Membrane Transporter/Ion Channel
- Piezo Channels(1)
- Aquaporins(1)
- AMPAR(57)
- ATPase(79)
- Calcium Channel(305)
- Cannabinoid Transporters(6)
- CFTR(43)
- Chloride Channels(13)
- GABA Receptor(191)
- Gardos Channel(0)
- Glucose Transporters(12)
- Glutamate (EAAT) Transporters(18)
- Glycine Receptors(2)
- GlyT(18)
- GTPase(21)
- Kv1.3(0)
- ICB(1)
- Lipophilic platinum complex(0)
- M2 ion Channel(0)
- MCT2(2)
- Monoamine transporter(17)
- Monocarboxylate Transporters(11)
- Multidrug Transporters(4)
- Na+/Ca2+ Exchanger(16)
- NKCC(7)
- NMDA Receptor(81)
- Nucleoside Transporters(4)
- Pannexin-1(2)
- P2X purinergic receptor(67)
- P-gp(11)
- Proton Pump(55)
- Potassium Channel(300)
- Sodium Channel(201)
- TRP Channel(157)
- TRPV1(10)
- URAT1(12)
- Other Channel Modulators(10)
- MCT(3)
- Kainate Receptors(12)
- Ionophore(3)
- sodium-hydrogen exchanger(3)
- Chloride Channel(26)
- CRAC Channel(13)
- EAAT2(6)
- HCN Channel(5)
- Na+/HCO3- Cotransporter(1)
- Na+/K+ ATPase(28)
- P-glycoprotein(48)
- iGluR(163)
- nAChR(100)
- VDAC(2)
- GlyR(1)
Products for Membrane Transporter/Ion Channel
- Cat.No. Product Name Information
-
GC49097
α-Conotoxin AuIB (trifluoroacetate salt)
GCCSYPPCFATNPDC
A conotoxin and an antagonist of α3β4 subunit-containing nAChRs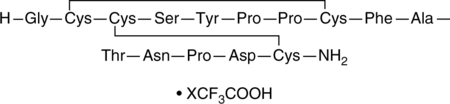
-
GC49140
α-Conotoxin ImI (trifluoroacetate salt)
α-CTx ImI, GCCSDPRCAWRC
A conotoxin and an antagonist of α7 nAChRs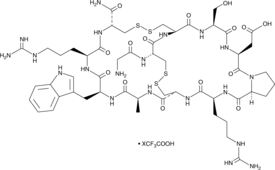
-
GC63269
α-Conotoxin PIA TFA

-
GC63723
α-Conotoxin PnIA TFA
-
GC10873
α-Spinasterol
Bessisterol, Hitodesterol
TRPV1 antagonist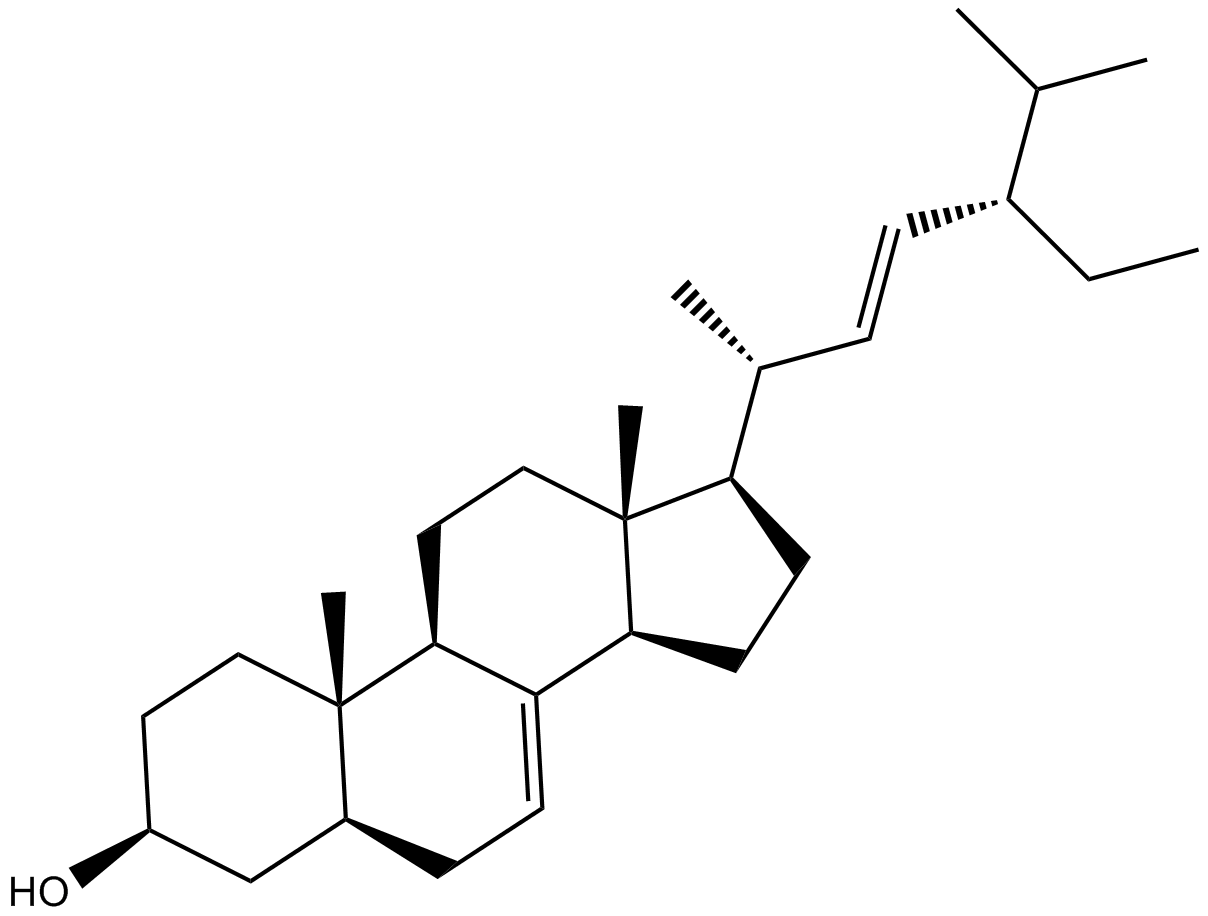
-
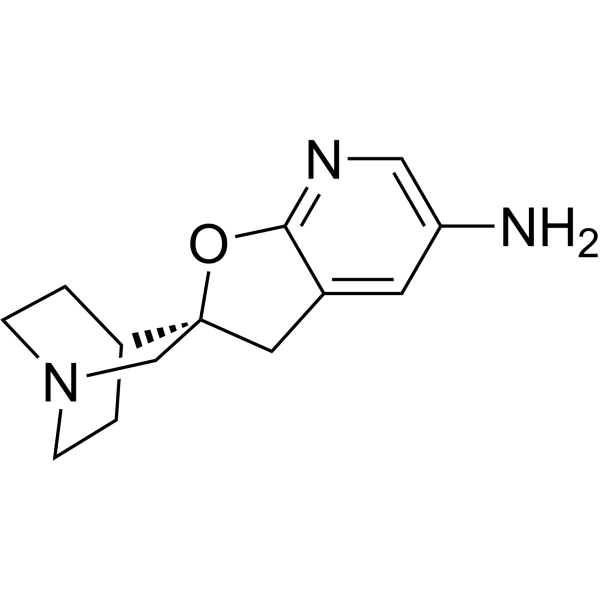
GC70179
α7 Nicotinic receptor agonist-1
Alpha-7 Nicotinic receptor agonist-1 (Preparation 5) is an alpha-7 nAChR agonist. It can be used for research on psychiatric disorders such as schizophrenia, mania or bipolar disorder, and anxiety disorders, as well as intellectual disabilities such as Alzheimer's disease, learning deficits, cognitive impairments, attention deficits, memory loss, Lewy body dementia and attention deficit hyperactivity disorder.
-
GC48998
β-Defensin-1 (human) (trifluoroacetate salt)
hBD-1
An antimicrobial peptide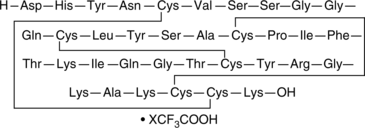
-
GC70650
γ-Acetylenic GABA hydrochloride
γ-Acetylenic GABA (GAG) drochloride is an irreversible inhibitor of GABA-transaminase.

-
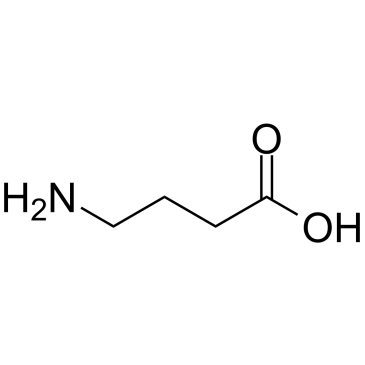
GC38010
γ-Aminobutyric acid
-
GC64508
γ-Aminobutyric acid-d6
γ-Aminobutyric acid-d6 (4-Aminobutyric acid-d6) is the deuterium labeled γ-Aminobutyric acid.

