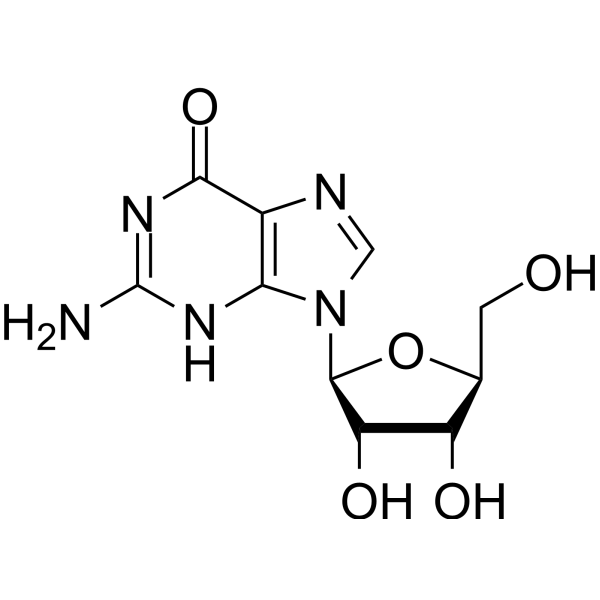
Nucleoside Antimetabolite/Analogue
Nucleoside analouges belong to the fmalily of antimetablits that resemble the neleosieds for uptake and metaolism that inibitr dna sytnse and couase cain termination. It is a durg target for cacner and viral infcations.
Products for Nucleoside Antimetabolite/Analogue
- Cat.No. Product Name Information
-
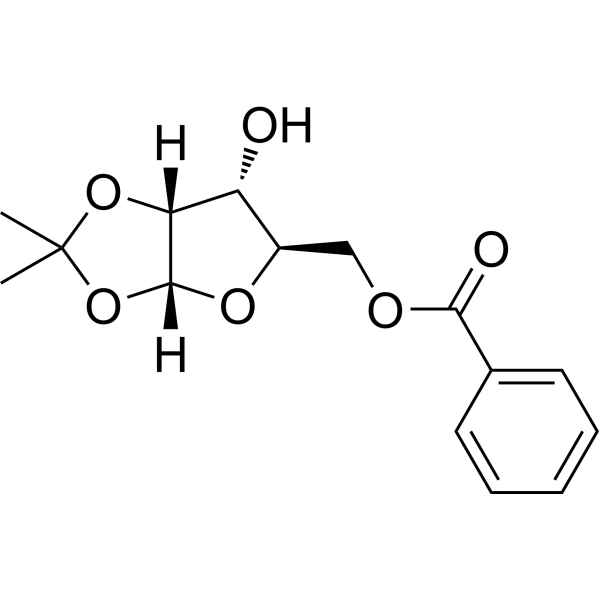
GC69793
(R)-5-O-Benzoyl-1,2-di-O-isopropylidene-alpha-D-xylofuranose
(R)-5-O-Benzoyl-1,2-di-O-isopropylidene-alpha-D-xylofuranose is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity and target inert lymphoid malignancies. The anticancer mechanism in this process depends on inhibiting DNA synthesis, inducing apoptosis (cell death), etc.
-
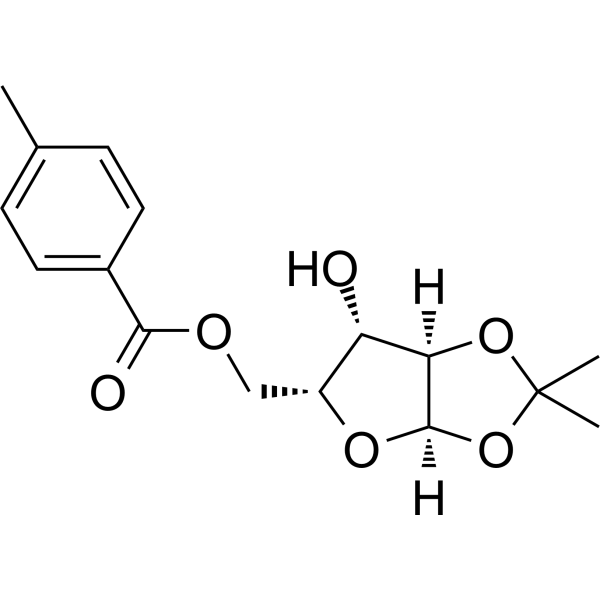
GC68484
1,2-O-Isopropylidene-5-O-p-toluoyl-a-D-xylofuranose
1,2-O-Isopropylidene-5-O-p-toluoyl-a-D-xylofuranose is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity and target malignant tumors in the inert lymphatic system. The anticancer mechanism in this process depends on inhibiting DNA synthesis, inducing apoptosis (cell death), etc.
-
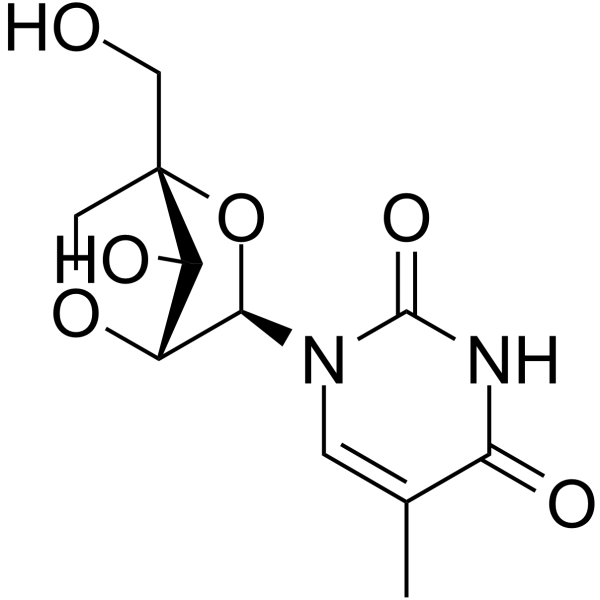
GC65551
1-(2'-O-4-C-Methylene-beta-D-ribofuranosyl)thymine
1-(2'-O-4-C-Methylene-beta-D-ribofuranosyl)thymine is a bicyclic nucleoside.
-
GC65038
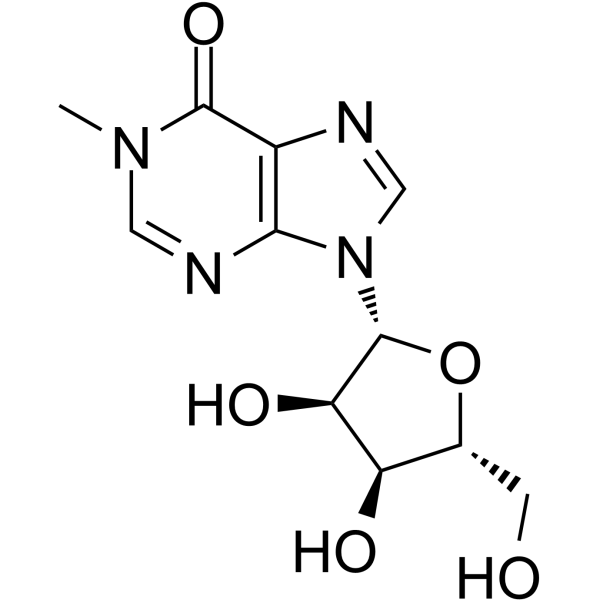
1-Methylinosine
1-Methylinosine is a modified nucleotide found at position 37 in tRNA 3' to the anticodon of eukaryotic tRNA.
-
GC65489
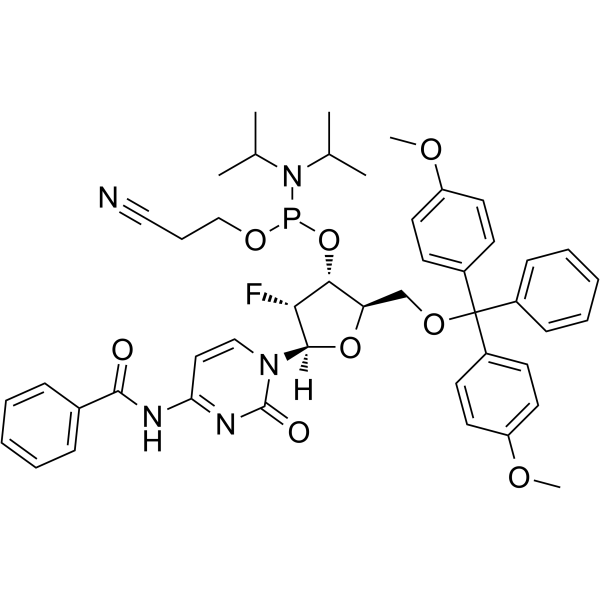
2'-F-Bz-dC Phosphoramidite
2'-F-Bz-dC Phosphoramidite can be used in the synthesis of oligoribonucleotides.
-
GC66651
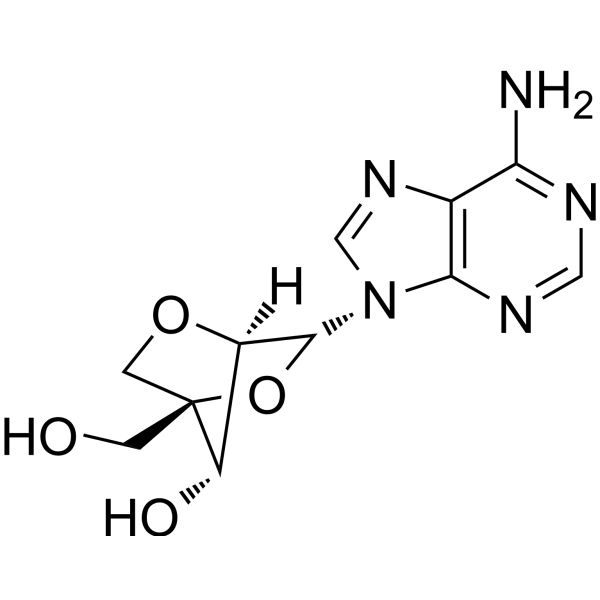
2'-O,4'-C-Methyleneadenosine
2'-O,4'-C-Methyleneadenosine (LNA-A) is a locked nucleic acid (LNA) and is also an adenosine analog.
-
GC66654
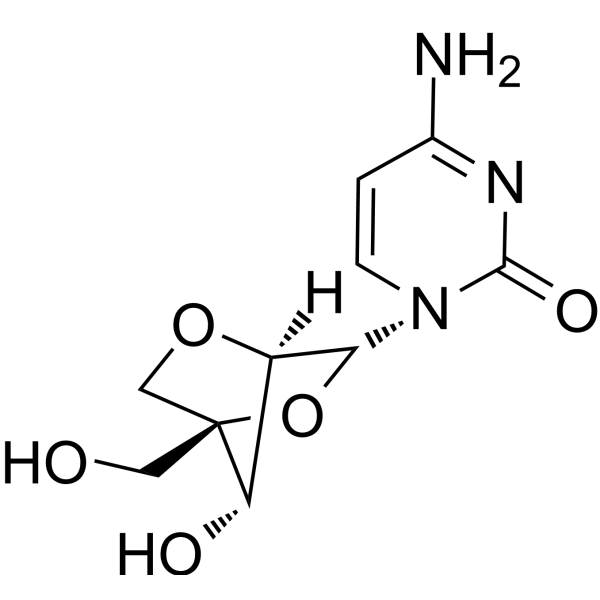
2'-O,4'-C-Methylenecytidine
2'-O,4'-C-Methylenecytidine (LNA-C(Bz)) is a bicyclic nucleoside analogue with fixed N-type conformation. 2'-O,4'-C-Methylenecytidine can be used to synthesize oligonucleotides. 2'-O,4'-C-Methylenecytidine forms duplexes with complementary DNA and RNA strands.
-
GC66655
2'-O,4'-C-Methyleneguanosine
2′-O,4′-C-Methyleneguanosine (LNA-G) is a reverse guanine analogue, where LNA (locked nucleic acid) is a nucleic acid analogue. LNA modification can be used in a variety of applications such as effective binding affinity to complementary sequences and greater nuclease resistance than natural nucleotides, offering great potential for applications in disease diagnosis and research. LNA-G is also available via KOD DNA polymerase, which allows the integration of LNA-G nucleotides into the DNA strand.
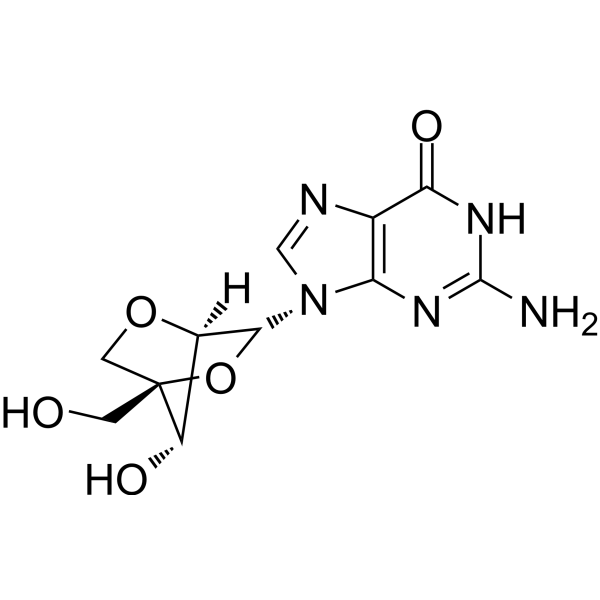
-
GC65170
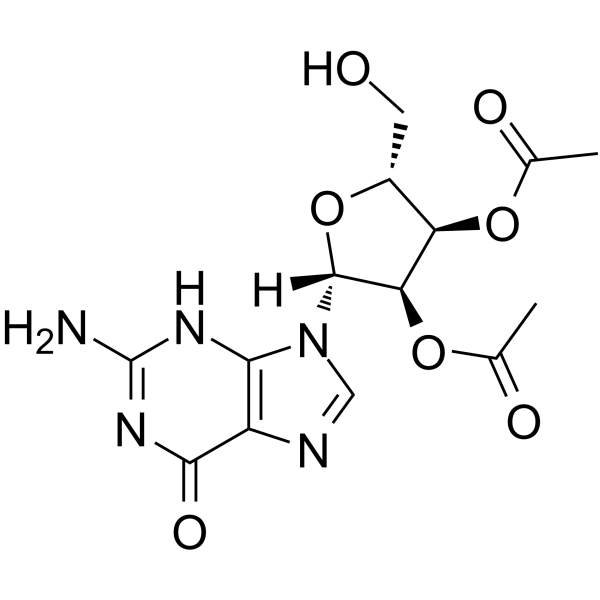
2′,3′-Di-O-acetylguanosine
2′,3′-Di-O-acetylguanosine is a nucleoside analog.
-
GC67384
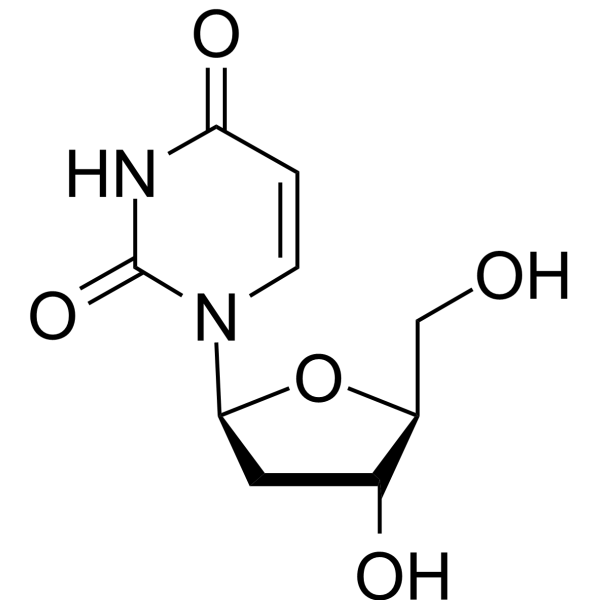
2′-Deoxy-β-L-uridine
2'-Deoxy-β-L-uridine is a nucledside analogue and a specific substrate for the viral enzyme, shows no stereospecificity against herpes simplex 1 (HSV1) thymidine kinase (TK). 2′-Deoxy-β-L-uridine exerts antiviral activity via the interation of 5'-triphosphates with the viral DNA polymerase.
-
GC61667
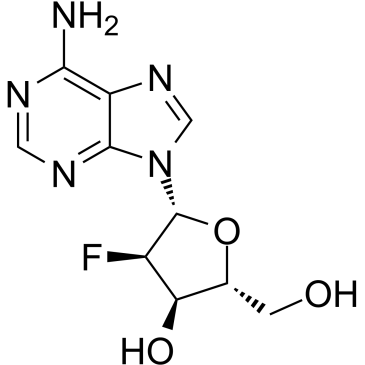
2′-Deoxy-2′-fluoroadenosine
2′-Deoxy-2′-fluoroadenosine can be used for thesynthesisof 2′-Deoxy-2′-fluoro-modified oligonucleotides hybridized with RNA. 2′-Deoxy-2′-fluoroadenosine can be cleaved efficiently by E. coli purine nucleoside phosphorylase (PNP) to the toxic agent 2-fluoroadenine (FAde). 2′-Deoxy-2′-fluoroadenosine shows excellent in vivo activity against tumors expressing E. coli PNP.
