PARP
Poly (ADP-ribose) polymerases (PARPs) is a large family of proteins with a conserved catalytic domain that catalyze an immediate DNA-damage-dependent post-translational modification of histones and other nuclear proteins leading to the survival of injured proliferating cells. So far, a total number of 18 human PARP proteins encoded by different genes have been identified, including PARP-1 to PARP-4, PARP-5a, PARP-5b, PARP-5c and PARP-6 to PARP-16. The general structural of PARP proteins has been revealed through the extensive study of the founding family member PARP-1, which is characterized by the presence of four functional domains, including a DNA-binding domain, a caspase-cleaved domain, an automodification domain and a catalytic domain.
Products for PARP
- Cat.No. Product Name Information
-
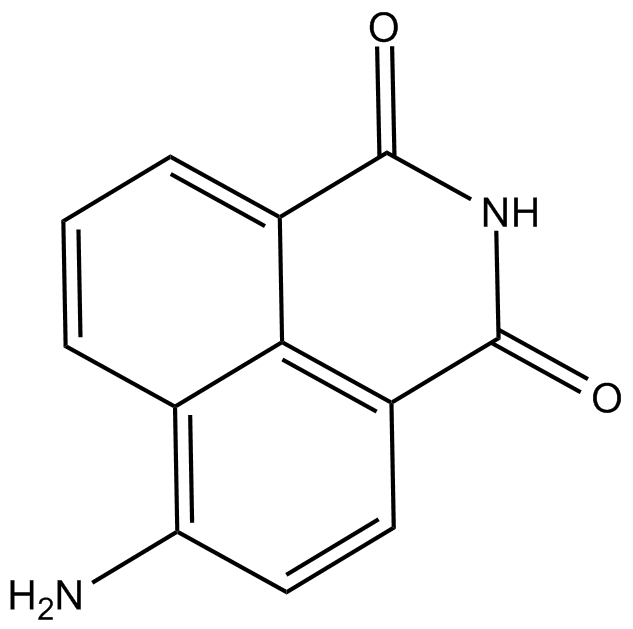
GC11761
4-amino-1,8-Naphthalimide
4-amino-1,8-Naphthalimide is a potent PARP inhibitor and potentiates the cytotoxicity of γ-radiation in cancer cells.
-
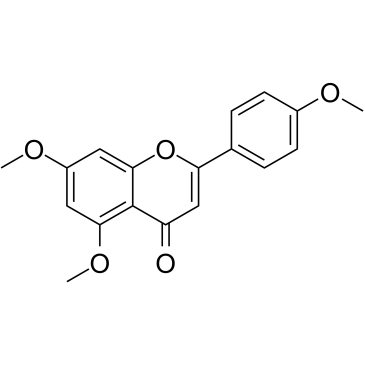
GC35150
5,7,4'-Trimethoxyflavone
5,7,4'-Trimethoxyflavone is isolated from Kaempferia parviflora (KP) that is a famous medicinal plant from Thailand. 5,7,4'-Trimethoxyflavone induces apoptosis, as evidenced by increments of sub-G1 phase, DNA fragmentation, annexin-V/PI staining, the Bax/Bcl-xL ratio, proteolytic activation of caspase-3, and degradation of poly (ADP-ribose) polymerase (PARP) protein.5,7,4'-Trimethoxyflavone is significantly effective at inhibiting proliferation of SNU-16 human gastric cancer cells in a concentration dependent manner.
-
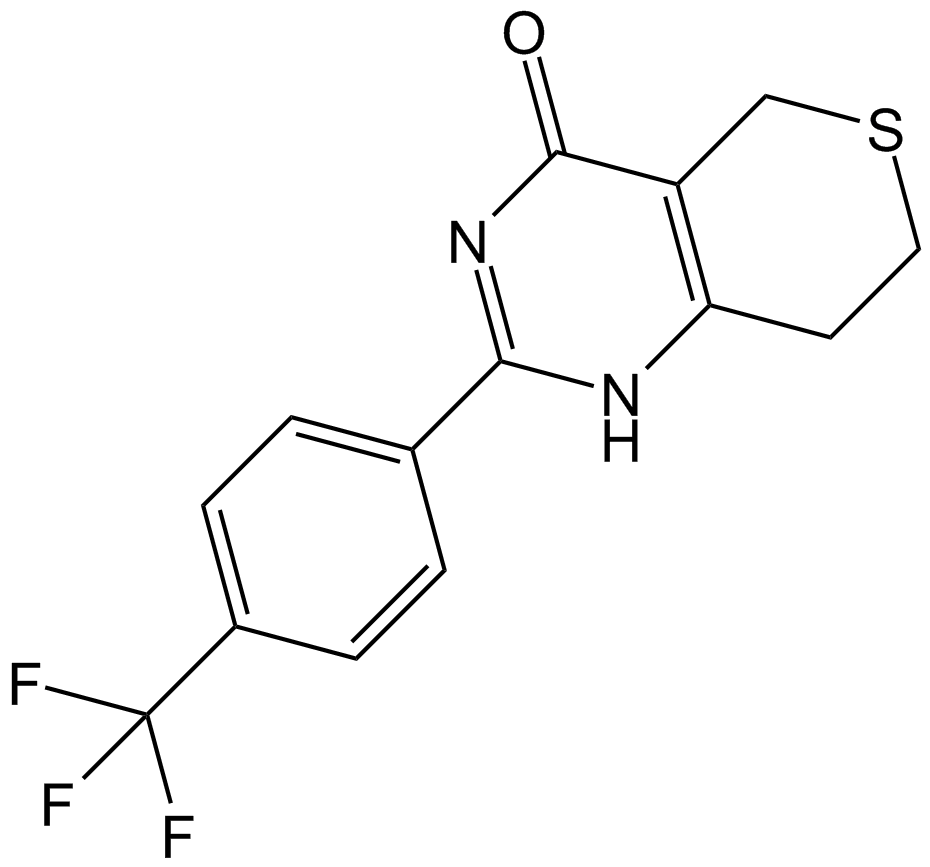
GC68161
5-AIQ
-
GC65899
AZ3391
AZ3391 is a potent inhibitor of PARP. AZ3391 is a quinoxaline derivative. PARP family of enzymes play an important role in a number of cellular processes, such as replication, recombination, chromatin remodeling, and DNA damage repair. AZ3391 has the potential for the research of diseases and conditions occurring in tissues in the central nervous system, such as the brain and spinal cord (extracted from patent WO2021260092A1, compound 23).
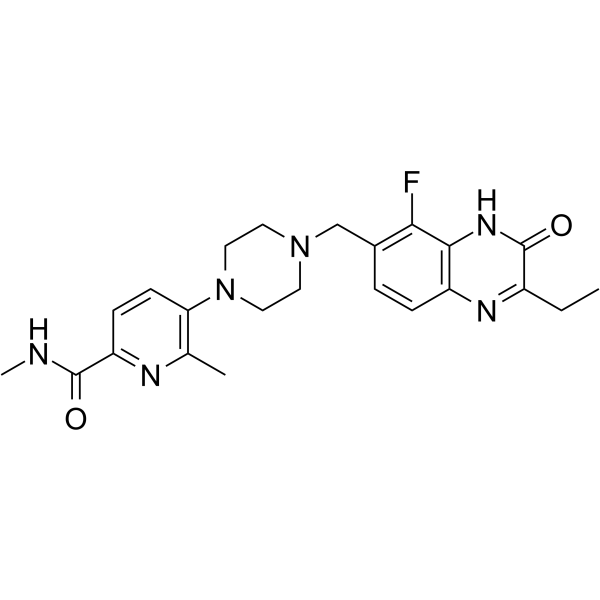
-
GC16725
AZ6102
TNKS1/2 inhibitor
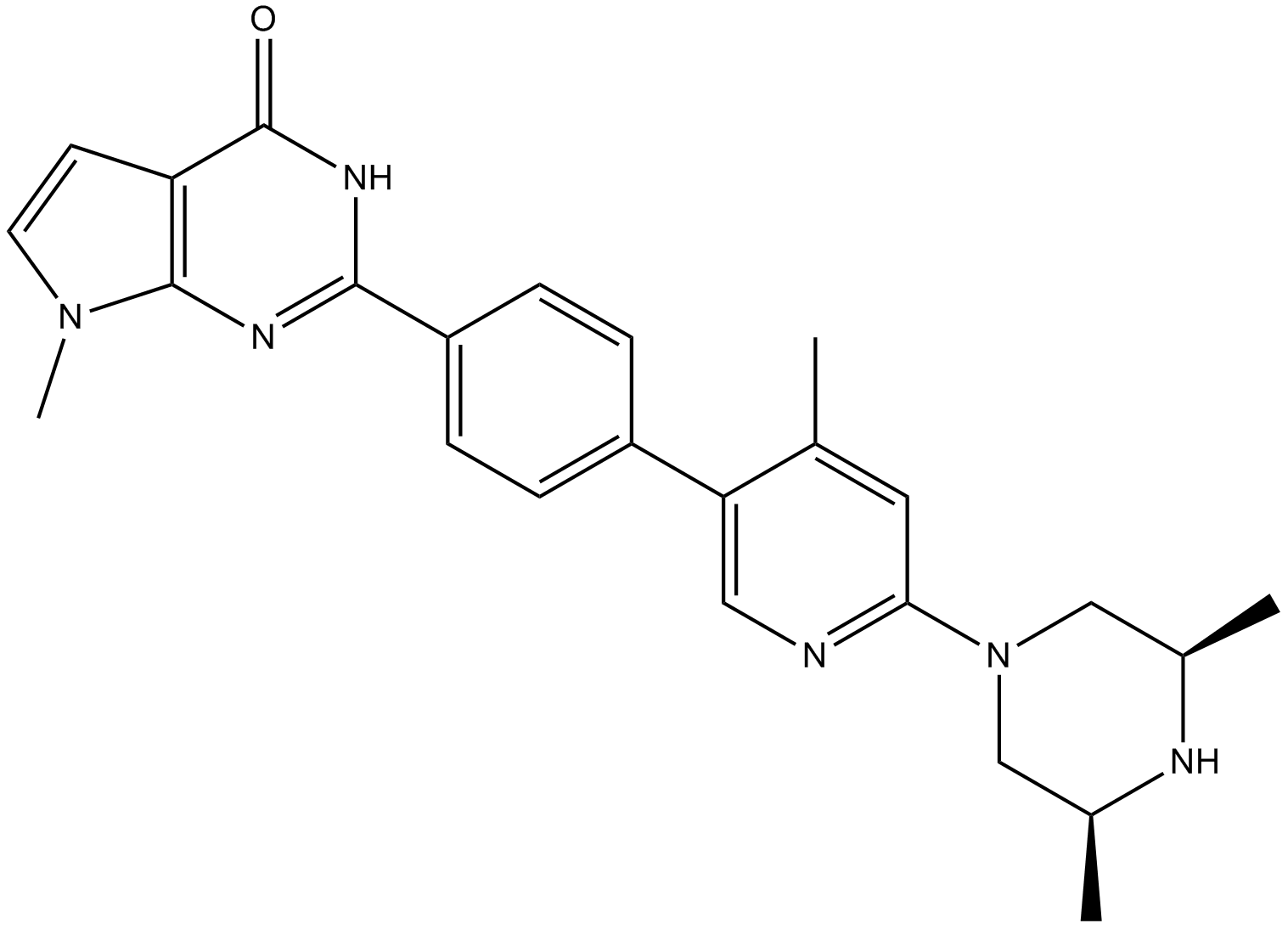
-
GC12844
Benzamide
poly (ADP-ribose) synthetase inhibitor
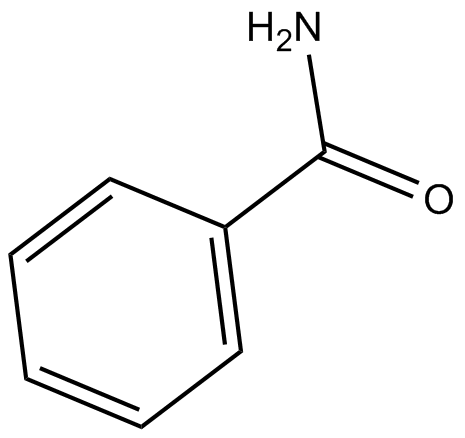
-
GC14380
BGP-15
PARP inhibitor
-
GC35547
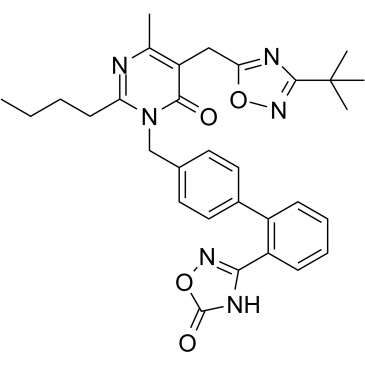
BR102375
BR102375 is a non-TZD peroxisome proliferator-activated receptor γ (PPAR γ) full agonist for the treatment of type 2 diabetes, reveals EC50 value of 0.28?μM and Amax ratio?of 98%.
