Fluorescent Probes
Products for Fluorescent Probes
- Cat.No. Product Name Information
-
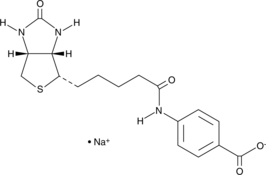
GC45258
(+)-Biotin 4-Amidobenzoic Acid (sodium salt)
(+)-Biotin 4-amidobenzoic acid is a substrate of biotinidase, which cleaves biotin amide to give biotin in vivo.
-
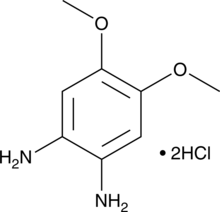
GC41784
1,2-Diamino-4,5-dimethoxybenzene (hydrochloride)
1,2-Diamino-4,5-dimethoxybenzene is an aromatic diamine that reacts with aldehydes to produce highly fluorescent benzimidazole derivatives with excitation/emission spectra of 361/448 nm.
-
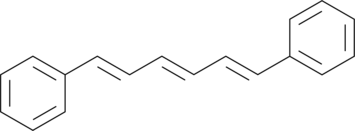
GC45306
1,6-Diphenyl-1,3,5-hexatriene
1,6-Diphenyl-1,3,5-hexatriene is a fluorescent probe.
-
GC41860
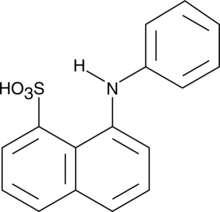
1,8-ANS
1,8-ANS is a fluorescent dye that binds with high affinity to hydrophobic surfaces of proteins.
-
GC41861
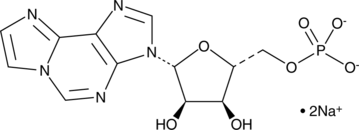
1,N6-Ethenoadenosine 5'-monophosphate (sodium salt)
1,N6-Ethenoadenosine 5'-monophosphate (1,N6-Etheno-AMP) sodium is a highly fluorescent analog of adenosine 5'-monophosphate (AMP).
-
GC45317
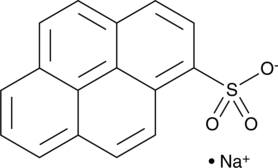
1-Pyrenesulfonic Acid (sodium salt)
-
GC41866
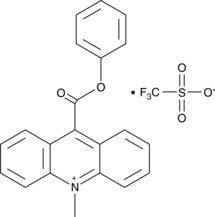
10-methyl-9-(phenoxycarbonyl) Acridinium (trifluoromethylsulfonate)
10-methyl-9-(phenoxycarbonyl) Acridinium is an acridinium ester that produces fluorescent 10-methyl-9-acridone upon oxidation with hydrogen peroxide, persulfates, and other oxidants in alkaline conditions.
-
GC42079
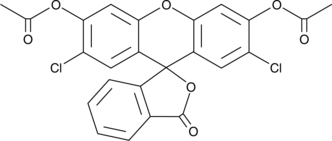
2',7'-Dichlorofluorescein diacetate
2',7'-Dichlorofluorescein diacetate is as a cell-permeable fluorogenic probe to quantify reactive oxygen species (ROS) and nitric oxide (NO).
-
GC42239
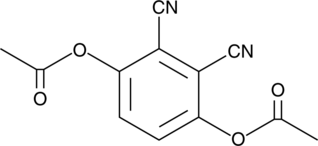
3,6-diacetoxy Phthalonitrile
3,6-diacetoxy Phthalonitrile is a cell-permeable fluorescent probe.
-
GC45348
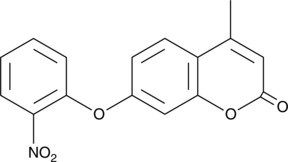
4-Methyl-7-(2-nitrophenoxy)-2H-chromen-2-one
-
GC45718
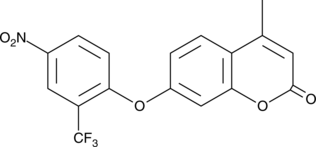
4-Methyl-7-(4-nitro-2-(trifluoromethyl)phenoxy)-2H-chromen-2-one
A fluorescent H2S probe
-
GC45349
4-Methyl-7-(4-nitrophenoxy)-2H-chromen-2-one
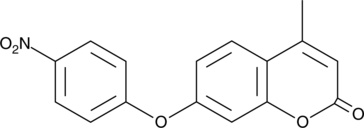
-
GC42445
4-Methylumbelliferyl β-D-Galactopyranoside-6-sulfate (sodium salt)
4-Methylumbelliferyl β-D-Galactopyranoside-6-sulfate (sodium salt) is a fluorescent dye.
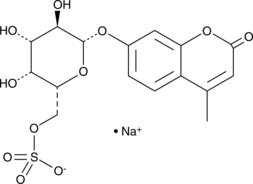
-
GC42446
4-Methylumbelliferyl β-D-N,N'-diacetylchitobioside
4-Methylumbelliferyl β-D-N,N'-diacetylchitobioside (4-μU-(GlcNAc)2) is a fluorogenic substrate for chitinases and chitobiosidases.
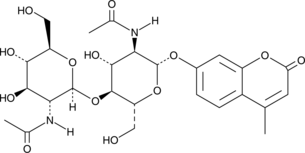
-
GC42441
4-Methylumbelliferyl 2-Acetamido-2-deoxy-α-D-galactopyranoside
4-Methylumbelliferyl 2-acetoamido-2-deoxy-α-D-galactopyranoside (4-MU-2-acetoamido-2-deoxy-α-Gal) is a fluorogenic substrate used to quantify α- and β-galactopyranosaminidase activity.
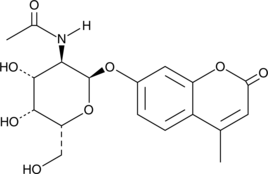
-
GC42442
4-Methylumbelliferyl 2-sulfamino-2-deoxy-α-D-Glucopyranoside (sodium salt)
4-Methylumbelliferyl 2-sulfamino-2-deoxy-α-D-Glucopyranoside (sodium salt) is a fluorogenic substrate of heparin sulphamidase, is desulfurized into 4-MU-α-GlcNH2.
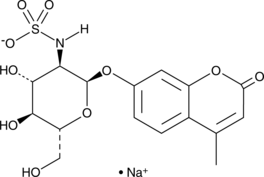
-
GC42443
4-Methylumbelliferyl 6-thio-Palmitate-β-D-Glucopyranoside
4-Methylumbelliferyl 6-thio-Palmitate-β-D-Glucopyranoside is a fluorogenic substrate for palmitoyl-protein thioesterase (PPT).
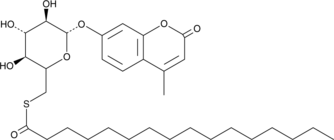
-
GC41299
4-Methylumbelliferyl Caprylate
4-Methylumbelliferyl caprylate (MUCAP) is a fluorogenic substrate for C8 esterase.
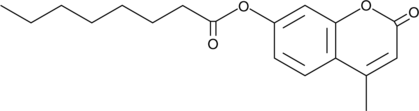
-
GC42444
4-Methylumbelliferyl Oleate
4-Methylumbelliferyl (4-MU) oleate is a fluorogenic substrate for acid and alkaline lipases.
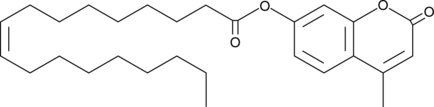
-
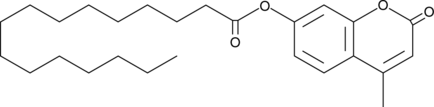
GC40904
4-Methylumbelliferyl Palmitate
Cholesterol ester storage disease and Wolman disease are recessive autosomal disorders caused by a deficiency in lysosomal acid lipase (LAL), also known as cholesteryl ester hydrolase.
-
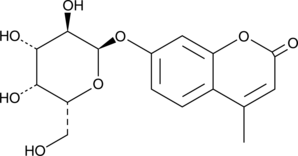
GC40510
4-Methylumbelliferyl-α-D-Galactopyranoside
4-Methylumbelliferyl-α-D-galactopyranoside (4-MU-α-Gal) is a fluorogenic substrate of α-galactosidase.
