TGF-β / Smad Signaling
Transforming growth factor-beta (TGF-beta) is a multifunctional cytokine that regulates proliferation, migration, differentiation, and survival of many different cell types. Deletion or mutation of different members of the TGF-β family have been shown to cause vascular remodeling defect and absence of mural cell formation, leading to embryonic lethality or severe vascular disorders. TGF-β induces smooth muscle differentiation via Notch or SMAD2 and SMAD3 signaling in ES cells or in a neural crest stem cell line. TGF-β binds to TGF-βRI and to induce phosphorylation of SMAD2/3, thereby inhibiting proliferation, tube formation, and migration of endothelial cells (ECs).
TGF-β is a pluripotent cytokine with dual tumour-suppressive and tumour-promoting effects. TGF-β induces the epithelial-to-mesenchymal transition (EMT) leading to increased cell plasticity at the onset of cancer cell invasion and metastasis.
Targets for TGF-β / Smad Signaling
Products for TGF-β / Smad Signaling
- Cat.No. Product Name Information
-
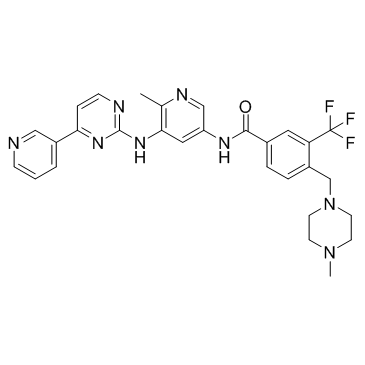
GC13890
(±)-Palmitoylcarnitine chloride
C16:0 Carnitine, CAR 16:0, DL-Carnitine hexadecanoyl ester, DL-Carnitine palmitoyl ester, Hexadecanoyl-DL-carnitine, DL-Hexadecanoylcarnitine, NSC 628323, DL-Palmitoylcarnitine
intermediate in mitochondrial fatty acid oxidation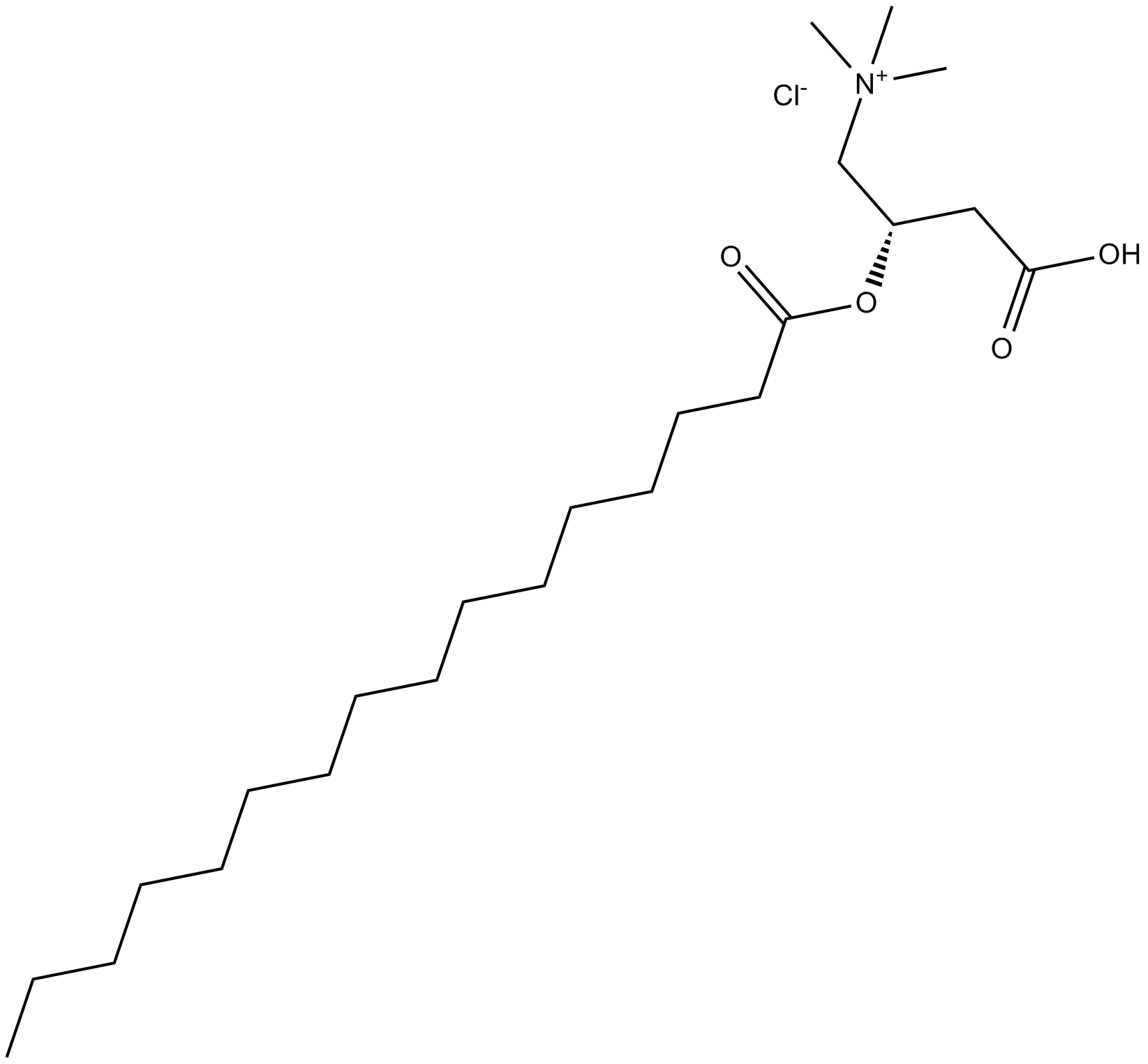
-
GC10603
(-)-epicatechin gallate
ECG
major catechin in green tea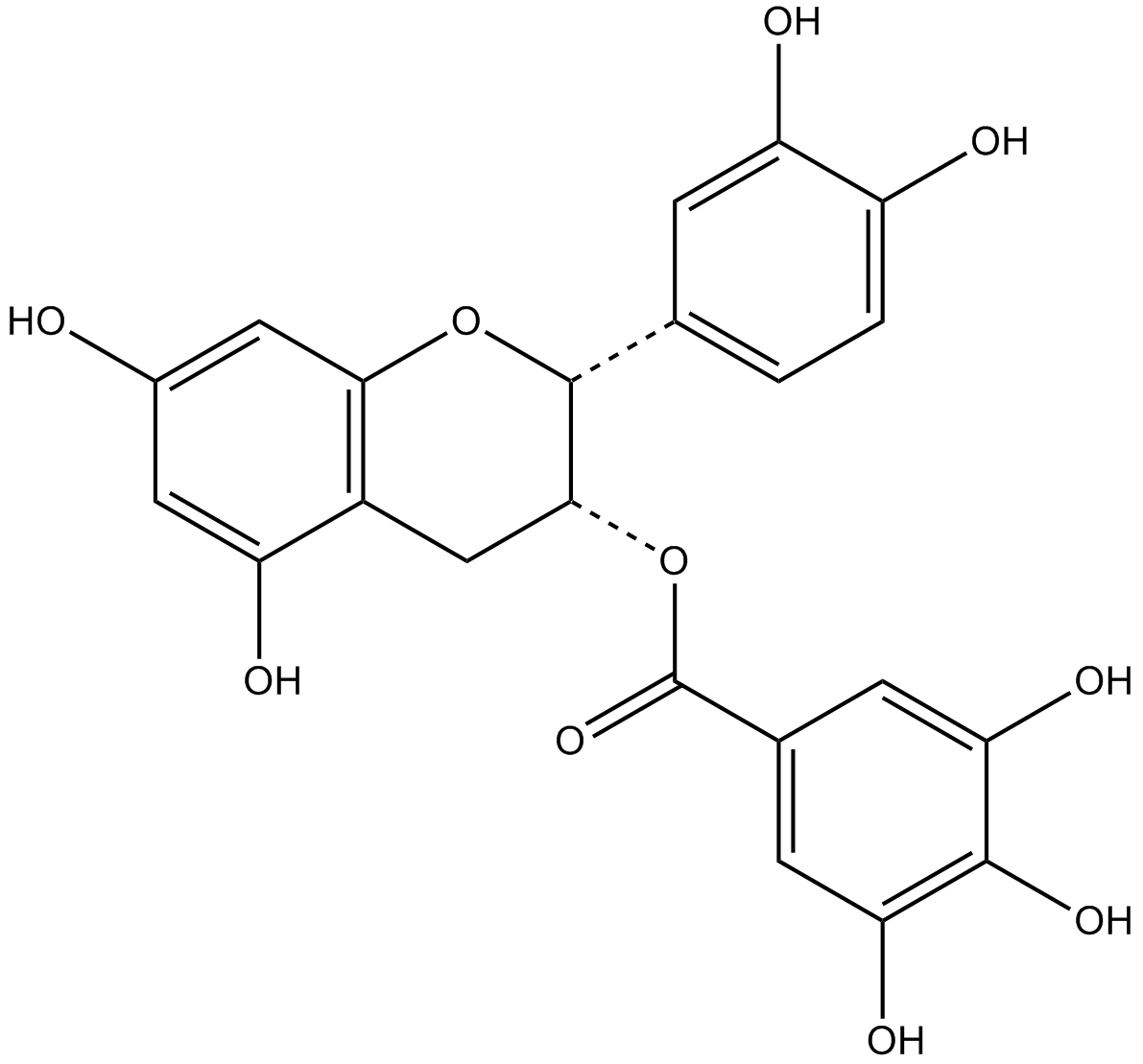
-
GC17242
(-)-epigallocatechin
(-)EGC, epi-Gallocatechin, NSC 674039
green tea epicatechin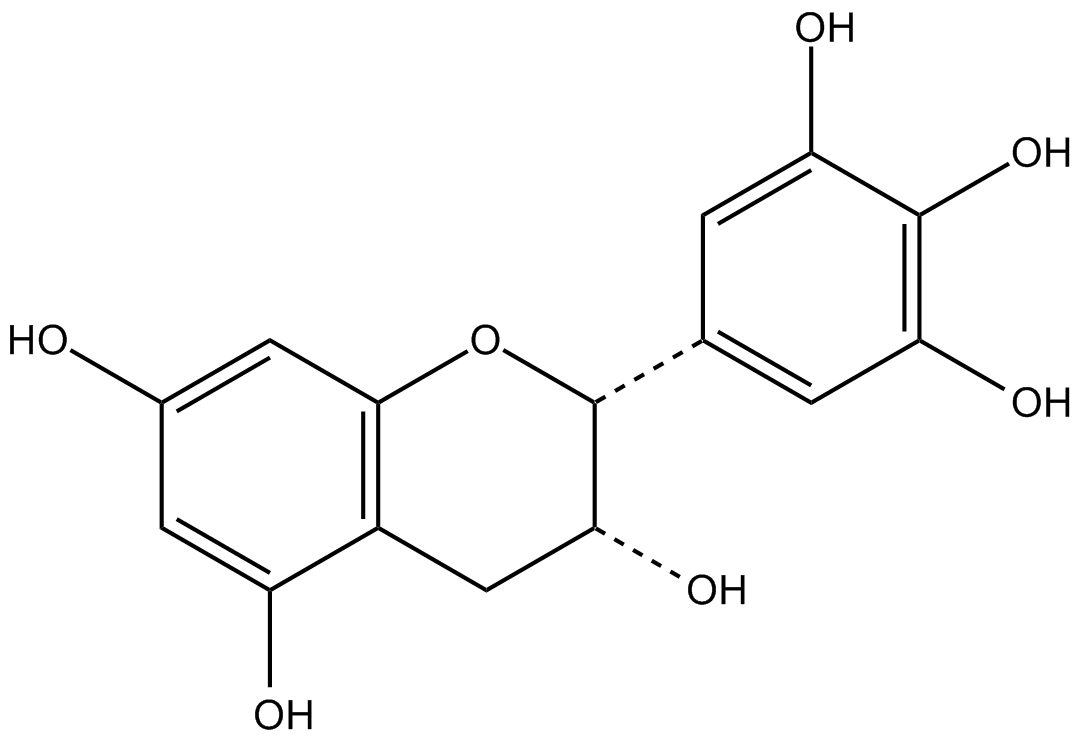
-
GC14049
(-)-Epigallocatechin gallate (EGCG)
EGCG
(-)-Epigallocatechin Gallate sulfate (EGCG) is a major polyphenol in green tea that inhibits cell proliferation and induces apoptosis.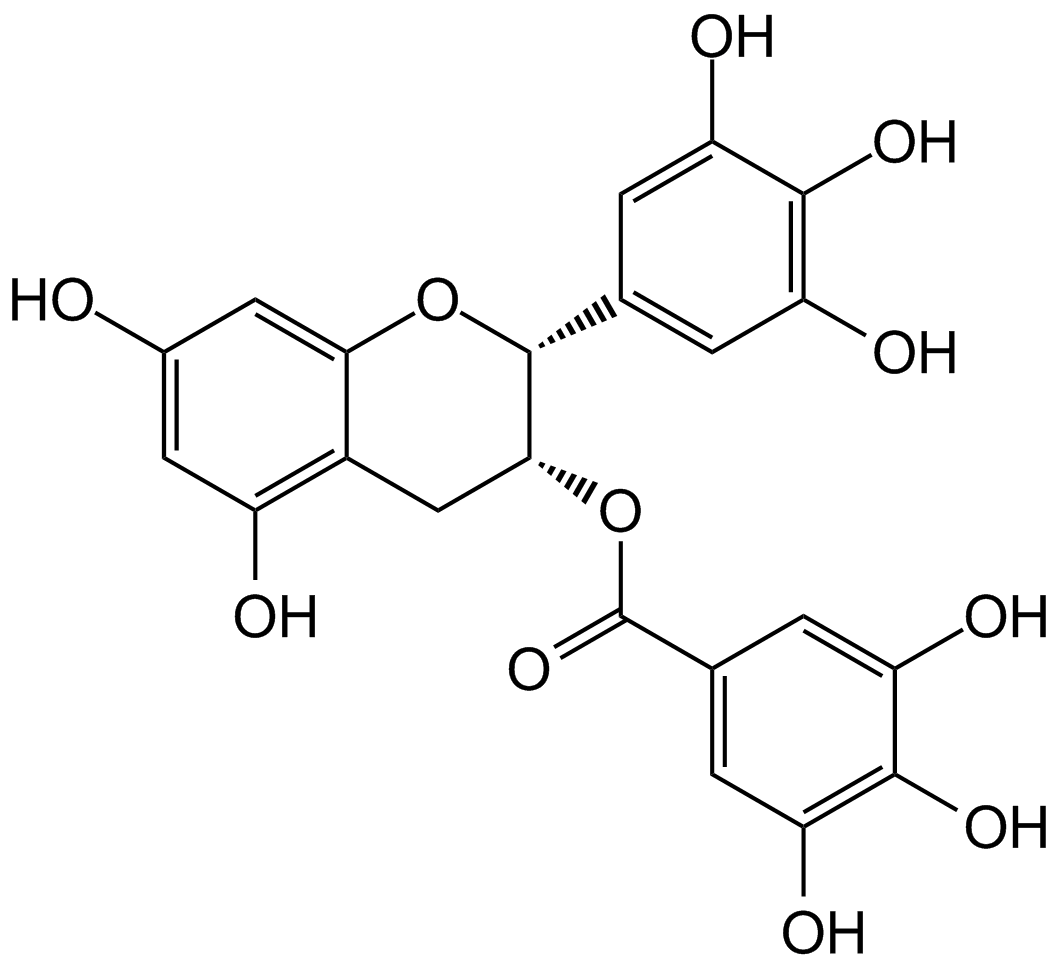
-
GC14012
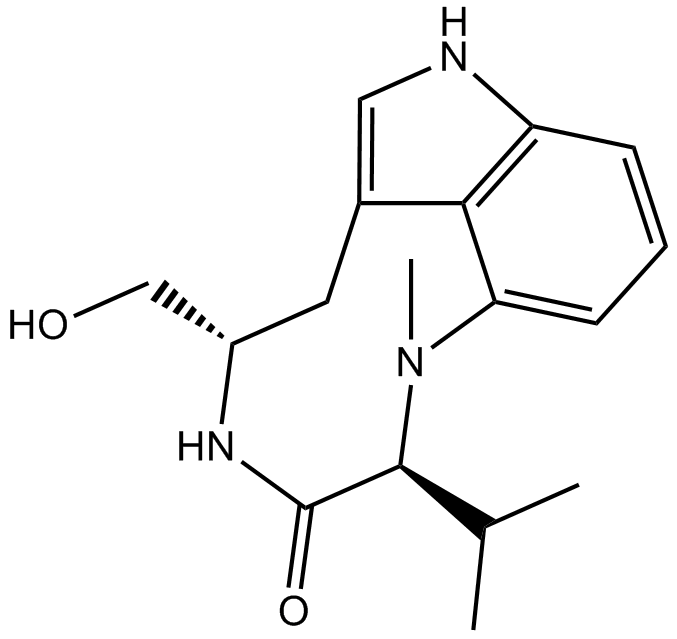
(-)-Indolactam V
A protein kinase C activator
-
GC70295
(S)-Ro 32-0432
(S)-Ro 32-0432 is a potent, selective, ATP-competitive and orally active PKC inhibitor.

