Peptides
Products for Peptides
- Cat.No. Product Name Information
-
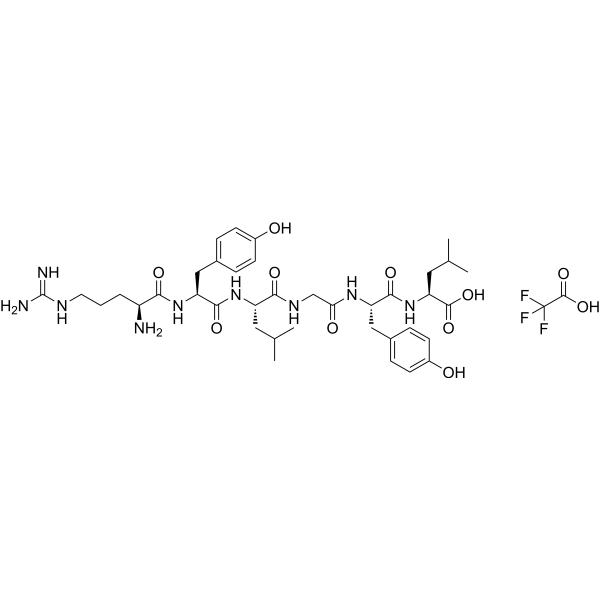
GC70180
α-Casein (90-95) (TFA)
Alpha-casein (90-95) TFA is a peptide fragment of alpha-casein.
-
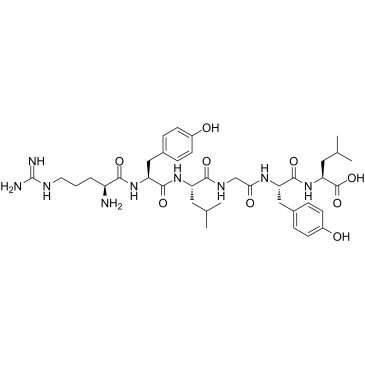
GC37976
α-Casein 90-95
α-Casein 90-95 is a peptide fragment of α-Casein.
-
GC62017
α-CGRP, rat TFA

-
GC62016
α-Conotoxin AuIB TFA

-
GA24163
α-Conotoxin EI (free acid)
α-Conotoxin El, Conus ermineus, free acid
Nicotinic acetylcholine receptor ligand.
-
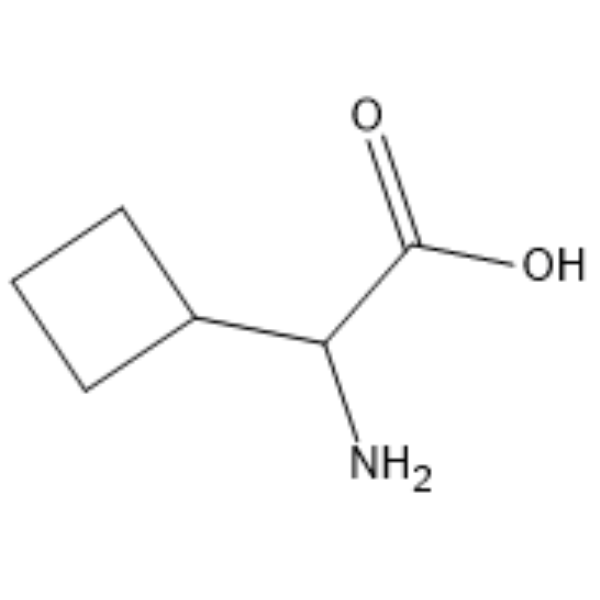
GC66247
α-Cyclobutylglycine
α-Cyclobutylglycine is a Glycine (HY-Y0966) derivative.
-
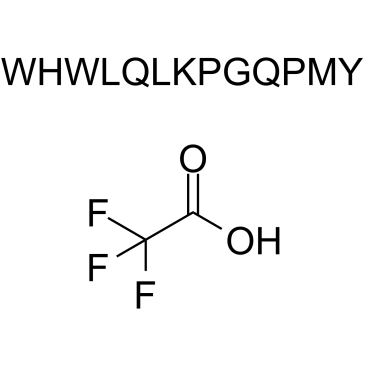
GC38042
α-Factor Mating Pheromone, yeast (TFA)
Mating Factor α TFA
-
GC63270
α-Glucosidase
α-Glucosidase (α-D-Glucosidase), a carbohydrate hydrolyzing enzyme, catalyzes the liberation of α-glucose from the non-reducing end of the substrate.

-
GC66189
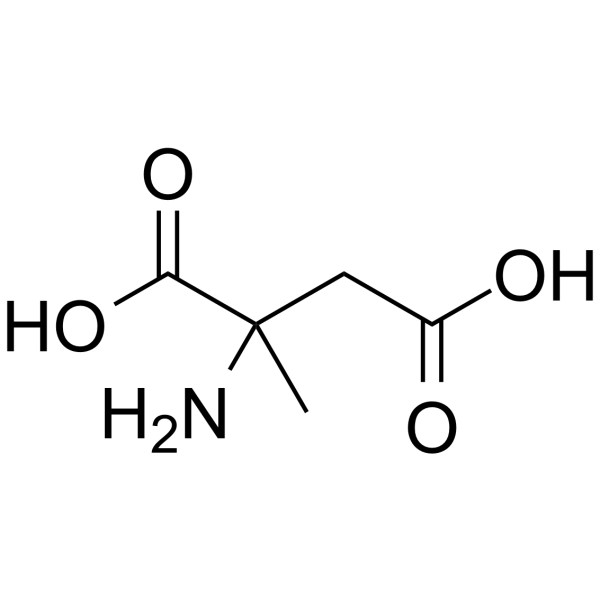
α-Methyl-DL-aspartic acid
α-Methyl-DL-aspartic acid is a specific inhibitor of argininosuccinate synthase (ASS), and also is the rate-limiting enzyme for the recycling of 1-citrulline to 1-arginine.
-
GC66256
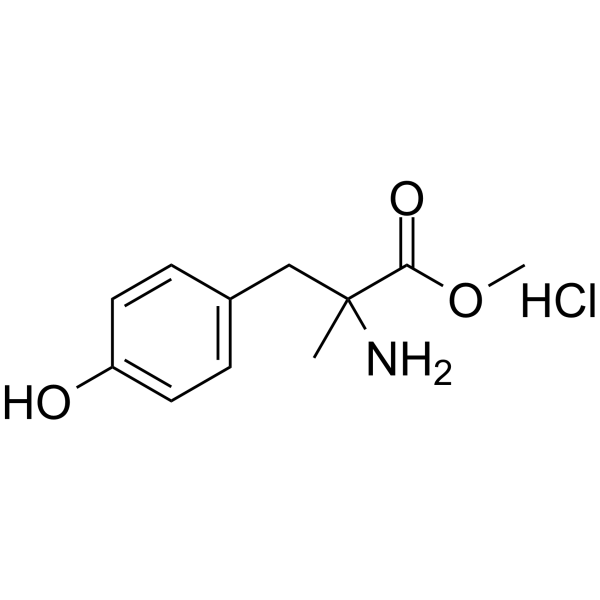
α-Methyltyrosine methyl ester hydrochloride
α-Methyltyrosine methyl ester hydrochloride is a competitive tyrosine hydroxylase inhibitor that inhibits the conversion of tyrosine to dopamine. α-Methyltyrosine methyl ester hydrochloride can be used as a tool for sympathetic nervous system research.
-
GC48292
α-MSH (human, mouse, rat, porcine, bovine, ovine) (trifluoroacetate salt)
α-Melanocyte-stimulating Hormone, Ac-SYSMEHFRWGKPV-NH2
α-MSH (α-Melanocyte-Stimulating Hormone) TFA, an endogenous neuropeptide, is an endogenous melanocortin receptor 4 (MC4R) agonist with anti-inflammatory and antipyretic activities.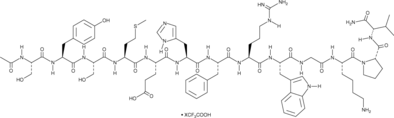
-
GC37978
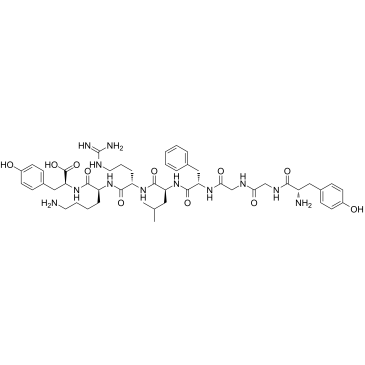
α-Neoendorphin 1-8
α-Neoendorphin 1-8 is a 8-amino acid peptide derived from the N-terminal of α-Neoendorphin.
-
GA24164
α-Synuclein (32-37) (human)
α-Synuclein (43-48) (human)

-
GC63271
α-Synuclein (61-75) (TFA)
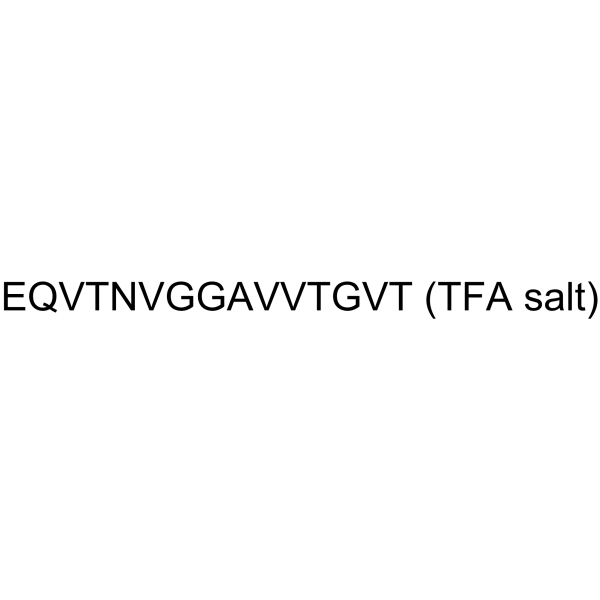
-
GC37975
α2β1 Integrin Ligand Peptide
α2β1 Integrin Ligand Peptide interacts with the α2β1 integrin receptor on the cell membrane and mediates extracellular signals into cells.

