DNA Damage/DNA Repair
- MTH1(4)
- PARP(61)
- ATM/ATR(27)
- DNA Alkylating(19)
- DNA Ligases(3)
- DNA Methyltransferase(18)
- DNA-PK(27)
- HDAC(99)
- Nucleoside Antimetabolite/Analogue(146)
- Telomerase(14)
- Topoisomerase(134)
- tankyrase(5)
- Antifolate(32)
- CDK(234)
- Checkpoint Kinase (Chk)(29)
- CRISPR/Cas9(9)
- Deubiquitinase(64)
- DNA Alkylator/Crosslinker(65)
- DNA/RNA Synthesis(410)
- Eukaryotic Initiation Factor (eIF)(23)
- IRE1(22)
- LIM Kinase (LIMK)(10)
- TOPK(6)
- Casein Kinase(55)
- DNA Intercalating Agents(7)
- DNA/RNA Oxidative Damage(12)
Products for DNA Damage/DNA Repair
- Cat.No. Product Name Information
-
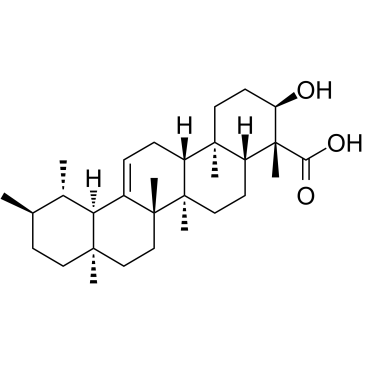
GC38000
β-Boswellic acid
A pentacyclic triterpene with diverse bioactivities
-
GC31365
γ-Oryzanol
γ-Oryzanol is a potent DNA methyltransferases (DNMTs) inhibitor in the striatum of mice.
-
GC38194
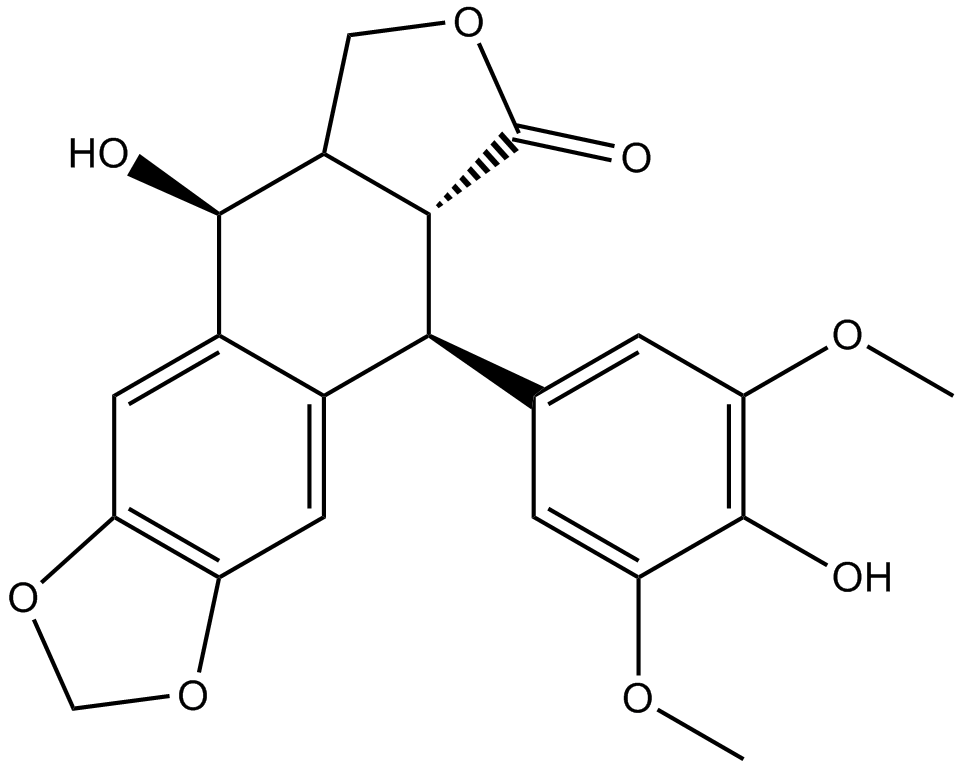
(±)-10-Hydroxycamptothecin
(±)-10-Hydroxycamptothecin is an indole alkaloid that inhibits the activity of topoisomerase I and has a broad spectrum of anticancer activity.
-
GC33107
(±)-BAY-1251152
(±)-BAY-1251152; (±)-VIP152
(±)-BAY-1251152 ((±)-BAY-1251152) is a racemic mixture of BAY-1251152. BAY-1251152 is a potent and highly selective PTEF/CDK9 inhibitor.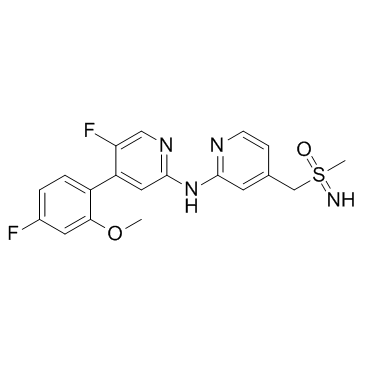
-
GC10867
(+)-Aphidicolin
ICI 69653, NSC 234714
Aphidicolin ((+)-Aphidicolin), a reversible inhibitor of eukaryotic nuclear DNA replication, can block the cell cycle at the pre-S phase.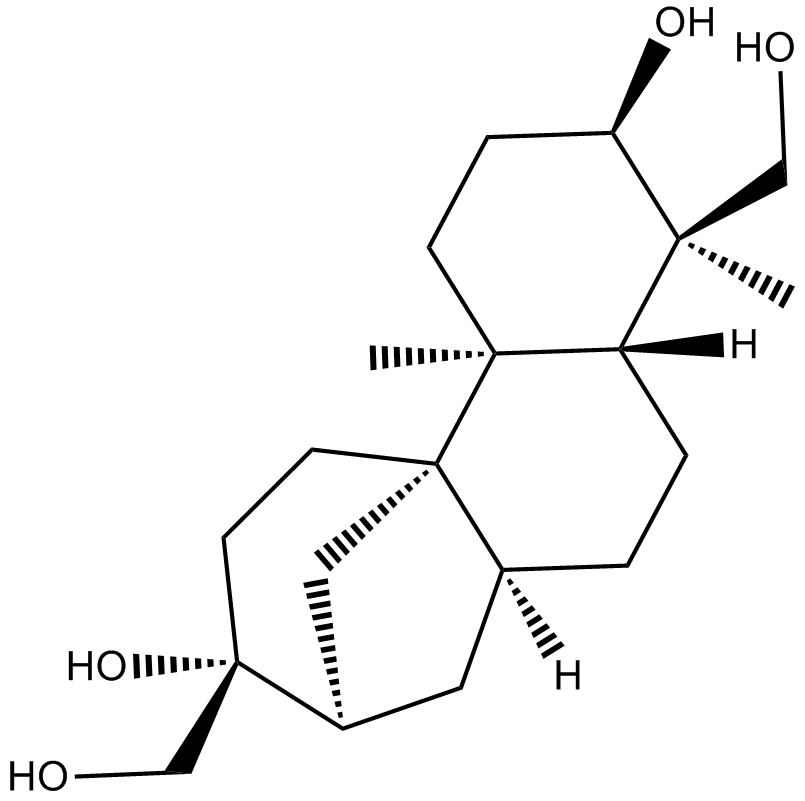
-
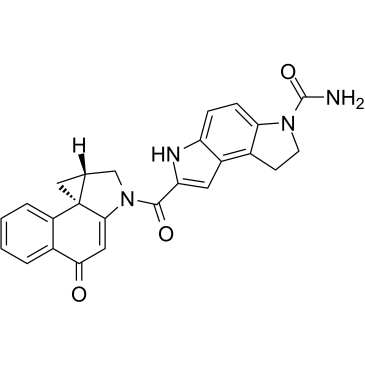
GC34955
(+)-CBI-CDPI1
(+)-CBI-CDPI1 is an enhanced functional analog of CC-1065. (+)-CBI-CDPI1 is a DNA alkylating agent. (+)-CBI-CDPI1 is an antibody drug conjugates (ADCs) toxin.
-
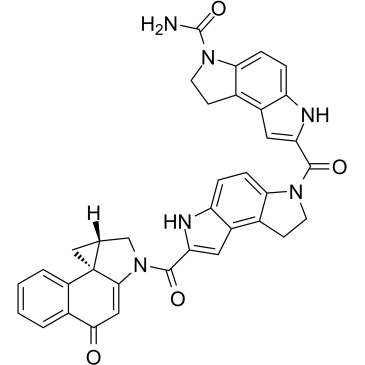
GC34956
(+)-CBI-CDPI2
(+)-CBI-CDPI2 is an enhanced functional analog of CC-1065. (+)-CBI-CDPI1 is a DNA alkylating agent. (+)-CBI-CDPI2 is an antibody drug conjugates (ADCs) toxin.
-
GC32429
(-)-BAY-1251152
(-)-BAY-1251152; (-)-VIP152
(-)-BAY-1251152 ((-)-BAY-1251152) is an enanthiomer of BAY-1251152 with rotation (-). BAY-1251152 is a potent and highly selective PTEF/CDK9 inhibitor.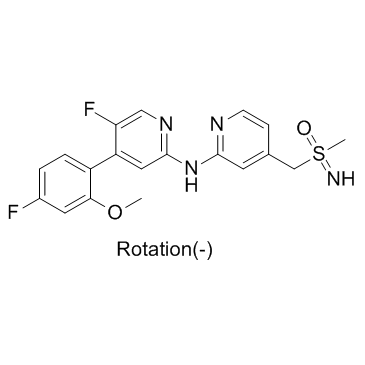
-
GC48635
(-)-Cryptopleurine
(R)-Cryptopleurine, NSC 19912
An alkaloid with diverse biological activities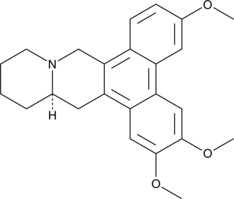
-
GC40076
(-)-Voacangarine
NSC 306219, (-)-Voacristine
(-)-Voacangarine is an indole alkaloid originally isolated from V.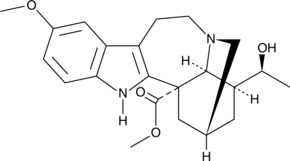
-
GC38377
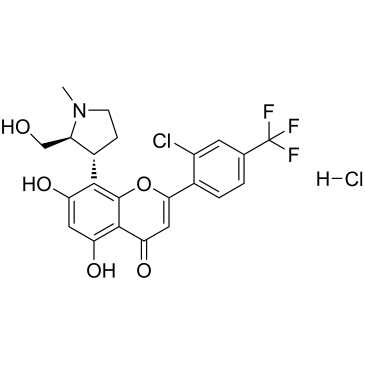
(2S,3R)-Voruciclib hydrochloride
(2S,3R)-Voruciclib hydrochloride is the enantiomer of Voruciclib hydrochloride. (2S,3R)-Voruciclib is an orally active CDK inhibitor.
