Caspase
Caspases (cysteine-dependent aspartate specific ptotease) are a family of cysteine proteases that play an essential role in cell apoptosis. Caspases contain a conserved QACXG pentapeptide active site motif and are characterized by specificity for aspartic acid in the P1 position. The synthesis of caspases involves the activation of inactive proenzymes, which contain an N-terminal peptide (prodomain) together with two subunits (one small and one large), following cleavage at specific aspartate cleavage sites. Caspases can be classified into three subfamilies, due to phylogenetic analysis, including an ICE subfamily (caspases-1, -4 and -5), a CED-3/CPP32 subfamily (caspases-3, -6, -7, -8, -9 and -10) and an ICH-1/Nedd2 subfamily (caspase-2).
Products for Caspase
- Cat.No. Product Name Information
-
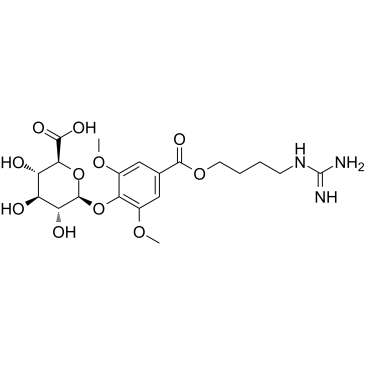
GC66048
δ-Secretase inhibitor 11
δ-Secretase inhibitor 11 (compound 11) is an orally active, potent, BBB-penetrated, non-toxic, selective and specific δ-secretase inhibitor, with an IC50 of 0.7 μM. δ-Secretase inhibitor 11 interacts with both the active site and allosteric site of δ-secretase. δ-Secretase inhibitor 11 attenuates tau and APP (amyloid precursor protein) cleavage. δ-Secretase inhibitor 11 ameliorates synaptic dysfunction and cognitive impairments in tau P301S and 5XFAD transgenic mouse models. δ-Secretase inhibitor 11 can be used for Alzheimer's disease research.
-
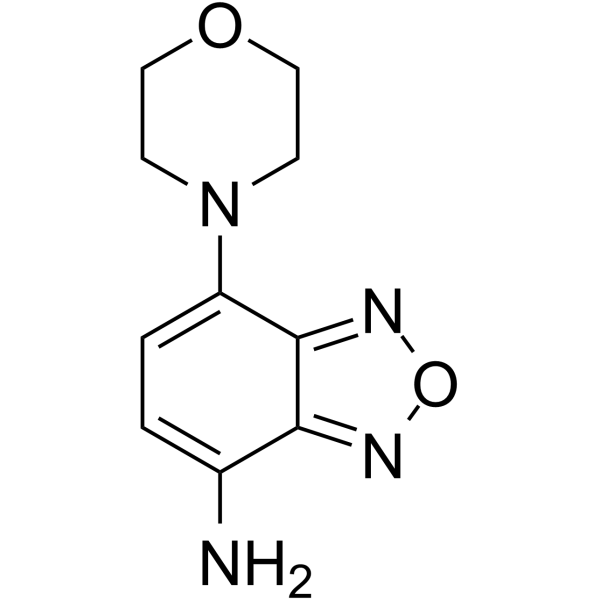
GC11988
15-acetoxy Scirpenol
mycotoxin that induce apoptotic cell death
-
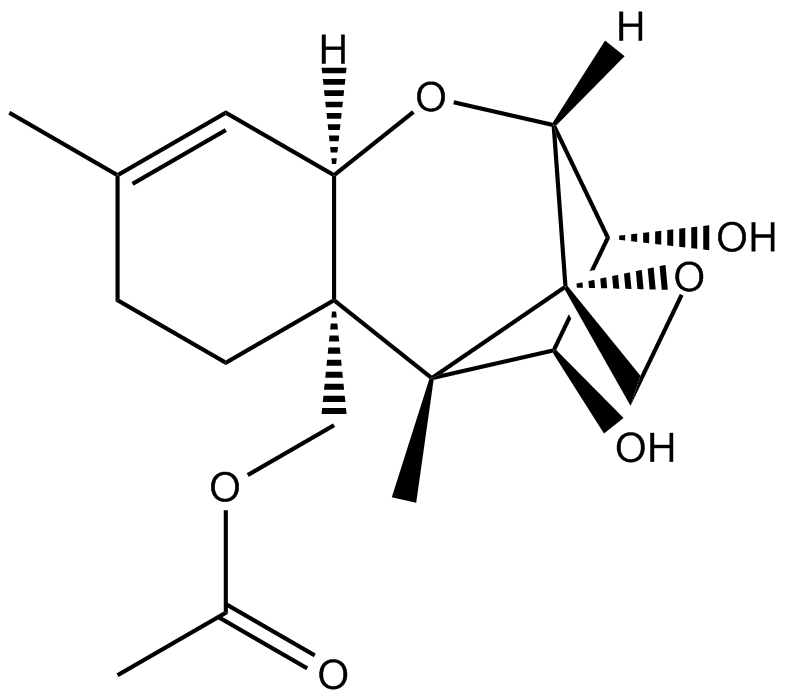
GC12545
2-HBA
indirect inducer of enzymes that catalyze detoxification reactions through the Keap1-Nrf2-ARE pathway.
-
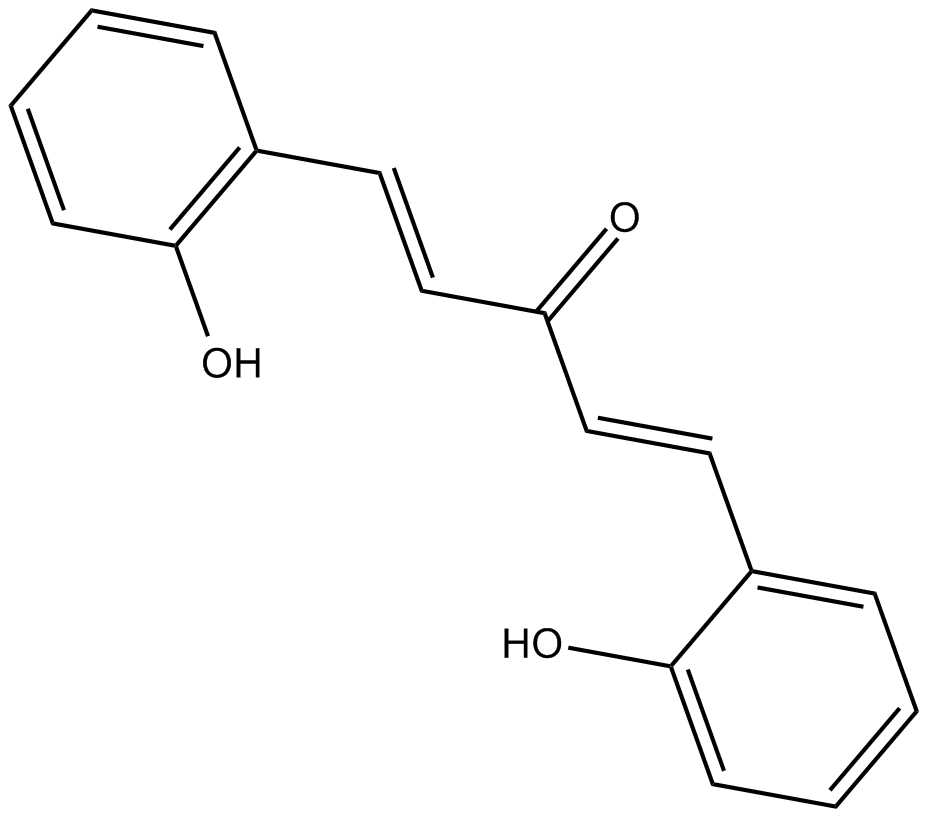
GC35150
5,7,4'-Trimethoxyflavone
5,7,4'-Trimethoxyflavone is isolated from Kaempferia parviflora (KP) that is a famous medicinal plant from Thailand. 5,7,4'-Trimethoxyflavone induces apoptosis, as evidenced by increments of sub-G1 phase, DNA fragmentation, annexin-V/PI staining, the Bax/Bcl-xL ratio, proteolytic activation of caspase-3, and degradation of poly (ADP-ribose) polymerase (PARP) protein.5,7,4'-Trimethoxyflavone is significantly effective at inhibiting proliferation of SNU-16 human gastric cancer cells in a concentration dependent manner.
