Ubiquitination/ Proteasome
- Autophagy(1089)
- DUB(20)
- E1 Activating(1)
- E2 conjugating(1)
- E2 Ligases(0)
- E3 Ligase(8)
- Proteasome(99)
- p97(12)
- Mitophagy(82)
- ULK(9)
Products for Ubiquitination/ Proteasome
- Cat.No. Product Name Information
-
GC30761
α-Hydroxylinoleic acid
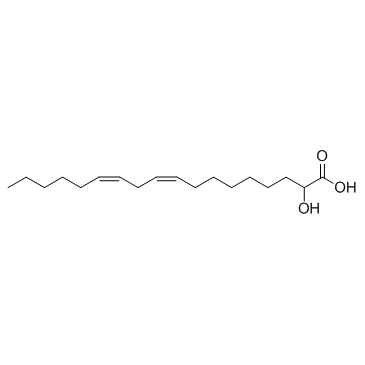
α-Hydroxylinoleic acid (α-Hydroxylinoleic acid) induces endoplasmic reticulum (ER) stress-mediated autophagy.
-
GC46008
(±)-Thalidomide-d4
N-Phthaloylglutamimide-d4
An internal standard for the quantification of (±)-thalidomide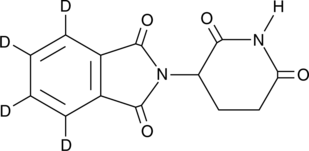
-
GC34960
(+)-Talarozole
(+)-Talarozole is a potent inhibitor of retinoic acid metabolism extracted from patent WO 1997049704 A1.
-
GC10603
(-)-epicatechin gallate
ECG
major catechin in green tea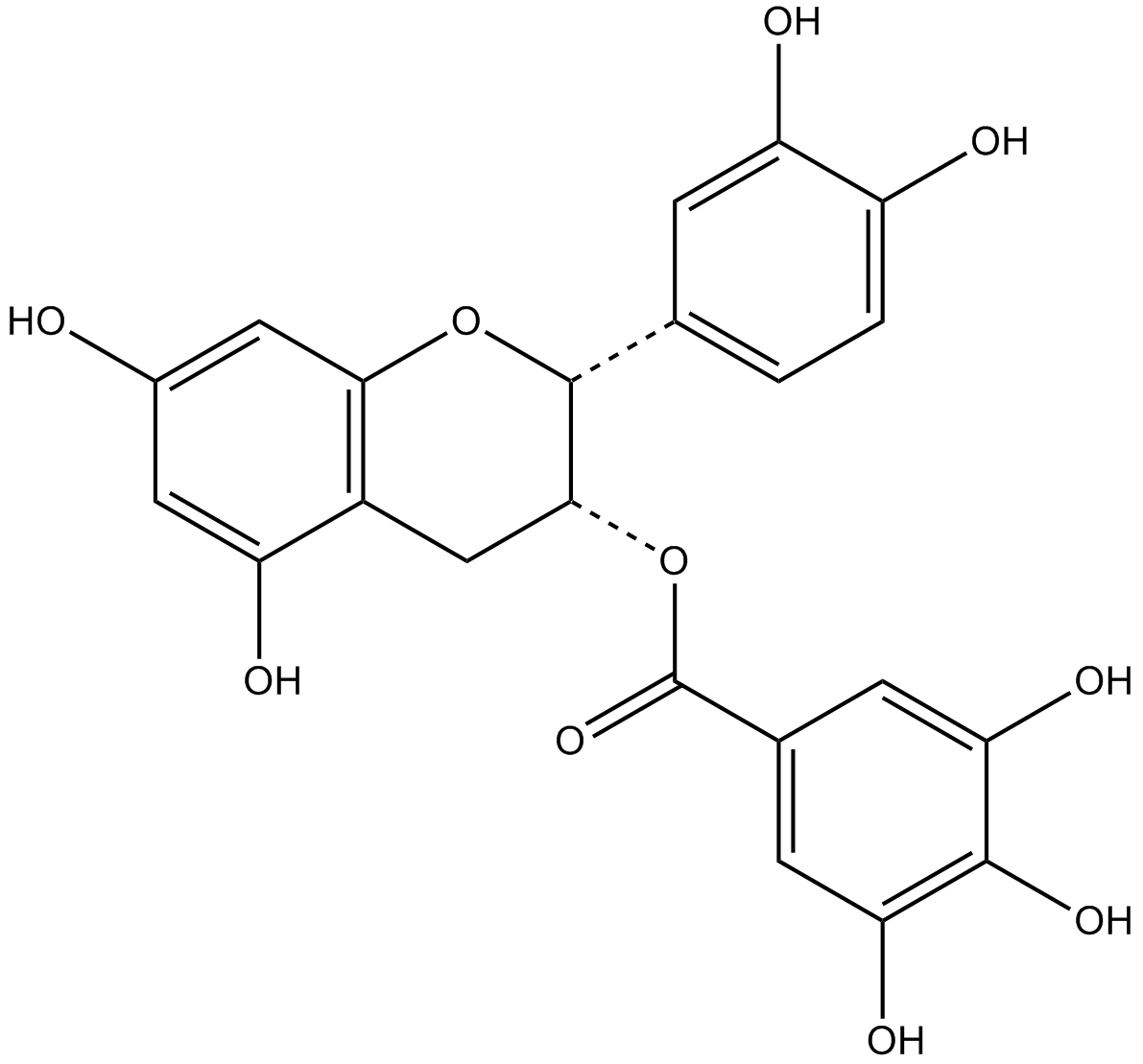
-
GC17242
(-)-epigallocatechin
(-)EGC, epi-Gallocatechin, NSC 674039
green tea epicatechin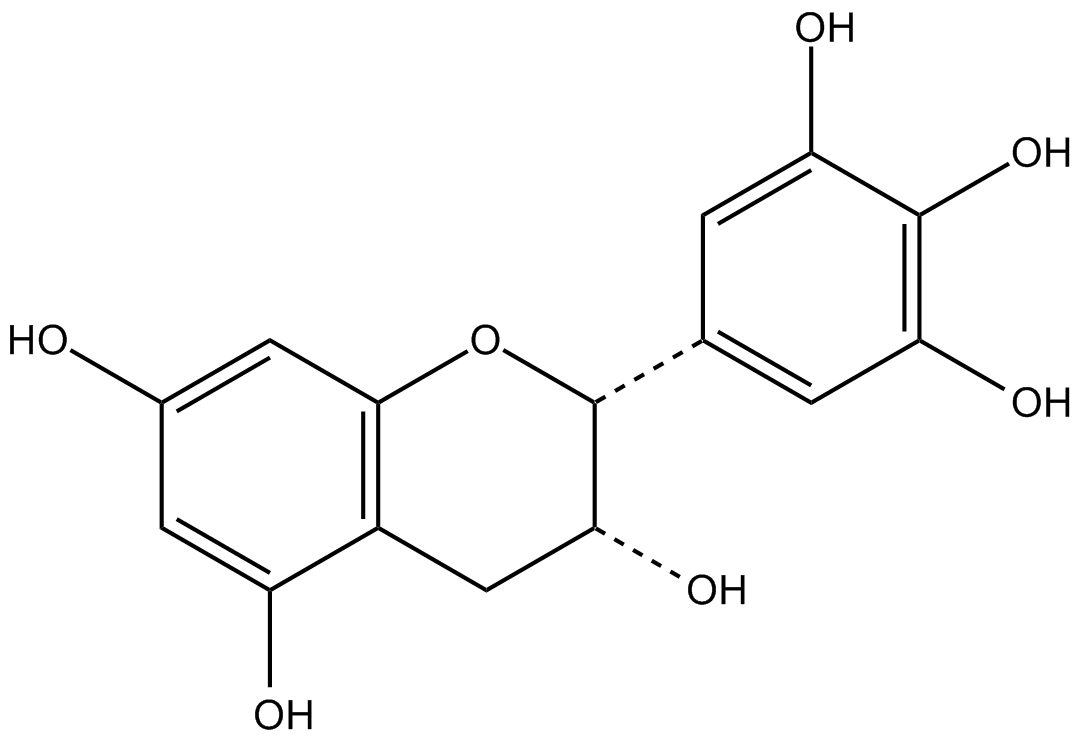
-
GC14049
(-)-Epigallocatechin gallate (EGCG)
EGCG
(-)-Epigallocatechin Gallate sulfate (EGCG) is a major polyphenol in green tea that inhibits cell proliferation and induces apoptosis.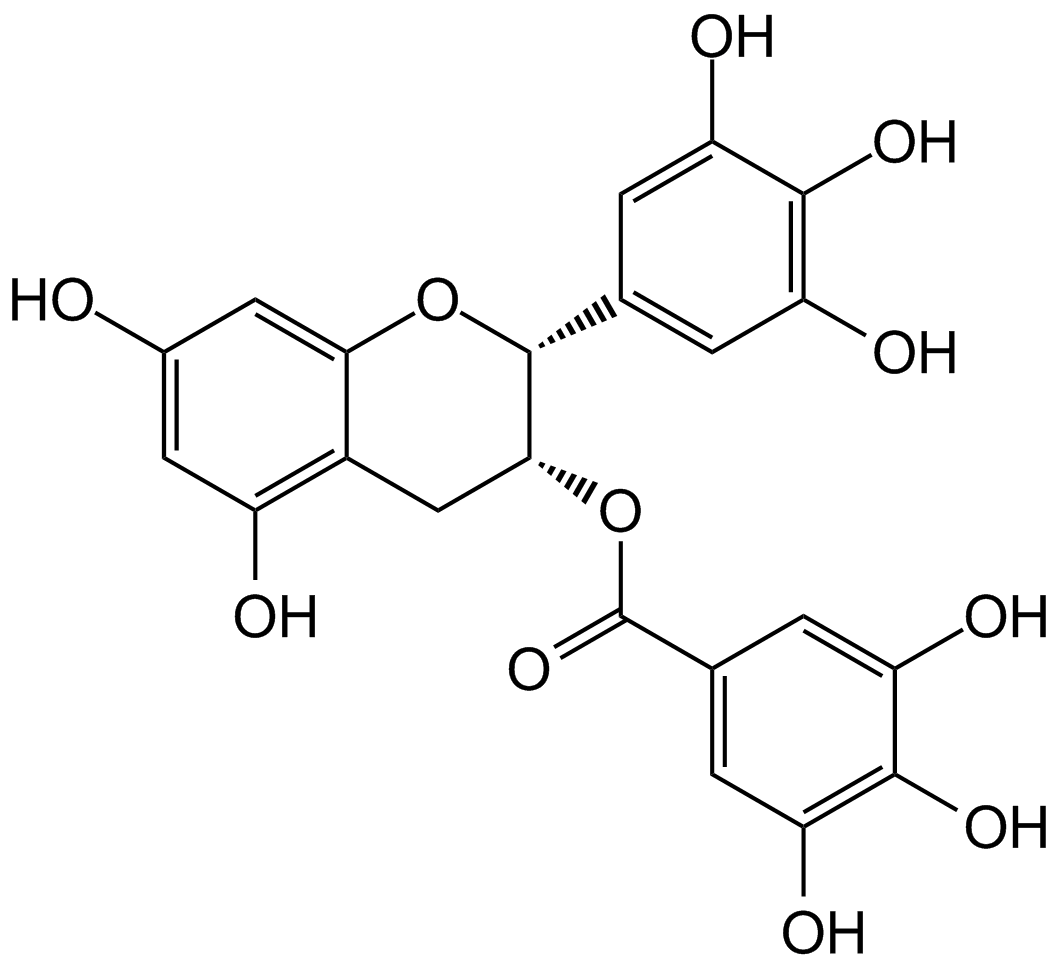
-
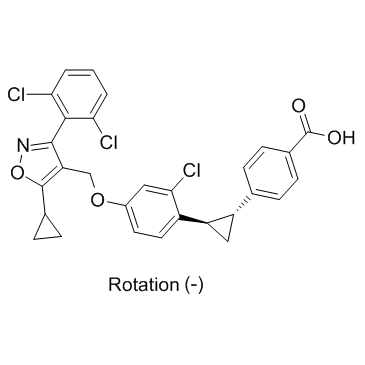
GC31386
(-)-PX20606 trans isomer ((-)-PX-102 trans isomer)
(-)-PX20606 trans isomer ((-)-PX-102 trans isomer) is a FXR agonist with EC50s of 18 and 29 nM for FXR in FRET and M1H assay, respectively.
-
GC65547
(1S,2R)-Alicapistat
(1S,2R)-ABT-957
(1S,2R)-Alicapistat ((1S,2R)-ABT-957) is an orally active selective inhibitor of human calpains 1 and 2 for the potential application of Alzheimer's disease (AD).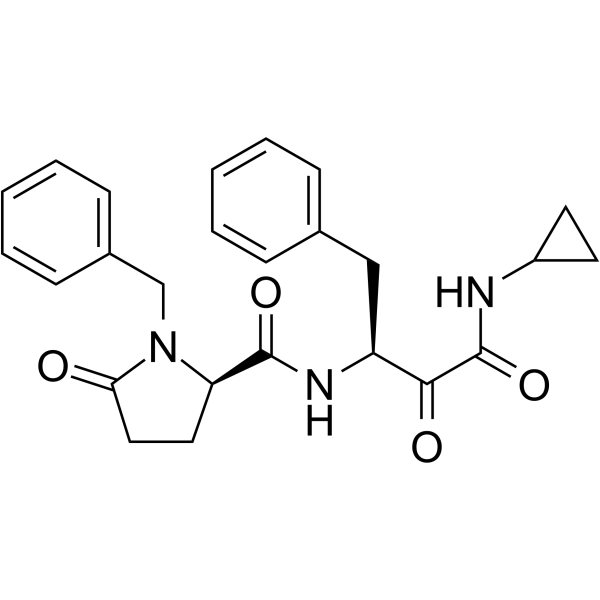
-
GC62193
(1S,2S)-Bortezomib
(1S,2S)-Bortezomib is an enantiomer of Bortezomib. Bortezomib is a cell-permeable, reversible, and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki of 0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is an anti-cancer agent and the first therapeutic proteasome inhibitor to be used in humans.
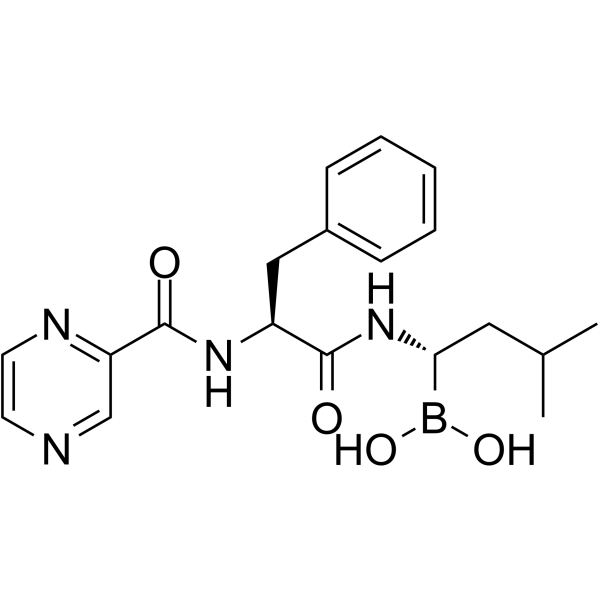
-
GC60395
(3S,5R)-Fluvastatin D6
(3S,5R)-XU 62-320 free acid D6
(3S,5R)-Fluvastatin D6 is the deuterium labeled (3S,5R)-Fluvastatin sodium.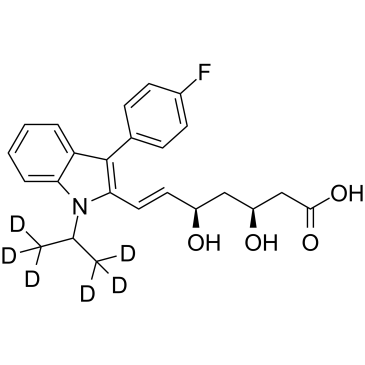
-
GC49189
(E/Z)-4-hydroxy Tamoxifen-d5
Afimoxifene-d5, 4-OHT-d5
An internal standard for the quantification of (E/Z)-4-hydroxy tamoxifen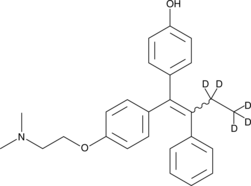
-
GC34096
(R)-(-)-Gossypol acetic acid (AT-101 (acetic acid))
(R)-(-)-Gossypol acetic acid (AT-101 (acetic acid)) (AT-101 (acetic acid)) is the levorotatory isomer of a natural product Gossypol. AT-101 is determined to bind to Bcl-2, Mcl-1 and Bcl-xL proteins with Kis of 260±30 nM, 170±10 nM, and 480±40 nM, respectively.
-
GC31665
(R)-BPO-27
(R)-BPO-27, the R enantiomer of BPO-27, is a potent, orally active and ATP-competitive CFTR?inhibitor with an?IC50 of 4 nM.
-
GC41233
(R)-MG132
(R)-MG132 is a proteasome inhibitor with an IC50 of 100 nM.
-
GC62528
(Rac)-Hesperetin
(Rac)-Hesperetin is the racemate of Hesperetin.
