PI3K/Akt/mTOR Signaling
- Akt(113)
- AMPK(73)
- CK2(8)
- DNA-PK(22)
- GSK-3(71)
- MELK(7)
- mTOR(104)
- PI3K(206)
- PI4K(14)
- PDK-1(14)
- PIKfyve(4)
- PKB(1)
- S6 Kinase(9)
Products for PI3K/Akt/mTOR Signaling
- Cat.No. Product Name Information
-
GC10350
TIC10 isomer
ONC201 isomer
Potent Akt/ERK inhibitor
-
GC17330
(+)-Usniacin
(+)-Usniacin is isolated from isolated from lichens, binds at the ATP-binding pocket of mTOR, and inhibits mTORC1/2 activity. (+)-Usniacin inhibits the phosphorylation of mTOR downstream effectors: Akt (Ser473), 4EBP1, S6K, induces autophay, with anti-cancer activity. (+)-Usniacin possesses antimicrobial activity against a number of planktonic gram-positive bacteria, including Staphylococcus aureus, Enterococcus faecalis, and Enterococcus faecium.
-
GC63524
(32-Carbonyl)-RMC-5552
(32-Carbonyl)-RMC-5552 is a potent mTOR inhibitor. (32-Carbonyl)-RMC-5552 inhibits mTORC1 and mTORC2 substrate (p-P70S6K-(T389), p-4E-BP1-(T37/36), AND p-AKT1/2/3-(S473)) phosphorylation with pIC50s of > 9, >9 and between 8 and 9, respectively (patent WO2019212990A1, example 2).
-
GC62122
(E)-Akt inhibitor-IV
(E)-AKTIV
(E)-Akt inhibitor-IV ((E)-AKTIV) is a PI3K-Akt inhibitor, with potent cytotoxic.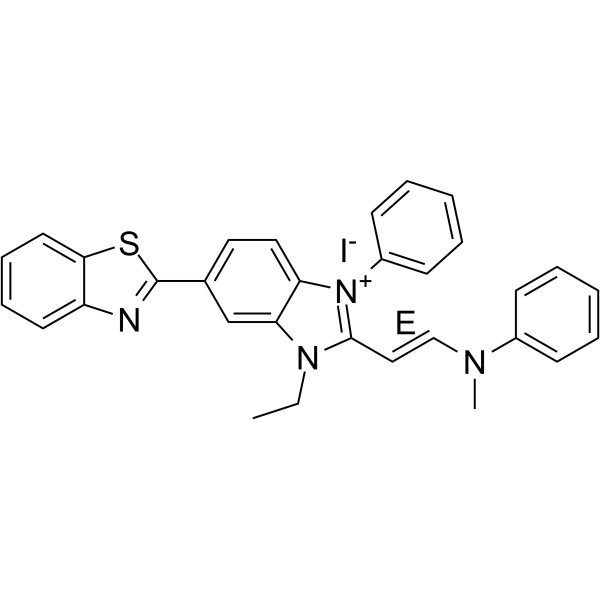
-
GC62736
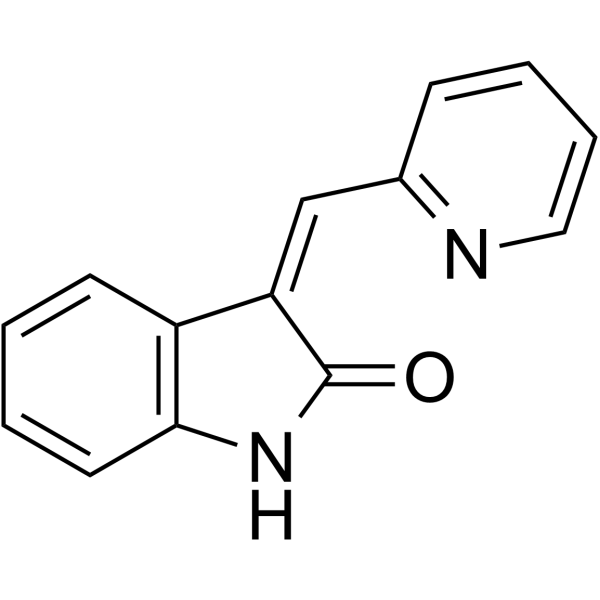
(E/Z)-GSK-3β inhibitor 1
(E/Z)-GSK-3β inhibitor 1 is a racemic compound of (E)-GSK-3β inhibitor 1 and (Z)-GSK-3β inhibitor 1 isomers.
-
GC63802
(R)-PS210
(R)-PS210, the R enantiomer of PS210 (compound 4h-eutomer), is a substrate-selective allosteric activator of PDK1 with an AC50 value of 1.8 μM. (R)-PS210 targets to the PIF-binding pocket of?PDK1. PIF: The protein kinase C-related kinase 2 (PRK2)-interacting fragment.
-
GC62450
(S)-PI3Kα-IN-4
(S)-PI3Kα-IN-4 is a potent inhibitor of PI3Kα, with an IC50 of 2.3 nM. (S)-PI3Kα-IN-4 shows 38.3-, 4.25-, and 4.93-fold selectivity for PI3Kα over PI3Kβ, PI3Kδ, and PI3Kγ, respectively. (S)-PI3Kα-IN-4 can be used for the research of cancer.
-
GC35037
1,3-Dicaffeoylquinic acid
1,5-DCQA, 1,3-Dicaffeoylquinic Acid
1,3-Dicaffeoylquinic acid is a caffeoylquinic acid derivative, and activates PI3K/Akt.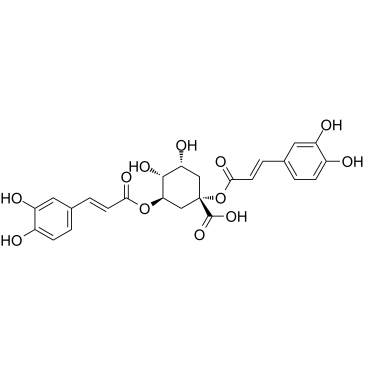
-
GC13907
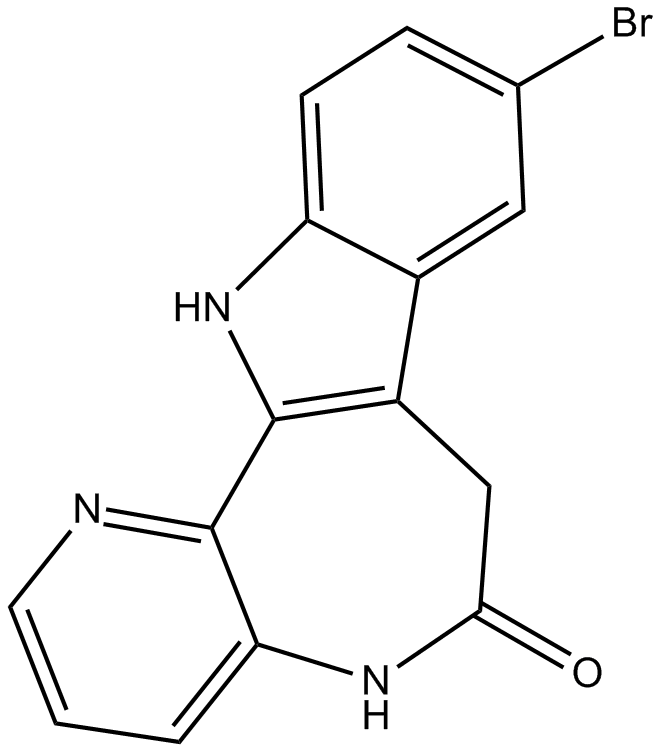
1-Azakenpaullone
GSK-3β inhibitor,potent and selective,cell-permeable
-
GC41992
1-Deoxynojirimycin (hydrochloride)
1-DNJ, 1-dNM, Moranoline
1-Deoxynojirimycin (1-dNM) (hydrochloride), produced by Bacillus species, is a glucose analog that potently inhibits α-glucosidase I and II.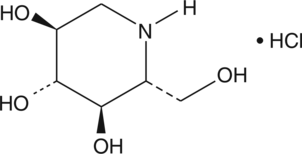
-
GC42018
1-O-Octadecyl-2-O-methyl-sn-glycerol
2Methyl1octadecylsnglycerol, PIA 7
1-O-Octadecyl-2-O-methyl-sn-glycerol is a metabolite of a phosphotidylinositol ether lipid analog (PIA).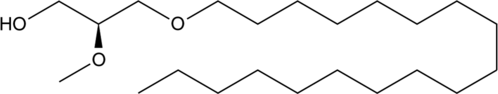
-
GC13006
10-DEBC hydrochloride
Akt Inhibitor X
Akt inhibitor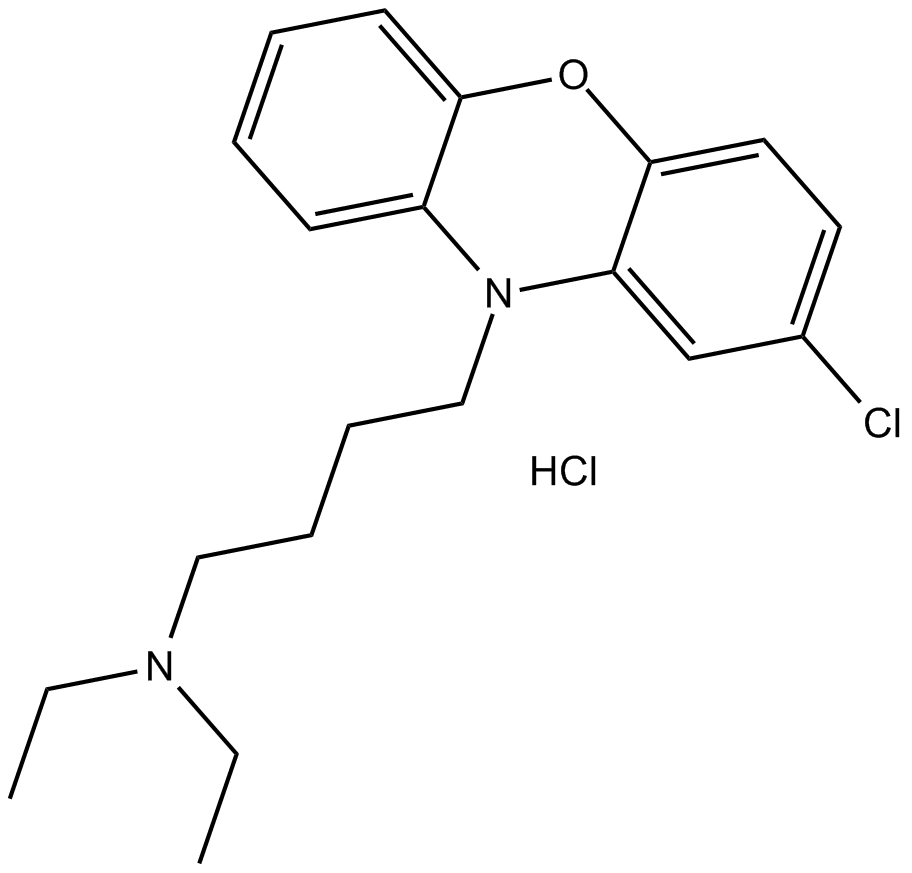
-
GC41979
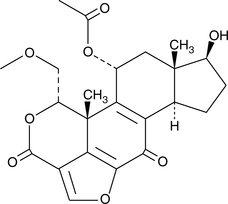
17β-hydroxy Wortmannin
17β-hydroxy Wortmannin is an analog of wortmannin.
-
GC40947
2,3-Dimethoxy-5-methyl-p-benzoquinone
Coenzyme Q0, CoQ0
2,3-Dimethoxy-5-methyl-p-benzoquinone (CoQ0) is a potent, oral active ubiquinone compound can be derived from Antrodia cinnamomea.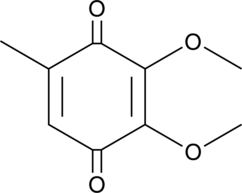
-
GC46531
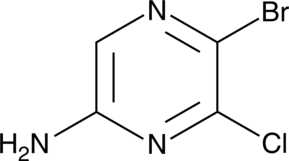
2-Amino-5-bromo-6-chloropyrazine
A heterocyclic building block
-
GC64670
24-Methylenecycloartanyl ferulate
24-Methylenecycloartanyl ferulate is a γ-oryzanol compound. 24-Methylenecycloartanyl ferulate promotes parvin-beta expression in human breast cancer cells. 24-Methylenecycloartanyl ferulate is a potential ATP-competitive Akt1 inhibitor (EC50= 33.3μM).

-
GC35124
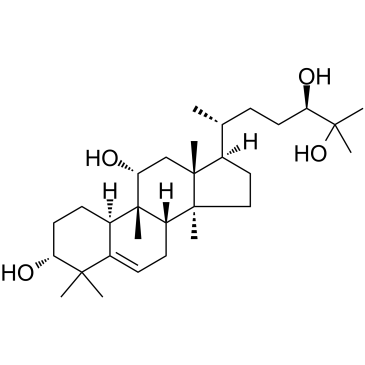
3α-Hydroxymogrol
3α-Hydroxymogrol is a triterpenoid isolated from Siraitia grosvenorii Swingle, acts as a potent AMPK activator, and enhances AMPK phosphorylation.
-
GC42237
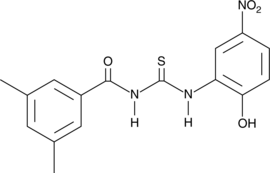
3,5-dimethyl PIT-1
PtdIns-(3,4,5)-P3 (PIP3) serves as an anchor for the binding of signal transduction proteins bearing pleckstrin homology (PH) domains such as phosphatidylinositol 3-kinase (PI3K) or PTEN.
-
GC13992
3-Guanidinopropionic Acid
β-GPA,PNU 10483
RGX-202 (3-Guanidinopropionic acid) is an oral small-molecule SLC6A8 transporter inhibitor. RGX-202 robustly inhibits creatine import in vitro and in vivo, reduces intracellular phosphocreatine and ATP levels, and induces tumor apoptosis. RGX-202 can be used for the research of cancer and duchenne muscular dystrophy.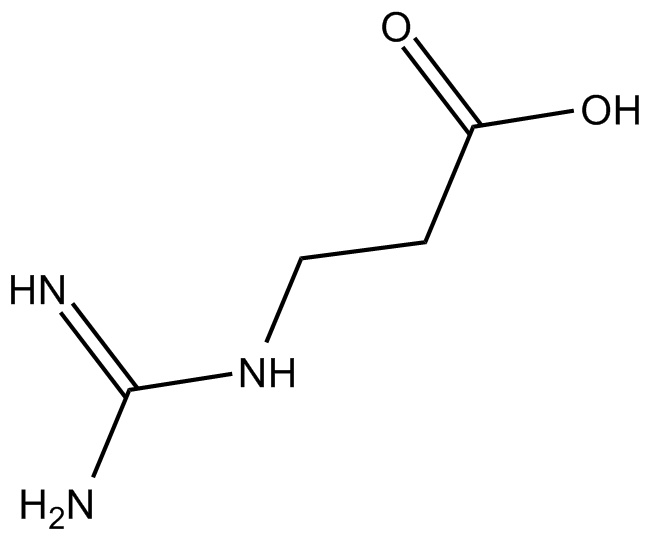
-
GC10710
3-Methyladenine
3-MA
3-Methyladenine is a classic autophagy inhibitor.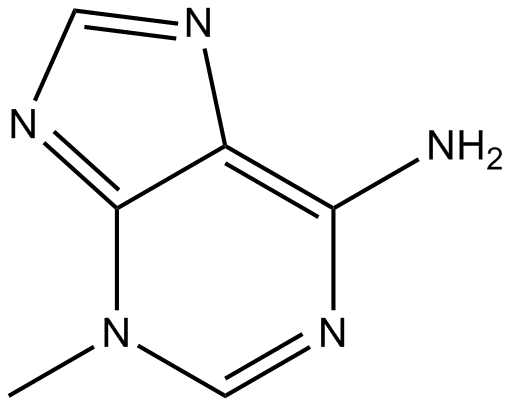
-
GC68539
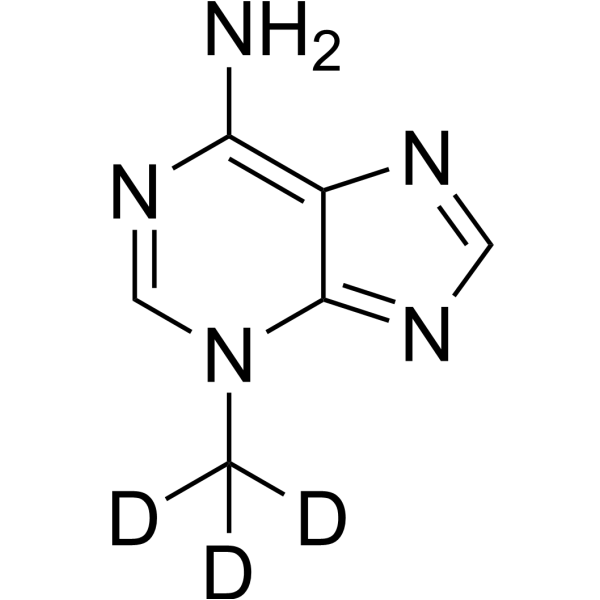
3-Methyladenine-d3
3-Methyladenine-d3 is the deuterated form of 3-Methyladenine. 3-Methyladenine (3-MA) is an inhibitor of PI3K. It is widely used as an autophagy inhibitor by inhibiting class III PI3K.
