BAY-598 |
| Catalog No.GC18159 |
A potent and selective SMYD2 inhibitor
Products are for research use only. Not for human use. We do not sell to patients.
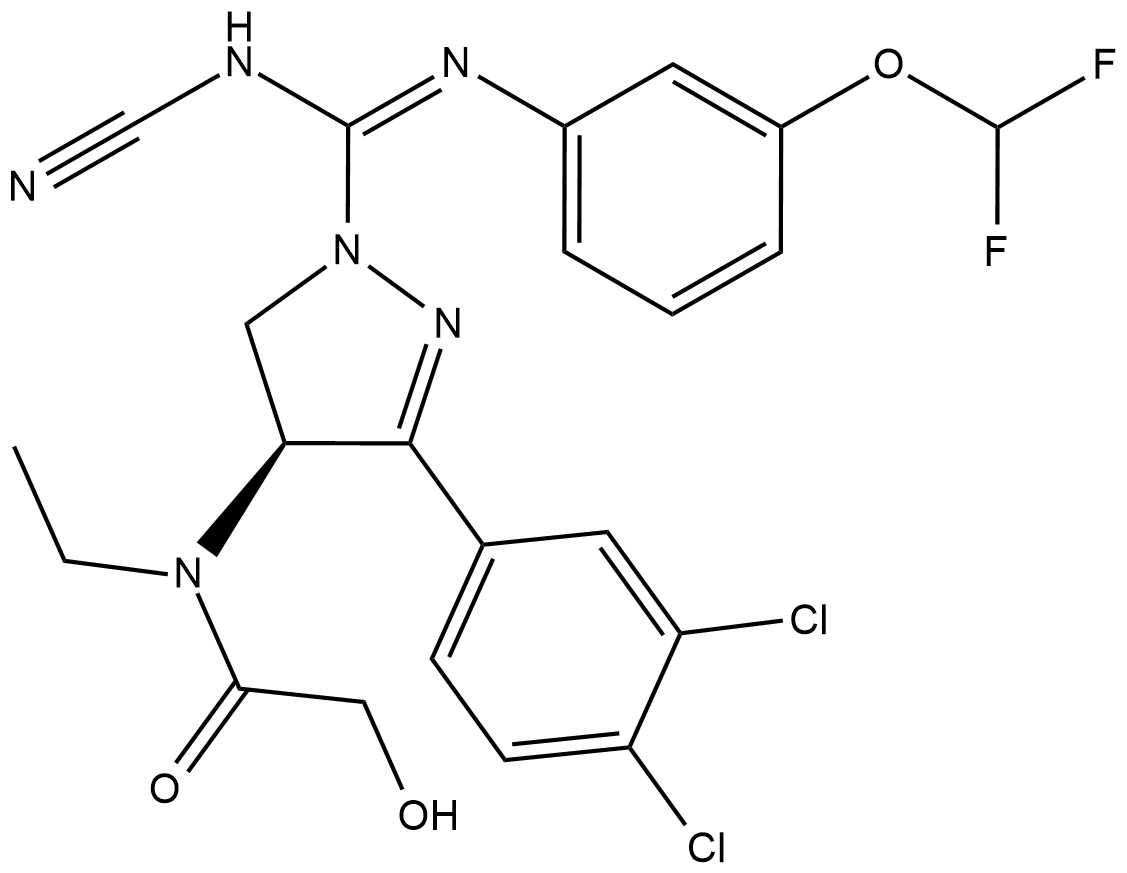
Cas No.: 1906919-67-2
Sample solution is provided at 25 µL, 10mM.
BAY-598 is a potent, peptide-competitive chemical probe for SET and MYND domain-containing protein 2 (SMYD2), a lysine methyl transferase inhibitor that dimethylates histone H3K36 and methylates histone H3K4. SMYD2 also methylates Lys-370 of p53, leading to decreased DNA-binding activity. SMYD2 is over-expressed in several cancers with poor prognosis. BAY-598 inhibits in vitro methylation of p53K370 with an IC50 value of 27 nM and in cells with an IC50 value < 1 μM. BAY-598 is more than 100-fold selective over other histone methyltransferases and non-epigenetic targets.
References
[1]. Reynoird N, et al. Coordination of stress signals by the lysine methyltransferase SMYD2 promotes pancreaticcancer. Genes Dev. 2016 Apr 1;30(7):772-85.
Average Rating: 5 (Based on Reviews and 4 reference(s) in Google Scholar.)
GLPBIO products are for RESEARCH USE ONLY. Please make sure your review or question is research based.
Required fields are marked with *