HIV
HIV (human immunodeficiency virus) is a lentivirus that infect immune system. It cause immune system failure and lead to AIDS (acquired immnudeficiency syndrome).
Products for HIV
- Cat.No. Product Name Information
-
GC14269
Cobicistat (GS-9350)
An inhibitor of CYP3A
-
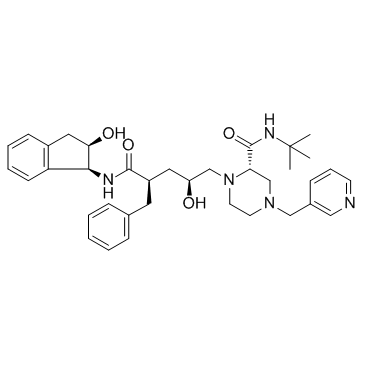
GC35731
Corydine
Corydine is a naturally occurring alkaloid which can be extracted from plants such as Croton echinocarpus leaves.
-
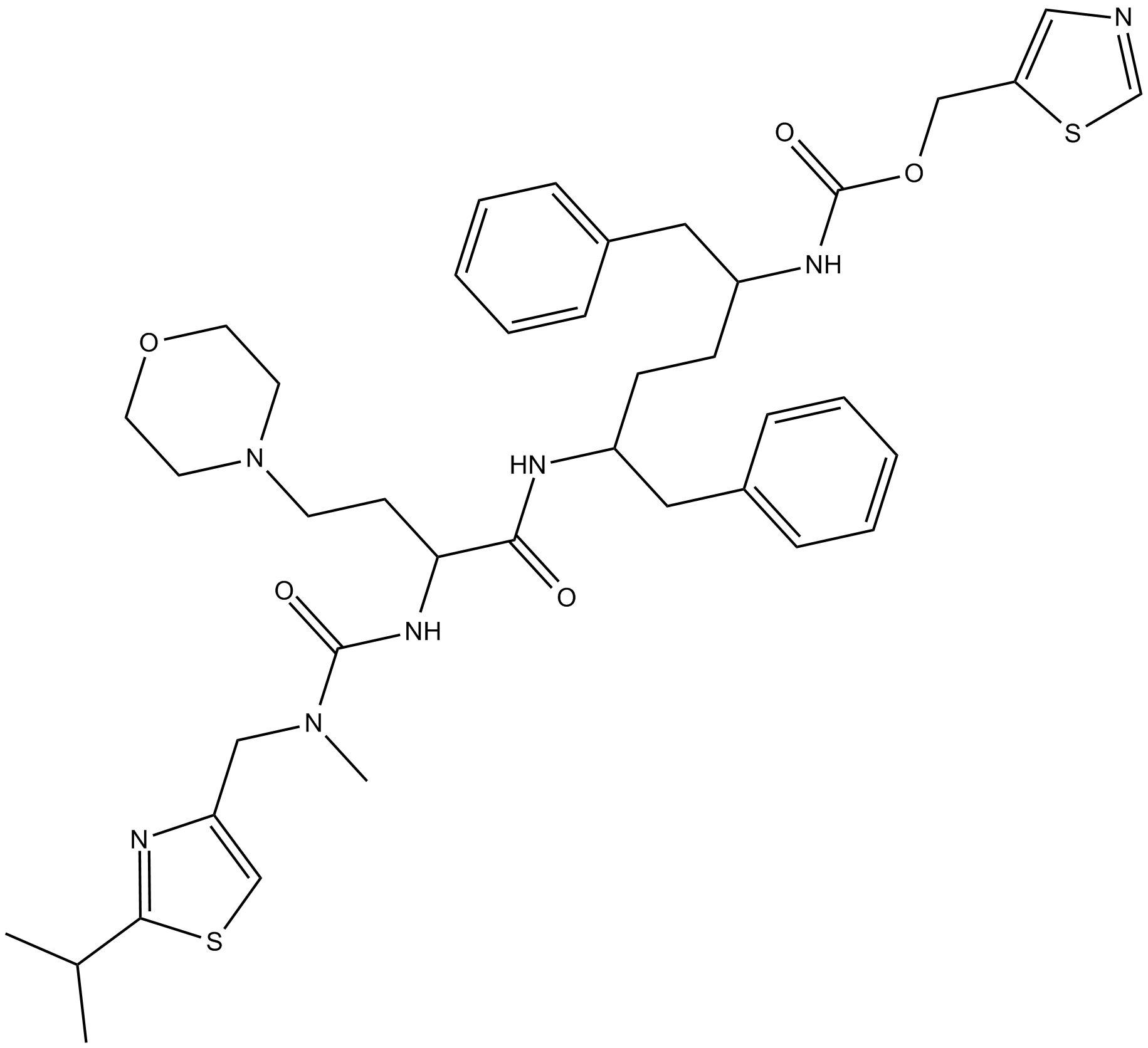
GC25306
CQ31
CQ31, a small molecule, selectively activates caspase activation and recruitment domain-containing 8 (CARD8).
-
GC64068
Cyclotriazadisulfonamide
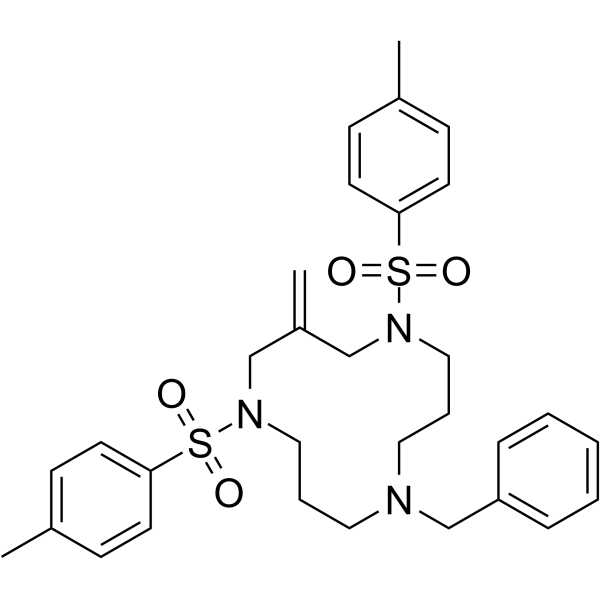
Cyclotriazadisulfonamide (CADA) is a specific CD4-targeted HIV entry inhibitors.
-
GC35790
Cys-TAT(47-57)
Cys-TAT(47-57) (Cys-[HIV-Tat (47-57)]) is an arginine rich cell penetrating peptide derived from the HIV-1 transactivating protein.

-
GC33982
D77
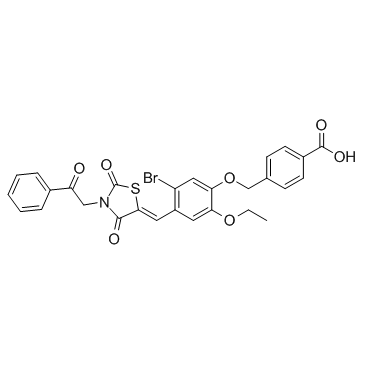
D77 is anti-HIV-1 inhibitor targeting the interaction between integrase and cellular LEDGF/p75. D77 inhibits HIV-1(IIIB) replication by EC50 value of 23.8 μg/ml in MT-4 cell (5.03 μg/ml for C8166 cells).
-
GC14452
Dapivirine (TMC120)
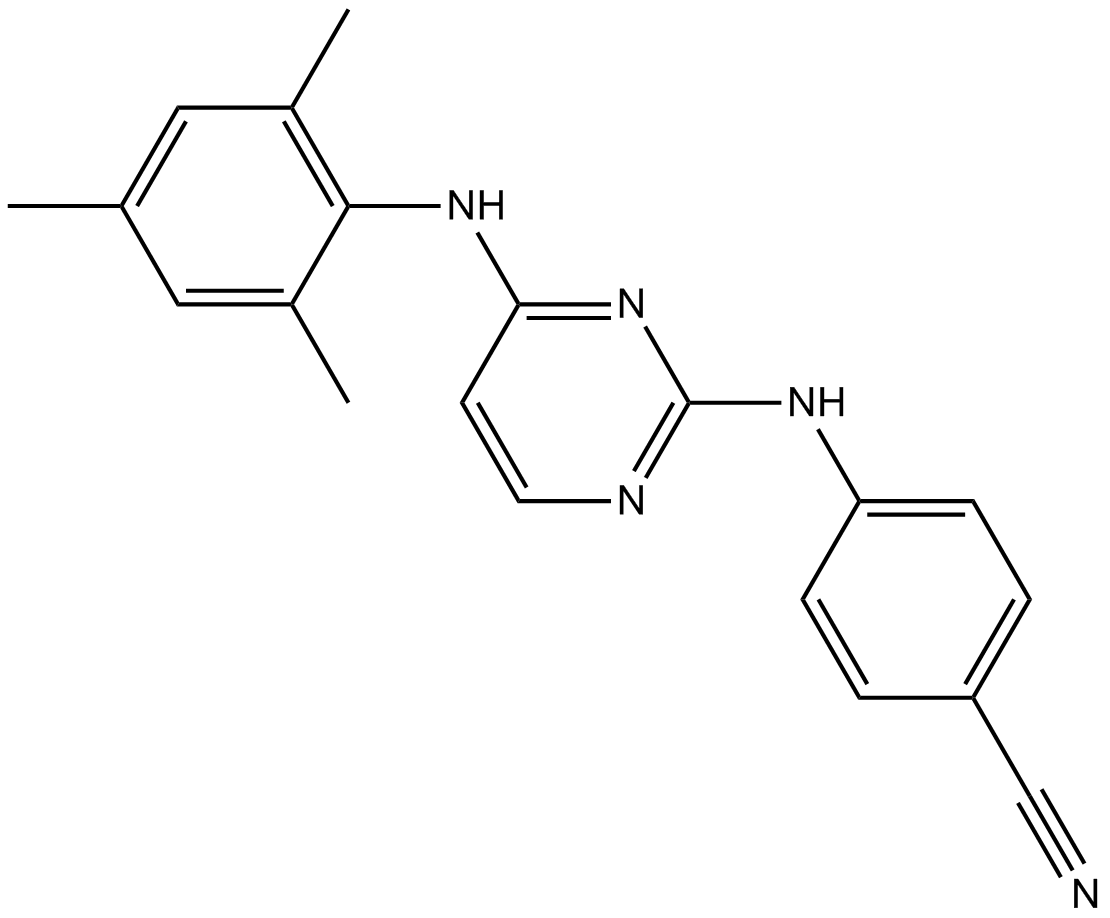
Dapivirine (TMC120) (TMC120), the prototype of diarylpyrimidines (DAPY), is an orally active and nonnucleoside reverse transcriptase inhibitor (NRTI).