-
GC48313
γ-Lindane
An insecticide and GABAA receptor antagonist

-
GC15513
ω-Agatoxin IVA
ω-Agatoxin IVA is a potent, selective P/Q type Ca2+ (Cav2.1) channel blocker with IC50s of 2 nM and 90 nM for P-type and Q-type Ca2+ channels, respectively.
-
GC12608
ω-Agatoxin TK
ω-Agatoxin TK, a peptidyl toxin of the venom of Agelenopsis aperta, is a potent and selective P/Q type Ca2+ channel blocker.
-
GC13886
ω-Conotoxin GVIA
ω-Conotoxin GVIA is a cone snail toxin that selectively blocks N-type channels in neurons .
-
GC18070
ω-Conotoxin MVIIC
wide spectrum blocker of N, P and Q type calcium channels
-
GC31248
β-Amino Acid Imagabalin Hydrochloride (PD-0332334)
PD-0332334
β-Amino Acid Imagabalin Hydrochloride (PD-0332334) (PD-0332334) is a ligand for the α2δ subunit of the voltage-dependent calcium channel.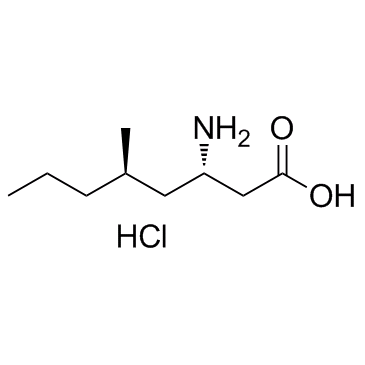
-
GC11449
β-CCB
benzodiazepine receptor ligand
-
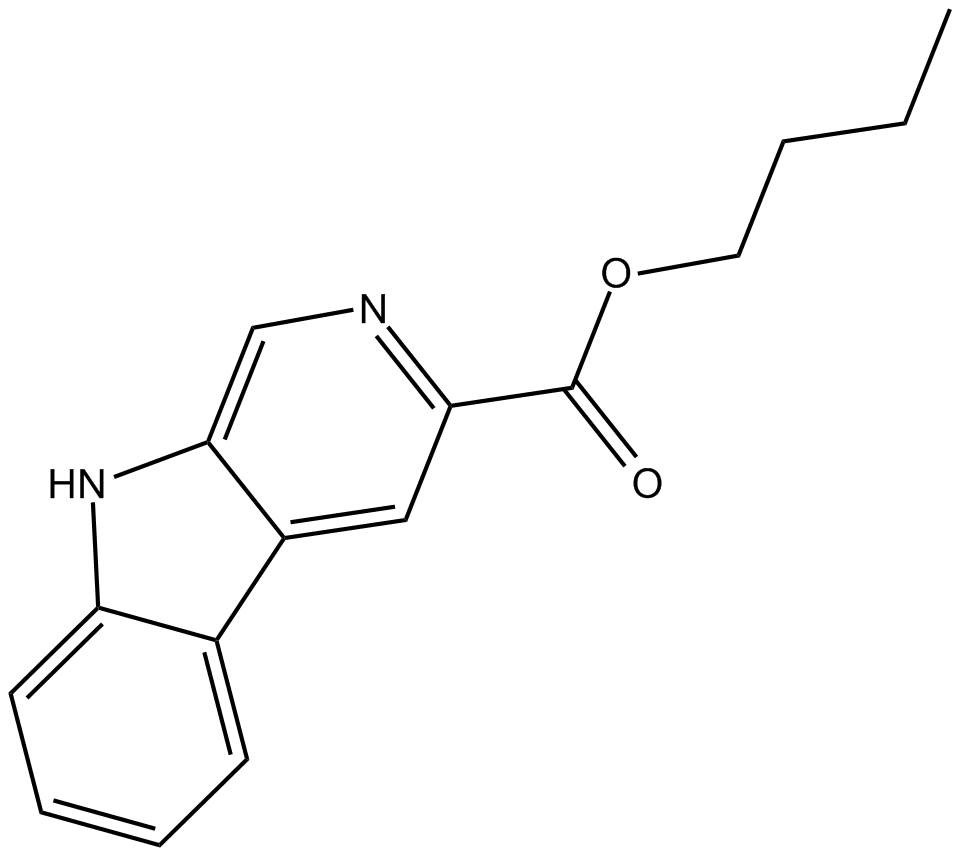
GC18033
γDGG
γDGG is a competitive AMPA receptor blocker.
-
GC70188
ω-Agatoxin IVA TFA
ω-Agatoxin IVA TFA is an effective selective blocker of P/Q-type Ca2+ (Cav2.1) channels, with IC50 values of 2 nM and 90 nM for P-type and Q-type Ca2+ channels, respectively. ω-Agatoxin IVA TFA (IC50, 30-225 nM) inhibits high potassium-induced glutamate release and calcium influx. It also blocks the release of serotonin and adrenaline induced by high potassium, without affecting L-type or N-type calcium channels.
-
GC62067
ω-Conotoxin GVIA TFA

-
GA24016
ω-Conotoxin MVIIA
ω-Conotoxin MVIIA (SNX-111), a peptide, is a potent and selective block of N-type calcium channels antagonist.

-
GC64899
ω-Conotoxin MVIIC TFA

-
GC49296
τ-Fluvalinate
tau-Fluvalinate
A pyrethroid acaricide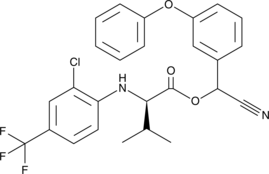
-
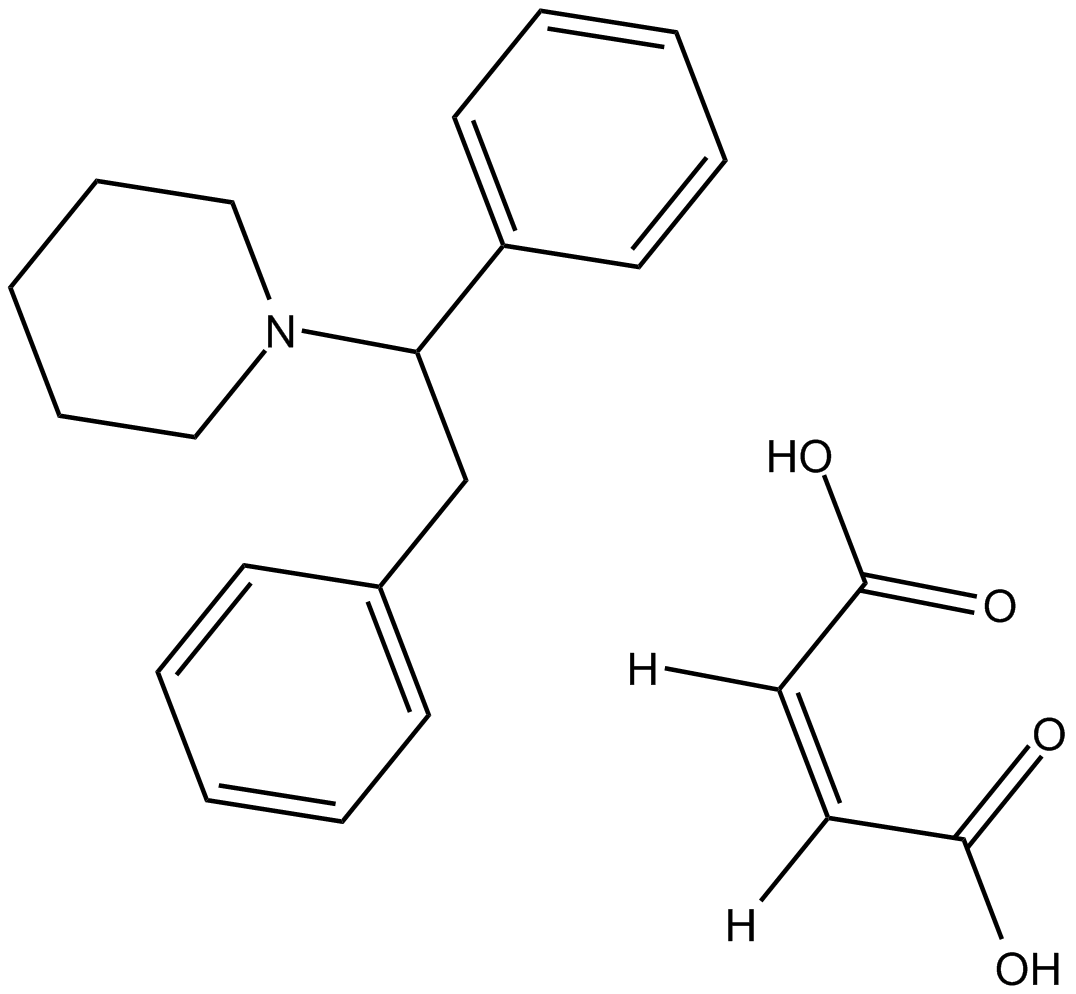
GC10237
(±)-1-(1,2-Diphenylethyl)piperidine maleate
NMDA receptor antagonist
-
GC72625
(±)-Coniine hydrochloride
(±)-Coniine drochloride (2-Propylpiperidine drochloride) is a potent nAChR agonist with an EC50 value of 0.3 mM.

-
GC14410
(±)-Epibatidine
CMI 545
nicotinic agonist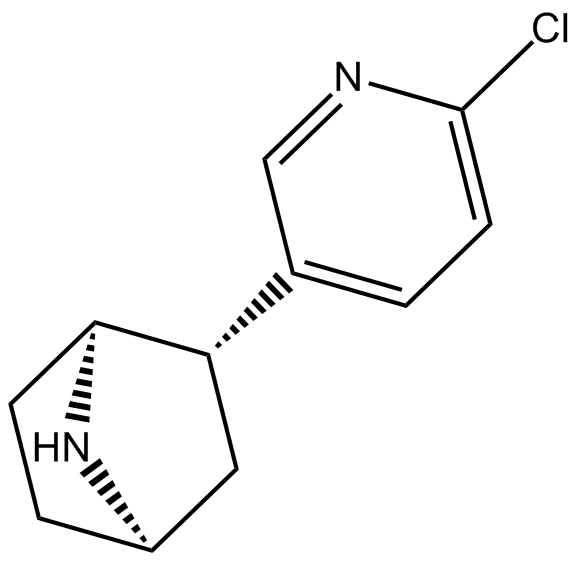
-
GC11138
(±)-HIP-A
excitatory amino acid transporter (EAAT) blocker

-
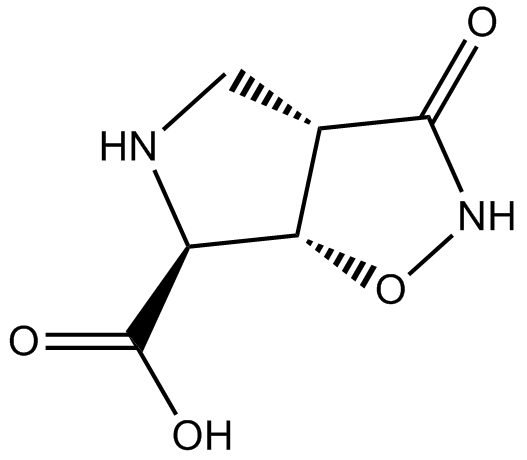
GC13735
(±)-HIP-B
excitatory amino acid transporter (EAAT) blocker
-
GC45985
(±)-Indoxacarb
DPX-JW062, DPX-MP062
A broad-spectrum insecticide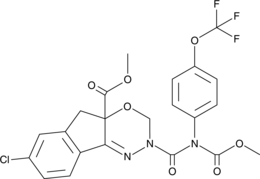
-
GC14834
(±)-Nipecotic acid
(±)-Nipecotic Acid, (R,S)-Nipecotic Acid, 3-Piperidine Carboxylic Acid, DL-Piperidine 3-Carboxylic Acid
GABA uptake inhibitor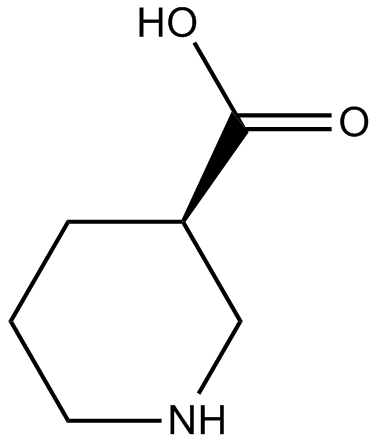
-
GC41676
(±)-Nornicotine
DL-Nornicotine, (R,S)-Nornicotine
(±)-Nornicotine is a metabolite of nicotine that acts as a neuronal nicotinic acetylcholine receptor (nAChR) agonist.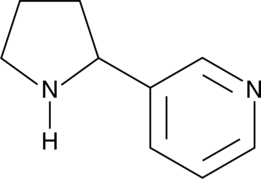
-
GC49482
(±)-Nornicotine-d4
DL-Nornicotine-d4, (R,S)-Nornicotine-d4
An internal standard for the quantification of (±)-nornicotine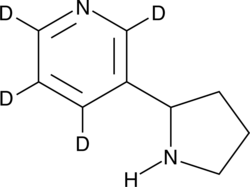
-
GC41677
(±)-SDZ-201 106
DPI 201-106
(±)-SDZ-201 106 (SDZ 201106) is a cardiotonic agent with a synergistic sarcolemmal and intracellular mechanism of action.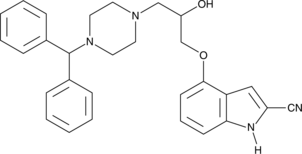
-
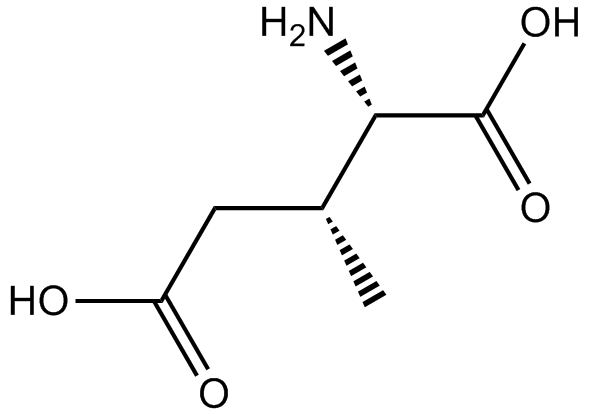
GC11982
(±)-threo-3-Methylglutamic acid
glutamate transport blocker
-
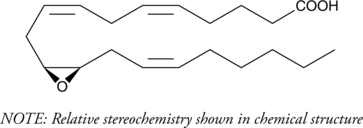
GC40466
(±)11(12)-EET
(±)11,12-EpETrE
(±)11(12)-EET is a fully racemic version of the R/S enantiomeric forms biosynthesized from arachidonic acid by cytochrome P450 enzymes.
-
GC40434
(±)16-HETE
(±)16-Hydroxyeicosatetraenoic Acid
Electrolyte and fluid transport in the kidney are regulated in part by arachidonic acid and its metabolites.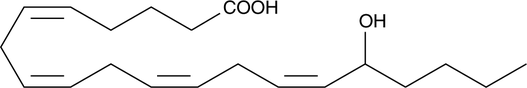
-
GC40270
(±)5(6)-DiHET
(±)5,6-DiHETrE
5(6)-DiHET is a fully racemic version of the enantiomeric forms biosynthesized from 5(6)-EET by epoxide hydrolases.
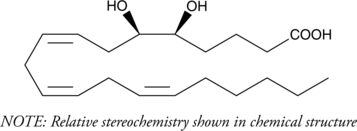
-
GC40438
(±)5(6)-EET
(±)5,6EpETrE
5(6)-EET is a fully racemic version of the enantiomeric forms biosynthesized from arachidonic acid by cytochrome P450 enzymes.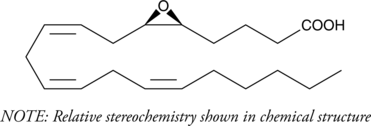
-
GN10612
(+)- Praeruptorin C

