DNA/RNA Synthesis
RNA synthesis, which is also called DNA transcription, is a highly selective process. Transcription by RNA polymerase II extends beyond RNA synthesis, towards a more active role in mRNA maturation, surveillance and export to the cytoplasm.
Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.DNA ligases discriminate against substrates containing RNA strands or mismatched base pairs at positions near the ends of the nickedDNA. Bleomycin (BLM) exerts its genotoxicity by generating free radicals, whichattack C-4′ in the deoxyribose backbone of DNA, leading to opening of the ribose ring and strand breakage; it is an S-independentradiomimetic agent that causes double-strand breaks in DNA.
First strand cDNA is synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H). Second strand cDNA synthesis is subsequently performed using DNA Polymerase I and RNase H. The remaining overhangs are converted into blunt ends using exonuclease/polymerase activity. After adenylation of the 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure is ligated to prepare the samples for hybridization. Cell cycle and DNA replication are the top two pathways regulated by BET bromodomain inhibition. Cycloheximide blocks the translation of mRNA to protein.
Targets for DNA/RNA Synthesis
Products for DNA/RNA Synthesis
- Cat.No. Product Name Information
-
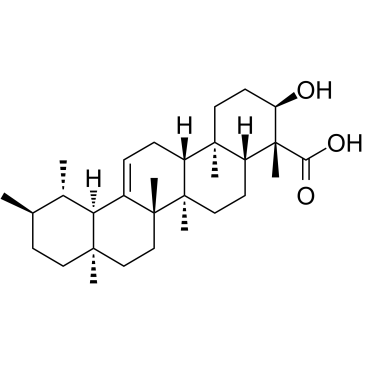
GC38000
β-Boswellic acid
A pentacyclic triterpene with diverse bioactivities
-
GC10867
(+)-Aphidicolin
ICI 69653, NSC 234714
Aphidicolin ((+)-Aphidicolin), a reversible inhibitor of eukaryotic nuclear DNA replication, can block the cell cycle at the pre-S phase.
-
GC48635
(-)-Cryptopleurine
(R)-Cryptopleurine, NSC 19912
An alkaloid with diverse biological activities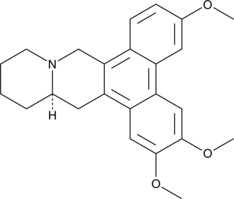
-
GC41695
(6R,S)-5,6,7,8-Tetrahydrofolic Acid (hydrochloride)
Tetrahydrofolate, THFA
(6R,S)-5,6,7,8-Tetrahydrofolic acid (THFA), the reduced form of folic acid, serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.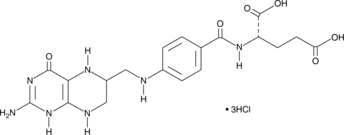
-
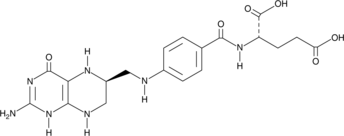
GC41088
(6S)-Tetrahydrofolic Acid
(6S)-Tetrahydrofolic acid is a diastereomer of tetrahydrofolic acid, a reduced form of folic acid that serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.
-
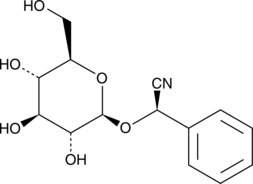
GC41633
(R)-Prunasin
(R)-Prunasin is a inhibitor of DNA Polymerase β.
-
GC41859
1,5,7-Triazabicyclo[4.4.0]dec-5-ene
TBD, Triazabicyclodecene
1,5,7-Triazabicyclo[4.4.0]dec-5-ene is a bicyclic guanidine base that can be used as a catalyst for a variety of base-mediated organic transformations, including Michael additions, Wittig reactions, Henry reactions, and transesterification reactions.![1,5,7-Triazabicyclo[4.4.0]dec-5-ene Chemical Structure 1,5,7-Triazabicyclo[4.4.0]dec-5-ene Chemical Structure](/media/struct/GC4/GC41859.png)
-
GC48909
1-Hydroxyanthraquinone
NSC 8640
An anthraquinone with genotoxic and carcinogenic activities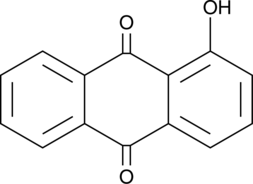
-
GC49872
10-Formyltetrahydrofolate (sodium salt) (technical grade)
10-CHO-FH4, 10-CHO-THF, N10-Formyltetrahydrofolate, 10-formyl H4PteGlu, 10-fTHF
10-Formyltetrahydrofolate (sodium salt) (technical grade) is a form of tetrahydrofolic acid that acts as a donor of formyl groups in anabolism.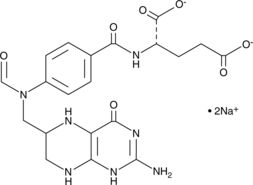
-
GC63796
116-9e
MAL2-11B
116-9e (MAL2-11B) is a Hsp70 co-chaperone DNAJA1 inhibitor.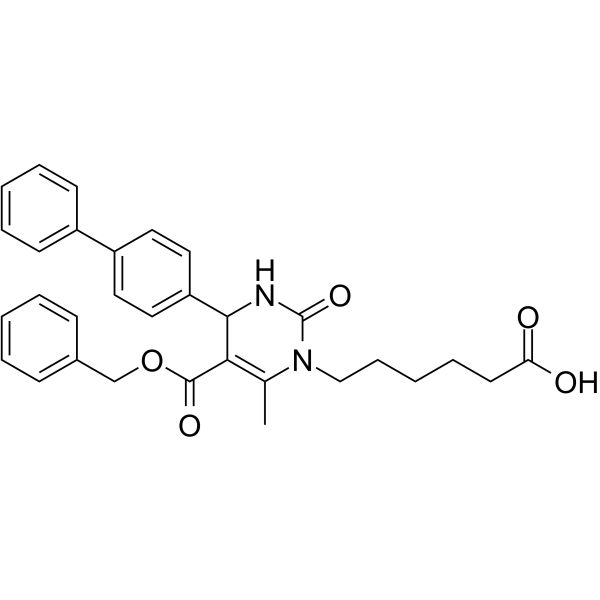
-
GC49759
13C17-Mycophenolic Acid
13C17-MPA
An internal standard for the quantification of mycophenolic acid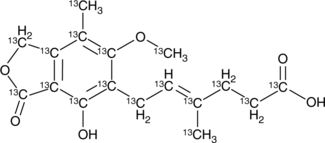
-
GC46474
18-Deoxyherboxidiene
RQN-18690A
A bacterial metabolite with antiangiogenic activity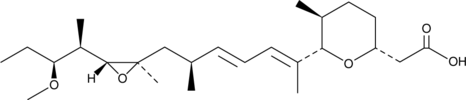
-
GC65489
2'-F-Bz-dC Phosphoramidite
2'-F-Bz-dC Phosphoramidite can be used in the synthesis of oligoribonucleotides.
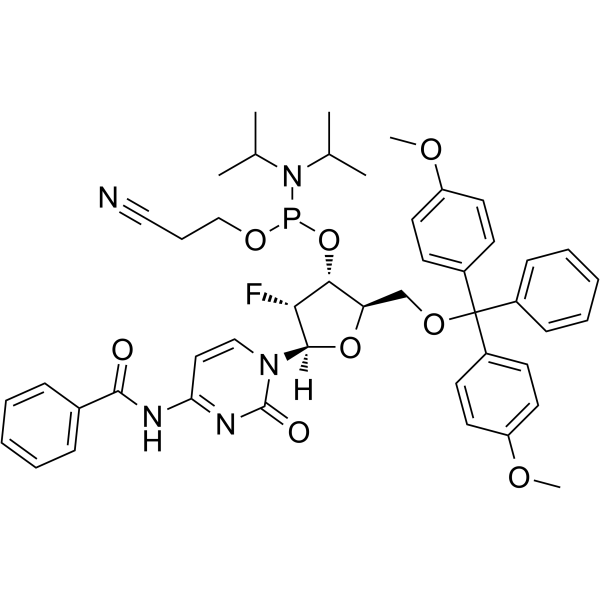
-
GC52183
2'-O-Methyl-5-methyluridine
2'-O-Methylribothymidine, 2'-O-Methylthymidine
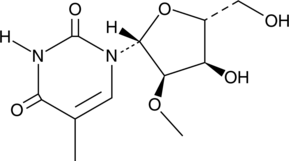
-
GC66713
2'-O-MOE-5-Me-C(Bz)
2'-O-MOE-5-Me-C (Bz) is a nucleotide for the stereoselective synthesis of nucleoside alkyl phosphonates.
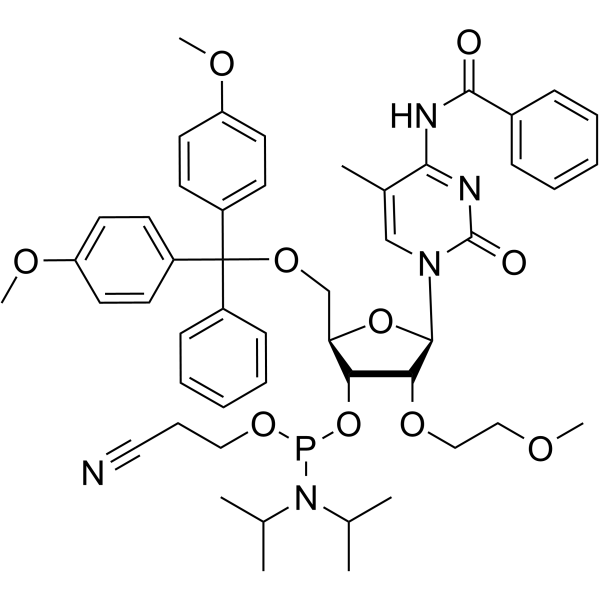
-
GC66657
2'-O-MOE-5-Me-rC
2'-O-MOE-5-Me-rC is an active compound. 2'-O-MOE-5-Me-rC can be used for oligonucleotide synthesis.
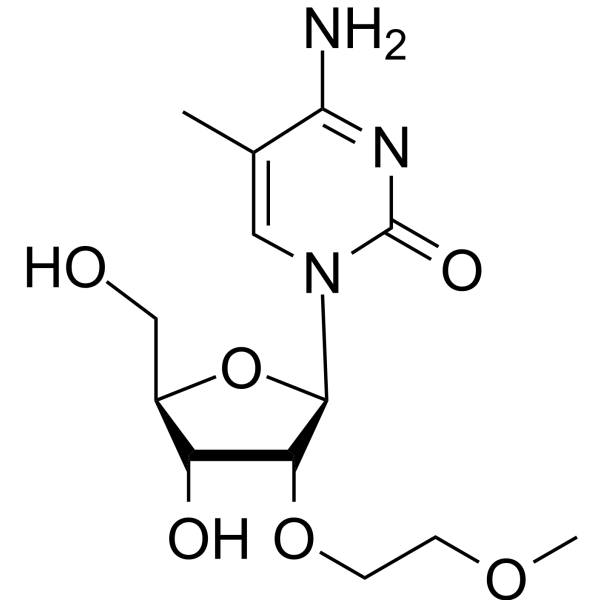
-
GC66091
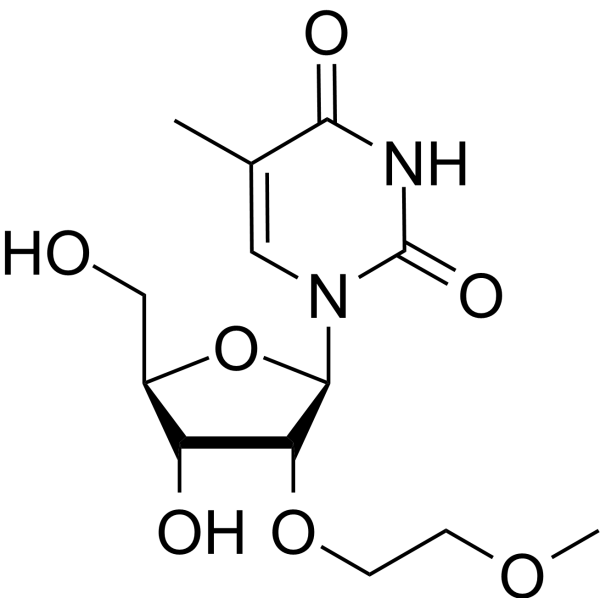
2'-O-MOE-5-Me-rU
2'-O-MOE-5-Me-rU is an active compound. 2'-O-MOE-5-Me-rU can be used for oligonucleotide synthesis.