-
GC18062
1,2-Dilauroyl-sn-glycerol
1,2-DLG
A longchain DAG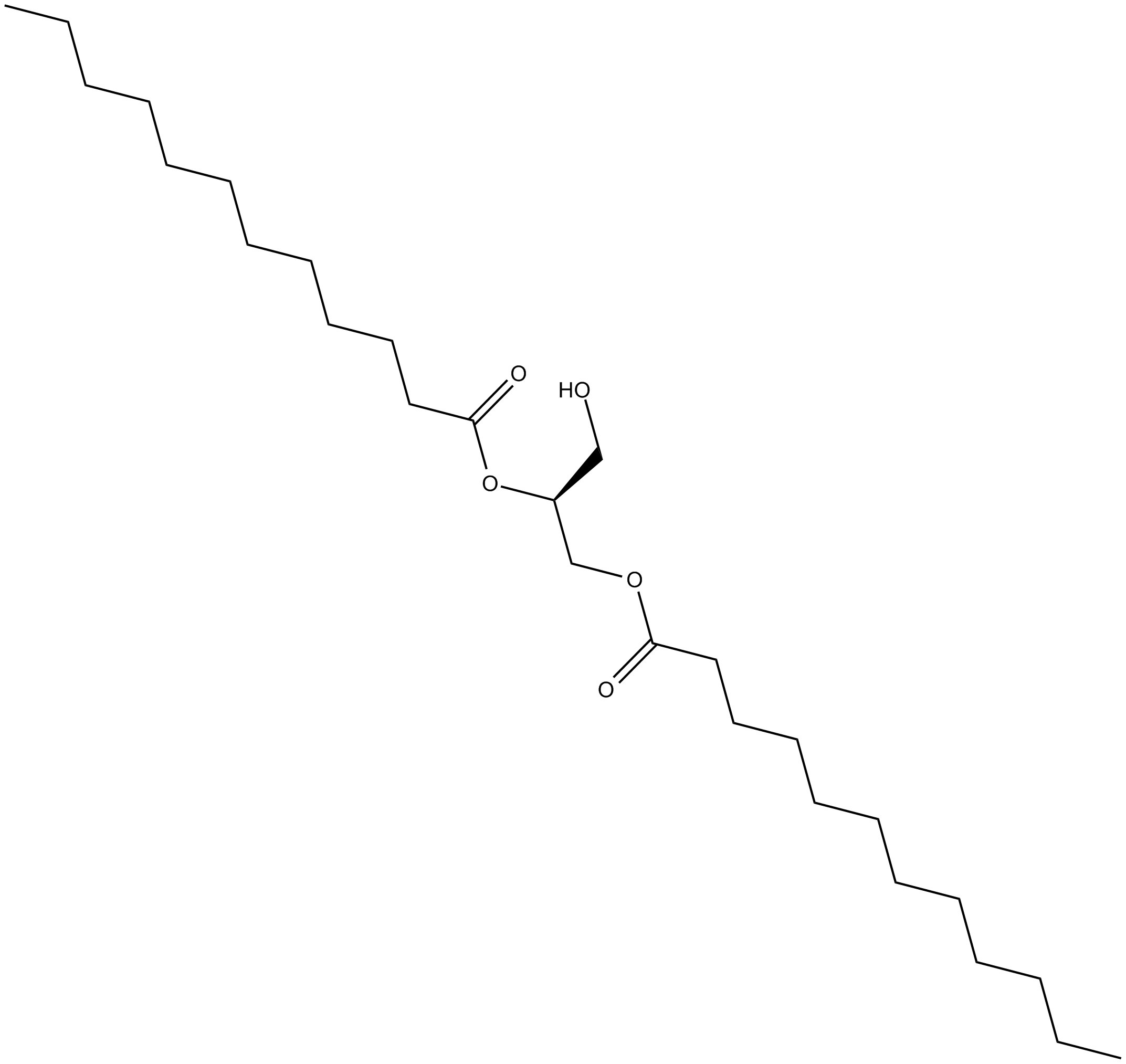
-
GC14134
1,2-Dimyristoyl-sn-glycerol
1,2-DMG
A longchain DAG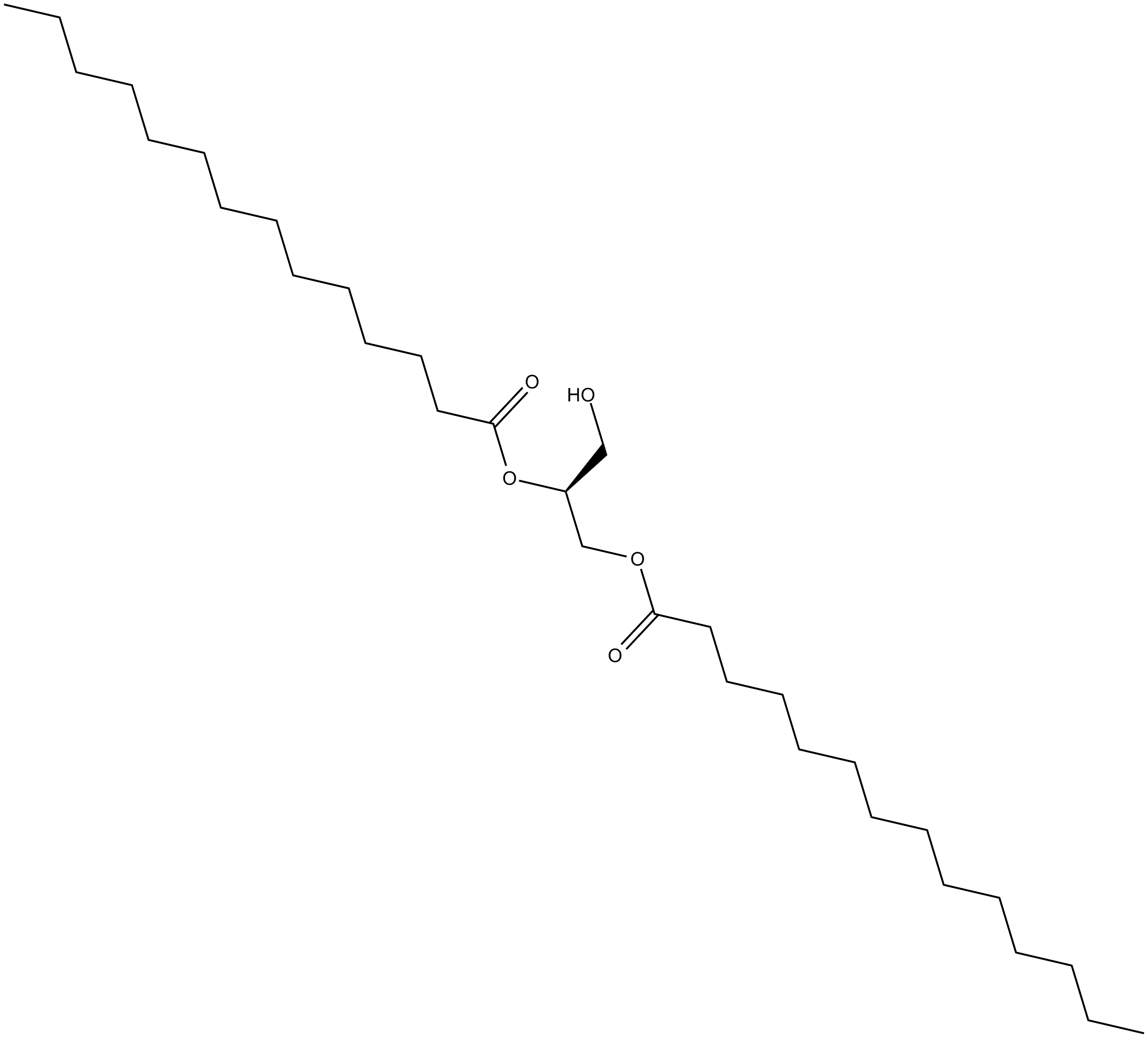
-
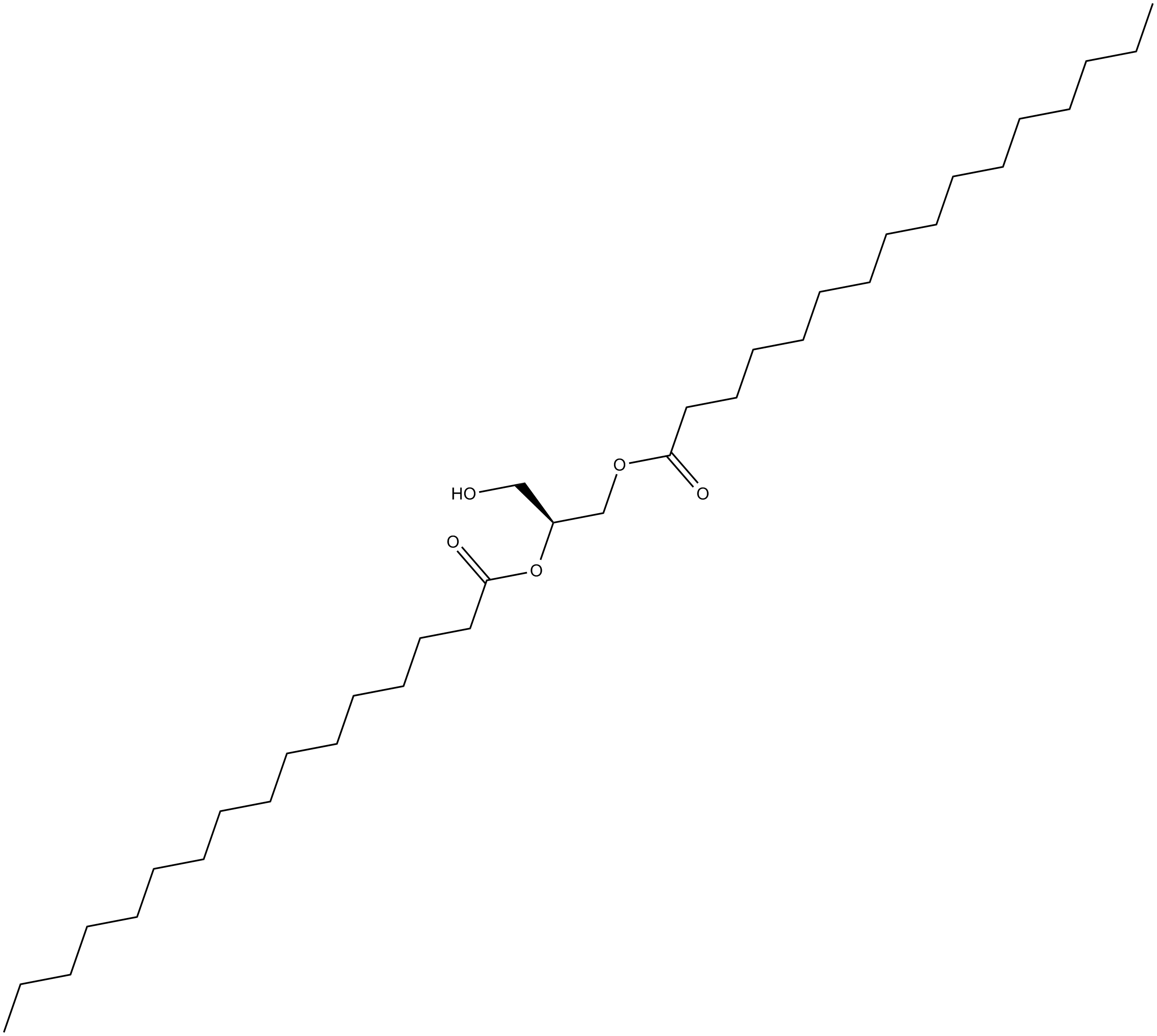
GC13877
1,2-Dipalmitoyl-sn-glycerol
1,2-DPG,NSC 269964
weak activator of PKC
-
GC12662
1,2-Distearoyl-sn-glycerol
1,2-Dioctadecanoyl-sn-glycerol,DSG
A diacylglycerol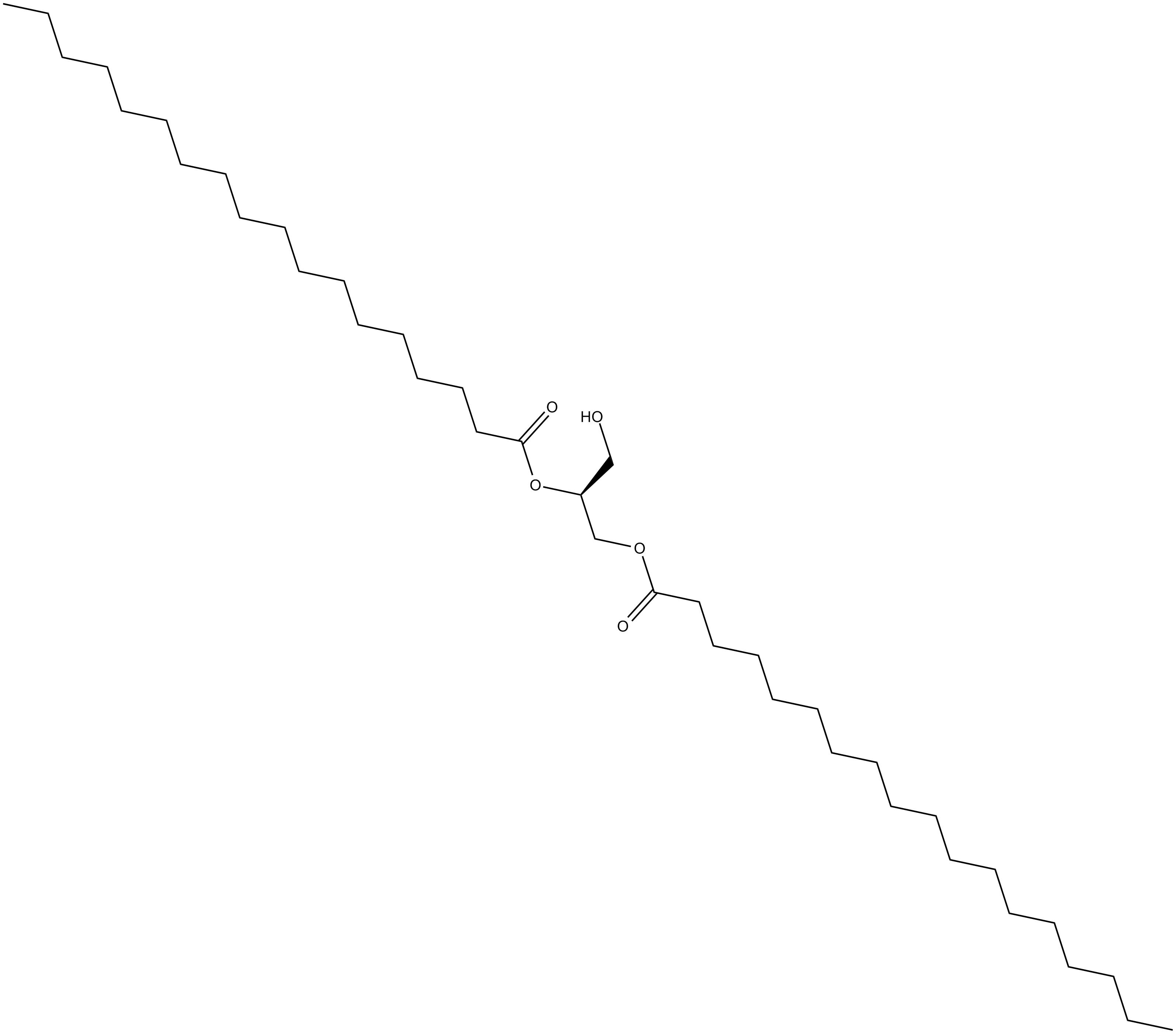
-
GC12172
1-Naphthyl PP1
1NaphthylPP1, PP1 Analog
Src family kinases inhibitor
-
GC41863
10,11-dehydro Curvularin
α,β-dehydro Curvularin
A natural mycotoxin
-
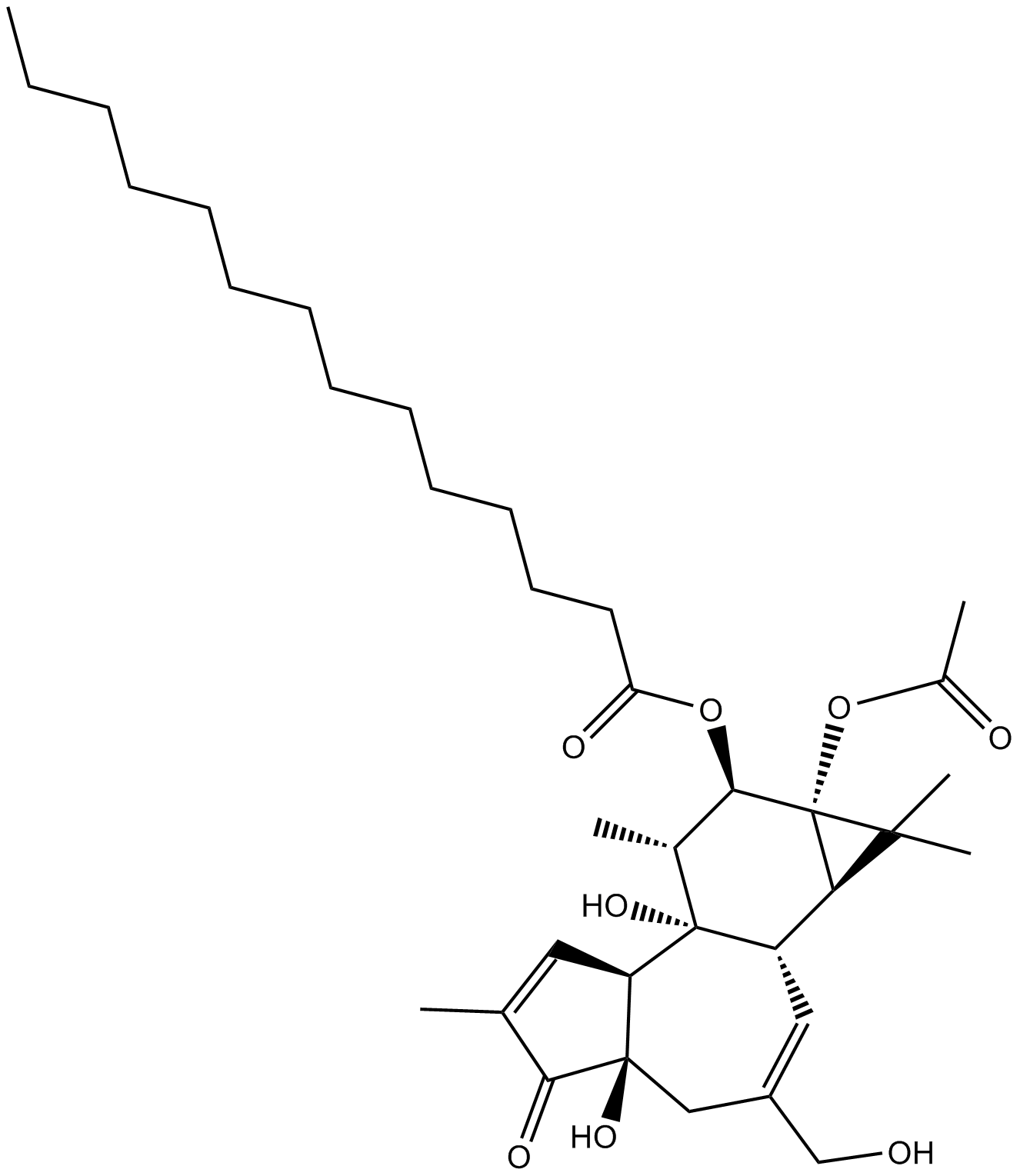
GN10444
12-O-tetradecanoyl phorbol-13-acetate
PMA; TPA; Phorbol myristate acetate
A PKC activator
-
GC13927
A 77-01
A77-01;A-77-01
A potent ALK5 inhibitor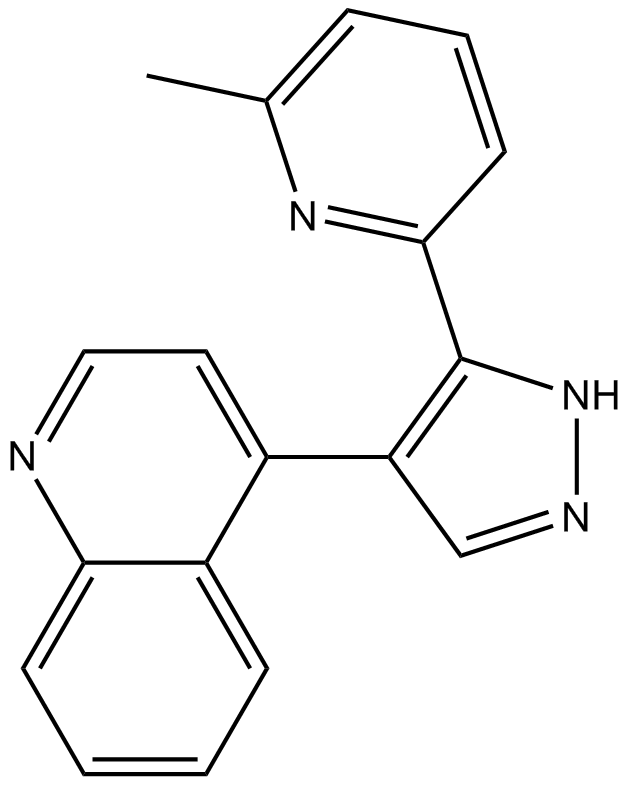
-
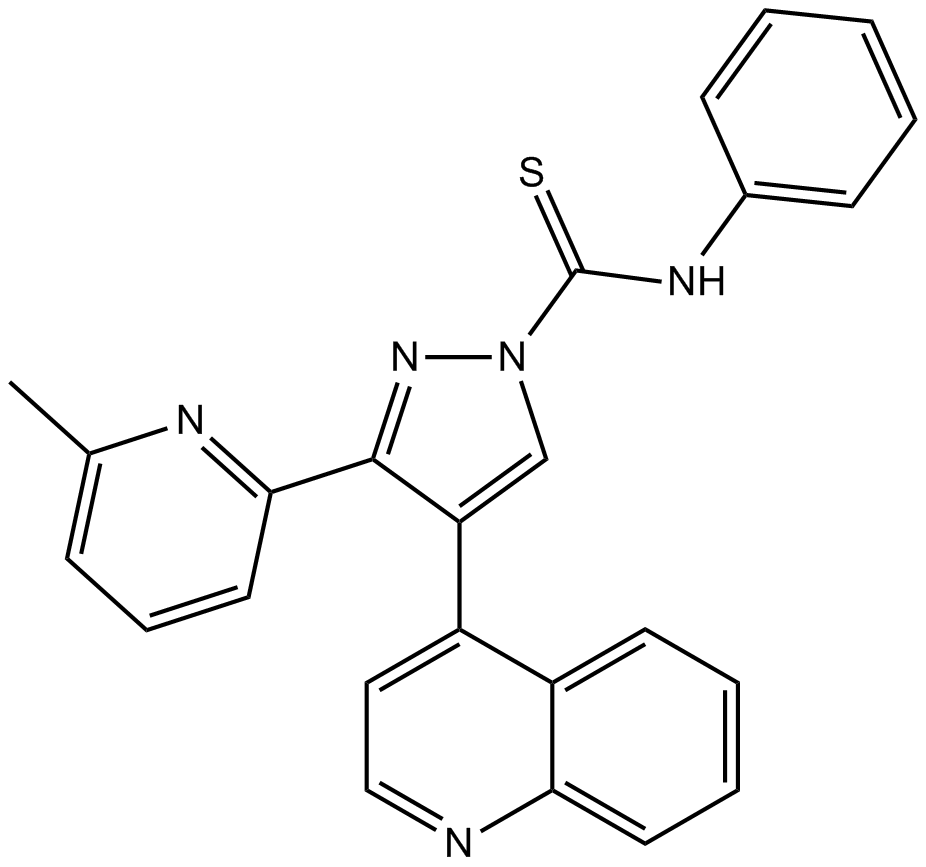
GC10166
A 83-01
A 83-01 is a potent inhibitor of TGF-β type I receptor ALK5 kinase, type I nodal receptor ALK4 and type I nodal receptor ALK7, with IC50 s of 12 nM,45 nM and 7.5 nM against the transcription induced by ALK5, ALK4 and ALK7, respectively .
