Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(15)
- Caspase(77)
- 14.3.3 Proteins(3)
- Apoptosis Inducers(71)
- Bax(15)
- Bcl-2 Family(136)
- Bcl-xL(13)
- c-RET(15)
- IAP(32)
- KEAP1-Nrf2(73)
- MDM2(21)
- p53(137)
- PC-PLC(6)
- PKD(8)
- RasGAP (Ras- P21)(2)
- Survivin(8)
- Thymidylate Synthase(12)
- TNF-α(141)
- Other Apoptosis(1144)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(6)
- PD-1/PD-L1 interaction(60)
- ASK1(4)
- PAR4(2)
- RIP kinase(47)
- FKBP(22)
Products for Apoptosis
- Cat.No. Nombre del producto Información
-
GC47042
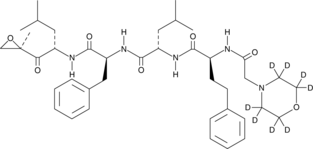
Carfilzomib-d8
Un estándar interno para la cuantificación de carfilzomib.
-
GN10733
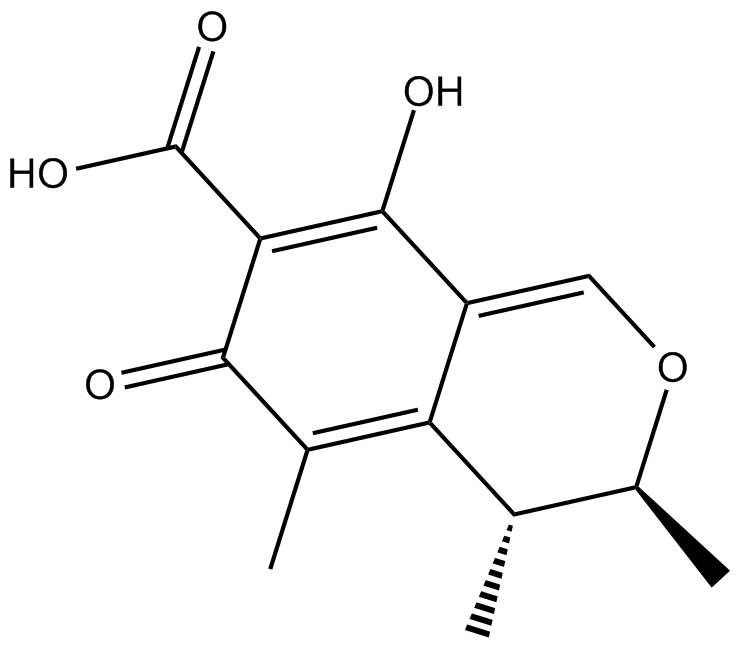
Carnosic acid
-
GC45679
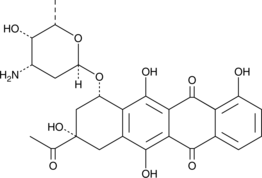
Carubicin
La carubicina (carminomicina) es un compuesto de origen microbiano.
-
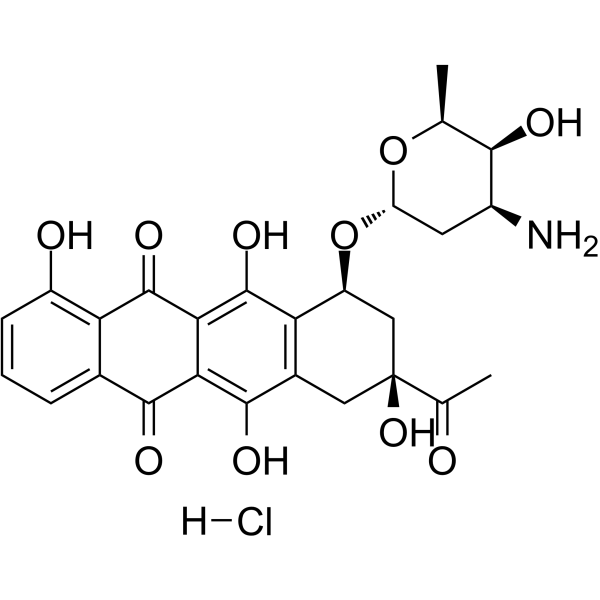
GC64110
Carubicin hydrochloride
El clorhidrato de carubicina es un compuesto derivado de microbios.
-
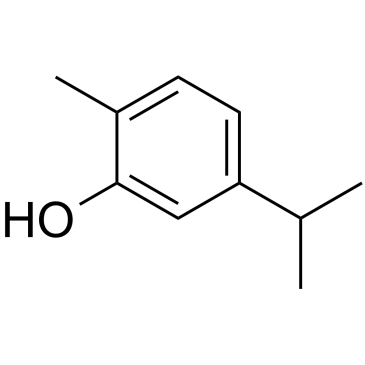
GC35612
Carvacrol
Carvacrol es un fenol monoterpenoide aislado de Thymus mongolicus Ronn.
-
GC62442
Casein Kinase inhibitor A51
El inhibidor de caseÍna quinasa A51 es un inhibidor potente y activo por vÍa oral de la caseÍna quinasa 1α (CK1α). El inhibidor de la caseÍna quinasa A51 induce la apoptosis de las células leucémicas y tiene potentes actividades antileucémicas.
-
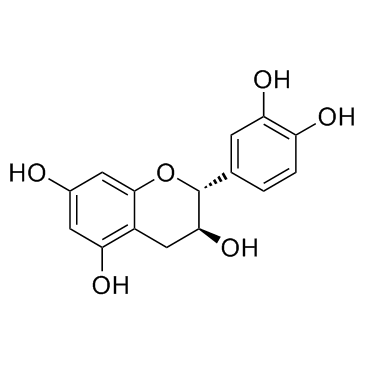
GC32841
Catechin ((+)-Catechin)
La catequina ((+)-catequina) ((+)-catequina ((+)-catequina)) inhibe la ciclooxigenasa-1 (COX-1) con una IC50 de 1,4 μM.
-
GN10543
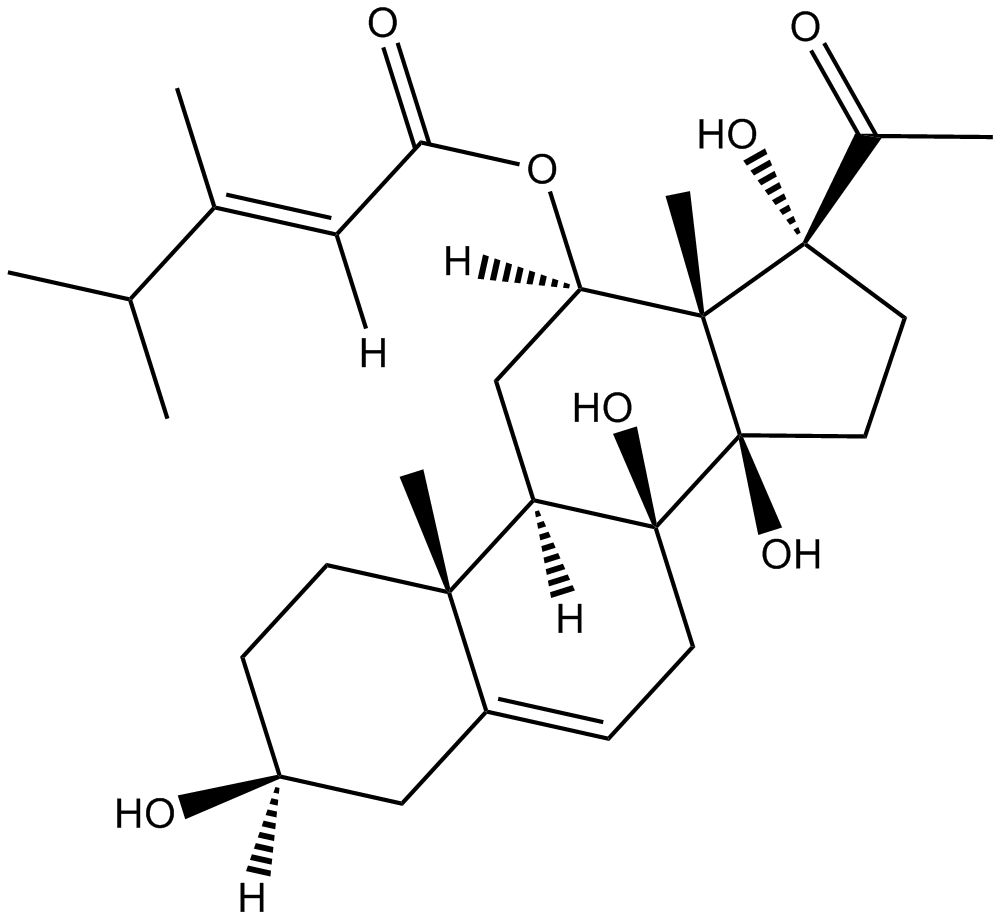
caudatin
-
GC43149
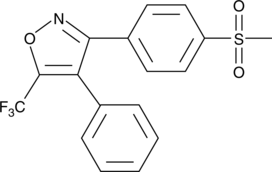
CAY10404
CAY10404 es un inhibidor potente y selectivo de la ciclooxigenasa-2 (COX-2) con una IC50 de 1 nM y un Índice de selectividad (SI; COX-1 IC50/COX-2 IC50) >500000.