-
GC34954
(+)-Borneol
D-Borneol, (1R)-(+)-Borneol, (+)-endo-Borneol

-
GC30933
(+)-Kavain
Kawain, NSC 112162
An Analytical Reference Standard
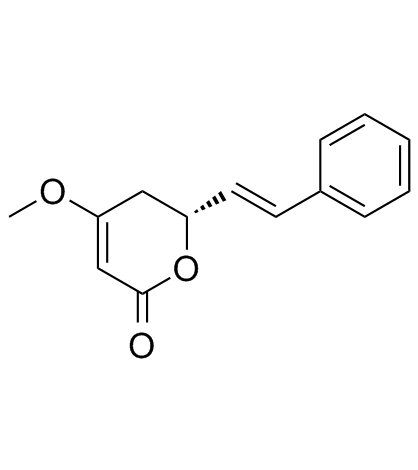
-
GC40808
(+)-Menthol
D-Menthol
(+)-Menthol is a monoterpene alcohol that has been found in Cannabis and has antifungal activity.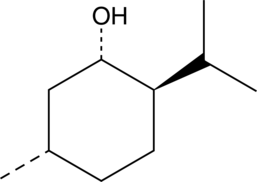
-
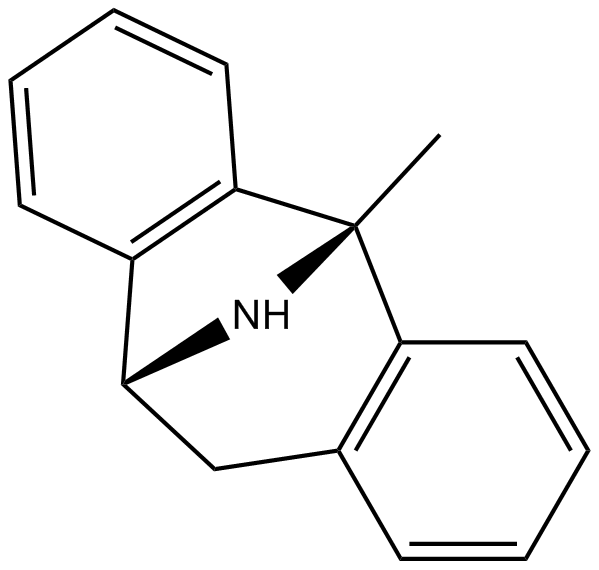
GC16616
(+)-MK 801
Dizocilpine maleate;Dizocilpine hydrogen maleate;(+)-MK 801;MK 801
-
GC11025
(+)-MK 801 Maleate
Dizocilpine Maleate
(+)-MK 801 Maleate (MK-801 maleate) is a potent, selective and non-competitive NMDA receptor antagonist with Kd of 37.2 nM in rat brain membranes.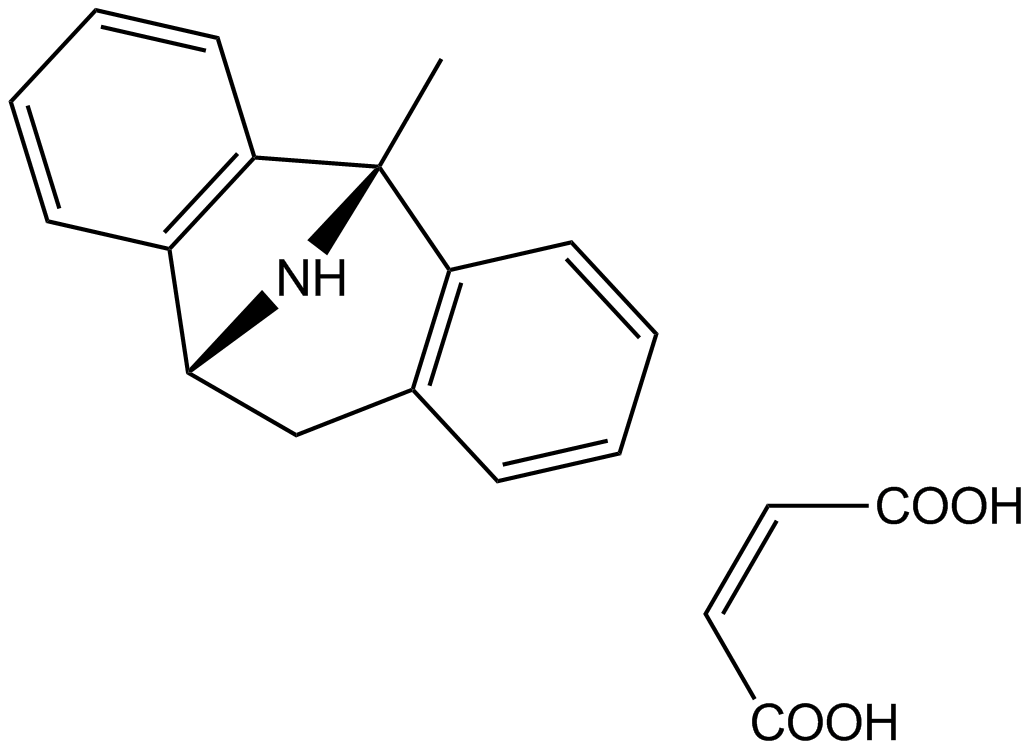
-
GC32130
(+)-SJ733 (SJ000557733)
SJ000557733
(+)-SJ733 (SJ000557733) is an anti-malaria agent which can also inhibit Na+-ATPase PfATP4.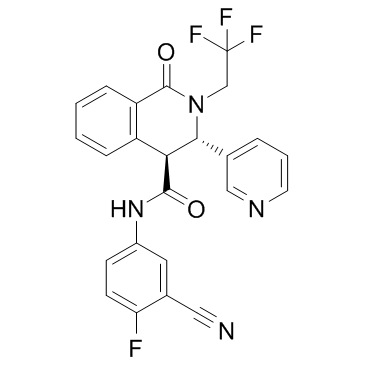
-
GC34959
(+)-Sparteine
Pachycarpin, Pachycarpine
(+)-Sparteine is a natural alkaloid acting as a ganglionic blocking agent.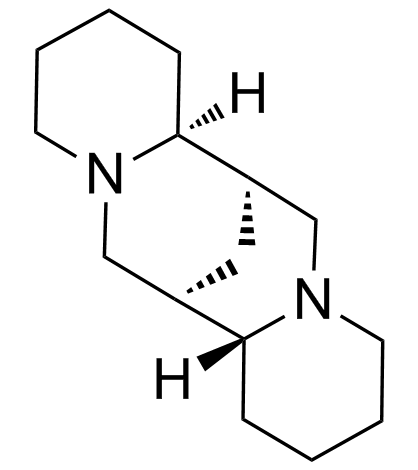
-
GC38677
(-)-α-Pinene
NSC 7727
(-)-α-Pinene is a monoterpene and shows sleep enhancing property through a direct binding to GABAA-benzodiazepine (BZD) receptors by acting as a partial modulator at the BZD binding site.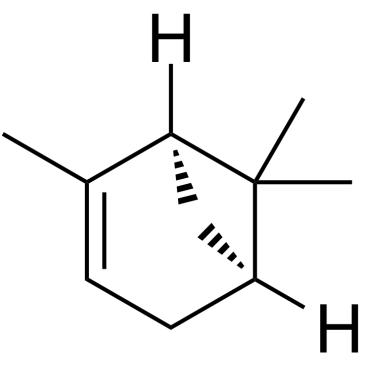
-
GC31084
(-)-(S)-B-973B
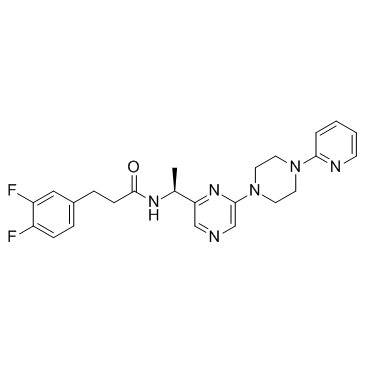
-
GC10065
(-)-Bicuculline methiodide
GABAA receptor antagonist
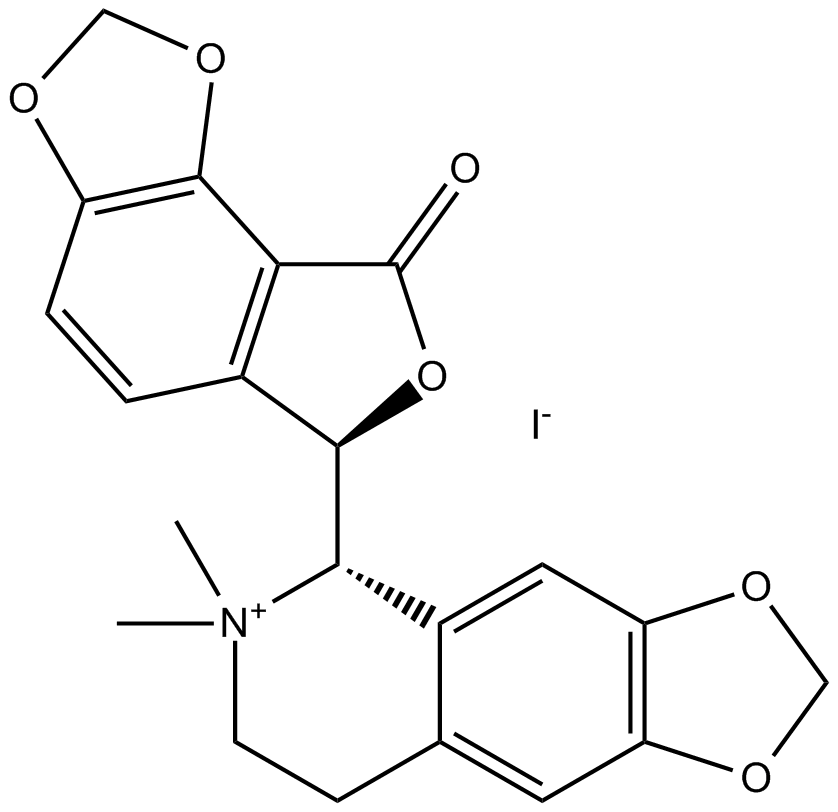
-
GC16857
(-)-Bicuculline methobromide
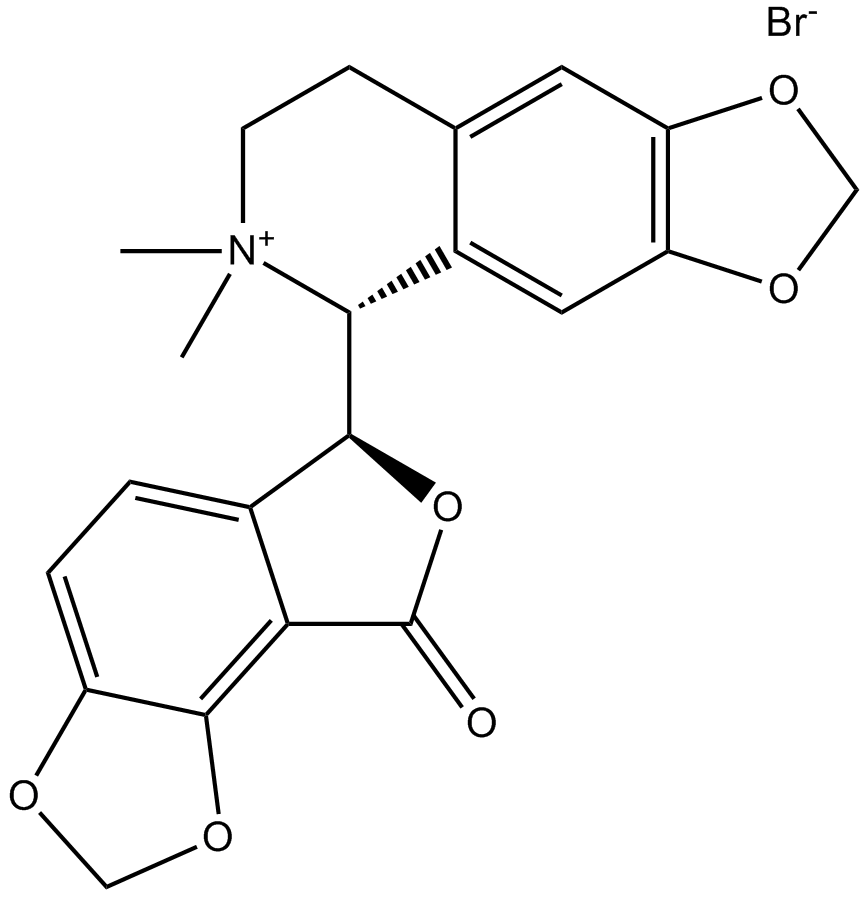
-
GC15636
(-)-Bicuculline methochloride
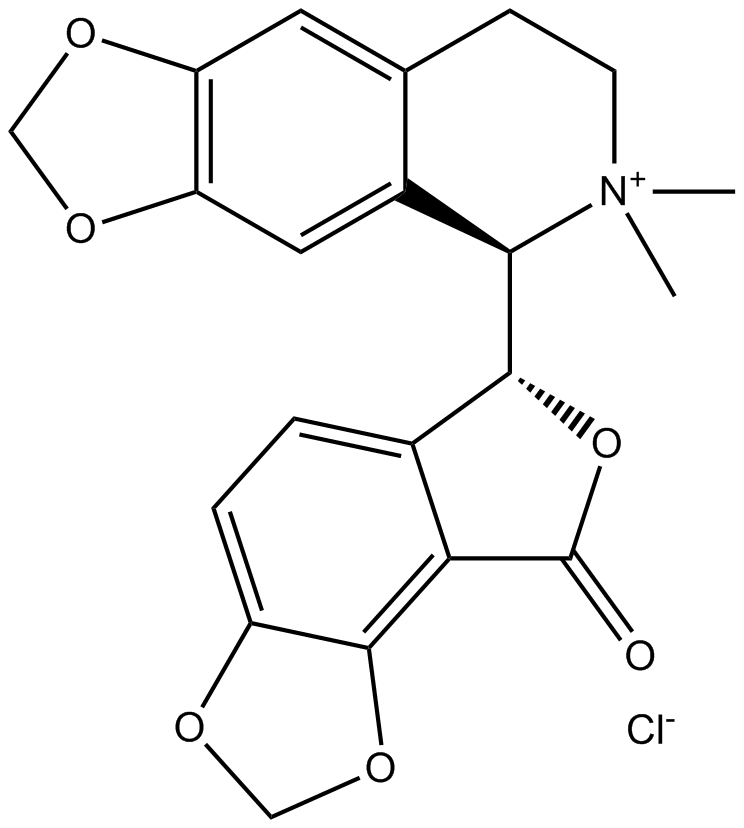
-
GC13430
(-)-Blebbistatin
(S)Blebbistatin
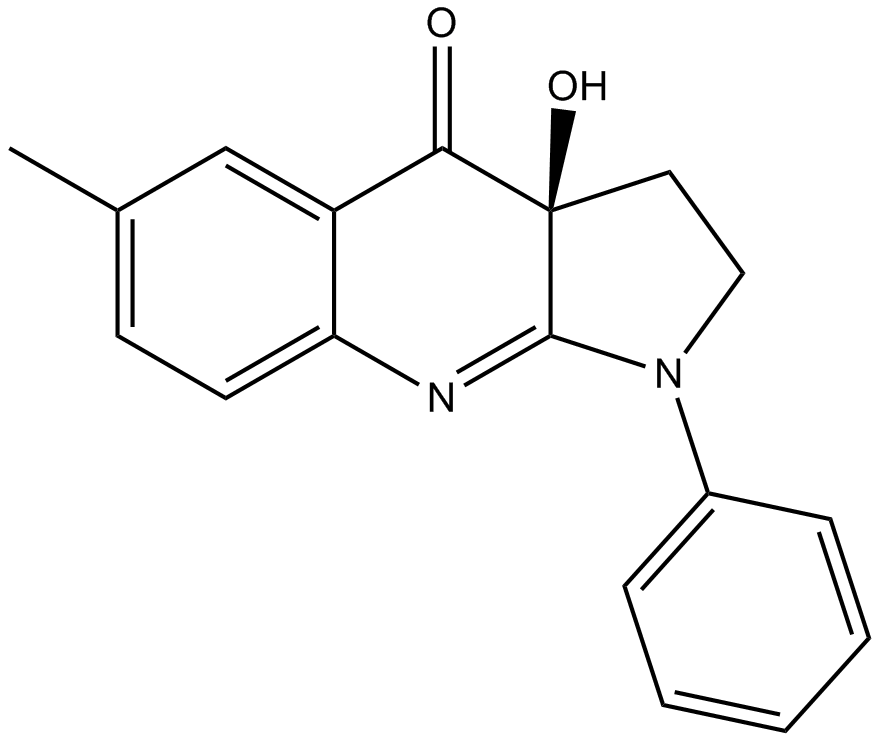
-
GC63940
(-)-Denudatin B
(-)-Denudatin B is an antiplatelet agent.

