PNGase F
In the intricate world of molecular biology, understanding the structure and function of proteins is paramount. Proteins, often adorned with various post-translational modifications, play crucial roles in virtually all biological processes. Among these modifications, glycosylation the attachment of sugar molecules to proteins, stands out due to its complexity and significance. PNGaseF (Peptide-N-Glycosidase F), a powerful enzyme that has become an indispensable tool for scientists studying glycoproteins. This article delves into the nature of PNGase F, its applications, and why it’s a game changer in the field of biochemistry and molecular biology.
PNGase F is a recombinant glycosidase cloned from Elizabethkingia miricola and overexpressed in E. coli. PNGase F has a molecular weight of 36kDa.It is active in a wide pH range and in the presence of many reagents. It can be used for protein trafficking, characterization of glycan structure, determining locations of glycosylation in proteins and identifying whether a protein is glycosylated. It is also used for the deglycosylation of glycoproteins. PNGase F catalyzes the cleavage of N-linked oligosaccharides between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins (see figure). PNGase F will not remove oligosaccharides containing α-(1,3)-linked core fucose commonly found on plant glycoproteins.
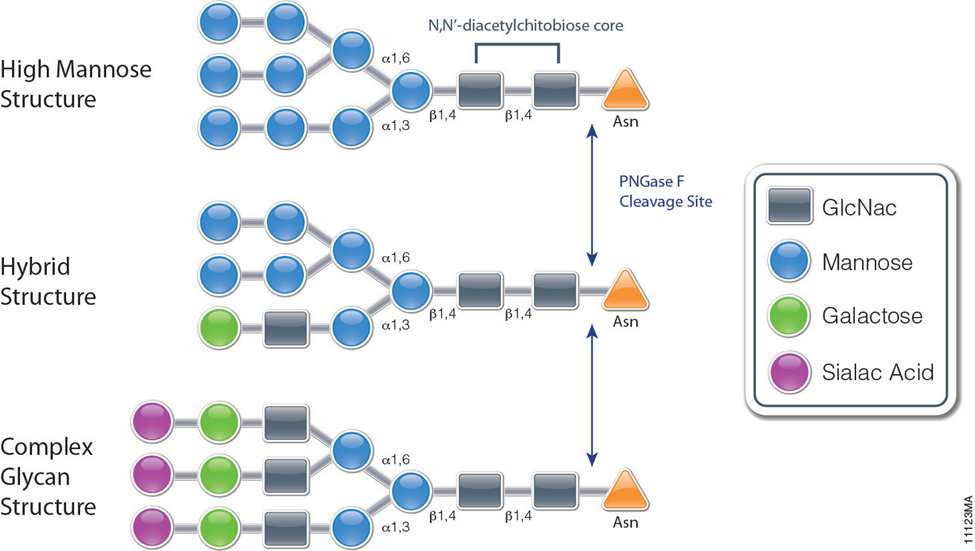
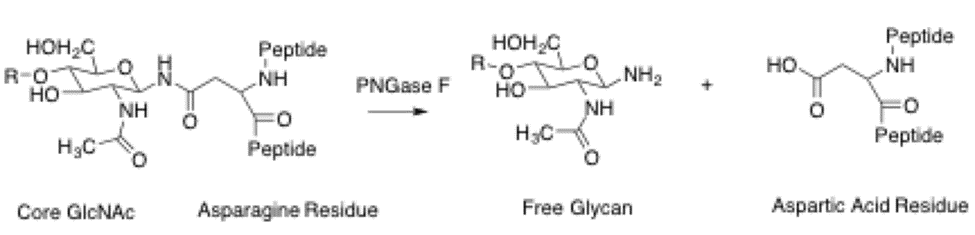
PNGase F catalyzes the cleavage of an internal glycosidic bond in an oligosaccharide. The asparagine residue from which the glycan is removed, is deaminated to aspartic acid. It requires a minimum of two GlcNAc oligosaccharide residues attached to the asparagine for catalysis to occur. This enzyme utilizes a catalytic triad of cysteine-histidine-aspartate in its active site, which is a common motif for amidases. This motif contains a nucleophile, a proton donor, and a positive charge to stabilize the tetrahedral intermediate. The crystal structure of PNGase from flavobacterium miningosepticum, with 1.8 Angstrom resolution, was found to be folded in two domains, each with an eight-stranded β Barrel, or jelly roll configuration. This structure is similar to lectins and glucanases suggesting similarities with lectins and other carbohydrate-binding proteins.