-
GC32767
3BDO
3-Benzyl-5-((2-nitrophenoxy)methyl)dihydrofuran-2(3H)-one
A butyrolactone derivative and autophagy inhibitor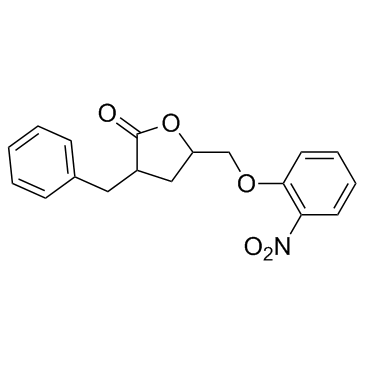
-
GC12078
3CAI
3-chloroacetyl Indole
AKT inhibitor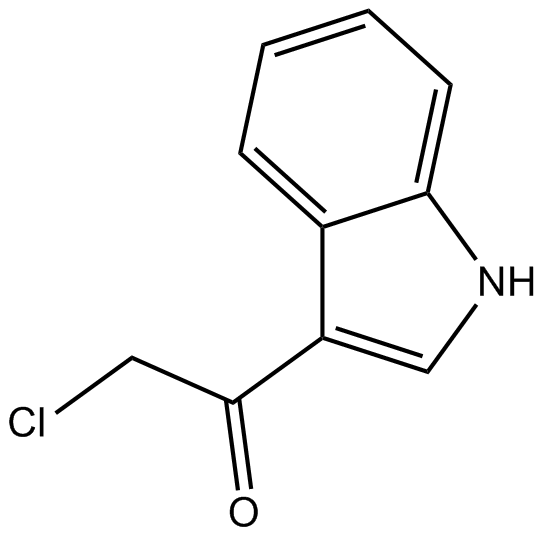
-
GC14306
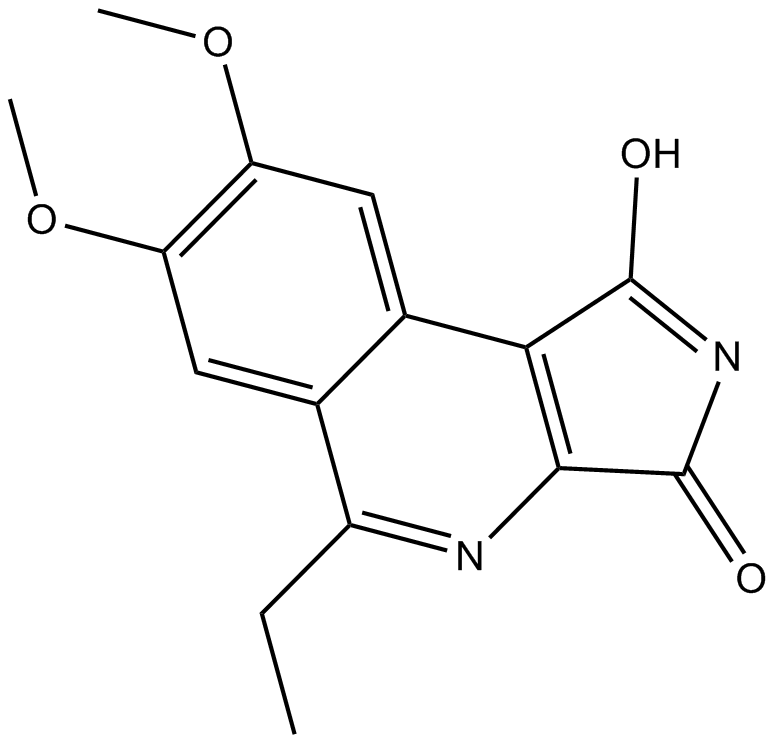
3F8
GSK-3β inhibitor
-
GC16960
4,5,6,7-Tetrabromobenzimidazole
Casein Kinase II Inhibitor XII,TBBz
4,5,6,7-Tetrabromobenzimidazole is a selective and ATP competitive CK2 (casein kinase 2) inhibitor.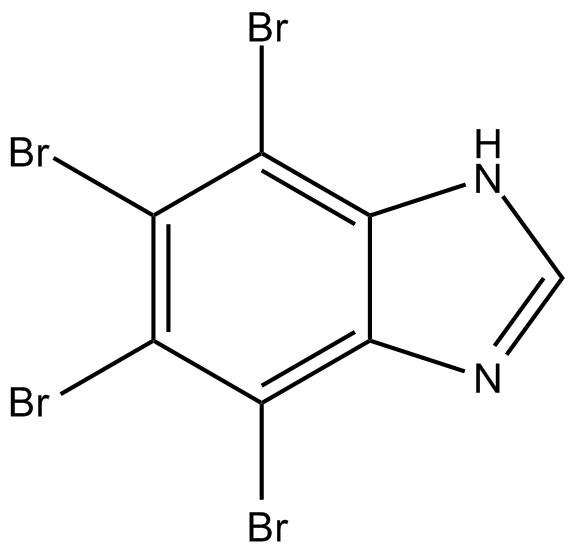
-
GC52091
5,7-Dichlorothiazolo[5,4-d]pyrimidine
A building block
![5,7-Dichlorothiazolo[5,4-d]pyrimidine Chemical Structure 5,7-Dichlorothiazolo[5,4-d]pyrimidine Chemical Structure](/media/struct/GC5/GC52091.png)
-
GC35162
5-Iodo-indirubin-3'-monoxime
5-Iodo-indirubin-3'-monoxime is a potent GSK-3β, CDK5/P25 and CDK1/cyclin B inhibitor, competing with ATP for binding to the catalytic site of the kinase, with IC50s of 9, 20 and 25 nM, respectively.
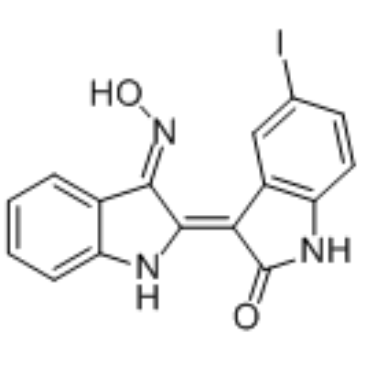
-
GN10093
6-gingerol
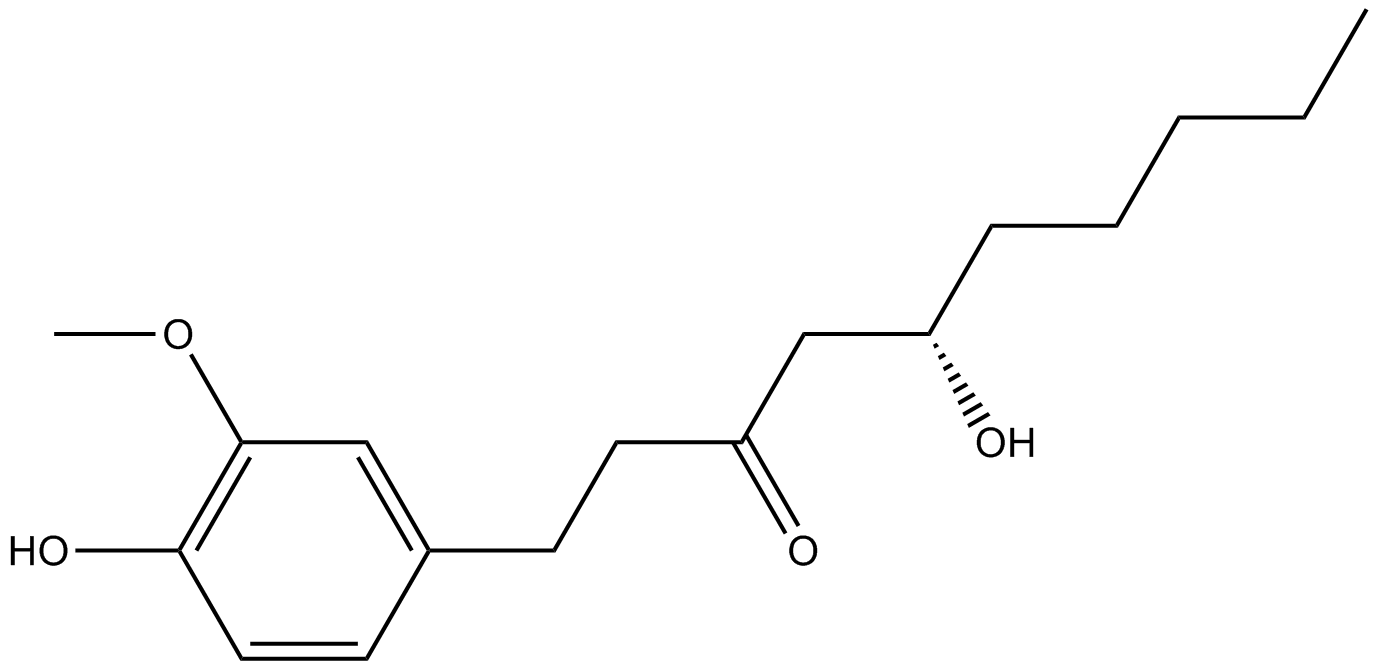
-
GC46724
6-Hydroxypyridin-3-ylboronic Acid
A heterocyclic building block
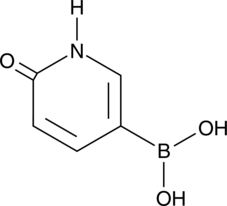
-
GC46738
7-Methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-amine
A building block
![7-Methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-amine Chemical Structure 7-Methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-amine Chemical Structure](/media/struct/GC4/GC46738.png)
-
GC31348
7-Methoxyisoflavone
5-methyl-7-Methoxyisoflavone
A synthetic isoflavone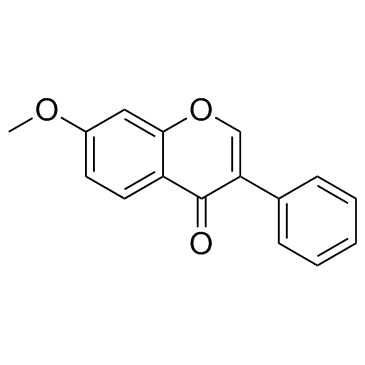
-
GC16157
740 Y-P
740YPDGFR; PDGFR 740Y-P
740 Y-P is an activator of PI3K with concentration of 20 μM [1].
-
GC39152
9-ING-41
Elraglusib
9-ING-41 is a maleimide-based ATP-competitive and selective glycogen synthase kinase-3β (GSK-3β) inhibitor with an IC50 of 0.71 μM. 9-ING-41 significantly leads to cell cycle arrest, autophagy and apoptosis in cancer cells. 9-ING-41 has anticancer activity and has the potential for enhancing the antitumor effects of chemotherapeutic drugs.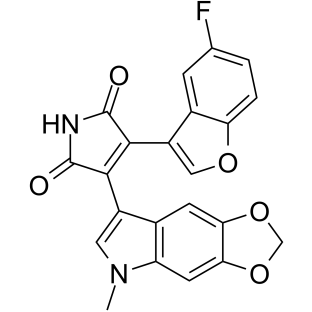
-
GC10336
A 1070722
GSK-3 inhibitor
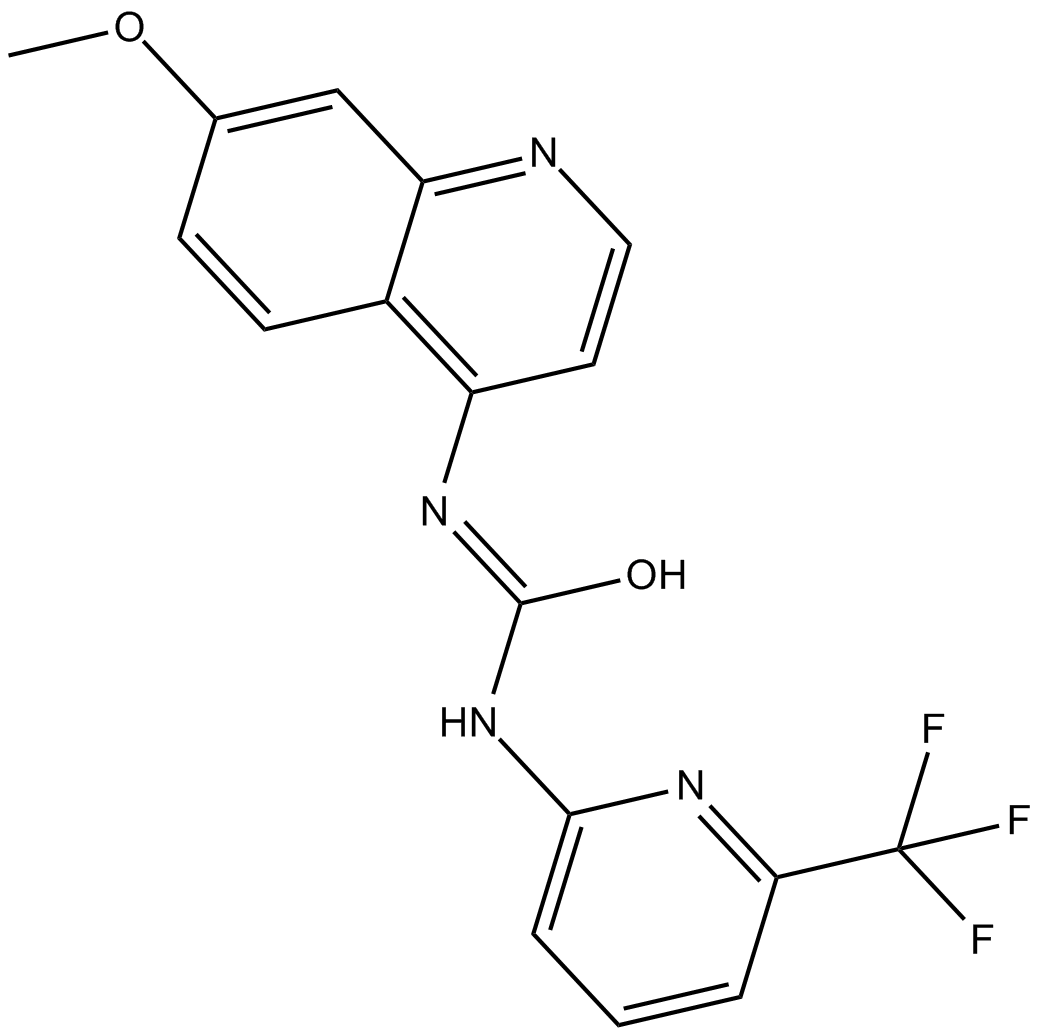
-
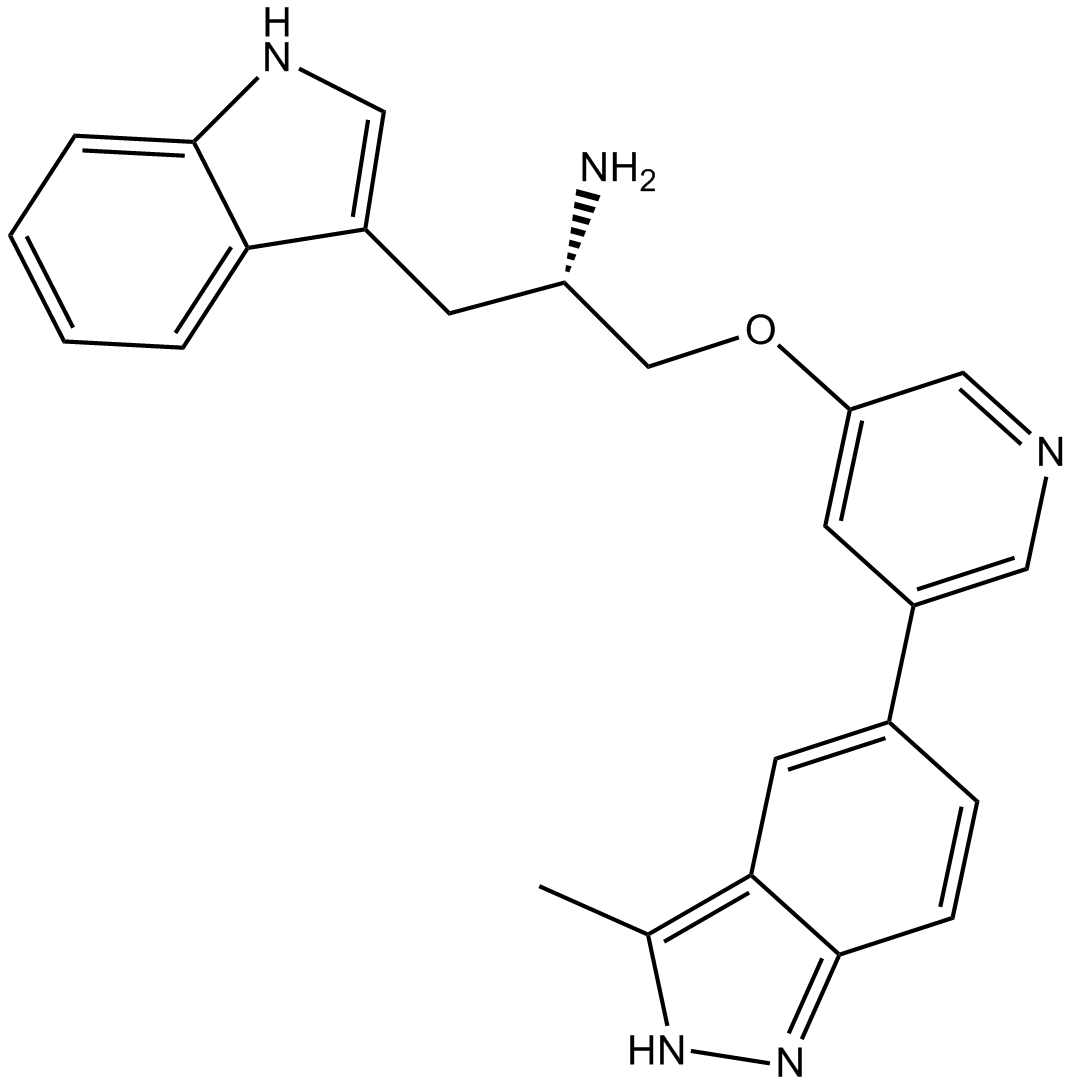
GC12502
A-443654
A pan Akt inhibitor
-
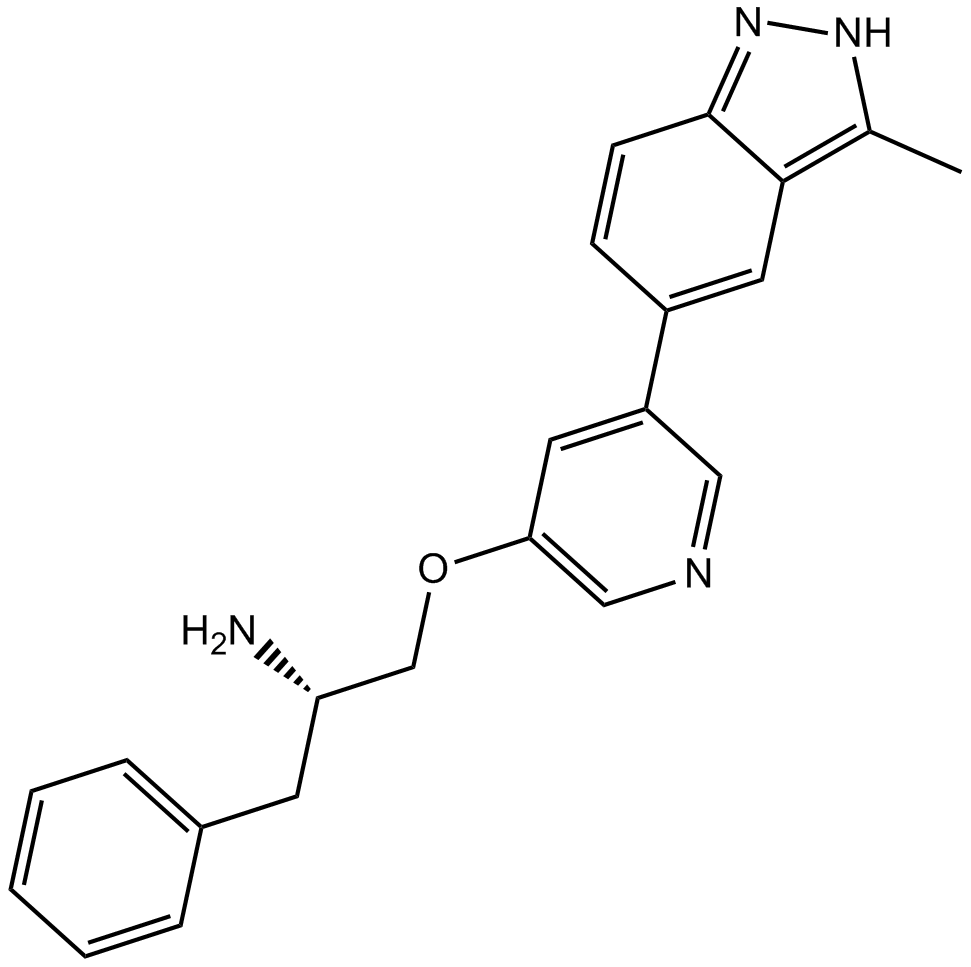
GC11875
A-674563
Akt1/PKA/CDK2 inhibitor,potent and selective
-
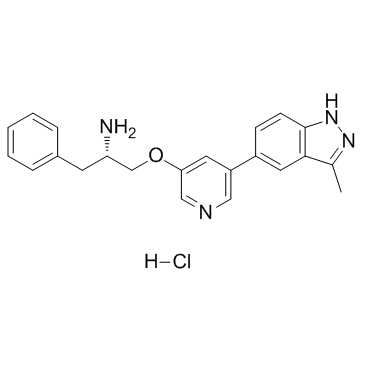
GC35212
A-674563 hydrochloride
A-674563 hydrochloride is a potent and selective Akt1 inhibitor with Ki of 11 nM.