-
GC14157
(-)-Lobeline hydrochloride
(–)-α-Lobeline, VUF 10751
nicotinic receptor partial agonist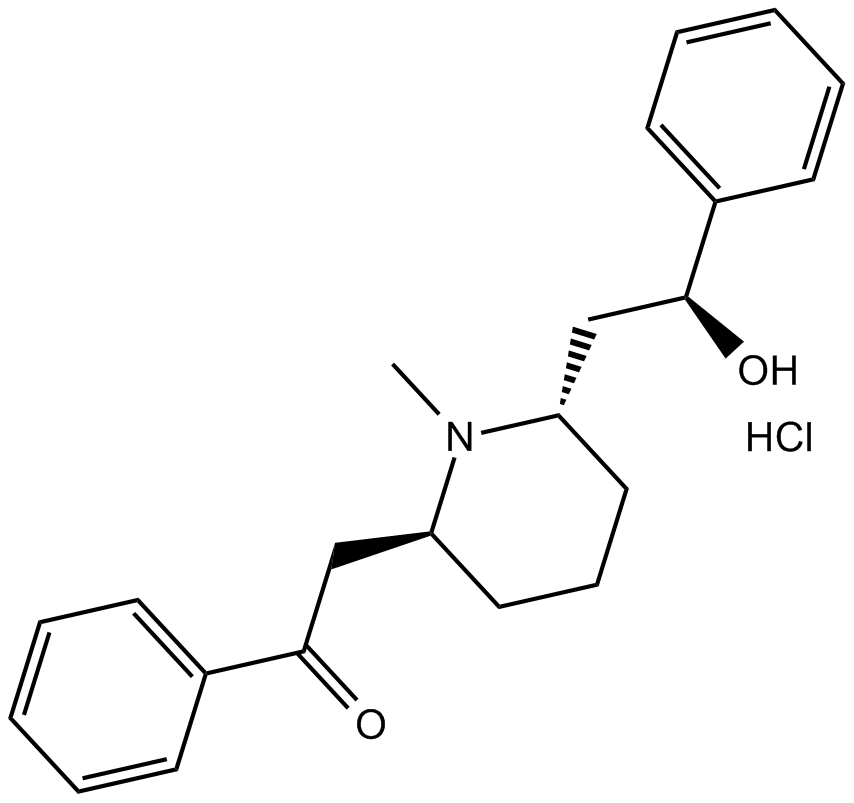
-
GC34951
(-)-Menthol
L-Menthol, (1R,2S,5R)-(-)-Menthol, NSC 62788
A monoterpene with diverse biological activities
-
GC15270
(-)-MK 801
(-)-Dizocilpine Maleate
(-)-MK 801 ((-)-MK-801 maleate) is a less active (-)-enantiomer of Dizocilpine.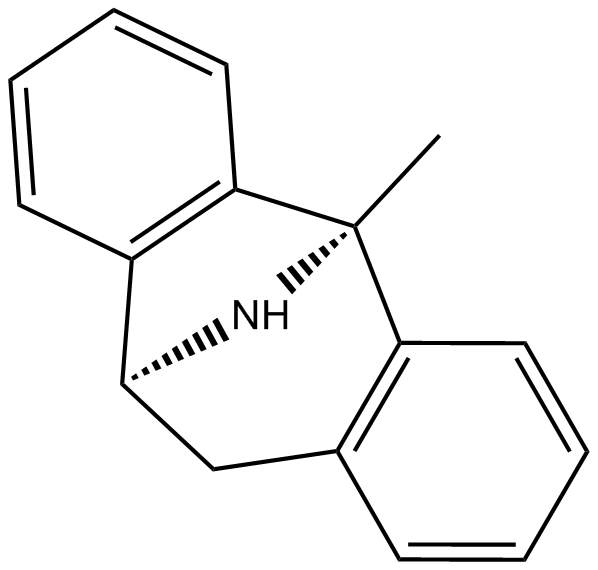
-
GN10445
(-)-pareruptorin A
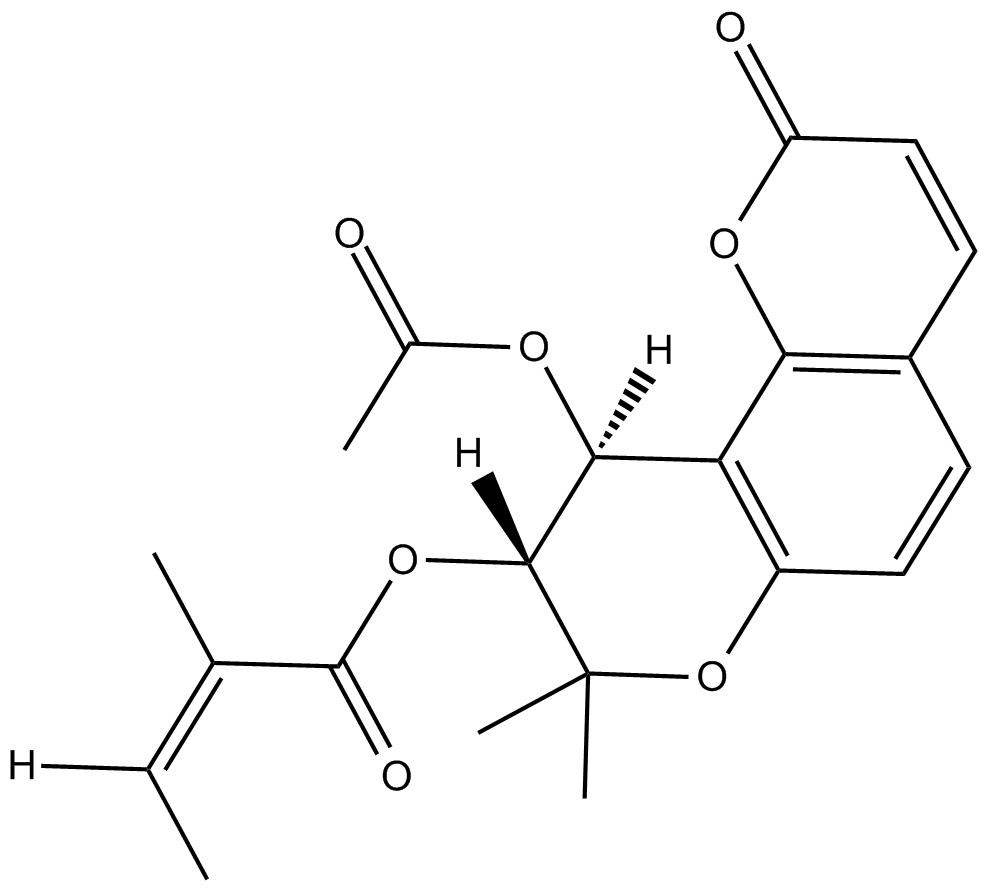
-
GC30889
(-)-Securinine
NSC 107413, L-Securinine
An alkaloid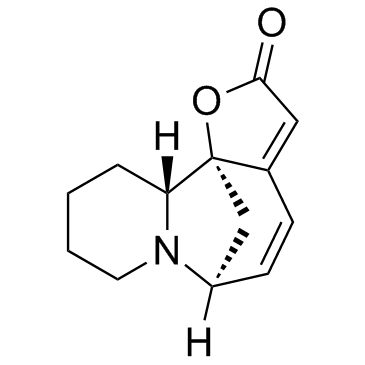
-
GC34952
(-)-Sparteine sulfate pentahydrate
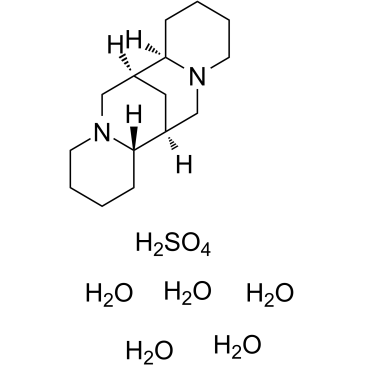
-
GC14484
(-)-Xestospongin C
XeC, Araguspongine E
IP3-dependent Ca2+ release inhibitor
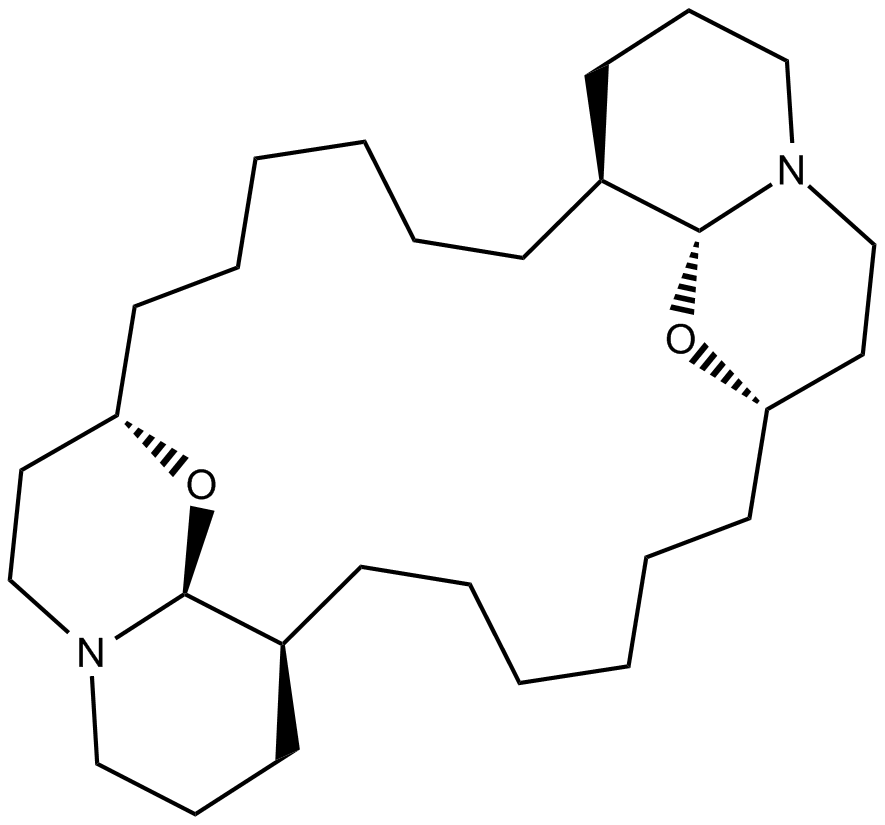
-
GC14044
(-)-[3R,4S]-Chromanol 293B
(-)-[3R,4S]-Chromanol 293B is a potent and selective inhibitor of the slow component of delayed rectifier K+ current (IKs).
![(-)-[3R,4S]-Chromanol 293B Chemical Structure (-)-[3R,4S]-Chromanol 293B Chemical Structure](/media/struct/GC1/GC14044.png)
-
GC64062
(1R,2R)-ML-SI3
(1R,2R)-ML-SI3 is a potent inhibitor of both TRPML1 and TRPML2 (IC50 values of 1.6 and 2.3 μM) and a weak inhibitor (IC50 12.5 μM) of TRPML3.
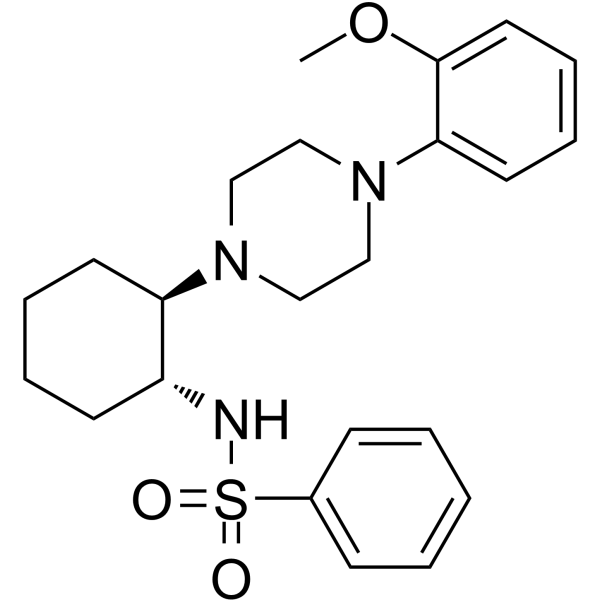
-
GC16915
(2R,3S)-Chlorpheg
NMDA receptor antagonist
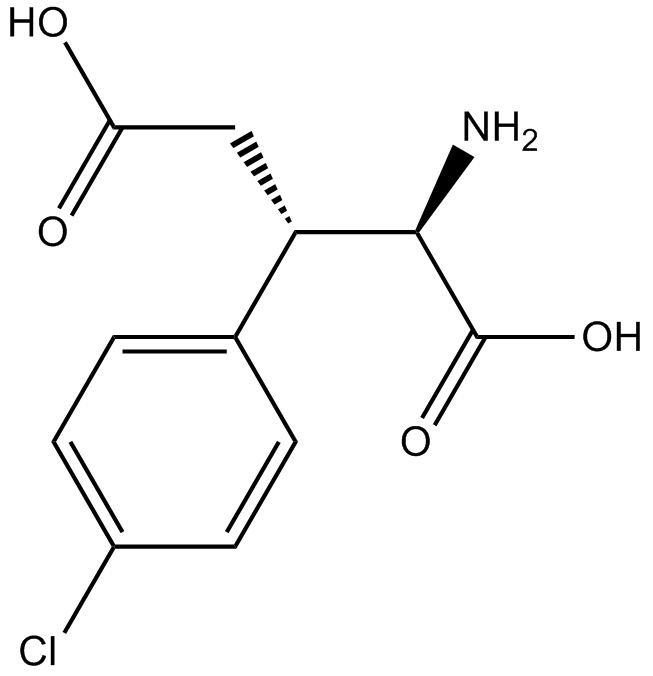
-
GC50536
(2R/S)-6-PNG
(±)-6-PN
CaV3.2 blocker; active in vivo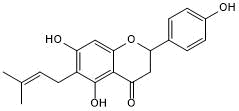
-
GC66255
(E)-3,4,5-Trimethoxycinnamic acid
TMCA
(E)-3,4,5-Trimethoxycinnamic acid (TMCA) is a cinnamic acid substituted by multi-methoxy groups. (E)-3,4,5-Trimethoxycinnamic acid is an orally active and potent GABAA/BZ receptor agonist. (E)-3,4,5-Trimethoxycinnamic exhibits favourable binding affinity to 5-HT2C and 5-HT1A receptor, with IC50 values of 2.5 and 7.6 μM, respectively. (E)-3,4,5-Trimethoxycinnamic acid shows anticonvulsant and sedative activity. (E)-3,4,5-Trimethoxycinnamic acid can be used for the research of insomnia, headache and epilepsy.
-
GC62560
(E/Z)-Sivopixant
(E/Z)-S-600918
(E/Z)-Sivopixant ((E/Z)-S-600918) is a potent P2X3 receptor antagonist with an IC50 of 4 nM.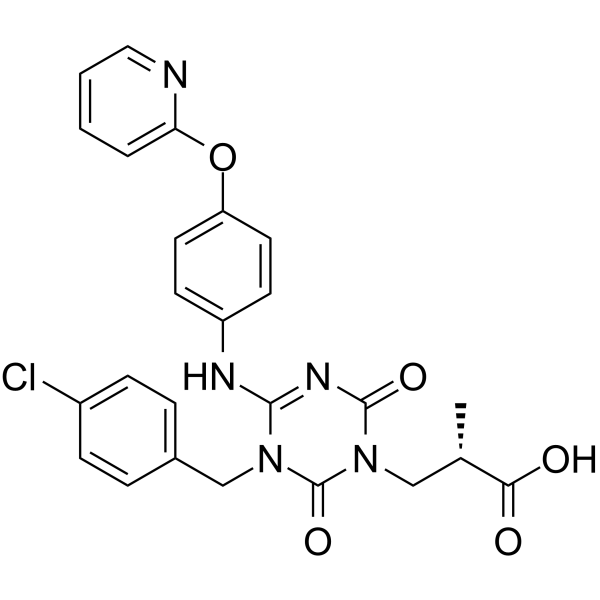
-
GC13895
(R)-(+)-Bay K 8644
NI 105, R 4407
L-type Ca2+-channel blocker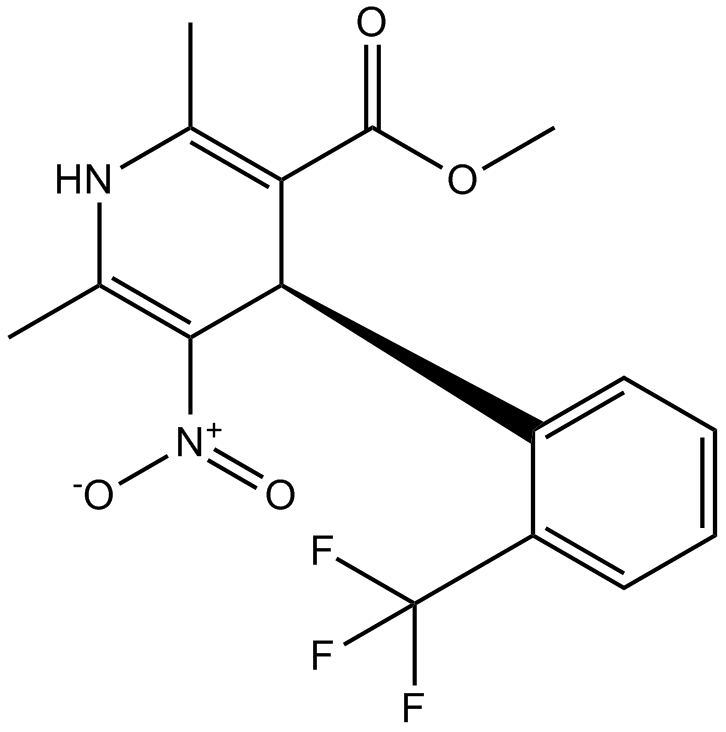
-
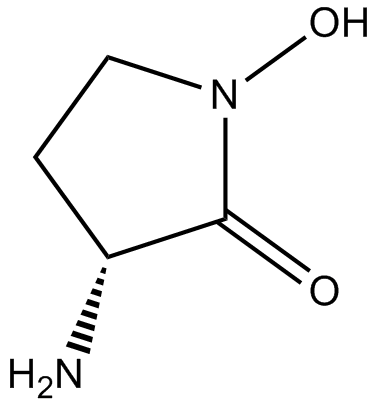
GC14085
(R)-(+)-HA-966
NMDA receptor antagonist/partial agonist
-
GC40696
(R)-(+)-Pulegone
NSC 15334, D-Pulegone
(R)-(+)-Pulegone, the major chemical constituent of Calamintha nepeta (L.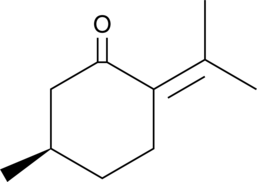
-
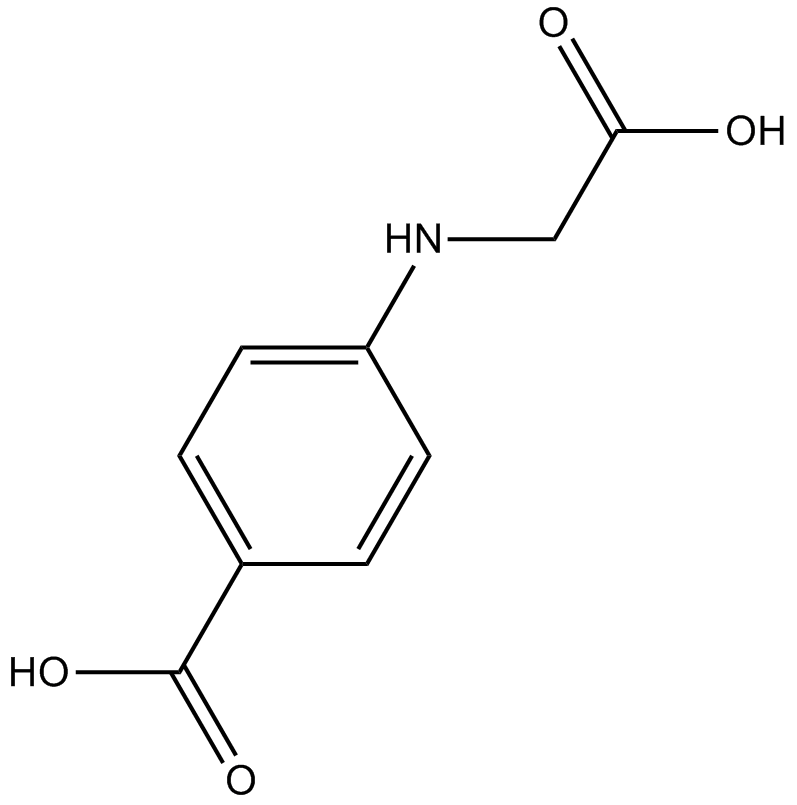
GC10168
(R)-4-Carboxyphenylglycine
NMDA receptor antagonist
-
GC11071
(R)-AMPA
inactive enantiomer of AMPA
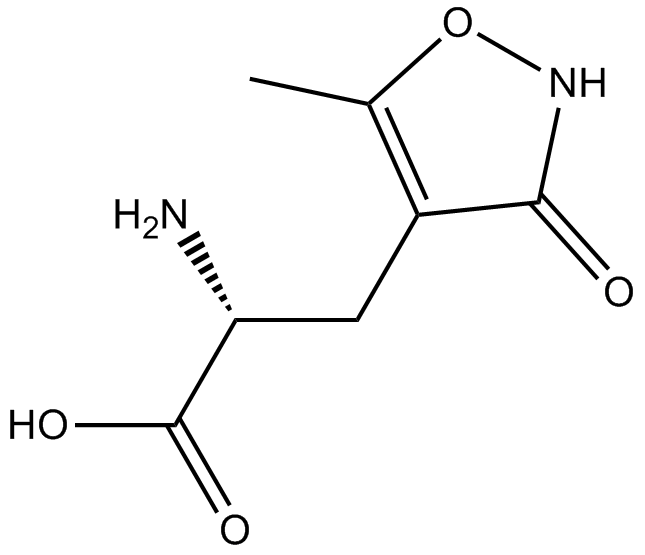
-
GC14111
(R)-baclofen
GABA receptor agonist
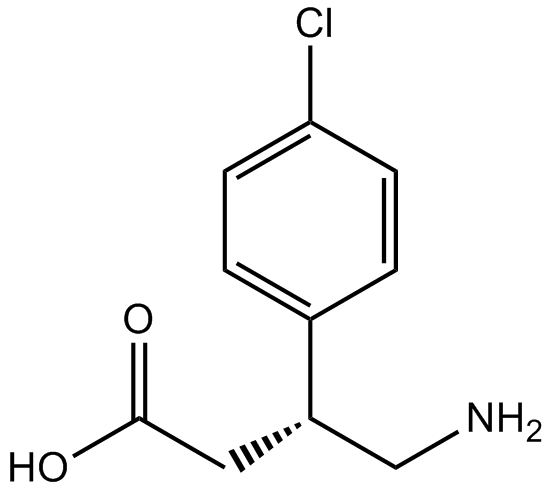
-
GC34986
(R)-Baclofen hydrochloride
(R)-Baclofen
(R)-Baclofen hydrochloride (Arbaclofen hydrochloride) is a selective GABAB receptor agonist.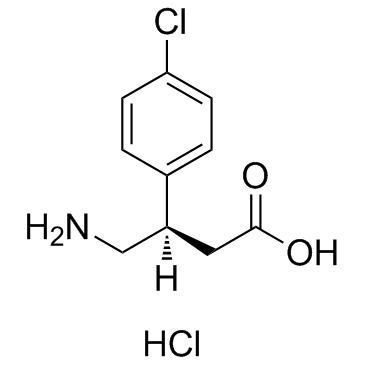
-
GC31665
(R)-BPO-27
(R)-BPO-27, the R enantiomer of BPO-27, is a potent, orally active and ATP-competitive CFTR?inhibitor with an?IC50 of 4 nM.
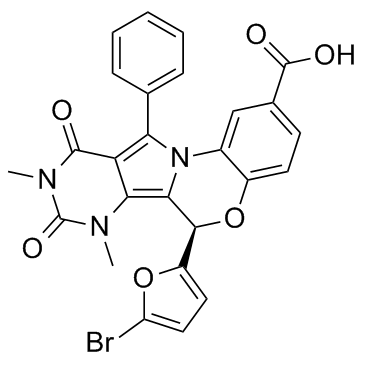
-
GC14236
(R)-CPP
NMDA antagonist
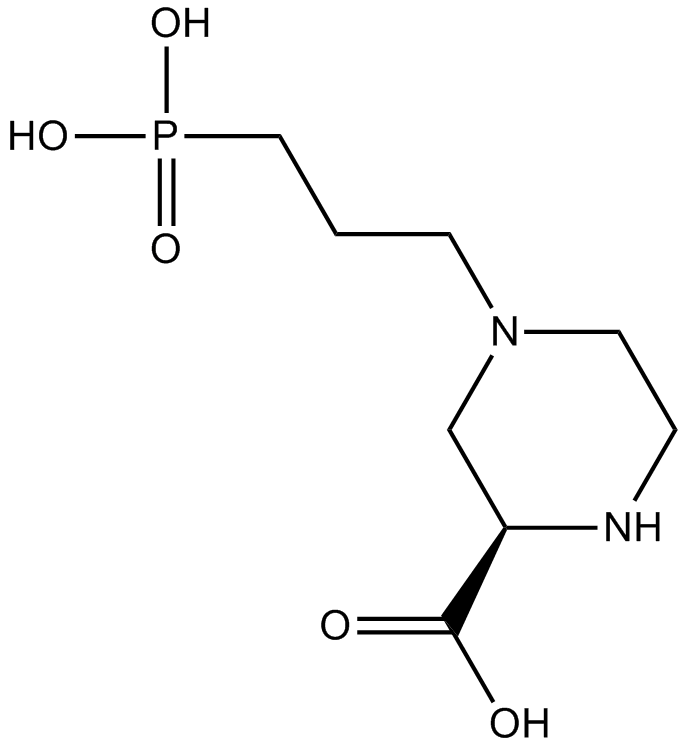
-
GC69812
(R)-Elexacaftor
(R)-VX-445
(R)-Elexacaftor is the enantiomer of Elexacaftor (Compound 1) and comes from Compound 37 in patent WO2018107100A1. It is a corrector of cystic fibrosis transmembrane conductance regulator (CFTR), with an EC50 value of 0.29 uM for CFTR dF508.
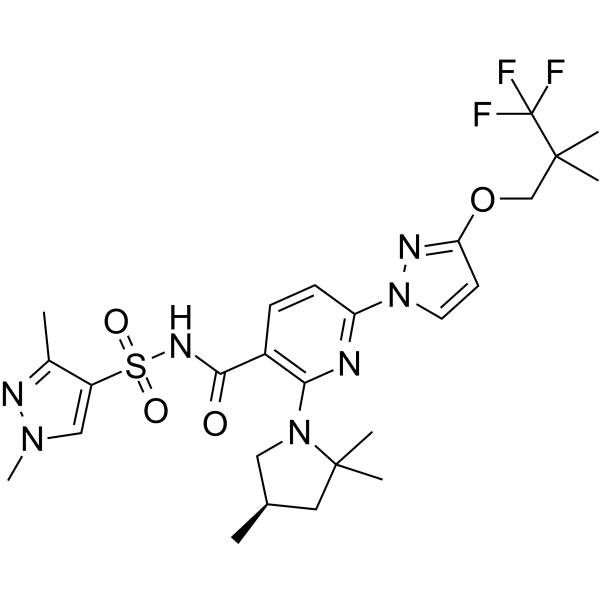
-
GC62739
(R)-Funapide
(R)-TV 45070; (R)-XEN402
(R)-Funapide ((R)-TV 45070) is the less active R-enantiomer of Funapide.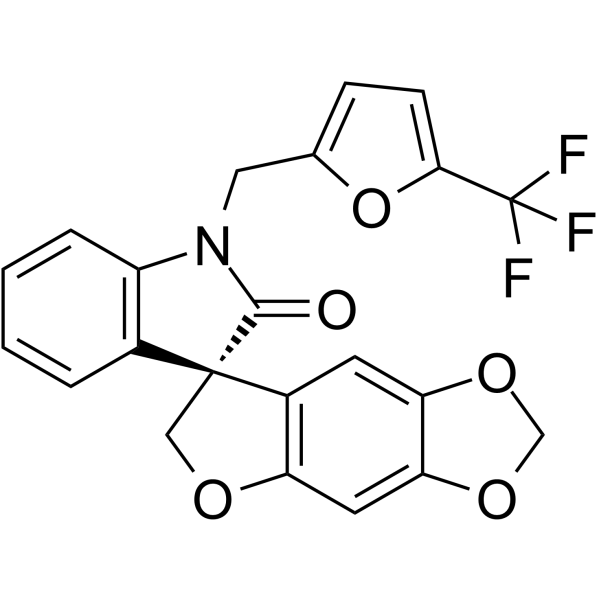
-
GC61847
(R)-Lanicemine
(R)-Lanicemine ((R)-AZD6765) is the less active R-enantiomer of Lanicemine.
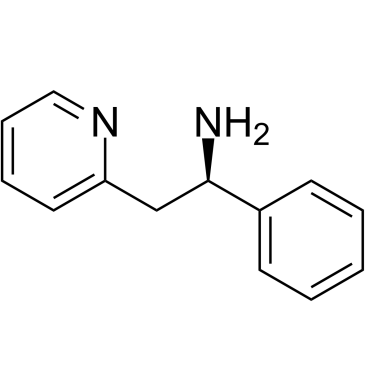
-
GC66419
(R)-Lercanidipine-d3 hydrochloride
(R)-lercanidipine D3 (hydrochloride) is a deuterium labeled (R)-Lercanidipine hydrochloride. (R)-Lercanidipine D3 (hydrochloride), the R-enantiomer of Lercanidipine, is a calcium channel blocker.
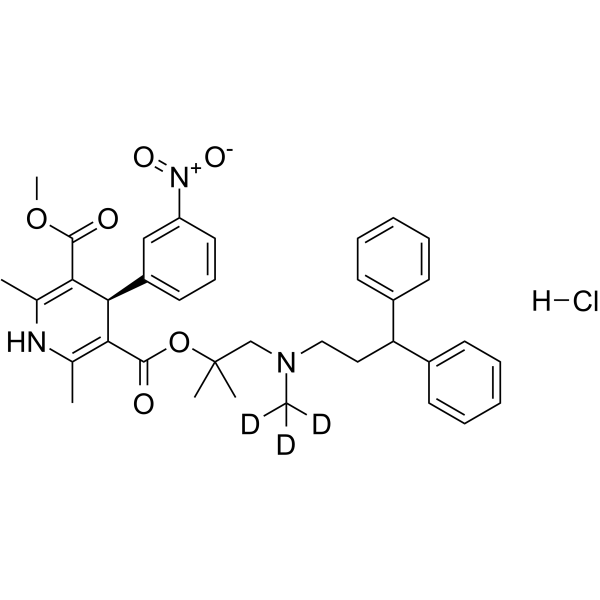
-
GC68475
(R)-Nicardipine
(R)-YC-93 free base

