Dyslipidemias
Products for Dyslipidemias
- Cat.No. Product Name Information
-
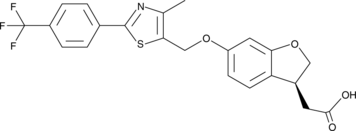
GC45210
α-Linolenic Acid (sodium salt)
α-Linolenic acid (ALA) is an essential fatty acid found in leafy green vegetables.
-
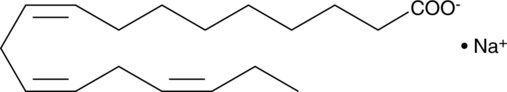
GC48283
α-Linolenic Acid-d14
An internal standard for the quantification of αLinolenic acid
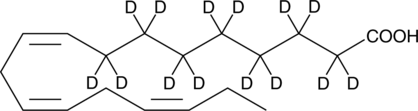
-
GC45602
α-Linolenic Acid-d5 MaxSpec• Standard

-
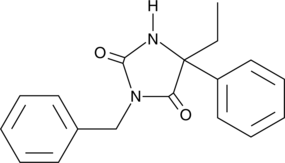
GC40954
(±)-N-3-Benzylnirvanol
(±)-N-3-Benzylnirvanol is a racemic mixture of (+)-N-3-benzylnirvanol and (-)-N-3-benzylnirvanol.
-
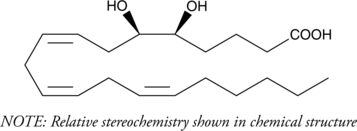
GC40270
(±)5(6)-DiHET
5(6)-DiHET is a fully racemic version of the enantiomeric forms biosynthesized from 5(6)-EET by epoxide hydrolases.
-
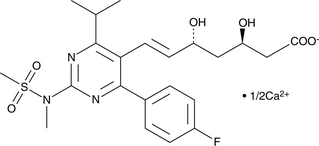
GC49690
(3R,5R)-Rosuvastatin (calcium salt)
A potential impurity found in bulk preparations of rosuvastatin
-
GC48719
(S)-Canadine
(S)-Canadine is an alkaloid and intermediate in the biosynthesis of berberine with insecticidal activity.

-
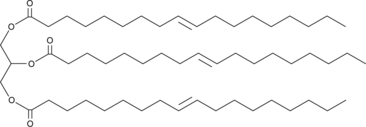
GC46040
1,2,3-Trielaidoyl-rac-glycerol
A triacylglycerol
-
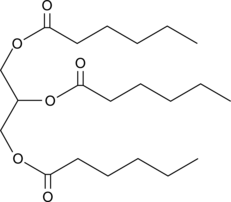
GC45285
1,2,3-Trihexanoyl-rac-glycerol
-
GC41846
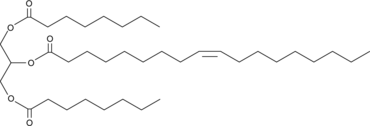
1,3-Dioctanoyl-2-Oleoyl-rac-glycerol
1,3-Dioctanoyl-2-oleoyl-rac-glycerol is a triacylglycerol that contains octanoic acid at the sn-1 and sn-3 positions and oleic acid at the sn-2 position.
-
GC40167
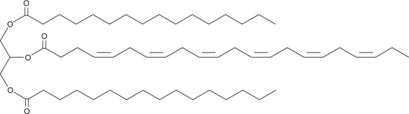
1,3-Dipalmitoyl-2-Docosahexaenoyl-rac-glycerol
1,3-Dipalmitoyl-2-docosahexaenoyl-rac-glycerol is a triacylglycerol that contains palmitic acid at the sn-1 and sn-3 positions and docosahexaenoic acid at the sn-2 position.
-
GC41850
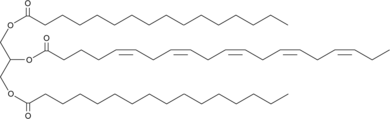
1,3-Dipalmitoyl-2-Eicosapentaenoyl-rac-glycerol
1,3-Dipalmitoyl-2-eicosapentaenoyl-rac-glycerol is a triacylglycerol that contains palmitic acid at the sn-1 and sn-3 positions and eicosapentaenoic acid at the sn-2 position.