-
GC11234
A-769662
A-769662;A769662
A direct activator of AMPK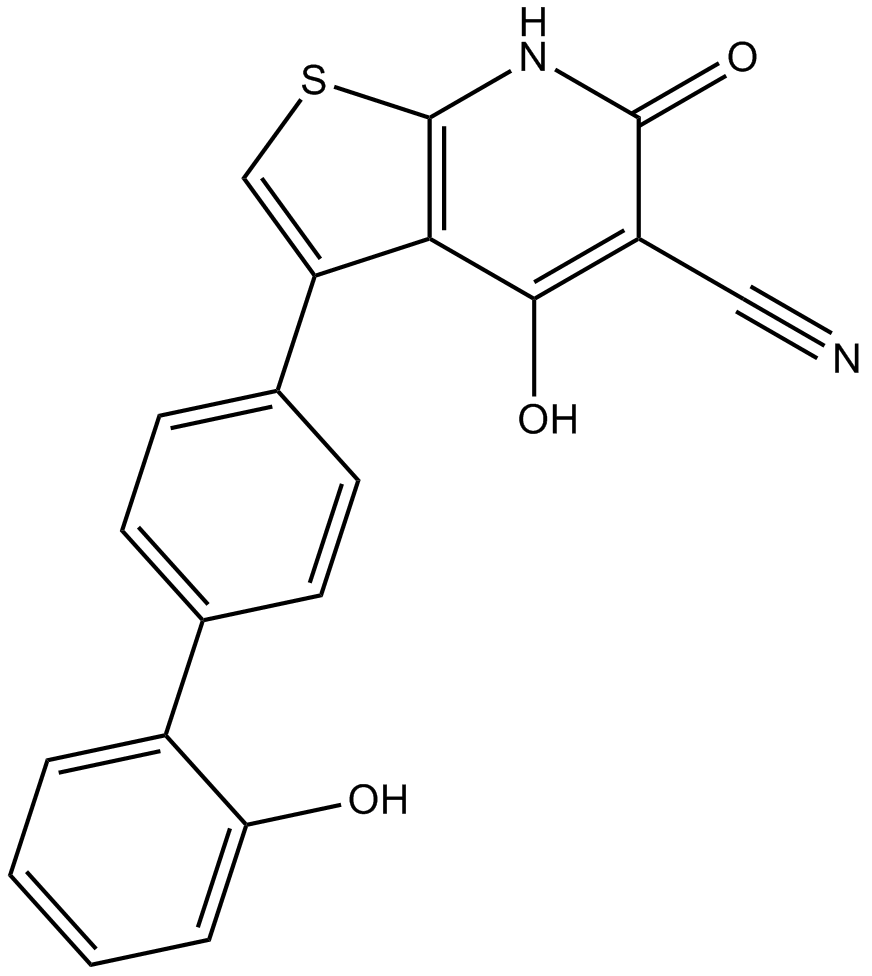
-
GC17550
A66
A 66; A-66
P110α inhibitor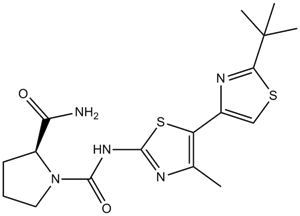
-
GC68592
ABC1183
ABC1183 is a selective dual inhibitor of GSK3 and CDK9 with oral activity. Its IC50 values for GSK3β, GSK3α, and CDK9/cyclin T1 are 657 nM, 327 nM, and 321 nM respectively. ABC1183 has anti-inflammatory properties as well as anti-tumor activity.

-
GC10860
Acalisib (GS-9820)
GS-9820
Acalisib (GS-9820) is a potent and selective PI3Kδ inhibitor with an IC50 of 12.7 nM.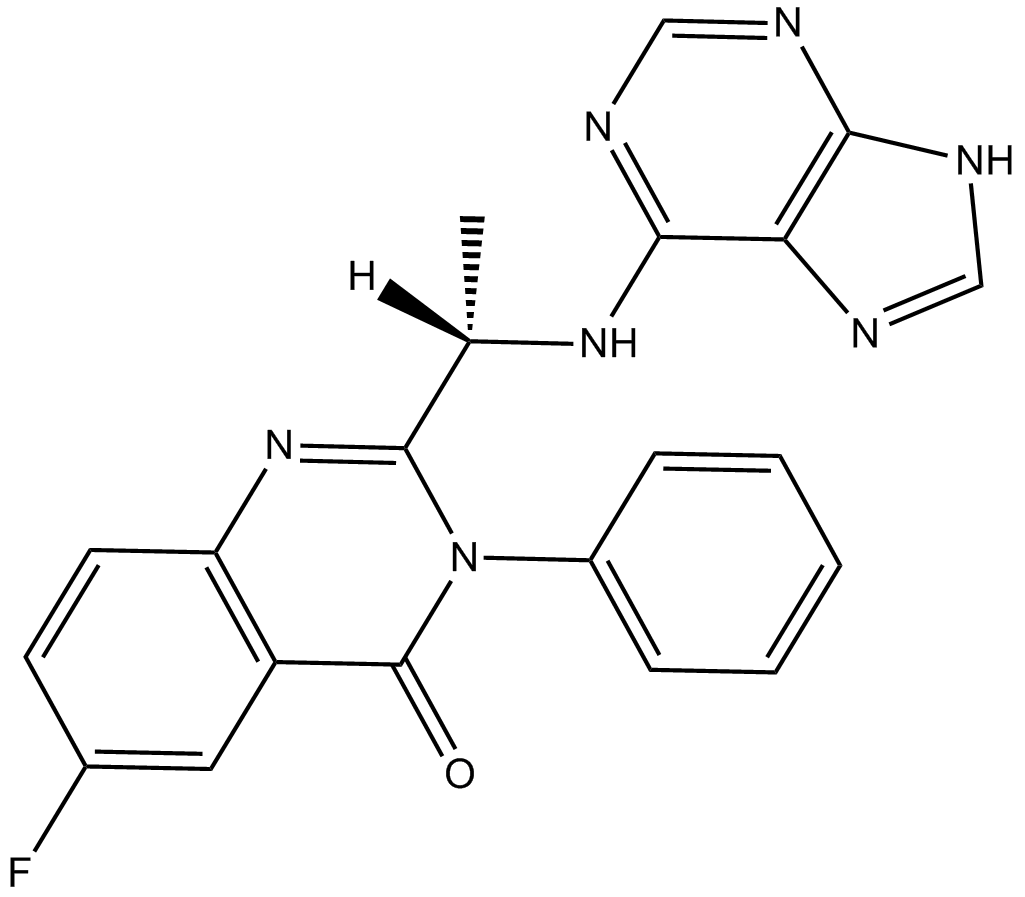
-
GC35242
Actein
Actein is a triterpene glycoside isolated from the rhizomes of Cimicifuga foetida. Actein suppresses cell proliferation, induces autophagy and apoptosis through promoting ROS/JNK activation, and blunting AKT pathway in human bladder cancer. Actein has little toxicity in vivo.
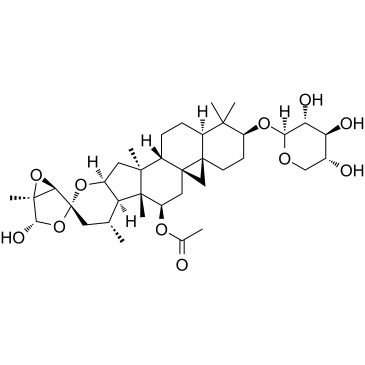
-
GC16475
Afuresertib
GSK2110183
pan-AKT inhibitor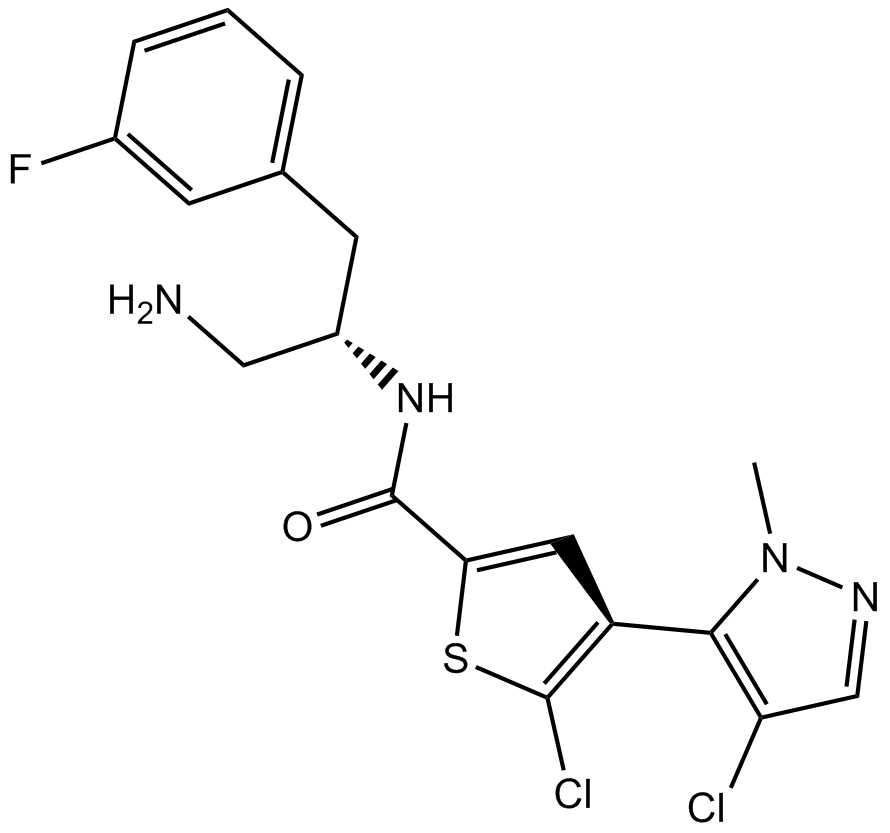
-
GC42747
Afuresertib (hydrochloride)
GSK2110183B
Afuresertib is a selective, orally bioavailable inhibitor of Akt1, 2, and 3 with Ki values of 0.08, 2, and 2.6 nM, respectively.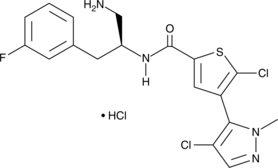
-
GC10518
AICAR
Acadesine, AICA-Riboside, NSC 105823
AICAR (also called acadesine) is a purine nucleoside.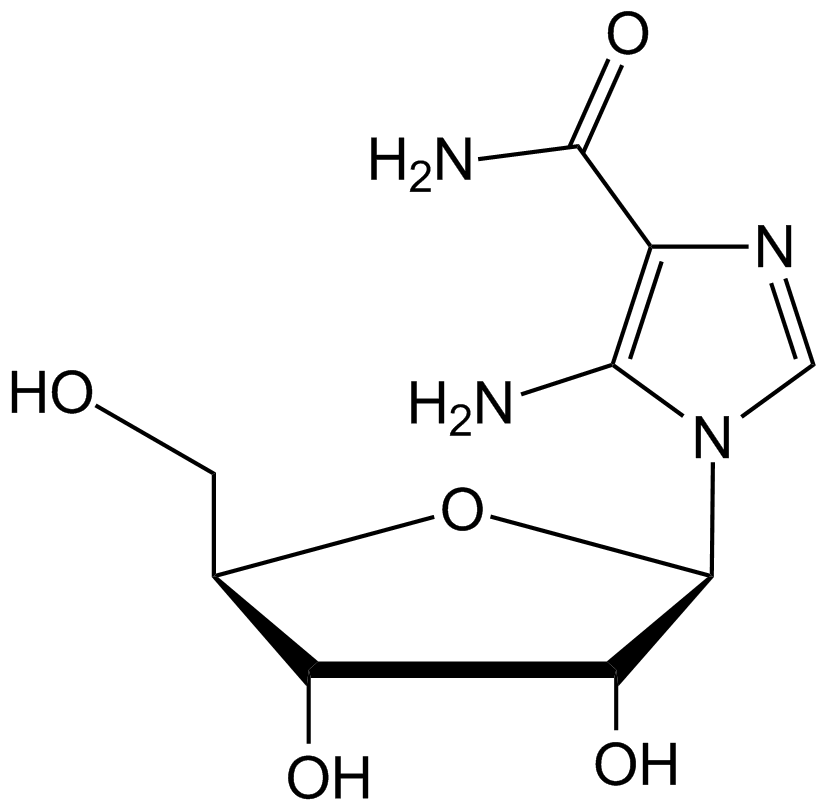
-
GC10580
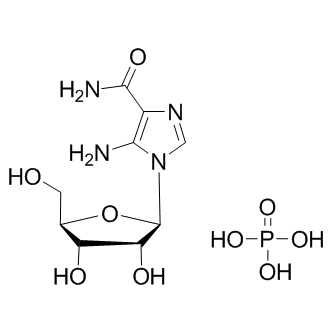
AICAR phosphate
AMPK activator
-
GC42759
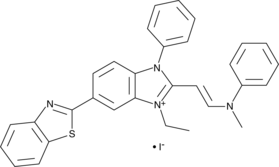
Akt Inhibitor IV
Akt inhibitor IV is an inhibitor of Akt activation that inhibits Akt-mediated nuclear export of Forkhead box class O transcription factor 1a (FOXO1a; IC50 = 625 nM) and reduces phosphorylation of Akt at Ser473 and Thr308 in a dose-dependent manner.
-
GC11589
AKT inhibitor VIII
A potent inhibitor of Akt1 and Akt2

-
GC16366
AKT Kinase Inhibitor
AKT Kinase Inhibitor is an Akt kinase inhibitor with anti-tumor activity[1].
