Other Apoptosis
Products for Other Apoptosis
- Cat.No. Product Name Information
-
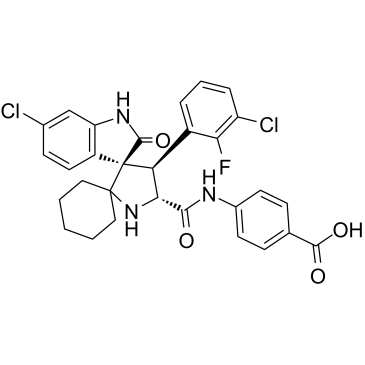
GC14503
IC261
CK1 inhibitor
-
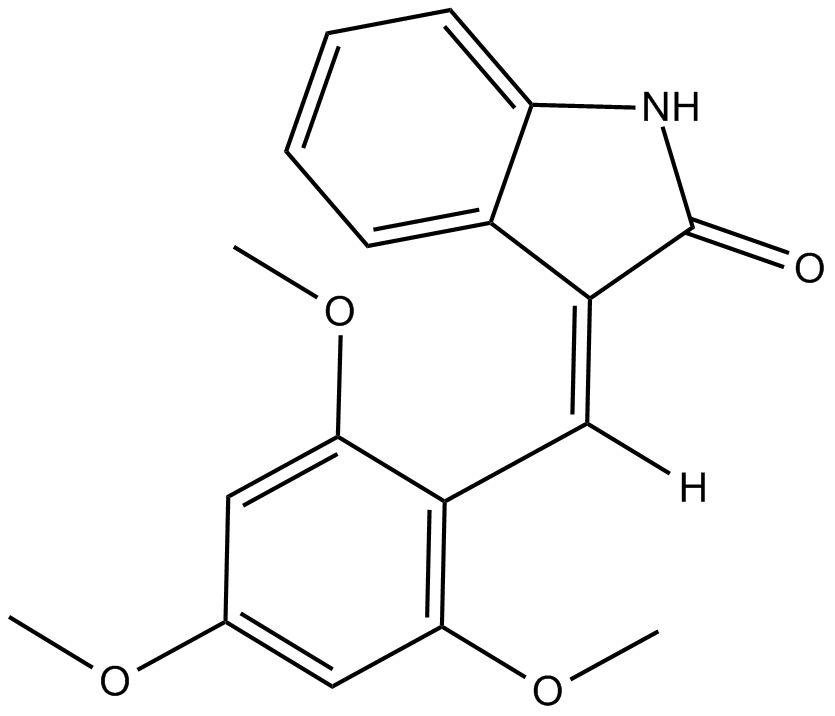
GC38096
Icaritin
Icaritin is a prenylflavonoid derivative obtained from the Epimedium genus.
-
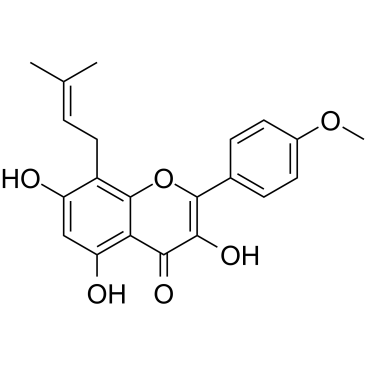
GC19198
iCRT3
iCRT3 is an inhibitor of both Wnt and β-catenin-responsive transcription.
-
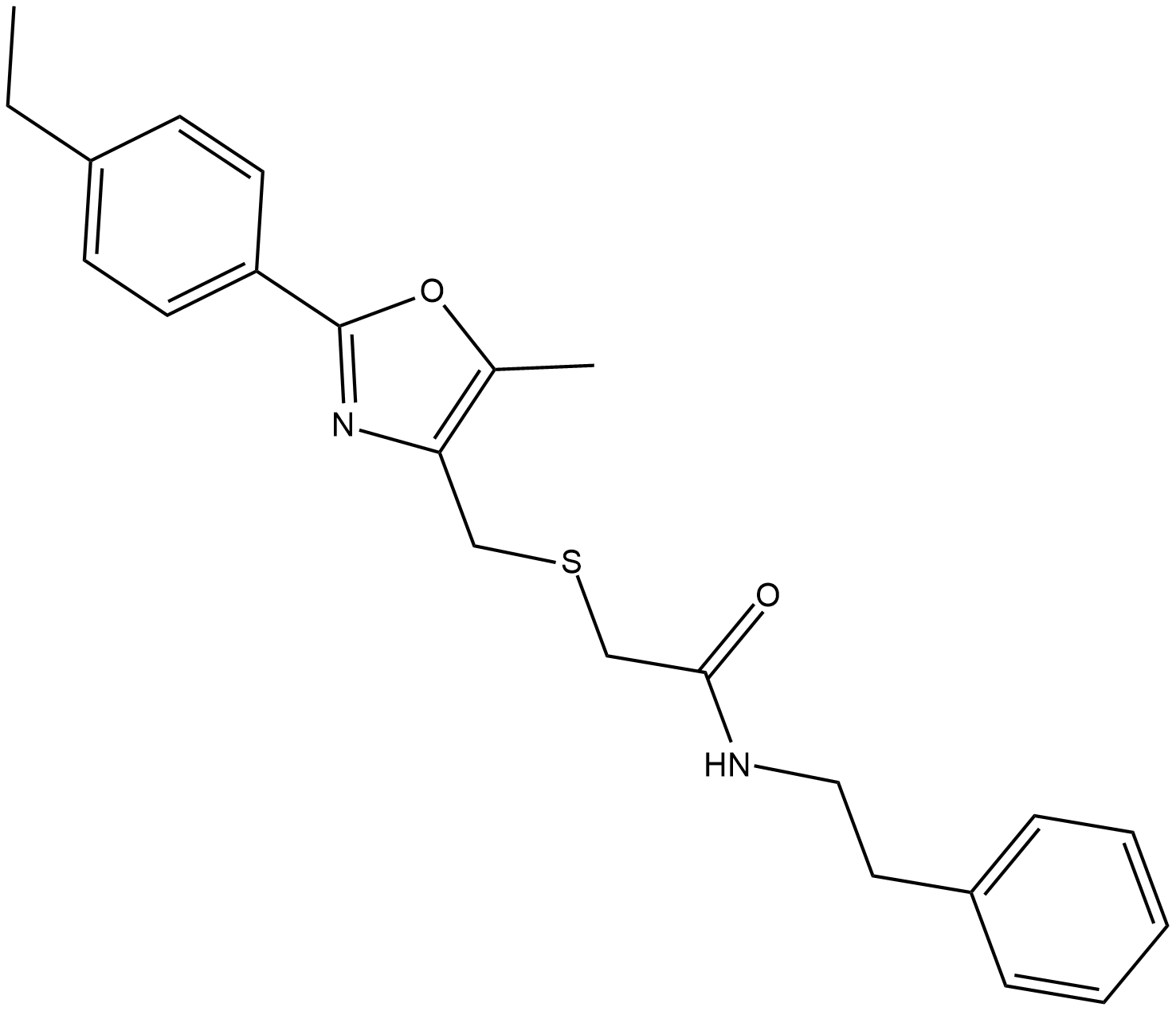
GC19411
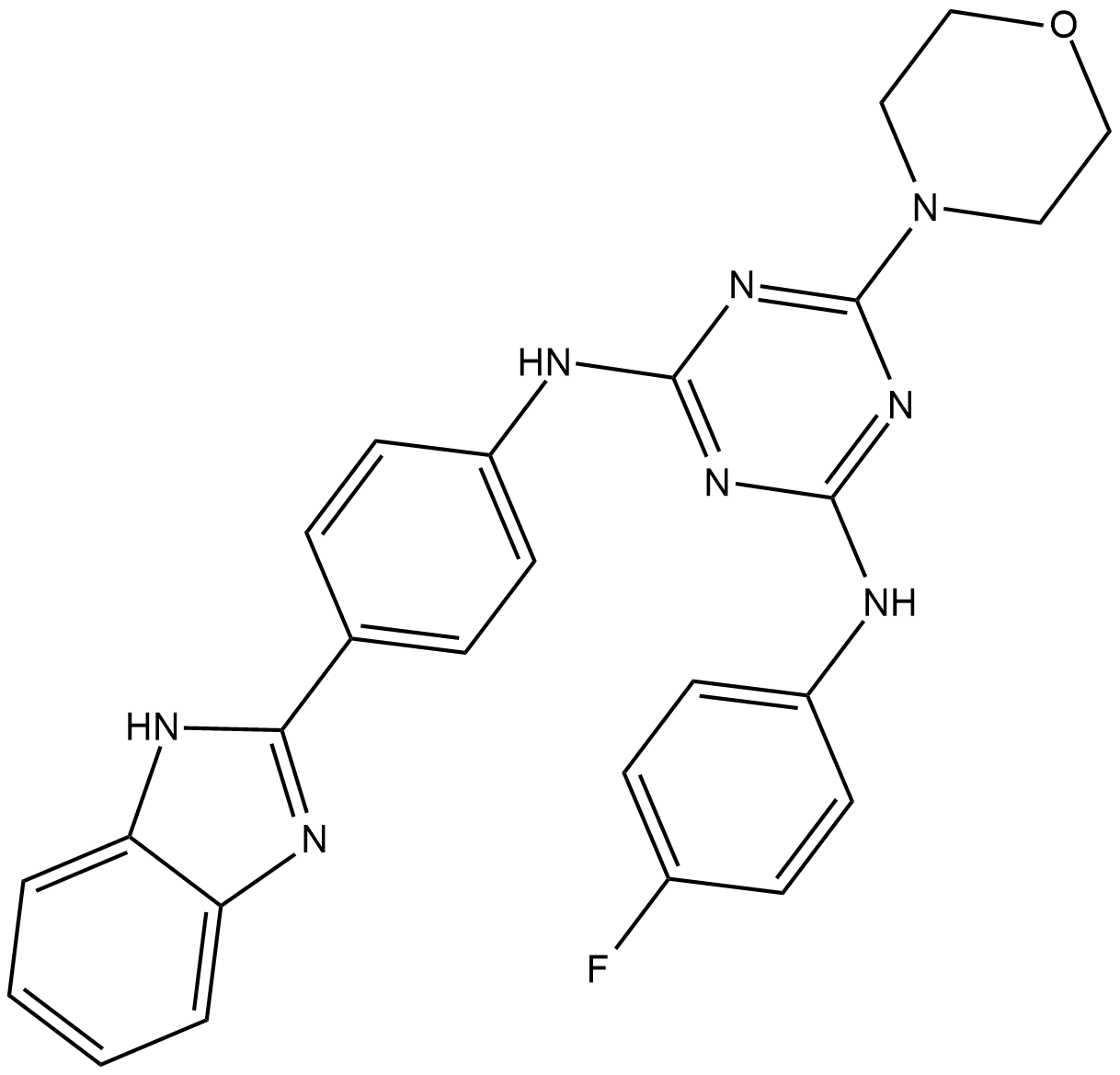
IITZ-01
IITZ-01 is a potent lysosomotropic autophagy inhibitor with single-agent antitumor activity, with an IC50 of 2.62 μM for PI3Kγ.
-
GC38566
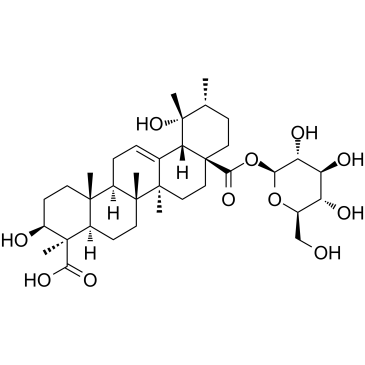
Ilexsaponin A
Ilexsaponin A, isolated from the root of Ilex pubescens, attenuates ischemia-reperfusion-induced myocardial injury through anti-apoptotic pathway.
-
GC19199
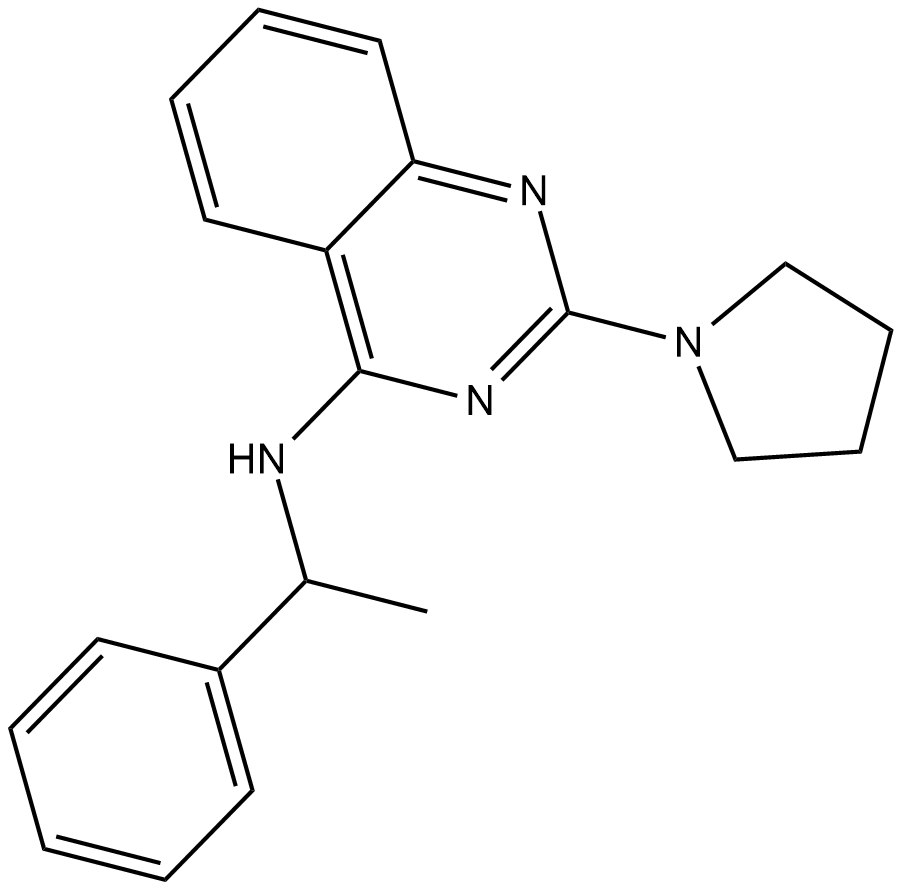
Importazole
Importazole is a small molecule inhibitor of the nuclear transport receptor importin-β.
-
GC19200
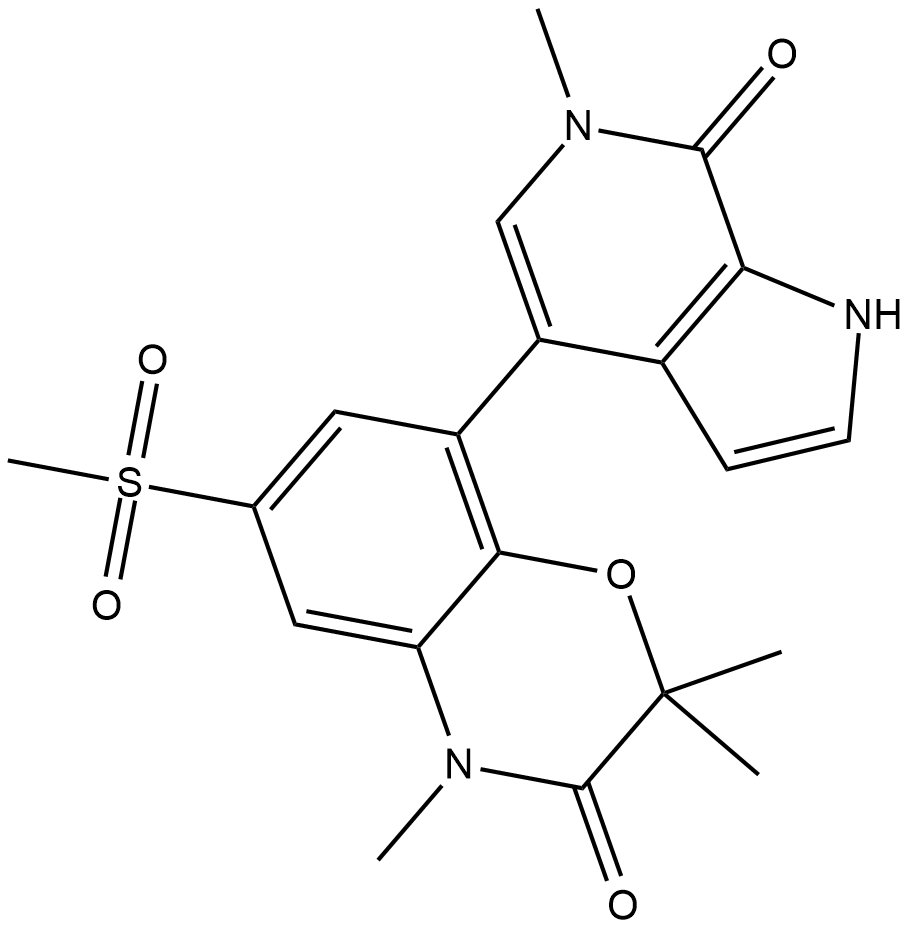
INCB-057643
INCB-057643 is a novel, orally bioavailable BET inhibitor.
-
GC17866
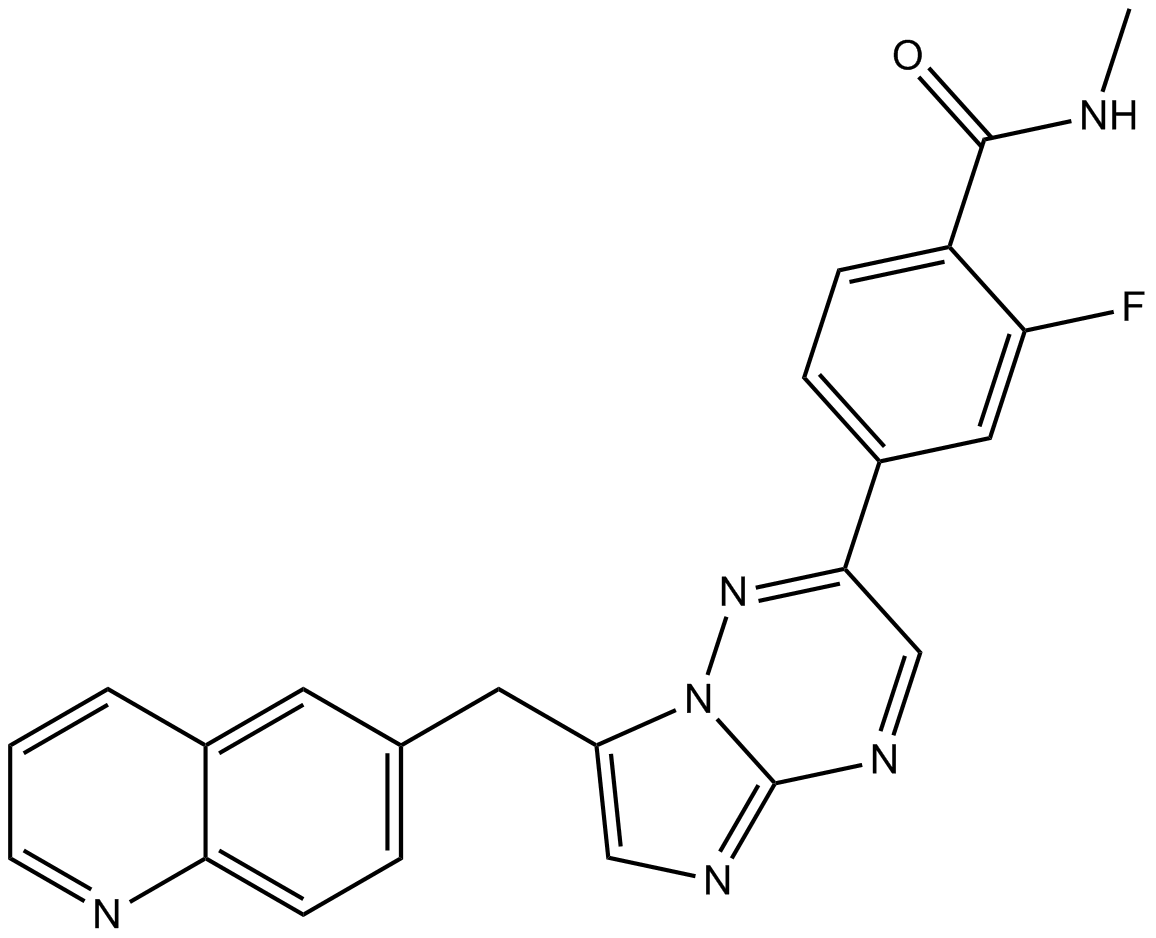
INCB28060
INCB28060 (INC280; INCB28060) is a potent, orally active, selective, and ATP competitive c-Met kinase inhibitor (IC50=0.13 nM). INCB28060 can inhibit phosphorylation of c-MET as well as c-MET pathway downstream effectors such as ERK1/2, AKT, FAK, GAB1, and STAT3/5. INCB28060 potently inhibits c-MET-dependent tumor cell proliferation and migration and effectively induces apoptosis. Antitumor activity. INCB28060 is largely metabolized by CYP3A4 and aldehyde oxidase.
-
GC38213
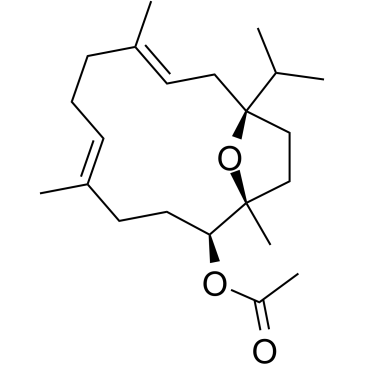
Incensole Acetate
Incensole acetate is a main constituent of Boswellia carterii resin, has neuroprotective effects against neuronal damage in traumatic and ischemic head injury.
