Immunology/Inflammation
The immune and inflammation-related pathway including the Toll-like receptors pathway, the B cell receptor signaling pathway, the T cell receptor signaling pathway, etc.
Toll-like receptors (TLRs) play a central role in host cell recognition and responses to microbial pathogens. TLR4 initially recruits TIRAP and MyD88. MyD88 then recruits IRAKs, TRAF6, and the TAK1 complex, leading to early-stage activation of NF-κB and MAP kinases [1]. TLR4 is endocytosed and delivered to intracellular vesicles and forms a complex with TRAM and TRIF, which then recruits TRAF3 and the protein kinases TBK1 and IKKi. TBK1 and IKKi catalyze the phosphorylation of IRF3, leading to the expression of type I IFN [2].
BCR signaling is initiated through ligation of mIg under conditions that induce phosphorylation of the ITAMs in CD79, leading to the activation of Syk. Once Syk is activated, the BCR signal is transmitted via a series of proteins associated with the adaptor protein B-cell linker (Blnk, SLP-65). Blnk binds CD79a via non-ITAM tyrosines and is phosphorylated by Syk. Phospho-Blnk acts as a scaffold for the assembly of the other components, including Bruton’s tyrosine kinase (Btk), Vav 1, and phospholipase C-gamma 2 (PLCγ2) [3]. Following the assembly of the BCR-signalosome, GRB2 binds and activates the Ras-guanine exchange factor SOS, which in turn activates the small GTPase RAS. The original RAS signal is transmitted and amplified through the mitogen-activated protein kinase (MAPK) pathway, which including the serine/threonine-specific protein kinase RAF followed by MEK and extracellular signal related kinases ERK 1 and 2 [4]. After stimulation of BCR, CD19 is phosphorylated by Lyn. Phosphorylated CD19 activates PI3K by binding to the p85 subunit of PI3K and produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) from PIP2, and PIP3 transmits signals downstream [5].
Central process of T cells responding to specific antigens is the binding of the T-cell receptor (TCR) to specific peptides bound to the major histocompatibility complex which expressed on antigen-presenting cells (APCs). Once TCR connected with its ligand, the ζ-chain–associated protein kinase 70 molecules (Zap-70) are recruited to the TCR-CD3 site and activated, resulting in an initiation of several signaling cascades. Once stimulation, Zap-70 forms complexes with several molecules including SLP-76; and a sequential protein kinase cascade is initiated, consisting of MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAPKK), and MAP kinase (MAPK) [6]. Two MAPK kinases, MKK4 and MKK7, have been reported to be the primary activators of JNK. MKK3, MKK4, and MKK6 are activators of P38 MAP kinase [7]. MAP kinase pathways are major pathways induced by TCR stimulation, and they play a key role in T-cell responses.
Phosphoinositide 3-kinase (PI3K) binds to the cytosolic domain of CD28, leading to conversion of PIP2 to PIP3, activation of PKB (Akt) and phosphoinositide-dependent kinase 1 (PDK1), and subsequent signaling transduction [8].
References
[1] Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature immunology, 2010, 11(5): 373-384.
[2] Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650.
[3] Packard T A, Cambier J C. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation[J]. F1000Prime Rep, 2013, 5(40.10): 12703.
[4] Zhong Y, Byrd J C, Dubovsky J A. The B-cell receptor pathway: a critical component of healthy and malignant immune biology[C]//Seminars in hematology. WB Saunders, 2014, 51(3): 206-218.
[5] Baba Y, Matsumoto M, Kurosaki T. Calcium signaling in B cells: regulation of cytosolic Ca 2+ increase and its sensor molecules, STIM1 and STIM2[J]. Molecular immunology, 2014, 62(2): 339-343.
[6] Adachi K, Davis M M. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells[J]. Proceedings of the National Academy of Sciences, 2011, 108(4): 1549-1554.
[7] Rincón M, Flavell R A, Davis R A. The Jnk and P38 MAP kinase signaling pathways in T cell–mediated immune responses[J]. Free Radical Biology and Medicine, 2000, 28(9): 1328-1337.
[8] Bashour K T, Gondarenko A, Chen H, et al. CD28 and CD3 have complementary roles in T-cell traction forces[J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2241-2246.
Targets for Immunology/Inflammation
- Cyclic GMP-AMP Synthase(1)
- Apoptosis(137)
- 5-Lipoxygenase(18)
- TLR(98)
- Papain(1)
- PGDS(1)
- PGE synthase(24)
- SIKs(10)
- IκB/IKK(60)
- AP-1(1)
- KEAP1-Nrf2(35)
- NOD1(1)
- NF-κB(214)
- Interleukin Related(120)
- 15-lipoxygenase(2)
- Others(10)
- Aryl Hydrocarbon Receptor(32)
- CD73(16)
- Complement System(46)
- Galectin(30)
- IFNAR(19)
- NO Synthase(69)
- NOD-like Receptor (NLR)(36)
- STING(83)
- Reactive Oxygen Species(384)
- FKBP(11)
- eNOS(4)
- iNOS(24)
- nNOS(20)
- Glutathione(37)
- Adaptive Immunity(144)
- Allergy(129)
- Arthritis(25)
- Autoimmunity(134)
- Gastric Disease(64)
- Immunosuppressants(27)
- Immunotherapeutics(3)
- Innate Immunity(411)
- Pulmonary Diseases(76)
- Reactive Nitrogen Species(43)
- Specialized Pro-Resolving Mediators(42)
- Reactive Sulfur Species(24)
Products for Immunology/Inflammation
- Cat.No. Nombre del producto Información
-
GC35412
Asperulosidic Acid
El Ácido asperulosÍdico (ASPA), un glucÓsido iridoide bioactivo, se extrae de las hierbas de Hedyotis diffusa Willd.
-
GC42860
Aspochalasin D
Aspochalasin D is a co-metabolite originally isolated from A.
-
GC46886
Aspyrone
A fungal metabolite with diverse biological activities
-
GC31350
Astaxanthin
La astaxantina, el carotenoide dietético rojo, es un antioxidante potente y eficaz por vÍa oral.
-
GC68702
Astegolimab
Astegolimab (MSTT 1041A; RG 6149) es un anticuerpo monoclonal humano IgG2 que puede bloquear la señalización de IL-33 al dirigirse al receptor ST2 de IL-33. Astegolimab tiene potencial para su uso en la investigación de enfermedad pulmonar obstructiva crónica (EPOC).

-
GC41640
Asterriquinol D dimethyl ether
El asterriquinol D dimetil éter es un metabolito fÚngico que puede inhibir las lÍneas celulares NS-1 del mieloma de ratÓn con una IC50 de 28 μg/mL.
-
GN10415
Astilbin
Taxifolin 3-O-rhamnoside
-
GC42863
Asukamycin
AM1042, Asukamycin A
Asukamycin is polyketide isolated from the S.
-
GC32457
Asymmetric dimethylarginine
ADMA, Asymmetric dimethylarginine
La dimetilarginina asimétrica es un inhibidor endÓgeno de la Óxido nÍtrico sintasa (NOS) y funciona como marcador de disfunciÓn endotelial en varios estados patolÓgicos.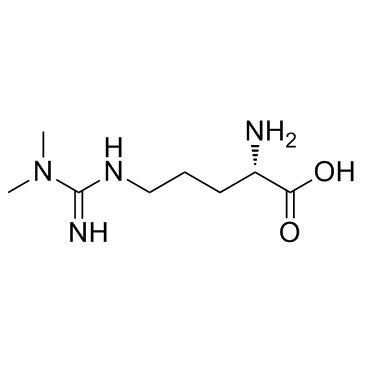
-
GC46091
Aszonapyrone A
La aszonapirona A es un metabolito producido por Aspergillus zonatus.
-
GC39554
AT2 receptor agonist C21
Compound 21
El agonista del receptor AT2 C21 es un agonista selectivo del receptor AT2 de la angiotensina II similar a un fÁrmaco con valores de Ki de 0,4 nM y >10 μM para el receptor AT2 y el receptor AT1, respectivamente.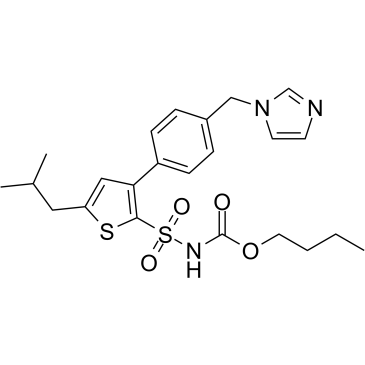
-
GC62334
AT791
AT791 es un inhibidor de TLR7 y TLR9 potente y biodisponible por vÍa oral.
