MAPK Signaling

Targets for MAPK Signaling
- ERK(77)
- MEK1/2(61)
- NKCC(5)
- MNK(7)
- PKA(53)
- p38(96)
- Rac(2)
- Raf(63)
- RasGAP (Ras- P21)(0)
- JNK(56)
- cAMP(33)
- Protein Kinase G(2)
- RSK(28)
- Other(759)
- MAPKAPK(1)
- MKK(2)
- KLF(3)
- MAP3K(20)
- MAP4K(20)
- MAPKAPK2 (MK2)(9)
- Mixed Lineage Kinase(9)
Products for MAPK Signaling
- Cat.No. Product Name Information
-
GC48920
β-Carboline-1-carboxylic Acid
1-Formic Acid-β-carboline
An alkaloid with diverse biological activities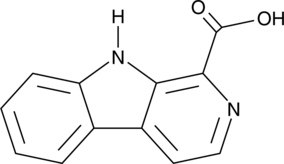
-
GC41656
(±)2-(14,15-Epoxyeicosatrienoyl) Glycerol
(±)214,15EG
2-Arachidonoyl glycerol (2-AG) is an endogenous central cannabinoid (CB1) receptor agonist that is present at relatively high levels in the central nervous system.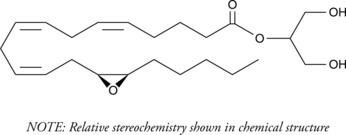
-
GC60393
(-)-Zuonin A
(-)-Zuonin A (D-Epigalbacin), a naturally occurring lignin, is a potent, selective JNKs inhibitor, with IC50s of 1.7 μM, 2.9 μM and 1.74 μM for JNK1, JNK2 and JNK3, respectively.
-
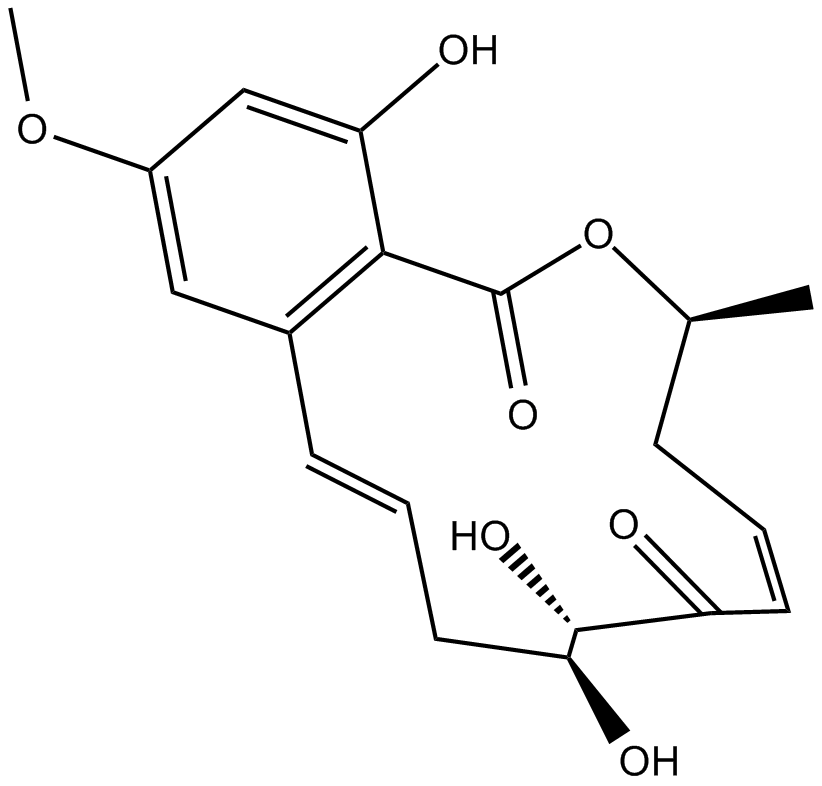
GC13944
(5Z)-7-Oxozeaenol
FR148083,L-783,279,LL-Z 1640-2
TAK1 mitogen-activated protein kinase kinase kinase (MAPKKK) inhibitor
-
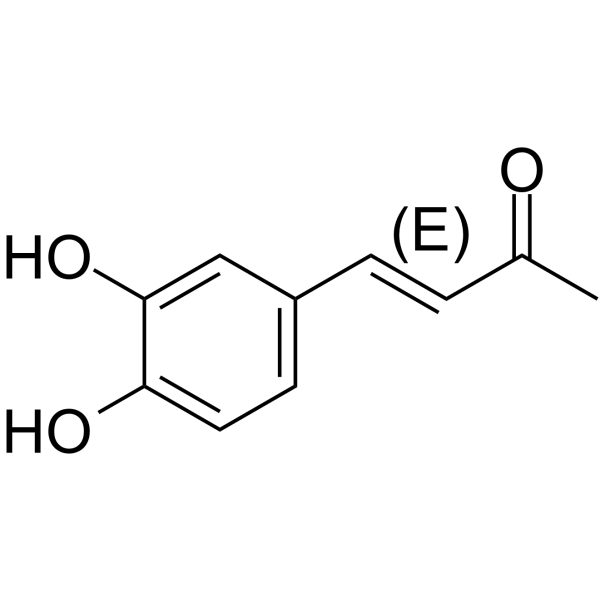
GC63903
(E)-Osmundacetone
(E)-Osmundacetone is the isomer of Osmundacetone.
-
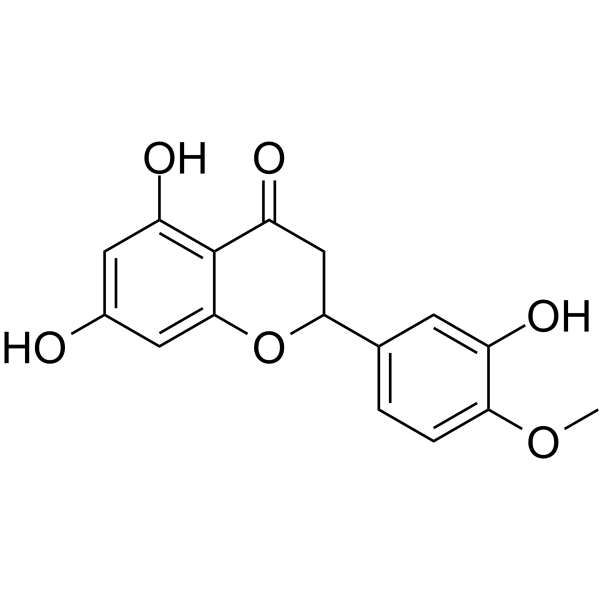
GC62528
(Rac)-Hesperetin
(Rac)-Hesperetin is the racemate of Hesperetin.
-
GC41740
(S)-p38 MAPK Inhibitor III
(S)-p38 MAP Kinase Inhibitor III, (S)-p38 Mitogen-activated Protein Kinase Inhibitor III
(S)-p38 MAPK inhibitor III is a methylsulfanylimidazole that inhibits p38 MAP kinase (IC50 = 0.90 μM in vitro).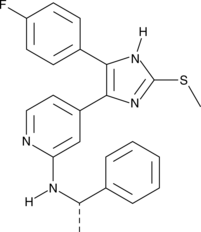
-
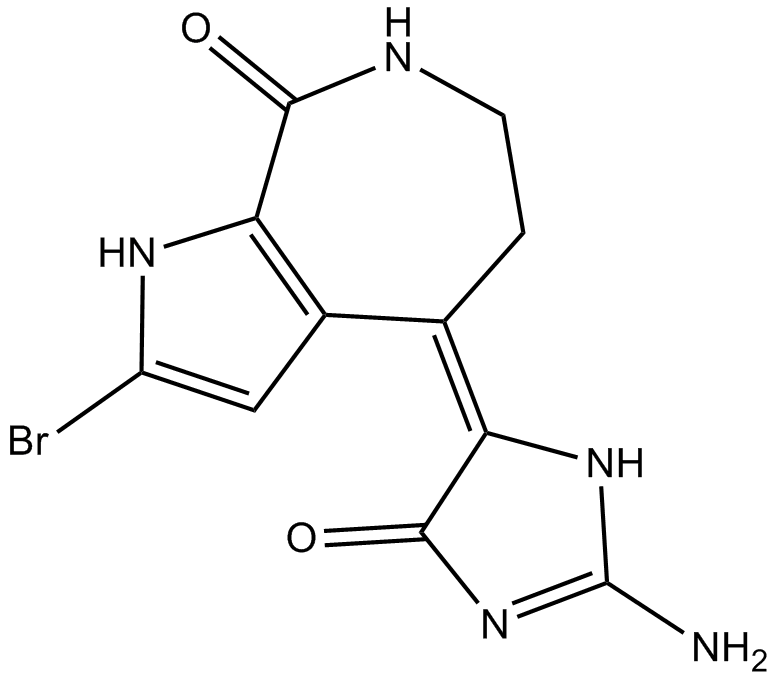
GC12851
10Z-Hymenialdisine
Pan kinase inhibitor
-
GC46554
2'-O-Monosuccinyladenosine-3',5'-cyclic monophosphate
2’-O-Succinyl-cAMP, 2’-O-Succinyl-3’,5’-cyclic AMP
2'-O-Monosuccinyladenosine-3',5'-cyclic monophosphate is a cAMP analog that can be covalently coupled to acetylcholinesterase.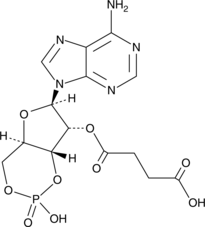
-
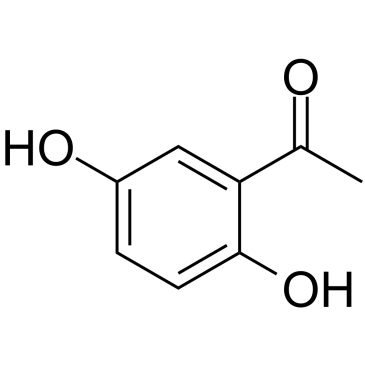
GC39325
2,5-Dihydroxyacetophenone
2,5-Dihydroxyacetophenone, isolated from Rehmanniae Radix Preparata, inhibits the production of inflammatory mediators in activated macrophages by blocking the ERK1/2 and NF-κB signaling pathways.
-
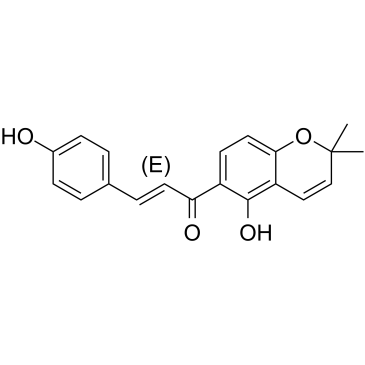
GC35132
4-Hydroxylonchocarpin
4-Hydroxylonchocarpin is a chalcone compound from an extract of Psoralea corylifolia.
-
GC48381
5'-pApA (sodium salt)
c-di-AMP Control, Cyclic di-AMP Negative Control
A linearized form of cyclic di-AMP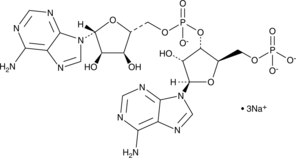
-
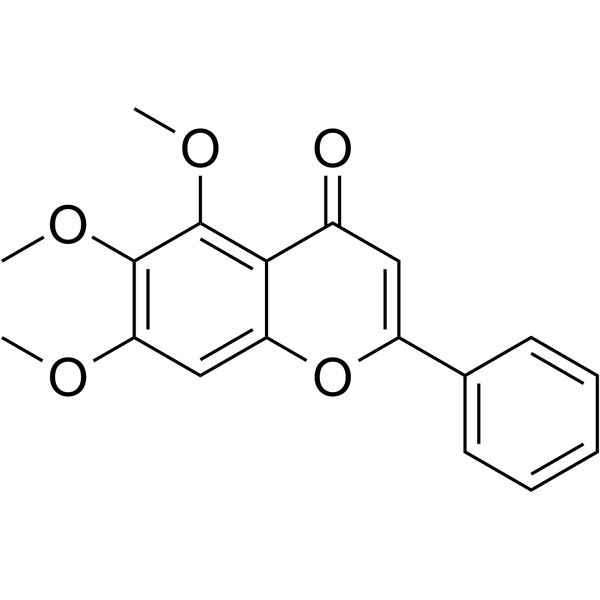
GC63700
5,6,7-Trimethoxyflavone
5,6,7-Trimethoxyflavone is a novel p38-α MAPK inhibitor with an anti-inflammatory effect.
-
GC41423
5-trans Prostaglandin E2
transDinoprostone, 5,6trans PGE2
5-trans PGE2 occurs naturally in some gorgonian corals and is a common impurity in commercial lots of PGE1.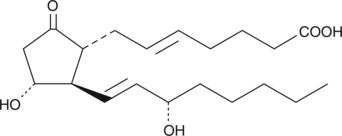
-
GC12834
6-Bnz-cAMP sodium salt
6-Bnz-cAMP
cAMP analog,PKA activator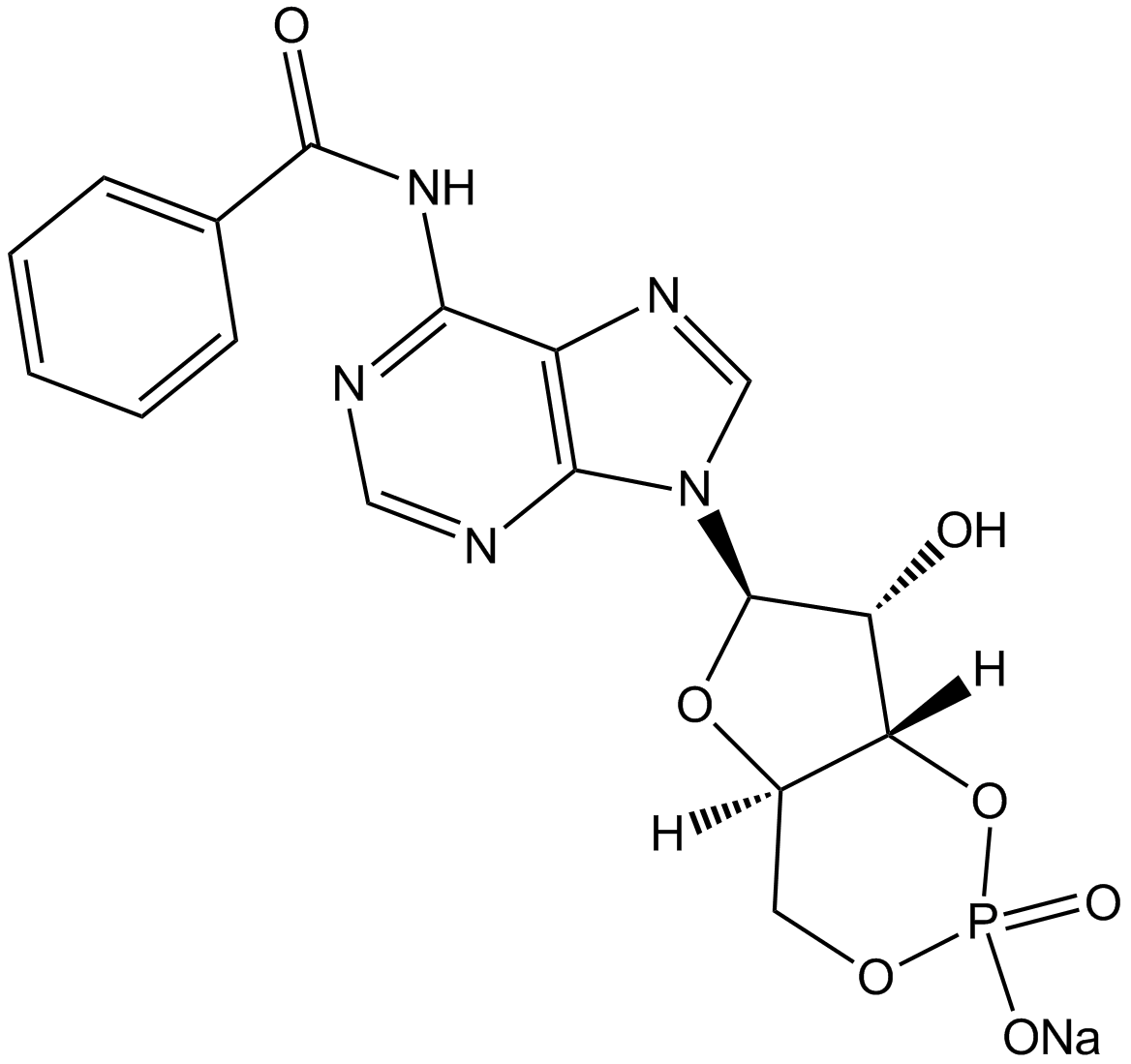
-
GC42616
7-oxo Staurosporine
BMY 41950, RK-1409
7-oxo Staurosporine is an antibiotic originally isolated from S.
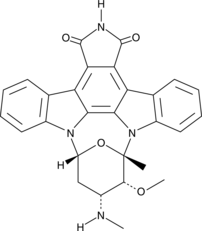
-
GC16929
8-Bromo-cAMP, sodium salt
8-Br-cAMP, 8-Bromoadenosine 3',5'-cyclic monophosphate, 8-bromo-cAMP
8-Bromo-cAMP (8-Bromo-cAMP, sodium salt) is a cell-permeable cAMP analogue that acts as a CAMP-dependent protein kinase activator (PKA activator).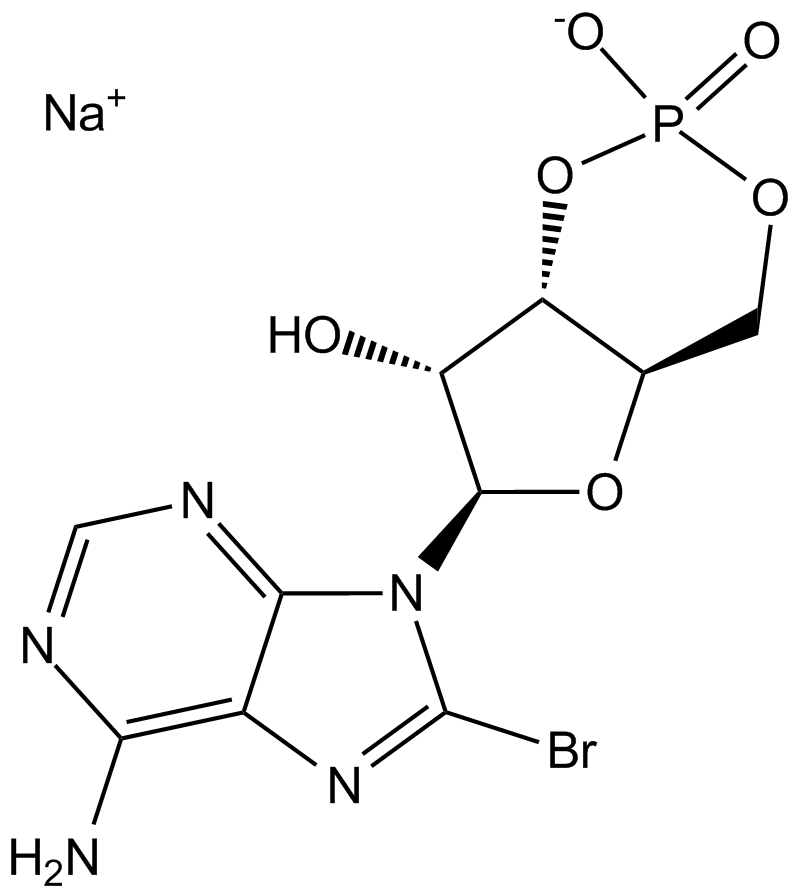
-
GC42622
8-bromo-Cyclic AMP
8-Bromoadenosine 3',5'-cyclic monophosphate, 8-Br-cAMP, 8-bromo-cAMP, NSC 171719
8-bromo-Cyclic AMP is a brominated derivative of cAMP that remains long-acting due to its resistance to degradation by cAMP phosphodiesterase.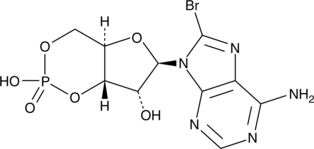
-
GC64460
8-Chloro-cAMP
8-Chloro-cAMP is a cAMP analogue that induces growth arrest, and modulates cAMP-dependent PKA activity. 8-Chloro-cAMP has anticancer activity.