-
GC33223
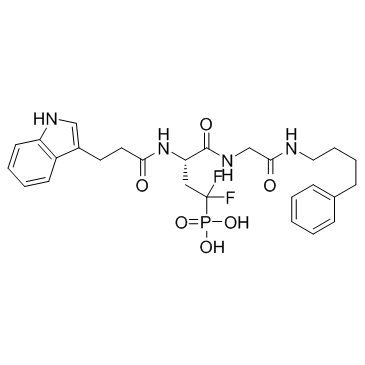
BRCA1-IN-1
BRCA1-IN-1 is a novel small-molecule-like BRCA1 inhibitor with IC50 and Ki of 0.53 μM and 0.71 μM, respecrively.
-
GC35550
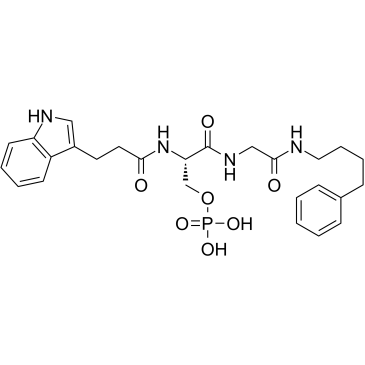
BRCA1-IN-2
BRCA1-IN-2 (compound 15) is a cell-permeable protein-protein interaction (PPI) inhibitor for BRCA1 with an IC50 of 0.31 μM and a Kd of 0.3 μM, which shows antitumor activities via the disruption of BRCA1 (BRCT)2/protein interactions.
-
GC64209
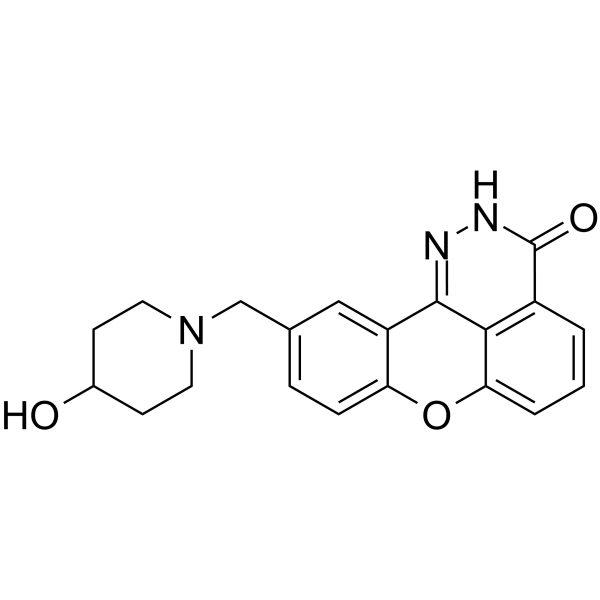
E7016
E7016 (GPI 21016) is an orally available PARP inhibitor. E7016 can enhance tumor cell radiosensitivity in vitro and in vivo through the inhibition of DNA repair. E7016 acts as a potential anticancer agent.
-
GC18172
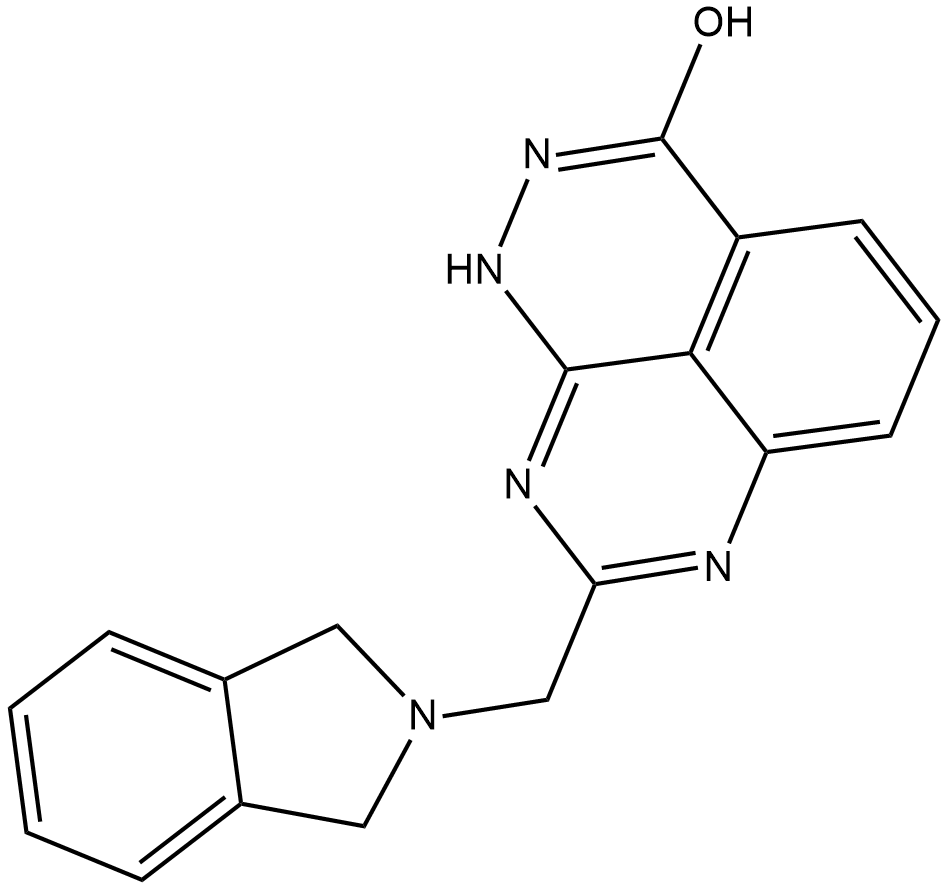
E7449
E7449 is an inhibitor of poly(ADP-ribose) polymerase 1 (PARP1) and PARP2 (IC50s = 1 and 1.2 nM, respectively) as well as tankyrase (TNKS) 1/2 (IC50s = 50-100 nM).
-
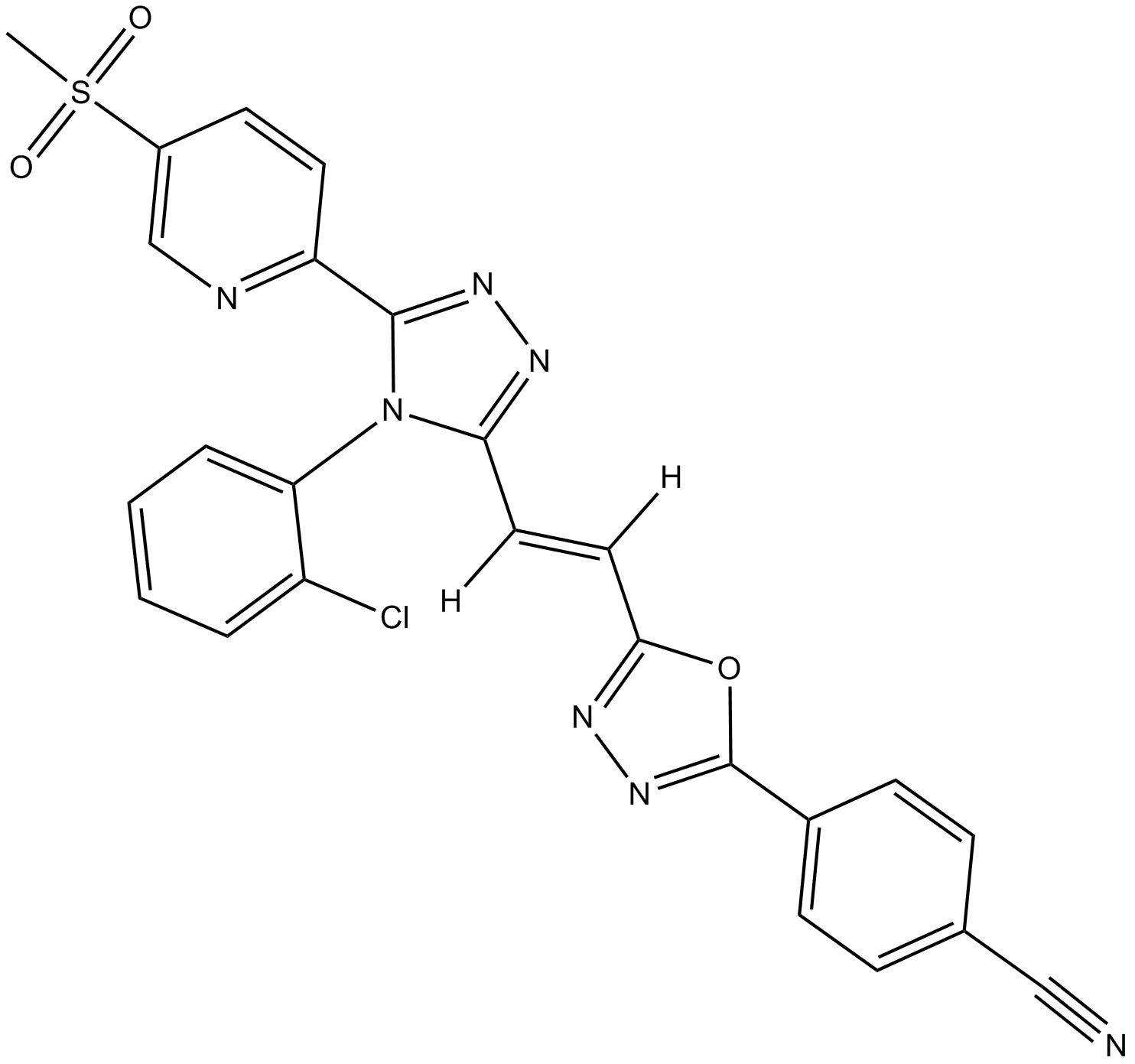
GC13541
G007-LK
tankyrase 1/2 inhibitor
-
GC19542
GeA-69
GeA-69 is a selective, highly cell permeable allosteric inhibitor

-
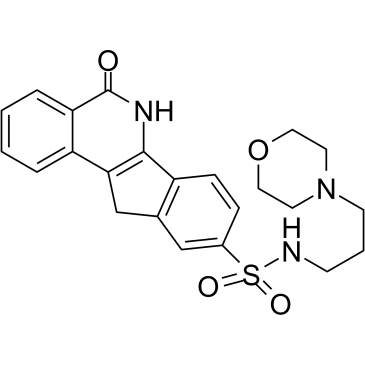
GC38385
INO-1001
-
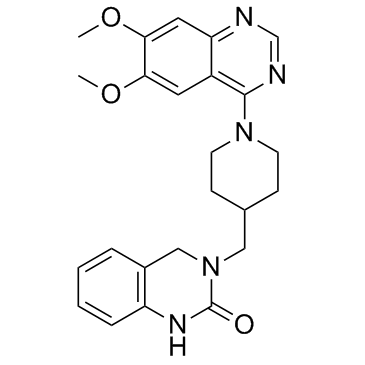
GC34195
K-756
K-756 is a direct and selective tankyrase (TNKS) inhibitor, which inhibits the ADP-ribosylation activity of TNKS1 and TNKS2 with IC50s of 31 and 36 nM, respectively.
