Mitophagy
Mitophagy is the selective degradation of mitochondria by autophagy.
Mitochondria are essential organelles that regulate cellular energy homeostasis and cell death. The removal of damaged mitochondria through autophagy, a process called mitophagy, is thus critical for maintaining proper cellular functions. Indeed, mitophagy has been recently proposed to play critical roles in terminal differentiation of red blood cells, paternal mitochondrial degradation, neurodegenerative diseases, and ischemia or drug-induced tissue injury.
Autophagy and mitophagy are important cellular processes that are responsible for breaking down cellular contents, preserving energy and safeguarding against accumulation of damaged and aggregated biomolecules.
Targets for Mitophagy
Products for Mitophagy
- Cat.No. Product Name Information
-
GC11720
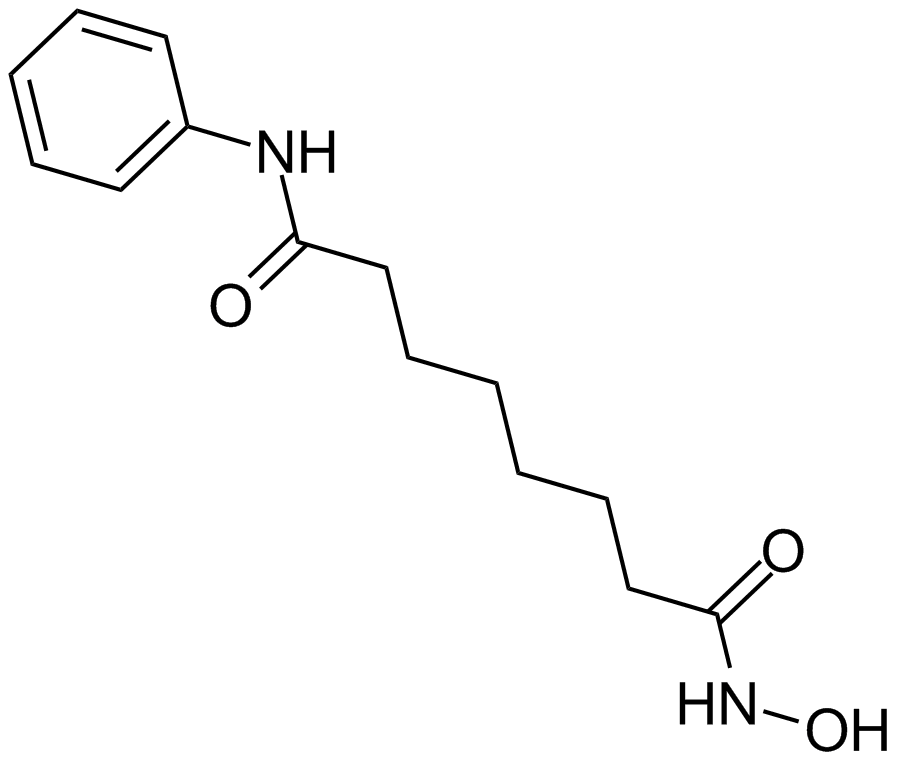
17-AAG (KOS953)
An inhibitor of Hsp90
-
GC10710
3-Methyladenine
3-Methyladenine is a classic autophagy inhibitor.
-
GC45356
5-Aminolevulinic Acid (hydrochloride)
-
GC16267
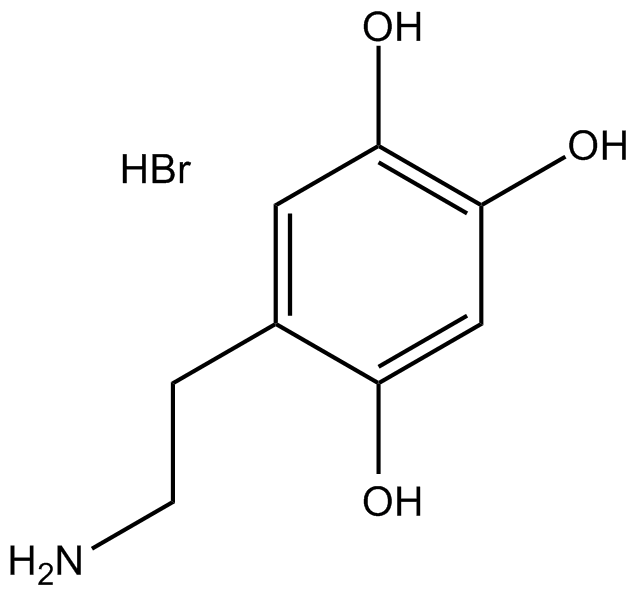
6-Hydroxydopamine hydrobromide
Oxidopamine (6-OHDA) hydrobromide is an antagonist of the neurotransmitter dopamine.
-
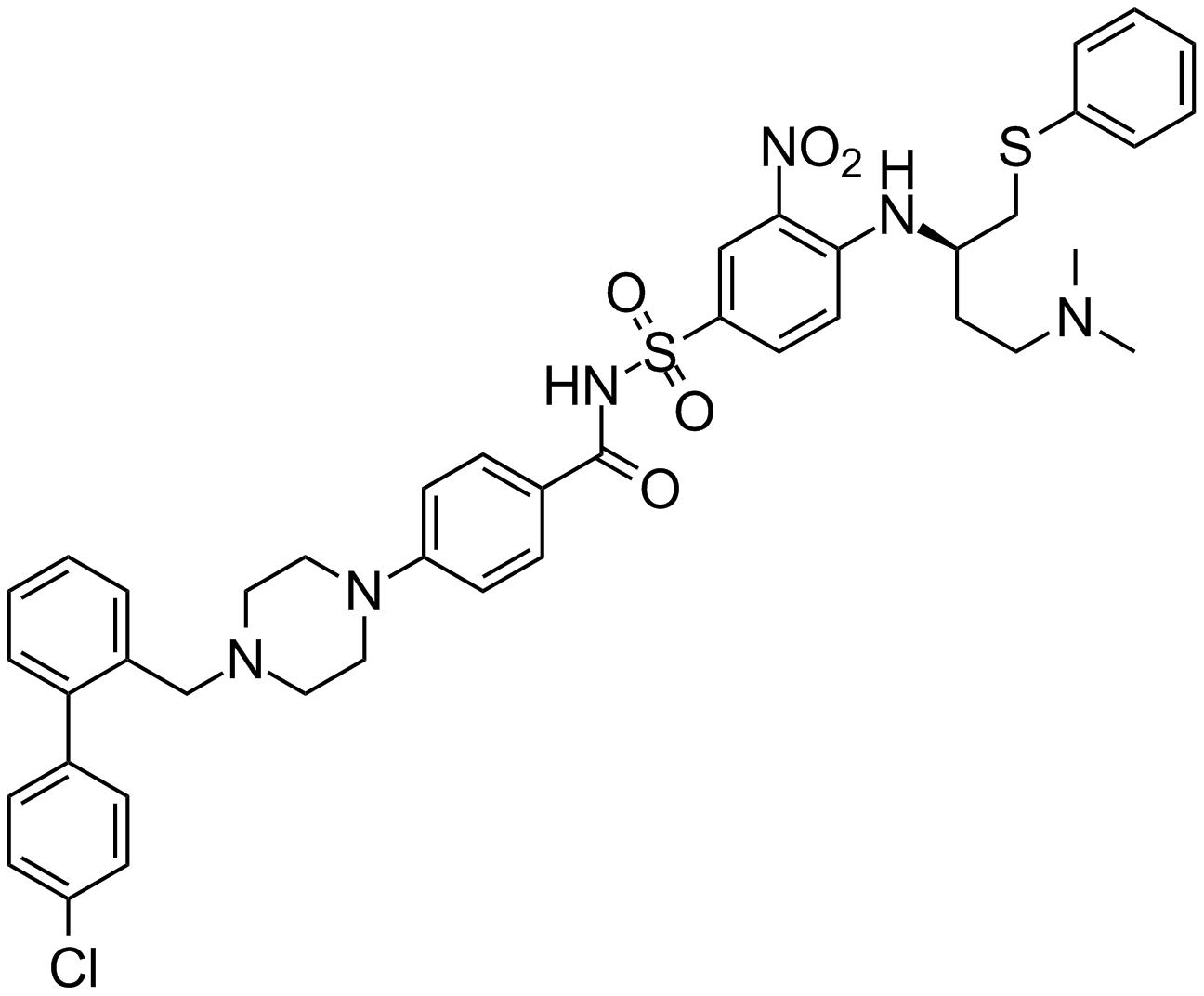
GC17234
ABT-737
An inhibitor of anti-apoptotic Bcl-2 proteins
-
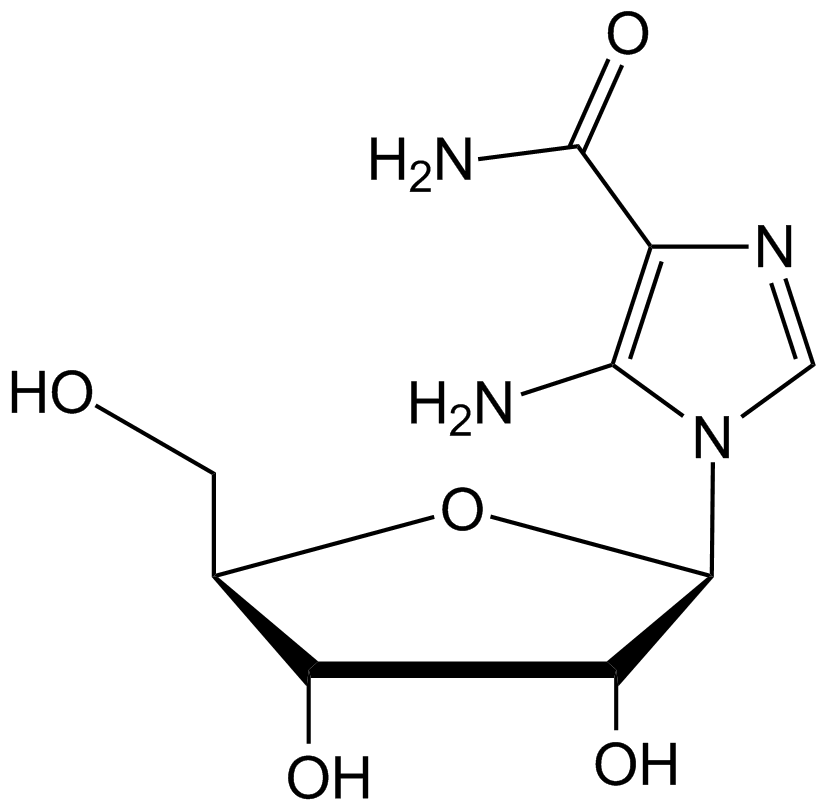
GC10518
AICAR
An activator of AMPK
-
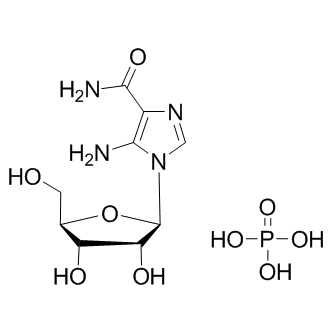
GC10580
AICAR phosphate
AMPK activator
-
GC15706
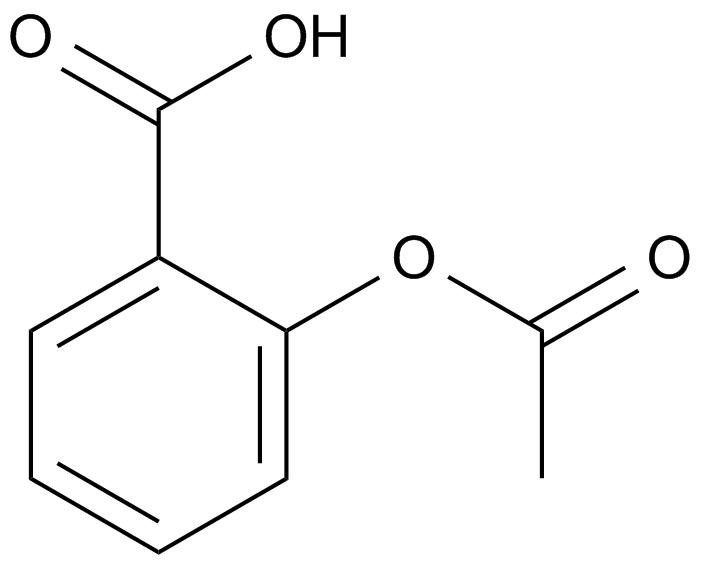
Aspirin (Acetylsalicylic acid)
A non-selective, irreversible COX inhibitor
-
GC62135
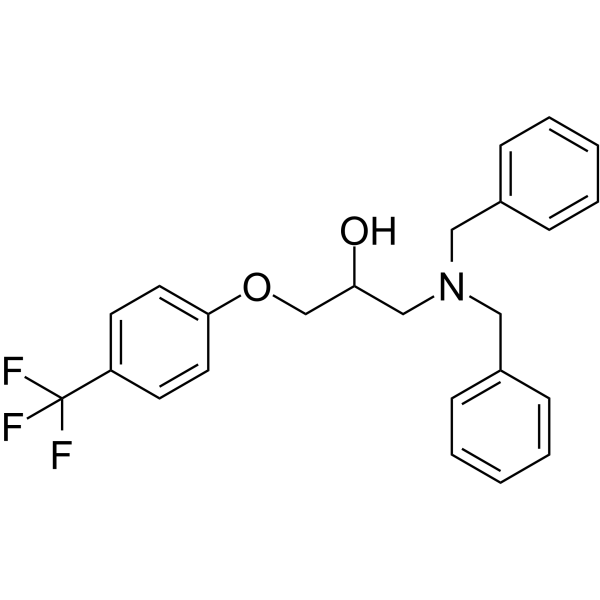
BC1618
BC1618, an orally active Fbxo48 inhibitory compound, stimulates Ampk-dependent signaling (via preventing activated pAmpkα from Fbxo48-mediated degradation).
