Other Apoptosis
Products for Other Apoptosis
- Cat.No. Product Name Information
-
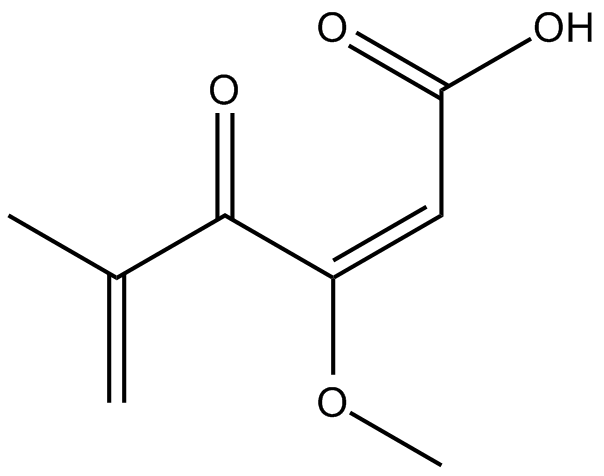
GC11837
MI-136
menin-MLL interaction inhibitor
-
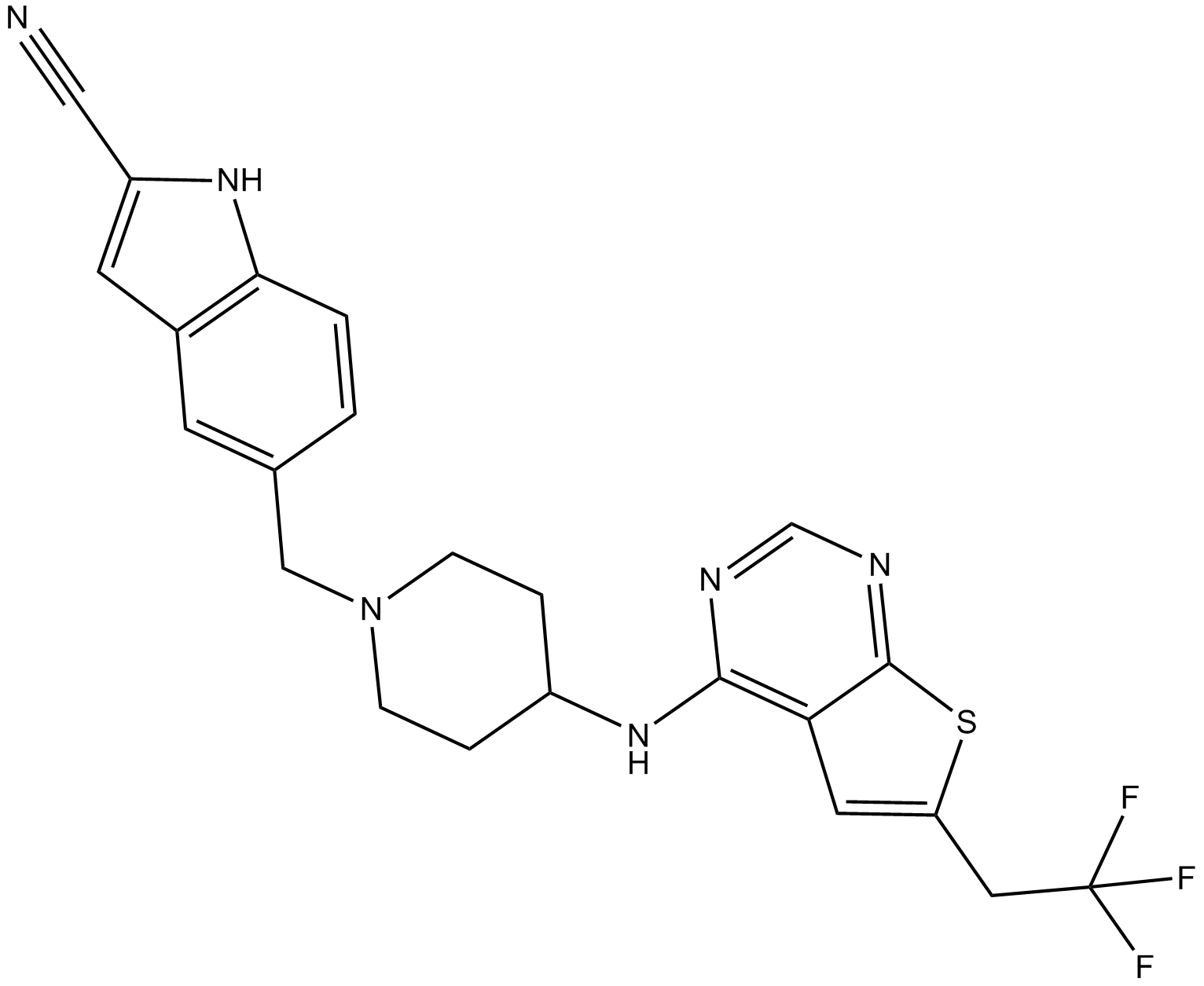
GC11418
MI-2
An inhibitor of menin-MLL fusion protein interactions
-
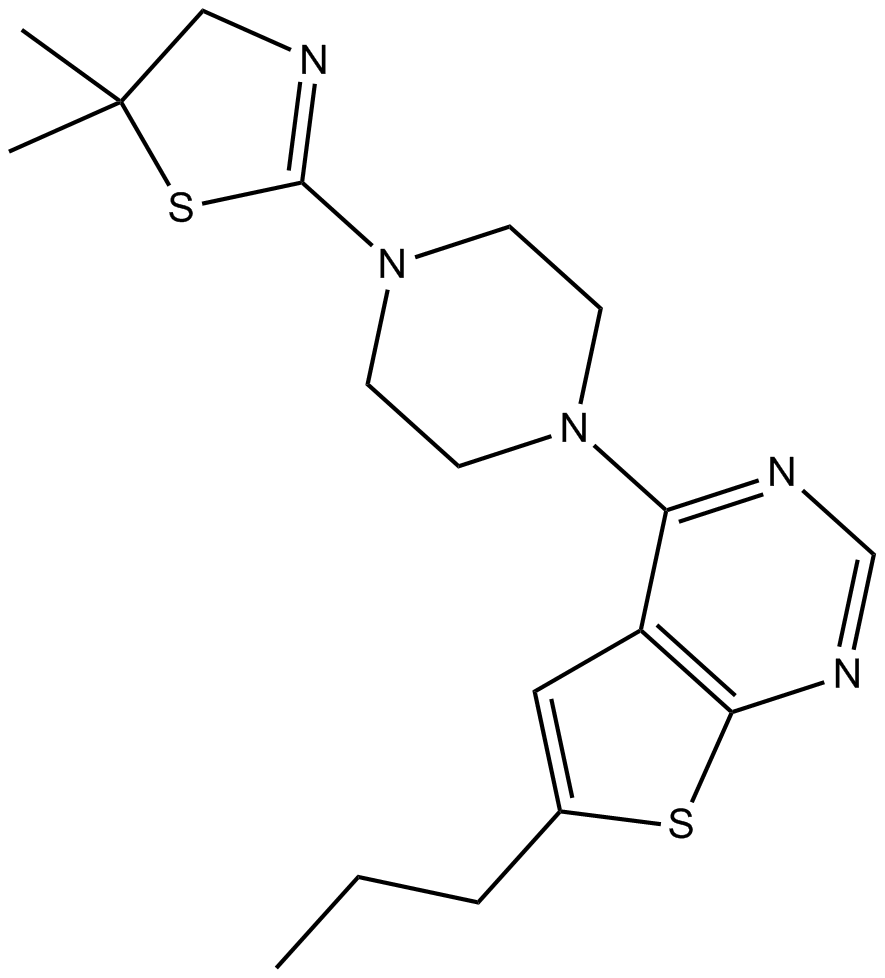
GC12199
MI-3
An inhibitor of menin-MLL fusion protein interactions
-
GC38817
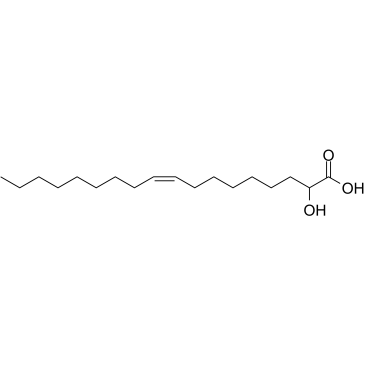
Minerval
Minerval (2-Hydroxyoleic acid) is a synthetic oleic acid (OA) derivative that binds to the plasma membrane and alters lipid organization. Minerval has anti-tumor effect.
-
GC36612
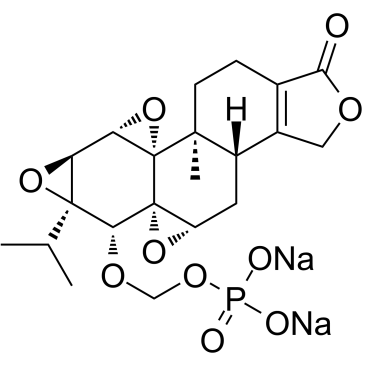
Minnelide
Minnelide is a prodrug of triptolide that shows potent antitumor activity in a number of tumor types, particularly in pancreatic cancer. Minnelide promotes apoptosis.
-
GC36613
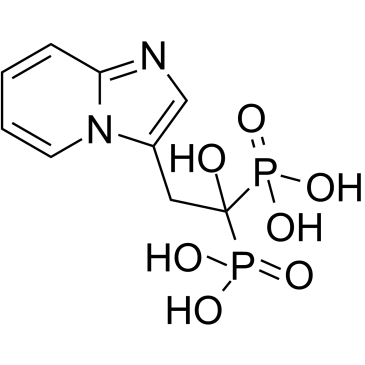
Minodronic acid
Minodronic acid (YM-529) is a third-generation bisphosphonate that directly and indirectly prevents proliferation, induces apoptosis, and inhibits metastasis of various types of cancer cells. Minodronic acid (YM-529) is an antagonist of purinergic P2X2/3 receptors involved in pain.
-
GC32929
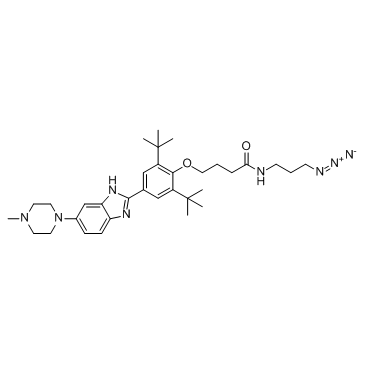
MIR96-IN-1
MIR96-IN-1 targets the Drosha site in the miR-96 (miRNA-96, microRNA-96) hairpin precursor, inhibiting its biogenesis, derepressing downstream targets, and triggering apoptosis in breast cancer cells. MIR96-IN-1 binds to RNAs with Kds of 1.3, 9.4, 3.4, 1.3 and 7.4 μM for RNA1, RNA2, RNA3, RNA4 and RNA5, respectively.
-
GC36619
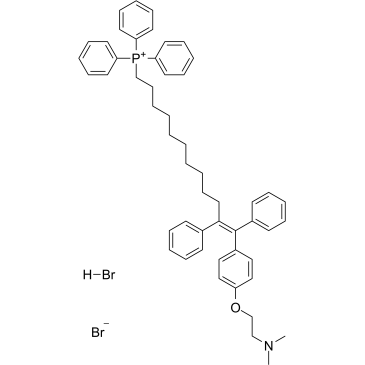
MitoTam bromide, hydrobromide
MitoTam bromide, hydrobromide, a Tamoxifen derivative, is an electron transport chain (ETC) inhibitor. MitoTam bromide, hydrobromide reduces mitochondrial membrane potential in senescent cells and affects mitochondrial morphology. MitoTam bromide, hydrobromide is an effective anticancer agent, suppresses respiratory complexes (CI-respiration) and disrupts respiratory supercomplexes (SCs) formation in breast cancer cells.
-
GC36620
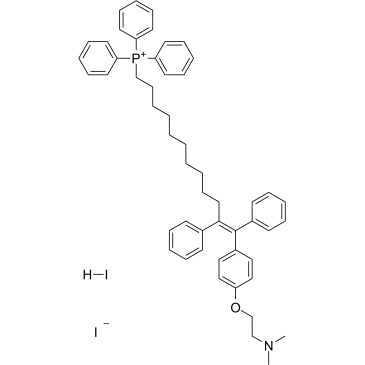
MitoTam iodide, hydriodide
MitoTam iodide, hydriodide is a Tamoxifen derivative, an electron transport chain (ETC) inhibitor, spreduces mitochondrial membrane potential in senescent cells and affects mitochondrial morphology.MitoTam iodide, hydriodide is an effective anticancer agent, suppresses respiratory complexes (CI-respiration) and disrupts respiratory supercomplexes (SCs) formation in breast cancer cells. MitoTam iodide, hydriodide causes apoptosis.
-
GC16976
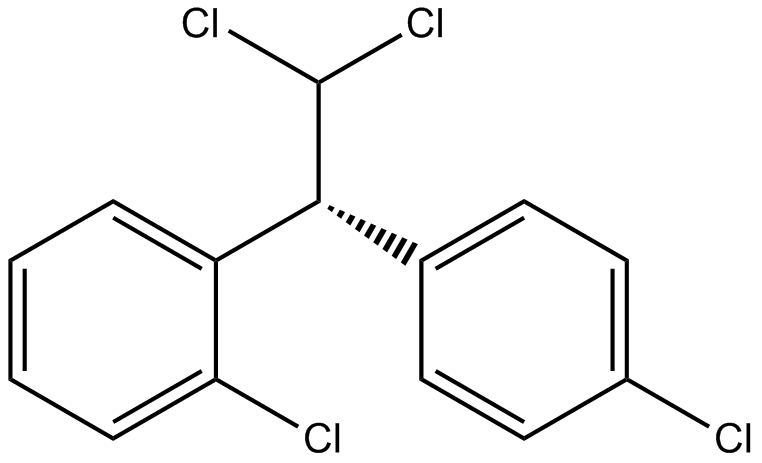
Mitotane (Lsodren)
Mitotane (Lsodren) (2,4′-DDD), an isomer of DDD and derivative of dichlorodiphenyltrichloroethane (DDT), is an antineoplastic agent, can be used to research adrenocortical carcinoma. Mitotane (Lsodren) exert its adrenocorticolytic effect at least in part through lipotoxicity induced by intracellular free cholesterol (FC) accumulation. Mitotane (Lsodren) can have direct pituitary effects on corticotroph cells. Mitotane (Lsodren) can induce CYP3A4 gene expression via steroid and xenobiotic receptor (SXR) activation, and has drug-drug interactions.
-
GC19248
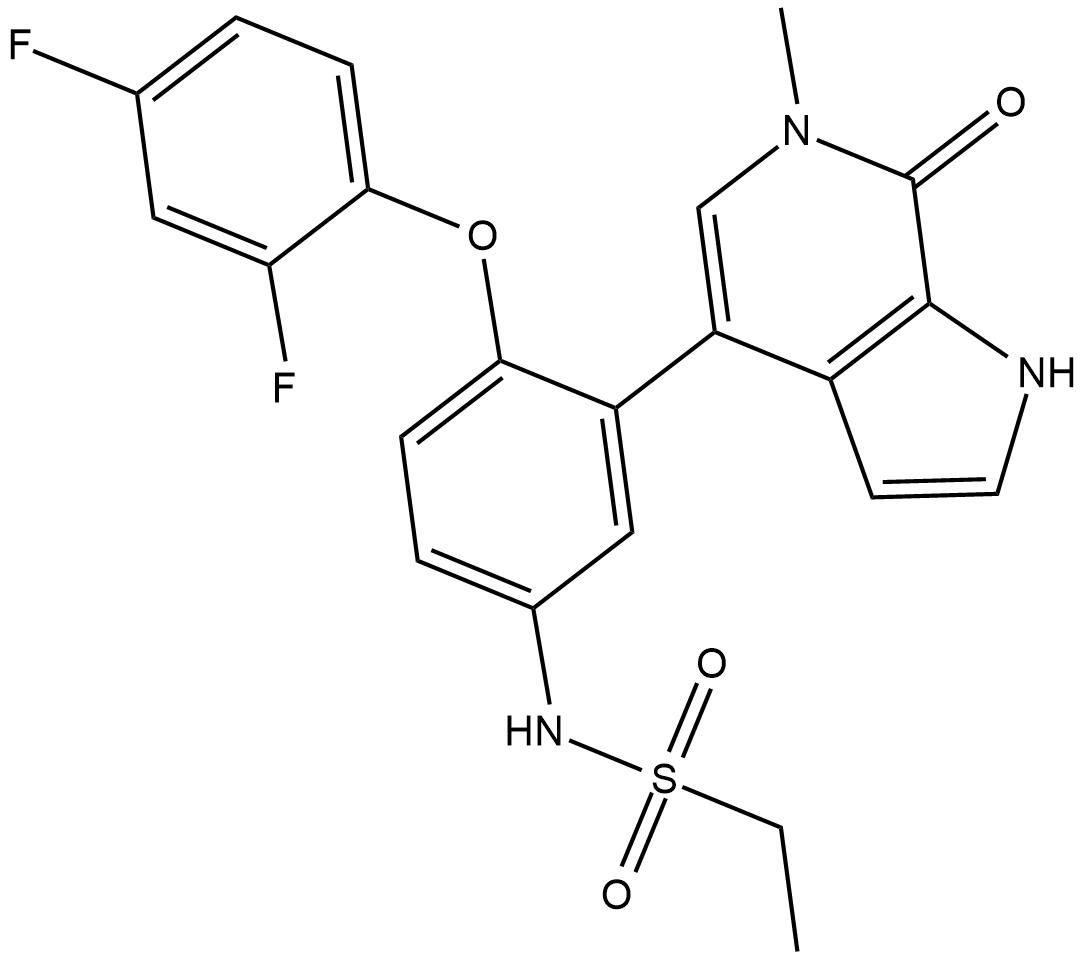
Mivebresib
Mivebresib is a potent and orally available bromodomain and extraterminal domain (BET) bromodomain inhibitor.
-
GC13630
MK-4101
Hedgehog (Hh) signaling pathway inhibitor

-
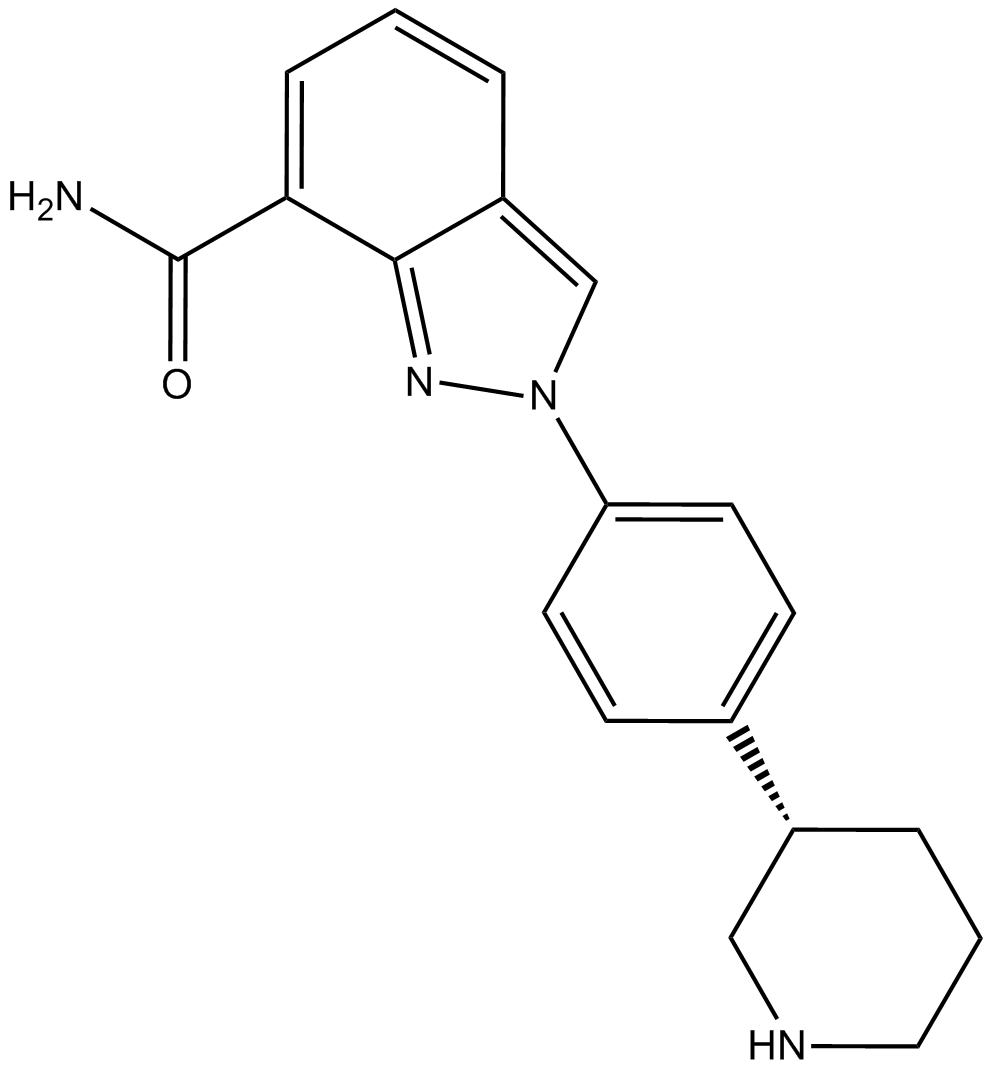
GC17802
MK-4827
An orally bioavailable PARP1/2 inhibitor
-
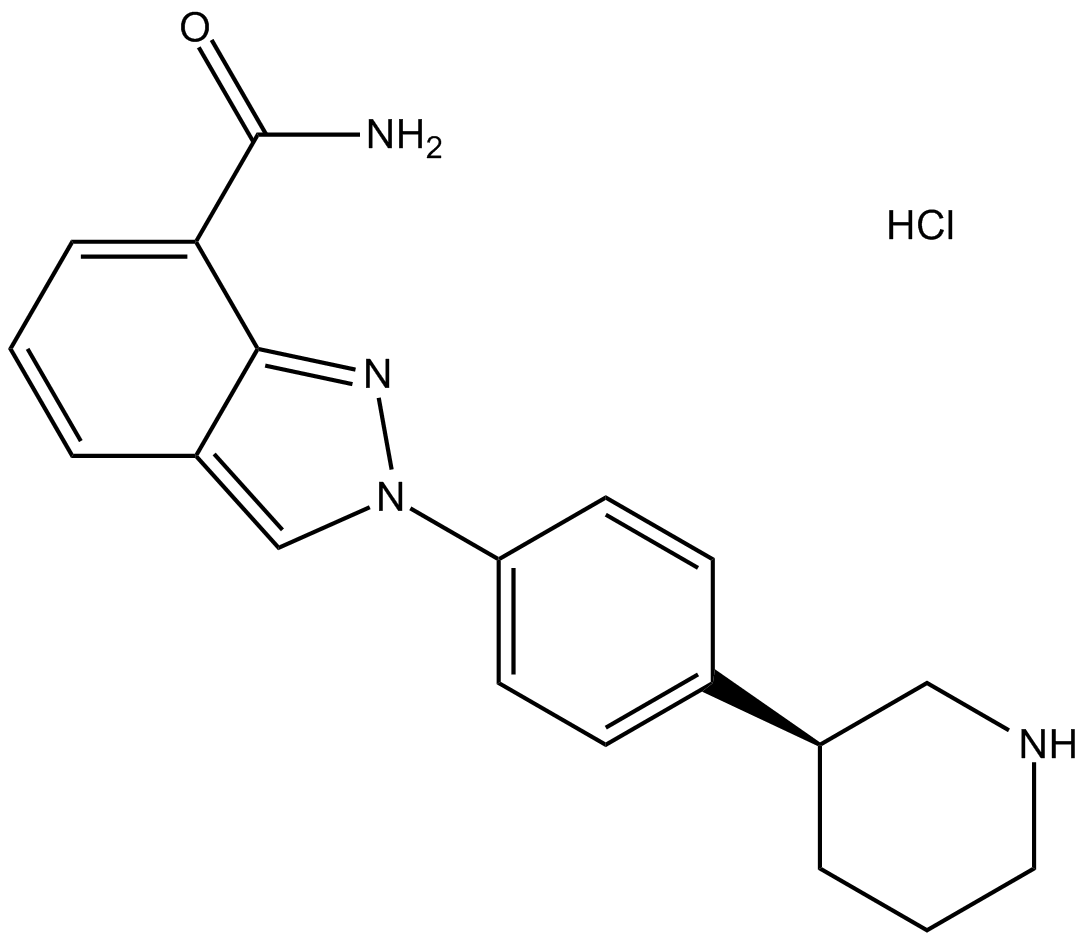
GC12756
MK-4827 hydrochloride
MK-4827 hydrochloride (MK-4827 hydrochloride) is a highly potent and orally bioavailable PARP1 and PARP2 inhibitor with IC50s of 3.8 and 2.1 nM, respectively. MK-4827 hydrochloride leads to inhibition of repair of DNA damage, activates apoptosis and shows anti-tumor activity.
-
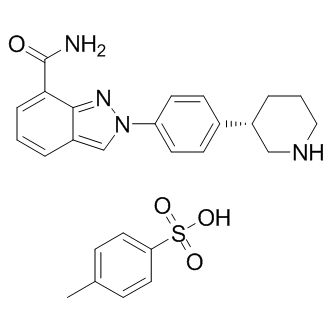
GC11537
MK-4827 tosylate
MK-4827 tosylate (MK-4827 tosylate) is a highly potent and orally bioavailable PARP1 and PARP2 inhibitor with an IC50 of 3.8 and 2.1 nM, respectively. MK-4827 tosylate leads to inhibition of repair of DNA damage, activates apoptosis and shows anti-tumor activity.
