Cell Cycle/Checkpoint
Cell Cycle
Cells undergo a complex cycle of growth and division that is referred to as the cell cycle. The cell cycle consists of four phases, G1 (GAP 1), S (synthesis), G2 (GAP 2) and M (mitosis). DNA replication occurs during S phase. When cells stop dividing temporarily or indefinitely, they enter a quiescent state called G0.
Targets for Cell Cycle/Checkpoint
- ATM/ATR(23)
- Aurora Kinase(17)
- Cdc42(4)
- Cdc7(3)
- Chk(14)
- c-Myc(20)
- CRM1(8)
- Cyclin-Dependent Kinases(77)
- E1 enzyme(1)
- G-quadruplex(11)
- Haspin(6)
- HMTase(1)
- Kinesin(23)
- Ksp(4)
- Microtubule/Tubulin(221)
- Mps1(15)
- Mitotic(7)
- RAD51(16)
- ROCK(65)
- Rho(13)
- PERK(11)
- PLK(33)
- PTEN(6)
- Wee1(8)
- PAK(21)
- Arp2/3 Complex(8)
- Dynamin(11)
- ECM & Adhesion Molecules(40)
- Cholesterol Metabolism(3)
- Endomembrane System & Vesicular Trafficking(26)
- G1(38)
- G2/M(26)
- G2/S(10)
- Genotoxic Stress(18)
- Inositol Phosphates(18)
- Proteolysis(99)
- Cytoskeleton & Motor Proteins(53)
- Cellular Chaperones(8)
Products for Cell Cycle/Checkpoint
- Cat.No. Product Name Information
-
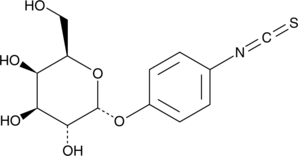
GC40702
α-D-Galactopyranosylphenyl isothiocyanate
α-D-Galactopyranosylphenyl isothiocyanate is a chemically activated form of galactose that has been used to prepare various neoglycoproteins, which consist of a glycosylated serum albumin substituted with either fluorescein or methotrexate.
-
GC41676
(±)-Nornicotine
DL-Nornicotine, (R,S)-Nornicotine
(±)-Nornicotine is a metabolite of nicotine that acts as a neuronal nicotinic acetylcholine receptor (nAChR) agonist.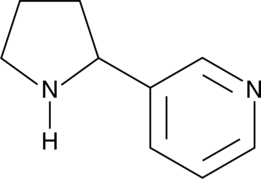
-
GC49482
(±)-Nornicotine-d4
DL-Nornicotine-d4, (R,S)-Nornicotine-d4
An internal standard for the quantification of (±)-nornicotine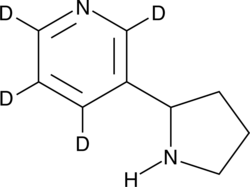
-
GC41684
(±)-trans-1,2-Bis(2-mercaptoacetamido)cyclohexane
BMC, Vectrase P
(±)-trans-1,2-Bis(2-mercaptoacetamido)cyclohexane (BMC) is a cyclohexane with two mercaptoacetamido groups.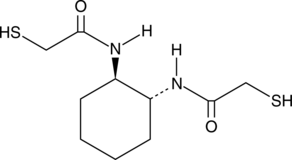
-
GC40802
(±)12(13)-DiHOME
Isoleukotoxin diol
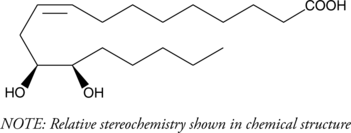
(±)12(13)-DiHOME is the diol form of (±)12(13)-EpOME, a cytochrome P450-derived epoxide of linoleic acid also known as isoleukotoxin.
-
GC41656
(±)2-(14,15-Epoxyeicosatrienoyl) Glycerol
(±)214,15EG
2-Arachidonoyl glycerol (2-AG) is an endogenous central cannabinoid (CB1) receptor agonist that is present at relatively high levels in the central nervous system.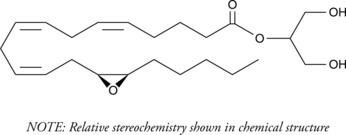
-
GC45890
(+)-Abscisic Acid-d6
(S)-(+)-Abscisic acid-d6; ABA-d6
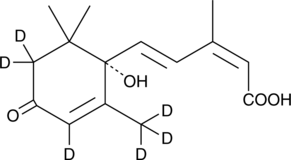
An internal standard for the quantification of (+)-abscisic acid
-
GC62728
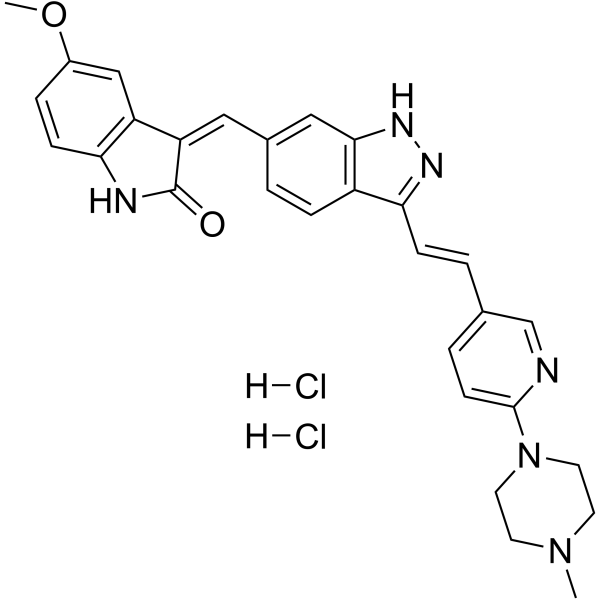
(1E)-CFI-400437 dihydrochloride
