Nucleotides/Nucleosides
Products for Nucleotides/Nucleosides
- Cat.No. Product Name Information
-
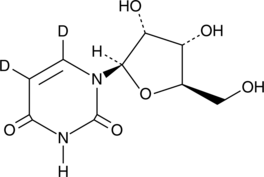
GC49514
2′-Deoxyuridine-d2
An internal standard for the quantification of 2’-deoxyuridine
-
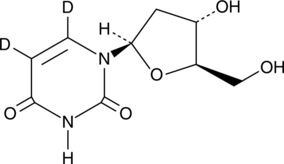
GC46540
2'-Deoxyadenosine-5'-triphosphate (sodium salt hydrate)
A purine nucleotide
-
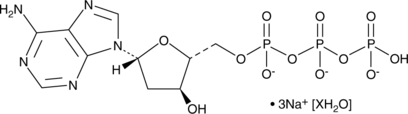
GC46554
2'-O-Monosuccinyladenosine-3',5'-cyclic monophosphate
2'-O-Monosuccinyladenosine-3',5'-cyclic monophosphate is a cAMP analog that can be covalently coupled to acetylcholinesterase.
-
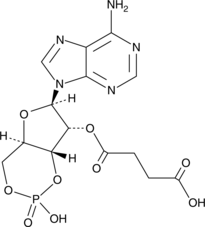
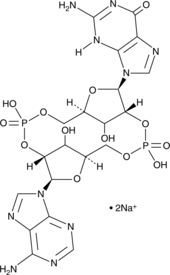
GC42080
2'2'-cGAMP (sodium salt)
2'2'-cGAMP is a synthetic dinucleotide (CDN) that contains non-canonical 2'5'-phosphodiester bonds.
-
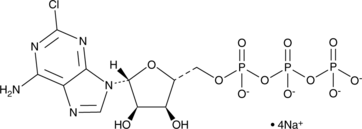
GC49718
2-Chloroadenosine 5-triphosphate (sodium salt)
An adenine nucleotide and ATP analog
-
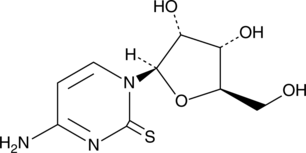
GC49348
2-Thiocytidine
A modified nucleobase
-
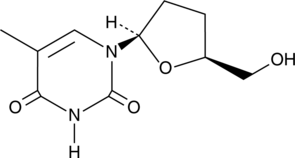
GC49122
3′-deoxy Thymidine
An antiviral nucleoside analog
-
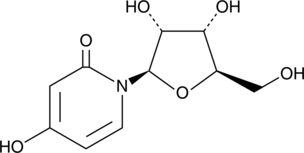
GC42260
3-Deazauridine
3-Deazauridine is a nucleoside analog.
-
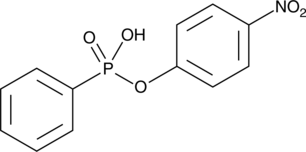
GC42460
4-Nitrophenyl Phenylphosphonate
4-Nitrophenyl phenylphosphonate is a substrate for 5'-nucleotide phosphodiesterases.
-
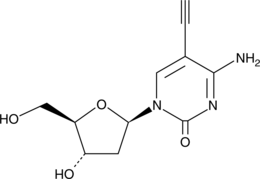
GC40641
5'-Ethynyl-2'-deoxycytidine
5'-Ethynyl-2'-deoxycytidine (EdC) is a nucleoside analog that inhibits replication of the herpes simplex virus-1 (HSV-1) KOS strain (ID50 = 0.2 μg/mL).
-
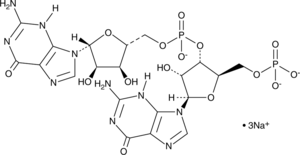
GC48375
5'-pGpG (sodium salt)
A linearized form of cyclic di-GMP
-
GC48662
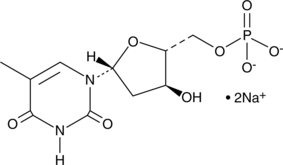
5'-Thymidylic Acid (sodium salt)
5'-Thymidylic Acid (sodium salt) is an endogenous metabolite.
