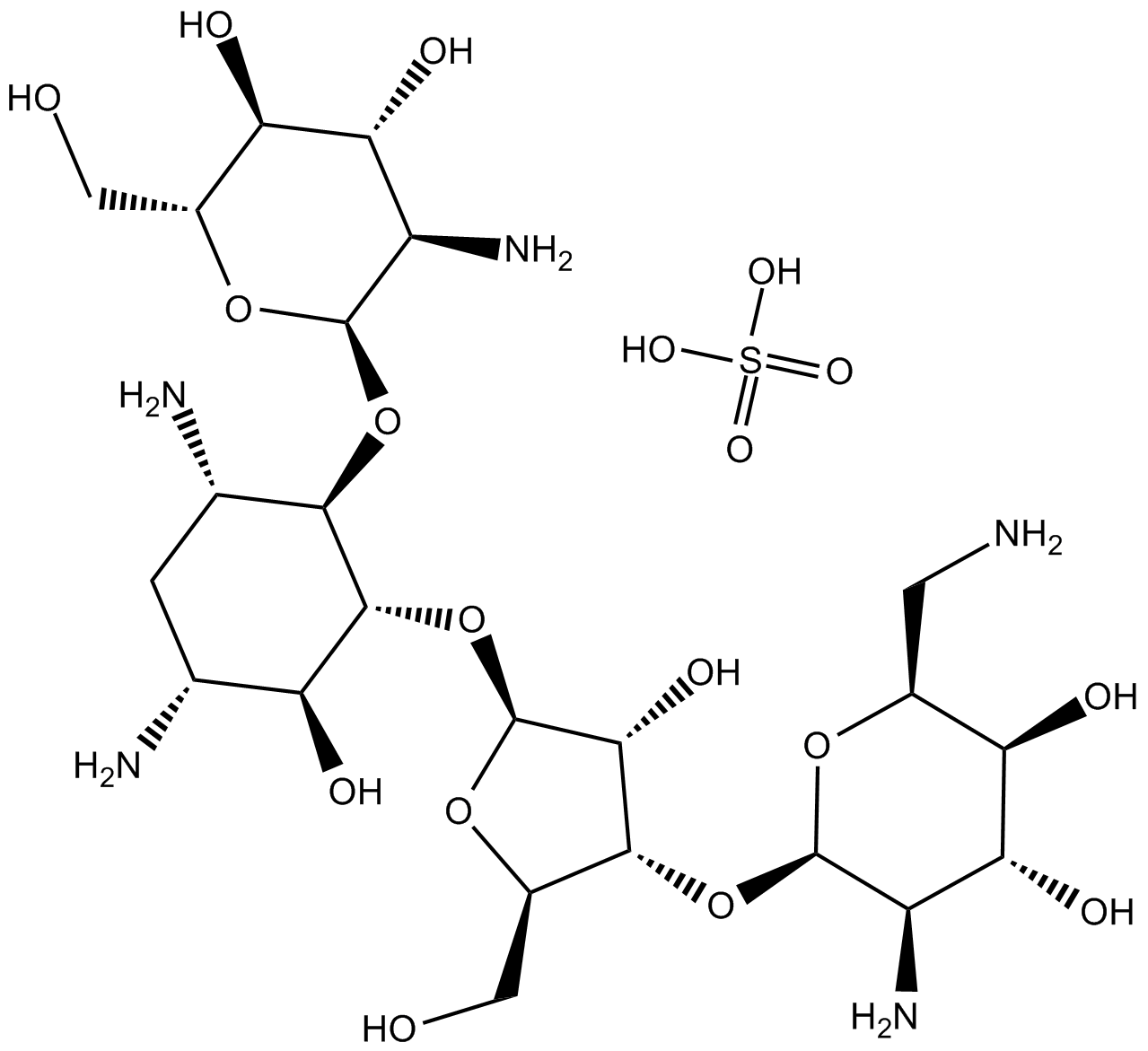
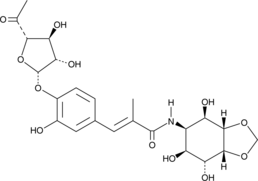
Hygromycin B
A naturally occurring bacterium called Streptomyces hygroscopicus, which is frequently found in soil, produces hygromycin B. The antibiotic is a derivative of the nucleoside adenosine and is categorized as an aminoglycoside. Its unique structure, which consists of a 4-aminoglucose ring linked to a hygromycin A moiety, is what makes it unique. Its antimicrobial qualities and value in research applications stem from its distinct structure. The molecular structure of Hygromycin B is shown in Figure 1.
