Infectious Disease
Products for Infectious Disease
- Cat.No. Product Name Information
-
GC52351
Citrullinated α-Enolase (R8 + R14) (1-19)-biotin Peptide
A biotinylated and citrullinated α-enolase peptide

-
GC66467
Claficapavir
Claficapavir (A1752) is a specific nucleocapsid protein (NC) inhibitor with an IC50 around 1 μM. Claficapavir strongly binds the HIV-1 NC (Kd=20 nM) thereby inhibiting the chaperone properties of NC and leading to good antiviral activity against the HIV-1.
-
GC49852
Clindamycin (hydrochloride hydrate)
Clindamycin (hydrochloride hydrate) is an oral protein synthesis inhibitory agent that has the ability to suppress the expression of virulence factors in Staphylococcus aureus at sub-inhibitory concentrations (sub-MICs).
-
GC43278
Clindamycin Sulfoxide
Clindamycin sulfoxide is an active metabolite of the antibiotic clindamycin.
-
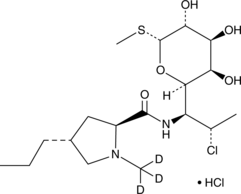
GC52171
Clindamycin-d3 (hydrochloride)
Clindamycin-d3 (hydrochloride) is the deuterium labeled Clindamycin.
-
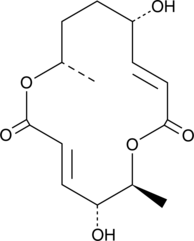
GC47105
Clonostachydiol
A fungal metabolite with anticancer and anthelmintic activities
-
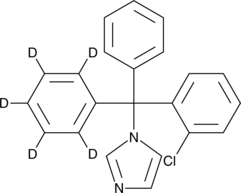
GC47109
Clotrimazole-d5
An internal standard for the quantification of clotrimazole
-
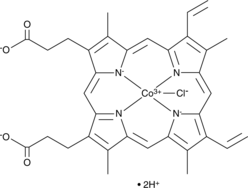
GC49096
Cobaltic Protoporphyrin IX (chloride)
An inducer of HO-1 activity
-
GC43298
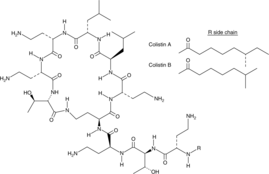
Colistin
Colistin is a complex antibiotic containing greater than 30 components, with the cyclic polypeptide antibiotics polymyxins E1 (colistin A) and E2 (colistin B) as the major components.
-
GC46122
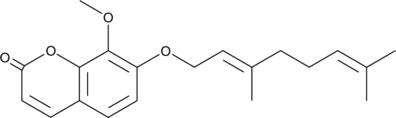
Collinin
A coumarin with diverse biological activities
-
GC43314
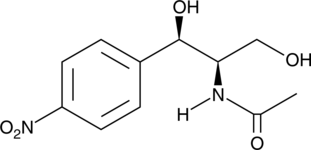
Corynecin I
Corynecin I is a chloramphenicol-like antibiotic originally isolated from Corynebacterium.
-
GC40079
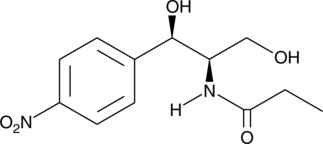
Corynecin II
Corynecin II is a chloramphenicol-like antibiotic originally isolated from Corynebacterium.
-
GC40083
Corynecin III
Corynecin III is a chloramphenicol-like antibiotic originally isolated from Corynebacterium.
-
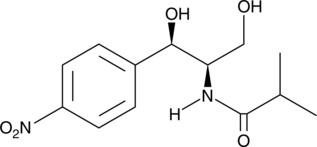
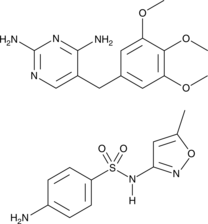
GC47121
Cotrimoxazole
A mixture of the antibiotics sulfamethoxazole and trimethoprom
-
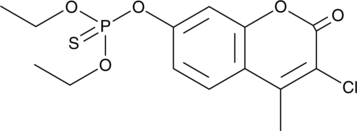
GC47122
Coumaphos
An organophosphate pesticide
-
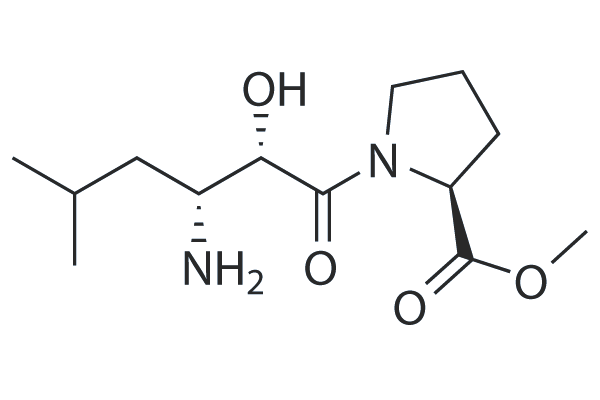
GC25306
CQ31
CQ31, a small molecule, selectively activates caspase activation and recruitment domain-containing 8 (CARD8).
-
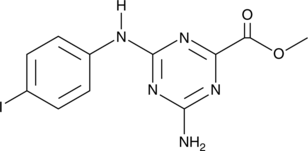
GC47128
CU-32
A cGAS inhibitor
-
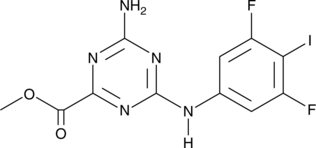
GC47129
CU-76
A cGAS inhibitor
-
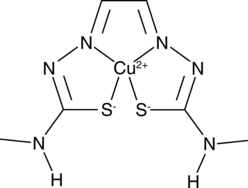
GC49557
Cu-GTSM
A copper-containing compound with diverse biological activities
-
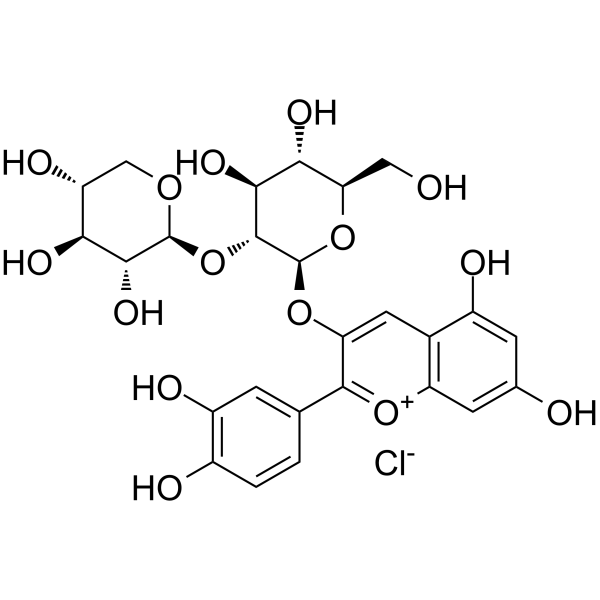
GC64348
Cyanidin 3-sambubioside chloride
Cyanidin 3-sambubioside chloride (Cyanidin-3-O-sambubioside chloride), a major anthocyanin, a natural colorant, and is a potent NO inhibitor.
