Immunology/Inflammation
The immune and inflammation-related pathway including the Toll-like receptors pathway, the B cell receptor signaling pathway, the T cell receptor signaling pathway, etc.
Toll-like receptors (TLRs) play a central role in host cell recognition and responses to microbial pathogens. TLR4 initially recruits TIRAP and MyD88. MyD88 then recruits IRAKs, TRAF6, and the TAK1 complex, leading to early-stage activation of NF-κB and MAP kinases [1]. TLR4 is endocytosed and delivered to intracellular vesicles and forms a complex with TRAM and TRIF, which then recruits TRAF3 and the protein kinases TBK1 and IKKi. TBK1 and IKKi catalyze the phosphorylation of IRF3, leading to the expression of type I IFN [2].
BCR signaling is initiated through ligation of mIg under conditions that induce phosphorylation of the ITAMs in CD79, leading to the activation of Syk. Once Syk is activated, the BCR signal is transmitted via a series of proteins associated with the adaptor protein B-cell linker (Blnk, SLP-65). Blnk binds CD79a via non-ITAM tyrosines and is phosphorylated by Syk. Phospho-Blnk acts as a scaffold for the assembly of the other components, including Bruton’s tyrosine kinase (Btk), Vav 1, and phospholipase C-gamma 2 (PLCγ2) [3]. Following the assembly of the BCR-signalosome, GRB2 binds and activates the Ras-guanine exchange factor SOS, which in turn activates the small GTPase RAS. The original RAS signal is transmitted and amplified through the mitogen-activated protein kinase (MAPK) pathway, which including the serine/threonine-specific protein kinase RAF followed by MEK and extracellular signal related kinases ERK 1 and 2 [4]. After stimulation of BCR, CD19 is phosphorylated by Lyn. Phosphorylated CD19 activates PI3K by binding to the p85 subunit of PI3K and produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) from PIP2, and PIP3 transmits signals downstream [5].
Central process of T cells responding to specific antigens is the binding of the T-cell receptor (TCR) to specific peptides bound to the major histocompatibility complex which expressed on antigen-presenting cells (APCs). Once TCR connected with its ligand, the ζ-chain–associated protein kinase 70 molecules (Zap-70) are recruited to the TCR-CD3 site and activated, resulting in an initiation of several signaling cascades. Once stimulation, Zap-70 forms complexes with several molecules including SLP-76; and a sequential protein kinase cascade is initiated, consisting of MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAPKK), and MAP kinase (MAPK) [6]. Two MAPK kinases, MKK4 and MKK7, have been reported to be the primary activators of JNK. MKK3, MKK4, and MKK6 are activators of P38 MAP kinase [7]. MAP kinase pathways are major pathways induced by TCR stimulation, and they play a key role in T-cell responses.
Phosphoinositide 3-kinase (PI3K) binds to the cytosolic domain of CD28, leading to conversion of PIP2 to PIP3, activation of PKB (Akt) and phosphoinositide-dependent kinase 1 (PDK1), and subsequent signaling transduction [8].
References
[1] Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature immunology, 2010, 11(5): 373-384.
[2] Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650.
[3] Packard T A, Cambier J C. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation[J]. F1000Prime Rep, 2013, 5(40.10): 12703.
[4] Zhong Y, Byrd J C, Dubovsky J A. The B-cell receptor pathway: a critical component of healthy and malignant immune biology[C]//Seminars in hematology. WB Saunders, 2014, 51(3): 206-218.
[5] Baba Y, Matsumoto M, Kurosaki T. Calcium signaling in B cells: regulation of cytosolic Ca 2+ increase and its sensor molecules, STIM1 and STIM2[J]. Molecular immunology, 2014, 62(2): 339-343.
[6] Adachi K, Davis M M. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells[J]. Proceedings of the National Academy of Sciences, 2011, 108(4): 1549-1554.
[7] Rincón M, Flavell R A, Davis R A. The Jnk and P38 MAP kinase signaling pathways in T cell–mediated immune responses[J]. Free Radical Biology and Medicine, 2000, 28(9): 1328-1337.
[8] Bashour K T, Gondarenko A, Chen H, et al. CD28 and CD3 have complementary roles in T-cell traction forces[J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2241-2246.
Targets for Immunology/Inflammation
- Cyclic GMP-AMP Synthase(2)
- Apoptosis(178)
- 5-Lipoxygenase(18)
- TLR(98)
- Papain(1)
- PGDS(1)
- PGE synthase(24)
- SIKs(10)
- IκB/IKK(60)
- AP-1(3)
- KEAP1-Nrf2(42)
- NOD1(1)
- NF-κB(224)
- Interleukin Related(147)
- 15-lipoxygenase(2)
- Others(10)
- Aryl Hydrocarbon Receptor(33)
- CD73(16)
- Complement System(52)
- Galectin(31)
- IFNAR(21)
- NO Synthase(75)
- NOD-like Receptor (NLR)(46)
- STING(86)
- Reactive Oxygen Species(402)
- FKBP(11)
- eNOS(4)
- iNOS(24)
- nNOS(20)
- Glutathione(37)
- Adaptive Immunity(144)
- Allergy(129)
- Arthritis(25)
- Autoimmunity(134)
- Gastric Disease(64)
- Immunosuppressants(27)
- Immunotherapeutics(3)
- Innate Immunity(411)
- Pulmonary Diseases(76)
- Reactive Nitrogen Species(43)
- Specialized Pro-Resolving Mediators(42)
- Reactive Sulfur Species(24)
Products for Immunology/Inflammation
- Cat.No. Product Name Information
-
GC50569
NLRP3-IN-2
NLRP3 inflammasome inhibitor

-
GC45194
α-(difluoromethyl)-DL-Arginine
DFMA, RMI 71897
Bacteria synthesize the cellular growth factor putrescine through a number of pathways.
-
GC65446
α-Amyrin acetate
α-Amyrin acetate, a natural triterpenoid, has anti-inflammatory activity, antispasmodic profile and the relaxant effect.
