Cancer Biology
- Cancer Biology Peptides(42)
- Ras-like GTPases(11)
- Leukemia(1)
- Cell Proliferation, Viability & Cytotoxicity(0)
- Others(7)
- Tumor Microenvironment(30)
- Cell Migration & Metastasis(19)
Produkte für Cancer Biology
- Bestell-Nr. Artikelname Informationen
-
GC49140
α-Conotoxin ImI (trifluoroacetate salt)
α-CTx ImI, GCCSDPRCAWRC
A conotoxin and an antagonist of α7 nAChRs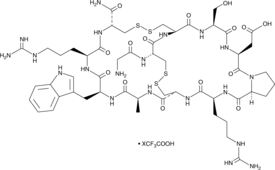
-
GC52338
α-Conotoxin PnIA (trifluoroacetate salt)
α-CtxPnIA, αPnIA
A peptide antagonist of α3β2-subunit containing and α7 nAChRs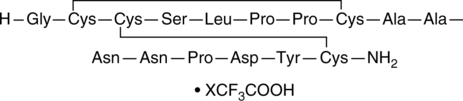
-
GC49838
α-Cortolone
20α-Cortolone, NSC 59872
A metabolite of cortisol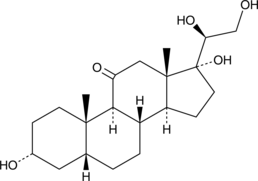
-
GC40702
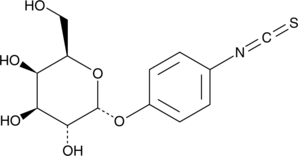
α-D-Galactopyranosylphenyl isothiocyanate
α-D-Galactopyranosylphenyl isothiocyanate is a chemically activated form of galactose that has been used to prepare various neoglycoproteins, which consist of a glycosylated serum albumin substituted with either fluorescein or methotrexate.
-
GC45207
α-hydroxy Farnesyl Phosphonic Acid
Hydroxyfarnesyl Phosphate
α-hydroxy Farnesyl phosphonic acid is a nonhydrolyzable analog of farnesyl pyrophosphate which acts as a competitive inhibitor of farnesyl transferase (FTase).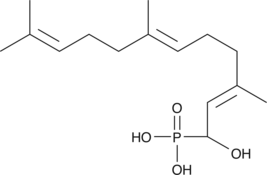
-
GC41499
α-Phellandrene
p-Mentha-1,5-diene, (±)-α-Phellandrene
α-Phellandrene is a cyclic monoterpene that has been found in various plants, including Cannabis, and has diverse biological activities.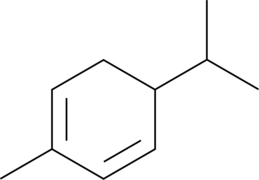
-
GC49467
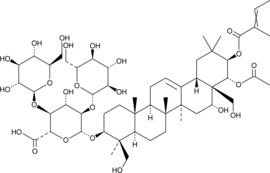
β-Aescin
A triterpenoid saponin with diverse biological activities
-
GC40789
β-Cembrenediol
β-CBD
β-Cembrenediol (β-CBT) is a natural product from tobacco plants that is found in cigarette smoke condensate.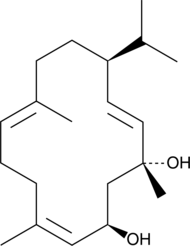
-
GC48299
β-D-Ribofuranose 1,2,3,5-tetraacetate
NSC 18738, 1,2,3,5-tetra-O-acetyl-β-D-Ribofuranose, 1’,2’,3’,5’-O-tetraacetyl-β-D-Ribofuranose
A precursor in the synthesis of nucleosides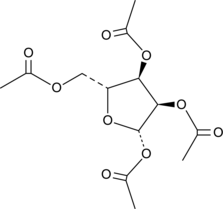
-
GC48998
β-Defensin-1 (human) (trifluoroacetate salt)
hBD-1
An antimicrobial peptide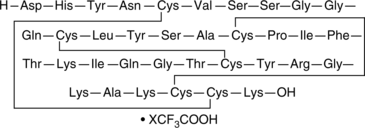
-
GC45604
β-Rubromycin
β-Rubromycin ist ein potenter und selektiver Inhibitor der RNA-gerichteten DNA-Polymerasen (reverse Transkriptase) des humanen ImmunschwÄchevirus-1 (HIV-1).
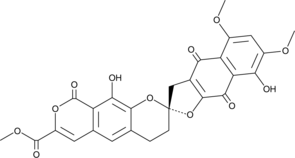
-
GC52404
γ-Glu-Phe (trifluoroacetate salt)
γ-Glutamylphenylalanine
A dipeptide with metabolism-altering activity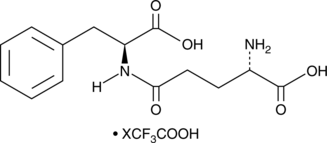
-
GC41109
δ12-Prostaglandin J2
Δ12PGJ2
δ12-Prostaglandin J2 (δ12-PGJ2) ist ein Cyclopentenon-Prostaglandin (PG) mit antiproliferativer Wirkung auf verschiedenes Tumorzellwachstum. δ12-Prostaglandin J2, ein natÜrlich vorkommendes Dehydratisierungsprodukt von Prostaglandin D2, kann Über Caspase-Aktivierung Apoptose in HeLa-Zellen induzieren.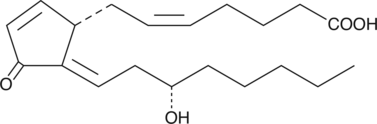
-
GC48532
ε-Rhodomycinone
NSC 196524
A bacterial metabolite and precursor to rhodomycin D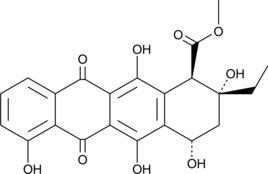
-
GC41669
(±)-Enterolactone
HPMF
(±)-Enterolacton ist ein bioaktiver phenolischer Metabolit, bekannt als SÄugetier-Lignan, das aus Nahrungs-Lignanen gewonnen wird. (±)-Enterolacton hat Östrogene Eigenschaften und Anti-Brustkrebs-AktivitÄt. (±)-Enterolacton ist ein Radiosensibilisator fÜr menschliche Brustkrebszelllinien durch beeintrÄchtigte DNA-Reparatur und erhÖhte Apoptose.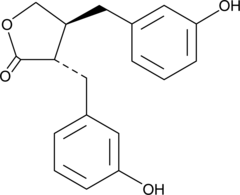
-
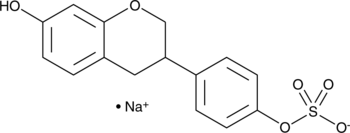
GC41671
(±)-Equol 4'-sulfate (sodium salt)
R,S-Equol 4'-Sulfate
(±)-Equol 4'-sulfate is a gut-mediated phase II metabolite of the isoflavonoid phytoestrogen (±)-equol.
