Viral Diseases
Products for Viral Diseases
- Cat.No. Product Name Information
-
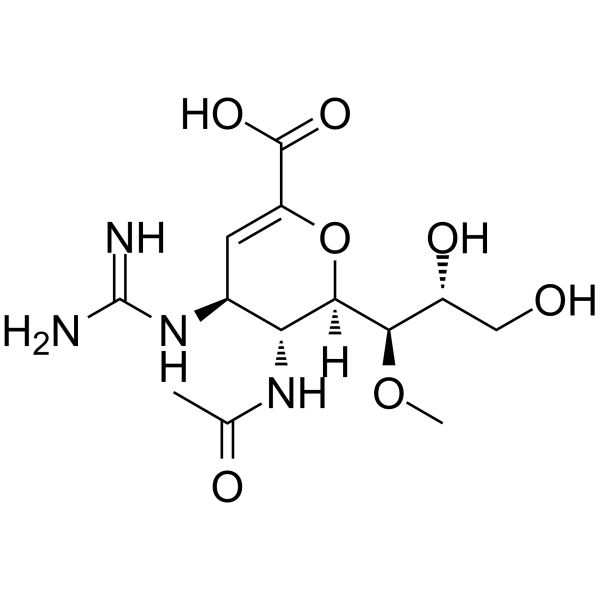
GC49467
β-Aescin
A triterpenoid saponin with diverse biological activities
-
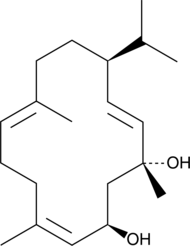
GC40789
β-Cembrenediol
β-Cembrenediol (β-CBT) is a natural product from tobacco plants that is found in cigarette smoke condensate.
-
GC41109
δ12-Prostaglandin J2
δ12-Prostaglandin J2 (δ12-PGJ2) is a cyclopentenone prostaglandin (PG) with anti-proliferative effect on various tumor cell growth. δ12-Prostaglandin J2, a naturally occurring dehydration product of prostaglandin D2, is able to induce apoptosis in HeLa cells via caspase activation.
-
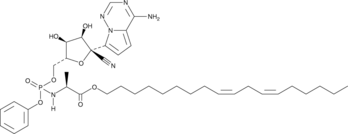
GC52224
(±)-MMT5-14
A derivative of remdesivir with antiviral activity
-
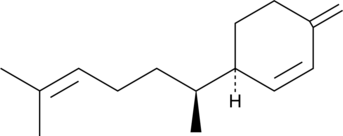
GC49502
(-)-β-Sesquiphellandrene
A sesquiterpene with antiviral and anticancer activities
-
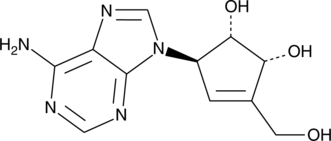
GC45251
(-)-Neplanocin A
S-Adenosylhomocysteine (SAH) hydrolase catalyzes the reversible hydrolysis of SAH to adenosine and homocysteine.
-
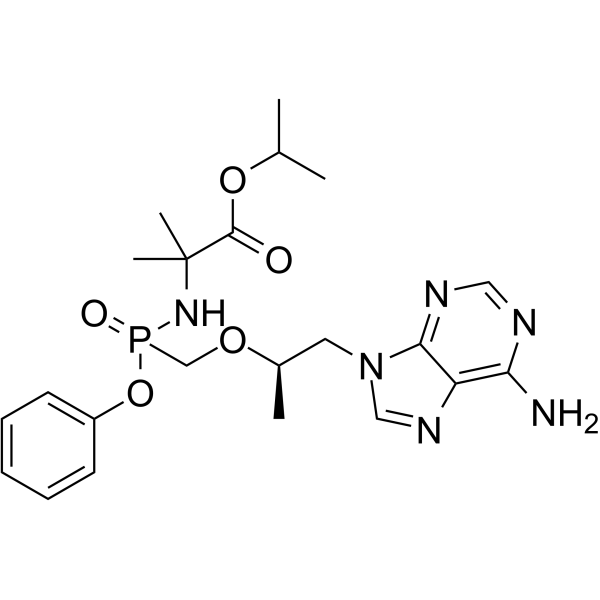
GC65363
(1R)-Tenofovir amibufenamide
(1R)-Tenofovir amibufenamide ((1R)-HS-10234) is the isomer of Tenofovir amibufenamide, is an orally active antiviral agent.
-
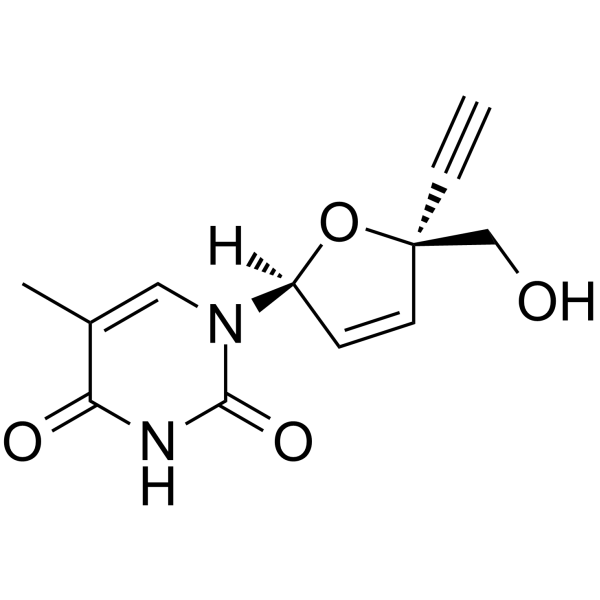
GC64260
(2S,5S)-Censavudine
(2S,5S)-Censavudine ((2S,5S)-OBP-601) is the (2S,5S)-enantiomer of Censavudine.
-
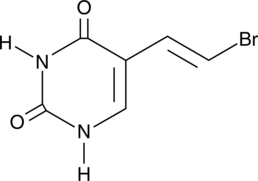
GC41702
(E)-5-(2-Bromovinyl)uracil
(E)-5-(2-Bromovinyl)uracil (BVU) is a pyrimidine base and an inactive metabolite of the antiviral agents sorivudine and (E)-5-(2-bromovinyl)-2'-deoxyuridine (BVDU) that may be regenerated to BVDU in vivo.
