Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Ziele für Chromatin/Epigenetics
- Bromodomain(43)
- Aurora Kinase(59)
- DNA Methyltransferase(32)
- HDAC(180)
- Histone Acetyltransferases(63)
- Histone Demethylases(95)
- Histone Methyltransferase(197)
- HIF(90)
- JAK(146)
- MBT Domains(0)
- PARP(105)
- Pim(26)
- Protein Ser/Thr Phosphatases(26)
- RNA Polymerase(5)
- Sirtuin(77)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(200)
- MicroRNA(20)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Produkte für Chromatin/Epigenetics
- Bestell-Nr. Artikelname Informationen
-
GC31365
γ-Oryzanol
γ-Oryzanol ist ein starker Inhibitor der DNA-Methyltransferasen (DNMTs) im Striatum von Mäusen.
-
GC45258
(+)-Biotin 4-Amidobenzoic Acid (sodium salt)
(+)-Biotin PABA, N-Biotinyl-4-aminobenzoic Acid
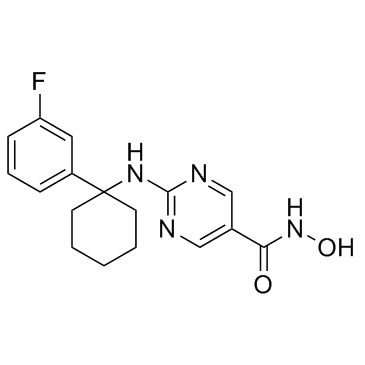
(+)-Biotin 4-amidobenzoic acid is a substrate of biotinidase, which cleaves biotin amide to give biotin in vivo.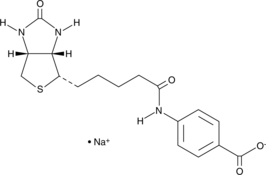
-
GC61595
(+)-JQ-1-aldehyde
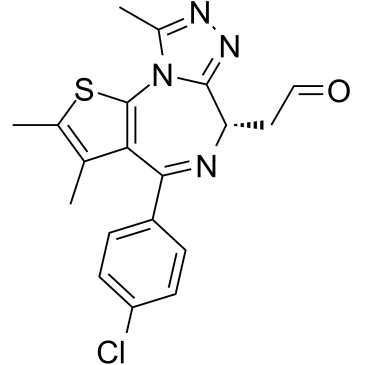
(+)-JQ-1-Aldehyd ist die Aldehydform von (+)-JQ1. (+)-JQ-1-Aldehyd kann als Vorstufe zur Synthese von PROTACs verwendet werden, die auf BET-BromodomÄnen abzielen.
-
GC34958
(+)-JQ1 PA
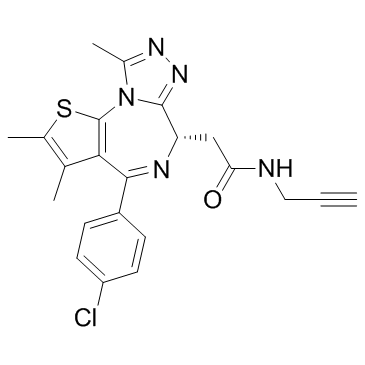
(+)-JQ1 PA ist ein Derivat der BromodomÄne und des extraterminalen (BET) Inhibitors JQ1 mit einem IC50 von 10,4 nM.
-
GC13822
(-)-JQ1
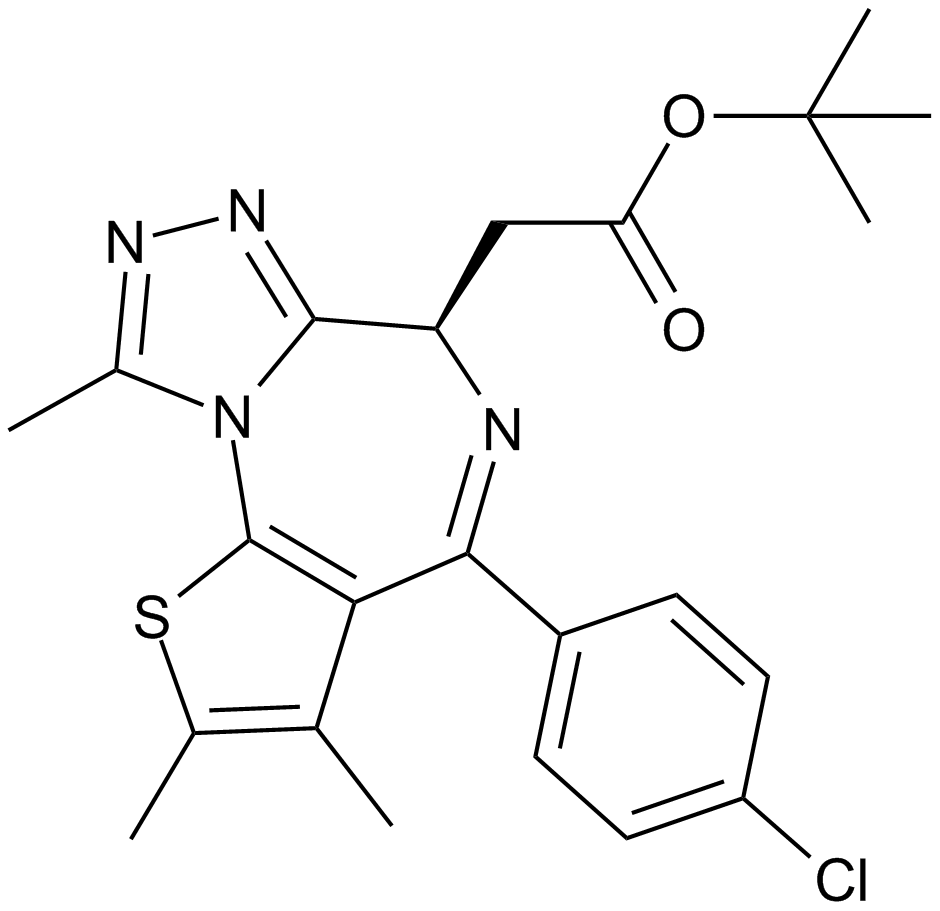
A selective inhibitor of BET bromodomains
-
GC34964
(1S,3R,5R)-PIM447 dihydrochloride
(1S,3R,5R)-LGH447 dihydrochloride
(1S,3R,5R)-PIM447 (Dihydrochlorid), ein PIM-Inhibitor, extrahiert aus Patent US 20100056576 A1, Verbindungsbeispiel 72, hat IC50-Werte von 0,095 μM fÜr Pim1, 0,522 μM fÜr Pim2 und 0,369 μM fÜr Pim3.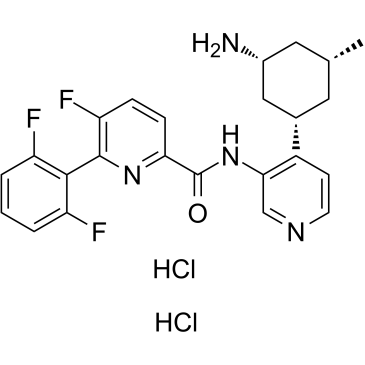
-
GC62130
(2R,5S)-Ritlecitinib
(2R,5S)-PF-06651600
(2R,5S)-Ritlecitinib ((2R,5S)-PF-06651600) ist ein potenter und selektiver JAK3-Inhibitor (IC50=144,8 nM), extrahiert aus Patent US20150158864A1, Beispiel 68.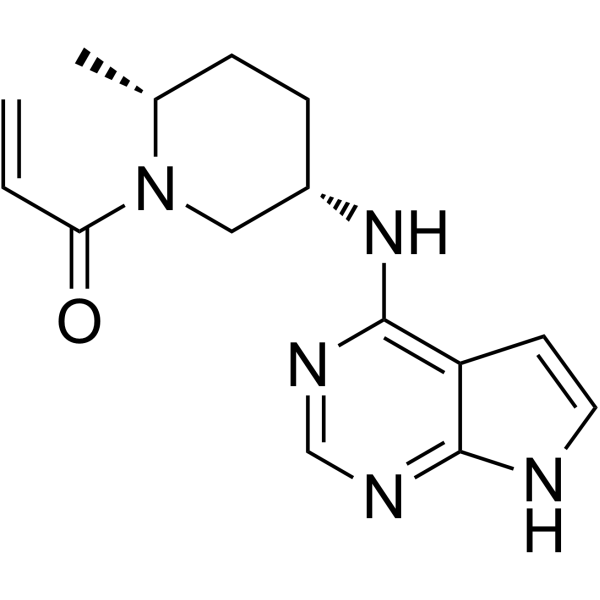
-
GC34971
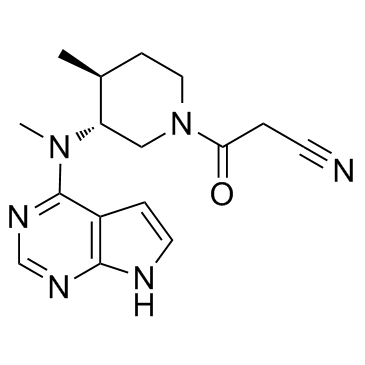
(3R,4S)-Tofacitinib
(3R,4S)-Tofacitinib ist ein weniger aktives Enantiomer von Tofacitinib.
-
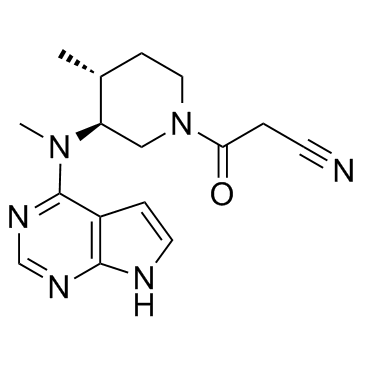
GC34972
(3S,4R)-Tofacitinib
(3S,4R)-Tofacitinib ist ein weniger aktives Enantiomer von Tofacitinib.
-
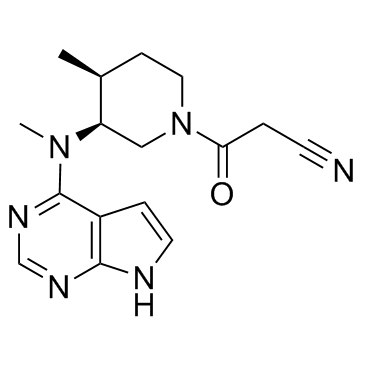
GC34973
(3S,4S)-Tofacitinib
(3S,4S)-Tofacitinib ist das weniger aktive S-Enantiomer von Tofacitinib.
-
GC41695
(6R,S)-5,6,7,8-Tetrahydrofolic Acid (hydrochloride)
Tetrahydrofolate, THFA
(6R,S)-5,6,7,8-Tetrahydrofolic acid (THFA), the reduced form of folic acid, serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.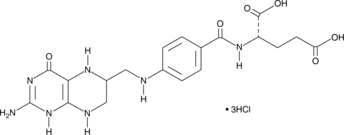
-
GC41088
(6S)-Tetrahydrofolic Acid
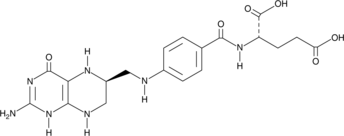
(6S)-Tetrahydrofolic acid is a diastereomer of tetrahydrofolic acid, a reduced form of folic acid that serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.
-
GC72591
(E,E)-RGFP966
(E,E)-RGFP966 is a selective and CNS permeable HDAC3 inhibitor that can be used for the research of Huntington’s disease.

-
GC61807
(E/Z)-AG490
(E/Z)-AG490 ((E/Z)-Tyrphostin AG490) ist eine racemische Verbindung der Isomere (E)-AG490 und (Z)-AG490. (E)-AG490 ist ein Tyrosinkinase-Inhibitor, der EGFR, Stat-3 und JAK2/3 hemmt.

-
GC63864
(E/Z)-Zotiraciclib hydrochloride
(E/Z)-TG02 hydrochloride; (E/Z)-SB1317 hydrochloride
(E/Z)-Zotiraciclib ((E/Z)-TG02)-Hydrochlorid ist ein potenter CDK2-, JAK2- und FLT3-Inhibitor.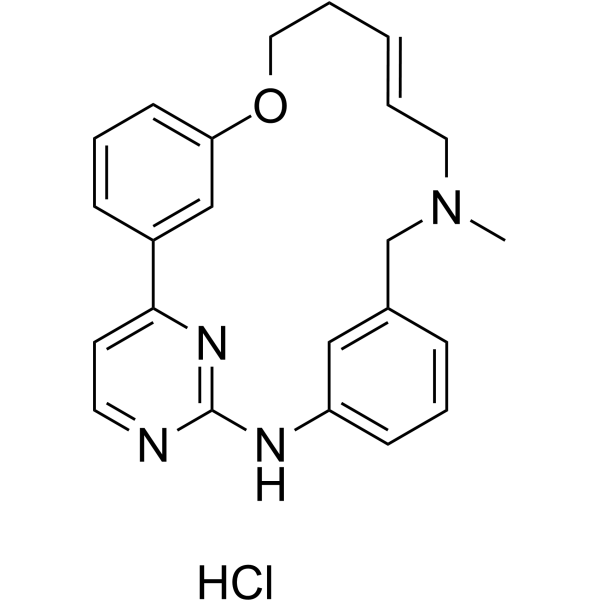
-
GC15104
(R)-(+)-Etomoxir sodium salt
(R)(+)Etomoxir
Etomoxir((R)-(+)-Etomoxir-Natriumsalz ist ein irreversibler Inhibitor der Carnitin-Palmitoyltransferase 1a (CPT-1a), hemmt die FettsÄureoxidation (FAO) durch CPT-1a und hemmt die Palmitat-β-Oxidation bei Mensch und Ratte und Meerschweinchen.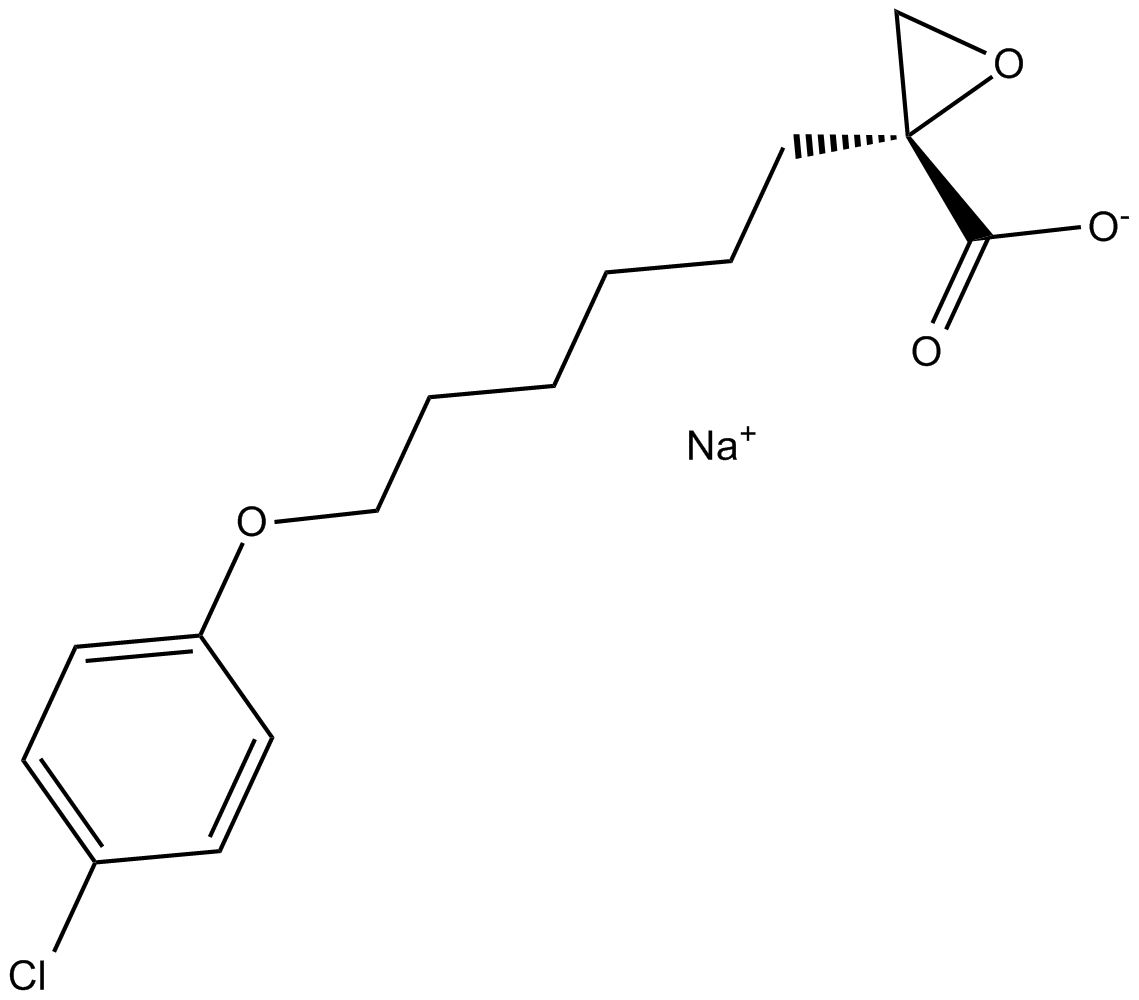
-
GC63791
(R)-(-)-JQ1 Enantiomer
(R)-(-)-JQ1 Enantiomer ist das Stereoisomer von (+)-JQ1. (+)-JQ1 verringert stark die Expression beider BRD4-Zielgene, wÄhrend (R)-(-)-JQ1-Enantiomer keine Wirkung hat.