-
GC66656
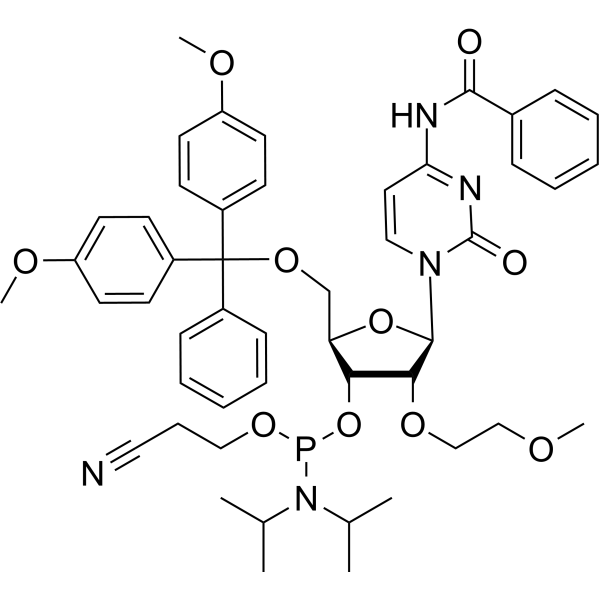
2'-O-MOE-rC
2'-O-MOE-rC is a 2'-O-MOE modified nucleoside. 2'-O-MOE-rC can be used for synthesis of DNA.
-
GC66652
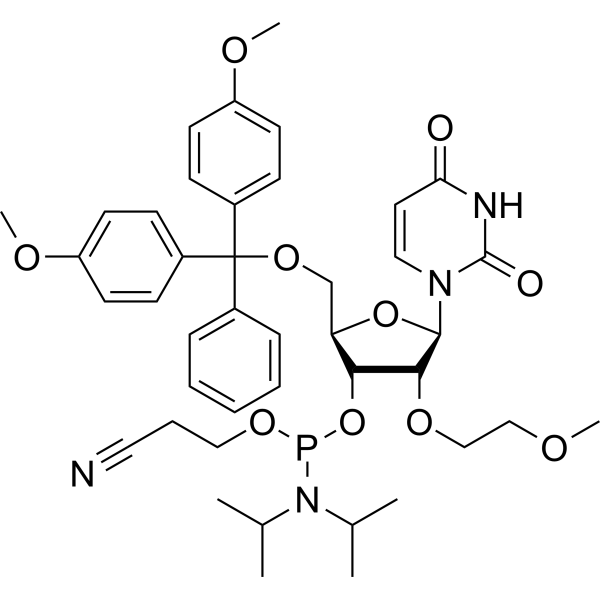
2'-O-MOE-U
2'-O-MOE-U is a phosphoramidite, can be used for oligonucleotide synthesis.
-
GC62530
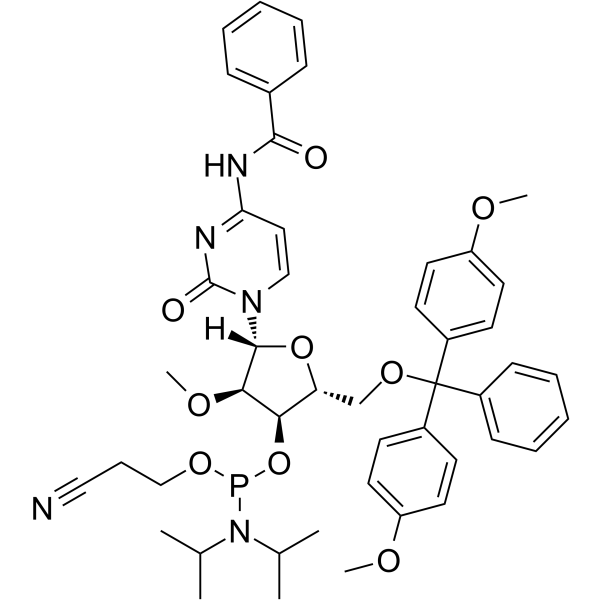
2’-O-Me-C(Bz) Phosphoramidite
2’-O-Me-C(Bz) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
-
GC62529
2’-OMe-A(Bz) Phosphoramidite
2’-OMe-A(Bz) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
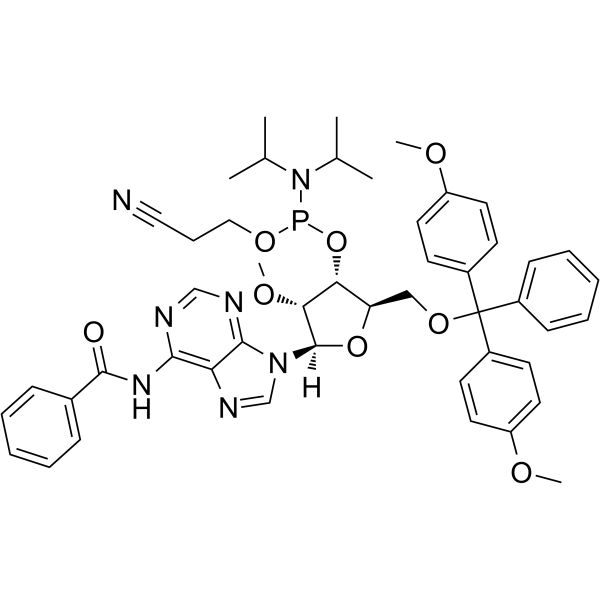
-
GC62531
2’-OMe-G(ibu) Phosphoramidite
2’-OMe-G(ibu) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
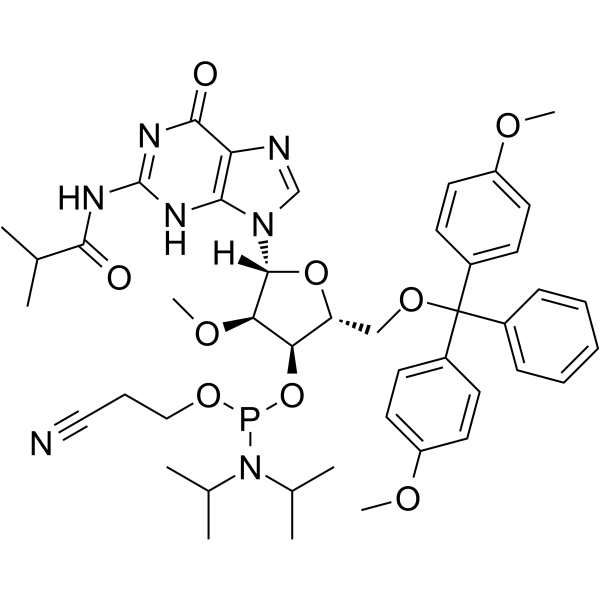
-
GC42061
2',3'-Dideoxyadenosine 5'-triphosphate
2',3'-Dideoxyadenosine 5'-triphosphate
2',3'-Dideoxyadenosine 5'-triphosphate (2',3'-Dideoxyadenosine 5'-triphosphate), an active metabolite of 2',3'-dideoxyinosine, is a chain-elongating inhibitor of DNA polymerase.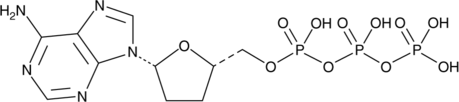
-
GC46519
2',3'-O-Isopropylideneguanosine
A building block
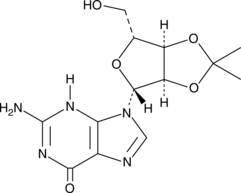
-
GC46540
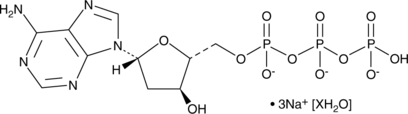
2'-Deoxyadenosine-5'-triphosphate (sodium salt hydrate)
A purine nucleotide
-
GC48440
2'-Deoxycytidine-5'-triphosphate (sodium salt)
dCTP
2'-Deoxycytidine-5'-triphosphate (sodium salt) (dCTP trisodium salt) is a nucleoside triphosphate that can be used for DNA synthesis.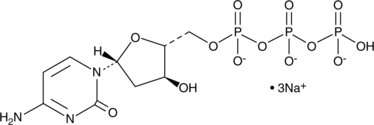
-
GC42151
2'-Deoxyguanosine 5'-monophosphate (sodium salt hydrate)
dGMP
2'-Deoxyguanosine 5'-monophosphate (dGMP) is used as a substrate of guanylate kinases to generate dGDP, which in turn is phosphorylated to dGTP, a nucleotide precursor used in DNA synthesis.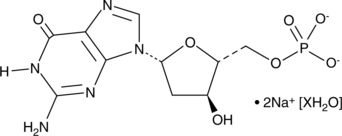
-
GC42080
2'2'-cGAMP (sodium salt)
Adenosine-Guanosine 2’,2’-cyclic monophosphate, cGAMP(2’-5’), 2’,2’-Cyclic GMP-AMP
2'2'-cGAMP is a synthetic dinucleotide (CDN) that contains non-canonical 2'5'-phosphodiester bonds.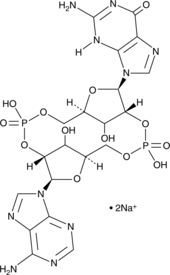
-
GC60459
2,4-D (2,4-Dichlorophenoxyacetic acid)
2,4D, Diclordon, Vidon 638
2,4-D (2,4-Dichlorophenoxyacetic acid) (2,4-D (2,4-Dichlorophenoxyacetic acid) ichlorophenoxyacetic acid) is a selective systemic herbicide for the control of broad-leaved weeds.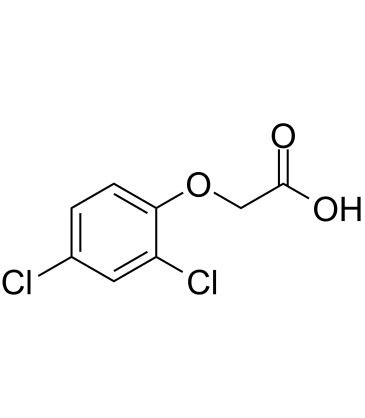
-
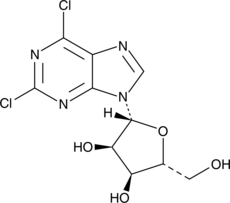
GC46524
2,6-Dichloropurine-9-β-D-riboside
2,6-Dichloropurine riboside
A building block
-
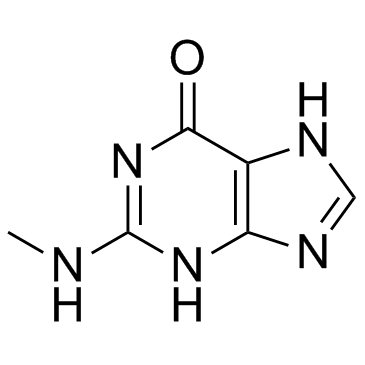
GC33496
2-(Methylamino)-1H-purin-6(7H)-one (N2-methylguanine)
2-(Methylamino)-1H-purin-6(7H)-one (N2-methylguanine) (N2-Methylguanine) is a modified nucleoside.
-
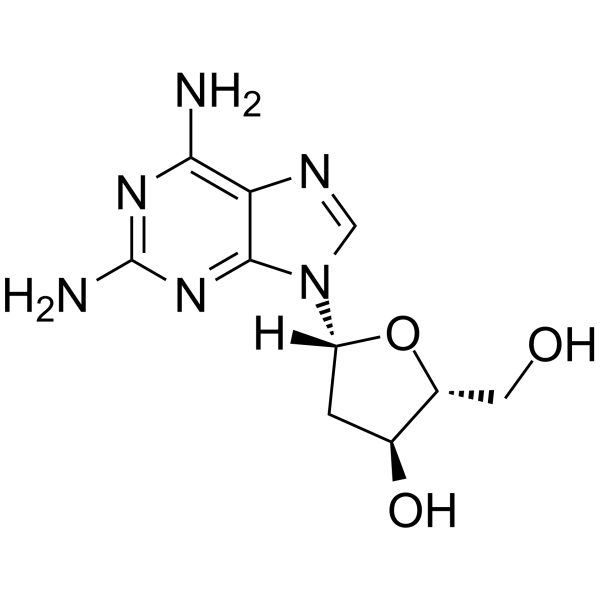
GC64983
2-Amino-2'-deoxyadenosine
2-Amino-2'-deoxyadenosine is a deoxyribonucleoside used for the oligonucleotide synthesis.