(1E)-CFI-400437 dihydrochloride is a potent PLK4 (IC50= 0.6 nM) inhibitor and selective against other members of the PLK family (>10 μM). (1E)-CFI-400437 dihydrochloride inhibits Aurora A, Aurora B, KDR and FLT-3 with IC50s of 0.37, 0.21, 0.48, and 0.18 μM, respectively. Antiproliferative activity.
-
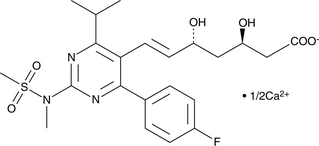
GC49690
(3R,5R)-Rosuvastatin (calcium salt)
A potential impurity found in bulk preparations of rosuvastatin
-
GC41268
(E)-2-Hexadecenal
trans-2-Hexadecenal
Sphingosine-1-phosphate (S1P), a bioactive lipid involved in many signaling processes, is irreversibly degraded by the membrane-bound S1P lyase.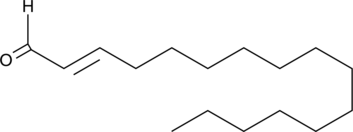
-
GC41701
(E)-2-Hexadecenal Alkyne
(E)-2-Hexadecenal alkyne is an alkyne version of the sphingolipid degradation product (E)-2-hexadecenal that can be used as a click chemistry probe.

-
GC46335
(E)-Fenpyroximate
Fenpyroximate, NNI-850
A phenoxypyrazole acaricide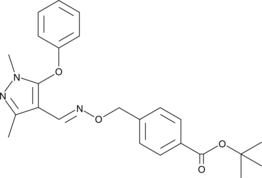
-
GC10419
(R)-CCG-1423
Rho inhibitor

-
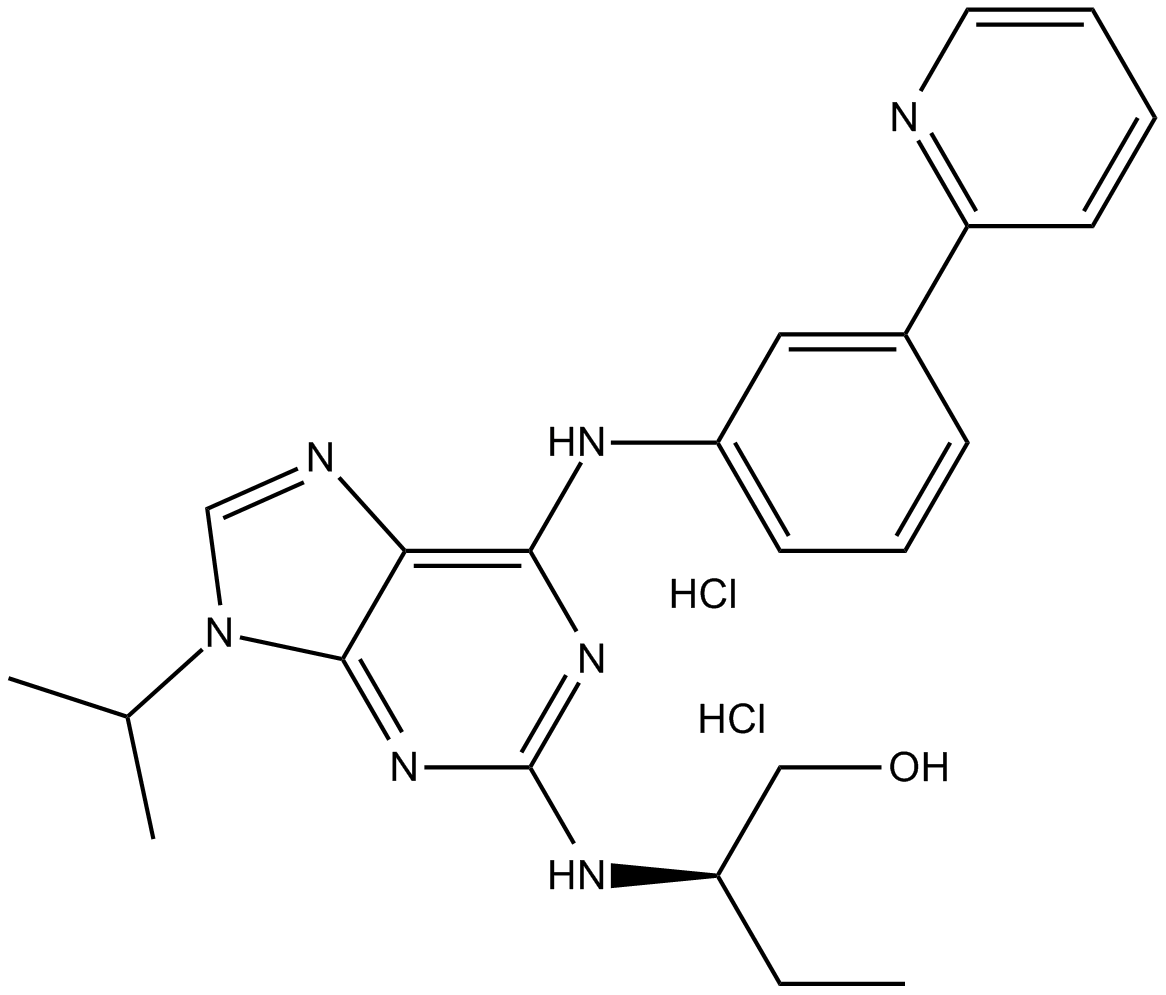
GC16429
(R)-DRF053 dihydrochloride
cdk/CK1 inhibitor,potent and ATP-competitive
-
GC41719
(R)-nitro-Blebbistatin
R(-)7Desmethyl8nitro Blebbistatin
(R)-nitro-Blebbistatin is a more stable form of (+)-blebbistatin, which is the inactive form of (-)-blebbistatin.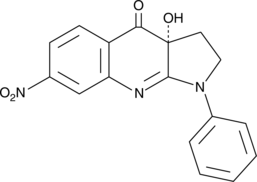
-
GC46347
(S)-(+)-Methoprene
Altosid, d-Methoprene, ZR 2458
An insect growth regulator and Met agonist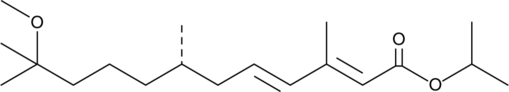
-
GC41557
(S)-3'-amino Blebbistatin
(-)-3'-amino Blebbistatin, m-amino Blebbistatin, meta-amino Blebbistatin
(S)-3'-amino Blebbistatin is a more stable and less phototoxic form of (-)-blebbistatin, which is a selective cell-permeable inhibitor of non-muscle myosin II ATPases.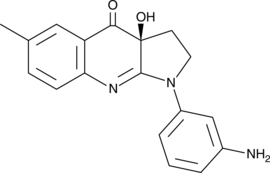
-
GC41484
(S)-3'-hydroxy Blebbistatin
(-)-3'-hydroxy Blebbistatin, meta-hydroxy-Blebbistatin, m-hydroxy-Blebbistatin
(S)-3'-hydroxy Blebbistatin is a more stable and less phototoxic form of (-)-blebbistatin, which is a selective cell-permeable inhibitor of non-muscle myosin II ATPases.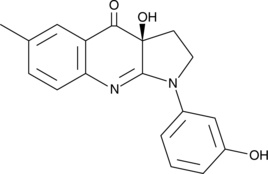
-
GC52192
(S)-4'-nitro-Blebbistatin
(-)-4'-nitro-Blebbistatin, p-nitro-Blebbistatin, para-nitro-Blebbistatin
(S)-4'-nitro-Blebbistatin is a non-cytotoxic, photostable, fluorescent and specific Myosin II inhibitor, usd in the study of the specific role of myosin II in physiological, developmental, and cell biological studies.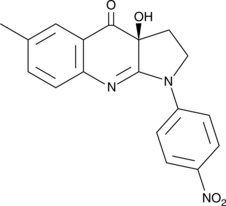
-
GC14497
(S)-CCG-1423
Rho inhibitor
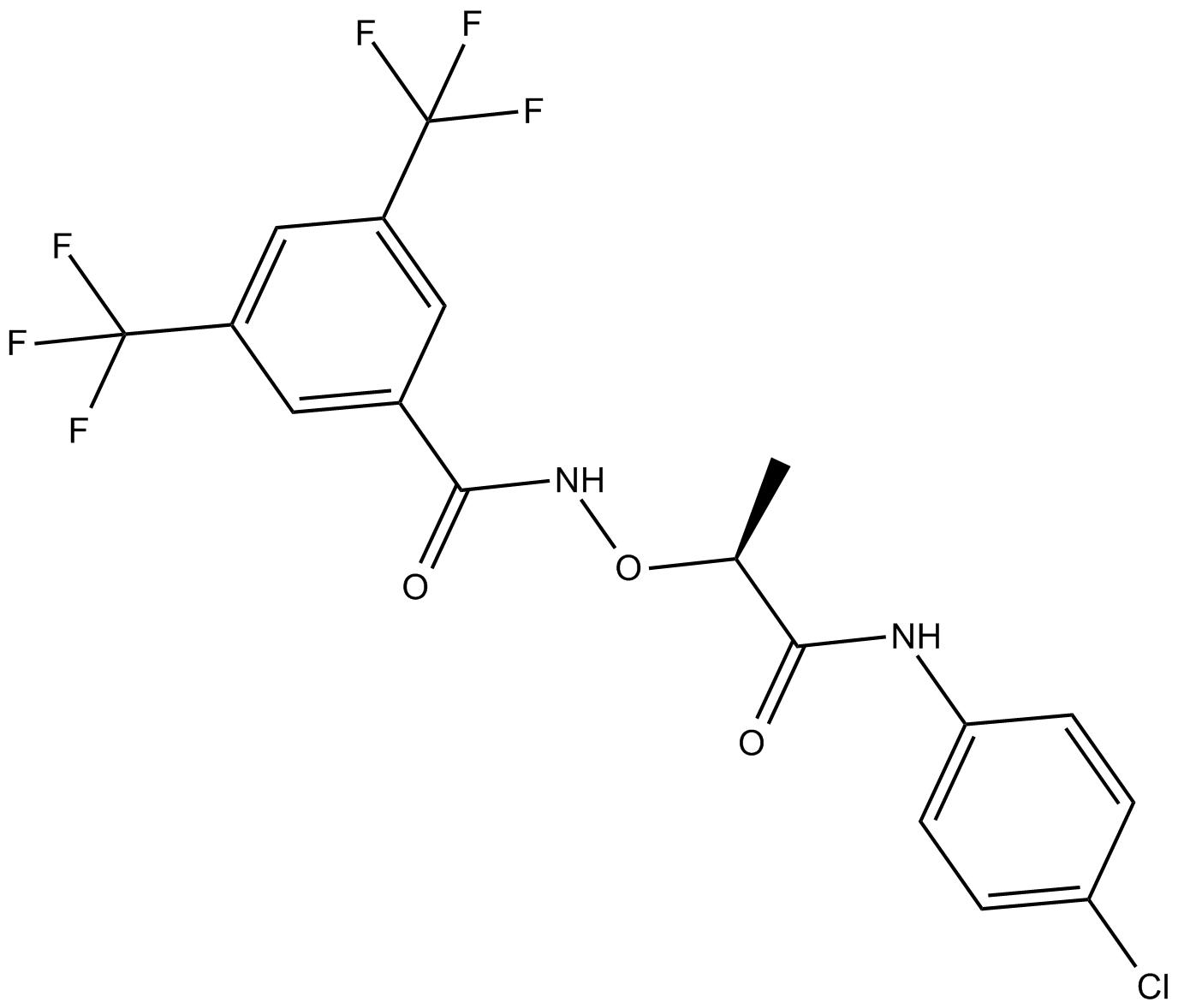
-
GC34999
(S)-Ceralasertib
(S)-AZD6738
(S)-Ceralasertib ((S)-AZD6738) is extracted from patent WO2011154737A1, Compound II, exhibits an IC50 of 2.578 nM.(S)-Ceralasertib is a potent and selective sulfoximine morpholinopyrimidine ATR inhibitor with excellent preclinical physicochemical and pharmacokinetic (PK) characteristics.(S)-Ceralasertib is developed improving aqueous solubility and eliminates CYP3A4 time-dependent inhibition.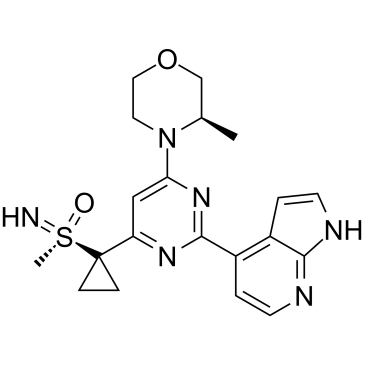
