Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(16)
- Caspase(54)
- 14.3.3 Proteins(1)
- Apoptosis Inducers(41)
- Bax(8)
- Bcl-2 Family(108)
- Bcl-xL(8)
- c-RET(8)
- IAP(26)
- KEAP1-Nrf2(68)
- MDM2(13)
- p53(108)
- PC-PLC(4)
- PKD(6)
- RasGAP (Ras- P21)(1)
- Survivin(6)
- Thymidylate Synthase(10)
- TNF-α(131)
- Other Apoptosis(884)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(5)
- PD-1/PD-L1 interaction(61)
- ASK1(3)
- PAR4(2)
- RIP kinase(49)
- FKBP(19)
Products for Apoptosis
- Cat.No. Product Name Information
-
GC10350
TIC10 isomer
ONC201 isomer
Potent Akt/ERK inhibitor
-
GC41183
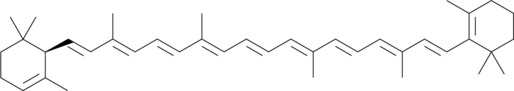
α-Carotene
all-trans-α-Carotene
α-Carotene is a precursor of vitamin A that has been found in various fruits and vegetables.
-
GC48292
α-MSH (human, mouse, rat, porcine, bovine, ovine) (trifluoroacetate salt)
α-Melanocyte-stimulating Hormone, Ac-SYSMEHFRWGKPV-NH2
α-MSH (α-Melanocyte-Stimulating Hormone) TFA, an endogenous neuropeptide, is an endogenous melanocortin receptor 4 (MC4R) agonist with anti-inflammatory and antipyretic activities.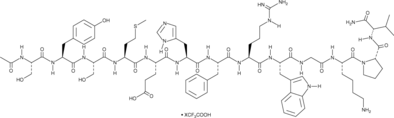
-
GC45213
α-NETA
Choline acetyltransferase (ChAT) mediates the synthesis of the neurotransmitter acetylcholine from acetyl-CoA and choline.
-
GC41499
α-Phellandrene
p-Mentha-1,5-diene, (±)-α-Phellandrene
α-Phellandrene is a cyclic monoterpene that has been found in various plants, including Cannabis, and has diverse biological activities.
-
GC63941
α-Solanine
α-solanine, a bioactive component and one of the major steroidal glycoalkaloids in potatoes, has been observed to inhibit growth and induce apoptosis in cancer cells.
-
GC67618
α-Tocopherol phosphate disodium
alpha-Tocopherol phosphate disodium; TocP disodium; Vitamin E phosphate disodium
α-Tocopherol phosphate (alpha-Tocopherol phosphate) disodium, a promising antioxidant, can protect against long-wave UVA1 induced cell death and scavenge UVA1 induced ROS in a skin cell model. α-Tocopherol phosphate disodium possesses therapeutic potential in the inhibition of apoptosis and increases the migratory capacity of endothelial progenitor cells under high-glucose/hypoxic conditions and promotes angiogenesis.
-
GC48920
β-Carboline-1-carboxylic Acid
1-Formic Acid-β-carboline
An alkaloid with diverse biological activities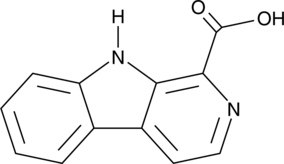
-
GC41623
β-Elemonic Acid
Elemadienonic Acid, 3-Oxotirucallenoic Acid, 3-oxo Tirucallic Acid
β-Elemonic acid is a triterpene isolated from Boswellia (Burseraceae) that exhibits anticancer activity.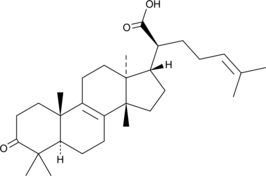
-
GC64619
β-Ionone
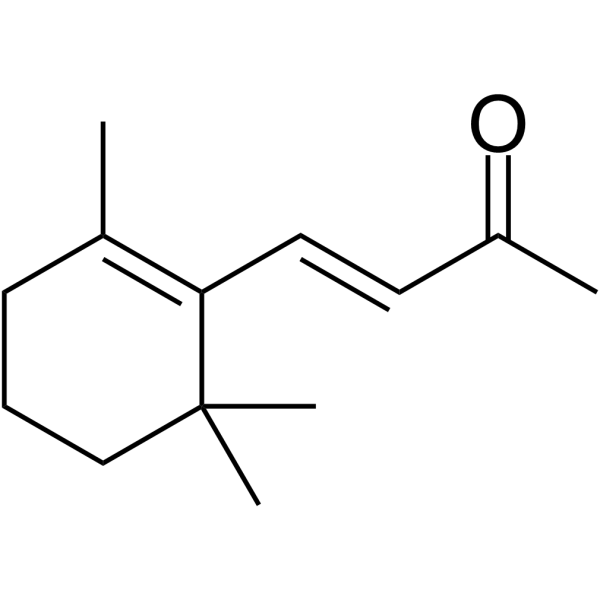
β-Ionone is effective in the induction of apoptosis in gastric adenocarcinoma SGC7901 cells. Anti-cancer activity.
-
GC66048
δ-Secretase inhibitor 11
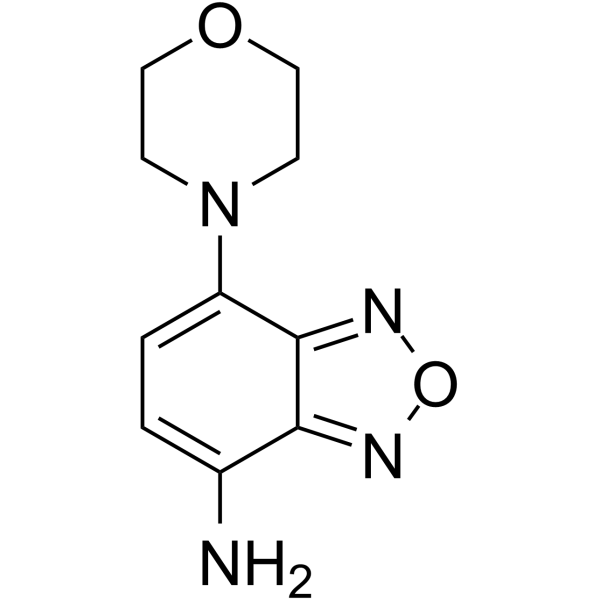
δ-Secretase inhibitor 11 (compound 11) is an orally active, potent, BBB-penetrated, non-toxic, selective and specific δ-secretase inhibitor, with an IC50 of 0.7 μM. δ-Secretase inhibitor 11 interacts with both the active site and allosteric site of δ-secretase. δ-Secretase inhibitor 11 attenuates tau and APP (amyloid precursor protein) cleavage. δ-Secretase inhibitor 11 ameliorates synaptic dysfunction and cognitive impairments in tau P301S and 5XFAD transgenic mouse models. δ-Secretase inhibitor 11 can be used for Alzheimer's disease research.