-
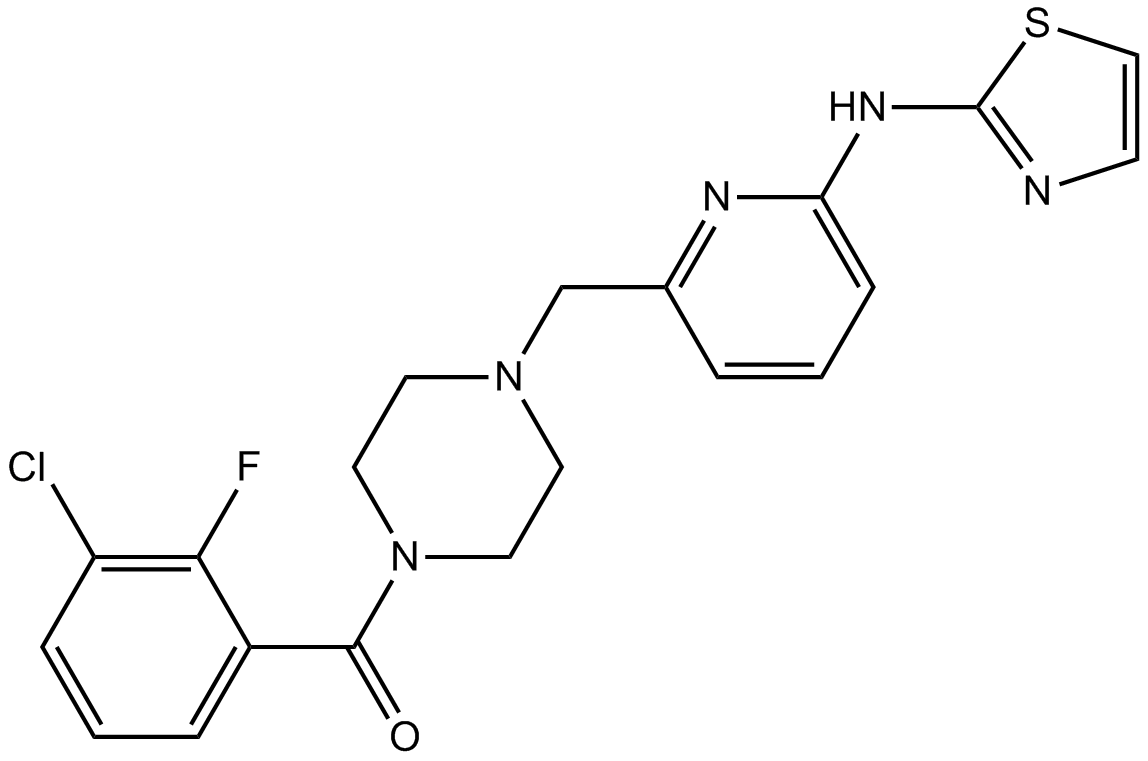
GC10442
MK-8745
Aurora A inhibitor,potent and selective
-
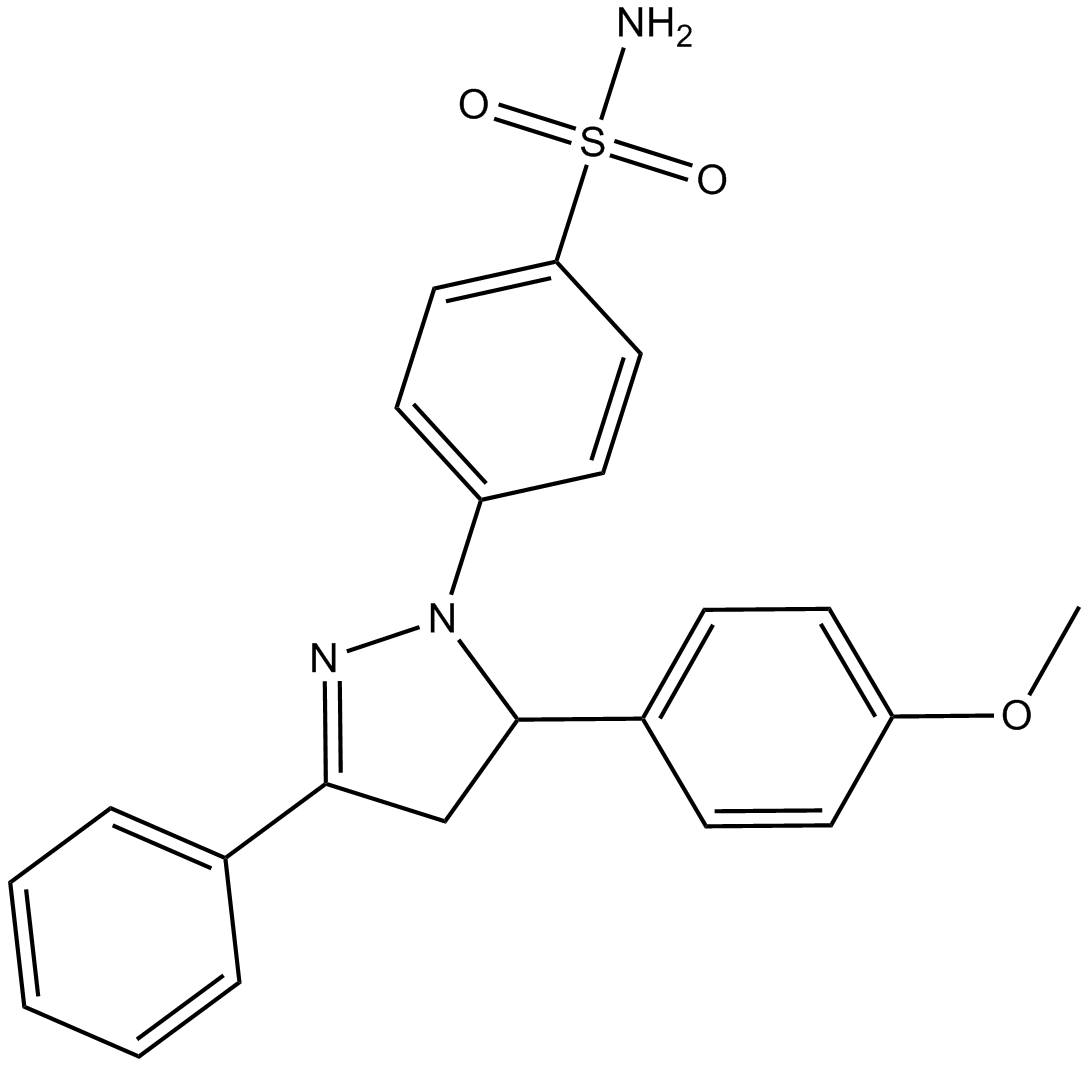
GC14800
ML 141
Cdc42 GTPase inhibitor
-
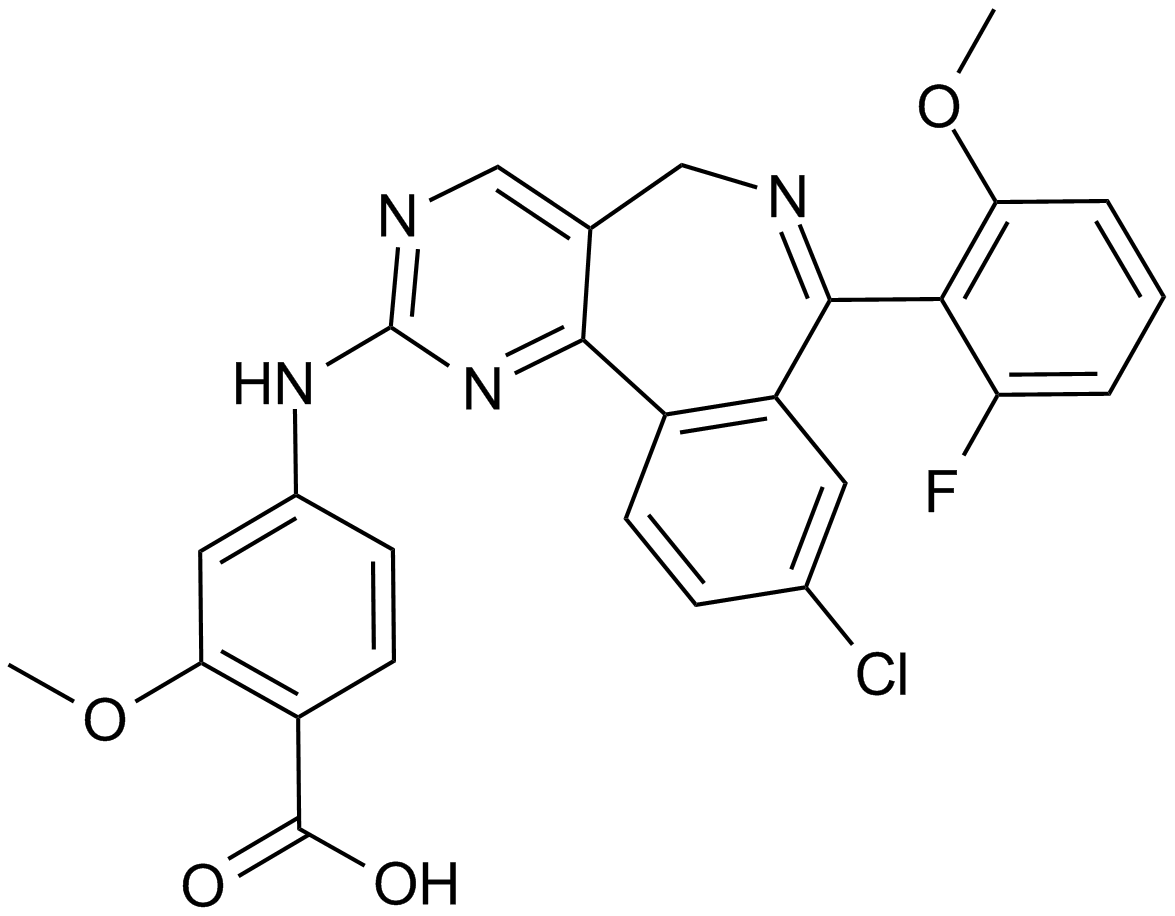
GC12690
MLN8237 (Alisertib)
An Aurora A kinase inhibitor
-
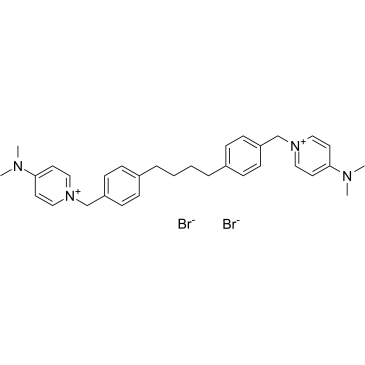
GC38406
MN58b
MN58b is a selective choline kinase α (CHKα) inhibitor, and results in inhibition of phosphocholine synthesis. MN58b reduces cell growth through the induction of apoptosis, and also has antitumoral activity.
-
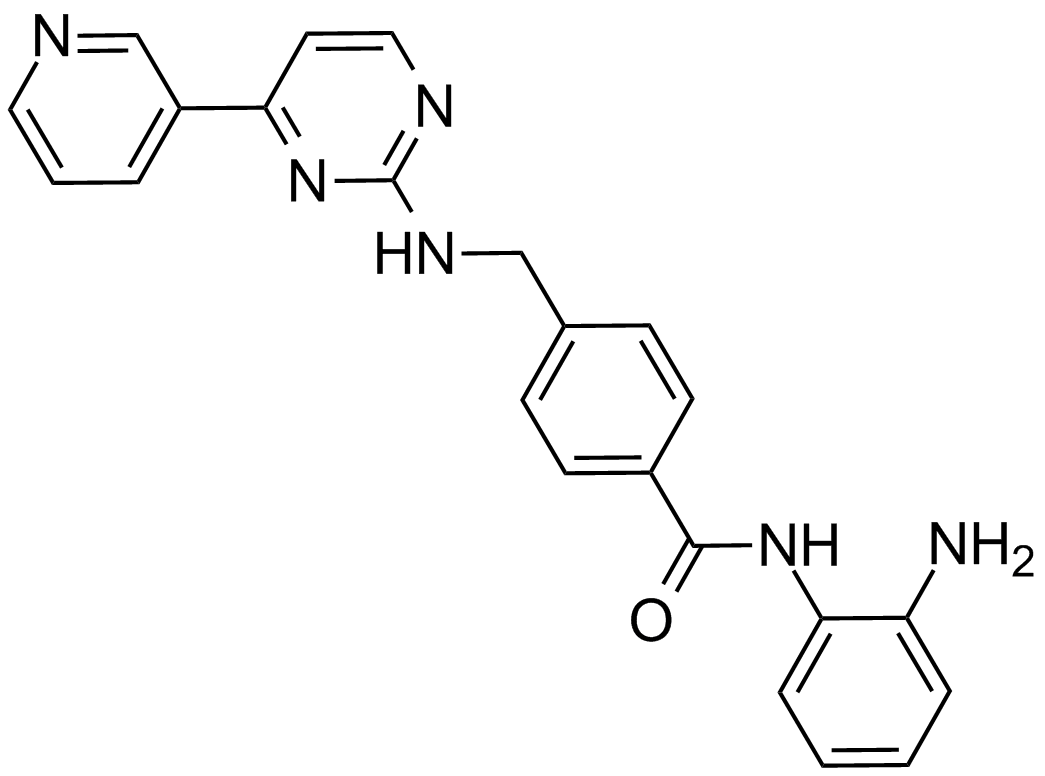
GC13055
Mocetinostat (MGCD0103, MG0103)
An orally available HDAC inhibitor
-
GC11655
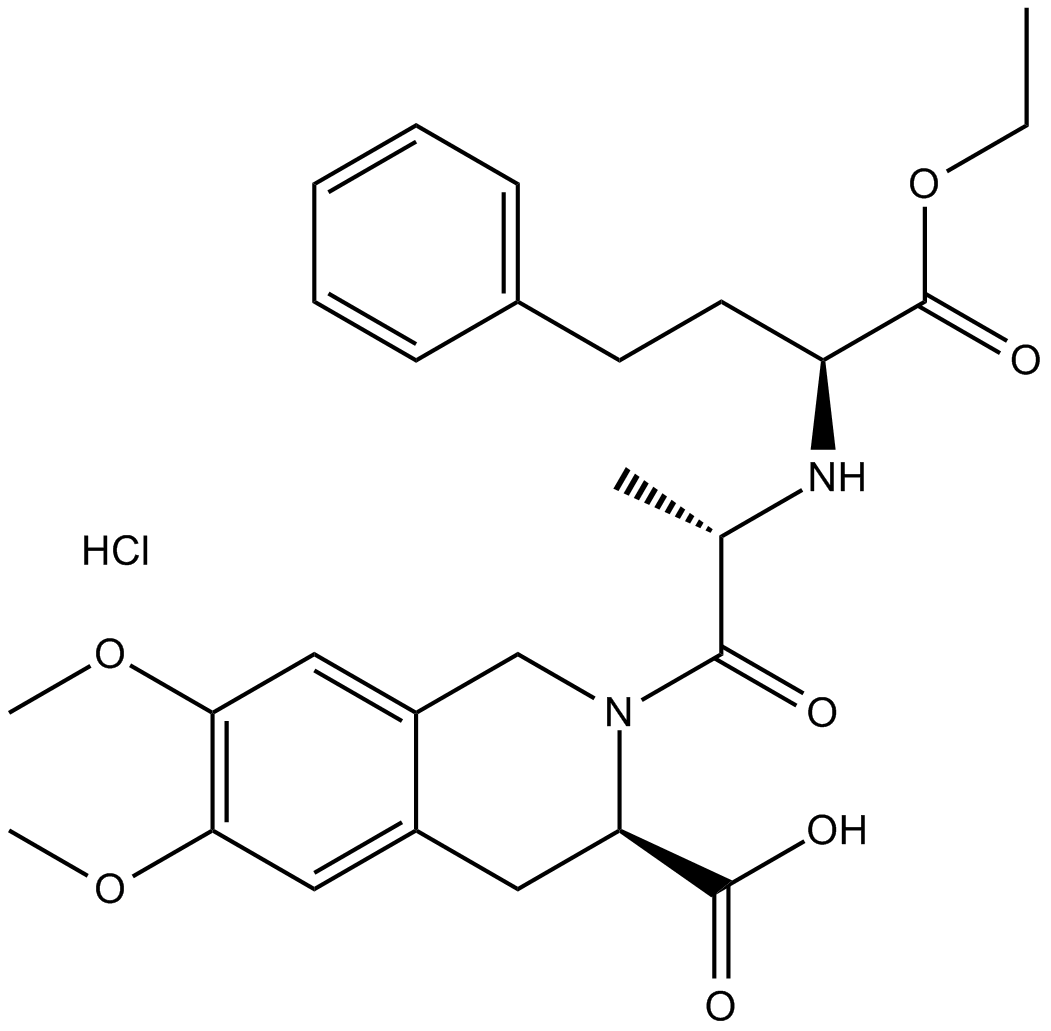
Moexipril HCl
Moexipril HCl (RS-10085) is an orally active inhibitor of angiotensin-converting enzyme (ACE), and becomes effective by being hydrolyzed to moexiprila (hydrochloride).
-
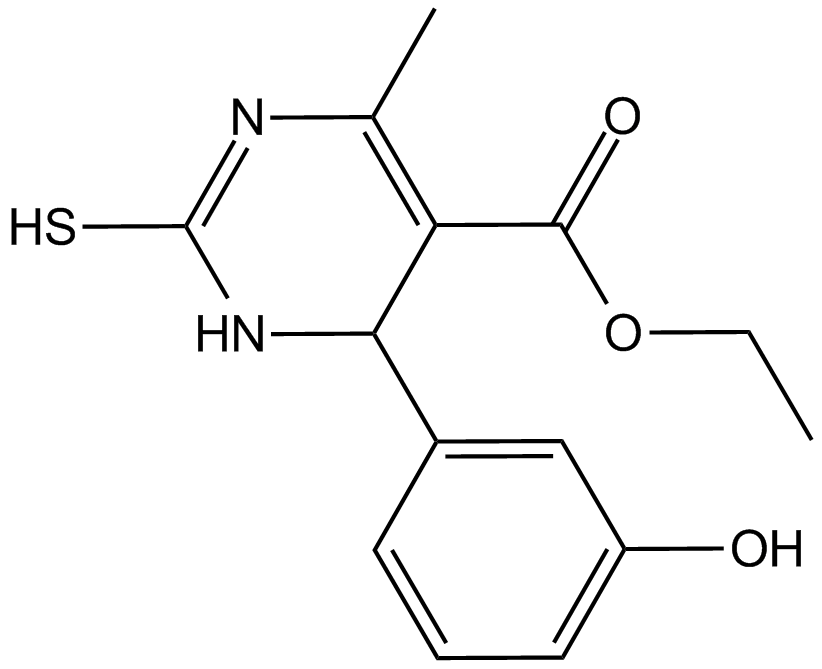
GC14929
Monastrol
Eg5 inhibitor
-
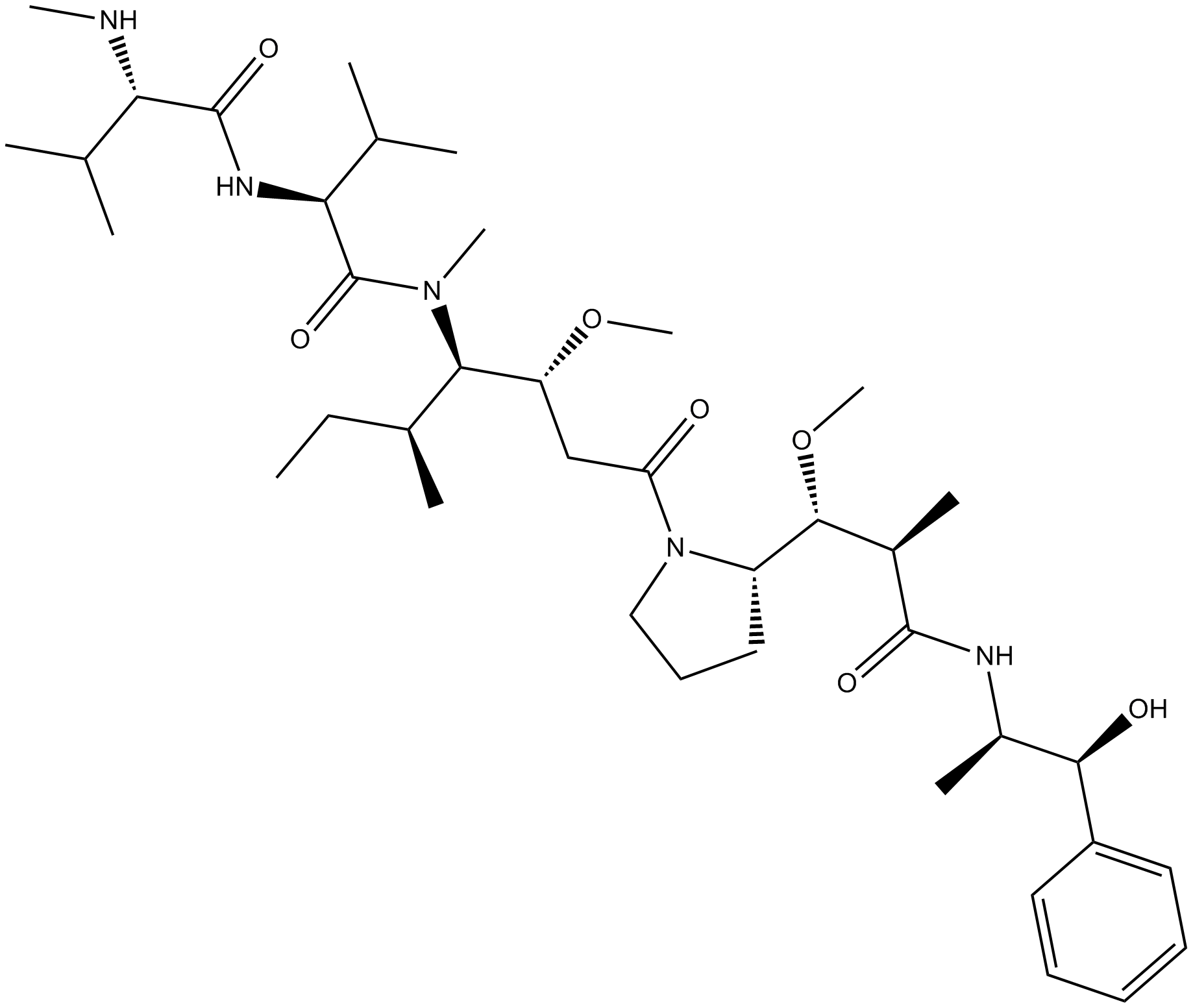
GC17276
Monomethyl auristatin E
A potent antimitotic compound
-
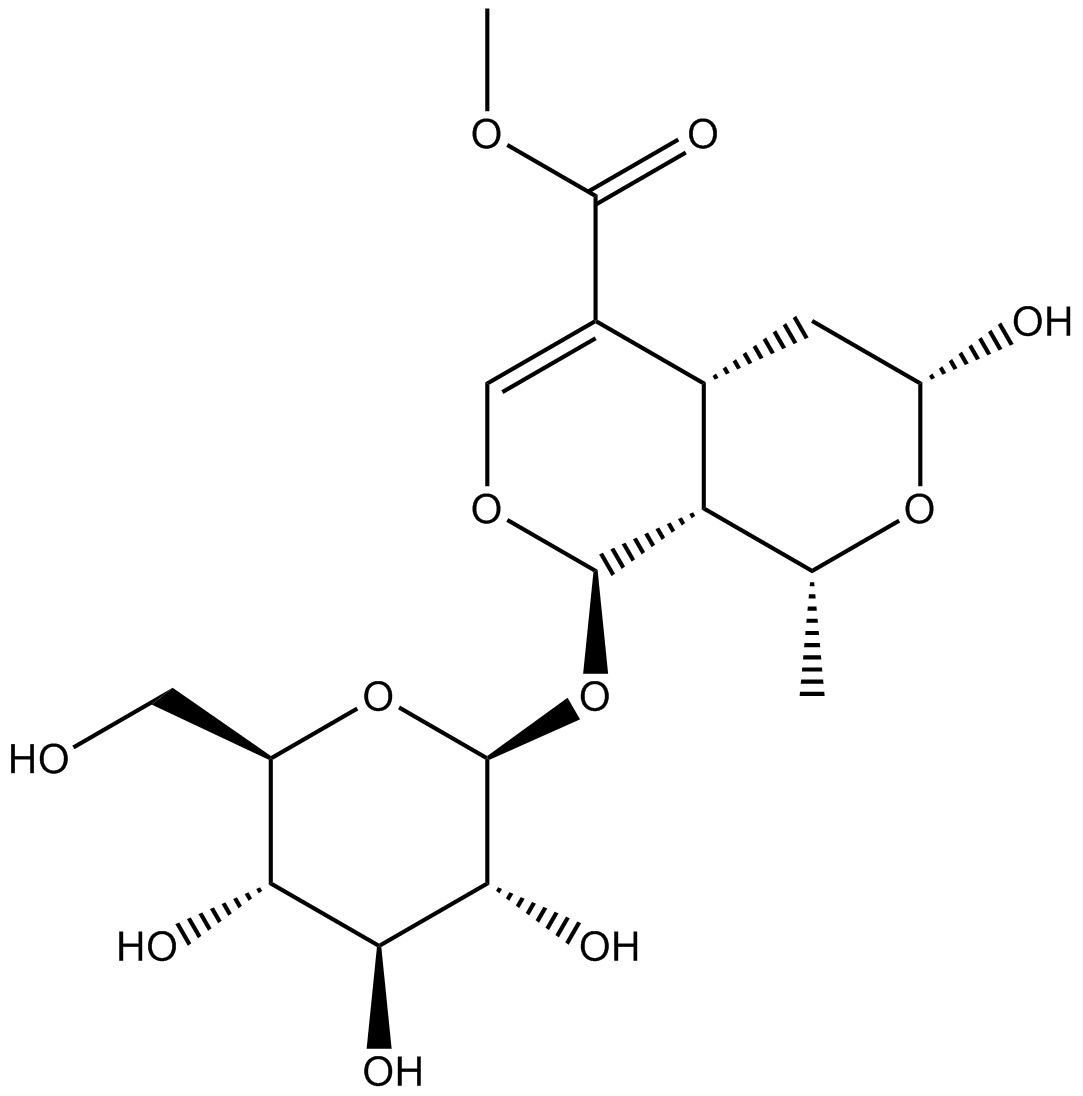
GN10360
Morroniside
-
GC10083
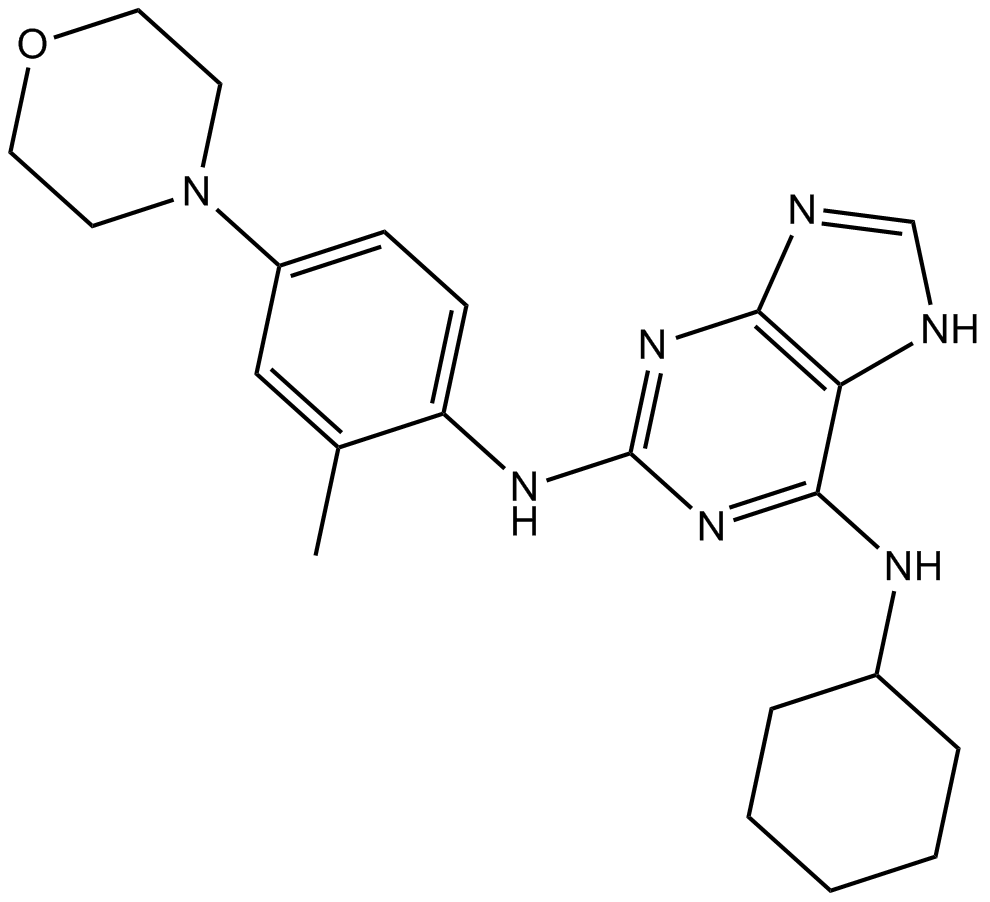
MPI-0479605
Mps1 inhibitor,selective and ATP competitive
-
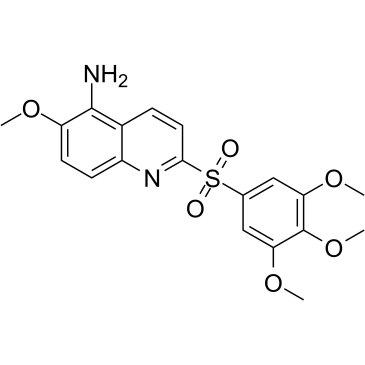
GC36652
MPT0B392
MPT0B392, an orally active quinoline derivative, induces c-Jun N-terminal kinase (JNK) activation, leading to apoptosis. MPT0B392 inhibits tubulin polymerization and triggers induction of the mitotic arrest, followed by mitochondrial membrane potential loss and caspases cleavage by activation of JNK and ultimately leads to apoptosis. MPT0B392 is demonstrated to be a novel microtubule-depolymerizing agent and enhances the cytotoxicity of sirolimus in sirolimus-resistant acute leukemic cells and the multidrug resistant cell line.
-
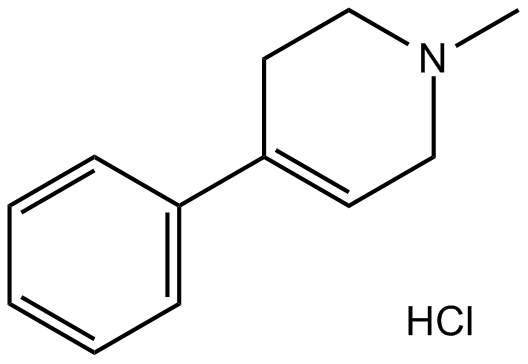
GC11097
MPTP hydrochloride
MPTP (1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine) is a neurotoxic agent that is a precusor of MPP+ which is toxic to dopaminergic neurons and causes Parkinsonism.
