Nucleic Acid Turnover/Signaling
Products for Nucleic Acid Turnover/Signaling
- Cat.No. Product Name Information
-
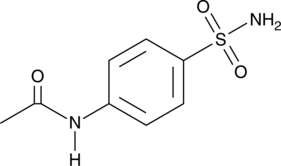
GC49337
4-Acetamidobenzenesulfonamide
A metabolite of asulam and sulfanilamide
-
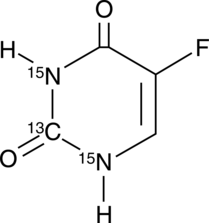
GC42545
5-Fluorouracil-13C,15N2
5-Fluorouracil-13C,15N2 is intended for use as an internal standard for the quantification of 5-flurouracil by GC- or LC-MS.
-
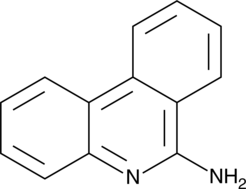
GC40479
6-Aminophenanthridine
6-Aminophenanthridine is an antiprion agent.
-
GC42578
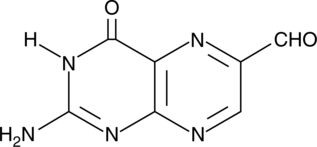
6-Formylpterin
Xanthine oxidase (XO) generates reactive oxygen species, including hydrogen peroxide (H2O2), as it oxidizes specific substrates in the presence of water and oxygen.
-
GC49235
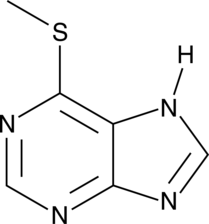
6-Methylmercaptopurine
A metabolite of 6-mercaptopurine
-
GC49488
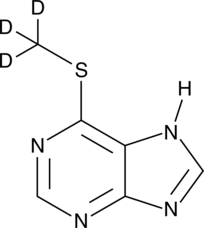
6-Methylmercaptopurine-d3
An internal standard for the quantification of 6-MMP
-
GC18426
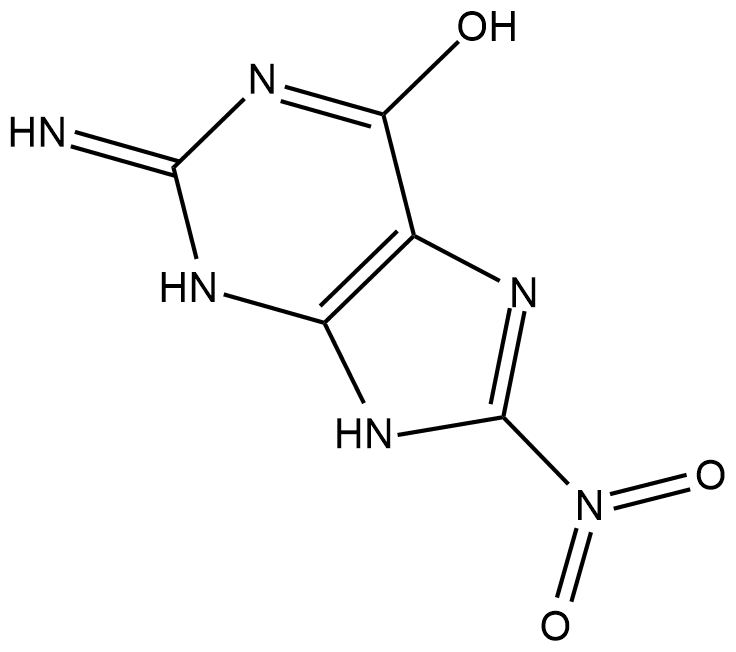
8-Nitroguanine
8-Nitroguanine is a nitrative guanine derivative formed by oxidative damage to the guanine base in DNA by reactive nitrogen species (RNS) during inflammation and in vitro by reaction of DNA with peroxynitrite and other RNS reagents.
-
GC49347
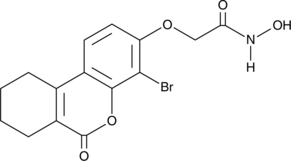
8Br-HA
An FHIT inhibitor
-
GC42644
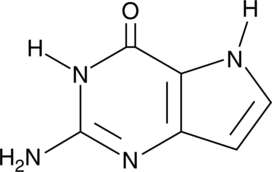
9-Deazaguanine
9-Deazaguanine is an analog of guanine that acts as an inhibitor of purine nucleoside phosphorylase (PNP; Kd = 160 nM).