-
GC46008
(±)-Thalidomide-d4
N-Phthaloylglutamimide-d4
An internal standard for the quantification of (±)-thalidomide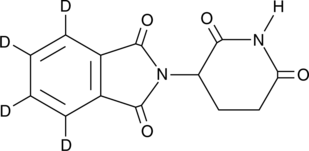
-
GC45618
(±)-trans-GK563
GK563
A GVIA iPLA2 inhibitor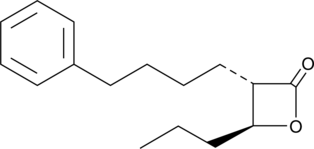
-
GC45270
(±)10(11)-EDP Ethanolamide
10,11-EDP-EA, 10,11-EDP epoxide, 10,11-epoxy Docosapentaenoic Ethanolamide
(±)10(11)-EDP ethanolamide is an ω-3 endocannabinoid epoxide and cannabinoid (CB) receptor agonist (EC50s = 0.43 and 22.5 nM for CB1 and CB2 receptors, respectively).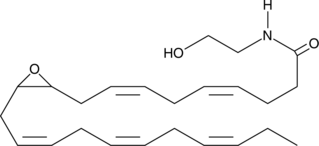
-
GC49268
(+)-δ-Cadinene
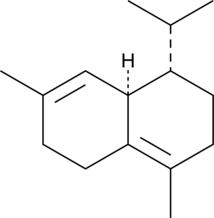
A sesquiterpene with antimicrobial and anticancer activities
-
GC18516
(+)-Aeroplysinin-1
NSC 170364
(+)-Aeroplysinin-1 is a metabolite originally isolated from the marine sponge V.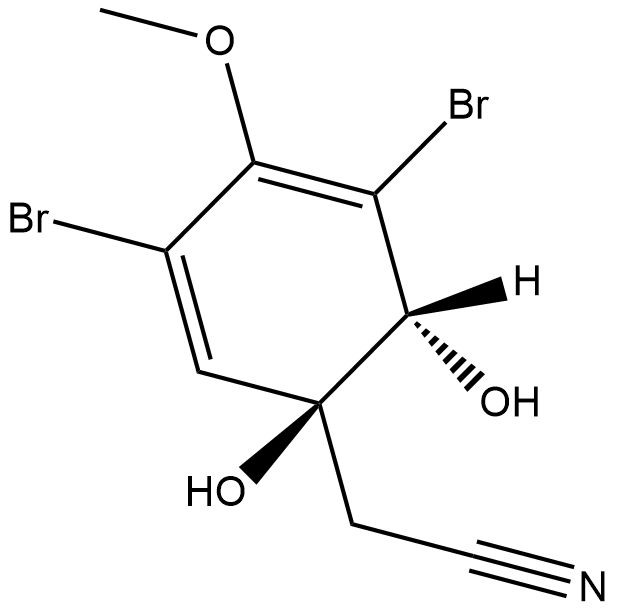
-
GC17008
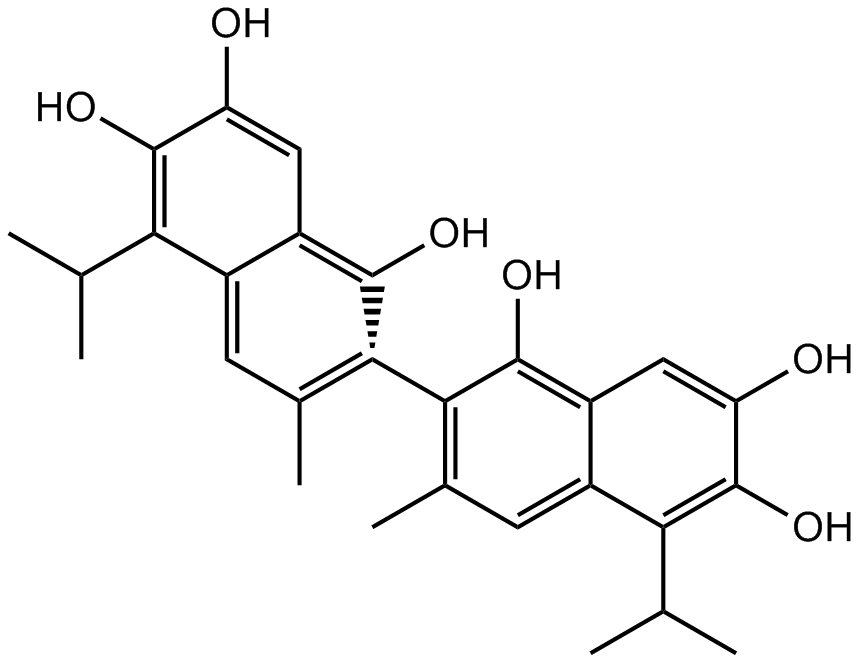
(+)-Apogossypol
inhibitor of Bcl-2 family proteins
-
GC45256
(+)-ar-Turmerone
(S)(+)arTurmerone
(+)-ar-Turmerone is an aromatic compound from the rhizomes of C.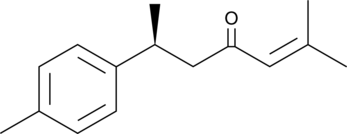
-
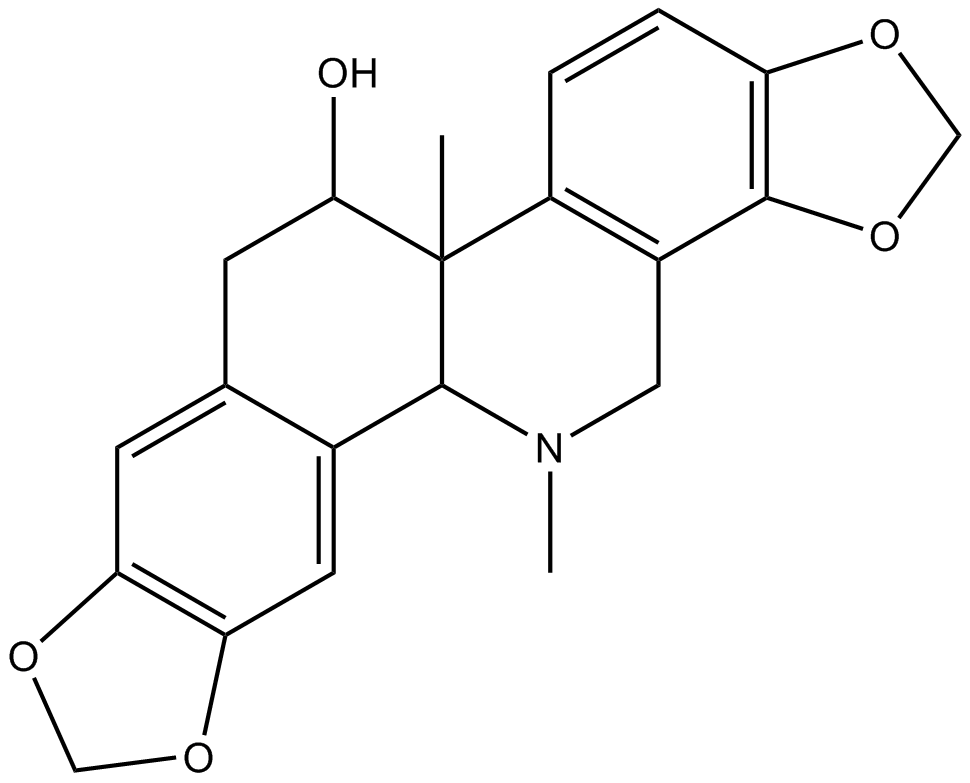
GN10654
(+)-Corynoline
(+)-Corynoline, 13-methyl-Chelidonine, CRL, (d)-Corynoline
-
GC31691
(+)-DHMEQ
(1R,2R,6R)-Dehydroxymethylepoxyquinomicin; (1R,2R,6R)-DHMEQ
(+)-DHMEQ is an activator of antioxidant transcription factor Nrf2.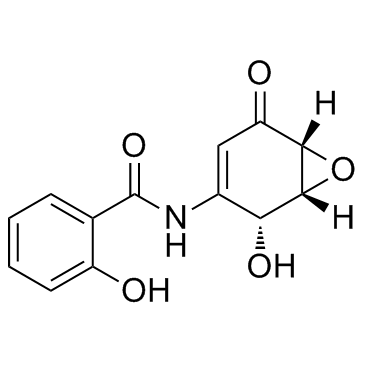
-
GC45265
(+)-Goniothalesdiol
(+)-Goniothalesdiol, isolated from the bark of the Malaysian tree G.

