Epigenetic Reader Domain
Epigenetic regulators of gene expression and chromatin state include so-called writers, erasers, and readers of chromatin modifications.Well-characterized examples of reader domains include bromodomains typically binding acetyllysine and chromatin organization modifier (chromo), malignant brain tumor (MBT), plant homeodomain (PHD), and Tudor domains generally associating with methyllysine. Research on epigenetic readers has been tremendously influenced by the discovery of selective inhibitors targeting the bromodomain and extraterminal motif (BET) family of acetyl-lysine readers. The human genome encodes 46 proteins containing 61 bromodomains clustered into eight families. Distinct experimental approaches are used to identify the first BET inhibitors, GSK 525762A and (+)-JQ-1.
The Polycomb group (PcG) protein, enhancer of zeste homologue 2 (EZH2), has an essential role in promoting histone H3 lysine 27 trimethylation (H3K27me3) and epigenetic gene silencing. This function of EZH2 is important for cell proliferation and inhibition of cell differentiation, and is implicated in cancer progression. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. In many types of cancers including lymphomas and leukemia, EZH2 is postulated to exert its oncogenic effects via aberrant histone and DNA methylation, causing silencing of tumor suppressor genes.
p300/CBP is not only a transcriptional adaptor but also a histone acetyltransferase.
Targets for Epigenetic Reader Domain
Products for Epigenetic Reader Domain
- Cat.No. Product Name Information
-
GC34078
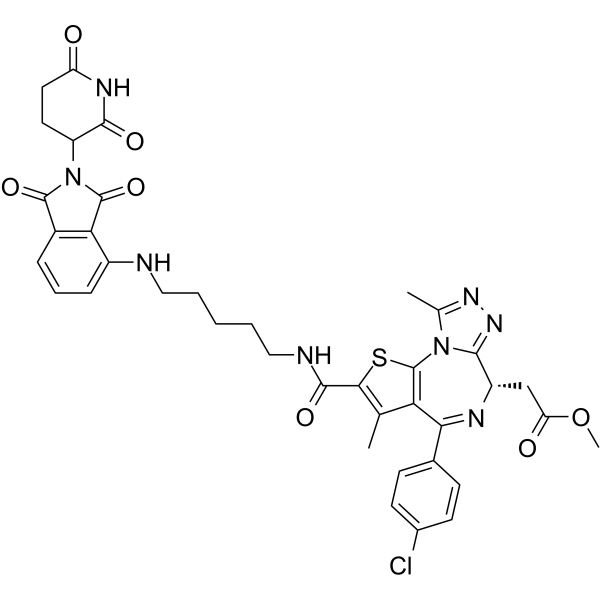
I-CBP112
I-CBP112 is a specific and potent acetyl-lysine competitive protein-protein interaction inhibitor, that inhibits the CBP/p300 bromodomains, enhances acetylation by p300.
-
GC43889
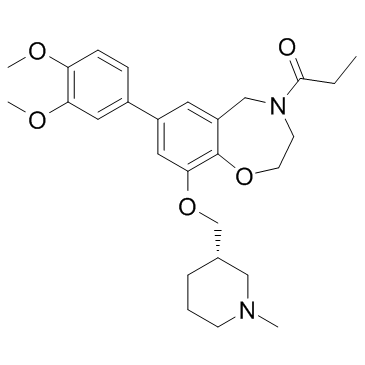
I-CBP112 (hydrochloride)
cAMP-responsive element-binding protein binding protein (CREBBP) and E1A-associated protein p300 (EP300) are transcriptional co-activators that modulate DNA replication, DNA repair, cell growth, transformation, and development.
-
GC33042
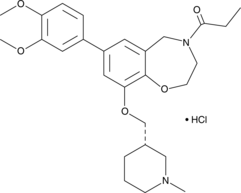
IACS-9571 (ASIS-P040)
IACS-9571 (ASIS-P040) is a potent and selective inhibitor of TRIM24 and BRPF1, with IC50 of 8 nM for TRIM24, and Kds of 31 nM and 14 nM for TRIM24 and BRPF1, respectively.
-
GC60925
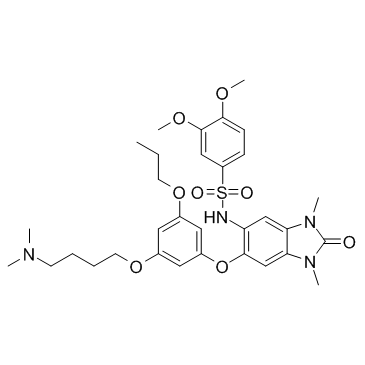
IACS-9571 hydrochloride
IACS-9571 (ASIS-P040) hydrochloride is a potent and selective inhibitor of TRIM24 and BRPF1, with an IC50 of 8 nM for TRIM24, and Kds of 31 nM and 14 nM for TRIM24 and BRPF1, respectively.
-
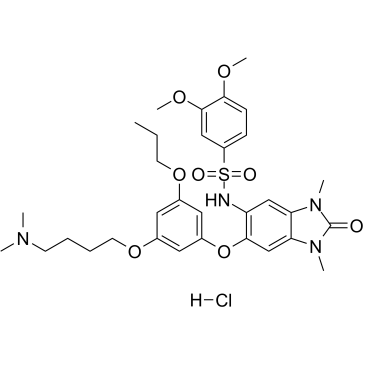
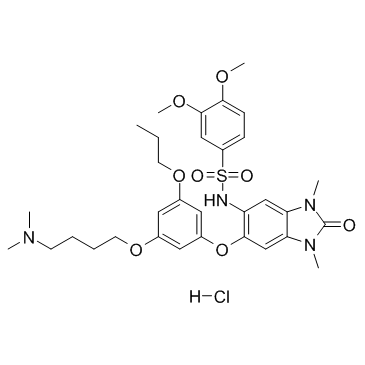
GC34437
IACS-9571 Hydrochloride (ASIS-P040 Hydrochloride)
-
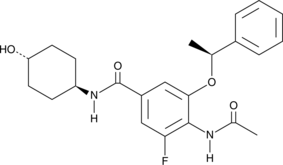
GC48548
iBET-BD2
A BD2 bromodomain inhibitor
-
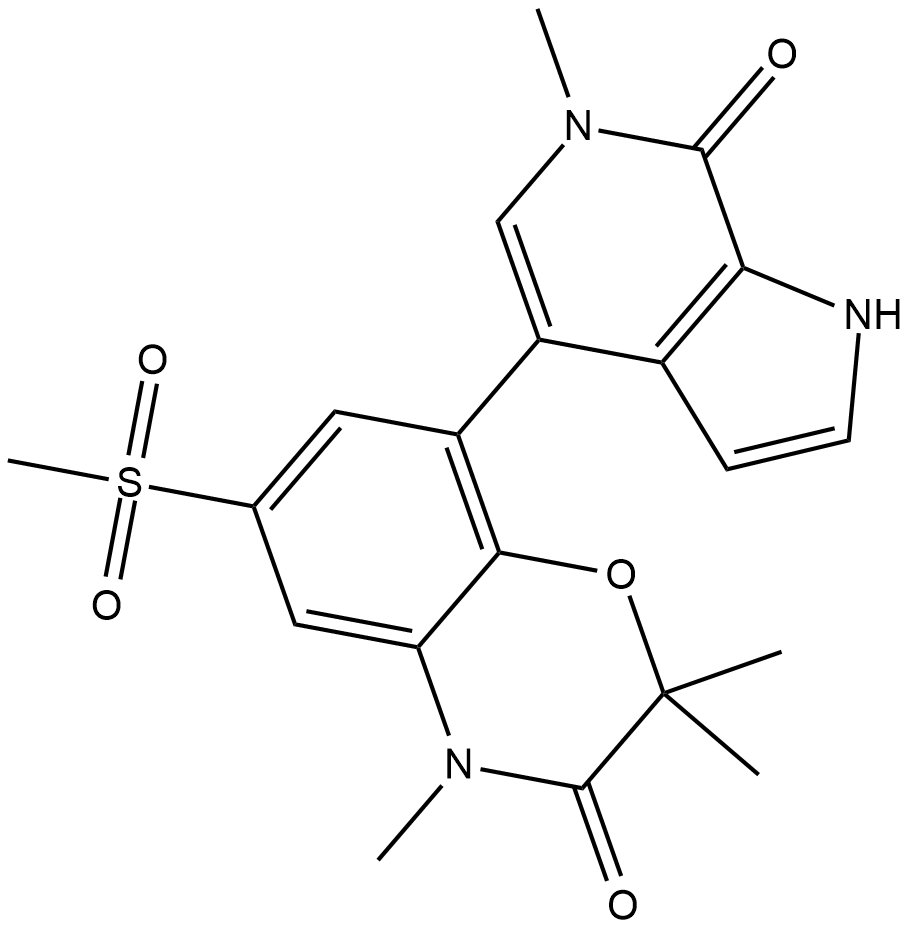
GC19200
INCB-057643
INCB-057643 is a novel, orally bioavailable BET inhibitor.
-
GC33026
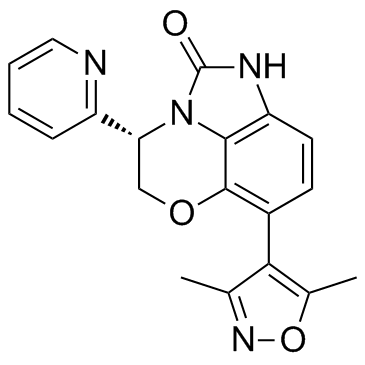
INCB054329
INCB054329 is a potent BET inhibitor.
-
GC30644
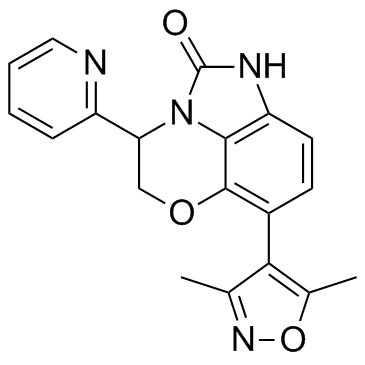
INCB054329 Racemate
INCB054329 Racemate is a BET protein inhibitor.
-
GC19210
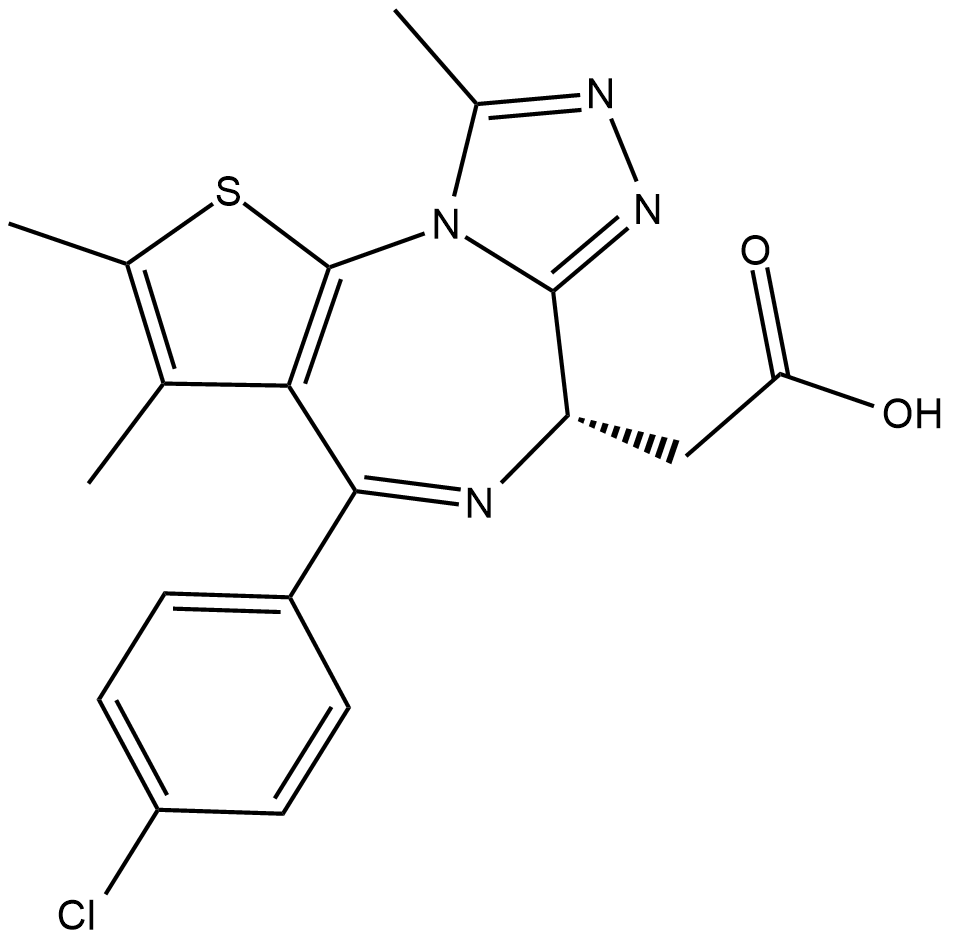
JQ-1 carboxylic acid
JQ-1 carboxylic acid is a highly potent, selective and cell-permeable BRD4 inhibitor with IC50s of 77 nM and 33 nM for BRD4(1) and BRD4(2), respectively.
-
GC31654
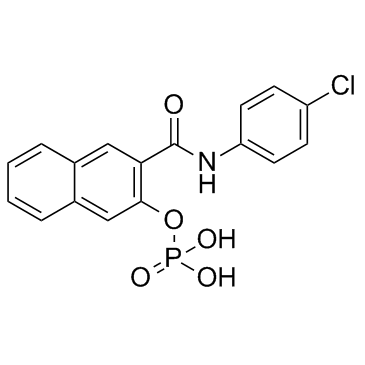
KG-501 (Naphthol AS-E phosphate)
KG-501 (Naphthol AS-E phosphate) is a CREB inhibitor, with an IC50 of 6.89 μM.