-
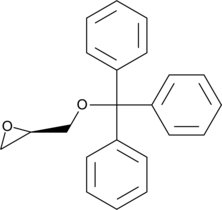
GC49167
(R)-(+)-Trityl glycidyl ether
A synthetic precursor
-
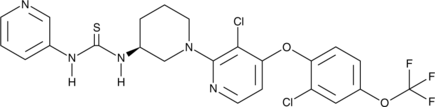
GC46352
(S)-DO271
An inactive control for DO264
-
GC49294
1-(4-Chlorobenzhydryl)piperazine
An inactive metabolite of meclizine and chlorcyclizine
-
GC46415
12-Bromododecanoic Acid
A halogenated form of lauric acid
-
GC49823
2′-C-β-Methylguanosine
An active nucleoside metabolite of BMS-986094
-
GC49514
2′-Deoxyuridine-d2
An internal standard for the quantification of 2’-deoxyuridine
-
GC41281
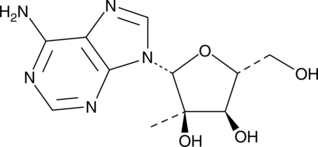
2'-C-Methyladenosine
2'-C-Methyladenosine is an inhibitor of hepatitis C virus (HCV) replication (IC50 = 0.3 μM in Huh-7 human hepatoma cells) that is not cytotoxic at concentrations up to 100 μM.
-
GC46533
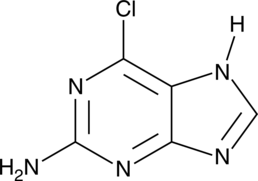
2-Amino-6-chloropurine
A precursor in the synthesis of nucleoside analogs
-
GC42123
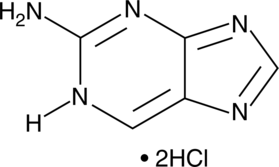
2-Aminopurine (hydrochloride)
2-Aminopurine (hydrochloride) is a fluorescent analog of guanosine.
-
GC68513
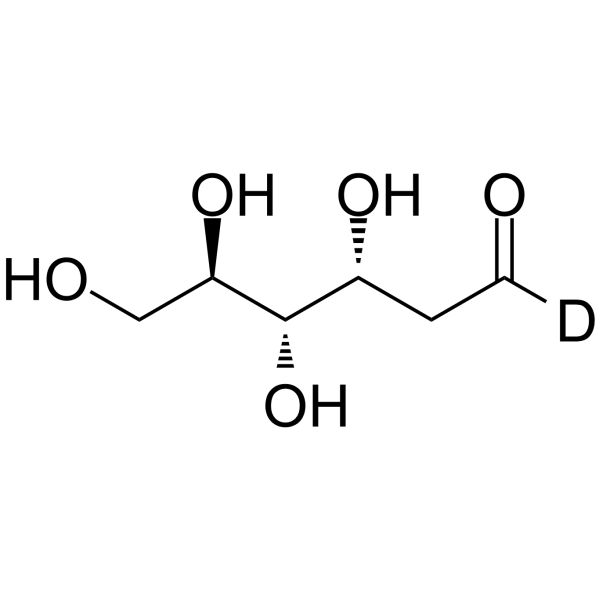
2-Deoxy-D-glucose-d
2-Deoxy-D-glucose-d is the deuterated form of 2-Deoxy-D-glucose. 2-Deoxy-D-glucose is a glucose analogue and a glycolysis inhibitor that works by inhibiting hexokinase, an enzyme involved in glucose metabolism.
-
GC40634
2-epi-Abamectin
2-epi-Abamectin is a degradation product of abamectin.
-
GC42166
2-hydroxy Myristic Acid
2-hydroxy Myristic acid is a hydroxy fatty acid that has been found in bovine, human, and horse milk, cow and buffalo cheeses, sea bass filet, seal oil, human vernix caseosa, and wool wax.
-
GC46553
2-Nonylquinolin-4(1H)-one
A quinolone alkaloid with diverse biological activities
-
GC52446
2-Nonylquinolin-4(1H)-one-d4
An internal standard for the quantification of 2-nonylquinolin-4(1H)-one
-
GC41387
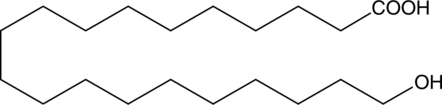
20-hydroxy Arachidic Acid
20-hydroxy Arachidic acid is a hydroxylated fatty acid that has been found in the suberin component of silver birch (B.
-
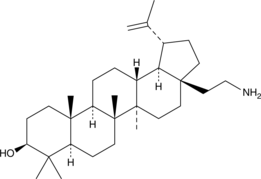
GC48503
28-Deoxybetulin methyleneamine
A derivative of betulin
-
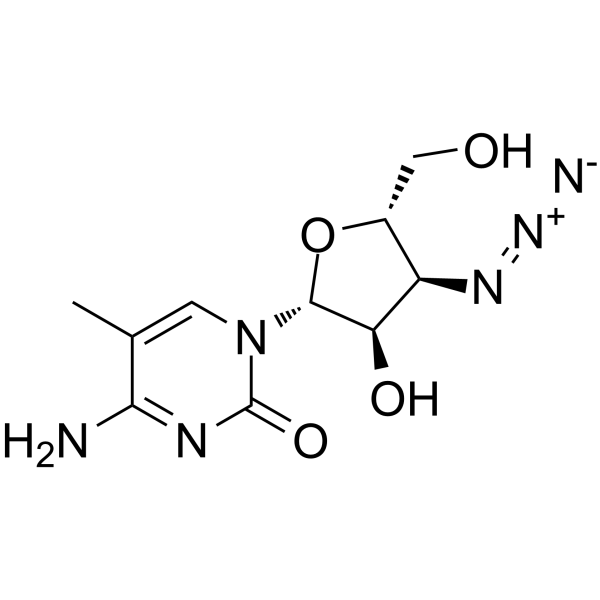
GC65085
3'-Azido-3'-deoxy-5-methylcytidine
3'-Azido-3'-deoxy-5-methylcytidine (CS-92) is a potent xenotropic murine leukemia-related retrovirus (XMRV) inhibitor with a CC50 of 43.5 μM in MCF-7 cells.
