- Cat.No. Product Name Information
-
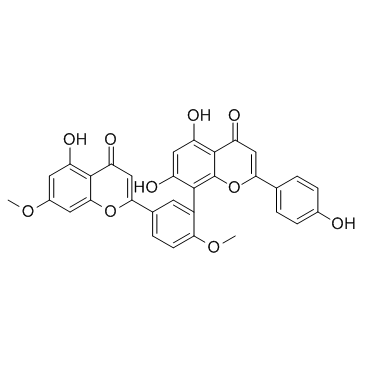
GC31717
Ginkgetin
A biflavonoid with diverse biological activities
-
GC14056
CFDA-SE
5(6)Carboxyfluorescein diacetate succinimidyl ester, 5(6)-CFDA N-succinmidyl ester
CFDA-SE, full name carboxycein diacetate, Succinimidyl Ester, with cell membrane permeability, is a fluorescent dye CFDA-SE widely used for viable cell tracing or cell proliferation detection.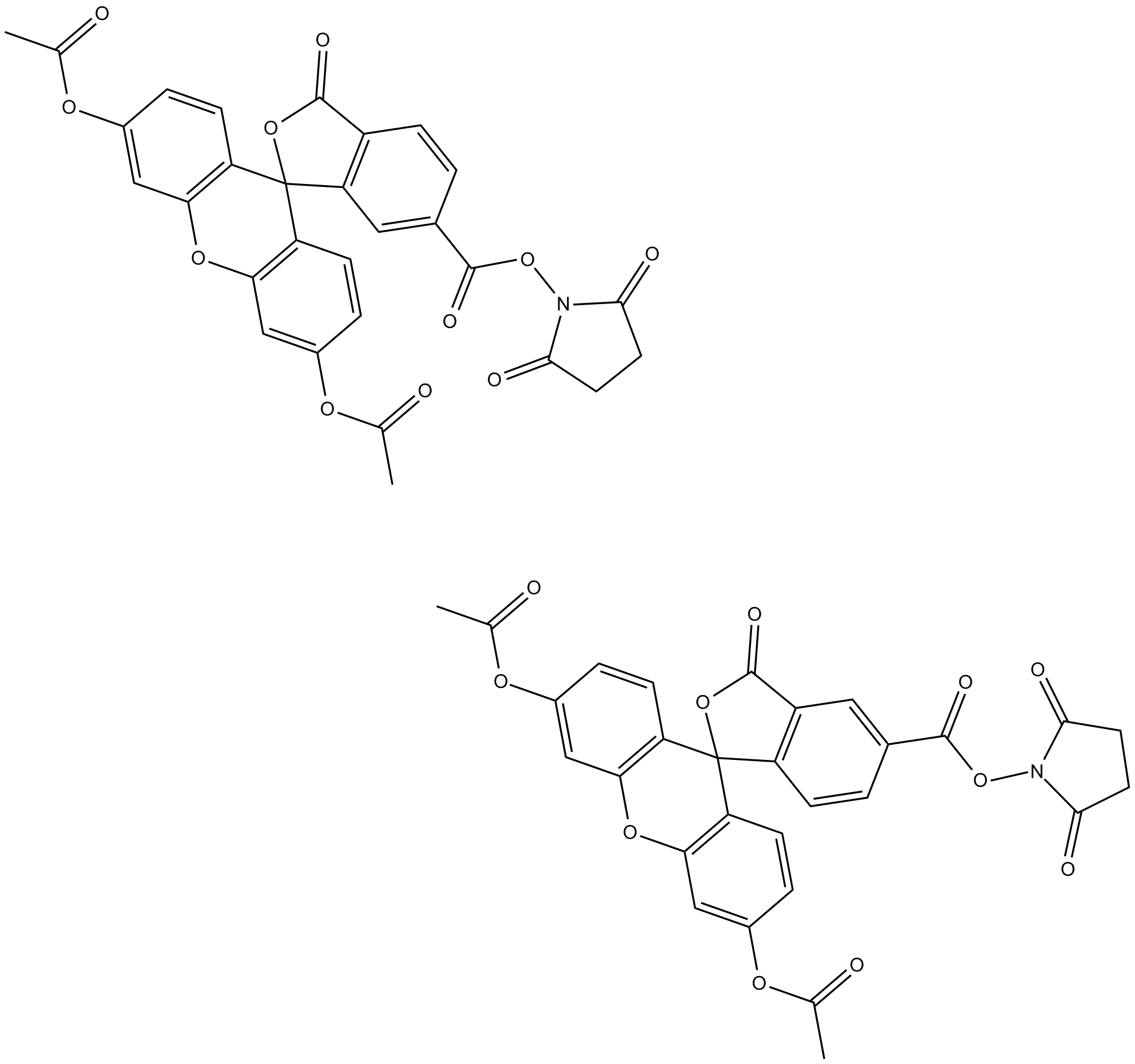
-
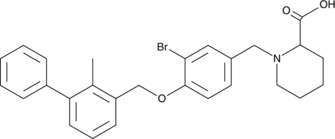
GC42957
BMS-8
An inhibitor of the PD-1/PD-L1 interaction
-
GC27560
Tetramethylrhodamine-Gemcitabine

-
GC17312
Jasplakinolide
NSC 613009
stabilizes pre-formed actin filaments and inhibits their disassembly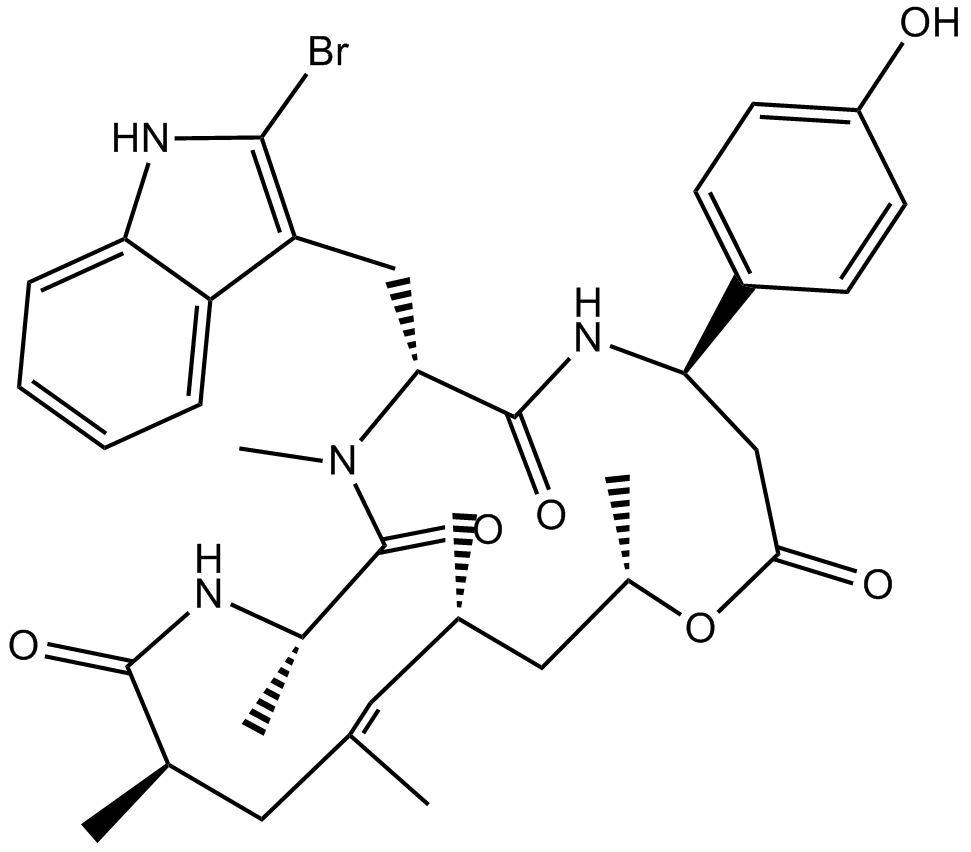
-
GC44254
MTSES
Sodium (2-Sulfonatoethyl)methanethiosulfonate
A negatively-charged MTS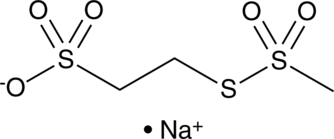
-
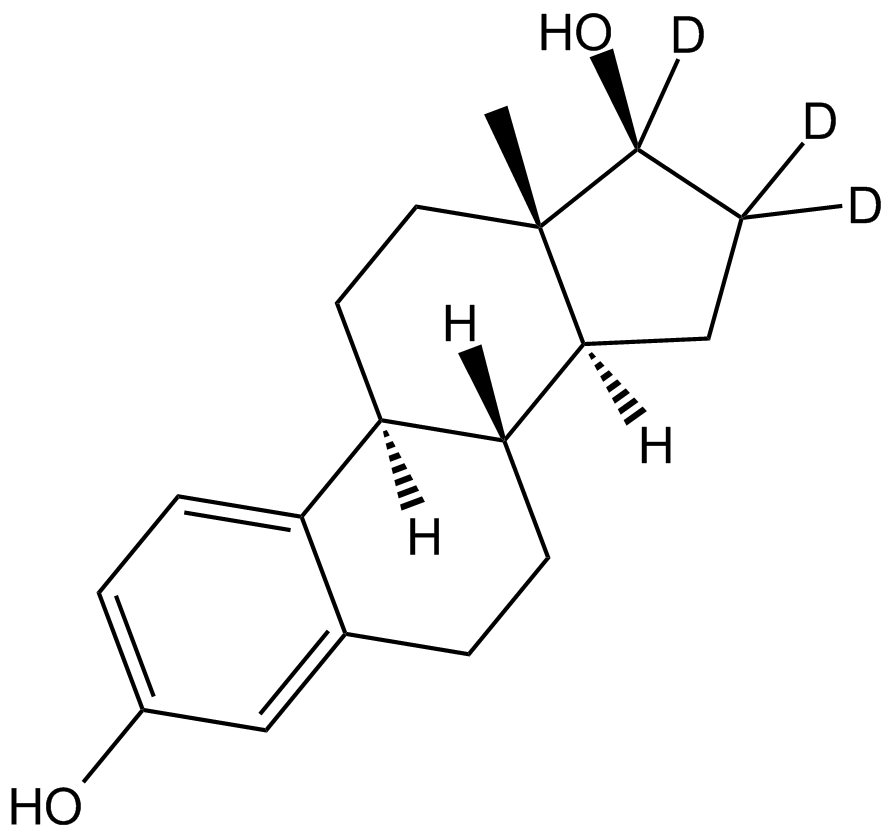
GC17258
β-Estradiol-d3
Deuterated β-estradiol
-
GC26230
Ammonium persulfate
Ammonium peroxydisulfate; PER; Ammonium peroxodisulfate; APS; AP
Catalyst for acrylamide gel polymerization.

-
GC16157
740 Y-P
740YPDGFR; PDGFR 740Y-P
PI 3-kinase activator,cell permeable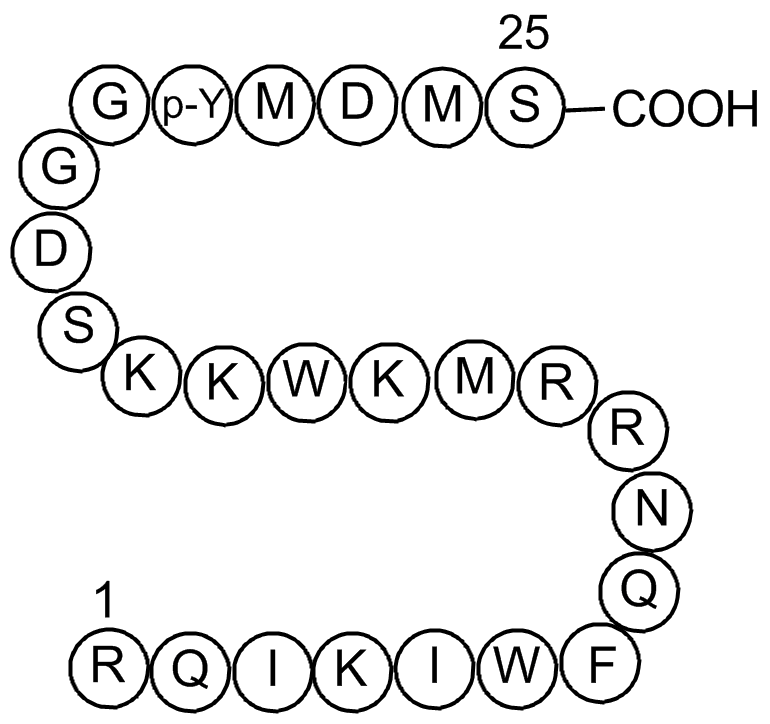
-
GC19778
Triton x
Triton x-100 is an oligomeric blend that can be used for biochemical research.

-
GC32111
Imazalil (Enilconazole)
Enilconazole, (±)-Imazalil
Imazalil (Enilconazole) (Enilconazole) is a fungicide, widely used in agriculture, particularly in the growing of citrus fruits, also used in veterinary medicine as a topical antimycotic.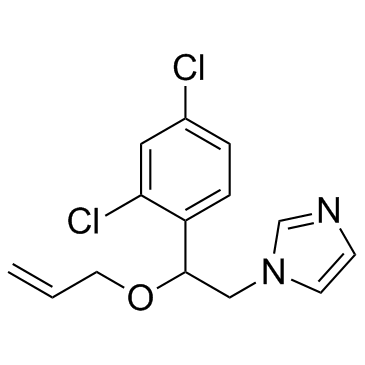
-
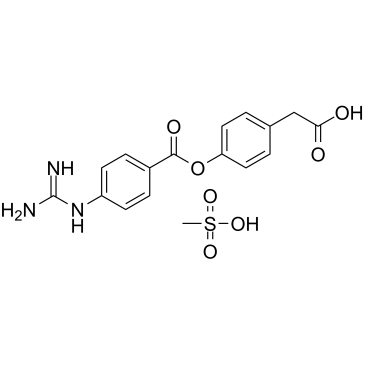
GC60856
FOY 251
FOY 251, an anti-proteolytic active metabolite Camostate, acts as a proteinase inhibitor.
-
GK10029
Dual Luciferase Reporter Gene Assay Kit
Firefly luciferase is widely used as a reporter for studying gene regulation and function, and for pharmaceutical screening.

-
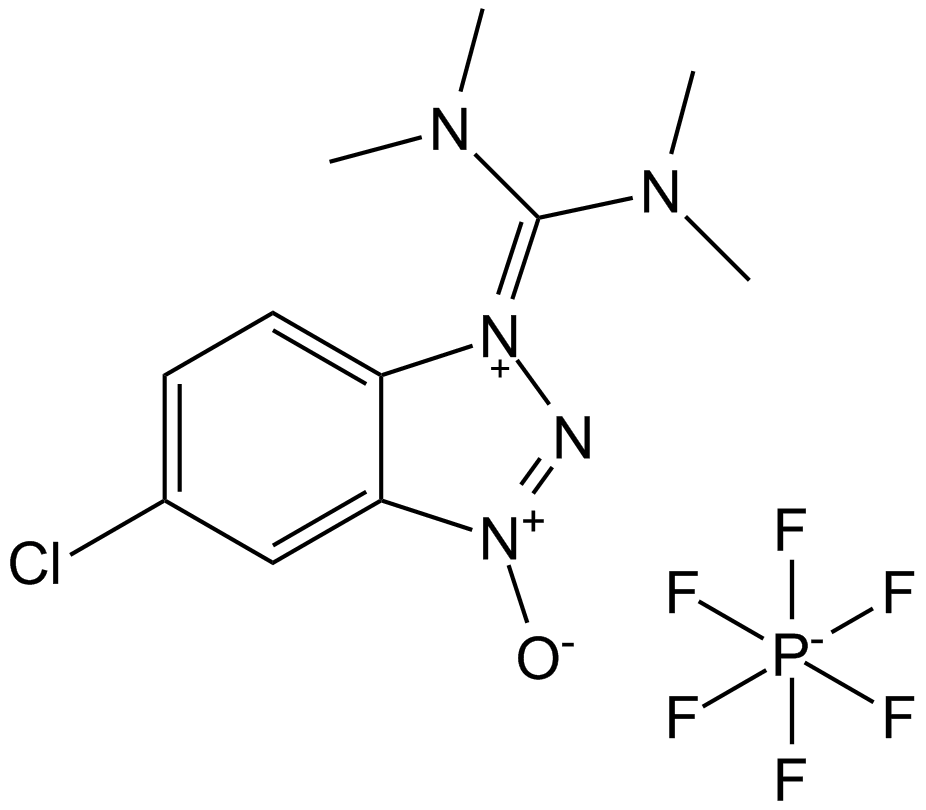
GA10910
HCTU
HCTU is a highly efficient aminium-based coupling reagent widely utilized in solid-phase and solution-phase peptide synthesis.
-
GC75345
Zosurabalpin
Abx MCP; RG6006
Zosurabalpin is a novel class of tethered macrocyclic peptide antibiotics active against various Acinetobacter baumannii complex (ABC) isolates with the MIC90 values of 0.06-0.5mg/L.