-
GC49821
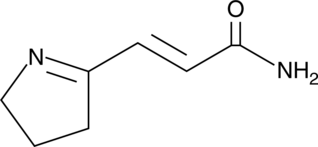
Cyclamidomycin
A bacterial metabolite with antibiotic activity
-
GC43339
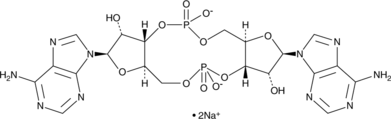
Cyclic di-AMP (sodium salt)
Cyclic di-AMP (c-di-AMP) is a second messenger produced by bacteria but not by mammals.
-
GC19526
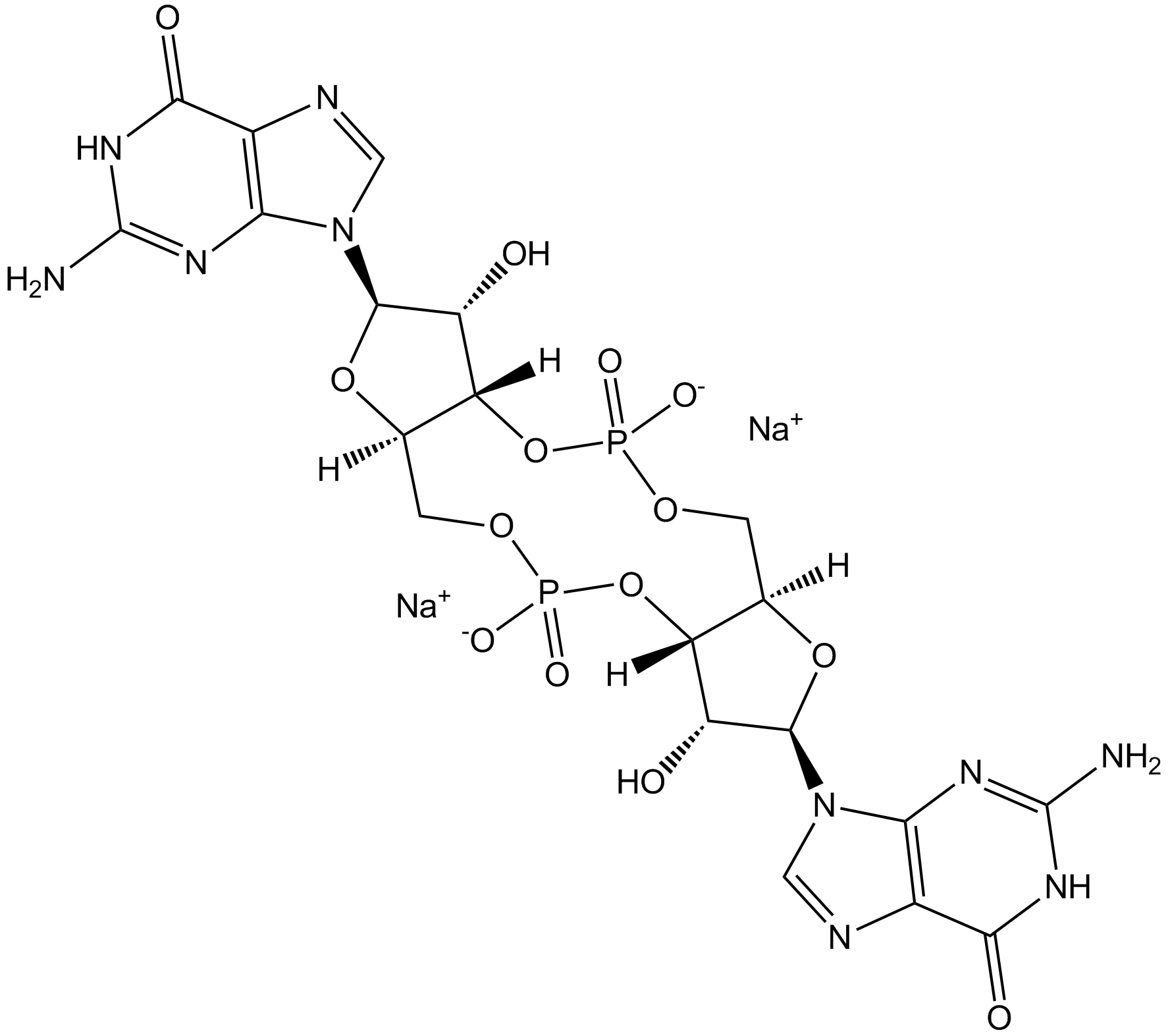
Cyclic di-GMP (sodium salt)
Cyclic di-GMP (sodium salt) is a STING agonist and a bacterial second messenger that coordinates different aspects of bacterial growth and behavior, including motility, virulence, biofilm formation, and cell cycle progression. Cyclic di-GMP (sodium salt) has anti-cancer cell proliferation activity and also induces elevated CD4 receptor expression and cell cycle arrest. Cyclic di-GMP (sodium salt) can be used in cancer research.
-
GC43340
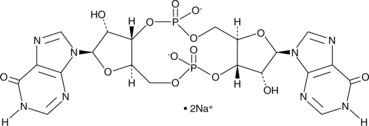
Cyclic di-IMP (sodium salt)
Cyclic di-IMP (sodium salt) (c-di-IMP) is a synthetic second messenger structurally related to the bacterial second messengers cyclic di-GMP and cyclic di-AMP.
-
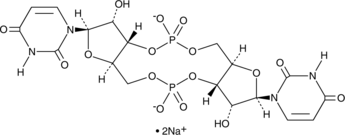
GC48397
Cyclic di-UMP (sodium salt)
A pyrimidine-containing CDN
-
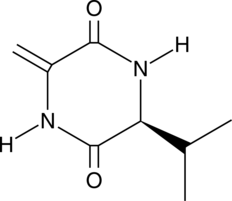
GC41176
Cyclo(δ-Ala-L-Val)
Cyclo(δ-Ala-L-Val) is a bacterial cyclic dipeptide characterized as a diketopiperazine.
-
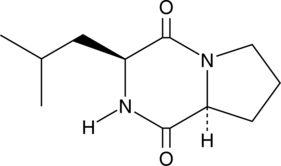
GC43342
Cyclo(L-Leu-L-Pro)
Cyclo(L-Leu-L-Pro) is a diketopiperazine metabolite that has been isolated from various bacterial and fungal species including Streptomyces.