-
GC41695
(6R,S)-5,6,7,8-Tetrahydrofolic Acid (hydrochloride)
Tetrahydrofolate, THFA
(6R,S)-5,6,7,8-Tetrahydrofolic acid (THFA), the reduced form of folic acid, serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.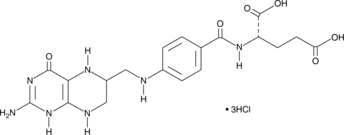
-
GC41088
(6S)-Tetrahydrofolic Acid
(6S)-Tetrahydrofolic acid is a diastereomer of tetrahydrofolic acid, a reduced form of folic acid that serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.
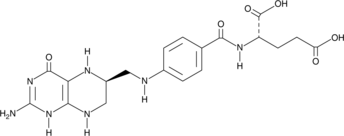
-
GC72591
(E,E)-RGFP966
(E,E)-RGFP966 is a selective and CNS permeable HDAC3 inhibitor that can be used for the research of Huntington’s disease.

-
GC62735
(E/Z)-GO289
(E/Z)-GO289 is a potent and selective casein kinase 2 (CK2) inhibitor (IC50=7 nM). (E/Z)-GO289 strongly lengthens circadian period. (E/Z)-GO289 exhibits cell type–dependent inhibition of cancer cell growth that correlated with cellular clock function.
-
GC64429
(E/Z)-Zotiraciclib citrate
(E/Z)-TG02 citrate; (E/Z)-SB1317 citrate
(E/Z)-Zotiraciclib citrate is a potent CDK2, JAK2, and FLT3 inhibitor.
-
GC63864
(E/Z)-Zotiraciclib hydrochloride
(E/Z)-TG02 hydrochloride; (E/Z)-SB1317 hydrochloride
(E/Z)-Zotiraciclib ((E/Z)-TG02) hydrochloride is a potent CDK2, JAK2, and FLT3 inhibitor.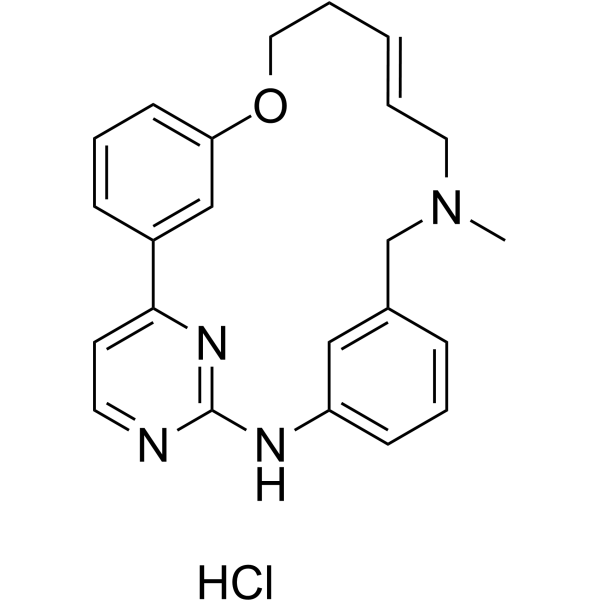
-
GC69793
(R)-5-O-Benzoyl-1,2-di-O-isopropylidene-alpha-D-xylofuranose
(R)-5-O-Benzoyl-1,2-di-O-isopropylidene-alpha-D-xylofuranose is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity and target inert lymphoid malignancies. The anticancer mechanism in this process depends on inhibiting DNA synthesis, inducing apoptosis (cell death), etc.
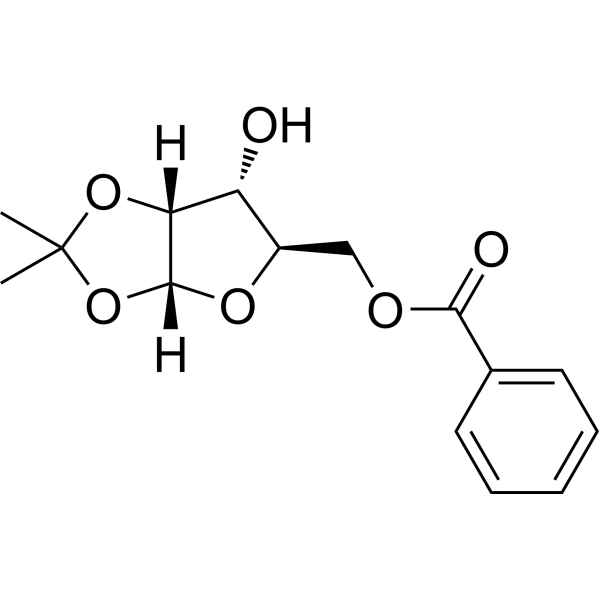
-
GC41716
(R)-CR8
Cyclin-dependent kinases (CDKs) are key regulators of cell cycle progression and are therefore promising targets for cancer therapy.
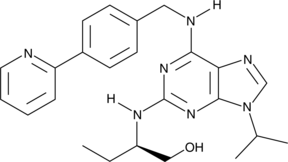
-
GC39281
(R)-CR8 trihydrochloride
CR8, (R)-Isomer trihydrochloride
(R)-CR8 (CR8) trihydrochloride, a second-generation analog of Roscovitine, is a potent CDK1/2/5/7/9 inhibitor.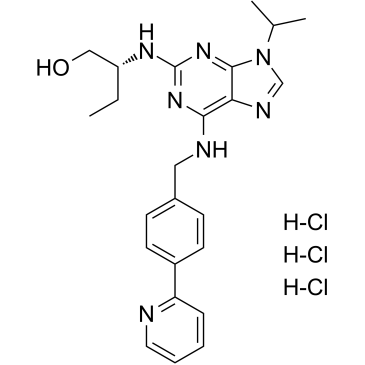
-
GC64210
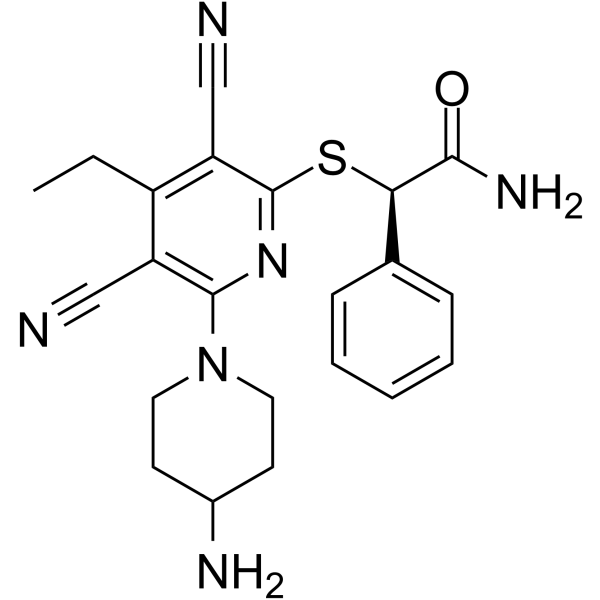
(R)-GSK-3685032
(R)-GSK-3685032 is the R-enantiomer of GSK-3685032. GSK-3685032 is a non-time-dependent, noncovalently, first-in-class reversible DNMT1-selective inhibitor, with an IC50 of 0.036 μM. GSK-3685032 induces robust loss of DNA methylation, transcriptional activation, and cancer cell growth inhibition.
-
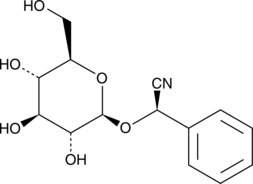
GC41633
(R)-Prunasin