-
GC50395
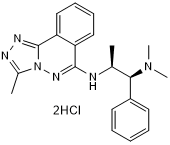
L Moses dihydrochloride
High affinity and selective PCAF bromodomain inhibitor
-
GC33183
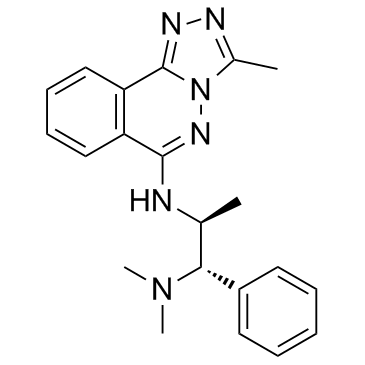
L-45 (L-Moses)
L-45 (L-Moses) (L-45) is the first potent, selective, and cell-active p300/CBP-associated factor (PCAF) bromodomain (Brd) inhibitor with a Kd of 126 nM.
-
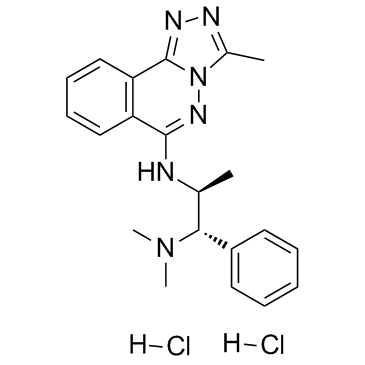
GC34377
L-45 dihydrochloride (L-Moses dihydrochloride)
-
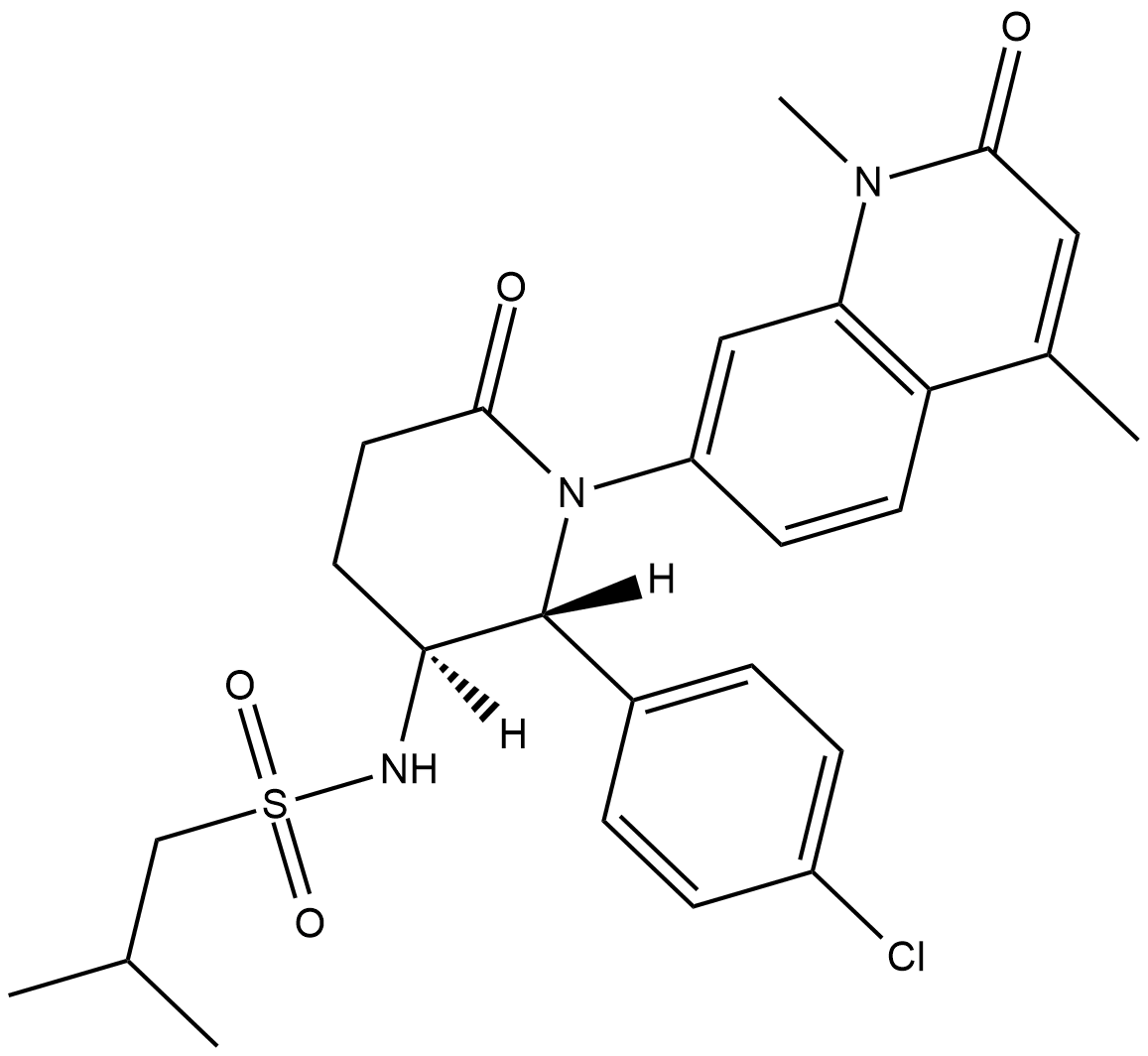
GC18731
LP99
LP99 is a potent inhibitor of the bromodomain containing proteins BRD7 and BRD9 that binds with Kd values of 99 and 909 nM, respectively, as determined by isothermal titration calorimetry.
-
GC62591
LT052
LT052 is a highly selective BET BD1 inhibitor with an IC50 of 87.7 nM.
-
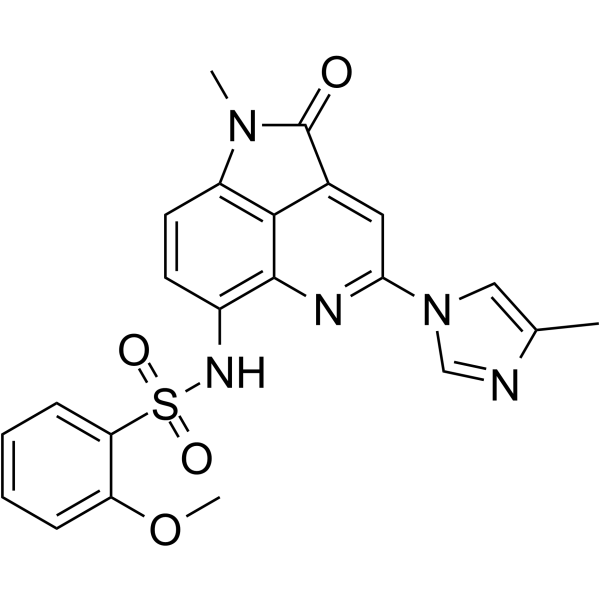
GC36521
M89
M-89 is a highly potent and specific menin inhibitor, with a Kd of 1.4 nM for binding to menin.
-
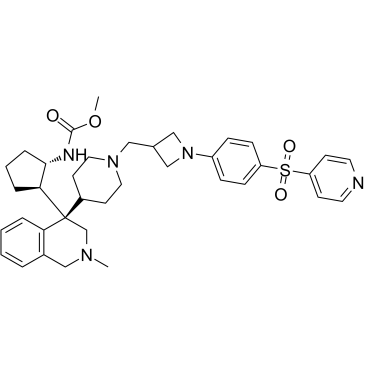
GC63064
Menin-MLL inhibitor 19
Menin-MLL inhibitor 19, a potent exo-aza spiro inhibitor of menin-mll interaction, example A17, extracted from patent WO2019120209A1.
-
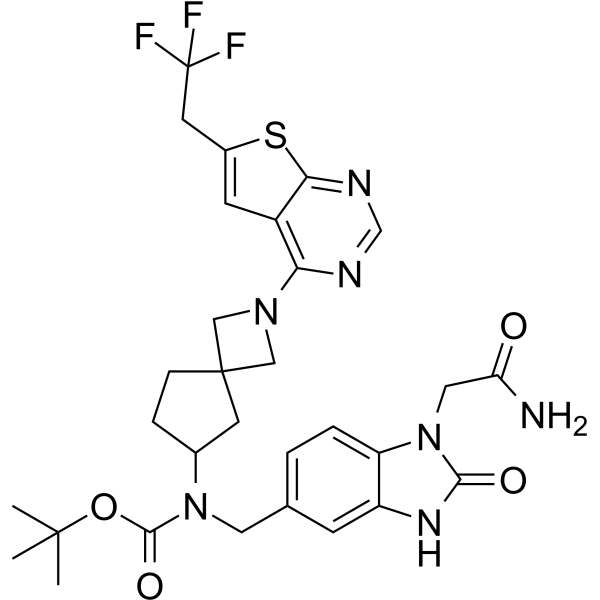
GC63538
Menin-MLL inhibitor 20
Menin-MLL inhibitor 20 is an irreversible menin-MLL interaction inhibitor with antitumor activities (WO2020142557A1, Intermediate 6).

-
GC11439
MG 149
HAT inhibitor
-
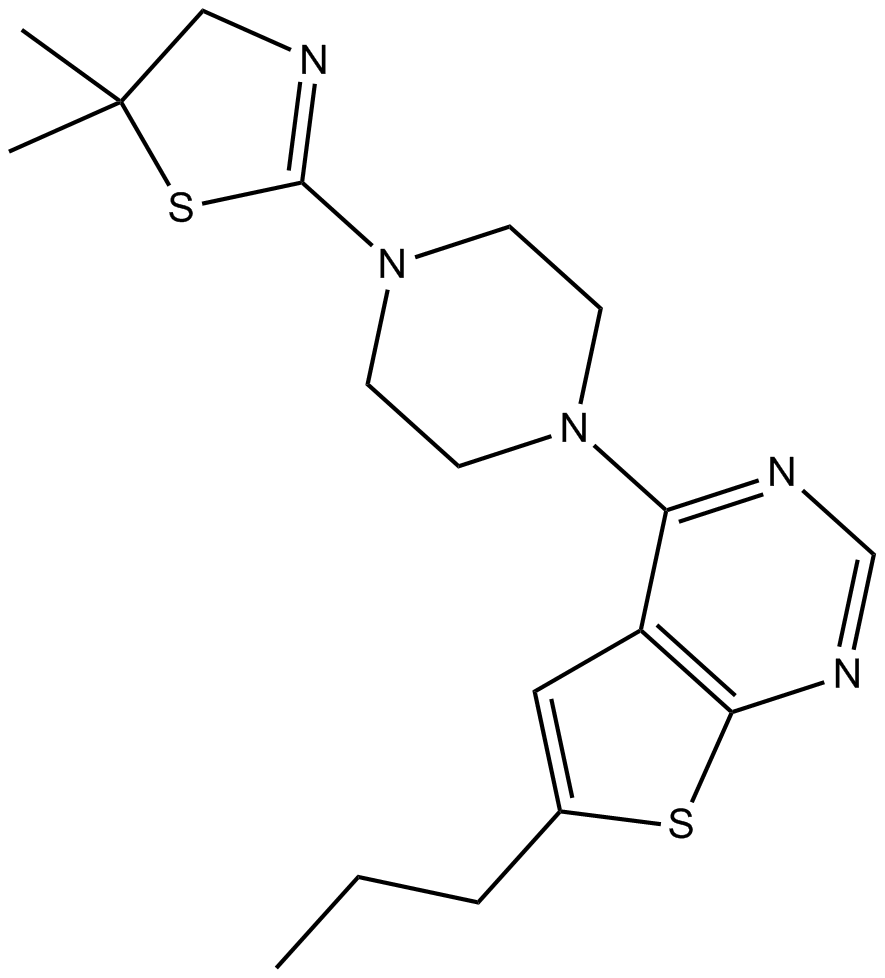
GC11418
MI-2
An inhibitor of menin-MLL fusion protein interactions
-
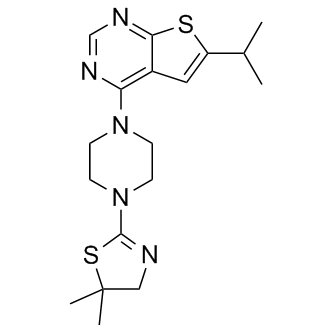
GC12199
MI-3
An inhibitor of menin-MLL fusion protein interactions
-
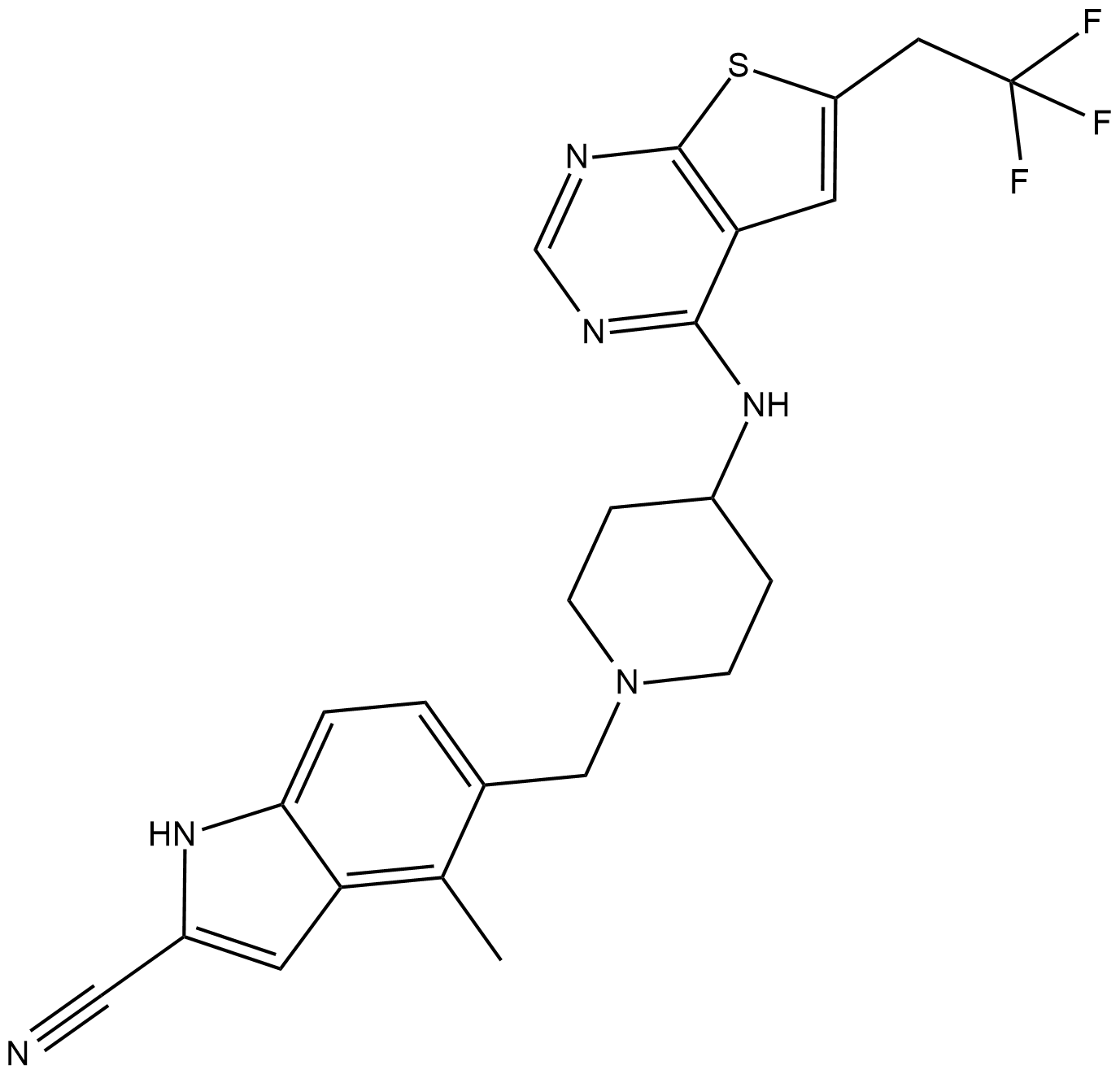
GC19246
MI-463
MI-463 is a highly potent and orally bioavailable small molecule inhibitor of the menin-mLL interaction.