-
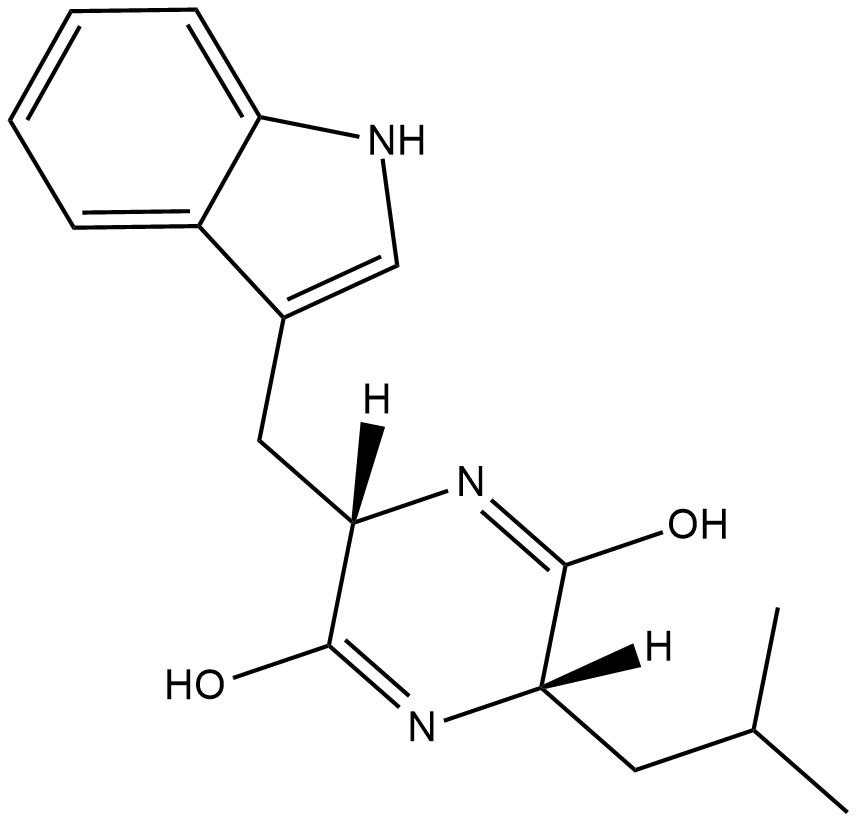
GC18488
Cyclo(L-Leu-L-Trp)
Cyclo(L-Leu-L-Trp) is a diketopiperazine metabolite originally isolated from Penicillium.
-
GC41555
Cyclo(L-Phe-L-Val)
Cyclo(L-Phe-L-Val) is a metabolite of the sponge bacterium Pseudoalteromonas sp.
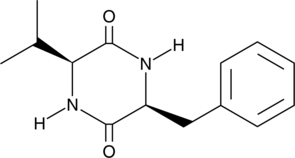
-
GC18268
Cyclo(L-Trp-L-Trp)
Cyclo(L-Trp-L-Trp) is a cyclic dipeptide that has been used as a substrate for indole prenyltransferases in the synthesis of mono- and diprenylated indolines.
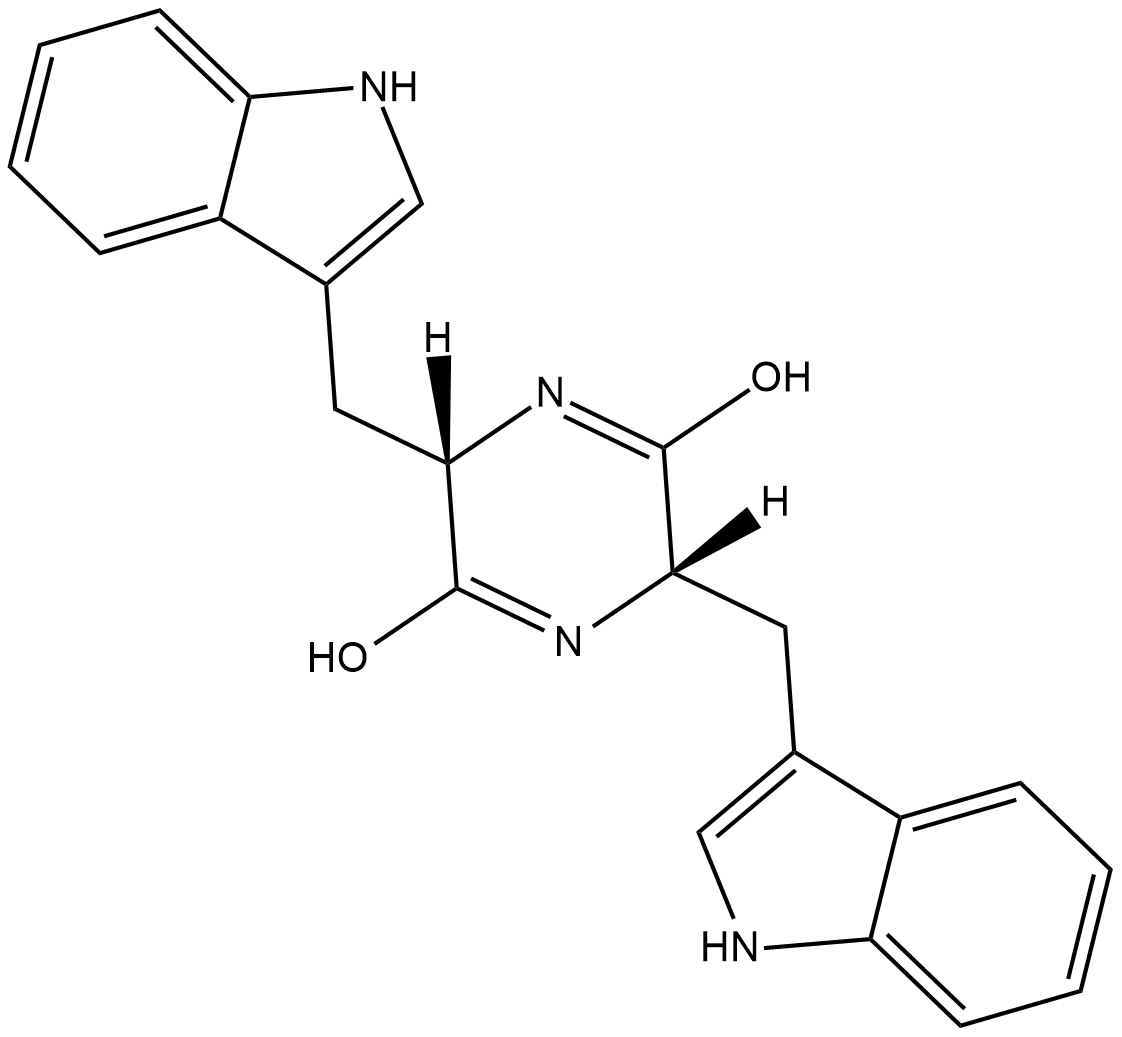
-
GC40569
Cyclopenin
Cyclopenin is an inhibitor of acetylcholinesterase (AChE; IC50 = 2.04 μM for human recombinant AChE) that is produced by Penicillium.
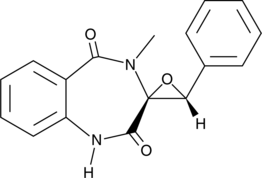
-
GC64068
Cyclotriazadisulfonamide
Cyclotriazadisulfonamide (CADA) is a specific CD4-targeted HIV entry inhibitors.
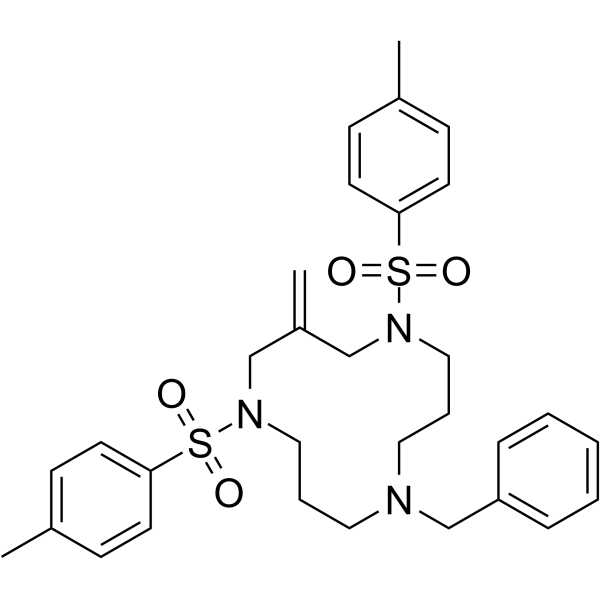
-
GC43368
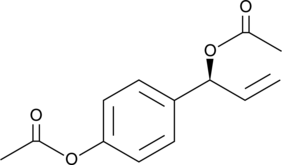
D,L-1′-Acetoxychavicol Acetate
D,L-1′-Acetoxychavicol acetate is a natural compound first isolated from the rhizomes of ginger-like plants.