-
GC10480
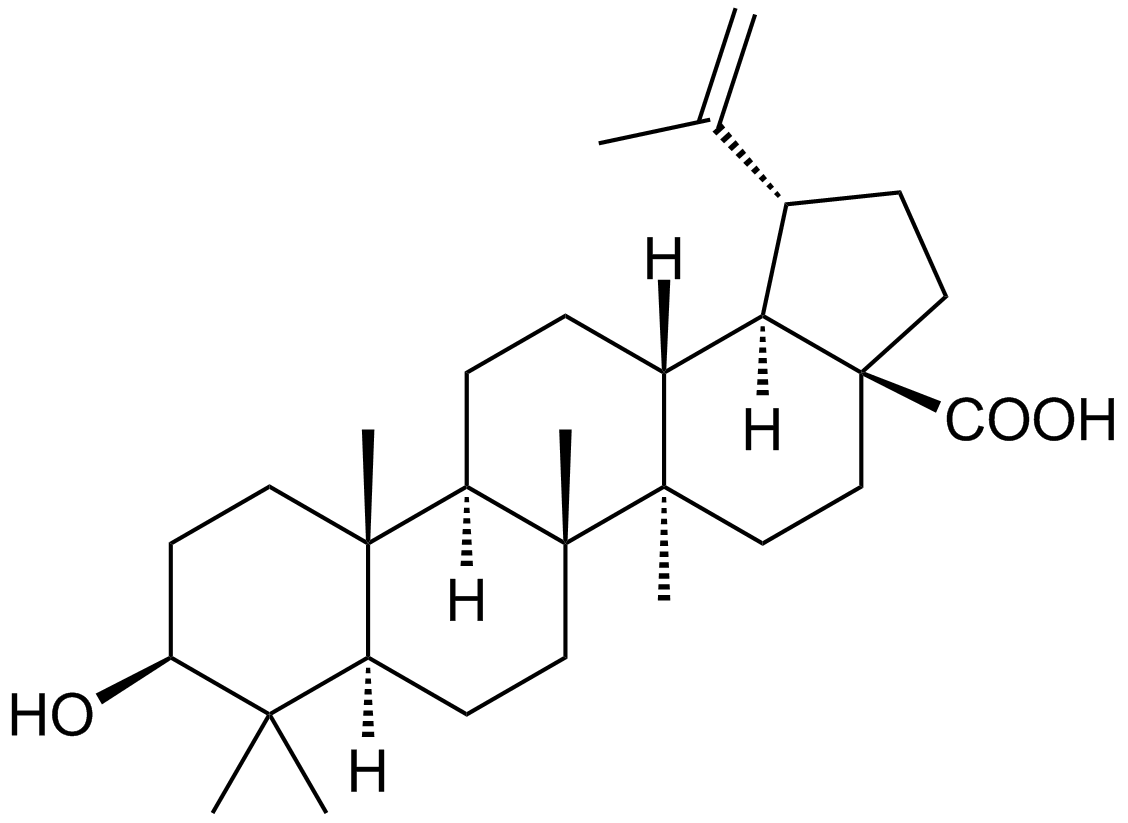
Betulinic acid
A plant triterpenoid similar to bile acids
-
GC17683
Brefeldin A
Brefeldin A (BFA) is a fungal macrocyclic lactone and a potent, reversible inhibitor of intracellular vesicle formation and protein trafficking between the endoplasmic reticulum (ER) and the Golgi apparatus.
-
GC17340
Carbamazepine
Inhibitor of neuronal voltage-gated Na+ channels
-
GC15083
Celastrol
A triterpenoid antioxidant
-
GC32078
Clioquinol (Iodochlorhydroxyquin)
Clioquinol (Iodochlorhydroxyquin) (Iodochlorhydroxyquin) is a topical antifungal agent with anticancer activity.
-
GC30360
Cresol (Cresol mixture of isomers)
-
GC14787
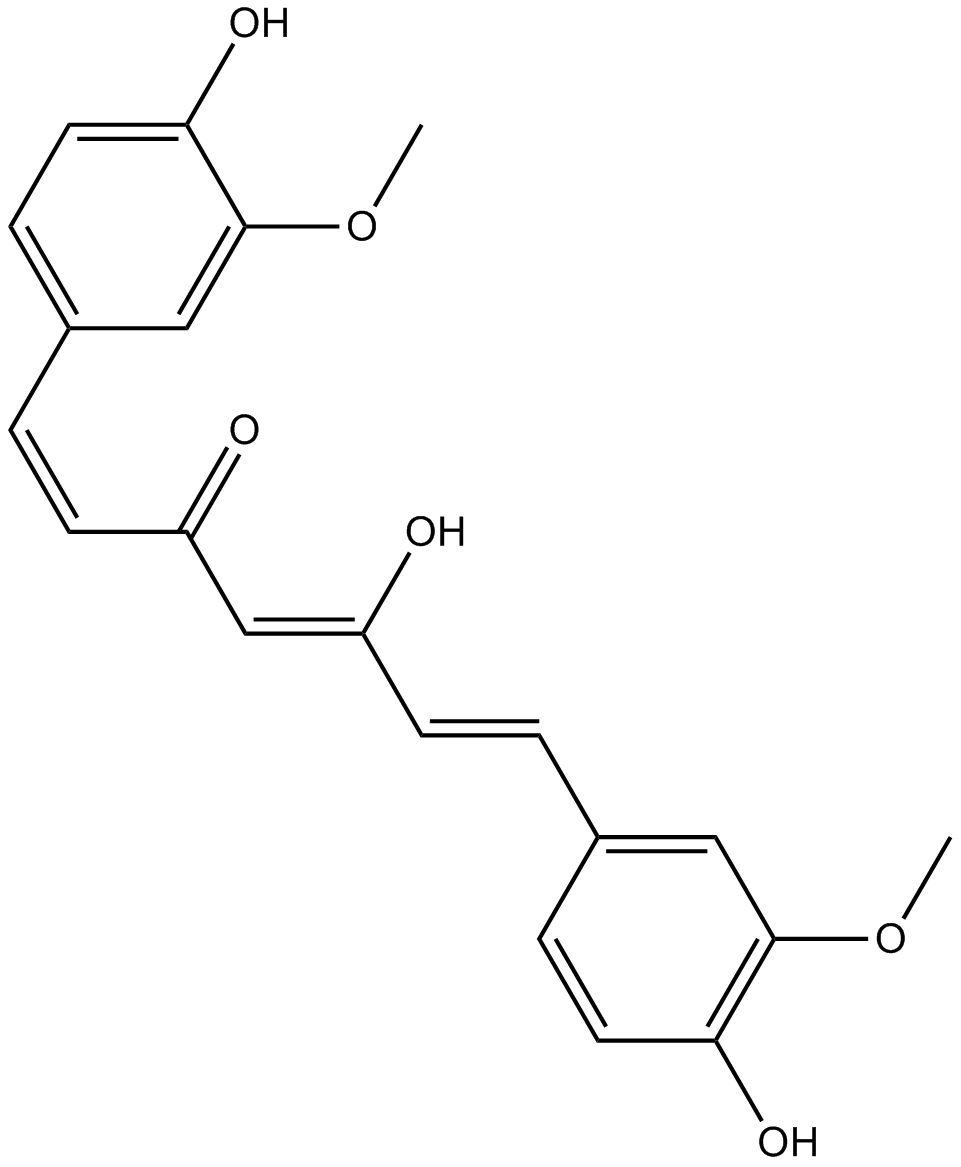
Curcumin
A yellow pigment with diverse biological activities
-
GC40226
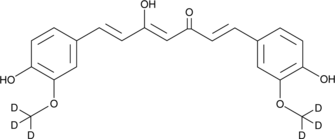
Curcumin-d6
Curcumin-d6 is intended for use as an internal standard for the quantification of curcumin by GC- or LC-MS.
-
GC13554
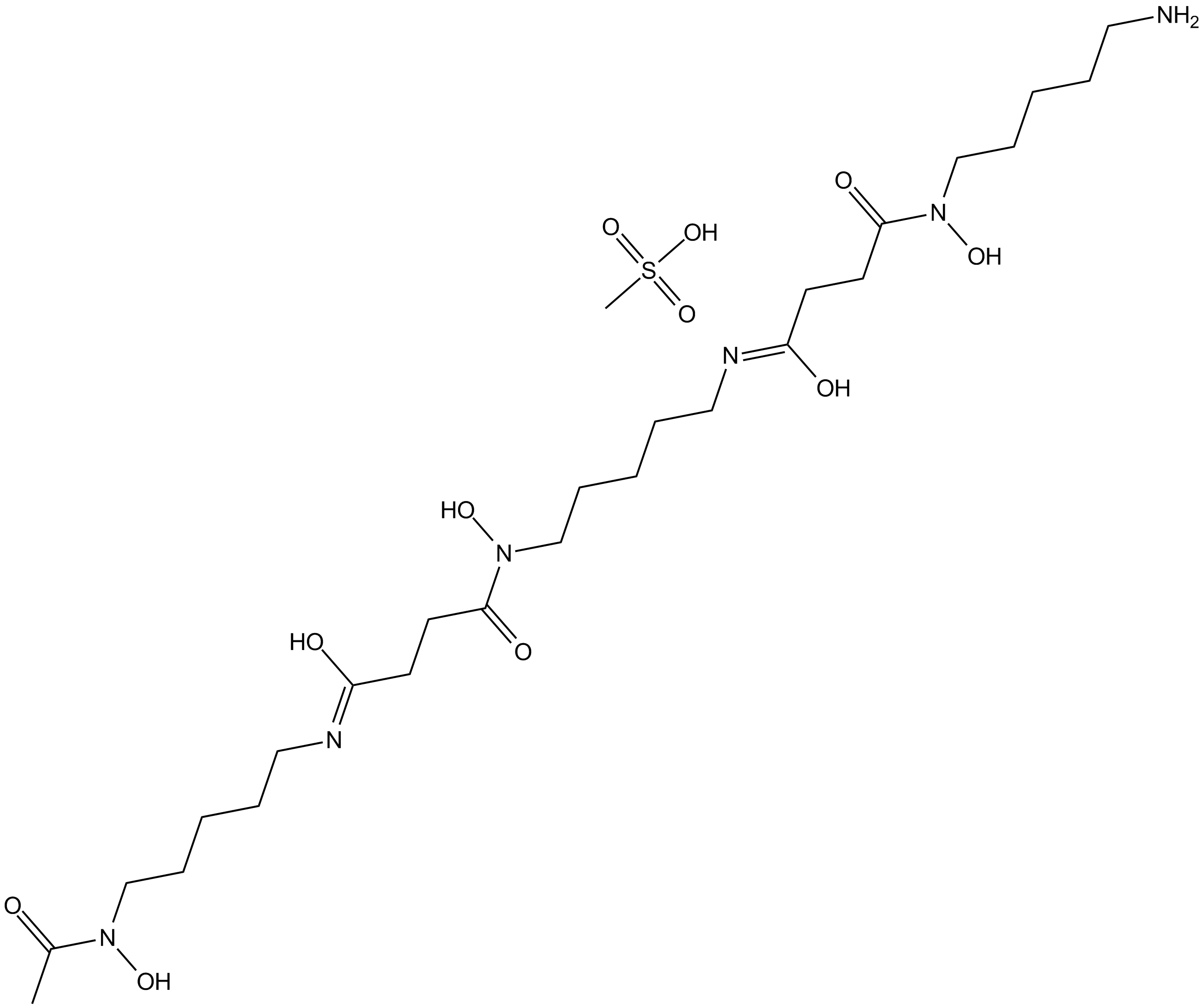
Deferoxamine mesylate
Deferoxamine mesylate is a drug that chelates iron by forming a stable complex that prevents the iron from entering into further chemical reactions, and is used for the treatment of chronic iron overload in patients with transfusion-dependent anemias.
-
GC40775
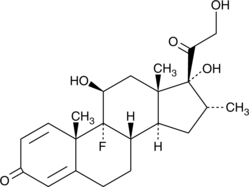
Dexamethasone
Dexamethasone, as one member of the glucocorticoid family, can protect against arthritis-related changes in cartilage structure and function, including matrix loss, inflammation and cartilage viability.
-
GC11237
Dexamethasone acetate
IL Receptor modulator
-
GC13443
Doxazosin Mesylate
α1-adrenergic receptor antagonist
-
GC16994
Doxorubicin
Doxorubicin(DOX), also known as adriamycin, is a compound of the anthracycline class that has the broadest spectrum of activity[1].
-
GC17567
Doxorubicin (Adriamycin) HCl
Doxorubicin (Hydroxydaunorubicin) hydrochloride, a cytotoxic anthracycline antibiotic, is an anti-cancer chemotherapy agent.
-
GC14968
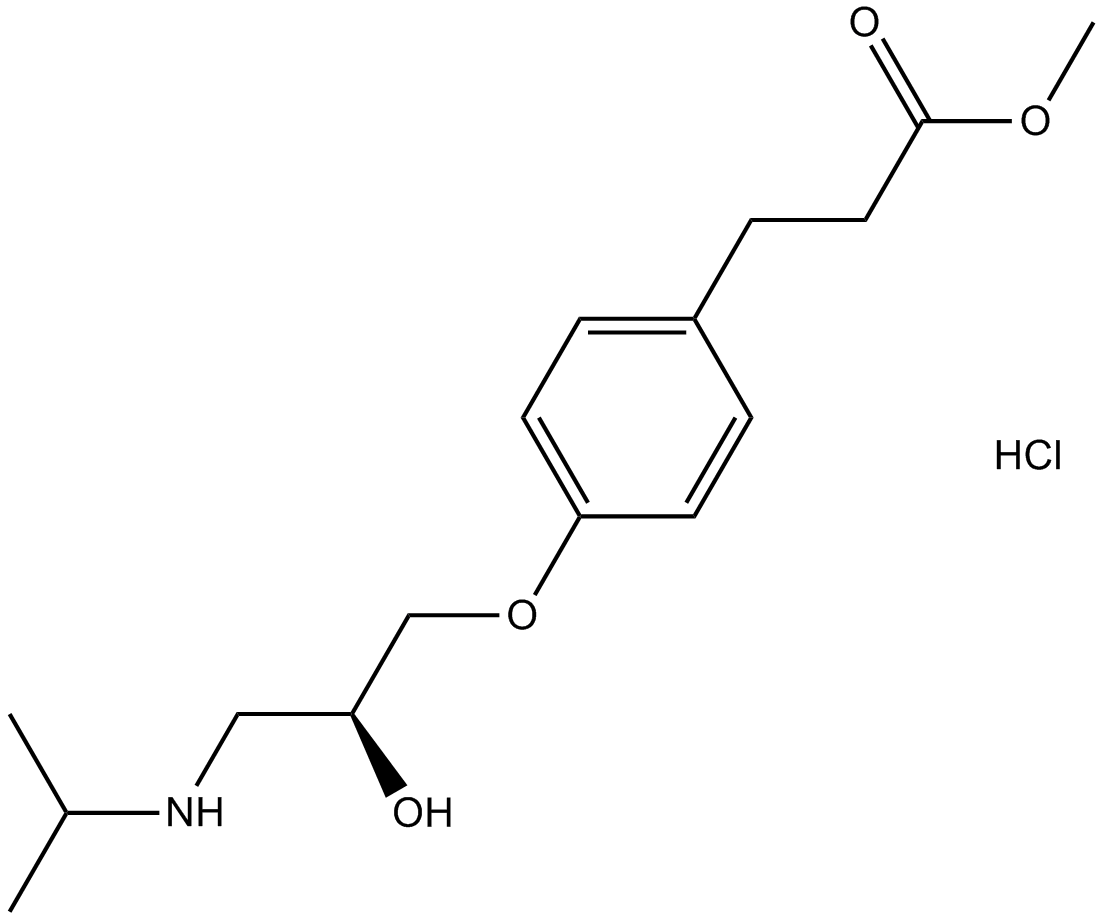
Esmolol HCl
Esmolol HCl is a beta adrenergic receptor blocker.