-
GC46351
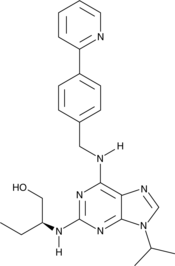
(S)-CR8
An inhibitor of cyclin-dependent kinases
-
GC41737
(S)-Glycyl-H-1152 (hydrochloride)
Rho Kinase Inhibitor IV
Two Rho-associated kinases (ROCK), ROCK-I and ROCK-II, act downstream of the G protein Rho to regulate cytoskeletal stability.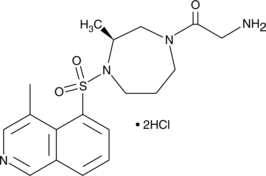
-
GC40145
(S)-Laudanosine
(+)-Laudanosine, L-Laudanosine, L-(+)-Laudanosine, NSC 35045
(S)-Laudanosine is the (S) enantiomer of laudanosine, a metabolite of the neuromuscular blocking agents atracurium and cisatracurium.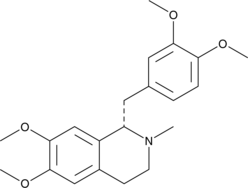
-
GC41739
(S)-nitro-Blebbistatin
S(-)7Desmethyl8nitro Blebbistatin
(S)-nitro-Blebbistatin is a more stable form of (-)-blebbistatin, which is a selective cell-permeable inhibitor of non-muscle myosin II ATPases.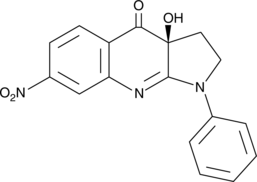
-
GC18275
1,2-Dihexadecyl-sn-glycero-3-PC
1,2-Dihexadecyl-sn-glycero-3-Phosphocholine
1,2-Dihexadecyl-sn-glycero-3-PC is a synthetic ether-linked phospholipid containing hexadecyl groups at the sn-1 and sn-2 positions.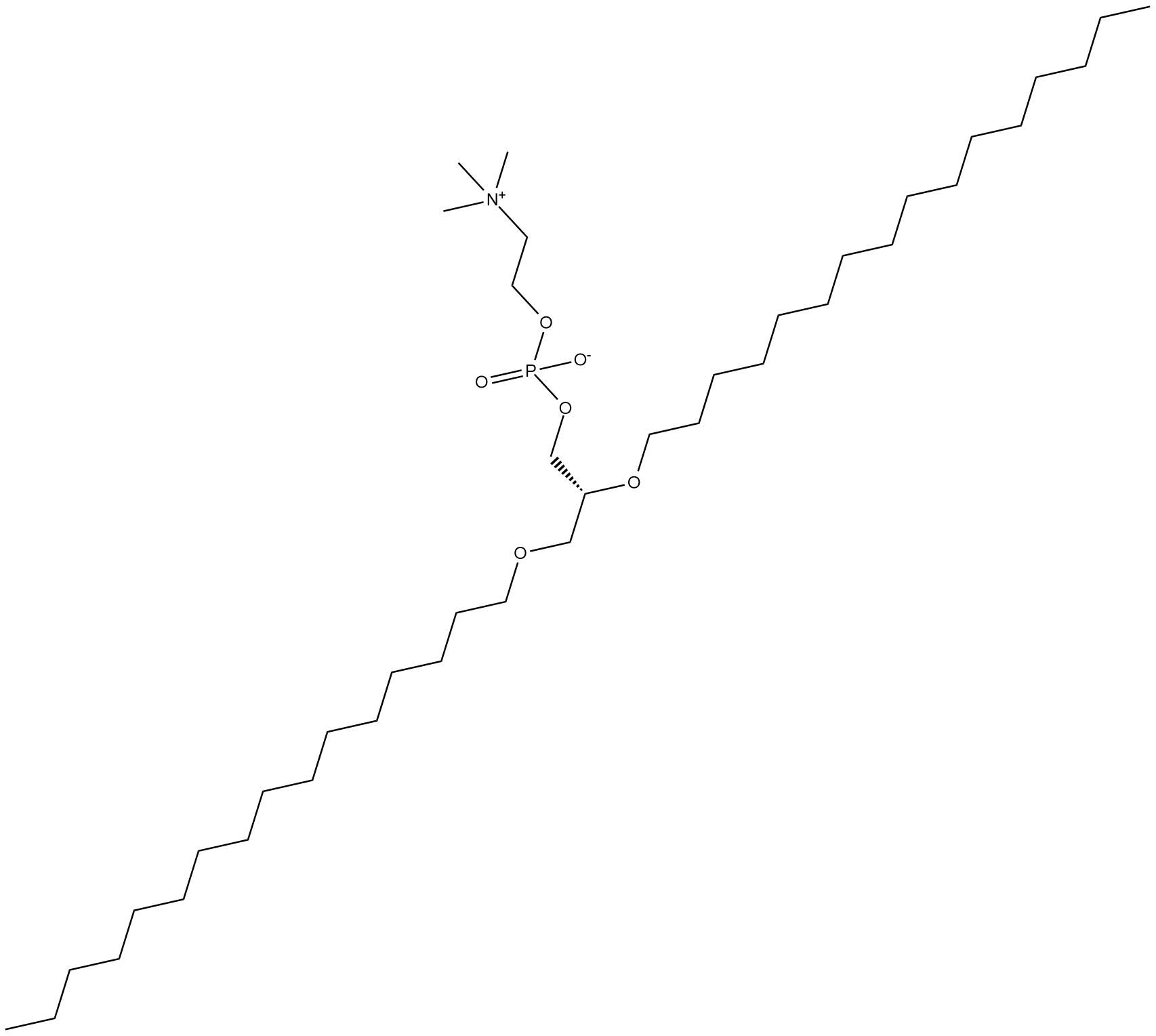
-
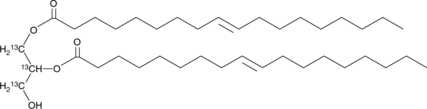
GC45783
1,2-Dioleoyl-rac-glycerol-13C3
An internal standard for the quantification of 1,2-dioleoyl-rac-glycerol
-
GC46379
1,2-Dioleoyl-sn-glycero-3-PS (sodium salt)
1,2-DOPS, 18:1/18:1-PS; PS(18:1/18:1), 1,2-Dioctadecenoyl-sn-glycero-3-Phosphoserine, 1,2-Dioctadecenoyl-sn-glycero-3-Phosphatidylserine
1,2-Dioleoyl-sn-glycero-3-PS (sodium salt) is a ubstitute for Phosphoserine/phosphatidylserine.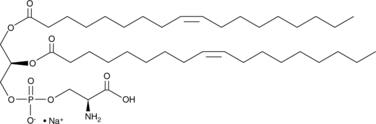
-
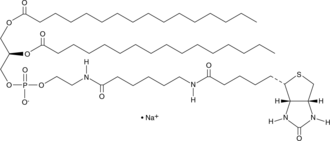
GC41823
1,2-Dipalmitoyl-sn-glycero-3-PE-N-(cap biotin) (sodium salt)
biotin-cap-DPPE
1,2-Dipalmitoyl-sn-glycero-3-PE-N-(cap biotin) is a biotinylated phospholipid.
-
GC46385
1,3,4,6-Tetra-O-acetyl-2-azido-2-deoxy-α-D-Mannopyranose
A ManNAc analog and building block

-
GC19528
1,4-Benzoquinone
p-Benzoquinone, NSC 36324, p-Quinone
A toxic metabolite of benzene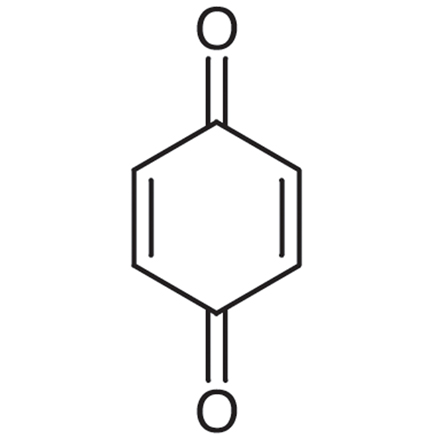
-
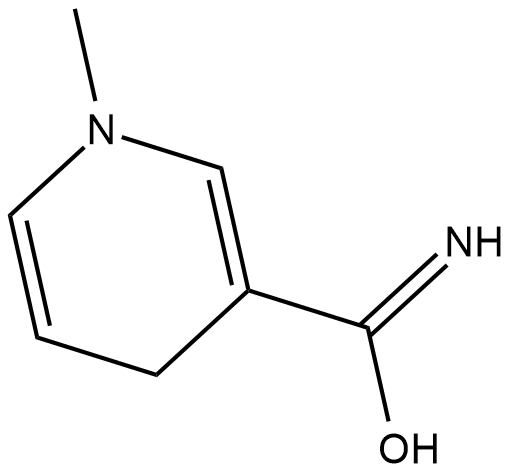
GC18726
1-Methyl-1,4-dihydronicotinamide
1-Methyl-1,4-dihydronicotinamide is a derivative of nicotinamide .
-
GC49736
10-acetyl Docetaxel
PNU 101383, 10-acetyl Taxotere
10-acetyl Docetaxel (10-Acetyl docetaxel) is an analog of Docetaxel, with anticancer activity. Docetaxel is a microtubule disassembly inhibitor, with antimitotic activity.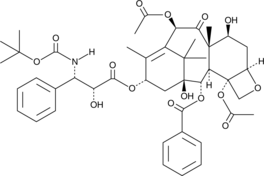
-
GC12954
10-DAB (10-Deacetylbaccatin)
NSC 251677
An inhibitor of microtubule assembly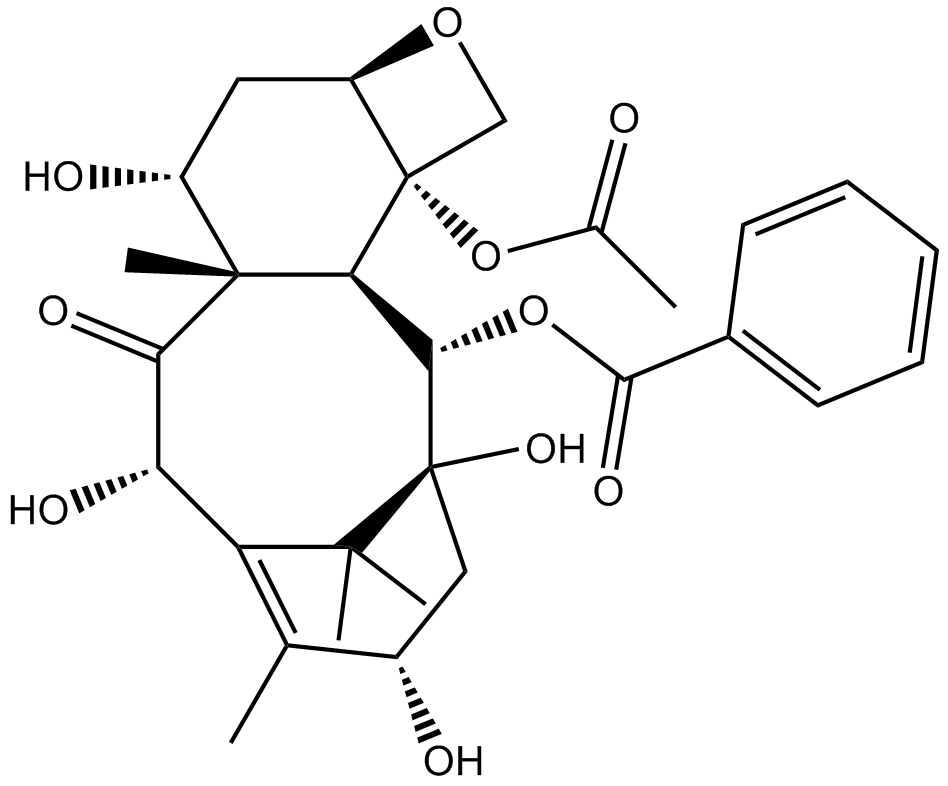
-
GC35044
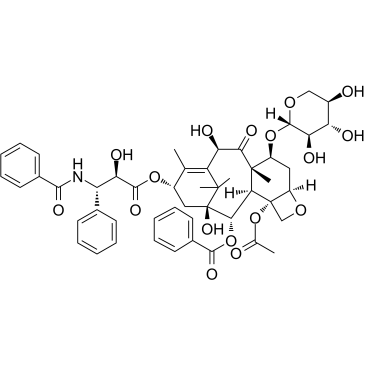
10-Deacetyl-7-xylosyl paclitaxel
10-Deacetyl-7-xylosyl paclitaxel is a Paclitaxel (a microtubule stabilizing agent; enhances tubulin polymerization) derivative with improved pharmacological features.