-
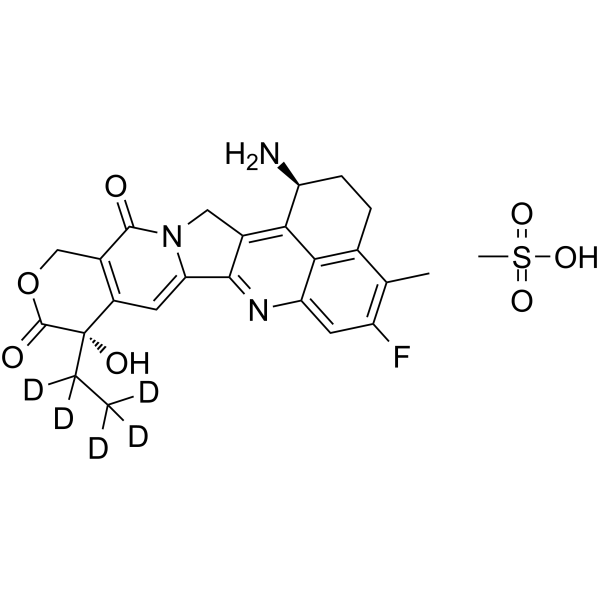
GC62184
Exatecan D5 mesylate
DX8951f-d5; Exatecan-d5 mesylate; Deuterated labeled Exatecan mesylate
-
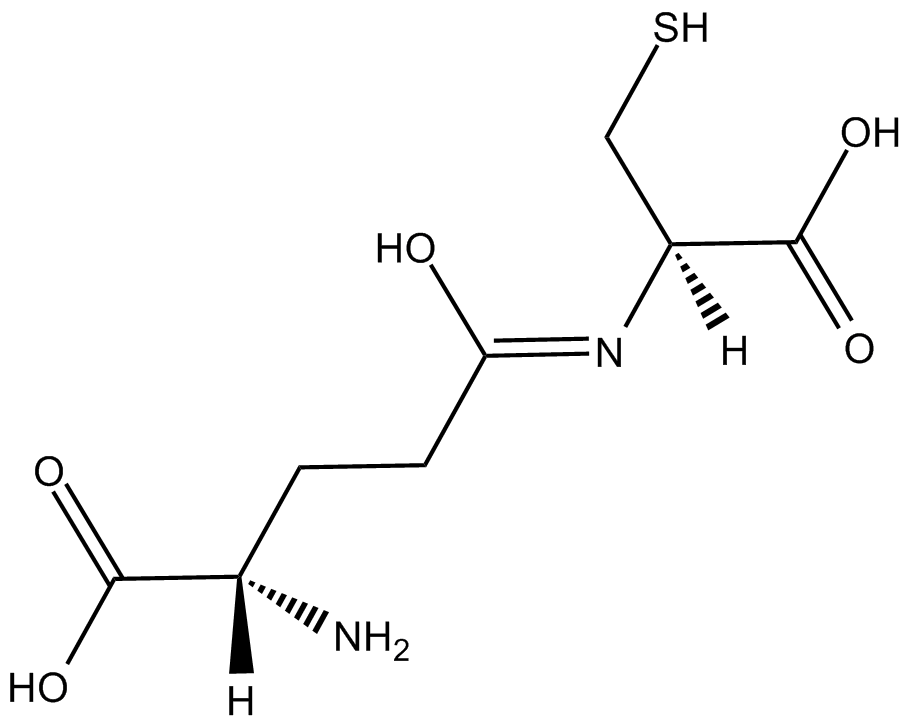
GC17170
gamma-Glu-Cys
gamma-Glu-Cys is a dipeptide produced by glutathione synthetase.
-
GC74394
DOTA-JR11
Satoreotide tetraxetan
DOTA-JR11 is a somatostatin receptor 2 (SSTR2)antagonist.
-
GC14731
MTT
Methylthialazole Tetrazolium, NSC 60102, Thiazolyl Blue, Thiazolyl Blue Tetrazolium Bromide
reagent used in the measurement of in vitro cell proliferation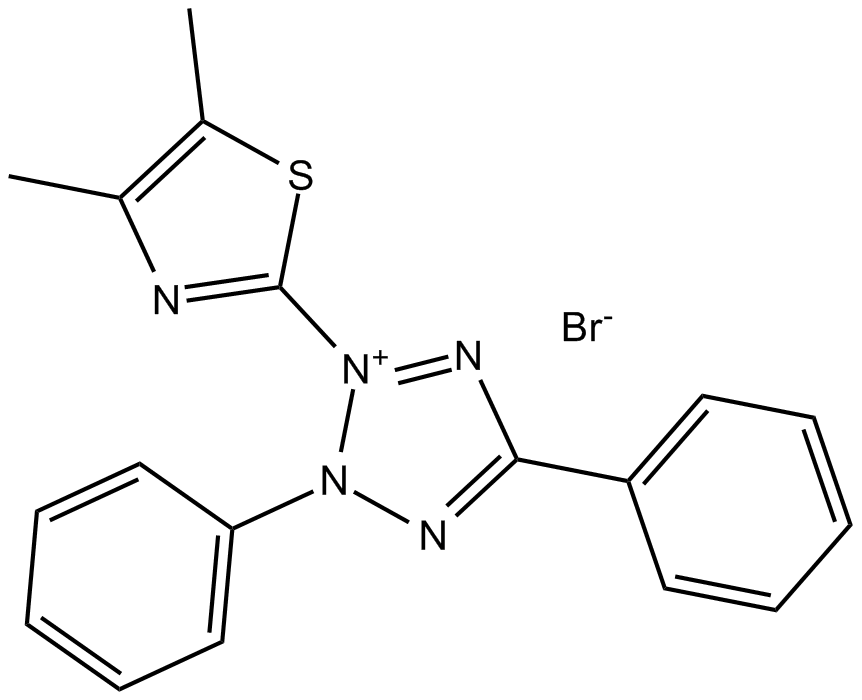
-
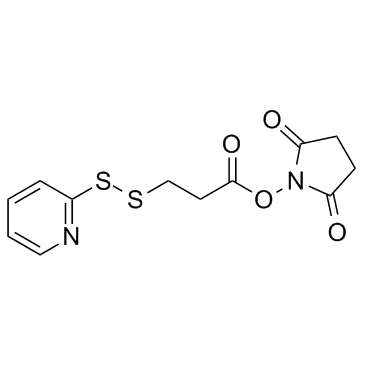
GC30018
SPDP (SPDP Crosslinker)
SPDP (SPDP Crosslinker) is a short-chain crosslinker that reacts with NHS ester groups and pyridyl disulfide reactive groups with cysteine thiols to form a cleavable (reducible) disulfide bond, thereby facilitating the coupling of amine and thiol groups.
-
GC19659
2-Phenoxyethanol
2-Phenoxyethanol has a broad spectrum of antimicrobial activity against various gram-negative and gram-positive bacteria.

-
GN10444
12-O-tetradecanoyl phorbol-13-acetate
PMA; TPA; Phorbol myristate acetate
An activator of ERK/MAPK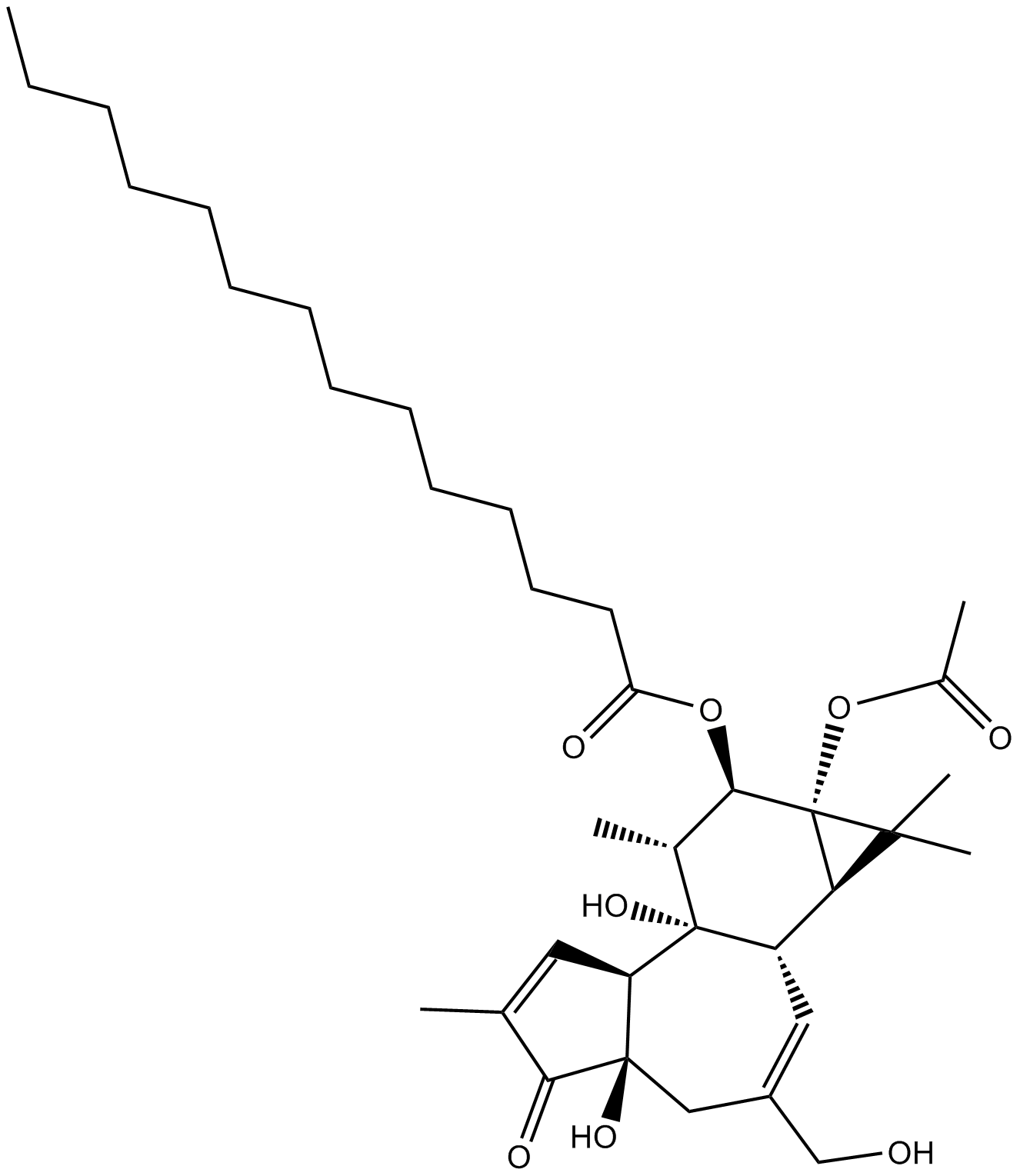
-
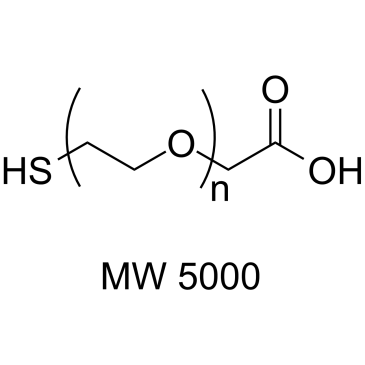
GC61333
Thiol-PEG-CH2COOH (MW 5000)
Thiol-PEG-CH2COOH (MW 5000) is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
-
GC65098
ARL67156 triethylamine
FPL 67156 triethylamine
ARL67156 (FPL 67156) triethylamine is a selective ecto-ATPase inhibitor.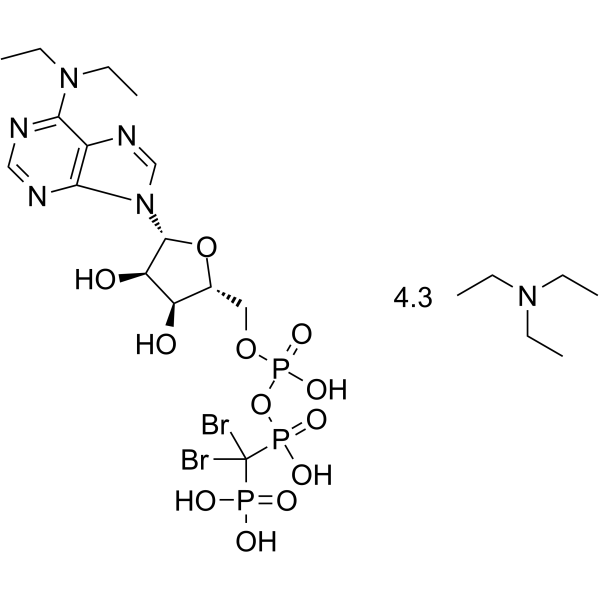
-
GC41283
N-acetyl-D-Glucosamine
GlcNAc, Marine Sweet, NAG, NSC 524344
N-acetyl-D-Glucosamine (GlcNAc) is a monosaccharide derivative of glucose with antitumor and anti-inflammatory activities.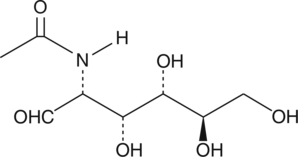
-
GC16952
SB 204990
ATP citrate lyase (ACLY) inhibitor
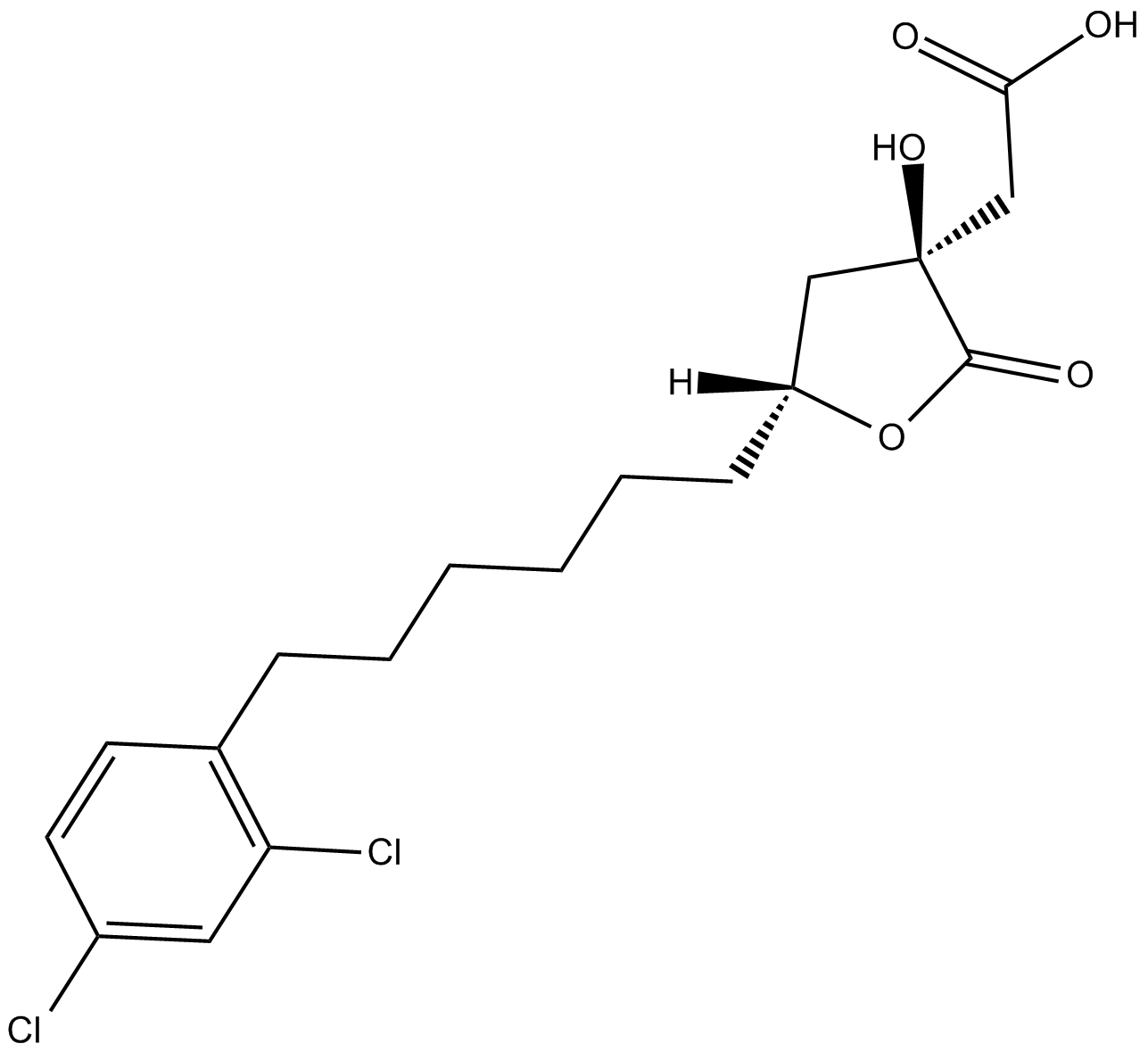
-
GC33902
Urethane (Carbamic acid ethyl ester)
Urethane (Carbamic acid ethyl ester) is an organic compound with an ester structure, which can be used for the synthesis of polyurethane materials, and has also been used as an anesthetic for intravenous injection.
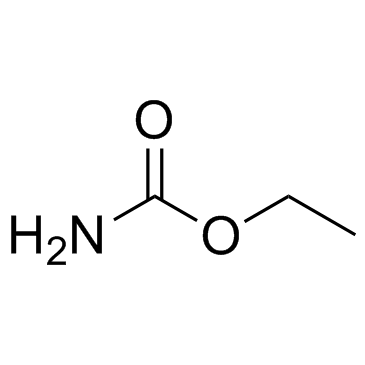
-
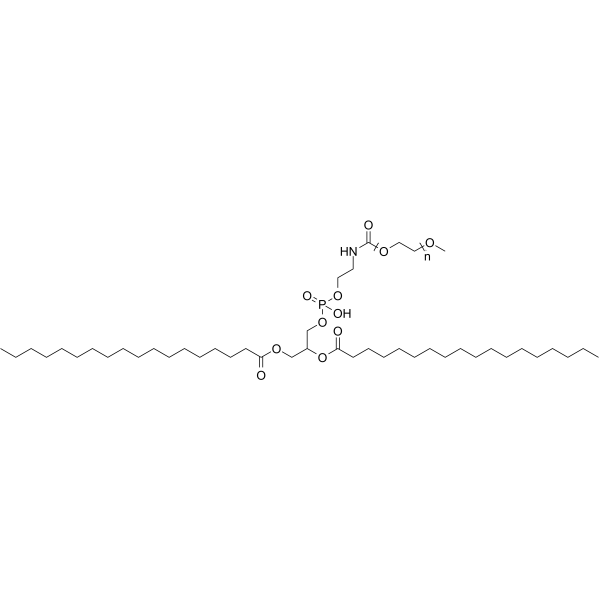
GC64095
MPEG-2000-DSPE
1,2-DSPE-MPEG(5000), Methyl-PEG2000-DSPE, MPEG-2000-DSPE