-
GC40527
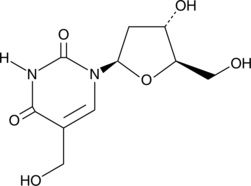
5-(Hydroxymethyl)-2'-deoxyuridine
5-(Hydroxymethyl)-2'-deoxyuridine is a nucleoside analog with anticancer and antiviral activities.
-
GC46681
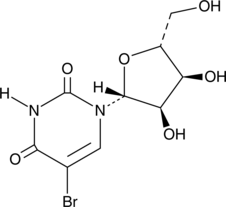
5-Bromouridine
A brominated uridine analog
-
GC45357
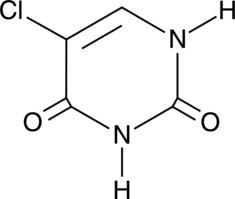
5-Chlorouracil
-
GC49245
5-Ethynyluridine
5-Ethynyluridine (5-EU) is a potent cell-permeable nucleoside can be used to label newly synthesized RNA.
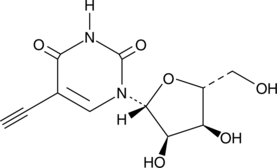
-
GC49551
6-Chloropurine Riboside
A nucleoside precursor

-
GC49561
7-Methylguanosine-d3
An internal standard for the quantification of 7-methylguanosine
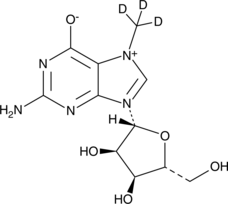
-
GC46805
Adefovir-d4
An internal standard for the quantification of adefovir
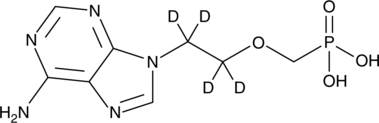
-
GC49004
Adenosine 3’-monophosphate (sodium salt hydrate)
A nucleotide
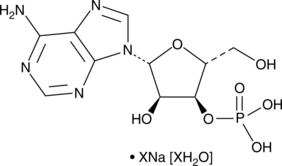
-
GC42732
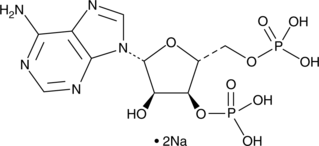
Adenosine 3',5'-diphosphate (sodium salt)
Adenosine 3',5'-diphosphate (sodium salt) is an hydroxysteroid sulfotransferases inhibitor.
-
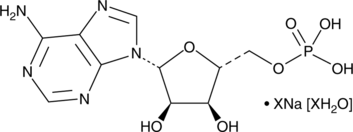
GC42733
Adenosine 5'-monophosphate (sodium salt hydrate)
Adenosine 5'-monophosphate (AMP) is a central nucleotide with functions in metabolism and cell signaling.
-
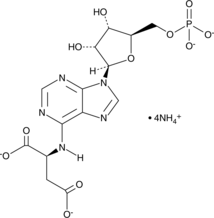
GC49231
Adenylosuccinic Acid (ammonium salt)
A purine nucleotide and an intermediate in the purine nucleotide cycle
-
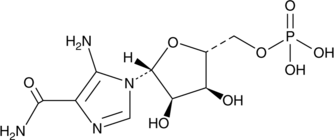
GC49110
AICA Ribonucleotide
An AMPK activator
-
GC19526
Cyclic di-GMP (sodium salt)
Cyclic di-GMP (sodium salt) is a STING agonist and a bacterial second messenger that coordinates different aspects of bacterial growth and behavior, including motility, virulence, biofilm formation, and cell cycle progression. Cyclic di-GMP (sodium salt) has anti-cancer cell proliferation activity and also induces elevated CD4 receptor expression and cell cycle arrest. Cyclic di-GMP (sodium salt) can be used in cancer research.
