Proteases
Proteases is a general term for a class of enzymes that hydrolyze protein peptide chains. According to the way they degrade polypeptides, they are divided into two categories: endopeptidases and telopeptidases. The former can cut the large molecular weight polypeptide chain from the middle to form prions and peptones with smaller molecular weights; the latter can be divided into carboxypeptidase and aminopeptidase, which respectively remove the peptide from the free carboxyl terminus or free amino terminus of the polypeptide one by one. Chain hydrolysis produces amino acids.
A general term for a class of enzymes that hydrolyze peptide bonds in proteins. According to the way they hydrolyze polypeptides, they can be divided into endopeptidases and exopeptidases. Endopeptidase cleaves the interior of the protein molecule to form smaller molecular weight peptones and peptones. Exopeptidase hydrolyzes peptide bonds one by one from the end of the free amino group or carboxyl group of protein molecules, and frees amino acids, the former is aminopeptidase and the latter is carboxypeptidase. Proteases can be classified into serine proteases, sulfhydryl proteases, metalloproteases and aspartic proteases according to their active centers and optimum pH. According to the optimum pH value of its reaction, it is divided into acidic protease, neutral protease and alkaline protease. The proteases used in industrial production are mainly endopeptidases.
Proteases are widely found in animal offal, plant stems and leaves, fruits and microorganisms. Microbial proteases are mainly produced by molds and bacteria, followed by yeast and actinomycetes.
Enzymes that catalyze the hydrolysis of proteins. There are many kinds, the important ones are pepsin, trypsin, cathepsin, papain and subtilisin. Proteases have strict selectivity for the reaction substrates they act on. A protease can only act on certain peptide bonds in protein molecules, such as the peptide bonds formed by the hydrolysis of basic amino acids catalyzed by trypsin. Proteases are widely distributed, mainly in the digestive tract of humans and animals, and are abundant in plants and microorganisms. Due to limited animal and plant resources, the industrial production of protease preparations is mainly prepared by fermentation of microorganisms such as Bacillus subtilis and Aspergillus terrestris.
Targets for Proteases
- Caspase(91)
- Aminopeptidase(20)
- ACE(74)
- Calpains(11)
- Carboxypeptidase(9)
- Cathepsin(75)
- DPP-4(18)
- Elastase(24)
- Gamma Secretase(46)
- HCV Protease(37)
- HSP(100)
- HIV Integrase(29)
- HIV Protease(34)
- MMP(207)
- NS3/4a protease(4)
- Serine Protease(11)
- Thrombin(48)
- Urokinase(2)
- Cysteine Protease(0)
- Other Proteases(15)
- Tyrosinases(46)
- 15-PGDH(1)
- Acetyl-CoA Carboxylase(14)
- Acyltransferase(56)
- Aldehyde Dehydrogenase (ALDH)(28)
- Aminoacyl-tRNA Synthetase(9)
- ATGL(1)
- Dipeptidyl Peptidase(48)
- Drug Metabolite(466)
- E1/E2/E3 Enzyme(83)
- Endogenous Metabolite(1709)
- FABP(30)
- Farnesyl Transferase(21)
- Glutaminase(16)
- Glutathione Peroxidase(15)
- Isocitrate Dehydrogenase (IDH)(26)
- Lactate Dehydrogenase(18)
- Lipoxygenase(233)
- Mitochondrial Metabolism(209)
- NEDD8-activating Enzyme(6)
- Neprilysin(12)
- PAI-1(13)
- Ser/Thr Protease(46)
- Tryptophan Hydroxylase(11)
- Xanthine Oxidase(18)
- MALT1(10)
- PCSK9(9)
Products for Proteases
- Cat.No. Product Name Information
-
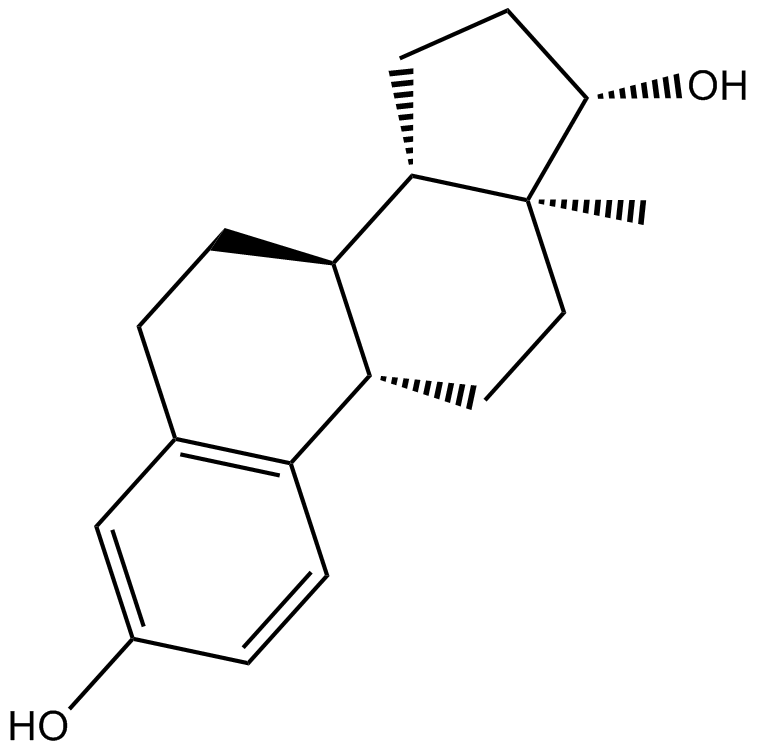
GC11282
β-Estradiol
β-Estradiol, Estradiol, 17β-Oestradiol, E2
Sex hormone
-
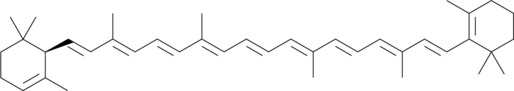
GC41183
α-Carotene
all-trans-α-Carotene
α-Carotene is a precursor of vitamin A that has been found in various fruits and vegetables.
-
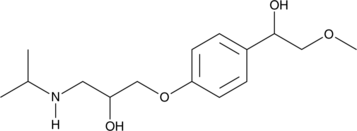
GC45208
α-hydroxy Metoprolol
α-hydroxy Metoprolol is an active metabolite of the β1-adrenergic receptor blocker metoprolol.
-
GC48948
α-Ketoglutaric Acid (sodium salt)
α-KGA, 2-Oxoglutaric Acid
α-Ketoglutaric Acid (sodium salt) (Alpha-Ketoglutaric acid Sodium) is an intermediate in the production of ATP or GTP in the Krebs cycle.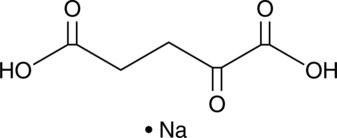
-
GC40718
α-Muricholic Acid
5β-Cholanic Acid-3α,6β,7α-triol
α-Muricholic acid is a murine-specific primary bile acid.
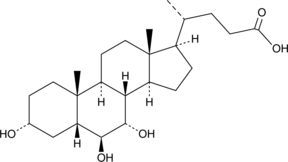
-
GC40480
α-Phenyl-α-(2-pyridyl)acetonitrile
NSC 16276, 2-Pyridylphenylacetonitrile
α-Phenyl-α-(2-pyridyl)thioacetamide, also known as antigastrin and SC-15396, is an inhibitor of gastric acid secretion.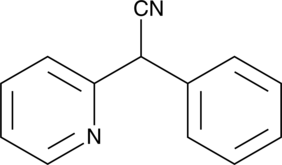
-
GC38287
α-Pyridone
α-Pyridone is an endogenous metabolite.
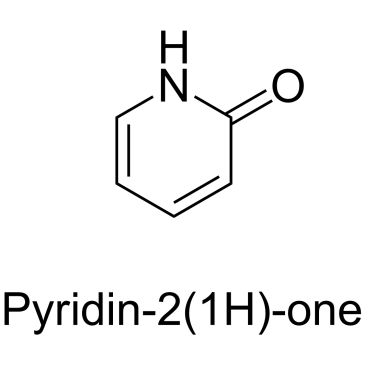
-
GC38000
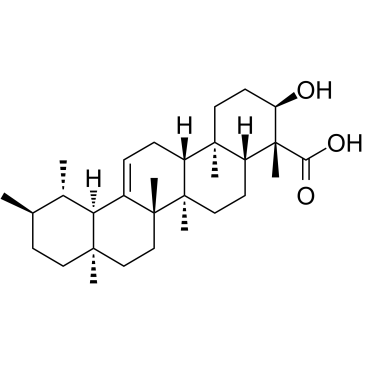
β-Boswellic acid
A pentacyclic triterpene with diverse bioactivities
-
GC71980
β-Caryophyllene-d2
β-Caryopllene-d2 is deuterium labeled β-Caryopllene.

-
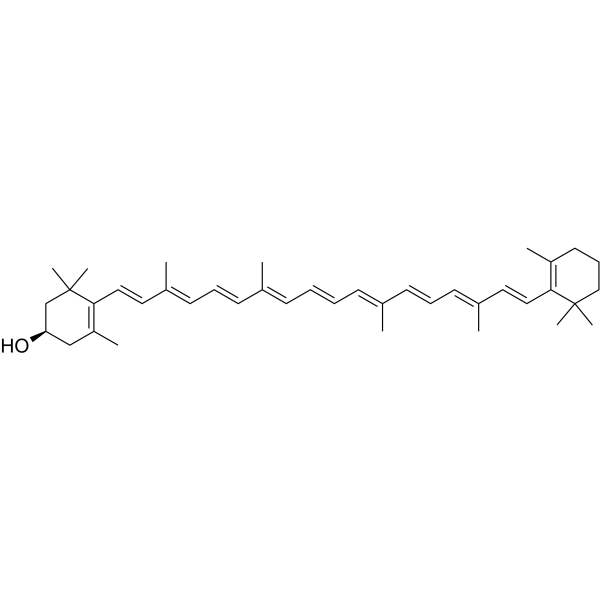
GC63275
β-Cryptoxanthin
β-Cryptoxanthin ((3R)-β-Cryptoxanthin), isolated from Satsuma mandarin orange, is an oxygenated carotenoid and a potent antioxidant.
-
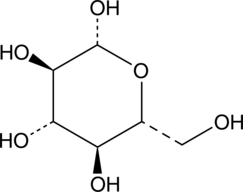
GC40777
β-D-Glucose
NSC 287045
D-Glucose, a naturally occurring monosaccharide found in plants, is the primary energy source for living organisms.