-
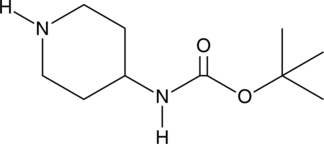
GC46608
4-(N-Boc-amino)piperidine
An organic building block
-
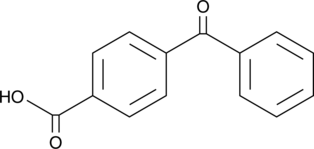
GC46609
4-(Phenylcarbonyl)benzoic Acid
A photooxidant
-
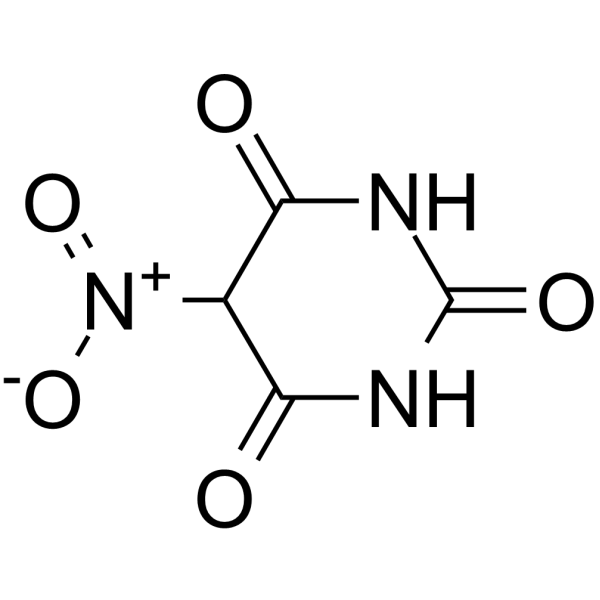
GC66324
5-Nitrobarbituric acid
5-Nitrobarbituric acid is a herpes simplex virus type-1 (HSV-1) inhibitor (IC50=1.7 μM).
-
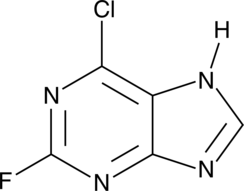
GC46721
6-Chloro-2-fluoropurine
A heterocyclic building block
-
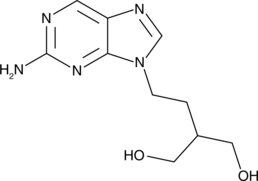
GC49749
6-Deoxypenciclovir
An inactive metabolite of famciclovir
-
GC46724
6-Hydroxypyridin-3-ylboronic Acid
A heterocyclic building block
-
GC52230
AA3-DLin
An ionizable cationic amino lipid
-
GC46767
Abacavir-d4
An internal standard for the quantification of abacavir
-
GC46797
Acyclovir-d4
An internal standard for the quantification of acyclovir
-
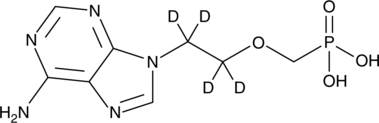
GC46805
Adefovir-d4
An internal standard for the quantification of adefovir
-
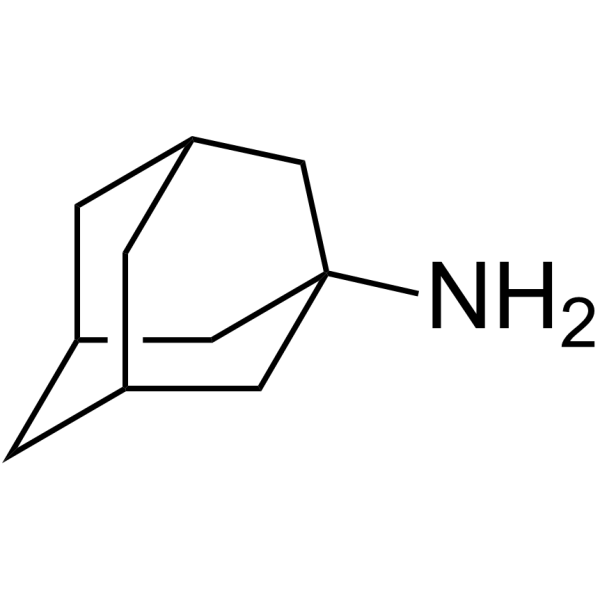
GC63814
Amantadine
Amantadine (1-Adamantanamine) is an orally avtive and potent antiviral agent with activity against influenza A viruses.
-
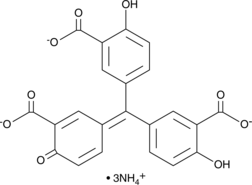
GC46895
Aurintricarboxylic Acid (ammonium salt)
A protein synthesis inhibitor with diverse biological activities
-
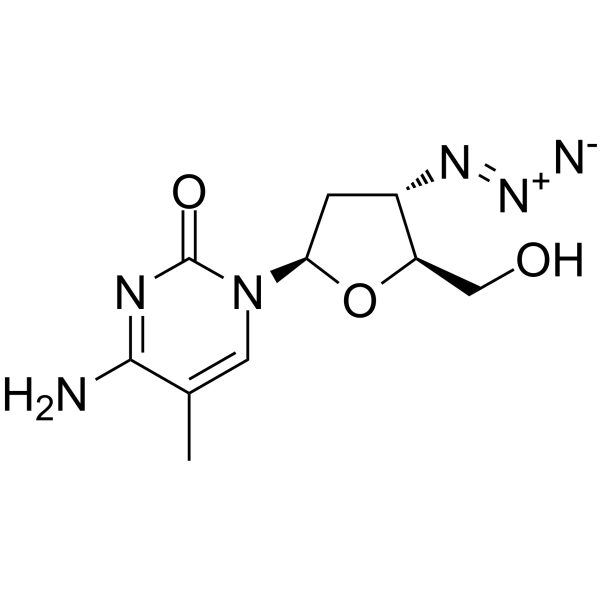
GC65963
AzddMeC
AzddMeC (CS-92) is an antiviral nucleoside analogue and a potent potent, selective and orally active HIV-1 reverse transcriptase and HIV-1 replication inhibitor. In HIV-1-infected human PBM cells and HIV-1-infected human macrophages, the EC50 values of AzddMeC are 9 nM and 6 nM, respectively.