MPEG-2000-DSPE is a phospholipid PEG conjugate, with both hydrophilicity and hydrophobility, MPEG-2000-DSPE is used as a component of liposomes.
-
GC40314
1,2-Dioleoyl-rac-glycerol
(±)1,2Diolein, racGlycerol 1,2dioleate
Diacylglycerol analog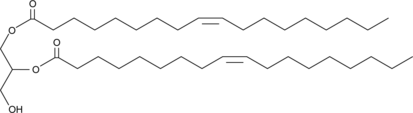
-
GC40412
2-Monostearin (2-Stearoyl-rac-glycerol)
MG(0:0/18:0/0:0), 18:0-MG, 2-Monostearin
2-Monostearin (2-Stearoyl-rac-glycerol) is a monoacylglycerol containing stearic acid at the sn-2 position.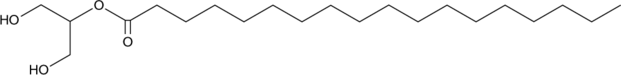
-
GC42444
4-Methylumbelliferyl Oleate
7-hydroxy-4-Methylcoumarin Oleate, 4-Methylumbelliferone Oleate, 4-MU Oleate, 4-MUO
4-Methylumbelliferyl Oleate is a fluorogenic substrate that releases 4-methylumbelliferone (4-MU; Ex/Em: 320/450nm) upon cleavage by lipases, features a pH-dependent excitation wavelength (330nm at pH 4.6, 370nm at pH 7.4, and 385nm at pH 10.4), and is widely used for detecting acid and alkaline lipase activity in biological samples.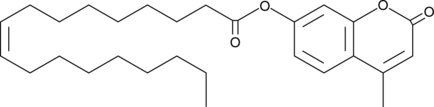
-
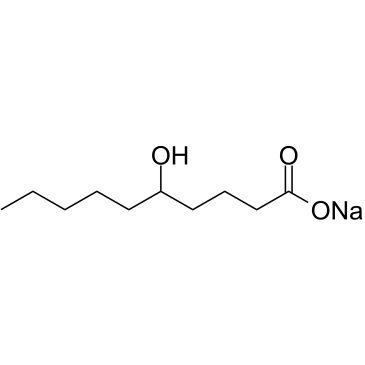
GC60029
5-Hydroxydecanoate sodium
5-Hydroxydecanoate sodium is a selective ATP-sensitive K+ (KATP) channel blocker with an IC50 value of 28.7μM.
-
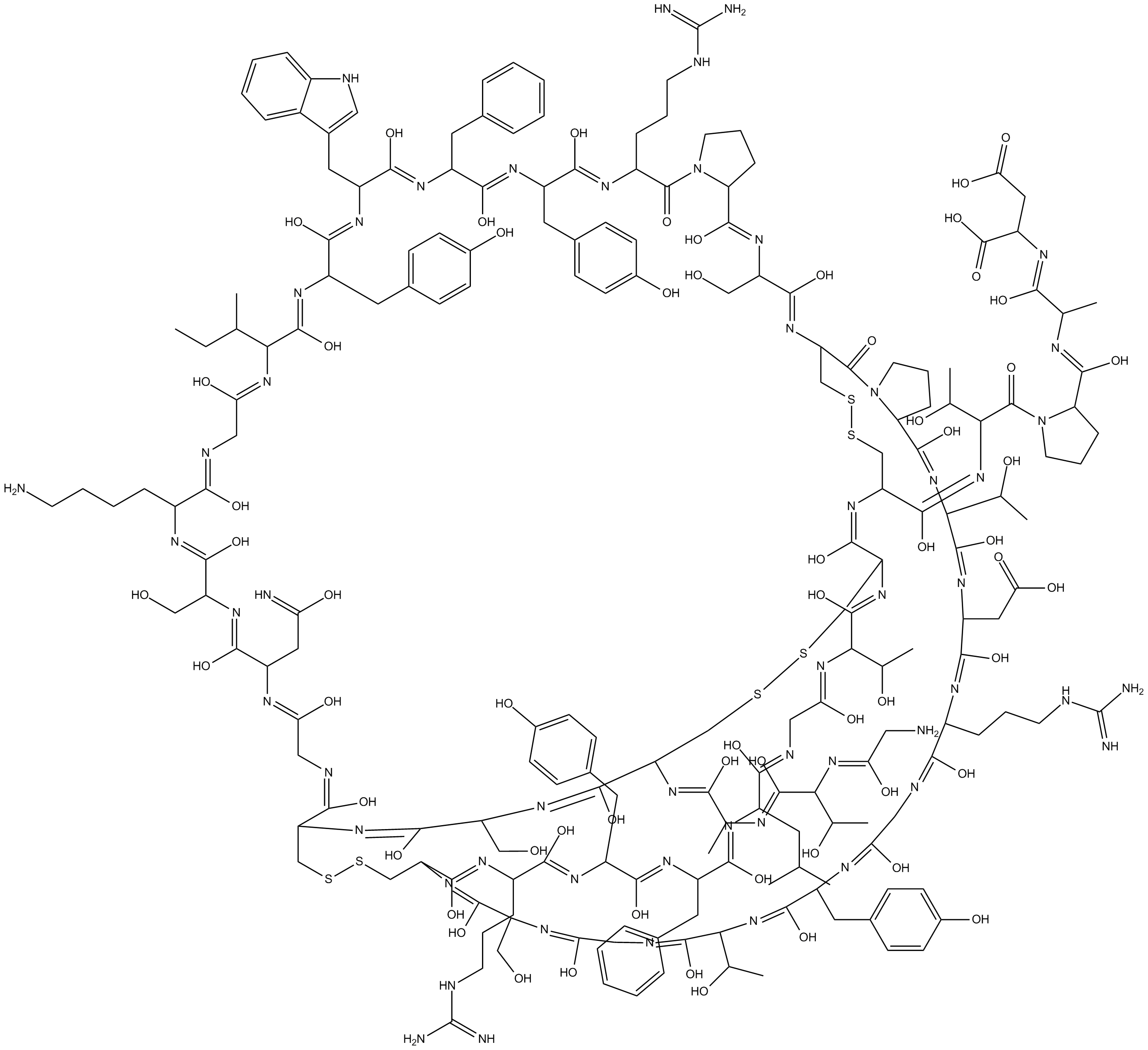
GC17168
APETx2
acid-sensing ion channel 3 (ASIC3) channel blocker
-
GC65272
APETx2 TFA
APETx2 is a peptide toxin derived from Anthopleura elegantissimaand serves as a selective, reversible inhibitor of the acid-sensing ion channel 3 (ASIC3).
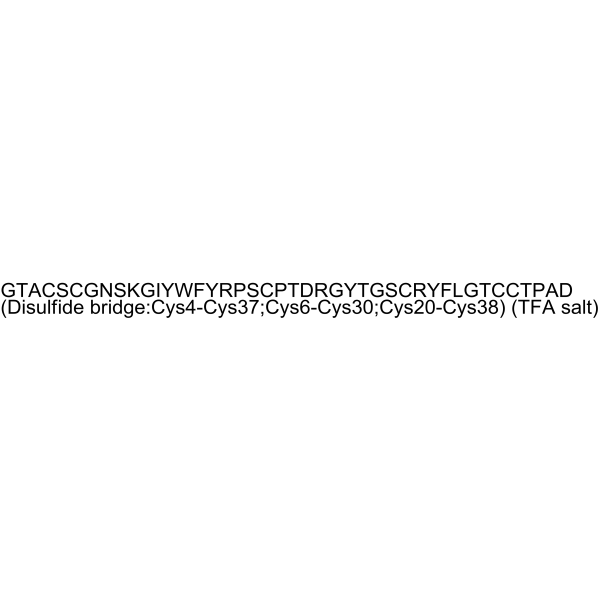
-
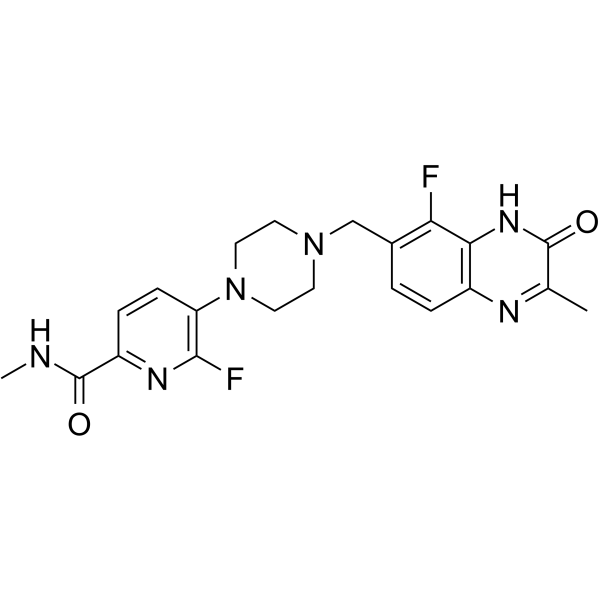
GC64099
AZD-9574
AZD-9574 is a novel, blood-brain barrier permeable PARP1 inhibitor that selectively inhibits PARP1 at single-strand break (SSB) sites.
-
GC34357
DBCO-(PEG)3-VC-PAB-MMAE
DBCO-(PEG)3-VC-PAB-MMAE is a drug-linker conjugate for ADC, formed by connecting Monomethylauristatin E (MMAE) to a DBCO-(PEG)3-vc-PAB linker.
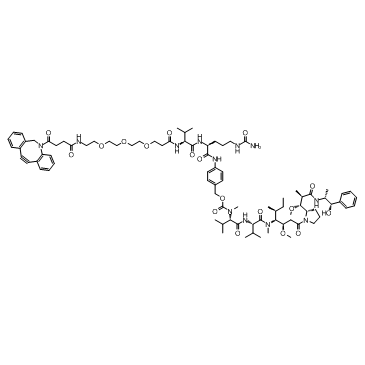
-
GN10422
Echinacoside
Kusaginin, NSC 603831, trans-Acteoside, trans-Verbascoside
Echinacoside is a phenylethanoid glycoside isolated from Cistanche tubulosa with potent antioxidant and anti-inflammatory effects.
-
GA22890
H-Ser-His-OH
H-Ser-His-OH is a short peptide with hydrolytic cleavage activity and an endogenous metabolite.