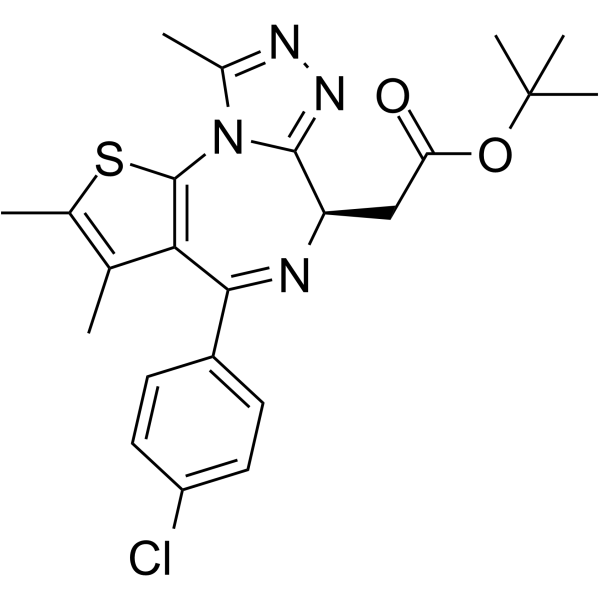
-
GC45908
(R)-BAY-598
An inhibitor of SMYD2
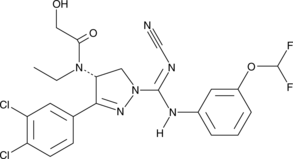
-
GC34443
(R)-BAY1238097
(R)-BAY1238097 ist das R-Isomer mit geringerer AktivitÄt von BAY1238097. BAY1238097 ist ein potenter und selektiver Inhibitor der BET-Bindung an Histone und hat eine starke antiproliferative AktivitÄt in verschiedenen AML- (akute myeloische LeukÄmie) und MM- (multiples Myelom) Modellen durch Herunterregulierung der c-Myc-Spiegel und seines nachgeschalteten Transkriptoms.
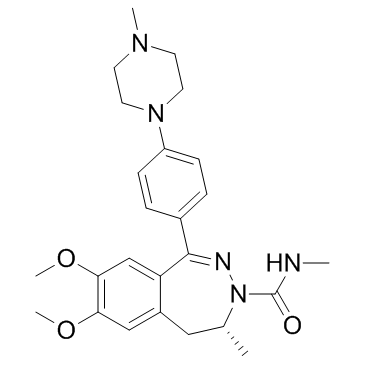
-
GC64210
(R)-GSK-3685032
(R)-GSK-3685032 ist das R-Enantiomer von GSK-3685032. GSK-3685032 ist ein nicht zeitabhÄngiger, nicht kovalenter, erster reversibler DNMT1-selektiver Inhibitor seiner Klasse mit einem IC50-Wert von 0,036 μM. GSK-3685032 induziert einen starken Verlust der DNA-Methylierung, Transkriptionsaktivierung und Wachstumshemmung von Krebszellen.
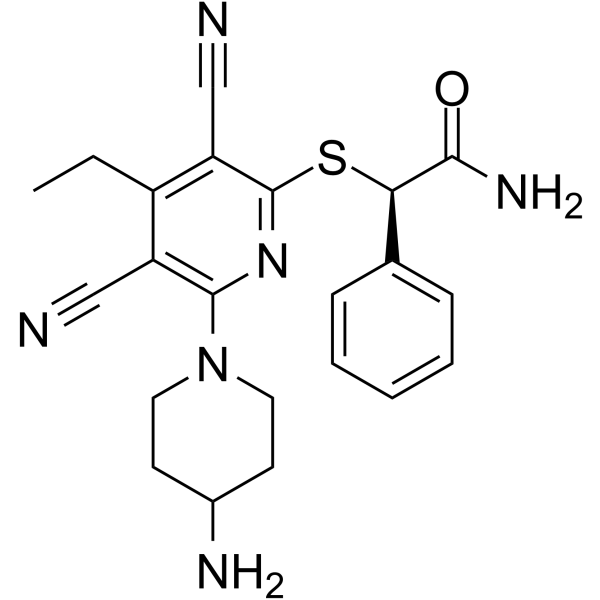
-
GC70906
(R)-HH2853
(R)-HH2853 is a mutant EZH2 inhibitor with an IC50 of <100 nM for EZH2-Y641F.

-
GC11340
(R)-PFI 2 hydrochloride
(-)PFI2
PFI-2 ((R)-(R)-PFI 2 Hydrochlorid) Hydrochlorid ist eine potente und selektive SET-DomÄne, die den Lysinmethyltransferase 7 (SETD7)-Inhibitor enthÄlt.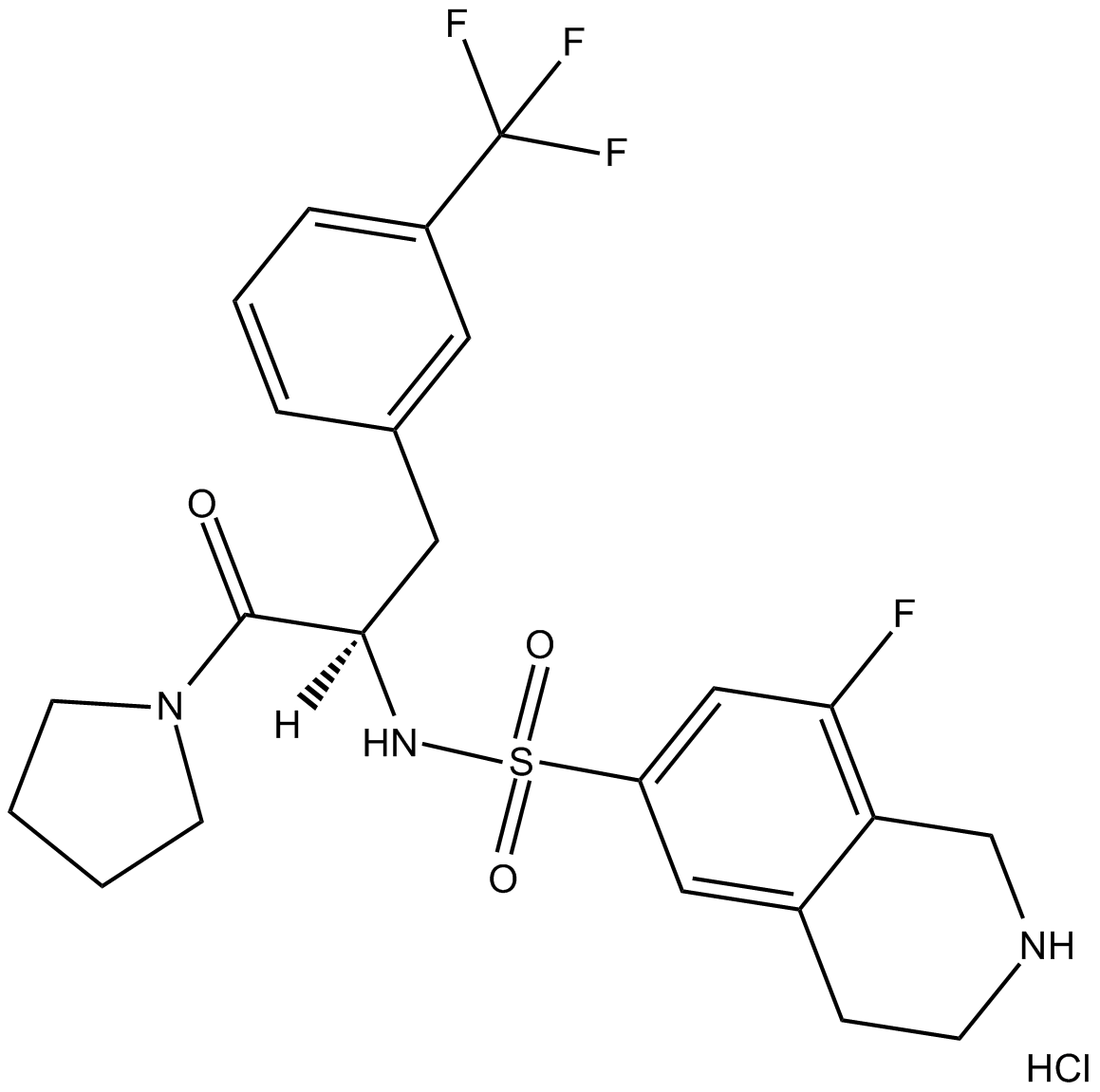
-
GC69794
(Rac)-Arnebin 1
(Rac)-β,β-Dimethylacrylalkannin; (Rac)-β,β-?Dimethylacrylshikonin
(Rac)-Arnebin 1 ((Rac)-β,β-Dimethylacrylalkannin) ist das Racemat von β,β-Dimethylacrylalkannin und/oder β,β-Dimethylacrylshikonin. Diese beiden Verbindungen sind Naphthochinone, die aus Pflanzen der Gattung Lithospermum isoliert werden können. β,β-Dimethylacrylshikonin besitzt antitumorale Aktivität.
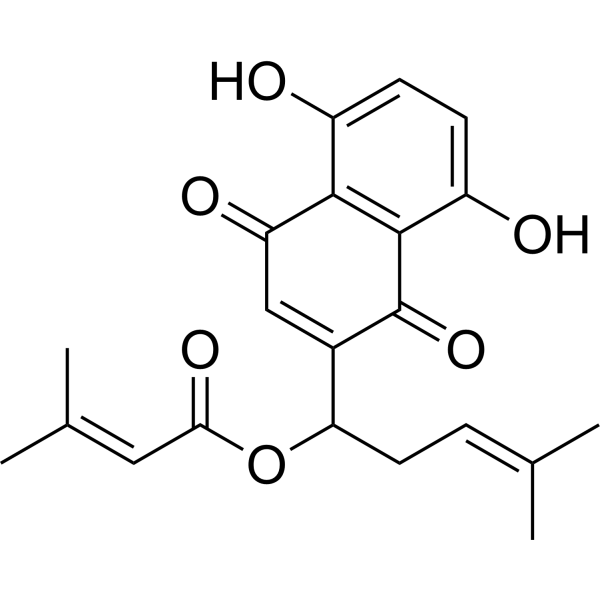
-
GC34446
(rac)-BAY1238097
(Rac)-BAY1238097 ist ein BET-Inhibitor mit einem IC50-Wert von 1,02 μM fÜr BRD4. Wird in der Krebsforschung verwendet.
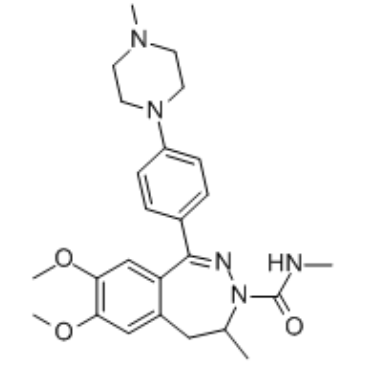
-
GC60004
(rel)-Tranylcypromine D5 hydrochloride
Tranylcypromin-d5 (SKF 385-d5)-Hydrochlorid ist ein mit Deuterium markiertes (rel)-Tranylcypromin-Hydrochlorid.
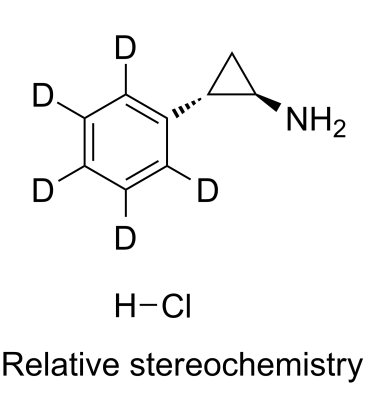
-
GC33198
(S)-JQ-35 (TEN-010)
TEN-010
(S)-JQ-35 (TEN-010) (TEN-010) ist ein Inhibitor der BromodomÄne und der Familie der extraterminalen (BET) BromodomÄne enthaltenden Proteine mit potenzieller antineoplastischer AktivitÄt.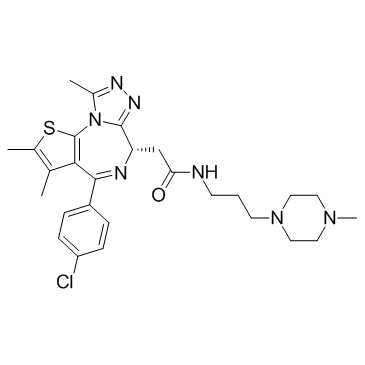
-
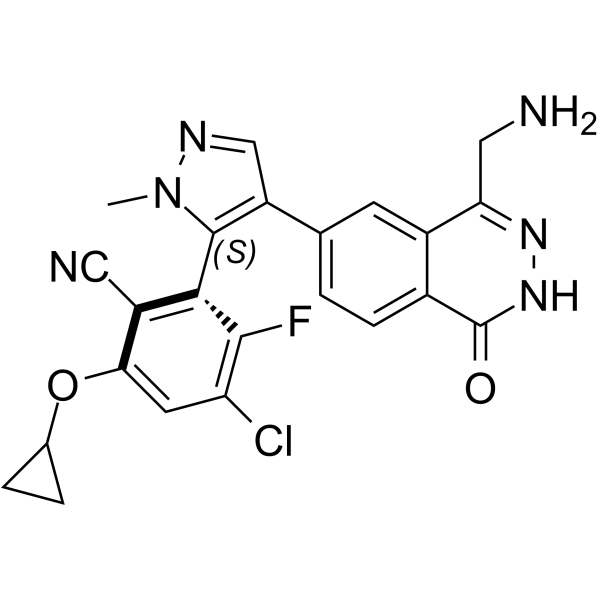
GC65045
(S)-MRTX-1719
(S)-MRTX-1719 (Beispiel 16-7) ist das S-Enantiomer von MRTX-1719. (S)-MRTX-1719 ist ein PRMT5/MTA-Komplex-Inhibitor mit einem IC50-Wert von 7070 nM.
