GTPase
Products for GTPase
- Cat.No. Product Name Information
-
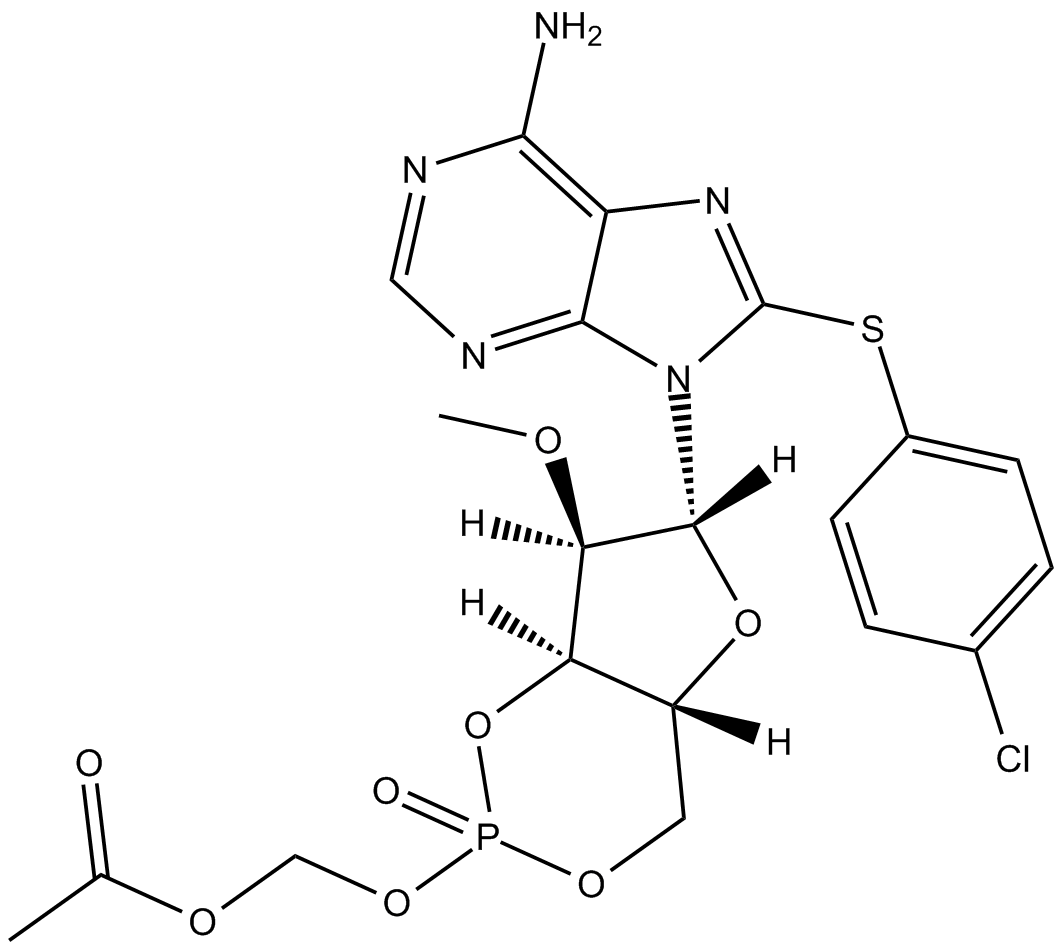
GC11483
8-CPT-2Me-cAMP, sodium salt
8-CPT-2Me-cAMP, sodium salt is a selective activator of exchange proteins activated by cAMP (Epac), the cAMP sensitive guanine nucleotide exchange factors (GEFs) for the small GTPases Rap1 and Rap2.