-
GC43150
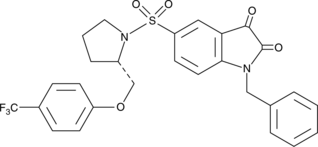
CAY10406
CAY10406 is a trifluoromethyl analog of an isatin sulfonamide compound that selectively inhibits caspases 3 and 7.
-
GC43154
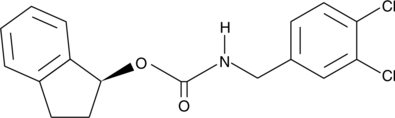
CAY10443
Mitochondrial release of cytochrome c triggers apoptosis via the assembly of a multimeric complex including caspase-9, Apaf-1, and other components, sometimes called the apoptosome.
-
GC43176
CAY10575
CAY10575 (Compuesto 8) es un inhibidor de IKK2 con un IC50 de 0,075 μM.
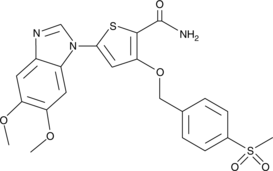
-
GC18530
CAY10616
Resveratrol is a natural polyphenolic antioxidant that has anti-cancer properties.
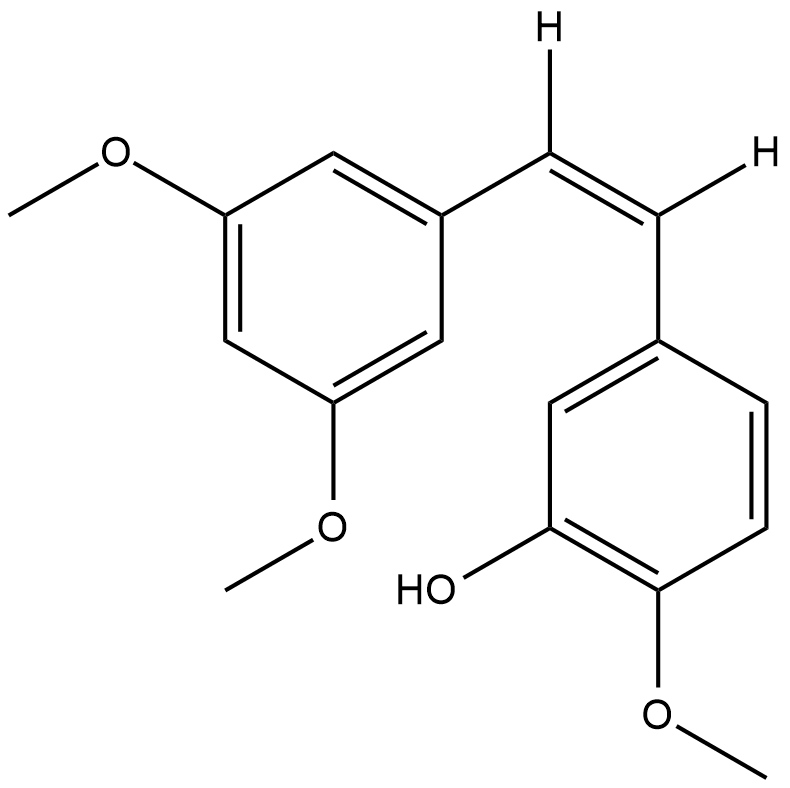
-
GC41317
CAY10625
Survivin is a cellular protein implicated in cell survival by interacting with and inhibiting the apoptotic function of several proteins including Smac/DIABLO, caspase-3, and caspase-7.
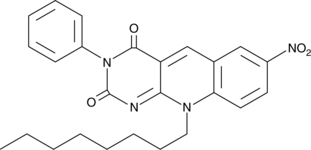
-
GC43189
CAY10681
Inactivation of the tumor suppressor p53 commonly coincides with increased signaling through NF-κB in cancer.
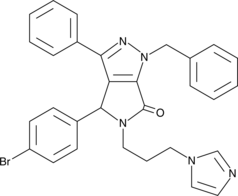
-
GC43190
CAY10682
(±)-Nutlin-3 blocks the interaction of p53 with its negative regulator Mdm2 (IC50 = 90 nM), inducing the expression of p53-regulated genes and blocking the growth of tumor xenografts in vivo.
-
GC40650
CAY10706
CAY10706 is a ligustrazine-curcumin hybrid that promotes intracellular reactive oxygen species accumulation preferentially in lung cancer cells.
-
GC43198
CAY10717
CAY10717 is a multi-targeted kinase inhibitor that exhibits greater than 40% inhibition of 34 of 104 kinases in an enzymatic assay at a concentration of 100 nM.
-
GC43203
CAY10726
CAY10726 is an arylurea fatty acid.
-
GC46113
CAY10744
A topoisomerase II-α poison
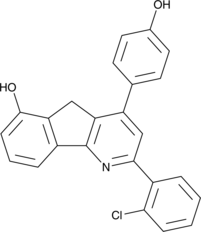
-
GC47053
CAY10746
A ROCK1 and ROCK2 inhibitor
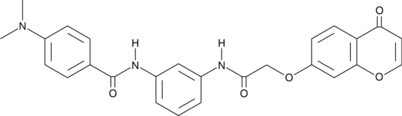
-
GC48392
CAY10747
An inhibitor of the Hsp90-Cdc37 protein-protein interaction
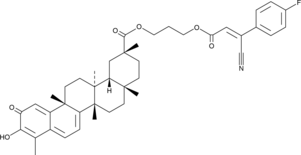
-
GC47055
CAY10749
CAY10749 (Compuesto 15) es un potente inhibidor PARP/PI3K con valores PIC50 de 8.22, 8.44, 8.25, 6.54, 8.13, 6.08 para PARP-1, PARP-2, PI3K⊵ ;, PI3K⋲ offlineefficient_models_2022q2.md.en_es_2021q4.mdCAY10749 is a highly effective anticancer compound targeted against a wide range of oncologic diseases.en_es_2021q4.md
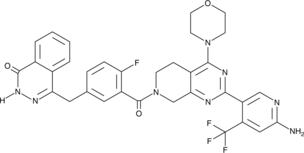
-
GC47057
CAY10755
A fungal metabolite with anticancer activity
-
GC47061
CAY10763
A dual inhibitor of IDO1 and STAT3 activation
-
GC47065
CAY10773
A derivative of sorafenib
-
GC49080
CAY10786
CAY10786 (Compuesto 43) es un antagonista de GPR52 con un IC50 de 0,63 μM.
-
GC52245
CAY10792
An anticancer agent
-
GC14634
CBL0137
curaxin that activates p53 and inhibits NF-κB
-
GC15394
CBL0137 (hydrochloride)
CBL0137 (clorhidrato) es un inhibidor de la histona chaperona, FACT. CBL0137 (clorhidrato) también puede activar p53 e inhibe NF-κB con EC50 de 0,37 y 0,47 µM, respectivamente.
-
GC61636
CBR-470-2
CBR-470-2, un anÁlogo sustituido con glicina, puede activar la seÑalizaciÓn de NRF2.
-
GC13648
CC-223
CC-223 (CC-223) es un inhibidor potente, selectivo y biodisponible por vÍa oral de la cinasa mTOR, con un valor IC50 para la cinasa mTOR de 16 nM. CC-223 inhibe tanto mTORC1 como mTORC2.