-
GC49838
α-Cortolone
20α-Cortolone, NSC 59872
A metabolite of cortisol
-
GC48279
α-D-Glucose-1-phosphate (sodium salt hydrate)
α-D-Glc 1-P
An intermediate in glycogen metabolism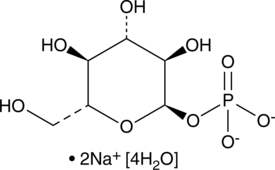
-
GC52253
α-Enolase (1-19)-biotin Peptide
Enolase-1 (1-19)-biotin
A biotinylated α-enolase peptide
-
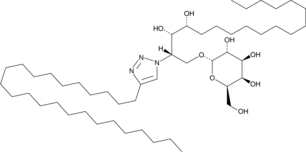
GC45206
α-GalCer analog 8
α-Galactosylceramide analog 8
α-Galactosylceramide analog 8 (α-GalCer analog 8) is a triazole derivative of α-galactosylceramide.
-
GC40262
α-Humulene
αCaryophyllene, (±)-αHumulene
α-Humulene is a sesquiterpene that has been found in C.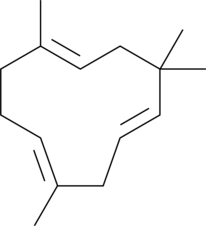
-
GC45601
α-Linolenic Acid ethyl ester-d5
ALAEE-d5, Ethyl α-Linolenate-d5, Ethyl Linolenate-d5, LAEE-d5, Linolenic Acid ethyl ester-d5
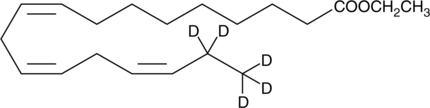
-
GC48292
α-MSH (human, mouse, rat, porcine, bovine, ovine) (trifluoroacetate salt)
α-Melanocyte-stimulating Hormone, Ac-SYSMEHFRWGKPV-NH2
α-MSH (α-Melanocyte-Stimulating Hormone) TFA, an endogenous neuropeptide, is an endogenous melanocortin receptor 4 (MC4R) agonist with anti-inflammatory and antipyretic activities.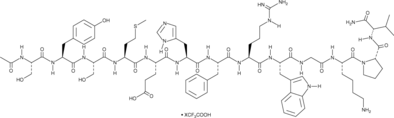
-
GC41499
α-Phellandrene
p-Mentha-1,5-diene, (±)-α-Phellandrene
α-Phellandrene is a cyclic monoterpene that has been found in various plants, including Cannabis, and has diverse biological activities.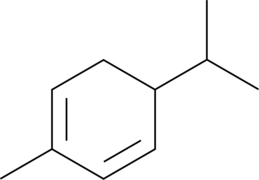
-
GC63941
α-Solanine
α-solanine, a bioactive component and one of the major steroidal glycoalkaloids in potatoes, has been observed to inhibit growth and induce apoptosis in cancer cells.
-
GC67618
α-Tocopherol phosphate disodium
alpha-Tocopherol phosphate disodium; TocP disodium; Vitamin E phosphate disodium
α-Tocopherol phosphate (alpha-Tocopherol phosphate) disodium, a promising antioxidant, can protect against long-wave UVA1 induced cell death and scavenge UVA1 induced ROS in a skin cell model. α-Tocopherol phosphate disodium possesses therapeutic potential in the inhibition of apoptosis and increases the migratory capacity of endothelial progenitor cells under high-glucose/hypoxic conditions and promotes angiogenesis.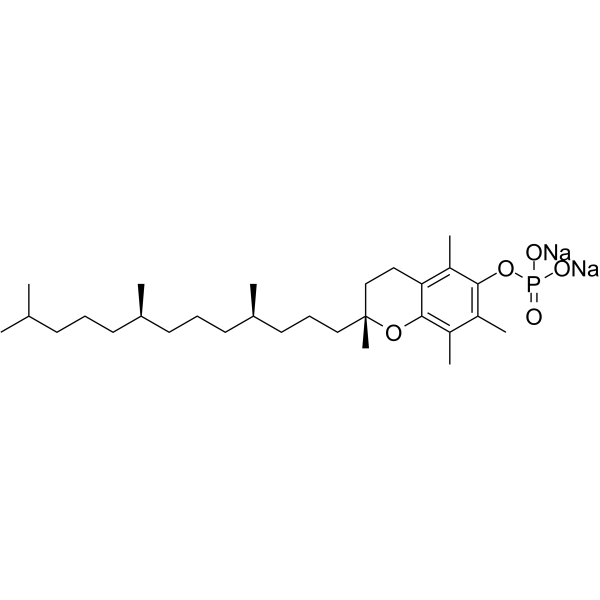
-
GC70953
α7 nAchR-JAK2-STAT3 agonist 1
α7 nAchR-JAK2-STAT3 agonist 1 is a potent α7 nAchR-JAK2-STAT3 agonist, with an IC50 value of 0.32 μM for nitric oxide (NO).

-
GC49467
β-Aescin
A triterpenoid saponin with diverse biological activities
-
GC70787
β-Aminoarteether
β-Aminoarteether (SM934 free base) is an Artemisinin derivative with orally active.

-
GC37999
β-Anhydroicaritin
β-Anhydroicaritin is isolated from Boswellia carterii Birdware, has important biological and pharmacological effects, such as antiosteoporosis, estrogen regulation and antitumor properties.
-
GC45225
β-Apooxytetracycline
β-Apo-Oxytetracycline, β-Apoterramycin
β-Apooxytetracycline is a potential impurity found in commercial preparations of oxytetracycline.
-
GC48920
β-Carboline-1-carboxylic Acid
1-Formic Acid-β-carboline
An alkaloid with diverse biological activities
-
GC66870
β-D-Glucan
β-D-glucan is a natural non-digestible polysaccharide and high biocompatibility that can be selectively recognized by recognition receptors such as Dectin-1 and Toll-like receptors as well as being easily internalized by murine or human macrophages, which is likely to attribute to a target delivery. β-d-glucan is an enteric delivery vehicle for probiotics.

-
GC48998
β-Defensin-1 (human) (trifluoroacetate salt)
hBD-1
An antimicrobial peptide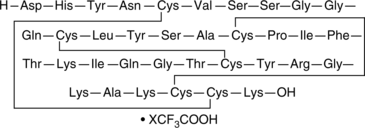
-
GC48298
β-Defensin-2 (human) (trifluoroacetate salt)
hBD-2
An antimicrobial peptide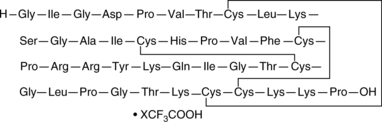
-
GC45230
β-Defensin-3 (human) (trifluoroacetate salt)
hBD-3
β-Defensin-3 is a peptide with antimicrobial properties that protects the skin and mucosal membranes of the respiratory, genitourinary, and gastrointestinal tracts.