-
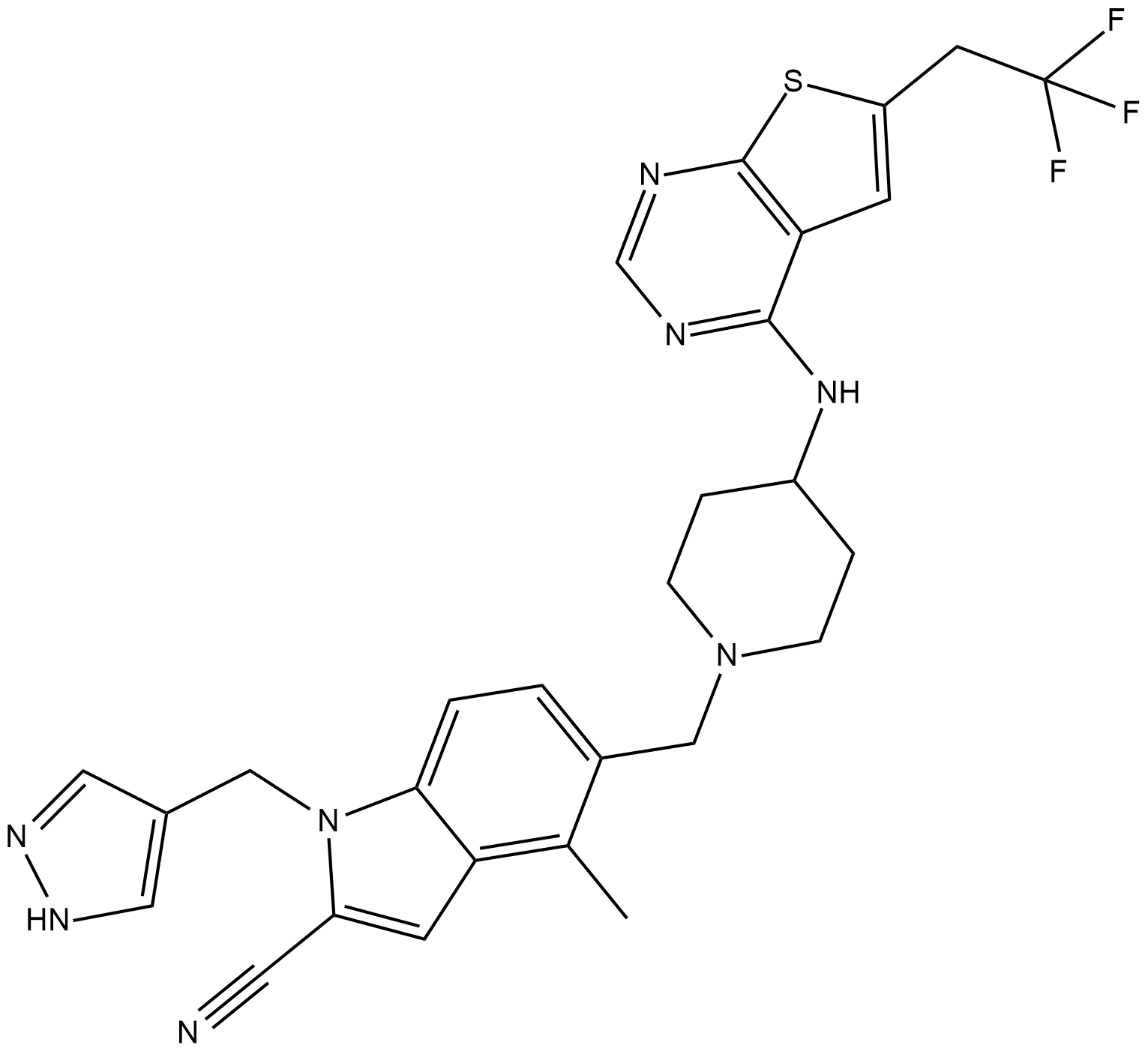
GC19247
MI-503
MI-503 is a highly potent and orally bioavailable small molecule inhibitor of the menin-mLL interaction.
-
GC32798
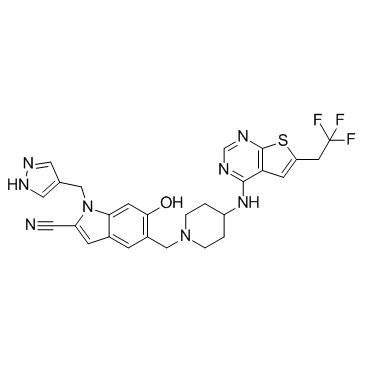
MI-538
MI-538 is an inhibitor of the interaction between menin and MLL fusion proteins with an IC50 of 21 nM.
-
GC19248
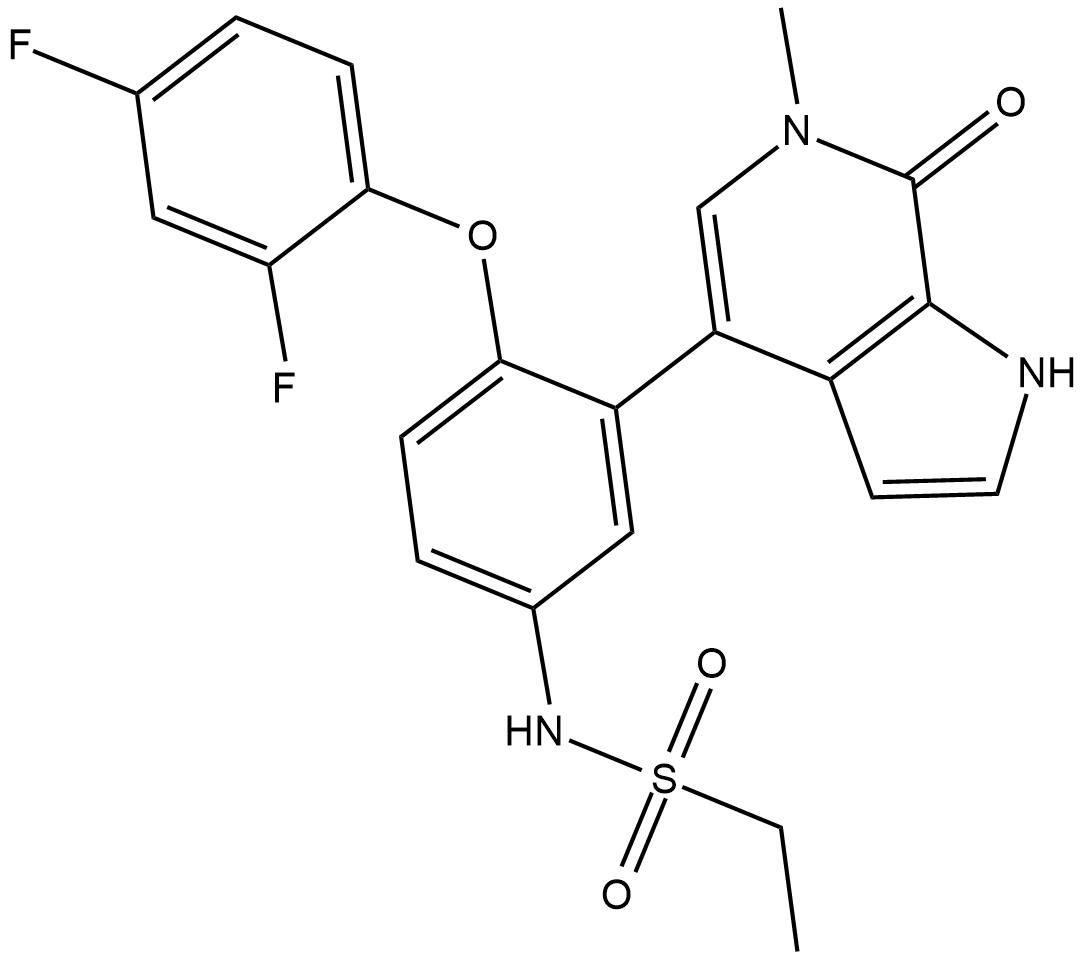
Mivebresib
Mivebresib is a potent and orally available bromodomain and extraterminal domain (BET) bromodomain inhibitor.
-
GC34675
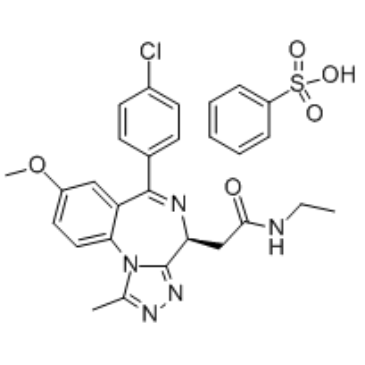
Molibresib besylate
Molibresib besylate (GSK 525762C; I-BET 762 besylate) is a BET bromodomain inhibitor with IC50 of 32.5-42.5 nM.
-
GC36657
MS31
MS31 is a potent, highly affinity and selective fragment-like methyllysine reader protein spindlin 1 (SPIN1) inhibitor. MS31 potently inhibits the interactions between SPIN1 and H3K4me3 (IC50=77 nM, AlphaLISA; 243 nM, FP). MS31 selectively binds Tudor domain II of SPIN1 (Kd=91 nM). MS31 potently inhibits binding of trimethyllysine-containing peptides to SPIN1. MS31 is not toxic to nontumorigenic cells.
-
GC39252
MS31 trihydrochloride
MS31 trihydrochloride is a potent, highly affinity and selective fragment-like methyllysine reader protein spindlin 1 (SPIN1) inhibitor. MS31 trihydrochloride potently inhibits the interactions between SPIN1 and H3K4me3 (IC50=77 nM, AlphaLISA; 243 nM, FP). MS31 trihydrochloride selectively binds Tudor domain II of SPIN1 (Kd=91 nM). MS31 trihydrochloride potently inhibits binding of trimethyllysine-containing peptides to SPIN1, and is not toxic to nontumorigenic cells.
-
GC44250
MS351
MS351 is an antagonist of chromobox 7 (CBX7) that acts by binding the CBX7 chromodomain.
-
GC64999
MS402
MS402 is a BD1-selective BET BrD inhibitor with Kis of 77 nM, 718 nM, 110 nM, 200 nM, 83 nM, and 240 nM for BRD4(BD1), BRD4(BD2), BRD3(BD1), BRD3(BD2), BRD2(BD1) and BRD2(BD2), respectively.

-
GC31881
MS417 (GTPL7512)
MS417 (GTPL7512) is a selective BET-specific BRD4 inhibitor, binds to BRD4-BD1 and BRD4-BD2 with IC50s of 30, 46 nM and Kds of 36.1, 25.4 nM, respectively, with weak selectivity at CBP BRD (IC50, 32.7 μM).
-
GC13148
MS436
BRD4 inhibitor
-
GC38474
MS645
MS645 is a bivalent BET bromodomains (BrD) inhibitor with a Ki of 18.4 nM for BRD4-BD1/BD2. MS645 spatially constrains bivalent inhibition of BRD4 BrDs resulting in a sustained repression of BRD4 transcriptional activity in solid-tumor cells.