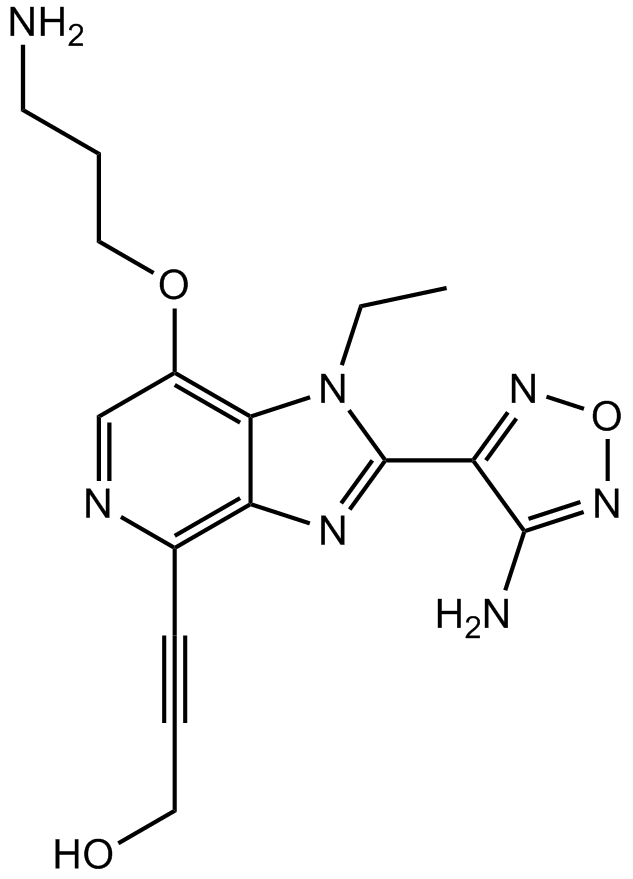
-
GC67700
AKT Kinase Inhibitor hydrochloride
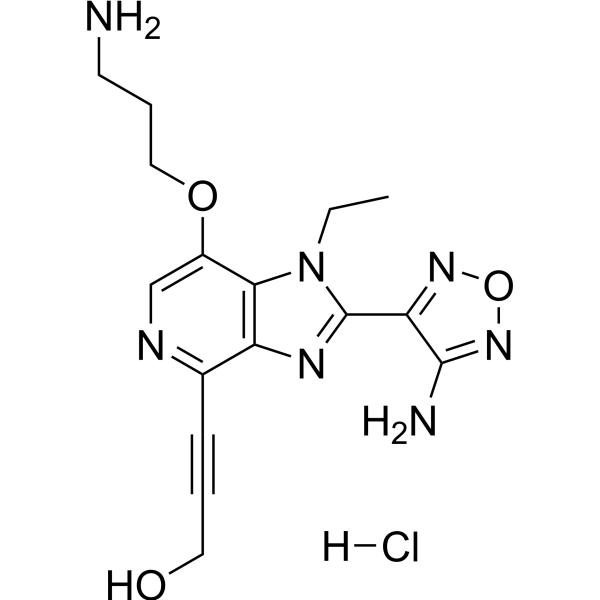
-
GC52443
Akt Substrate (trifluoroacetate salt)
RPRAATF
An Akt peptide substrate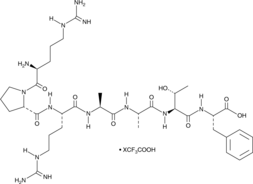
-
GC33308
AKT-IN-1
AKT-IN-1 is an allosteric AKT inhibitor with an IC50 of 1.042 μM.
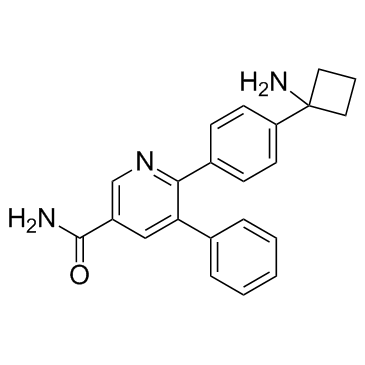
-
GC35275
AKT-IN-3
AKT-IN-3 (compound E22) is a potent, orally active low hERG blocking Akt inhibitor, with 1.4 nM, 1.2 nM and 1.7 nM for Akt1, Akt2 and Akt3, respectively. AKT-IN-3 (compound E22) also exhibits good inhibitory activity against other AGC family kinases, such as PKA, PKC, ROCK1, RSK1, P70S6K, and SGK. AKT-IN-3 (compound E22) induces apoptosis and inhibits metastasis of cancer cells.
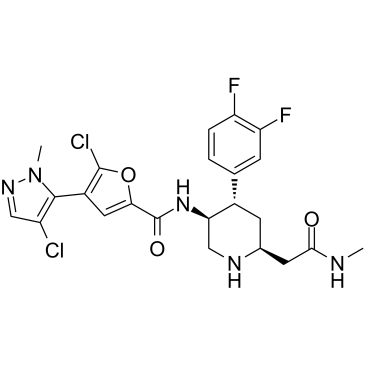
-
GC62190
AKT-IN-6
AKT-IN-6 (Example 13) is a potent Akt inhibitor. AKT-IN-6 inhibits Akt1, Akt2 and Akt3 with IC50s < 500nM, respectively. (patent WO2013056015A1).
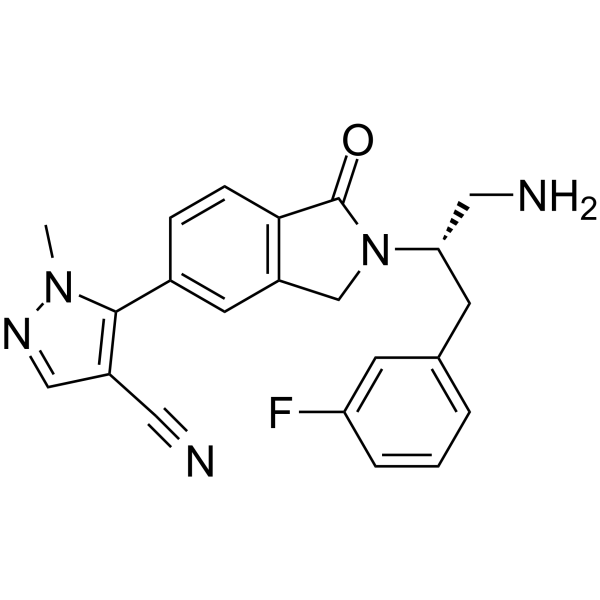
-
GC32939
Akt1 and Akt2-IN-1
Akt1 and Akt2-IN-1 is an allosteric inhibitor of Akt1 (IC50=3.5 nM) and Akt2 (IC50=42 nM), with potent and balanced activity.
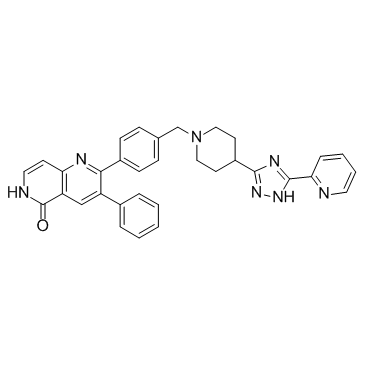
-
GC66024
Aldometanib
LXY-05-029
Aldometanib (LXY-05-029) is an orally active aldolase inhibitor. Aldometanib can activate lysosomal adenosine monophosphate-activated protein kinase (AMPK) and decreases blood glucose. Aldometanib can be used for the research of metabolic homeostasis.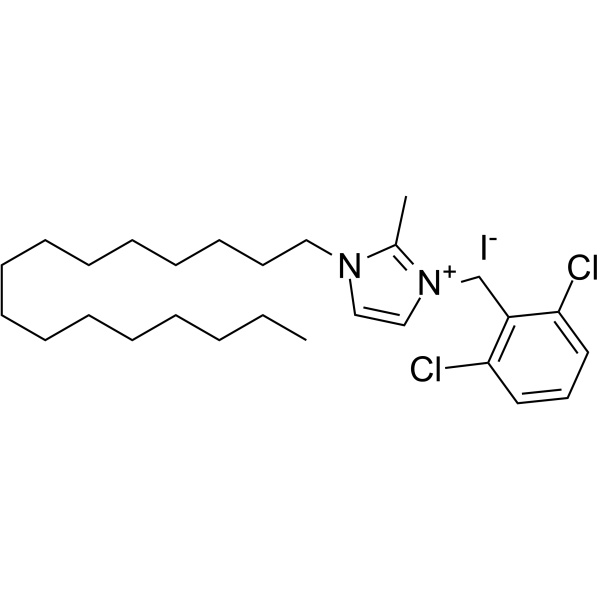
-
GC68640
ALM301
ALM301 is a highly specific AKT inhibitor with oral activity. Its IC50 values for AKT1, AKT2 and AKT3 are 0.13 μM, 0.09 μM and 2.75 μM, respectively. ALM301 inhibits the phosphorylation of AKT and regulates downstream signaling in vitro. It can inhibit cancer cell proliferation and tumor growth.

-
GC15841
Alsterpaullone
9-Nitropaullone,NSC 705701
CDKs and GSK3β inhibitor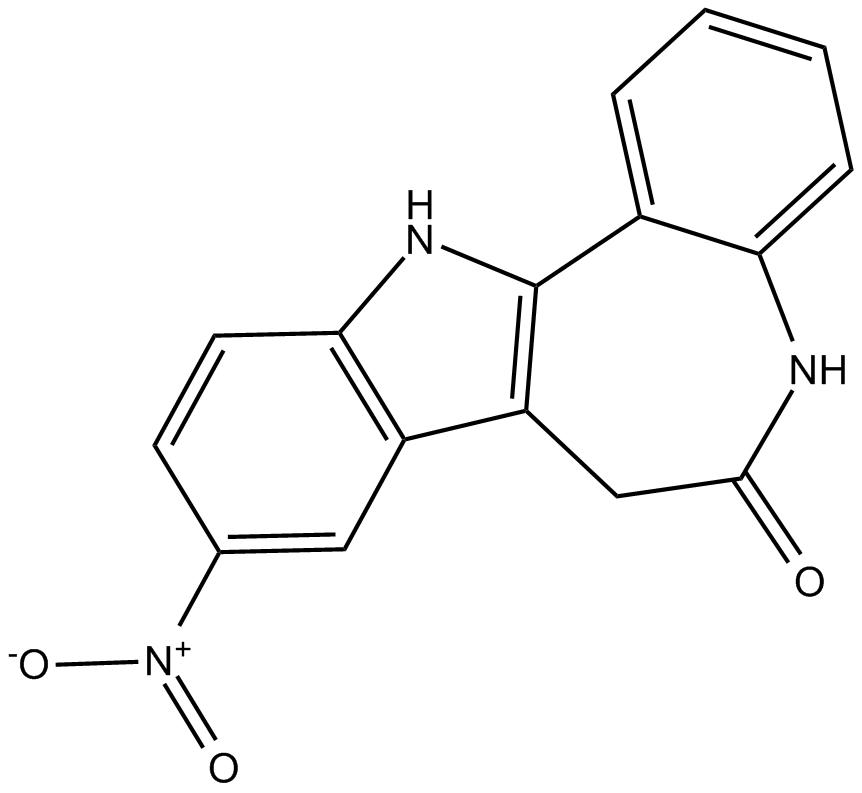
-
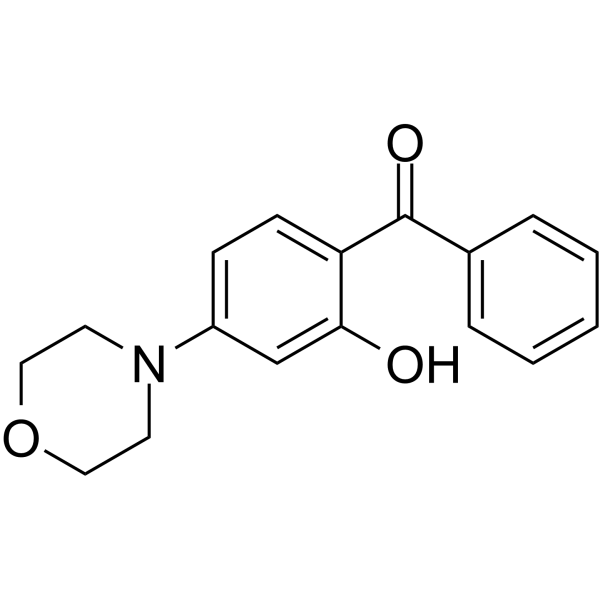
GC64818
AMA-37
AMA-37, an Arylmorpholine analog, is ATP-competitive DNA-PK inhibitor, with IC50 values of 0.27 μM (DNA-PK), 32 μM (p110α), 3.7 μM (p110β), and 22 μM (p110γ), respectively.
-
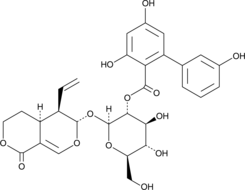
GC42776
Amarogentin
A secoiridoid glycoside with diverse biological activities
-
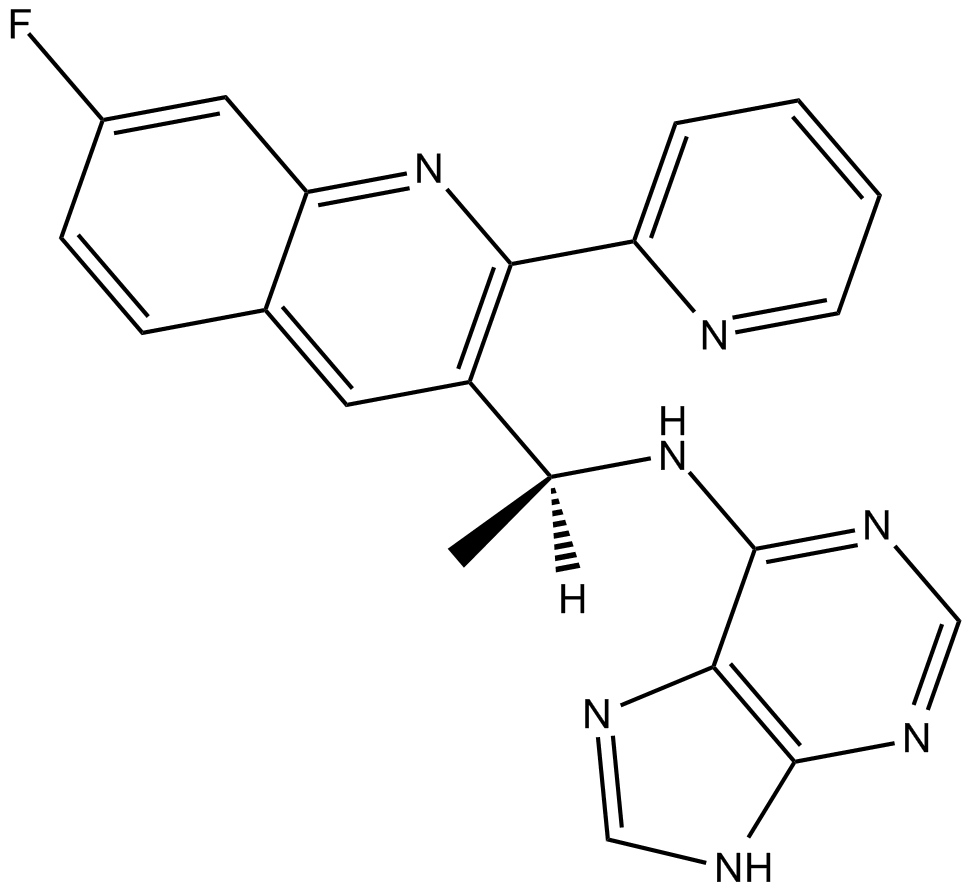
GC13913
AMG319
PI3Kδ inhibitor
-
GC11190
AMPK activator
D942
AMPK activator is a cell penetrant AMPK activator and partially inhibits the mitochondrial complex I. In multiple myeloma cells, AMPK activator inhibits cell growth.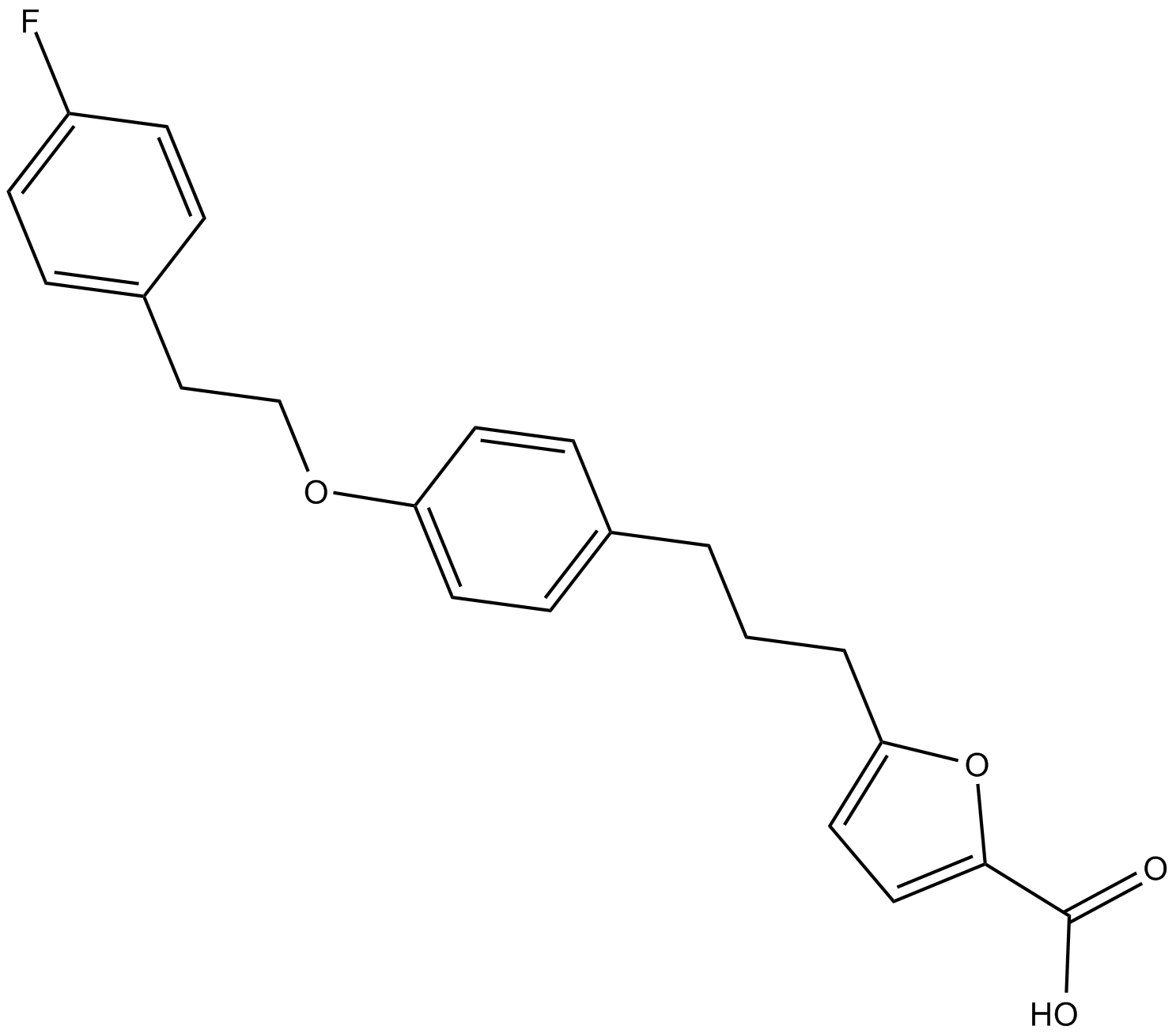
-
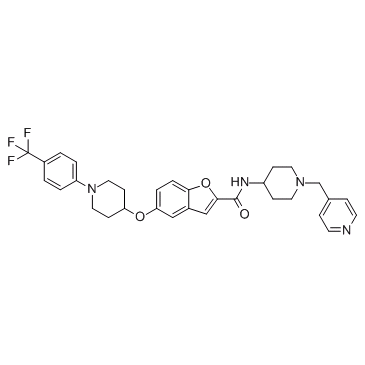
GC35327
AMPK activator 1
AMPK activator 1 is an AMPK activator extracted from patent WO2013116491A1, compound No.1-75, has an EC50 of <0.1μM.