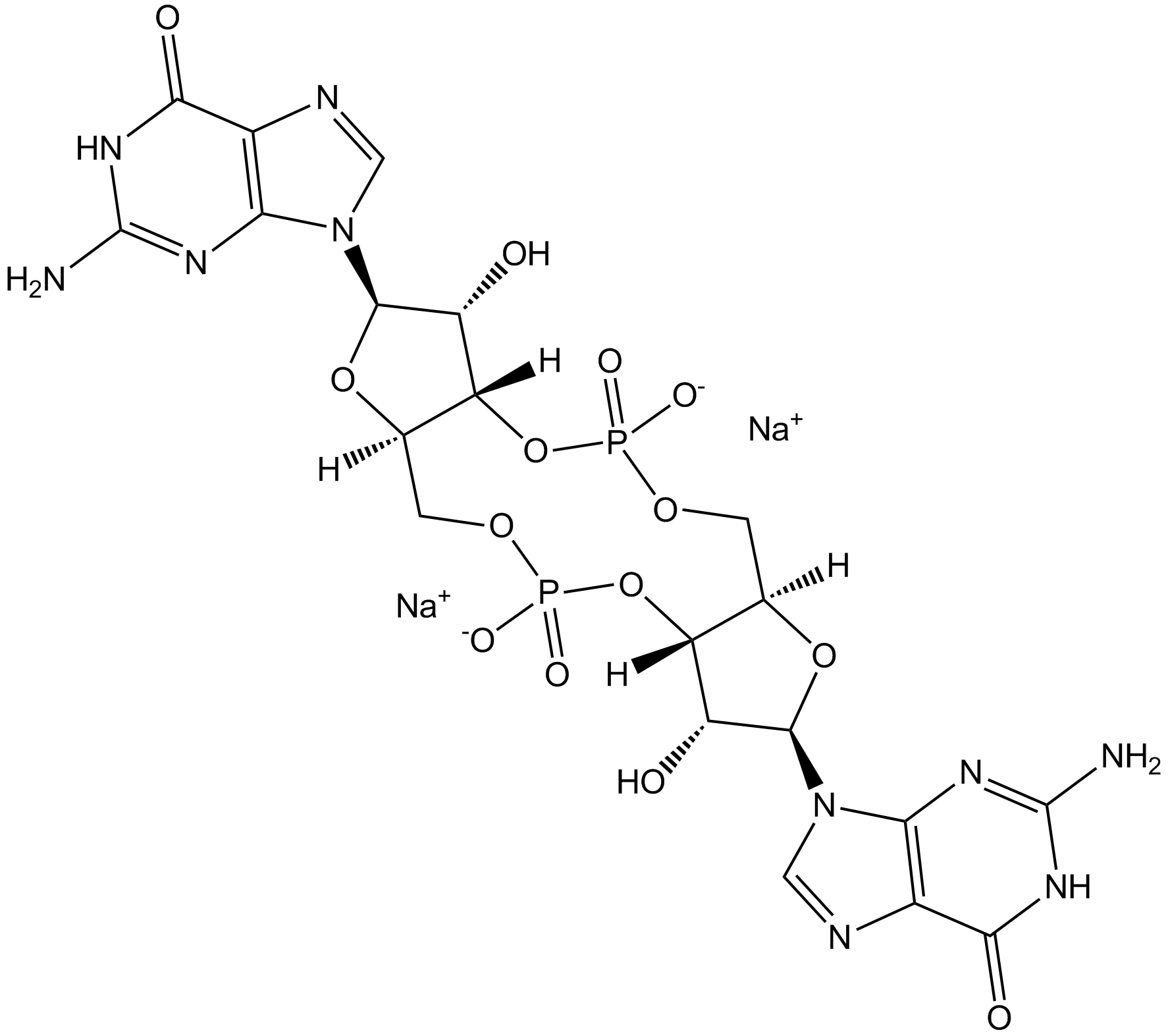
-
GC48397
Cyclic di-UMP (sodium salt)
A pyrimidine-containing CDN
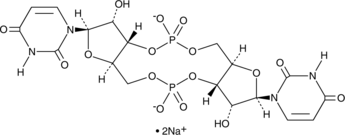
-
GC49104
Cytidine 3’-monophosphate
A ribonucleotide
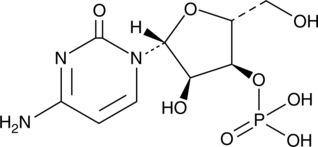
-
GC47162
Cytidine 5'-triphosphate (sodium salt hydrate)
A pyrimidine nucleoside triphosphate
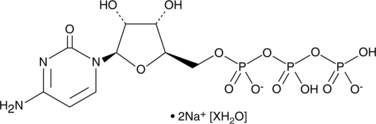
-
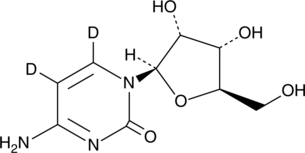
GC49526
Cytidine-d2
An internal standard for the quantification of cytidine
-
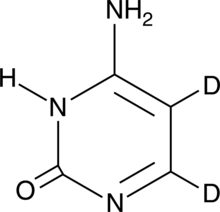
GC49464
Cytosine-d2
An internal standard for the quantification of cytosine
-
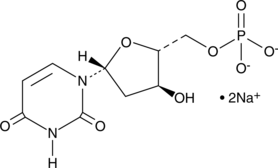