(R)-Prunasin is a inhibitor of DNA Polymerase β.
-
GC34124
(rel)-MC180295
(rel)-MC180295
(rel)-MC180295 ((rel)-(rel)-MC180295) is a potent and selective CDK9-Cyclin T1 inhibitor, with an IC50 of 5 nM, at least 22-fold more selective for CDK9 over other CDKs. (rel)-MC180295 also inhibits GSK-3α and GSK-3β. (rel)-MC180295 ((rel)-(rel)-MC180295) has potent anti-tumor effect.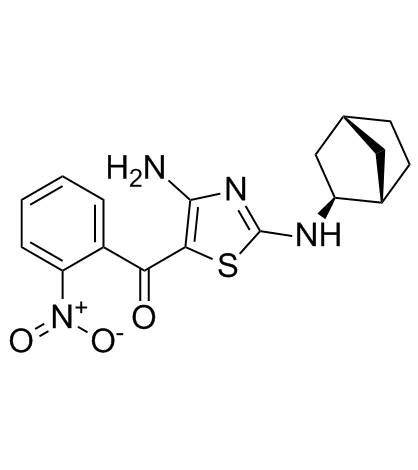
-
GC10098
(S)-10-Hydroxycamptothecin
ChEMBL 273862, NSC 107124
inhibitor of topoisomerase I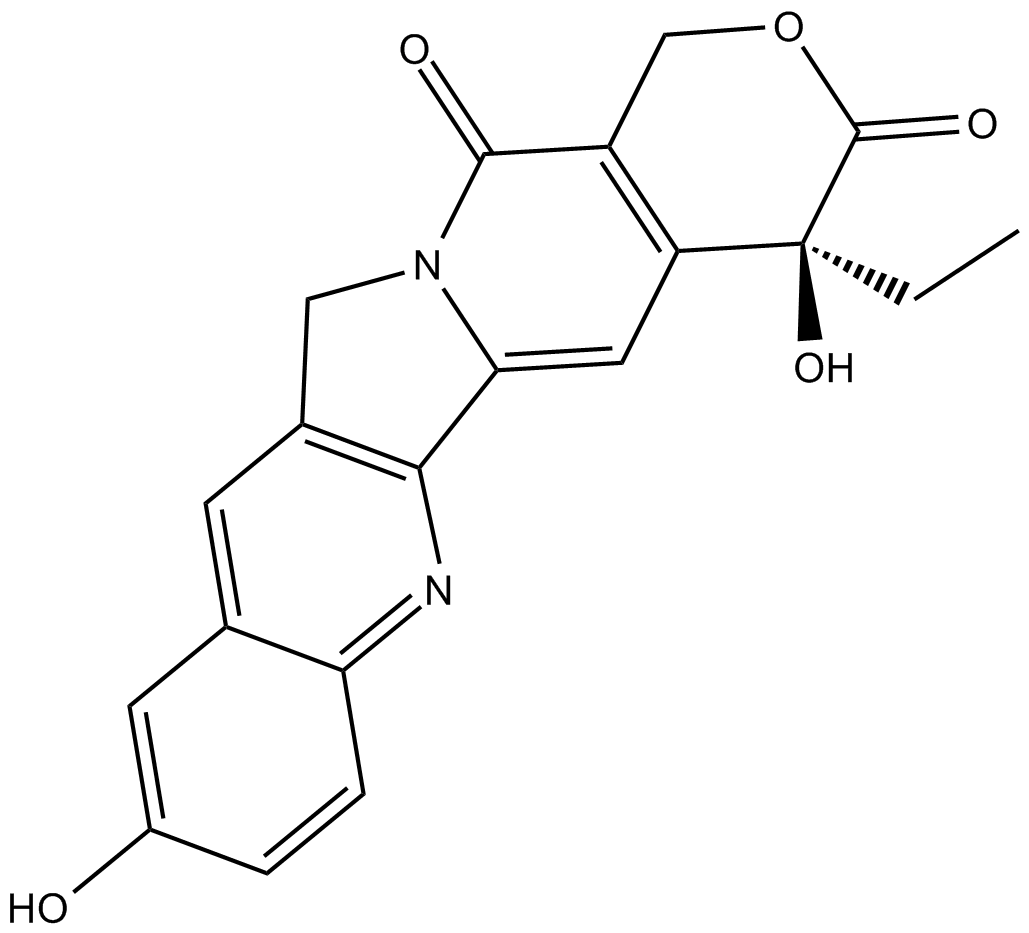
-
GC34999
(S)-Ceralasertib
(S)-AZD6738
(S)-Ceralasertib ((S)-AZD6738) is extracted from patent WO2011154737A1, Compound II, exhibits an IC50 of 2.578 nM.(S)-Ceralasertib is a potent and selective sulfoximine morpholinopyrimidine ATR inhibitor with excellent preclinical physicochemical and pharmacokinetic (PK) characteristics.(S)-Ceralasertib is developed improving aqueous solubility and eliminates CYP3A4 time-dependent inhibition.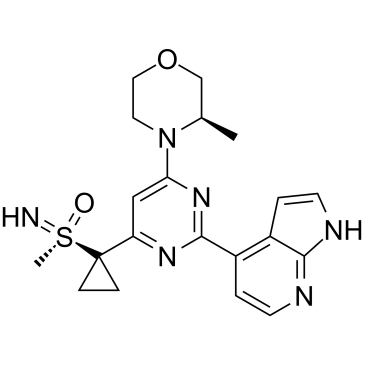
-
GC46351
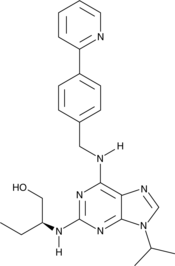
(S)-CR8
An inhibitor of cyclin-dependent kinases
-
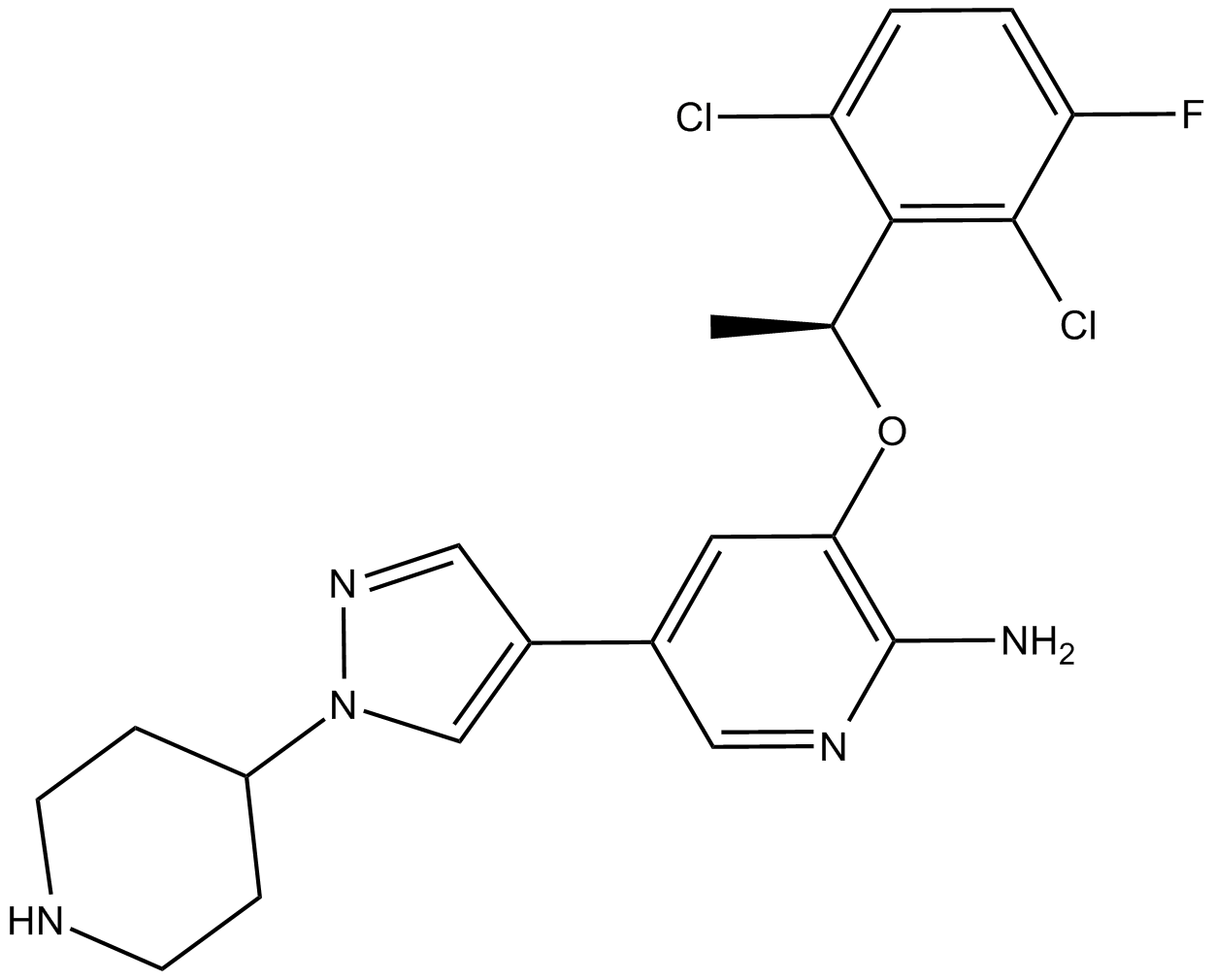
GC13136
(S)-Crizotinib
Potent MTH1 inhibitor
-
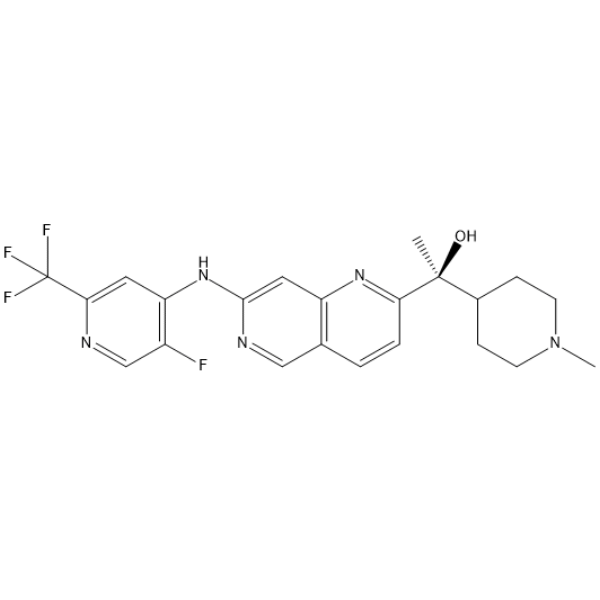
GC65877
(S)-GFB-12811
(S)-GFB-12811 (compound 596) is a potent and selective CDK5 inhibitor, with an IC50 value less than 10 nM. (S)-GFB-12811 can be used in the research of cell cycle progression, neuronal development, tumorigenesis.
-
GC65997
(S)-LY3177833 hydrate
(S)-LY3177833 ((S)-Example 2) hydrate is an orally active CDC7 kinase inhibitor. (S)-LY3177833 hydrate shows broad in vitro anticancer activity.
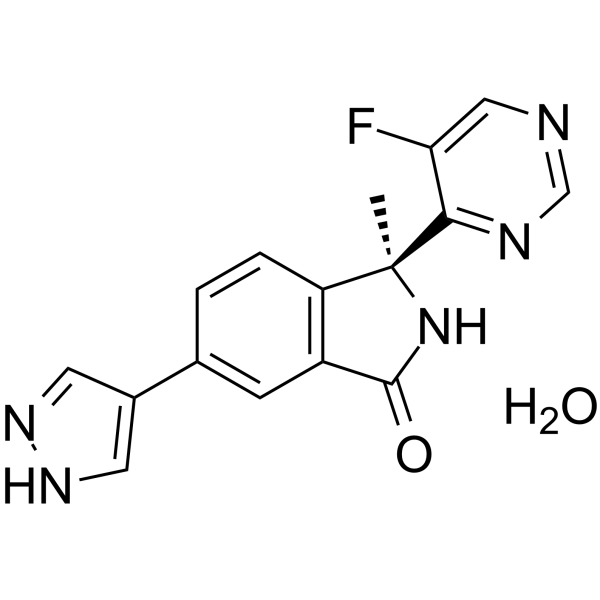
-
GC60421
(S)-Seco-Duocarmycin SA
(S)-Seco-Duocarmycin SA is a DNA alkylator, cytotoxic to cancer cells, and acts as a ADC cytotoxin for antibody-drug conjugates.
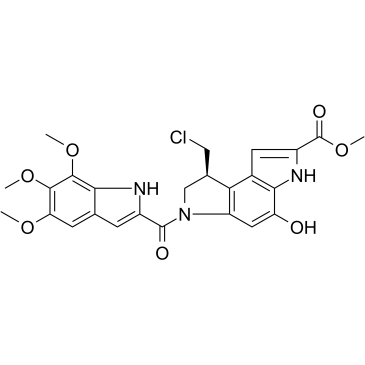
-
GC39842
(Z)-4EGI-1
(Z)-4EGI-1 is the Z-isomer of 4EGI-1 and is an inhibitor of eIF4E/eIF4G interaction and of translation initiation. (Z)-4EGI-1 effectively binds to eIF4E with an IC50 of 43.5 μM and a Kd value of 8.74 μM. (Z)-4EGI-1 has anticancer activity.
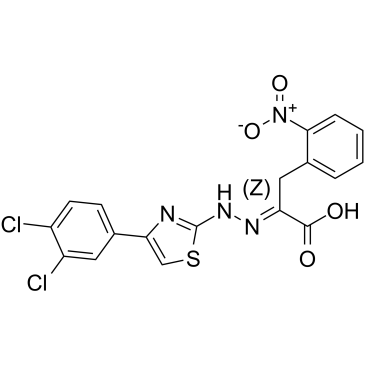
-
GC68484
1,2-O-Isopropylidene-5-O-p-toluoyl-a-D-xylofuranose
1,2-O-Isopropylidene-5-O-p-toluoyl-a-D-xylofuranose is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity and target malignant tumors in the inert lymphatic system. The anticancer mechanism in this process depends on inhibiting DNA synthesis, inducing apoptosis (cell death), etc.