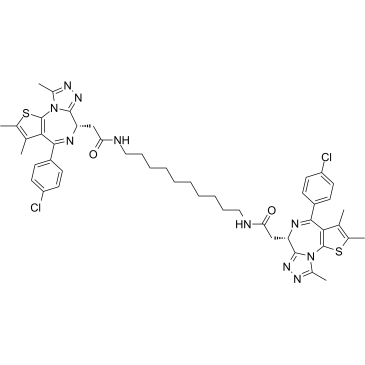
-
GC18729
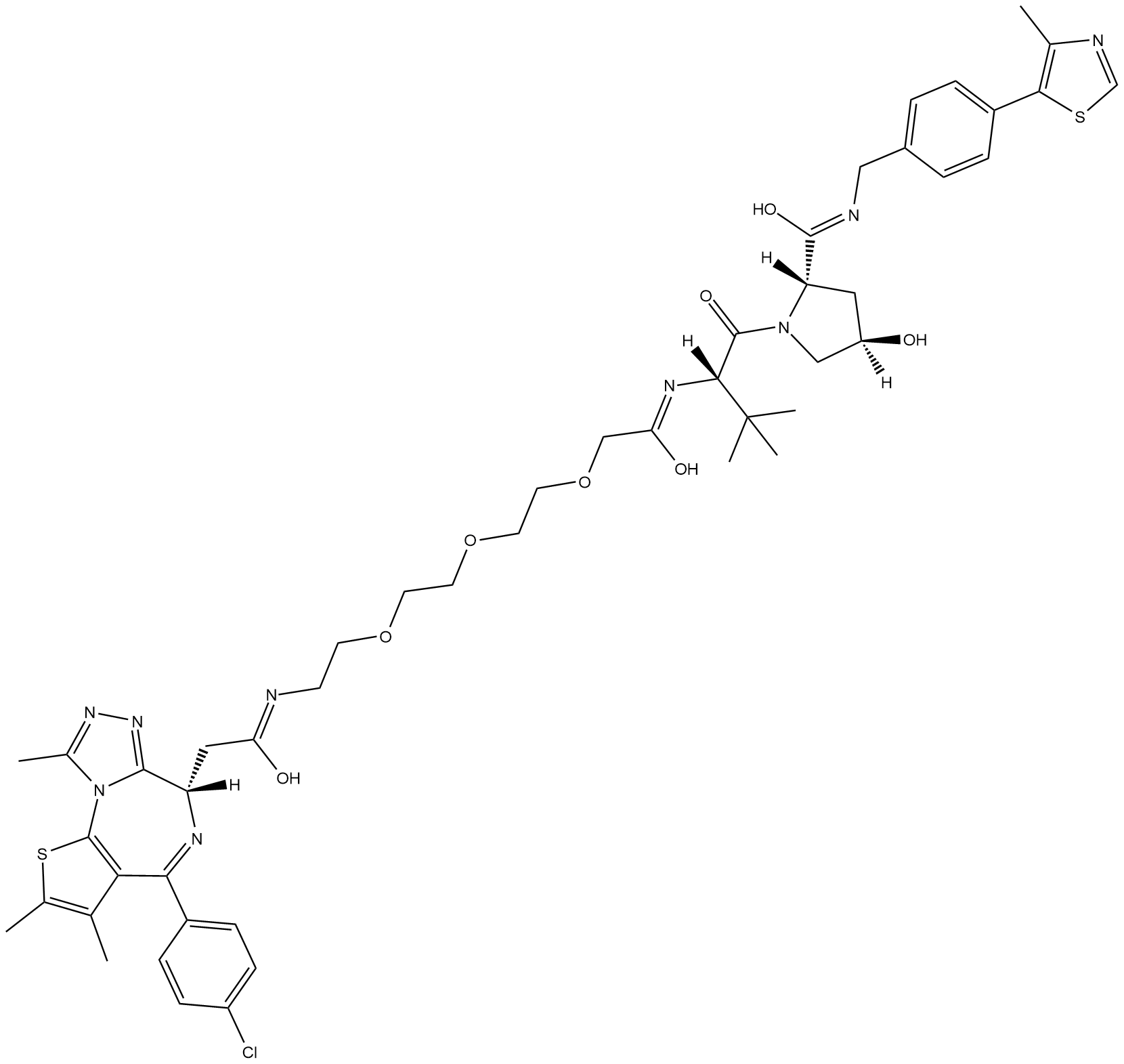
MZ1
MZ1 is a hybrid compound that drives the selective proteasomal degradation of bromodomain-containing protein 4 (BRD4).
-
GC33102
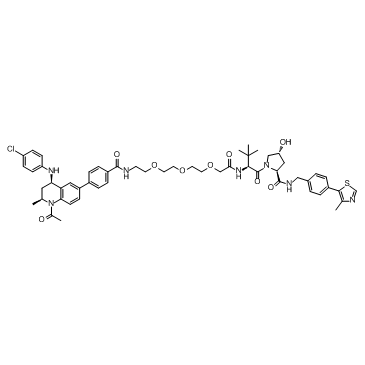
MZP-54
MZP-54 is a PROTAC connected by ligands for von Hippel-Lindau and BRD3/4, with a Kd of 4 nM for Brd4BD2.
-
GC33363
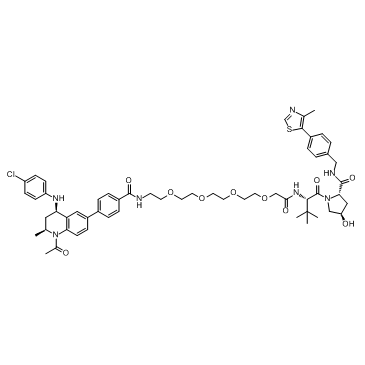
MZP-55
MZP-55 is a PROTAC connected by ligands for von Hippel-Lindau and BRD3/4, with a Kd of 8 nM for Brd4BD2.
-
GC61841
Naphthol AS-E
Naphthol AS-E is a potent and cell-permeable inhibitor of KIX-KID interaction. Naphthol AS-E directly binds to the KIX domain of CBP (Kd:8.6 ?M), blocks the interaction between the KIX domain and the KID domain of CREB with IC50 of 2.26 ?M. Naphthol AS-E can be used for cancer research.
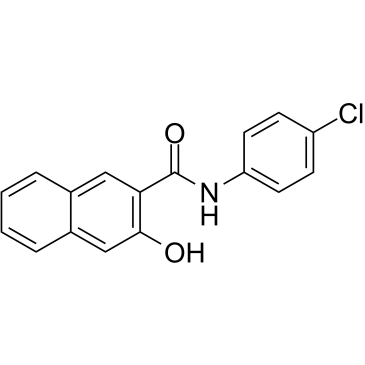
-
GC62620
NHWD-870
NHWD-870 is a potent, orally active and selective BET family bromodomain inhibitor and only binds bromodomains of BRD2, BRD3, BRD4 (IC50=2.7 nM), and BRDT. NHWD-870 has potent tumor suppressive efficacies and suppresses cancer cell-macrophage interaction. NHWD-870 increases tumor apoptosis and inhibits tumor proliferation.
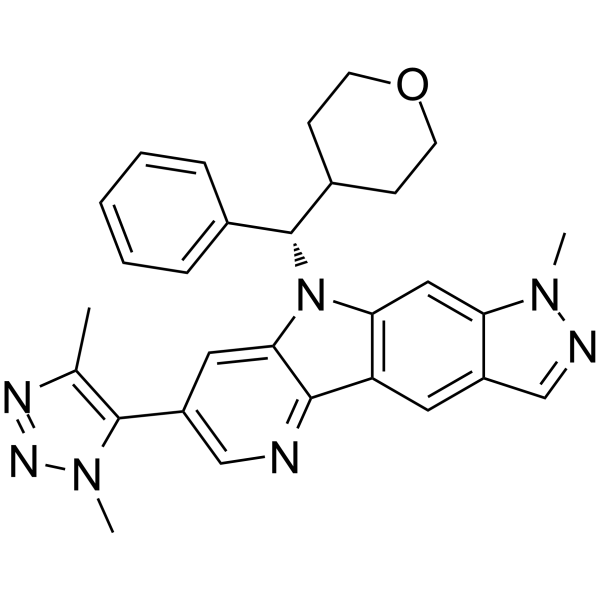
-
GC36734
NI-42
An inhibitor of the BRPF1 bromodomain
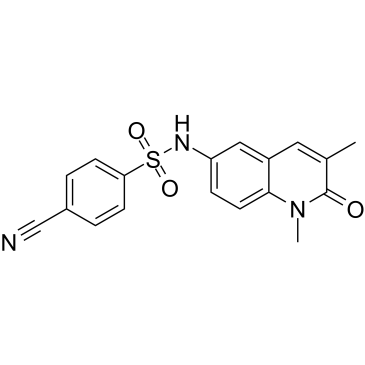
-
GC16763
NI-57
bromodomains of BRPF proteins inhibitor
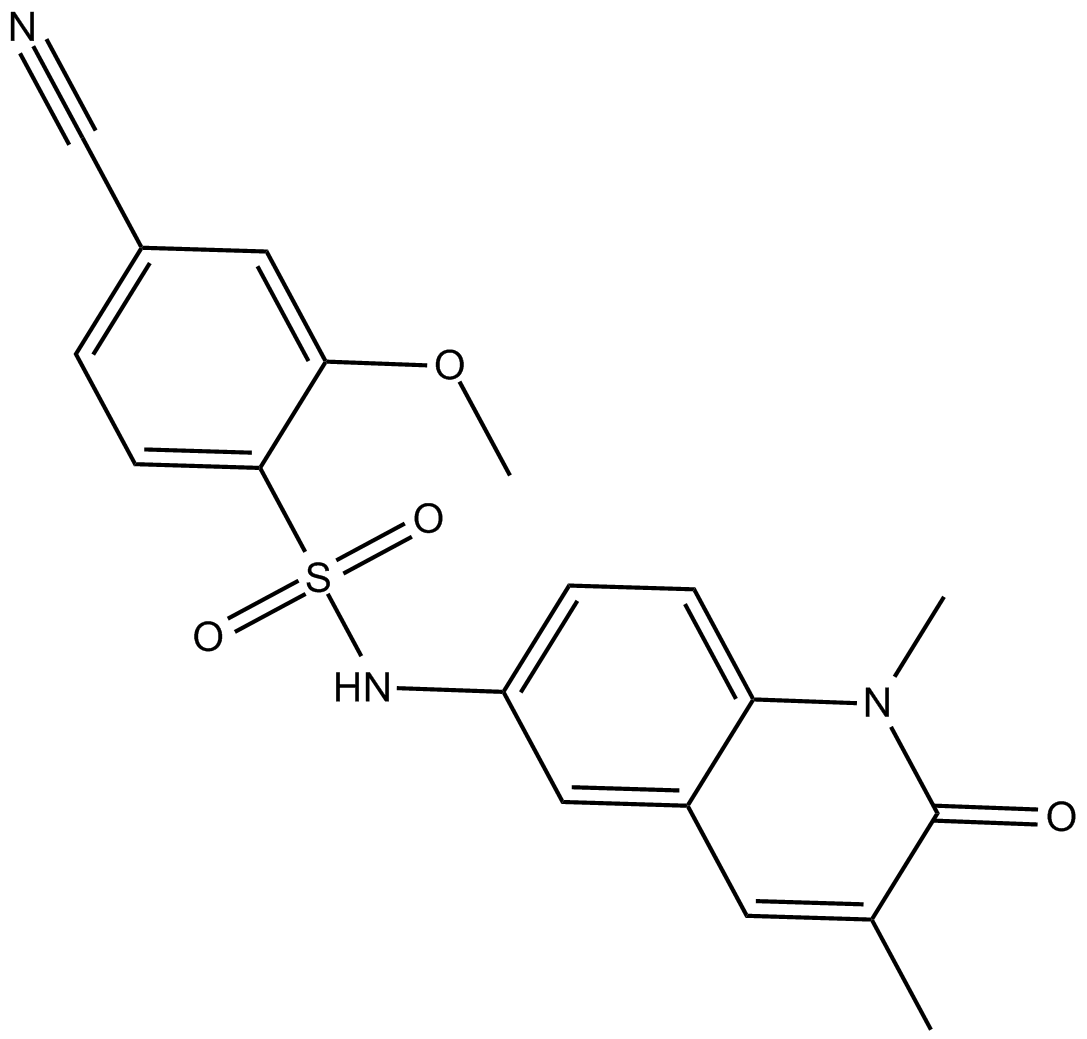
-
GC14103
NSC228155
EGFR activator
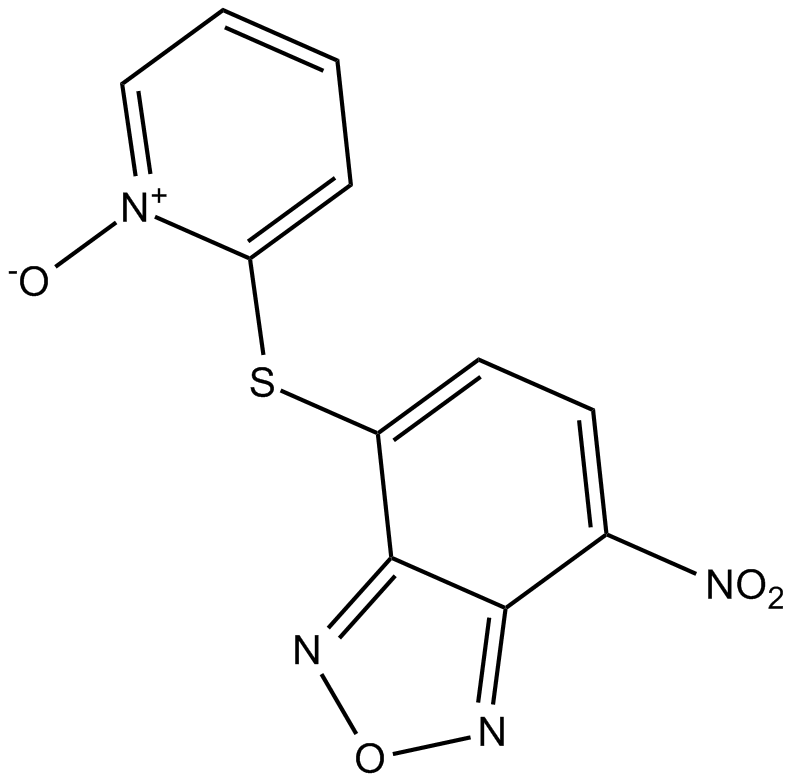
-
GC44477
NVS-CECR2-1
NVS-CECR2-1 is a potent inhibitor of CECR2 (cat eye syndrome chromosome region, candidate 2), a component of chromatin complexes that regulate gene expression controlling development.
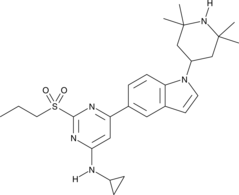
-
GC69601
OARV-771
OARV-771 is a VHL-based BET degrader (PROTAC) with improved cell permeability. OARV-771 shows DC50 values of 6, 1, and 4 nM for Brd4, Brd2, and Brd3 respectively.
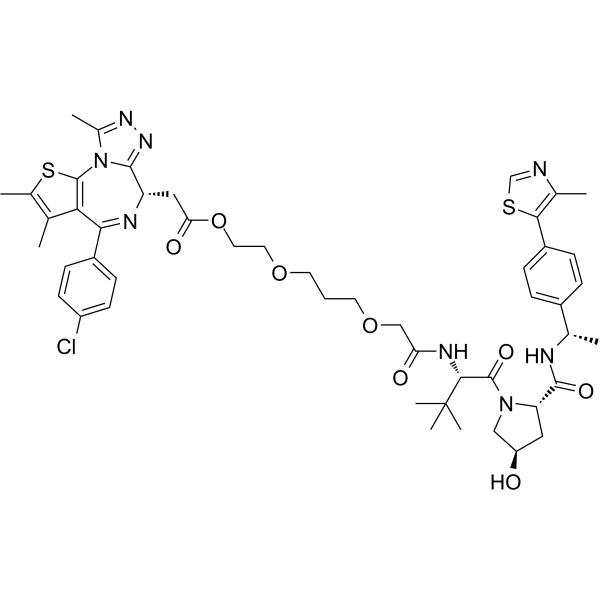
-
GC66067
ODM-207
ODM-207 (BET-IN-4) is a potent BET bromodomain protein (BRD4) inhibitor, with an IC50 of ≤ 1 μM.