-
GC49057
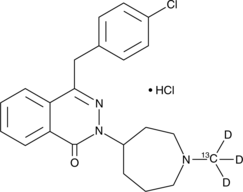
Azelastine-13C-d3 (hydrochloride)
An internal standard for the quantification of azelastine
-
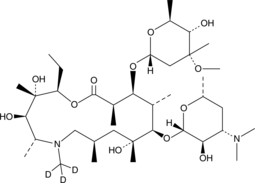
GC46903
Azithromycin-d3
An internal standard for the quantification of azithromycin
-
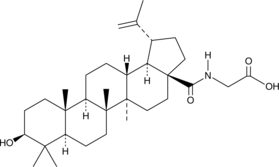
GC48458
Betulinic glycine amide
A derivative of betulinic acid
-
GC52326
Biotin-PEG4-LL-37 (human) (trifluoroacetate salt)
A biotinylated and pegylated form of LL-37

-
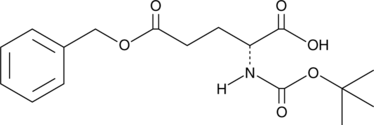
GC46937
Boc-Glu-OBzl
An amino acid-containing building block
-
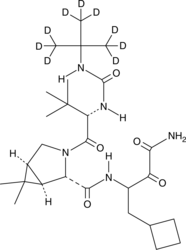
GC46936
Boceprevir-d9
An internal standard for the quantification of boceprevir
-
GA21221
Bz-Nle-Lys-Arg-Arg-AMC
Bz-Nle-Lys-Arg-Arg-AMC is a fluorogenic tetra-peptide substrate for yellow fever virus (YFV) non-structural 3 (NS3).

-
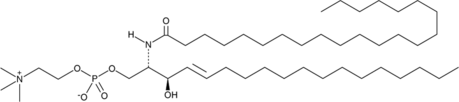
GC43071
C22 Sphingomyelin (d18:1/22:0)
C22 Sphingomyelin is a naturally occurring form of sphingomyelin.
-
GC68449
CA inhibitor 1

-
GC66051
Cabotegravir sodium
Cabotegravir (GSK-1265744) sodium is a orally active and long-acting HIV integrase inhibitor and organic anion transporter 1/3 (OAT1/OAT3) inhibitor with IC50 values of 2.5 nM, 0.41 μM and 0.81 μM for HIVADA, OAT3 and OAT1, respectively. Cabotegravir sodium is primarily metabolized by uridine diphosphate glucuronosyltransferase (UGT) 1A1, with low potential to interact with other antiretroviral drugs (ARVs). Cabotegravir sodium can be used to research AIDS.

-
GC18322
CAY10567
Akt1, 2, and 3 are serine/threonine protein kinases in the phosphatidylinositol 3 (PI3)-kinase signalling pathway that play a critical role in the regulation of cell proliferation and survival.
-
GC43196
CAY10704
CAY10704 is a potent inhibitor of hepatitis C virus (HCV) infection (EC50 = 17 nM) that displays low cytotoxicity of virally-infected human hepatoma Huh7.5.1 cells (CC50 = 21.3 μM).
-
GC64261
Censavudine
Censavudine (OBP-601; BMS-986001), a nucleoside analog, is a nucleoside reverse transcriptase inhibitor.
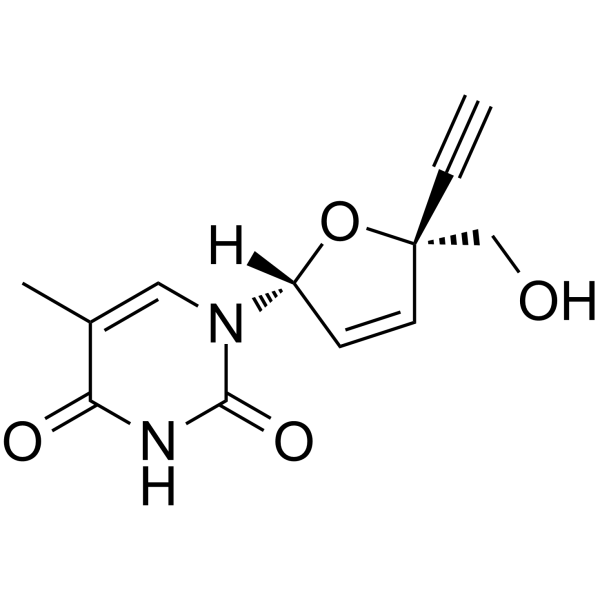
-
GC45885
Chloroquine-d5 (phosphate)
An internal standard for the quantification of chloroquine
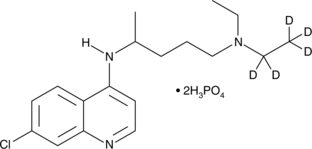
-
GC66467
Claficapavir
Claficapavir (A1752) is a specific nucleocapsid protein (NC) inhibitor with an IC50 around 1 μM. Claficapavir strongly binds the HIV-1 NC (Kd=20 nM) thereby inhibiting the chaperone properties of NC and leading to good antiviral activity against the HIV-1.
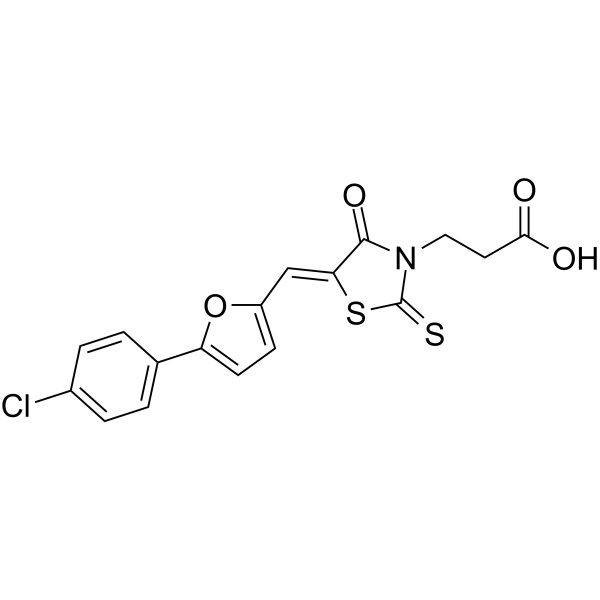
-
GC25306
CQ31
CQ31, a small molecule, selectively activates caspase activation and recruitment domain-containing 8 (CARD8).