-
GC35210
A 83-01 sodium salt
3-(6-Methylpyridin-2-yl)-N-phenyl-4-(quinolin-4-yl)-1H-pyrazole-1-carbothioamide, sodium salt
A potent inhibitor of TGF-β type I receptor ALK5 kinase

-
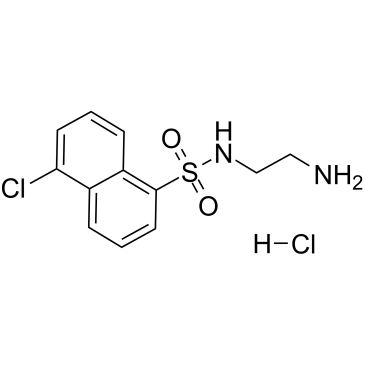
GC60037
A-3 hydrochloride
A-3 hydrochloride is a potent, cell-permeable, reversible, ATP-competitive non-selective antagonist of various kinases.
-
GC15579
Adaphostin
NSC 680410
P210bcr/abl tyrosine kinase inhibitor
-
GC16475
Afuresertib
GSK2110183
pan-AKT inhibitor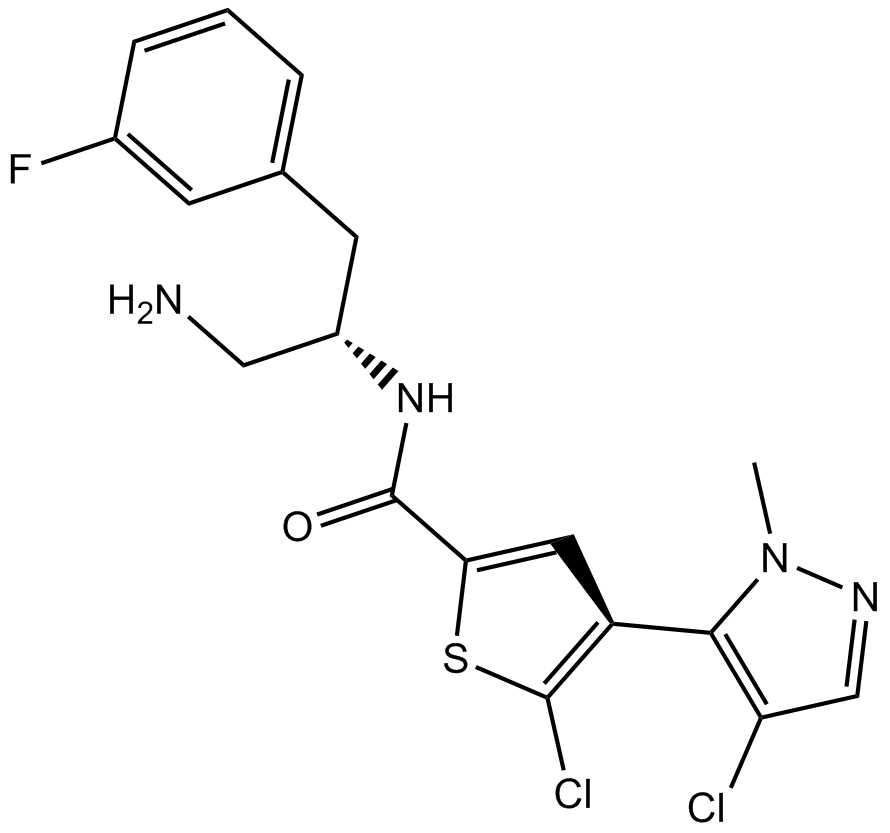
-
GC42747
Afuresertib (hydrochloride)
GSK2110183B
Afuresertib is a selective, orally bioavailable inhibitor of Akt1, 2, and 3 with Ki values of 0.08, 2, and 2.6 nM, respectively.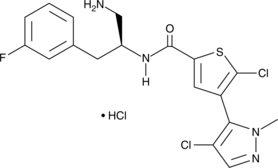
-
GC65294
ALK2-IN-2
ALK2-IN-2 is a potent and selective inhibitor of activin receptor-like kinase 2 (ALK2) with an IC50 of 9 nM, and over 700-fold selectivity against ALK3.
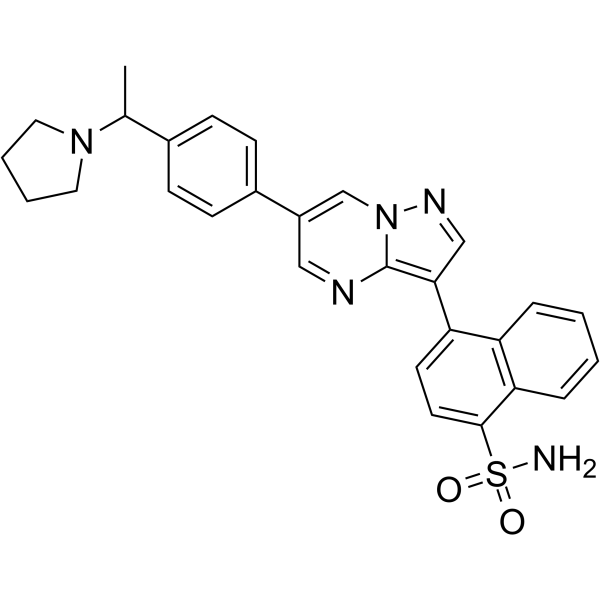
-
GC11050
ALK5 Inhibitor II (hydrochloride)
E 616452,RepSox,SJN 2511
ALK5 inhibitor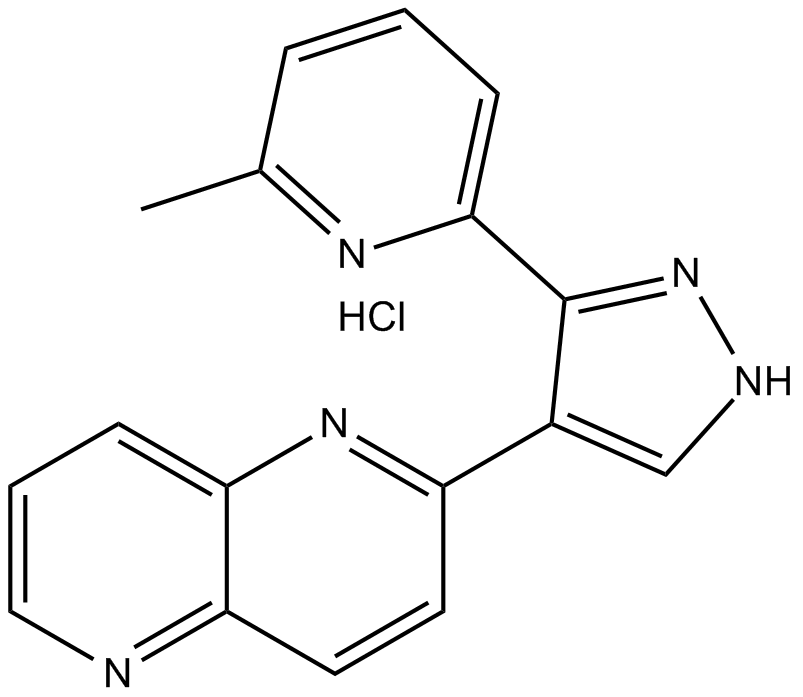
-
GC68679
Aprinocarsen sodium
ISIS 3521 sodium
Aprinocarsen (ISIS 3521) sodium is a specific antisense oligonucleotide inhibitor of protein kinase C-alpha (PKC-alpha). Aprinocarsen sodium is a 20-mer oligonucleotide that can regulate cell differentiation and proliferation. It can inhibit the growth of human tumor cell lines in nude mice. Aprinocarsen sodium has value as a chemotherapy compound for human cancer.

-
GC34300
AR-13324 analog mesylate
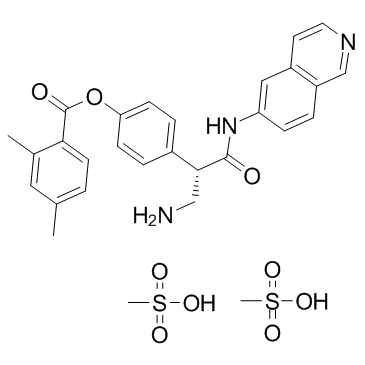
-
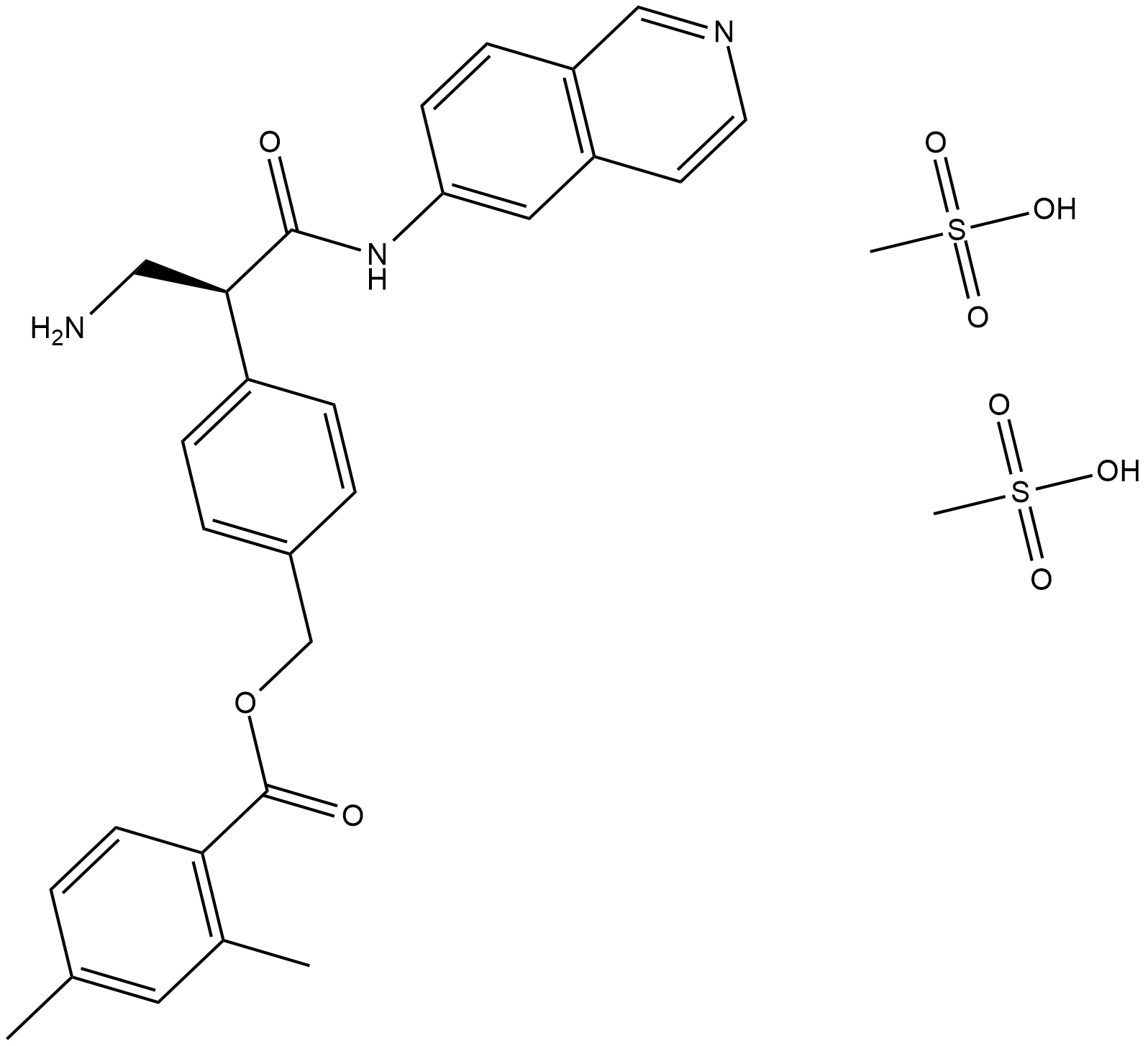
GC19034
AR-13324 mesylate
AR-13324 is a potent inhibitor of ROCK I and ROCK II that has been shown to induce morphologic changes in cultured human and porcine TM cells at low concentration.
-
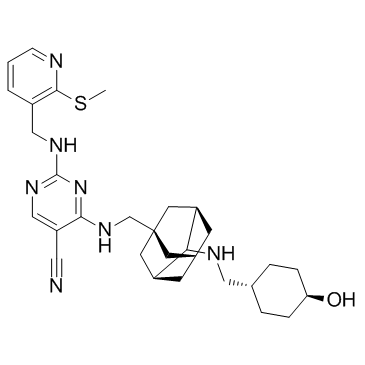
GC31959
AS2521780
AS2521780 is a novel PKCθ selective inhibitor with an IC50 of 0.48 nM.
-
GC32703
Asciminib (ABL001)
ABL001
Asciminib (ABL001) (ABL001) is a potent and selective allosteric BCR-ABL1 inhibitor, which inhibits Ba/F3 cells grown with an IC50 of 0.25 nM.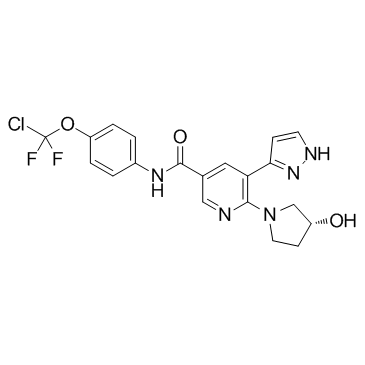
-
GC64462
Asciminib hydrochloride
Asciminib (ABL001) hydrochloride is a potent and selective allosteric BCR-ABL1 inhibitor, which inhibits Ba/F3 cells grown with an IC50 of 0.25 nM.

-
GN10534
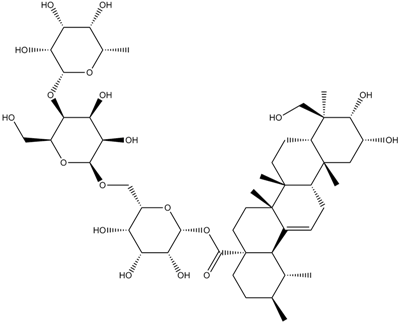
Asiaticoside
Ba 2742, NSC 36002, NSC 166062
-
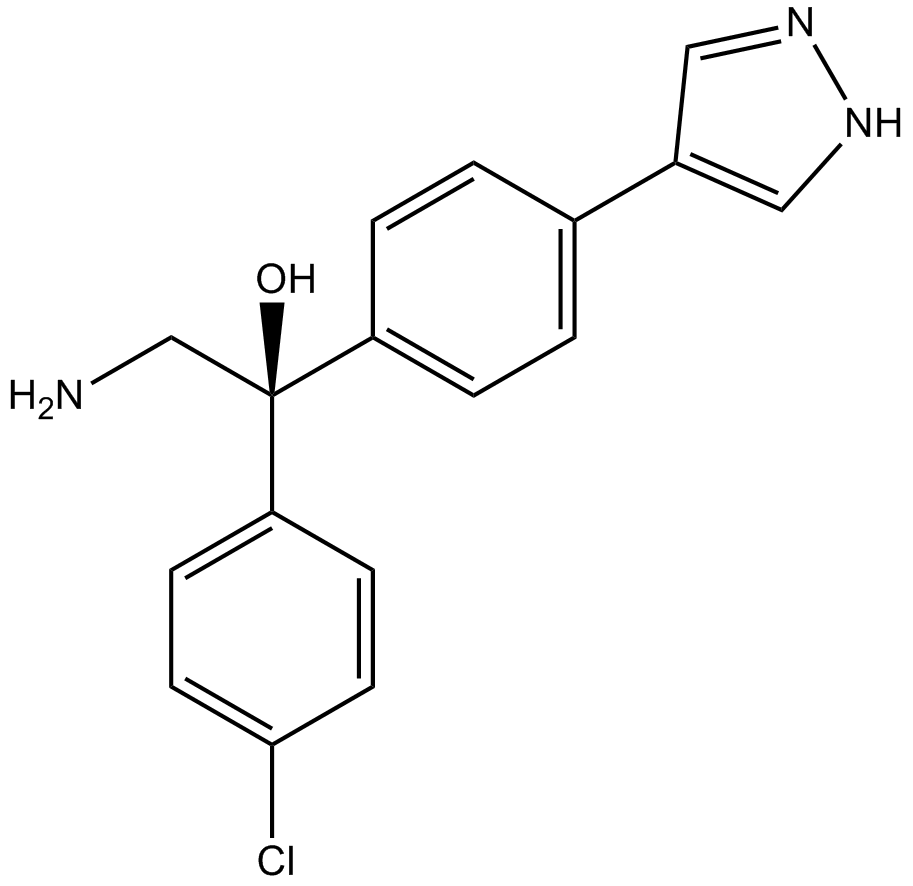
GC17712
AT13148
Multi-AGC kinase inhibitor,ATP-competitive
-
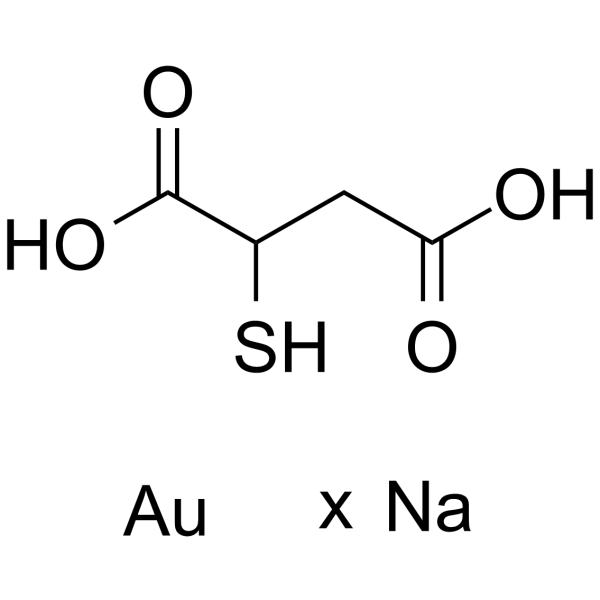
GC64815
Aurothiomalate sodium
Aurothiomalate sodium is a potent and selective oncogenic PKCι signaling inhibitor.
