HSP
Heat shock proteins (HSPs) are a family of highly conserved proteins that are expressed, either constitutively or regulated inductively, in response to a wide range of physiological and environmental insults, such as heat shock. According to molecular weight, mammalian HSPs are classified into four subfamilies, including HSP90 (90 kDa), HSP70 (70 kDa), HSP60 (60 kDa) and small HSPs (15 to 30 kDa), among which HSP90 and HSP70 are two of the most studied stress-inducible HSPs. HSP90 is a highly abundant chaperone protein characterized by containing three relevant domains, including the N-terminal domain, the charged middle linker region and the C-terminal dimerization domain; while HSP70 is an ATP-dependent chaperone protein characterized by containing two distinct functional domains, including a peptide binding domain (PBD) and the amino-terminal ATPase domain (ABD). Collectively HSPs play a fundamental role in maintaining the stability of other cellular proteins.
Products for HSP
- Cat.No. Product Name Information
-
GC63796
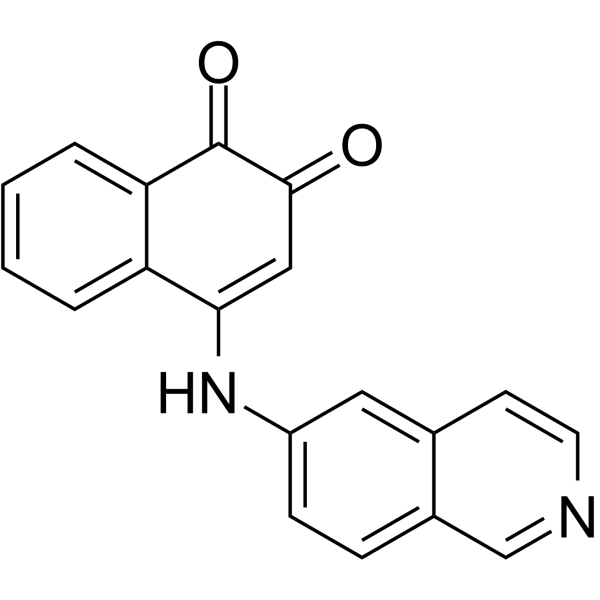
116-9e
116-9e (MAL2-11B) is a Hsp70 co-chaperone DNAJA1 inhibitor.
-
GC11720
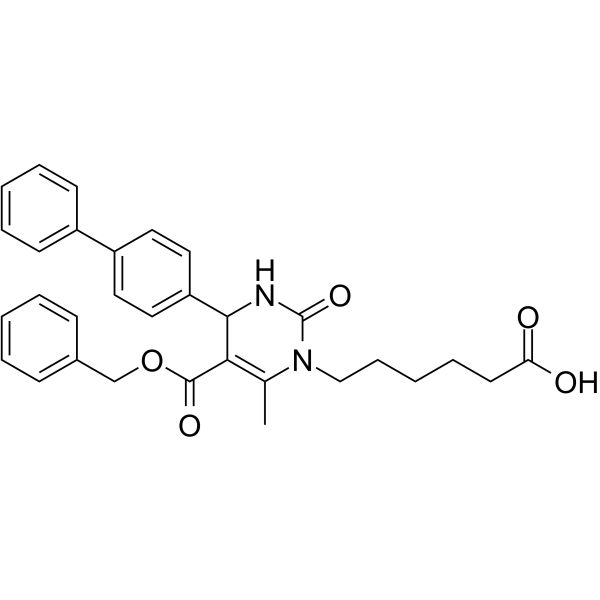
17-AAG (KOS953)
An inhibitor of Hsp90
-
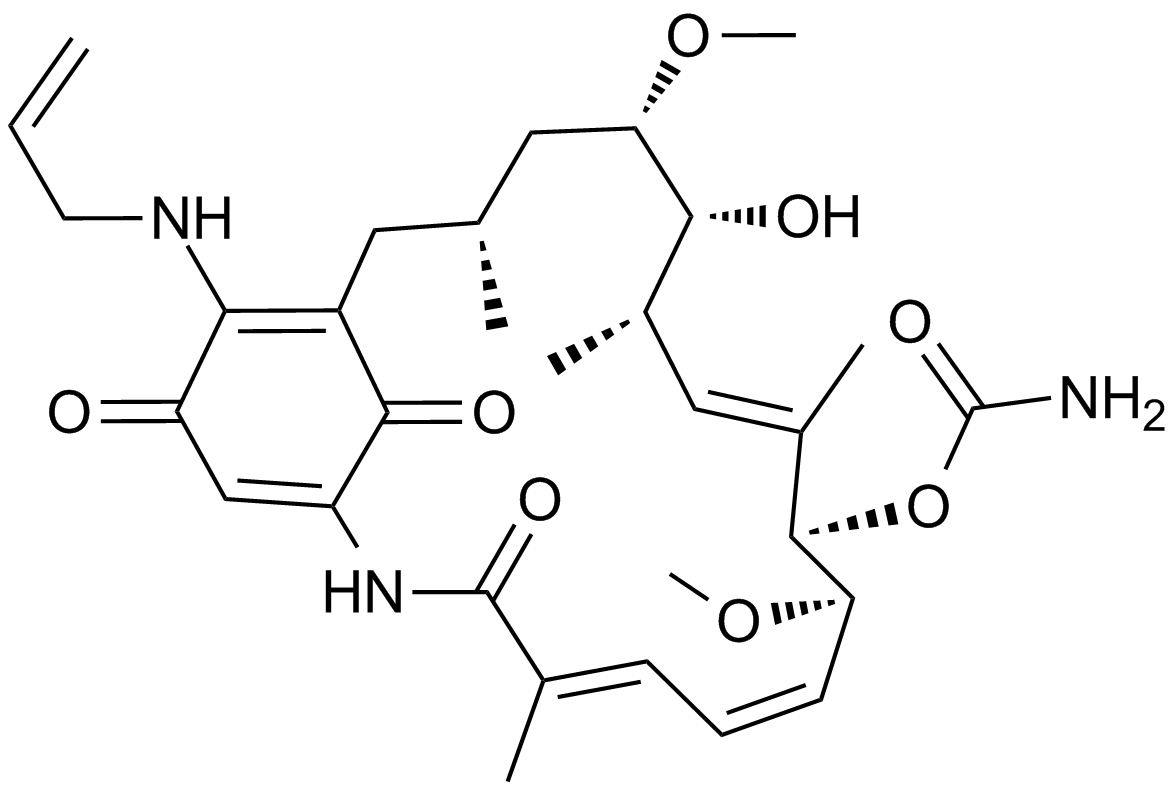
GC17210
17-AAG Hydrochloride
Hsp90 inhibitor,geldanamycin analogue
-
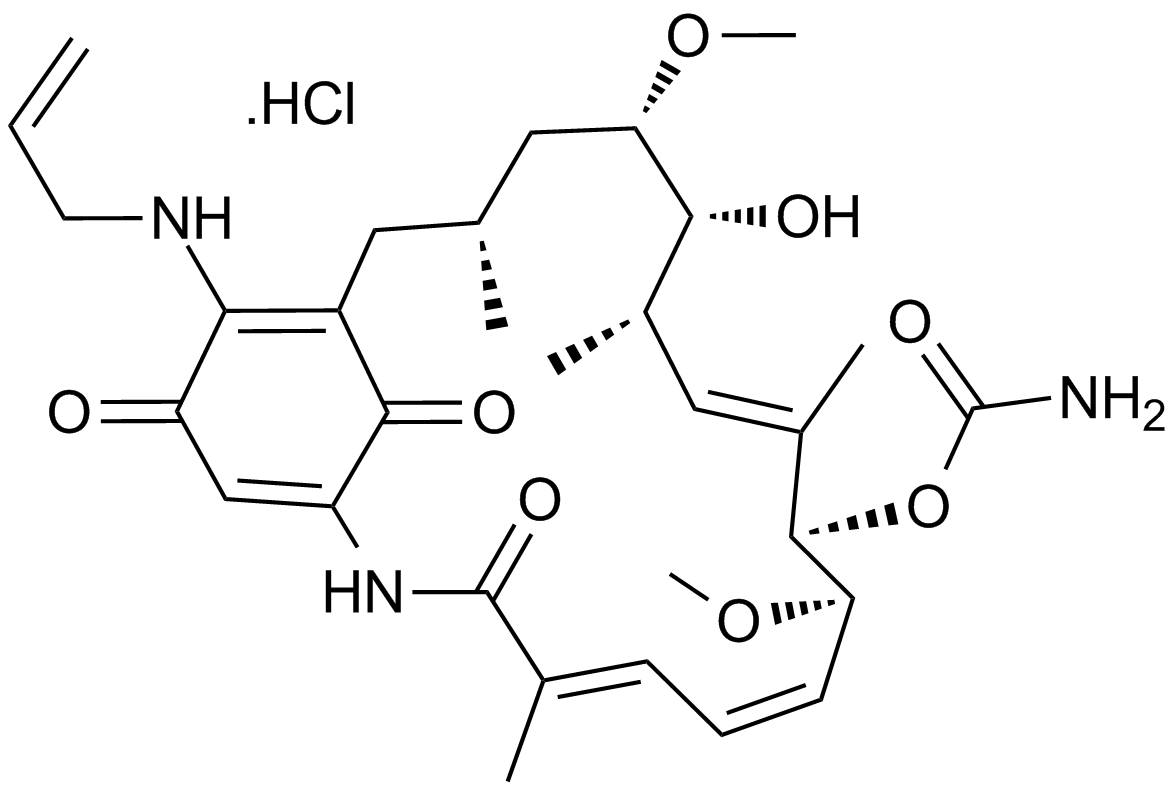
GC41955
17-DMAG
17-DMAG (17-DMAG) is a potent inhibitor of Hsp90, binding to Hsp90 with an EC50 of 62 ± 29 nM.
-
GC13044
17-DMAG (Alvespimycin) HCl
17-DMAG (Alvespimycin) HCl (17-DMAG hydrochloride; KOS-1022; BMS 826476) is a potent inhibitor of Hsp90, binding to Hsp90 with EC50 of 62±29 nM.

-
GC61582
3-Phenyltoxoflavin
3-Phenyltoxoflavin, a derivative of Toxoflavin, is an Hsp90 inhibitor, with a Kd of 585 nM for the interaction of Hsp90-TPR2A. 3-Phenyltoxoflavin has anti-cancer activity.
-
GC61563
Aminohexylgeldanamycin
Aminohexylgeldanamycin (AHGDM), a Geldanamycin derivative, is a potent HSP90 inhibitor. Aminohexylgeldanamycin shows antiangiogenic and antitumor activities.
-
GC39477
AMP-PCP disodium
AMP-PCP disodium is an ATP analogue and can bind to Hsp90 N-terminal domain with a Kd value of 3.8 μM. AMP-PCP disodium binding favors the formation of the active homodimer of Hsp90.
-
GC65035
Apatorsen
Apatorsen?is?an?antisense?oligonucleotide?designed?to?bind?to?Hsp27?mRNA,?resulting?in?the?inhibition of?the?production?of?Hsp27?protein.

-
GC14411
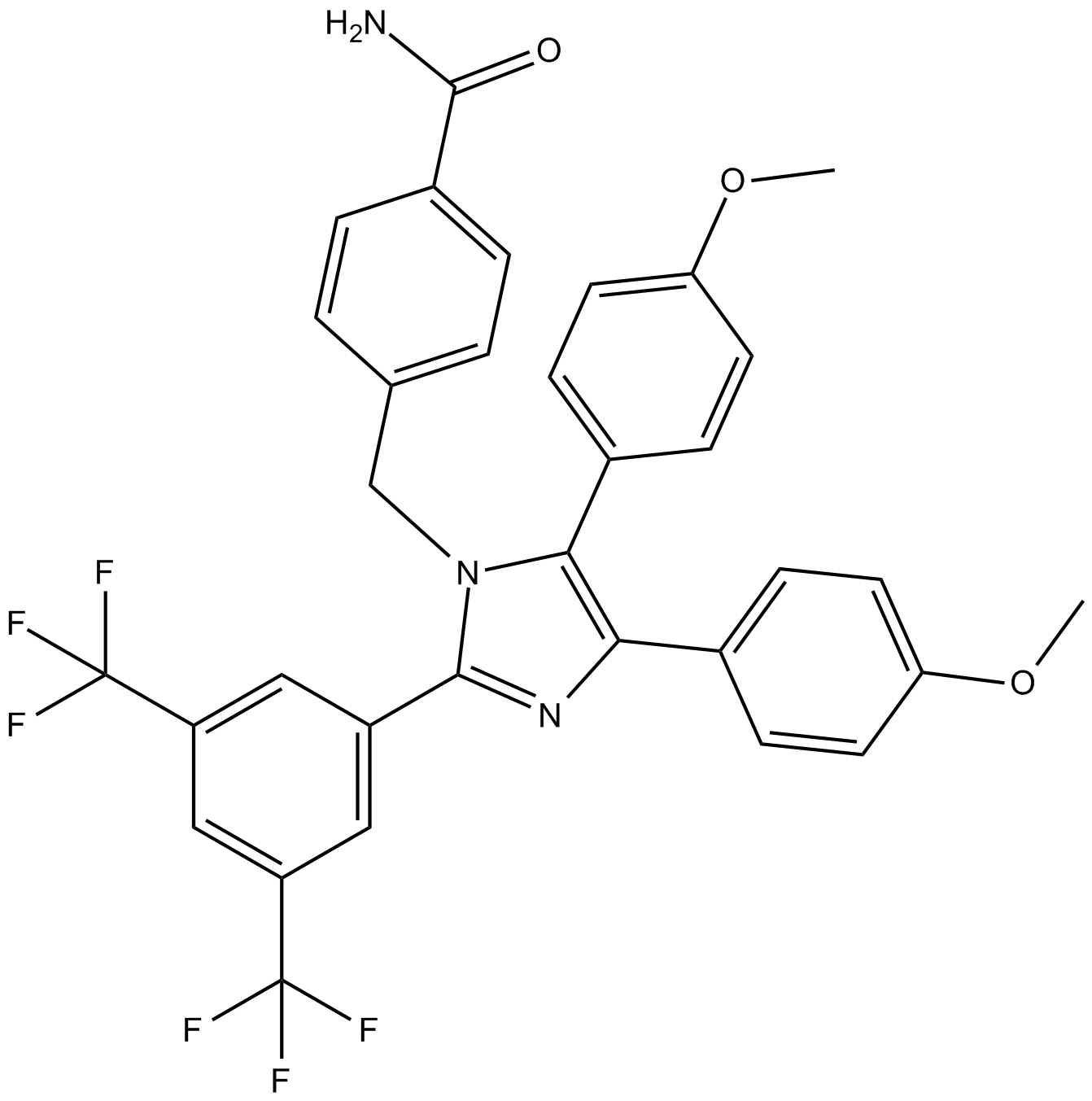
Apoptozole
inhibitor of heat shock protein 70 (Hsp70)
-
GC31103
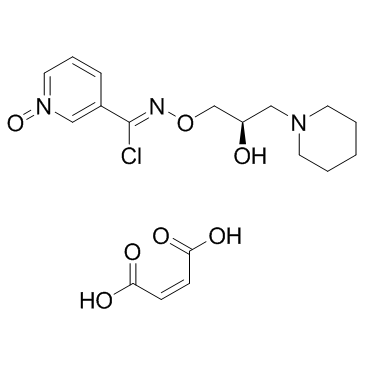
Arimoclomol maleate (BRX-220)
Arimoclomol maleate (BRX-220) (BRX-220) is a co-inducer of heat shock proteins (HSP).