-
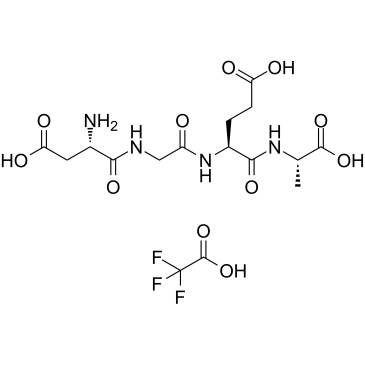
GC38873
α2β1 Integrin Ligand Peptide TFA
-
GC37984
β-Amyloid (1-42), rat
Amyloid β-peptide (1-42) (rat/mouse)
β-Amyloid (1-42), rat is a 42-aa peptide, shows cytotoxic effect on acute hippocampal slices, and used in the research of Alzheimer's disease.
-
GC61988
β-amyloid (12-28) (TFA)
Amyloid β-Protein (12-28) (TFA); Amyloid Beta-Peptide (12-28) (human) TFA; β-Amyloid protein fragment(12-28) TFA

-
GC37986
β-Amyloid 1-17
β-Amyloid 1-17 is a peptide of β-Amyloid, stabilizes the fibres and plays a role in Aβ fibre formation.

-
GC37987
β-Amyloid 1-20
β-Amyloid 1-20 consists of amino acids 1 to 20 of beta amyloid protein.

-
GC37990
β-Amyloid 1-34
β-Amyloid 1-34 is a β-Amyloid peptide consists of 34 amino acid.

-
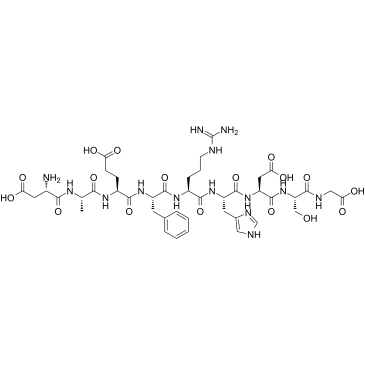
GC37993
β-Amyloid 1-9
β-Amyloid 1-9, an N-terminal fragment of beta amyloid, consists of amino acid residues 1 to 9.
-
GC37985
β-Amyloid 11-22
β-Amyloid 11-22 is a peptide fragment of β-Amyloid.
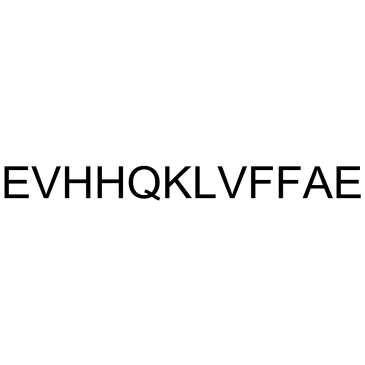
-
GC37988
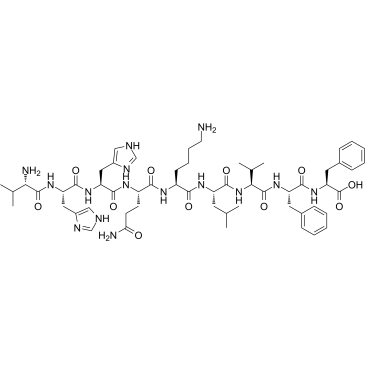
β-Amyloid 12-20
β-Amyloid 12-20 is a peptide fragment of β-Amyloid.
-
GC37989
β-Amyloid 13-27
β-Amyloid 13-27 is a peptide consisting of amino acid of 13 to 27 of beta amyloid protein.
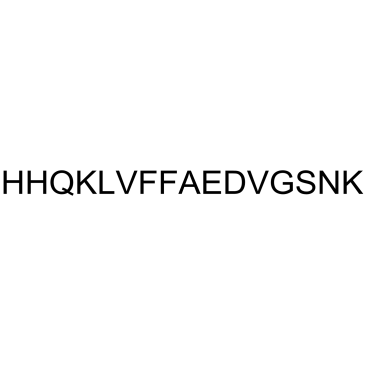
-
GC39466
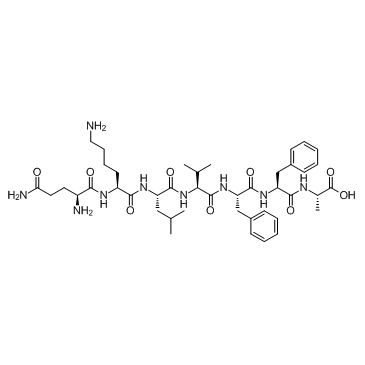
β-Amyloid 15-21

-
GC37991
β-Amyloid 15-21
-
GC37992
β-Amyloid 18-28
β-Amyloid 18-28 is a peptide fragment of β-Amyloid.
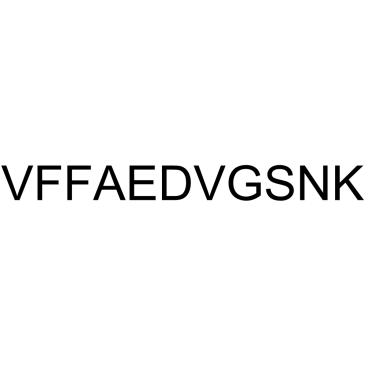
-
GC37994
β-Amyloid 22-40
β-Amyloid 22-40 is a peptide fragment of β-Amyloid.
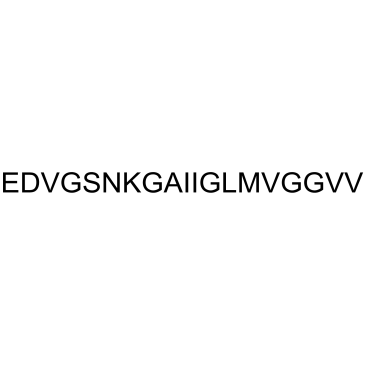
-
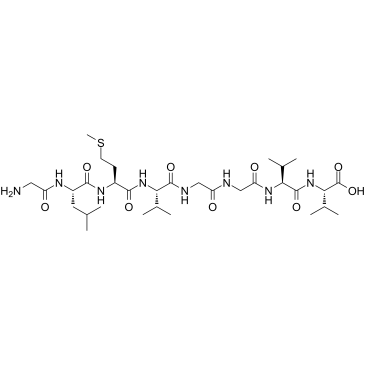
GC37995
β-Amyloid 33-40
β-Amyloid 33-40 is a peptide consisting of amino acid of 33 to 40 of beta amyloid protein.
-
GC37996
β-Amyloid 35-42
β-Amyloid 35-42 is a peptide consisting of amino acid of 35 to 42 of beta amyloid protein.
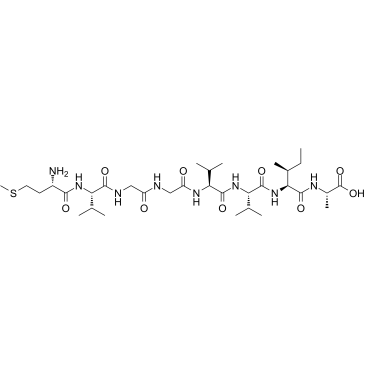
-
GC37997
β-Amyloid 4-10
β-Amyloid 4-10 is an epitope for the polyclonal anti-Aβ(1-42) antibody, reduces amyloid deposition in a transgenic Alzheimer disease mouse model.
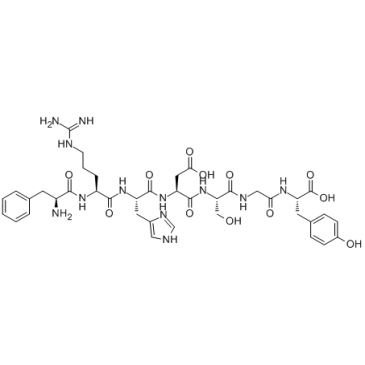
-
GC37998
β-Amyloid Protein Precursor 770 135-155
β-Amyloid Protein Precursor 770 135-155 is a peptide of amyloid precursor protein isoform (APP 770).

-
GC38001
β-Casomorphin (1-3), amide
β-Casomorphin (1-3), amide is a peptide fragment of β-Casomorphin with 3 amino acid.
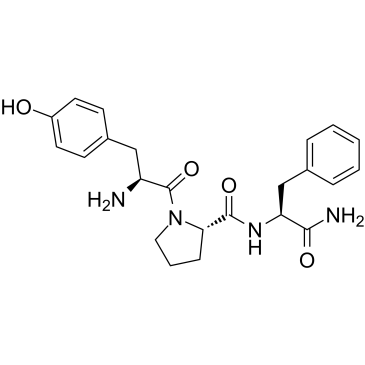
-
GC38002
β-Casomorphin (1-5), amide, bovine
β-Casomorphin (1-5), amide, bovine is a peptide of bovine β-Casomorphin.
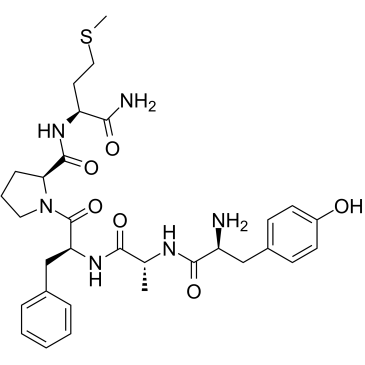
-
GC38003
β-Casomorphin (1-5), bovine
β-Casomorphin (1-5), bovine is a peptide of bovine β-Casomorphin.
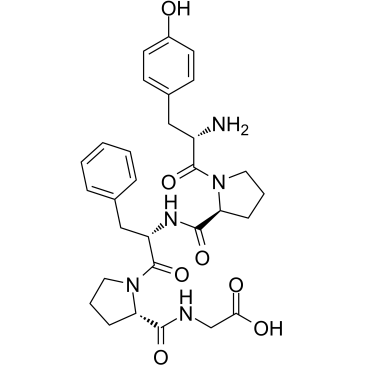
-
GC38004
β-Casomorphin (1-6), bovine
β-Casomorphin (1-6), bovine is a opioid-like bioactive peptide of β-Casomorphin.
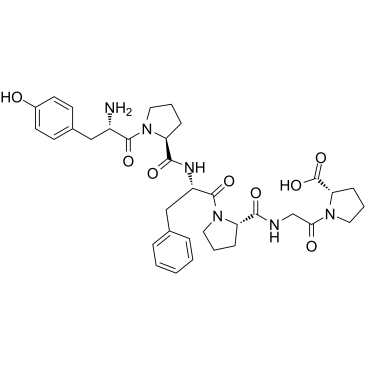
-
GC60391
β-Casomorphin, bovine TFA
β-Casomorphin-7 (bovine) (TFA); Bovine β-casomorphin-7 TFA