-
GC62530
2’-O-Me-C(Bz) Phosphoramidite
2’-O-Me-C(Bz) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
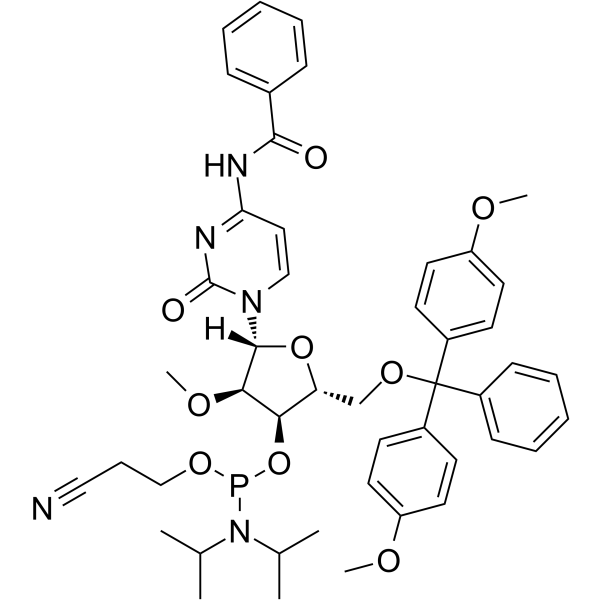
-
GC62529
2’-OMe-A(Bz) Phosphoramidite
2’-OMe-A(Bz) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
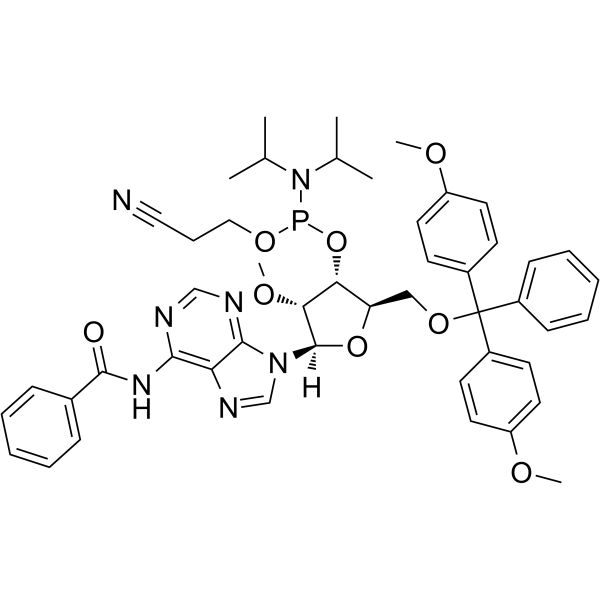
-
GC62531
2’-OMe-G(ibu) Phosphoramidite
2’-OMe-G(ibu) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
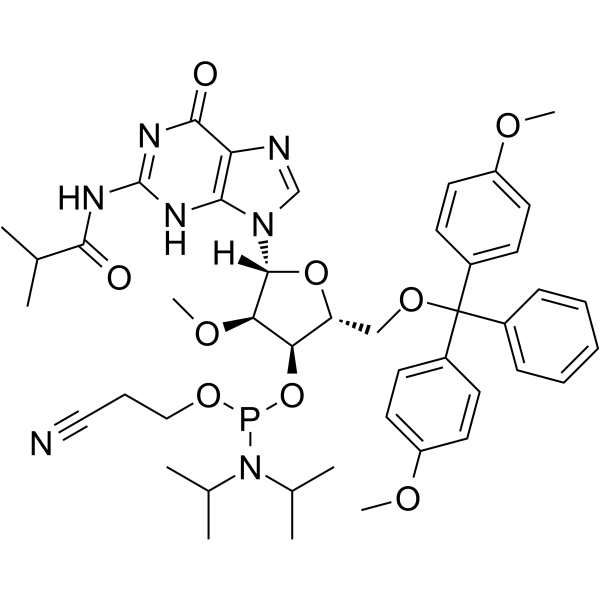
-
GC48440
2'-Deoxycytidine-5'-triphosphate (sodium salt)
2'-Deoxycytidine-5'-triphosphate (sodium salt) (dCTP trisodium salt) is a nucleoside triphosphate that can be used for DNA synthesis.
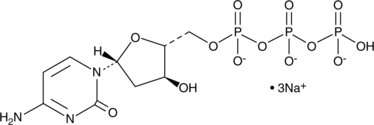
-
GC33430
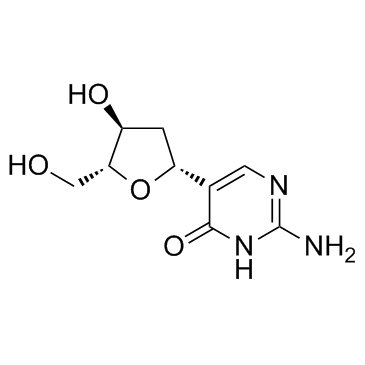
2'-Deoxypseudoisocytidine
2'-Deoxypseudoisocytidine is a nucleoside analogue.
-
GC35072
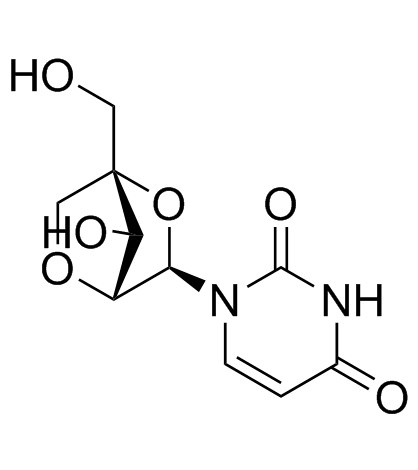
2'-O,4'-C-Methyleneuridine
2'-O,4'-C-Methyleneuridine (Compound 15a) is a bicyclic nucleoside.
-
GC64983
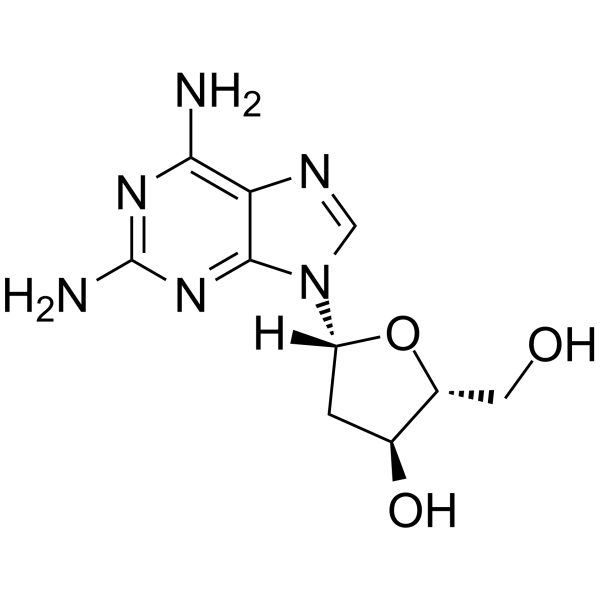
2-Amino-2'-deoxyadenosine
2-Amino-2'-deoxyadenosine is a deoxyribonucleoside used for the oligonucleotide synthesis.
-
GC65083
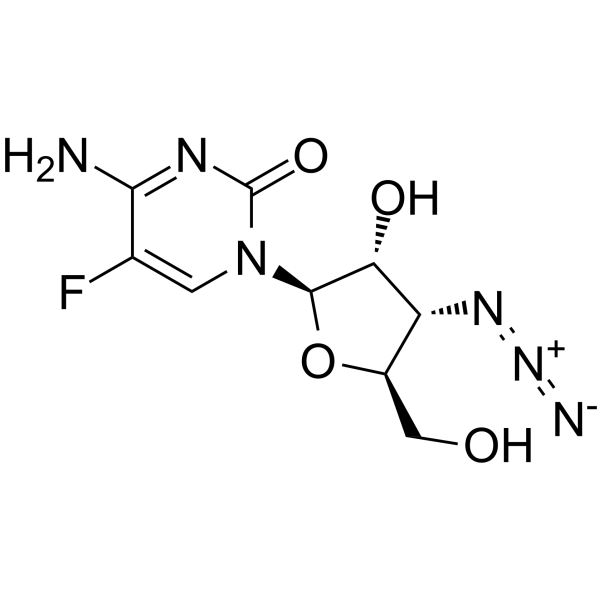
3'-Azido-3'-deoxy-5-fluorocytidine
3'-Azido-3'-deoxy-5-fluorocytidine (Compound 12) is a cytidine derivative.
-
GC64985
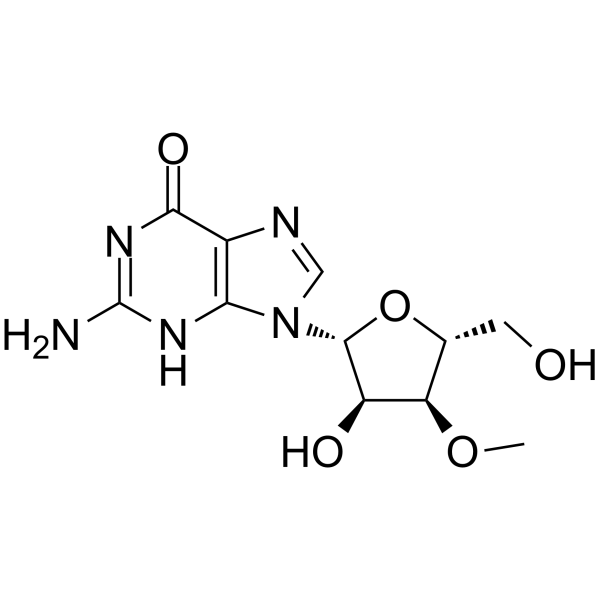
3'-O-Methylguanosine
3'-O-Methylguanosine is a methylated nucleoside analogs and a RNA chain terminator.
-
GC35105
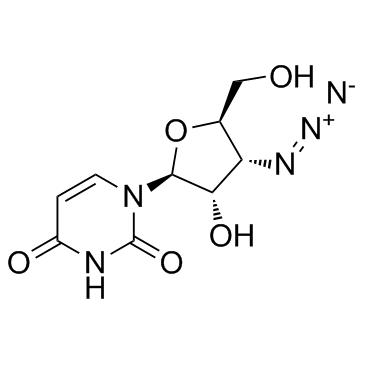
3'-Azido-3'-deoxy-beta-L-uridine
3'-Azido-3'-deoxy-beta-L-uridine (Compound 25) is a nucleoside derivative.
-
GC60023
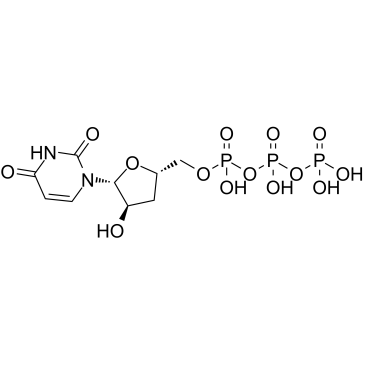
3'-Deoxyuridine-5'-triphosphate
3'-Deoxyuridine-5'-triphosphate (3'-dUTP) is a nucleotide analogue that inhibits DNA-dependent RNA polymerases I and II.
-
GC61862
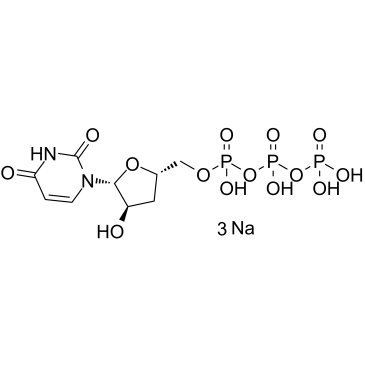
3'-Deoxyuridine-5'-triphosphate trisodium
3'-Deoxyuridine-5'-triphosphate trisodium (3'-dUTP trisodium) is a nucleotide analogue that inhibits DNA-dependent RNA polymerases I and II.
-
GC65084
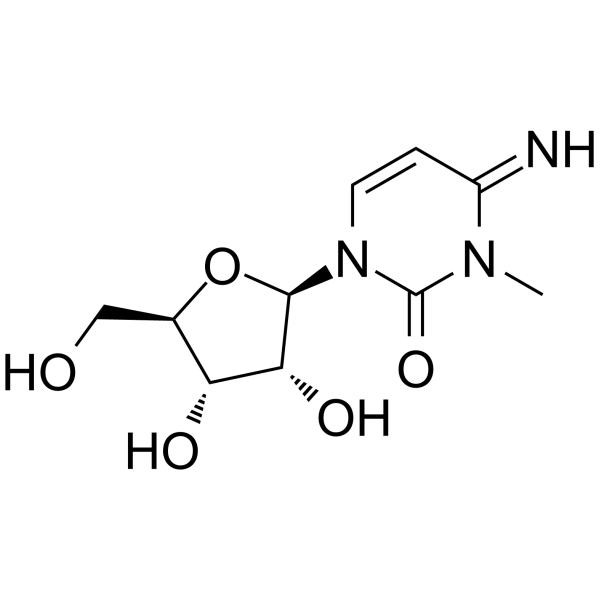
3-Methylcytidine
3-Methylcytidine, a urinary nucleoside, can be used as a biomarker of four different types of cancer: lung cancer, gastric cancer, colon cancer, and breast cancer.
-
GC68557
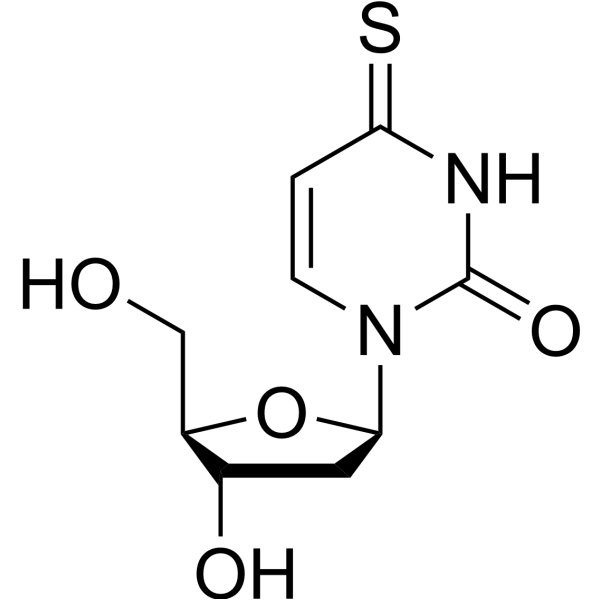
4-Thio-2'-deoxyuridine
4-Thio-2'-deoxyuridine is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity, targeting inert lymphatic system malignancies. The anticancer mechanism in this process depends on inhibiting DNA synthesis and inducing cell apoptosis.
-
GC42474
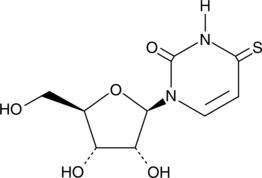
4-Thiouridine
4-Thiouridine (4-SU) is a photoactivatable ribonucleoside analog that is widely used for RNA analysis, including short-range RNA-RNA crosslinking and nascent RNA labeling.
-
GC65520
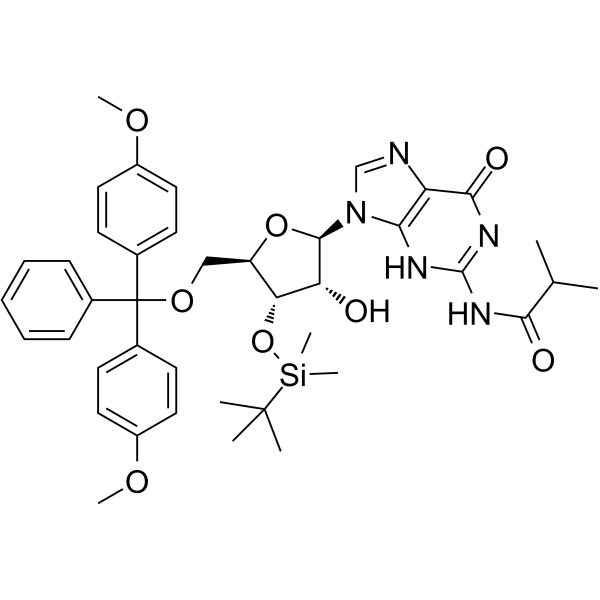
5'-DMT-3'-TBDMS-ibu-rG
5'-DMT-3'-TBDMS-ibu-rG is is a modified nucleoside.