-
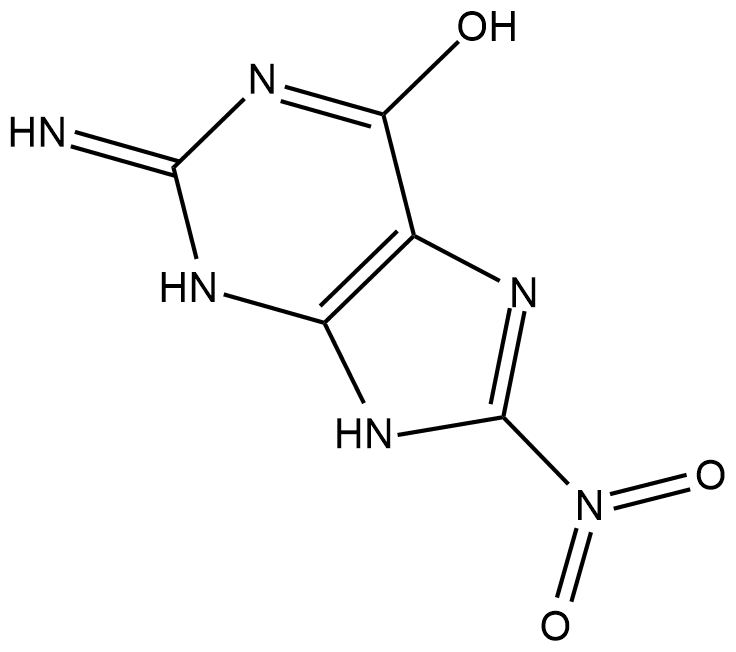
GC18426
8-Nitroguanine
8-Nitroguanine is a nitrative guanine derivative formed by oxidative damage to the guanine base in DNA by reactive nitrogen species (RNS) during inflammation and in vitro by reaction of DNA with peroxynitrite and other RNS reagents.
-
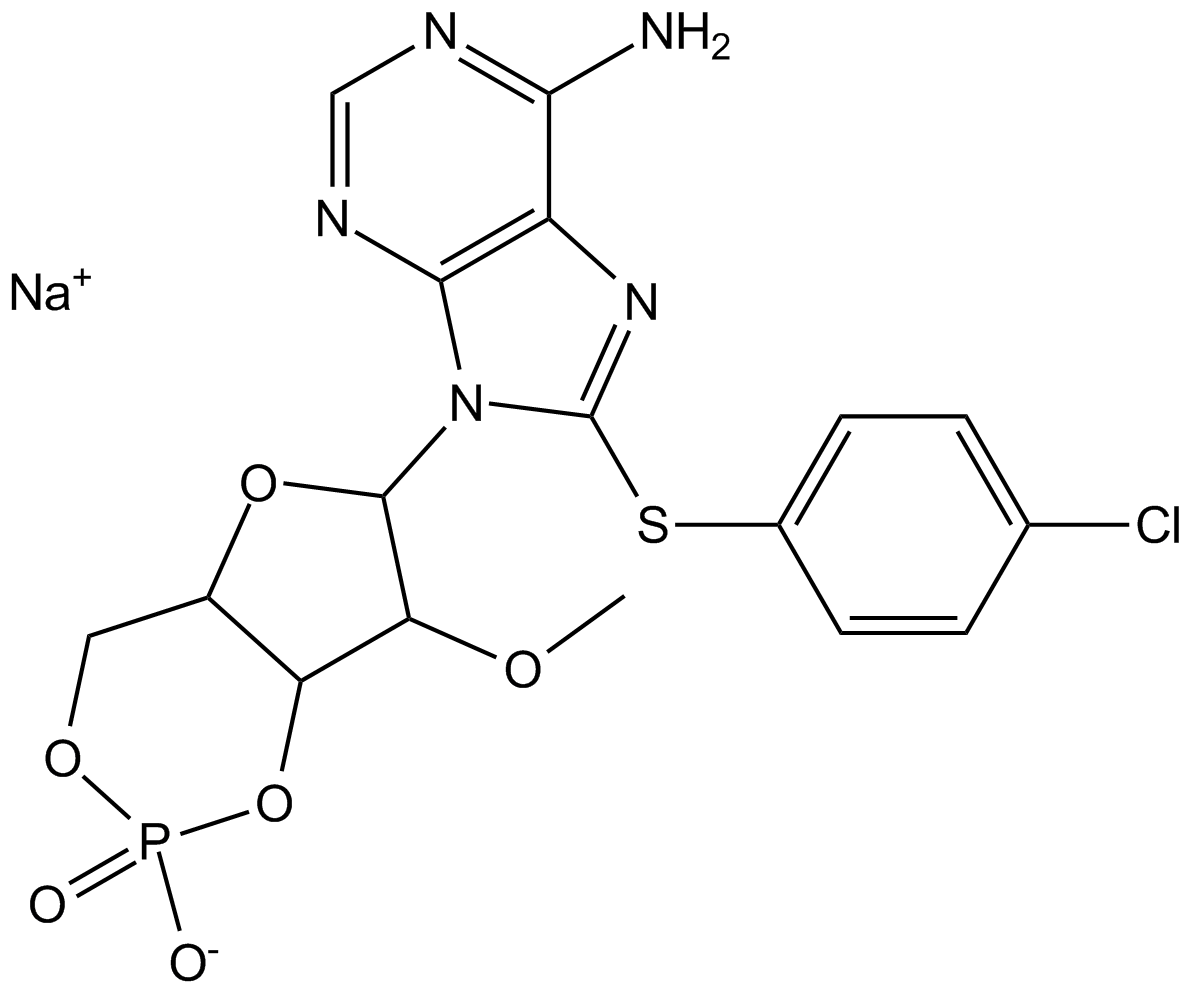
GC16199
8-pCPT-2-O-Me-cAMP-AM
Epac activator
-
GC43306
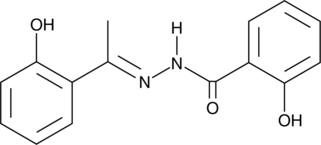
Compound C108
Compound C108 is an inhibitor of the GTPase-activating protein (SH3 domain)-binding protein 2 (G3BP2), which is involved in breast tumor initiation.