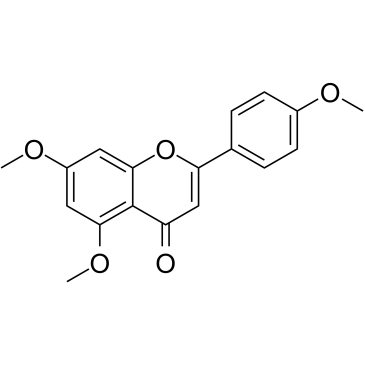
-
GC17602
Ac-DEVD-AFC
fluorogenic substrate for activated caspase-3
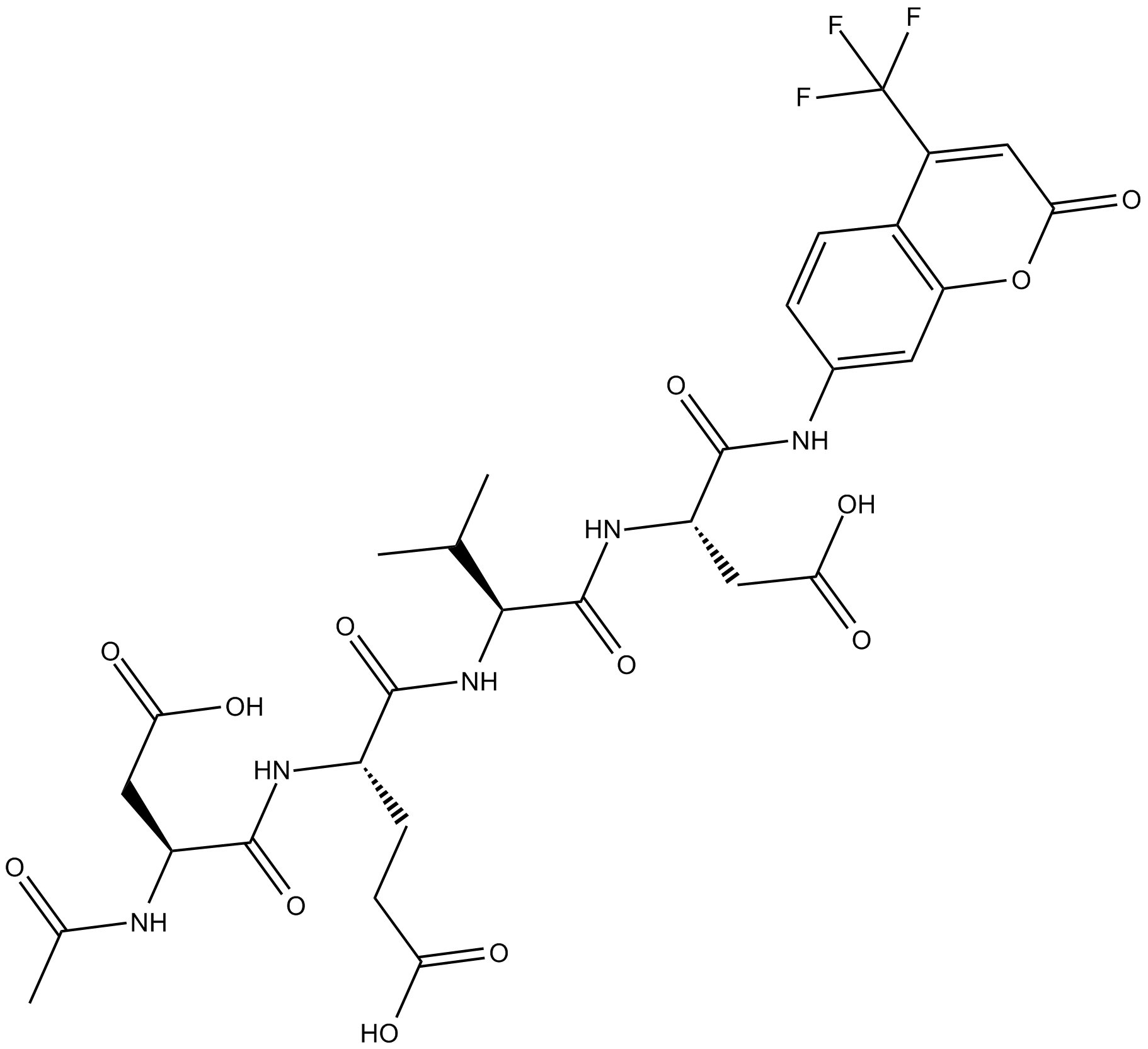
-
GC32695
Ac-DEVD-CHO
Ac-DEVD-CHO is a specific Caspase-3 inhibitor with a Ki value of 230 pM.
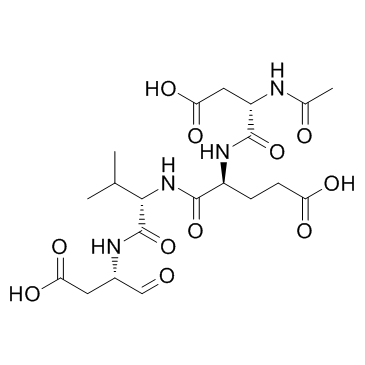
-
GC10951
Ac-DEVD-CMK
cell-permeable, and irreversible inhibitor of caspase
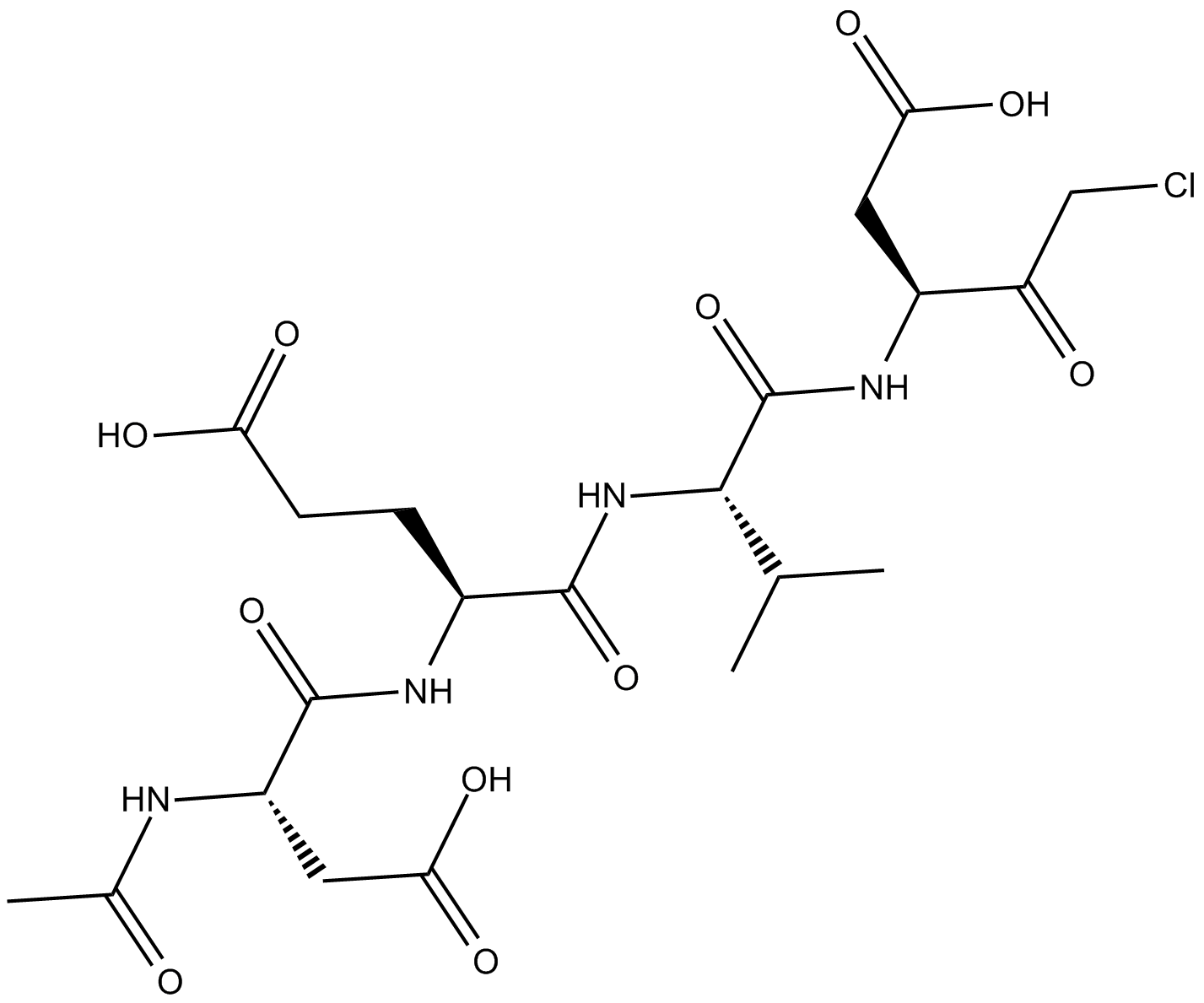
-
GC35388
Aristolactam I
Aristololactam I (AL-I), is the main metabolite of aristolochic acid I (AA-I), participates in the processes that lead to renal damage.
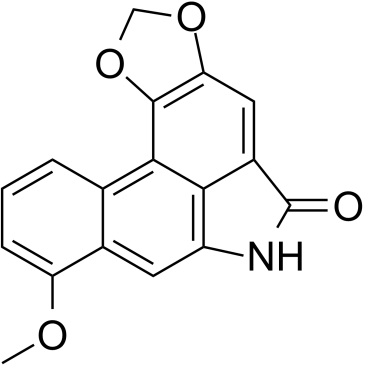
-
GC35395
Arnicolide D
Arnicolide D is a sesquiterpene lactone isolated from Centipeda minima. Arnicolide D modulates the cell cycle, activates the caspase signaling pathway and inhibits the PI3K/AKT/mTOR and STAT3 signaling pathways. Arnicolide D inhibits Nasopharyngeal carcinoma (NPC) cell viability in a concentration- and time-dependent manner.
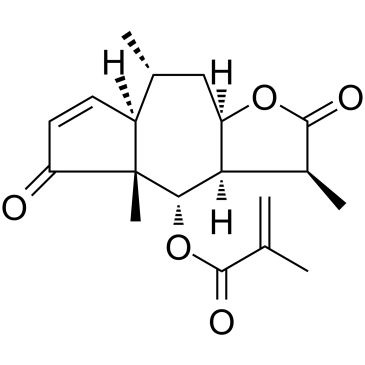
-
GC18476
Biotin-VAD-FMK
Biotin-VAD-FMK is a biotin-conjugated form of the pan-caspase inhibitor Z-VAD(OH)-FMK .