-
GC39169
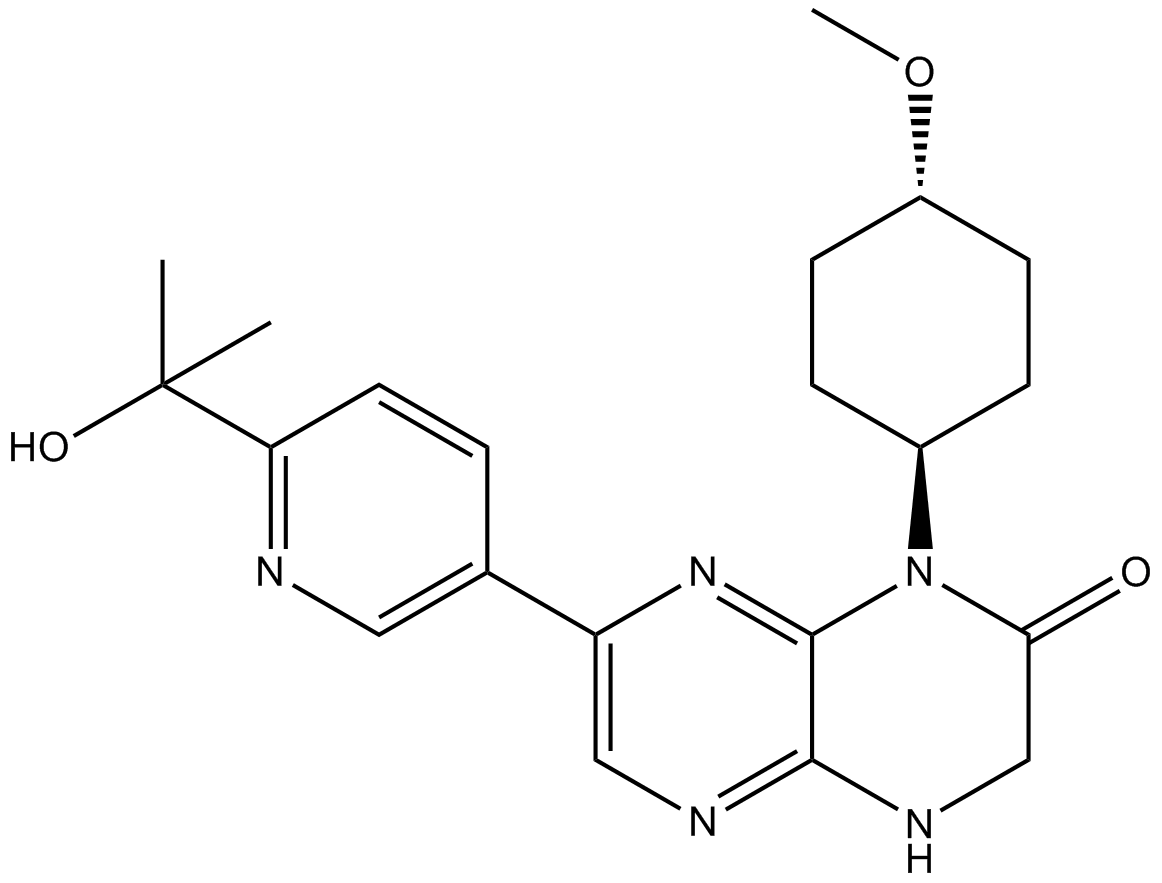
CC-92480
CC-92480 (CC-92480), un fÁrmaco modulador de ubiquitina ligasa de cereblon E3 (CELMoD), actÚa como un pegamento molecular. CC-92480 muestra una alta afinidad por cereblon, lo que resulta en una potente actividad antimieloma.
-
GC19088
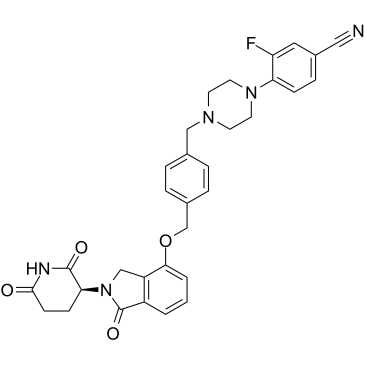
CC122
CC122 (CC 122) es un modulador de cerebro activo por vÍa oral.
-
GC61532
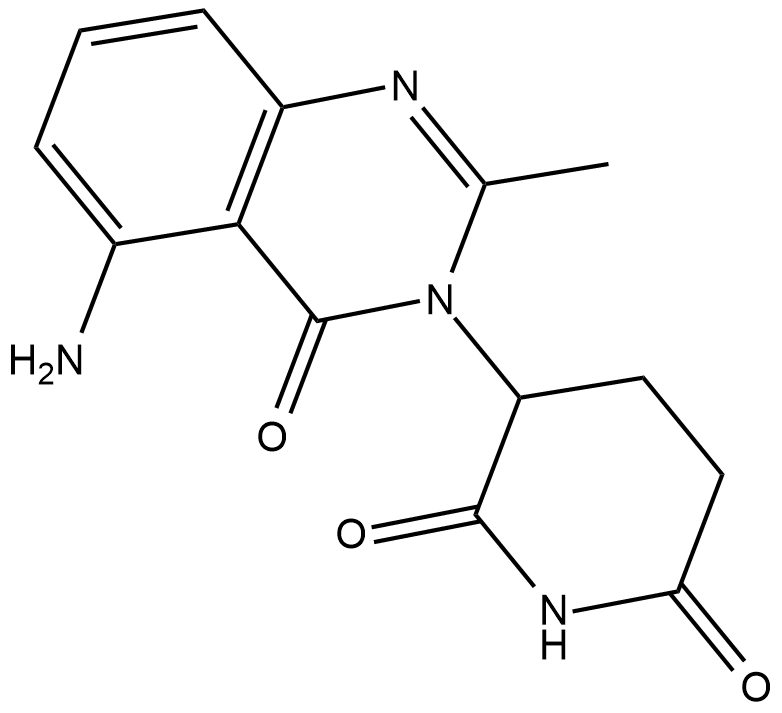
CCI-007
CCI-007 es una molécula pequeÑa con actividad citotÓxica contra la leucemia infantil con reordenamientos de MLL, con valores de IC50 de 2,5-6,2 μM en células sensibles.
-
GC12891
CCT007093
CCT007093 es un inhibidor eficaz de la proteÍna fosfatasa 1D (PPM1D Wip1). La inhibiciÓn de Wip1 puede activar la vÍa mTORC1 y mejorar la proliferaciÓn de hepatocitos después de la hepatectomÍa.
-
GC14566
CCT137690
An inhibitor of Aurora kinases and FLT3
-
GC62561
CCT369260
CCT369260 (compuesto 1) es un inhibidor del linfoma 6 de células B activo por vÍa oral (BCL6) con actividad antitumoral. CCT369260 (compuesto 1) exhibe un IC50 de 520 nM.
-
GC33337
CDC801
CDC801 es una fosfodiesterasa 4 (PDE4) potente y activa por vÍa oral y factor de necrosis tumoral-α (TNF-α) inhibidor con IC50 de 1,1 μM y 2,5 μM, respectivamente.
-
GC39555
CDDO-2P-Im
CDDO-2P-Im es un anÁlogo de CDDO-imidazolida con efecto quimiopreventivo. CDDO-2P-Im puede reducir el tamaÑo y la gravedad de los tumores de pulmÓn en el modelo de cÁncer de pulmÓn de ratÓn.
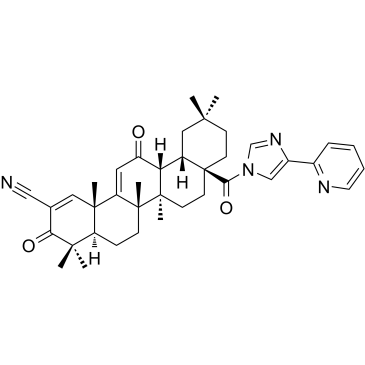
-
GC39556
CDDO-3P-Im
CDDO-3P-Im es un anÁlogo de CDDO-imidazolida con efecto quimiopreventivo. CDDO-3P-Im puede reducir el tamaÑo y la gravedad de los tumores de pulmÓn en el modelo de cÁncer de pulmÓn de ratÓn. CDDO-3P-Im es un inhibidor de necroptosis activo por vÍa oral que se puede utilizar para la investigaciÓn de isquemia/reperfusiÓn (I/R).