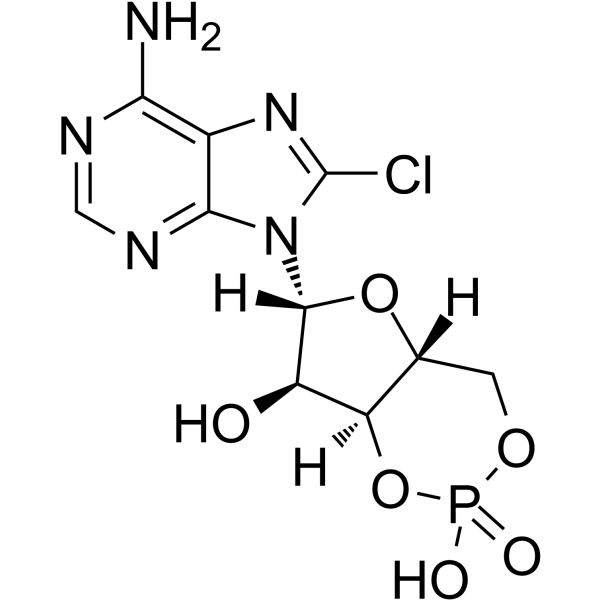
-
GC15352
8-CPT-Cyclic AMP (sodium salt)
8-(p-Chlorophenylthio)-cAMP,8-CPT-cAMP
8-CPT-Cyclic AMP (8-CPT-cAMP) sodium is a selective activator of cyclic AMP-dependent protein kinase (PKA).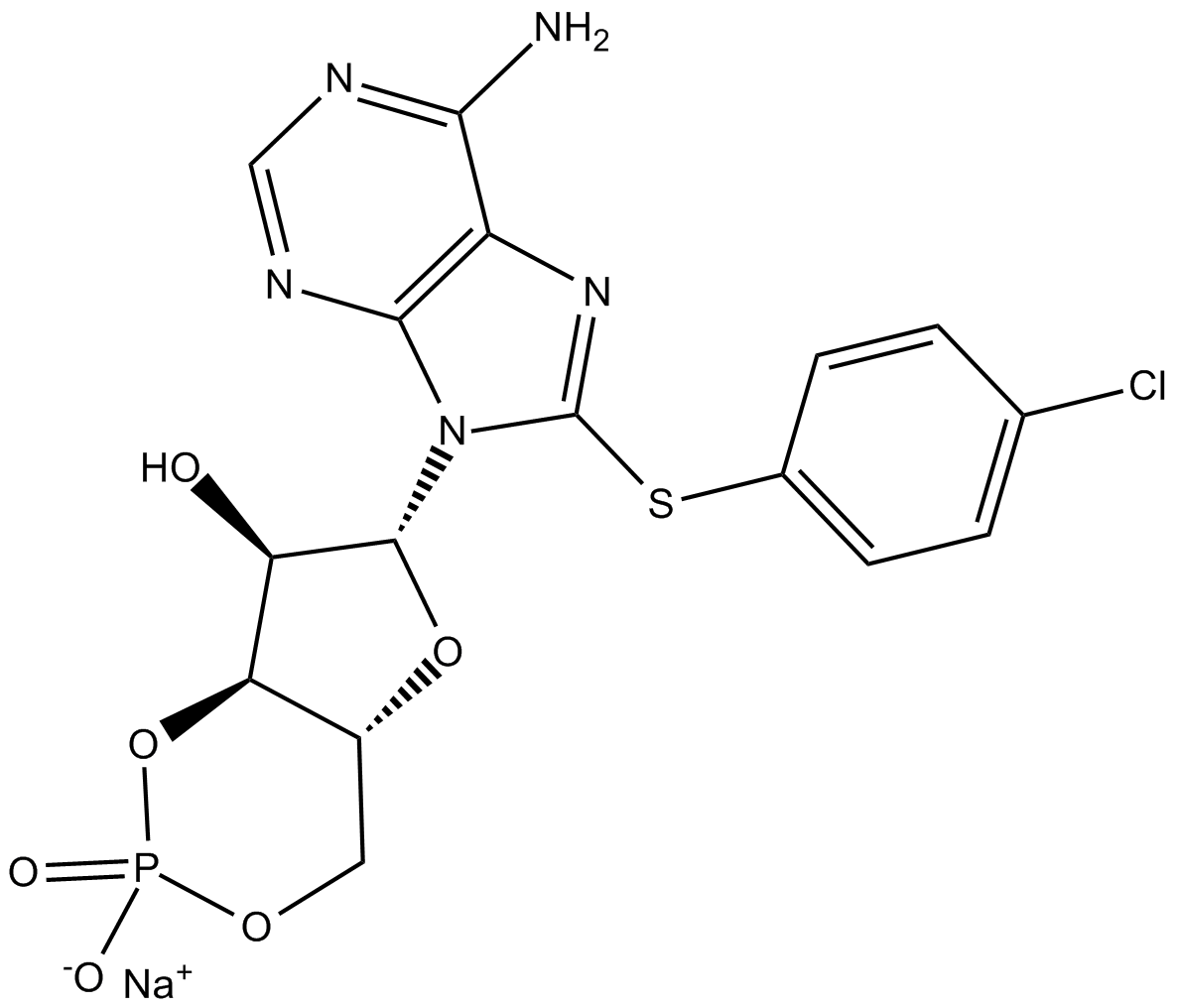
-
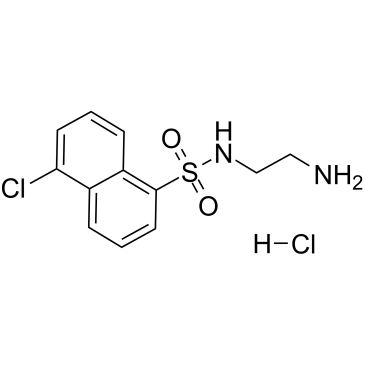
GC60037
A-3 hydrochloride
A-3 hydrochloride is a potent, cell-permeable, reversible, ATP-competitive non-selective antagonist of various kinases.
-
GA20623
Ac-muramyl-Ala-D-Glu-NH₂
MDP, N-Acetylmuramoyl dipeptide
Ac-muramyl-Ala-D-Glu-NH₂ (MDP) is a synthetic immunoreactive peptide, consisting of N-acetyl muramic acid attached to a short amino acid chain of L-Ala-D-isoGln.
-
GC68597
ACA-28
ACA-28 (compound 2a) is an effective ERK MAPK signaling regulator. ACA-28 induces apoptosis through excessive activation of ERK and selectively inhibits cancer cell growth. ACA-28 inhibits the growth of melanoma cells (SK-MEL-28) and normal melanocytes (NHEM), with IC50 values of 5.3 and 10.1 μM, respectively.
-
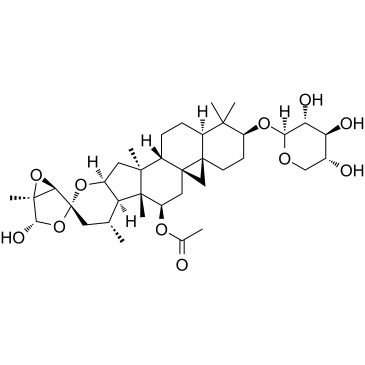
GC35242
Actein
Actein is a triterpene glycoside isolated from the rhizomes of Cimicifuga foetida. Actein suppresses cell proliferation, induces autophagy and apoptosis through promoting ROS/JNK activation, and blunting AKT pathway in human bladder cancer. Actein has little toxicity in vivo.
-
GC19019
Acumapimod
BCT-197
Acumapimod (BCT197) is an orally active p38 MAP kinase inhibitor, with an IC50 of less than 1 uM for p38α.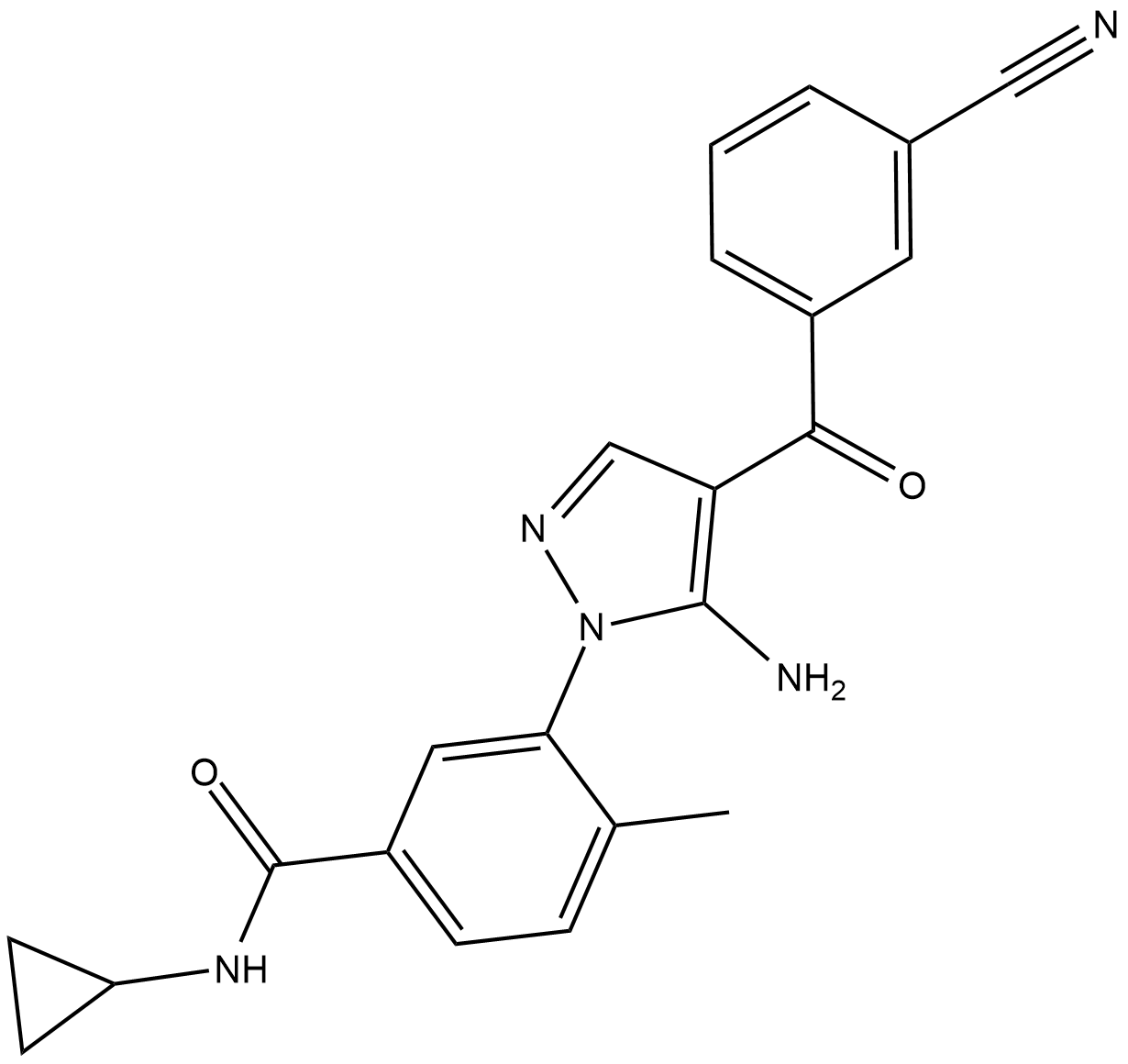
-
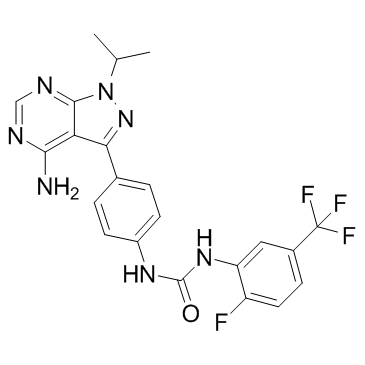
GC32797
AD80
AD80, a multikinase inhibitor, inhibits RET, RAF,SRCand S6K, with greatly reduced mTOR activity.