-
GC39527
2-O-Methylcytosine
2-O-Methylcytosine, an O-alkylated analogue a DNA adduct, is the damaged nucleobase.
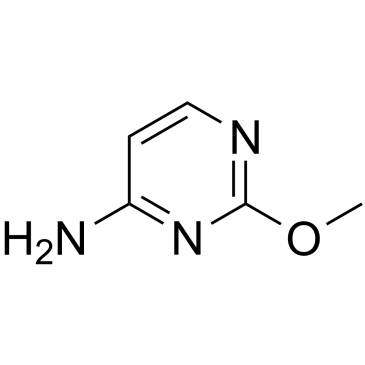
-
GC49348
2-Thiocytidine
α-2-Thioribocytidine
A modified nucleobase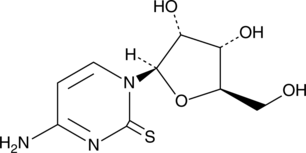
-
GC42197
2-Thiouridine
1-β-D-ribofuranosyl-2-thiouracil, s2U
2-Thiouridine (s2U) is a modified nucleobase found in tRNAs that is known to stabilize U:A pairs and modestly destabilize U:G wobble pairs.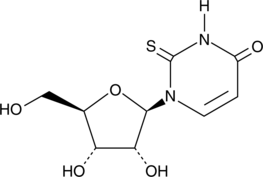
-
GC68542
3'Ome-m7GpppAmpG ammonium
3'Ome-m7GpppAmpG ammonium is a trinucleotide cap analog containing locked nucleic acid (LNA) molecules. It has significant translation efficiency and can be used as a potential molecular biology tool in the fields of mRNA vaccines and mRNA transfection, such as protein production, gene therapy, and anti-cancer immunity.

-
GC40618
3',4',7-Trihydroxyisoflavone
3'-hydroxy Daidzein, 3’,4’,7-THIF
3',4',7-Trihydroxyisoflavone, a major metabolite of Daidzein, is an ATP-competitive inhibitor of Cot (Tpl2/MAP3K8) and MKK4. 3',4',7-Trihydroxyisoflavone has anticancer, anti-angiogenic, chemoprotective, and free radical scavenging activities.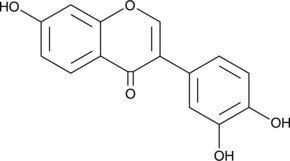
-
GC60023
3'-Deoxyuridine-5'-triphosphate
3'-dUTP
3'-Deoxyuridine-5'-triphosphate (3'-dUTP) is a nucleotide analogue that inhibits DNA-dependent RNA polymerases I and II.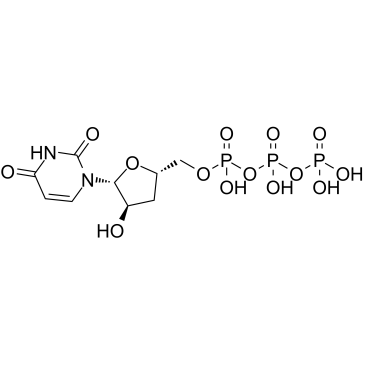
-
GC61862
3'-Deoxyuridine-5'-triphosphate trisodium
3'-dUTP trisodium
3'-Deoxyuridine-5'-triphosphate trisodium (3'-dUTP trisodium) is a nucleotide analogue that inhibits DNA-dependent RNA polymerases I and II.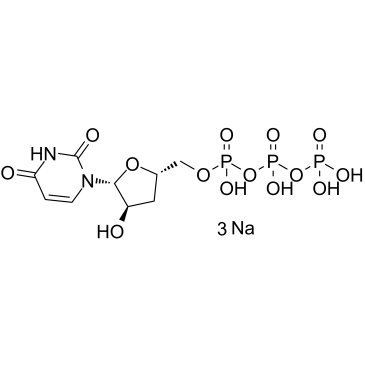
-
GC45332
3'-Dephosphocoenzyme A
depCoA, Dephospho-CoA
An intermediate in the biosynthesis of CoA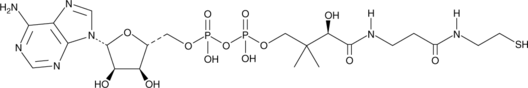
-
GC34452
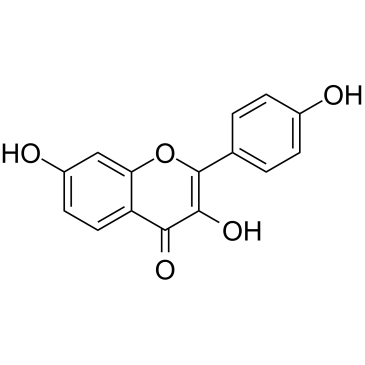
3,7,4'-Trihydroxyflavone
3,7,4'-Trihydroxyflavone, isolated from Rhus javanica var.
-
GC13510
3-AP
3-Aminopyridine-2-Carboxyaldehyde Thiosemicarbazone,NSC 663249,Triapine™
ribonucleotide reductase inhibitor and iron chelator with antitumor activity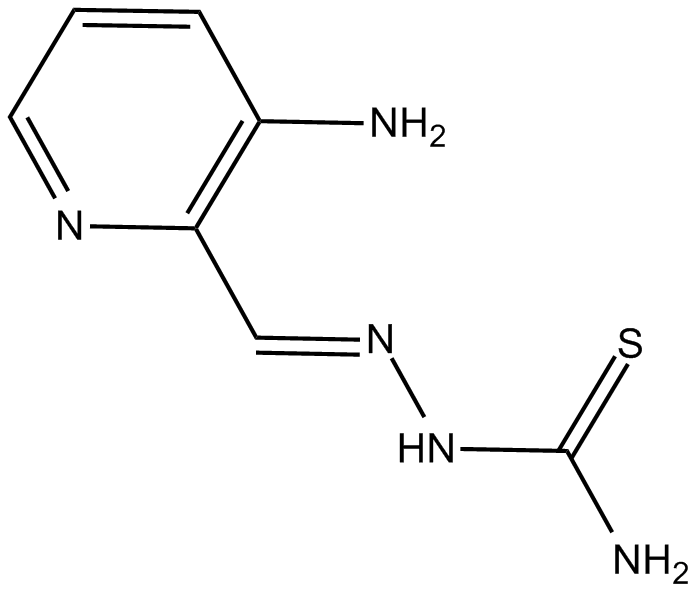
-
GC48457
3-keto Fusidic Acid
3-keto FA, 3-Oxofusidic Acid
An active metabolite of fusidic acid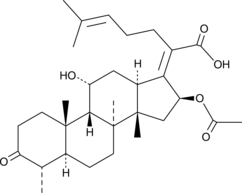
-
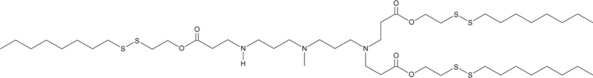
GC52391
306-O12B-3
An ionizable cationic lipidoid
-
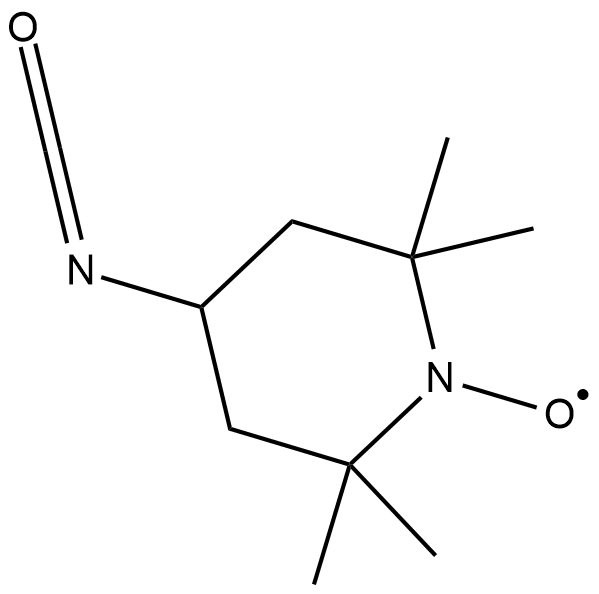
GC18853
4-isocyanato TEMPO
4-isocyanato TEMPO is a spin labeling reagent used to label the 2'-position in RNA.
-
GC61683
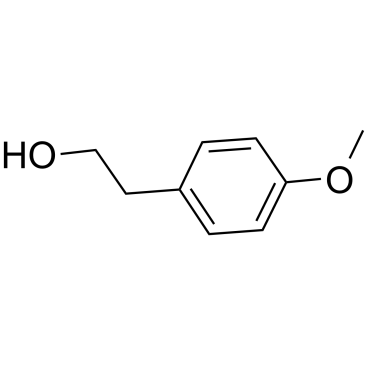
4-Methoxyphenethyl alcohol
4-Methoxyphenethyl alcohol, an aromatic alcohol, is the major component in the anise-like odour produced by A.
-
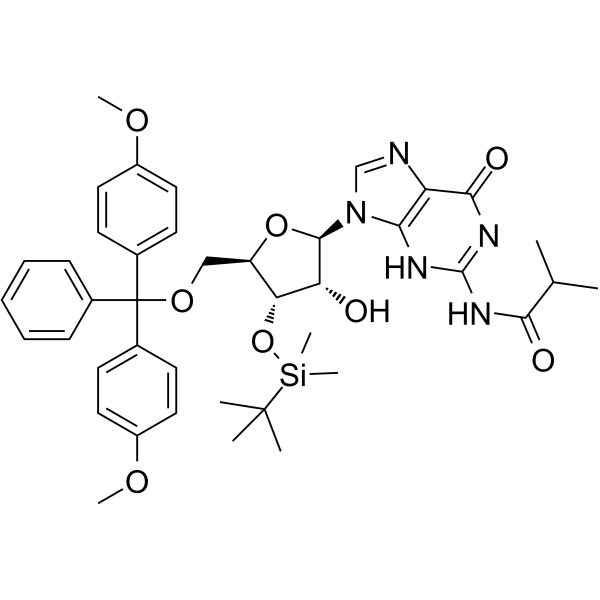
GC65520
5'-DMT-3'-TBDMS-ibu-rG
5'-DMT-3'-TBDMS-ibu-rG is is a modified nucleoside.
-
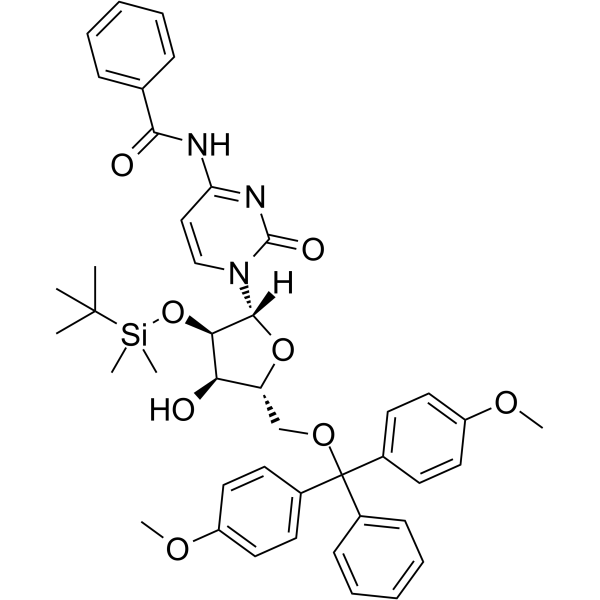
GC65402
5'-O-DMT-2'-O-TBDMS-Bz-rC
5'-O-DMT-2'-O-TBDMS-Bz-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
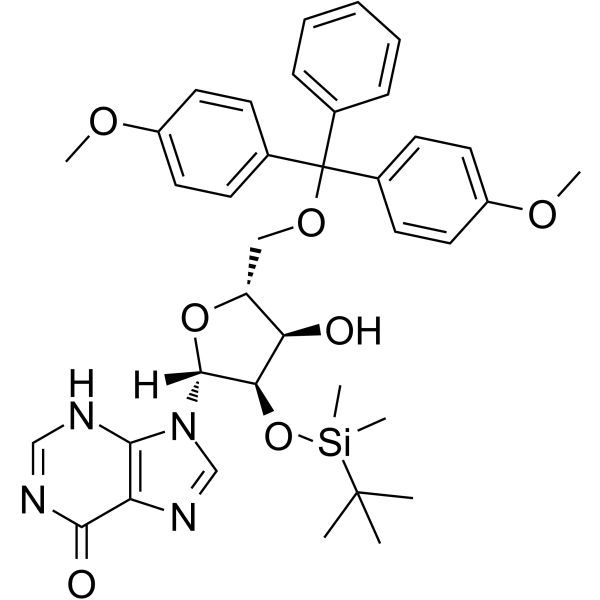
GC65164
5'-O-DMT-2'-O-TBDMS-rI
5'-O-DMT-2'-O-TBDMS-rI is a modified nucleoside.