-
GC13634
(S)-PFI-2 (hydrochloride)
(+)-PFI-2
Negative control of (R)-PFI 2 hydrochloride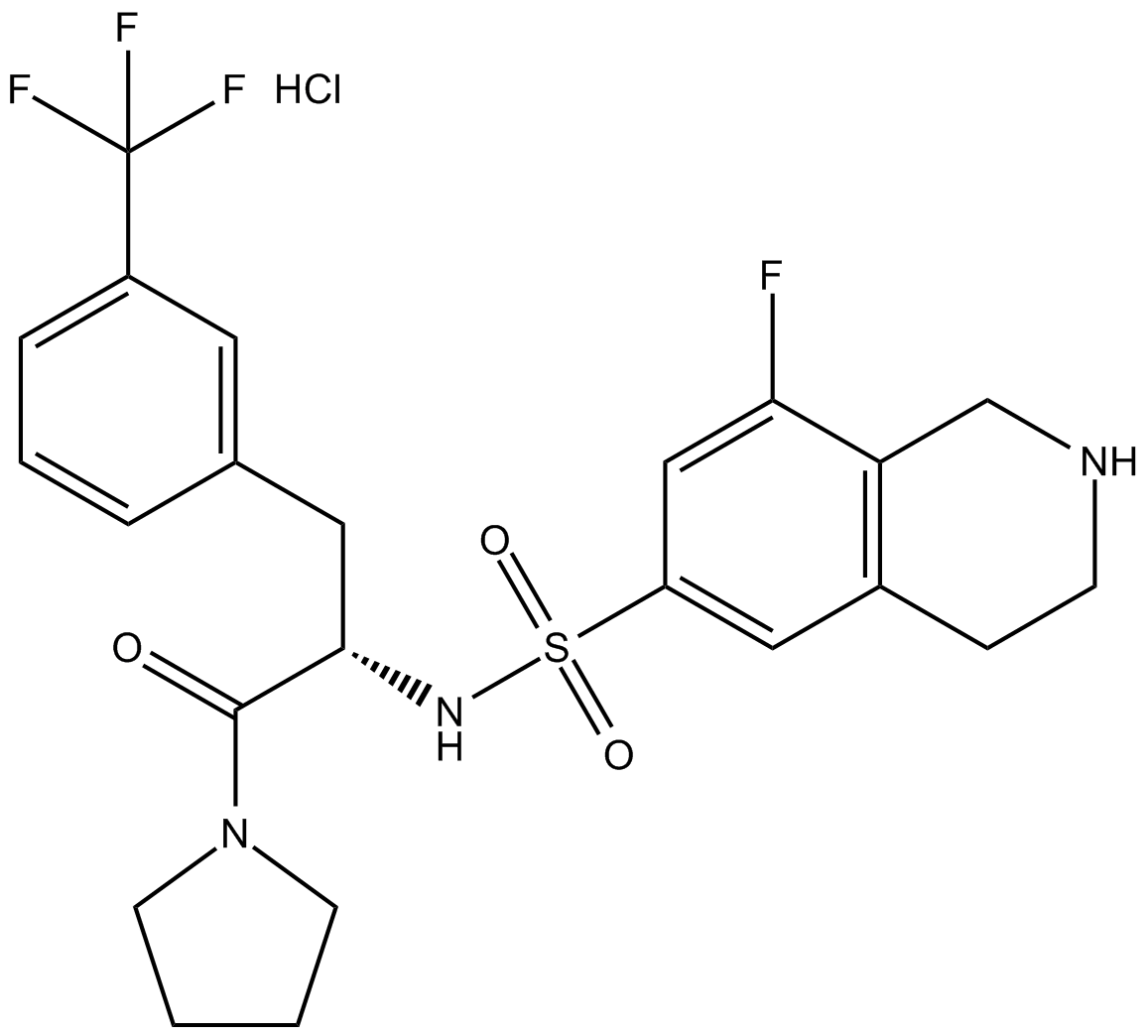
-
GC65133
(S,R,S)-AHPC-C5-COOH
VH032-C5-COOH
(S,R,S)-AHPC-C5-COOH (VH032-C5-COOH) ist ein synthetisiertes E3-Ligase-Ligand-Linker-Konjugat, das den VH032-VHL-basierten Liganden und einen Linker zur Bildung von PROTACs enthÄlt. VH-032 ist ein selektiver und potenter Inhibitor der VHL/HIF-1α-Wechselwirkung mit einem Kd von 185 nM und hat das Potenzial fÜr die Untersuchung von AnÄmie und ischÄmischen Erkrankungen.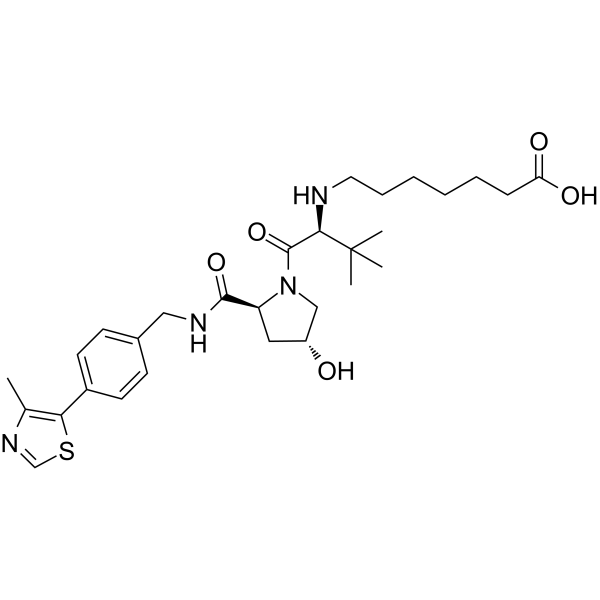
-
GC17055
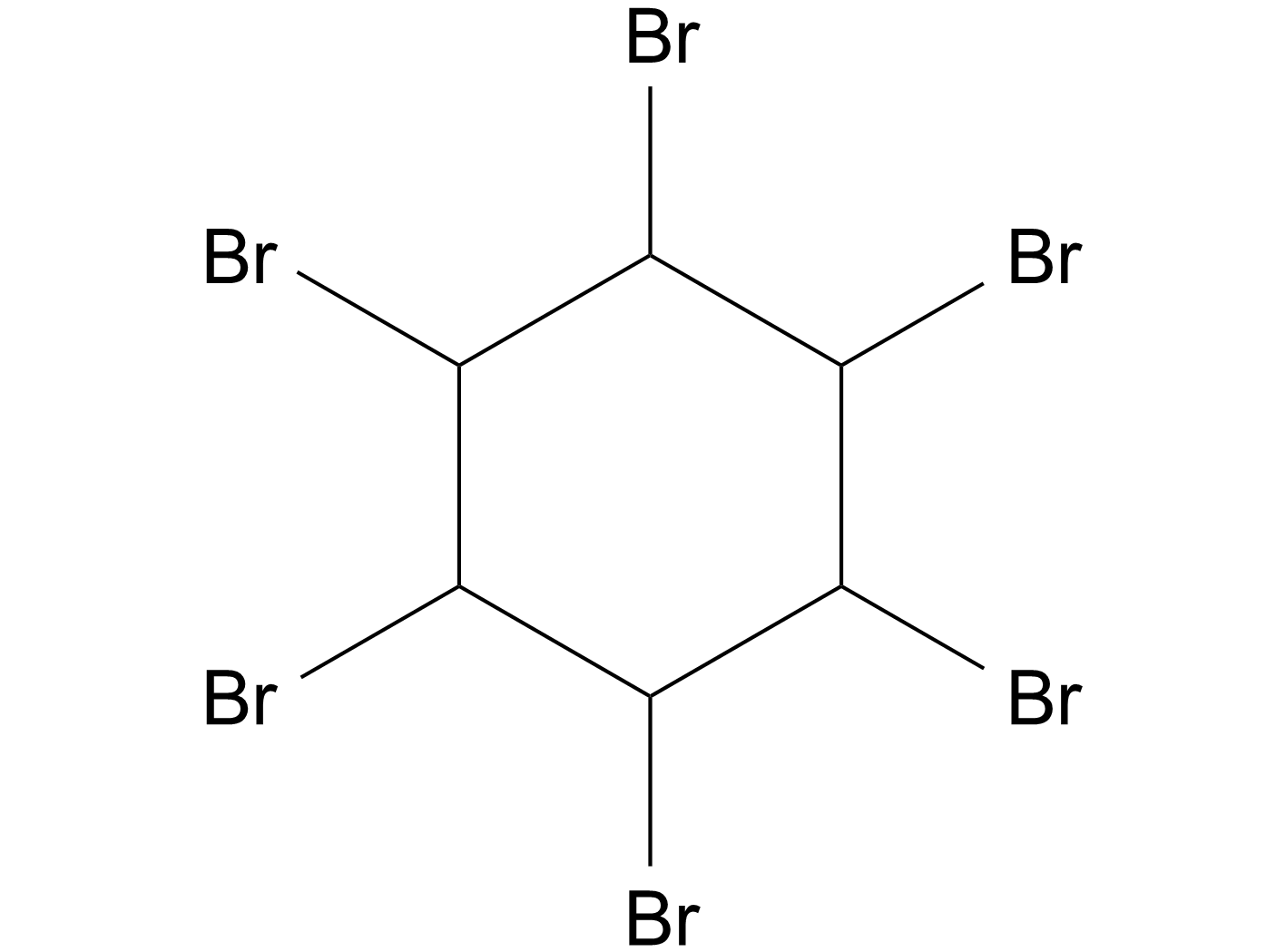
1,2,3,4,5,6-Hexabromocyclohexane
1,2,3,4,5,6-六溴环己烷(1,2,3,4,5,6-Hexabromocyclohexane)直接抑制JAK2激酶结构域内的口袋,抑制自身磷酸化。
-
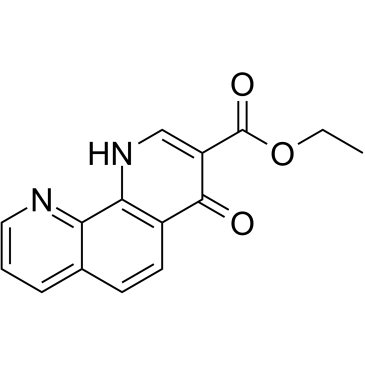
GC61949
1,4-DPCA ethyl ester
1,4-DPCA-Ethylester ist der Ethylester von 1,4-DPCA und kann Faktor-inhibierendes HIF (FIH) hemmen.
-
GC40468
1,5-Isoquinolinediol
NSC 65585
1,5-Isochinolindiol ist ein potenter PARP-Hemmer mit einem IC50-Wert von 0,18-0,37 μM.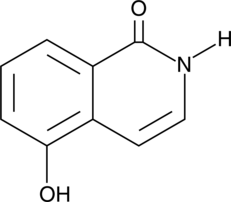
-
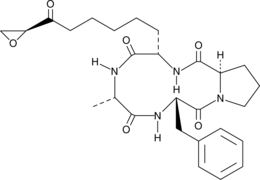
GC41984
1-Alaninechlamydocin
1-Alaninechlamydocin, ein zyklisches Tetrapeptid, ist ein potenter HDAC-Inhibitor (IC50 = 6,4 nM). 1-Alaninechlamydocin induziert G2/M-Zellzyklusarrest und Apoptose in MIA-PaCa-2-Zellen.
-
GC39214
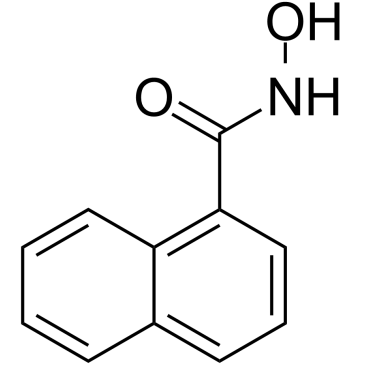
1-Naphthohydroxamic acid
1-NaphthohydroxamsÄure (Verbindung 2) ist ein potenter und selektiver HDAC8-Inhibitor mit einem IC50 von 14 μM. 1-NaphthohydroxamsÄure ist selektiver fÜr HDAC8 als Klasse-I-HDAC1 und Klasse-II-HDAC6 (IC50 > 100 μM). 1-NaphthohydroxamsÄure erhÖht die globale Histon-H4-Acetylierung nicht und verringert auch nicht die intrazellulÄre HDAC-GesamtaktivitÄt. 1-NaphthohydroxamsÄure kann die Tubulinacetylierung induzieren.
-
GC46508
2',2'-Difluoro-2'-deoxyuridine
dFdU
An active metabolite of gemcitabine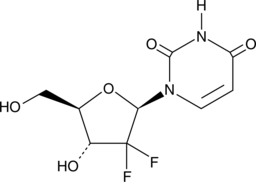
-
GC12775
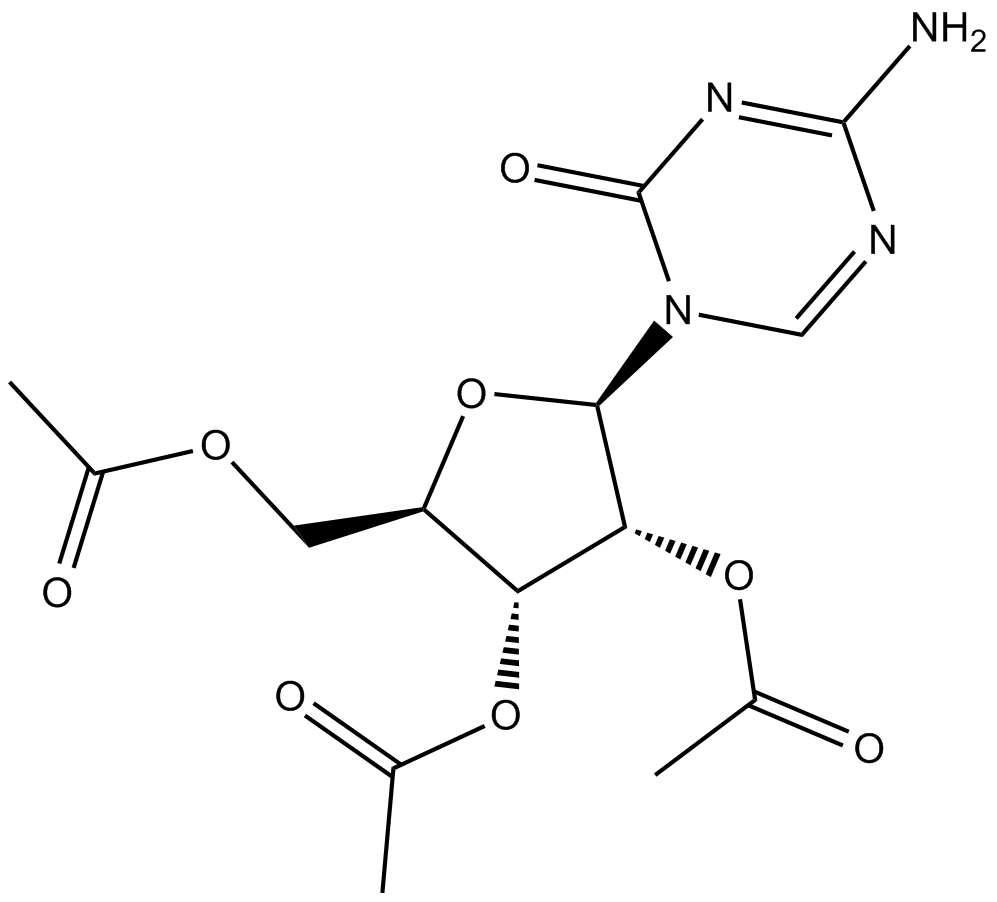
2',3',5'-triacetyl-5-Azacytidine
2',3',5'-Triacetyl-5-Azacytidin ist ein oral wirksames Prodrug von 5-Azacytidin. 5-Azacytidin ist ein Inhibitor der DNA-Methyltransferase.
-
GC16195
2,4-DPD
2,4-Diethylpyridine dicarboxylate
Diethylpyridin-2,4-dicarb ist ein potenter Prolyl-4-hydroxylase-gerichteter Proinhibitor.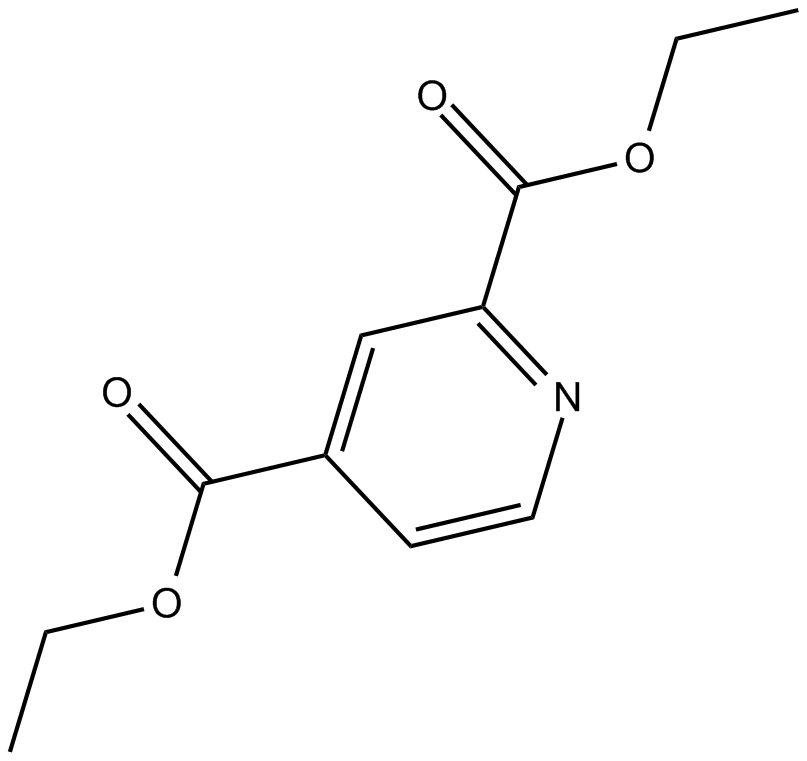
-
GC11873
2,4-Pyridinedicarboxylic Acid
2,4-PyridindicarbonsÄure (2,4-PyridindicarbonsÄure) ist ein Breitband-Inhibitor der 2OG-Oxygenase, einschließlich der JmjC-DomÄnen-enthaltenden Familie der Histon-Demethylasen (JHDMs). 2,4-PyridindicarbonsÄure ist eine Zielchemikalie im Bereich der biobasierten Kunststoffe.