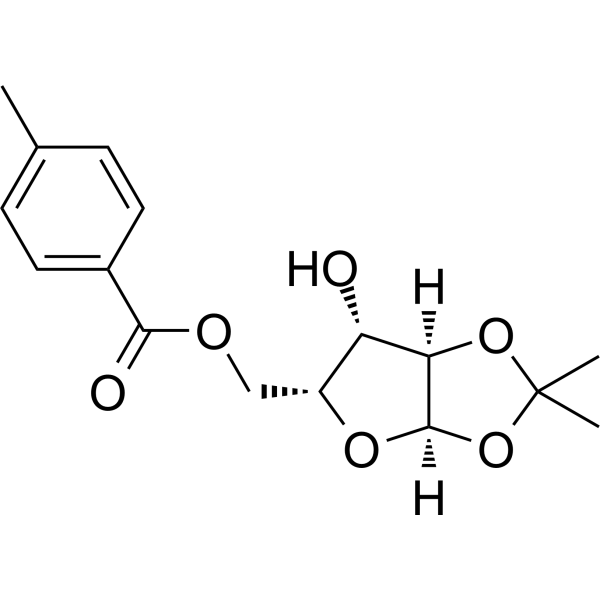
-
GC19528
1,4-Benzoquinone
p-Benzoquinone, NSC 36324, p-Quinone
A toxic metabolite of benzene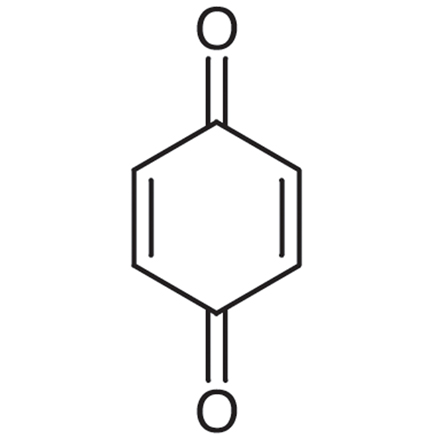
-
GC41859
1,5,7-Triazabicyclo[4.4.0]dec-5-ene
TBD, Triazabicyclodecene
1,5,7-Triazabicyclo[4.4.0]dec-5-ene is a bicyclic guanidine base that can be used as a catalyst for a variety of base-mediated organic transformations, including Michael additions, Wittig reactions, Henry reactions, and transesterification reactions.![1,5,7-Triazabicyclo[4.4.0]dec-5-ene Chemical Structure 1,5,7-Triazabicyclo[4.4.0]dec-5-ene Chemical Structure](/media/struct/GC4/GC41859.png)
-
GC65551
1-(2'-O-4-C-Methylene-beta-D-ribofuranosyl)thymine
1-(2'-O-4-C-Methylene-beta-D-ribofuranosyl)thymine is a bicyclic nucleoside.
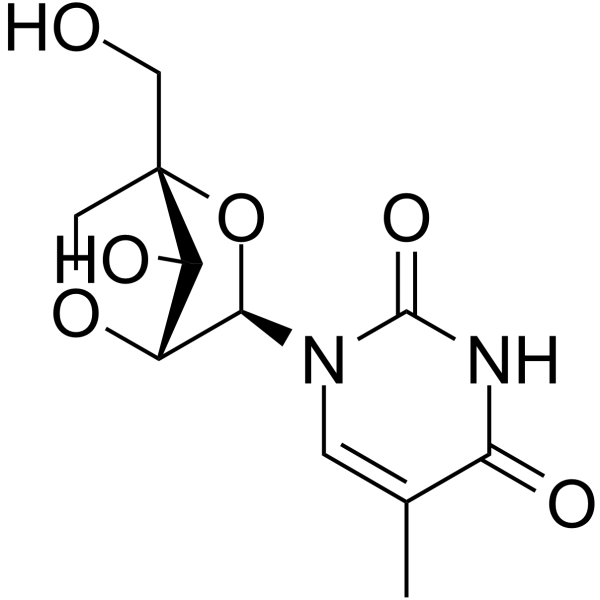
-
GC71294
1-(2-Deoxy-β-D-threo-pentofuranosyl)thymine
1-(2-Deoxy-β-D-threo-pentofuranosyl)tmine is a tmidine analog.

-
GC49470
1-Ethyl-1-nitrosourea (hydrate)
ENU, Ethylnitrosourea, N-Ethyl-N-nitrosourea, N-Nitroso-N-ethylurea
A DNA alkylating agent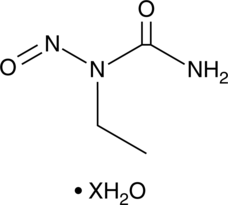
-
GC48909
1-Hydroxyanthraquinone
NSC 8640
An anthraquinone with genotoxic and carcinogenic activities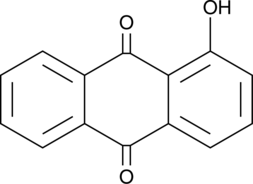
-
GC65038
1-Methylinosine
1-methyl Inosine, N1-Methylinosine
1-Methylinosine is a modified nucleotide found at position 37 in tRNA 3' to the anticodon of eukaryotic tRNA.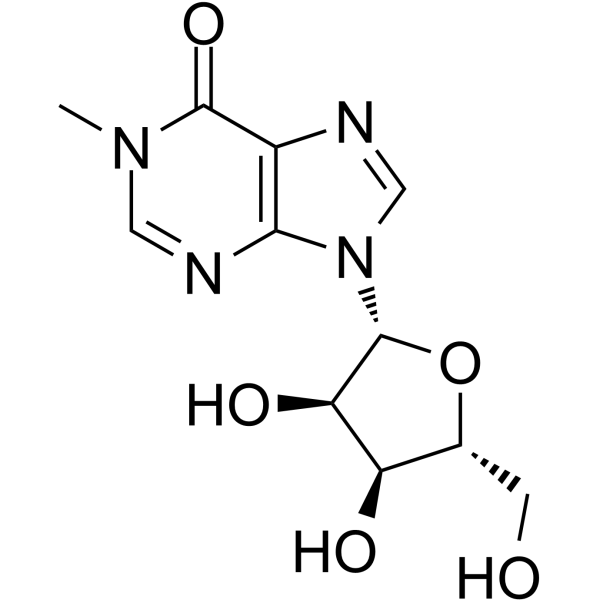
-
GC49872
10-Formyltetrahydrofolate (sodium salt) (technical grade)
10-CHO-FH4, 10-CHO-THF, N10-Formyltetrahydrofolate, 10-formyl H4PteGlu, 10-fTHF
10-Formyltetrahydrofolate (sodium salt) (technical grade) is a form of tetrahydrofolic acid that acts as a donor of formyl groups in anabolism.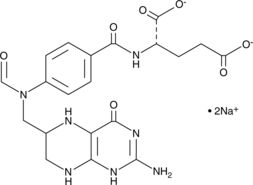
-
GC63796
116-9e
MAL2-11B
116-9e (MAL2-11B) is a Hsp70 co-chaperone DNAJA1 inhibitor.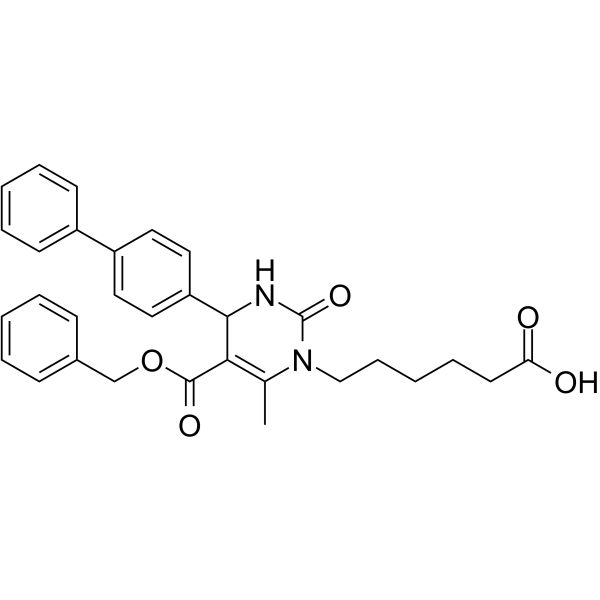
-
GC71326
12R-LOX-IN-2
12R-LOX-IN-2 (compound 7b) is an inhibitor of 12R-lipoxygenase (12R-LOX).

-
GC49759
13C17-Mycophenolic Acid
13C17-MPA
An internal standard for the quantification of mycophenolic acid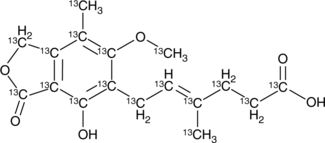
-
GC46474
18-Deoxyherboxidiene
RQN-18690A
A bacterial metabolite with antiangiogenic activity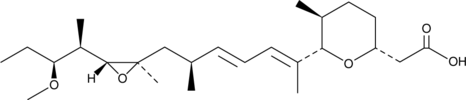
-
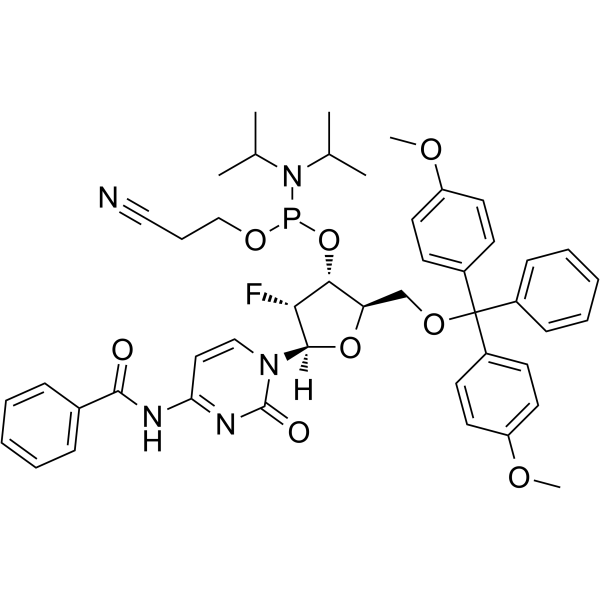
GC65489
2'-F-Bz-dC Phosphoramidite
2'-F-Bz-dC Phosphoramidite can be used in the synthesis of oligoribonucleotides.