-
GC17040
DAPTA
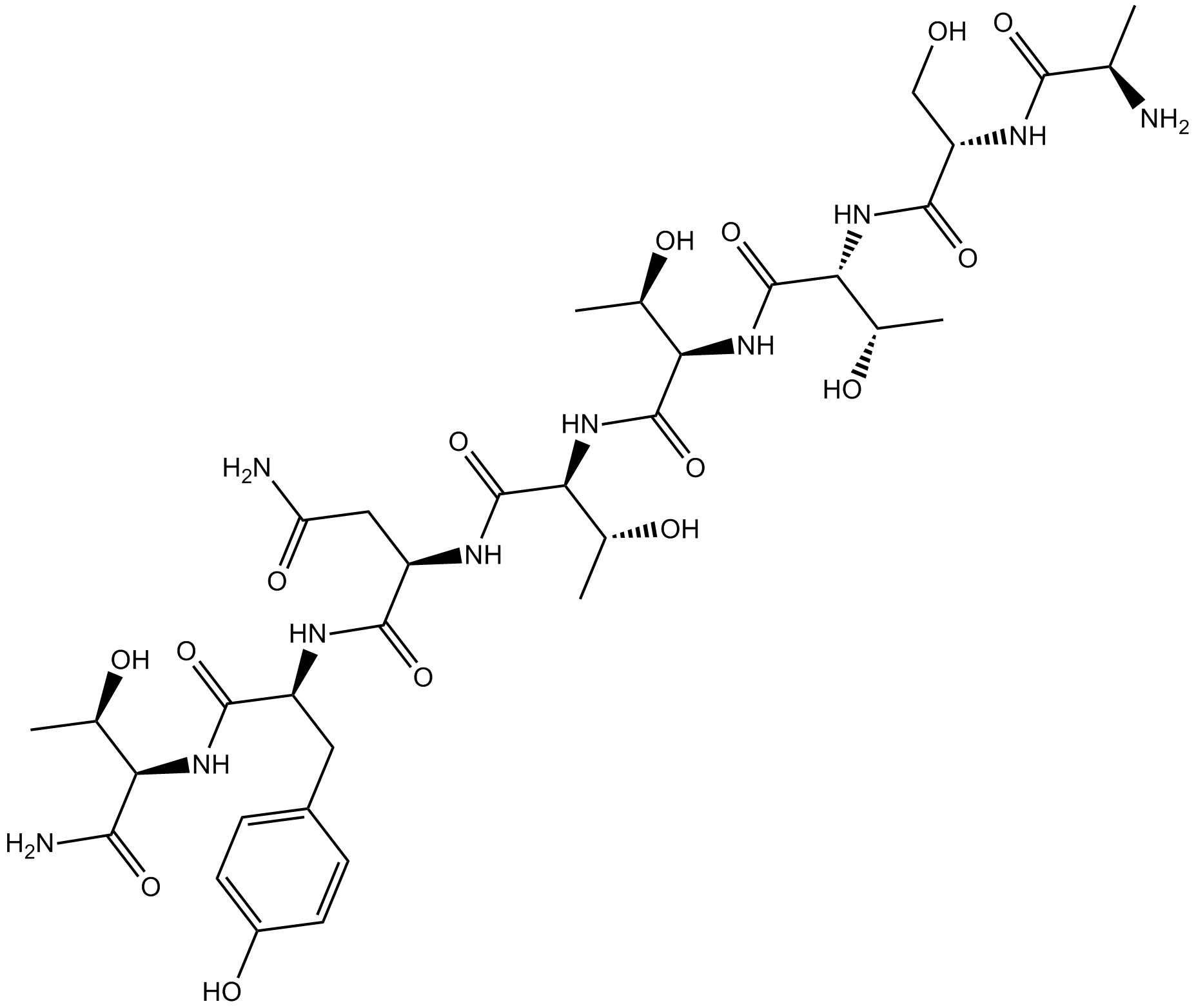
Chemokine receptor 5 (CCR5) antagonist
-
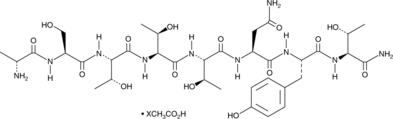
GC48359
DAPTA (acetate)
A CCR5 receptor antagonist
-
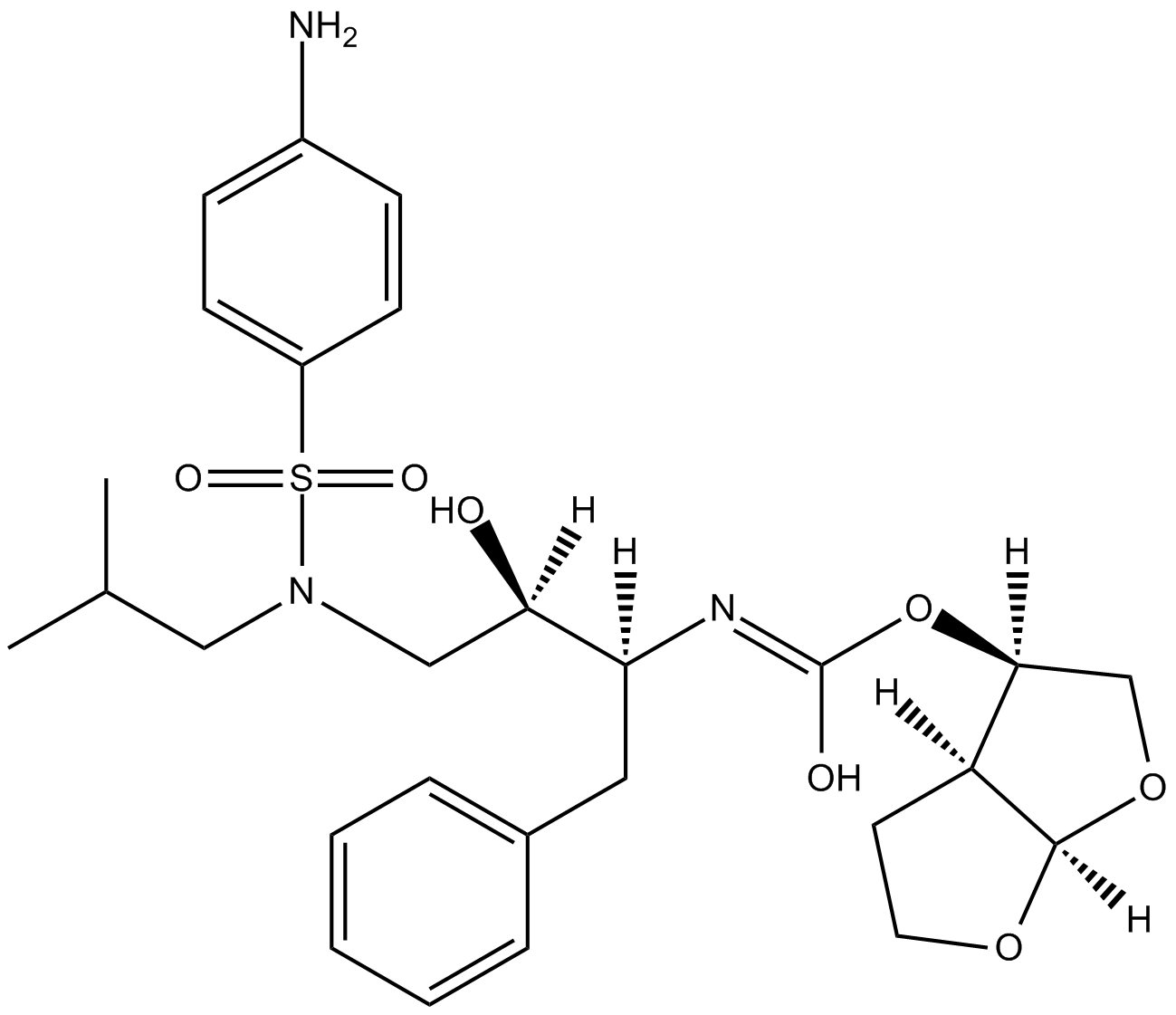
GC16602
Darunavir
An HIV-1 protease inhibitor
-
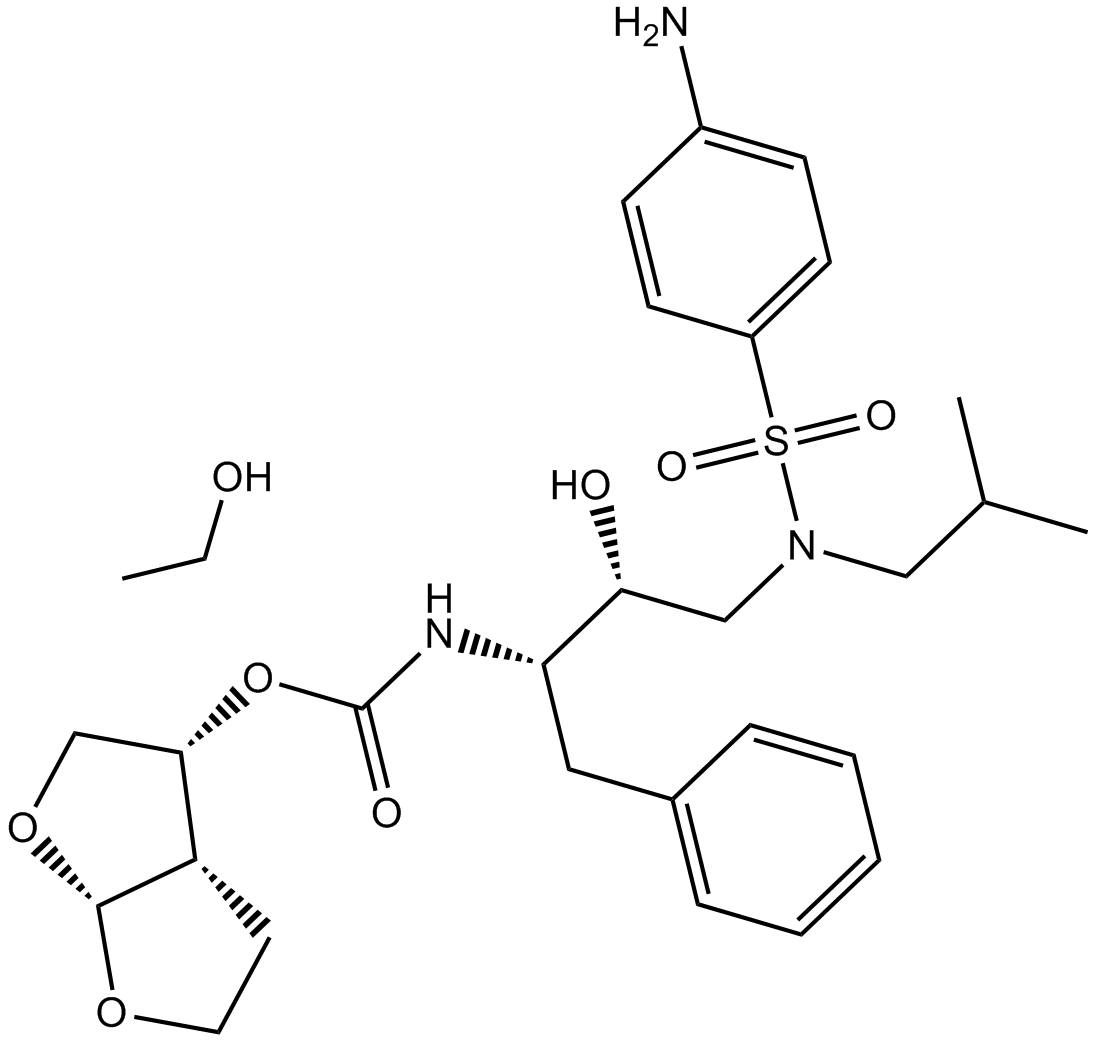
GC17942
Darunavir Ethanolate
nonpeptidic HIV protease inhibitor
-
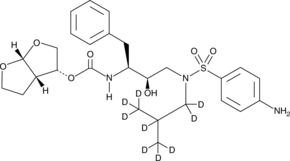
GC47173
Darunavir-d9
An internal standard for the quantification of darunavir
-
GC35823
DDX3-IN-1