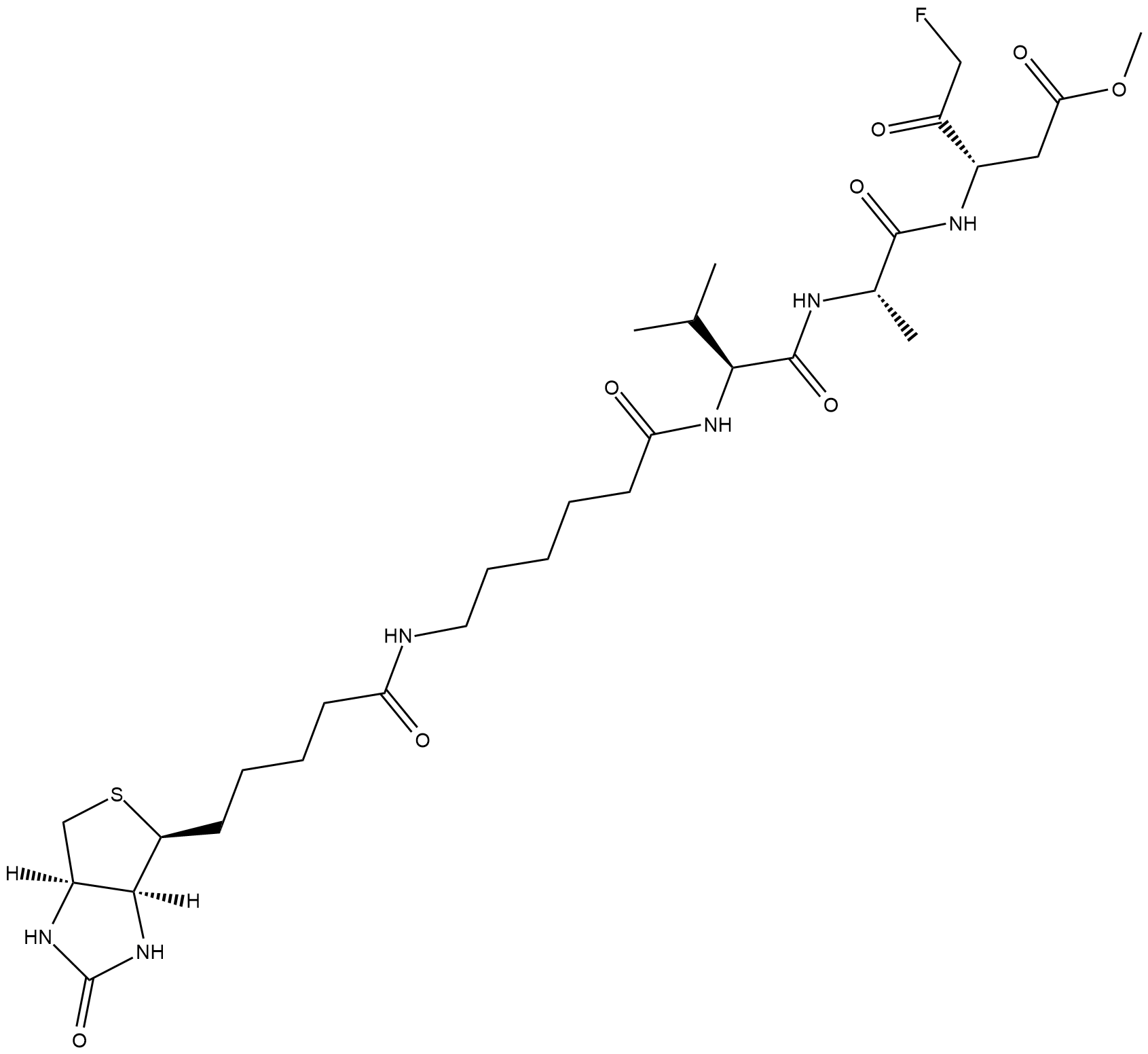
-
GC31886
Chelidonic acid
A pyran with diverse biological activities
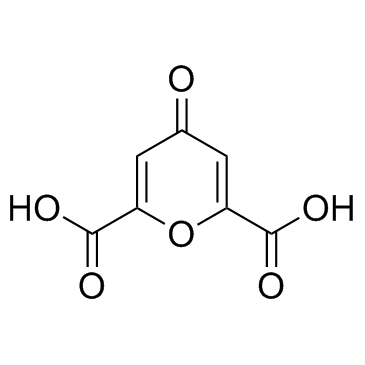
-
GC11908
Cisplatin
Cisplatin is one of the best and first metal-based chemotherapeutic drugs, which is used for wide range of solid cancers such as testicular, ovarian, bladder, lung, cervical, head and neck cancer, gastric cancer and some other cancers.

-
GC38452
Dehydrotrametenolic acid
Dehydrotrametenolic acid is a sterol isolated from the sclerotium of Poria cocos.
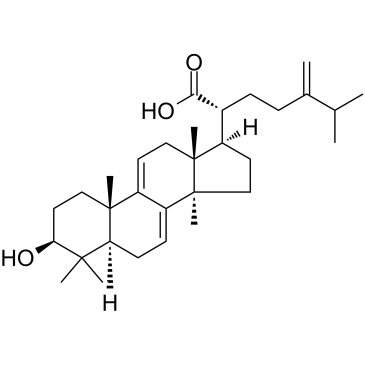
-
GC10661
Destruxin B
insecticidal and phytotoxic activity;induces apoptosis
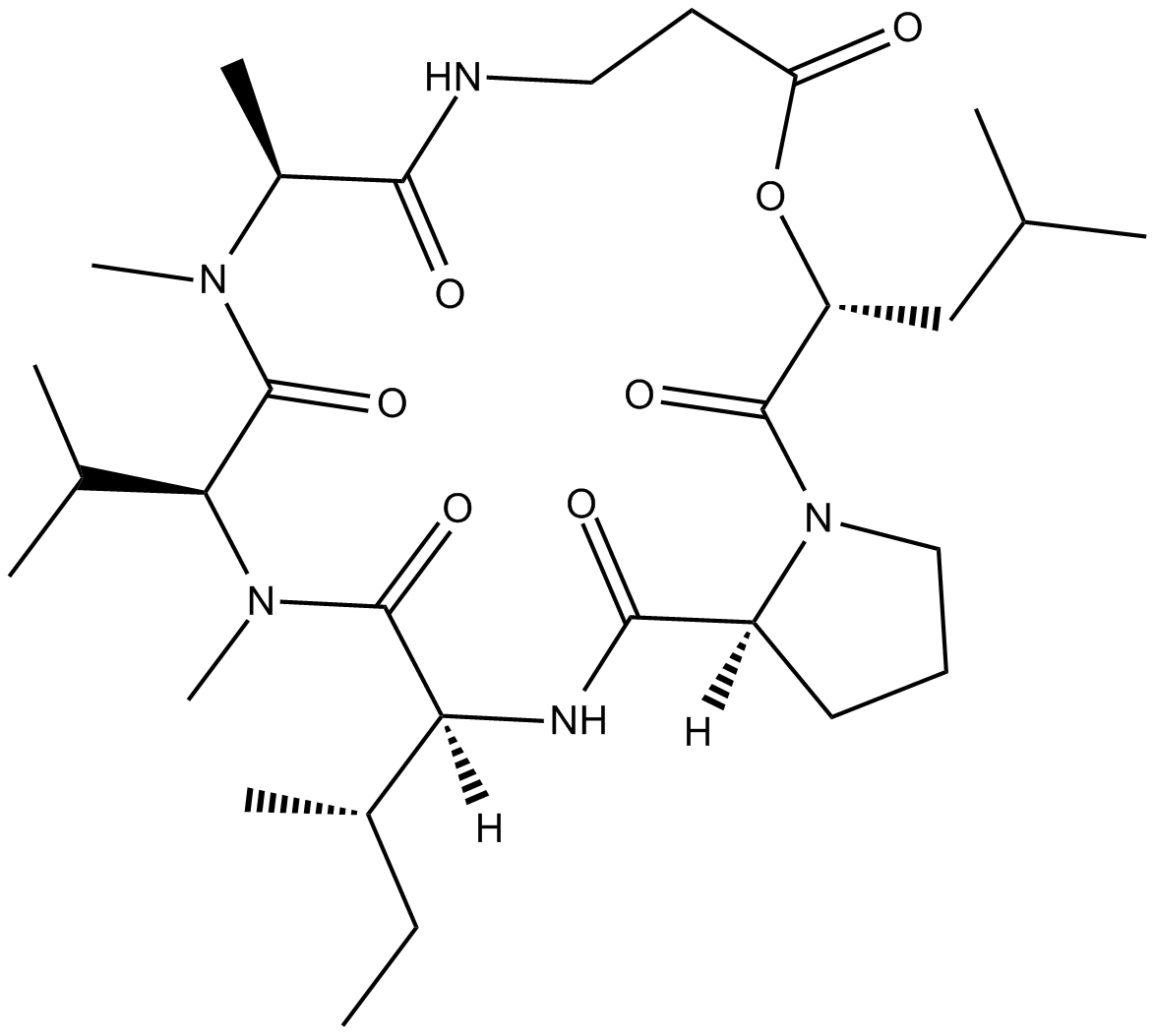
-
GC35908
Duocarmycin A
Duocarmycin A, which is one of well-known antitumor antibiotics, is a DNA alkylator and efficiently alkylates adenine N3 at the 3′ end of AT-rich sequences in the DNA. Duocarmycin A, as a chemotherapeutic agent, results HLC-2 cells typically apoptotic changes, including chromatin condensation, sub-G1 accumulation in DNA histogram pattern, and decrease in procaspase-3 and 9 levels.