-
GC66651
2'-O,4'-C-Methyleneadenosine
LNA-A
2'-O,4'-C-Methyleneadenosine (LNA-A) is a locked nucleic acid (LNA) and is also an adenosine analog.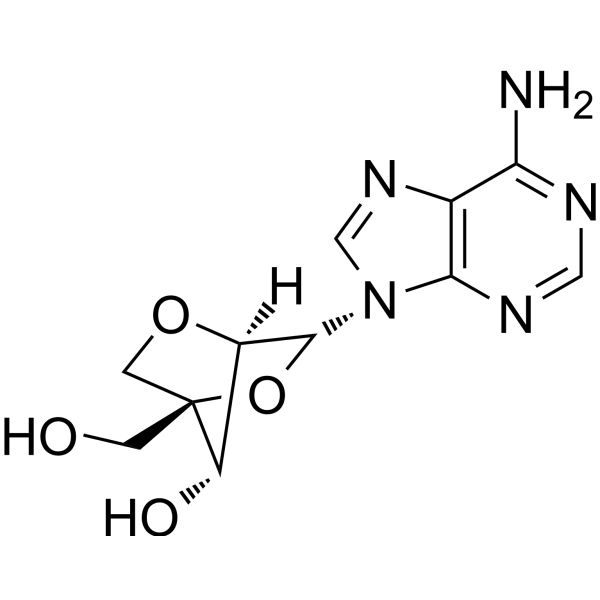
-
GC66654
2'-O,4'-C-Methylenecytidine
LNA-C(Bz)
2'-O,4'-C-Methylenecytidine (LNA-C(Bz)) is a bicyclic nucleoside analogue with fixed N-type conformation. 2'-O,4'-C-Methylenecytidine can be used to synthesize oligonucleotides. 2'-O,4'-C-Methylenecytidine forms duplexes with complementary DNA and RNA strands.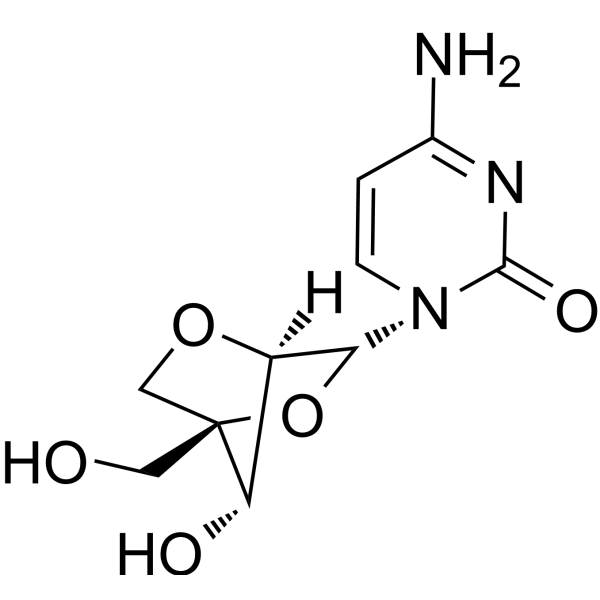
-
GC66655
2'-O,4'-C-Methyleneguanosine
LNA-G
2′-O,4′-C-Methyleneguanosine (LNA-G) is a reverse guanine analogue, where LNA (locked nucleic acid) is a nucleic acid analogue. LNA modification can be used in a variety of applications such as effective binding affinity to complementary sequences and greater nuclease resistance than natural nucleotides, offering great potential for applications in disease diagnosis and research. LNA-G is also available via KOD DNA polymerase, which allows the integration of LNA-G nucleotides into the DNA strand.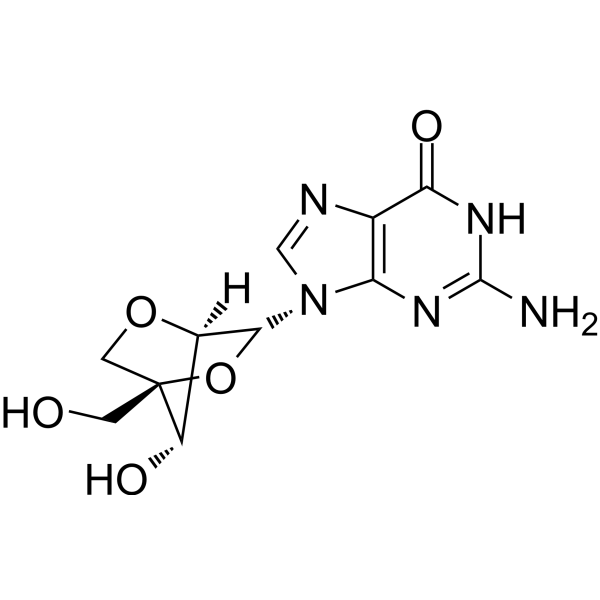
-
GC52183
2'-O-Methyl-5-methyluridine
2'-O-Methylribothymidine, 2'-O-Methylthymidine
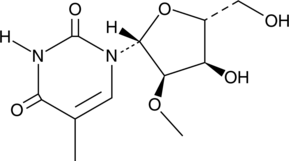
-
GC66713
2'-O-MOE-5-Me-C(Bz)
2'-O-MOE-5-Me-C (Bz) is a nucleotide for the stereoselective synthesis of nucleoside alkyl phosphonates.
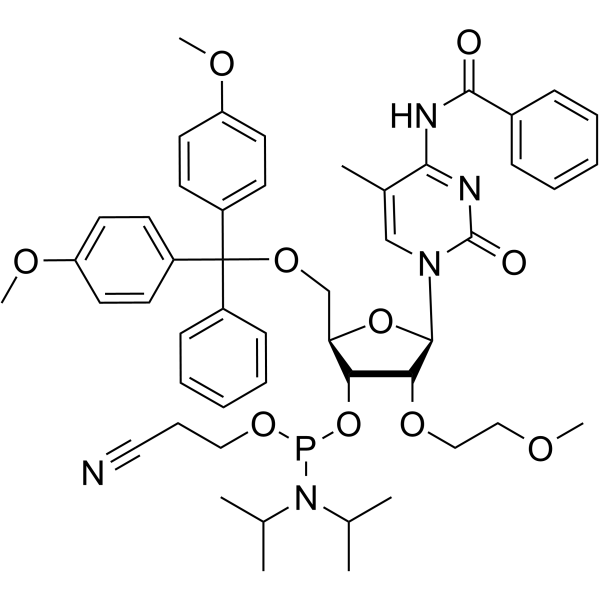
-
GC66657
2'-O-MOE-5-Me-rC
2'-O-MOE-5-Me-rC is an active compound. 2'-O-MOE-5-Me-rC can be used for oligonucleotide synthesis.
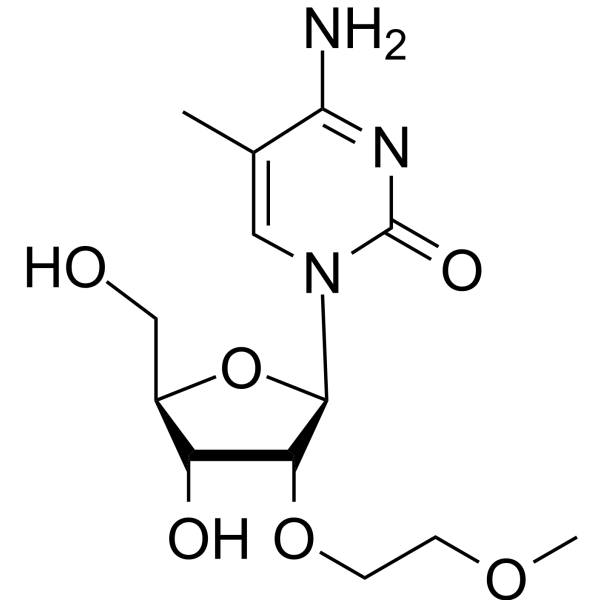
-
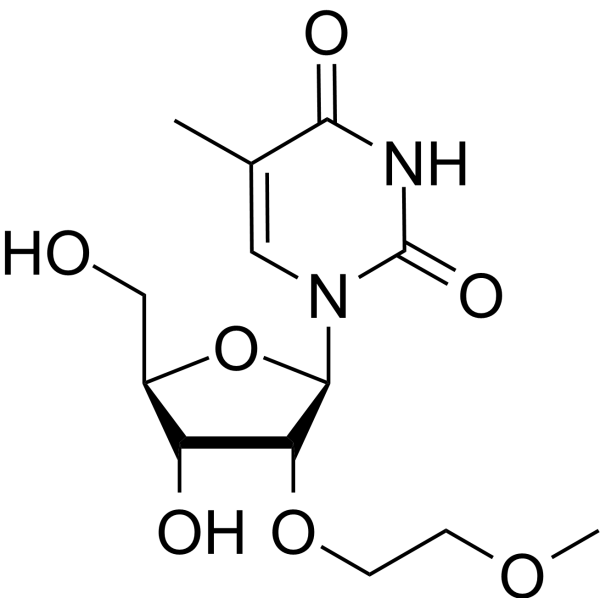
GC66091
2'-O-MOE-5-Me-rU
2'-O-MOE-5-Me-rU is an active compound. 2'-O-MOE-5-Me-rU can be used for oligonucleotide synthesis.
-
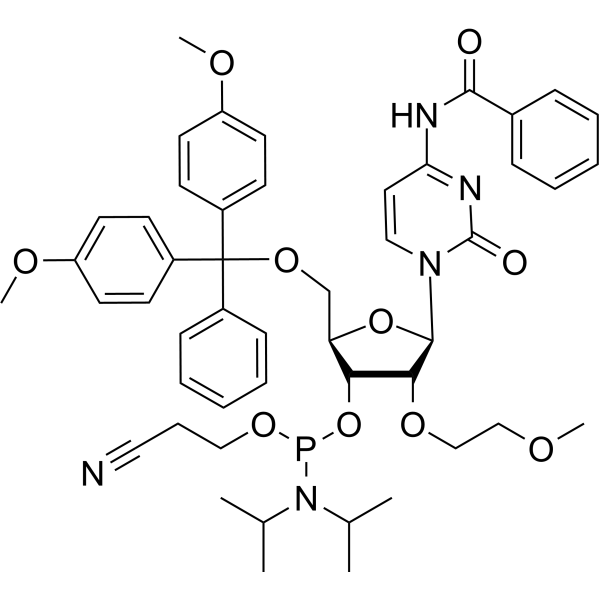
GC66656
2'-O-MOE-rC
2'-O-MOE-rC is a 2'-O-MOE modified nucleoside. 2'-O-MOE-rC can be used for synthesis of DNA.
-
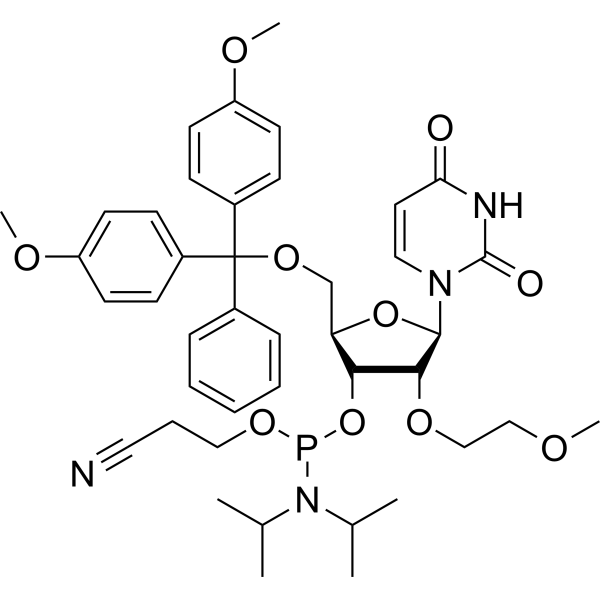
GC66652
2'-O-MOE-U
2'-O-MOE-U is a phosphoramidite, can be used for oligonucleotide synthesis.