-
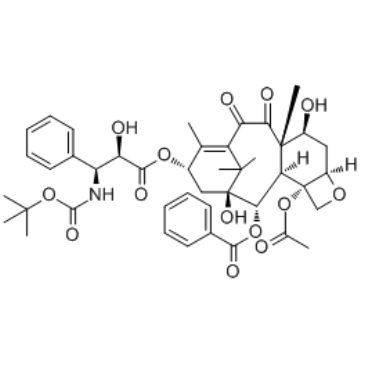
GC35045
10-Oxo Docetaxel
A docetaxel degradation product
-
GC17295
10058-F4
C-Myc-Max dimerization inhibitor

-
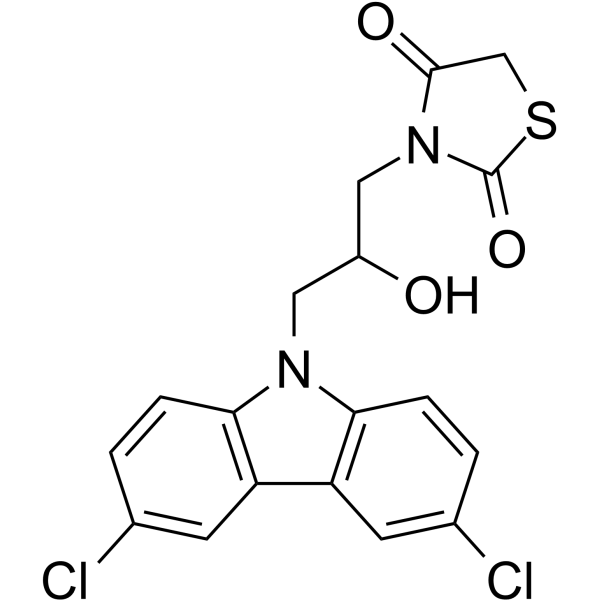
GC63576
10074-A4
10074-A4 is a c-Myc inhibitor. 10074-A4 could bind to c-Myc370-409 at different sites along the peptide chain. 10074-A4 has anticancer effects.
-
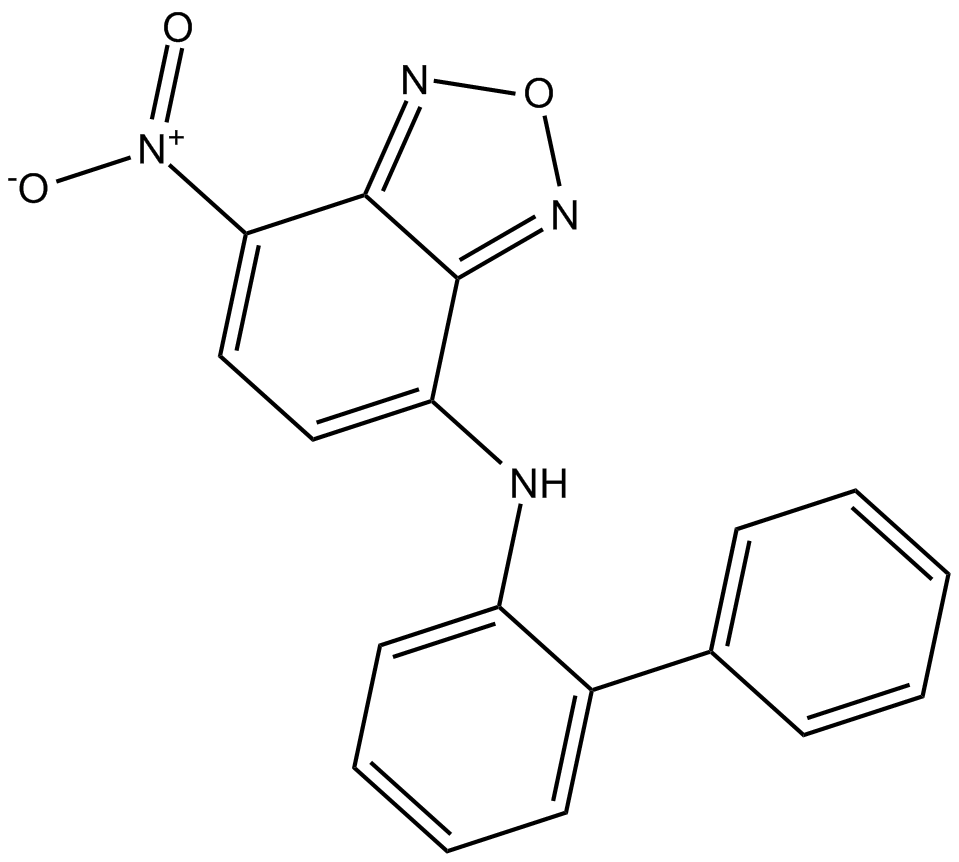
GC14918
10074-G5
c-Myc inhibitor
-
GC40448
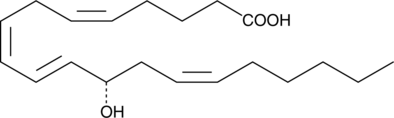
12(S)-HETE
12(S)-Hydroxyeicosatetraenoic Acid
12(S)-HETE is the predominant lipoxygenase product of mammalian platelets.
-
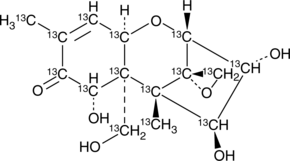
GC46434
13C15-Nivalenol
An internal standard for the quantification of nivalenol
-
GC49390
13C6-4-Nitroaniline
13C6-p-Nitroaniline
An internal standard for the quantification of 4-nitroaniline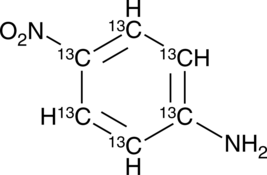
-
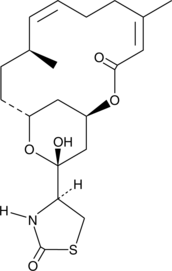
GC41110
16-epi Latrunculin B
16-epi Latrunculin B, first isolated from the Red Sea sponge N.
-
GC48423
19-O-Acetylchaetoglobosin A
Chaetoglobosin A Acetate
A fungal metabolite with actin polymerization inhibitory and cytotoxic activities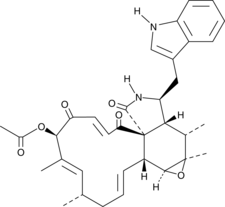
-
GC42151
2'-Deoxyguanosine 5'-monophosphate (sodium salt hydrate)
dGMP
2'-Deoxyguanosine 5'-monophosphate (dGMP) is used as a substrate of guanylate kinases to generate dGDP, which in turn is phosphorylated to dGTP, a nucleotide precursor used in DNA synthesis.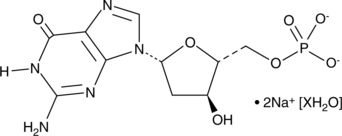
-
GC40947
2,3-Dimethoxy-5-methyl-p-benzoquinone
Coenzyme Q0, CoQ0
2,3-Dimethoxy-5-methyl-p-benzoquinone (CoQ0) is a potent, oral active ubiquinone compound can be derived from Antrodia cinnamomea.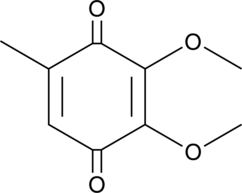
-
GC49671
2,3-Oxidosqualene
(3R,S)-Oxidosqualene, Squalene 2,3-oxide
An intermediate in the biosynthesis of sterols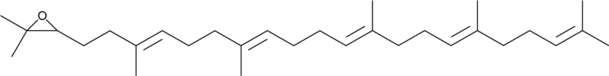
-
GC45324
2,5-dimethyl Celecoxib
DMC
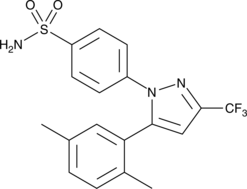
-
GC49362
2-Amino-3,8-dimethylimidazo-[4,5-f]-quinoxaline
MeIQx
A food-derived carcinogen![2-Amino-3,8-dimethylimidazo-[4,5-f]-quinoxaline Chemical Structure 2-Amino-3,8-dimethylimidazo-[4,5-f]-quinoxaline Chemical Structure](/media/struct/GC4/GC49362.png)
-
GC52029
2-Aminoflubendazole
Hydrolyzed Flubendazole
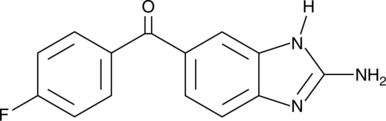
-
GC40675
2-deoxy-Artemisinin
2-deoxy-Artemisinin is an inactive metabolite of the antimalarial agent artemisinin.