To confirm the GNTI reduction in fruits we performed immunological assays by treating protein extracts with PNGase F, an enzyme that releases asparagine-bound-N -glycans when alpha 1,3-fructose is missing. Glycoproteins of GNTI-RNAi plants (or GNTI mutants) are PNGase F-sensitive, as indicated by the vINV shift (four complex N- glycans in wild type) and inverse loss of ConA binding to most glycoproteins. No shifts were detected for wild type and only minor ones for MANII-RNAi extracts (compare α- cgly to ConA), confirming the presence of core focuses on most protein-bound N-glycans of MANII-RNAi fruits.
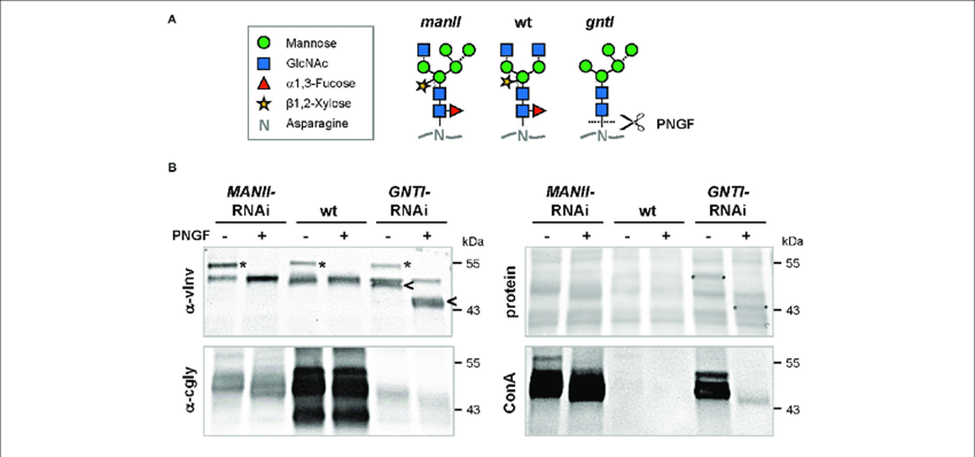
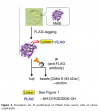
Figure 1: Peptide:N-glycosidase F (PNGase F) treatment verifies the presence of core fucoses on MANII-RNAi glycans.
Peptide:N-glycosidase F (PNGase F) treatment verifies the presence of core fucoses on MANII-RNAi glycans. (A) N-glycan structures that may be produced in wild-type (wt), manII, and gntI mutant plants (known as hgl1 and cgl1 in Arabidopsis, respectively) with indicated potential release by PNGase F. This endoglycosidase can cleave asparagine-bound N-glycans when coreα1,3-fucose is missing. In the cell wall or vacuole, terminal sugar residues can be liberated by hexosaminidases (GlcNAc) and α-mannosidases (dotted lines). (B) Immunoblots of untreated (-) and PNGase F-treated (+) fruit extracts from MANII-RNAi, Micro-Tom wild-type (wt), and GNTI-RNAi (#20) plants were developed with α-vINV, α-cgly (α-PHA-L), or the lectin ConA (detecting mannose-terminating N-glycans). The Ponceau S-stained blot (protein) is shown as the loading reference.
In GNTI-RNAi, PNGase-F treatment results in a shift of vacuolar invertase (vINV, arrowheads) and loss of ConA binding. This is not the case for MANII-RNAi or wt samples, confirming the presence of core fucoses on most glycoproteins. Note that the upper band detected by α-vINV (stars) shifts in all PNGase F-treated extracts (protein with high mannose precursor), indicating completeness of the enzyme treatment. Apparent molecular masses are indicated in kDa (PageRuler Prestained Protein Ladder, Fermentas).