-
GC13414
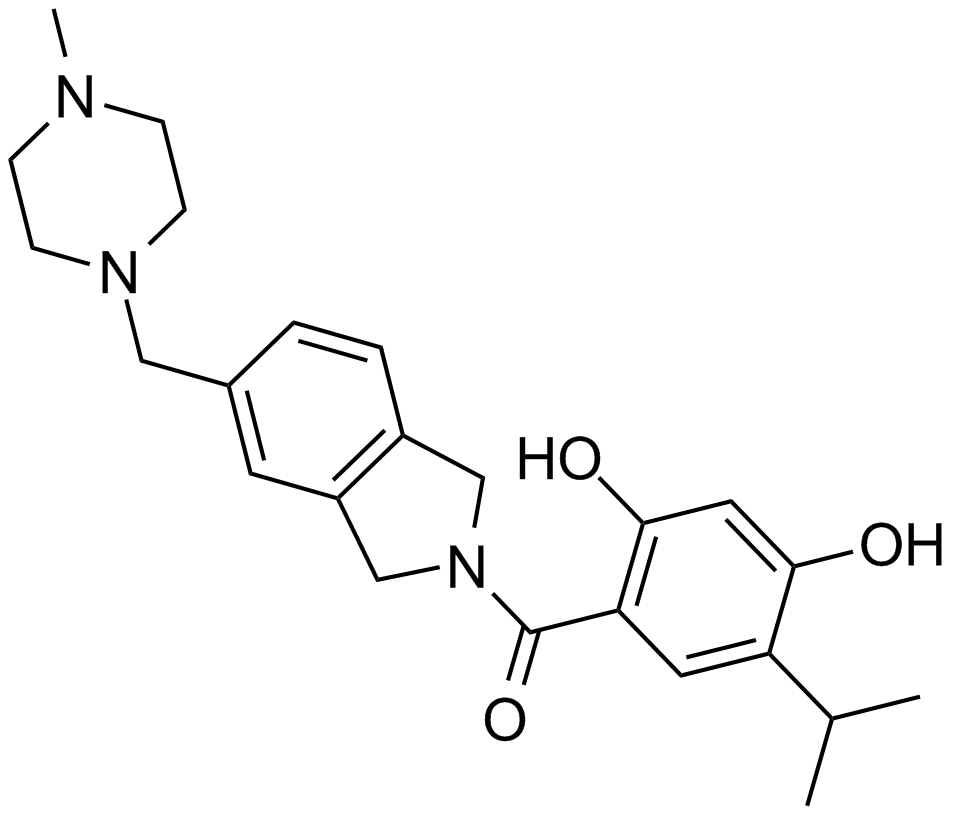
AT13387
AT13387 (AT13387) is a long-acting second-generation Hsp90 inhibitor with a Kd of 0.71 nM.
-
GC15295
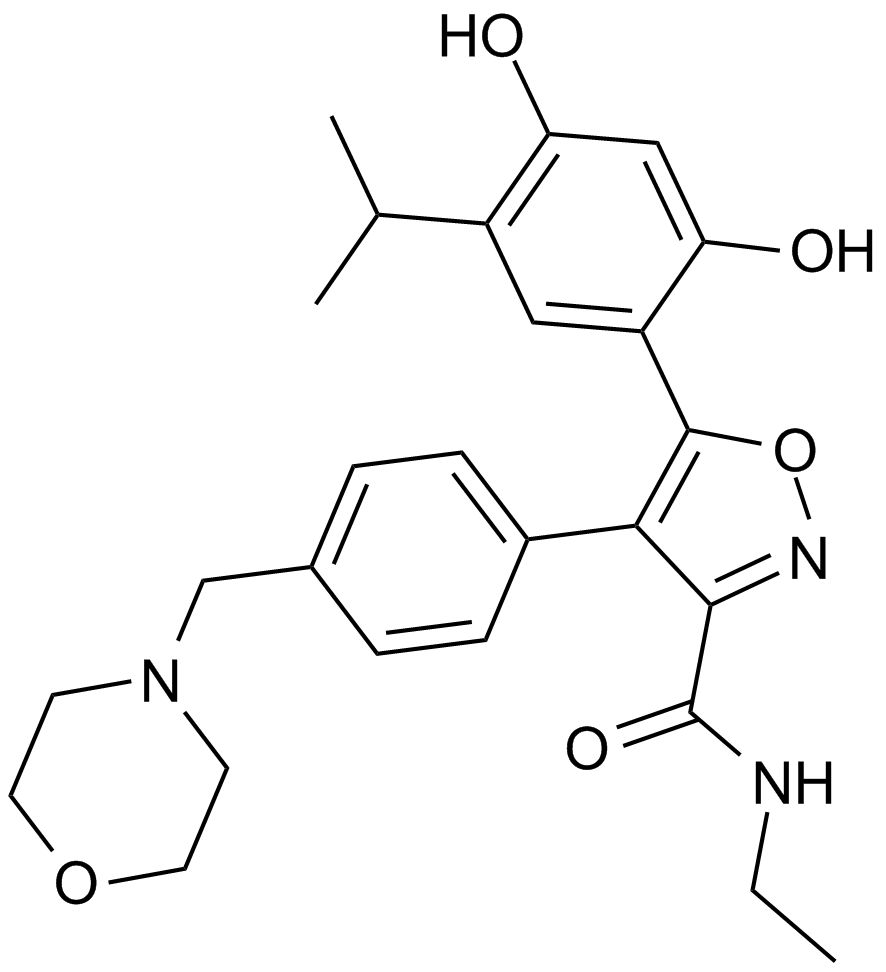
AUY922 (NVP-AUY922)
An Hsp90 inhibitor
-
GC41532
Benzisoxazole Hsp90 Inhibitor
Heat shock protein 90 (Hsp90) is a molecular chaperone that modulates intracellular signaling and protein folding, trafficking, and turnover.
-
GC12218
BIIB021
A potent, orally-available Hsp90 inhibitor
-
GC32620
Bimoclomol
Bimoclomol is a heat shock protein (HSP) coinducer, used for treatment of cardiovascular diseases.
-
GC50153
BIX
BIX, a selective inducer of immunoglobulin heavy chain binding protein (BiP)/GRP78, is an effective ER (endoplasmic reticulum) stress inhibitor.
-
GC65318
BOLD-100
BOLD-100 is a ruthenium-based anticancer agent. BOLD-100 also is an inhibitor of stress-induced GRP78 upregulation, disrupting endoplasmic reticulum (ER) homeostasis and inducing ER stress and unfolded protein response (UPR). BOLD-100 interferes with the complex interplay between ER-stress response, lysosome dynamics, and autophagy execution.

-
GC10223
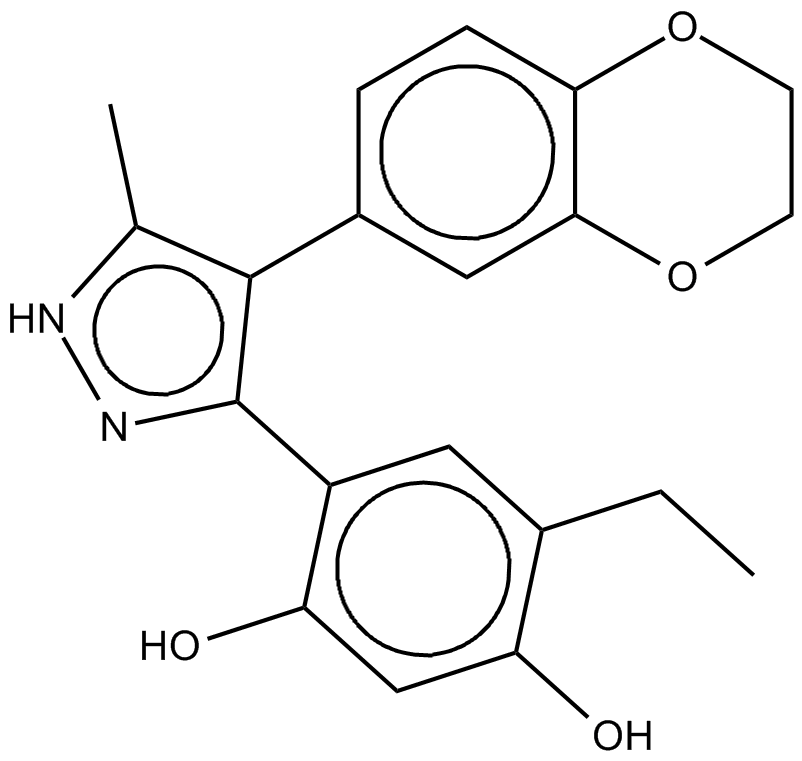
CCT018159
inhibitor of the ATPase activity of Hsp90
-
GC19094
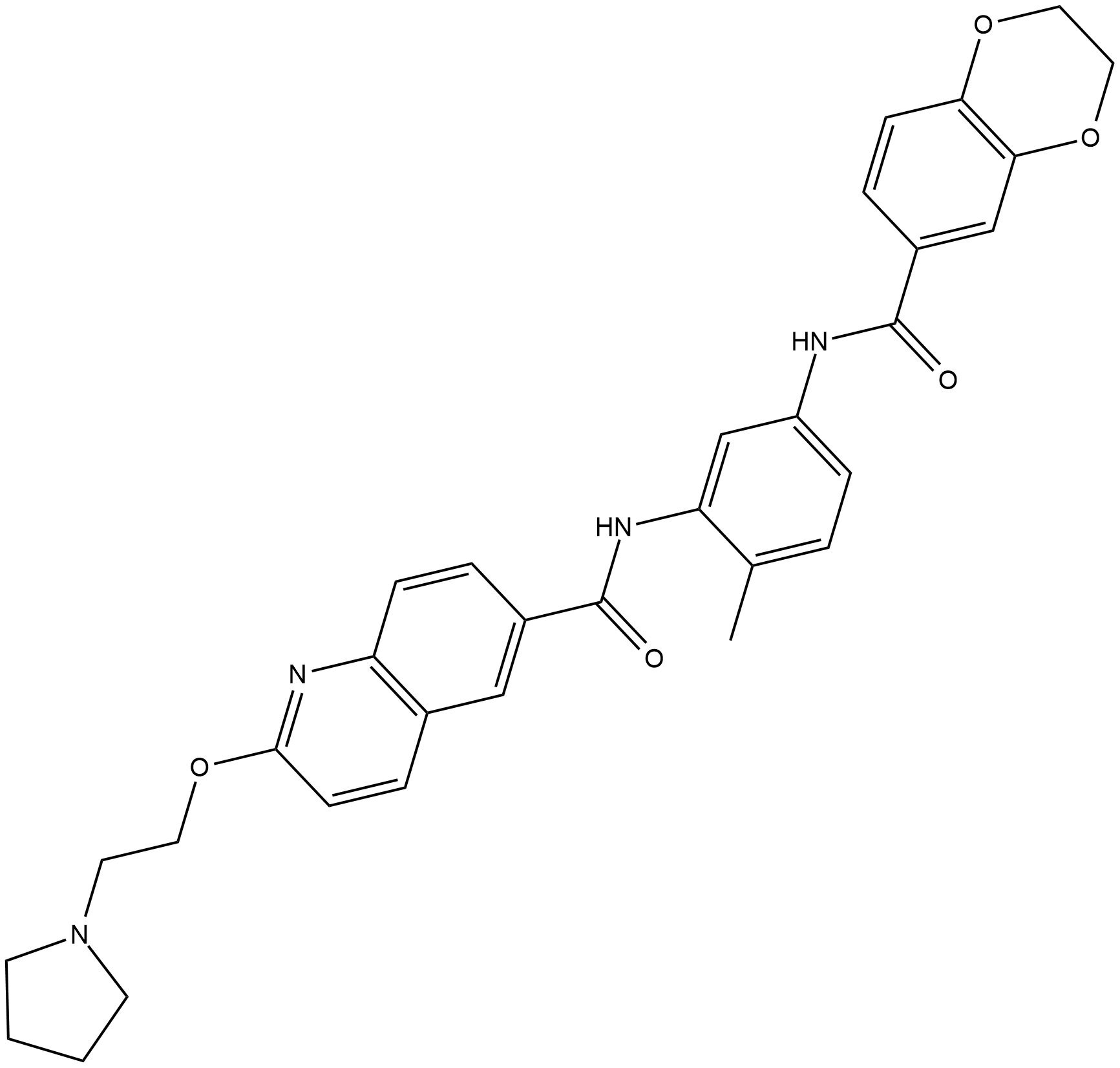
CCT251236
CCT251236 is an orally available pirin ligand from a heat shock transcription factor 1 (hsf1) phenotypic screen with an IC50 of 19 nM for inhibition of HSF1-mediated HSP72 induction.
-
GC17822
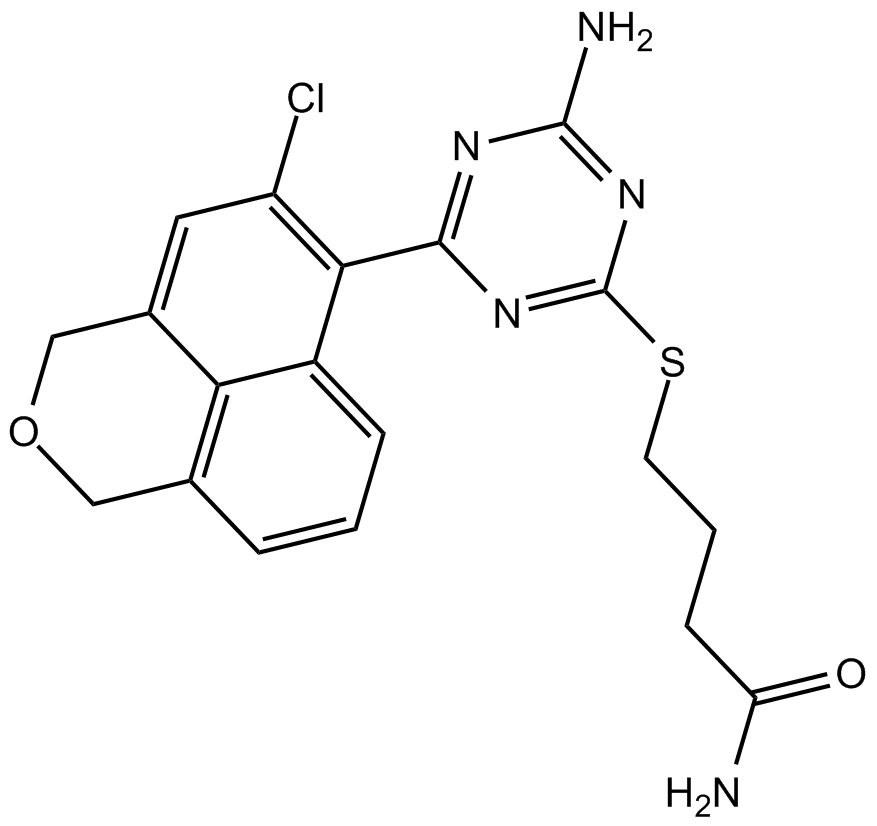
CH5138303
Hsp90 inhibitor, orally available
-
GC34539
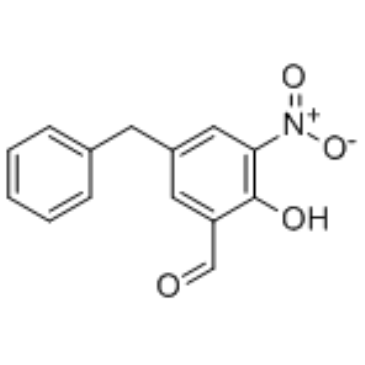
Col003
Col003 is a selective and potent inhibitor of?Hsp47 and competitively binds to the collagen binding site on Hsp47 (IC50=1.8 μM).
-
GC18832
Conglobatin
Conglobatin is a dimeric macrolide dilactone originally isolated from S.