-
GC65396
5'-O-DMT-ibu-dC
5'-O-DMT-ibu-dC can be used in the synthesis of oligodeoxyribonucleotides.
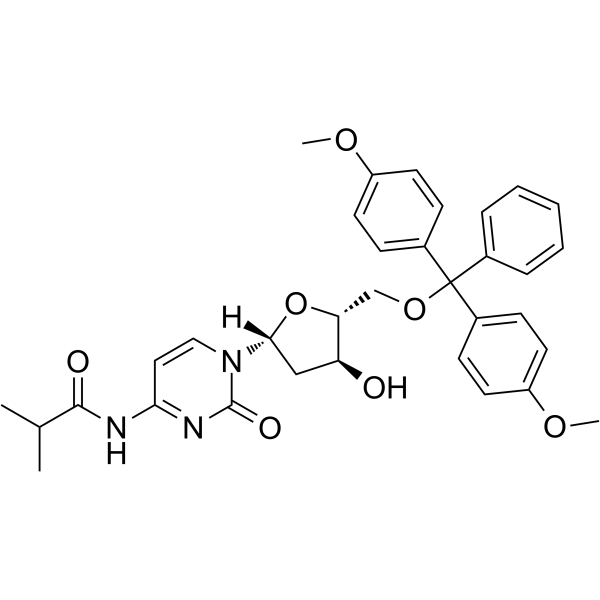
-
GC64982
5'-O-DMT-N2-DMF-dG
5'-O-DMT-2'-O-TBDMS-rI is a modified nucleoside.
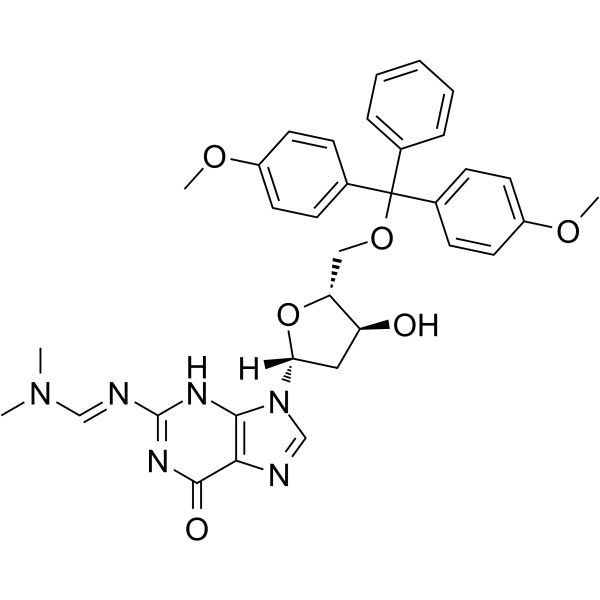
-
GC65482
5'-O-DMT-N6-ibu-dA
5'-O-DMT-N6-ibu-dA can be used in the synthesis of oligodeoxyribonucleotides.
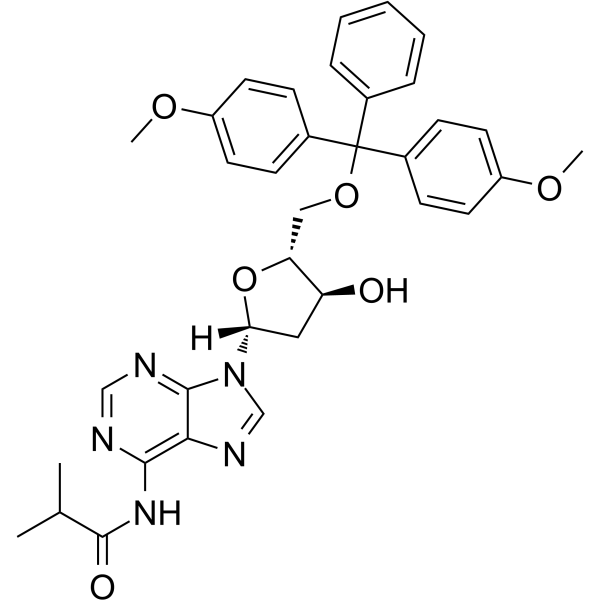
-
GC65562
5'-O-DMT-N6-Me-2'-dA
5'-O-DMT-N6-Me-2'-dA is a nucleoside with protective and modification effects.
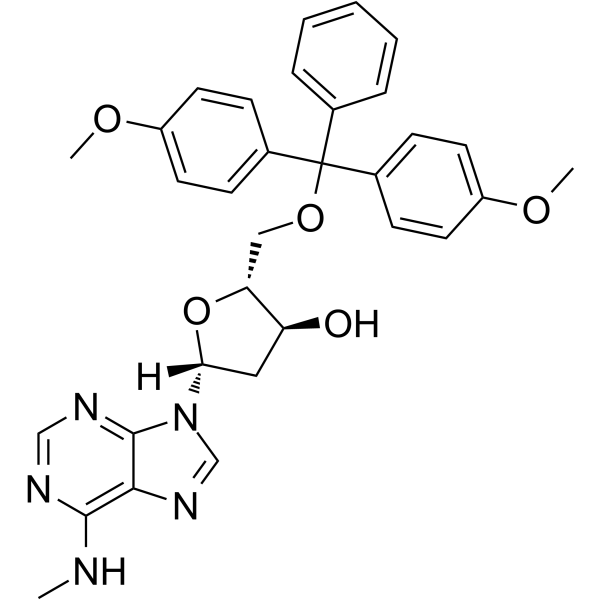
-
GC65401
5'-O-TBDMS-dU
5'-O-TBDMS-dU can be used in the synthesis of oligoribonucleotides.
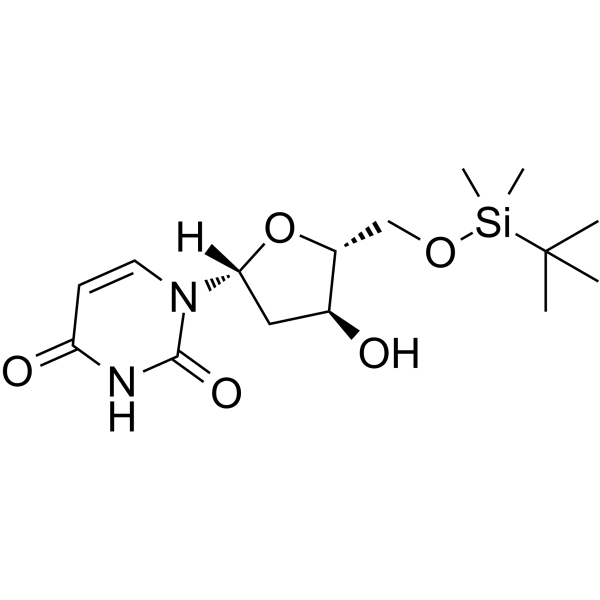
-
GC62157
5’-O-DMT-2’-O-TBDMS-Ac-rC
5’-O-DMT-2’-O-TBDMS-Ac-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
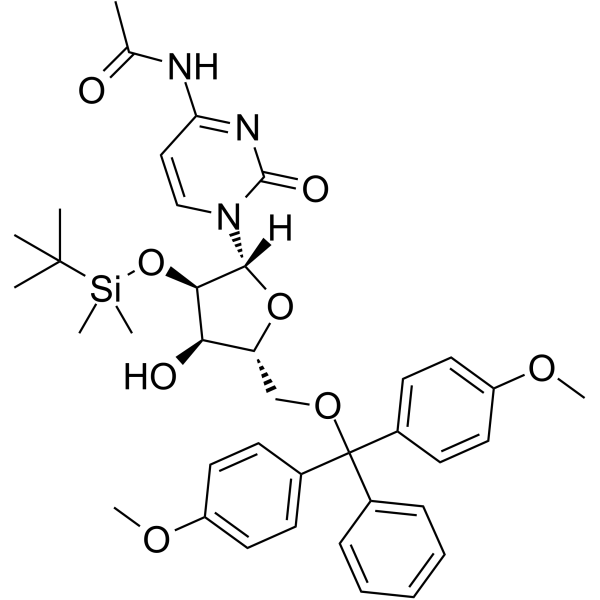
-
GC62149
5’-O-DMT-2’-TBDMS-Uridine
5’-O-DMT-2’-TBDMS-Uridine is a deoxyribonucleoside used for the oligonucleotide synthesis.
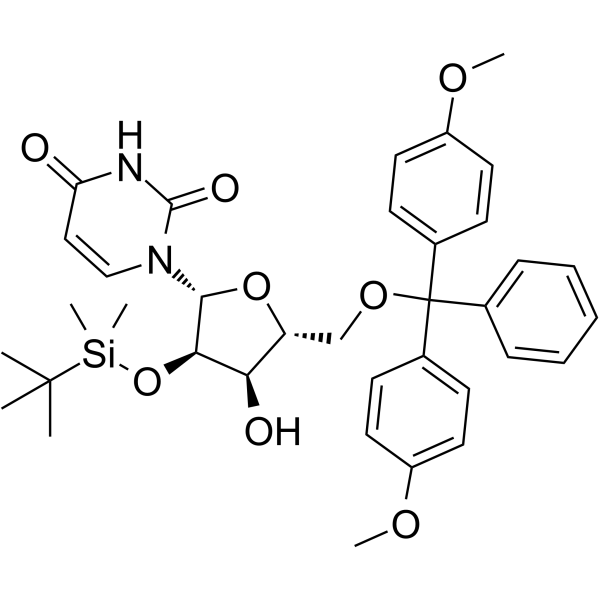
-
GC62532
5’-O-DMT-3’-O-TBDMS-Ac-rC
5’-O-DMT-3’-O-TBDMS-Ac-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
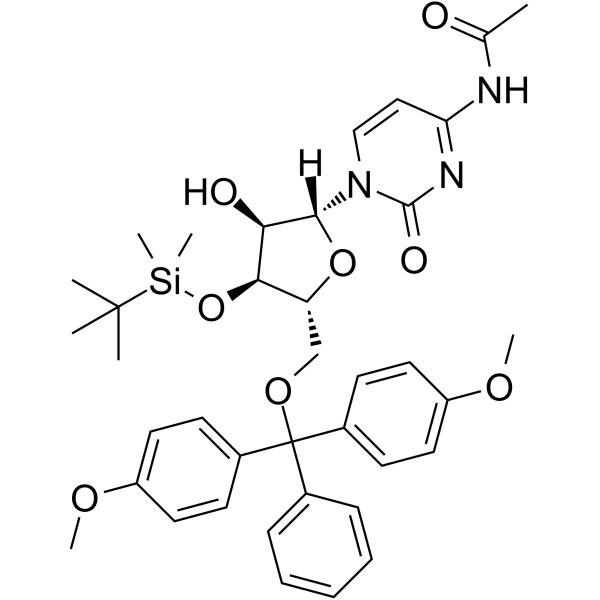
-
GC62573
5’-O-DMT-Bz-rC
5'-O-DMT-Bz-Rc is a modified nucleoside and can be used to synthesize DNA or RNA.
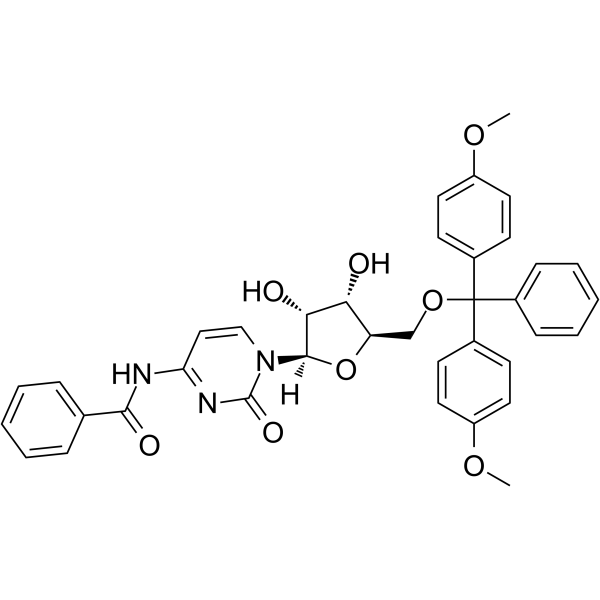
-
GC62574
5’-O-DMT-N4-Ac-2’-F-dC
5’-O-DMT-N4-Ac-2’-F-dC is a modified nucleoside and can be used to synthesize DNA or RNA.