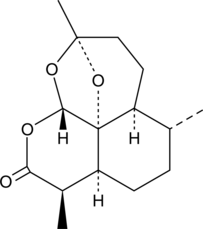
-
GC15084
2-Methoxyestradiol (2-MeOE2)
2Hydroxyestradiol 2methyl ether, 2ME2, NSC 659853, Panzem
2-Methoxyestradiol (2-MeOE2/2-Me) is an HIF-1α inhibitor.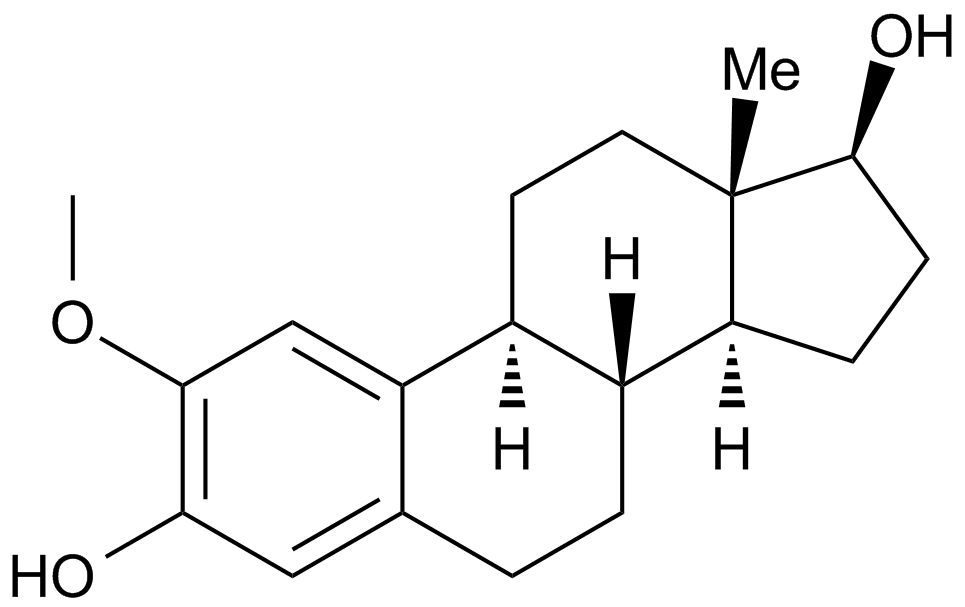
-
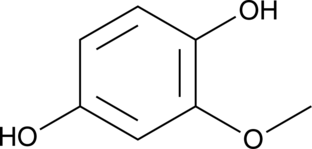
GC52140
2-Methoxyhydroquinone
o-Methoxyhydroquinone, MHQ, MOHQ
-
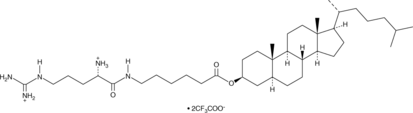
GC42161
2H-Cho-Arg (trifluoroacetate salt)
2H-Cho-Arg is a steroid-based cationic lipid that contains a 2H-cholesterol skeleton coupled to an L-arginine head group and can be used to facilitate gene transfection.
-
GC40618
3',4',7-Trihydroxyisoflavone
3'-hydroxy Daidzein, 3’,4’,7-THIF
3',4',7-Trihydroxyisoflavone, a major metabolite of Daidzein, is an ATP-competitive inhibitor of Cot (Tpl2/MAP3K8) and MKK4. 3',4',7-Trihydroxyisoflavone has anticancer, anti-angiogenic, chemoprotective, and free radical scavenging activities.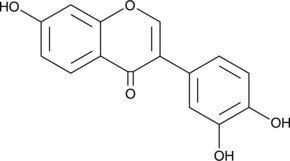
-
GC45332
3'-Dephosphocoenzyme A
depCoA, Dephospho-CoA
An intermediate in the biosynthesis of CoA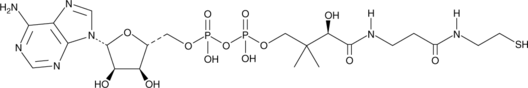
-
GC42239
3,6-diacetoxy Phthalonitrile
ADB, 1,4-Diacetoxy-2,3-dicyanobenzene, 2,3-Dicyano-1,4-hydroquinone diacetate
3,6-diacetoxy Phthalonitrile is a cell-permeable fluorescent probe.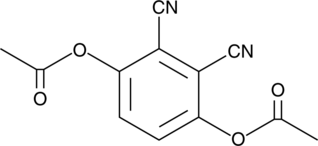
-
GC12314
3-(4-Pyridyl)indole
Rho Kinase Inhibitor III,ROCK Inhibitor III,Rockout
ROCK-I inhibitor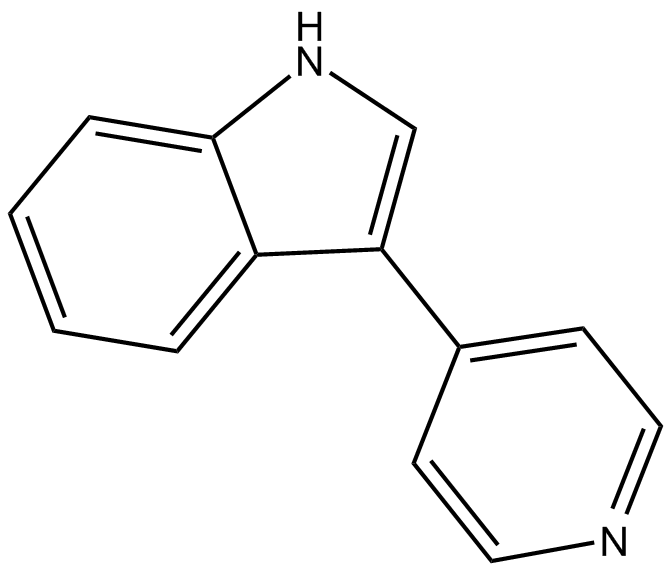
-
GC46582
3-Acetyldeoxy Nivalenol-13C17
3-AcDON-13C17, DON 3-acetate-13C17, Deoxy Nivalenol-3-acetate-13C17
An internal standard for the quantification of 3-acetyldeoxy nivalenol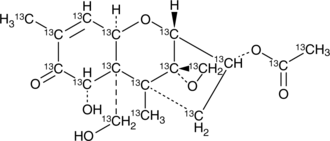
-
GC52129
3-Amino-5-hydroxybenzoic Acid
AHBA
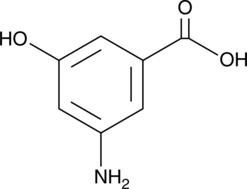
-
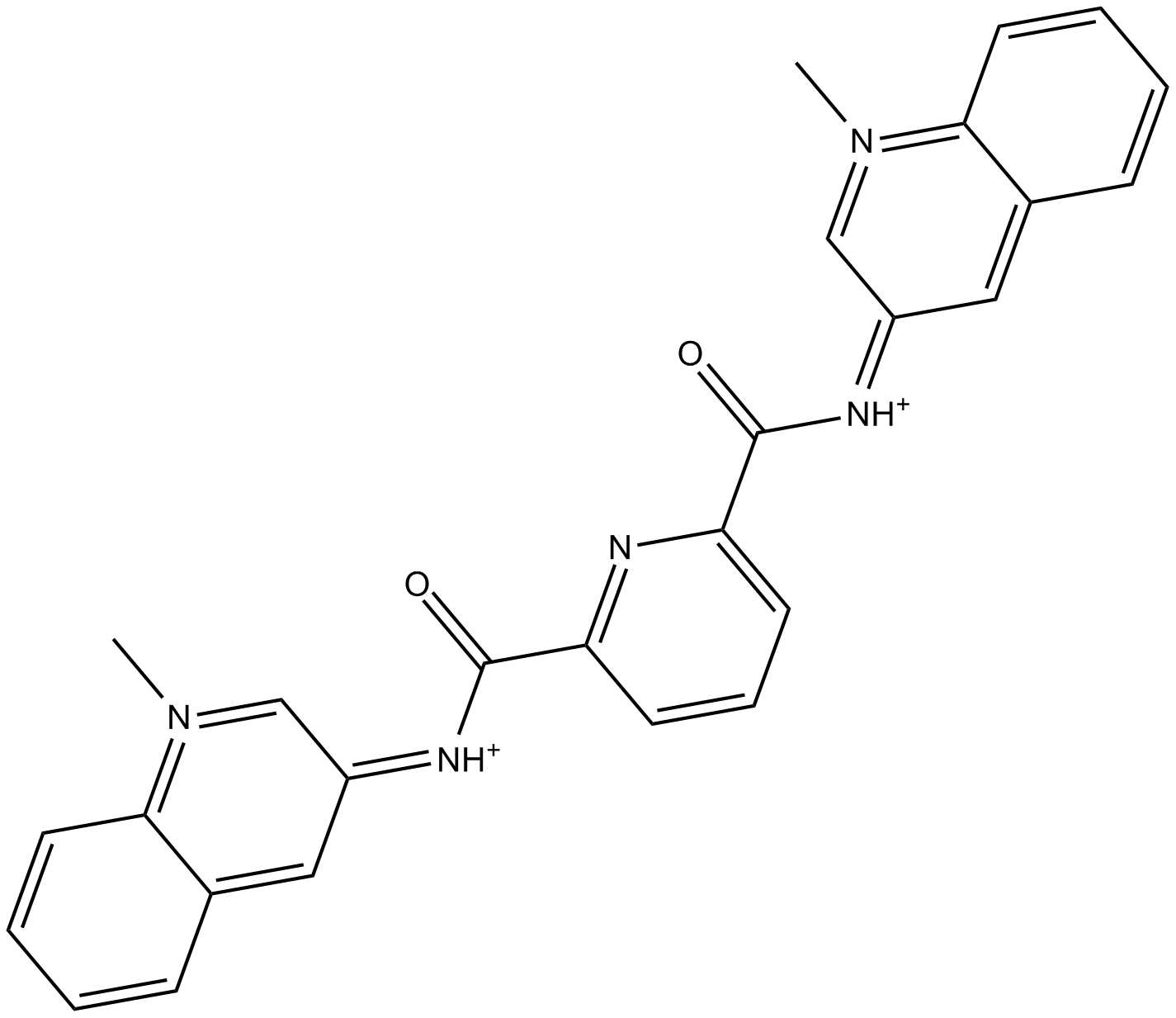
GC15389
360A
-
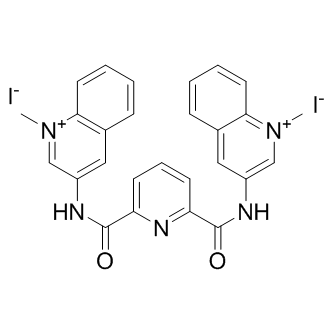
GC10115
360A iodide
-
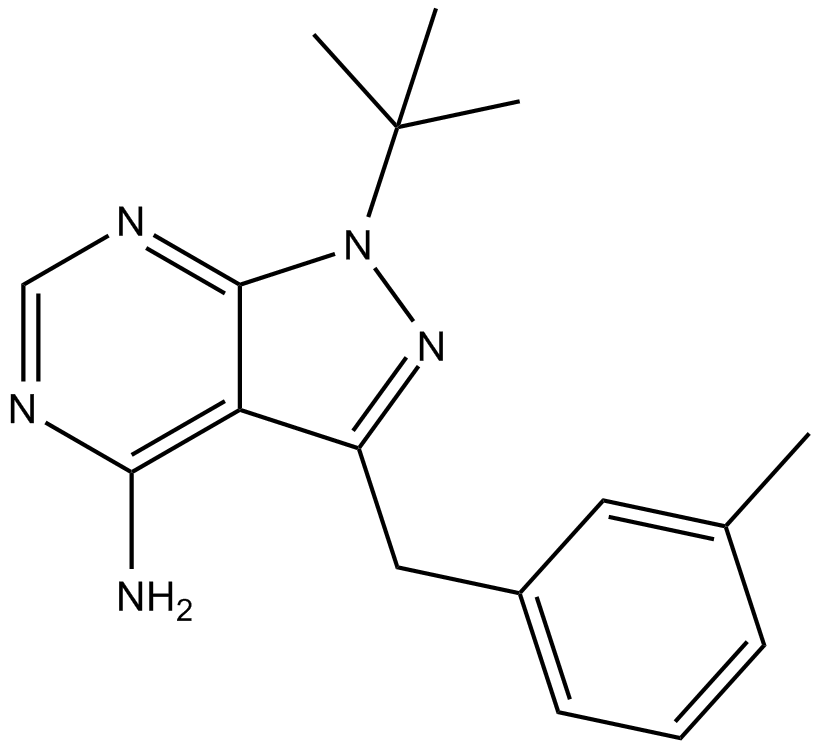
GC14186
3MB-PP1
polo-like kinase 1 (Plk1) allele inhibitor,ATP-competitive