-
GC15617
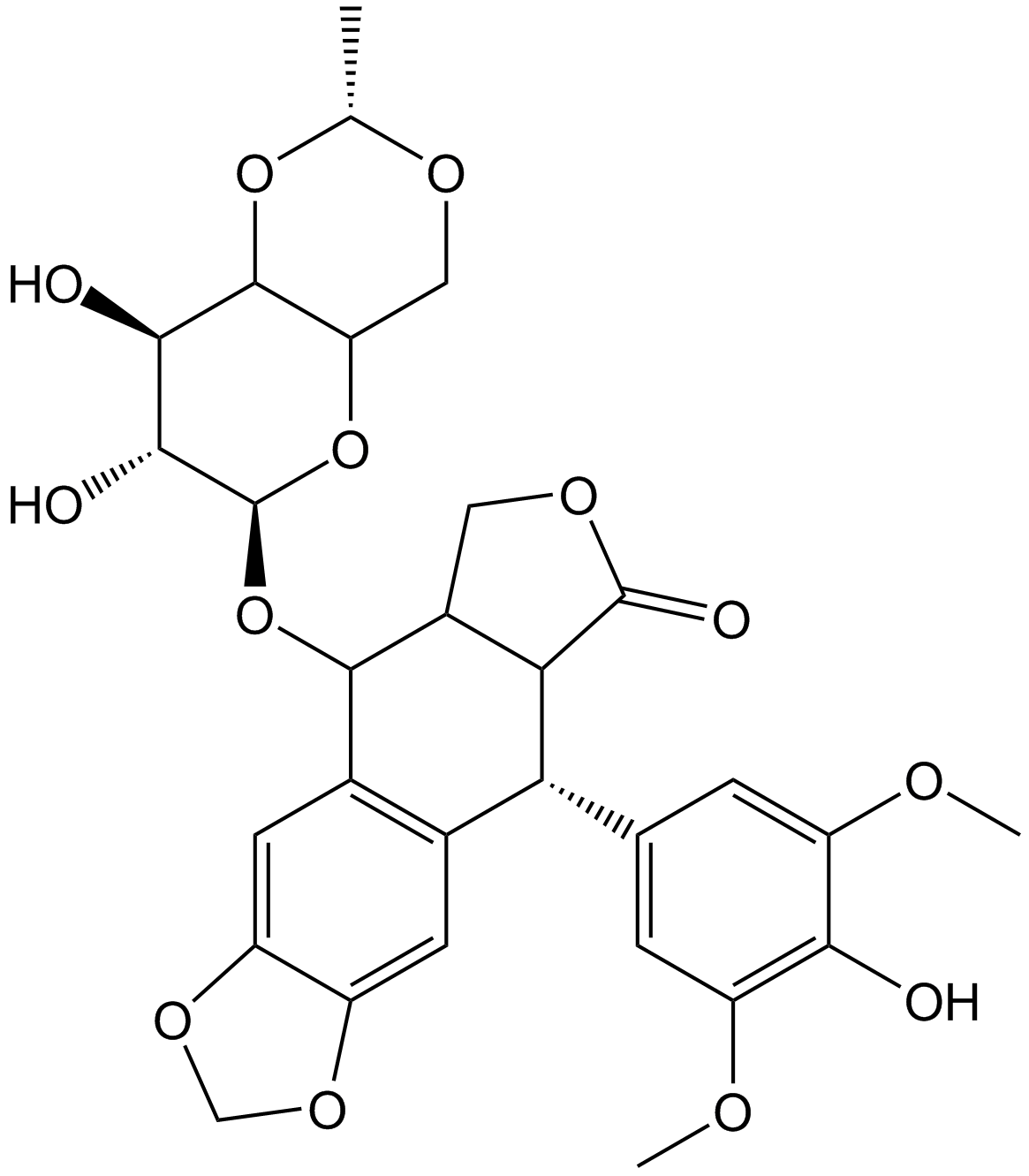
Etoposide
Topo II inhibitor
-
GN10156
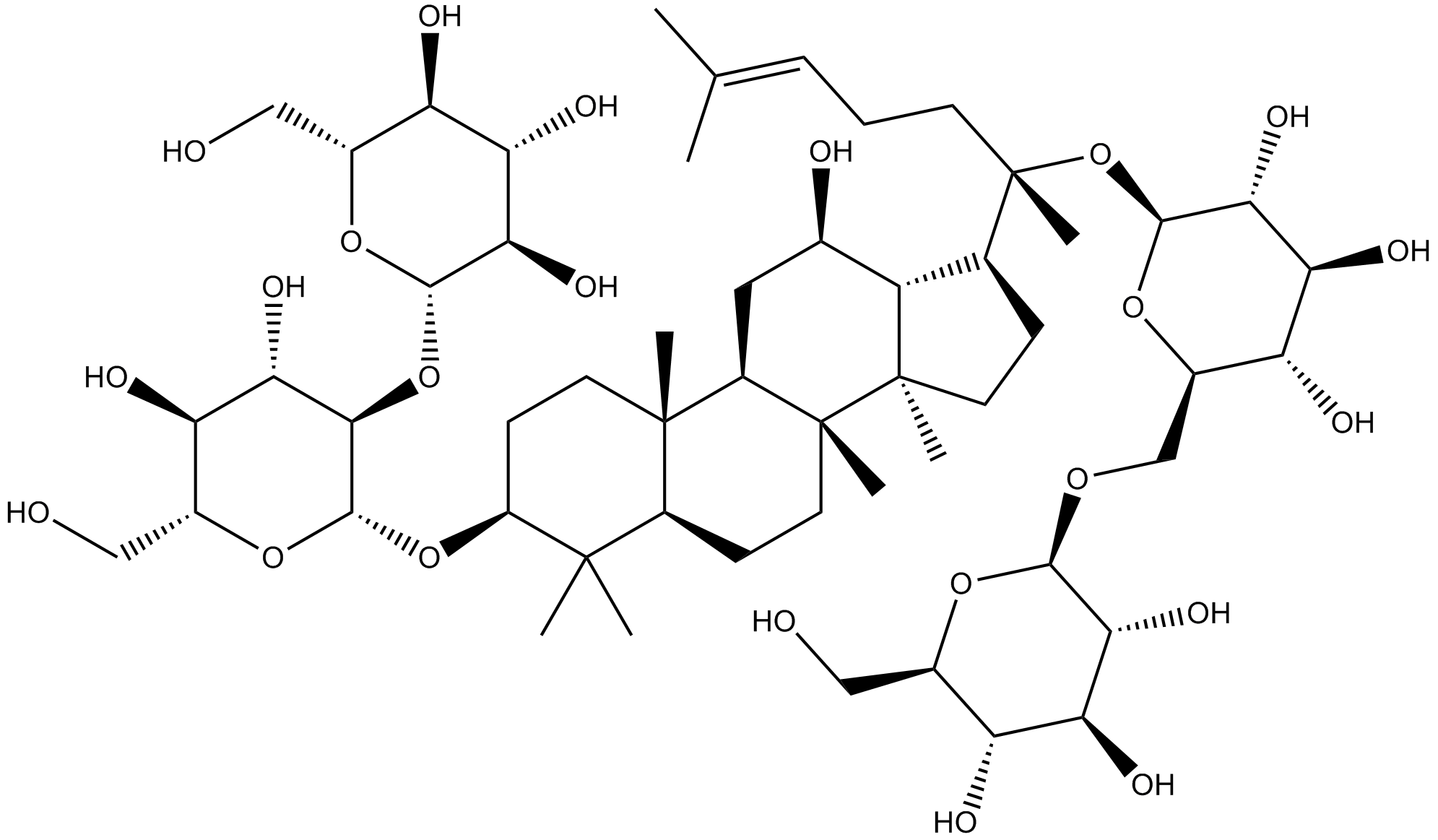
Ginsenoside Rb1
-
GC18049
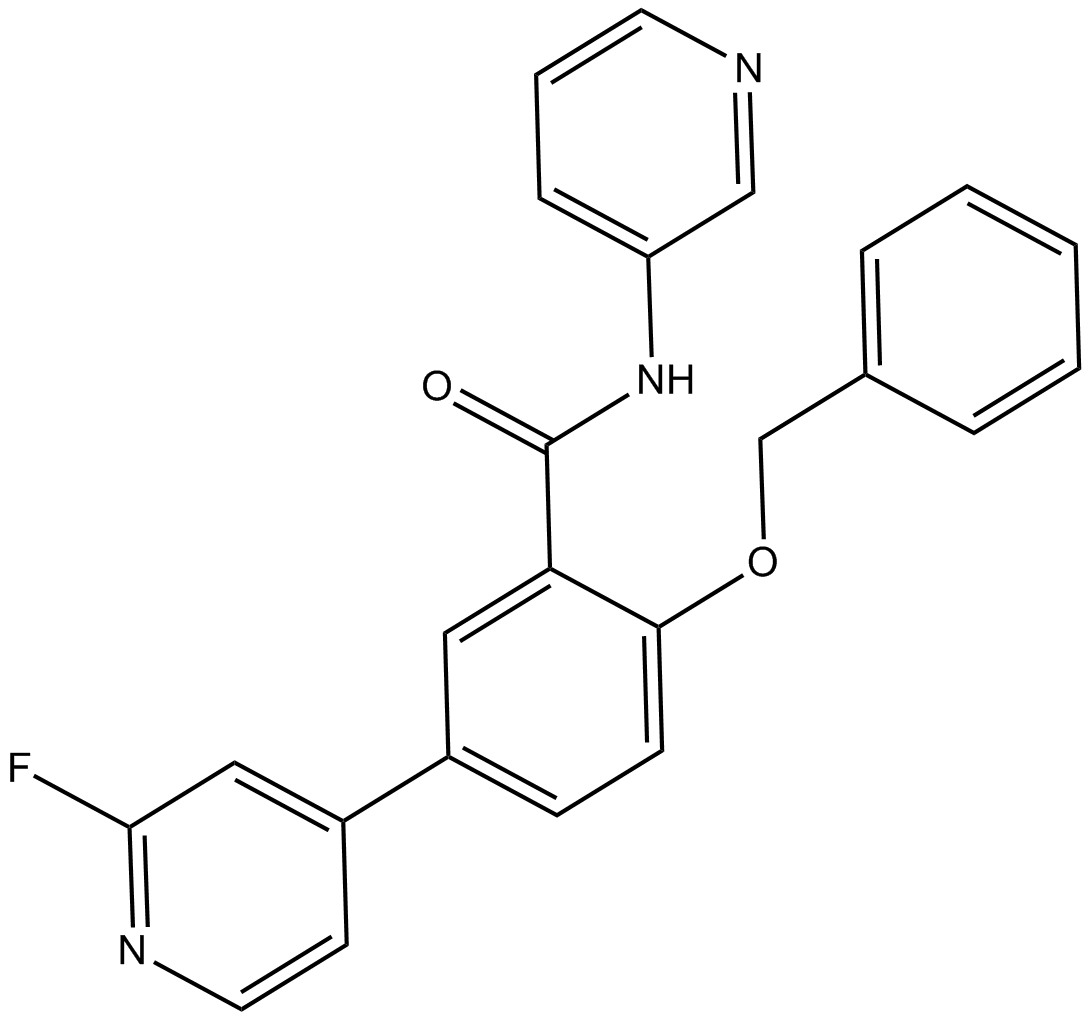
GSK2578215A
LRRK2 inhibitor
-
GA10942
H-D-Gln-OH
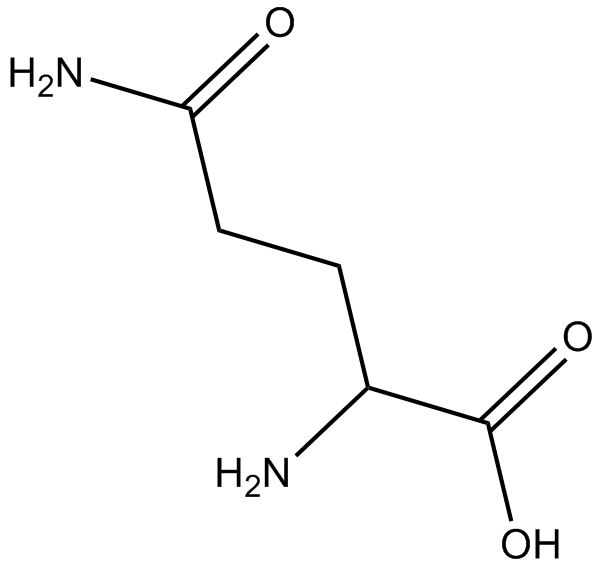
-
GC14591
Hemin chloride
Hemin chloride is an iron-containing porphyrin.