-
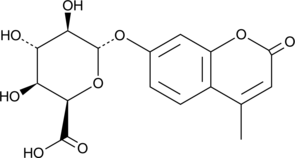
GC42449
4-Methylumbelliferyl-α-L-Iduronide (free acid)
4-Methylumbelliferyl-α-L-iduronide (free acid) is a fluorogenic substrate for α-L-iduronidase, an enzyme found in cell lysosomes that is involved in the degradation of glycosaminoglycans such as dermatan sulfate and heparin sulfate.
-
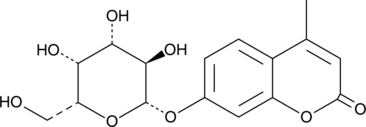
GC49114
4-Methylumbelliferyl-β-D-Galactoside
4-Methylumbelliferyl-β-D-Galactoside is a fluorescent substrate for β-galactosidase which, when cleaved, produces a water-soluble blue fluorescent coumarin fluorophore that can be detected using a fluoroenzymeter or fluorometer.
-
GC42452
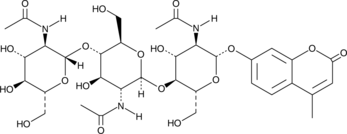
4-Methylumbelliferyl-β-D-N,N',N''-Triacetylchitotrioside
4-Methylumbelliferyl-β-D-N,N',N''-triacetylchitotrioside is a fluorogenic substrate for chitinases and chitotriosidases.
-
GC42447
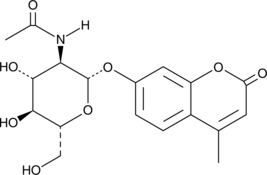
4-Methylumbelliferyl-2-acetamido-2-deoxy-β-D-Glucopyranoside
4-Methylumbelliferyl-2-acetamido-2-deoxy-β-D-glucopyranoside is a fluorogenic substrate for β-hexosaminidases.
-
GC42448
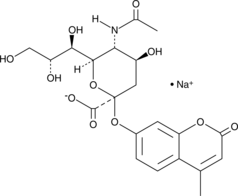
4-Methylumbelliferyl-N-acetyl-α-D-Neuraminic Acid (sodium salt)
4-Methylumbelliferyl-N-acetyl-α-D-Neuraminic Acid (sodium salt) is a fluorescent substrate used for neuraminidase activity assay.
-
GC11870
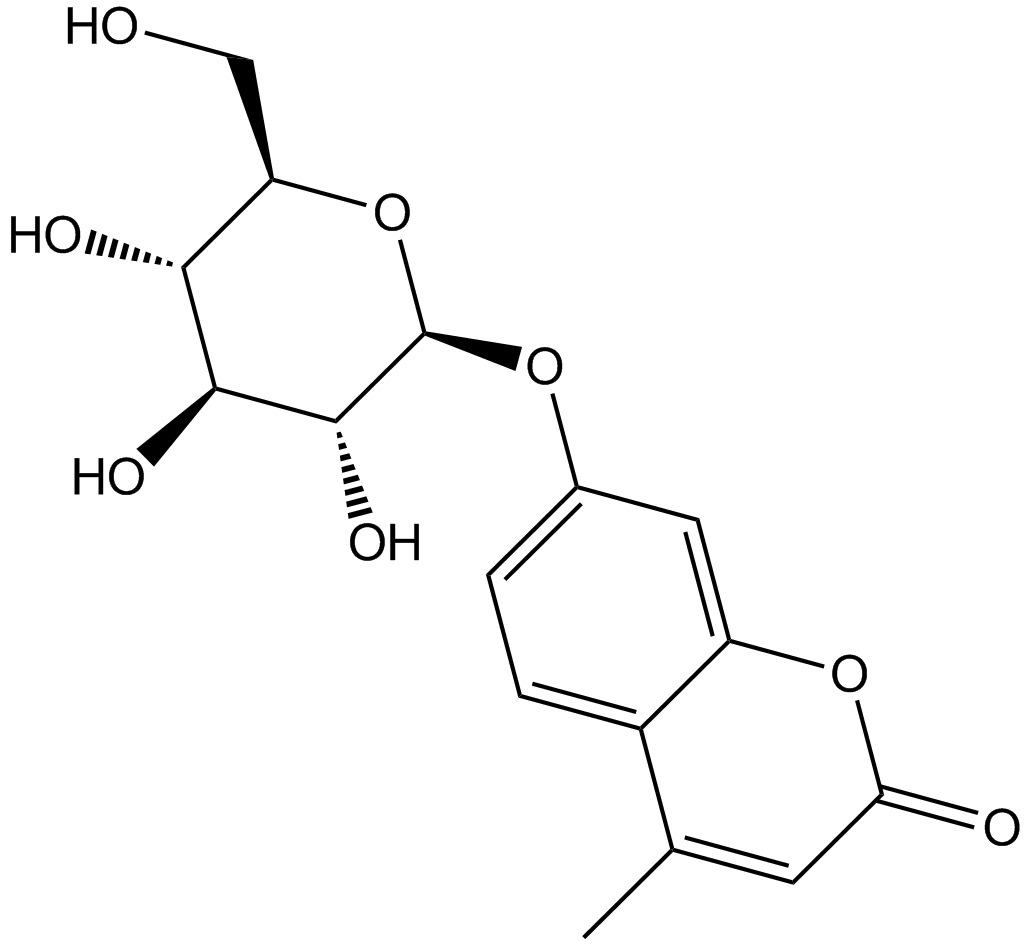
4-Methylumbelliferyl-β-D-Glucopyranoside
4-Methylumbelliferyl-β-D-Glucopyranoside (4-MUG) is a fluorogenic substrate of β-glucosidase and β-glucocerebrosidase (also known as glucosylceramidase).
-
GC49339
5(6)-Carboxy-2′,7′-dichlorofluorescein diacetate
An oxidant-sensitive fluorescent probe

-
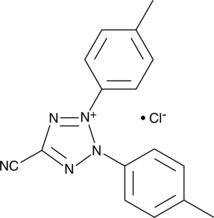
GC42502
5-Cyano-2,3-di-(p-tolyl)tetrazolium (chloride)
5-Cyano-2,3-di-(p-tolyl)tetrazolium (chloride) is a redox-sensitive tetrazolium salt used primarily to detect metabolic activity in microorganisms.
-
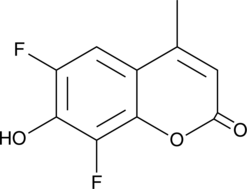
GC49504
6,8-Difluoro-7-hydroxy-4-methylcoumarin
A fluorophore
-
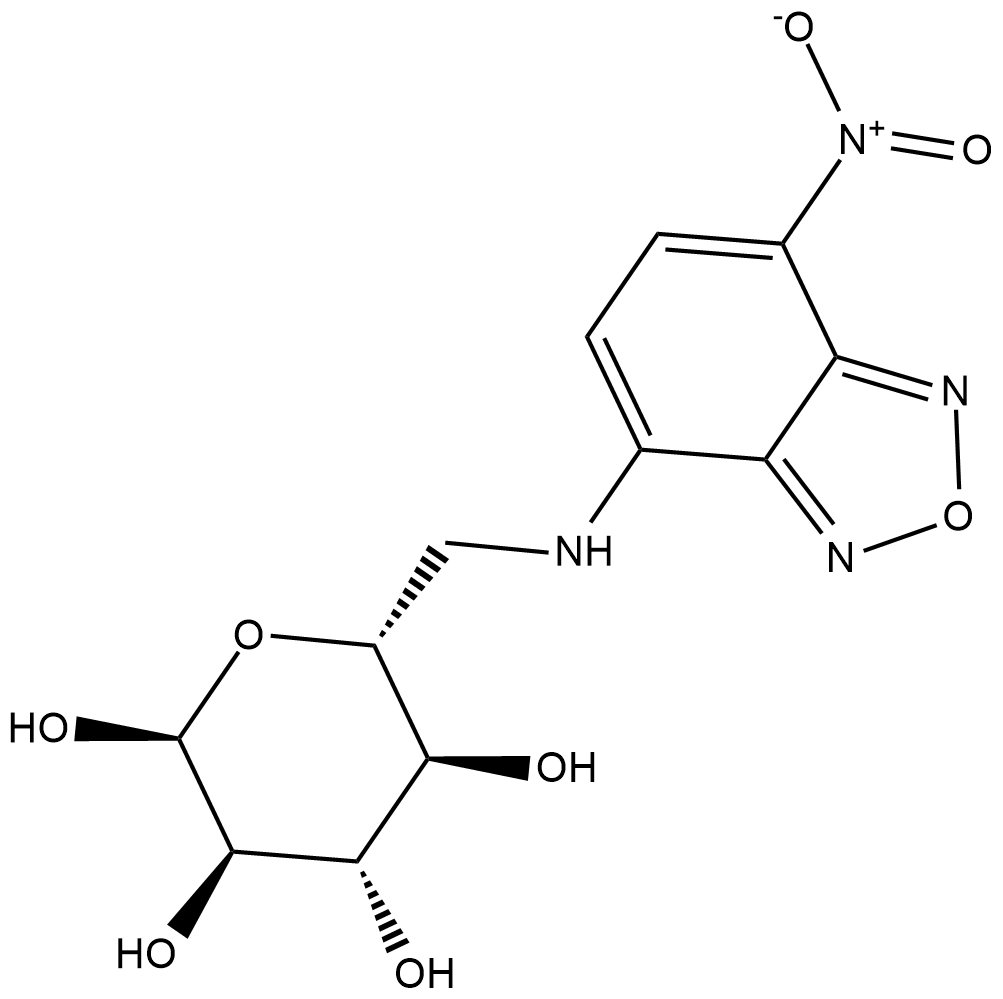
GC18870
6-NBDG
6-NBDG is a non-hydrolyzable fluorescent glucose analog that is used to monitor glucose uptake and transport in living cells.