DDX3-IN-1 (Compound 16f) is a DEAD-box polypeptide 3 (DDX3) inhibitor with CC50s of 50 and 36 μM for HIV and HCV, respectively.
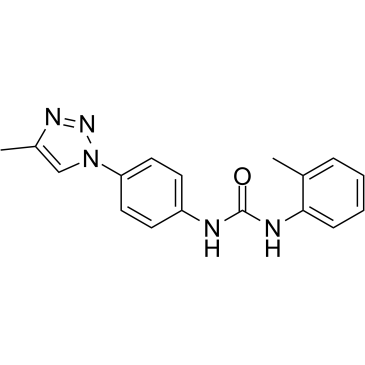
-
GC64391
DDX3-IN-2
DDX3-IN-2 is an active DEADbox polypeptide 3 (DDX3) inhibitor with an IC50 value of 0.3 μM.
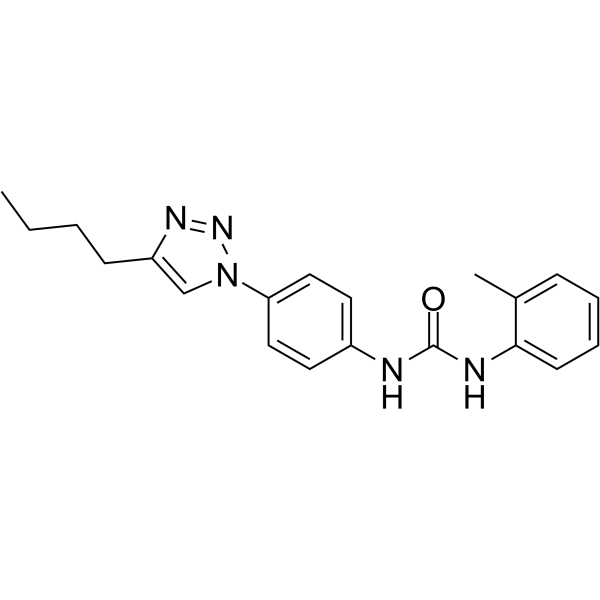
-
GC64270
Decanoyl-RVKR-CMK TFA
Decanoyl-RVKR-CMK (DecRVKRcmk) TFA inhibits over-expressed gp160 processing and HIV-1 replication.
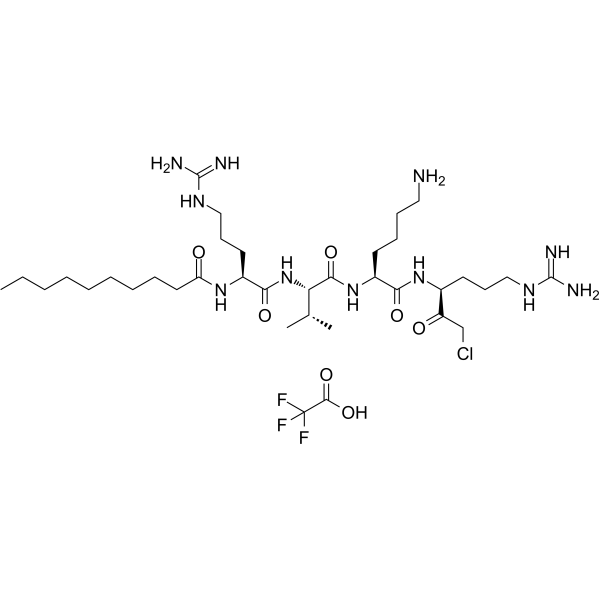
-
GC35835
Delavirdine
Delavirdine (U 90152) is a potent, highly specific and orally active non-nucleoside reverse transcriptase inhibitor (NNRTI).