-
GC65402
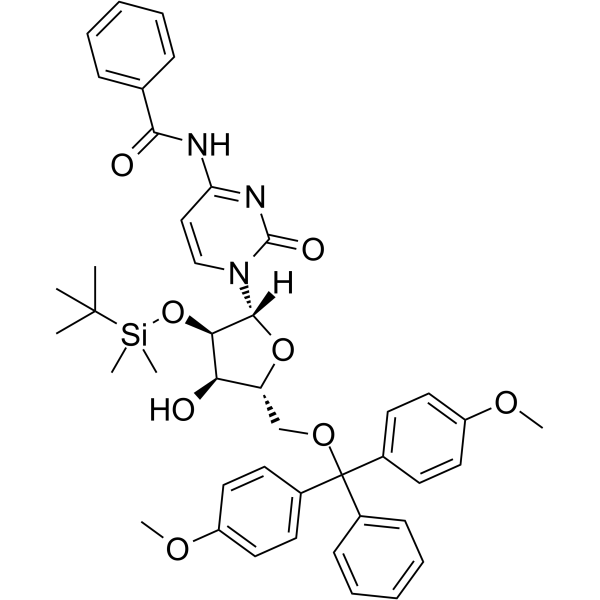
5'-O-DMT-2'-O-TBDMS-Bz-rC
5'-O-DMT-2'-O-TBDMS-Bz-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC65164
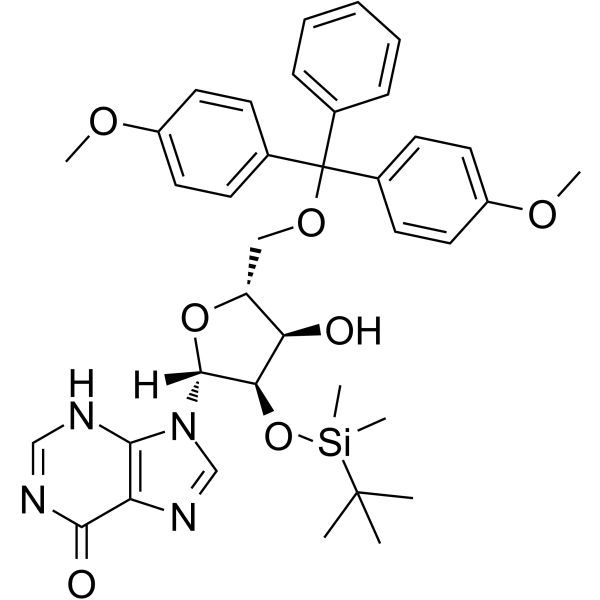
5'-O-DMT-2'-O-TBDMS-rI
5'-O-DMT-2'-O-TBDMS-rI is a modified nucleoside.
-
GC65396
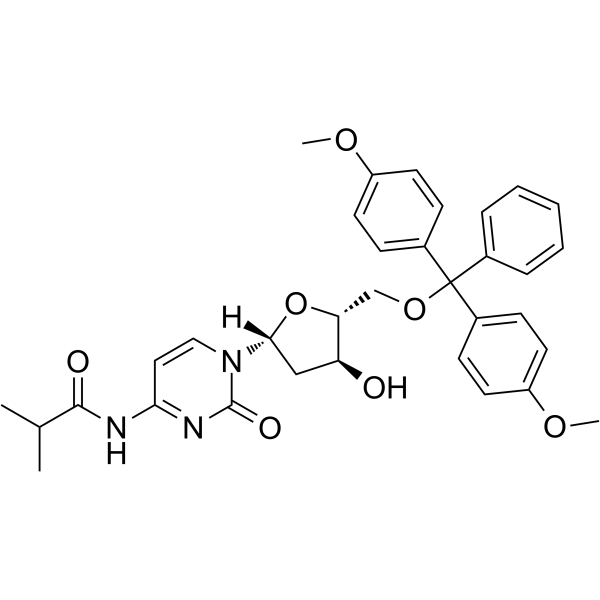
5'-O-DMT-ibu-dC
5'-O-DMT-ibu-dC can be used in the synthesis of oligodeoxyribonucleotides.
-
GC64982
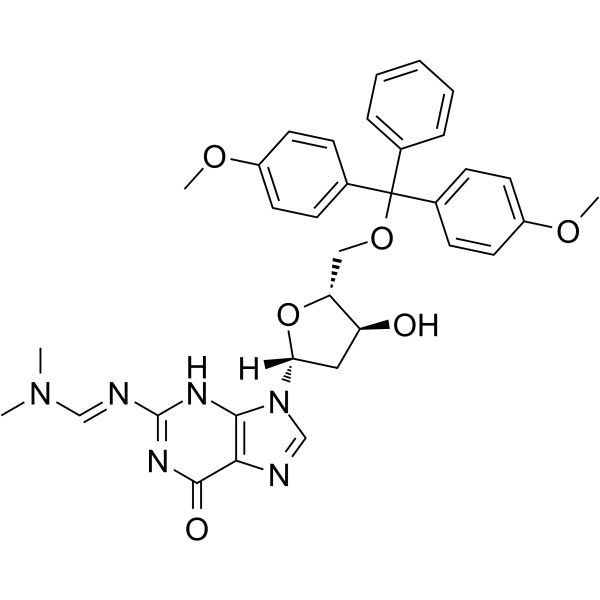
5'-O-DMT-N2-DMF-dG
5'-O-DMT-2'-O-TBDMS-rI is a modified nucleoside.
-
GC65482
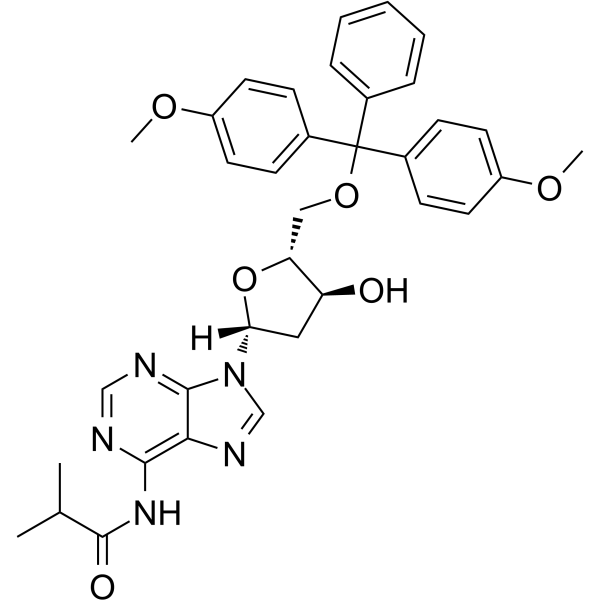
5'-O-DMT-N6-ibu-dA
5'-O-DMT-N6-ibu-dA can be used in the synthesis of oligodeoxyribonucleotides.
-
GC65562
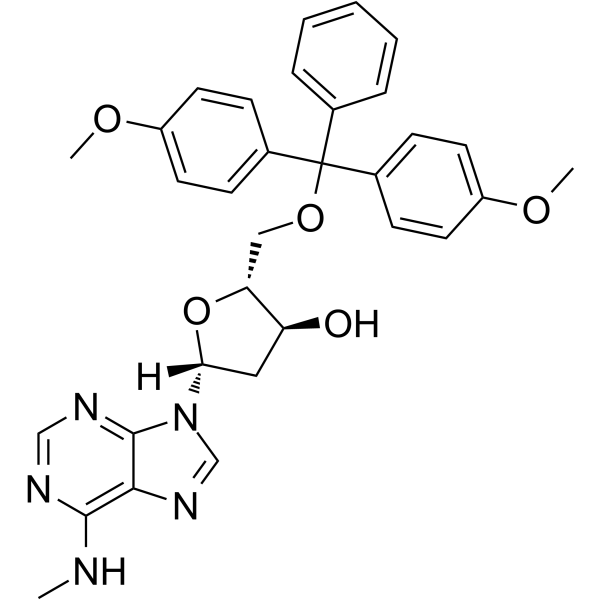
5'-O-DMT-N6-Me-2'-dA
5'-O-DMT-N6-Me-2'-dA is a nucleoside with protective and modification effects.
-
GC65401
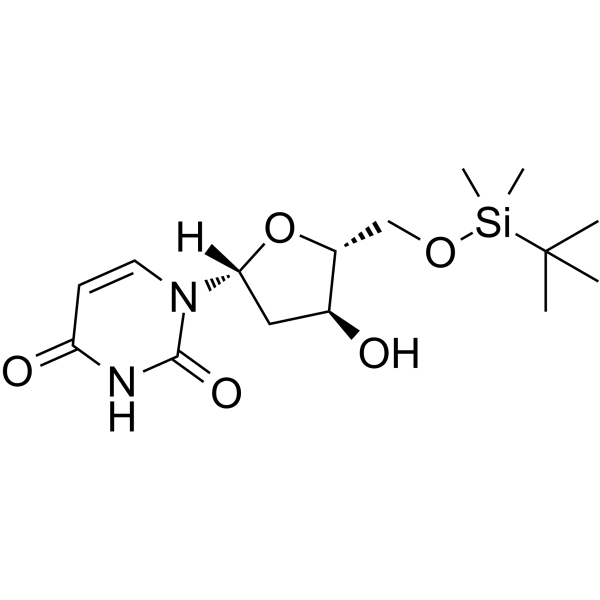
5'-O-TBDMS-dU
5'-O-TBDMS-dU can be used in the synthesis of oligoribonucleotides.
-
GC62157
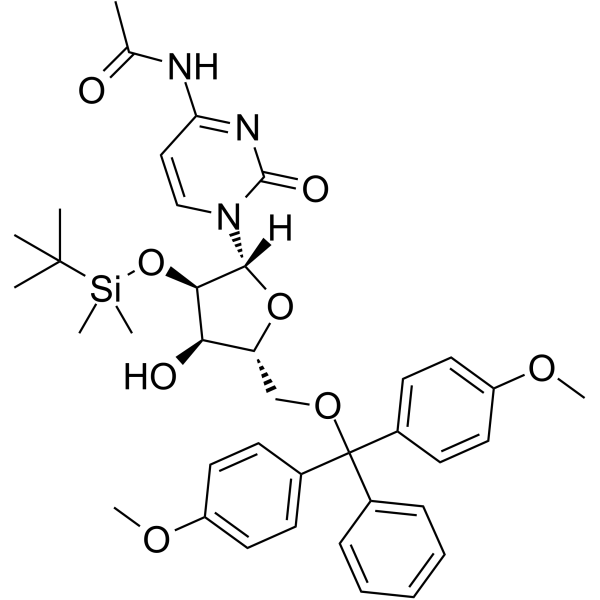
5’-O-DMT-2’-O-TBDMS-Ac-rC
5’-O-DMT-2’-O-TBDMS-Ac-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62149
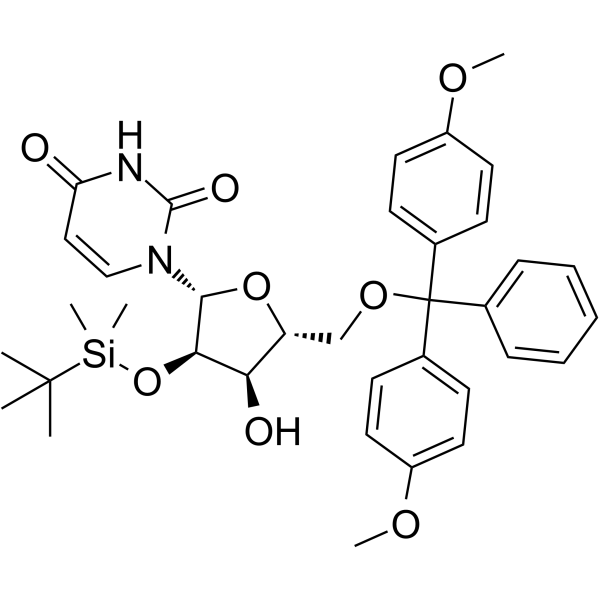
5’-O-DMT-2’-TBDMS-Uridine
5’-O-DMT-2’-TBDMS-Uridine is a deoxyribonucleoside used for the oligonucleotide synthesis.
-
GC62532
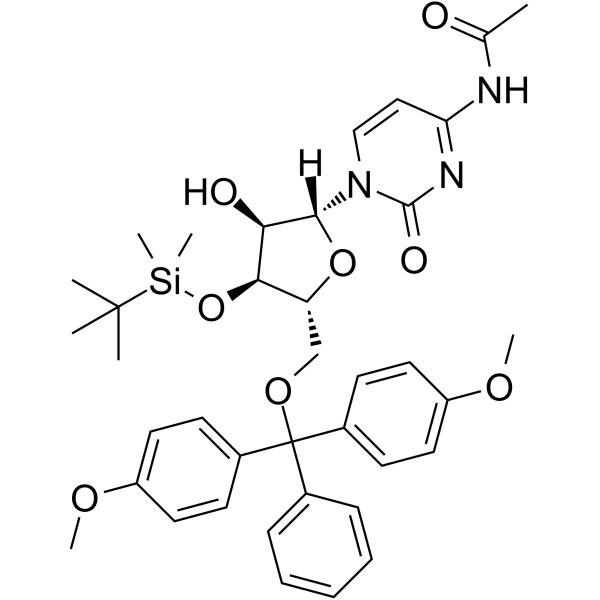
5’-O-DMT-3’-O-TBDMS-Ac-rC
5’-O-DMT-3’-O-TBDMS-Ac-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62573
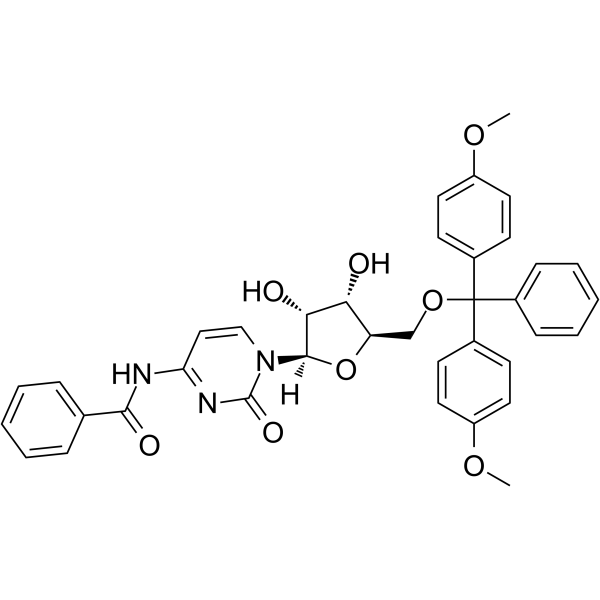
5’-O-DMT-Bz-rC
5'-O-DMT-Bz-Rc is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62574
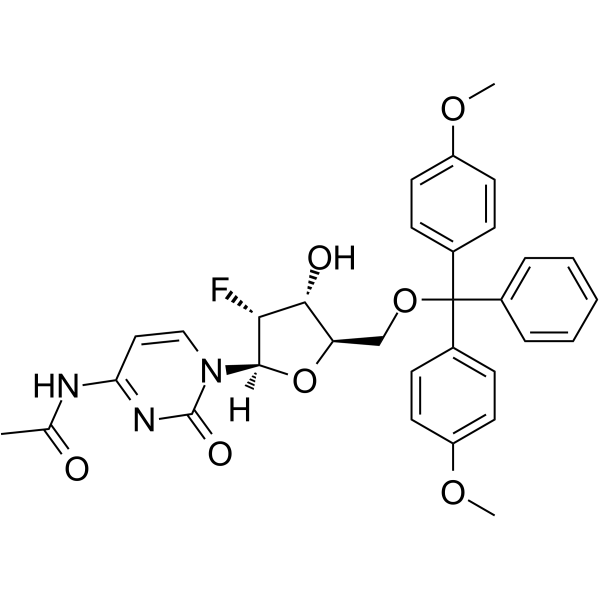
5’-O-DMT-N4-Ac-2’-F-dC
5’-O-DMT-N4-Ac-2’-F-dC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62575
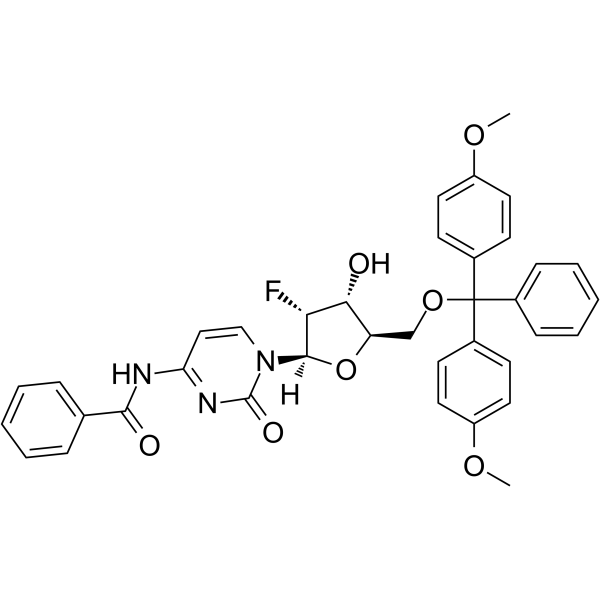
5’-O-DMT-N4-Bz-2’-F-dC
5’-O-DMT-N4-Bz-2’-F-dC is a nucleoside with protective and modification effects.