-
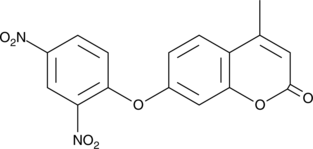
GC45363
7-(2,4-Dinitrophenoxy)-4-methyl-2H-chromen-2-one
-
GC45986
7-Amino-4-methyl-3-coumarinylacetic Acid
7-Amino-4-methyl-3-coumarinylacetic Acid is a fluorescent protein labelling agent.
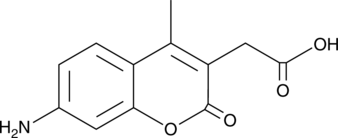
-
GC45707
7-Azido-4-methylcoumarin
A fluorescent H2S probe
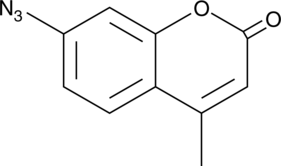
-
GC40882
7-Diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin
7-Diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin is a maleimide derivative, acting as a blue fluorescent thiol-reactive dye.
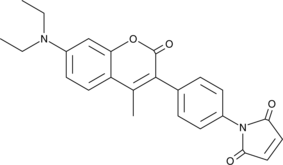
-
GC49320
7-Ethoxy-4-(trifluoromethyl)coumarin
A fluorogenic substrate for CYPs
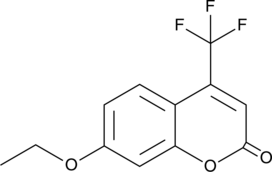
-
GC49312
7-Fluoro-2,1,3-benzoxadiazole-4-sulfonate (ammonium salt)
7-Fluoro-2,1,3-benzoxadiazole-4-sulfonate (ammonium salt) is a fluorescent label.
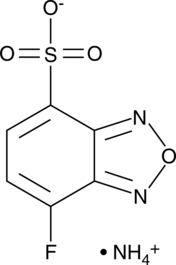
-
GC49248
7-Methoxy-4-(trifluoromethyl)coumarin
A fluorogenic substrate for CYPs
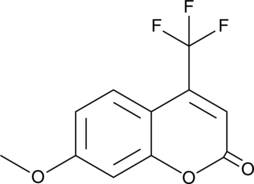
-
GC42620
8-Aminopyrene-1,3,6-trisulfonic Acid (sodium salt)
8-Aminopyrene-1,3,6-trisulfonic Acid (sodium salt) is a water-soluble anionic fluorescent dye.
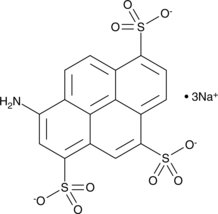
-
GC40844
9-(2,2-Dicyanovinyl)julolidine
9-(2,2-Dicyanovinyl)julolidine (9-(2,2-Dicyanovinyl)julolidine), a molecular rotor and unique fluorescent dye, binds to tubulin and actin, and increases its fluorescence intensity drastically upon polymerization.
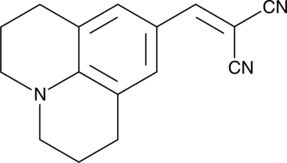
-
GC42643
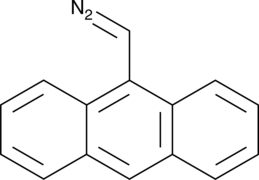
9-Anthryldiazomethane
9-Anthryldiazomethane is a fluorescent probe that has been used for the detection of fatty acids, arachidonic acid metabolites, prostaglandins, leukotrienes, and thromboxanes.
-
GC42683
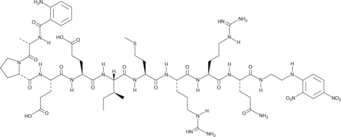
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp is a fluorescence-quenched peptide substrate for human neutrophil elastase (kcat/Km = 531 mM-1s-1).
-
GC42684
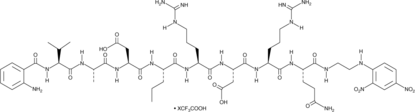
Abz-Val-Ala-Asp-Nva-Arg-Asp-Arg-Gln-EDDnp (trifluoroacetate salt)
Abz-Val-Ala-Asp-Nva-Arg-Asp-Arg-Gln-EDDnp is a fluorescence-quenched peptide substrate for human proteinase 3 (kcat/Km = 1,570 mM-1s-1).
-
GC42685
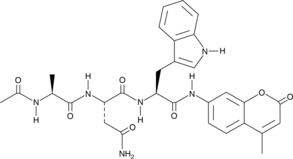
Ac-ANW-AMC
Ac-ANW-AMC is a fluorogenic substrate for the β5i/LMP7 subunit of the 20S immunoproteasome.
-
GC42689
Ac-DNLD-AMC
Ac-WLA-AMC is a fluorogenic substrate of caspase-3.