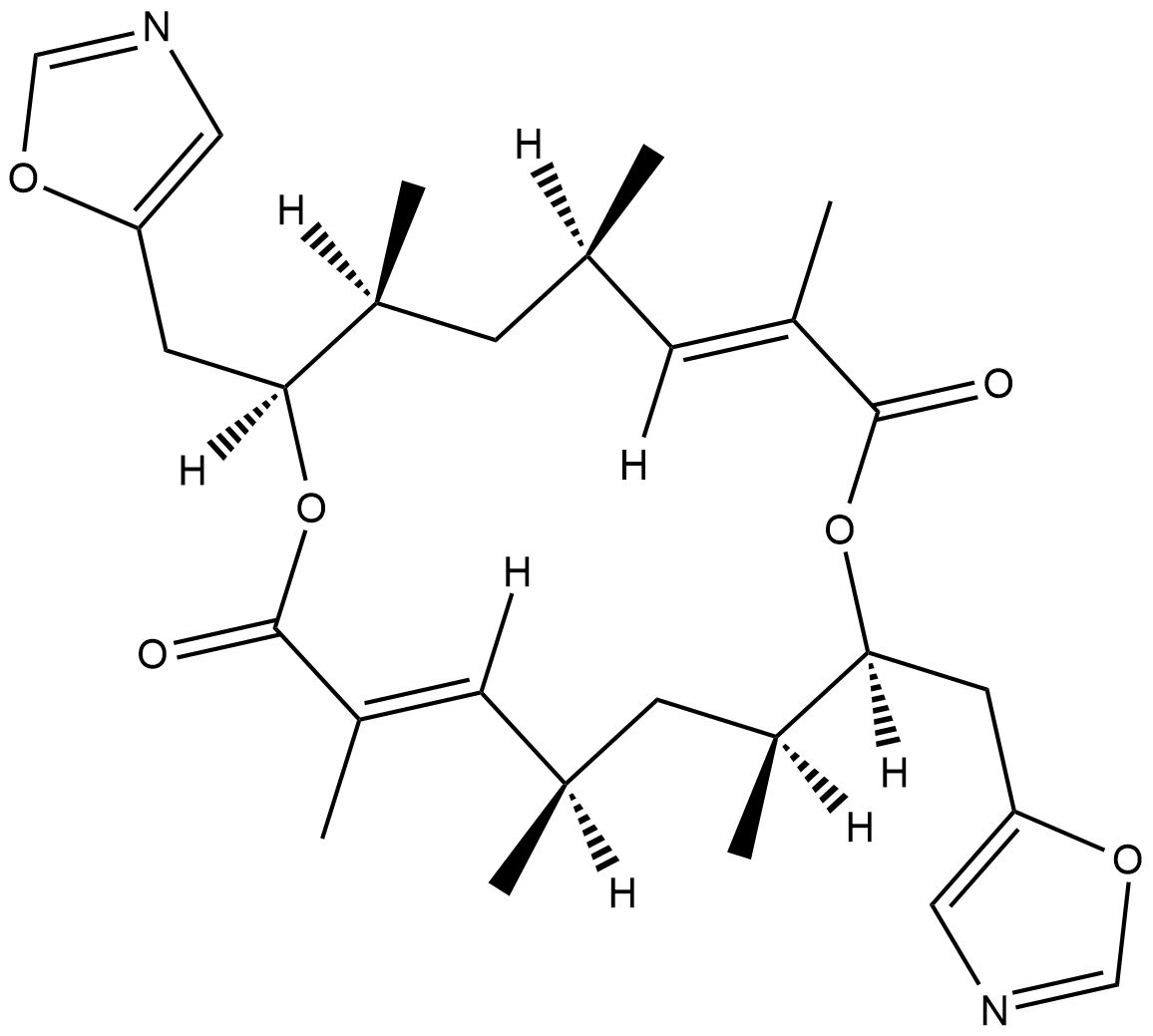
-
GC35757
Cucurbitacin D
Cucurbitacin D is an active component in Cucurbita texana, disrupts interactions between Hsp90 and two co-chaperones, Cdc37 and p23. Cucurbitacin D prevents Hsp90 client (Her2, Raf, Cdk6, pAkt) maturation without induction of the heat shock response. Anti-cancer activity.
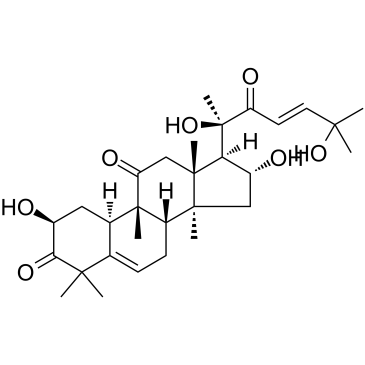
-
GC62305
DDO-5936
DDO-5936 is a potent and specific Hsp90-Cdc37 PPI inhibitor. DDO-5936 can be used for the research of colorectal cancer.
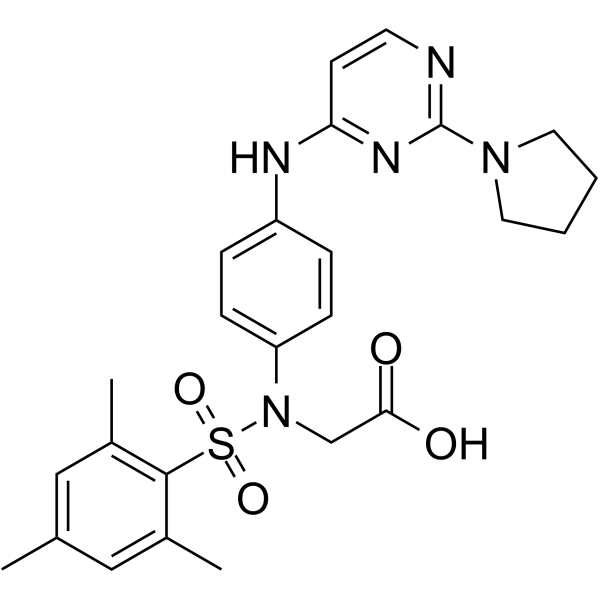
-
GC19121
Debio 0932
Debio 0932 is an orally active HSP90 inhibitor, with IC50s of 100 and 103 nM for HSP90α and HSP90β, respectively.
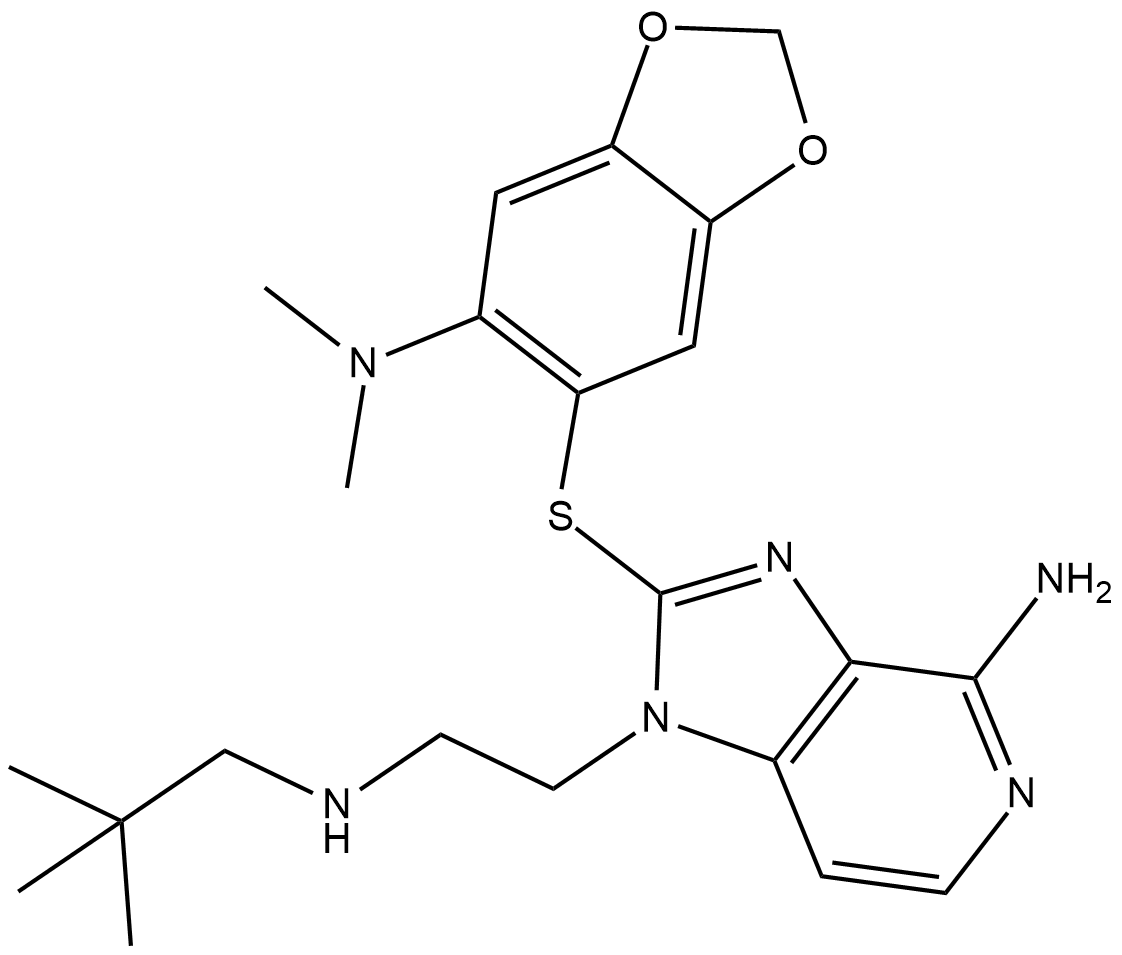
-
GC38223
Dihydroberberine
-
GC62120
DTHIB
DTHIB is a direct and selective heat shock factor 1 (HSF1) inhibitor with a Kd of 160 nM for DTHIB binding to the HSF1 DNA binding domain (DBD). DTHIB inhibits HSF1 cancer gene signature (HSF1 CaSig) and selectively stimulates degradation of nuclear HSF1. DTHIB has potently anticancer activities and can be used for prostate cancer research.
-
GC14776
EC 144
High affinity, potent and selective Hsp90 inhibitor
-
GC13885
Elesclomol (STA-4783)
An apoptosis inducer
-
GC32965
Ethoxyquin
An antioxidant used in animal feed
-
GC43649
Falcarinol
Falcarinol is a C17-polyacetylene produced by the Apiaceae family that has antimicrobial properties due to its inhibition of fatty acid biosynthesis.
-
GC32008
Feretoside
Feretoside, a phenolic compound extracted from the barks of E.
-
GC17655
Ganetespib (STA-9090)
-
GC10881
Gedunin
naturally occurring Hsp90 inhibitor
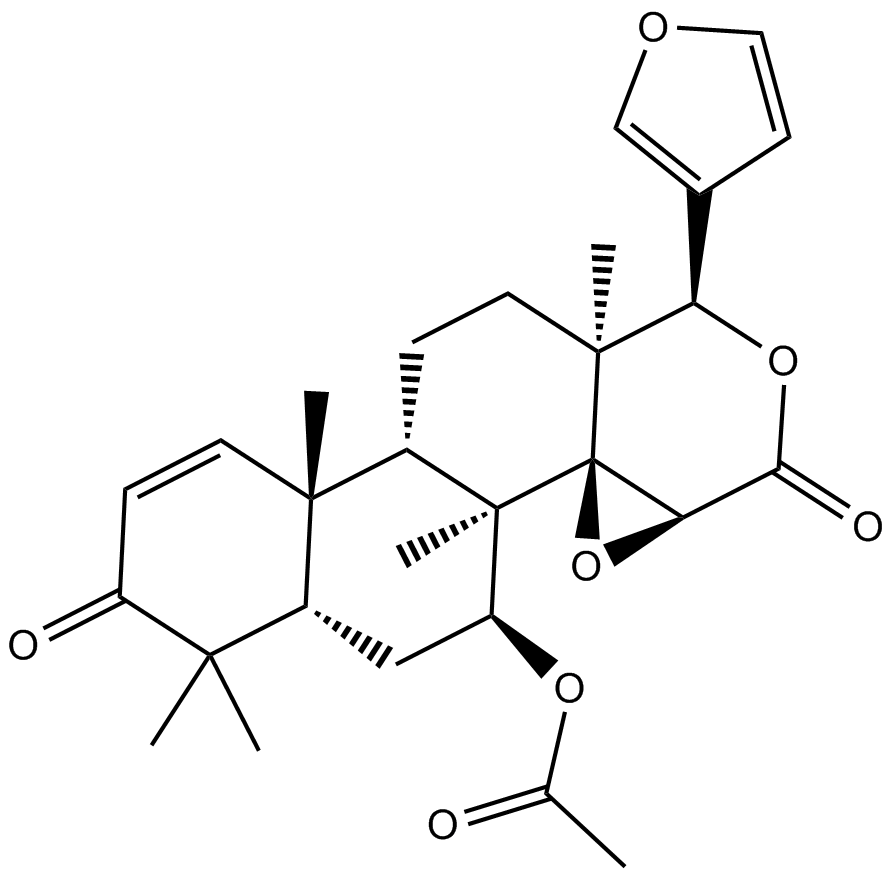
-
GC16361
Geldanamycin
Binds Hsp90 and GRP94, inhibits growth of cancer cells
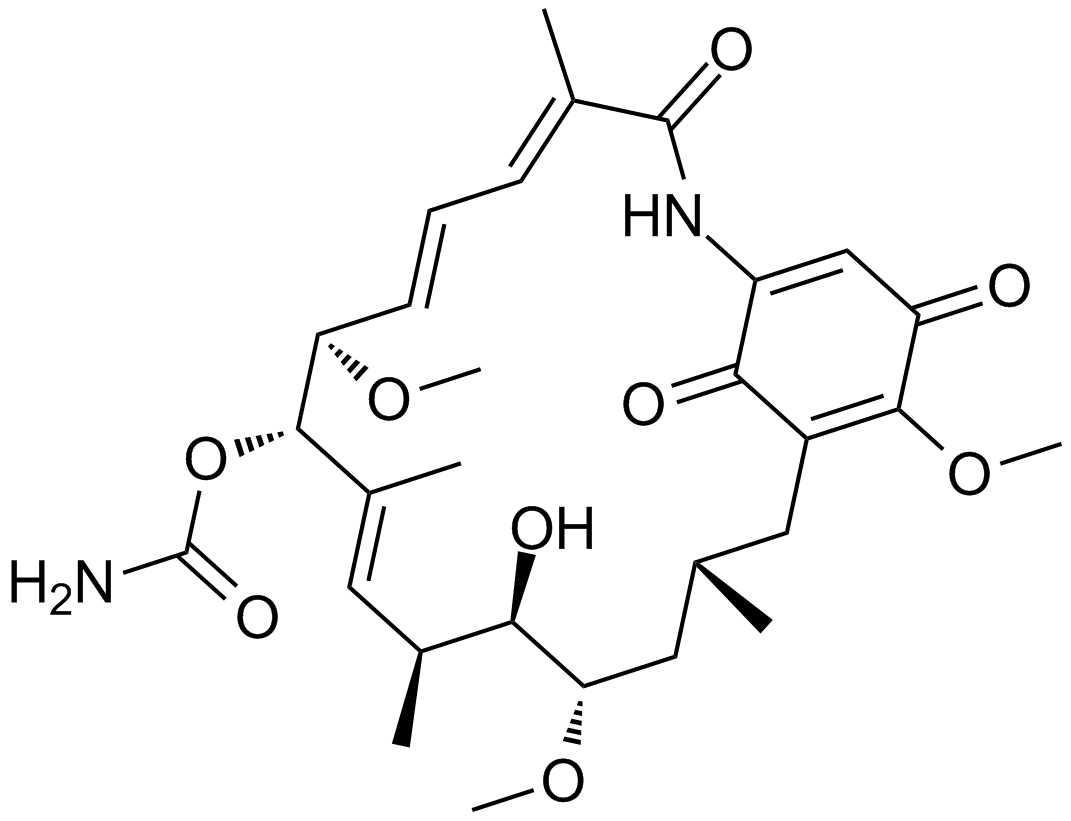
-
GC64795
Geldanamycin-FITC
Geldanamycin-FITC, a Geldanamycin fluorescent probe, can be used in a fluorescence polarization assay for HSP90 inhibitors.