-
GC49285
Adenosine 5’-methylenediphosphate (hydrate)
Adenosine 5'-(α,β-methylene)diphosphate, AMP-CP, APCP, 5'-APCP
An inhibitor of ecto-5’-nucleotidase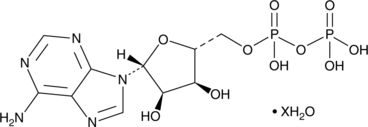
-
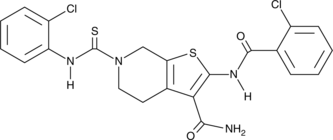
GC46808
ADTL-EI1712
A dual ERK1 and ERK5 inhibitor
-
GC10204
AEG 3482
inhibitor of Jun kinase (JNK)-dependent apoptosis
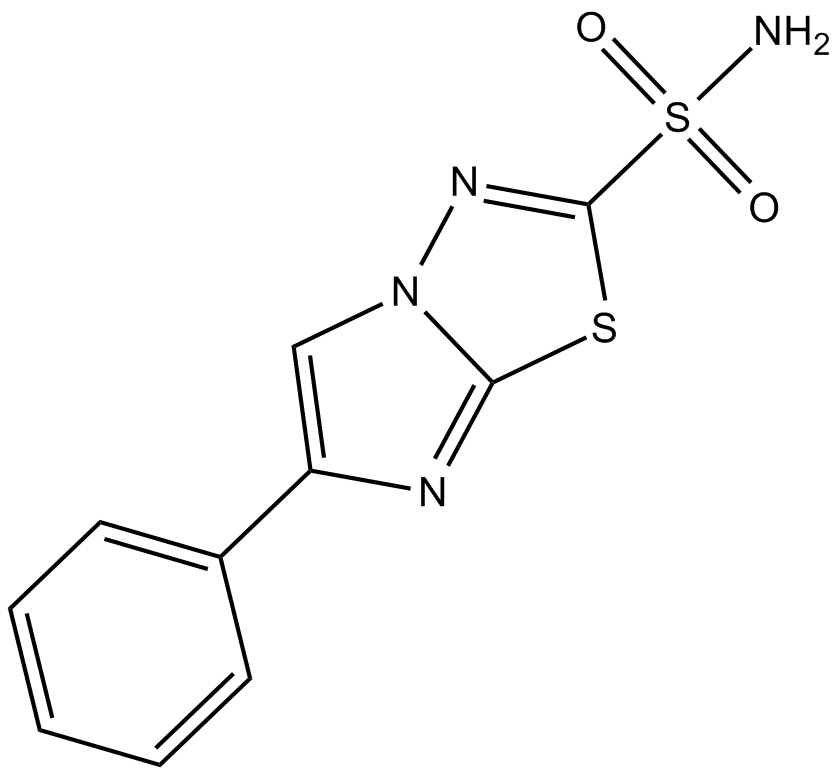
-
GC17265
AG-126
Tyrphostin AG-126
ERK1 (p44) and ERK2 (p42) inhibitor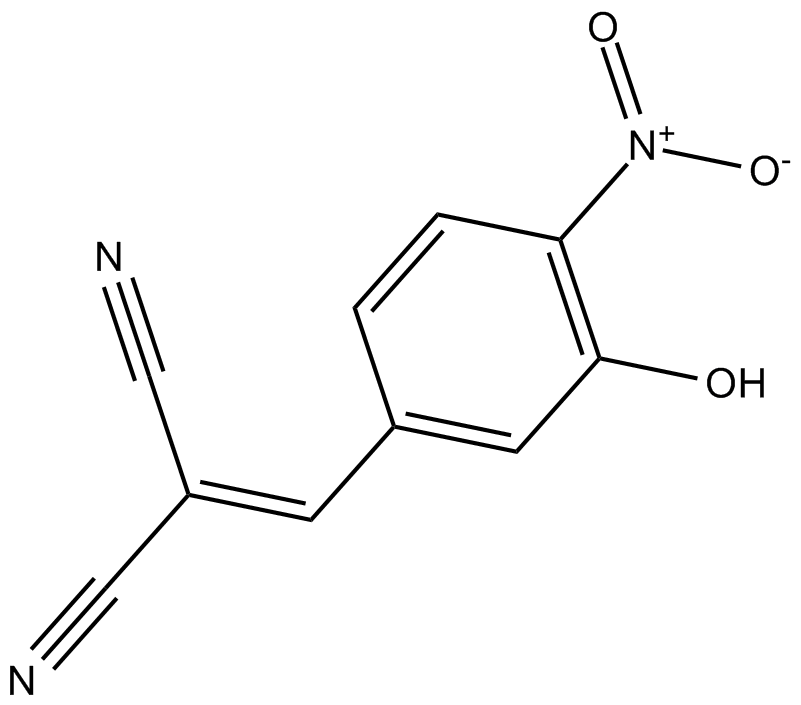
-
GC12646
AL 8697
p38α inhibitor,potent and selective
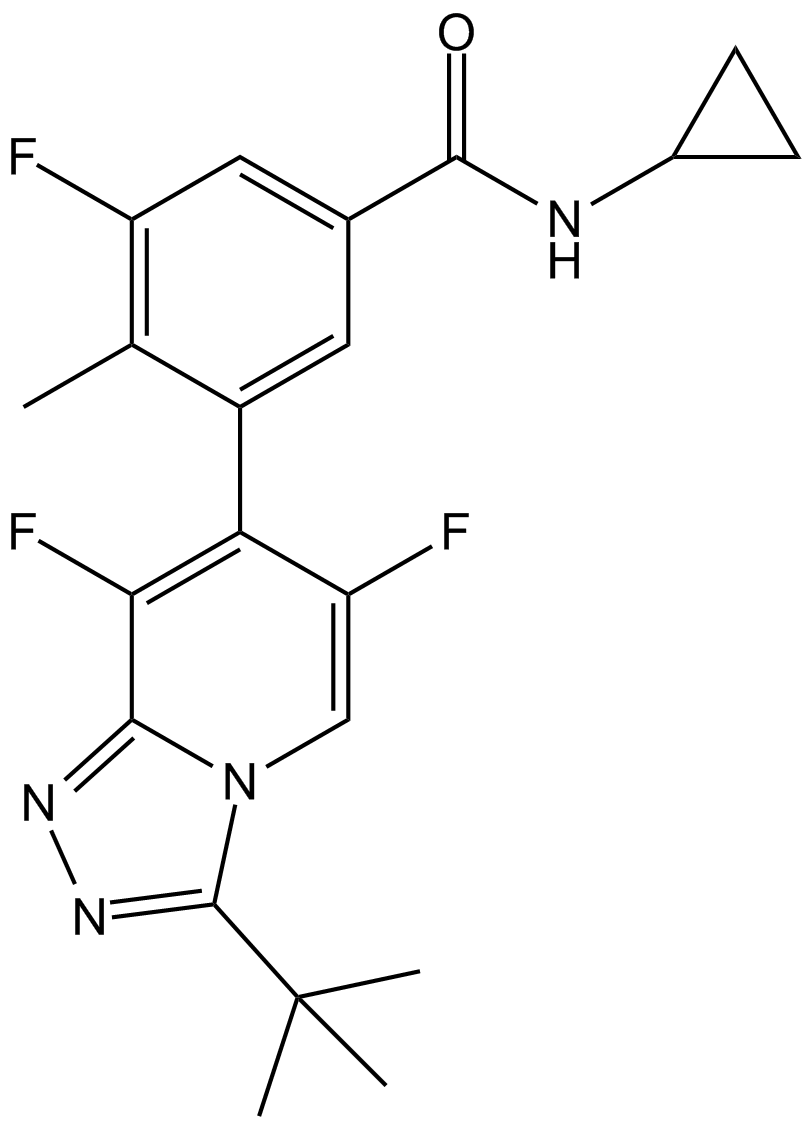
-
GC14899
AMG 548
P38α inhibitor,potent and selective
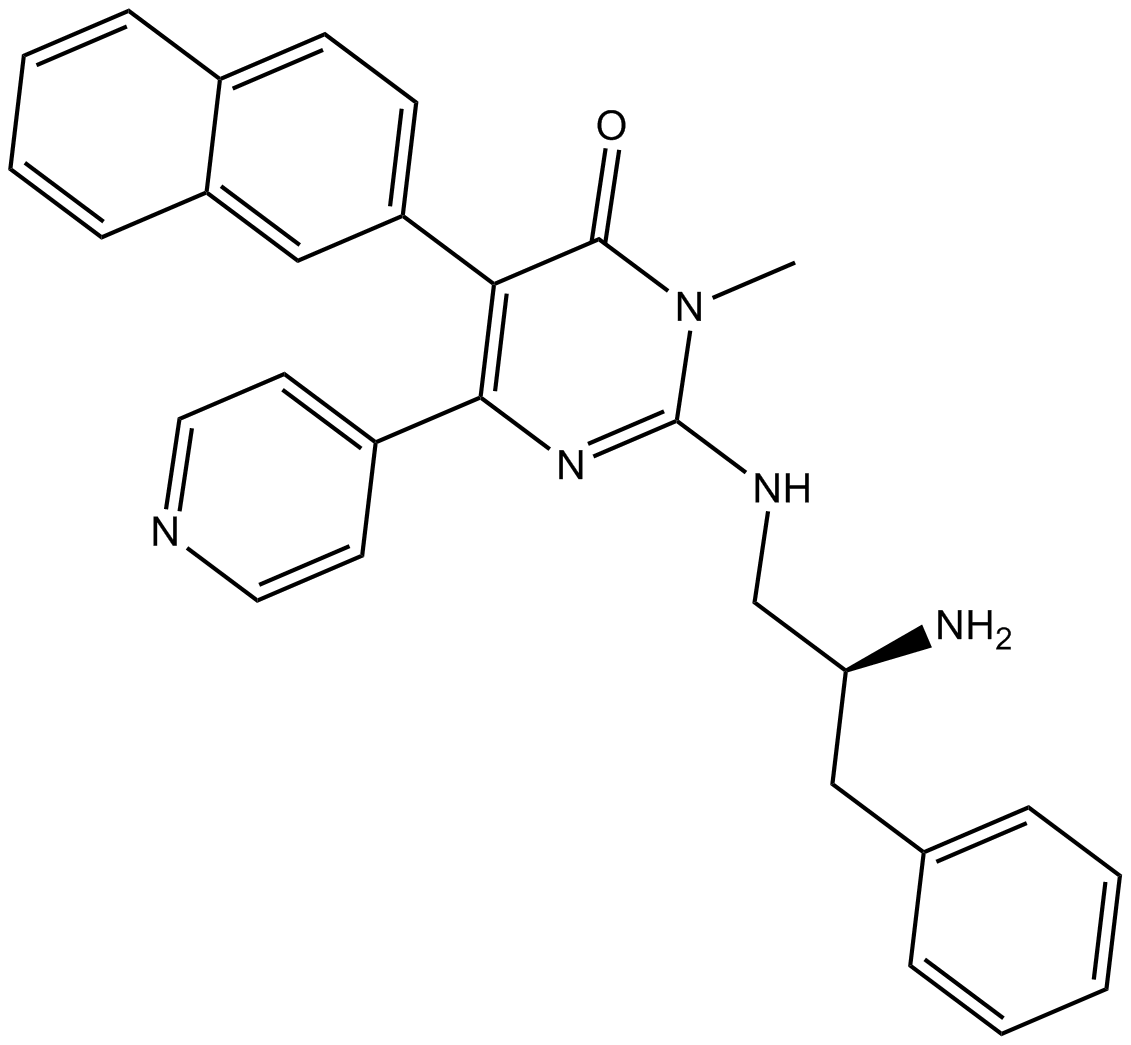
-
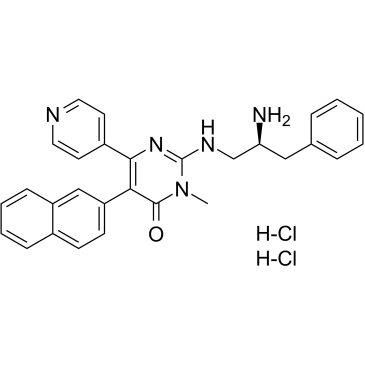
GC38518
AMG-548 dihydrochloride
AMG-548 dihydrochloride, an orally active and selective p38α inhibitor (Ki=0.5 nM), shows slightly selective over p38β (Ki=36 nM) and >1000 fold selective against p38γ and p38δ.
-
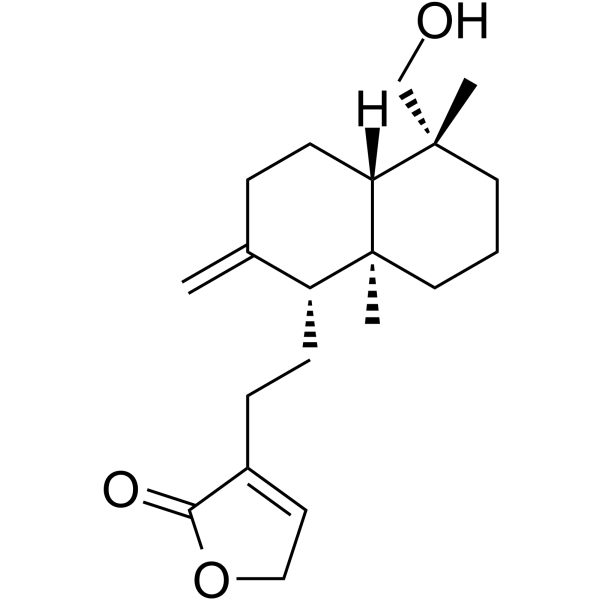
GC65917
Andrograpanin
Andrograpanin, a bioactive compound from Andrographis paniculata, exhibits anti-inflammatory and anti-infectious properties.
-
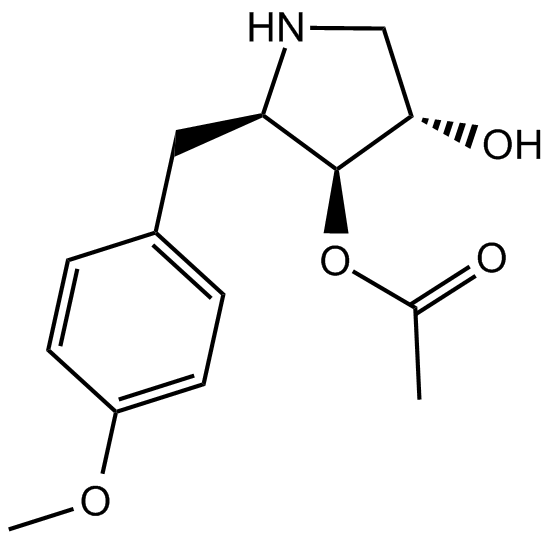
GC11559
Anisomycin
Flagecidin, NSC 76712, Wuningmeisu C
JNK agonist, potent and specific
-
GC68671
Anti-inflammatory agent 35
Anti-inflammatory agent 35 (compound 5a27) is an orally effective curcumin analogue with anti-inflammatory activity. It can block the signaling of mitogen-activated protein kinase (MAPK) and nuclear translocation of NF-kB. Anti-inflammatory agent 35 also inhibits neutrophil infiltration and proinflammatory cytokine production in vitro. In vivo studies have shown that Anti-inflammatory agent 35 significantly alleviates acute lung injury (ALI) induced by lipopolysaccharide (LPS).

-
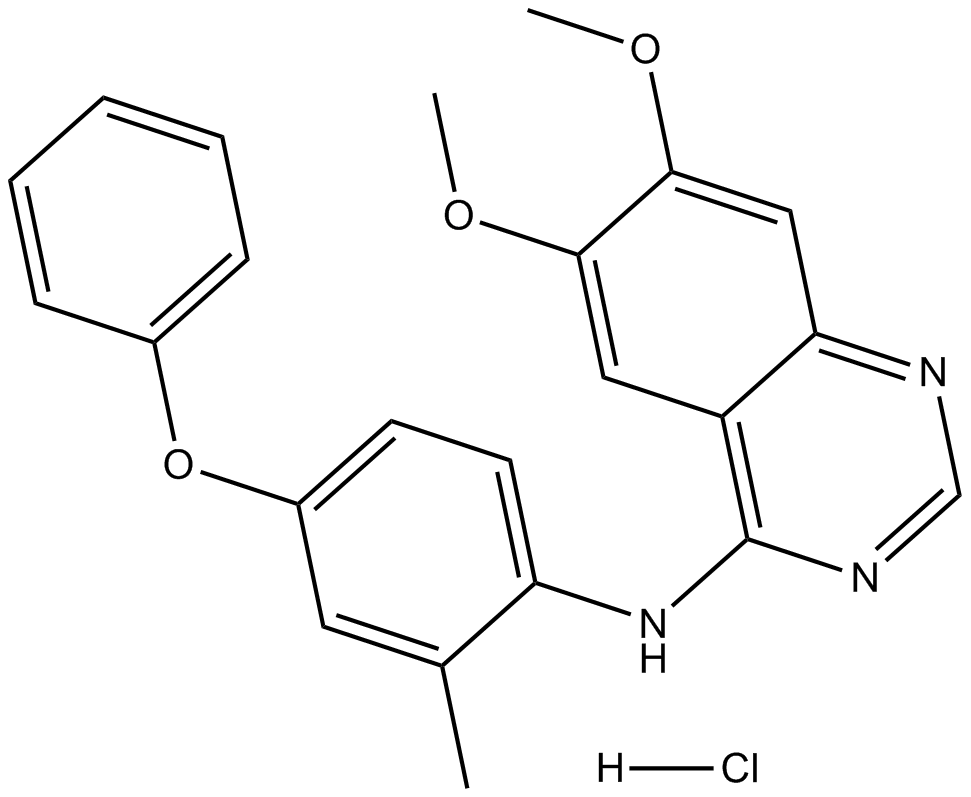
GC15240
APS-2-79
APS-2-79 is a KSR-dependent MEK antagonist. APS-2-79 inhibits ATPbiotin binding to KSR2 within the KSR2-MEK1 complexe with an IC50 of 120 nM. APS-2-79 makes the stabilization of the KSR inactive state antagonizes oncogenic Ras-MAPK signaling.
-
GC32692
APTO-253 (LOR-253)
APTO-253
APTO-253 (LOR-253) (LOR-253) is a small molecule that inhibits c-Myc expression, stabilizes G-quadruplex DNA, and induces cell cycle arrest and apoptosis in acute myeloid leukemia cells.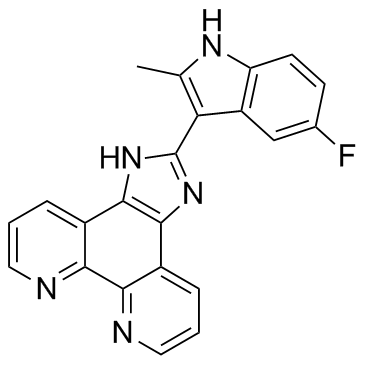
-
GN10063
Arctigenin
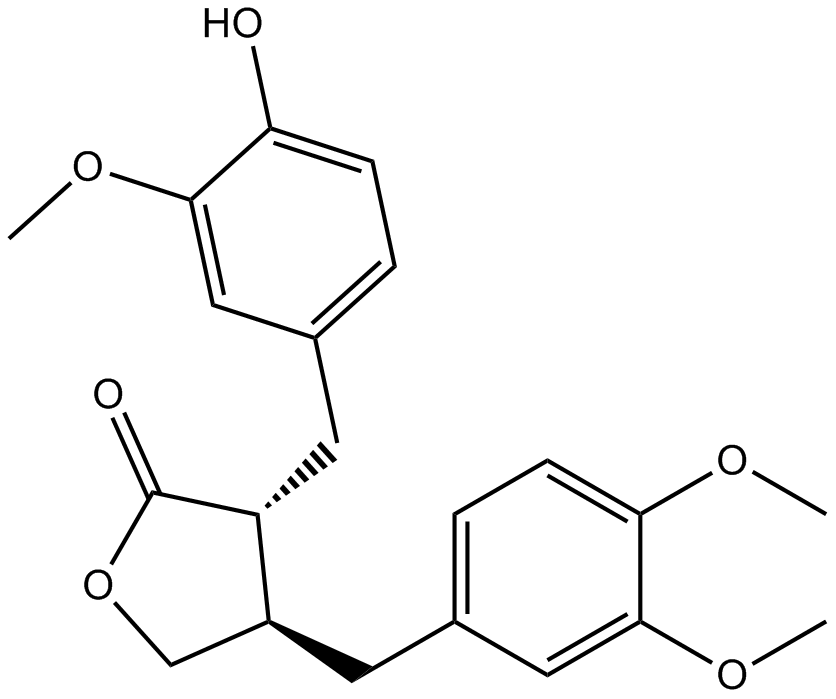
-
GC12882
AS 602801
AS-602801
A selective, orally bioavailable JNK inhibitor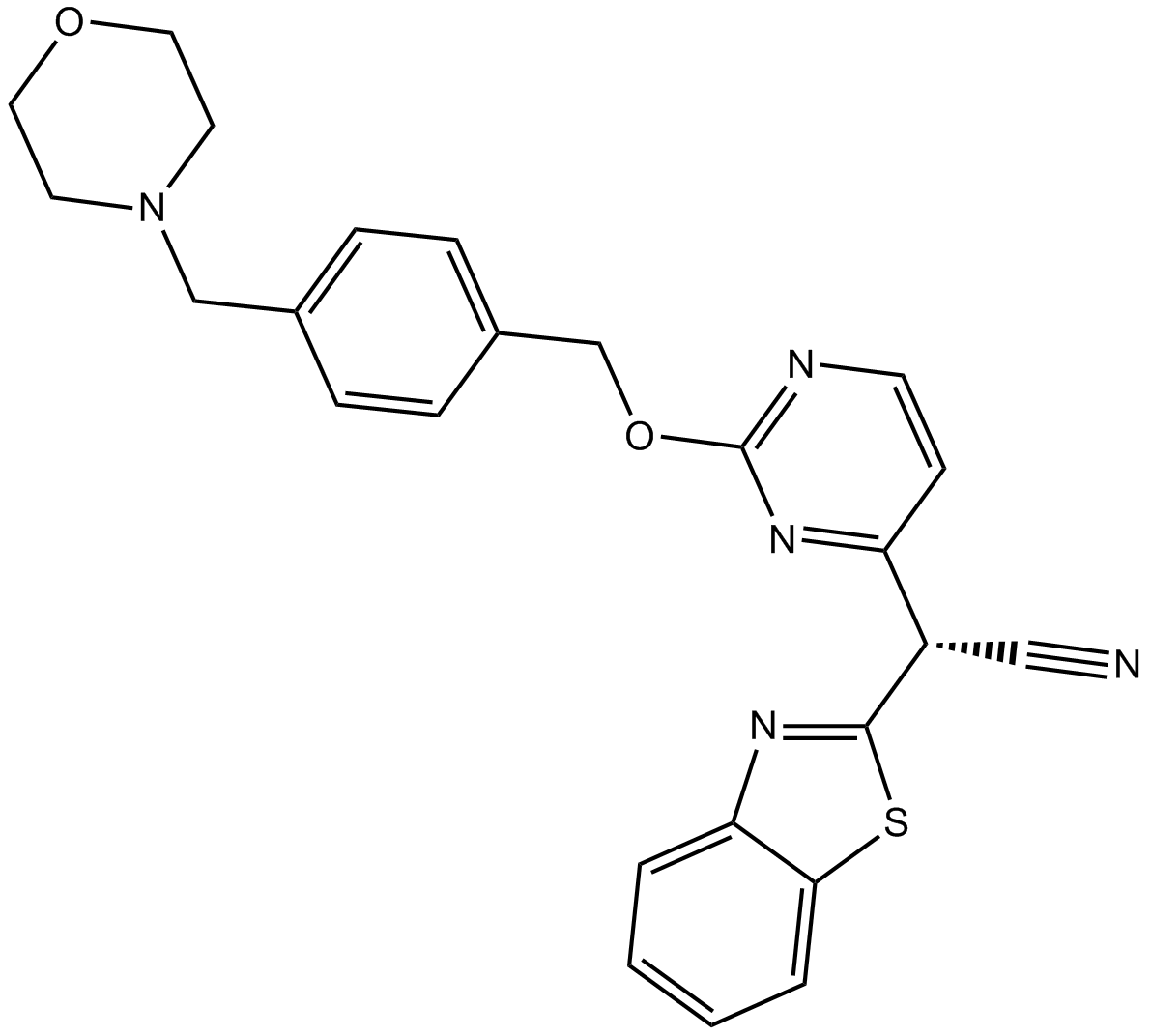
-
GC10010
AS601245
AS-601245, c-Jun N-terminal Kinase Inhibitor V
A c-Jun NH2-terminal protein kinase inhibitor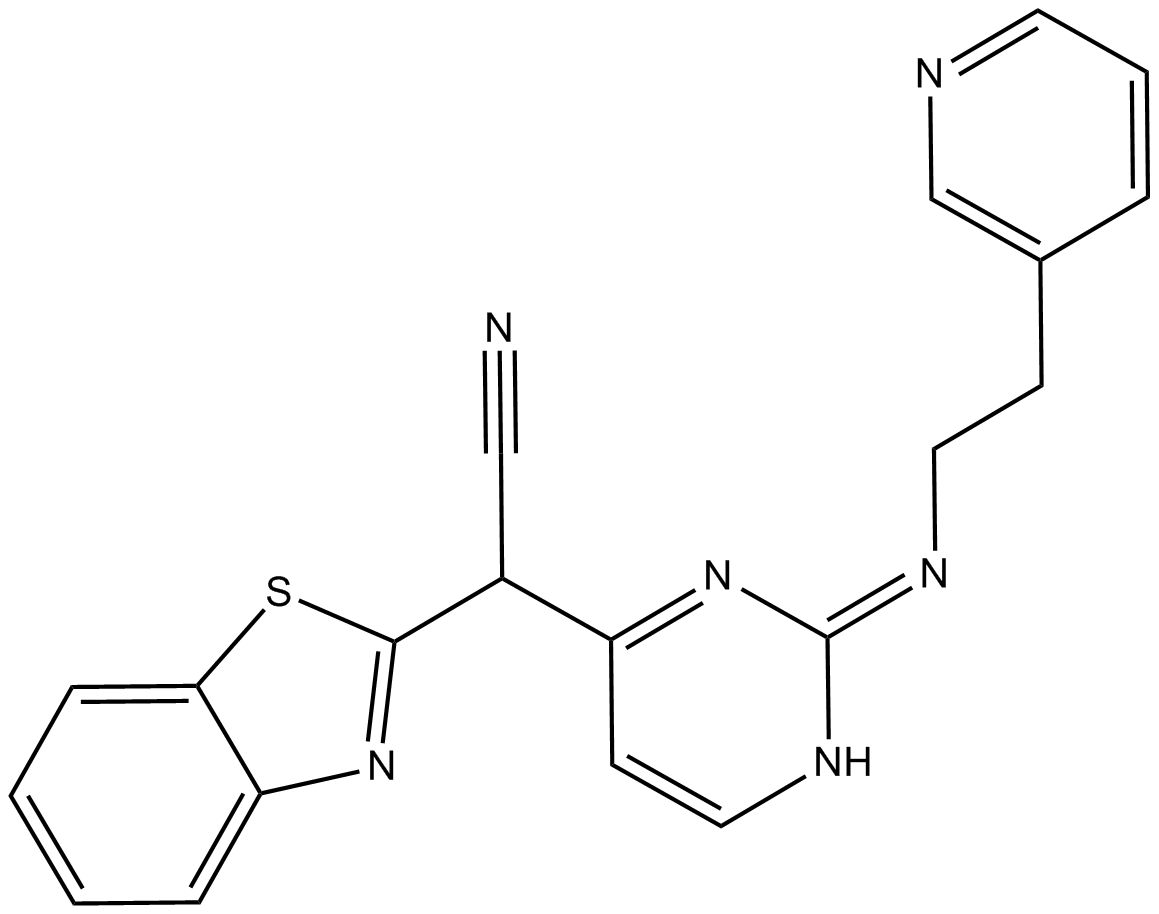
-
GC19041
ASK1-IN-1
ASK1-IN-1 is a potent, orally available and selective ATP-competitive inhibitor of apoptosis signal-regulating kinase 1 (ASK1) with an IC50 of 2.87 nM.