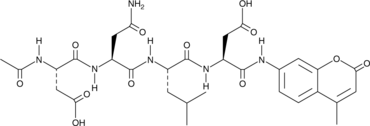
-
GC11094
Ac-IETD-AFC
A fluorogenic substrate for caspase-8
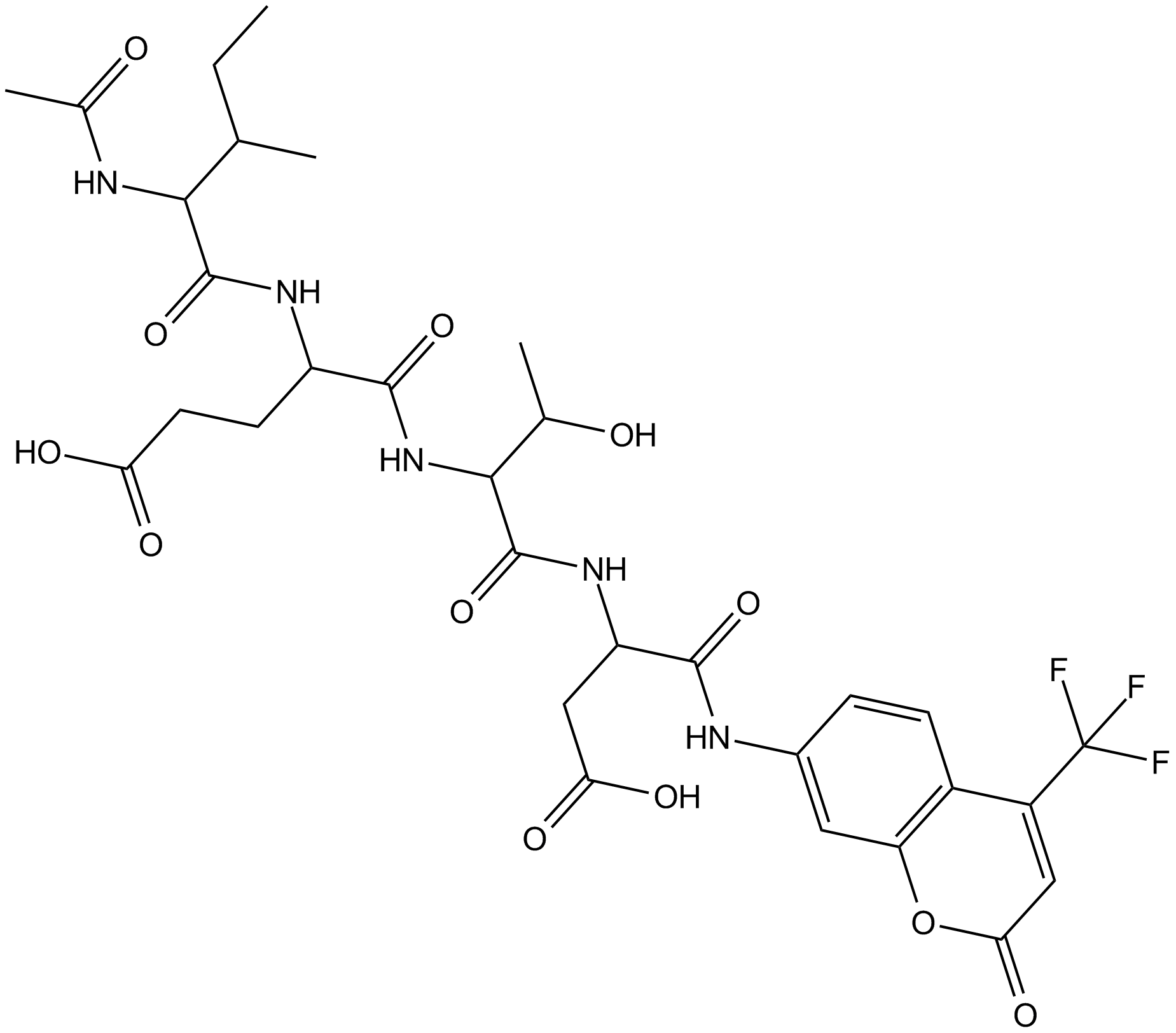
-
GC49725
Ac-IETD-AFC (trifluoroacetate salt)
A fluorogenic substrate for caspase-8
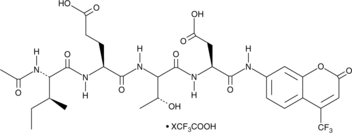
-
GC18226
Ac-LEHD-AMC (trifluoroacetate salt)
Ac-LEHD-AMC (trifluoroacetate salt) is a fluorogenic substrate for caspase-9 (Excitation: 341 nm; Emission: 441 nm).
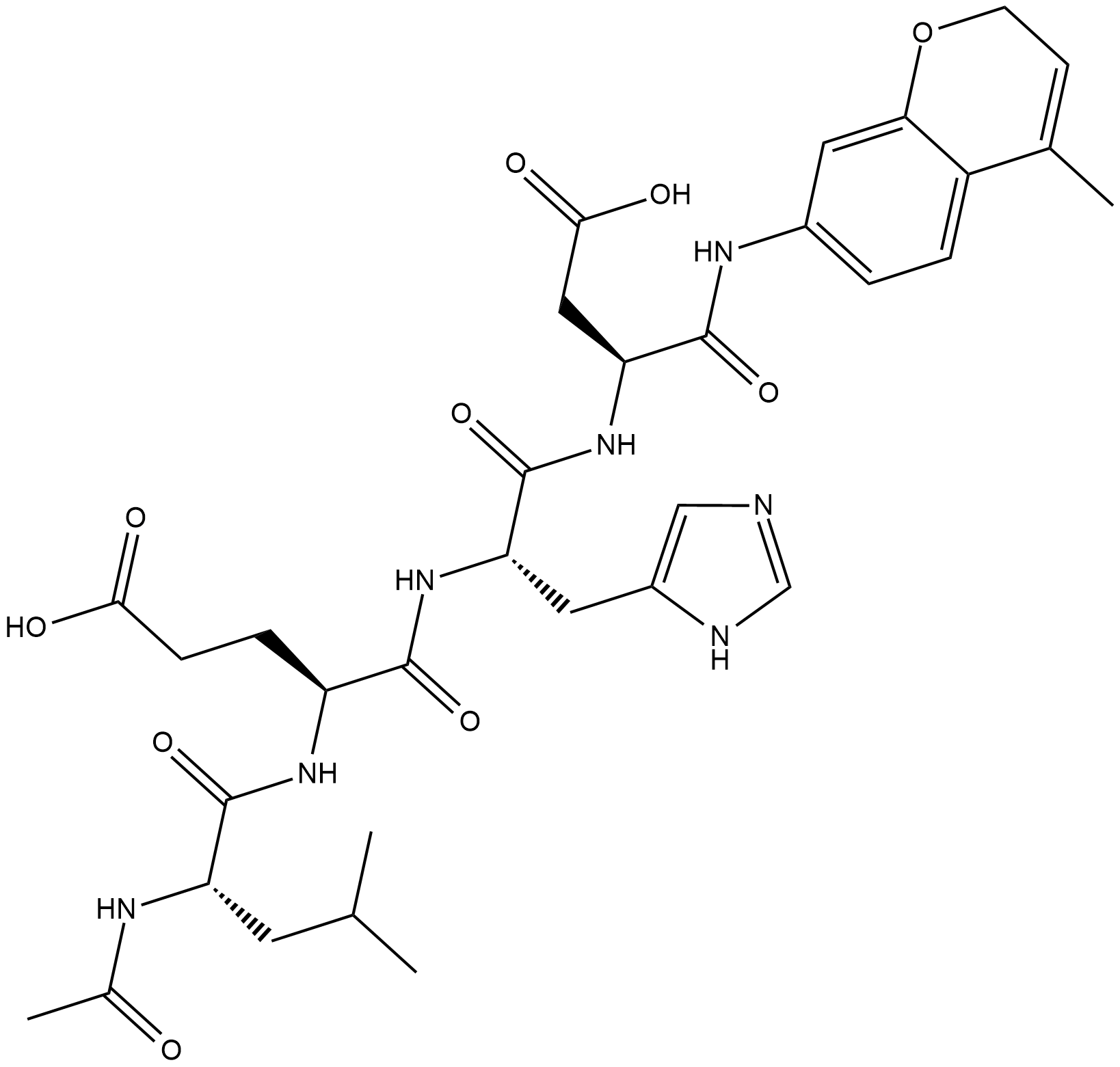
-
GC40556
Ac-LETD-AFC
Ac-LETD-AFC is a fluorogenic substrate that can be cleaved specifically by caspase-8.