-
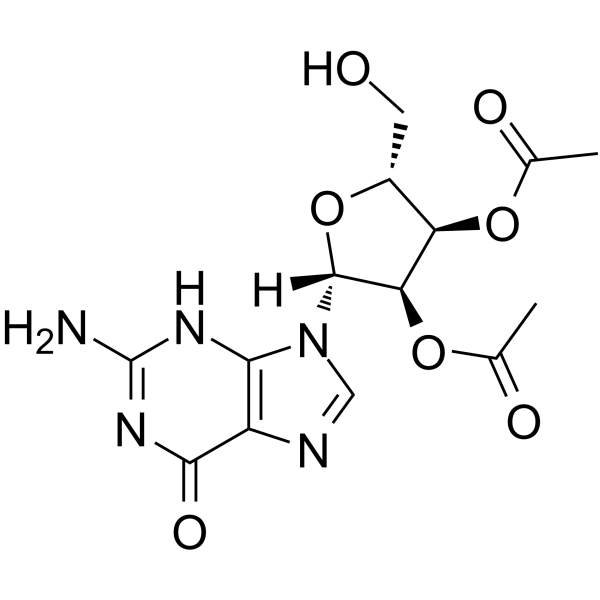
GC65170
2′,3′-Di-O-acetylguanosine
2′,3′-Di-O-acetylguanosine is a nucleoside analog.
-
GC67384
2′-Deoxy-β-L-uridine
2'-Deoxy-β-L-uridine is a nucledside analogue and a specific substrate for the viral enzyme, shows no stereospecificity against herpes simplex 1 (HSV1) thymidine kinase (TK). 2′-Deoxy-β-L-uridine exerts antiviral activity via the interation of 5'-triphosphates with the viral DNA polymerase.
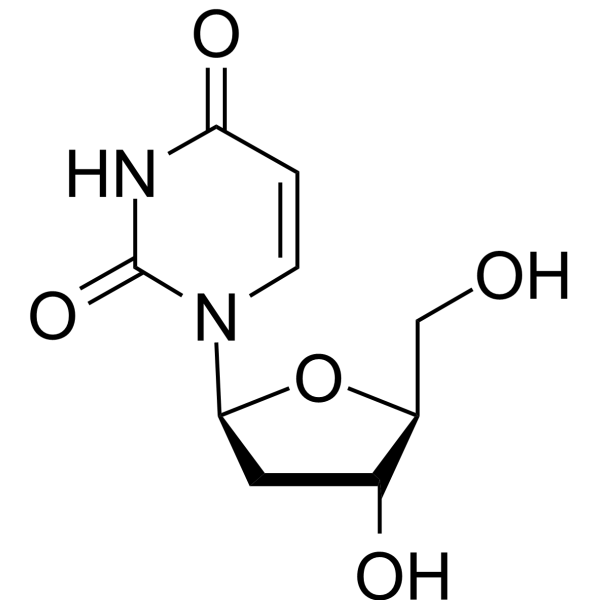
-
GC61667
2′-Deoxy-2′-fluoroadenosine
2′-Deoxy-2′-fluoroadenosine can be used for thesynthesisof 2′-Deoxy-2′-fluoro-modified oligonucleotides hybridized with RNA. 2′-Deoxy-2′-fluoroadenosine can be cleaved efficiently by E. coli purine nucleoside phosphorylase (PNP) to the toxic agent 2-fluoroadenine (FAde). 2′-Deoxy-2′-fluoroadenosine shows excellent in vivo activity against tumors expressing E. coli PNP.
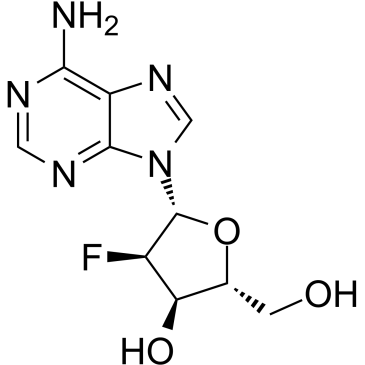
-
GC62530
2’-O-Me-C(Bz) Phosphoramidite
2’-O-Me-C(Bz) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
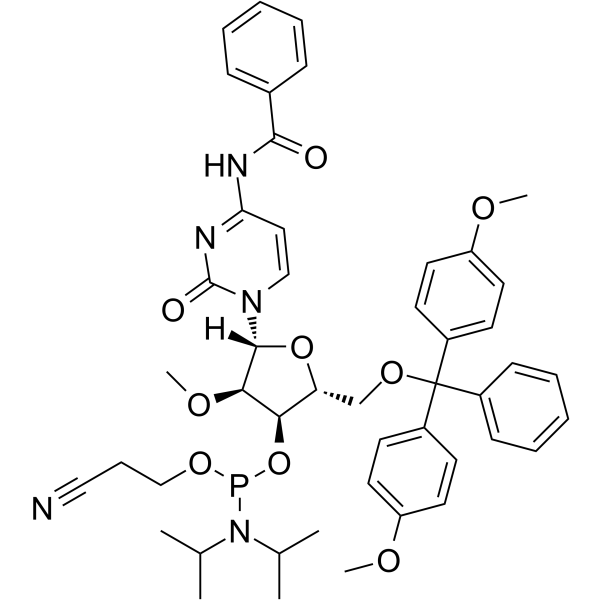
-
GC62529
2’-OMe-A(Bz) Phosphoramidite
2’-OMe-A(Bz) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
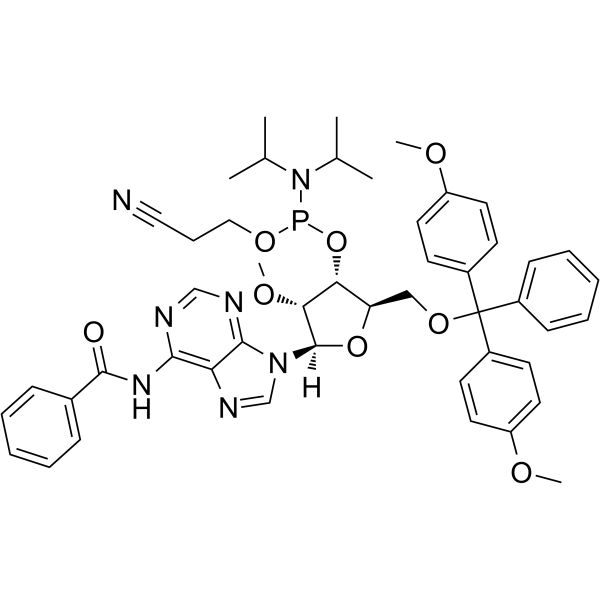
-
GC62531
2’-OMe-G(ibu) Phosphoramidite
2’-OMe-G(ibu) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
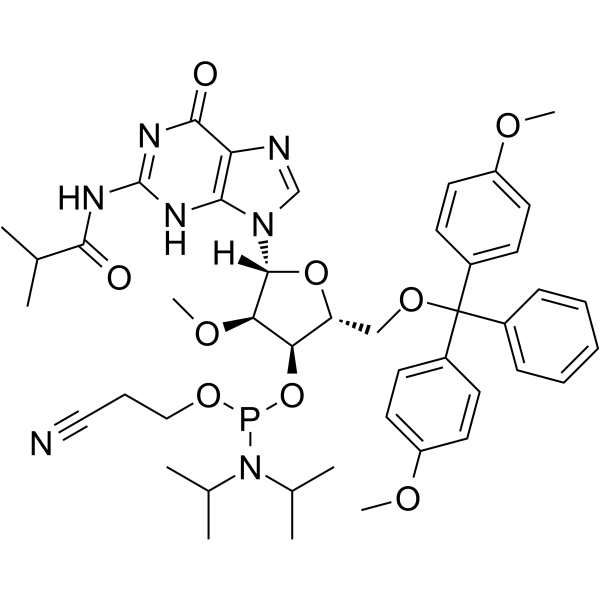
-
GC46508
2',2'-Difluoro-2'-deoxyuridine
dFdU
An active metabolite of gemcitabine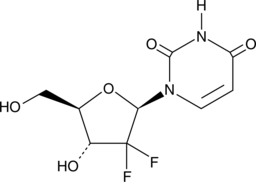
-
GC42061
2',3'-Dideoxyadenosine 5'-triphosphate
2',3'-Dideoxyadenosine 5'-triphosphate
2',3'-Dideoxyadenosine 5'-triphosphate (2',3'-Dideoxyadenosine 5'-triphosphate), an active metabolite of 2',3'-dideoxyinosine, is a chain-elongating inhibitor of DNA polymerase.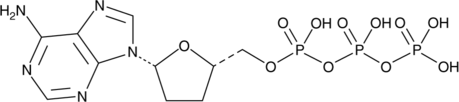
-
GC46519
2',3'-O-Isopropylideneguanosine
A building block
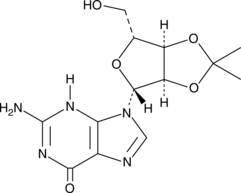
-
GC46540
2'-Deoxyadenosine-5'-triphosphate (sodium salt hydrate)
A purine nucleotide
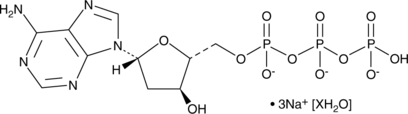
-
GC48440
2'-Deoxycytidine-5'-triphosphate (sodium salt)
dCTP
2'-Deoxycytidine-5'-triphosphate (sodium salt) (dCTP trisodium salt) is a nucleoside triphosphate that can be used for DNA synthesis.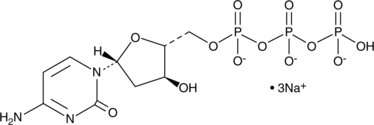
-
GC42151
2'-Deoxyguanosine 5'-monophosphate (sodium salt hydrate)
dGMP
2'-Deoxyguanosine 5'-monophosphate (dGMP) is used as a substrate of guanylate kinases to generate dGDP, which in turn is phosphorylated to dGTP, a nucleotide precursor used in DNA synthesis.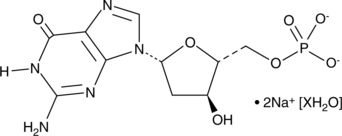
-
GC33430
2'-Deoxypseudoisocytidine
2'-Deoxypseudoisocytidine is a nucleoside analogue.
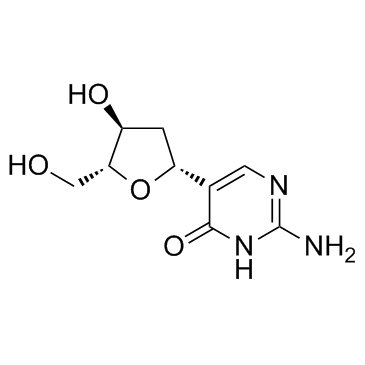
-
GC35072
2'-O,4'-C-Methyleneuridine
2'-O,4'-C-Methyleneuridine (Compound 15a) is a bicyclic nucleoside.