-
GC62576
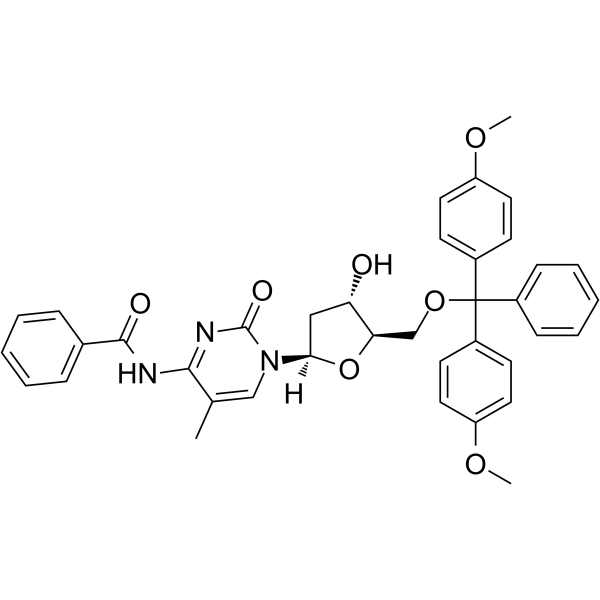
5’-O-DMT-N4-Bz-5-Me-dC
5’-O-DMT-N4-Bz-5-Me-dC is a modified nucleoside.
-
GC62577
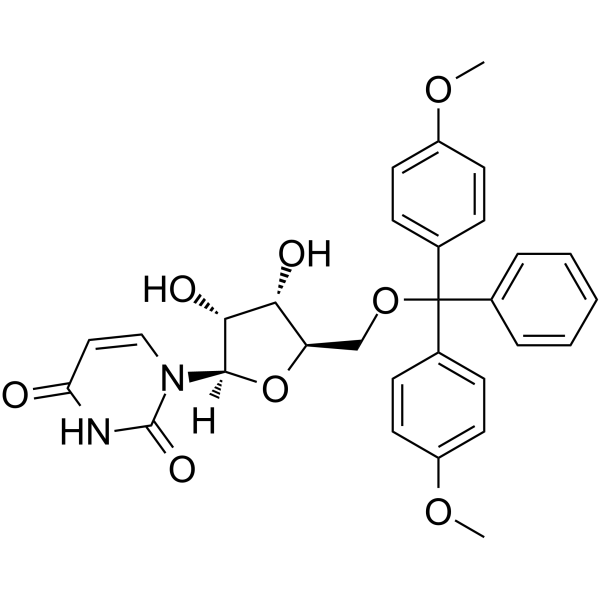
5’-O-DMT-rU
5’-O-DMT-rU is a modified nucleoside and can be used to synthesize RNA.
-
GC62578
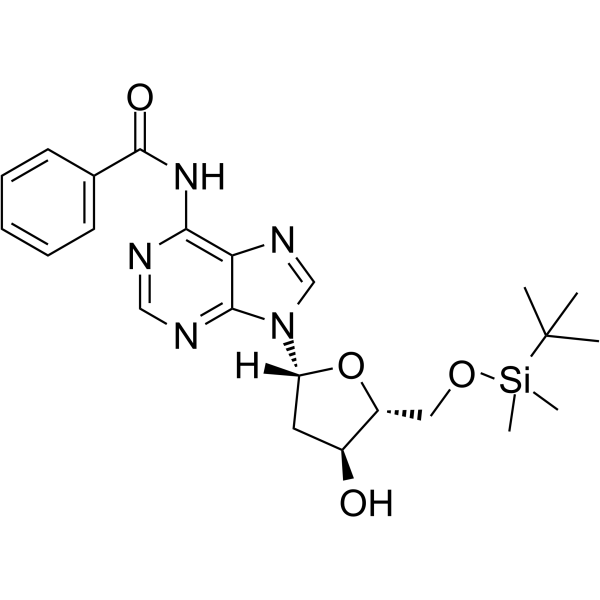
5’-O-TBDMS-Bz-dA
5’-O-TBDMS-Bz-dA is a nucleoside with protective and modification effects.
-
GC62579
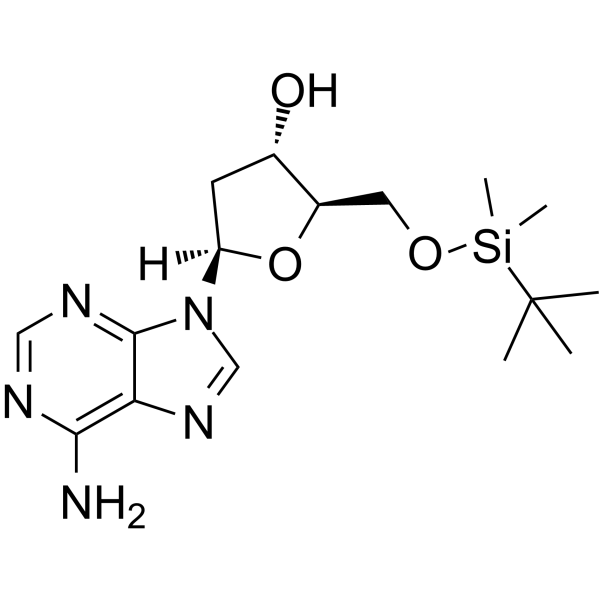
5’-O-TBDMS-dA
5’-O-TBDMS-dA is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62580
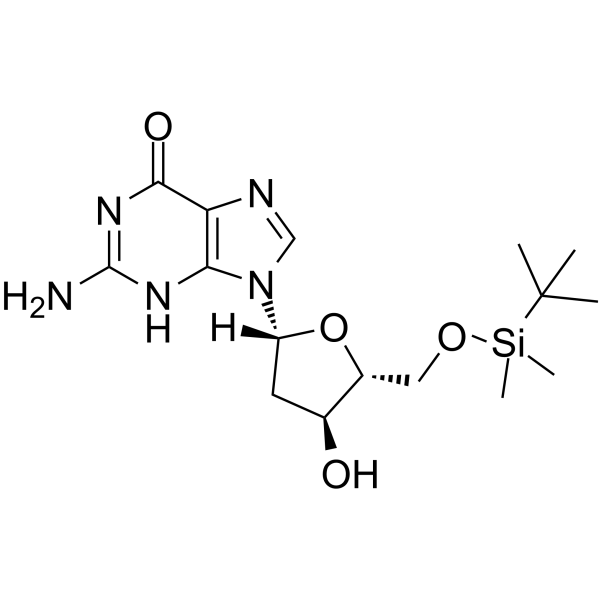
5’-O-TBDMS-dG
5’-O-TBDMS-dG is a modified nucleoside.
-
GC62581
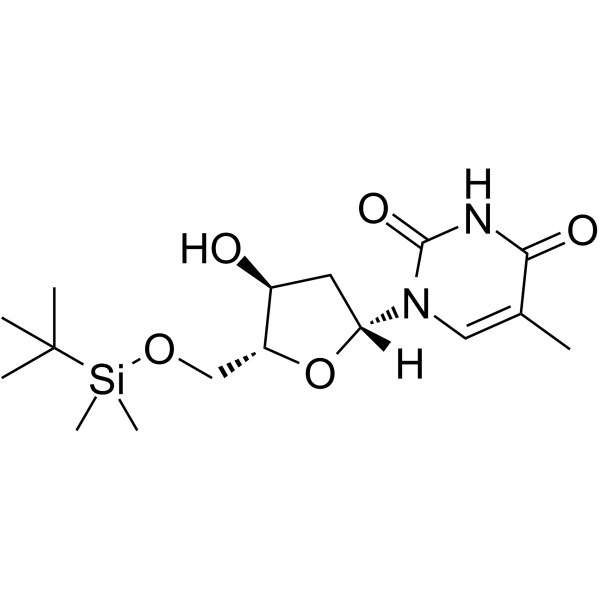
5’-O-TBDMS-dT
5’-O-TBDMS-dT is a nucleoside with protective and modification effects.
-
GC61432
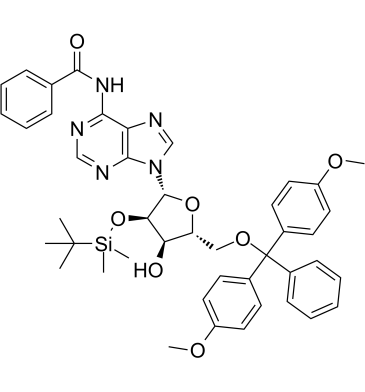
5'-O-DMT-2'-O-TBDMS-N-Bz-Adenosine
5'-O-DMT-2'-O-TBDMS-N-Bz-Adenosine?is an adenosine derivative and can be used as an intermediate for oligonucleotides synthesis.
-
GC64952
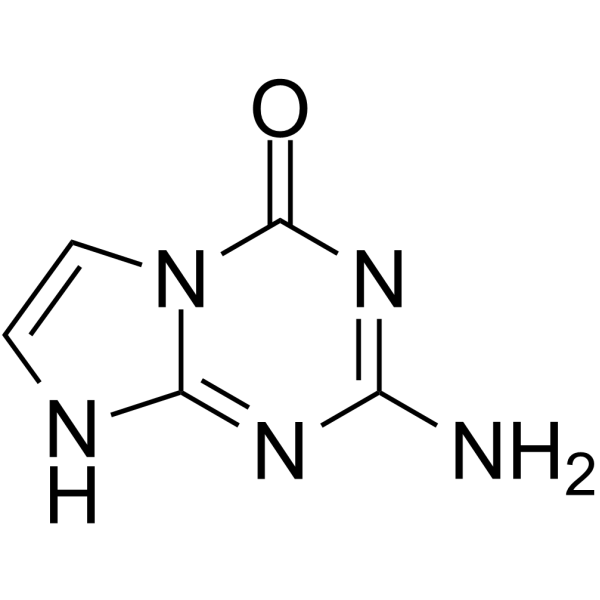
5-Aza-7-deazaguanine
5-Aza-7-deazaguanine is a substrate for wild-type (WT) E. coli purine nucleoside phosphorylase and its Ser90Ala mutant in the synthesis of base-modified nucleosides.
-
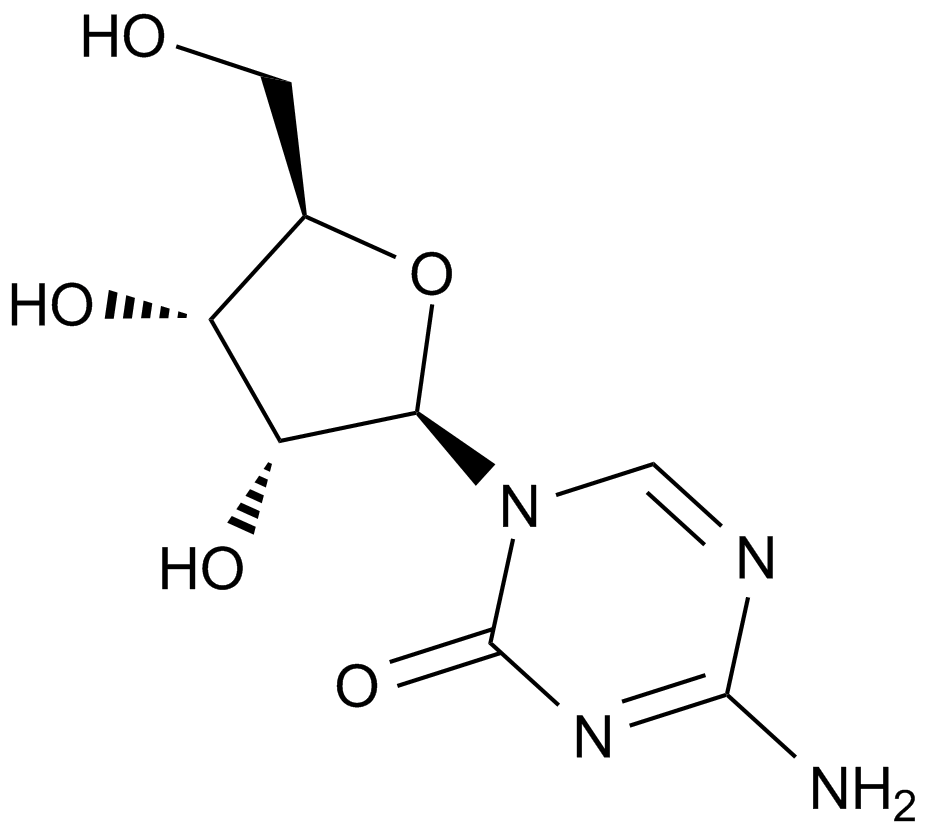
GC10946
5-Azacytidine
A DNA methyltransferase inhibitor
-
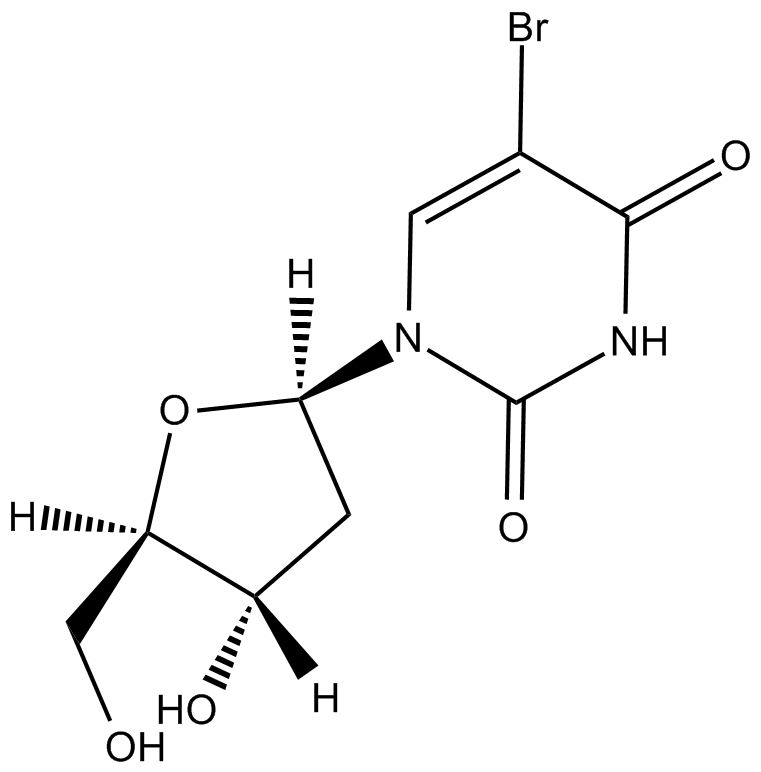
GC11940
5-BrdU
Synthetic thymidine analog
-
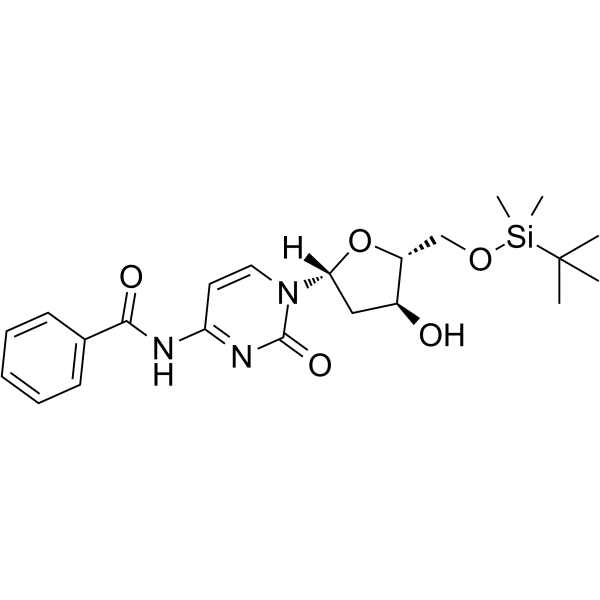
GC62547
5-O-TBDMS-N4-Benzoyl-2-deoxycytidine
5-O-TBDMS-N4-Benzoyl-2-deoxycytidine is a modified nucleoside.
-
GC30548
6-Amino-5-nitropyridin-2-one
6-Amino-5-nitropyridin-2-one is a pyridine base and used as a nucleobase of hachimoji DNA, in which it pairs with 5-aza-7-deazaguanine.