-
GC13136
(S)-Crizotinib
Potent MTH1 inhibitor
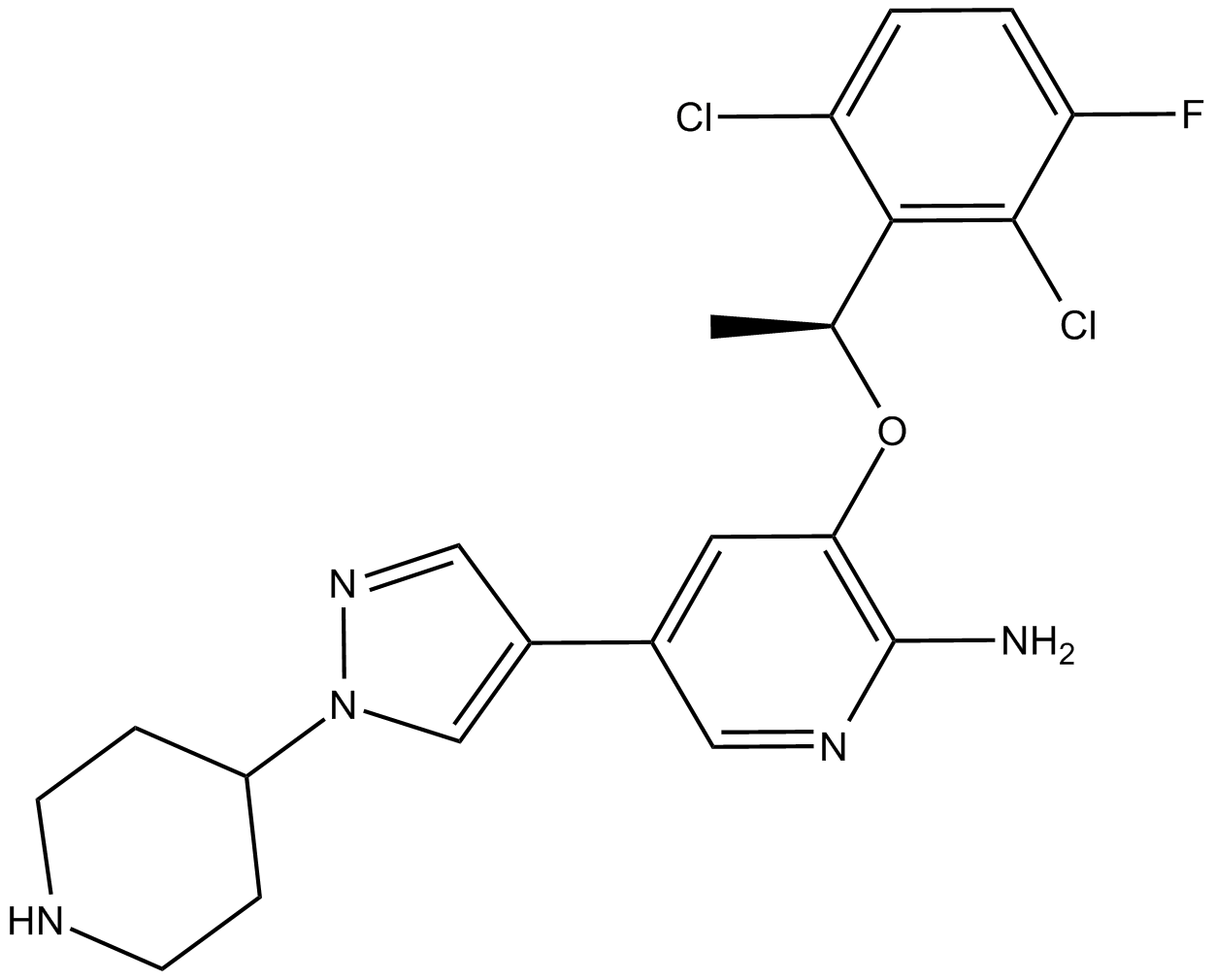
-
GC14820
(S)-Naproxen
CG 3117, (S)-Naproxen
(S)-Naproxen is a COX-1 and COX-2 inhibitor with IC50s of 8.72 and 5.15 μM, respectively in cell assay.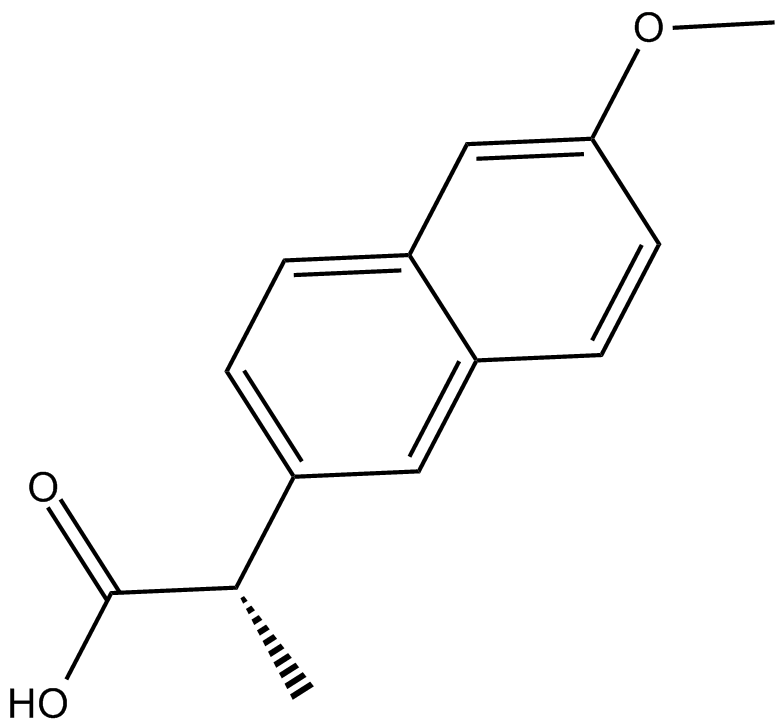
-
GC15015
(±)-Bay K 8644
SQ 28,873
L-type Ca2+-channel activator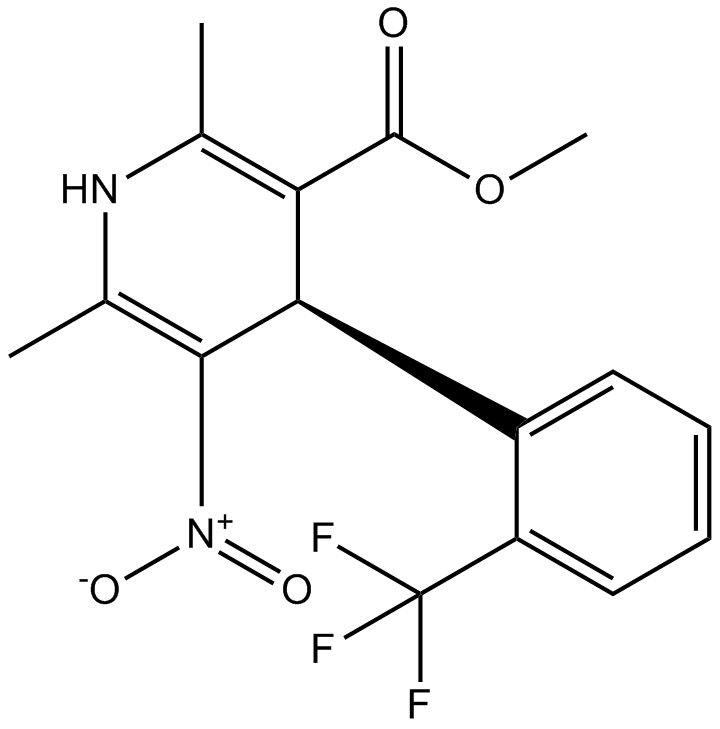
-
GC35068
1-Monomyristin
MG(14:0/0:0/0:0), 1-Monomyristin
A monoacylglycerol
-
GC17295
10058-F4
C-Myc-Max dimerization inhibitor

-
GC14918
10074-G5
c-Myc inhibitor
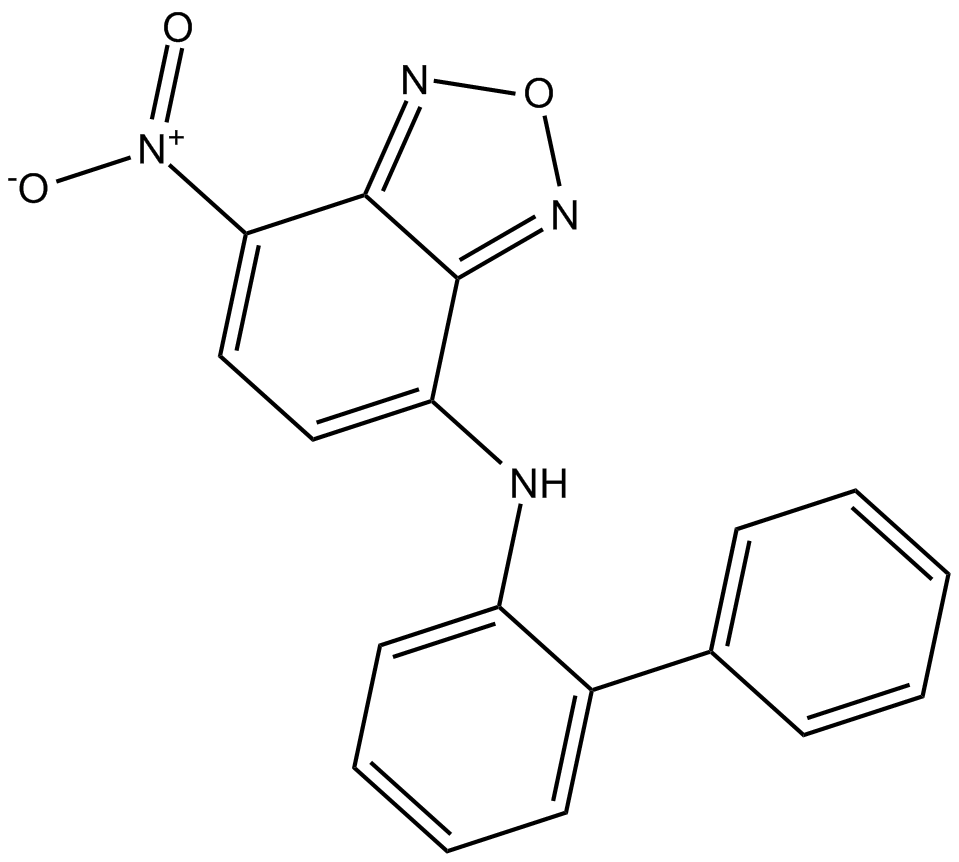
-
GC11720
17-AAG (KOS953)
BMS 722782, CP 127374, KOS 953, NSC 330507, Tanespimycin
17-AAG(Geldanamycin), a natural benzoquinone ansamycin antibiotic, is the first established inhibitor of Hsp90.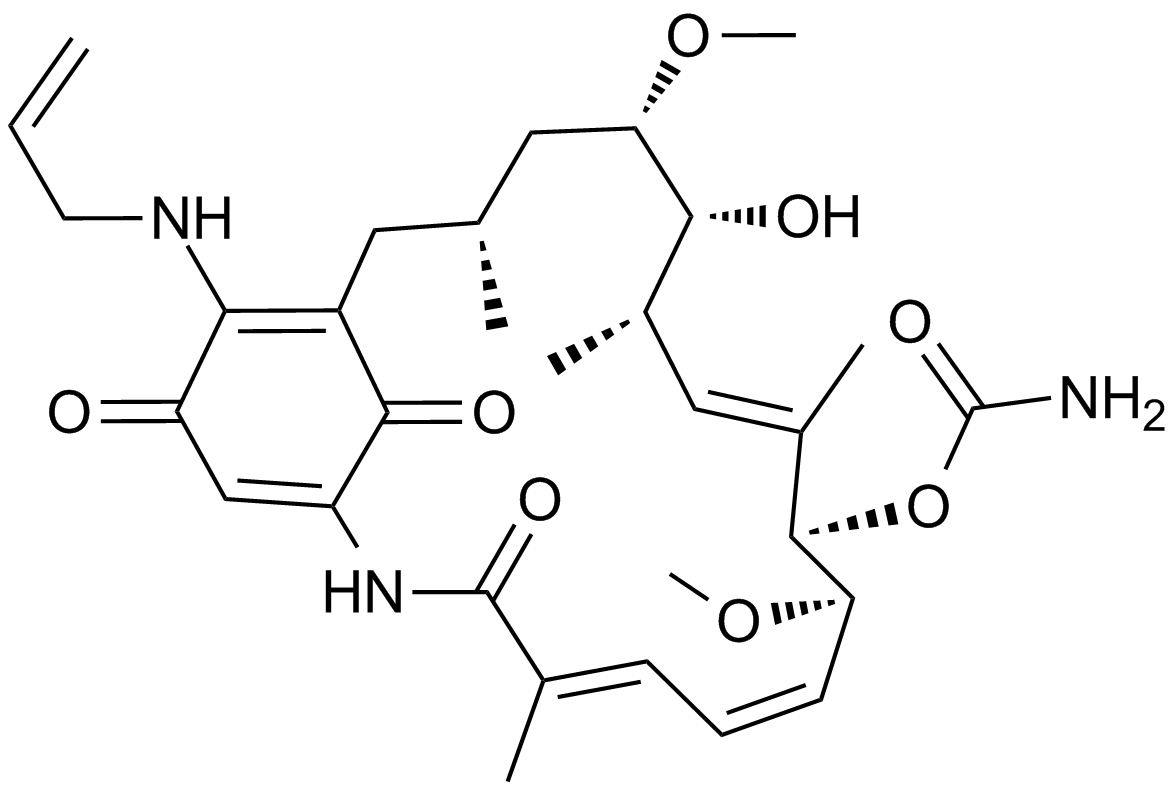
-
GC40947
2,3-Dimethoxy-5-methyl-p-benzoquinone
Coenzyme Q0, CoQ0
2,3-Dimethoxy-5-methyl-p-benzoquinone (CoQ0) is a potent, oral active ubiquinone compound can be derived from Antrodia cinnamomea.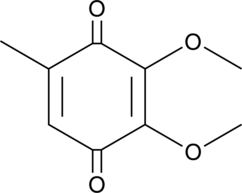
-
GC46057
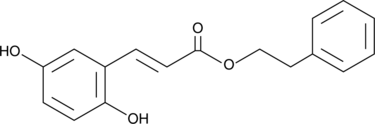
2,5-Dihydroxycinnamic Acid phenethyl ester
An inhibitor of 5-LO
-
GC15084
2-Methoxyestradiol (2-MeOE2)
2Hydroxyestradiol 2methyl ether, 2ME2, NSC 659853, Panzem
2-Methoxyestradiol (2-MeOE2/2-Me) is an HIF-1α inhibitor.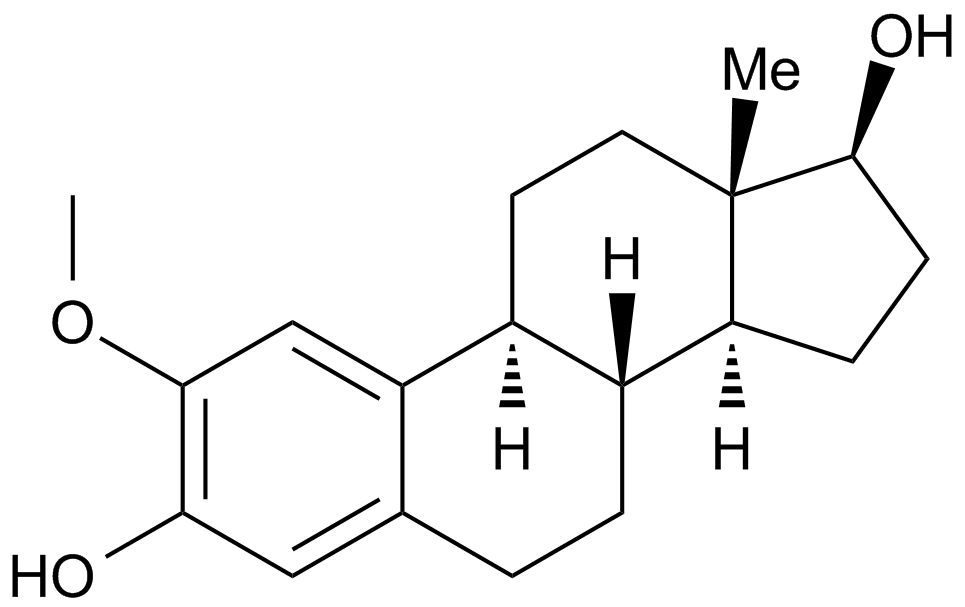
-
GC30011
20-Deoxyingenol
A diterpenoid with antioxidant and osteoprotective activities
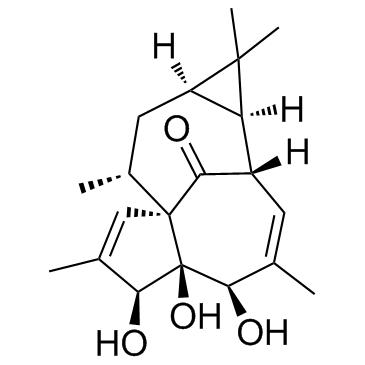
-
GC68503
20S Proteasome activator 1
20S Proteasome activator 1 is an effective activator of the 20S protease, with IC50 values of 0.3 μM, 0.7 μM and 1.8 μM for trypsin-like sites, chymotrypsin-like sites and caspase-like sites respectively. It can be translated in cellular systems to prevent accumulation of pathogenic A53T mutant alpha-synuclein. 20S Proteasome activator 1 can be used for research on neurodegenerative diseases.
-
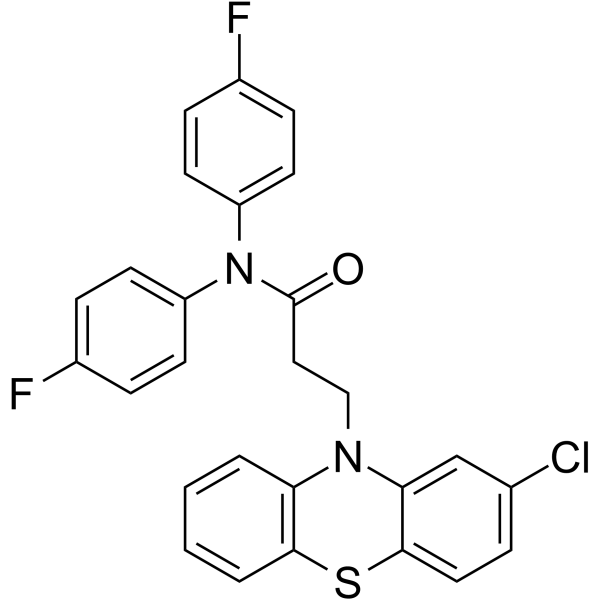
GC64472
20S Proteasome-IN-1
20S Proteasome-IN-1 is a 26S proteasome inhibitor extracted from patent WO2006128196A2 compound 2.
-
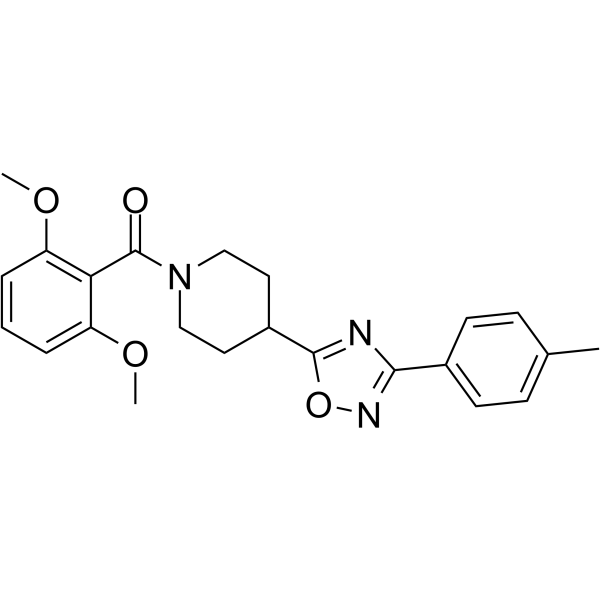
GC35112
3'-Hydroxypterostilbene
3'-Hydroxypterostilbene is a Pterostilbene analogue. 3'-Hydroxypterostilbene inhibits the growth of COLO 205, HCT-116 and HT-29 cells with IC50s of 9.0, 40.2 and 70.9 ?M, respectively. 3'-Hydroxypterostilbene significantly down-regulates PI3K/Akt and MAPKs signaling pathways and effectively inhibits the growth of human colon cancer cells by inducing apoptosis and autophagy. 3'-Hydroxypterostilbene can be used for the research of cancer.