-
GC45231
β-Defensin-4 (human) (trifluoroacetate salt)
hBD-4 (human)
β-Defensin-4 is a peptide with antimicrobial properties that protects the skin and mucosal membranes of the respiratory, genitourinary, and gastrointestinal tracts.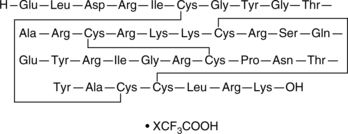
-
GC41623
β-Elemonic Acid
Elemadienonic Acid, 3-Oxotirucallenoic Acid, 3-oxo Tirucallic Acid
β-Elemonic acid is a triterpene isolated from Boswellia (Burseraceae) that exhibits anticancer activity.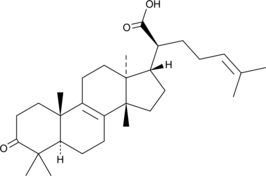
-
GC49769
β-Glucogallin
1-O-Galloyl-β-D-glucose
A plant metabolite and an aldose reductase 2 inhibitor
-
GC64619
β-Ionone
β-Ionone is effective in the induction of apoptosis in gastric adenocarcinoma SGC7901 cells. Anti-cancer activity.
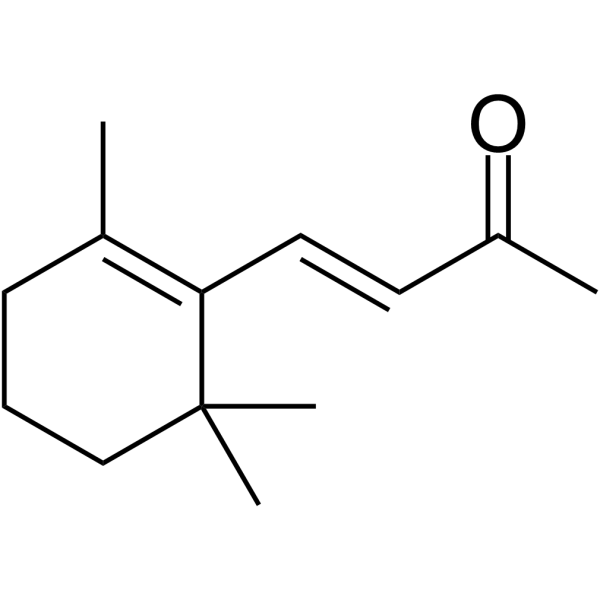
-
GC41502
β-Myrcene
NSC 406264
β-Myrcene is a terpene that has been found in Cannabis and has antioxidative properties.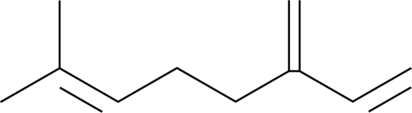
-
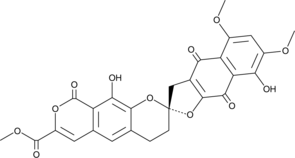
GC45604
β-Rubromycin
β-Rubromycin is a potent and selective inhibitor of human immunodeficiency virus-1 (HIV-1) RNA-directed DNA polymeras (reverse transcriptase).
-
GC52400
γ-Glu-Ala (trifluoroacetate salt)
γ-Glutamylalanine, γ-L-Glutamyl-L-alanine
A dipeptide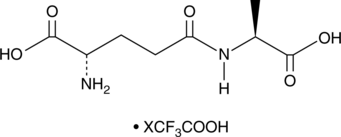
-
GC48312
γ-Glu-Cys (ammonium salt)
γ-Glutamylcysteine
An intermediate in GSH synthesis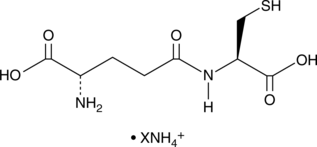
-
GC45238
δ14-Triamcinolone acetonide
14,15-dehydro Triamcinolone acetonide, Triamcinolone acetonide Impurity B
δ14-Triamcinolone acetonide is a potential impurity found in commercial preparations of triamcinolone acetonide.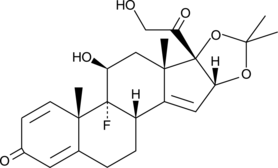
-
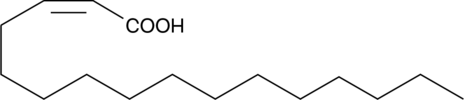
GC40307
δ2-cis-Hexadecenoic Acid
One of the first organisms in which quorum sensing was observed were Myxobacteria, a group of gram-negative bacteria, found mainly in soil and also common to marine and freshwater systems.
-
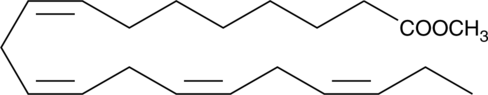
GC41393
ω-3 Arachidonic Acid methyl ester
ω-3 Fatty acids, represented primarily by docosahexaenoic acid, eicosapentaenoic acid, and α-linoleate, are essential dietary nutrients required for normal growth and development.
-
GC45713
(±)-α-Tocopherol Acetate
all-rac-α-Tocopherol Acetate, DL-α-Tocopherol Acetate, DL-Vitamin E acetate
(±)-α-Tocopherol Acetate ((±)-Vitamin E acetate), is a orally active synthetic form of vitamin E.
-
GC67191
(±)-α-Tocopherol nicotinate
(±)-α-Tocopherol nicotinate, vitamin E - nicotinate, is an orally active fat-soluble antioxidant that prevents lipid peroxidation in cell membranes. (±)-α-Tocopherol nicotinate is hydrolysed in the blood to α -tocopherol and niacin and may be used in studies of related vascular diseases.