-
GC45274
(+)-Pinoresinol
NSC 35444
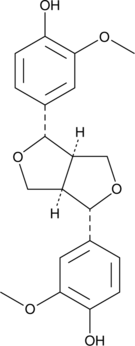
-
GC18749
(+)-Rugulosin
NSC 160880, NSC 249990, Rugulosin A
(+)-Rugulosin is a pigment and mycotoxin produced by certain fungi.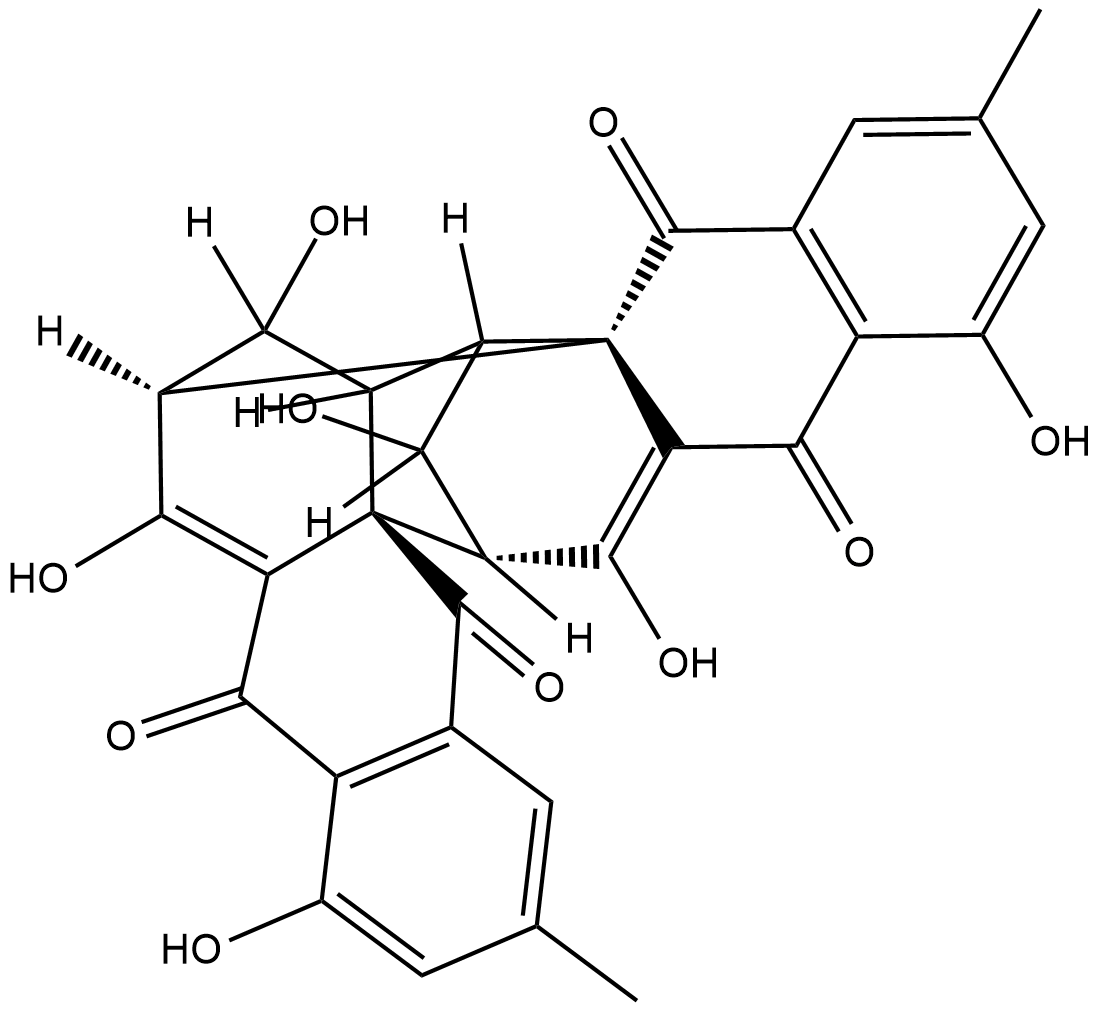
-
GC41345
(-)-α-Bisabolol
DL-α-Bisabolol
(-)-α-Bisabolol ((-)-α-Bisabolol), a monocyclic sesquiterpene alcohol, exerts antioxidant, anti-inflammatory, and anti-apoptotic activities.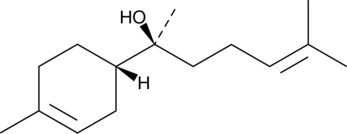
-
GC49502
(-)-β-Sesquiphellandrene
A sesquiterpene with antiviral and anticancer activities
-
GC45244
(-)-(α)-Kainic Acid (hydrate)
A potent central nervous system stimulant for induction of seizures

-
GC45246
(-)-Chaetominine
(-)-Chaetominine
(-)-Chaetominine is a cytotoxic alkaloid originally isolated from Chaetomium sp.
-
GC40698
(-)-Perillyl Alcohol
(L)-Perillyl Alcohol, (S)-Perillic Alcohol, (S)-(–)-Perillyl Alcohol
(-)-Perillyl Alcohol is a monoterpene found in lavender, inhibits farnesylation of Ras, upregulates the mannose-6-phosphate receptor and induces apoptosis. Anti-cancer activity.
-
GC40076
(-)-Voacangarine
NSC 306219, (-)-Voacristine
(-)-Voacangarine is an indole alkaloid originally isolated from V.
-
GC62193
(1S,2S)-Bortezomib
(1S,2S)-Bortezomib is an enantiomer of Bortezomib. Bortezomib is a cell-permeable, reversible, and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki of 0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is an anti-cancer agent and the first therapeutic proteasome inhibitor to be used in humans.
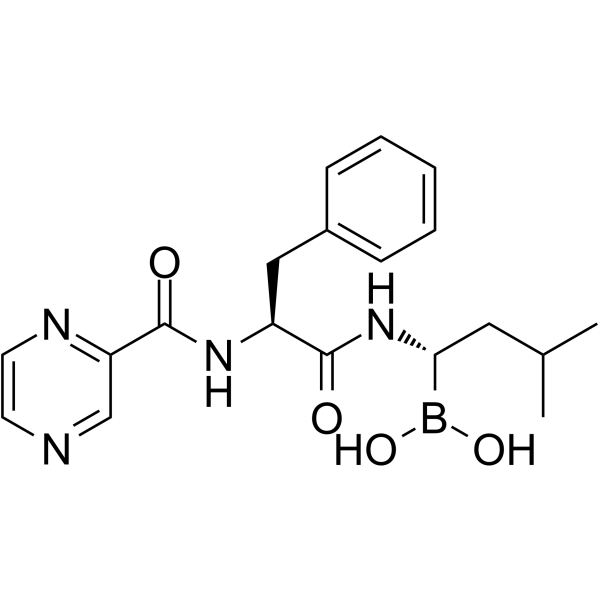
-
GC34965
(20S)-Protopanaxatriol
20(S)-APPT, 20(S)-PPT
An active ginsenoside metabolite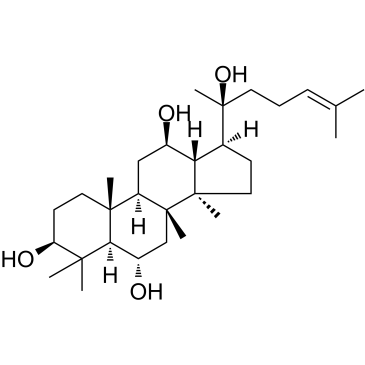
-
GC60397
(5Z,2E)-CU-3
(5Z,2E)-CU-3 is a potent and selective inhibitor against the α-isozyme of DGK with an IC50 value of 0.6 μM, competitively inhibits the affinity of DGKα for ATP with a Km value of 0.48 mM. (5Z,2E)-CU-3 targets the catalytic region, but not the regulatory region of DGKα. (5Z,2E)-CU-3 has antitumoral and proimmunogenic effects, enhances the apoptosis of cancer cells and the activation of T cells.
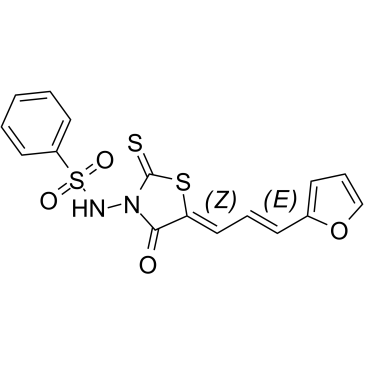
-
GC60398
(6R)-FR054
(6R)-FR054 is a less active isomer of FR054.
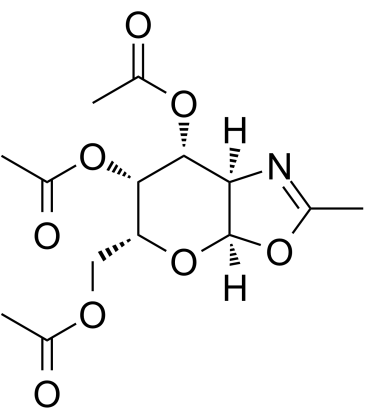
-
GC50482
(D)-PPA 1
PD-1/PD-L1 interaction inhibitor

-
GC69009
(D)-PPA 1 TFA
(D)-PPA 1 TFA is an anti-hydrolysis D-peptide antagonist. It is an effective PD-1/PD-L1 inhibitor with an affinity of 0.51 μM for binding to PD-1, and it works both in vitro and in vivo.

-
GA20156
(D-Ser(tBu)⁶,Azagly¹⁰)-LHRH (free base)

-
GC41700
(E)-2-(2-Chlorostyryl)-3,5,6-trimethylpyrazine
CSTMP
(E)-2-(2-Chlorostyryl)-3,5,6-trimethylpyrazine (CSTMP) is a stilbene derivative with antioxidant and anticancer activities.
-
GC41268
(E)-2-Hexadecenal
trans-2-Hexadecenal
Sphingosine-1-phosphate (S1P), a bioactive lipid involved in many signaling processes, is irreversibly degraded by the membrane-bound S1P lyase.
-
GC41701
(E)-2-Hexadecenal Alkyne
(E)-2-Hexadecenal alkyne is an alkyne version of the sphingolipid degradation product (E)-2-hexadecenal that can be used as a click chemistry probe.