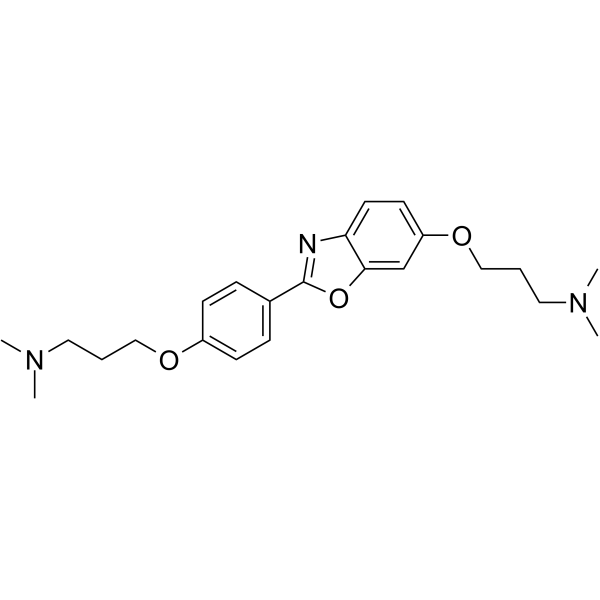
-
GC46887
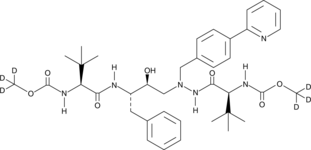
Atazanavir-d6
An internal standard for the quantification of atazanavir
-
GC12537
ATB-337
ACS 15,S-Diclofenac
ATB-337 es una molécula hÍbrida de un donante de H2S y el AINE diclofenaco.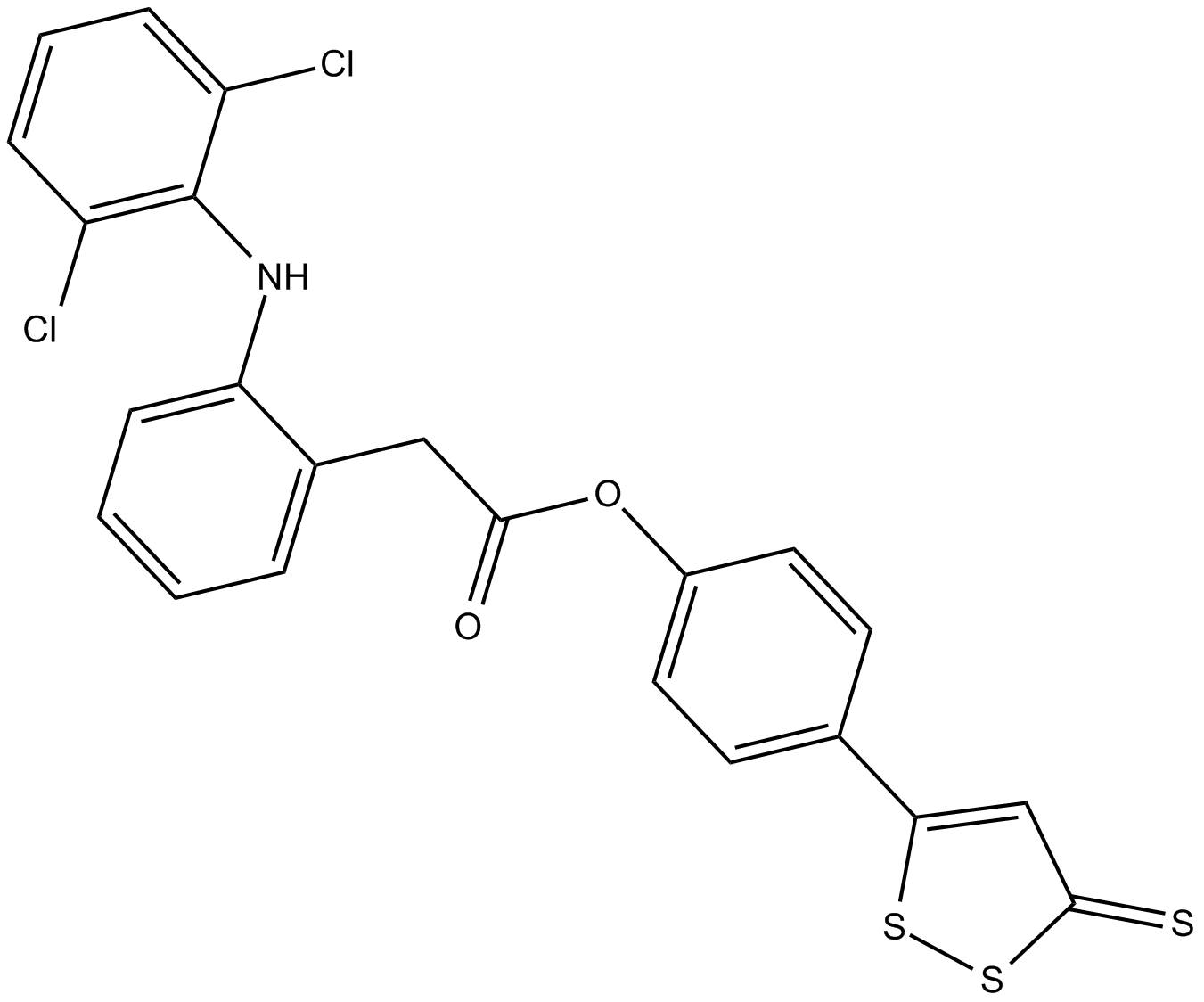
-
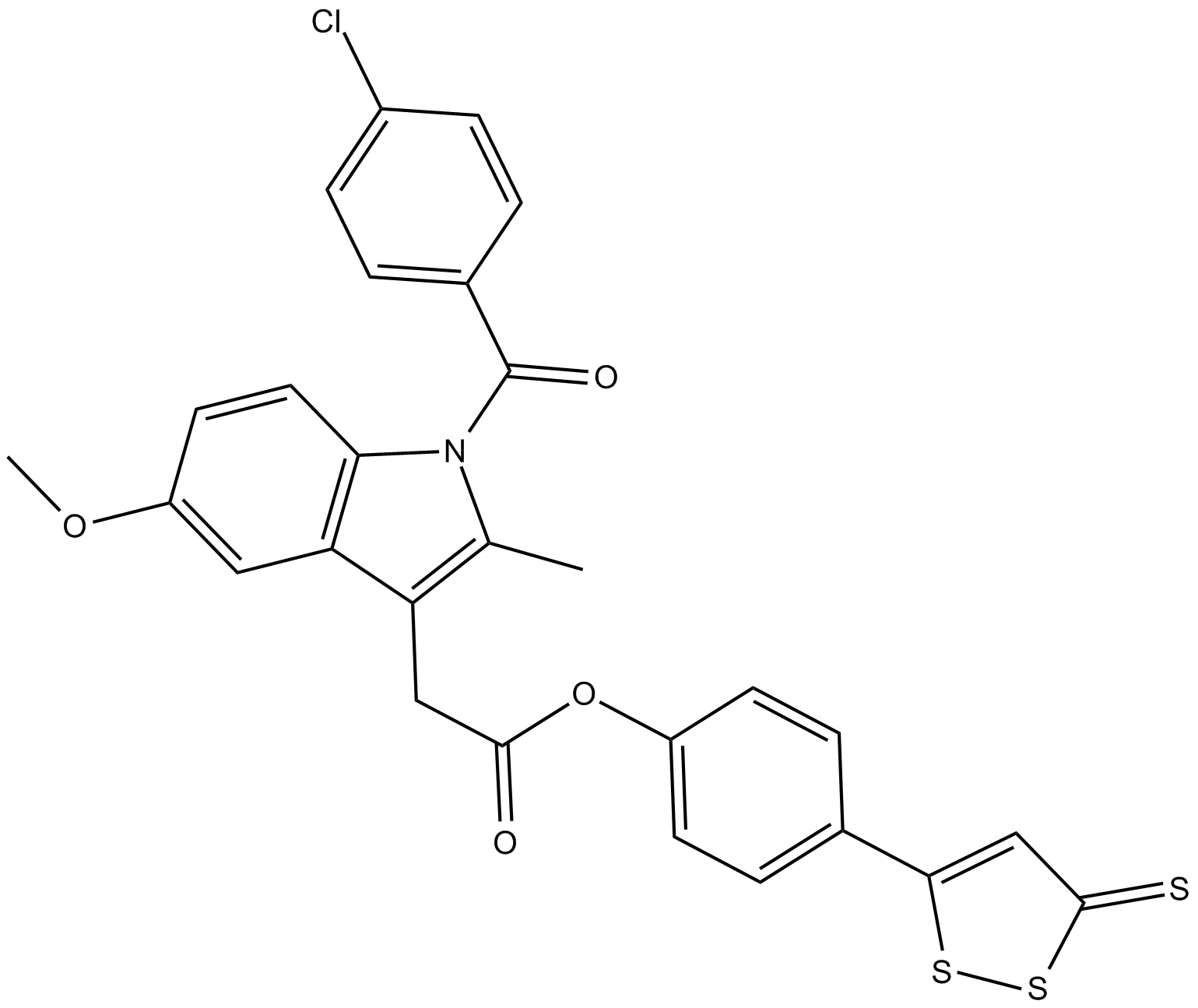
GC16245
ATB-343
hybrid molecule of an H2S donor and the NSAID indomethacin
-
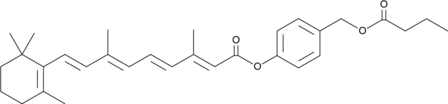
GC46892
ATRA-BA Hybrid
A prodrug form of all-trans retinoic acid and butyric acid
-
GN10627
Atractylenolide I
-
GC48925
Aureonitol
A fungal metabolite
-
GC41490
Aureusimine B
Phevalin
Aureusimine B, also known as phevalin, is a natural pyrazinone produced by certain fungi and by Staphylococcus spp., including S.
-
GC46895
Aurintricarboxylic Acid (ammonium salt)
ATA
A protein synthesis inhibitor with diverse biological activities
-
GC40005
Aurodox
1-methyl-Mocimycin
Aurodox is a polyketide antibiotic originally isolated from S.
-
GC49646
Aurothioglucose (hydrate)
Gold Thioglucose
A TrxR inhibitor
-
GC42877
AUY954
AUY954 is an orally bioavailable and selective agonist of the sphingosine-1-phosphate receptor 1 (S1P1; EC50 = 1.2 nM for stimulating GTPγS binding to S1P1 in CHO cells).
-
GC32486
AVE-3085
AVE-3085 es un potente potenciador de la sintasa de óxido nítrico endotelial, utilizado para el tratamiento de enfermedades cardiovasculares.
-
GC42880
Avenanthramide-C methyl ester
Avenanthramide-C methyl ester is an inhibitor of NF-κB activation that acts by blocking the phosphorylation of IKK and IκB (IC50 ~ 40 μM).
-
GC45388
Averantin
(–)-Averantin
La averantina es el metabolito menor del hongo Cercospora arachidicola.
-
GC42881
Avermectin B1a aglycone
Avermectin B1a aglycone is an aglycone form of the anthelmintic and insecticide avermectin B1a.
-
GC42882
Avermectin B1a monosaccharide
Avermectin B1a monosaccharide is a macrolide anthelmintic and monosaccharide form of avermectin B1a.
-
GC45984
Avilamycin A
An antibiotic
-
GC48511
Avrainvillamide
CJ-17,665
La avrainvillamida ((+)-Avrainvillamida) es un alcaloide natural con efectos antiproliferativos, se une a la chaperona nuclear nucleofosmina, una proteÍna oncogénica propuesta que se sobreexpresa en muchos tumores humanos diferentes.
-
GC42885
AX 048
The group IVA phospholipase A2 (PLA2), known as calcium-dependent cytosolic PLA2 (cPLA2), selectively releases arachidonic acid from membrane phospholipids, playing a central role in initiating the synthesis of prostaglandins and leukotrienes.
-
GC35440
AX-024
AX-024 es un inhibidor primero en su clase disponible por vÍa oral de la interacciÓn TCR-Nck que inhibe selectivamente la activaciÓn de células T desencadenada por TCR con un IC50 ~ 1 nM.
-
GC19046
AX-024 hydrochloride
El clorhidrato de AX-024 es un inhibidor primero en su clase disponible por vÍa oral de la interacciÓn TCR-Nck que inhibe selectivamente la activaciÓn de células T desencadenada por TCR con un IC50 ~1 nM.