GC49225
Deoxyuridine 5′-monophosphate (sodium salt)
Deoxyuridine 5′-monophosphate (sodium salt) is reductively methylated to dTMP (2'-deoxythymidine 5'-monophosphate) by bisubstrate enzyme thymidylate synthase (TS).
-
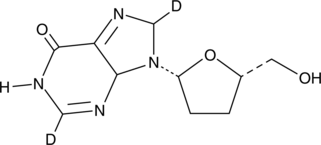
GC49022
Didanosine-d2
An internal standard for the quantification of didanosine
-
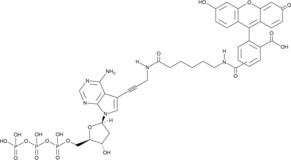
GC49455
Fluorescein-12-dATP
A fluorescently labeled form of dATP
-
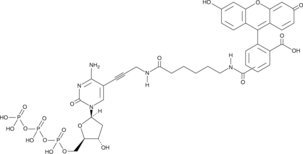
GC49746
Fluorescein-12-dCTP
A fluorescently labeled form of dCTP
-
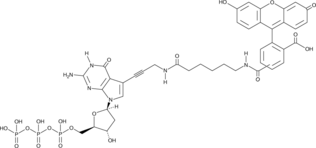
GC49456
Fluorescein-12-dGTP
A fluorescently labeled form of dGTP