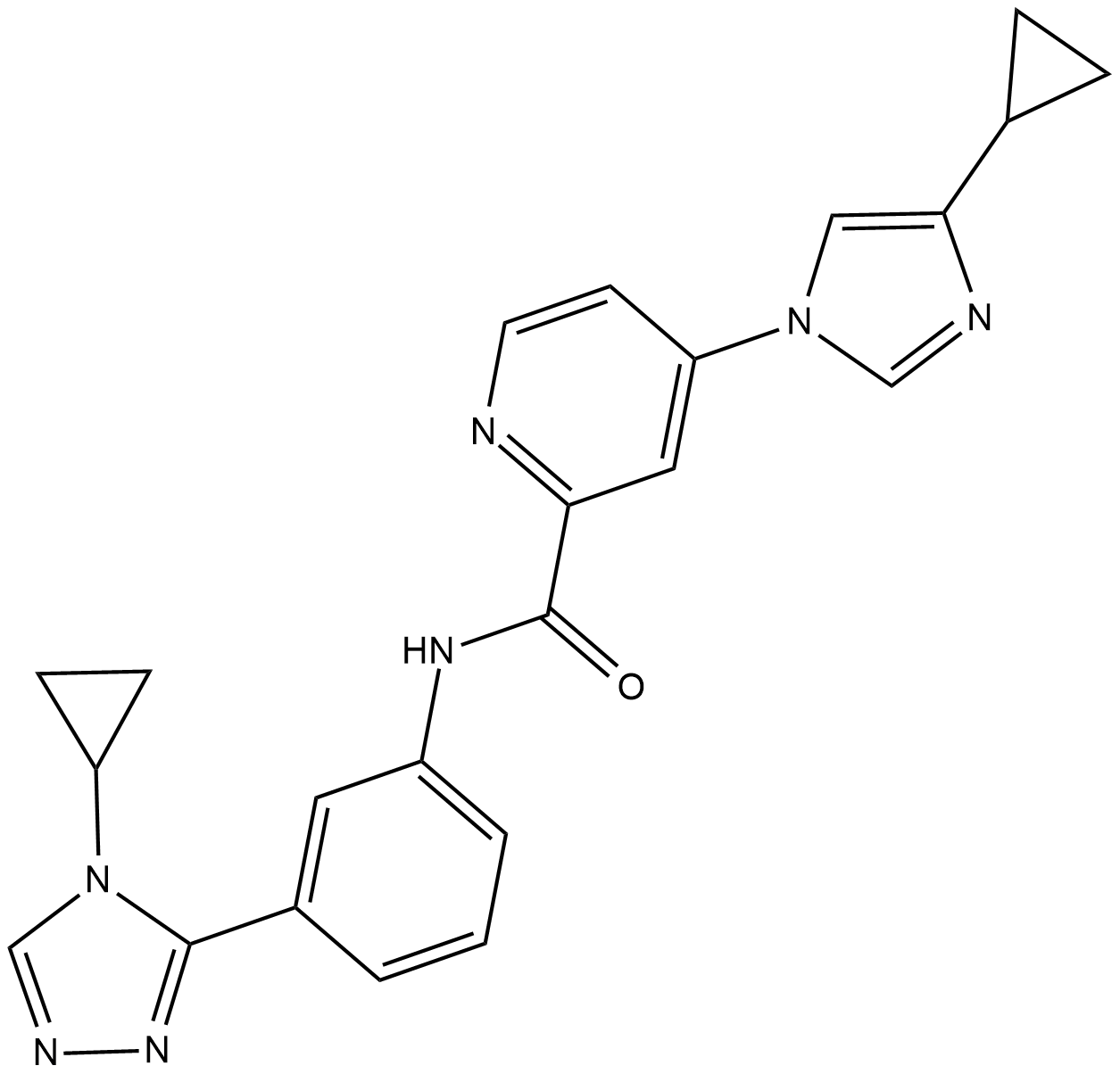
-
GC62426
ASK1-IN-2
ASK1-IN-2 is a potent and orally active inhibitor of apoptosis signal-regulating kinase 1 (ASK1), with an IC50 of 32.8 nM.
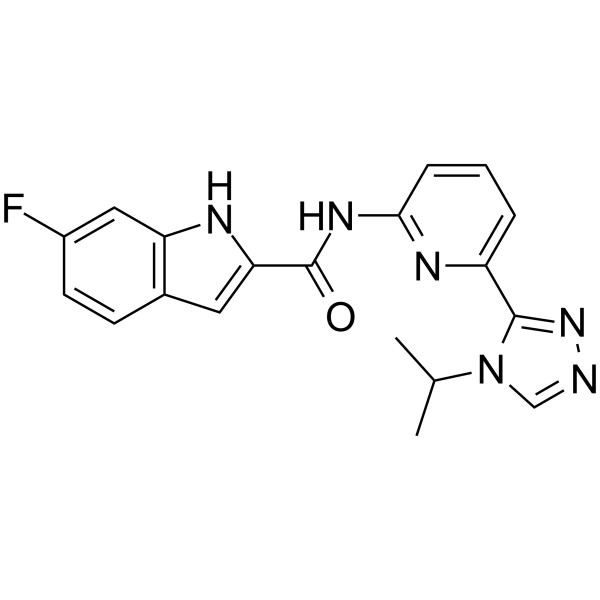
-
GC35412
Asperulosidic Acid
Asperulosidic Acid (ASPA), a bioactive iridoid glycoside, is extracted from the herbs of Hedyotis diffusa Willd.
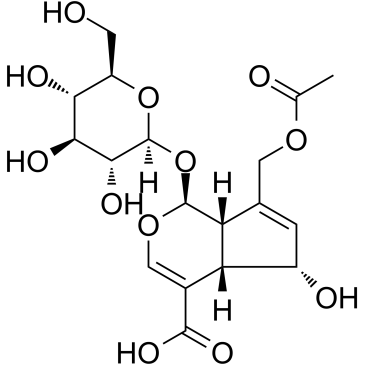
-
GN10494
Astragaloside IV
AS-IV, AST-IV
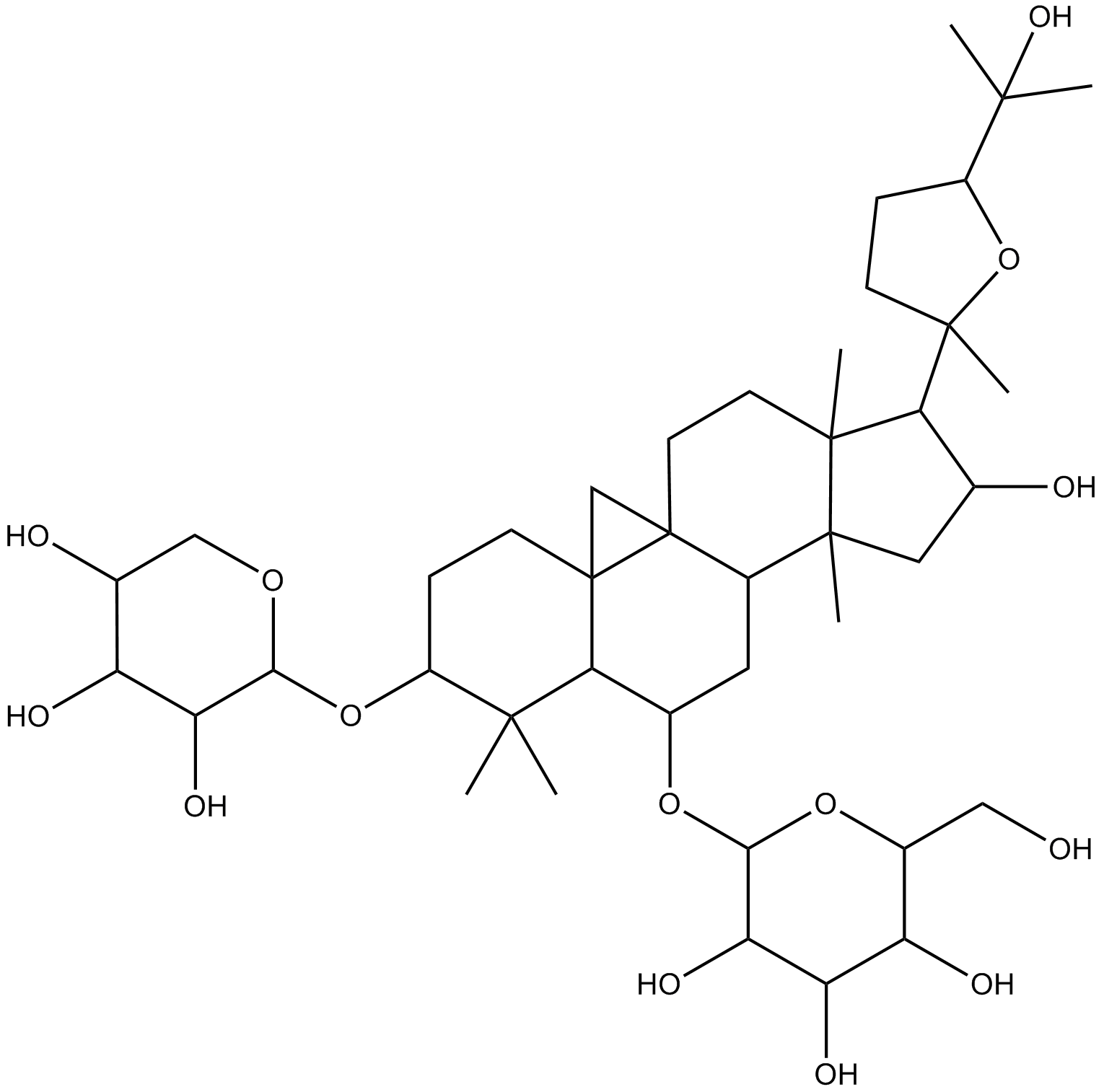
-
GC17712
AT13148
Multi-AGC kinase inhibitor,ATP-competitive
-
GC12297
AT7867
Akt1/2/3 and p70S6K/PKA inhibitor
-
GC10918
AT7867 dihydrochloride
A potent and orally bioavailable pan-Akt inhibitor
-
GC31667
AX-15836
AX-15836 is a potent and selective ERK5 inhibitor with an IC50 of 8 nM.
-
GC15055
AZ 628
AZ-628; AZ628
Raf kinases,potent and ATP-competitive

-
GC19479
AZ304
A dual inhibitor

-
GC19048
AZD-0364
AZD0364
AZD-0364 (AZD0364) is a potent and selective ERK2 inhibitor extracted from patent WO2017080979A1, compound example 18, has an IC50 of 0.6 nM.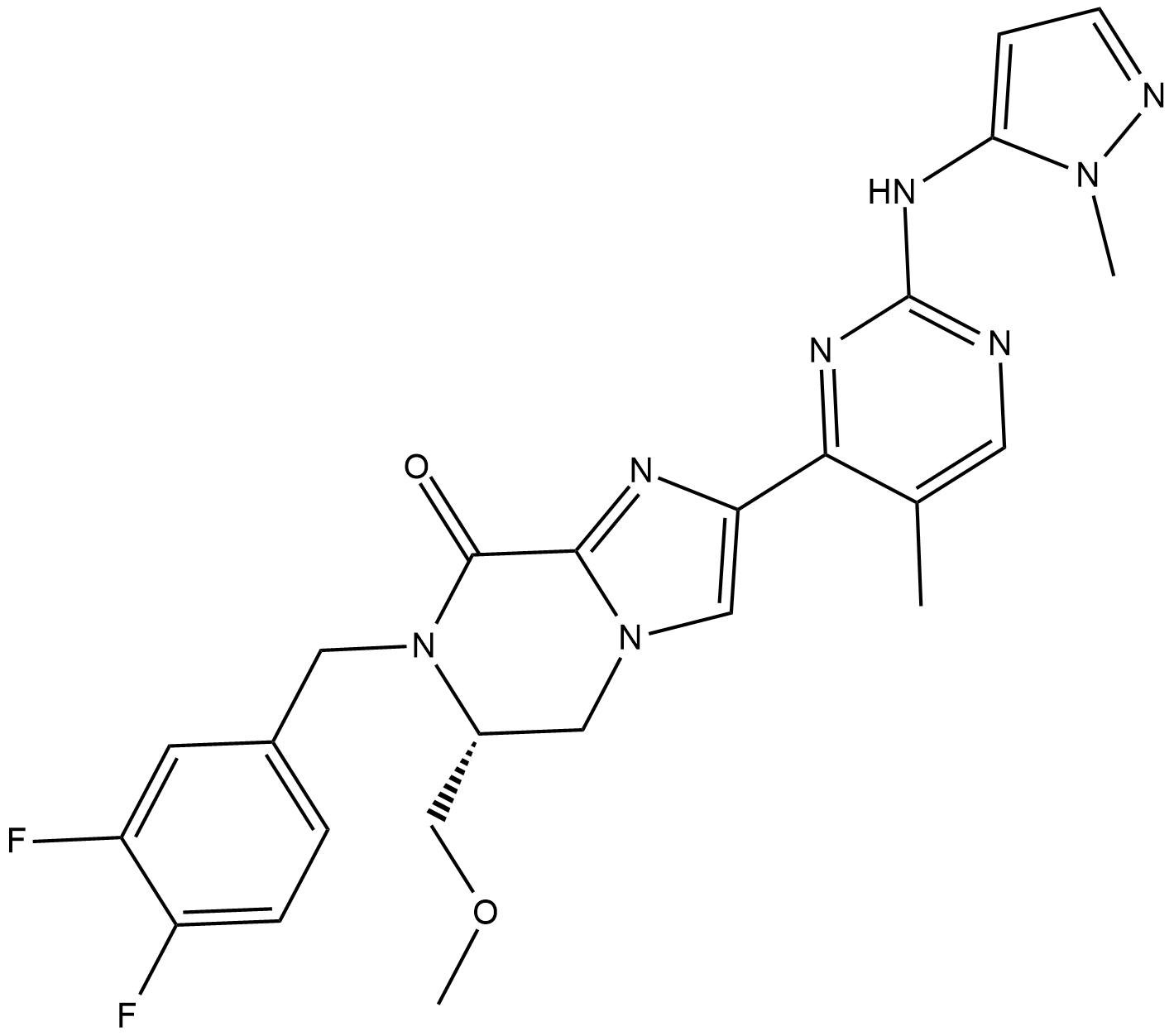
-
GC17030
AZD6244(Selumetinib)
AZD6244; ARRY-142886
A highly selective inhibitor of MEK1/2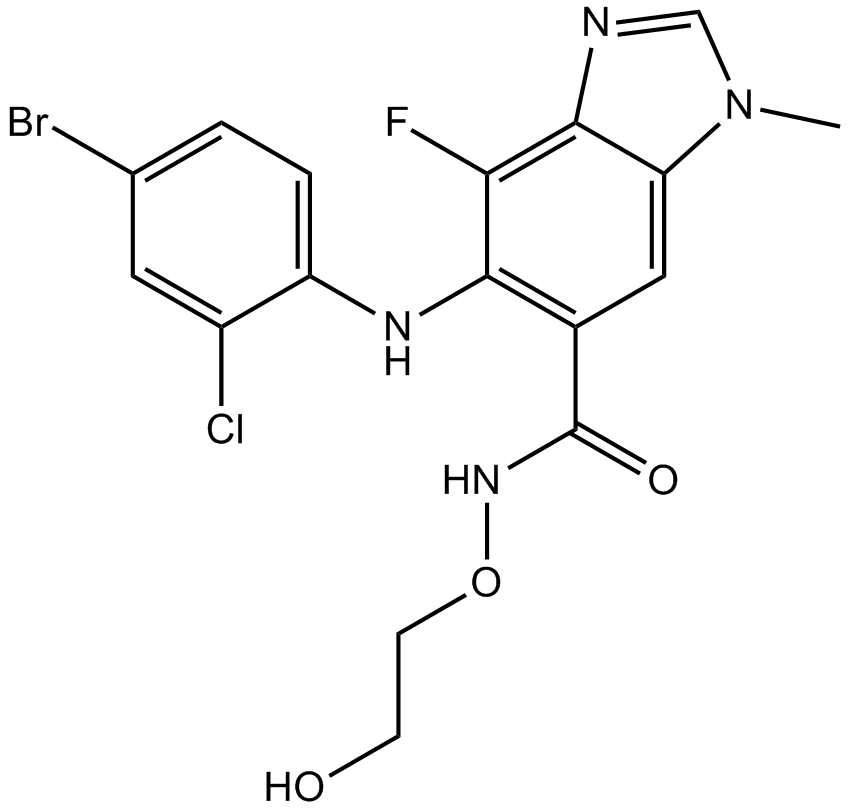
-
GC31924
AZD7624
AZD7624 is an inhaled p38 inhibitor, with potent anti-inflammatory activity.