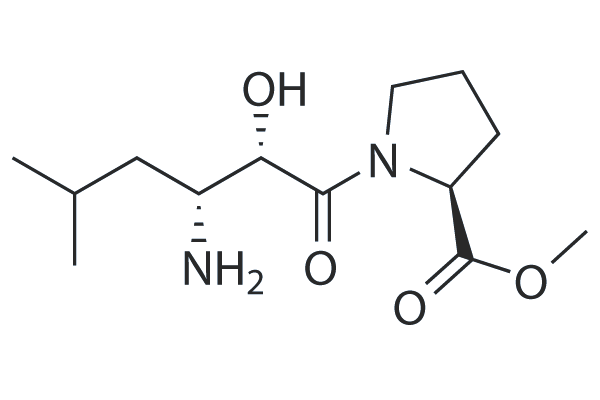
-
GC47128
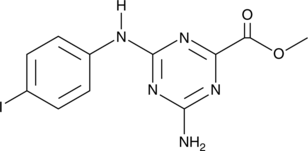
CU-32
A cGAS inhibitor
-
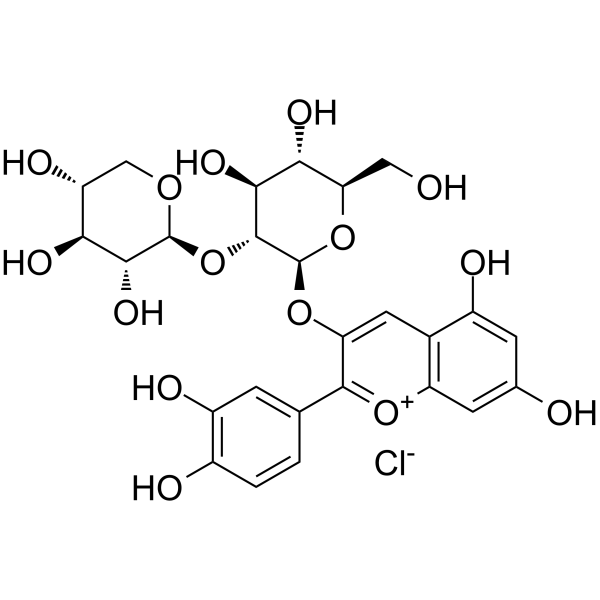
GC64348
Cyanidin 3-sambubioside chloride
Cyanidin 3-sambubioside chloride (Cyanidin-3-O-sambubioside chloride), a major anthocyanin, a natural colorant, and is a potent NO inhibitor.
-
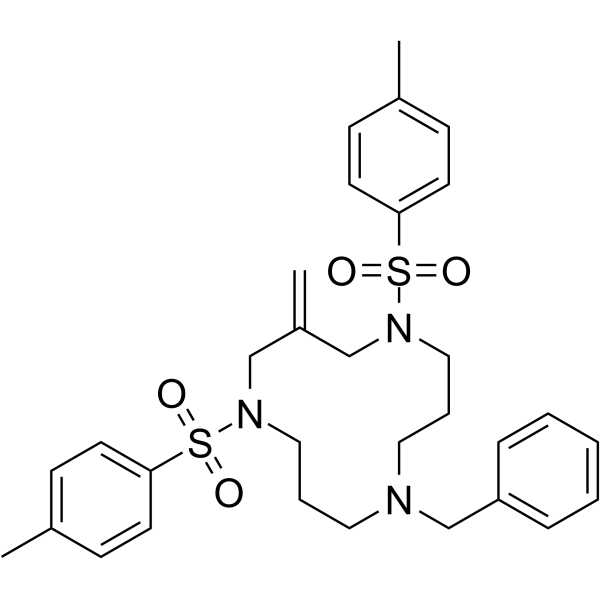
GC64068
Cyclotriazadisulfonamide
Cyclotriazadisulfonamide (CADA) is a specific CD4-targeted HIV entry inhibitors.
-
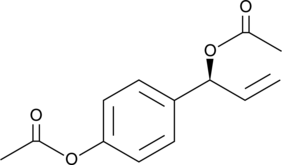
GC43368
D,L-1′-Acetoxychavicol Acetate
D,L-1′-Acetoxychavicol acetate is a natural compound first isolated from the rhizomes of ginger-like plants.
-
GC47266
D-Ornithine lactam
A building block
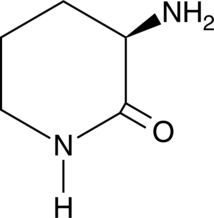
-
GC45883
Daclatasvir-d6
An internal standard for the quantification of daclatasvir
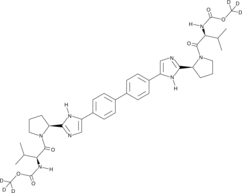
-
GC64391
DDX3-IN-2
DDX3-IN-2 is an active DEADbox polypeptide 3 (DDX3) inhibitor with an IC50 value of 0.3 μM.
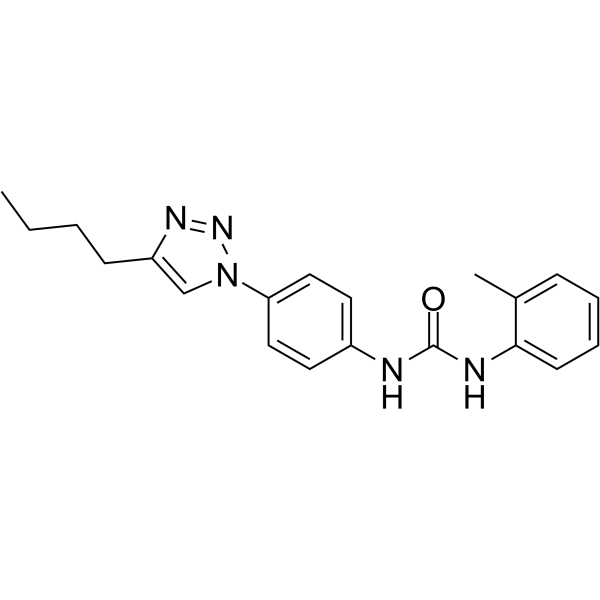
-
GC64270
Decanoyl-RVKR-CMK TFA
Decanoyl-RVKR-CMK (DecRVKRcmk) TFA inhibits over-expressed gp160 processing and HIV-1 replication.