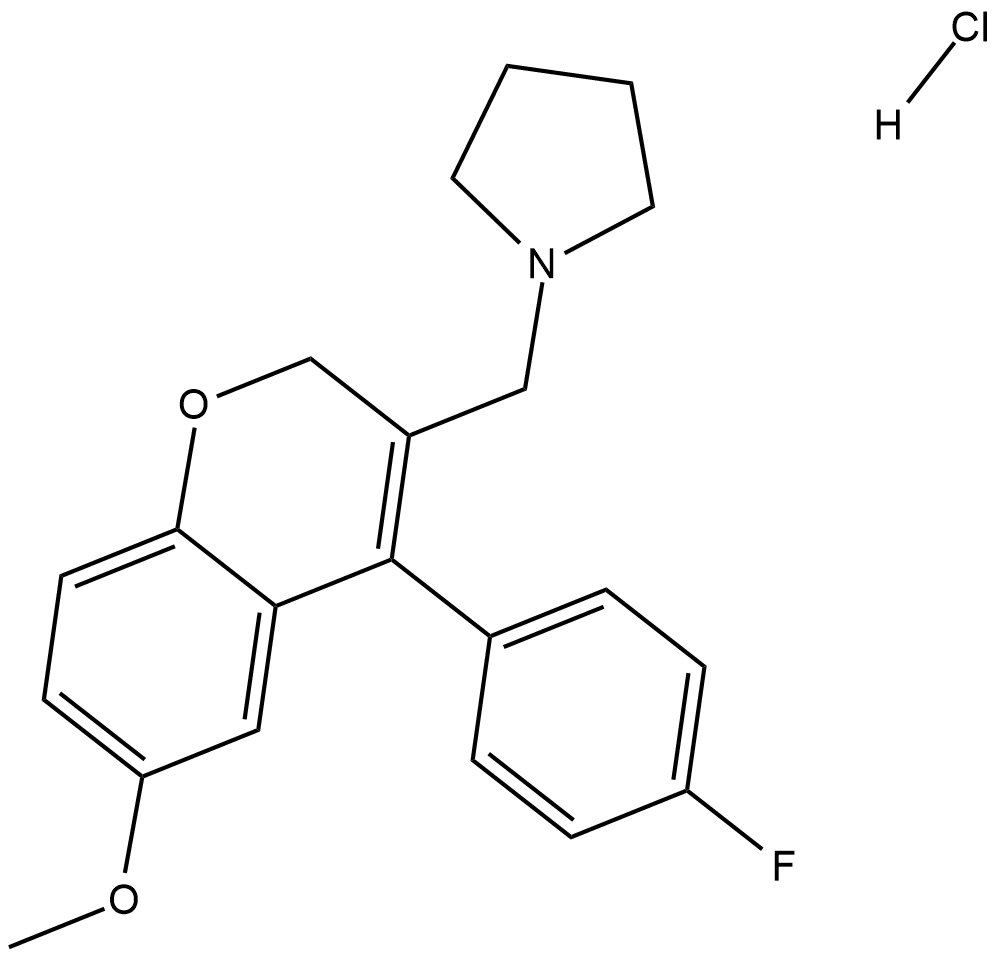
-
GC65283
AXC-715 trihydrochloride
T785 trihydrochloride
El trihidrocloruro de AXC-715 (T785) es un agonista dual de TLR7/TLR8, extraÍdo de la patente WO2020168017 A1.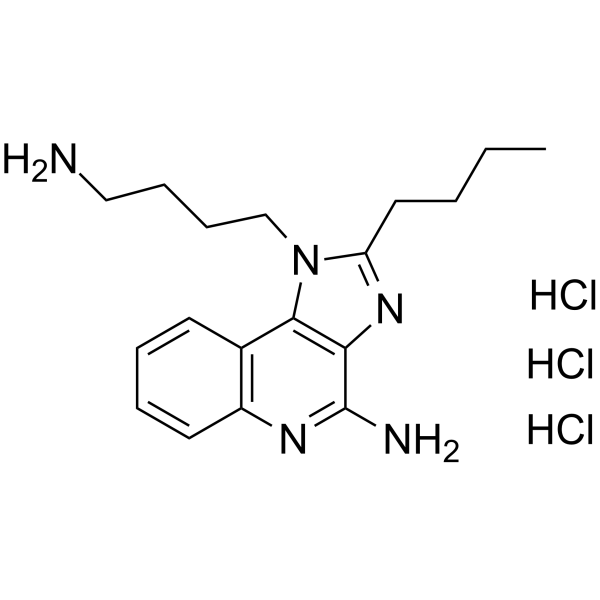
-
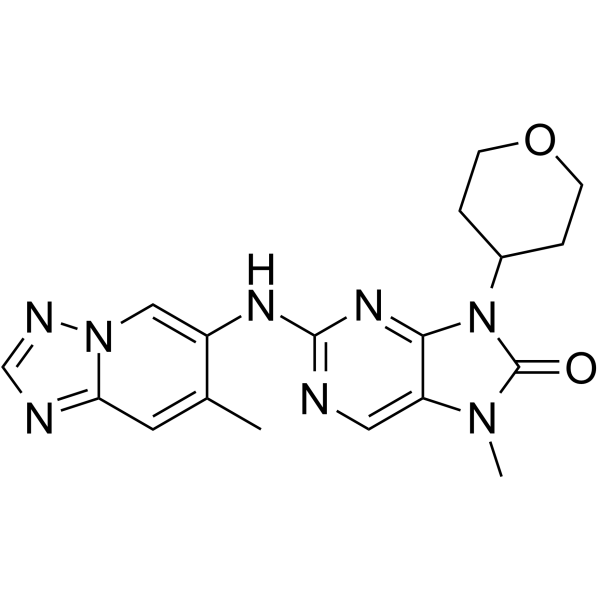
GC64938
AZD-7648
AZD-7648 es un potente inhibidor selectivo de DNA-PK activo por vÍa oral con una IC50 de 0,6 nM. AZD-7648 induce apoptosis y muestra actividad antitumoral.
-
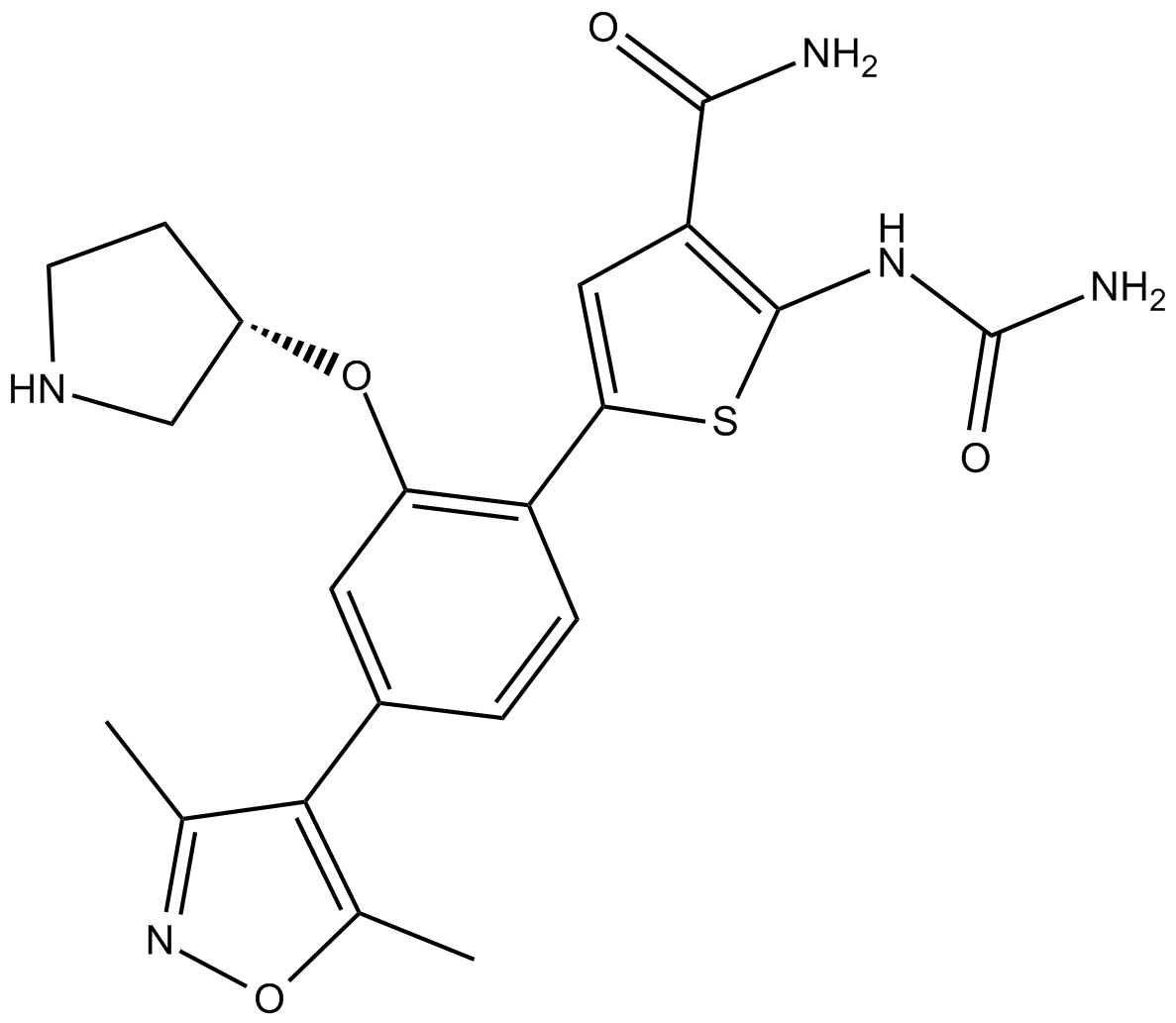
GC10135
AZD3264
AZD3264 es un inhibidor selectivo de IkB-quinasa IKK2.
-
GC62488
AZD8848
AZD8848 es un agonista antifÁrmaco selectivo del receptor 7 tipo toll (TLR7) desarrollado para la investigaciÓn del asma y la rinitis alérgica.
-
GC49057
Azelastine-13C-d3 (hydrochloride)
An internal standard for the quantification of azelastine
-
GC42891
azido-FTY720
Azido-FTY720 es un anÁlogo fotoactivable de FTY720.
-
GC46903
Azithromycin-d3
La azitromicina-d3 (CP 62993-d3) es la azitromicina marcada con deuterio.
-
GC46904
Azoxystrobin
ICI-A 5504
La azoxistrobina es un fungicida de β-metoxiacrilato de amplio espectro.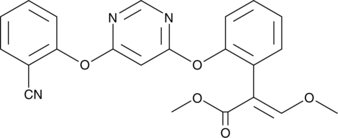
-
GC60616
AZT triphosphate
3'-Azido-3'-deoxythymidine-5'-triphosphate
El trifosfato de AZT (3'-azido-3'-desoxitimidina-5'-trifosfato) es un metabolito trifosfato activo de la zidovudina (AZT).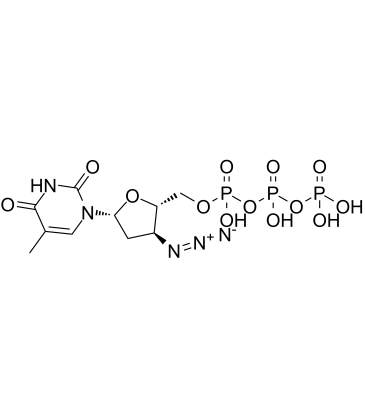
-
GC60617
AZT triphosphate TEA
3'-Azido-3'-deoxythymidine-5'-triphosphate TEA
AZT trifosfato TEA (3'-azido-3'-desoxitimidina-5'-trifosfato TEA) es un metabolito trifosfato activo de zidovudina (AZT).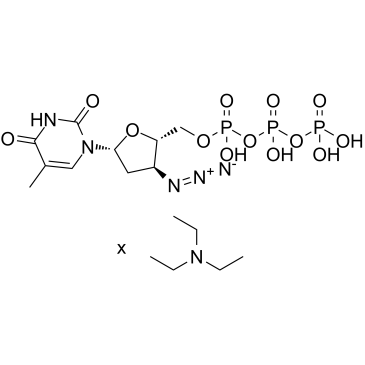
-
GC45795
Aztreonam-d6
SQ 26,776-d6
An internal standard for the quantification of aztreonam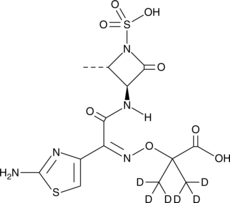
-
GC39280
B022
B022 es un inhibidor potente y selectivo de la cinasa inductora de NF-κB (NIK) (Ki de 4,2 nM; IC50 = 15,1 nM).