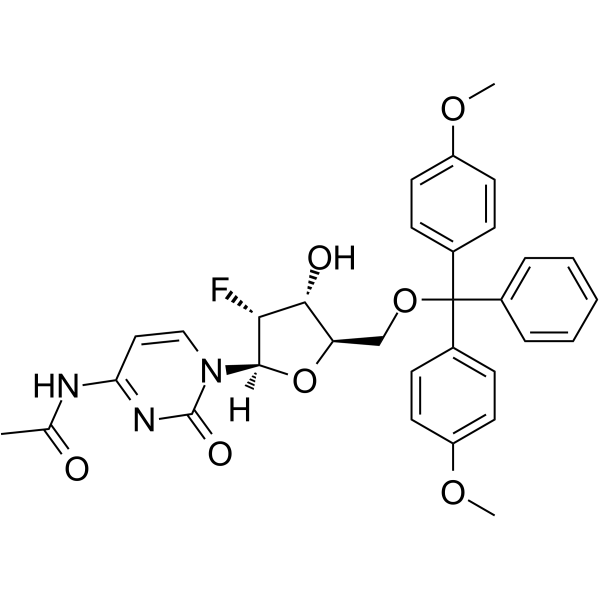
-
GC62575
5’-O-DMT-N4-Bz-2’-F-dC
5’-O-DMT-N4-Bz-2’-F-dC is a nucleoside with protective and modification effects.
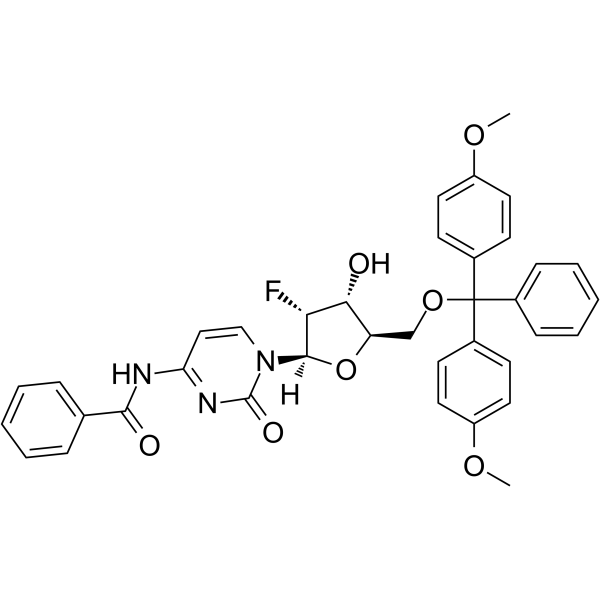
-
GC62576
5’-O-DMT-N4-Bz-5-Me-dC
5’-O-DMT-N4-Bz-5-Me-dC is a modified nucleoside.
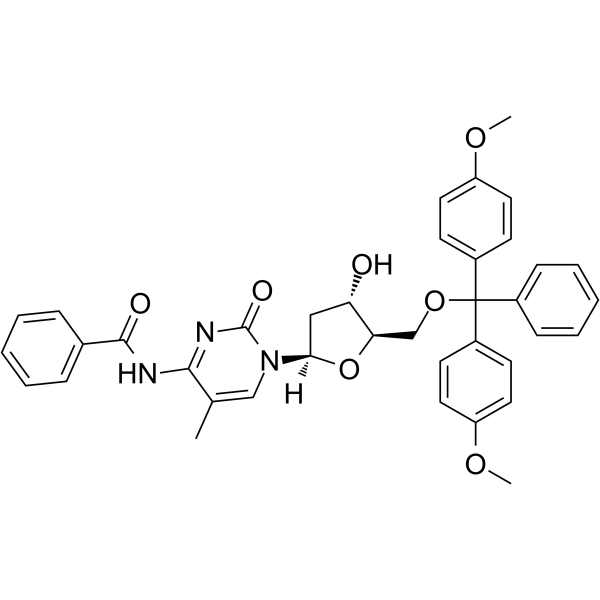
-
GC62812
5’-O-DMT-PAC-dA
5’-O-DMT-PAC-dA can be used in the synthesis of oligoribonucleotides.
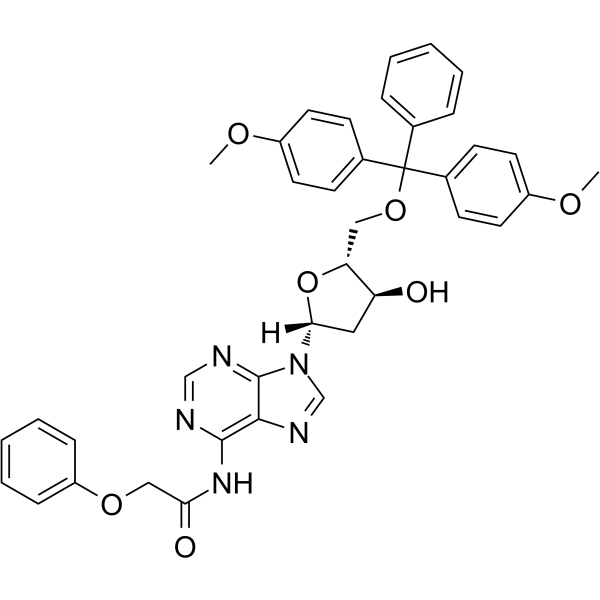
-
GC62813
5’-O-DMT-rI
5'-O-DMT-Ri can be used in the synthesis of oligoribonucleotides.
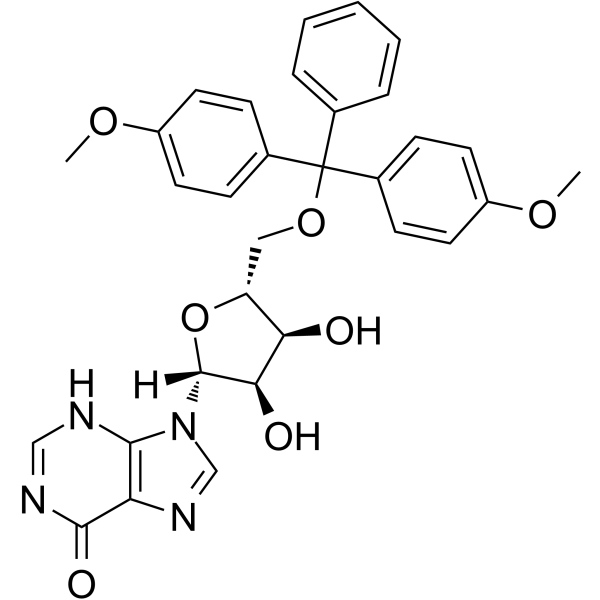
-
GC62577
5’-O-DMT-rU
5’-O-DMT-rU is a modified nucleoside and can be used to synthesize RNA.
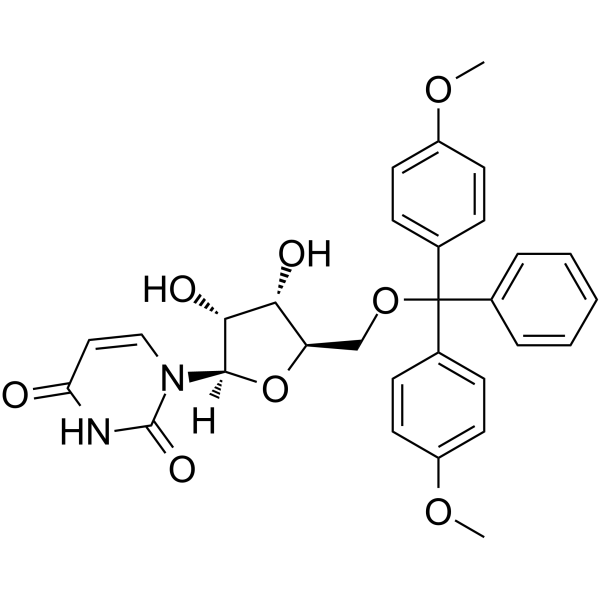
-
GC62578
5’-O-TBDMS-Bz-dA
5’-O-TBDMS-Bz-dA is a nucleoside with protective and modification effects.
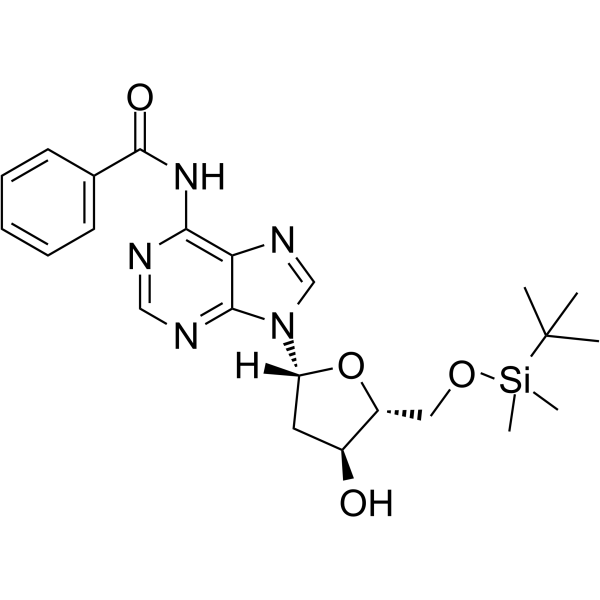
-
GC62579
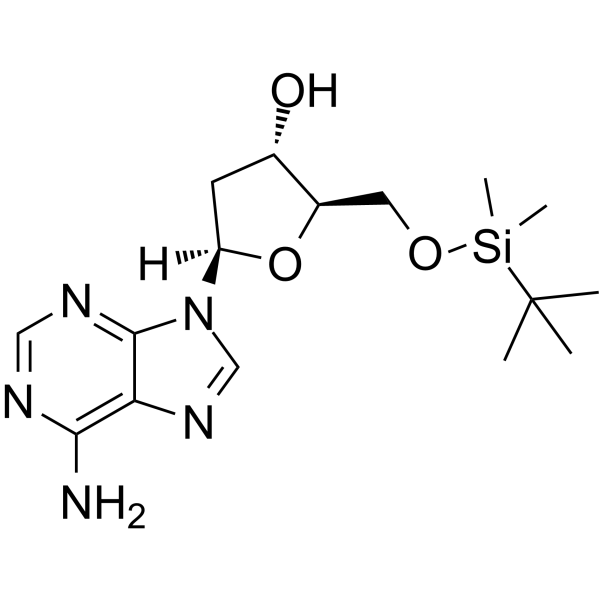
5’-O-TBDMS-dA
5’-O-TBDMS-dA is a modified nucleoside and can be used to synthesize DNA or RNA.
-
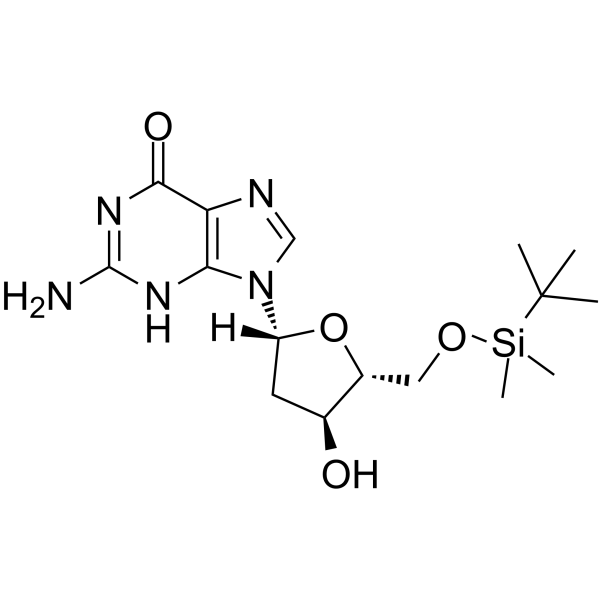
GC62580
5’-O-TBDMS-dG
5’-O-TBDMS-dG is a modified nucleoside.