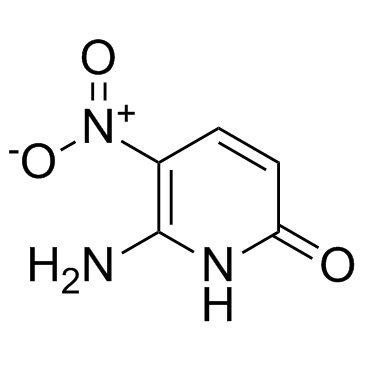
-
GC60031
6-Azathymine
6-Azathymine, a 6-nitrogen analog of thymine, is a potent D-3-aminoisobutyrate-pyruvate aminotransferase inhibitor.
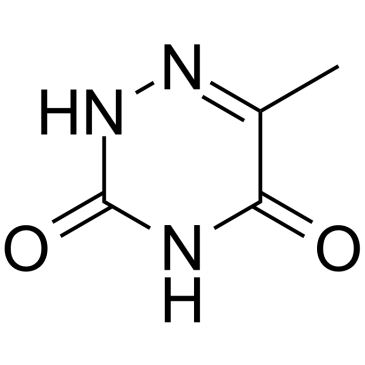
-
GC32975
6-Mercaptopurine hydrate
An inhibitor of purine synthesis and interconversion
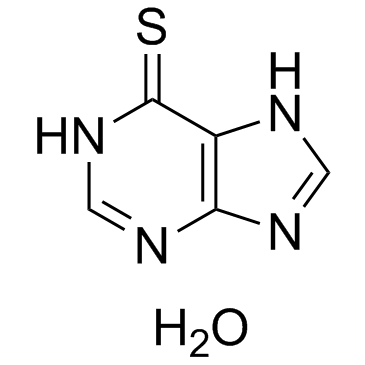
-
GC65082
6-O-Methyl Guanosine
6-O-Methyl Guanosine is a modified nucleoside.
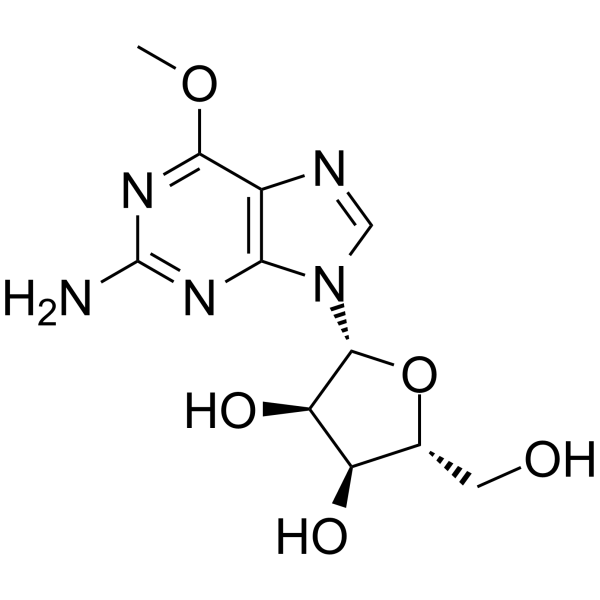
-
GC35171
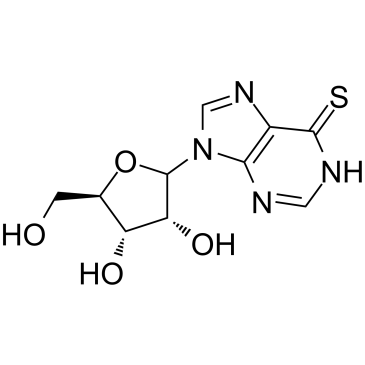
6-Thioinosine
6-Thioinosine (6TI) is a purine antimetabolite, acts as an anti-adipogenesis agent, downregulates mRNA levels of PPAR γ and C/EBPα, as well as PPAR γ target protein such as LPL, CD36, aP2, and LXRα.
-
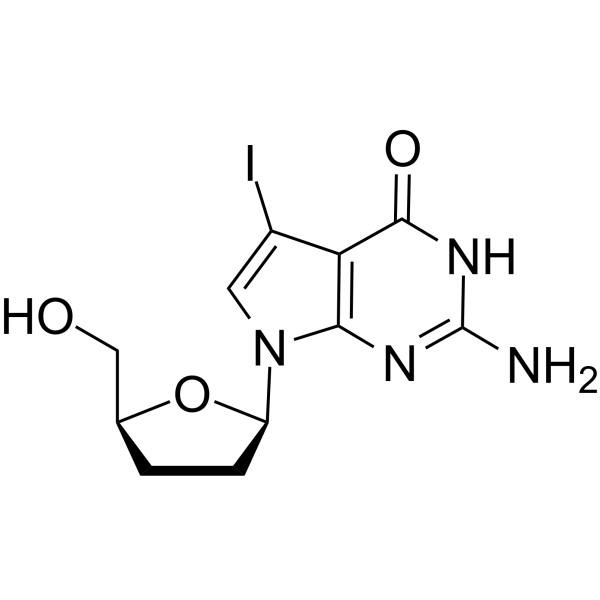
GC66060
7-Iodo-2',3'-dideoxy-7-deaza-guanosine
7-Iodo-2',3'-dideoxy-7-deaza-guanosine is a dideoxynucleoside that can be used in DNA synthesis and sequencing reactions.
-
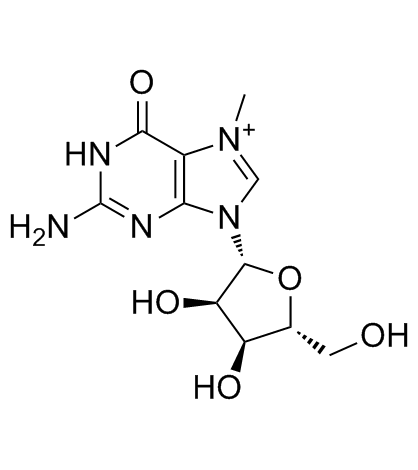
GC30531
7-Methylguanosine
A nucleotide analog
-
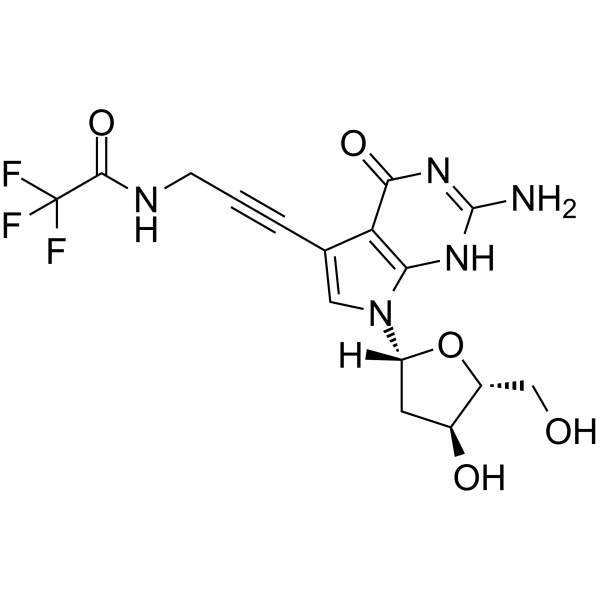
GC65525
7-TFA-ap-7-Deaza-dG
5'-O-TBDMS-dG is a modified nucleoside.
-
GC13805
Abacavir
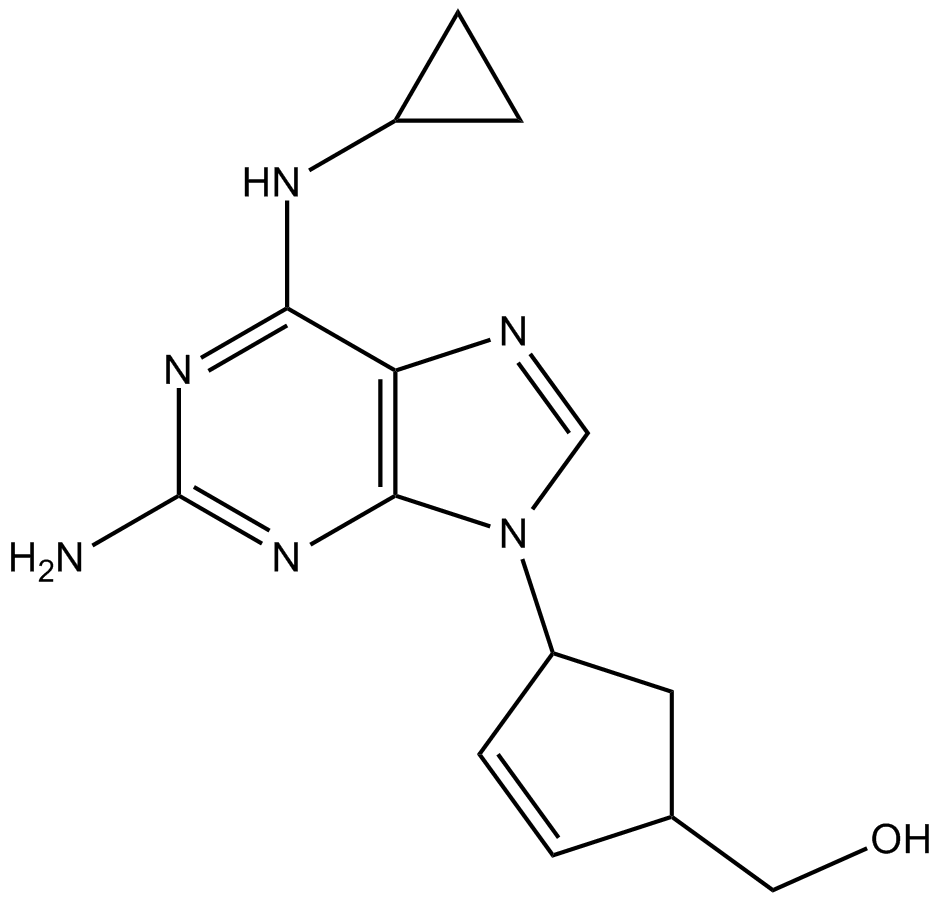
-
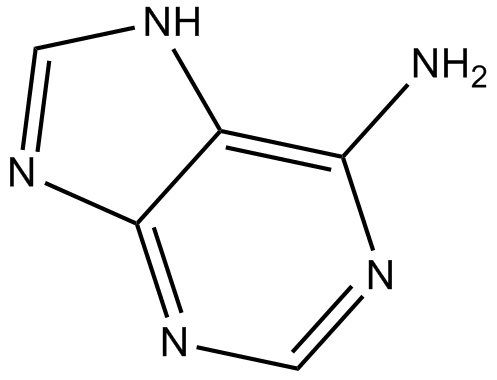
GC13432
Adenine
High affinity adenine receptor agonist
-
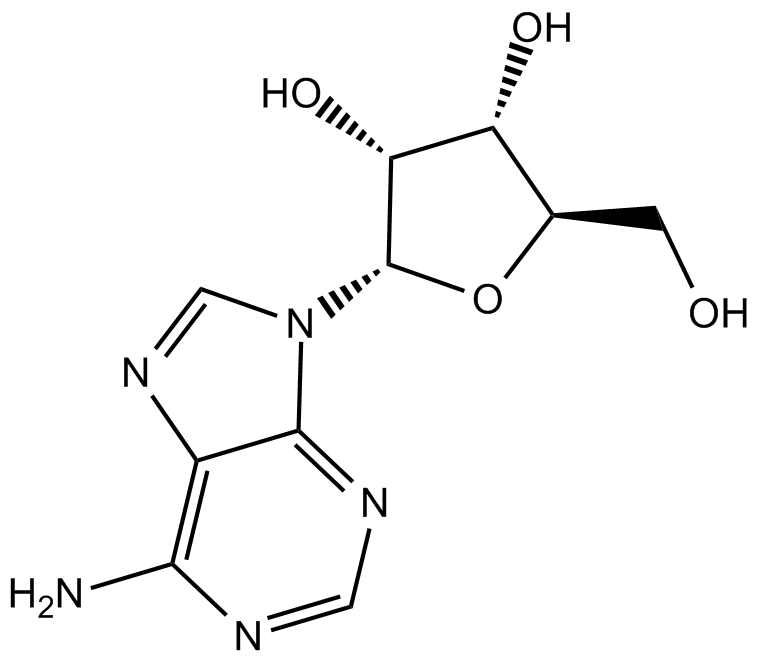
GC14106
Adenosine
nucleoside
-
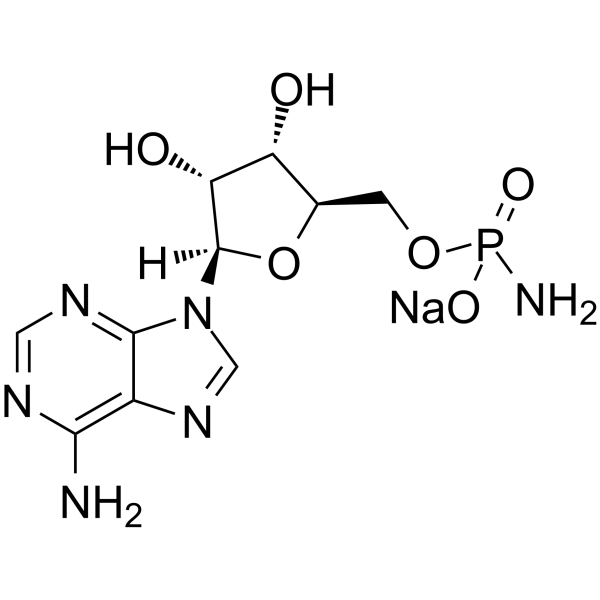
GC65462
Adenosine 5′-monophosphoramidate sodium
Adenosine 5′-monophosphoramidate sodium is an adenosine derivative and can be used as an intermediate for nucleotide synthesis.
-
GC11843
Azaguanine-8
Azaguanine-8 is a purine analogue that shows antineoplastic activity. Azaguanine-8 functions as an antimetabolite and easily incorporates into ribonucleic acids, interfering with normal biosynthetic pathways, thus inhibiting cellular growth.