-
GC36308
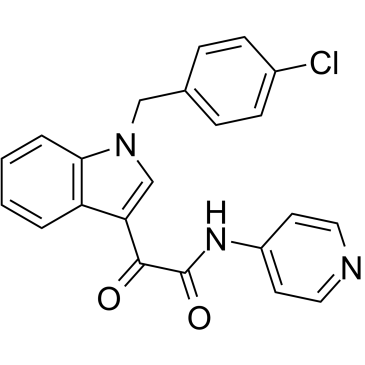
Indibulin
An inhibitor of microtubule assembly
-
GC12975
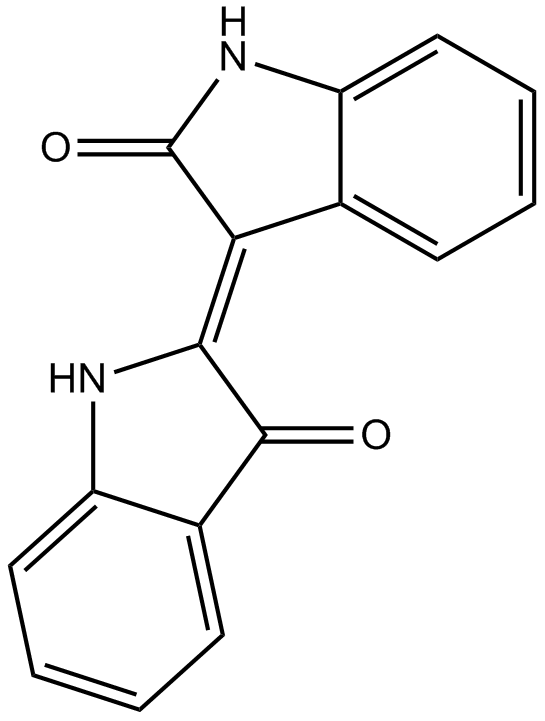
Indirubin
A cyclin-dependent kinases and GSK-3β inhibitor
-
GC10334
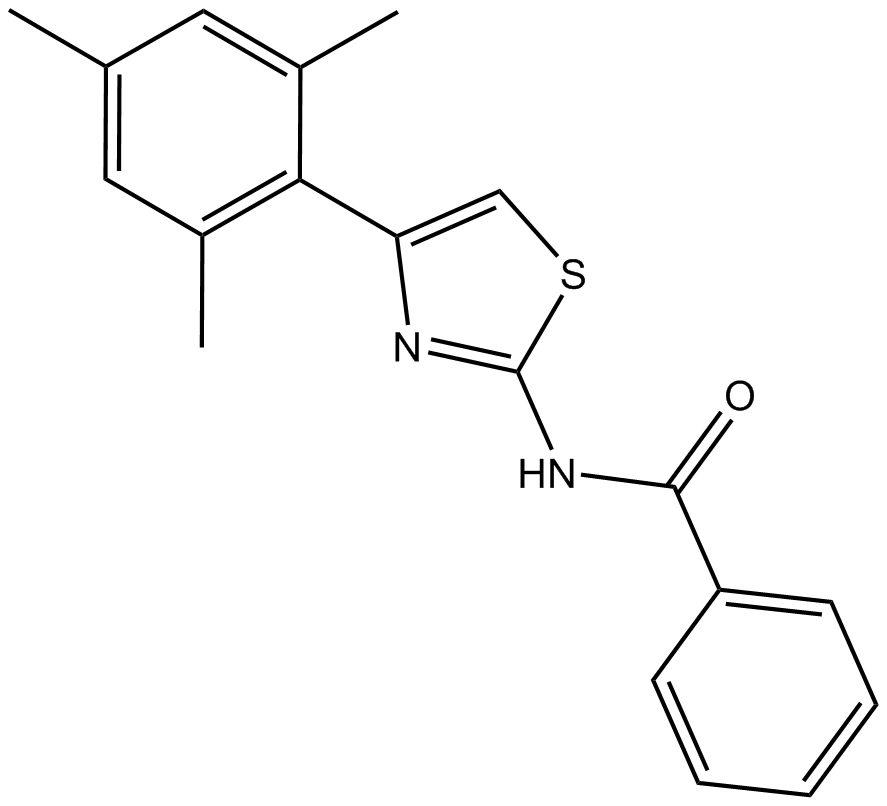
INH6
Hec1/Nek2 inhibitor, potent
-
GC15148
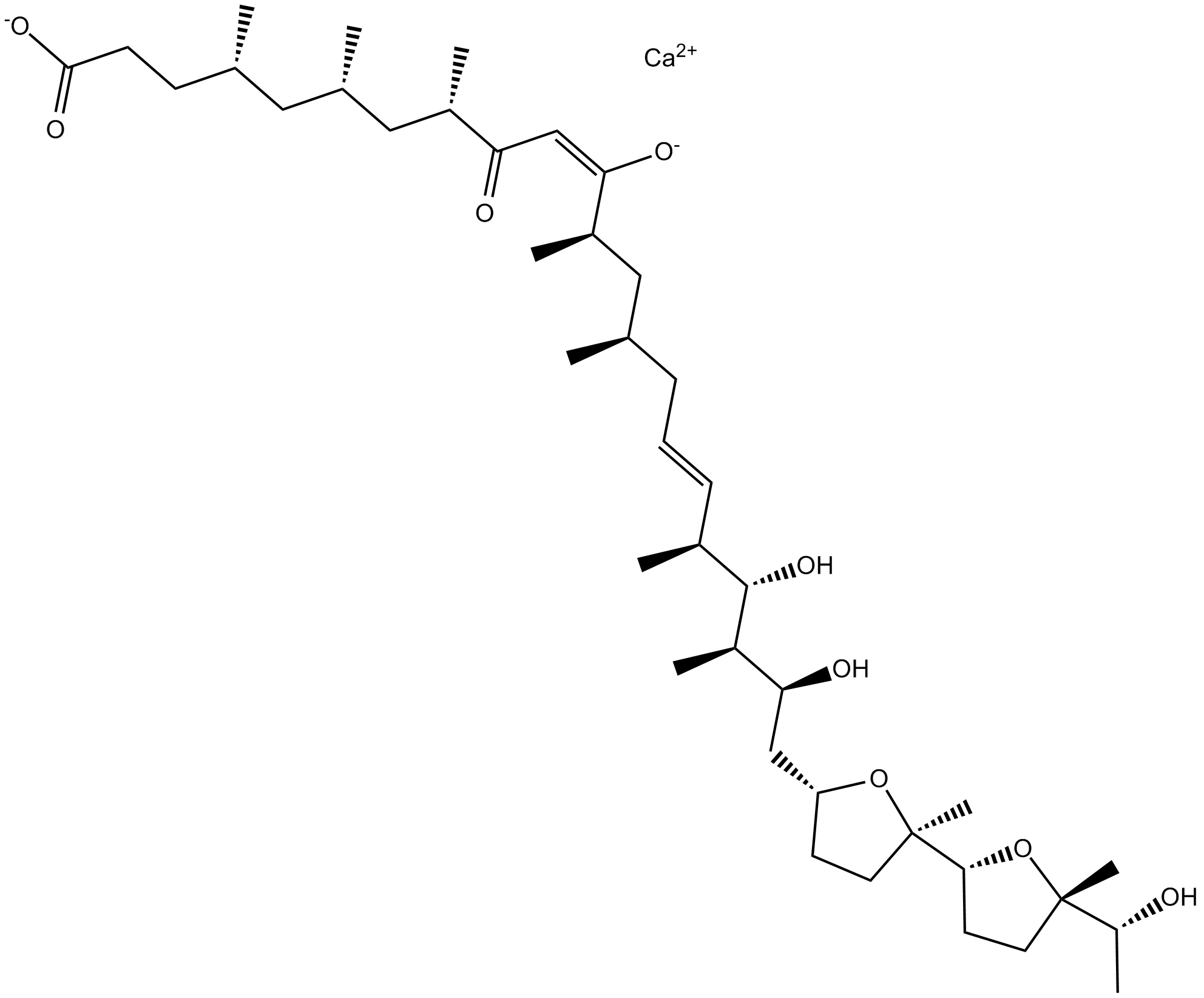
Ionomycin calcium salt
Lonomycin is a selective calcium ionophore derived from S. conglobatus that mobilizes intracellular calcium stores.
-
GC15446
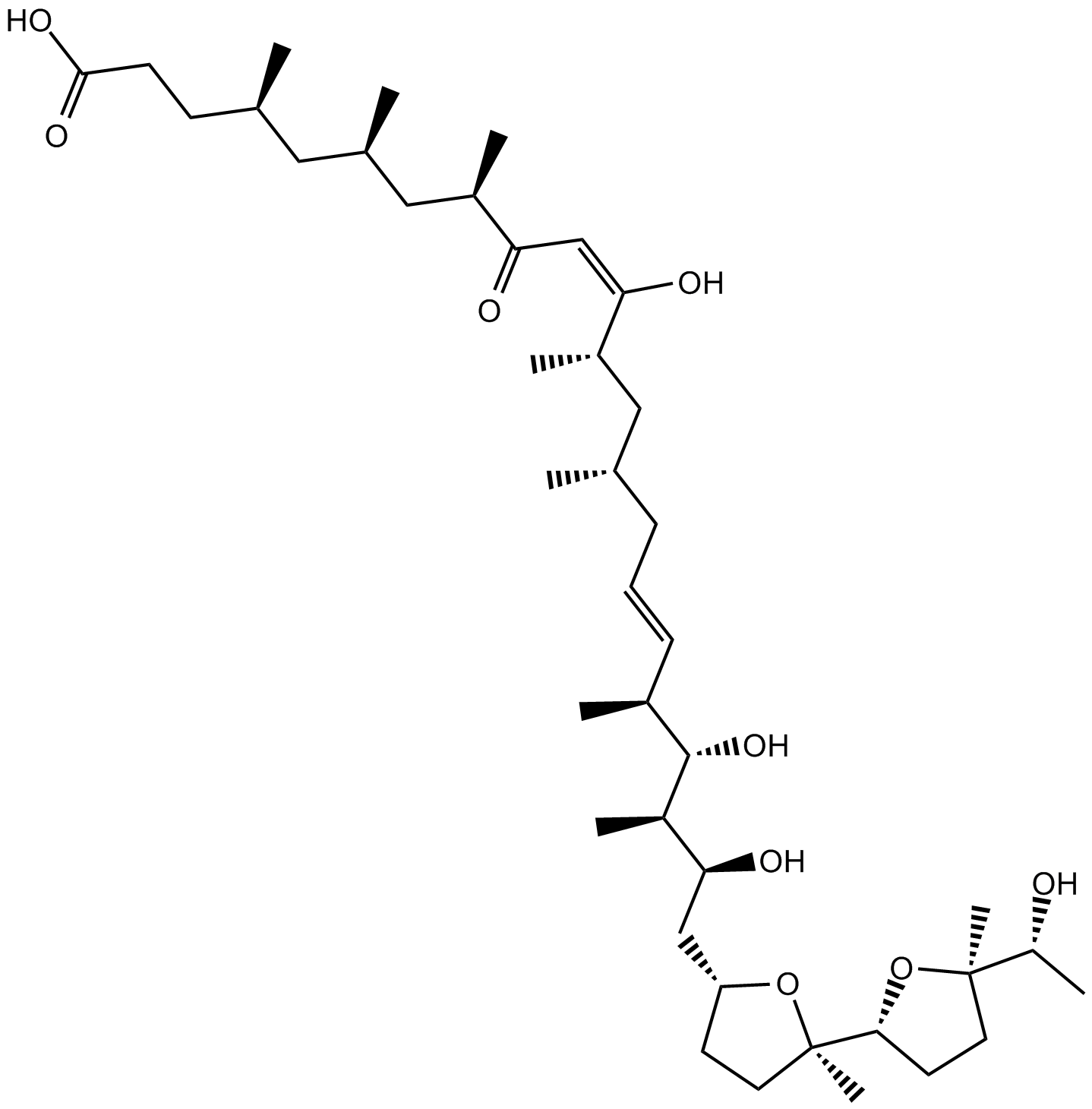
Ionomycin free acid
Ionomycin free acid (SQ23377) is a potent, selective calcium ionophore and an antibiotic produced by Streptomyces conglobatus.
-
GC46027
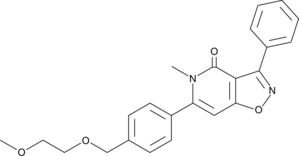
IP7e
An activator of Nurr1 signaling
-
GC10467
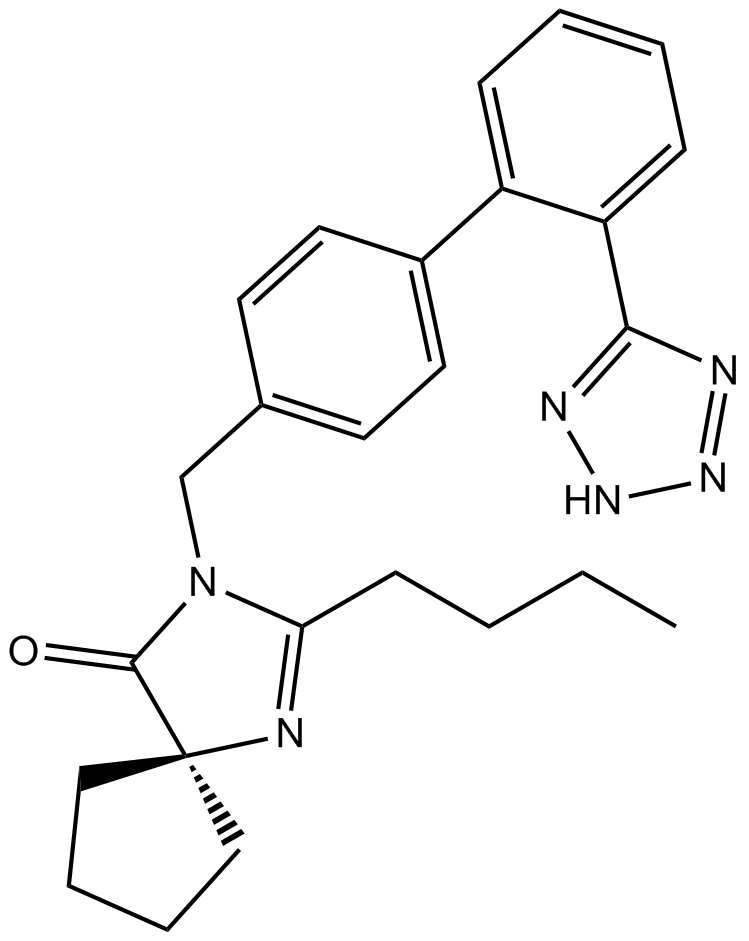
Irbesartan
Angiotensin II inhibitor
-
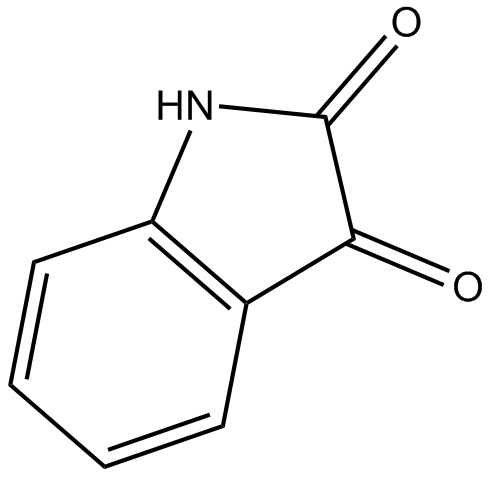
GC12962
Isatin
endogenous monoamine oxidase inhibitor
-
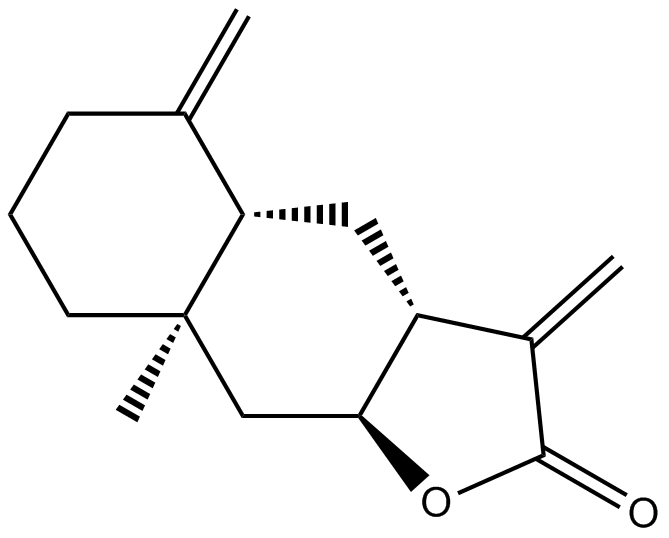
GN10666
Isoalantolactone
-
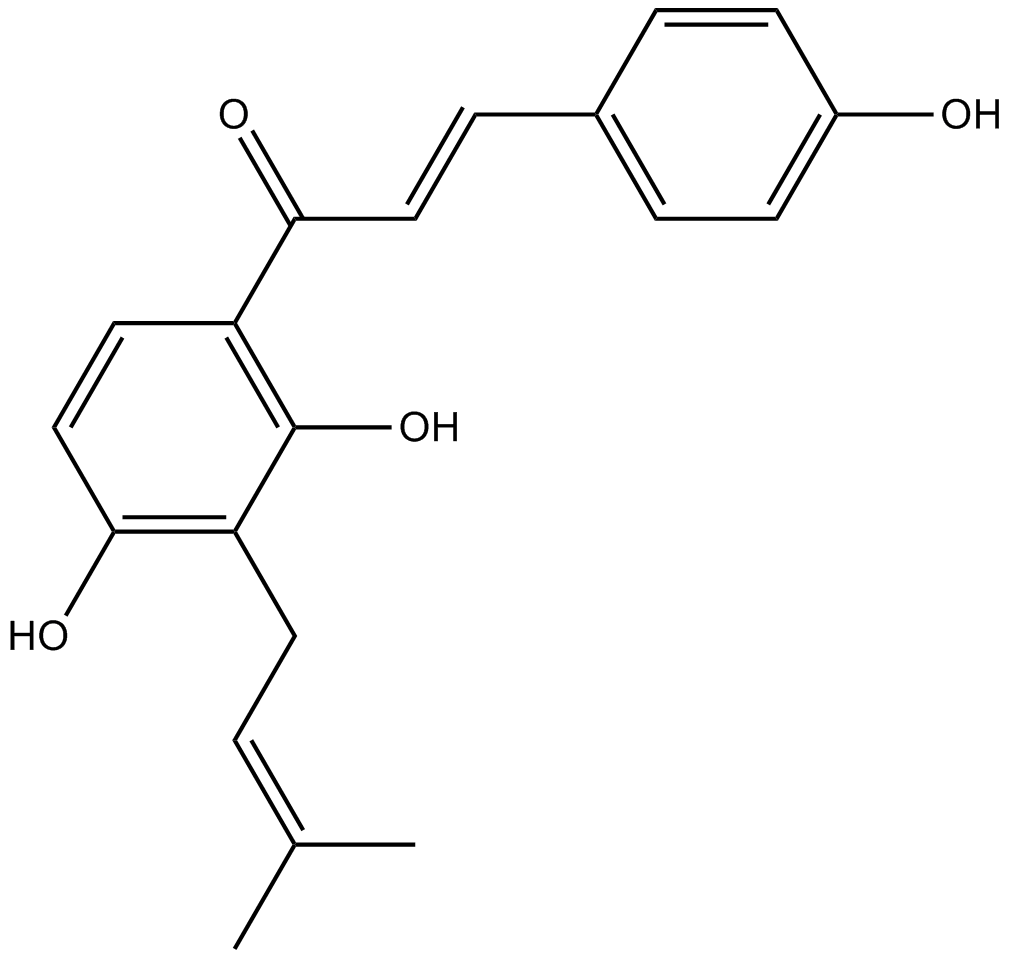
GC13795
Isobavachalcone
A chalcone and flavonoid with diverse biological activities
-
GC38220
Isoliensinine
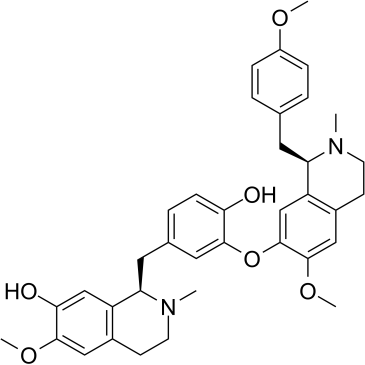
-
GN10088
Isoliquiritigenin
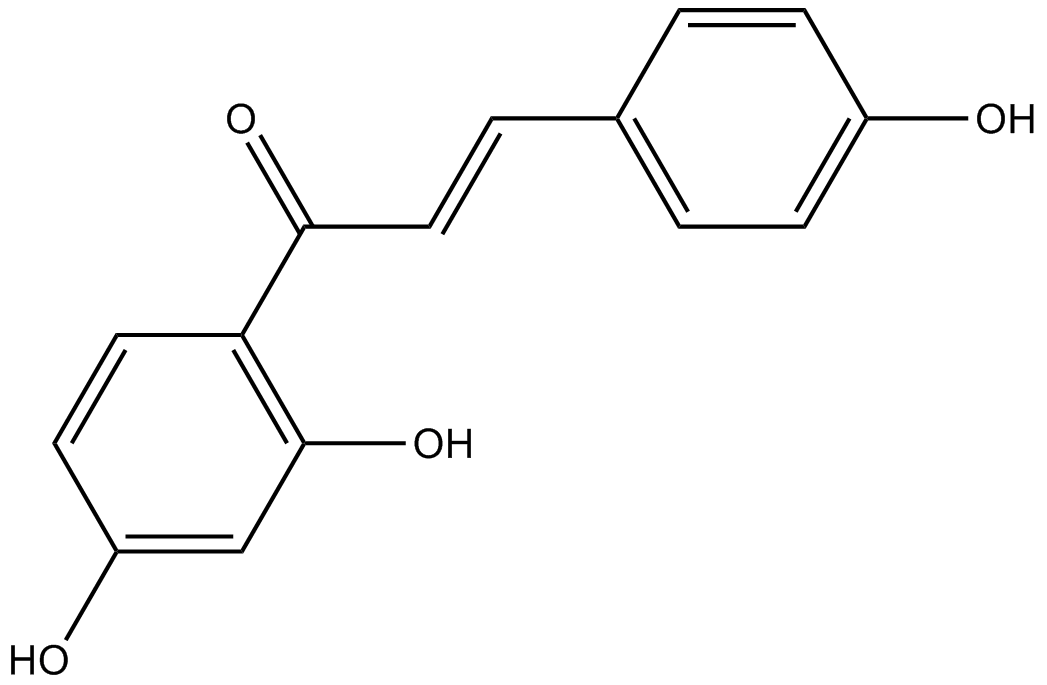
-
GC36343
Isosilybin A
Isosilybin A, a flavonolignan isolated from silymarin, has anti-prostate cancer (PCA) activity. Isosilybin A inhibits proliferation and induces G1 phase arrest and apoptosis in cancer cells, which activates apoptotic machinery in PCA cells via targeting Akt-NF-κB-androgen receptor (AR) axis.
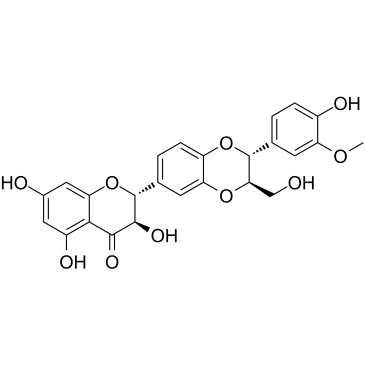
-
GC36344
Isosilybin B
Isosilybin B, a flavonolignan isolated from silymarin, has anti-prostate cancer (PCA) activity via inhibiting proliferation and inducing G1 phase arrest and apoptosis. Isosilybin B causes androgen receptor (AR) degradation.
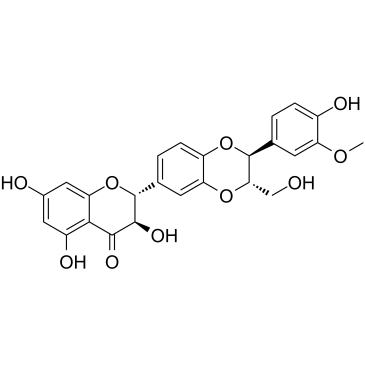
-
GC13394
Ispinesib (SB-715992)
Ispinesib (SB-715992) is a specific inhibitor of kinesin spindle protein (KSP), with a Ki app of 1.7 nM.