-
GC41719
(R)-nitro-Blebbistatin
R(-)7Desmethyl8nitro Blebbistatin
(R)-nitro-Blebbistatin is a more stable form of (+)-blebbistatin, which is the inactive form of (-)-blebbistatin.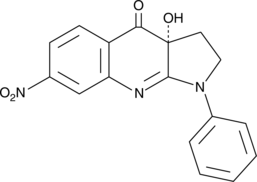
-
GC67767
(R)-Olacaftor
(R)-VX-440
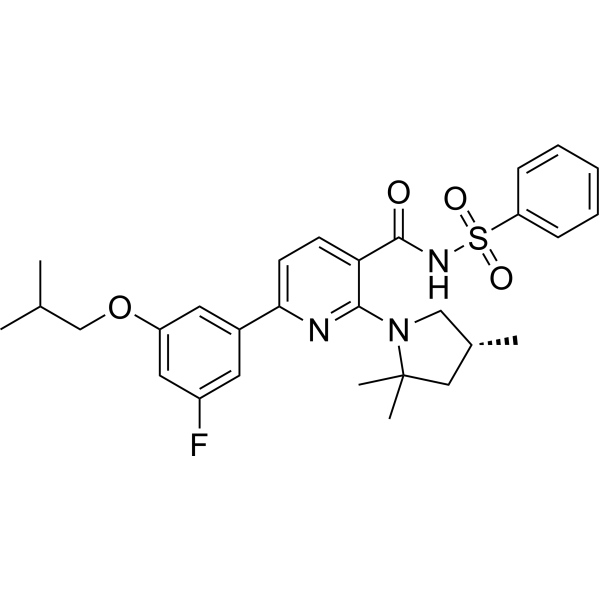
-
GC62513
(R)-Posenacaftor sodium
(R)-PTI-801 sodium
(R)-Posenacaftor (R)-PTI-801) sodium is the R enantiomer of Posenacaftor.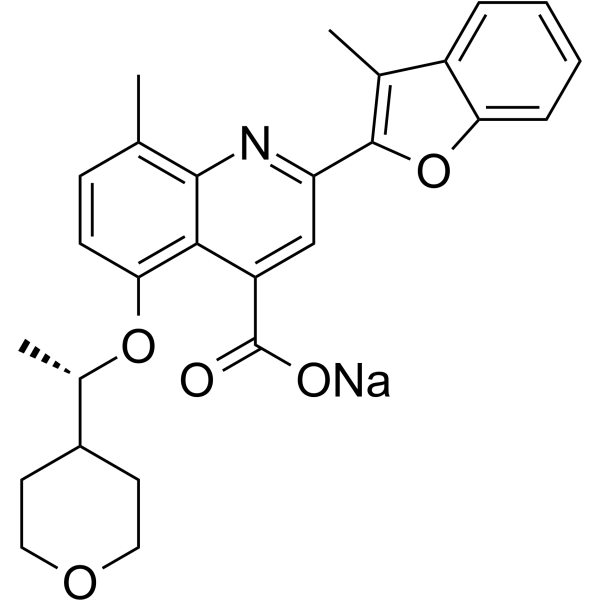
-
GC69839
(R)-Tegoprazan
(R)-CJ-12420; (R)-RQ-00000004
(R)-Tegoprazan ((R)-CJ-12420; example 3) is a benzimidazole derivative and an effective inhibitor of gastric H+/K+-ATPase. Its IC50 for canine kidney Na+/K+-ATPase is 98 nM. (R)-Tegoprazan has potential in the research of gastrointestinal diseases.
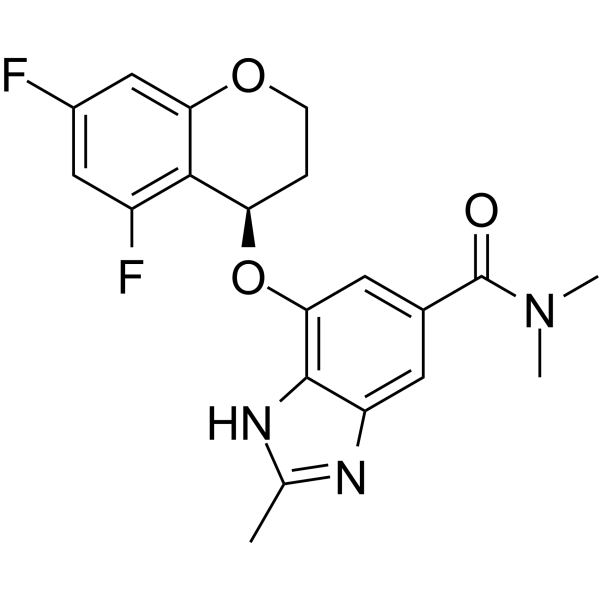
-
GC70934
(R)-Vanzacaftor
(R)-Vanzacaftor ((R)-VX-121) is a regulator of cystic fibrosis transmembrane conduction regulator (CFTR).