-
GC52010
(±)-10-hydroxy-12(Z),15(Z)-Octadecadienoic Acid
αHYA, (±)-10-hydroxy-12(Z),15(Z)-ODE
An oxylipin gut microbiota metabolite
-
GC52013
(±)-10-hydroxy-12(Z)-Octadecenoic Acid
10-hydroxy-cis-12-Octadecenoic Acid
An oxylipin and metabolite of linoleic acid
-
GC52421
(±)-10-hydroxy-12(Z)-Octadecenoic Acid-d5
10-hydroxy-cis-12-Octadecenoic Acid-d5
An internal standard for the quantification of (±)-10-hydroxy-12(Z)-octadecenoic acid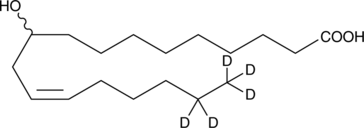
-
GC40112
(±)-Climbazole-d4
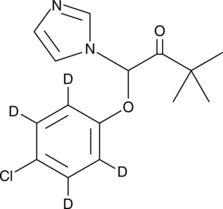
(±)-Climbazole-d4 is intended for use as an internal standard for the quantification of climbazole by GC- or LC-MS.
-
GC50708
(±)-ML 209
An RORγt antagonist
-
GC39271
(±)-Naringenin
SDihydrogenistein, NSC 11855, NSC 34875, Salipurol
A citrusderived flavonoid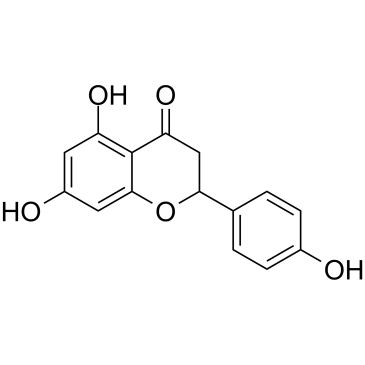
-
GC41212
(±)10(11)-EpDPA
(±)10,11-EDP, (±)10,11-EpDPE, (±)10,11-epoxy DPA, (±)10,11-epoxy Docosapentaenoic Acid
Cytochrome P450 metabolism of polyunsaturated fatty acids produces numerous bioactive epoxide regioisomers.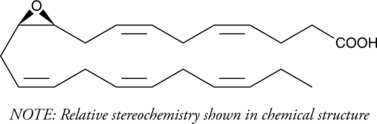
-
GC40466
(±)11(12)-EET
(±)11,12-EpETrE
(±)11(12)-EET is a fully racemic version of the R/S enantiomeric forms biosynthesized from arachidonic acid by cytochrome P450 enzymes.
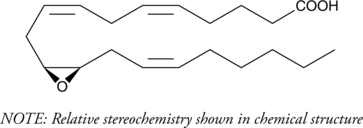
-
GC40467
(±)11-HETE
(±)11-Hydroxyeicosatetraenoic Acid
(±)11-HETE is one of the six monohydroxy fatty acids produced by the non-enzymatic oxidation of arachidonic acid.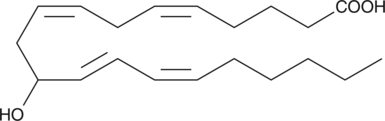
-
GC40802
(±)12(13)-DiHOME
Isoleukotoxin diol
(±)12(13)-DiHOME is the diol form of (±)12(13)-EpOME, a cytochrome P450-derived epoxide of linoleic acid also known as isoleukotoxin.
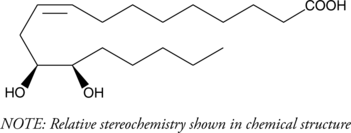
-
GC41191
(±)13(14)-EpDPA
(±)13,14-EDP, (±)13,14-EpDPE, (±)13,14-epoxy DPA, (±)13,14-epoxy Docosapentaenoic Acid
Cytochrome P450 metabolism of polyunsaturated fatty acids produces numerous bioactive epoxide regioisomers.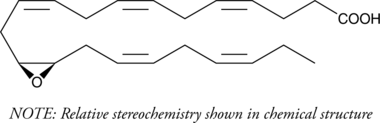
-
GC40355
(±)13-HpODE
13-Hydroperoxylinoleic acid; Linoleic acid 13-hydroperoxide
(±)13-HpODE is a racemic mixture of hydroperoxides derived non-enzymatically from linoleic acid through the action of reactive oxygen species.
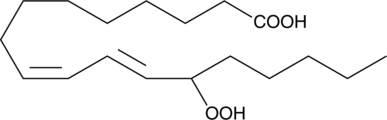
-
GC41288
(±)17(18)-EpETE-Ethanolamide
17,18-EEQ-EA, (±)17,18-EEQ-Ethanolamide, (±)17(18)-EpETE-EA, 17,18-epoxy-Eicosatetraenoic Acid Ethanolamide
(±)17(18)-EpETE-Ethanolamide is an ω-3 endocannabinoid epoxide.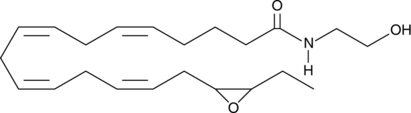
-
GC40362
(±)18-HEPE
(±)18-HEPE is produced by non-enzymatic oxidation of EPA.
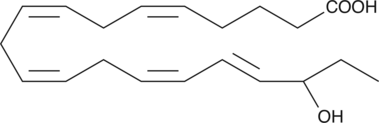
-
GC41655
(±)19(20)-EDP Ethanolamide
19,20-DHEA epoxide, 19,20-epoxy Docosapentaenoic Acid Ethanolamide, 19,20-EDP-EA, 19,20-EDP epoxide
(±)19(20)-EDP ethanolamide is an ω-3 endocannabinoid epoxide and cannabinoid (CB) receptor agonist (EC50s = 108 and 280 nM for CB1 and CB2, respectively).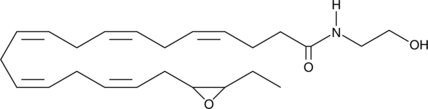
-
GC40270
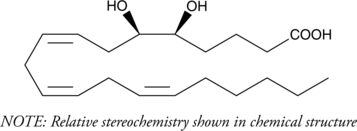
(±)5(6)-DiHET
(±)5,6-DiHETrE
5(6)-DiHET is a fully racemic version of the enantiomeric forms biosynthesized from 5(6)-EET by epoxide hydrolases.
