Anti Reverse Cap Analog (ARCA)
A crucial instrument in the study of molecular biology and messenger RNA-based investigations is the Anti-Reverse Cap Analogue (ARCA). This artificially modified nucleotide analogue has several advantages over conventional cap analogues when it comes to capping the 5'-end of RNA molecules . The expanding significance of messenger RNA (mRNA)in the expression of genes, protein production and RNA-based treatments has significantly accelerated the research of RNA biochemistry in recent years. Chemically modified cap analogues have been demonstrated to offer promising anti-cancer capabilities after extensive investigation. Several altered cap analogues were put under trial for cap-dependent translation inhibition in vitro via competing for the eukaryotic translation initiation factor 4E (eIF-4E) binding site, against native capped-mRNA within the cell. Some of them have intriguing biological characteristics. An appealing class of analogues has the methylation guanine moiety's exocyclic amine group changed. Previously it has been demonstrated, in vitro, that enhanced translation inhibition results from a single modification at the N2 site of the first guanine moiety.
The 5' cap structures, considered crucial for stability, translation effectiveness along cellular recognition, is a major component of mRNA processing. By interacting with the eIF-4E, mRNA capping significantly improves translation efficiency and intracellular mRNA stability . It is common practice to cap in vitro transcribed (IVT) mRNA using a cap analogue, which may be added either during or after transcription.
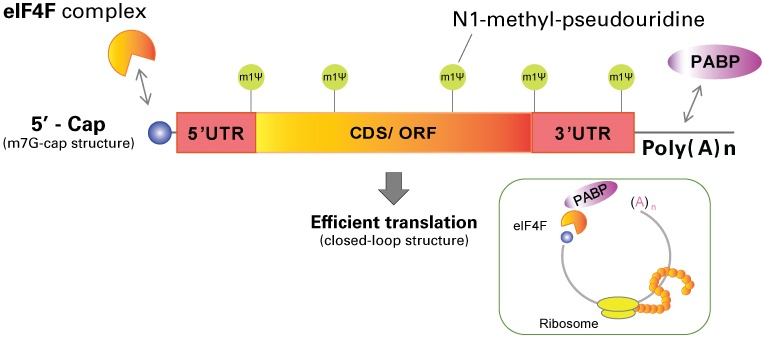
Figure 1: An overview of the transcribed mRNA, in vitro, with cap structure.
These analogues do have drawbacks, too, such as limited translational effectiveness and hydrolysis vulnerability. The ARCA was developed as a possible solution to these problems, as deregulated translational regulation is an important characteristic of many malignancies. Increased cell division and malignancies are brought on by the overexpression of eIF-4E, which has a significant impact on cell proliferation and phenotypic . Guanosine containing a methyl group attached to the 7th position makes up the 5′ cap; this altered nucleoside is frequently referred to as m7G while this initial 5’ cap structure is called Cap-0. The first nucleotide on the 5′ end of mRNA (at +1 position, where transcription begins), is connected to the cap by triphosphate linkage (PPP), modified in eukaryotes and is called Cap-1 .
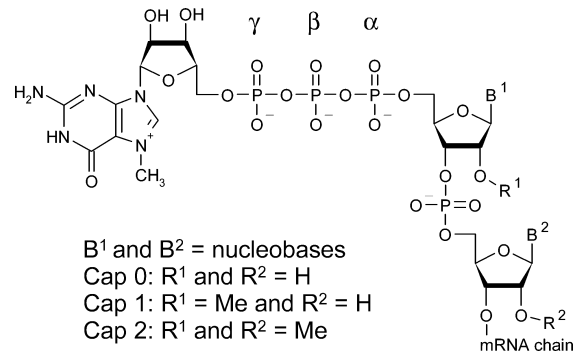
Figure 2: A general structural description of eukaryotic mRNA cap structure.
The mRNA, however, can be reversely capped, which causes quick breakdown and subpar translation for this, ARCA guarantees proper capping orientation and prevents reverse 5′ cap incorporation . However, only inserted with the right orientation, ARCA can create mRNAs with twice the translation efficiency compared to those started with conventional mCAP. ARCA as an altered nucleotide analogue offers several benefits when during in vitro transcription it is added to the 5' end of the newly synthesised RNA transcripts. ARCA, in contrast to conventional cap analogues, has a special reverse orientation that gives it resistance against 5' to 3' exonuclease action. This feature greatly improves the long-term stability of ARCA-capped RNA strands, making them useful for a variety of research applications. Its reversed cap structure, which uses a 3'-5' linkage as opposed to the typical 5'-5' linkage seen in mRNA caps, is what makes ARCA unique . Because of its unusual orientation, ARCA is resistant to intracellular decapping enzymes, preventing the breakdown of mRNA.
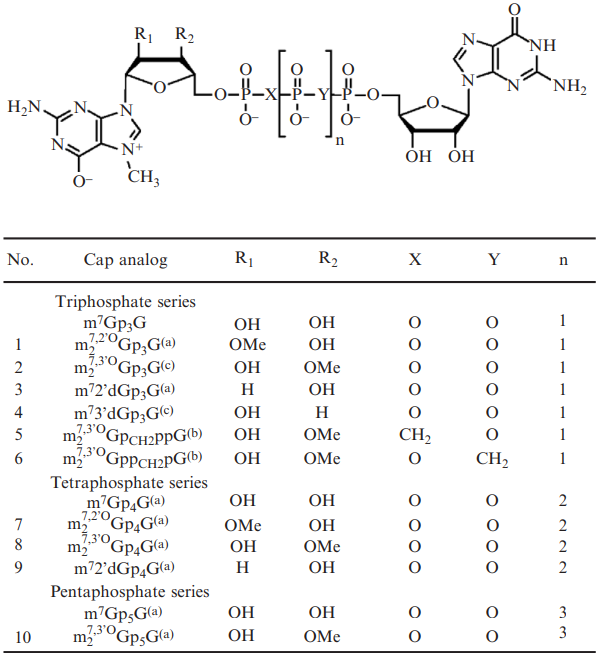
Figure 3: Some structures of the ARCA analogues.