-
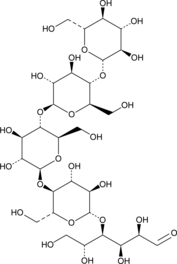
GC49792
D-(+)-Cellopentaose
D-(+)-Cellopentaose is a penta oligosaccharide of cellulose.
-
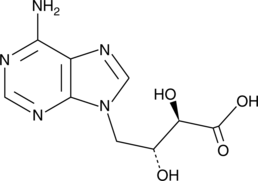
GC40294
D-Eritadenine
D-Eritadenine is an adenosine analog and a potent, reversible inhibitor of S-adenosylhomocysteine hydrolase (SAAH; IC50 = 7 nM).
-
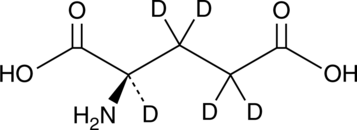
GC49378
D-Glutamic Acid-d5
An internal standard for the quantification of D-glutamic acid
-
GC47246
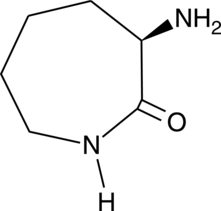
D-Lysine lactam
A chiral building block
-
GC47266
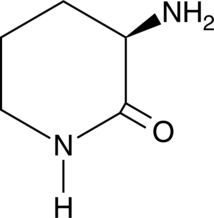
D-Ornithine lactam
A building block
-
GC47270
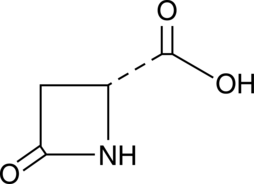
D-Pyroaspartic Acid
A synthetic intermediate
-
GC46124
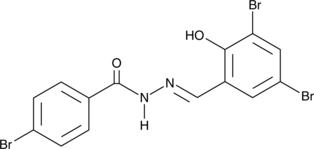
D13
An acylhydrazone antifungal
-
GC45883
Daclatasvir-d6
An internal standard for the quantification of daclatasvir
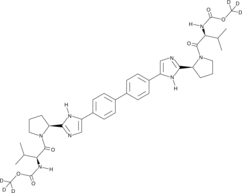
-
GC43386
DDD85646
DDD85646 is a moderately bioavailable pyrazole sulphonamide inhibitor of T.
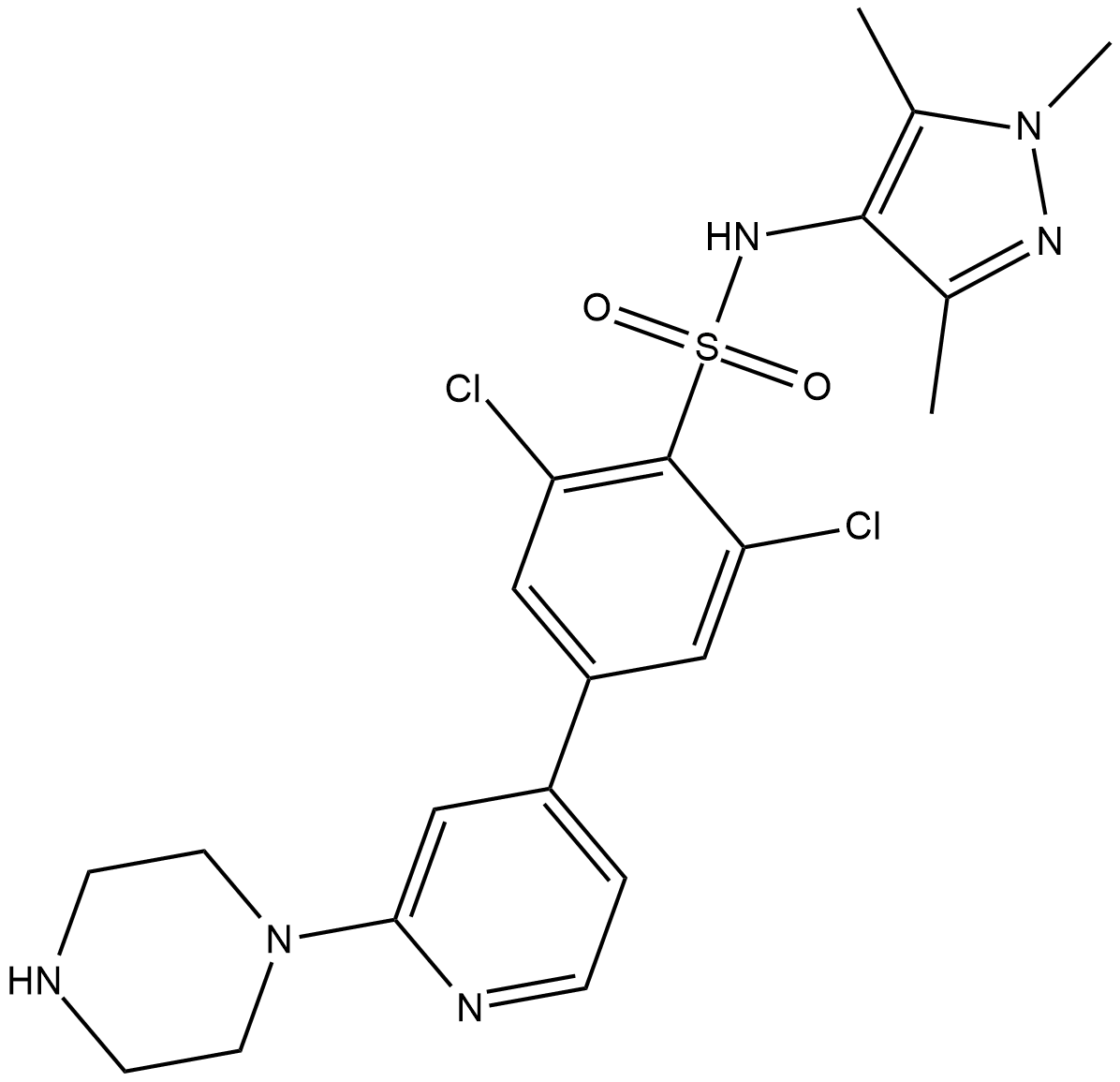
-
GC64391
DDX3-IN-2
DDX3-IN-2 is an active DEADbox polypeptide 3 (DDX3) inhibitor with an IC50 value of 0.3 μM.