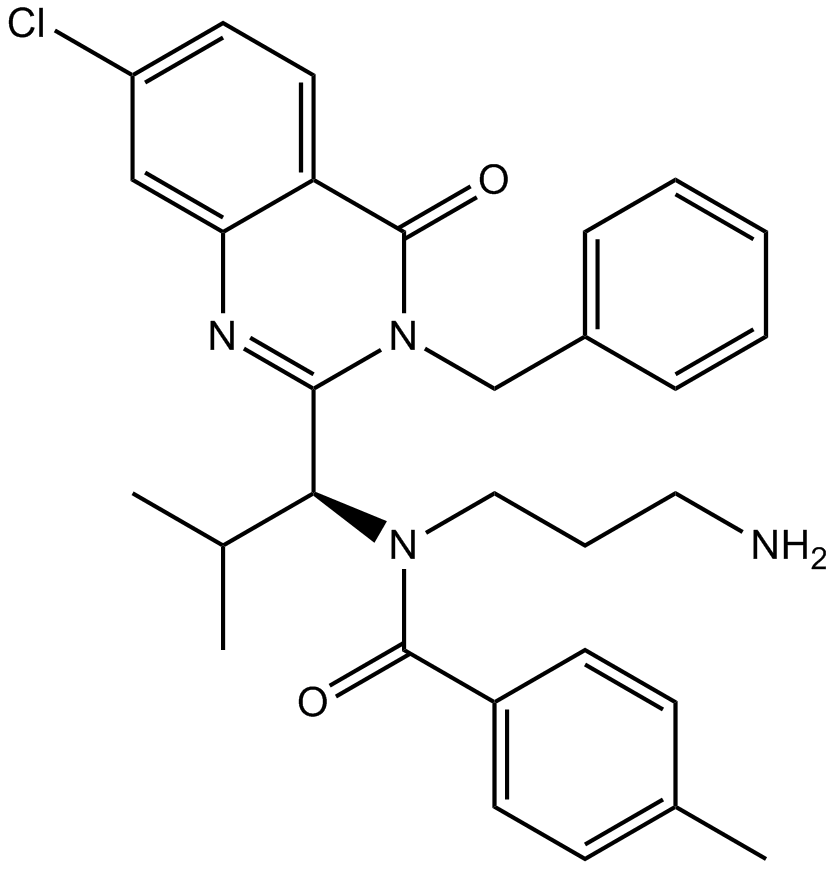
-
GC16869
Ixabepilone
A broad-spectrum anticancer agent
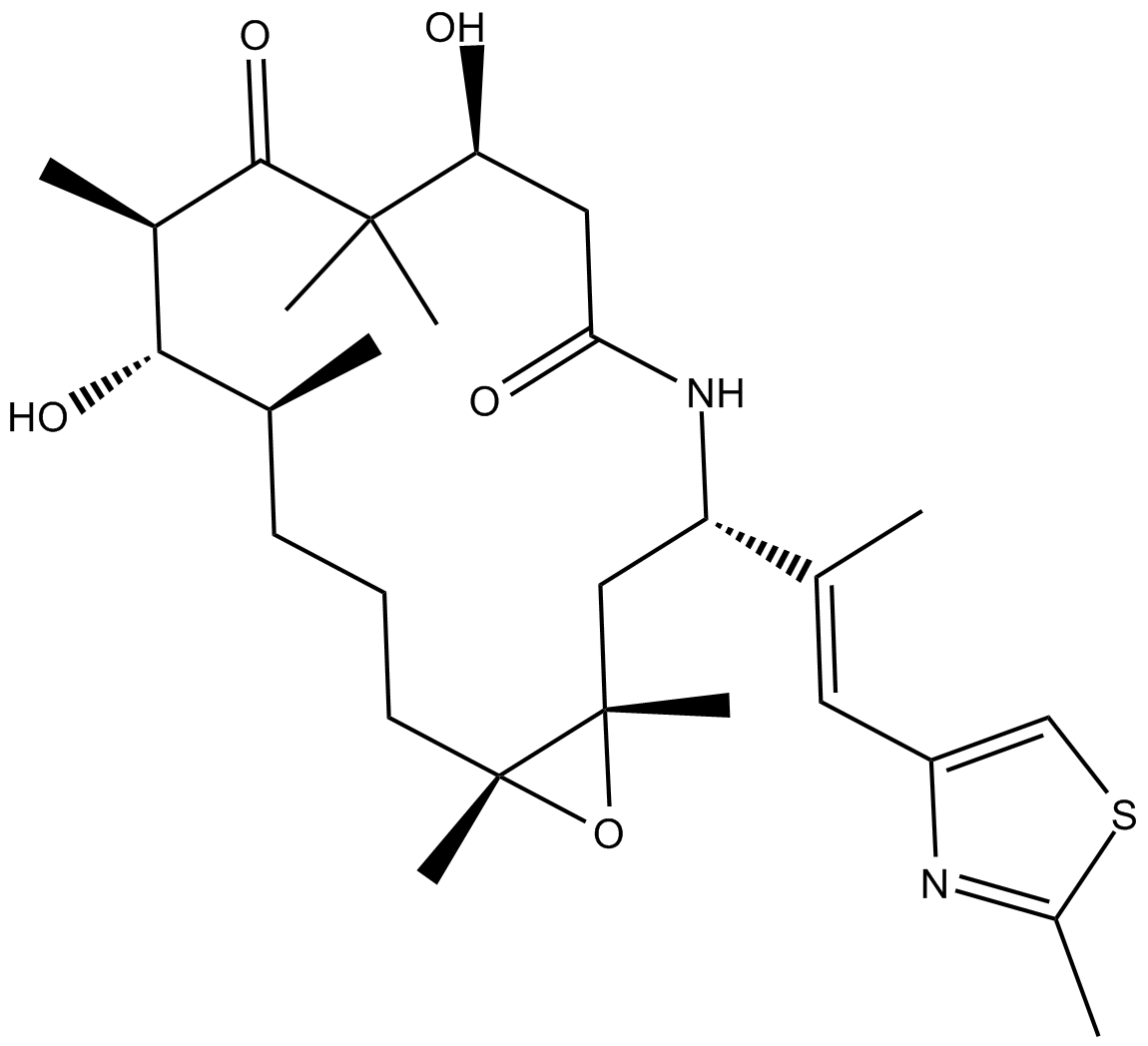
-
GN10645
Jaceosidin
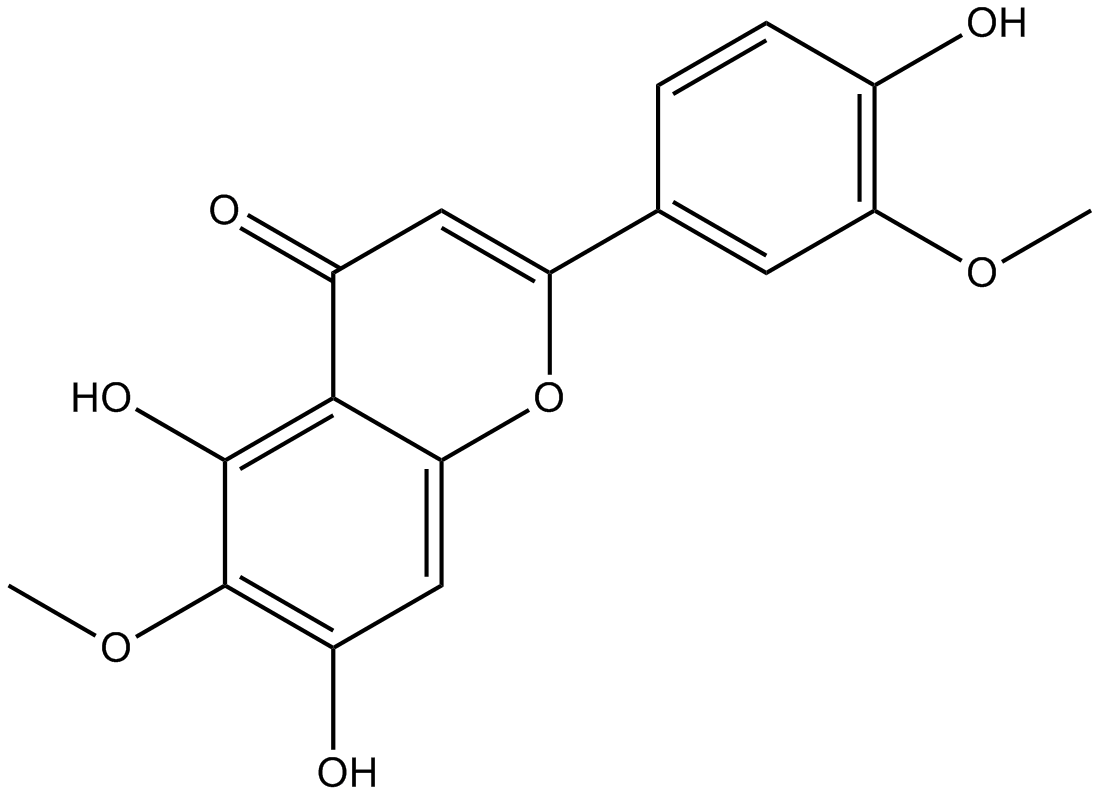
-
GC34634
JG-98
JG-98, an allosteric heat shock protein 70 (Hsp70) inhibitor, which binds tightly to a conserved site on Hsp70 and disrupts the Hsp70-Bag3 interaction. JG-98 shows anti-cancer activities affecting both cancer cells and tumor-associated macrophages.
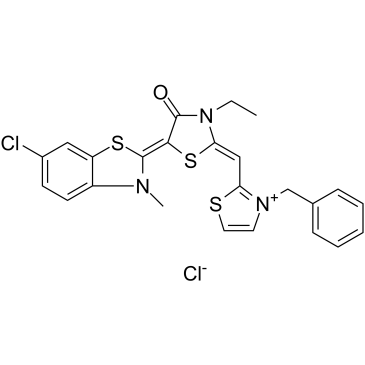
-
GC15603
JIB-04
Jumonji histone demethylase inihibitor
-
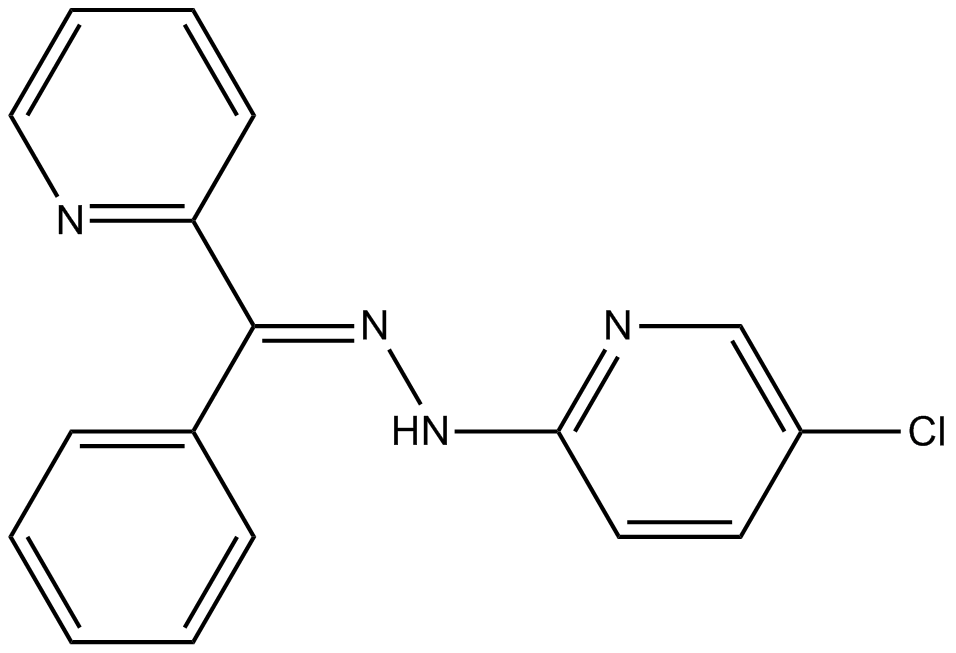
GC12117
JNJ-26854165 (Serdemetan)
An antagonist of MDM2 action
-
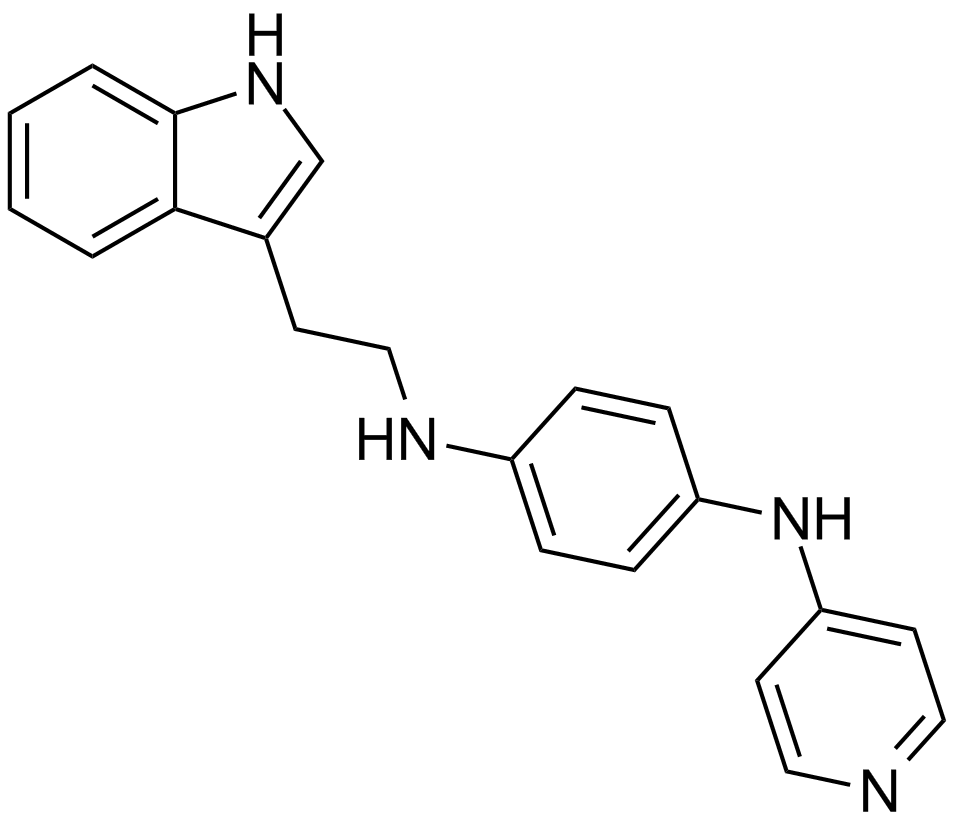
GC12612
JNJ-7706621
A dual inhibitor of CDKs and Aurora kinases
-
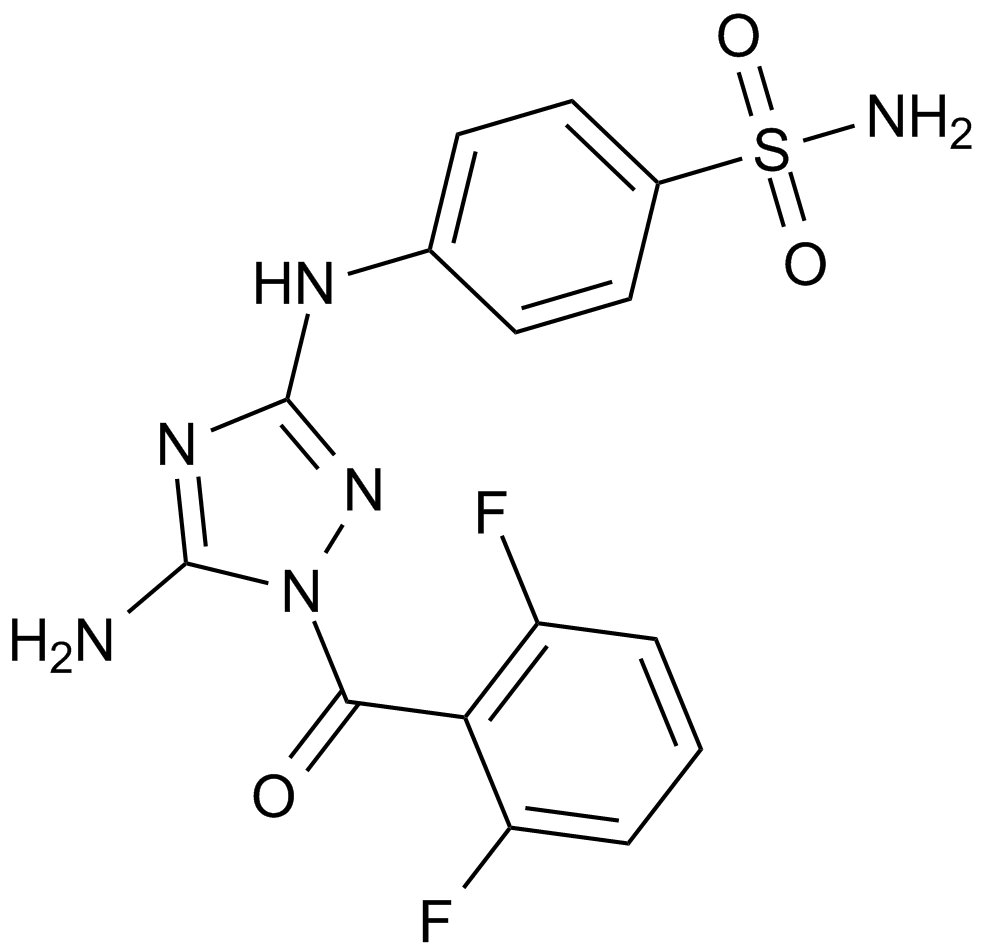
GC38082
JTE-013
A selective S1P2 receptor antagonist
-
GC16167
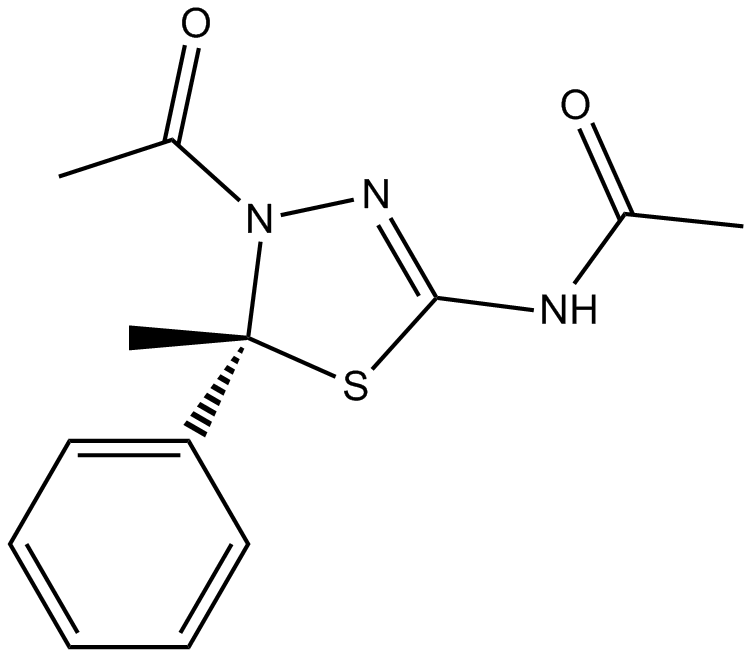
K 858
K858 Racemic is an ATP-uncompetitive inhibitor of kinesin Eg5 with an IC50 of 1.3 μM.
-
GC15281
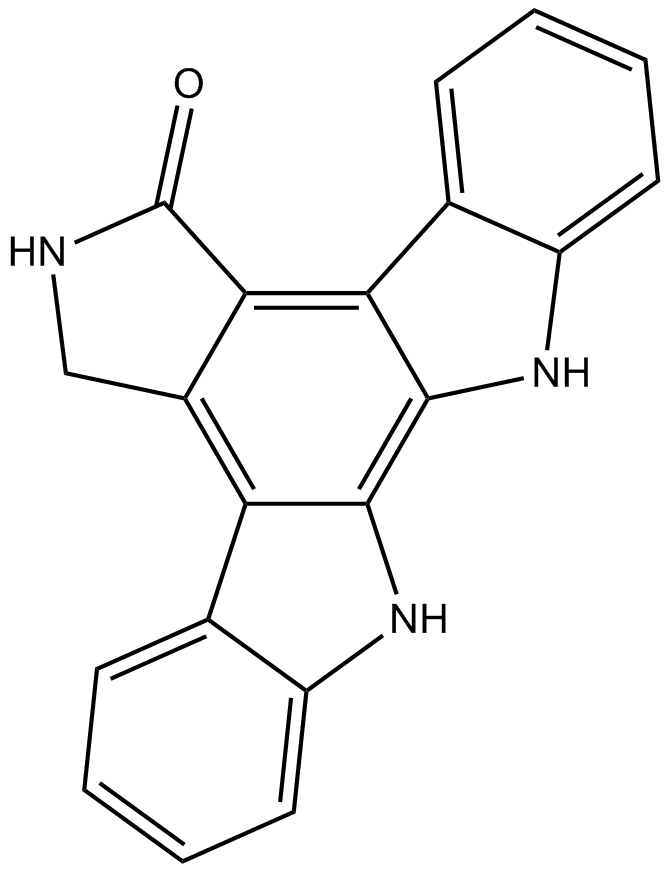
K-252c
Protein kinase inhibitor
-
GC12247
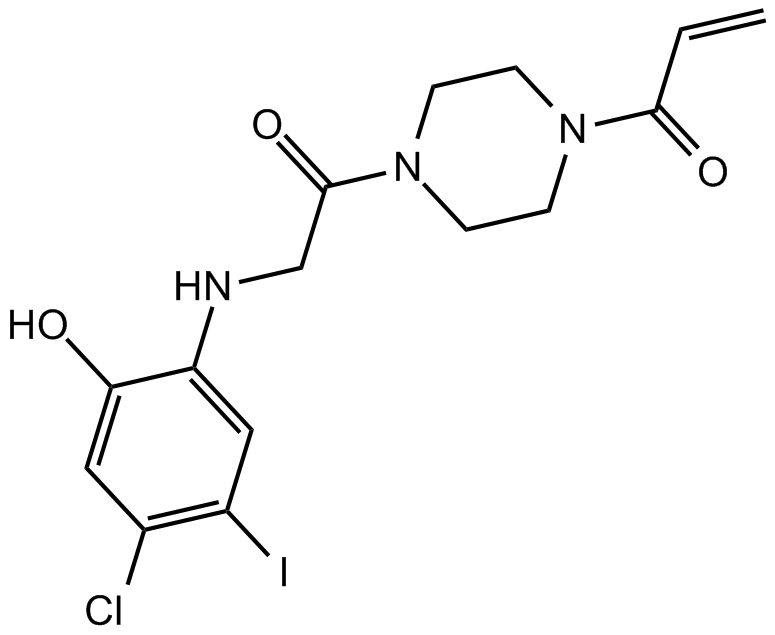
K-Ras(G12C) inhibitor 12
allosteric inhibitor of K-Ras(G12C)
-
GC38209
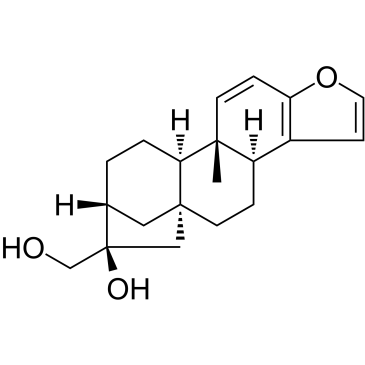
Kahweol
A natural diterpene with antiinflammatory and antiangiogenic properties
-
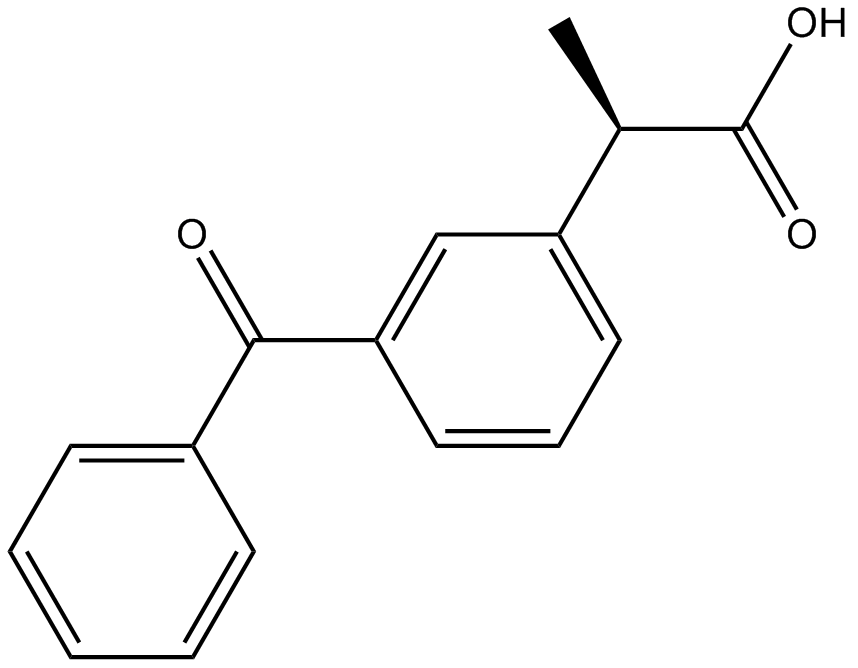
GC13339
Ketoprofen
Dual COX1/2 inhibitor
-
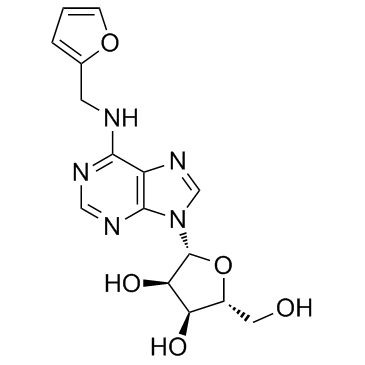
GC36393
Kinetin riboside
Kinetin riboside, a cytokinin analog, can induce apoptosis in cancer cells. It inhibits the proliferation of HCT-15 cells with an IC50 of 2.5 μM.