-
GC65907
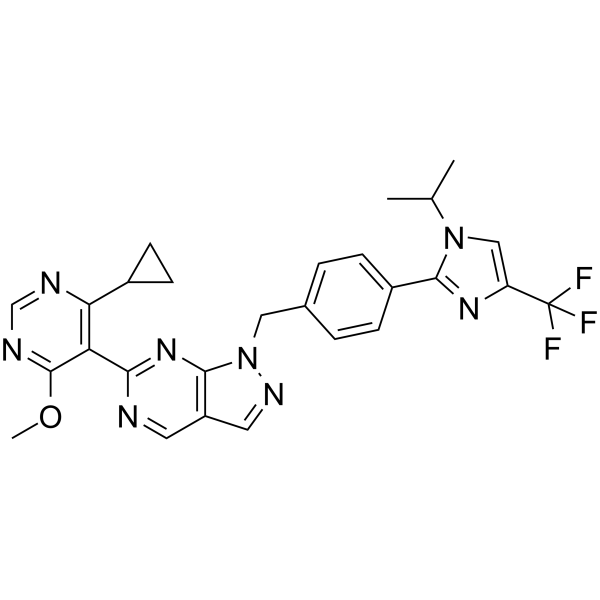
KSQ-4279
KSQ-4279 (USP1-IN-1, Formula I) is a USP1 and PARP inhibitor (extracted from patent WO2021163530).
-
GC13419
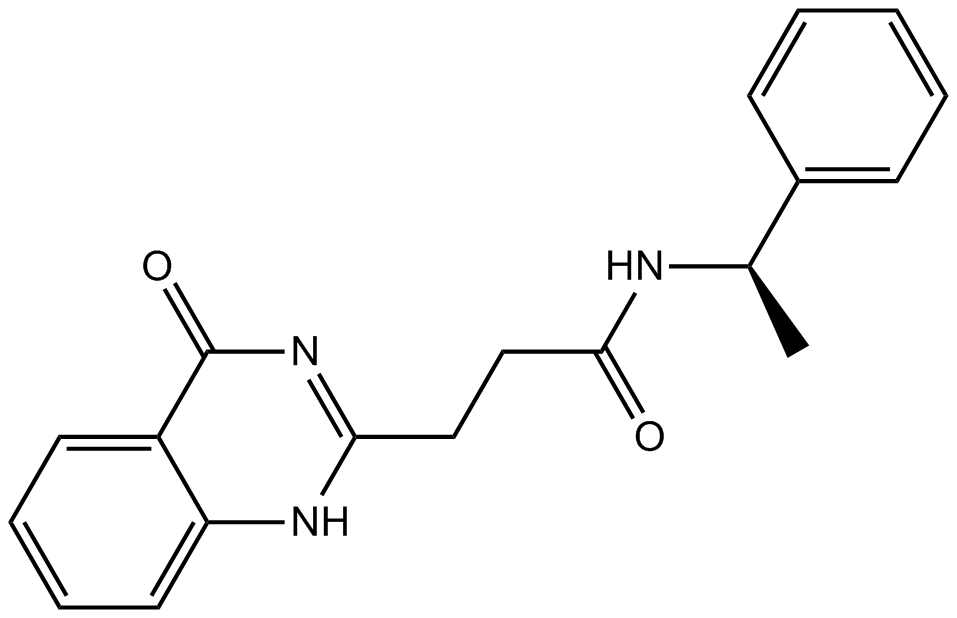
ME0328
PARP inhibitor,potent and selective
-
GC12756
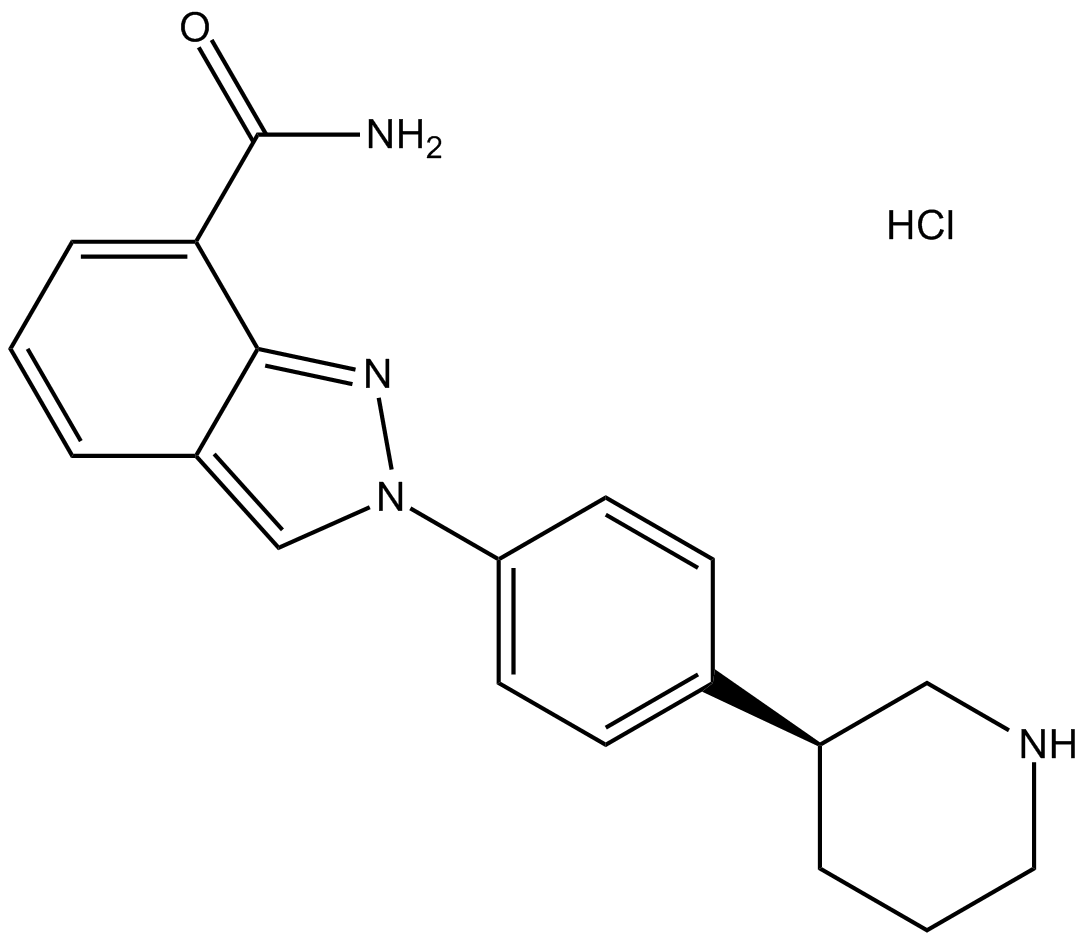
MK-4827 hydrochloride
MK-4827 hydrochloride (MK-4827 hydrochloride) is a highly potent and orally bioavailable PARP1 and PARP2 inhibitor with IC50s of 3.8 and 2.1 nM, respectively. MK-4827 hydrochloride leads to inhibition of repair of DNA damage, activates apoptosis and shows anti-tumor activity.
-
GC11537
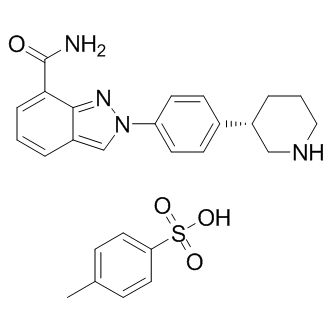
MK-4827 tosylate
MK-4827 tosylate (MK-4827 tosylate) is a highly potent and orally bioavailable PARP1 and PARP2 inhibitor with an IC50 of 3.8 and 2.1 nM, respectively. MK-4827 tosylate leads to inhibition of repair of DNA damage, activates apoptosis and shows anti-tumor activity.
-
GC16914
MN 64
tankyrase inhibitor
-
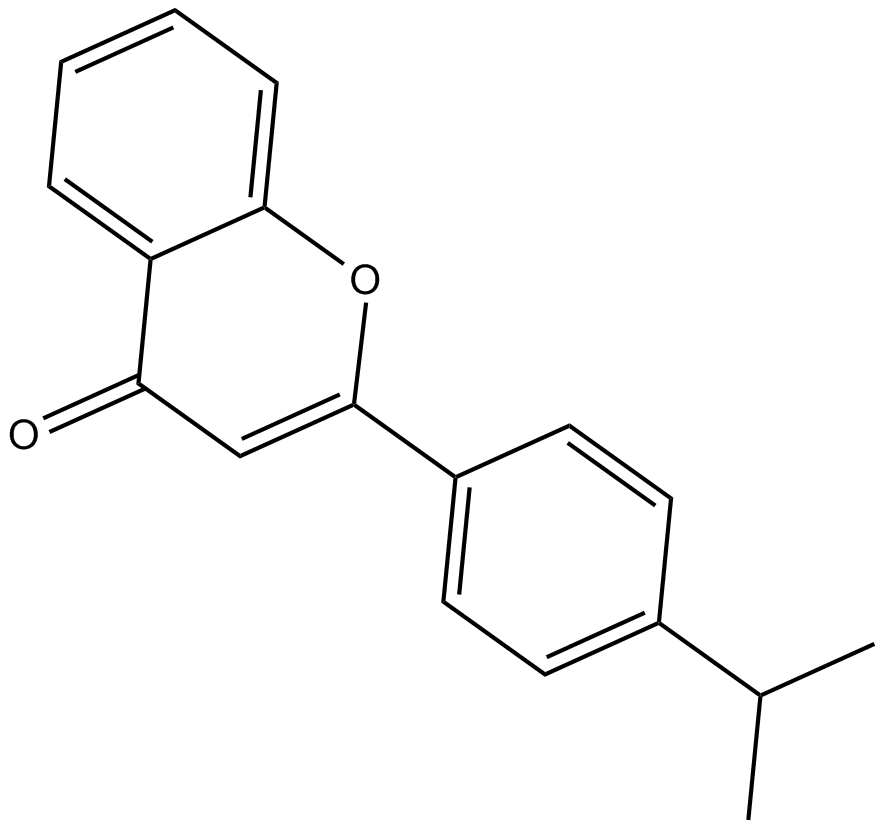
GC65202
Nesuparib
Nesuparib is a potent inhibitor of PARP.
