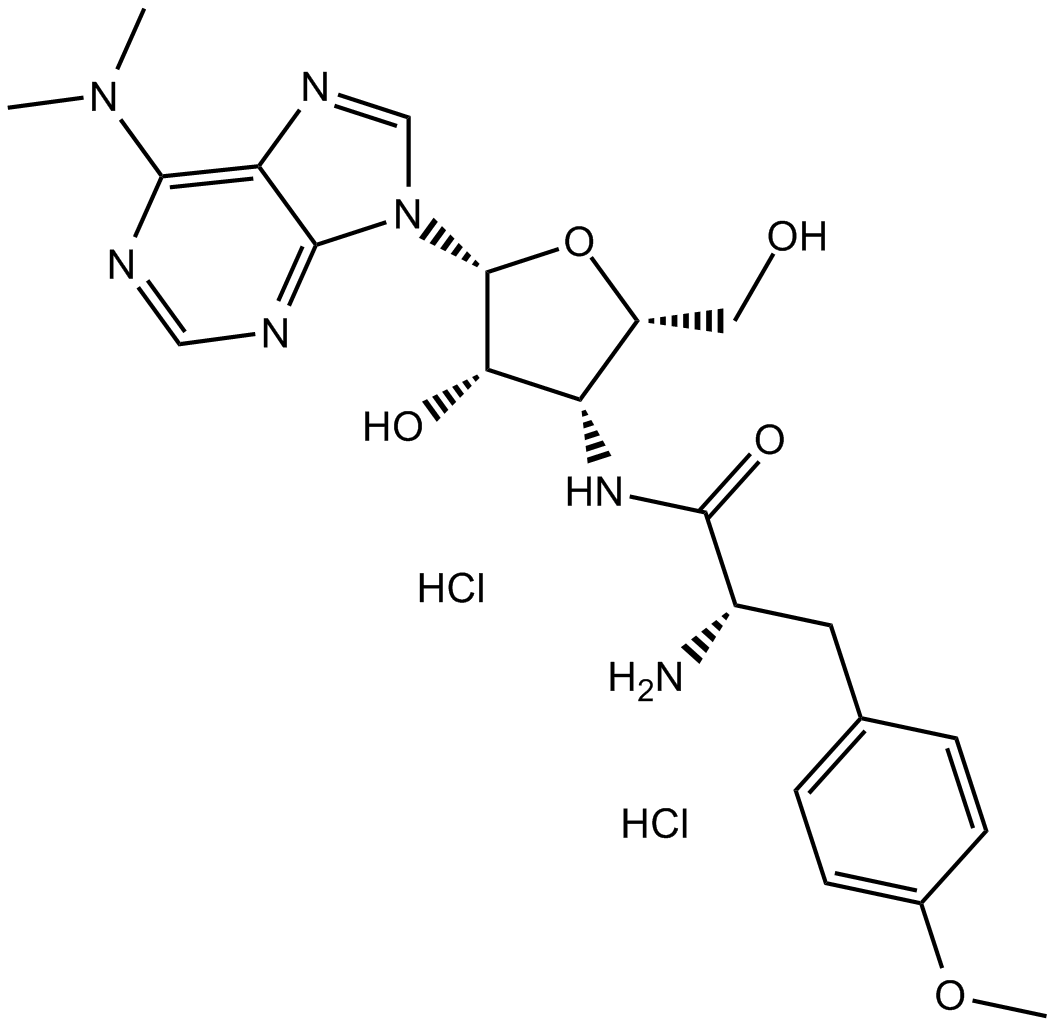
Puromycin dihydrochloride (Synonyms: CL13900) |
| Catalog No.GC16384 |
Puromycin dihydrochloride is produced by Streptomyces alboniger, a grampositive actinomycete, through a series of enzymatic reactions
Products are for research use only. Not for human use. We do not sell to patients.
Cas No.: 58-58-2
Sample solution is provided at 25 µL, 10mM.
Puromycin dihydrochloride is produced by Streptomyces alboniger, a grampositive actinomycete, through a series of enzymatic reactions.[1] Puromycin dihydrochloride included a nucleoside covalently bound to an amino acid, mimicking the 30 end of aminoacylated tRNAs that participate in delivery of amino acids to elongating ribosomes.[2] It inhibits the growth of animal cells and blocks protein synthesis by binding to 80S ribosomes at low doses.[3]
In vitro study determined the optimal concentration of Puromycin dihydrochloride for selecting EGFPac-transfected cells by performing a Puromycin dihydrochloride resistance test. The puromycin-resistant gene (termed pac) encoding puromycin N-acetyl transferase was isolated from Streptomyces aboniger. If pac is introduced and expressed in animal cells, the cells can survive in the presence of Puromycin dihydrochloride. Results ahowed that it could successfully produce a somatically cloned transgenic piglet using recombinant cells obtained after gene transfer of a transgene (carrying both EGFP and pac expression units) and subsequent in vitro selection with a low concentration (2 mg/ml) of puromycin.[3]
In vivo study was conducted to determine the surface sensing of translation (SUnSET) technique could be used to measure the protein synthesis in whole tissues. Since there is currently an intense interest in identifying the molecular mechanisms that regulate skeletal muscle protein synthesis. It allows for the visualization and quantification of protein synthesis and eliminates the need for generating radioactive tissues/animals. This study also determined that the surface sensing of translation could detect relatively acute changes in protein synthesis in the absence of changes in rRNA as well as detect not only increases but also decreases in protein synthesis in vivo. [4]
References:
[1]. Tercero JA, Espinosa JC, Lacalle RA, Jiménez A. The biosynthetic pathway of the aminonucleoside antibiotic puromycin, as deduced from the molecular analysis of the pur cluster of Streptomyces alboniger. J Biol Chem 1996;271(3):1579–90.
[2]. Aviner R. et al. The science of puromycin: From studies of ribosome function to applications in biotechnology. Comput Struct Biotechnol J. 2020 Apr 24;18:1074-1083.
[3]. Watanabe S, Iwamoto M, et al. A novel method for the production of transgenic cloned pigs: electroporation-mediated gene transfer to non-cultured cells and subsequent selection with puromycin. Biol Reprod. 2005 Feb;72(2):309-15.
[4]. Goodman CA, Mabrey DM, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011 Mar;25(3):1028-39.
Average Rating: 5 (Based on Reviews and 35 reference(s) in Google Scholar.)
GLPBIO products are for RESEARCH USE ONLY. Please make sure your review or question is research based.
Required fields are marked with *