-
GC45354
4β-Hydroxywithanolide E
NSC 212509
A withanolide with anti-inflammatory and anticancer activities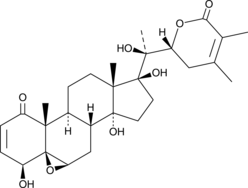
-
GC17271
4'-Demethylepipodophyllotoxin
(-)-4′-Demethylepipodophyllotoxin
An inhibitor of tubulin polymerization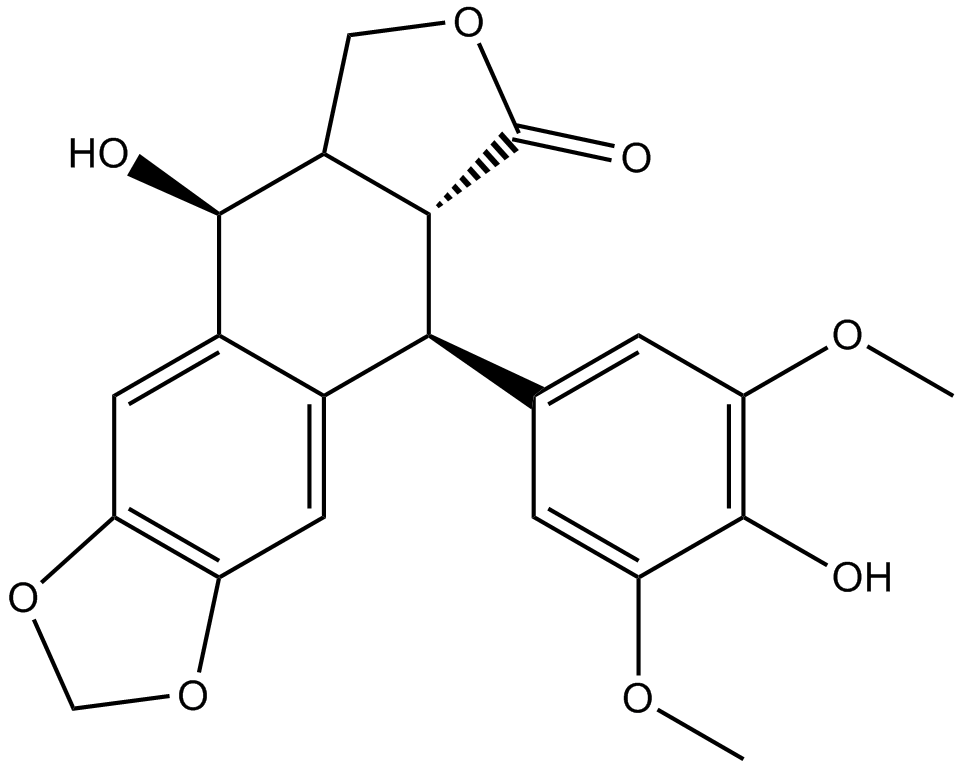
-
GC46606
4-(4,4,5,5-Tetramethyl-1,3,2-dioxaboran-2yl)aniline
4-Aminophenylboronic Acid pinacol ester
A heterocyclic building block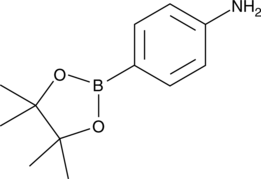
-
GC46635
4-deoxy Nivalenol-13C15
Vomitoxin-13C15, Deoxynivalenol-13C15, DON-13C15
An internal standard for the quantification of 4-deoxy nivalenol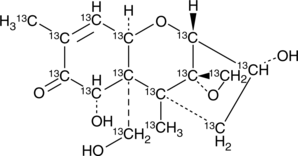
-
GC18359
4-Epianhydrochlortetracycline (hydrochloride)
4-Epianhydrochlortetracycline is a derivative of tetracycline .
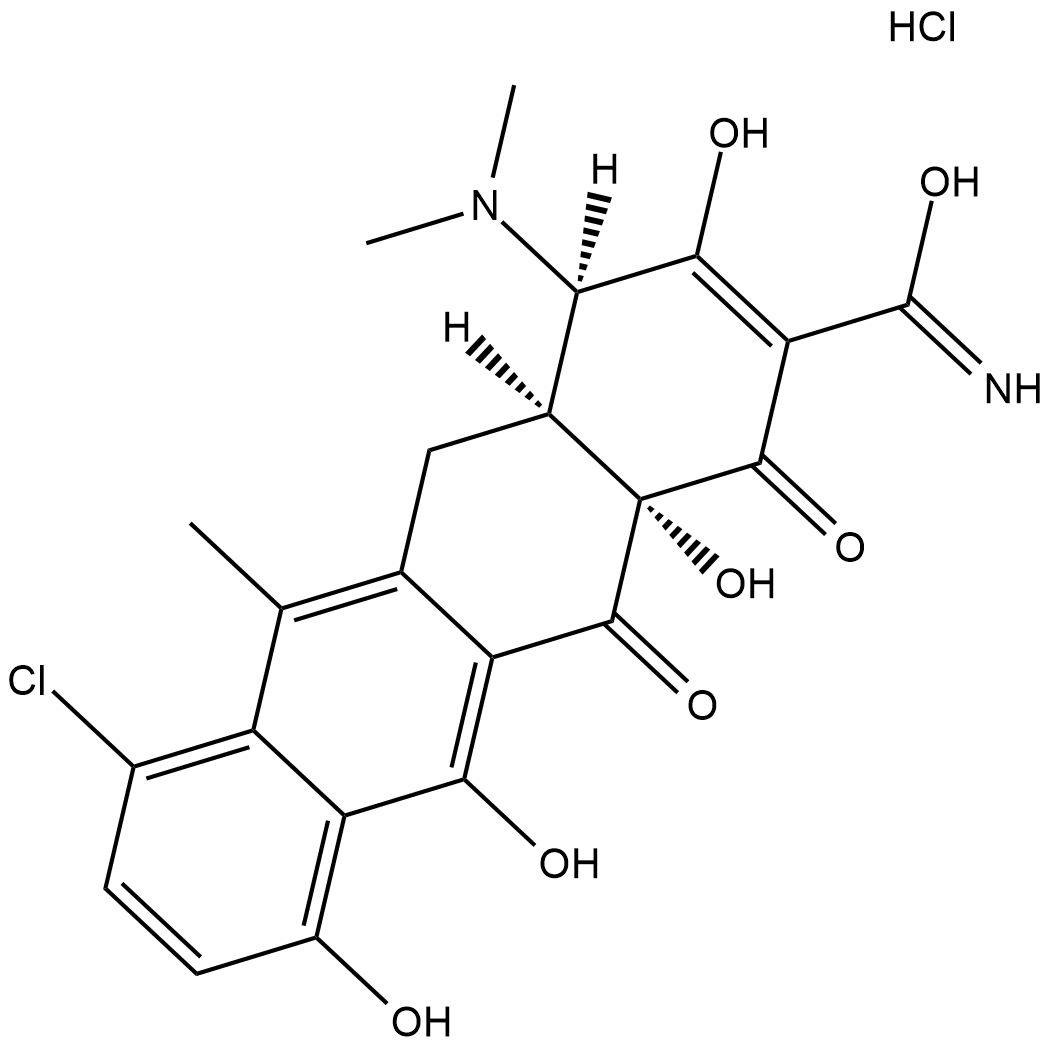
-
GC42449
4-Methylumbelliferyl-α-L-Iduronide (free acid)
4-Methylumbelliferyl-α-L-Idopyranosiduronic Acid, 4-MU-α-IdoA, MU-α-IdoA
4-Methylumbelliferyl-α-L-iduronide (free acid) is a fluorogenic substrate for α-L-iduronidase, an enzyme found in cell lysosomes that is involved in the degradation of glycosaminoglycans such as dermatan sulfate and heparin sulfate.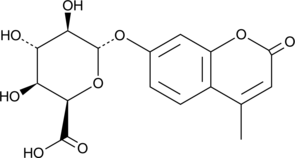
-
GC49244
4-oxo Isotretinoin
Ro 22-6595
An active metabolite of isotretinoin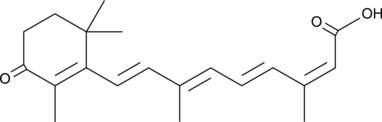
-
GC52365
4-tert-Octylphenol monoethoxylate
NSC 5259, OP1EO
An alkylphenolethoxylate and a degradation product of non-ionic surfactants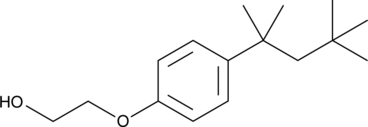
-
GC52227
5-(3',4'-Dihydroxyphenyl)-γ-Valerolactone
(±)-δ-(3,4-Dihydroxyphenyl)-γ-Valerolactone, 5-(3',4'-Dihydroxyphenyl)-γ-VL
An active metabolite of various polyphenols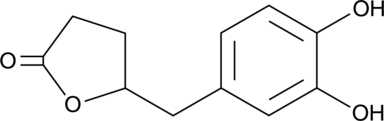
-
GC46681
5-Bromouridine
(–)-5-Bromouridine, BrU, BrUrd, NSC 38296
A brominated uridine analog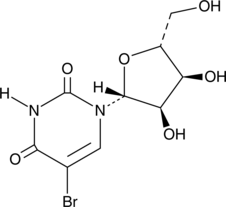
-
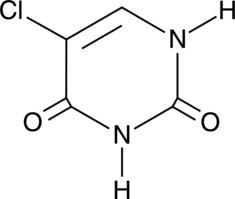
GC45357
5-Chlorouracil
-
GC41156