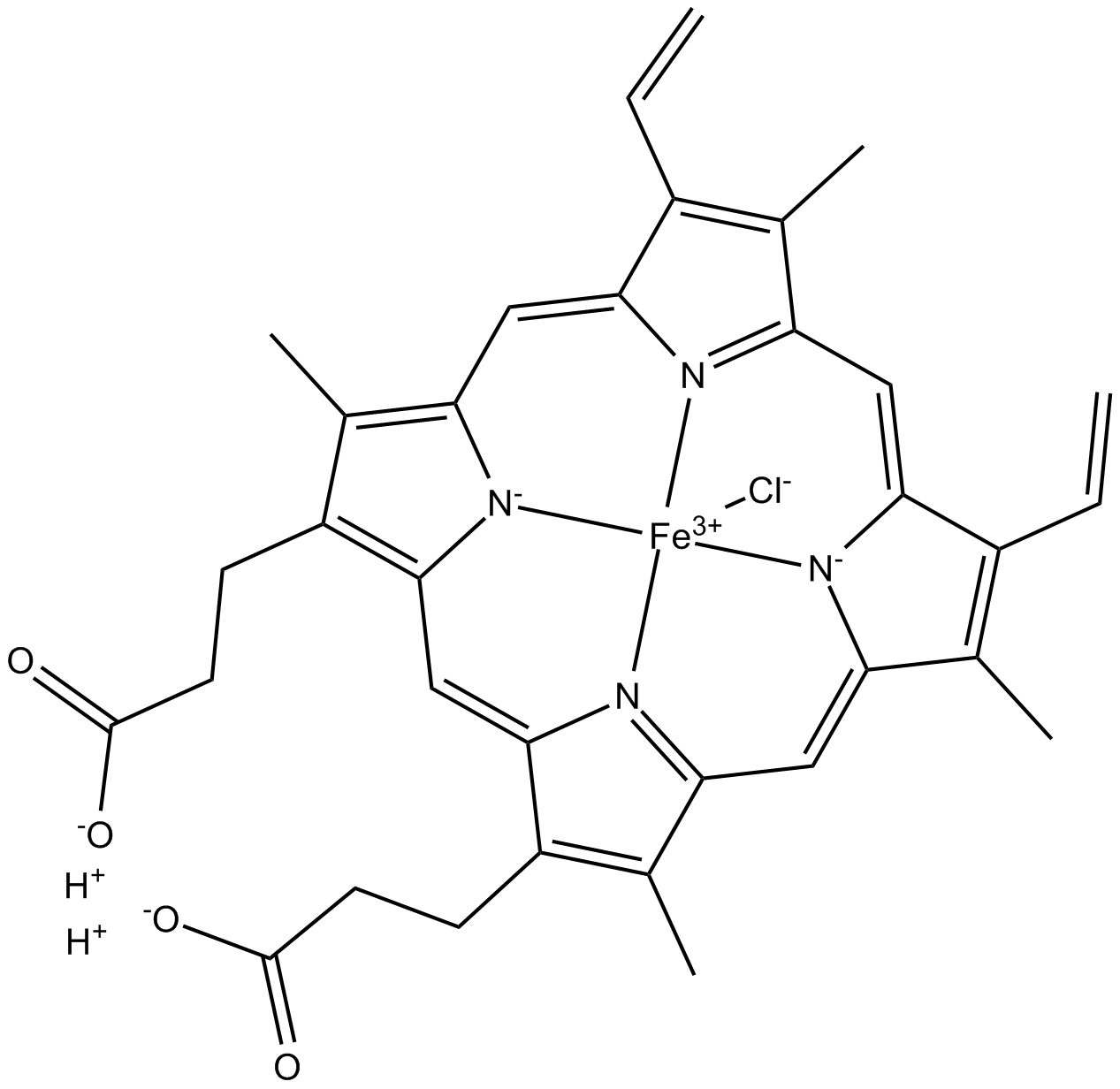
-
GC16105
Iohexol
contrast agent
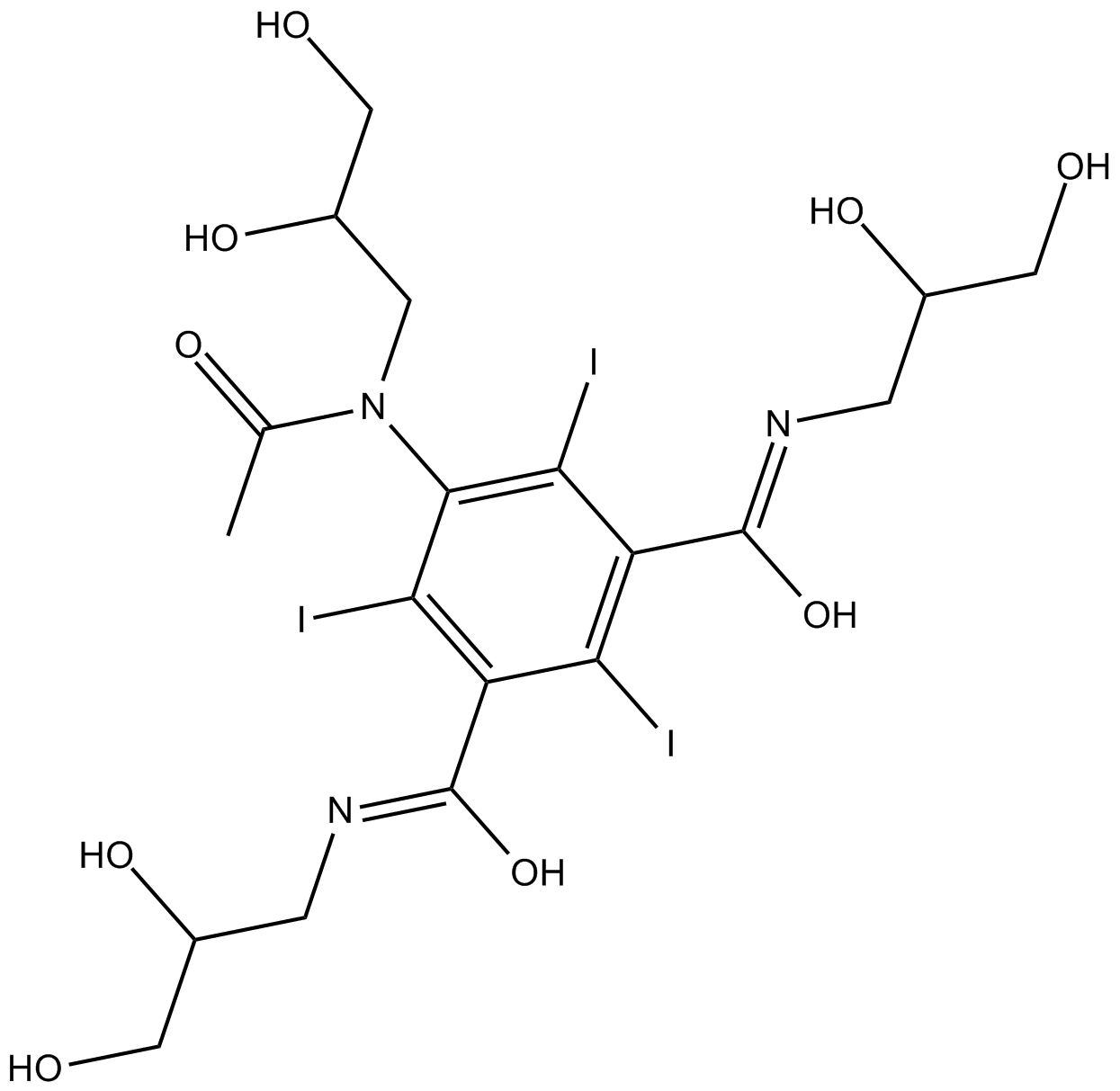
-
GC12017
Isoniazid
antibiotic used for the treatment of tuberculosis
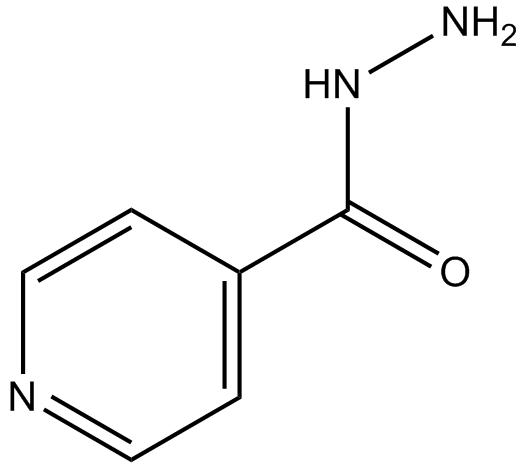
-
GC64098
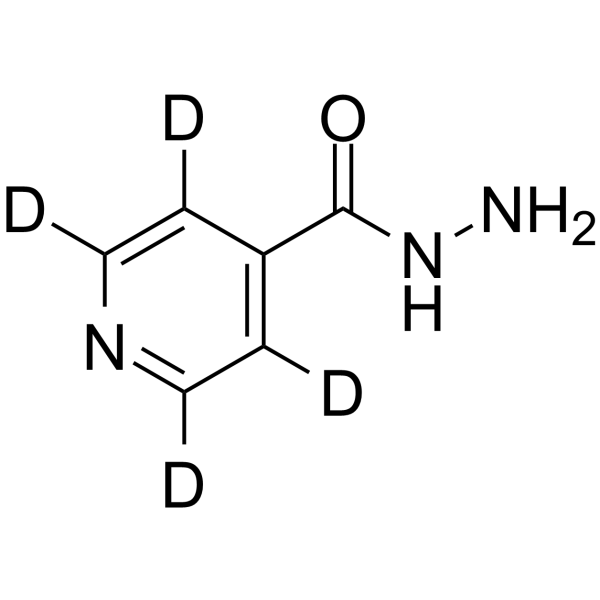
Isoniazid-d4
-
GC12339
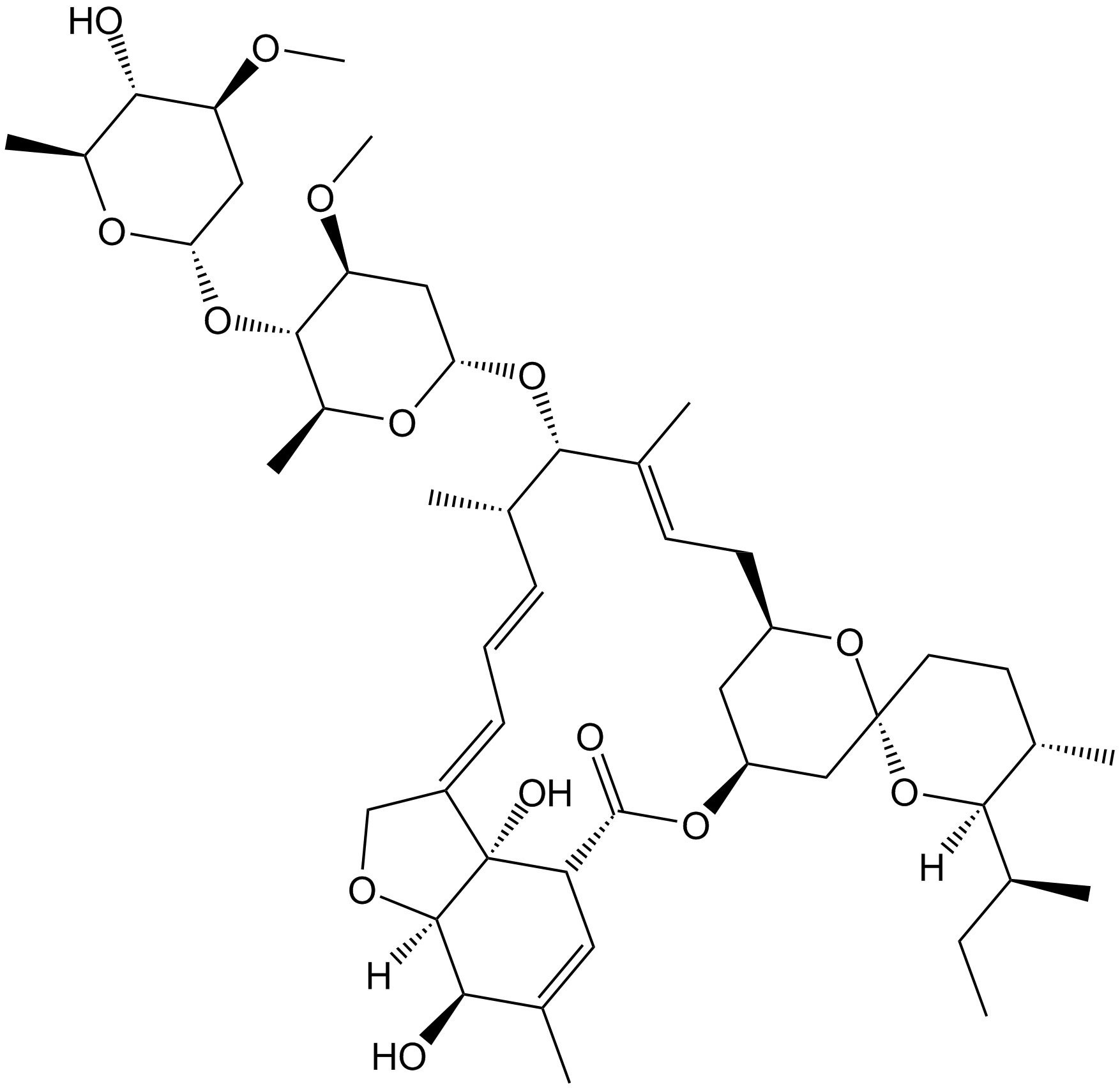
Ivermectin
NAChR/purinergic P2X4 receptor modulator
-
GC38150
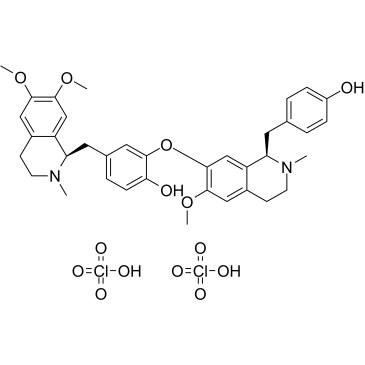
Liensinine Diperchlorate
Liensinine Diperchlor?ate is a major isoquinoline alkaloid, extracted from the seed embryo of Nelumbo nucifera Gaertn.
-
GC17874
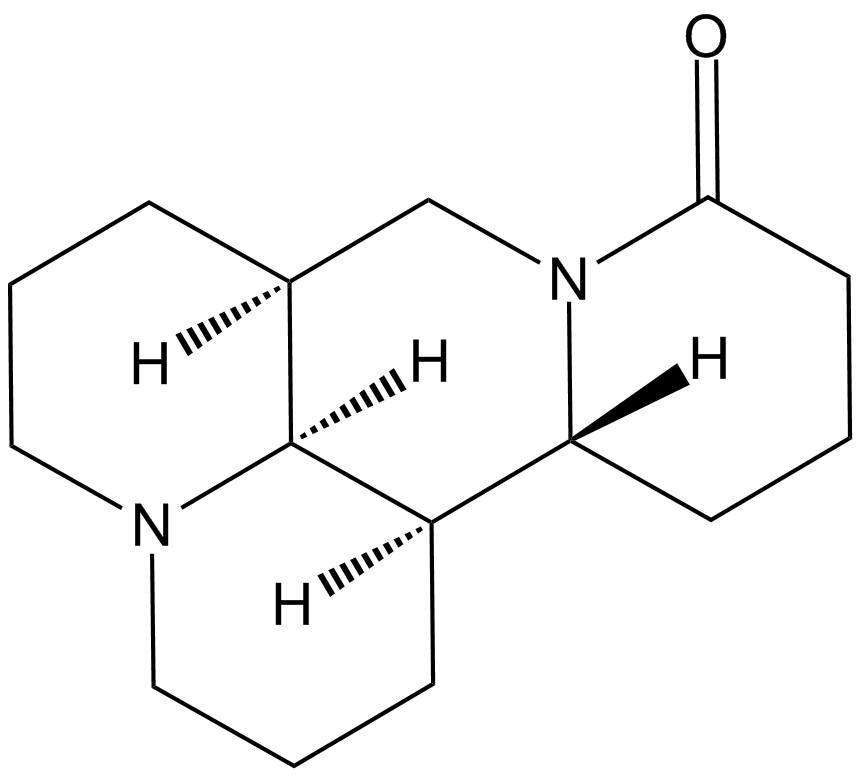
Matrine
An alkaloid with diverse biological activities
-
GC10200
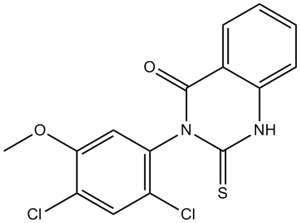
Mdivi 1
A mitochondrial division inhibitor
-
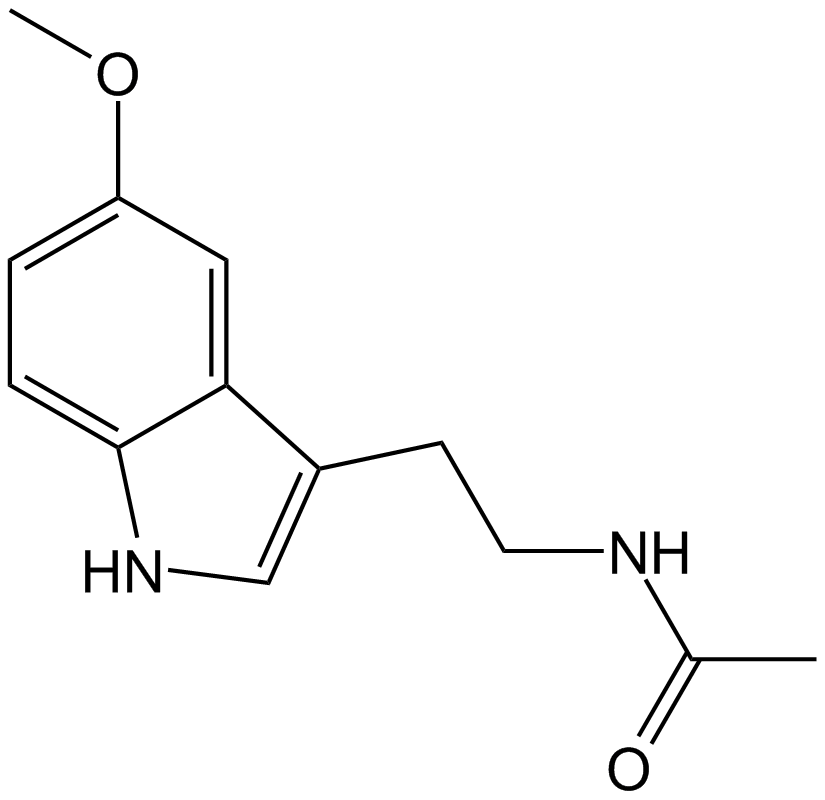
GC15618
Melatonin
Melatonin receptors agonist
-
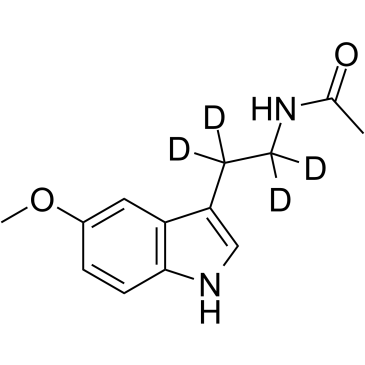
GC60241
Melatonin D5
An internal standard for the quantification of melatonin
-
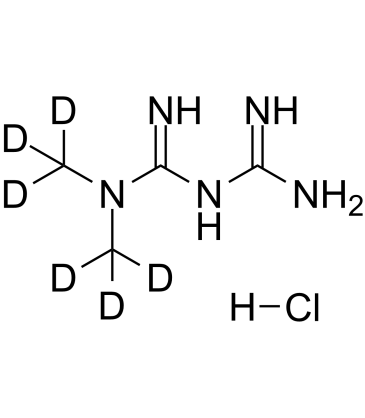
GC61045
Metformin D6 hydrochloride
An internal standard for the quantification of metformin
-
GC17443
Metformin HCl
Metformin HCl (1,1-Dimethylbiguanide hydrochloride) inhibits the mitochondrial respiratory chain in the liver, leading to activation of AMPK, enhancing insulin sensitivity for type 2 diabetes research.