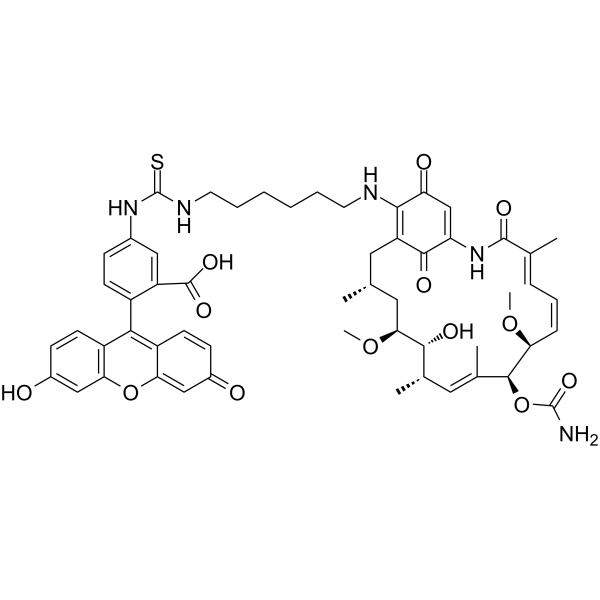
-
GC64988
Grp94 Inhibitor-1
Grp94 Inhibitor-1 is a potent, selective Grp94 inhibitor with an IC50 value of 2 nM, and over 1000-fold selectivity to Grp94 against Hsp90α.
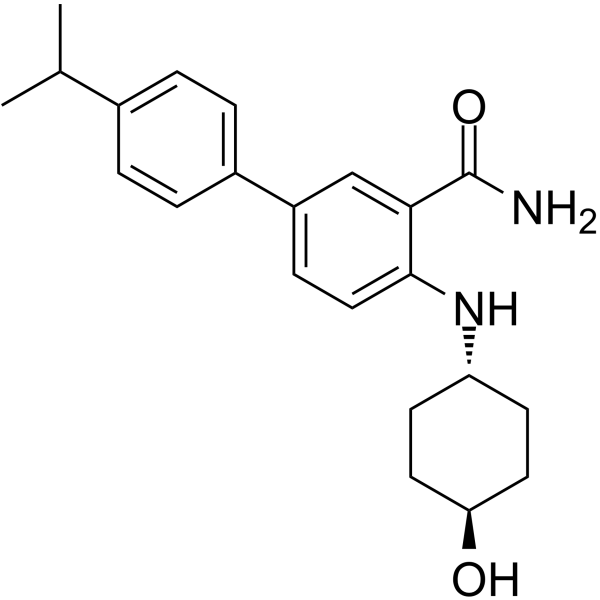
-
GC19188
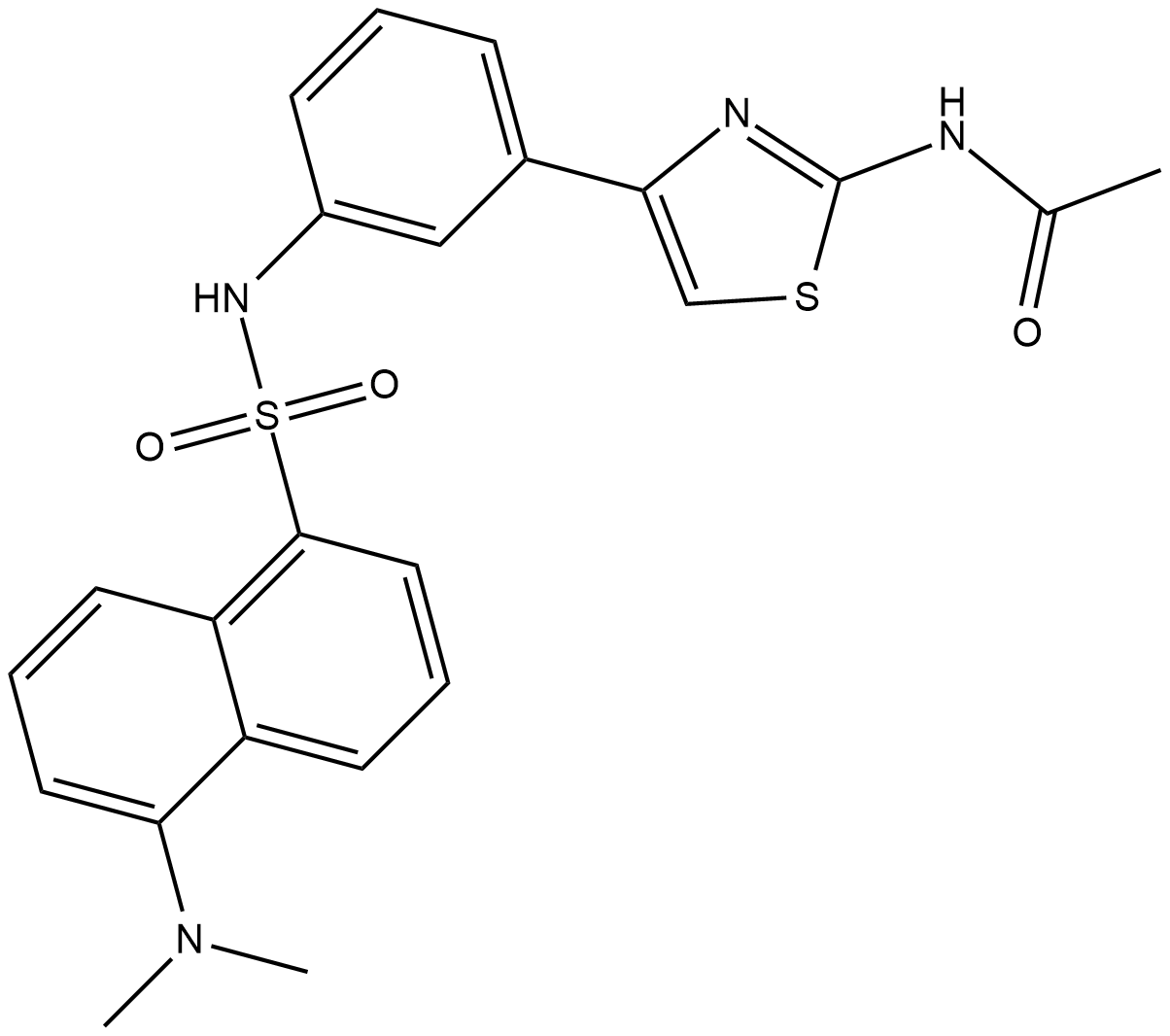
HA15
HA15 is a molecule that targets specifically BiP/GRP78/HSPA5.
-
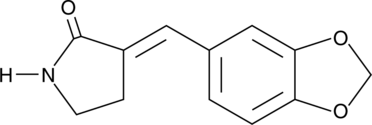
GC40724
Heat Shock Protein Inhibitor II
Heat shock protein (Hsp) inhibitor II is the active form of Hsp inhibitor I and a benzylidene lactam compound that prevents the synthesis of inducible Hsps, such as Hsp105, Hsp72, and Hsp40.
-
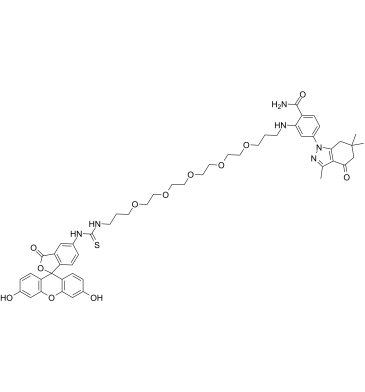
GC61784
HS-27
HS-27, a fluorescently-tethered Hsp90 inhibitor, assays surface Hsp90 expression on intact tissue specimens. HS-27 is made up of the core elements of SNX-5422, an Hsp90 inhibitor, tethered via a PEG linker to a fluorescein derivative (fluorescein isothiocyanate or FITC), that binds to ectopically expressed Hsp90. HS-27 has potential use in a see-and-treat paradigm in breast cancer.
-
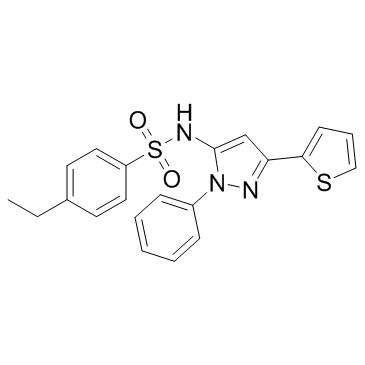
GC32437
HSF1A
HSF1A is a cell-permeable activator of heat shock transcription factor 1 (HSF1).
-
GC38503
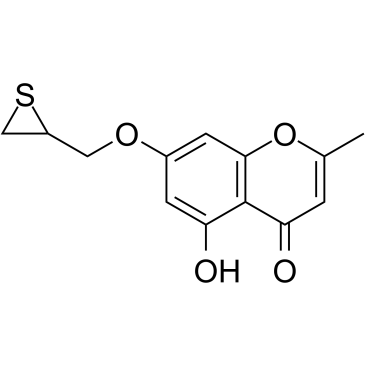
HSP27 inhibitor J2
HSP27 inhibitor J2 (J2) is an inhibitor of HSP27, which significantly induces abnormal HSP27 dimer formation and inhibits HSP27 macropolymer production[1-3].
-
GC32869
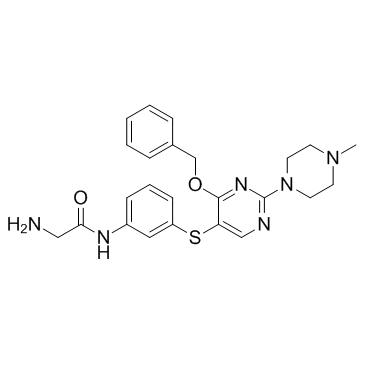
HSP70-IN-1
HSP70-IN-1 is a heat shock protein (HSP) inhibitor; inhibits the growth of Kasumi-1 cells with an IC50 of 2.3 μM.
-
GC68450
Hsp90-IN-17 hydrochloride

-
GC11973
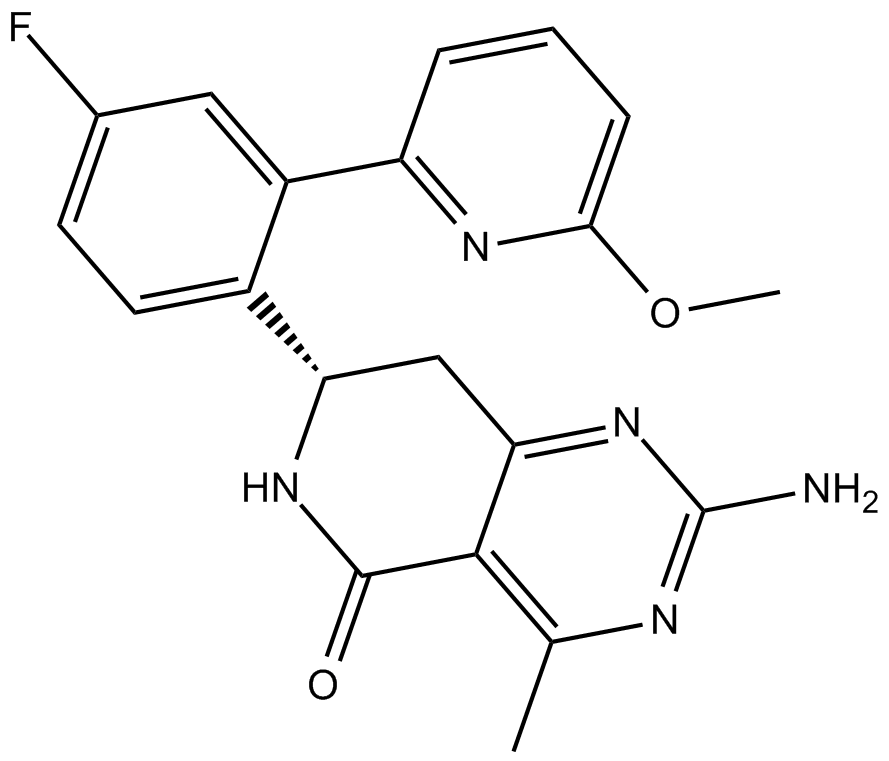
HSP990 (NVP-HSP990)
HSP990 (NVP-HSP990) is a potent and selective Hsp90 inhibitor, with IC50 values of 0.6, 0.8, and 8.5 nM for Hsp90α, Hsp90β, and Grp94, respectively.
-
GC63282
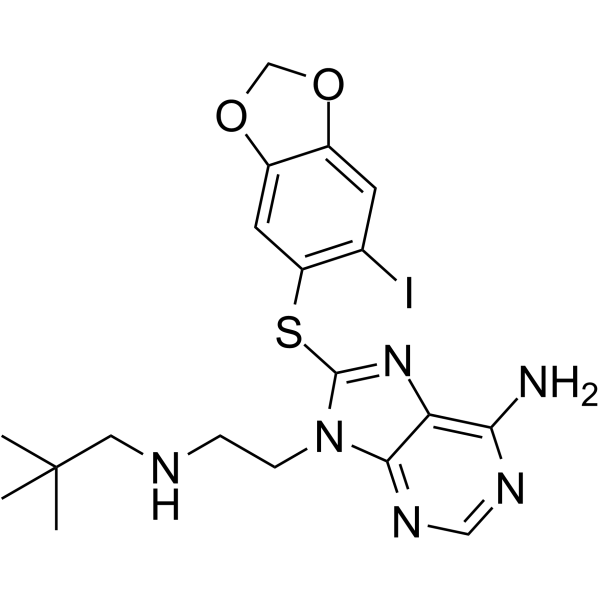
Icapamespib
Icapamespib (PU-HZ151) is a potent HSP90 inhibitor with an EC50 of 5?nM.
-
GC11391
IPI-504 (Retaspimycin hydrochloride)
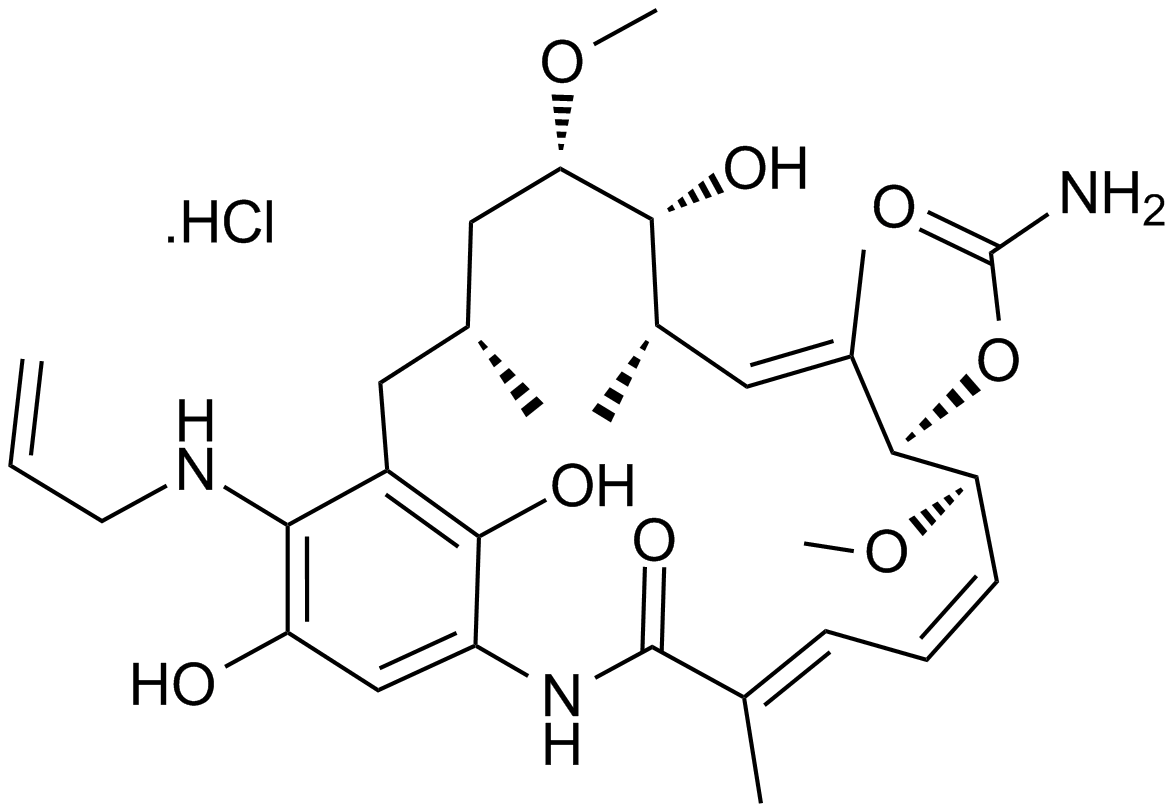
-
GC34634
JG-98
JG-98, an allosteric heat shock protein 70 (Hsp70) inhibitor, which binds tightly to a conserved site on Hsp70 and disrupts the Hsp70-Bag3 interaction. JG-98 shows anti-cancer activities affecting both cancer cells and tumor-associated macrophages.