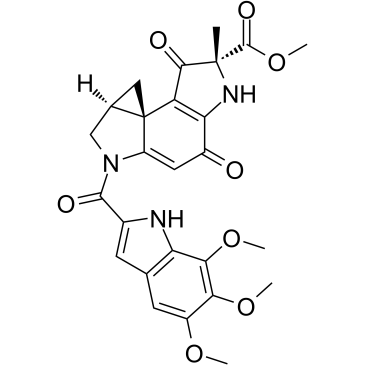
-
GC35980
Emricasan
A pan-caspase inhibitor

-
GC31390
EP1013 (F1013)
EP1013 (F1013) (F1013) is a broad-spectrum caspase selective inhibitor, used in the research of type 1 diabetes.
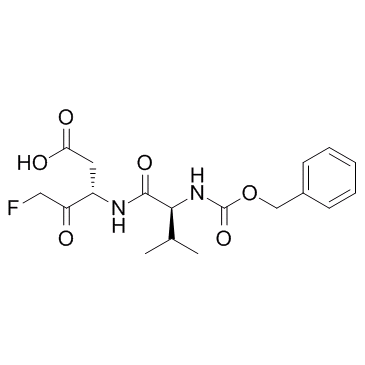
-
GC32998
Ginsenoside Rh4
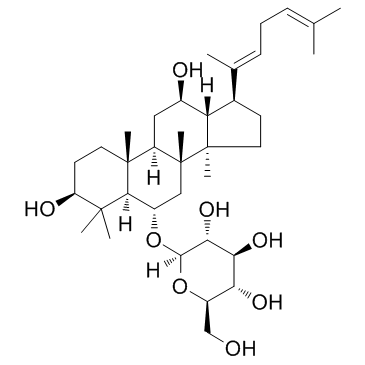
-
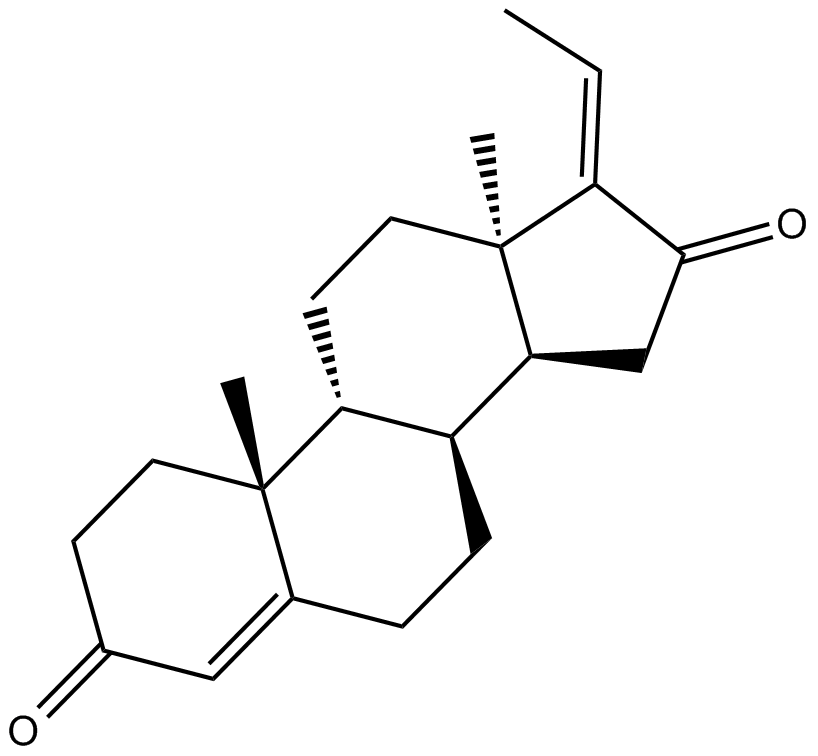
GC17658
Guggulsterone
Broad spectrum steroid receptor ligand
-
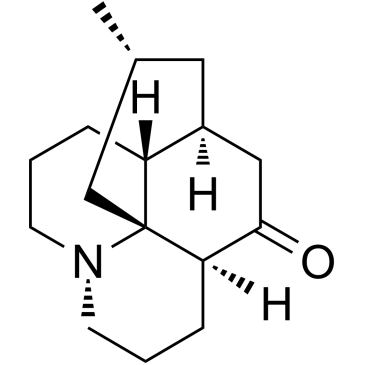
GC36516
Lycopodine
Lycopodine, a pharmacologically important bioactive component derived from Lycopodium clavatumspores, triggers apoptosis by modulating 5-lipoxygenase, and depolarizing mitochondrial membrane potential in refractory prostate cancer cells without modulating p53 activity. Lycopodine inhibits proliferation of HeLa cells through induction of apoptosis via caspase-3 activation.
-
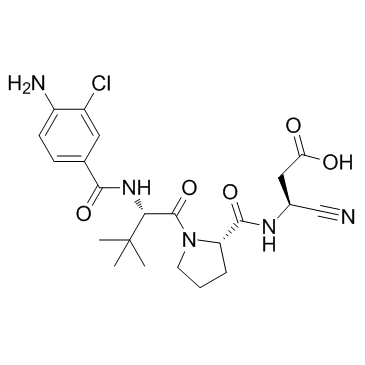
GC33309
ML132 (NCGC 00185682)
ML132 (NCGC 00185682) (NCGC-00183434) is a selective caspase 1 inhibitor with an IC50 of 34.9 nM.
-
GC36652
MPT0B392
MPT0B392, an orally active quinoline derivative, induces c-Jun N-terminal kinase (JNK) activation, leading to apoptosis. MPT0B392 inhibits tubulin polymerization and triggers induction of the mitotic arrest, followed by mitochondrial membrane potential loss and caspases cleavage by activation of JNK and ultimately leads to apoptosis. MPT0B392 is demonstrated to be a novel microtubule-depolymerizing agent and enhances the cytotoxicity of sirolimus in sirolimus-resistant acute leukemic cells and the multidrug resistant cell line.
-
GC44409
Nivalenol
Nivalenol is a type B trichothecene mycotoxin produced by Fusarium that is commonly found in contaminated foods.
-
GC12639
NS3694
inhibits apoptosome formation and caspase activation
-
GC36855
Paris saponin VII
Paris saponin VII (Chonglou Saponin VII) is a steroidal saponin isolated from the roots and rhizomes of Trillium tschonoskii Maxim. Paris saponin VII-induced apoptosis in K562/ADR cells is associated with Akt/MAPK and the inhibition of P-gp. Paris saponin VII attenuates mitochondrial membrane potential, increases the expression of apoptosis-related proteins, such as Bax and cytochrome c, and decreases the protein expression levels of Bcl-2, caspase-9, caspase-3, PARP-1, and p-Akt. Paris saponin VII induces a robust autophagy in K562/ADR cells and provides a biochemical basis in the treatment of leukemia.
-
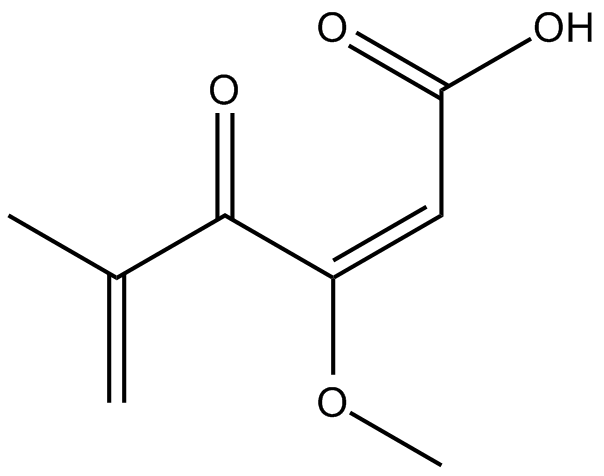
GC15880
Penicillic Acid
mycotoxin
-
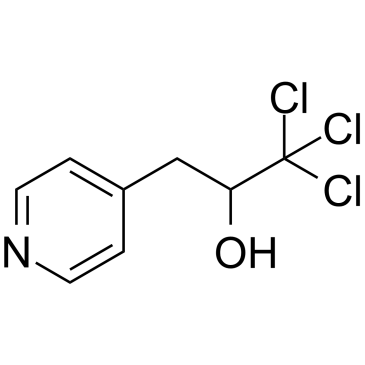
GC38064
PETCM
-
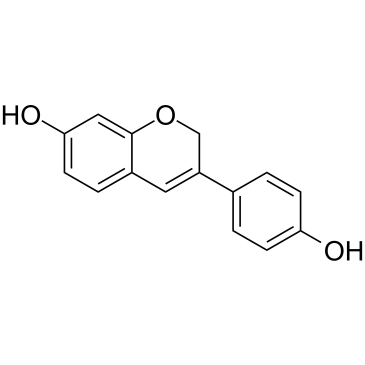
GC36895
Phenoxodiol
A phenol with anticancer activity
-
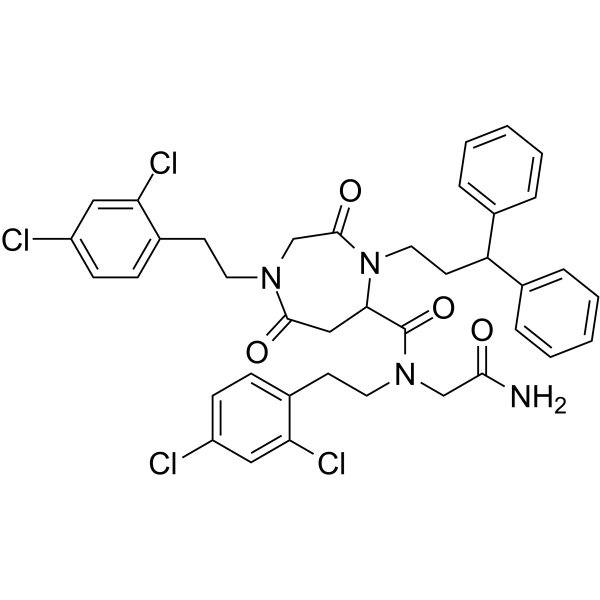
GC65474
QM31
QM31 (SVT016426), a cytoprotective agent, is a selective inhibitor of Apaf-1. QM31 inhibits the formation of the apoptosome (IC50=7.9μM), the caspase activation complex composed by Apaf-1, cytochrome c, dATP and caspase-9. QM31 exerts mitochondrioprotective functions and interferes with the intra-S-phase DNA damage checkpoint.