-
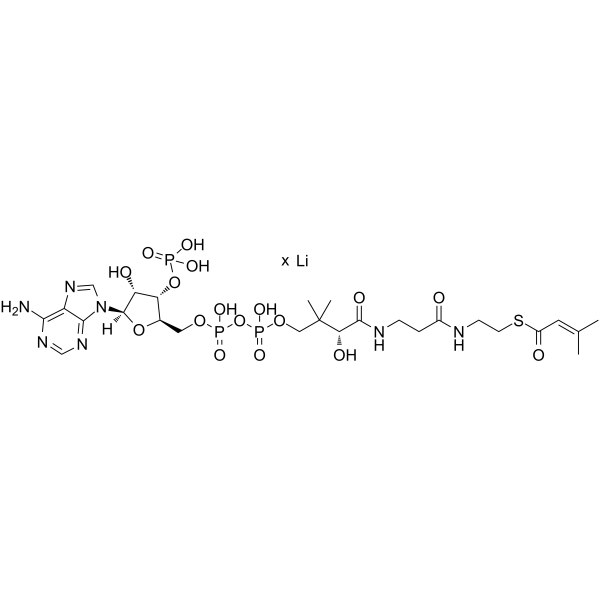
GC66842
β-Methylcrotonyl coenzyme A lithium
β-Methylcrotonyl coenzyme A lithium is an intermediate in leucine metabolism and can be used as a substrate to study the specificity and kinetics of β-methylcrotonyl coenzyme A carboxylase (MCCase).
-
GC40719
β-Muricholic Acid
5β-Cholanic Acid-3α,6β,7β-triol, β-MCA
β-Muricholic acid is a naturally occurring trihydroxy hydrophilic bile acid, as a biliary cholesterol-desaturating agent and facilitates the dissolution of cholesterol gallstones.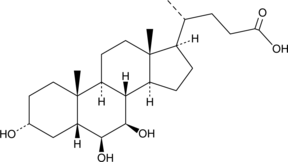
-
GC67484
β-Nicotinamide adenine dinucleotide reduced dipotassium
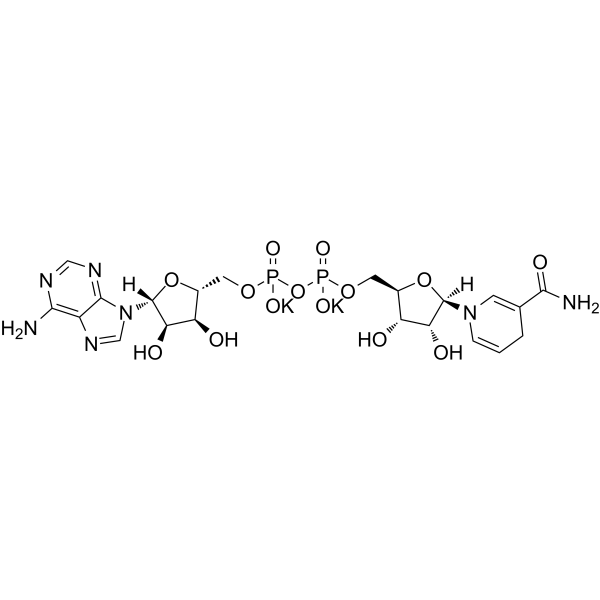
β-Nicotinamide adenine dinucleotide reduced dipotassium is an orally active reduced coenzyme. β-Nicotinamide adenine dinucleotide reduced dipotassium is a donor of ADP-ribose units in ADP-ribosylaton reactions and a precursor of cyclic ADP-ribose. β-Nicotinamide adenine dinucleotide reduced dipotassium plays a role as a regenerative electron donor in cellular energy metabolism, including glycolysis, β-oxidation and the tricarboxylic acid (TCA) cycle.
-
GC70185
β-Sitostenone
δ4-Sitosterol-3-one; β-Rosasterol oxide
Beta-sitostenone is a sterol isolated from Cochlospermum vitifolium. It can inhibit the activity of tyrosinase and has anti-melanogenesis and anti-tumor activities.

-
GC38010
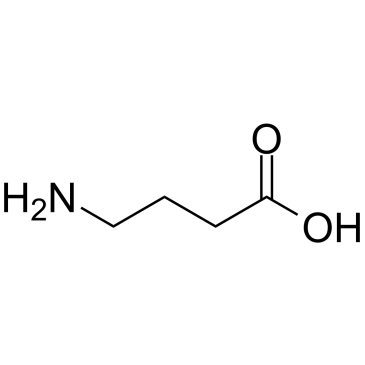
γ-Aminobutyric acid
-
GC63279
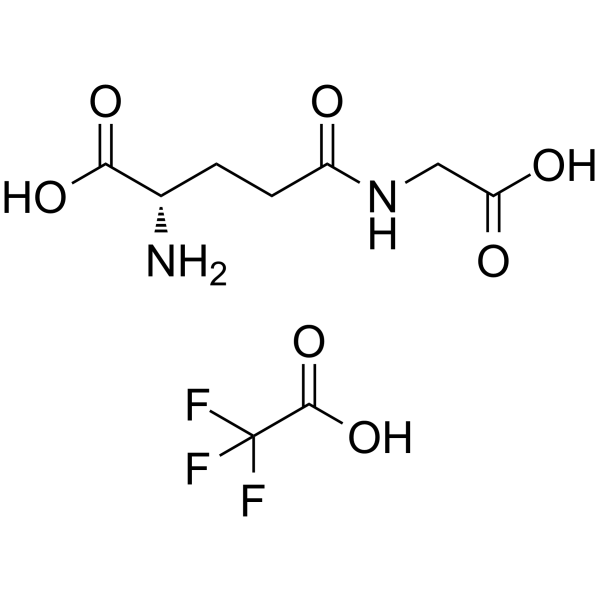
γ-Glu-Gly TFA
-
GC38011
γ-Secretase modulator 4
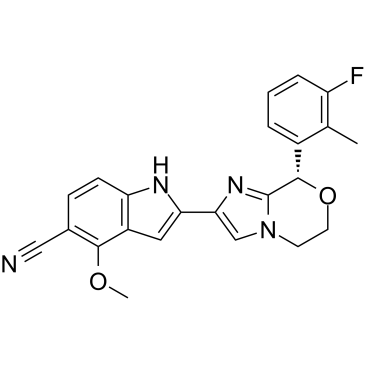
-
GC66048
δ-Secretase inhibitor 11
δ-Secretase inhibitor 11 (compound 11) is an orally active, potent, BBB-penetrated, non-toxic, selective and specific δ-secretase inhibitor, with an IC50 of 0.7 μM. δ-Secretase inhibitor 11 interacts with both the active site and allosteric site of δ-secretase. δ-Secretase inhibitor 11 attenuates tau and APP (amyloid precursor protein) cleavage. δ-Secretase inhibitor 11 ameliorates synaptic dysfunction and cognitive impairments in tau P301S and 5XFAD transgenic mouse models. δ-Secretase inhibitor 11 can be used for Alzheimer's disease research.
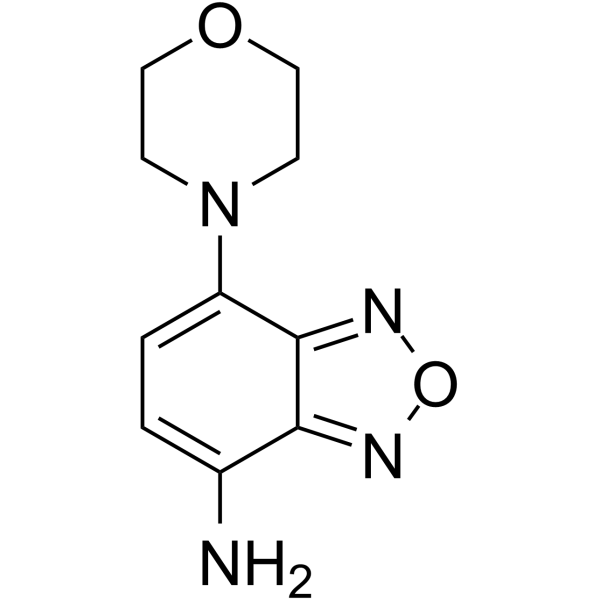
-
GC15975
α-Estradiol
Alfatradiol, α-Estradiol, 17-epi Estradiol, NSC 20293, 17α-Oestradiol
α-Estradiol is a weak estrogen and a 5α-reductase inhibitor which is used as a topical medication in the treatment of androgenic alopecia.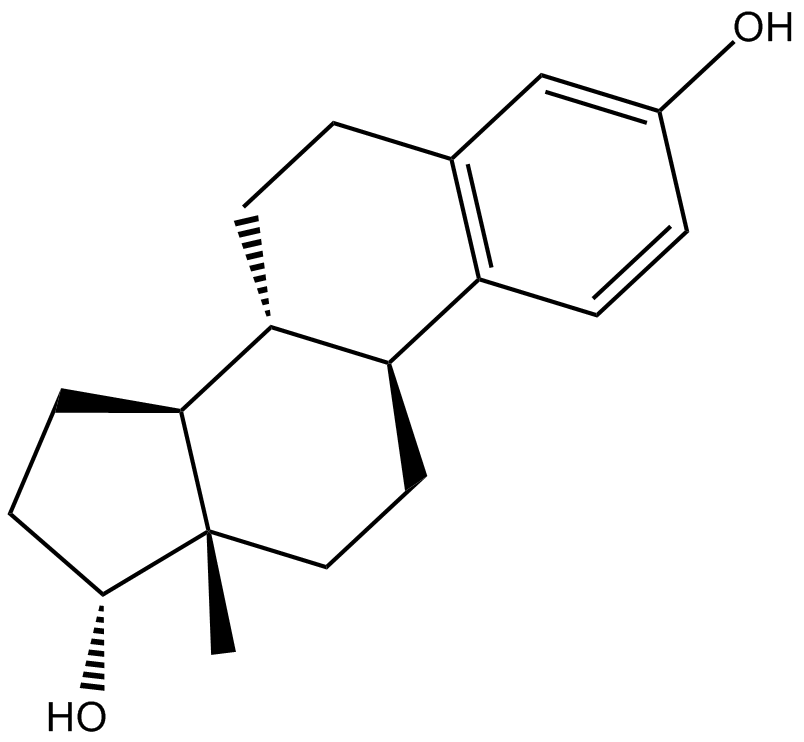
-
GC30187
γ-Glu-Phe (γ-Glutamylphenylalanine)
γ-Glutamylphenylalanine
γ-Glu-Phe (γ-Glutamylphenylalanine) (γ-Glutamylphenylalanine) is synthesized by Bacillus amyloliquefaciens (GBA) and Aspergillus oryzae (GAO).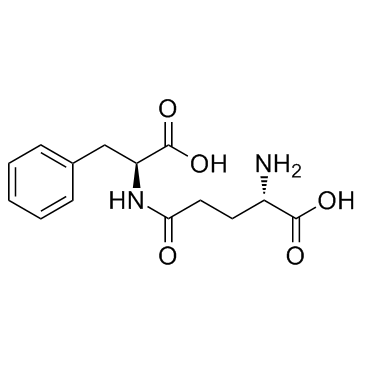
-
GC41552
ω-3 Arachidonic Acid
ω3 AA
ω-3 Arachidonic acid is a rare PUFA found in trace amounts in dietary sources.
