Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Targets for Chromatin/Epigenetics
- Bromodomain(43)
- Aurora Kinase(59)
- DNA Methyltransferase(32)
- HDAC(180)
- Histone Acetyltransferases(63)
- Histone Demethylases(95)
- Histone Methyltransferase(197)
- HIF(90)
- JAK(146)
- MBT Domains(0)
- PARP(105)
- Pim(26)
- Protein Ser/Thr Phosphatases(26)
- RNA Polymerase(5)
- Sirtuin(77)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(200)
- MicroRNA(20)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Products for Chromatin/Epigenetics
- Cat.No. Product Name Information
-
GC31365
γ-Oryzanol
γ-Oryzanol is a potent DNA methyltransferases (DNMTs) inhibitor in the striatum of mice.
-
GC45258
(+)-Biotin 4-Amidobenzoic Acid (sodium salt)
(+)-Biotin PABA, N-Biotinyl-4-aminobenzoic Acid
(+)-Biotin 4-amidobenzoic acid is a substrate of biotinidase, which cleaves biotin amide to give biotin in vivo.
-
GC61595
(+)-JQ-1-aldehyde
(+)-JQ-1-aldehyde is the aldehyde form of (+)-JQ1. (+)-JQ-1-aldehyde can be uesd as a precursor to synthesize PROTACs, which targets BET bromodomains.
-
GC34958
(+)-JQ1 PA
A clickable form of (+)-JQ1
-
GC13822
(-)-JQ1
A selective inhibitor of BET bromodomains
-
GC34964
(1S,3R,5R)-PIM447 dihydrochloride
(1S,3R,5R)-LGH447 dihydrochloride
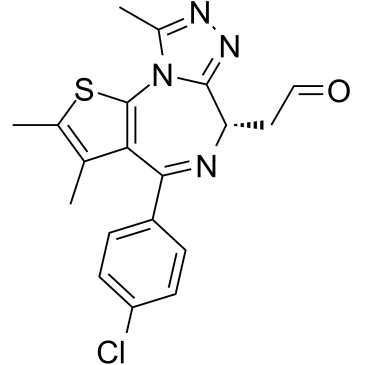
(1S,3R,5R)-PIM447 (dihydrochloride) an PIM inhibitor extracted from patent US 20100056576 A1, compound example 72, has IC50 values of 0.095 μM for Pim1, 0.522 μM for Pim2 and 0.369 μM for Pim3.
-
GC62130
(2R,5S)-Ritlecitinib
(2R,5S)-PF-06651600
(2R,5S)-Ritlecitinib ((2R,5S)-PF-06651600) is a potent and selective JAK3 inhibitor (IC50=144.8 nM) extracted from patent US20150158864A1, example 68.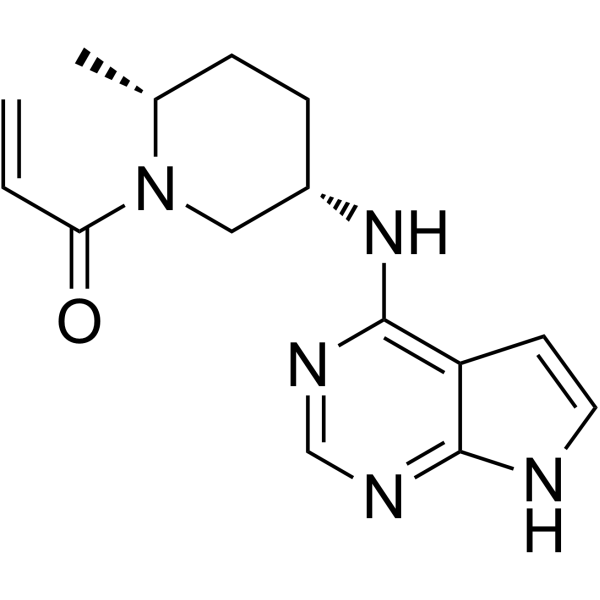
-
GC34971
(3R,4S)-Tofacitinib
(3R,4S)-Tofacitinib is an less active enantiomer of Tofacitinib.
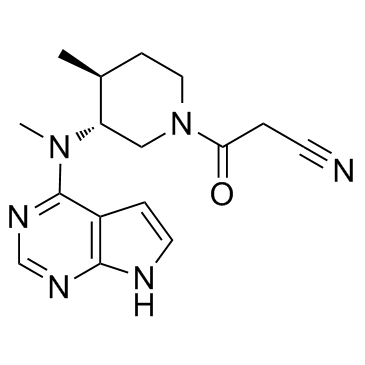
-
GC34972
(3S,4R)-Tofacitinib
(3S,4R)-Tofacitinib is an less active enantiomer of Tofacitinib.
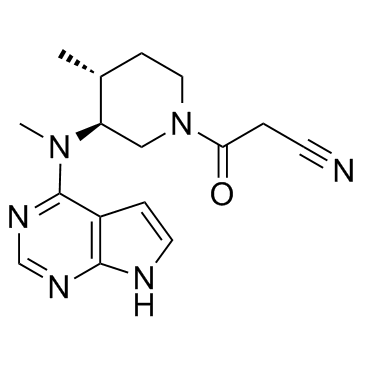
-
GC34973
(3S,4S)-Tofacitinib
(3S,4S)-Tofacitinib is the less active S-enantiomer of Tofacitinib.
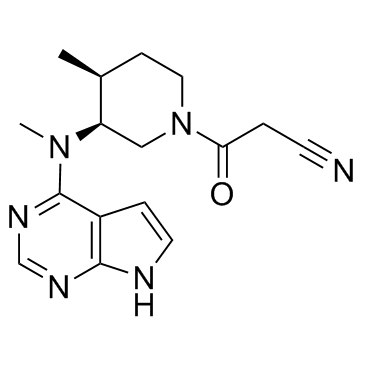
-
GC41695
(6R,S)-5,6,7,8-Tetrahydrofolic Acid (hydrochloride)
Tetrahydrofolate, THFA
(6R,S)-5,6,7,8-Tetrahydrofolic acid (THFA), the reduced form of folic acid, serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.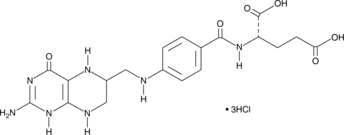
-
GC41088
(6S)-Tetrahydrofolic Acid
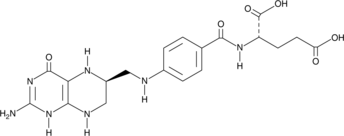
(6S)-Tetrahydrofolic acid is a diastereomer of tetrahydrofolic acid, a reduced form of folic acid that serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.
-
GC72591
(E,E)-RGFP966
(E,E)-RGFP966 is a selective and CNS permeable HDAC3 inhibitor that can be used for the research of Huntington’s disease.

-
GC61807
(E/Z)-AG490
(E/Z)-AG490 ((E/Z)-Tyrphostin AG490) is a racemic compound of (E)-AG490 and (Z)-AG490 isomers. (E)-AG490 is a tyrosine kinase inhibitor that inhibits EGFR, Stat-3 and JAK2/3.

-
GC63864
(E/Z)-Zotiraciclib hydrochloride
(E/Z)-TG02 hydrochloride; (E/Z)-SB1317 hydrochloride
(E/Z)-Zotiraciclib ((E/Z)-TG02) hydrochloride is a potent CDK2, JAK2, and FLT3 inhibitor.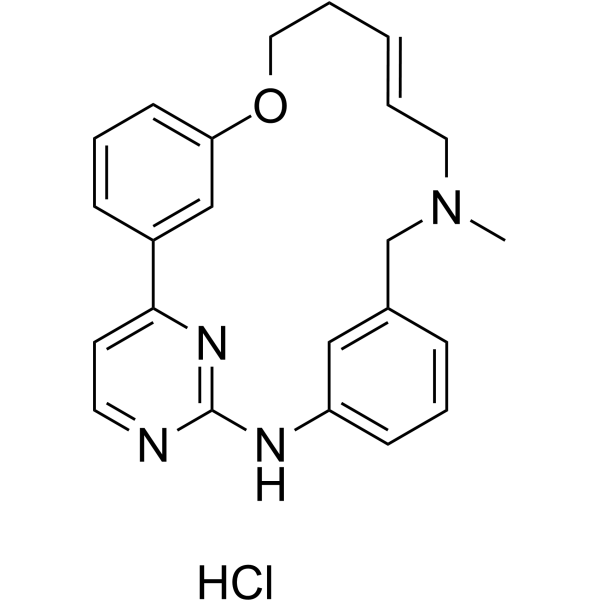
-
GC15104
(R)-(+)-Etomoxir sodium salt
(R)(+)Etomoxir
Etomoxir((R)-(+)-Etomoxir) sodium salt is an irreversible inhibitor of carnitine palmitoyltransferase 1a (CPT-1a), inhibits fatty acid oxidation (FAO) through CPT-1a and inhibits palmitate β-oxidation in human, rat and guinea pig.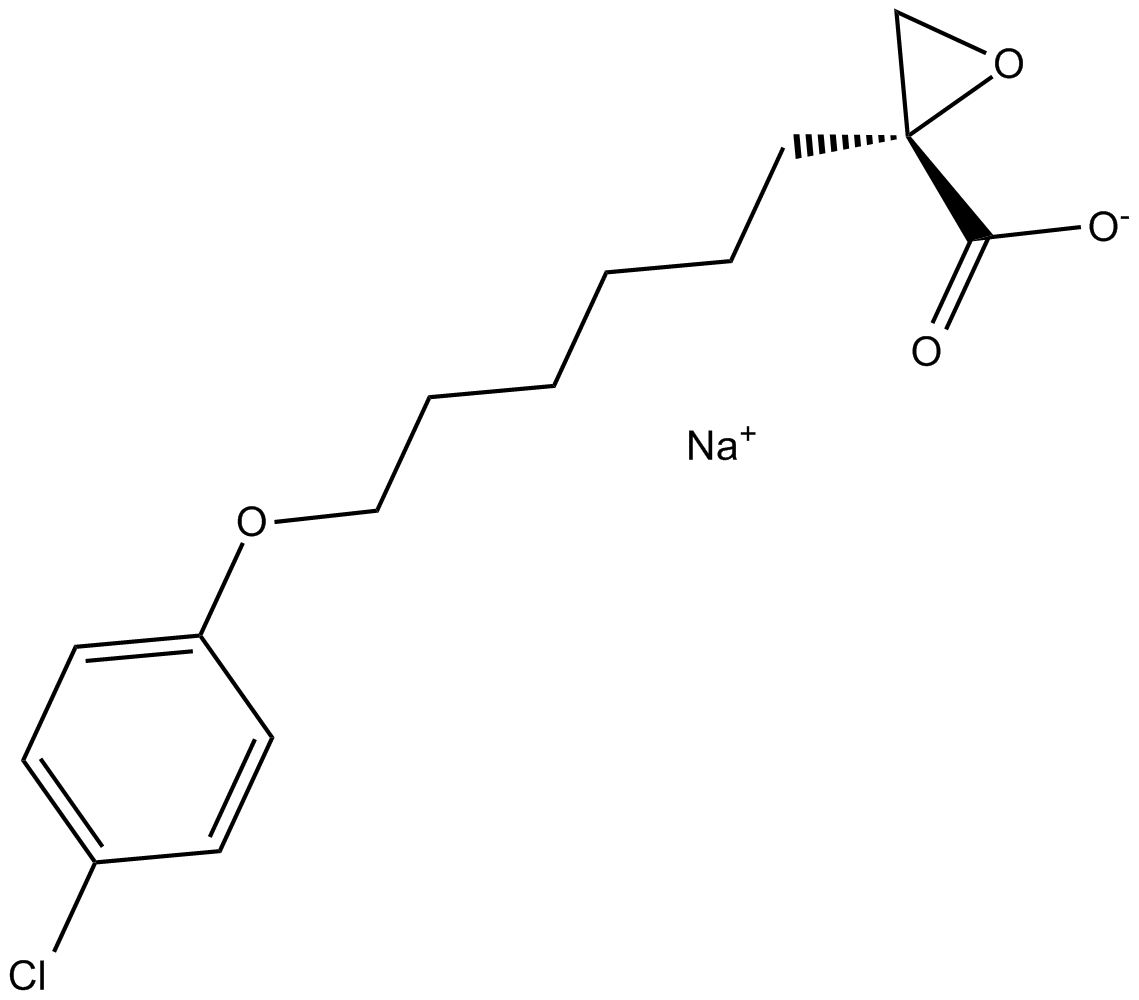
-
GC63791
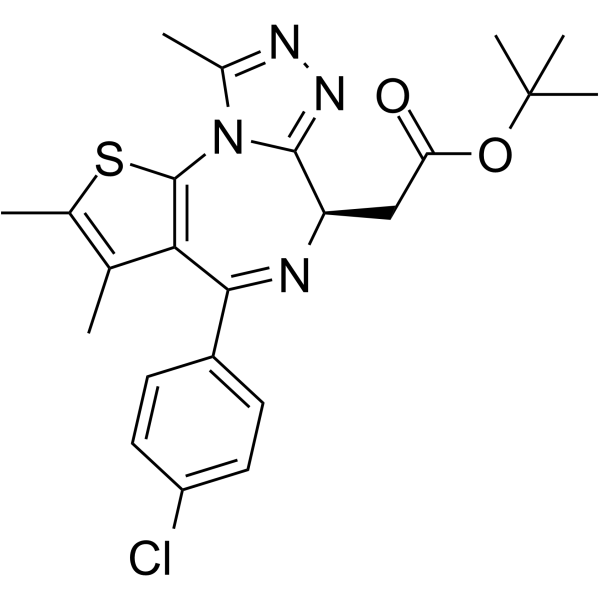
(R)-(-)-JQ1 Enantiomer
(R)-(-)-JQ1 Enantiomer is the stereoisomer of (+)-JQ1. (+)-JQ1 potently decreases expression of both BRD4 target genes, whereas (R)-(-)-JQ1 Enantiomer has no effect.
-
GC45908
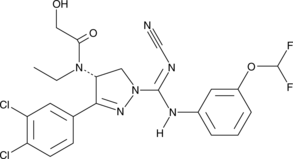
(R)-BAY-598
An inhibitor of SMYD2
-
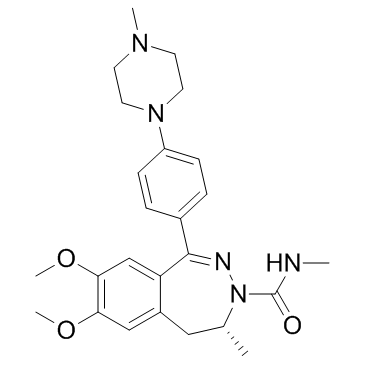
GC34443
(R)-BAY1238097
(R)-BAY1238097 is the R-isomer with lower activity of BAY1238097. BAY1238097 is a potent and selective inhibitor of BET binding to histones and has strong anti-proliferative activity in different AML (acute myeloid leukemia) and MM (multiple myeloma) models through down-regulation of c-Myc levels and its downstream transcriptome.