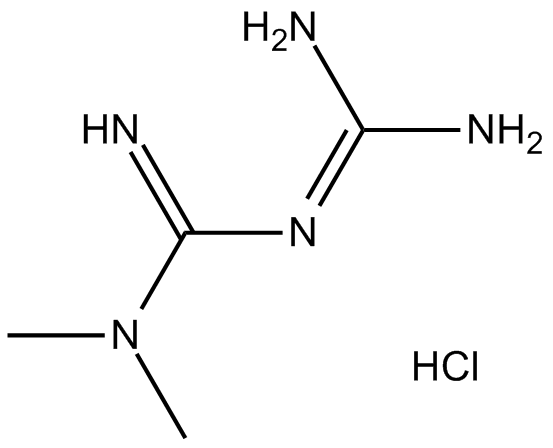
-
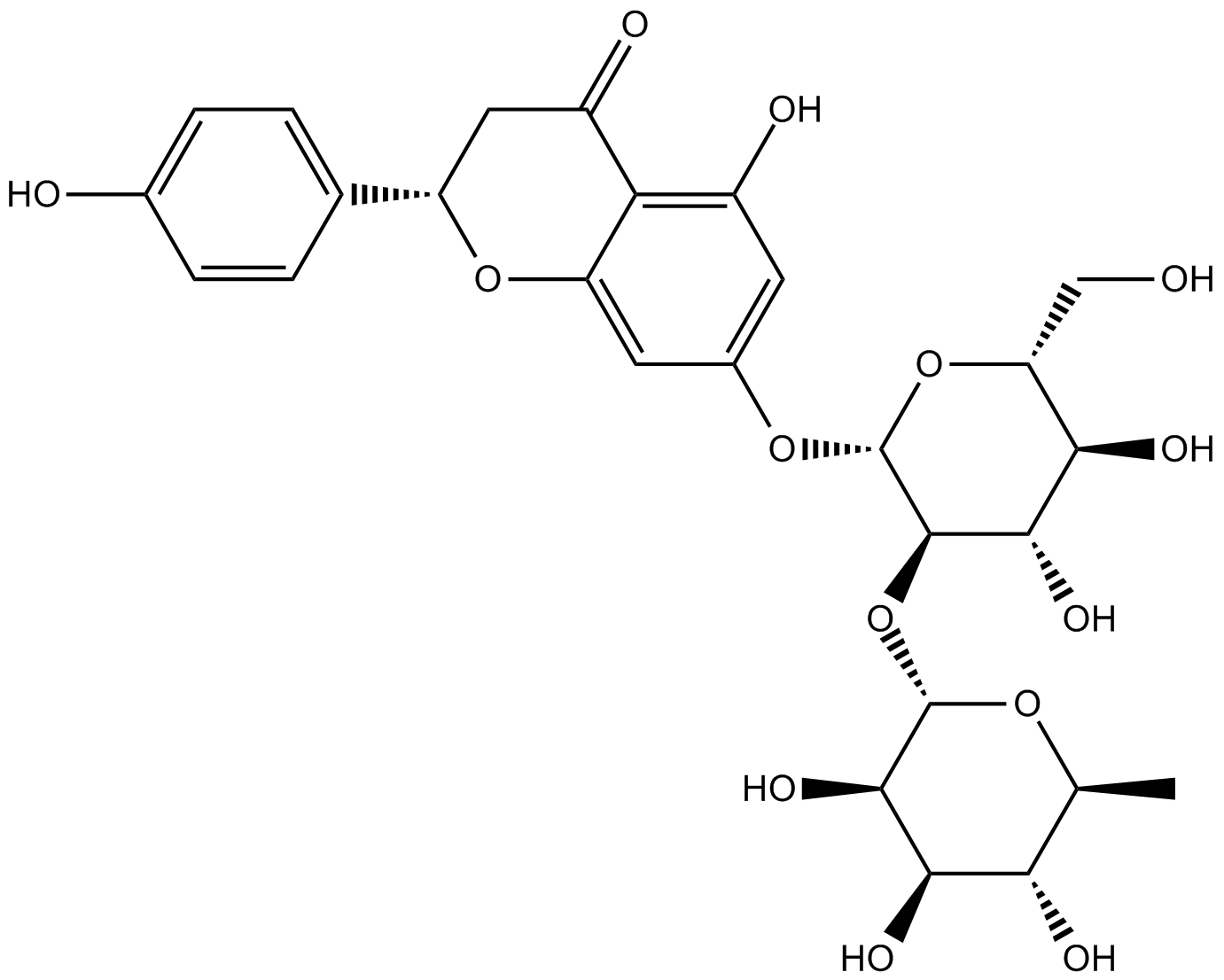
GN10257
Naringin
-
GC12495
Olanzapine
Olanzapine (LY170053) is a selective, orally active monoaminergic antagonist with high affinity binding to serotonin H1, 5HT2A/2C, 5HT3, 5HT6 (Ki=7, 4, 11, 57, and 5 nM, respectively), dopamine D1-4 (Ki=11 to 31 nM), muscarinic M1-5 (Ki=1.9-25 nM), and adrenergic α1 receptor (Ki=19 nM).
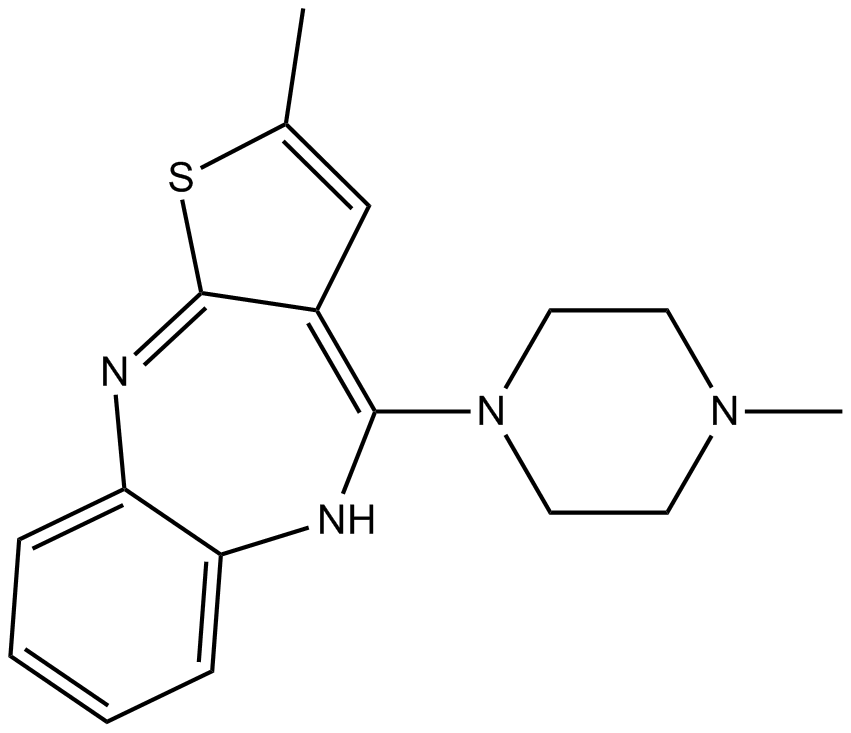
-
GC61809
Olanzapine D3
Olanzapine D3 (LY170053-d3) is the deuterium labeled Olanzapine.
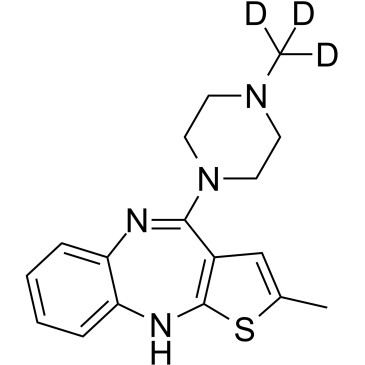
-
GC17580
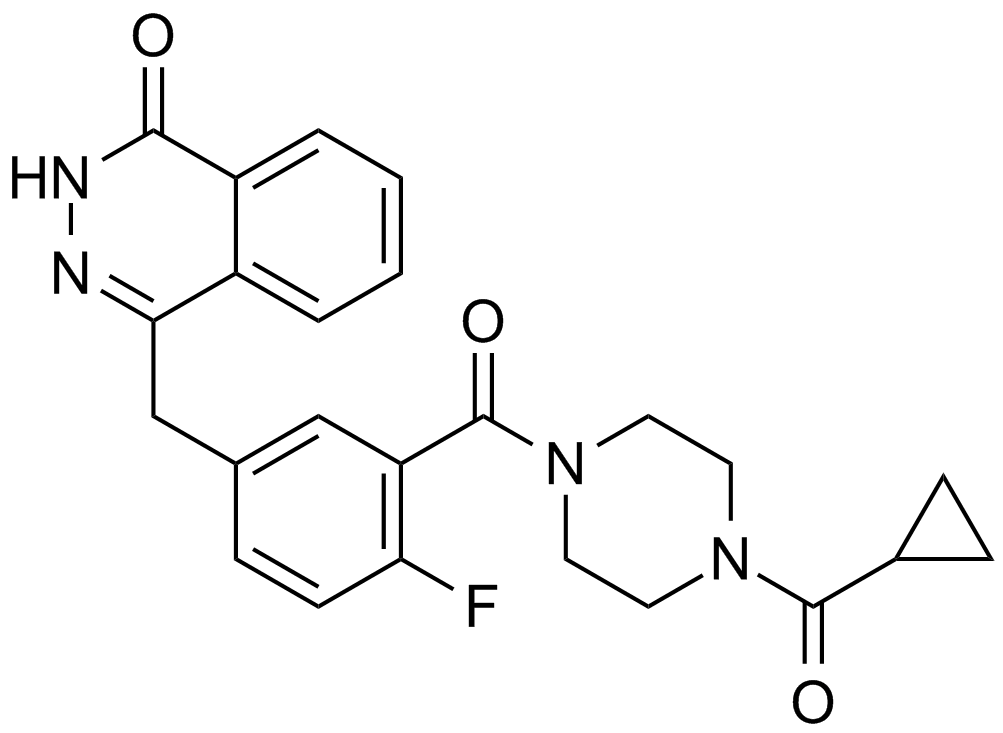
Olaparib (AZD2281, Ku-0059436)
A PARP inhibitor
-
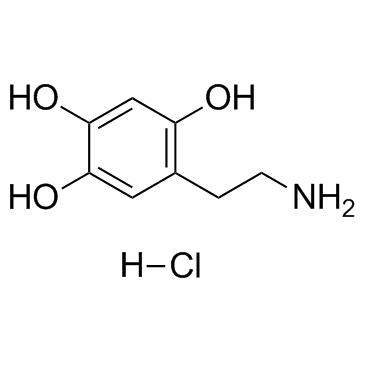
GC30871
Oxidopamine hydrochloride (6-Hydroxydopamine hydrochloride)
Oxidopamine (6-OHDA) hydrochloride is an antagonist of the neurotransmitter dopamine.
-
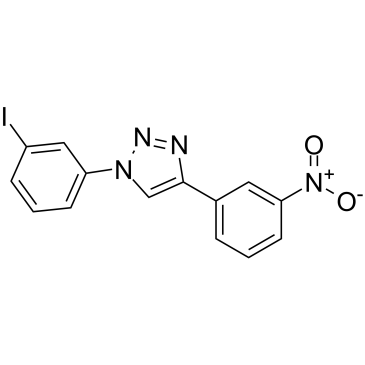
GC38562
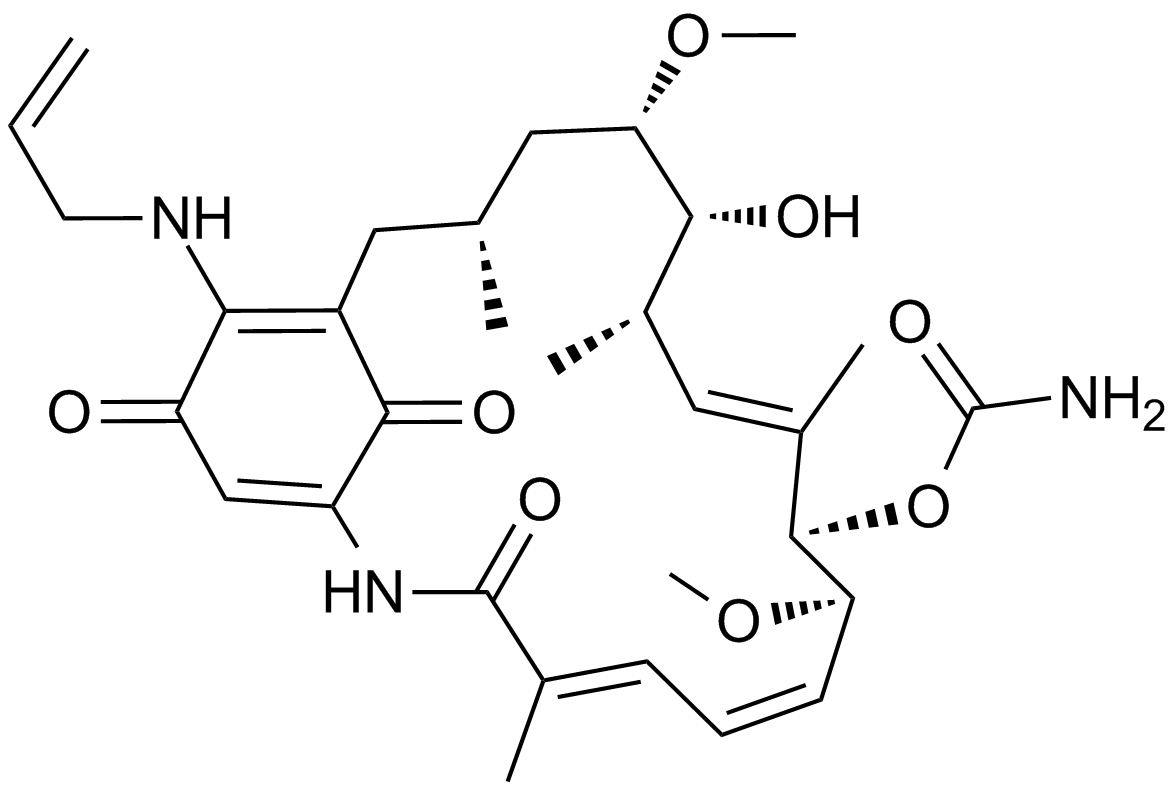
P62-mediated mitophagy inducer
P62-mediated mitophagy inducer is a mitophagy regulator which activates mitophagy without recruiting Parkin or collapsing ΔΨm and retains activity in cells devoid of a fully functional PINK1/Parkin pathway.