-
GA22902
H-Ser-Tyr-OH
H-Ser-Tyr-OH is a dipeptide composed of L-serine and L-tyrosine.

-
GC41513
Nε-(1-Carboxymethyl)-L-lysine
CML
Nε-(1-Carboxymethyl)-L-lysine is a unique post-translational modification (PTM) of proteins that is generated by the non-enzymatic glycation of lysine residues.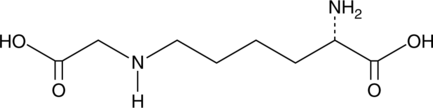
-
GA23321
Ophthalmic Acid
Ophthalmic Acid is an analog of glutathione (GSH) and a marker of oxidative stress and hepatic glutathione depletion.

-
GC10164
Orbifloxacin
CP 104,354
Synthetic broad-spectrum fluoroquinolone antibiotic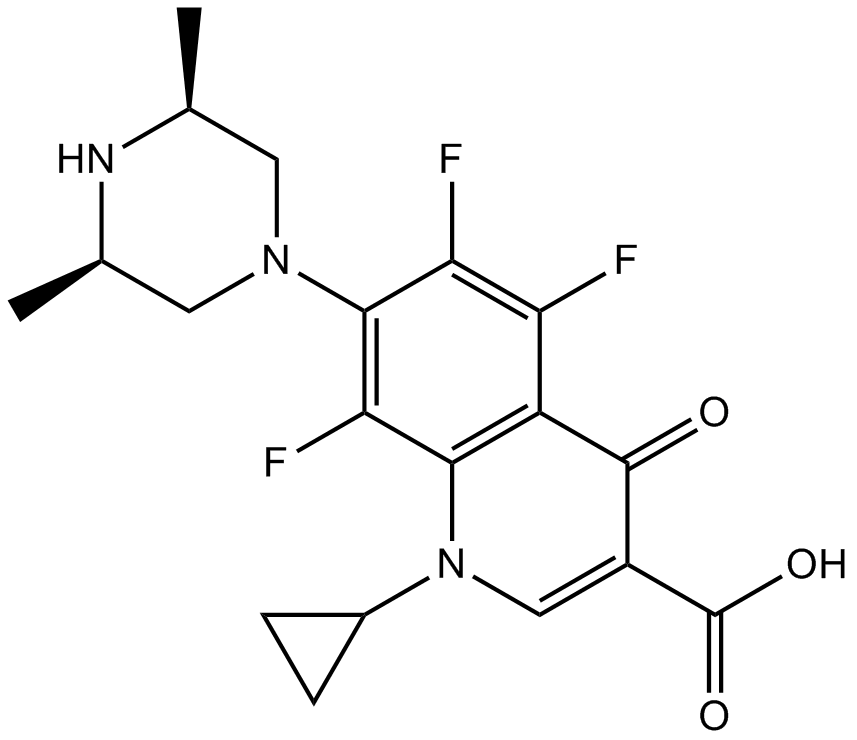
-
GC13561
PA-824
Pretomanid
Anti-tuberculosis drug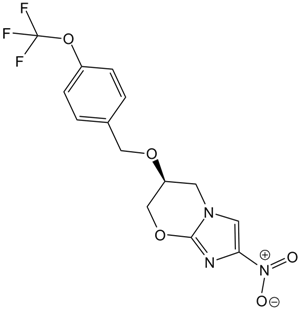
-
GC30112
PAT-1251
PAT-1251; GB2064
PAT-1251 is a lysyl oxidase-like 2 (LOXL2; IC50=0.71μM) inhibitor.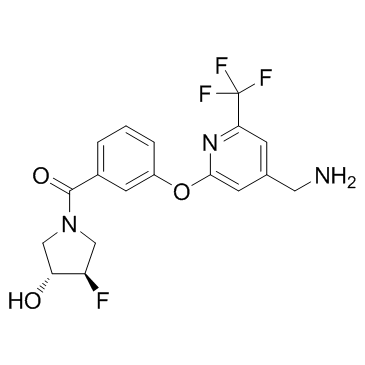
-
GC31949
PAT-1251 Hydrochloride
PAT-1251 is a lysyl oxidase-like 2 (LOXL2; IC50=0.71μM) inhibitor.
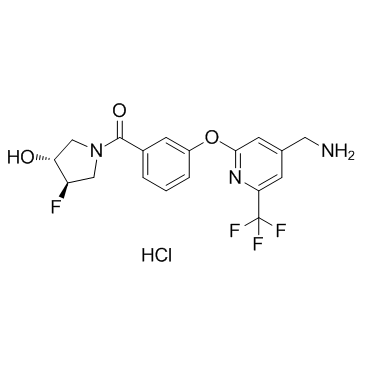
-
GC65250
QTZ
QTZ is a bioluminescent substrate used for in vivo imaging.

-
GC12877
SR3335
ML 176
SR3335 is a highly potent and selective inverse agonist of retinoic acid receptor-related orphan receptor alpha (RORα), with a Ki value of 220nM.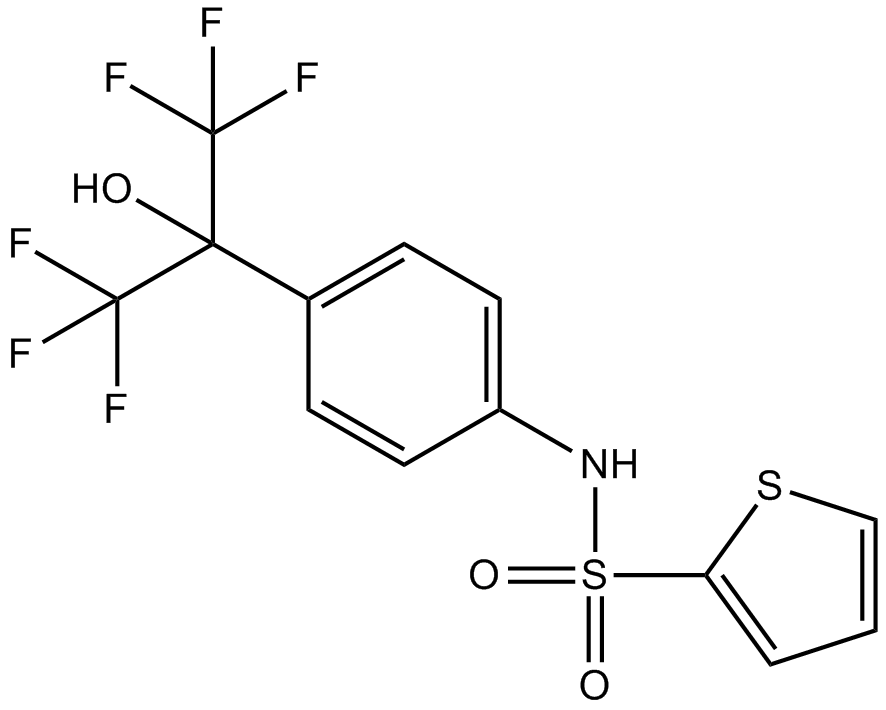
-
GA23548
Suc-Ala-Phe-Lys-AMC
Suc-Ala-Phe-Lys-AMC is a highly sensitive fluorogenic substrate.

-
GC30282
NBD-F (4-Fluoro-7-nitrobenzofurazan)
NBD-F (4-Fluoro-7-nitrobenzofurazan) (4-Fluoro-7-nitrobenzofurazan) is a pro-fluorescent reagent which is developed for amino acid analysis.
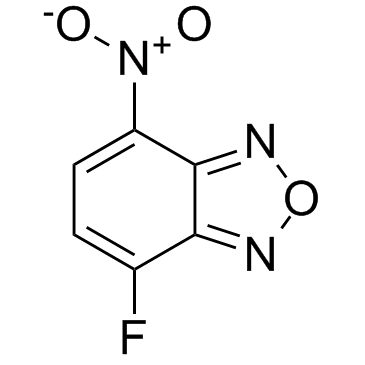
-
GC27559
Human Glu-Plasminogen
Human Glu-Plasminogen can be isolated and purified from human plasma.

-
GB55969
4CzIPN

-
GA20063
(D-Ala2)-GIP (human)
(D-Ala2)-GIP (human) is an analog of glucose-dependent insulinotropic polypeptide that activates the GIP receptor (EC50=630pM) and is commonly used in research on type 2 diabetes treatment.

-
GC67423
Propargyl-PEG6-SH
Propargyl-PEG6-SH is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.

-
GC66755
HS-PEG5-CH2CH2N3
HS-PEG5-CH2CH2N3 is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.

-
GC27558
Human Lys-Plasminogen
Human Lys-Plasminogen can be purified from homogeneous glu-plasminogen by activation with Plasmin.

-
GC26296
Squalene
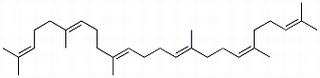
-
GA20486
Ac-Arg-pNA . HCl
Ac-Arg-pNA . HCl is a commonly used substrate for trypsin, which is utilized for inhibitor screening or enzyme concentration determination.

-
GC27557
Magnoloside F
Magnoloside F is a natural compound that can be isolated from the Magnolia officinalis, and has antioxidant activity.

