CDK
<div class="colum_1 clearer"><p>CDKs (Cyclin-dependent kinases) are serine-threonine kinases first discovered for their role in regulating the cell cycle. They are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells. CDKs are relatively small proteins, with molecular weights ranging from 34 to 40 kDa, and contain little more than the kinase domain. In fact, yeast cells can proliferate normally when their CDK gene has been replaced with the homologous human gene. By definition, a CDK binds a regulatory protein called a cyclin. Without cyclin, CDK has little kinase activity; only the cyclin-CDK complex is an active kinase.</p><p>There are around 20 Cyclin-dependent kinases (CDK1-20) known till date. CDK1, 4 and 5 are involved in cell cycle, and CDK 7, 8, 9 and 11 are associated with transcription.</p><p>CDK levels remain relatively constant throughout the cell cycle and most regulation is post-translational. Most knowledge of CDK structure and function is based on CDKs of <i>S. pombe</i> (Cdc2), <i>S. cerevisia</i> (CDC28), and vertebrates (CDC2 and CDK2). The four major mechanisms of CDK regulation are cyclin binding, CAK phosphorylation, regulatory inhibitory phosphorylation, and binding of CDK inhibitory subunits (CKIs).</p></div>
Targets for CDK
Products for CDK
- Cat.No. Product Name Information
-
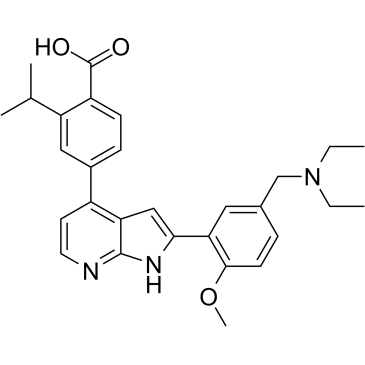
GC36062
FMF-04-159-2
FMF-04-159-2 is a covalent CDK14 inhibitor. FMF-04-159-2 inhibits CDK14 and CDK2 with IC50s of 39.6 nM and 256 nM in NanoBRET assay, respectively.
-
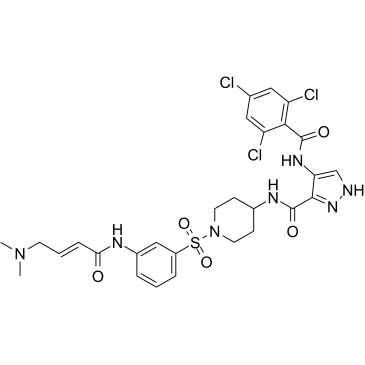
GC50719
FMF-04-159-R
-
GC33049
FN-1501
FN-1501 is a potent inhibitor of FLT3 and CDK, with IC50s of 2.47, 0.85, 1.96, and 0.28 nM for CDK2/cyclin A, CDK4/cyclin D1, CDK6/cyclin D1 and FLT3, respectively. FN-1501 has anticancer activity.
-
GN10696
Garcinone C
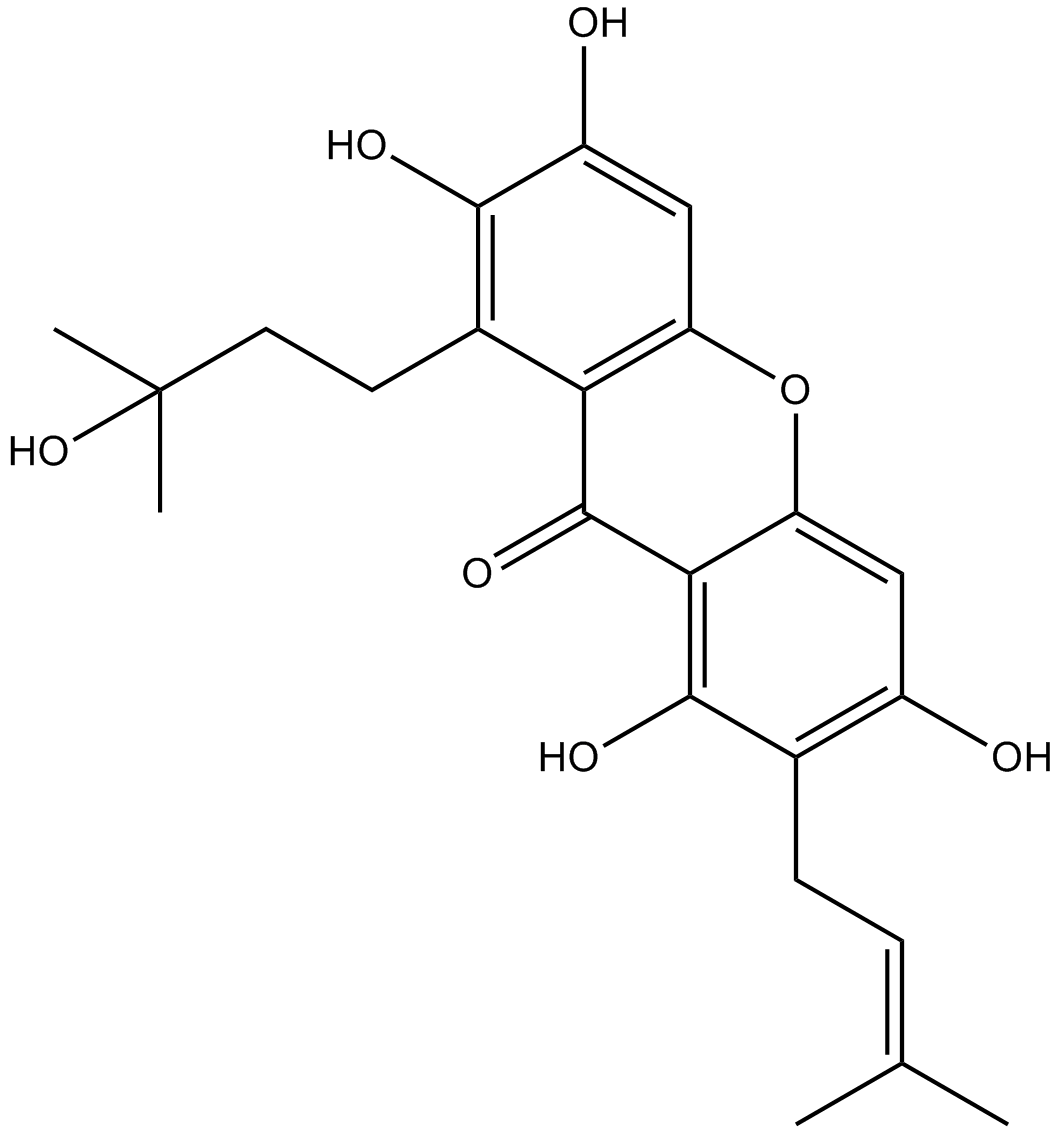
-
GC64926
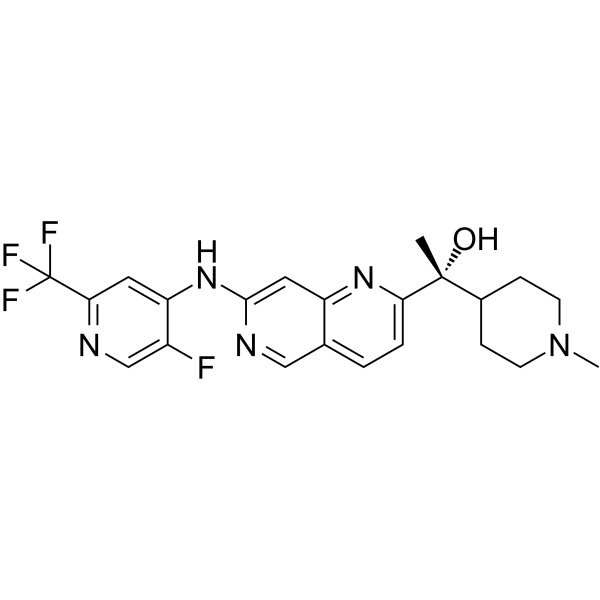
GFB-12811
GFB-12811 is a high selective and orally active CDK5 inhibitor with an IC50 of 2.3 nM.
-
GC14987
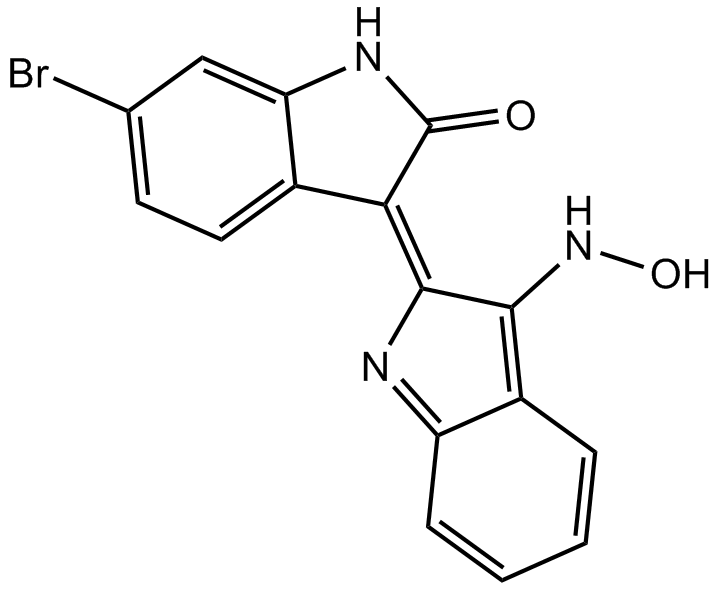
GSK-3 Inhibitor IX (BIO)
GSK-3 Inhibitor IX (BIO) (6-Bromoindirubin-3'-oxime; BIO) is a potent, selective, reversible and ATP-competitive inhibitor of GSK-3α/β and CDK1-cyclinB complex with IC50s of 5 nM/320 nM/80 nM for (GSK-3α/β)/CDK1/CDK5, respectively.
-
GC62423
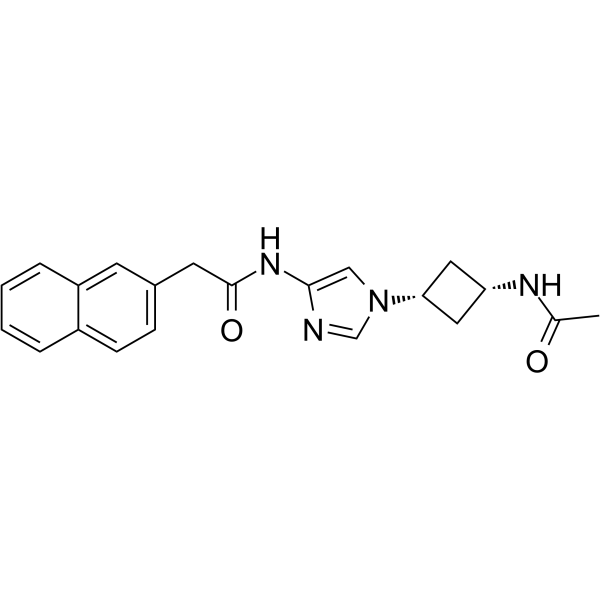
GSK-3/CDK5/CDK2-IN-1
GSK-3/CDK5/CDK2-IN-1, an imidazole derivative, is an inhibitor of cdk5, cdk2, and GSK-3 extracted from patent WO2002010141A1, example 9a.
-
GC63854
HQ461
HQ461 is a molecular glue that promotes CDK12-DDB1 interaction to trigger cyclin K degradation. HQ461-mediated degradation of cyclin K impairs CDK12 function, resulting in decreased CDK12 substrate phosphorylation, downregulation of DNA damage response genes, and cell death.
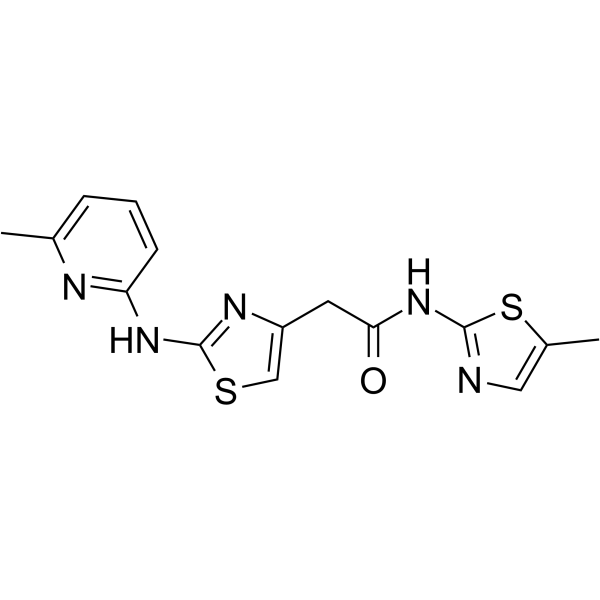
-
GC62572
hSMG-1 inhibitor 11e
hSMG-1 inhibitor 11e is a potent and selective hSMG-1 kinase inhibitor with an IC50 of 900-fold selectivity over mTOR (IC50 of 45 nM), PI3Kα/γ (IC50s of 61 nM and 92 nM) and CDK1/CDK2 (IC50s of 32 μM and 7.1 μM).
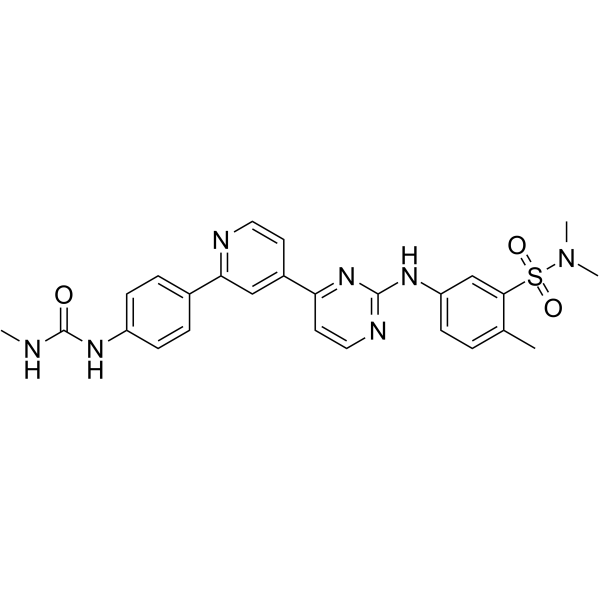
-
GC61925
hSMG-1 inhibitor 11j
hSMG-1 inhibitor 11j, a pyrimidine derivative, is a potent and selective inhibitor of hSMG-1, with an IC50 of 0.11 nM. hSMG-1 inhibitor 11j exhibits >455-fold selectivity for hSMG-1 over mTOR (IC50=50 nM), PI3Kα/γ (IC50=92/60 nM) and CDK1/CDK2 (IC50=32/7.1 μM). hSMG-1 inhibitor 11j can be used for the research of cancer.
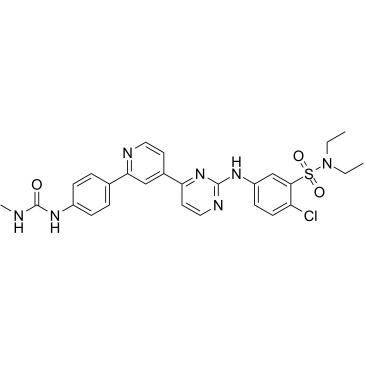
-
GC36312
Indirubin-3'-monoxime-5-sulphonic acid
Indirubin-3'-monoxime-5-sulphonic acid is a potent and selective inhibitor of CDK1, CDK5, and GSK-3β with IC50s of 5 nM, 7 nM, and 80 nM, respectively.
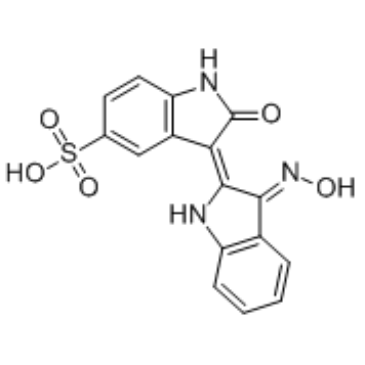
-
GC36313
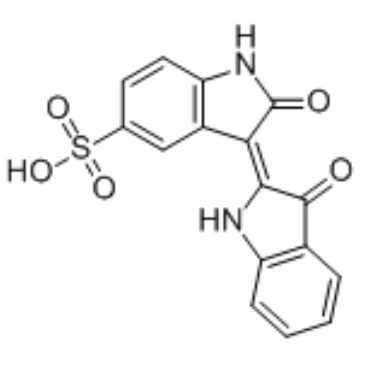
Indirubin-5-sulfonate
Indirubin-5-sulfonate is a cyclin-dependent kinase (CDK) inhibitor, with IC50 values of 55 nM, 35 nM, 150 nM, 300 nM and 65 nM for CDK1/cyclin B, CDK2/cyclin A, CDK2/cyclin E, CDK4/cyclin D1, and CDK5/p35, respectively. Indirubin-5-sulfonate also shows inhibitory activity against GSK-3β.
-
GC63023
Ipivivint
Ipivivint (compound 38) is a potent CDC-like kinase (CLK) inhibitor with EC50s of 1 nM, 7 nM for CLK2 and CLK3, respectively. Ipivivint inhibits Wnt pathway (EC50=13 nM).
-
GC63460
IV-361
IV-361 is an orally active and selective CDK7 inhibitor (Ki≤50 nM). IV-361 has anti-cancer activity (US20190256531A1).
-
GC65471
JH-XI-10-02
JH-XI-10-02 is a PROTAC connected by ligands for Cereblon and CDK. JH-XI-10-02 is a highly potent and selective PROTAC CDK8 degrader, with an IC50 of 159 nM. JH-XI-10-02 causes proteasomal degradation, does not affect CDK8 mRNA levels. JH-XI-10-02 shows no effect on CDK19.
-
GC65577
JH-XVI-178
JH-XVI-178 is a highly potent and selective inhibitor of CDK8/19 that displays low clearance and moderate oral pharmacokinetic properties.