-
GC46000
(•)-Drimenol
NSC 169775
A sesquiterpene alcohol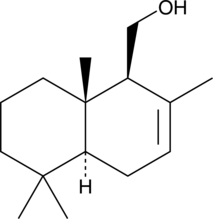
-
GC48635
(-)-Cryptopleurine
(R)-Cryptopleurine, NSC 19912
An alkaloid with diverse biological activities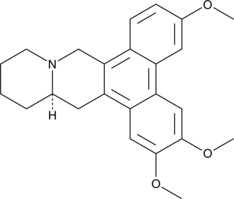
-
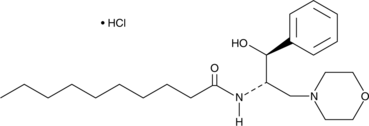
GC45250
(-)-L-threo-PDMP (hydrochloride)
(-)-L-threo-PDMP is an enhancer of ganglioside biosynthesis.
-
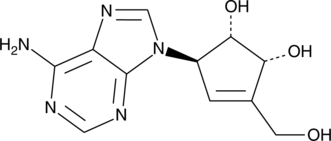
GC45251
(-)-Neplanocin A
S-Adenosylhomocysteine (SAH) hydrolase catalyzes the reversible hydrolysis of SAH to adenosine and homocysteine.
-
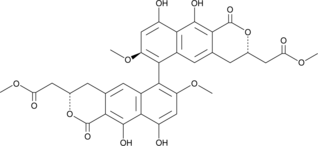
GC45253
(-)-Viriditoxin
(-)-Viriditoxin is a mycotoxin originally isolated from A.
-
GC48508
(4-Carboxybutyl-d4)triphenylphosphonium (bromide)
TPP-d4, 5-Triphenylphosphoniovaleric Acid-d4
An internal standard for the quantification of (4-carboxybutyl)triphenylphosphonium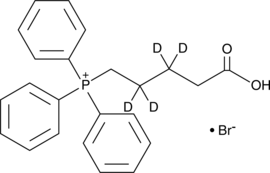
-
GC46038
(5E)-7-Oxozeaenol
LL-Z 1640-2
A resorcylic acid lactone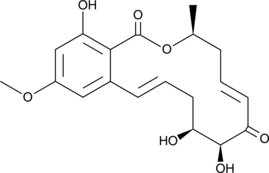
-
GC41702
(E)-5-(2-Bromovinyl)uracil
BVU
(E)-5-(2-Bromovinyl)uracil (BVU) is a pyrimidine base and an inactive metabolite of the antiviral agents sorivudine and (E)-5-(2-bromovinyl)-2'-deoxyuridine (BVDU) that may be regenerated to BVDU in vivo.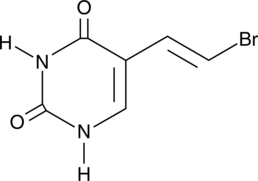
-
GC49003
(E)-Ajoene
NSC 614554
A disulfide with diverse biological activities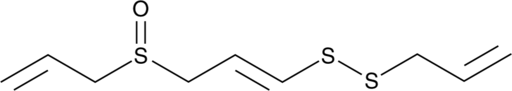
-
GC41620
(R)-(-)-Mellein
Ochracin
(R)-(-)-Mellein ist ein aus KulturflÜssigkeiten dieses Aspergillus isoliertes Antibiotikum.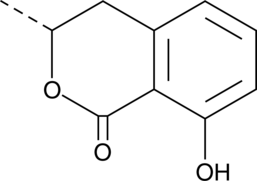
-
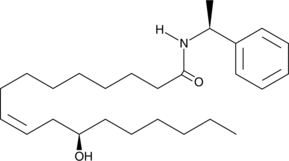
GC41741
(S)-α-Methylbenzyl Ricinoleamide
(S)-α-Methylbenzyl ricinoleamide is a fatty acid amide derived from ricinoleic acid and methyl benzylamine.
-
GC18787
(±)-Dunnione
NSC 95403
(±)-Dunnione is a naturally occurring naphthoquinone with diverse biological activities.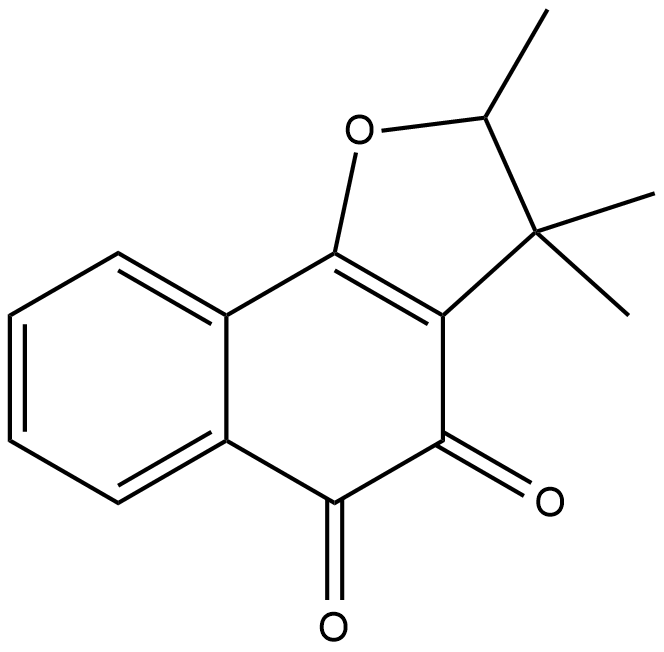
-
GC46039
1,2,3,4-Tetrahydrostaurosporine
AFN-941
A mutant EGFR inhibitor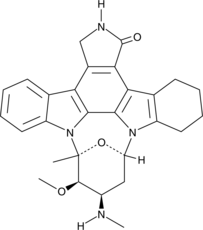
-
GA20357
1,2-Dimyristoyl-rac-glycerol
1,2-Dimyristin, TG(14:0/14:0/0:0)
A diacylglycerol
-
GC41823
1,2-Dipalmitoyl-sn-glycero-3-PE-N-(cap biotin) (sodium salt)
biotin-cap-DPPE
1,2-Dipalmitoyl-sn-glycero-3-PE-N-(cap biotin) is a biotinylated phospholipid.