-
GC12791
3,3'-Diindolylmethane
DIM
A phytochemical with antiradiation and chemopreventative effects
-
GC10710
3-Methyladenine
3-MA
3-Methyladenine is a classic autophagy inhibitor.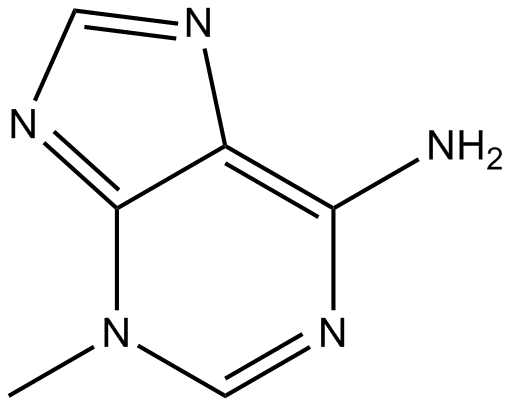
-
GC32767
3BDO
3-Benzyl-5-((2-nitrophenoxy)methyl)dihydrofuran-2(3H)-one
A butyrolactone derivative and autophagy inhibitor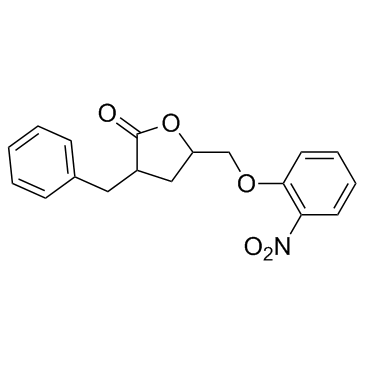
-
GC39658
4,4'-Dimethoxychalcone
4,4'-Dimethoxychalcone acts as a natural autophagy inducer with anti-ageing properties.
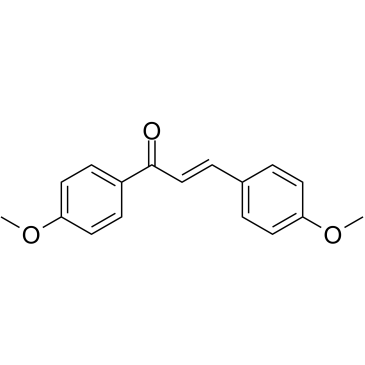
-
GC42414
4-hydroxy Tolbutamide
4-hydroxy Tolbutamide is a cytochrome P450 2C8 (CYP2C8) and CYP2C9 metabolite of tolbutamide, a first-generation potassium channel blocker.
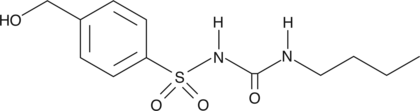
-
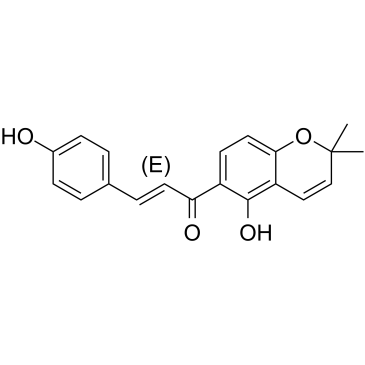
GC35132
4-Hydroxylonchocarpin
4-Hydroxylonchocarpin is a chalcone compound from an extract of Psoralea corylifolia.
-
GC70208
4-Hydroxytolbutamide-d9
4-droxytolbutamide-d9 is the deuterium labeled 4-droxytolbutamide.

-
GC13104
4E1RCat
eIF4E/eIF4G Interaction Inhibitor II
Dual inhibitor of eIF4E:eIF4G and eIF4E:4E-BP1 interaction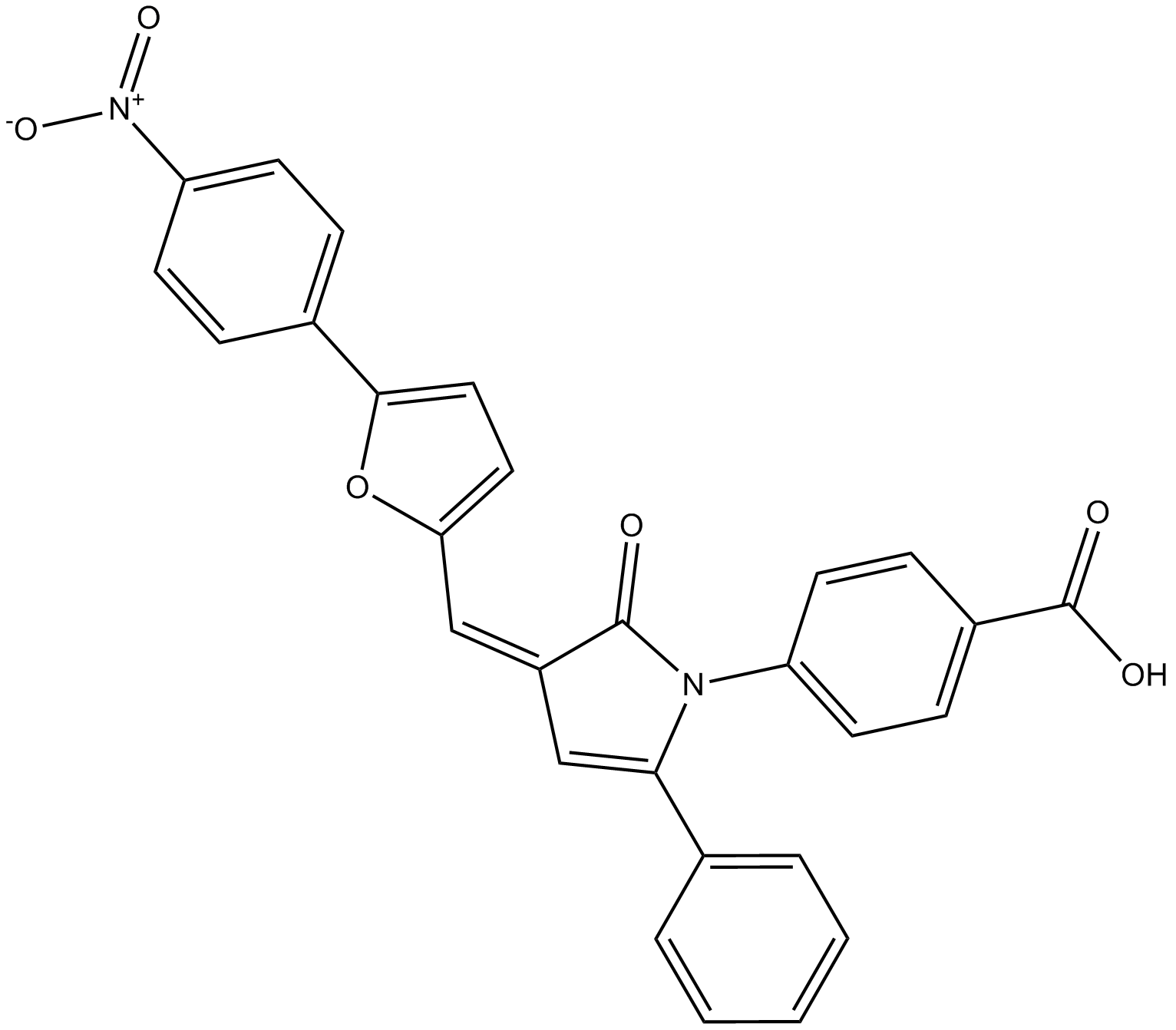
-
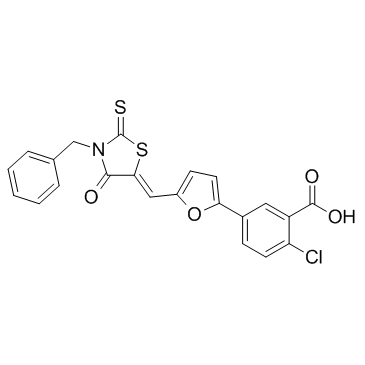
GC30785
4E2RCat
4E2RCat is an inhibitor of eIF4E-eIF4G interaction with an IC50 of 13.5 μM.
-
GC10468
4EGI-1
eIF4E/eIF4G Interaction Inhibitor
Competitive eIF4E/eIF4G interaction inhibitor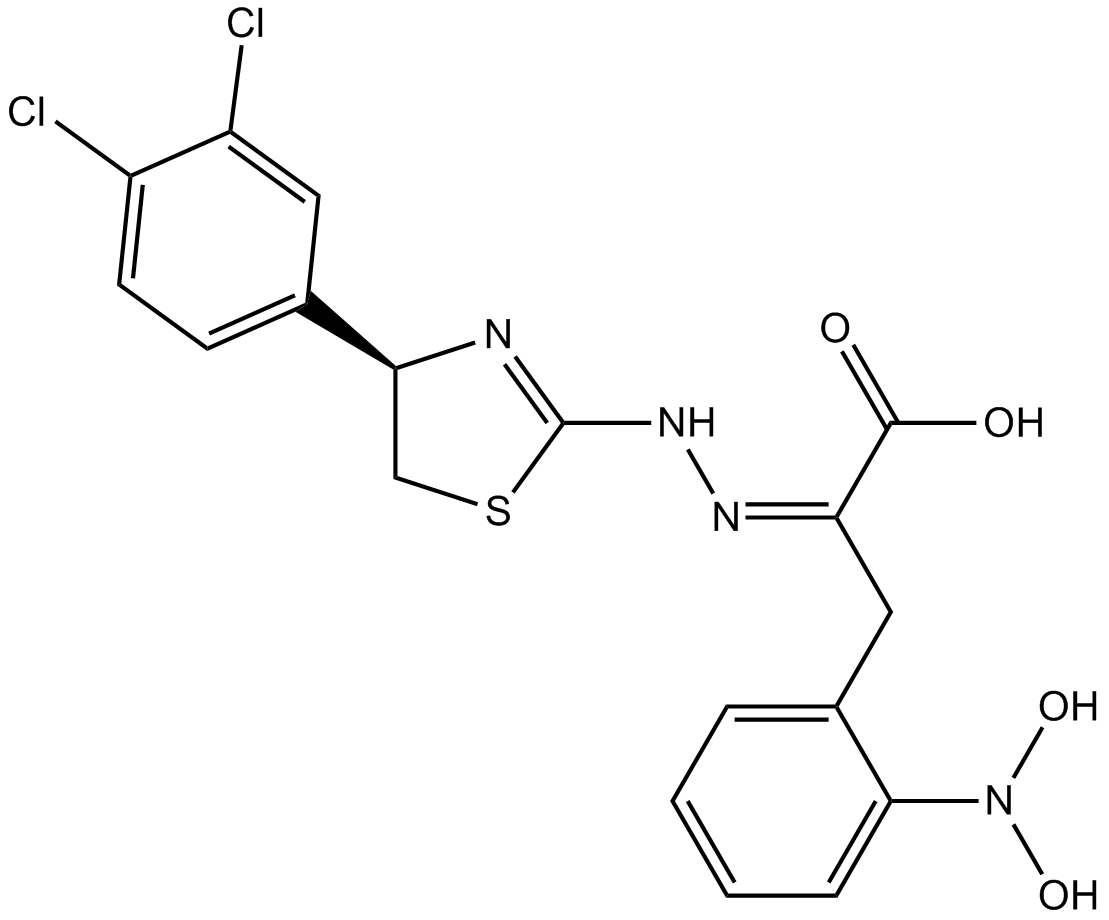
-
GC25019
5-amino-2,4-dimethylpyridine (5A-DMP)
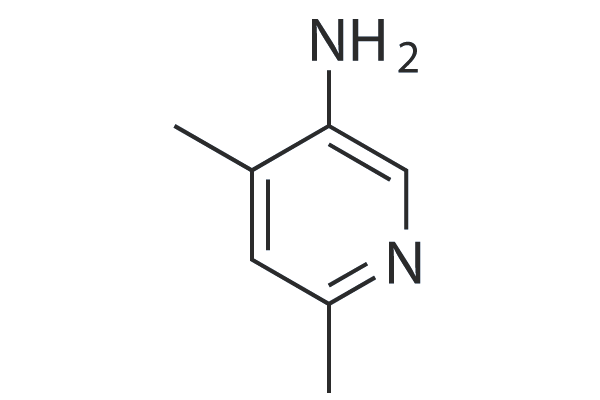
-
GC65668
5-Amino-8-hydroxyquinoline
5A8HQ
5-Amino-8-hydroxyquinoline (5A8HQ), a potential anticancer candidate, has promising proteasome inhibitory activity.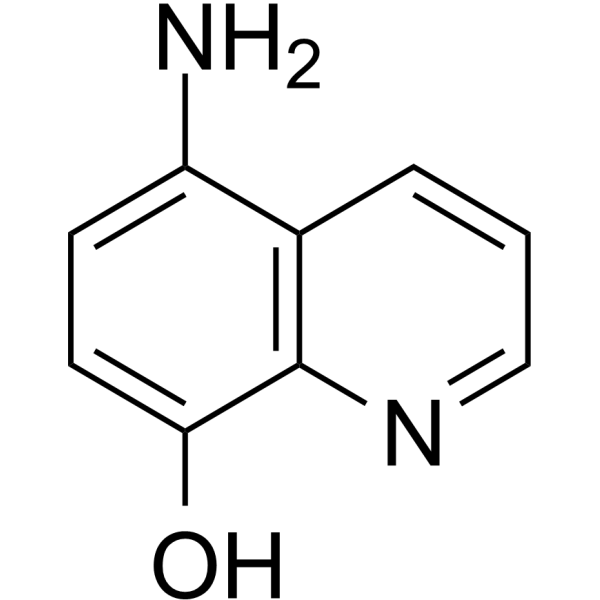
-
GC45356
5-Aminolevulinic Acid (hydrochloride)
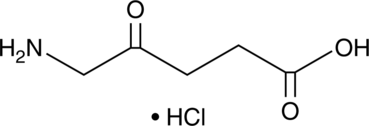
-
GC10946
5-Azacytidine
Antibiotic U 18496, 5AzaC, Ladakamycin, Mylosar, NSC 102816, NSC 103627, U 18496, WR 183027