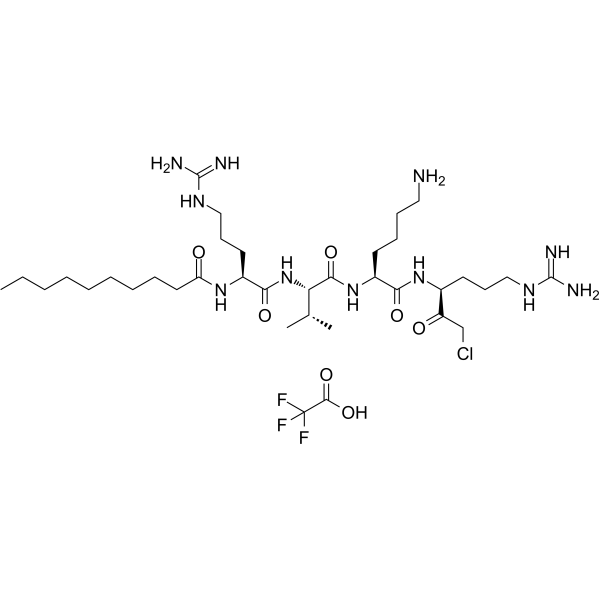
-
GC46130
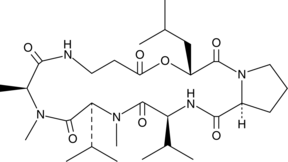
Destruxin B2
A mycotoxin with antiviral, insecticidal, and phytotoxic activities
-
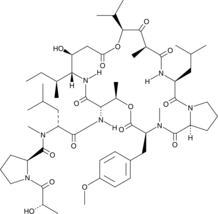
GC49153
Didemnin B
Didemnin B is a cyclic depsipeptide produced by marine tunicates that specifically binds the GTP-bound conformation of EEF1A.
-
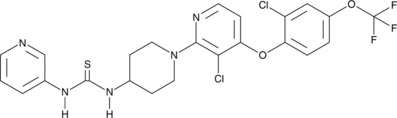
GC45995
DO264
An ABHD12 inhibitor
-
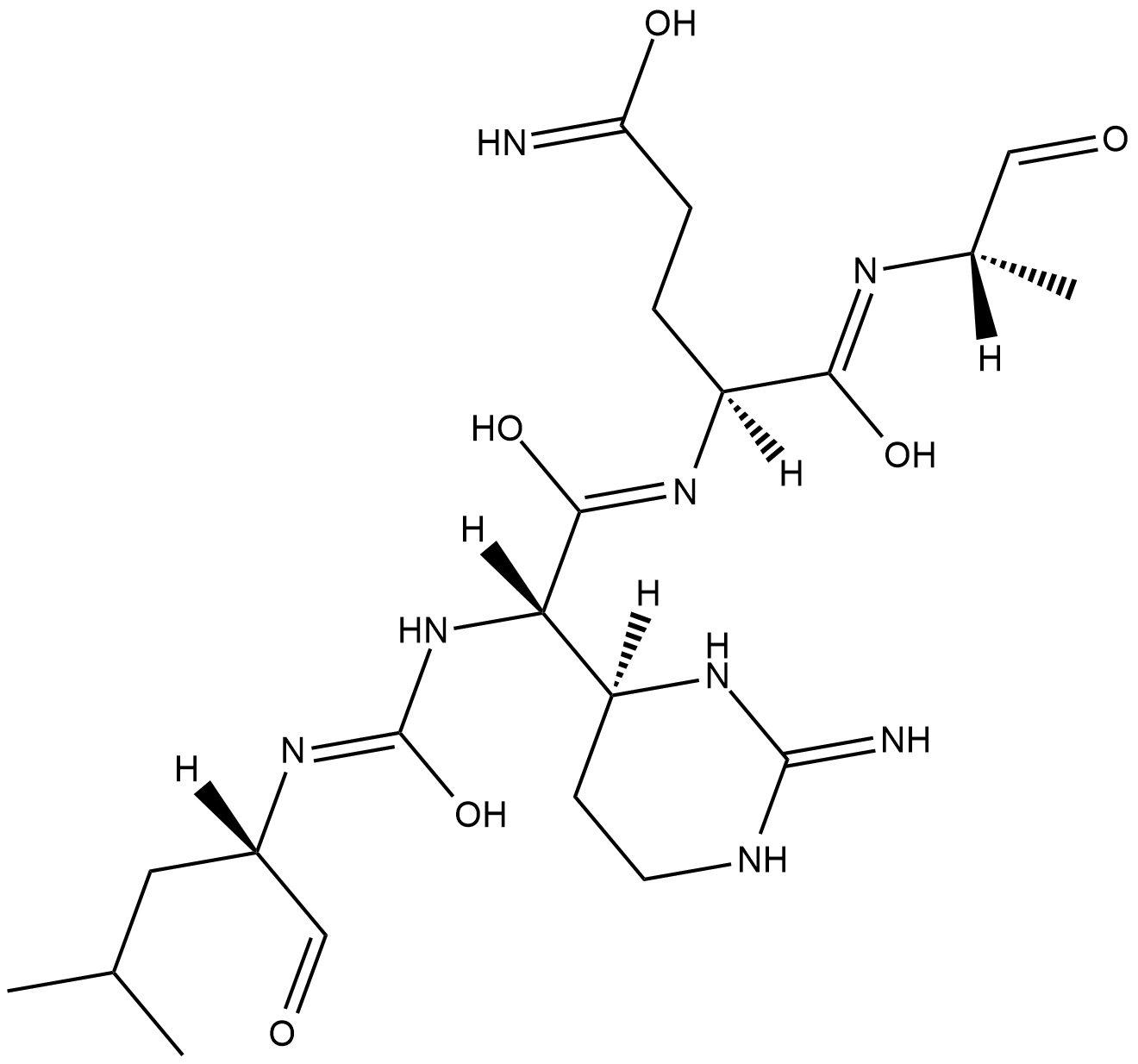
GC18191
Elastatinal
Elastatinal is a potent inhibitor of pancreatic elastase (Ki = 240 nM) that is produced by various species of Actinomycetes.
-
GC63987
Eleutheroside B1
Eleutheroside B1, a coumarin compound, has a wide spectrum of anti-human influenza virus efficacy, with an IC50 value of 64-125μg/ml.
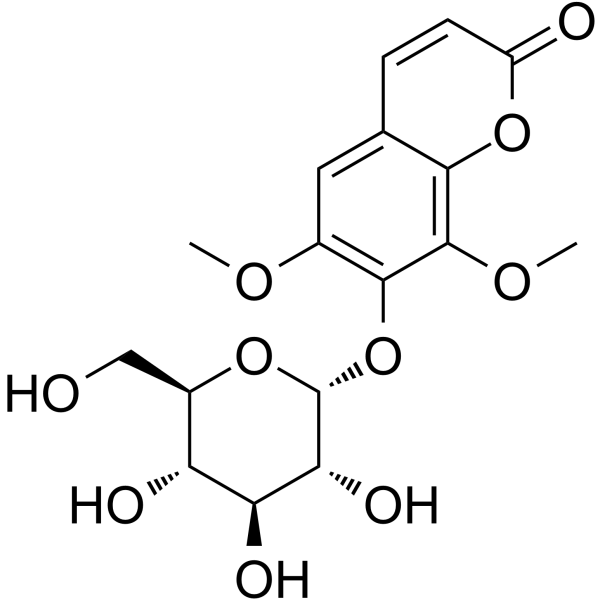
-
GC64484
Emivirine
Emivirine (MKC-442) is a non-nucleoside reverse transcriptase inhibitors (NNRTIs) with Ki values of 0.20 and 0.01 μM for dTTP- and dGTP-dependent DNA or RNA polymerase activity, respectively.