-
GC12612
JNJ-7706621
A dual inhibitor of CDKs and Aurora kinases
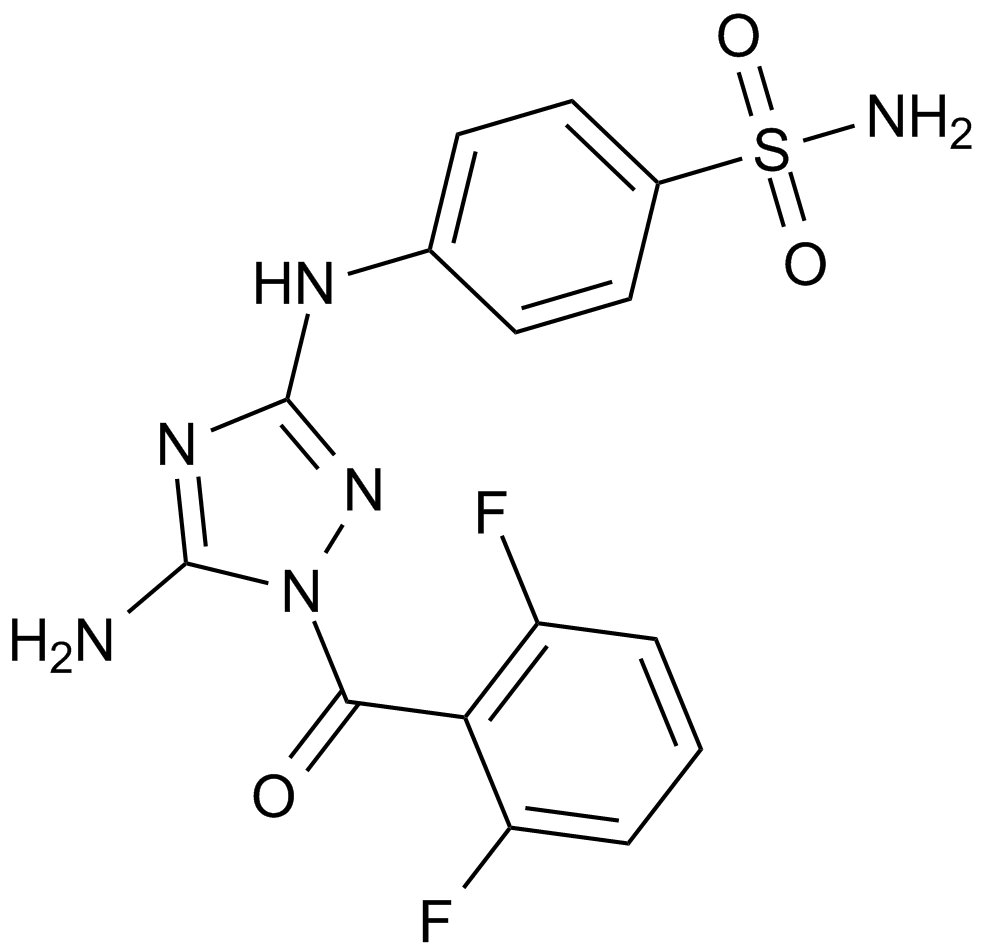
-
GC34204
JSH-150
JSH-150 is a highly selective and potent CDK9 inhibitor with an IC50 of 1 nM.
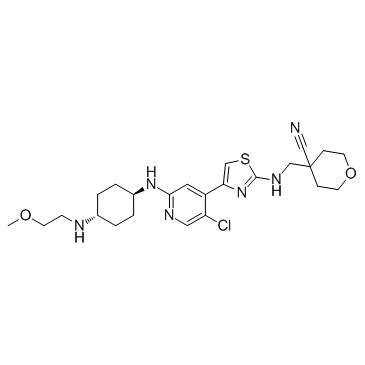
-
GC14230
K03861
K03861 (K03861) is a type II CDK2 inhibitor with Kd of 8.2 nM. K03861 (K03861) inhibits CDK2 activity by competing with binding of activating cyclins.
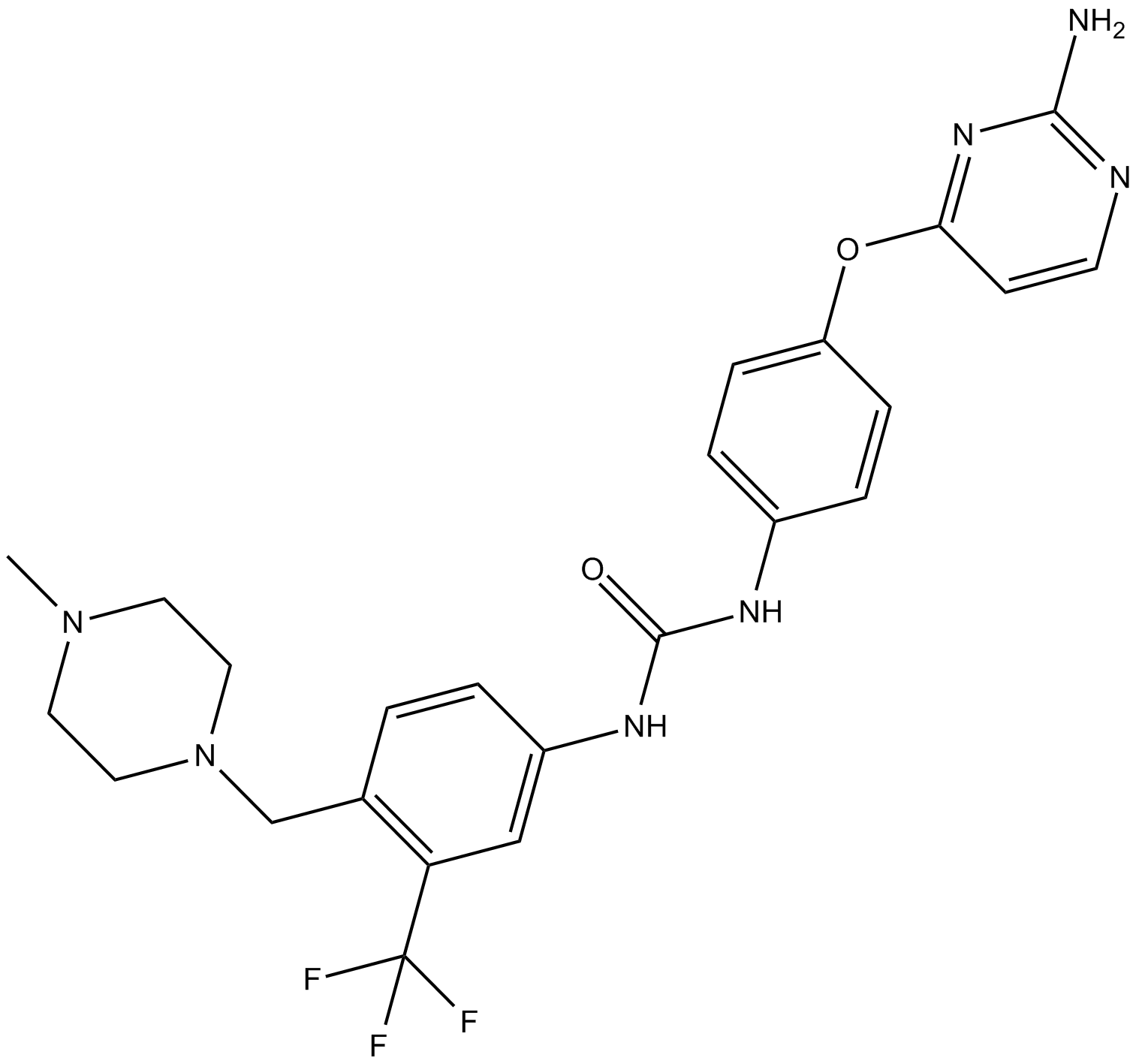
-
GC62392
KB-0742 dihydrochloride
KB-0742 dihydrochloride is a potent, selective and orally active CDK9 inhibitor with an IC50 of 6 nM for CDK9/cyclin T1. KB-0742 dihydrochloride is selective for CDK9/cyclin T1 with >50-fold selectivity over other CDK kinases. KB-0742 dihydrochloride has potent anti-tumor activity.
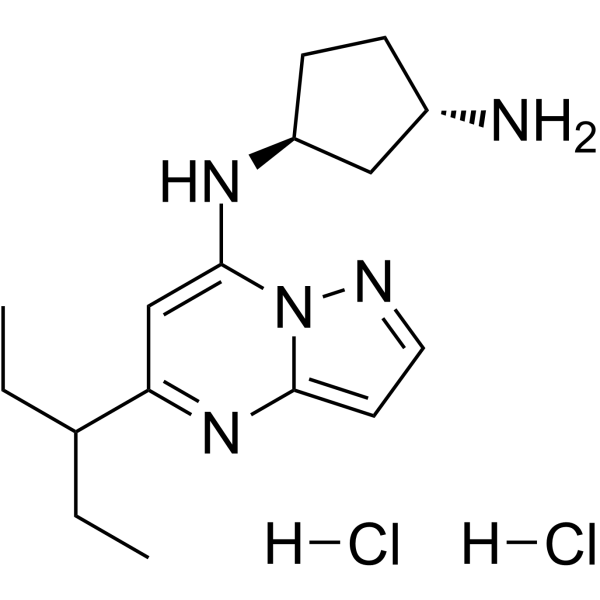
-
GC14182
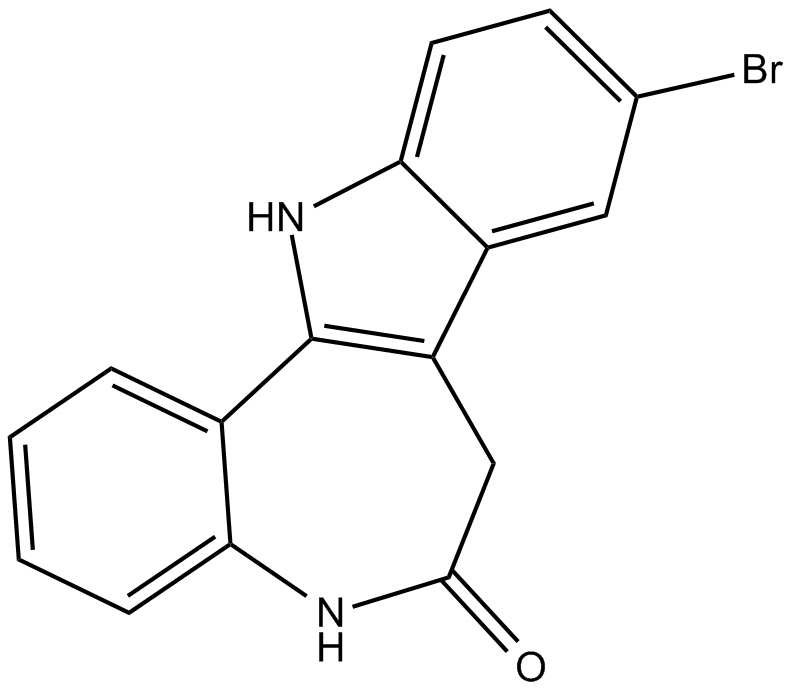
Kenpaullone
CDK1/cyclin B and GSK-3β inhibitor
-
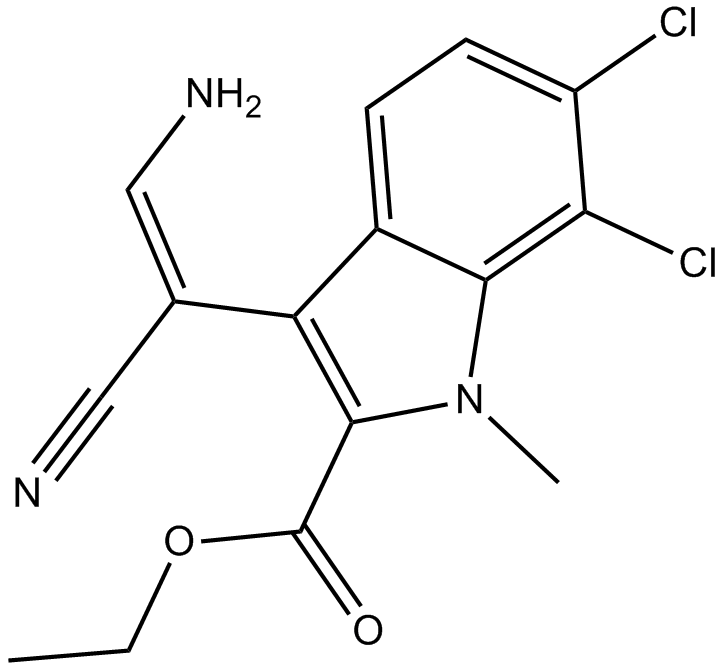
GC11774
KH CB19
inhibitor of CLK1 and CLK4, potent and selective
-
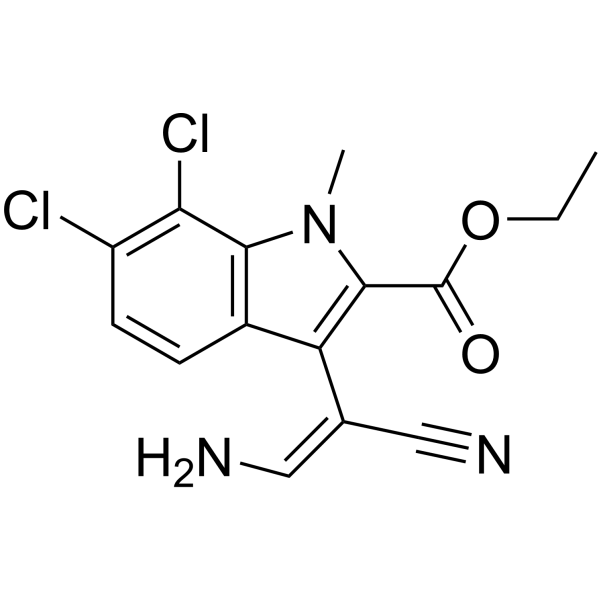
GC63943
KH-CB20
KH-CB20, an E/Z mixture, is a potent and selective inhibitor of CLK1 and the closely related isoform CLK4, with an IC50 of 16.5 nM for CLK1.
-
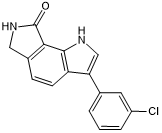
GC50532
KuWal151
Potent and selective CLK inhibitor
-
GC17067
LDC000067
CDK9 inhibitor, novel and highly specific
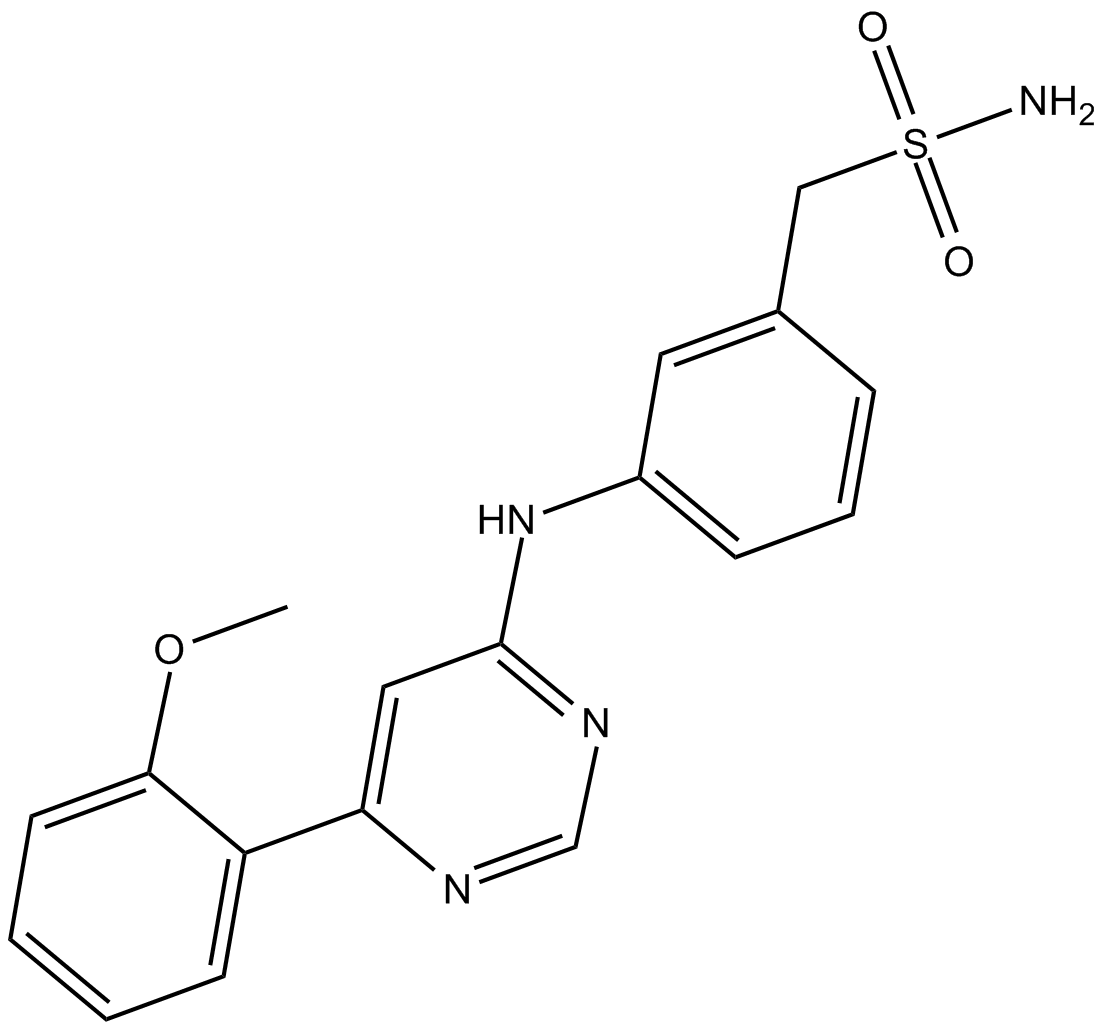
-
GC19219
LDC4297
LDC4297 is a potent and selective CDK7 inhibitor with an IC50 of 0.13 nM.
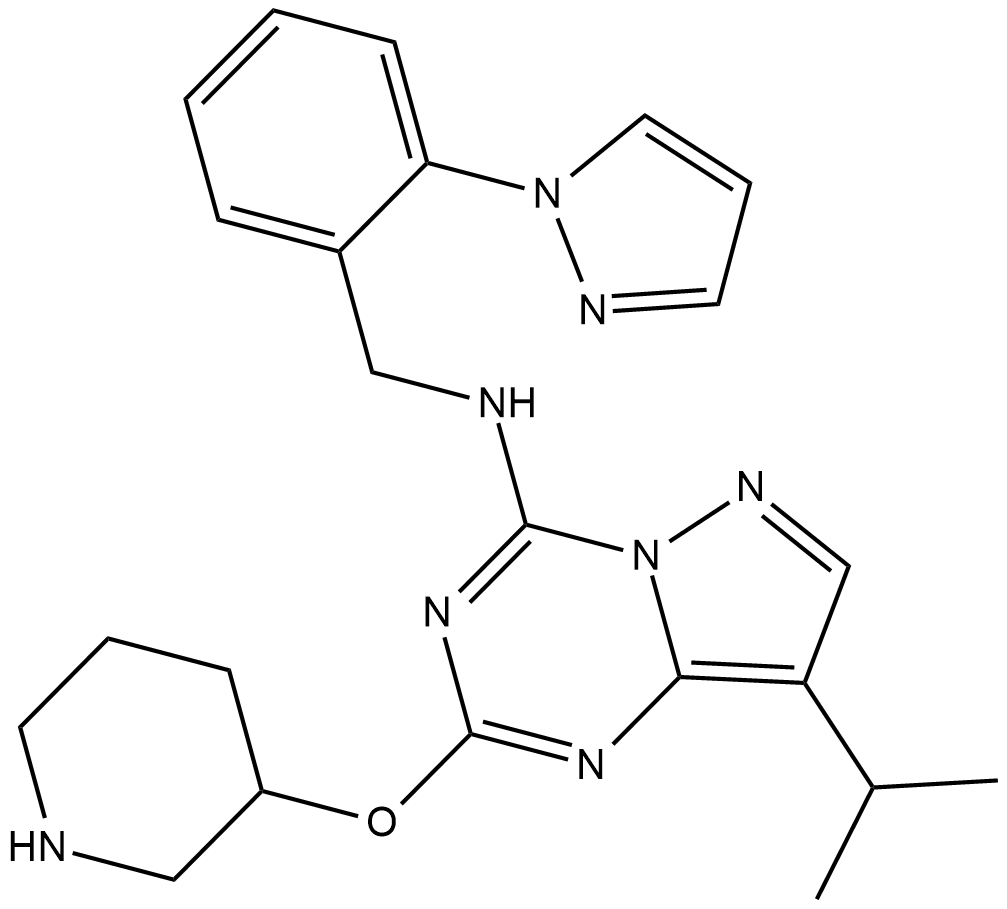
-
GC66446
LDC4297 hydrochloride
LDC4297 hydrochloride is a selective inhibitor of CDK7 with an IC50 value of 0.13 nM. LDC4297 hydrochloride inhibits human cytomegalovirus (HCMV) replication with an EC50 value of 24.5 nM. LDC4297 hydrochloride shows broad antiviral activities to Herpesviridae, Adenoviridae, Poxviridae, Retroviridae and Orthomyxoviridae with EC50 values of 0.02-1.21 μM. LDC4297 hydrochloride can be used for the research of infection.