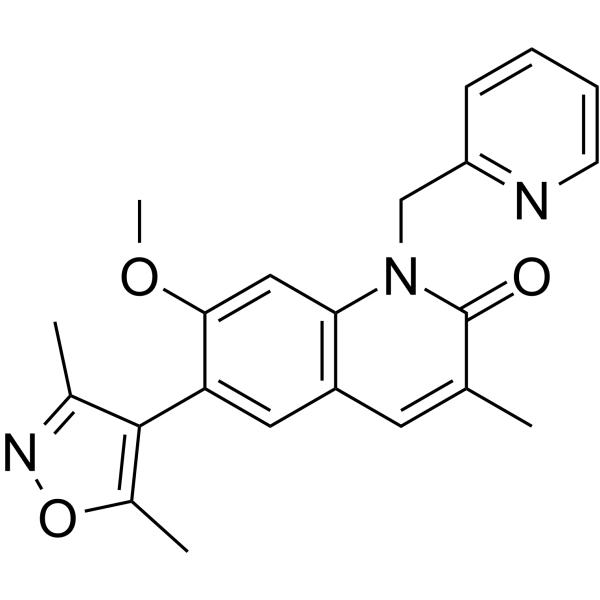
-
GC17482
OF-1
BRPF1B and BRPF2 bromodomain inhibitor
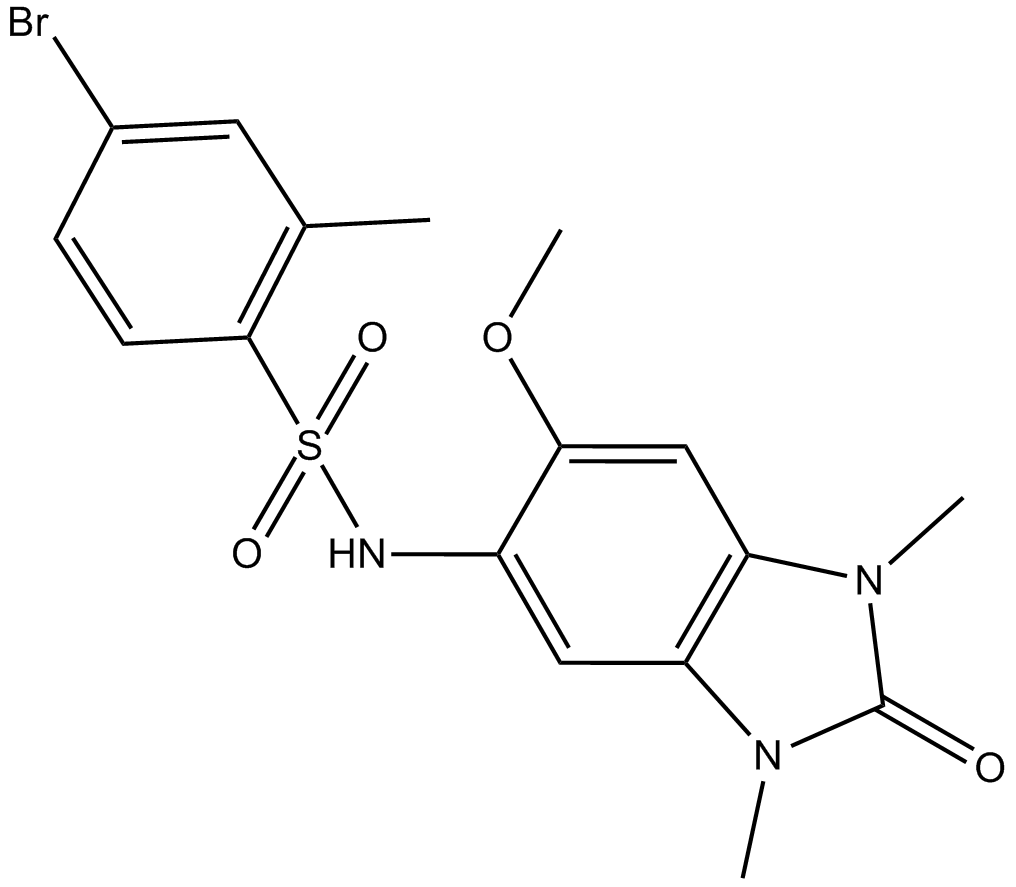
-
GC17973
OTX-015
A BRD2, BRD3, and BRD4 inhibitor
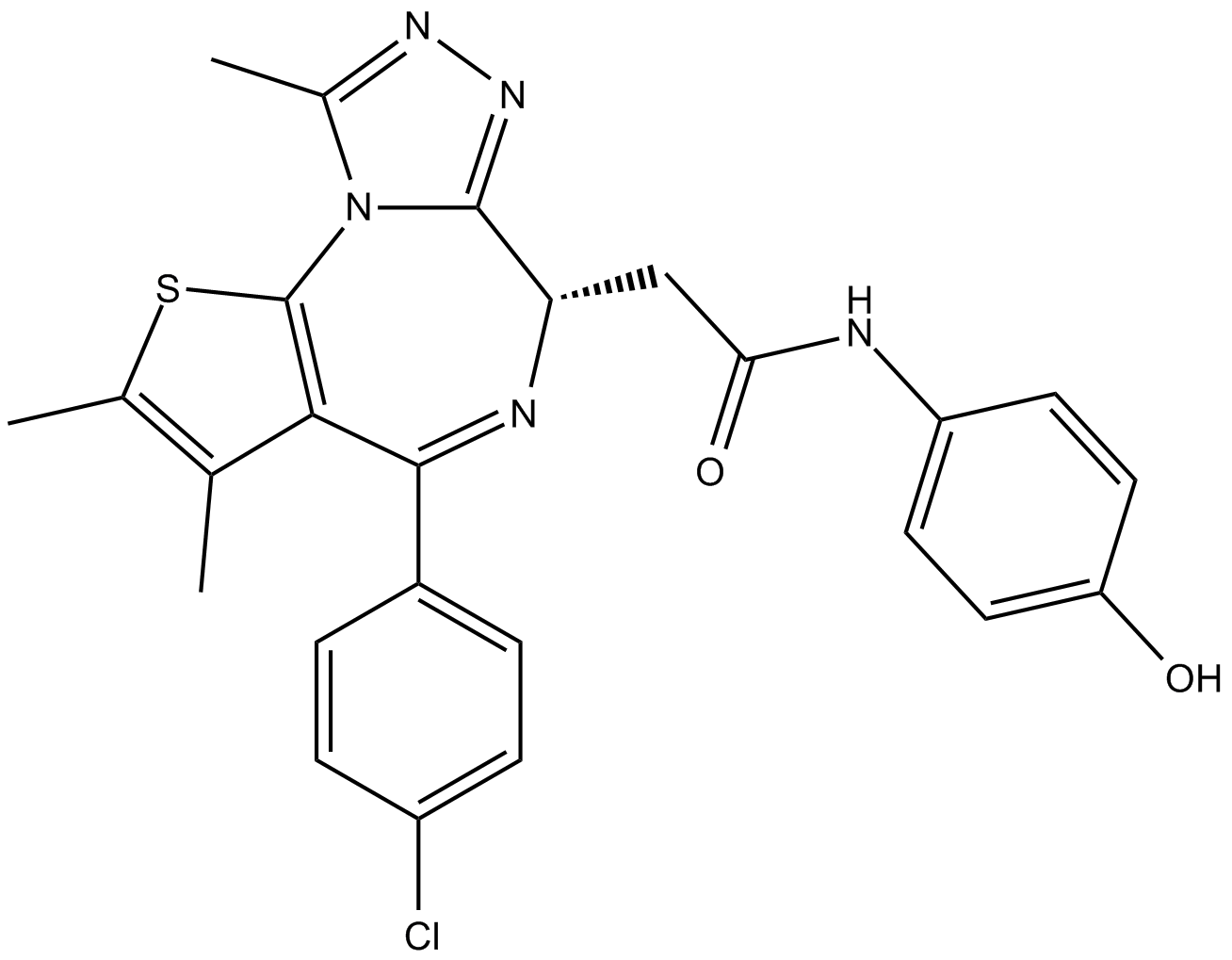
-
GC39417
OXFBD04
OXFBD04 is a potent and selective BRD4 inhibitor with an IC50 of 166?nM. OXFBD04 is a potent BET bromodomain ligand with additional modest affinity for the CREBBP bromodomain. OXFBD04 has anti-cancer activity.
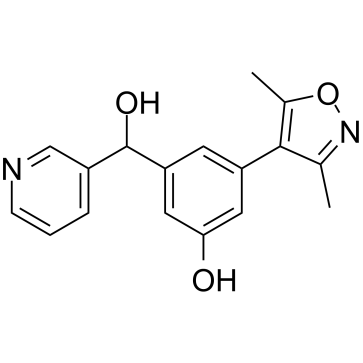
-
GC36887
PF-CBP1 hydrochloride
PF-CBP1 hydrochloride is a highly selective inhibitor of the CREB binding protein bromodomain (CBP BRD).
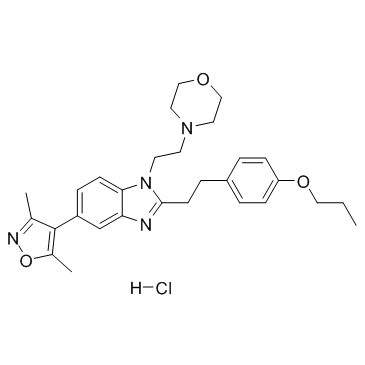
-
GC14361
PFI 3
inhibitor of polybromo 1 and SMARCA4
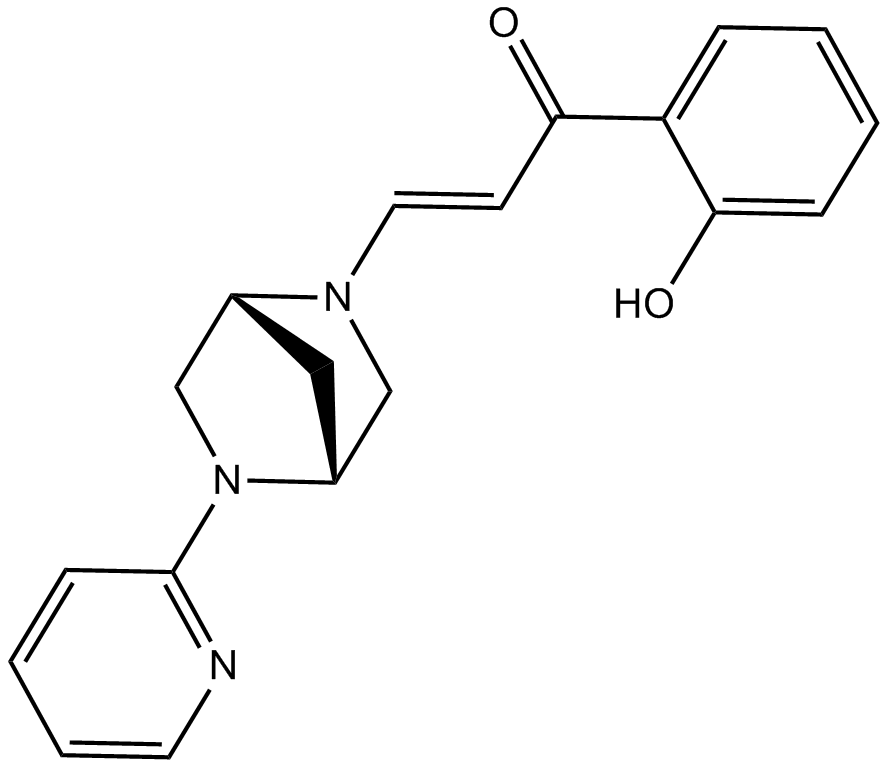
-
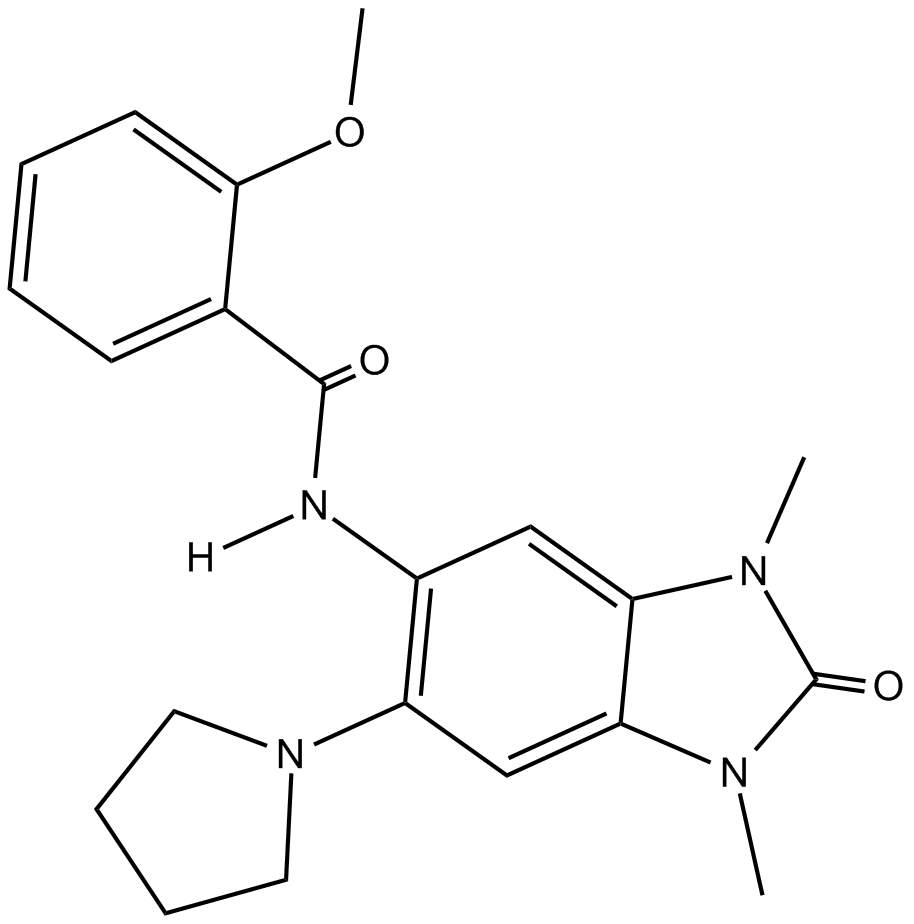
GC10979
PFI 4
Potent and selective BRPF1 Bromodomain inhibitor
-
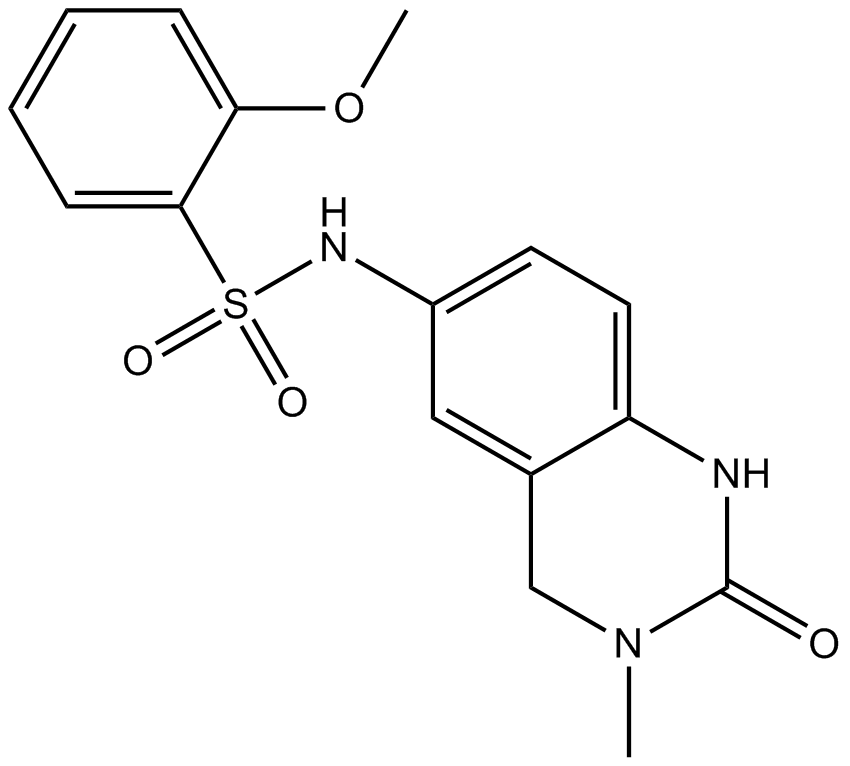
GC15375
PFI-1 (PF-6405761)
A BET bromodomain inhibitor
-
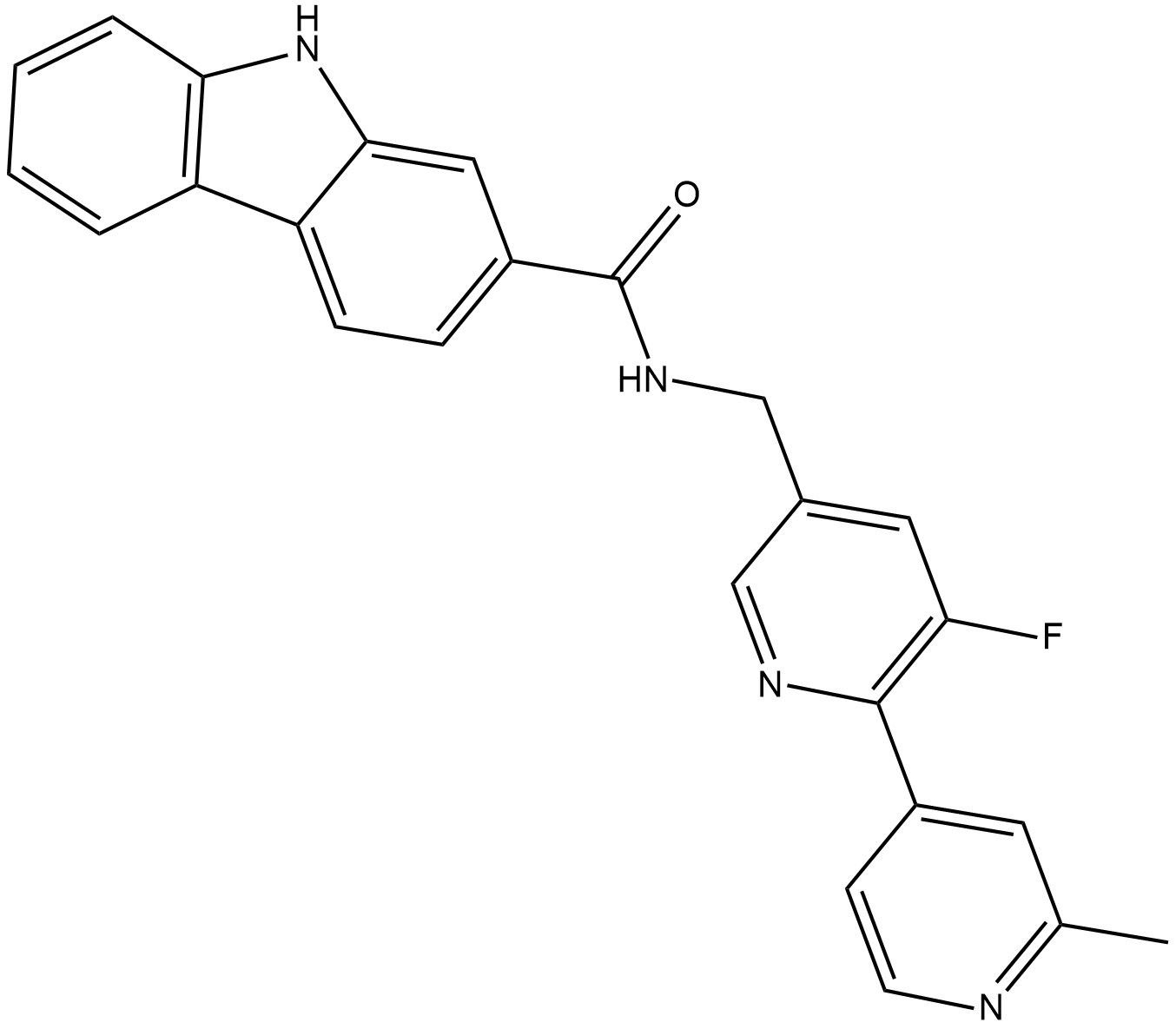
GC19298
PLX51107
PLX51107 is a potent and selective BET inhibitor, with Kds of 1.6, 2.1, 1.7, and 5 nM for BD1 and 5.9, 6.2, 6.1, and 120 nM for BD2 of BRD2, BRD3, BRD4, and BRDT, respectively; PLX51107 also interacts with the bromodomains of CBP and EP300 (Kd, in the 100 nM range).
-
GC36943
PNZ5
PNZ5 is a potent and isoxazole-based pan-BET inhibitor with high selectivity and potency similar to the well-established (+)-JQ1, with a KD of 5.43 nM for BRD4(1).

-
GC33109
PROTAC BET Degrader-1
PROTAC BET Degrader-1 is a PROTAC connected by ligands for Cereblon and BET, decreasing BRD2, BRD3, and BRD4 protein levels at low concentration.
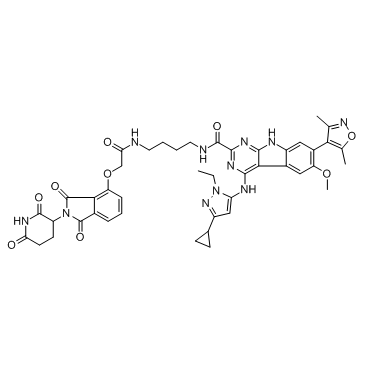
-
GC32980
PROTAC BET degrader-2