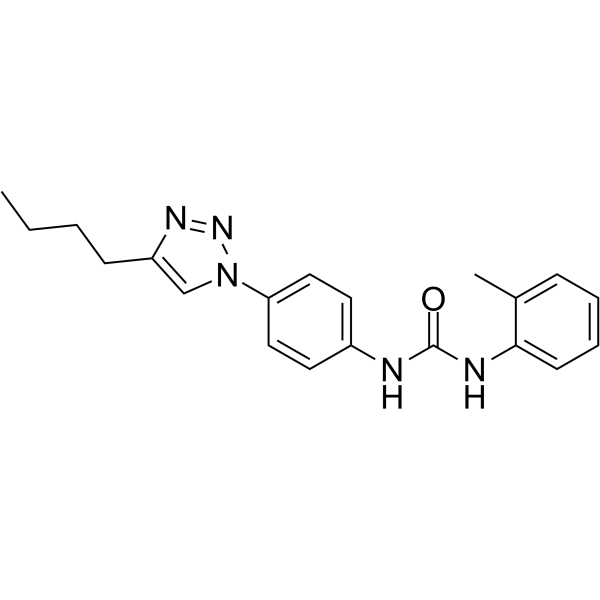
-
GC52191
Deacetylanisomycin
Deacetylanisomycin is a potent growth regulator in plants and an inactive derivative of Anisomycin.
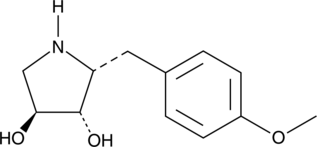
-
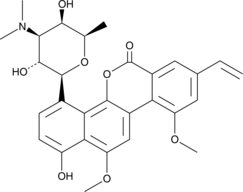
GC43390
Deacetylravidomycin
Deacetylravidomycin is a microbial metabolite that has been found in Streptomyces and has light-dependent antibiotic and anticancer activities.
-
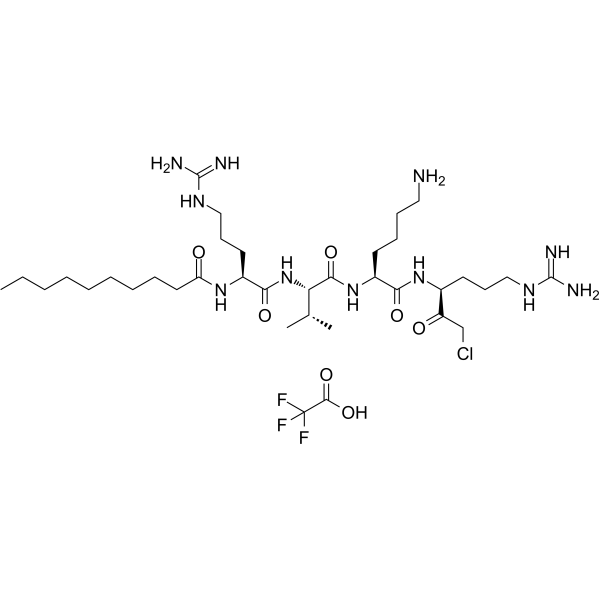
GC64270
Decanoyl-RVKR-CMK TFA
Decanoyl-RVKR-CMK (DecRVKRcmk) TFA inhibits over-expressed gp160 processing and HIV-1 replication.
-
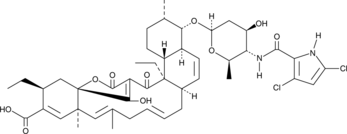
GC43394
Decatromicin B
Decatromicin B is a bacterial metabolite that has been found in Actinomadura.
-
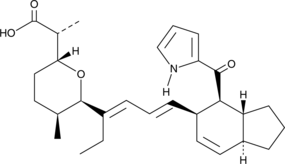
GC43395
Deethylindanomycin
Deethylindanomycin is a polyether antibiotic that has been found in S.
-
GC43398
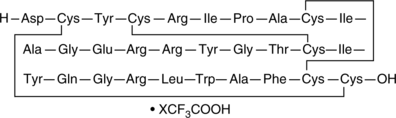
Defensin HNP-3 (human) (trifluoroacetate salt)
Defensin HNP-3 is a peptide secreted by human polymorphonuclear leukocytes (PMNs) that has antimicrobial properties.
-
GC43408
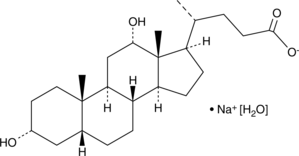
Deoxycholic Acid (sodium salt hydrate)
Deoxycholic acid (cholanoic acid) sodium hydrate,a bile acid, is a by-product of intestinal metabolism, that activates the G protein-coupled bile acid receptorTGR5.
-
GC47187
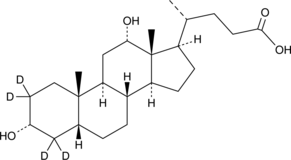
Deoxycholic Acid-d4
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
GC18869
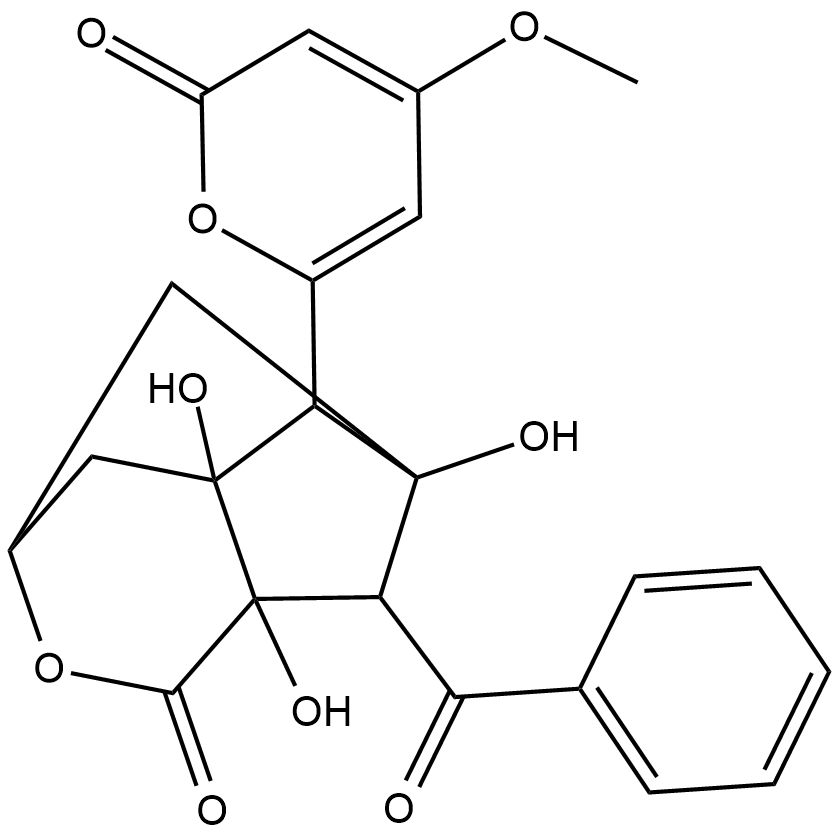
Deoxyenterocin
Deoxyenterocin is a bacterial metabolite originally isolated from Streptomyces that has diverse biological activities, including antibiotic, antiviral, and antioxidant properties.
-
GC41092
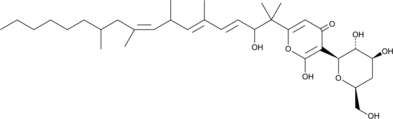
Deoxyfusapyrone
Deoxyfusapyrone is an α-pyrone fungal metabolite originally isolated from F.
-
GC40654
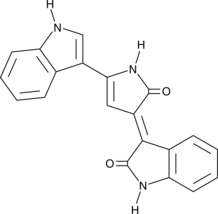
Deoxyviolacein
Deoxyviolacein is a bacterial metabolite and byproduct in the biosynthesis of the bisindole alkaloid violacein that has anticancer, antibacterial, and antifungal properties.