5-Azacytidine (also known as 5-AzaC), a compound belonging to a class of cytosine analogues, is a DNA methyl transferase (DNMT) inhibitor that exerts potent cytotoxicity against multiple myeloma (MM) cells, including MM.1S, MM.1R, RPMI-8266, RPMI-LR5, RPMI-Dox40 and Patient-derived MM, with the half maximal inhibition concentration IC50 values of 1.5 μmol/L, 0.7 μmol/L, 1.1 μmol/L, 2.5 μmol/L, 3.2 μmol/L and 1.5 μmol/L respectively.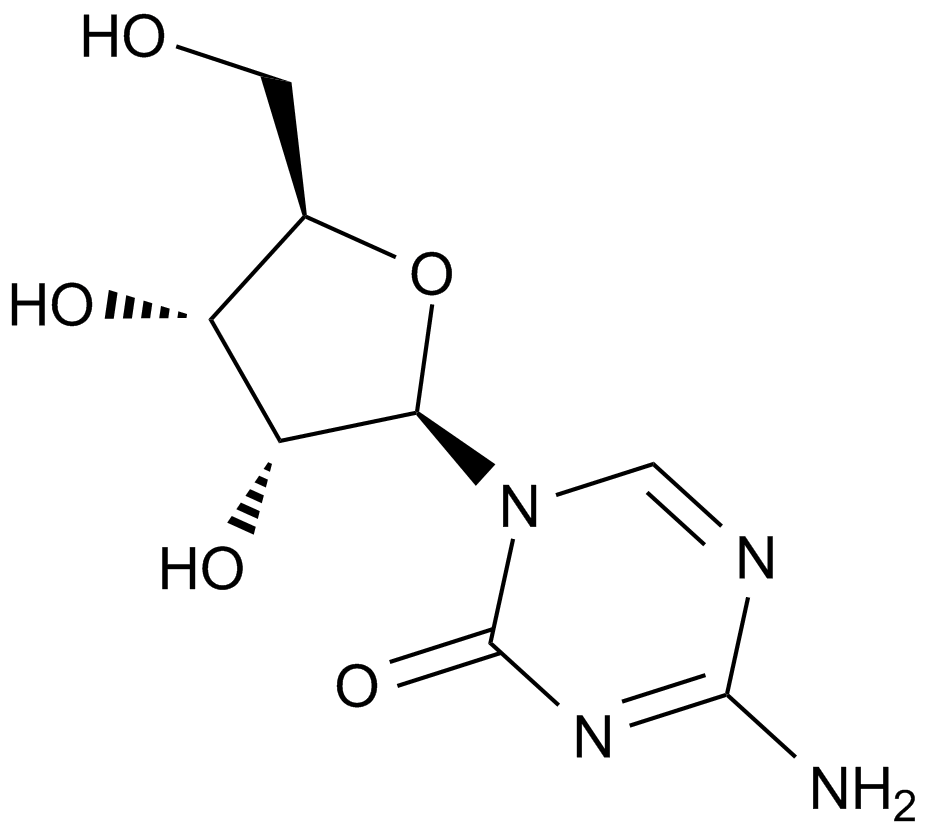
-
GN10241
5-Methoxypsoralen
5-Methoxypsoralen, 5-MOP, Heraclin, NSC 95437
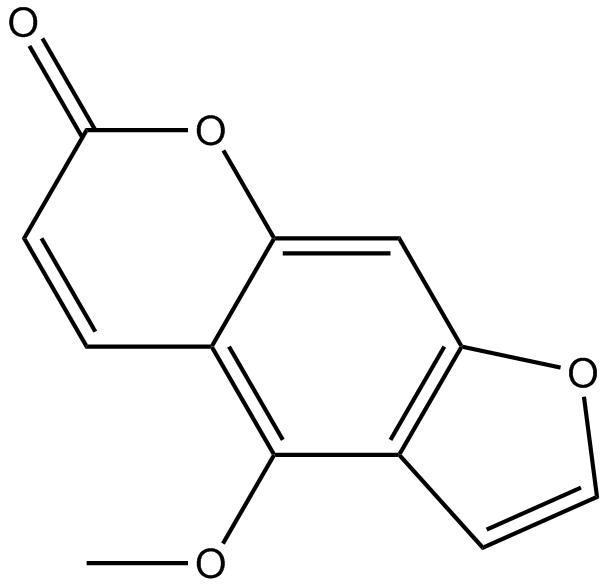
-
GC16267
6-Hydroxydopamine hydrobromide
6-hydroxy Dopamine, Oxidopamine, 2,4,5-Trihydroxyphenethylamine
Oxidopamine (6-OHDA) hydrobromide is an antagonist of the neurotransmitter dopamine.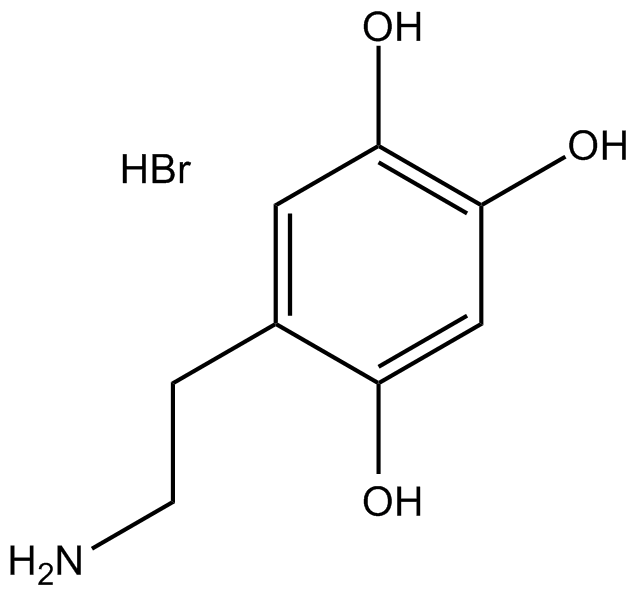
-
GN10228
6-Shogaol
A bioactive component of ginger
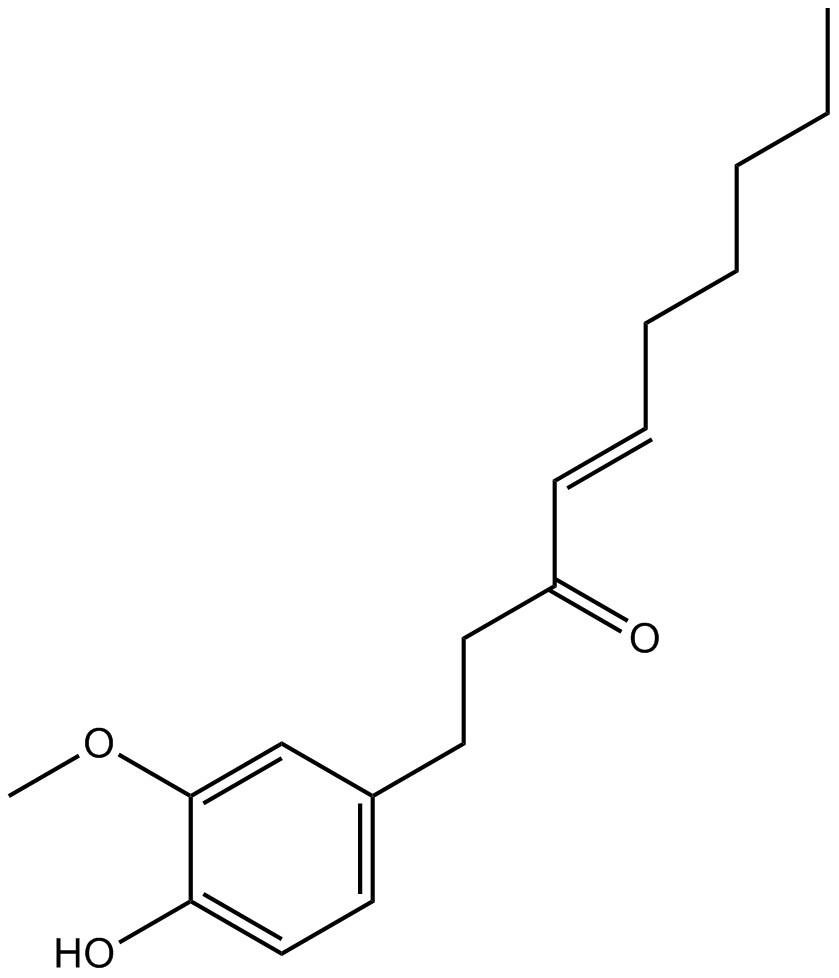
-
GC12739
A 484954
A eukaryotic elongation factor-2 (eEF-2) kinase inhibitor
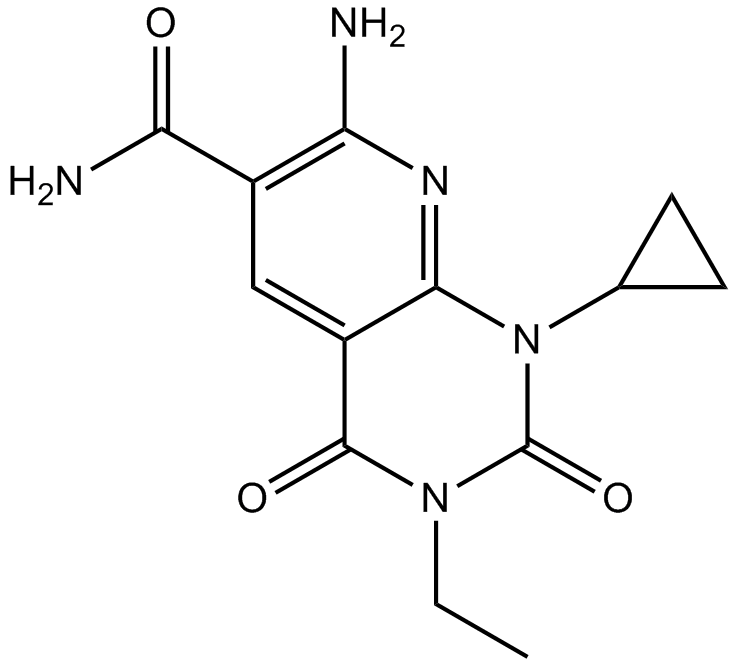
-
GC17710
A-317491
A 317491;A317491
A P2X3 and P2X2/3 receptor antagonist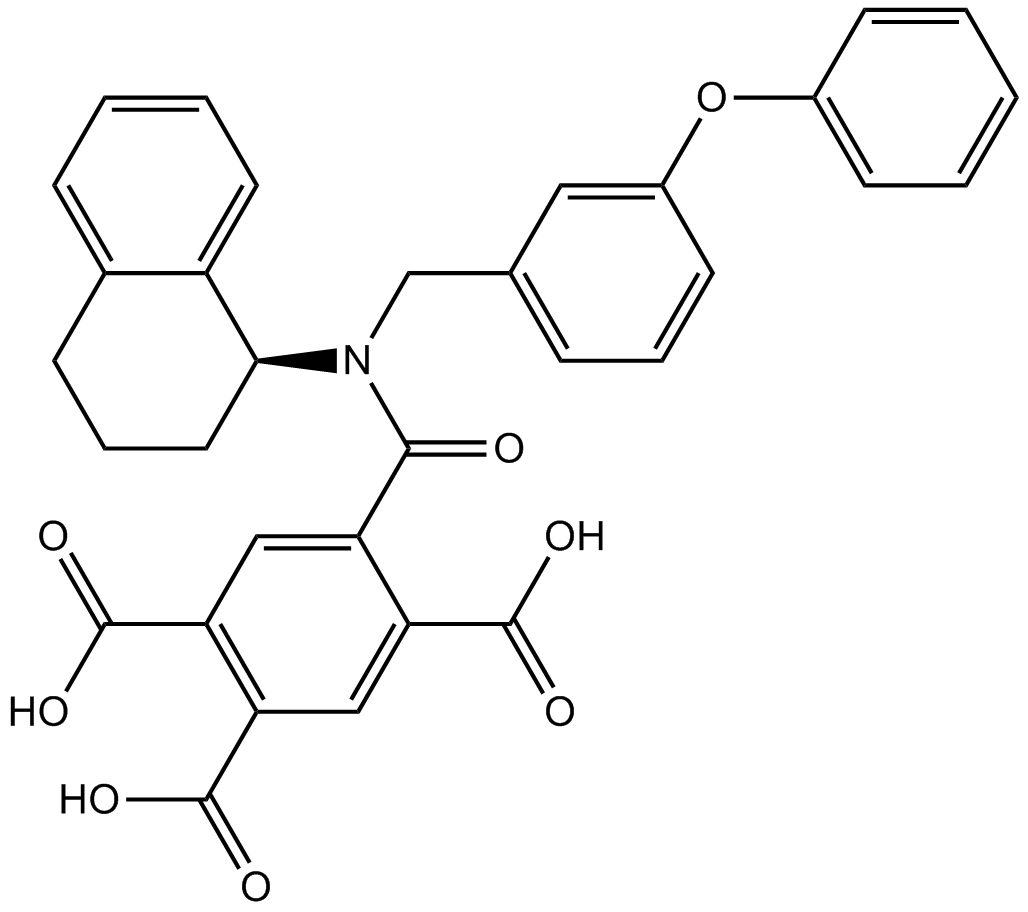
-
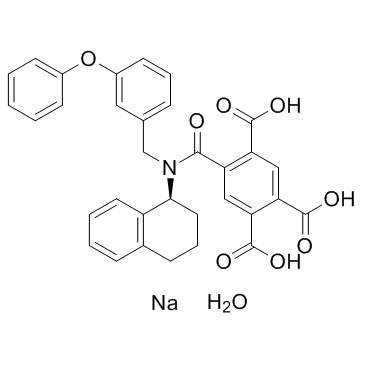
GC35211
A-317491 sodium salt hydrate
A-317491 sodium salt hydrate is a potent, selective and non-nucleotide antagonist of P2X3 and P2X2/3 receptors, with Kis of 22, 22, 9, and 92 nM for hP2X3, rP2X3, hP2X2/3, and rP2X2/3, respectively.
-
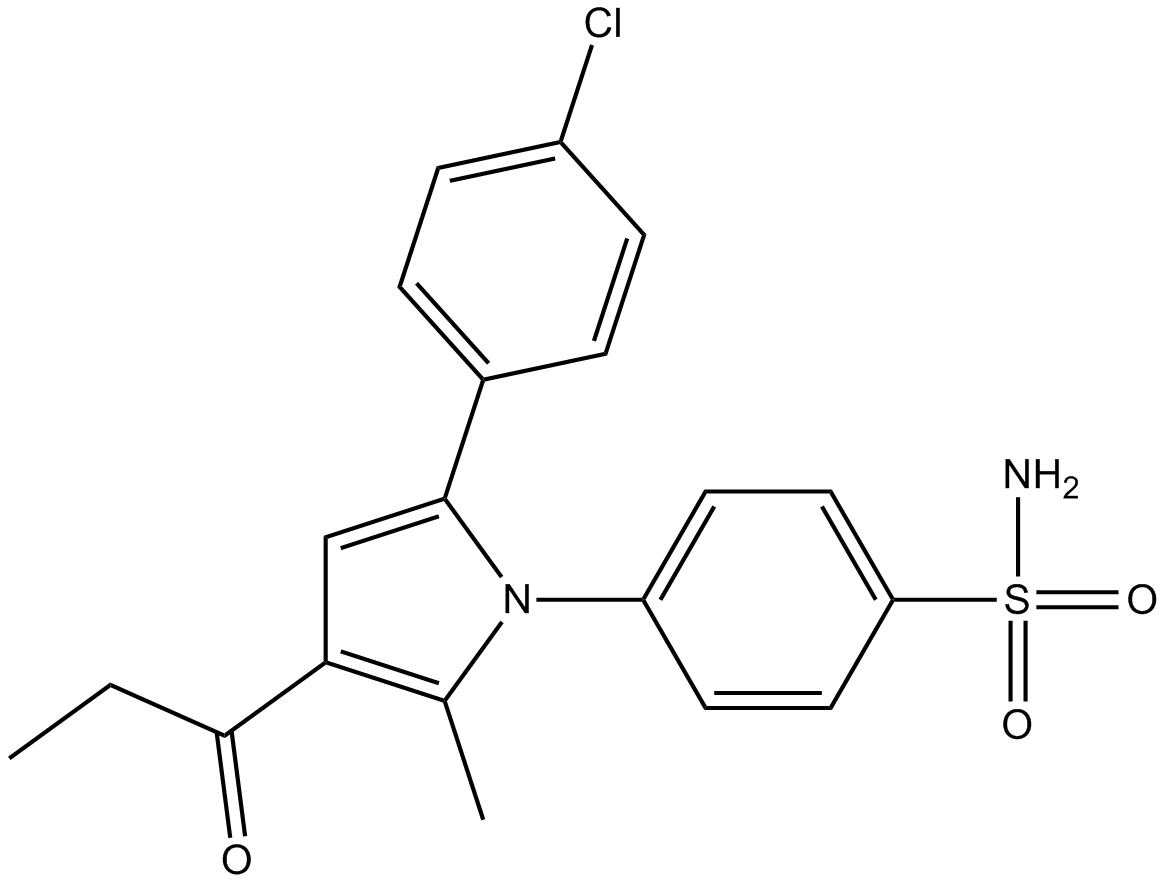
GC15014
A-867744
A positive allosteric modulator of α7 nAChRs
-
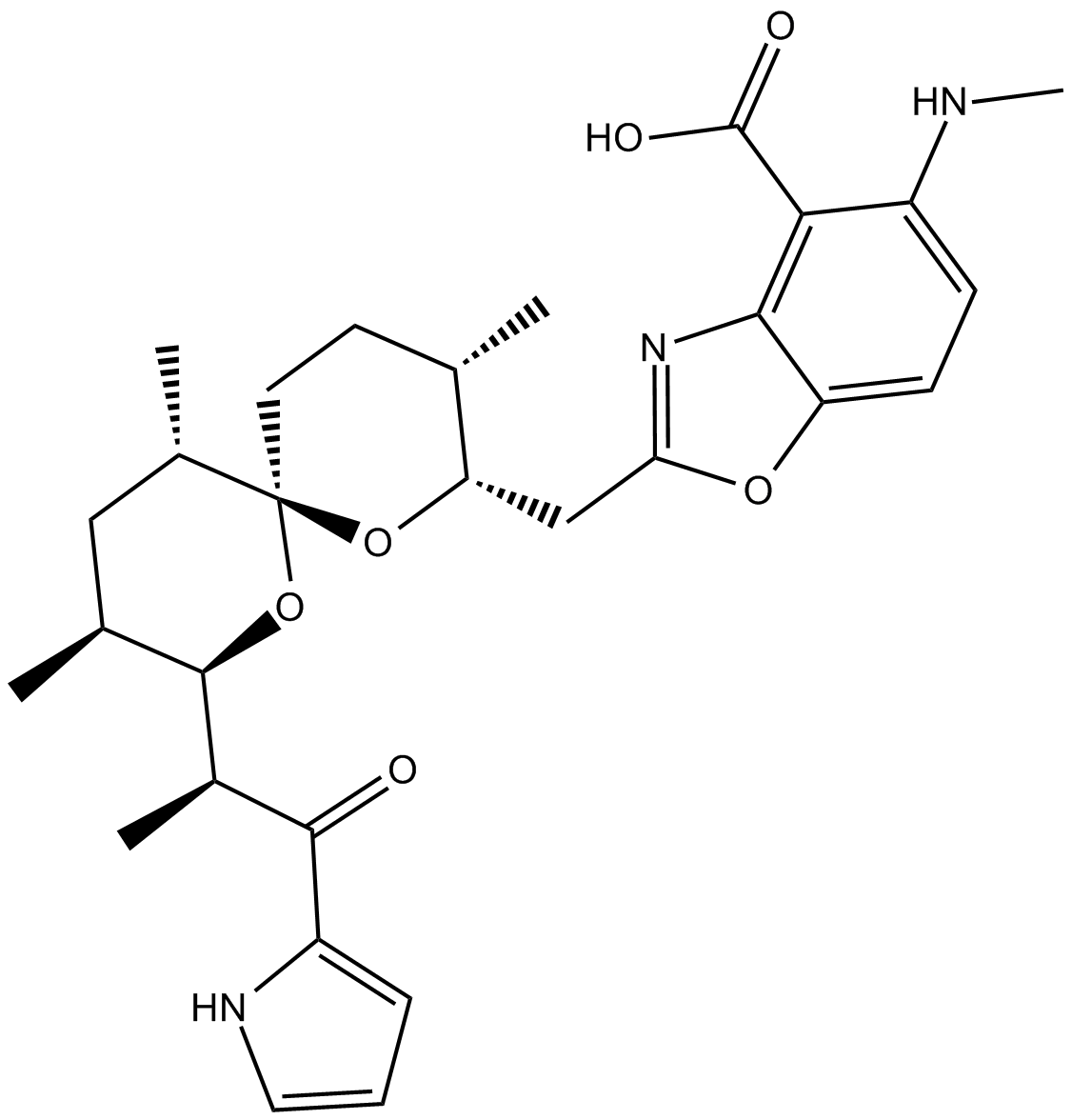
GC11200
A23187
Calcimycin
A23187, free acid is a Ca2+ ionophore
-
GC14069
ABT-199
GDC 0199, Venetoclax
Venetoclax (ABT-199, GDC-0199) is a selective inhibitor of Bcl-2 with a K i of 0.01 nM in cell-free assays.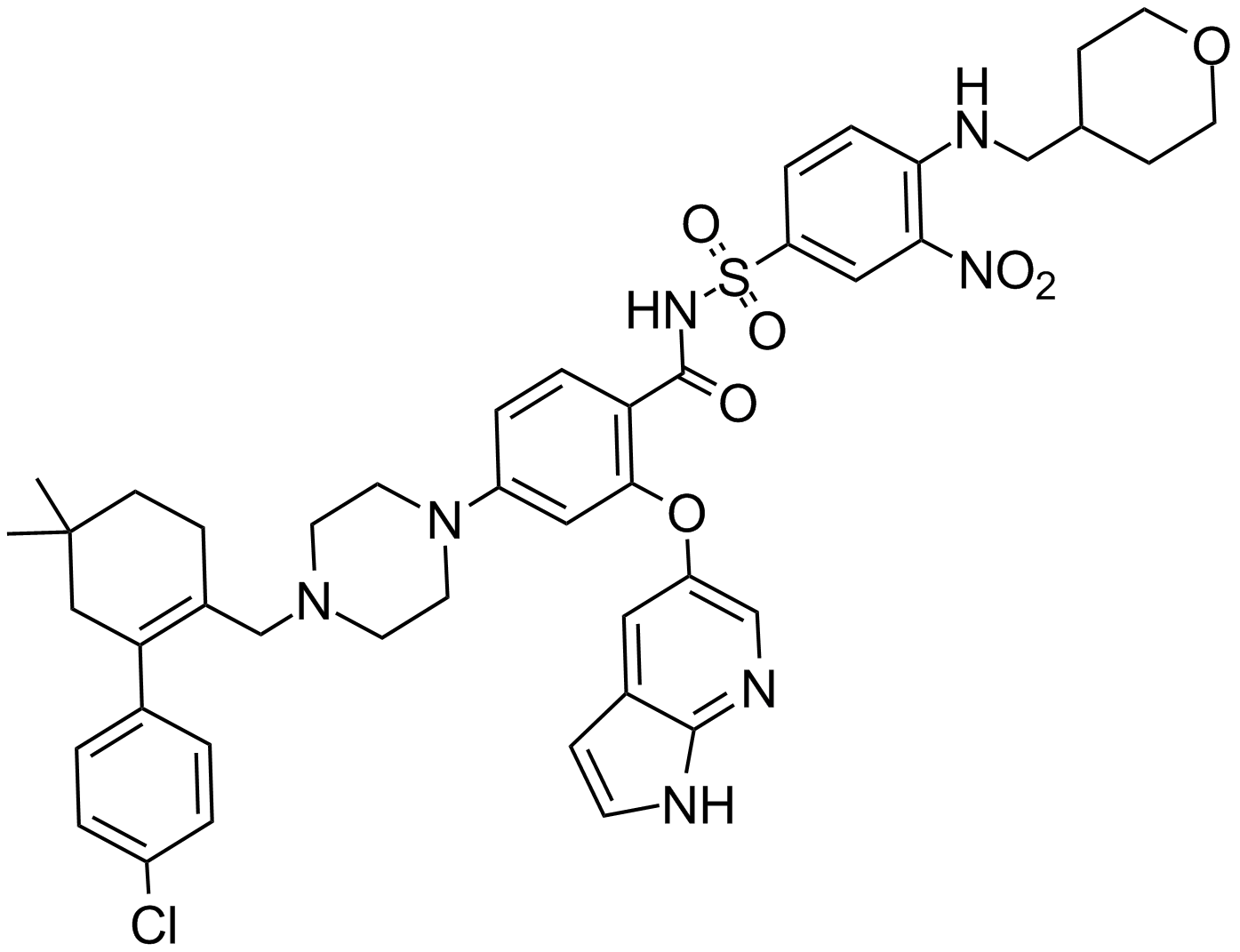
-
GC17234
ABT-737
ABT 737, ABT737
An inhibitor of anti-apoptotic Bcl-2 proteins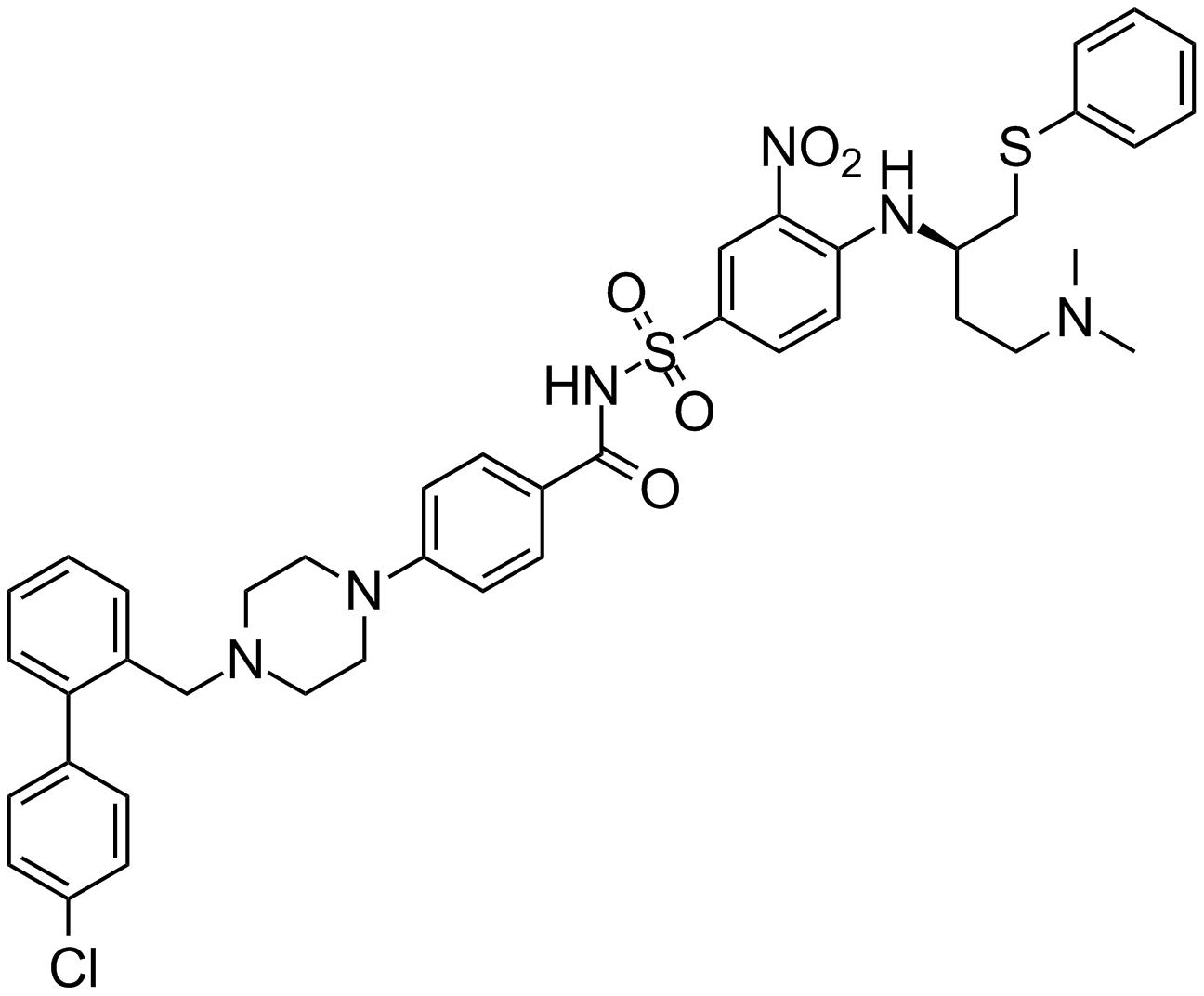
-
GC15875
ABT-751 (E7010)
ABT-751 (E7010)(E 7010) is a novel bioavailable tubulin-binding and antimitotic sulfonamide agent with IC50 of about 1.5 and 3.4 μM in neuroblastoma and non-neuroblastoma cell lines, respectively.
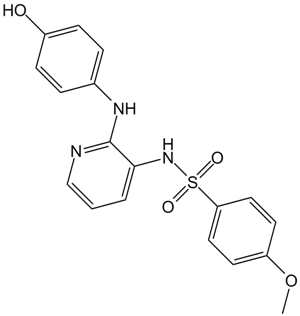
-
GC12422