-
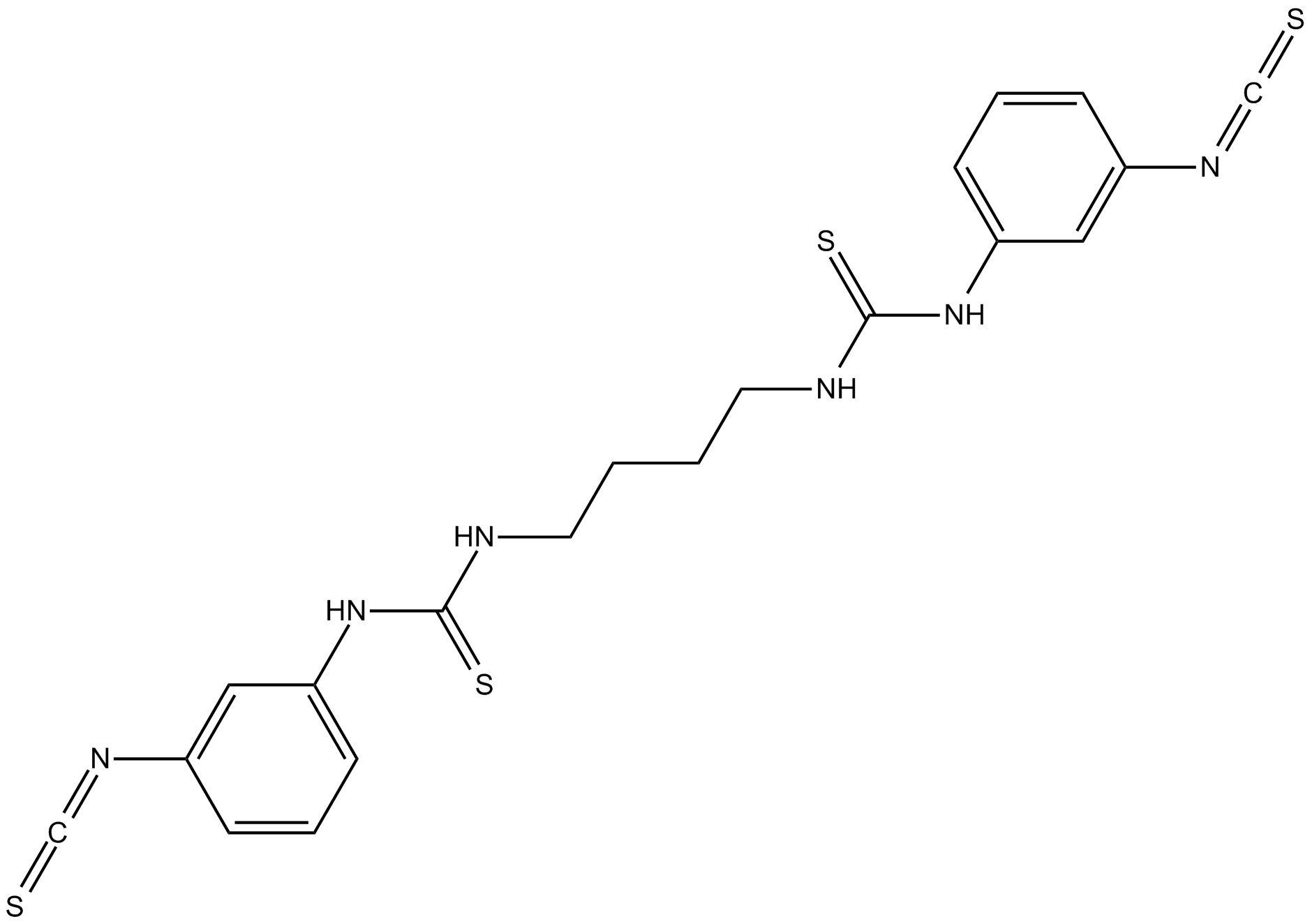
GC11078
MRS 2578
P2Y6 receptor antagonist,potent and selective
-
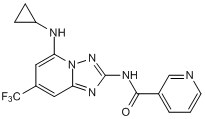
GC50223
MSC 2032964A
Potent and selective ASK1 inhibitor; orally bioavailable
-
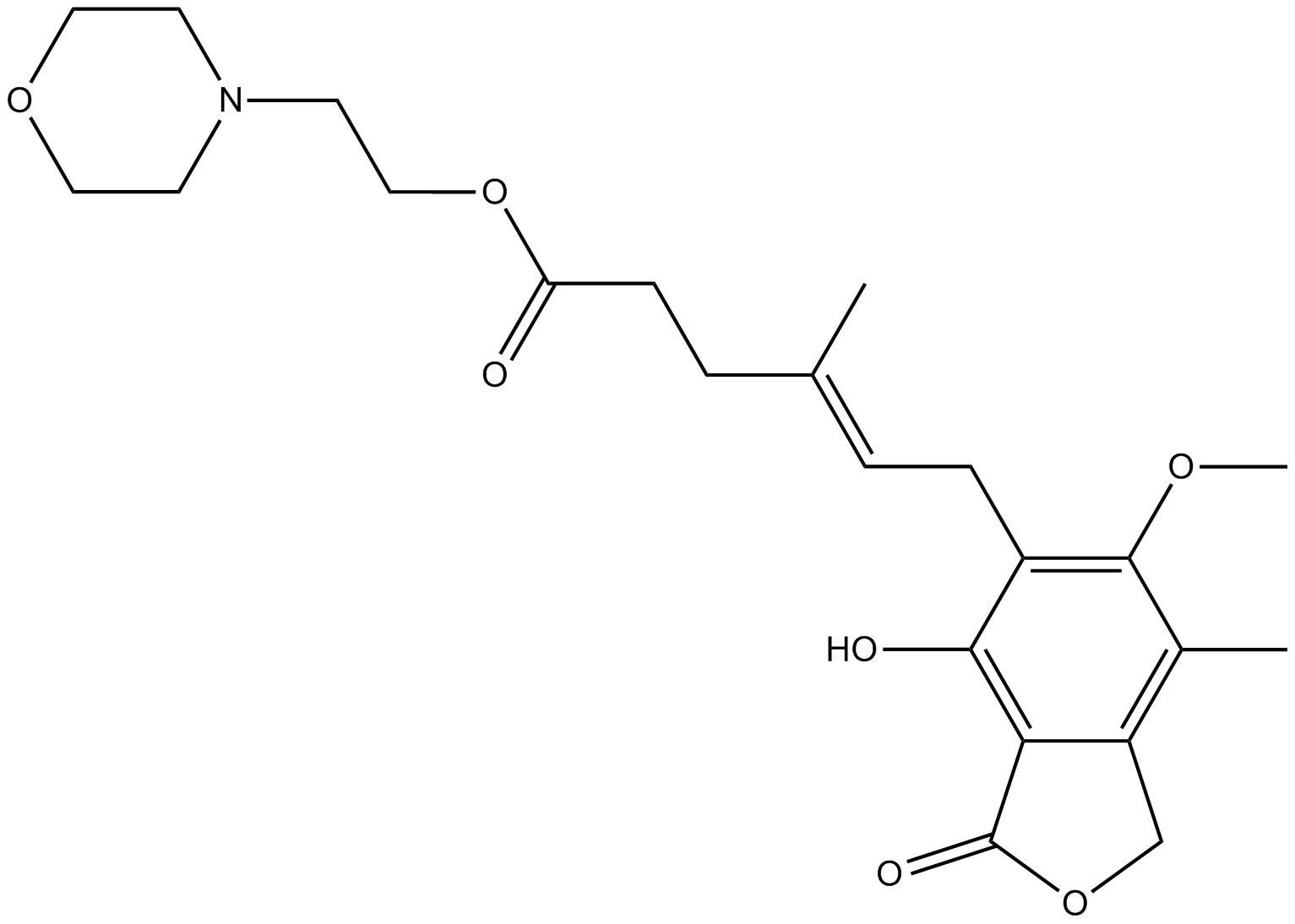
GC13614
Mycophenolate Mofetil
A prodrug form of mycophenolic acid
-
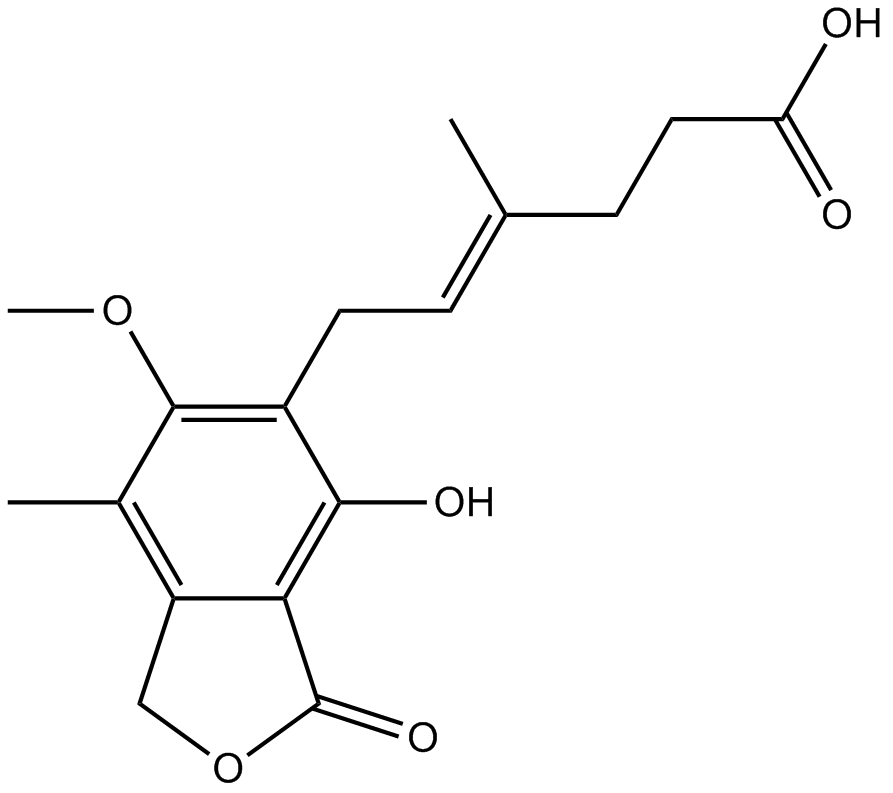
GC17121
Mycophenolic acid
Dehydrogenase inhibitor
-
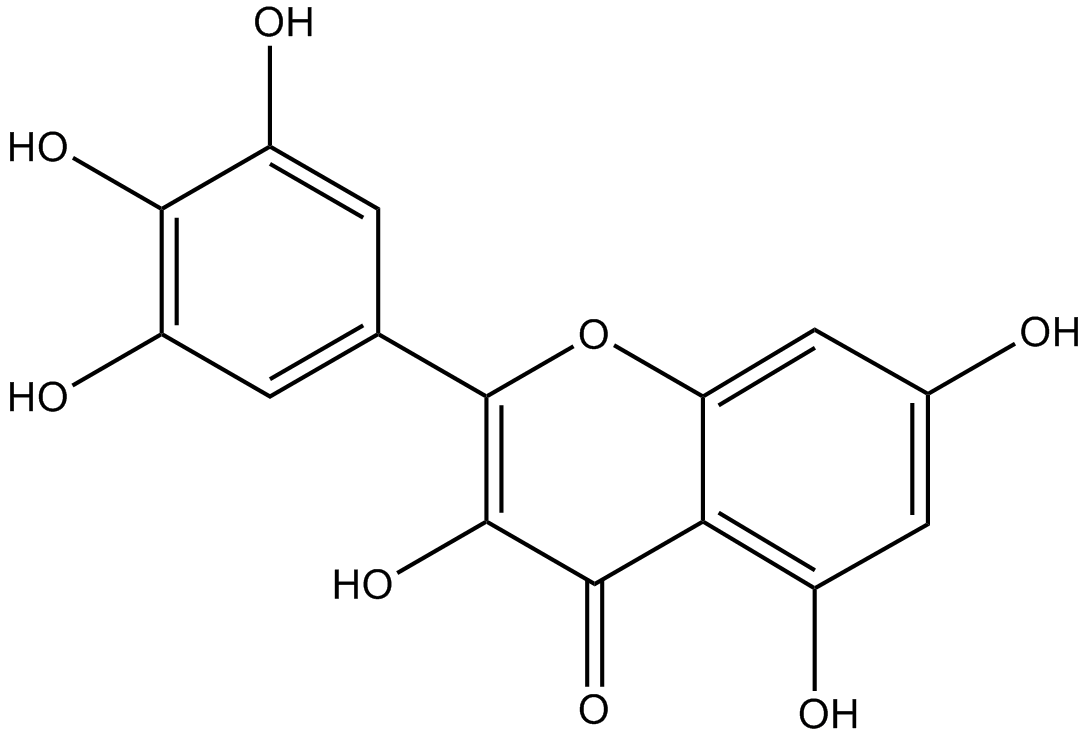
GN10634
Myricetin
-
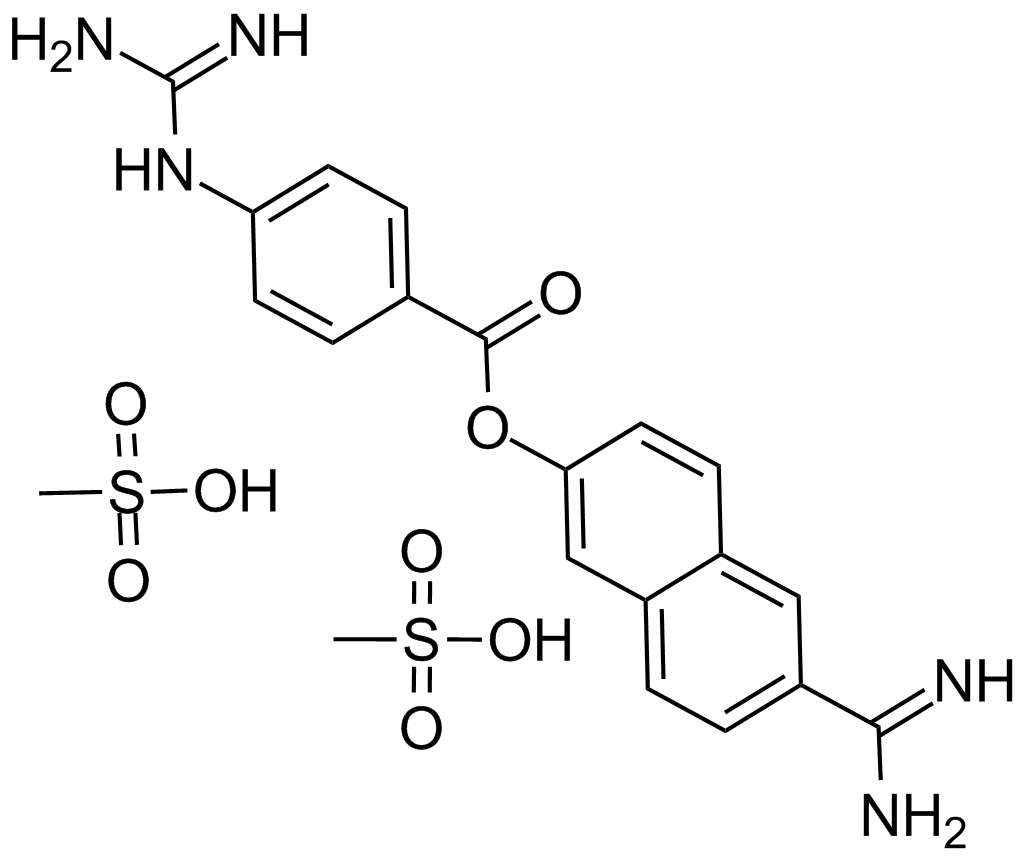
GC10613
Nafamostat Mesylate(FUT-175)
A serine protease inhibitor
-
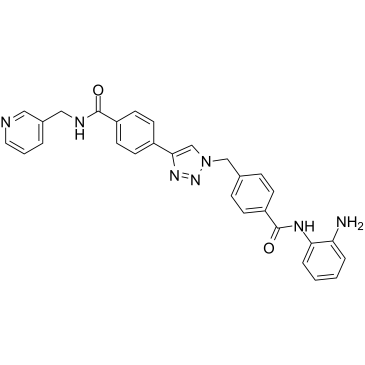
GC36690
Nampt-IN-3
Nampt-IN-3 (Compound 35) simultaneously inhibit nicotinamide phosphoribosyltransferase (NAMPT) and HDAC with IC50s of 31 nM and 55 nM, respectively. Nampt-IN-3 effectively induces cell apoptosis and autophagy and ultimately leads to cell death.
-
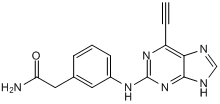
GC50112
NCL 00017509
NCL 00017509 (compound 6) is a potent and irreversible Nek2 ((Never in mitosis gene a)-related kinase 2) inhibitior.
-
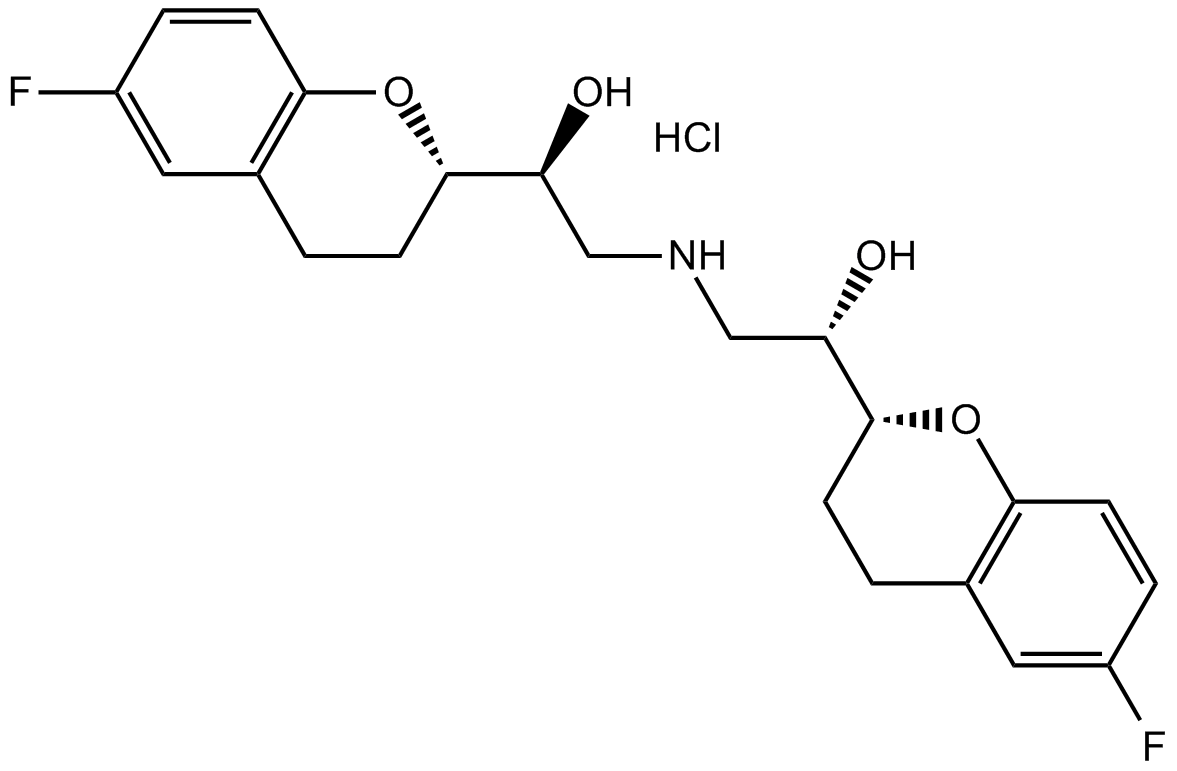
GC13906
Nebivolol hydrochloride
Highly selective β1-adrenoceptor antagonist
-
GC10150
Necrosulfonamide
Necrosulfonamide (NSA) is a specific inhibitor of MLKL (mixed lineage kinase domain-like protein)[1].
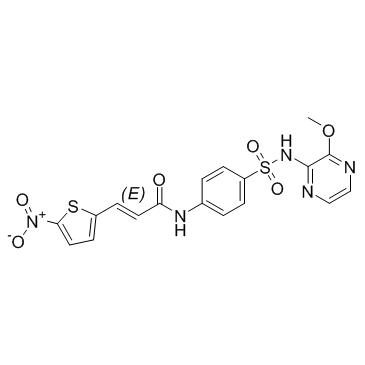
-
GN10103
Neferine

-
GC10591
Nelarabine
Prodrug of ara-G for T-LBL/T-ALL
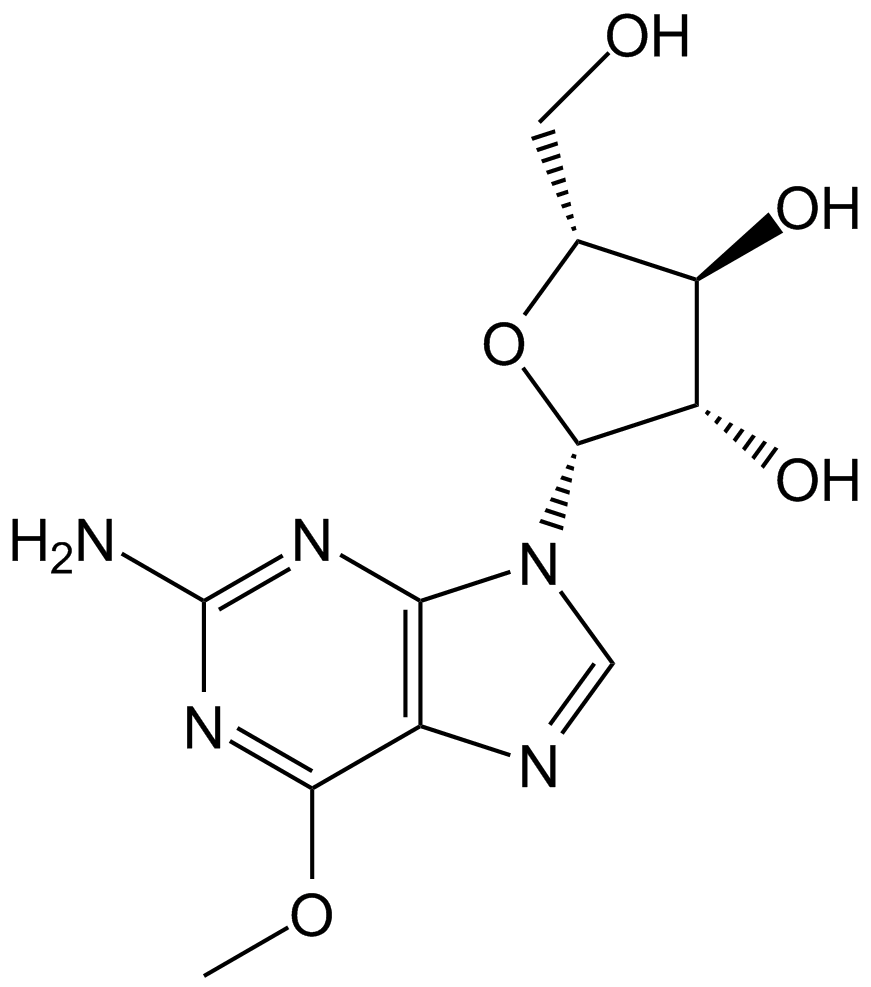
-
GC30085
Neobavaisoflavone
A natural isoflavone
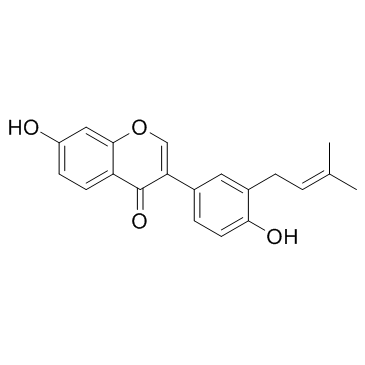
-
GC40669
Nerol
Nerol is a monoterpene and isomer of geraniol that has been found in a variety of plants, including Cannabis.
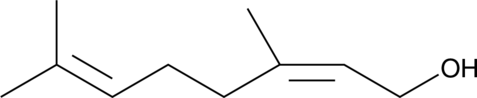
-
GC36743
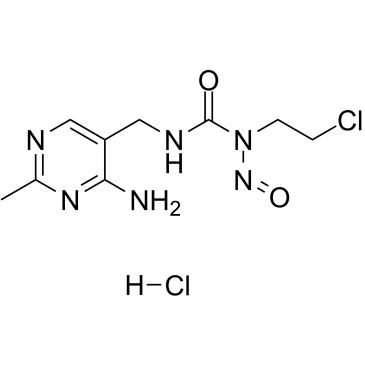
Nimustine hydrochloride
Nimustine hydrochloride (ACNU) is a DNA cross-linking and DNA alkylating agent, which induces DNA replication blocking lesions and DNA double-strand breaks and inhibits DNA synthesis, commonly used in chemotherapy for glioblastomas.
-
GC36747
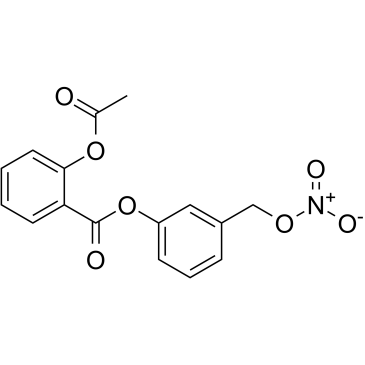
Nitroaspirin
Nitroaspirin (NCX 4016) is a nitric oxide (NO) donor and a nitro-derivative of Aspirin, which combines with Nitroaspirin to inhibit cyclooxygenase. Nitroaspirin (NCX 4016) has antithrombotic and anti-platelet properties and acts as a direct and irreversible inhibitor of COX-1. Nitroaspirin (NCX 4016) causes significant induction of cell cycle arrest and apoptosis in Cisplatin-resistant human ovarian cancer cells via down-regulation of EGFR/PI3K/STAT3 signaling and modulation of Bcl-2 family proteins.
-
GC44409
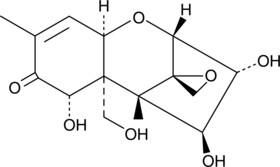
Nivalenol
Nivalenol is a type B trichothecene mycotoxin produced by Fusarium that is commonly found in contaminated foods.
-
GC19263
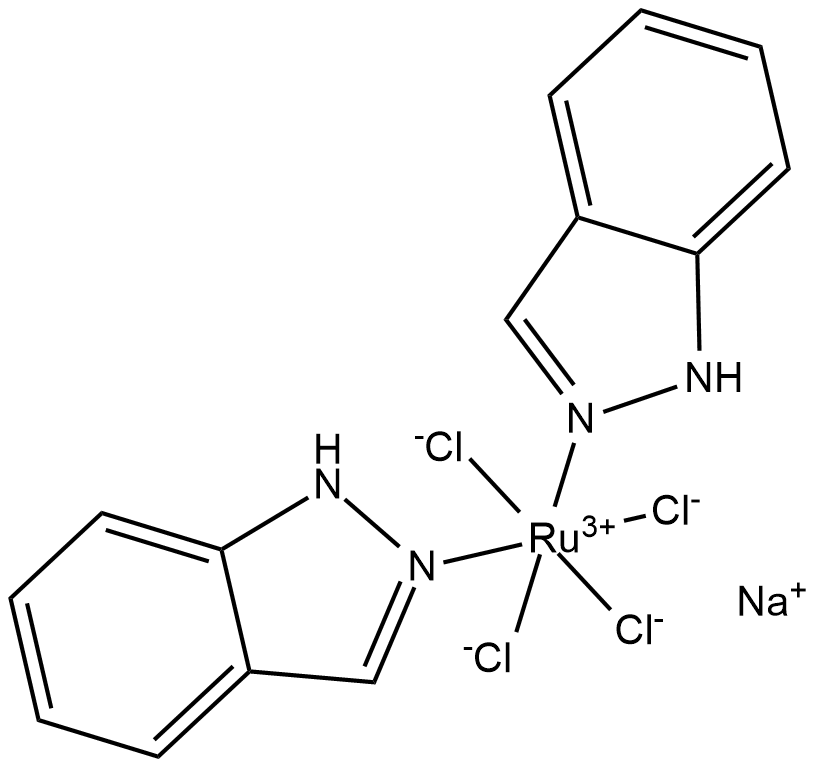
NKP-1339
NKP-1339(IT-139) is a ruthenium(iii) coordination anticancer compound based on target to transferrin.