-
GC40259
(±)-β-Tocopherol
DL-β-Tocopherol, rac-β-Tocopherol
(±)-β-Tocopherol is a lipid-soluble form of vitamin E with antioxidant activity.
-
GC40260
(±)-γ-Tocopherol
7,8-Dimethyltocol, all-rac-γ-Tocopherol, DL-γ-Tocopherol
(±)-γ-Tocopherol is a form of vitamin E with antioxidant and anti-inflammatory properties.
-
GC41661
(±)-4-hydroxy Propranolol β-D-Glucuronide
(±)-4-hydroxy Propranolol β-D-glucuronide is a metabolite of (±)-4-hydroxy propranolol, which is a metabolite of propranolol.
-
GC34961
(±)-BI-D
(±)-BI-D is a potent ALLINI(An allosteric IN inhibitor) that binds integrase at the LEDGF/p75 binding site.
-
GC46284
(±)-Cotinine-d3
An internal standard for the quantification of cotinine
-
GC41670
(±)-Epinephrine (hydrochloride)
(±)-Adrenaline, DL-Adrenaline, DL-Epinephrine
(±)-Epinephrine is a natural neurotransmitter that is released from the adrenal medulla and activates adrenoceptors (Kis = 15, 735, and 3,970 nM for α1A-, β2-, and β1-adrenergic receptors, respectively).
-
GC41671
(±)-Equol 4'-sulfate (sodium salt)
R,S-Equol 4'-Sulfate
(±)-Equol 4'-sulfate is a gut-mediated phase II metabolite of the isoflavonoid phytoestrogen (±)-equol.
-
GC41315
(±)-Ketoprofen Glucuronide
rac-Ketoprofen Acyl-β-D-glucuronide, (R,S)-Ketoprofen Glucuronide
(±)-Ketoprofen glucuronide is a phase II metabolite of the non-steroidal anti-inflammatory drug (NSAID) ketoprofen.
-
GC38369
(±)-Leucine

-
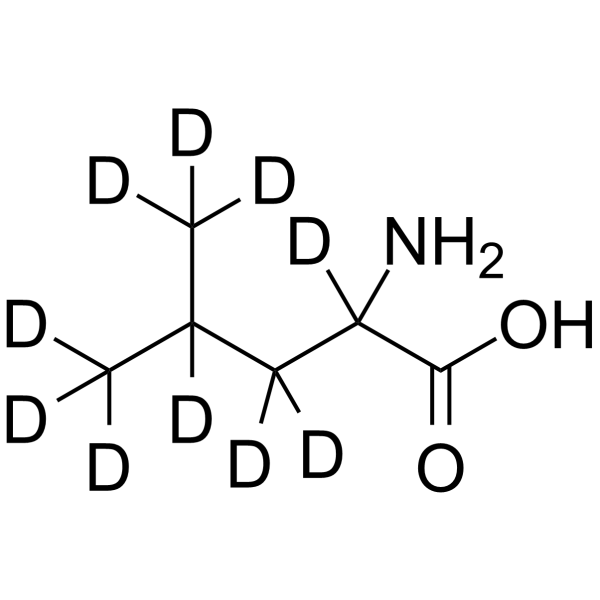
GC65599
(±)-Leucine-d10
-
GC49875
(±)-N-desmethyl Venlafaxine (hydrochloride)
Wy 45494
A minor active metabolite of venlafaxine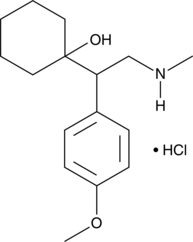
-
GC39271
(±)-Naringenin
SDihydrogenistein, NSC 11855, NSC 34875, Salipurol
A citrusderived flavonoid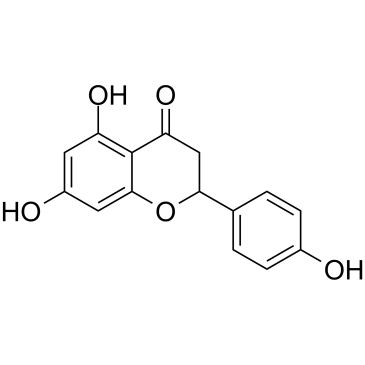
-
GC13890
(±)-Palmitoylcarnitine chloride
C16:0 Carnitine, CAR 16:0, DL-Carnitine hexadecanoyl ester, DL-Carnitine palmitoyl ester, Hexadecanoyl-DL-carnitine, DL-Hexadecanoylcarnitine, NSC 628323, DL-Palmitoylcarnitine
intermediate in mitochondrial fatty acid oxidation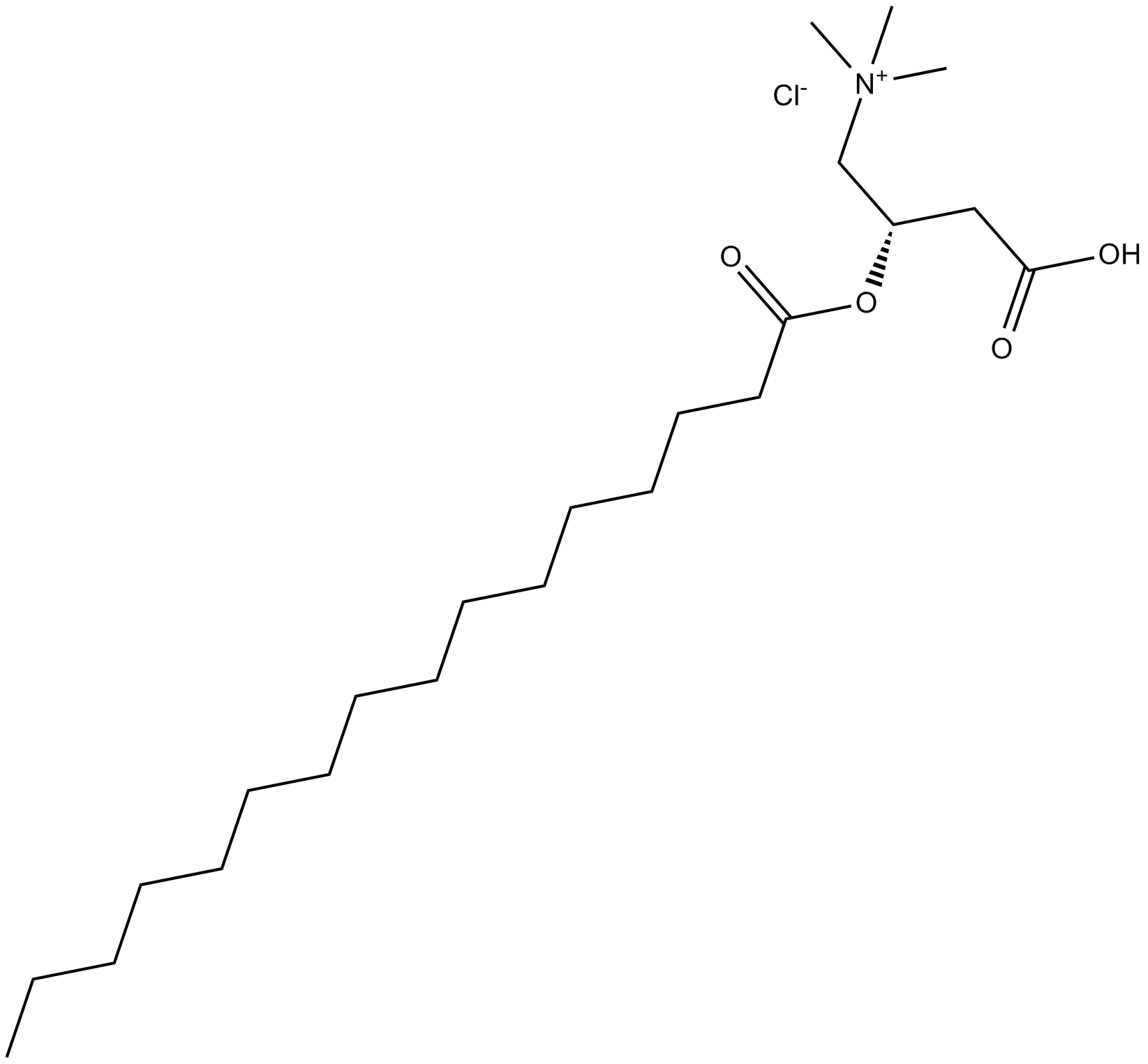
-
GC40229
(±)-Warfarin-d5
(±)-Warfarin-d5 is intended for use as an internal standard for the quantification of warfarin by GC- or LC-MS.

-
GC41649
(±)13-HODE cholesteryl ester
(±)13-HODE cholesteryl ester was originally extracted from atherosclerotic lesions and shown to be produced by Cu2+-catalyzed oxidation of LDL.

-
GC19444
(±)20-HDHA
20hydroxy Docosahexaenoic Acid, (±)20HDoHE
An autoxidation product of DHA and potential marker of oxidative stress
-
GC40828
(±)5-HETE lactone
(±)5-Hydroxyeicosatetraenoic Acid lactone
(±)5-HETE lactone is a cyclic ester formed by acid-catalyzed nucleophilic addition of the C-5 hydroxyl to the C-1 carboxyl of (±)5-HETE.
-
GC40442
(±)8-HETE
(±)8-Hydroxyeicosatetraenoic Acid
(±)8-HETE is one of the six monohydroxy fatty acids produced by the non-enzymatic oxidation of arachidonic acid.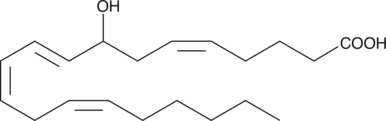
-
GC40801
(±)9(10)-DiHOME
Leukotoxin diol
Leukotoxin is the 9(10) epoxide of linoleic acid, generated by neutrophils during the oxidative burst.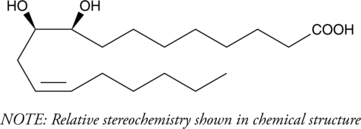
-
GC41666
(±)9-HODE cholesteryl ester
(±)9-HODE cholesteryl ester was originally extracted from atherosclerotic lesions and shown to be produced by Cu2+-catalyzed oxidation of LDL.
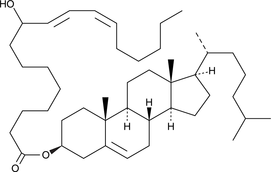
-
GC30586
(±) Anabasine
(±) Anabasine is a biphasic muscle relaxant.