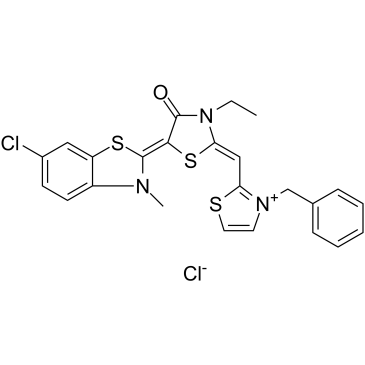
-
GC11930
KNK437
stress-induced HSP synthesis inhibitor
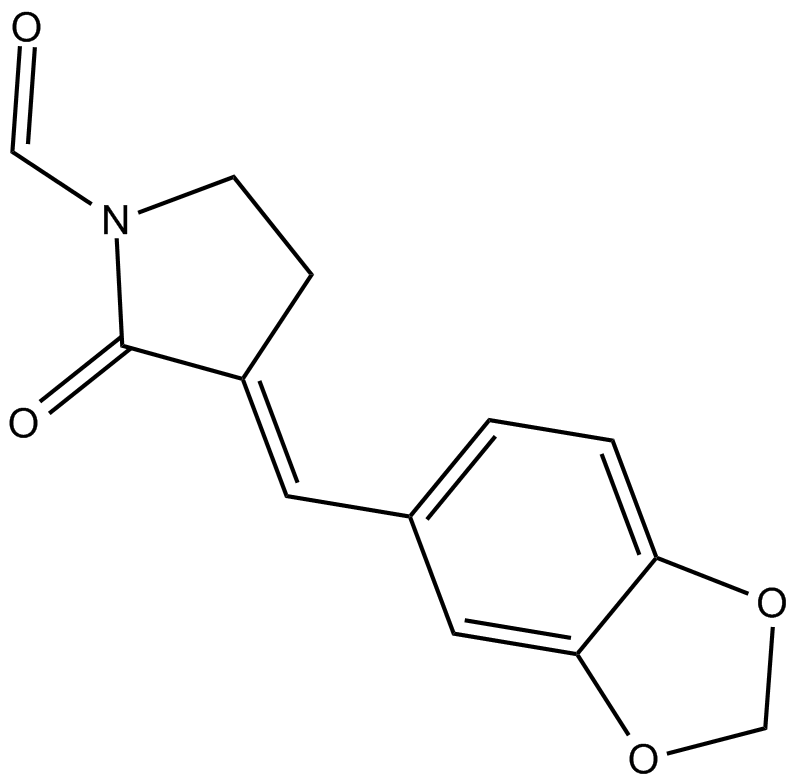
-
GC64370
Kongensin A
Kongensin A is a natural product isolated from Croton kongensis.
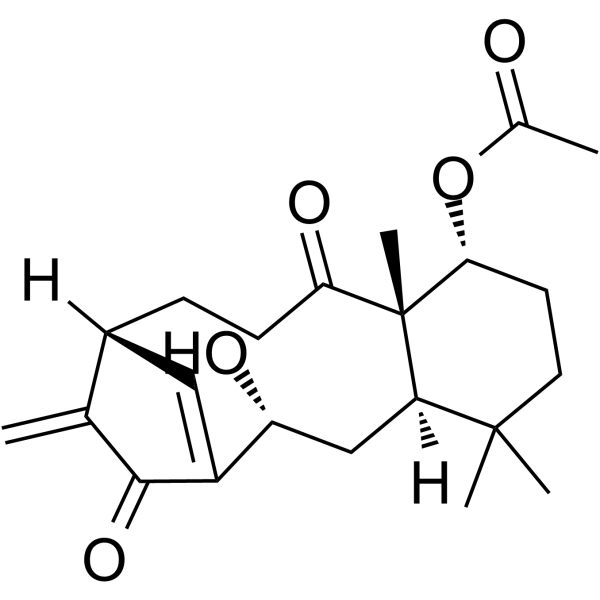
-
GC19215
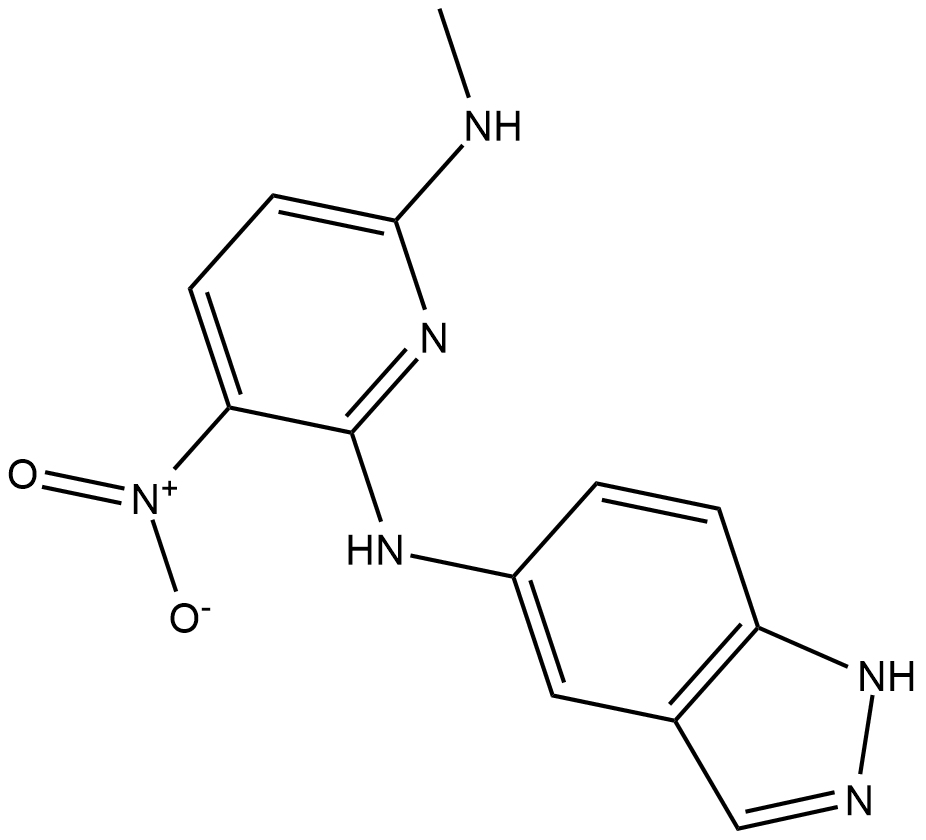
KRIBB11
KRIBB11 is an inhibitor of Heat shock factor 1 (HSF1), with IC50 of 1.2 uM.
-
GC31118
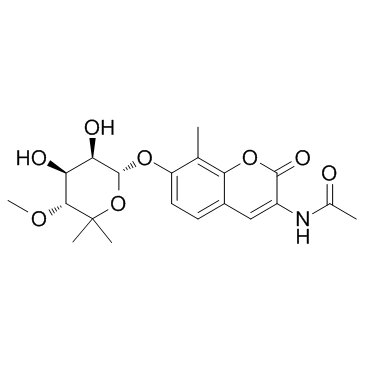
KU-32
KU-32 is a novel, novobiocin-based Hsp90 inhibitor that can protect against neuronal cell death.
-
GC14636
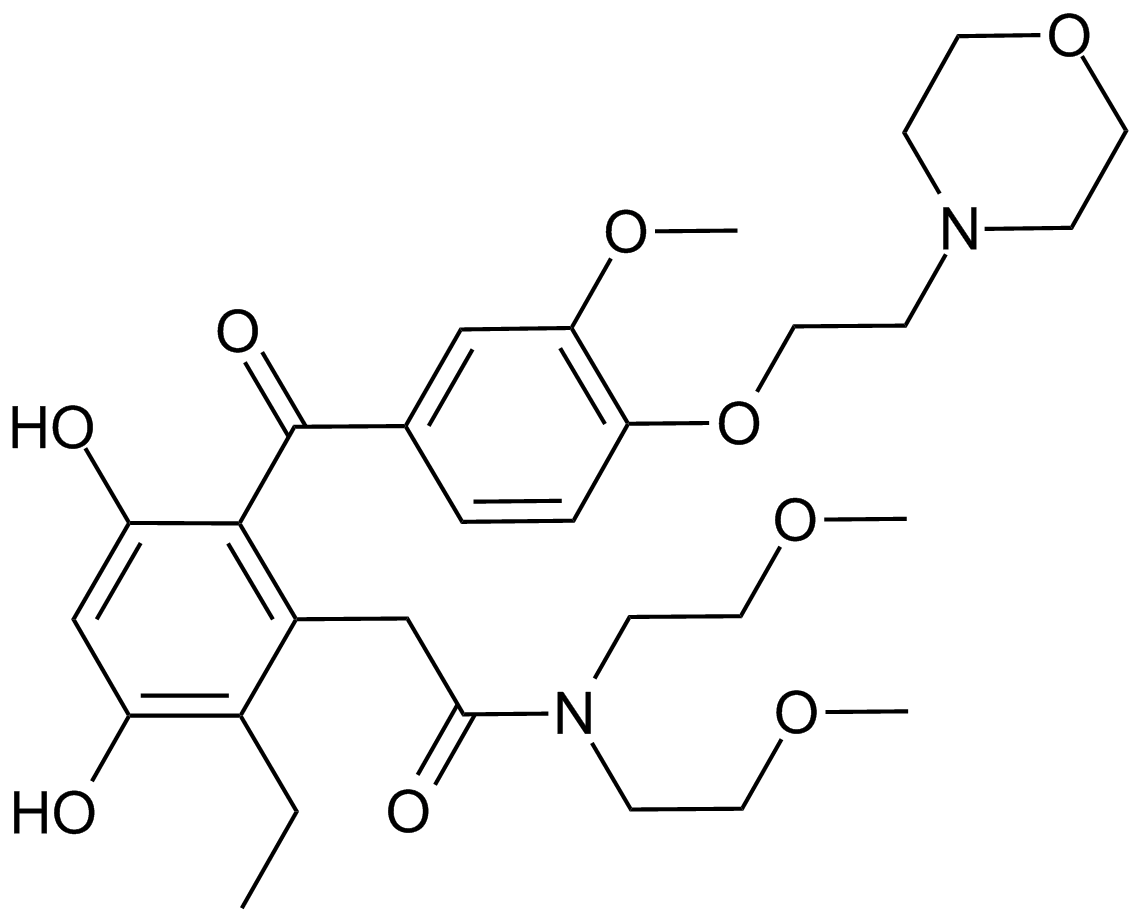
KW-2478
An Hsp90 inhibitor
-
GC15042
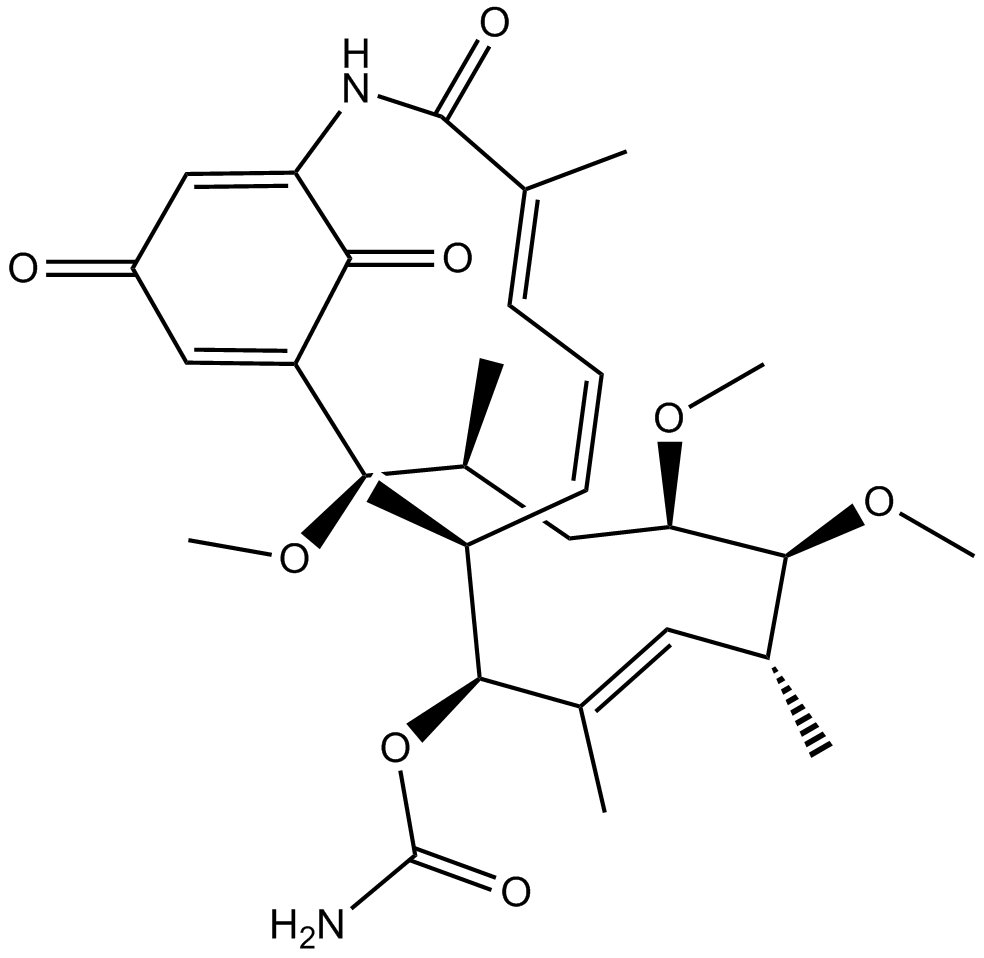
Macbecin I
Macbecin I is a stable HSP90 inhibitor by binding to the ATP-binding site with an IC50 of 2 μM and a Kd of 0.24 μM. Macbecin I exhibits antitumor and cytocidal activities.