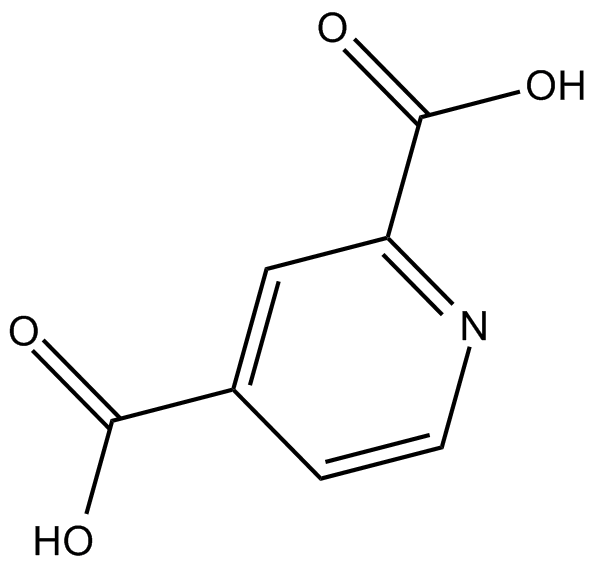
-
GC42075
2,4-Pyridinedicarboxylic Acid (hydrate)
2,4-Dicarboxylpyridine, 2,4-PDCA
2,4-Pyridinedicarboxylic Acid (2,4-PDCA) is a compound that structurally mimics 2-oxoglutarate (2-OG, also known as α-ketoglutarate) and chelates zinc, thus affecting a range of enzymes.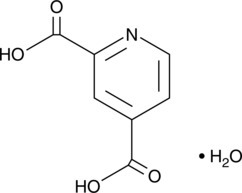
-
GC62233
2,6-Dichloro-N-(2-(cyclopropanecarboxamido)pyridin-4-yl)benzamide
GDC-046 ist ein potenter, selektiver und oral bioverfÜgbarer TYK2-Inhibitor mit Kis von 4,8, 0,7, 0,7 und 0,4 nM fÜr TYK2, JAK1, JAK2 bzw. JAK3.
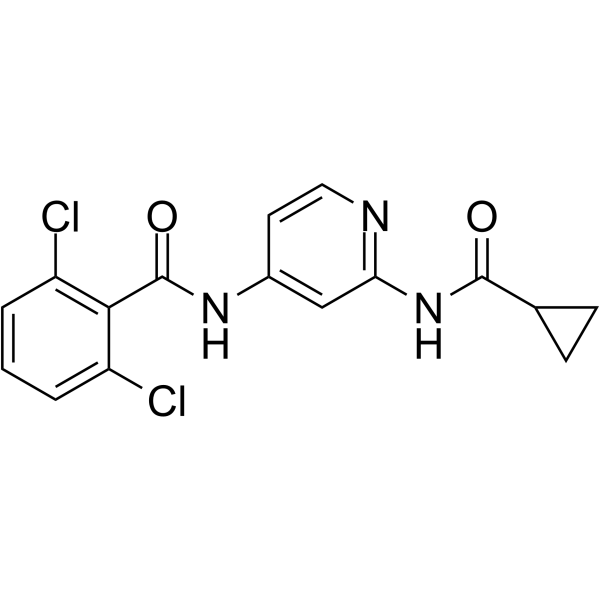
-
GC46503
2-(1,8-Naphthyridin-2-yl)phenol
2-NP
2-(1,8-Naphthyridin-2-yl)phenol ist ein selektiver VerstÄrker der STAT1-Transkription. 2-(1,8-Naphthyridin-2-yl)phenol kann die FÄhigkeit von IFN-γ verstÄrken; um die Proliferation menschlicher Brustkrebs- und Fibrosarkomzellen zu hemmen.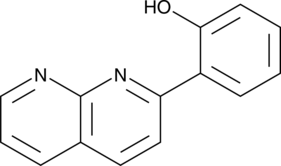
-
GC10408
2-D08
2-D08 ist ein zellgÄngiger, mechanistisch einzigartiger Inhibitor der Protein-SUMOylierung. 2-D08 hemmt auch Axl mit einem IC50 von 0,49 nM.
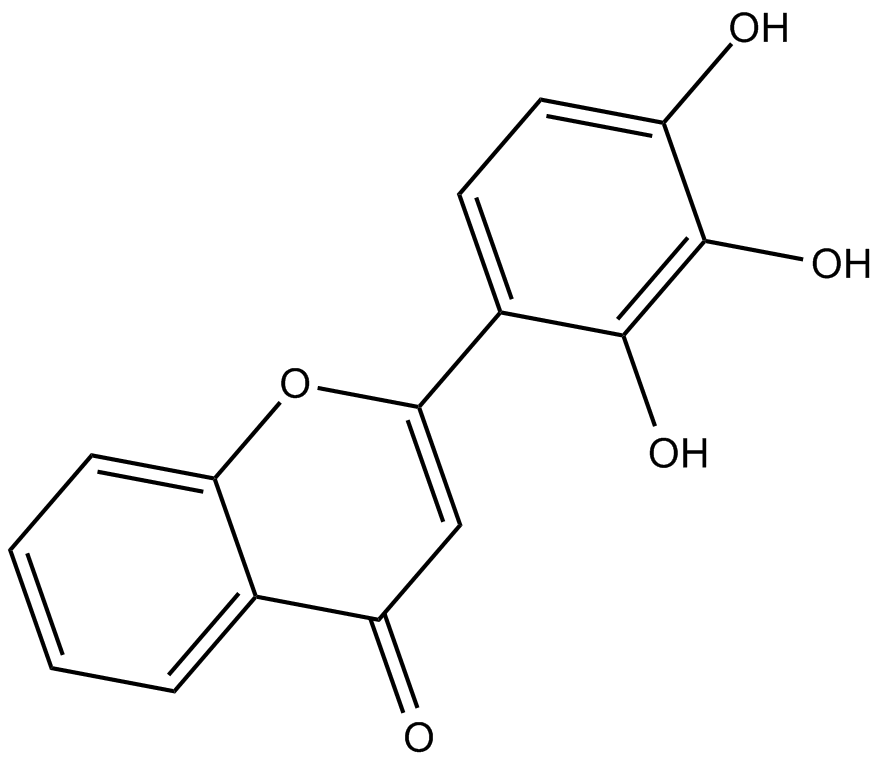
-
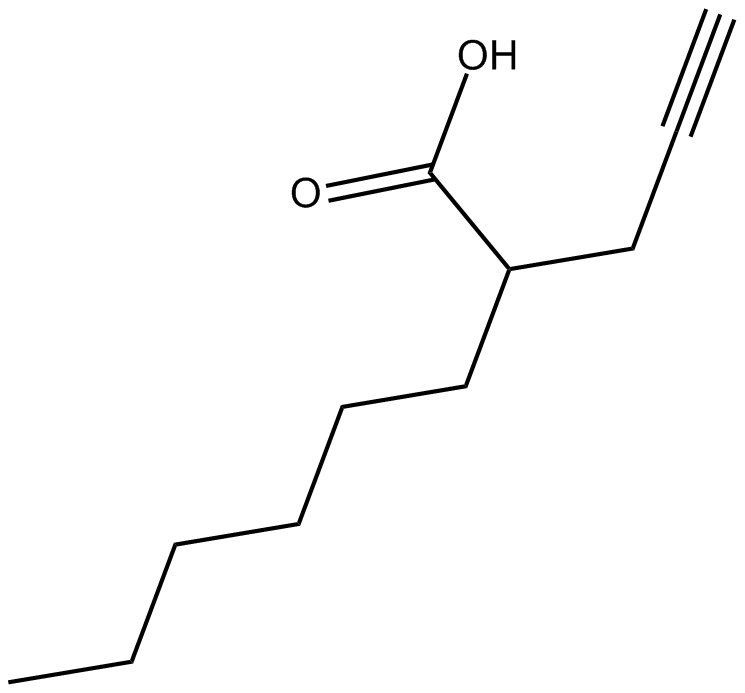
GC11249
2-hexyl-4-Pentynoic Acid
Potent and robust HDACs inhibitor
-
GC15084
2-Methoxyestradiol (2-MeOE2)
2Hydroxyestradiol 2methyl ether, 2ME2, NSC 659853, Panzem
2-Methoxyestradiol (2-MeOE2/2-Me) is an HIF-1α inhibitor.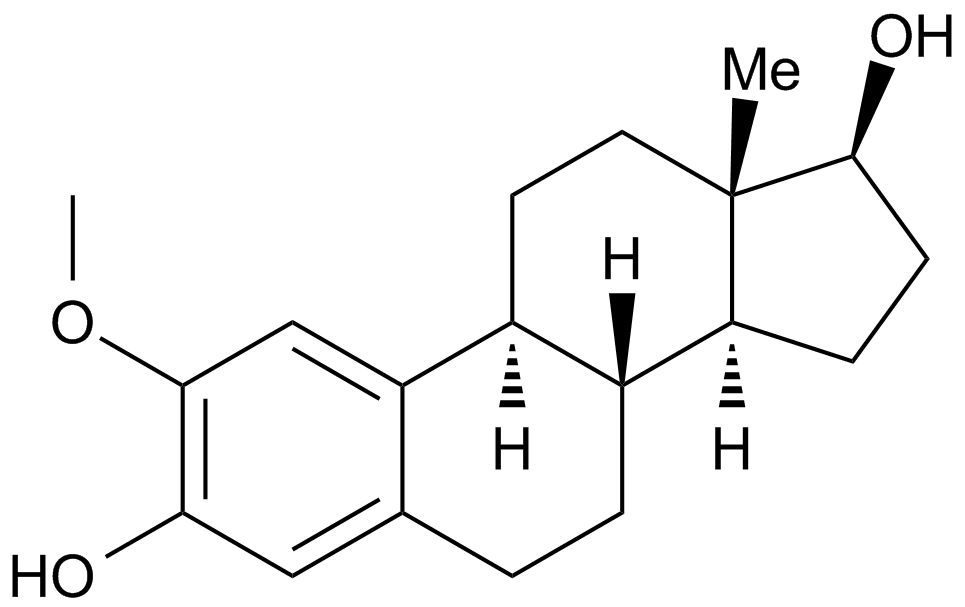
-
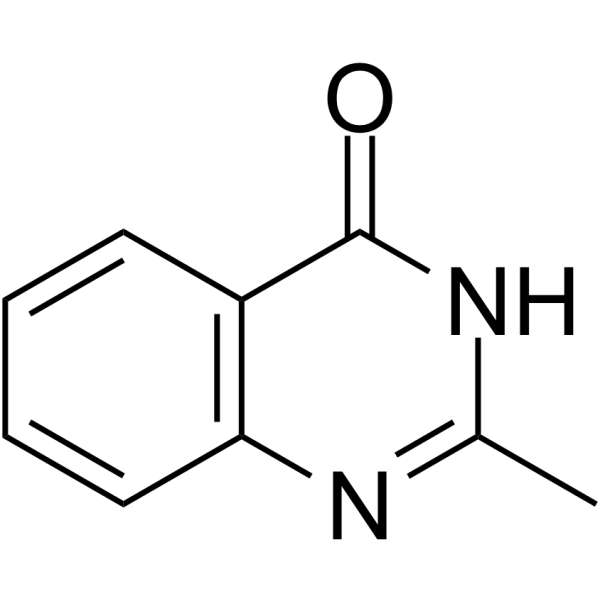
GC62781
2-Methylquinazolin-4-ol
2-Methylchinazolin-4-ol ist ein potenter kompetitiver Poly(ADP-Ribose)-Synthetase-Inhibitor mit einem Ki von 1,1 μM.
-
GC14282
3-acetyl-11-keto-β-Boswellic Acid
3-O-acetyl-11-keto-β-Boswellic acid,AKBA
3-Acetyl-11-keto-β-BoswelliasÄure (Acetyl-11-keto-β-BoswelliasÄure) ist eine aktive Triterpenoidverbindung aus dem Extrakt von Boswellia serrate und ein neuartiger Nrf2-Aktivator.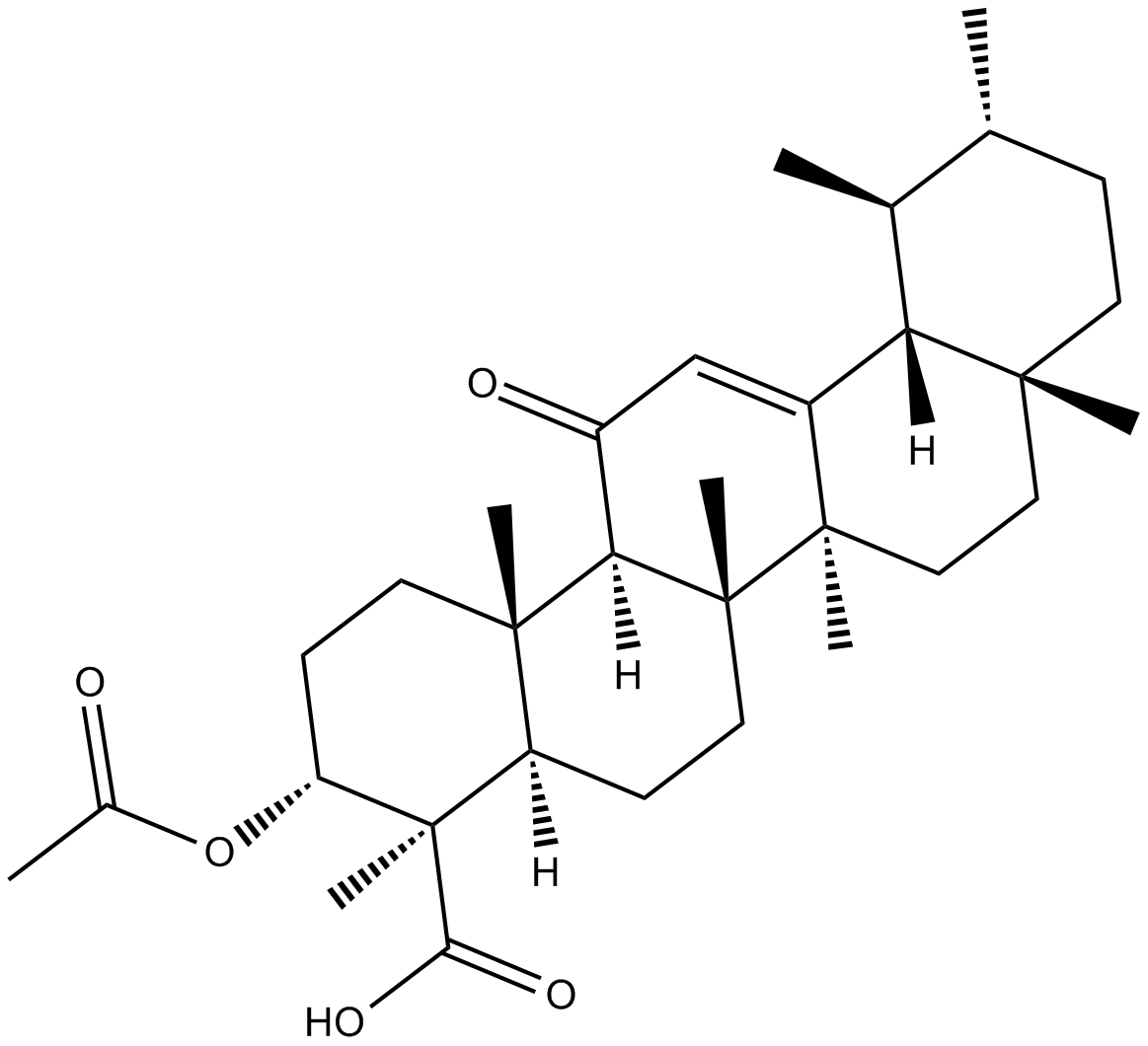
-
GC17907
3-Deazaneplanocin A (DZNep) hydrochloride
2,3DMMC
Ein Inhibitor der Lysin-Methyltransferase EZH2.