-
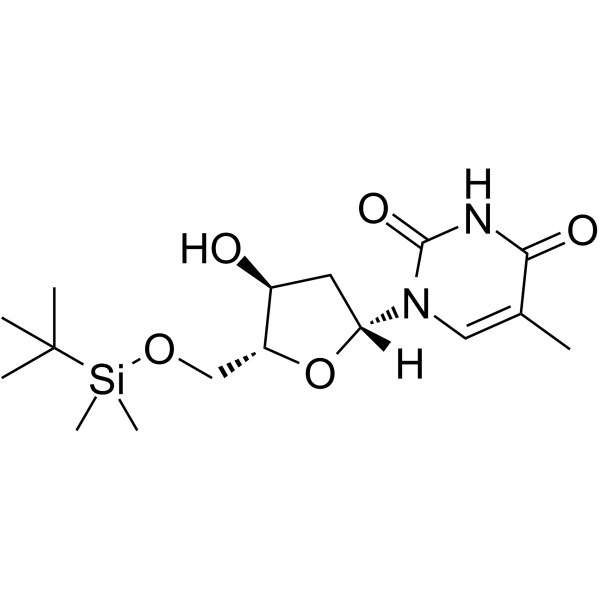
GC62581
5’-O-TBDMS-dT
5’-O-TBDMS-dT is a nucleoside with protective and modification effects.
-
GC40641
5'-Ethynyl-2'-deoxycytidine
EdC, 2'-deoxy-5-Ethynylcytidine
5'-Ethynyl-2'-deoxycytidine (EdC) is a nucleoside analog that inhibits replication of the herpes simplex virus-1 (HSV-1) KOS strain (ID50 = 0.2 μg/mL).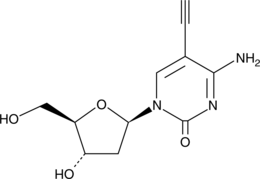
-
GC61726
5'-O-DMT-N4-Ac-dC
5'-O-DMT-N4-Ac-dC (N4-Acetyl-2'-deoxy-5'-O-DMT-cytidine, compound 7), a deoxynucleoside, can be used to synthesize of dodecyl phosphoramidite which is the raw material for dod‐DNA (amphiphilic DNA containing an internal hydrophobic region consisting of dodecyl phosphotriester linkages) synthesis.
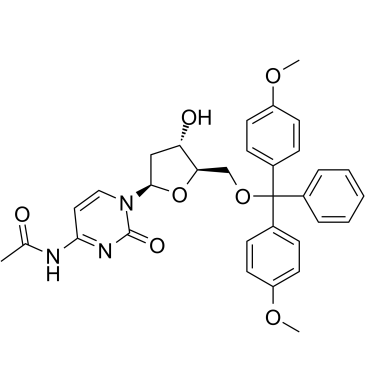
-
GC48662
5'-Thymidylic Acid (sodium salt)
Deoxythymidine-5’-monophosphate, Thymidine monophosphate, Thymidine-5’-monophosphate, dTMP
5'-Thymidylic Acid (sodium salt) is an endogenous metabolite.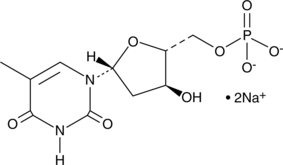
-
GC46681
5-Bromouridine
(–)-5-Bromouridine, BrU, BrUrd, NSC 38296
A brominated uridine analog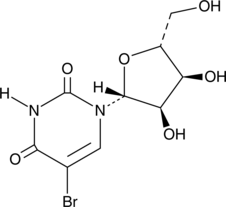
-
GC49245
5-Ethynyluridine
5-Ethynyluridine (5-EU) is a potent cell-permeable nucleoside can be used to label newly synthesized RNA.
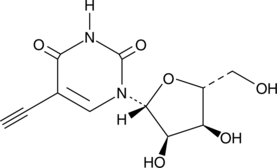
-
GC62389
5-Iminodaunorubicin
5-Iminodaunorubicin is a quinone-modified anthracycline that retains antitumor activity. 5-Iminodaunorubicin produces protein-concealed DNA strand breaks in cancer cells.
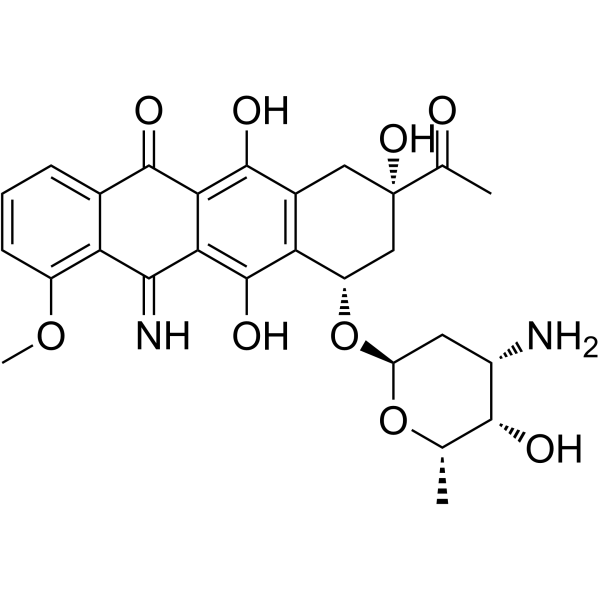
-
GC66658
5-Me-dC(Ac) amidite
5-Me-dC(Ac) amidite is used for synthesizing DNA or RNA.
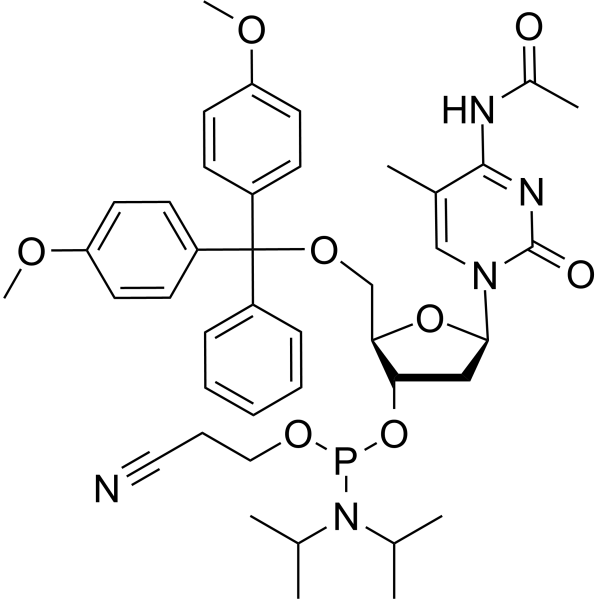
-
GC31187
5-Methoxyflavone
5-Methoxyflavone, belonged to Flavonoid family, is a DNA polymerase-beta inhibitor and neuroprotective agent against beta-amyloid toxicity.
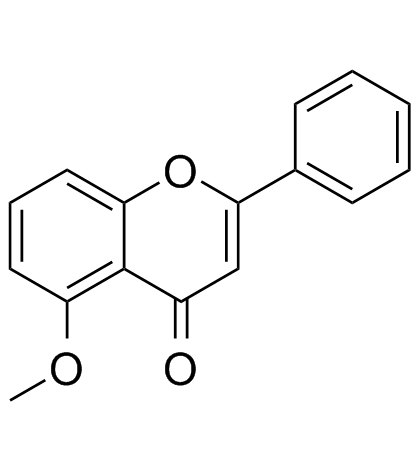
-
GC67671
5-Methylcytidine 5′-triphosphate trisodium
5-Methyl-CTP trisodium
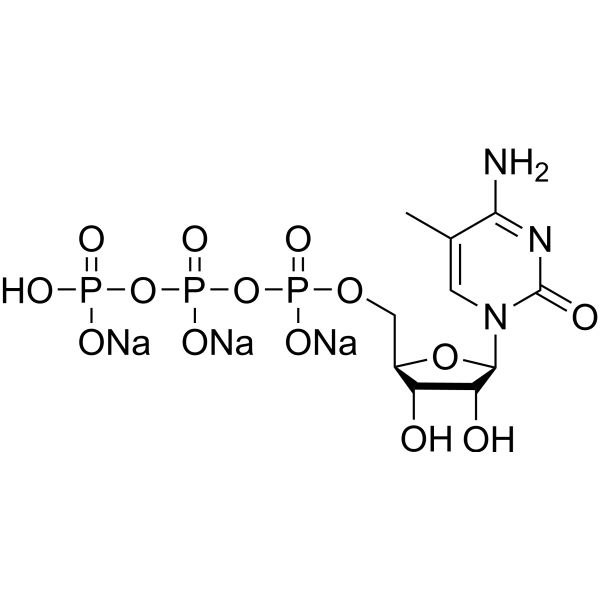
-
GC35166
5-Methylcytosine
5-Methylcytosine is a well-characterized DNA modification, and is also predominantly in abundant non-coding RNAs in both prokaryotes and eukaryotes.
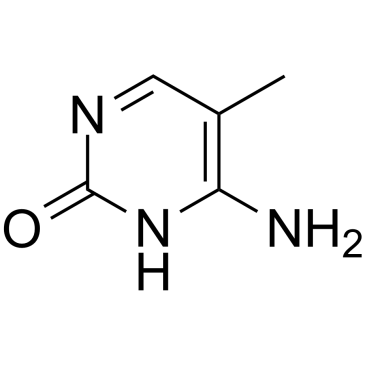
-
GC62547
5-O-TBDMS-N4-Benzoyl-2-deoxycytidine
5-O-TBDMS-N4-Benzoyl-2-deoxycytidine is a modified nucleoside.
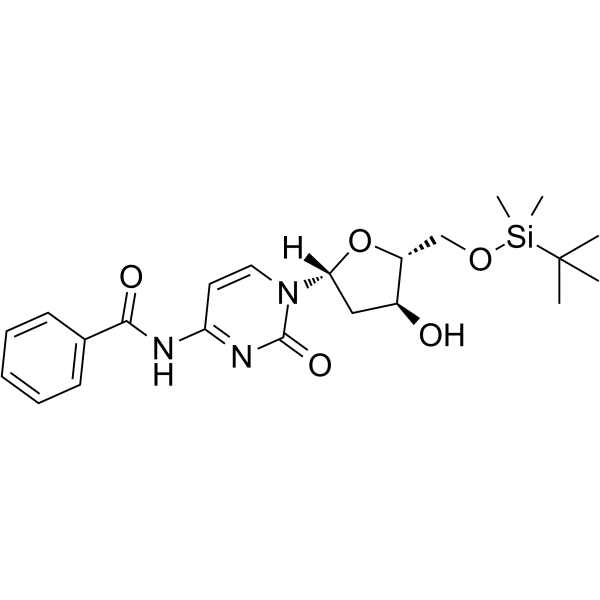
-
GC46713
5-Phospho-D-ribose 1-diphosphate (sodium salt hydrate)
Phosphoribosyl Pyrophosphate, P-rib-PP, PRPP
An intermediate in several biochemical pathways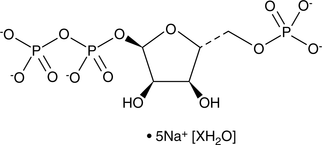
-
GC60031
6-Azathymine
6-Azathymine, a 6-nitrogen analog of thymine, is a potent D-3-aminoisobutyrate-pyruvate aminotransferase inhibitor.
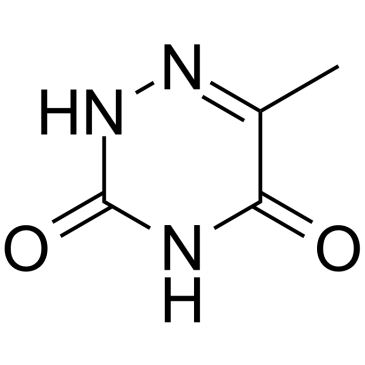
-
GC50249
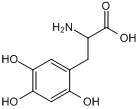
6-Hydroxy-DL-DOPA
6-Hydroxy-DL-DOPA is a selective and effective allosteric inhibitor of the RAD52 ssDNA binding domain. 6-Hydroxy-DL-DOPA can be used for the research of cancer.