-
GC49285
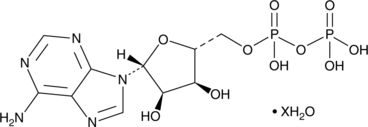
Adenosine 5’-methylenediphosphate (hydrate)
An inhibitor of ecto-5’-nucleotidase
-
GC49039
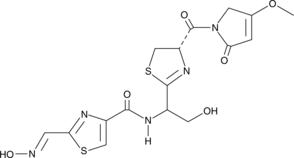
Althiomycin
A thiazole antibiotic
-
GC42783
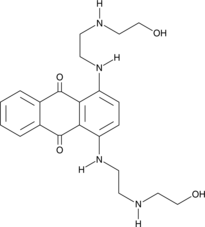
Ametantrone
Ametantrone (NSC 196473) is an antitumor agent that intercalates into DNA and induces topoisomerase II (TOP2)-mediated DNA break.
-
GC46895
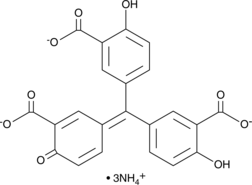
Aurintricarboxylic Acid (ammonium salt)
A protein synthesis inhibitor with diverse biological activities
-
GC18322
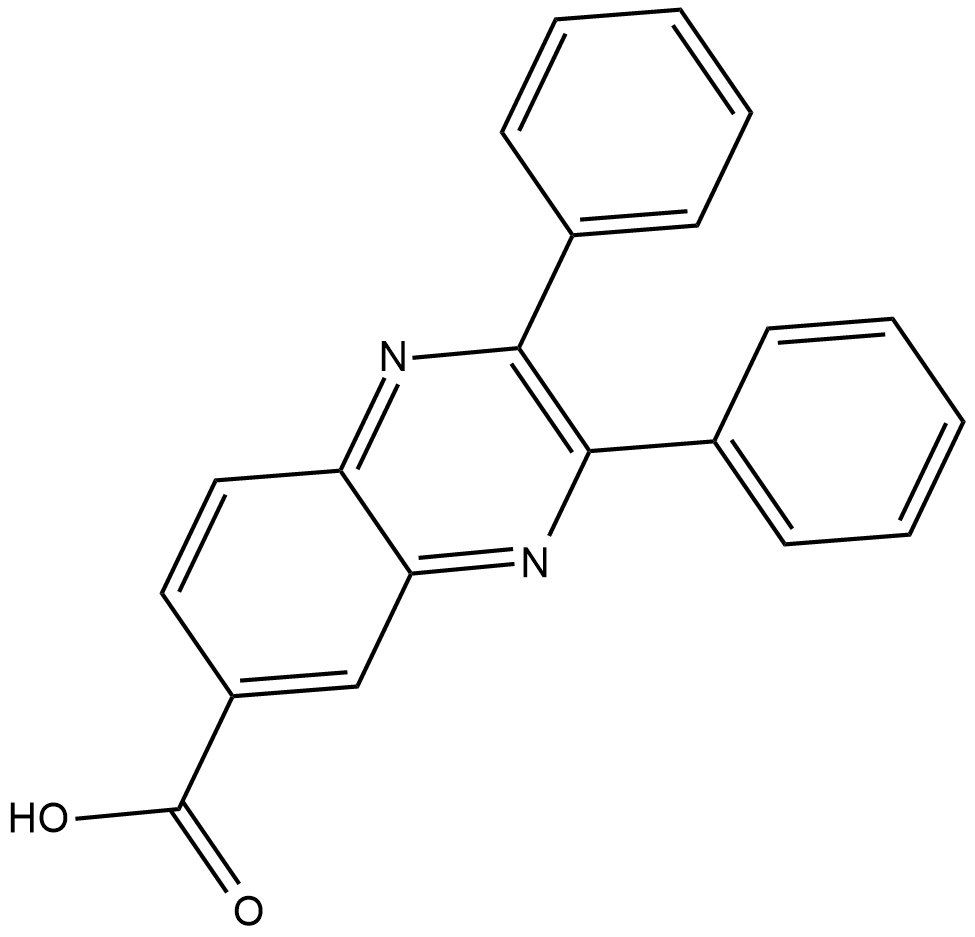
CAY10567
Akt1, 2, and 3 are serine/threonine protein kinases in the phosphatidylinositol 3 (PI3)-kinase signalling pathway that play a critical role in the regulation of cell proliferation and survival.
-
GC40219
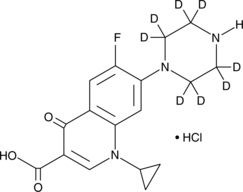
Ciprofloxacin-d8 (hydrochloride)
Ciprofloxacin-d8 is intended for use as an internal standard for the quantification of ciprofloxacin by GC- or LC-MS.