-
GC12515
NMS-1286937
NMS-1286937 is a potent, selective and orally available PLK1 inhibitor, with an IC50 of 2 nM.
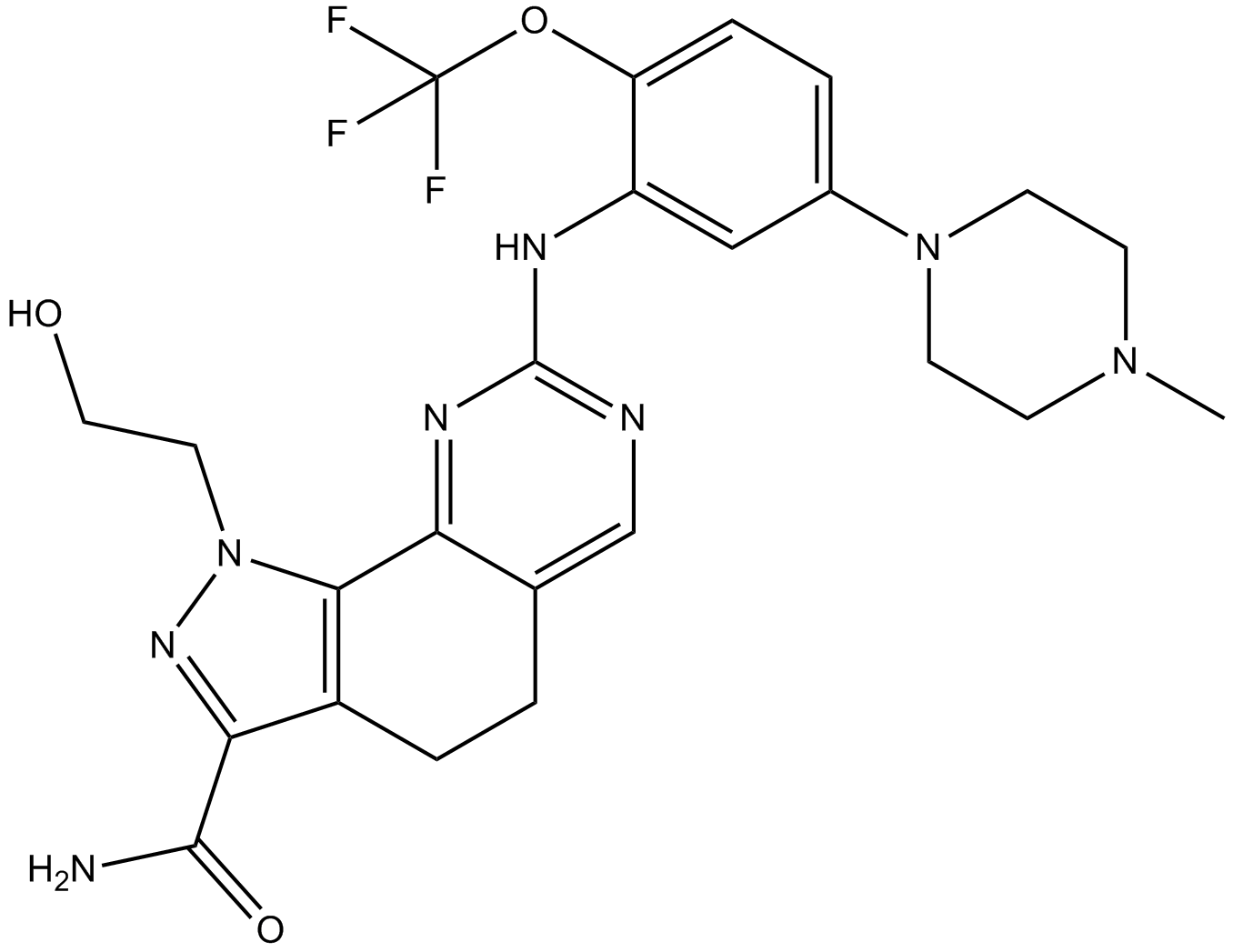
-
GN10325
Nobiletin
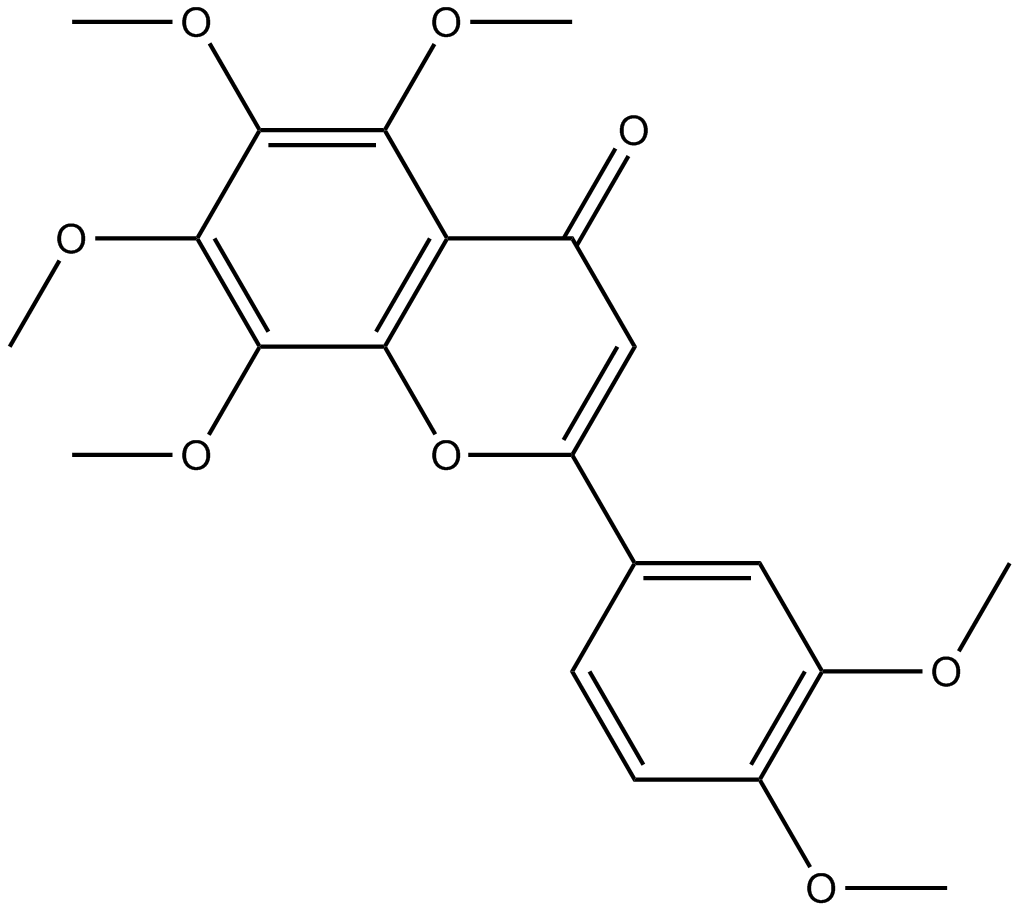
-
GC14075
Nocodazole
A tubulin production inhibitor,anti-neoplastic agent
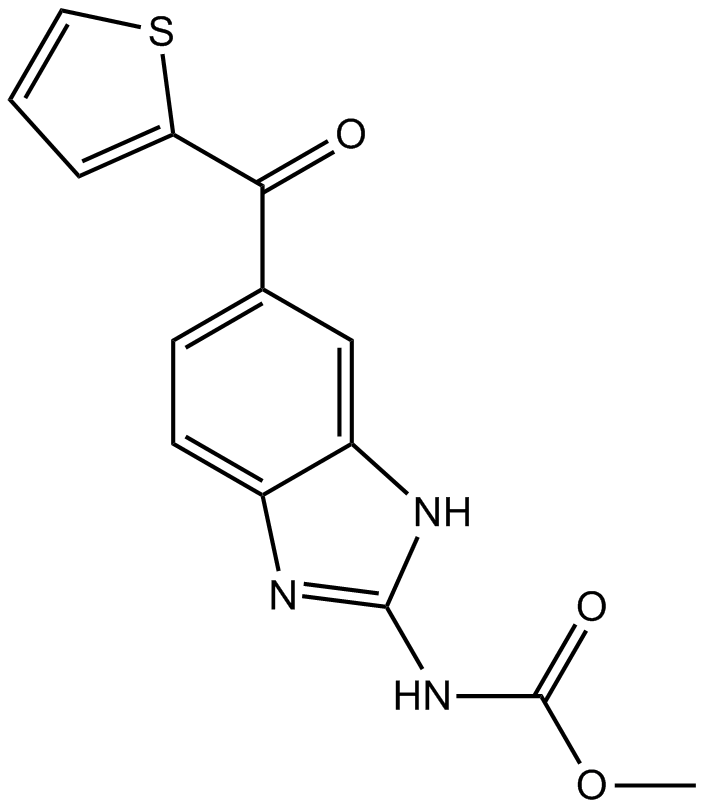
-
GC12090
Nonactin
Monovalent cation ionophore that displays selectivity for K+ and NH4+
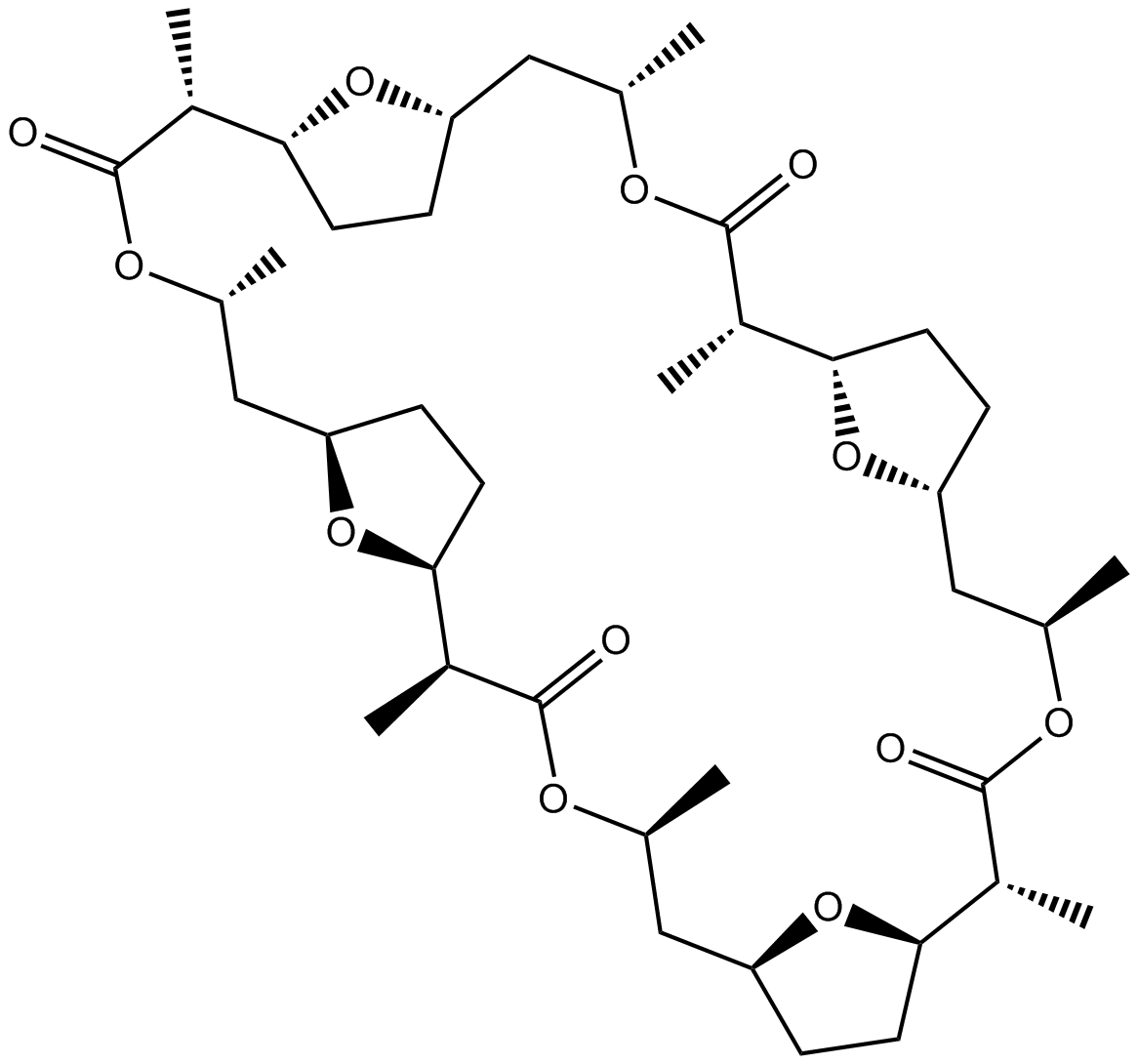
-
GC10628
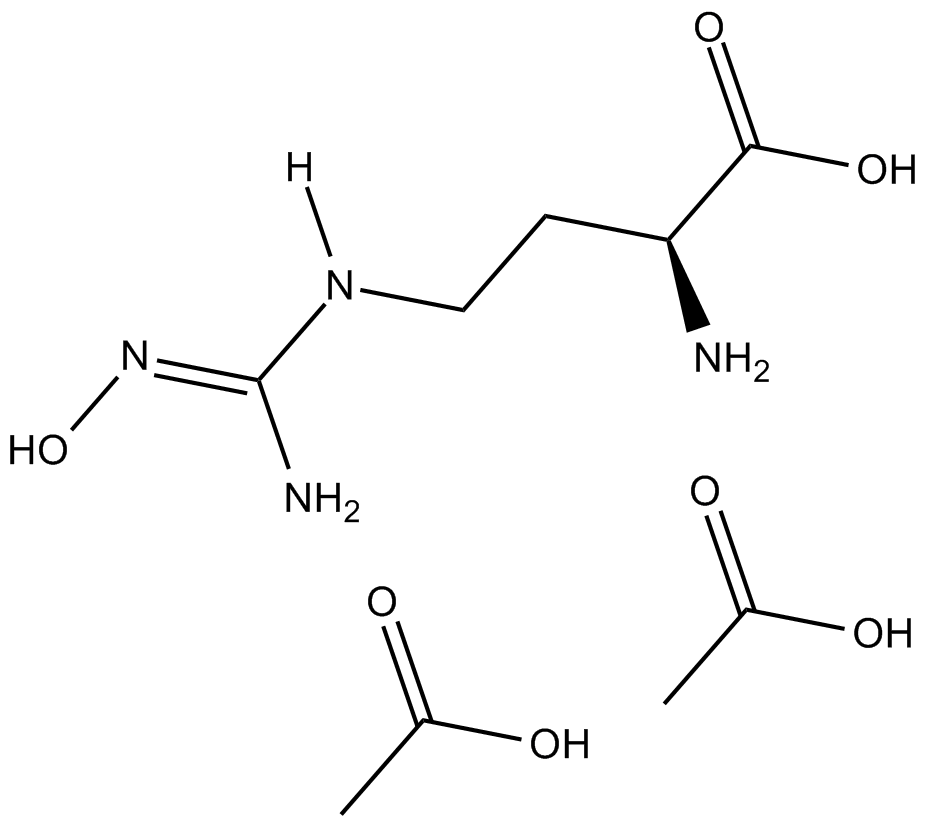
nor-NOHA (acetate)
nor-NOHA (acetate) (alpha-amino acid N(omega) -Hydroxy-Nor-L-arginine) is a potent and reversible selective arginase inhibitor,
-
GC11989
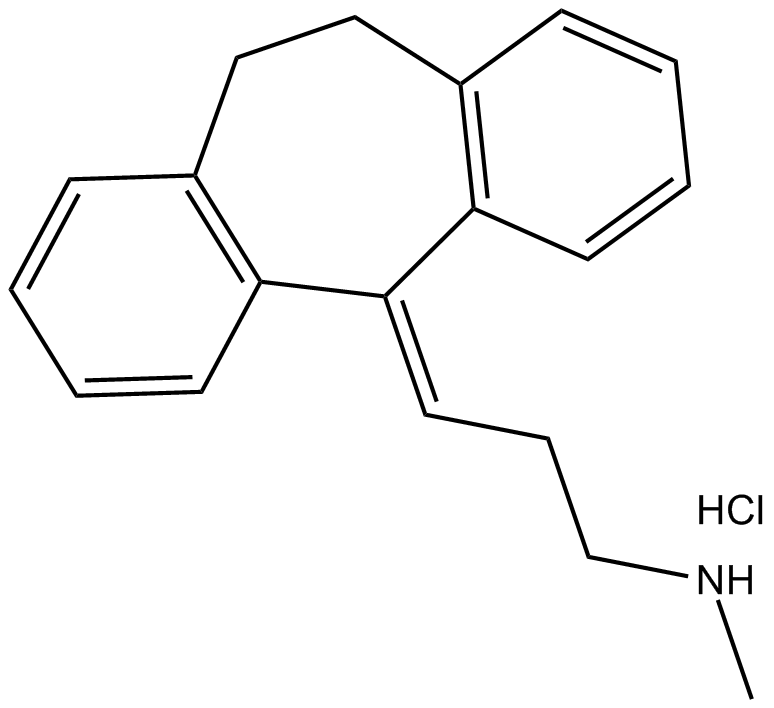
Nortriptyline (hydrochloride)
norepinephrine and serotonin transporters blocker
-
GC14488
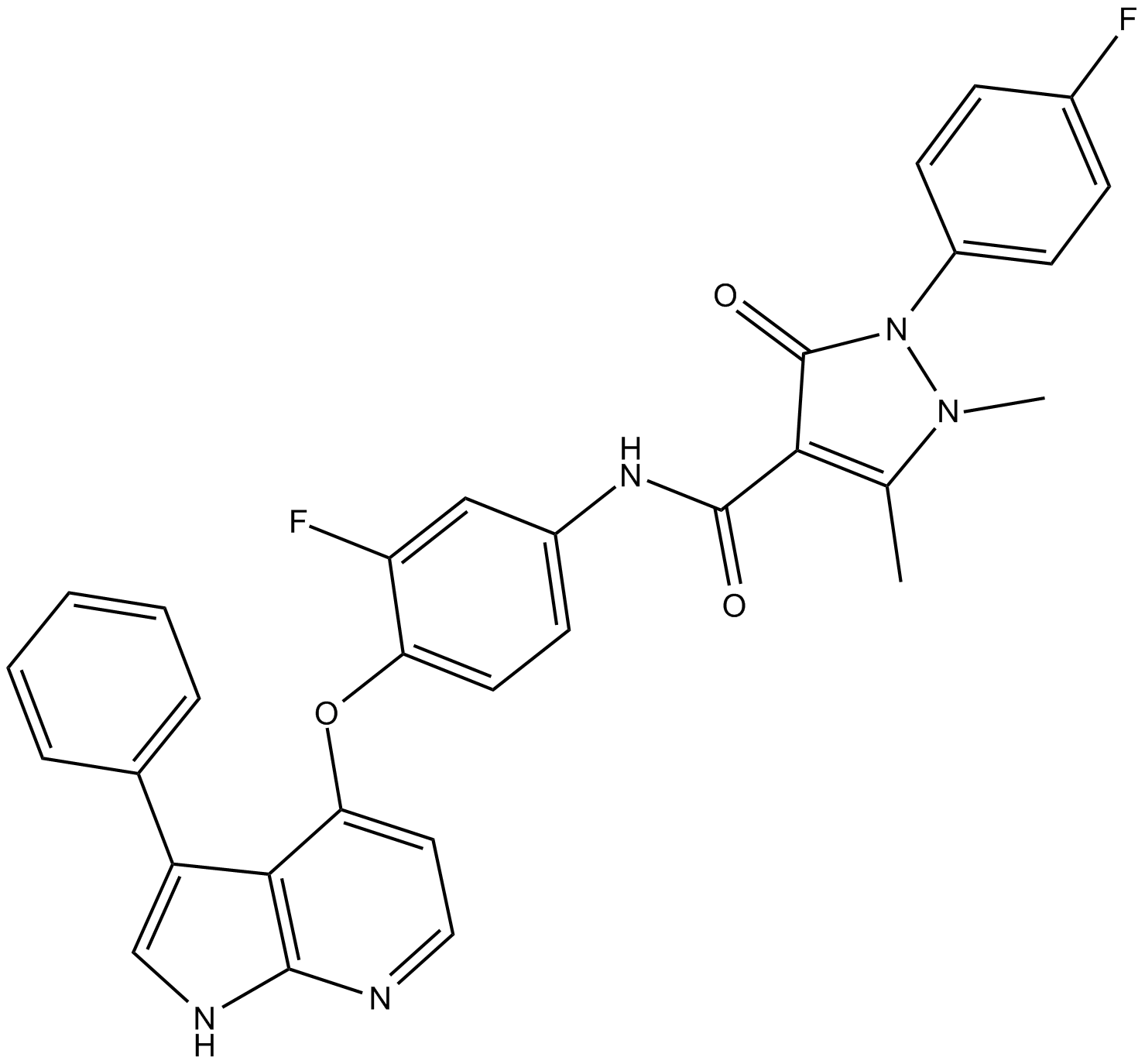
NPS-1034
MET inhibitor
-
GC17577
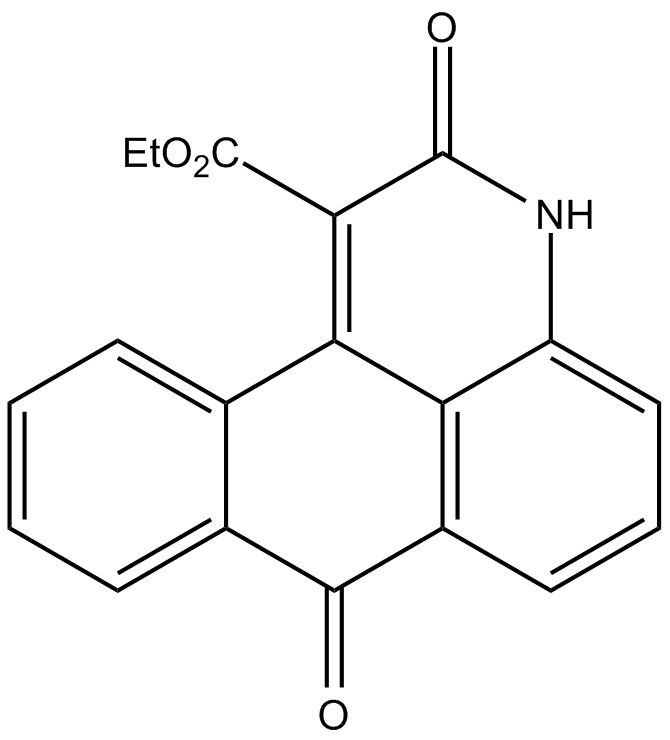
NQDI 1
A selective inhibitor of ASK1
-
GC10069
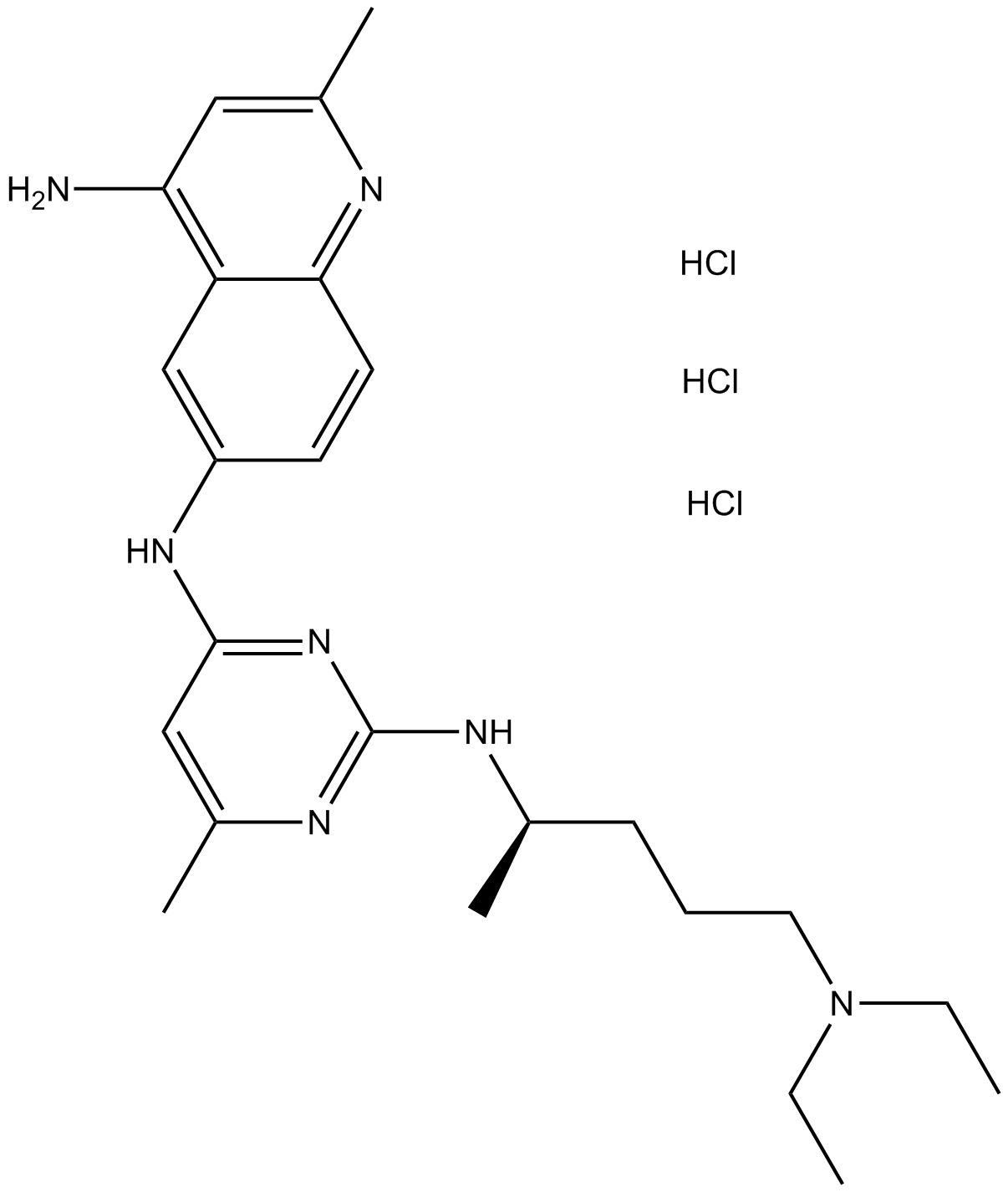
NSC 23766 trihydrochloride
Selective inhibitor of Rac1-GEF interaction.