-
GC41203
(±)7(8)-EpDPA
(±)7,8-EDP, (±)7,8-EpDPE, (±)7,8-epoxy DPA, (±)7,8-epoxy Docosapentaenoic Acid
Docosahexaenoic acid is the most abundant ω-3 fatty acid in neural tissues, especially in the brain and retina.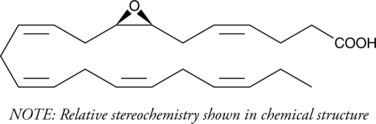
-
GC40801
(±)9(10)-DiHOME
Leukotoxin diol
Leukotoxin is the 9(10) epoxide of linoleic acid, generated by neutrophils during the oxidative burst.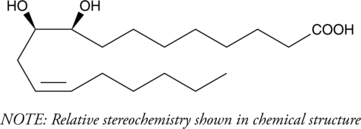
-
GC46000
(•)-Drimenol
NSC 169775
A sesquiterpene alcohol
-
GC40809
(+)-β-Citronellol
(R)-Citronellol, (+)-Citronellol, (+)-(R)-Citronellol, (R)-(+)-β-Citronellol
(+)-β-Citronellol (D-Citronellol) is an alcoholic monoterpene found in geranium essential oil.
-
GC49268
(+)-δ-Cadinene
A sesquiterpene with antimicrobial and anticancer activities
-
GC45263
(+)-D-threo-PDMP (hydrochloride)
D-PDMP
(+)-D-threo-PDMP is a ceramide analog and is one of the four possible stereoisomers of PDMP.
-
GC31691
(+)-DHMEQ
(1R,2R,6R)-Dehydroxymethylepoxyquinomicin; (1R,2R,6R)-DHMEQ
(+)-DHMEQ is an activator of antioxidant transcription factor Nrf2.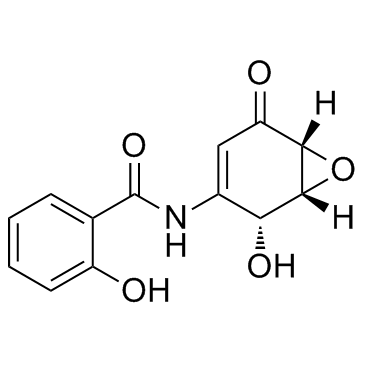
-
GC45266
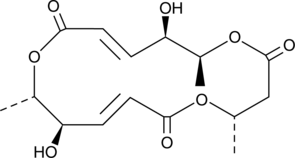
(+)-Macrosphelide A
(+)-Macrosphelide A is a fungal metabolite originally isolated from Microsphaeropsis.
-
GC40266
(+)-Praeruptorin A
(+)-Praeruptorin A is a coumarin derivative originally isolated from P.

-
GC18749
(+)-Rugulosin
NSC 160880, NSC 249990, Rugulosin A
(+)-Rugulosin is a pigment and mycotoxin produced by certain fungi.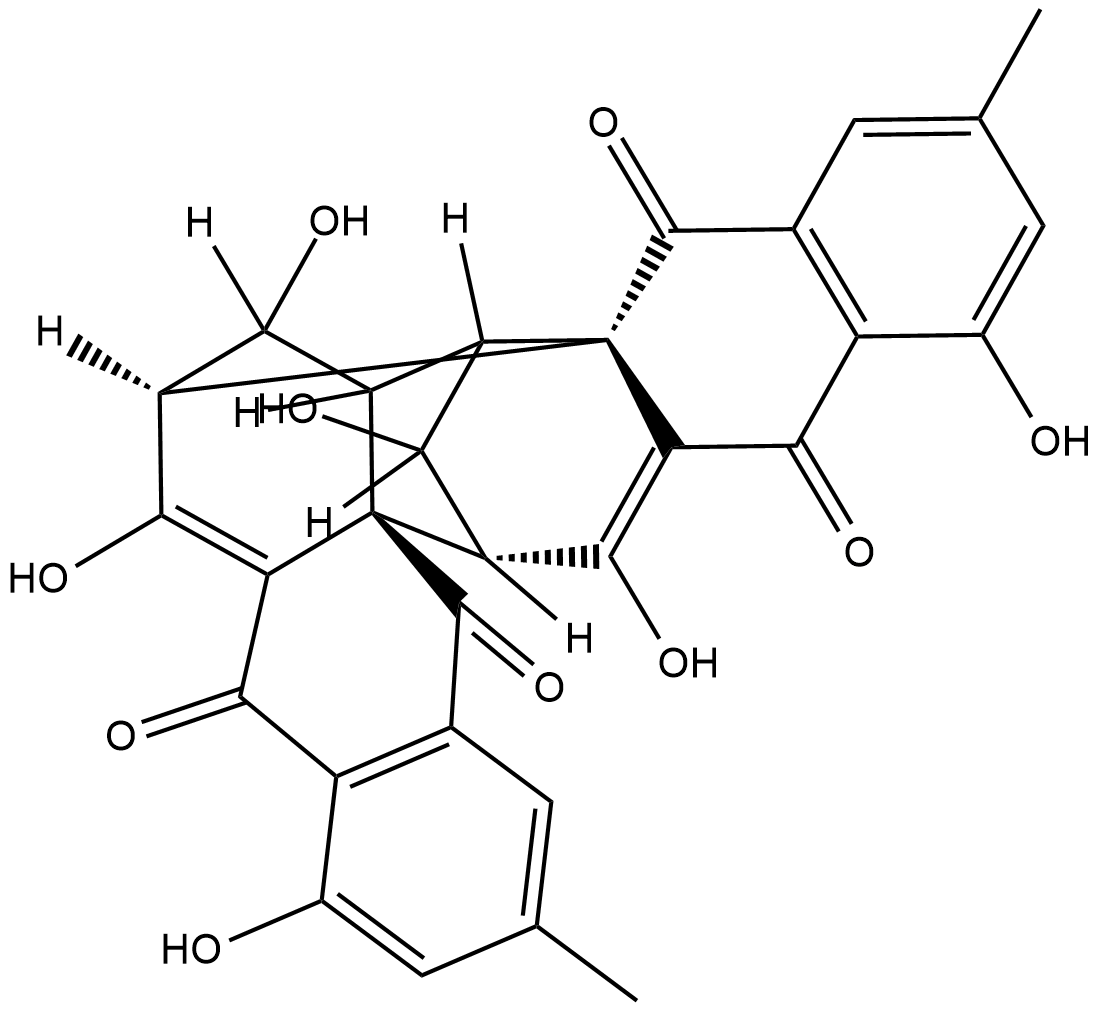
-
GC63969
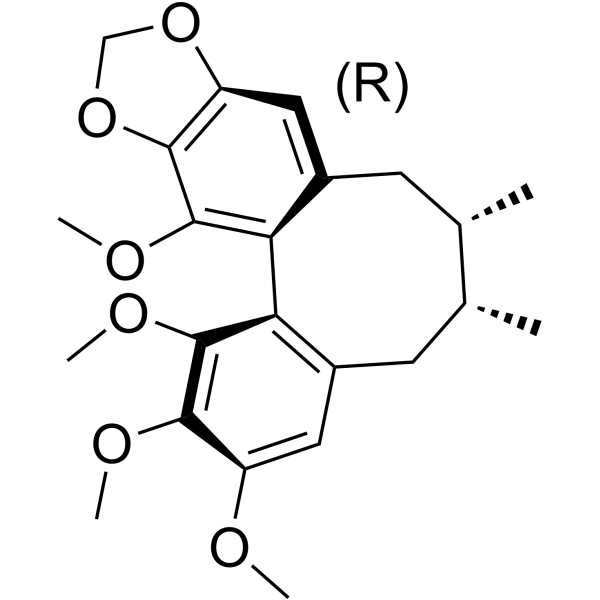
(+)-Schisandrin B
(+)-Schisandrin B is an enantiomer of Schisandrin B.