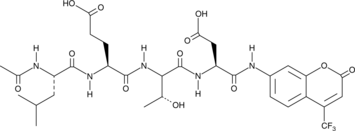
-
GC15171
Ac-LEVD-AFC
A fluorogenic substrate for caspase-4
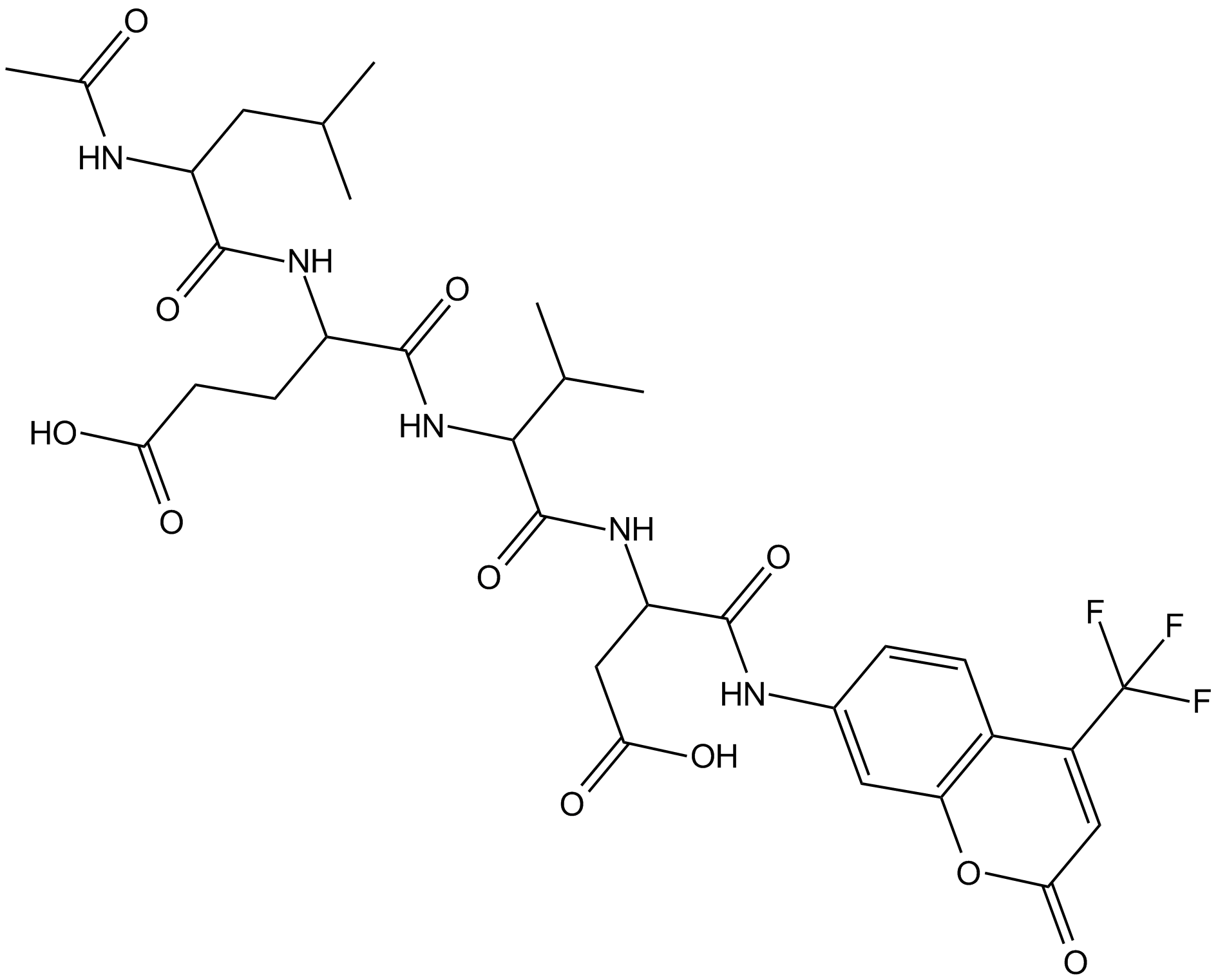
-
GC48382
Ac-QPKK(Ac)-AMC
A fluorogenic substrate for SIRT1, SIRT2, and SIRT3

-
GC13400
Ac-VDVAD-AFC
Ac-VDVAD-AFC is a caspase-specific fluorescent substrate. Ac-VDVAD-AFC can measure caspase-3-like activity and caspase-2 activity and can be used for the research of tumor and cancer.
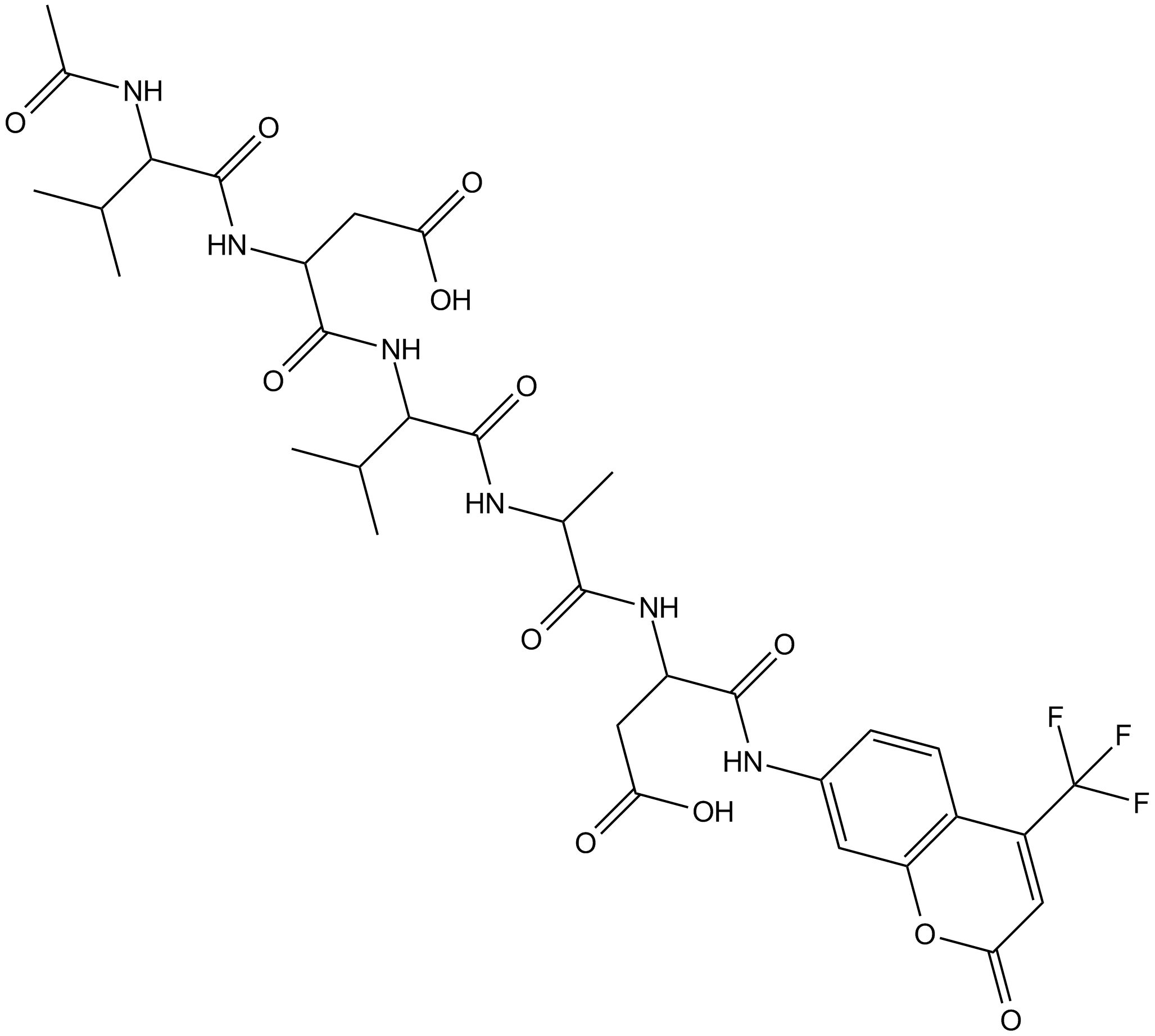
-
GC48974
Ac-VEID-AMC (ammonium acetate salt)
A caspase-6 fluorogenic substrate
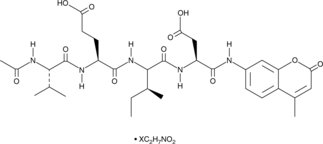
-
GC42720
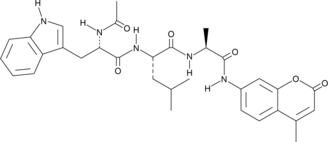
Ac-WLA-AMC
Ac-WLA-AMC is a fluorogenic substrate for the β5c subunit of the 20S proteasome.