-
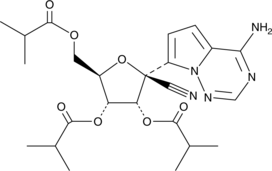
GC49165
GS-441524 tris-isobutyryl ester
A prodrug form of GS-441524
-
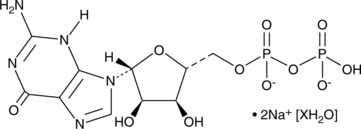
GC49033
Guanosine 5’-diphosphate (sodium salt hydrate)
A purine nucleotide
-
GC49703
Guanosine 5’-[γ-thio]triphosphate (lithium salt)
A hydrolysis-resistant analog of GTP
![Guanosine 5’-[γ-thio]triphosphate (lithium salt) Chemical Structure Guanosine 5’-[γ-thio]triphosphate (lithium salt) Chemical Structure](/media/struct/GC4/GC49703.png)
-
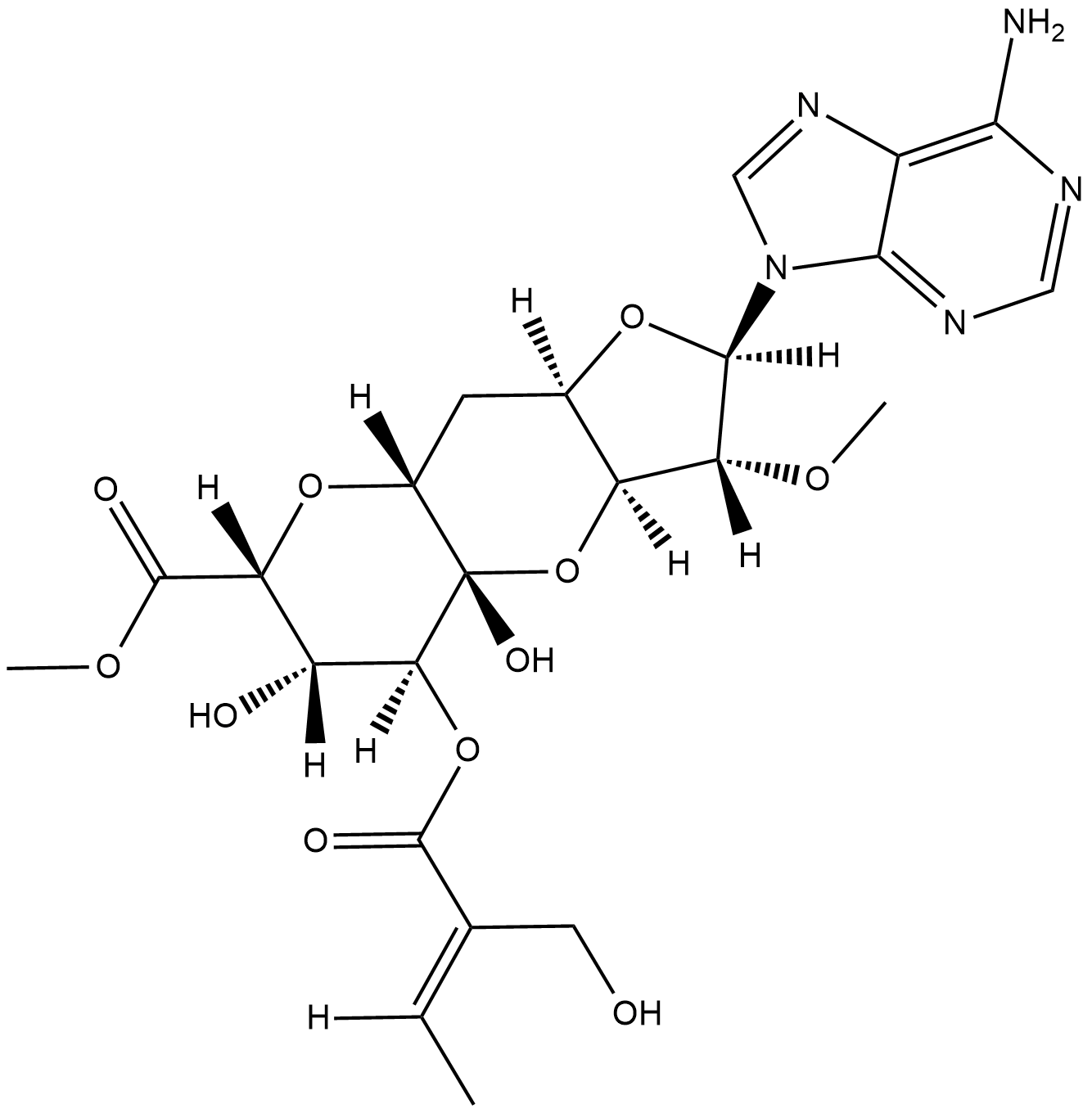
GC18796
Herbicidin A
Herbicidin A is an adenine nucleoside antibiotic originally isolated from S.