-
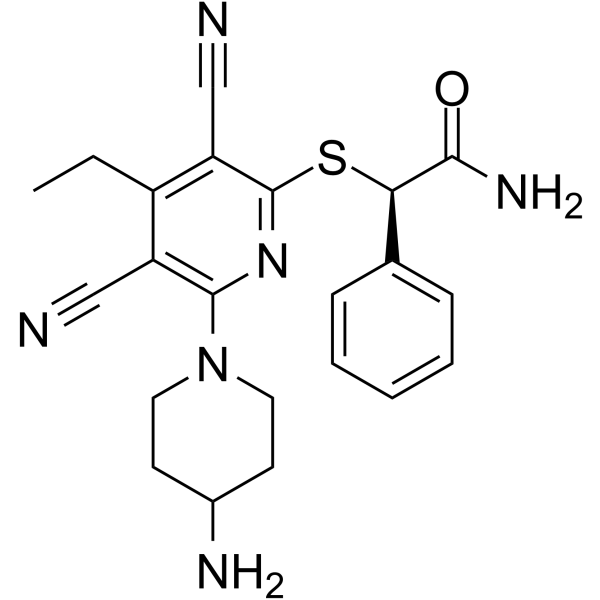
GC64210
(R)-GSK-3685032
(R)-GSK-3685032 is the R-enantiomer of GSK-3685032. GSK-3685032 is a non-time-dependent, noncovalently, first-in-class reversible DNMT1-selective inhibitor, with an IC50 of 0.036 μM. GSK-3685032 induces robust loss of DNA methylation, transcriptional activation, and cancer cell growth inhibition.
-
GC70906
(R)-HH2853
(R)-HH2853 is a mutant EZH2 inhibitor with an IC50 of <100 nM for EZH2-Y641F.

-
GC11340
(R)-PFI 2 hydrochloride
(-)PFI2
PFI-2 ((R)-(R)-PFI 2 hydrochloride) hydrochloride is a potent and selective SET domain containing lysine methyltransferase 7 (SETD7) inhibitor.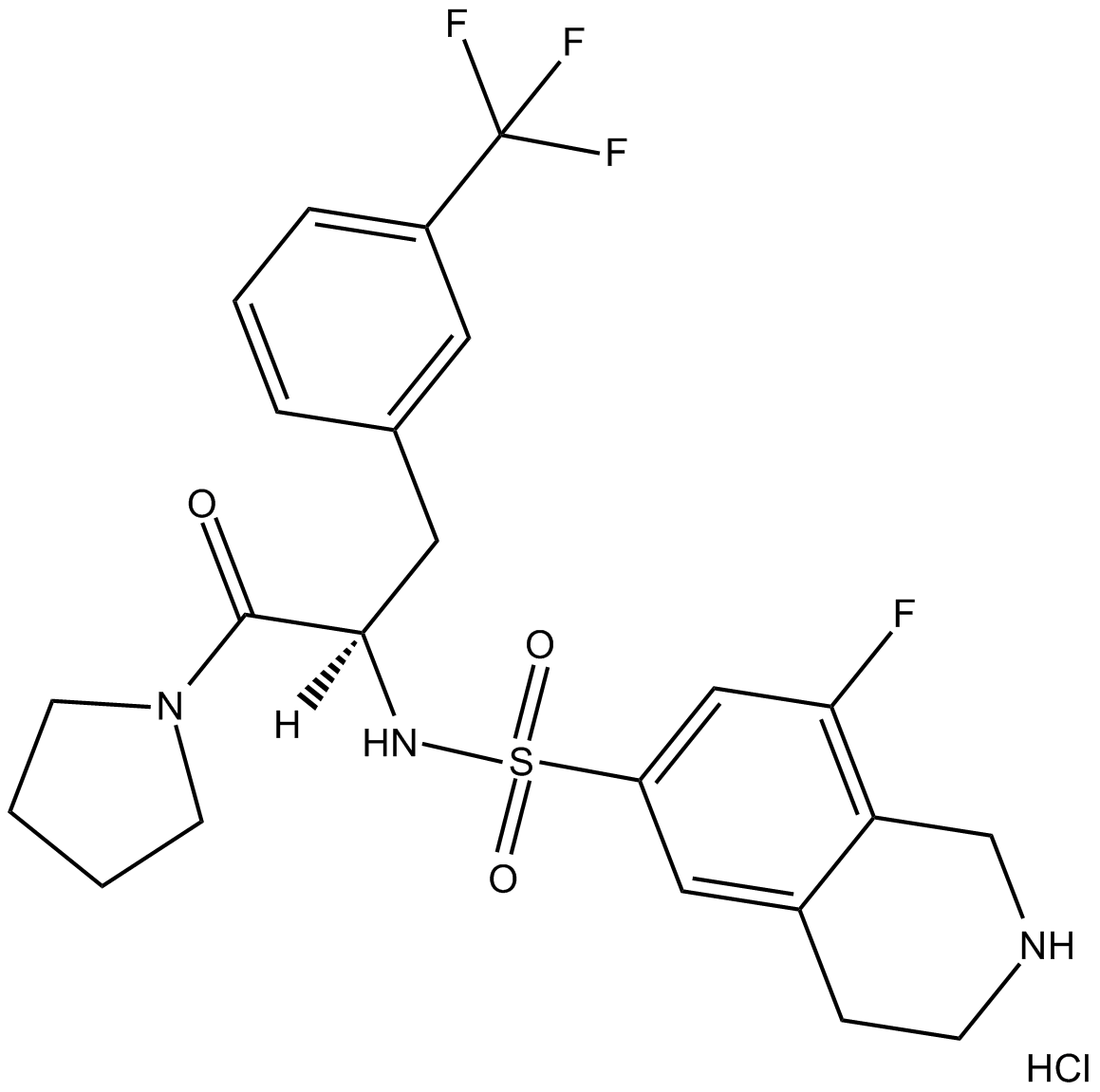
-
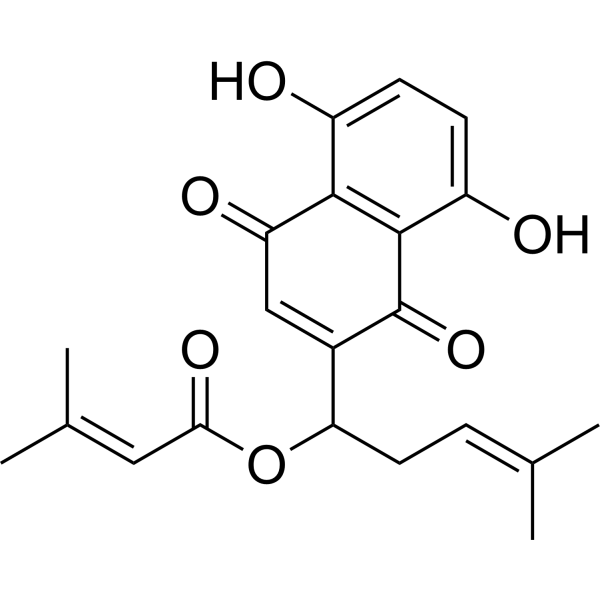
GC69794
(Rac)-Arnebin 1
(Rac)-β,β-Dimethylacrylalkannin; (Rac)-β,β-?Dimethylacrylshikonin
(Rac)-Arnebin 1 is the racemic form of β,β-Dimethylacrylalkannin and/or β,β-Dimethylacrylshikonin. These compounds are naphthoquinones isolated from plants in the Lithospermum genus, with β,β-Dimethylacrylshikonin exhibiting anti-tumor activity.
-
GC34446
(rac)-BAY1238097
(Rac)-BAY1238097 is a BET inhibitor, with an IC50 of 1.02 μM for BRD4. Used in cancer research.
-
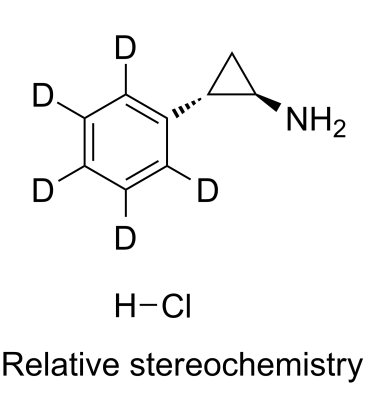
GC60004
(rel)-Tranylcypromine D5 hydrochloride
Tranylcypromine-d5 (SKF 385-d5) hydrochloride is a deuterium labeled (rel)-Tranylcypromine hydrochloride.
-
GC33198
(S)-JQ-35 (TEN-010)
TEN-010
(S)-JQ-35 (TEN-010) (TEN-010) is an inhibitor of the Bromodomain and Extra-Terminal (BET) family bromodomain-containing proteins with potential antineoplastic activity.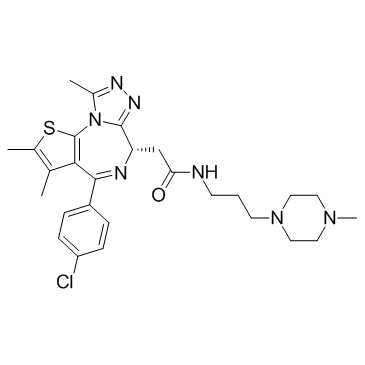
-
GC65045
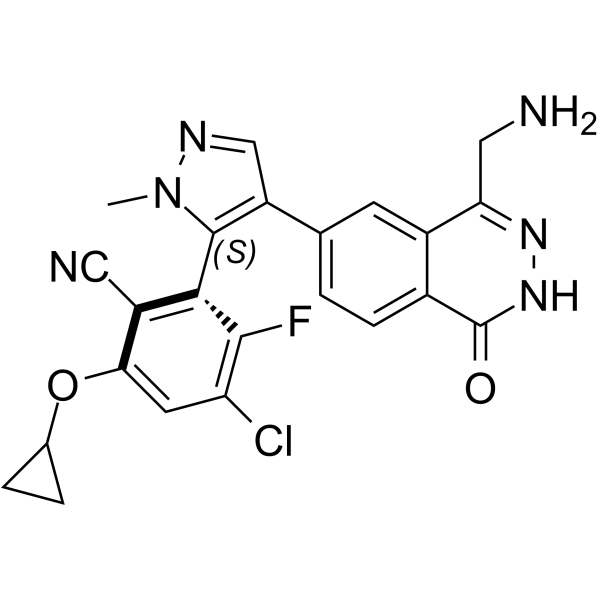
(S)-MRTX-1719
(S)-MRTX-1719 (example 16-7) is the S-enantiomer of MRTX-1719. (S)-MRTX-1719 is a PRMT5/MTA complex inhibitor, with an IC50 of 7070 nM.
-
GC13634
(S)-PFI-2 (hydrochloride)
(+)-PFI-2
Negative control of (R)-PFI 2 hydrochloride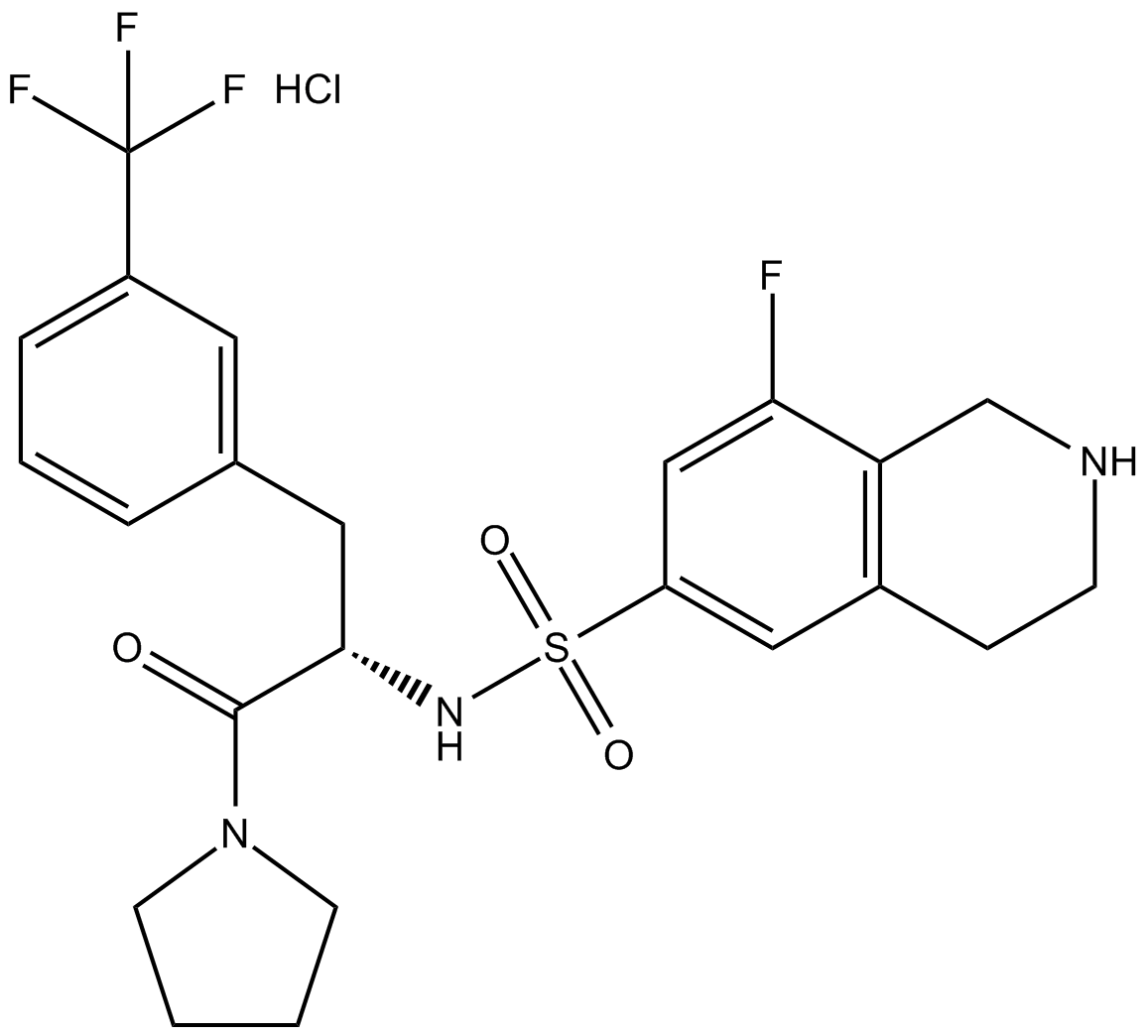
-
GC65133
(S,R,S)-AHPC-C5-COOH
VH032-C5-COOH
(S,R,S)-AHPC-C5-COOH (VH032-C5-COOH) is a synthesized?E3 ligase ligand-linker conjugate, contains the VH032 VHL-based ligand and a linker to form PROTACs. VH-032 is a selective and potent inhibitor of VHL/HIF-1α interaction with a Kd of 185 nM, has the potential for the study of anemia and ischemic diseases.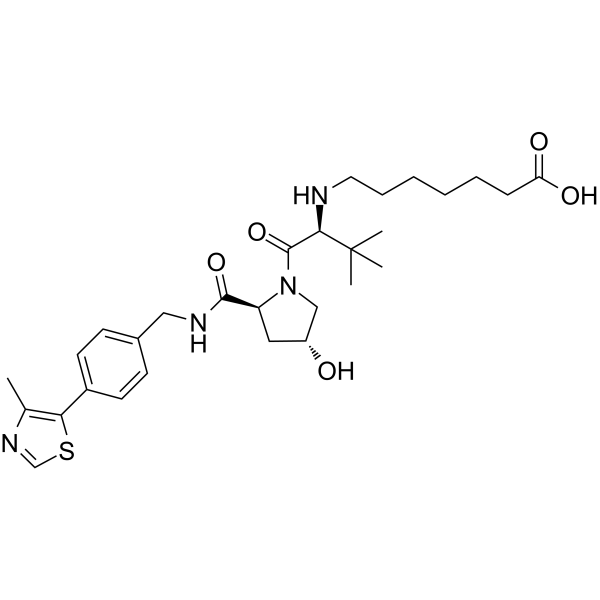
-
GC17055
1,2,3,4,5,6-Hexabromocyclohexane
1,2,3,4,5,6-六溴环己烷(1,2,3,4,5,6-Hexabromocyclohexane)直接抑制JAK2激酶结构域内的口袋,抑制自身磷酸化。