ABT-888 (Veliparib)
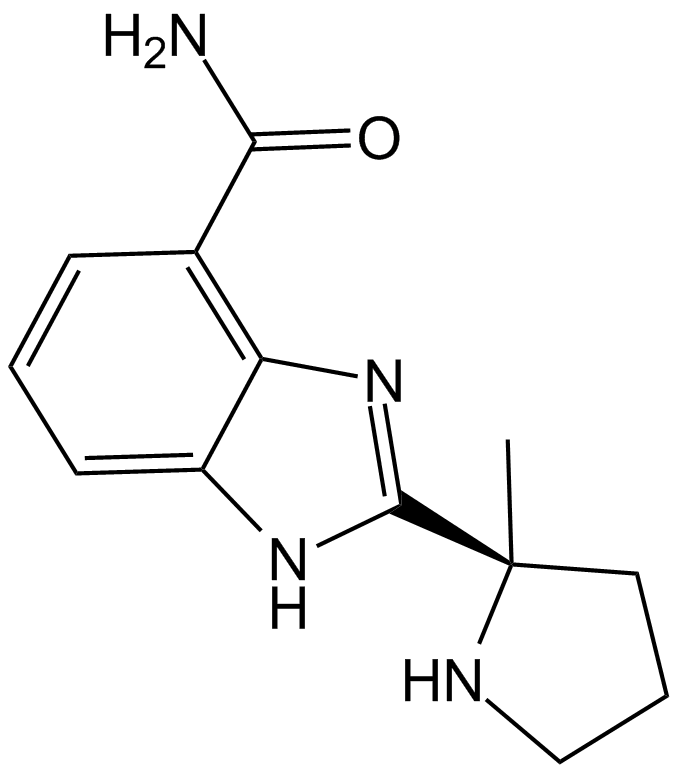
-
GC10871
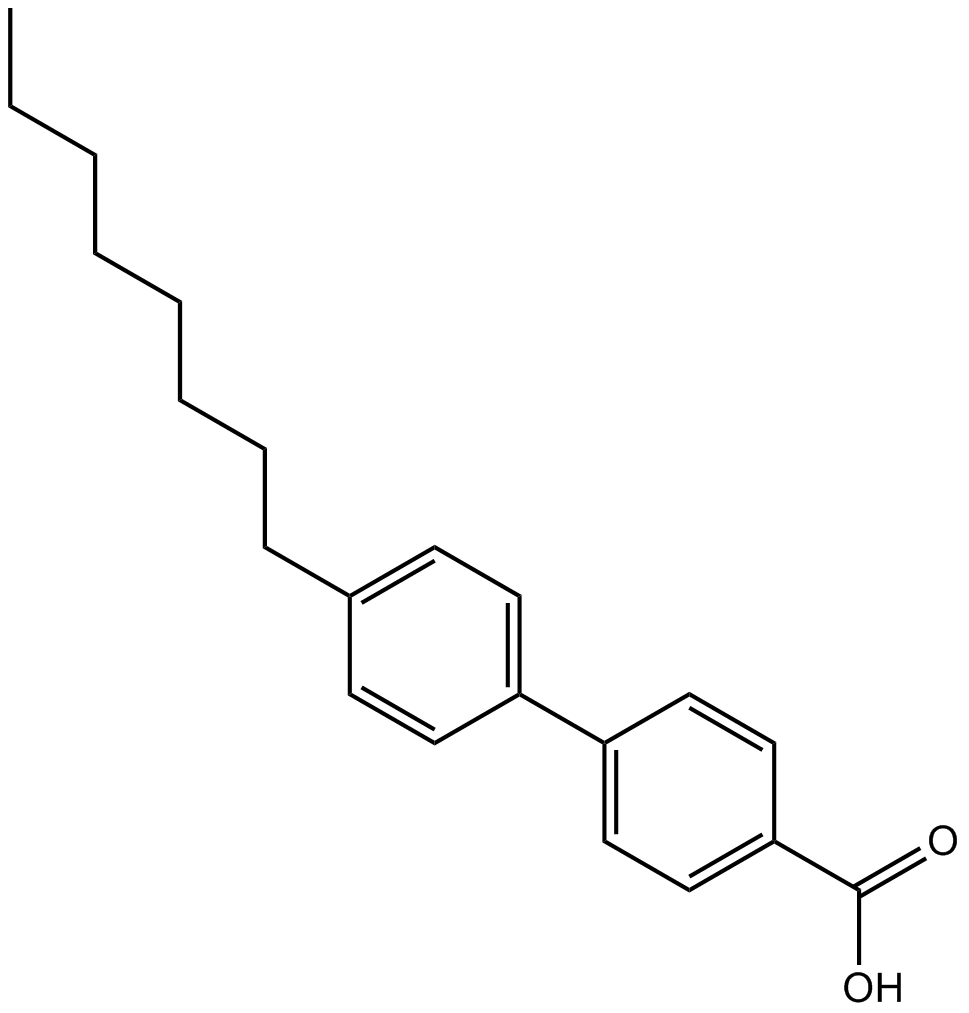
AC 55649
RARβ2 agonist, potent and selective
-
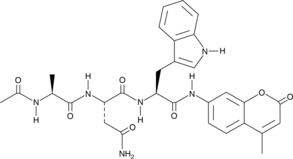
GC42685
Ac-ANW-AMC
Ac-Ala-Asn-Trp-AMC
Ac-ANW-AMC is a fluorogenic substrate for the β5i/LMP7 subunit of the 20S immunoproteasome.
-
GC42710
Ac-PAL-AMC
Acetyl-Pro-Ala-Leu-7-amino-4-Methylcoumarin, Ac-Pro-Ala-Leu-AMC
Ac-PAL-AMC is a fluorogenic substrate for the β1i/LMP2 subunit of the 20S immunoproteasome.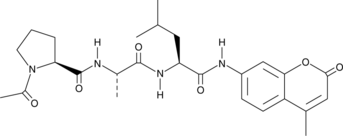
-
GC42720
Ac-WLA-AMC
Acetyl-Trp-Leu-Ala-7-amino-4-Methylcoumarin, Ac-Trp-Leu-Ala-AMC
Ac-WLA-AMC is a fluorogenic substrate for the β5c subunit of the 20S proteasome.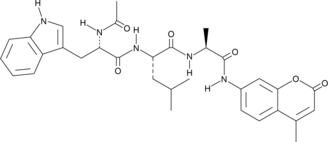
-
GC35228
Aceglutamide
Aceglutamide, Acetylglutamine, N-acetyl Gln, NSC 186896
Aceglutamide (α-N-Acetyl-L-glutamine) is a psychostimulant and nootropic, used to improve memory and concentration.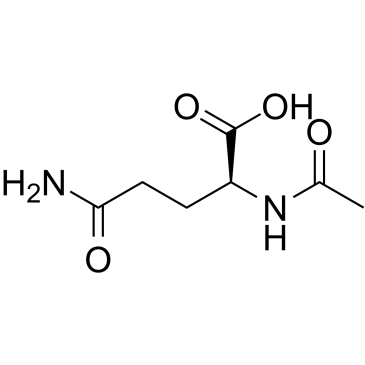
-
GC11567
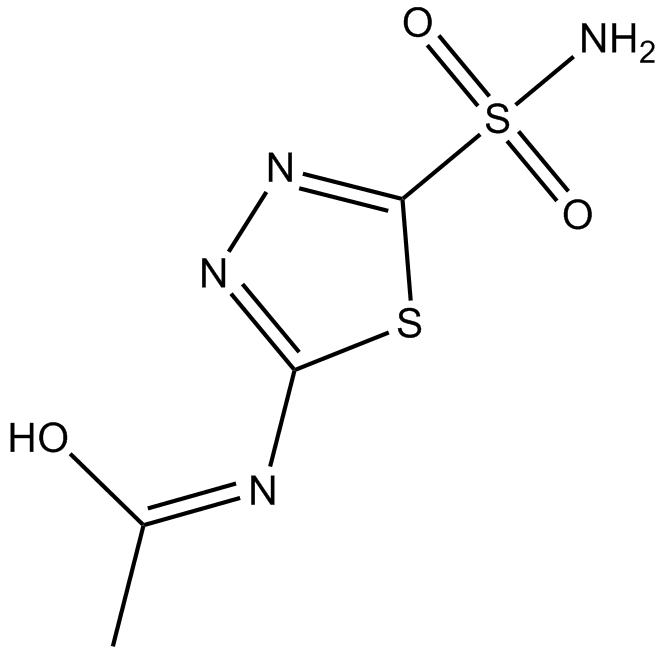
Acetazolamide
L-579,486, NSC 145177
carbonic anhydrase (CA) inhibitor
-
GC14142
Acetylcholine Chloride
ACh
Major transmitter at many nervous sites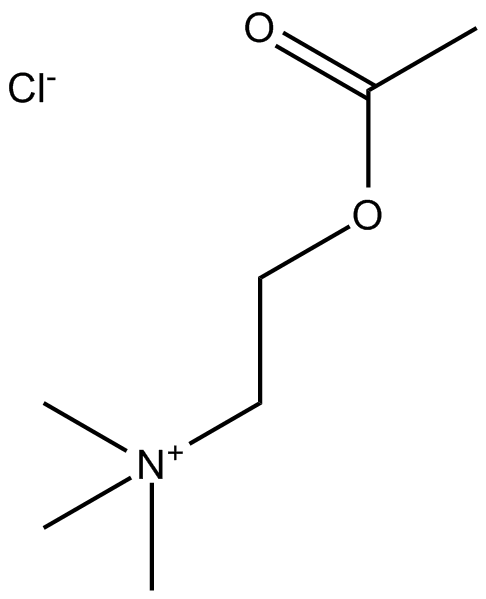
-
GC17094
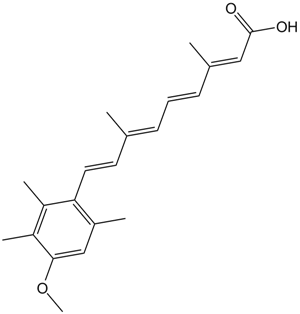
Acitretin
all-trans Acitretin, Ro 10-1670, Ro 10-1670/000
Metabolite of etretinate
-
GC17113
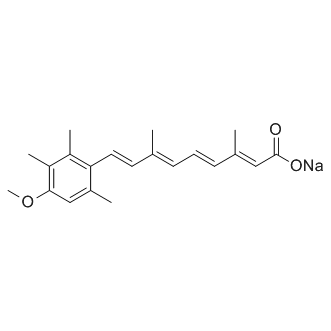
Acitretin sodium
Second-generation, systemic retinoid
-
GC10102
Aclacinomycin A
NSC 208734
An anthracycline with antibiotic and anticancer activities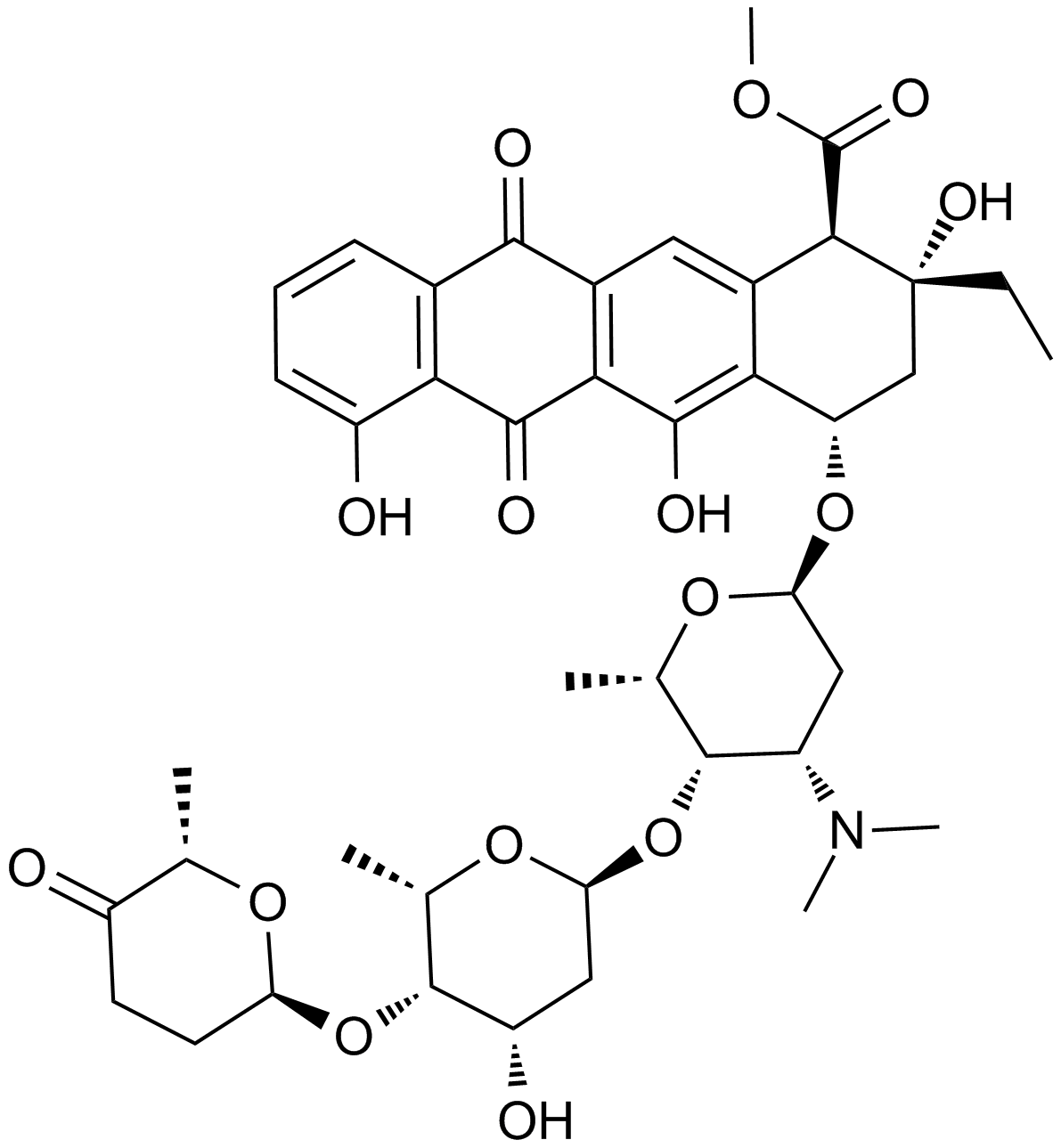
-
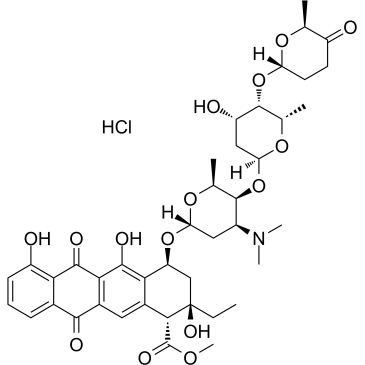
GC35238
Aclacinomycin A hydrochloride
Aclacinomycin A hydrochloride (Aclarubicin hydrochloride), a fluorescent molecule and the first described non-peptidic inhibitor showing discrete specificity for the CTRL (chymotrypsin-like) activity of the 20S proteasome. Aclacinomycin A hydrochloride is also a dual inhibitor of topoisomerase I and II. An effective anthracycline chemotherapeutic agent for hematologic cancers and solid tumors.