-
GC40264
(+)-Valencene
NSC 148969
(+)-Valencene is a sesquiterpene that has been found in C.
-
GC49502
(-)-β-Sesquiphellandrene
A sesquiterpene with antiviral and anticancer activities
-
GC32705
(-)-DHMEQ (Dehydroxymethylepoxyquinomicin)
Dehydroxymethylepoxyquinomicin
(-)-DHMEQ (Dehydroxymethylepoxyquinomicin) (Dehydroxymethylepoxyquinomicin) is a potent, selective and irreversible NF-κB inhibitor that covalently binds to a cysteine residue.
-
GC14049
(-)-Epigallocatechin gallate (EGCG)
EGCG
(-)-Epigallocatechin Gallate sulfate (EGCG) is a major polyphenol in green tea that inhibits cell proliferation and induces apoptosis.
-
GC45248
(-)-FINO2
(-)-FINO2 is a ferroptosis-inducing peroxide compound that indirectly inhibits glutathione peroxidase 4 (GPX4) and oxidizes iron.
-
GC46245
(-)-G-Lactone
A bicyclic γ-lactone
-
GC38316
(-)-Limonene
(±)-Dipentene, DL-Limonene, NSC 844, NSC 21446
(-)-Limonene ((S)-(-)-Limonene) is a monoterpene found in many pine-needle oils and in turpentine.
-
GC46247
(-)-Mycousnine
Mycousunin
A microbial metabolite with antibacterial and antifungal activities
-
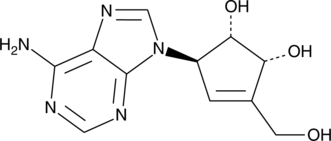
GC45251
(-)-Neplanocin A
S-Adenosylhomocysteine (SAH) hydrolase catalyzes the reversible hydrolysis of SAH to adenosine and homocysteine.
-
GC45272
(-)-Rasfonin
TT-1
-
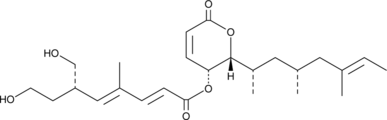
GC40803
(25S)-δ7-Dafachronic Acid
UPF1404
During unfavorable environmental conditions, C.
-
GC52442
(D)-PPA 1 (trifluoroacetate salt)
DPPA-1, NYSKPTDRQYHF
An inhibitor of the PD-1-PD-L1 protein-protein interaction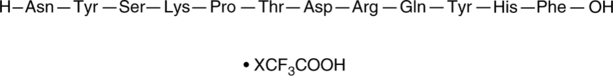
-
GC41700
(E)-2-(2-Chlorostyryl)-3,5,6-trimethylpyrazine
CSTMP
(E)-2-(2-Chlorostyryl)-3,5,6-trimethylpyrazine (CSTMP) is a stilbene derivative with antioxidant and anticancer activities.
-
GC61668
(E)-3,4-Dimethoxycinnamic acid
(E)-3,4-Dimethoxycinnamic acid is the less active isomer of 3,4-Dimethoxycinnamic acid.
-
GC41702
(E)-5-(2-Bromovinyl)uracil
BVU
(E)-5-(2-Bromovinyl)uracil (BVU) is a pyrimidine base and an inactive metabolite of the antiviral agents sorivudine and (E)-5-(2-bromovinyl)-2'-deoxyuridine (BVDU) that may be regenerated to BVDU in vivo.
-
GC49003
(E)-Ajoene
NSC 614554
A disulfide with diverse biological activities
-
GC41703
(E)-C-HDMAPP (ammonium salt)
(E)5hydroxy4methylpent3enyl pyrophosphate
Synthetic and natural alkyl phosphates, also known as phosphoantigens, stimulate the proliferation of γδ-T lymphocytes.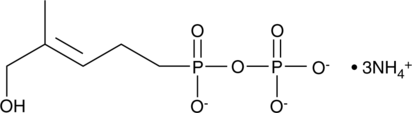
-
GC39747
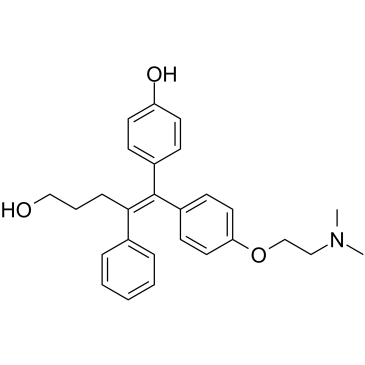
(E/Z)-GSK5182
(E/Z)-GSK5182 is a racemic compound of (E)-GSK5182 and (Z)-GSK5182 isomers.
-
GC61564
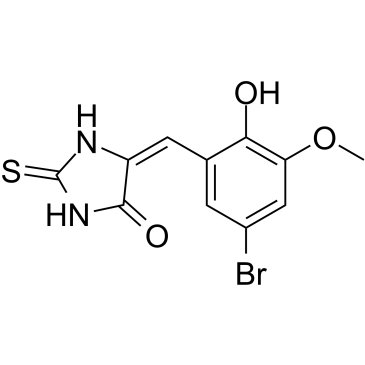
(E/Z)-IT-603
(E/Z)-IT-603 is a mixture of E-IT-603 and Z-IT-603 (IT-603).