-
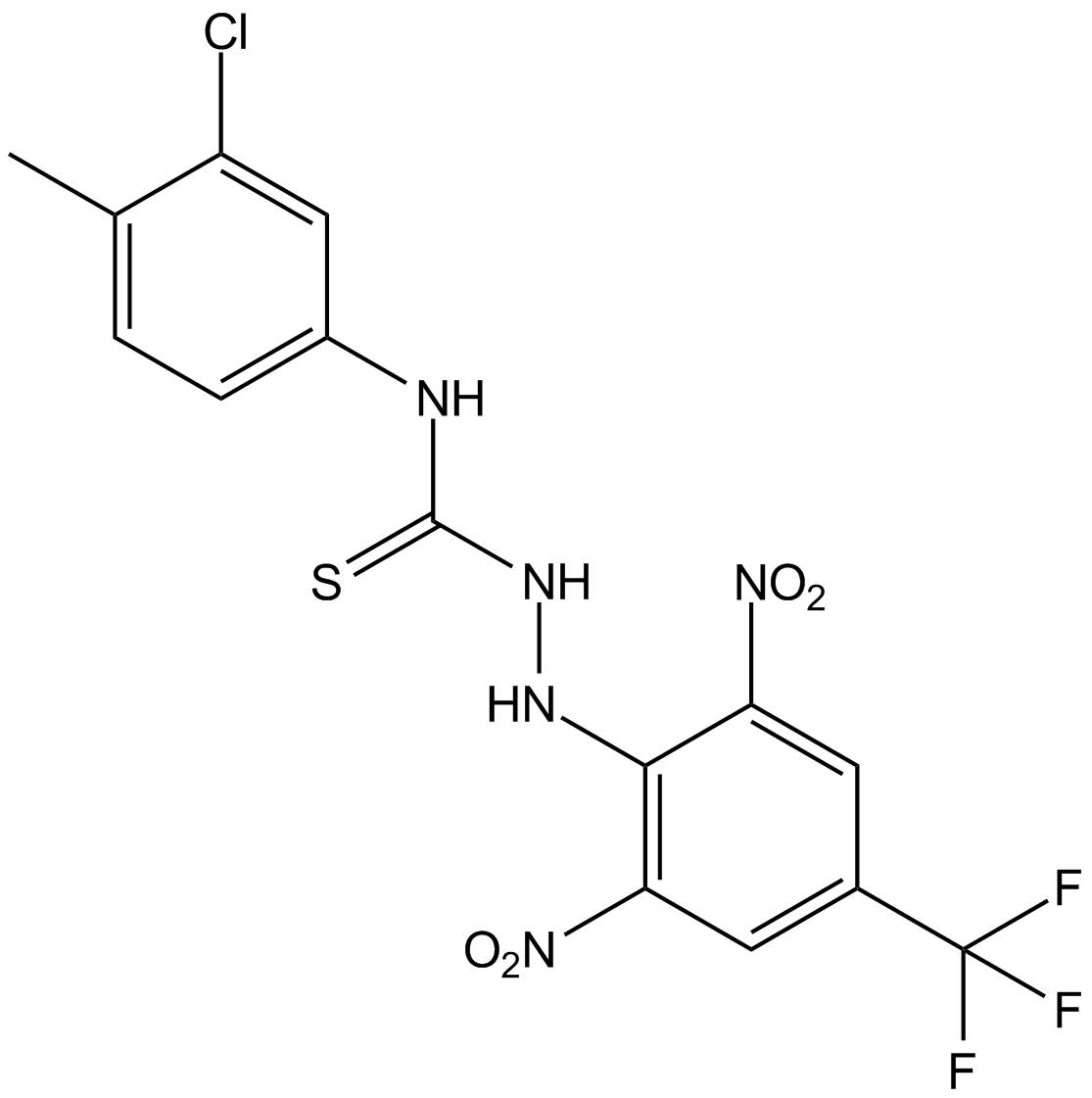
GC16922
Kobe0065
Ras inhibitor
-
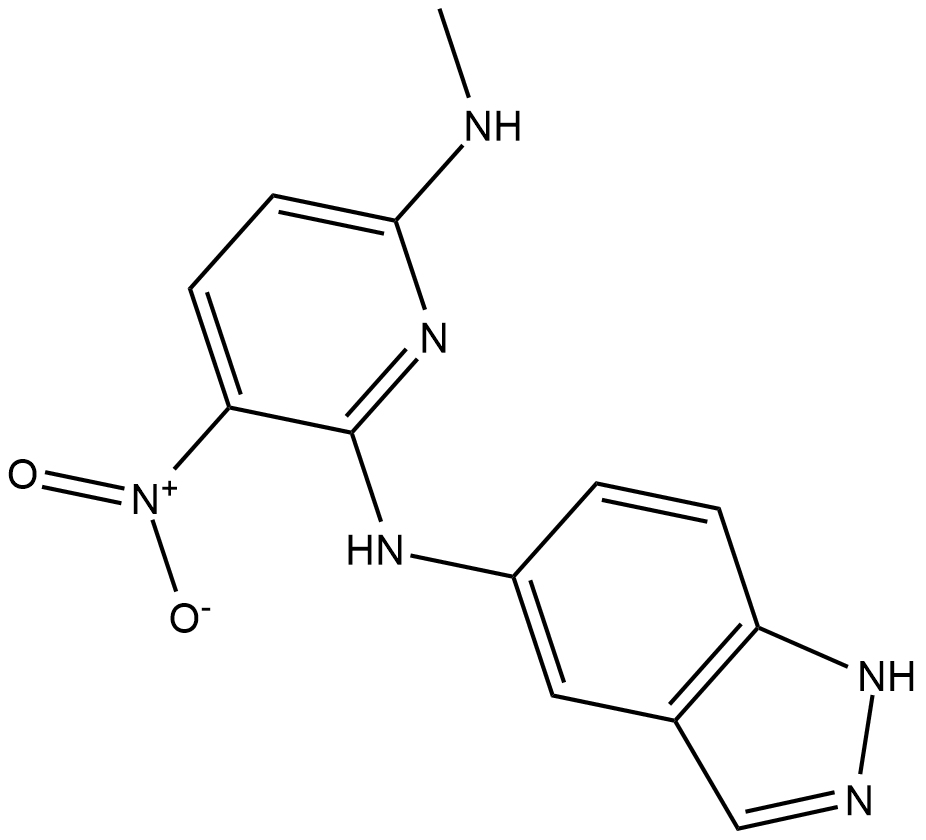
GC19215
KRIBB11
KRIBB11 is an inhibitor of Heat shock factor 1 (HSF1), with IC50 of 1.2 uM.
-
GC17346
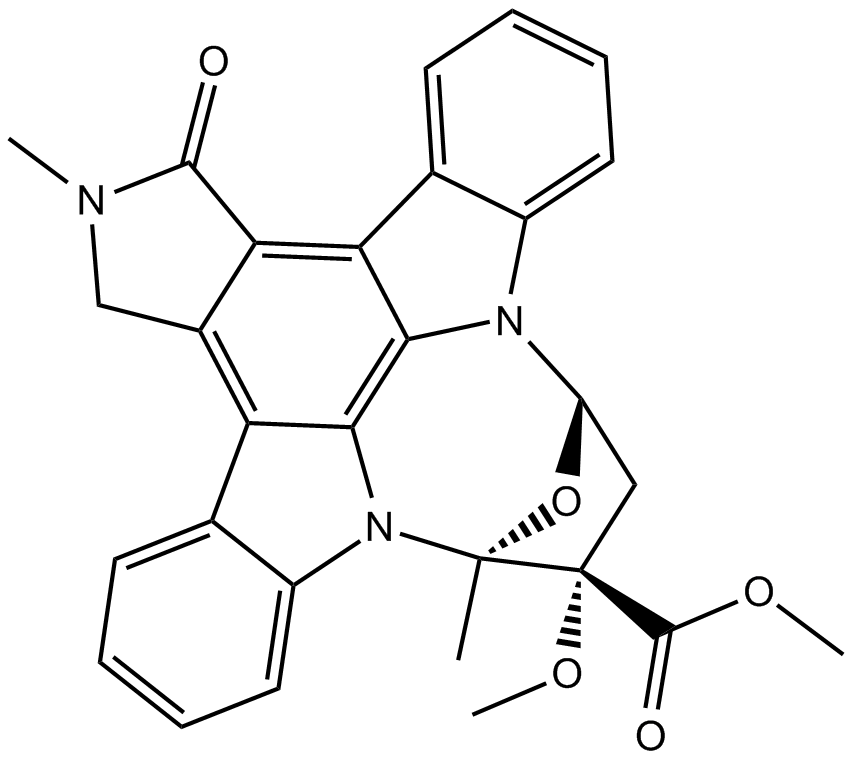
KT 5823
A protein kinase G inhibitor
-
GC14592
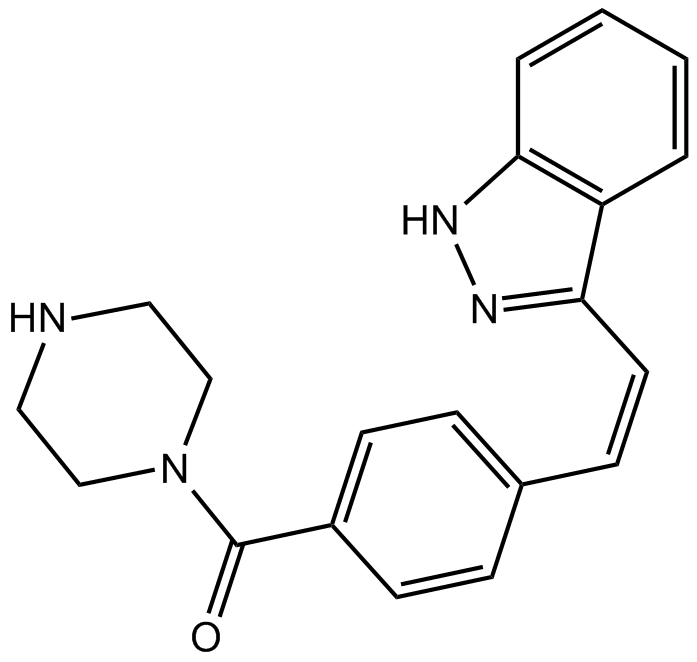
KW 2449
A multi-kinase inhibitor
-
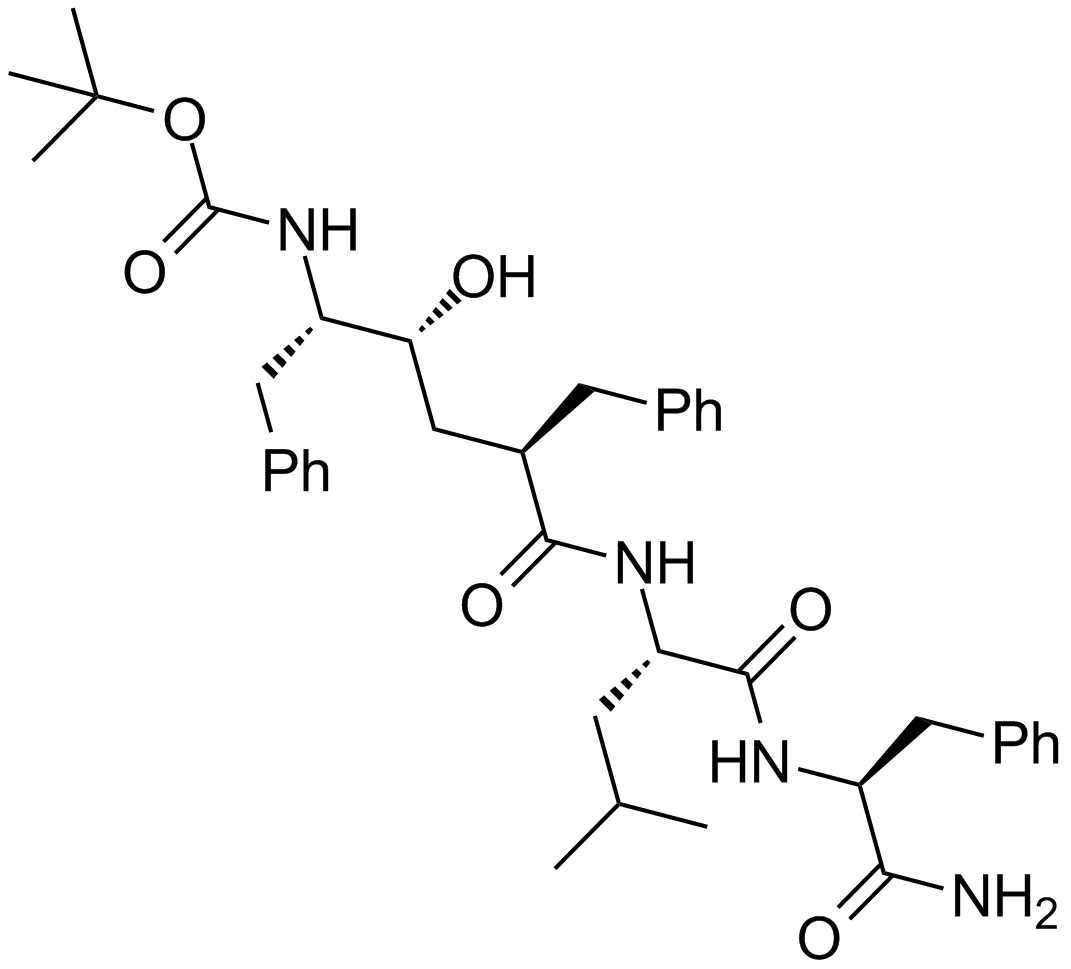
GC15364
L-685,458
A γ-secretase inhibitor
-
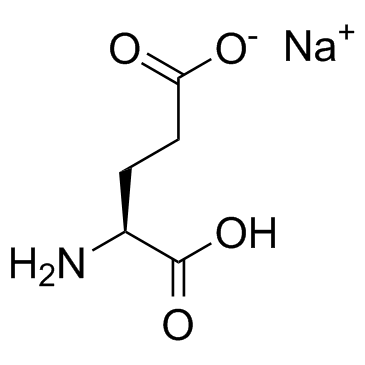
GC30788
L-Glutamic acid monosodium salt (Monosodium glutamate)
L-Glutamic acid monosodium salt (Monosodium glutamate) acts as an excitatory transmitter and an agonist at all subtypes of glutamate receptors (metabotropic, kainate, NMDA, and AMPA).
-
GC30266
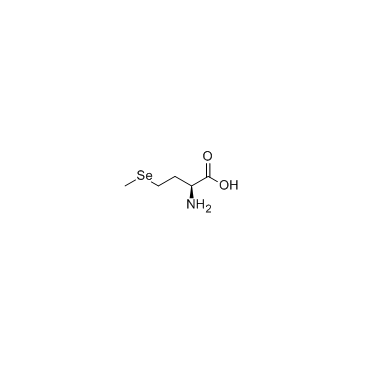
L-SelenoMethionine
A naturally occurring amino acid with antioxidant properties
-
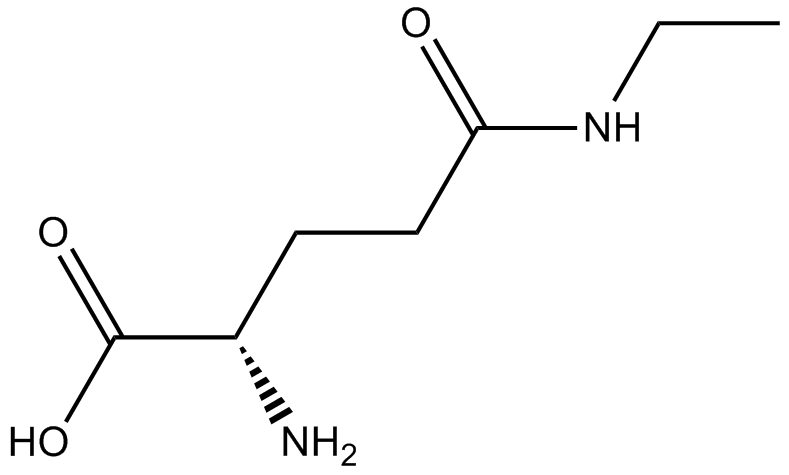
GN10456
L-Theanine
-
GC15550
Lacidipine
L-type calcium channel blocker
-
GC10474
Laquinimod (ABR-215062)
Laquinimod (ABR-215062) (ABR-215062), an orally available carboxamide derivative, is a potent immunomodulator which prevents neurodegeneration and inflammation in the central nervous system.
-
GC36424
Larotaxel
Larotaxel (XRP9881) is a taxane analogue with preclinical activity against taxane-resistant breast cancer. Larotaxel (XRP9881) exerts its cytotoxic effect by promoting tubulin assembly and stabilizing microtubules, ultimately leading to cell death by apoptosis. It presents the ability to cross the blood brain barrier and has a much lower affinity for P-glycoprotein 1 than Docetaxel.
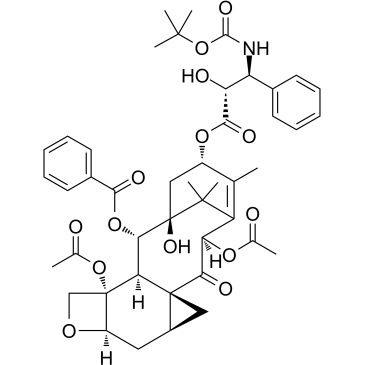
-
GC11784
LCZ696
LCZ696 (LCZ696), comprised Valsartan and Sacubitril (AHU377) in 1:1 molar ratio, is a first-in-class, orally bioavailable, and dual-acting angiotensin receptor-neprilysin (ARN) inhibitor for hypertension and heart failure.