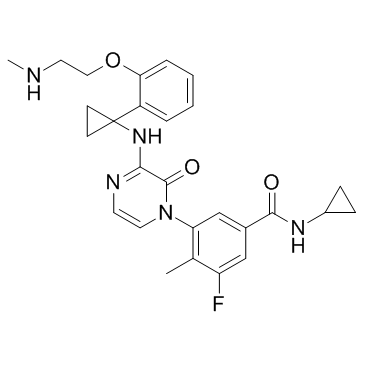
-
GC14643
AZD8330
ARRY-424704; ARRY-704; AZD-8330; ARRY424704; ARRY704; AZD8330
MEK 1/2 inhibitor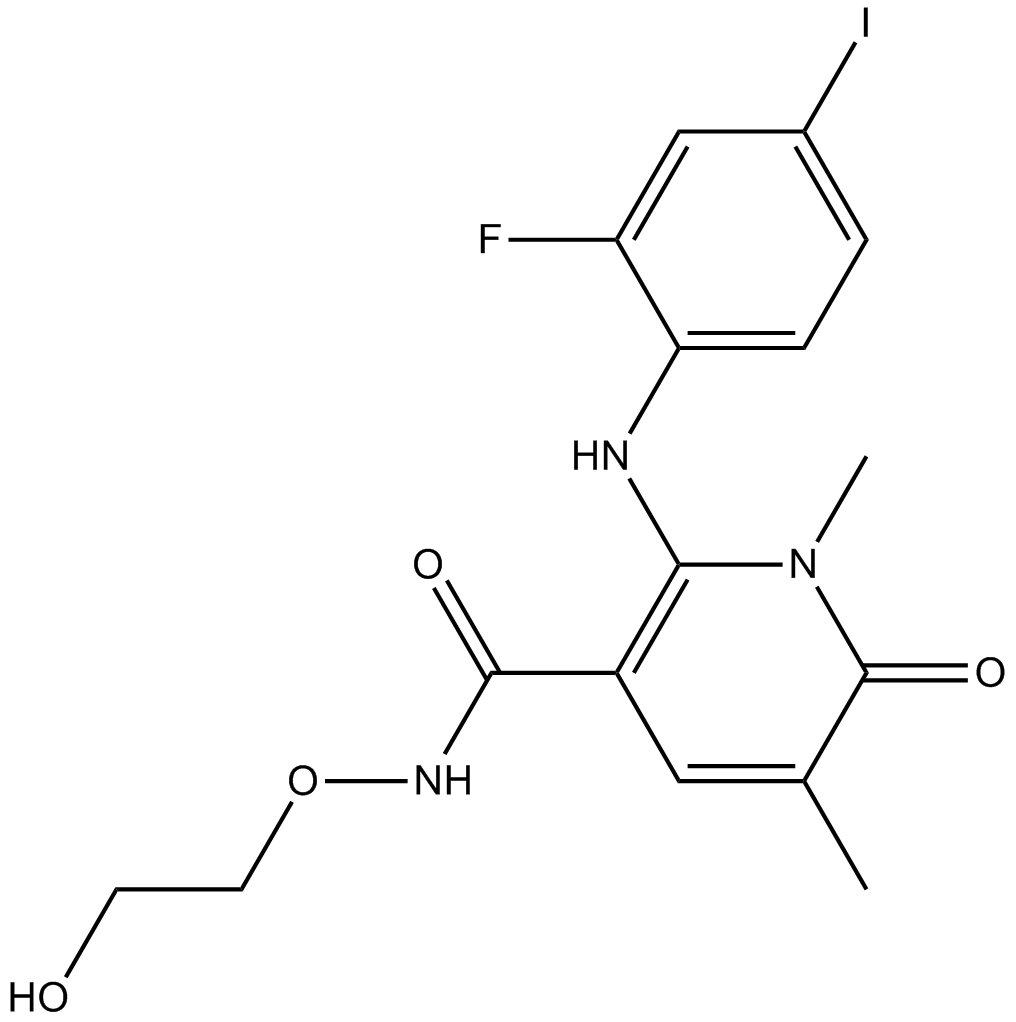
-
GC38738
Azosemide
Azosemide, a sulfonamide loop diuretic, is a potent NKCC1 inhibitor with IC50s of 0.246??M and 0.197??M for hNKCC1A and NKCC1B, respectively.
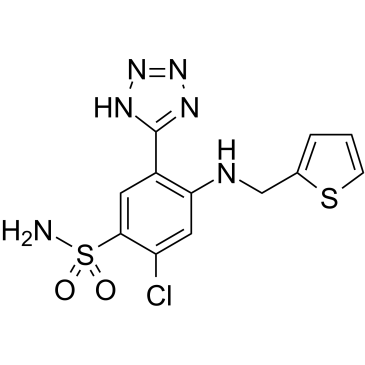
-
GC10534
B-Raf IN 1
B-RAF Inhibitor 1
B-Raf inhibitor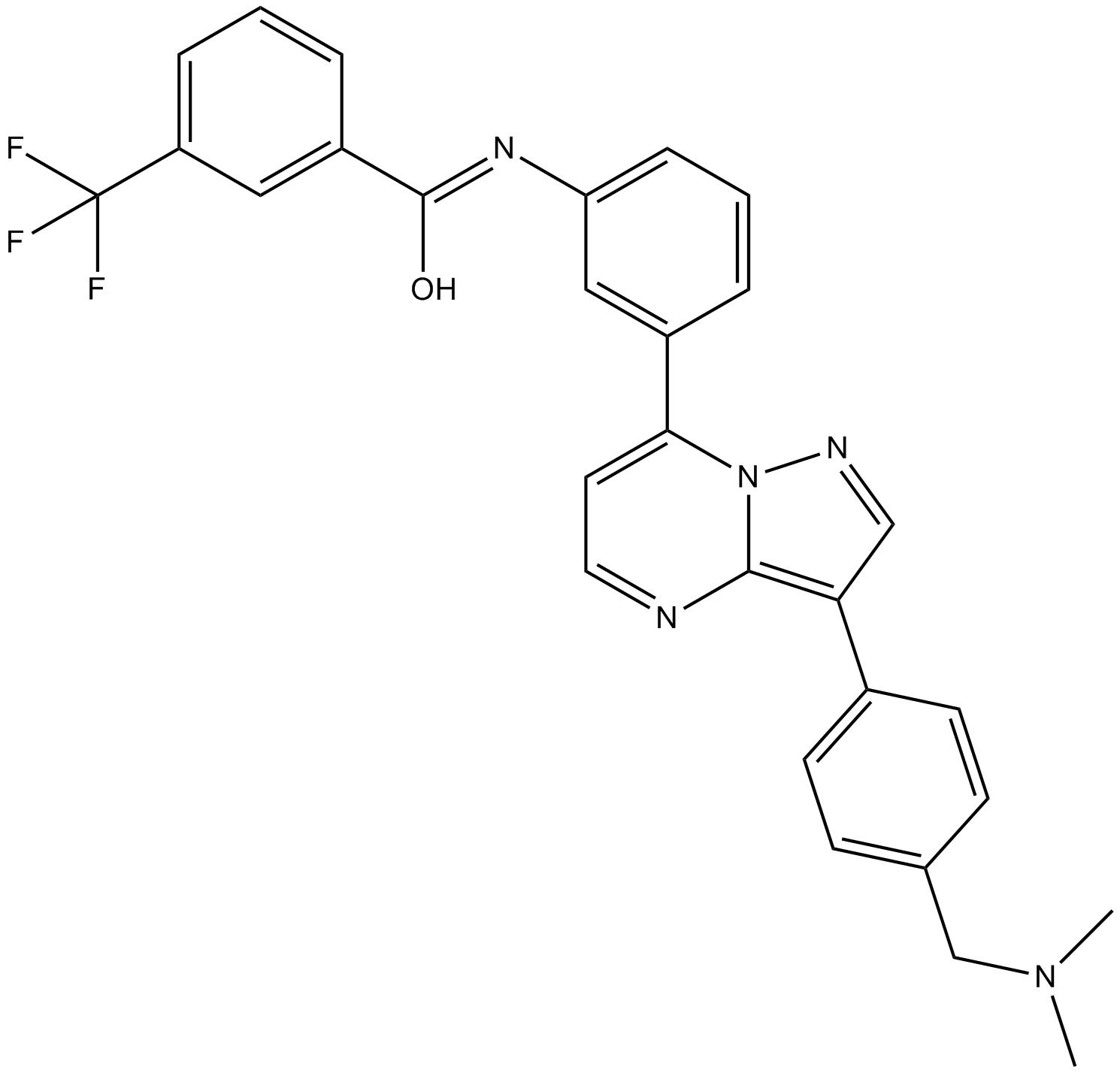
-
GC65978
B-Raf IN 10
B-Raf IN 10 (Compound C09) is a BRAF inhibitor with an IC50 between 50 and 100 nM. B-Raf IN 10 shows antitumor activity.
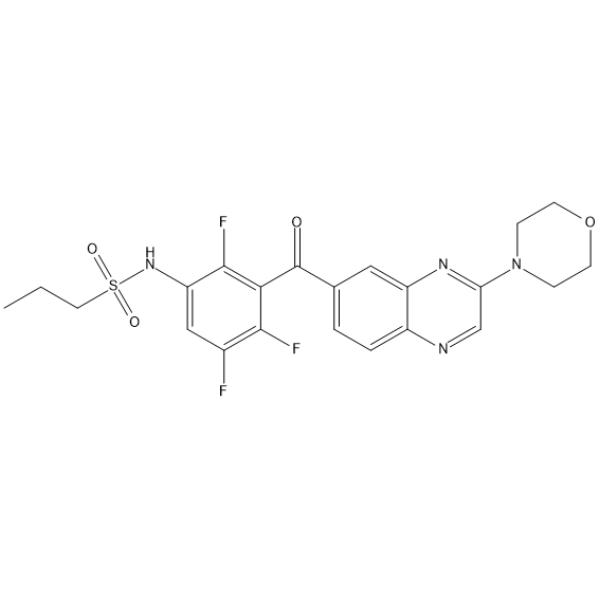
-
GC67800
B-Raf IN 11
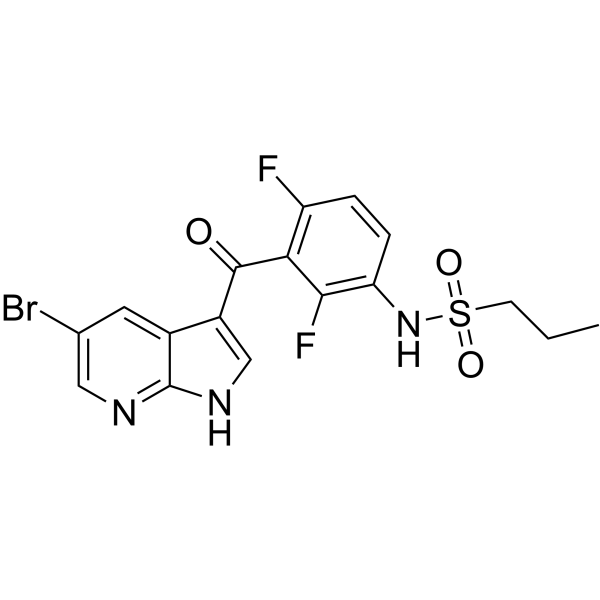
-
GC64550
B-Raf IN 2
B-Raf IN 2 is a potent and selective BRAF inhibitor extracted from patent WO2021116055A1, compound Ia. B-Raf IN 2 can be used for the research of cancer.
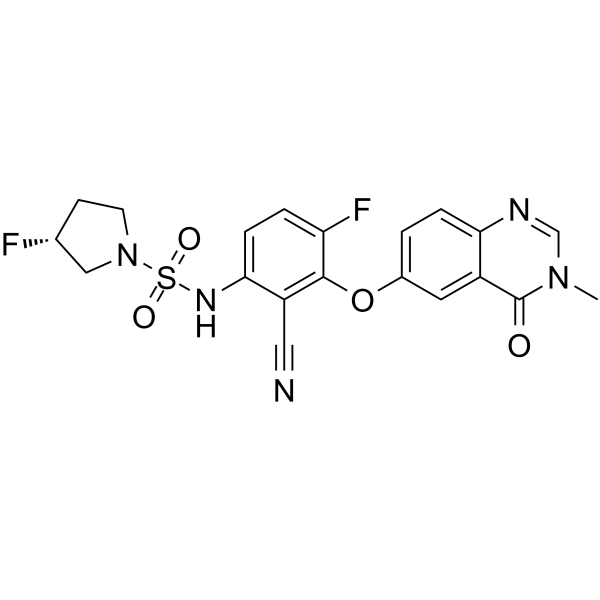
-
GC12151
B-Raf inhibitor
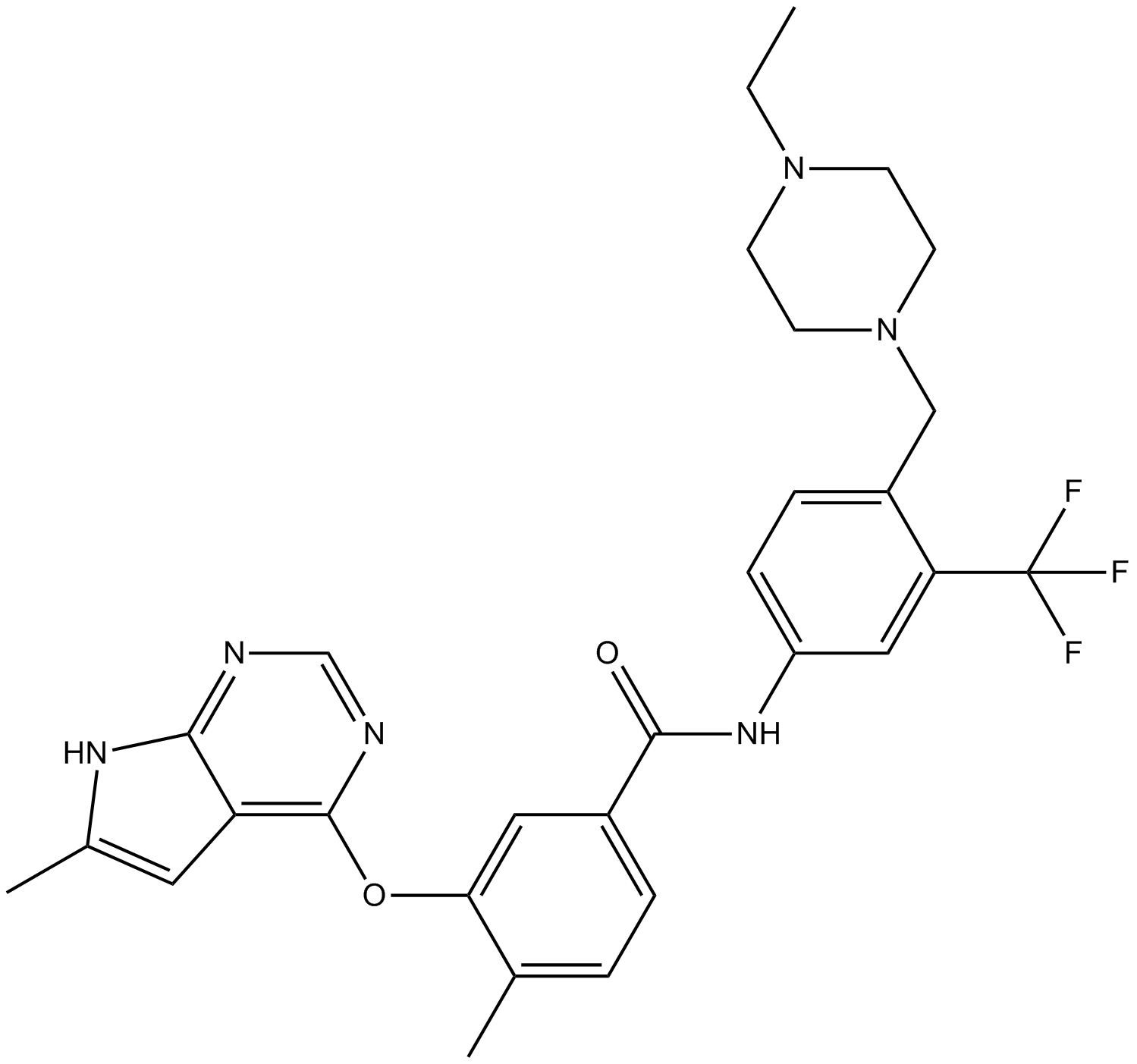
-
GC11599
B-Raf inhibitor 1
B-Raf inhibitor 1 is a potent Raf kinase inhibitor with Kis of 1 nM, 1 nM, and 0.3 nM for B-RafWT, B-RafV600E, and C-Raf, respectively.