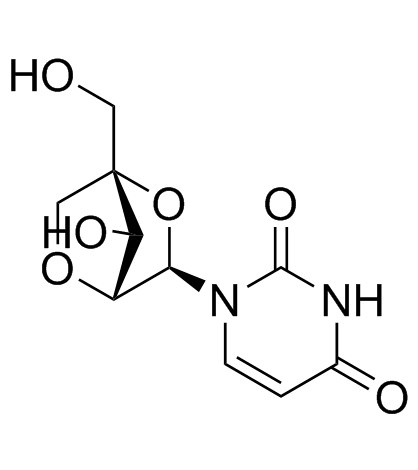
-
GC42080
2'2'-cGAMP (sodium salt)
Adenosine-Guanosine 2’,2’-cyclic monophosphate, cGAMP(2’-5’), 2’,2’-Cyclic GMP-AMP
2'2'-cGAMP is a synthetic dinucleotide (CDN) that contains non-canonical 2'5'-phosphodiester bonds.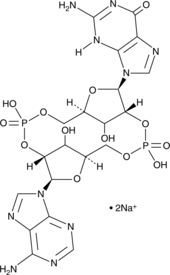
-
GC64399
2,4,6-Trihydroxybenzoic acid
2,4,6-Trihydroxybenzoic acid, the flavonoid metabolite, is a CDK inhibitor. 2,4,6-Trihydroxybenzoic acid can be used for the research of cancer.
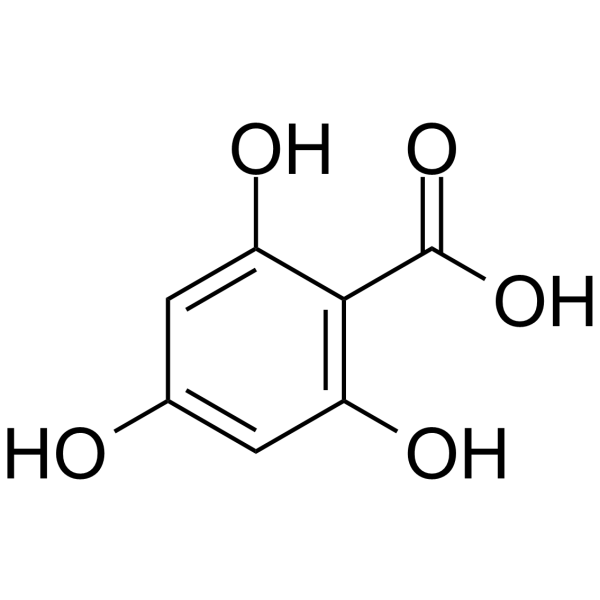
-
GC60459
2,4-D (2,4-Dichlorophenoxyacetic acid)
2,4D, Diclordon, Vidon 638
2,4-D (2,4-Dichlorophenoxyacetic acid) (2,4-D (2,4-Dichlorophenoxyacetic acid) ichlorophenoxyacetic acid) is a selective systemic herbicide for the control of broad-leaved weeds.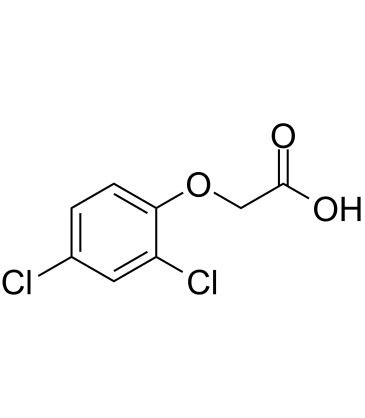
-
GC71530
2,4-D-13C6
2,4-D-13C6 is the 13C-labeled 2,4-D.

-
GC46524
2,6-Dichloropurine-9-β-D-riboside
2,6-Dichloropurine riboside
A building block
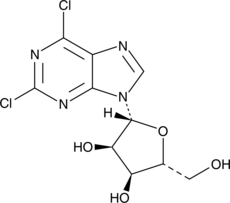
-
GC33496
2-(Methylamino)-1H-purin-6(7H)-one (N2-methylguanine)
2-(Methylamino)-1H-purin-6(7H)-one (N2-methylguanine) (N2-Methylguanine) is a modified nucleoside.
-
GC64983
2-Amino-2'-deoxyadenosine
2-Amino-2'-deoxyadenosine is a deoxyribonucleoside used for the oligonucleotide synthesis.
-
GC71560
2-Iodoacetamide-d4
2-Iodoacetamide-d4 is the deuterium labeled 2-Iodoacetamide.

-
GC71208
2-Methyladenosine
2-Metladenosine is an adenosine analogue.

-
GC39527
2-O-Methylcytosine
2-O-Methylcytosine, an O-alkylated analogue a DNA adduct, is the damaged nucleobase.
-
GC49348
2-Thiocytidine
α-2-Thioribocytidine
A modified nucleobase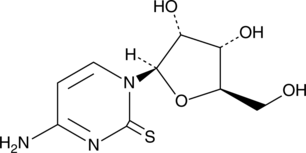
-
GC42197
2-Thiouridine
1-β-D-ribofuranosyl-2-thiouracil, s2U
2-Thiouridine (s2U) is a modified nucleobase found in tRNAs that is known to stabilize U:A pairs and modestly destabilize U:G wobble pairs.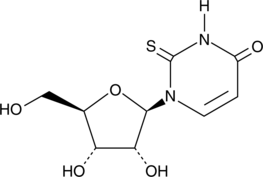
-
GC65083
3'-Azido-3'-deoxy-5-fluorocytidine
3'-Azido-3'-deoxy-5-fluorocytidine (Compound 12) is a cytidine derivative.
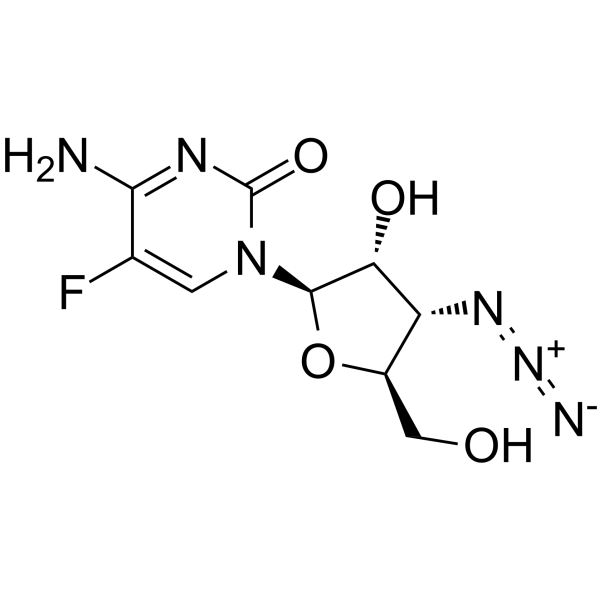
-
GC71204
3'-O-(2-Methoxyethyl)guanosine
3’-O-(2-Methoxyetl)guanosine is a guanosine analogue.

-
GC64985
3'-O-Methylguanosine
3'-O-Methylguanosine is a methylated nucleoside analogs and a RNA chain terminator.
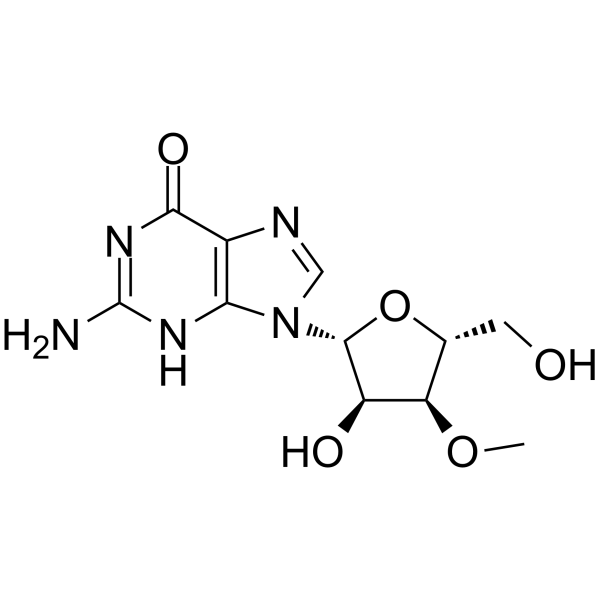
-
GC68542
3'Ome-m7GpppAmpG ammonium
3'Ome-m7GpppAmpG ammonium is a trinucleotide cap analog containing locked nucleic acid (LNA) molecules. It has significant translation efficiency and can be used as a potential molecular biology tool in the fields of mRNA vaccines and mRNA transfection, such as protein production, gene therapy, and anti-cancer immunity.

-
GC40618
3',4',7-Trihydroxyisoflavone
3'-hydroxy Daidzein, 3’,4’,7-THIF
3',4',7-Trihydroxyisoflavone, a major metabolite of Daidzein, is an ATP-competitive inhibitor of Cot (Tpl2/MAP3K8) and MKK4. 3',4',7-Trihydroxyisoflavone has anticancer, anti-angiogenic, chemoprotective, and free radical scavenging activities.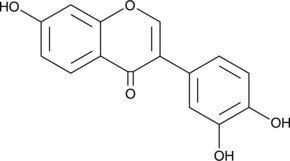
-
GC35105
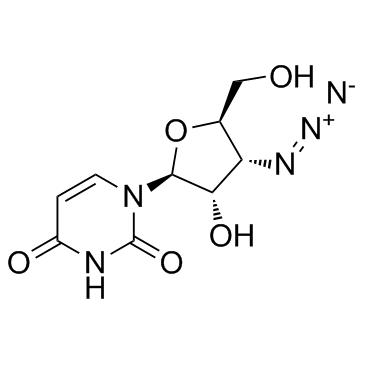
3'-Azido-3'-deoxy-beta-L-uridine
3'-Azido-3'-deoxy-beta-L-uridine (Compound 25) is a nucleoside derivative.
-
GC60023
3'-Deoxyuridine-5'-triphosphate
3'-dUTP
3'-Deoxyuridine-5'-triphosphate (3'-dUTP) is a nucleotide analogue that inhibits DNA-dependent RNA polymerases I and II.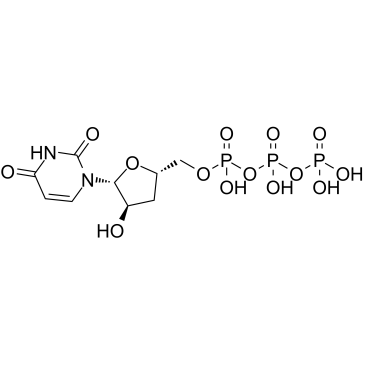
-
GC61862
3'-Deoxyuridine-5'-triphosphate trisodium
3'-dUTP trisodium
3'-Deoxyuridine-5'-triphosphate trisodium (3'-dUTP trisodium) is a nucleotide analogue that inhibits DNA-dependent RNA polymerases I and II.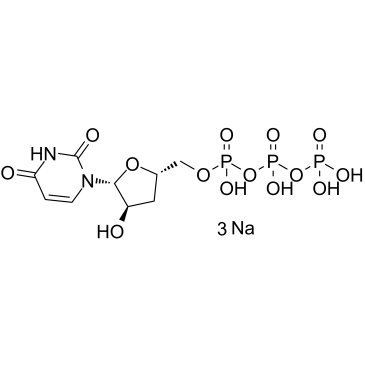
-
GC45332
3'-Dephosphocoenzyme A
depCoA, Dephospho-CoA
An intermediate in the biosynthesis of CoA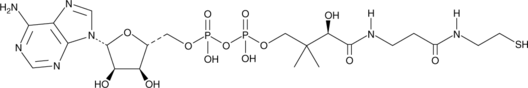
-
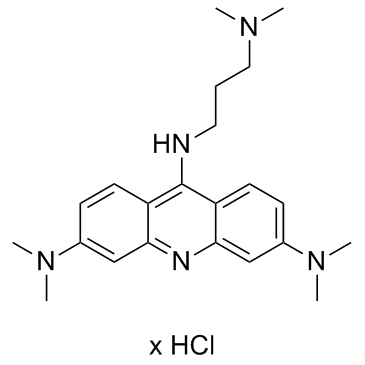
GC34384
3,6-DMAD hydrochloride
3,6-DMAD hydrochloride, an acridine derivative, is a potent IRE1α-XBP1s pathway inhibitor. 3,6-DMAD hydrochloride promotes IL-6 secretion via the IRE1α-XBP1s pathway. 3,6-DMAD hydrochloride inhibits IRE1α oligomerization and endoribonuclease (RNase) activity. 3,6-DMAD hydrochloride can be used for research of cancer.