-
GC13326
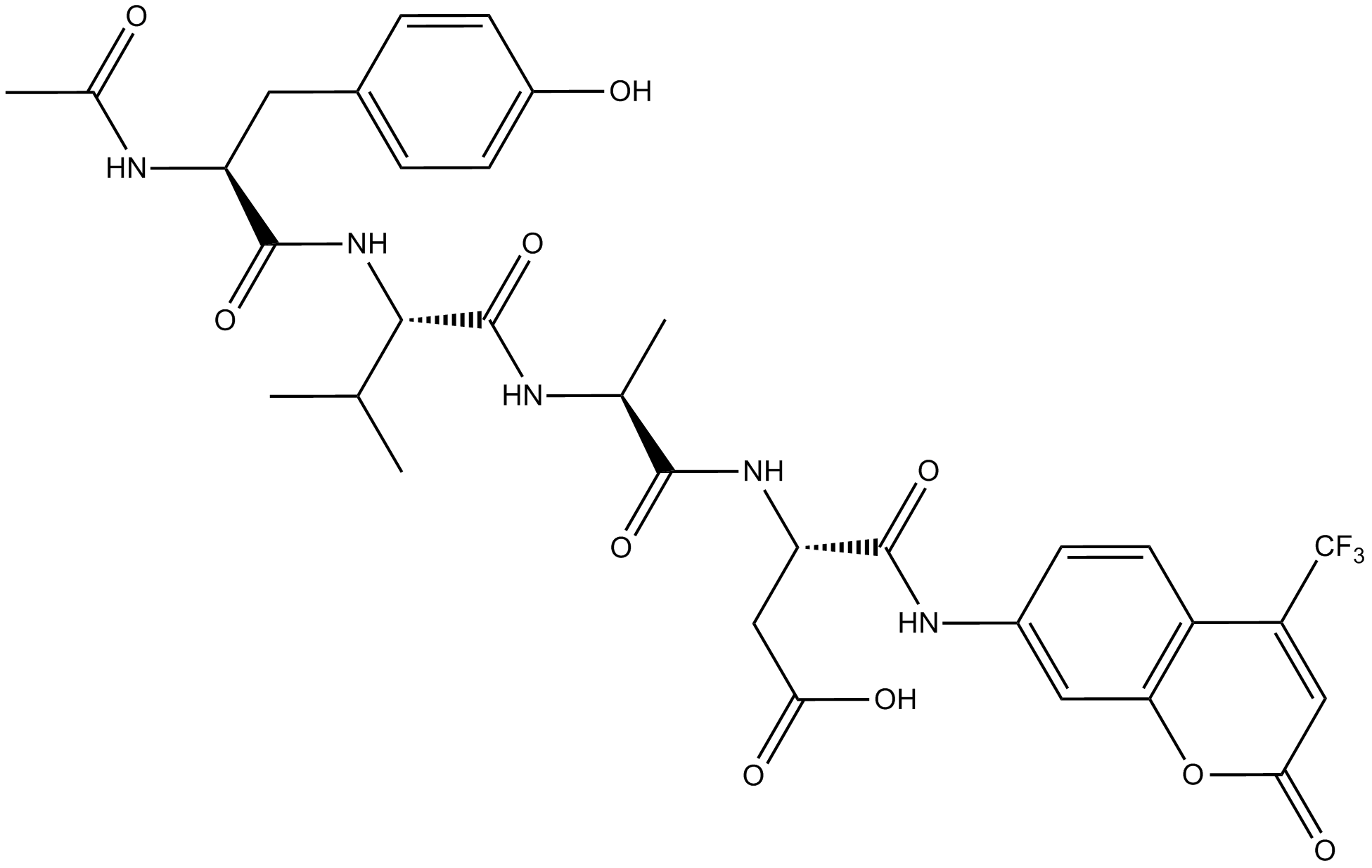
Ac-YVAD-AFC
Fluorogenic peptide substrate for caspase-1
-
GC41242
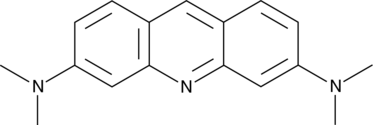
Acridine Orange
Acridine Orange is a cell-permeable fluorescent dye that stains organisms (bacteria, parasites, viruses, etc.
-
GC49331
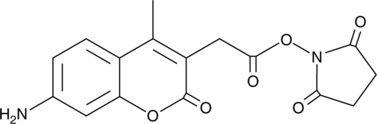
AMCA N-succinimidyl ester
AMCA N-succinimidyl ester is a bioReagent, suitable for fluorescence, can be used for amine-reactive labeling.
-
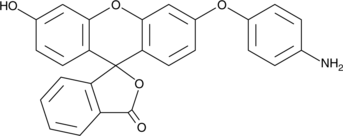
GC42825
APF
APF is an aromatic amino-fluorescein derivative and fluorescent probe for highly reactive radicals.
-
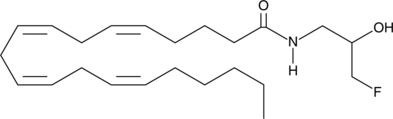
GC42853
ARN1203
Arachidonoyl ethanolamide, also known as anandamide, is an endogenous ligand of the cannabinoid receptors.
-
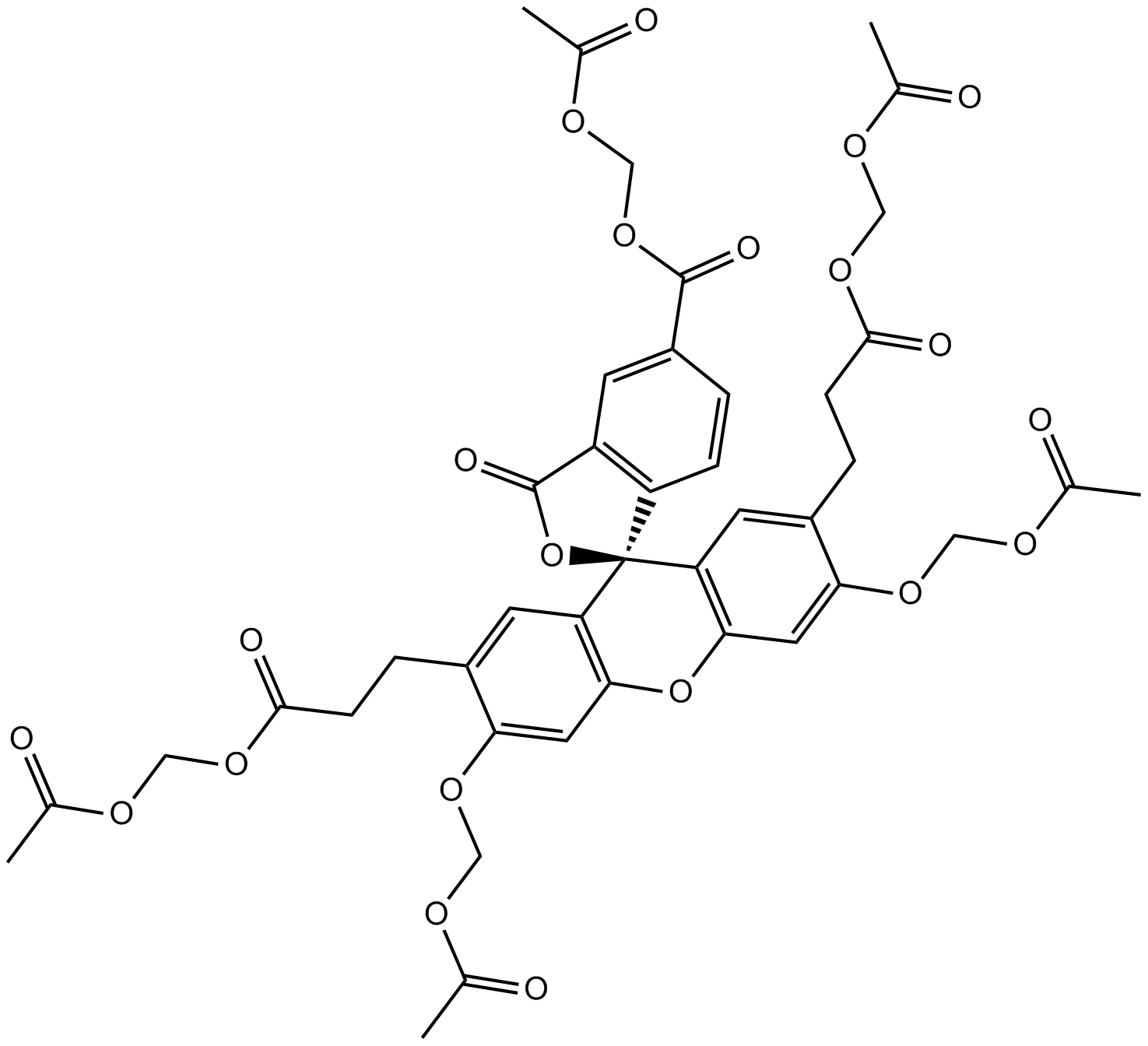
GC14603
BCECF-AM
Intracellular ratiometric pH indicator
-
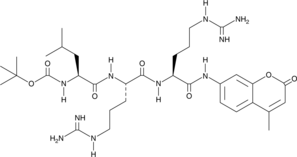
GC42958
Boc-LRR-AMC
Boc-LRR-AMC is a fluorogenic substrate for the trypsin-like activity of the 26S proteasome or 20S proteolytic core.