-
GC16285
Amikacin
Antibiotic BBK 8, BAY 416551, BBK 8, Lukadin, Potentox
aminoglycoside antibiotic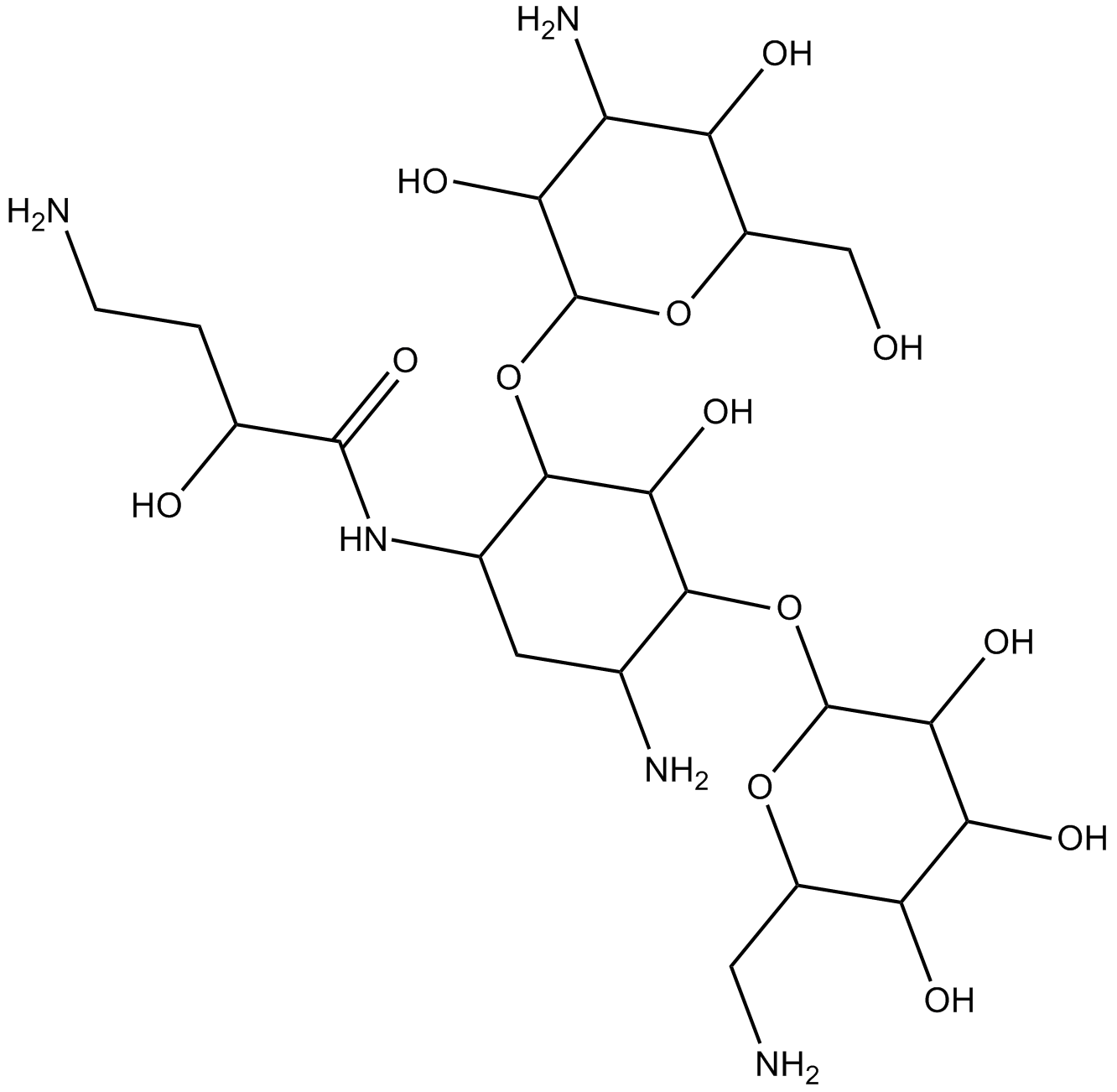
-
GC11093
Amikacin disulfate
Semisynthetic aminoglycoside antibiotic
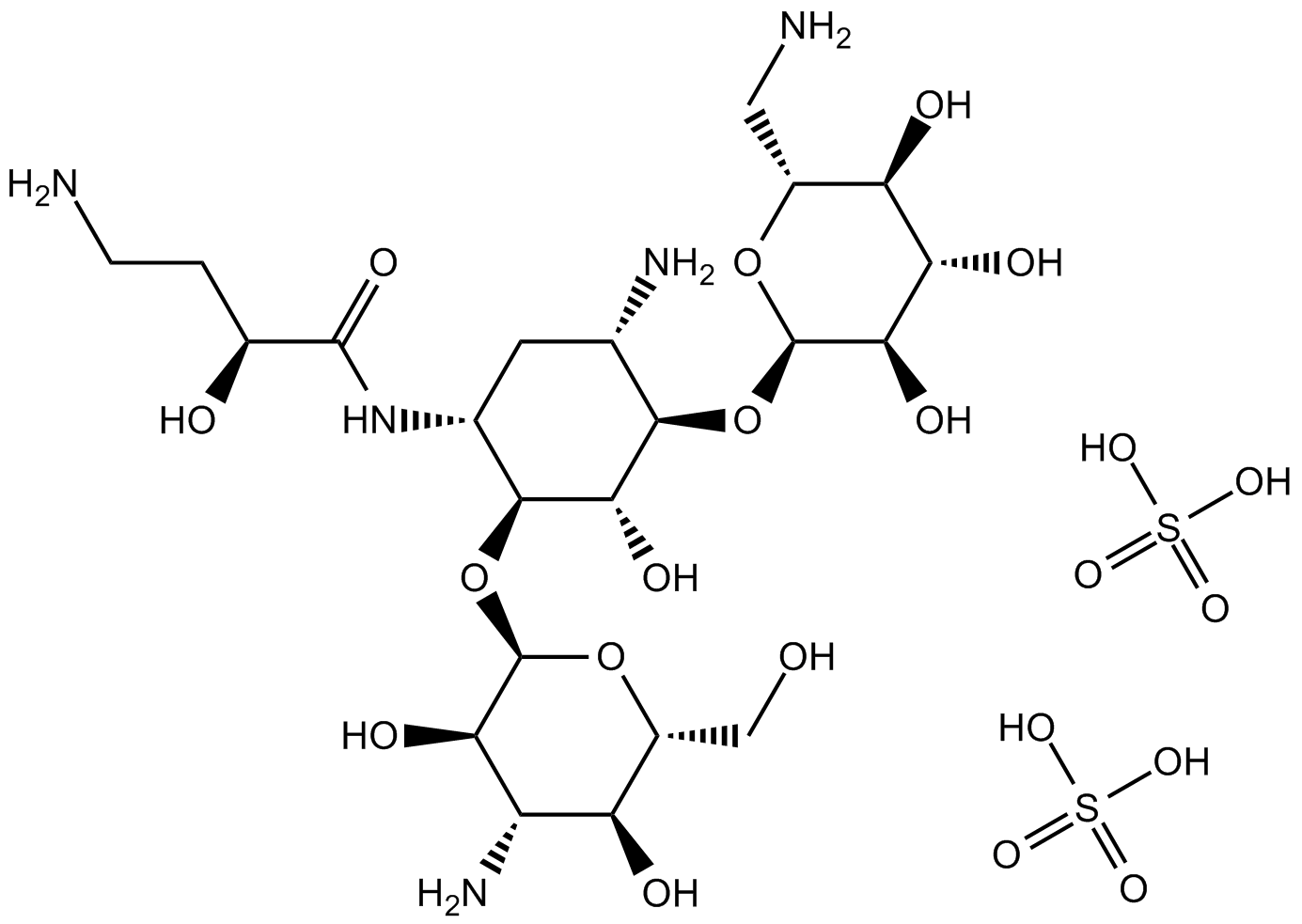
-
GC15685
Amikacin hydrate
Aminoglycoside antibiotic
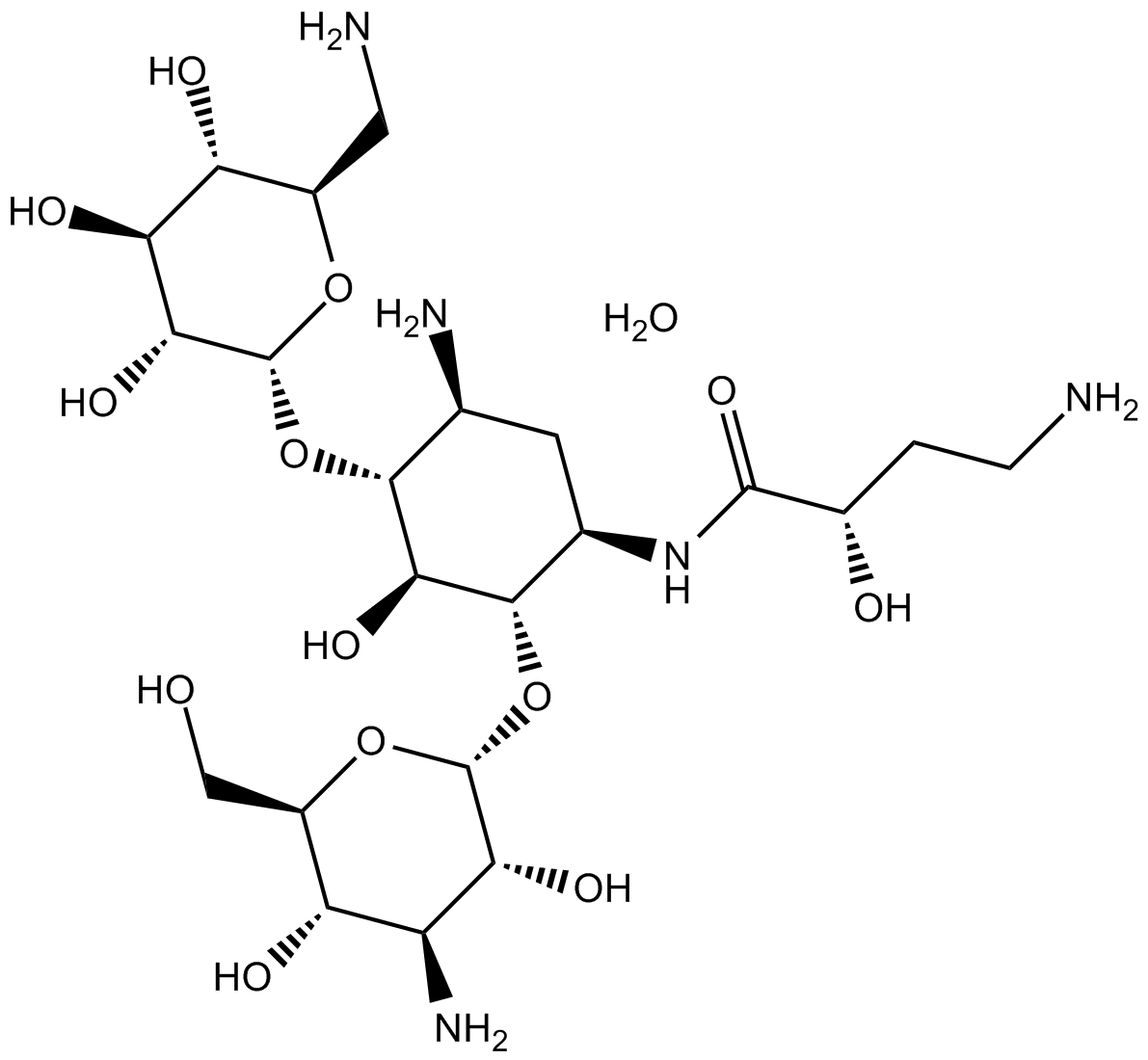
-
GC14411
Apoptozole
Apoptosis Activator VII
inhibitor of heat shock protein 70 (Hsp70)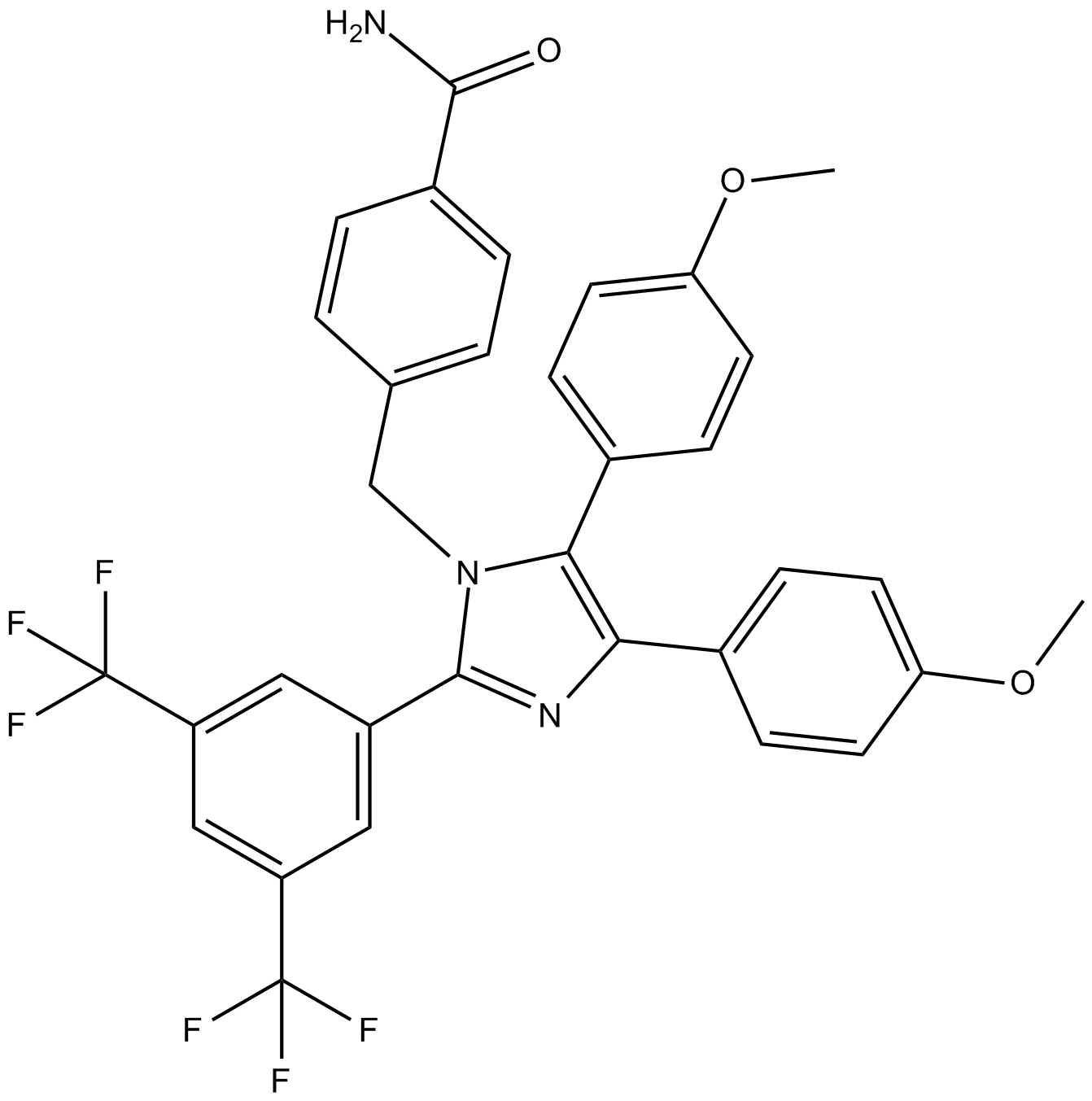
-
GC32685
ARV-771
ARV-771, a pan-BET-PROTAC, potently degrades BRD2/3/4 with a DC50<5nM.
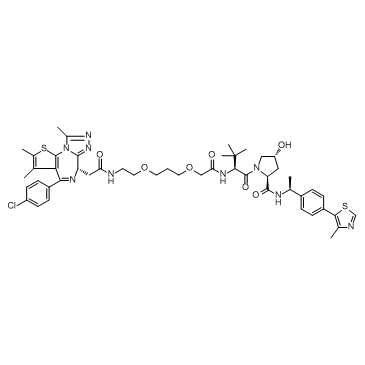
-
GC17903
Atovaquone
BW 566C, BW 556C-80
unique naphthoquinone with broad-spectrum antiprotozoal activity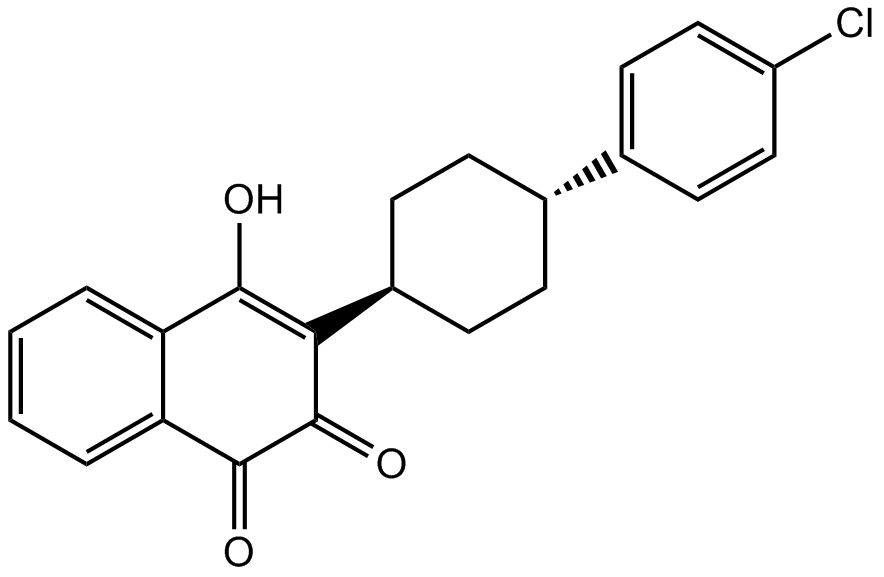
-
GC12948
CID-1067700
ML-282
competitive inhibitor of nucleotide binding by Ras-related GTPases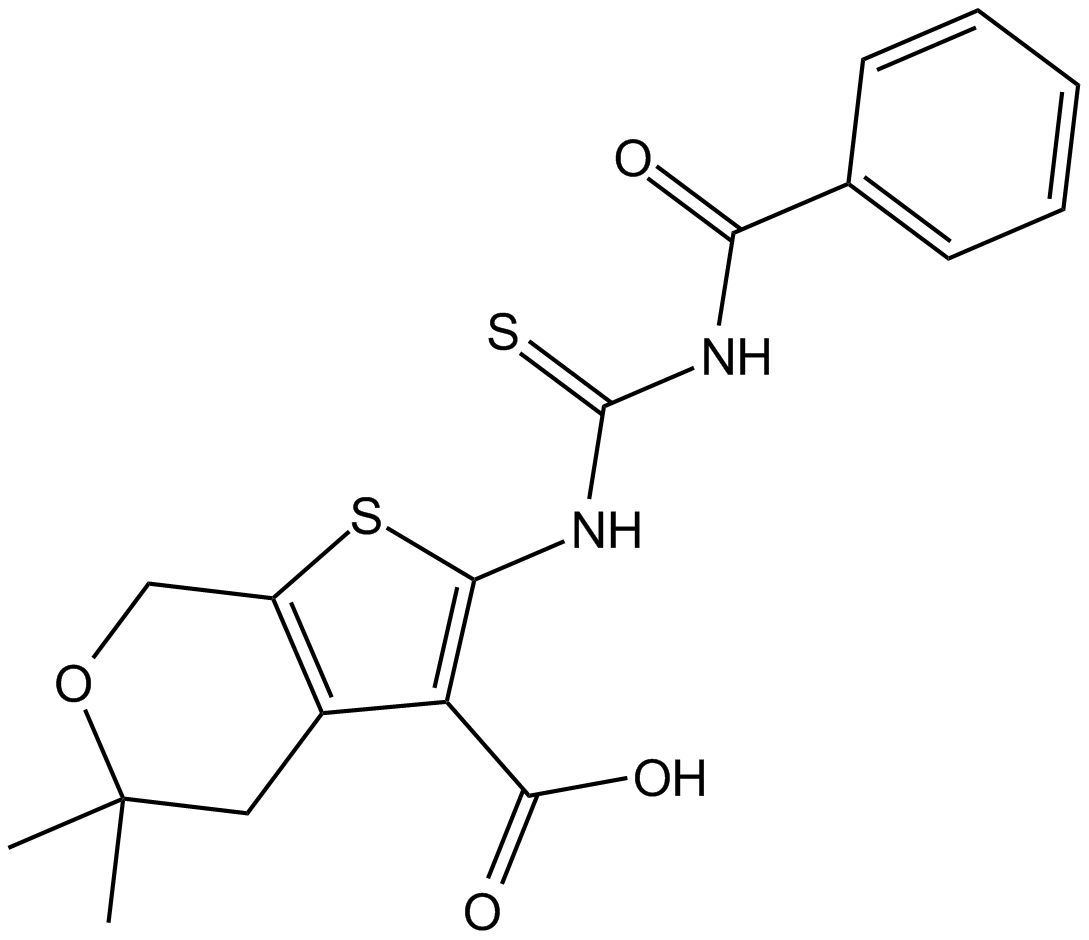
-
GC15484
Deguelin
(-)cisDeguelin
Deguelin is one of four major naturally occurring rotenoids isolated from root extracts and is best recognized as a NADH: ubiquinone oxidoreductase (complex I) inhibitor, resulting in significant alterations in mitochondrial function.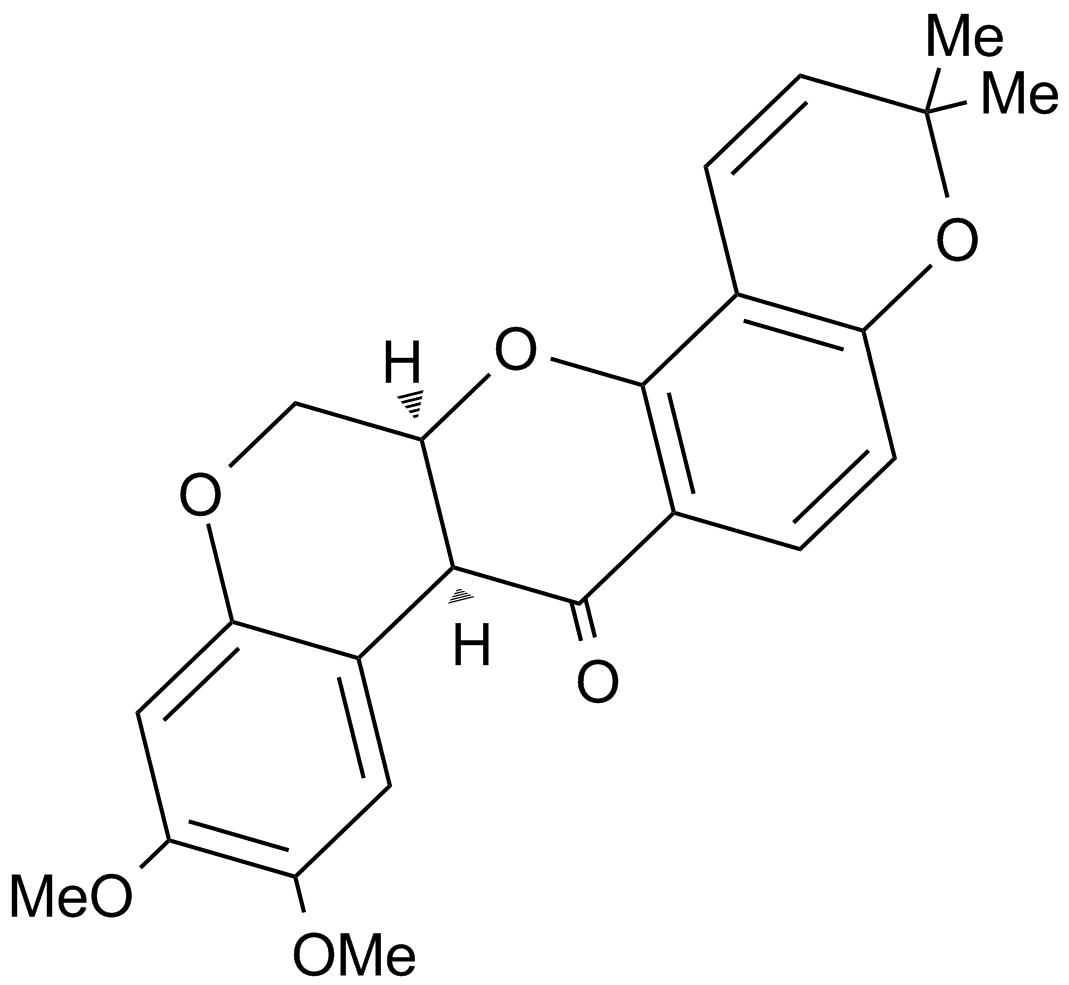
-
GP10119
Gap 27
Ser-Arg-Pro-Thr-Glu-Lys-Thr-Ile-Phe-Ile-Ile
Gap 27 is a peptide (Ser-Arg-Pro-Thr-Glu-Lys-Thr-Ile-Phe-Ile-Ile) derived from connexin43 (Cx43) and is a selective gap junction blocker.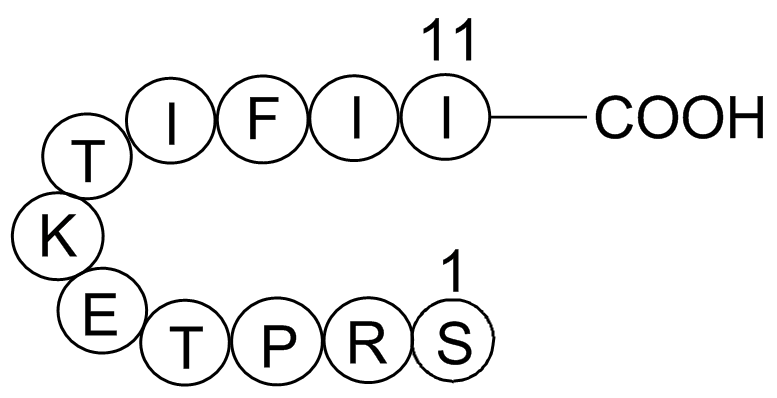
-
GC16460
KIN-59
5’-O-Tritylinosine
thymidine phosphorylase (Tpase) inhibitor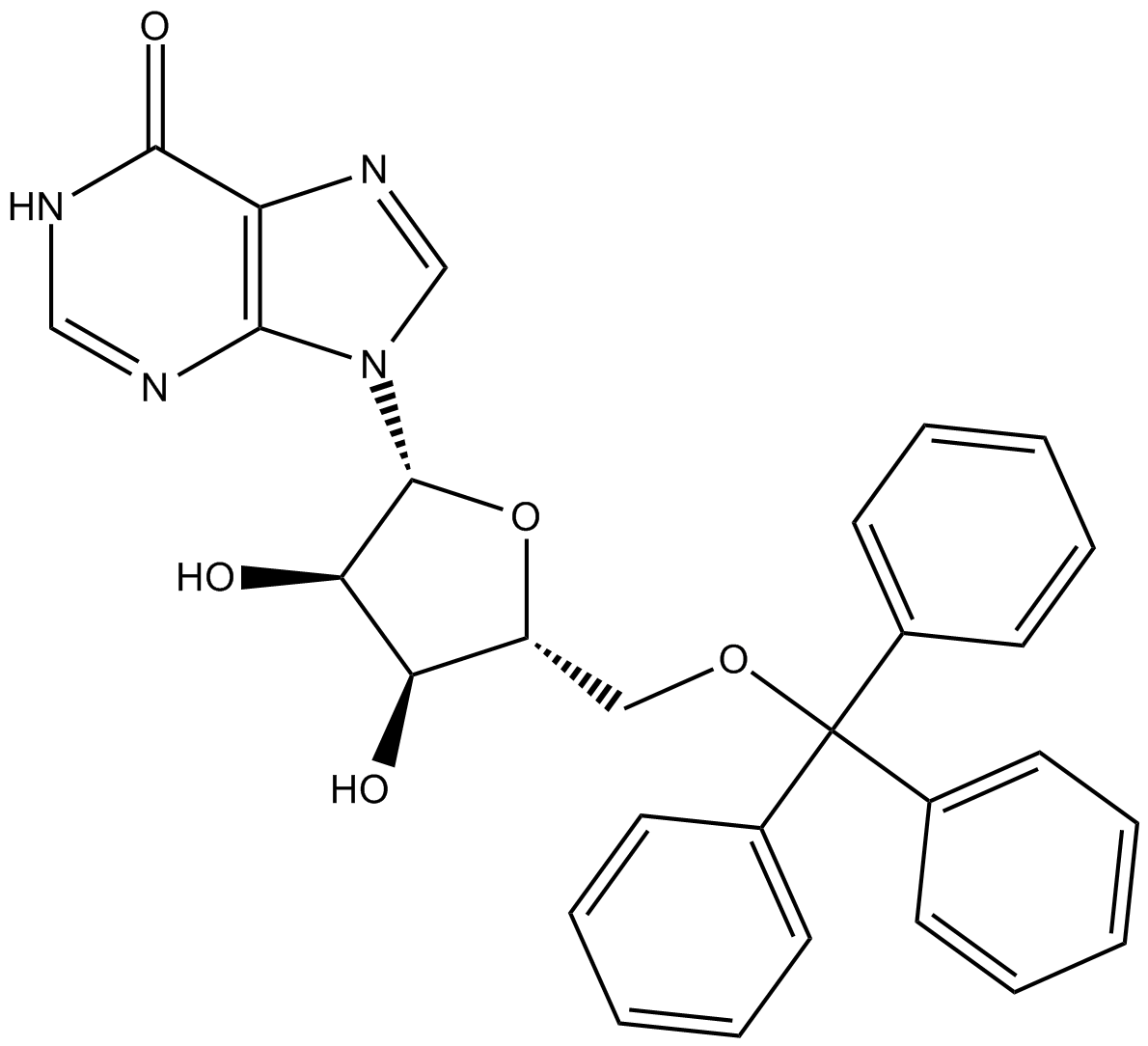
-
GC11439
MG 149
Tip60 HAT inhibitor
HAT inhibitor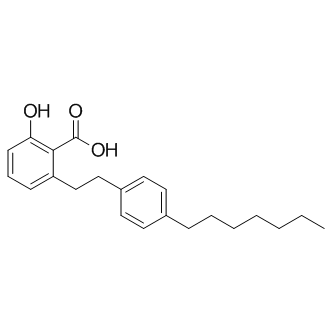
-
GC50176
PMX 53
3D53
PMX 53 is a potent, cell-permeable, and orally active C5a receptor (CD88) antagonist with an IC50 value of 20nM.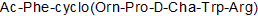