PROTAC BET degrader-2 is a PROTAC connected by ligands for Cereblon and BET with an IC50 value of 9.6 nM in cell growth inhibition in the RS4;11 cells and capable of achieving tumor regression.
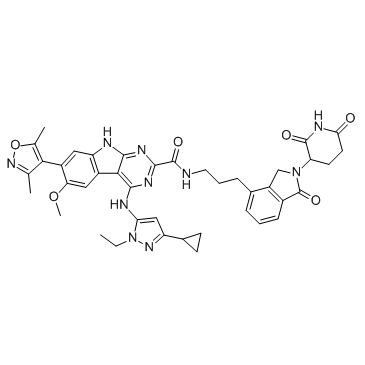
-
GC34325
PROTAC BET degrader-3
PROTAC BET Degrader-3 is a PROTAC connected by ligands for von Hippel-Lindau and BET.
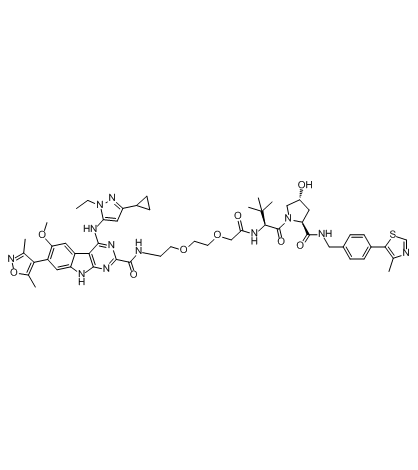
-
GC61212
PROTAC BRD4 Degrader-5
PROTAC BRD4 Degrader-5 is a PROTAC connected by ligands for von Hippel-Lindau and BRD4. PROTAC BRD4 Degrader-5 can potent degrade BRD4 in HER2 positive and negative breast cancer cell lines.
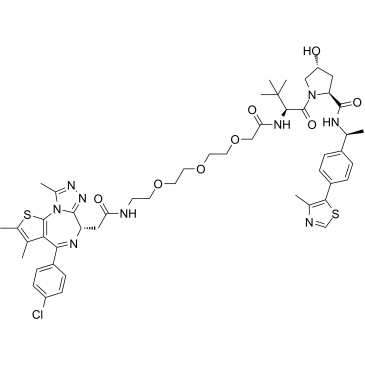
-
GC62606
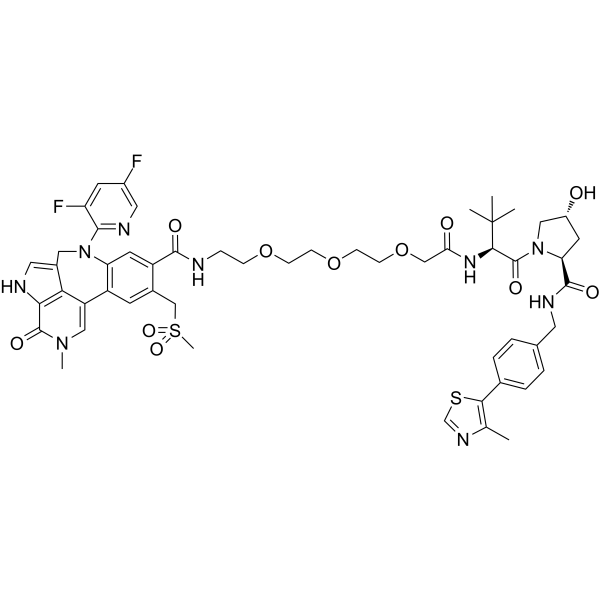
PROTAC BRD4 Degrader-8
PROTAC BRD4 Degrader-8 is a PROTAC connected by ligands for von Hippel-Lindau and BRD4, with IC50s of 1.1 nM and 1.4 nM for BRD4 BD1 and BD2, respectively. PROTAC BRD4 Degrader-8 is capable of potently degrading the BRD4 protein in PC3 prostate cancer cells.
-
GC39180
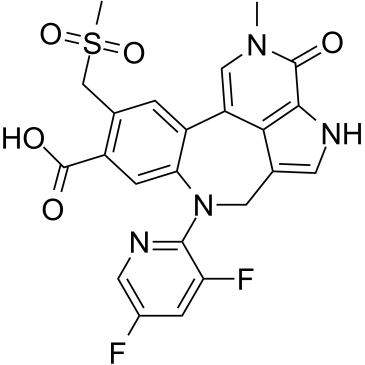
PROTAC BRD4 ligand-1
PROTAC BRD4 ligand-1 is a potent BET inhibitor and a ligand for target BRD4 protein for PROTACT GNE-987.
-
GC33372
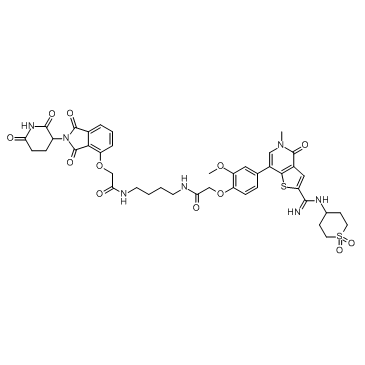
PROTAC BRD9 Degrader-1
PROTAC BRD9 Degrader-1 is a PROTAC connected by ligands for Cereblon and BRD9 (IC50=13.5 nM), which can be used as a selective probe useful for the study of BAF complex biology.
-
GC33217
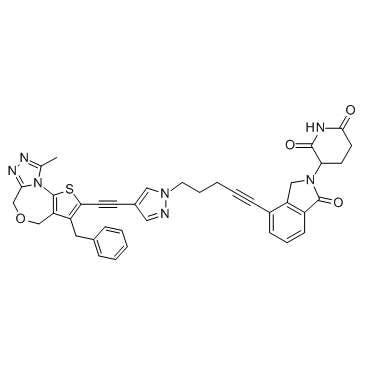
QCA570
QCA570 is a PROTAC connected by ligands for Cereblon and BET, with an IC50 of 10 nM for BRD4 BD1 Protein.
-
GC14199
RVX-208
RVX-208 (RVX-208) is an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. The IC50s are 87 μM and 0.51 μM for BD1 and BD2, respectively.