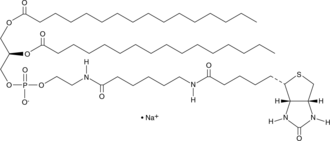
-
GC49294
1-(4-Chlorobenzhydryl)piperazine
N-(p-Chlorobenzhydryl)-piperazine, Norchlorcyclizine, NSC 86164
An inactive metabolite of meclizine and chlorcyclizine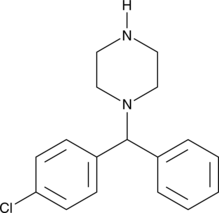
-
GC41986
1-Arachidonoyl Lysophosphatidic Acid (ammonium salt)
1-Arachidonoyl LPA, 1-Arachidonoyl-sn-glycero-3-phosphate, 1-Eicosatetraenoyl-sn-glycero-3-PA, 20:4 Lyso PA, LPA(20:4), PA(20:4/0:0)
1-Arachidonoyl lysophosphatidic acid is a phospholipid containing arachidonic acid at the sn-1 position.
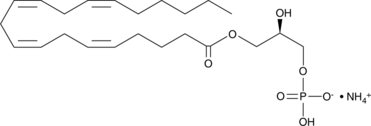
-
GC42001
1-Myristoyl-2-hydroxy-sn-glycero-3-PE
14:0 Lyso-PE, 1-Myristoyl-2-hydroxy-sn-glycero-3-Phosphoethanolamine
1-Myristoyl-2-hydroxy-sn-glycero-3-PE is a naturally occurring lysophospholipid.
-
GC18651
1-Palmitoyl-2-hydroxy-sn-glycero-3-PE
1-Hexadecanoyl-sn-glycero-3-Phosphoethanolamine
1-Palmitoyl-2-hydroxy-sn-glycero-3-PE is a naturally-occurring lysophospholipid.
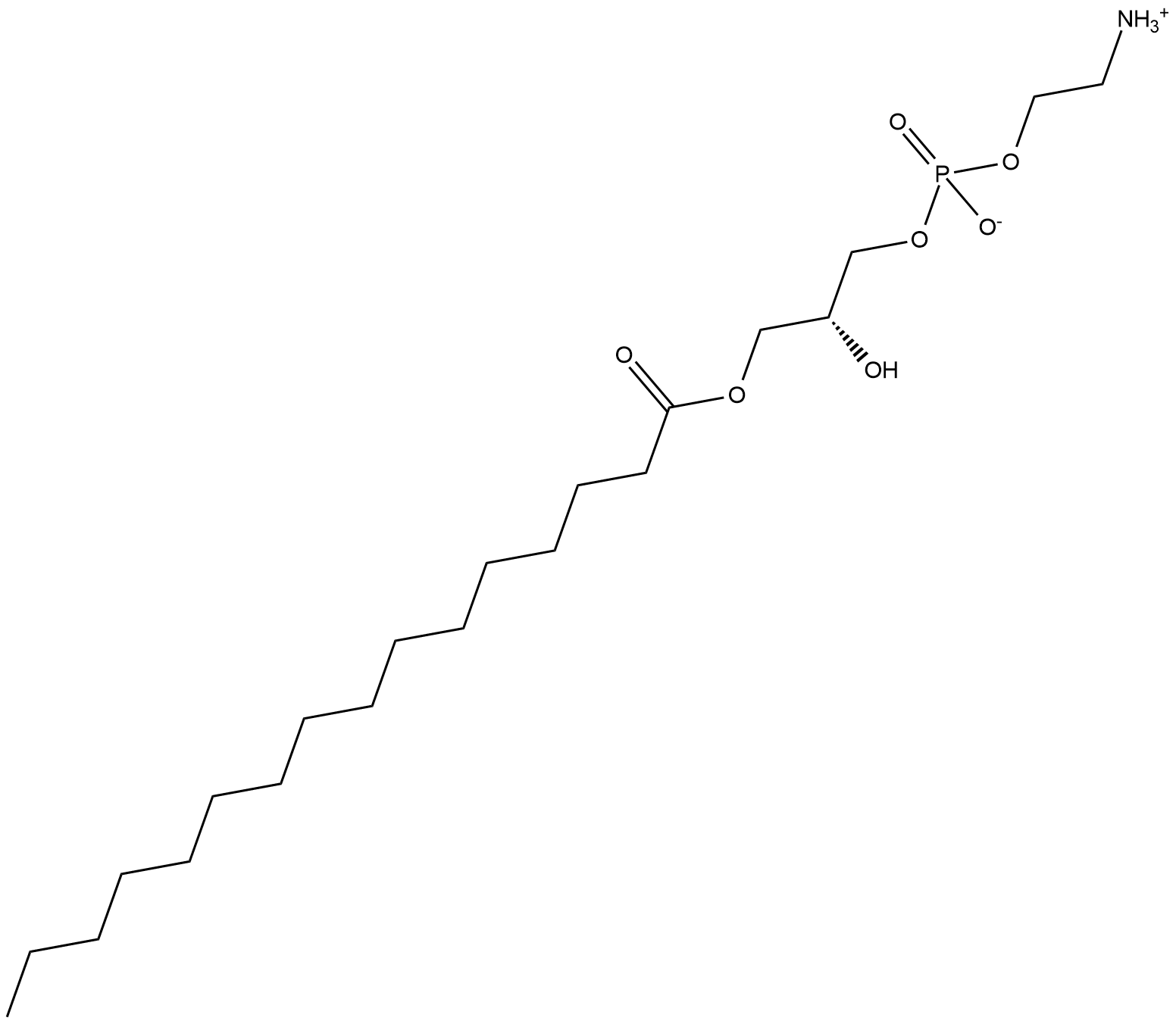
-
GC46496
1-Stearoyl-2-Arachidonoyl-d8-sn-glycero-3-PC
18:0/20:4-d8-PC, PC(18:0/20:4-d8), SAPCd8, 1-Stearoyl-2-Arachidonoyl-d8-sn-glycero-3-Phosphatidylcholine, 1-Stearoyl-2-Arachidonoyl-d8-sn-glycero-3-Phosphocholine
An internal standard for the quantification of 1-stearoyl-2-arachidonoyl-sn-glycero-3-PC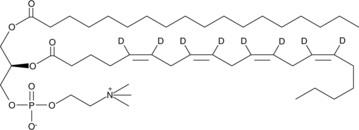
-
GC41331
1-Stearoyl-2-hydroxy-sn-glycero-3-PC
C18:0-PC, Lyso-PC, 1-Octadecanoyl-2-hydroxy-sn-glycero-3-phosphatidylcholine, PC(18:0/0:0), 1-Stearoyl-2-hydroxy-sn-glycero-3-Phosphocholine, 1-Stearoyl-2-lyso-sn-glycero-3-PC
1-Stearoyl-2-hydroxy-sn-glycero-3-PC is a saturated 18:0 lysophosphatidylcholine found in plasma and oxidized LDL that is thought to play a role in inflammatory diseases and atherosclerosis.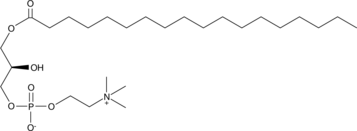