-
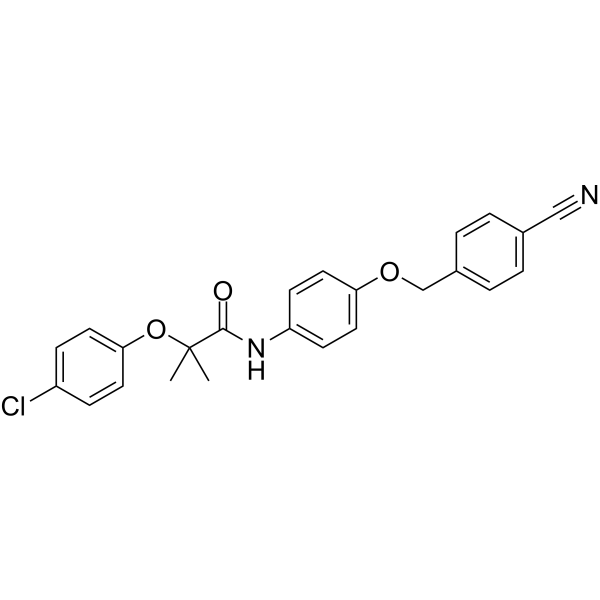
GC62841
AMPK activator 4
AMPK activator 4 is a potent AMPK activator without inhibition of mitochondrial complex I.
-
GC67961
AMPK-IN-3
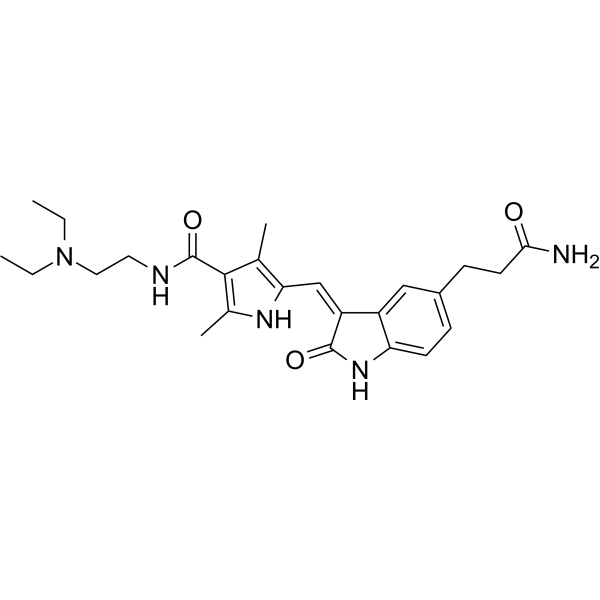
-
GC16534
Ampkinone
AMPK activator
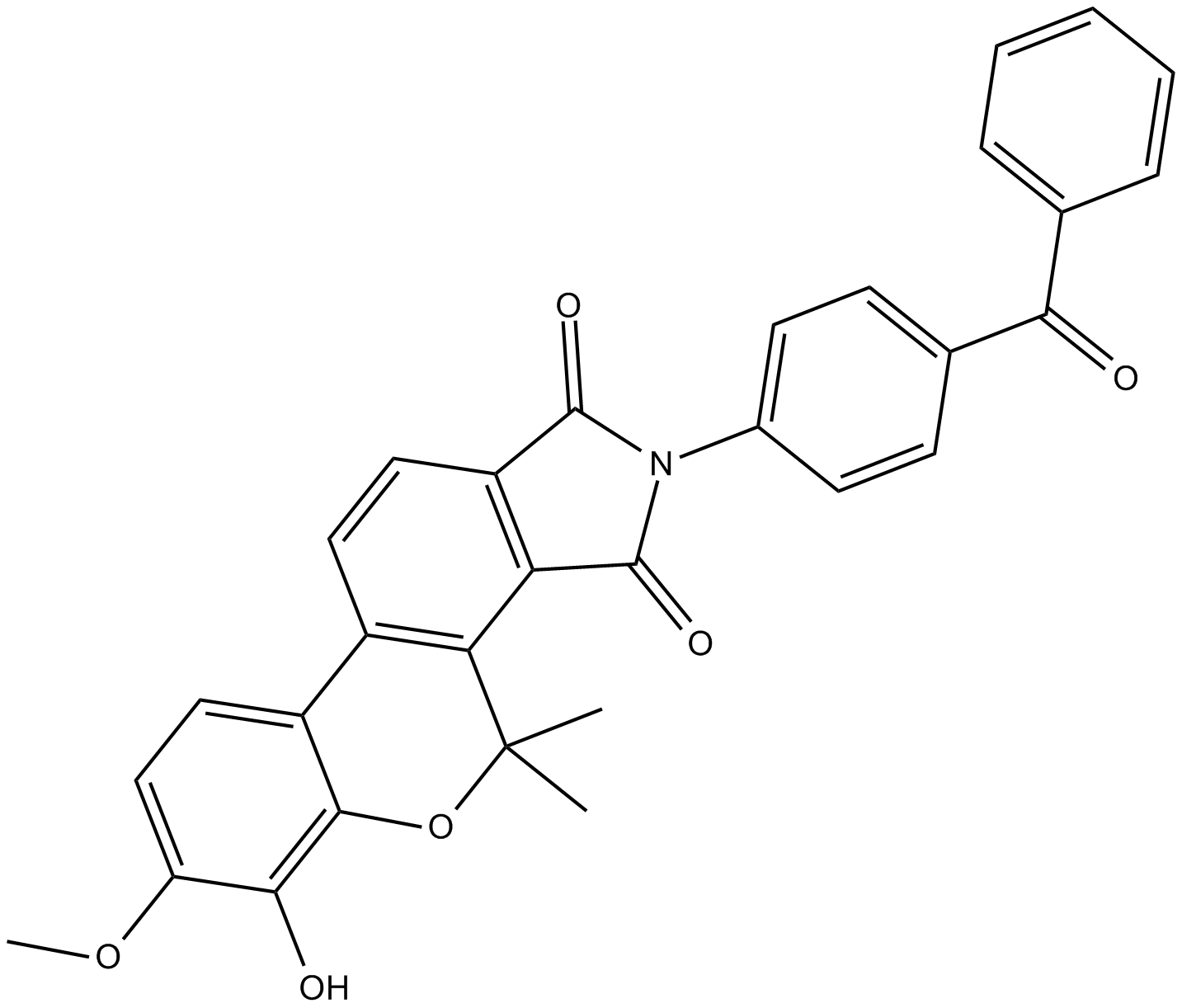
-
GC18374
API-1
NSC 177223
API-1 is an inhibitor of Akt that reduces the level of phosphorylated Akt (IC50 = ~0.8 uM in OVCAR3 cells) by binding to Akt and blocking its translocation to the cell membrane.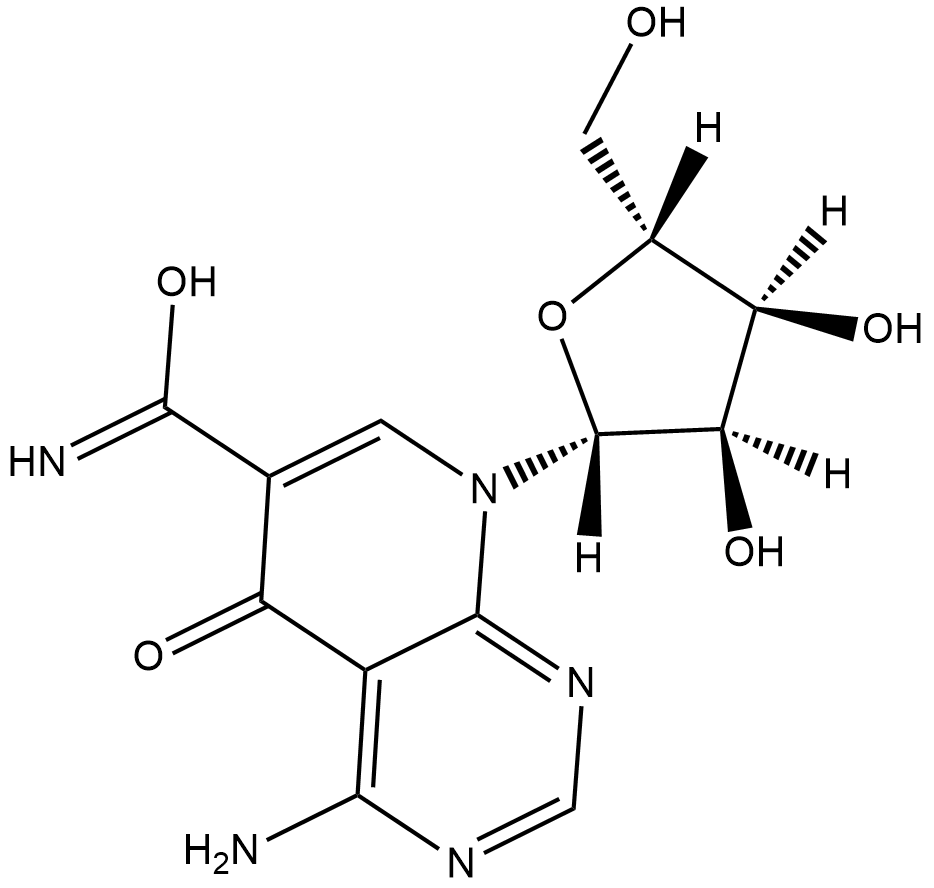
-
GC31661
APY0201
APY0201 is a potent PIKfyve inhibitor, which inhibits the conversion of PtdIns3P to PtdIns(3,5)P2 in the presence of in the presence of [33P]ATP with an IC50 of 5.2 nM.
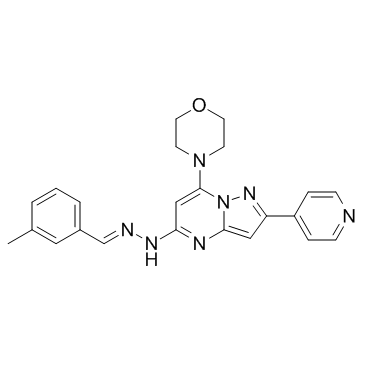
-
GC15425
AR-A014418
AR-AO 14418;AR 0133418;AR 014418;GSK 3β inhibitor
A selective inhibitor of GSK3β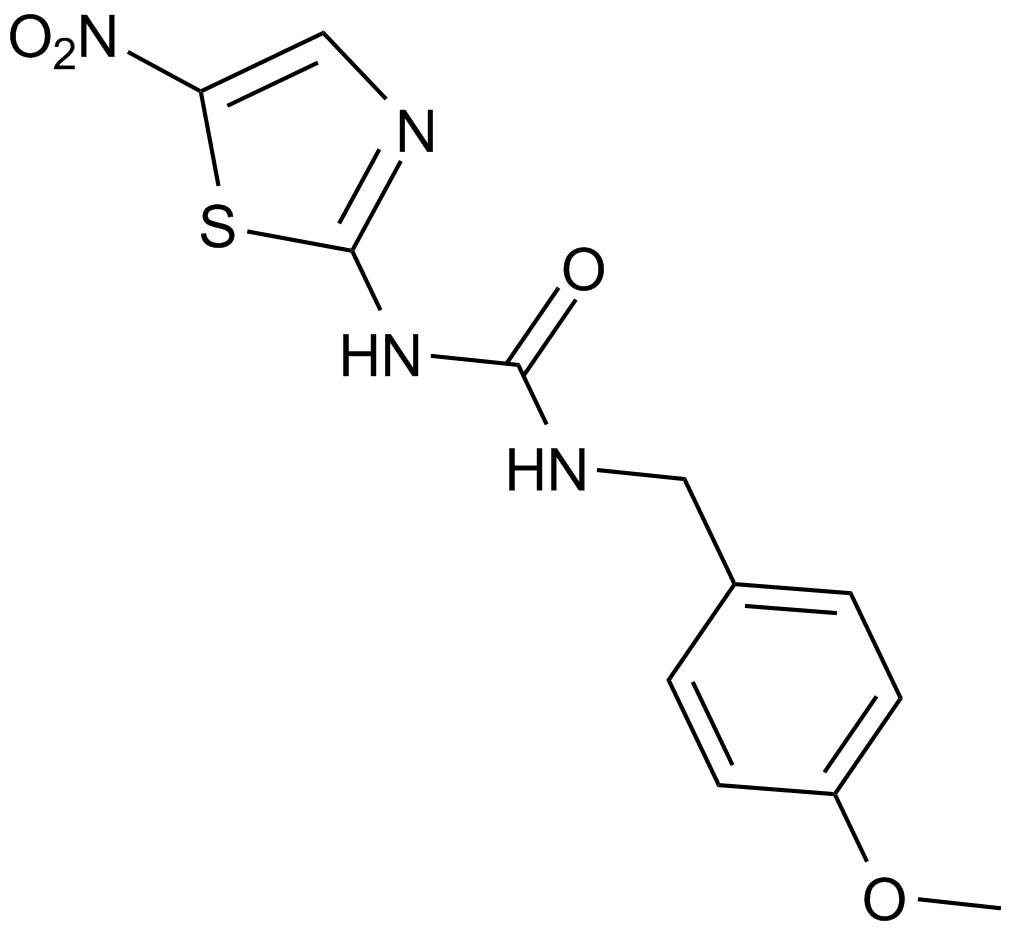
-
GC35395
Arnicolide D
ARD
Arnicolide D is a sesquiterpene lactone isolated from Centipeda minima. Arnicolide D modulates the cell cycle, activates the caspase signaling pathway and inhibits the PI3K/AKT/mTOR and STAT3 signaling pathways. Arnicolide D inhibits Nasopharyngeal carcinoma (NPC) cell viability in a concentration- and time-dependent manner.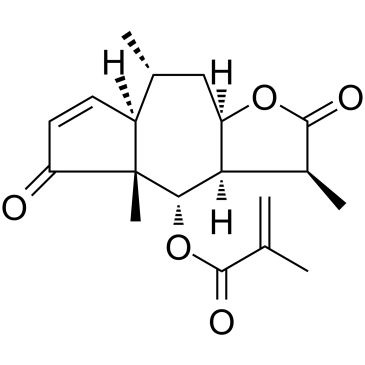
-
GC19036
ARQ-092
Miransertib
ARQ-092 (ARQ-092) is a potent, orally active, selective and allosteric Akt inhibitor with IC50s of 2.7 nM, 14 nM and 8.1 nM for Akt1, Akt2, Akt3, respectively.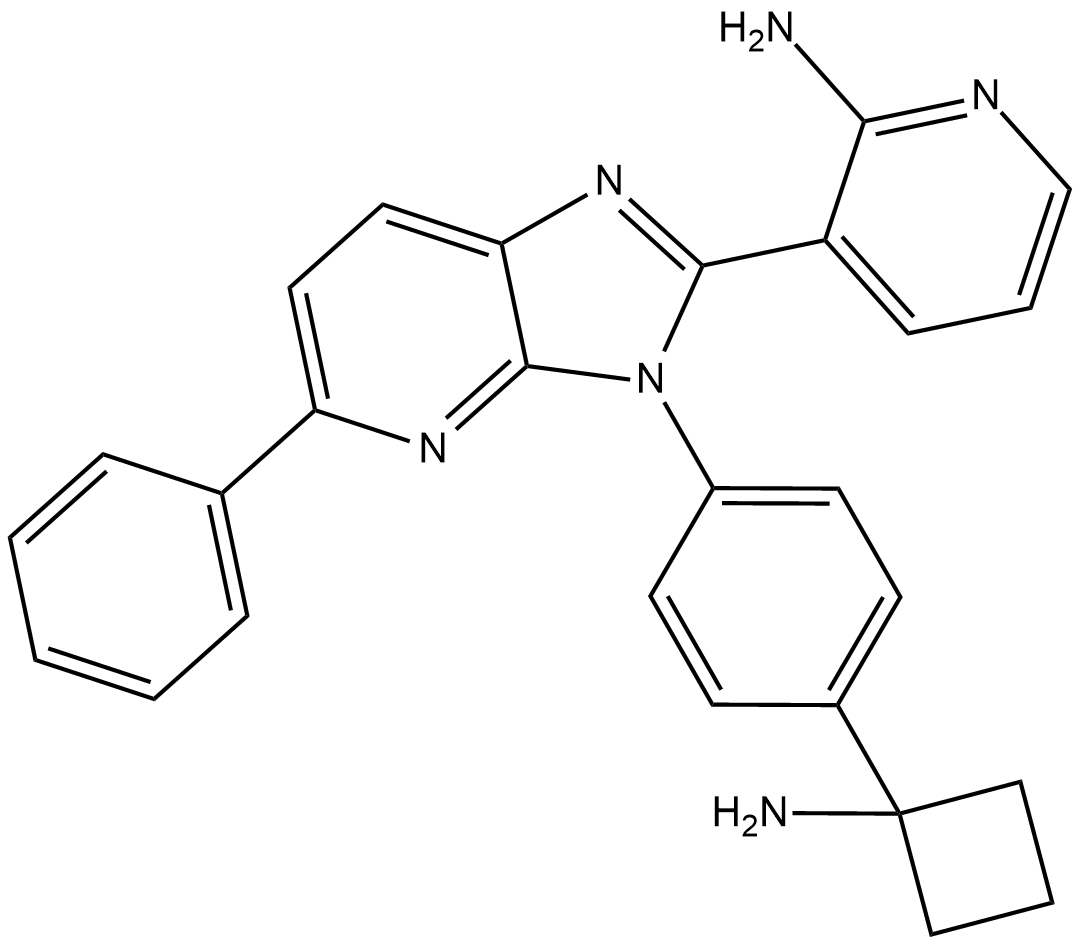
-
GN10647
Artemisinine
NSC 369397
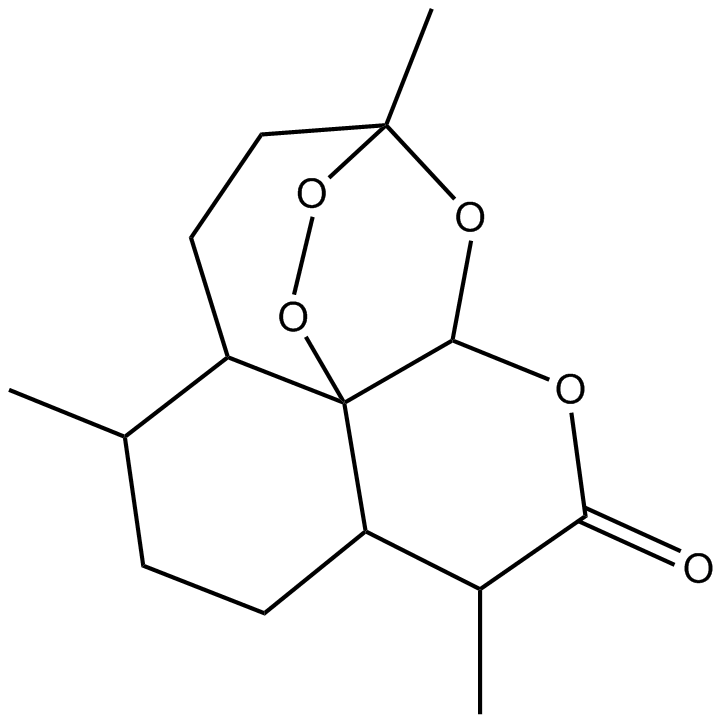
-
GC11910