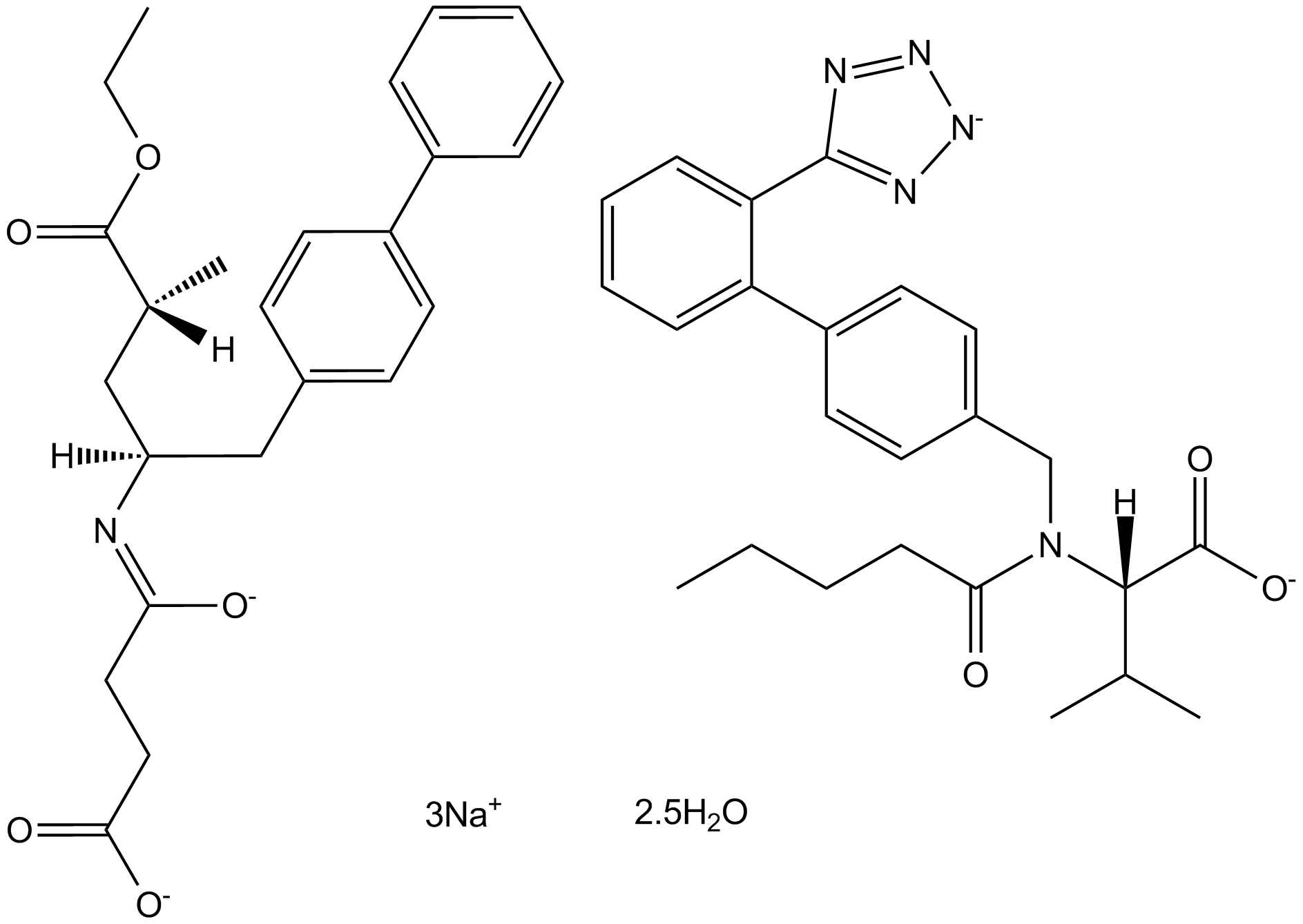
-
GC17067
LDC000067
CDK9 inhibitor, novel and highly specific
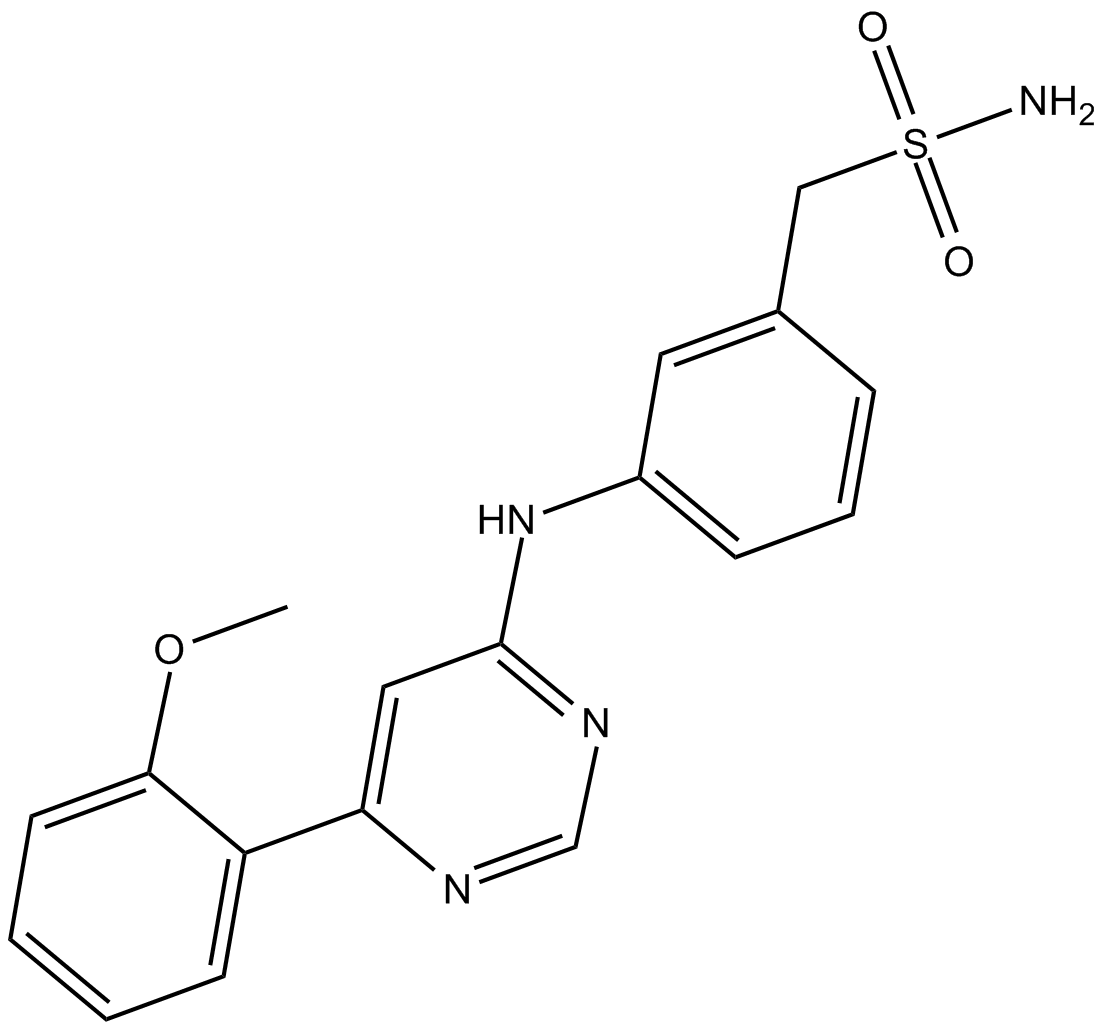
-
GC10510
LDN 57444
An inhibitor of UCH-L1
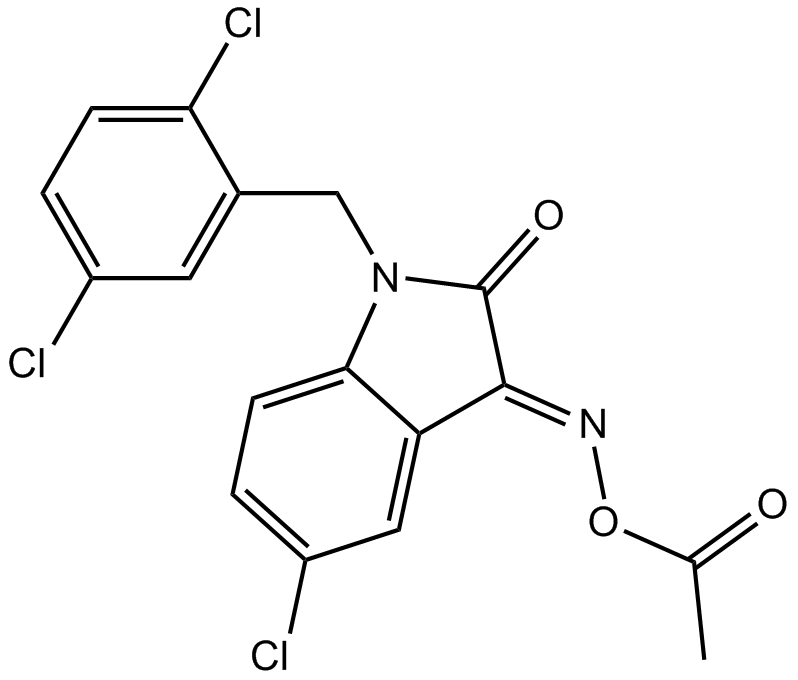
-
GC36434
Leachianone A
Leachianone A, isolated from Radix Sophorae, has anti-malarial, anti-inflammatory, and cytotoxic potent.
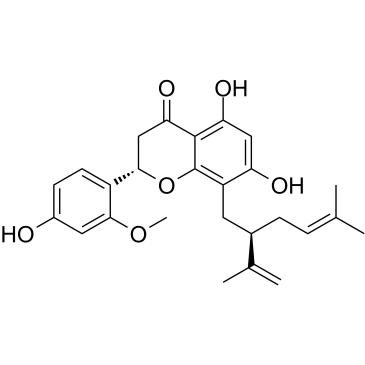
-
GC14976
Lenalidomide (CC-5013)
An analog of thalidomide
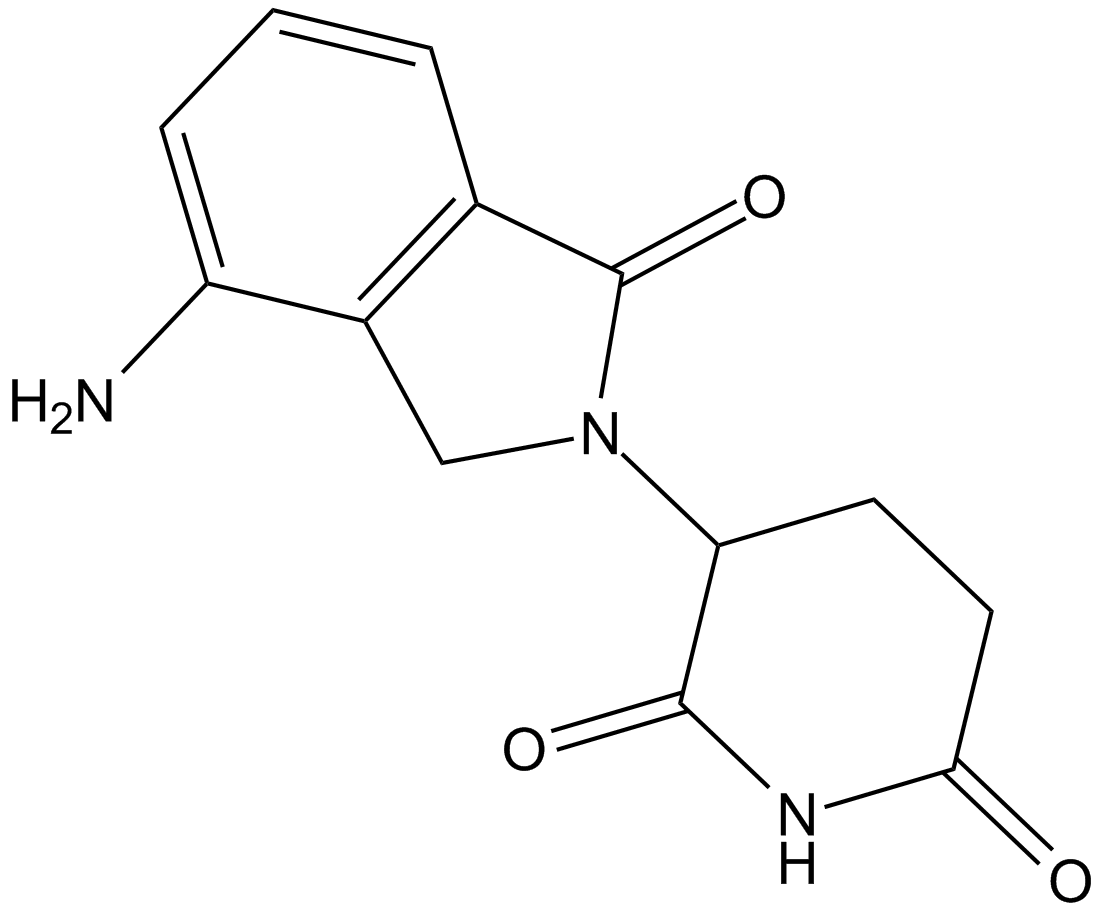
-
GC38196
Licofelone
A dual inhibitor of COX1/COX2 and 5LO
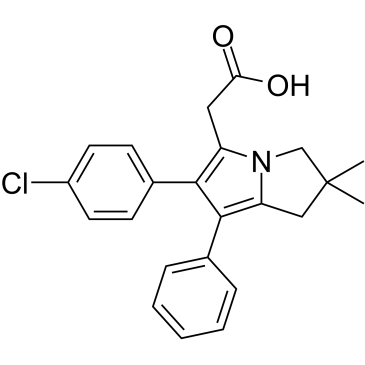
-
GC36456
Licoricidin
Licoricidin (LCD) is isolated from Glycyrrhiza uralensis Fisch, possesses anti-cancer activities.
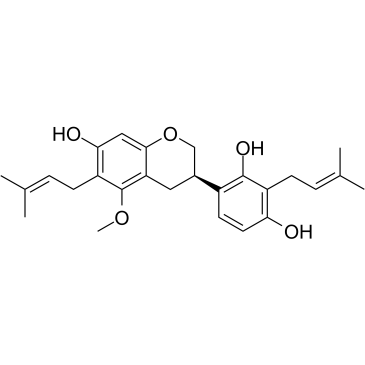
-
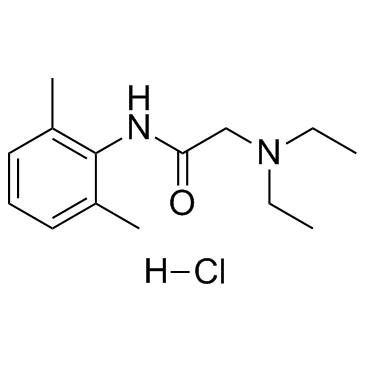
GC33831
Lidocaine hydrochloride (Lignocaine hydrochloride)
Lidocaine hydrochloride (Lignocaine hydrochloride) (Lignocaine hydrochloride) inhibits sodium channels involving complex voltage and using dependence.
-
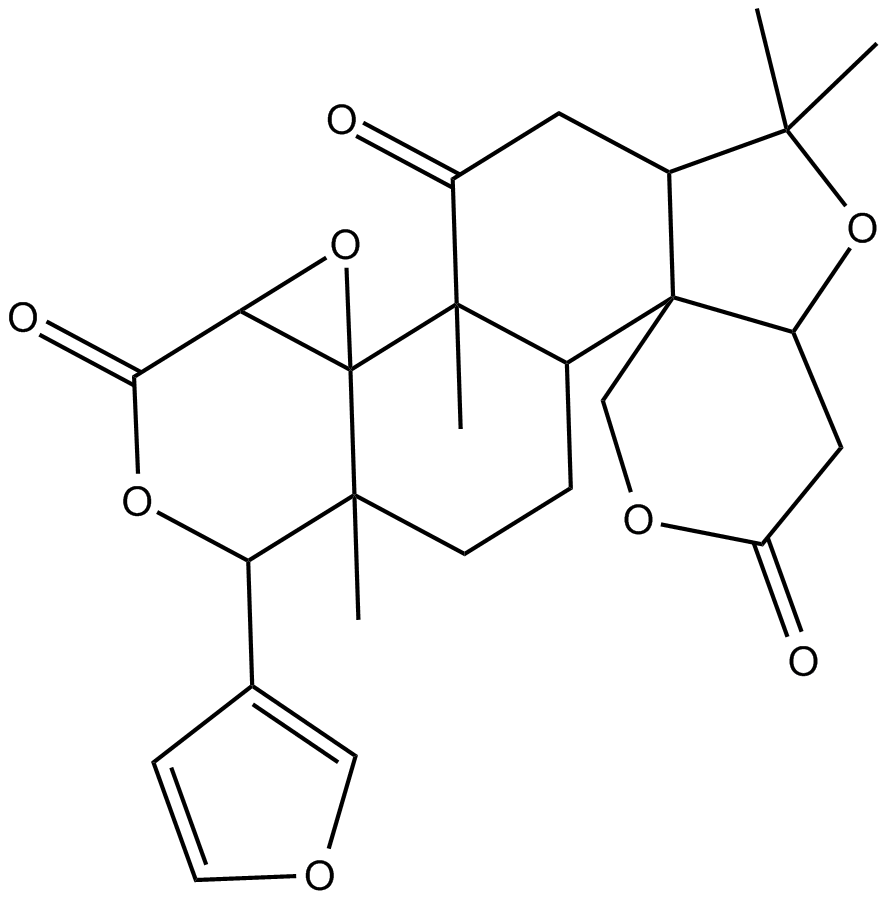
GC16589
Limonin
A limonoid
-
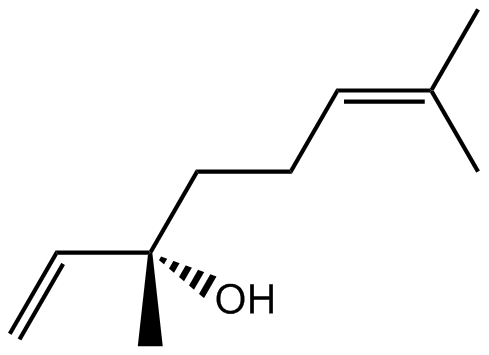
GN10122
Linalool
-
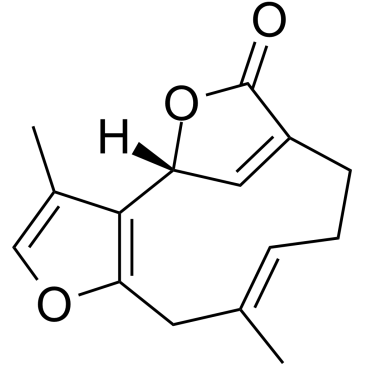
GC38086
Linderalactone
-
GC17958
Linifanib (ABT-869)
Linifanib (ABT-869) (ABT-869) is a potent and orally active multi-target inhibitor of VEGFR and PDGFR family with IC50s of 4, 3, 66, and 4 nM for KDR, FLT1, PDGFRβ, and FLT3, respectively. Linifanib (ABT-869) shows prominent antitumor activity. Linifanib (ABT-869) has much less activity against unrelated RTKs, soluble tyrosine kinases, or serine/threonine kinases. Linifanib (ABT-869) is a specific miR-10b inhibitor that blocks miR-10b biogenesis.
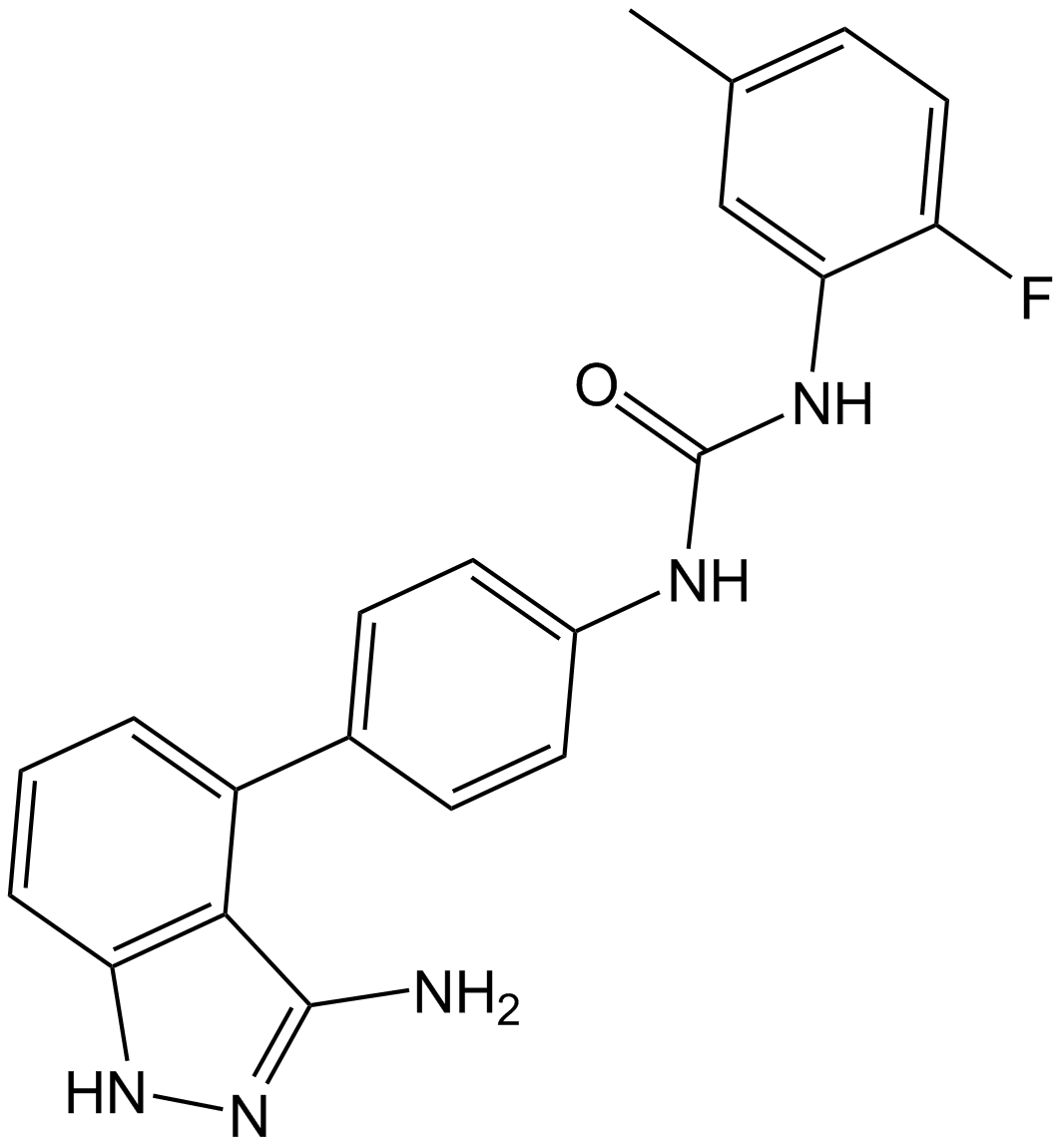
-
GC17057
Lithocholic Acid
Activator of vitamin D receptor,PXR and FXR
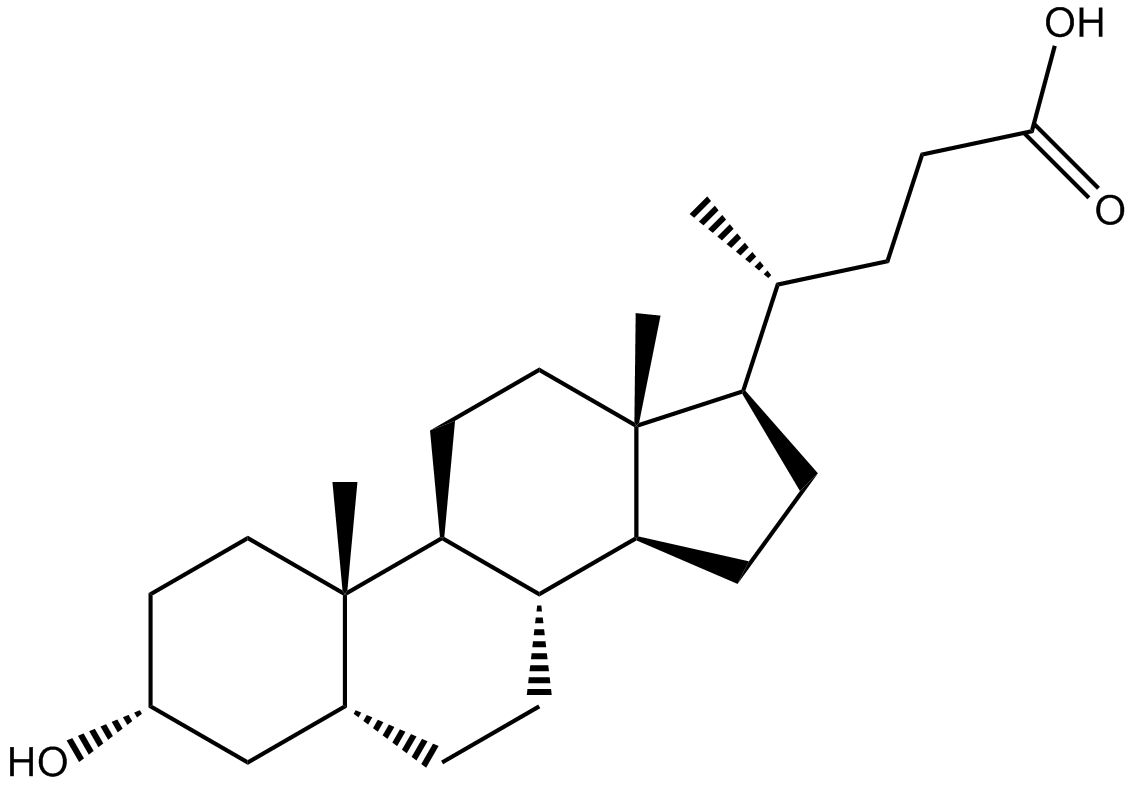
-
GC16039
LJH685
RSK (p90 ribosomal S6 kinase) inhibitor
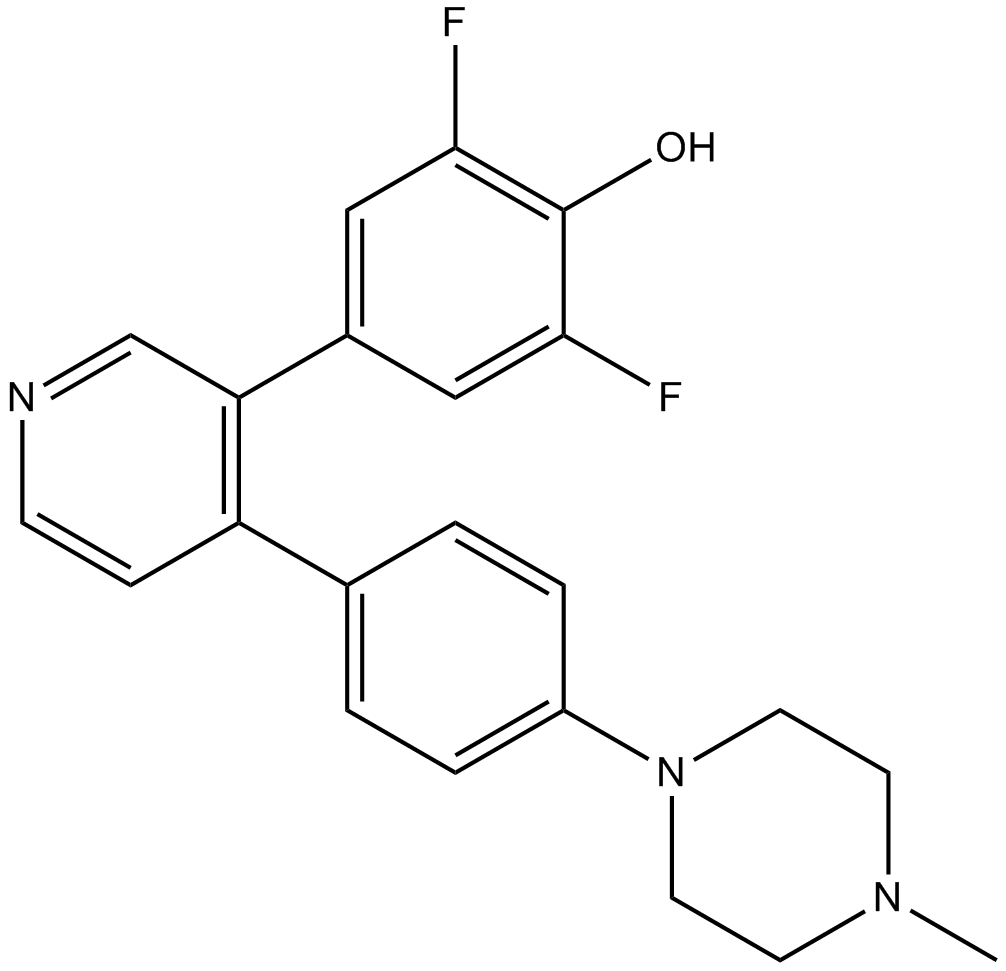
-
GN10405
Loganin
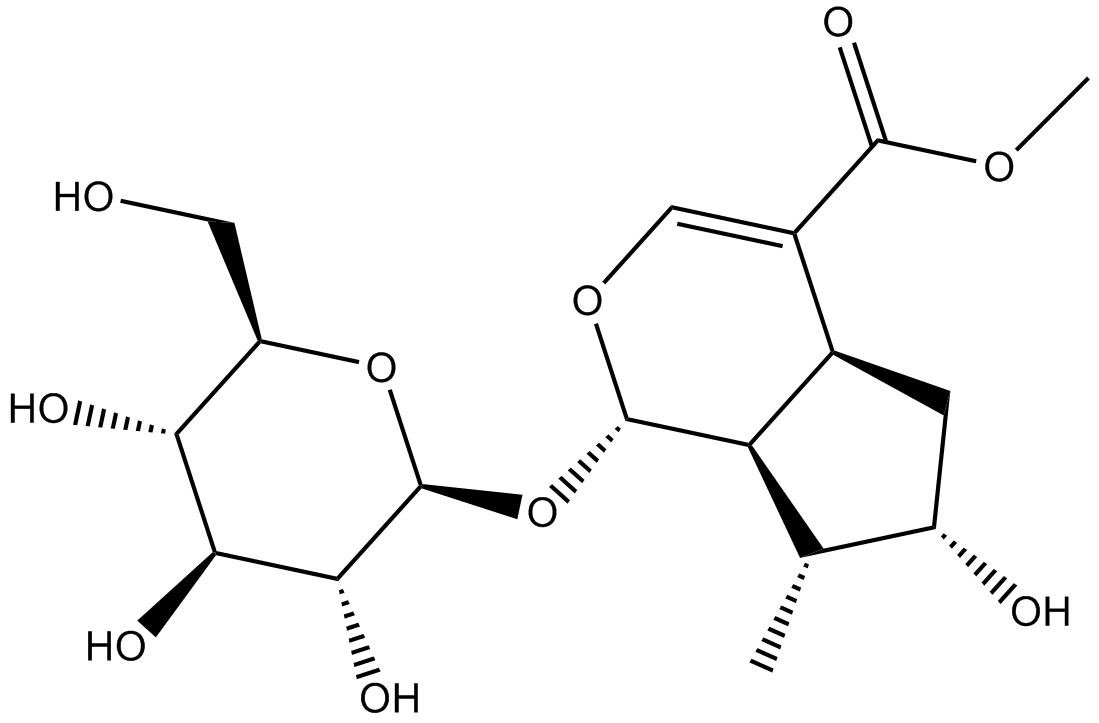
-
GC18404
Lometrexol
Glycinamide ribonucleotide formyltransferase (GART) is a folate-dependent enzyme required for de novo purine synthesis.