More than 25 years ago, scientists used bacterial or bacteriophage RNA polymerases (i.e. T7 or SP6) to transcribe a DNA template to create capped mRNA in a lab setting . Following a standard procedure involving four NTPs and a dinucleotide cap analogue m7G. Usually, the 3’ OH group of GTP attacks the alpha-phosphate of the following NTP of the DNA template. Whereas transcription mostly starts with an attack of the 3' OH group of the cap dinucleotide, if a cap analogue is present in high concentration in the reaction mixture. As a result, m7GpppGpNp(Np)n-shaped capped mRNA transcripts are created . It was found that the cap dinucleotide's guanosine (Guo) or methylated guanosine (m7-Guo) moieties can both be the target of this attack at the 3' OH. Due to this, up to 50% of the transcripts had their caps in reversed orientation. The H-bonding patterns of Guo and m7-Guo were identical, which made it possible for both of them to pair with the dC in the template making it difficult for the viral polymerase to distinguish between them. According to Jemielity et al. when the pH of the reaction mixture was elevated from 6.5 to 7.5, the proportion of appropriately capped transcripts improved from 52% to 65%, however, 28% of these transcripts were reverse capped even at pH 8.4. It is difficult to correctly interpret data due to these reverse-capped transcripts in mRNA mixes diminished their total translational activity and might impact the results of biological experiments.
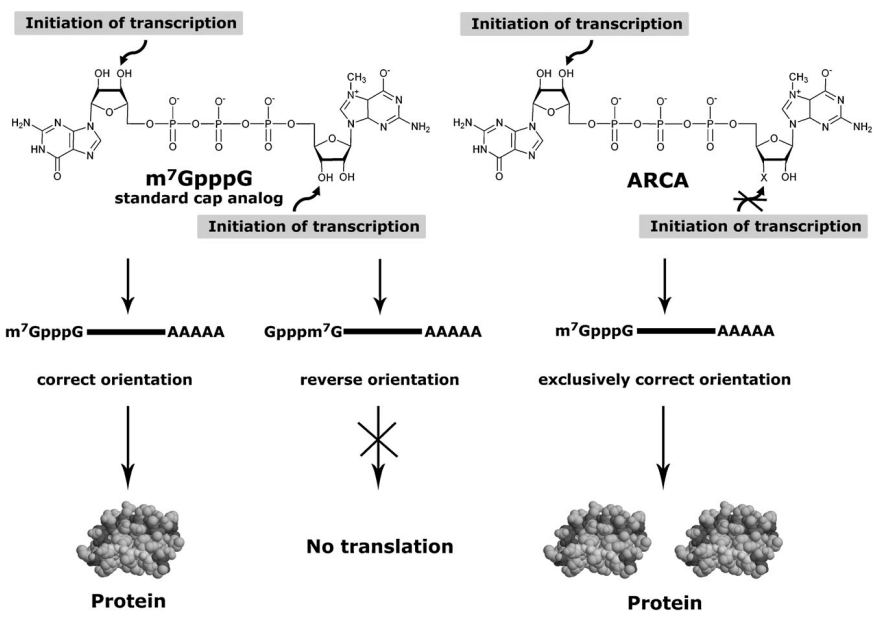
The m7-Guo moiety's 3'-OH group was changed in ARCA analogues by either 3'-O-CH3 or 3'-H alterations. This modification ensured that ARCAs were only integrated into mRNA in the proper orientation, where transcription is started by the Guo 3'-OH. The in vitro translational efficiency was doubled due to ARCA-capped mRNAs compared to those capped with m7G . The empirical evidence suggests that ARCA replacements have no impact on the translation machinery's ability to recognise caps. All triphosphate ARCA complexes with eIF4E had KAS (equilibrium association affinity) constants comparable to those of m7G. Additionally, the 2'- and 3'-OH groups in the crystalline structures of the cap analogues attached to eIF4E showed that they are accessible to the solution and contribute little to cap recognition . This makes them an ideal location for alterations not affecting the binding to eIF-4E.

Figure 4: The co-crystal structure: A: of m7G bound to eIF4E where 2' and 3'-OH groups are accessible to the solution but have no effect on cap binding B: Essential amino acids for recognising the cap.
In gene regulation research, like reporter assays, functional genomics and in vitro translation, ARCA-capped mRNA finds widely applications. Even during lengthy testing times, the stability offered by ARCA guarantees repeatable and trustworthy results. Success of mRNA vaccines, like those for COVID-19, has stoked demand for ARCA-capped mRNA due to its medicinal uses involving its improved translational efficacy and sustained expression. This has resulted in more powerful and long-lasting immune responses . In addition to vaccines, ARCA is being investigated for a number of RNA-based drugs, such as mRNA treatments for genetic conditions, tumour immunotherapy and regenerative medicine. In research that involves high-throughput functional genomics, the effective synthesis of capped RNA libraries are used for like CRISPR/Cas9 screening and RNA interference.
ARCA has a lot of potential, but before it can be widely used in therapeutic settings, issues related to scalability, affordability and inherent toxicity must be resolved. Additionally, research is being done to enhance ARCA's translational effectiveness and tailor it for particular mRNA-based therapeutics.










コメント