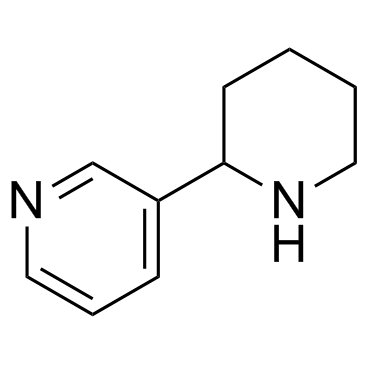
-
GC61647
(+)-Longifolene
(+)-Longifolene is a sesquiterpenoid and a metabolite in rabbits.
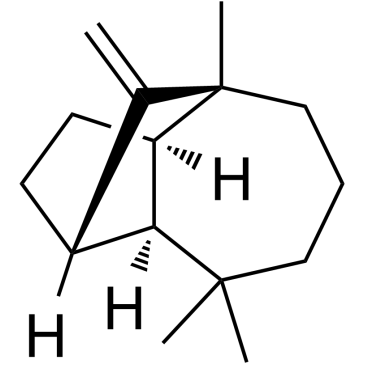
-
GC10675
(+,-)-Octopamine HCl
β,4-Dihydroxyphenethylamine, Epirenor, Norfen, NSC 108685, (±)-4-Octopamine, (±)-p-Octopamine
Octopamine ((±)-p-Octopamine) hydrochloride, a biogenic monoamine structurally related to noradrenaline, acts as a neurohormone, a neuromodulator and a neurotransmitter in invertebrates.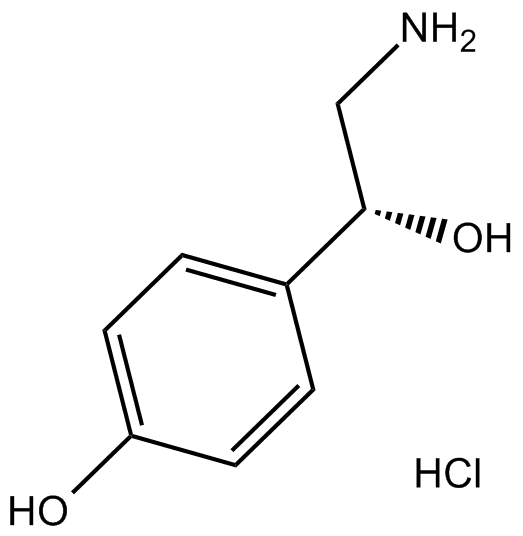
-
GC45245
(-)-Caryophyllene oxide
BCP oxide, β-Caryophyllene epoxide, β-Caryophyllene oxide
(-)-Caryophyllene oxide is a bicyclic sesquiterpene and a metabolite of β-caryophyllene that has been found in C.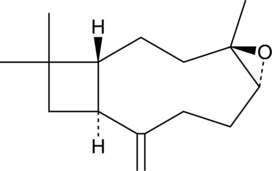
-
GC17470
(-)-Cotinine
NIH 10498
α3/α6β2 nAChR activator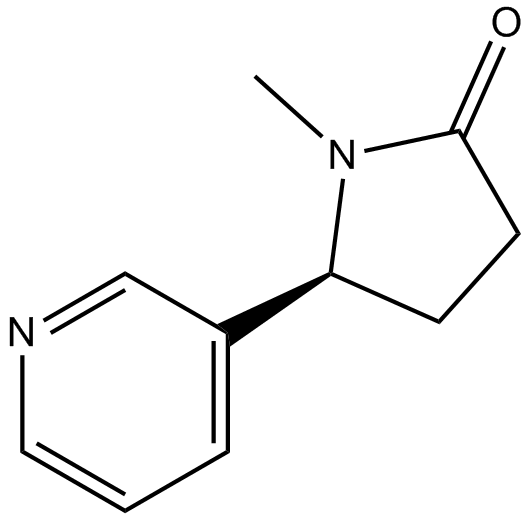
-
GC17242
(-)-epigallocatechin
(-)EGC, epi-Gallocatechin, NSC 674039
green tea epicatechin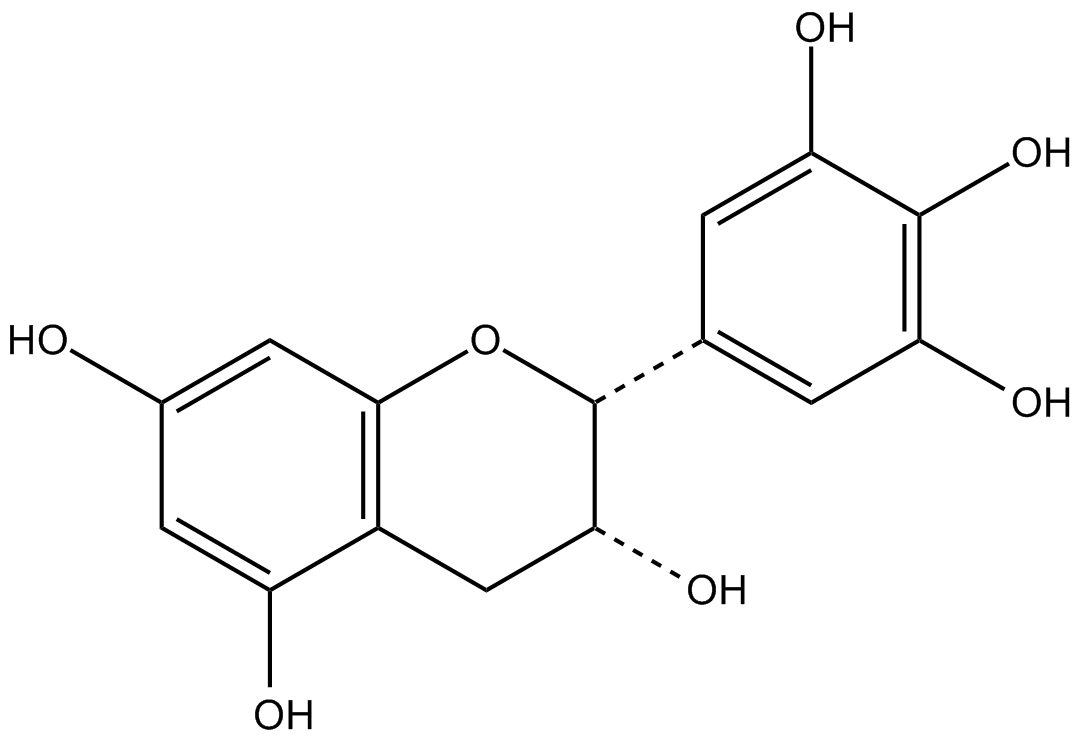
-
GC14049
(-)-Epigallocatechin gallate (EGCG)
EGCG
(-)-Epigallocatechin Gallate sulfate (EGCG) is a major polyphenol in green tea that inhibits cell proliferation and induces apoptosis.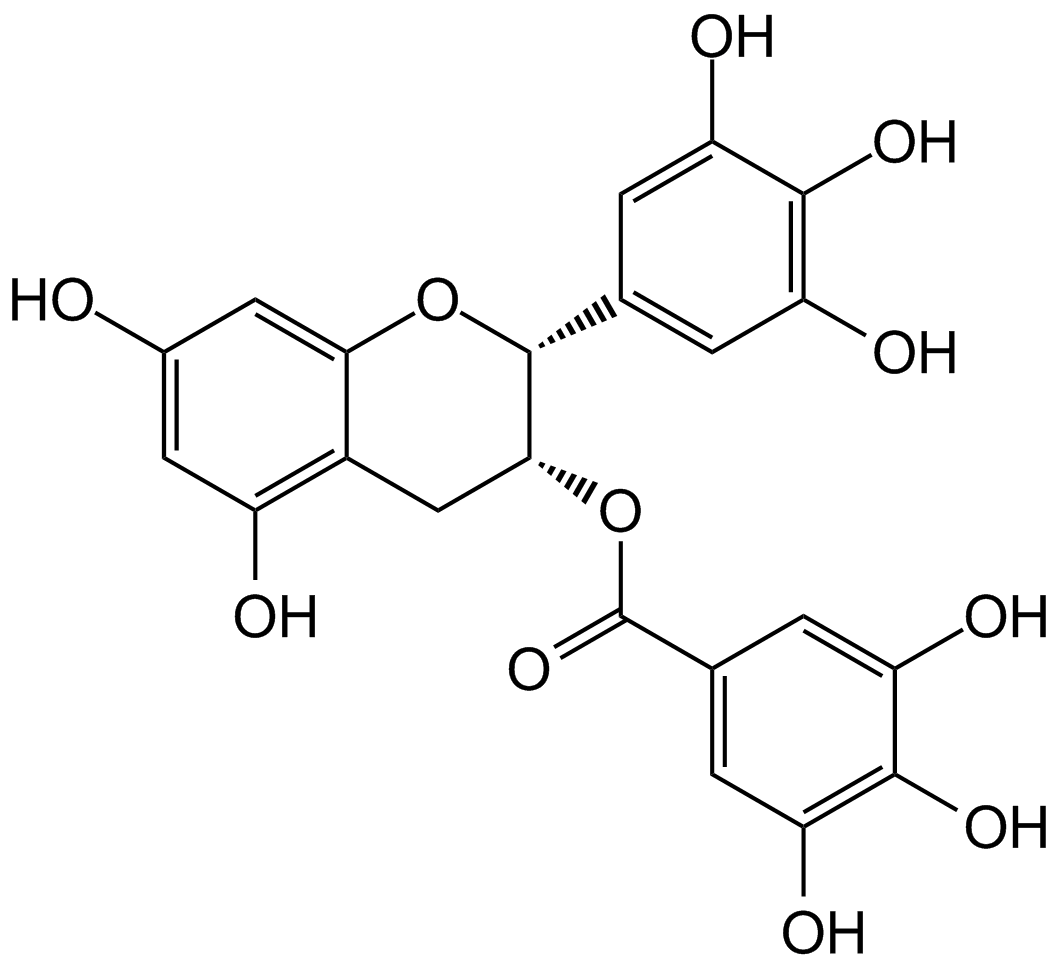
-
GC34951
(-)-Menthol
L-Menthol, (1R,2S,5R)-(-)-Menthol, NSC 62788
A monoterpene with diverse biological activities
-
GC45252
(-)-Sitagliptin Carbamoyl Glucuronide
(R)-Sitagliptin Carbamoyl Glucuronide
(-)-Sitagliptin carbamoyl glucuronide is a minor phase II metabolite of the dipeptidyl peptidase 4 (DPP-4) inhibitor (-)-sitagliptin.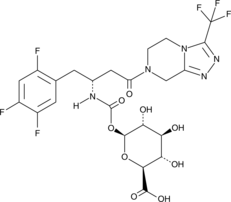
-
GC18622
(2'S)-Nicotine-1-oxide
Nicotine-N-oxide
(2'S)-Nicotine-1-oxide is a metabolite of nicotine .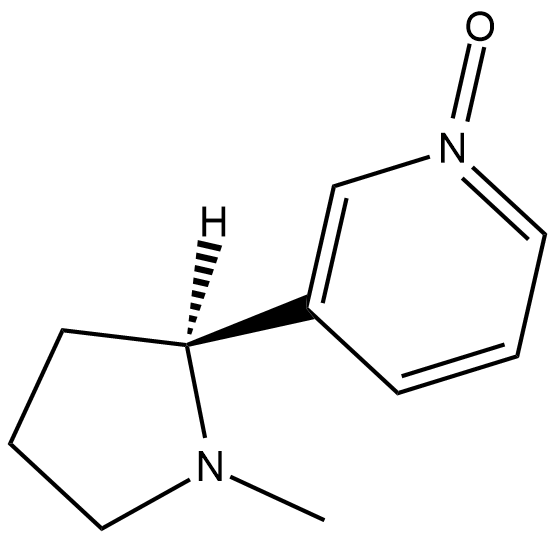
-
GC38299
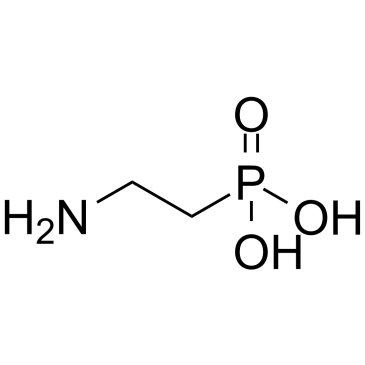
(2-Aminoethyl)phosphonic acid
(2-Aminoethyl)phosphonic acid is an endogenous metabolite.