-
GC34980
(E)-Ferulic acid
(E)-Ferulic acid is a isomer of Ferulic acid which is an aromatic compound, abundant in plant cell walls. (E)-Ferulic acid causes the phosphorylation of β-catenin, resulting in proteasomal degradation of β-catenin and increases the expression of pro-apoptotic factor Bax and decreases the expression of pro-survival factor survivin. (E)-Ferulic acid shows a potent ability to remove reactive oxygen species (ROS) and inhibits lipid peroxidation. (E)-Ferulic acid exerts both anti-proliferation and anti-migration effects in the human lung cancer cell line H1299.
-
GC34981
(E)-Flavokawain A
Flavokavain A, 4-methoxy Flavokawain B
(E)-Flavokawain A, a chalcone extracted from Kava, has anticarcinogenic effect. (E)-Flavokawain A induces apoptosis in bladder cancer cells by involvement of bax protein-dependent and mitochondria-dependent apoptotic pathway and suppresses tumor growth in mice.
-
GC61437
(E)-Methyl 4-coumarate
trans-4-Coumaric Acid methyl ester, trans-p-Coumaric Acid methyl ester, trans-para-Coumaric Acid methyl ester
(E)-Methyl 4-coumarate (Methyl 4-hydroxycinnamate), found in several plants, such as green onion (Allium cepa) or noni (Morinda citrifolia L.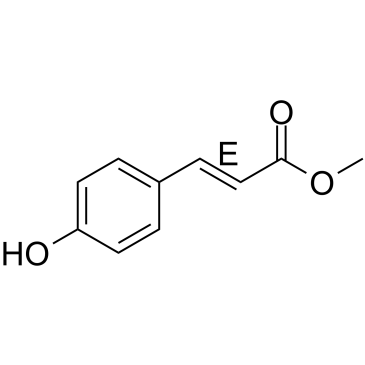
-
GC34125
(E)-[6]-Dehydroparadol
(E)-[6]-Dehydroparadol, an oxidative metabolite of [6]-Shogaol, is a potent Nrf2 activator. (E)-[6]-Dehydroparadol can inhibit the growth and induce the apoptosis of human cancer cells.
![(E)-[6]-Dehydroparadol Chemical Structure (E)-[6]-Dehydroparadol Chemical Structure](/media/struct/GC3/GC34125.png)
-
GC49189
(E/Z)-4-hydroxy Tamoxifen-d5
Afimoxifene-d5, 4-OHT-d5
An internal standard for the quantification of (E/Z)-4-hydroxy tamoxifen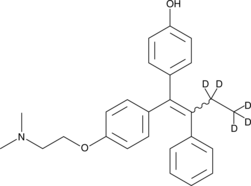
-
GN10783
(R) Ginsenoside Rh2
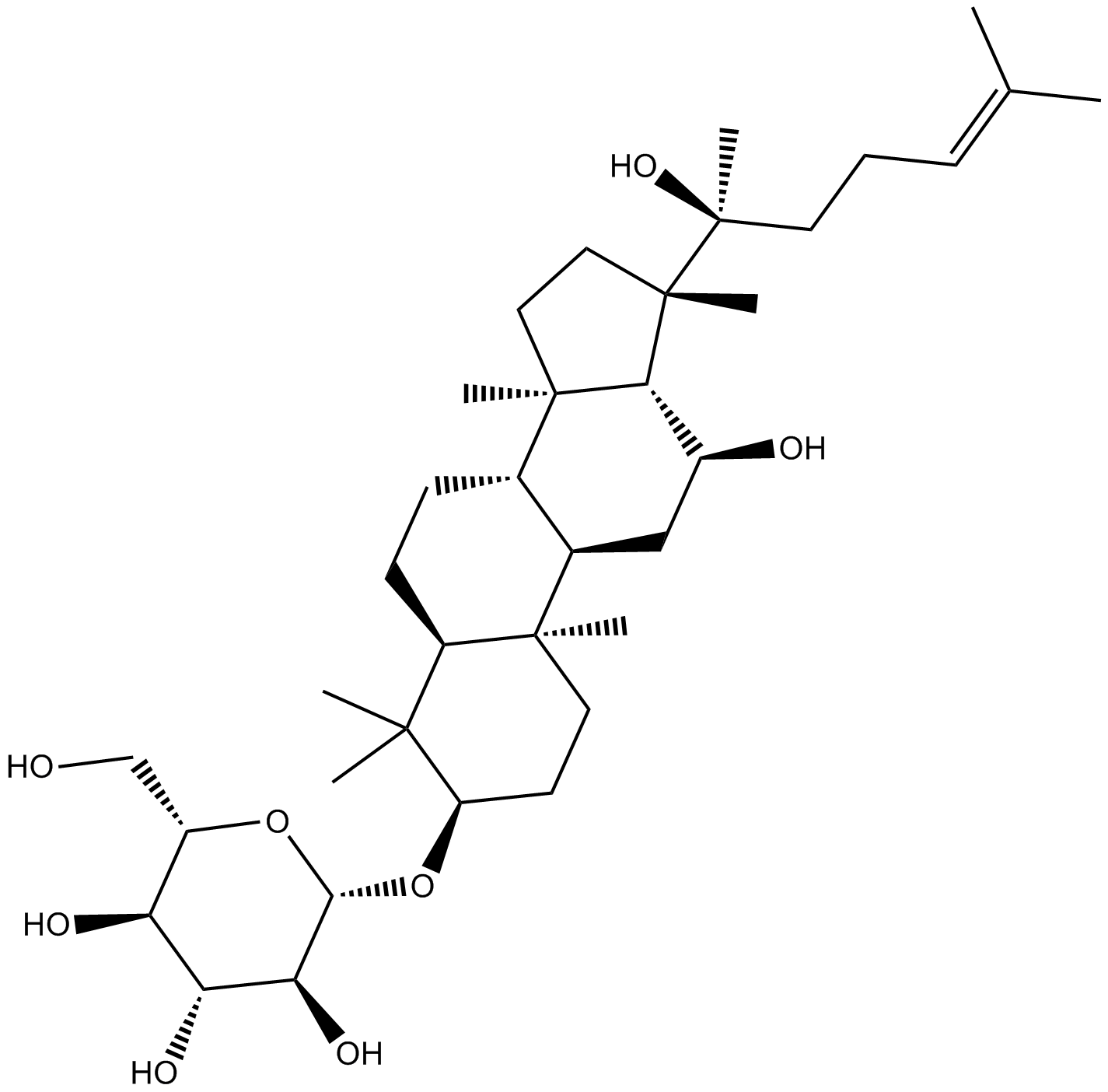
-
GC15104
(R)-(+)-Etomoxir sodium salt
(R)(+)Etomoxir
Etomoxir((R)-(+)-Etomoxir) sodium salt is an irreversible inhibitor of carnitine palmitoyltransferase 1a (CPT-1a), inhibits fatty acid oxidation (FAO) through CPT-1a and inhibits palmitate β-oxidation in human, rat and guinea pig.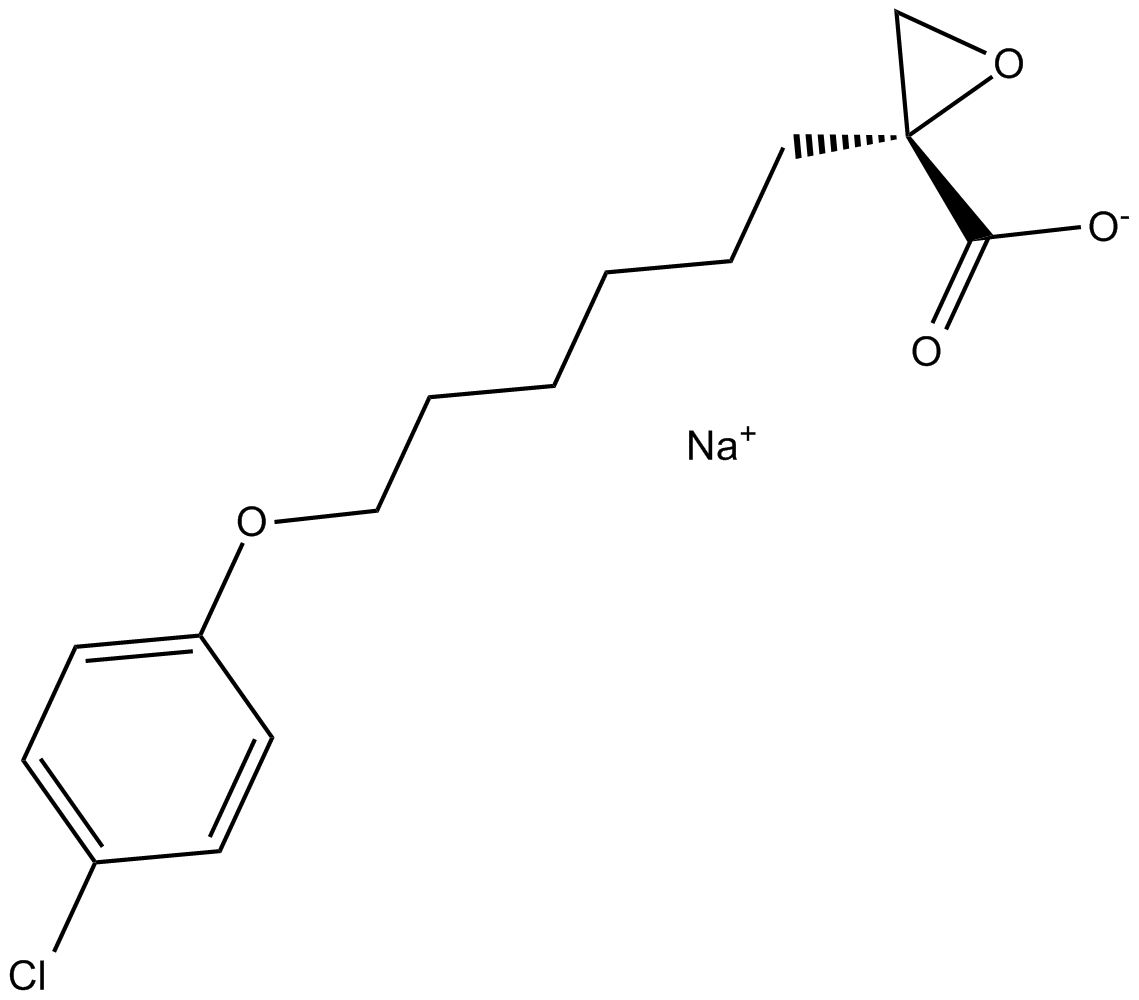
-
GC34096
(R)-(-)-Gossypol acetic acid (AT-101 (acetic acid))
(R)-(-)-Gossypol acetic acid (AT-101 (acetic acid)) (AT-101 (acetic acid)) is the levorotatory isomer of a natural product Gossypol. AT-101 is determined to bind to Bcl-2, Mcl-1 and Bcl-xL proteins with Kis of 260±30 nM, 170±10 nM, and 480±40 nM, respectively.