-
GC41721
(R)-α-Lipoic Acid
(R)-(+)-Lipoic Acid
(R)-α-Lipoic acid is the naturally occurring enantiomer of lipoic acid, a cyclic disulfide antioxidant.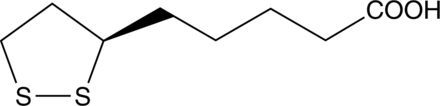
-
GC49167
(R)-(+)-Trityl glycidyl ether
(R)-Trityl Glycidol
A synthetic precursor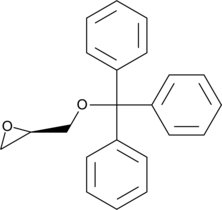
-
GC13030
(R)-(-)-Ibuprofen
(-)-Ibuprofen
Inhibitor of Cox-1 and Cox-2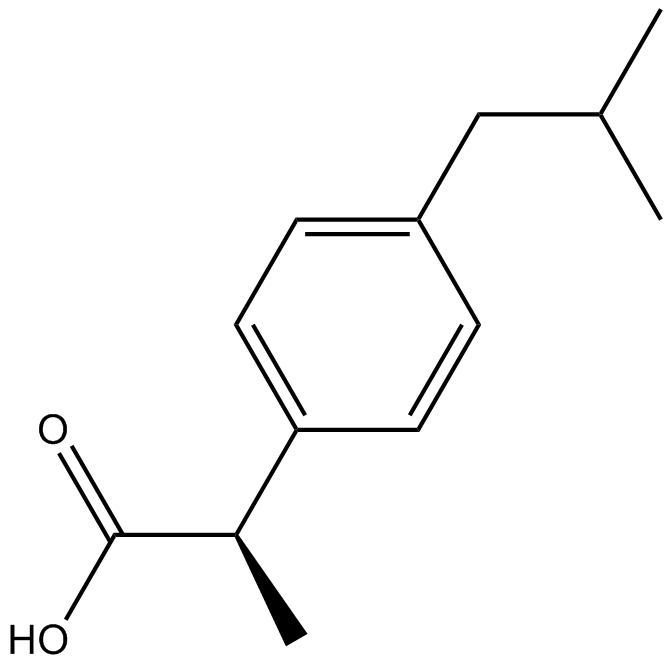
-
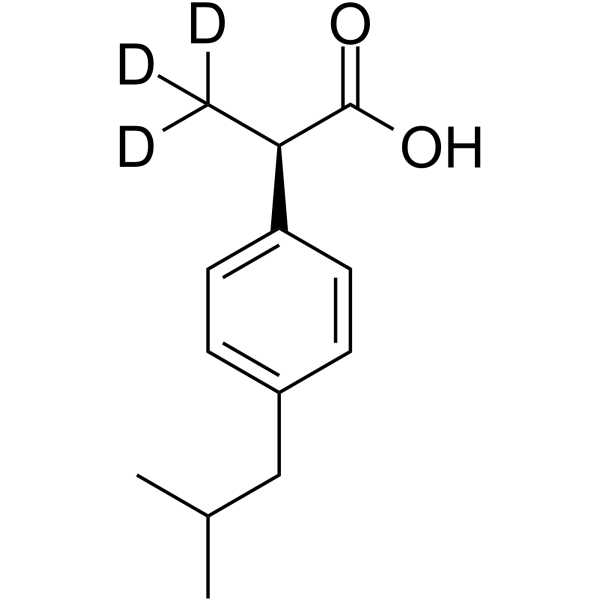
GC69823
(R)-(-)-Ibuprofen-d3
(R)-Ibuprofen-d3
(R)-(-)-Ibuprofen-d3 is the deuterated form of (R)-(-)-Ibuprofen. (R)-(-)-Ibuprofen is the R enantiomer of Ibuprofen, which has no effect on COX and can inhibit NF-κB activation. (R)-(-)-Ibuprofen has anti-inflammatory properties and can be used in pain relief research.
-
GC41620
(R)-(-)-Mellein
Ochracin
(R)-(-)-Mellein is an antibiotic isolated from culture fluids of this Aspergillus.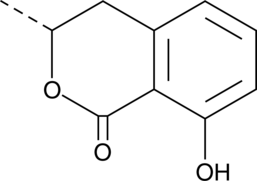
-
GC41712
(R)-3-hydroxy Myristic Acid
(R)-3-hydroxy Tetradecanoic Acid
Lipopolysaccharides (LPS) are components of the cell walls of Gram-negative bacteria.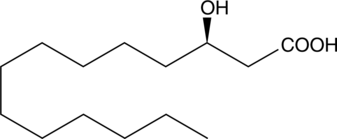
-
GC65610
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone is a diarylheptanoid that can be found in Alpinia officinarum.

-
GC65373
(R)-IL-17 modulator 4
(R)-IL-17 modulator 4 is the R-configure of IL-17 modulator 4.