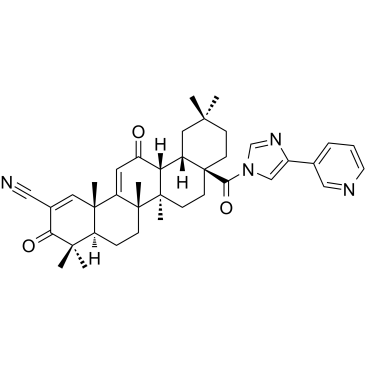
-
GC35629
CDDO-dhTFEA
CDDO-dhTFEA (RTA dh404) es un compuesto triterpenoide de oleanano sintético que activa Nrf2 de forma potente e inhibe el factor de transcripciÓn proinflamatorio NF-κB.
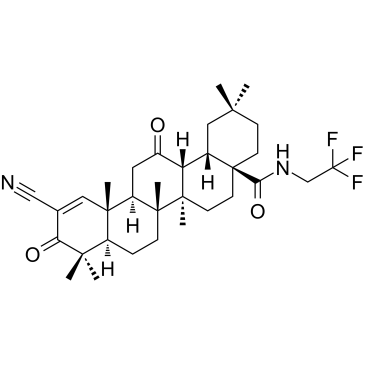
-
GC35630
CDDO-EA
CDDO-EA es un activador del elemento de respuesta antioxidante/factor 2 relacionado con NF-E2 (Nrf2/ARE).

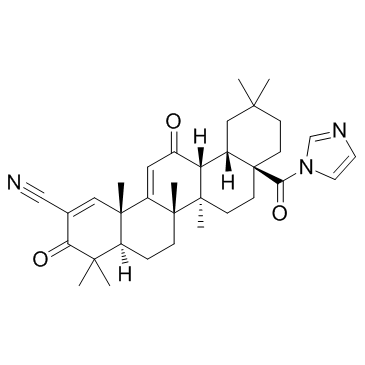
-
GC32723
CDDO-Im (RTA-403)
CDDO-Im (RTA-403) (RTA-403) es un activador de Nrf2 y PPAR, con Kis de 232 y 344 nM para PPARα y PPARγ.
-
GC16625
CDDO-TFEA
Nrf2 activator
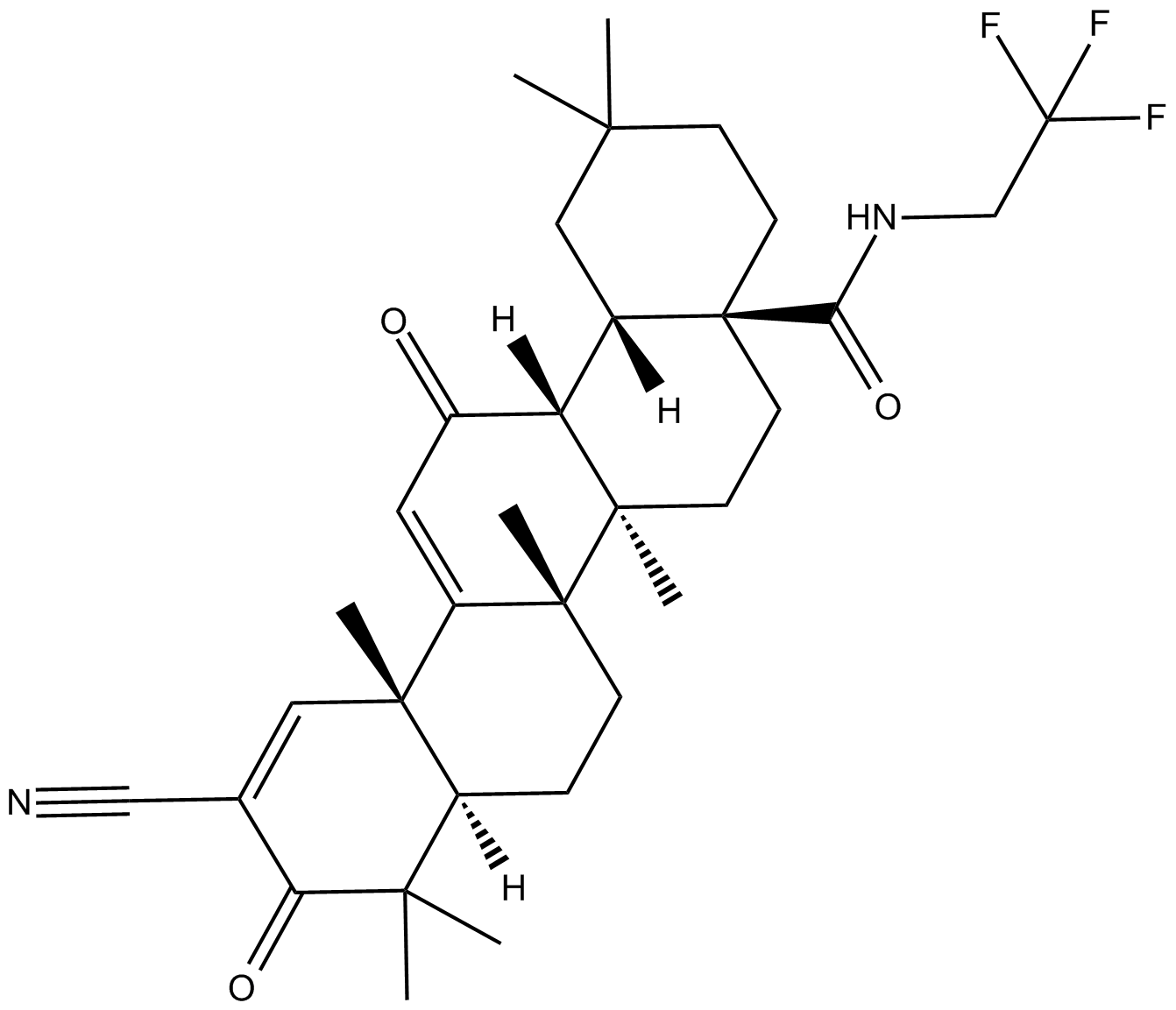
-
GC43217
CDK/CRK Inhibitor
CDK/CRK inhibitor is an inhibitor of cyclin-dependent kinases (CDK) and CDK-related kinases (CRK) with IC50 values ranging from 9-839 nM in vitro.
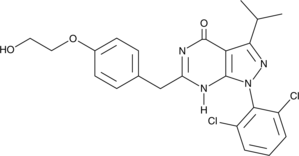
-
GC62596
CDK7-IN-3
CDK7-IN-3 (CDK7-IN-3) es un inhibidor de CDK7 no covalente, altamente selectivo, activo por vÍa oral con una KD de 0,065 nM. CDK7-IN-3 muestra una inhibiciÓn pobre de CDK2 (Ki=2600 nM), CDK9 (Ki=960 nM), CDK12 (Ki=870 nM). CDK7-IN-3 induce apoptosis en células tumorales y tiene actividad antitumoral.
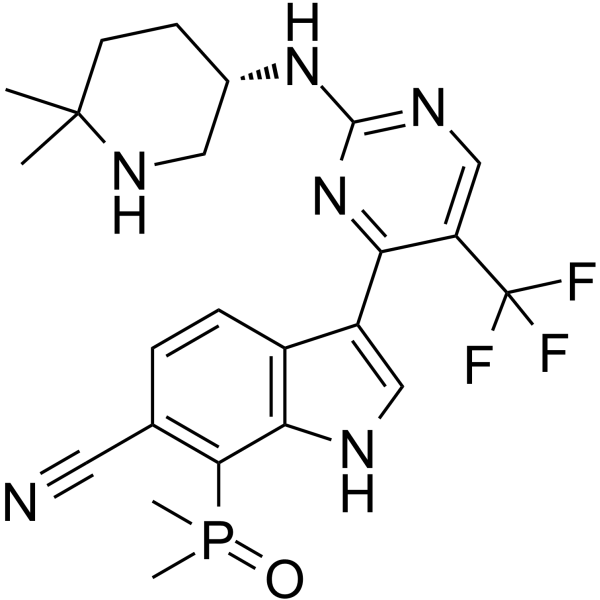
-
GC35636
CDK9-IN-7
CDK9-IN-7 (compuesto 21e) es un inhibidor de CDK9/ciclina T selectivo, muy potente y activo por vÍa oral (IC50=11 nM), que muestra una mayor potencia que otras CDK (CDK4/ciclinaD=148 nM; CDK6/ciclinaD= 145 nM). CDK9-IN-7 muestra actividad antitumoral sin toxicidad evidente. CDK9-IN-7 induce la apoptosis de las células NSCLC, detiene el ciclo celular en la fase G2 y suprime las propiedades de tallo del NSCLC.