-
GC49115
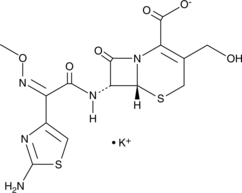
Desacetylcefotaxime (potassium salt)
An active metabolite of cefotaxime
-
GC43417
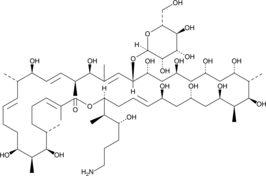
Desertomycin A
Desertomycin A is fungal metabolite and the major component of the desertomycin antibiotic complex that has been found in S.
-
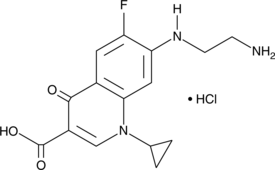
GC43418
Desethylene Ciprofloxacin (hydrochloride)
Desethylene ciprofloxacin is a major metabolite of ciprofloxacin.
-
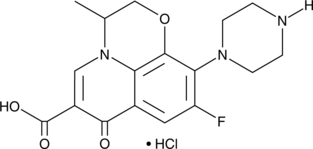
GC49100
Desmethyl Ofloxacin (hydrochloride)
A metabolite of ofloxacin
-
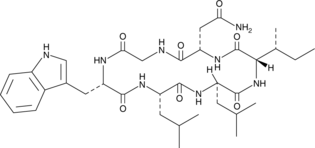
GC43424
Desotamide
Desotamide is a cyclic hexapeptide antibiotic originally isolated from Streptomyces.
-
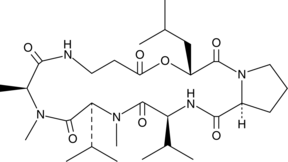
GC46130
Destruxin B2
A mycotoxin with antiviral, insecticidal, and phytotoxic activities
-
GC45992
Diallyl Tetrasulfide
An organosulfur compound with diverse biological activities
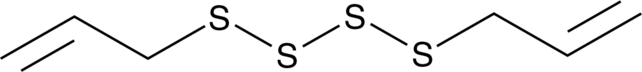
-
GC49153
Didemnin B
Didemnin B is a cyclic depsipeptide produced by marine tunicates that specifically binds the GTP-bound conformation of EEF1A.
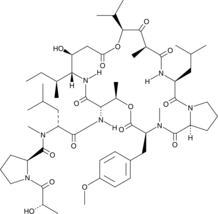
-
GC45662
Dieugenol
A neolignan with antioxidative and antiparasitic activities
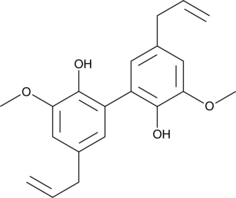
-
GC47220
Difloxacin-d3
An internal standard for the quantification of difloxacin
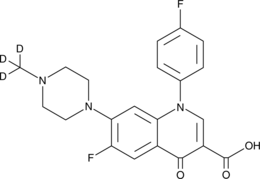
-
GC40063
Dihydroaeruginoic Acid
Dihydroaeruginoic acid is an antibiotic originally isolated from P.
-
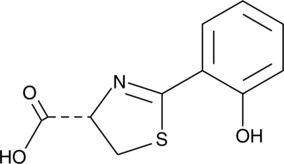
GC47228
Dihydronovobiocin
A coumarin antibiotic
-
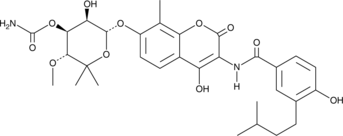
GC41162
Dihydropleuromutilin
Dihydropleuromutilin is a semisynthetic antibiotic and derivative of pleuromutilin.
-
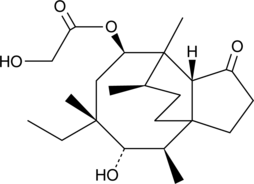
GC43467
Dimethyldioctadecylammonium (bromide)
Dimethyldioctadecylammonium (DDA) is a cationic amphipathic lipid.
-
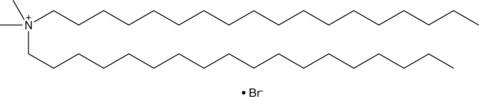
GC52082
Dimidium (bromide)
A trypanocide
-
GC43488
DL-erythro/threo Sphinganine (d16:0)
DL-erythro/threo Sphinganine (d16:0) is a sphingolipid that is decreased in rats following long-term, low-dose administration of dimethoate and is used as a biomarker for dimethoate exposure.
-
GC49383
DL-Phenyl-d5-alanine
An internal standard for the quantification of DL-phenylalanine
-
GC52120
DMPE-MPEG(2000)
A PEGylated form of DMPE
-
GC45995
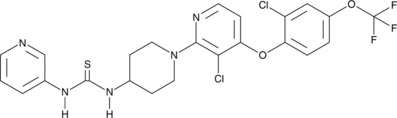
DO264
An ABHD12 inhibitor
-
GC47257
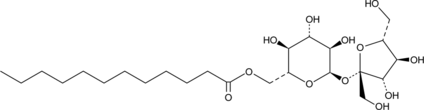
Dodecanoyl D-Sucrose
A nonionic surfactant
-
GC52095
Dodecyltrimethylammonium (bromide)
A cationic surfactant

-
GC43562
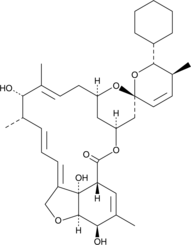
Doramectin aglycone
Doramectin aglycone is an acid degradation product of doramectin, a disaccharide-containing anthelmintic that potentiates glutamate- and GABA-gated chloride channel opening in nematodes, and its acid-catalyzed hydrolysis product doramectin monosaccharide.
-
GC43563
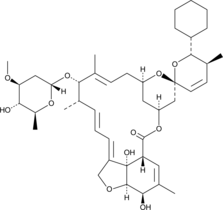
Doramectin monosaccharide
Doramectin monosaccharide is an acid degradation product of doramectin, a disaccharide-containing anthelmintic that potentiates glutamate- and GABA-gated chloride channel opening in nematodes.
-
GC46135
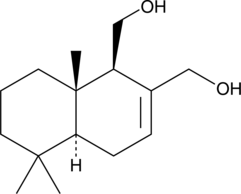
Drimendiol
A sesquiterpene
-
GC43572
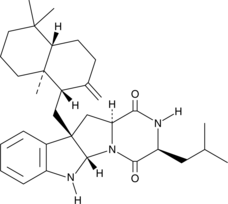
Drimentine A
Drimentine A is a terpenylated diketopiperazine antibiotic originally isolated from Actinomycete bacteria.
-
GC40082
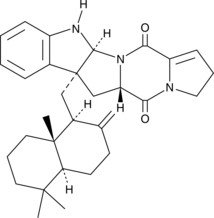
Drimentine B
Drimentine B is a terpenylated diketopiperazine antibiotic originally isolated from Actinomycete bacteria.
-
GC40059
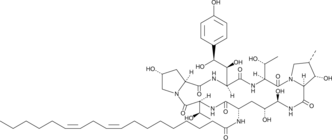
Echinocandin B
Echinocandin B is an echinocandin antifungal originally isolated from Aspergillus.