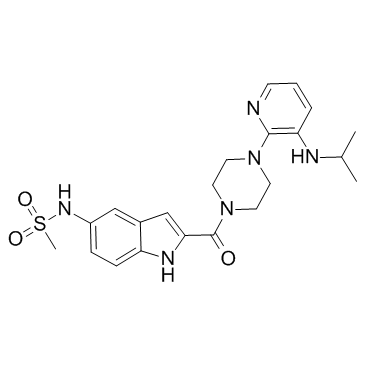
-
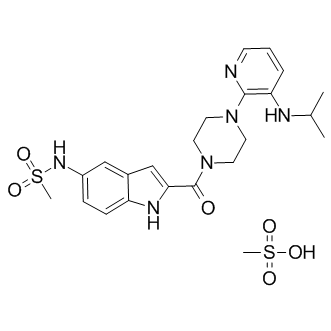
GC15452
Delavirdine mesylate
Non-nucleoside reverse transcriptase inhibitor
-
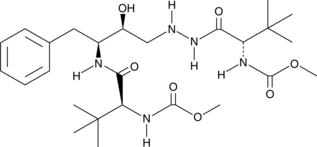
GC45650
Des(benzylpyridyl) Atazanavir
A metabolite of atazanavir
-
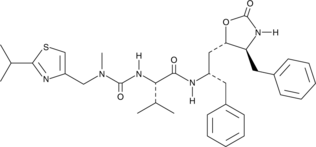
GC52038
Desthiazolylmethyl Ritonavir
A ritonavir degradation product
-
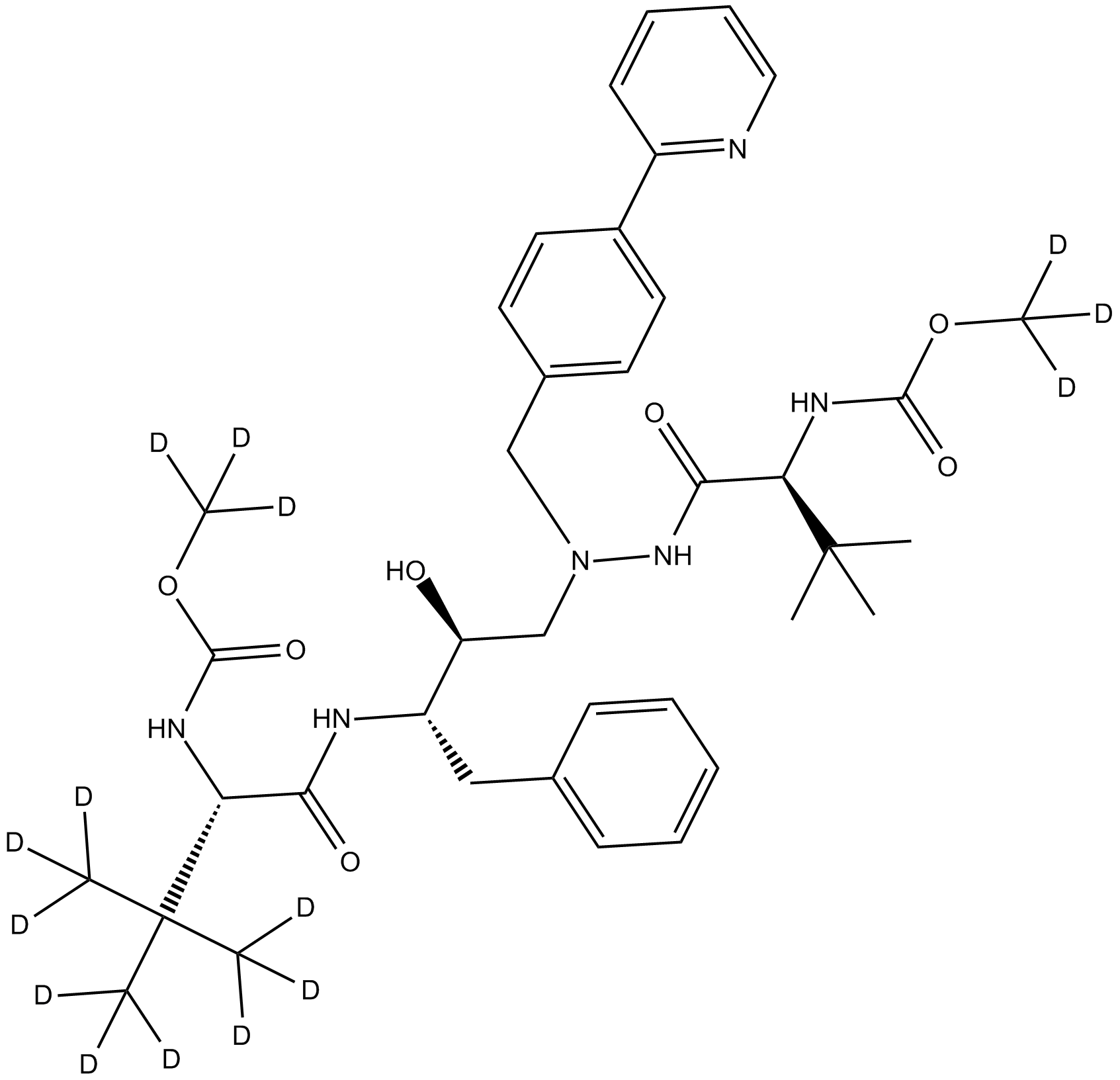
GC15984
Deuterated Atazanivir-D3-1
HIV-1 protease inhibitor
-
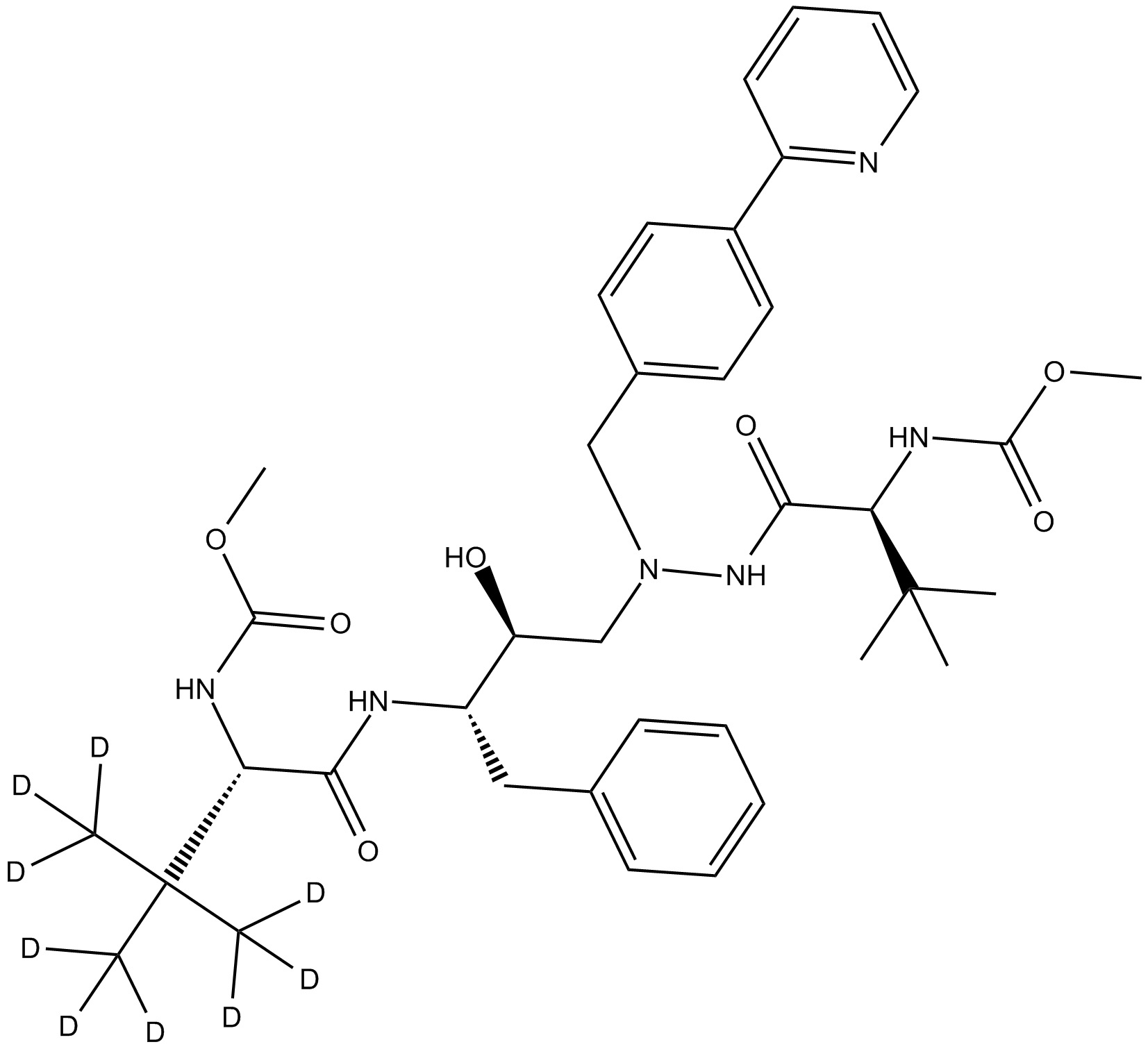
GC11772
Deuterated Atazanivir-D3-2
HIV-1 protease inhibitor
-
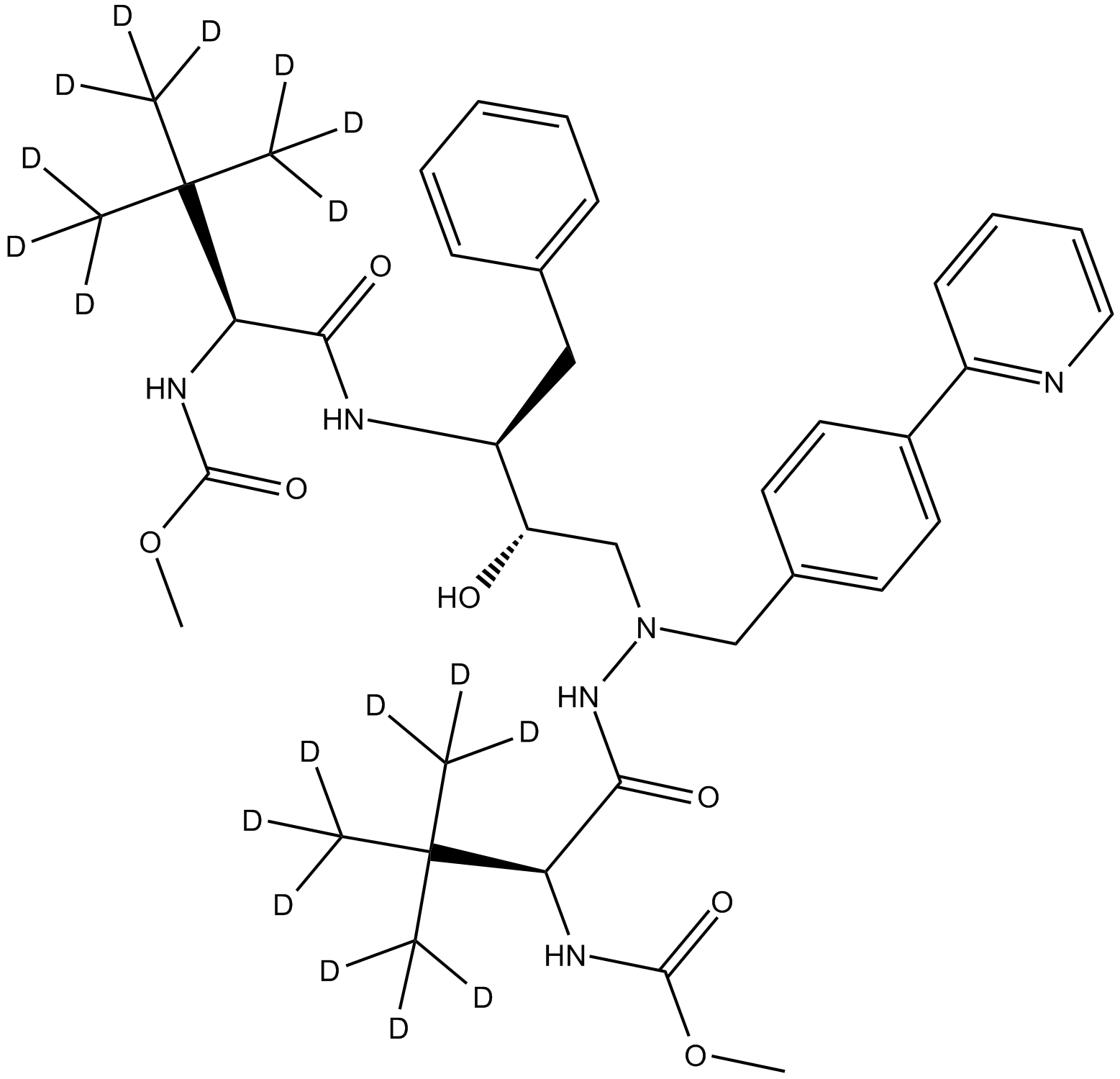
GC15050
Deuterated Atazanivir-D3-3
HIV-1 protease inhibitor
-
GC10520
Dextran sulfate sodium salt (M.W 200000)
Dextran sulfate sodium salt is a is a polymer of anhydroglucose with the molecular weight range of 35000-45000.