5-Octyl D-glutamate
5-Octyl ester D-glutamatic acid
5-Octyl D-glutamate, also known as 5-octyl ester D-glutamate, is a stable, cell-permeable molecule that generates free D-glutamate upon hydrolysis of the ester bond by cytoplasmic esterases.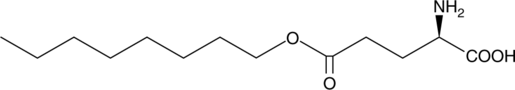
-
GC41423
5-trans Prostaglandin E2
transDinoprostone, 5,6trans PGE2
5-trans PGE2 occurs naturally in some gorgonian corals and is a common impurity in commercial lots of PGE1.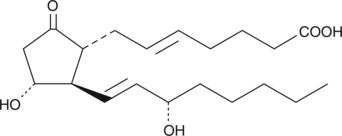
-
GC42586
6α-hydroxy Paclitaxel
6α-hydroxy Taxol
6α-hydroxy Paclitaxel is a primary metabolite of the anticancer compound paclitaxel, produced by the action of the cytochrome P450 isoform CYP2C8.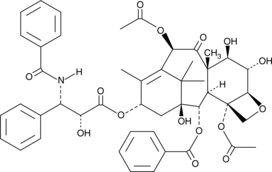
-
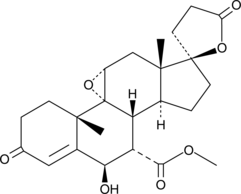
GC45969
6β-hydroxy Eplerenone
A major metabolite of eplerenone
-
GC46721
6-Chloro-2-fluoropurine
NSC 37363
A heterocyclic building block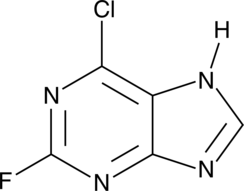
-
GC48721
6-O-Demethyl Griseofulvin
6-Demethylgriseofulvin
A metabolite of griseofulvin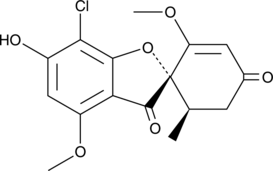
-
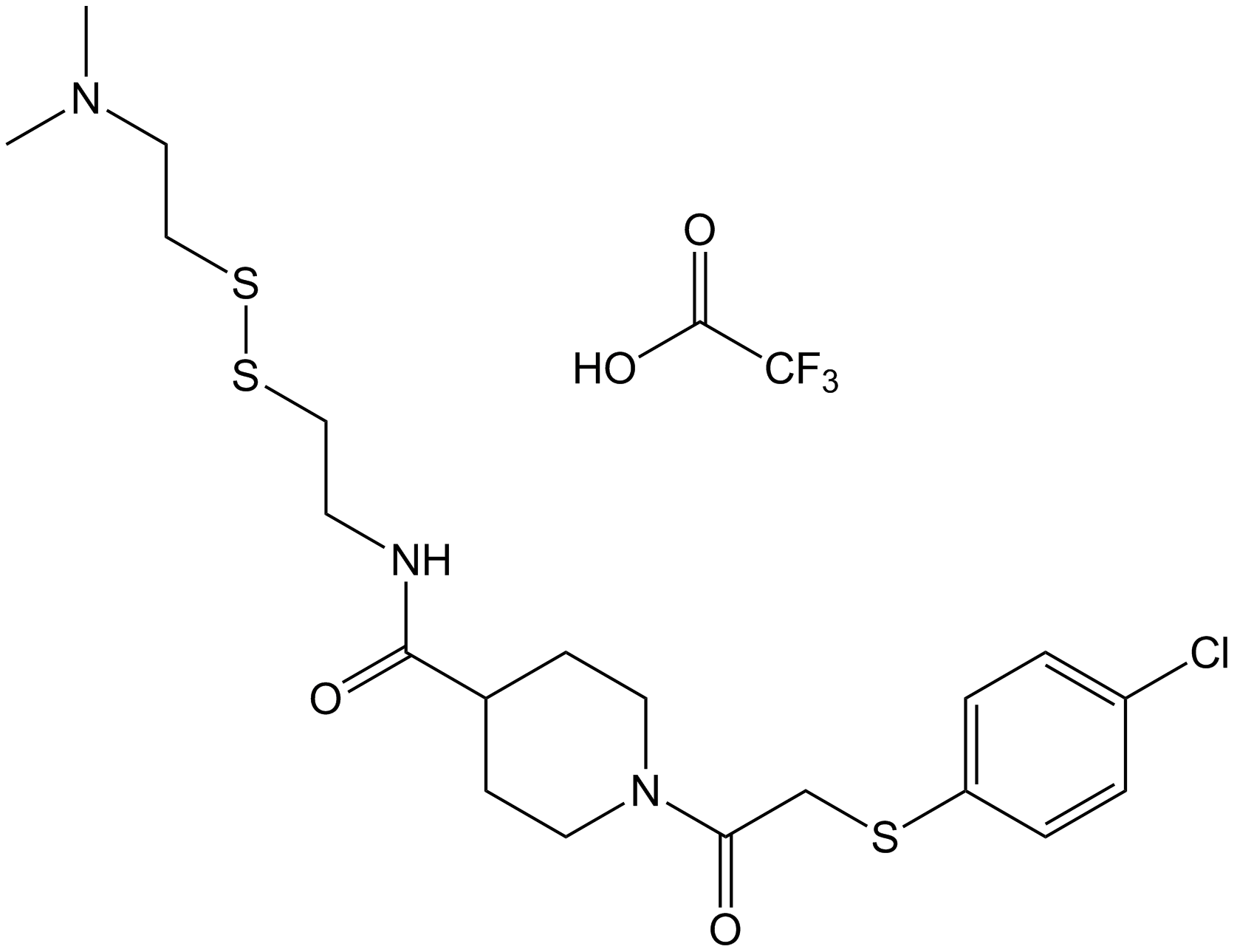
GC15478
6H05
K-Ras inhibitor
-
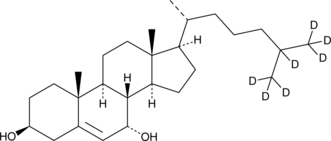
GC40202
7α-hydroxy Cholesterol-d7
7α-hydroxycholesterol-d7
7α-hydroxy Cholesterol-d7 is intended for use as an internal standard for the quantification of 7α-hydroxy cholesterol by GC- or LC-MS.
-
GC46733
7,12-Dimethylbenz[a]anthracene
DMBA
7,12-Dimethylbenz[a]anthracene has carcinogenic activity as a polycyclic aromatic hydrocarbon (PAH). 7,12-Dimethylbenz[a]anthracene is used to induce tumor formation in various rodent models.![7,12-Dimethylbenz[a]anthracene Chemical Structure 7,12-Dimethylbenz[a]anthracene Chemical Structure](/media/struct/GC4/GC46733.png)
-
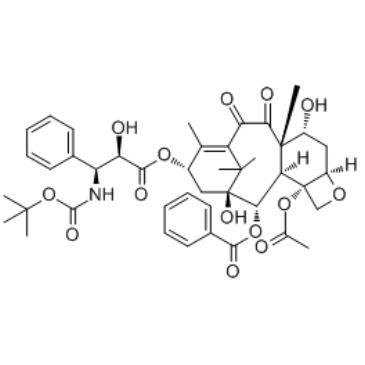
GC35188
7-Epi-10-oxo-docetaxel
7-Epi-10-oxo-docetaxel (Docetaxel Impurity 2) is a impurity of docetaxel detected by high performance liquid chromatography (HPLC).