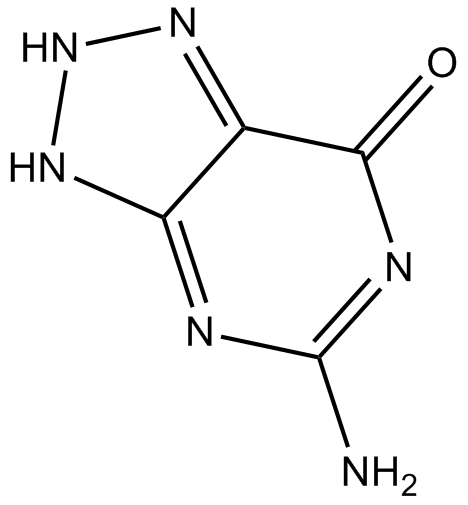
-
GC15866
Capecitabine
DNA, RNA and protein synthesis inhibitor
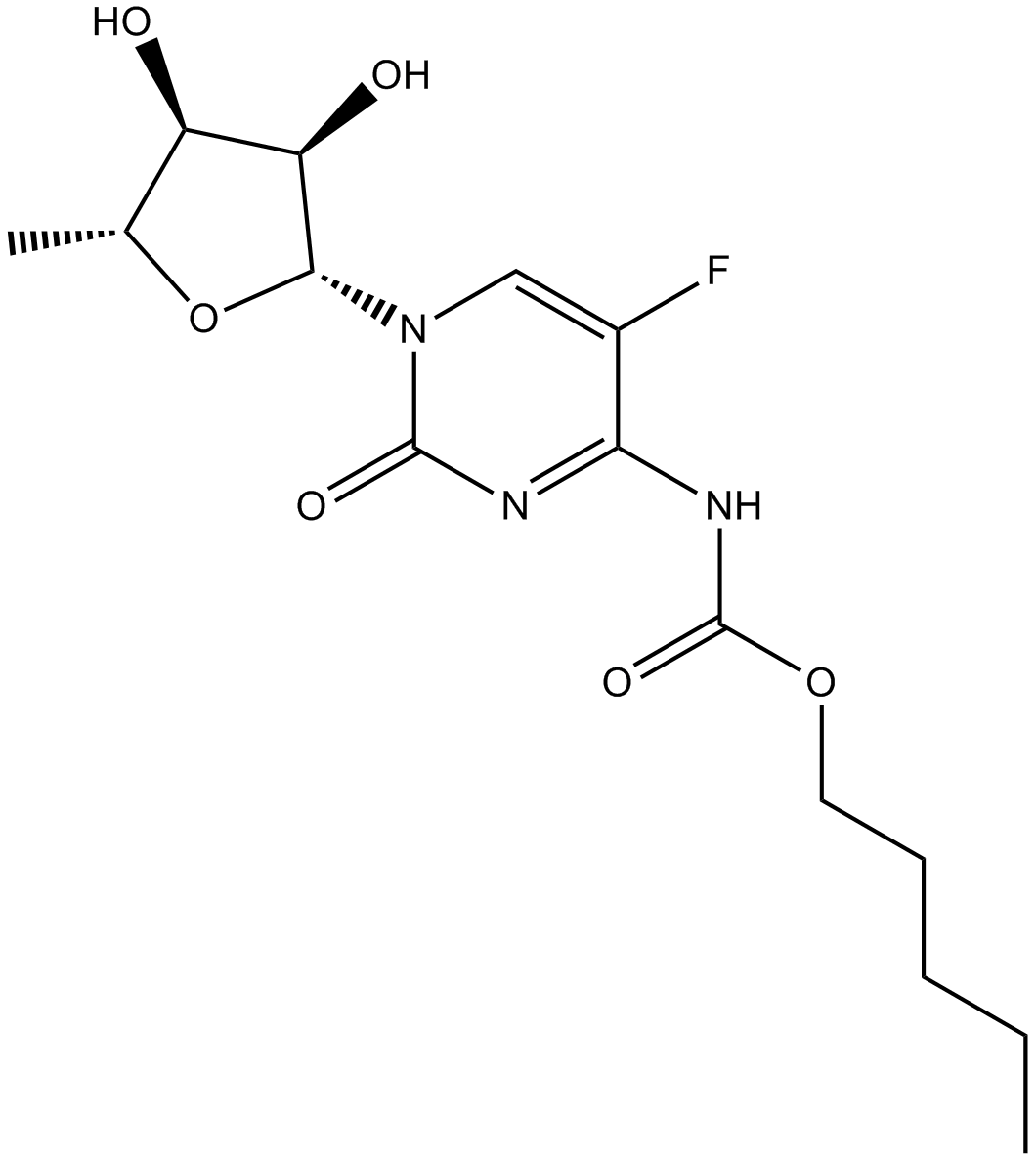
-
GC12748
Carmofur
Cytostatic derivative of fluorouracilm,antineoplatic agent
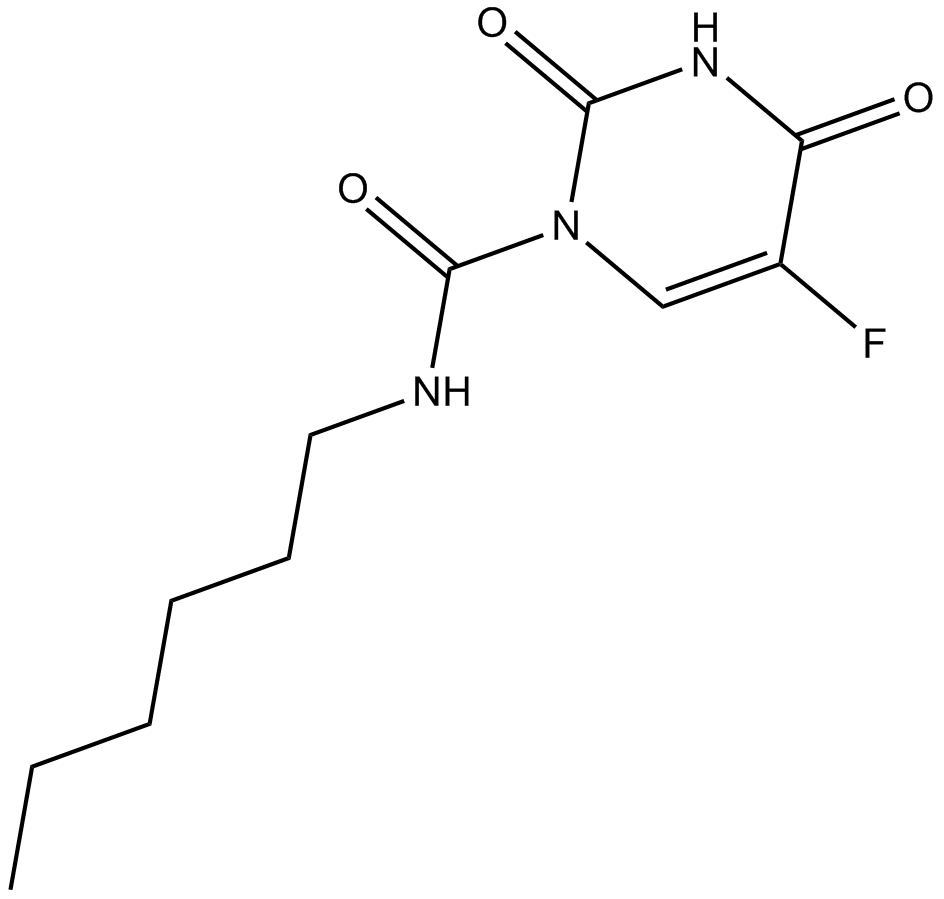
-
GC64261
Censavudine
Censavudine (OBP-601; BMS-986001), a nucleoside analog, is a nucleoside reverse transcriptase inhibitor.
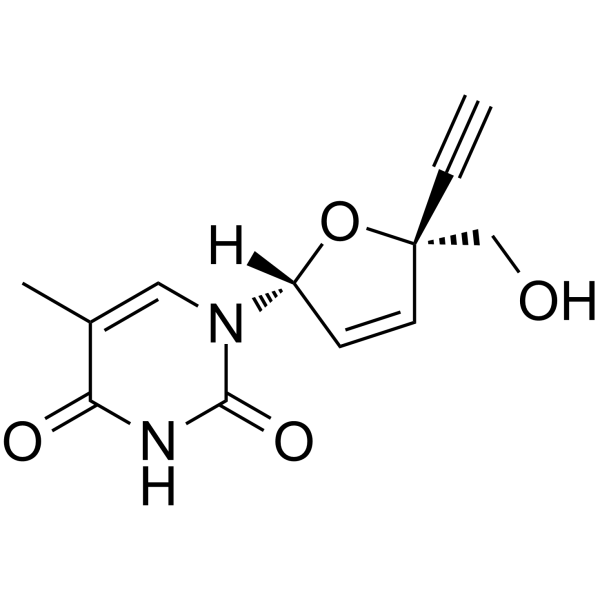
-
GC35693
CI 972 (anhydrous)
CI 972 (anhydrous) is a potent, orally active, and competitive inhibitor of purine nucleoside phosphorylase (PNP) (Ki=0.83 μM) under development as a T cell-selective immunosuppressive agent.
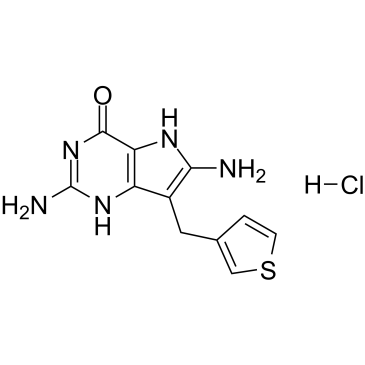
-
GC15219
Clofarabine
Antimetabolite,inhibit DNA polymerase and ribonucleotide reductase
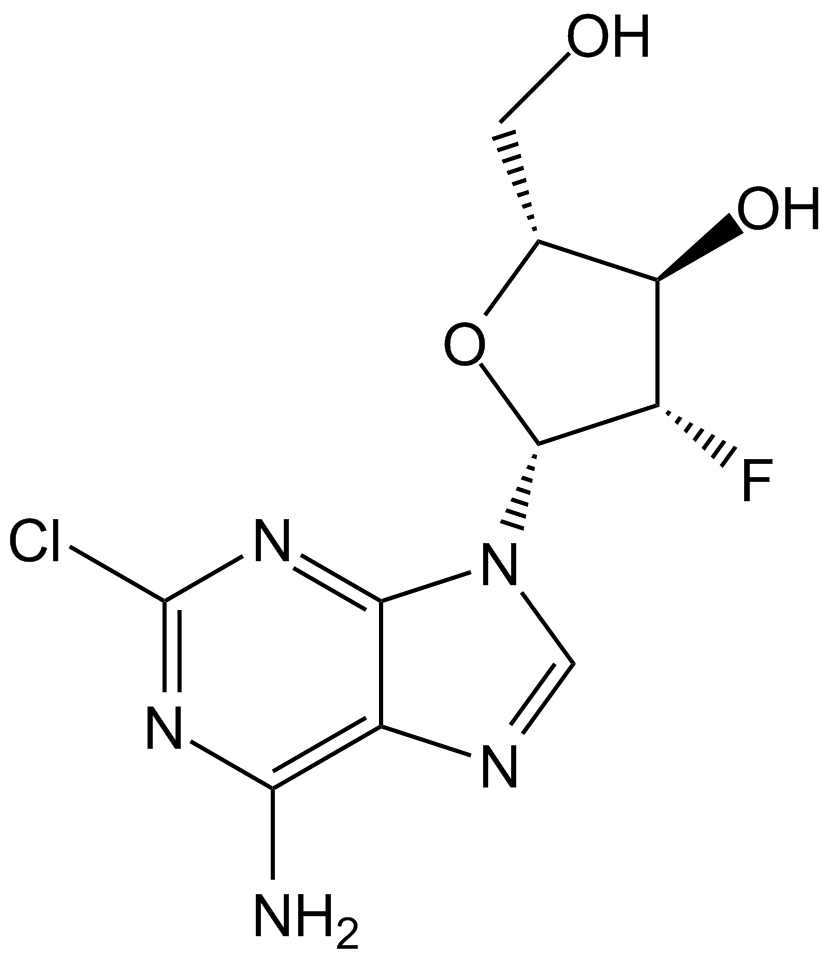
-
GC33177
CNDAC
CNDAC is a major metabolite of oral drug sapacitabine, and a nucleoside analog.
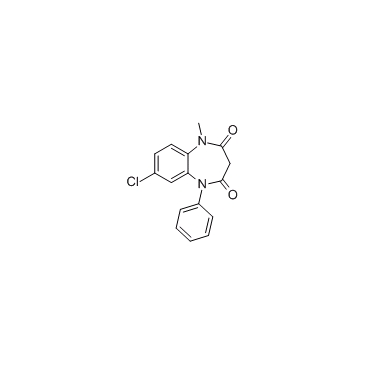
-
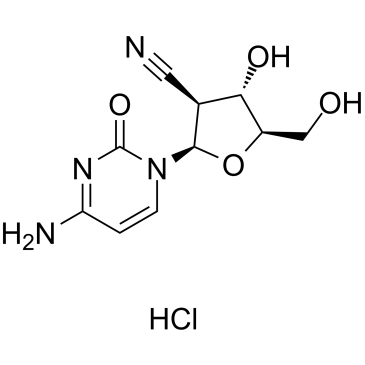
GC38382
CNDAC hydrochloride
CNDAC hydrochloride is a metabolite of the orally active agent sapacitabine, and a nucleoside analog.
-
GC13070
Cytarabine
Cytotoxic agent, blocks DNA synthesis
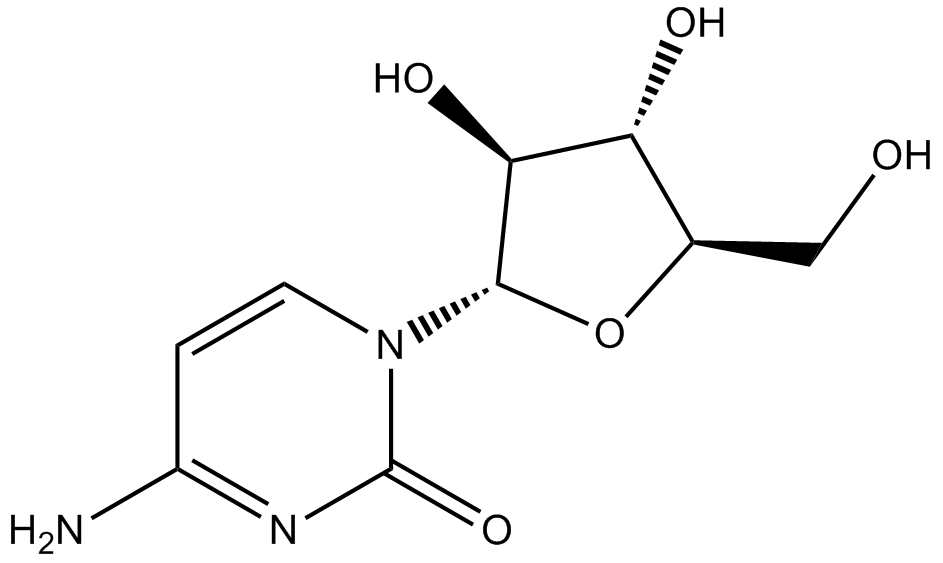
-
GC14961
Cytarabine hydrochloride
DNA synthsis inhibitor
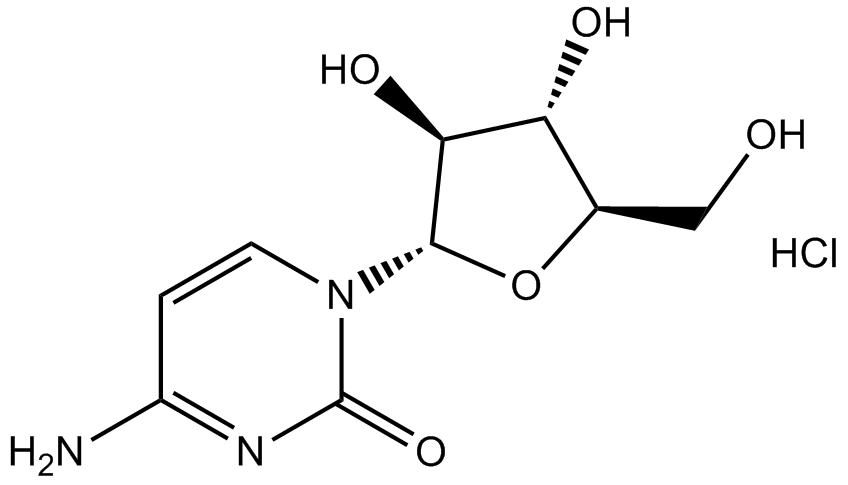
-
GC13729
Cytidine
pyrimidine nucleoside
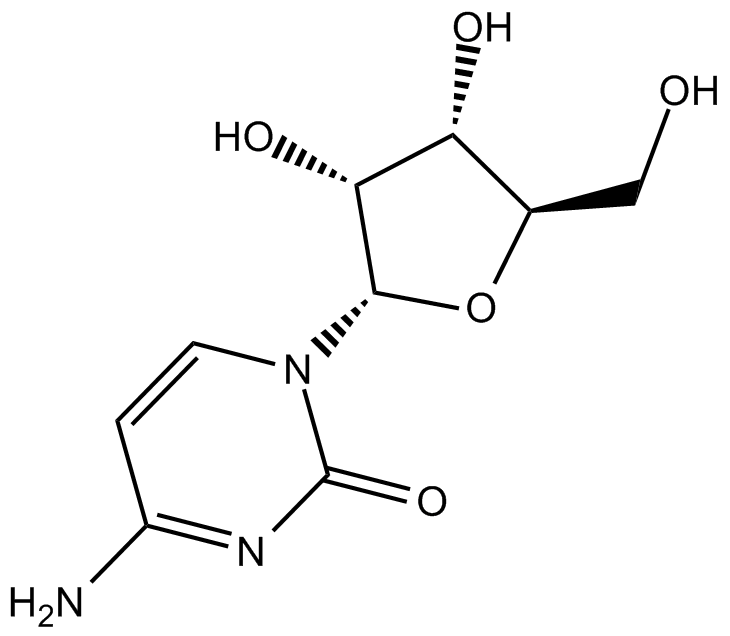
-
GC14485
Dacarbazine
Antineoplastic( malignant melanoma and sarcomas)
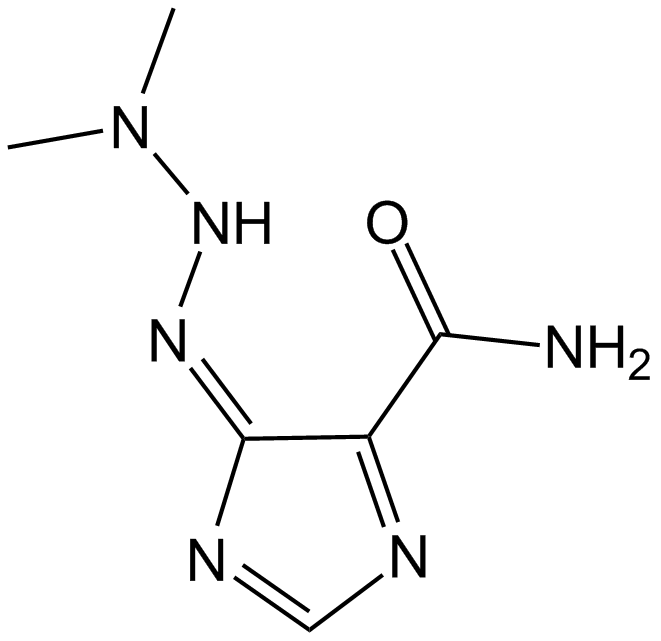
-
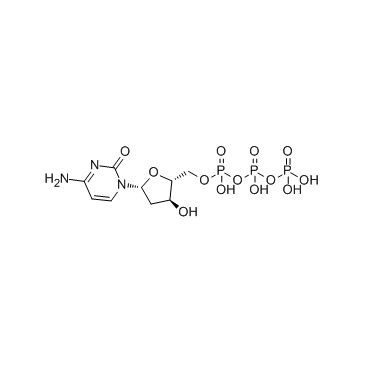
GC34123
Deoxycytidine triphosphate (dCTP)
Deoxycytidine triphosphate (dCTP) (dCTP) is a nucleoside triphosphate that can be used for DNA synthesis.
