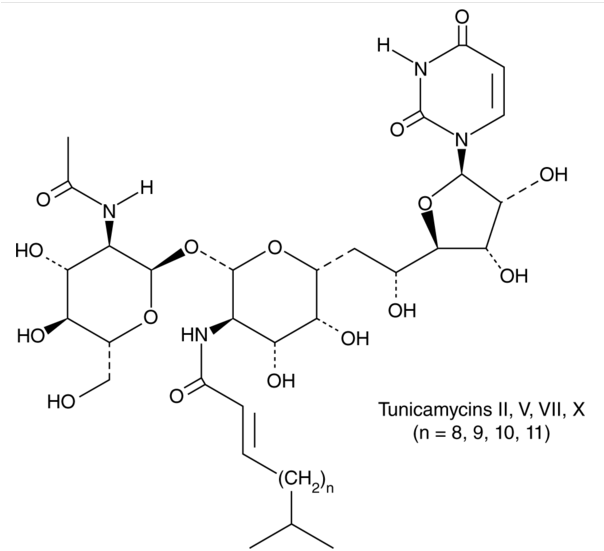
Tunicamycin
Numerous bacteria, including Streptomyces clavuligerus and Streptomyces lysosuperificus, are producers of tunicamycin. The antibacterial, antiviral, and anticancer properties of tunicamycins, which are naturally occurring nucleoside derivatives, are ascribed to their suppression of enzymatic processes involving polyisoprenyl phosphate and UDP-GlcNAc or UDP-MurNAc-pentapeptide. The diverse biological actions of tunicamycins make them promising candidates for use as therapeutic agents in the treatment of cancer or infectious disorders. A lipid chain, an undecodialdose skeleton, and a GlcNAc fragment connected by a 1,1-β,α-trehalose-type glycosidic bond make up the distinct structural makeup of tunicamycins. The molecular structure on tunicamycin is shown in Figure 1.
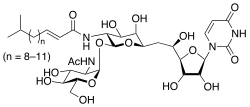
Figure 1: The molecular structure of Tunicamycin.
N- glycosylation, a frequent post-translational modification, significantly affects the stability and folding of proteins. N-glycosylation is an intricate process involving several different enzymes that takes place in the endoplasmic reticulum (ER). GlcNAc-1-P-transferase (GPT) is necessary to start the process of N-glycosylation in the ER. Tunicamycin is a naturally occurring substance that is used in research because it causes ER stress and inhibits N-glycosylation . LLO in humans is initiated by the enzyme GlcNAc-1-P-transferase (GPT), which belongs to the polyprenyl-phosphate N-acetylhexoamine 1-phosphate transferase (PNPT) family. Comprehending the structure of GPT is crucial in order to confirm its involvement in N-glycosylation. Thus far, the apo-form (PDB:5LEV and 6FM9) and UDP-GlcNAc-bound structures of GPT (PDB:6FWZ) have been identified by X-ray crystallography. Additionally, the binding structures of tunicamycin and GPT have been identified (PDB:5O5E, 6BW5, and 6BW6), meaning that a molecular explanation for the inhibitory action of tunicamycin against GPT is also possible .
Transmembrane helices (TMH) in the ER membrane make up GPT (Fig. 2A). There are five cytoplasmic loops (CLs) connecting TMH to one another. Particularly TMH1 and TMH2, as well as TMH9b and TMH10, have loops that are significantly more exposed to the ER lumen than other loops; these loops are essential for binding to substrates like tunicamycin and UDP-GlcNAc, and Fig. 2B shows these loops as "Loop A" and "Loop E," respectively .
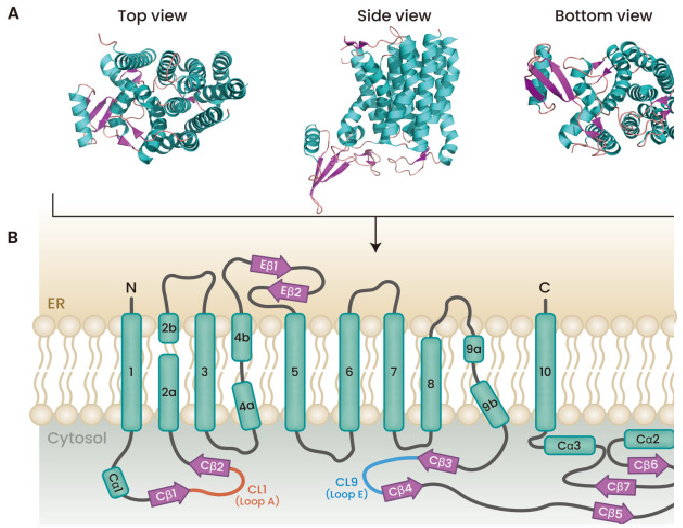
Figure 2: Structure of GPT from different views (PDB ID: 5LEV).