-
GC13002
Thiostrepton
Bryamycin, NSC 81722, NSC 170365, Thiactin, Thiostreptin A
Antibiotic that inhibits bacterial protein synthesis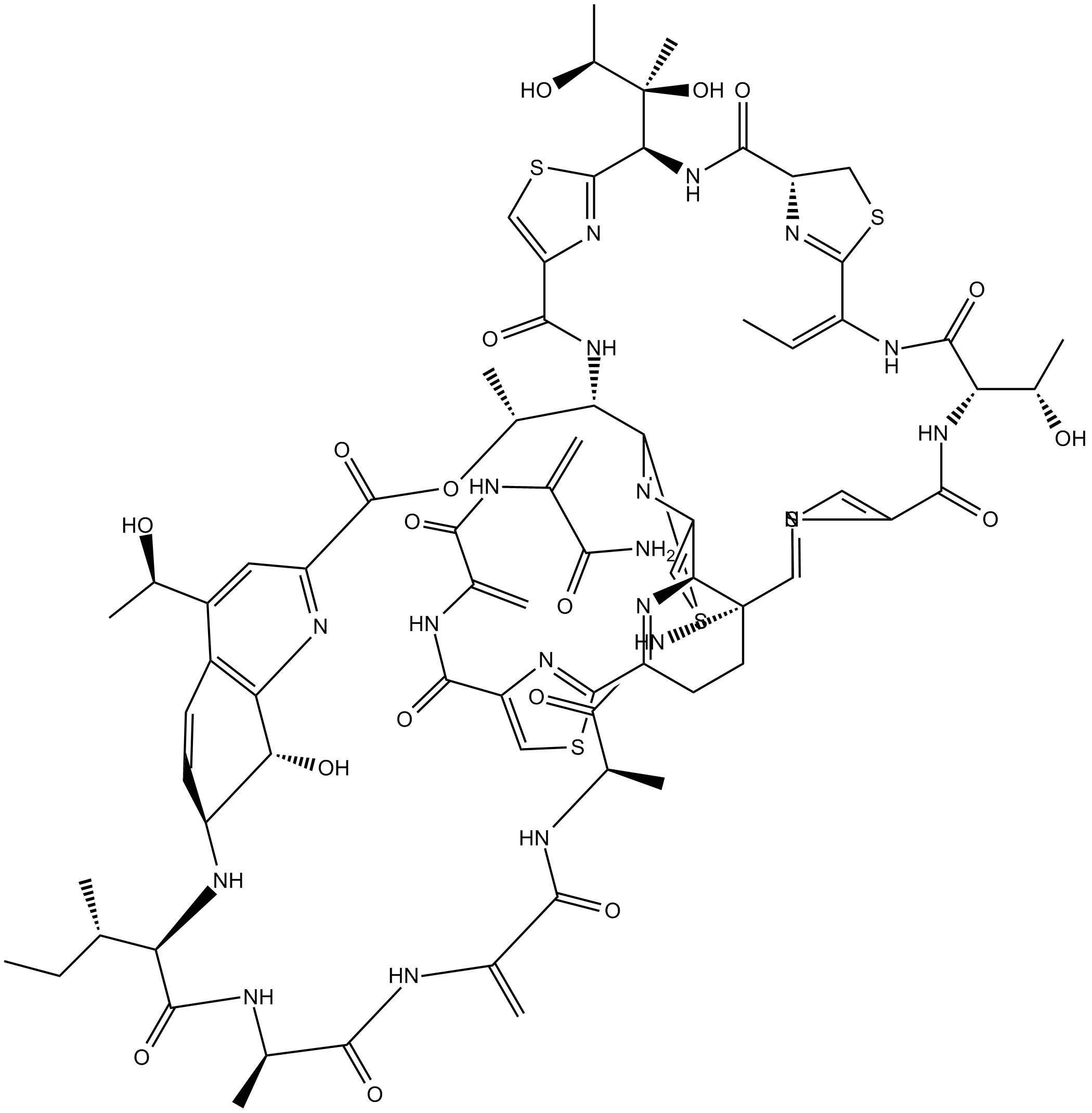
-
GC45067
Tpl2 Kinase Inhibitor
c-Cot Kinase Inhibitor, MAP3K8 Kinase Inhibitor, Tumor Progression Locus 2 Kinase Inhibitor
Tpl2 Kinase Inhibitor (Compound 1) is a potent and selective Tpl2 (COT kinase, MAP3K8) inhibitor, plays an important role in the regulation of the inflammatory response and the progression of some cancers.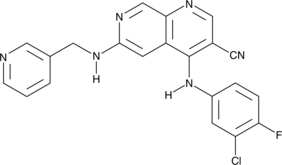
-
GC13107
YM155
Sepantronium bromide
YM155, a small imidazolium-based compound, potently inhibits survivin promoter activity with an IC50 value of 0.54nM.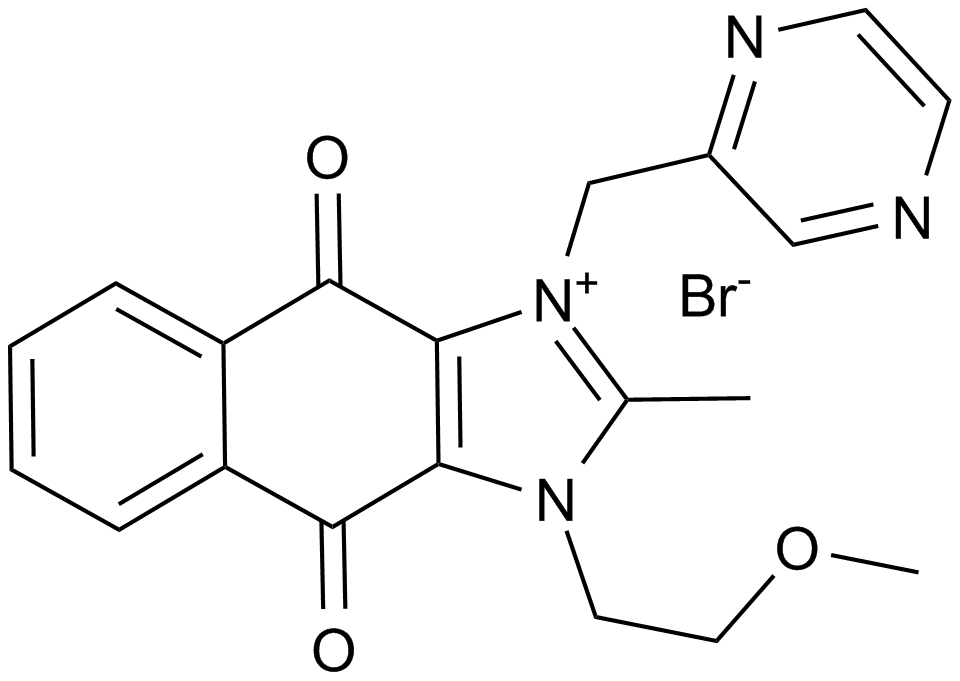
-
GC16186
4-P-PDOT
AH 024
Melatonin receptor antagonist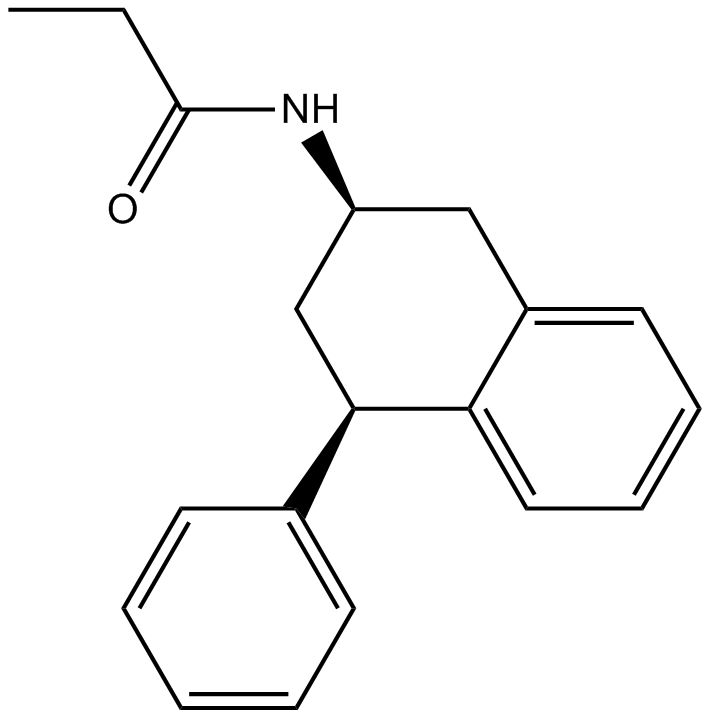
-
GN10228
6-Shogaol
A bioactive component of ginger
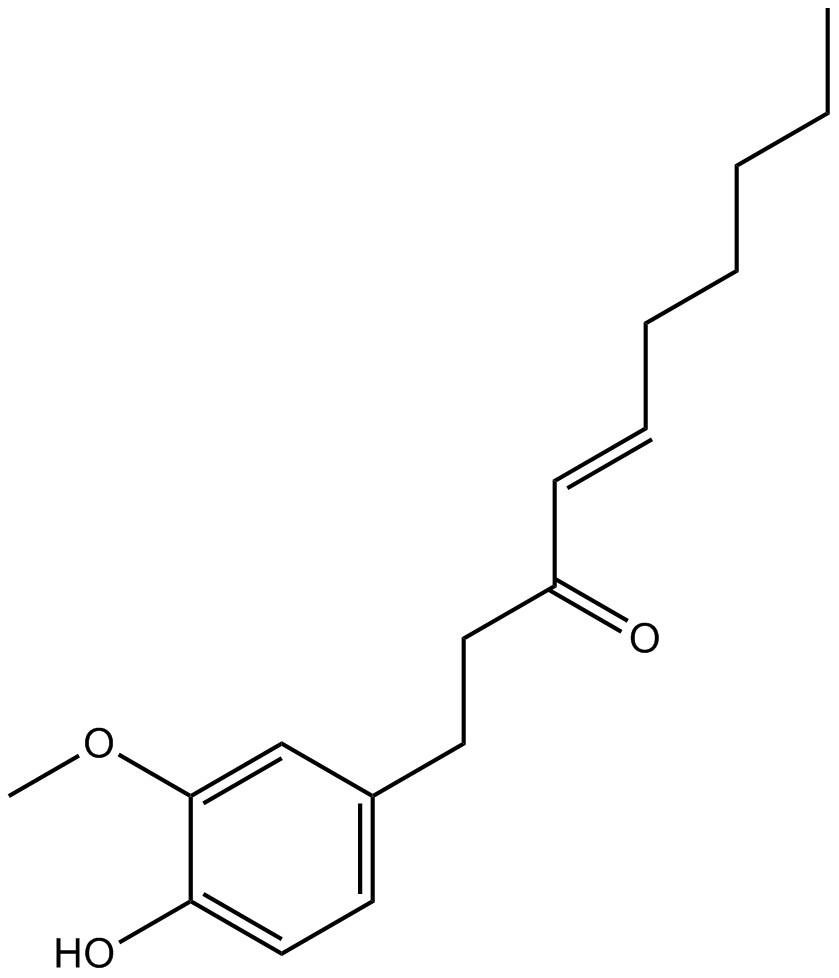
-
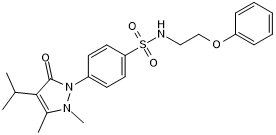
GC50572
BC-LI-0186
BC-LI-0186 is a potent and highly selective inhibitor of the interaction between leucyl-tRNA synthetase (LRS) and Ras-related GTP-binding protein D (RagD), with an IC₅₀ value of 46.11nM.
-
GC33340
BDP9066
BDP9066 is a selective inhibitor of myotonic dystrophy-associated Cdc42-binding kinase (MRCK), with an IC50 of 64nM for MRCKβ and Ki values of 0.0136nM and 0.0233nM for MRCKα/β in SCC12 cells.
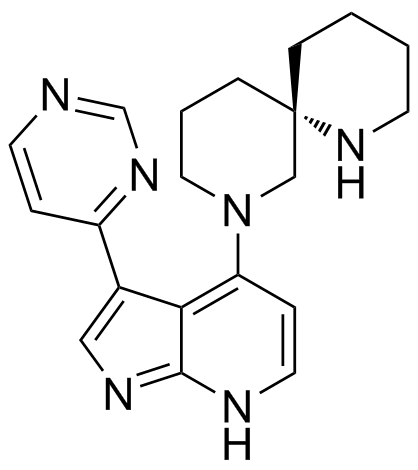
-
GC18354
Bisindolylmaleimide X (hydrochloride)
BIM X, Ro 31-8425
A PKC inhibitor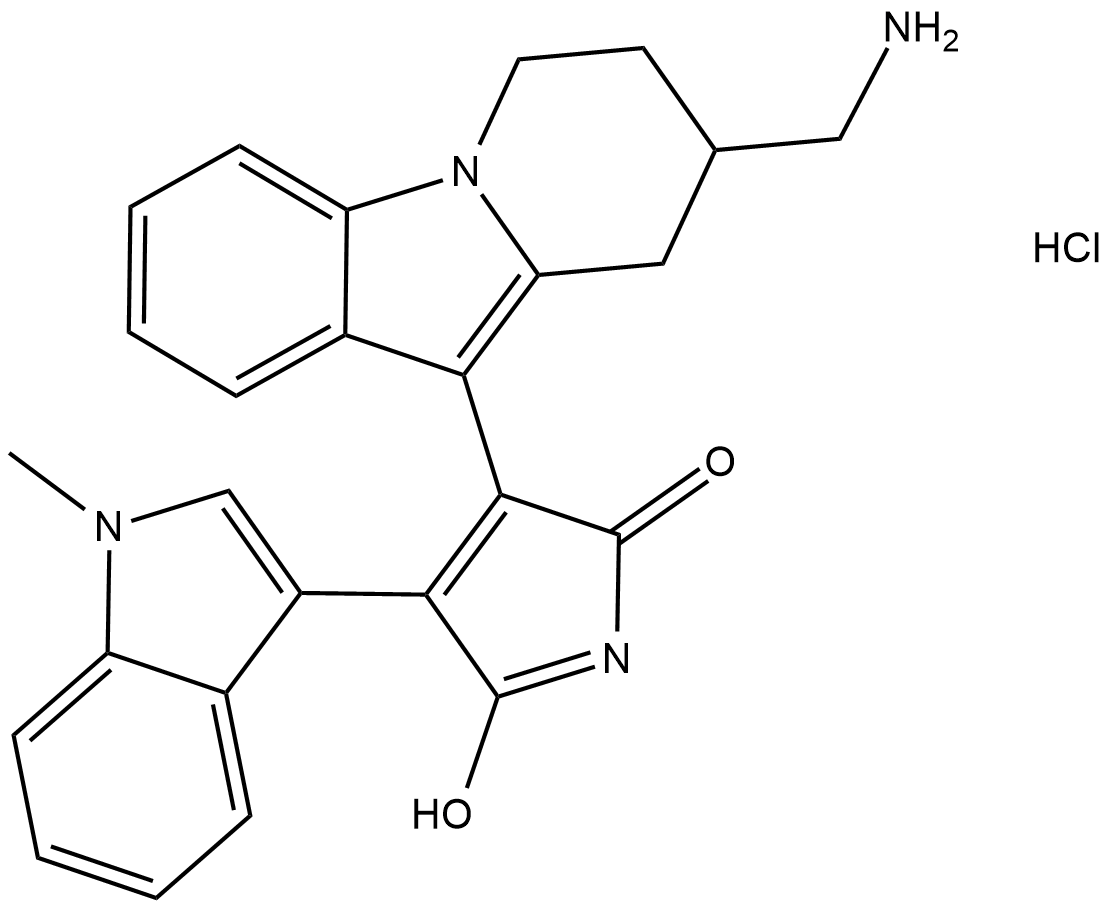
-
GC68793
BODIPY FL C5-Ceramide
BODIPY FL C5-Ceramide is a Golgi-specific green fluorescent dye, with Ex/Em=505nm/512nm.