-
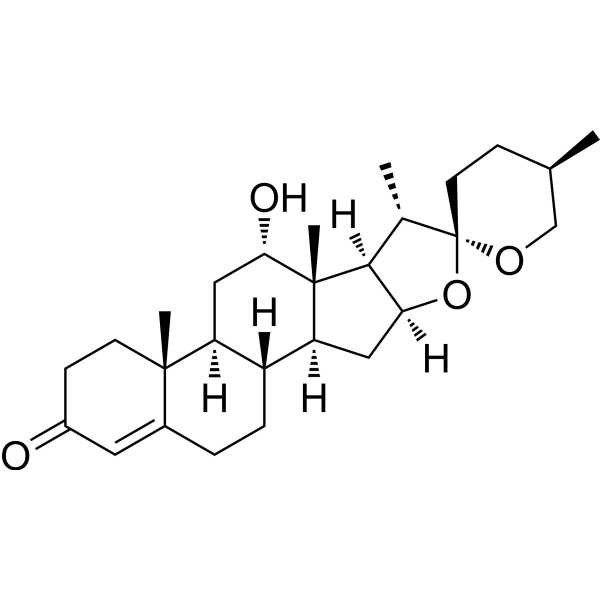
GC68509
(25R)-12α-Hydroxyspirost-4-en-3-one
(25R)-12α-Hydroxyspirost-4-en-3-one is a secondary metabolite produced by Nocardia globerula from Hecogenin.
-
GC38265
(2R,3R)-2,3-Dihydroxysuccinic acid
(2R,3R)-2,3-Dihydroxysuccinic acid (L-(+)-Tartaric acid) is an endogenous metabolite.

-
GC62731
(2R,3R)-Butane-2,3-diol
(2R,3R)-Butane-2,3-diol is an endogenous metabolite.

-
GC38296
(2S,3R,4S,5R)-2-Amino-3,4,5,6-tetrahydroxyhexanal hydrochloride
(2S,3R,4S,5R)-2-Amino-3,4,5,6-tetrahydroxyhexanal hydrochloride is an endogenous metabolite.

-
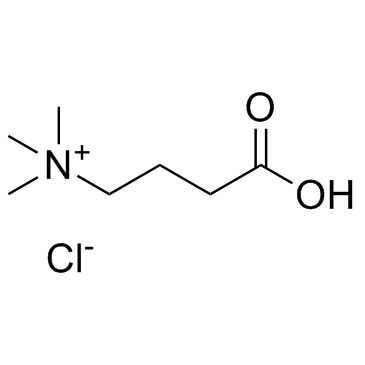
GC33797
(3-Carboxypropyl)trimethylammonium chloride
(3-Carboxypropyl)trimethylammonium chloride is angiopathic substance produced as an intermediary metabolite by gut microbiota that feed on carnitine in dietary red meat.
-
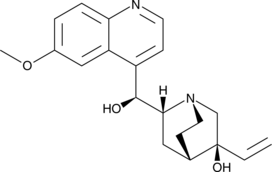
GC41694
(3S)-hydroxy Quinidine
(3S)-hydroxy Quinidine is an active quinidine metabolite.
-
GC38144
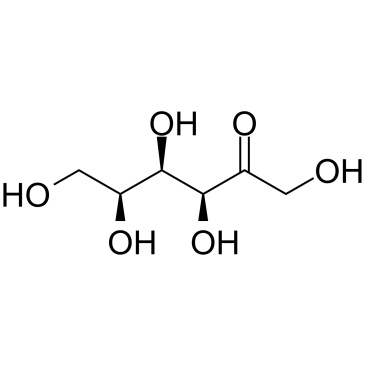
(3S,4R,5S)-1,3,4,5,6-Pentahydroxyhexan-2-one
A monosaccharide
-
GC38283
(3S,4S,5R)-1,3,4,5,6-Pentahydroxyhexan-2-one
(3S,4S,5R)-1,3,4,5,6-Pentahydroxyhexan-2-one (D-(-)-Tagatose) is a rare monosaccharide found in nature with prebiotic characteristics.

-
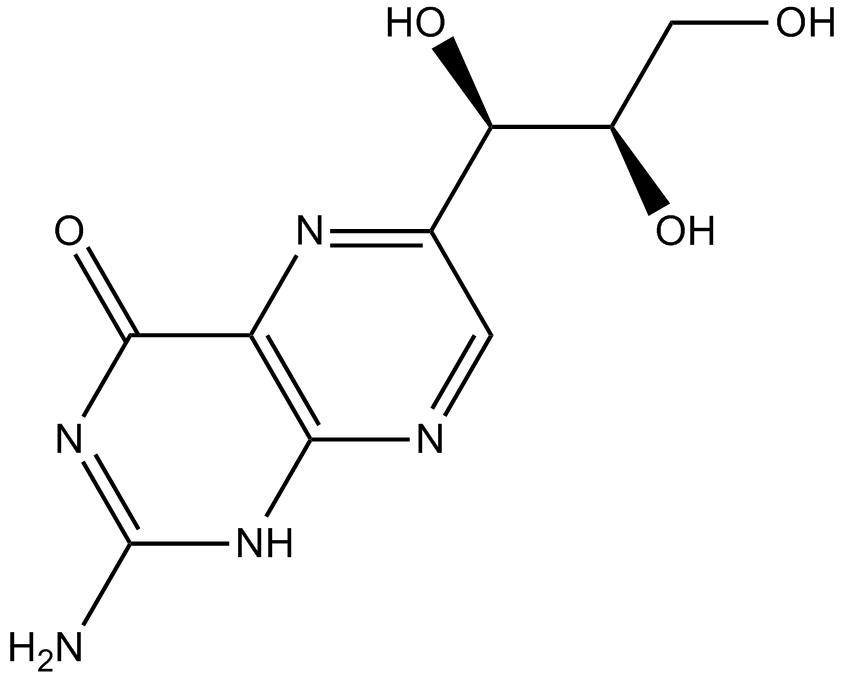
GC12395
(D)-(+)-Neopterin
(D)-(+)-Neopterin (D-(+)-(D)-(+)-Neopterin), a catabolic product of guanosine triphosphate (GTM), serves as a marker of cellular immune system activation.
-
GC60399
(E)-10-Hydroxynortriptyline
10(E)-hydroxy Nortriptyline
(E)-10-Hydroxynortriptyline (E-10-OH-NT) is a metabolite of Nortriptyline.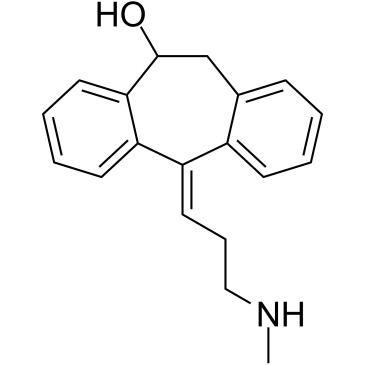
-
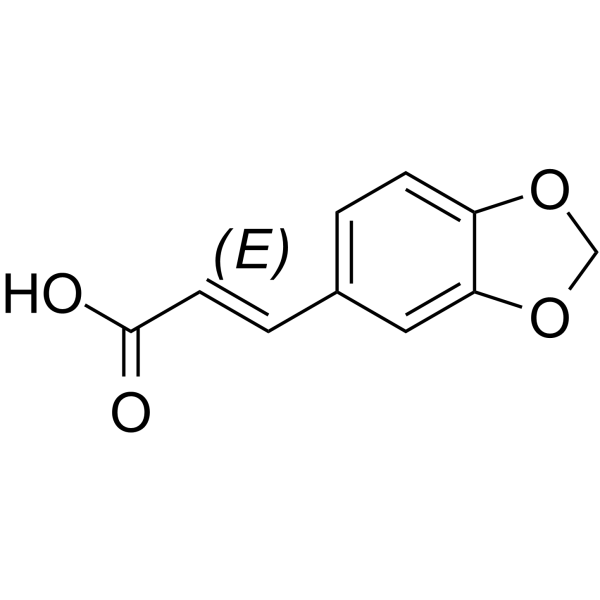
GC65239
(E)-3,4-(Methylenedioxy)cinnamic acid
(E)-3,4-(Methylenedioxy)cinnamic acid is a cinnamic acid derivative obtained from the stem bark of Brombya platynema.
-
GC38684
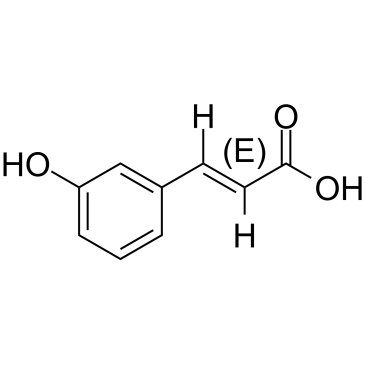
(E)-m-Coumaric acid
(E)-m-Coumaric acid (3-Hydroxycinnamic acid) is an aromatic acid that highly abundant in food.