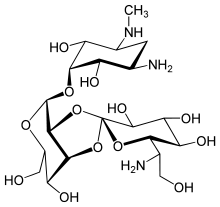
Figure 1: The molecular structure of Hygromycin B.
Among the most popular and clinically significant groups of ribosome-targeting antibiotics are aminoglycosides. An unusual aminoglycoside antibiotic with distinct structural and functional characteristics is hygromycin B. Unlike its effects reported in the structure of the isolated 30S ribosomal subunit in association with the medication, the antibiotic attaches to the mRNA decoding center in the small (30S) ribosomal subunit of the 70S ribosome and generates a localized conformational shift. Hygromycin B binding results in a different conformational shift in the ribosome than other aminoglycosides. Furthermore, hygromycin B potently blocks the spontaneous reverse translocation of tRNAs and mRNA on the ribosome, in contrast to other aminoglycosides . Comparing Hygromycin B to other aminoglycosides reveals that it has distinct structural and functional characteristics. It forms a third ring with a dual ether connectivity between its second and fourth rings. The antibiotic hygromycin B has unique features since it lacks a glucosamine ring I and instead has a 2-DOS core as its first ring .
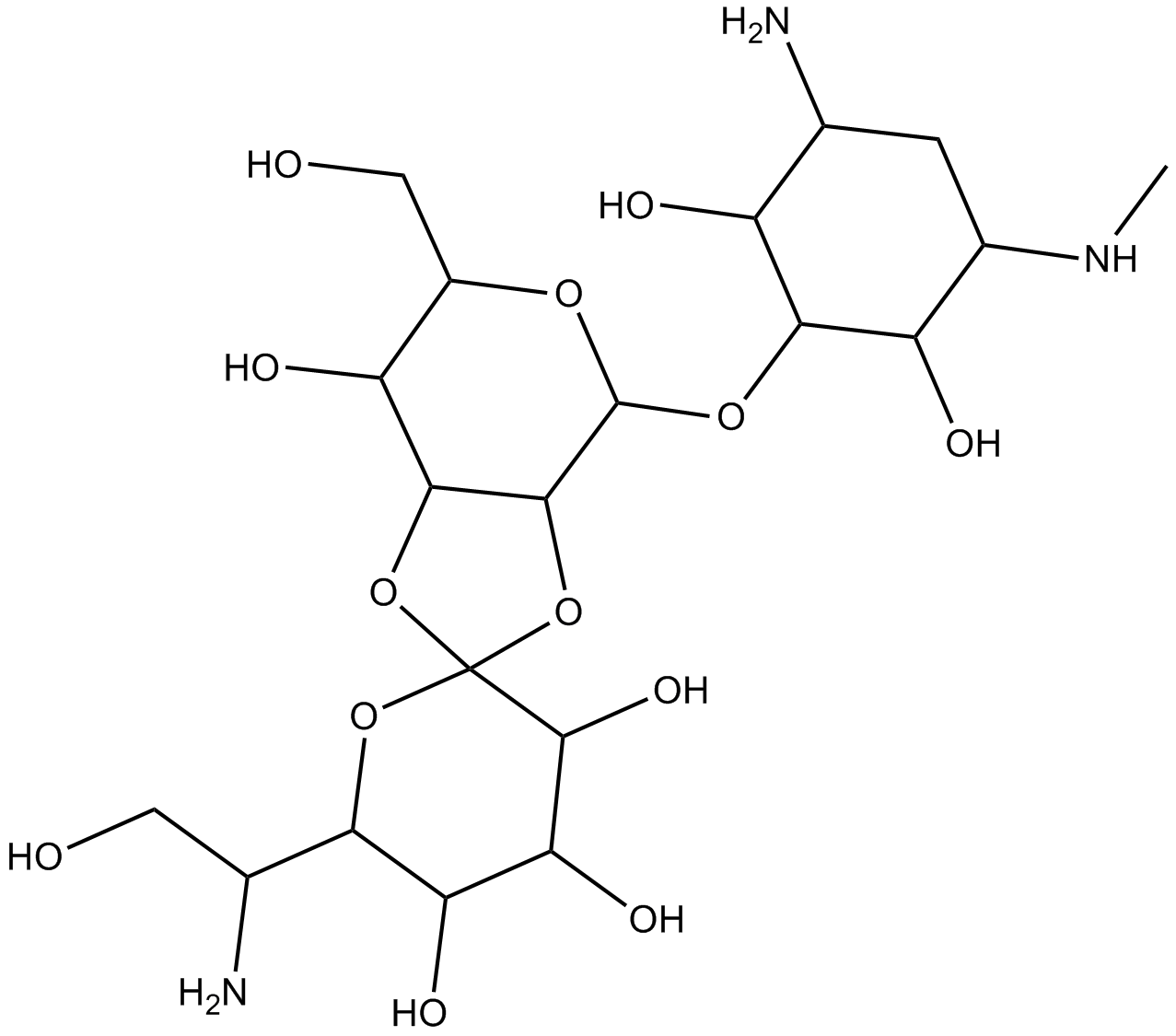
Hygromycin B binds to a single location in RNA helix h44 of the small ribosomal subunit in the 70S ribosome (Figure 2), which is consistent with the drug's monophasic manner of inhibition of translation. It is probable that all aminoglycosides containing 2-DOS target helix h44 in the small subunit. A few angstroms separate the hygromycin B binding site in h44 from the aminoglycoside binding sites. Furthermore, hygromycin B's stiffness prevents it from fitting into the binding pockets in h44 or h69, where other 2-DOS aminoglycosides bind .