-
GC12301
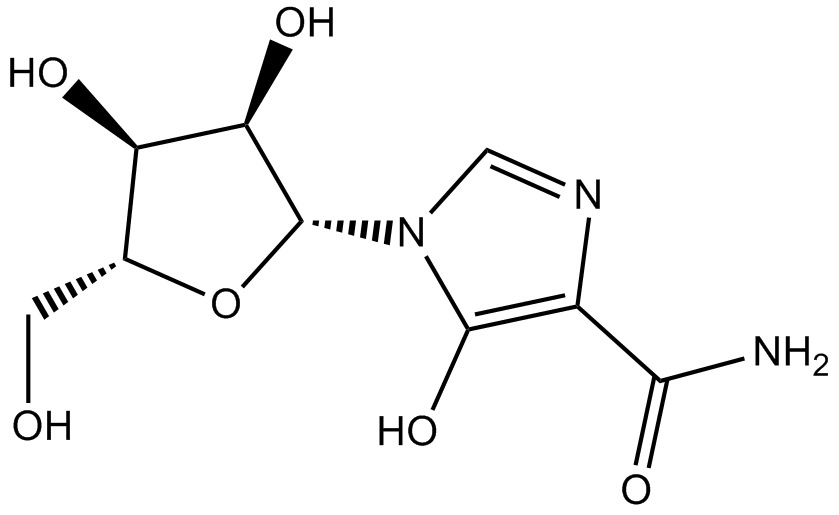
Mizoribine
DNA/RNA synthesis inhibitor
-
GC17751
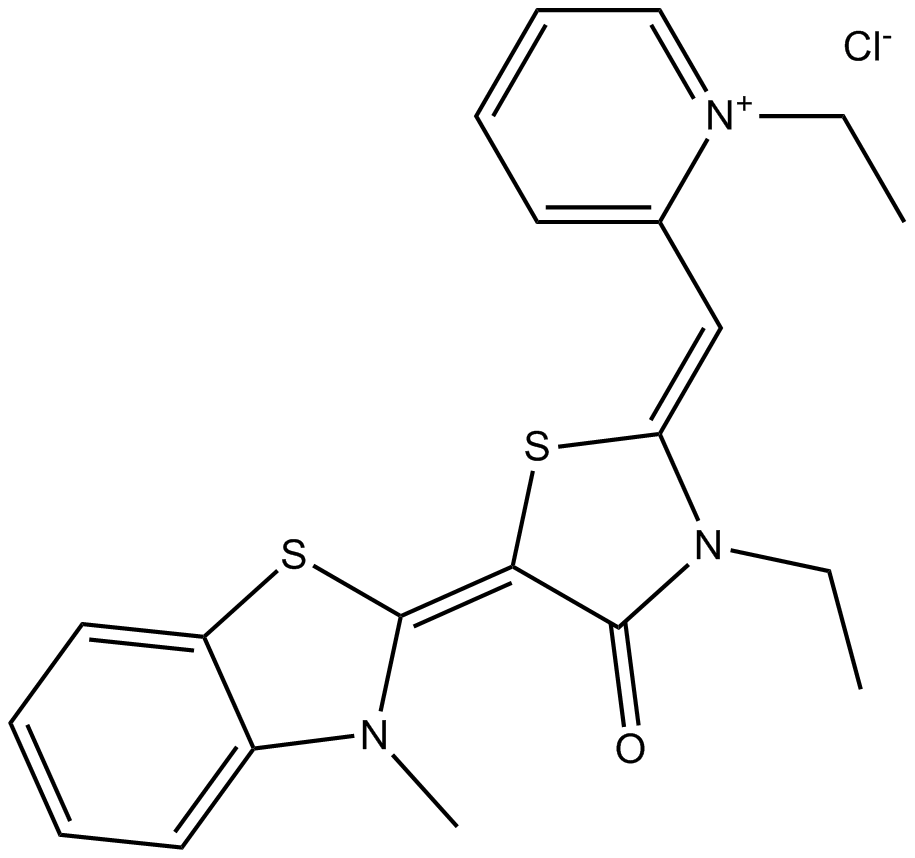
MKT 077
mortalin-2 (mot-2) inhibitor
-
GC18170
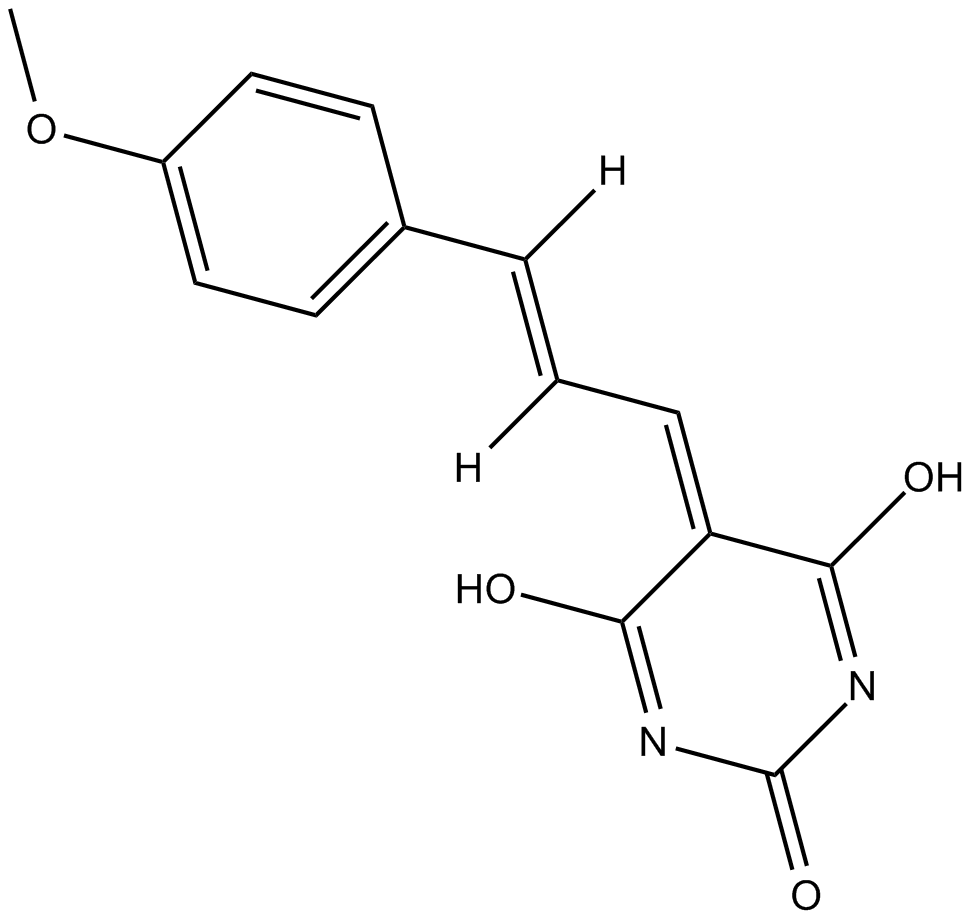
ML-346
ML-346 is an activator of the heat shock response that induces the expression of the heat shock proteins HSP70, HSP40, and HSP27.
-
GC14322
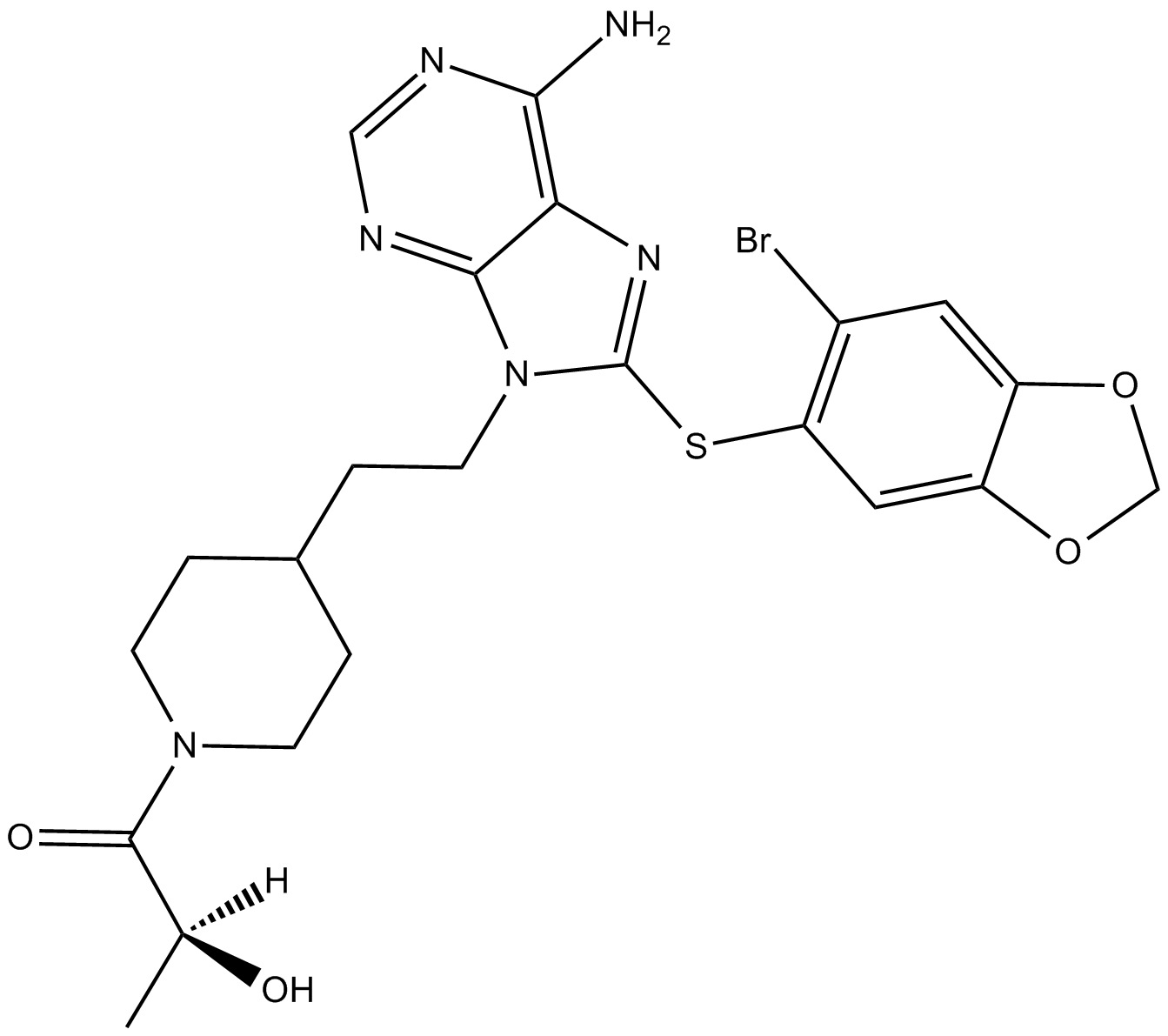
MPC-3100
An inhibitor of Hsp90
-
GC64714
NCT-58
NCT-58 is a potent inhibitor of C-terminal HSP90. NCT-58 does not induce the heat shock response (HSR) due to its targeting of the C-terminal region and elicits anti-tumor activity via the simultaneous downregulation of HER family members as well as inhibition of Akt phosphorylation. NCT-58 kills Trastuzumab-resistant breast cancer stem-like cells. NCT-58 induces apoptosis in HER2-positive breast cancer cells.
-
GC45926
Nelfinavir-d3
An internal standard for the quantification of nelfinavir
-
GC13105
NMS-E973
Hsp90 inhibitor,potent and selective
-
GC11179
NVP-BEP800
An Hsp90 inhibitor
-
GC64655
NXP800
NXP800 (CCT361814) is a potent heat shock factor 1 (HSF1) inhibitor. NXP800 has?the?potential?for?cancer research.
-
GC36834
p5 Ligand for Dnak and DnaJ
p5 Ligand for Dnak and DnaJ is a nonapeptide, which corresponds to the main binding site for the 23-residue part of the presequence of mitochondrial aspartate aminotransferase.
-
GC63852
Palmitic acid-d3
-
GC67916
Palmitic acid-d4

-
GC11912
PF-04929113 (SNX-5422)
Hsp90 inhibitor,potent and selective

-
GC69777
PU-H54
PU-H54 is a selective inhibitor of Grp94, which can be used for the study of breast cancer. The Hsp90 chaperone family includes Hsp90α, Hsp90β, Grp94 and Trap-1, which play important roles in malignant tumors.
-
GC15143
PU-H71
Hsp90 inhibitor,potent and selective
-
GC37040
PU-H71 hydrochloride
-
GC13307
PU-WS13
Grp94-specific Hsp90 inhibitor
-
GC11403
Radicicol
An inhibitor of Hsp90
-
GC10327
Retaspimycin
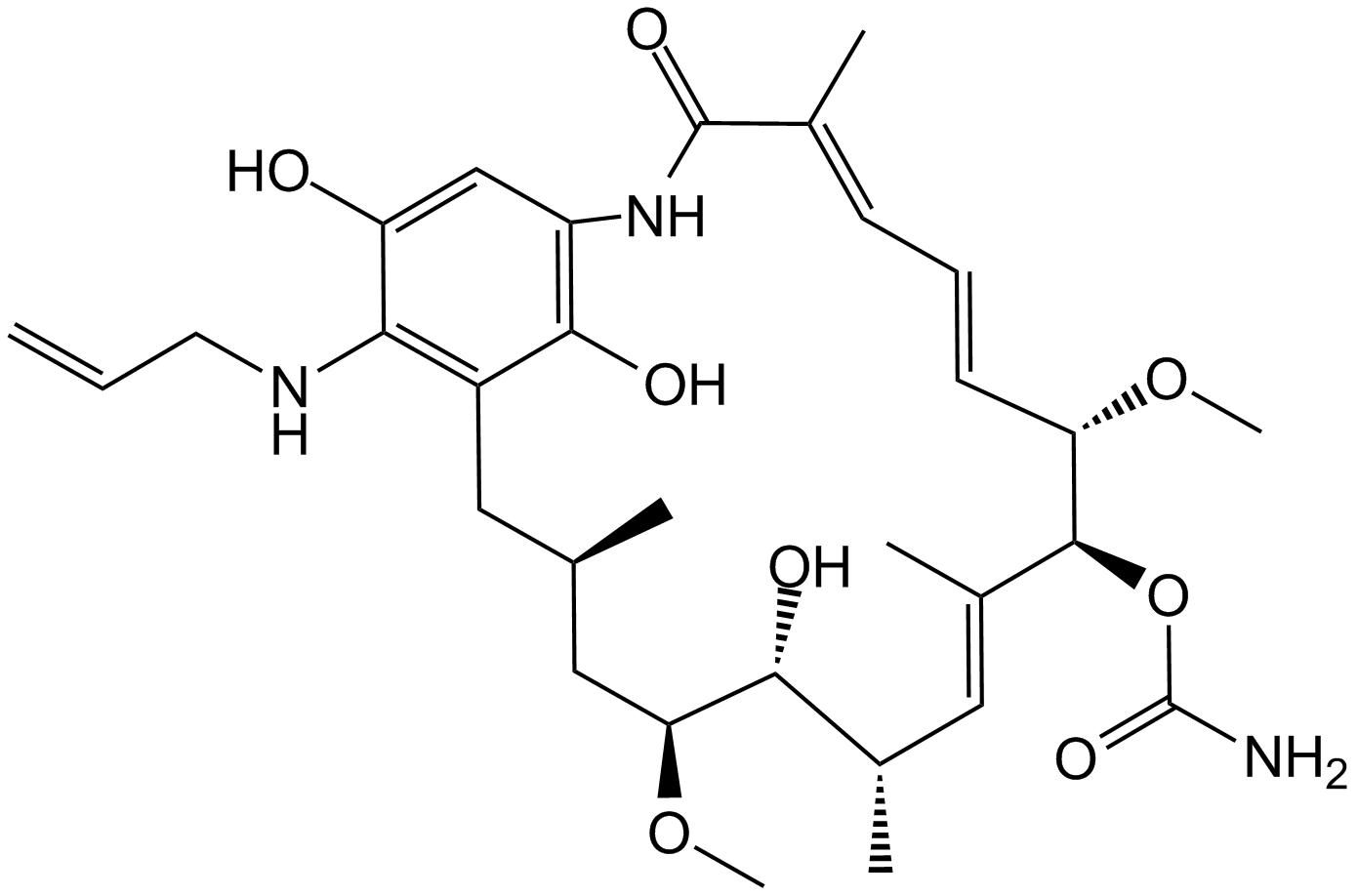
-
GC19548
Rocaglamide
Rocaglamide is a potent NF-κB activation inhibitor.

-
GC33426
Rocaglamide (Rocaglamide A)
Rocaglamide (Rocaglamide A) is isolated from the genus Aglaia (family Meliaceae).