-
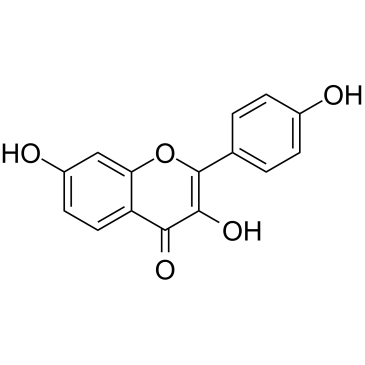
GC34452
3,7,4'-Trihydroxyflavone
3,7,4'-Trihydroxyflavone, isolated from Rhus javanica var.
-
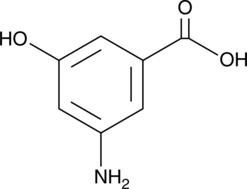
GC52129
3-Amino-5-hydroxybenzoic Acid
AHBA
-
GC13510
3-AP
3-Aminopyridine-2-Carboxyaldehyde Thiosemicarbazone,NSC 663249,Triapine™
ribonucleotide reductase inhibitor and iron chelator with antitumor activity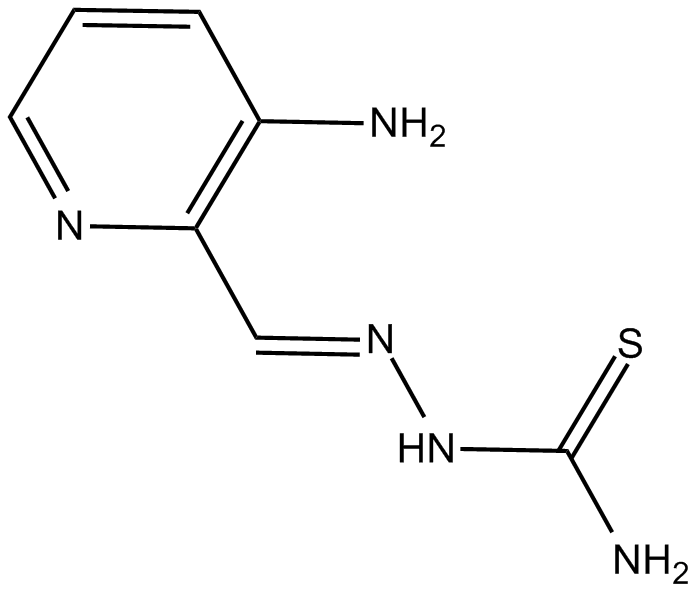
-
GC48457
3-keto Fusidic Acid
3-keto FA, 3-Oxofusidic Acid
An active metabolite of fusidic acid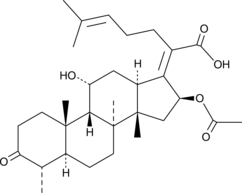
-
GC65084
3-Methylcytidine
3-Methylcytidine, a urinary nucleoside, can be used as a biomarker of four different types of cancer: lung cancer, gastric cancer, colon cancer, and breast cancer.
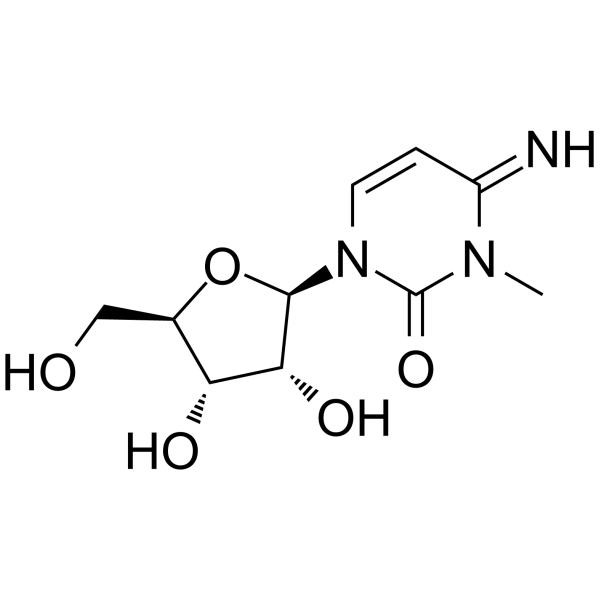
-
GC52391
306-O12B-3
An ionizable cationic lipidoid
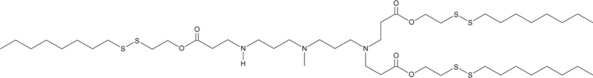
-
GC15389
360A
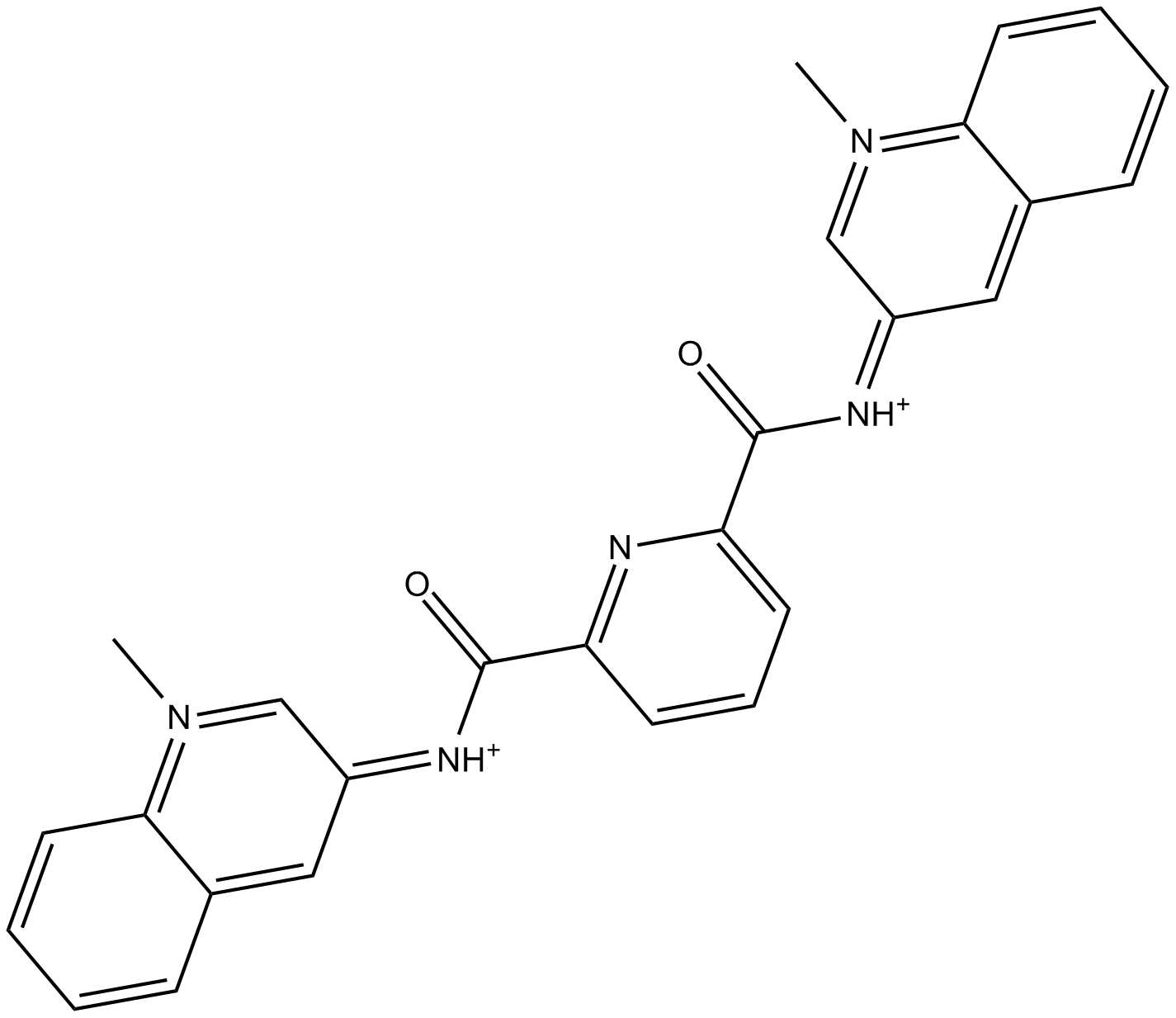
-
GC10115
360A iodide
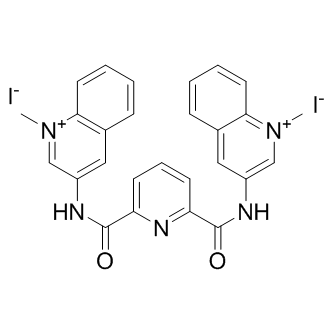
-
GC14493
4μ8C
4u8C
IRE1 Rnase inhibitor, potent and non-toxic
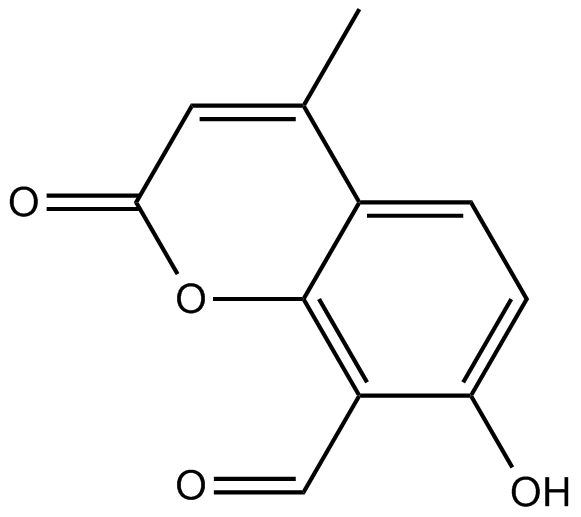
-
GC17271
4'-Demethylepipodophyllotoxin
(-)-4′-Demethylepipodophyllotoxin
An inhibitor of tubulin polymerization