-
GC12193
6-Thio-dG
β-TGdR; 6-Thio-2'-Deoxyguanosine
6-Thio-dG is a nucleoside analogue that can be incorporated into de novo-synthesized telomeres by telomerase.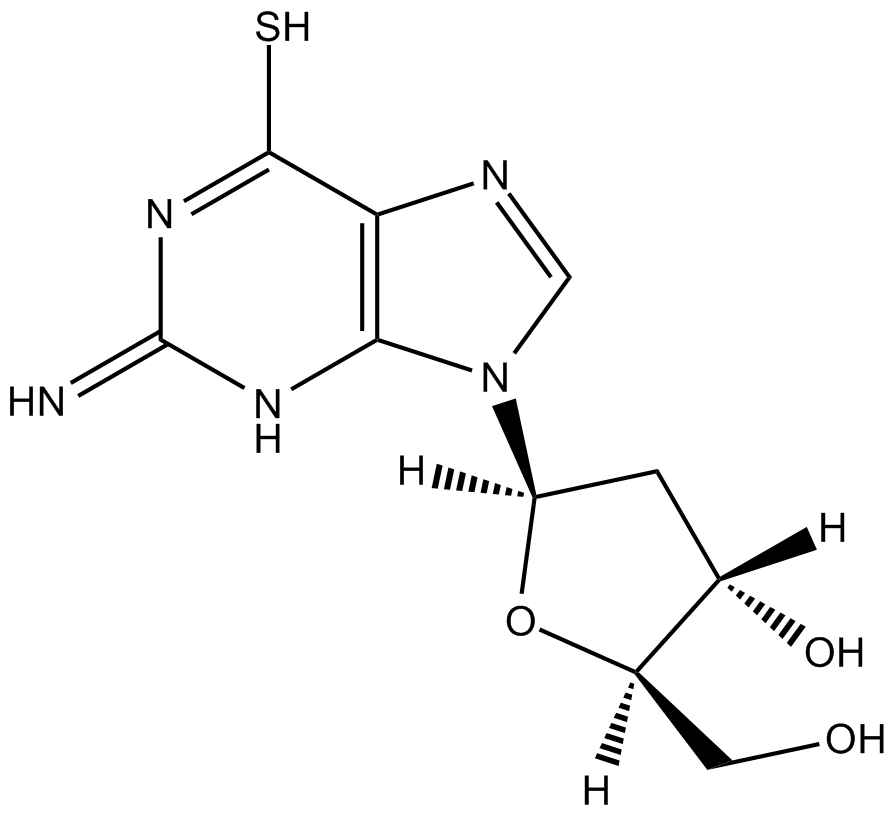
-
GC33577
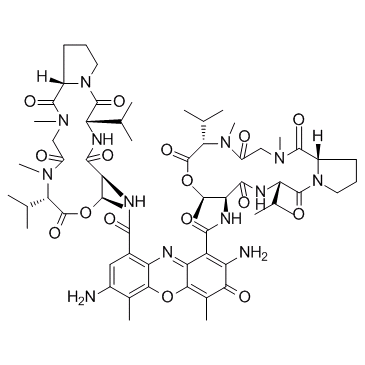
7-Aminoactinomycin D (7-AAD)
7-AAD, NSC 239759
7-Aminoactinomycin D is a fluorescent derivative of actinomycin D that selectively binds to the GC region of DNA.
-
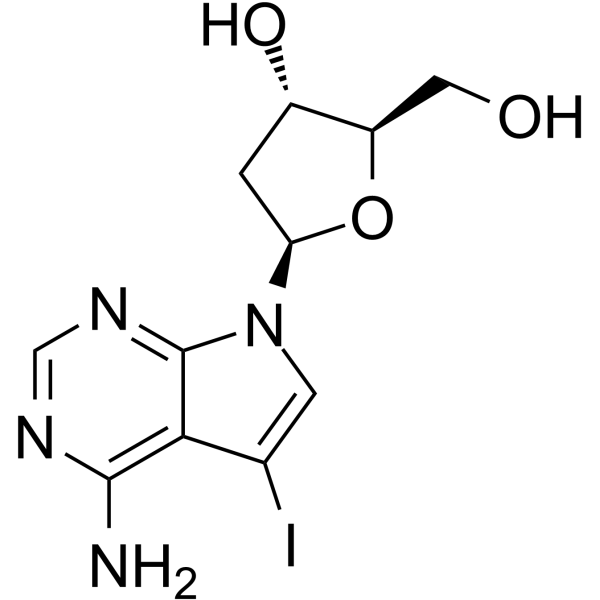
GC65408
7-Deaza-2'-deoxy-7-iodoadenosine
7-Deaza-2'-deoxy-7-iodoadenosine is a modified oligonucleotide containing 7-Deazaadenine.
-
GC65675
7-Iodo-7-deaza-2'-deoxyguanosine
7-Deaza-7-Iodo-2'-deoxyguanosine
7-Iodo-7-deaza-2'-deoxyguanosine (7-Deaza-7-Iodo-2'-deoxyguanosine) is a deoxyguanosine derivative that can be used in DNA synthesis and sequencing reactions.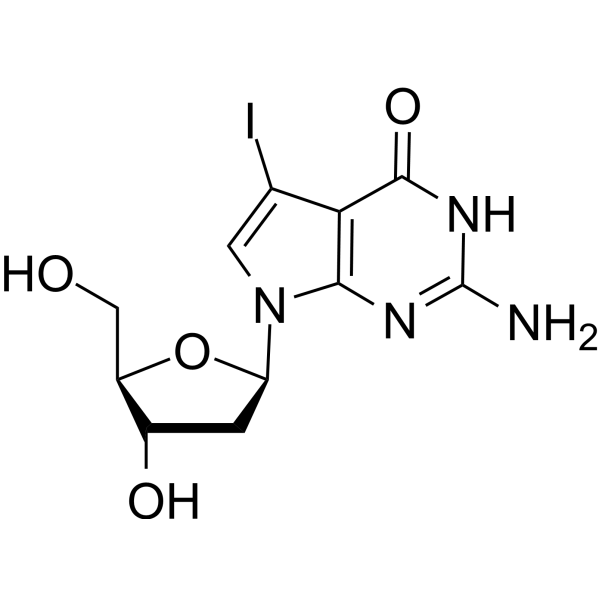
-
GC49561
7-Methylguanosine-d3
m7G-d3, 7-MeGua-d3
An internal standard for the quantification of 7-methylguanosine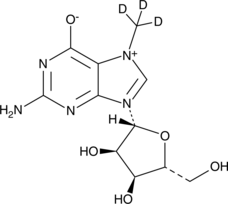
-
GC66058
7-TFA-ap-7-Deaza-dA
7-TFA-ap-7-Deaza-dA is a modified nucleoside. 7-TFA-ap-7-Deaza-dA can be used in the synthesis of deoxyribonucleic acid or nucleic acid.
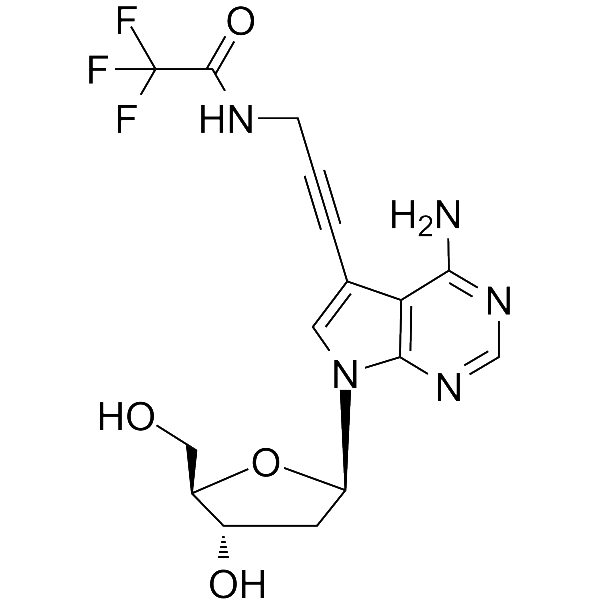
-
GC65525
7-TFA-ap-7-Deaza-dG
5'-O-TBDMS-dG is a modified nucleoside.
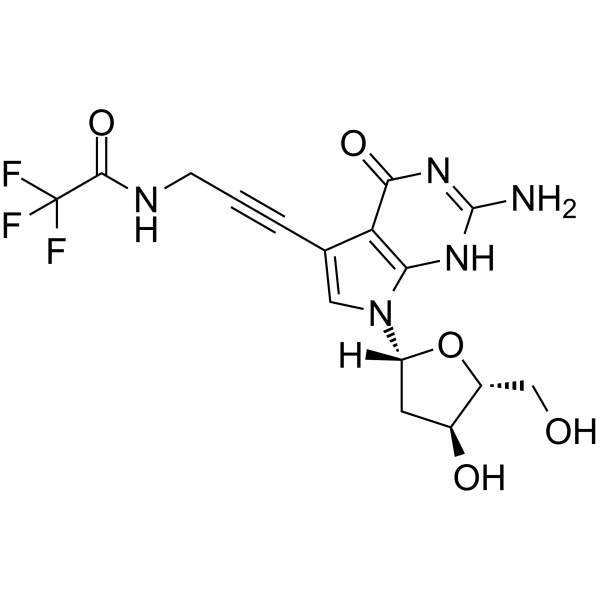
-
GC12777
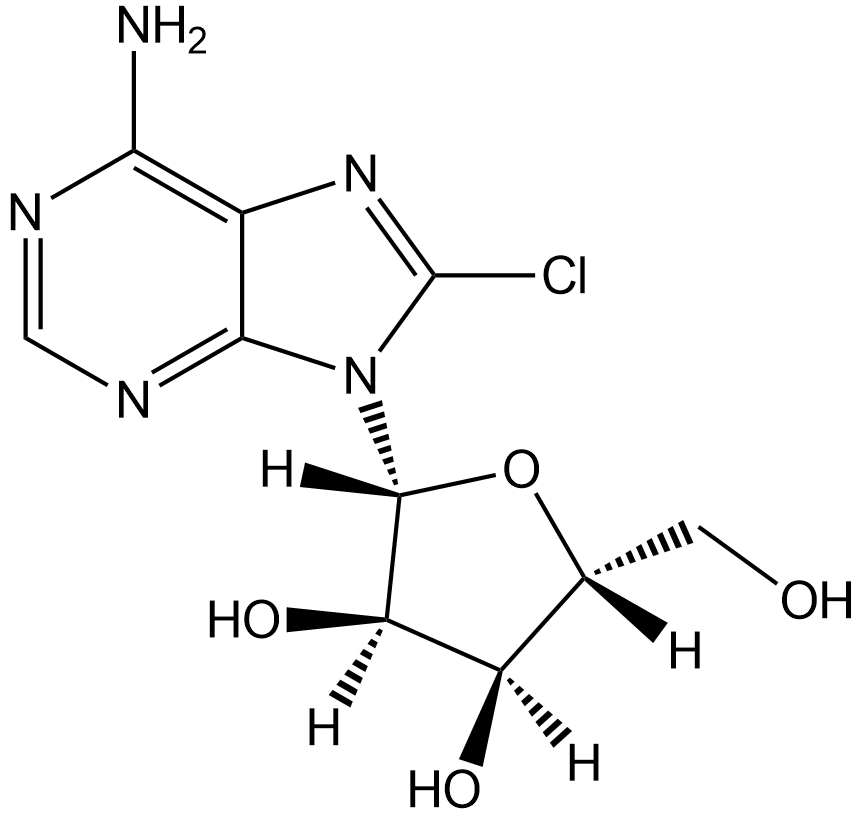
8-Chloroadenosine
Nucleoside analog, inhibits RNA synthesis
-
GC61763
Ac-rC Phosphoramidite
Ac-rC Phosphoramidite is used for the oligoribonucleotide phosphorodithioate modification (PS2-RNA).
-
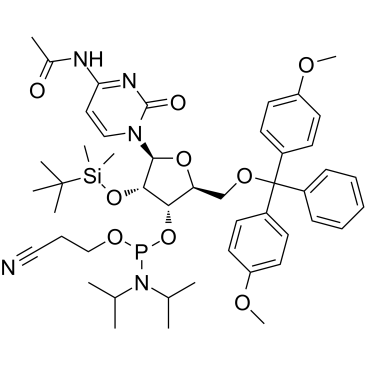
GC34090
Acelarin (NUC-1031)
Acelarin (NUC-1031) (NUC-1031) is a ProTide transformation and enhancement of the widely-used nucleoside analogue, gemcitabine.