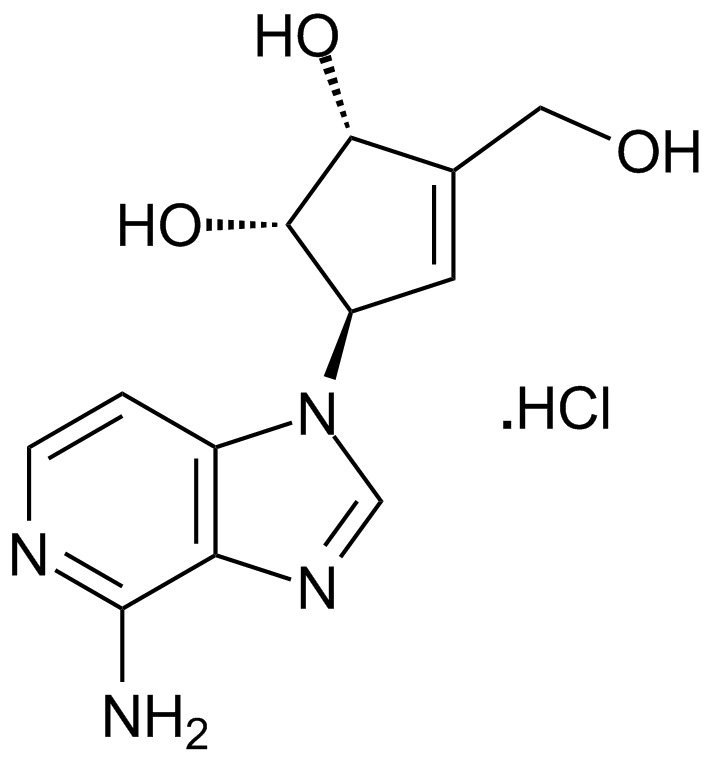
-
GC13145
3-Deazaneplanocin,DZNep
DZNep, NSC 617989
An inhibitor of lysine methyltransferase EZH2
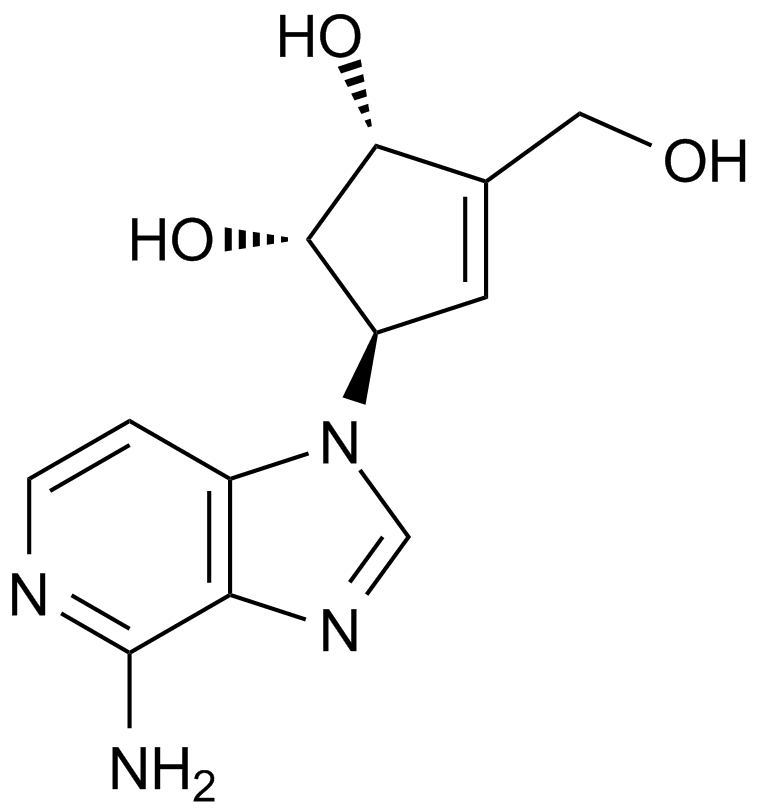
-
GC18509
3-Methylcytidine (methosulfate)
3-Methylcytidine (methosulfate) is a cytidine derivative used as an internal standard for HPLC.
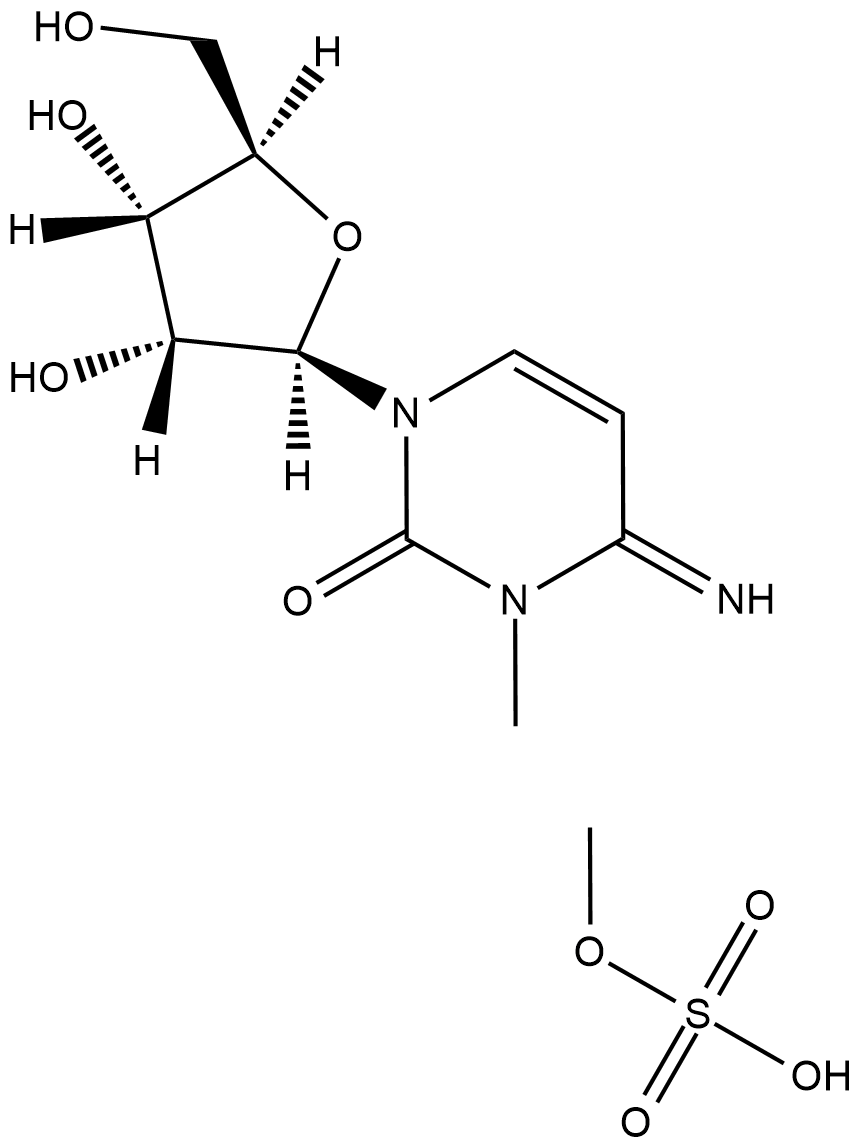
-
GC19013
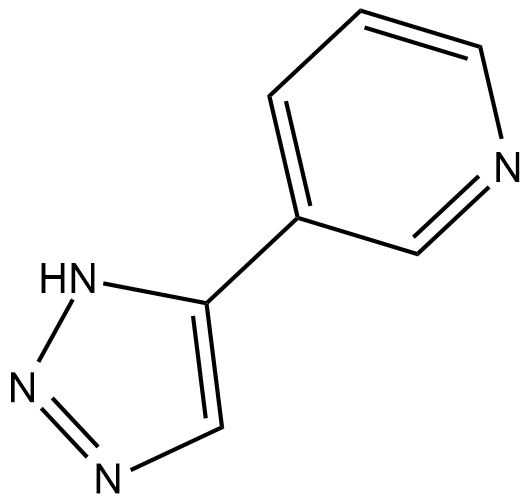
3-TYP
3-TYP hemmt SIRT3 mit einem IC50-Wert von 16 nM und ist gegenüber SIRT1 und SIRT2 mit IC50-Werten von 88 nM bzw. 92 nM potenter.
-
GC62805
4’-Methoxychalcone
4’-Methoxychalcon reguliert die Adipozytendifferenzierung durch PPARγ-Aktivierung.

-
GC17922
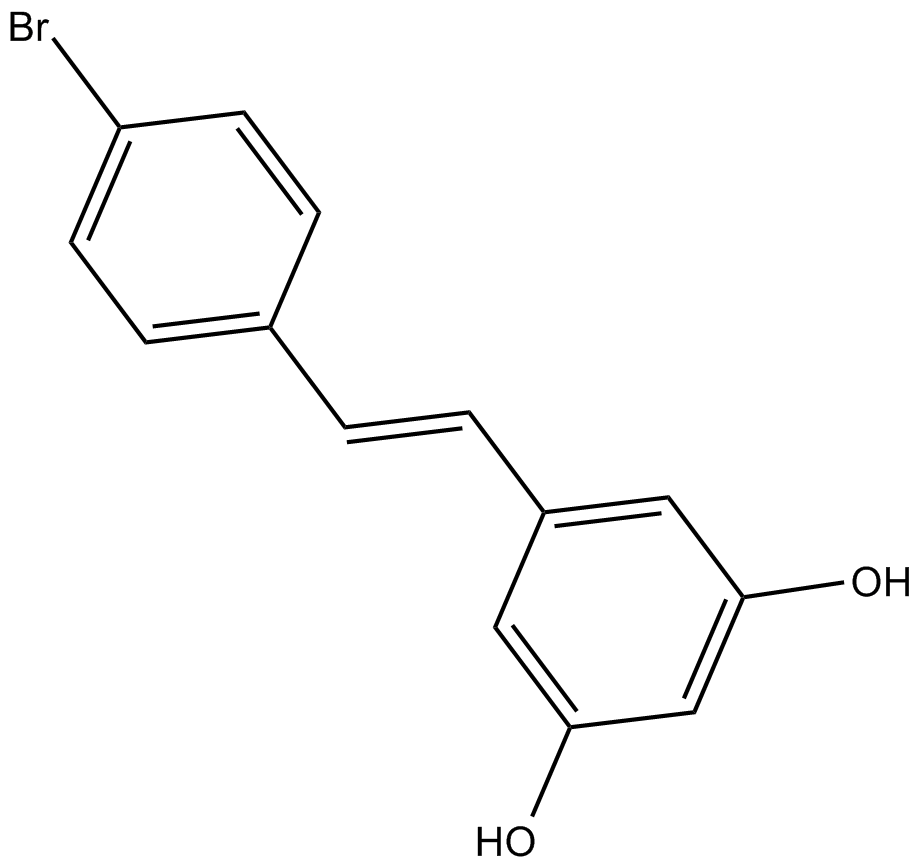
4'-bromo-Resveratrol
4'-Brom-Resveratrol ist ein potenter und dualer Inhibitor von Sirtuin-1 und Sirtuin-3. 4'-Brom-Resveratrol hemmt das Wachstum von Melanomzellen durch die Umprogrammierung des mitochondrialen Stoffwechsels. 4'-Brom-Resveratrol verleiht Melanomzellen durch eine metabolische Umprogrammierung antiproliferative Wirkungen und beeinflusst den Zellzyklus und die Apoptose-Signalgebung.