-
GC52071
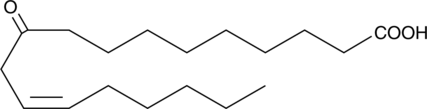
10-oxo-12(Z)-Octadecenoic Acid
A metabolite of linoleic acid and an activator of TRPV1
-
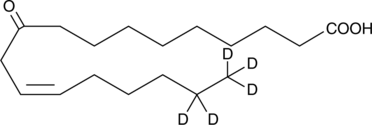
GC52428
10-oxo-12(Z)-Octadecenoic Acid-d5
An internal standard for the quantification of 10-oxo-12(Z)-octadecenoic acid
-
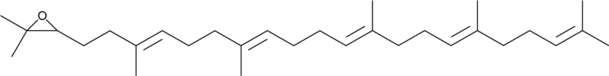
GC49671
2,3-Oxidosqualene
An intermediate in the biosynthesis of sterols
-
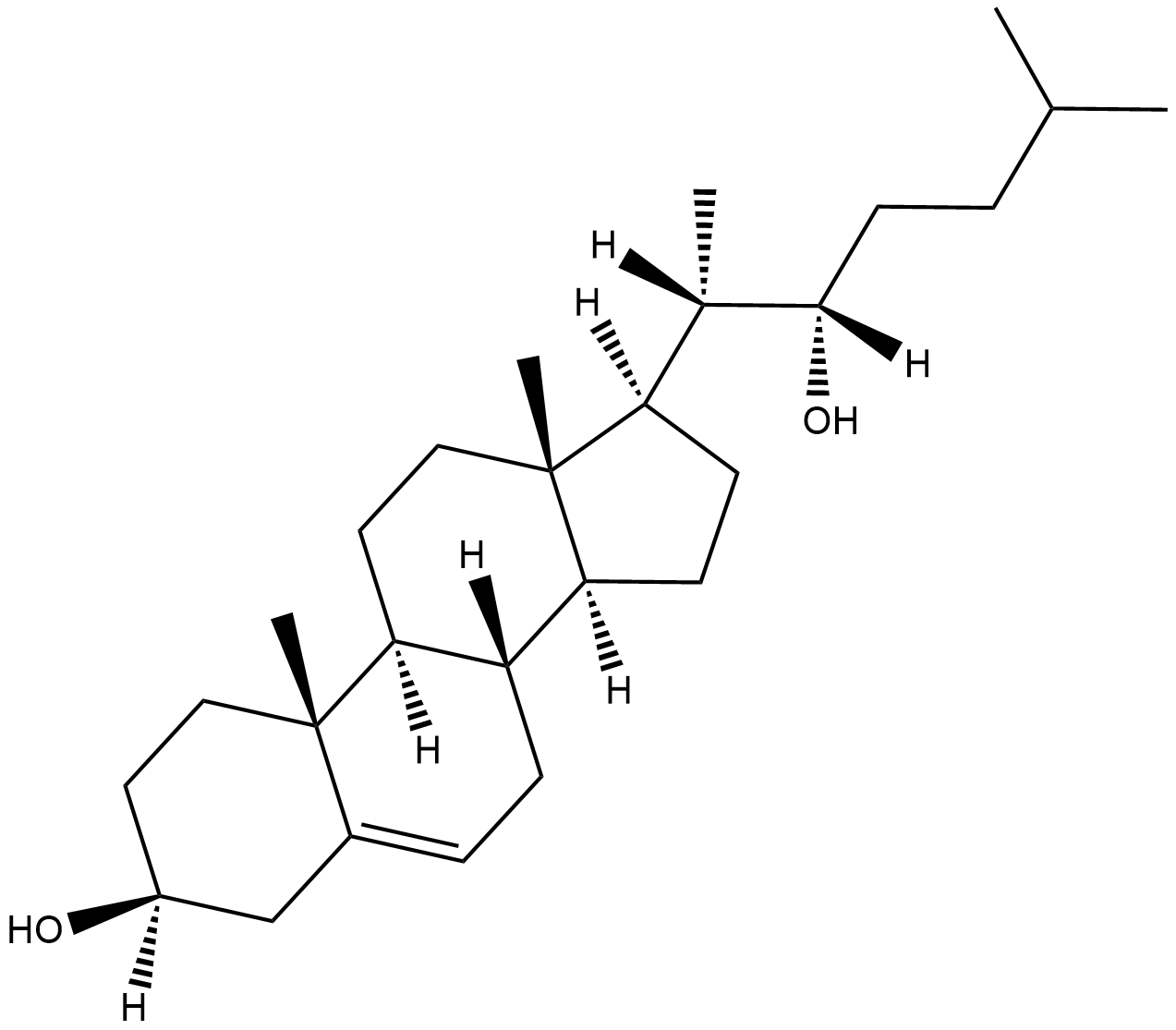
GC18270
22(S)-hydroxy Cholesterol
A synthetic oxysterol and LXR modulator
-
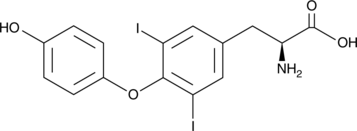
GC49427
3,5-Diiodo-L-thyronine
3,5-Diiodo-L-thyronine is an endogenous metabolite.