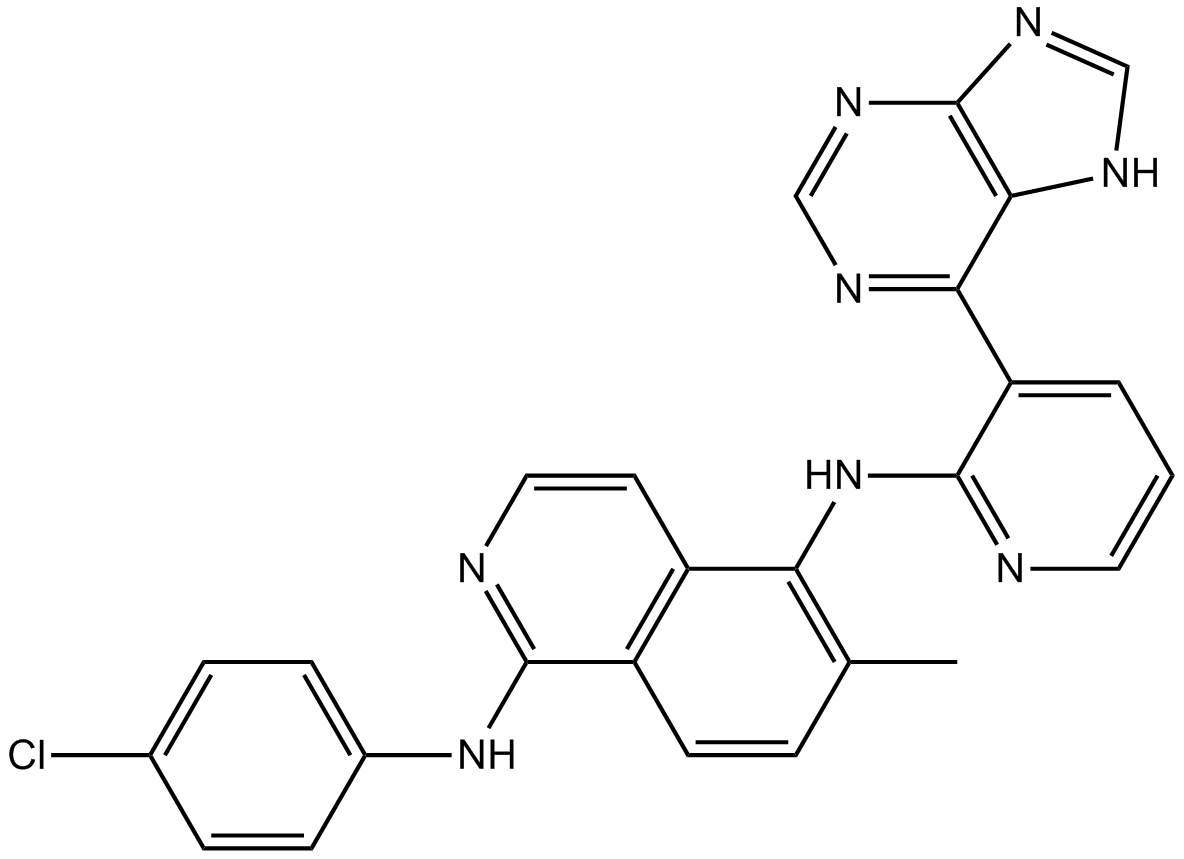
-
GC14907
B-Raf inhibitor 1 dihydrochloride
B-B-Raf inhibitor 1 dihydrochloride is a potent Raf kinase inhibitor with Kis of 1 nM, 1 nM, and 0.3 nM for B-RafWT, B-RafV600E, and C-Raf, respectively.
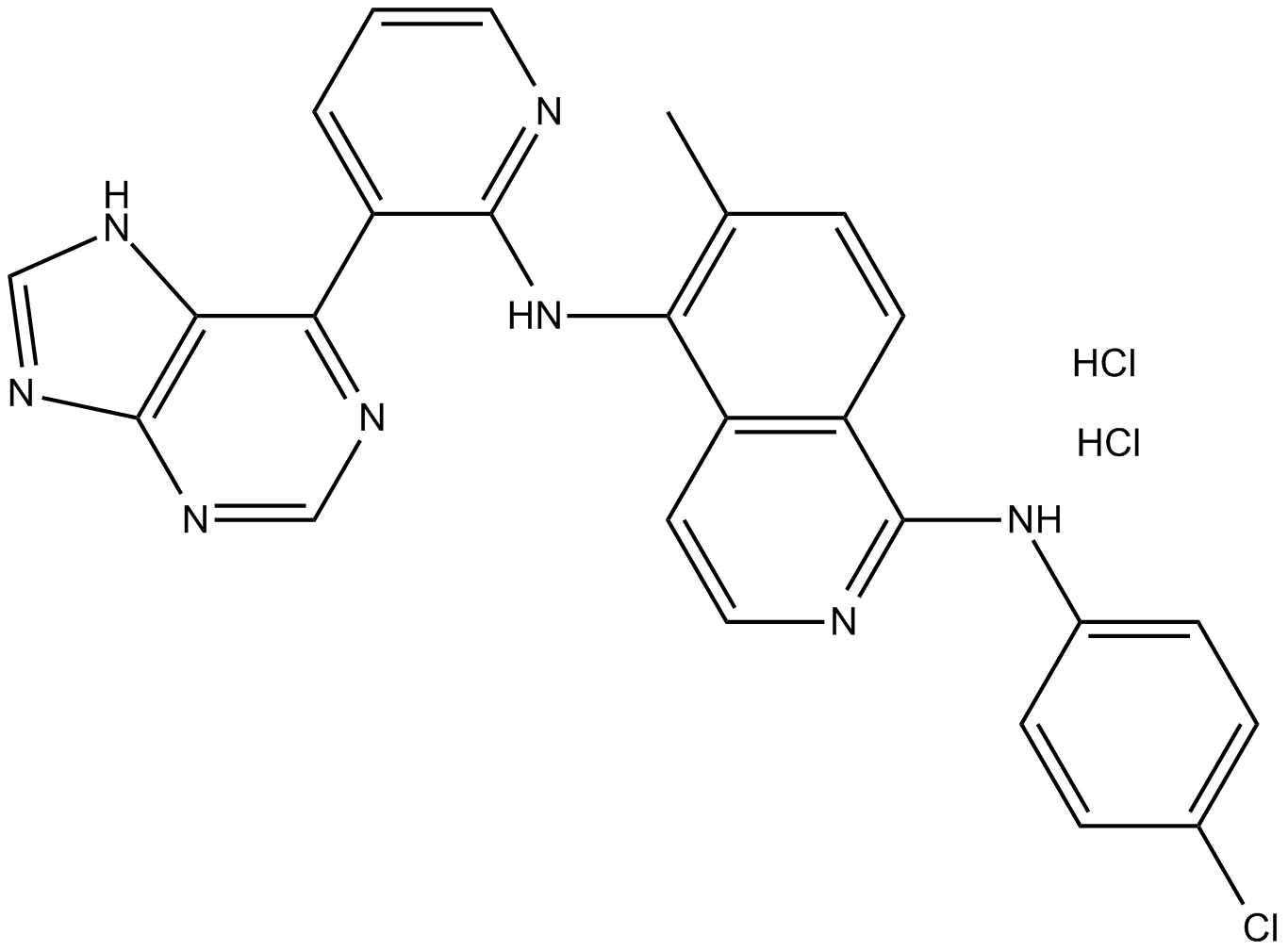
-
GN10590
Bakuchiol
(S)-(+)-Bakuchiol
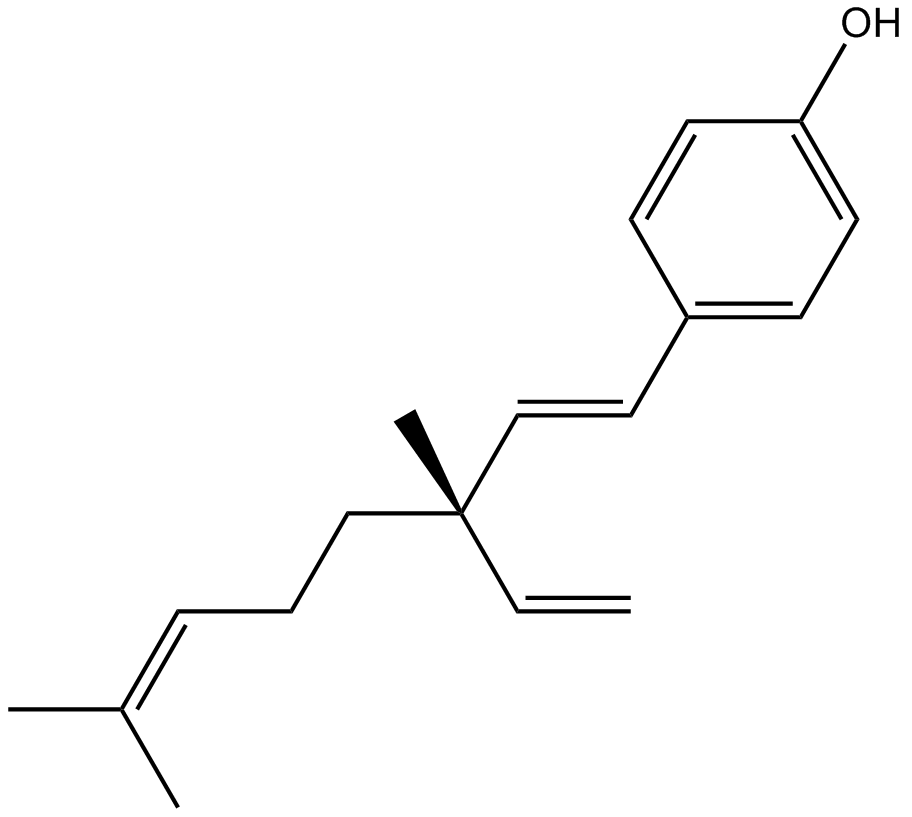
-
GC33305
Balamapimod (MKI 833)
MKI 833
Balamapimod (MKI 833) (MKI 833) is a reversible Ras/Raf/MEK inhibitor with potential anti-tumor activity.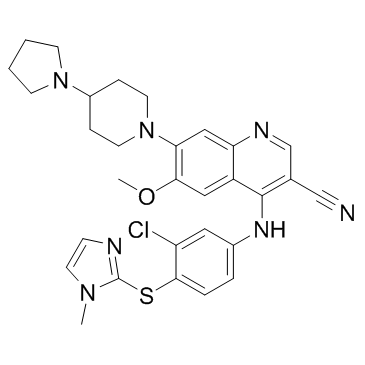
-
GC34342
BAY885
An ERK5 inhibitor
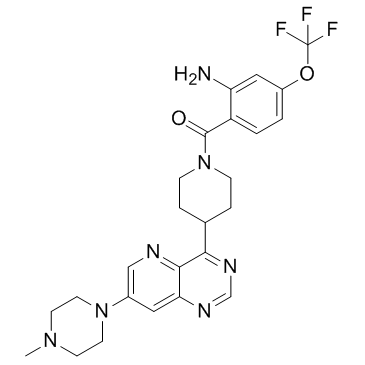
-
GC32950
Belvarafenib
GDC-5573, HM95573
An inhibitor of B-RAF and C-RAF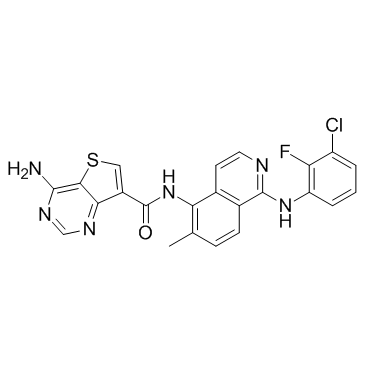
-
GC35488
Belvarafenib TFA
HM95573 TFA; GDC-5573 TFA; RG6185 TFA
Belvarafenib TFA (HM95573 TFA) is a potent and pan RAF (Rapidly Accelerated Fibrosarcoma) inhibitor, with IC50s of 56 nM, 7 nM and 5 nM for B-RAF, B-RAFv600E and C-RAF respectively.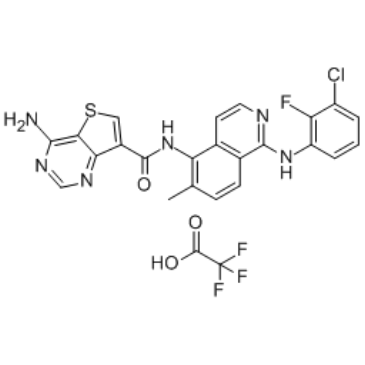
-
GC19066
BGB-283
BGB-283 is a novel and potent Raf Kinase and EGFR inhibitor with IC50 values of 23 and 29 nM for recombinant BRafV600E and EGFR, respectively.