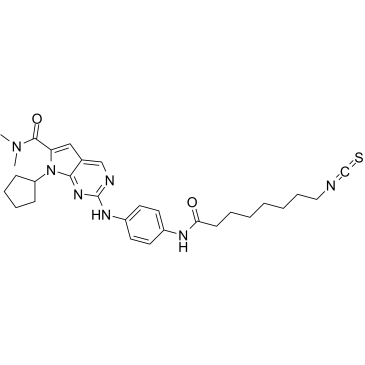
-
GC19096
CDKI-73
CDKI-73 (LS-007) es un inhibidor de CDK9 muy eficaz y activo por vÍa oral, con valores de Ki de 4 nM, 4 nM y 3 nM para CDK9, CDK1 y CDK2, respectivamente.
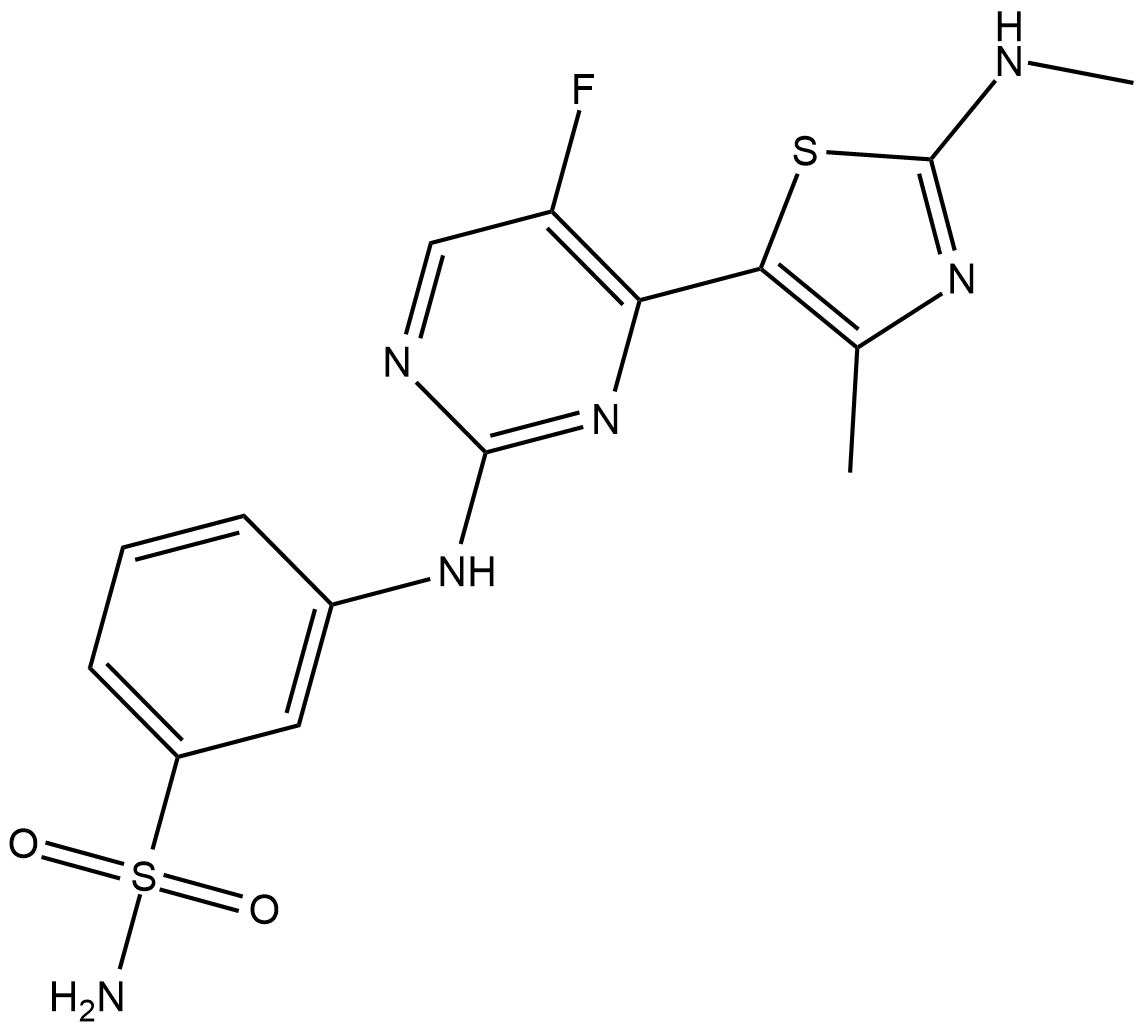
-
GC61865
Cearoin
La cearoÍna aumenta la autofagia y la apoptosis a través de la producciÓn de ROS y la activaciÓn de ERK.
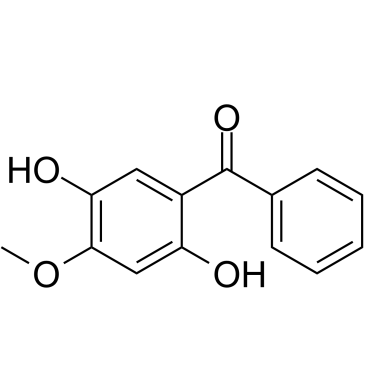
-
GC15083
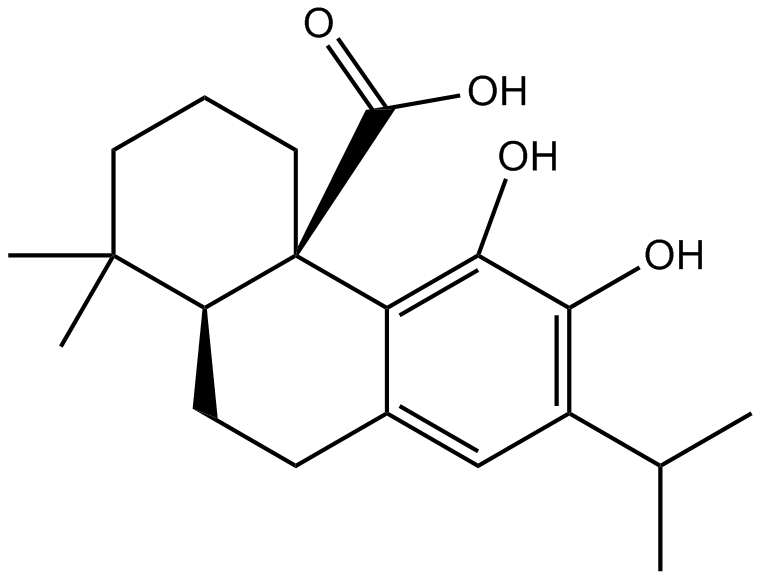
Celastrol
A triterpenoid antioxidant
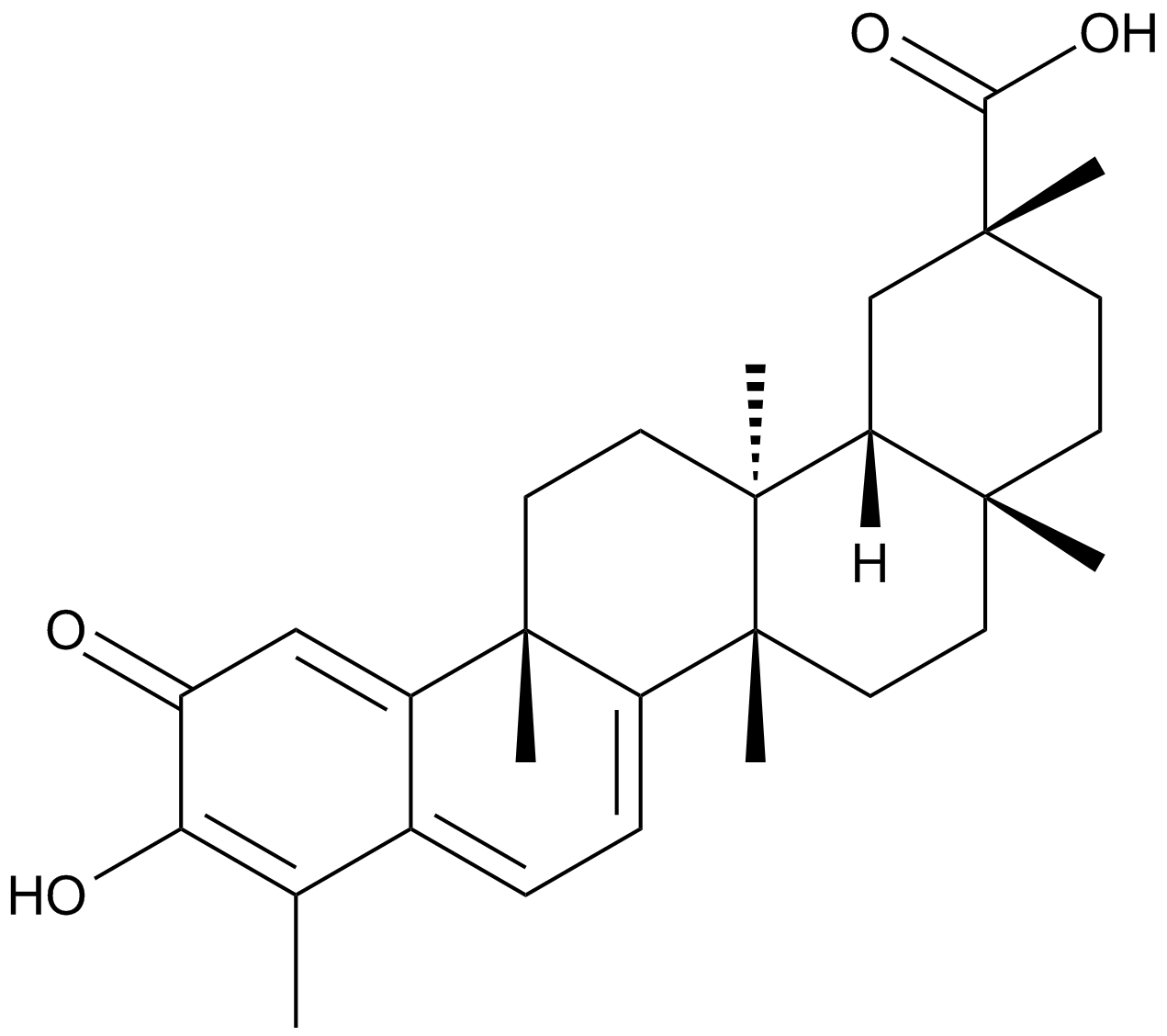
-
GC49152
Celecoxib Carboxylic Acid
An inactive metabolite of celecoxib
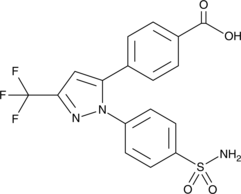
-
GC47070
Celecoxib-d7
An internal standard for the quantification of celecoxib
-
GC18392
Cellocidin
Cellocidin is an antibiotic originally isolated from S.
-
GN10113
Cepharanthine
-
GC52489
Ceramide (hydroxy) (bovine spinal cord)
A sphingolipid
-
GC52485
Ceramide (non-hydroxy) (bovine spinal cord)
A sphingolipid
-
GC52486
Ceramide Phosphoethanolamine (bovine)
A sphingolipid
-
GC43229
Ceramide Phosphoethanolamines (bovine)
Ceramide phosphoethanolamine (CPE) is an analog of sphingomyelin that contains ethanolamine rather than choline as the head group.
-
GC47073
Ceramides (hydroxy)
A mixture of hydroxy fatty acid-containing ceramides
-
GC43230
Ceramides (non-hydroxy)
Ceramides are generated from sphingomyelin through activation of sphingomyelinases or through the de novo synthesis pathway, which requires the coordinated action of serine palmitoyl transferase and ceramide synthase.
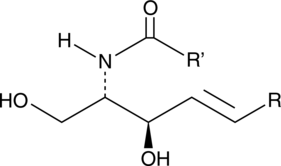
-
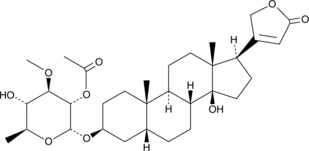
GC49706
Cerberin
A cardiac glycoside with cytotoxic and cardiac modulatory activities
-
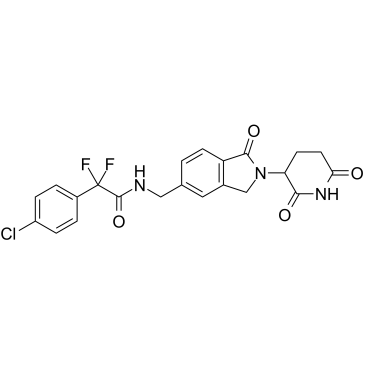
GC60688
Cereblon modulator 1
Cereblon modulator 1 (CC-90009) es un modulador de ligasa E3 de cereblon (CRBN) selectivo para GSPT1, el primero en su clase, que actÚa como un pegamento molecular.
-
GC65487
Certolizumab pegol