-
GC61484
β-Casomorphin, human TFA
Human β-casomorphin 7 TFA
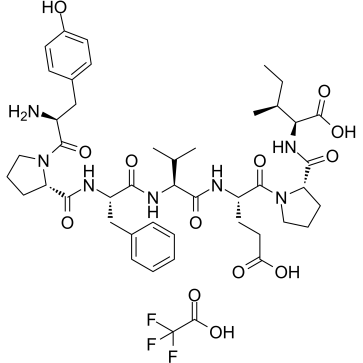
-
GC34944
β-CGRP, human TFA
Human β-CGRP TFA; CGRP-II (Human) (TFA)

-
GC48998
β-Defensin-1 (human) (trifluoroacetate salt)
hBD-1
An antimicrobial peptide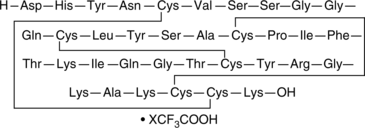
-
GC48298
β-Defensin-2 (human) (trifluoroacetate salt)
hBD-2
An antimicrobial peptide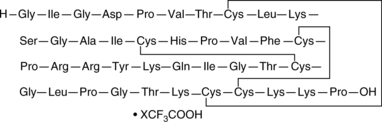
-
GC45230
β-Defensin-3 (human) (trifluoroacetate salt)
hBD-3
β-Defensin-3 is a peptide with antimicrobial properties that protects the skin and mucosal membranes of the respiratory, genitourinary, and gastrointestinal tracts.
-
GC45231
β-Defensin-4 (human) (trifluoroacetate salt)
hBD-4 (human)
β-Defensin-4 is a peptide with antimicrobial properties that protects the skin and mucosal membranes of the respiratory, genitourinary, and gastrointestinal tracts.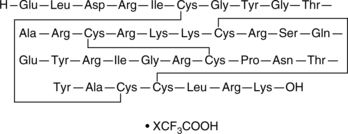
-
GC45234
β-Endorphin (1-27) (human) (trifluoroacetate salt)
β-Endorphin (1-27) is an endogenous peptide that binds to μ-, δ-, and κ-opioid receptors (Kis = 5.31, 6.17, and 39.82 nM, respectively, in COS-1 cells expressing rat receptors).

-
GC45236
β-Endorphin (rat)
β-Endorphin (β-EP) is an endogenous opioid neuropeptide with diverse biological activities.

-
GC38005
β-Endorphin, equine
β-Endorphin, equine is an endogenous opioid peptide, which binds at high affinity to both μ/δ opioid receptors.
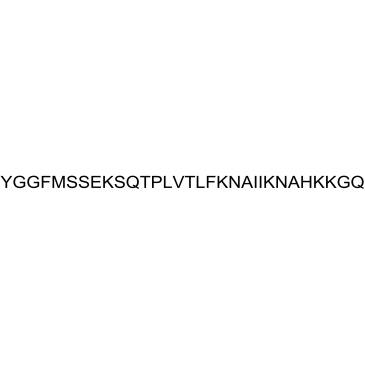
-
GC38030
β-Endorphin, equine (TFA)
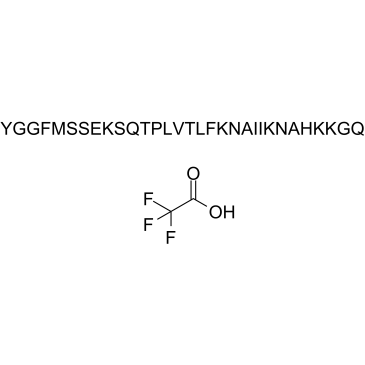
-
GC38007
β-Melanocyte Stimulating Hormone (MSH), human TFA
Beta-MSH (1-22) (human) TFA
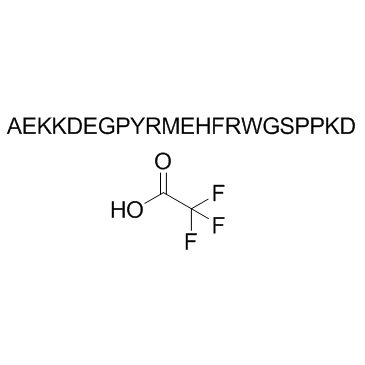
-
GC61462
γ-1-Melanocyte Stimulating Hormone (MSH), amide
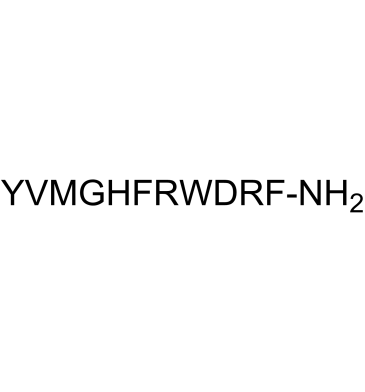
-
GC38009
γ-2-MSH (41-58), amide
γ-2-MSH (41-58), amide is derived from γ-2-MSH.
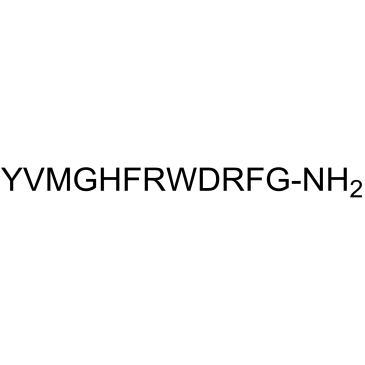
-
GC48312
γ-Glu-Cys (ammonium salt)
γ-Glutamylcysteine
An intermediate in GSH synthesis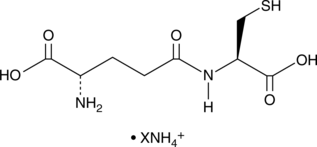
-
GC62081
γ-Glu-Phe TFA
γ-Glutamylphenylalanine TFA