-
GC65610
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone is a diarylheptanoid that can be found in Alpinia officinarum.

-
GC41716
(R)-CR8
Cyclin-dependent kinases (CDKs) are key regulators of cell cycle progression and are therefore promising targets for cancer therapy.
-
GC39281
(R)-CR8 trihydrochloride
CR8, (R)-Isomer trihydrochloride
(R)-CR8 (CR8) trihydrochloride, a second-generation analog of Roscovitine, is a potent CDK1/2/5/7/9 inhibitor.
-
GC41719
(R)-nitro-Blebbistatin
R(-)7Desmethyl8nitro Blebbistatin
(R)-nitro-Blebbistatin is a more stable form of (+)-blebbistatin, which is the inactive form of (-)-blebbistatin.
-
GC60407
(R)-Verapamil D7 hydrochloride
(R)-(+)-Verapamil D7 hydrochloride
(R)-Verapamil D7 hydrochloride ((R)-(+)-Verapamil D7 hydrochloride) is a deuterium labeled (R)-Verapamil hydrochloride. (R)-Verapamil hydrochloride ((R)-(+)-Verapamil hydrochloride) is a P-Glycoprotein inhibitor. (R)-Verapamil hydrochloride blocks MRP1 mediated transport, resulting in chemosensitization of MRP1-overexpressing cells to anticancer drugs.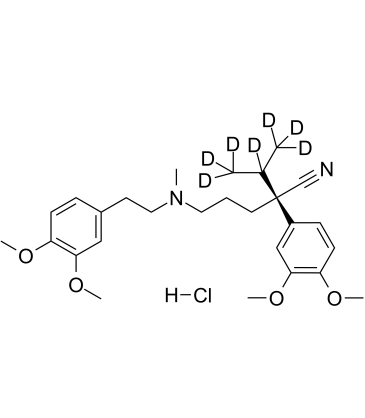
-
GC60408
(R)-Verapamil hydrochloride
(R)-Verapamil hydrochloride ((R)-(+)-Verapamil hydrochloride) is a P-Glycoprotein inhibitor. (R)-Verapamil hydrochloride blocks MRP1 mediated transport, resulting in chemosensitization of MRP1-overexpressing cells to anticancer drugs.
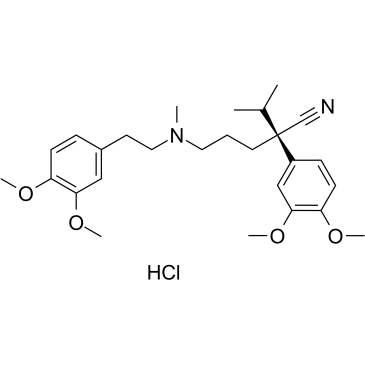
-
GC19541
(rac)-Antineoplaston A10
(rac)-Antineoplaston A10 is the racemate of Antineoplaston A10

-
GC69795
(Rac)-BIO8898
(Rac)-BIO8898 is a CD40-CD154 co-stimulatory interaction inhibitor. (Rac)-BIO8898 inhibits the binding of CD154 to CD40-Ig, with an IC50 of 25 μM.
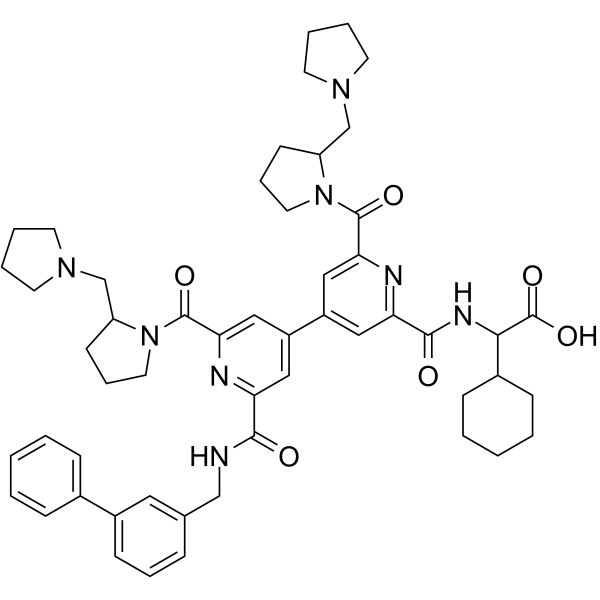
-
GC62528
(Rac)-Hesperetin
(Rac)-Hesperetin is the racemate of Hesperetin.
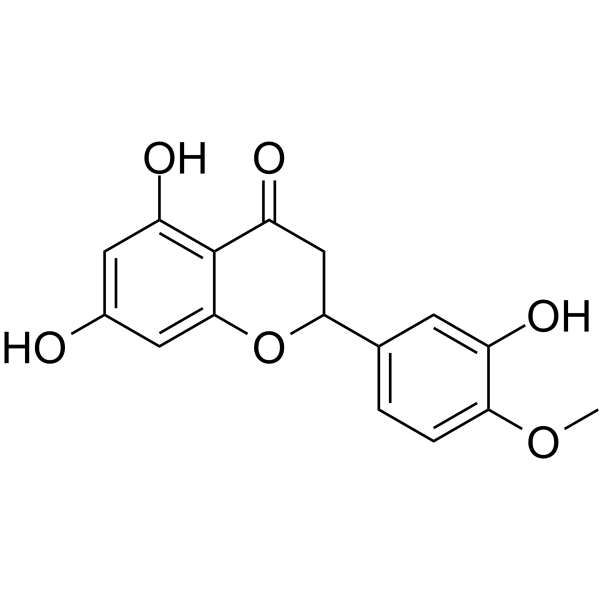
-
GC61750
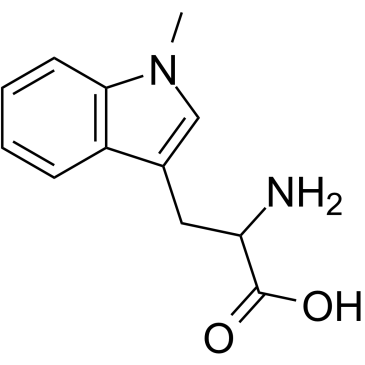
(Rac)-Indoximod
(Rac)-Indoximod (1-Methyl-DL-tryptophan) is an indoleamine 2,3-dioxygenase (IDO) inhibitor.