-
GC11641
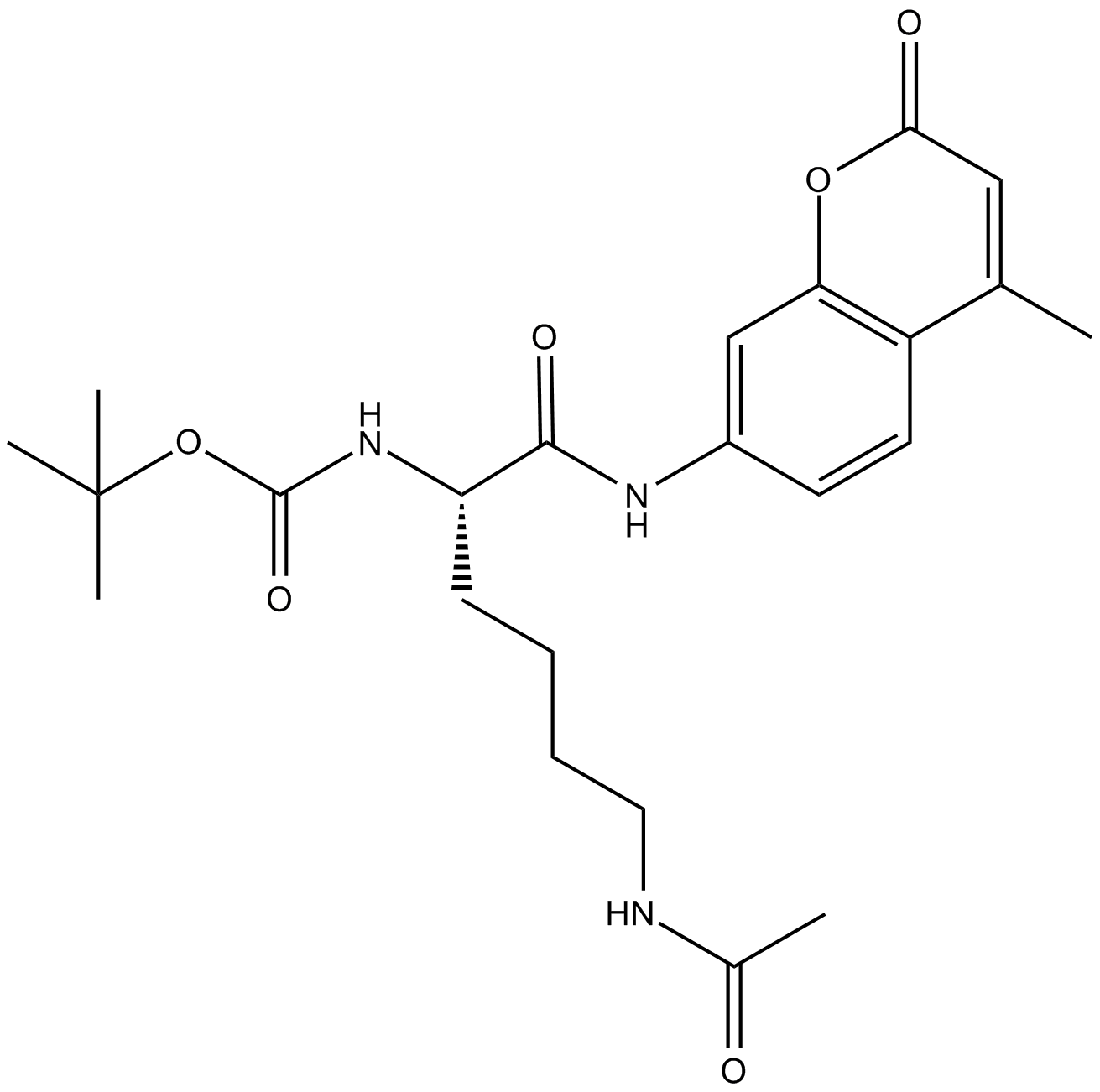
Boc-Lys(Ac)-AMC
GPCR G2A/GPR132 agonist
-
GC42959
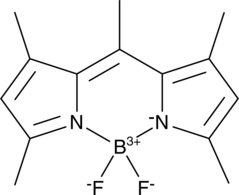
BODIPY 493/503
BODIPY 493/503, a lipophilic fluorescence dye, emits bright green fluorescence has been used extensively for lipid droplet labeling (Ex/Em: 493/503 nm).
-
GC45796
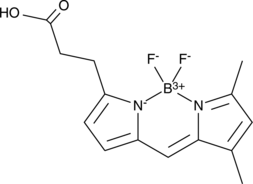
BODIPY 503/512
BODIPY 503/512 is a potent fluorescent dye.
-
GC42960
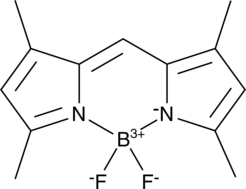
BODIPY 505/515
BODIPY 505/515, a lipophilic fluorescence dye, emits fluorescence has been used extensively for lipid droplet labeling (Ex/Em: 505/515 nm).
-
GC42961
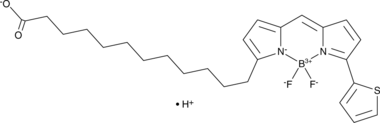
BODIPY 558/568 C12
BODIPY 558/568 C12 is a fatty acid-conjugated fluorescent probe for lipid droplets.
-
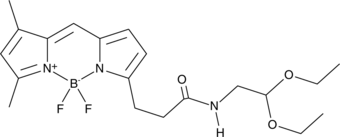
GC46939
BODIPY-aminoacetaldehyde diethyl acetal
A stable precursor to the fluorescent ALDH substrate BAAA
-
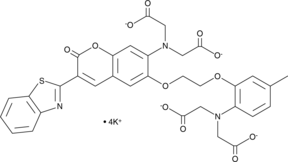
GC42984
BTC (potassium salt)
BTC (potassium salt) is a low affinity calcium indicator (Kd approximately 7-26 μM) featuring many desirable properties for cellular calcium imaging, including long excitation wavelengths (400/485 nm), low sensitivity to Mg2+, and accuracy of ratiometric measurement.
-
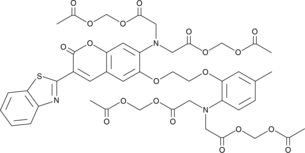
GC42985
BTC AM
BTC AM is a cell-permeable acetoxy-methyl ester of BTC.
-
GC43002
Bz-FVR-AMC
Bz-FVR-AMC is a fluorogenic peptide substrate for bovine cathepsins S and H, Xenopus endopeptidase XSCEP1, and esterase E-II from B.
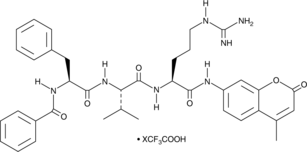
-
GA21221
Bz-Nle-Lys-Arg-Arg-AMC
Bz-Nle-Lys-Arg-Arg-AMC is a fluorogenic tetra-peptide substrate for yellow fever virus (YFV) non-structural 3 (NS3).

-
GC43095
C-6 NBD
C-6 NBD is a fluorescent probe for the study of fatty acids and sterols.
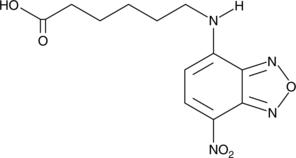
-
GC43021
C12FDG
C12FDG releases luciferin by enzymolysis of β-galactosidase, and the fluorescence of luciferin can be used to quantify the activity of β-galactosidase. The maximum excitation/emission of fluorescein is 485/530 nm.
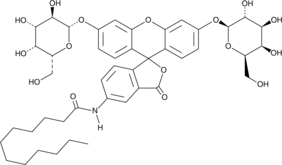
-
GC43115
Cal Green™ 1 (potassium salt)
Cal Green™ 1 is a cell-impermeant fluorescent calcium indicator that is characterized by high quantum yield and low phototoxicity.