-
GC63178
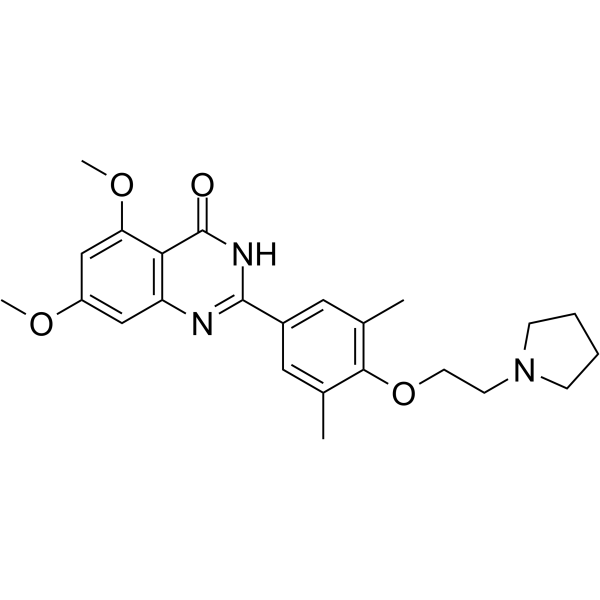
RVX-297
RVX-297 is a potent, orally active BET bromodomain inhibitor with selectivity for BD2.
-
GC19328
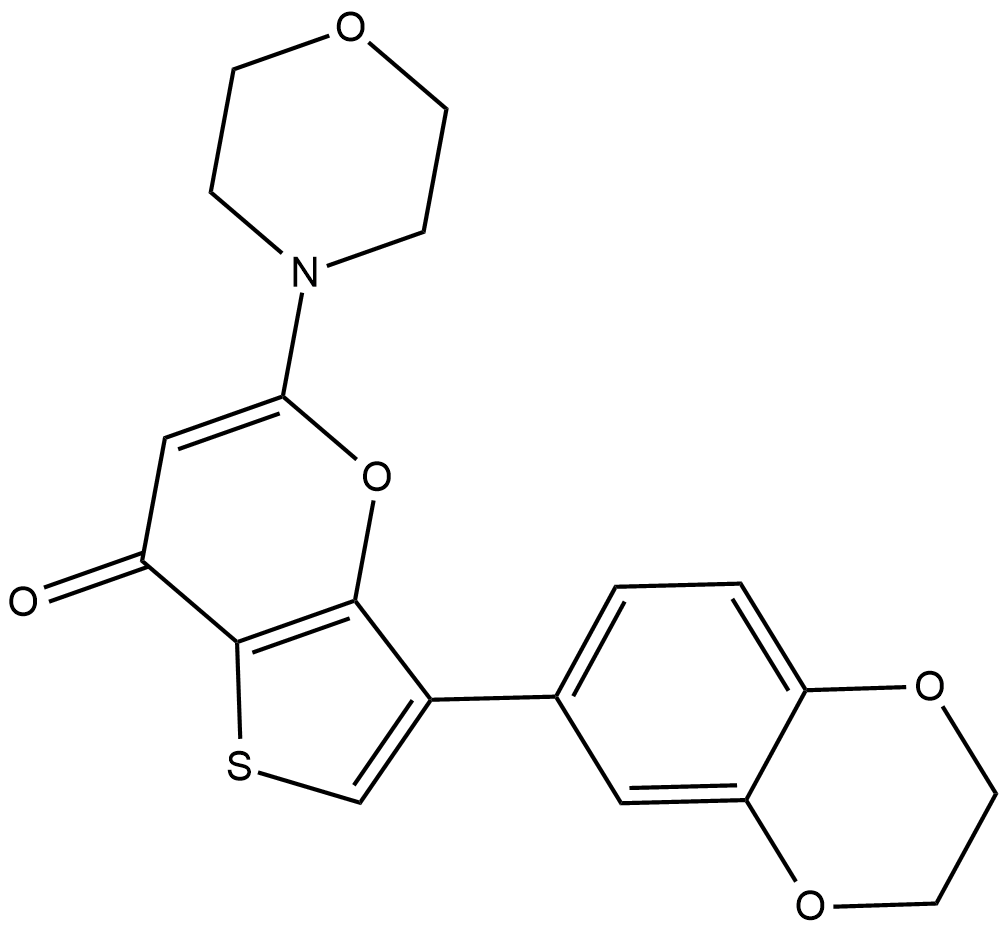
SF2523
SF2523 is a highly selective and potent inhibitor of PI3K with IC50s of 34 nM, 158 nM, 9 nM, 241 nM and 280 nM for PI3Kα, PI3Kγ, DNA-PK, BRD4 and mTOR, respectively.
-
GC10174
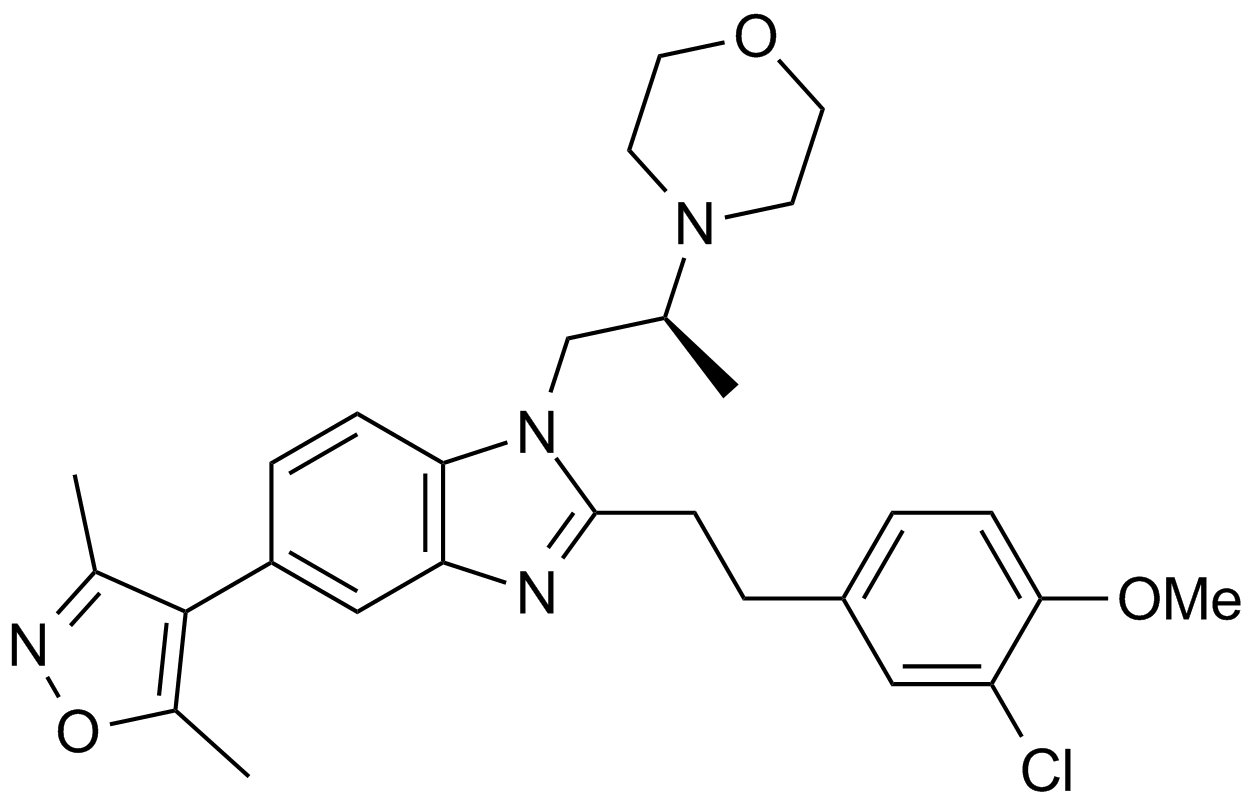
SGC-CBP30
A potent inhibitor of CREBBP/EP300 bromodomains
-
GC34778
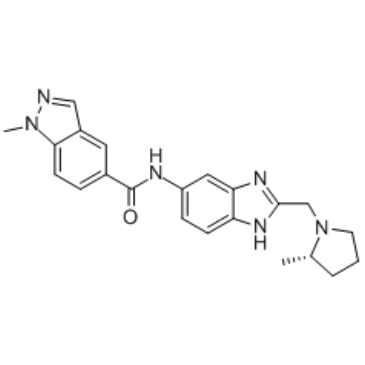
SGC-iMLLT
An inhibitor of MLLT1 and MLLT3 YEATS domains
-
GC63887
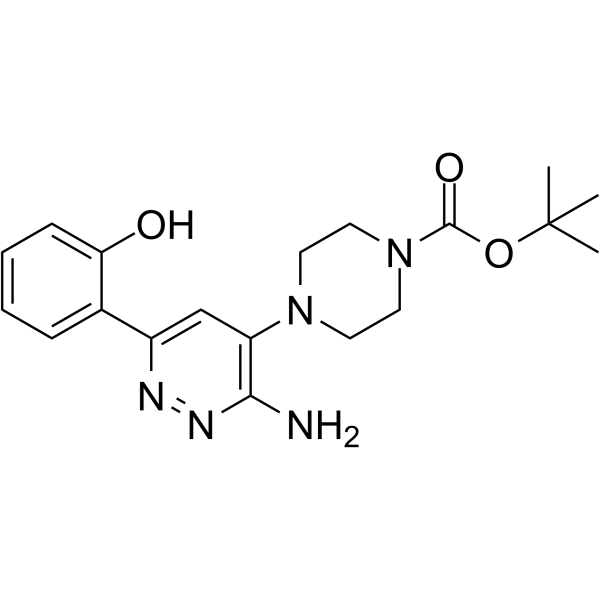
SGC-SMARCA-BRDVIII
SGC-SMARCA-BRDVIII is a potent and selective inhibitor of SMARCA2/4 and PB1(5), with Kds of 35 nM, 36 nM, and 13 nM, respectively.
-
GC65574
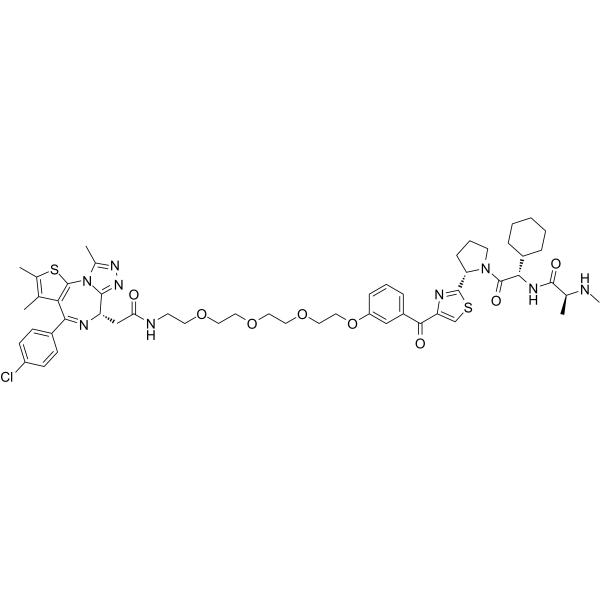
SNIPER(BRD)-1
SNIPER(BRD)-1, consists of an IAP antagonist LCL-161 derivative and a BET inhibitor, (+)-JQ-1, connected by a linker. SNIPER(BRD)-1 induces the degradation of BRD4 via the ubiquitin-proteasome pathway. SNIPER(BRD)-1 also degrades cIAP1 , cIAP2 and XIAP with IC50s of 6.8 nM, 17 nM, and 49nM, respectively.
-
GC32863
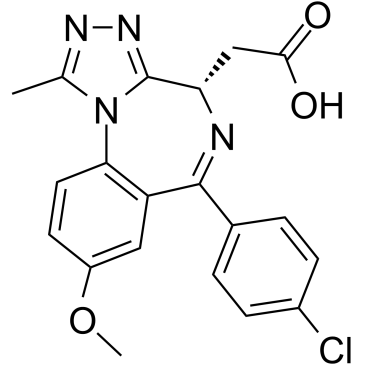
Target Protein-binding moiety 4
Target Protein-binding moiety 4 (Molibresib carboxylic acid) is an I-BET762-based warhead ligand for conjugation reactions of PROTAC targeting on BET. Target Protein-binding moiety 4 (Molibresib carboxylic acid) is a BRD4 inhibitor with a pIC50 of 5.1.
-
GC50340
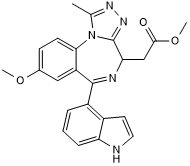
TC AC 28
High affinity BET bromodomain ligand
-
GC34319
TD-428
TD-428 is a PROTAC connected by ligands for Cereblon and BRD4. TD-428 is a highly specific BRD4 degrader with a DC50?of?0.32?nM. TD-428 is a BET PROTAC, which comprises TD-106 (a CRBN ligand) linked to JQ1 (a BET inhibitor). TD-428 efficiently induce BET protein degradation.
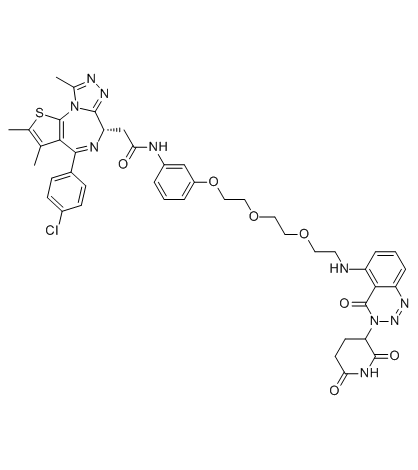
-
GC70037
TMX1
TMX1 is a BRD4 covalent molecular glue degrader. TMX1 selectively recruits DCAF16 to BRD4BD2, leading to the degradation of BRD4.
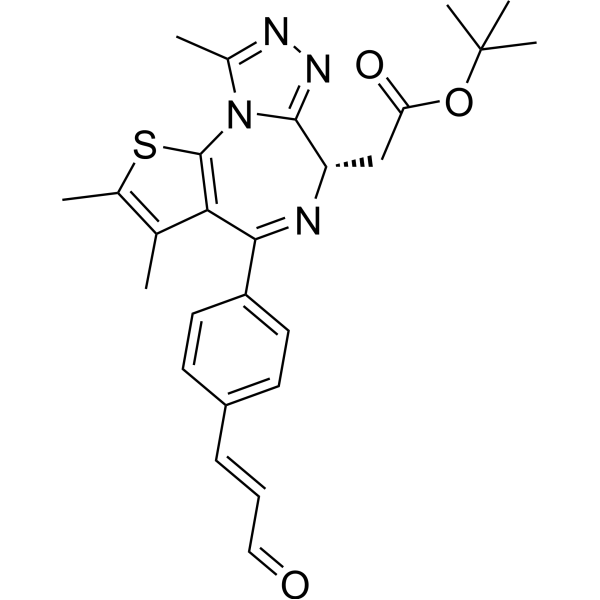
-
GC50542
TP 238
CECR2 and BPTF/FALZ inhibitor
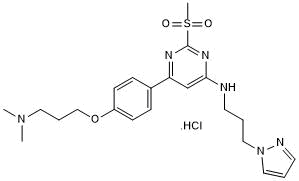
-
GC40628
TP-472
TP-472 is an inhibitor of bromodomain BRD9 (Kd = 33 nM; EC50 = 320 nM in a NanoBRET assay).