-
GC46607
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone
NNK
A tobacco-specific nitrosamine carcinogen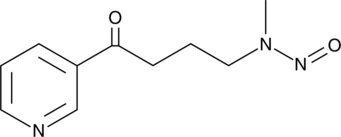
-
GC11761
4-amino-1,8-Naphthalimide
4-Aminonaphthalimide,4-ANI
4-Amino-1,8-Naphthalimid ist ein potenter PARP-Hemmer und potenziert die ZytotoxizitÄt von ⋳-Strahlung in Krebszellen.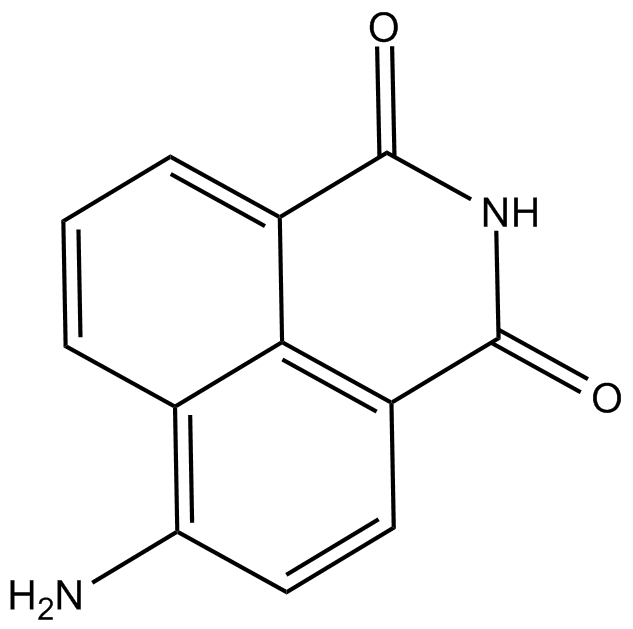
-
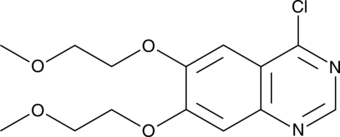
GC46628
4-Chloro-6,7-bis(2-methoxyethoxy)quinazoline
A building block and synthetic intermediate
-
GC17005
4-iodo-SAHA
ISAHA
4-Iod-SAHA (1k) ist ein oral aktiver Klasse-I- und Klasse-II-Histondeacetylase (HDAC)-Hemmer mit EC50-Werten von 1,1, 0,95, 0,12, 0,24, 0,85 und 1,3 μM fÜr Skbr3-, HT29-, U937-, JA16- und HL60-Zelllinien , beziehungsweise. 4-Iodo-SAHA (1k) kann fÜr die Krebsforschung verwendet werden.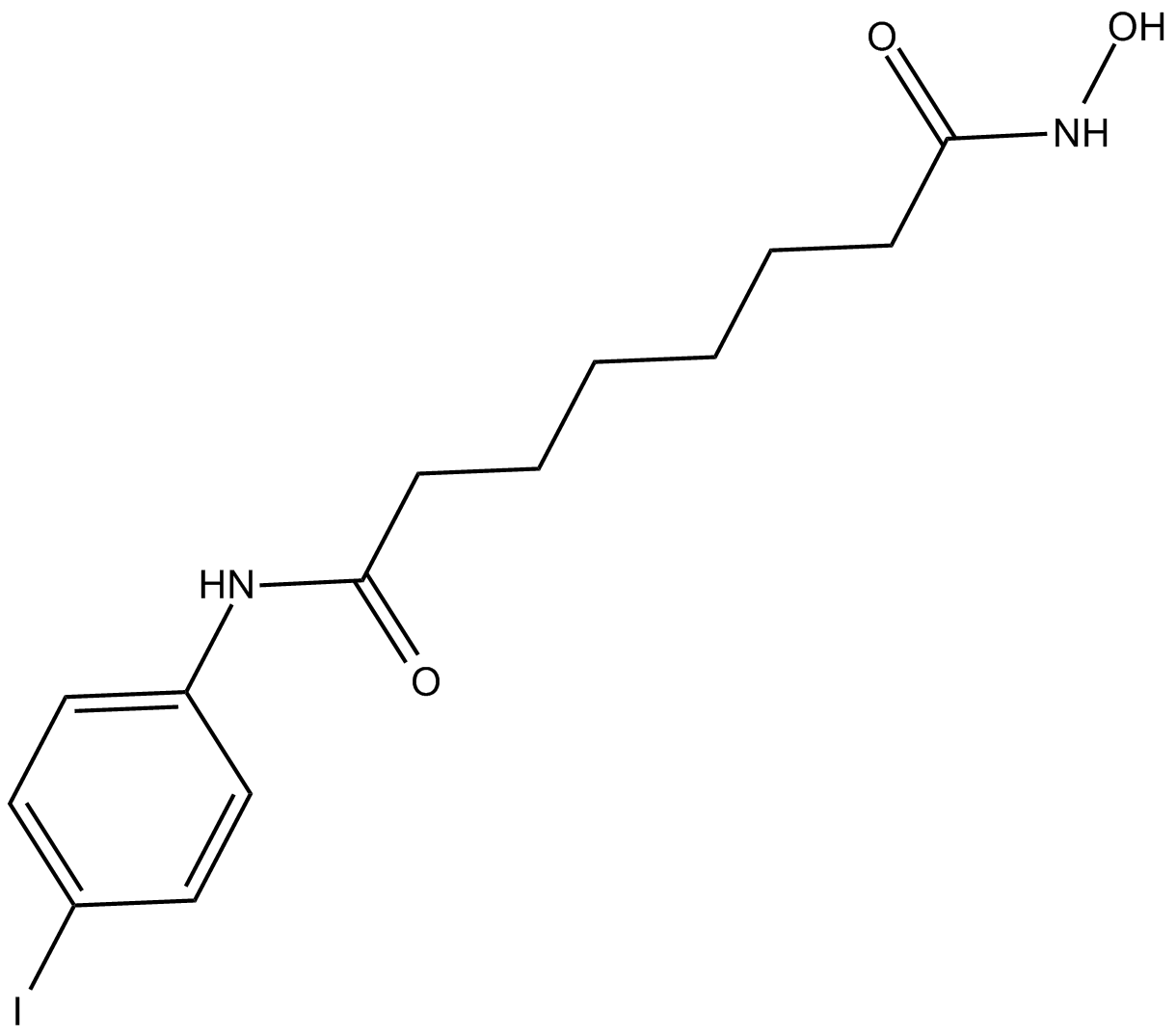
-
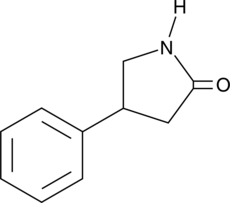
GC46675
4-Phenyl-2-pyrrolidinone
A precursor and synthetic intermediate
-
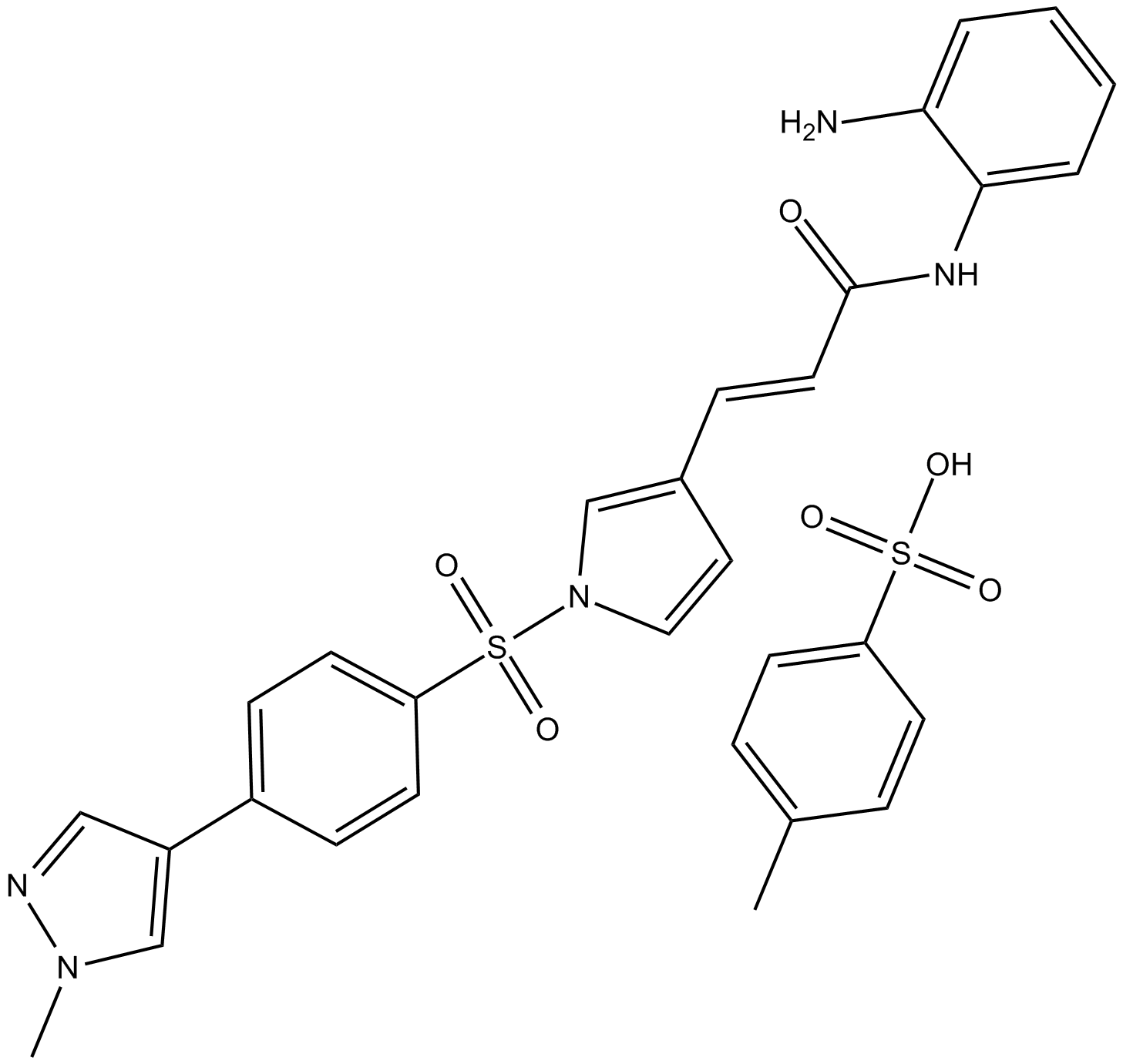
GC12667
4SC-202
4SC-202 (4SC-202) ist ein selektiver HDAC-Inhibitor der Klasse I mit einem IC50-Wert von 1,20 μM, 1,12 μM und 0,57 ⋼M fÜr HDAC1, HDAC2 bzw. HDAC3. Es zeigt auch eine hemmende AktivitÄt gegen Lysin-spezifische Demethylase 1 (LSD1).
-
GC35150
5,7,4'-Trimethoxyflavone
5,7,4'-Trimethoxyflavon wird aus Kaempferia parviflora (KP) isoliert, einer berühmten Heilpflanze aus Thailand. 5,7,4'-Trimethoxyflavon induziert Apoptose, wie durch Inkremente der Sub-G1-Phase, DNA-Fragmentierung, Annexin-V/PI-Färbung, das Bax/Bcl-xL-Verhältnis, proteolytische Aktivierung von Caspase-3 und Abbau von Poly belegt wird (ADP-Ribose)-Polymerase (PARP)-Protein.5,7,4'-Trimethoxyflavon ist bei der konzentrationsabhängigen Hemmung der Proliferation von menschlichen SNU-16-Magenkrebszellen signifikant wirksam.
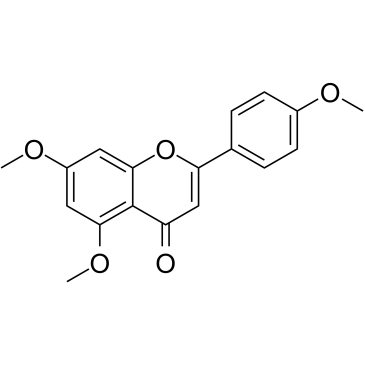
-
GN10629
5,7-dihydroxychromone
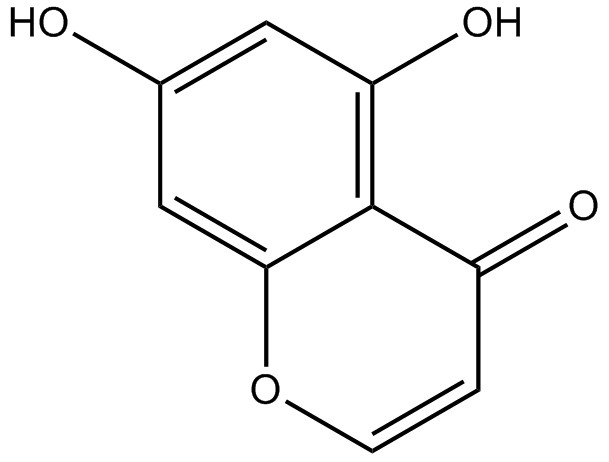
-
GC10898
5-(N,N-dimethyl)-Amiloride (hydrochloride)
DMA,L-591,605,MK-685
NHE1, NHE2, and NHE3 inhibitor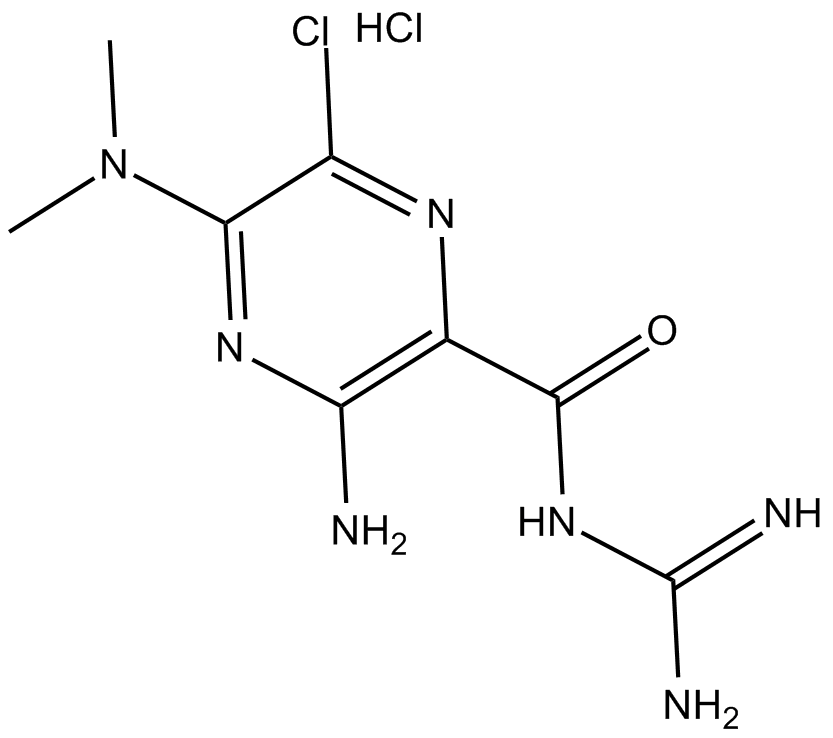
-
GC68161
5-AIQ
5-Aminoisoquinolin-1-one
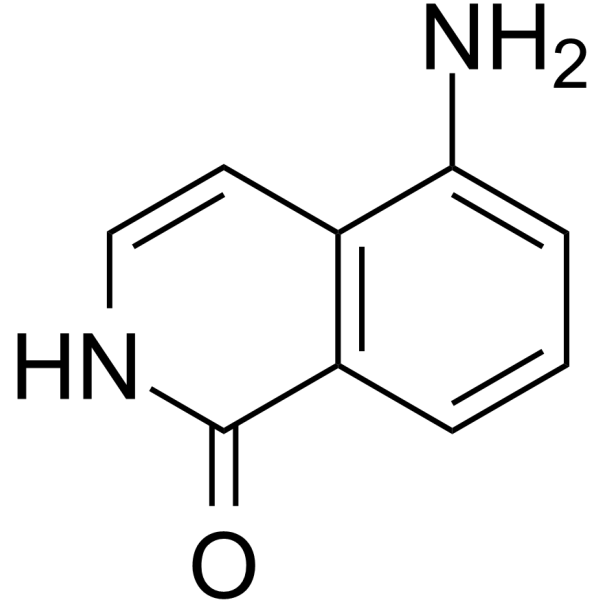
-
GC10946
5-Azacytidine
Antibiotic U 18496, 5AzaC, Ladakamycin, Mylosar, NSC 102816, NSC 103627, U 18496, WR 183027
5-Azacytidine (also known as 5-AzaC), a compound belonging to a class of cytosine analogues, is a DNA methyl transferase (DNMT) inhibitor that exerts potent cytotoxicity against multiple myeloma (MM) cells, including MM.1S, MM.1R, RPMI-8266, RPMI-LR5, RPMI-Dox40 and Patient-derived MM, with the half maximal inhibition concentration IC50 values of 1.5 μmol/L, 0.7 μmol/L, 1.1 μmol/L, 2.5 μmol/L, 3.2 μmol/L and 1.5 μmol/L respectively.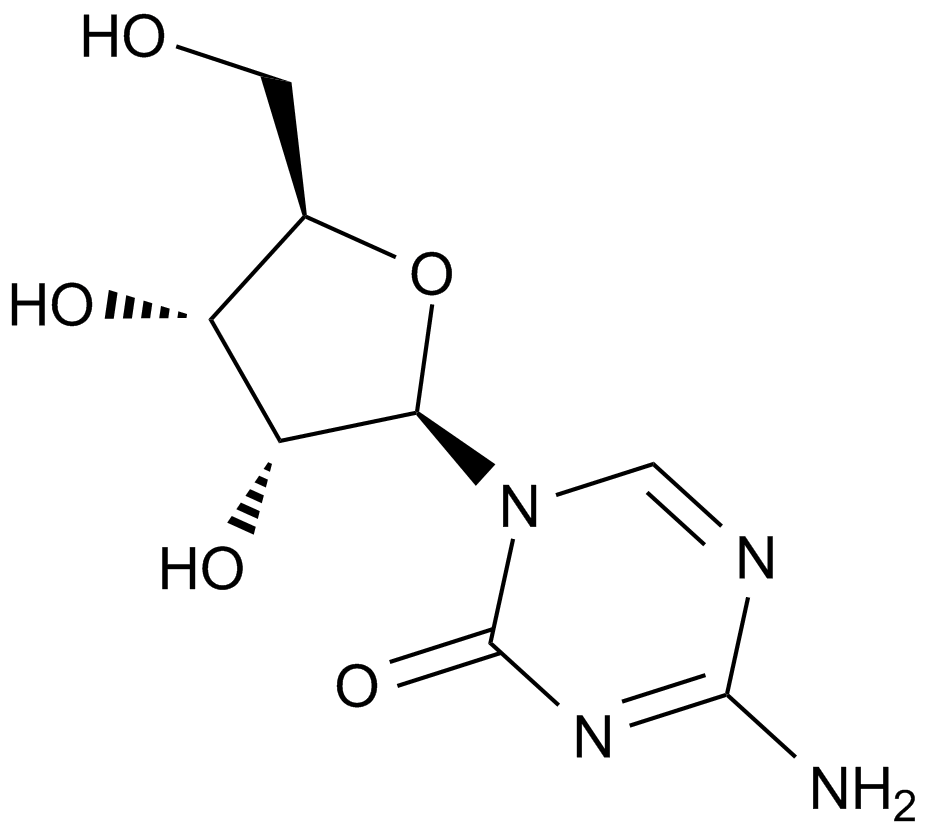
-
GC61662
5-Fluoro-2'-deoxycytidine
5-Fluor-2'-desoxycytidin, ein Fluorpyrimidin-Nukleosid-Analogon, ist ein DNA-Methyltransferase (DNMT)-Inhibitor. 5-Fluor-2'-desoxycytidin ist ein tumorselektives Prodrug des potenten Thymidylatsynthase-Inhibitors 5-Fluor-2'-dUMP.