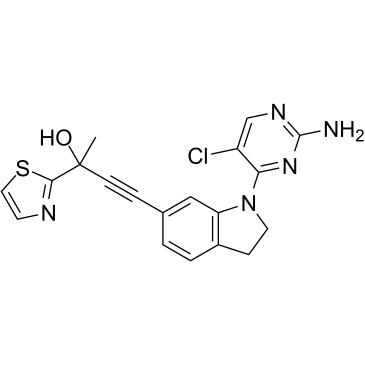
-
GC18580
B355252
B355252, una molécula pequeña de fenoxi tiofeno sulfonamida, es un potente agonista del receptor de NGF.
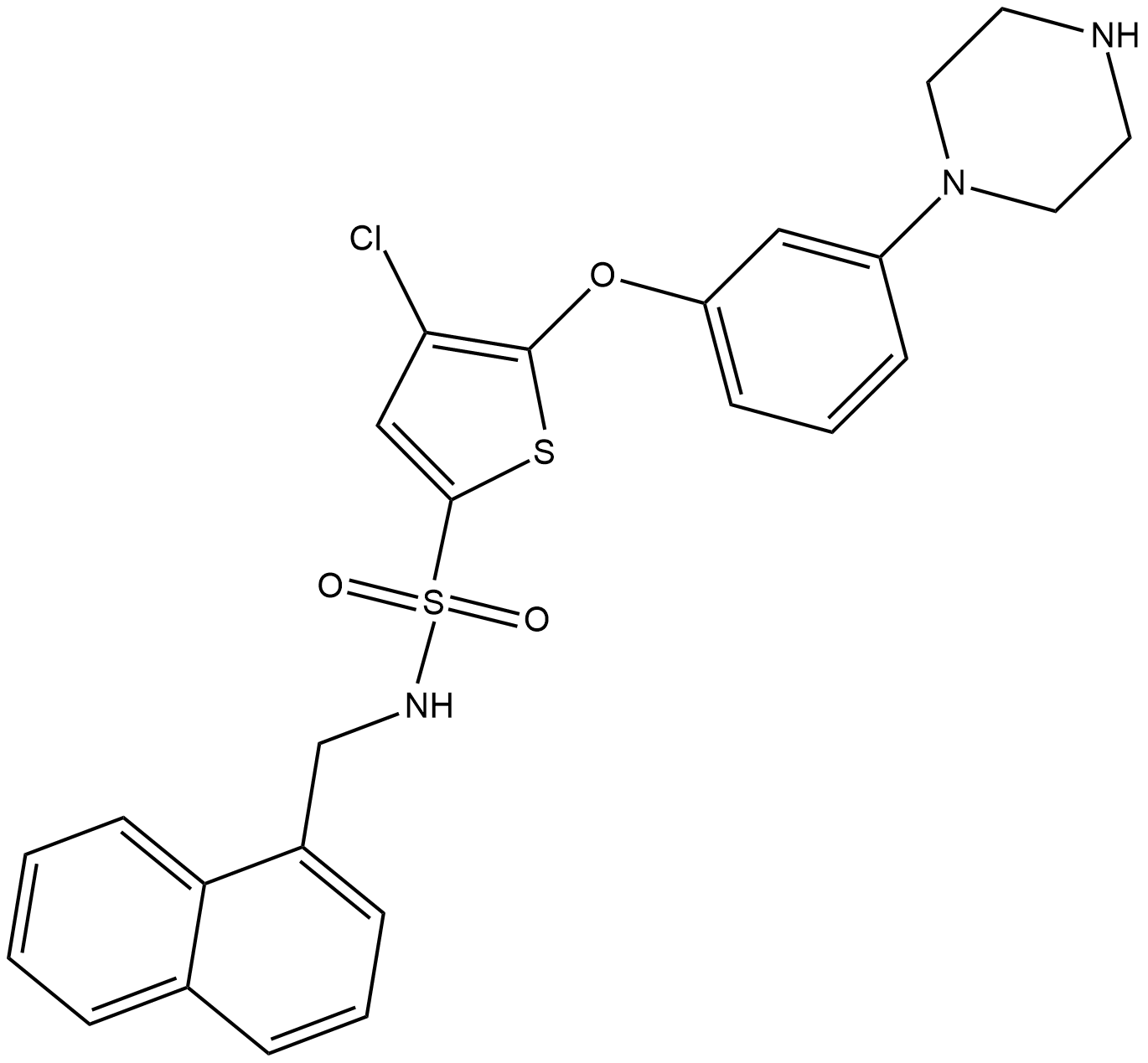
-
GC42895
Bacillosporin C
Bacillosporin C is an oxaphenalenone dimer originally isolated from T.
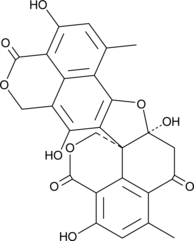
-
GC49793
Bacitracin A (technical grade)
NSC 45737
A polypeptide antibiotic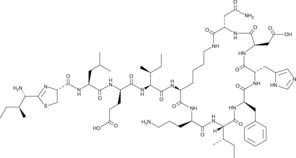
-
GC46905
Bacitracin Complex
A mixture of bacitracin polypeptides in complex with copper
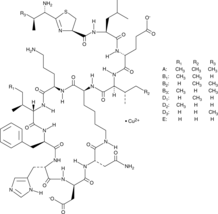
-
GC45938
Bacopaside X
Bacopaside VII, Jujubogenin isomer of Bacopasaponin C
El bacopasido X se encuentra en Bacopa monnieri y muestra una afinidad de uniÓn hacia el receptor D1.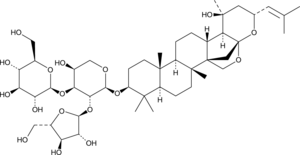
-
GC49302
Bactenecin (bovine) (trifluoroacetate salt)
H-Arg-Leu-Cys-Arg-Ile-Val-Val-Ile-Arg-Val-Cys-Arg-OH, RLCRIVVIRVCR-OH
A cationic peptide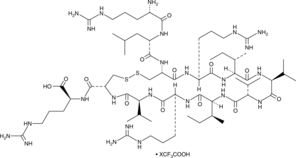
-
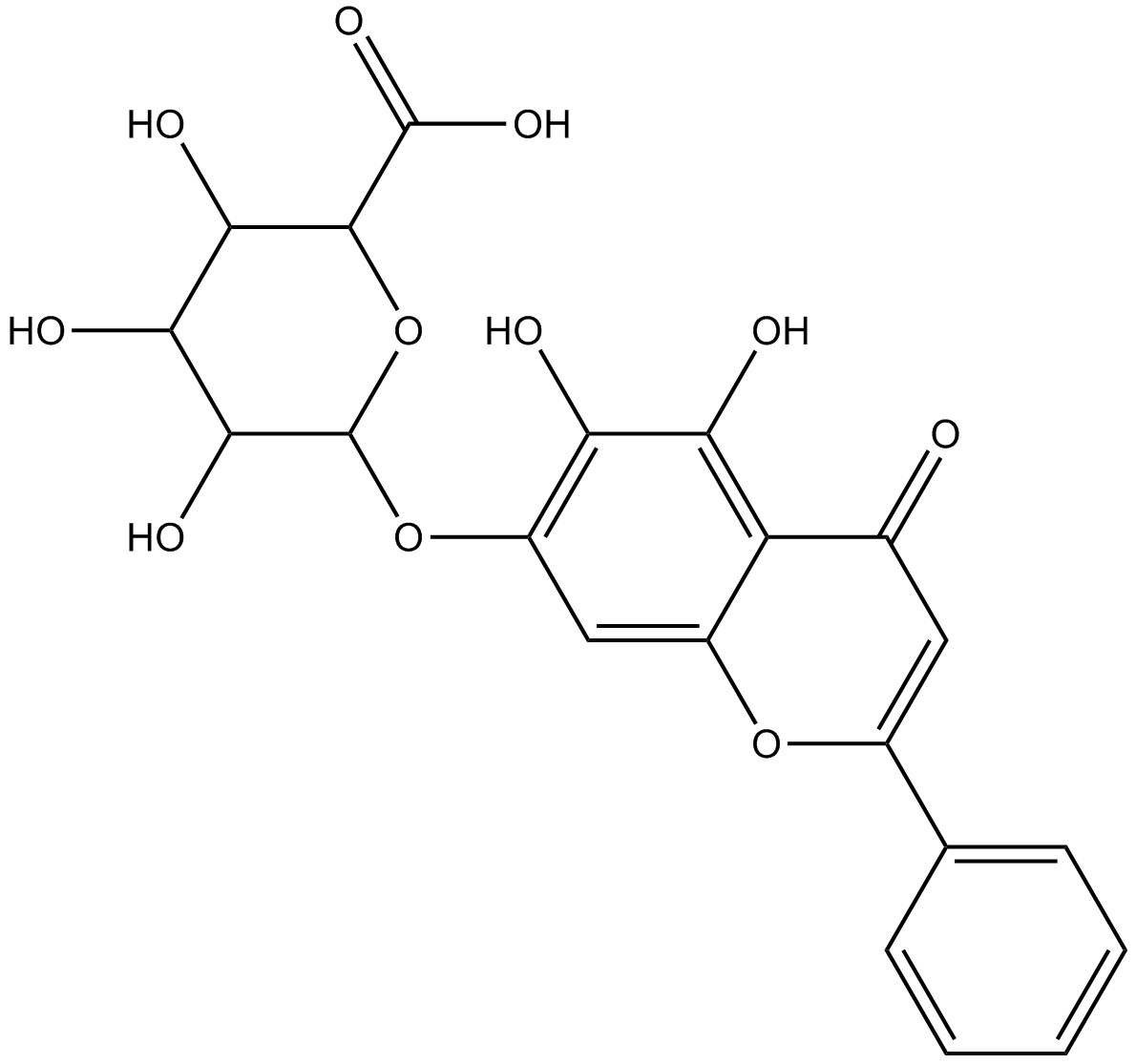
GN10018
Baicalin
Baicalein 7-glucuronide
Un flavonoide con diversas actividades biológicas.
-
GC52344
Bak BH3 (72-87) (human) (trifluoroacetate salt)
A Bak-derived peptide
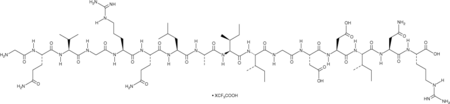
-
GC18126
Balsalazide
La balsalazida podrÍa suprimir la carcinogénesis asociada a la colitis mediante la modulaciÓn de la vÍa IL-6/STAT3.