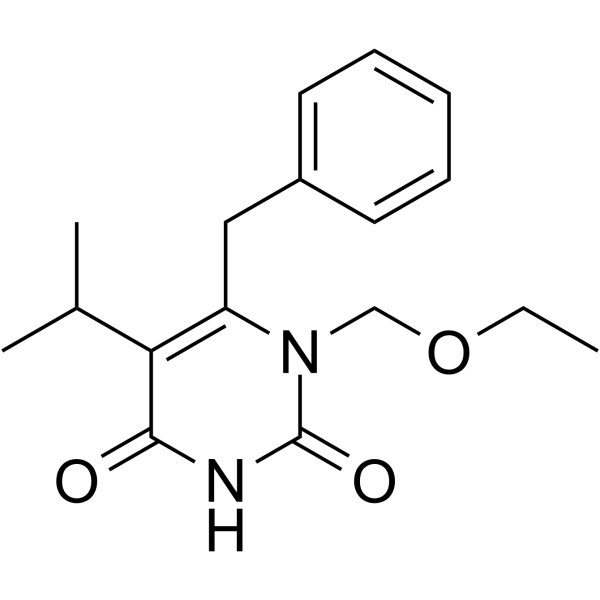
-
GC52261
Entecavir-d2
An internal standard for the quantification of entecavir
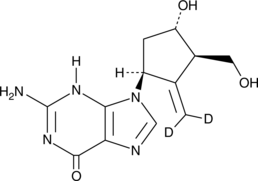
-
GC47295
Enviroxime
An antiviral agent
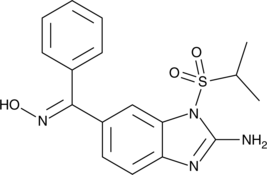
-
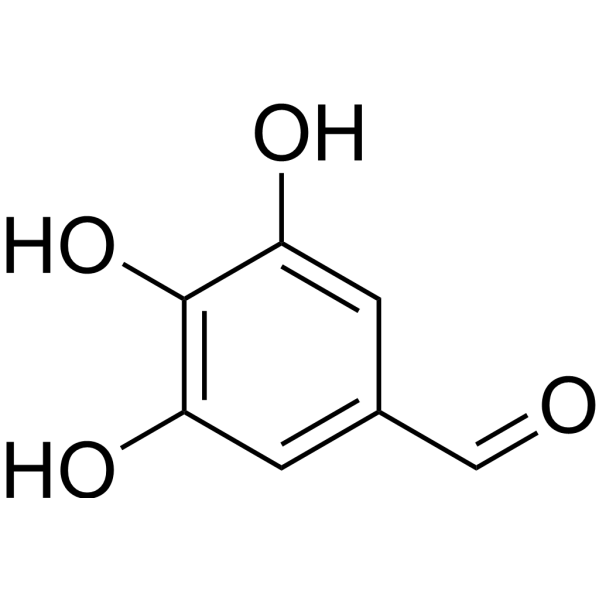
GC67733
Gallic aldehyde
-
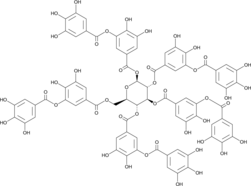
GC43726
Gallotannin
Gallotannin is a polyphenol of gallic acid that has been found in various plants and has antioxidant, anti-inflammatory, antiviral, and antiproliferative biological activities.
-
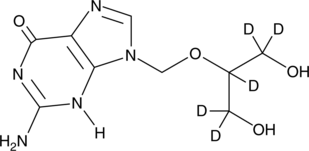
GC47391
Ganciclovir-d5
An internal standard for the quantification of ganciclovir
-
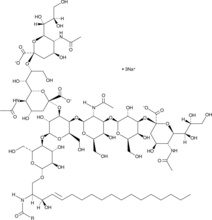
GC52505
Ganglioside GT1b (bovine) (sodium salt)
A sphingolipid
-
GC43735
Ganglioside GT1b Mixture (sodium salt)
Ganglioside GT1b is a trisialoganglioside that is characterized by having two sialic residues linked to the inner galactose unit.
-
GC48464
GC376 (sodium salt)
An inhibitor of 3C- and 3C-like proteases
-
GC52315
GK241
An sPLA2 (Type IIA) inhibitor
-
GC49437
Gliotoxin-13C13
An internal standard for the quantification of gliotoxin
-
GC64233
Gomisin M2
Gomisin M2 ((+)-Gomisin M2) is a lignan isolated from the fruits of Schisandra rubriflora with anti-HIV activity (EC50 of 2.4 μM).
-
GC49165
GS-441524 tris-isobutyryl ester
A prodrug form of GS-441524
-
GC49284
GSK-A1
A PI4KIIIα inhibitor
-
GC65043
Haemanthamine
Haemanthamine is a crinine-type alkaloid isolated from the Amaryllidaceae plants with potent anticancer activity.
-
GC49915
Hexa-D-Arginine (trifluoroacetate salt)
A furin inhibitor
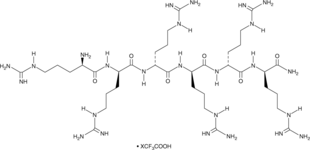
-
GC68023
HIV-1 inhibitor-48
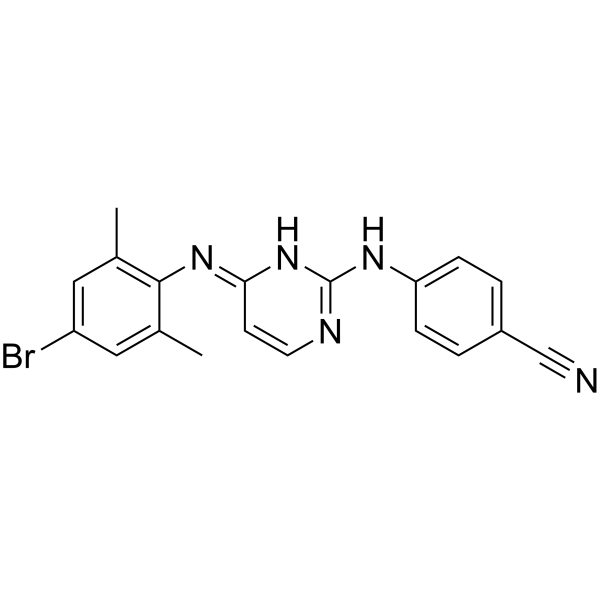
-
GC65256
HIV-1 inhibitor-6
HIV-1 inhibitor-6 (compound 9), a diheteroarylamide-based compound, is a potent HIV-1 pre-mRNA alternative splicing inhibitor.