-
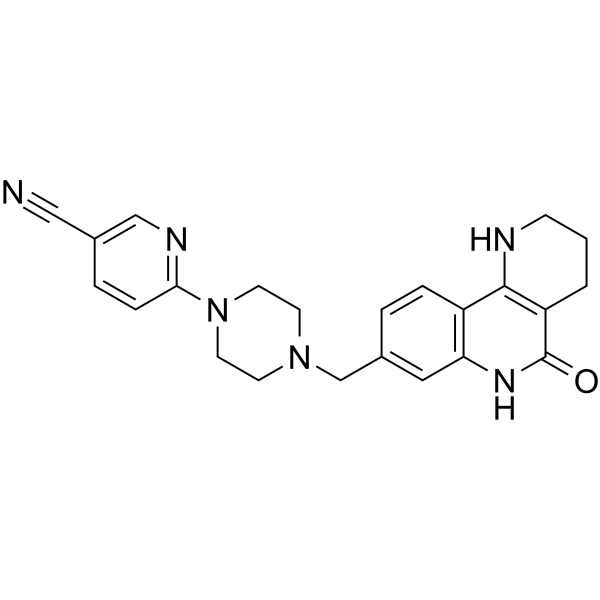
GC34120
Niraparib R-enantiomer (MK 4827 (R-enantiomer))
Niraparib R-enantiomer (MK-4827 R-enantiomer) is an excellent PARP1 inhibitor with IC50 of 2.4 nM.
-
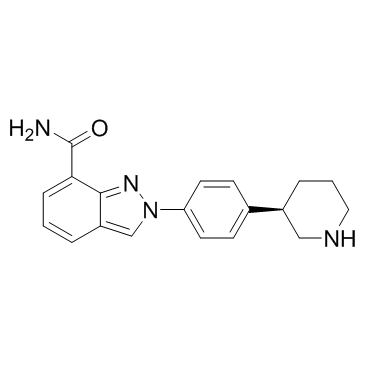
GC19264
NMS-P118
NMS-P118 is a potent, orally available, and highly selective PARP-1 Inhibitor for cancer therapy.
-
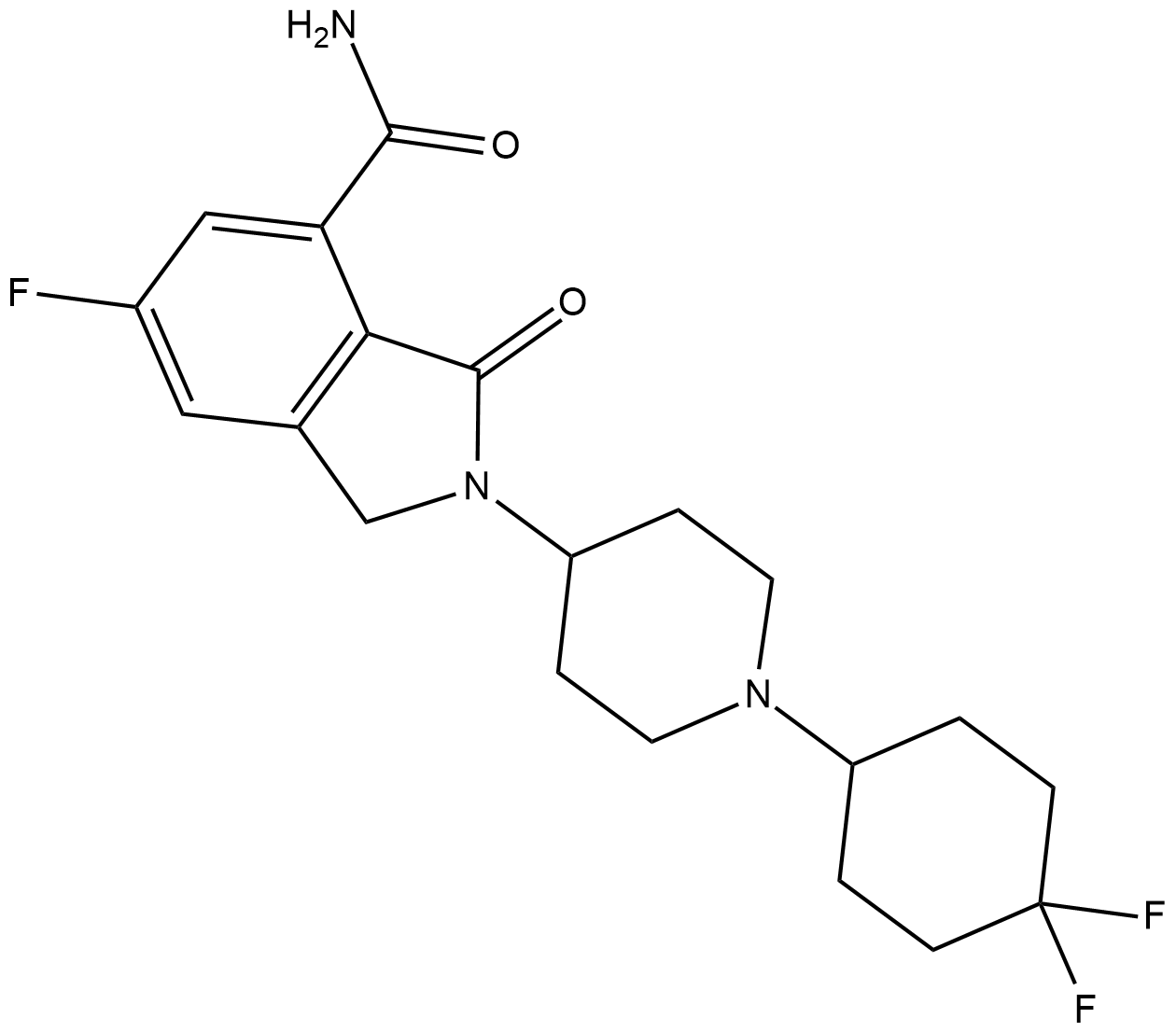
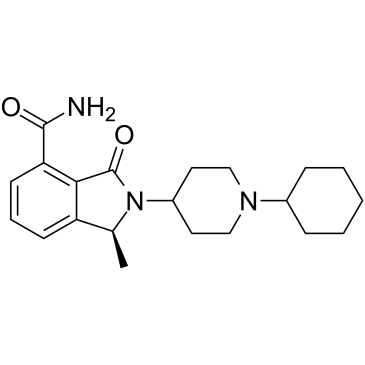
GC36751
NMS-P515
NMS-P515 is a potent, orally active and stereospecific PARP-1 inhibitor, with a Kd of 16 nM and an IC50 of 27 nM (in Hela cells). Anti-tumor activity.
-
GC17555
NVP-TNKS656
TNKS2 inhibitor
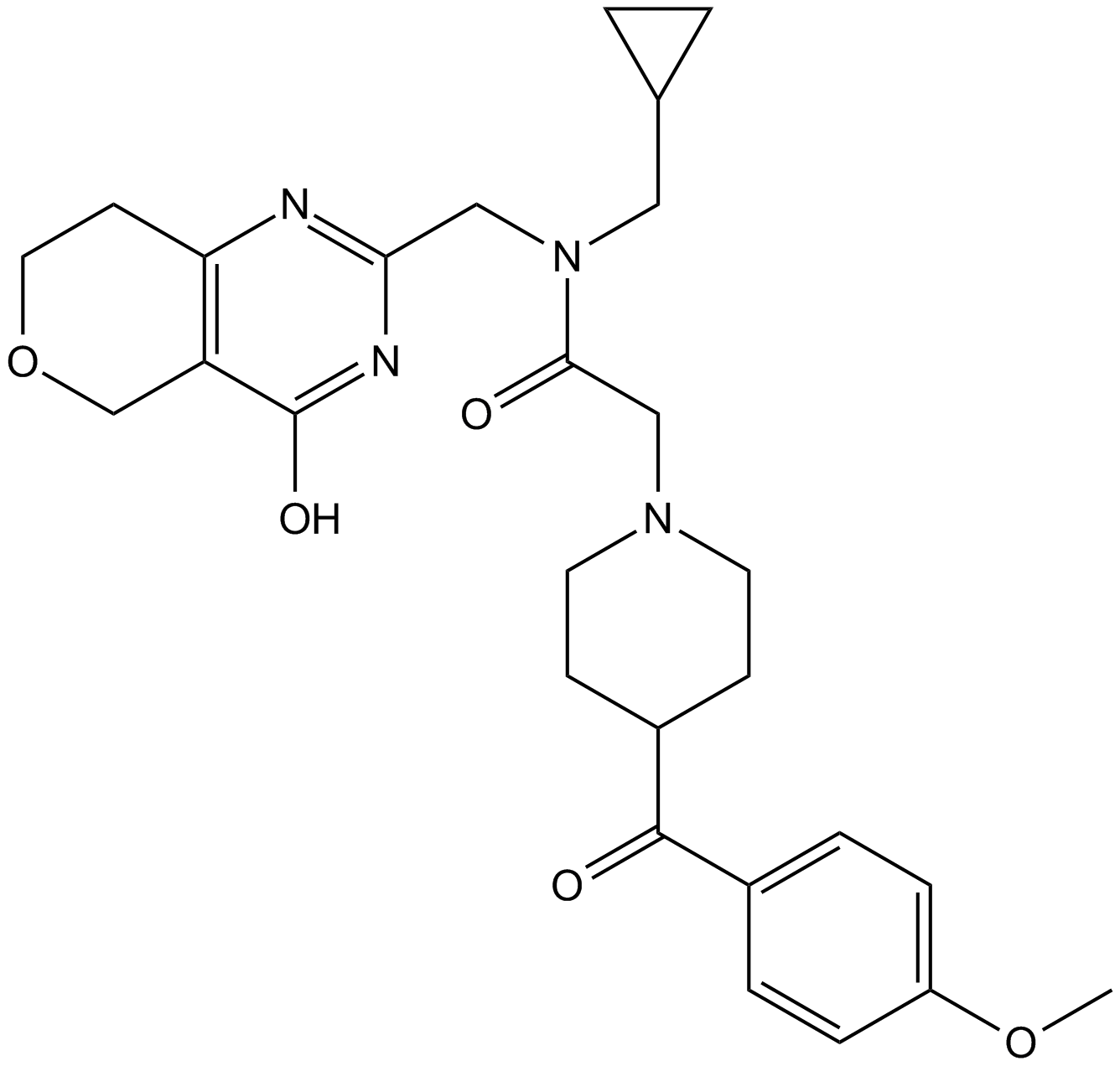
-
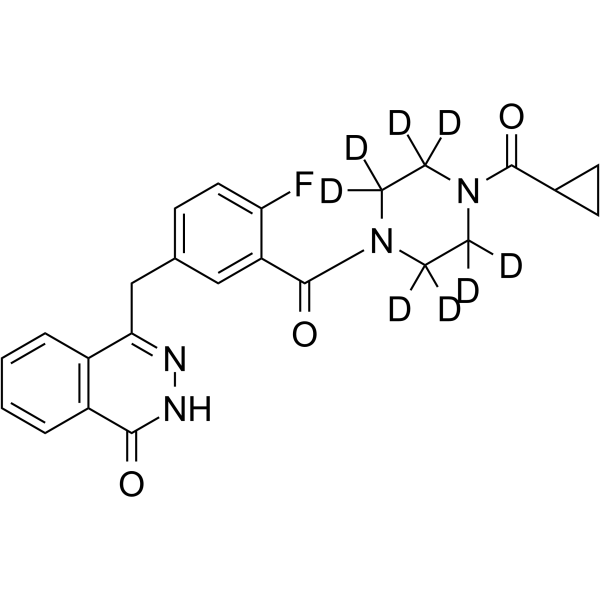
GC69618
Olaparib-d8
Olaparib-d8 is the deuterated form of Olaparib (AZD2281). Olaparib is an orally effective PARP inhibitor that inhibits PARP-1 and PARP-2 with IC50 values of 5 and 1 nM, respectively. Olaparib is also an activator of autophagy and mitophagy.