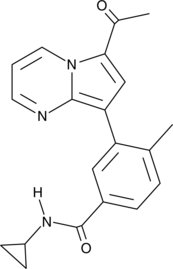
-
GC32880
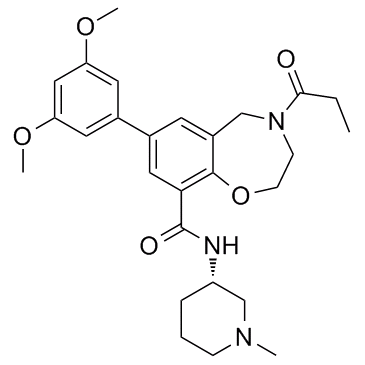
TPOP146
TPOP146 is a selective CBP/P300 benzoxazepine bromodomain inhibitor with Kd values of 134 nM and 5.02 μM for CBP and BRD4.
-
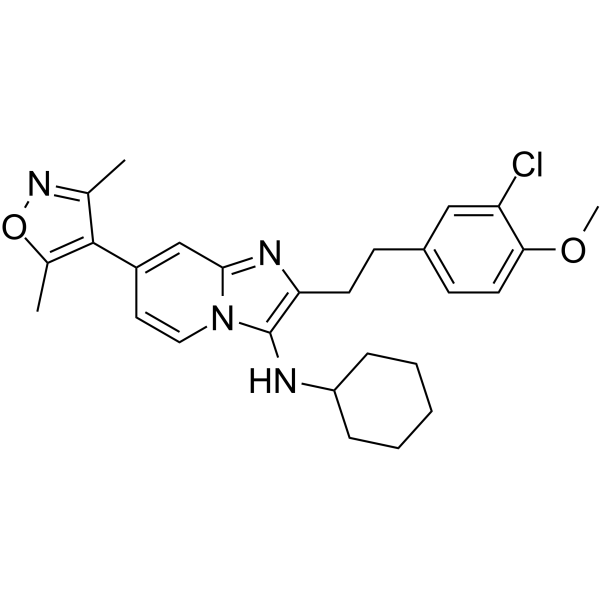
GC62405
UMB298
UMB298 is a potent and selective CBP/P300 bromodomain inhibitor.
-
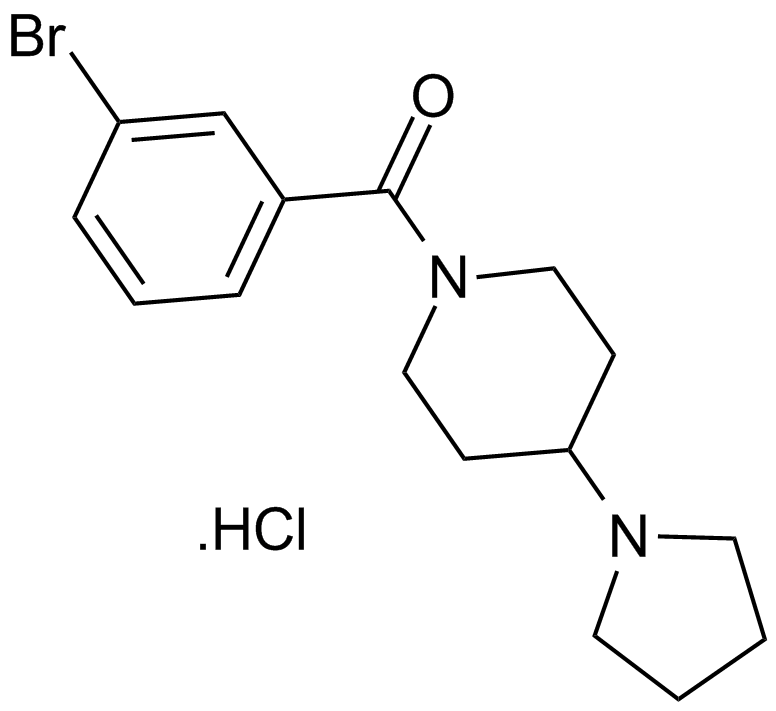
GC12419
UNC 926 hydrochloride
An antagonist of L3MBTL1, L3MBTL3, and L3MBTL4
-
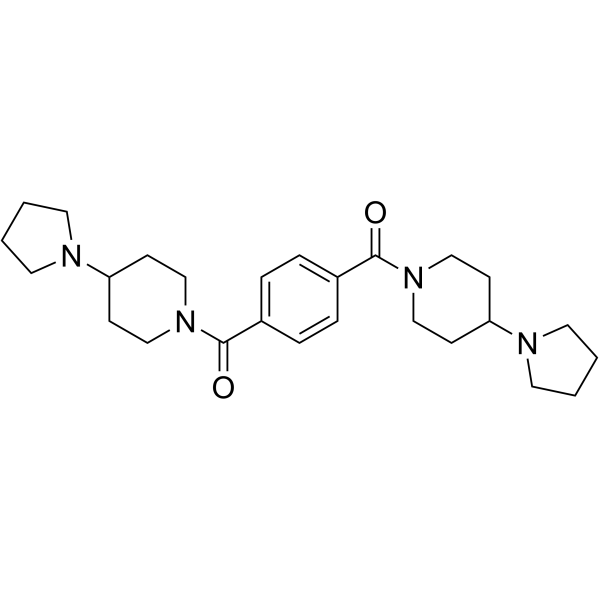
GC68025
UNC1021
-
GC64040
VTP50469 fumarate
VTP50469 fumarate is a potent, highly selective and orally active Menin-MLL interaction inhibitor with a Ki of 104 pM. VTP50469 fumarate has potently anti-leukemia activity.
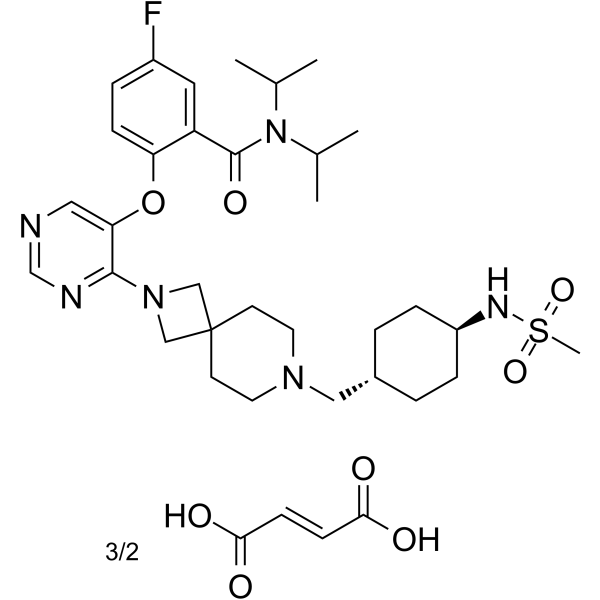
-
GC65135
VZ185
VZ185 is a potent, fast, and selective von Hippel-Lindau based dual degrader probe of BRD9 and BRD7 with DC50s of 4.5 and 1.8 nM, respectively. VZ185 is cytotoxic in EOL-1 and A-402 cells, with EC50s of 3 nM and 40 nM, respectively.
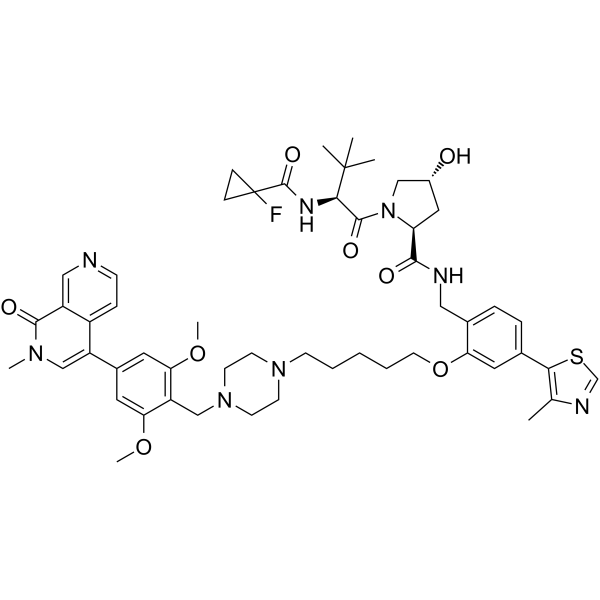
-
GC33285
Y06036
Y06036 is a potent and selective BET inhibitor, which binds to the BRD4(1) bromodomain with Kd value of 82 nM. Antitumor activity.

-
GC33264
Y06137
Y06137 is a potent and selective BET inhibitor for treatment of castration-resistant prostate cancer (CRPC). Y06137 binds to the BRD4(1) bromodomain with a Kd of 81 nM.
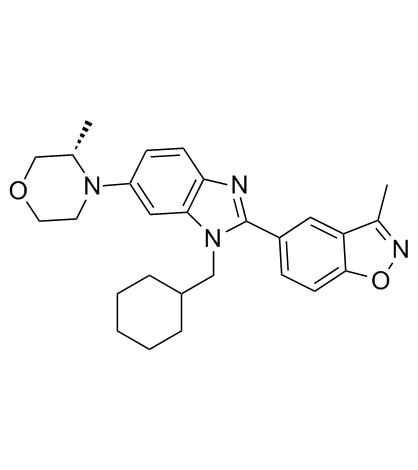
-
GC64518
Ziftomenib
Ziftomenib (KO-539) is a menin-MLL interaction inhibitor with antitumor activities (WO2017161028A1, compound 151).
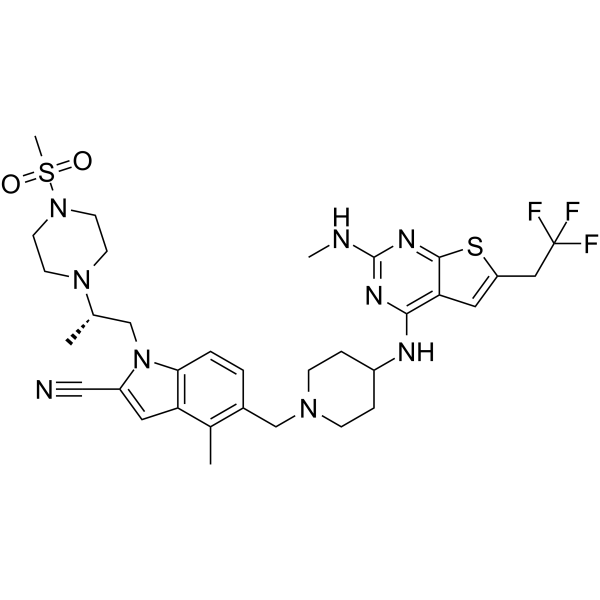
-
GC19455
ZL0420
ZL0420 is a potent and selective BRD4 inhibitor

-
GC65204
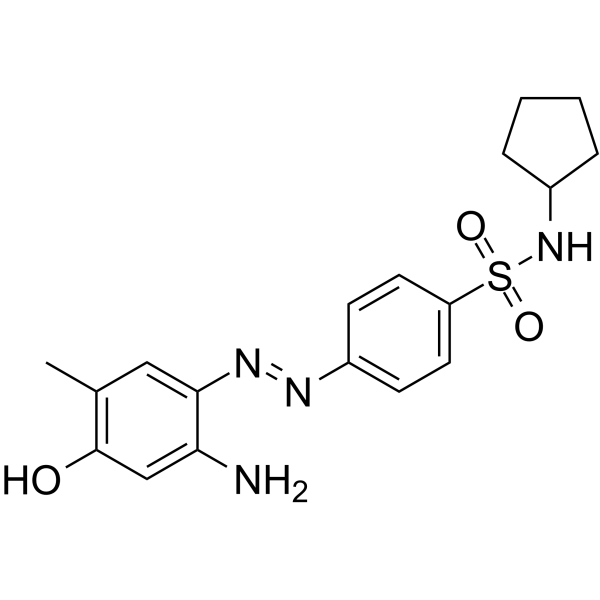
ZL0454
ZL0454 is a potent and selective Bromodomain-containing protein 4 (BRD4) inhibitor with an IC50 of 49 and 32 nM for BD1 and BD2.
-
GC64529
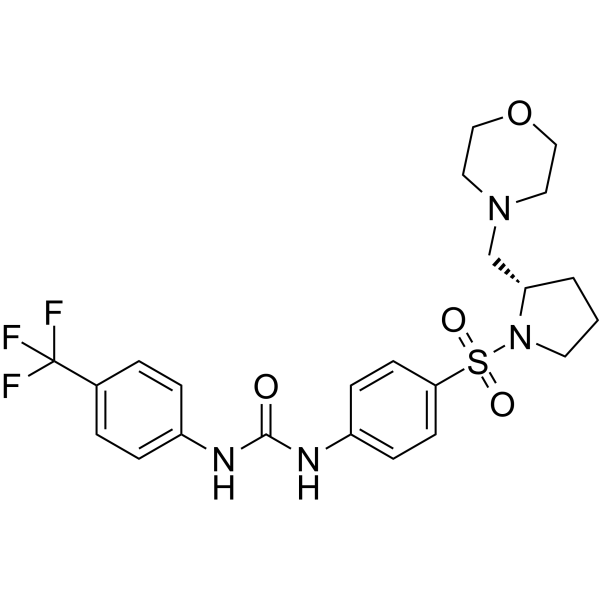
ZL0590
ZL0590 is a potent, orally active BRD4 BD1-selective inhibitor with an IC50 of 90 nM for human BRD4 BD1.
-
GC65269
ZXH-3-26
ZXH-3-26 is a PROTAC connected by ligands for Cereblon and BRD4 with a DC50/5h of 5 nM. The DC50/5h refers to half-maximal degradation after 5 hours of treatment of ~ 5 nM.