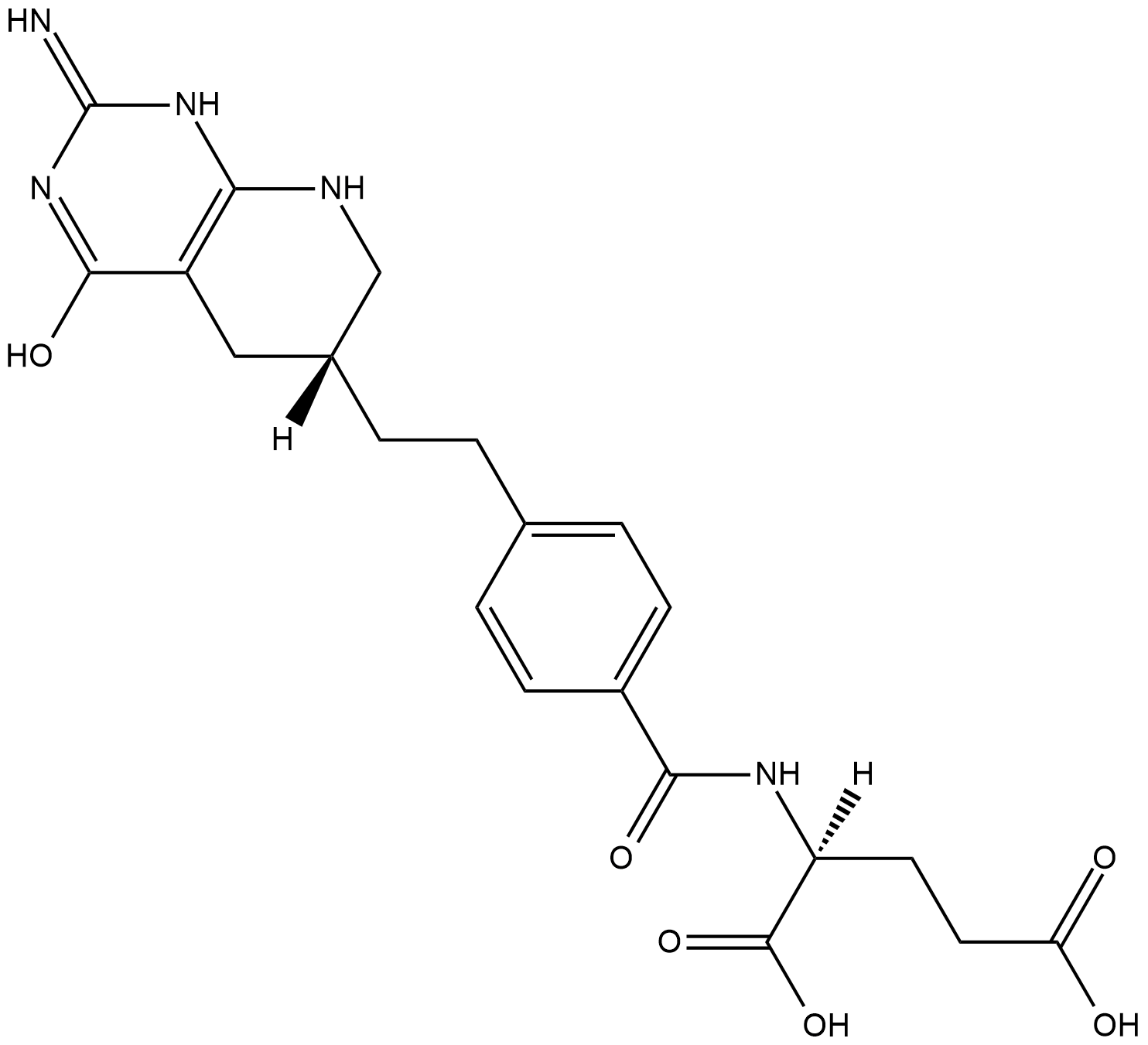
-
GC17865
Lomustine
Antineoplastic drug
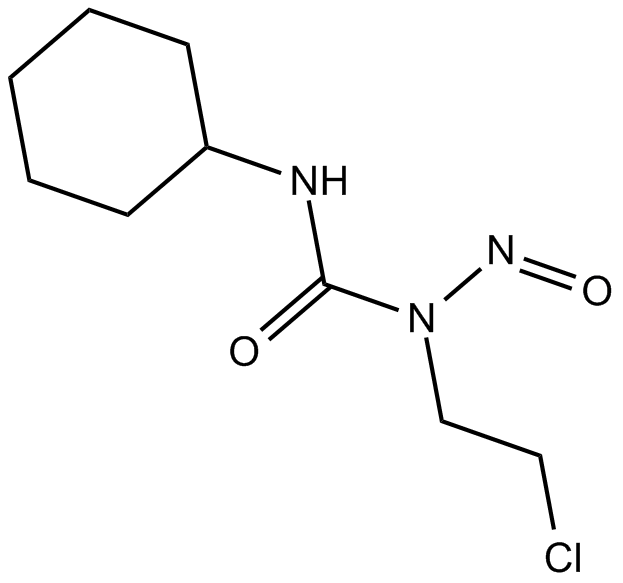
-
GC11532
Lonidamine
hexokinase inactivator
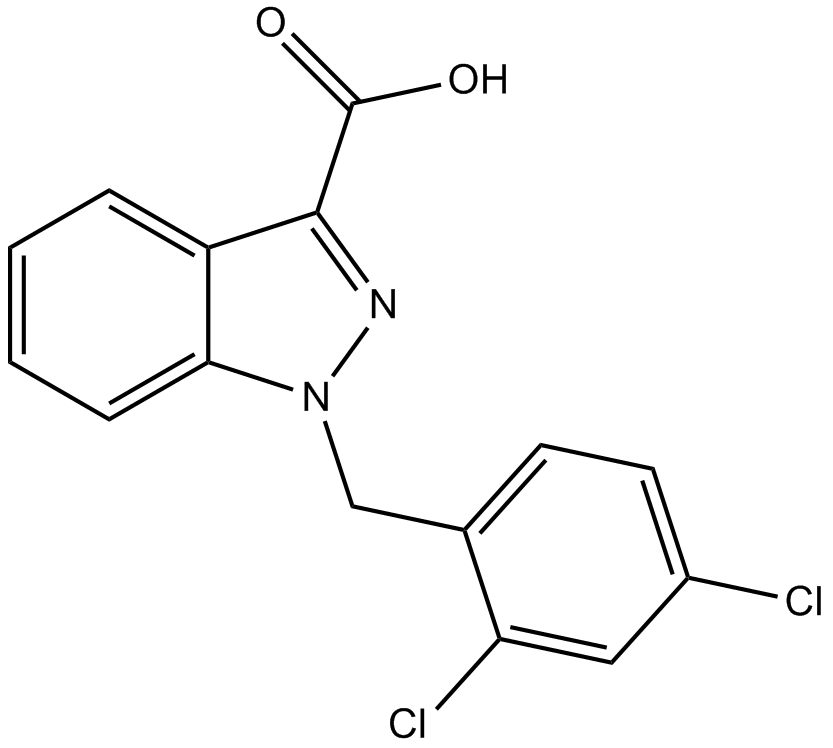
-
GC15282
LOXO-101 (Larotrectinib) sulfate
LOXO-101 (Larotrectinib) sulfate (LOXO-101 sulfate; ARRY-470 sulfate) is an ATP-competitive oral, selective inhibitor of the tropomyosin-related kinase (TRK) family receptors, with low nanomolar 50% inhibitory concentrations against all three isoforms (TRKA, B, and C).
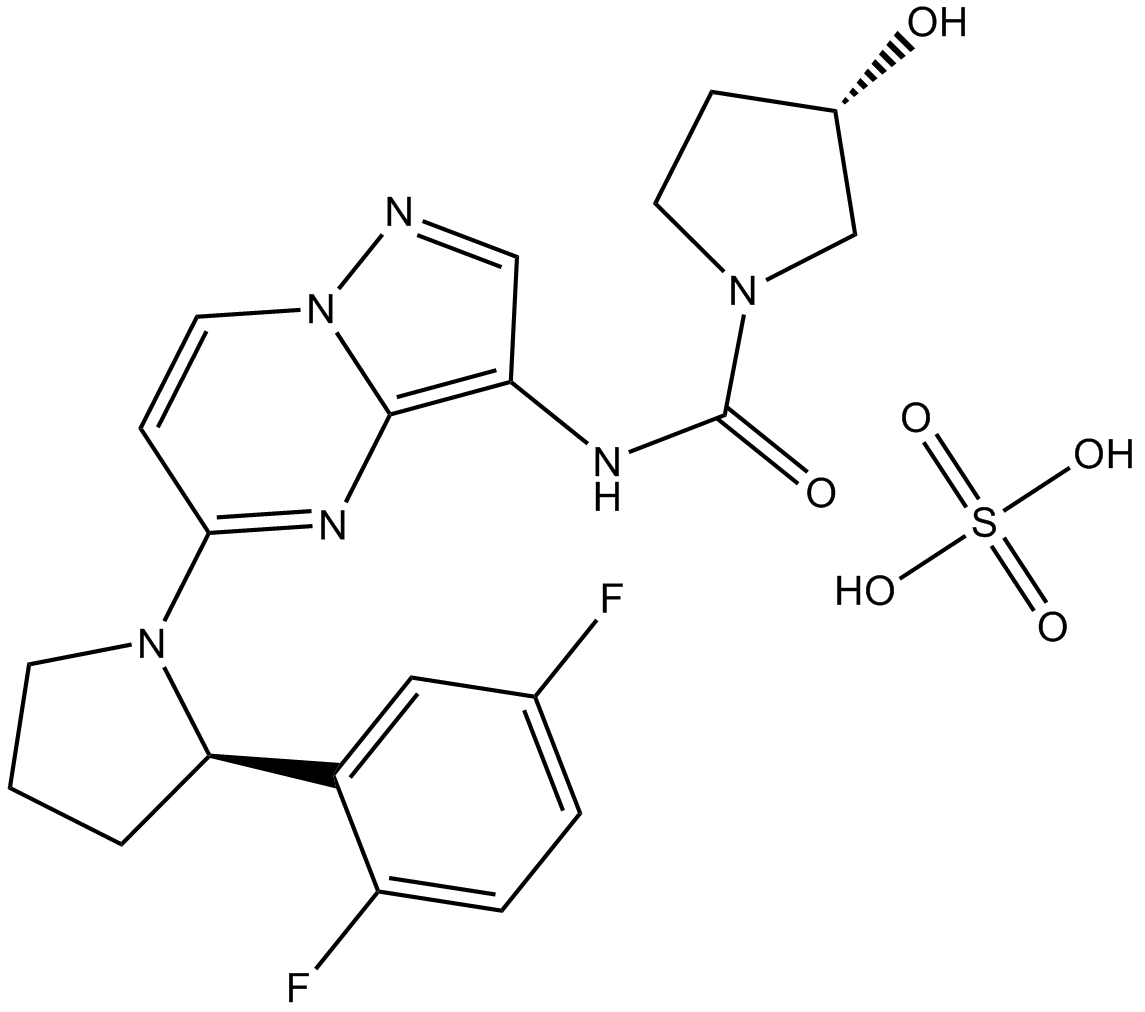
-
GC10809
LRRK2-IN-1
A selective LRRK2 inhibitor

-
GC36488
Lucidenic acid B
Lucidenic acid B is a natural compound isolated from Ganoderma lucidum, induces apoptosis of cancer cells, and causes the activation of caspase-9 and caspase-3, and cleavage of PARP. Lucidenic acid B does not affect the cell cycle profile, or the number of necrotic cells.
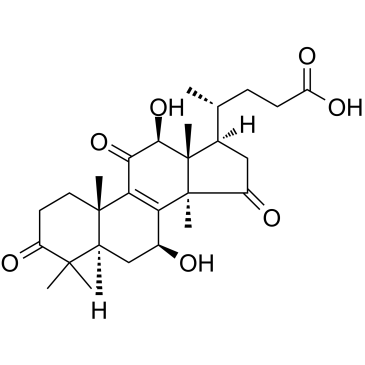
-
GN10142
Lupeol
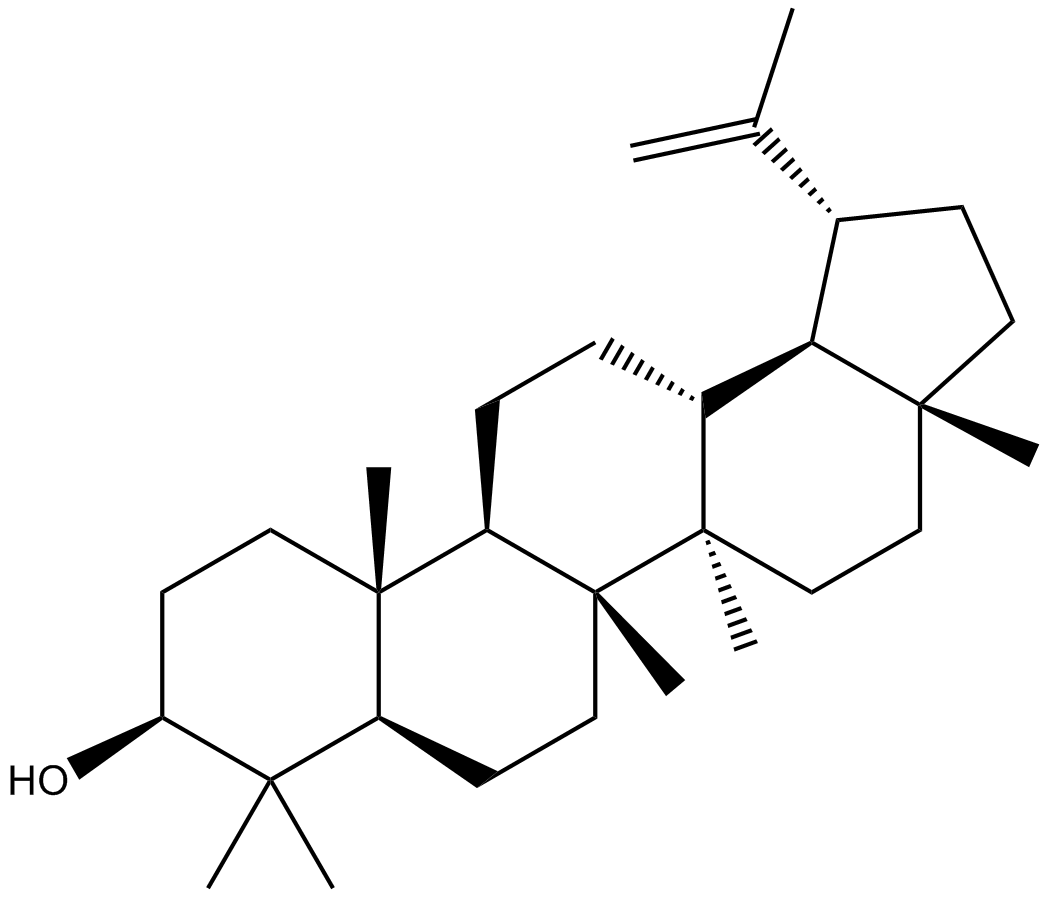
-
GN10370
Luteolin
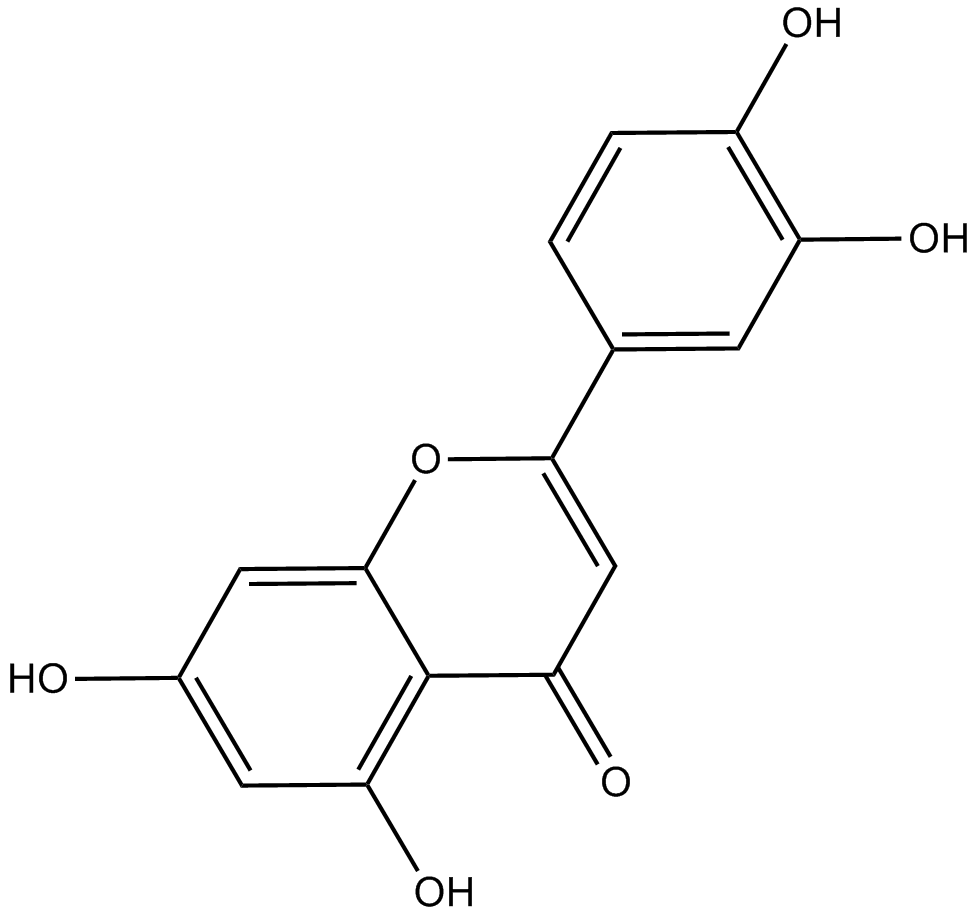
-
GC32724
LW6 (HIF-1α inhibitor)
LW6 (HIF-1α inhibitor) (HIF-1α inhibitor) is a novel HIF-1 inhibitor with an IC50 of 4.4 μM. LW6 (HIF-1α inhibitor) decreases HIF-1α protein expression without affecting HIF-1β expression.