-
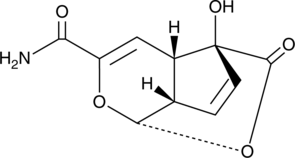
GC47279
Echinosporin
A bacterial metabolite with antibacterial and anticancer activities
-
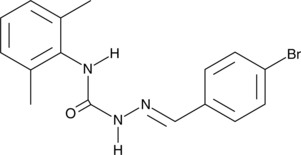
GC43588
EGA
EGA is an inhibitor of endosomal trafficking.
-
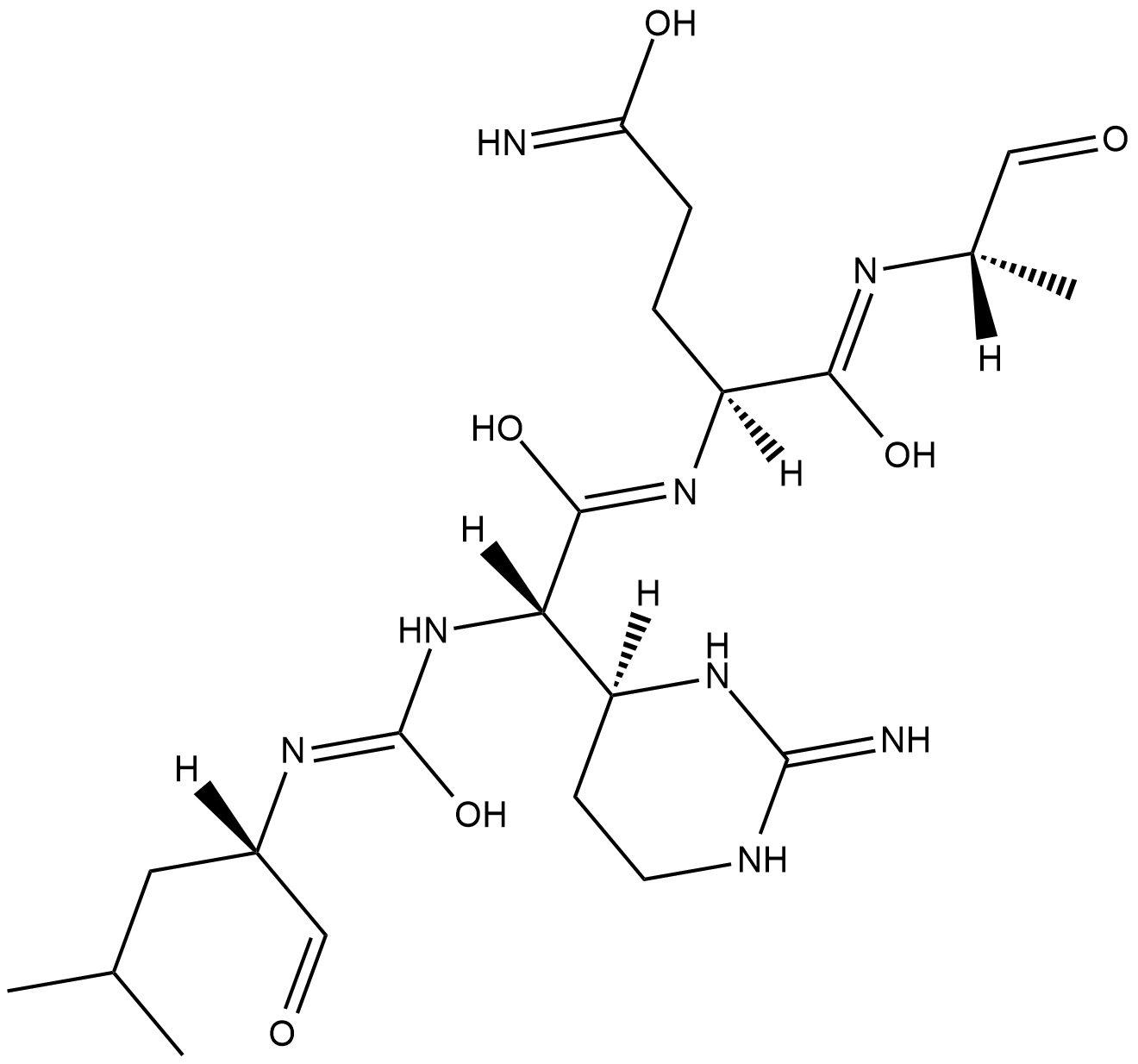
GC18191
Elastatinal
Elastatinal is a potent inhibitor of pancreatic elastase (Ki = 240 nM) that is produced by various species of Actinomycetes.
-
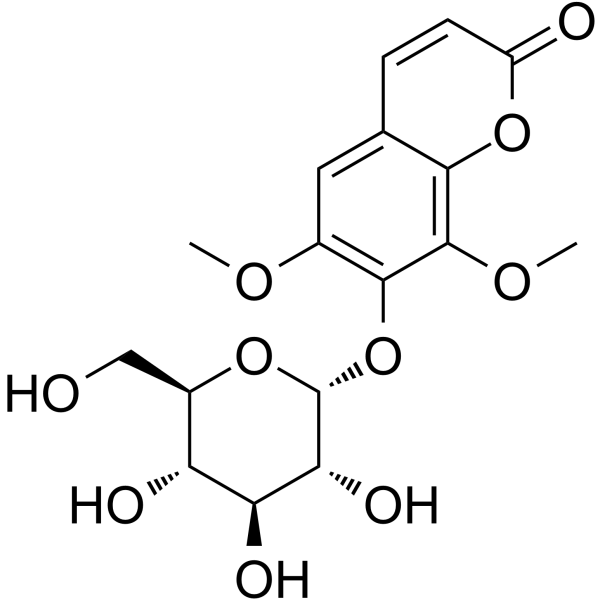
GC63987
Eleutheroside B1
Eleutheroside B1, a coumarin compound, has a wide spectrum of anti-human influenza virus efficacy, with an IC50 value of 64-125μg/ml.
-
GC48546
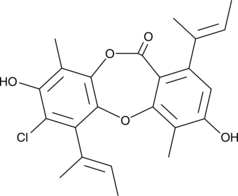
Emeguisin A
A fungal metabolite
-
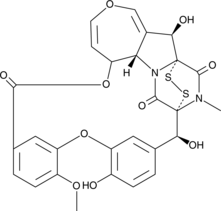
GC46137
Emestrin
A mycotoxin
-
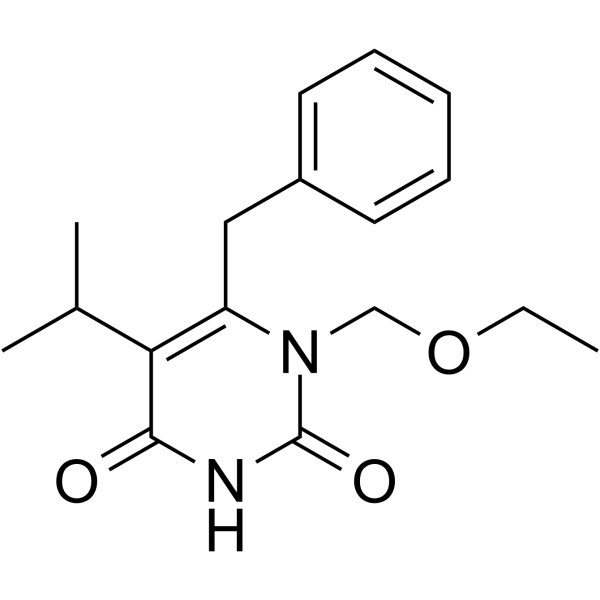
GC64484
Emivirine
Emivirine (MKC-442) is a non-nucleoside reverse transcriptase inhibitors (NNRTIs) with Ki values of 0.20 and 0.01 μM for dTTP- and dGTP-dependent DNA or RNA polymerase activity, respectively.