-
GC16866
Actinomycin D
Cosmegen, Dactinomycin, Meractinomycin, NCI C04682, NSC 3053, Oncostatin K
Actinomycin D (dactinomycin) is a natural chromopeptide isolated from Streptomyces species, and has one heterocyclic chromophore and two cyclic pentapeptide lactone rings. [1]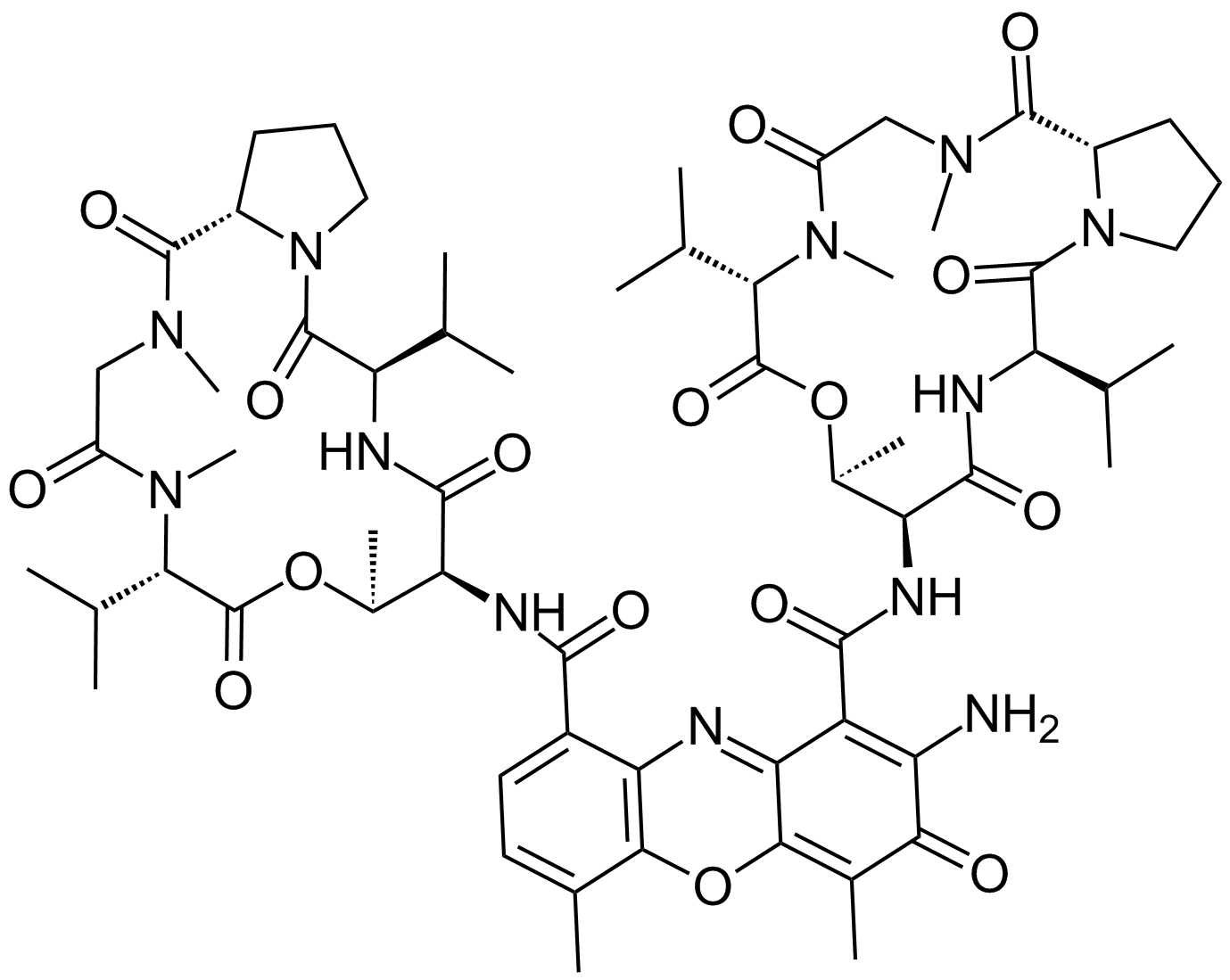
-
GC35247
ACX-362E
ACX-362E; GLS-362E
ACX-362E (ACX-362E) is a first-in-class, orally active DNA polymerase IIIC (pol IIIC) inhibitor, with a Ki of 0.325 μM for the DNA pol IIIC from C.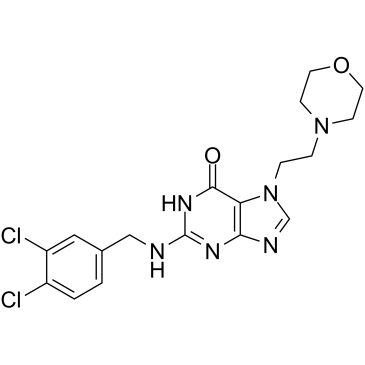
-
GC17186
Acyclovir
ACV, BW 248U, NSC 645011
Antiviral agent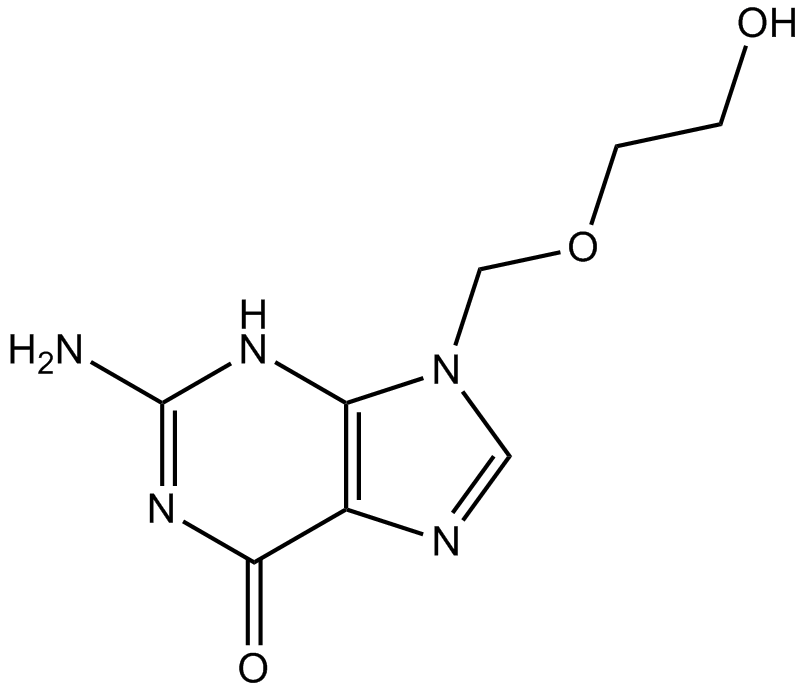
-
GC13432
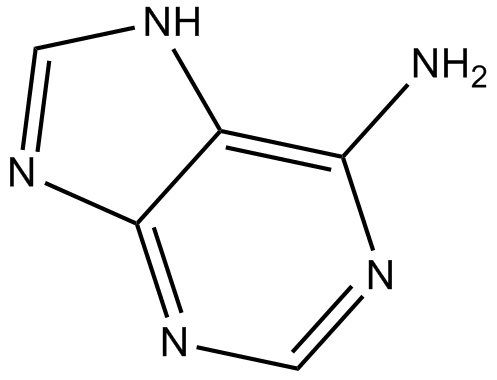
Adenine
High affinity adenine receptor agonist
-
GC11825
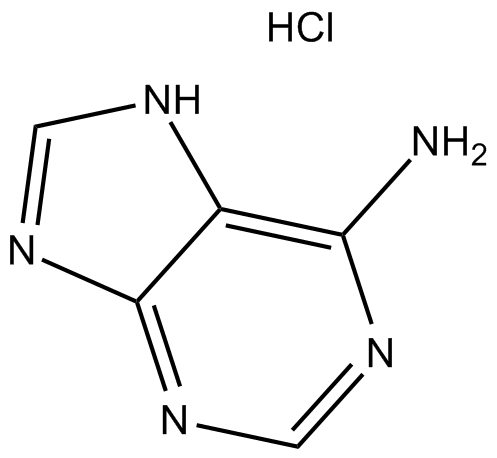
Adenine HCl
Adenine HCl (6-Aminopurine hydrochloride), a purine, is one of the four nucleobases in the nucleic acid of DNA. Adenine HCl acts as a chemical component of DNA and RNA. Adenine HCl also plays an important role in biochemistry involved in cellular respiration, the form of both ATP and the cofactors (NAD and FAD), and protein synthesis.
-
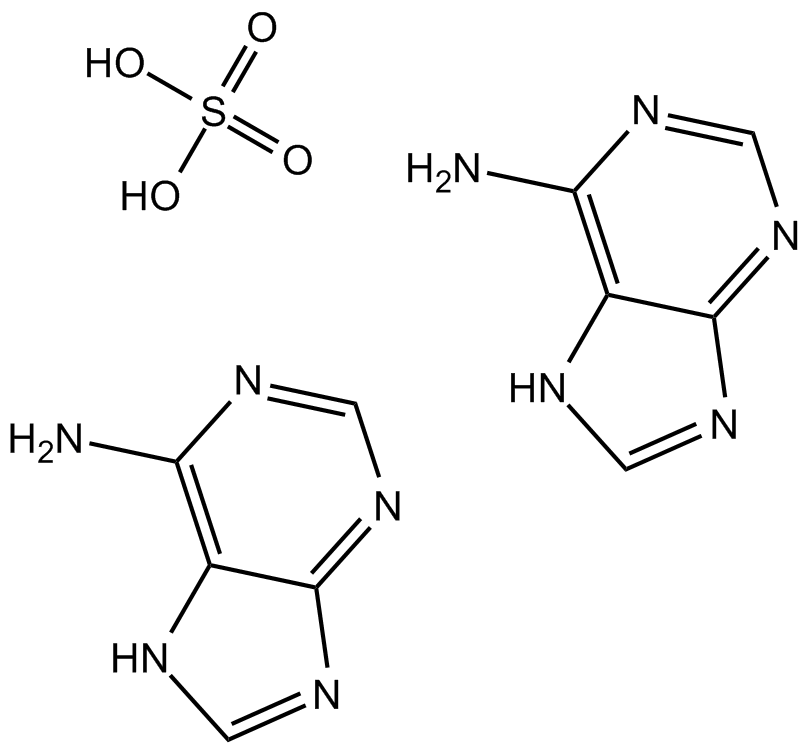
GC17278
Adenine sulfate
Adenine sulfate (6-Aminopurine hemisulfate), a purine, is one of the four nucleobases in the nucleic acid of DNA. Adenine sulfate acts as a chemical component of DNA and RNA. Adenine sulfate also plays an important role in biochemistry involved in cellular respiration, the form of both ATP and the cofactors (NAD and FAD), and protein synthesis.
-
GC68624
Adenine-13C
Adenine-8-C13; 9H-Purin-6-amine-8-13C
Adenine-13C is a 13C-labeled Adenine. Adenine (6-Aminopurine) is a purine and one of the four nucleobases in DNA. Adenine is a chemical component of both DNA and RNA. It plays an important role in cellular respiration, the formation of ATP, coenzymes NAD and FAD, as well as protein synthesis.

-
GC67946
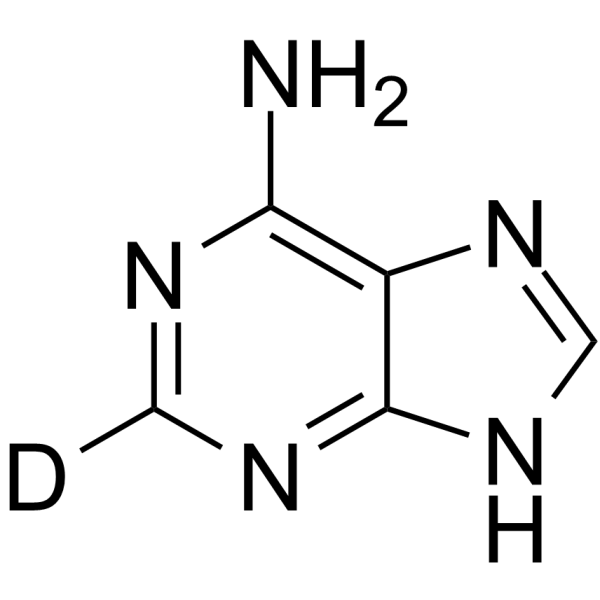
Adenine-d1
6-Aminopurine-d1; Vitamin B4-d1