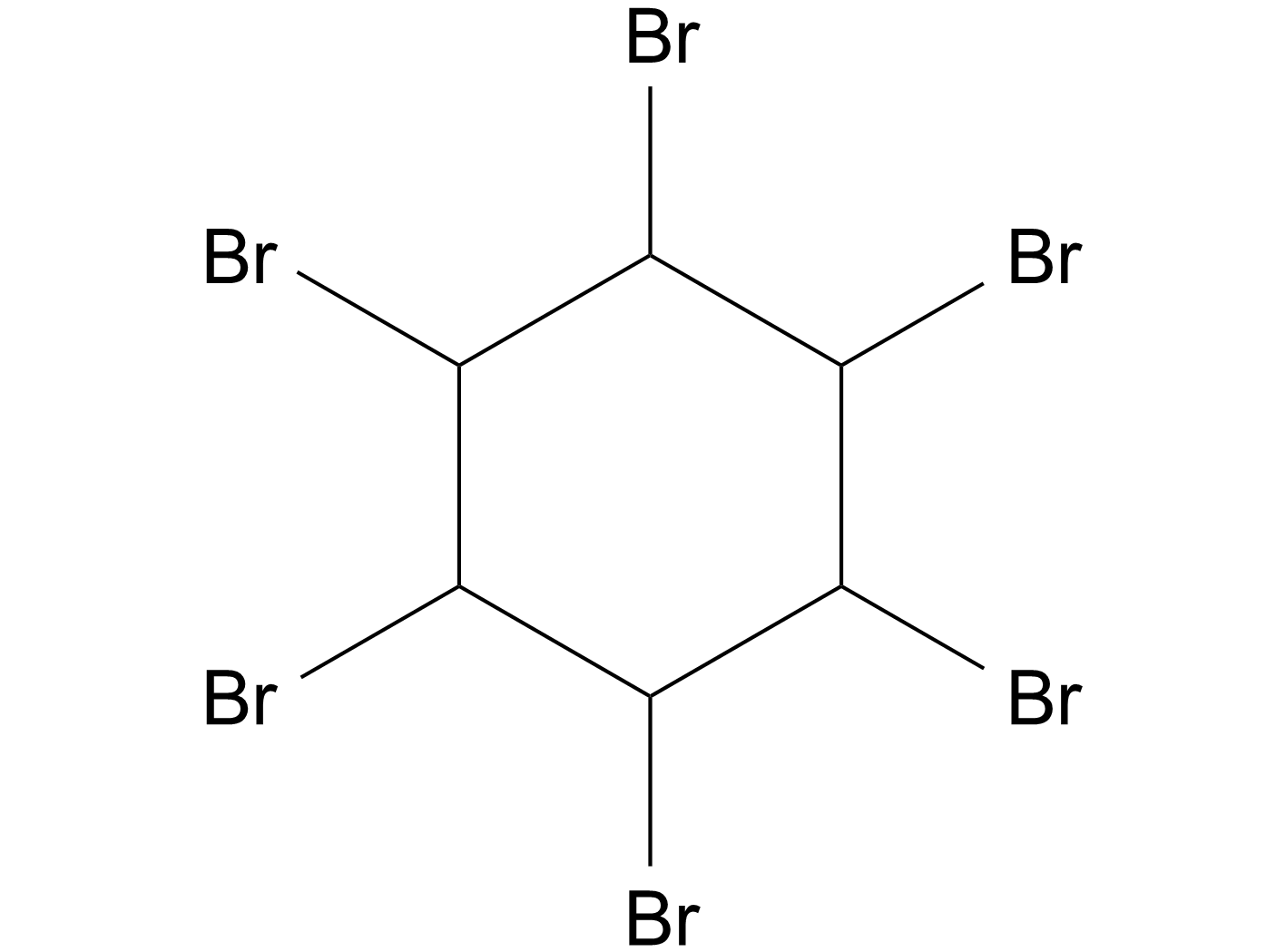
-
GC61949
1,4-DPCA ethyl ester
1,4-DPCA ethyl ester is the ethyl ester of 1,4-DPCA and can inhibit factor inhibiting HIF (FIH).
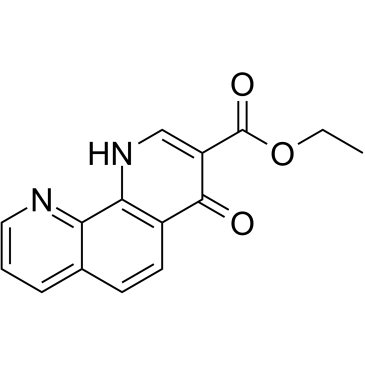
-
GC40468
1,5-Isoquinolinediol
NSC 65585
The poly(ADP-ribose) polymerases (PARPs) form a family of enzymes with roles in DNA repair and apoptosis.
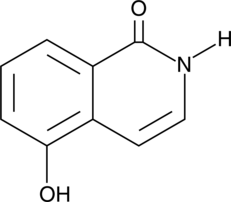
-
GC41984
1-Alaninechlamydocin
1-Alaninechalmydocin is a fungal metabolite originally isolated from a Great Lakes-derived Tolypocladium sp.
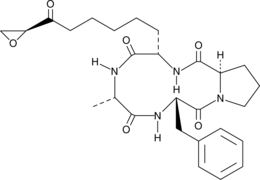
-
GC39214
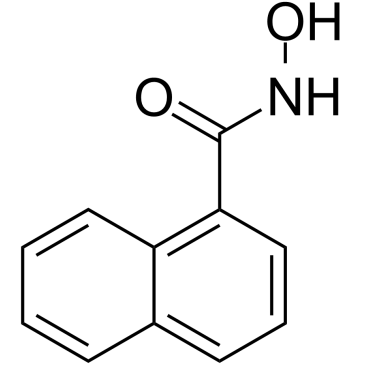
1-Naphthohydroxamic acid
1-Naphthohydroxamic acid (Compound 2) is a potent and selective HDAC8 inhibitor with an IC50 of 14 μM. 1-Naphthohydroxamic acid is more selectively for HDAC8 than class I HDAC1 and class II HDAC6 (IC50 >100 μM). 1-Naphthohydroxamic acid does not increase global histone H4 acetylation and also does not reduce total intracellular HDAC activity.1-Naphthohydroxamic acid can induce tubulin acetylation.
-
GC46508
2',2'-Difluoro-2'-deoxyuridine
dFdU
An active metabolite of gemcitabine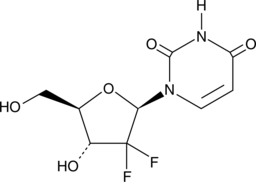
-
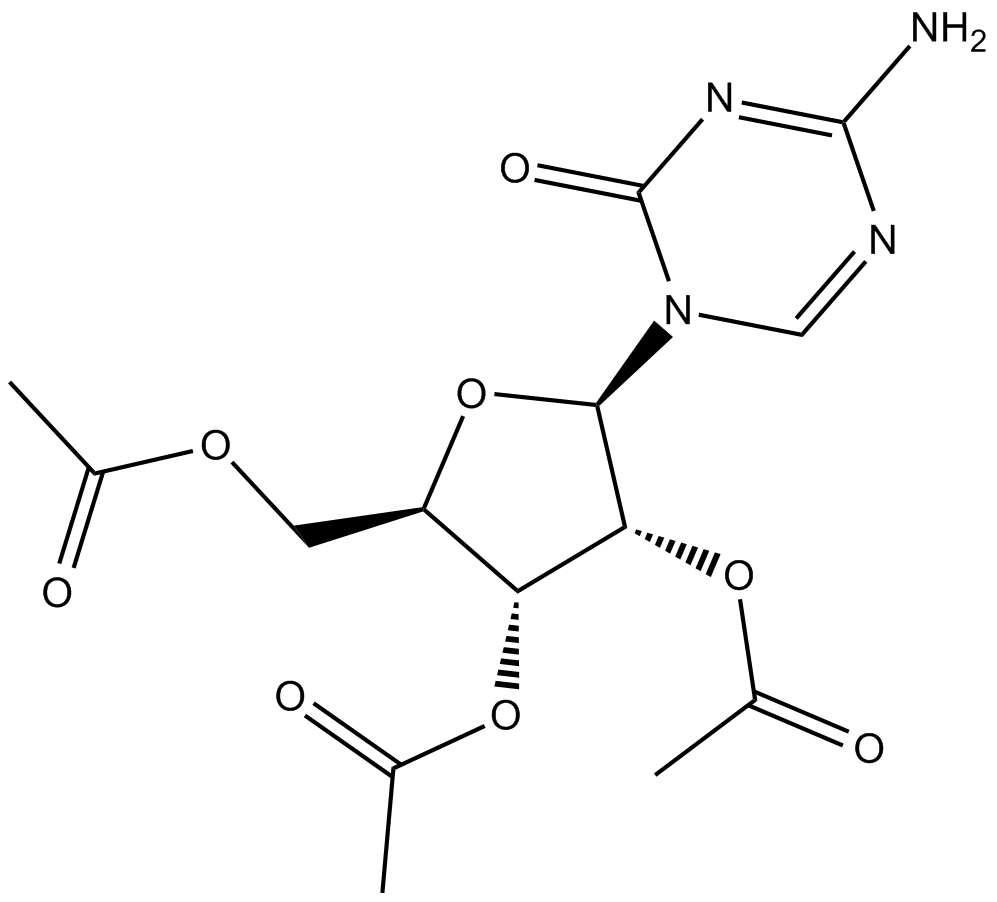
GC12775
2',3',5'-triacetyl-5-Azacytidine
prodrug form of 5-azacytidine, a DNA methyltransferase inhibitor
-
GC16195
2,4-DPD
2,4-Diethylpyridine dicarboxylate
Diethyl pyridine-2,4-dicarb is a potent prolyl 4-hydroxylase-directed proinhibitor.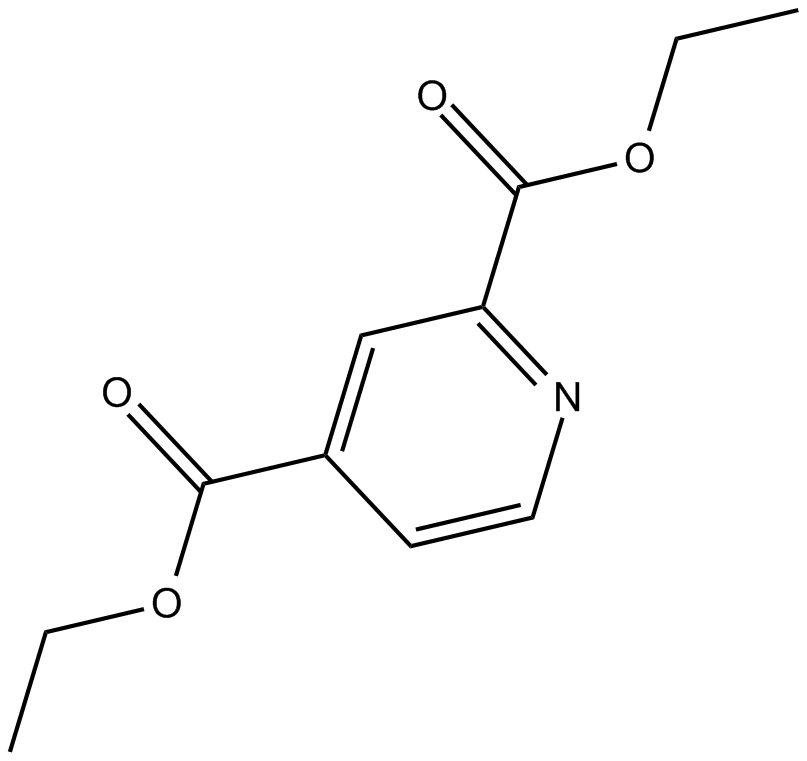
-
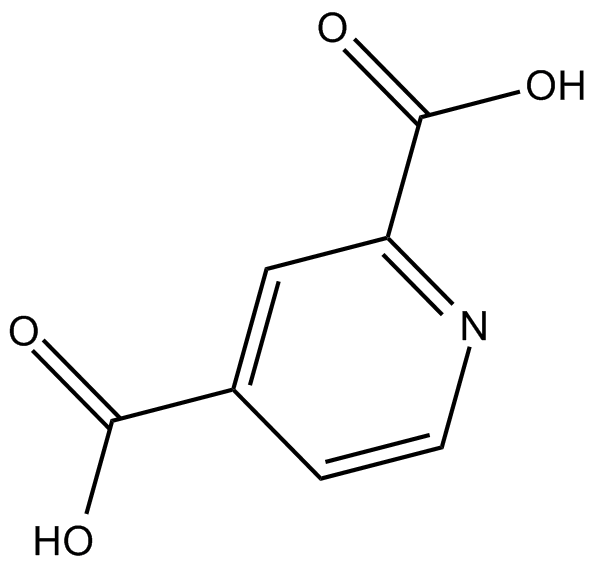
GC11873
2,4-Pyridinedicarboxylic Acid
2,4-Pyridinedicarboxylic Acid (2,4 pyridine dicarboxylic acid) is a broad-spectrum inhibitor of 2OG oxygenase, including JmjC domain-containing family of histone demethylases (JHDMs). 2,4-Pyridinedicarboxylic Acid is a target chemical in the field of bio-based plastics.
-
GC42075
2,4-Pyridinedicarboxylic Acid (hydrate)
2,4-Dicarboxylpyridine, 2,4-PDCA
2,4-Pyridinedicarboxylic Acid (2,4-PDCA) is a compound that structurally mimics 2-oxoglutarate (2-OG, also known as α-ketoglutarate) and chelates zinc, thus affecting a range of enzymes.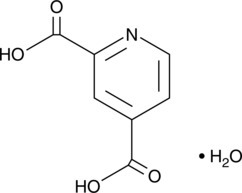
-
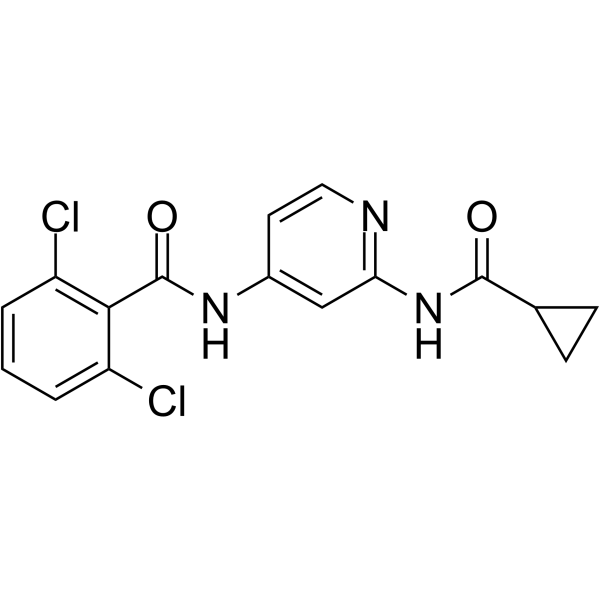
GC62233
2,6-Dichloro-N-(2-(cyclopropanecarboxamido)pyridin-4-yl)benzamide
GDC-046 is a potent, selective, and orally bioavailable TYK2 inhibitor with Kis of 4.8, 0.7, 0.7, and 0.4 nM for TYK2, JAK1, JAK2, and JAK3, respectively.