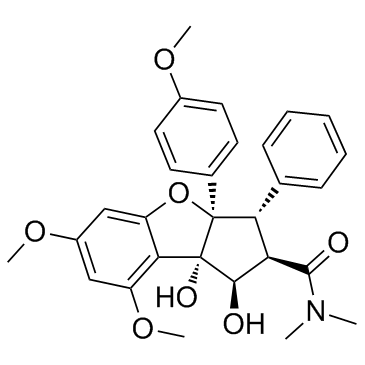
-
GC64685
SNX-0723
SNX-0723 is a potent Hsp90 Inhibitor with anti-Plasmodium activity.
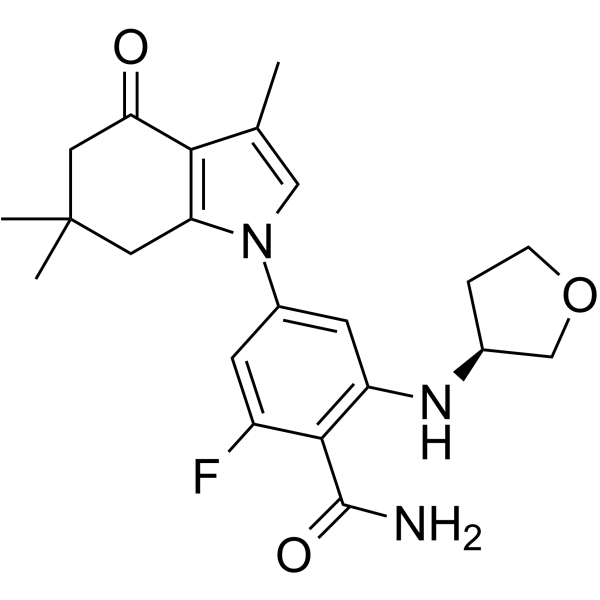
-
GC13946
SNX-2112
A potent Hsp90 inhibitor
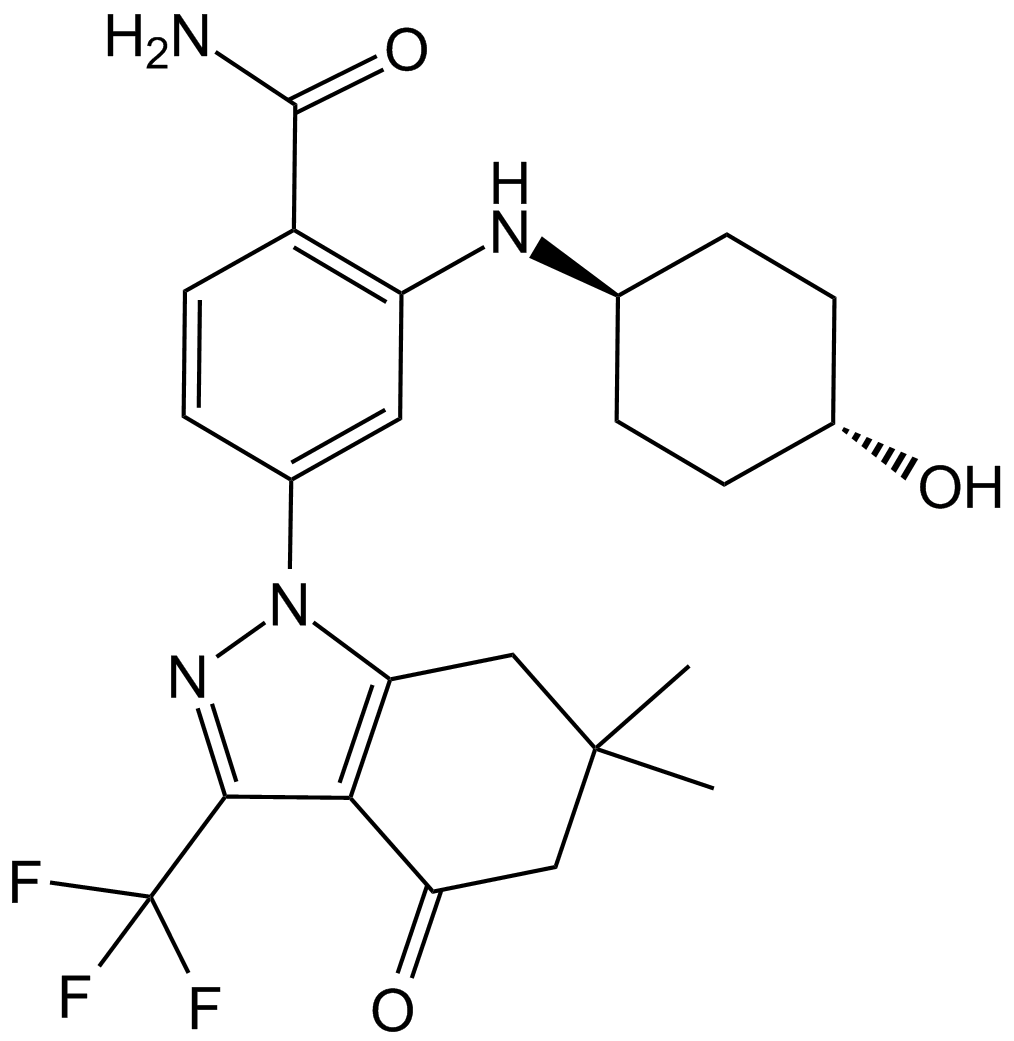
-
GC33047
SNX-5422 Mesylate (PF-04929113 (Mesylate))
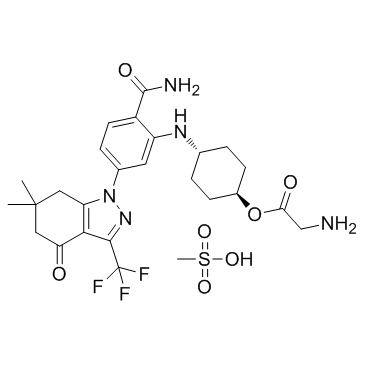
-
GC17901
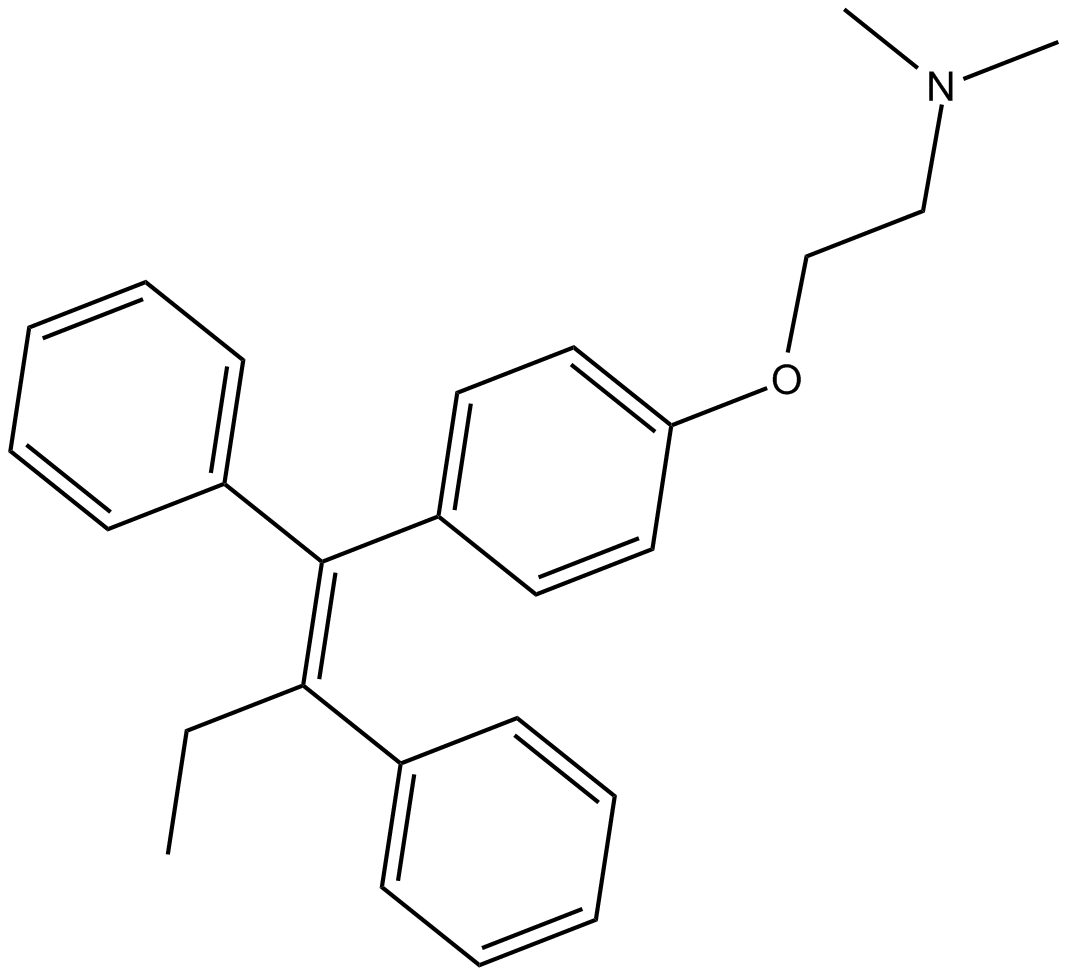
Tamoxifen
Tamoxifen(TAM) serves as a selective estrogen receptor regulator (SERM), inhibiting estrogen's effects in breast cells while potentially stimulating estrogen activity in cells found in different tissues.
-
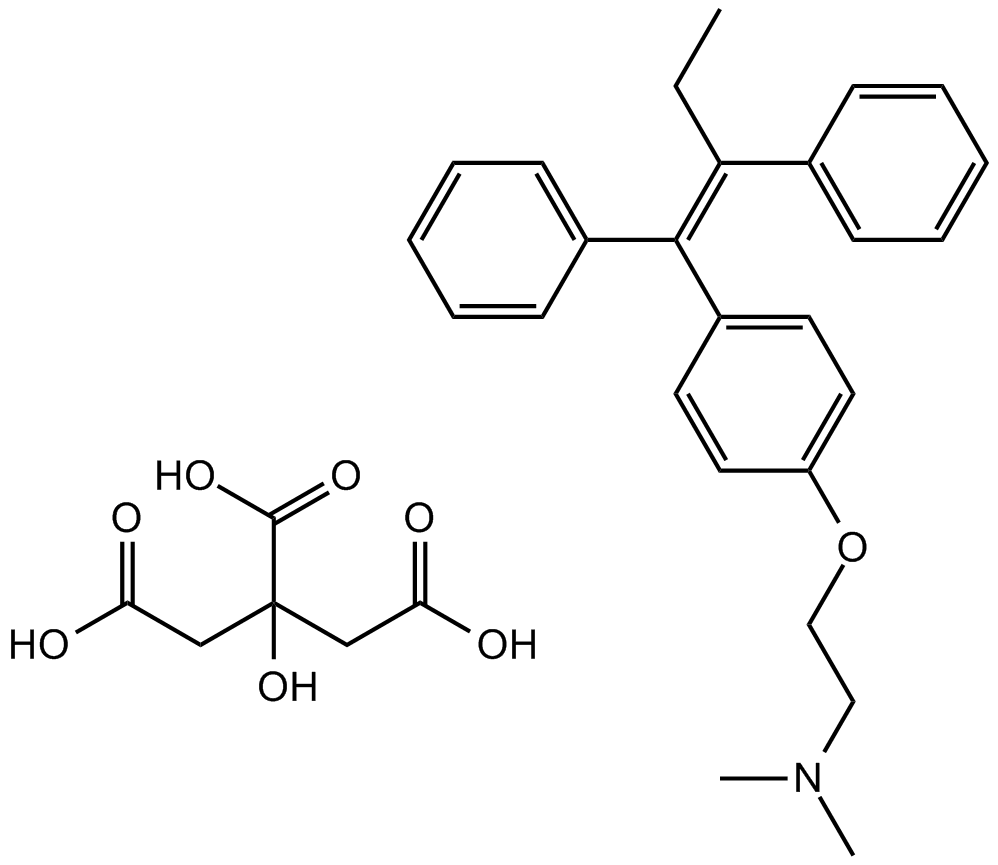
GC11669
Tamoxifen Citrate
Antiestrogen drug
-
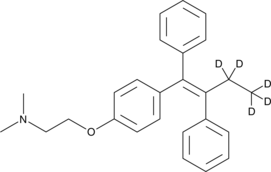
GC48124
Tamoxifen-d5
An internal standard for the quantification of tamoxifen
-
GC33286
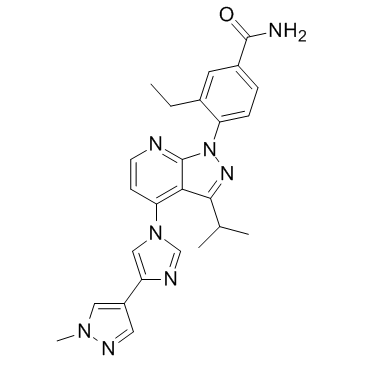
TAS-116
TAS-116 (TAS-116) is an oral bioavailable, ATP-competitive, highly specific HSP90α/HSP90β inhibitor (Kis of 34.7 nM and 21.3 nM, respectively) without inhibiting other HSP90 family proteins such as GRP94. TAS-116 demonstrates less ocular toxicity.
-
GC33663
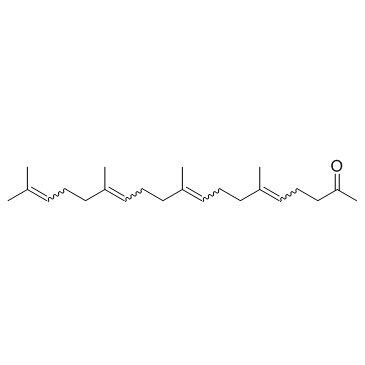
Teprenone (Geranylgeranylacetone)
Teprenone (Geranylgeranylacetone) is an anti-ulcer drug, and works as an inducer of heat shock proteins (HSPs).