-
GC11561
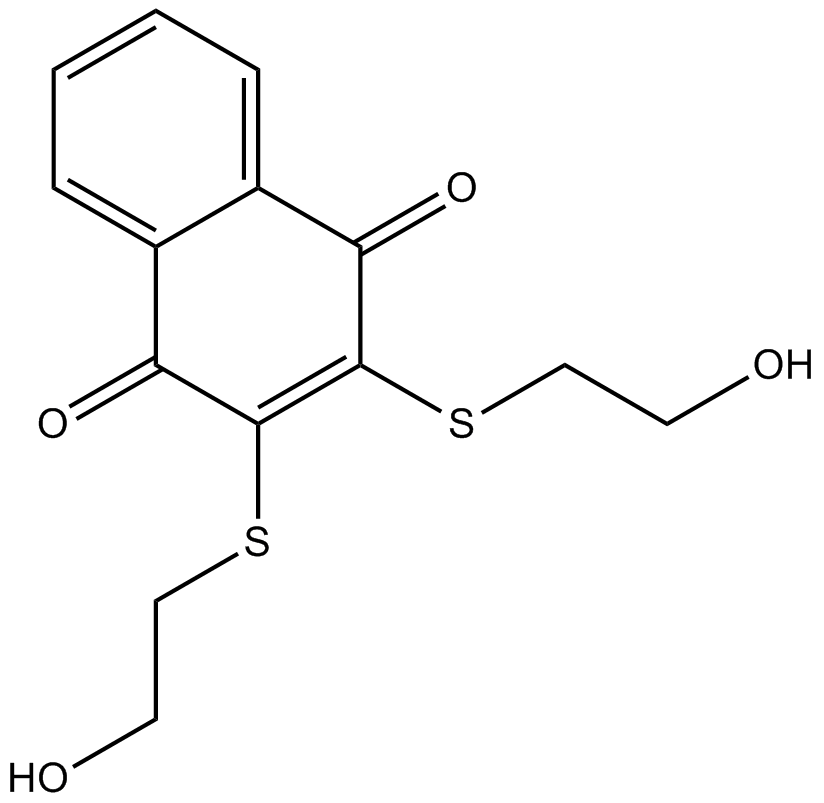
NSC 95397
Cdc25 dual specificity phosphatases inhibitor
-
GC16151
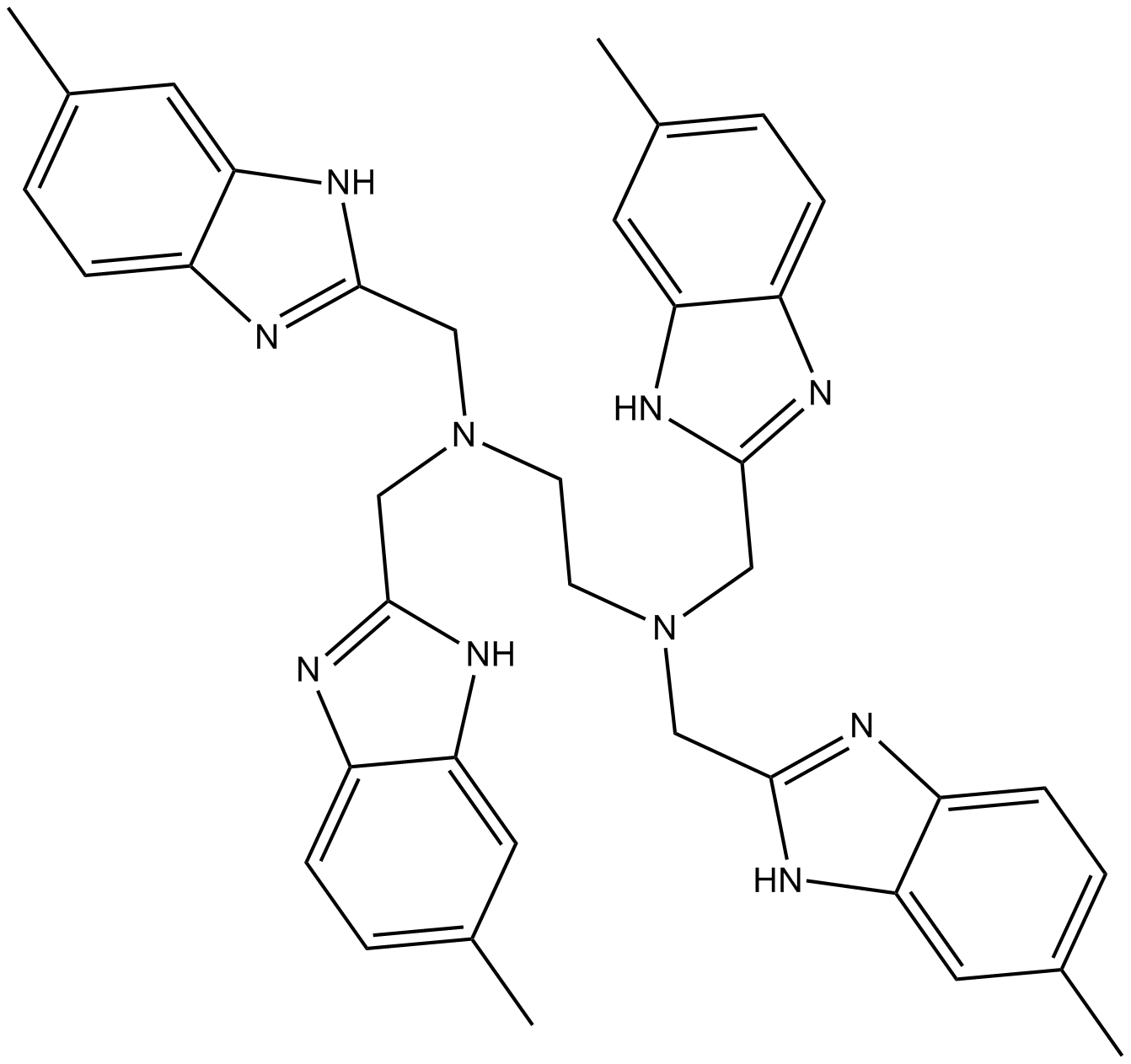
NSC348884
nucleophosmin inhibitor
-
GC12332
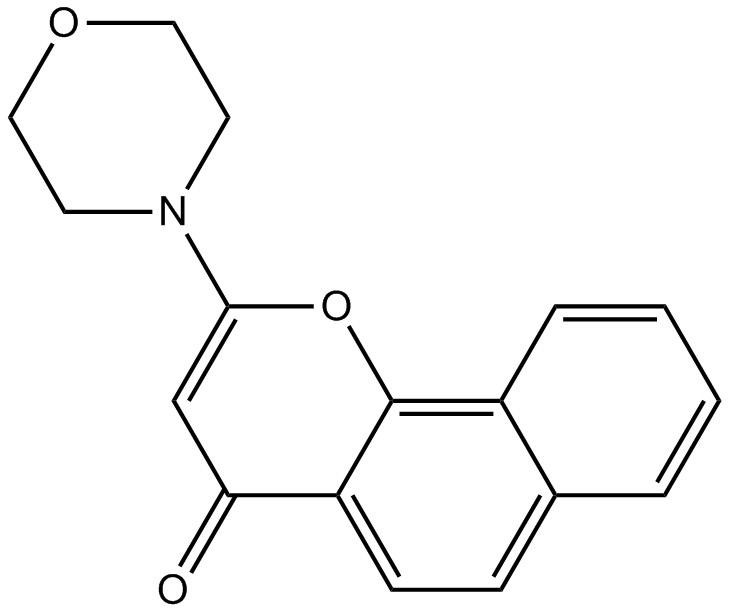
NU 7026
DNPK inhibitor,ATP-competitive and potent
-
GC10470
Nutlin-3a chiral
An inhibitor of the p53-Mdm2 interaction
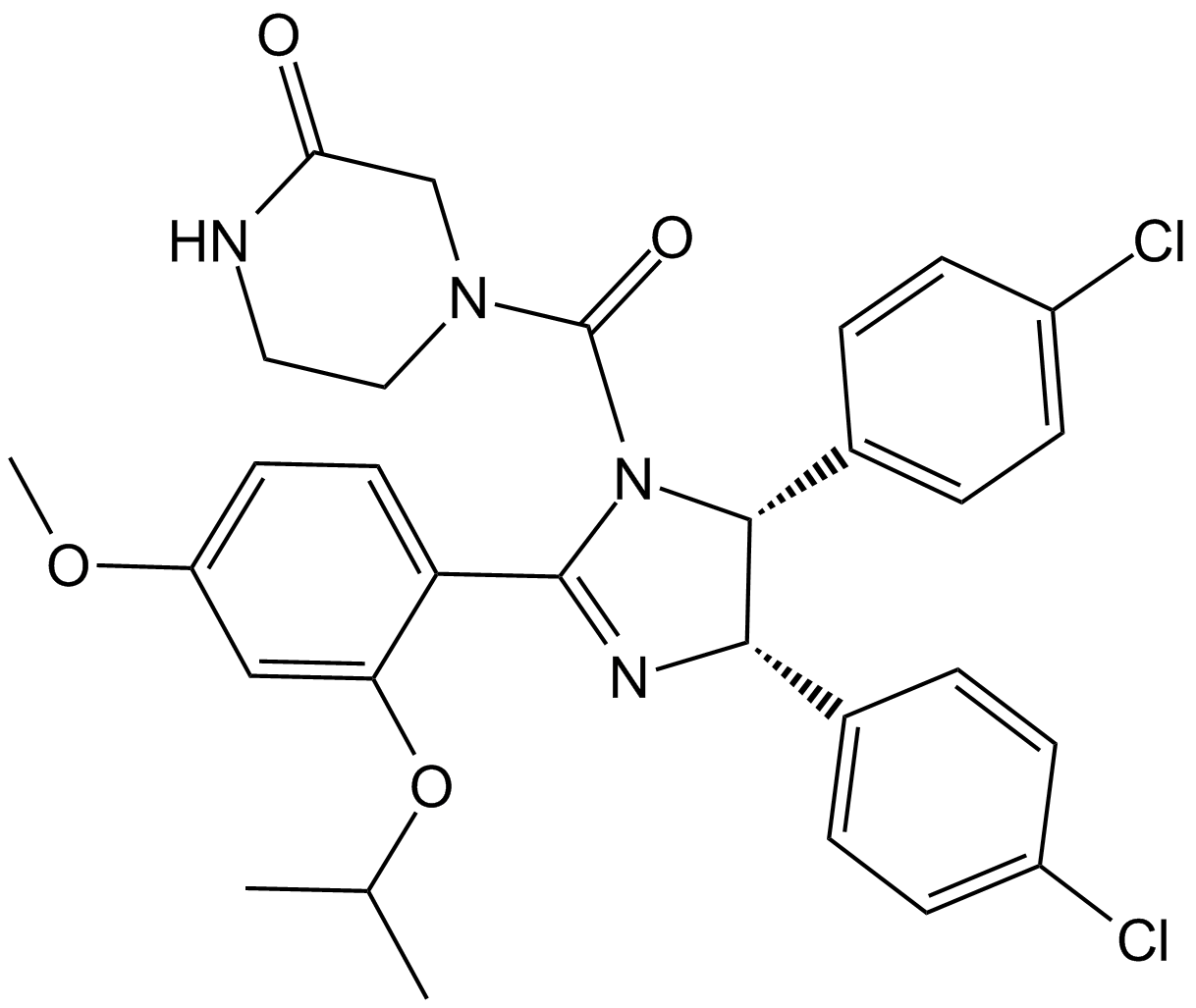
-
GC16145
NVP-BGT226
BGT226 (NVP-NVP-BGT226) is a PI3K (with IC50s of 4 nM, 63 nM and 38 nM for PI3Kα, PI3Kβ and PI3Kγ) /mTOR dual inhibitor which displays potent growth-inhibitory activity against human head and neck cancer cells.
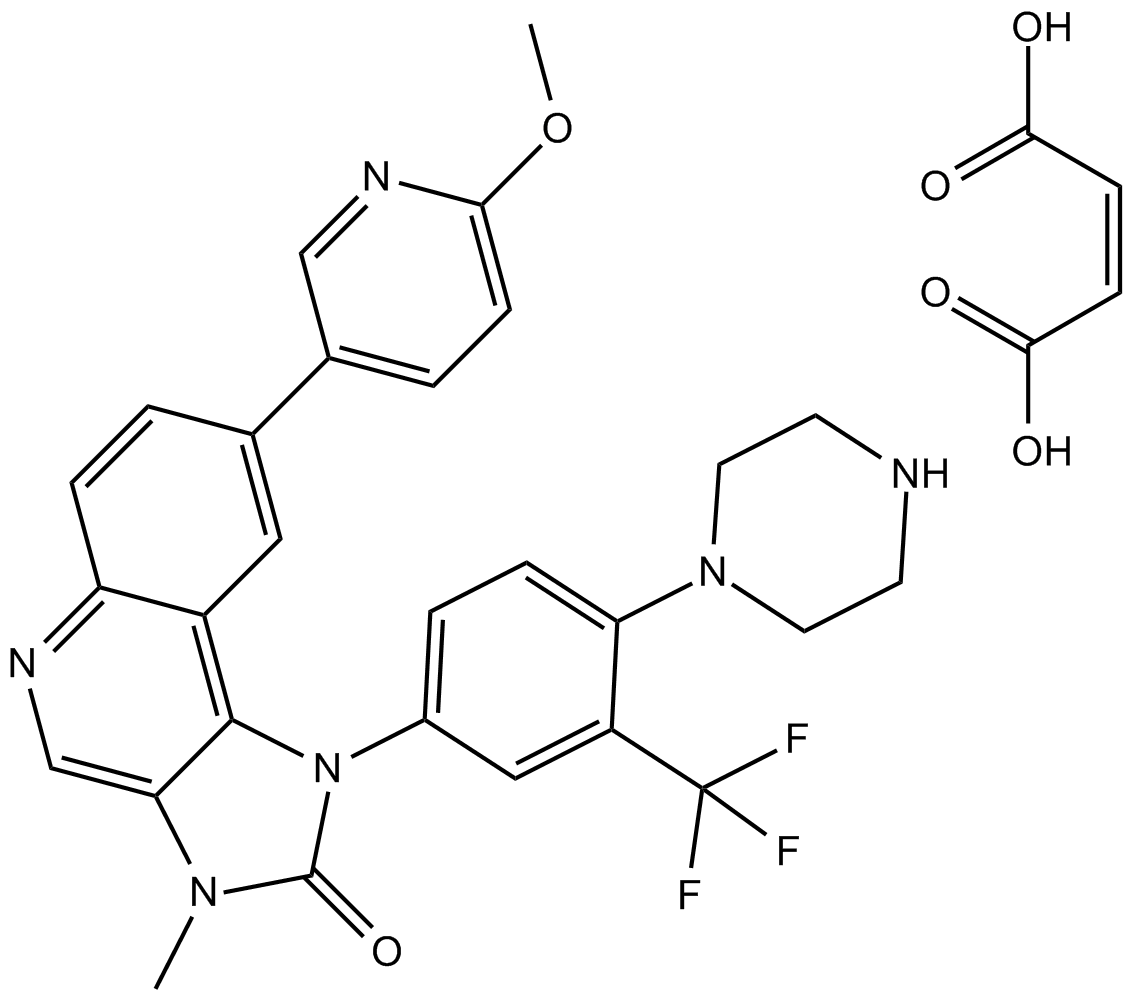
-
GC17555
NVP-TNKS656
TNKS2 inhibitor
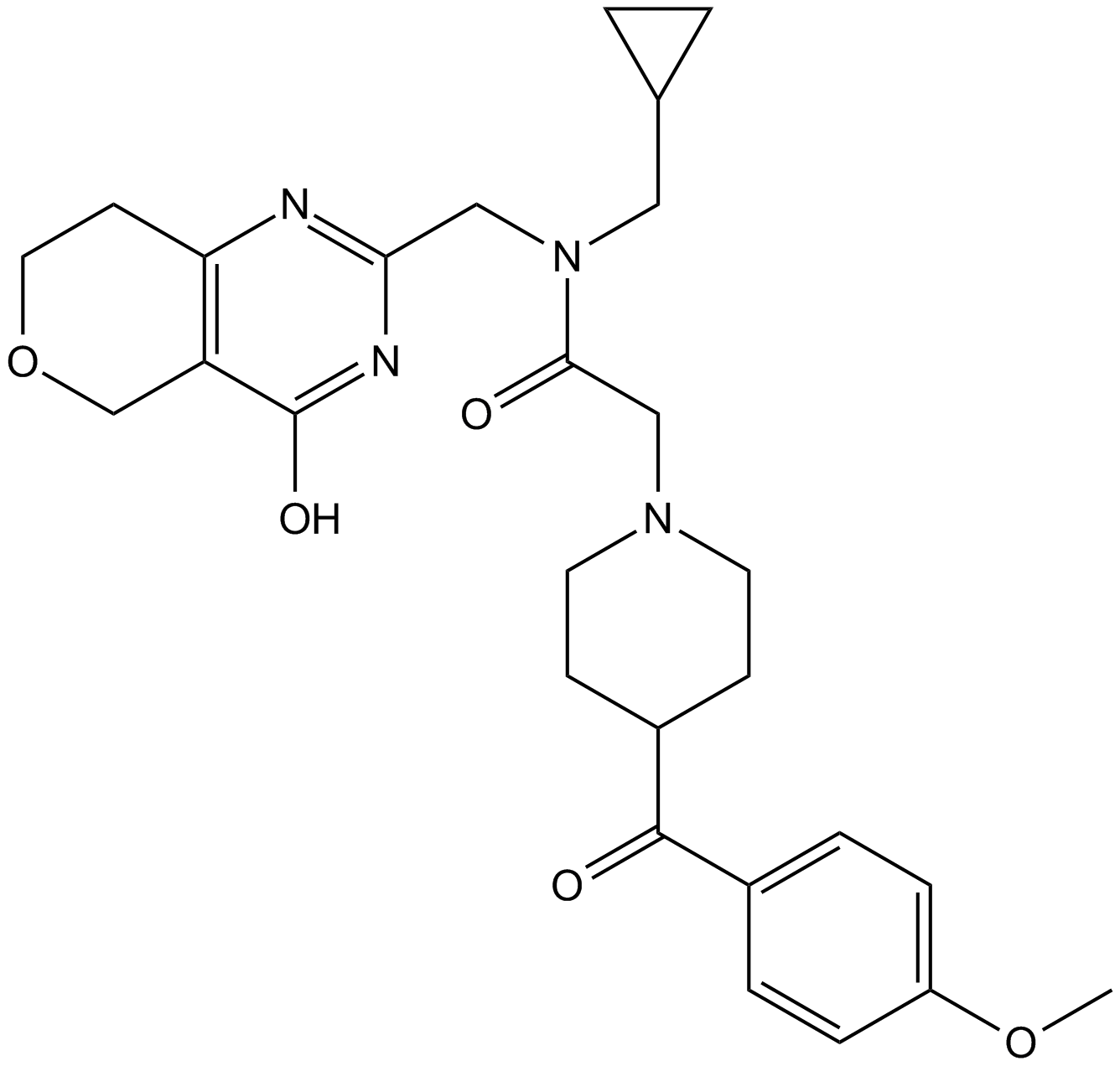
-
GN10776
Obacunone
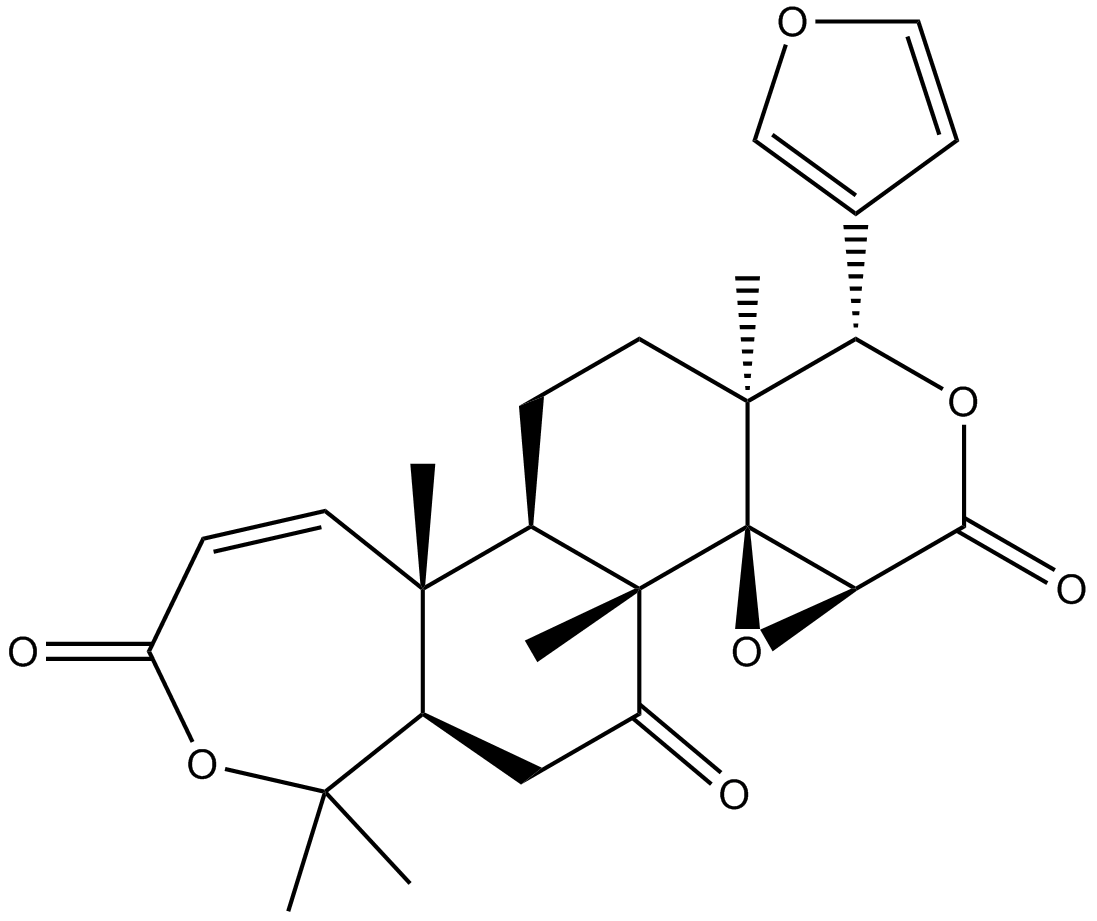
-
GN10144
Oleandrin
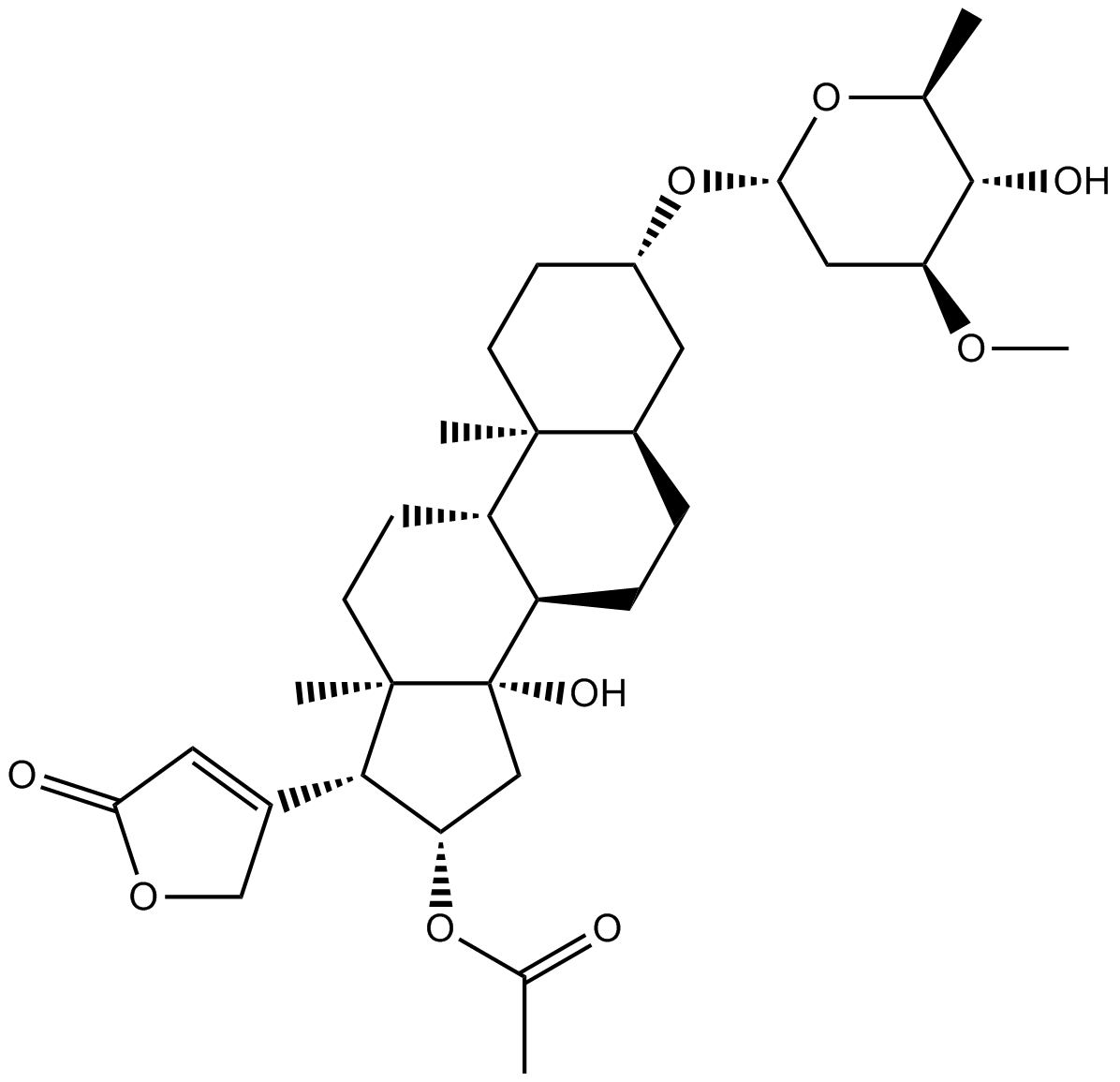
-
GC30110
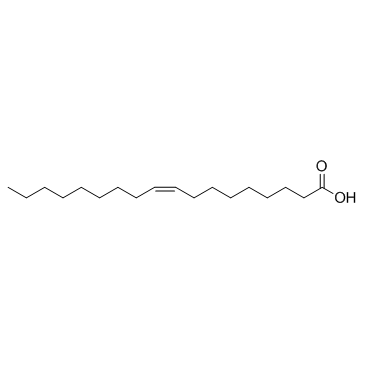
Oleic acid (9-cis-Octadecenoic acid)
Oleic acid (9-cis-Octadecenoic acid) (9-cis-Octadecenoic acid) is an abundant monounsaturated fatty acid.