Certolizumab pegol (Certolizumab) es un fragmento de anticuerpo monoclonal humanizado recombinante, polietilenglicolilado y de unión a antígenos que se dirige selectivamente y neutraliza el factor de necrosis tumoral-α (TNF-α).
-
GC11543
Cesium chloride
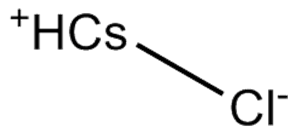
-
GC11710
CFM 4
CFM 4 es un potente antagonista molecular pequeÑo de la uniÓn de CARP-1/APC-2. CFM 4 evita la uniÓn de CARP-1 con APC-2, provoca la detenciÓn del ciclo celular G2M e induce la apoptosis con un intervalo de IC50 de 10-15 μM. CFM 4 también suprime el crecimiento de células de cÁncer de mama humano resistentes a los medicamentos.
-
GC35668
CG-200745
CG-200745 (CG-200745) es un potente inhibidor pan-HDAC activo por vÍa oral que tiene la fracciÓn de Ácido hidroxÁmico para unirse al zinc en la parte inferior de la bolsa catalÍtica. CG-200745 inhibe la desacetilaciÓn de histona H3 y tubulina. CG-200745 induce la acumulaciÓn de p53, promueve la transactivaciÓn dependiente de p53 y mejora la expresiÓn de las proteÍnas MDM2 y p21 (Waf1/Cip1). CG-200745 mejora la sensibilidad de las células resistentes a la gemcitabina a la gemcitabina y al 5-fluorouracilo (5-FU; ). CG-200745 induce apoptosis y tiene efectos antitumorales.
-
GC10666
CGP 57380
CGP 57380 es un compuesto de pirazolo-pirimidina permeable a las células que actÚa como un inhibidor selectivo de Mnk1 con IC50 de 2,2 μM, pero no tiene actividad inhibidora contra p38, JNK1, ERK1/2, PKC o quinasas similares a Src.
-
GC43234
Chaetoglobosin A
La quetoglobosina A, el principio activo del extracto de Penicillium aquamarinium, es miembro de la familia de las citocalasanas.
-
GC18536
Chartreusin
Chartreusin is an antibiotic originally isolated from S.
-
GN10463
Chelerythrine
-
GC13065
Chelerythrine Chloride
Potent inhibitor of PKC and Bcl-xL
-
GC31886
Chelidonic acid
El Ácido quelidÓnico es un componente de Chelidonium majus L.
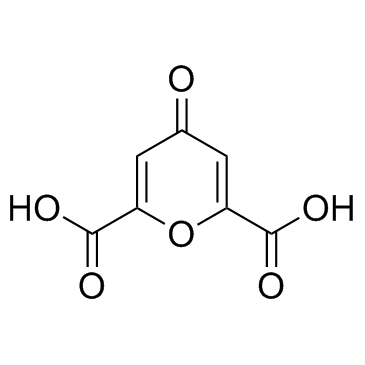
-
GC40878
Chelidonine
Chelidonine es un alcaloide de isoquinolina, se puede aislar de Chelidonium majus L.