-
GC60407
(R)-Verapamil D7 hydrochloride
(R)-(+)-Verapamil D7 hydrochloride
(R)-Verapamil D7 hydrochloride ((R)-(+)-Verapamil D7 hydrochloride) is a deuterium labeled (R)-Verapamil hydrochloride. (R)-Verapamil hydrochloride ((R)-(+)-Verapamil hydrochloride) is a P-Glycoprotein inhibitor. (R)-Verapamil hydrochloride blocks MRP1 mediated transport, resulting in chemosensitization of MRP1-overexpressing cells to anticancer drugs.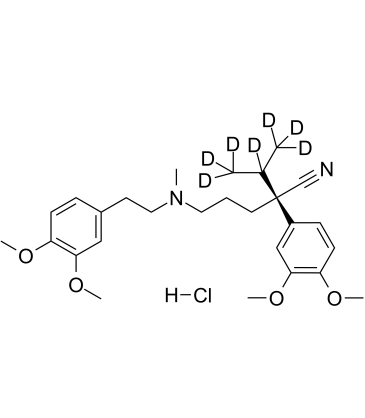
-
GC60408
(R)-Verapamil hydrochloride
(R)-Verapamil hydrochloride ((R)-(+)-Verapamil hydrochloride) is a P-Glycoprotein inhibitor. (R)-Verapamil hydrochloride blocks MRP1 mediated transport, resulting in chemosensitization of MRP1-overexpressing cells to anticancer drugs.
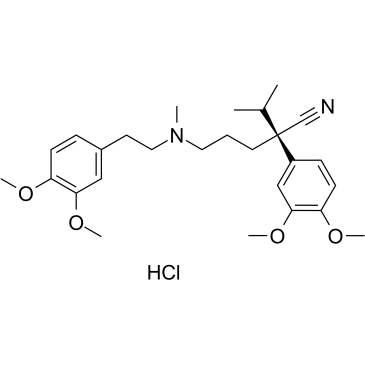
-
GC70292
(Rac)-AMG8379
(Rac)-AMG8379 ((Rac)-AMG8380) is a racemate of AMG8379. AMG8379 is a potent, orally active and selective sulfonamide antagonist of NaV1.7, with IC50s of 8.5 and 18.6 nM for hNaV1.7 and mNaV1.7, respectively .