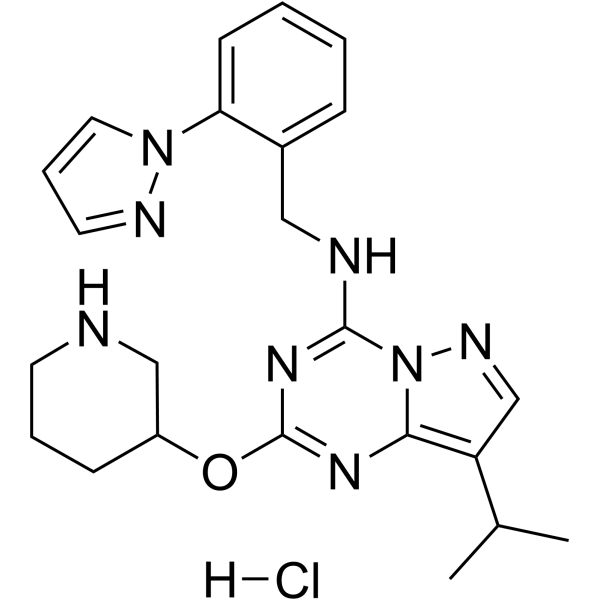
-
GC10842
LEE011
LEE011 (LEE01) is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
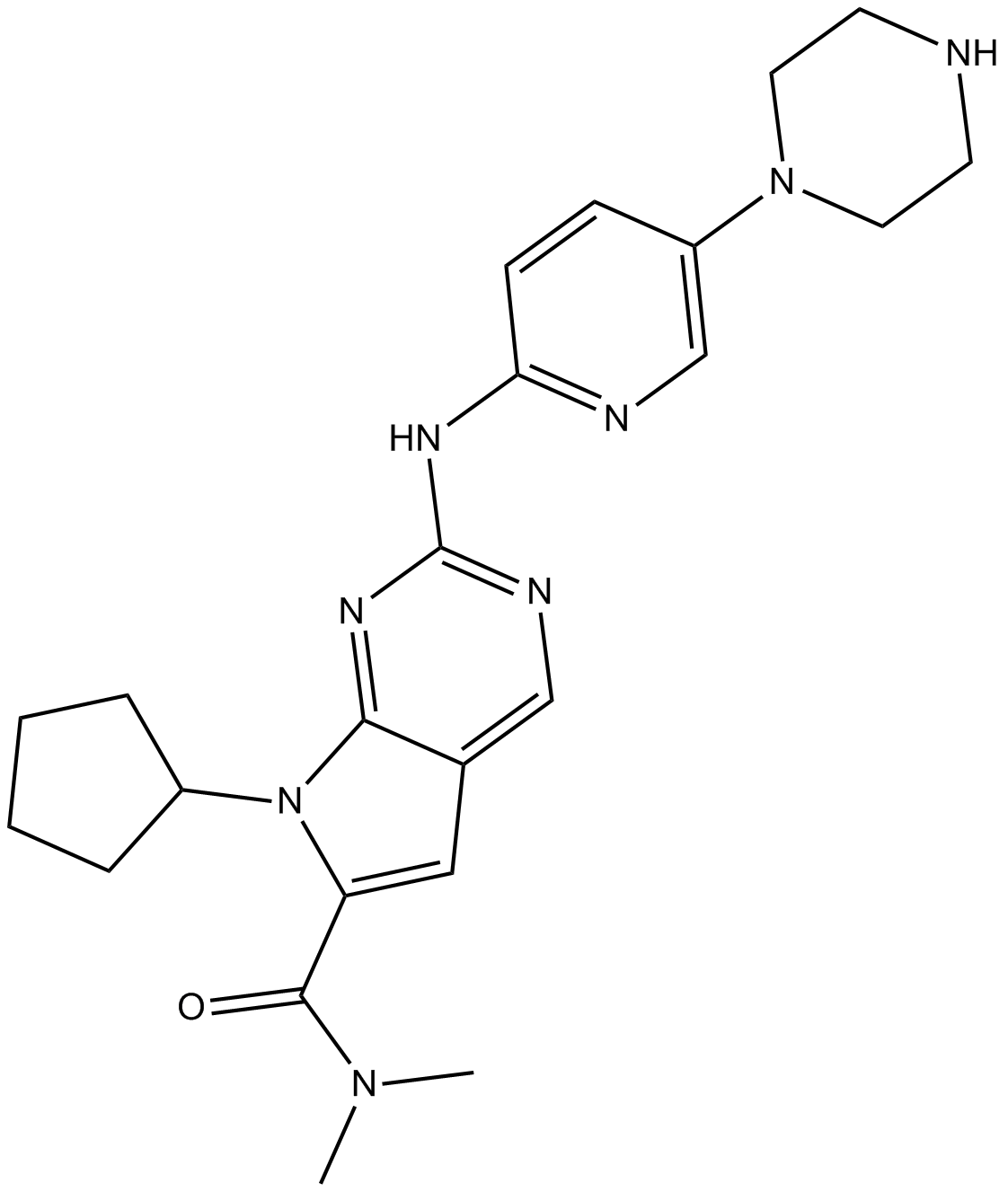
-
GC15922
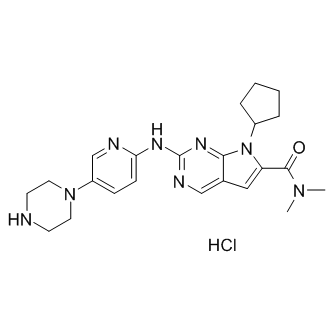
LEE011 hydrochloride
LEE011 hydrochloride (LEE011 hydrochloride) is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
-
GC15377
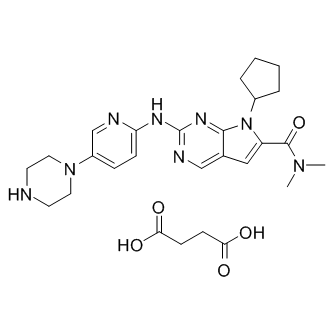
LEE011 succinate
LEE011 succinate (LEE011 succinate) is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
-
GC34163
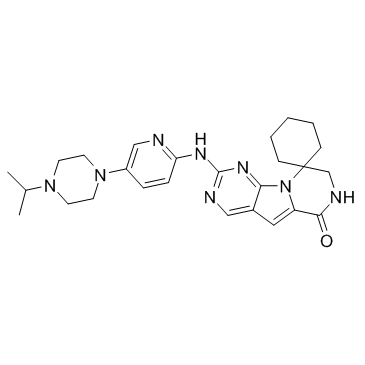
Lerociclib (G1T38)
Lerociclib (G1T38) (G1T38) is a potent and selective inhibitor of CDK4/6, with IC50s of 1 nM, 2 nM for CDK4/CyclinD1 and CDK6/CyclinD3, respectively.
-
GC34100
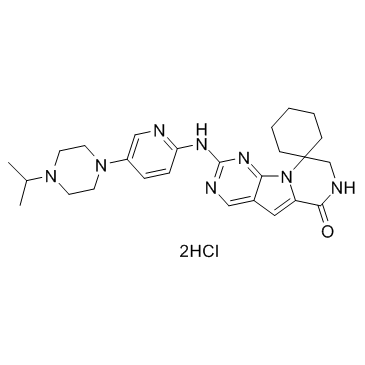
Lerociclib dihydrochloride (G1T38 dihydrochloride)
Lerociclib dihydrochloride (G1T38 dihydrochloride) (G1T38 dihydrochloride) is a potent and selective inhibitor of CDK4/CDK6, with IC50s of 1 nM and 2 nM for CDK4/CyclinD1 and CDK6/CyclinD3, respectively.
-
GC34650
Longdaysin
Longdaysin is a inhibitor of the Wnt/β-catenin signaling pathway, which exerts antitumor effect through blocking CK1δ/ε-dependent Wnt signaling. Longdaysin inhibits CK1α, CK1δ, CDK7, and ERK2 with IC50s of? 5.6 ?M, 8.8 ?M, 29 ?M, and 52 ?M, respectively.
-
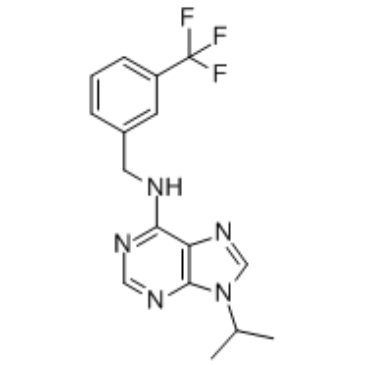
GC61008
LSN3106729 hydrochloride
-
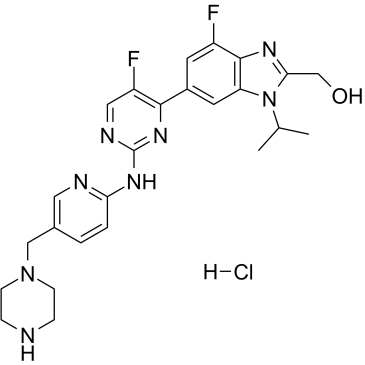
GC16822
LY2835219
LY2835219 (LY2835219 methanesulfonate) is a selective CDK4/6 inhibitor with IC50s of 2 nM and 10 nM for CDK4 and CDK6, respectively.
-
GC11823
LY2835219 free base
A dual inhibitor of Cdk4 and Cdk6

-
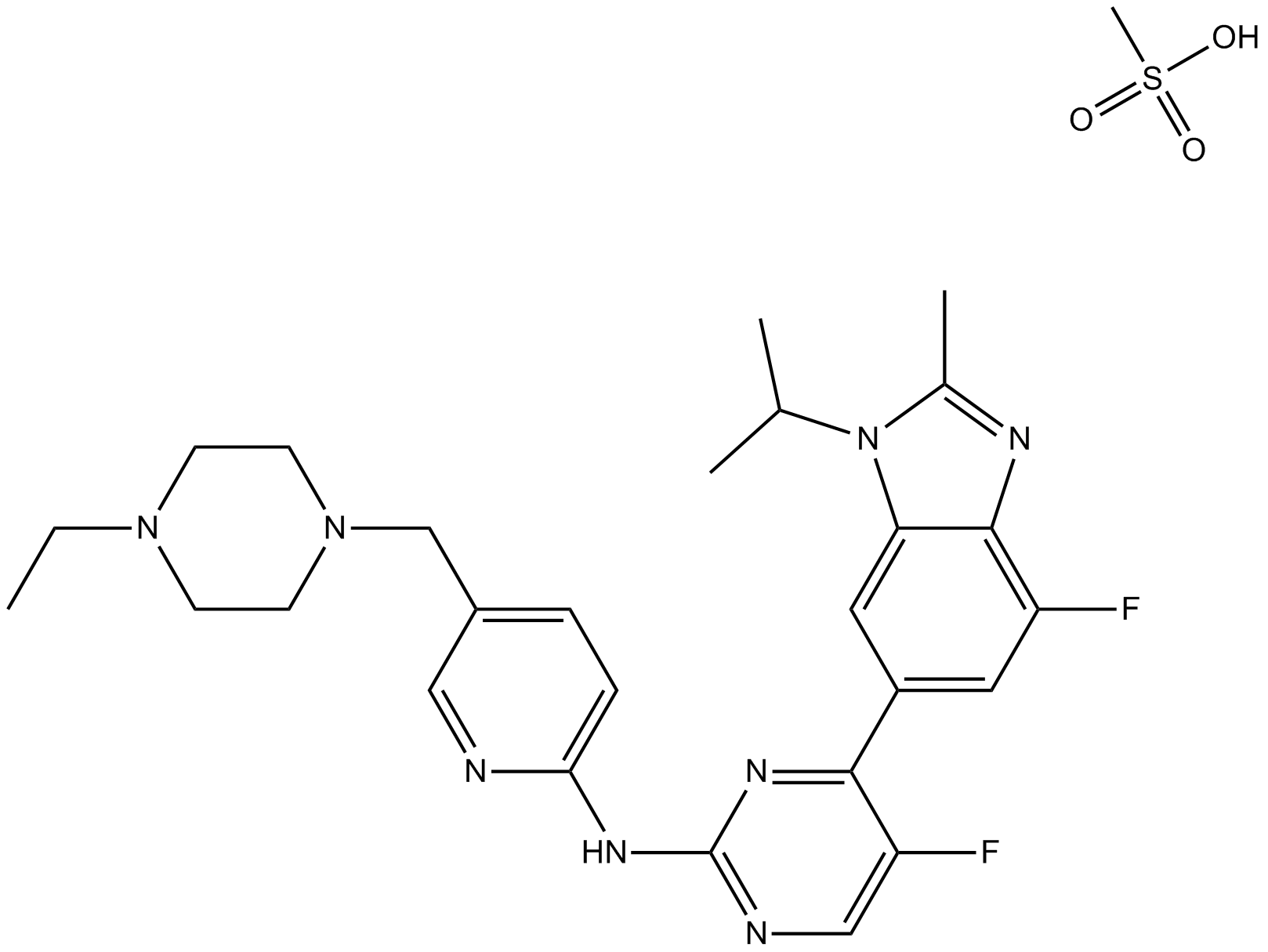
GC11971
LY2857785
CDK9 inhibitor
-
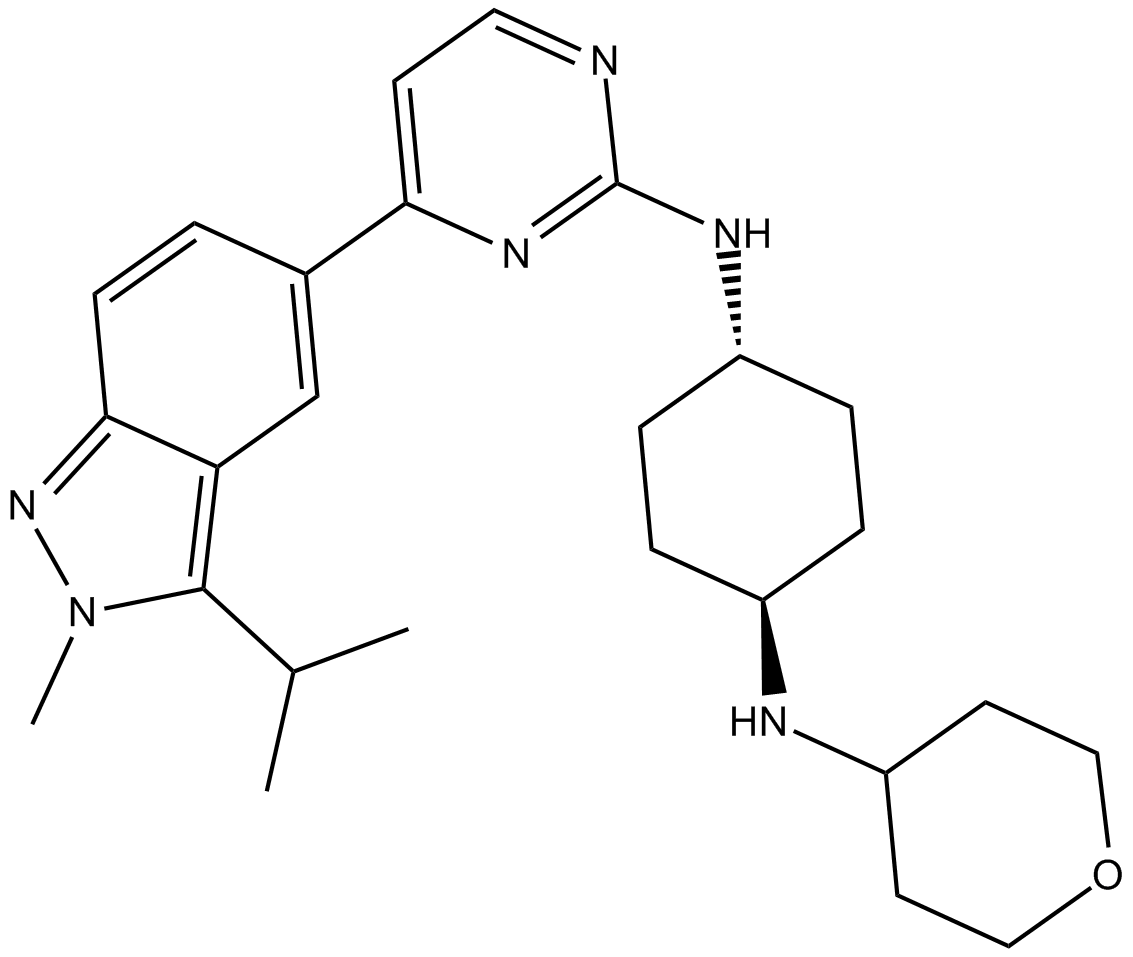
GC25593
LY3143921 hydrate
LY3143921 hydrate is an orally administered ATP-competitive CDC7 inhibitor.
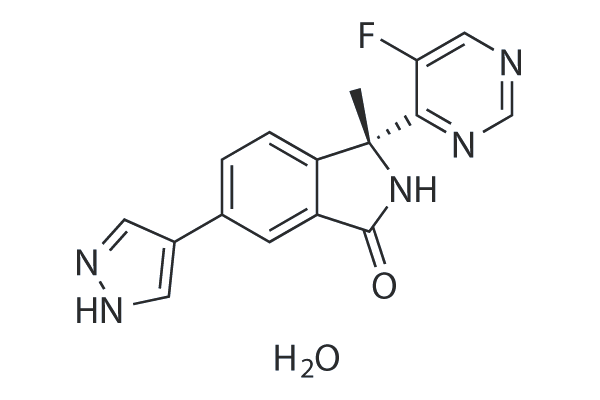
-
GC19233
LY3177833
LY3177833 is an CDC7 and pMCM2 inhibitor extracted from patent US 20140275131and patent WO 2014143601 A1 compound example 4; has IC50 values of 3.3 nM and 290 nM, respectively.