-
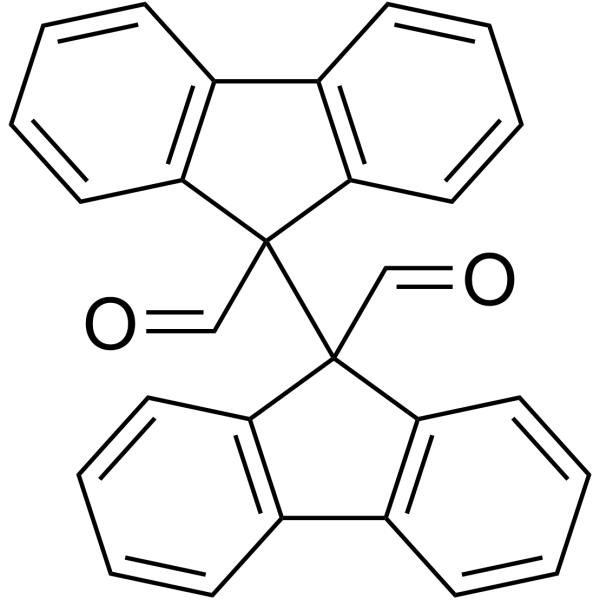
GC63914
Raptinal
Raptinal, a agent that directly activates caspase-3, initiates intrinsic pathway caspase-dependent apoptosis. Raptinal is able to rapidly induce cancer cell death by directly activating the effector caspase-3, bypassing the activation of initiator caspase-8 and caspase-9.
-
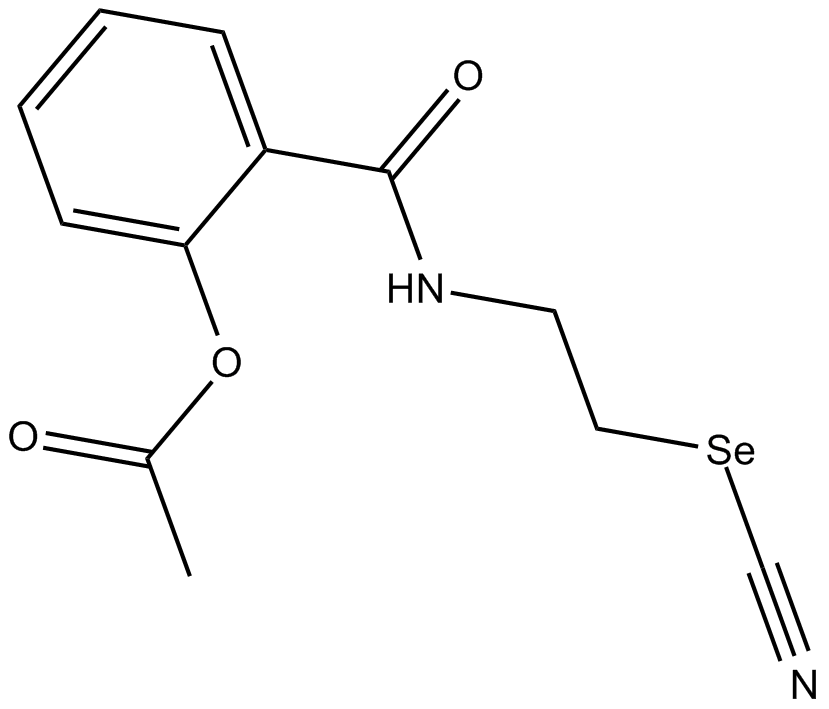
GC15267
Se-Aspirin
nonsteroidal anti-inflammatory drug
-
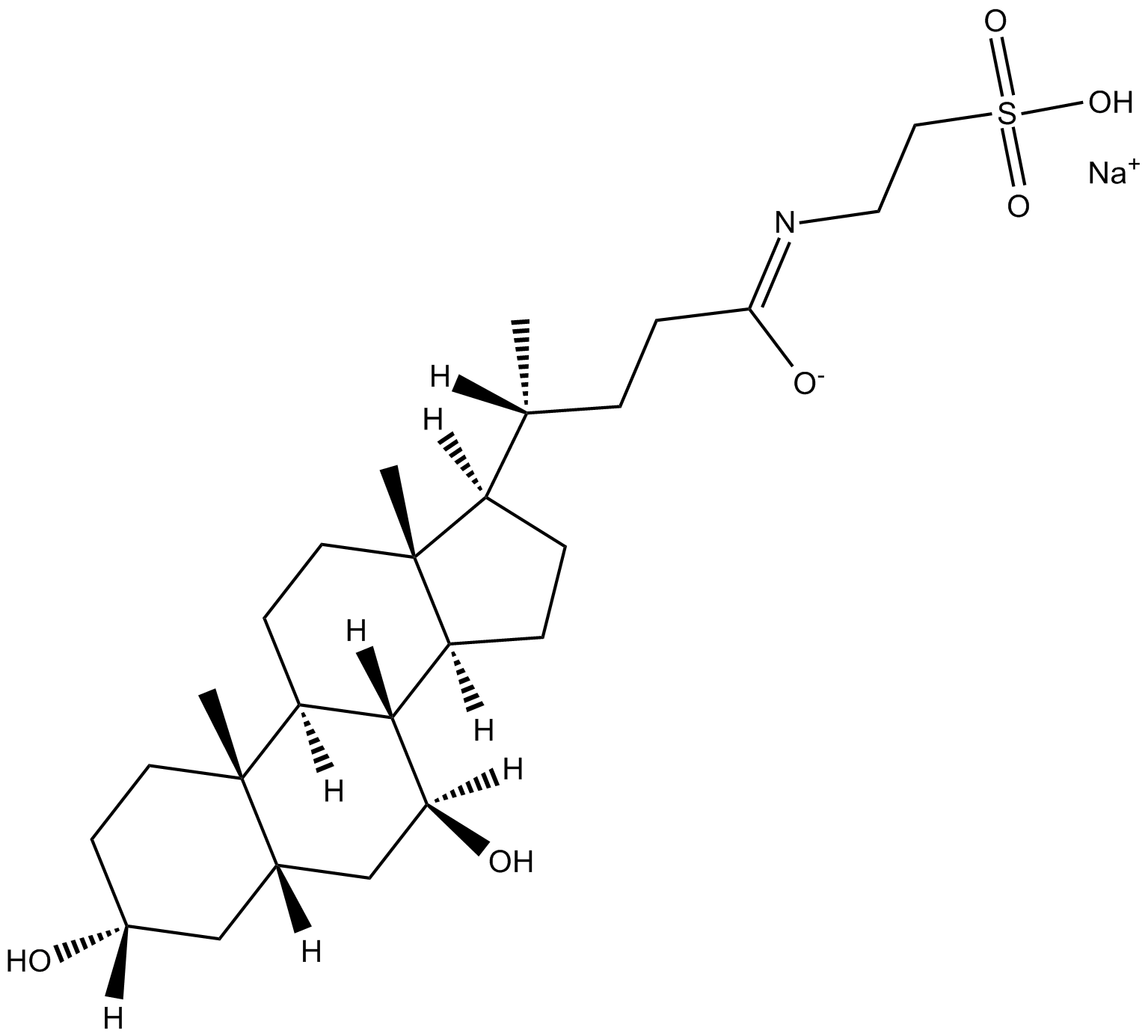
GC17425
Sodium Tauroursodeoxycholate (TUDC)
Tauroursodeoxycholate (Tauroursodeoxycholic acid; TUDCA) sodium is an endoplasmic reticulum (ER) stress inhibitor. Tauroursodeoxycholate significantly reduces expression of apoptosis molecules, such as caspase-3 and caspase-12. Tauroursodeoxycholate also inhibits ERK.
-
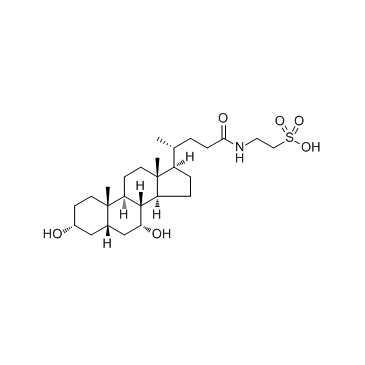
GC33825
Taurochenodeoxycholic acid (12-Deoxycholyltaurine)
Taurochenodeoxycholic acid (12-Deoxycholyltaurine) (12-Deoxycholyltaurine) is one of the main bioactive substances of animals' bile acid.
-
GC37745
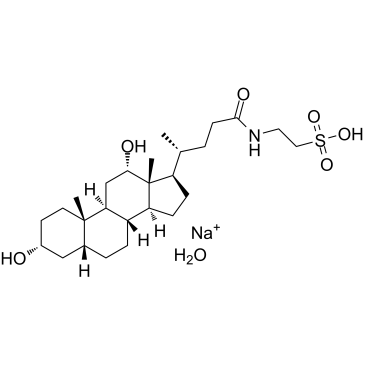
Taurodeoxycholic acid sodium hydrate
Taurodeoxycholic acid sodium hydrate (Sodium taurodeoxycholate monohydrate), a bile acid, is an amphiphilic surfactant molecule synthesized from cholesterol in the liver. It activates S1PR2 pathway in addition to the TGR5 pathway.