-
GC38250
Dextran sulfate sodium salt (MW 16000-24000)
A sulfated polysaccharide

-
GC38252
Dextran sulfate sodium salt (MW 35000-45000)
A sulfated polysaccharide

-
GC38253
Dextran sulfate sodium salt (MW 4500-5500)
A sulfated polysaccharide

-
GC38251
Dextran sulfate sodium salt (MW 450000-550000)
A sulfated polysaccharide

-
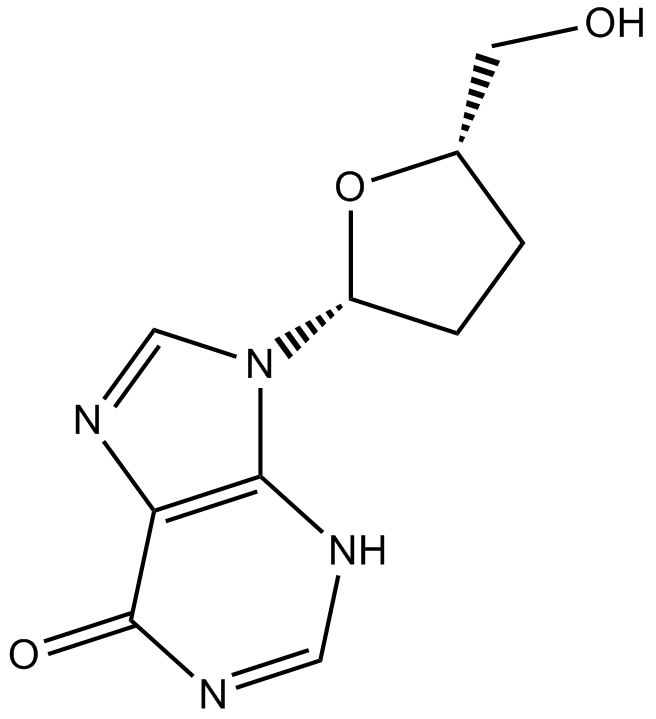
GC15330
Didanosine
Reverse transcriptase inhibitor
-
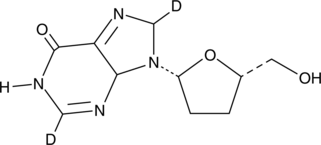
GC49022
Didanosine-d2
An internal standard for the quantification of didanosine
-
GC18456
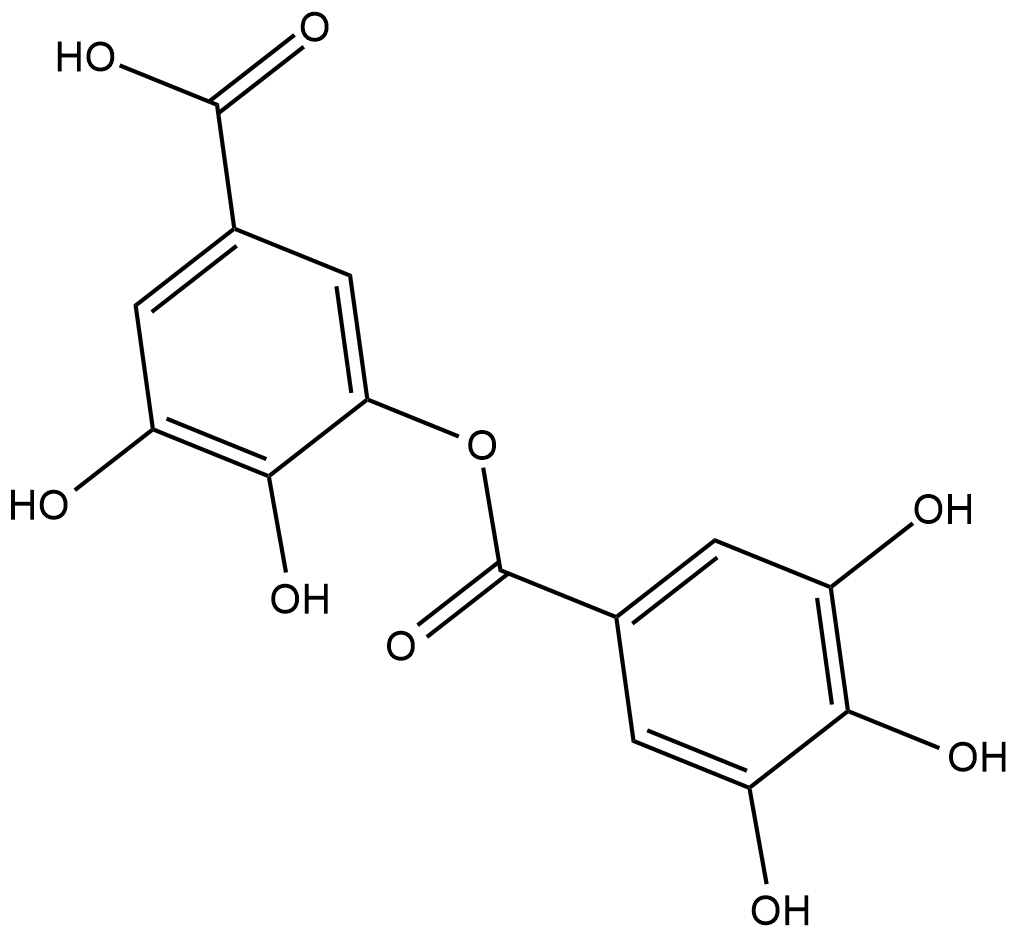
Digallic Acid
Digallic acid is a natural polyphenolic that can be produced by hydrolysis of gallotannins.
-
GC38662
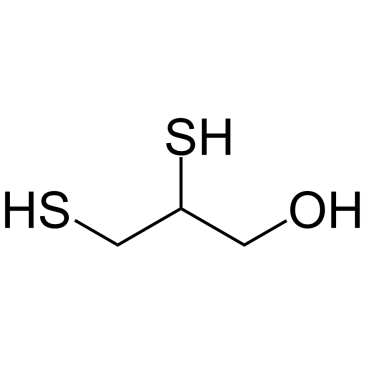
Dimercaprol
Dimercaprol (2,3-Dimercapto-1-propanol) is an anti-heavy metal-poisoning drug, which exhibits anti-HIV activity.
-
GC39734
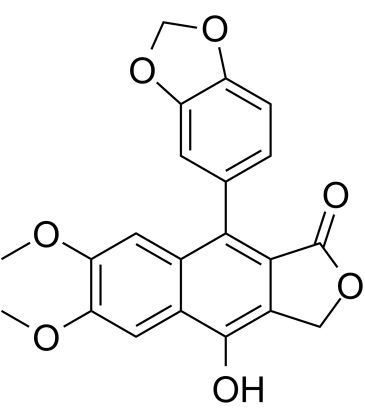
Diphyllin
Diphyllin is an arylnaphthalene lignan isolated from Justicia procumbens and is a potent HIV-1 inhibitor with an IC50 of 0.38 μM.
-
GC32142
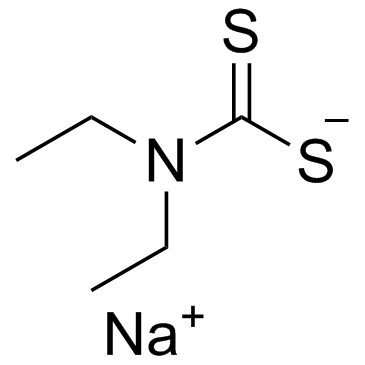
Ditiocarb sodium (Sodium diethyldithiocarbamate)
Ditiocarb sodium (Sodium diethyldithiocarbamate) (Sodium diethyldithiocarbamate) is an accelerator of the rate of copper cementation.
-
GC43498
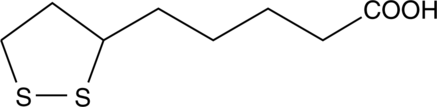
DL-α-Lipoic Acid
DL-α-Lipoic Acid is an antioxidant, which is an essential cofactor of mitochondrial enzyme complexes.