-
GC42562
5-Methyl-2'-deoxycytidine
5Methyldeoxycytidine
5-Methyl-2'-desoxycytidin in einzelstrÄngiger DNA kann in cis wirken, um die De-novo-DNA-Methylierung zu signalisieren.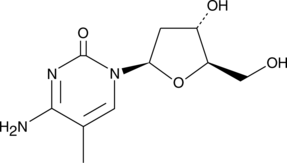
-
GC66055
5-Phenylpentan-2-one
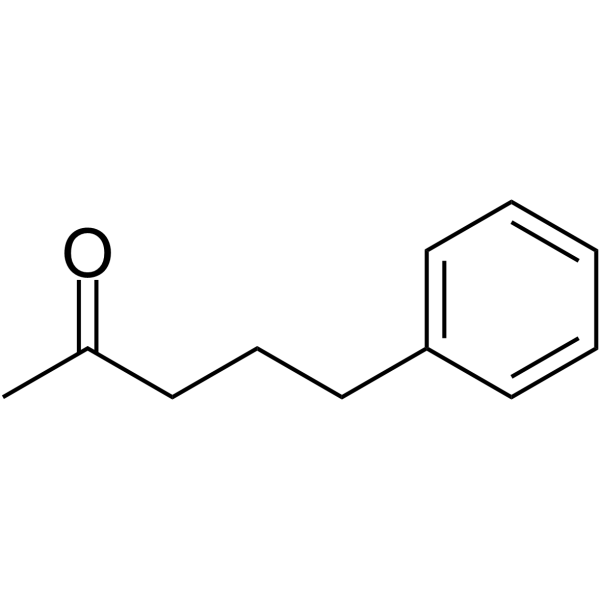
5-Phenylpentan-2-on ist ein potenter Inhibitor der Histon-Deacetylase (HDACs). 5-Phenylpentan-2-on kann fÜr die Erforschung von StÖrungen des Harnstoffzyklus verwendet werden.
-
GC45772
6(5H)-Phenanthridinone
NSC 11021, NSC 40943, NSC 61083
An inhibitor of PARP1 and 2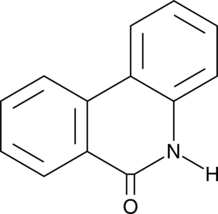
-
GC35175
6-Demethoxytangeretin
6-Demethoxytangeretin ist ein aus Citrus depressa isoliertes Zitrusflavonoid.
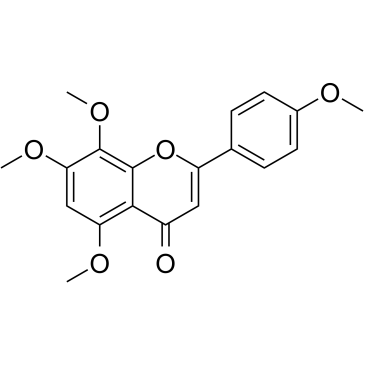
-
GC35180
6-Methyl-5-azacytidine
6-Methyl-5-azacytidin ist ein potenter DNMT-Inhibitor.
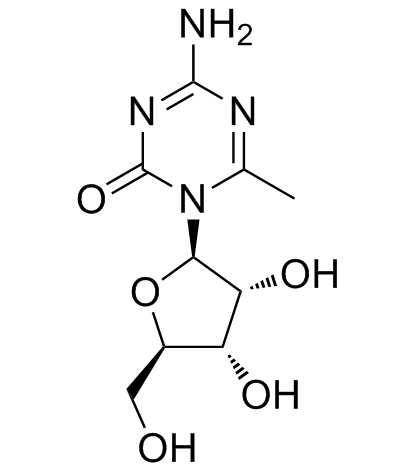
-
GC62279
653-47
653-47, ein Potenzierer, potenziert signifikant die cAMP-Response-Element-Bindungsprotein (CREB)-InhibitoraktivitÄt von 666-15. 653-47 ist ebenfalls ein sehr schwacher CREB-Inhibitor mit IC50 von 26,3 μM.
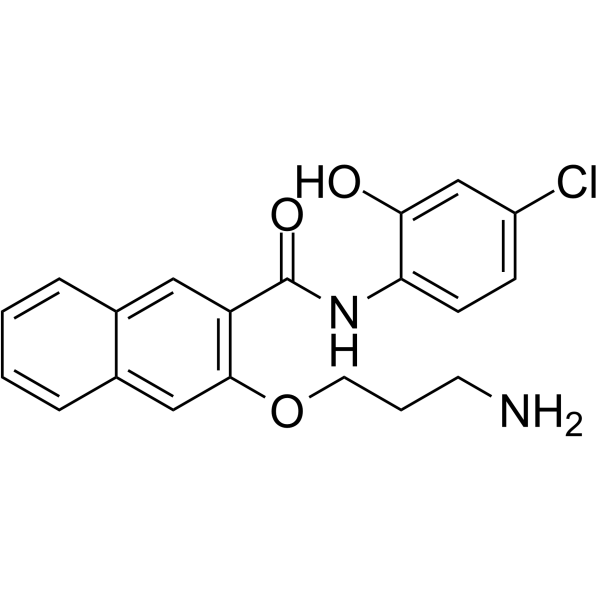
-
GC62144
653-47 hydrochloride
653-47-Hydrochlorid, ein Potentiator, potenziert die HemmaktivitÄt des cAMP-Response-Element-Bindungsproteins (CREB) von 666-15 signifikant. 653-47 Hydrochlorid ist auch ein sehr schwacher CREB-Inhibitor mit IC50 von 26,3 μM.
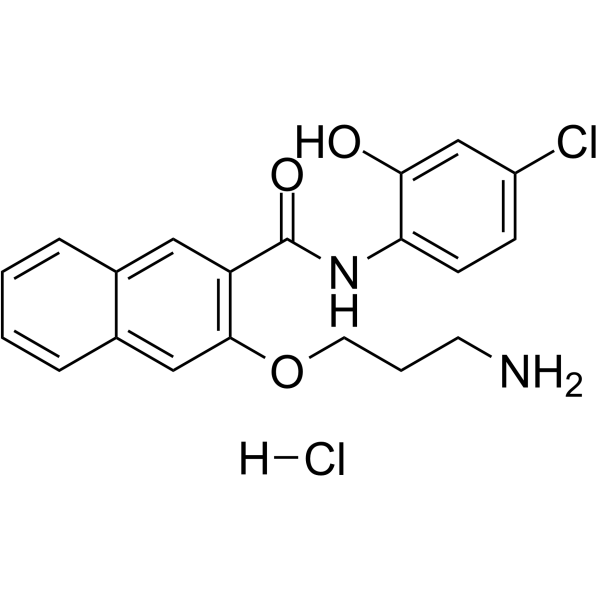
-
GC32689
666-15
Compound 3i
666-15 ist ein potenter und selektiver CREB-Inhibitor mit einem IC50 von 81 nM. 666-15 unterdrÜckt das Tumorwachstum in einem Brustkrebs-Xenotransplantatmodell.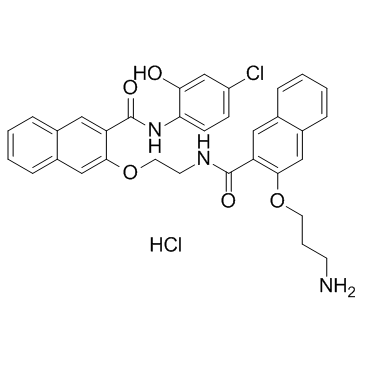
-
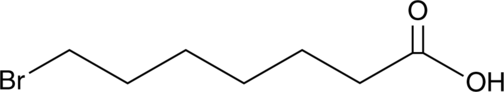
GC46736
7-Bromoheptanoic Acid
A building block
-
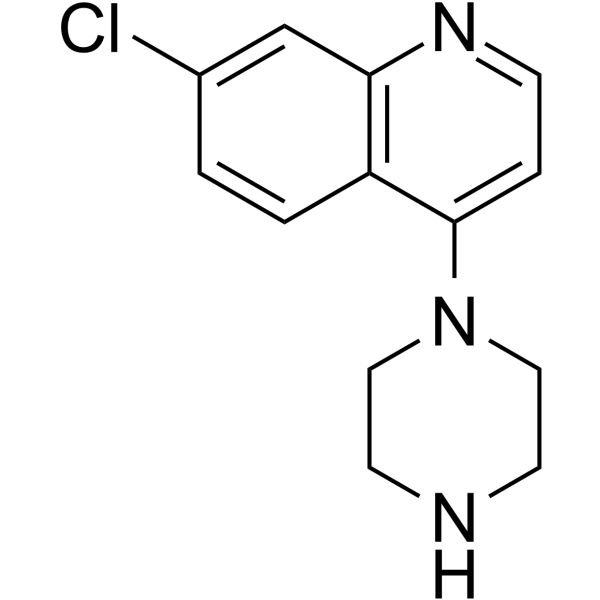
GC62651
7-Chloro-4-(piperazin-1-yl)quinoline
7-Chlor-4-(piperazin-1-yl)chinolon ist ein wichtiges GerÜst in der medizinischen Chemie.
-
GC48986
9-hydroxy Stearic Acid
9-HSA, 9-hydroxy Octadecanoic Acid
A hydroxy fatty acid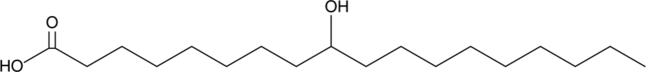
-
GC16015
A 366
A 366 ist ein potenter, hochselektiver, Peptid-kompetitiver Histon-Methyltransferase-G9a-Inhibitor mit IC50-Werten von 3,3 und 38 nM für G9a bzw. GLP (EHMT1). A 366 zeigt eine \u003e1000-fache Selektivität gegenüber 21 anderen Methyltransferasen. A 366 ist auch ein potenter, nanomolarer Inhibitor der Spindlin1-H3K4me3-Interaktion (IC50\u003d182,6 nM). A 366 zeigt eine hohe Affinität zum menschlichen Histamin-H3-Rezeptor (Ki \u003d 17 nM) und zeigt Subtyp-Selektivität unter Untergruppen der histaminergen und dopaminergen Rezeptorfamilien.
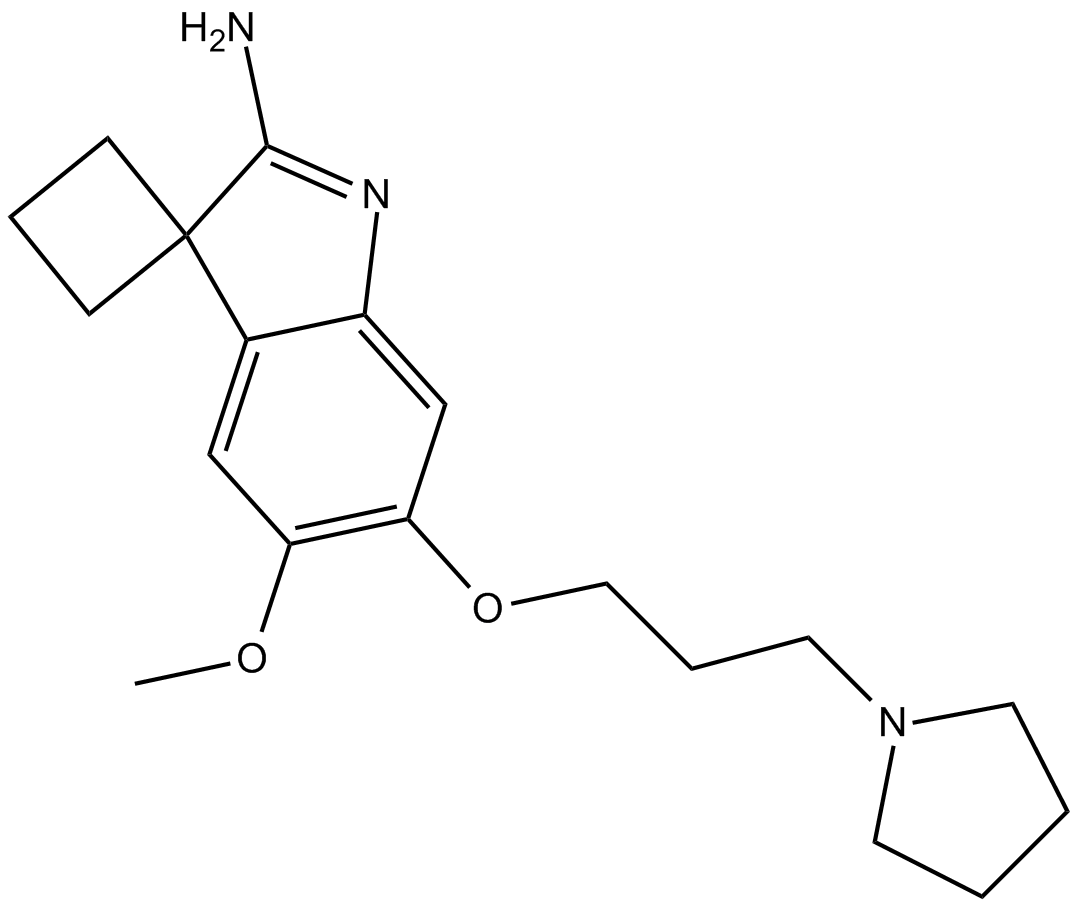
-
GC32861
A-196
A-196 ist ein potenter und selektiver Inhibitor von SUV420H1 und SUV420H2 mit IC50-Werten von 25 nM bzw. 144 nM. A-196 hemmt SUV4-20 biochemisch substratkompetitiv. A-196 ist die erste chemische Sonde ihrer Klasse von SUV4-20 zur Untersuchung der Rolle von Histon-Methyltransferasen bei der genomischen IntegritÄt.
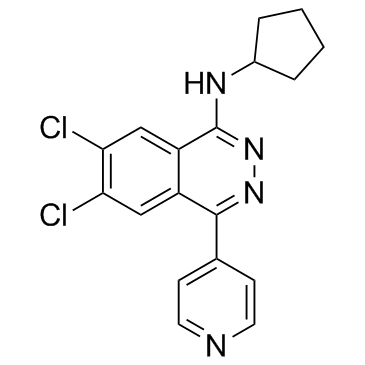
-
GC33187
A-395 (A395)
A-395 (A395) ist ein Antagonist der Protein-Protein-Interaktionen des polycomb repressiven Komplexes 2 (PRC2), der den trimeren PRC2-Komplex (EZH2-EED-SUZ12) mit einer IC50 von 18 nM stark hemmt.
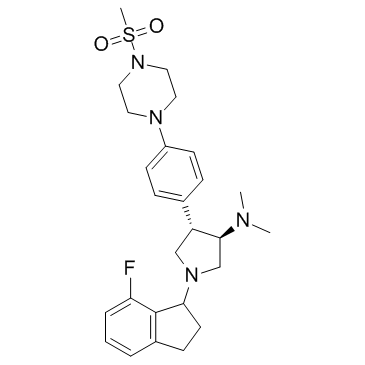
-
GC32677
A-485
A-485 ist ein potenter und selektiver katalytischer Inhibitor von p300/CBP mit IC50-Werten von 9,8nM und 2,6nM fÜr p300 bzw. CBP-Histonacetyltransferase (HAT).
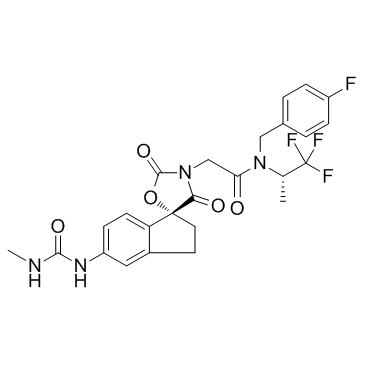
-
GC30503
A-893
A-893 ist ein zellaktiver Inhibitor der Methyltransferase SMYD2 mit einem IC50 von 2,8 nM.
-
GC12390
A-966492
A PARP1 and PARP2 inhibitor
-
GC33280
A1874
A1874 ist ein Nutlin-basiertes (MDM2-Ligand) und BRD4-abbauendes PROTAC mit einem DC50 von 32 nM (induziert BRD4-Abbau in Zellen). Wirksam bei der Hemmung der Proliferation vieler Krebszelllinien.
-
GC35216
AAPK-25
AAPK-25 ist ein potenter und selektiver Aurora/PLK-Doppelinhibitor mit Anti-Tumor-AktivitÄt, der eine mitotische VerzÖgerung verursachen und Zellen in einer Prometaphase anhalten kann, was durch die Phosphorylierung des Biomarkers Histon H3Ser10 widergespiegelt wird, gefolgt von einem Anstieg der Apoptose. AAPK-25 zielt auf Aurora-A, -B und -C mit Kd-Werten im Bereich von 23-289 nM sowie PLK-1, -2 und -3 mit Kd-Werten im Bereich von 55-456 nM ab.