-
GN10457
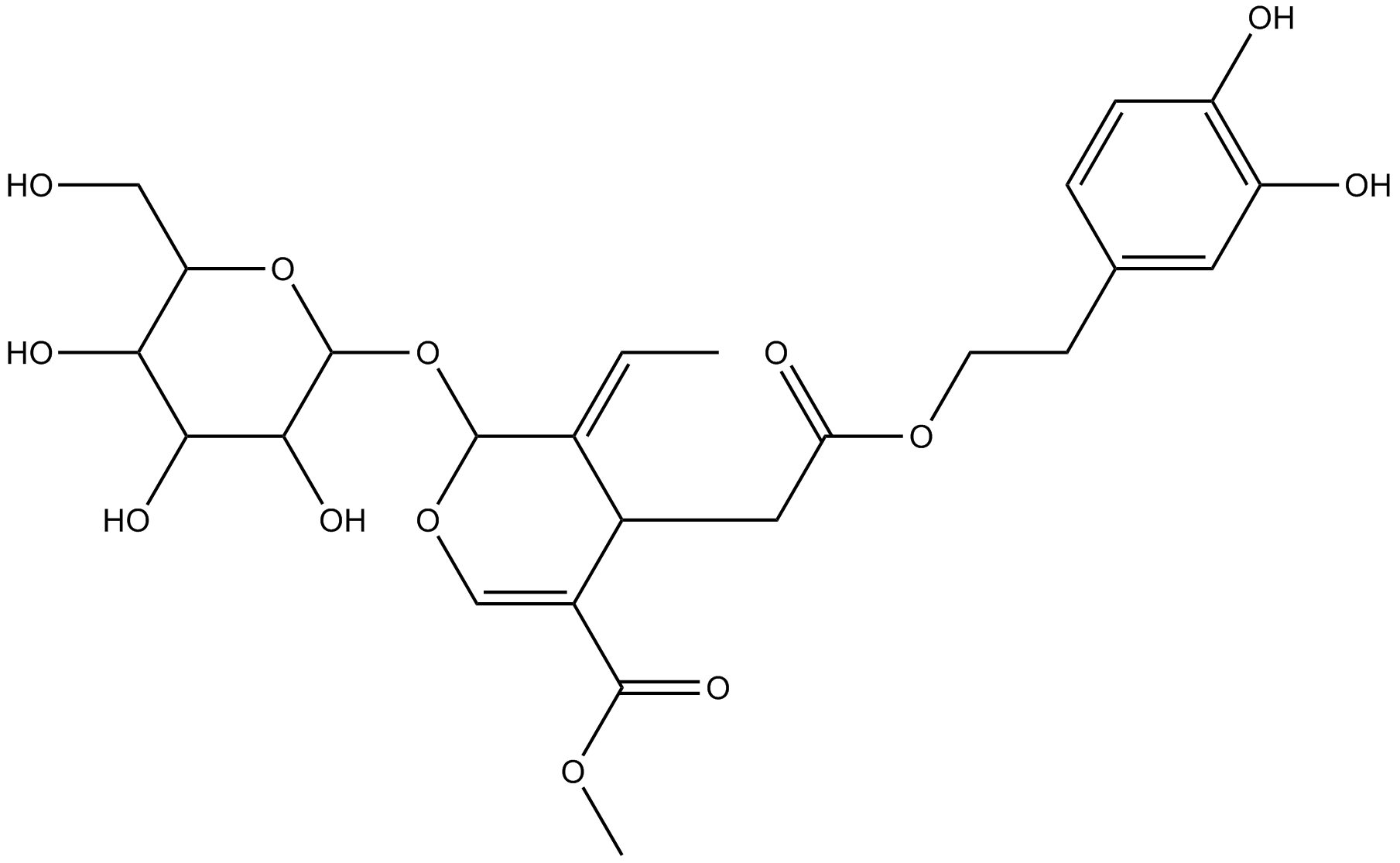
Oleuropein
A polyphenol
-
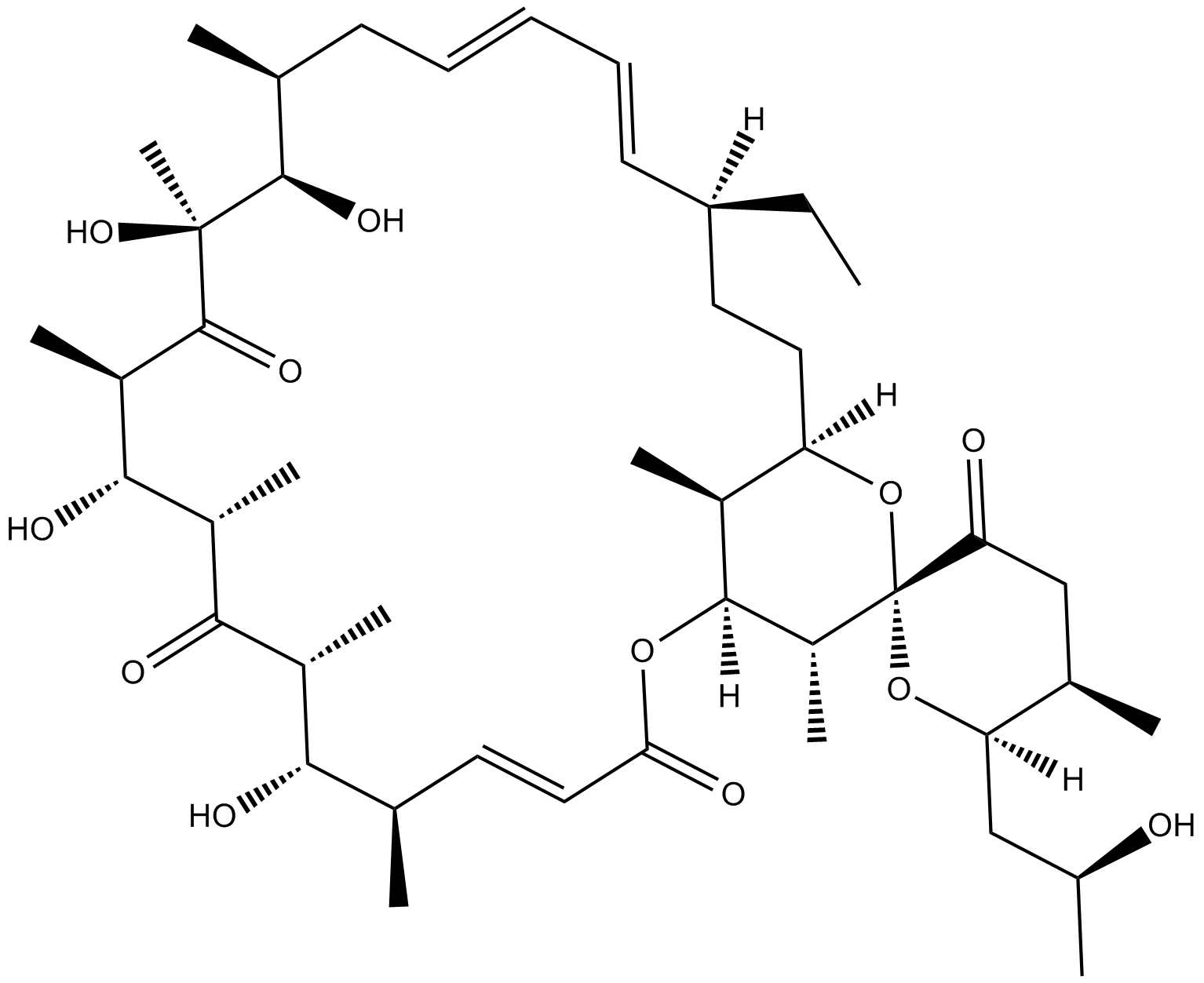
GC16409
Oligomycin B
mitochondrial F1FO ATP synthase inhibitor
-
GC13693
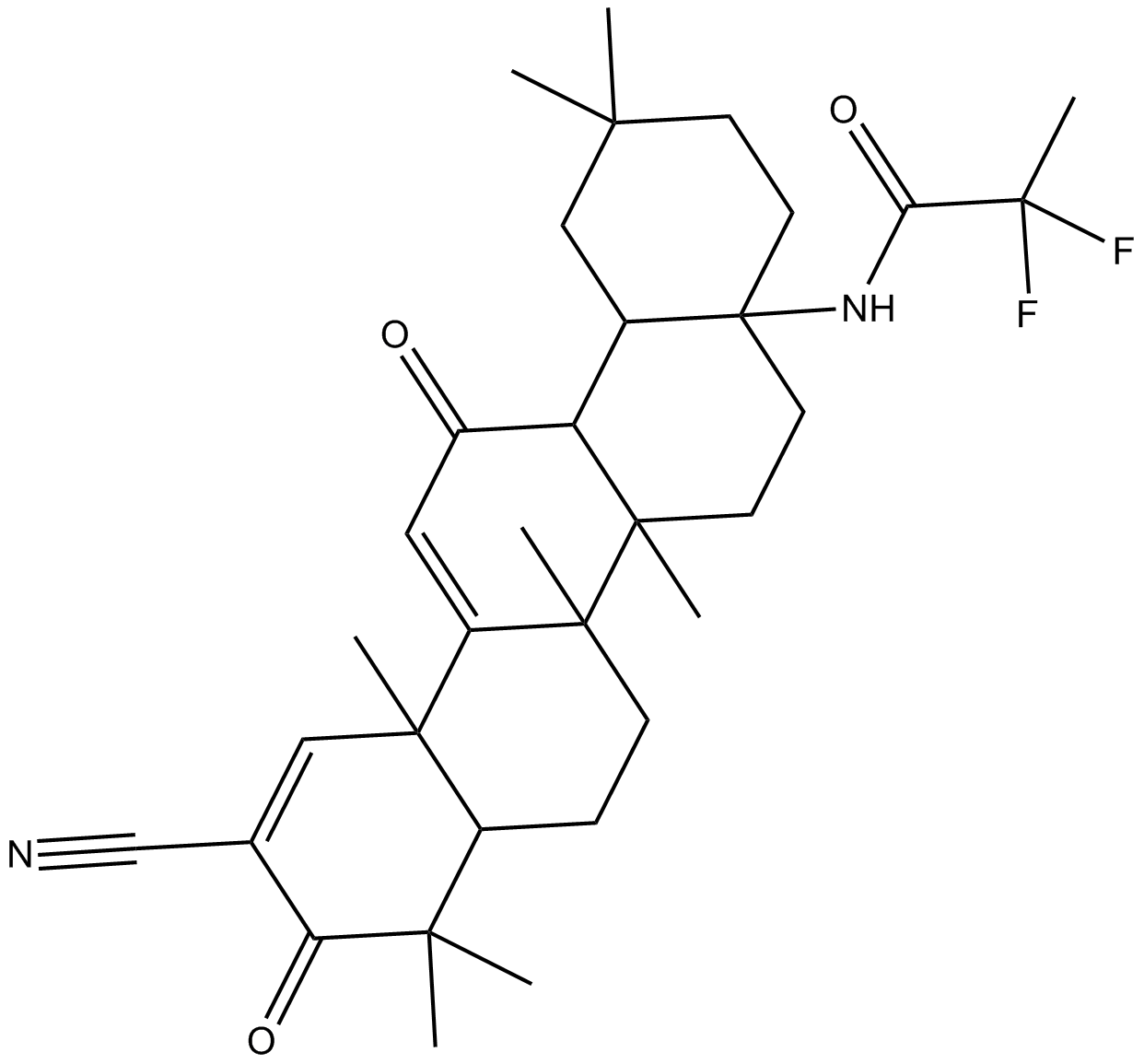
Omaveloxolone (RTA-408)
Omaveloxolone (RTA-408) (RTA 408) is an antioxidant inflammation modulator (AIM), which activates Nrf2 and suppresses nitric oxide (NO).
-
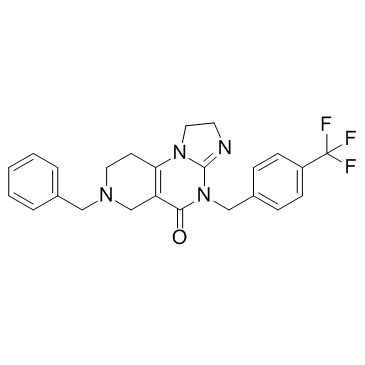
GC34095
ONC212
A GPR132 agonist
-
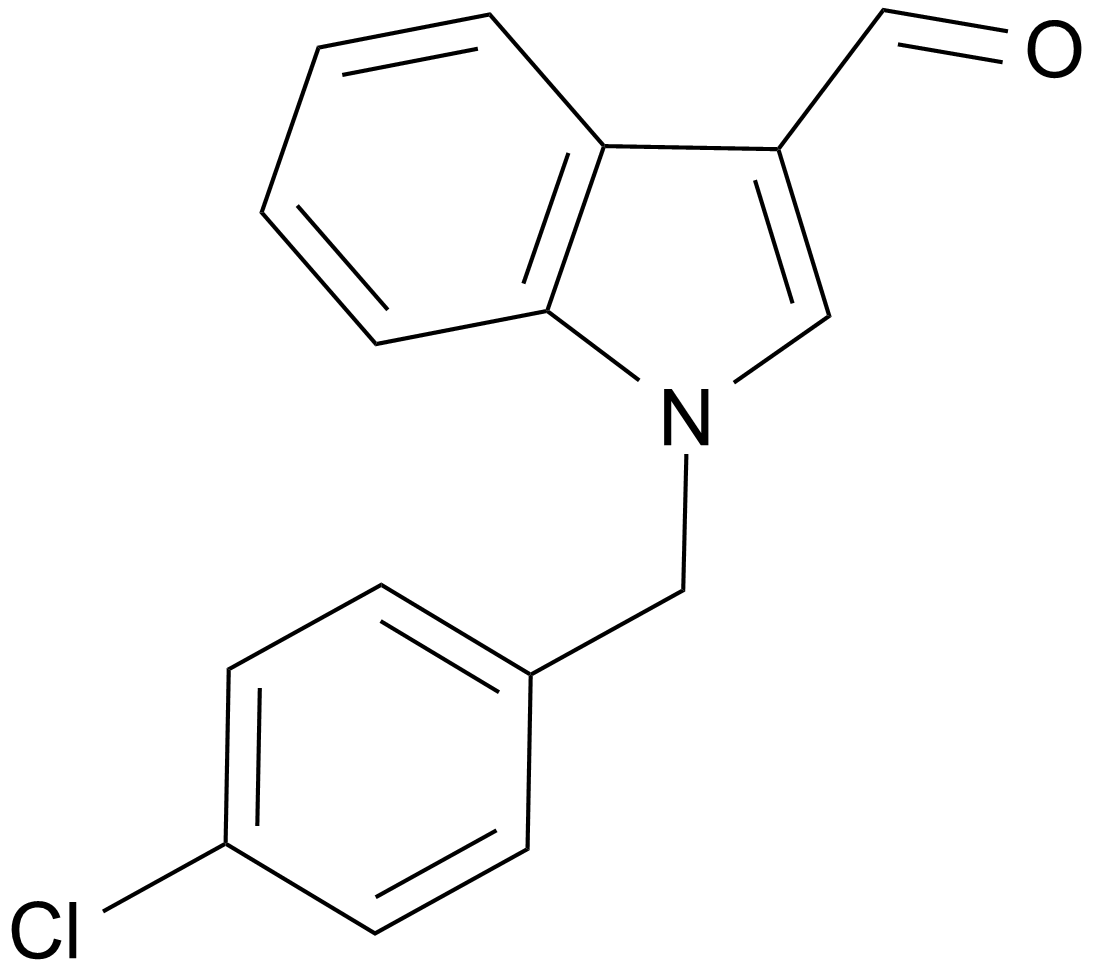
GC14860
Oncrasin 1
An anticancer agent
-
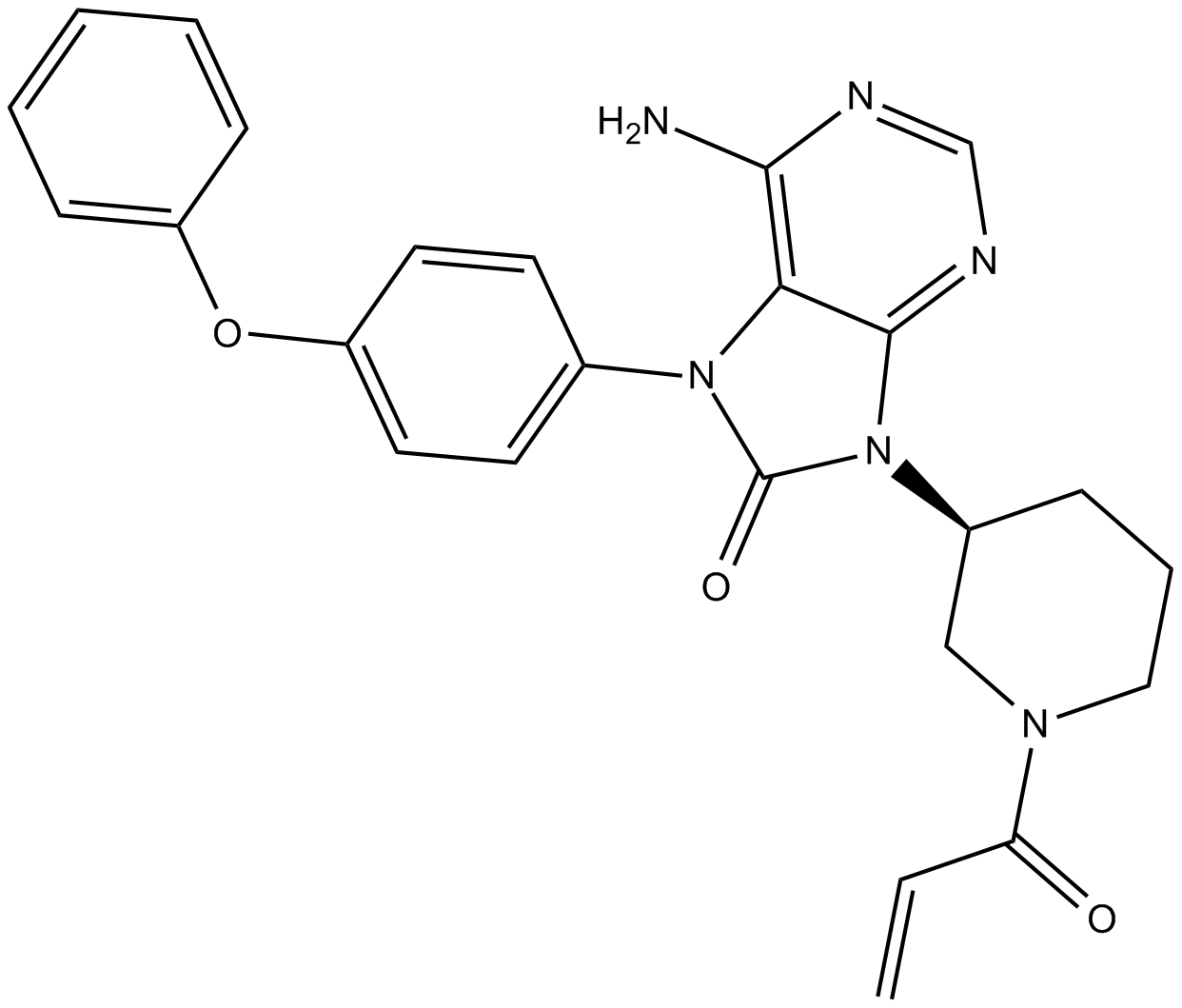
GC13219
ONO-4059
ONO-4059 is the analog of ONO-4059, ONO-4059 is a highly potent and selective Btk inhibitor.
-
GC17318
Orlistat
Orlistat is an irreversible inhibitor of lipases in the pancreas and stomach.
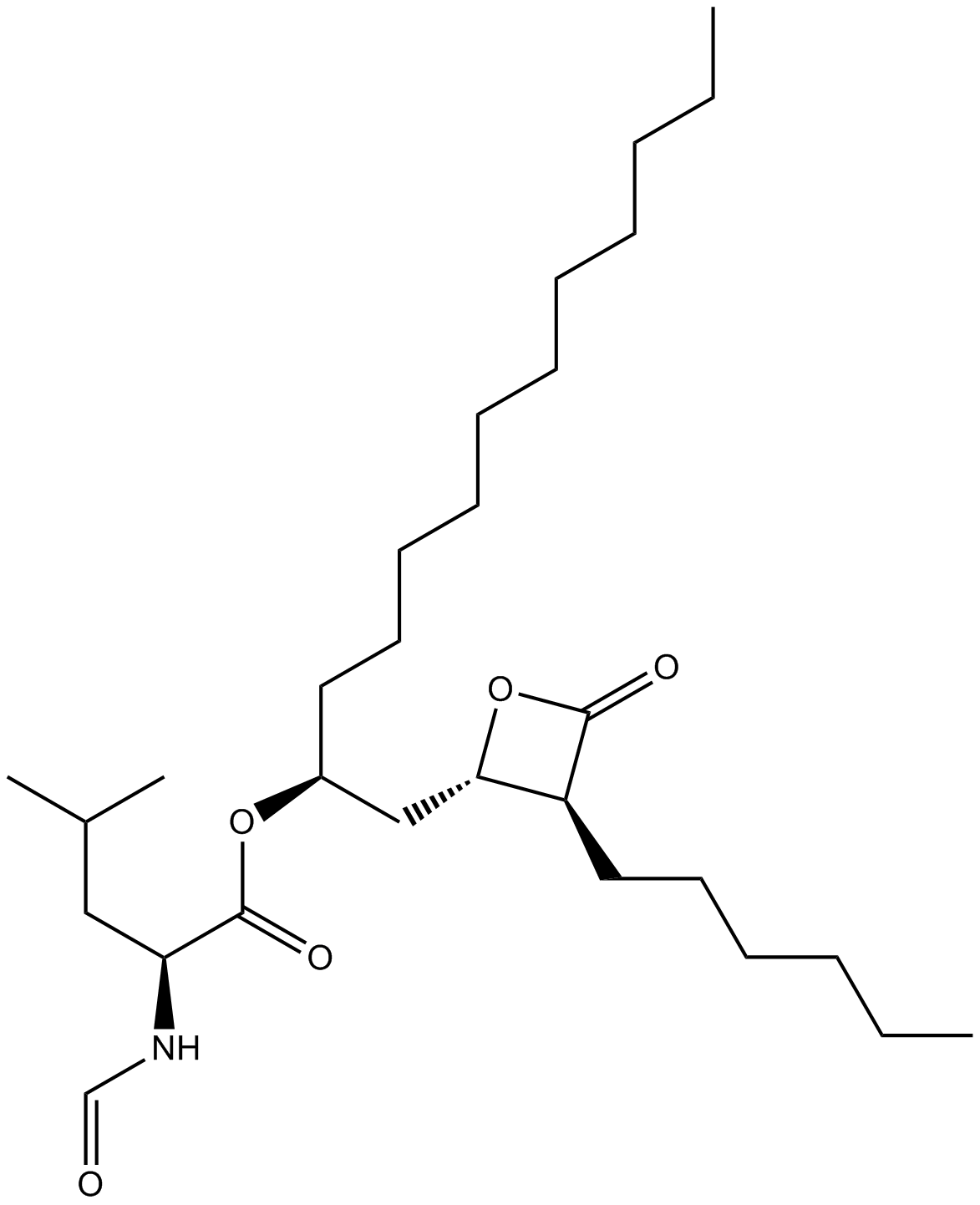
-
GC11528
Orotic acid
Pyrimidinecarboxylic acid
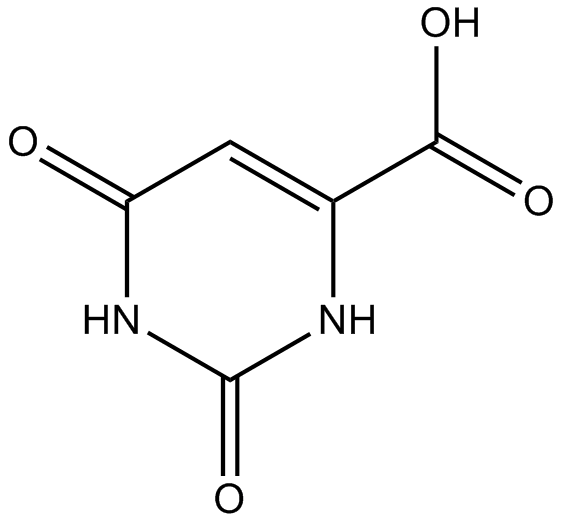
-
GN10732
Oroxin B
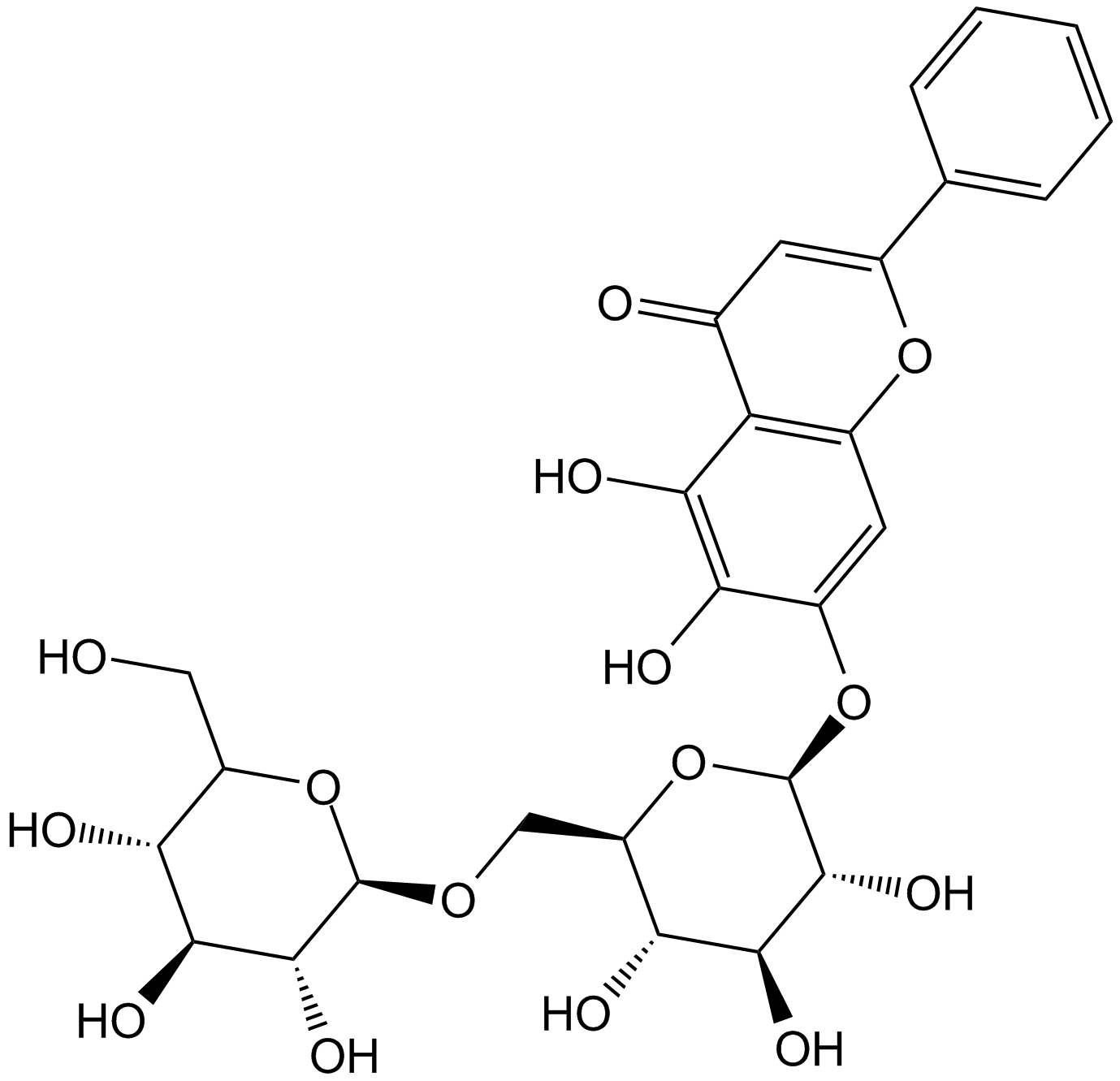
-
GC31596
Osajin (CID 95168)
Osajin (CID 95168) is the major bioactive isoflavone present in the fruit of Maclura pomifera with antitumor, antioxidant and anti-inflammatory activities.
