Cell Migration & Metastasis
Products for Cell Migration & Metastasis
- Cat.No. Product Name Information
-
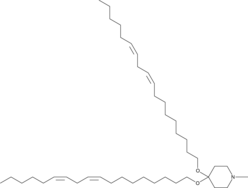
GC48998
β-Defensin-1 (human) (trifluoroacetate salt)
An antimicrobial peptide
-
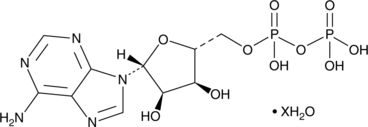
GC49285
Adenosine 5’-methylenediphosphate (hydrate)
An inhibitor of ecto-5’-nucleotidase
-
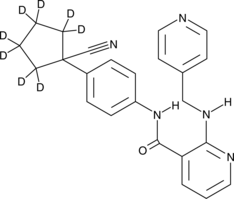
GC49799
Apatinib-d8
An internal standard for the quantification of apatinib
-
GC52194
Dimethylamino Parthenolide
-
GC49563
Fenhexamid
Fenhexamid, a botryticide, is a sterol biosynthesis inhibitor.
-
GC49089
FR900359
A cyclic depsipeptide and an inhibitor of Gαq, Gα11, and Gα14
-
GC49229
Ganodermanontriol
Ganodermanontriol, a sterol isolated from Ganoderma lucidum, induces anti-inflammatory activity in tert-butyl hydroperoxide (t-BHP)-damaged hepatic cells through the expression of HO-1. Ganodermanontriol exhibits hepatoprotective activity.
-
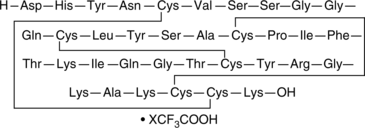
GC49882
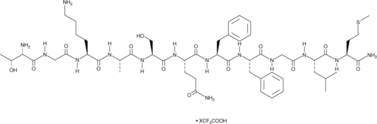
Hemokinin 1 (human) (trifluoroacetate salt)
A peptide agonist of NK1 receptors
-
GC49915
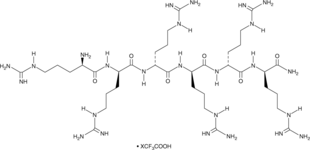
Hexa-D-Arginine (trifluoroacetate salt)
A furin inhibitor
-
GC52472
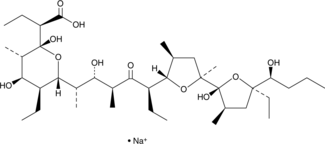
Inostamycin A (sodium salt)
A bacterial metabolite with anticancer activity
-
GC49560
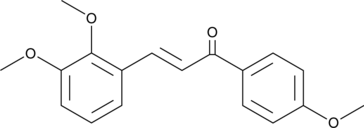
L6H21
L6H21, a Chalcone derivative, is an orally active, potent and specific myeloid differentiation 2 (MD-2) inhibitor.
-
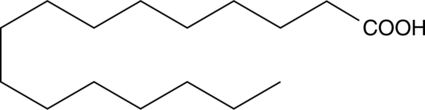
GC49024
Palmitic Acid MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
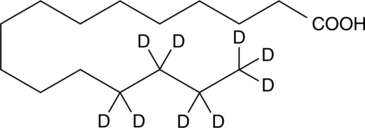
GC49023
Palmitic Acid-d9 MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
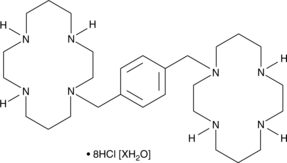
GC49441
Plerixafor (hydrochloride hydrate)
An antagonist of CXCR4
-
GC52484
Purified Ganglioside Mixture (bovine) (ammonium salt)
A mixture of purified bovine gangliosides

-
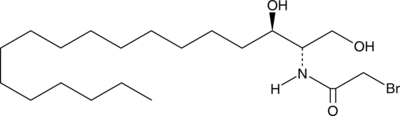
GC49668
SABRAC
An inhibitor of acid ceramidase
-
GC52460
tcY-NH2 (trifluoroacetate salt)
A peptide antagonist of PAR4
-
GC52084
YSK05
YSK05 is a pH-sensitive cationic lipid. YSK05 improves the intracellular trafficking of non-viral vectors. YSK05-MEND shows significantly good gene silencing activity and hemolytic activity. YSK05 overcomes the suppression of endosomal escape by PEGylation. YSK05 effectively enhances siRNA delivery both in vitro and in vivo.