-
GC34181
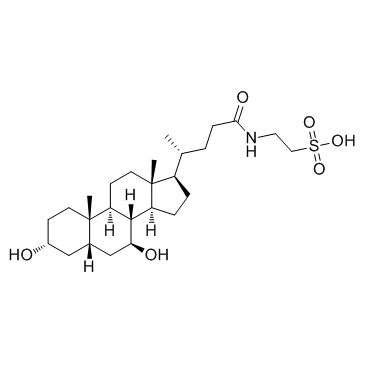
Tauroursodeoxycholate (TUDCA)
Tauroursodeoxycholate (TUDCA) (Tauroursodeoxycholic acid) is an endoplasmic reticulum (ER) stress inhibitor. Tauroursodeoxycholate (TUDCA) significantly reduces expression of apoptosis molecules, such as caspase-3 and caspase-12. Tauroursodeoxycholate (TUDCA) also inhibits ERK.
-
GC34831
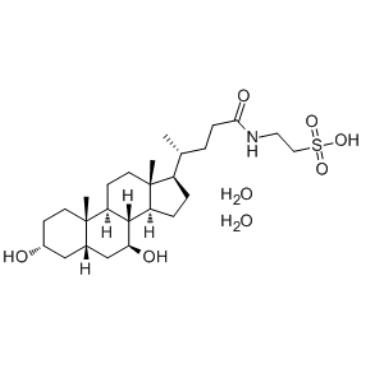
Tauroursodeoxycholate dihydrate
Tauroursodeoxycholate (Tauroursodeoxycholic acid; TDUCA) dihydrate is an endoplasmic reticulum (ER) stress inhibitor. Tauroursodeoxycholate significantly reduces expression of apoptosis molecules, such as caspase-3 and caspase-12. Tauroursodeoxycholate also inhibits ERK.
-
GC10978
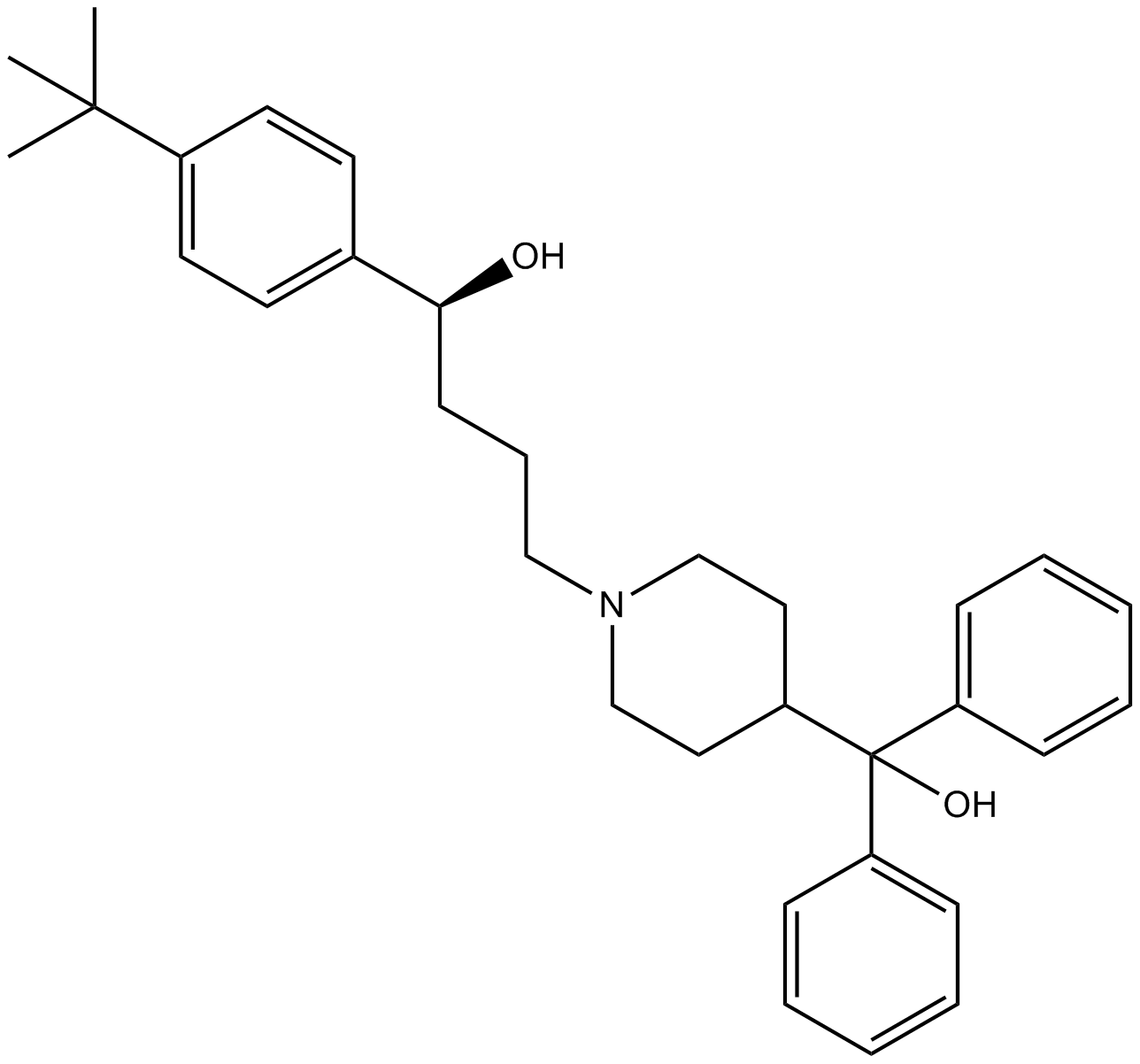
Terfenadine
Histamine H1-receptor antagonist
-
GC37780
Thevetiaflavone
Thevetiaflavone could upregulate the expression of Bcl?2 and downregulate that of Bax and caspase?3.
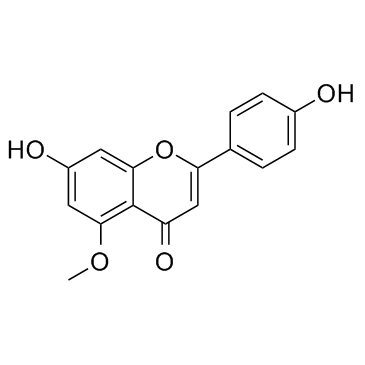
-
GC12223
Z-Asp-CH2-DCB
caspase inhibitor
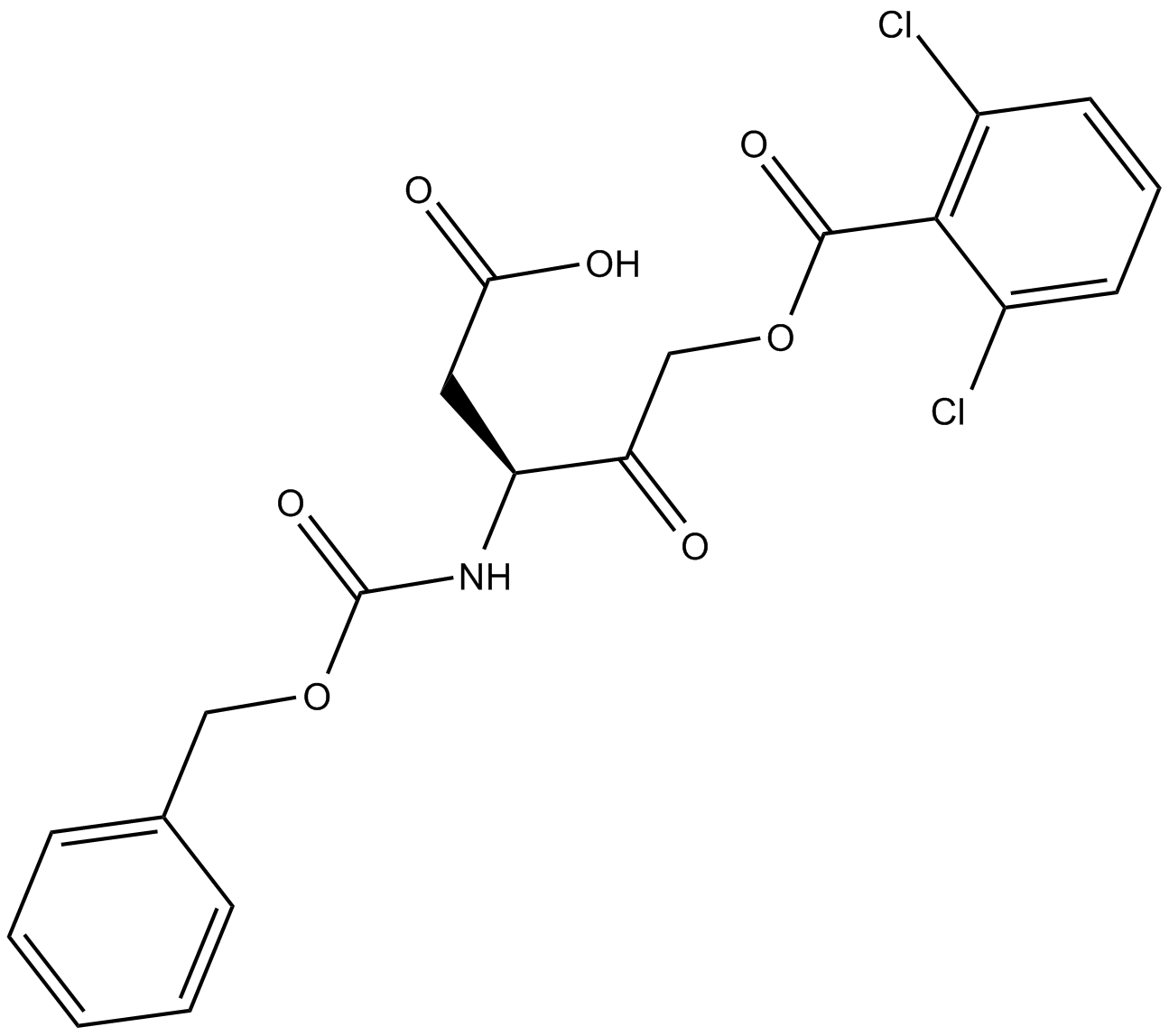
-
GC66403
Z-DEVD-AMC
Z-DEVD-AMC is a selective caspase-3 substrate that can be measured by fluorescence spectrometry. AMC can be used as a fluorescence reference standard for AMC-based enzyme substrates including AMC-based caspase substrates.