-
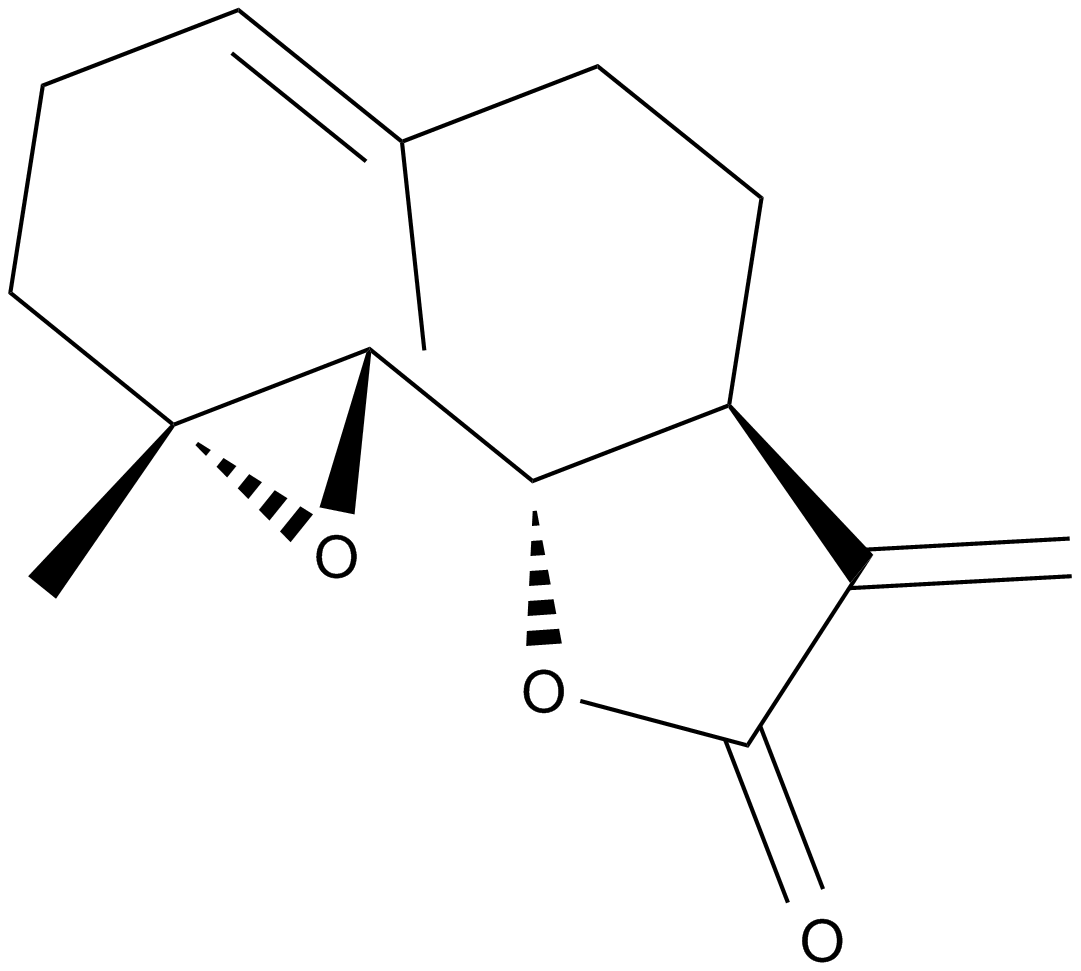
GN10357
Parthenolide
-
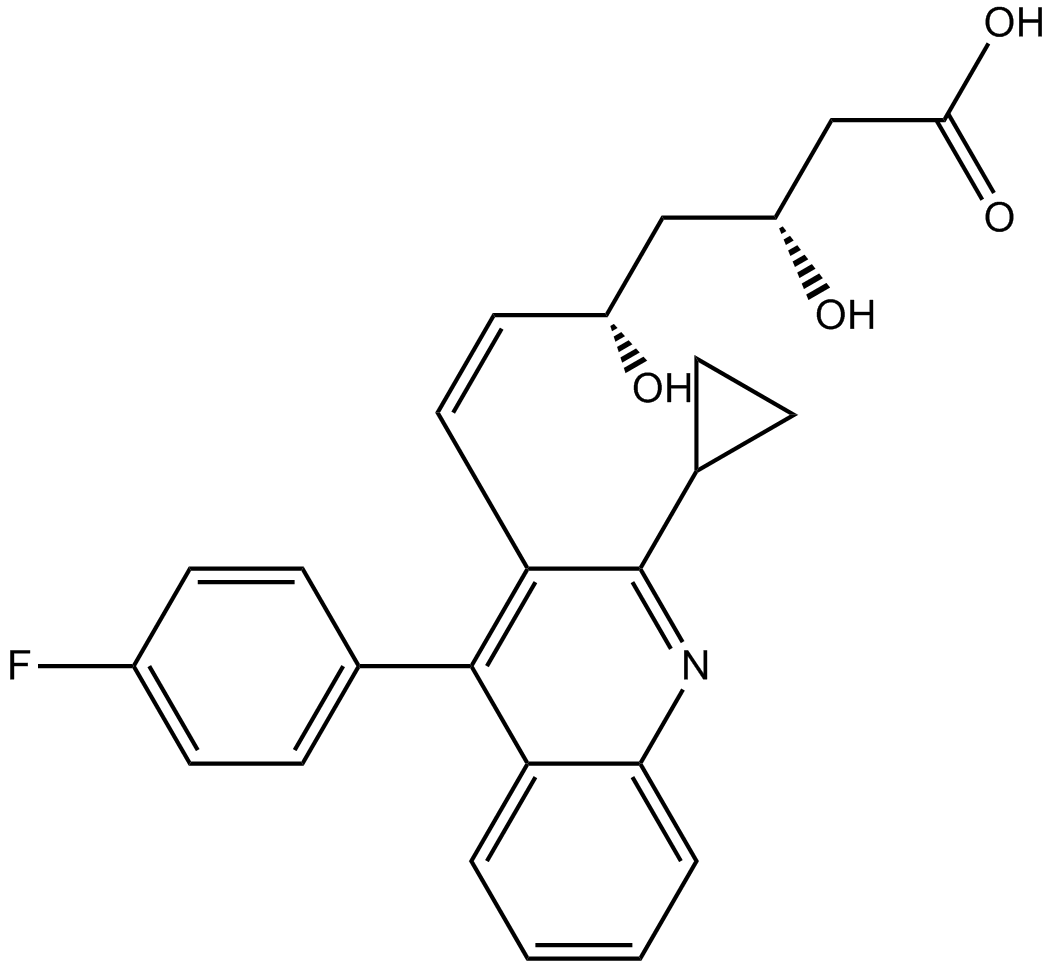
GC11332
Pitavastatin
HMG-CoA reductase inhibitor
-
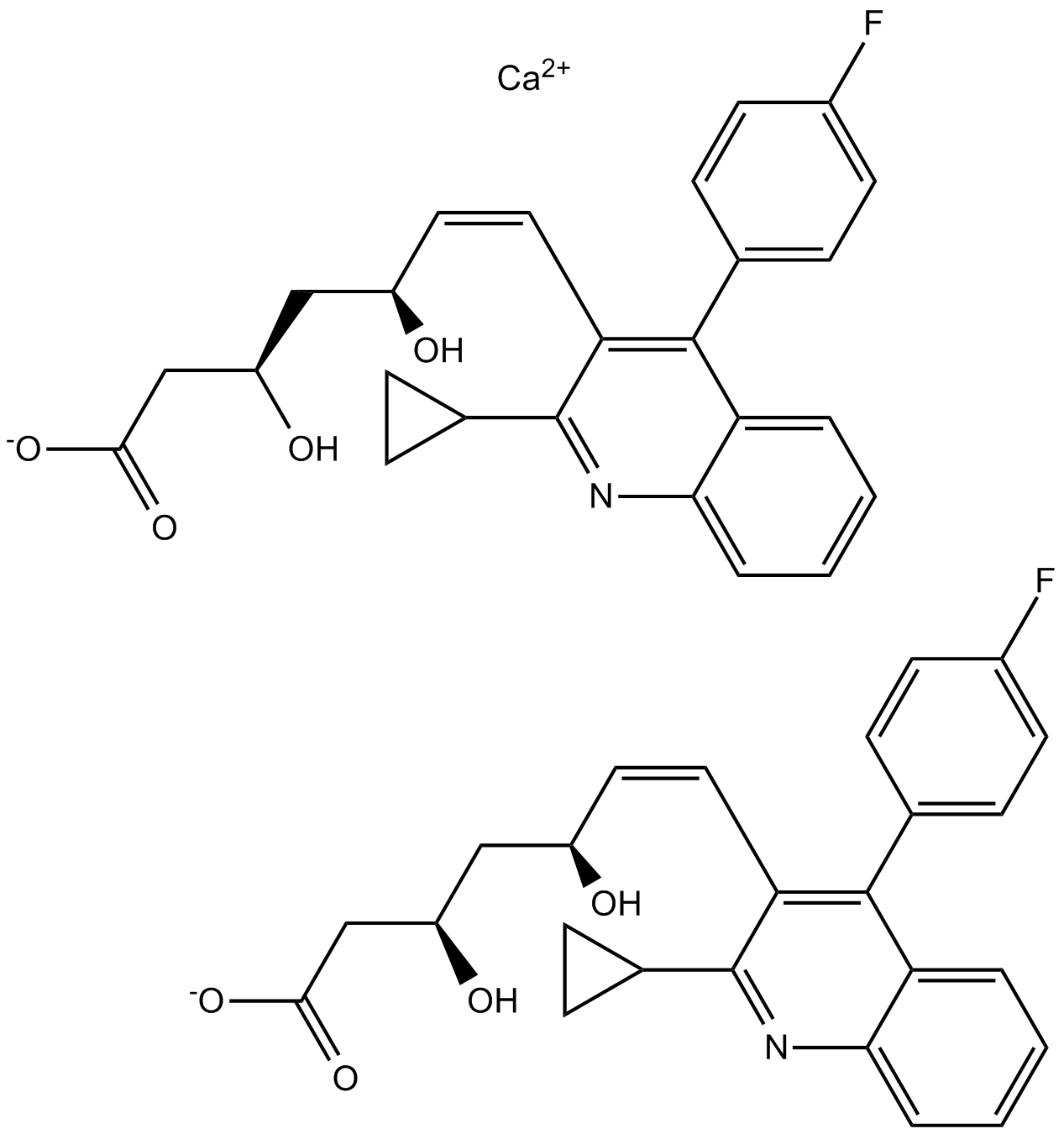
GC12116
Pitavastatin Calcium
Enzyme HMGCR inhibitor
-
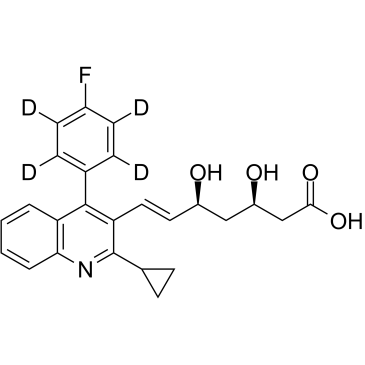
GC36930
Pitavastatin D4
Pitavastatin D4 (NK-104 D4) is deuterium labeled Pitavastatin.
-
GN10331
Polydatin

-
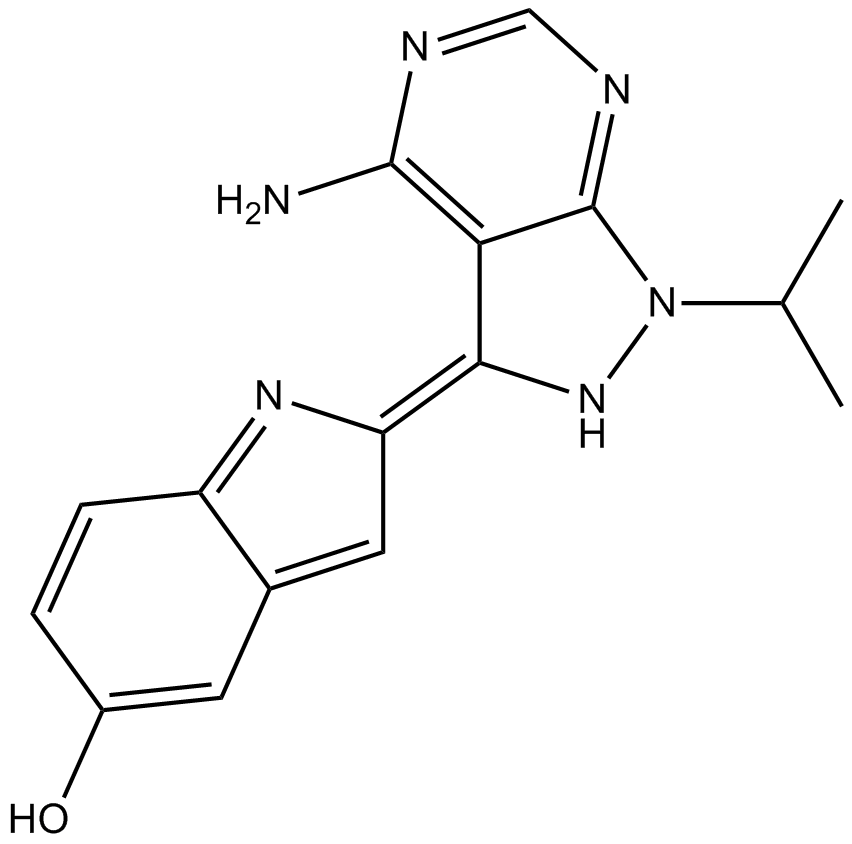
GC15904
PP242
PP242 (PP 242) is a selective and ATP-competitive mTOR inhibitor with an IC50 of 8 nM. PP242 inhibits both mTORC1 and mTORC2 with IC50s of 30 nM and 58 nM, respectively.
-
GN10266
Quercetin
Quercetin is an important dietary flavonoid, present in vegetables, fruits, seeds, nuts, tea and red wine.