-
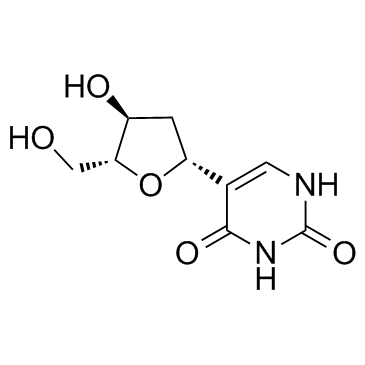
GC33494
Deoxypseudouridine
Deoxypseudouridine is a nucleoside analog.
-
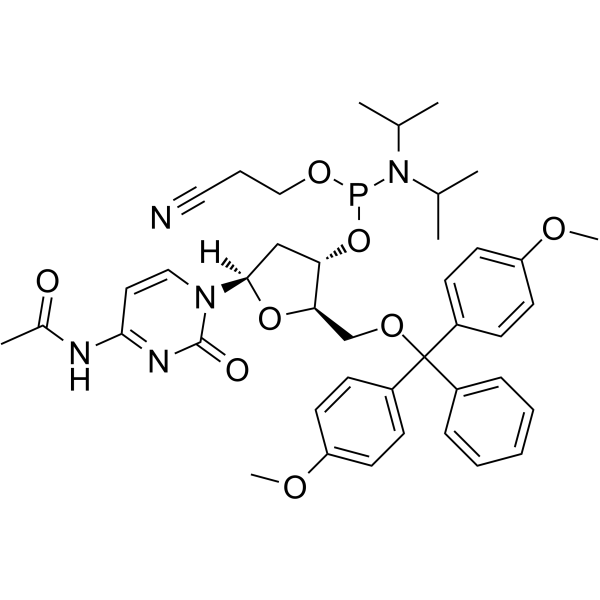
GC62150
DMT-dC(ac) Phosphoramidite
DMT-dC(ac) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
-
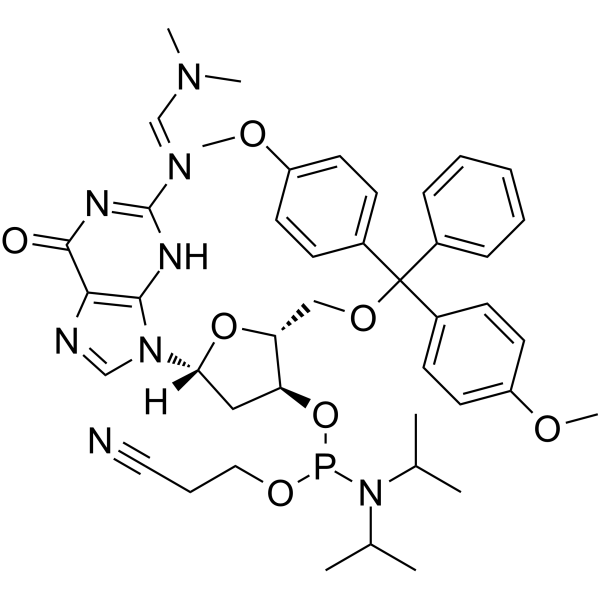
GC62538
DMT-dG(dmf) Phosphoramidite
DMT-dG(dmf) Phosphoramidite is a phosphinamide monomer that can be used in the preparation of oligonucleotides
-
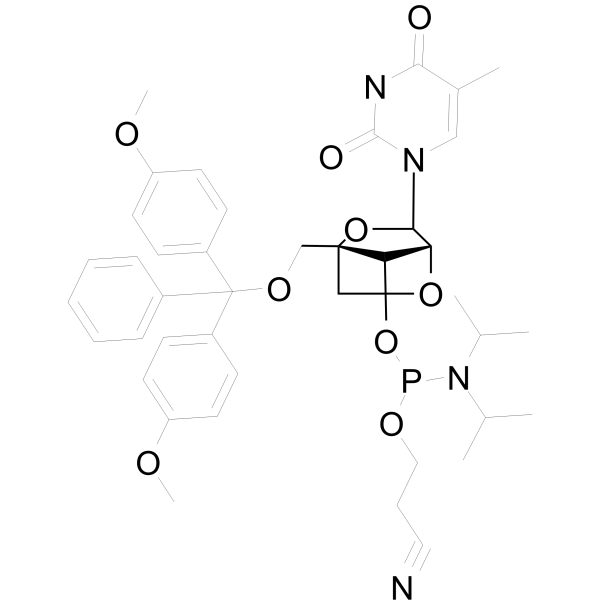
GC66711
DMTr-LNA-5MeU-3-CED-phosphoramidite
DMTr-LNA-5MeU-3-CED-phosphoramidite is a nucleoside derivative.
-
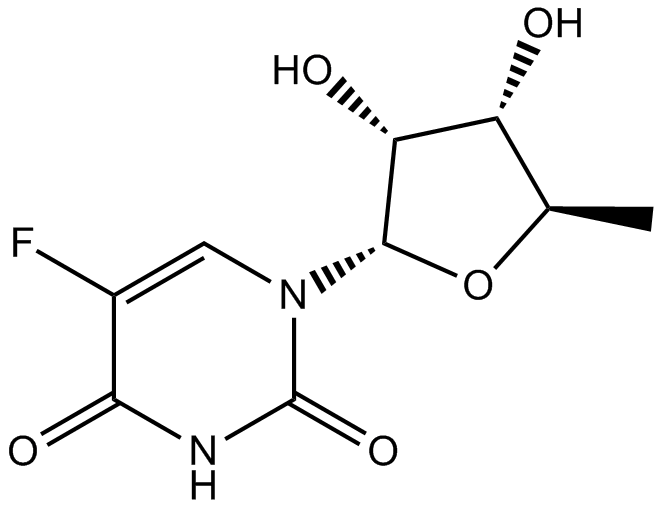
GC11099
Doxifluridine
oral prodrug of the antineoplastic agent 5-fluorouracil (5-FU)
-
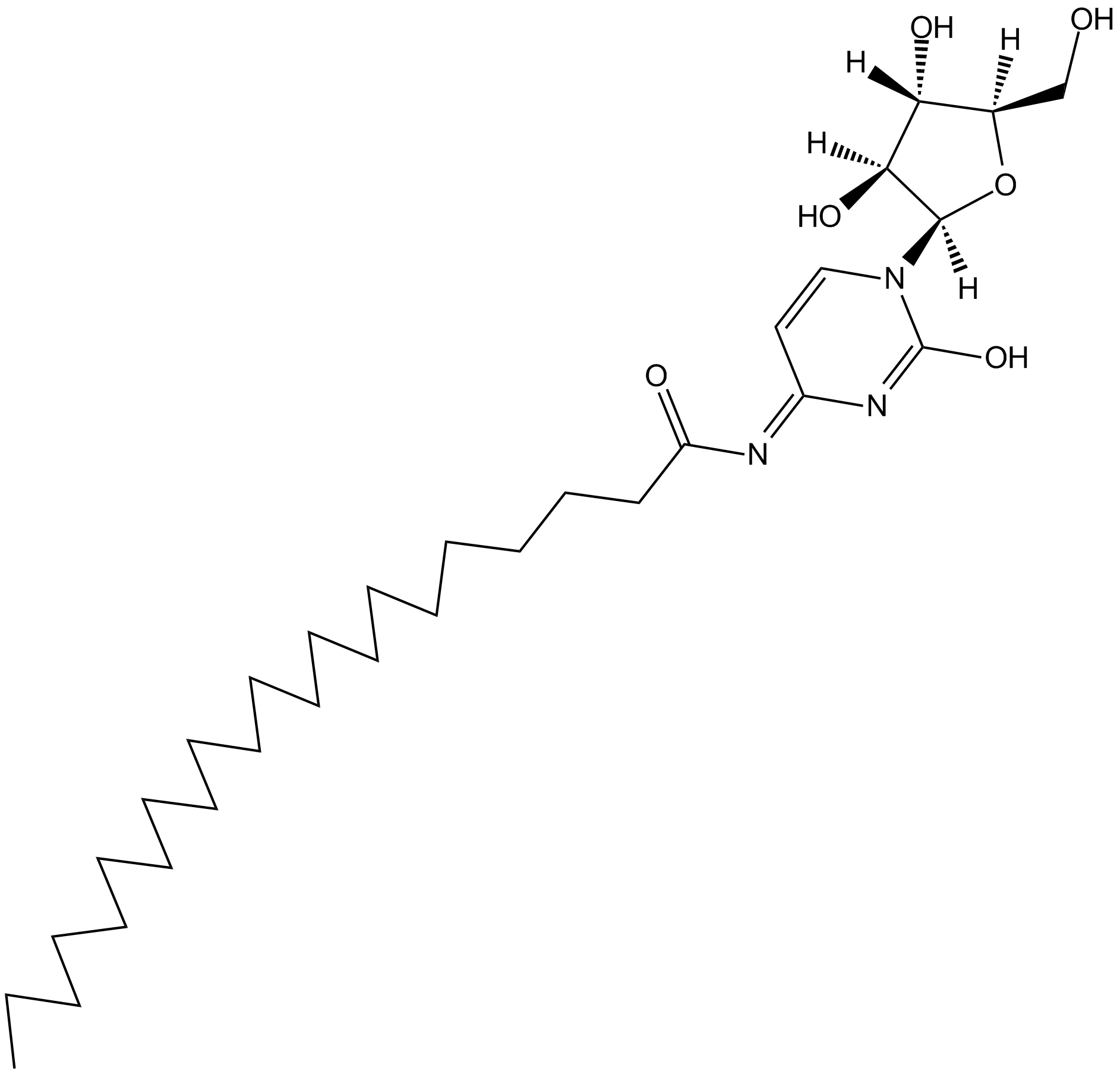
GC11654
Enocitabine
nucleoside analog used as chemotherapy
-
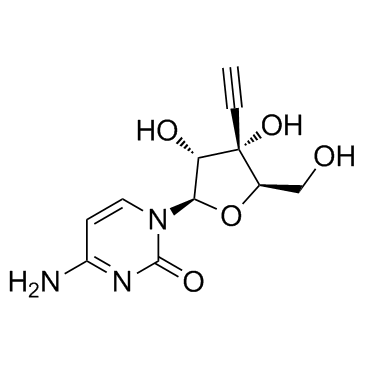
GC34112
Ethynylcytidine (ECyD)
Ethynylcytidine (ECyD) (ECyD), a nucleoside analog and a potent inhibitor of RNA synthesis, inhibits RNA polymerases I, II and II. Ethynylcytidine (ECyD) has robust antitumor activity in a wide range of models of cancer.
-
GC18014
Floxuridine
Antineoplastic antimetabolite
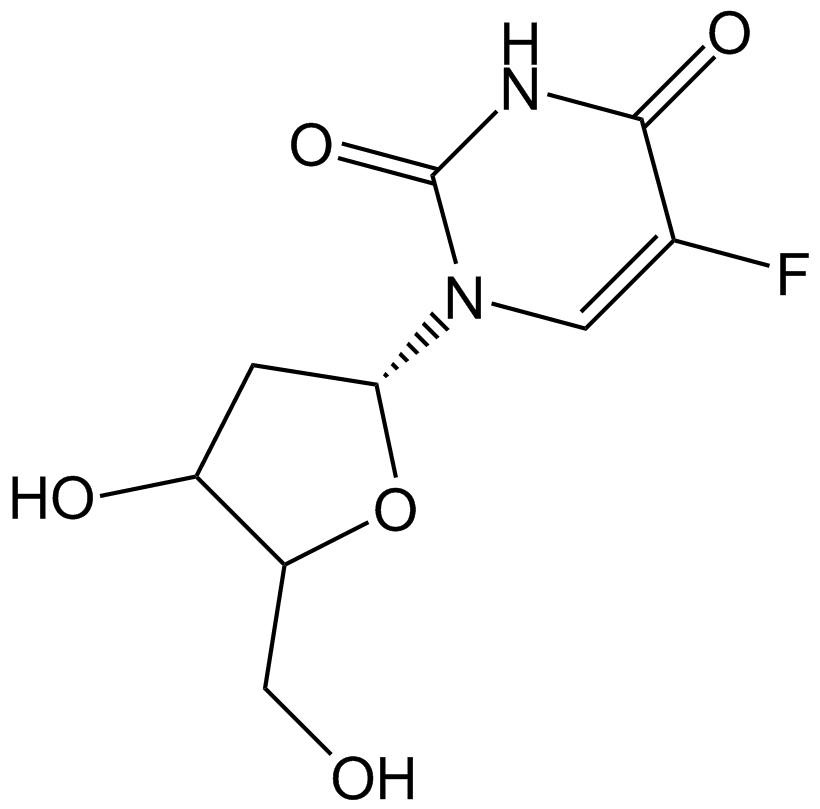
-
GC14144
Fludarabine
DNA synthsis inhibitor
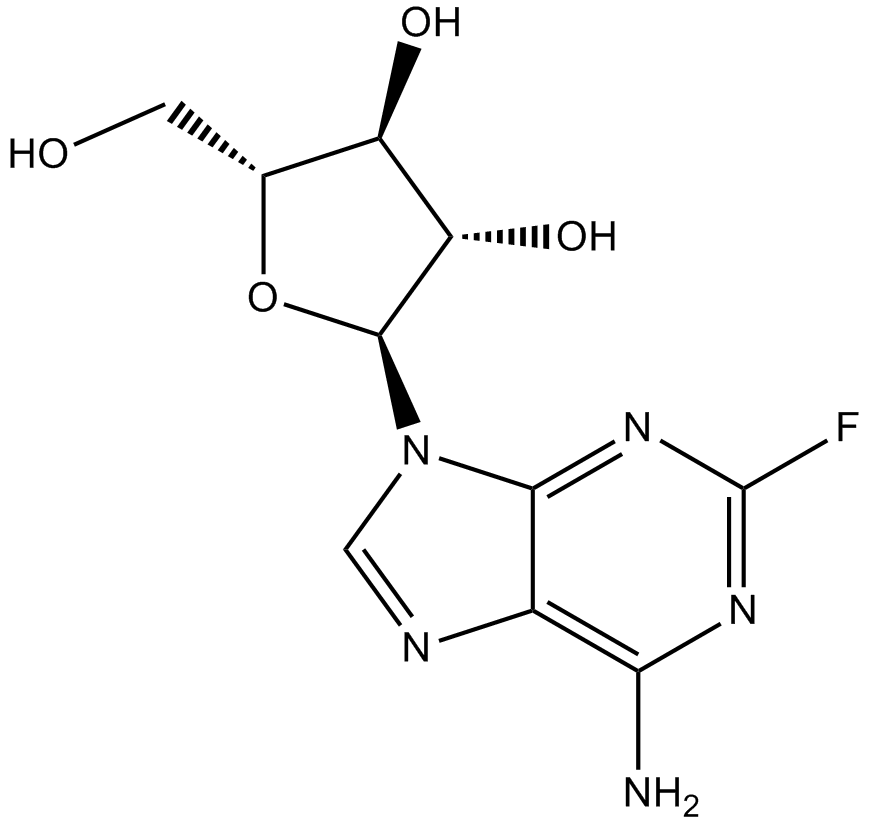
-
GC15134
Fludarabine Phosphate (Fludara)
Fludarabine (phosphate) is an analogue of adenosine and deoxyadenosine, which is able to compete with dATP for incorporation into DNA and inhibit DNA synthesis.