-
GC46503
2-(1,8-Naphthyridin-2-yl)phenol
2-NP
2-(1,8-Naphthyridin-2-yl)phenol is a selective enhancer of STAT1 transcription. 2-(1,8-Naphthyridin-2-yl)phenol can enhance the ability of IFN-γ to inhibit the proliferation of human breast cancer and fibrosarcoma cells.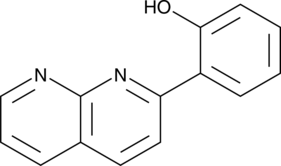
-
GC10408
2-D08
Sumoylation inhibitor
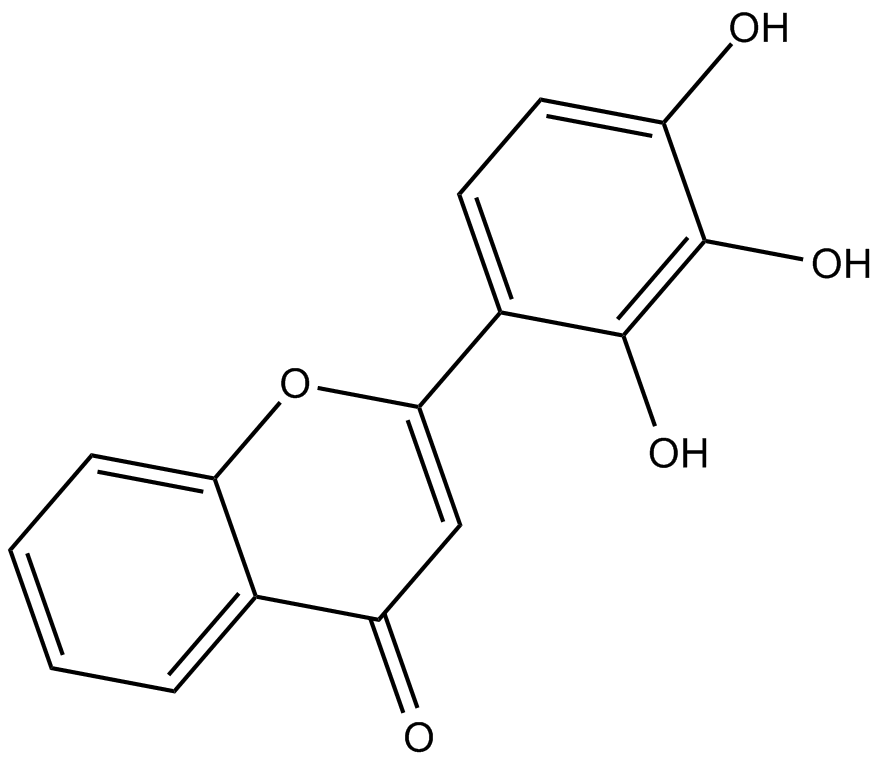
-
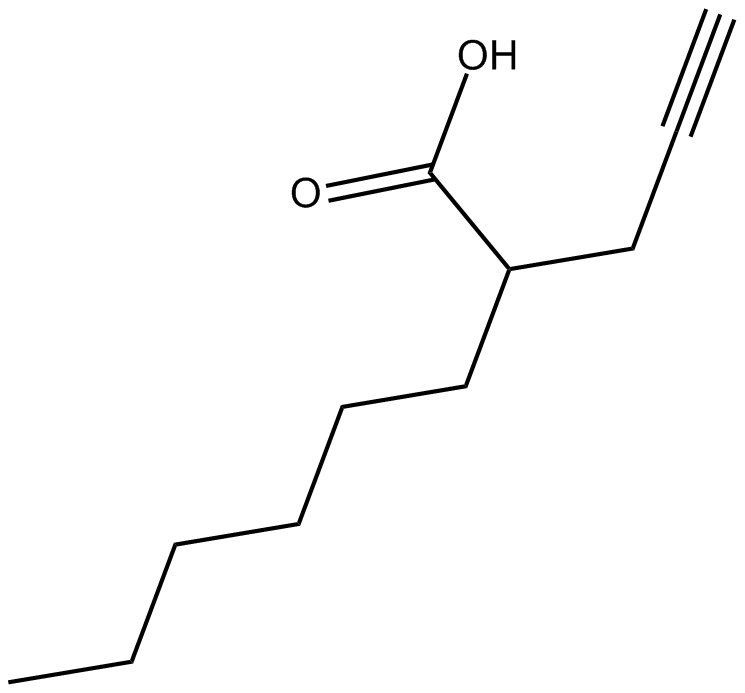
GC11249
2-hexyl-4-Pentynoic Acid
Potent and robust HDACs inhibitor
-
GC15084
2-Methoxyestradiol (2-MeOE2)
2Hydroxyestradiol 2methyl ether, 2ME2, NSC 659853, Panzem
2-Methoxyestradiol (2-MeOE2/2-Me) is an HIF-1α inhibitor.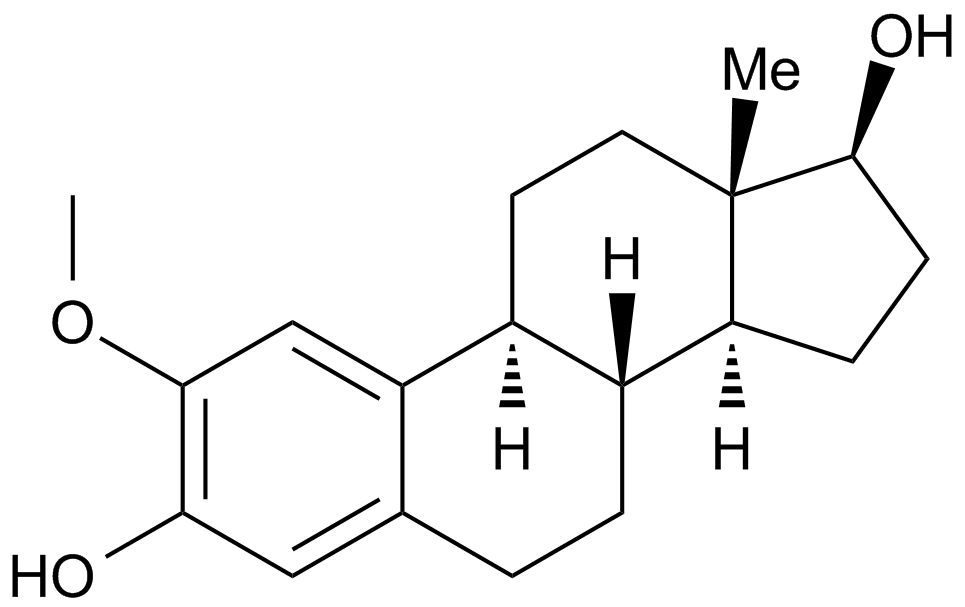
-
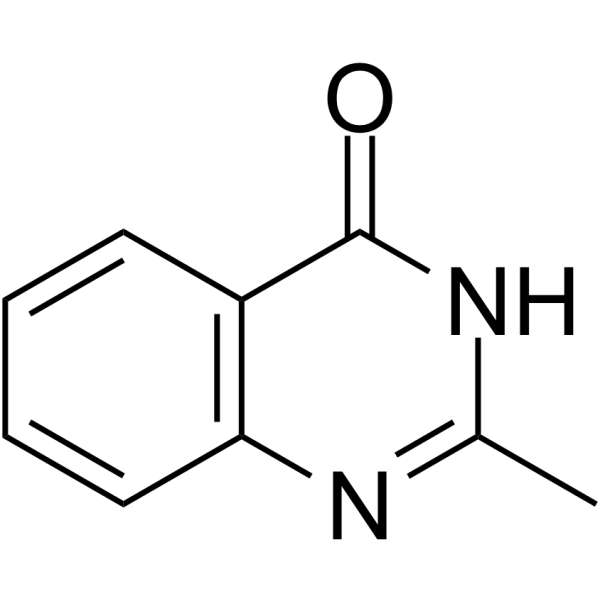
GC62781
2-Methylquinazolin-4-ol
2-Methylquinazolin-4-ol is a potent competitive poly(ADP-ribose) synthetase inhibitor, with a Ki of 1.1 μM.
-
GC14282
3-acetyl-11-keto-β-Boswellic Acid
3-O-acetyl-11-keto-β-Boswellic acid,AKBA
3-acetyl-11-keto-β-Boswellic Acid (Acetyl-11-keto-β-boswellic acid) is an active triterpenoid compound from the extract of Boswellia serrate and a novel Nrf2 activator.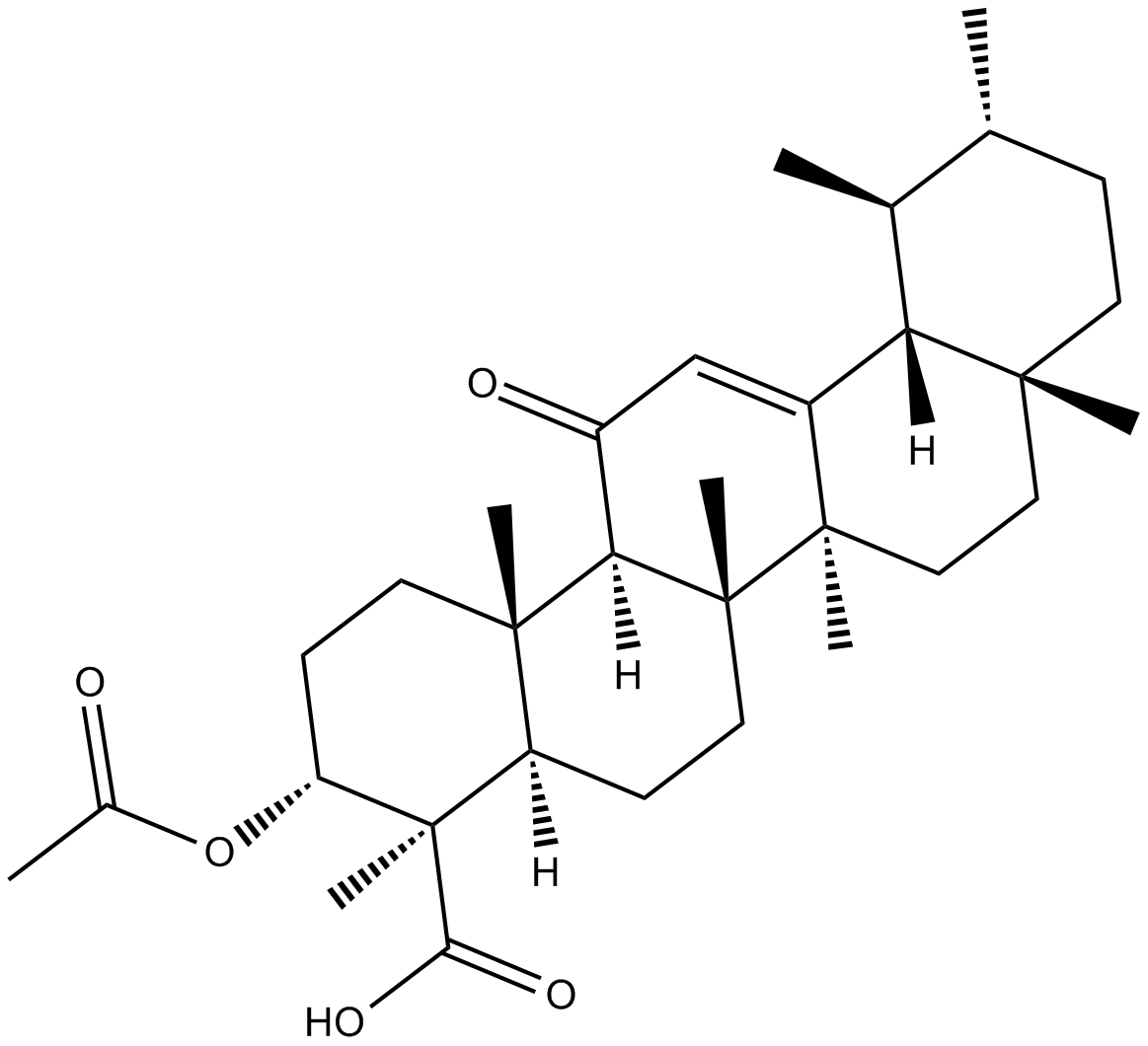
-
GC17907
3-Deazaneplanocin A (DZNep) hydrochloride
2,3DMMC
3-Deazaneplanocin A (DZNep) hydrochloride is an adenosine analogue and is a competitive S-adenosylhomocysteine hydrolase inhibitor with a Ki of 50 pM in cell-free tests.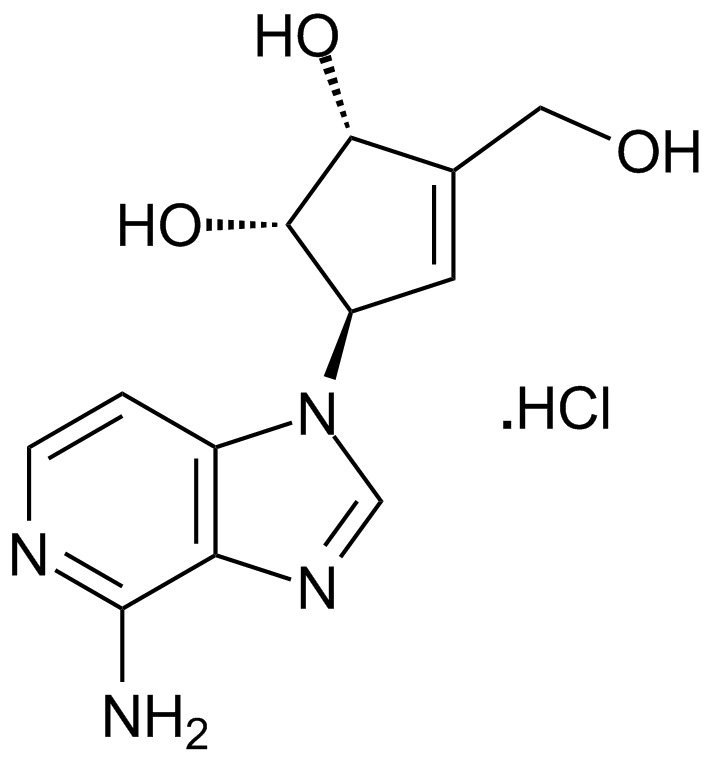
-
GC13145
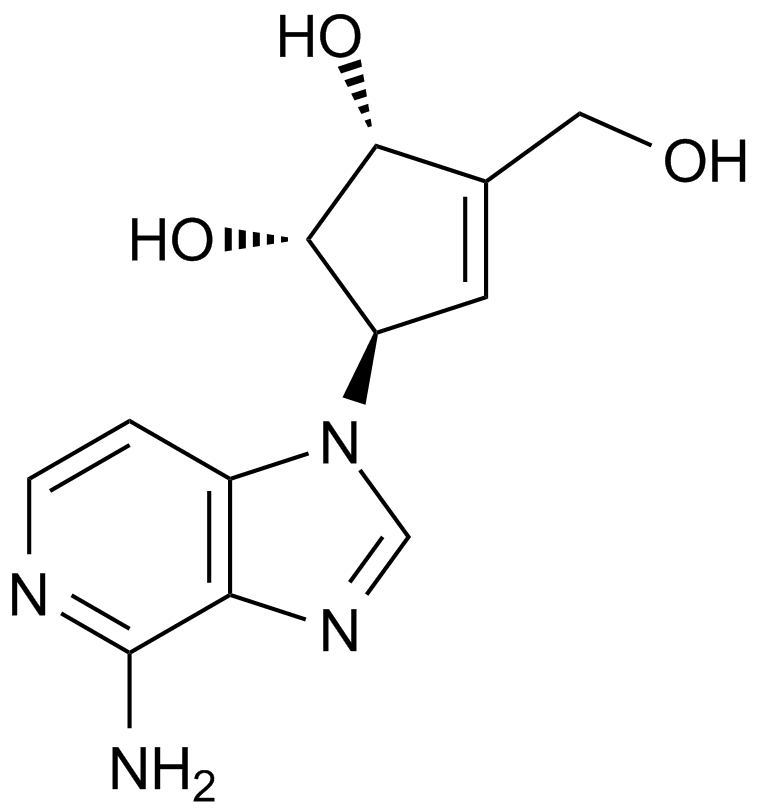
3-Deazaneplanocin,DZNep
DZNep, NSC 617989
An inhibitor of lysine methyltransferase EZH2
-
GC18509