-
GC10098
(S)-10-Hydroxycamptothecin
ChEMBL 273862, NSC 107124
inhibitor of topoisomerase I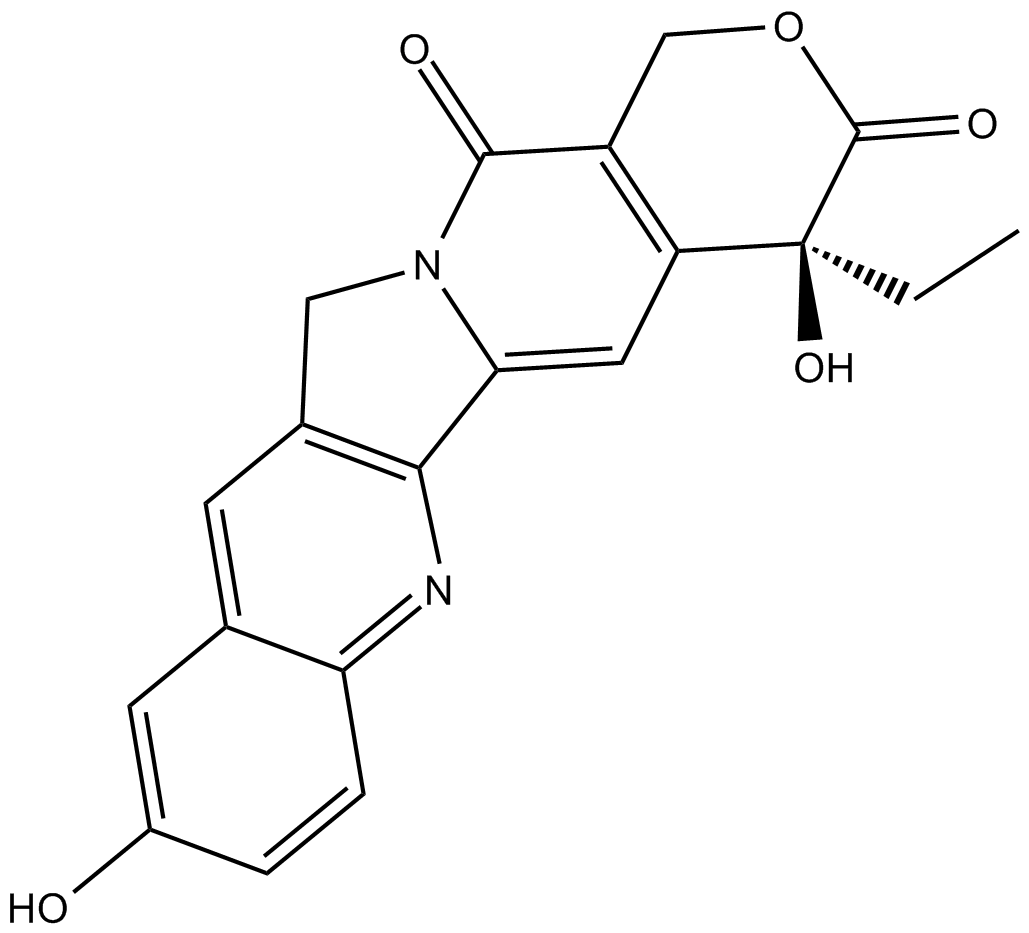
-
GC41557
(S)-3'-amino Blebbistatin
(-)-3'-amino Blebbistatin, m-amino Blebbistatin, meta-amino Blebbistatin
(S)-3'-amino Blebbistatin is a more stable and less phototoxic form of (-)-blebbistatin, which is a selective cell-permeable inhibitor of non-muscle myosin II ATPases.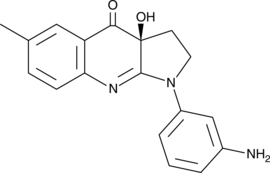
-
GC41484
(S)-3'-hydroxy Blebbistatin
(-)-3'-hydroxy Blebbistatin, meta-hydroxy-Blebbistatin, m-hydroxy-Blebbistatin
(S)-3'-hydroxy Blebbistatin is a more stable and less phototoxic form of (-)-blebbistatin, which is a selective cell-permeable inhibitor of non-muscle myosin II ATPases.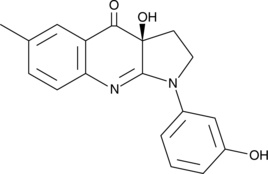
-
GC52192
(S)-4'-nitro-Blebbistatin
(-)-4'-nitro-Blebbistatin, p-nitro-Blebbistatin, para-nitro-Blebbistatin
(S)-4'-nitro-Blebbistatin is a non-cytotoxic, photostable, fluorescent and specific Myosin II inhibitor, usd in the study of the specific role of myosin II in physiological, developmental, and cell biological studies.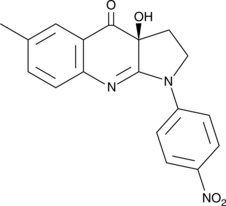
-
GC35001
(S)-Gossypol acetic acid
(S)-(+)-Gossypol acetic acid
(S)-Gossypol is the isomer of a natural product Gossypol. (S)-Gossypol binds to the BH3-binding groove of Bcl-xL and Bcl-2 proteins with high affinity.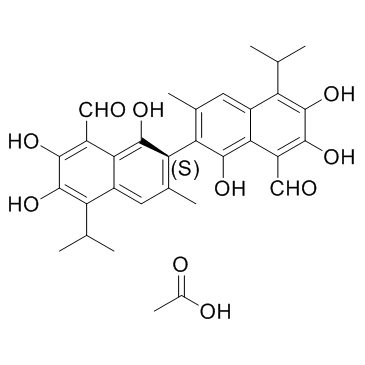
-
GC41739
(S)-nitro-Blebbistatin
S(-)7Desmethyl8nitro Blebbistatin
(S)-nitro-Blebbistatin is a more stable form of (-)-blebbistatin, which is a selective cell-permeable inhibitor of non-muscle myosin II ATPases.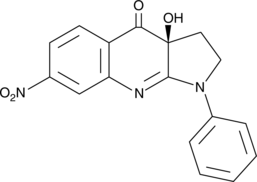
-
GC60425
(S)-Verapamil D7 hydrochloride
(S)-(-)-Verapamil D7 hydrochloride
(S)-Verapamil D7 hydrochloride ((S)-(-)-Verapamil D7 hydrochloride) is a deuterium labeled (S)-Verapamil hydrochloride. (S)-Verapamil hydrochloride (S(-)-Verapamil hydrochloride) inhibits leukotriene C4 (LTC4) and calcein transport by MRP1. (S)-Verapamil hydrochloride leads to the death of potentially resistant tumor cells.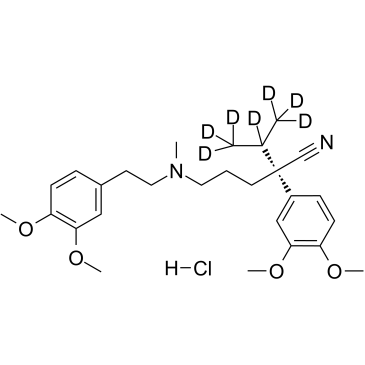
-
GC60008
(S)-Verapamil hydrochloride
(S)-Verapamil hydrochloride (S(-)-Verapamil hydrochloride) inhibits leukotriene C4 (LTC4) and calcein transport by MRP1. (S)-Verapamil hydrochloride leads to the death of potentially resistant tumor cells.
-
GC18787
(±)-Dunnione
NSC 95403
(±)-Dunnione is a naturally occurring naphthoquinone with diverse biological activities.
-
GC11965
(±)-Huperzine A
Hup A, (-)-Selagine
A neuroprotective AChE inhibitor
-
GC16375
(±)-Jasmonic Acid methyl ester
(±)-Methyl Jasmonate
(±)-Jasmonic Acid methyl ester is an endogenous metabolite.
-
GC14154
(±)-Nutlin-3
Nutlin 3b
MDM2 antagonist, potent and selective
-
GC46379
1,2-Dioleoyl-sn-glycero-3-PS (sodium salt)
1,2-DOPS, 18:1/18:1-PS; PS(18:1/18:1), 1,2-Dioctadecenoyl-sn-glycero-3-Phosphoserine, 1,2-Dioctadecenoyl-sn-glycero-3-Phosphatidylserine
1,2-Dioleoyl-sn-glycero-3-PS (sodium salt) is a ubstitute for Phosphoserine/phosphatidylserine.
-
GC19528
1,4-Benzoquinone
p-Benzoquinone, NSC 36324, p-Quinone
A toxic metabolite of benzene
-
GC42018
1-O-Octadecyl-2-O-methyl-sn-glycerol
2Methyl1octadecylsnglycerol, PIA 7
1-O-Octadecyl-2-O-methyl-sn-glycerol is a metabolite of a phosphotidylinositol ether lipid analog (PIA).