-
GC12910
Canertinib (CI-1033)
HER family tyrosine kinase inhibitor
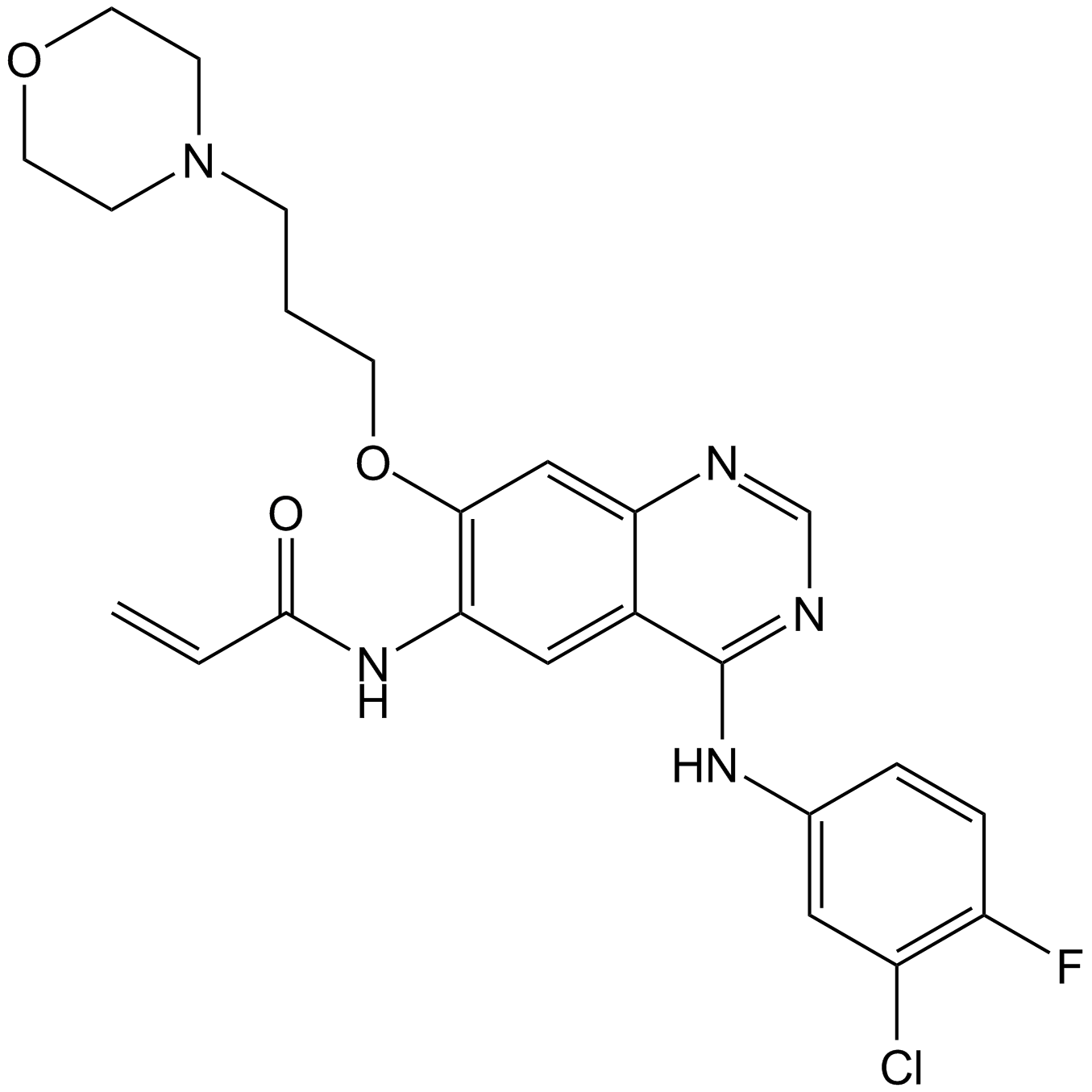
-
GC10803
DAMGO
μ opioid receptor agonist
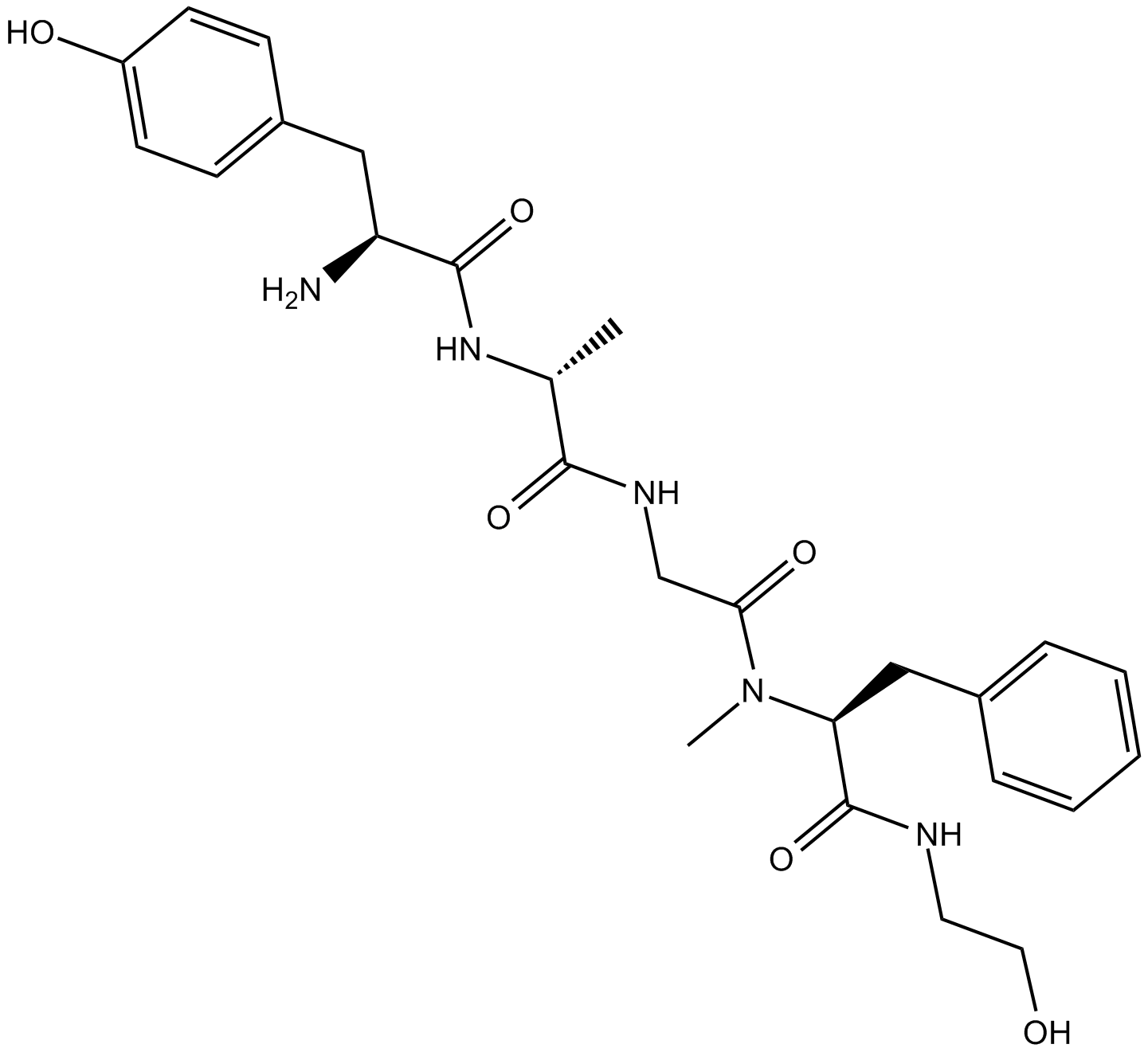
-
GC35805
DAMGO (TFA)
DAMGO (TFA) is a μ-opioid receptor (μ-OPR ) selective agonist with a Kd of 3.46 nM for native μ-OPR.
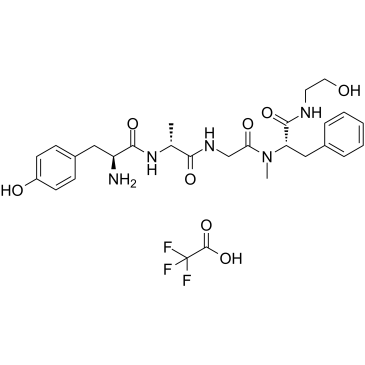
-
GC38102
Epithalon
Epithalon is an anti-aging agent and a telomerase activator.
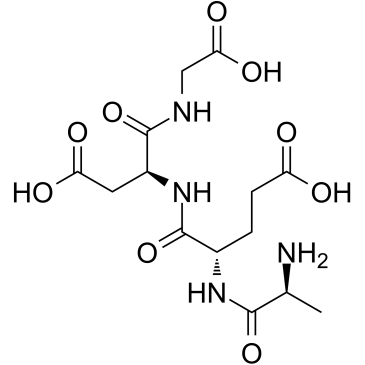
-
GC12932
EPZ5676
EPZ5676 is a novel, highly potent and selective DOT1L histone methyltransferase inhibitor with a Ki value of less than 80pM and an IC50 of 0.8nM against DOT1L, showing selectivity that is 37,000-fold higher than against other protein methyltransferases.
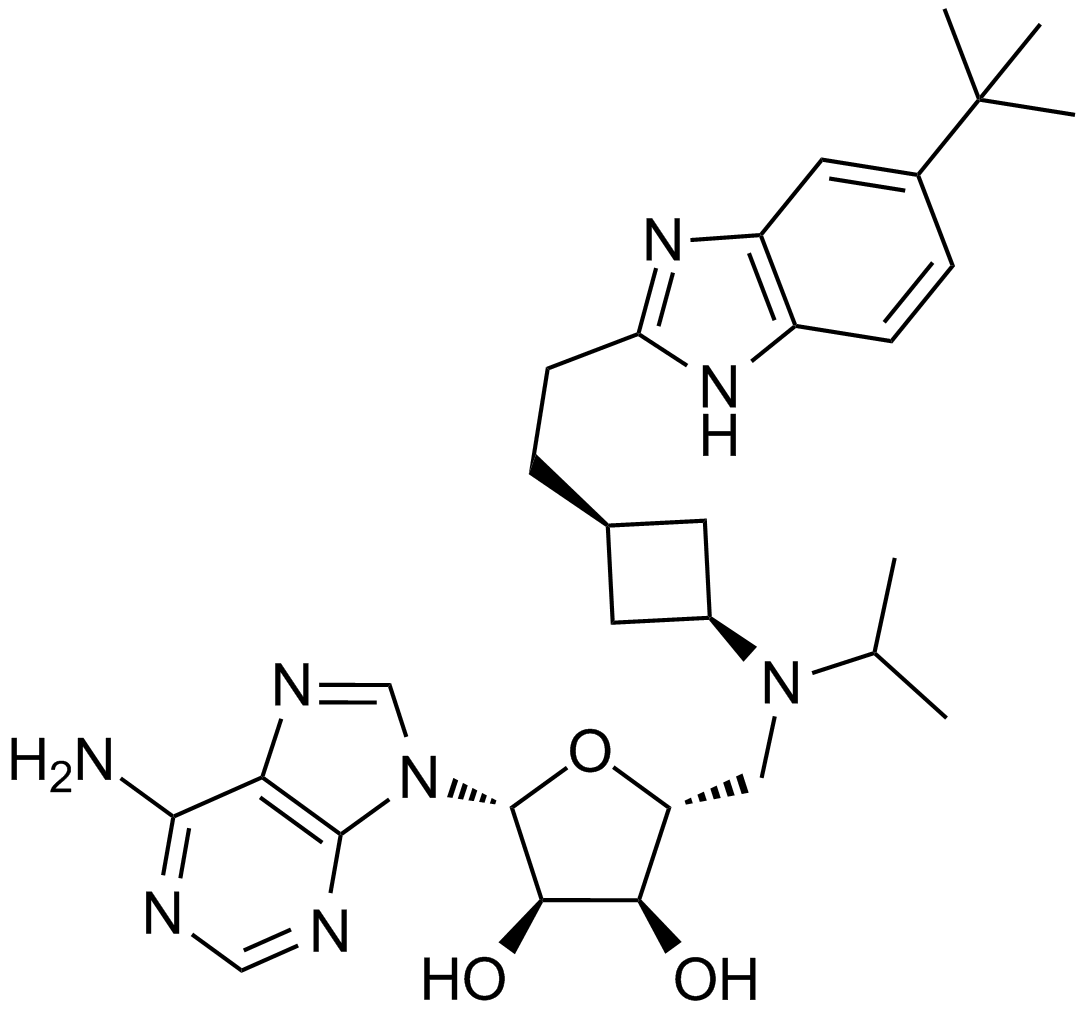
-
GN10151
Geraniin
NSC 359346
Geraniin, a TNF-α releasing inhibitor with an IC50 value of 43μM, exhibits numerous activities including anticancer, anti-inflammatory, and anti-hyperglycemic activities.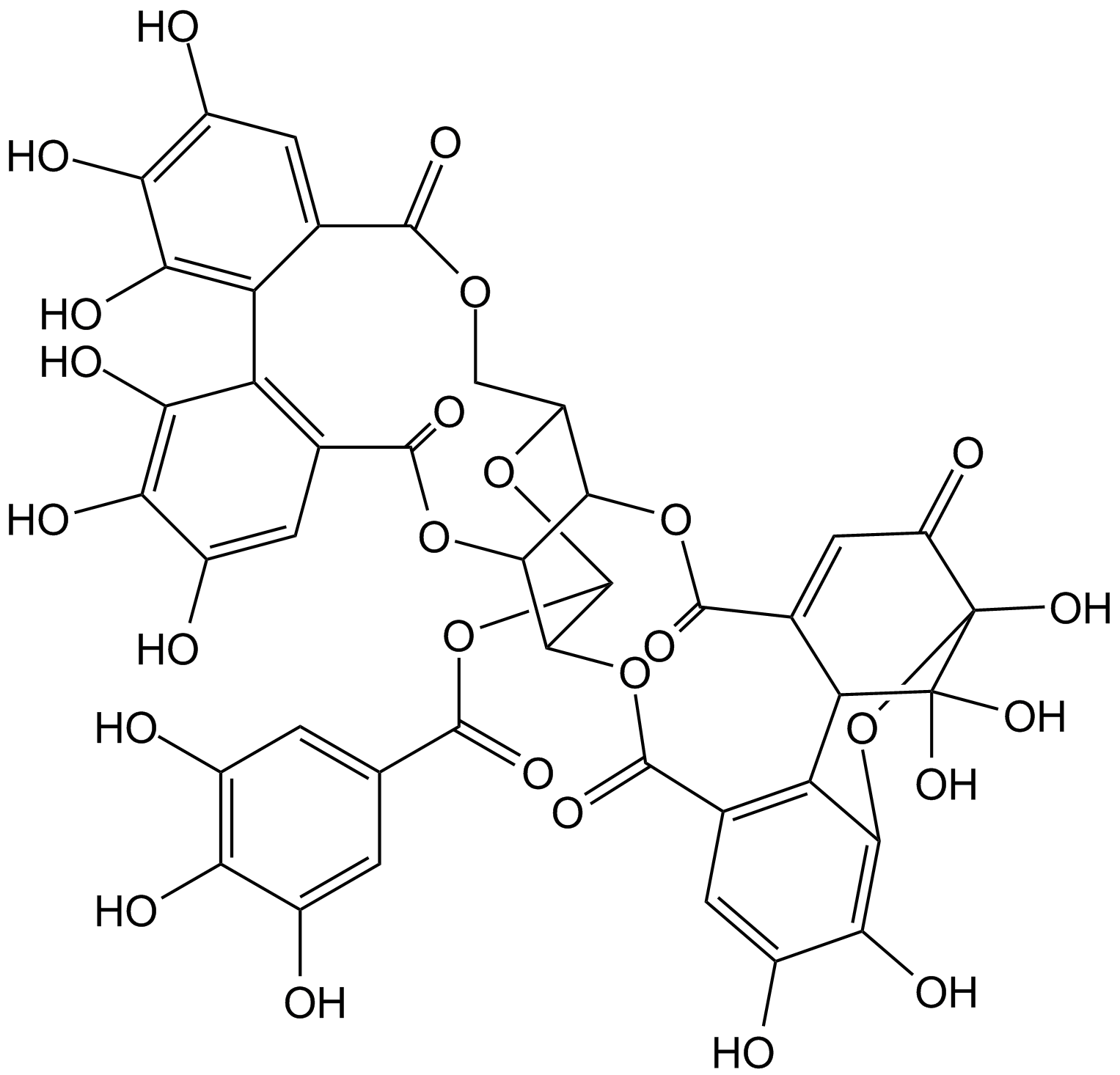
-
GC39387
GSK2798745
GSK2798745, a clinical candidate, was identified as an inhibitor of the transient receptor potential vanilloid 4 (TRPV4) ion channel with IC50 of 1.8 and 1.6nM for hTRPV4 and rTRPV4, respectively.
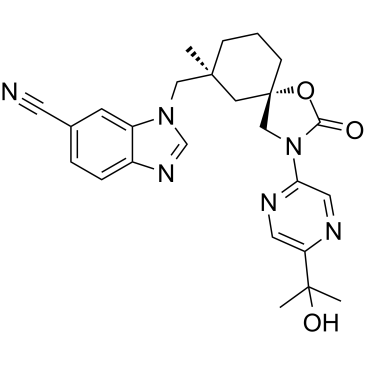
-
GC63022
Inupadenant
EOS-850
Inupadenant is an orally active, highly selective A2A receptor antagonist, which maintains full potency at the high adenosine levels found in tumors, exhibiting strong antitumor activity.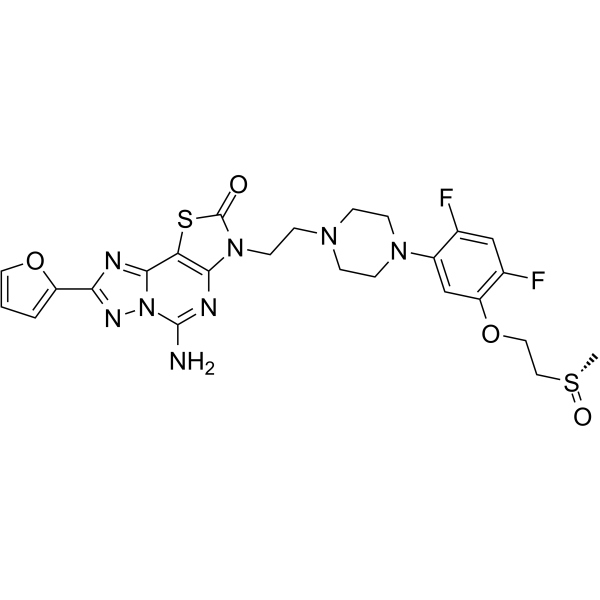
-
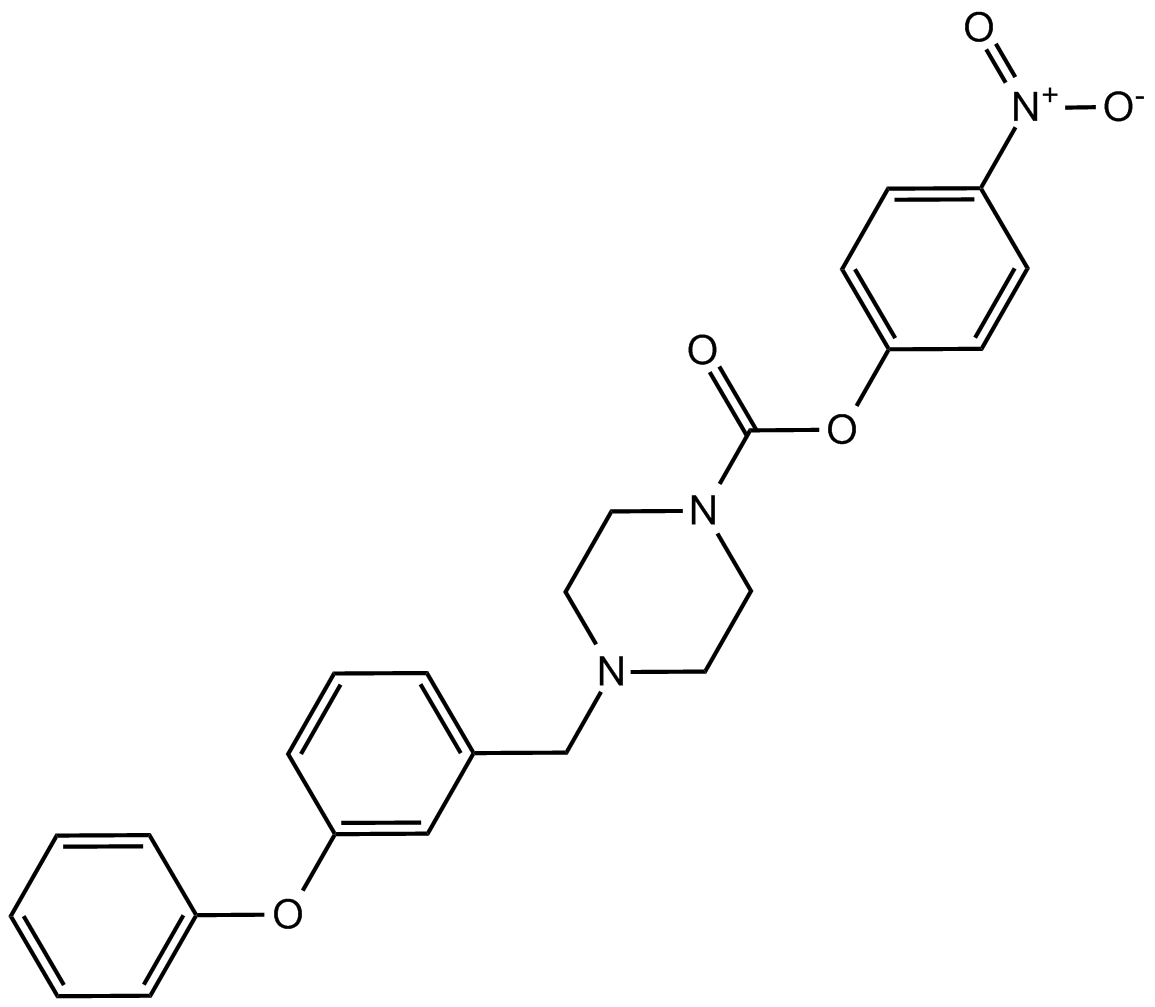
GC12693
JZL 195
Dual inhibitor of fatty acid amide hydrolase (FAAH) and monoacylglycerol lipase (MAGL)
-
GB10076
Lysozyme chloride
Lysozyme chloride is an orally active bacteriolytic enzyme that specifically hydrolyzes the β(1-4) glycosidic bonds between N-acetylmuramic acid and N-acetylglucosamine in the peptidoglycan layer of bacterial cell walls, thereby lysing Gram-positive bacteria.