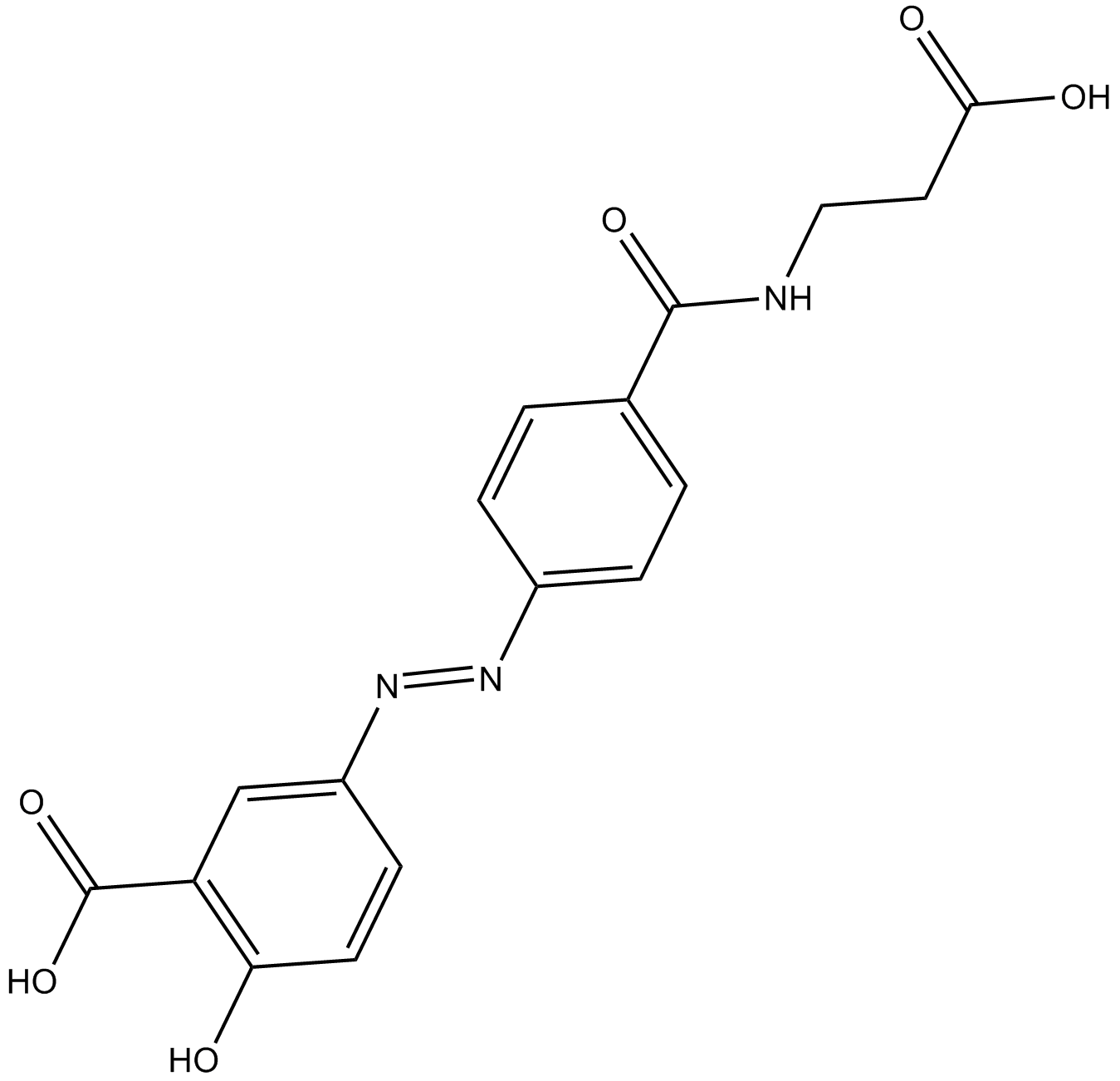
-
GC35466
Balsalazide sodium hydrate
El hidrato de sodio de balsalazida podrÍa suprimir la carcinogénesis asociada a la colitis a través de la modulaciÓn de la vÍa IL-6/STAT3.
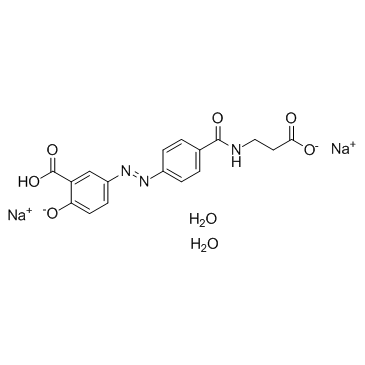
-
GC17574
BAPTA
BAPTA es un quelante selectivo del calcio.
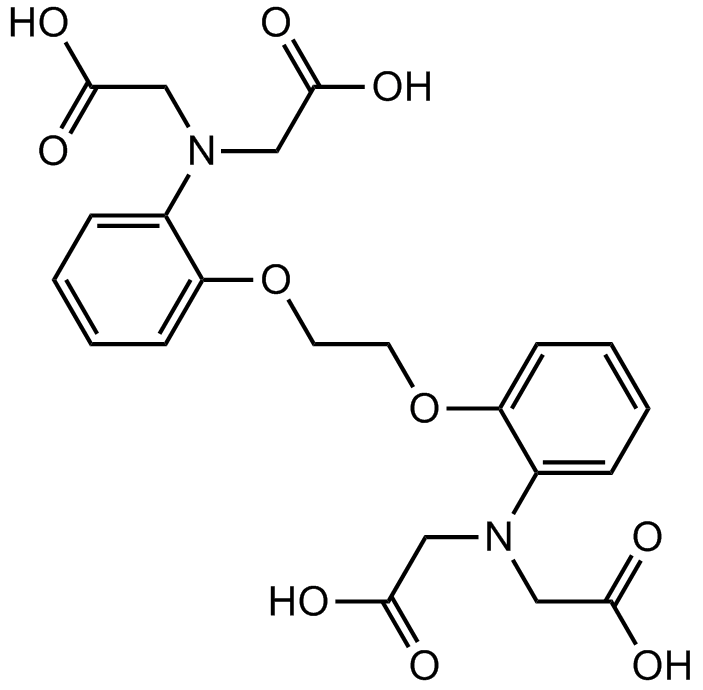
-
GC18313
BAR501 Impurity
BAR501 impurity is an impurity found in the preparation of BAR501 that acts as an agonist of the G protein-coupled bile acid-activated receptor (GP-BAR1).
-
GC66331
Basiliximab
CHI 621
Basiliximab (CHI 621) es un anticuerpo anti-receptor de interleucina-2 monoclonal murino/humano IgG1 recombinante quimérico. Basiliximab se puede utilizar para la investigaciÓn del trasplante renal.
-
GC52476
Bax Inhibitor Peptide V5 (trifluoroacetate salt)
BIP V5, VPMLK
A Bax inhibitor
-
GC10345
Bay 11-7085
BAY 11-7085 (BAY 11-7083) es un inhibidor de la activaciÓn de NF-κB y la fosforilaciÓn de IκBα; estabiliza el IκBα con una IC50 de 10 μM.
-
GC13035
Bay 11-7821
BAY 11-7082
Un inhibidor selectivo e irreversible de NF-κB.
-
GC42897
BAY 61-3606 (hydrochloride)
BAY 61-3606 is a cell-permeable, reversible inhibitor of spleen tyrosine kinase (Syk; Ki = 7.5 nM; IC50 = 10 nM).
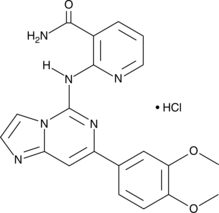
-
GC35474
Bay 65-1942 free base
La base libre Bay 65-1942 es un inhibidor de IKKβ selectivo y competitivo con ATP.
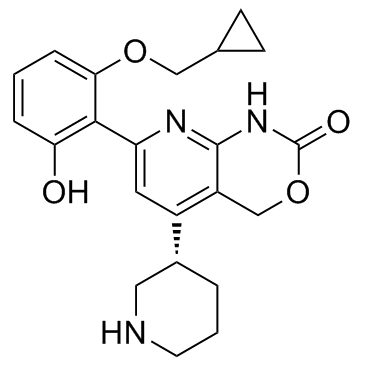
-
GC16303
Bay 65-1942 HCl salt
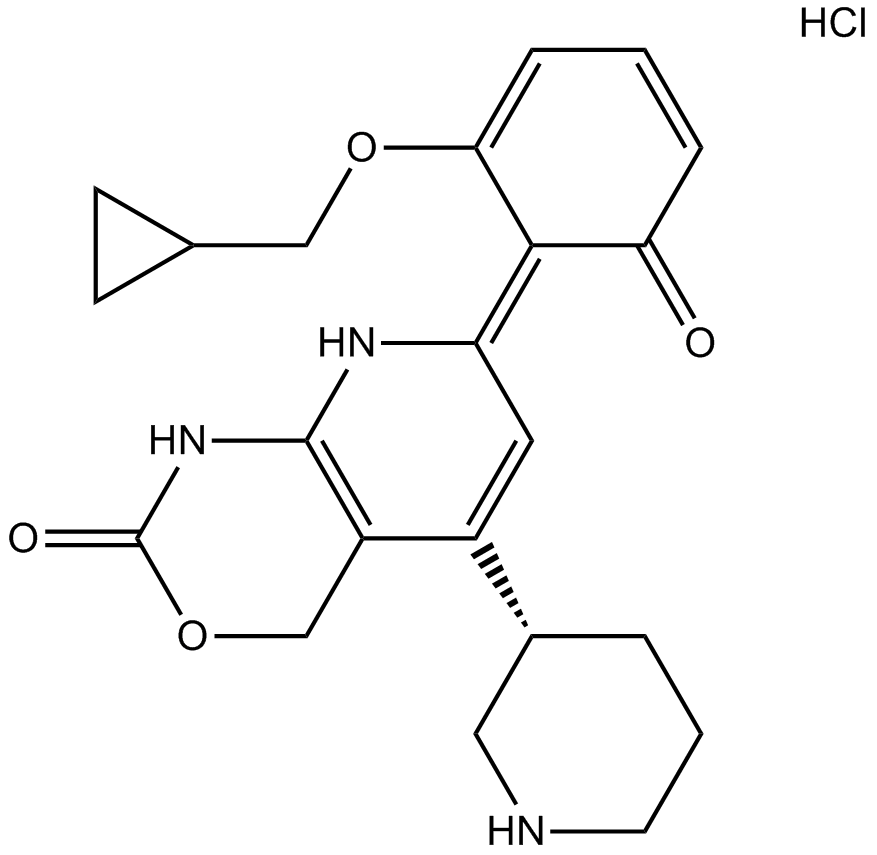
-
GC35475
Bay 65-1942 R form
La forma R de Bay 65-1942 es la forma R menos activa de Bay 65-1942.
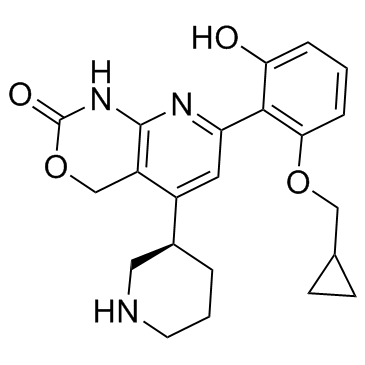
-
GC60624
BAY-985
BAY-985 es un inhibidor dual de TBK1 e IKKε altamente potente, oralmente activo y selectivo competitivo con ATP con IC50 de 2/30 y 2 nM para TBK1 (ATP bajo/alto) e IKKε, respectivamente. Eficacia antitumoral.