-
GC14179
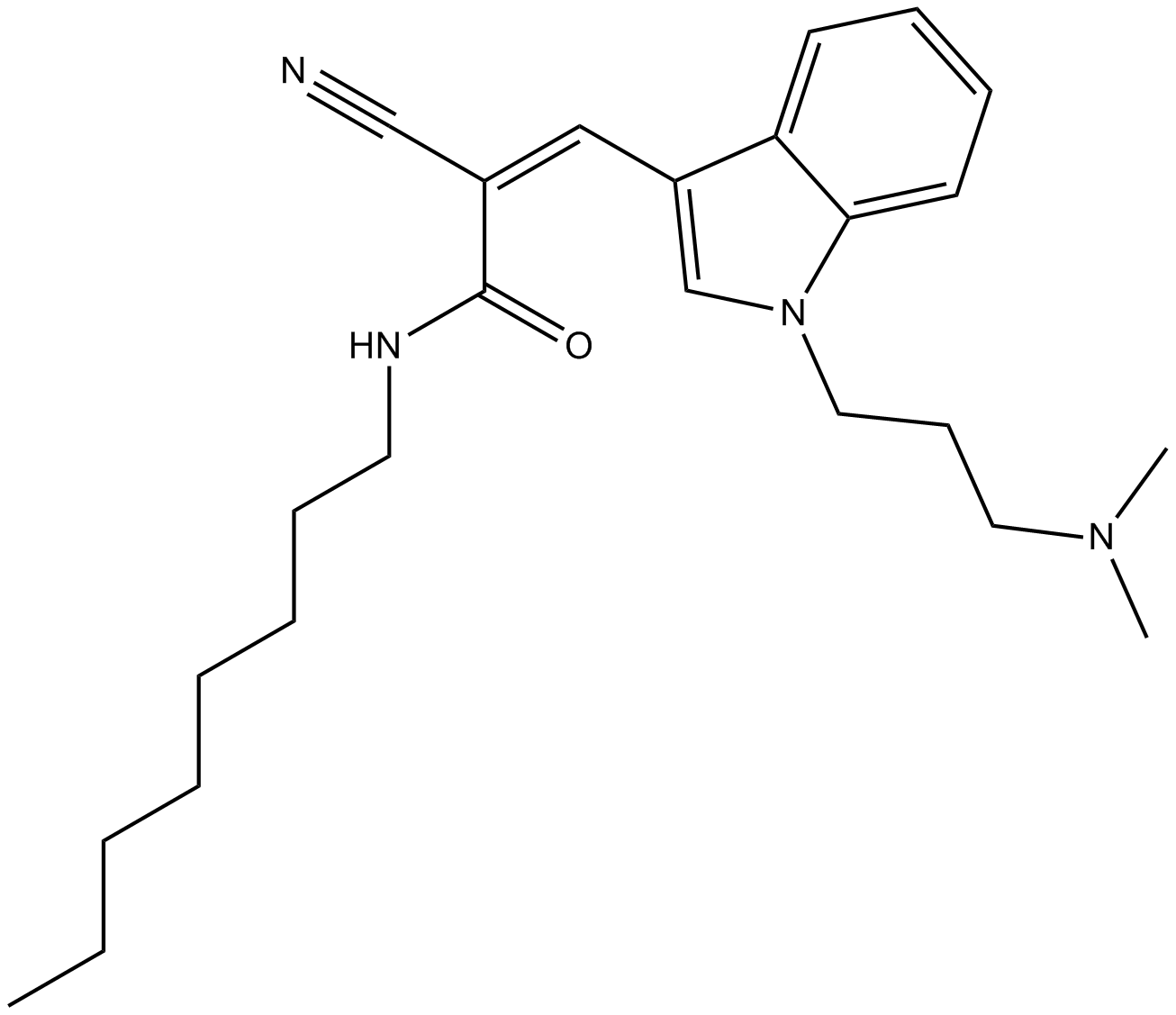
Dynole 34-2
Dynamin I inhibitor
-
GC49089
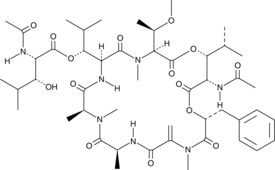
FR900359
A cyclic depsipeptide and an inhibitor of Gαq, Gα11, and Gα14
-
GC18260