-
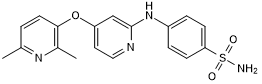
GC50589
AZ 12799734
Potent TGF-βRI inhibitor
-
GC35442
Azaindole 1
ROCK-IN-2 ,TC-S 7001
Azaindole 1 (Azaindole 1; TC-S 7001) is an orally active and ATP-competitive ROCK inhibitor with IC50s of 0.6 and 1.1nM for human ROCK-1 and ROCK-2, respectively.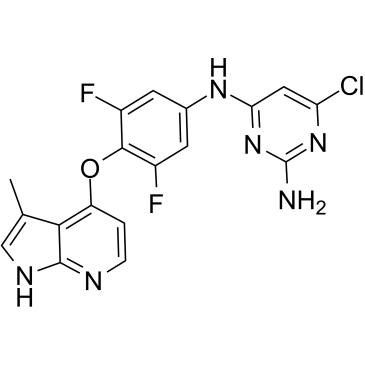
-
GC35462
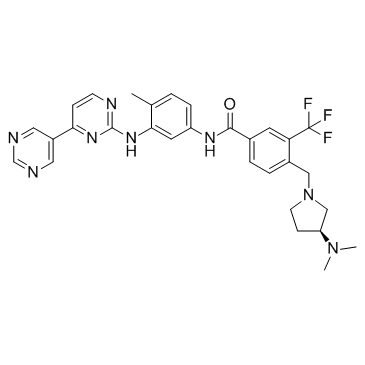
Bafetinib
INNO-406
Bcr-Abl/Lyn tyrosine kinase inhibitor
-
GC33343
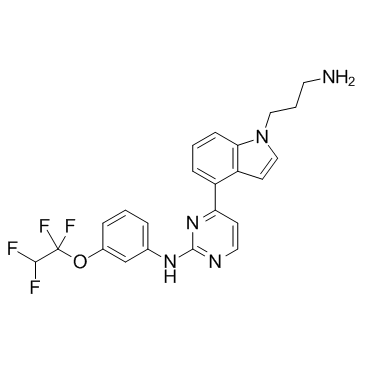
BCR-ABL-IN-1
BCR-ABL-IN-1 is an inhibitor of BCR-ABL tyrosine kinase, with a pIC50 of 6.46, and may be used in the research of chronic myelogenous leukemia.
-
GC33368
BCR-ABL-IN-2
BCR-ABL-IN-2 is an inhibitor of BCR-ABL1 tyrosine kinase, with IC50s of 57 nM, 773 nm for ABL1native and ABL1T315I, respectively.

-
GC32868
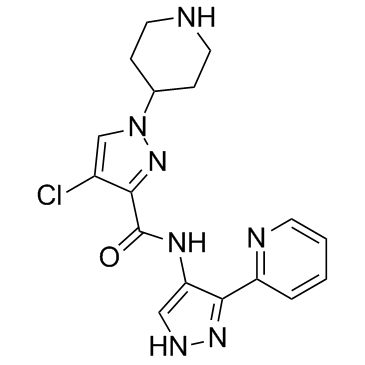
BDP5290
BDP5290 is a potent inhibitor of ROCK and MRCK with IC50 values of 17nM, 230nM, 123nM and 100nM for MRCKβ, ROCK1, ROCK2, respectively.
-
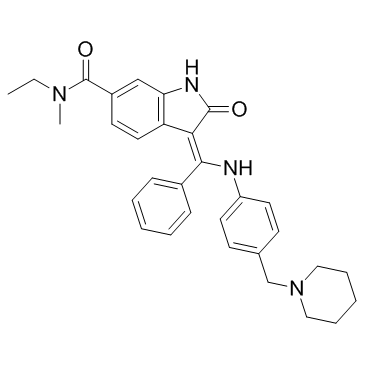
GC34493
BIBF0775
BIBF0775 is a potent and selective transforming growth factor β (TGFβ) type I receptor (Alk5) inhibitor with an IC50 of 34 nM.
-
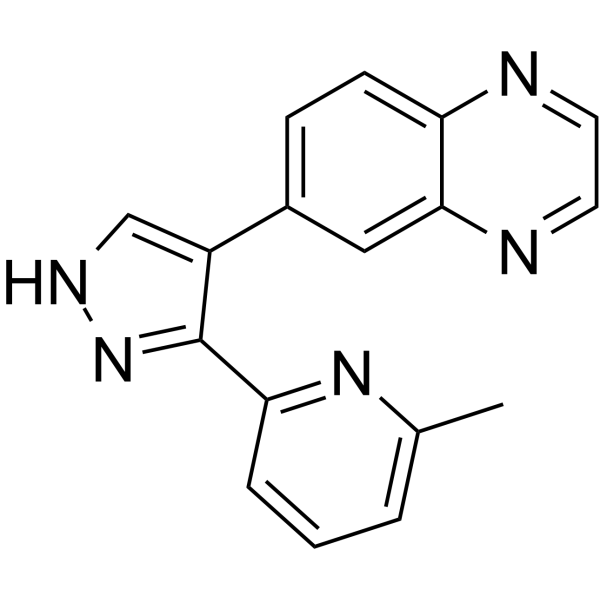
GC62868
BIO-013077-01
BIO-013077-01 is a pyrazole TGF-β inhibitor.
-
GC12135
Bisindolylmaleimide II
BIM II
protein kinase C (PKC) inhibitor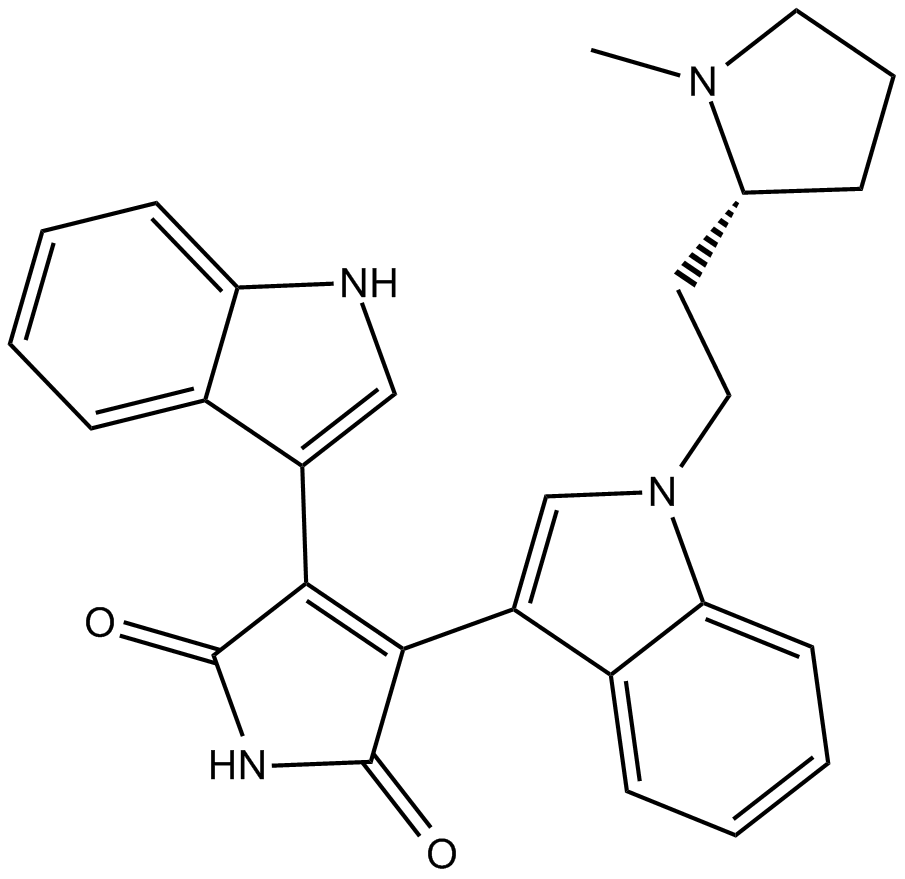
-
GC14716
Bisindolylmaleimide IV
Arcyriarubin A,BIM IV
protein kinase C (PKC) inhibitor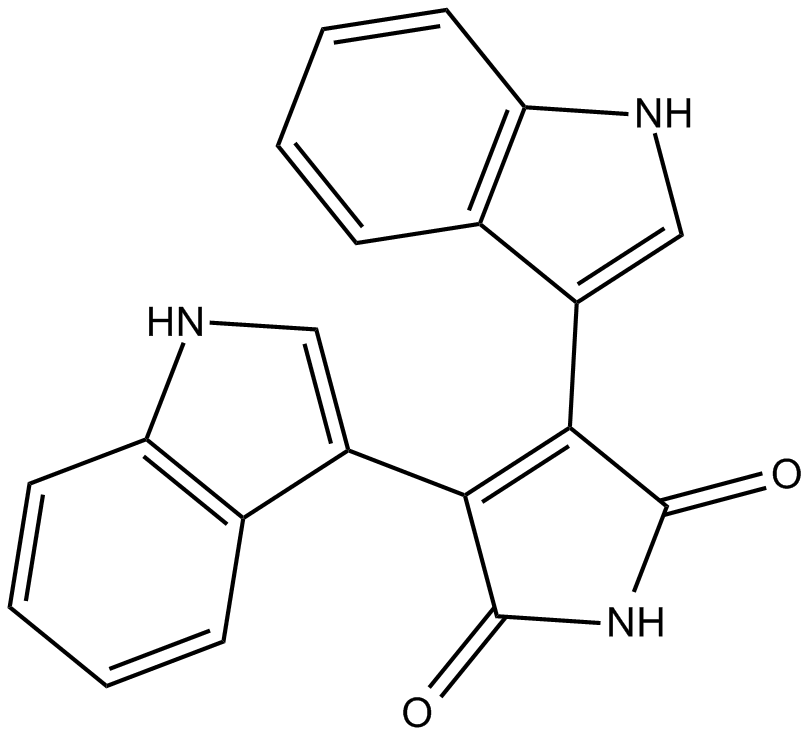
-
GC17239
Bisindolylmaleimide V
BIM V, Ro 316045
negative control for protein kinase C (PKC)-inhibitory activity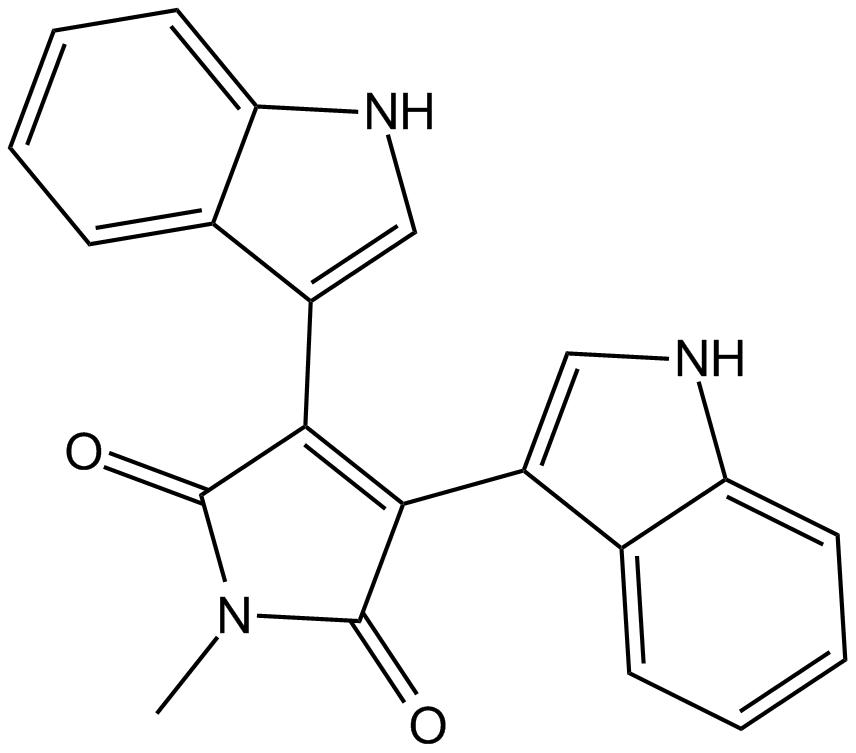
-
GC13226
Bisindolylmaleimide VIII (acetate)
BIM VIII,Ro 31-7549
A protein kinase C (PKC) inhibitor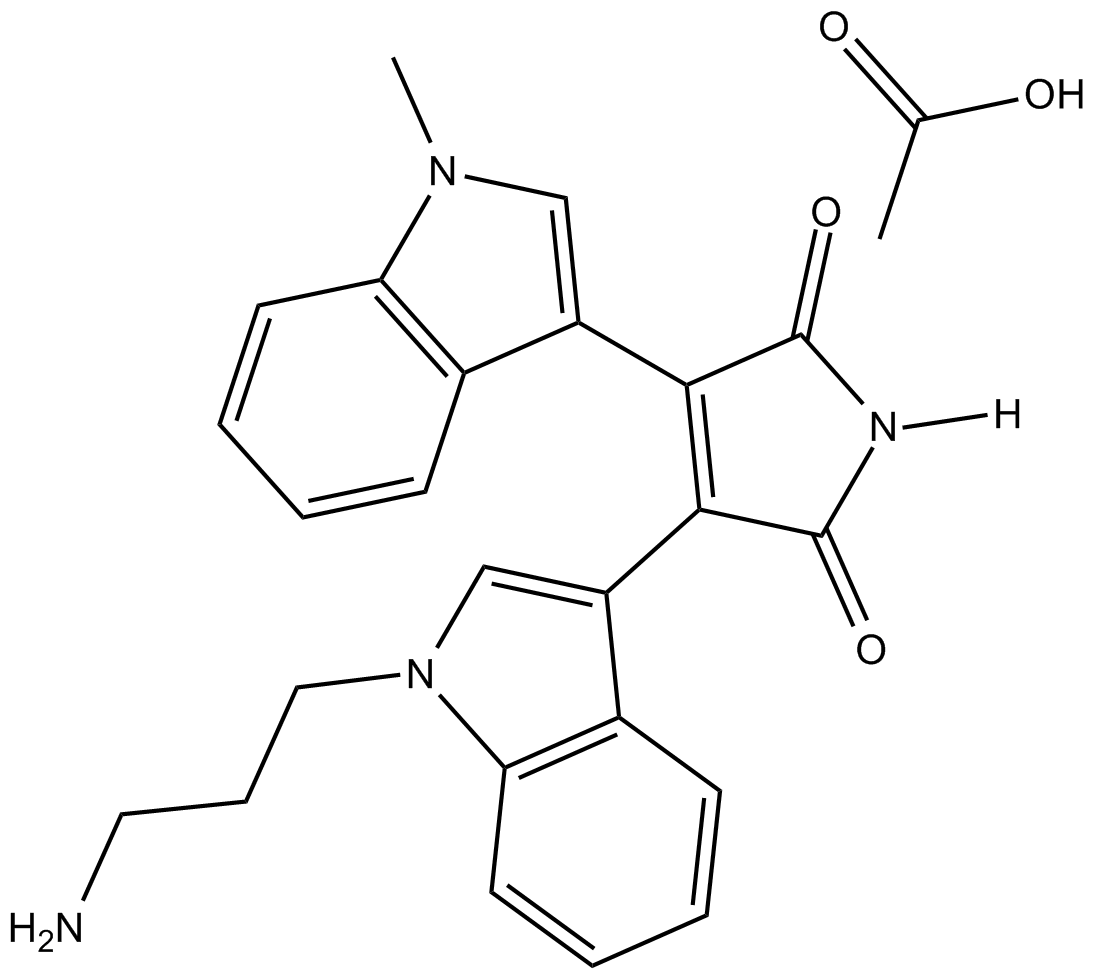
-
GC18354
Bisindolylmaleimide X (hydrochloride)
BIM X, Ro 31-8425
Bisindolylmaleimide X is a cell-permeable, reversible, ATP-competitive protein kinase C (PKC) inhibitor (IC50 = 15 nM, rat brain PKC).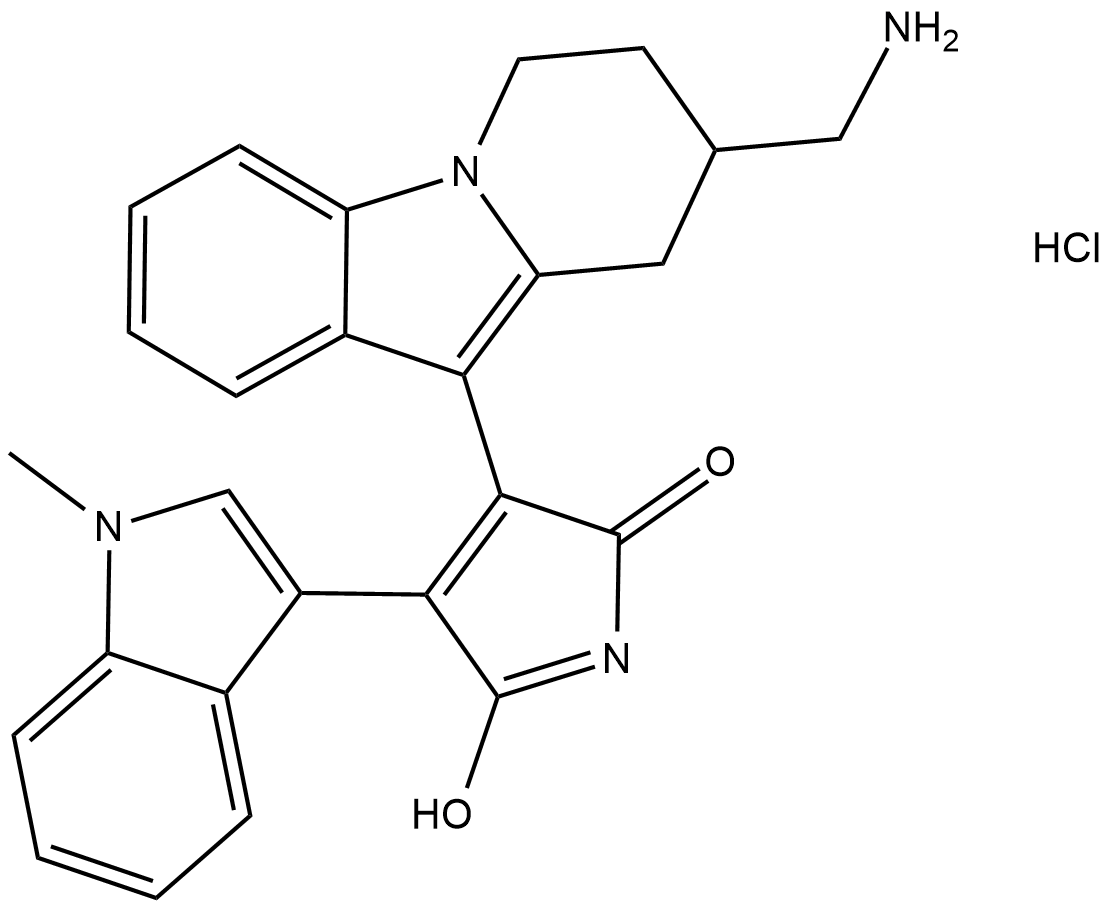
-
GC35530
BJE6-106
B106
BJE6-106 (B106) is a potent, selective 3rd generation PKCδ inhibitor with an IC50 of 0.05 μM and targets selectivity over classical PKC isozyme PKCα (IC50=50 μM).