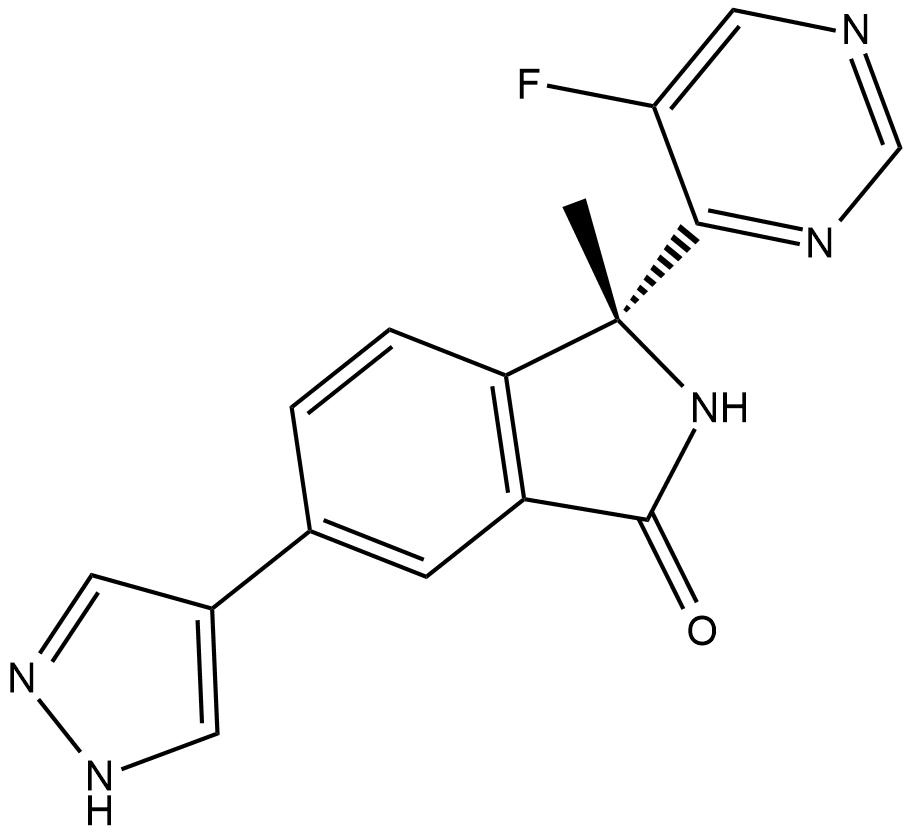
-
GC25594
LY3405105
LY3405105 is an orally active CDK7 inhibitor with an IC50 of 92.8 nM. LY3405105 shows potential antineoplastic activity.
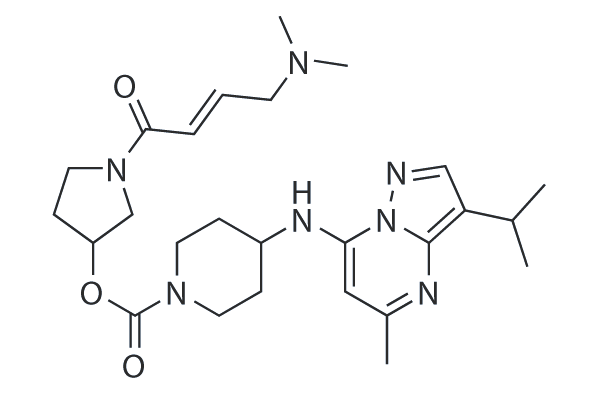
-
GC32970
MBQ-167
MBQ-167 is a dual Rac/Cdc42 inhibitor, with IC50s of 103 nM for Rac 1/2/3 and 78 nM for Cdc42 in MDA-MB-231 cells, respectively.
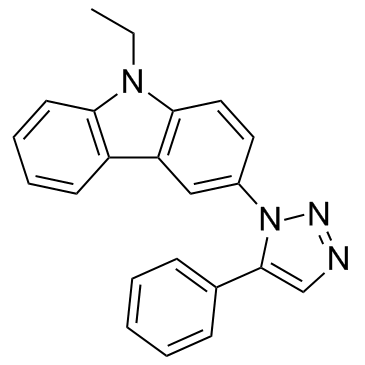
-
GC50239
ML 315 hydrochloride
Inhibitor of Clk and DYRK kinases
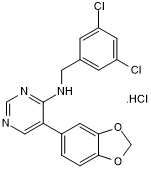
-
GC14162
ML167
Clk4 inhibitor,highly selective
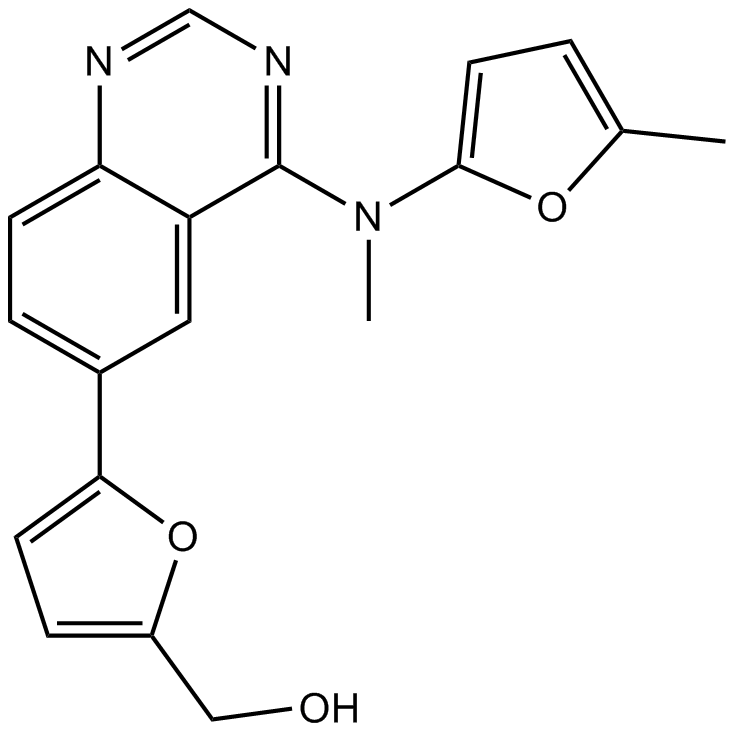
-
GC32746
MSC2530818
A Cdk8 inhibitor
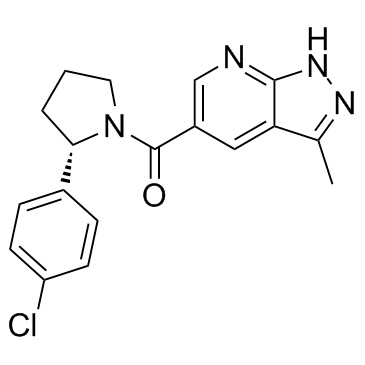
-
GC64393
NCT02
NCT02 is a cyclin K degrader. NCT02 induces ubiquitination of cyclin K (CCNK) and proteasomal degradation of CCNK and its complex partner CDK12. NCT02 has the potential for the research of metastatic colorectal cancer (CRC).
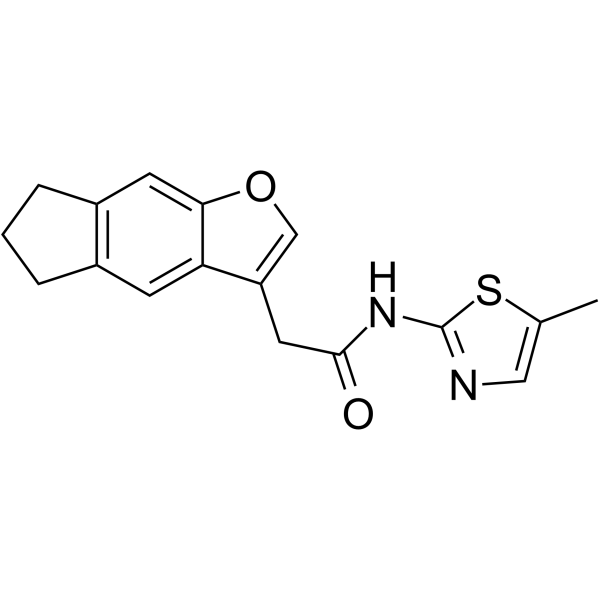
-
GC36732
NG 52
NG 52 is a potent, cell-permeable, selective, ATP-compatible and orally active Cdc28p and Pho85p kinase inhibitor with IC50s of 7 μM and 2 μM, respectively. NG 52 also inhibits the activity of phosphoglycerate kinase 1 (PGK1) with an IC50 of 2.5 μM. NG 52 is inactive against yeast kinases Kin28p, Srb10, and Cak1p.
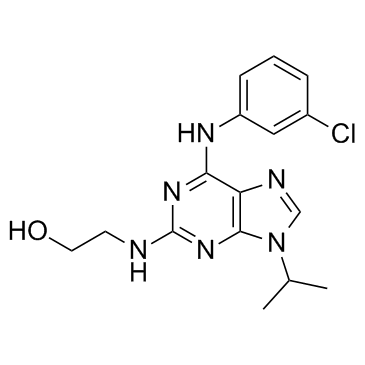
-
GC63874
NSC 107512
NSC 107512 is a potent inhibitor of cyclin-dependent kinase 9 (CDK9). NSC 107512 is a class of sangivamycin-like molecules (SLM). NSC 107512 inhibits growth and induces apoptosis of multiple myeloma tumors.
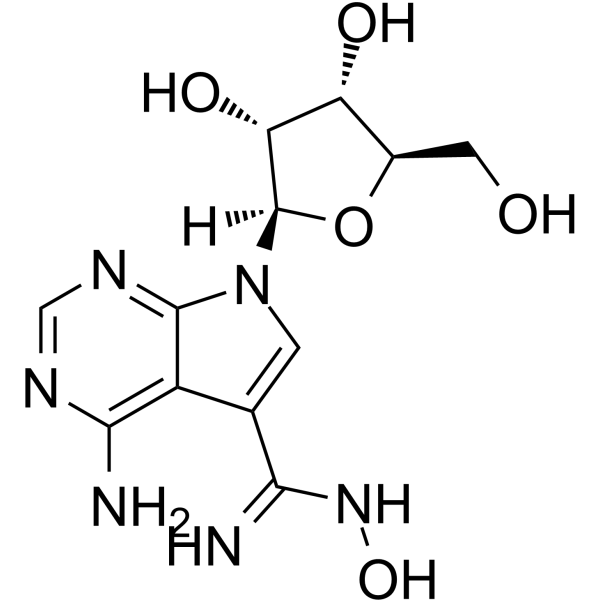
-
GC19267
NU2058
NU2058 is a guanine-based CDK inhibitor with IC50 of 17 uM and 26 uM for CDK2 and CDK1.
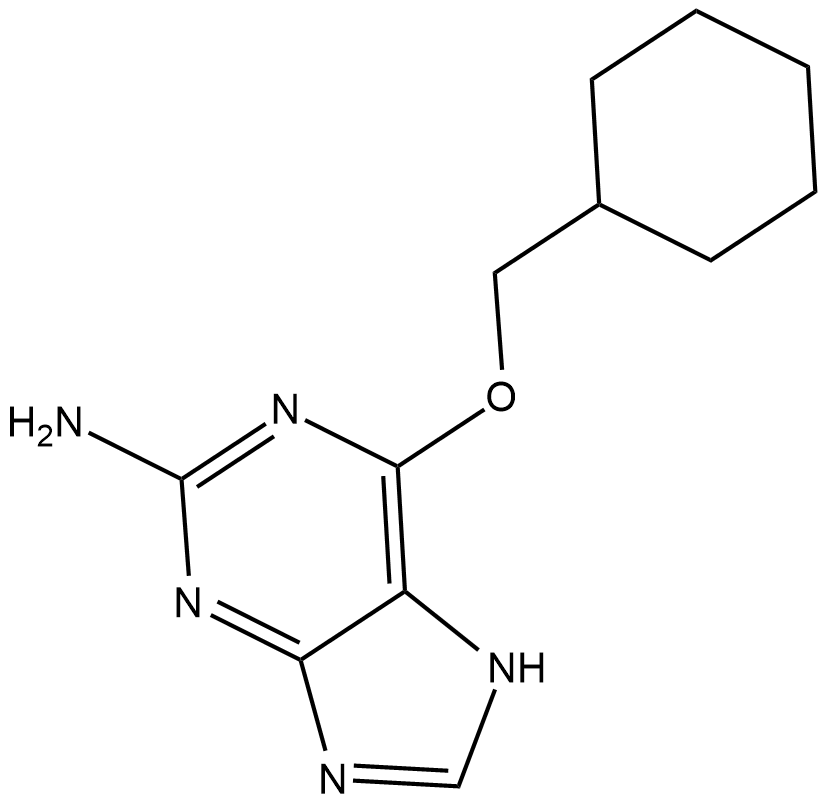
-
GC34692
NU6140
A Cdk2 inhibitor
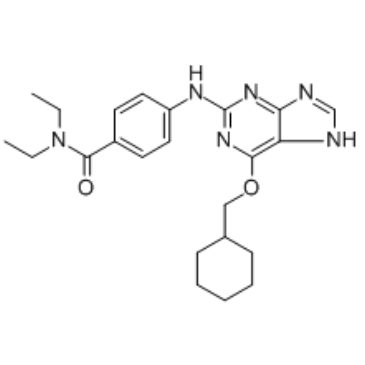
-
GC32829
NU6300
NU6300 is a covalent, irreversible and ATP-competitive CDK2 inhibitor with an IC50 value of 0.16 μM. NU6300 can be used for the research of eukaryotic cell cycle- and transcription-related.
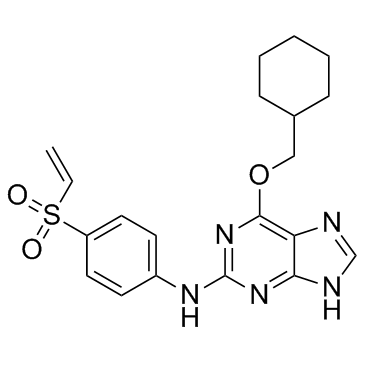
-
GC34056
NVP-2
NVP-2 is a potent and selective ATP-competitive cyclin dependent kinase 9 (CDK9) probe, inhibits CDK9/CycT activity with an IC50 of 0.514 nM. NVP-2 displays inhibitory effcts on CDK1/CycB, CDK2/CycA and CDK16/CycY kinases with IC50 values of 0.584 ?M, 0.706 ?M, and 0.605 ?M, respectively. NVP-2 induces cell apoptosis.
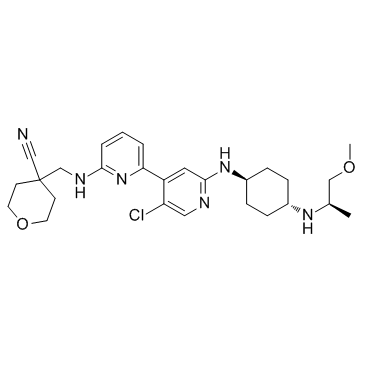
-
GC16959
NVP-LCQ195
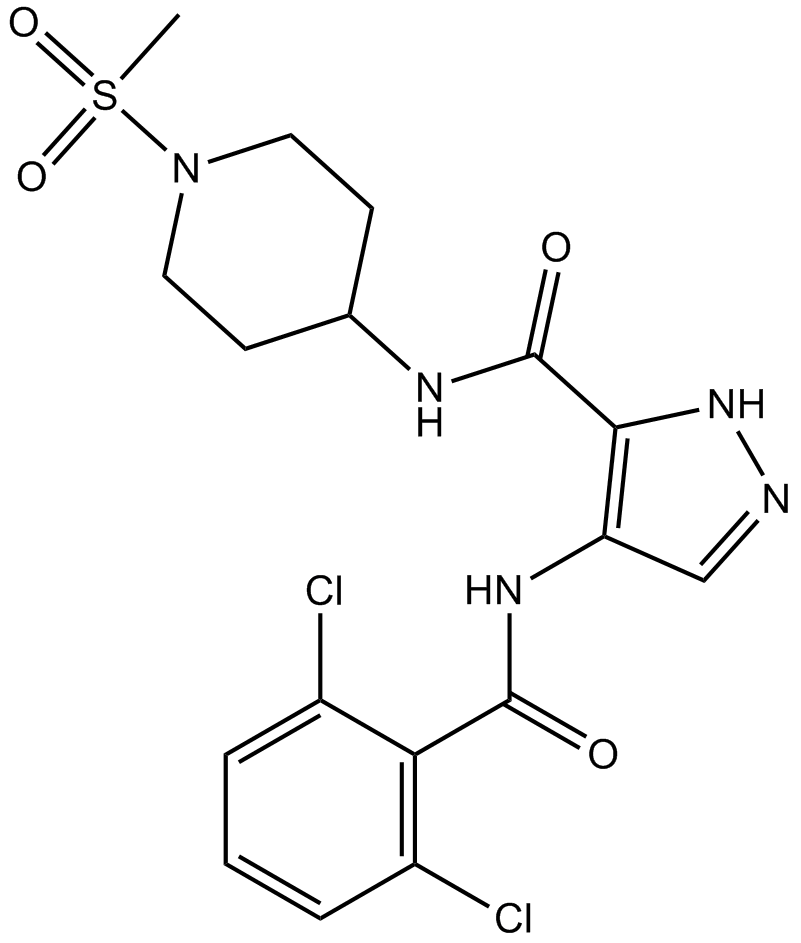
-
GC33353
ON-013100
ON-013100, an antineoplastic drug, acts a mitotic inhibitor that could inhibit Cyclin D1 expression.