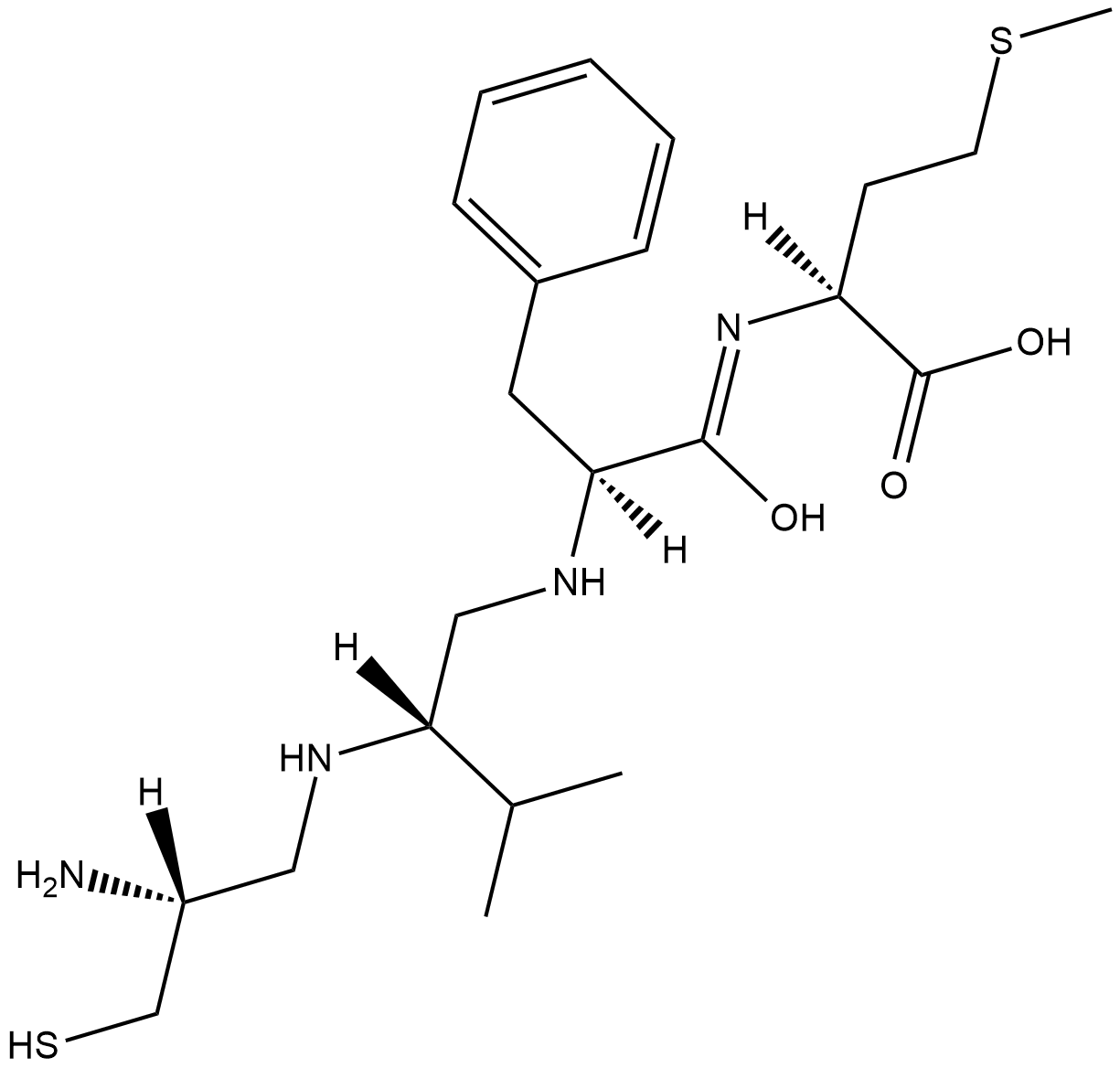
FTase Inhibitor I
FTase inhibitor I is a potent inhibitor of farnesyltransferase (FTase; IC50 = 21 nM) that is greater than 30-fold selective for FTase over geranylgeranyl transferase (GGTase; IC50 = 790 nM).