Tunicamycin is one of the inhibitors of PNPT superfamily enzymes; it also inhibits GPT activity, stops N-glycosylation, and causes ER stress in humans. Although tunicamycin's effects have long been known, its exact mode of action is still unknown. Nonetheless, the discovery of the complex structure involving GPT and tunicamycin allows for an investigation of the molecular underpinnings of the drug's mechanism of action .
The regions that TMH4, 5, 6, and 8 make up the binding region of tunicamycin are almost the same as the regions where UDP-GlcNAc binds to (Figure 3). Tunicamycin's uridine moiety is covered by CL1 (Loop A), and its GlcNAc component—more precisely, UDP-GlcNAc—is covered by Loop E. Similar to the interaction between GPT and UDP-GlcNAc, the uridine moiety of tunicamycin and GPT had a similar relationship. The uridine moiety of tunicamycin's hydrogen bond network and its π-π stacking interaction with F249 were found to be identical to those of UDP-GlcNAc (Figure 3). However, there are notable differences between tunicamycin and UDP-GlcNAc concerning the GlcNAc moiety. H302 and R303 both assisted in binding to UDP-GlcNAc, but only R303 contributed to binding to the GlcNAc moiety of tunicamycin. Nevertheless, Loop E approaches GlcNAc in the same way as UDP-GlcNAc due to the interaction between tunicamycin and R303 (Figure 3).
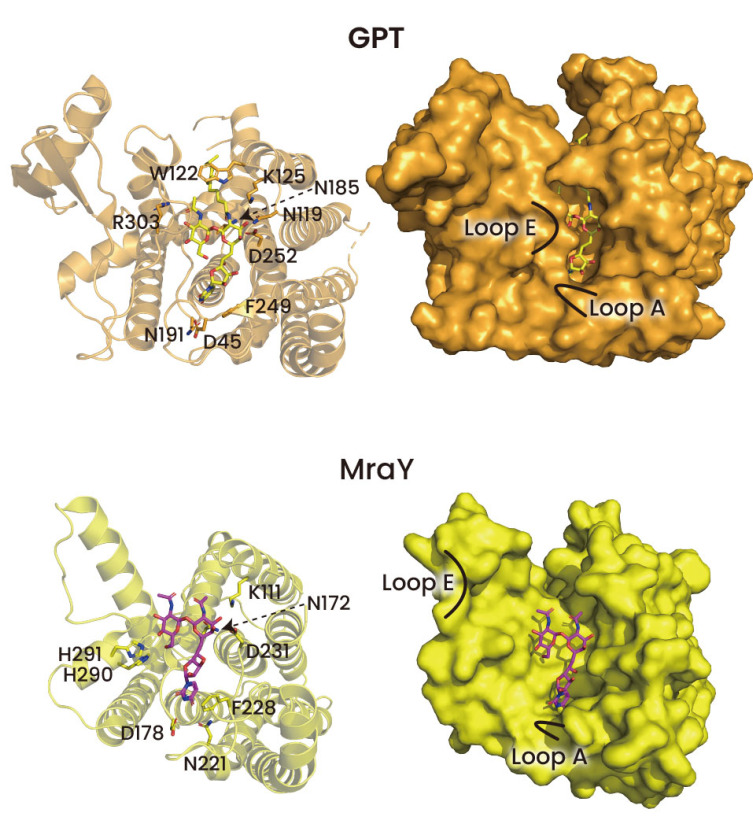
Figure 3: To demonstrate the binding of tunicamycin to GPT and MraY, a surface model and cartoon were utilized (PDB ID: 6BW5 and 5JNQ). Tunicamycin is indicated by yellow magenta in GPT and MraY. The structure of MraY is complicated, and it did not display the lipid tail of tunicamycin.