-
GC17002
γ1-MSH
γ1-MSH is a melanocortin MC3 receptor agonist, with a Ki of 34 nM for the rat MC3 receptor.
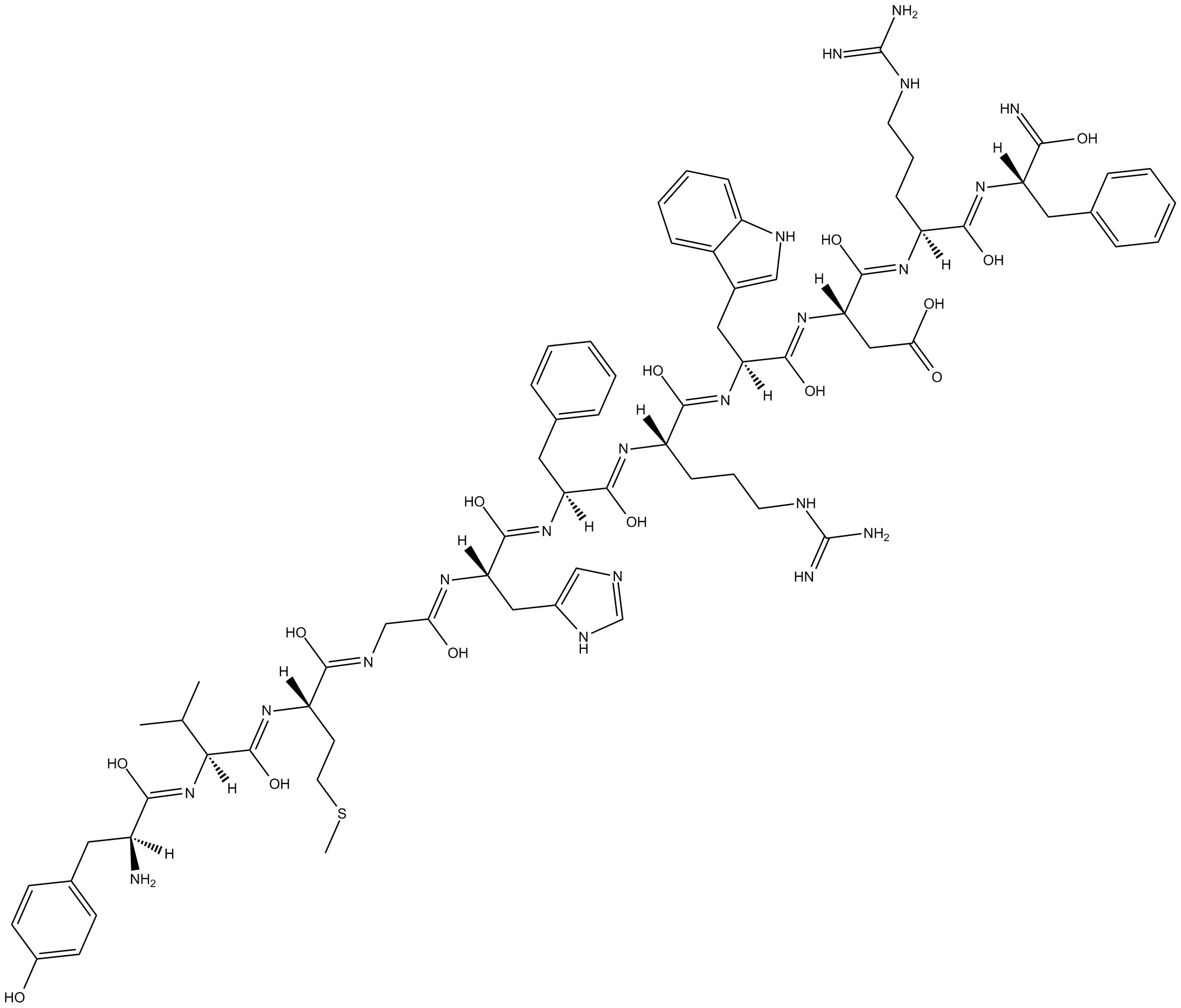
-
GC70187
δ-Sleep Inducing Peptide acetate
Delta-Sleep Inducing Peptide acetate
Delta-Sleep Inducing Peptide acetate is a neuropeptide that has antioxidant and anti-anxiety effects.
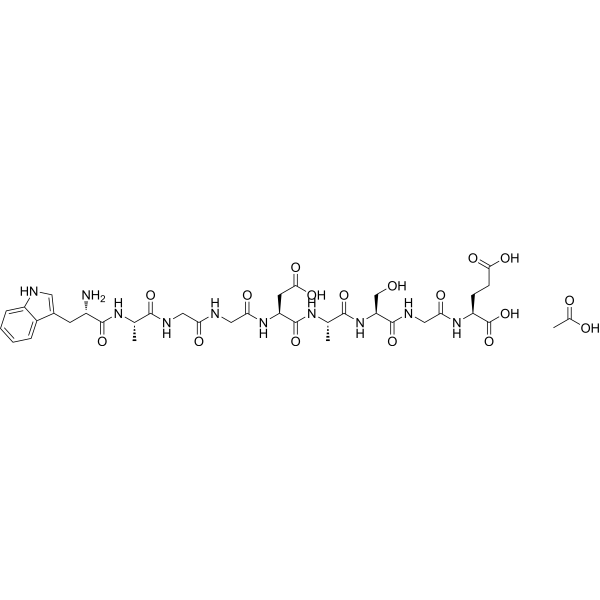
-
GC15513
ω-Agatoxin IVA
ω-Agatoxin IVA is a potent, selective P/Q type Ca2+ (Cav2.1) channel blocker with IC50s of 2 nM and 90 nM for P-type and Q-type Ca2+ channels, respectively.
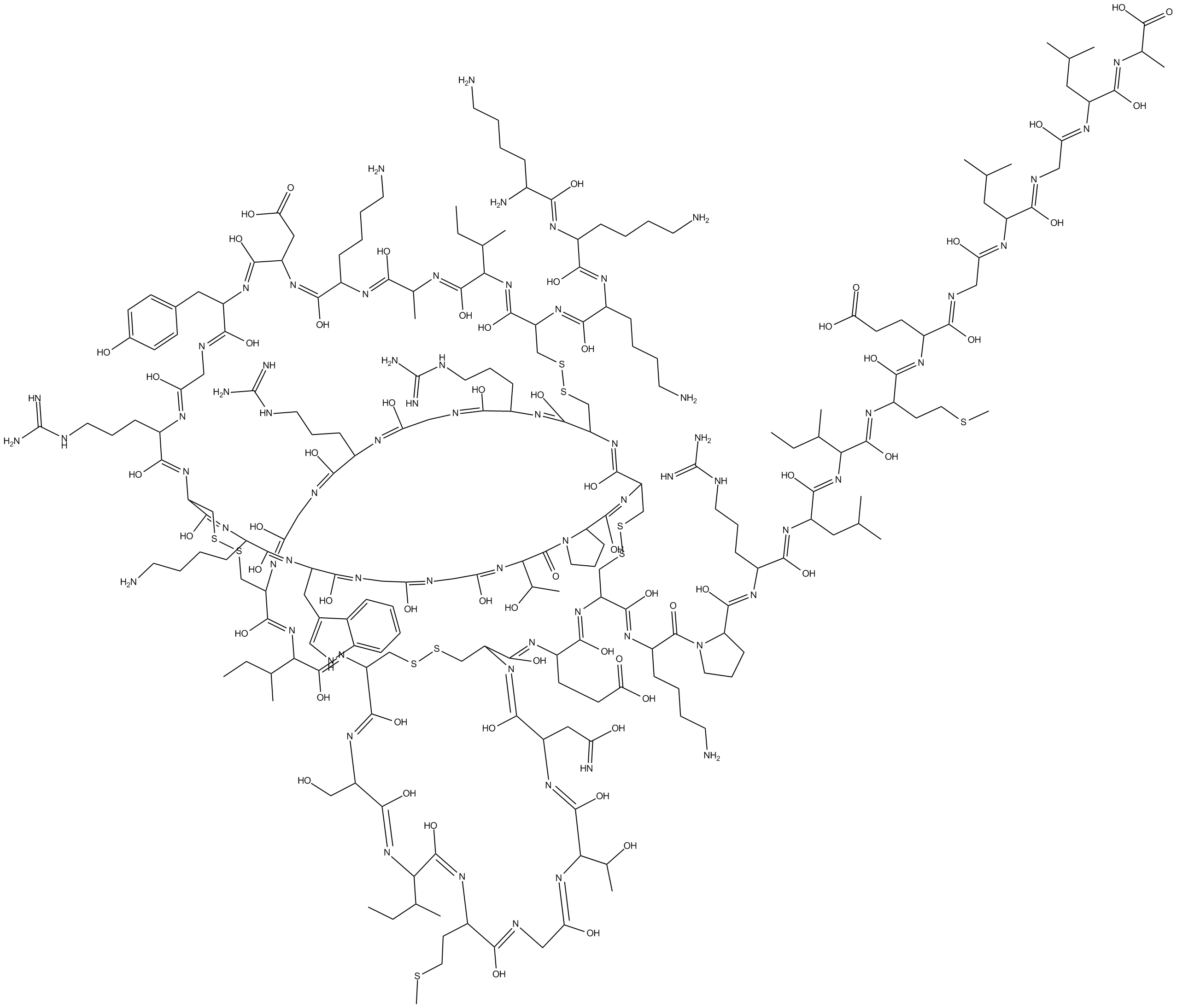
-
GC12608
ω-Agatoxin TK
ω-Agatoxin TK, a peptidyl toxin of the venom of Agelenopsis aperta, is a potent and selective P/Q type Ca2+ channel blocker.
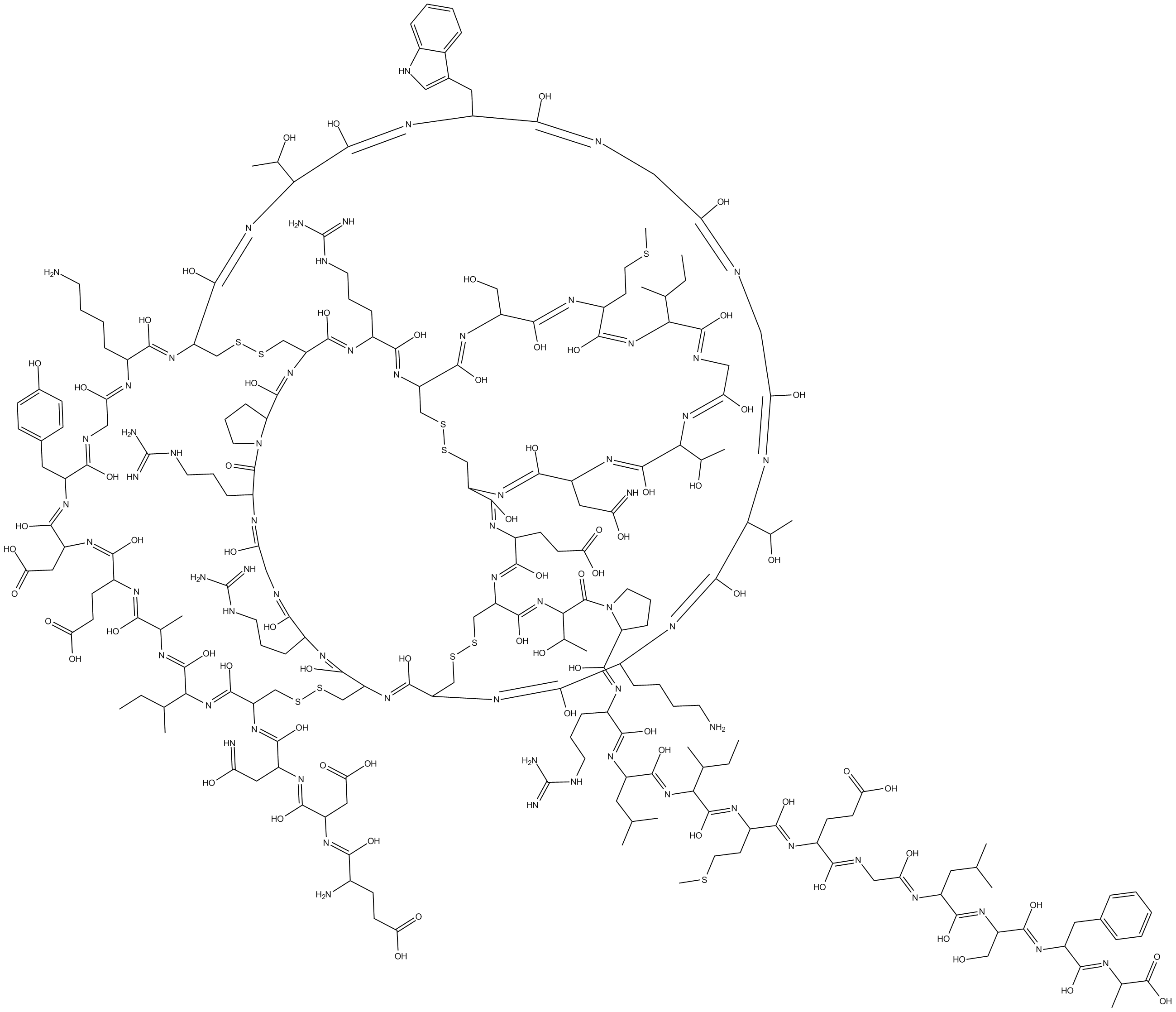
-
GC13886
ω-Conotoxin GVIA
ω-Conotoxin GVIA is a cone snail toxin that selectively blocks N-type channels in neurons .
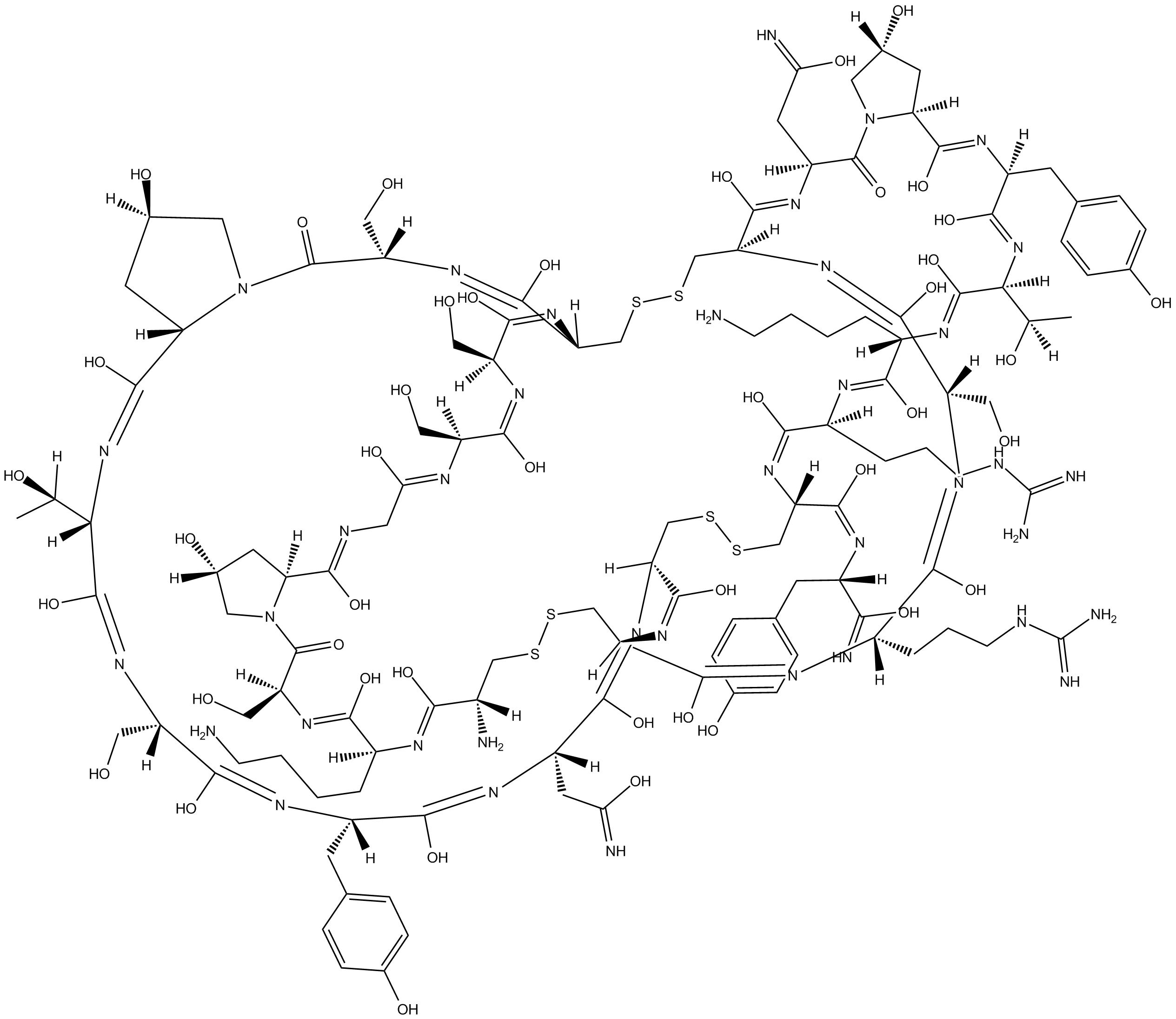
-
GC18070
ω-Conotoxin MVIIC
wide spectrum blocker of N, P and Q type calcium channels
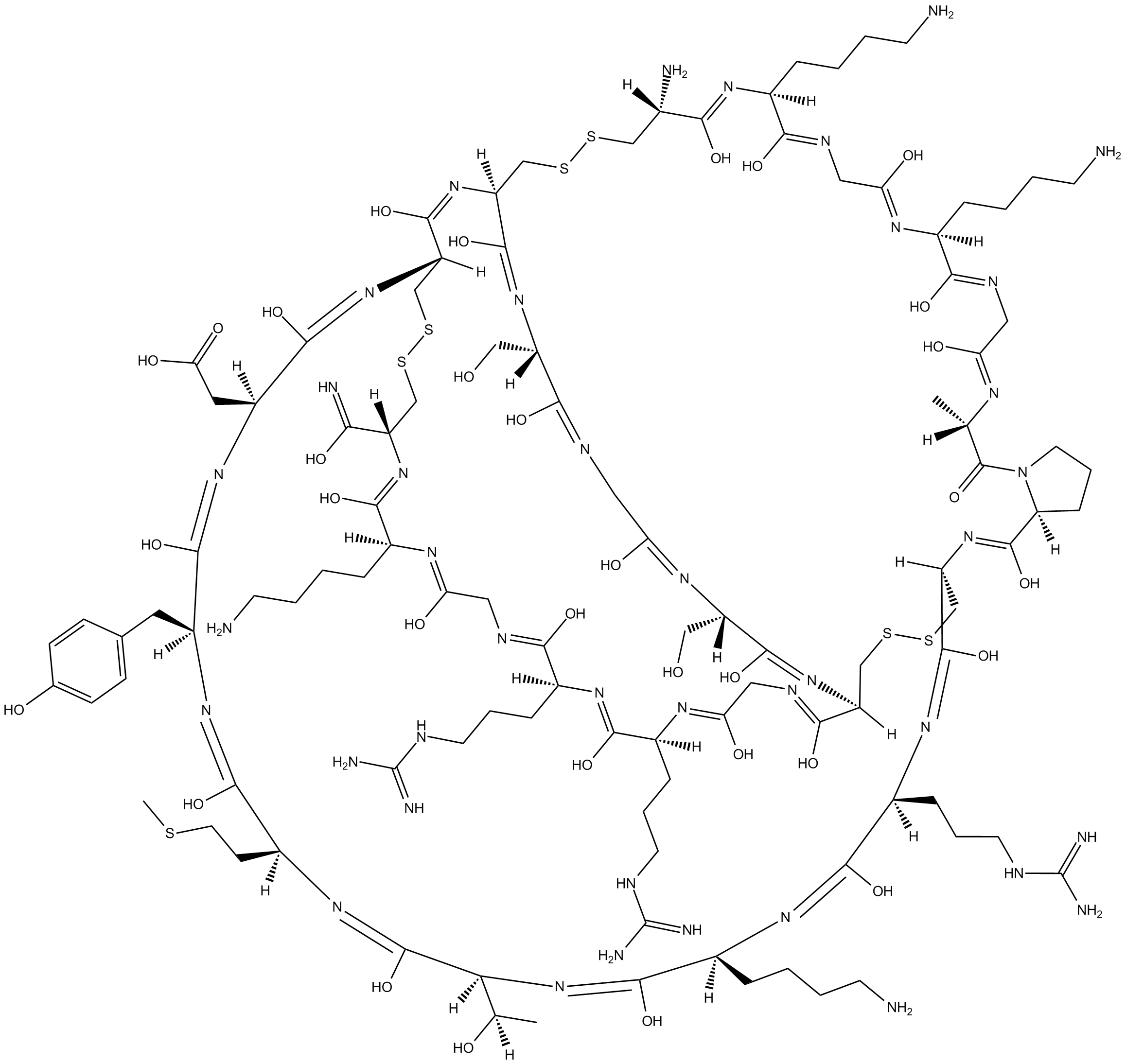
-
GC15519
α-CGRP (human)
Endogenous calcitonin gene-related peptide receptor (CGRP) agonist