-
GC50593
BMP signaling agonist sb4
Potent BMP4 agonist
-
GC13343
Bosutinib (SKI-606)
SKI 606
Bosutinib (SKI-606) is an oral Src/Abl tyrosine kinase inhibito with IC50 of 1.2 nM and 1 nM, respectively.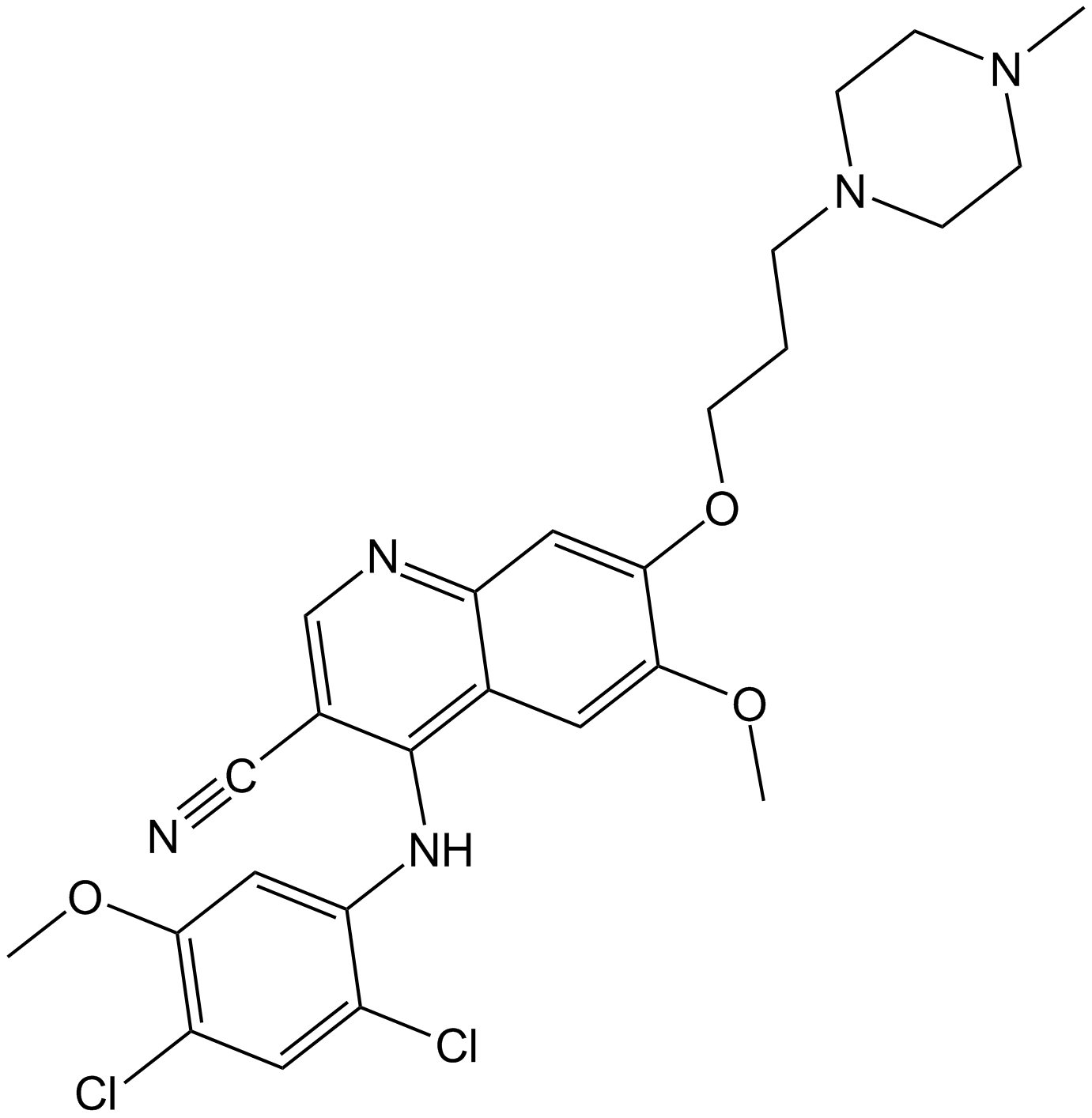
-
GC40080
Bosutinib-d8
SKI-606 D8
Bosutinib-d8 is intended for use as an internal standard for the quantification of bosutinib by GC- or LC-MS.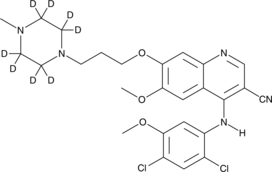
-
GC17670
Bryostatin 1
NSC 339555
PKC activator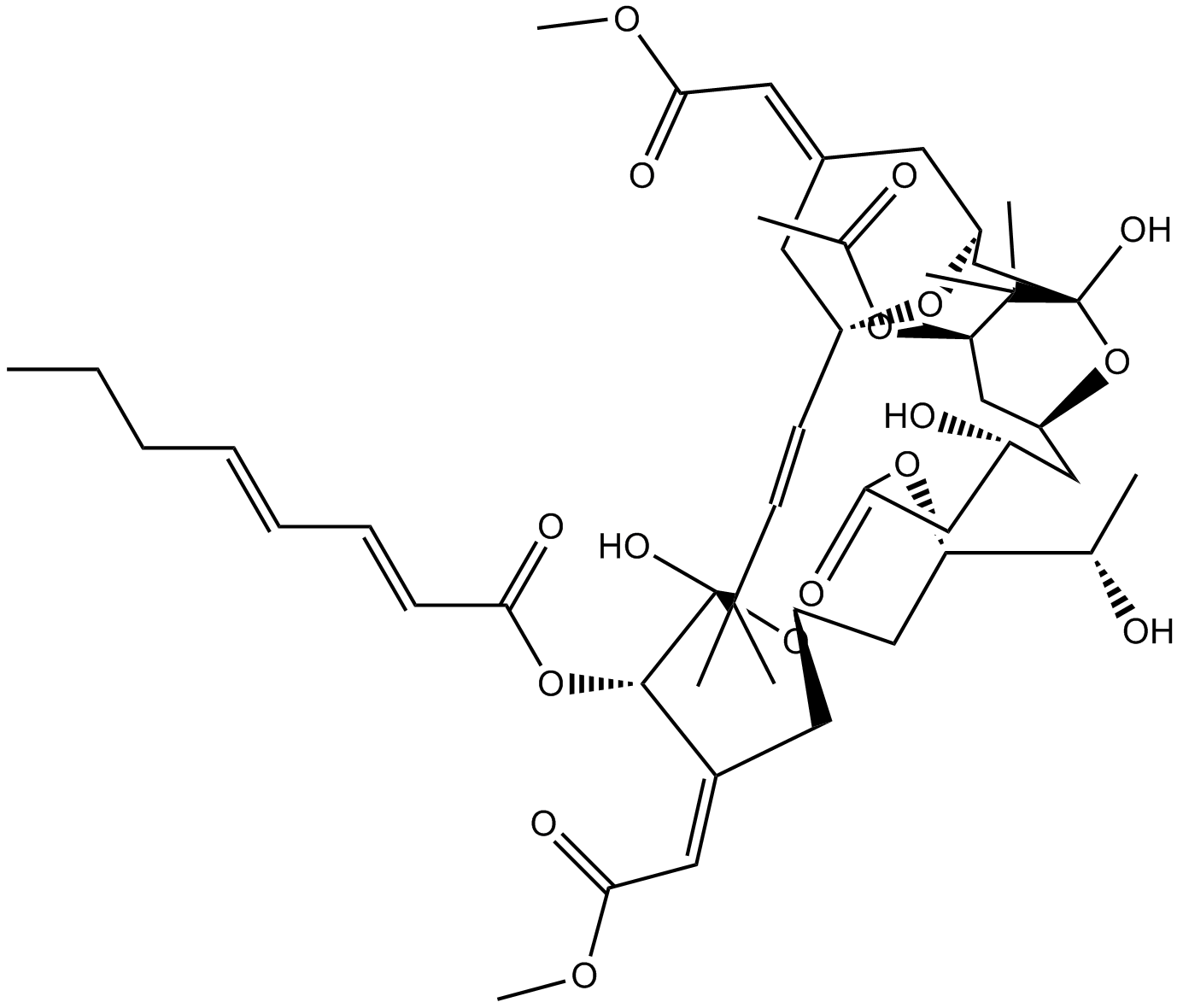
-
GC12842
Bryostatin 2
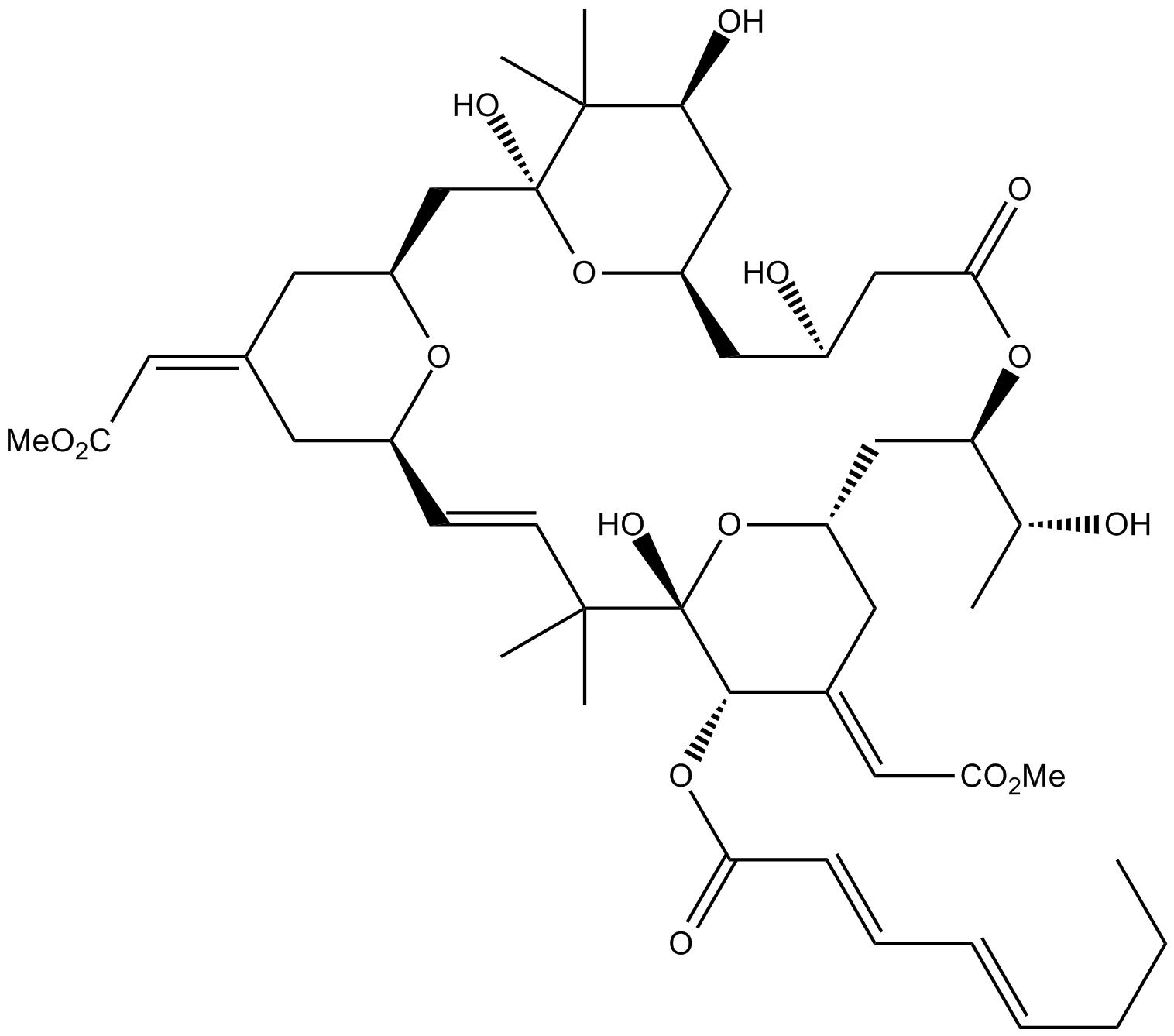
Protein kinase C (PKC) activator
-
GC12695
Bryostatin 3
protein kinase C activator
-
GC16539
Butaprost
15deoxy16Shydroxy17cyclobutyl PGE1 methyl ester, TR 4979
A selective agonist for the EP2 receptors
-
GC11605
C-1
C-1 is a potent protein kinase inhibitor, with IC50s of 4 μM, 8 μM, 12 μM and 240 μM for cGMP-dependent protein kinase (PKG), cAMP-dependent protein kinase (PKA), protein kinase C (PKC) and MLC-kinase, respectively. C-1 also used as a ROCK inhibitor.

-
GC43105
C8 Ceramide (d18:1.8:0)
N-octanoyl-D-erythro-Sphingosine
C8 Ceramide (d18:1.8:0) (N-Octanoyl-D-erythro-sphingosine) is a cell-permeable analog of naturally occurring ceramides.
-
GC11159
Calphostin C
Cladochrome E, PKF 115584, UCN 1028C
protein kinase C inhibitor
-
GC68833
Carotuximab
TRC105; DE-122
Carotuximab (TRC105) is an IgG1 monoclonal antibody that can block endoglin (CD105) and its downstream Smad signaling pathway. Carotuximab has immunomodulatory and anti-tumor effects.

-
GC62612
CC-90005
CC-90005 is a potent, selective and orally active inhibitor of protein kinase C-θ (PKC-θ), with an IC50 of 8 nM.
-
GC38898
CCG-222740
CCG-222740 is an orally active and selective Rho/myocardin-related transcription factor (MRTF) pathway inhibitor. CCG-222740 is also a potent inhibitor of alpha-smooth muscle actin protein expression. CCG-222740 effectively reduces fibrosis in skin and blocks melanoma metastasis.