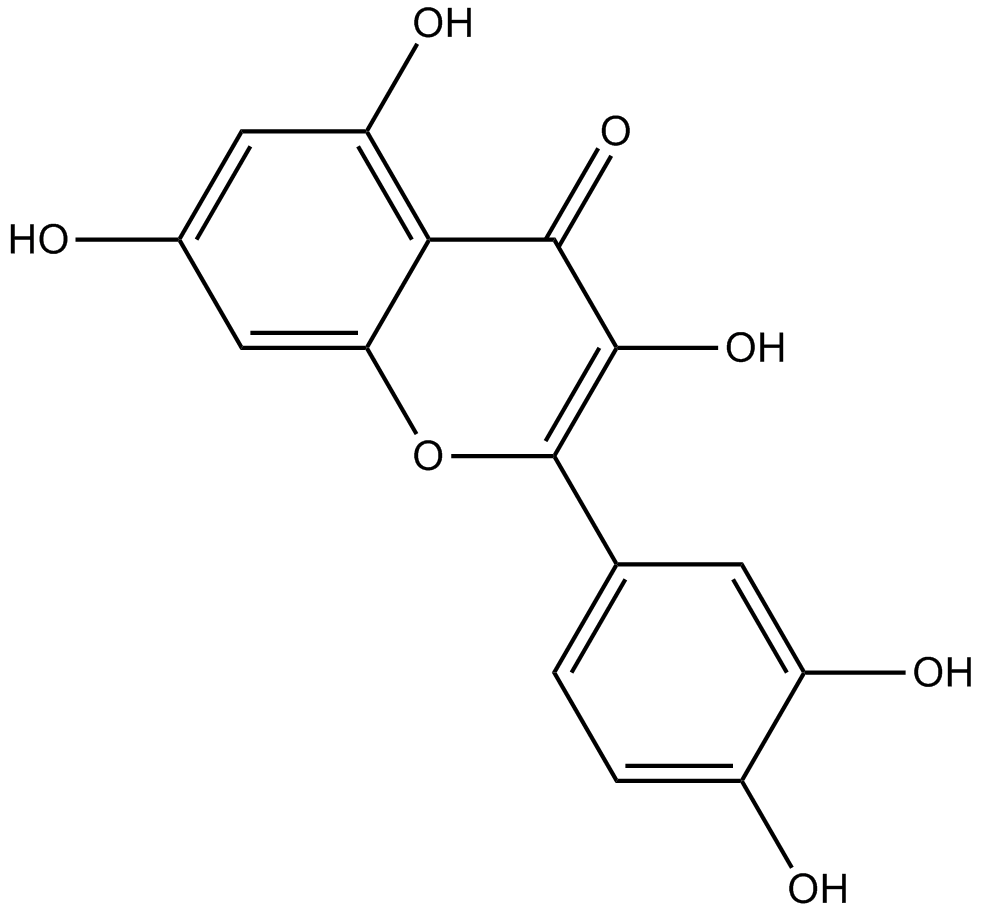
-
GC61227
Quercetin D5
Quercetin D5 is a deuterium labeled Quercetin. Quercetin, a natural flavonoid, is a stimulator of recombinant SIRT1 and also a PI3K inhibitor with IC50 of 2.4 μM, 3.0 μM and 5.4 μM for PI3K γ, PI3K δ and PI3K β, respectively.
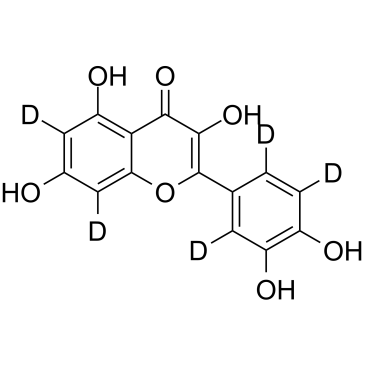
-
GC61229
Quinacrine dihydrochloride
Quinacrine (Mepacrine) dihydrochloride is an orally bioavailable antimalarial agent, which possess anticancer effect both in vitro and vivo.
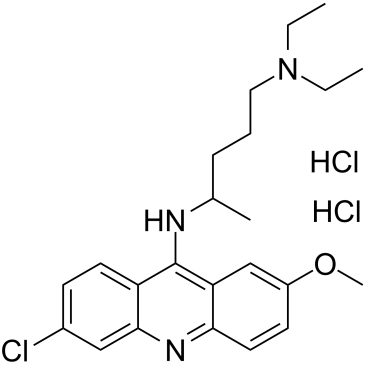
-
GC32768
Quinacrine dihydrochloride (Mepacrine dihydrochloride)
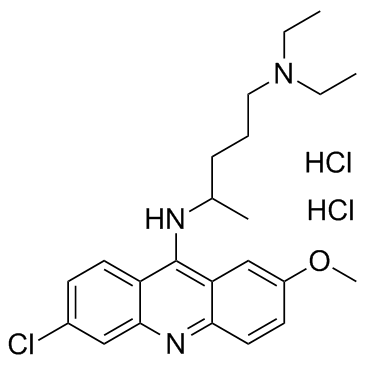
-
GC14553
Resveratrol
Resveratrol (trans-Resveratrol; SRT501) is a phytoalexin
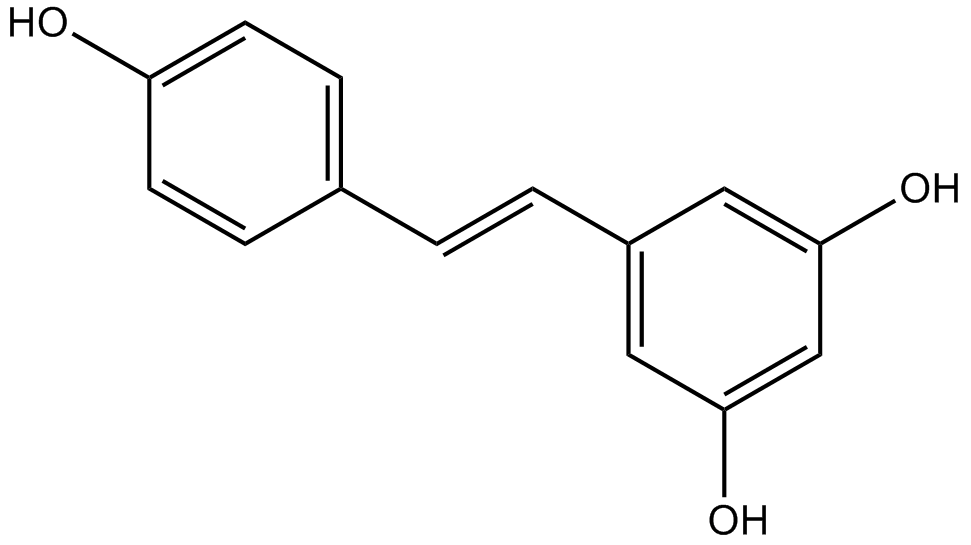
-
GC14191
Ruxolitinib (INCB018424)
Ruxolitinib (INCB018424), as an inhibitor, inhibits Janus-associated kinases (JAKs)JAK1 and JAK2 with IC50 values of 3.3 nM and 2.8 nM, respectively.
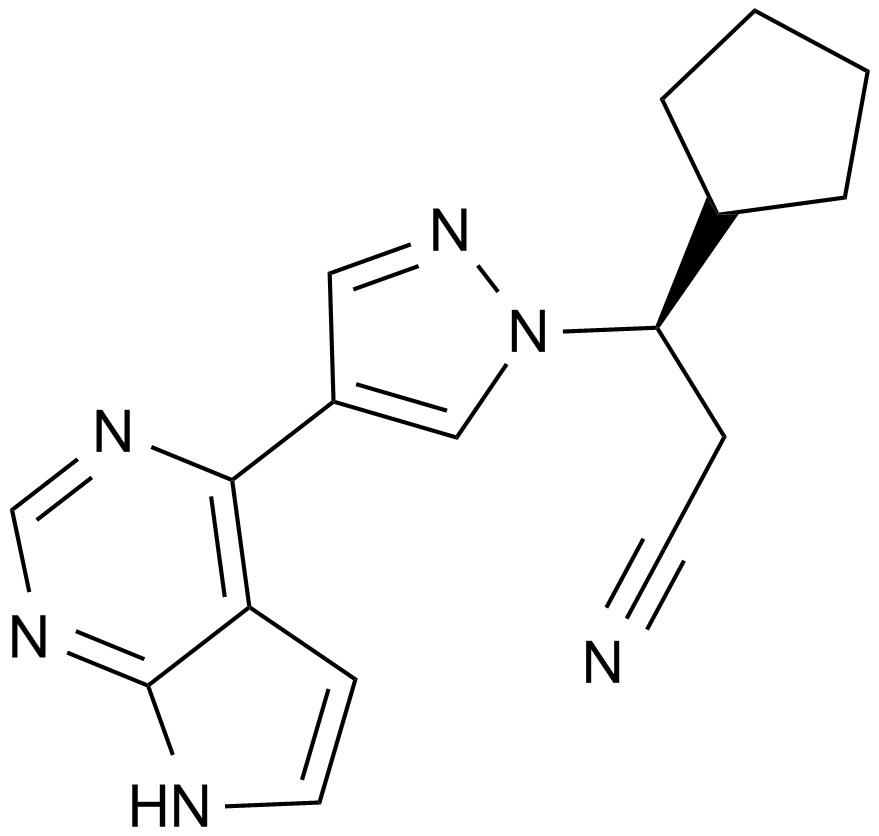
-
GC16124
Ruxolitinib phosphate
A potent and selective JAK1/JAK2 inhibitor
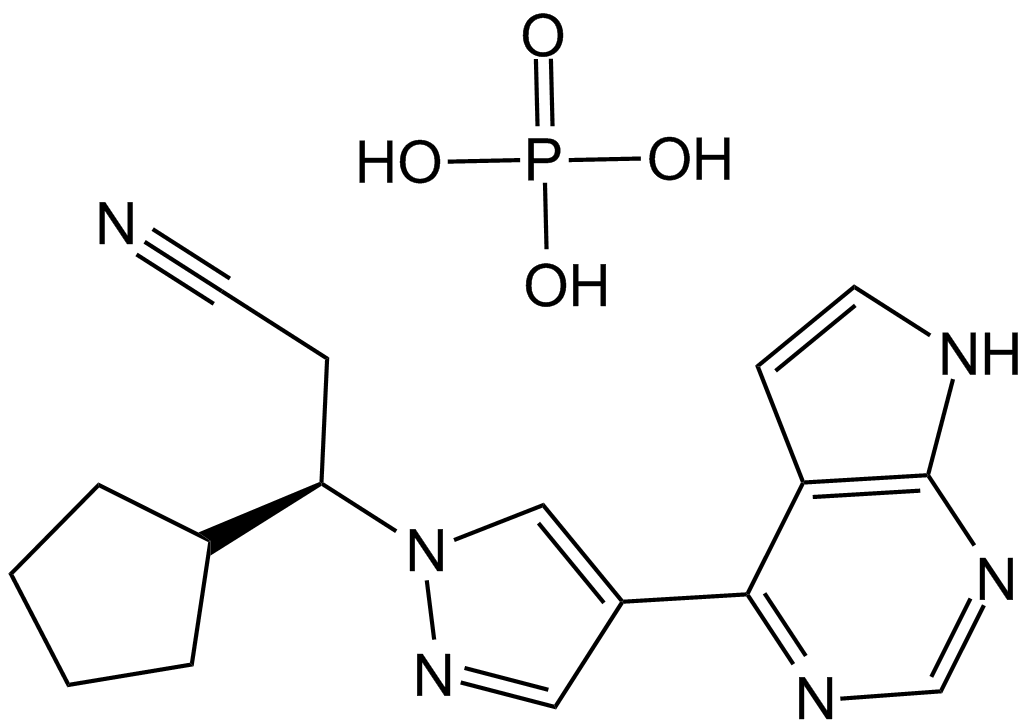
-
GC16508
Salicylic acid
COX inhibitor
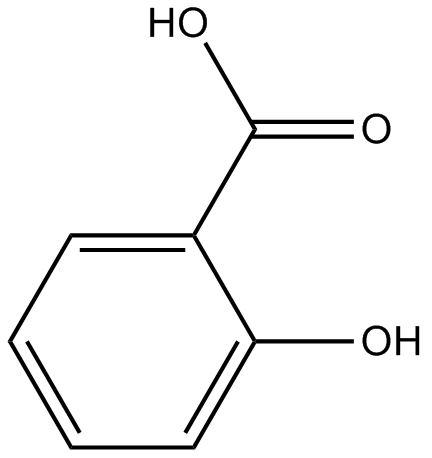
-
GC64032
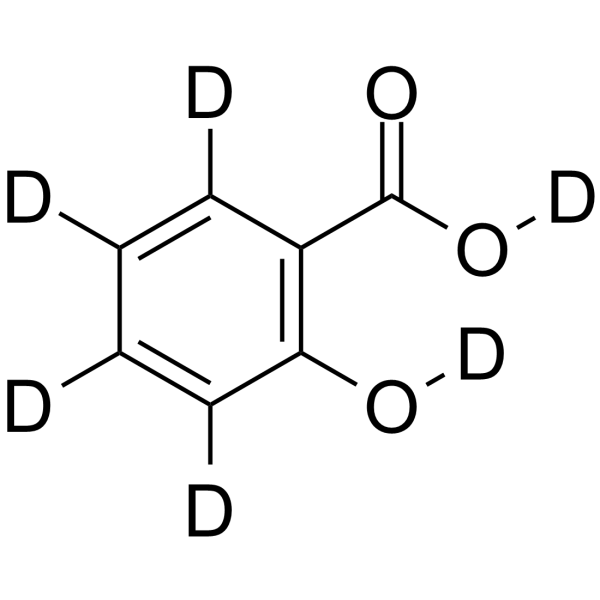
Salicylic acid-d6
Salicylic acid-D6 (2-Hydroxybenzoic acid-D6) is a deuterium labeled Salicylic acid.