-
GC18205
3,5-Diiodothyroacetic Acid
3,5-Diiodothyroacetic acid (diac) is the acetic acid variant of thyroxine.
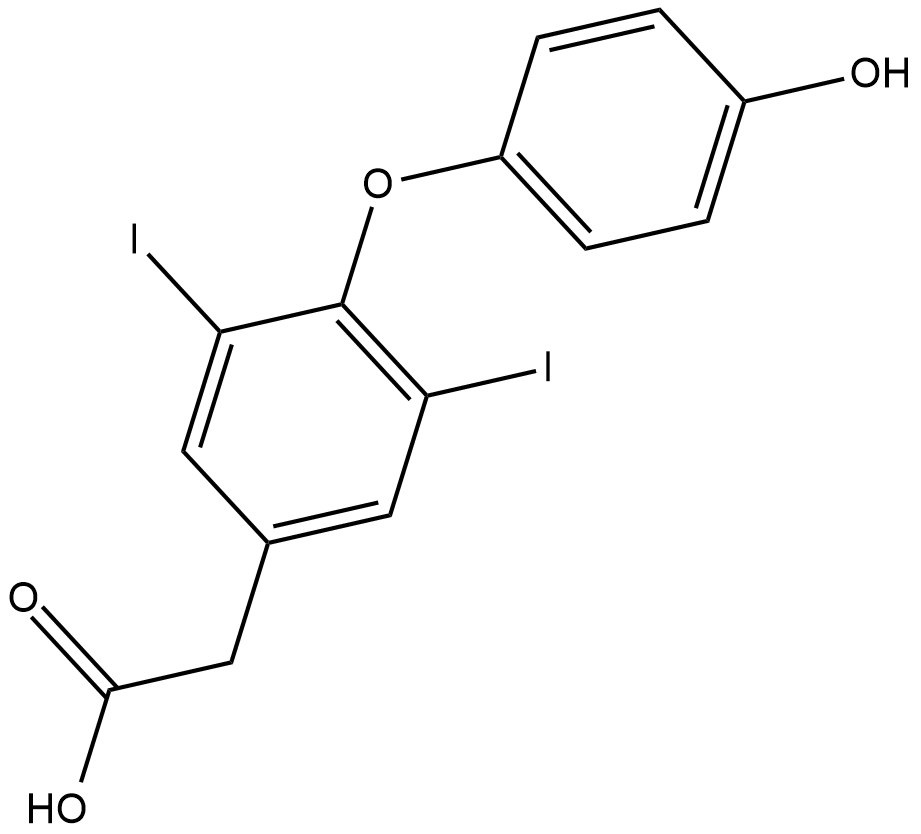
-
GC52324
3-(3-Hydroxyphenyl)propionic Acid sulfate
A metabolite of certain phenols and glycosides
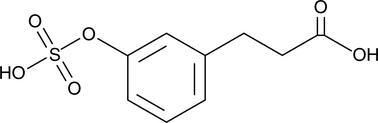
-
GC52149
306-O12B
An ionizable cationic lipidoid
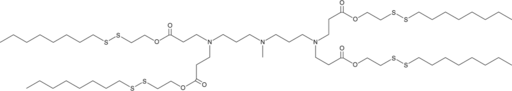
-
GC52056
6(Z),9(Z),12(Z),15(Z)-Hexadecatetraenoic Acid ethyl ester
6(Z),9(Z),12(Z),15(Z)-Hexadecatetraenoic Acid ethyl ester is an orally active n-1PUFA.