-
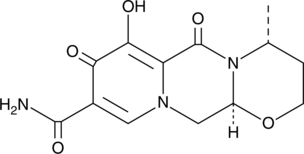
GC49288
Dolutegravir M1
A metabolite of dolutegravir
-
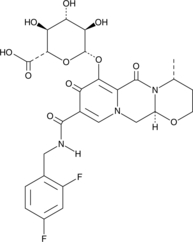
GC49289
Dolutegravir O-β-D-Glucuronide
A metabolite of dolutegravir
-
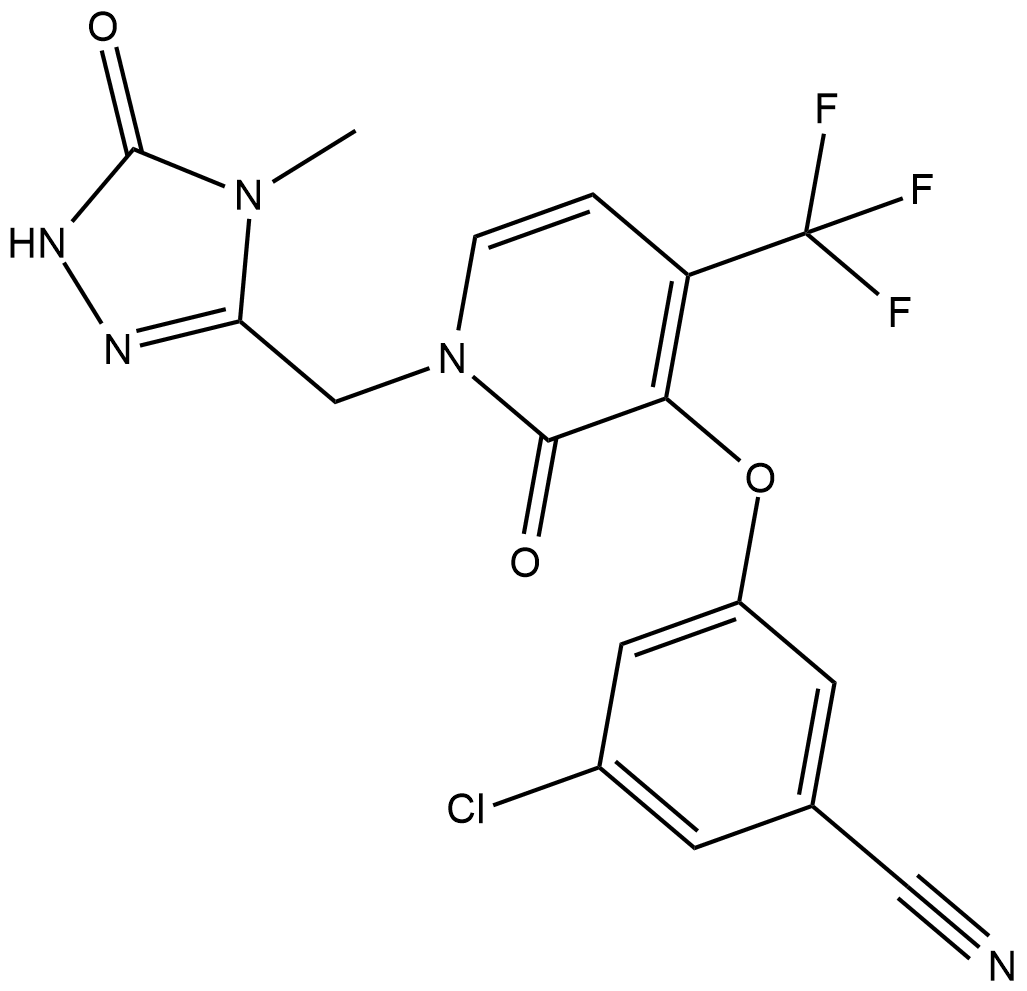
GC19126
Doravirine
Doravirine is a novel non-nucleoside inhibitor of HIV-1 reverse transcriptase with potent activity against wild-type virus (95% inhibitory concentration 19 nM, 50% human serum).
-
GC17567
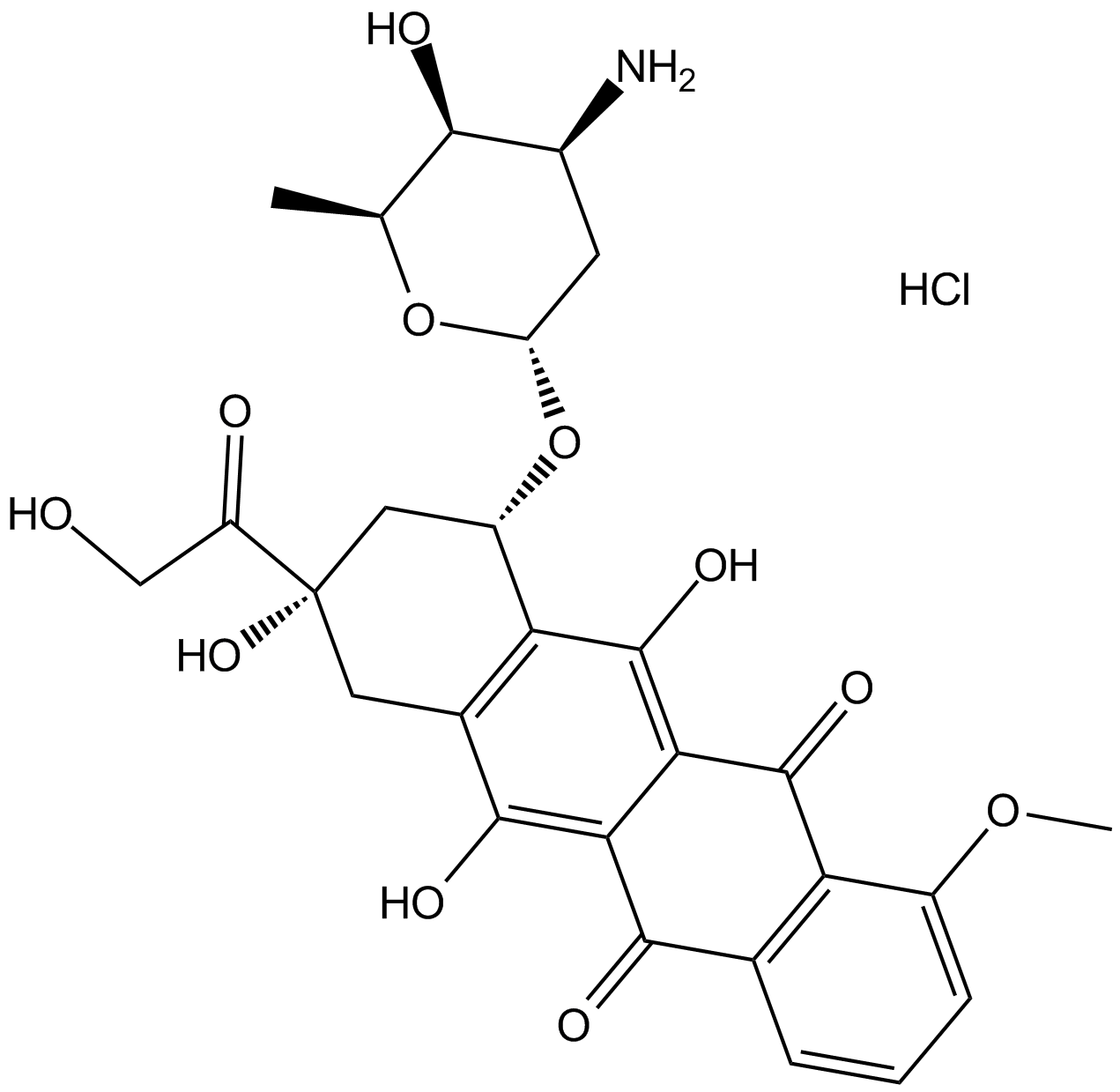
Doxorubicin (Adriamycin) HCl
Doxorubicin (Hydroxydaunorubicin) hydrochloride, a cytotoxic anthracycline antibiotic, is an anti-cancer chemotherapy agent.
-
GC33968
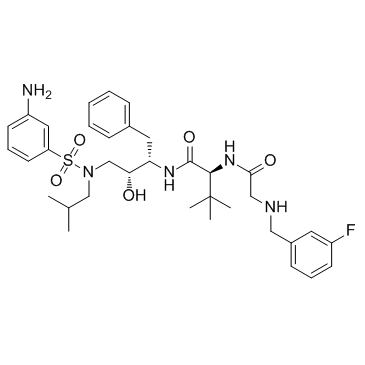
DPC-681 (DPH-153893)
DPC-681 (DPH-153893) is a potent and selective inhibitor of HIV protease with IC90s for wild-type HIV-1 of 4 to 40 nM.
-
GC17015
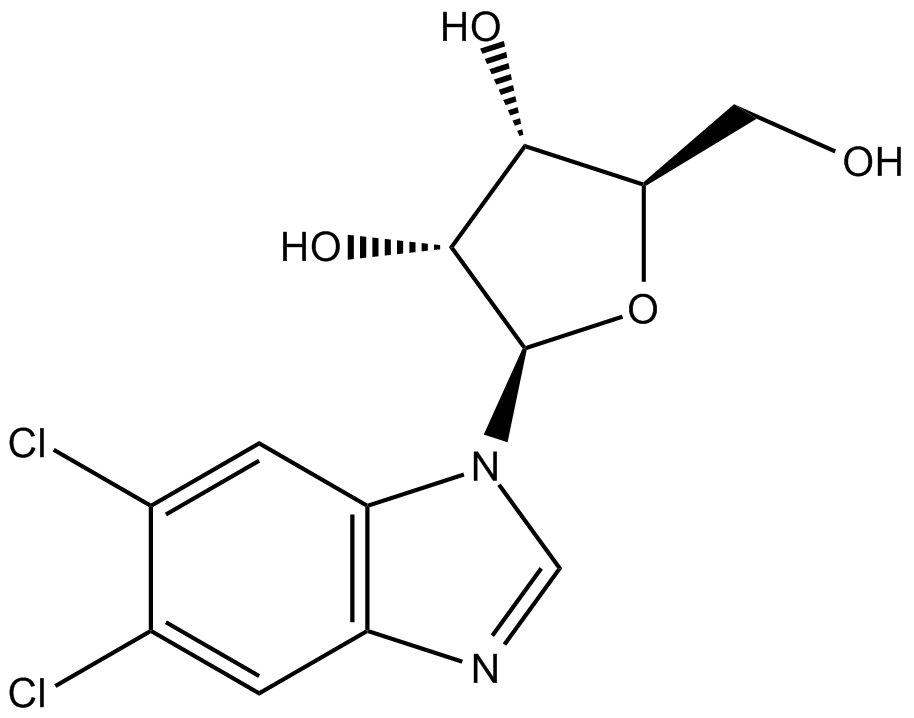
DRB
DRB is a nucleoside analog that inhibits several carboxyl-terminal domain (CTD) kinases including casein kinase II and CDKs. DRB trigger p53-dependent apoptosis of human colon adenocarcinoma cells without inducing genotoxic stress to healthy cells.