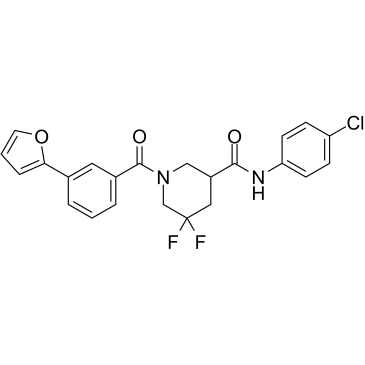
-
GC35651
Cenisertib
AS-703569; R-763
Cenisertib (AS-703569) is an ATP-competitive multi-kinase inhibitor that blocks the activity of Aurora-kinase-A/B, ABL1, AKT, STAT5 and FLT3. Cenisertib induces major growth-inhibitory effects by blocking the activity of several different molecular targets in neoplastic mast cells (MC). Cenisertib inhibits tumor growth in xenograft models of pancreatic, breast, colon, ovarian, and lung tumors and leukemia.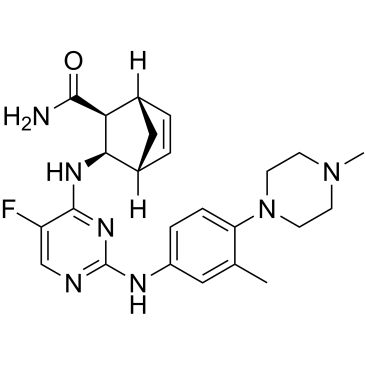
-
GC18521
Cercosporin
CGP 049090, NSC 153111
Cercosporin is a pigmented phytotoxin isolated from the fungus C.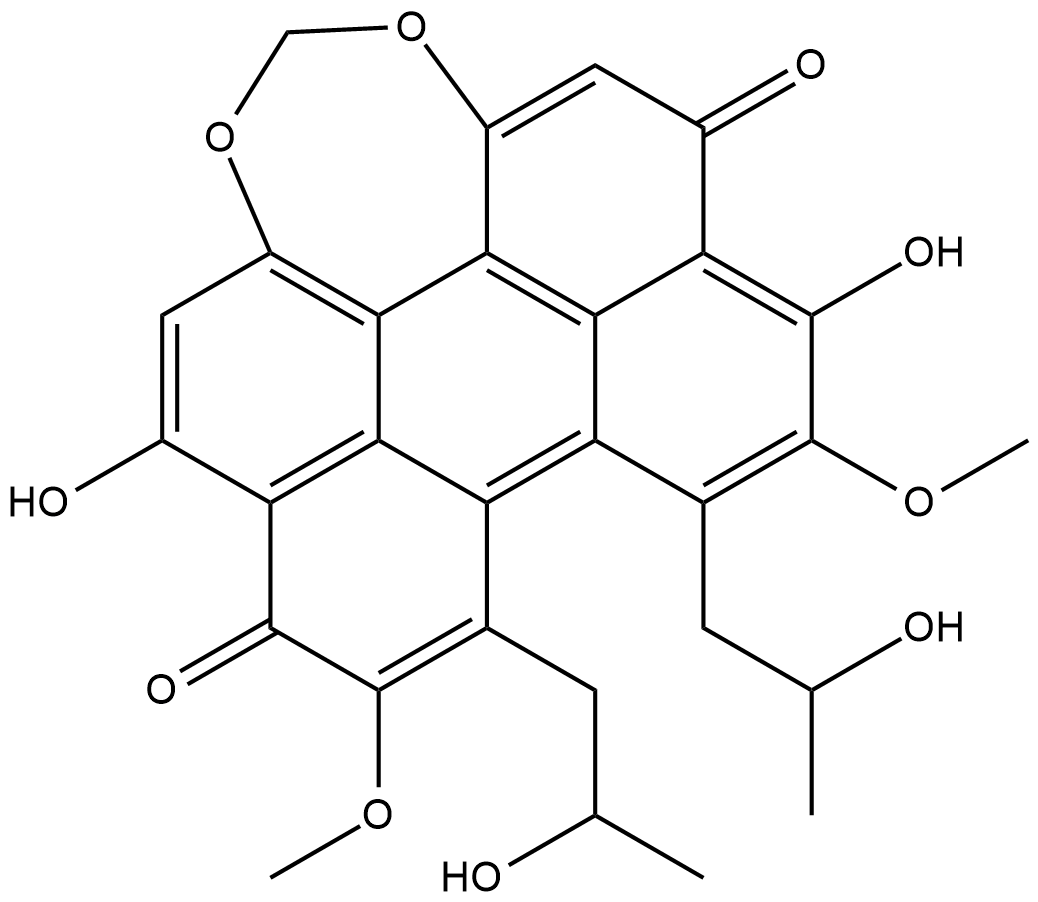
-
GC10687
CGP 53353
DAPH-7
PKCβII inhibitor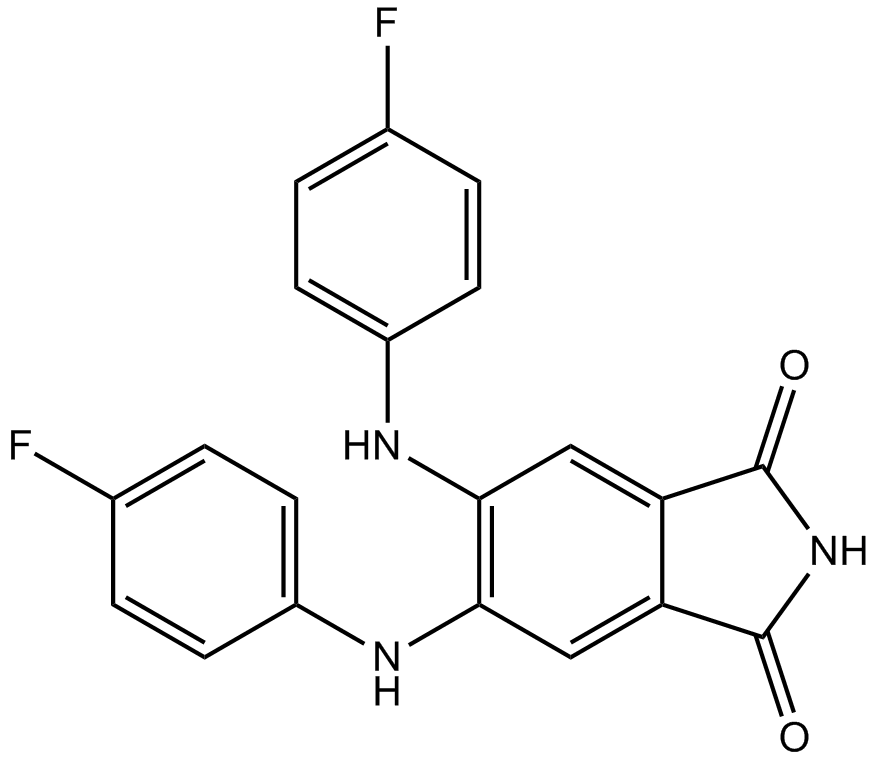
-
GC14650
CGP60474
A CDK inhibitor
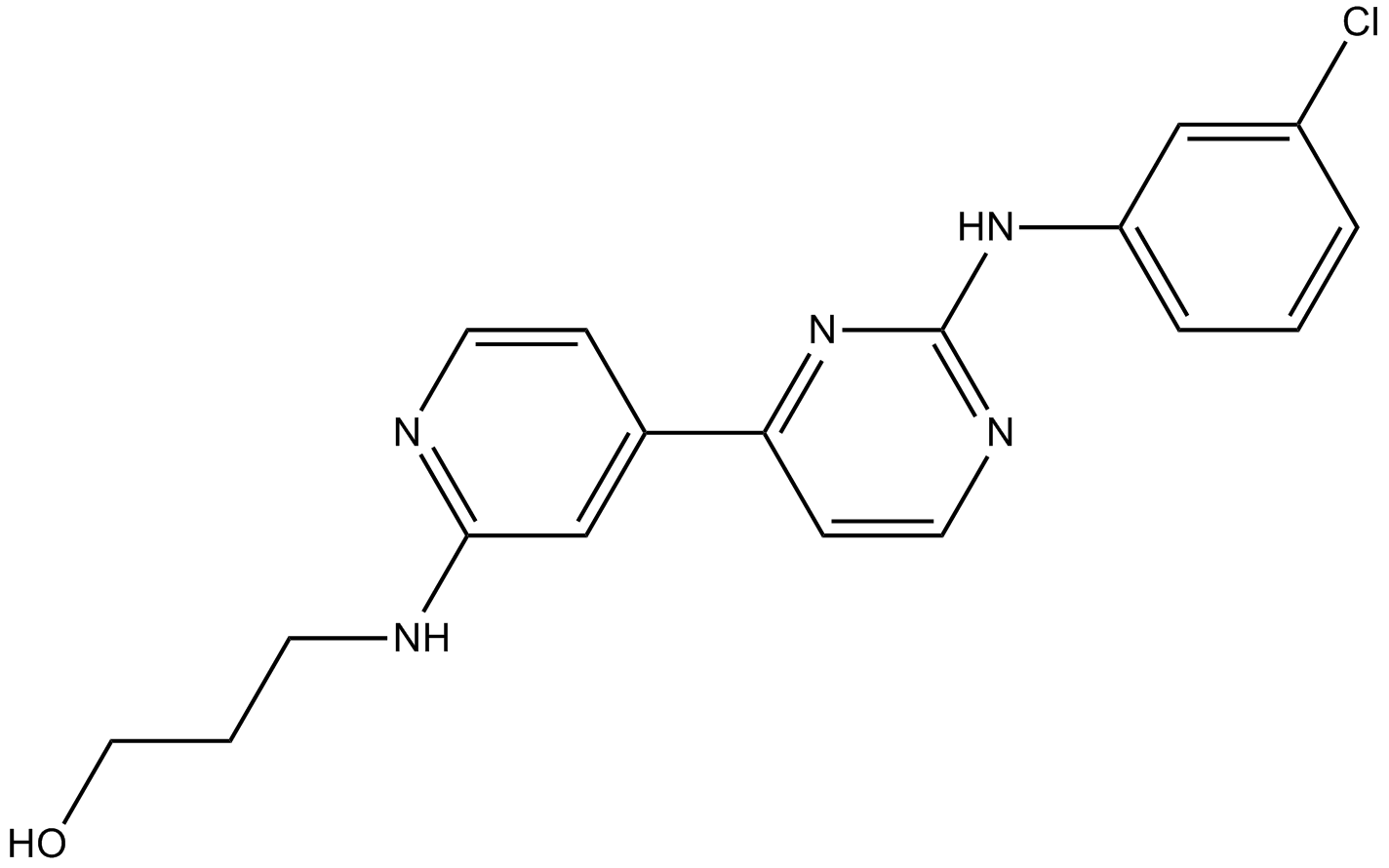
-
GC32250
Chebulinic acid
Eutannin, NSC 69862
An ellagitannin with diverse biological activities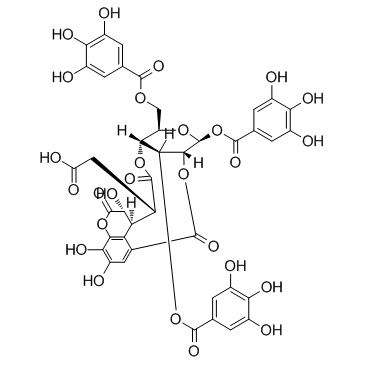
-
GN10463
Chelerythrine
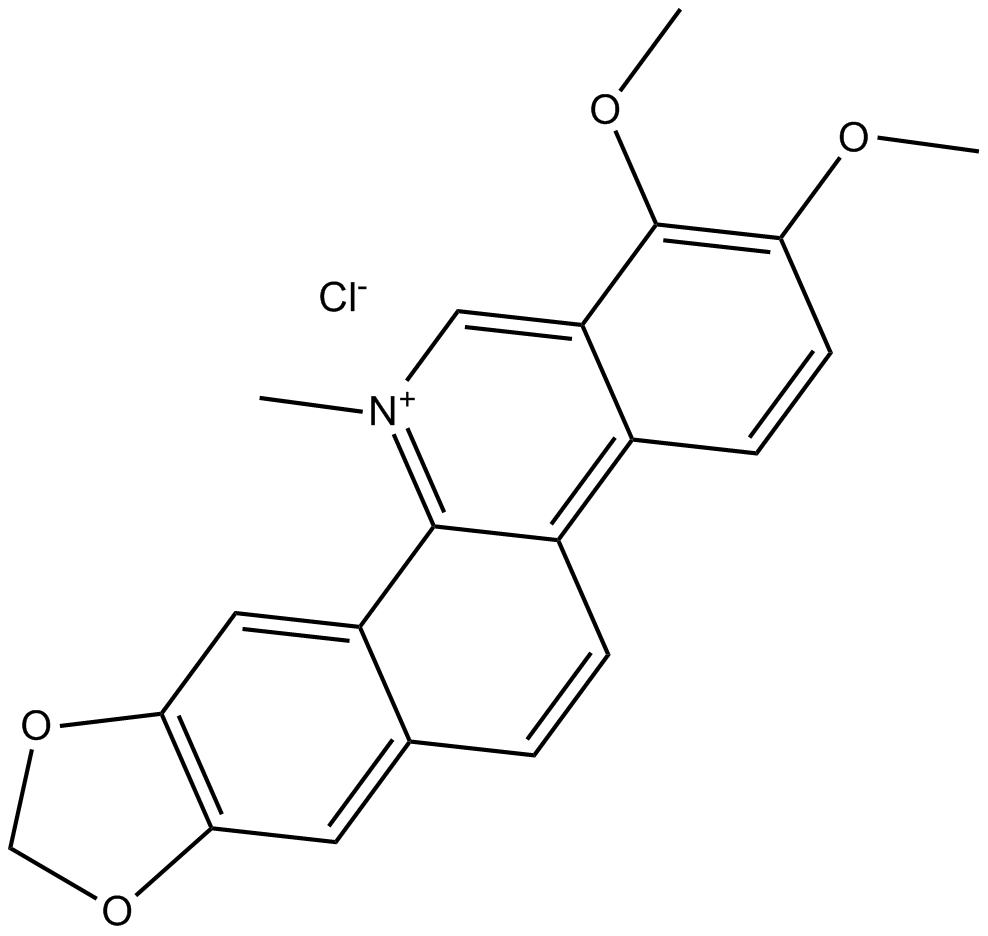
-
GC13065
Chelerythrine Chloride
Broussonpapyrine chloride, NSC 646662
Potent inhibitor of PKC and Bcl-xL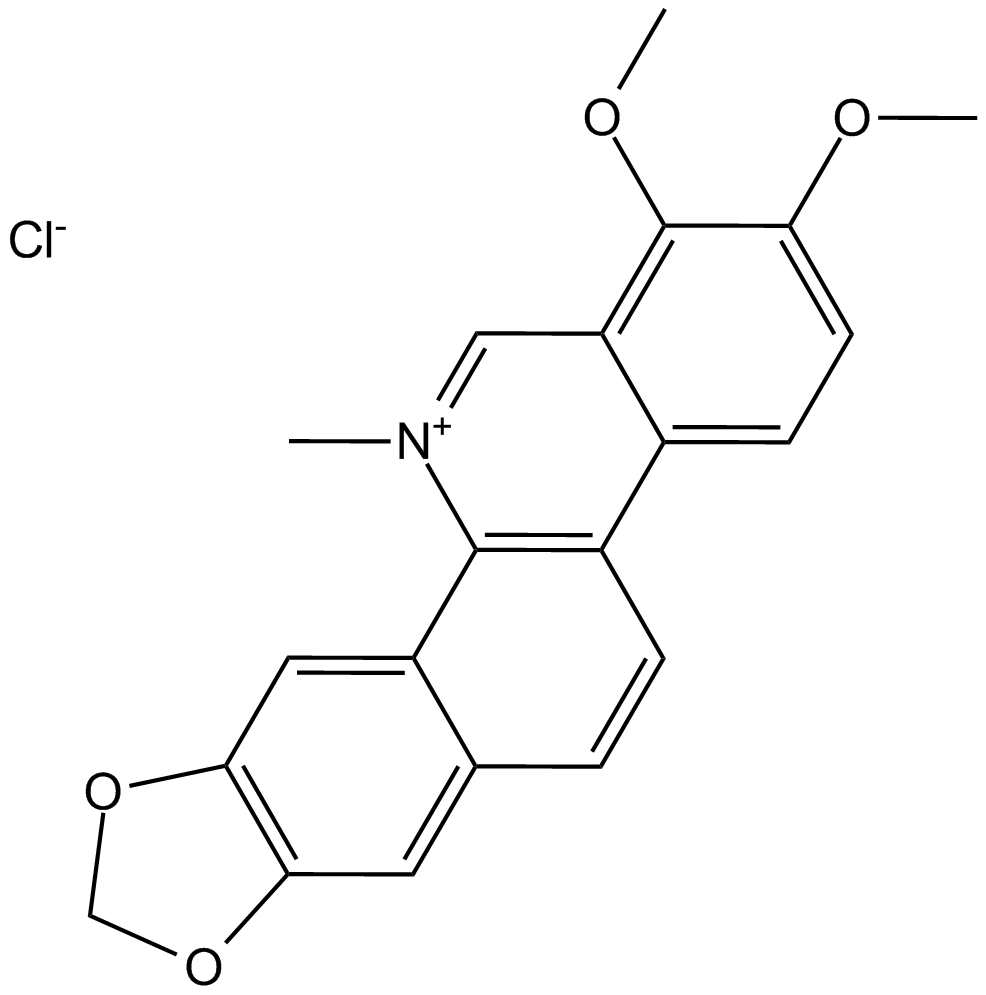
-
GC35682
CHMFL-ABL/KIT-155
CHMFL-ABL-KIT-155
CHMFL-ABL/KIT-155 (CHMFL-ABL-KIT-155; compound 34) is a highly potent and orally active type II ABL/c-KIT dual kinase inhibitor (IC50s of 46 nM and 75 nM, respectively), and it also presents significant inhibitory activities to BLK (IC50=81 nM), CSF1R (IC50=227 nM), DDR1 (IC50=116 nM), DDR2 (IC50=325 nM), LCK (IC50=12 nM) and PDGFRβ (IC50=80 nM) kinases. CHMFL-ABL/KIT-155 (CHMFL-ABL-KIT-155) arrests cell cycle progression and induces apoptosis.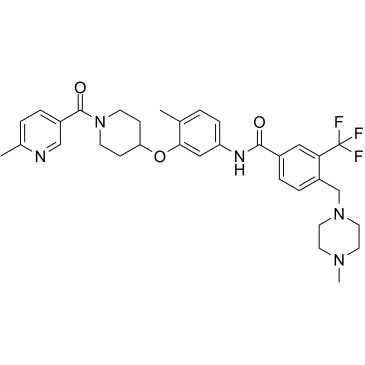
-
GC70882
Chromenone 1
Chromenone 1 is a potent osteogenic bone morphogenetic protein (BMP) potentiator.

-
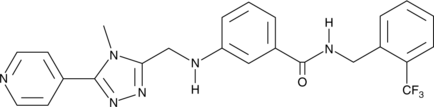
GC43286
CMPD101
A GRK2 and GRK3 inhibitor
-
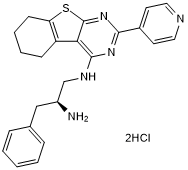
GC50704
CRT 0066854 hydrochloride
CRT 0066854 hydrochloride is a potent and selective atypical PKCs inhibitor.
-
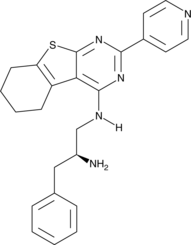
GC45414
CRT0066854
-
GC35753
CT-721
CT-721 is a potent and time-dependent Bcr-Abl kinase inhibitor with an IC50 of 21.3 nM for wild-type Bcr-Abl kinase, and possesses anti-chronic myeloid leukemia (CML) activities.
-
GC33351
CZC-8004 (CZC-00008004)
Dianilinopyrimidine-01
CZC-8004 (CZC-00008004) (CZC-00008004), an aminopyrimidine, is a pan-kinase inhibitor. CZC-8004 (CZC-00008004) can bind a range of tyrosine kinases, including EGFR and VEGFR2 with IC50 values of 650 and 437 nM, respectively.
-
GC17591
D-erythro-Sphingosine (synthetic)
(-)Sphingosine, DerythroSphingosine C18
D-erythro-sphingosine (Erythrosphingosine) is a very potent activator of p32-kinase with an EC50 value of 8μM.
-
GC72448
Dalutrafusp alfa
Dalutrafusp alfa (AGEN-1423; GS-1423) is a specific bifunctional antibody against CD73 and TGF-β, which is involved in the immunosuppressive pathway.

-
GN10336
Daphnetin
7,8-Dihydroxycoumarin, NSC 633563
-
GC38186
Daphnoretin
NSC 291852
A coumarin with diverse biological activities
-
GC35812
Dasatinib hydrochloride
A potent and dual AblWT/Src inhibitor
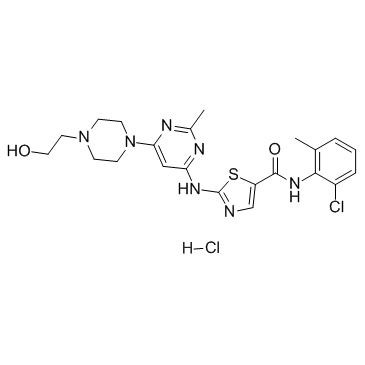
-
GC15884
Dasatinib Monohydrate
Inhibitor of ABL, SRC, KIT, PDGFR, and other tyrosine kinases.