-
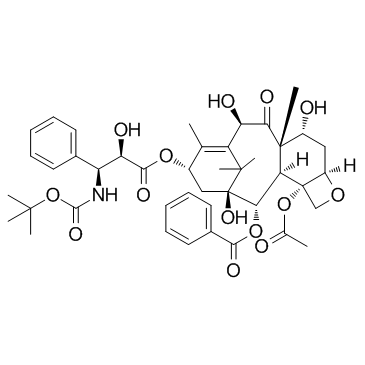
GC35189
7-Epi-docetaxel
7-Epi-10-oxo-docetaxel (Docetaxel Impurity C; 7-Epitaxotere) is a impurity of docetaxel.
-
GN10625
7-Epitaxol
7-epi Taxol
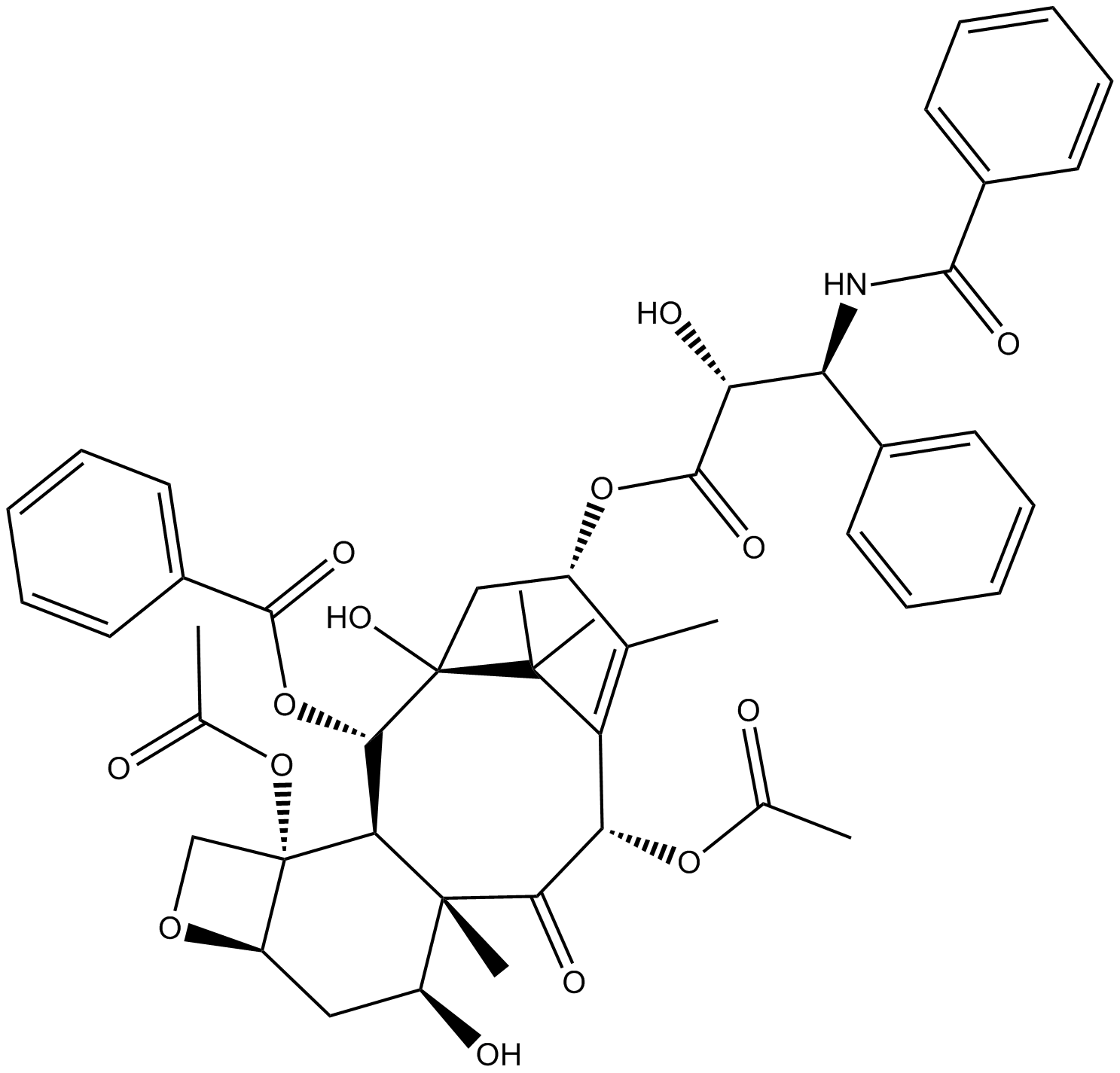
-
GC42610
7-hydroxy Pestalotin
LL-P880β
7-hydroxy Pestalotin is a fungal metabolite originally isolated from Penicillium.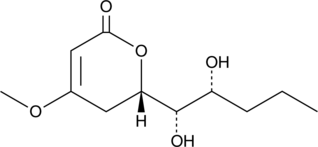
-
GC42616
7-oxo Staurosporine
BMY 41950, RK-1409
7-oxo Staurosporine is an antibiotic originally isolated from S.
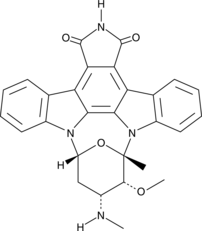
-
GC35197
7-xylosyltaxol
A taxane with microtubule disruptor and anticancer activities

-
GC52126
8-chloro Caffeine
NSC 6277
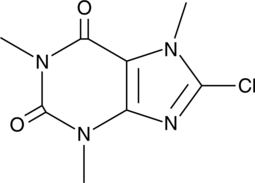
-
GC40844
9-(2,2-Dicyanovinyl)julolidine
DCVJ, 9-Julolidine Methylene Malononitrile, NSC 160064
9-(2,2-Dicyanovinyl)julolidine (9-(2,2-Dicyanovinyl)julolidine), a molecular rotor and unique fluorescent dye, binds to tubulin and actin, and increases its fluorescence intensity drastically upon polymerization.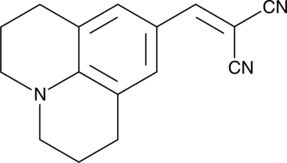
-
GC48986
9-hydroxy Stearic Acid
9-HSA, 9-hydroxy Octadecanoic Acid
A hydroxy fatty acid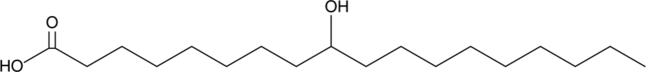
-
GC42665
AAF-CMK (trifluoroacetate salt)
NAlaAlaPheCMK, Tripeptidyl Peptidase Inhibitor II
Tripeptidyl peptidase II (TPPII) is a serine peptidase of the subtilisin-type which removes tripeptides from the free NH2 terminus of oligopeptides.
-
GC35216
AAPK-25
AAPK-25 is a potent and selective Aurora/PLK dual inhibitor with anti-tumor activity, which can cause mitotic delay and arrest cells in a prometaphase, reflecting by the biomarker histone H3Ser10 phosphorylation and followed by a surge in apoptosis. AAPK-25 targets Aurora-A, -B, and -C with Kd values ranging from 23-289 nM, as well as PLK-1, -2, and -3 with Kd values ranging from 55-456 nM.
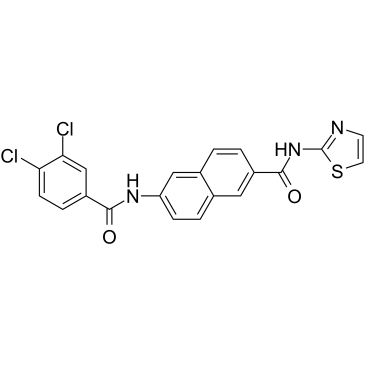
-
GC25025
Abraxane
Nab-Paclitaxel
Abraxane (Nab-Paclitaxel), a novel solvent-free taxane with the binding ratio of Paclitaxel to human serum albumin of 1:9, is an anti-microtubule drug that promotes microtubule aggregation in tubulin dimer and inhibits microtubule depolymerization to stabilize the microtubule system.
-
GC15875
ABT-751 (E7010)
ABT-751 (E7010)(E 7010) is a novel bioavailable tubulin-binding and antimitotic sulfonamide agent with IC50 of about 1.5 and 3.4 μM in neuroblastoma and non-neuroblastoma cell lines, respectively.
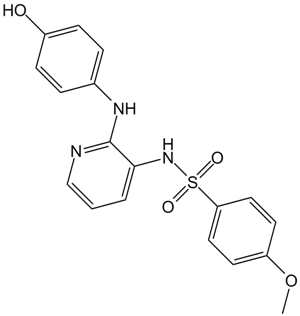
-
GC42683
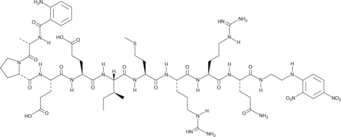
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp is a fluorescence-quenched peptide substrate for human neutrophil elastase (kcat/Km = 531 mM-1s-1).
-
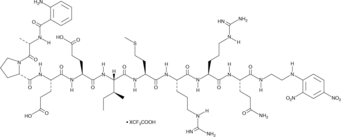
GC52499
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp (trifluoroacetate salt)
A sensitive substrate for neutrophil elastase
-
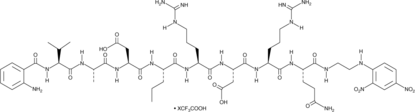
GC42684
Abz-Val-Ala-Asp-Nva-Arg-Asp-Arg-Gln-EDDnp (trifluoroacetate salt)
Abz-Val-Ala-Asp-Nva-Arg-Asp-Arg-Gln-EDDnp is a fluorescence-quenched peptide substrate for human proteinase 3 (kcat/Km = 1,570 mM-1s-1).
-
GA20494
Ac-Asp-Glu-Val-Asp-pNA
Ac-Asp-Glu-Val-Asp-pNA
The cleavage of the chromogenic caspase-3 substrate Ac-DEVD-pNA can be monitored at 405 nm.