-
GC46498
1-Stearoyl-d35-2-hydroxy-sn-glycero-3-PC
S-Lyso-PC-d35, 1-Stearoyl-d35-2-hydroxy-sn-glycero-3-Phosphocholine, 1-Stearoyl-d35-2-lyso-sn-glycero-3-PC
An internal standard for the quantification of 1-stearoyl-2-hydroxy-sn-glycero-3-PC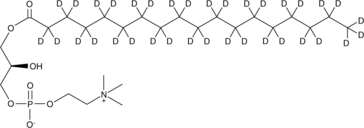
-
GC40176
1-Stearoyl-rac-glycerol
18:0-MG, MG(18:0/0:0/0:0), 1-Monostearin, NSC 3875
1-Stearoyl-rac-glycerol is a monoacylglycerol that contains stearic acid at the sn-1 position.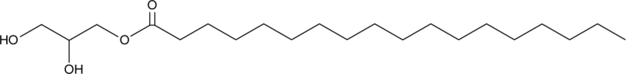
-
GC46412
11β-Prostaglandin F2α-d4
9α,11βPGF2αd4, 11βPGF2αd4, 11epi PGF2αd4
An internal standard for the quantification of 11β-PGF F2α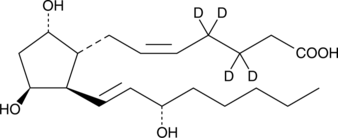
-
GC18637
11β-Prostaglandin F2α
9α,11βPGF2α, 11βPGF2α, 11epi PGF2α
11β-Prostaglandin F2α (11β-PGF2α) is the primary plasma metabolite of PGD2 in vivo.
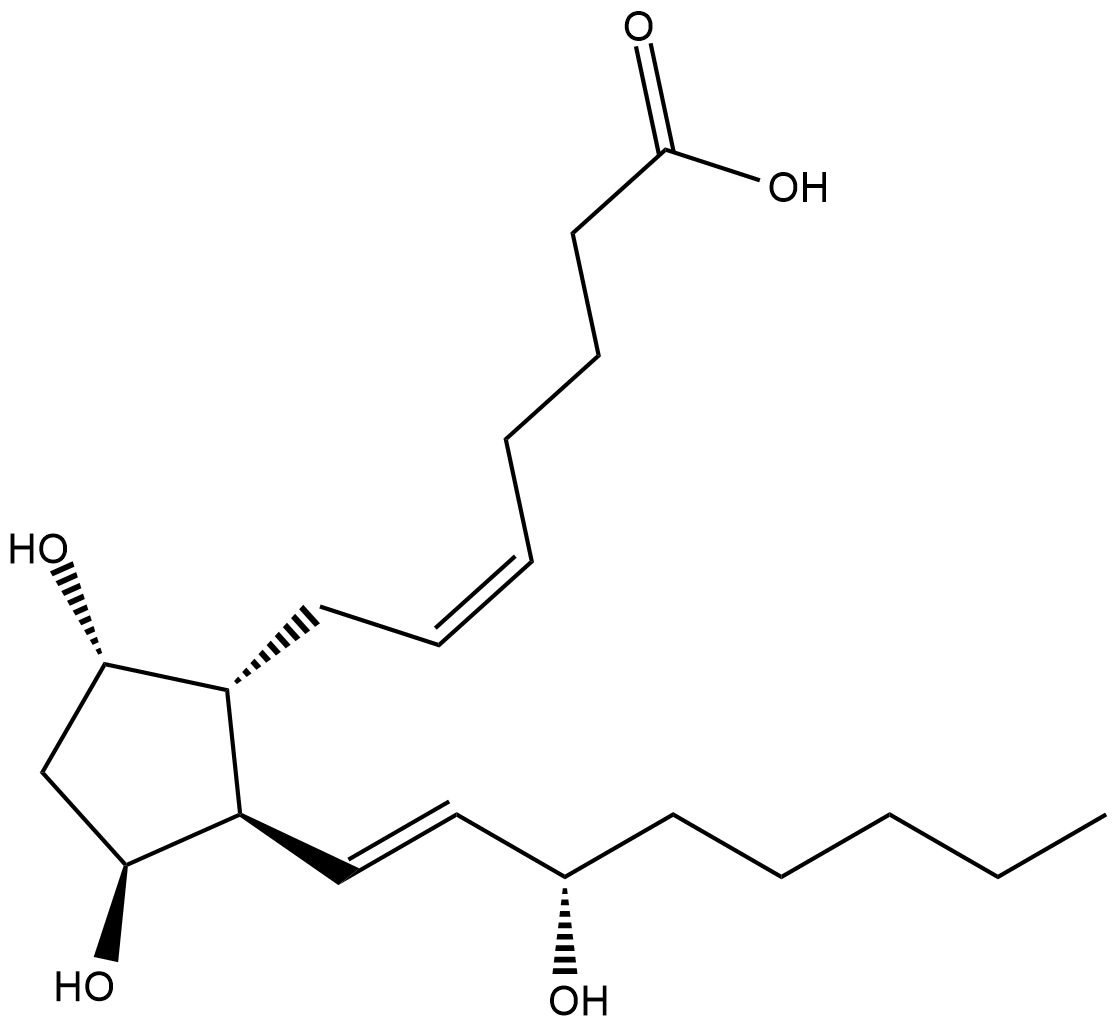
-
GC49310
12-hydroxy Stearic Acid
12-HSA, NSC 2385
12-Hydroxystearinsäure ist ein strukturell einfacher und kostengünstiger Organogelierer mit niedrigem Molekulargewicht, und seine Metallsalze und Derivate finden in vielen wichtigen Anwendungen eine Rolle.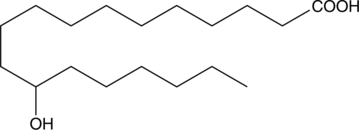
-
GC49759
13C17-Mycophenolic Acid
13C17-MPA
An internal standard for the quantification of mycophenolic acid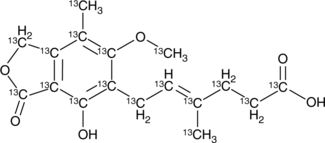
-
GC41950
16α-hydroxy Estrone
16α-hydroxy E1, 16αOHE1
16α-Hydroxy Estrone (16αOHE) ist ein Hauptmetabolit von Estradiol.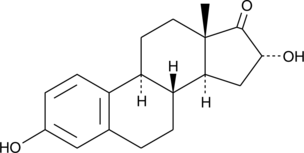
-
GC41942
16,16-dimethyl Prostaglandin A2
16,16dimethyl PGA2
16,16-dimethyl PGA2 is a metabolism-resistant analog of PGA2 with a prolonged in vivo half-life.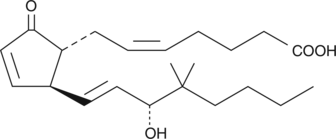
-
GC41300
17β-hydroxy Exemestane
17β-hydroxy Exemestane is the primary active metabolite of exemestane.