-
GC10481
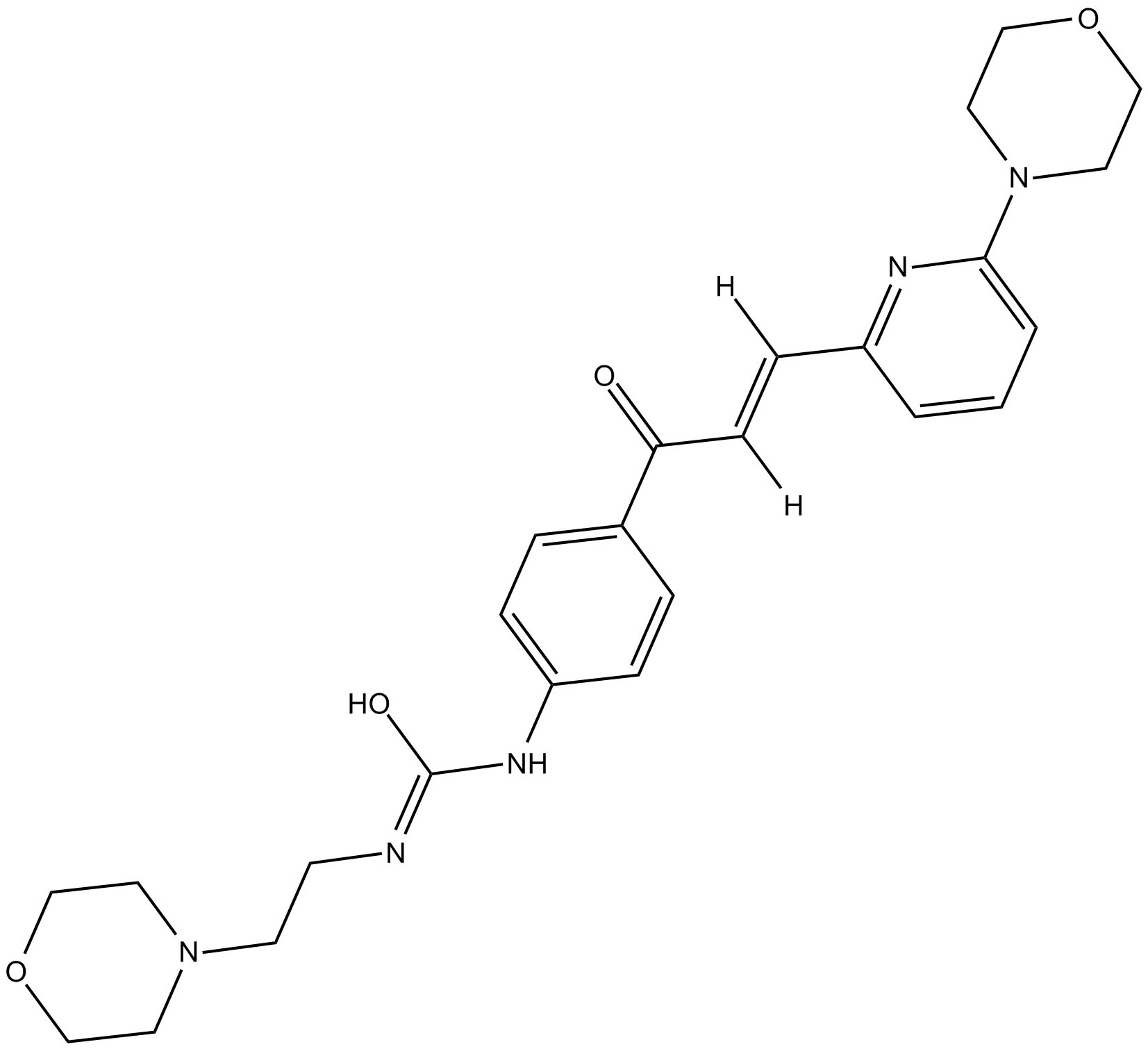
TRC 051384
HSP70 inducer
-
GC16966
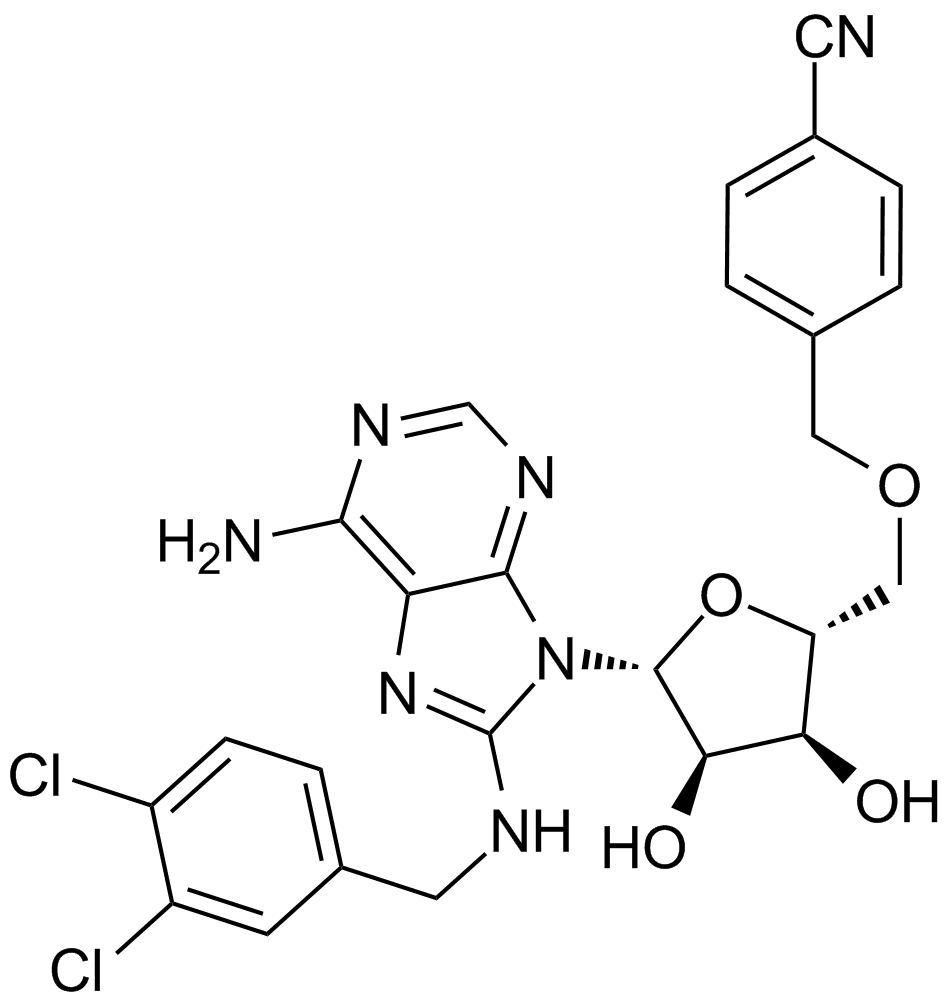
VER 155008
An Hsp70 inhibitor
-
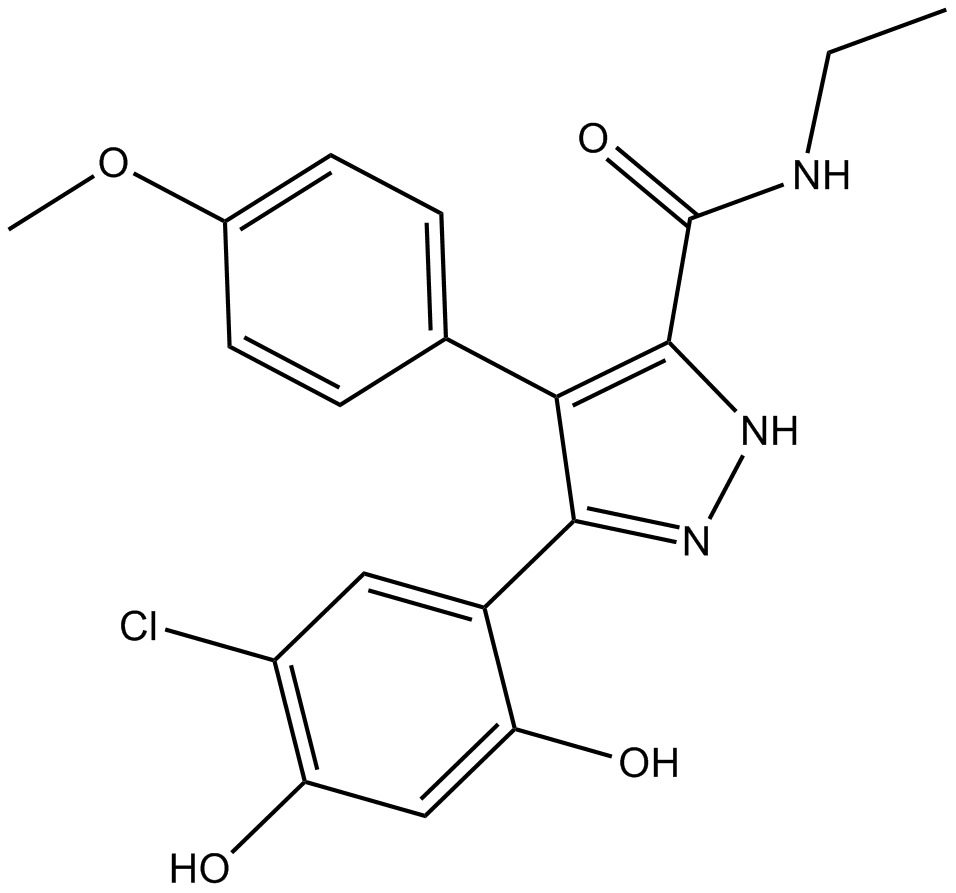
GC13556
VER-49009
HSP90 inhibitor, potent and selective
-
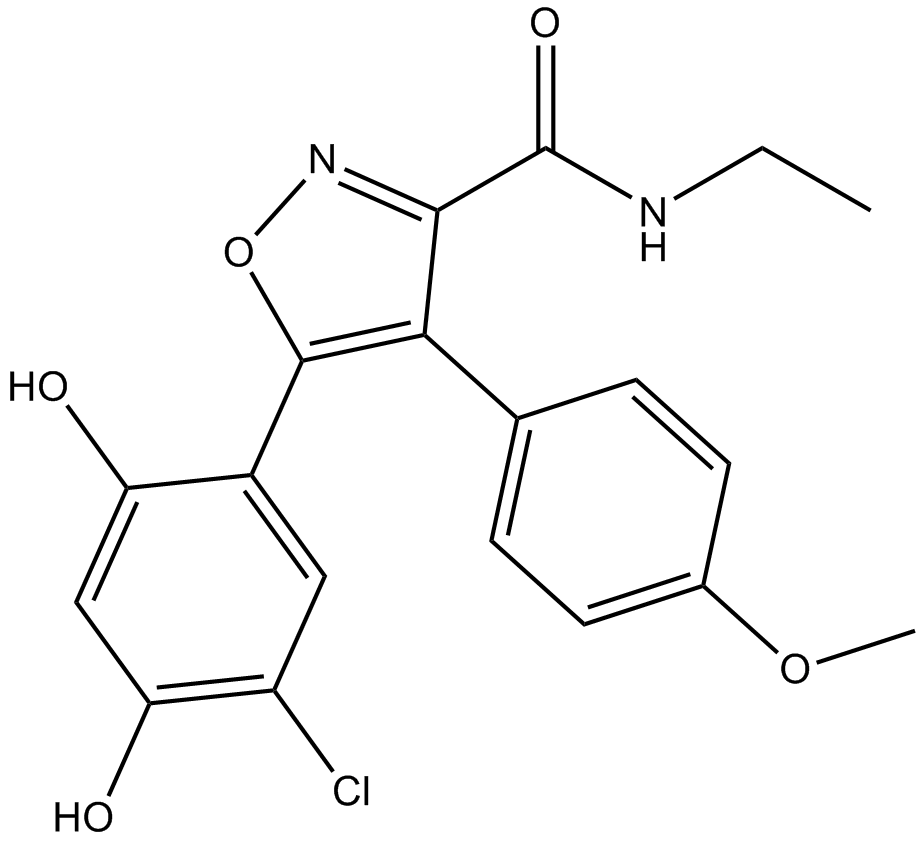
GC10501
VER-50589
HSP90 inhibitor, potent
-
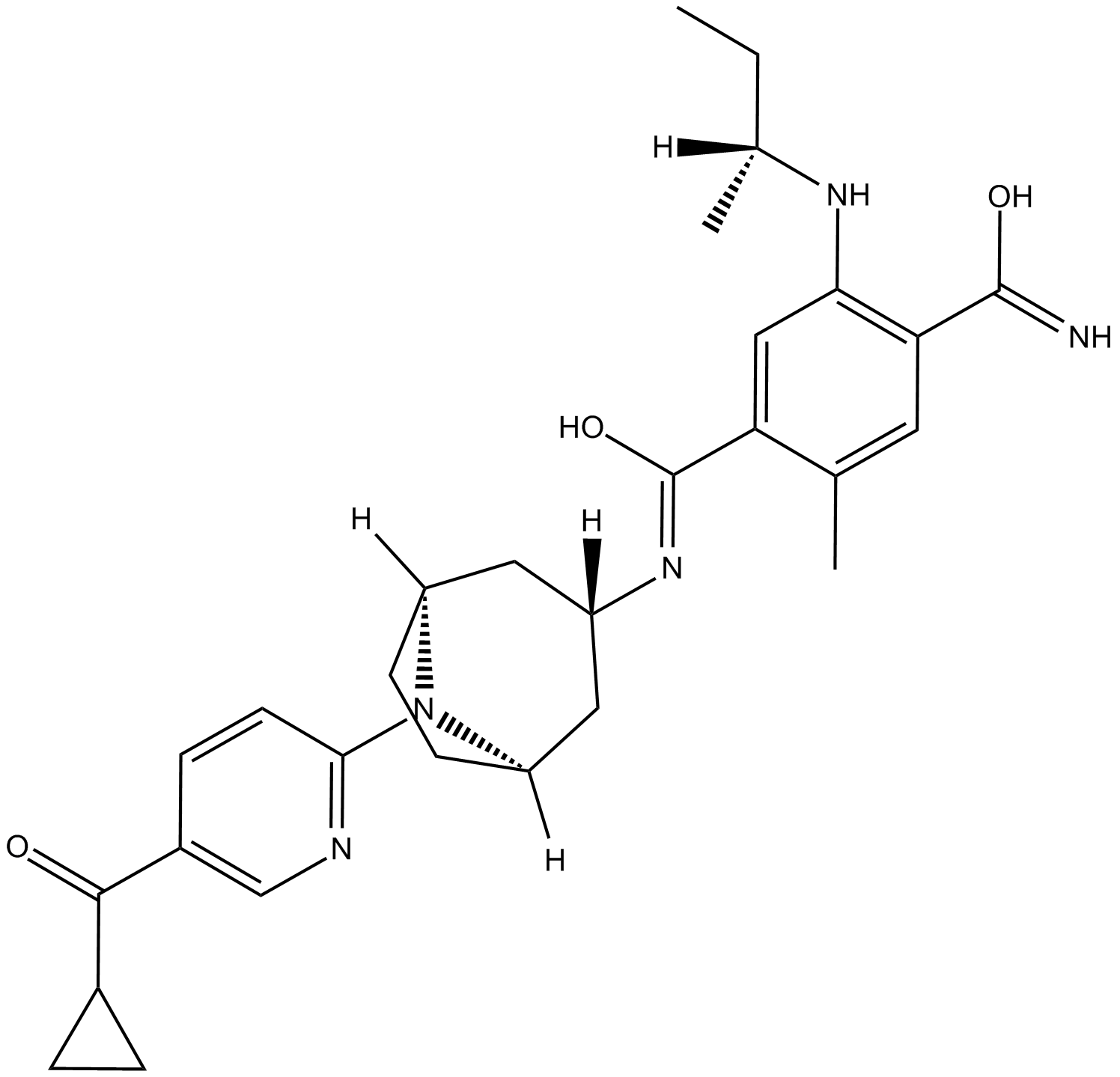
GC16892
XL-888
An orally bioavailable Hsp90 inhibitor
-
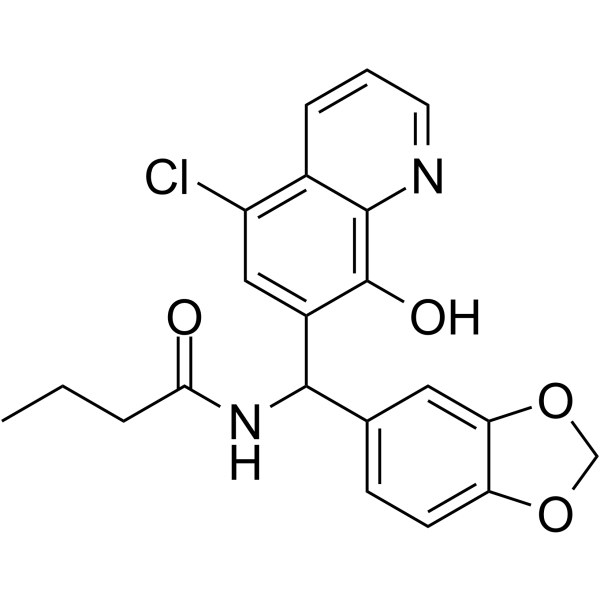
GC62388
YUM70
YUM70 is a potent and selective inhibitor of glucose-regulated protein 78 (GRP78), with an IC50 of 1.5 μM for inhibiting GRP78 ATPase activity of the full-length protein. YUM70 induces endoplasmic reticulum (ER) stress-mediated apoptosis in pancreatic cancer. YUM70 also has in vivo efficacy in a pancreatic cancer xenograft model.
-
GC66059
YZ129
YZ129 is an inhibitor of the HSP90-calcineurin-NFAT pathway against glioblastoma, directly binding to heat shock protein 90 (HSP90) with an IC50 of 820 nM on NFAT nuclear translocation. YZ129-induced GBM cell-cycle arrest at the G2/M phase promotes apoptosis and inhibited tumor cell proliferation and migration.