Overall, the interactions between the uridine and GlcNAc moieties, which are similar to those between UDP-GlcNAc and GlcNAc, are very different from those between the tunicamine moiety of tunicamycin and GPT. The interaction formed in the tunicamine moiety of tunicamycin may only be different from that of UDP-GlcNAc as the pyrophosphate moiety of UDP-GlcNAc is substituted by the tunicamine moiety of tunicamycin (Fig. 4). This is due to the fact that pyrophosphate and tunicamine have quite different structures. First, as can be seen in Figure 3, the tunicamine moiety is not in contact with R301, which is in contact with UDP-GlcNAc's pyrophosphate. But as was already established, even in the case that R301 and the pyrophosphate in UDP-GlcNAc do not connect, Loop E approaches GlcNAc because R303 interacts with the GlcNAc of tunicamycin. The most prominent alteration in the tunicamine moiety is that the Mg2+ ion that is involved in the binding of GPT and UDP-GlcNAc was not seen in the binding of GPT and tunicamycin (Figs. 3 and 4). The placement of the Mg2+ ion in the binding structures of UDP-GlcNAc and GPT and the tunicamine moiety in the binding structures of GPT and tunicamycin are identical (Fig. 4). In this case, Mg2+ did not act as a mediator in the interactions between the tunicamine moiety and N185 and D252. This structural alteration contradicts previous expectations regarding tunicamycin's need on Mg2+ when linked to GPT . It's noteworthy to note that MraY and tunicamycin interactions are Mg2+ ion-dependent, in contrast to GPT. This was corroborated by the discovery of a mutation that altered MraY's Asp residue, which interacts with the Mg2+ ion, to Ala. This mutation led to a significant decrease in the binding affinity of tunicamycin to MraY, but not to GPT . These findings imply that Mg2+ contributes differently to GPT and MraY's tunicamycin binding.
The complex interaction between the tunicamycin and GPT structures shows that the latter decreases GPT activity. Since tunicamycin's molecular structure resembles that of GPT substrates DolP and UDP-GlcNAc, it can attach to the GPT active site similarly to those substrates. Furthermore, the structure of the GPT-bound tunicamycin is remarkably similar to that of UDP-GlcNAc. These protein structures show how the GPT substrates DolP and UDP-GlcNAc are in competition with tunicamycin to reduce GPT activity. The Kd values of pure GPT protein and tunicamycin are roughly 6 nM, indicating that the drug can effectively inhibit the activity of GPT through competitive interactions with UDP-GlcNAc and DolP .
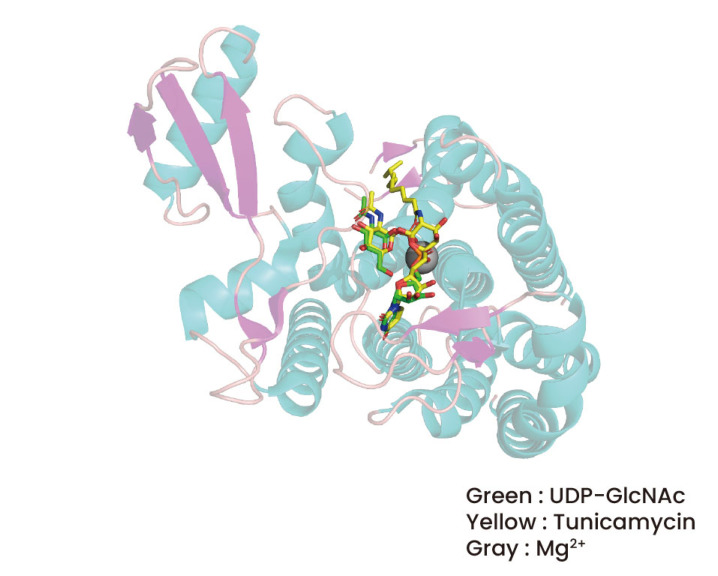
Figure 4: The comparison of tunicamycin binding and UDP-GlcNAc binding to GPT.
GPT functions as a "off-target" of tunicamycin, which is why it hasn't been utilized in clinical practice despite the fact that tunicamycin is a naturally occurring chemical with enticing antibacterial properties. Therefore, in order to establish tunicamycin as a next-generation antibiotic, it must be developed in a way that allows it to selectively act on MraY and not GPT. Based on the previously discovered structures of GPT and MraY, tricainemycin compounds with high selectivity exclusively for MraY are constantly being developed .
Studies have shown that tunicamycin has anticancer properties in addition to antibiotics . It was established that tunicamycin exhibited anticancer properties in a number of malignancies, including head and neck cancer and stomach cancer. Tunicamycin efficiently inhibits N-glycosylation in cancer cells, which causes significant ER stress and anticancer effects. Because of this, even though tunicamycin is now difficult to utilize as an anticancer drug due to its toxicity, there is a good chance that it will develop into a novel anticancer agent because it has demonstrated high anticancer potential. It is important to look into the anticancer effects of tunicamycin in patients who are resistant to anticancer therapy since anticancer medications that inhibit N-glycosylation and have anticancer benefits have not yet been used in cancer patients.
In summary, tunicamycin is an intriguing substance with an intriguing past that has significantly advanced our knowledge of glycoproteins, cellular stress responses, and cellular processes. Its uses across a wide range of scientific fields are still growing, making it a vital resource for scientists trying to solve the cellular secrets of life.










Comments