-
GC63511
(Rac)-CP-601927 hydrochloride
(Rac)-CP-601927 hydrochloride is the racemate of CP-601927.
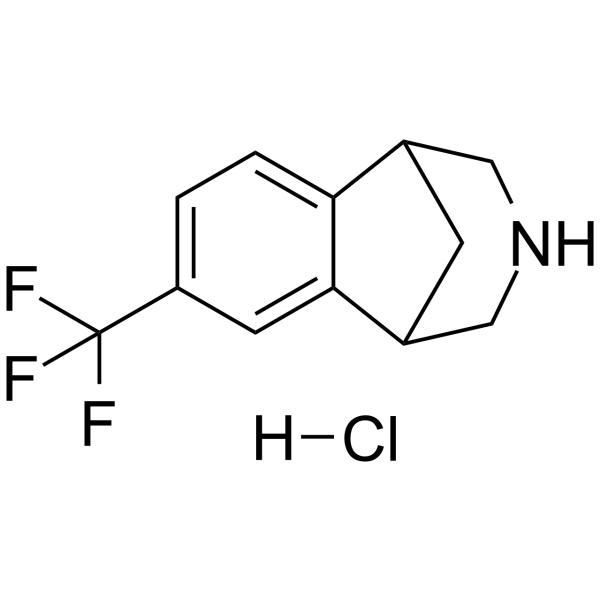
-
GC61446
(Rac)-Lanicemine
(Rac)-Lanicemine ((Rac)-AZD6765) is the racemate of Lanicemine.
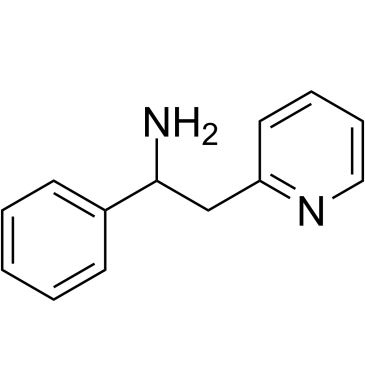
-
GC61873
(Rac)-MEM 1003
(Rac)-MEM 1003 is the racemate of MEM 1003.
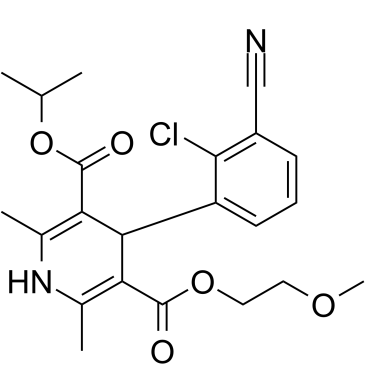
-
GC60413
(Rac)-NMDAR antagonist 1
(Rac)-NMDAR antagonist 1 is the racemate of NMDAR antagonist 1.
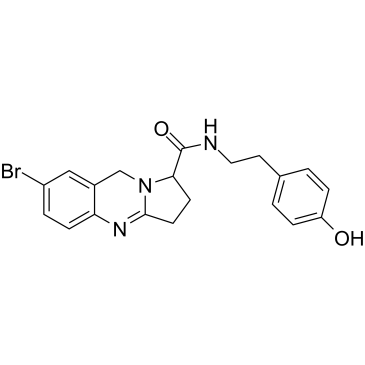
-
GC12488
(RS)-(Tetrazol-5-yl)glycine
Tet-Glycine, LY285265, DL-(Tetrazol-5-yl)glycine
NMDA receptor agonist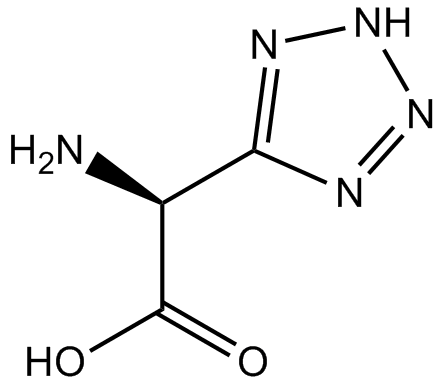
-
GC13631
(RS)-AMPA
AMPA agonist
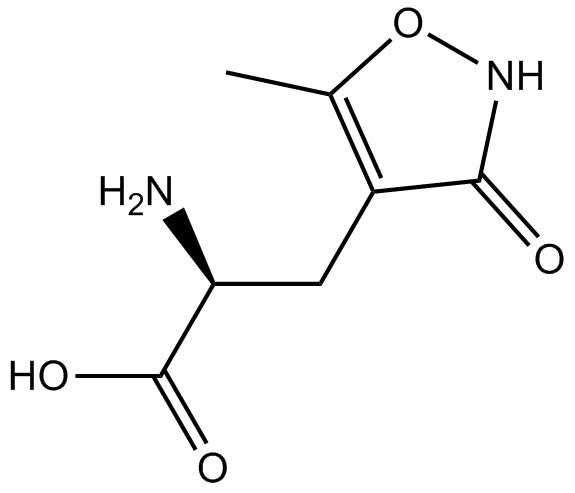
-
GC13032
(RS)-AMPA hydrobromide
AMPAR agonist