-
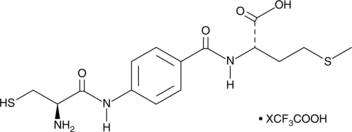
GC52503
FTase Inhibitor II (trifluoroacetate salt)
A potent, selective inhibitor of farnesyltransferase
-
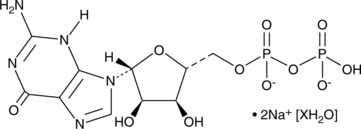
GC49033
Guanosine 5’-diphosphate (sodium salt hydrate)
A purine nucleotide
-
GC49703
Guanosine 5’-[γ-thio]triphosphate (lithium salt)
A hydrolysis-resistant analog of GTP
![Guanosine 5’-[γ-thio]triphosphate (lithium salt) Chemical Structure Guanosine 5’-[γ-thio]triphosphate (lithium salt) Chemical Structure](/media/struct/GC4/GC49703.png)
-
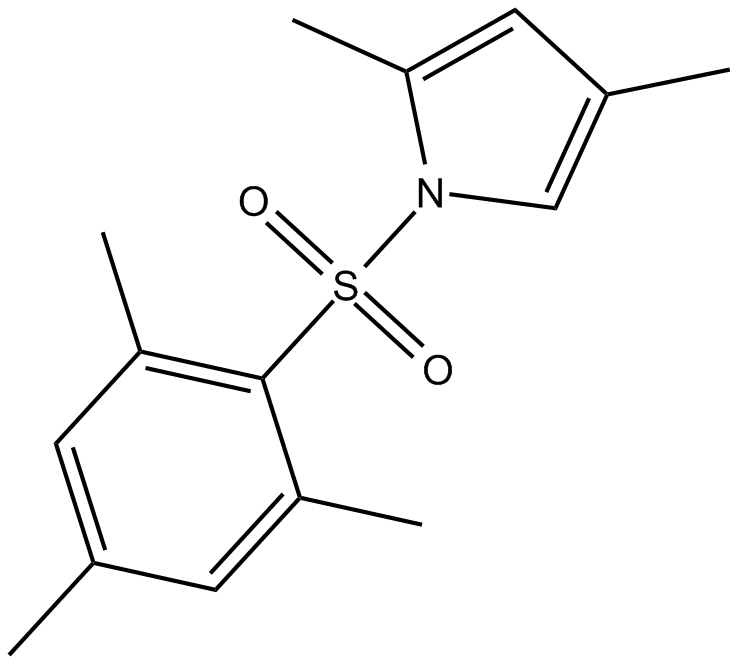
GC17852
Gue 1654
Oxoeicosanoid receptor 1 (OXE-R) modulator
-
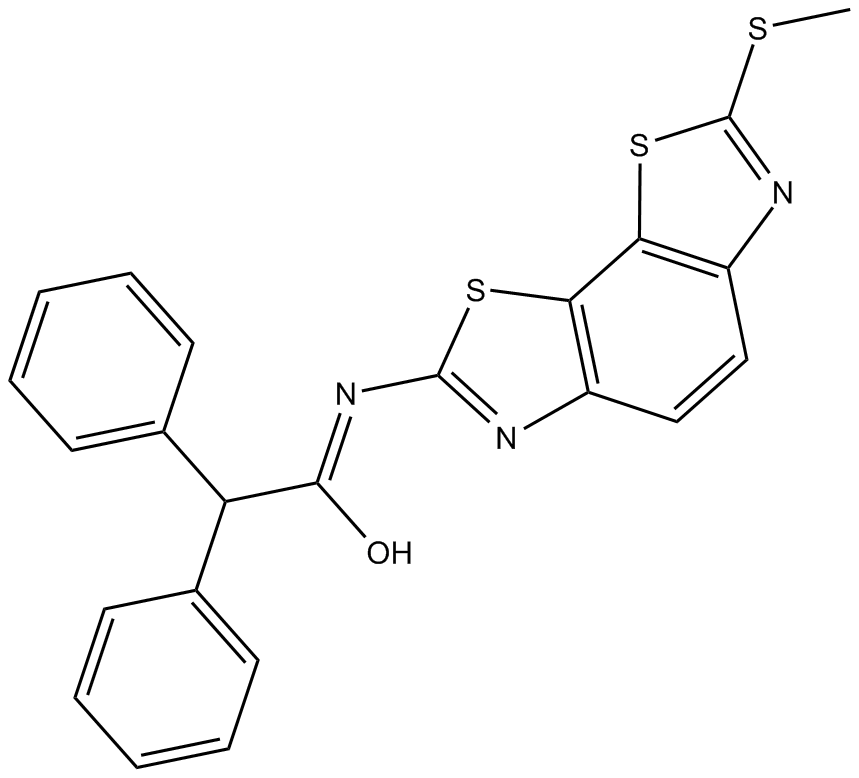
GC15722
HJC 0350
EPAC2 antagonist, potent and selective
-
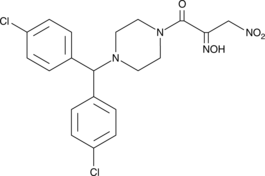
GC47477
JKE-1674
A GPX4 inhibitor and active metabolite of ML-210
-
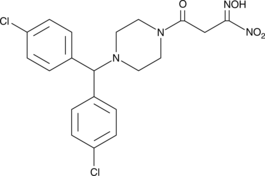
GC47478
JKE-1716
A GPX4 inhibitor
-