-
GC67906
OM-153

-
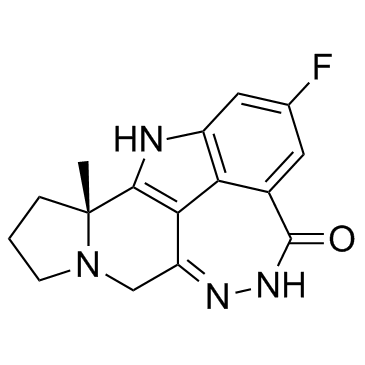
GC34071
Pamiparib (BGB-290)
Pamiparib (BGB-290) (BGB-290) is an orally active, potent, highly selective PARP inhibitor, with IC50 values of 0.9 nM and 0.5 nM for PARP1 and PARP2, respectively. Pamiparib (BGB-290) has potent PARP trapping, and capability to penetrate the brain, and can be used for the research of various cancers including the solid tumor.
-
GC36855
Paris saponin VII
Paris saponin VII (Chonglou Saponin VII) is a steroidal saponin isolated from the roots and rhizomes of Trillium tschonoskii Maxim. Paris saponin VII-induced apoptosis in K562/ADR cells is associated with Akt/MAPK and the inhibition of P-gp. Paris saponin VII attenuates mitochondrial membrane potential, increases the expression of apoptosis-related proteins, such as Bax and cytochrome c, and decreases the protein expression levels of Bcl-2, caspase-9, caspase-3, PARP-1, and p-Akt. Paris saponin VII induces a robust autophagy in K562/ADR cells and provides a biochemical basis in the treatment of leukemia.
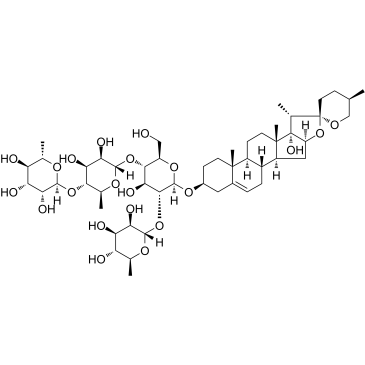
-
GC64579
PARP-1-IN-2
PARP-1-IN-2 (compound 11g) is a potent and BBB-penetrated PARP1 inhibitor, with an IC50 of 149 nM. PARP1-IN-2 shows significantly potent anti-proliferative activity against Human lung adenocarcinoma epithelial cell line A549. PARP1-IN-2 can induce A549 cells apoptosis.
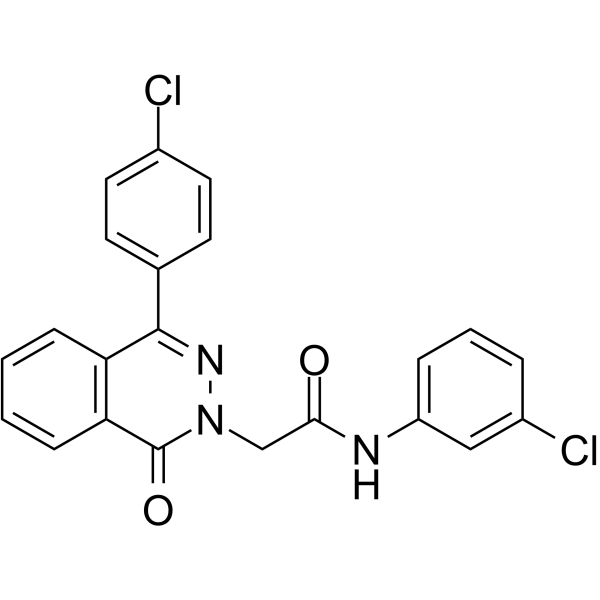
-
GC65927
PARP-2-IN-1
PARP-2-IN-1 is a potent and selective PARP-2 inhibitor with an IC50 of 11.5 nM.
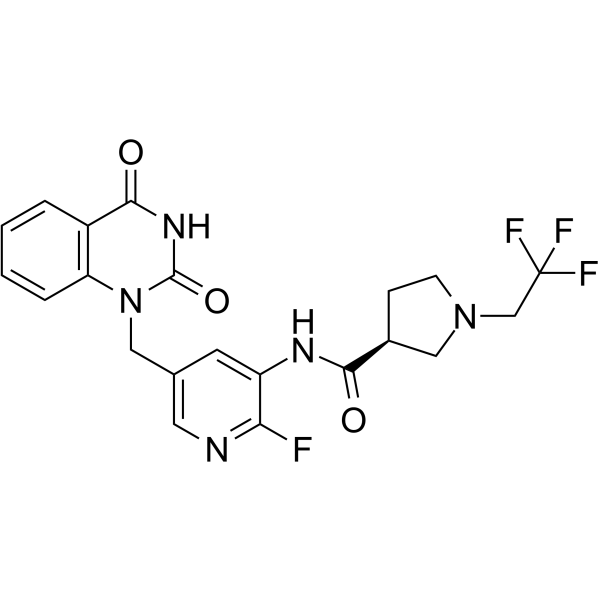
-
GC68006
PARP1-IN-11
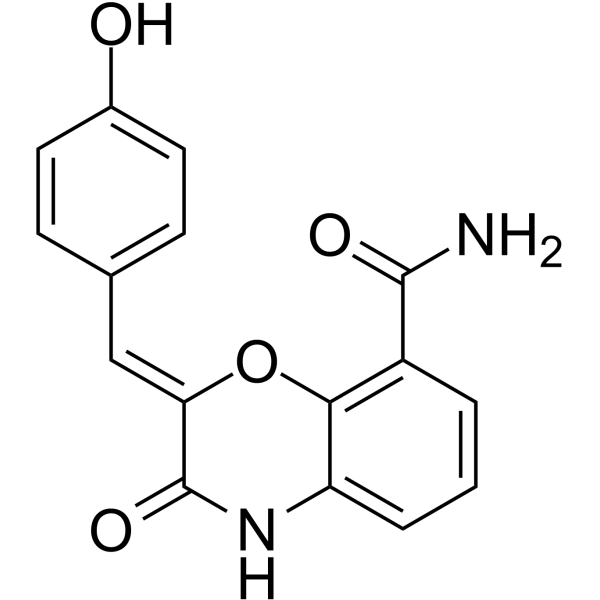
-
GC69660
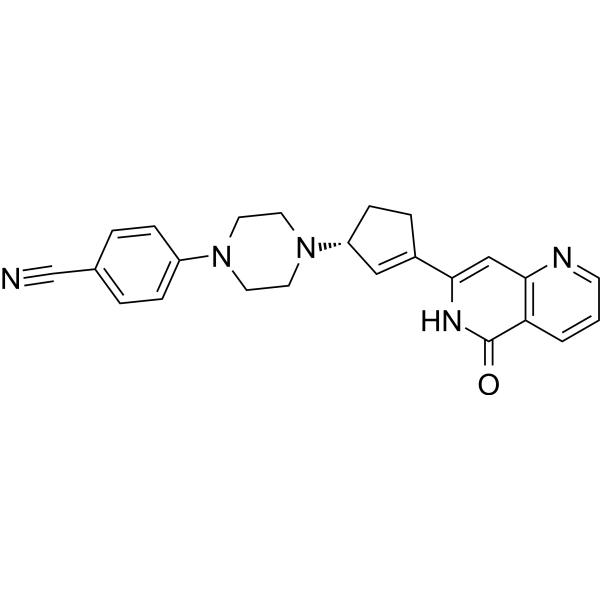
PARP1-IN-7
PARP1-IN-7 is an inhibitor of poly ADP-ribose polymerase-1 (PARP1), used as an anticancer agent.
-
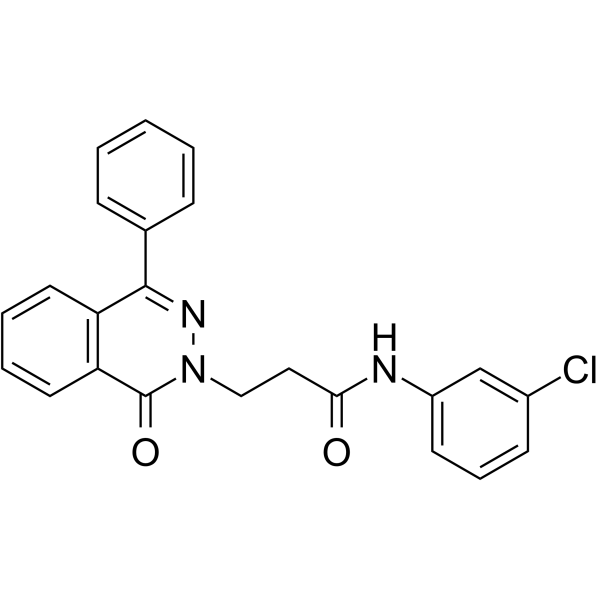
GC64578
PARP1-IN-8
PARP1-IN-8 (compound 11c) is a potent and BBB-penetrated PARP1 inhibitor, with an IC50 of 97 nM. PARP1-IN-8 shows significantly potent anti-proliferative activity against Human lung adenocarcinoma epithelial cell line A549.
-
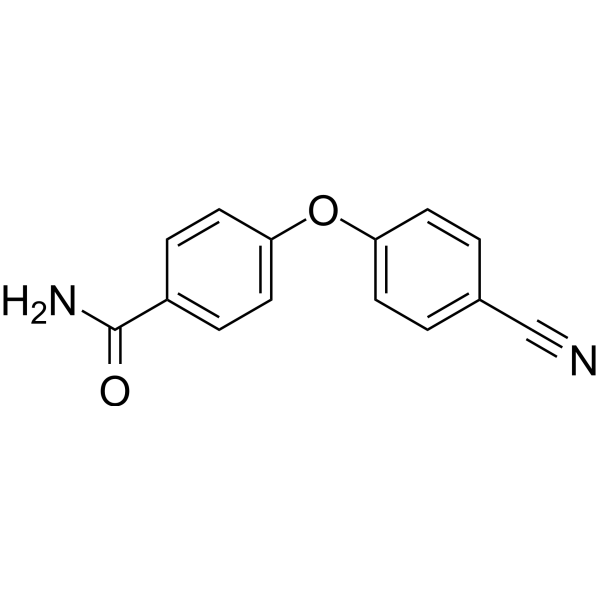
GC69657
PARP10-IN-2
PARP10-IN-2 is an effective inhibitor of mono-ADP-ribosyltransferase PARP10, with an IC50 of 3.64 μM for human PARP10. It also inhibits PARP2 and PARP15, with IC50 values of 27 μM and 11 μM for human PARP2 and human PARP15, respectively.