-
GC62746
(RS)-AMPA monohydrate
(±)-AMPA monohydrate
(RS)-AMPA ((±)-AMPA) monohydrate is a glutamate analogue and a potent and selective excitatory neurotransmitter L-glutamic acid agonist.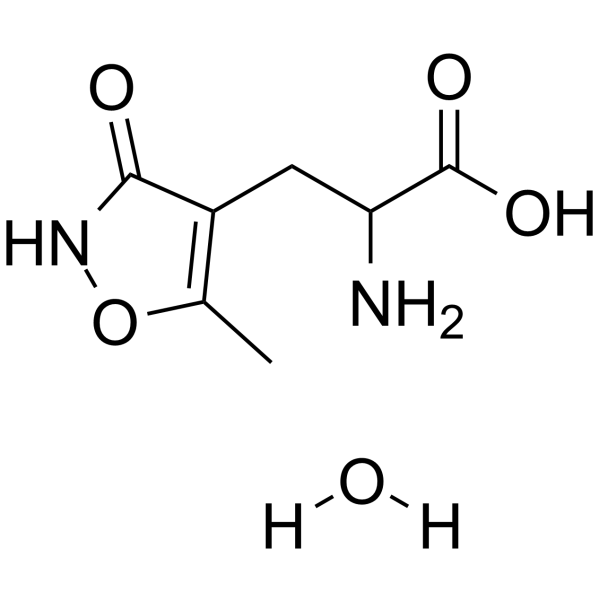
-
GC10476
(RS)-CPP
NMDA antagonist
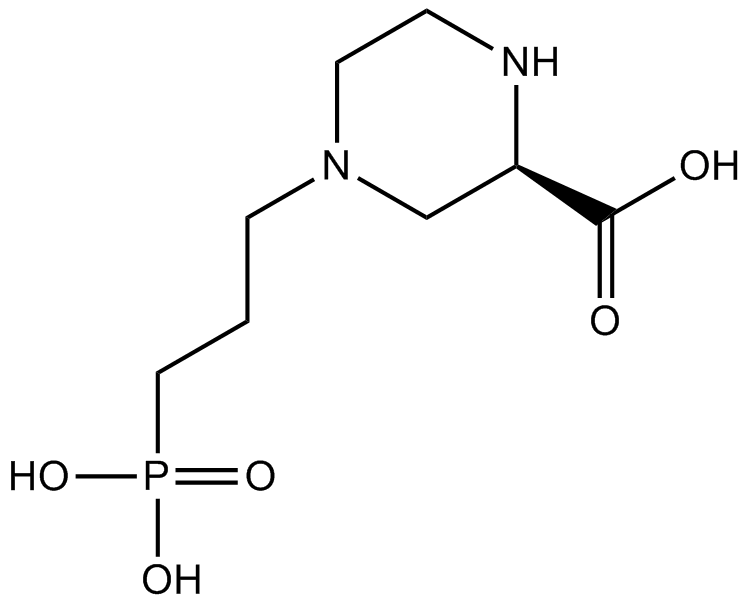
-
GC11889
(S)-(-)-5-Fluorowillardiine
AMPA receptor agonist
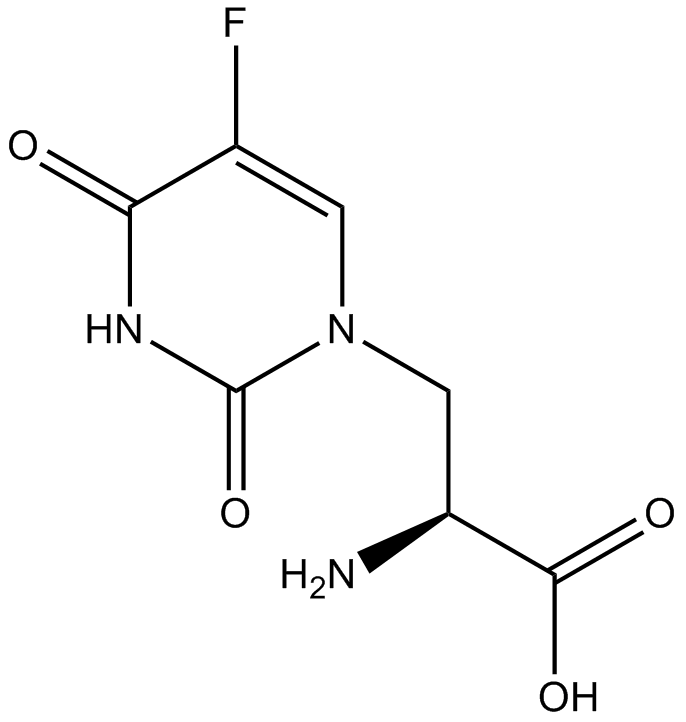
-
GC34998
(S)-(-)-5-Fluorowillardiine hydrochloride
(5S)-Fluorowillardiine hydrochloride; (S)-5-Fluorowillardiine hydrochloride
(S)-(-)-5-Fluorowillardiine hydrochloride is a potent and specific AMPAR agonist.
-
GC15768
(S)-(-)-5-Iodowillardiine
hGluR5 kainate receptor agonist