-
GC46757
9(Z),11(E)-Conjugated Linoleic Acid (sodium salt)
An isomer of linoleic acid
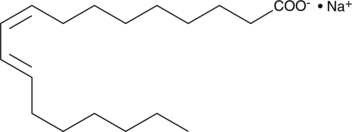
-
GC49393
all-trans-13,14-Dihydroretinol
A metabolite of all-trans retinoic acid

-
GC40024
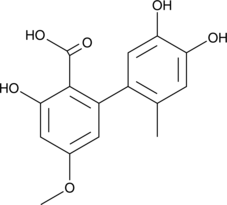
Altenusin
Altenusin is a polyphenol fungal metabolite originally isolated from the fungus Alternaria that has diverse biological activities.
-
GC40636
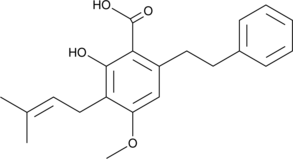
Amorfrutin A
Amorfrutin A is an isoprenoid-substituted benzoic acid natural product found in the fruit of A.
-
GC42791
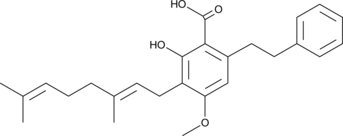
Amorfrutin B
Amorfrutin B is a partial agonist of the peroxisome proliferator-activated receptor γ (PPARγ; Ki = 19 nM and EC50 = 73 nM) that was first isolated from A.
-
GC46892
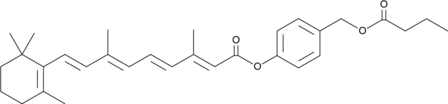
ATRA-BA Hybrid
A prodrug form of all-trans retinoic acid and butyric acid
-
GC49387
Berberine-d6 (chloride)
An internal standard for the quantification of berberine
-
GC49885
BQ-3020 (trifluoroacetate salt)
A peptide ETB receptor antagonist
-
GC46104
Butyric Acid-d7
An internal standard for the quantification of sodium butyrate
-
GC43136
Campestanol
Campestanol is a phytosterol found in vegetables, fruits, nuts, and seeds.
-
GC40203
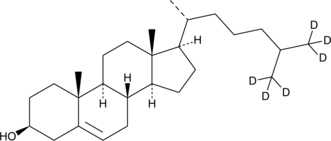
Cholesterol-d6
Cholesterol-d6 is intended for use as an internal standard for the quantification of cholesterol by GC- or LC-MS.
-
GC47085
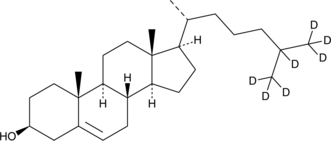
Cholesterol-d7
An internal standard for the quantification of cholesterol
-
GC47204
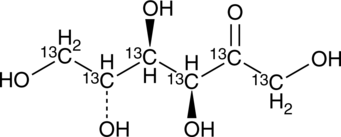
D-Fructose-13C6
An internal standard for the quantification of D-fructose
-
GC47254
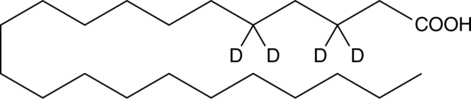
Docosanoic Acid-d4
An internal standard for the quantification of docosanoic acid
-
GC43646
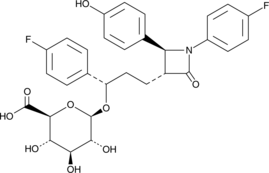
Ezetimibe Hydroxy Glucuronide
Ezetimibe hydroxy glucuronide is a minor phase II glucuronide metabolite of ezetimibe .