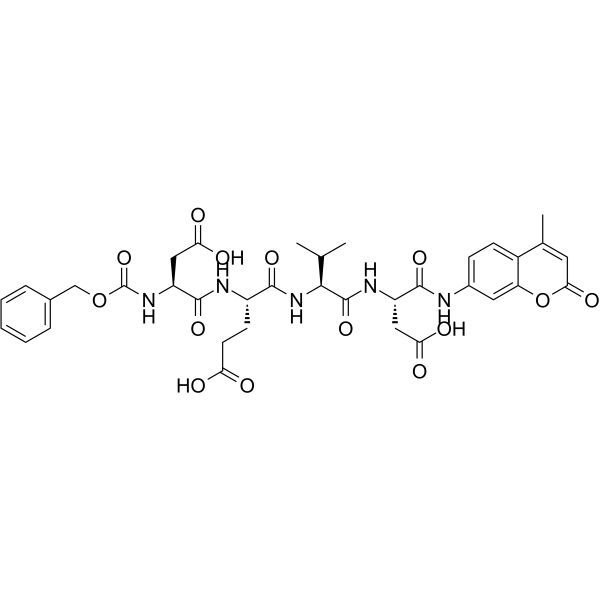
-
GC12407
Z-IETD-FMK
Caspase-8 inhibitor
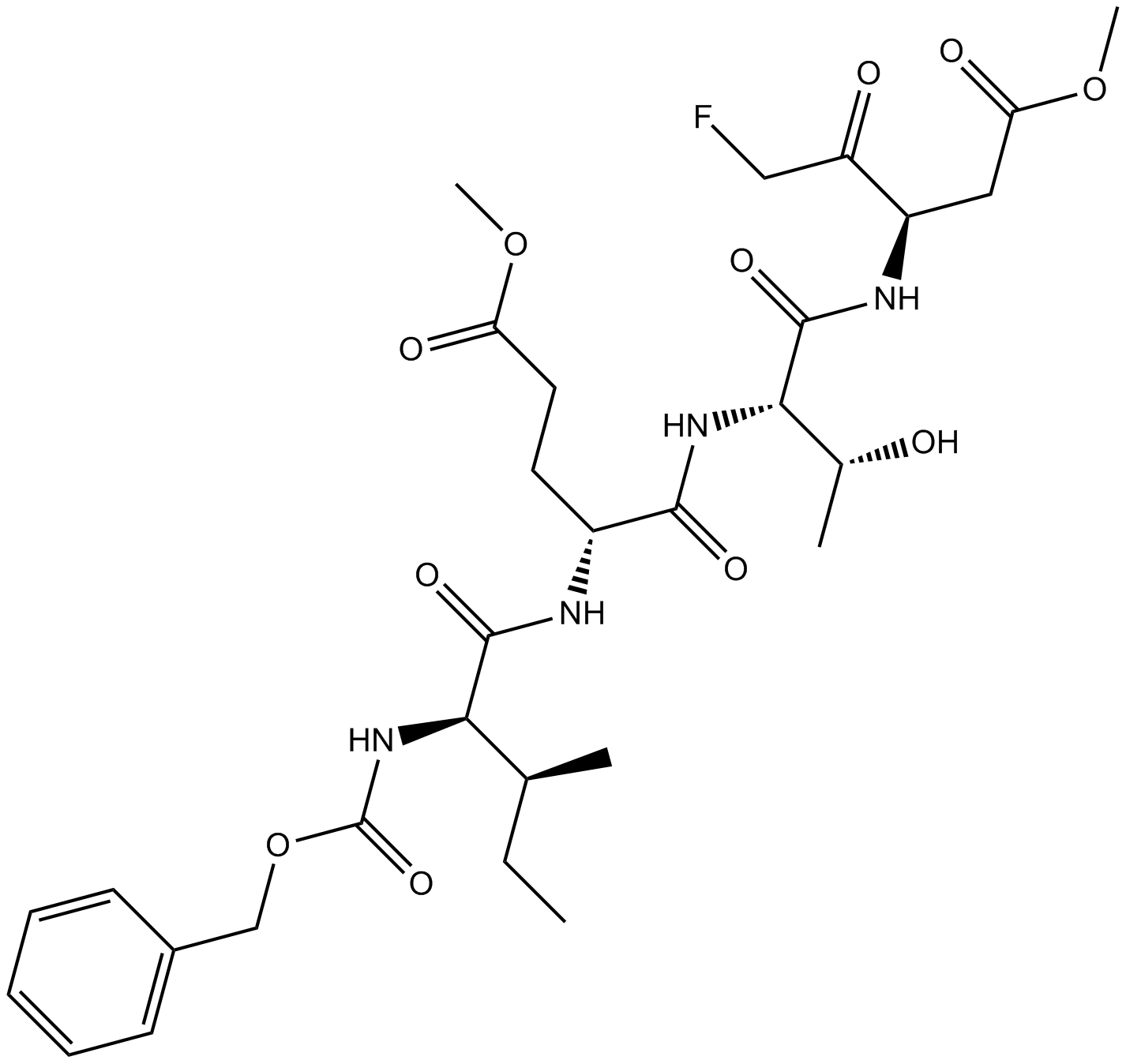
-
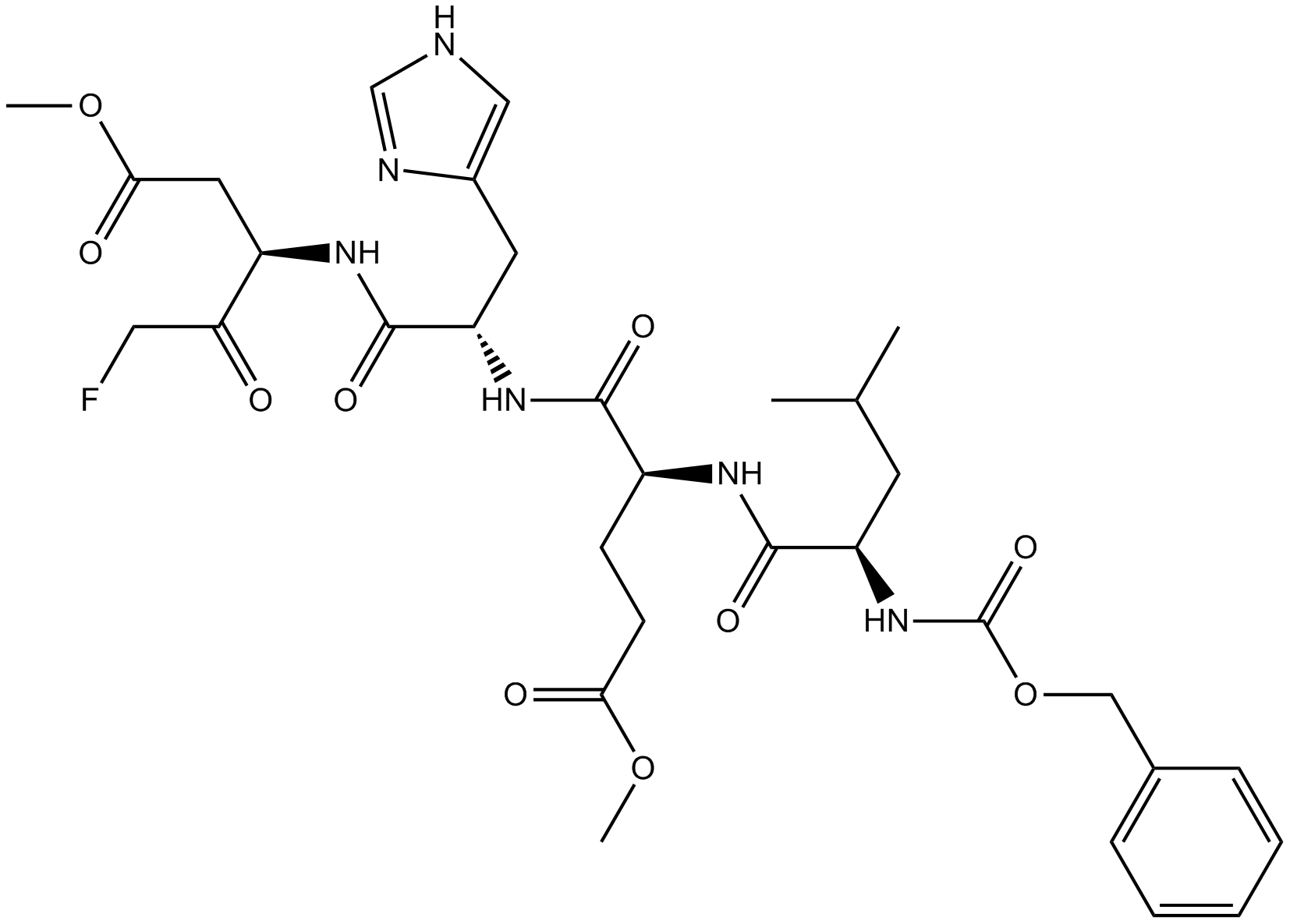
GC16990
Z-LEHD-FMK
Irreversible Caspase-9 inhibitor.