AS-041164
PI3Kγ inhibitor
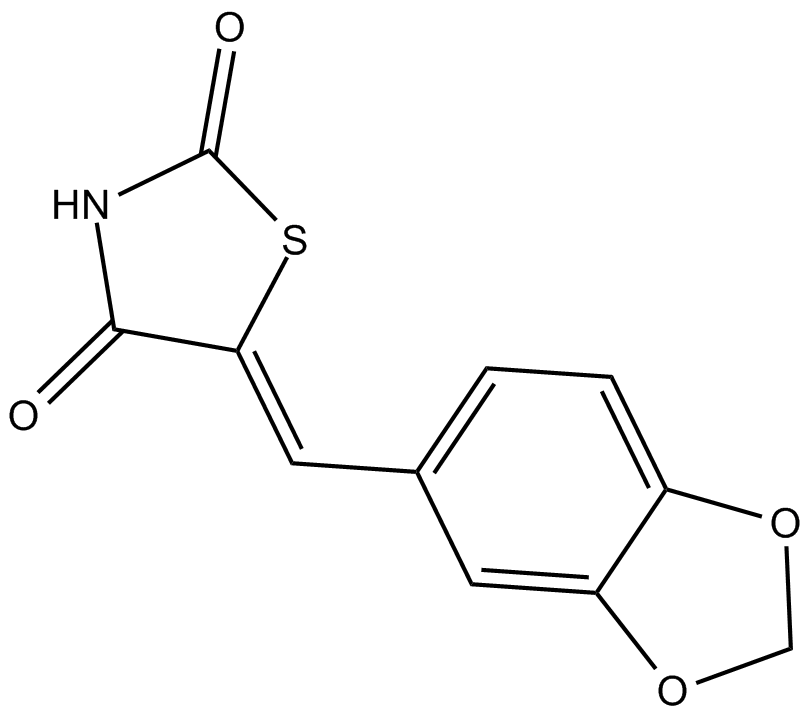
-
GC13901
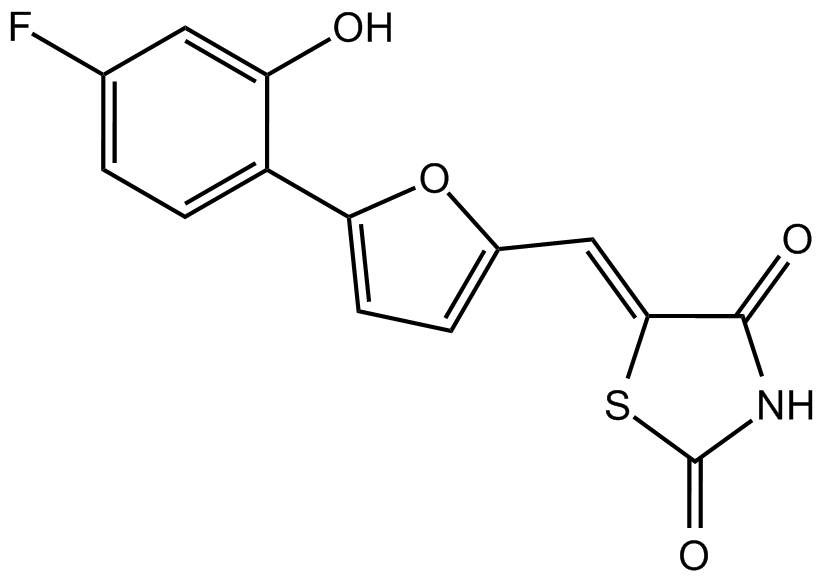
AS-252424
PI3Kγ inhibitor,novel and potent
-
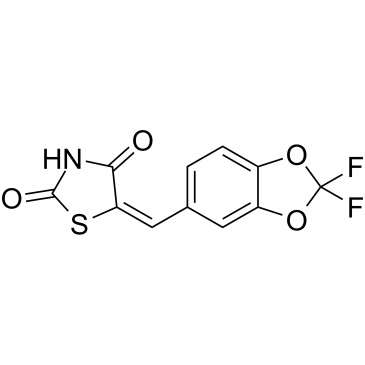
GC38061
AS-604850
A selective inhibitor of PI3Kγ
-
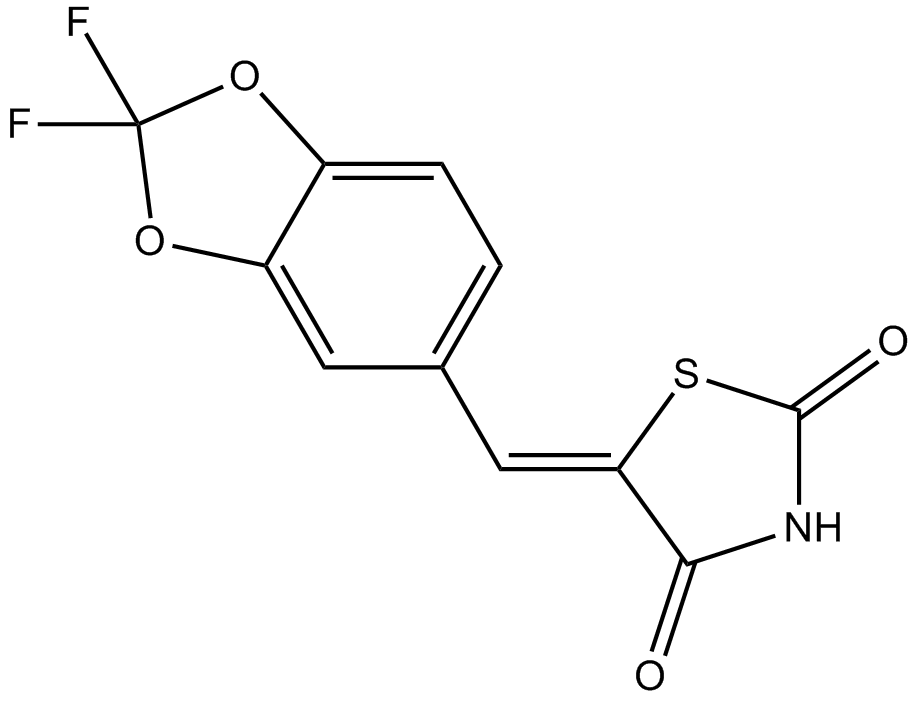
GC12080
AS-604850
PI3Kγ inhibitor,selective and ATP-competitve
-
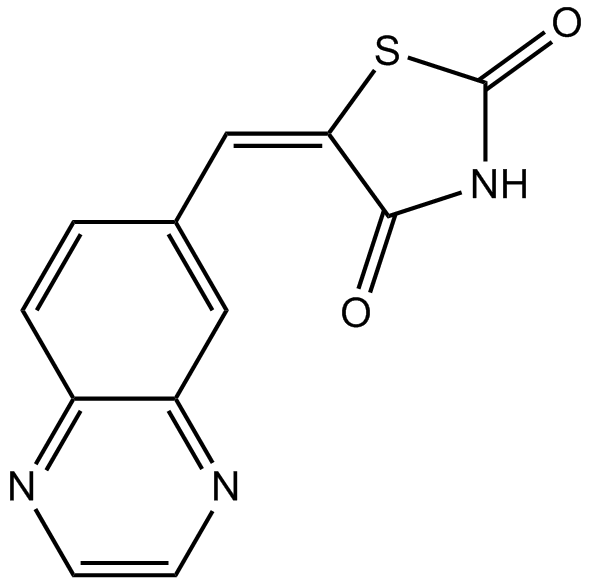
GC12474
AS-605240
Potent and selective PI 3-Kγ inhibitor
-
GC17712
AT13148
Multi-AGC kinase inhibitor,ATP-competitive
-
GC12297
AT7867
Akt1/2/3 and p70S6K/PKA inhibitor
-
GC10918
AT7867 dihydrochloride
A potent and orally bioavailable pan-Akt inhibitor
-
GC62335
AZ2
AZ2 is a highly selective PI3Kγ inhibitor (The pIC50 value for PI3Kγ is 9.3).
-
GC64938
AZD-7648
AZD-7648 is a potent, orally active, selective DNA-PK inhibitor with an IC50 of 0.6 nM. AZD-7648 induces apoptosis and shows antitumor activity.
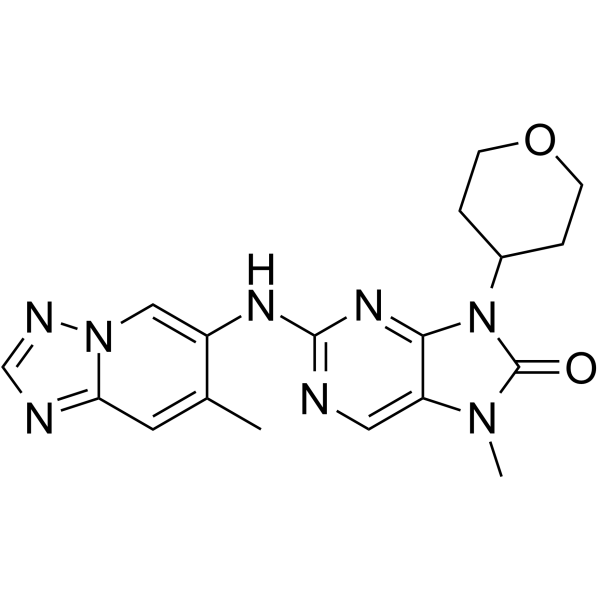
-
GC16568
AZD1080
GSK3βinhibitor

-
GC13029
AZD2014
AZD 2014; AZD-2014
AZD2014 (AZD2014) is an ATP competitive mTOR inhibitor with an IC50 of 2.81 nM. AZD2014 inhibits both mTORC1 and mTORC2 complexes.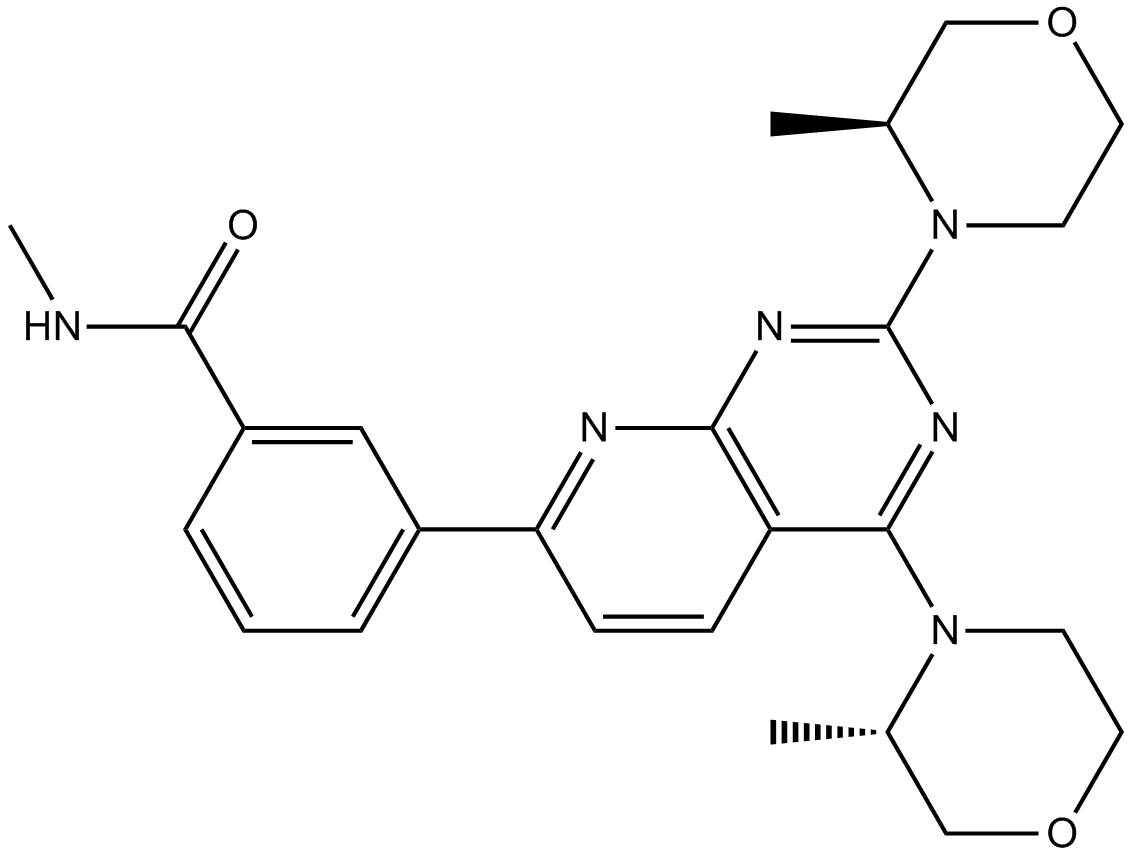
-
GC14736
AZD2858
GSK-3 inhibitor,potent and selective

-
GC65149
AZD3458
AZD3458 is a potent and remarkably selective PI3Kγ inhibitor with pIC50s of 9.1, 5.1, <4.5, and 6.5 for PI3Kγ, PI3Kα, PI3Kβ, and PI3Kδ, respectively.
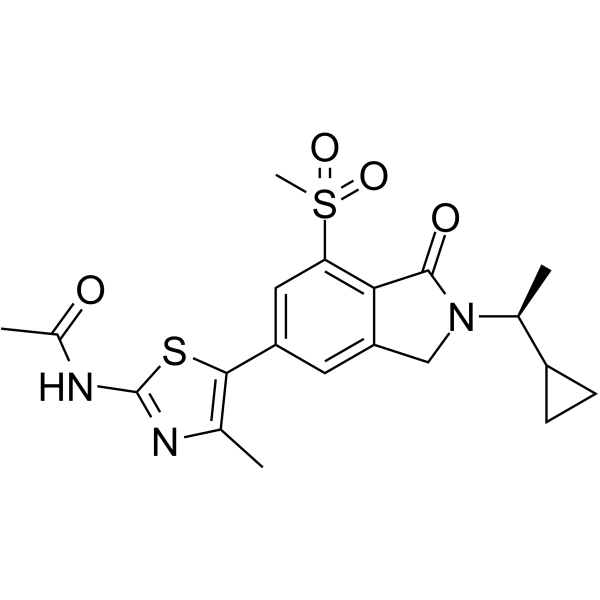
-
GC11752
AZD5363
AZD5363 (AZD5363) is an orally active and potent pan-AKT kinase inhibitor with IC50 of 3, 7 and 7 nM for Akt1,Akt2 and Akt3, respectively.