-
GC47337
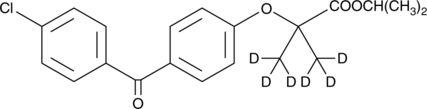
Fenofibrate-d6
An internal standard for the quantification of fenofibrate
-
GC47367
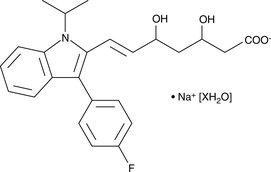
Fluvastatin (sodium salt hydrate)
An HMG-CoA reductase inhibitor
-
GC52047
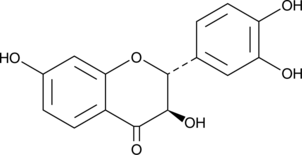
Fustin
-
GC48778
Gadoleic Acid
A monounsaturated fatty acid

-
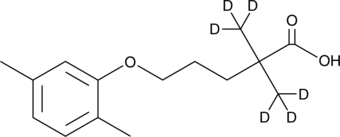
GC47397
Gemfibrozil-d6
An internal standard for the quantification of gemfibrozil
-
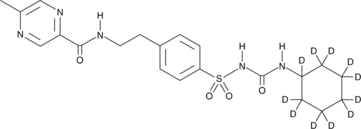
GC47404
Glipizide-d11
An internal standard for the quantification of glipizide
-
GC43764
GLP-2 (human) (trifluoroacetate salt)
Glucagon-like peptide-2 (GLP-2) is an endogenous peptide hormone formed in L cells of the small and large intestine by cleavage of proglucagon in response to nutrient ingestion.

-
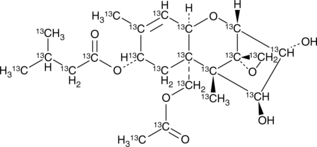
GC47436
HT-2 Toxin-13C22
An internal standard for the quantification of HT-2 toxin
-
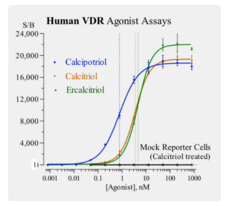
GC52504
Human Vitamin D Receptor Reporter Assay System
A nuclear receptor cell-based reporter assay
-
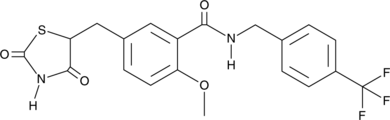
GC48521
KRP 297
A dual agonist of PPARα and PPARγ
-
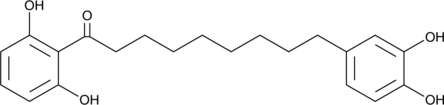
GC46028
Malabaricone C
A diarylnonanoid with diverse biological activities
-
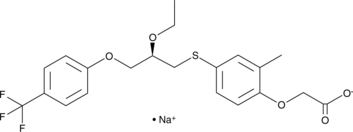
GC46168
MBX-8025 (sodium salt)
A PPARδ agonist
-
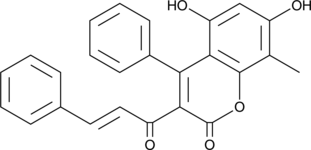
GC45506
MD001
-
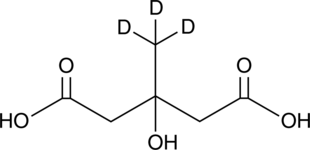
GC49486
Meglutol-d3
An internal standard for the quantification of meglutol
-
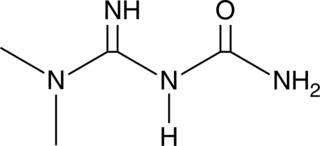
GC49879
Metformin hydroxy analog 2
An oxidation product of metformin
-
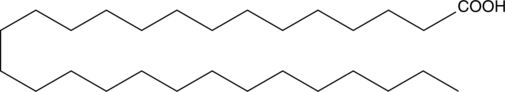
GC47815
Octacosanoic Acid
A very long-chain saturated fatty acid
-
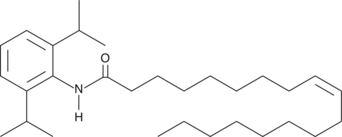
GC44497
Oleic Acid-2,6-diisopropylanilide
AcylCoA:cholesterol acyltransferase (ACAT) is an intracellular cholesteryl ester synthase tied closely to the absorption of dietary cholesterol.