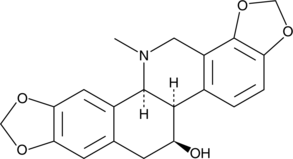
-
GC43236
Chevalone B
Chevalone B is a meroterpenoid originally isolated from the fungus E.
-
GC43237
Chevalone C
Chevalone C, un metabolito fÚngico meroterpenoide, muestra actividad antipalÚdica con un valor IC50 de 25,00 μg/mL.
-
GC64993
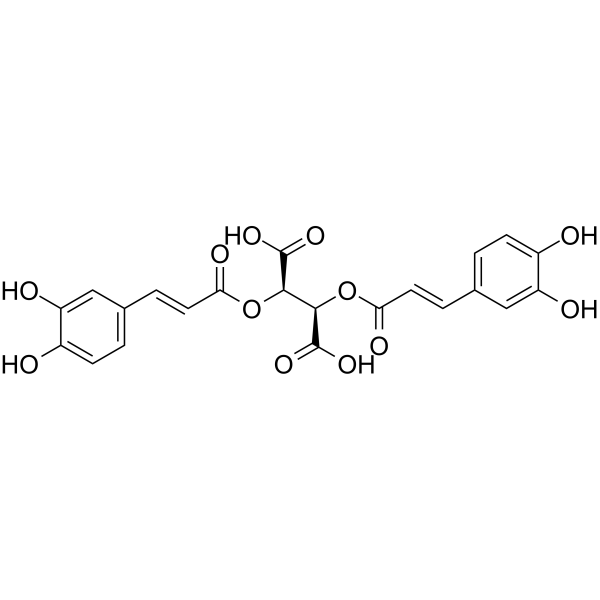
Chicoric acid
El Ácido chicorico (Ácido cicÓrico), un Ácido dicaffeyltartarico activo por vÍa oral, induce la generaciÓn de especies reactivas de oxÍgeno (ROS).
-
GC15739
CHIR-124
CHIR-124 es un inhibidor potente y selectivo de Chk1 con IC50 de 0,3 nM, y también se dirige potentemente a PDGFR y FLT3 con IC50 de 6,6 nM y 5,8 nM.
-
GC43239
Chk2 Inhibitor
El inhibidor de Chk2 (compuesto 1) es un inhibidor potente y selectivo de la quinasa de punto de control 2 (Chk2), con IC50 de 13,5 nM y 220,4 nM para Chk2 y Chk1, respectivamente. El inhibidor de Chk2 puede provocar un fuerte efecto de radioprotecciÓn mediado por Chk2 dependiente de ataxia telangiectasia mutada (ATM).
-
GC45717
Chlamydocin
La clamidocina, un metabolito fÚngico, es un inhibidor de HDAC muy potente, con una IC50 de 1,3 nM. La clamidocina exhibe potentes actividades antiproliferativas y anticancerÍgenas. La clamidocina induce la apoptosis al activar la caspasa-3.
-
GC17969
CHM 1
An inhibitor of tubulin polymerization
-
GC35682
CHMFL-ABL/KIT-155
CHMFL-ABL/KIT-155 (CHMFL-ABL-KIT-155; compuesto 34) es un inhibidor dual de la cinasa ABL/c-KIT de tipo II altamente potente y activo por vÍa oral (CI50 de 46 nM y 75 nM, respectivamente), y también presenta actividades inhibitorias significativas para BLK (IC50=81 nM), CSF1R (IC50=227 nM), DDR1 (IC50=116 nM), DDR2 (IC50=325 nM), LCK (IC50=12 nM) y PDGFRβ (IC50= 80 nM) quinasas. CHMFL-ABL/KIT-155 (CHMFL-ABL-KIT-155) detiene la progresiÓn del ciclo celular e induce la apoptosis.
-
GC64028
Chrysosplenol D
El crisosplenol D es un metoxiflavonoide que induce la apoptosis mediada por ERK1/2 en células de cÁncer de mama humano triple negativas.
-
GC13408
CI994 (Tacedinaline)
An inhibitor of HDAC1, -2, and -3
-
GC13589
CID 755673
CID 755673 es un potente inhibidor de PKD con IC50 de 182 nM, 280 nM y 227 nM para PKD1, PKD2 y PKD3, respectivamente.
-
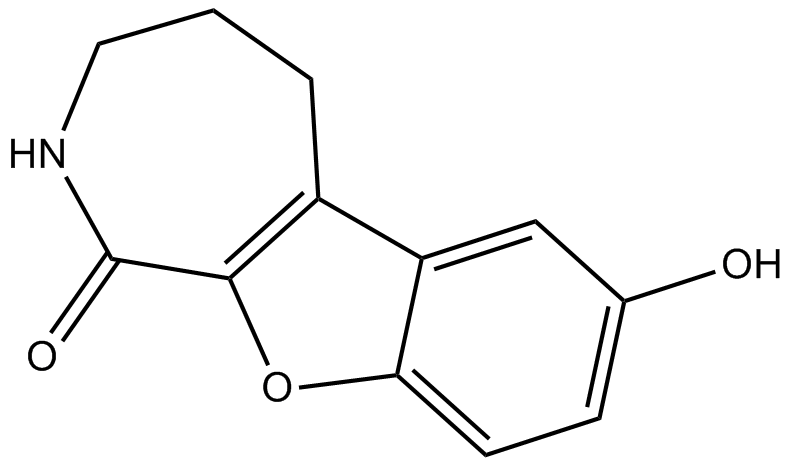
GC19436
CID-5721353
CID-5721353 es un inhibidor de BCL6 con un valor IC50 de 212 μM, lo que corresponde a una Ki de 147 μM.
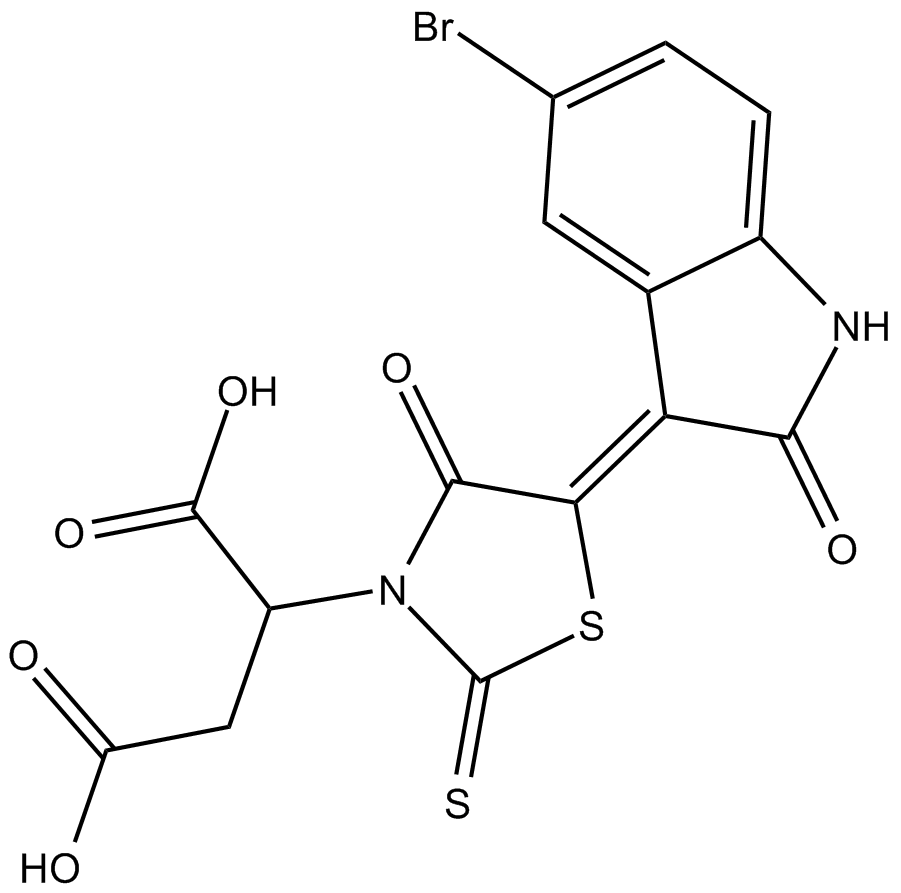
-
GC32997
Cinchonine ((8R,9S)-Cinchonine)
Cinchonine ((8R,9S)-Cinchonine) es un compuesto natural presente en la corteza de Cinchona. La cinconina ((8R,9S)-cinchonina) activa la apoptosis inducida por estrés del retÍculo endoplÁsmico en células de cÁncer de hÍgado humano.