-
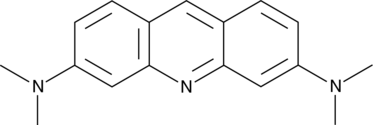
GC41242
Acridine Orange
NSC 194350
Acridine Orange is a cell-penetrating, nucleic acid-selective fluorescent dye.
-
GC35242
Actein
Actein is a triterpene glycoside isolated from the rhizomes of Cimicifuga foetida. Actein suppresses cell proliferation, induces autophagy and apoptosis through promoting ROS/JNK activation, and blunting AKT pathway in human bladder cancer. Actein has little toxicity in vivo.
-
GC16866
Actinomycin D
Cosmegen, Dactinomycin, Meractinomycin, NCI C04682, NSC 3053, Oncostatin K
Actinomycin D (dactinomycin) is a natural chromopeptide isolated from Streptomyces species, and has one heterocyclic chromophore and two cyclic pentapeptide lactone rings. [1]
-
GC19019
Acumapimod
BCT-197
Acumapimod (BCT197) is an orally active p38 MAP kinase inhibitor, with an IC50 of less than 1 uM for p38α.
-
GC10610
Adapalene
CD 271
RARβ and RARγ agonist
-
GC14379
Adapalene sodium salt
Retinoic acid receptor agonist
-
GC14106
Adenosine
NSC 7652
nucleoside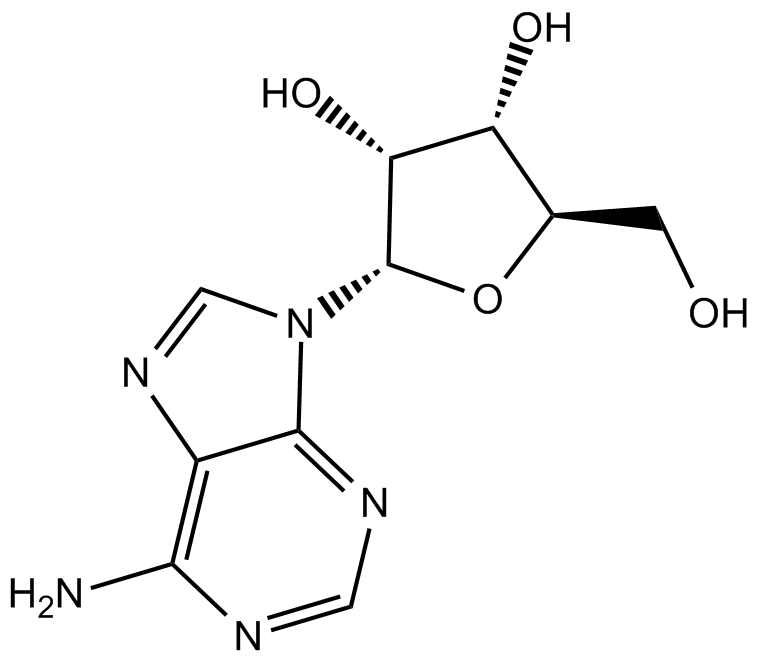
-
GC19729
Adenosine 5′-diphosphoribose sodium
ADP ribose sodium
Adenosine 5′-diphosphoribose sodium (ADP ribose sodium) is a nicotinamide adenine nucleotide (NAD+) metabolite.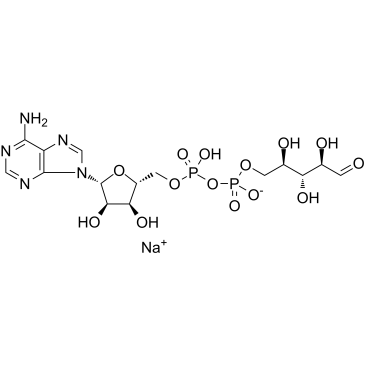
-
GC10707
Afatinib dimaleate
Afatinib dimaleate, BIBW 2992, BIBW 2992MA2
An inhibitor of EGFR and ErbB2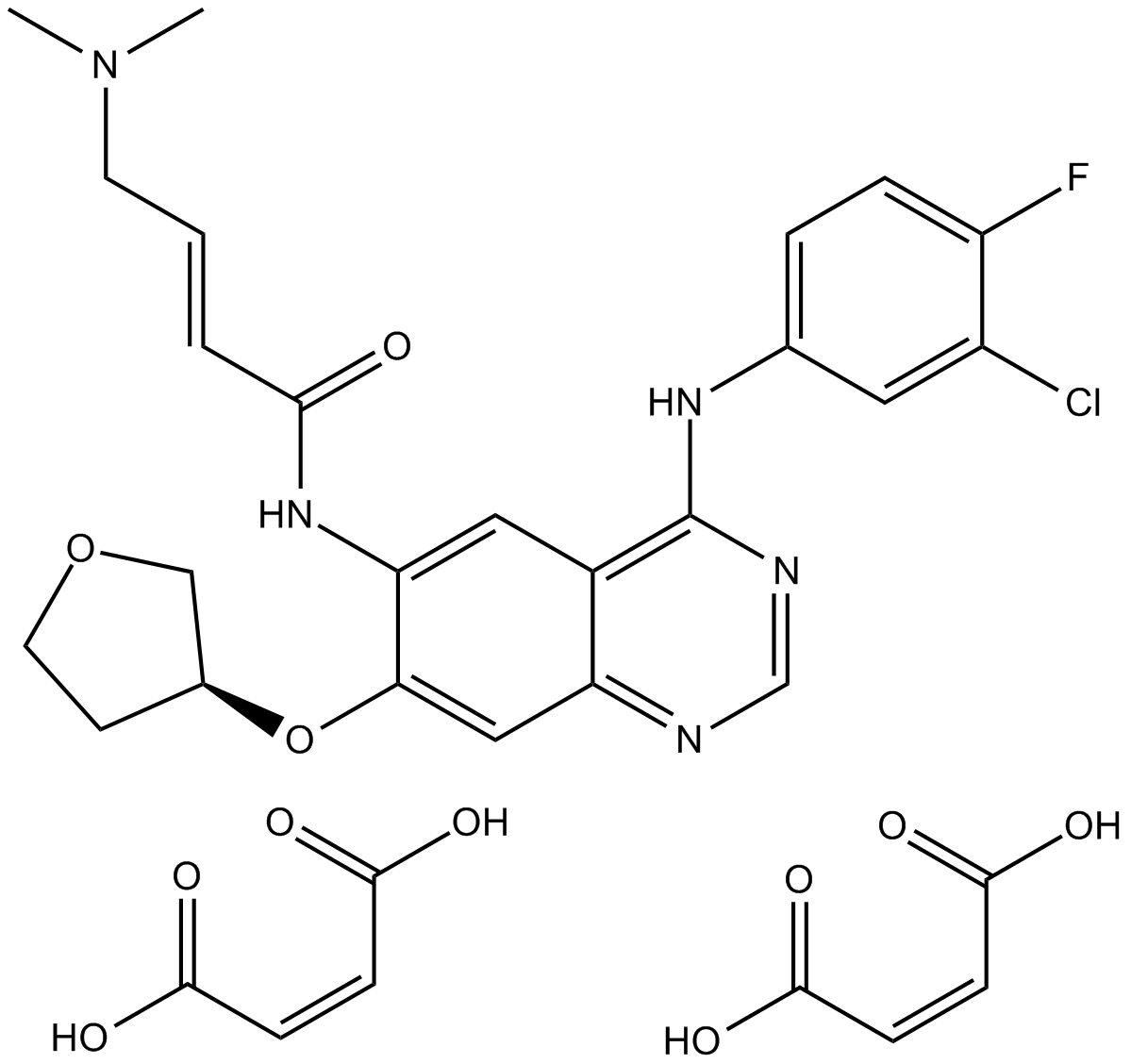
-
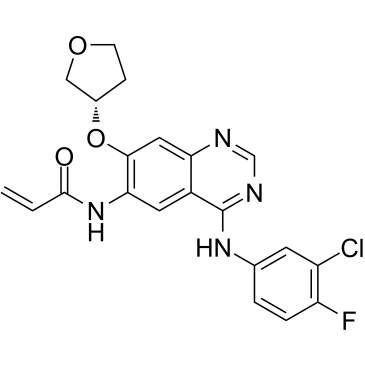
GC60567
Afatinib impurity 11
Afatinib impurity 11 is an impurity of Afatinib. Afatinib is an irreversible EGFR family inhibitor with IC50s of 0.5 nM, 0.4 nM, 10 nM and 14 nM for EGFRwt, EGFRL858R, EGFRL858R/T790M and HER2, respectively.
-
GC35262
Afzelin
Kaempferin, Kaempferol 3-O-rhamnoside, Kaempferol 3-O-α-L-rhamnopyranoside
A polyphenolic glycoside flavone with diverse biological activities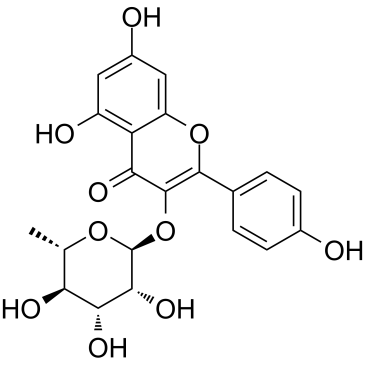
-
GC13697
AG-1024
AGS 200, Tyrphostin AG1024
Selective IGF-1R inhibitor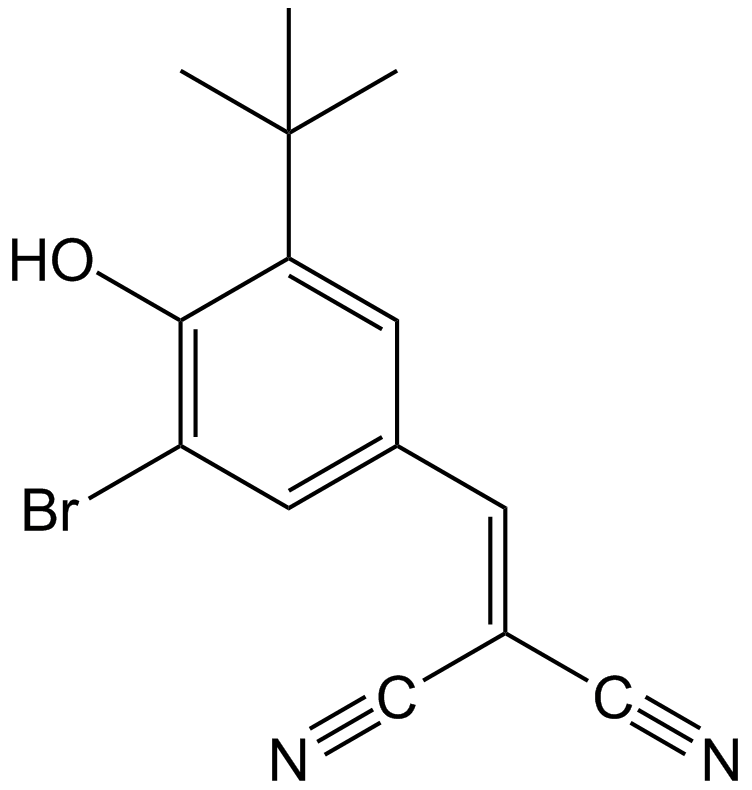
-
GC13854
AG-490 (Tyrphostin B42)
Tyrphostin AG-490
AG-490 (Tyrphostin B42) (Tyrphostin AG-490 (Tyrphostin B42)) is a tyrosine kinase inhibitor that inhibits EGFR, Stat-3 and JAK2/3.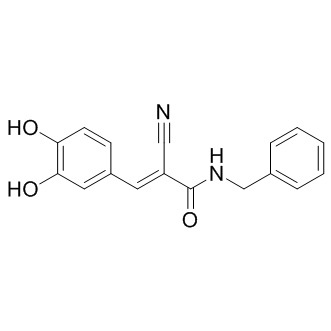
-
GC32817
AGN 193109
AGN 193109 is a retinoid analog, and acts as a specific and highly effective antagonist of retinoic acid receptors (RARs), with Kds of 2 nM, 2 nM, and 3 nM for RARα, RARβ, and RARγ, respectively.
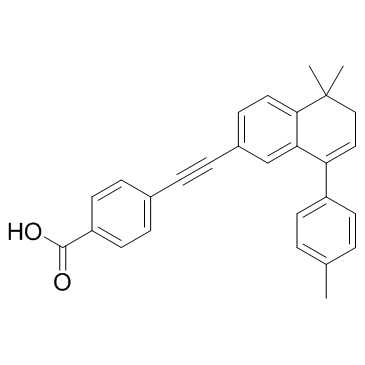
-
GC33315
AGN 194078
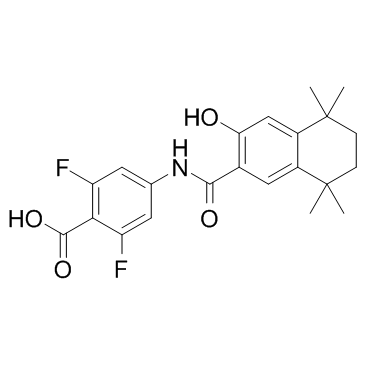
AGN 194078 is a selective RARα agonist with a Kd and EC50 of 3 and 112 nM, respectively.