-
GC47824
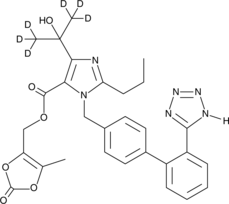
Olmesartan Medoxomil-d6
An internal standard for the quantification of olmesartan medoxomil
-
GC49236
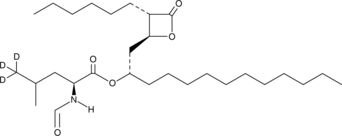
Orlistat-d3
An internal standard for the quantification of orlistat
-
GC44529
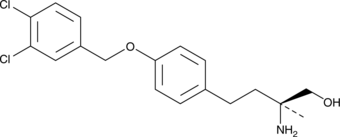
P053
P053 is an inhibitor of ceramide synthase 1 (CerS1; IC50s = 0.54 and 0.46 μM for human and mouse CerS1, respectively).
-
GC47864
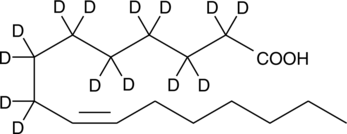
Palmitoleic Acid-d14
An internal standard for the quantification of palmitoleic acid
-
GC44645
Pioglitazone (potassium salt)
Pioglitazone (U 72107) potassium is an orally active and selective PPARγ (peroxisome proliferator-activated receptor) agonist with high affinity binding to the PPARγ ligand-binding domain with EC50 of 0.93 μM and 0.99 μM for human and mouse PPARγ, respectively.
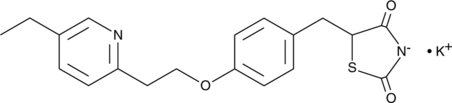
-
GC41339
Pioglitazone Ketone
Pioglitazone ketone is an active metabolite of the PPARγ agonist pioglitazone.
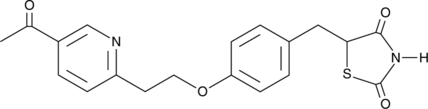
-
GC44660
Platensimycin
Platensimycin (PTM) is an antibiotic produced by S.
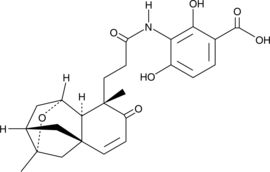
-
GC40220
Pravastatin-d3 (sodium salt)
Pravastatin-d3 is intended for use as an internal standard for the quantification of pravastatin by GC- or LC-MS.
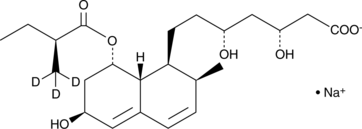
-
GC18762
Remogliflozin A
Remogliflozin A is a potent and selective inhibitor of SGLT2 (sodium-glucose cotransporter 2) with Kis of 12.4 and 26 nM for human and rat SGLT2, respectively.