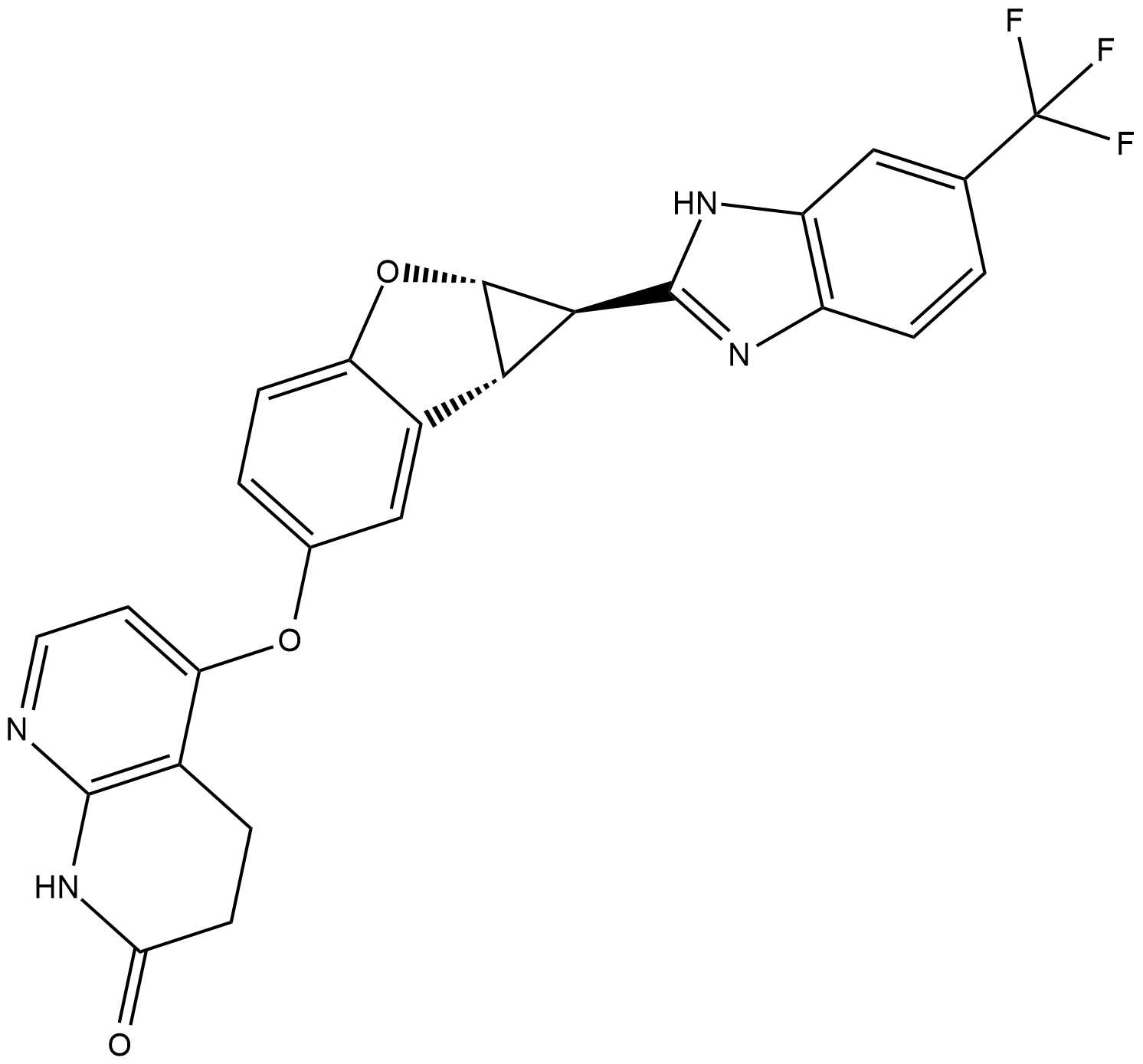
-
GC12904
BI 78D3
JNK Inhibitor X, c-Jun N-terminal Kinase Inhibitor X
Competitive JNK inhibitor
-
GC17828
BI-847325
dual inhibitor of MEK and Aurora kinases

-
GC18178
BI-882370
A RAF inhibitor
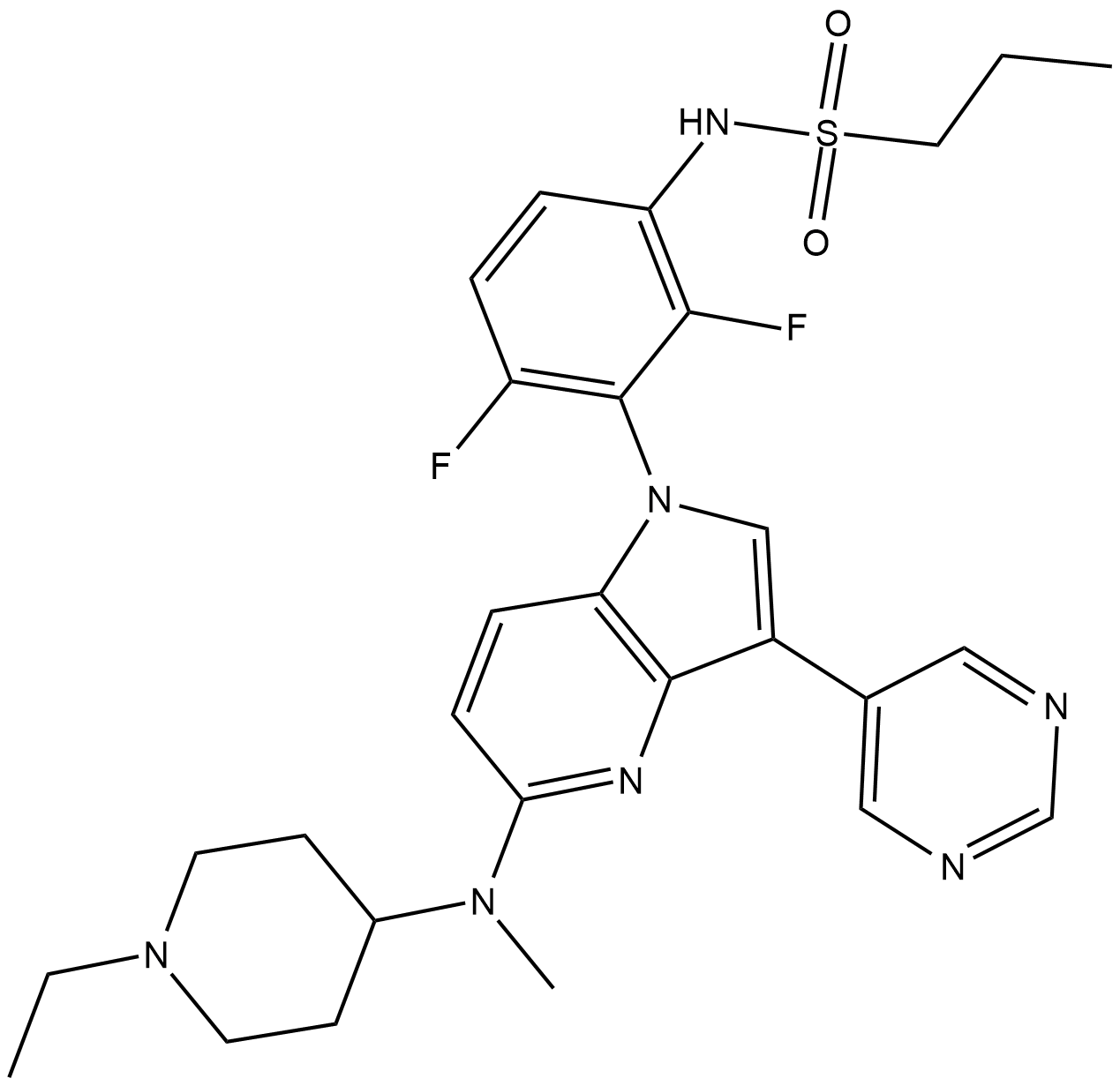
-
GC13468
BI-D1870
P90 RSK inhibitor,ATP-competitive and cell-permeable

-
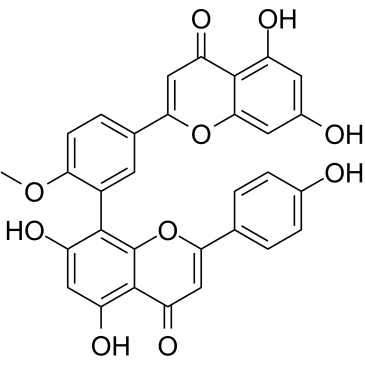
GC35518
Bilobetin
A biflavonoid with diverse biological activities
-
GC11497
BIRB 796 (Doramapimod)
BIRB796
BIRB 796 (Doramapimod) (BIRB 796) is an orally active, highly potent p38 MAPK inhibitor, which has an IC50 for p38α=38 nM, for p38β=65 nM, for p38γ=200 nM, and for p38δ=520 nM.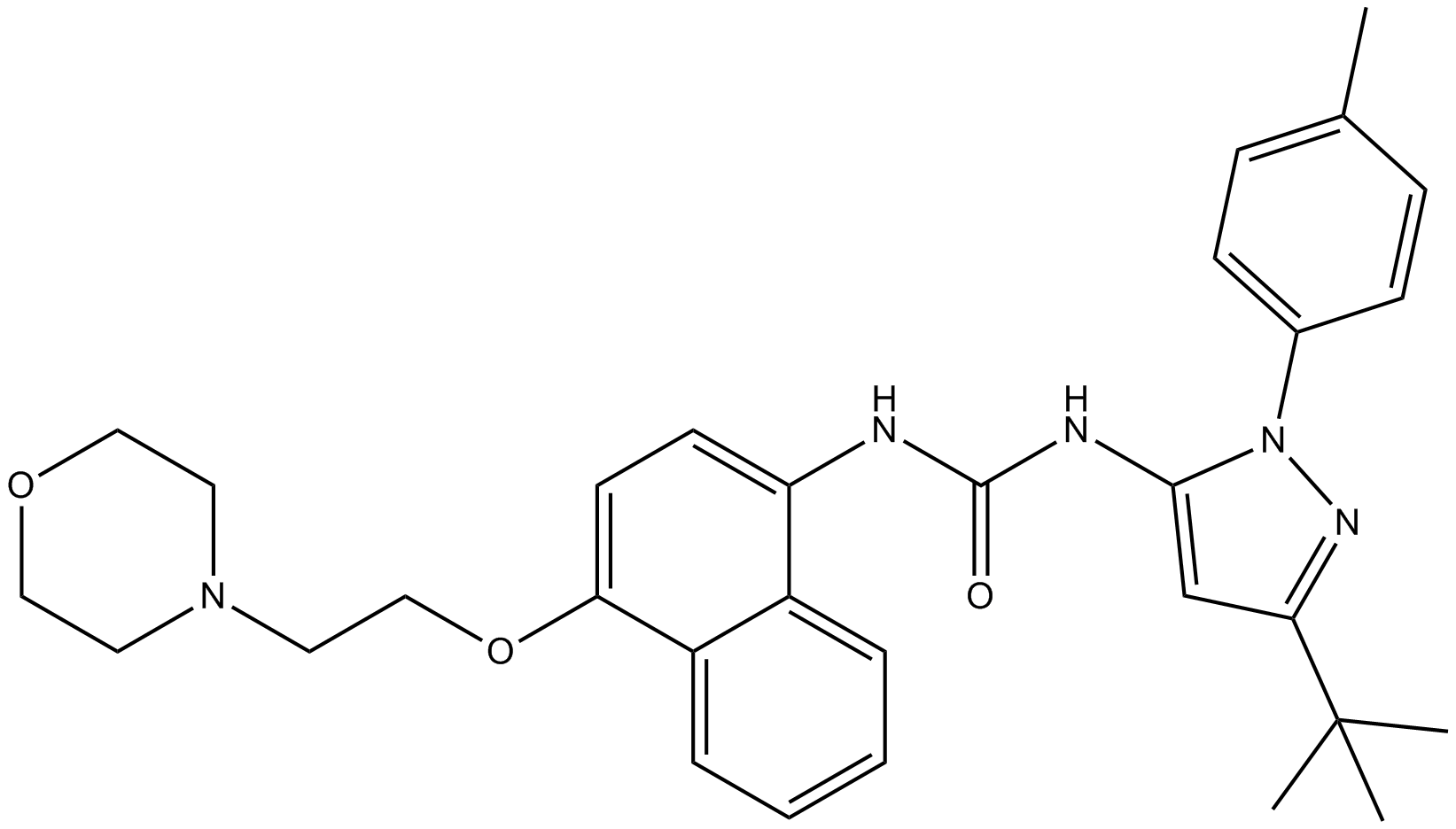
-
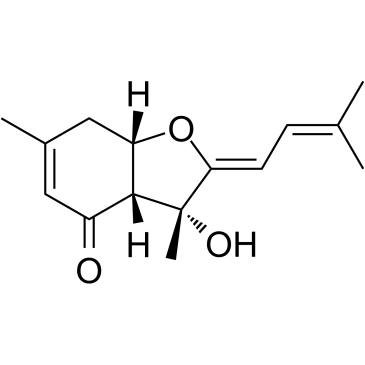
GC35525
Bisabolangelone
Bisabolangelone, a sesquiterpene derivative, is isolated from the roots of Osterici Radix.
-
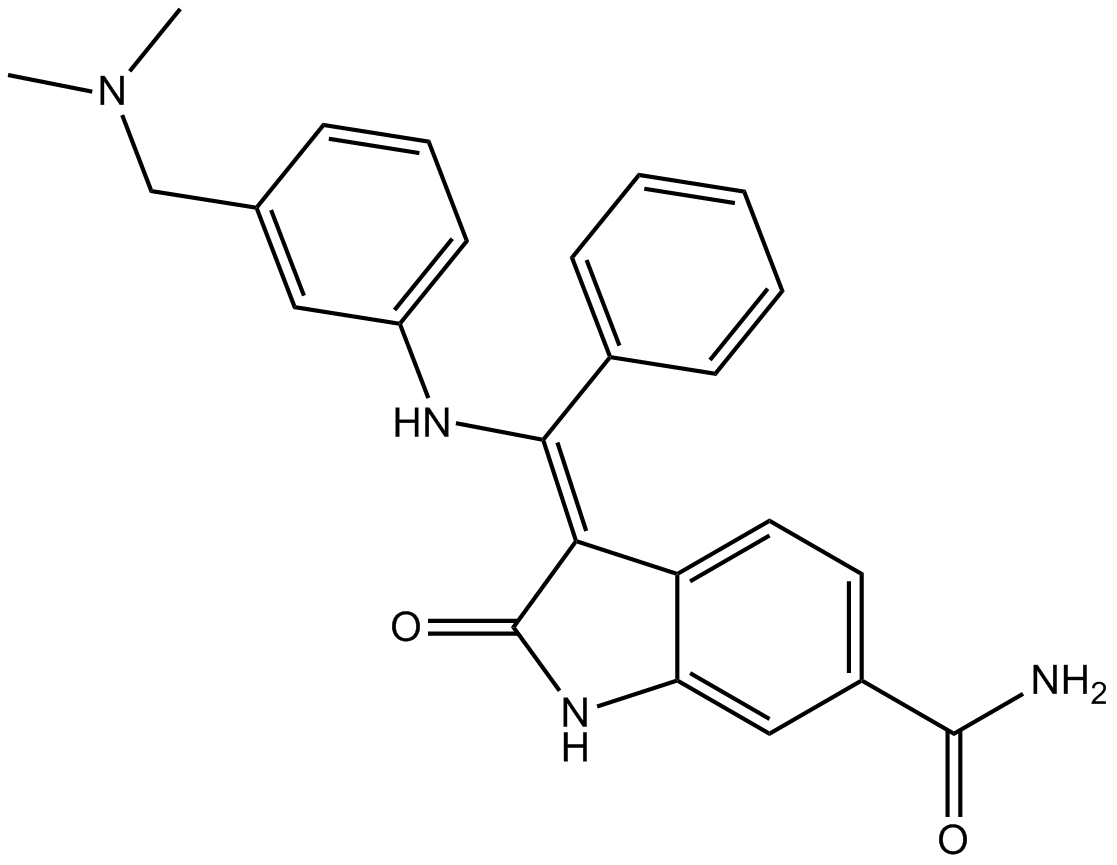
GC15693
BIX 02188
MEK5 inhibitor,potent and selective
-
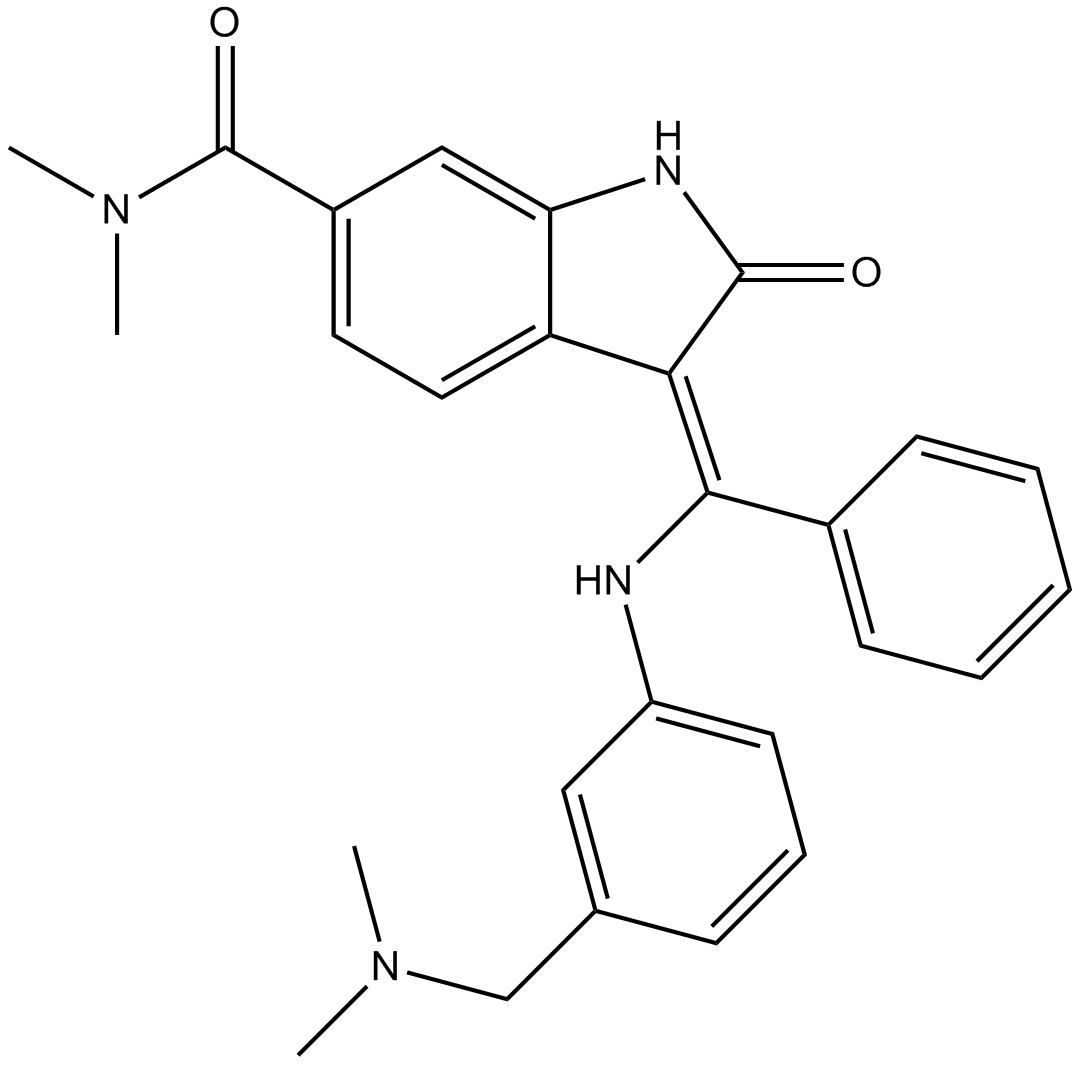
GC12220
BIX 02189
Selective MEK5 inhibitor
-
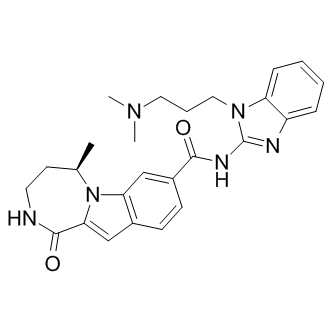
GC12982
BIX 02565
RSK2 inhibitor
-
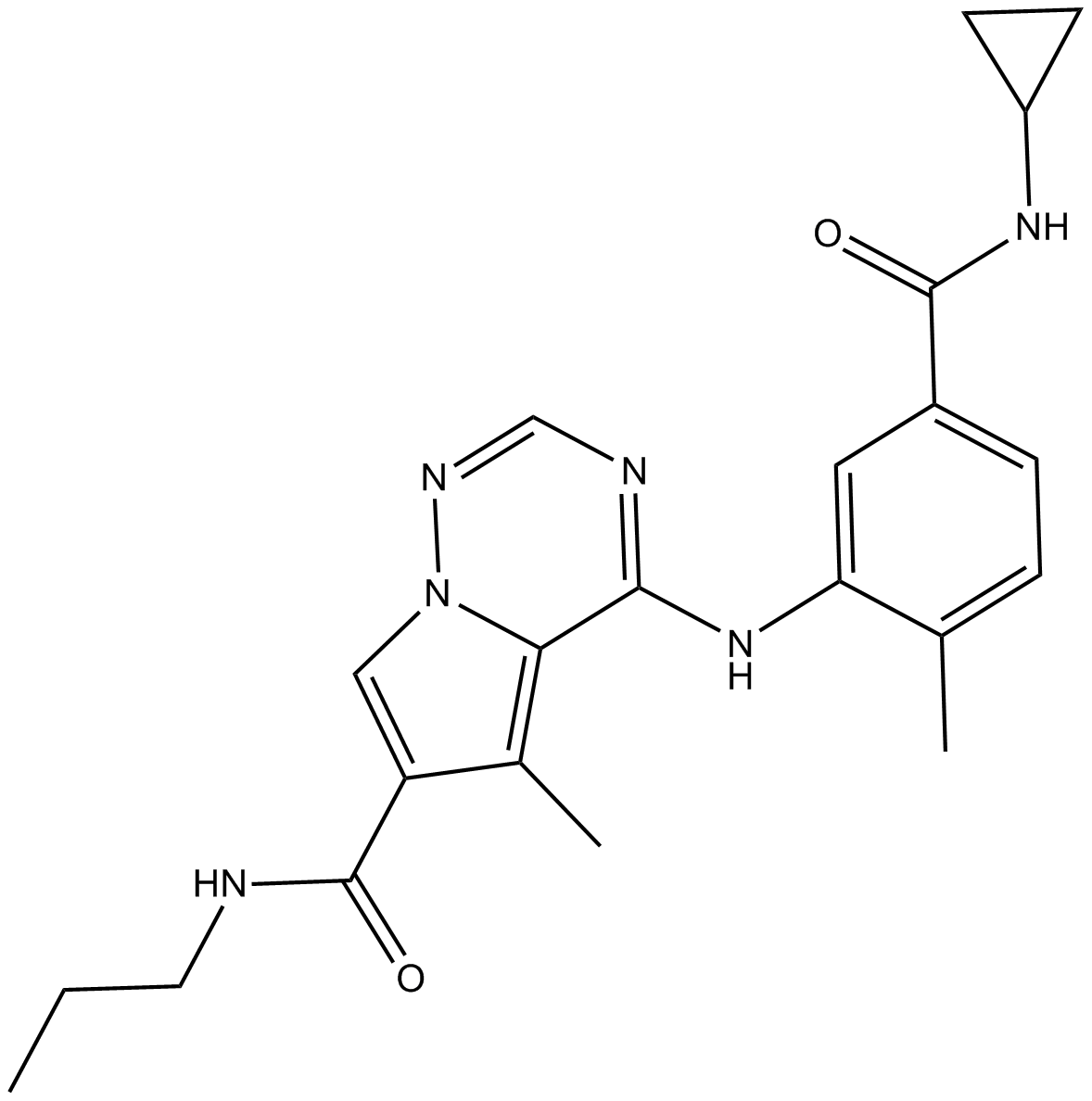
GC16997
BMS-582949
p38 MAPK inhibitor
-
GC10593
BMS-582949 hydrochloride
p38 MAPK inhibitor

-
GC42968
BPIQ-II (hydrochloride)
PD 158294
BPIQ-II is a linear imidazoloquinazoline that potently inhibits the tyrosine kinase activity of the epidermal growth factor receptor (EGFR; IC50 = 8 pM).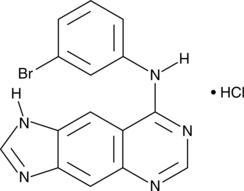
-
GC12482
BRAF inhibitor
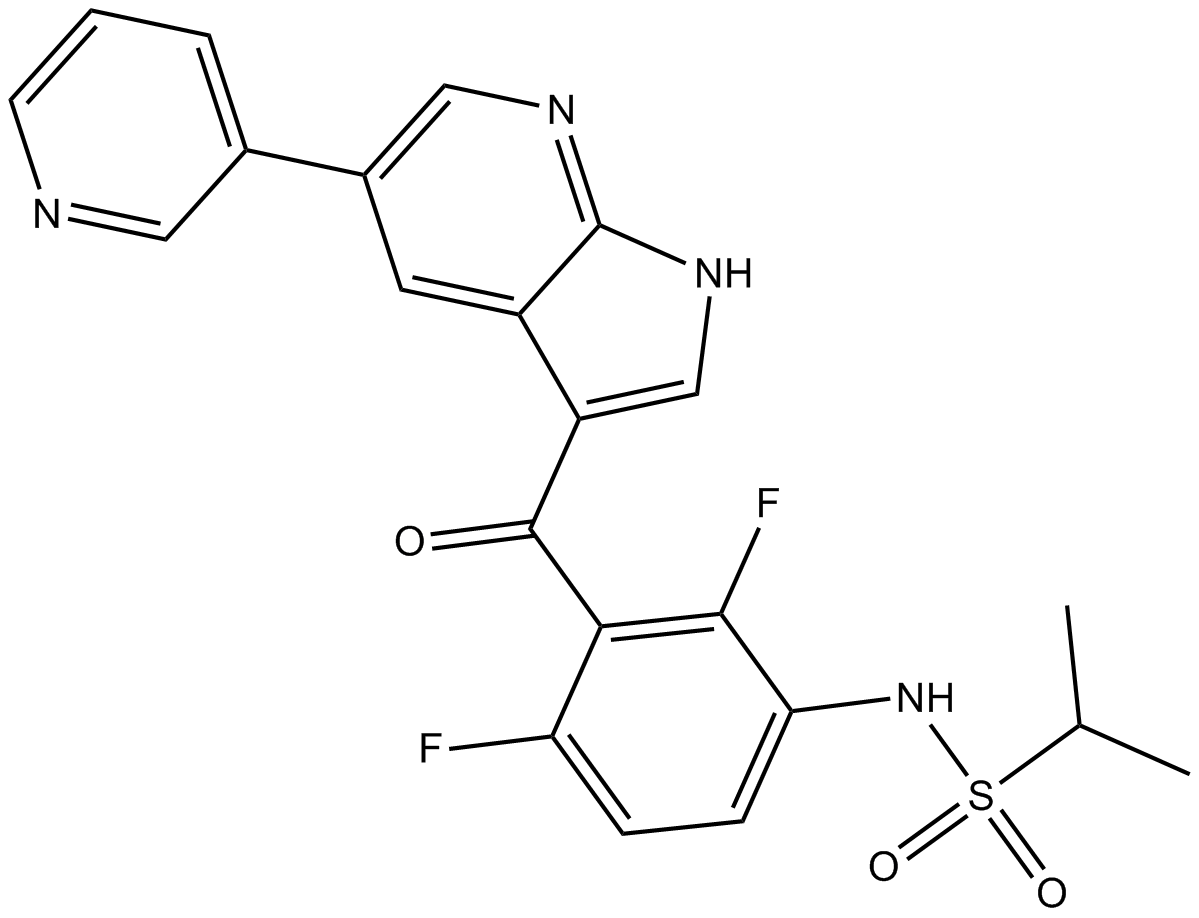
-
GC16779
BRD 7389
P90 RSK inhibitor
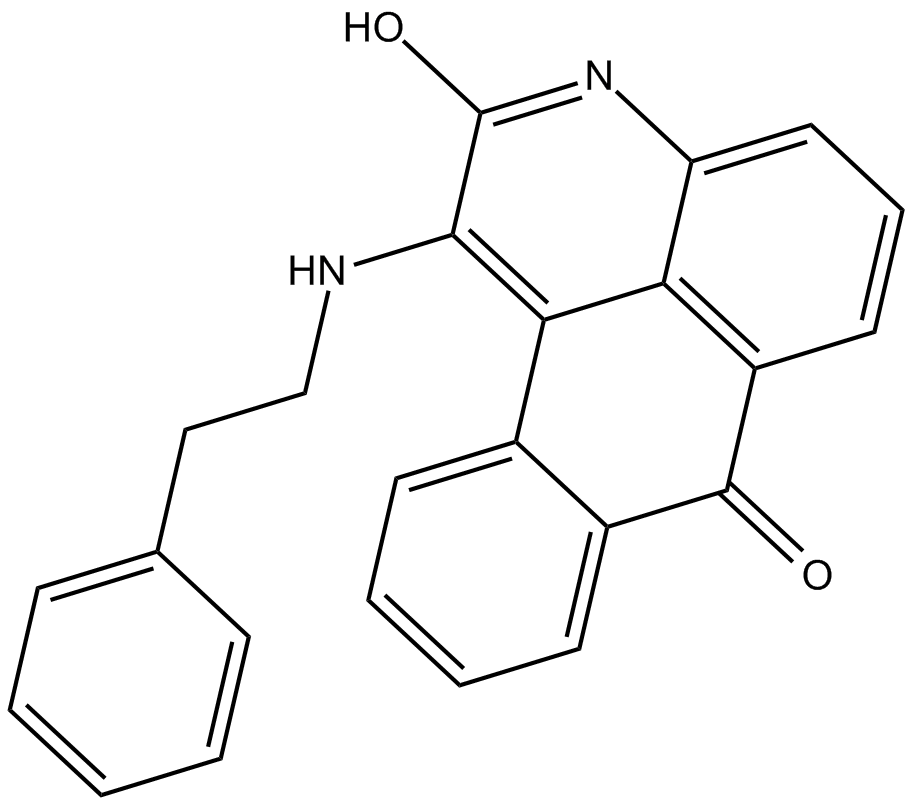
-
GC68811
BSJ-04-122
BSJ-04-122 is a covalent dual inhibitor of MKK4/7. It inhibits both MKK4 and MKK7 with IC50 values of 4 nM and 181 nM, respectively. BSJ-04-122 can be used in cancer research.

-
GC35565
Bucladesine calcium salt
Bucladesine calcium salt salt (Dibutyryl-cAMP calcium salt;DC2797 calcium salt) is a cell-permeable cyclic AMP (cAMP) analog and selectively activates cAMP dependent protein kinase (PKA) by increasing the intracellular level of cAMP.
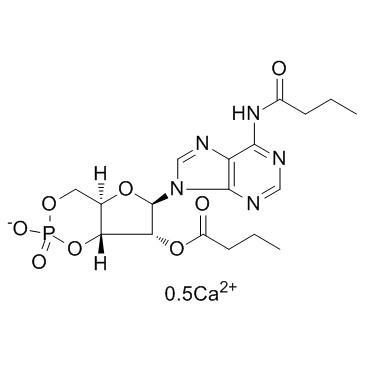
-
GC15524
Bumetanide