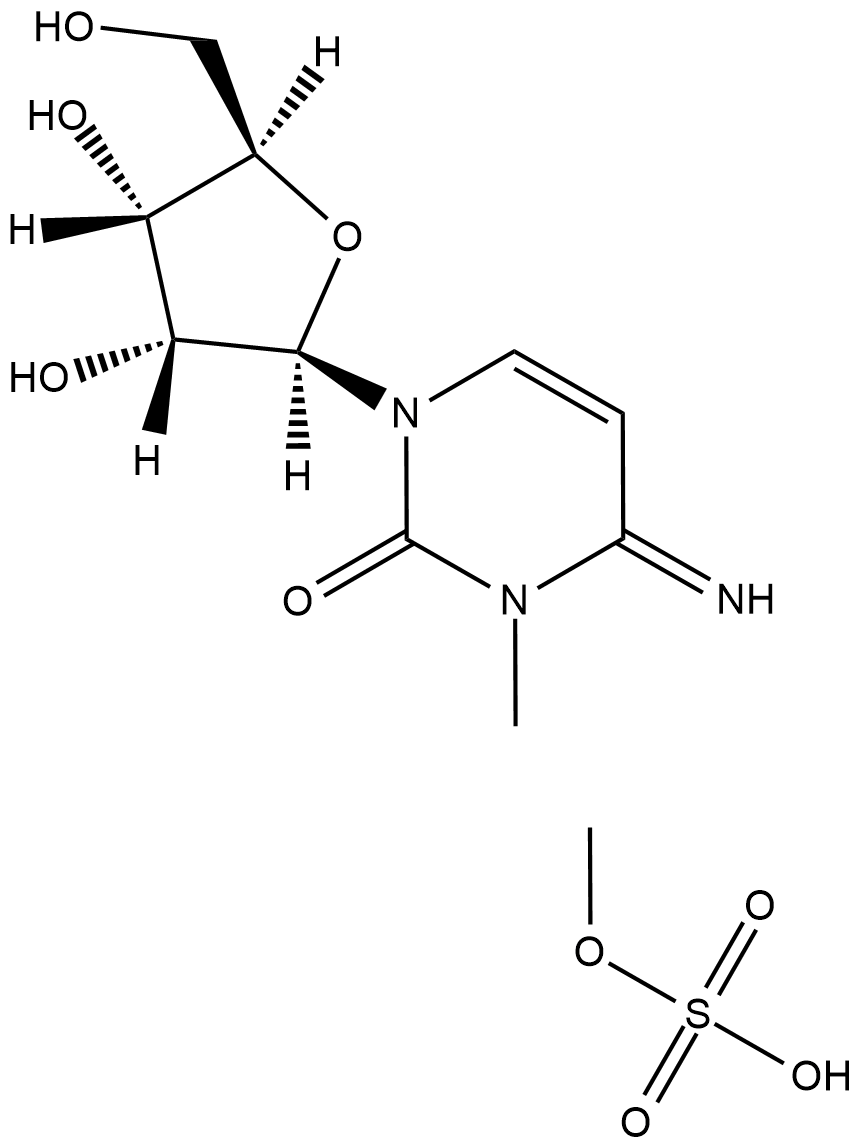
3-Methylcytidine (methosulfate)
3-Methylcytidine (methosulfate) is a cytidine derivative used as an internal standard for HPLC.
-
GC19013
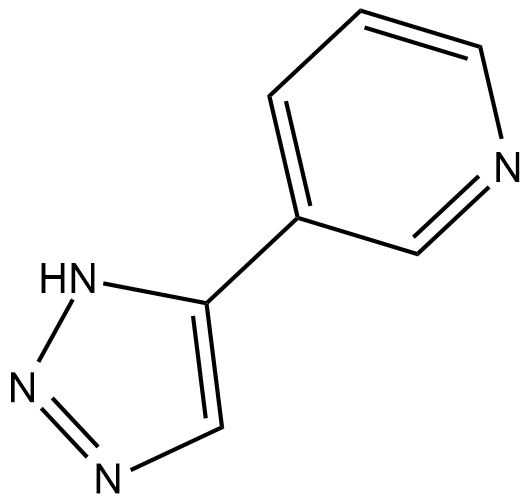
3-TYP
3-TYP inhibit SIRT3 with an IC50 of 16 nM, and is more potent over SIRT1 and SIRT2 with IC50 of 88 nM and 92 nM, respectively.
-
GC62805
4’-Methoxychalcone
4’-Methoxychalcone regulates adipocyte differentiation through PPARγ activation.

-
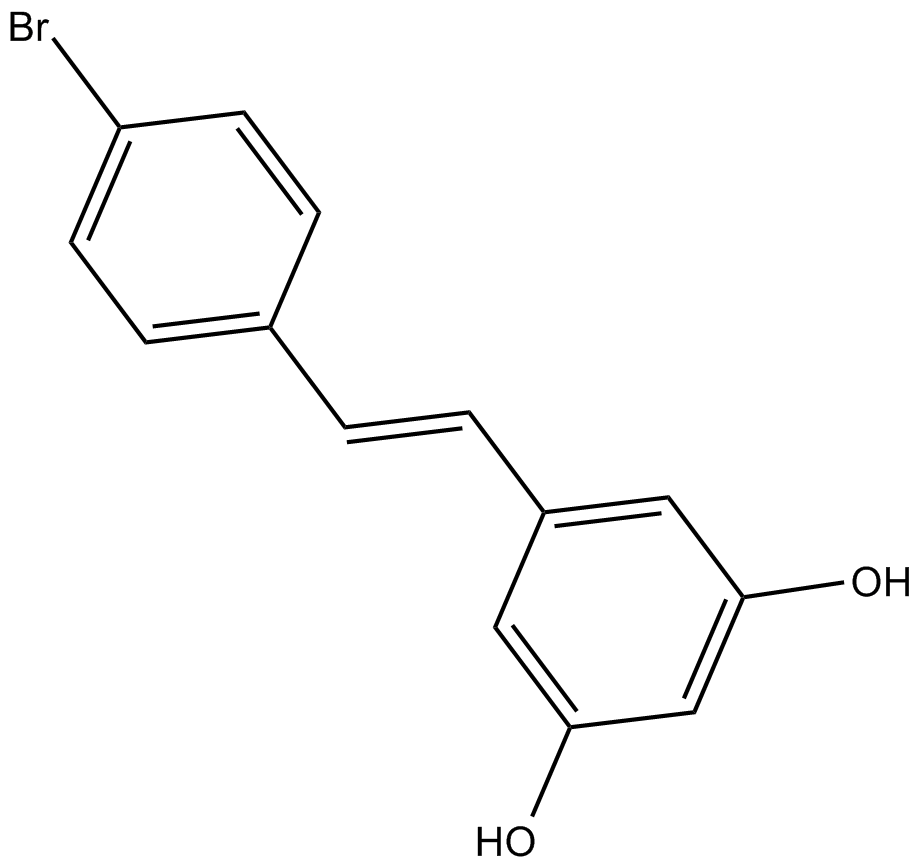
GC17922
4'-bromo-Resveratrol
Sirt1 and Sirt3 inhibitor
-
GC46607
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone
NNK
A tobacco-specific nitrosamine carcinogen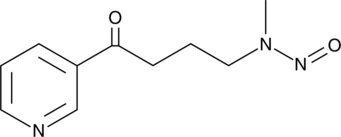
-
GC11761
4-amino-1,8-Naphthalimide
4-Aminonaphthalimide,4-ANI
4-amino-1,8-Naphthalimide is a potent PARP inhibitor and potentiates the cytotoxicity of γ-radiation in cancer cells.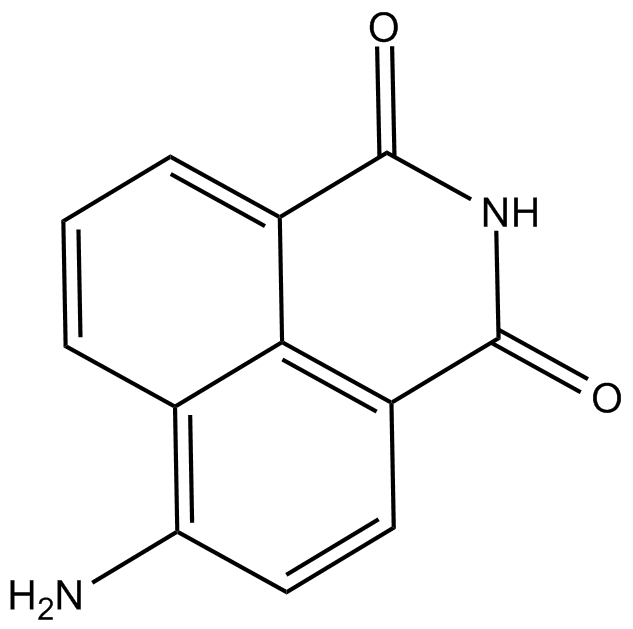
-
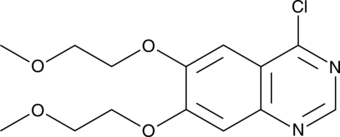
GC46628
4-Chloro-6,7-bis(2-methoxyethoxy)quinazoline
A building block and synthetic intermediate
-
GC17005
4-iodo-SAHA
ISAHA
class I and class II histone deacetylase (HDAC) inhibitor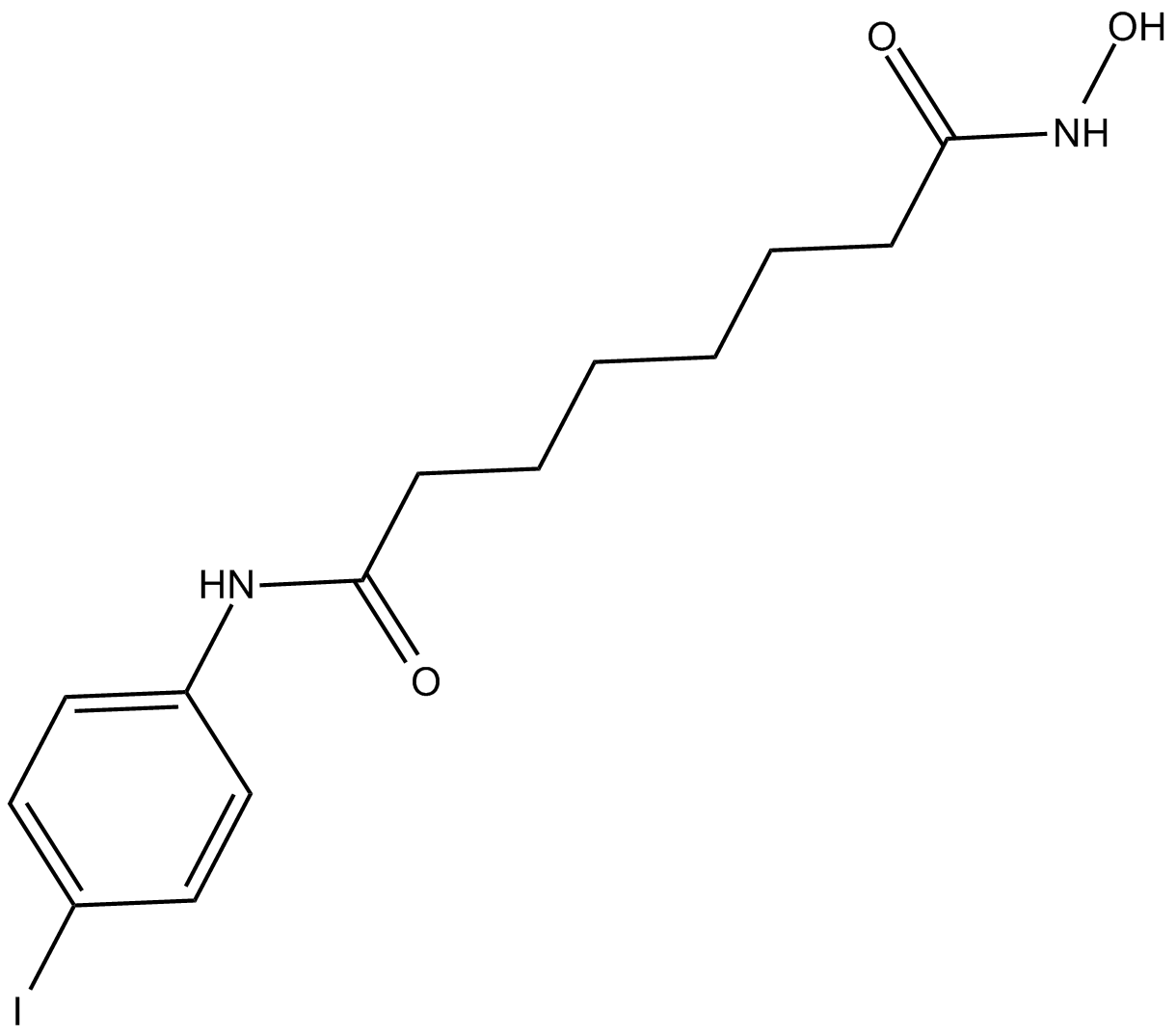
-
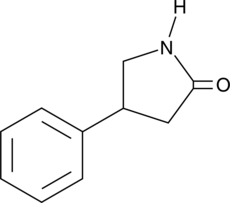
GC46675
4-Phenyl-2-pyrrolidinone
A precursor and synthetic intermediate
-
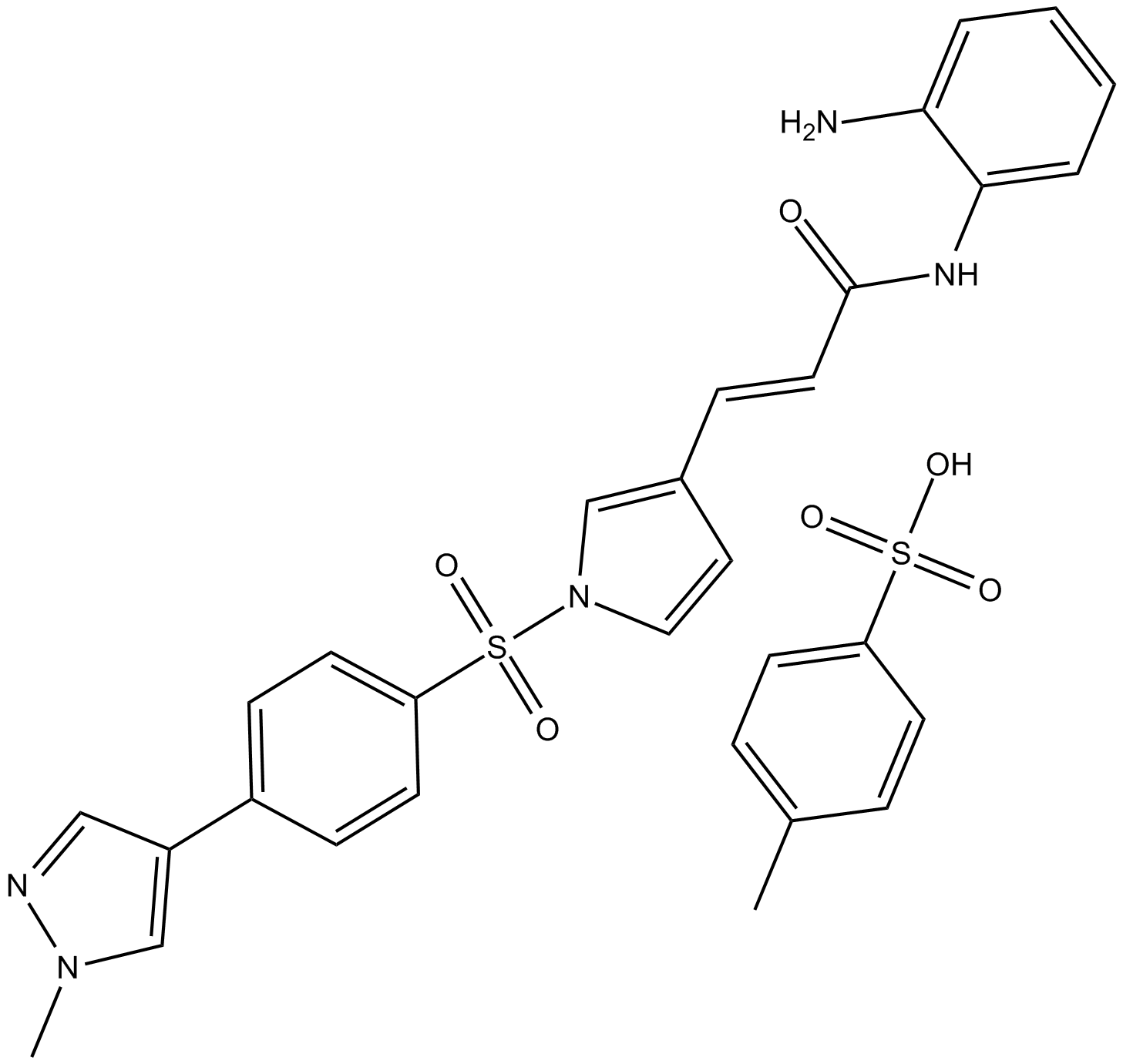
GC12667
4SC-202
4SC-202 (4SC-202) is a selective class I HDAC inhibitor with IC50 of 1.20 μM, 1.12 μM, and 0.57 μM for HDAC1, HDAC2, and HDAC3, respectively. It also displays inhibitory activity against Lysine specific demethylase 1 (LSD1).