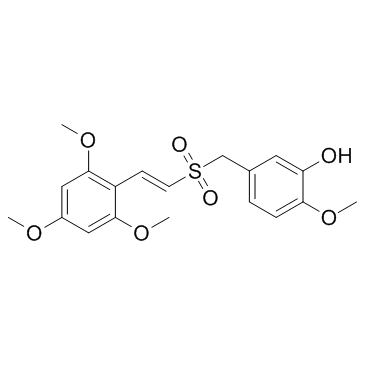
-
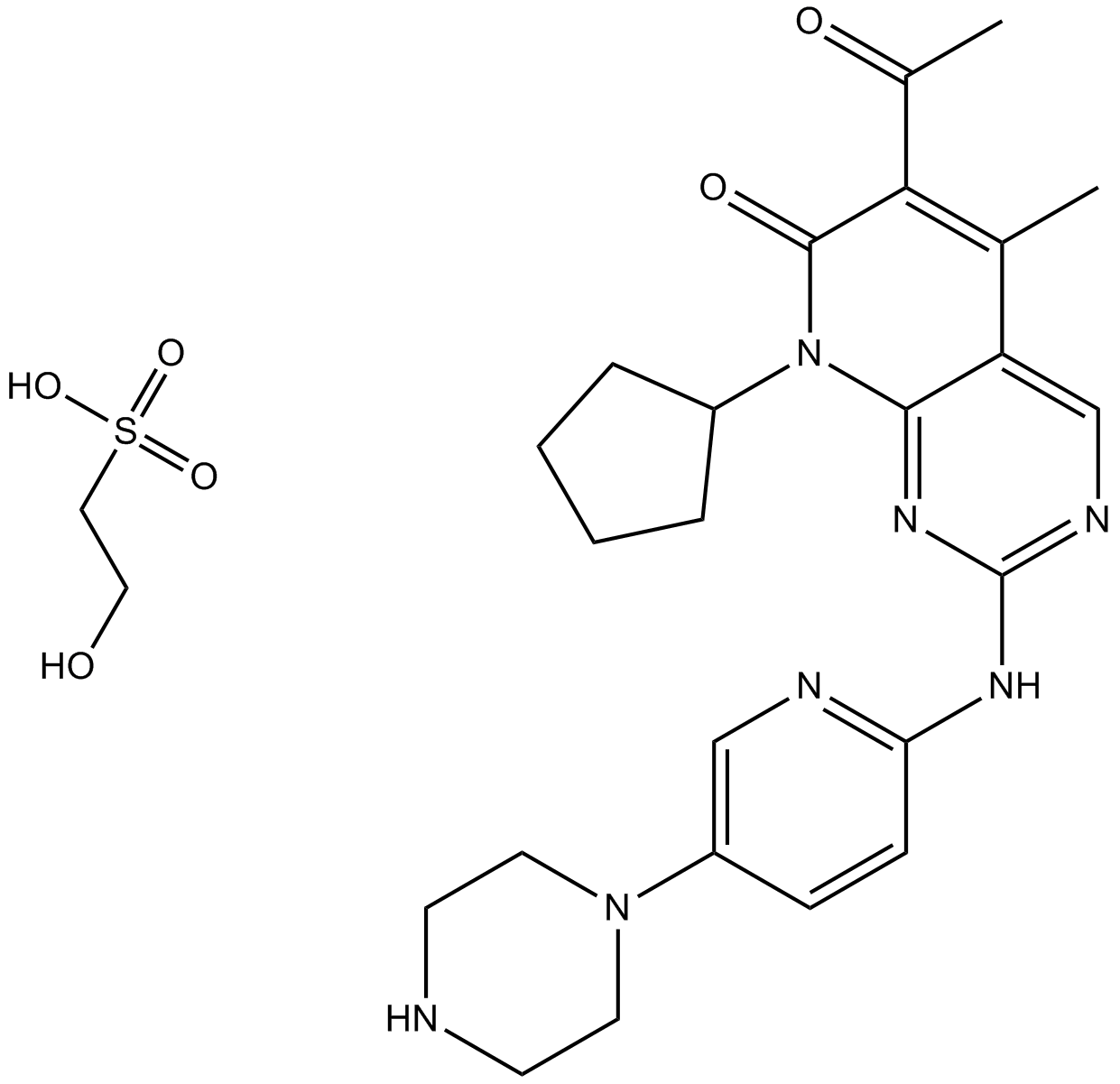
GC15607
ON123300
multi-targeted kinase inhibitor,inhibits CDK4, Ark5, PDGFRβ, FGFR1, RET, and Fyn
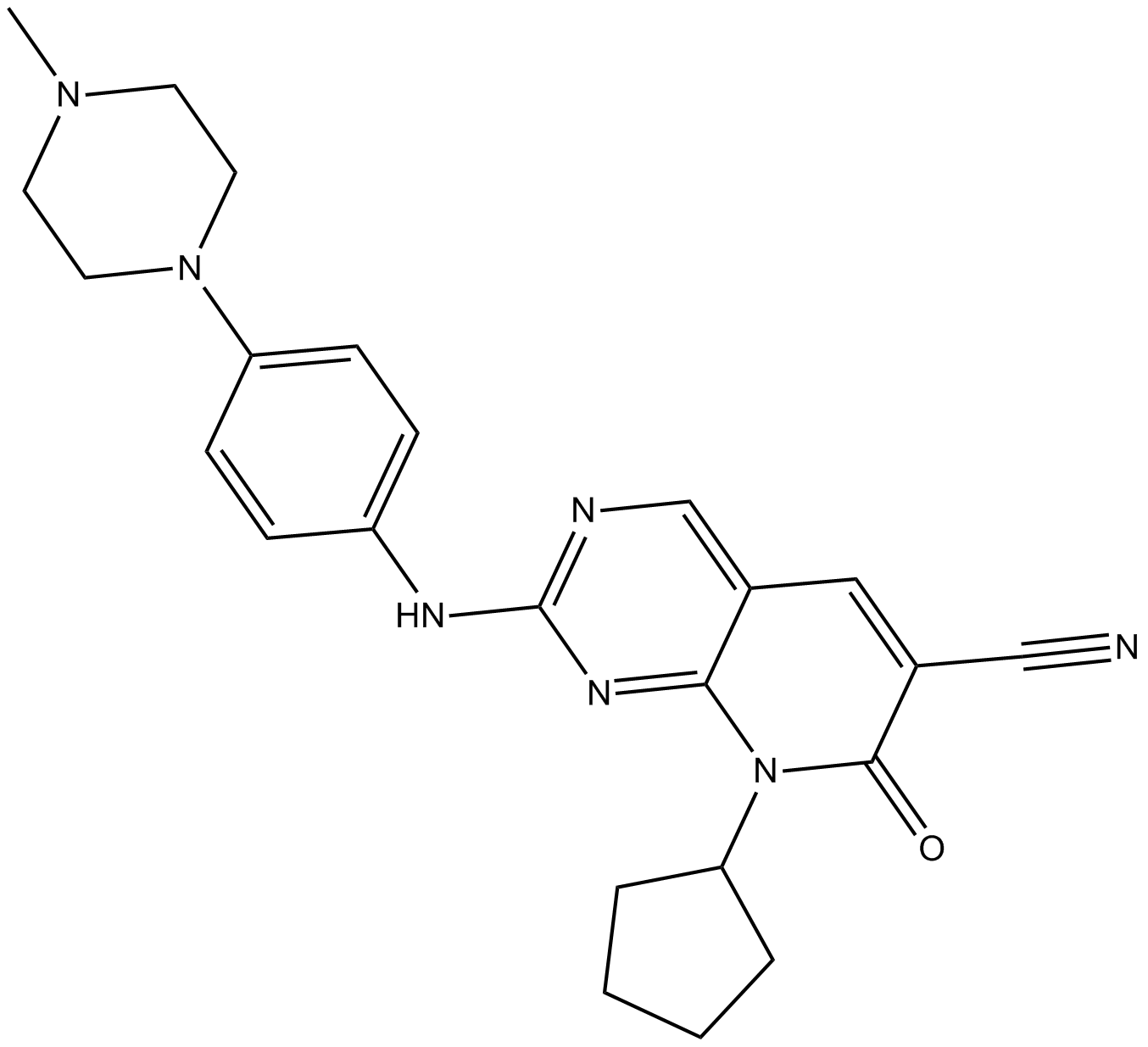
-
GC16511
OTS964
OTS964 is an orally active, high affinity and selective TOPK (T-lymphokine-activated killer cell-originated protein kinase) inhibitor with an IC50 of 28 nM. OTS964 is also a potent inhibitor of the cyclin-dependent kinase CDK11, which binds to CDK11B with a Kd of 40 nM.
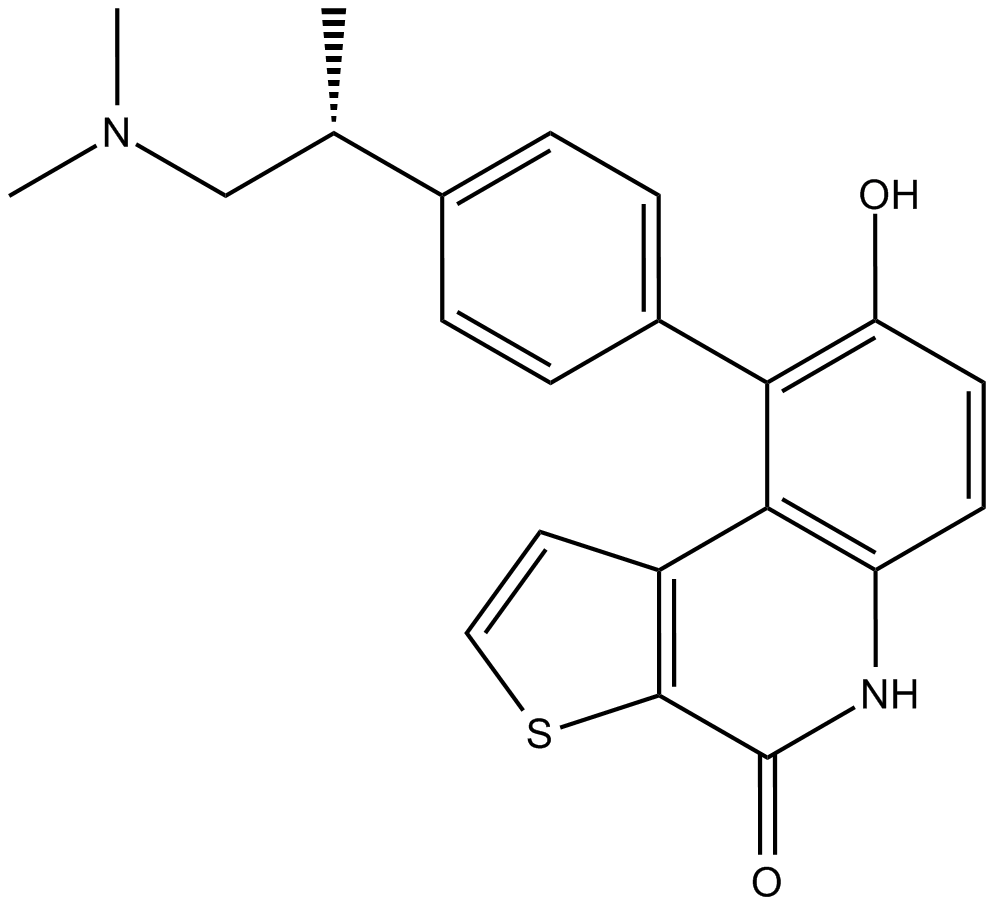
-
GC12011
P276-00
P276-00 (P276-00) is a potent cyclin-dependent kinase (CDK) inhibitor, which inhibits CDK9-cyclinT1, CDK4-cyclin D1, and CDK1-cyclinB with IC50s of 20 nM, 63 nM, and 79 nM, respectively.P276-00 (P276-00) shows antitumor activity on cisplatin-resistant cells.
-
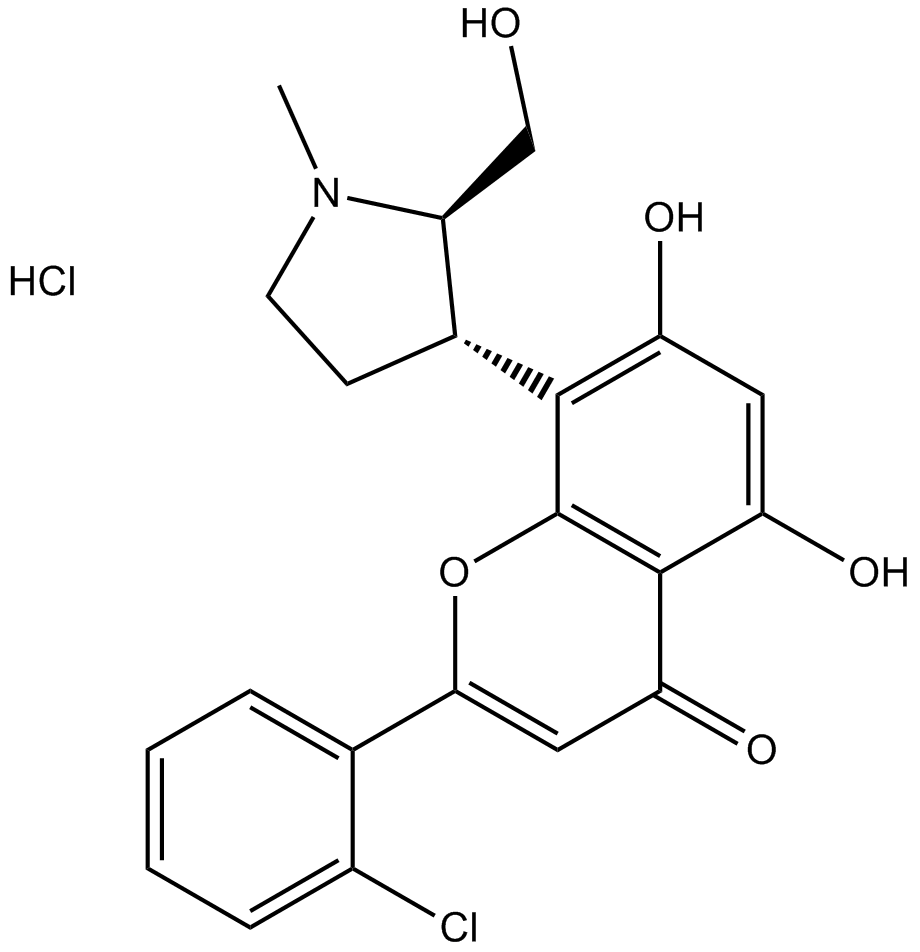
GC15421
Palbociclib (PD0332991) Isethionate
Palbociclib (PD 0332991) isethionate is an orally active selective CDK4 and CDK6 inhibitor with IC50 values of 11 and 16 nM, respectively. Palbociclib (PD0332991) Isethionate has potent anti-proliferative activity and induces cell cycle arrest in cancer cells, which can be used in the research of HR-positive and HER2-negative breast cancer and hepatocellular carcinoma.
-
GC17935
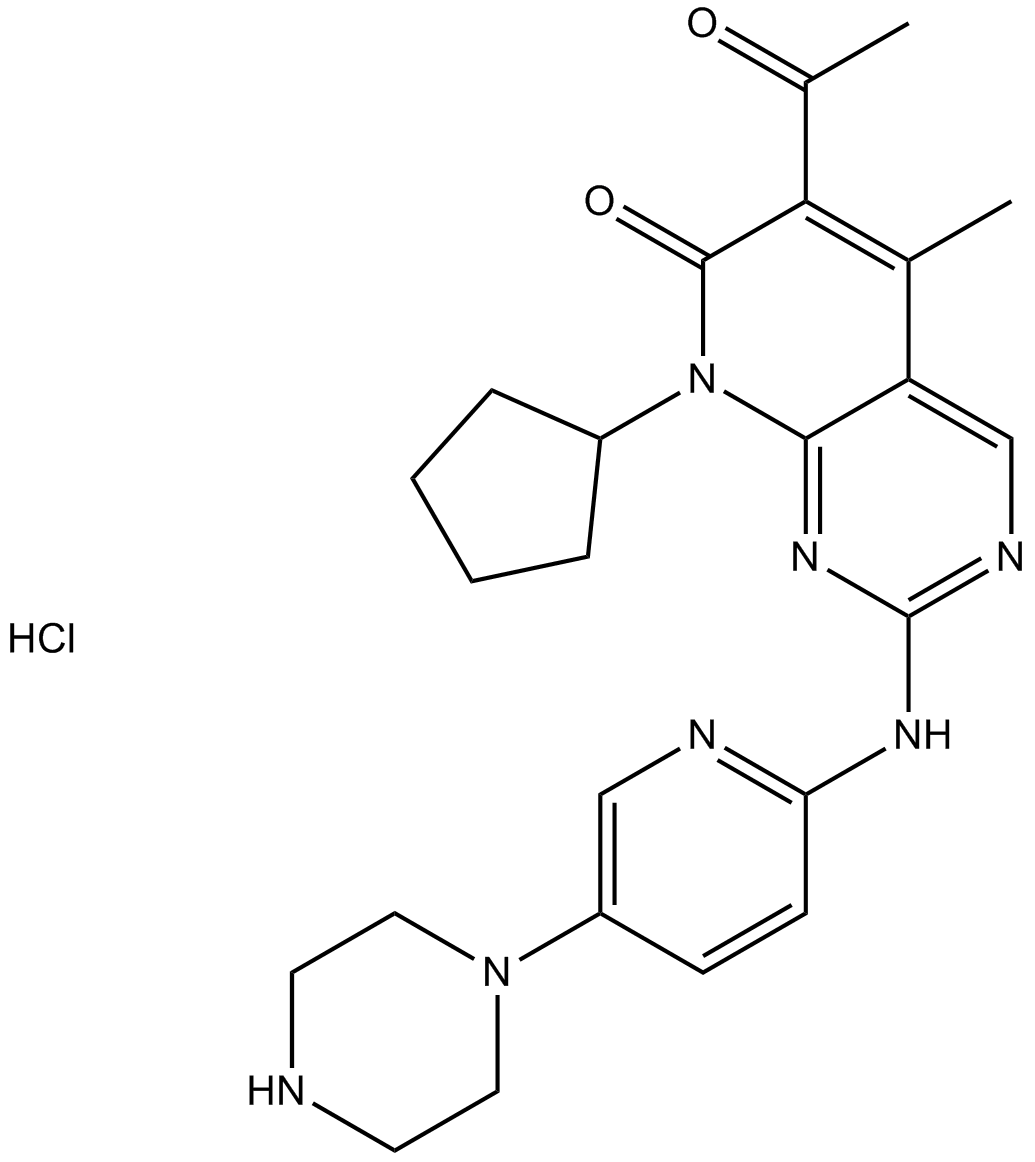
PD 0332991 (Palbociclib) HCl
Palbociclib (PD 0332991) monohydrochloride is an orally active selective CDK4 and CDK6 inhibitor with IC50 values of 11 and 16 nM, respectively. PD 0332991 (Palbociclib) HCl has potent anti-proliferative activity and induces cell cycle arrest in cancer cells, which can be used in the research of HR-positive and HER2-negative breast cancer and hepatocellular carcinoma.
-
GC40092
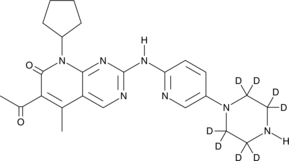
PD 0332991-d8
Palbociclib D8 (PD 0332991 D8) is a deuterium labeled Palbociclib. Palbociclib is a selective and orally active CDK4 and CDK6 inhibitor with IC50s of 11 and 16 nM, respectively. Palbociclib has the potential for ER-positive and HER2-negative breast cancer research.
-
GC32832
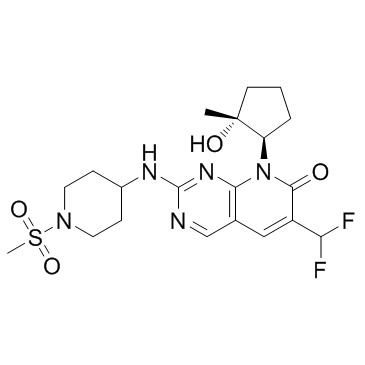
PF-06873600
-
GC62660
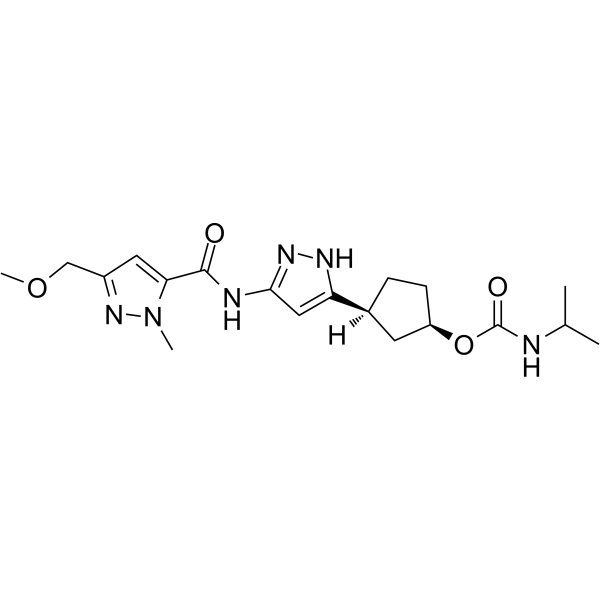
PF-07104091
PF-07104091 is a CDK2/cyclin E1 inhibitor, a selective ATP-site inhibitor targeting the cyclin-bound activated state of the kinase [1].