-
GC12598
AGN 194310
VTP-194310
pan-RAR antagonist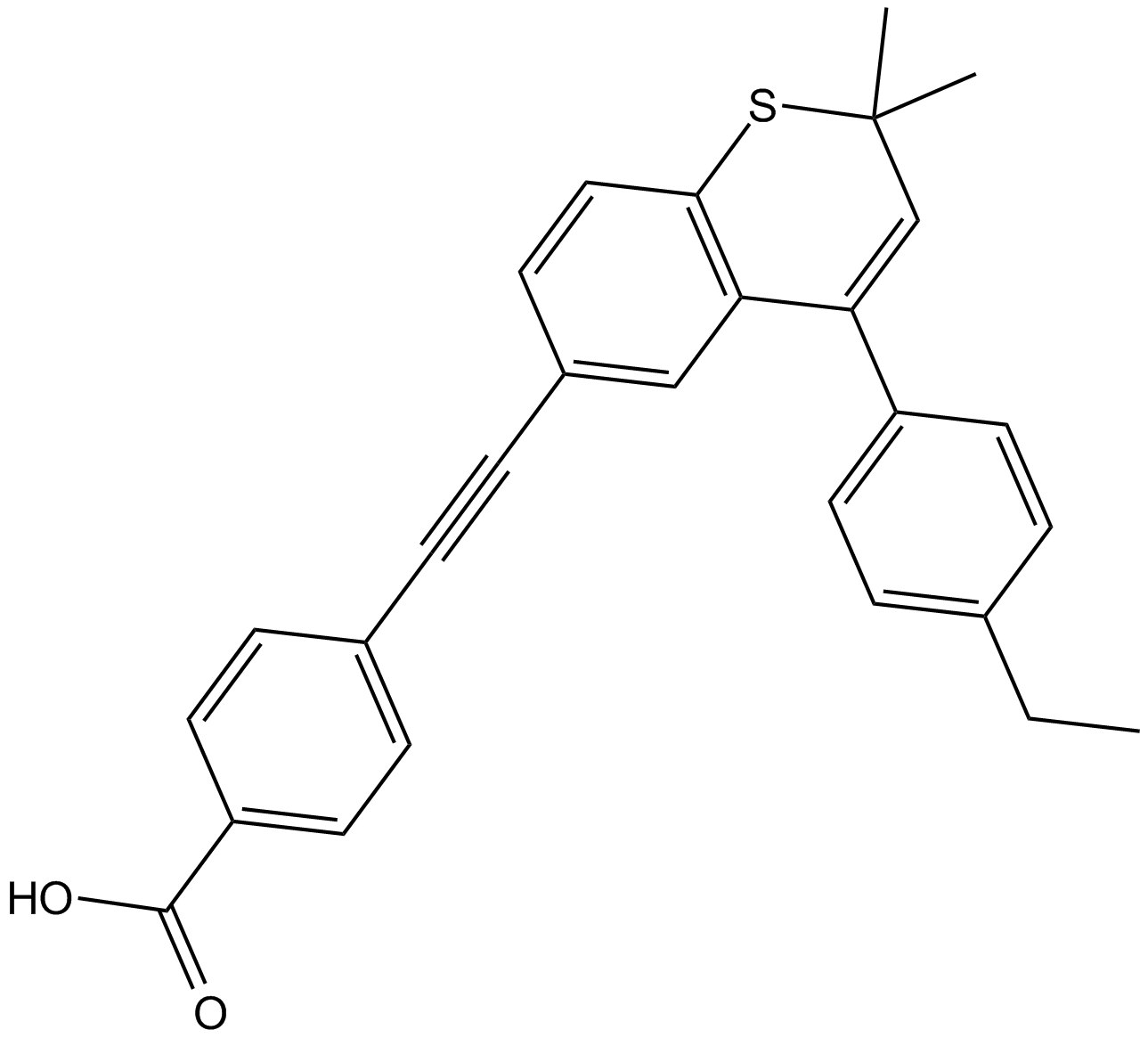
-
GC35265
AGN 195183
IRX-5183; VTP-195183; NRX-195183
AGN 195183 (IRX-5183) is a potent and selective agonist of RARα (Kd=3 nM) with improved binding selectivity relative to AGN 193836. AGN 195183 has no activity on RARβ/γ.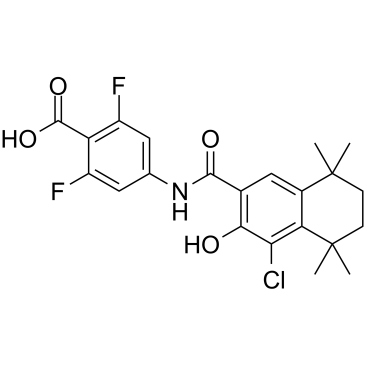
-
GC35266
AGN 196996
AGN 196996 is a potent and selective RARα antagonist with Ki value of 2 nM; little binding affinity for RARβ(Ki=1087 nM) and RARγ(Ki=8523 nM).
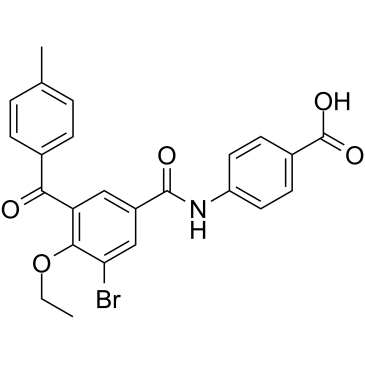
-
GC35267
AGN 205327
AGN 205327 is a potent synthetic RARs agonist with EC50 of 3766/734/32 nM for RARα/β/γ respectively; no inhibition on RXR.
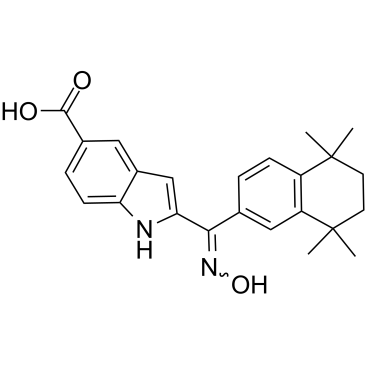
-
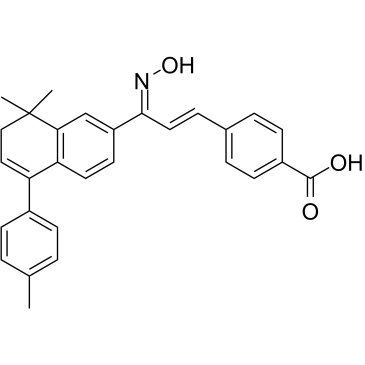
GC35268
AGN 205728
AGN 205728 is a potent and selective RARγ antagonist with Ki/IC95 values of 3 nM/ 0.6 nM; no inhibiton on RARα and RARβ.
-
GC10518
AICAR
Acadesine, AICA-Riboside, NSC 105823
AICAR (also called acadesine) is a purine nucleoside.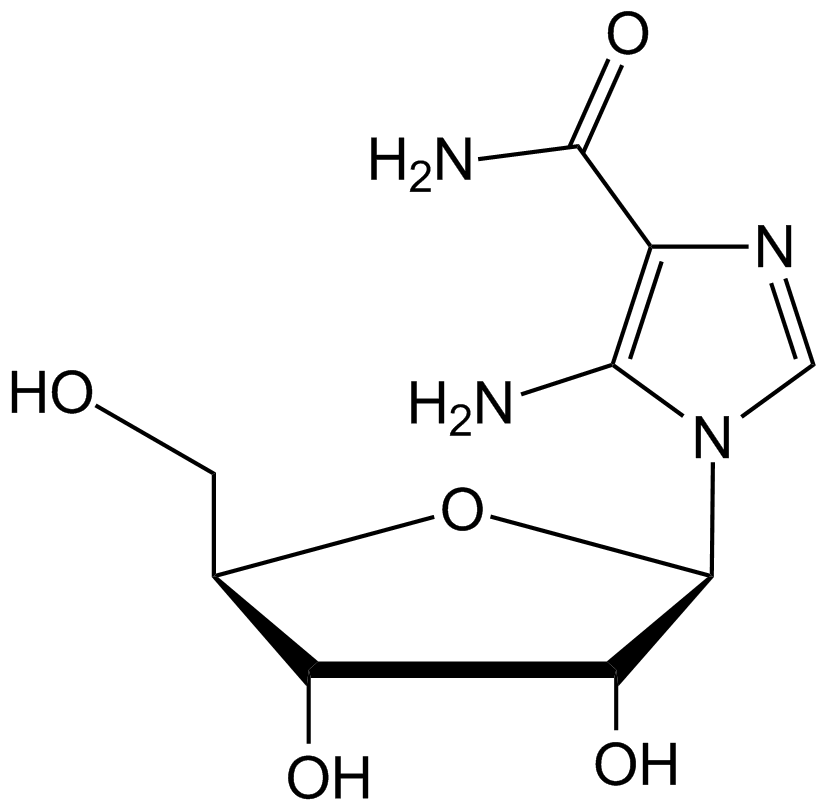
-
GC10580
AICAR phosphate
AMPK activator
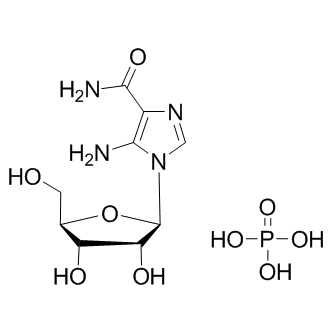
-
GC12646
AL 8697
p38α inhibitor,potent and selective
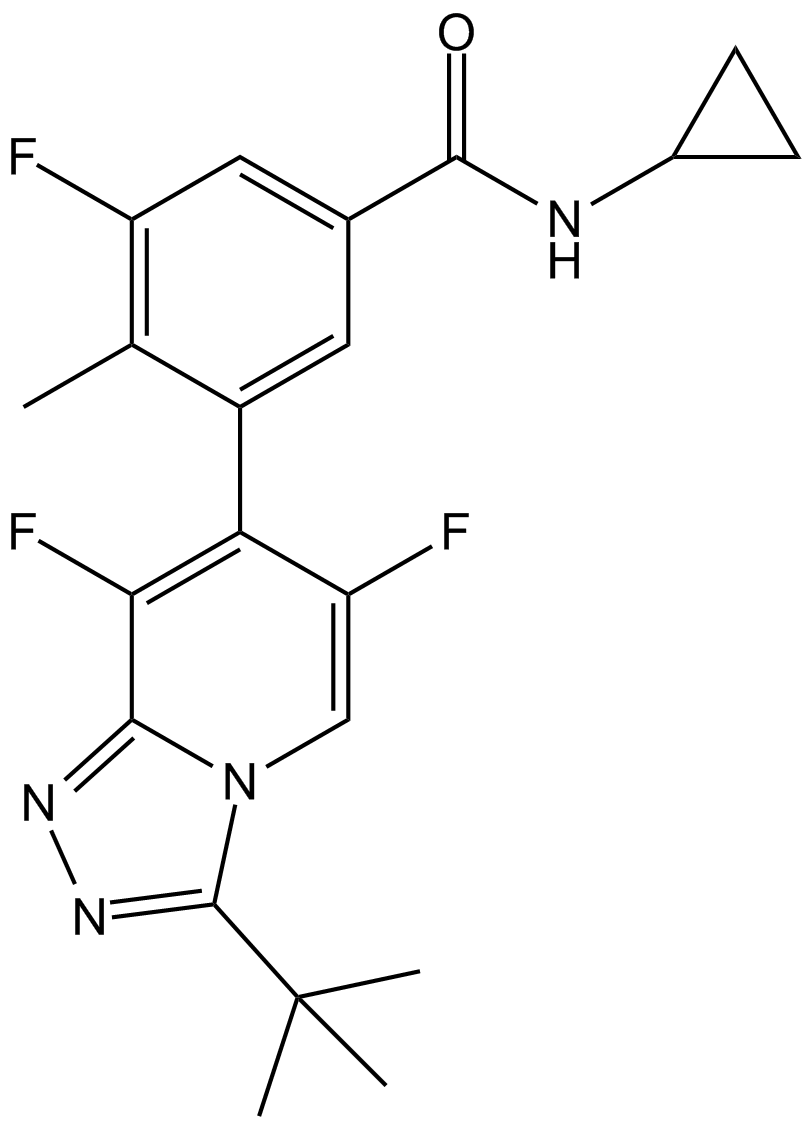
-
GC33743
Alicapistat (ABT-957)
ABT-957