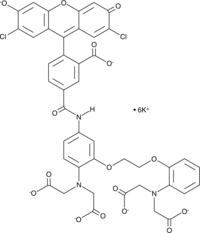
-
GC43116
Cal Green™ 1 AM
Cal Green™ 1 AM is a cell-permeant fluorescent calcium indicator (Excitation 506 nm; Emission 531 nm).
-
GC18604
Calcein Blue AM
Calcein Blue AM is a fluorogenic dye that is used to assess cell viability.
-
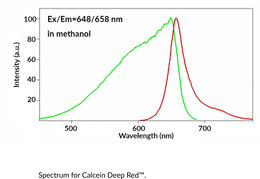
GC43118
Calcein Deep Red™
Calcein Deep Red? is a membrane-impermeant fluorescent dye.
-
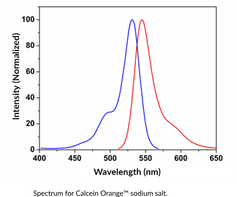
GC43120
Calcein Orange™ (sodium salt)
Calcein Orange is a membrane-impermeant fluorescent dye.
-
GC43121
Calcein Orange™ Diacetate
Calcein Orange Diacetate is a fluorogenic dye that is used to assess cell viability.

-
GC43122
Calcein Red™ (sodium salt)
Calcein Red? is a membrane-impermeant fluorescent dye.
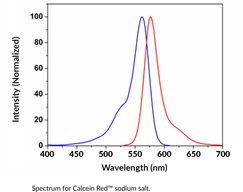
-
GC43123
Calcein Red™ AM
Calcein Red? AM is a fluorogenic dye that is used to assess cell viability.
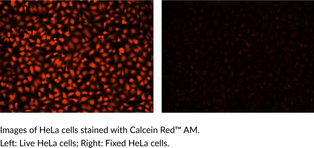
-
GC43124
Calcein UltraBlue™ (sodium salt)
Calcein UltraBlue? is a membrane-impermeant fluorescent dye.

-
GC43204
CAY10730
CAY10730 is a turn-on fluorescent probe for the detection of nitroreductase activity, a marker of hypoxia.
-
GC43205
CAY10731
CAY10731 is a fluorescent probe for hydrogen sulfide (H2S).
-
GC43206
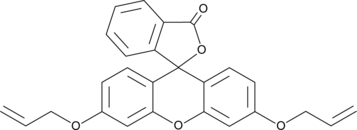
CAY10732
CAY10732 functions as a fluorescent agent designed to identify the presence of carbon monoxide.
-
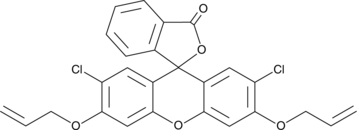
GC40030
CAY10733
CAY10733 is a fluorescent probe for the detection of carbon monoxide.
-
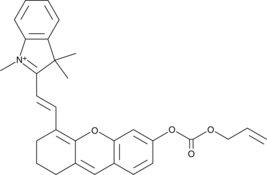
GC45405
CAY10737
-
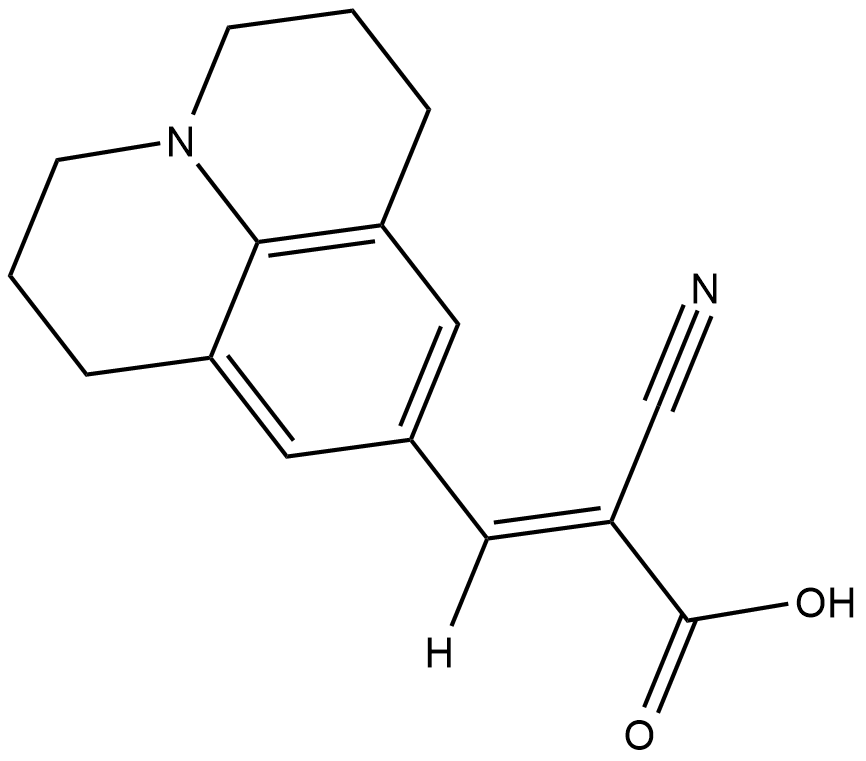
GC18179
CCVJ
CCVJ is a fluorescent molecular rotor.
-
GC43275
Citrulline-specific Probe-Rhodamine
Protein arginine deiminases (PADs) catalyze the posttranslational modification of arginine residues on proteins to form citrulline, which plays a large role in regulating gene expression.
-
GC43290
Coelenterazine 400a
Coelenterazine 400a (Bisdeoxycoelenterazine), a derivative of Coelenterazine, is a Renilla luciferase (RLuc) substrate.
-
GC49345
Coelenterazine hcp
A synthetic bioluminescent luciferin
-
GC49454
Complex 3
A fluorescent copper complex with anticancer activity
-
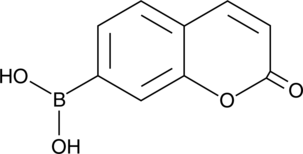
GC43316
Coumarin Boronic Acid
Coumarin boronic acid (CBA) is a fluorescent probe that can be used to detect peroxynitrite, hypochlorous acid, and hydrogen peroxide.
-
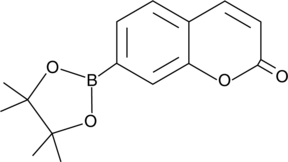
GC43317
Coumarin Boronic Acid pinacolate ester
Coumarin boronic acid pinacolate ester (CBE) is a more soluble form of coumarin boronic acid that can be used to detect peroxynitrite, hypochlorous acid, and hydrogen peroxide.
-
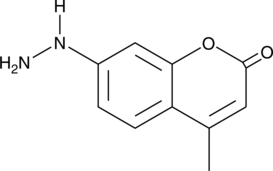
GC40950
Coumarin hydrazine
Coumarin hydrazine is an aromatic hydrazine-containing fluorophore that reacts with aldehydes or ketones for fluorescent labeling.
-
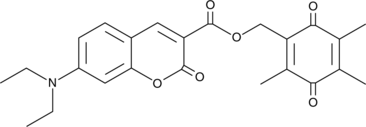
GC47123
Coumarin-Quinone Conjugate
A fluorescent substrate for NADH:ubiquinone oxidoreductases
-
GC43322
CRANAD 28
CRANAD 28 is a curcumin derivative and non-conjugated fluorescent affinity probe for the detection of amyloid-β (Aβ) in vitro and in vivo that has excitation/emission spectra of 498/578 nm, respectively.
-
GC47136
Cy5 (triethylamine salt)
A cyanine-containing fluorochrome
-
GC47137
Cy5-SE (triethylamine salt)
An amine-reactive fluorescent probe
-
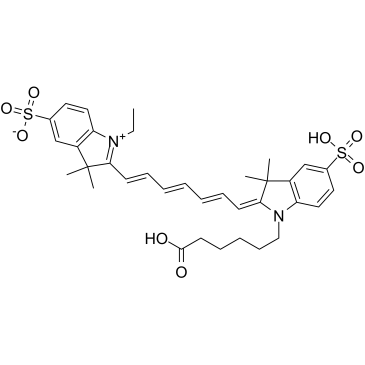
GC35772
CY7
Cy7 (Sulfo-Cyanine7) is a fluorescence labeling agent (Ex=750 nm, Em=773 nm).