-
GC62661
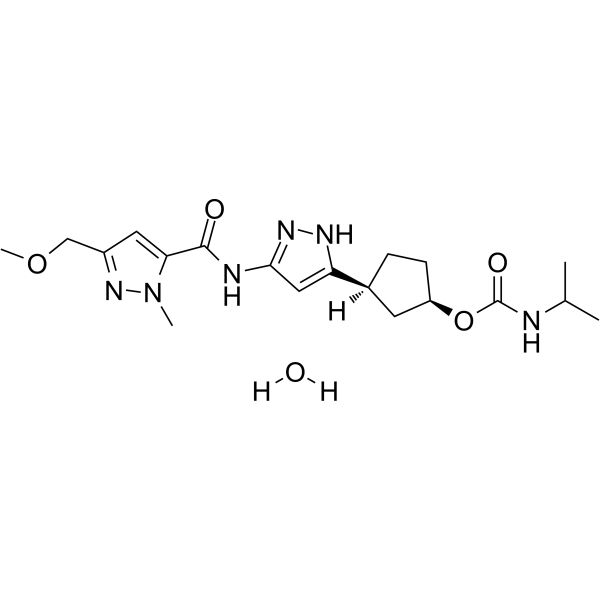
PF-07104091 hydrate
PF-07104091 hydrate is a potent and selective CDK2/cyclin E1 and GSK3β inhibitor, with Kis of 1.16 and 537.81 nM, respectively.
-
GC11324
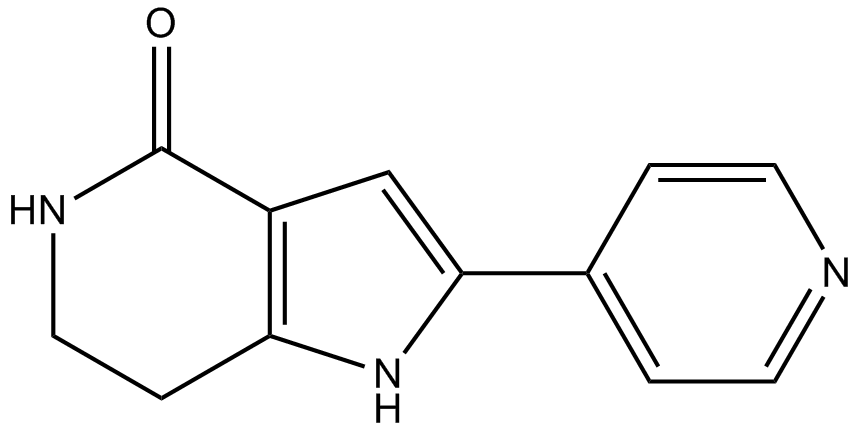
PHA-767491
A potent Cdc7 kinase inhibitor
-
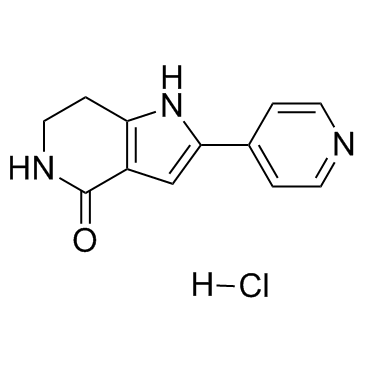
GC36892
PHA-767491 hydrochloride
A potent Cdc7 kinase inhibitor
-
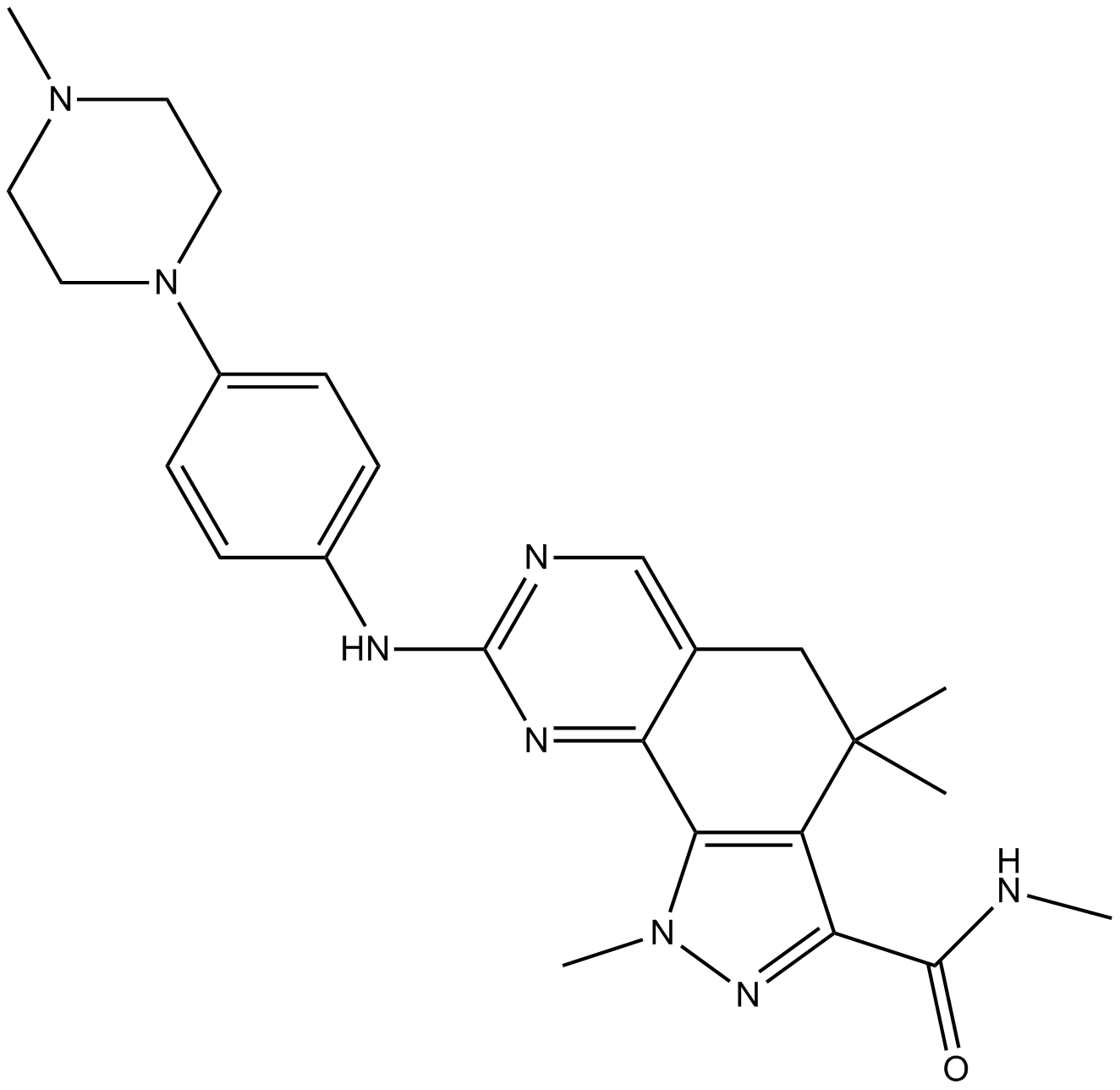
GC15588
PHA-848125
PHA-848125 (PHA-848125) is a potent, ATP-competitive and dual inhibitor of CDK and Tropomyosin receptor kinase (TRK), with IC50s of 45, 150, 160, 363, 398 nM and 53 nM for cyclin A/CDK2, cyclin H/CDK7, cyclin D1/CDK4, cyclin E/CDK2, cyclin B/CDK1 and TRKA, respectively.
-
GC39421
PNU112455A hydrochloride
A Cdk2 and Cdk5 inhibitor
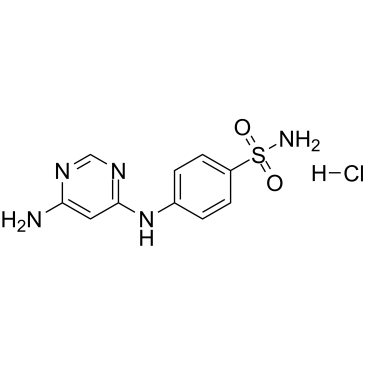
-
GC69747
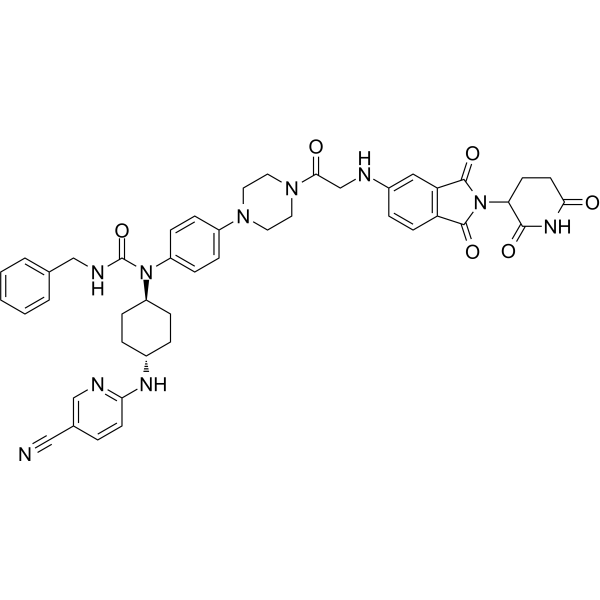
PROTAC CDK12/13 Degrader-1
PROTAC CDK12/13 Degrader-1 (7f) is a highly efficient and selective dual degrader of cell cycle-dependent kinases CDK12/CDK13, with DC50 values of 2.2 nM and 2.1 nM, respectively. PROTAC CDK12/13 Degrader-1 has anti-proliferative activity and can be used for breast cancer research.
-
GC62666
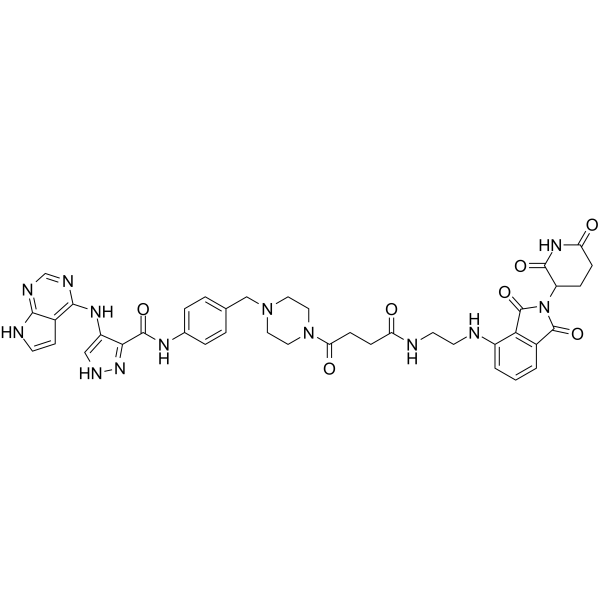
PROTAC CDK2/9 Degrader-1
PROTAC CDK2/9 Degrader-1 (Compound F3) is a potent dual degrader for CDK2 (DC50=62 nM) and CDK9 (DC50=33 nM). PROTAC CDK2/9 Degrader-1 suppresses prostate cancer PC-3 cell proliferation (IC50=0.12 ?M) by effectively blocking the cell cycle in S and G2/M phases. PROTAC CDK2/9 Degrader-1 is a PROTAC by tethering CDK inhibitor with Cereblon ligand.
-
GC32845
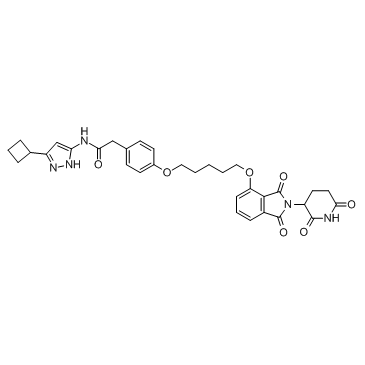
PROTAC CDK9 Degrader-1
PROTAC CDK9 Degrader-1 is a PROTAC connected by ligands for Cereblon and CDK as a selective CDK9 degrader.
-
GC14707
Purvalanol A
potent, and cell-permeable CDK inhibitor
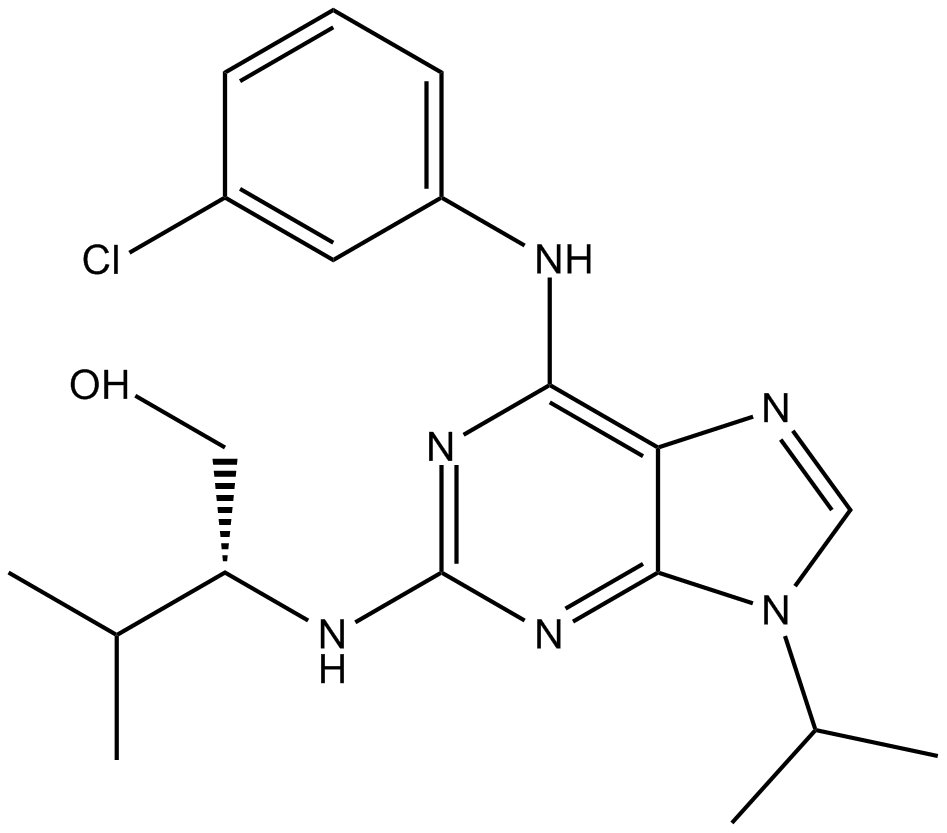
-
GC16268
Purvalanol B
CDK1/CDK2/CDK4 inhibitor
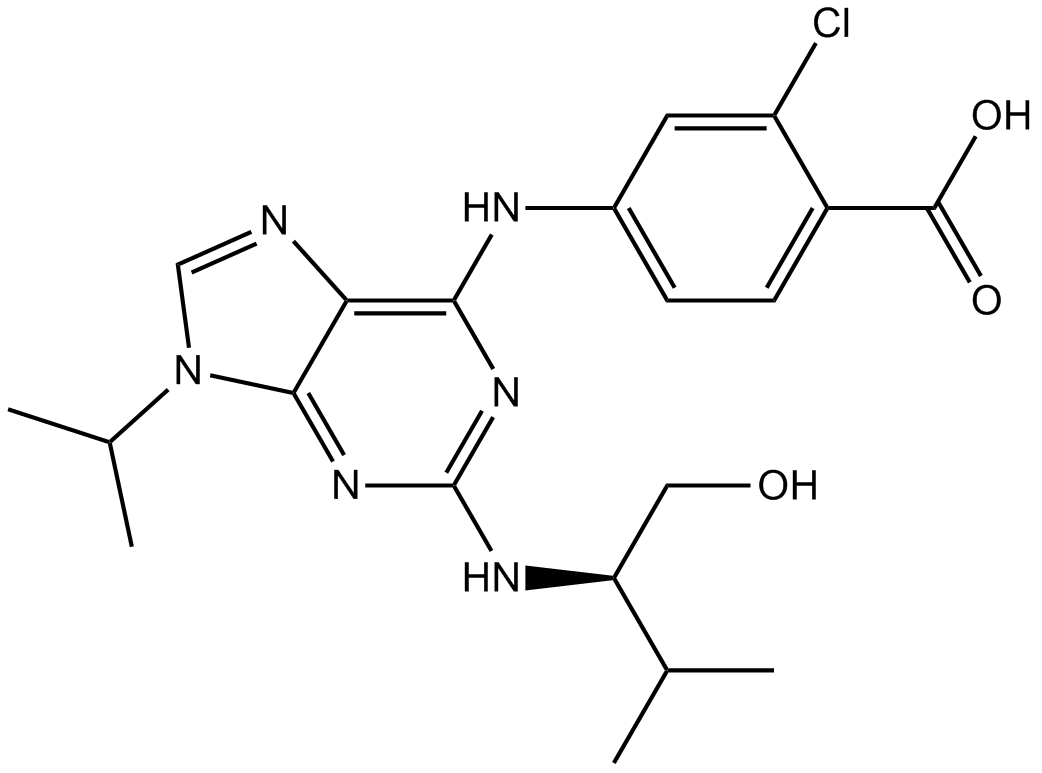
-
GC17400
R547
CDK1/2/4 inhibitor,ATP-competitive
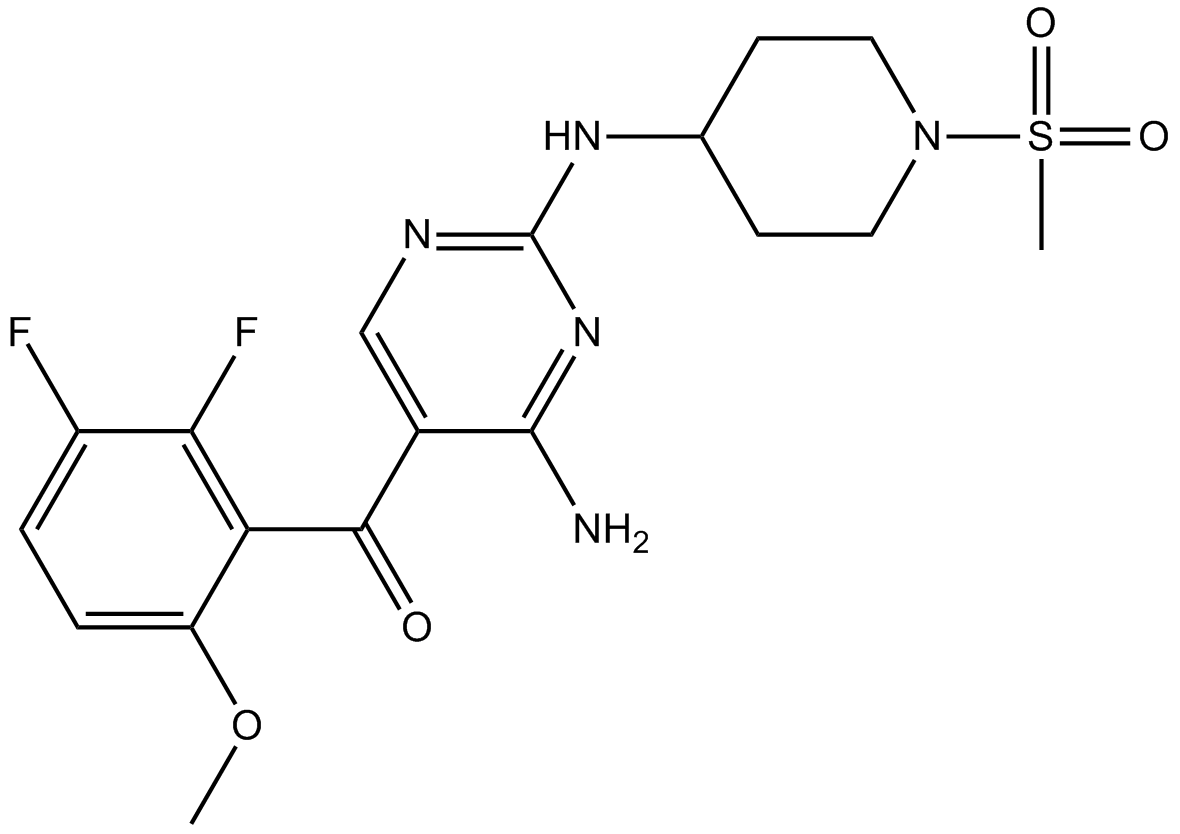
-
GC37522
RGB-286638
A multi-kinase inhibitor
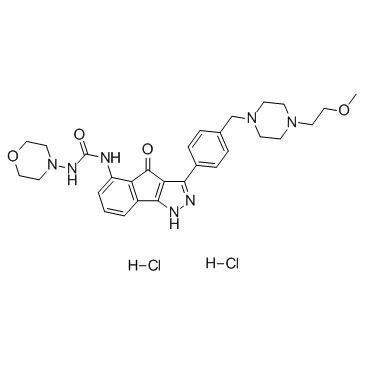
-
GC37523
RGB-286638 free base
A multi-kinase inhibitor
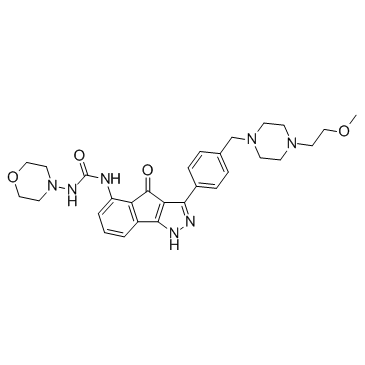
-
GC60324
Ribociclib D6
Ribociclib D6 (LEE011 D6) is a deuterium labeled Ribociclib. Ribociclib is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
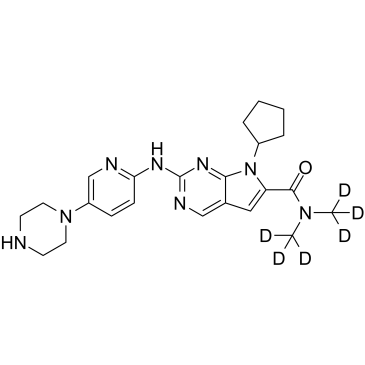
-
GC62638
Ribociclib D6 hydrochloride