Alicapistat (ABT-957) (ABT-957) is an orally active selective inhibitor of human calpains 1 and 2 for the potential application of Alzheimer's disease (AD).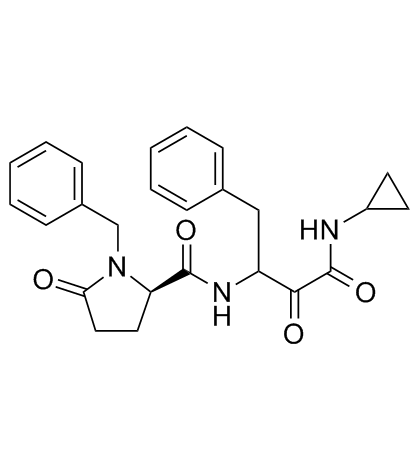
-
GC15451
Aliskiren
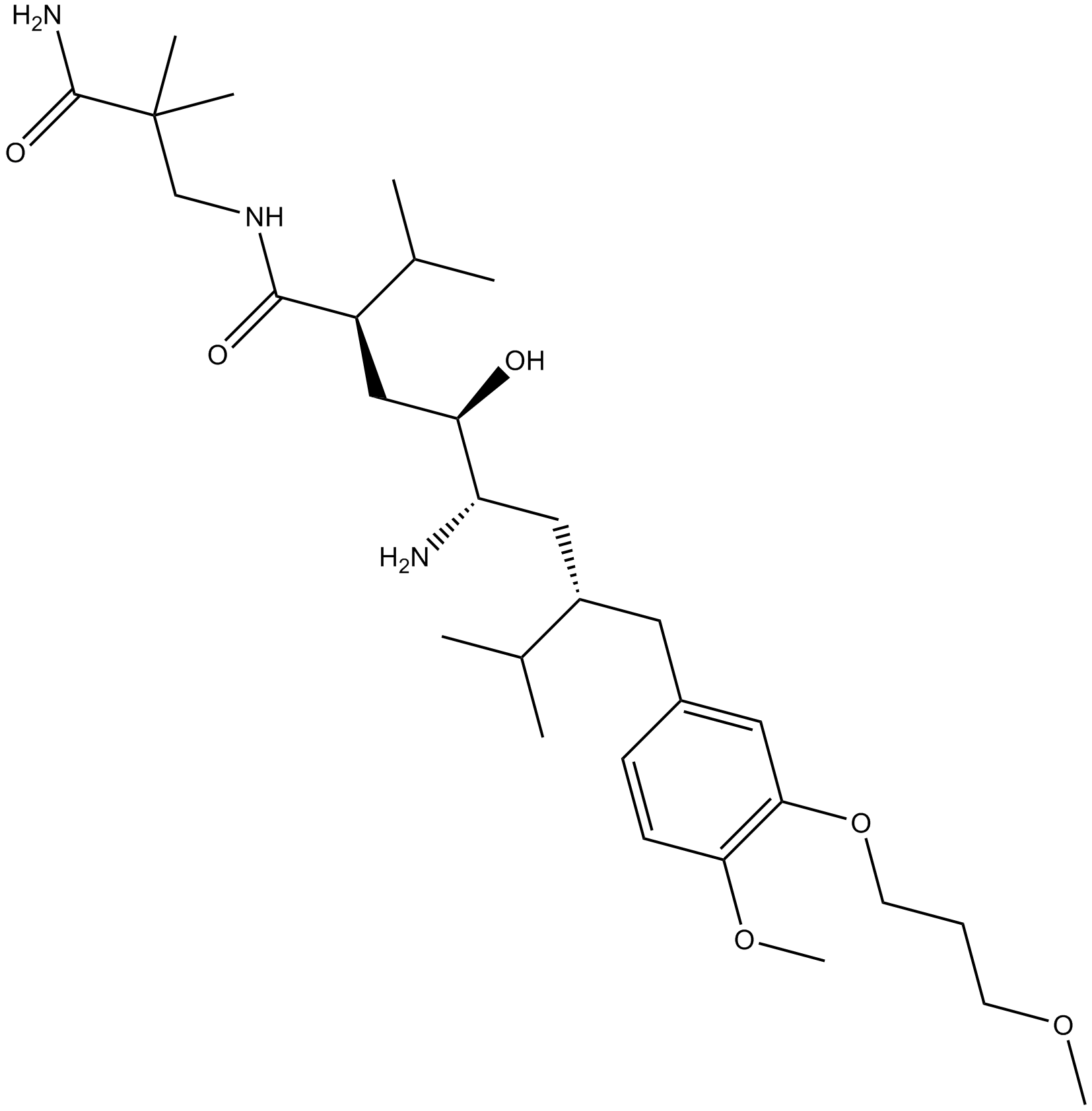
-
GC13223
Aliskiren Hemifumarate
CGP 60536, SPP 100
renin inhibitor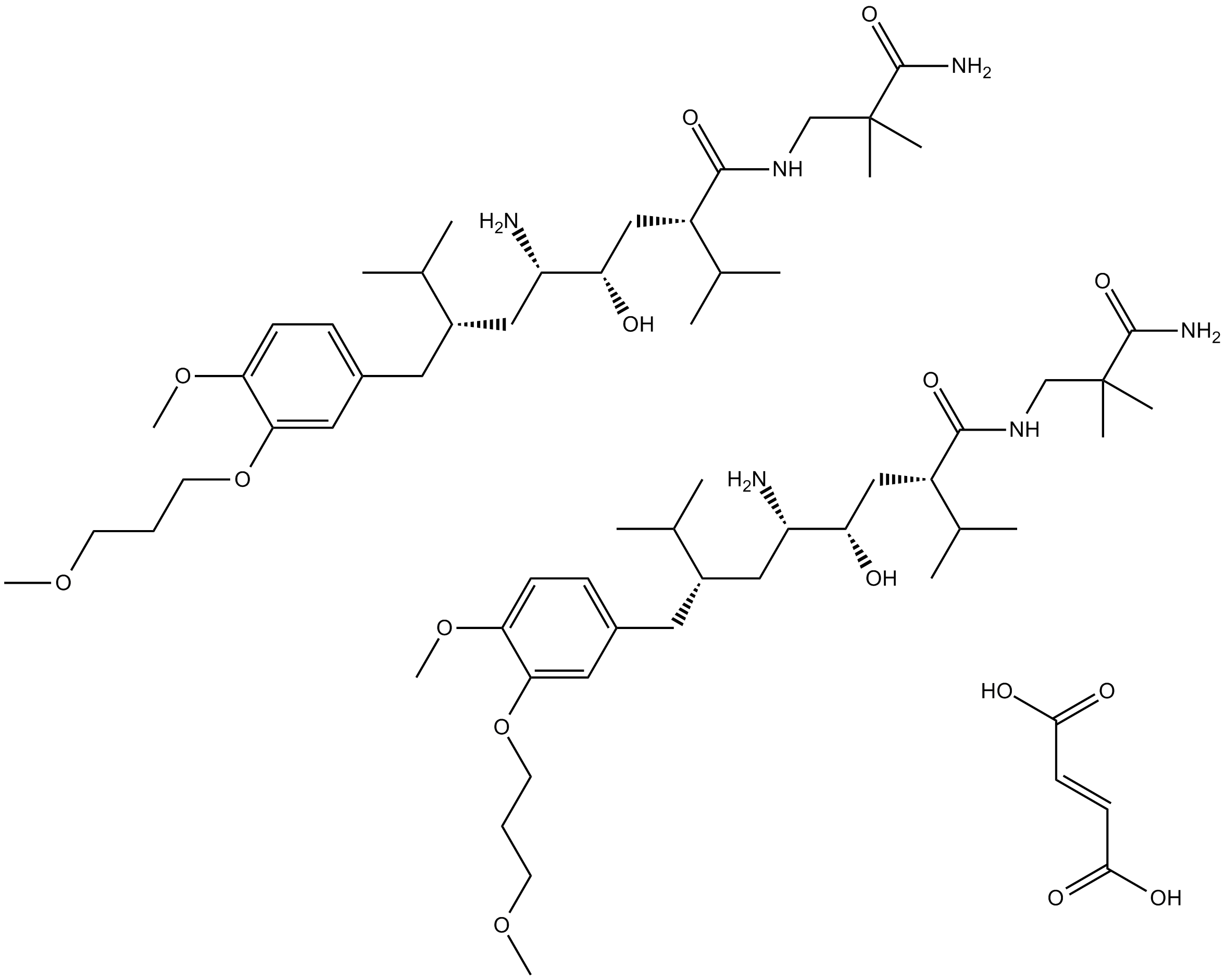
-
GN10407
Alisol A
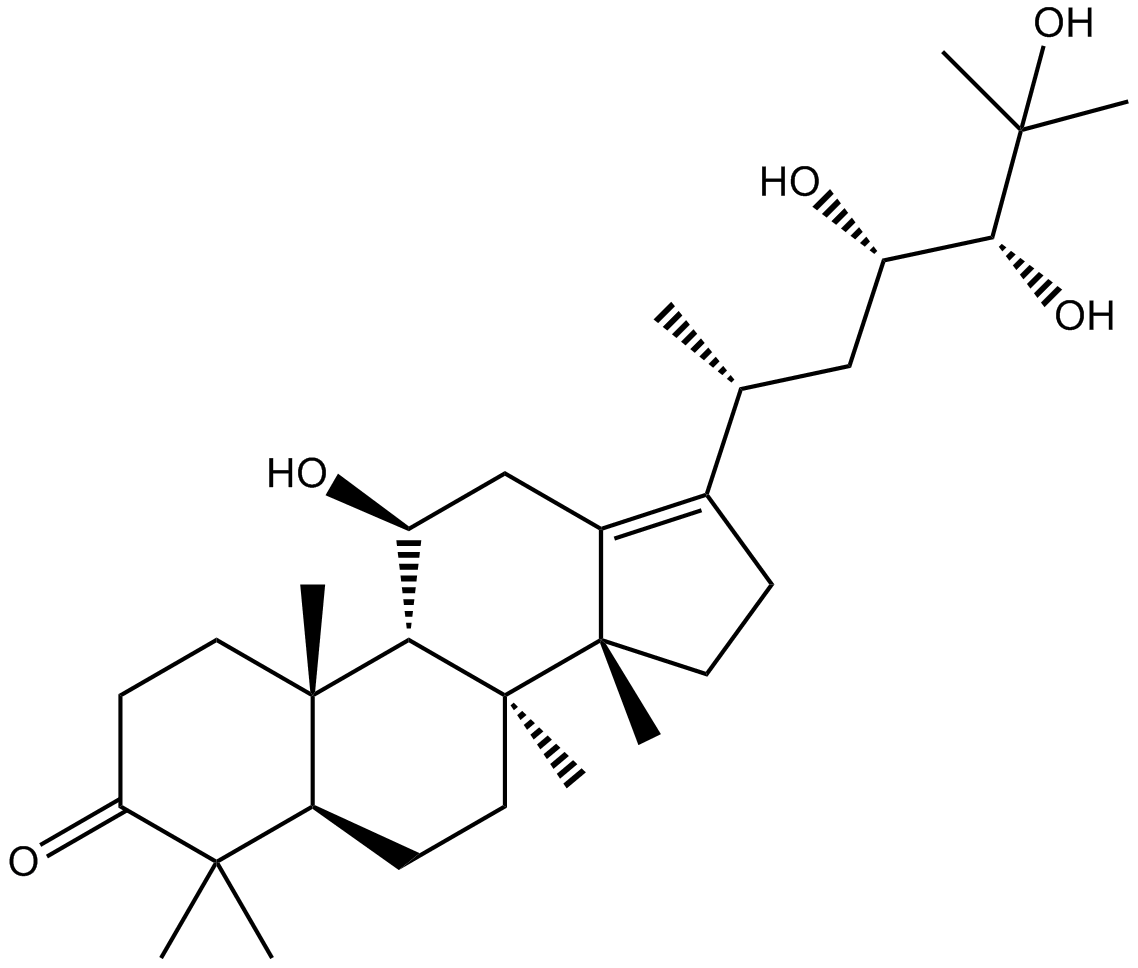
-
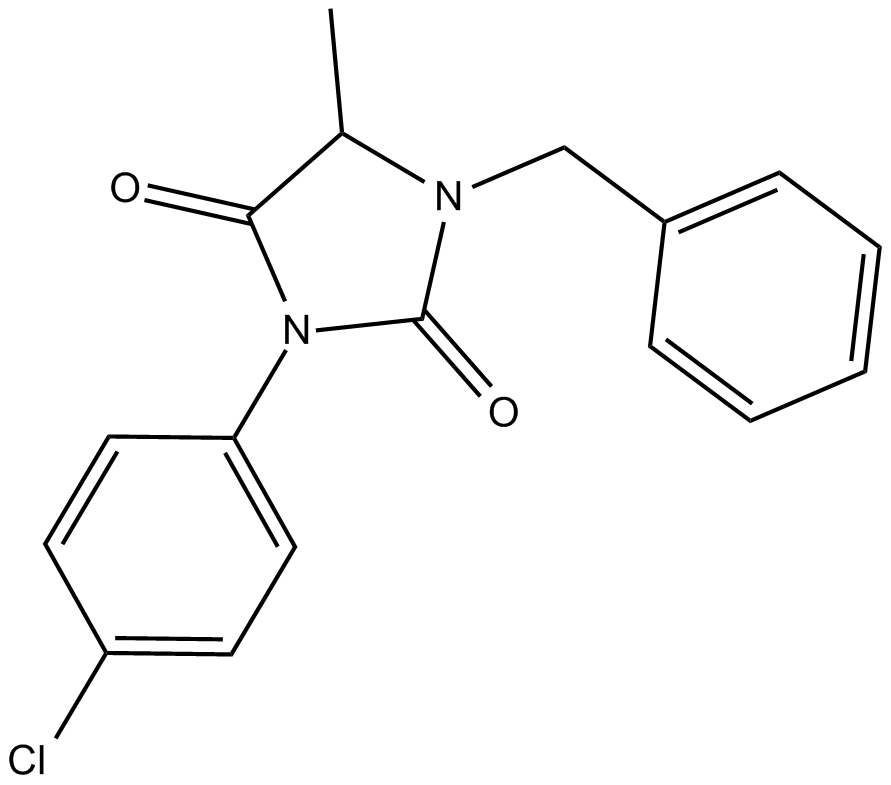
GC14749
ALLO-1
ALLO-1, an autophagy receptor, is essential for autophagosome formation around paternal organelles and directly binds to the worm LC3 homologue LGG-1 through its LC3-interacting region (LIR) motif.
-
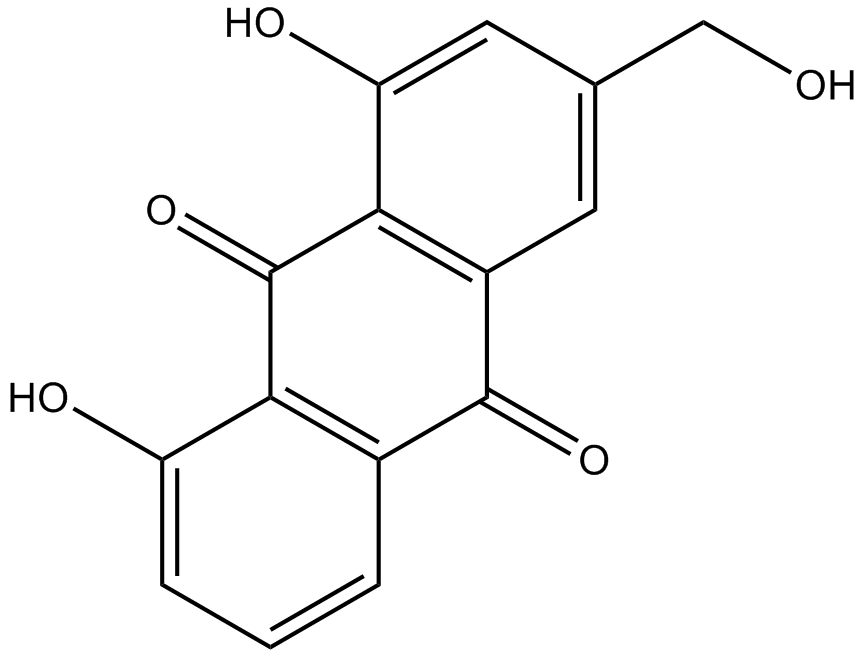
GN10516
Aloeemodin
3-Hydroxymethylchrysazine, NSC 38628, Rhabarberone
-
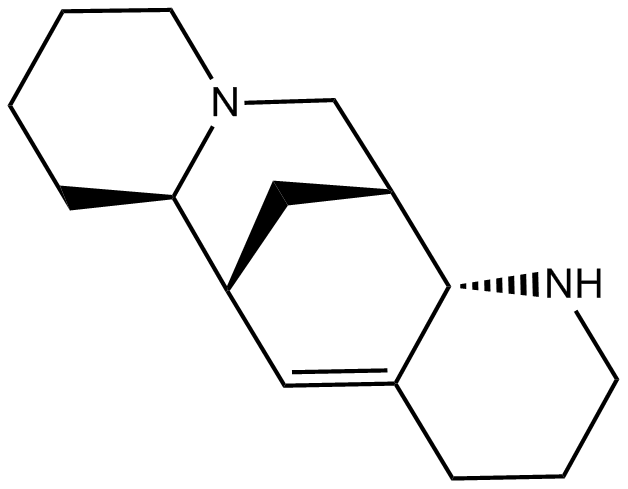
GC14314
Aloperine
An alkaloid
-
GC13789
AM 114
PS-IX; AM114
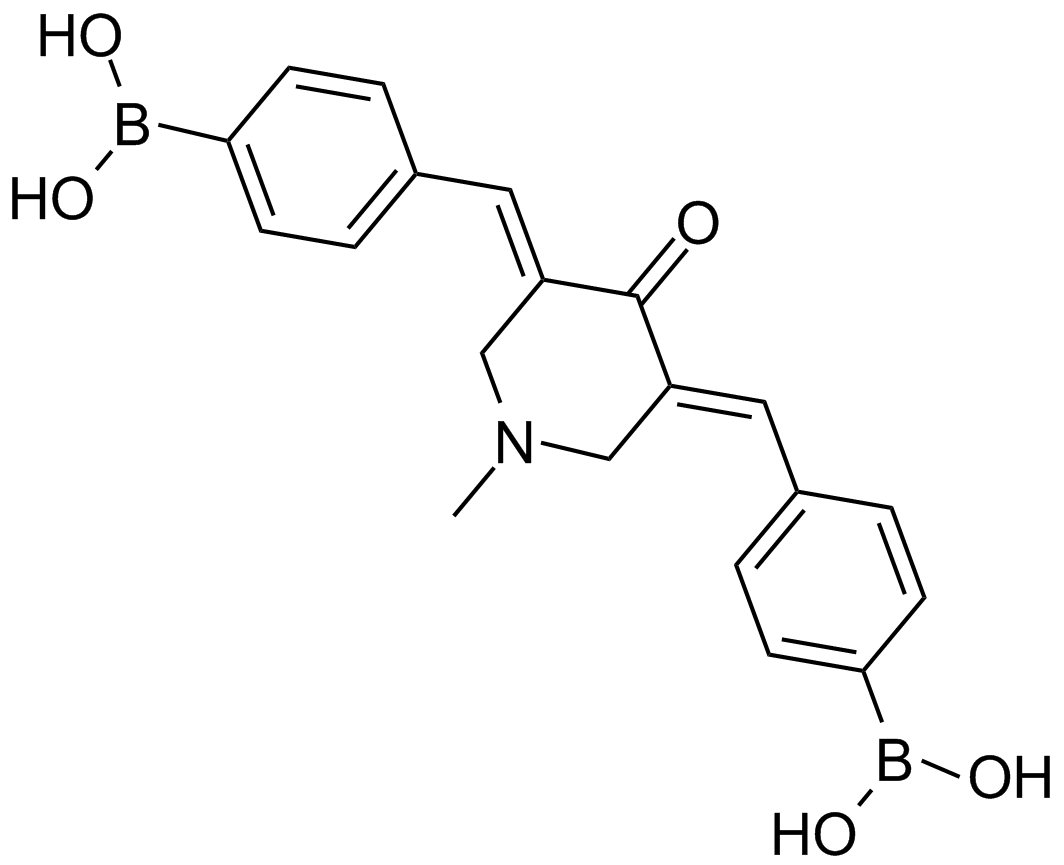
-
GC13664
AM580
CD336, NSC 608001, Ro 406055
Selective RARα agonist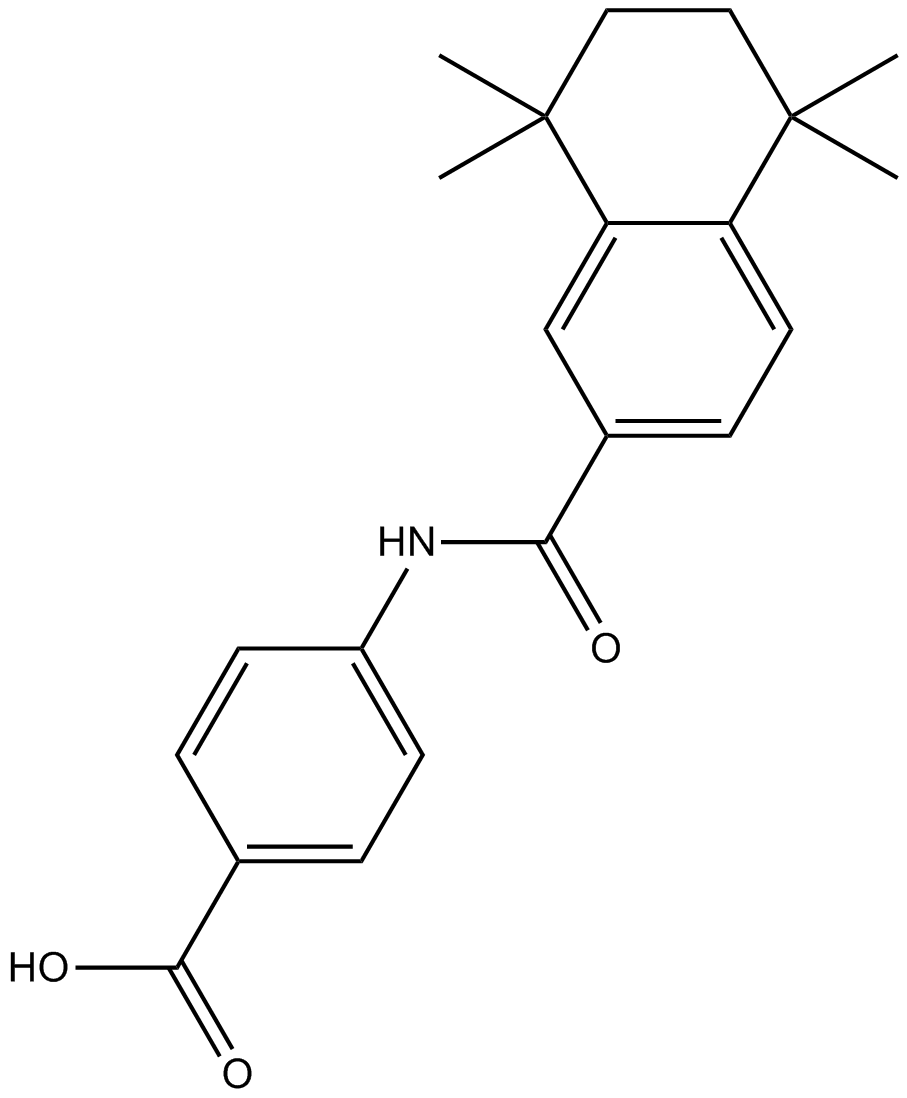
-
GC35322
Amiodarone
Amiodarone is an antiarrhythmic drug for inhibition of ATP-sensitive potassium channel with an IC50 of 19.1 μM.
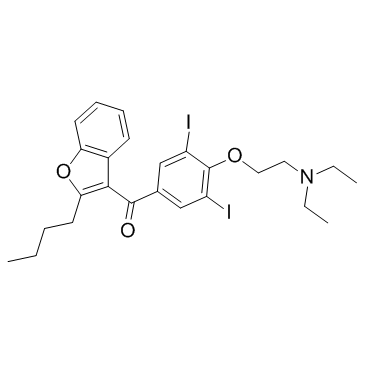
-
GC11432
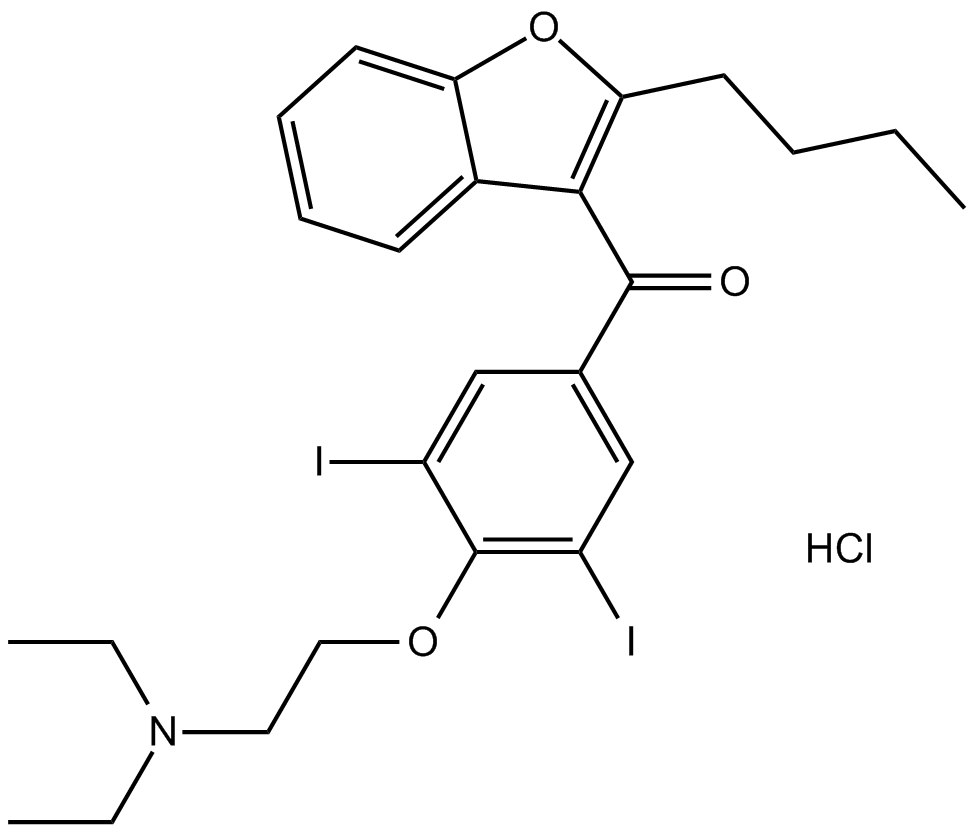
Amiodarone HCl
Amiodarone HCl, a benzofuran-based Class III antiarrhythmic agent, inhibits WT outwardIhERG tails with an IC50 of ~45 nM.