-
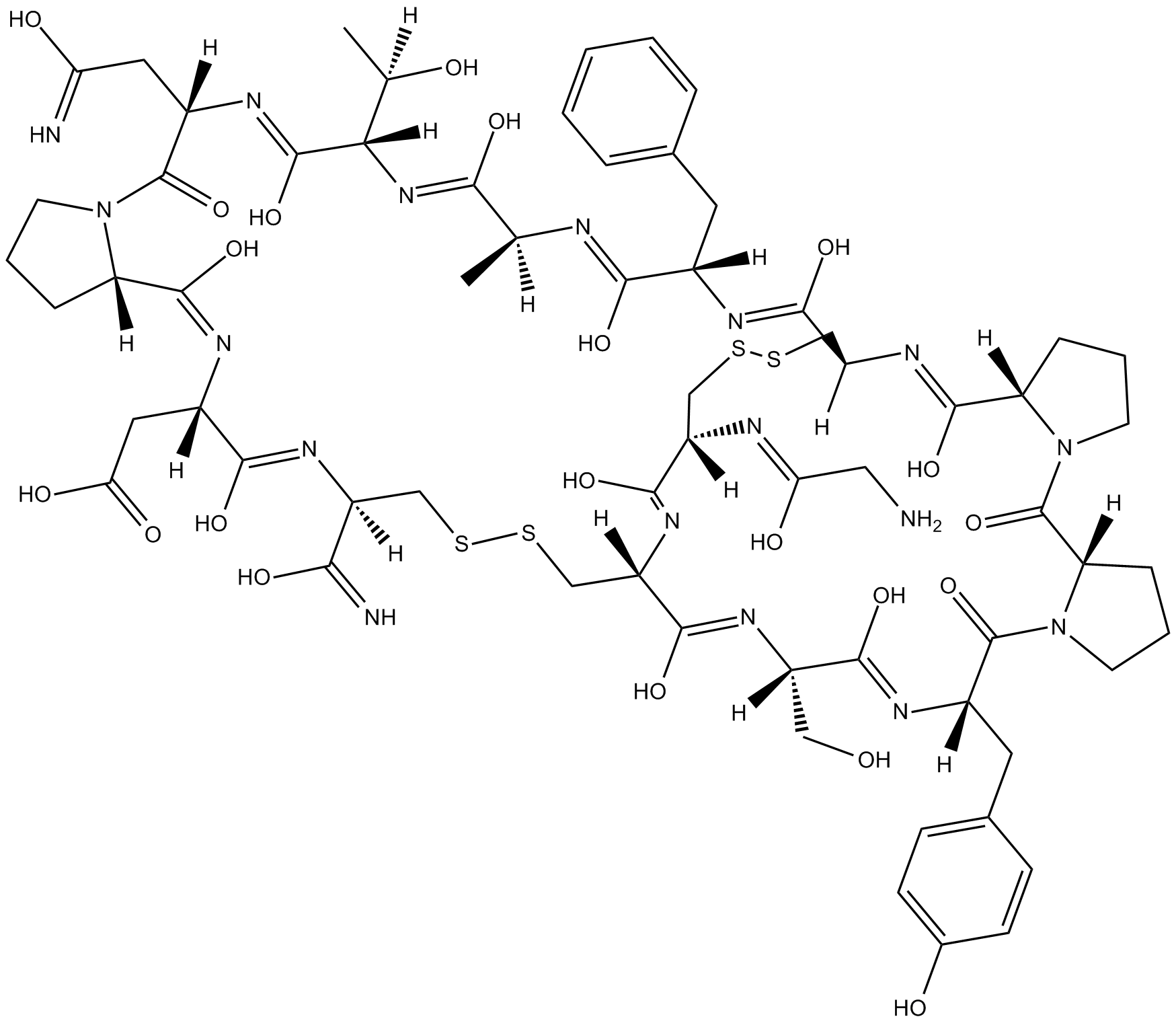
GC10872
α-Conotoxin AuIB
Selective antagonist of α3β4 nicotinic acetylcholine receptors
-
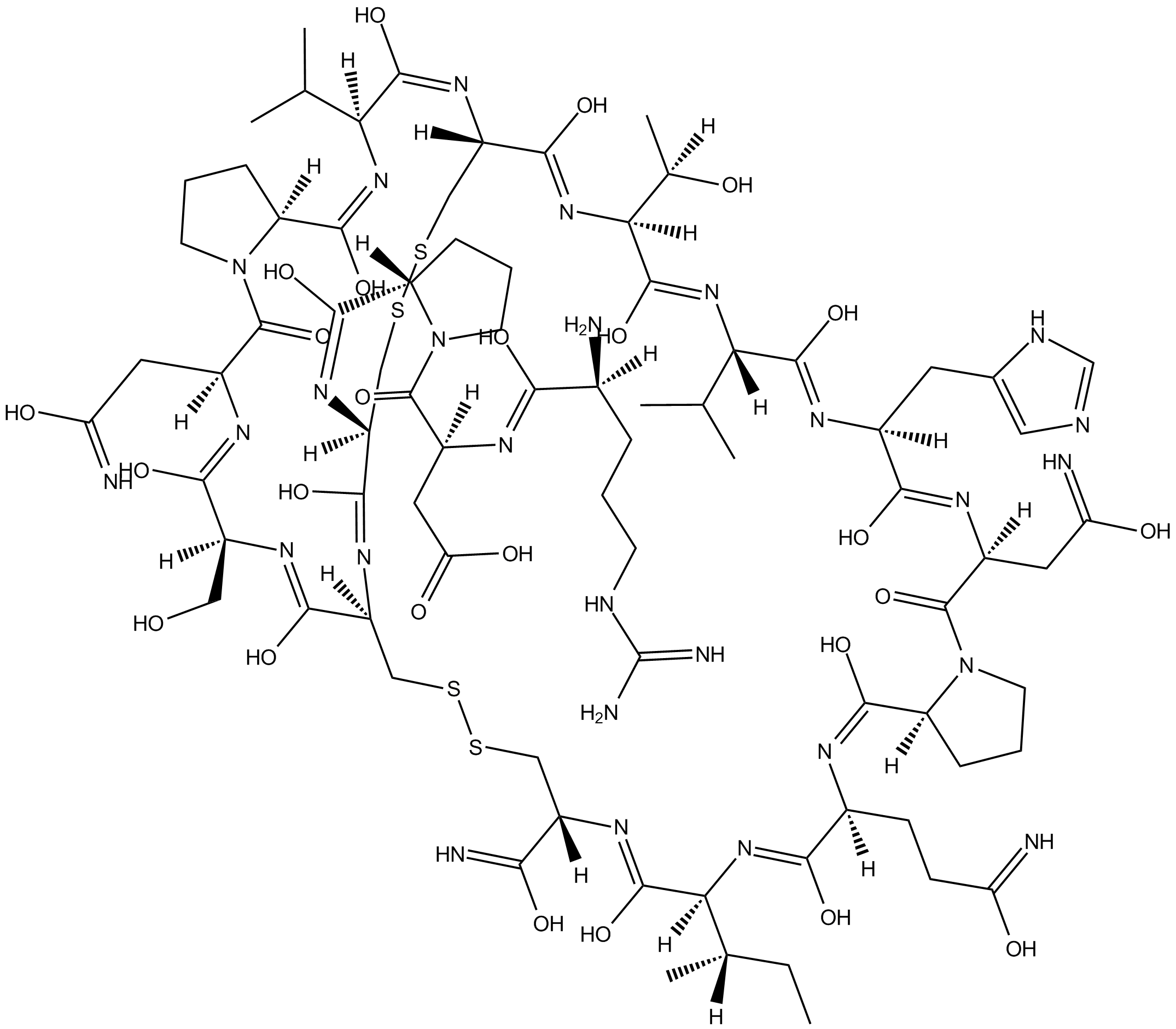
GC14296
α-Conotoxin PIA
Selective antagonist of α6-containing nicotinic receptors
-
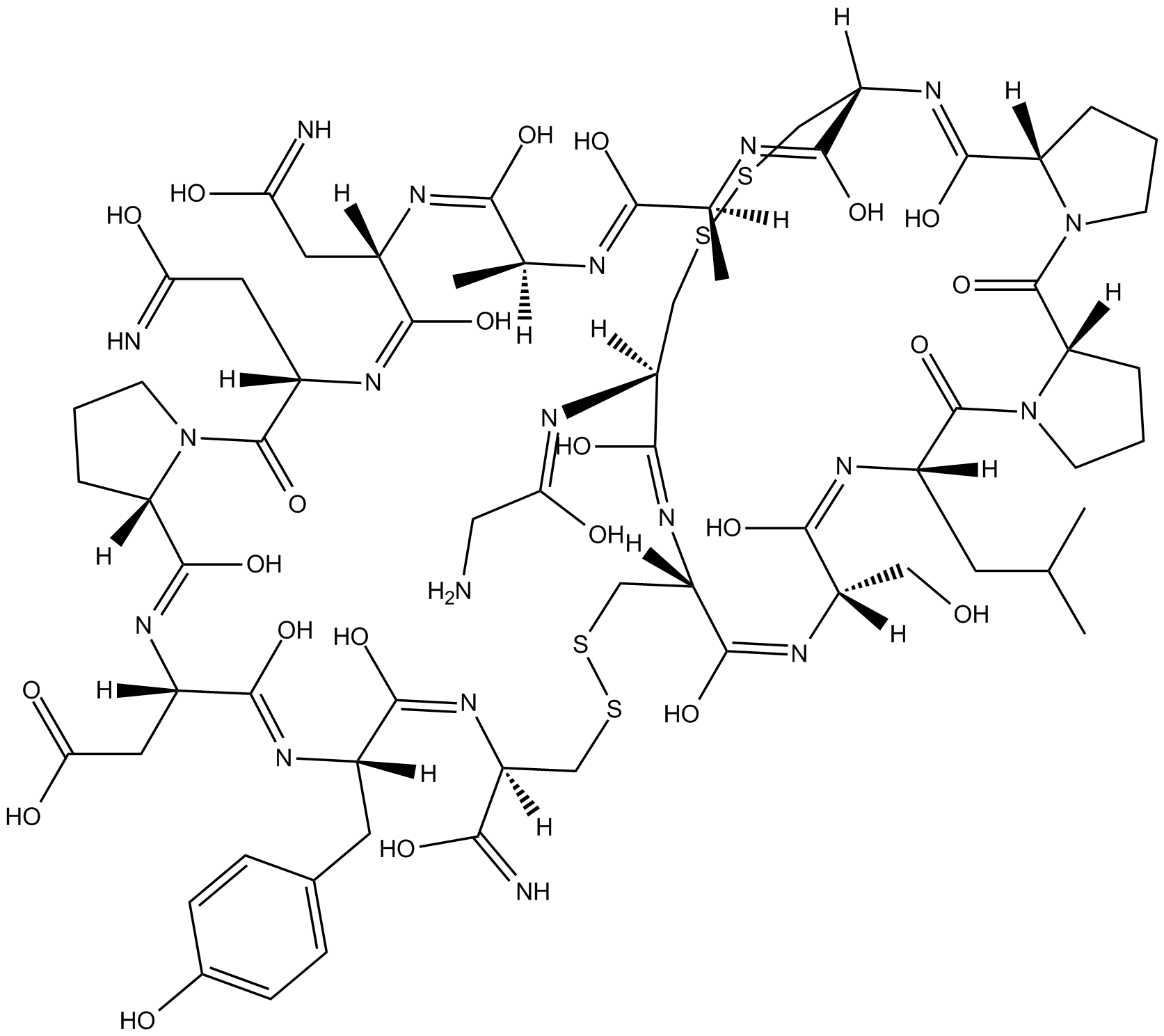
GC10368
α-Conotoxin PnIA
α-Conotoxin PnIA, a potent and selective antagonist of the mammalian α7 nAChR, has the potential for the research of neurological conditions such as neuropathic pain and Alzheimer's disease.
-
GC30587
α-Factor Mating Pheromone, yeast (Mating Factor α)
α-Factor Mating Pheromone, yeast (Mating Factor α) is a tridecapeptide secreted by S.
-
GC11346
α-helical CRF 9-41
Antagonist of corticotropin releasing factor receptor