-
GC45307
17(R)-Resolvin D3
Aspirin-triggered Resolvin D3, 17-epi-Resolvin D3, AT-RvD3, 17(R)-RvD3
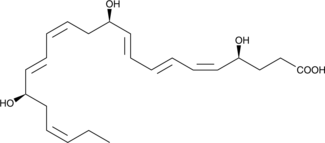
-
GC46510
2,3,4-Tri-O-acetyl-β-D-Glucuronide methyl ester
A synthetic intermediate
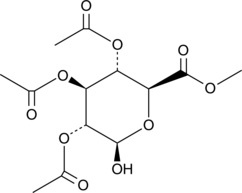
-
GC40947
2,3-Dimethoxy-5-methyl-p-benzoquinone
Coenzyme Q0, CoQ0
2,3-Dimethoxy-5-methyl-p-benzochinon (CoQ0) ist eine potente, oral aktive Ubichinonverbindung, die aus Antrodia cinnamomea gewonnen werden kann.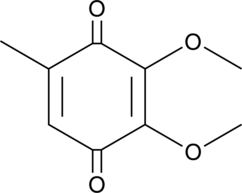
-
GC48417
2,3-Indolobetulinic glycine amide
A derivative of betulinic acid
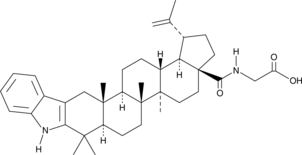
-
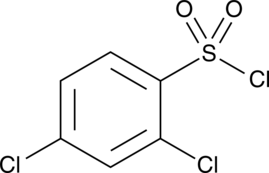
GC46522
2,4-Dichlorobenzenesulfonyl chloride
A heterocyclic building block
-
GC46502
2-(1-(Thiophen-2-yl)ethylidene)hydrazinecarbothioamide
NSC 707, 1-(Thiophen-2-yl)ethanone thiosemicarbazone
An antimicrobial agent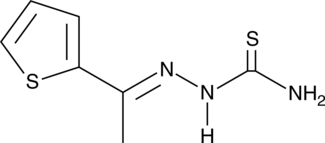
-
GC46530
2-Amino-3,4-dimethyl-3H-imidazo[4,5-f]quinoline
MelQ
A mutagenic heterocyclic amine![2-Amino-3,4-dimethyl-3H-imidazo[4,5-f]quinoline Chemical Structure 2-Amino-3,4-dimethyl-3H-imidazo[4,5-f]quinoline Chemical Structure](/media/struct/GC4/GC46530.png)
-
GC49362
2-Amino-3,8-dimethylimidazo-[4,5-f]-quinoxaline
MeIQx
A food-derived carcinogen![2-Amino-3,8-dimethylimidazo-[4,5-f]-quinoxaline Chemical Structure 2-Amino-3,8-dimethylimidazo-[4,5-f]-quinoxaline Chemical Structure](/media/struct/GC4/GC49362.png)
-
GC52029
2-Aminoflubendazole
Hydrolyzed Flubendazole
2-Aminoflubendazol ist der Metabolit von Benzimidazolen.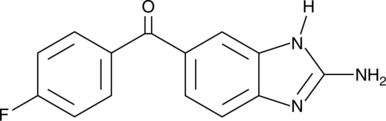
-
GC46539
2-Chloroadenine
6-amino-2-chloropurine, 2-chloro-6-aminopurine, NSC 7362
A heterocyclic building block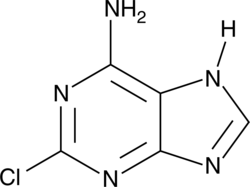
-
GC48943
2-deoxy-2-fluoro-D-Glucose
FDG, 2-FG, Fluorodeoxyglucose, 2-fluoro-2-deoxy-D-Glucose
A glucose derivative with anticancer activity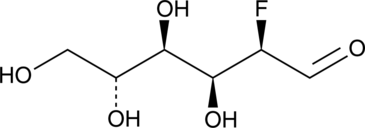
-
GC49223
2-deoxy-D-Glucose-13C6
2-DG-13C6
An internal standard for the quantification of 2-deoxy-D-glucose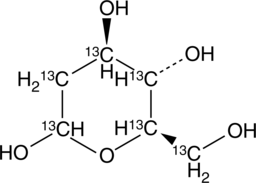
-
GC49172
2-hydroxy Estrone
Catecholestrone, 2-hydroxy E1, 2-OHE1
2-Hydroxyöstron (Catecholestrone) ist ein spezifischer rezeptorvermittelter antiöstrogener Wirkstoff.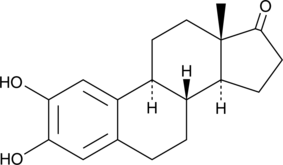
-
GC42170
2-hydroxy Stearic Acid
NSC 907, (±)-α-hydroxy Octadecanoic Acid, (±)-α-hydroxy Stearic Acid