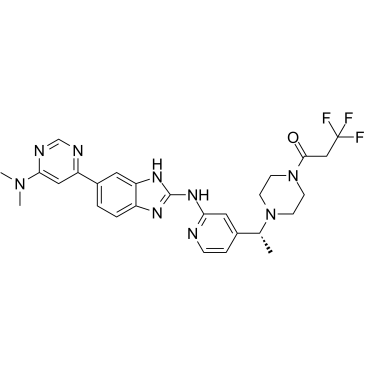
-
GC12232
BAY-X 1005
BAY X 1005; DG-031
BAY-X 1005 (BAY X 1005; DG-031) es un inhibidor de la proteÍna activadora de la 5-lipoxigenasa (FLAP) selectivo y activo por vÍa oral.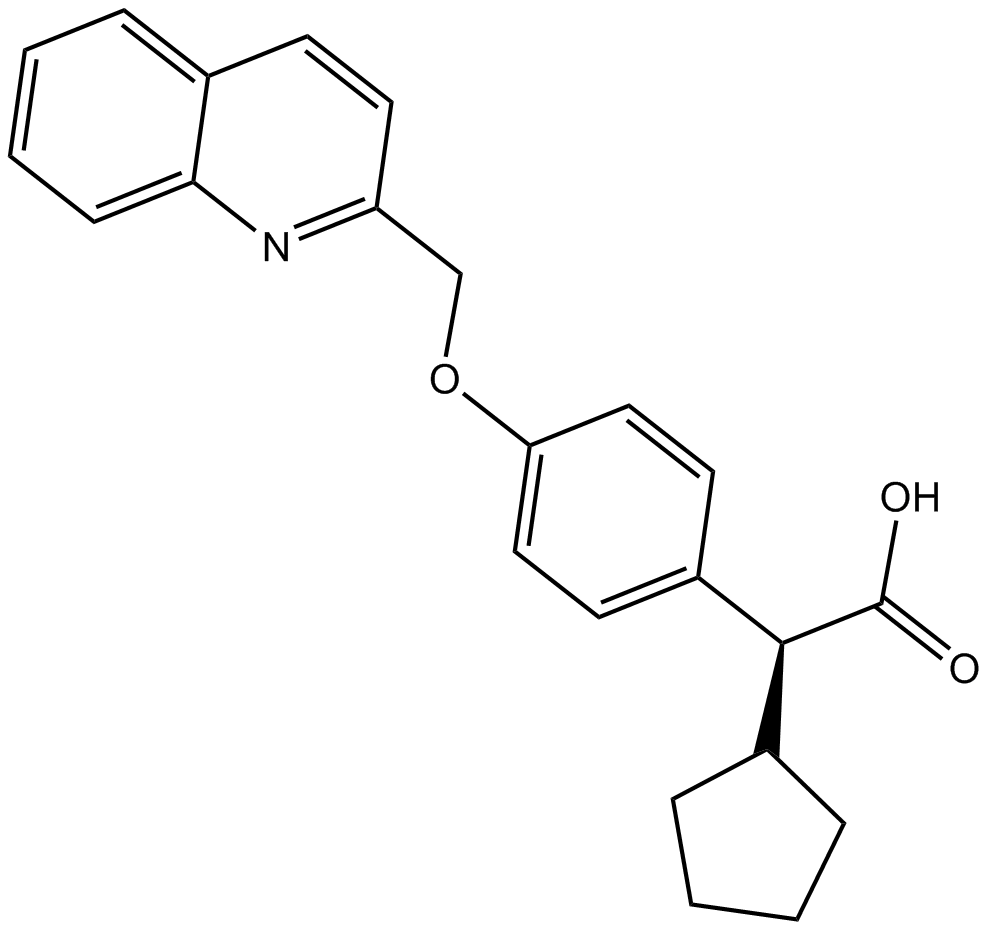
-
GC18487
BC-1215
BC-1215 es un inhibidor de la proteÍna 3 de la caja F (Fbxo3).
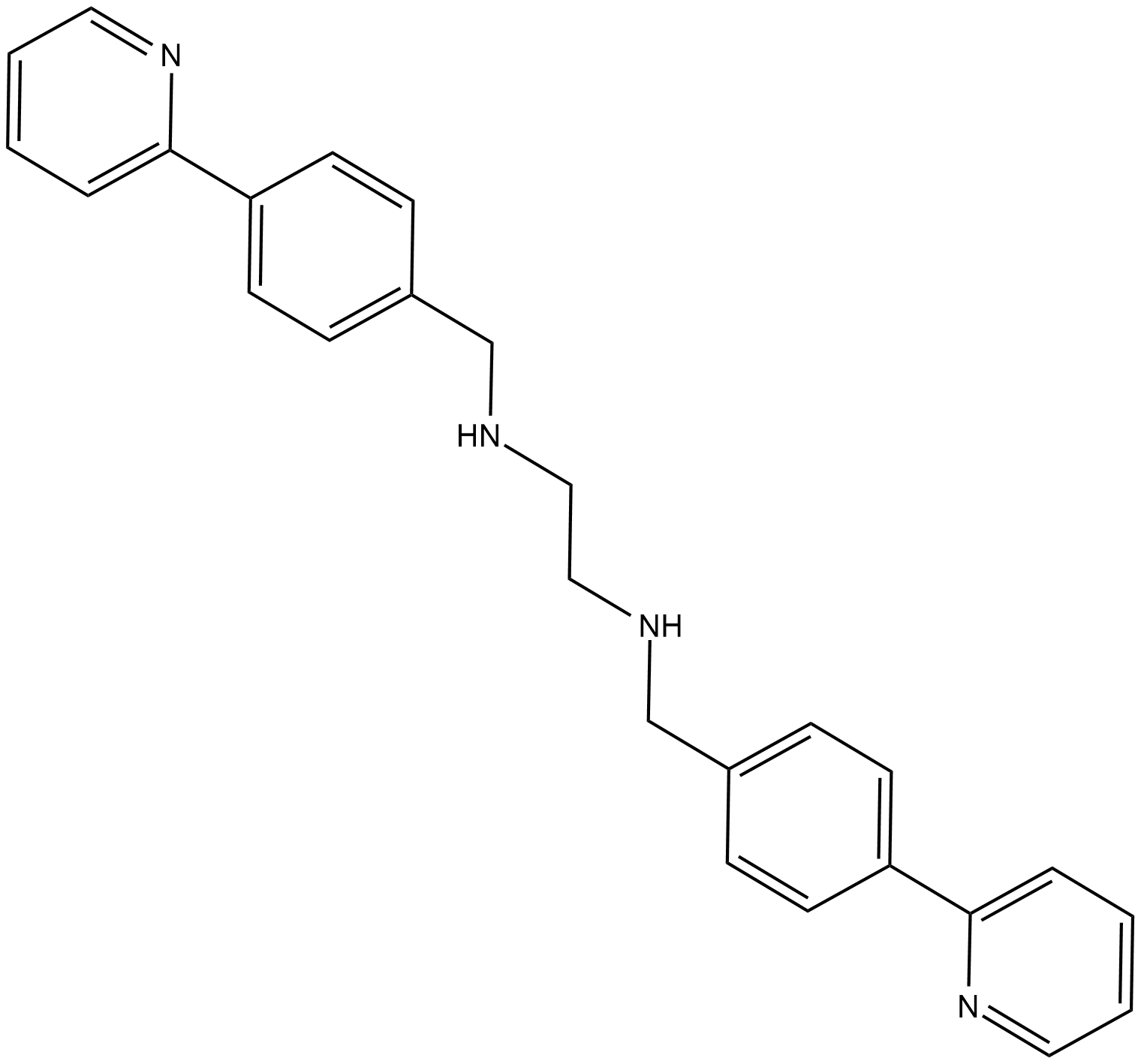
-
GC41583
BCN-E-BCN
BCN-E-BCN is a strained cycloalkyne probe for detecting proteins that have been sulfenylated, the first intermediate step in protein oxidation.
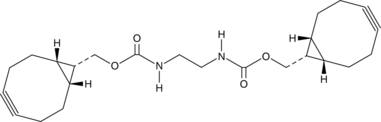
-
GC35481
BCX 1470
BCX 1470 inhibe la actividad esterolÍtica del factor D (IC50=96 nM) y C1s (IC50=1,6 nM), 3,4 y 200 veces mejor, respectivamente, que la de la tripsina.
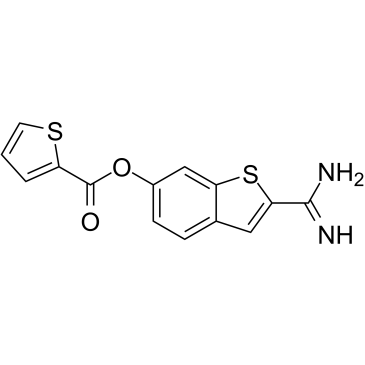
-
GC35482
BCX 1470 methanesulfonate
El metanosulfonato BCX 1470 inhibe la actividad esterolÍtica del factor D (IC50=96 nM) y C1s (IC50=1,6 nM), 3,4 y 200 veces mejor, respectivamente, que la de la tripsina.
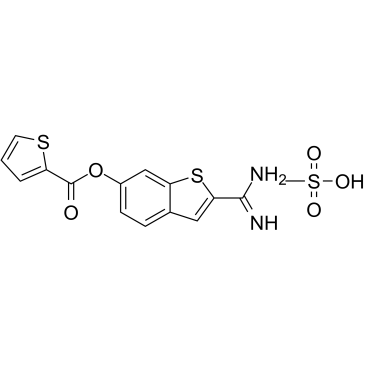
-
GC46908
BE-24566B
L-755,805
A fungal metabolite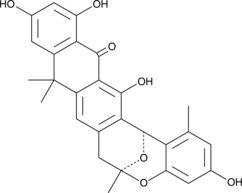
-
GC46910
Beauvericin A
A cyclodepsipeptide with diverse biological activities
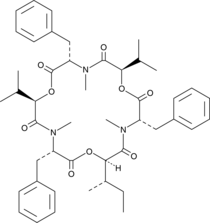
-
GC49038
Benanomicin A
A microbial metabolite with antifungal, fungicidal, and antiviral activities
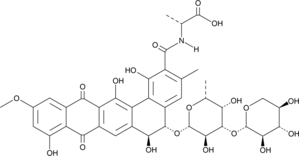
-
GC52468
Benanomicin B
Antibiotic BU 3608C, Pradimicin C
A microbial metabolite with antifungal, fungicidal, and antiviral activities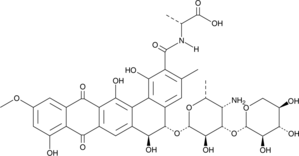
-
GC49040
Benanomicin B (formate)
A microbial metabolite with antifungal, fungicidal, and antiviral activities
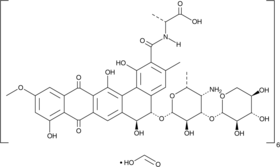
-
GC49042
Benastatin A
A bacterial metabolite with diverse biological activities
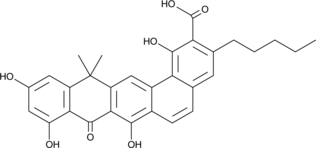
-
GC49043
Benastatin B
A bacterial metabolite with diverse biological activities
-
GC49044
Benastatin C
A bacterial metabolite with diverse biological activities
-
GC64354
Bendamustine
La bendamustina (SDX-105 base libre), un anÁlogo de purina, es un agente de entrecruzamiento del ADN. La bendamustina activa la respuesta al estrés por daÑo del ADN y la apoptosis. La bendamustina tiene potentes propiedades alquilantes, anticancerÍgenas y antimetabolitos.
-
GC34046
Bendazol
Bendazol es un fÁrmaco hipotensor que también puede aumentar la actividad de la NO sintasa en los glomérulos renales y tÚbulos colectores.