-
GC35150
5,7,4'-Trimethoxyflavone
5,7,4'-Trimethoxyflavone is isolated from Kaempferia parviflora (KP) that is a famous medicinal plant from Thailand. 5,7,4'-Trimethoxyflavone induces apoptosis, as evidenced by increments of sub-G1 phase, DNA fragmentation, annexin-V/PI staining, the Bax/Bcl-xL ratio, proteolytic activation of caspase-3, and degradation of poly (ADP-ribose) polymerase (PARP) protein.5,7,4'-Trimethoxyflavone is significantly effective at inhibiting proliferation of SNU-16 human gastric cancer cells in a concentration dependent manner.
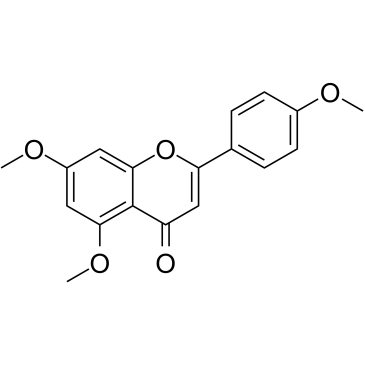
-
GN10629
5,7-dihydroxychromone
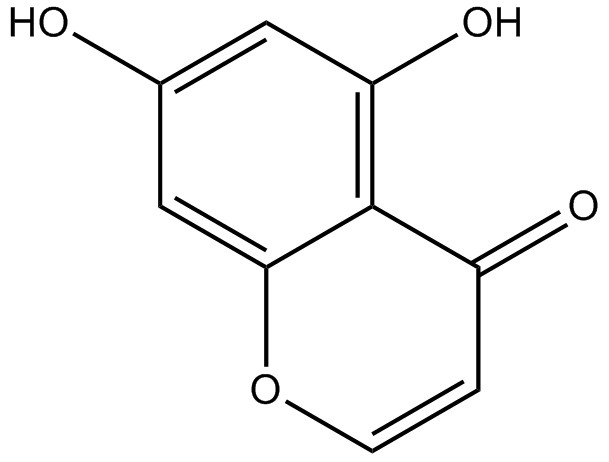
-
GC10898
5-(N,N-dimethyl)-Amiloride (hydrochloride)
DMA,L-591,605,MK-685
NHE1, NHE2, and NHE3 inhibitor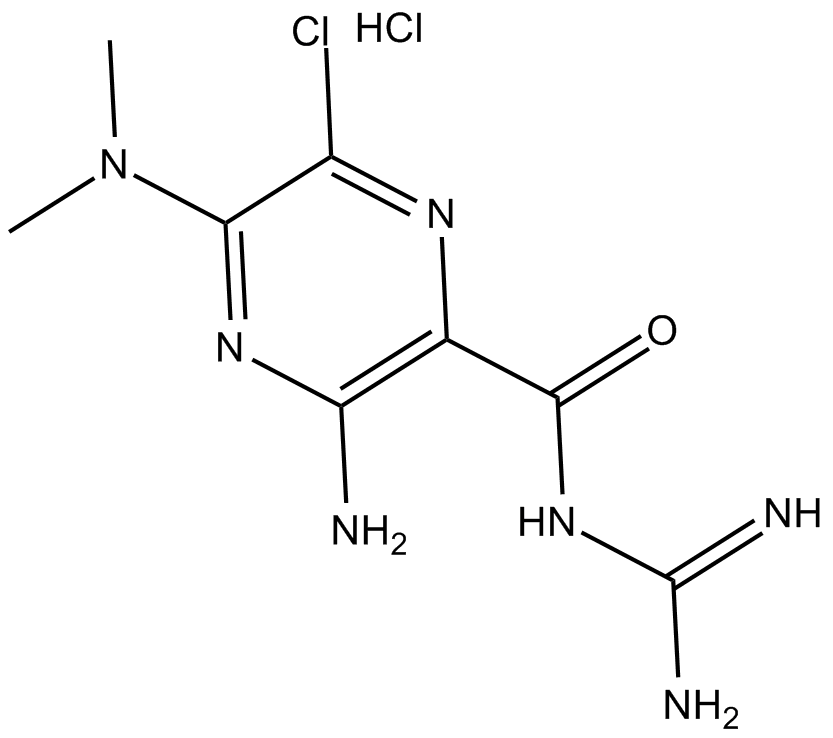
-
GC68161
5-AIQ
5-Aminoisoquinolin-1-one
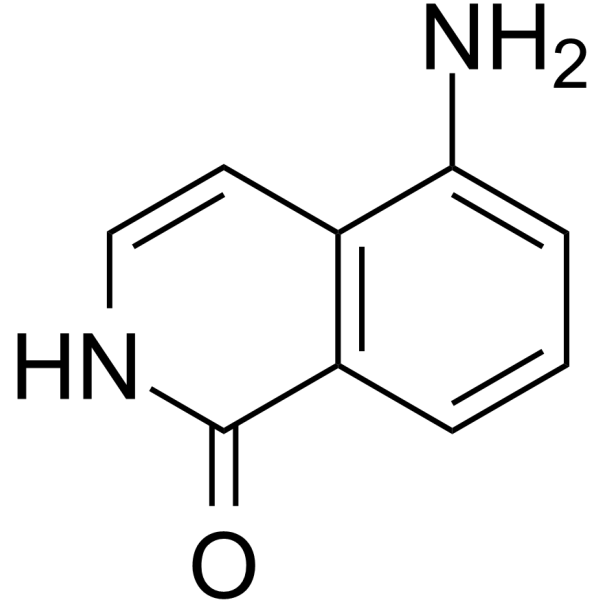
-
GC10946
5-Azacytidine
Antibiotic U 18496, 5AzaC, Ladakamycin, Mylosar, NSC 102816, NSC 103627, U 18496, WR 183027
5-Azacytidine (also known as 5-AzaC), a compound belonging to a class of cytosine analogues, is a DNA methyl transferase (DNMT) inhibitor that exerts potent cytotoxicity against multiple myeloma (MM) cells, including MM.1S, MM.1R, RPMI-8266, RPMI-LR5, RPMI-Dox40 and Patient-derived MM, with the half maximal inhibition concentration IC50 values of 1.5 μmol/L, 0.7 μmol/L, 1.1 μmol/L, 2.5 μmol/L, 3.2 μmol/L and 1.5 μmol/L respectively.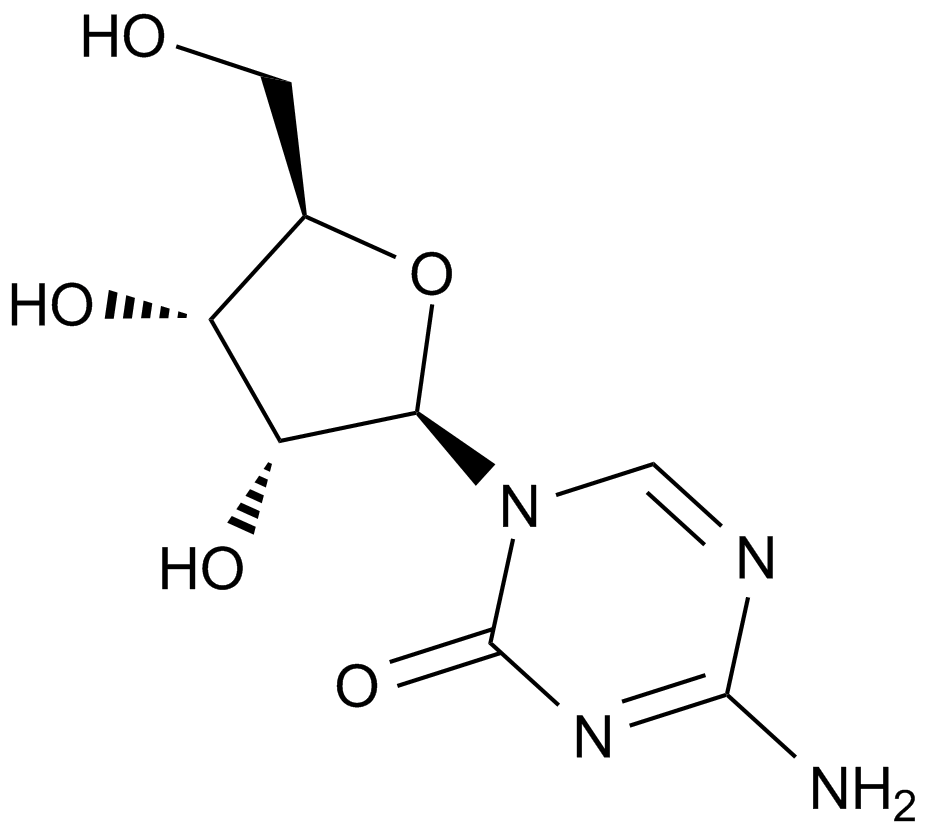
-
GC61662
5-Fluoro-2'-deoxycytidine
5-Fluoro-2'-deoxycytidine, a fluoropyrimidine nucleoside analogue, is a DNA methyltransferase (DNMT) inhibitor. 5-Fluoro-2'-deoxycytidine is a tumor-selective prodrug of the potent thymidylate synthase inhibitor 5-fluoro-2′-dUMP.

-
GC42562
5-Methyl-2'-deoxycytidine
5Methyldeoxycytidine
5-Methyl-2'-deoxycytidine is a pyrimidine nucleoside that when incorporated into single-stranded DNA can act in cis to signal de novo DNA methylation.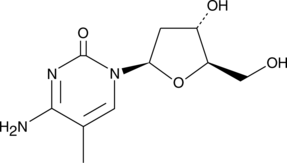
-
GC66055
5-Phenylpentan-2-one
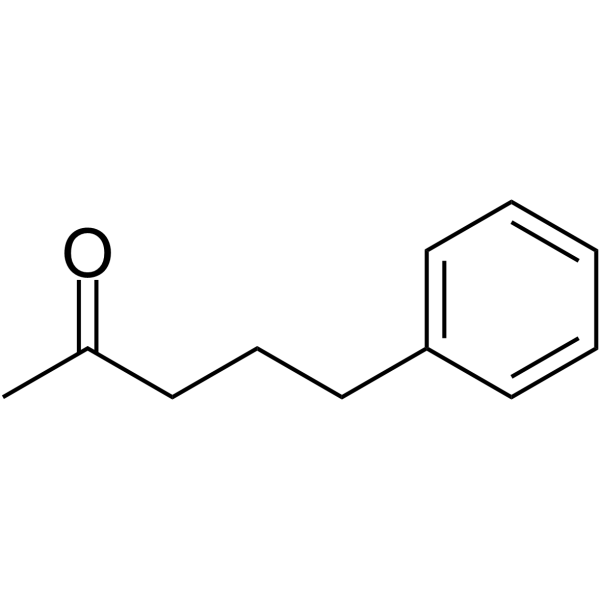
5-Phenylpentan-2-one is a potent histone deacetylases (HDACs) inhibitor. 5-Phenylpentan-2-one can be used for urea cycle disorder research.
-
GC45772
6(5H)-Phenanthridinone
NSC 11021, NSC 40943, NSC 61083
An inhibitor of PARP1 and 2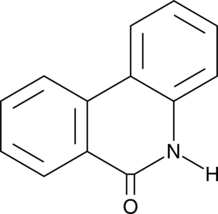
-
GC35175
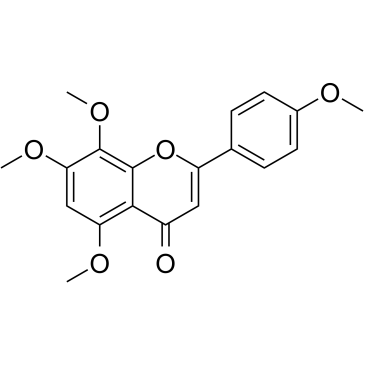
6-Demethoxytangeretin
6-Demethoxytangeretin is a citrus flavonoid isolated from Citrus depressa.