2-hydroxy Stearic acid is a naturally occurring hydroxylated fatty acid that has been found in humans and U.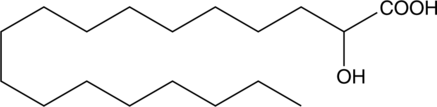
-
GC40944
2-hydroxy-6-Methylbenzoic Acid
2,6-Cresotic Acid, 6-Methylsalicylic Acid, 6-MSA, NSC 403256, 6-hydroxy-o-Toluic Acid
2-hydroxy-6-Methylbenzoic acid is a constituent of G.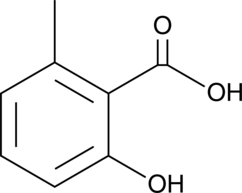
-
GC13460
2-Hydroxyestradiol
2-HE2,NSC 61711
2-Hydroxyestradiol, ein Metabolit von 17β-Estradiol mit minimaler Östrogener AktivitÄt, besitzt antioxidative Wirkungen und reagiert mit DNA unter Bildung stabiler Addukte und Übt GenotoxizitÄt aus.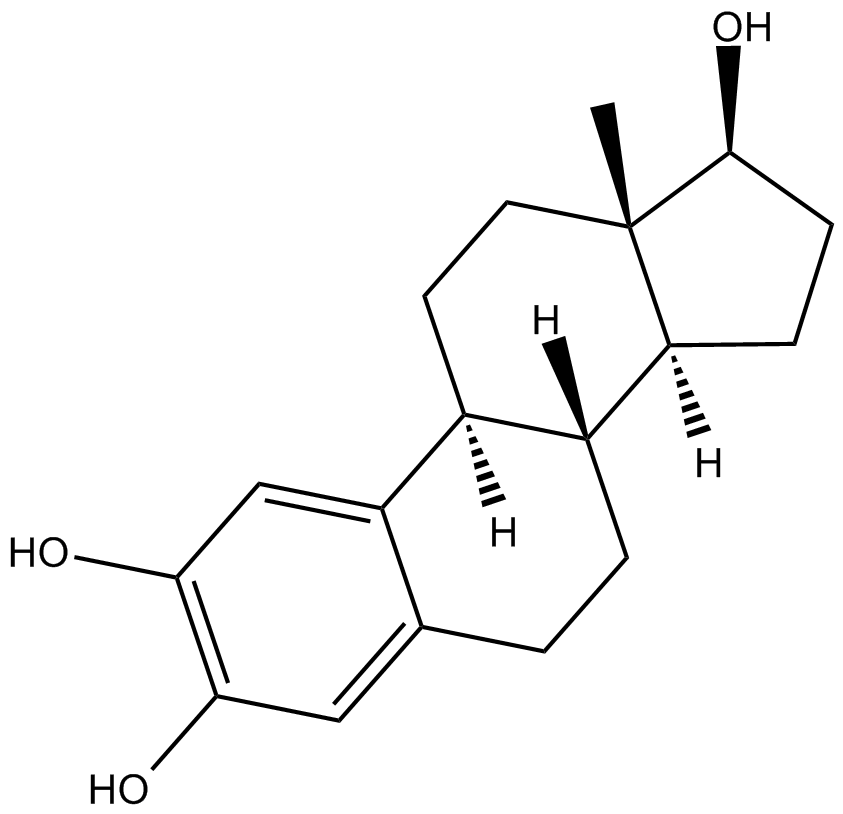
-
GC42203
3,4',5-Trismethoxybenzophenone
Resveratrol is a potent phenolic antioxidant found in natural sources that has antiproliferative activity.
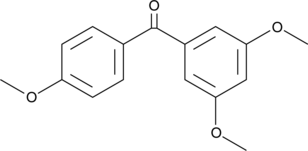
-
GC49361
3,6-Dimethoxy-9H-xanthen-9-one
3,6-Dimethoxyxanthone
A synthetic intermediate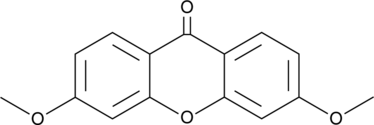
-
GC52324
3-(3-Hydroxyphenyl)propionic Acid sulfate
3-HPPA sulfate
A metabolite of certain phenols and glycosides
-
GC48481
3-Acetyl Betulin
3-O-Acetyl Betulin
A derivative of betulin
-
GC48487
3-Acetyl Betulinaldehyde
3-O-Acetylbetulin Aldehyde, Acetylbetulinaldehyde
A triterpene
-
GC42260
3-Deazauridine
NSC 126849
3-Deazauridine is a nucleoside analog.
-
GC45337
3-Hydroxyterphenyllin
NSC 299113
3-Hydroxyterphenyllin ist ein Metabolit von Aspergillus candidus. 3-Hydroxyterphenyllin unterdrÜckt die Proliferation und verursacht ZytotoxizitÄt gegenÜber A2780/CP70- und OVCAR-3-Zellen. 3-Hydroxyterphenyllin induziert S-Phasen-Arrest und Apoptose. 3-Hydroxyterphenyllin hat das Potenzial fÜr die Erforschung von Eierstockkrebs.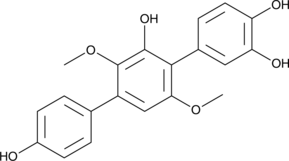
-
GC49874
3-Methoxycatechol
1,2-Dihydroxy-3-methoxybenzene, 3-Methoxypyrocatechol, NSC 66525, Pyrogallol 1-monomethyl ether
A lignan-derived phenol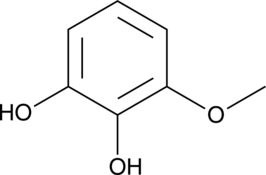
-
GC48488
3-Oxobetulin Acetate
28-O-acetyl-3-Oxobetulin, 3-oxo-28-O-Acetylbetulin
A derivative of betulin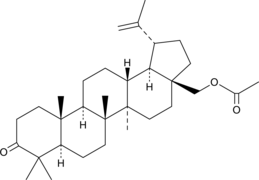
-
GC46606
4-(4,4,5,5-Tetramethyl-1,3,2-dioxaboran-2yl)aniline
4-Aminophenylboronic Acid pinacol ester
A heterocyclic building block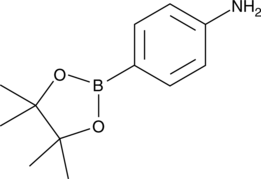
-
GC40045
4-(Dimethylamino)-1-methylpyridinium (iodide)
4-(Dimethylamino)-1-methylpyridinium is a monoquaternary pyridinium salt with anticholinesterase and antiproliferative activities.