-
GC41865
10'-Desmethoxystreptonigrin
10'-Desmethoxystreptonigrin is an antibiotic originally isolated from Streptomyces and a derivative of the antibiotic streptonigrin.
-
GC49736
10-acetyl Docetaxel
PNU 101383, 10-acetyl Taxotere
10-acetyl Docetaxel (10-Acetyl docetaxel) is an analog of Docetaxel, with anticancer activity. Docetaxel is a microtubule disassembly inhibitor, with antimitotic activity.
-
GC64726
10-Formyl-5,8-dideazafolic acid
10-Formyl-5,8-dideazafolic acid is a thymidylate synthase inhibitor.

-
GC49872
10-Formyltetrahydrofolate (sodium salt) (technical grade)
10-CHO-FH4, 10-CHO-THF, N10-Formyltetrahydrofolate, 10-formyl H4PteGlu, 10-fTHF
10-Formyltetrahydrofolate (sodium salt) (technical grade) is a form of tetrahydrofolic acid that acts as a donor of formyl groups in anabolism.
-
GC70375
12-HETE-d8
12-HETE-d8 is the deuterium labeled 12-HETE.

-
GC35057
14-Deoxyandrographolide
14-DAG
A diterpene lactone with diverse biological properties
-
GC11988
15-acetoxy Scirpenol
4-Deacetylanguidin,NSC 267030
mycotoxin that induce apoptotic cell death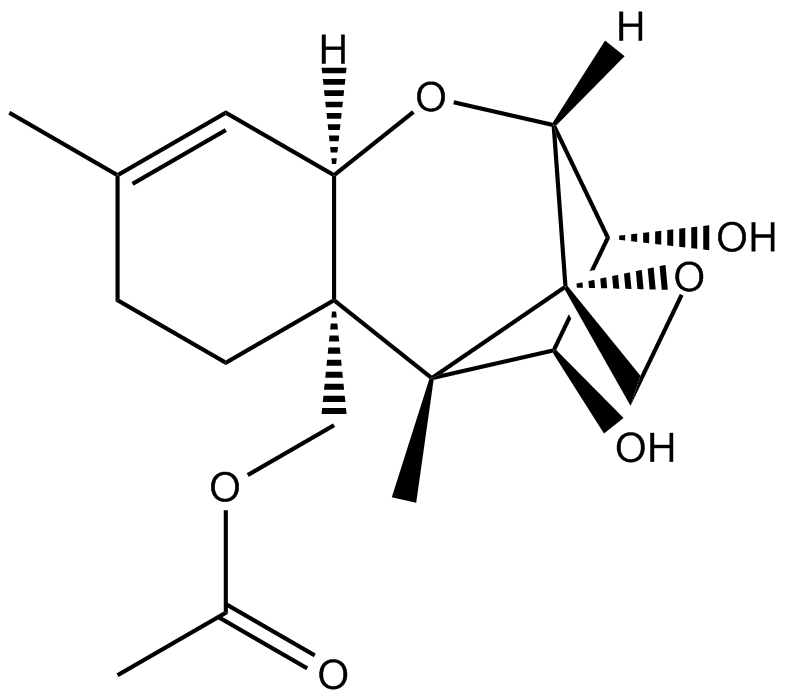
-
GC41938
15-Lipoxygenase Inhibitor 1
15LO Inhibitor 1
15-Lipoxygenase Inhibitor 1 is a selective inhibitor of 15-lipoxygenase, with an IC50 of 18 μM. 15-Lipoxygenase Inhibitor 1 has IC50s of 19.5 μM and 19.1 μM for soybean 15-lipoxygenase (SLO) and human 15-lipoxygenase-1 (15-LOX-1), respectively. 15-Lipoxygenase Inhibitor 1 has potential for the research of prostate cancer.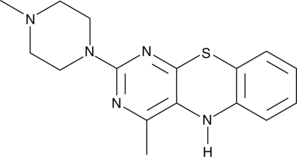
-
GC46451
16F16
A PDI inhibitor

-
GC11720
17-AAG (KOS953)
BMS 722782, CP 127374, KOS 953, NSC 330507, Tanespimycin
17-AAG(Geldanamycin), a natural benzoquinone ansamycin antibiotic, is the first established inhibitor of Hsp90.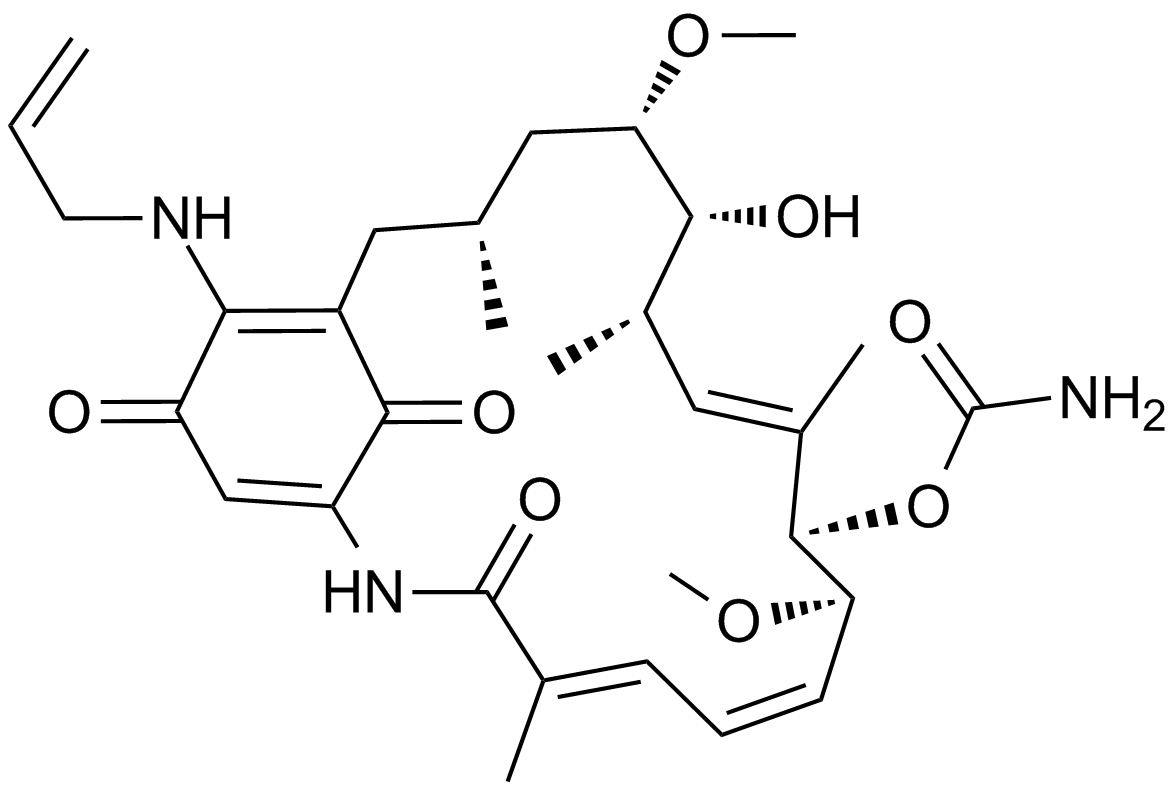
-
GC13044
17-DMAG (Alvespimycin) HCl
17-DMAG (Alvespimycin) HCl (17-DMAG hydrochloride; KOS-1022; BMS 826476) is a potent inhibitor of Hsp90, binding to Hsp90 with EC50 of 62±29 nM.

-
GC41983
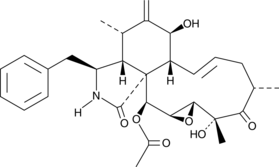
19,20-Epoxycytochalasin D
19,20-Epoxycytochalasin D is a fungal metabolite that has been found in the endophytic fungus Nemania sp.
-
GC48423
19-O-Acetylchaetoglobosin A
Chaetoglobosin A Acetate
A fungal metabolite with actin polymerization inhibitory and cytotoxic activities
-
GC39296
1G244
1G244 is a potent DPP8/9 inhibitor with IC50s of 12 nM and 84 nM, respectively. 1G244 does not inhibit DPPIV and DPPII. 1G244 induces apoptosis in multiple myeloma cells and has anti-myeloma effects.
-
GC46508
2',2'-Difluoro-2'-deoxyuridine
dFdU
An active metabolite of gemcitabine
-
GC41612
2'-O-Methylguanosine
2'-O-Methylguanosine is a modified nucleoside that is produced in tRNAs by the action of tRNA guanosine-2'-O-methyltransferase, using S-adenosyl-L-methionine as a substrate.