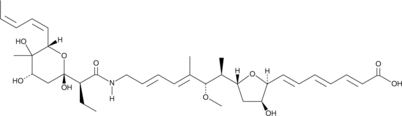
GC44020
L-681,217
L-681,217 is an antibiotic originally isolated from S.
-
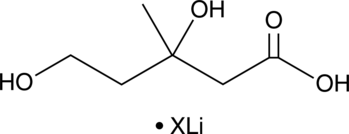
GC52250
Mevalonate (lithium salt)
An intermediate in the mevalonate pathway
-
GC13243
MitMAB
MitMAB, an organic building block, is a cationic surfactant with asymmetrical structure.
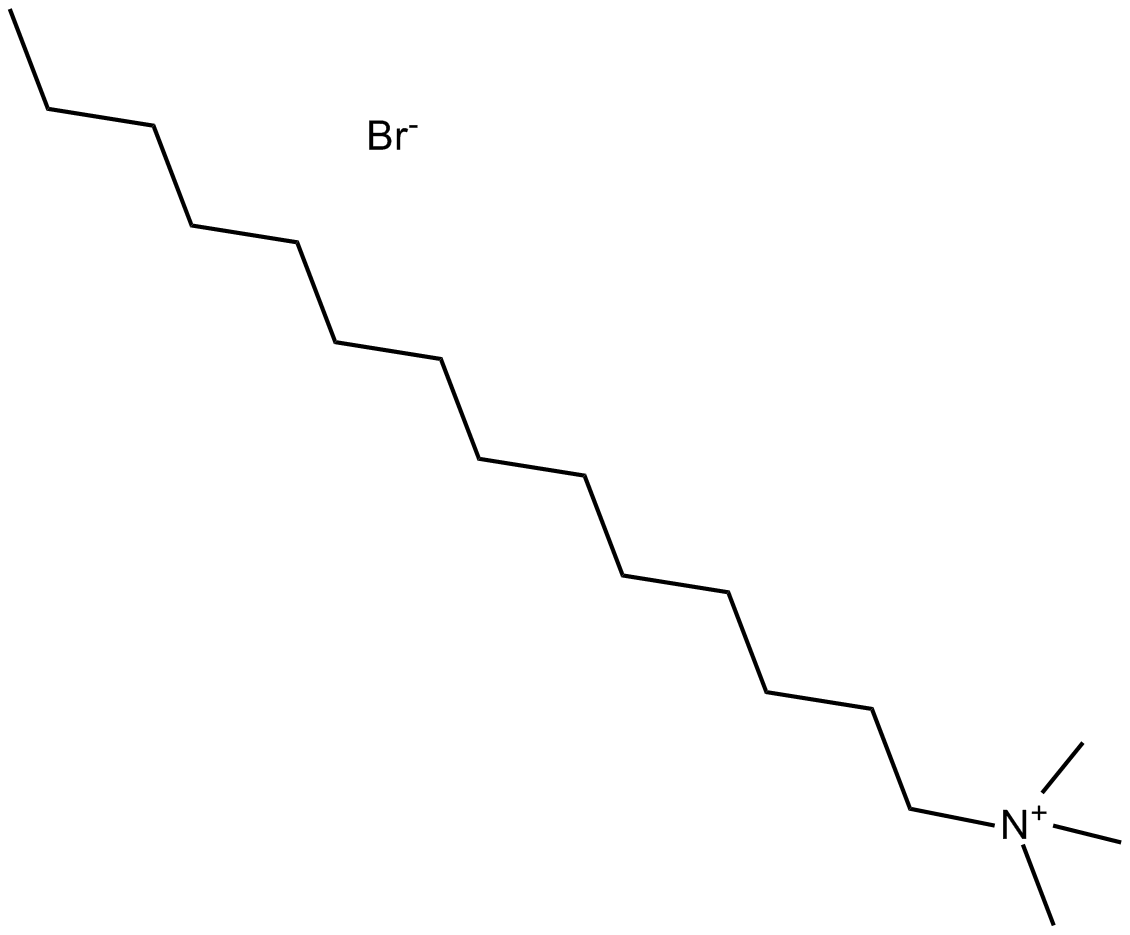
-
GC18445
N-Deacetylcolchicine
An inhibitor of microtubule polymerization