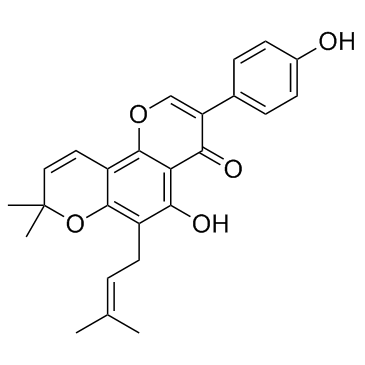
-
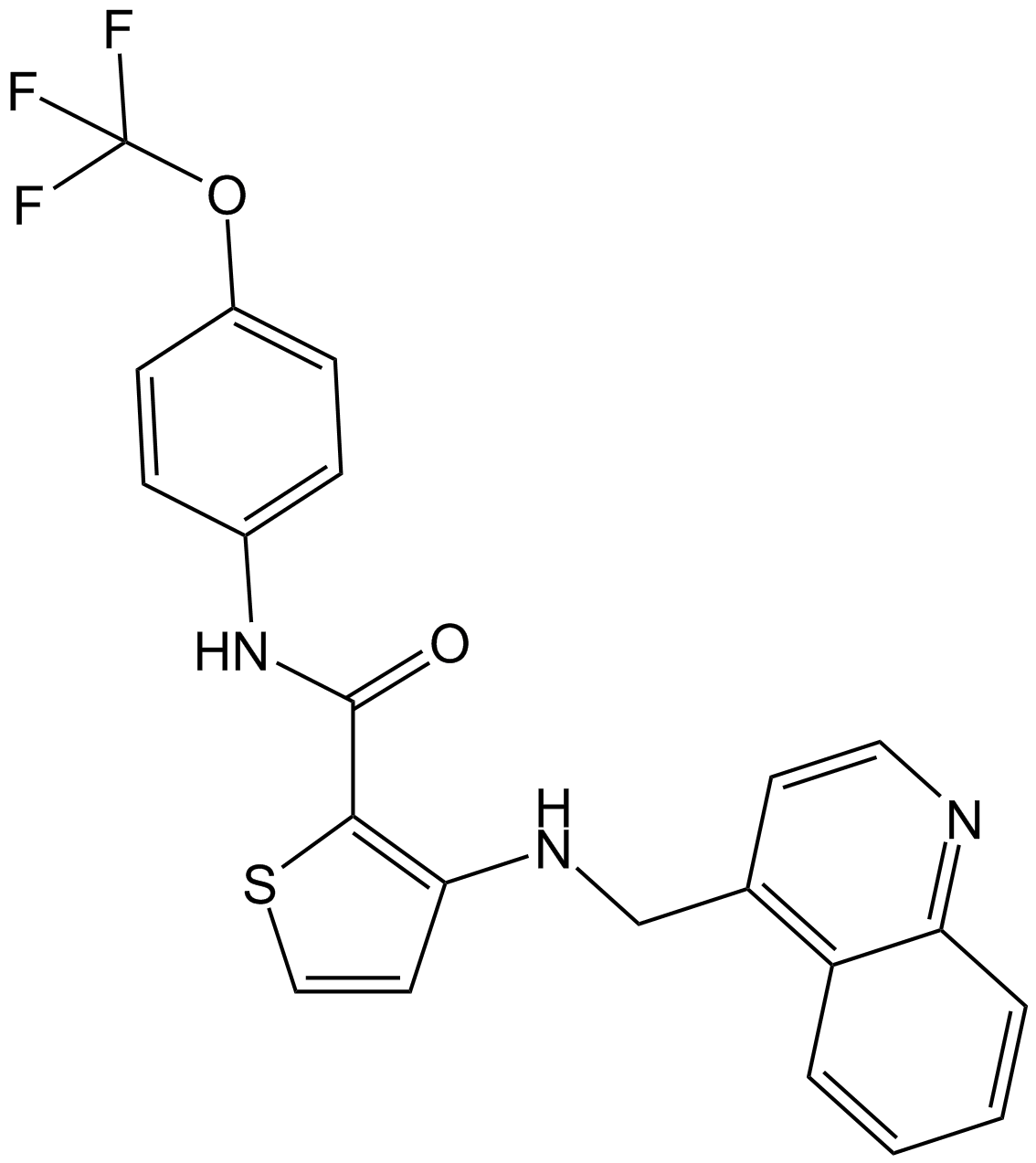
GC14957
OSI-930
Inhibitor of Kit, KDR, Flt, CSF-1R, c-Raf and Lck
-
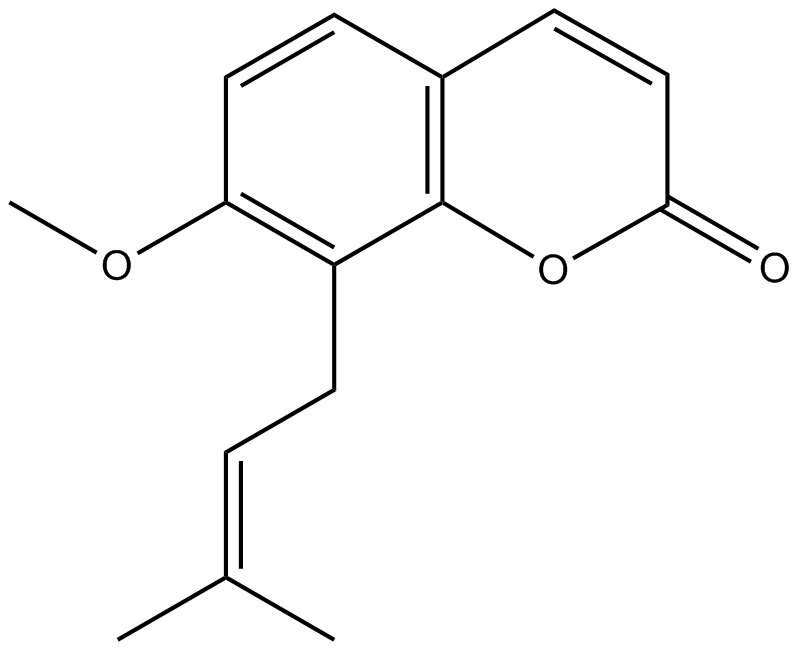
GN10333
Osthole
-
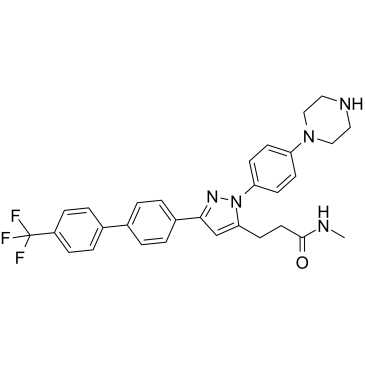
GC36821
OSU-T315
OSU-T315 (ILK-IN-1) is a small Integrin-linked kinase (ILK) inhibitor with an IC50 of 0.6 μM, inhibiting PI3K/AKT signaling by dephosphorylation of AKT-Ser473 and other ILK targets (GSK-3β and myosin light chain). OSU-T315 abrogates AKT activation by impeding AKT localization in lipid rafts and triggers caspase-dependent apoptosis in an ILK-independent manner. OSU-T315 causes cell death through apoptosis and autophagy.
-
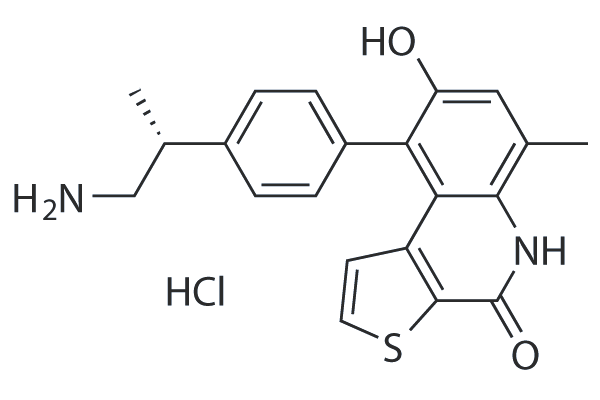
GC25691
OTS514 hydrochloride
OTS514 is a highly potent TOPK(T-LAK cell-originated protein kinase) inhibitor with an IC50 value of 2.6 nM. OTS514 induces cell cycle arrest and apoptosis.
-
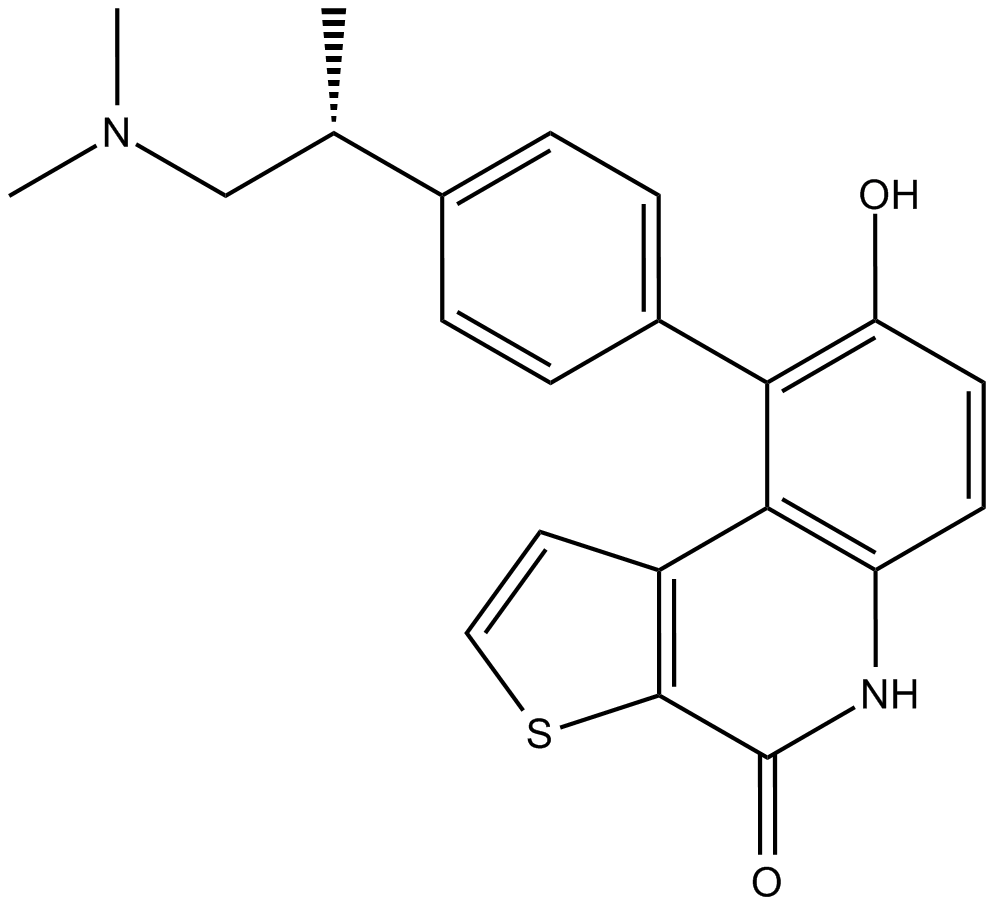
GC16511
OTS964
OTS964 is an orally active, high affinity and selective TOPK (T-lymphokine-activated killer cell-originated protein kinase) inhibitor with an IC50 of 28 nM. OTS964 is also a potent inhibitor of the cyclin-dependent kinase CDK11, which binds to CDK11B with a Kd of 40 nM.
-
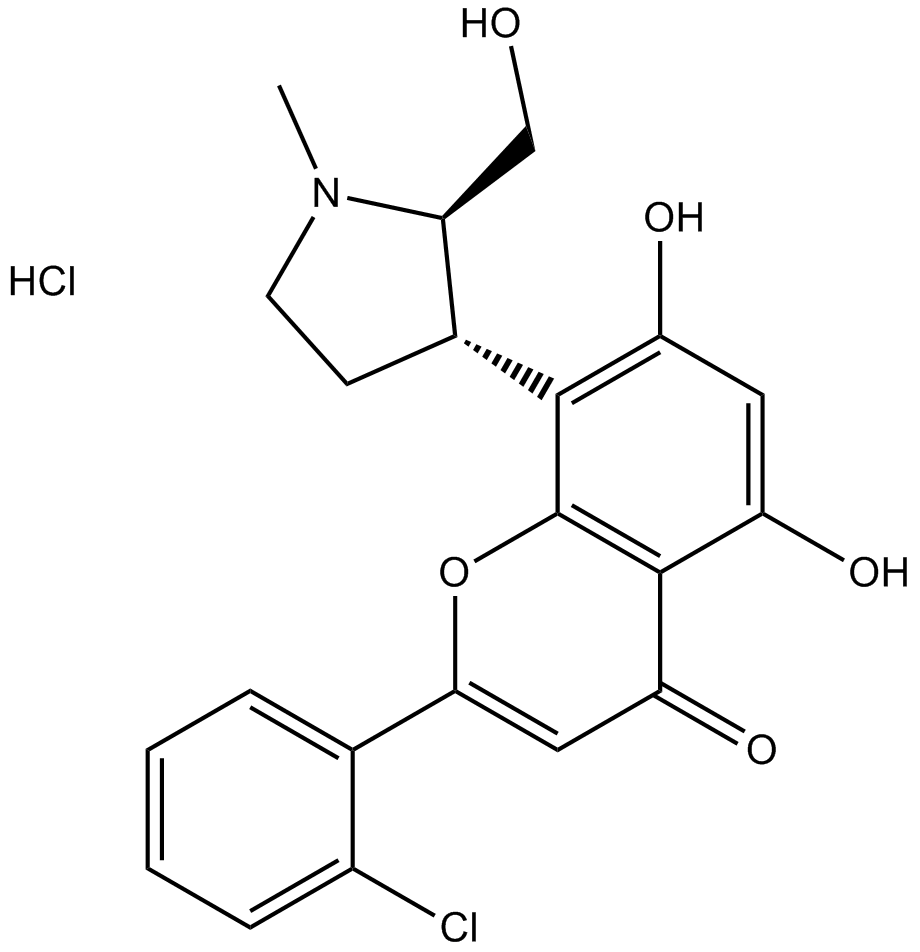
GC12011
P276-00
P276-00 (P276-00) is a potent cyclin-dependent kinase (CDK) inhibitor, which inhibits CDK9-cyclinT1, CDK4-cyclin D1, and CDK1-cyclinB with IC50s of 20 nM, 63 nM, and 79 nM, respectively.P276-00 (P276-00) shows antitumor activity on cisplatin-resistant cells.
-
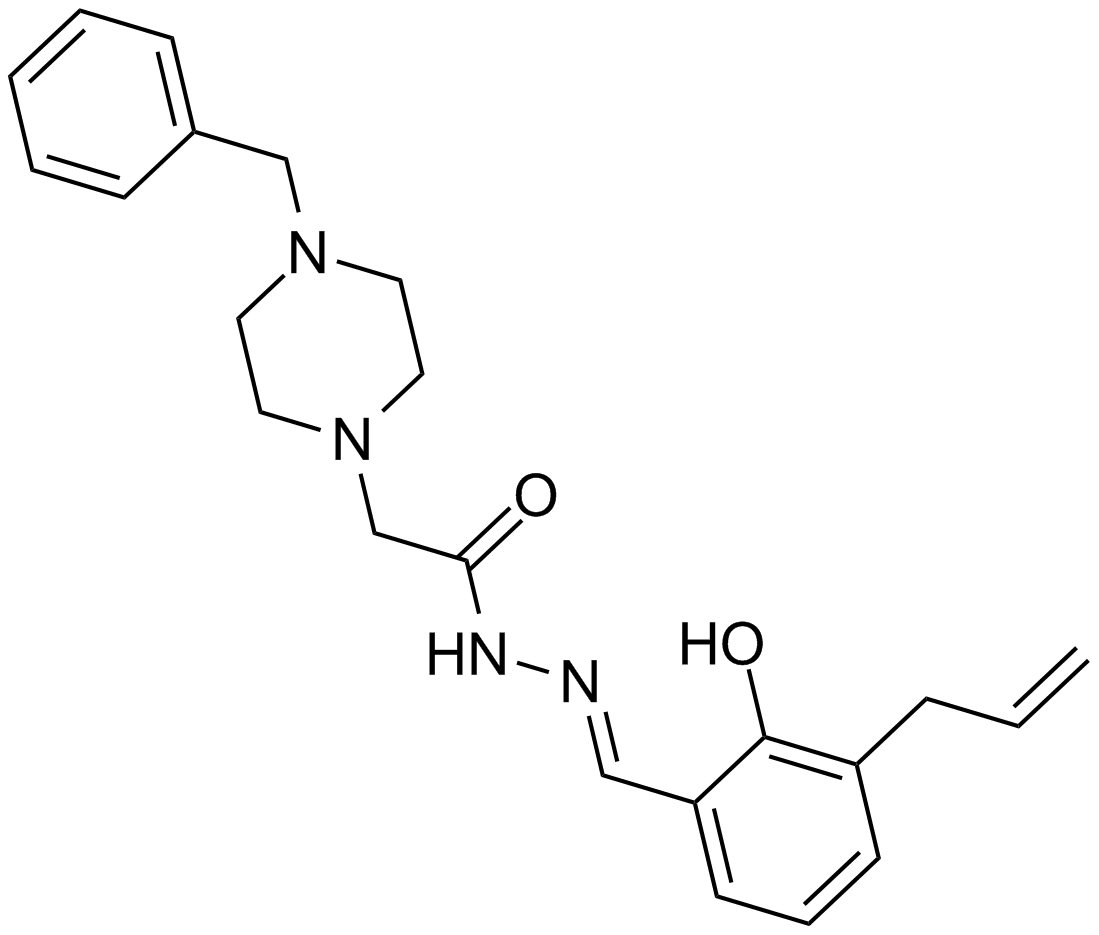
GC11993
PAC-1
An in vitro procaspase-3 activator
-
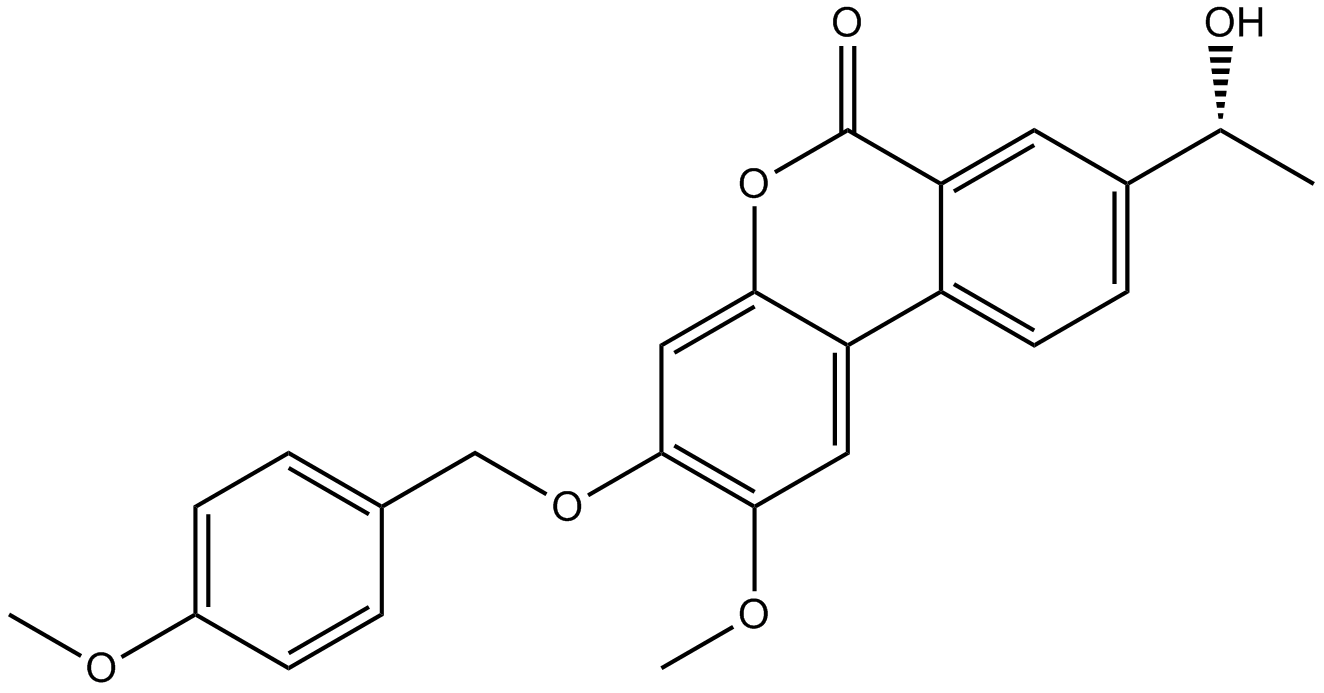
GC12773
Palomid 529
PI3K/Akt/mTOR inhibitor
-
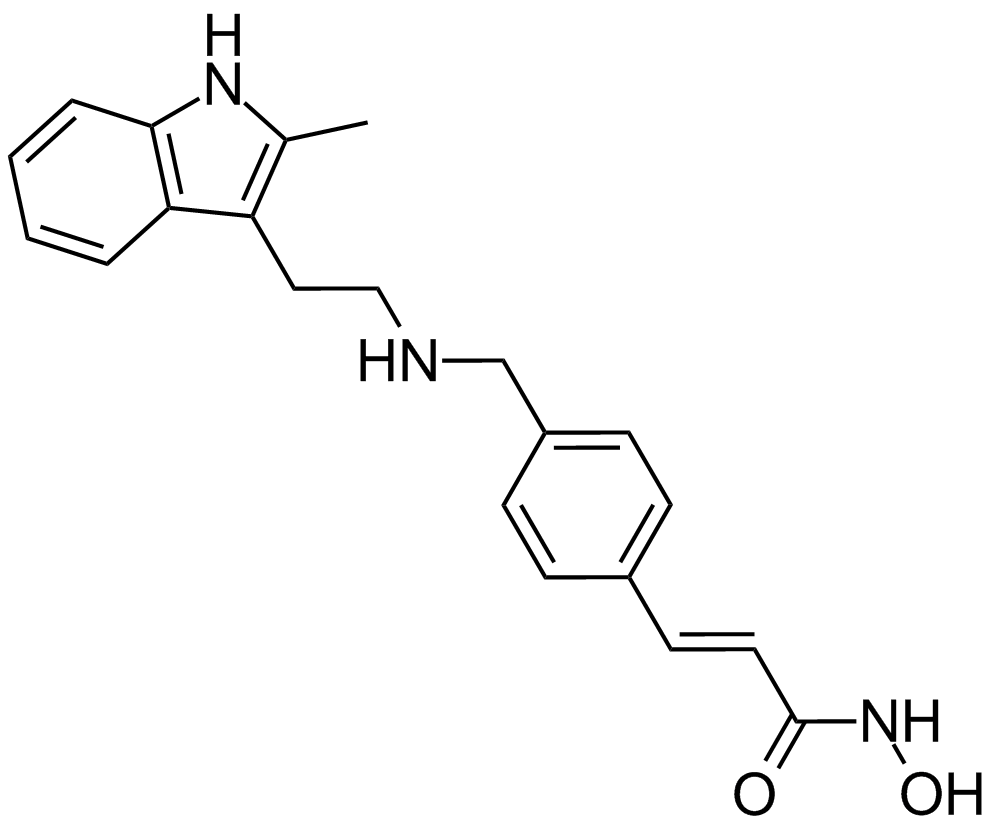
GC12257
Panobinostat (LBH589)
A pan-HDAC inhibitor
-
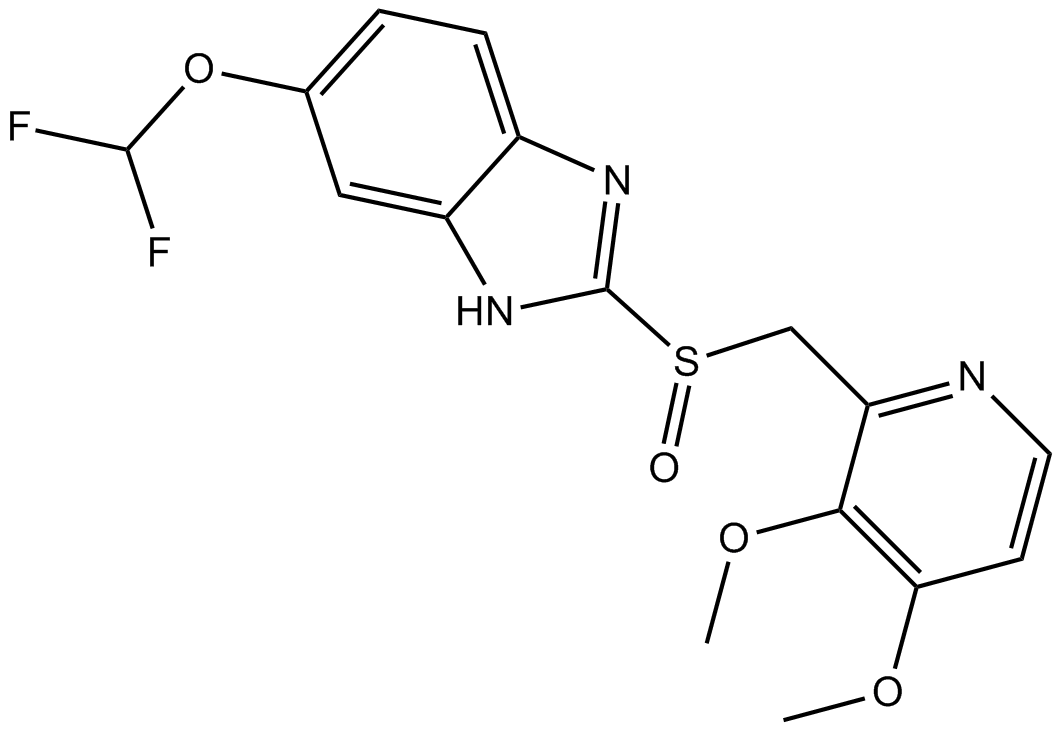
GC12718
Pantoprazole
H+/K+-ATPase inhibitor
-
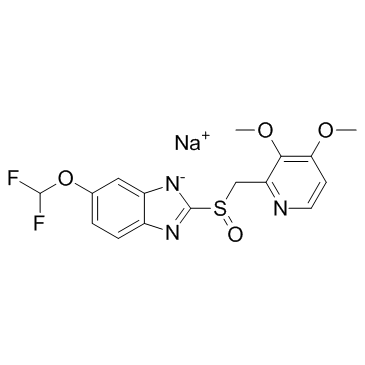
GC36847
Pantoprazole sodium
Pantoprazole sodium (BY10232 sodium) is an orally active and potent proton pump inhibitor (PPI).
-
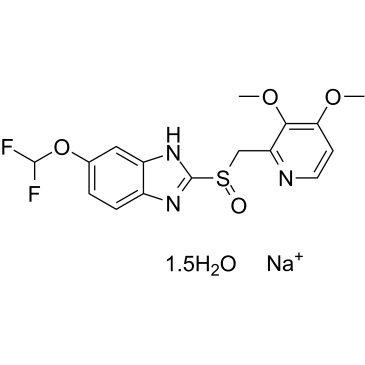
GC36848
Pantoprazole sodium hydrate
Pantoprazole sodium hydrate (BY10232 sodium hydrate) is an orally active and potent proton pump inhibitor (PPI).
-
GN10357
Parthenolide
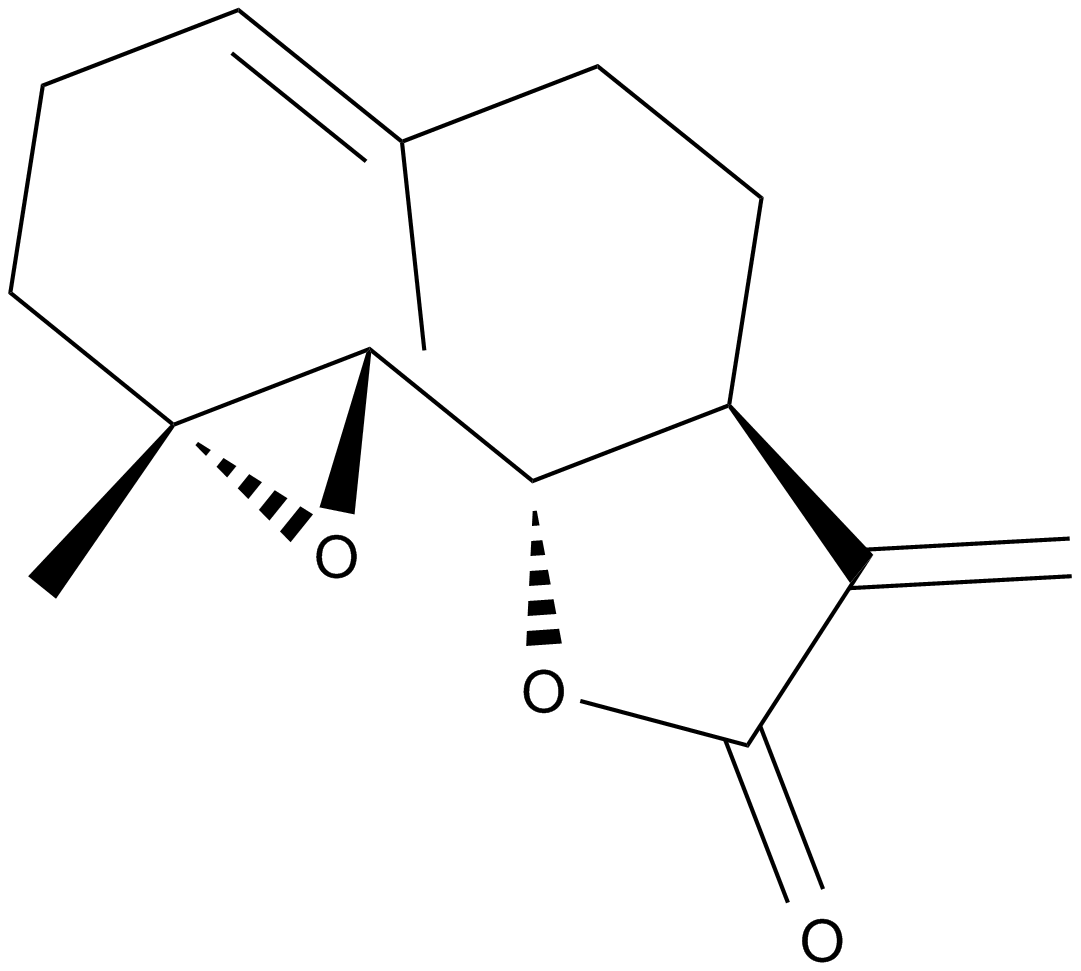
-
GC40483
Patulin
Patulin is a mycotoxin produced by a variety of molds commonly found in rotting apples, including Aspergillus and Penicillium.
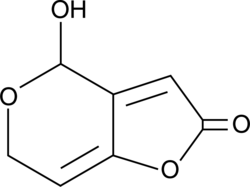
-
GC33002
PBOX 6
PBOX 6 is a pyrrolo-1,5-benzoxazepine (PBOX) compound, acts as a microtubule-depolymerizing agent and an apoptotic agent.