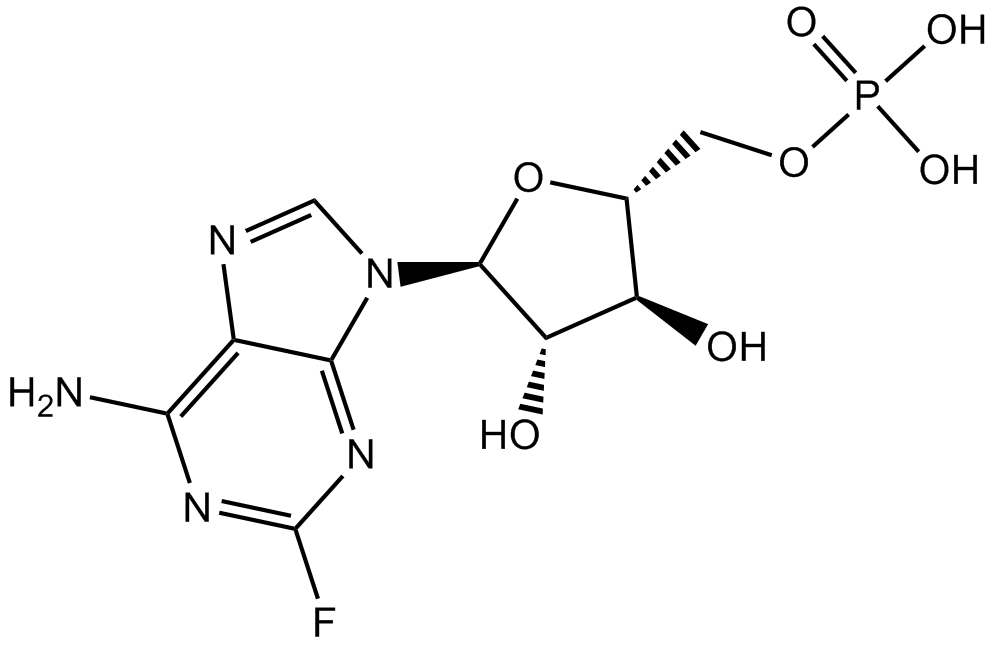
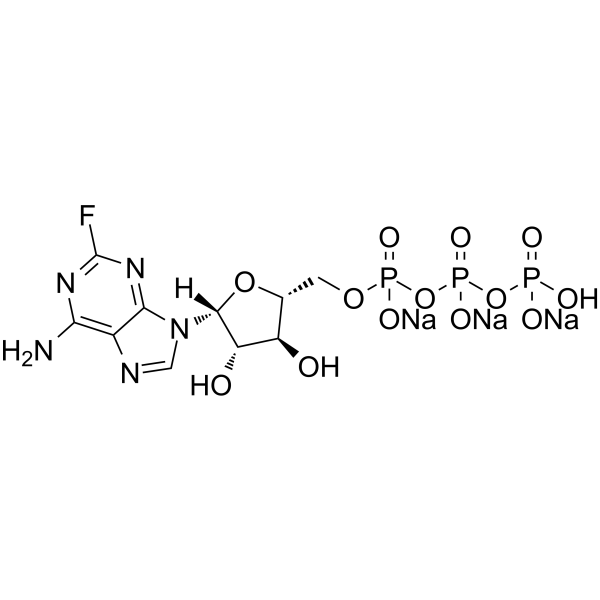
-
GC66430
Fludarabine triphosphate trisodium
Fludarabine triphosphate (F-ara-ATP) trisodium, the active metabolite of Fludarabine, is a potent, noncompetitive and specific inhibitor of DNA primase, with an IC50 of 2.3 μM and a Ki of 6.1 μM. Fludarabine triphosphate trisodium inhibits DNA synthesis by blocking DNA primase and primer RNA formation. Fludarabine triphosphate trisodium inhibits ribonucleotide reductase and DNA polymerase and ultimately leads to cellular apoptosis.
-
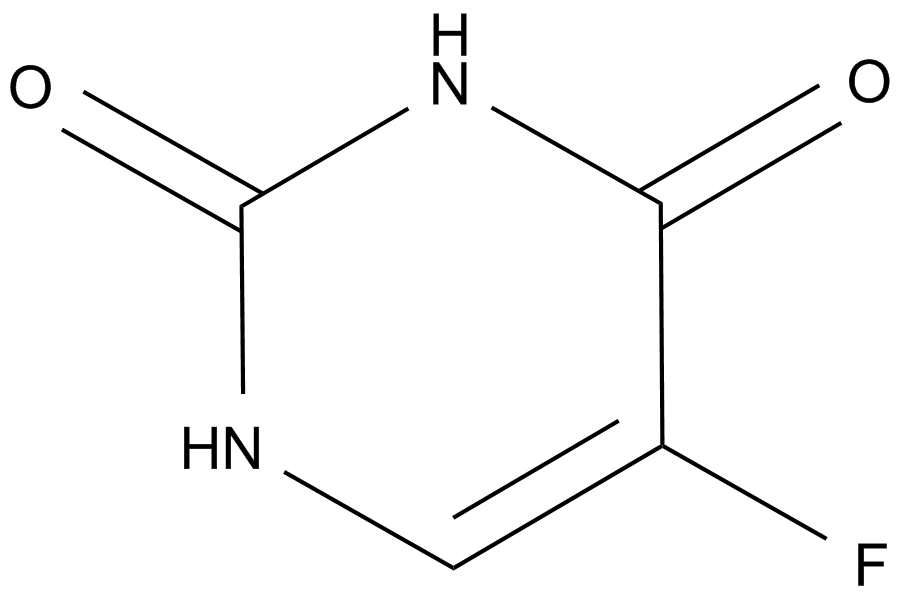
GC14466
Fluorouracil (Adrucil)
A prodrug form of FdUMP
-
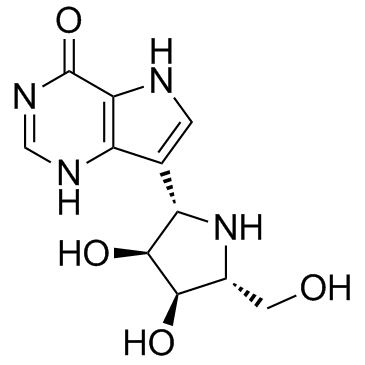
GC33029
Forodesine (BCX-1777 freebase)
Forodesine (BCX-1777 freebase) (BCX-1777) is a highly potent and orally active purine nucleoside phosphorylase (PNP) inhibitor with IC50 values ranging from 0.48 to 1.57 nM for human, mouse, rat, monkey and dog PNP. Forodesine (BCX-1777 freebase) is a potent human lymphocyte proliferation inhibitor. Forodesine (BCX-1777 freebase) could induce apoptosis in leukemic cells by increasing the dGTP levels.
-
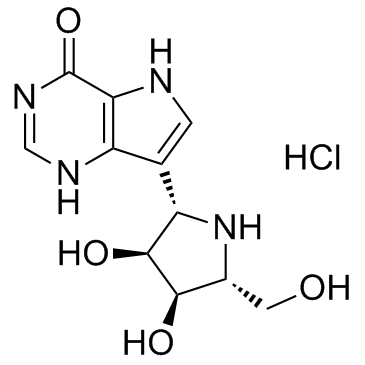
GC32708
Forodesine hydrochloride (BCX-1777)
Forodesine hydrochloride (BCX-1777) (BCX-1777 hydrochloride) is a highly potent and orally active purine nucleoside phosphorylase (PNP) inhibitor with IC50 values ranging from 0.48 to 1.57 nM for human, mouse, rat, monkey and dog PNP. Forodesine hydrochloride (BCX-1777) is a potent human lymphocyte proliferation inhibitor. Forodesine hydrochloride (BCX-1777) could induce apoptosis in leukemic cells by increasing the dGTP levels.
-
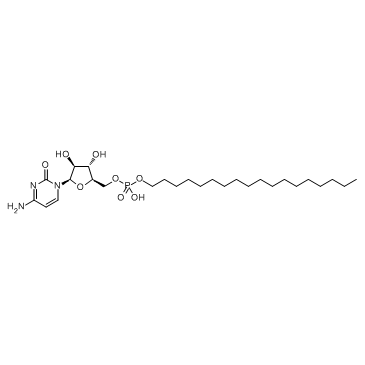
GC36071
Fosteabine
Fosteabine is an oral and prodrug analogue of cytarabine which is resistant to deoxycytidine deaminase.
-
GC13831
FT-207 (NSC 148958)
FT-207 (NSC 148958) (FT 207; NSC 148958) is a chemotherapeutic 5-FU prodrug used in the treatment of cancers; is a component of tegafur-uracil.
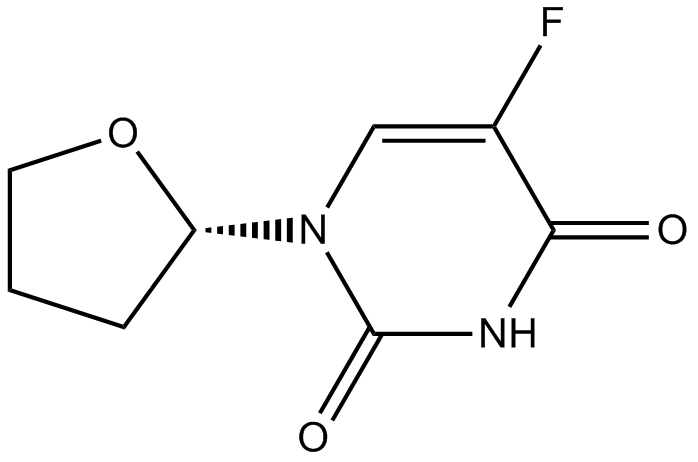
-
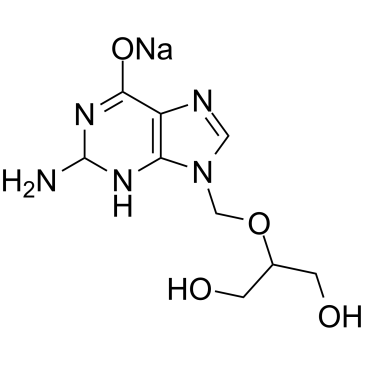
GC60865
Ganciclovir sodium
Ganciclovir (BW 759) sodium, a nucleoside analogue, is an orally active antiviral agent with activity against CMV.
-
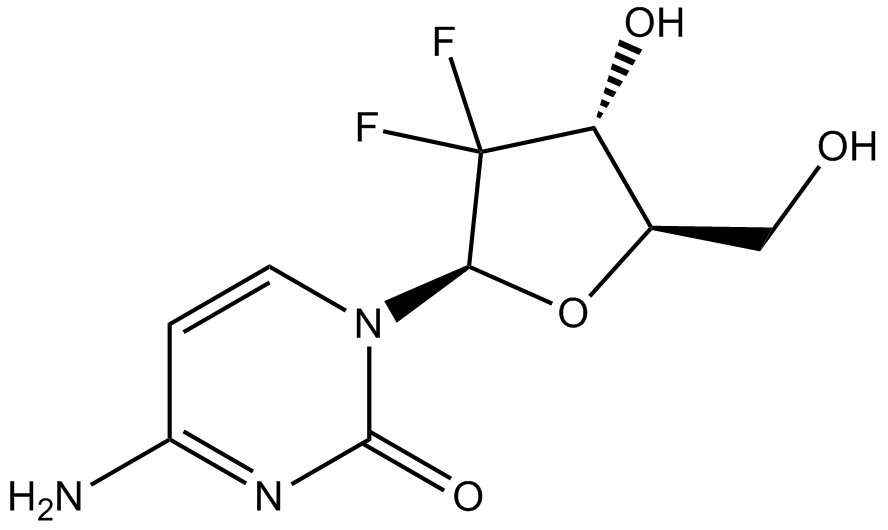
GC16805
Gemcitabine
An inhibitor of DNA synthesis
-
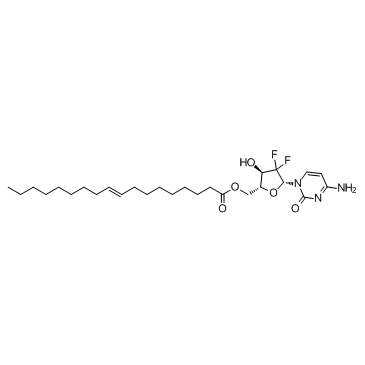
GC36130
Gemcitabine elaidate
A prodrug form of gemcitabine
-
GC63777
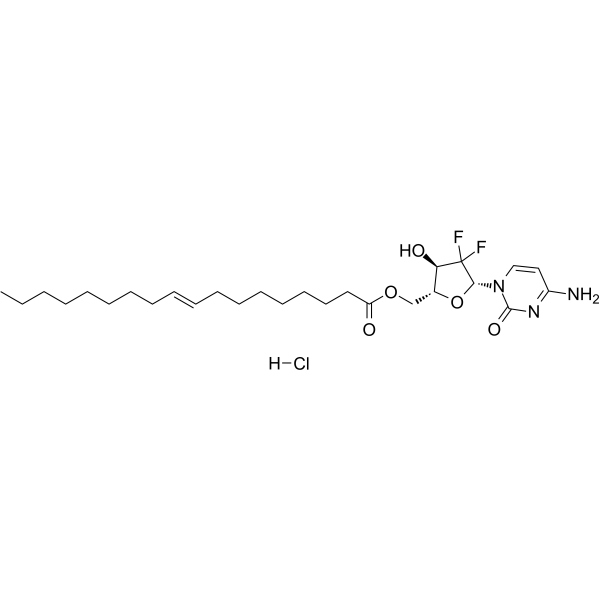
Gemcitabine elaidate hydrochloride
Gemcitabine elaidate (CP-4126) hydrochloride is lipophilic pro-drug of Gemcitabine. Gemcitabine elaidate hydrochloride is converted to Gemcitabine by esterases in order to be phosphorylated. Gemcitabine elaidate hydrochloride exhibits anti-tumor activity.
-
GC14447
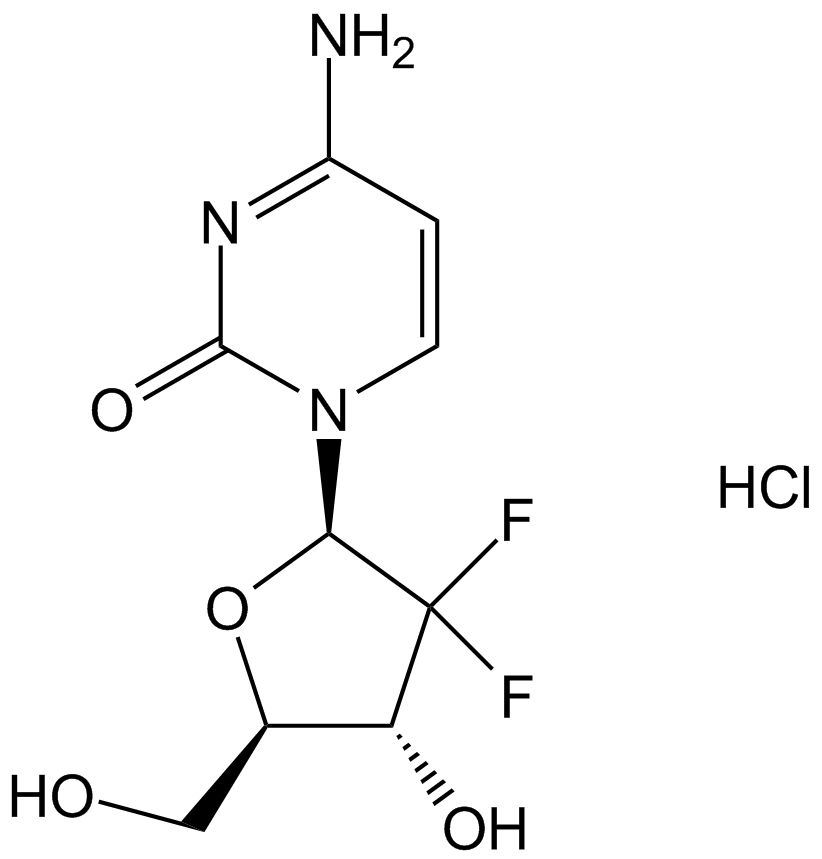
Gemcitabine HCl
Gemcitabine Hydrochloride (LY 188011 Hydrochloride) is a pyrimidine nucleoside analog antimetabolite and an antineoplastic agent. Gemcitabine Hydrochloride inhibits DNA synthesis and repair, resulting in autophagyand apoptosis.
-
GC30554
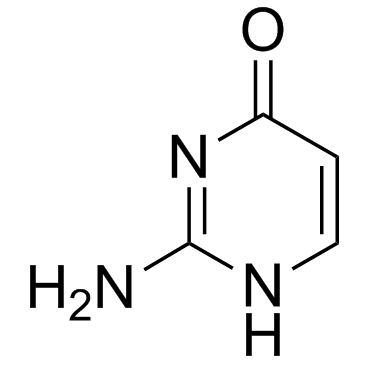
Isocytosine
Isocytosine is a non-natural nucleobase and an isomer of cytosine.
-
GC33662
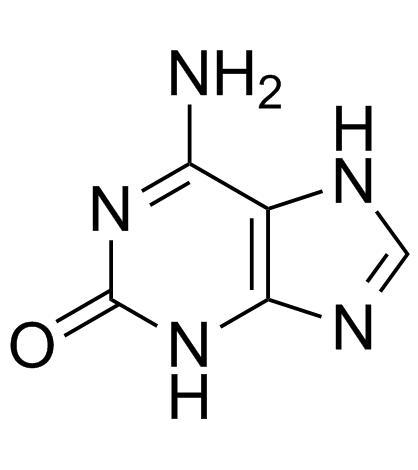
Isoguanine
An isomer of guanine
-
GC66622
L-Guanosine
L-Guanosine is the L-configuration of Guanosine . Guanosine is a purine nucleoside with anti-herpesvirus activity.