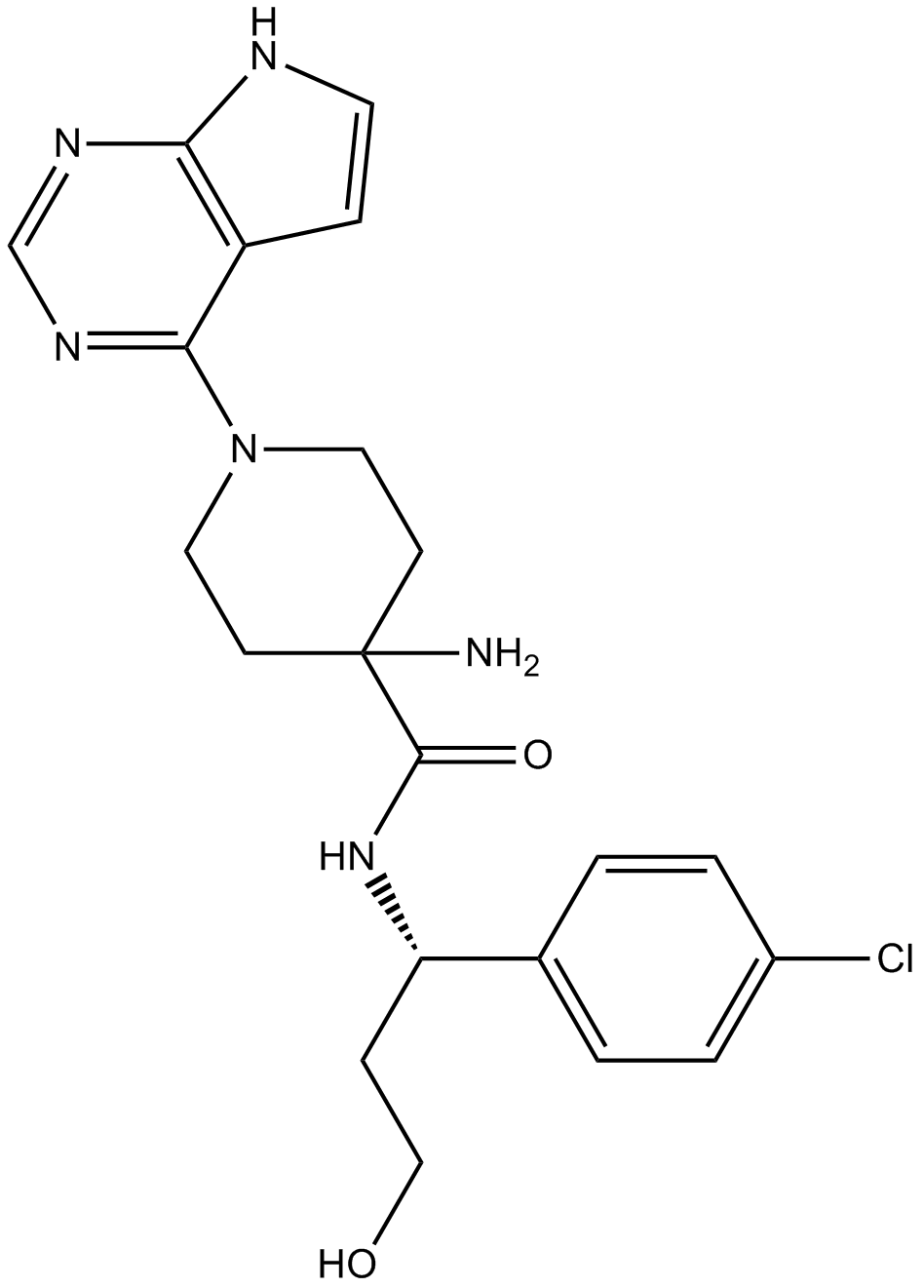
-
GC16851
AZD6482
KIN-193
PI3Kβ inhibitor,potent and selective
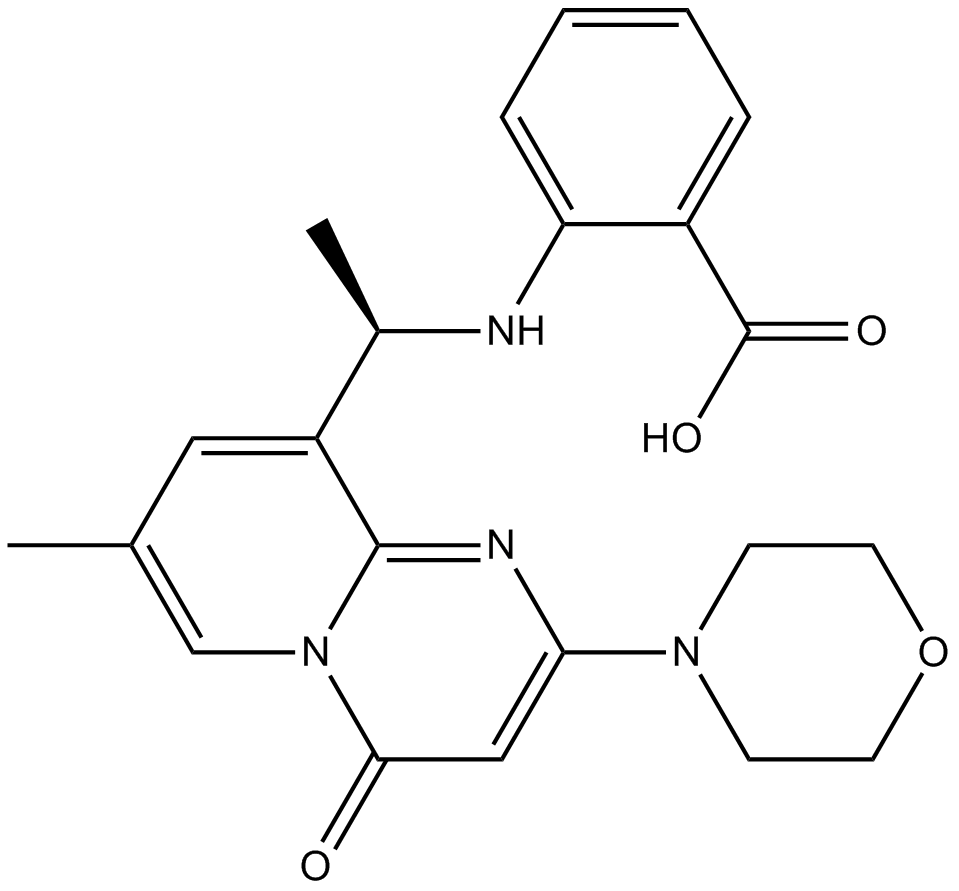
-
GC16380
AZD8055
CCG-168
AZD8055 is a new ATP-competitive mTOR inhibitor with an IC50 of 0.8 nmol/L and a Ki of 1.3 nmol/L.
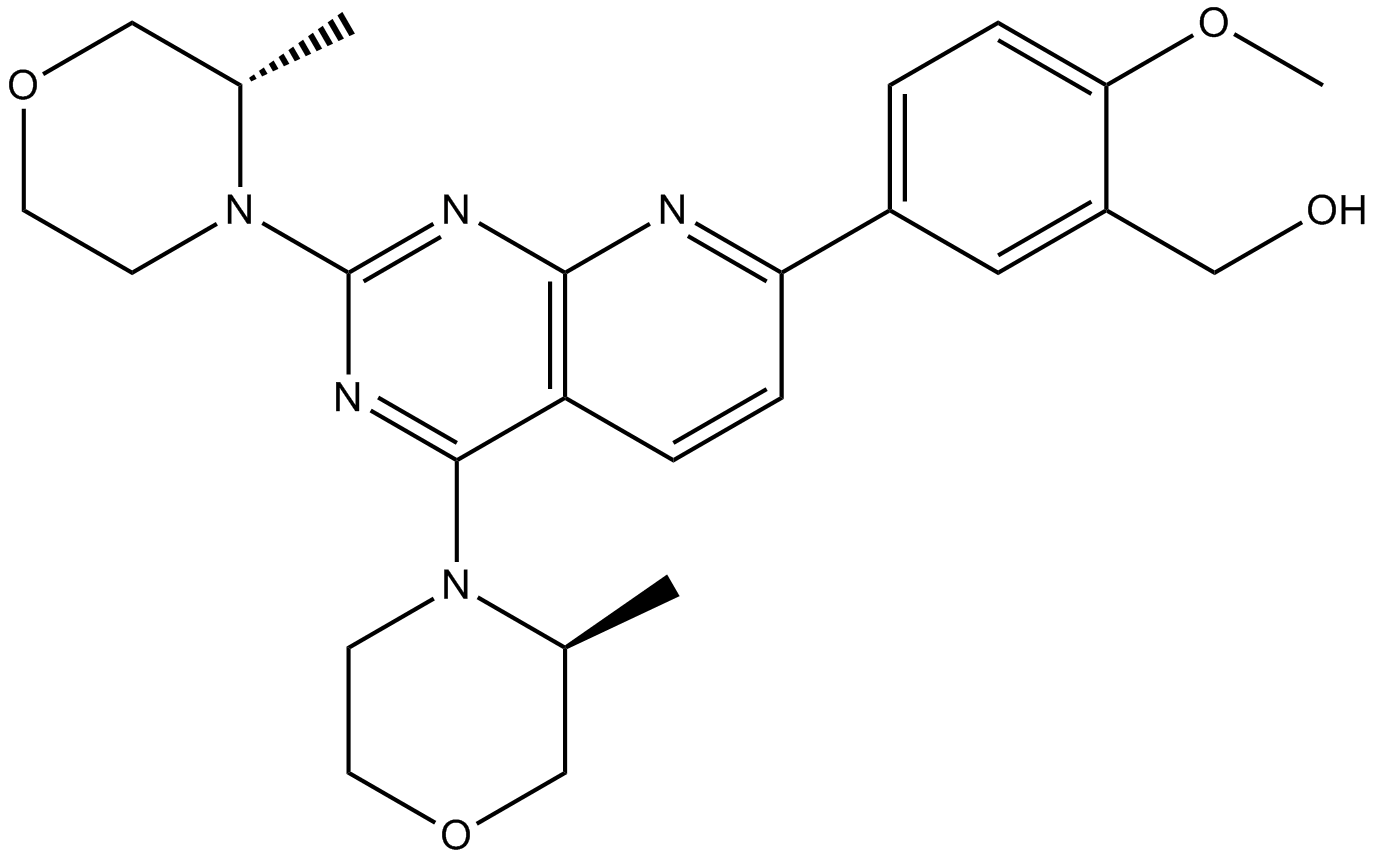
-
GC65507
AZD8154
AZD8154 is a novel inhaled selective PI3Kγδ dual inhibitor targeting airway inflammatory disease.
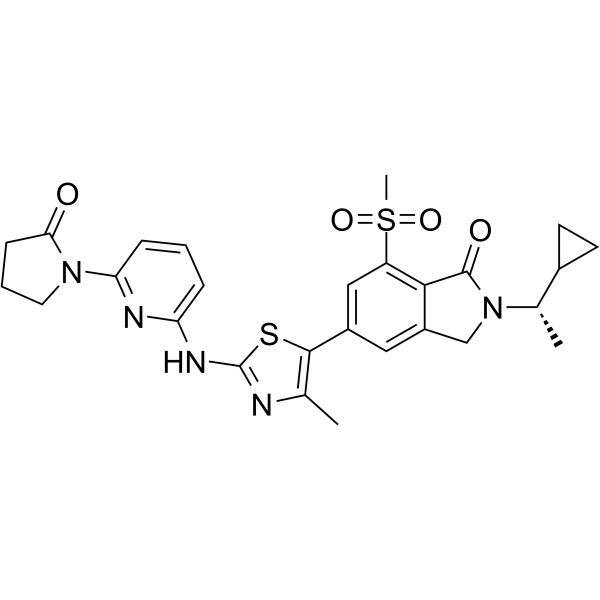
-
GC12576
AZD8186
Potent and selective inhibitor of PI3Kβ and PI3Kδ
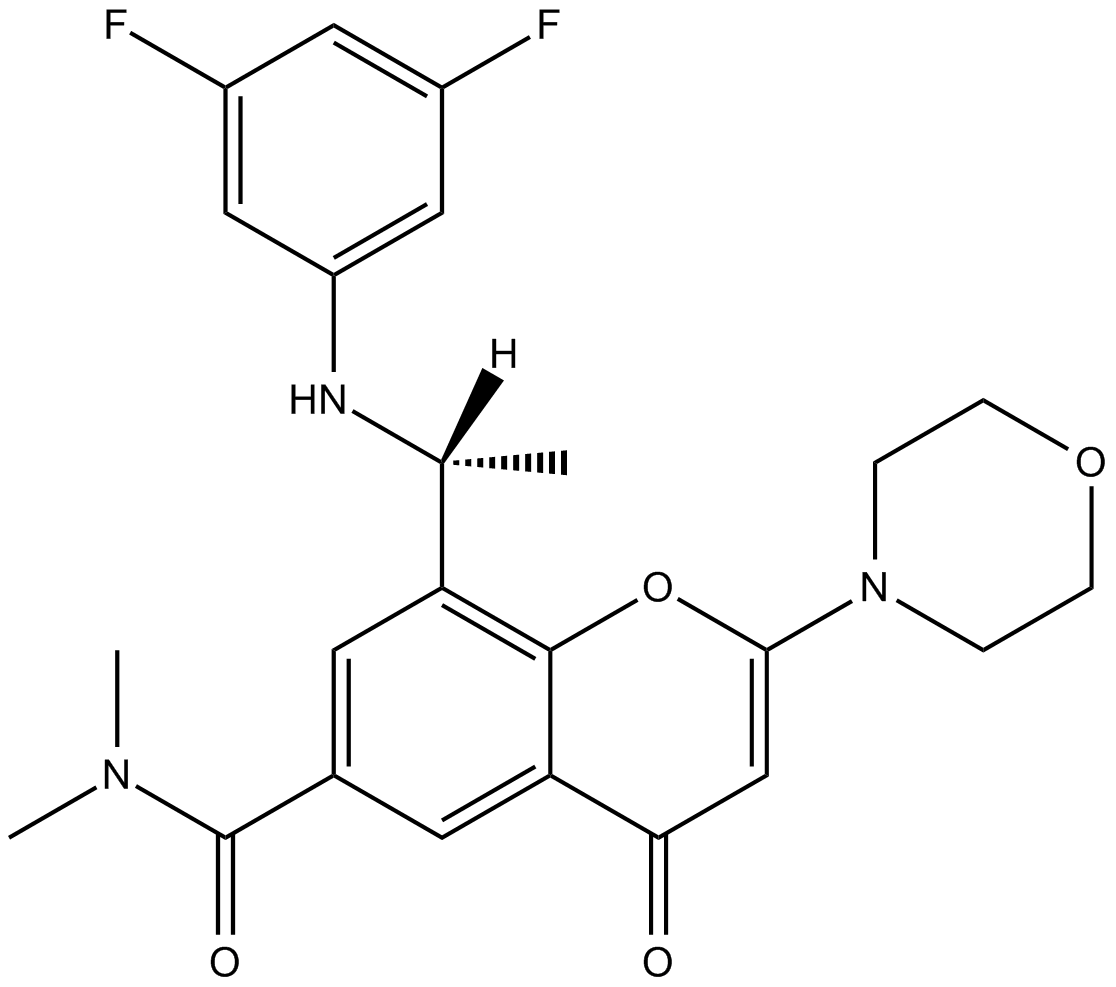
-
GC11248
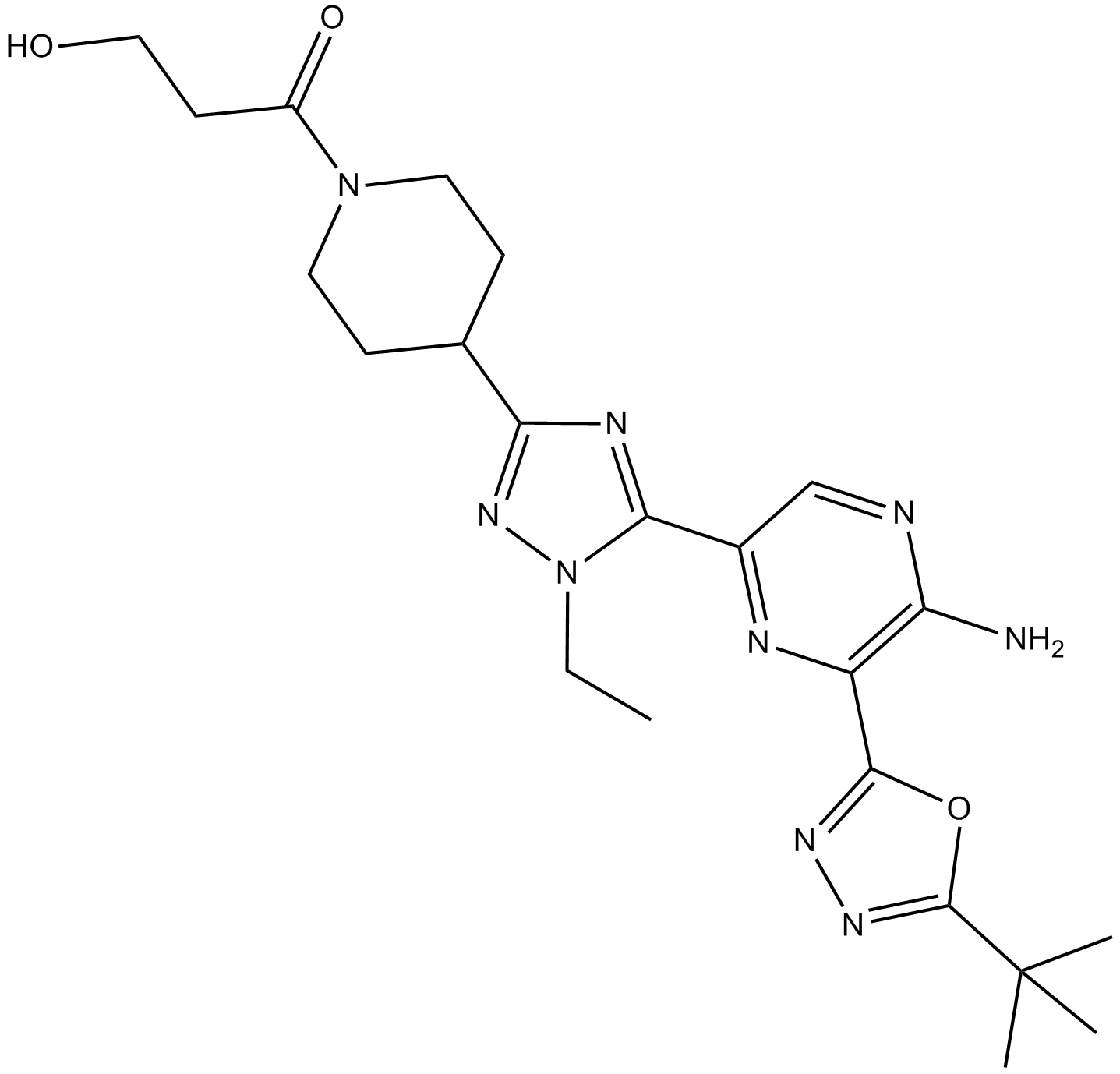
AZD8835
PI3Kα and PI3Kδ inhibitor
-
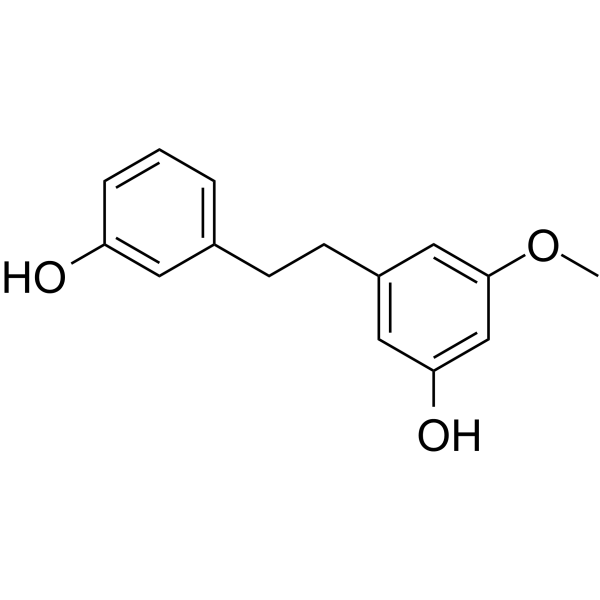
GC64377
Batatasin III
Batatasin III, a stilbenoid, inhibits cancer migration and invasion by suppressing epithelial to mesenchymal transition (EMT) and FAK-AKT signals. Batatasin III has anti-cancer activities.