-
GC10331
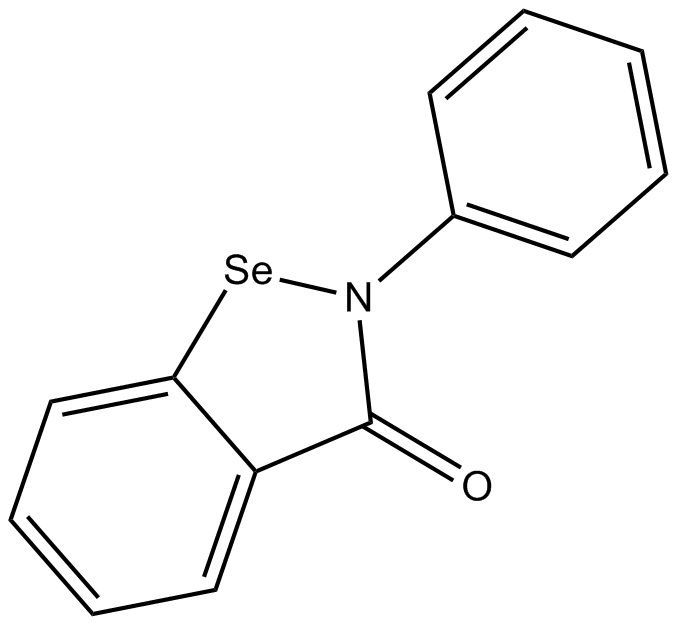
Ebselen
A peroxynitrite scavenger
-
GC12389
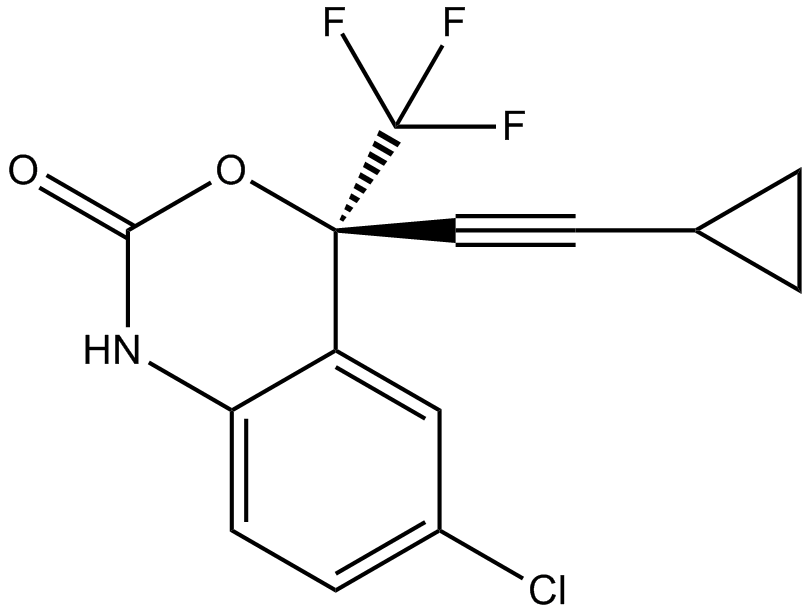
Efavirenz
Reverse transcriptase inhibitor
-
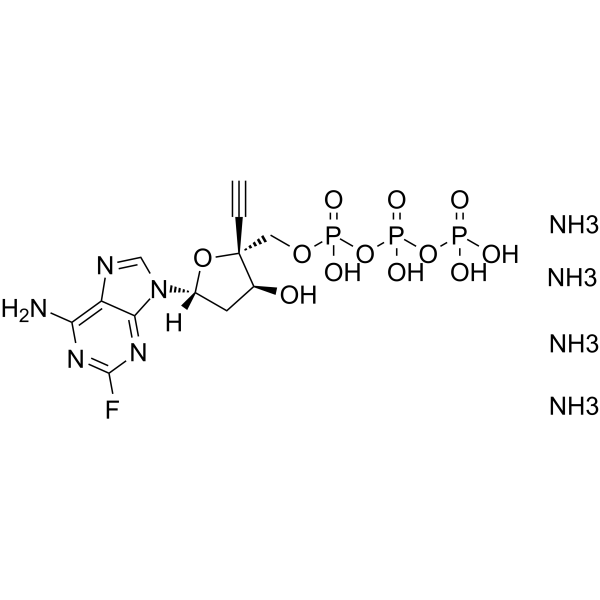
GC62607
EFdA-TP tetraammonium
EFdA-TP tetraammonium is a potent nucleoside reverse transcriptase (RT) inhibitor.
-
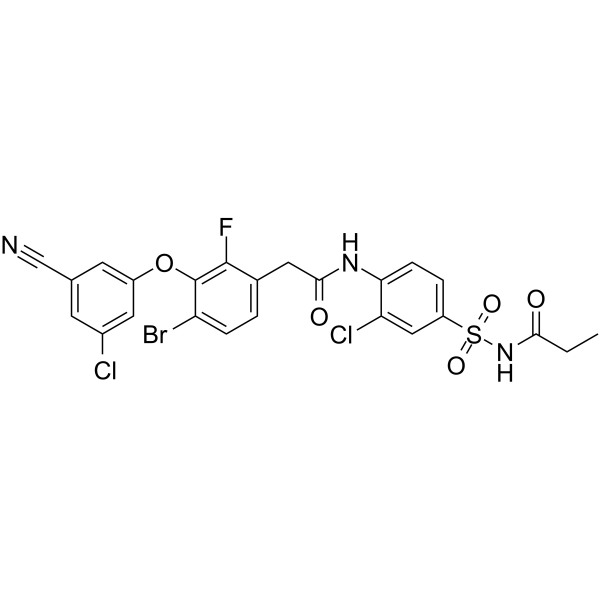
GC62295
Elsulfavirine
Elsulfavirine is a reverse transcriptase inhibitors for HIV-1 infection and is a new anti-HIV drug.
-
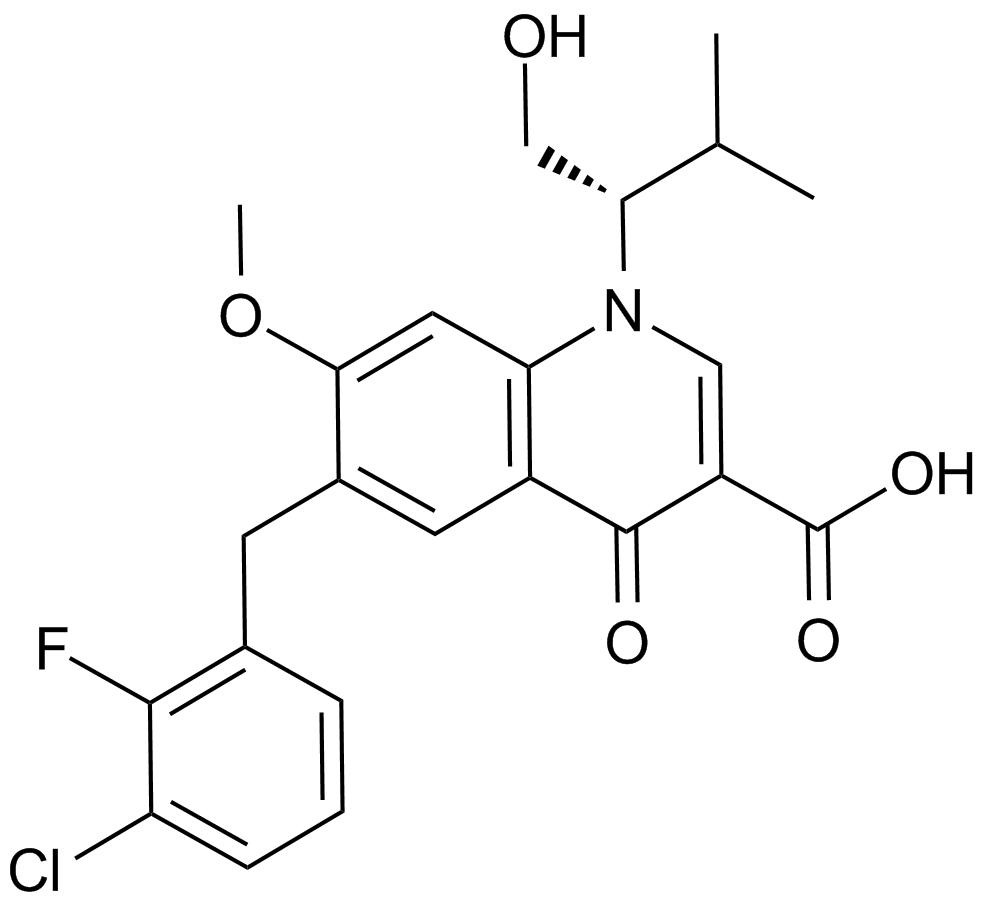
GC14996
Elvitegravir (GS-9137)
An inhibitor of HIV-1 integrase
-
GC64484
Emivirine
Emivirine (MKC-442) is a non-nucleoside reverse transcriptase inhibitors (NNRTIs) with Ki values of 0.20 and 0.01 μM for dTTP- and dGTP-dependent DNA or RNA polymerase activity, respectively.