-
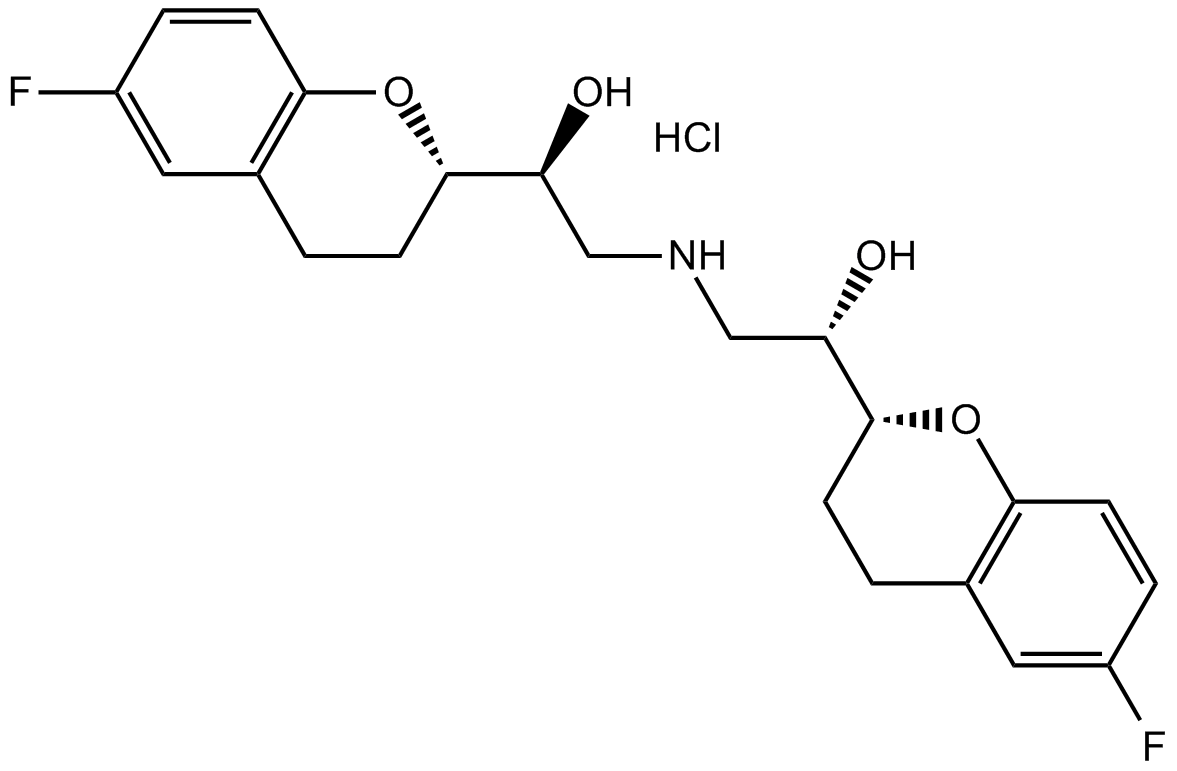
GC13906
Nebivolol hydrochloride
Highly selective β1-adrenoceptor antagonist
-
GC44451
Norfluoxetine (hydrochloride)
Desmethylfluoxetine
An active metabolite of fluoxetine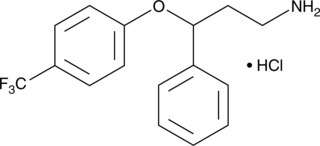
-
GC26095
Paclitaxel-SMCC
Paclitaxel-SMCC; Paclitaxel with SMCC linker; Paclitaxel ADC conjugate.; SMCC-Taxol; SMCC taxol, Taxol-SMCC, Taxol SMCC
Paclitaxel-SMCC is a paclitaxel derivative with a SMCC linker.
-
GC13776
Piericidin A
AR 054, Shaoguanmycin B, SN 198E
mitochondrial complex I inhibitor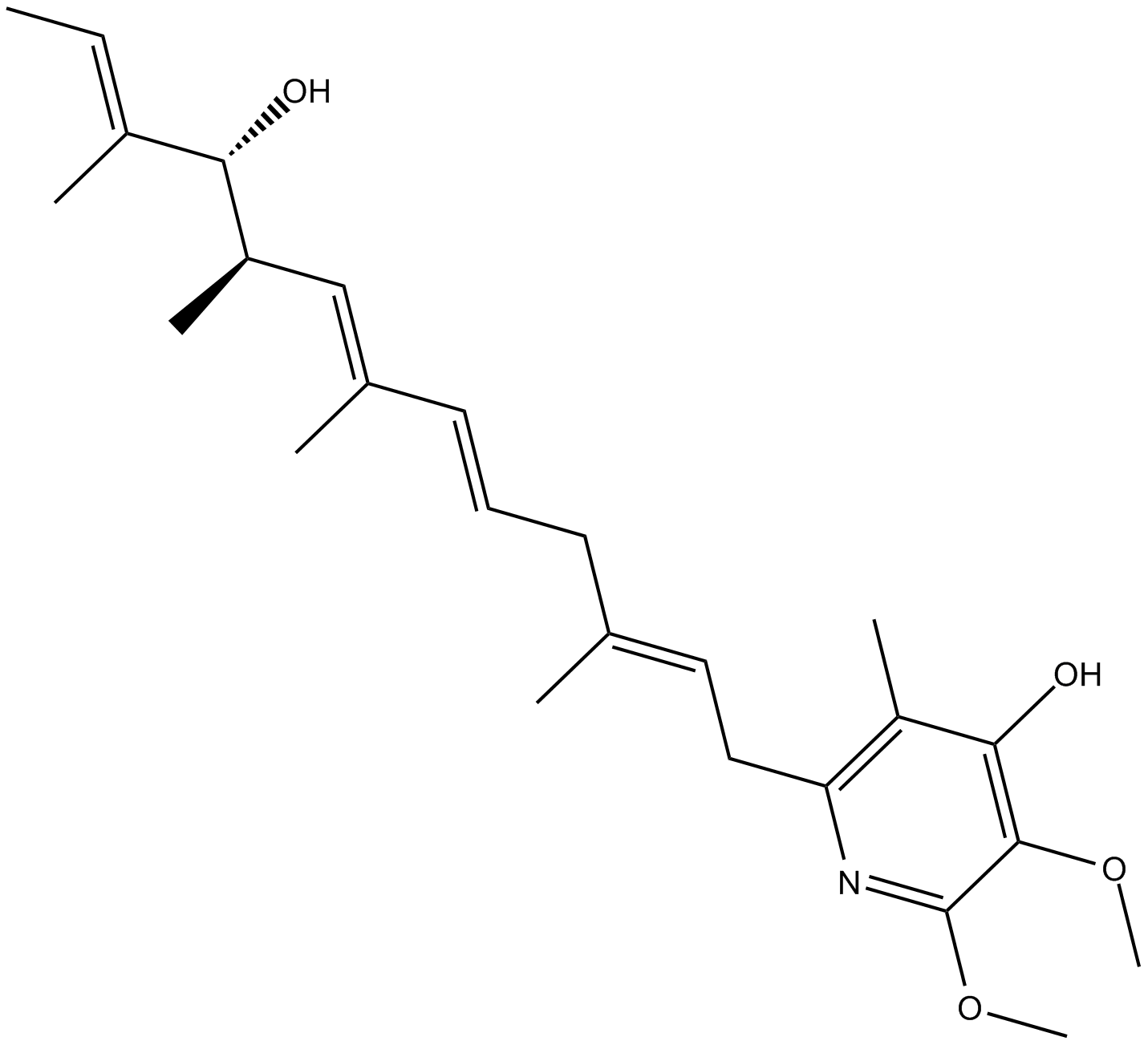
-
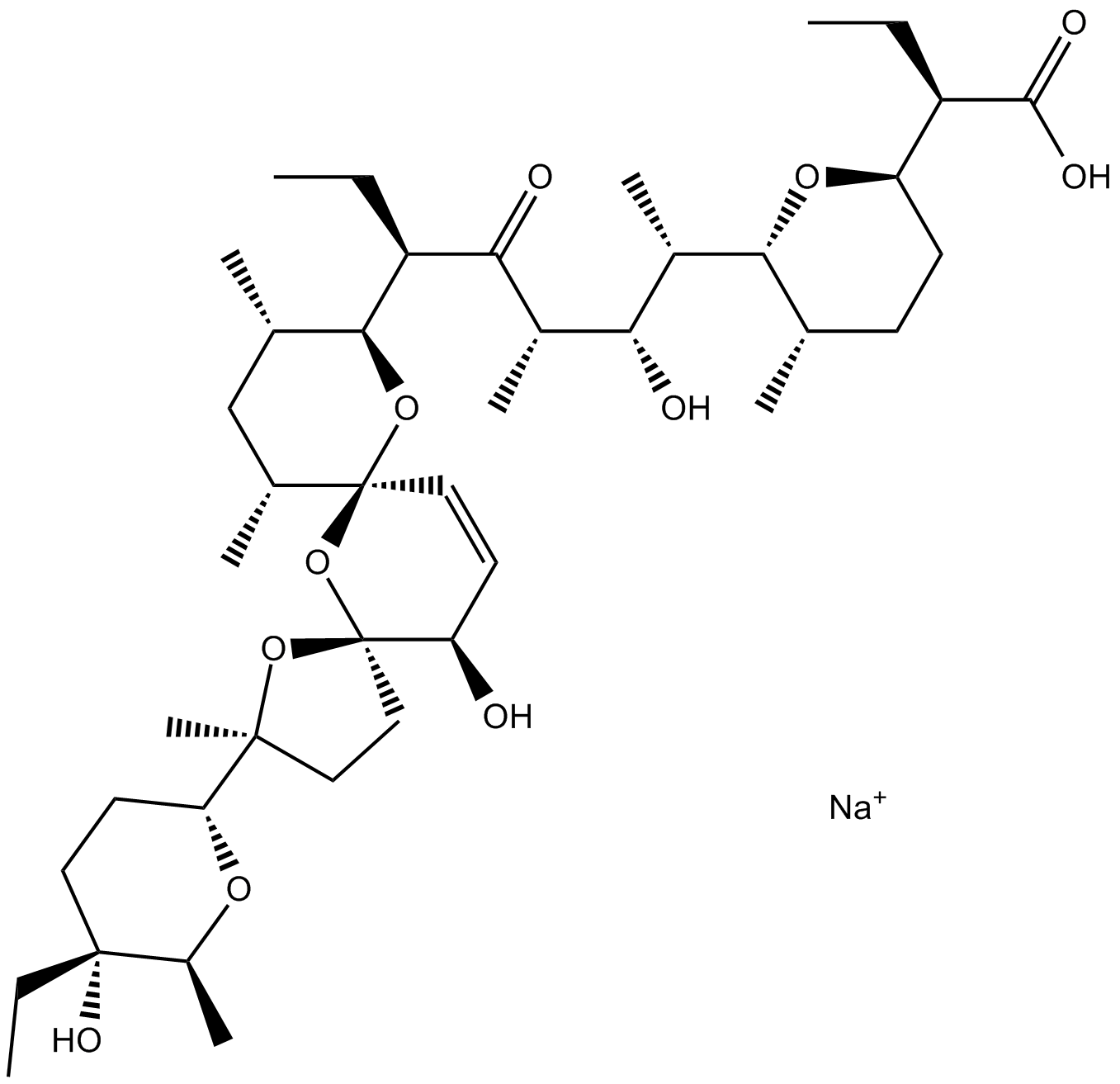
GC18107
Salinomycin sodium salt
Salinomycin sodium salt is a macrocyclic polyether ionophore antibiotic produced by the fermentation of Streptomyces albus.
-
GC32774
Temoporfin (m-THPC)
Foscan, KW 2345, m-THPC
A light-activated chlorin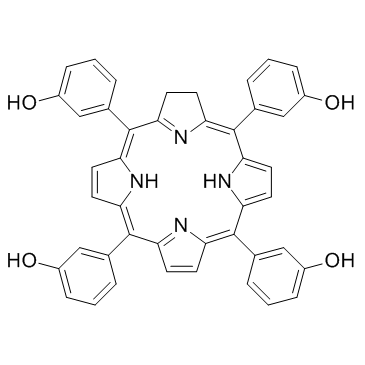
-
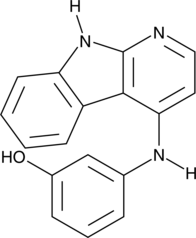
GC40927
Tilfrinib
Tilfrinib is a selective Brk/PTK6 inhibitor with an IC₅₀ value of 3.15nM for Brk.
-
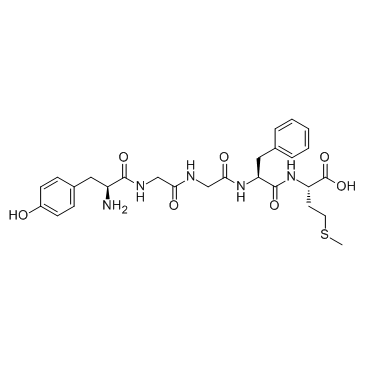
GC30816
Tyr-Gly-Gly-Phe-Met-OH (Met-Enkephalin)
Tyr-Gly-Gly-Phe-Met-OH (Met-Enkephalin) regulates human immune function and inhibits tumor growth via binding to the opioid receptor.
-
GC17203
Varespladib (LY315920)
Varespladib
Varespladib (LY315920) is a potent and selective inhibitor of group IIA, nonpancreatic secretory PLA2 (sPLA2) with an IC50 value of 9nM.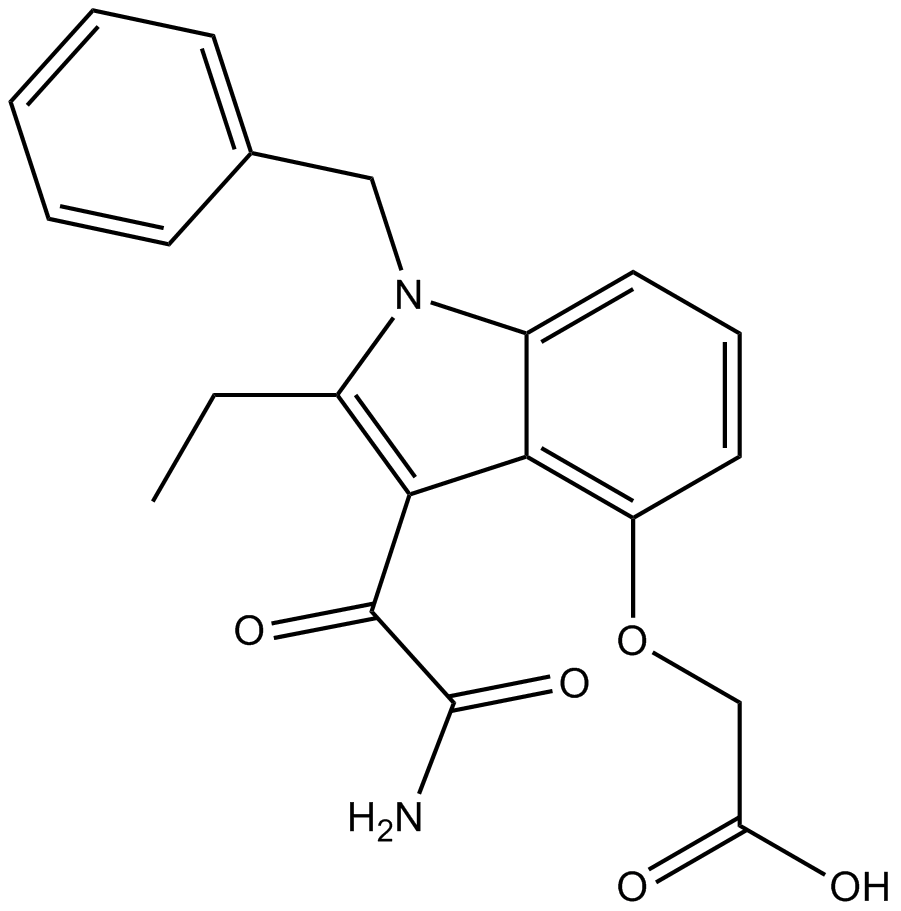
-
GC10911
Venlafaxine hydrochloride
Wy 45030 hydrochloride
Dual serotonin/noradrenalin re-uptake inhibitor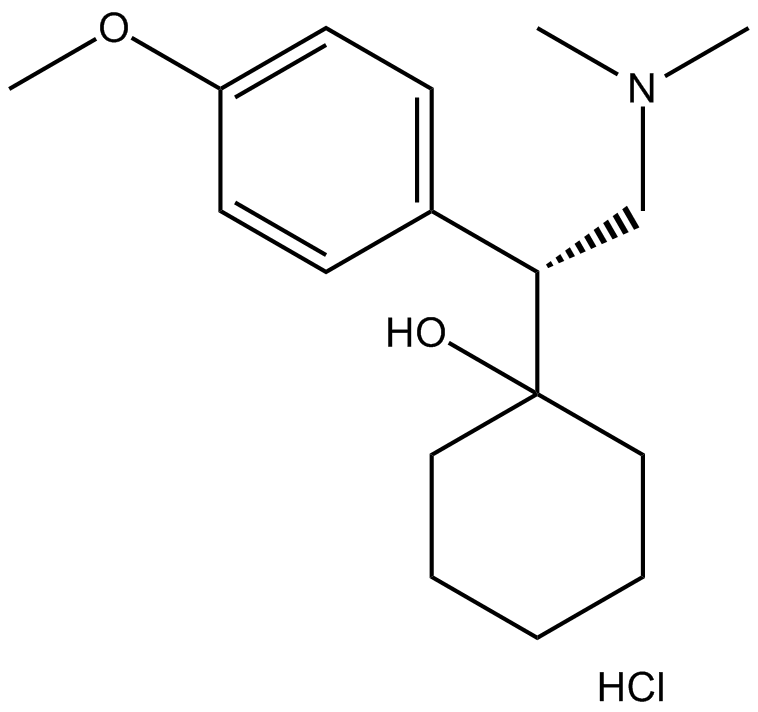
-
GC14567
Simplyblue Coomassie G-250 Staining Solution
Acid Blue 90,CBBG,Coomassie Brilliant Blue G-250,NSC 328382
fast, sensitive, and safe Coomassie G-250 staining of proteins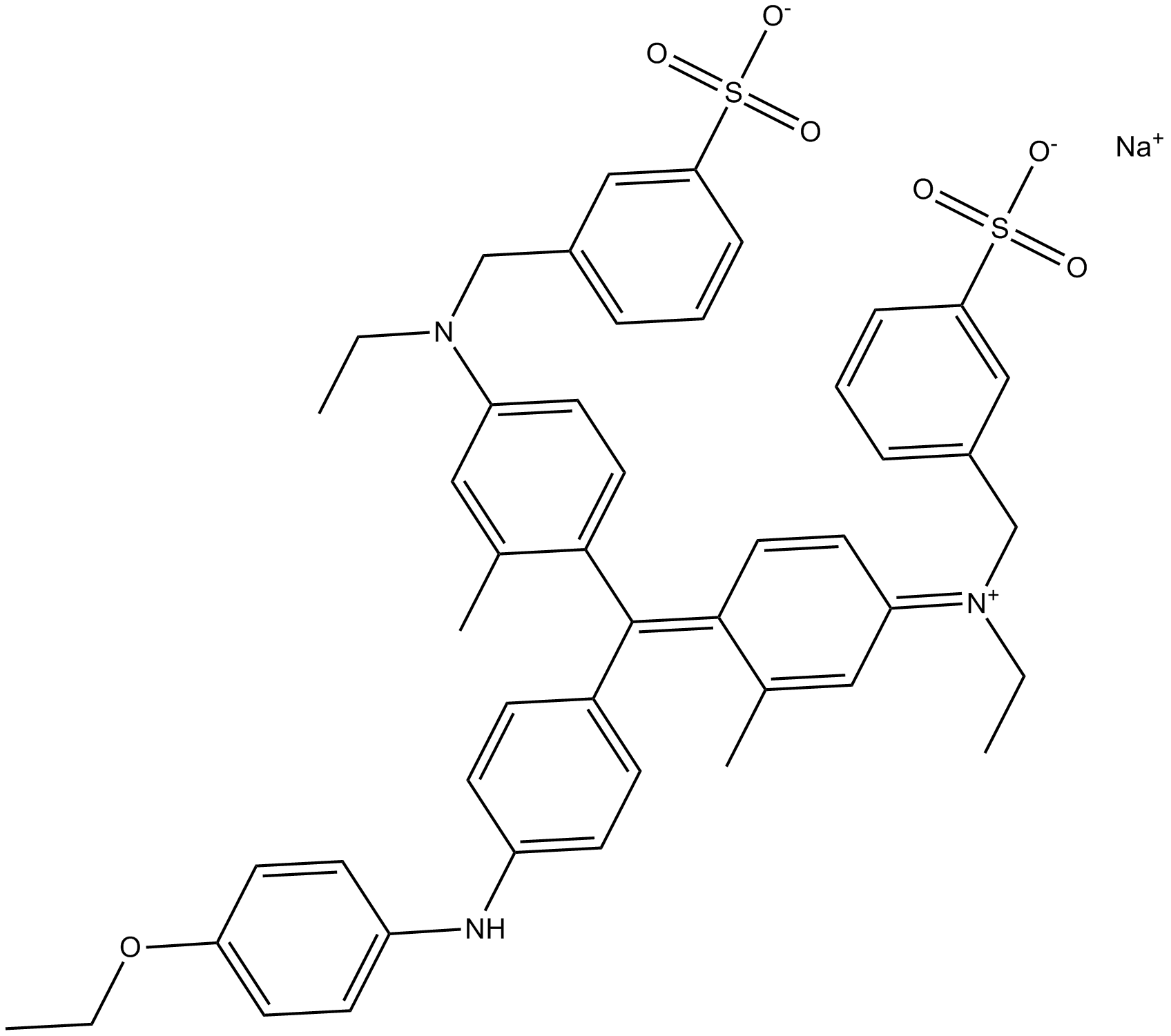
- GlpBio - Master of Small Molecules | Compounds - Peptides - Kits.
- Login | Register
- sales@glpbio.com
- (909) 407-4943
-
LanguageEN - English
-
Research Areas
-
Signaling Pathways
- Proteases
- Apoptosis
- Chromatin/Epigenetics
- Metabolism
- MAPK Signaling
- Tyrosine Kinase
- DNA Damage/DNA Repair
- PI3K/Akt/mTOR Signaling
- Microbiology & Virology
- Cell Cycle/Checkpoint
- Ubiquitination/ Proteasome
- JAK/STAT Signaling
- TGF-β / Smad Signaling
- Angiogenesis
- GPCR/G protein
- Stem Cell
- Cancer Biology
- Endocrinology and Hormones
- Immunology/Inflammation
- Neuroscience
- Membrane Transporter/Ion Channel
- Other Signal Transduction
- Product Type
-
Signaling Pathways
- Screening Library
- Kits
- Tools
- Reporter mRNA
- mRNA Customize
- Contact us
- Blog