-
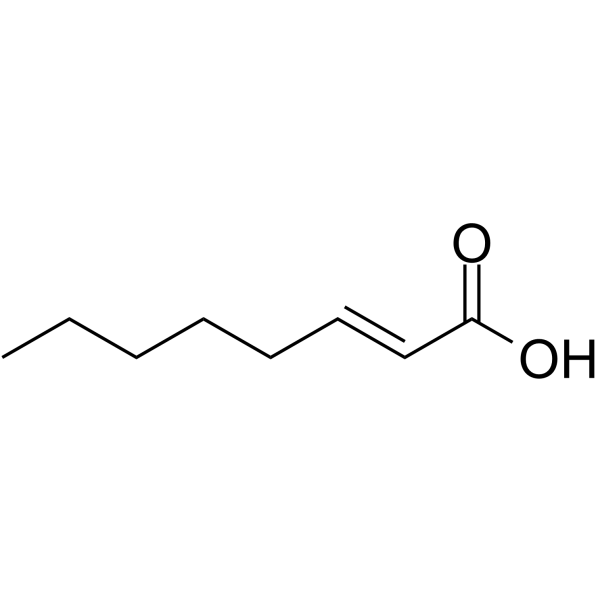
GC62734
(E)-Oct-2-enoic acid
(E)-Oct-2-enoic acid is an endogenous metabolite.
-
GC40286
(E,Z)-2-propyl-2-Pentenoic Acid
2-propyl-2-Pentenoate, 2-propylpenten-2-oic Acid, 2-ene-VPA
(E,Z)-2-propyl-2-Pentenoic acid is a bioactive metabolite of valproic acid that exhibits the same profile and potency of anticonvulsant activity in animal models as its parent compound without any observed teratogenicity and hepatotoxicity.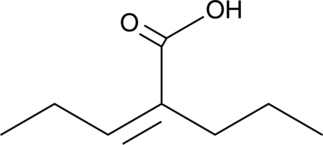
-
GC49189
(E/Z)-4-hydroxy Tamoxifen-d5
Afimoxifene-d5, 4-OHT-d5
An internal standard for the quantification of (E/Z)-4-hydroxy tamoxifen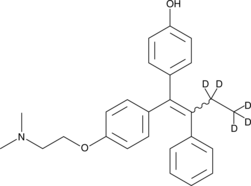
-
GC67476
(E/Z)-Sulindac sulfide
(E/Z)-Sulindac sulfide is a potent γ-secretase modulator (GSM). (E/Z)-Sulindac sulfide selectively reduces Aβ42 production in favor of shorter Aβ species. (E/Z)-Sulindac sulfide can be used for researching Alzheimer's disease.
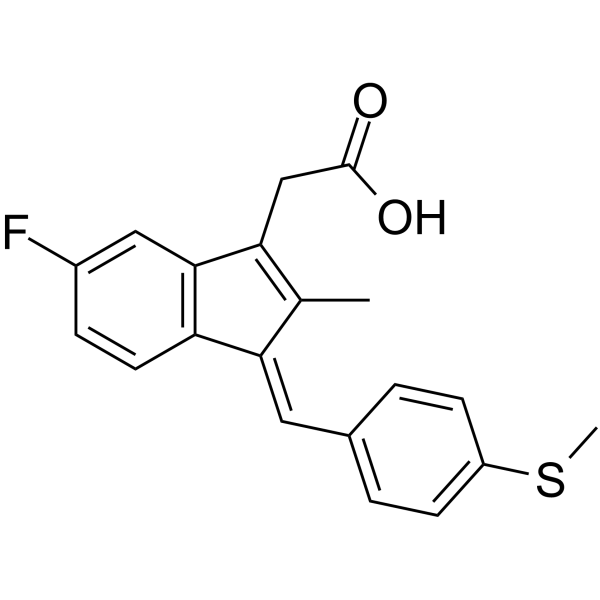
-
GC60404
(Ethoxymethyl)benzene
(Ethoxymethyl)benzene is an endogenous metabolite.
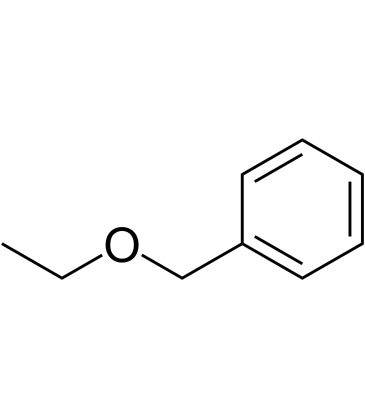
-
GA11210
(H-Cys-OH)2
(–)-Cystine, NSC 13203
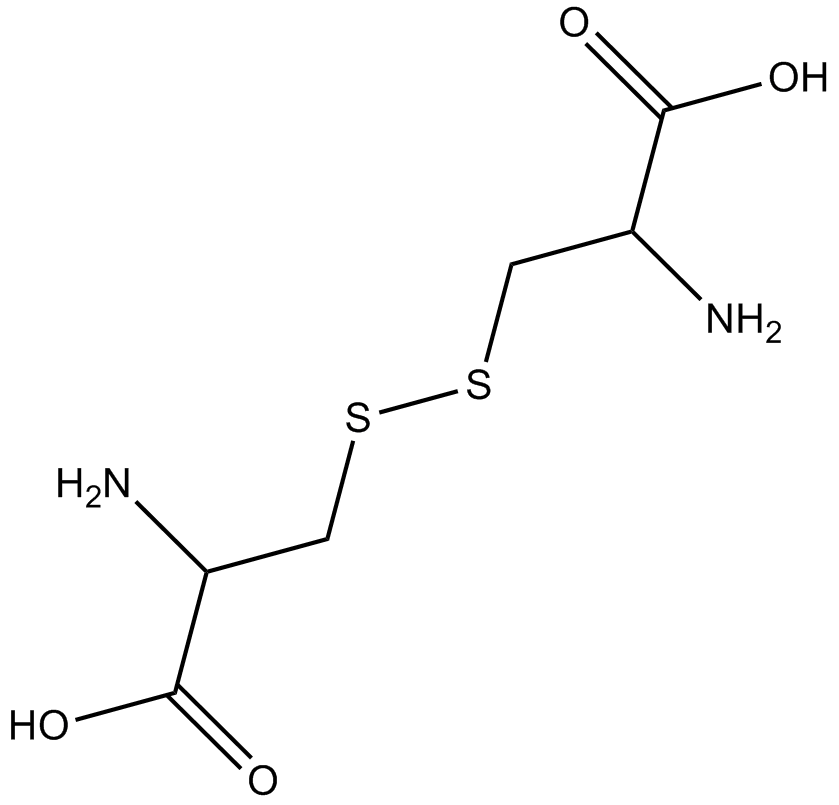
-
GN10783
(R) Ginsenoside Rh2
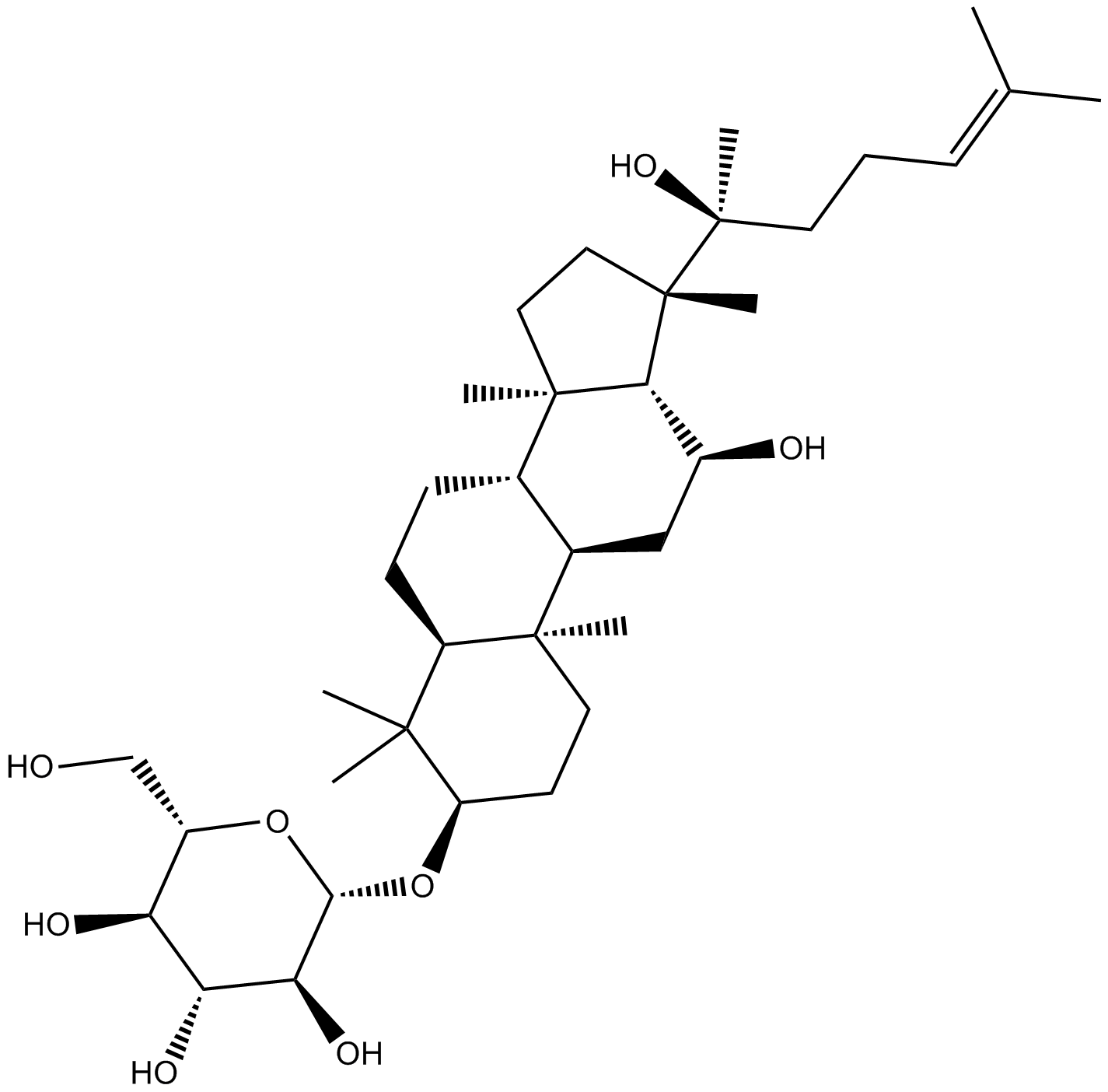
-
GC41721
(R)-α-Lipoic Acid
(R)-(+)-Lipoic Acid
(R)-α-Lipoic acid is the naturally occurring enantiomer of lipoic acid, a cyclic disulfide antioxidant.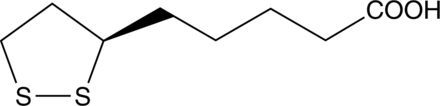
-
GC34442
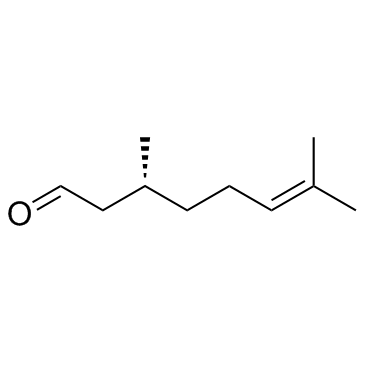
(R)-(+)-Citronellal
(R)-(+)-Citronellal, isolated from citrus, lavender and eucalyptus oils, is a monoterpenoid and main component of citronellal oil with a distinct lemon scent.