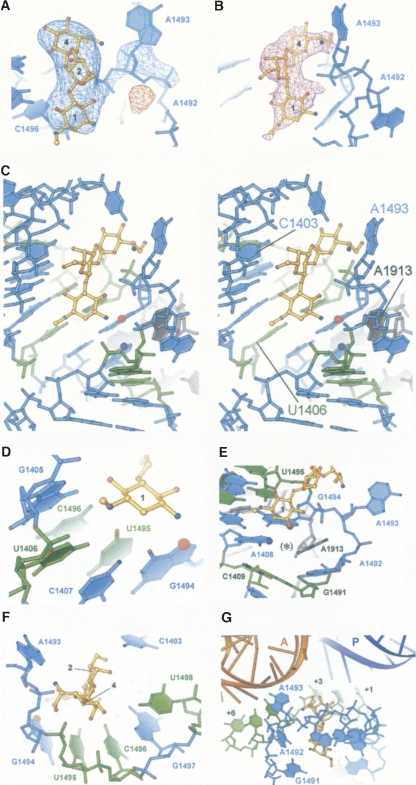
Figure 2: Interactions of hygromycin B with h44 of the ribosome.
Hygromycin B binds to the same location inside h44 in the 70S ribosome, as demonstrated by the structure of the 30S subunit in drug-bound complex . But the antibiotic's conformation, how it interacts with 16S ribosomal RNA, and how it affects h44's conformation are very different from what is seen in the 30S subunit structure (Figure 3). Rotating 180° in comparison to the 30S subunit structural model, the antibiotic's initial ring creates a network of hydrogen bonds that crosses the main groove of nucleotides C1404, G1405, U1406, and G1494–C1496 to the phosphate of G1494. C1403 and the second ring form a hydrogen bond. In contrast to the previous structure, the fourth ring in the current structure is rotated from the P site toward the A site, where it makes hydrogen bonds with both the phosphate of U1495 and A1493 (Figure 3). Hygromycin B alters the conformations of three globally conserved adenosines in the current 70S ribosome structure: A1913 in 23S rRNA and A1492 and A1493 in 16S rRNA . A1913 of 23S rRNA and A1492 of 16S rRNA are reoriented, and the medication causes A1493 of 16S rRNA to flip out of h44 (Figure 3). The structure of the 30S subunit in combination with hygromycin did not exhibit this conformational change.
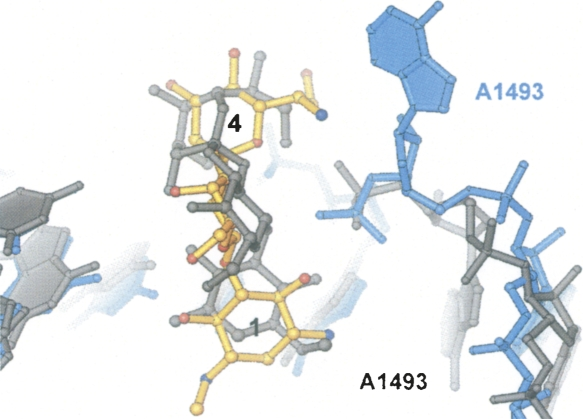
Figure 3: Superposition of the hygromycin B and h44 conformations observed in the present structure of hygromycin B bound to 70S ribosome with the structure of hygromycin B bound to 30S ribosomal subunit.