-
GC34242
β-Amyloid (1-42), rat TFA
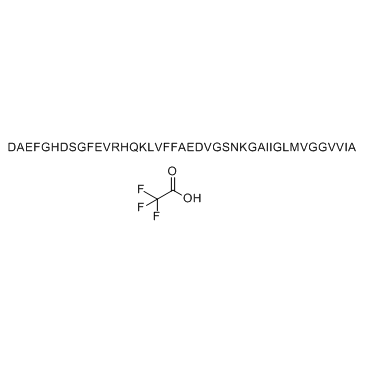
-
GC31146
β-Amyloid (10-35), amide
β-Amyloid (10-35), amide is composed of 26 aa (10-35 residues of the Aβ peptide) and is the primary component of the amyloid plaques of Alzheimer's disease.

-
GC31129
β-Amyloid 1-16 (Amyloid β-Protein (1-16))
Amyloid β-Protein (1-16)
β-Amyloid 1-16 (Amyloid β-Protein (1-16)) is a β-Amyloid protein fragment involved in metal binding.
-
GC31171
β-Amyloid 1-28 (Amyloid β-Protein (1-28))
Amyloid β-Protein (1-28)
β-Amyloid 1-28 (Amyloid β-Protein (1-28)) is a β-Amyloid protein fragment involved in metal binding.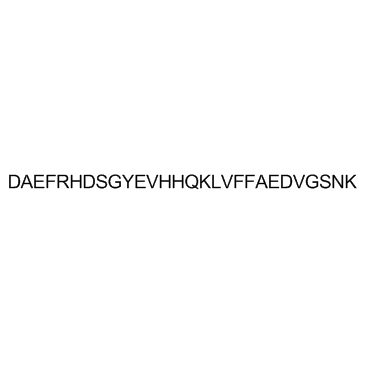
-
GC34391
β-Amyloid 15-21 (Beta-Amyloid (15-21))
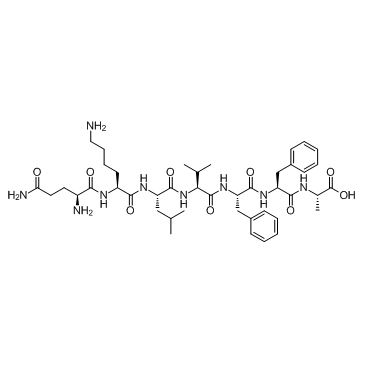
-
GC30325
β-Amyloid 22-35 (Amyloid β-Protein (22-35))
β-Amyloid 22-35 (Amyloid β-Protein 22-35), the residues 22-35 fragment ofβ-amyloid protein, has a cytotoxic effect on cultured neurons from the rat hippocampus in serum-free medium.