-
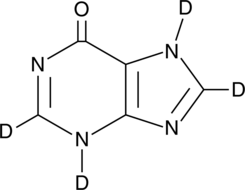
GC49479
Hypoxanthine-d4
An internal standard for the quantification of hypoxanthine
-
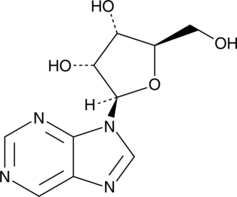
GC47760
Nebularine
A purine ribonucleoside with diverse biological activities
-
GC49395
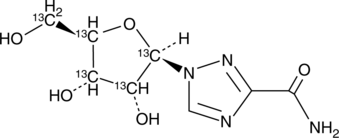
Ribavirin-13C5
An internal standard for the quantification of ribavirin
-
GC44836
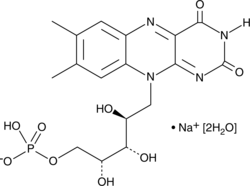
Riboflavin 5'-monophosphate (sodium salt hydrate)
Riboflavin 5'-monophosphate (FMN) is a coenzyme that is tightly bound to enzymes catalyzing oxidation and reduction reactions in a variety of biosynthetic pathways.
-
GC49529
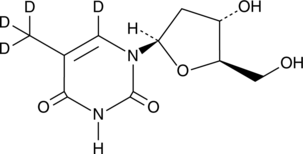
Thymidine-d4
An internal standard for the quantification of thymidine
-
GC49328
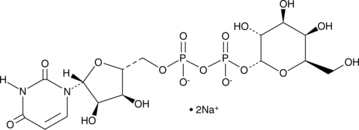
UDP-α-D-Galactose (sodium salt)
A nucleotide sugar
-
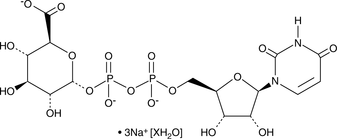
GC48991
UDP-Glucuronic Acid (sodium salt hydrate)
A nucleoside diphosphate sugar
-
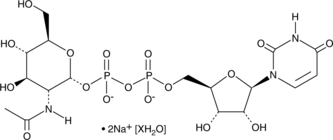
GC48990
UDP-N-acetyl-D-Glucosamine (sodium salt hydrate)
A nucleotide sugar
-
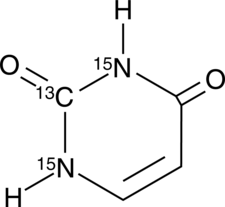
GC49132
Uracil-13C,15N2
An internal standard for the quantification of uracil
-
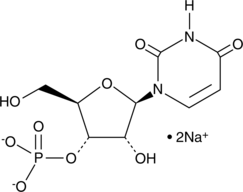
GC45127
Uridine-3'-monophosphate (sodium salt)
Uridine-3'-monophosphate is a nucleoside used in the biochemical synthesis of ribothymidine-3'-phosphate.
-
GC49297
Uridine-5’-monophosphate (sodium salt hydrate)
A ribonucleotide
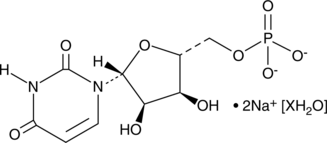
-
GC48228
Uridine-5'-diphosphoglucuronic Acid (sodium salt hydrate)
A nucleoside diphosphate sugar
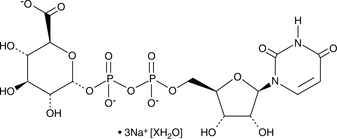
-
GC49528
Uridine-d2
Uridine-d2 is the deuterium labeled Uridine.