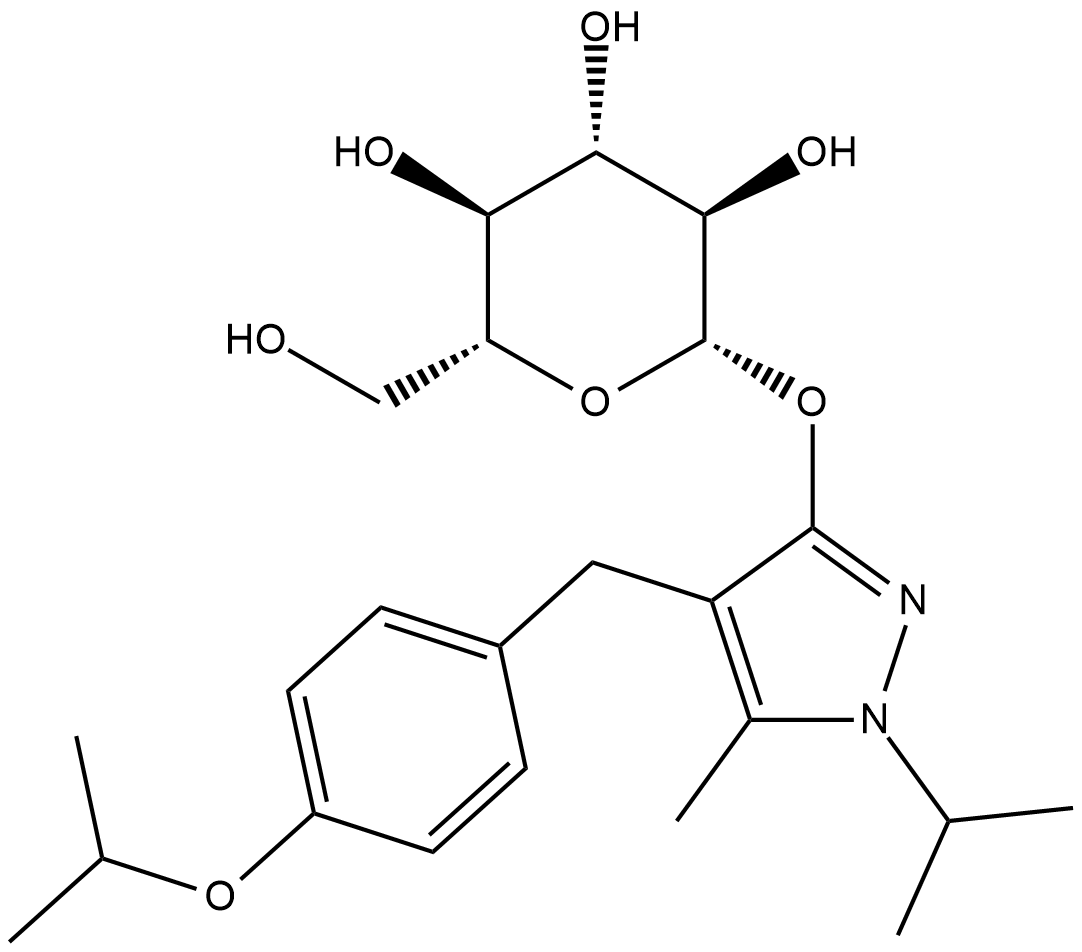
-
GC44811
Remogliflozin etabonate
Remogliflozin etabonate is a prodrug form of the sodium-glucose transporter 2 (SGLT2) inhibitor remogliflozin A.
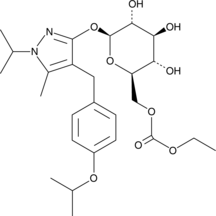
-
GC48054
Rimonabant-d10
An internal standard for the quantification of rimonabant
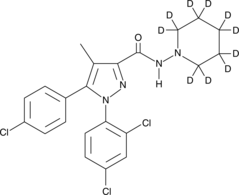
-
GC49333
SH-42
An inhibitor of DHCR24

-
GC50196
Simvastatin - d6
Deuterated simvastatin
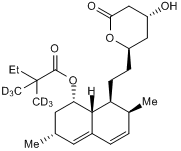
-
GC52390
Spexin 1 (human, mouse, rat, bovine) (acetate)
An endogenous peptide and agonist of GAL2 and GAL3
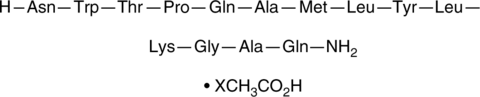
-
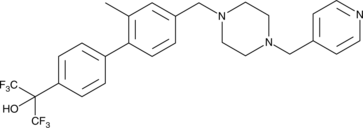
GC45567
SR 1903
-
GC52125
T-1095
T-1095 is a selective and orally active Na+-glucose cotransporter (SGLT) inhibitor with IC50s of 22.8 ?M and 2.3 ?M for human SGLT1 and SGLT2, respectively.

-
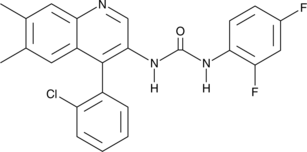
GC45060
TMP-153
Acyl-CoA:cholesterol acyltransferase (ACAT) catalyses the esterification of excess cellular cholesterol with fatty acids and is important for intestinal cholesterol absorption, hepatic lipoprotein secretion, and cholesterol accumulation in vascular tissues.
-
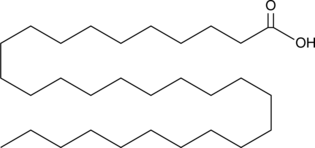
GC49141
Triacontanoic Acid
A very long-chain saturated fatty acid
-
GC49389
Tritylolmesartan Medoxomil
An intermediate in the synthesis of olmesartan medoxomil
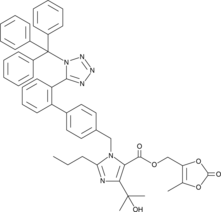
-
GC48272
ZLY032
A dual FFAR1 and PPARδ agonist