-
GC15949
Benfotiamine
Benzoylthiamine monophosphate
A lipid-soluble form of vitamin B1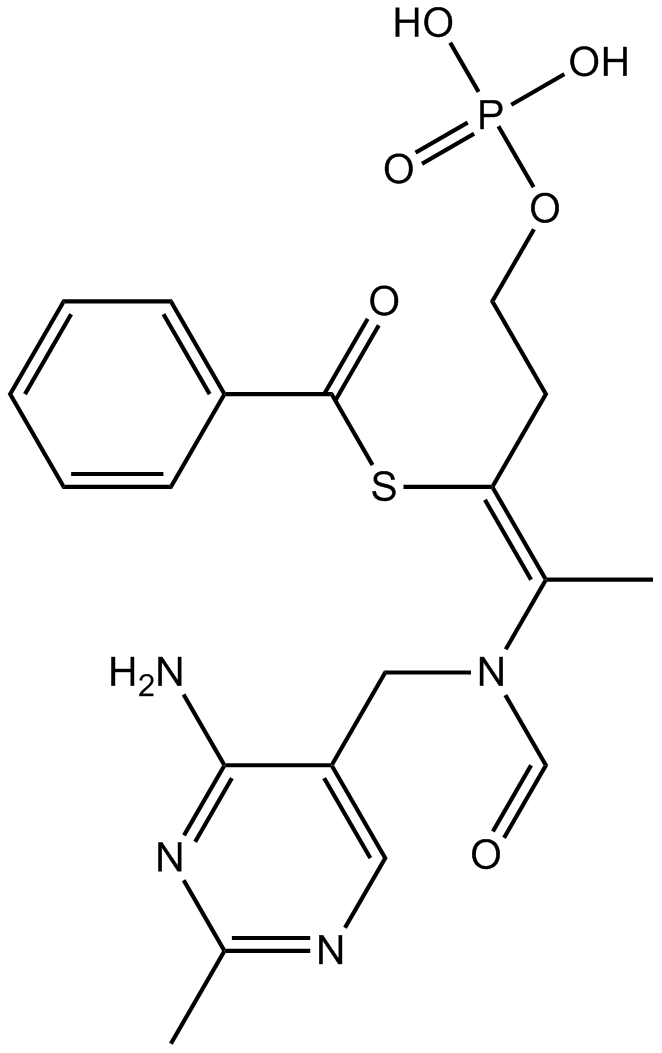
-
GC49836
Benoxaprofen
LRCL 3794, NSC 299582
El benoxaprofeno (LRCL 3794) es un compuesto antiinflamatorio y antipirético potente y de acciÓn prolongada.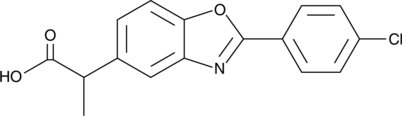
-
GC39346
Benralizumab
MEDI-563; BIW-8405
Benralizumab (MEDI-563) es un anticuerpo monoclonal citolÍtico dirigido por el receptor de la interleucina-5 α (IL-5Rα) que induce la depleciÓn directa, rÁpida y casi completa de los eosinÓfilos a través de una citotoxicidad mediada por células dependiente de anticuerpos mejorada.
-
GC35494
Benzoyloxypaeoniflorin
La benzoiloxipaeoniflorina, aislada de la raÍz de Paeonia suffruticosa, es un inhibidor de la tirosinasa contra la tirosinasa del hongo con IC50 de 0,453 mM.
-
GC15058
Benzydamine HCl
El clorhidrato de bencidamina es un fÁrmaco antiinflamatorio no esteroideo de acciÓn local con propiedades analgésicas y anestésicas locales; se une selectivamente a la prostaglandina sintetasa y tiene una notable actividad antibacteriana in vitro.
-
GC60636
Benzyl butyl phthalate
El ftalato de bencilo butilo, un miembro de los ésteres de Ácido ftÁlico (PAE), puede desencadenar la migraciÓn e invasiÓn de células de hemangioma (HA) a través de la regulaciÓn positiva de Zeb1. El ftalato de butilo de bencilo activa el receptor de hidrocarburo de arilo (AhR) en las células de cÁncer de mama para estimular la seÑalizaciÓn de SPHK1/S1P/S1PR3 y mejora la formaciÓn de células madre de cÁncer de mama iniciadoras de metÁstasis (BCSC).
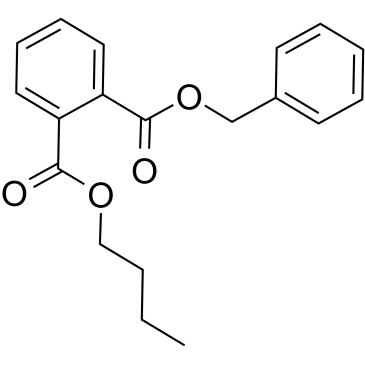
-
GN10443
Berbamine
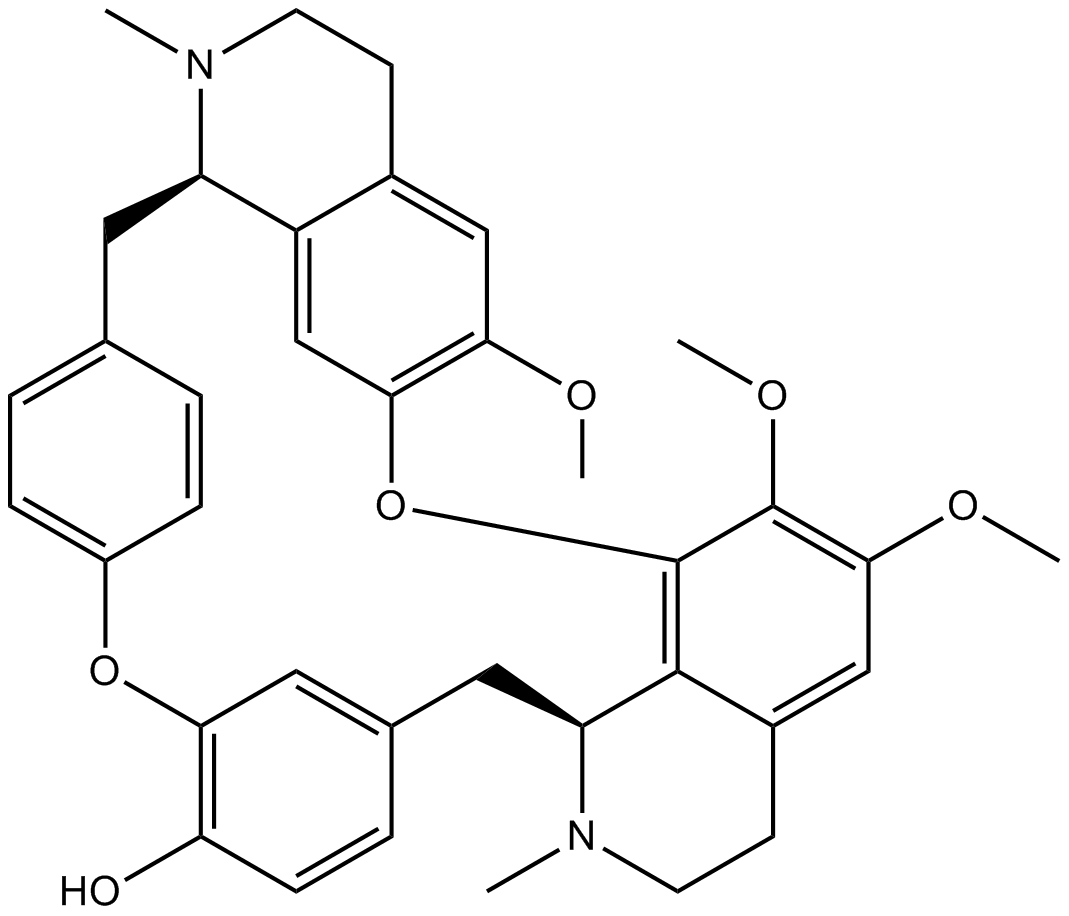
-
GN10358
Berbamine hydrochloride
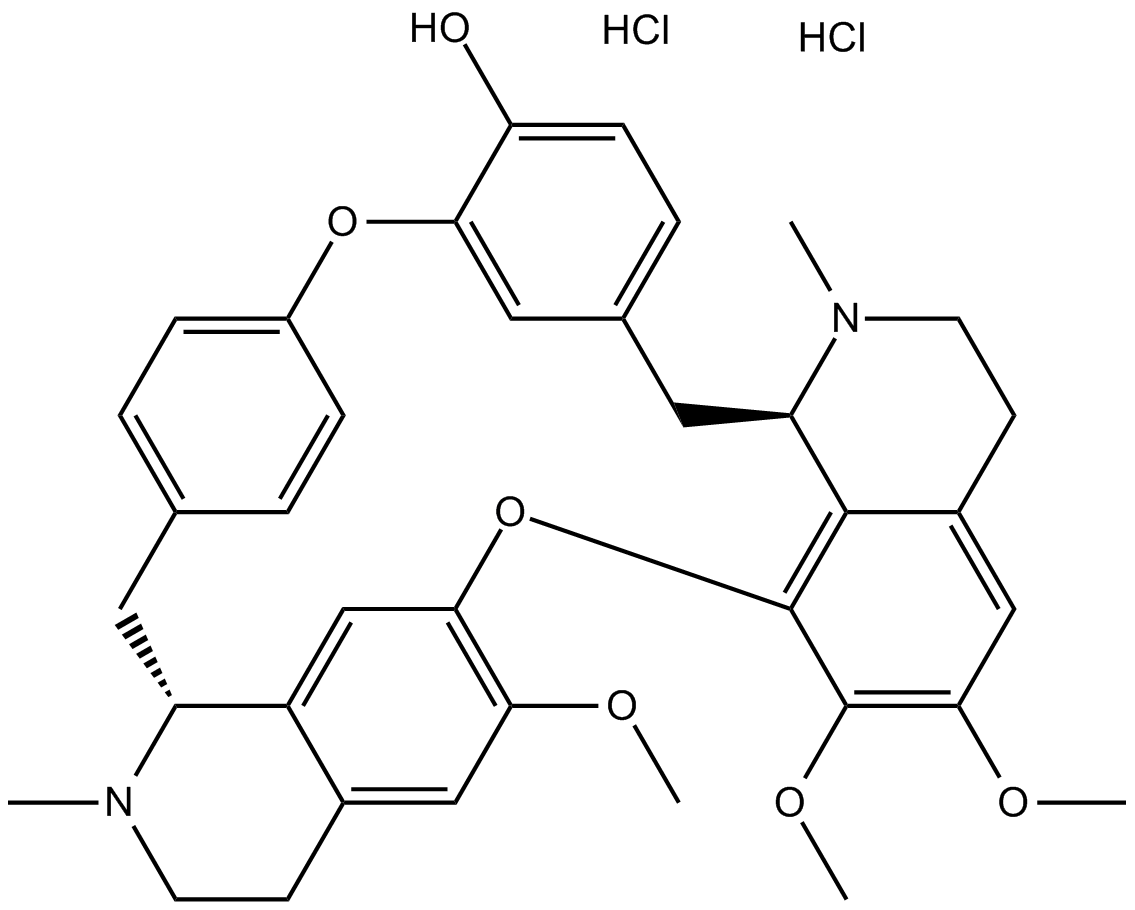
-
GN10221
Berberine
La berberina (Amarillo Natural 18) es un alcaloide aislado de la medicina herbal china Huanglian, como antibiótico. La berberina (Amarillo Natural 18) induce la generación de especies reactivas de oxígeno (ROS) e inhibe la topoisomerasa del ADN.