-
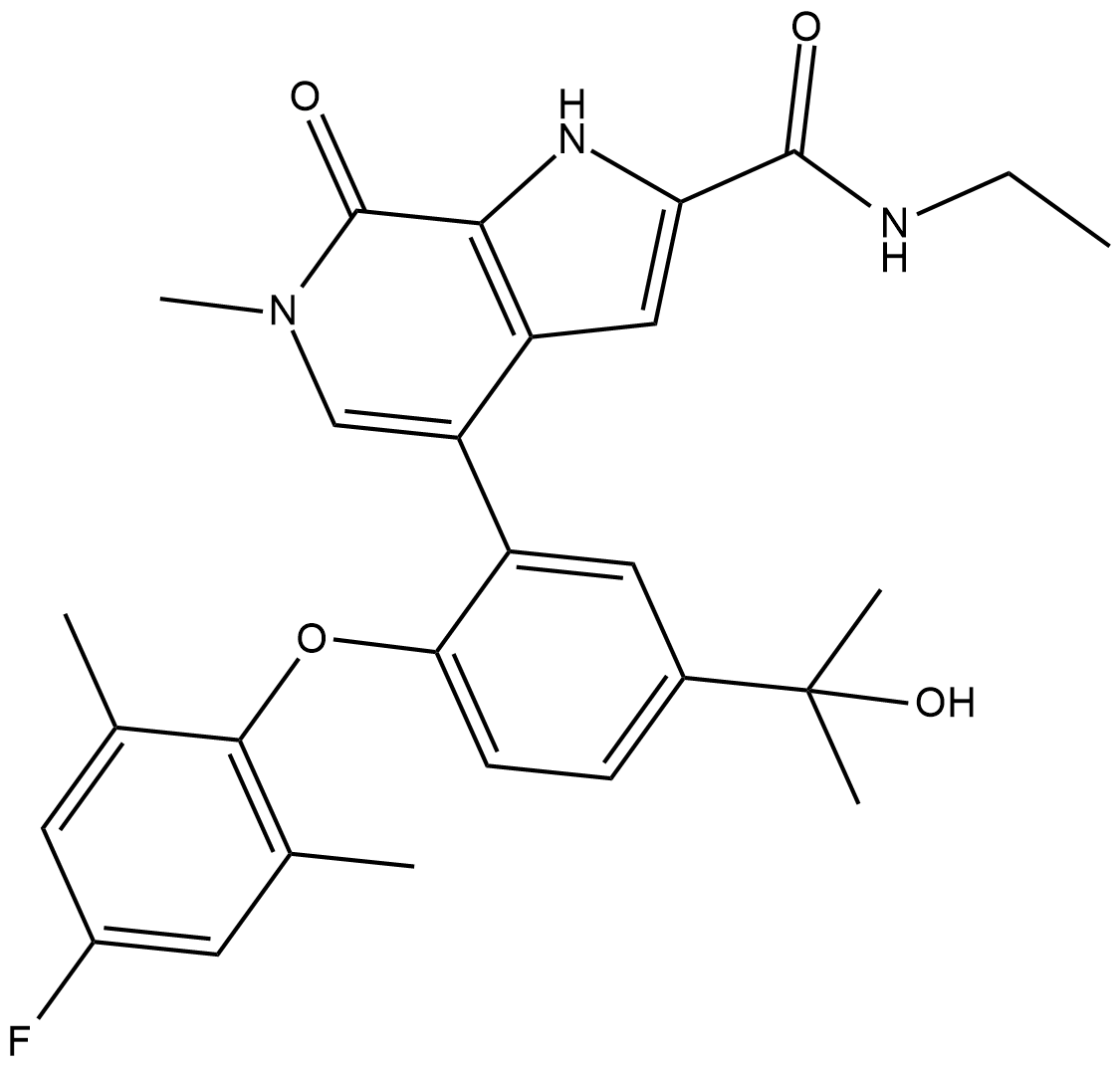
GC19409
ABBV-744
ABBV-744 ist ein erstklassiger, oral aktiver und selektiver Inhibitor der BDII-DomÄne von Proteinen der BET-Familie mit IC50-Werten im Bereich von 4 bis 18 nM fÜr BRD2, BRD3, BRD4 und BRDT.
-
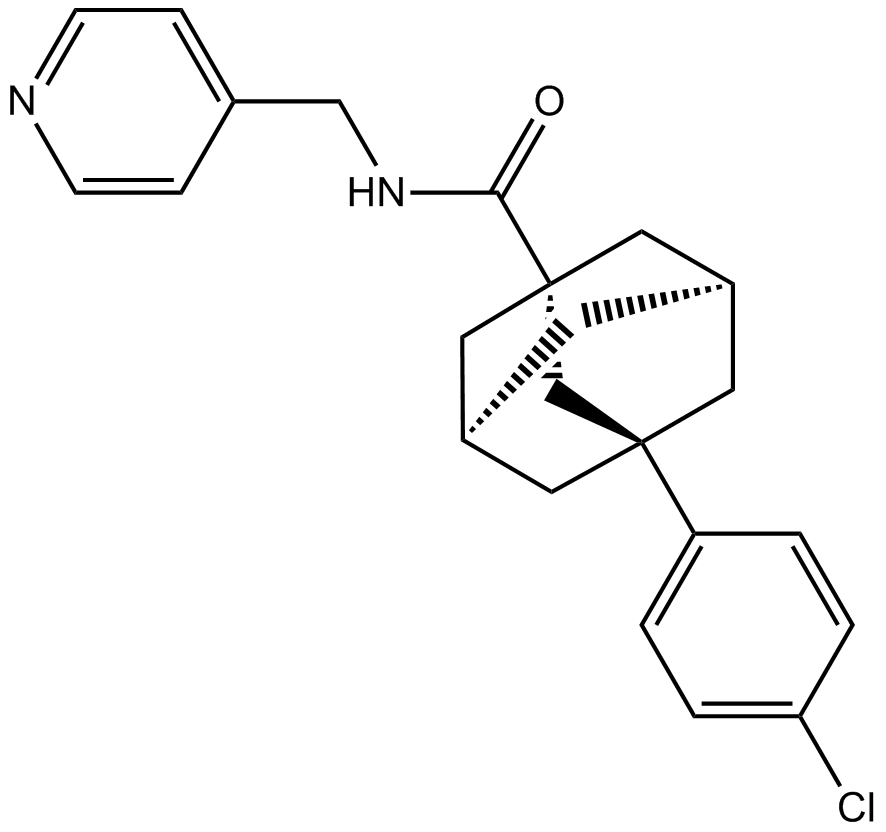
GC10154
ABC294640
ABC294640 (ABC294640) ist ein selektiver, kompetitiver Sphingosinkinase 2 (SK2)-Inhibitor mit einem Ki von 9,8 μM.
-
GC32023
Abrocitinib (PF-04965842)
PF-04965842
Abrocitinib (PF-04965842) (PF-04965842) ist ein potenter, oral aktiver und selektiver JAK1-Inhibitor mit IC50-Werten von 29 und 803 nM fÜr JAK1 bzw. JAK2.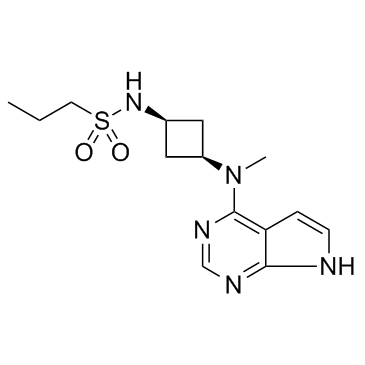
-
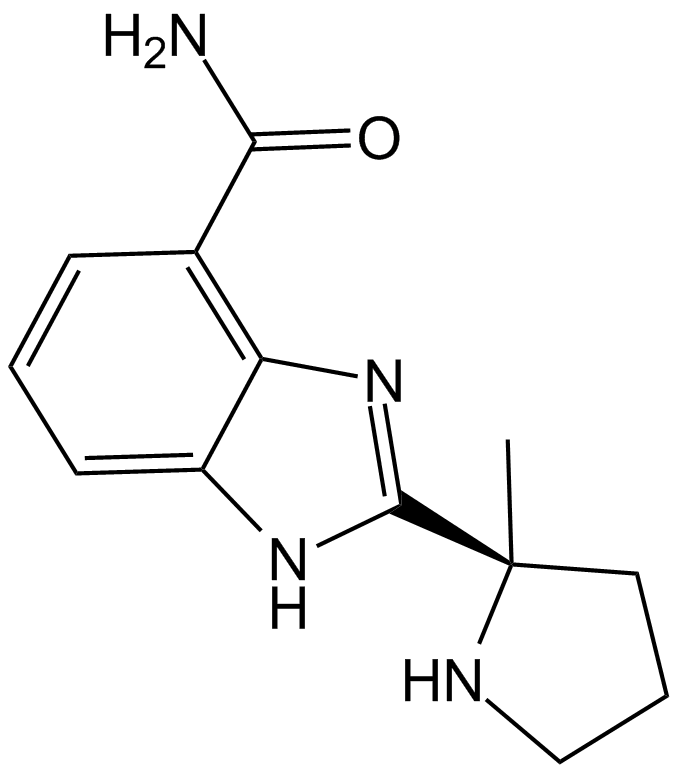
GC12422
ABT-888 (Veliparib)
-
GA20481
Ac-Arg-Gly-Lys(Ac)-AMC
Ac-Arg-Gly-Lys(Ac)-AMC ist ein Substrat fÜr HDAC.

-
GA20605
Ac-Lys-AMC
Ac-Lys-AMC (Hexanamid), auch MAL genannt, ist ein fluoreszierendes Substrat fÜr Histon-Deacetylase-HDACs.

-
GC48382
Ac-QPKK(Ac)-AMC
Ac-Gln-Pro-Lys-Lys-(Ac)-AMC, Ac-Gln-Pro-Lys-Lys-(Ac)-7-amino-4-methylcoumarin, p53317-320 Substrate (Ac-QPKK(Ac)-AMC)
A fluorogenic substrate for SIRT1, SIRT2, and SIRT3
-
GC35227
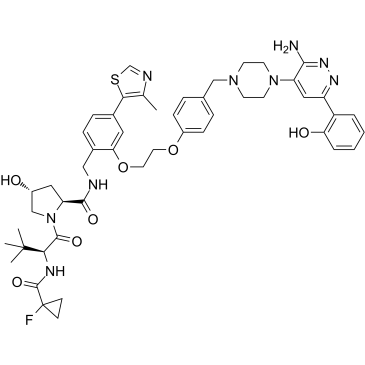
ACBI1
ACBI1 ist ein potenter und kooperativer SMARCA2-, SMARCA4- und PBRM1-Abbaustoff mit DC50-Werten von 6, 11 bzw. 32 nM. ACBI1 ist ein PROTAC-Degrader. ACBI1 zeigt antiproliferative AktivitÄt. ACBI1 induziert Apoptose.
-
GC12917
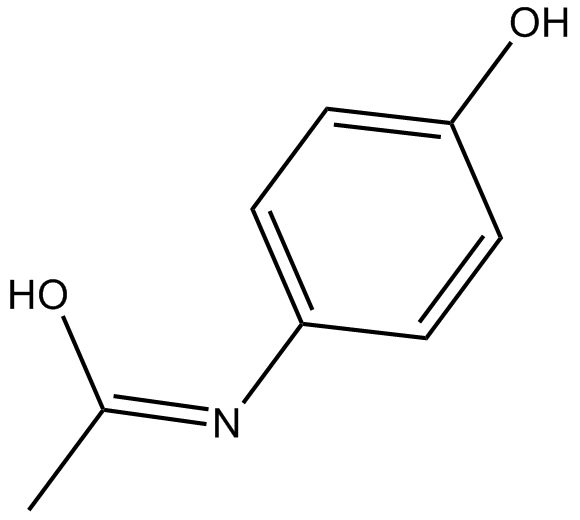
Acetaminophen
4-Acetamidophenol, APAP, 4'-Hydroxyacetanilide, NSC 3991, NSC 109028, Paracetamol
Ein COX-Inhibitor
-
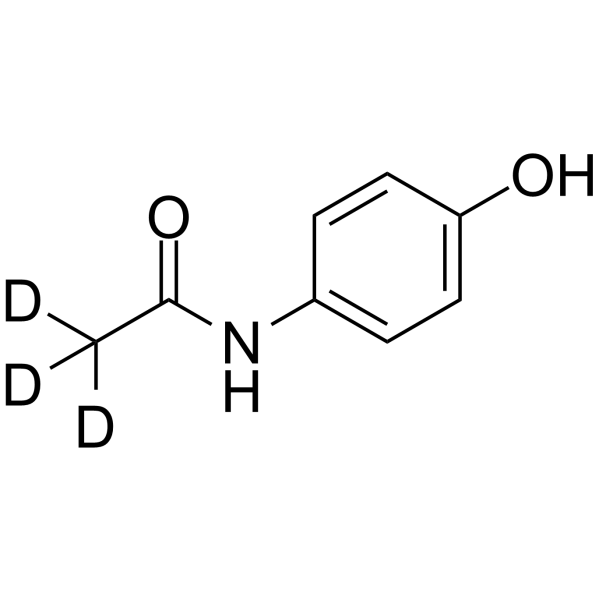
GC64137
Acetaminophen-d3
-
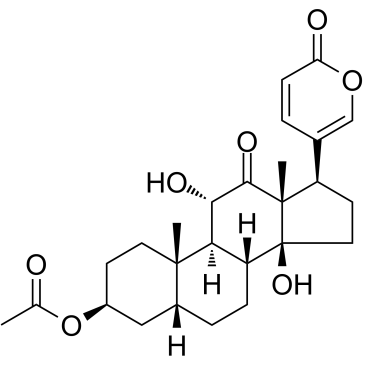
GC35230
Acetylarenobufagin
Acetylarenobufagin ist ein Modulator des steroidalen Hypoxie-induzierbaren Faktors 1 (HIF-I).
-
GC32083
Acriflavine
Acriflavine ist ein Fluoreszenzfarbstoff zur Markierung von RNA mit hohem Molekulargewicht.

-
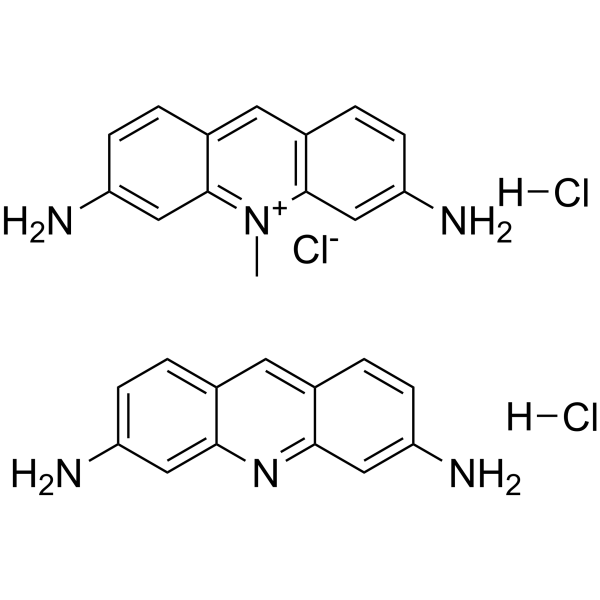
GC62354
Acriflavine hydrochloride
Acriflavinhydrochlorid (Acriflaviniumchloridhydrochlorid) ist ein fluoreszierender Acridinfarbstoff, der zur Markierung von NukleinsÄure verwendet werden kann.
-
GC34458
ACY-1083
ACY-1083 ist ein selektiver und gehirngängiger HDAC6-Inhibitor mit einer IC50 von 3 nM und ist 260-fach selektiver für HDAC6 als alle anderen Klassen von HDAC-Isoformen. ACY-1083 kehrt die durch Chemotherapie verursachte periphere Neuropathie effektiv um.
-
GC10417
ACY-241
Citarinostat
ACY-241 (ACY241) ist ein potenter, oral aktiver und hochselektiver HDAC6-Inhibitor der zweiten Generation mit einem IC50-Wert von 2,6 nM (IC50-Werte von 35 nM, 45 nM, 46 nM und 137 nM fÜr HDAC1, HDAC2, HDAC3 bzw. HDAC8). ). ACY-241 hat Antikrebswirkungen.
-
GC19020
ACY-738
ACY-738 ist ein potenter, selektiver und oral bioverfÜgbarer HDAC6-Inhibitor mit einem IC50 von 1,7 nM; ACY-738 hemmt auch HDAC1, HDAC2 und HDAC3 mit IC50-Werten von 94, 128 und 218 nM.

-
GC30782
ACY-775
ACY-775 ist ein potenter und selektiver Inhibitor der Histon-Deacetylase 6 (HDAC6) mit einem IC50 von 7,5nM.