The ability to profile and determine the two-dimensional localization and histopathology of multiple N-glycans in tissues has been developed using a MALDI-IMS approach. The goal of the studies was to present new data for analysis of N-glycan distribution in different FFPE hepatocellular carcinoma tissues, to highlight the capabilities and limitations of the method, how it compares with other glycan analysis approaches, and identify areas of improvement. Initial MALDI-IMS of a commercially available FFPE HCC tissue of complex histopathology demonstrated the specific release of N-glycans following PNGaseF digestion and the ability of released glycans to distinguish tissue subtypes and pathologies. Two serial sections were prepared identically with the exception that one tissue received PNGaseF while the other was shielded from enzyme application to serve as a negative control. Average spectra of each tissue region revealed a robust signal increase following PNGaseF digestion (Figure 2a) compared to the control tissue (Figure 2b). Putative glycan structures were annotated using GlycoWorkbench based on mass accuracy and previous studies, but no information is available regarding specific anomeric linkages. All glycan structures reported herein are the [M + Na]+ adducts unless otherwise noted. Three representative glycans, Hex5dHex1HexNAc4NeuAc1 (m/z 2100.759, blue), Hex5HexNAc4 (m/z 1663.582, red) and Hex9HexNAc2 (m/z 1905.612, green) were selected and shown in an overlay image (Figure 2c). The three glycans map to the tissue histopathology marked on the H & E stain (Figure 2d), with three evident tissue morphologies; necrosis (outlined in red), HCC tissue (outlined in green), and fibroconnective tissue (outlined in blue). The three N-glycans in Figure 2c were selected to illustrate their regiospecific distribution of them concerning the different histopathologies. Interestingly, high mannose glycans were elevated in the tumor tissue, while fucosylated or sialylated/fucosylated complex diantennary glycans were elevated in the fibroconnective tissue.
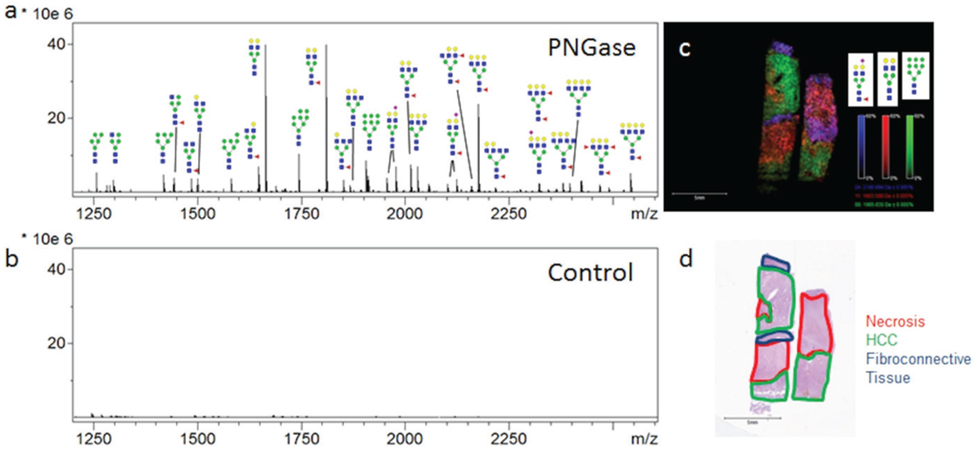
Figure 2: . PNGaseF Releases N-linked Glycans from HCC Tissue in a Spatially Defined Manner.
PNGaseF Releases N-linked Glycans from HCC Tissue in a Spatially Defined Manner. A HCC tissue slide (Biochain) containing two serial slices of HCC tissue was analyzed by N-glycan MALDI-IMS (a); One section was shielded from enzyme application to serve as a negative control (b); Three representative ions were selected in FlexImaging and displayed as an overlay image (c). Hex5dHex1HexNAc4NeuAc (m/z = 2100.759, blue), Hex5HexNAc4 (m/z = 1663.582, red) and Hex9HexNAc2 (m/z = 1905.612, green) each display unique localization within the tissue slice (c); As highlighted in the H & E stain of the slide (d), indicated regions were identified to be fibroconnective tissue (blue), necrotic tissue (red), and HCC tissue (green). Tri- and Tetra-antennary compositions are shown with representative glycan structures. Glycan structures with single fucoses are shown as being attached to the core N-acetylglucosamine for consistency.
PNGase F has revolutionized the study of glycoproteins, offering a window into the complex world of protein glycosylation. Its ability to precisely remove N-linked glycans has made it an essential tool in the arsenal of molecular biologists and biochemists. As our understanding of glycosylation deepens, the applications of PNGase F will undoubtedly expand, further unlocking the secrets of biological systems and advancing the development of glycan-based therapeutics.











Comments