-
GC17766
BAY 80-6946 (Copanlisib)
BAY 80-6946
BAY 80-6946 (Copanlisib) (BAY 80-6946) is a potent, selective and ATP-competitive pan-class I PI3K inhibitor, with IC50s of 0.5 nM, 0.7 nM, 3.7 nM and 6.4 nM for PI3Kα, PI3Kδ, PI3Kβ and PI3Kγ, respectively. BAY 80-6946 (Copanlisib) has more than 2,000-fold selectivity against other lipid and protein kinases, except for mTOR. BAY 80-6946 (Copanlisib) has superior antitumor activity.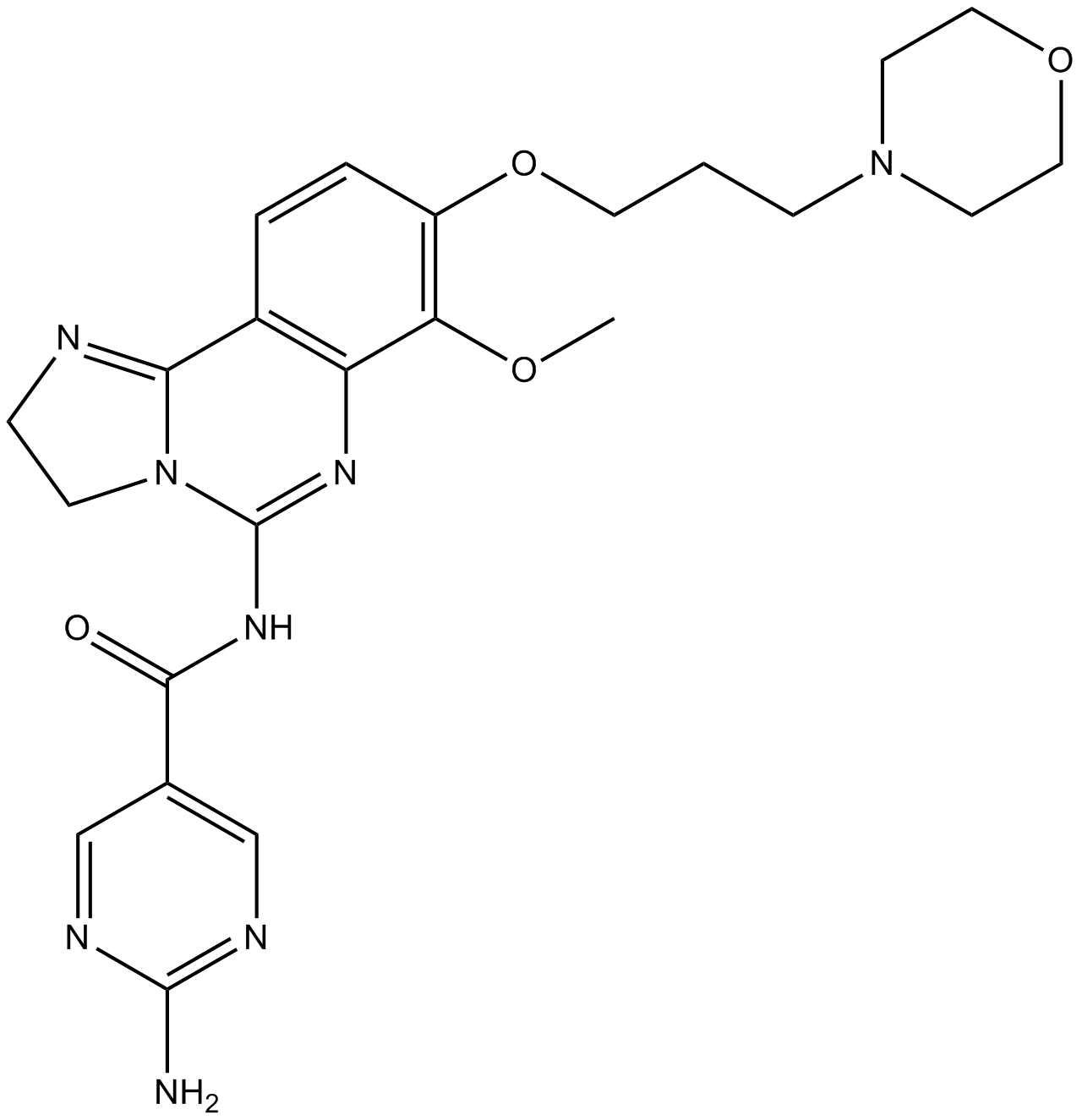
-
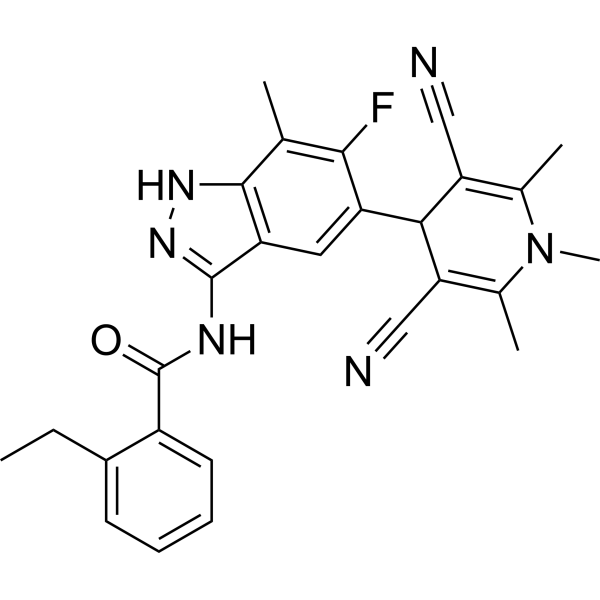
GC67772
BAY-3827
-
GC62164
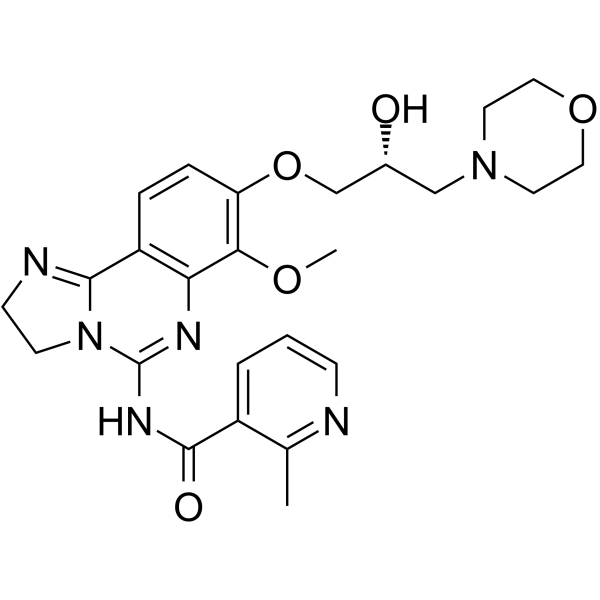
BAY1082439
BAY1082439 is an orally bioavailable, selective PI3Kα/β/δ inhibitor. BAY1082439 also inhibits mutated forms of PIK3CA. BAY1082439 is highly effective in inhibiting Pten-null prostate cancer growth.
-
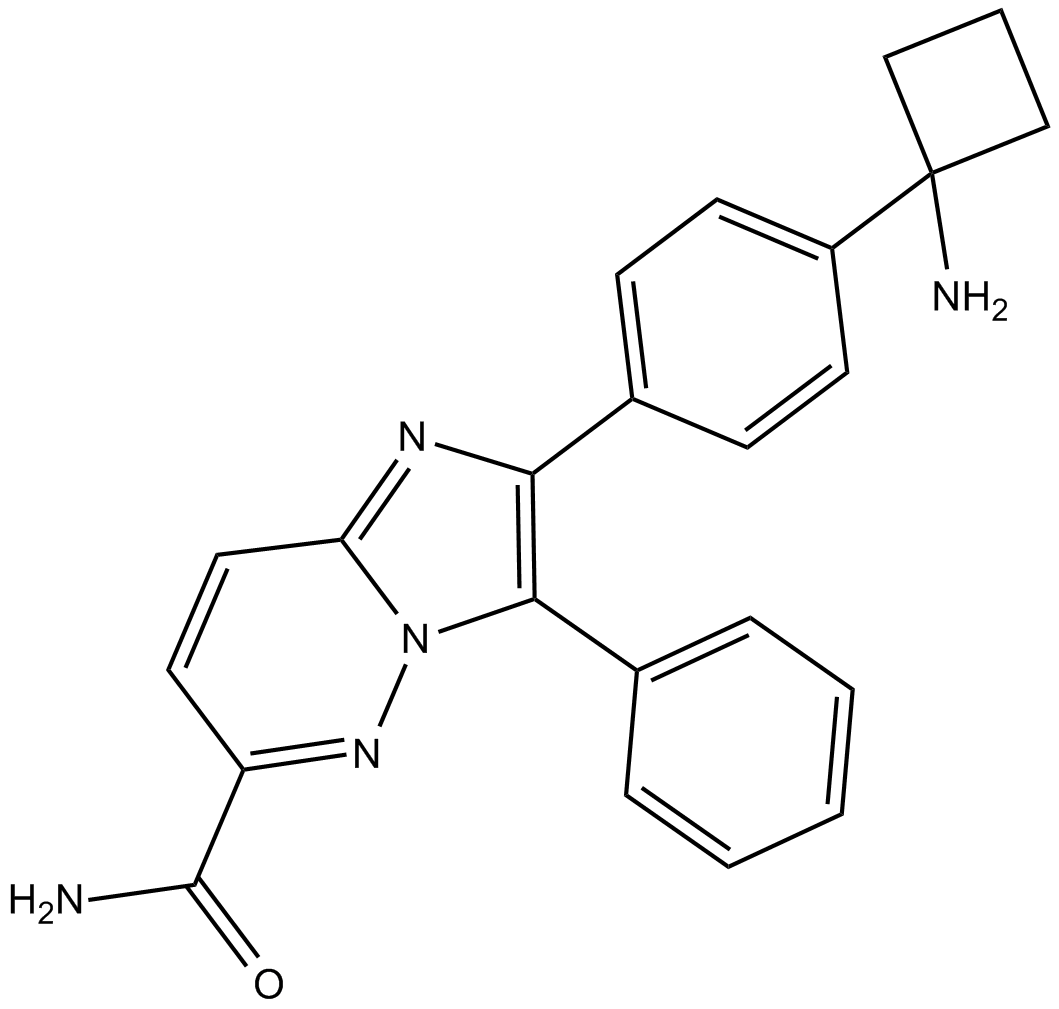
GC19484
BAY1125976
A selective allosteric Akt1/Akt2 inhibitor
-
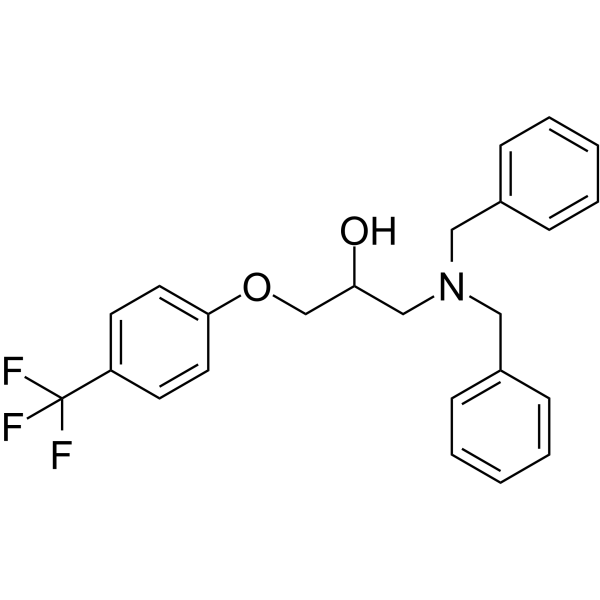
GC62135
BC1618
BC1618, an orally active Fbxo48 inhibitory compound, stimulates Ampk-dependent signaling (via preventing activated pAmpkα from Fbxo48-mediated degradation).
-
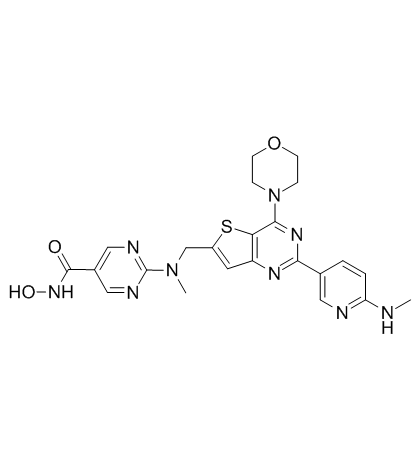
GC33145
BEBT-908 (PI3Kα inhibitor 1)
BEBT-908 (PI3Kα inhibitor 1) (PI3Kα inhibitor 1) is a selective PI3Kα inhibitor extracted from patent US/20120088764A1, Compound 243, has an IC50<0.1 μM, PI3Kα inhibitor 1 also inhibits HDAC (0.1 μM≤IC50≤1 μM) .
-
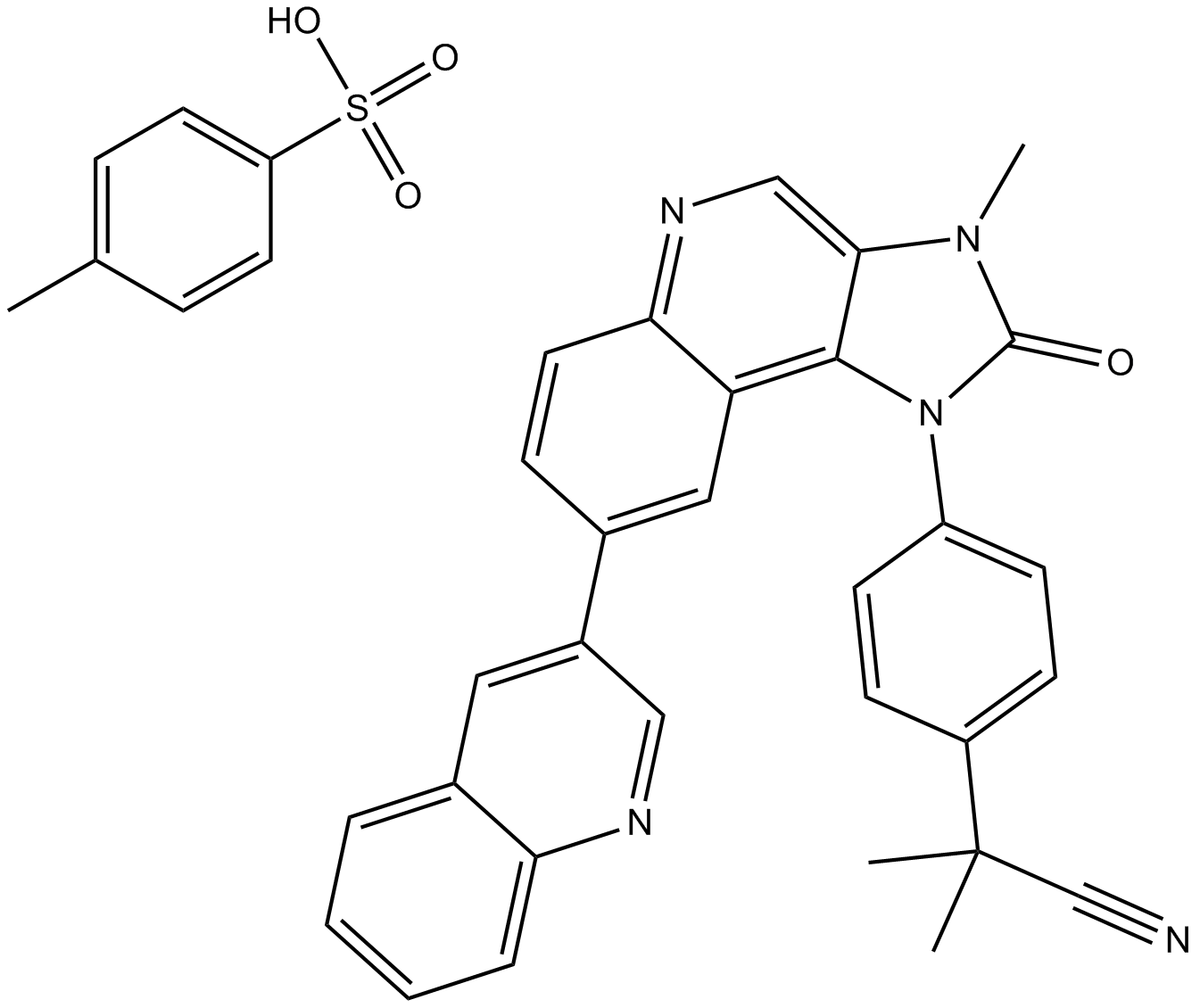
GC13271
BEZ235 Tosylate