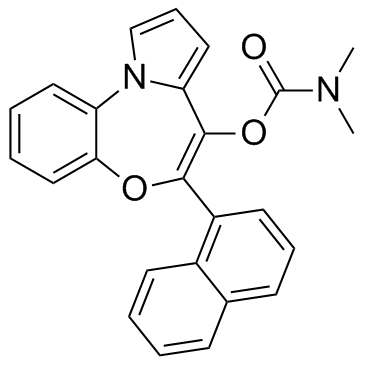
-
GC18008
PCI-34051
A potent, selective HDAC8 inhibitor
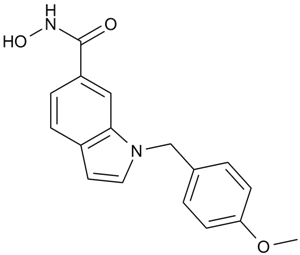
-
GC15142
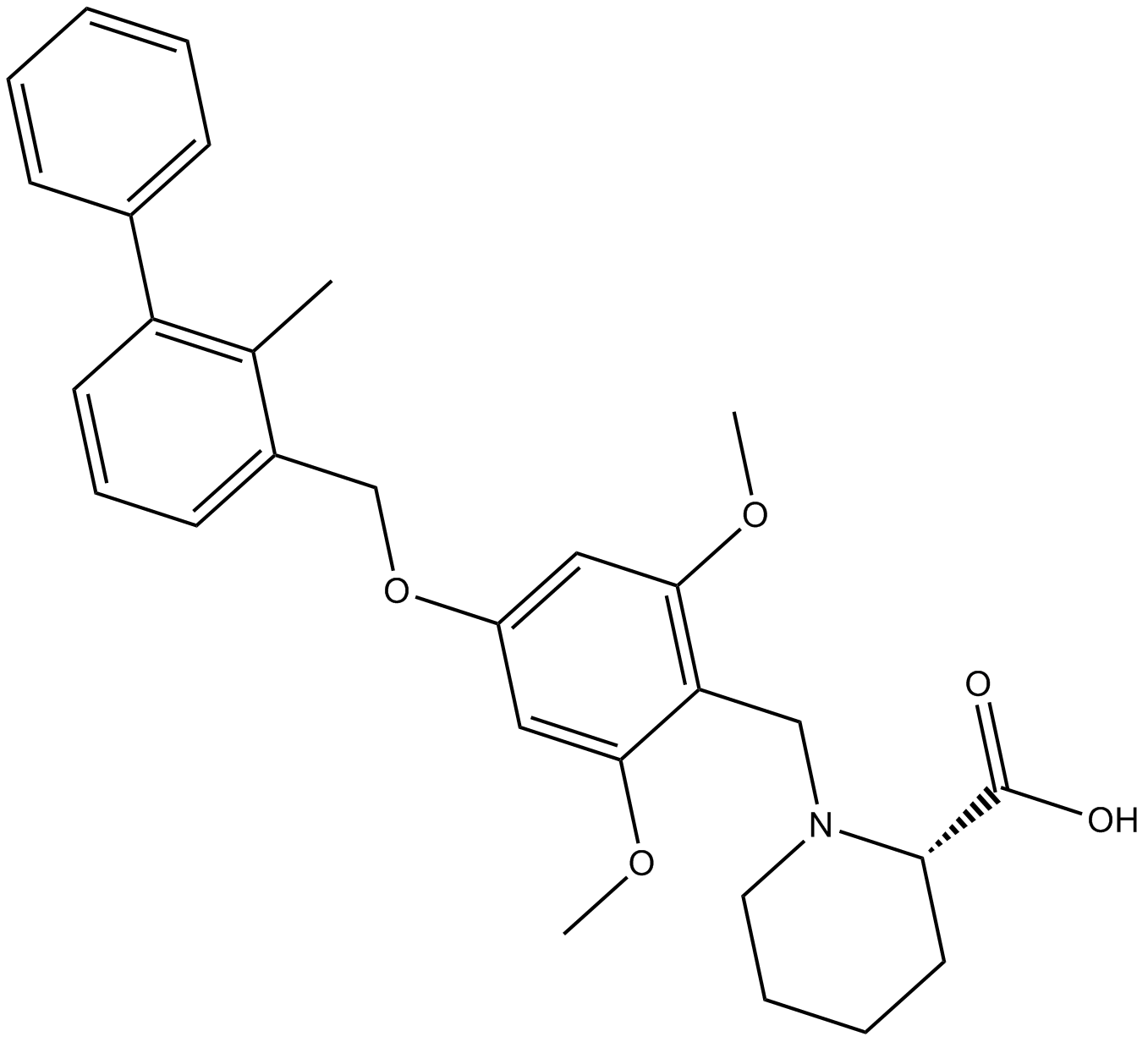
PD-1/PD-L1 inhibitor 1 (BMS-1)
PD-1/PD-L1 inhibitor 1 (BMS-1) is an inhibitor of the PD-1/PD-L1 protein/protein interaction (IC50 between 6 and 100 nM).
-
GC16762
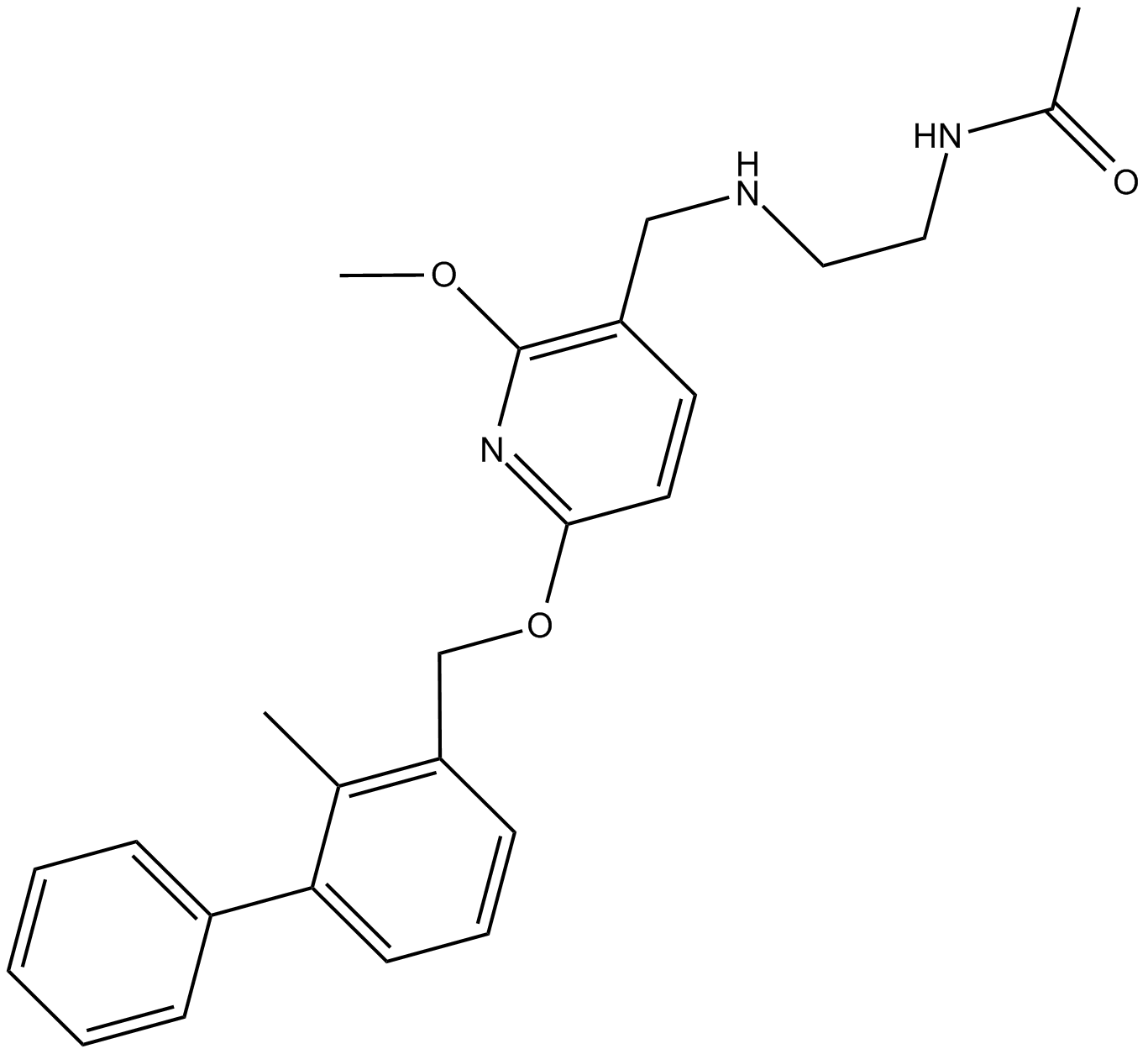
PD-1/PD-L1 inhibitor 2
PD-1/PD-L1 inhibitor 2 is a potent and nonpeptidic PD-1/PD-L1 complex inhibitor with an IC50 of 18 nM and a KD of 8 μM. PD-1/PD-L1 inhibitor 2 binds to PD-L1 and blocks human PD-1/PD-L1 interaction. PD-1/PD-L1 inhibitor 2 has antitumor activity.
-
GC32853
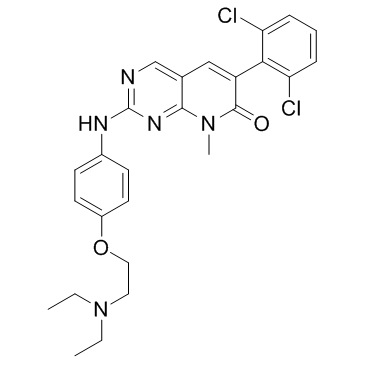
PD0166285
PD0166285, a substrate of P-gp, is a WEE1 inhibitor and a weak Myt1 inhibitor with IC50 values of 24 and 72 nM, respectively. PD0166285 exhibits an IC50 of 3.433 μM for Chk1.
-
GC10397
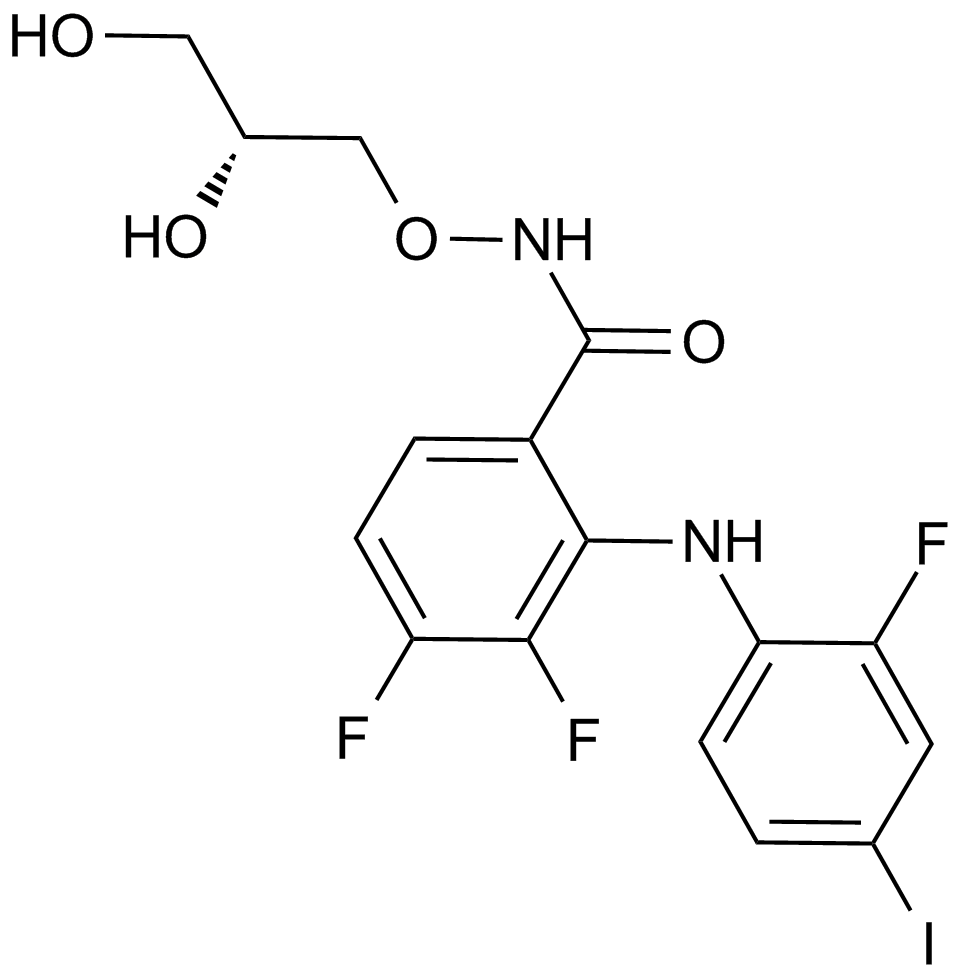
PD0325901
A MEK inhibitor that sustains stem cell renewal
-
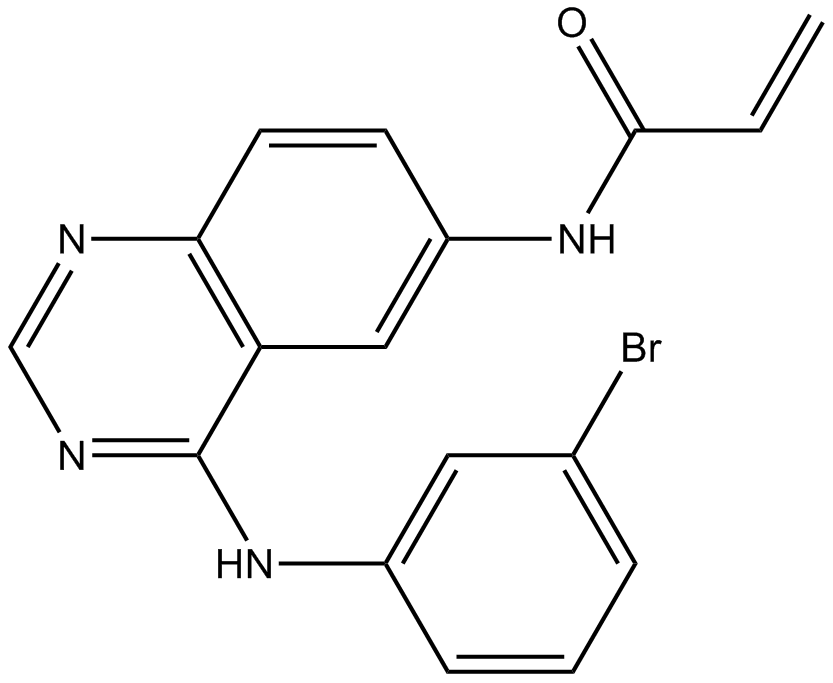
GC11015
PD168393
EGFR inhibitor
-
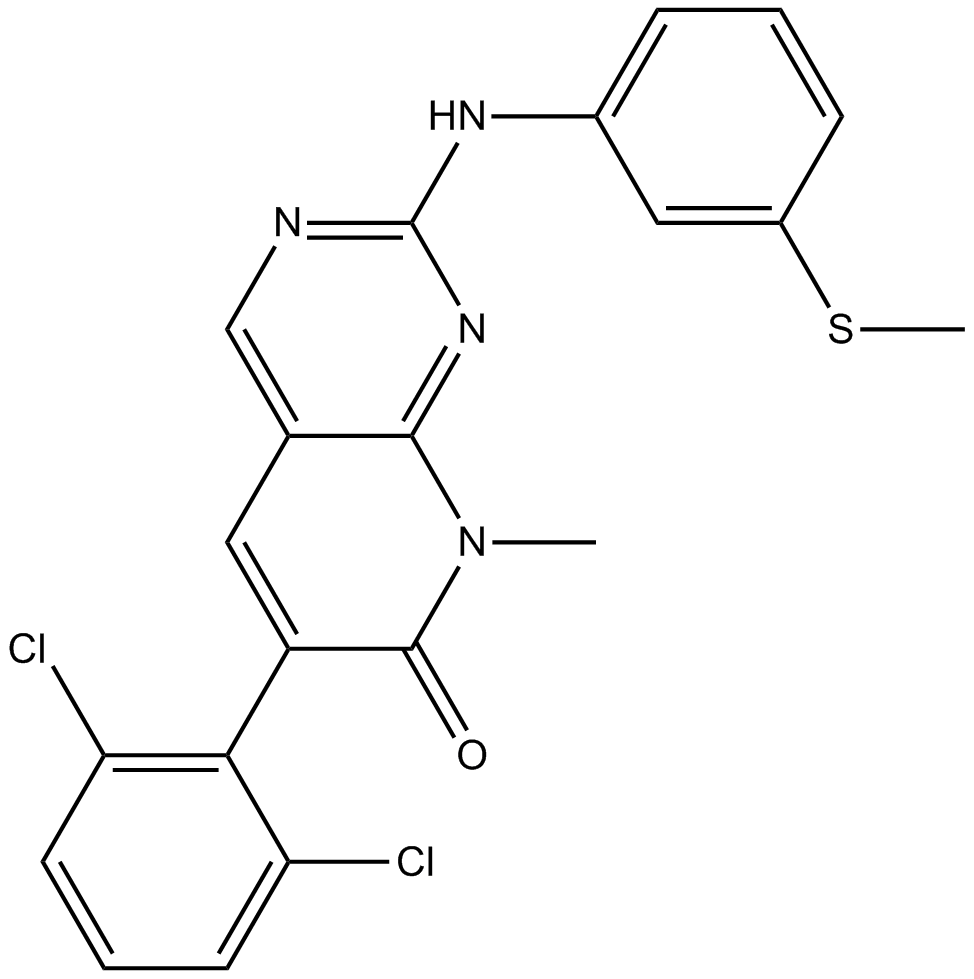
GC13592
PD173955
Dual Src/Abl kinase inhibitor, ATP-competitive,
-
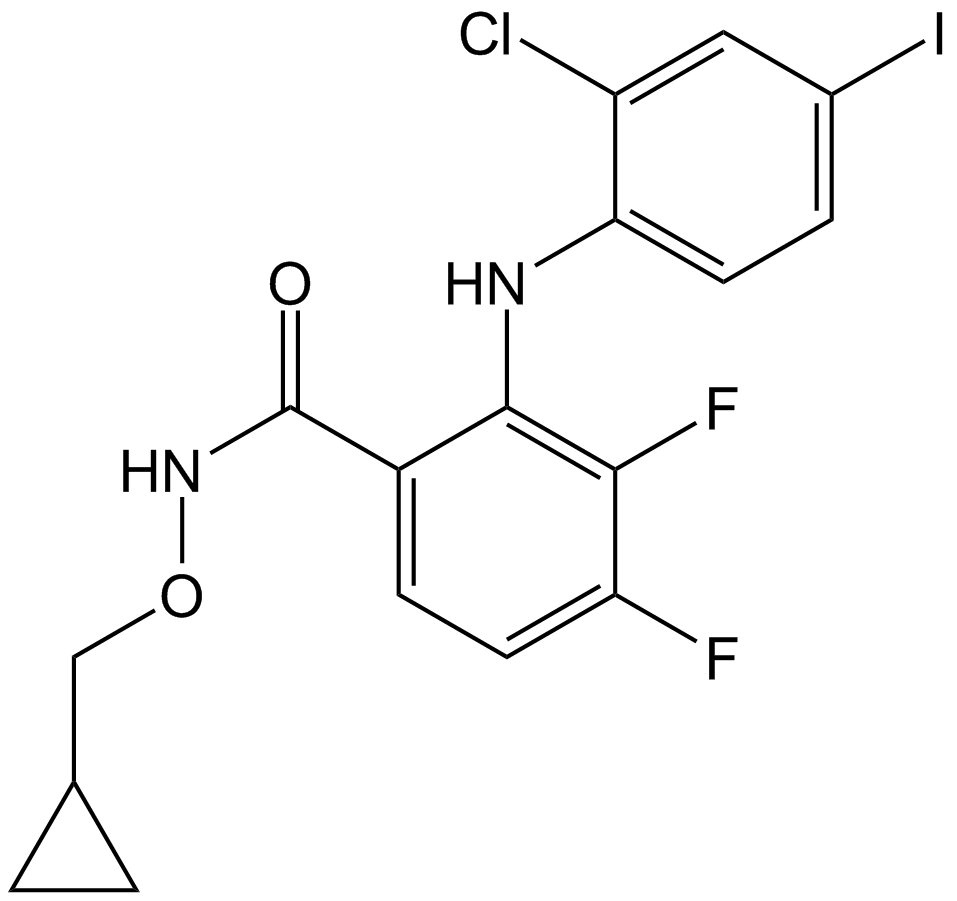
GC12989
PD184352 (CI-1040)
PD184352 (CI-1040) (PD 184352) is an orally active, highly specific, small-molecule inhibitor of MEK with an IC50 of 17 nM for MEK1.
-
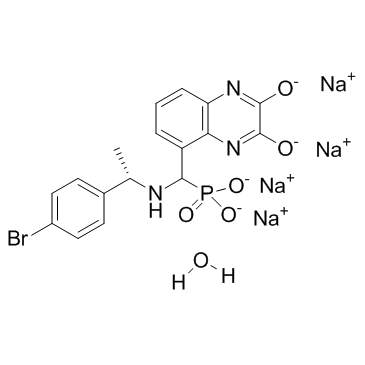
GC36865
PEAQX tetrasodium hydrate
PEAQX (NVP-AAM077) tetrasodium hydrate is a potent, selective and orally active NMDA antagonist, with IC50 values of 270 nM and 29600 nM for hNMDAR 1A/2B and hNMDAR 1A/2B, respectively.
-
GC11610
Pemetrexed
An antifolate with anticancer activity
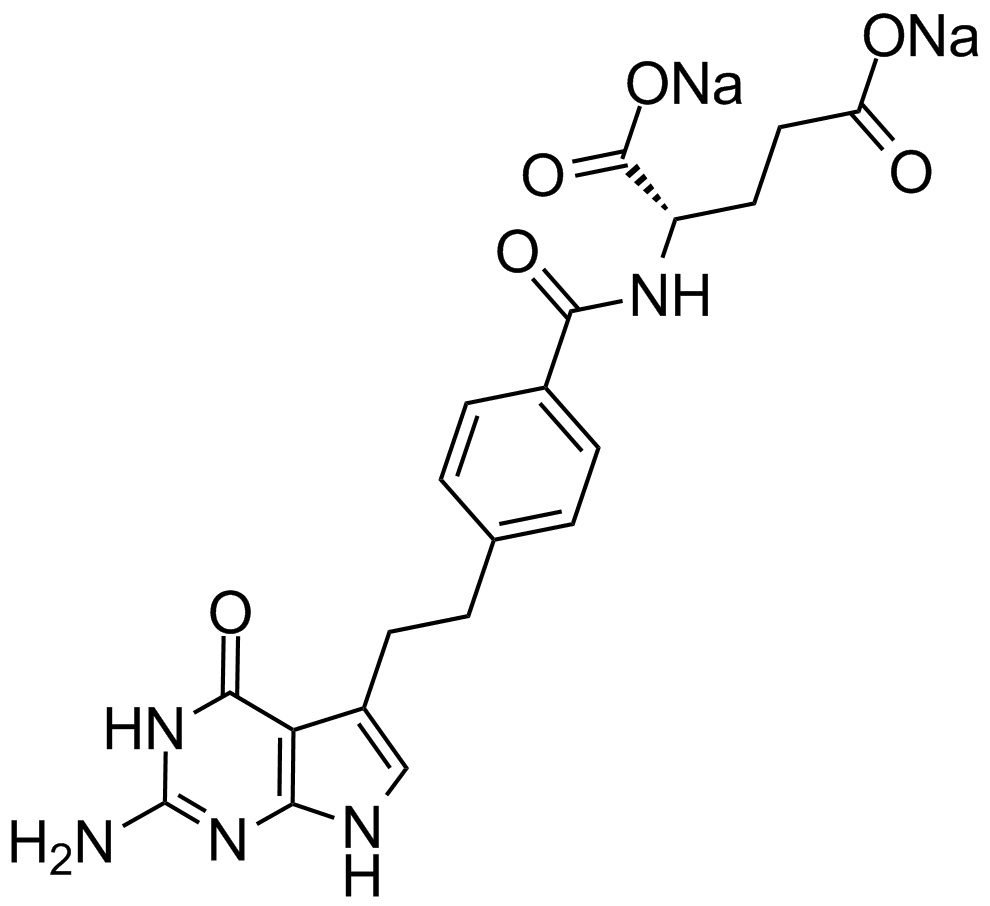
-
GC17694
Pemetrexed disodium hemipenta hydrate
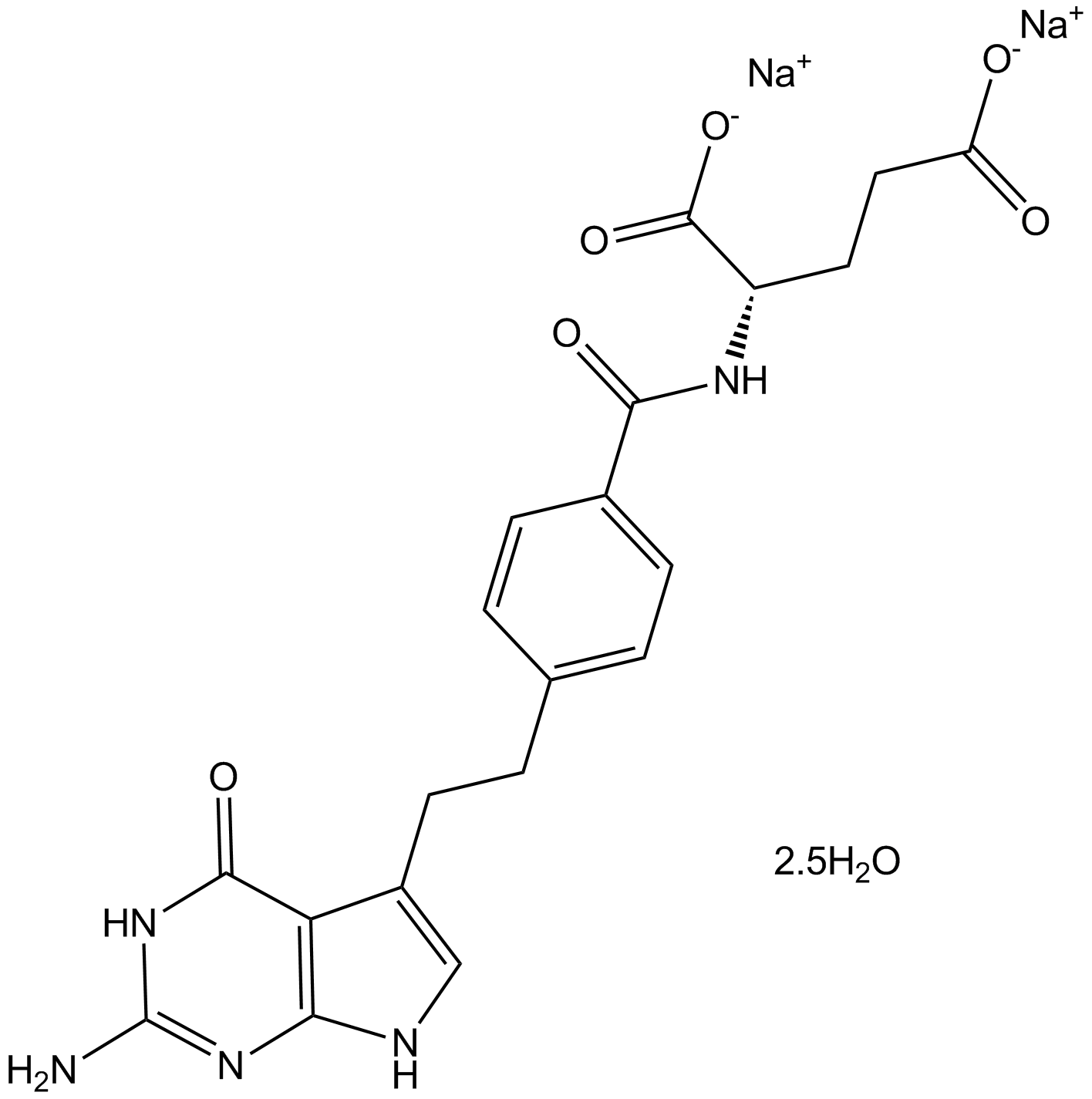
-
GC15880
Penicillic Acid
mycotoxin