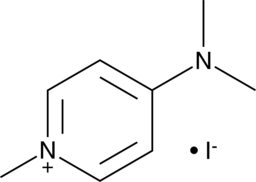
-
GC48824
4-hydroxy Estrone
4-hydroxy E1, 4-OHE1
4-Hydroxy-Estron (4-OHE1), ein Estron-Metabolit, hat eine starke neuroprotektive Wirkung gegen oxidative Neurotoxizität.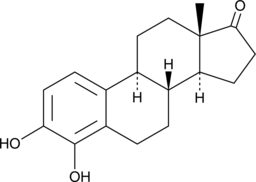
-
GC42411
4-hydroxy Nonenal Alkyne
Click Tag 4HNE Alkyne
4-hydroxy Nonenal (4-HNE) is a major aldehyde produced during the lipid peroxidation of ω-6 polyunsaturated fatty acids, such as arachidonic acid and linoleic acid.
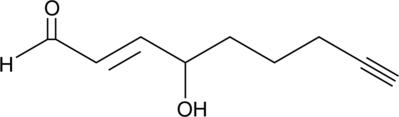
-
GC41530
4-oxo Docosahexaenoic Acid
4-oxo DHA
4-oxo Docosahexaenoic acid (4-oxo DHA) is a putative metabolite of DHA with antiproliferative and PPARγ agonist activity.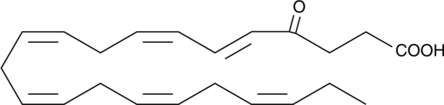
-
GC49244
4-oxo Isotretinoin
Ro 22-6595
An active metabolite of isotretinoin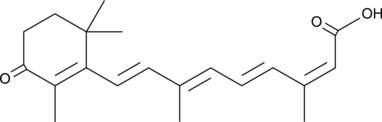
-
GC40053
5α,6α-epoxy Cholestanol
NSC 18176
An oxysterol and a metabolite of cholesterol produced by oxidation
-
GC18826
5'-chloro-5'-Deoxyadenosine (hydrate)
5’-ClDA, 5'-Deoxy-5'-chloroadenosine
5'-chloro-5'-Deoxyadenosine (hydrate) is a nucleoside analog used as a substrate in polyketide biosynthesis.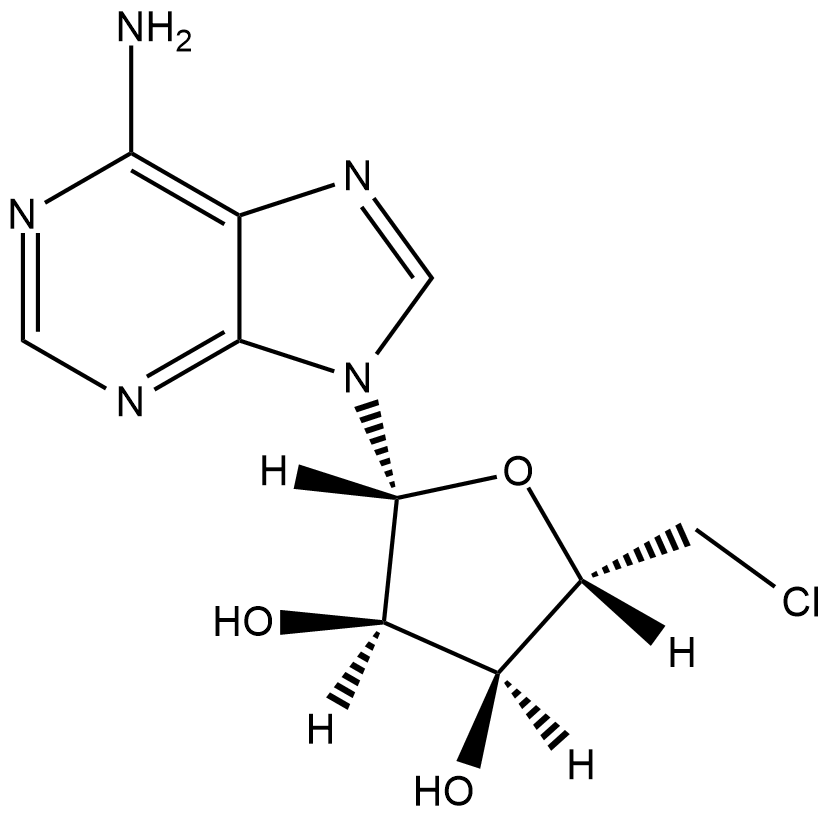
-
GC42503
5'-deoxy-5-Fluorocytidine
5'-DFCR
5'-deoxy-5-Fluorocytidine is an intermediate metabolite of the DNA synthesis inhibitor capecitabine.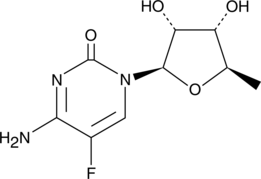
-
GC49499
5(Z)-Dodecenoic Acid
C12:1 n-7, C12:1(5Z), cis-5-Dodecenoic Acid
5(Z)-DodecensÄure ist ein endogener Metabolit mit hemmenden AktivitÄten gegen COX-I und COX-II.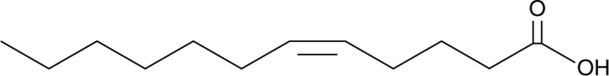
-
GC18542
5,6-dihydro-5-Fluorouracil
5-DHFU, 5-Fluorodihydropyrimidine-2,4-dione, 5-Fluorodihydrouracil, 5-Fluoro-5,6-dihydrouracil, 5-FUH2
5,6-dihydro-5-Fluorouracil (5-FUH2) is formed by the hydrogenation of 5-fluorouracil via the enzyme dihydropyrimidine dehydrogenase (DPD).
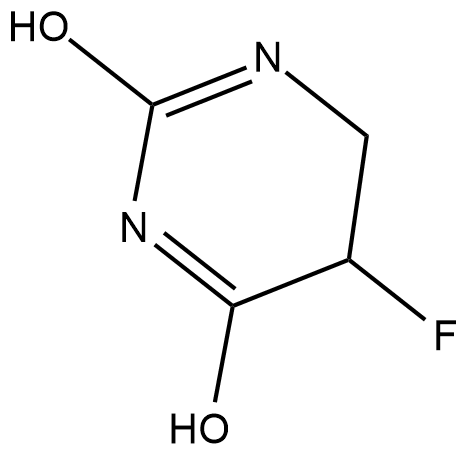
-
GC40527
5-(Hydroxymethyl)-2'-deoxyuridine
5-(Hydroxymethyl)-2'-deoxyuridine is a nucleoside analog with anticancer and antiviral activities.