-
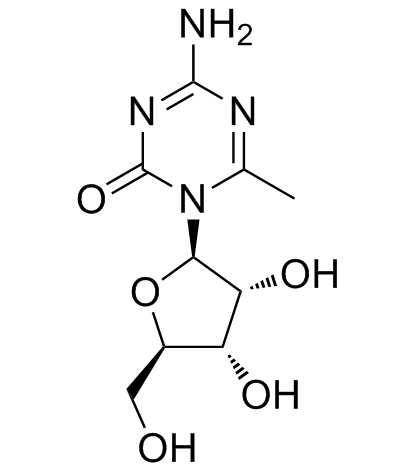
GC35180
6-Methyl-5-azacytidine
6-Methyl-5-azacytidine is a potent DNMT inhibitor.
-
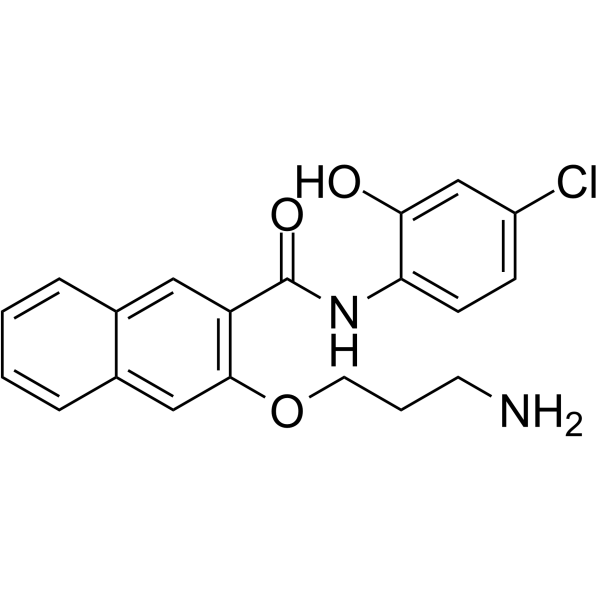
GC62279
653-47
653-47, a potentiator, significantly potentiates the cAMP-response element binding protein (CREB) inhibitory activity of 666-15. 653-47 is also a very weak CREB inhibitor with IC50 of 26.3 μM.
-
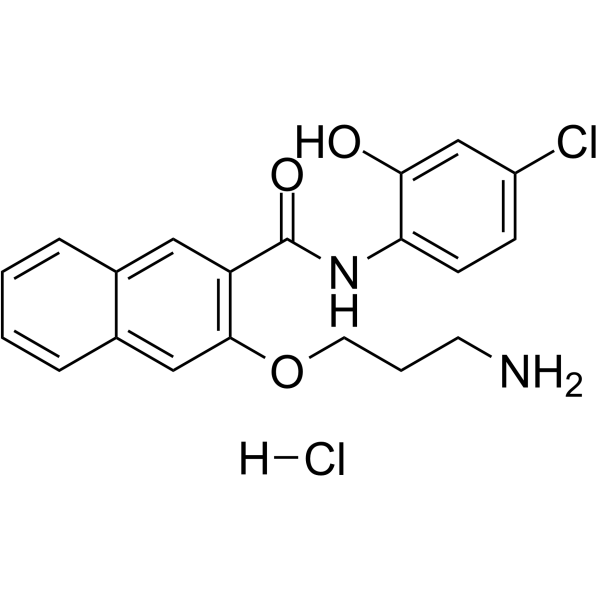
GC62144
653-47 hydrochloride
653-47 hydrochloride, a potentiator, significantly potentiates the cAMP-response element binding protein (CREB) inhibitory activity of 666-15. 653-47 hydrochloride is also a very weak CREB inhibitor with IC50 of 26.3 μM.
-
GC32689
666-15
Compound 3i
An inhibitor of CREB-mediated gene transcription
-
GC46736
7-Bromoheptanoic Acid
A building block
-
GC62651
7-Chloro-4-(piperazin-1-yl)quinoline
7-Chloro-4-(piperazin-1-yl)quinolone is an important scaffold in medicinal chemistry.
-
GC48986
9-hydroxy Stearic Acid
9-HSA, 9-hydroxy Octadecanoic Acid
A hydroxy fatty acid
-
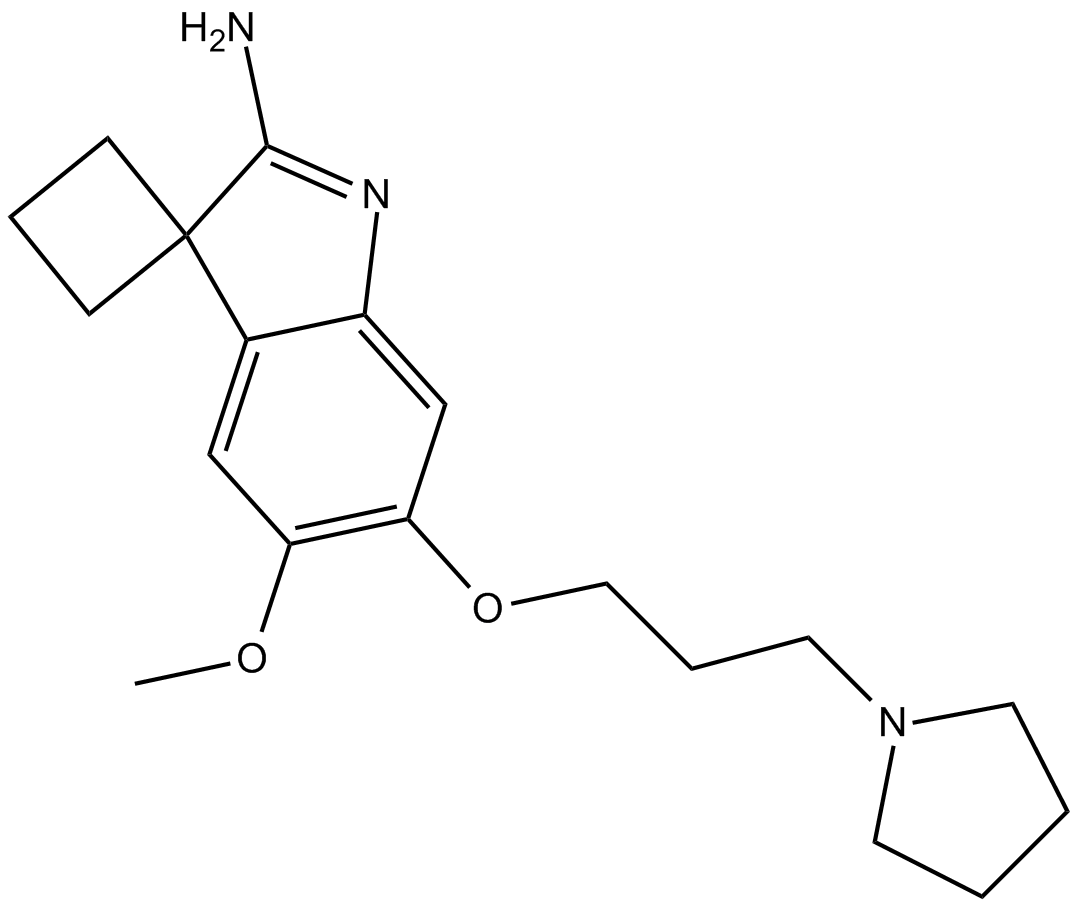
GC16015
A 366
G9a/GLP histone lysine methyltransferase inhibitor
-
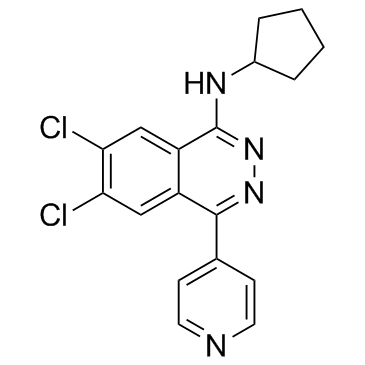
GC32861
A-196
A selective inhibitor of SUV420H1 and SUV420H2
-
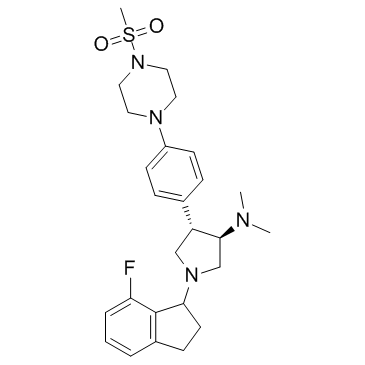
GC33187
A-395 (A395)
A-395 (A395) is an antagonist of polycomb repressive complex 2 (PRC2) protein-protein interactions that potently inhibits the trimeric PRC2 complex (EZH2-EED-SUZ12) with an IC50 of 18 nM.
-
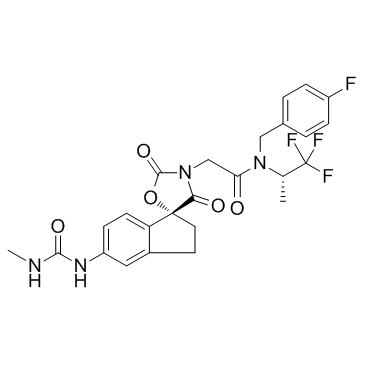
GC32677
A-485
A p300/CBP inhibitor
-
GC30503
A-893
A-893 is a cell-active inhibitor of Methyltransferase SMYD2, with an IC50 of 2.8 nM.
-
GC12390
A-966492
A PARP1 and PARP2 inhibitor
-
GC33280
A1874
A1874 is a nutlin-based (MDM2 ligand) and BRD4-degrading PROTAC with a DC50 of 32 nM (induce BRD4 degradation in cells). Effective in inhibiting many cancer cell lines proliferation.
-
GC35216
AAPK-25
AAPK-25 is a potent and selective Aurora/PLK dual inhibitor with anti-tumor activity, which can cause mitotic delay and arrest cells in a prometaphase, reflecting by the biomarker histone H3Ser10 phosphorylation and followed by a surge in apoptosis. AAPK-25 targets Aurora-A, -B, and -C with Kd values ranging from 23-289 nM, as well as PLK-1, -2, and -3 with Kd values ranging from 55-456 nM.
-
GC19409
ABBV-744
ABBV-744 is a BDII-selective BET bromodomain inhibitor
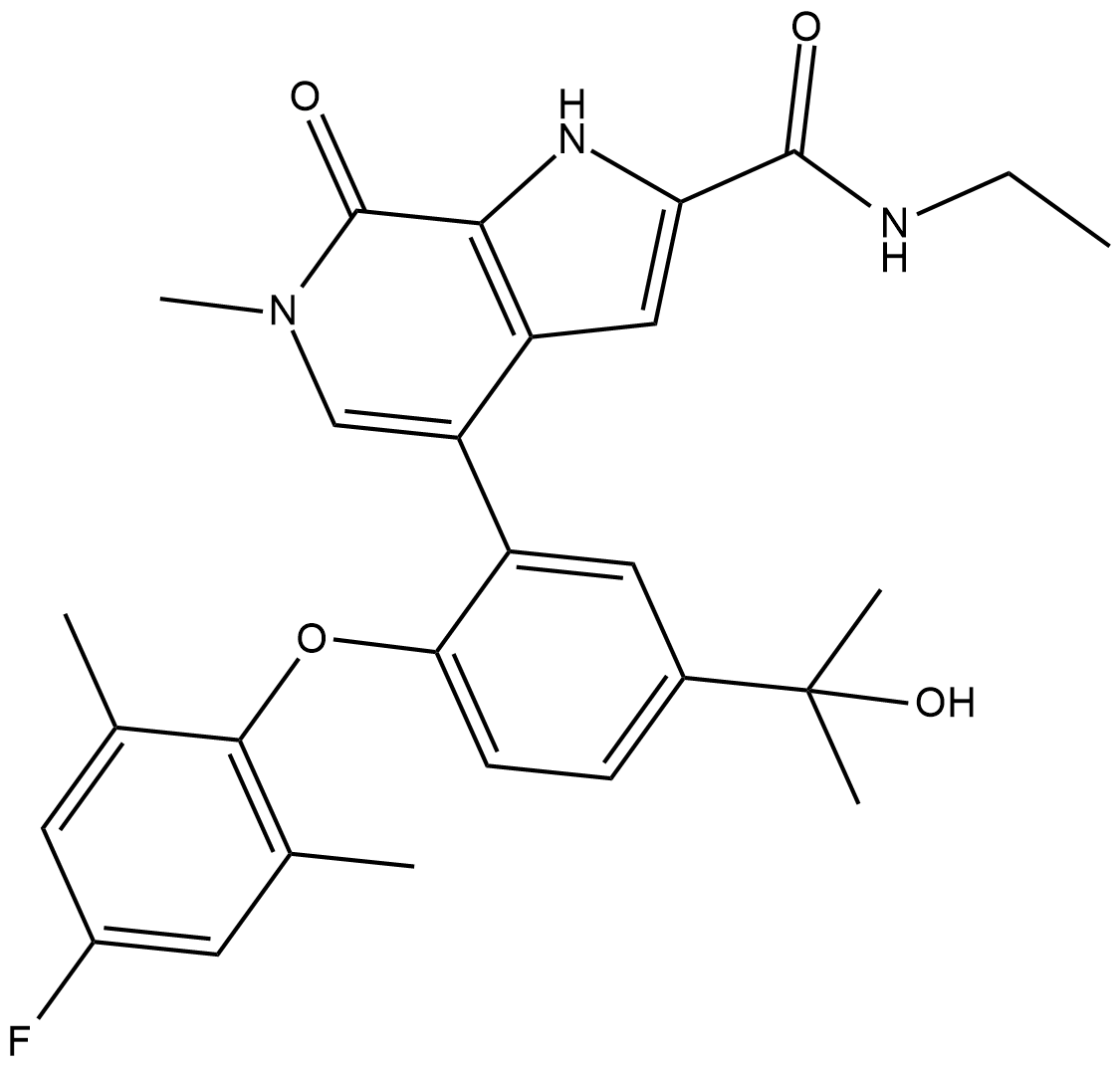
-
GC10154
ABC294640
ABC294640 (ABC294640) is a selective, competitive sphingosine kinase 2 (SK2) inhibitor with Ki of 9.8 μM.