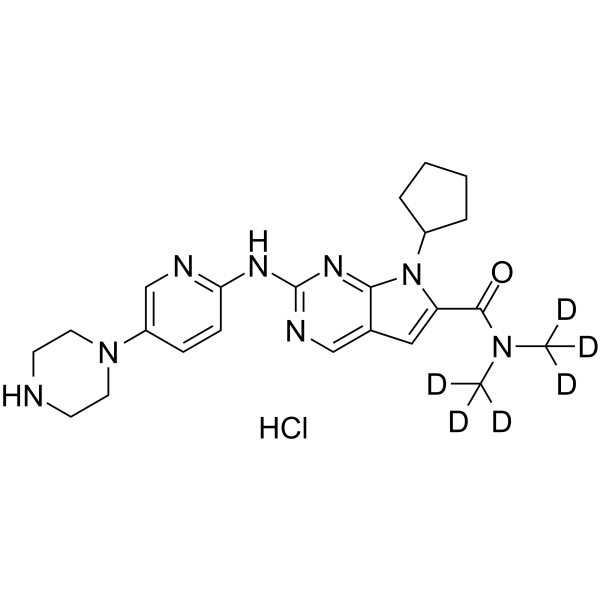
-
GC37529
Ribociclib succinate hydrate
Ribociclib succinate hydrate (LEE011 succinate hydrate) is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
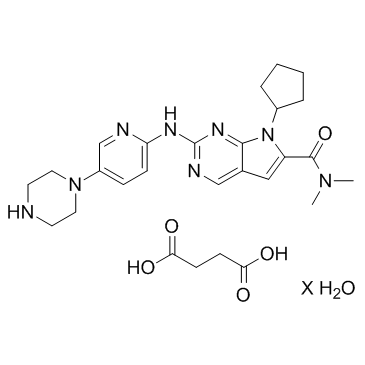
-
GC12348
Ro 3306
An ATP-competitive, potent CDK1 inhibitor
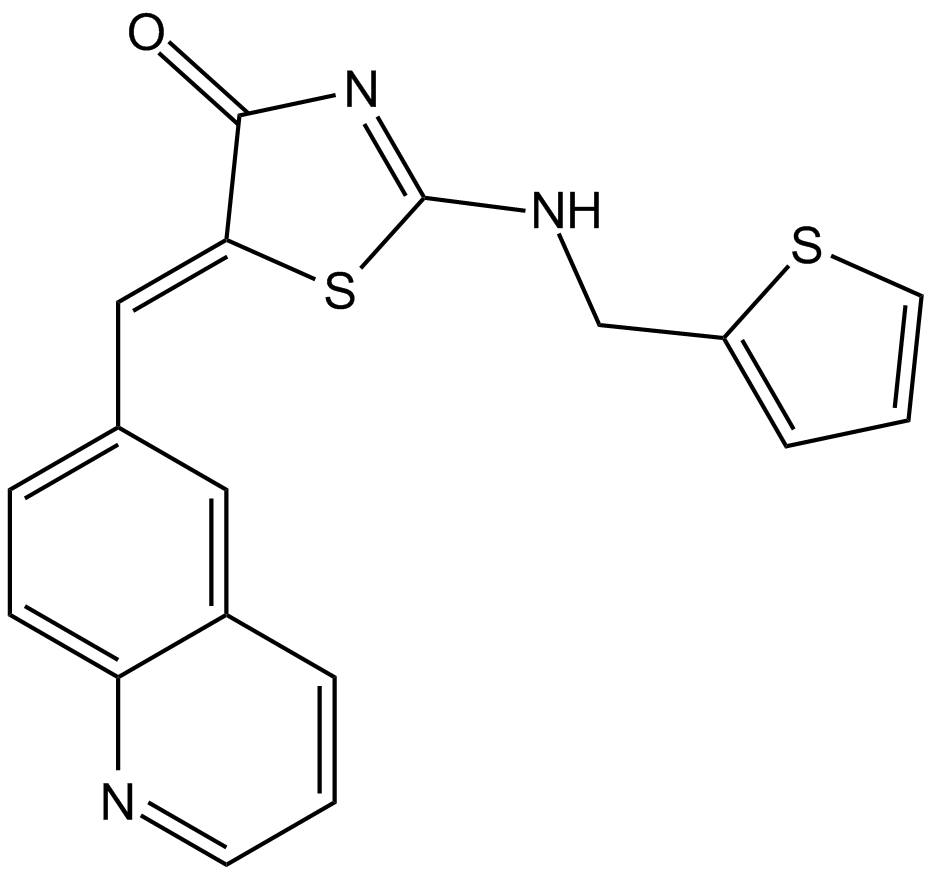
-
GC33360
Roniciclib (BAY 1000394)
Roniciclib (BAY 1000394) is an orally bioavailable pan-cyclin dependent kinase (CDK) inhibitor, with IC50s of 5-25 nM for CDK1, CDK2, CDK3, CDK4, CDK7 and CDK9.
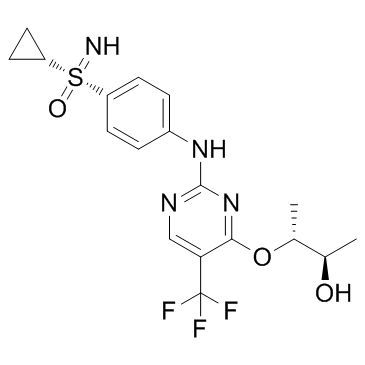
-
GC11401
Roscovitine (Seliciclib,CYC202)
Roscovitine (Seliciclib,CYC202) (Roscovitine) is an orally bioavailable and selective CDKs inhibitor with IC50s of 0.2 μM, 0.65 μM, and 0.7 μM for CDK5, Cdc2, and CDK2, respectively.
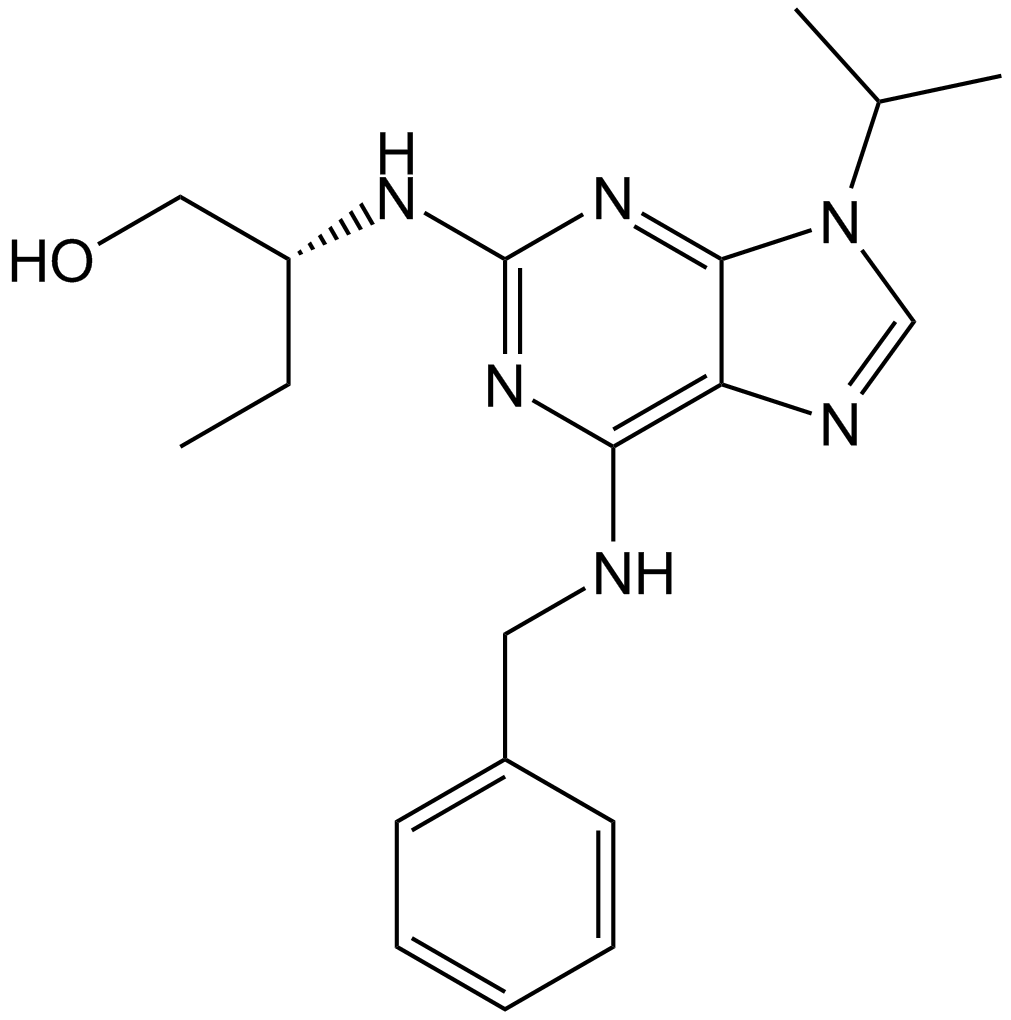
-
GC32735
Samuraciclib hydrochloride (ICEC0942 hydrochloride)
CT7001 hydrochloride (ICEC0942 hydrochloride) (CT7001 hydrochloride) is a potent, selective, ATP-competitive and orally active CDK7 inhibitor, with an IC50 of 41 nM. CT7001 hydrochloride (ICEC0942 hydrochloride) displays 45-, 15-, 230- and 30-fold selectivity over CDK1, CDK2 (IC50 of 578 nM), CDK5 and CDK9, respectively. CT7001 hydrochloride (ICEC0942 hydrochloride) inhibits the growth of breast cancer cell lines with GI50 values between 0.2-0.3 μM. CT7001 hydrochloride (ICEC0942 hydrochloride) has anti-tumor effects.
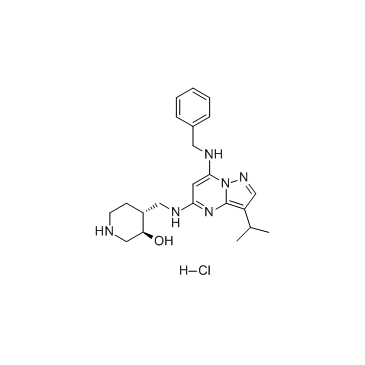
-
GC63736
Samuraciclib hydrochloride hydrate
Samuraciclib (CT7001) hydrochloride hydrate is a potent, selective, ATP-competitive and orally active CDK7 inhibitor, with an IC50 of 41 nM. Samuraciclib hydrochloride hydrate displays 45-, 15-, 230- and 30-fold selectivity over CDK1, CDK2 (IC50 of 578 nM), CDK5 and CDK9, respectively. Samuraciclib hydrochloride hydrate inhibits the growth of breast cancer cell lines with GI50 values between 0.2-0.3 ?M. Samuraciclib hydrochloride hydrate has anti-tumor effects.
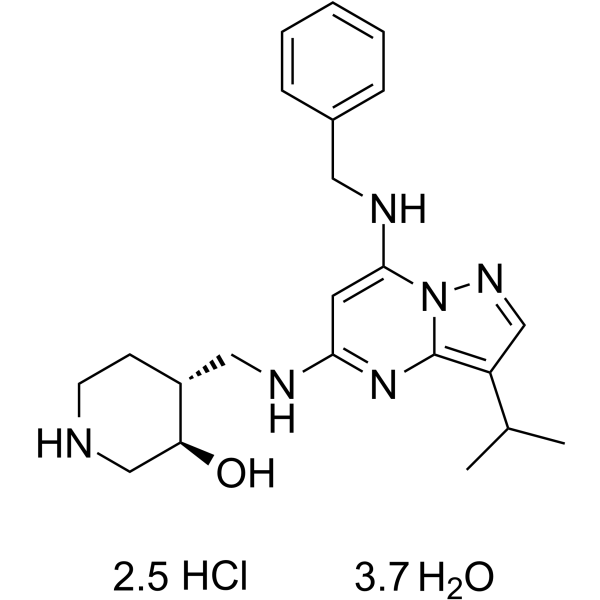
-
GC62319
Samuraciclib trihydrochloride
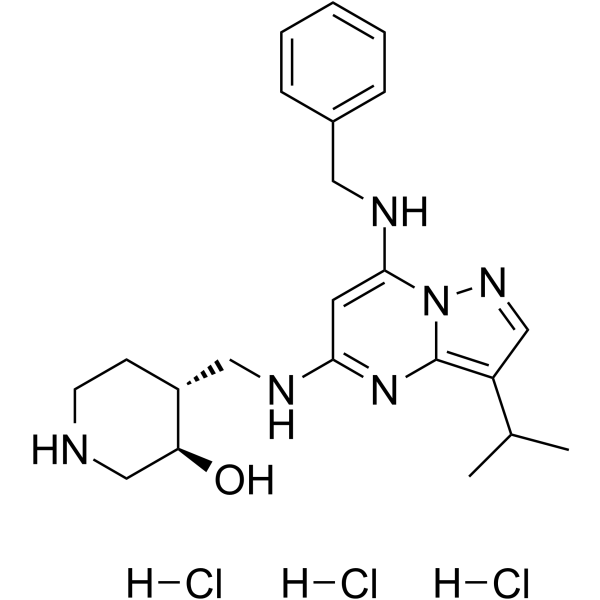
-
GC11022
SB 218078
checkpoint kinase 1 (Chk1) inhibitor
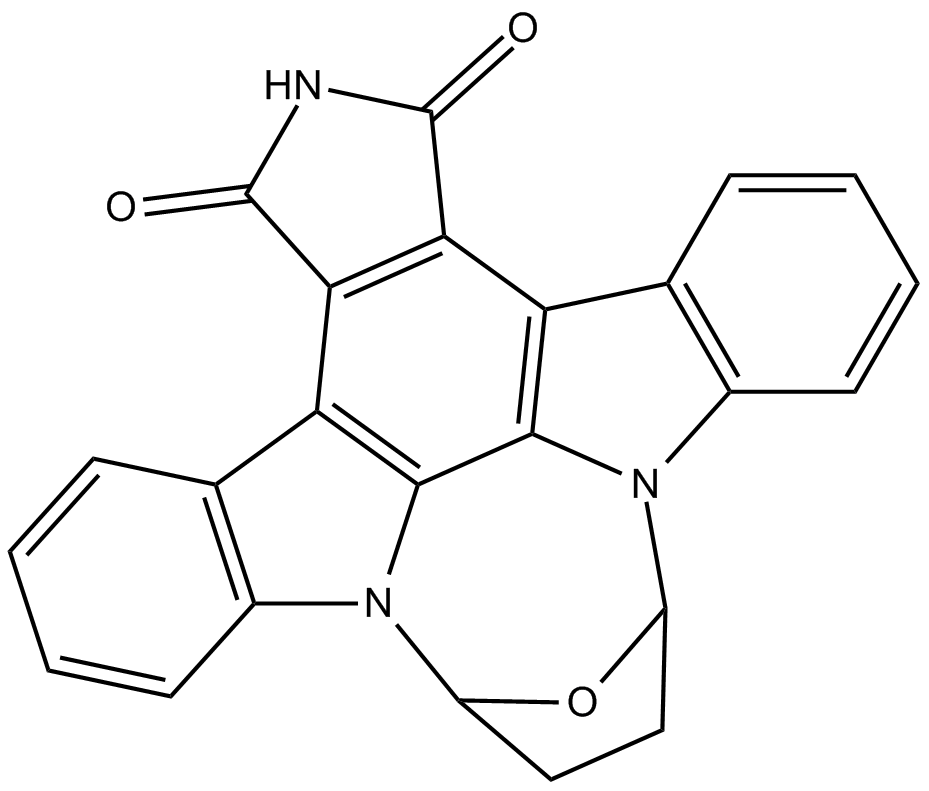
-
GC12064
SB1317
A multi-kinase inhibitor
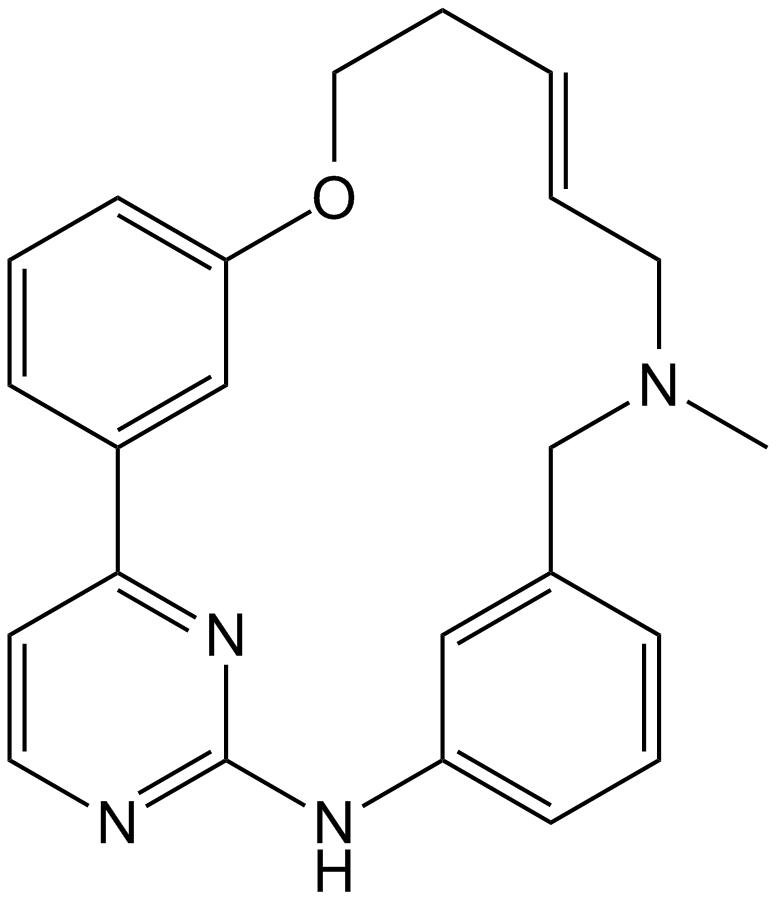
-
GC33137
SEL120-34A
SEL120-34A is a potent, selective, orally available, ATP-competitive CDK8 inhibitor, with IC50s of 4.4 nM and 10.4 nM for CDK8/CycC and CDK19/CycC, respectively, with antitumor activity.