-
GC31137
β-Amyloid 29-40 (Amyloid beta-protein(29-40))
β-Amyloid 29-40 (Amyloid beta-protein(29-40)) is a fragment of Amyloid-β peptide.

-
GC31179
β-Amyloid 31-35
β-Amyloid 31-35 is the shortest sequence of native Amyloid-β peptide that retains neurotoxic activity.
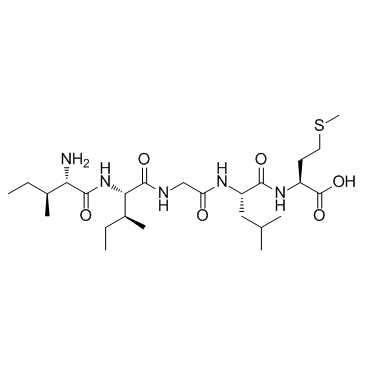
-
GC33736
β-Casomorphin, bovine (β-Casomorphin-7 (bovine))
β-Casomorphin, bovine (β-Casomorphin-7 (bovine)) (β-Casomorphin-7 (bovine) ) is a opioid peptide with an IC50 of 14 μM in an Opioid receptors binding assay.
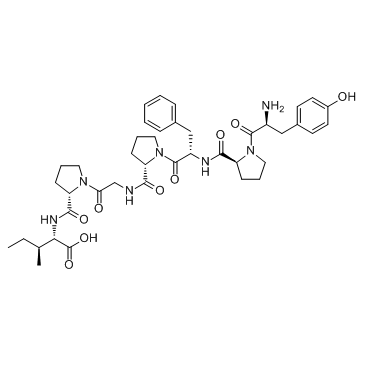
-
GC33784
β-Casomorphin, human (Human β-casomorphin 7)
Human β-casomorphin 7
is an opioid peptide, acts as an agonist of opioid receptor.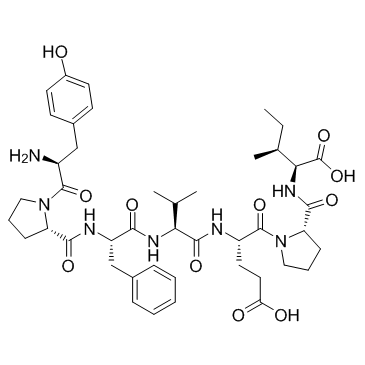
-
GC33585
β-catenin peptide
β-catenin peptide is a 8-aa peptide, and can promote thymocyte positive selection.
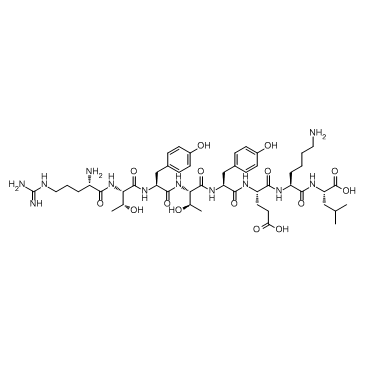
-
GC33595
β-CGRP, human (Human β-CGRP)
β-CGRP, human (Human β-CGRP) (Human β-CGRP) is one of calcitonin peptides, acts via the complex of calcitonin-receptor-like receptor (CRLR) and receptor-activity-modifying protein (RAMP), with IC50s of 1 nM and 300 nM for CRLR/RAMP1 and CRLR/RAMP2 in cells.

-
GC33693
β-Melanocyte Stimulating Hormone (MSH), human (Beta-MSH (1-22) (human))
β-Melanocyte Stimulating Hormone (MSH), human (Beta-MSH (1-22) (human)), a 22-residue peptide, acts as an endogenous melanocortin-4 receptor (MC4-R) agonist.

-
GC31172
δ-Sleep Inducing Peptide (Delta-Sleep Inducing Peptide)
δ-Sleep Inducing Peptide (Delta-Sleep Inducing Peptide) is a neuropeptide, with antioxidant and anxiolytic properties.
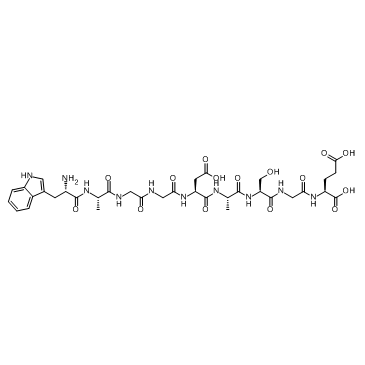
-
GC30187
γ-Glu-Phe (γ-Glutamylphenylalanine)
γ-Glutamylphenylalanine
γ-Glu-Phe (γ-Glutamylphenylalanine) (γ-Glutamylphenylalanine) is synthesized by Bacillus amyloliquefaciens (GBA) and Aspergillus oryzae (GAO).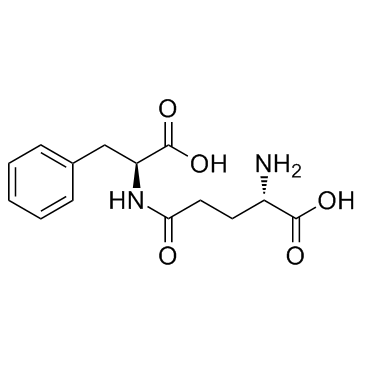
-
GC62067
ω-Conotoxin GVIA TFA

-
GA24016
ω-Conotoxin MVIIA
ω-Conotoxin MVIIA (SNX-111), a peptide, is a potent and selective block of N-type calcium channels antagonist.

-
GA24056
(β-Ala²?)-Glucagon trifluoroacetate salt
The peptide is an impurity of glucagon.