-
GC38262
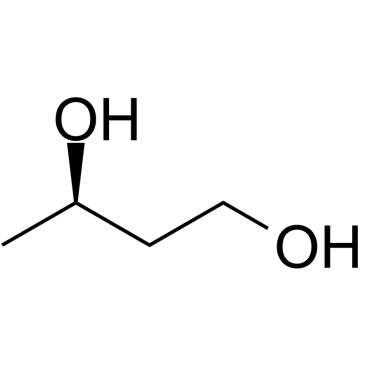
(R)-(-)-1,3-Butanediol
(R)-(-)-1,3-Butanediol is used to regulate the metabolism of carbohydrate and lipid.
-
GC62737
(R)-(-)-O-Desmethyl Venlafaxine D6

-
GC30210
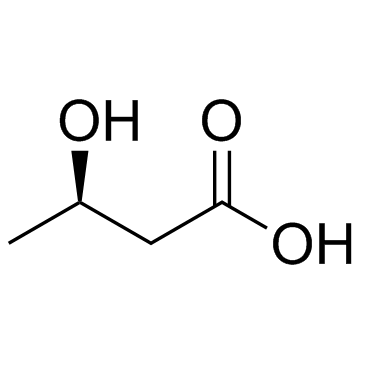
(R)-3-Hydroxybutanoic acid
(R)-3-Hydroxybutanoic acid is a metabolite, and converted from acetoacetic acid catalyzed by 3-hydroxybutyrate dehydrogenase.
-
GC61759
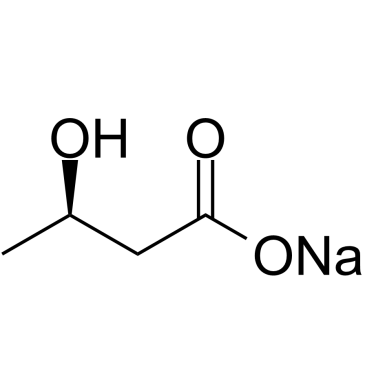
(R)-3-Hydroxybutanoic acid sodium
(R)-3-Hydroxybutanoic acid sodium ((R)-3-Hydroxybutyric acid) is a metabolite converted from acetoacetic acid catalyzed by 3-hydroxybutyrate dehydrogenase.
-
GC71626
(R)-3-Hydroxybutanoic acid-13C2 sodium
(R)-3-droxybutanoic acid-13C2 (sodium) is the 13C labeled (R)-3-droxybutanoic acid (sodium) .

-
GC30661
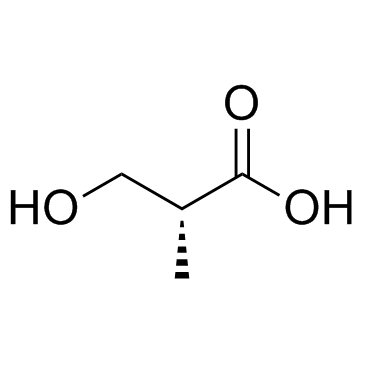
(R)-3-Hydroxyisobutyric acid
(R)-3-Hydroxyisobutyric acid is an intermediate in the pathways of l-valine and thymine and plays an important role in the diagnosis of the very rare inherited metabolic diseases 3-hydroxyisobutyric aciduria and methylmalonic semialdehyde dehydrogenase deficiency.
-
GC38282
(R)-5-Oxopyrrolidine-2-carboxylic acid
(R)-5-Oxoproline, (+)-Pyroglutamic Acid
(R)-5-Oxopyrrolidine-2-carboxylic acid is an endogenous metabolite.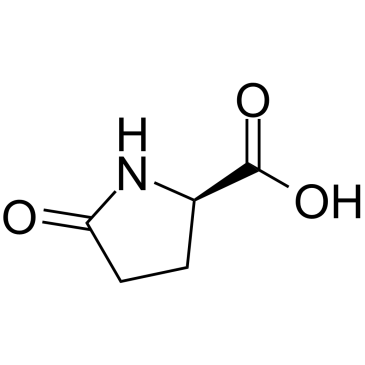
-
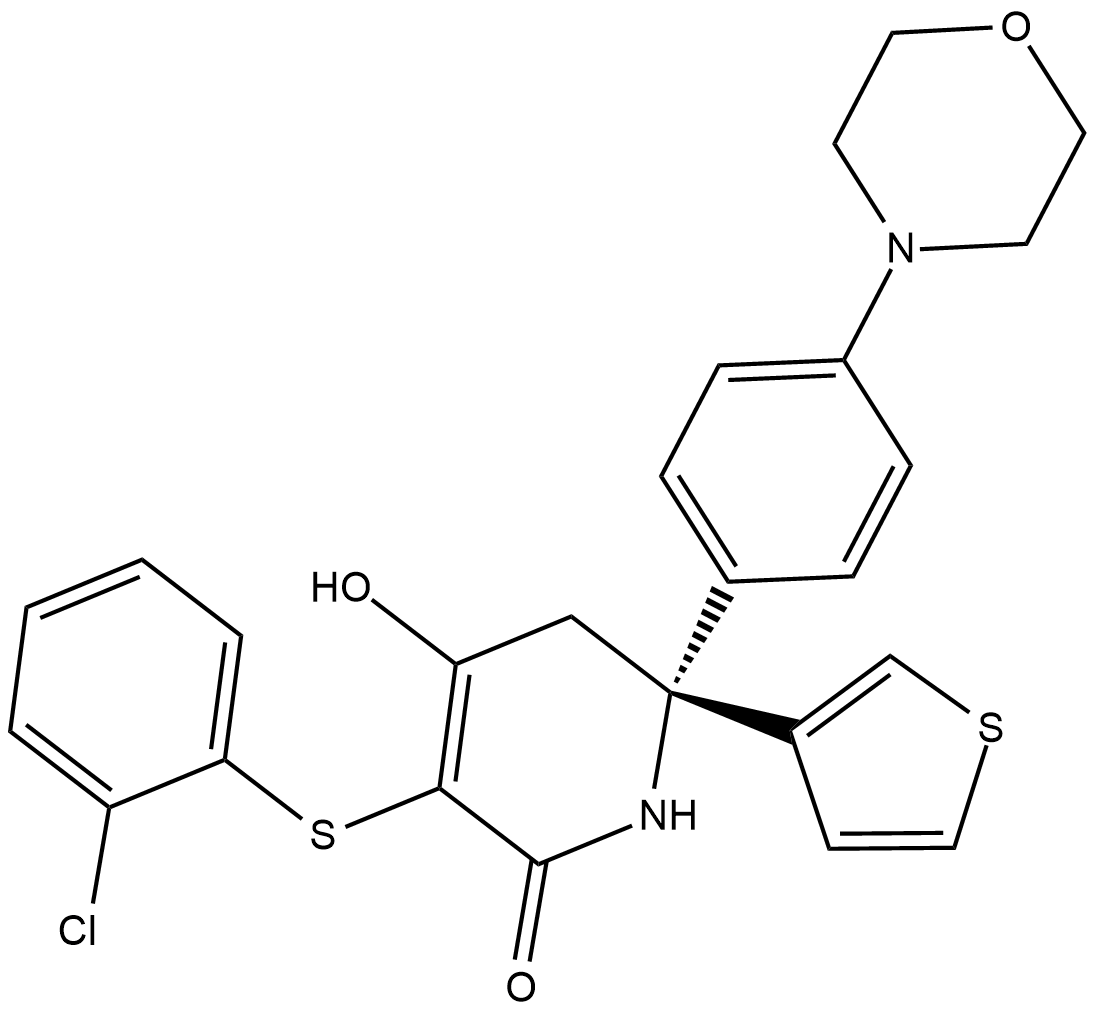
GC19012
(R)-GNE-140
?A TRK kinase (TKI) inhibitor
-
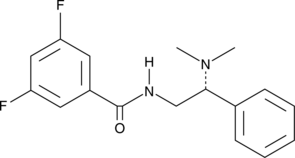
GC52290
(R)-HTS-3
An inhibitor of LPCAT3
-
GC61858
(R)-MLN-4760
(R)-MLN-4760, the R-enantiomer of MLN-4760, is an ACE2 inhibitor, with an IC50 of 8.4 μM.

-
GC38364
(R)-Ornithine hydrochloride

-
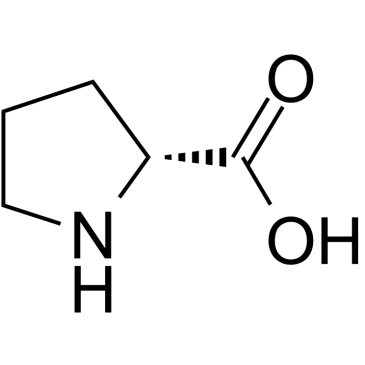
GC38363
(R)-pyrrolidine-2-carboxylic acid
(R)-pyrrolidine-2-carboxylic acid is an endogenous metabolite.
-
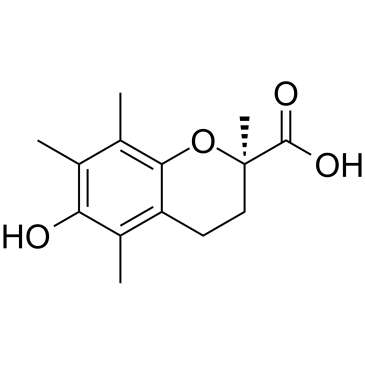
GC38720
(R)-Trolox
(R)-Trolox is a vitamin E analogue and a competitive tyrosinase inhibitor with a Ki value of 0.83 mM and a ID50 value of 1.88 mM. The (R)-Trolox has stronger tyrosinase affinity than the (S) enantiomer (Ki value of 0.61 mM).