-
GC12258
2,3-DCPE hydrochloride
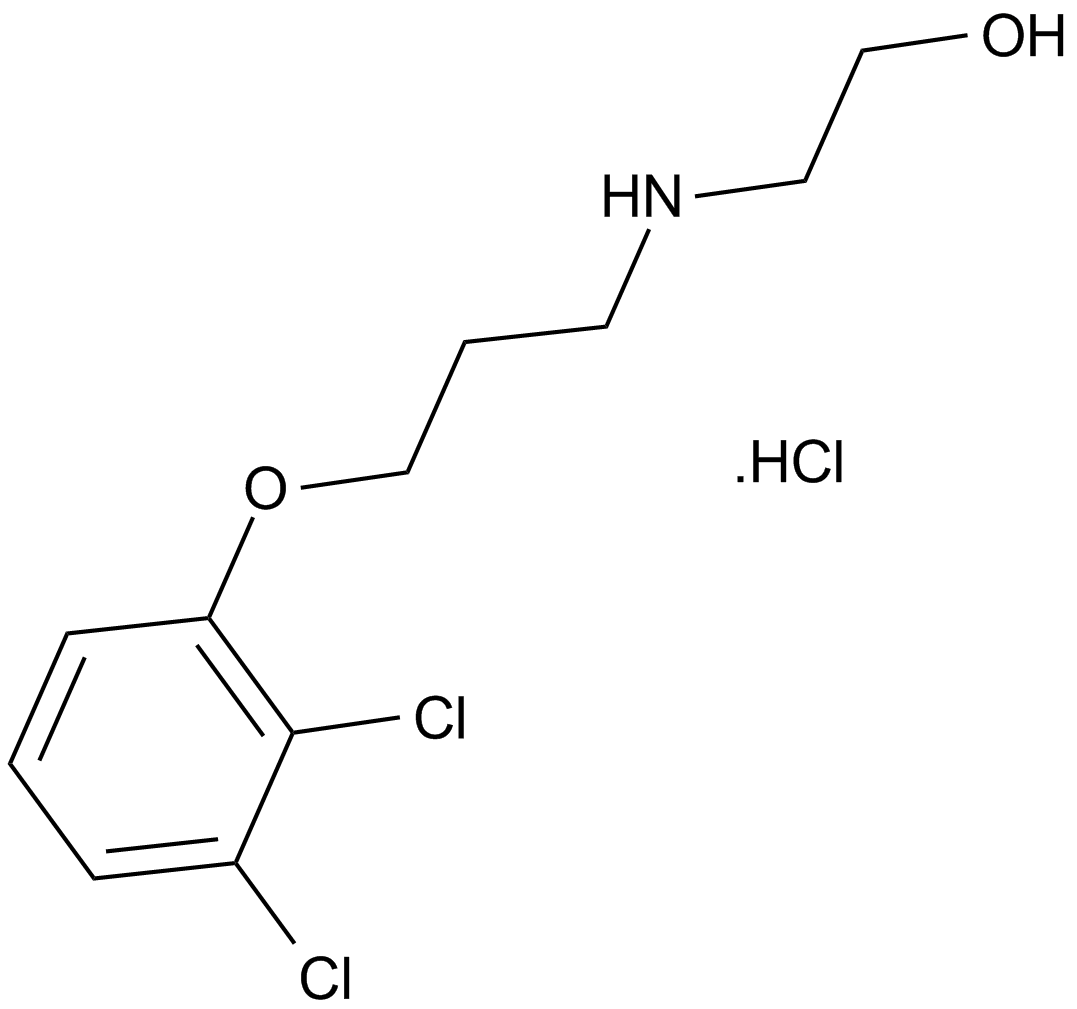
-
GC40947
2,3-Dimethoxy-5-methyl-p-benzoquinone
Coenzyme Q0, CoQ0
2,3-Dimethoxy-5-methyl-p-benzoquinone (CoQ0) is a potent, oral active ubiquinone compound can be derived from Antrodia cinnamomea.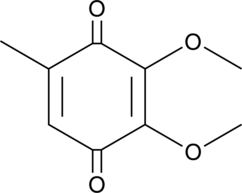
-
GC68452
2,4,6-Triiodophenol

-
GC46057
2,5-Dihydroxycinnamic Acid phenethyl ester
An inhibitor of 5-LO
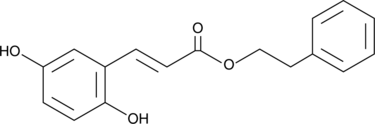
-
GC45324
2,5-dimethyl Celecoxib
DMC
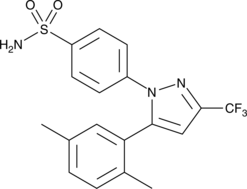
-
GN10065
2-Atractylenolide
2-Atractylenolide
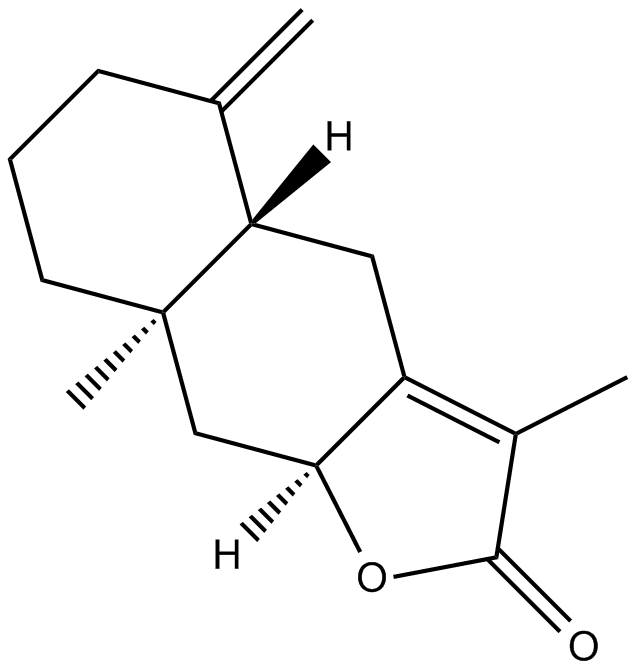
-
GC40675
2-deoxy-Artemisinin
2-deoxy-Artemisinin is an inactive metabolite of the antimalarial agent artemisinin.
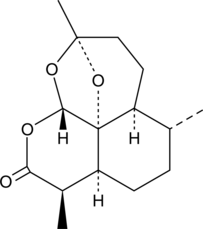
-
GC17430
2-Deoxy-D-glucose
2-DG
2-Deoxy-D-glucose (2DG), is a glucose analogue, act as competitive glycolytic inhibitor.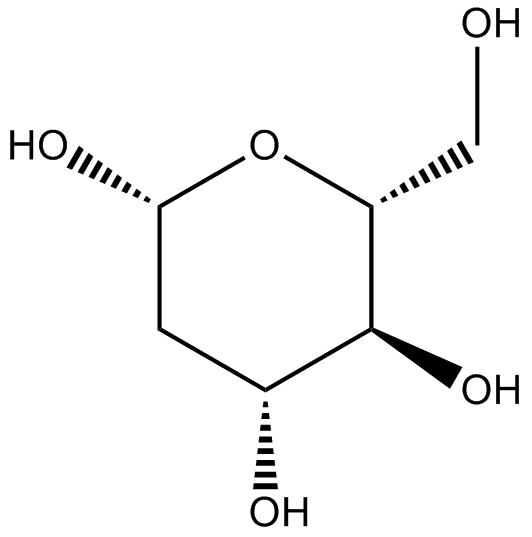
-
GC49223
2-deoxy-D-Glucose-13C6
2-DG-13C6
An internal standard for the quantification of 2-deoxy-D-glucose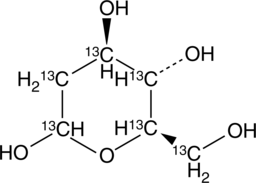
-
GC46545
2-Fluoroadenine
F-Ade, NSC 27364
A heterocyclic building block
-
GC12545
2-HBA
Bis(2-hydroxybenzylidene)acetone
indirect inducer of enzymes that catalyze detoxification reactions through the Keap1-Nrf2-ARE pathway.
-
GC38318
2-Methoxycinnamaldehyde
2-Methoxycinnamaldehyde (o-Methoxycinnamaldehyde) is a natural compound of Cinnamomum cassia, with antitumor activity. 2-Methoxycinnamaldehyde inhibits proliferation and induces apoptosis by mitochondrial membrane potential (ΔΨm) loss, activation of both caspase-3 and caspase-9. 2-Methoxycinnamaldehyde effectively inhibits platelet-derived growth factor (PDGF)-induced HASMC migration.