-
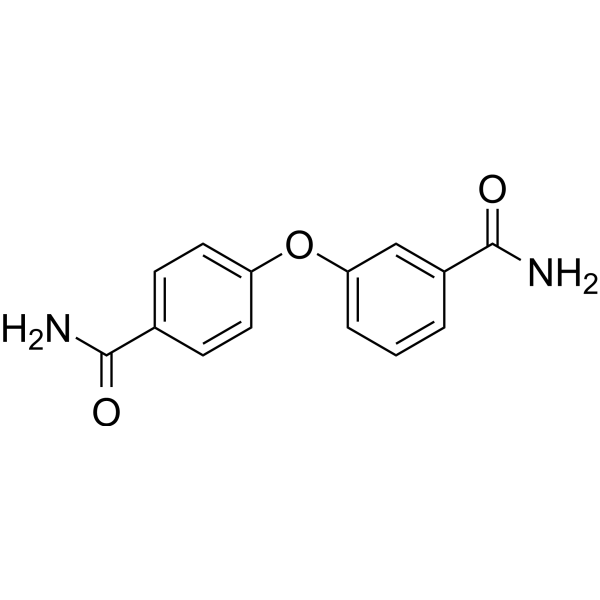
GC69658
PARP10-IN-3
PARP10-IN-3 is a selective mono-ADP-ribosyltransferase PARP10 inhibitor with an IC50 of 480 nM for human PARP10. It also inhibits PARP2 and PARP15, with IC50 values of 1.7 μM for both human PARP2 and human PARP15.
-
GC68035
PARP10/15-IN-1
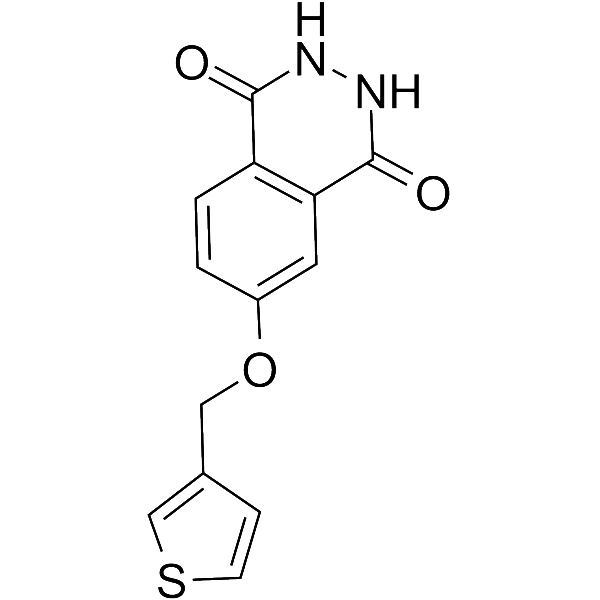
-
GC69655
PARP10/15-IN-2
PARP10/15-IN-2 (Compound 8h) is an effective dual inhibitor of PARP10 and PARP15, with IC50 values of 0.15 μM and 0.37 μM, respectively. It can enter cells and prevent apoptosis.
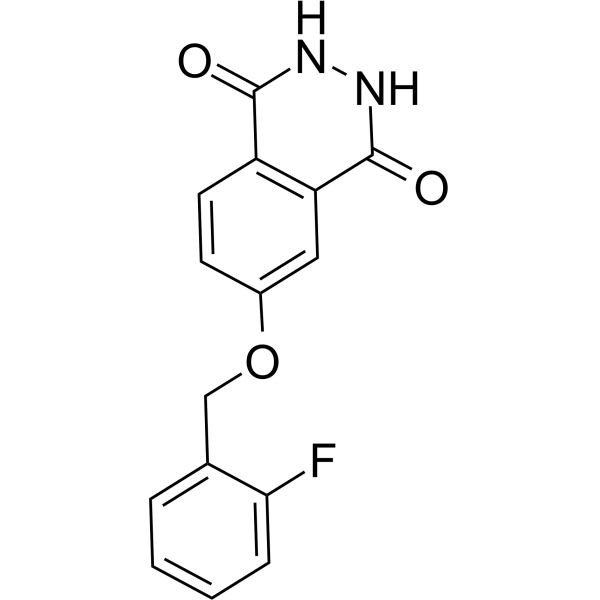
-
GC69656
PARP10/15-IN-3
PARP10/15-IN-3 (Compound 8a) is an effective dual inhibitor of PARP10 and PARP15, with IC50 values of 0.14 μM and 0.40 μM, respectively. It can enter cells and prevent apoptosis.
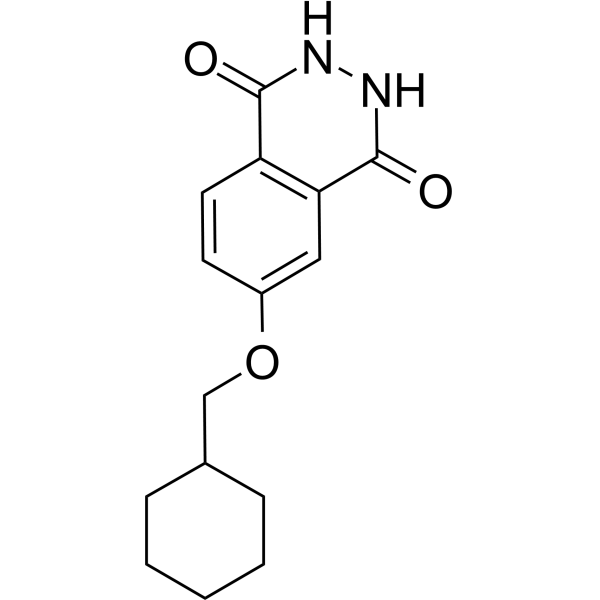
-
GC69659
PARP11 inhibitor ITK7
PARP11 inhibitor ITK7 (ITK7) is an effective and selective inhibitor of PARP11. It can effectively inhibit PARP11 with an IC50 value of 14 nM. PARP11 inhibitor ITK7 can be used for research on cellular localization.
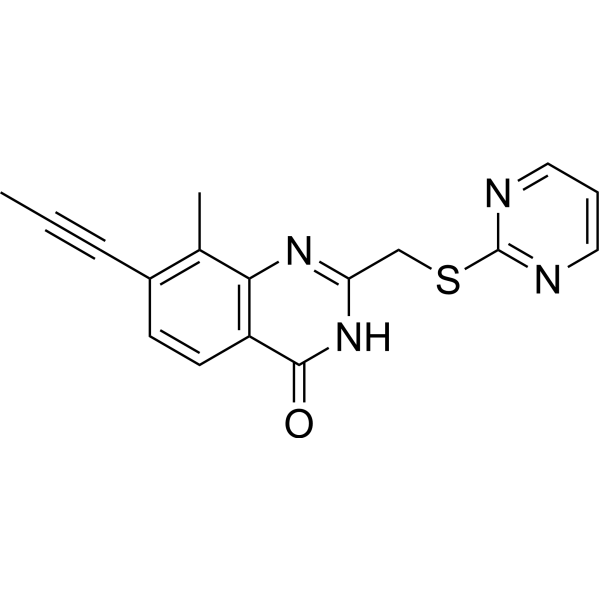
-
GC69661
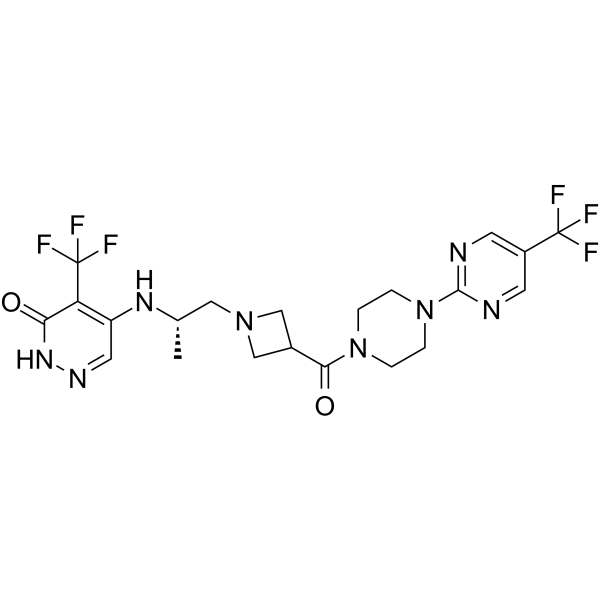
PARP7-IN-14
PARP7-IN-14 (I-1) is an effective selective PARP7 inhibitor with an IC50 value of 7.6 nM. PARP7-IN-14 has anti-cancer activity.
-
GC14251
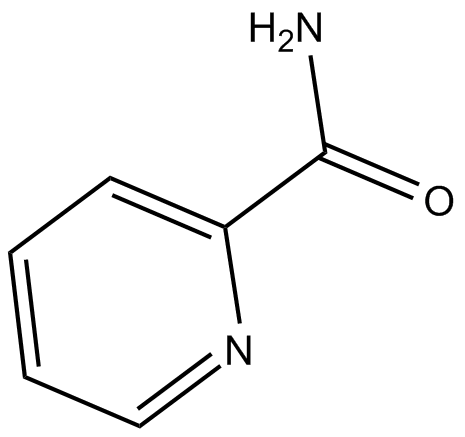
Picolinamide
poly (ADP-ribose) synthetase inhibitor
-
GC65147
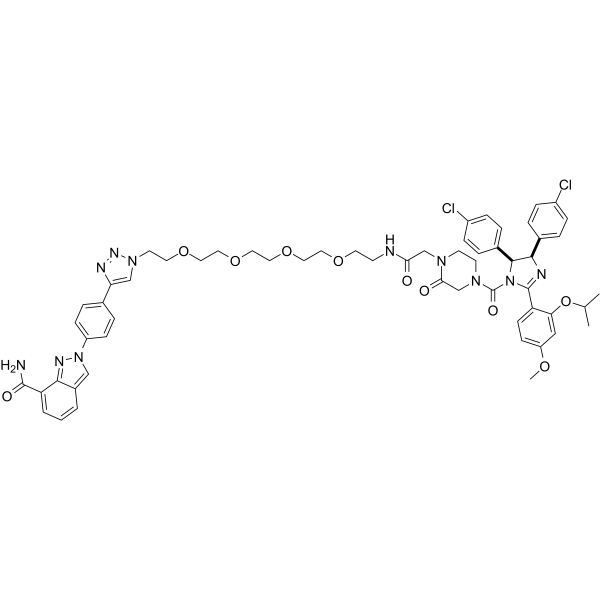
PROTAC PARP1 degrader
PROTAC PARP1 degrader is a PARP1 degrader based on MDM2 E3 ligand. It induces significant PARP1 cleavage and programmed cell death. PROTAC PARP1 degrader at 10 μM at 24 h inhibits MDA-MB-231 cell line with an IC50 of 6.12 μM.