Despite the poorly ordered adenine base of A1493, binding of hygromycin B orients it between the locations that would be occupied by A-site and P-site tRNAs (Figure 4). When hygromycin B attaches, nucleotides A1913 in 23S rRNA and A1492 in 16S rRNA experience a relatively minor conformational change. While the conformations of A1492 and A1913 are different from those in the vacant 70S ribosome structure, they both retain base stacking with A1913 at the tip of H69 in the 50S subunit in the hygromycin B complex. This contact acts as an intersubunit bridge in the apo–70S ribosome structure.
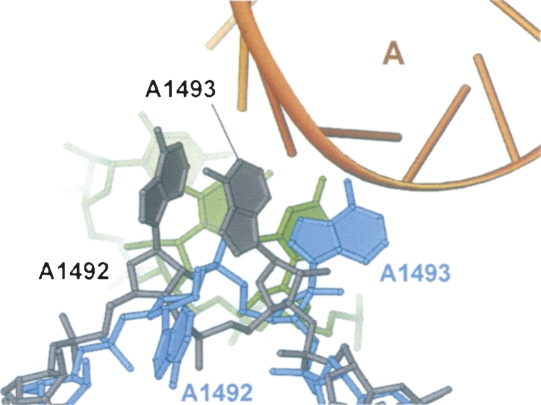
Figure 4: Superposition of the h44 conformations observed in the present structure of the E. coli 70S ribosome in complex with hygromycin B (rRNA in light blue) and in the structure of the T. thermophilus 70S ribosome in complex with tRNAs, mRNA, and paromomycin (rRNA in gray).
With the current framework, interpreting two sets of mutations is more difficult. Mutations in base paired nucleotides C1409–G1491 cause drug resistance even though these nucleotides do not directly interact with hygromycin B. By undermining the helix's stability or altering the shape of the hygromycin B binding pocket, breaking this base pair may confer resistance. Moreover, hygromycin B does not come into direct touch with U1498, which makes it difficult to understand why M. smegmatis has the resistance mutation U1498C. Notably, all three domains of life share universally conserved nucleotides necessary for hygromycin B sensitivity and/or those implicated in hygromycin B binding . This explains why hygromycin B is generally harmful and lacks specificity.
It is believed that hygromycin B mostly inhibits translation by interfering with translocation, which is the coordinated movement of mRNA and tRNA on the ribosome after each peptide bond is formed. hygromycin B inhibits translocation with repeated turnovers and reduces the rate of translocation in a single round by more than 300 times. Translocation is the movement of tRNA substrates from the A (aminoacyl-) and P (peptidyl-) sites on the ribosome to the P site and E (Exit) sites, respectively. It is catalyzed by elongation factor G (EF-G) in a GTP-dependent reaction. Additionally, translocation involves the shifting of mRNA in the 5′ direction by one codon, which opens the A site for a new round of mRNA decoding and aminoacyl–tRNA binding. According to current theories, translocation happens in a number of kinetically and structurally discrete phases, with the peptidyl-tRNA and deacylated tRNA moving in relation to the 50S subunit first and the 30S subunit second . Interestingly, the ribosome can still accomplish translocation on its own without EF-G, but much more slowly. Moreover, in factor-dependent and spontaneous events, the ribosome can facilitate back-translocation, also known as reverse translocation. Remarkably, aminoglycosides have remarkably distinct effects on the rate of the spontaneous reverse translocation reaction (Figure 5). Hygromycin B significantly alters the extent of the response by reducing the final population of the PRE complex, which in turn causes a threefold decrease in the rate of reverse translocation (Figure 5). Neomycin, gentamicin, and paromomycin, on the other hand, have no influence on the size of the reaction and increase the observed rate of reverse translocation by a factor of two to three .
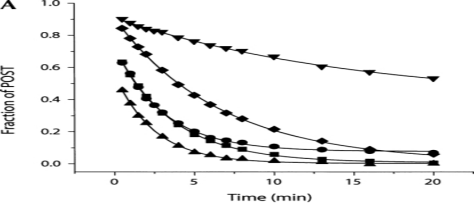
Figure 5: Effect of the aminoglycosides on reverse translocation
Antibiotics may prevent translocation through a variety of methods. By stabilizing tRNAs in their specific binding locations on the ribosome, they can raise the translocation energy barrier. Antibiotics can also cause conformational changes that would obstruct translocation or impede the ribosome's necessary conformational adjustments for translocation to occur.
A useful antibiotic, hygromycin B has grown to be an essential resource in the domains of microbiology, biotechnology, and molecular biology. Hygromycin B's function in genetic engineering and biotechnology is expected to grow as technology develops, opening up new avenues for the production of genetically modified organisms and the improvement of scientific understanding. But while handling this antibiotic in a lab setting, researchers need to constantly exercise caution and follow best practices to ensure safety.










Kommentare