-
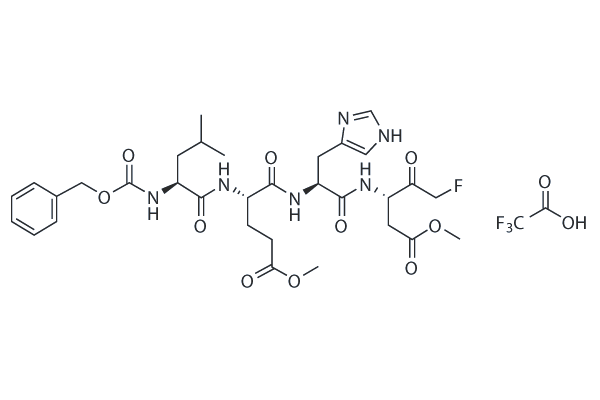
GC26092
Z-LEHD-FMK TFA
Z-LEHD-FMK TFA (Caspase-9 Inhibitor) is a cell-permeable, competitive and irreversible inhibitor of enzyme caspase-9, which helps in cell survival.
-
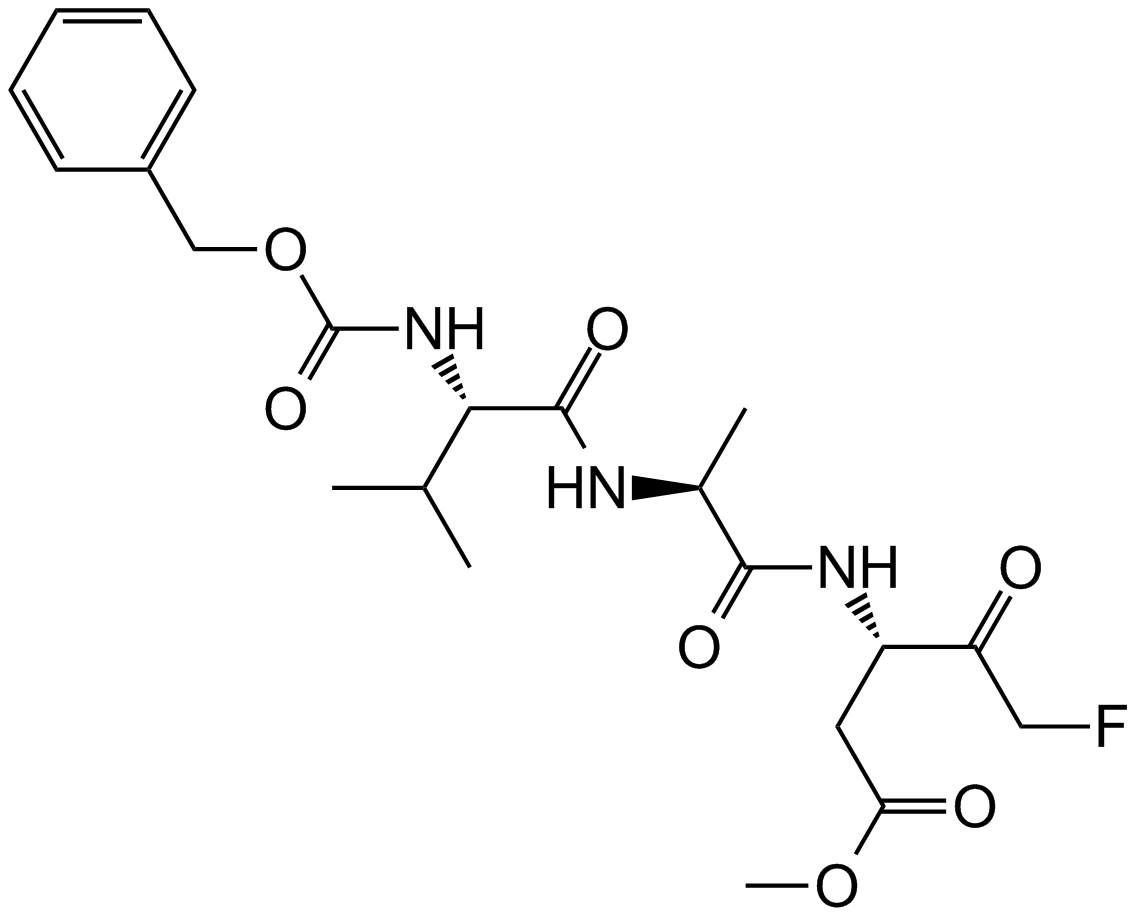
GC12861
Z-VAD-FMK
Z-VAD-FMK (Z-Val-Ala-Asp(OMe)-FMK) is a cell-permeable and irreversible pan-caspase inhibitor. Z-VAD-FMK is an ubiquitin carboxy-terminal hydrolase L1 (UCHL1) inhibitor. Z-VAD-FMK irreversibly modifies UCHL1 by targeting the active site of UCHL1.
-
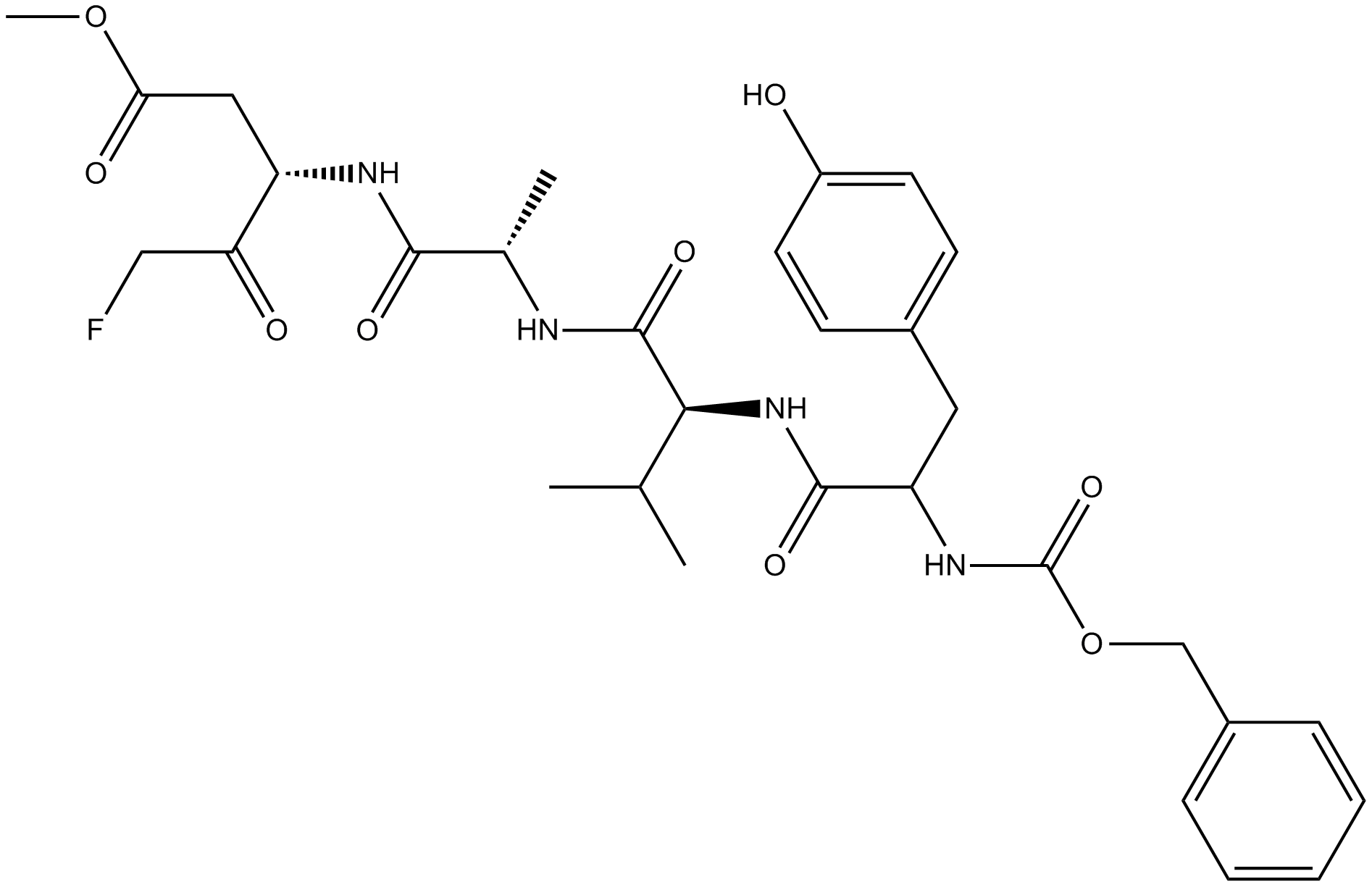
GC14188
Z-YVAD-FMK
Caspase-1 inhibitor
-
GC37974
ZYZ-488
ZYZ-488 is a competitive apoptotic protease activating factor-1 (Apaf-1) inhibitor. ZYZ-488 inhibits the activation of binding protein procaspase-9 and procaspase-3.