-
GC47128
CU-32
A cGAS inhibitor
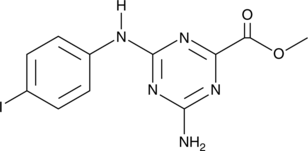
-
GC47129
CU-76
A cGAS inhibitor
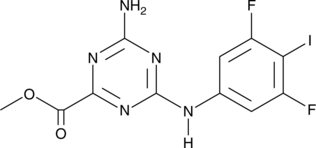
-
GC49100
Desmethyl Ofloxacin (hydrochloride)
A metabolite of ofloxacin
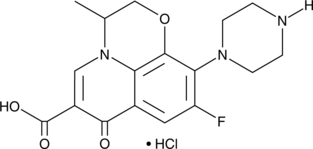
-
GC47228
Dihydronovobiocin
A coumarin antibiotic
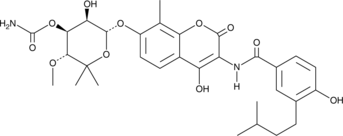
-
GC45881
Docosanoic Acid-d43
An internal standard for the quantification of docosanoic acid
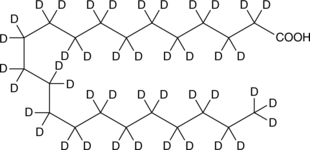
-
GC49288
Dolutegravir M1
A metabolite of dolutegravir
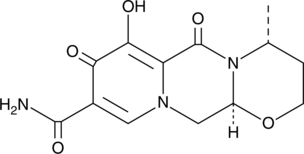
-
GC49289
Dolutegravir O-β-D-Glucuronide
A metabolite of dolutegravir
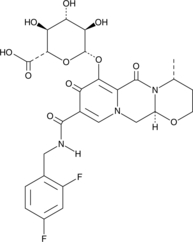
-
GC49101
Enrofloxacin-d5
An internal standard for the quantification of enrofloxacin
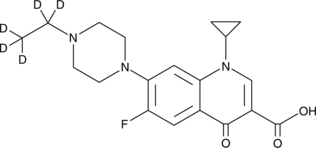
-
GC49165
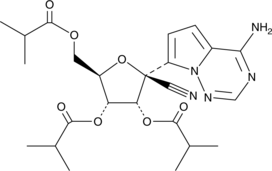
GS-441524 tris-isobutyryl ester
A prodrug form of GS-441524
-
GC47552
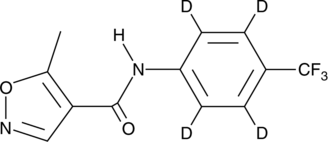
Leflunomide-d4
An internal standard for the quantification of leflunomide
-
GC61002
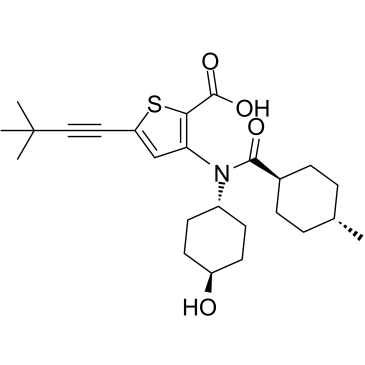
Lomibuvir
An HCV polymerase inhibitor
-
GC47649
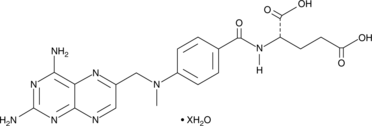
Methotrexate (hydrate)
A folic acid and aminopterin derivative
-
GC18769
MST-312
MST312 is a telomerase inhibitor (IC50 = 0.67 μM in a TRAP assay).
-
GC40081
Nargenicin
Nargenicin is a macrolide antibiotic that selectively inhibits the growth of S.
-
GC44591
Pemetrexed (sodium salt hydrate)
Pemetrexed (sodium salt hydrate) is a novel antifolate, the Ki values of the pentaglutamate of LY231514 are 1.3, 7.2, and 65 nM for inhibits thymidylate synthase (TS), dihydrofolate reductase (DHFR), and glycinamide ribonucleotide formyltransferase (GARFT), respectively.
-
GC49053
Pseudomonic Acid (lithium salt)
An antibiotic
-
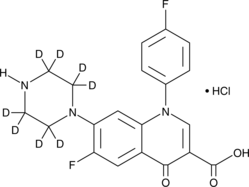
GC49188
Sarafloxacin-d8 (hydrochloride)
An internal standard for the quantification of sarafloxacin
-
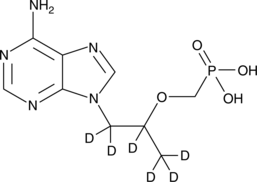
GC48143
Tenofovir-d6
An internal standard for the quantification of tenofovir
-
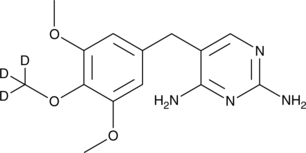
GC48205
Trimethoprim-d3
An internal standard for the quantification of trimethoprim