-
GC68266
(((9H-Fluoren-9-yl)methoxy)carbonyl)-L-cysteine
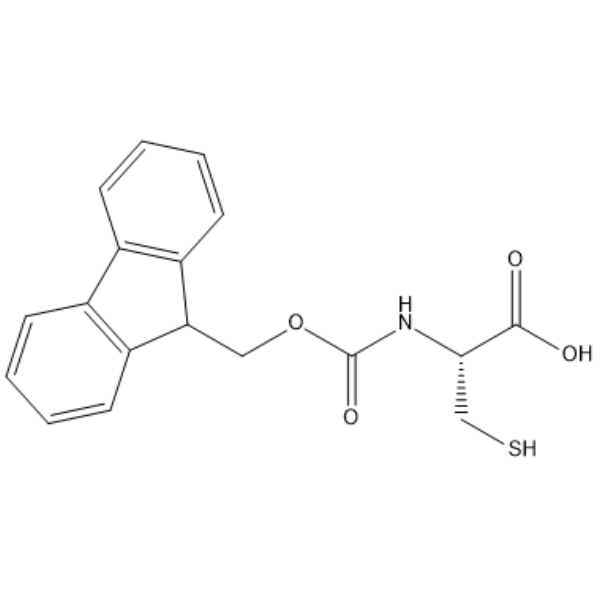
-
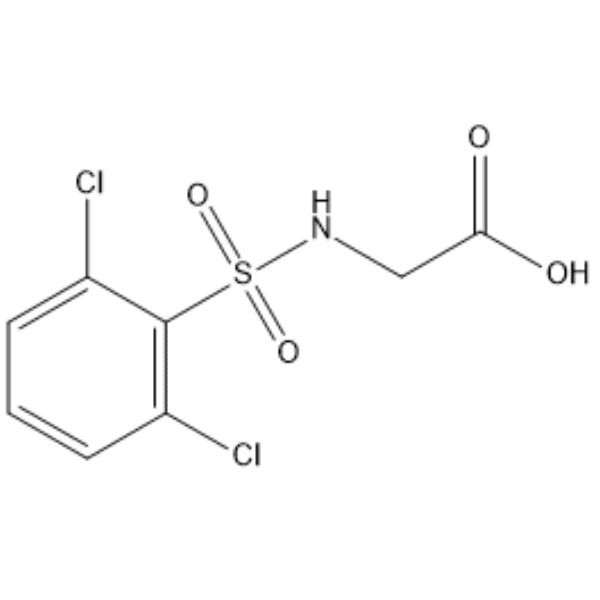
GC66647
((2,6-Dichlorophenyl)sulfonyl)glycine
((2,6-Dichlorophenyl)sulfonyl)glycine is a Glycine derivative.
-
GC66669
((3-Chlorophenyl)sulfonyl)phenylalanine
((3-Chlorophenyl)sulfonyl)phenylalanine is a phenylalanine analogue, contains both an amino group and a carboxyl group in its molecule.
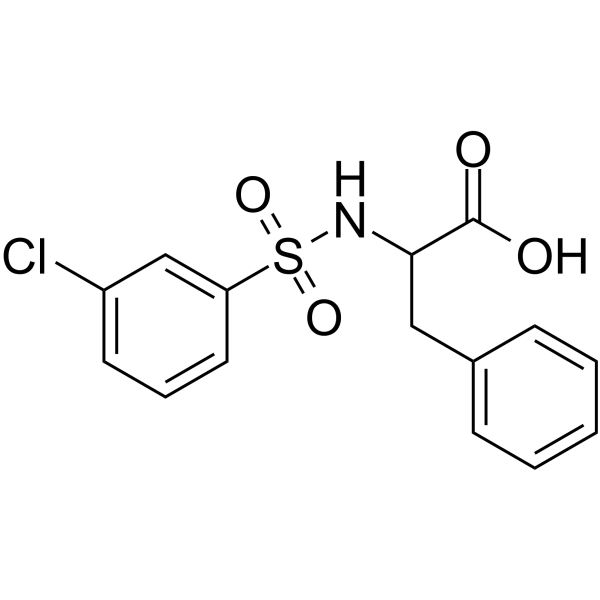
-
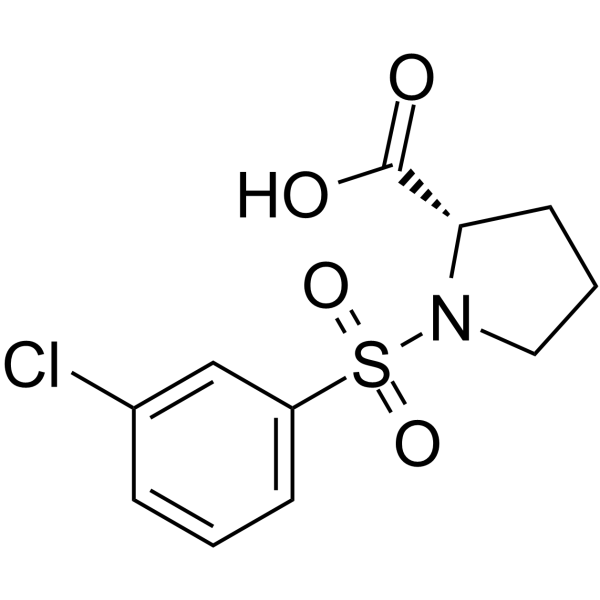
GC66623
((3-Chlorophenyl)sulfonyl)proline
((3-Chlorophenyl)sulfonyl)proline is a proline derivative containing amino and carboxyl groups.
-
GC66672
((4'-Chloro-[1,1'-biphenyl]-4-yl)sulfonyl)glycine
((4'-Chloro-[1,1'-biphenyl]-4-yl)sulfonyl)glycine is a synthetic amino acid.
![((4'-Chloro-[1,1'-biphenyl]-4-yl)sulfonyl)glycine Chemical Structure ((4'-Chloro-[1,1'-biphenyl]-4-yl)sulfonyl)glycine Chemical Structure](/media/struct/GC6/GC66672.png)
-
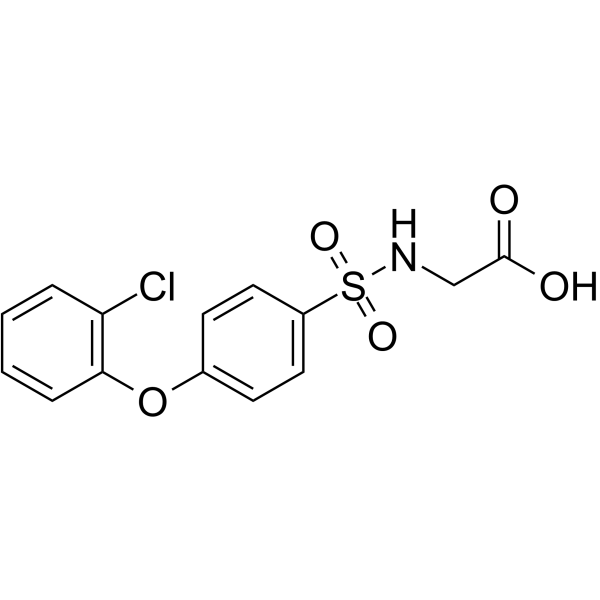
GC66625
((4-(2-Chlorophenoxy)phenyl)sulfonyl)glycine
((4-(2-Chlorophenoxy)phenyl)sulfonyl)glycine used for scientific research and chemical synthesis intermediates.
-
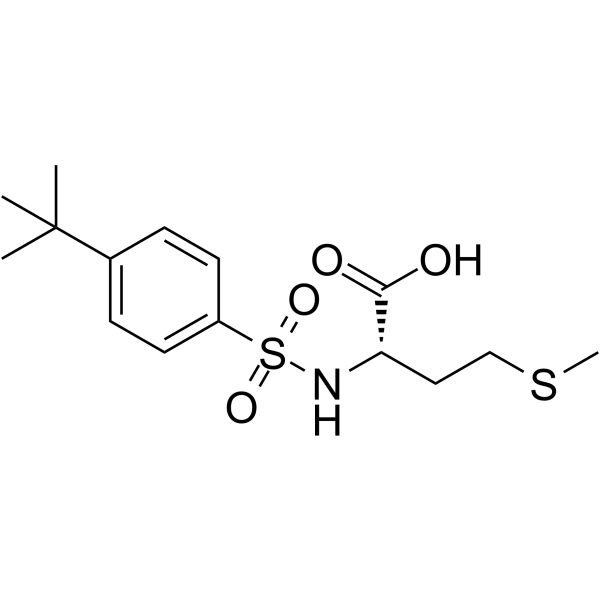
GC66671
((4-(tert-Butyl)phenyl)sulfonyl)methionine
((4-(tert-Butyl)phenyl)sulfonyl)methionine can be used for peptide synthesis.
-
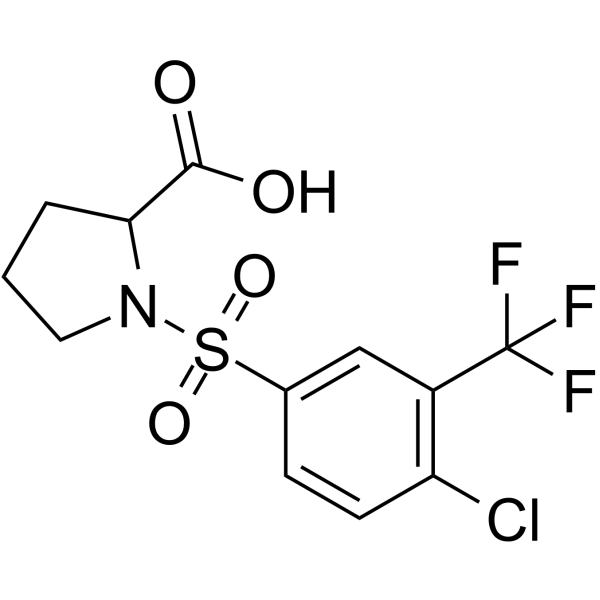
GC66661
((4-Chloro-3-(trifluoromethyl)phenyl)sulfonyl)proline
((4-Chloro-3-(trifluoromethyl)phenyl)sulfonyl)proline is a proline derivative, contains both an amino group and a carboxyl group in its molecule.
-
GC66664
((4-Chloro-3-(trifluoromethyl)phenyl)sulfonyl)valine
((4-Chloro-3-(trifluoromethyl)phenyl)sulfonyl)valine is a Valine derivative.
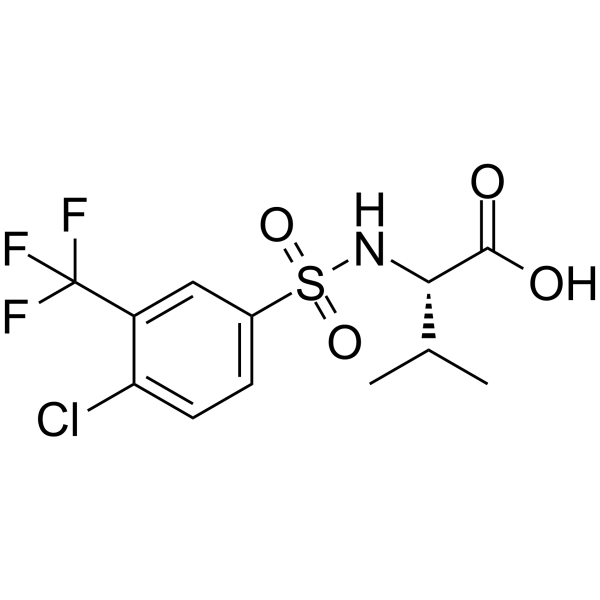
-
GA24018
(2-(1,3-Dithiolan-2-yl)-Trp²?)-Liraglutide trifluoroacetate salt
The peptide is an impurity of Liraglutide.

-
GA24019
(2-(2-Trifluoromethyl-1,3-dithiolan-2-yl)-Trp²?)-Liraglutide trifluoroacetate salt
The peptide is an impurity of Liraglutide.

-
GA24020
(2-Hydroxy-Trp²?)-Liraglutide trifluoroacetate salt
The peptide is an impurity of Liraglutide.

-
GC68059
(2R,2'R)-4,4'-Disulfanediylbis(2-aminobutanoic acid)
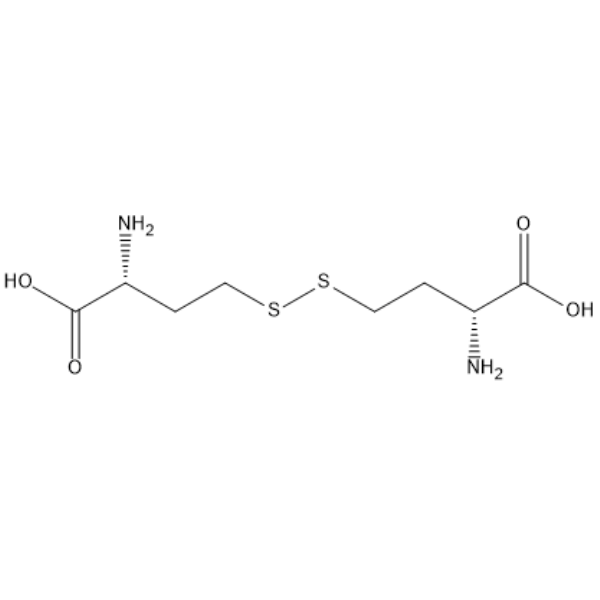
-
GC68248
(2R,3S)-3-Phenylisoserine hydrochloride
Benzenepropanoic acid, β-amino-α-hydroxy-, hydrochloride, (αR,βS)-

-
GC66180
(2S)-2-[(tert-Butoxycarbonyl)amino]-3-(3,4-difluorophenyl)propionic acid
(2S)-2-[(tert-Butoxycarbonyl)amino]-3-(3,4-difluorophenyl)propionic acid is a phenylalanine derivative.
![(2S)-2-[(tert-Butoxycarbonyl)amino]-3-(3,4-difluorophenyl)propionic acid Chemical Structure (2S)-2-[(tert-Butoxycarbonyl)amino]-3-(3,4-difluorophenyl)propionic acid Chemical Structure](/media/struct/GC6/GC66180.png)
-
GC68261
(2S)-2-[(tert-Butoxycarbonyl)amino]-3-(3-fluorophenyl)propionic acid
(S)-2-[(tert-Butoxycarbonyl)amino]-3-(3-fluorophenyl)propionic acid; tert-Butoxycarbonyl-L-3-fluorophenylalanine
![(2S)-2-[(tert-Butoxycarbonyl)amino]-3-(3-fluorophenyl)propionic acid Chemical Structure (2S)-2-[(tert-Butoxycarbonyl)amino]-3-(3-fluorophenyl)propionic acid Chemical Structure](/media/struct/GC6/GC68261.png)
-
GC68251
(2S)-4-(benzyloxy)-2-{[(9H-fluoren-9-ylmethoxy)carbonyl]amino}-4-oxobutanoic acid
![(2S)-4-(benzyloxy)-2-{[(9H-fluoren-9-ylmethoxy)carbonyl]amino}-4-oxobutanoic acid Chemical Structure (2S)-4-(benzyloxy)-2-{[(9H-fluoren-9-ylmethoxy)carbonyl]amino}-4-oxobutanoic acid Chemical Structure](/media/struct/default.png)