-
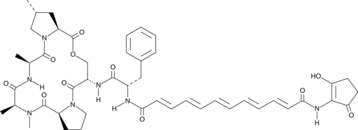
GC43612
Enopeptin A
Enopeptin A, originally isolated from a culture broth of Streptomyces sp.
-
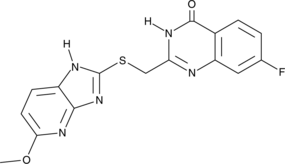
GC52334
ENPP1 Inhibitor 4e
An ENPP1 inhibitor
-
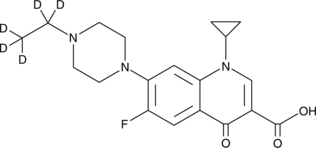
GC49101
Enrofloxacin-d5
An internal standard for the quantification of enrofloxacin
-
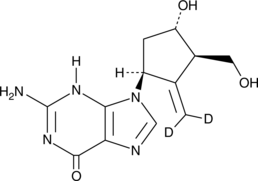
GC52261
Entecavir-d2
An internal standard for the quantification of entecavir
-
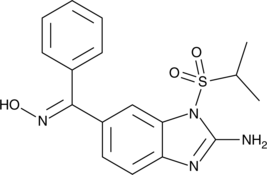
GC47295
Enviroxime
An antiviral agent
-
GC41326
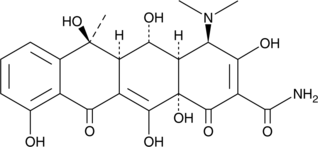
Epioxytetracycline
Epioxytetracycline, the degradation product of Oxytetracycline (OTC), can be found in swine manure compost and wastewater.
-
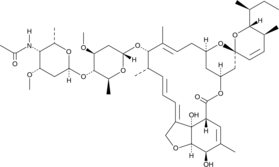
GC40845
Eprinomectin B1a
Eprinomectin B1a is a major component (>90%) of the antiparasitic compound eprinomectin, which also contains eprinomectin B1b and belongs to the avermectin family of insecticides and anthelmintics.
-
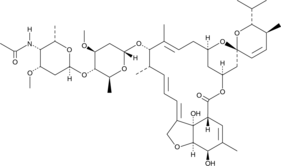
GC43622
Eprinomectin B1b
Eprinomectin B1b is a minor component and belongs to the avermectin family of insecticides and anthelmintics.
-
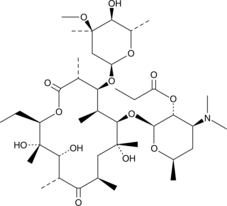
GC48372
Erythromycin 2'-Propionate
An antibiotic
-
GC43626
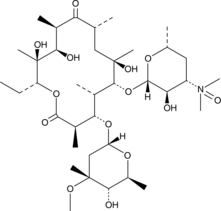
Erythromycin A N-oxide
Erythromycin A N-oxide is a potential impurity found in commercial preparations of erythromycin.
-
GC18499
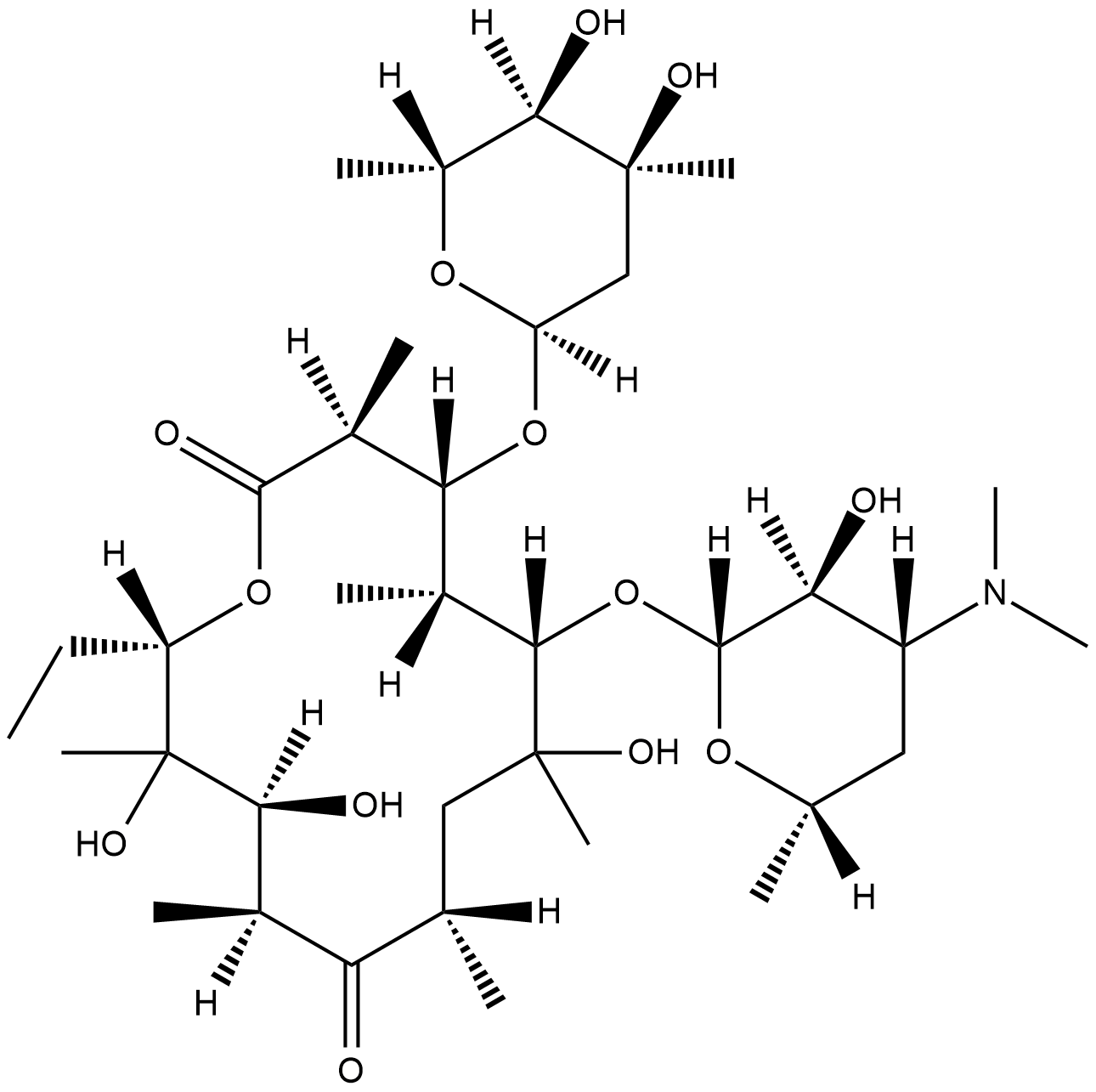
Erythromycin C
Erythromycin C is an intermediate in the biosynthesis of erythromycin .
-
GC43627
Erythromycin lactobionate
A macrolide antibiotic