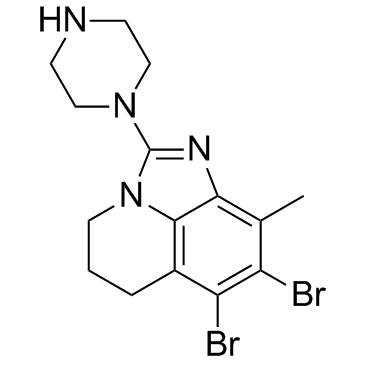
-
GC32898
SEL120-34A HCl
SEL120-34A HCl is a potent, selective, orally available, ATP-competitive CDK8 inhibitor, with IC50s of 4.4 nM and 10.4 nM for CDK8/CycC and CDK19/CycC, respectively, with antitumor activity.
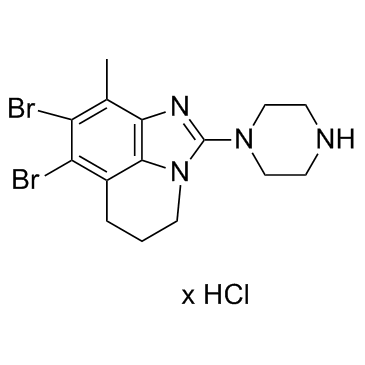
-
GC34355
SEL120-34A monohydrochloride
SEL120-34A monohydrochloride is an ATP-competitive and selective CDK8 inhibitor, inhibits kinase activities of CDK8/CycC and CDK19/CycC complexes with IC50s of 4.4 nM and 10.4 nM, respectively, with a Kd of 3 nM for CDK8. SEL120-34A monohydrochloride weakly inhibits CDK9 (calculated IC50=1070 nM), but shows no obvious activity against CDK1, 2, 4, 6, 5, 7. SEL120-34A monohydrochloride inhibits phosphorylation of STAT1 S727 and STAT5 S726. Has anti-tumor activity.
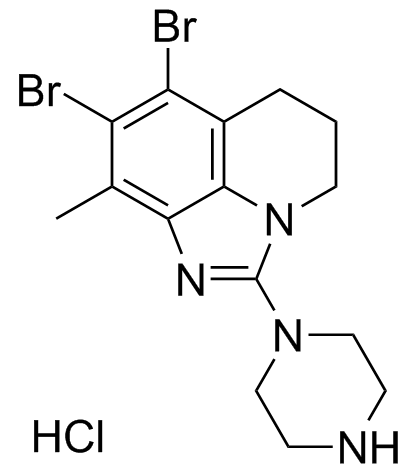
-
GC37627
Senexin A
Senexin A is a CDK8 inhibitor with an IC50 of 280 nM.
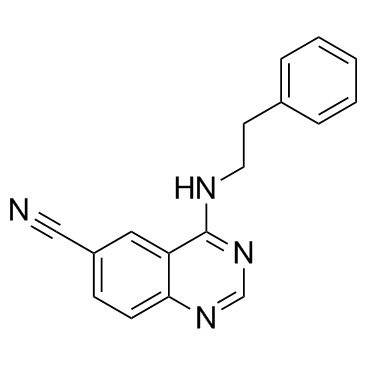
-
GC19326
Senexin B
Senexin B is a potent, highly water-soluble and bioavailable CDK8/19 inhibitor, with Kds of 140 nM for CDK8 and 80 nM for CDK19.
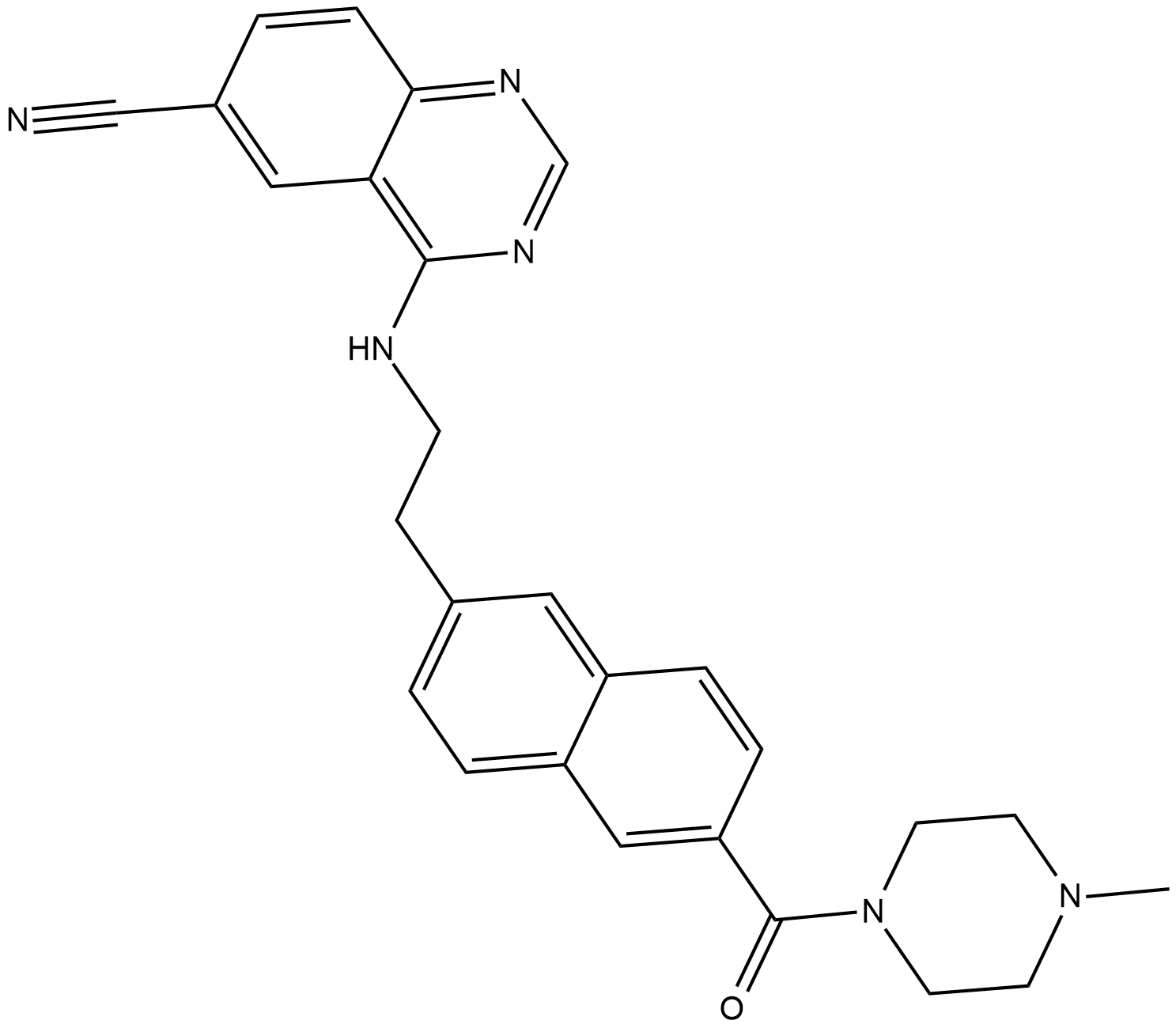
-
GC32930
Simurosertib (TAK-931)
Simurosertib (TAK-931) (TAK-931) is an orally active, selective and ATP-competitive cell division cycle 7 (CDC7) kinase inhibitor, with an IC50 of <0.3 nM. Simurosertib (TAK-931) has anti-cancer activity.
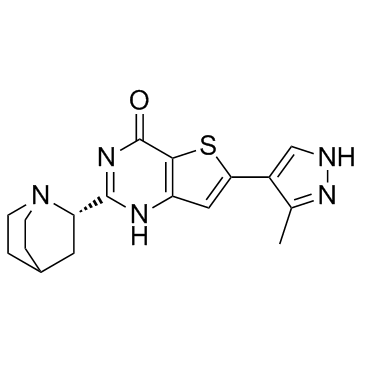
-
GC11396
SNS-032 (BMS-387032)
SNS-032 (BMS-387032) (BMS-387032) is a potent and selective inhibitor ofCDK2, CDK7, and CDK9 withIC50sof 38 nM, 62 nM and 4 nM, respectively. SNS-032 (BMS-387032) has antitumor effect.
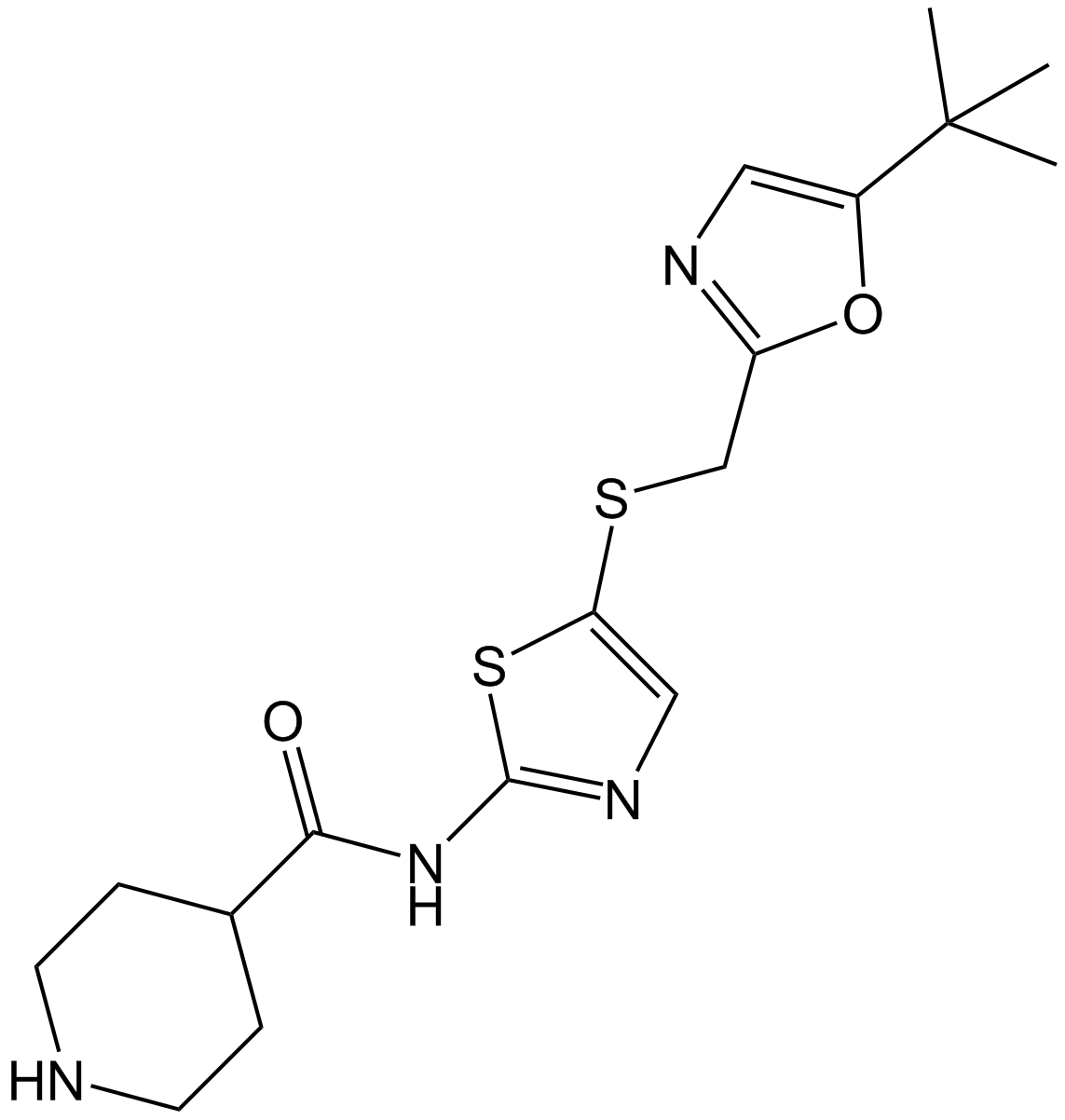
-
GC39175
SR-4835
SR-4835 is a potent, highly selective and ATP competitive dual inhibitor of CDK12/CDK13 (CDK12: IC50=99 nM, Kd=98 nM; CDK13: Kd=4.9 nM). SR-4835 acts in synergy with DNA-damaging chemotherapy and PARP inhibitors and provokes triple-negative breast cancer (TNBC) cell death.
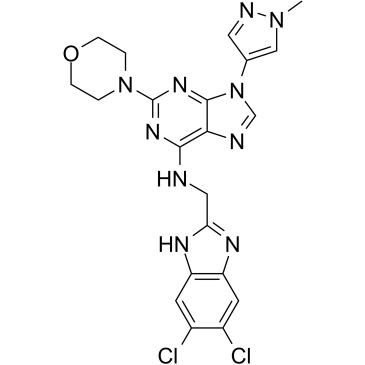
-
GC63201
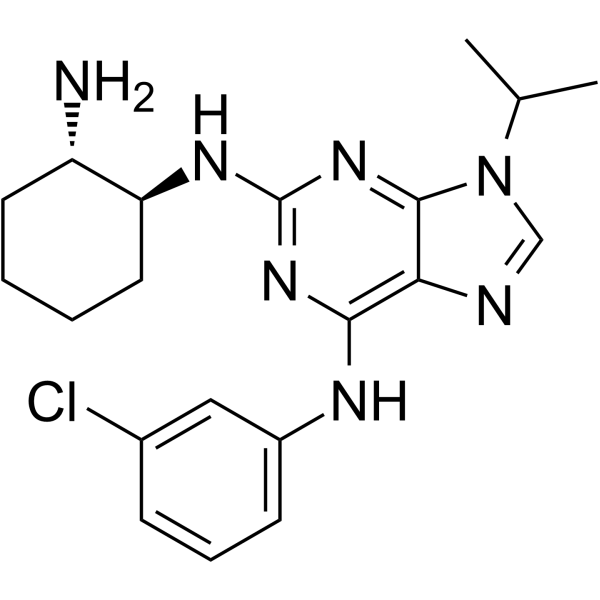
SRI-29329
SRI-29329 is a specific CLK inhibitor, with IC50 values of 78 nM, 16 nM and 86 nM for CLK1, CLK2 and CLK4, respectively.
-
GC15571
SU 9516
A pro-apoptotic Cdk2/cyclin A inhibitor

-
GC37709
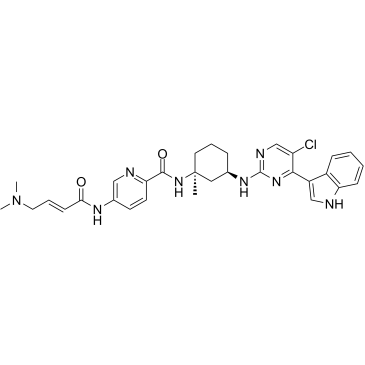
SY-1365
SY-1365 (SY-1365) is a potent and first-in-class selective CDK7 inhibitor, with a Ki of 17.4 nM. SY-1365 exhibits anti-proliferative and apoptotic effects in solid tumor cell lines. SY-1365 possesses anti-tumor activity in hematological and multiple aggressive solid tumors.
-
GC65395
T025
T025 is an orally active and highly potent inhibitor of Cdc2-like kinase (CLKs), with Kd values of 4.8, 0.096, 6.5, 0.61, 0.074, 1.5 and 32 nM for CLK1, CLK2, CLK3, CLK4, DYRK1A, DYRK1B and DYRK2, respectively.

-
GC69987
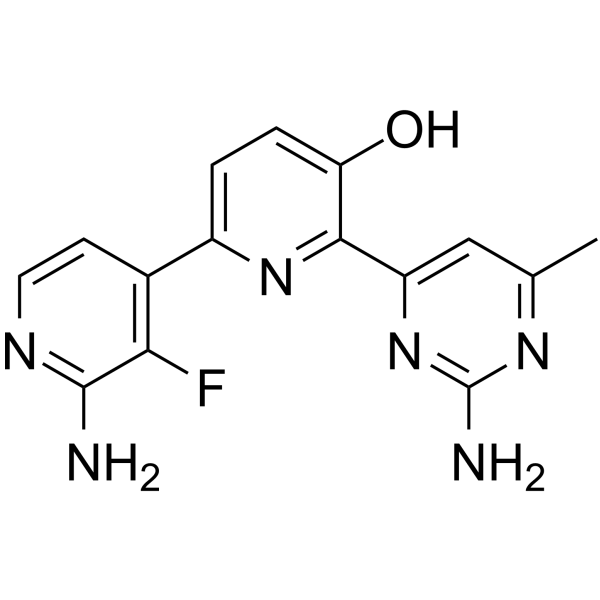
Tanuxiciclib
Tanuxiciclib is a cell cycle cyclin-dependent kinase (CDK) inhibitor.
-
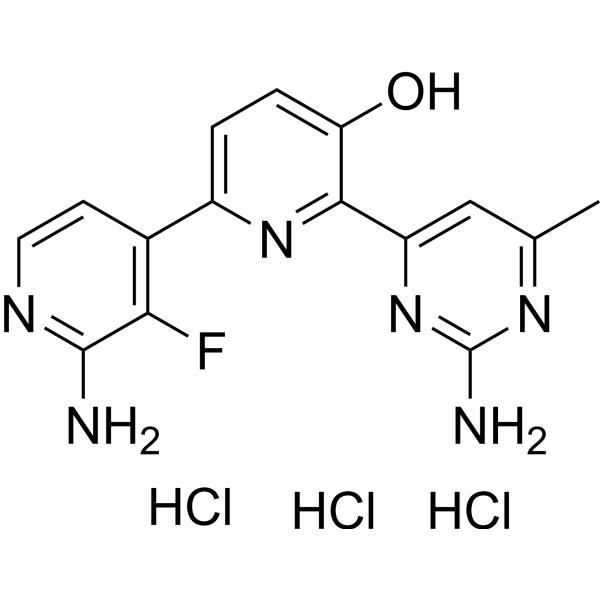
GC69988
Tanuxiciclib trihydrochloride
Tanuxiciclib trihydrochloride is a cell cycle cyclin-dependent kinase (CDK) inhibitor.
-
GC39162
TCMDC-135051
TCMDC-135051 is a highly selective and potent protein kinase PfCLK3 inhibitor with low off-target toxicity.