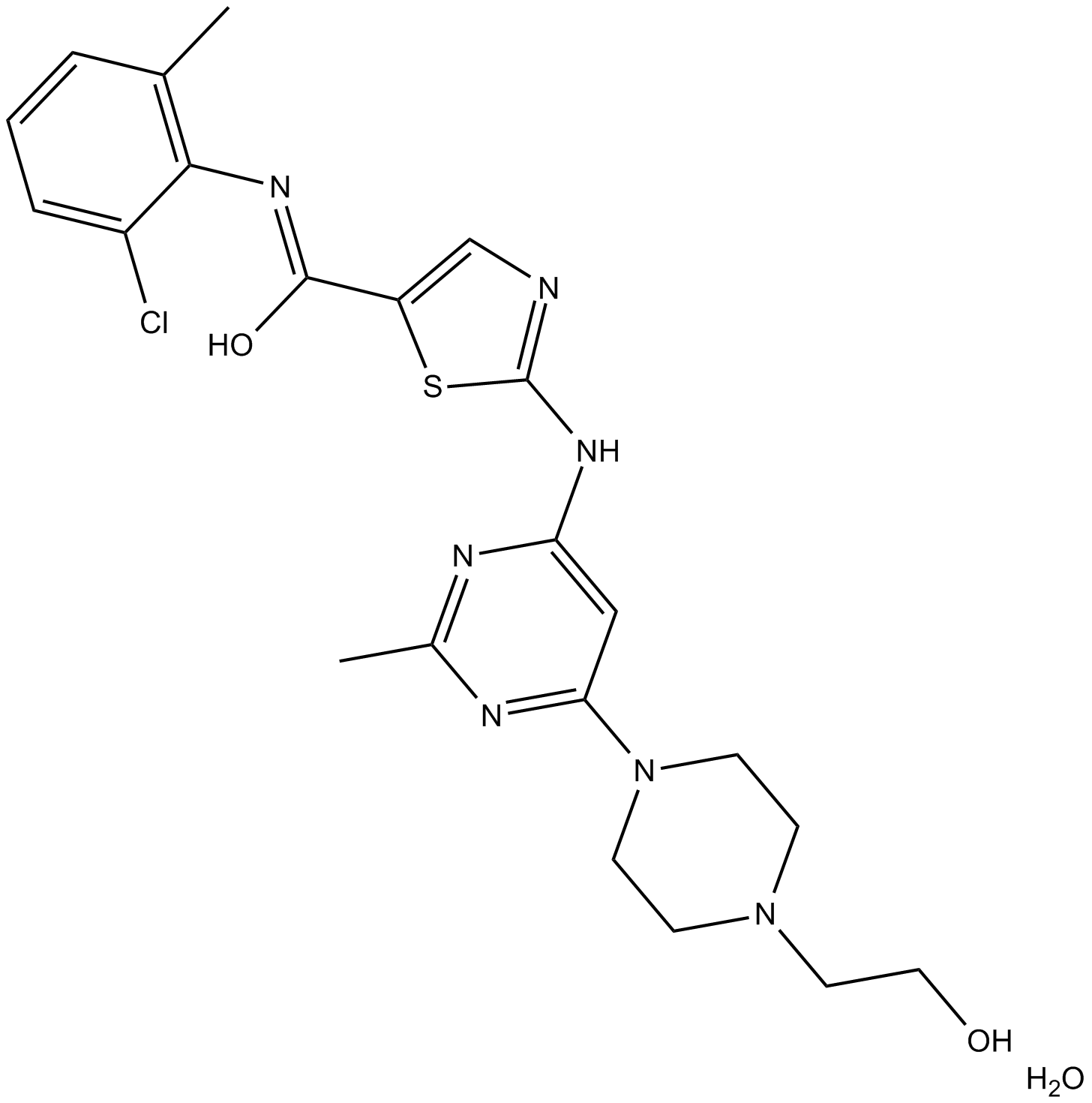
-
GC14007
DCC-2036 (Rebastinib)
DCC-2036
DCC-2036 (Rebastinib) (DCC-2036) is an orally active, non-ATP-competitiveBcr-Abl inhibitor for Abl1WT and Abl1T315I with IC50s of 0.8 nM and 4 nM, respectively. DCC-2036 (Rebastinib) also inhibits SRC, KDR, FLT3, and Tie-2, and has low activity to seen towards c-Kit.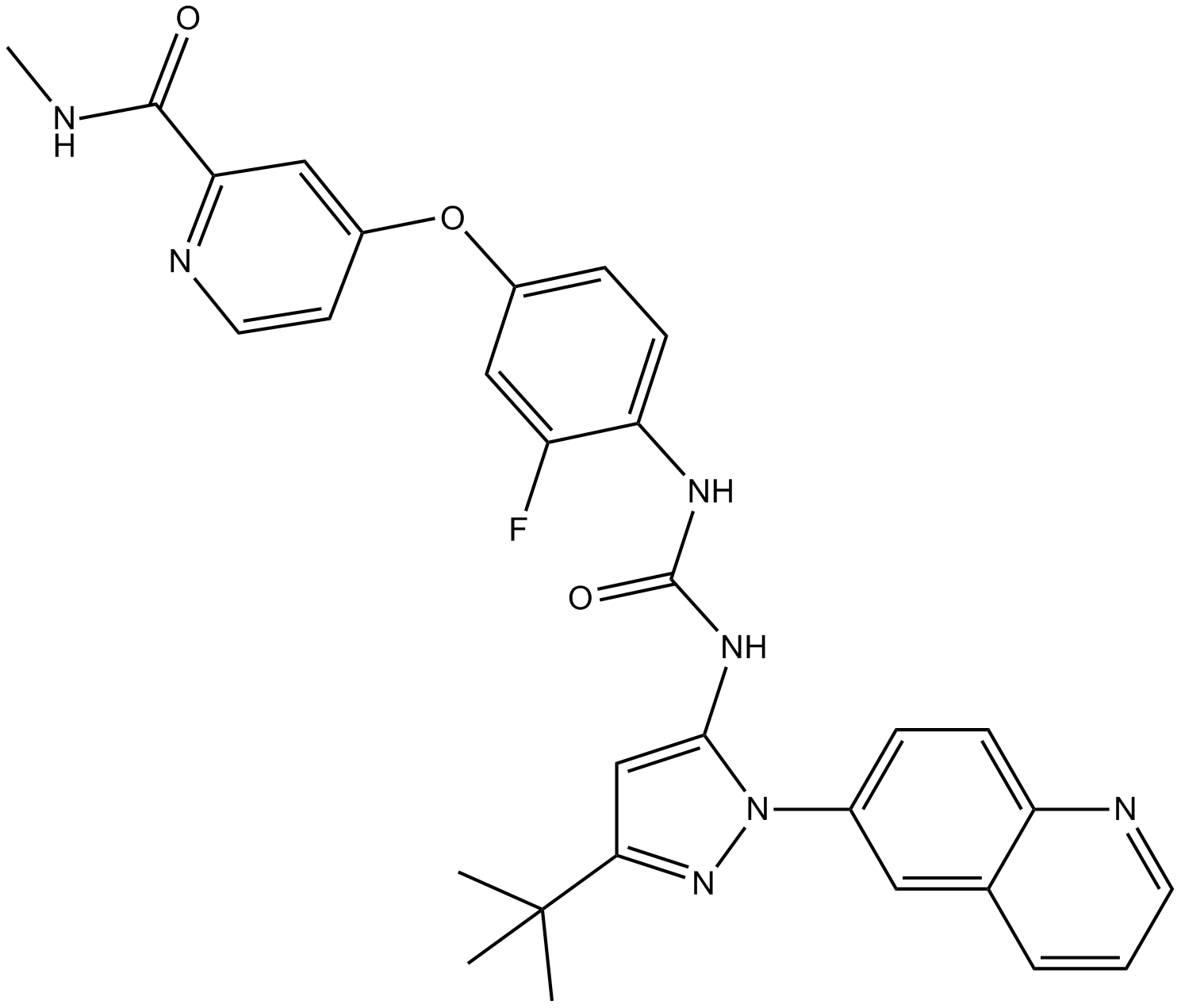
-
GC38388
DCPLA-ME
DCPLA methyl ester
DCPLA-ME, the methyl ester form of DCPLA, is a potent PKCε activator for use in the treatment of neurodegenerative diseases.
-
GC31892
Decursin ((+)-Decursin)
(+)-Decursin
Decursin ((+)-Decursin) ((+)-Decursin ((+)-Decursin)) is a potent anti-tumor agent.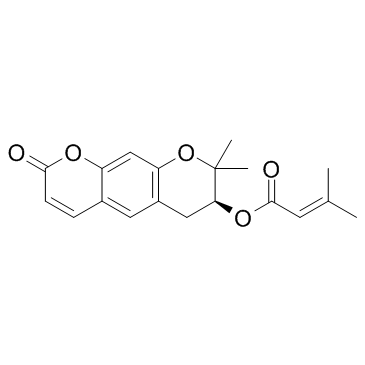
-
GC38085
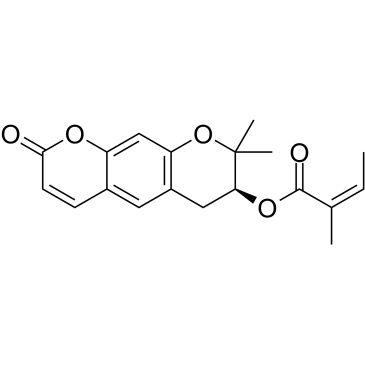
Decursinol angelate
Decursinol angelate, a cytotoxic and protein kinase C (PKC) activating agent from the root of Angelica gigas, possesses anti-tumor and anti-inflammatory activities.
-
GC16354
Dequalinium Chloride
NSC 166454
anti-tumor agent and PKC inhibitor
-
GC38482
Desmethylglycitein
6-hydroxy Daidzein, 6,7,4’-THIF
Desmethylglycitein (4',6,7-Trihydroxyisoflavone), a metabolite of daidzein, sourced from Glycine max with antioxidant, and anti-cancer activities.Desmethylglycitein binds directly to CDK1 and CDK2 in vivo, resulting in the suppresses CDK1 and CDK2 activity. Desmethylglycitein is a direct inhibitor of protein kinase C (PKC)α, against solar UV (sUV)-induced matrix matrix metalloproteinase 1 (MMP1). Desmethylglycitein binds to PI3K in an ATP competitive manner in the cytosol, where it inhibits the activity of PI3K and downstream signaling cascades, leading to the suppression of adipogenesis in 3T3-L1 preadipocytes.
-
GC17767
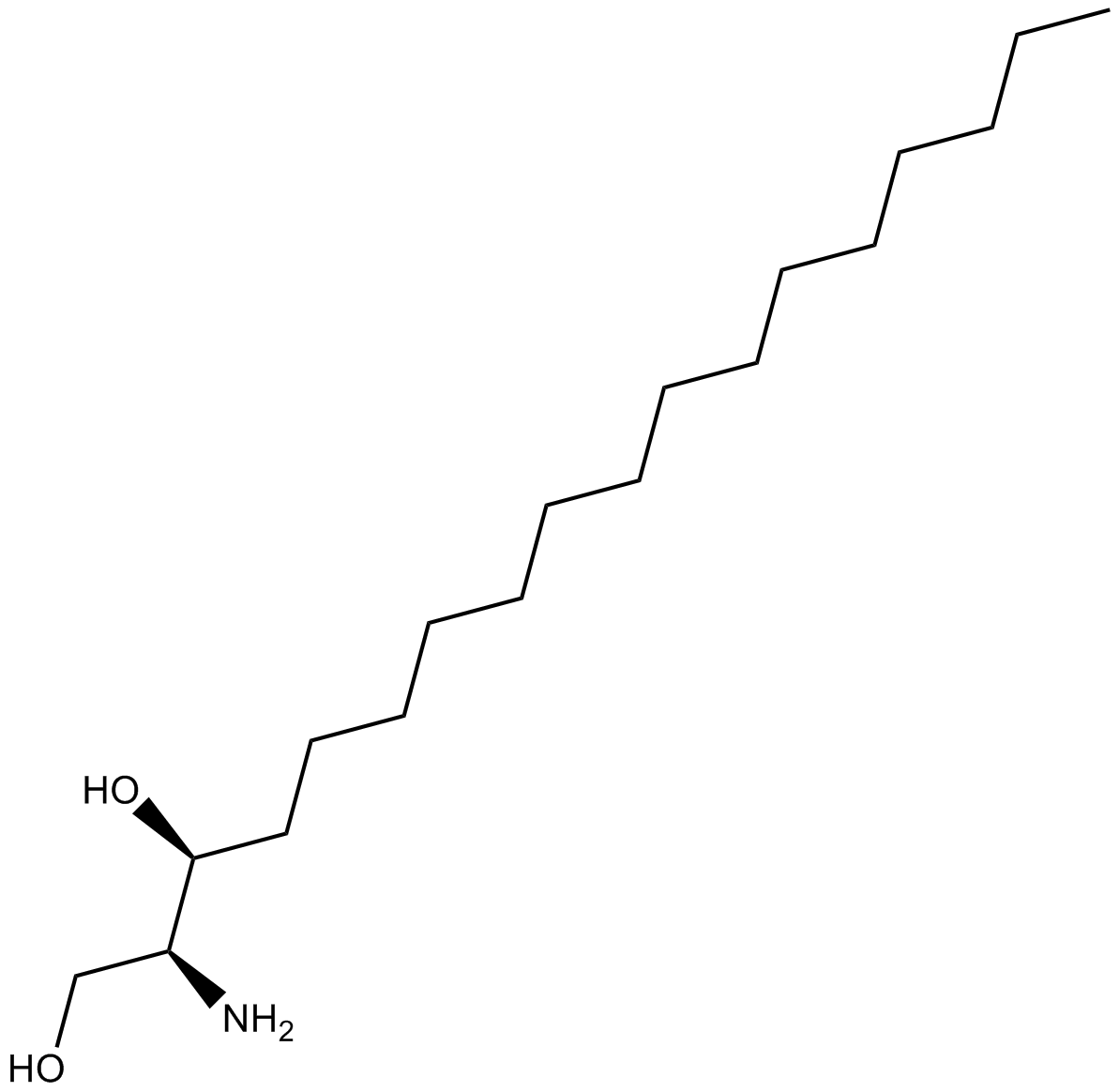
Dihydrosphingosine
Dihydrosphingosine is a potent inhibitor of PKC and phospholipase A2 (PLA2).
-
GC34060
Disitertide (P144)
Disitertide (P144) (P144) is a peptidic transforming growth factor-beta 1 (TGF-β1) inhibitor specifically designed to block the interaction with its receptor. Disitertide (P144) (P144) is also a PI3K inhibitor and an apoptosis inducer.

-
GC68333
Disitertide diammonium
P144 diammonium

-
GC60782
Disitertide TFA
P144 TFA
Disitertide (P144) TFA is a peptidic transforming growth factor-beta 1 (TGF-β1) inhibitor specifically designed to block the interaction with its receptor. Disitertide (P144) TFA is also a PI3K inhibitor and an apoptosis inducer.
-
GC14298
DMH-1
BMP Inhibitor II, DorsoMorphin Homolog 1, VU036482
Selective BMP ALK2 receptor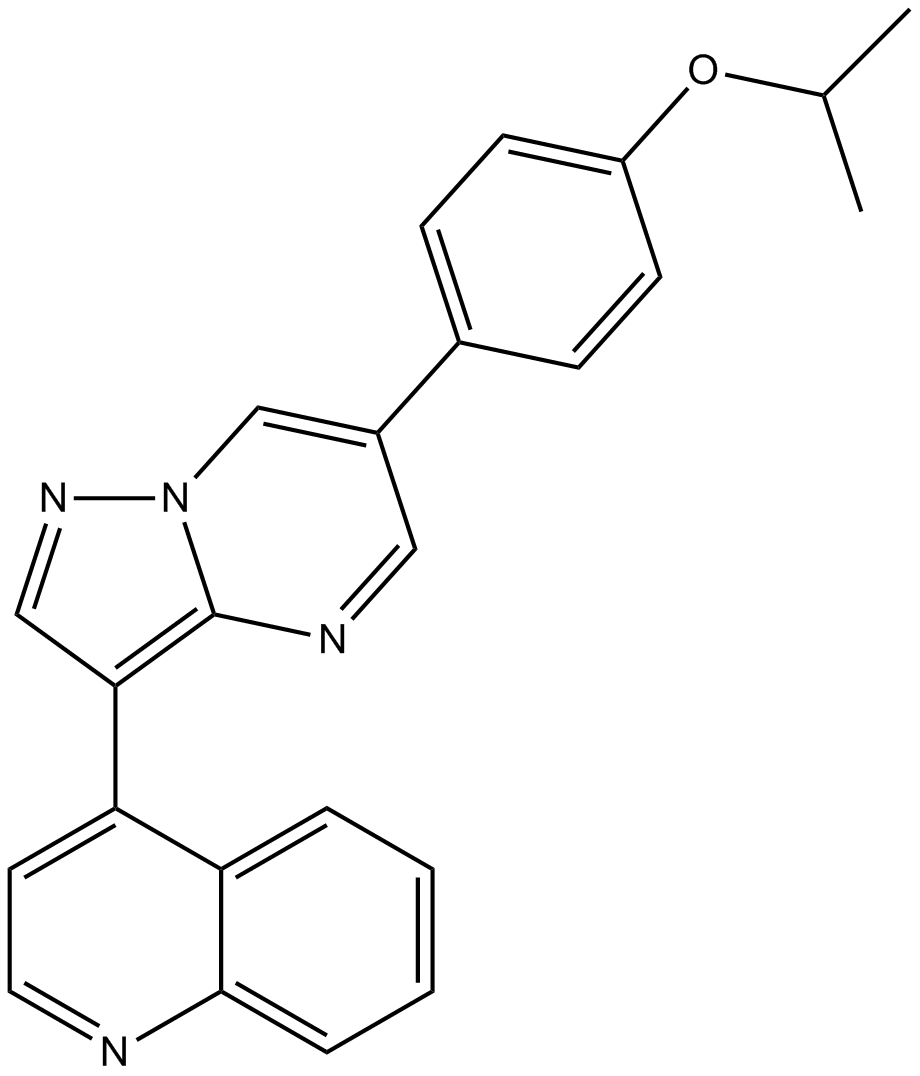
-
GC17243
Dorsomorphin (Compound C)
Compound C
Dorsomorphin (Compound C) (Compound C) is a selective and ATP-competitive AMPK inhibitor (Ki=109 nM in the absence of AMP). Dorsomorphin (Compound C) (BML-275) selectively inhibits BMP type I receptors ALK2, ALK3, and ALK6. Dorsomorphin (Compound C) induces autophagy.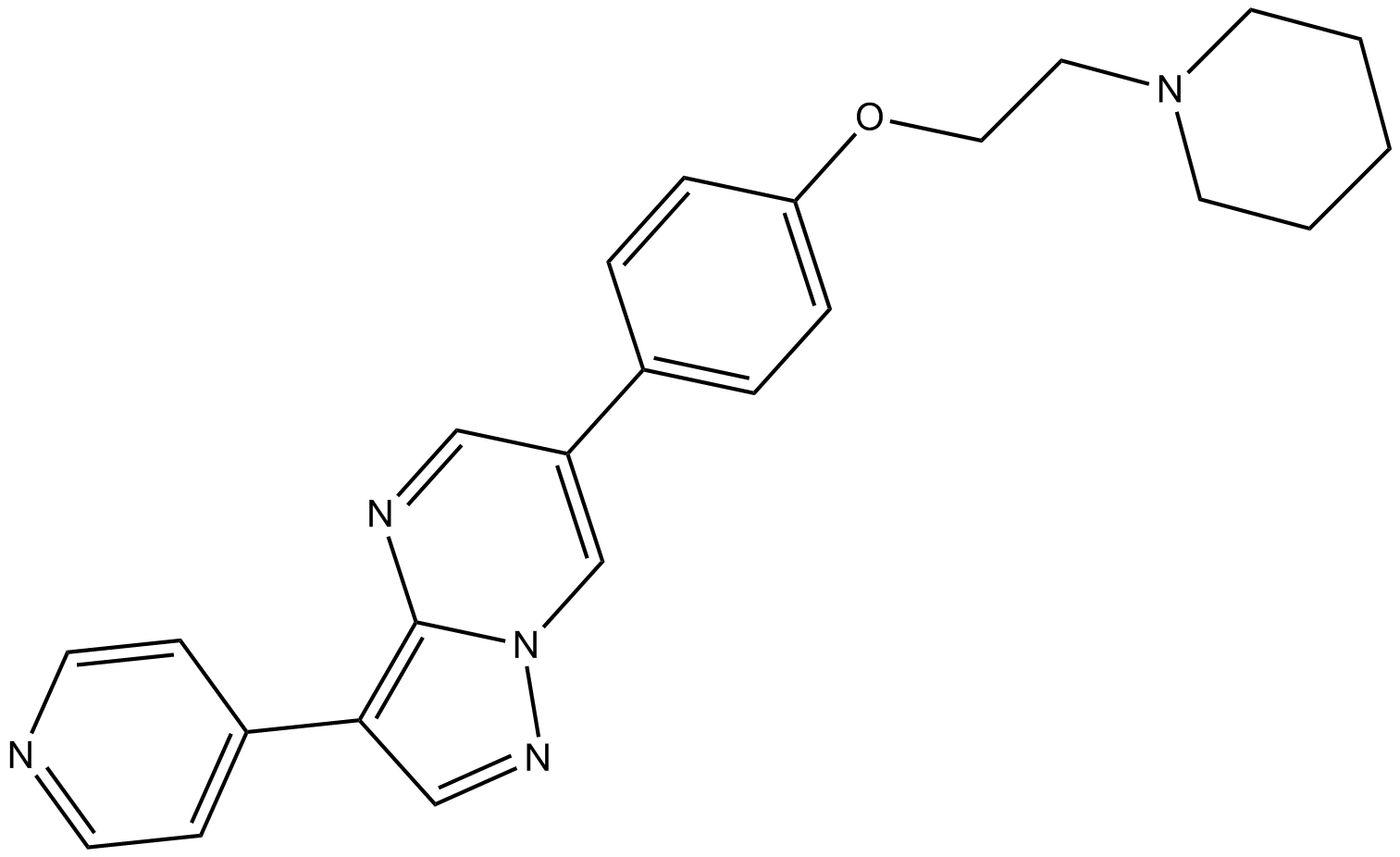