-
GC69804
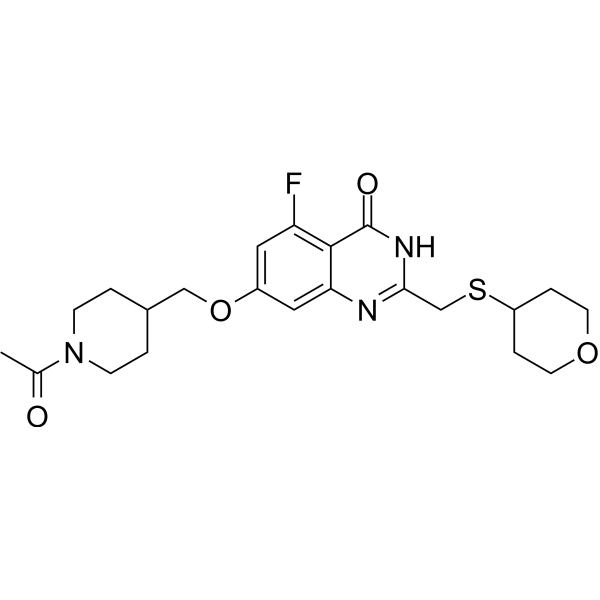
RBN-3143
RBN-3143 is an effective NAD+ competitive catalytic PARP14 inhibitor with an IC50 value of 4 nM. RBN-3143 inhibits PARP14-mediated ADP-ribosylation and stabilizes PARP14 in cell lines. RBN-3143 is used for research on lung inflammation.
-
GC19505
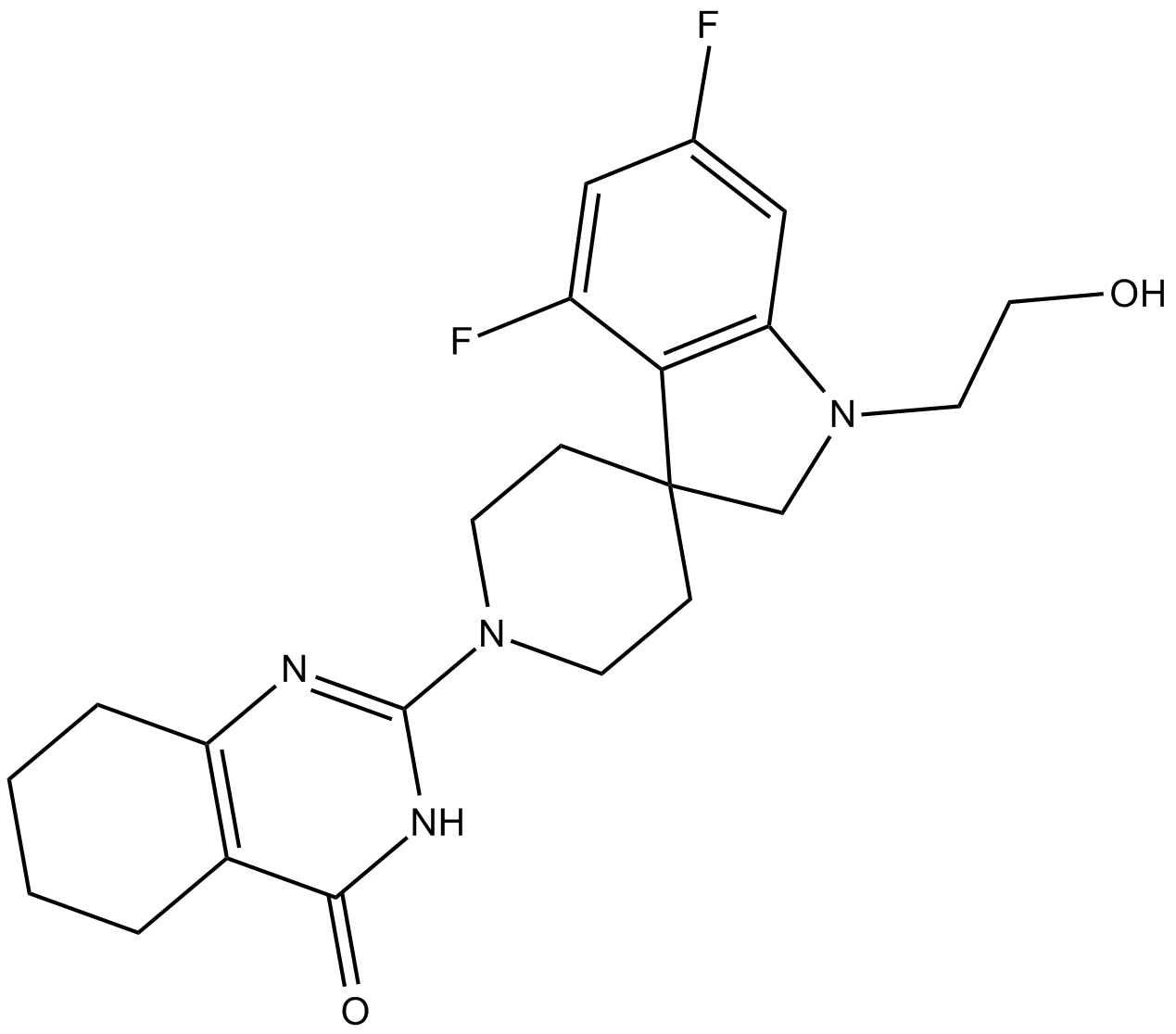
RK-287107
RK-287107 is a potent and specific tankyrase inhibitor with IC50s of 14.3 and 10.6 nM for tankyrase-1 and tankyrase-2, respectively
-
GC13249
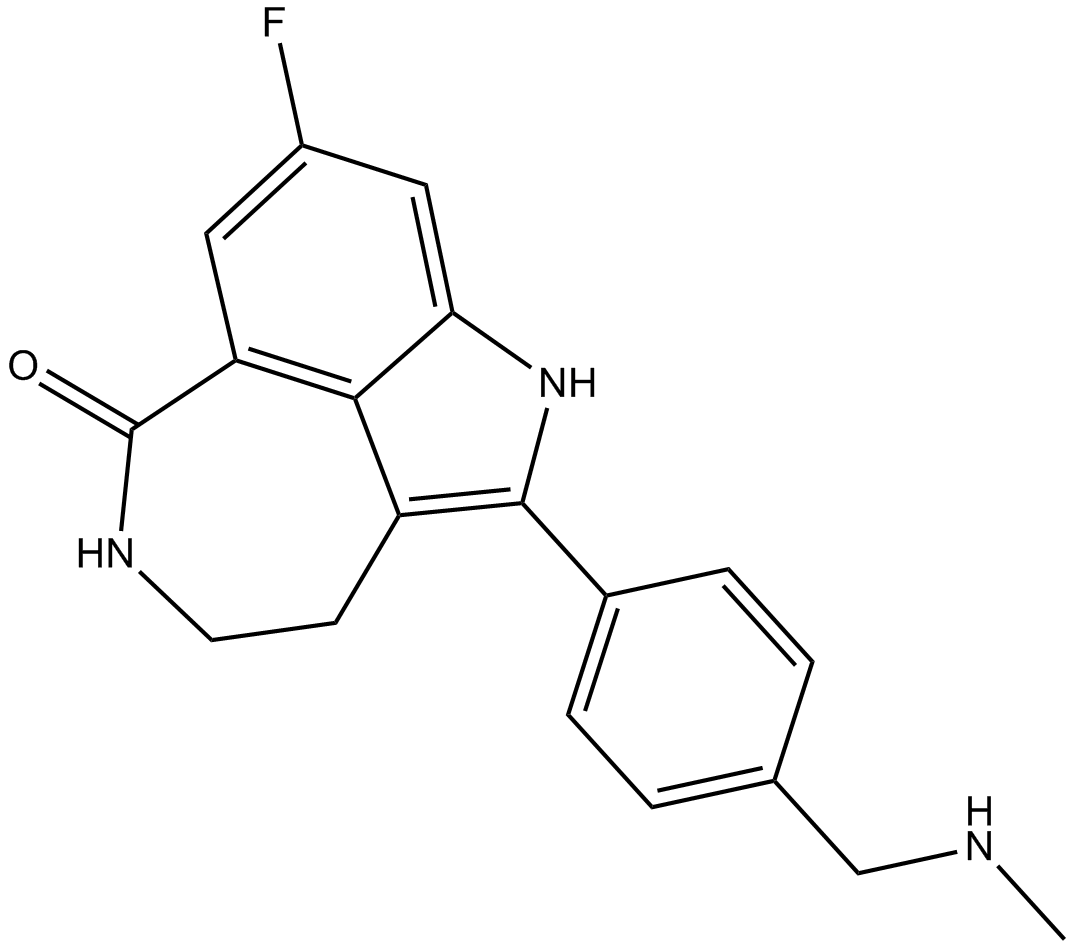
Rucaparib (free base)
Rucaparib (free base) (AG014699) is an orally active, potent inhibitor of PARP proteins (PARP-1, PARP-2 and PARP-3) with a Ki of 1.4 nM for PARP1. Rucaparib (free base) is a modest hexose-6-phosphate dehydrogenase (H6PD) inhibitor. Rucaparib (free base) has the potential for castration-resistant prostate cancer (CRPC) research.
-
GC32793
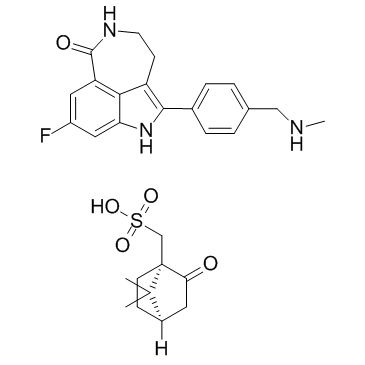
Rucaparib Camsylate
Rucaparib (AG014699) monocamsylate is an orally active, potent inhibitor of PARP proteins (PARP-1, PARP-2 and PARP-3) with a Ki of 1.4 nM for PARP1. Rucaparib Camsylate is a modest hexose-6-phosphate dehydrogenase (H6PD) inhibitor. Rucaparib Camsylate has the potential for castration-resistant prostate cancer (CRPC) research.