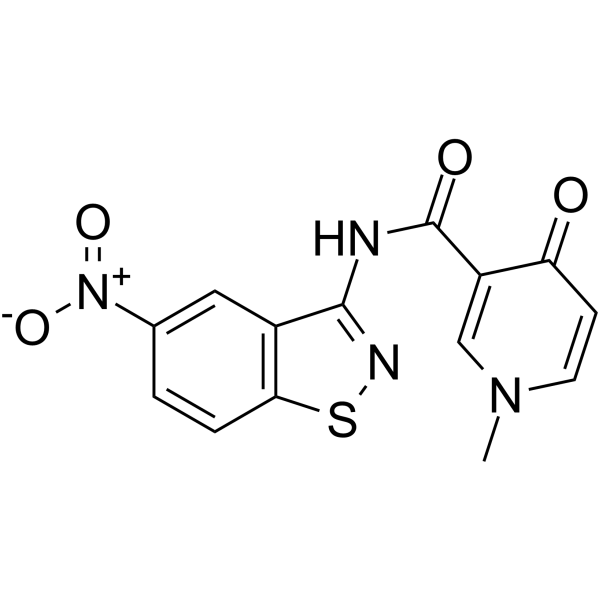
-
GC65158
HIV-1 inhibitor-8
HIV-1 inhibitor-8 is an orally active, low-toxicity and potent HIV?1 non-nucleoside reverse transcriptase inhibitor (NNRTI).
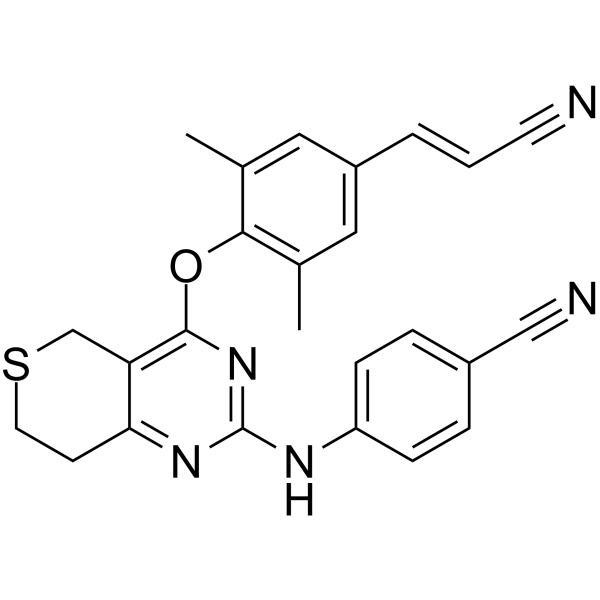
-
GC65134
HIV-IN-6
HIV-IN-6 is a HIV-Ⅰ viral replication inhibitor by targeting Src family kinases (SFK) that interact with Nef protein of the virus, such as Hck.

-
GC47445
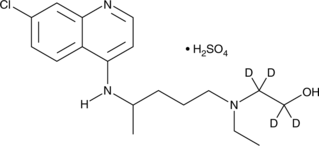
Hydroxychloroquine-d4 (sulfate)
An internal standard for the quantification of hydroxychloroquine
-
GC65034
Ibalizumab
Ibalizumab (TMB-355) is a humanised IgG4 monoclonal antibody that prevents HIV cell entry by binding to CD4 receptor.

-
GC47452
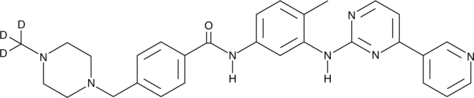
Imatinib-d3
Imatinib-d3 (STI571-d3) hydrochloride is the deuterium labeled Imatinib. Imatinib (STI571) is an orally bioavailable tyrosine kinases inhibitor that selectively inhibits BCR/ABL, v-Abl, PDGFR and c-kit kinase activity. Imatinib (STI571) works by binding close to the ATP binding site, locking it in a closed or self-inhibited conformation, therefore inhibiting the enzyme activity of the protein semicompetitively. Imatinib also is an inhibitor of SARS-CoV and MERS-CoV.
-
GC40101
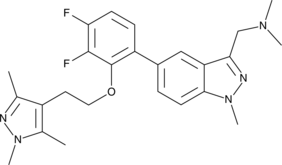
IMP-1088
An inhibitor of N-myristoyltransferase
-
GC47474
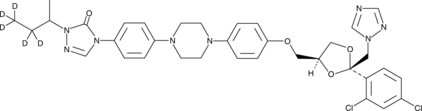
Itraconazole-d5
An internal standard for the quantification of itraconazole
-
GC52403
Ivermectin-d2
An internal standard for the quantification of ivermectin
-
GC65968
JNJ4796
JNJ4796 is an oral active fusion inhibitor of influenza virus, neutralizing influenza A group 1 viruses by inhibiting hemagglutinin (HA)-mediated fusion. JNJ4796 mimics the functionality of the broadly neutralizing antibodies (bnAbs).
-
GC52272
Jun9-72-2
A SARS-CoV-2 PLpro inhibitor

-
GC50716
K 22
An antiviral agent
-
GC64253
KIN101
KIN101 is a potent RNA viral inhibitor with IC50s of 2 μM, >5 μM for influenza virus and Dengue virus (DNV), respectively.
-
GC44005
KIN1400
KIN1400 is a small molecule activator of the RIG-1-like receptor (RLR) pathway that has antiviral activity.
-
GC65069
Laninamivir
Laninamivir (R 125489) is a potent influenza neuraminidase (NA) inhibitor with IC50s of 0.90 nM, 1.83 nM and 3.12 nM for avian H12N5 NA (N5), pH1N1 N1 NA (p09N1) and A/RI/5+/1957 H2N2 N2 (p57N2), respectively.