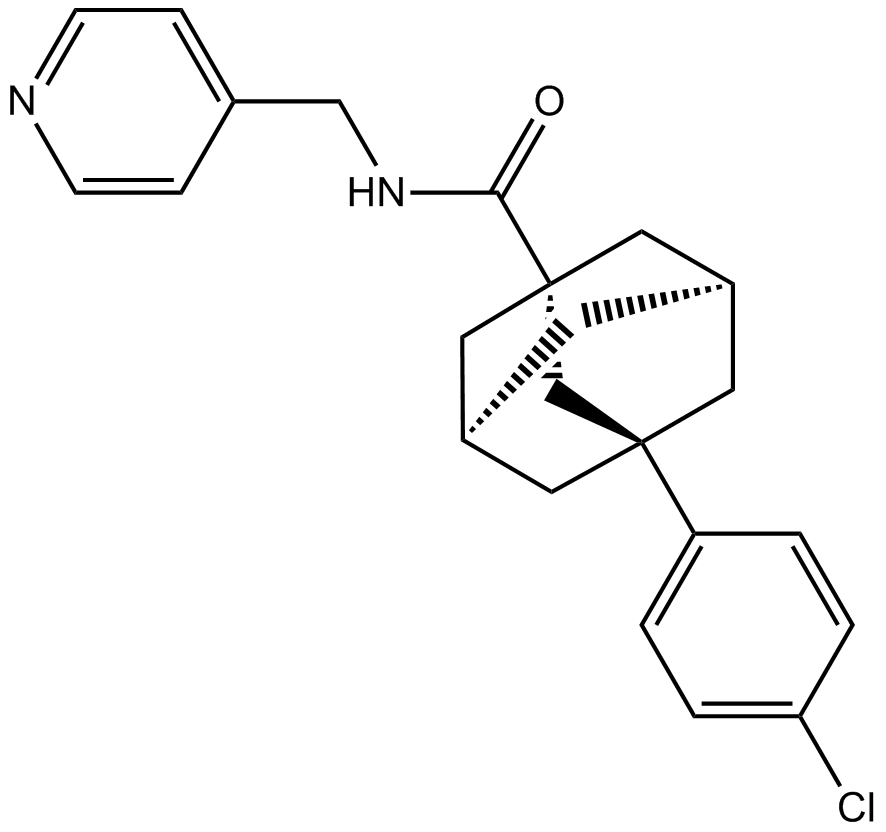
-
GC32023
Abrocitinib (PF-04965842)
PF-04965842
Abrocitinib (PF-04965842) (PF-04965842) is a potent, orally active and selective JAK1 inhibitor, with IC50s of 29 and 803 nM for JAK1 and JAK2, respectively.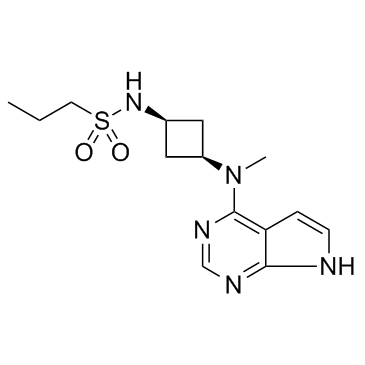
-
GC12422
ABT-888 (Veliparib)
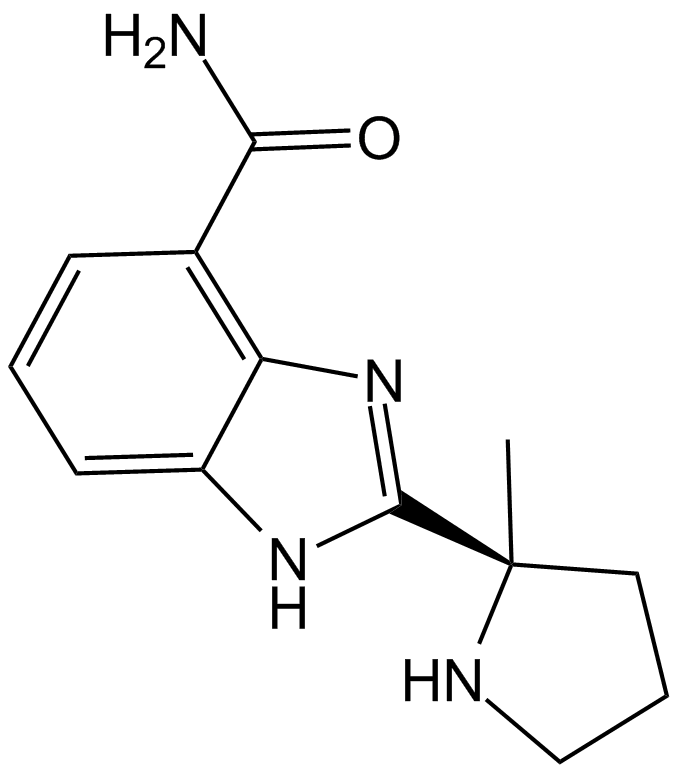
-
GA20481
Ac-Arg-Gly-Lys(Ac)-AMC
Ac-RGK(Ac)-AMC, fluorogenic substrate for assaying histone deacetylase (HDAC) activity in a two-step enzymatic reaction. The assay consists of the initial lysine deacetylation by HDAC followed by the release of the fluorescent group by trypsin.

-
GA20605
Ac-Lys-AMC
Ac-Lys-AMC is cleaved by trypsin.

-
GC48382
Ac-QPKK(Ac)-AMC
Ac-Gln-Pro-Lys-Lys-(Ac)-AMC, Ac-Gln-Pro-Lys-Lys-(Ac)-7-amino-4-methylcoumarin, p53317-320 Substrate (Ac-QPKK(Ac)-AMC)
A fluorogenic substrate for SIRT1, SIRT2, and SIRT3
-
GC35227
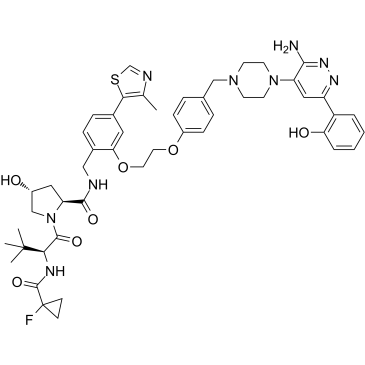
ACBI1
ACBI1 is a potent and cooperative SMARCA2, SMARCA4 and PBRM1 degrader with DC50s of 6, 11 and 32 nM, respectively. ACBI1 is a PROTAC degrader. ACBI1 shows anti-proliferative activity. ACBI1 induces apoptosis.
-
GC12917
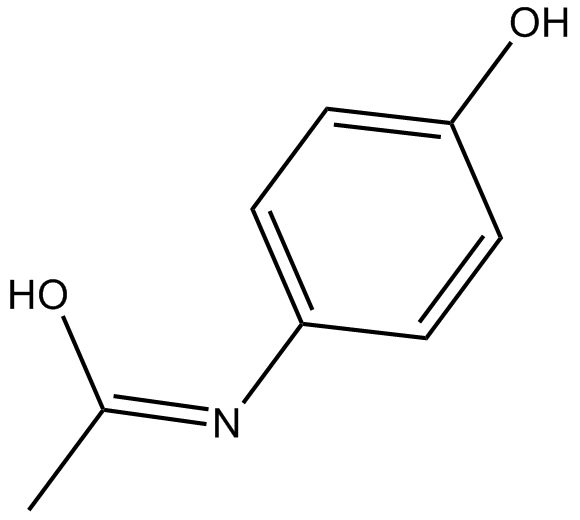
Acetaminophen
4-Acetamidophenol, APAP, 4'-Hydroxyacetanilide, NSC 3991, NSC 109028, Paracetamol
A COX inhibitor
-
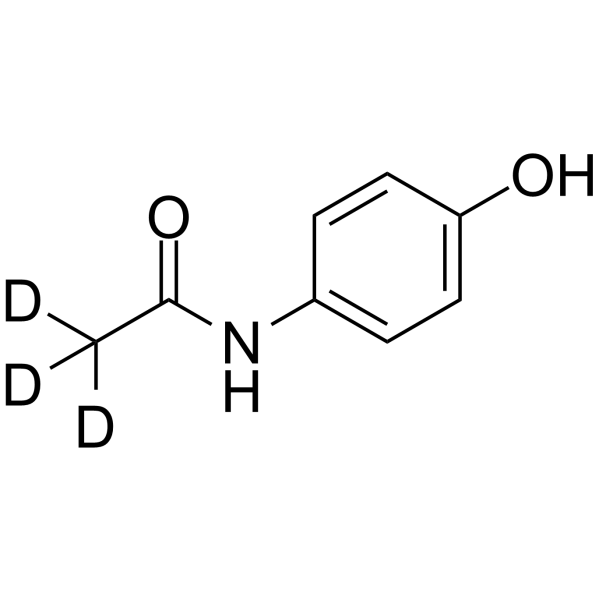
GC64137
Acetaminophen-d3
-
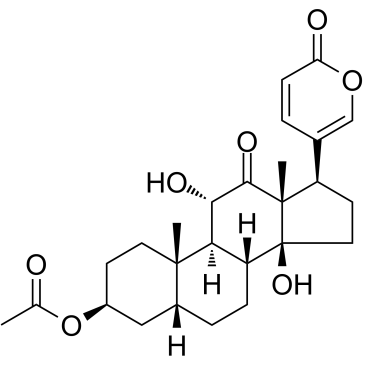
GC35230
Acetylarenobufagin
Acetylarenobufagin is a steroidal hypoxia inducible factor-1 (HIF-I) modulator.
-
GC32083
Acriflavine
Acriflavine is a fluorescent dye for labeling high molecular weight RNA.

-
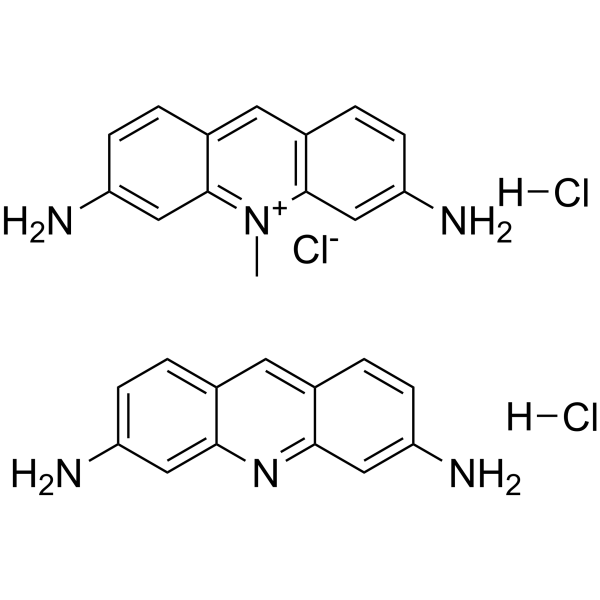
GC62354
Acriflavine hydrochloride
Acriflavine hydrochloride (Acriflavinium chloride hydrochloride) is a fluorescent acridine dye that can be used to label nucleic acid.
-
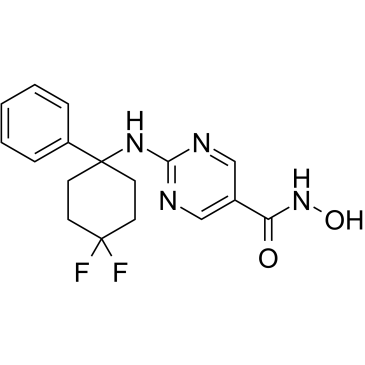
GC34458
ACY-1083
ACY-1083 is a selective and brain-penetrating HDAC6 inhibitor with an IC50 of 3 nM and is 260-fold more selective for HDAC6 than all other classes of HDAC isoforms. ACY-1083 effectively reverses chemotherapy-induced peripheral neuropathy.
-
GC10417
ACY-241
Citarinostat
ACY-241 (ACY241) is a second generation potent, orally active and high-selective HDAC6 inhibitor with an IC50 of 2.6 nM (IC50s of 35 nM, 45 nM, 46 nM and 137 nM for HDAC1, HDAC2, HDAC3 and HDAC8, respectively). ACY-241 has anticancer effects.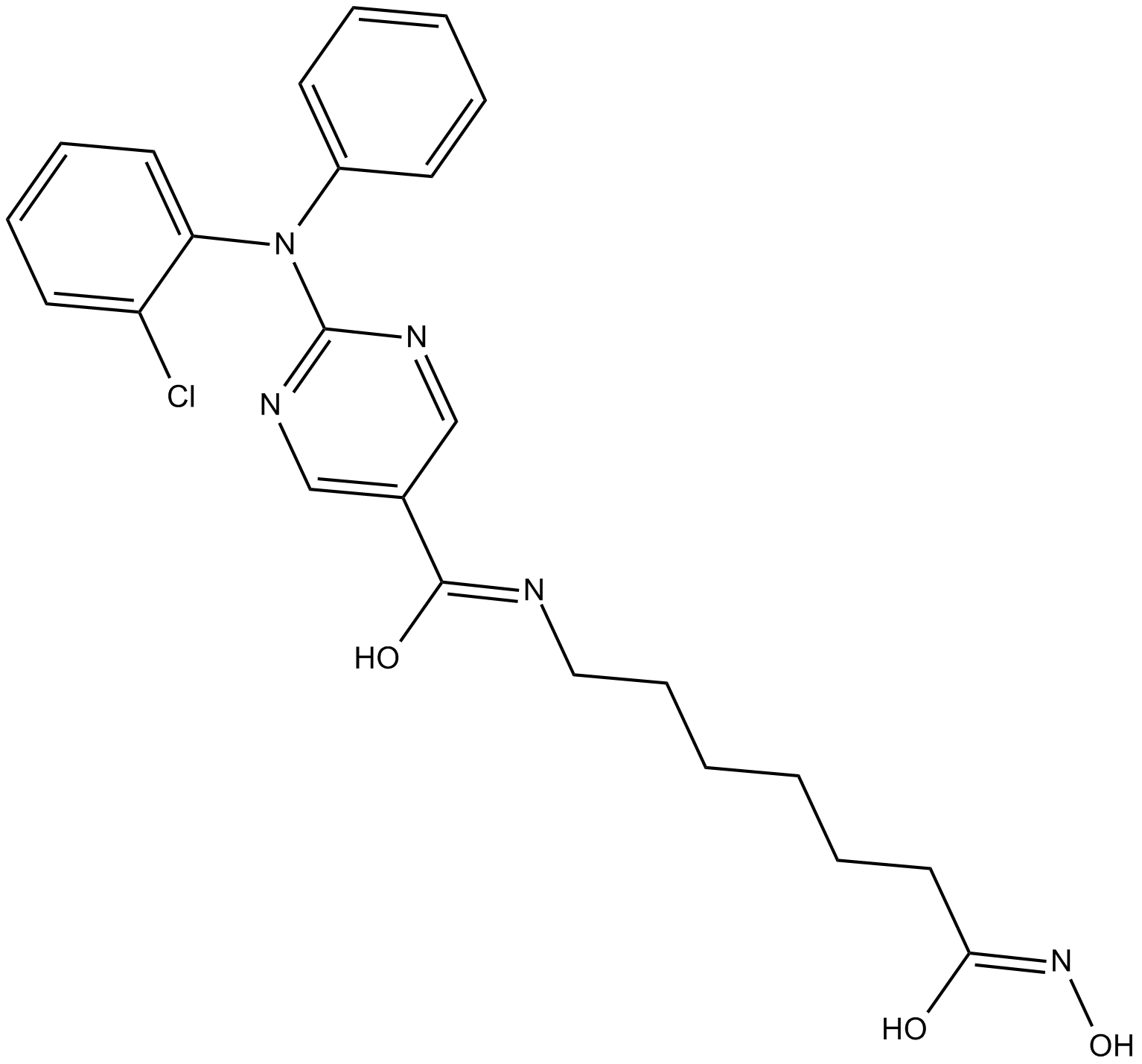
-
GC19020
ACY-738
ACY-738 is a potent, selective and orally-bioavailable HDAC6 inhibitor, with an IC50s of 1.7 nM; ACY-738 also inhibits HDAC1, HDAC2, and HDAC3, with IC50s of 94, 128, and 218 nM.

-
GC30782
ACY-775
An HDAC6 inhibitor