-
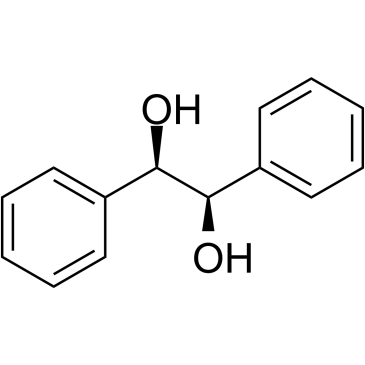
GC39832
(R,R)-(+)-Hydrobenzoin
(R,R)-(+)-Hydrobenzoin is a organocatalysts.
-
GC41722
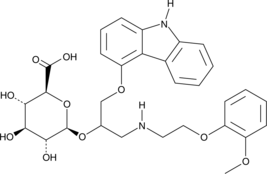
(R,S)-Carvedilol Glucuronide
(R,S)-Carvedilol glucuronide is a racemic mixture of the carvedilol metabolites (R)-carvedilol glucuronide and (S)-carvedilol glucuronide.
-
GC34417
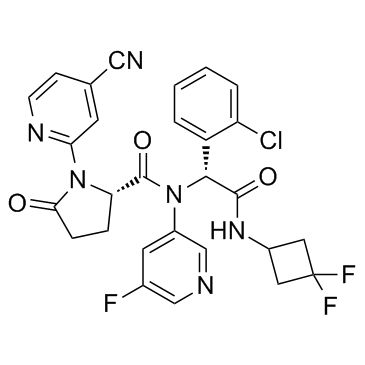
(R,S)-Ivosidenib ((R,S)-AG-120)
-
GC60410
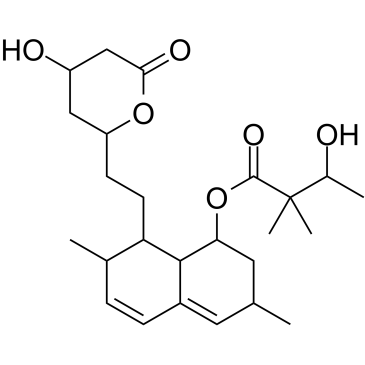
(Rac)-3′-Hydroxy simvastatin
(Rac)-3′-Hydroxy simvastatin is a metabolite of Simvastatin.
-
GC68414
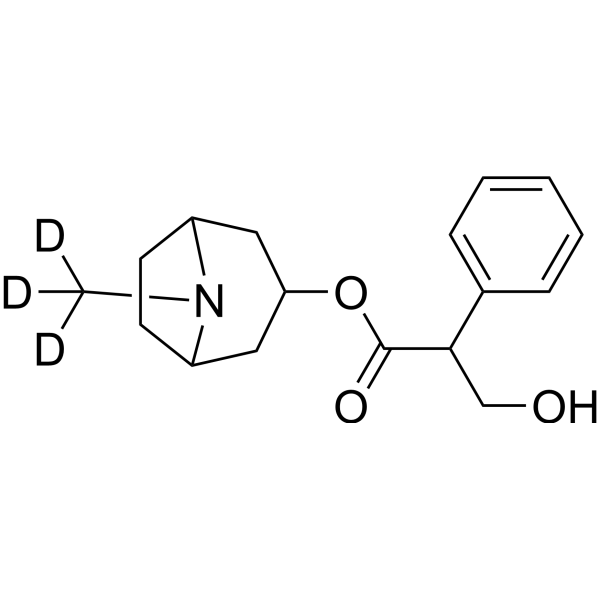
(Rac)-Atropine-d3
(Rac)-Tropine tropate-d3; (Rac)-Hyoscyamine-d3
-
GC67993
(Rac)-Cotinine-d4
(±)-Cotinine-d4; (Rac)-NIH-10498-d4
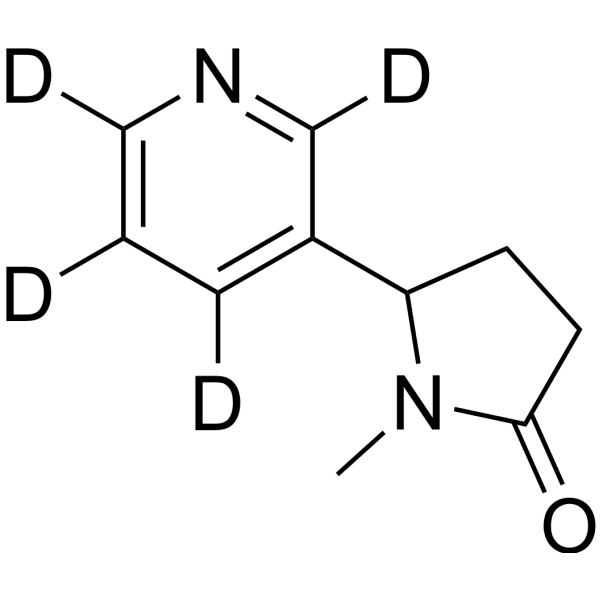
-
GC62744
(Rac)-OSMI-1
(Rac)-OSMI-1 is the racemate of OSMI-1.
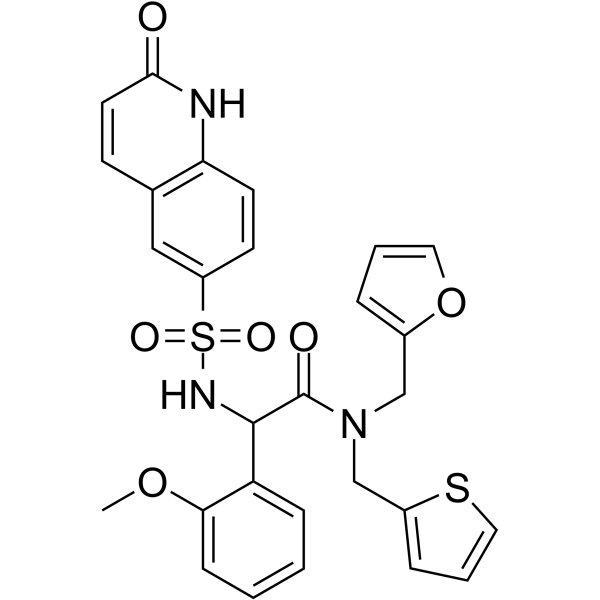
-
GC39833
(S)-(+)-1,2-Propanediol
(S)-(+)-1,2-Propanediol is an endogenous metabolite.
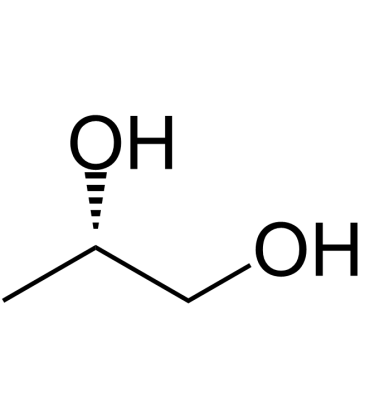
-
GC62747
(S)-(-)-Citronellal
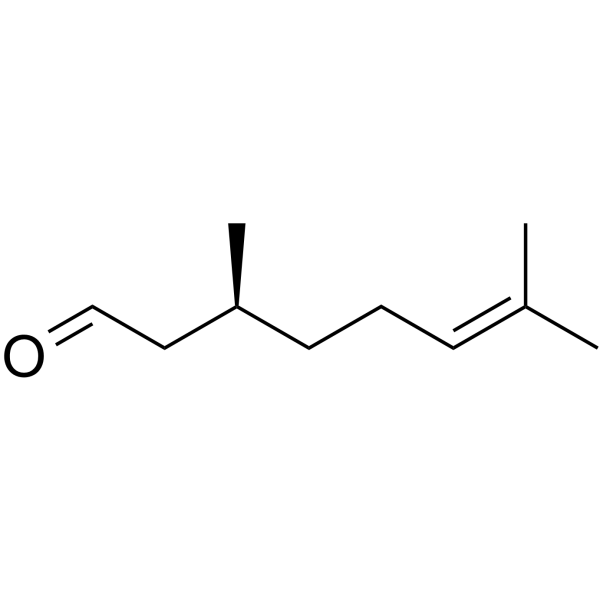
-
GC38371
(S)-(-)-Phenylethanol
(S)-(-)-Phenylethanol is an endogenous metabolite.
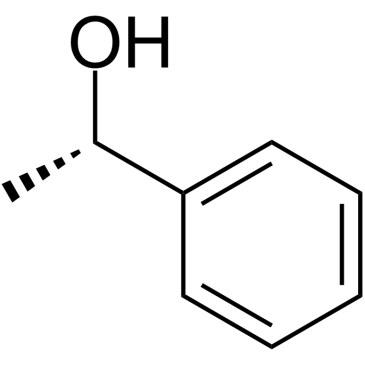
-
GC62748
(S)-2-Amino-3-(4-hydroxy-3,5-diiodophenyl)propanoic acid dihydrate
(S)-2-Amino-3-(4-hydroxy-3,5-diiodophenyl)propanoic acid dihydrate is an endogenous metabolite.
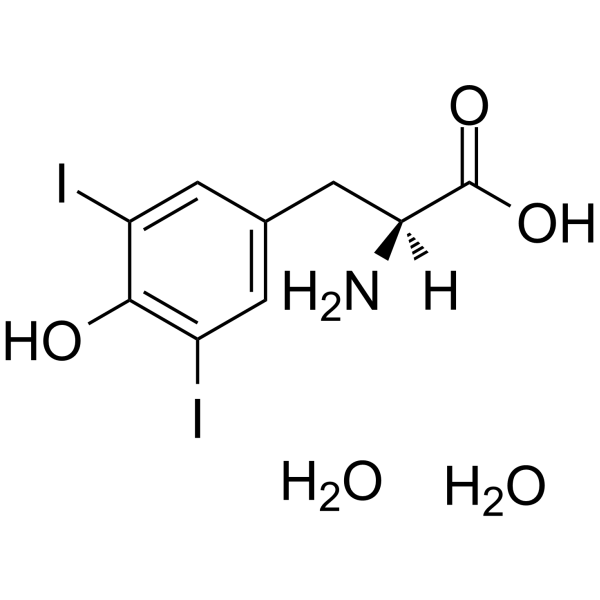
-
GC31630
(S)-2-Hydroxy-3-phenylpropanoic acid
(S)-2-Hydroxy-3-phenylpropanoic acid is a product of phenylalanine catabolism.
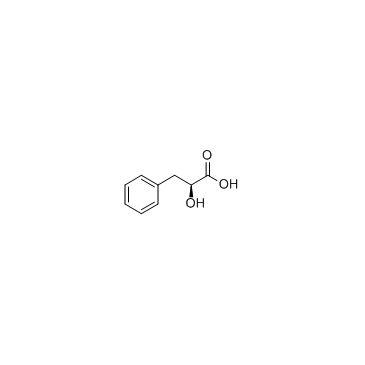
-
GC64746
(S)-2-Hydroxybutanoic acid
(S)-2-Hydroxybutanoic acid is the S-enantiomer of?2-Hydroxybutanoic acid.