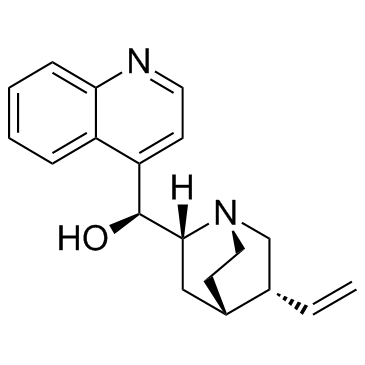
-
GC60708
Cinchonine hydrochloride
El clorhidrato de cinconina ((8R,9S)-clorhidrato de cinconina) es un alcaloide natural presente en la corteza de cinconina, con actividad antipalÚdica. El clorhidrato de cinconina activa la apoptosis inducida por estrés del retÍculo endoplÁsmico (ER) en células de cÁncer de hÍgado humano.
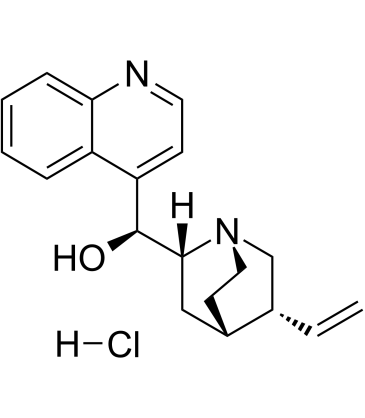
-
GC52269
Cinnabarinic Acid-d4
An internal standard for the quantification of cinnabarinic acid
-
GC40986
Cinnamamide
Cinnamamide is an amide form of of trans-cinnamic acid and a metabolite of Streptomyces.
-
GN10189
Cinobufagin
-
GC11908
Cisplatin
Cisplatin actualmente es uno de los mejores y primeros medicamentos de quimioterapia que base de metales, y se han utilizado ampliamente en cáncer sólido, como dídimo, vejiga, pulmón, cuello uterino, cabeza y cuello, estómago, etc.

-
GC17491
CITCO
CITCO, un derivado de imidazotiazol, es un agonista selectivo del receptor de androstano constitutivo (CAR). CITCO inhibe el crecimiento y la expansiÓn de las células madre de tumores cerebrales (BTSC) y tiene una EC50 de 49nM sobre el receptor X de pregnano (PXR) y no tiene actividad sobre otros receptores nucleares.
-
GC35703
Citicoline
La citicolina (citidina difosfato-colina) es un intermediario en la sÍntesis de fosfatidilcolina, un componente de las membranas celulares.
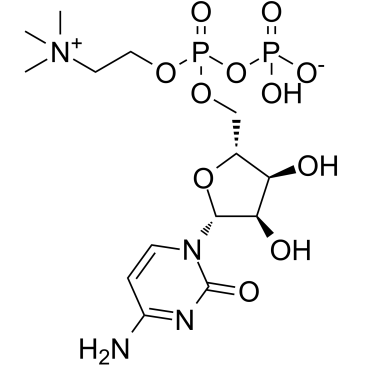
-
GC31186
Citicoline sodium salt
El citicolina sal sódica es un intermediario en la síntesis de fosfatidilcolina, que es un componente de las membranas celulares y también ejerce efectos neuroprotectores.
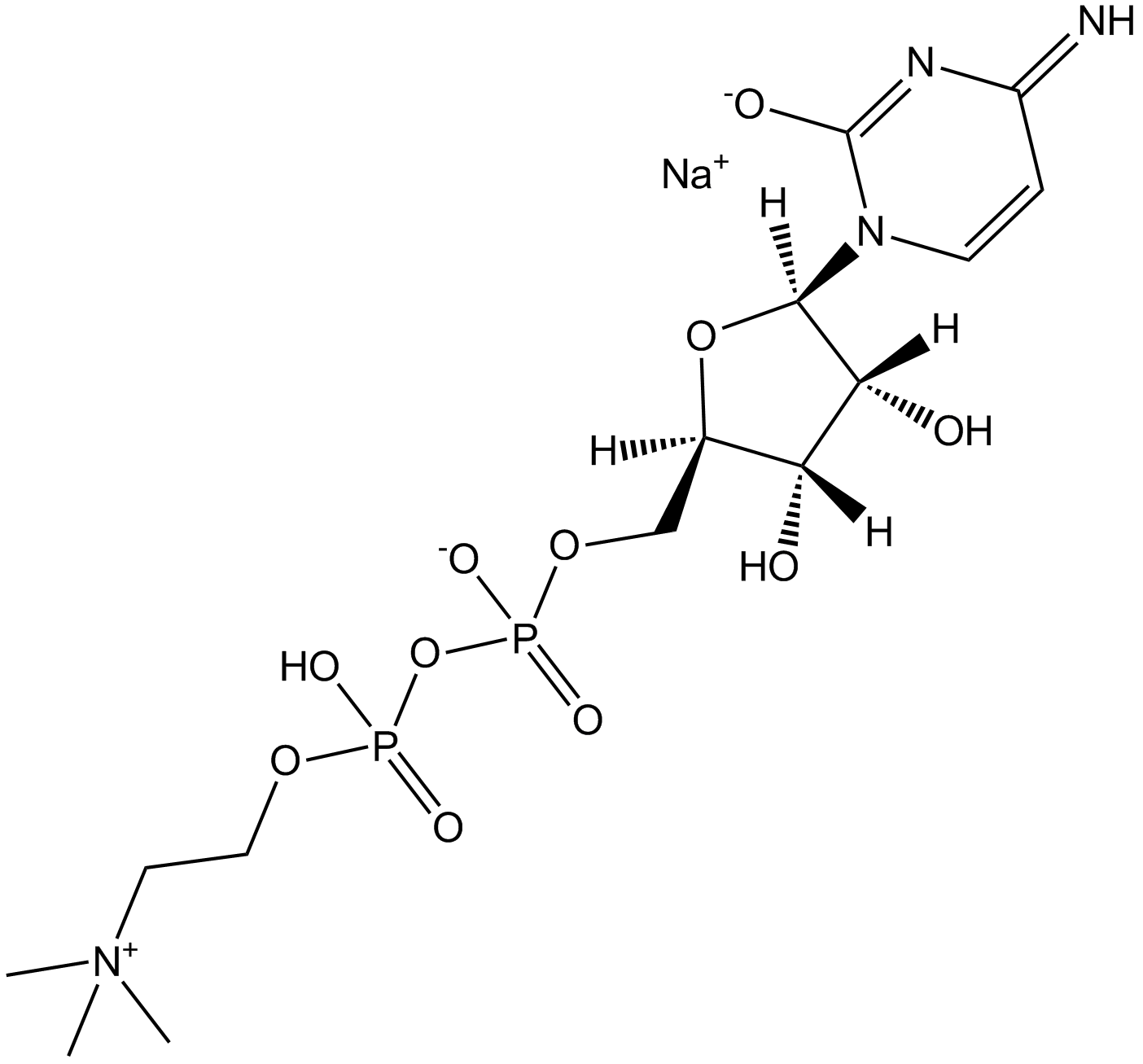
-
GC43273
Citreoindole
Citreoindole is a diketopiperazine metabolite isolated from a hybrid cell fusion of two strains of P.
-
GC41514
Citreoviridin
Citreoviridina, una toxina de Penicillium citreoviride NRRL 2579, inhibe la Na+/K+-ATPasa sinaptosÓmica cerebral mientras que en los microsomas, tanto la Na+/K+-ATPasa como la Mg2+-ATPasa son significativamente estimulados de una manera dependiente de la dosis.
-
GC14203
Citric acid
El Ácido cÍtrico es un conservante natural y potenciador de la acidez de los alimentos.
-
GC68051
Citric acid-d4

-
GC16661
Citrinin
Un micotoxina que induce apoptosis.