-
GC12560
Dorsomorphin (Compound C) 2HCl
BML-275 2HCl,Compound C 2HCl
Dorsomorphin (Compound C) 2HCl (BML-275 dihydrochloride; Compound C dihydrochloride) is a potent, selective and ATP-competitive AMPK inhibitor, with a Ki of 109 nM. Dorsomorphin (Compound C) 2HCl inhibits BMP pathway by targeting the type I receptors ALK2, ALK3, and ALK6. Dorsomorphin (Compound C) 2HCl induces autophagy.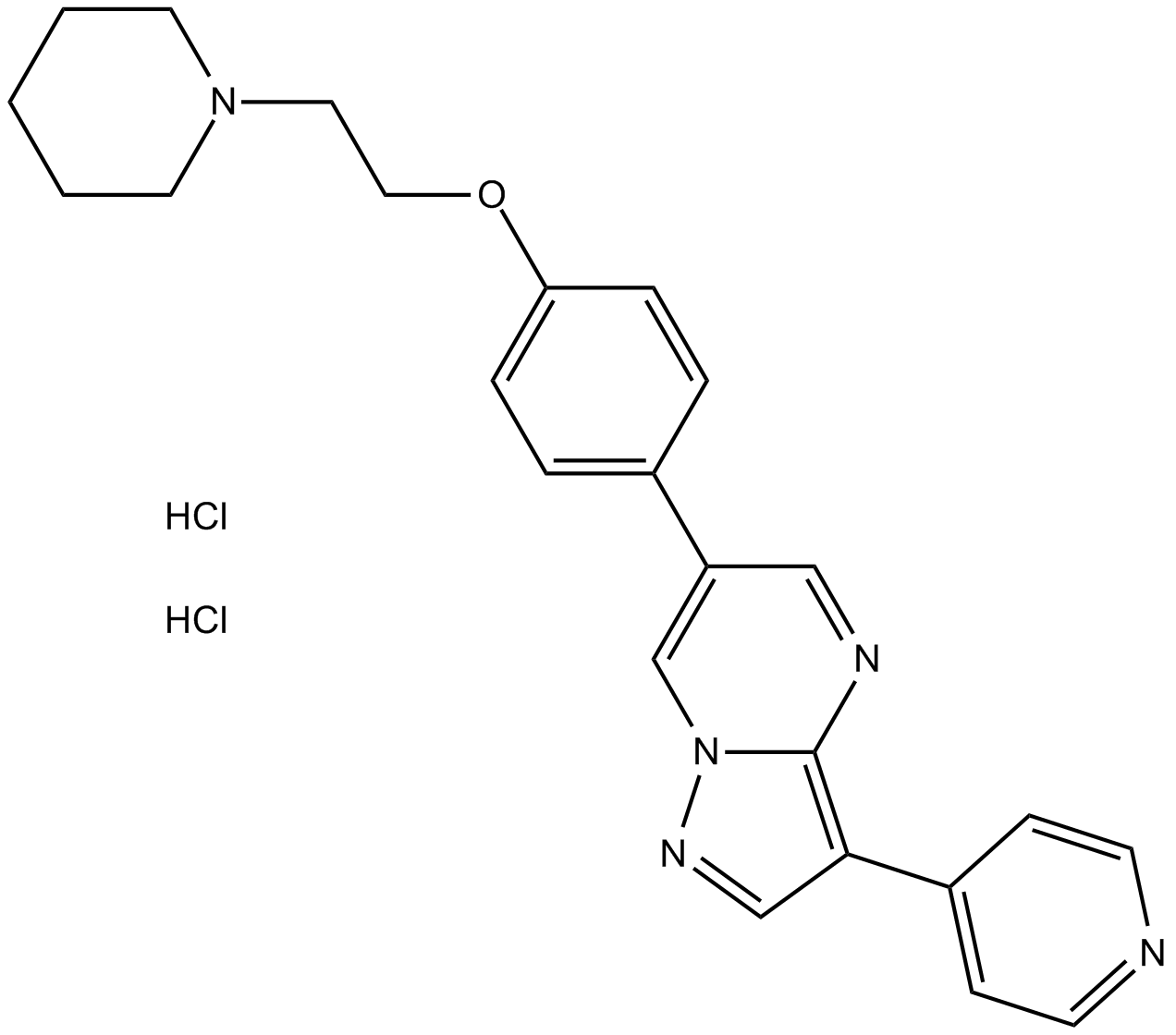
-
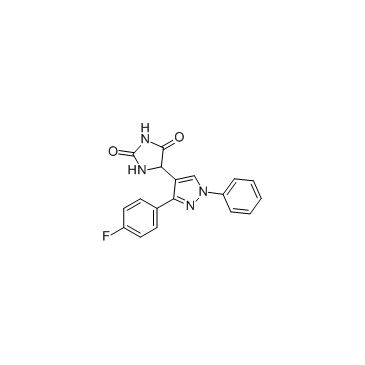
GC35897
DPH
A potent cell permeable c-Abl activator
-
GC69052
Elezanumab
Elezanumab (ABT-555; AE12-1Y-QL) is a human monoclonal antibody that selectively targets repulsive guidance molecule A (RGMa). Elezanumab effectively inhibits RGMa-mediated BMP signaling through the SMAD1/5/8 pathway, with an IC50 of approximately 97 pM. Elezanumab promotes neuronal regeneration and neuroprotection in models of nerve injury and demyelination by binding to the N-terminal of RGMa, blocking BMP signaling, and lacking cross-reactivity with RGMc. Elezanumab has activity in promoting neuronal regeneration and neuroprotection without affecting iron metabolism.

-
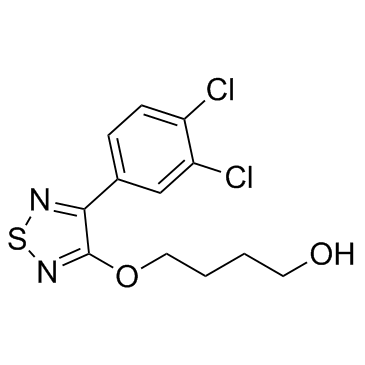
GC32914
EMT inhibitor-1
EMT inhibitor-1 is an inhibitor of of Hippo, TGF-β, and Wnt signaling pathways with antitumor activities.
-
GC11499
Enzastaurin (LY317615)
LY317615
Enzastaurin (LY317615) (LY317615) is a potent and selective PKCβ inhibitor with an IC50 of 6 nM, showing 6- to 20-fold selectivity over PKCα, PKCγ and PKCε.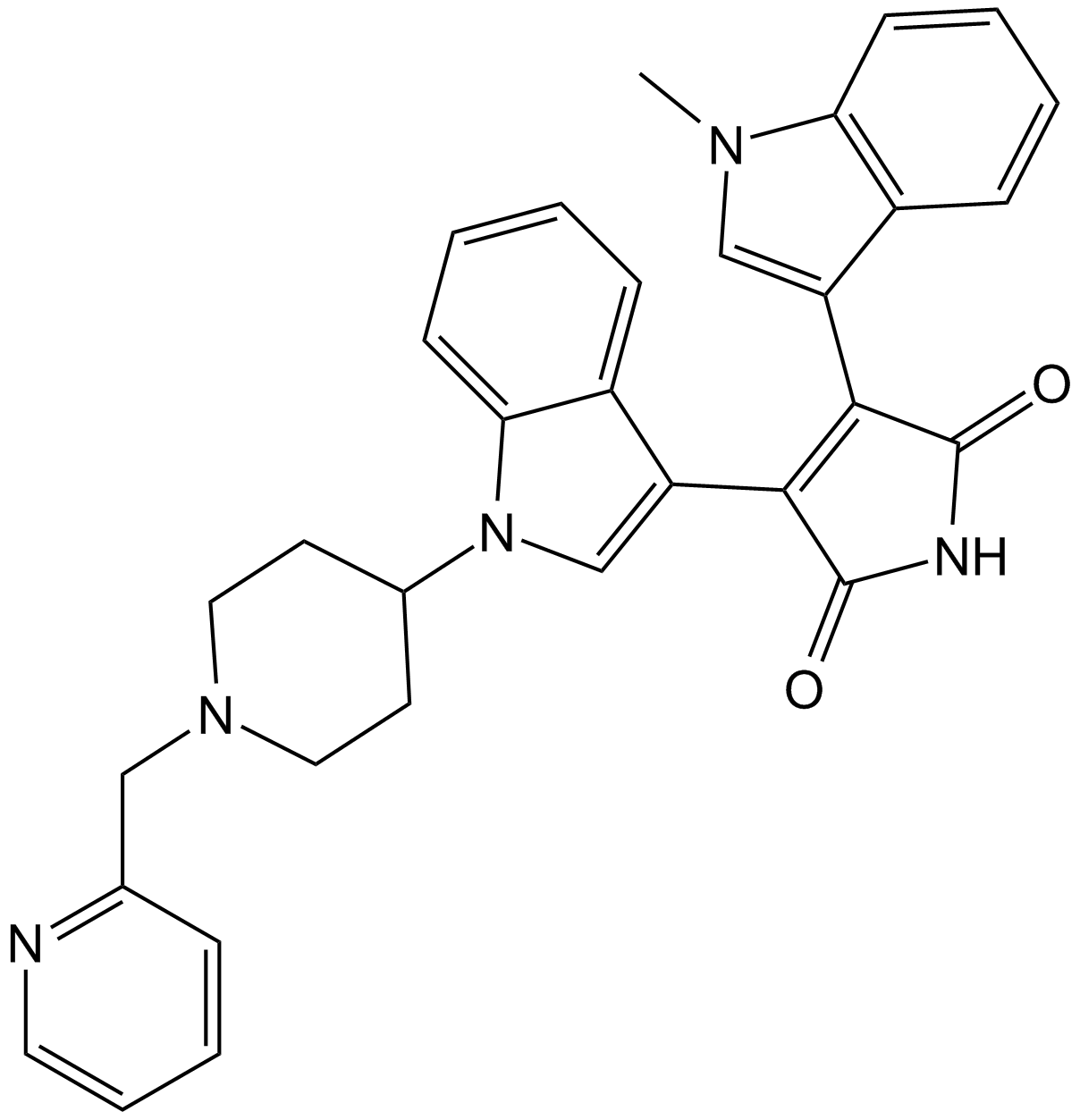
-
GC65329
EW-7195
EW-7195 is a potent and selective ALK5 (TGFβR1) inhibitor with an IC50 of 4.83 nM. EW-7195 has >300-fold selectivity for ALK5 over p38α. EW-7195 efficiently inhibits TGF-β1-induced Smad signaling, epithelial-to-mesenchymal transition (EMT) and breast tumour metastasis to the lung.
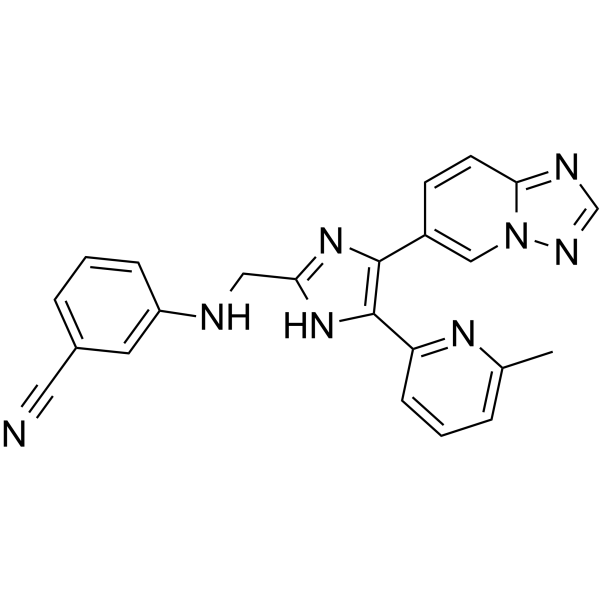
-
GC13354
EW-7197
Vactosertib, TEW-7197
EW-7197 (EW-7197) is a potent, orally active and ATP-competitive activin receptor-like kinase 5 (ALK5) inhibitor with an IC50 of 12.9 nM. EW-7197 also inhibits ALK2 and ALK4 (IC50 of 17.3 nM) at nanomolar concentrations. EW-7197 has potently antimetastatic activity and anticancer effect.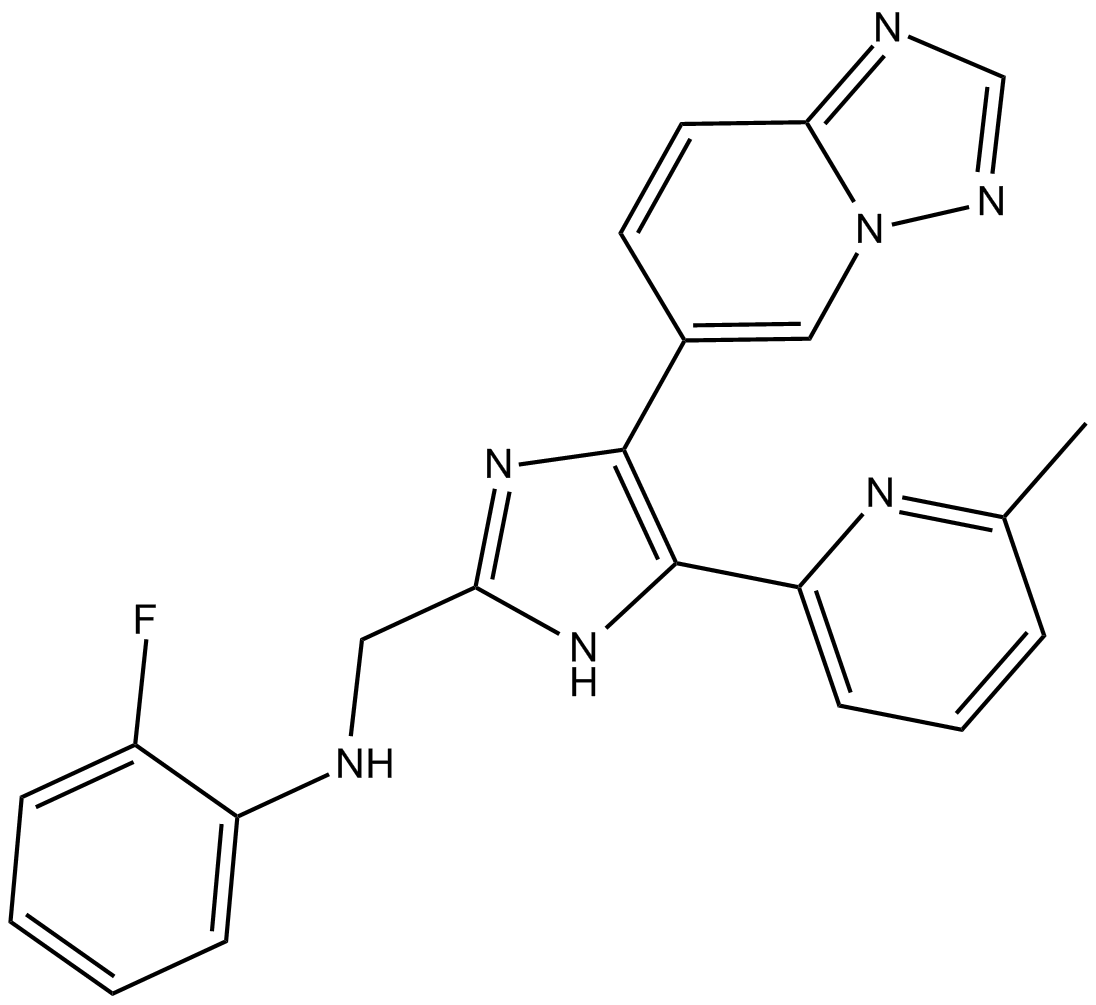
-
GC13869
Fasudil
Calcium antagonist

-
GC14289
Fasudil (HA-1077) HCl
Fasudil (HA-1077; AT877) Hydrochloride is a nonspecific RhoA/ROCK inhibitor and also has inhibitory effect on protein kinases, with an Ki of 0.33 μM for ROCK1, IC50s of 0.158 μM and 4.58 μM, 12.30 μM, 1.650 μM for ROCK2 and PKA, PKC, PKG, respectively. Fasudil (HA-1077) HCl is also a potent Ca2+ channel antagonist and vasodilator.
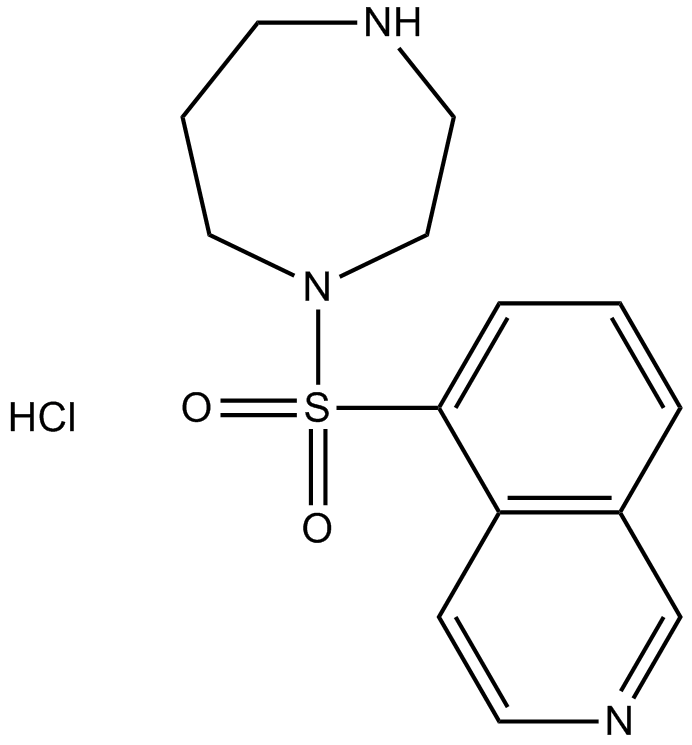
-
GC32867
Flumatinib (HHGV678)
HH-GV-678
Flumatinib (HHGV678) (HHGV678) is an orally available, selective inhibitor of Bcr-Abl. Flumatinib (HHGV678) inhibits c-Abl, PDGFRβ and c-Kit with IC50s of 1.2 nM, 307.6 nM and 665.5 nM, respectively.