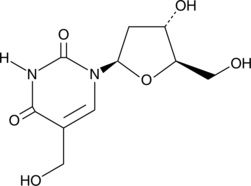
-
GC46033
5-Heneicosylresorcinol
An alkylresorcinol

-
GC52172
5-hydroxy Indole-3-acetic Acid-d6
5-HIAA-d6, 5-Hydroxyindoleacetic Acid-d6, 5-hydroxy IAA-d6
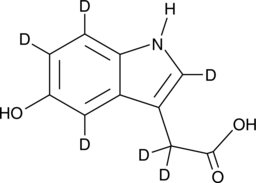
-
GC64610
5-O-Caffeoylshikimic acid
5-O-Caffeoylshikimic-SÄure kann in der Studie fÜr NSCLC verwendet werden.
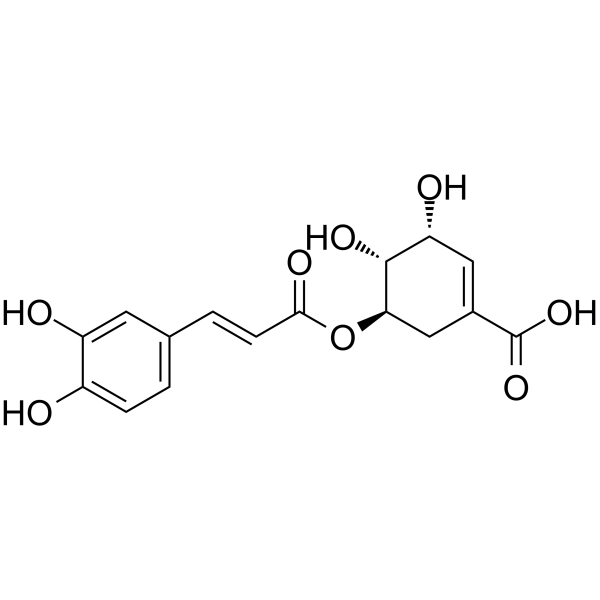
-
GC41310
5-Octyl-α-ketoglutarate
αKG octyl ester
In addition to its role in the Krebs cycle, α-ketoglutarate (2-oxoglutarate) has roles as a substrate or modulator of enzymes.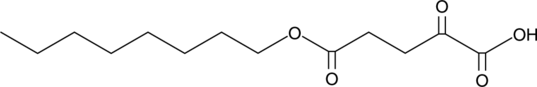
-
GC49629
6β-hydroxy Prednisolone
A metabolite of prednisolone
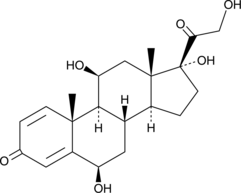
-
GC46718
6,7-Dihydro-5H-quinolin-8-one
8-Aza-1-tetralone, 5,6-Dihydro-7(7H)-quinolinone
A synthetic intermediate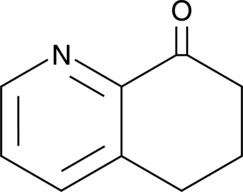
-
GC49551
6-Chloropurine Riboside
NSC 4910
A nucleoside precursor
-
GC48721
6-O-Demethyl Griseofulvin
6-Demethylgriseofulvin
A metabolite of griseofulvin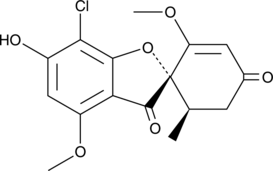
-
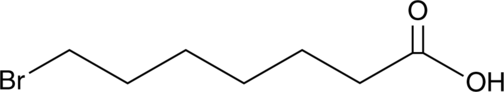
GC46736
7-Bromoheptanoic Acid
A building block
-
GC49051
7-hydroxy Methotrexate
NSC 380962, 7-hydroxy MTX
7-Hydroxy-Methotrexat ist ein Hauptmetabolit von Methotrexat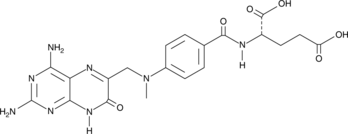
-
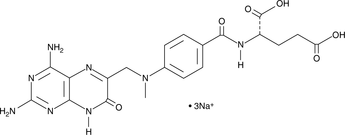
GC42608
7-hydroxy Methotrexate (sodium salt)
7-hydroxy MTX
7-hydroxy Methotrexate (7-hydroxy MTX) is a phase I metabolite of MTX, which is converted by hepatic aldehyde oxidases.
-
GC52239
7-Ketositosterol
7-oxo-β-Sitosterol, 7-Oxositosterol
A phytosterol and phytosterol oxidation product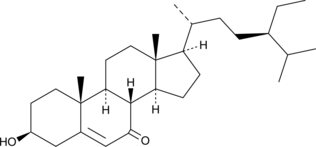
-
GC49561
7-Methylguanosine-d3
m7G-d3, 7-MeGua-d3
An internal standard for the quantification of 7-methylguanosine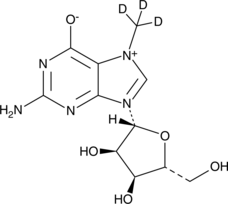
-
GC52142
80-O16B
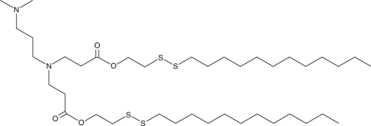
-
GC49347
8Br-HA
An FHIT inhibitor
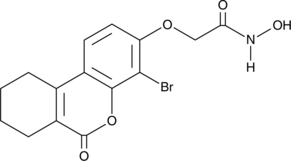
-
GC41642
9(E),11(E),13(E)-Octadecatrienoic Acid
β-Eleostearic Acid, β-ESA
9(E),11(E),13(E)-Octadecatrienoic acid (β-ESA) is a conjugated polyunsaturated fatty acid that is found in plant seed oils and in mixtures of conjugated linolenic acids synthesized by the alkaline isomerization of linolenic acid.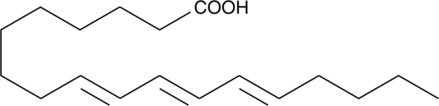
-
GC42644
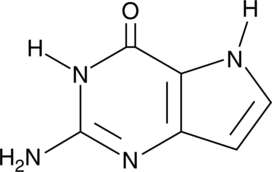
9-Deazaguanine
NSC 344522
9-Deazaguanine is an analog of guanine that acts as an inhibitor of purine nucleoside phosphorylase (PNP; Kd = 160 nM).