-
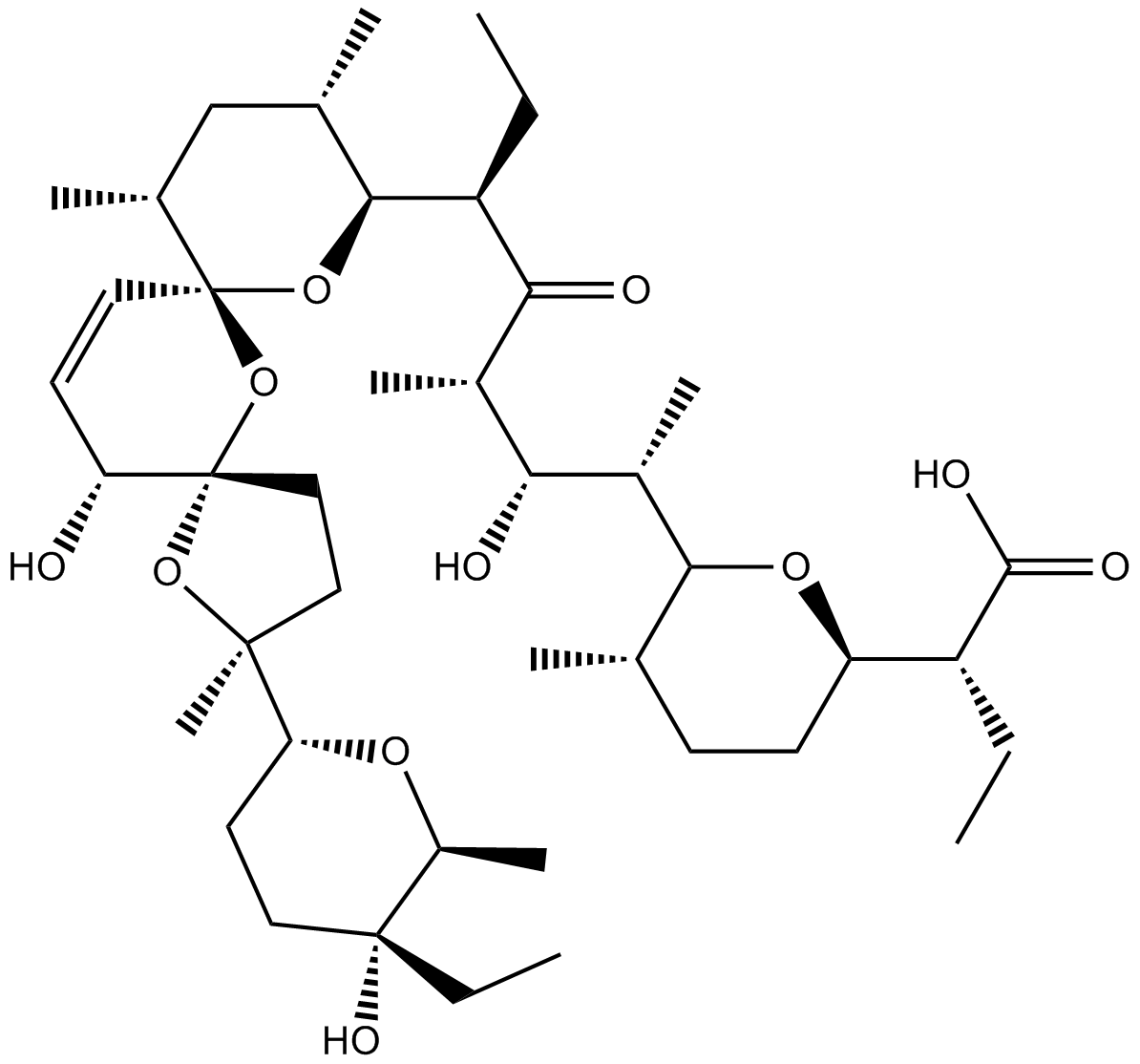
GC14882
Salinomycin
A selective cancer stem cell inhibitor
-
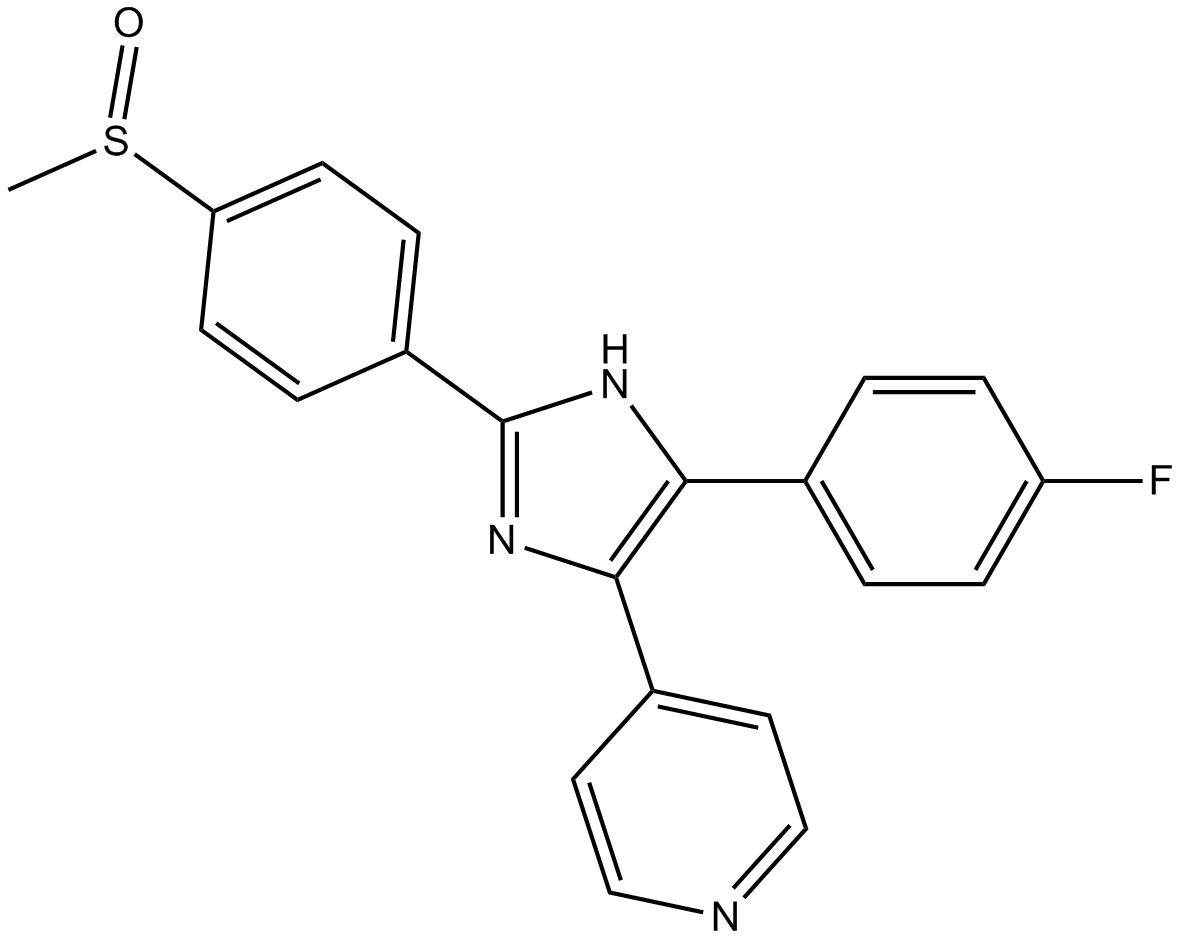
GC13595
SB 203580
SB 203580 is a specific inhibitor of p38-MAPK (Mitogen-activated Protein Kinase) pathway [1,2].
-
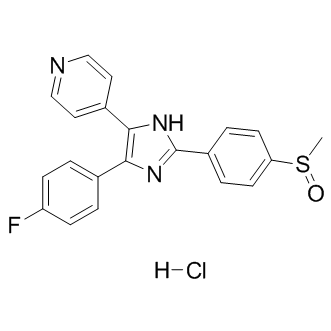
GC13001
SB 203580 hydrochloride
Adezmapimod (SB 203580) hydrochloride is a selective and ATP-competitive p38 MAPK inhibitor with IC50s of 50 nM and 500 nM for SAPK2a/p38 and SAPK2b/p38β2, respectively.
-
GC10585
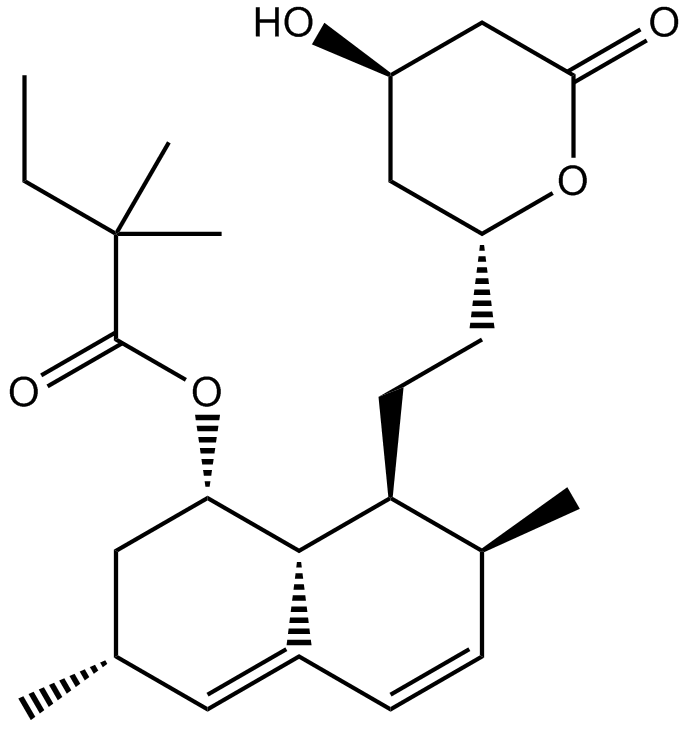
Simvastatin (Zocor)
Simvastatin (Zocor) (MK 733) is a competitive inhibitor of HMG-CoA reductase with a Ki of 0.2 nM.
-
GC31964
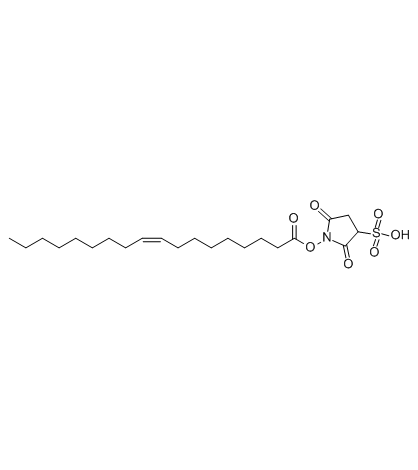
Sulfosuccinimidyl oleate
Sulfosuccinimidyl oleate (Sulfo-N-succinimidyl oleate) is a long chain fatty acid that inhibits fatty acid transport into cells.
-
GC34817
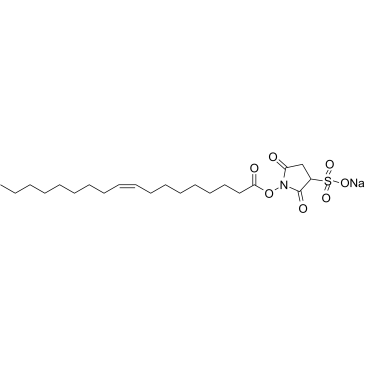
Sulfosuccinimidyl oleate sodium
A long chain fatty acid that inhibits fatty acid transport into cells
-
GC17651
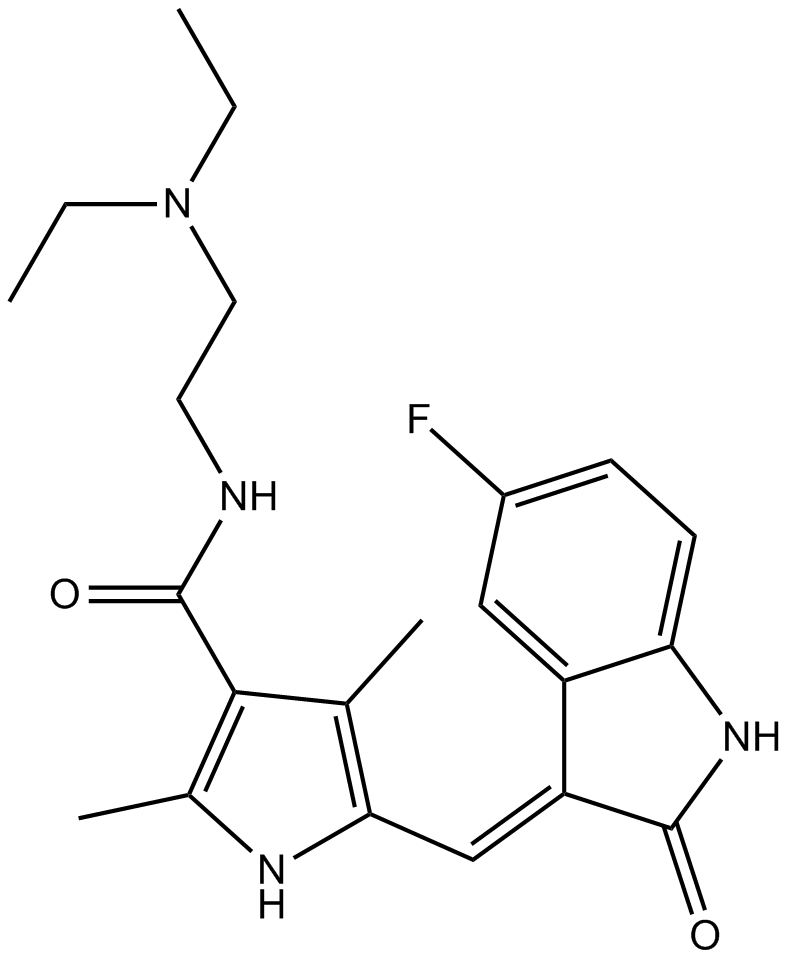
Sunitinib
RTK inhibitor
-
GC48118
Sunitinib-d10
An internal standard for the quantification of sunitinib
-
GC45099
U-0126
A MEK inhibitor
-
GC15951
URB597
An inhibitor of FAAH
-
GC11424
Valproic acid
HDAC1 inhibitor
-
GC15794
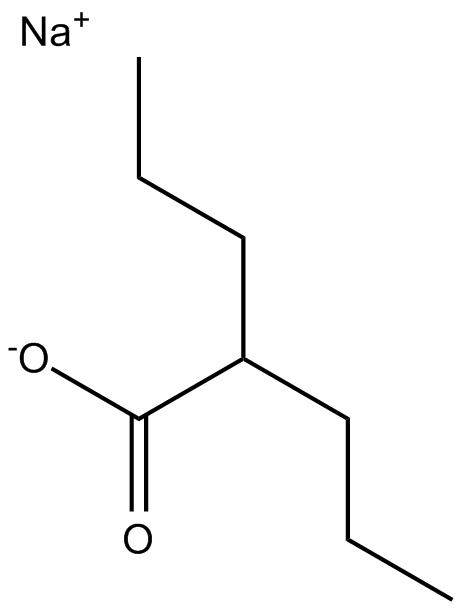
Valproic acid sodium salt (Sodium valproate)
A class I HDAC inhibitor
-
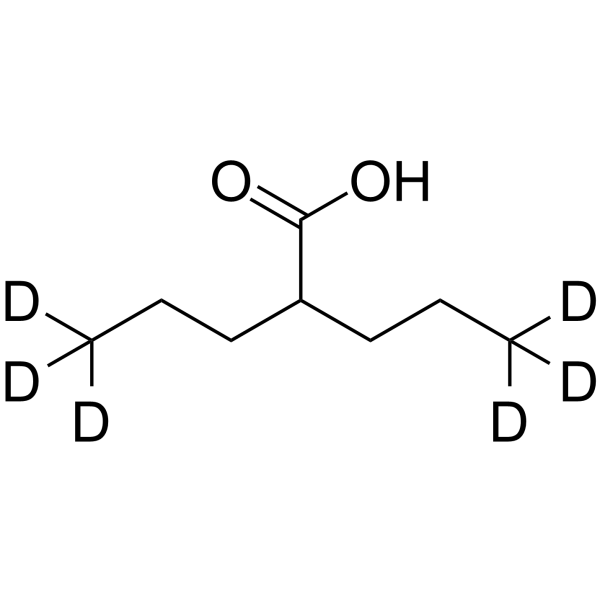
GC64378
Valproic acid-d6
Valproic acid-d6 (VPA-d6) is the deuterium labeled Valproic acid. Valproic acid (VPA; 2-Propylpentanoic Acid) is an HDAC inhibitor, with IC50 in the range of 0.5 and 2 mM, also inhibits HDAC1 (IC50, 400 μM), and induces proteasomal degradation of HDAC2. Valproic acid activates Notch1 signaling and inhibits proliferation in small cell lung cancer (SCLC) cells. Valproic acid sodium salt is used in the treatment of epilepsy, bipolar disorder and prevention of migraine headaches.
-
GC17390
Vorinostat (SAHA, MK0683)
An HDAC inhibitor