-
GC67962
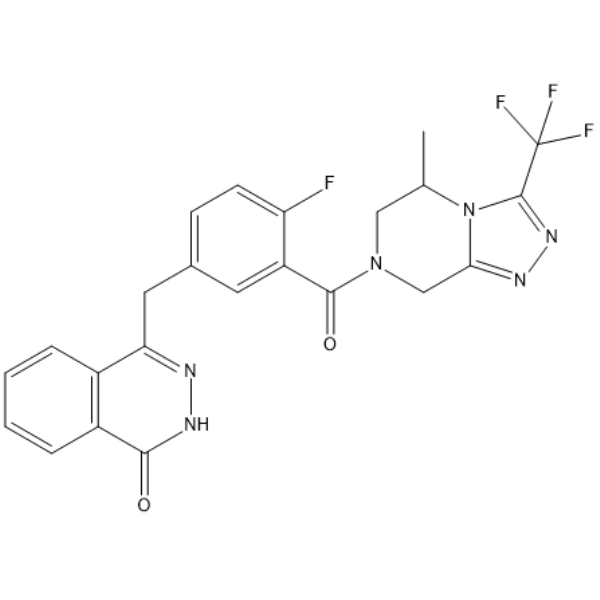
Simmiparib
-
GC69907
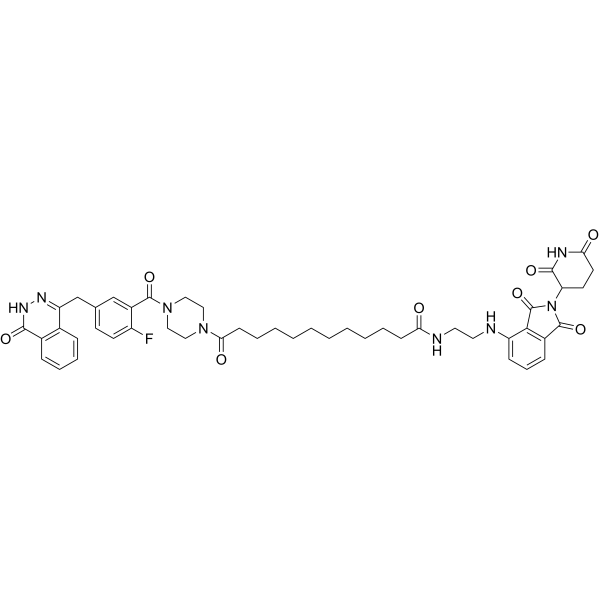
SK-575
SK-575 is an efficient and specific protein-hydrolyzing targeted chimeric molecule (PROTAC) PARP1 degrader with an IC50 of 2.30 nM. SK-575 can effectively inhibit the growth of cancer cells carrying BRCA1/2 mutations.
-
GC37728
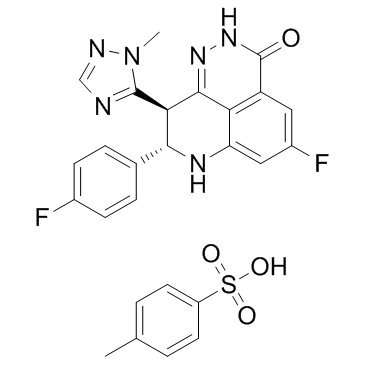
Talazoparib tosylate
Talazoparib tosylate (BMN 673ts) is a novel, potent and orally available PARP1/2 inhibitor with an IC50 of 0.57 nM for PARP1.
-
GC15041
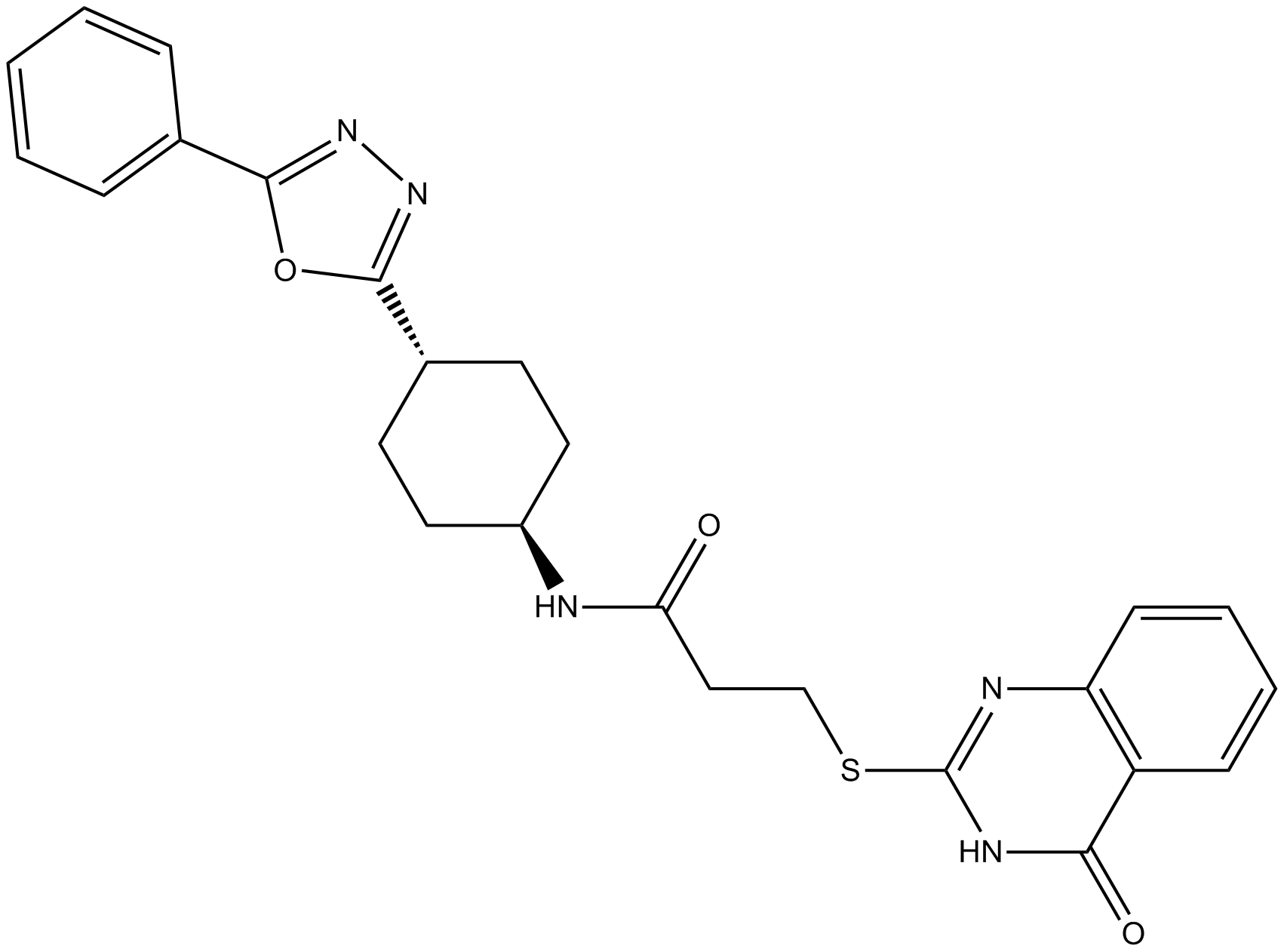
Tankyrase Inhibitors (TNKS) 22
Tankyrase inhibitor
-
GC11548
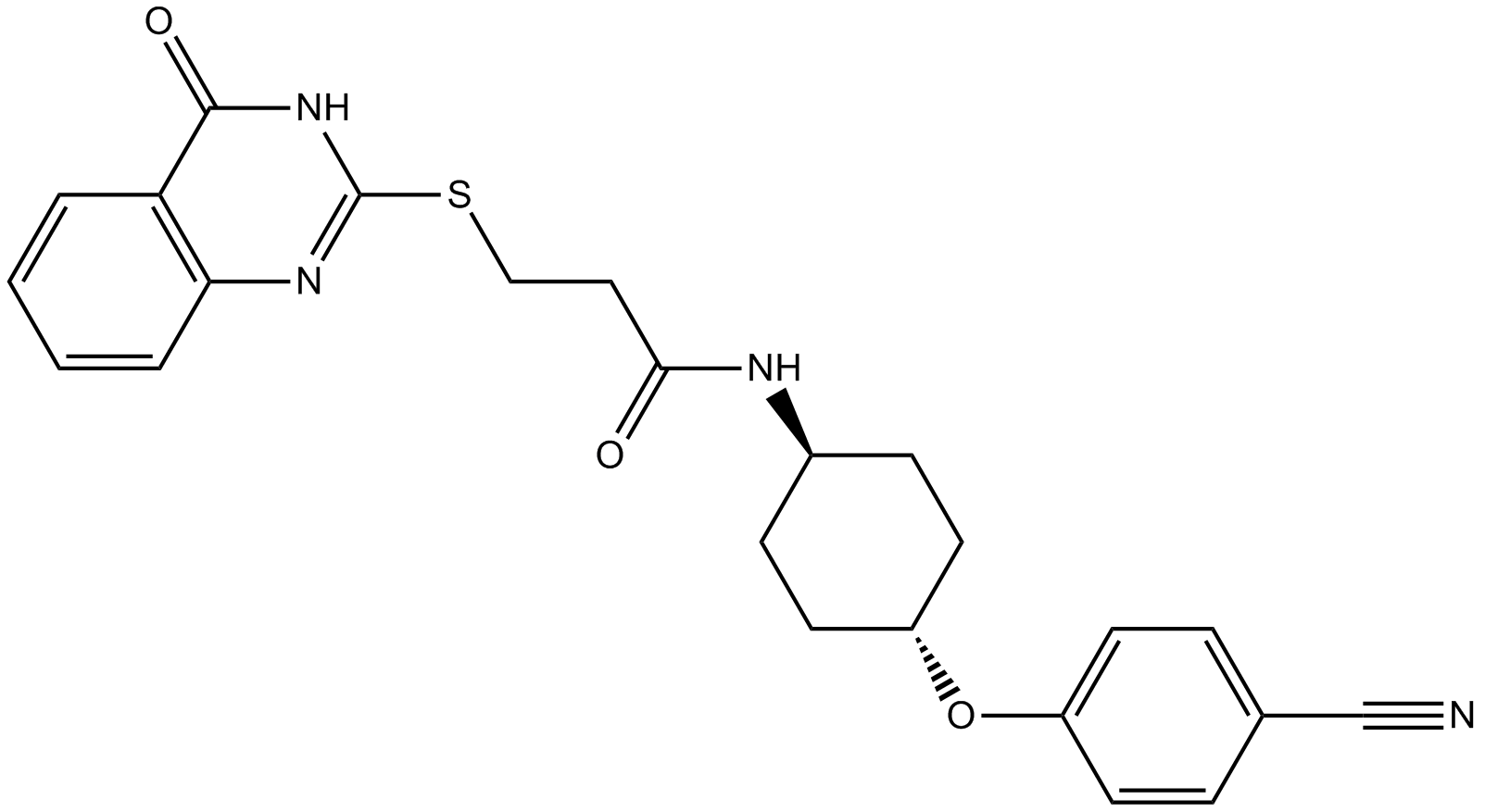
Tankyrase Inhibitors (TNKS) 49
Tankyrase inhibitor
-
GC37735
Tankyrase-IN-2
A TNKS1/2 inhibitor
-
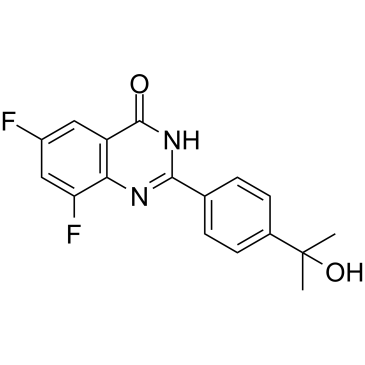
GC33358
WD2000-012547
WD2000-012547 is a selective poly(ADP-ribose)-polymerase (PARP-1) inhibitor with a pKi of 8.221.
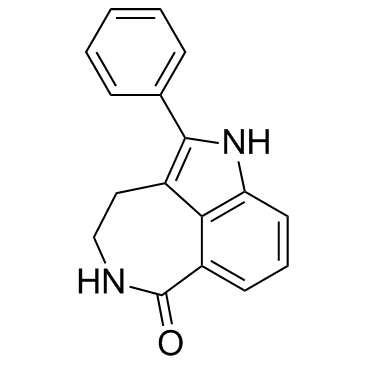
-
GC12781
XAV-939
XAV-939 selectively inhibits β-catenin-mediated transcription.