-
GC12578
(R)-Lisofylline
(−)-Lisofylline,(R)-LSF
anti-inflammatory agent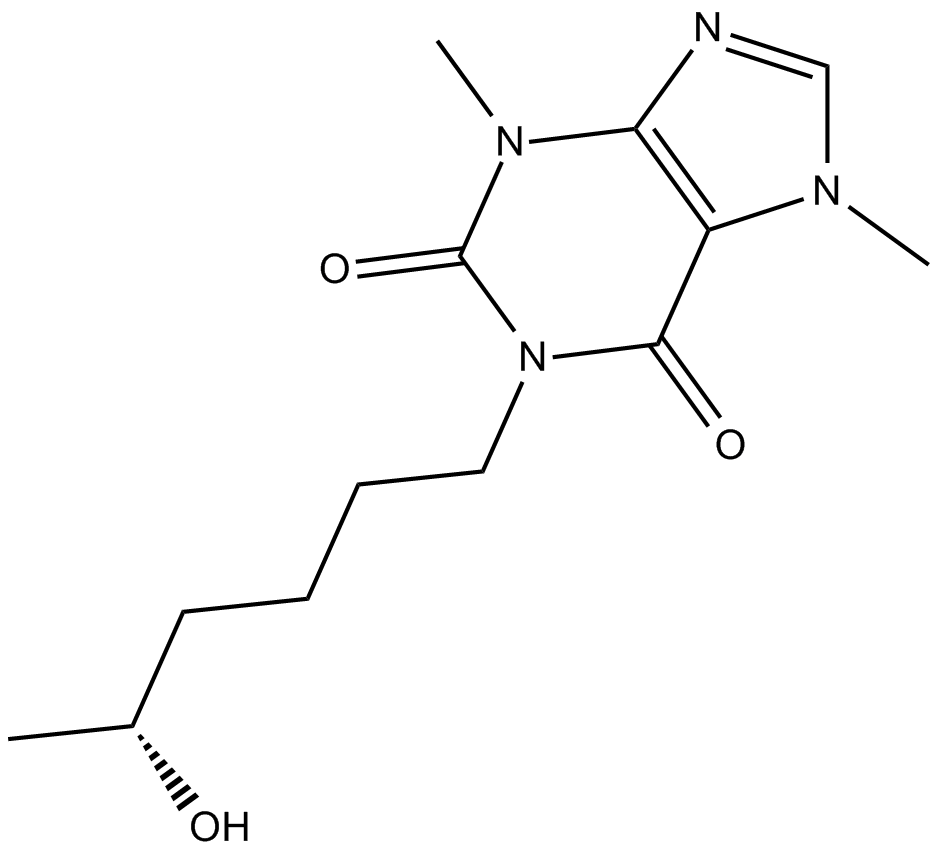
-
GC52185
(R,S)-Anatabine-d4
(±)-Anatabine-d4

-
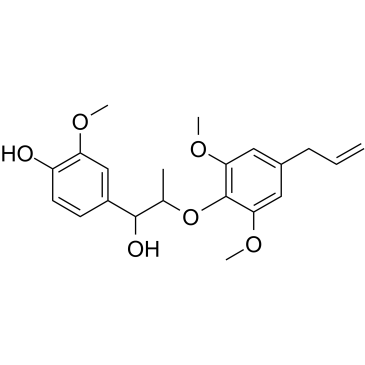
GC39321
(Rac)-Myrislignan
(Rac)-Myrislignan is the racemate of Myrislignan.
-
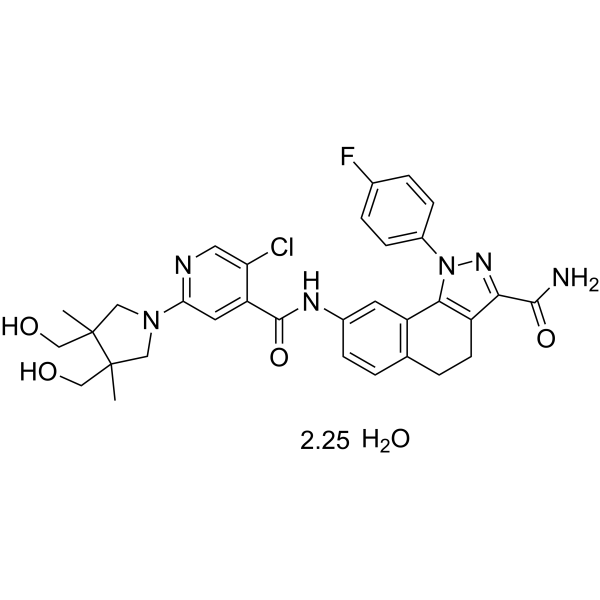
GC66334
(Rac)-PF-184 hydrate
(Rac)-PF-184 hydrate is a potent inhibitory factor-κB kinase 2 (IKK-2) inhibitor with an IC50 of 37 nM. (Rac)-PF-184 hydrate has anti-inflammatory effects.
-
GC69799
(Rac)-ZLc-002
(Rac)-ZLc-002 is an inhibitor that interacts with the binding protein between nNOS and nitric oxide synthase 1 (NOS1AP). It inhibits inflammatory pain and chemotherapy-induced neuropathic pain, and synergistically reduces tumor cell viability with Paclitaxel.
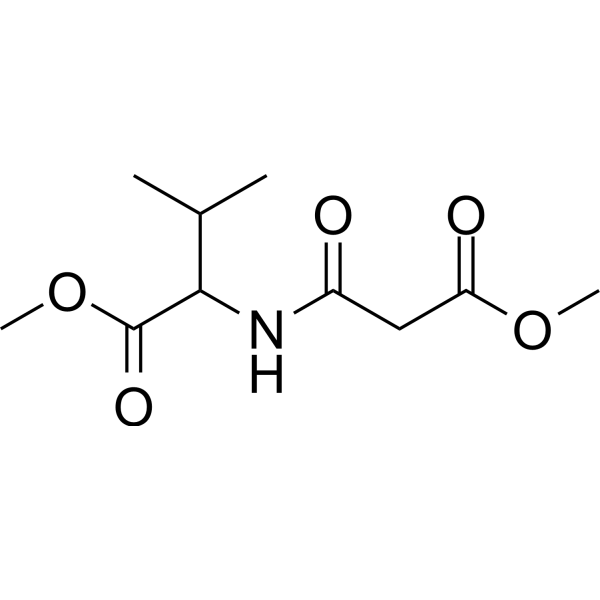
-
GC46345
(S)-(-)-Perillaldehyde
(–)-Perillaldehyde, L-Perillaldehyde, (S)-Perillaldehyde
(S)-(-)-Perillaldehyde is a major component in the essential oil containing in Perillae Herba.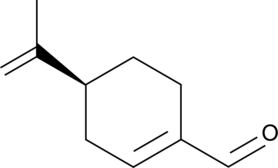
-
GC49028
(S)-3-Thienylglycine
L-R-(3-Thienyl)glycine, L-α-3-Thienylglycine
A thienyl-containing amino acid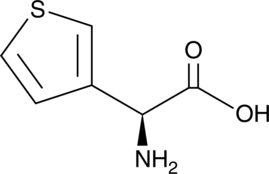
-
GC52192
(S)-4'-nitro-Blebbistatin
(-)-4'-nitro-Blebbistatin, p-nitro-Blebbistatin, para-nitro-Blebbistatin
(S)-4'-nitro-Blebbistatin is a non-cytotoxic, photostable, fluorescent and specific Myosin II inhibitor, usd in the study of the specific role of myosin II in physiological, developmental, and cell biological studies.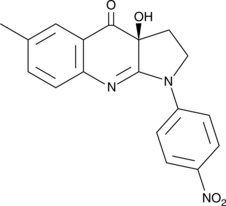
-
GC48719
(S)-Canadine
(–)-Canadine, (S)-Tetrahydroberberine
(S)-Canadine is an alkaloid and intermediate in the biosynthesis of berberine with insecticidal activity.
-
GC46352
(S)-DO271
An inactive control for DO264