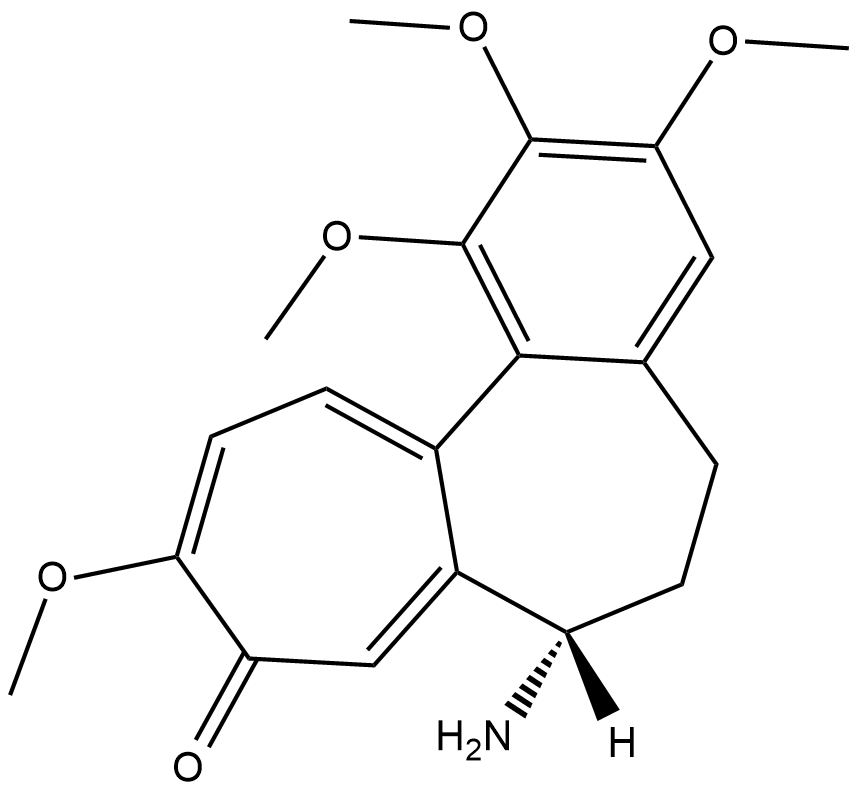
-
GC11880
OctMAB
Dynamin inhibitor
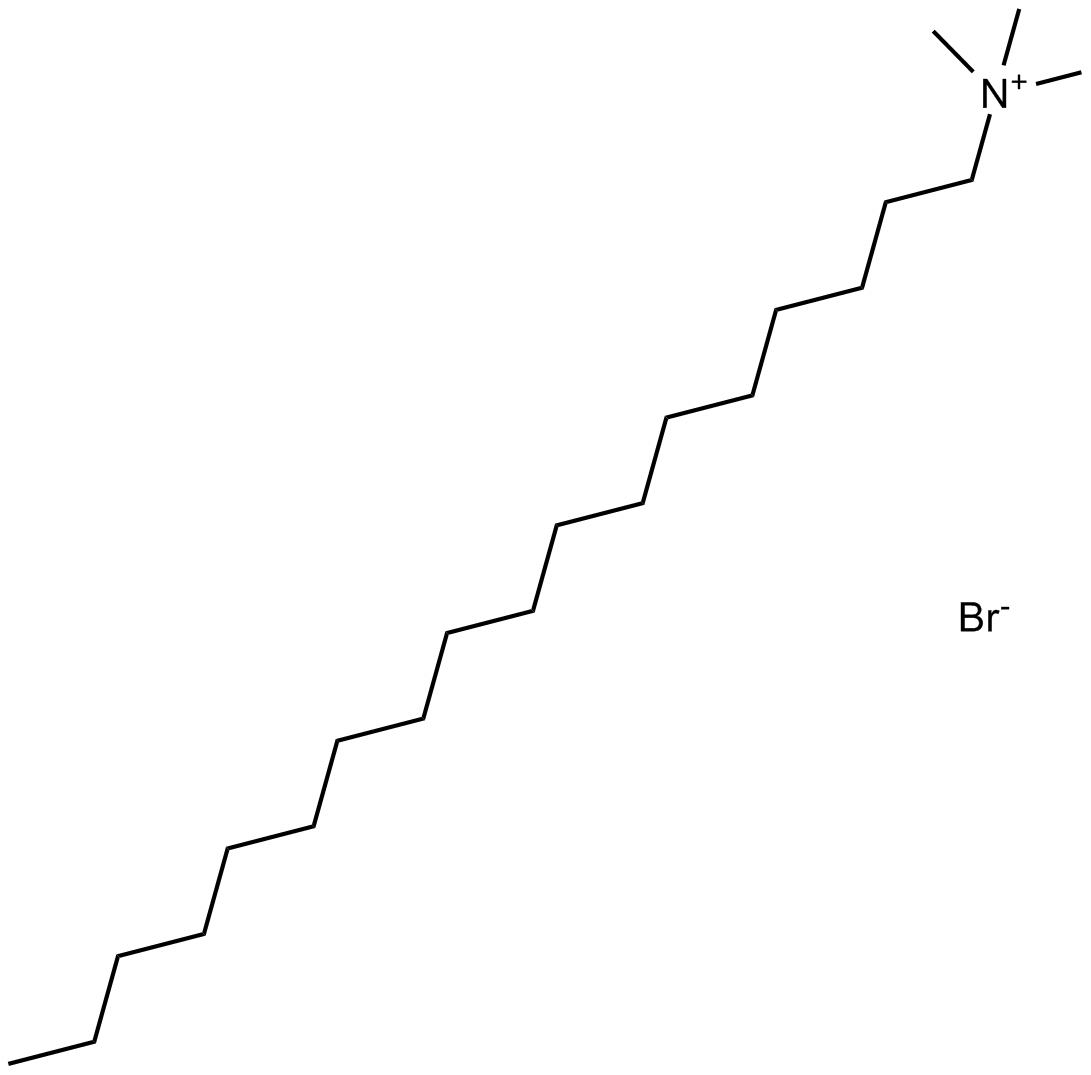
-
GC44506
Oligomycin D
Oligomycin D is a macrolide antibiotic produced by several species of Streptomyces that inhibits the mitochondrial F1FO-ATPase and is used to uncouple oxidative phosphorylation from electron transport.
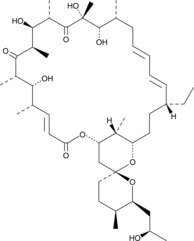
-
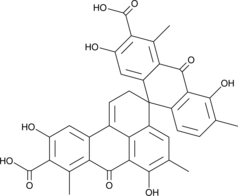
GC48094
Spiro-Oxanthromicin A
A polyketide