PF-1593, Ro 10-6338
NKCC cotransporter inhibitor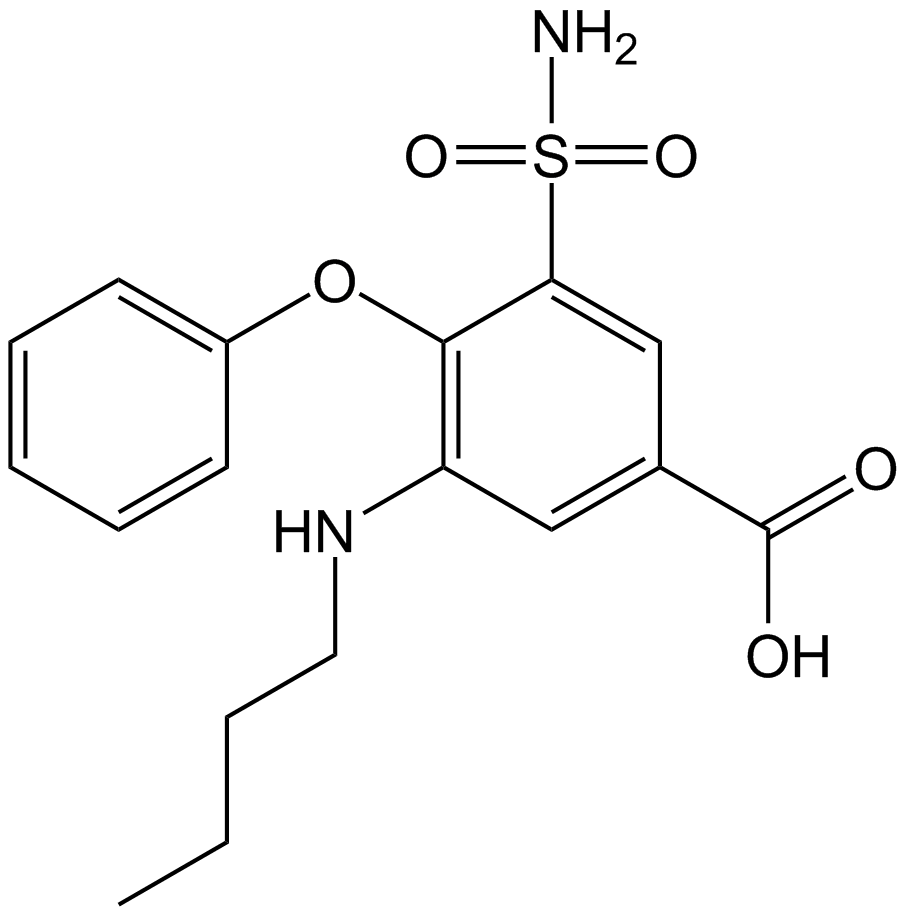
-
GC11605
C-1
C-1 is a potent protein kinase inhibitor, with IC50s of 4 μM, 8 μM, 12 μM and 240 μM for cGMP-dependent protein kinase (PKG), cAMP-dependent protein kinase (PKA), protein kinase C (PKC) and MLC-kinase, respectively. C-1 also used as a ROCK inhibitor.

-
GC10693
c-JUN peptide
JNK/c-Jun interaction inhibitor

-
GC43065
C2 Phytoceramide (t18:0/2:0)
N-Acetyl Phytosphingosine, C2:0 Phytoceramide, Cer(t18:0/2:0), Ceramide (t18:0/2:0), NAPS
C2 Phytoceramide is a bioactive semisynthetic sphingolipid that inhibits formyl peptide-induced oxidant release (IC50 = 0.38 μM) in suspended polymorphonuclear cells.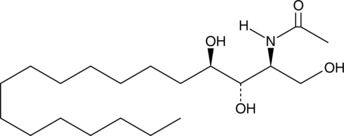
-
GC40352
Cafestol
Cafestol is a natural diterpene which is abundant in unfiltered coffee.
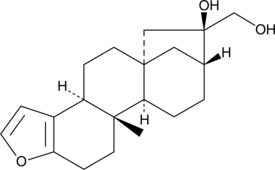
-
GC10941
cAMPS-Rp, triethylammonium salt
Rp-cAMPS
cAMPS-Rp, triethylammonium salt, a cAMP analog, is a potent, competitive cAMP-induced activation of cAMP-dependent PKA I and II (Kis of 12.5 μM and 4.5 μM, respectively) antagonist.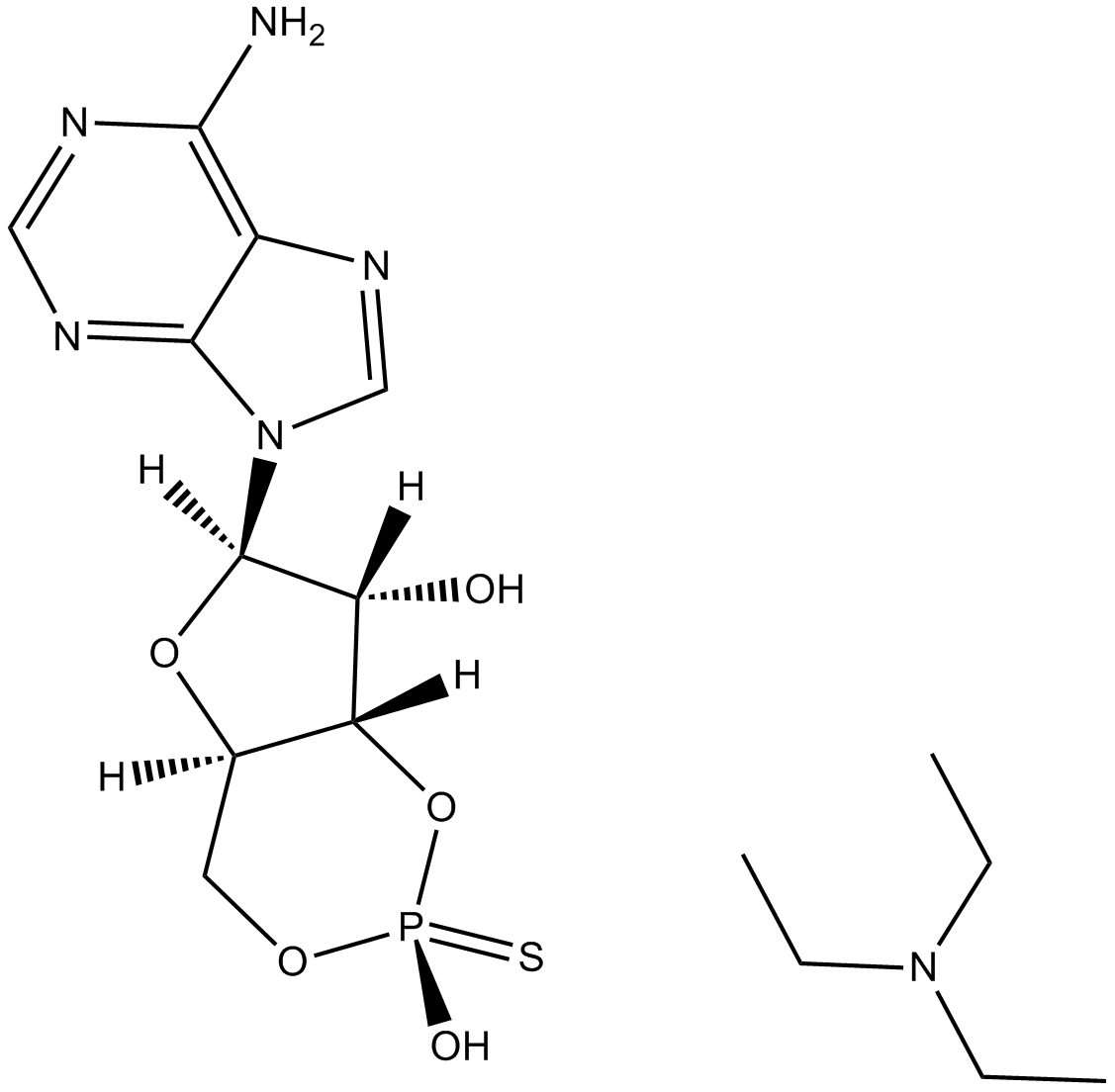
-
GC12706
cAMPS-Sp, triethylammonium salt
cAMPS-Sp, triethylammonium salt, a cAMP analog, is a potent, competitive cAMP-induced activation of cAMP-dependent PKA I and II (Kis of 12.5 μM and 4.5 μM, respectively) antagonist.

-
GN10016
Carnosol
NSC 39143
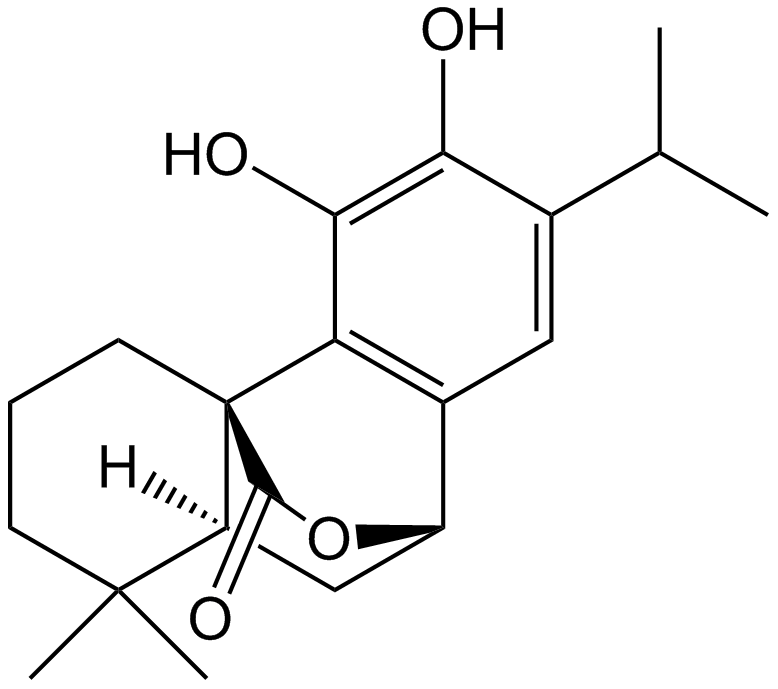
-
GC47045
Carvedilol-d5
An internal standard for the quantification of carvedilol
-
GC43167
CAY10561
Pyrazolylpyrrole ERK Inhibitor
The extracellular signal-regulated kinase (ERK) signal transduction pathway regulates a diverse array of cellular processes.
-
GC40650
CAY10706
CAY10706 is a ligustrazine-curcumin hybrid that promotes intracellular reactive oxygen species accumulation preferentially in lung cancer cells.
-
GC43198
CAY10717
CAY10717 is a multi-targeted kinase inhibitor that exhibits greater than 40% inhibition of 34 of 104 kinases in an enzymatic assay at a concentration of 100 nM.
-
GC50400
CC 401 dihydrochloride
High affinity JNK inhibitor; also inhibits HCMV replication
-
GC13529
CC-401
JNK inhibitor,ATP-competitive