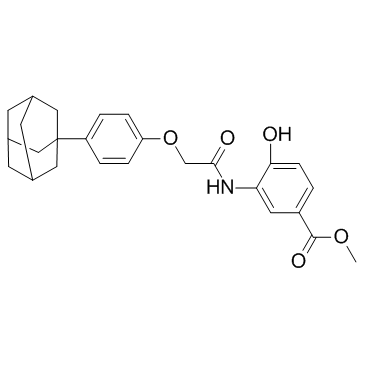
-
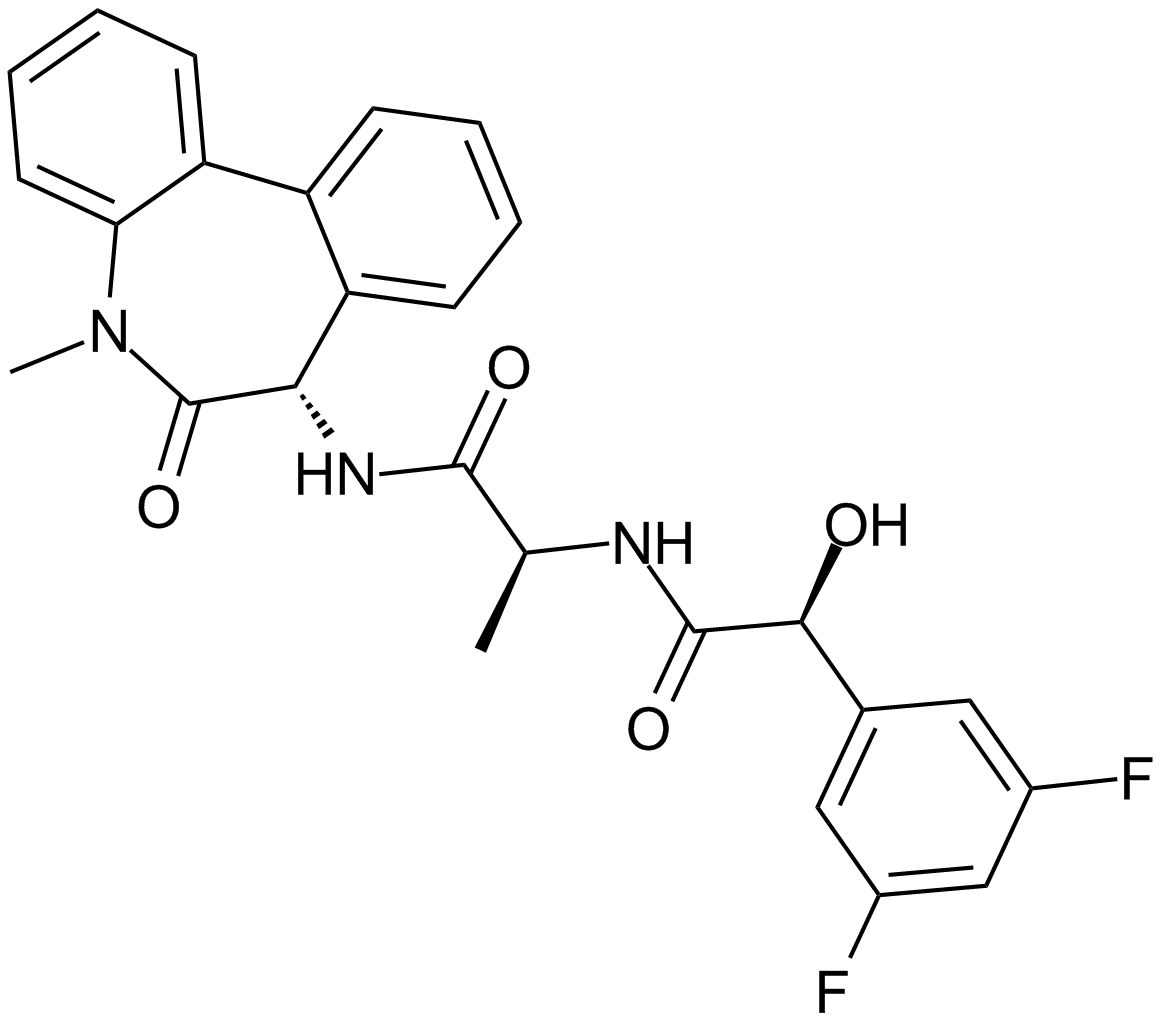
GC12191
LY-411575
A γ-secretase inhibitor
-
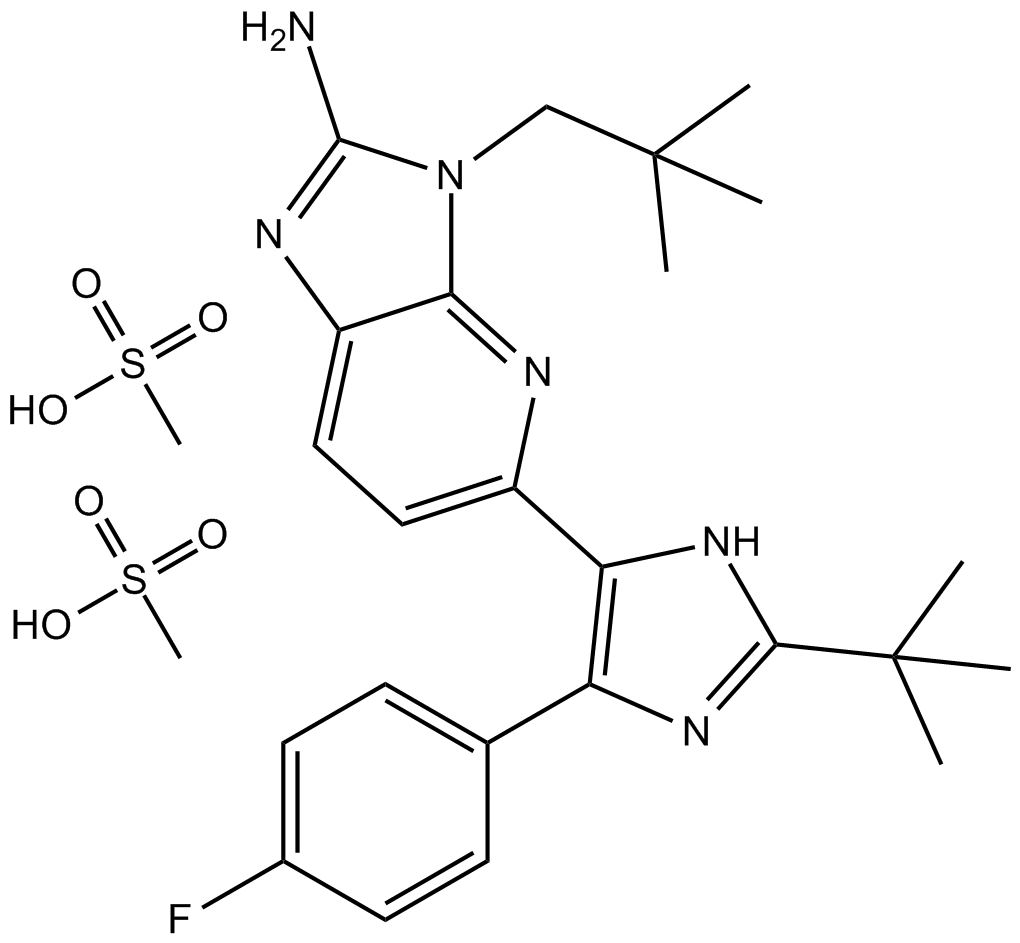
GC16835
LY2228820
LY2228820 (LY2228820 dimesylate) is a selective, ATP-competitive inhibitor of p38 MAPK α/β with IC50s of 5.3 and 3.2 nM, respectively.
-
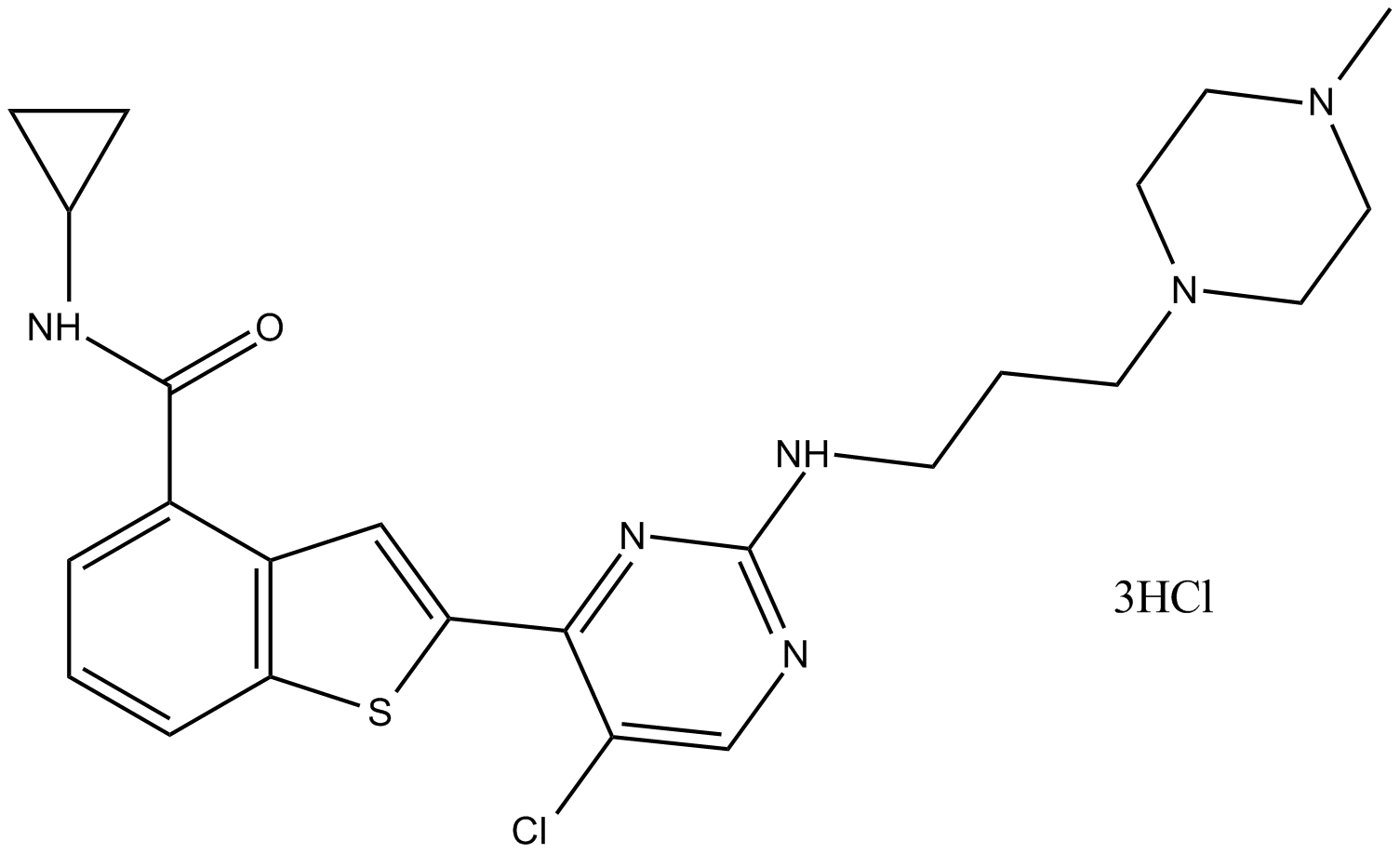
GC10093
LY2409881
LY2409881 is a selective IκB kinase β (IKK2) inhibitor with an IC50 of 30 nM.
-
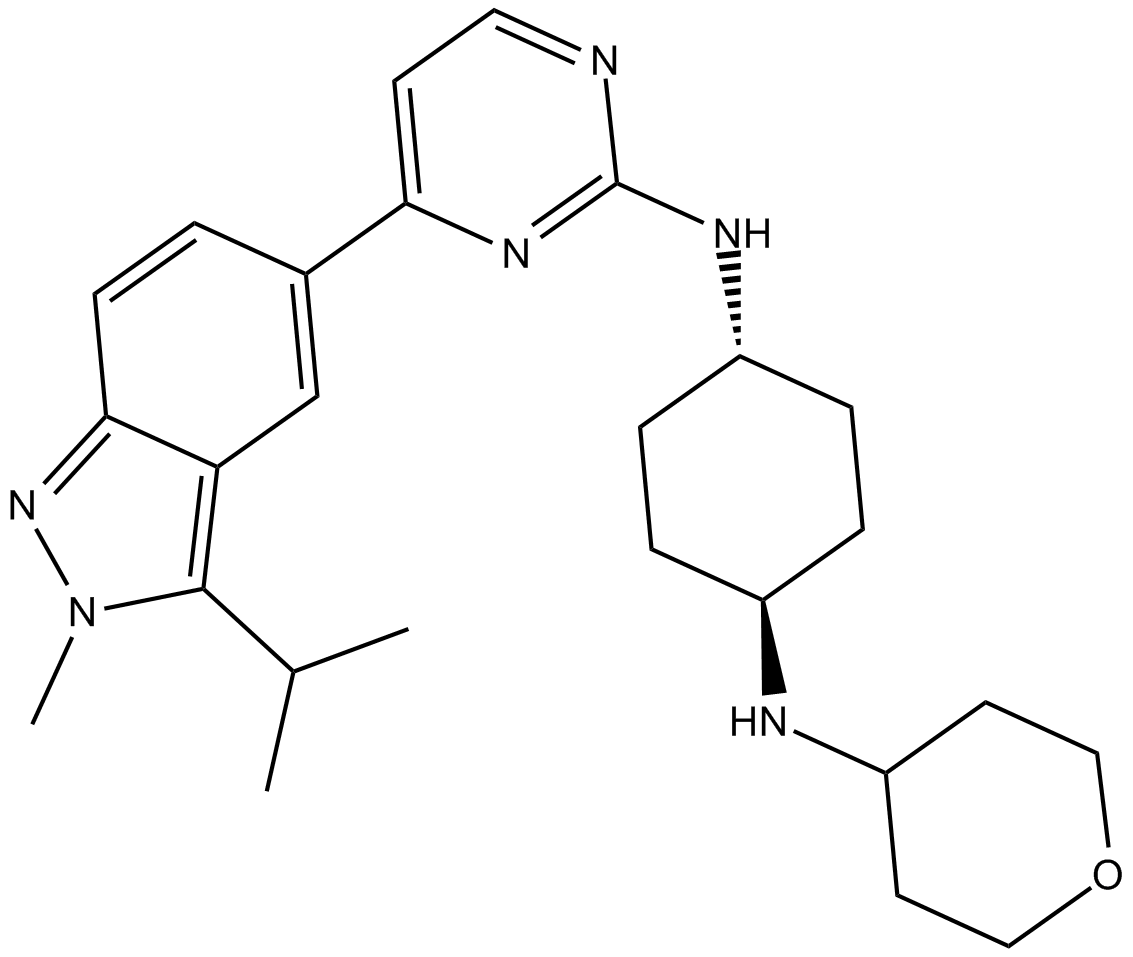
GC11971
LY2857785
CDK9 inhibitor
-
GC36516
Lycopodine
Lycopodine, a pharmacologically important bioactive component derived from Lycopodium clavatumspores, triggers apoptosis by modulating 5-lipoxygenase, and depolarizing mitochondrial membrane potential in refractory prostate cancer cells without modulating p53 activity. Lycopodine inhibits proliferation of HeLa cells through induction of apoptosis via caspase-3 activation.
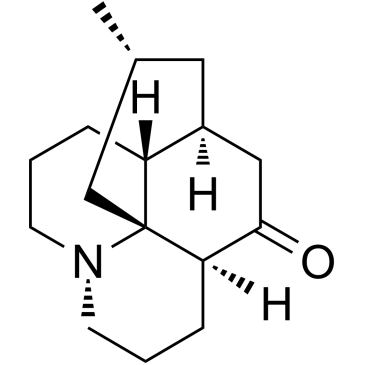
-
GC17785
Macitentan
Endothelin (ET)(A) and ET(B) receptor antagonist
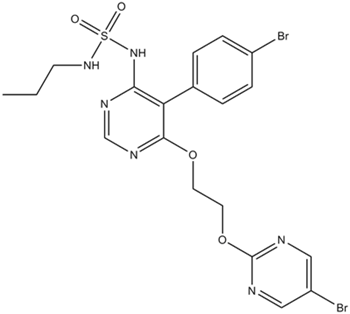
-
GC45723
Macitentan-d4
An internal standard for the quantification of macitentan
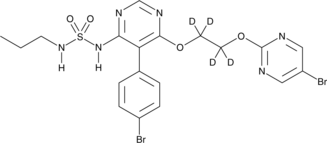
-
GN10173
Madecassoside
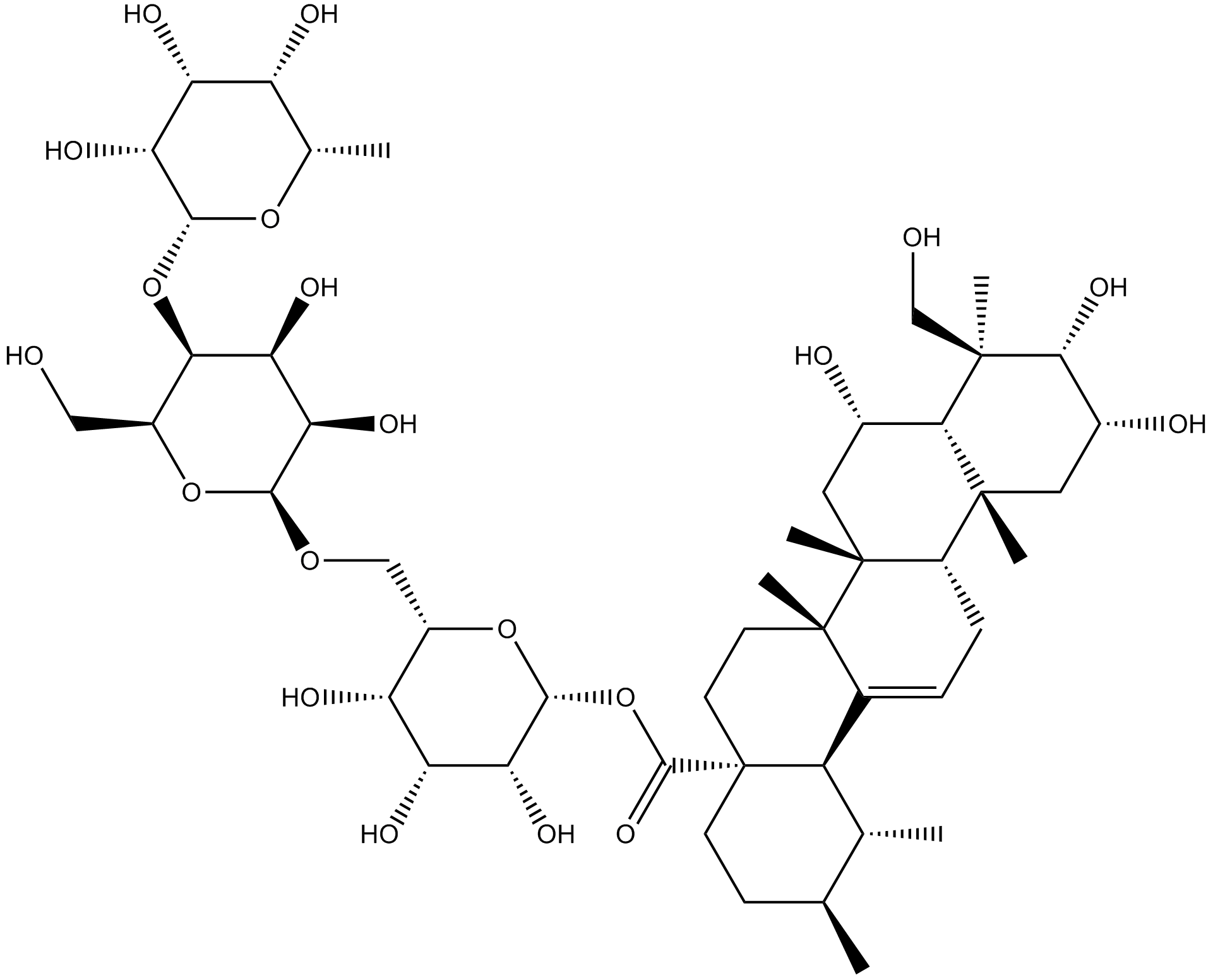
-
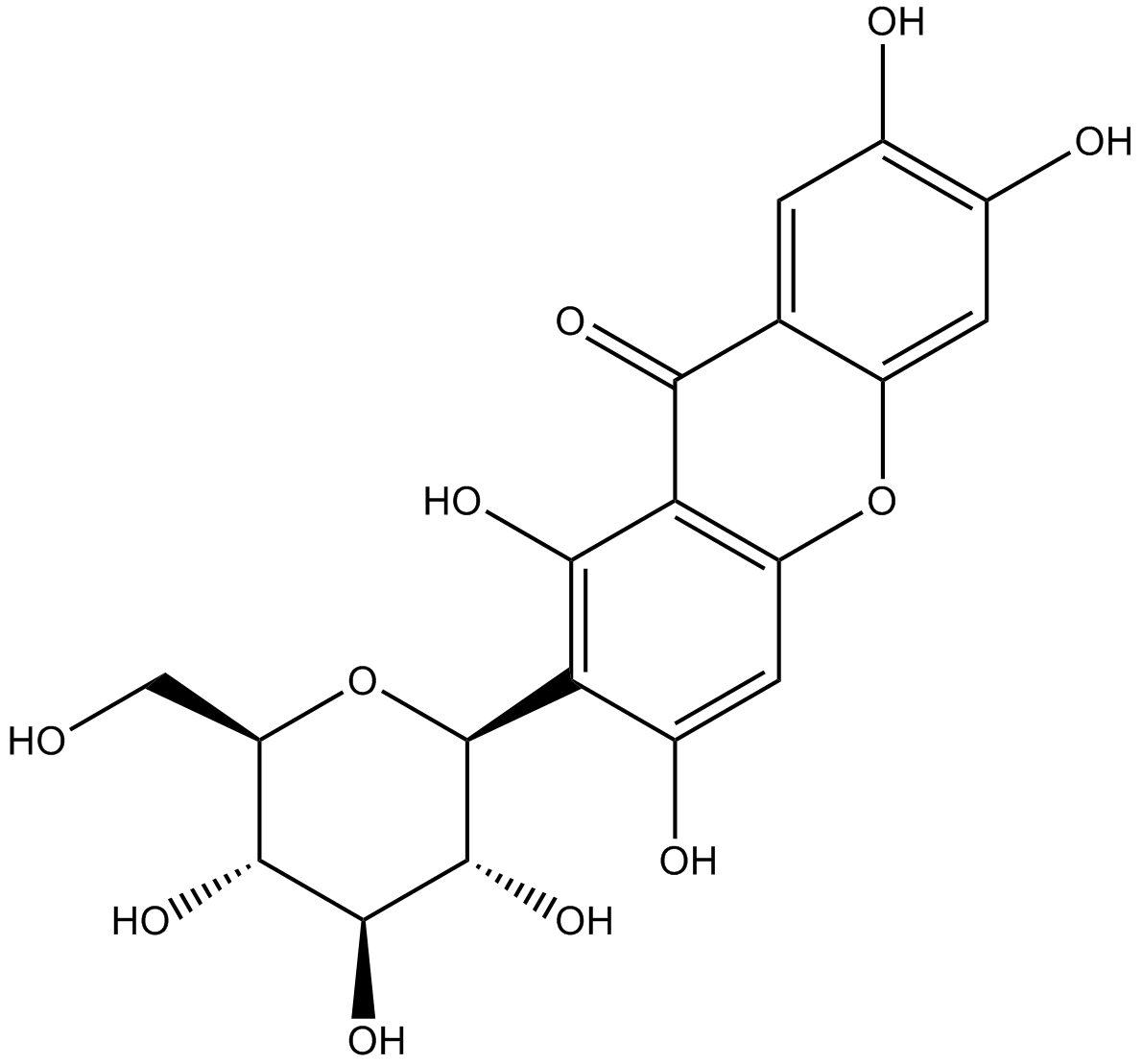
GN10491
Mangiferin
-
GC10265
Manumycin A
farnesyltransferase inhibitor
-
GC36542
MARK4 inhibitor 1
MARK4 inhibitor 1 is a potent microtubule affinity-regulating kinase 4 (MARK4) inhibitor, with an IC50 of 1.54 μM. MARK4 inhibitor 1 inhibits cancer cell proliferation, metastasis and induces apoptosis.