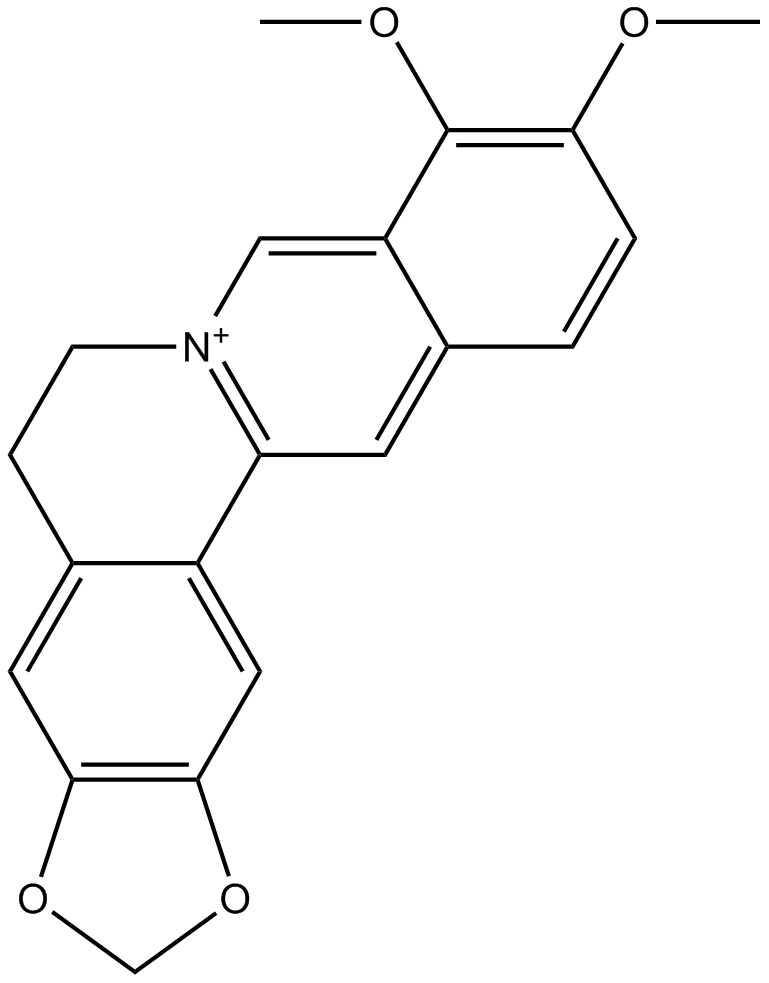
-
GC35497
Berberine chloride hydrate
El cloruro de berberina hidratado (Natural Yellow 18 cloruro hidrato) es un alcaloide que actÚa como antibiÓtico. El hidrato de cloruro de berberina induce la generaciÓn de especies reactivas de oxÍgeno (ROS) e inhibe la topoisomerasa del ADN. Propiedades antineoplÁsicas.
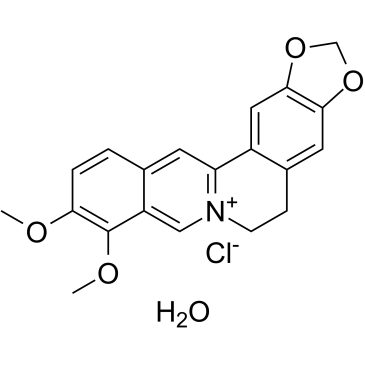
-
GN10208
Berberine hydrochloride
BBR, Umbellatine, NSC 163088, NSC 646666
Berberine hydrochloride is an isoquinoline alkaloid derived from the Ranunculaceae medicinal plant Coptis chinensis. It has various pharmacological activities such as anti-tumor, anti-inflammatory, and hypoglycemic activities.
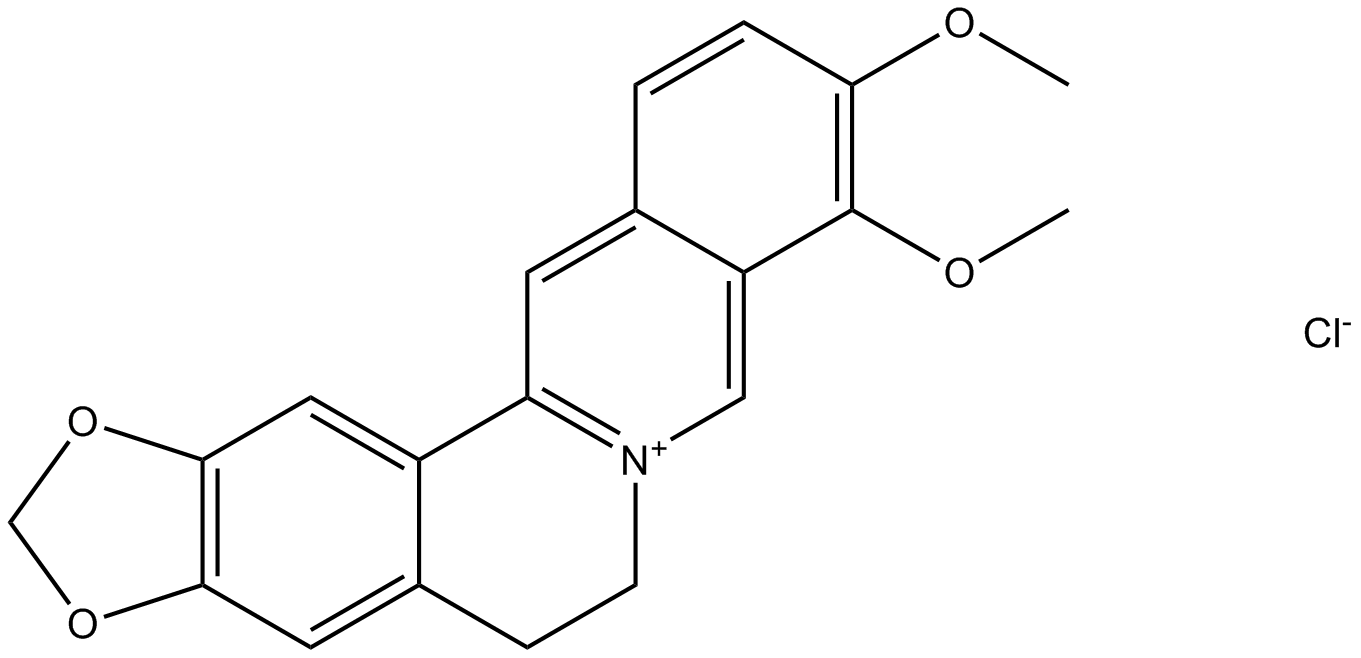
-
GN10523
Berberine Sulfate
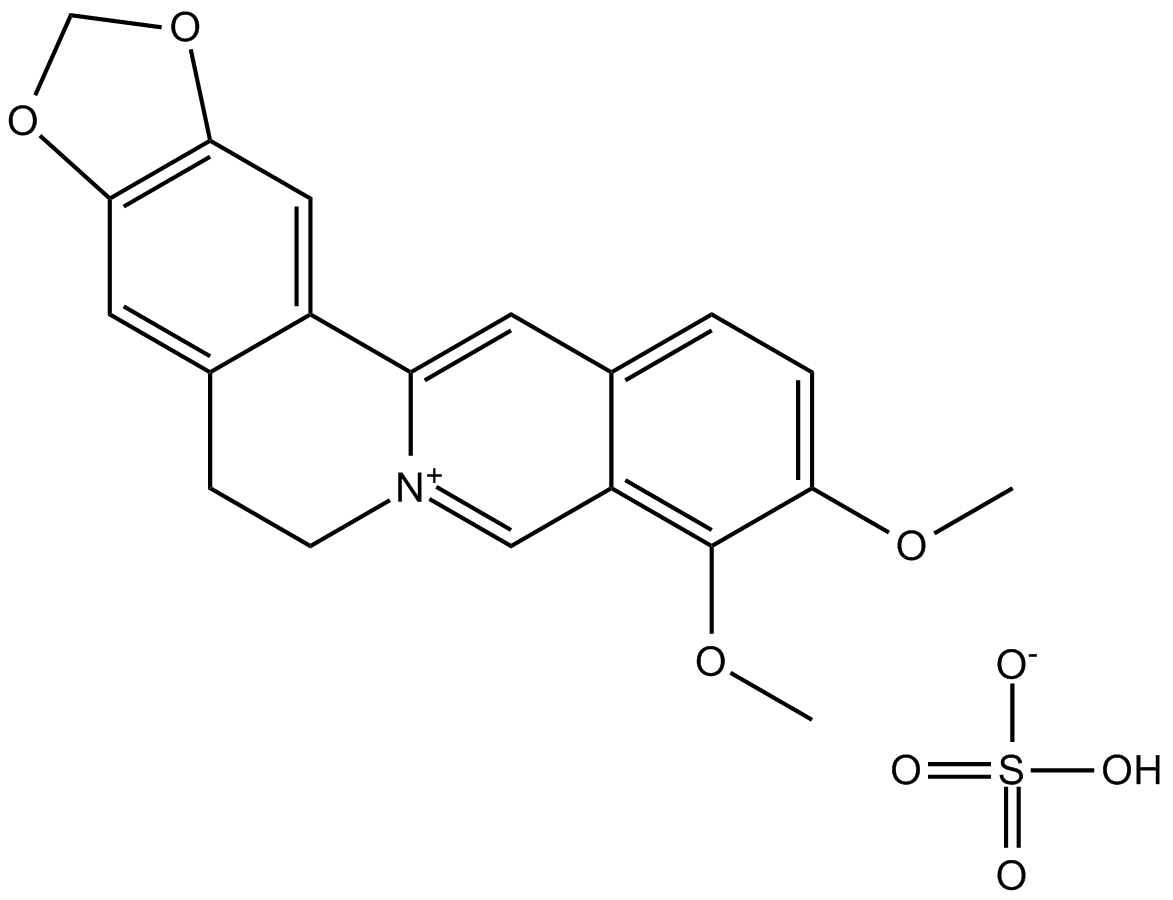
-
GC49387
Berberine-d6 (chloride)
BBR-d6, Umbellatine-d6
An internal standard for the quantification of berberine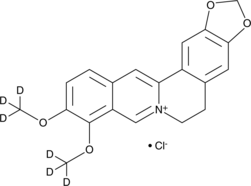
-
GC46098
Berkeleylactone E
A macrolide antibiotic
-
GC42925
Berteroin
5-Methylthiopentyl isothiocyanate
BerteroÍna, un anÁlogo de sulforafano de origen natural, entre otros, un agente antimetastÁsico.
-
GC46922
Betamethasone 21-phosphate (sodium salt hydrate)
A synthetic glucocorticoid
-
GC52329
Betamethasone-d5
β-Methasone-d5, SCH 4831-d5
An internal standard for the quantification of betamethasone
-
GC10480
Betulinic acid
Lupatic Acid, NSC 113090
A plant triterpenoid similar to bile acids
-
GC48504
Betulinic Aldehyde oxime
Betulin 28-oxime
A derivative of betulin