-
GC36546
Masitinib mesylate
Masitinib mesylate (AB-1010 mesylate) is a potent, orally bioavailable, and selective inhibitor of c-Kit (IC50=200 nM for human recombinant c-Kit). It also inhibits PDGFRα/β (IC50s=540/800 nM), Lyn (IC50= 510 nM for LynB), Lck, and, to a lesser extent, FGFR3 and FAK. Masitinib mesylate (AB-1010 mesylate) has anti-proliferative, pro-apoptotic activity and low toxicity.
-
GC32895
Maytansinol (Ansamitocin P-0)
Maytansinol (Ansamitocin P-0) inhibits microtubule assembly and induces microtubule disassembly in vitro.
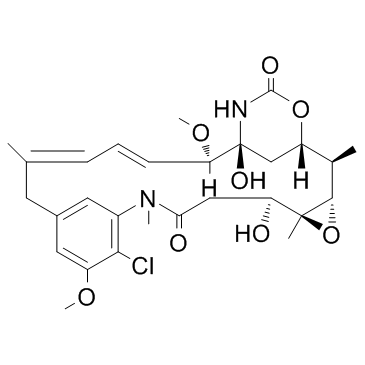
-
GC36552
MBM-17
MBM-17 is a potent NIMA-related kinase 2 (Nek2) inhibitor with an IC50 of 3 nM. It effectively inhibits the proliferation of cancer cells by inducing cell cycle arrest and apoptosis. MBM-55 shows antitumor activities, and no obvious toxicity to mice.
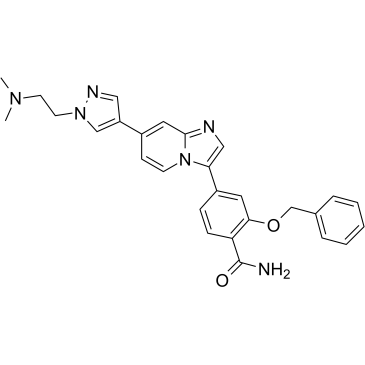
-
GC36553
MBM-55
MBM-55 is a potent NIMA-related kinase 2 (Nek2) inhibitor with an IC50 of 1 nM. MBM-55 shows a 20-fold or greater selectivity in most kinases with the exception of RSK1 (IC50=5.4 nM) and DYRK1a (IC50=6.5 nM). MBM-55 effectively inhibits the proliferation of cancer cells by inducing cell cycle arrest and apoptosis. MBM-55 shows antitumor activities, and no obvious toxicity to mice.
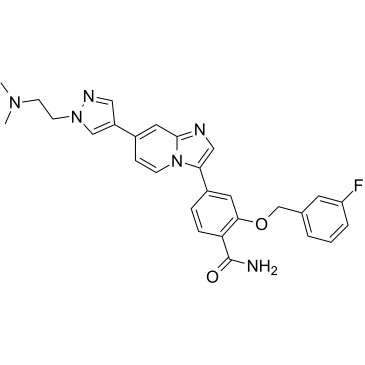
-
GC10200
Mdivi 1
A mitochondrial division inhibitor
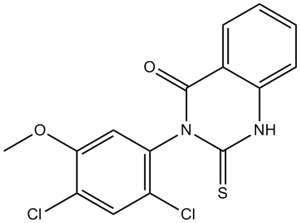
-
GC17649
Mebendazole
broad-spectrum anthelmintic that inhibits intestinal microtubule synthesis
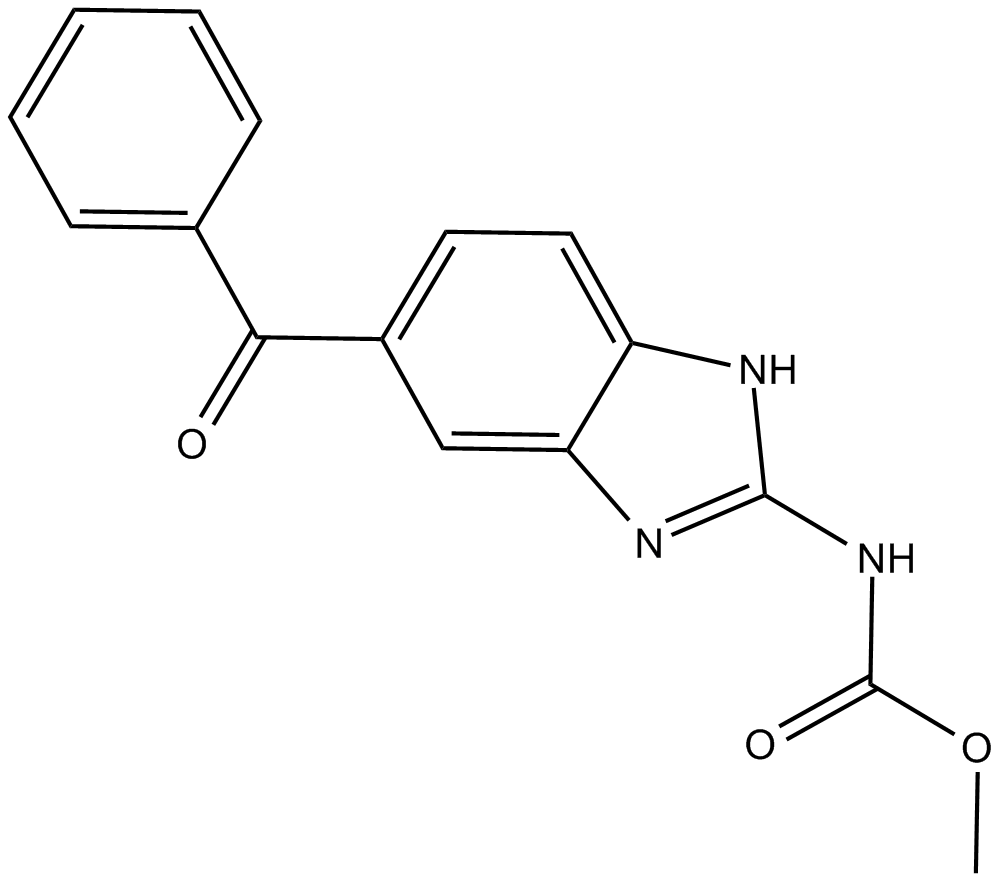
-
GC14312
Meisoindigo
Apoptosis inducer;potential agent for AML
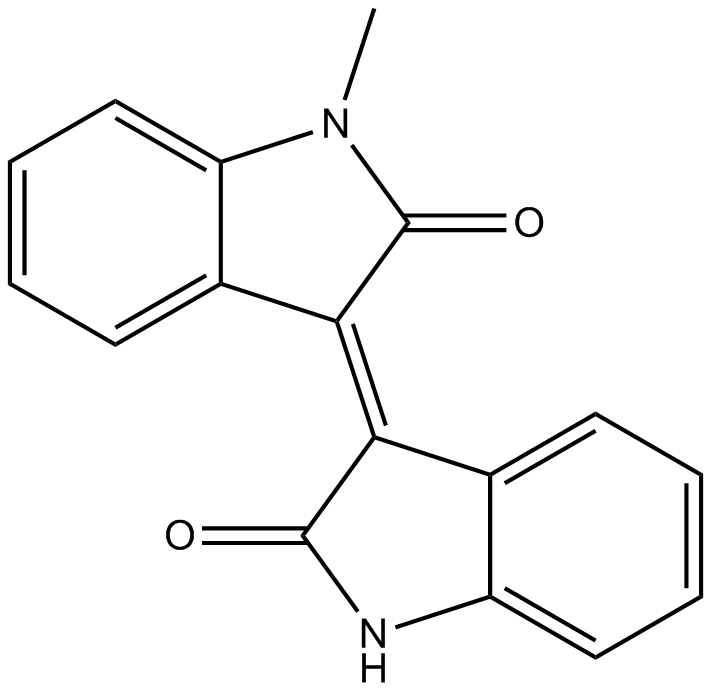
-
GC11762
Meloxicam (Mobic)
Meloxicam (Mobic) is a non-steroidal antiinflammatory agent, inhibits COX activity, with IC50s of 0.49 μM and 36.6 μM for COX-2 and COX-1, respectively.
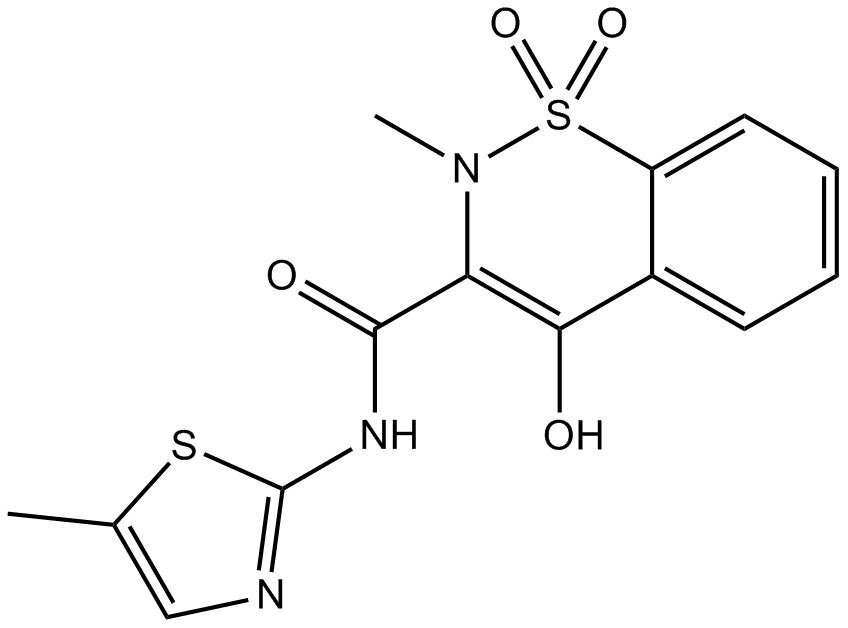
-
GC12393
Melphalan
DNA alkylating agent
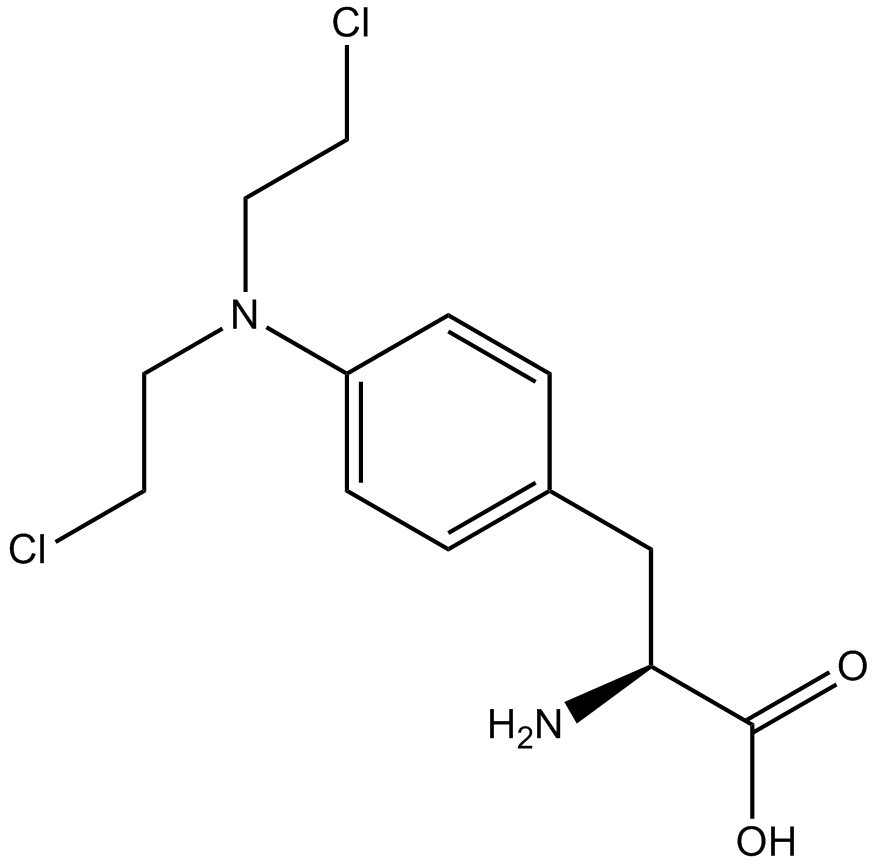
-
GC10405
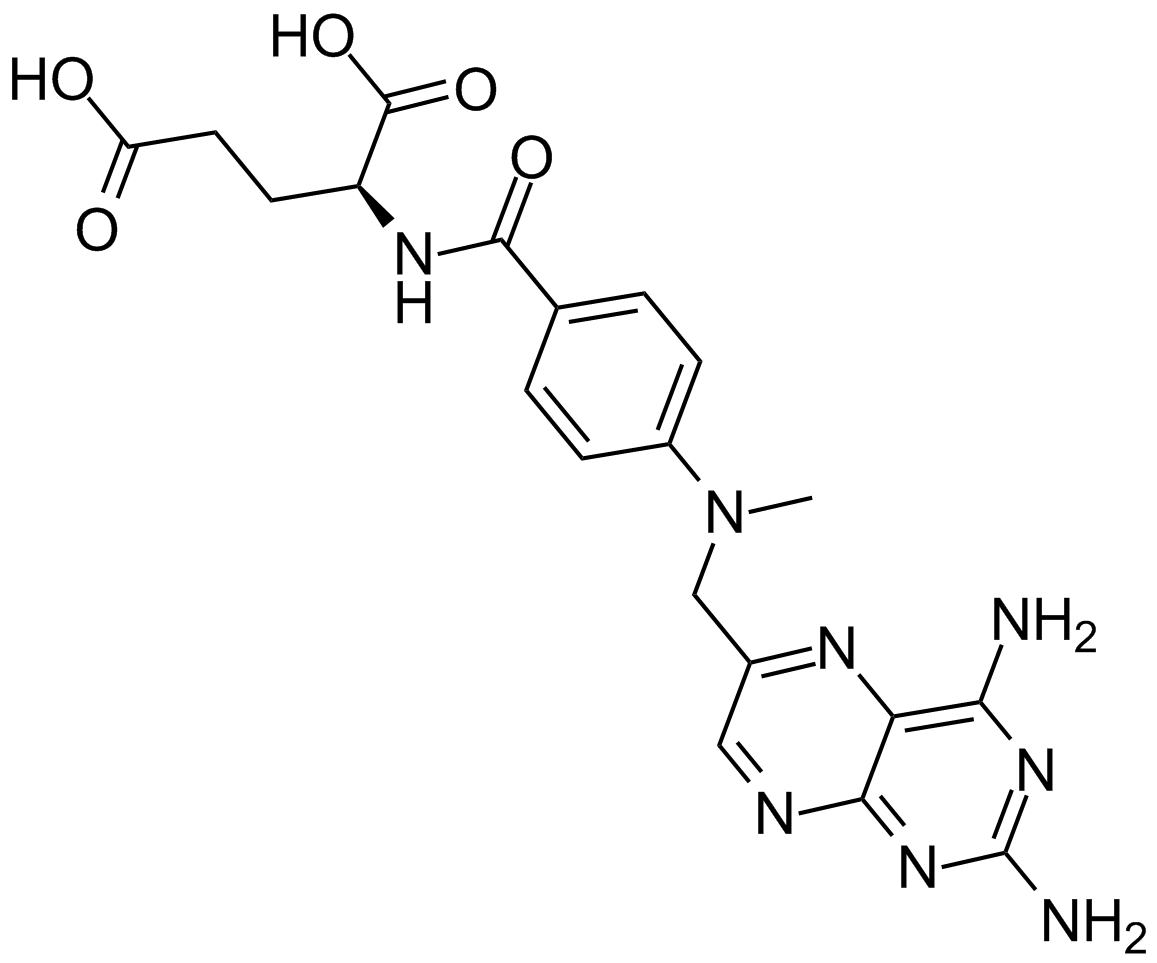
Methotrexate
Methotrexate, a folate antagonist, is a potent anti-inflammatory agent when used weekly in low concentrations, the anti-phlogistic action of which is due to increased adenosine release at inflamed sites.
-
GC31783
Methyl 3,4-dihydroxybenzoate (Protocatechuic acid methyl ester)
Methyl 3,4-dihydroxybenzoate (Protocatechuic acid methyl ester) (Protocatechuic acid methyl ester; Methyl protocatechuate) is a major metabolite of antioxidant polyphenols found in green tea.

-
GN10639
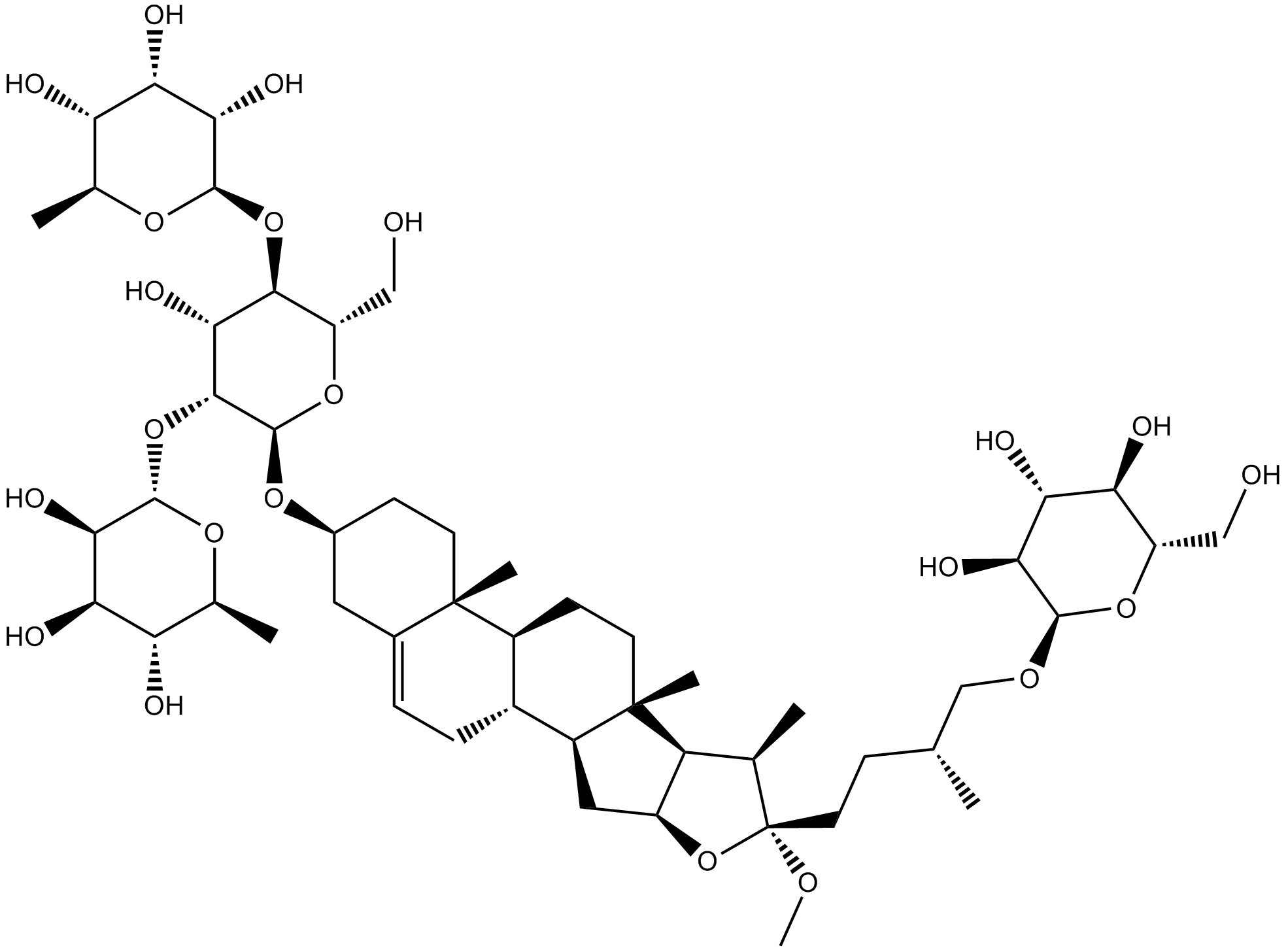
Methyl protodioscin
-
GC15775
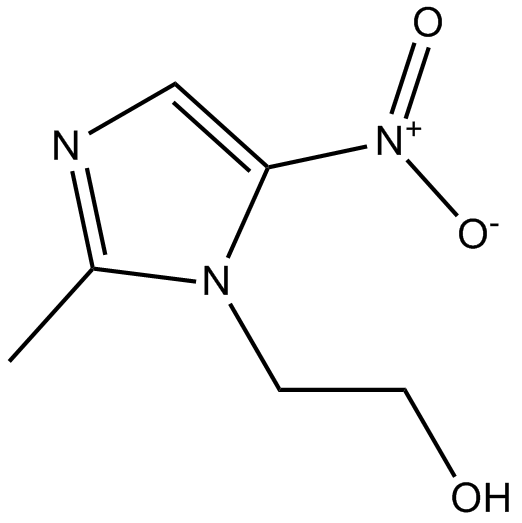
Metronidazole
nitroimidazole antibiotic
-
GC11895
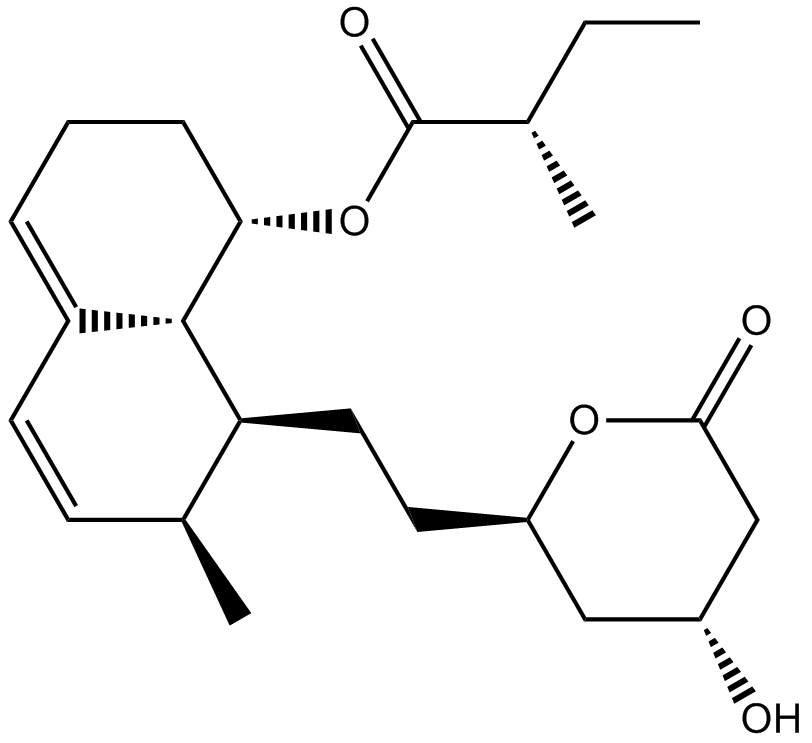
Mevastatin
HMG-CoA reductase inhibitor
-
GC11439
MG 149
HAT inhibitor
-
GC13598
MGCD-265
MGCD-265 is a potent and oral active inhibitor of c-Met and VEGFR2 tyrosine kinases, with IC50s of 29 nM and 10 nM, respectively. MGCD-265 has significant antitumor activity.
-
GC36605
MI-1061
MI-1061 is a potent, orally bioavailable, and chemically stable MDM2 (MDM2-p53 interaction) inhibitor (IC50=4.4 nM; Ki=0.16 nM). MI-1061 potently activates p53 and induces apoptosis in the SJSA-1 xenograft tumor tissue in mice. Anti-tumor activity.