-
GC13653
Emtricitabine
Reverse transcriptase inhibitor
-
GC43600
Emtricitabine S-oxide
Emtricitabine S-oxide is a potential impurity found in commercial preparations of emtricitabine.
-
GC11162
Enfuvirtide
HIV fusion inhibitor

-
GC38109
Enfuvirtide acetate
Enfuvirtide (T20; DP178) acetate is an anti-HIV-1 fusion inhibitor peptide.
-
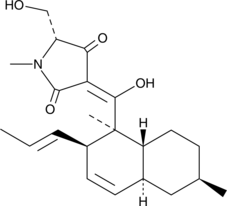
GC40224
Epiequisetin
Epiequisetin is a fungal metabolite and an epimer of equisetin.
-
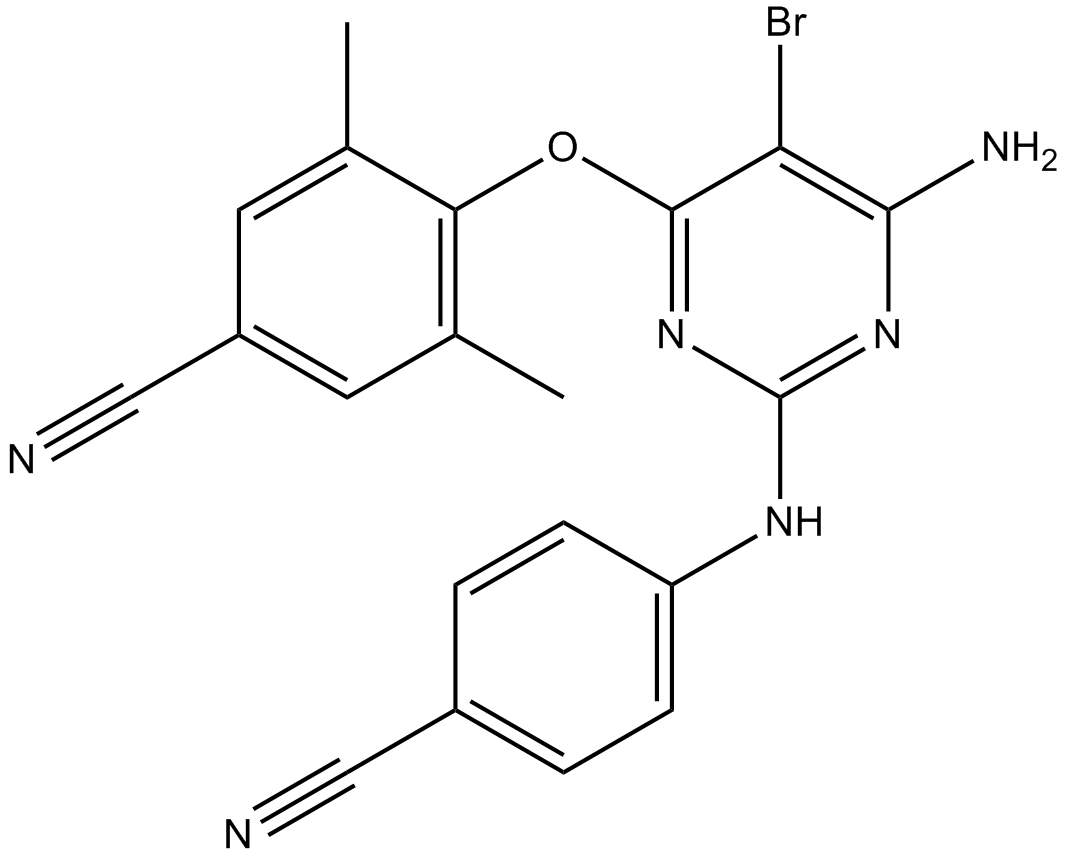
GC15996
Etravirine (TMC125)
Etravirine (TMC125) is a non-nucleoside reverse transcriptase inhibitor (NNRTI) used for the treatment of HIV.
-
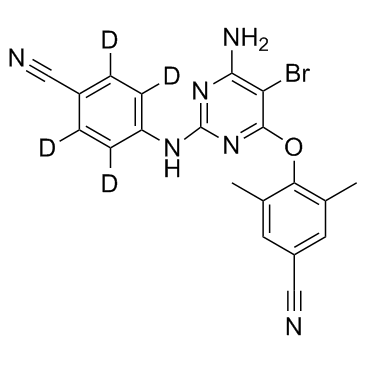
GC33536
Etravirine D4 (TMC-125 D4)
Etravirine D4 (TMC-125 D4) (TMC-125 D4) is the deuterium labeled Etravirine.
-
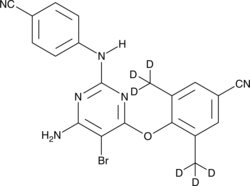
GC45825
Etravirine-d6
An internal standard for the quantification of etravirine
-
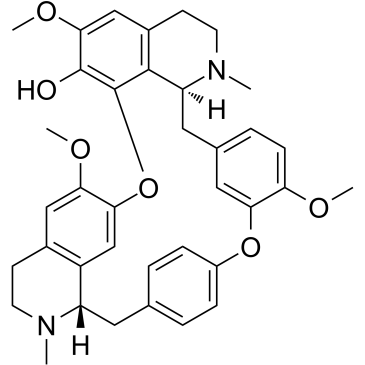
GC38437
Fangchinoline
An alkaloid with diverse biological activities
-
GC14289
Fasudil (HA-1077) HCl
Fasudil (HA-1077; AT877) Hydrochloride is a nonspecific RhoA/ROCK inhibitor and also has inhibitory effect on protein kinases, with an Ki of 0.33 μM for ROCK1, IC50s of 0.158 μM and 4.58 μM, 12.30 μM, 1.650 μM for ROCK2 and PKA, PKC, PKG, respectively. Fasudil (HA-1077) HCl is also a potent Ca2+ channel antagonist and vasodilator.
-
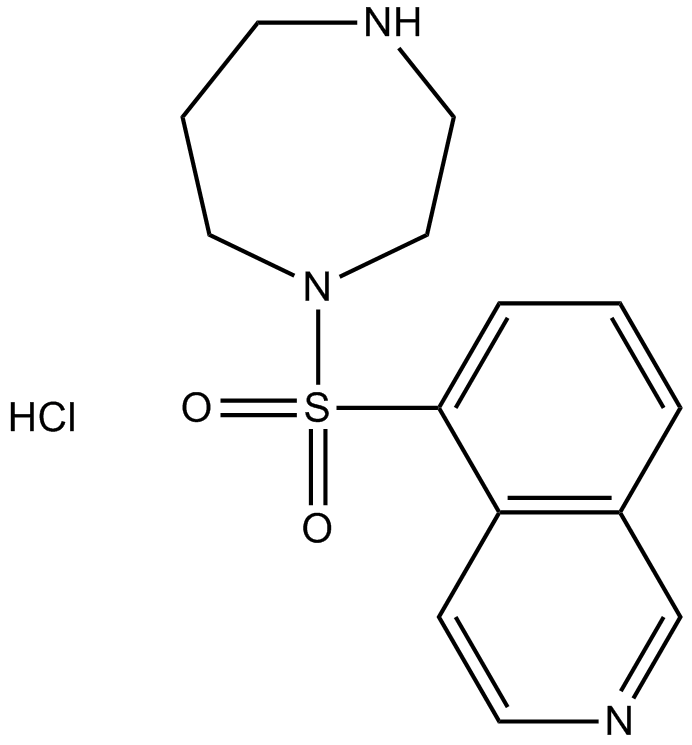
GC36033
FC131 TFA
FC131 TFA is a CXCR4 antagonist, inhibits [125I]-SDF-1 binding to CXCR4, with an IC50 of 4.5 nM.
-
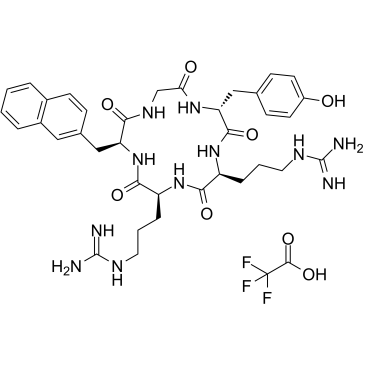
GC43659
Feglymycin
Feglymycin is a 13-amino acid peptide originally isolated from Streptomyces that has antibacterial and antiviral activities.
-
GC60840
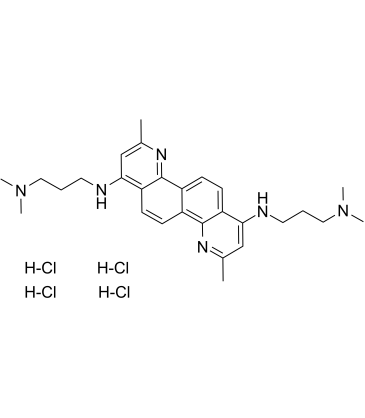
FGI-106 tetrahydrochloride
FGI-106 tetrahydrochloride is a potent and broad-spectrum inhibitor with inhibitory activity against multiple viruses.
-
GC16063
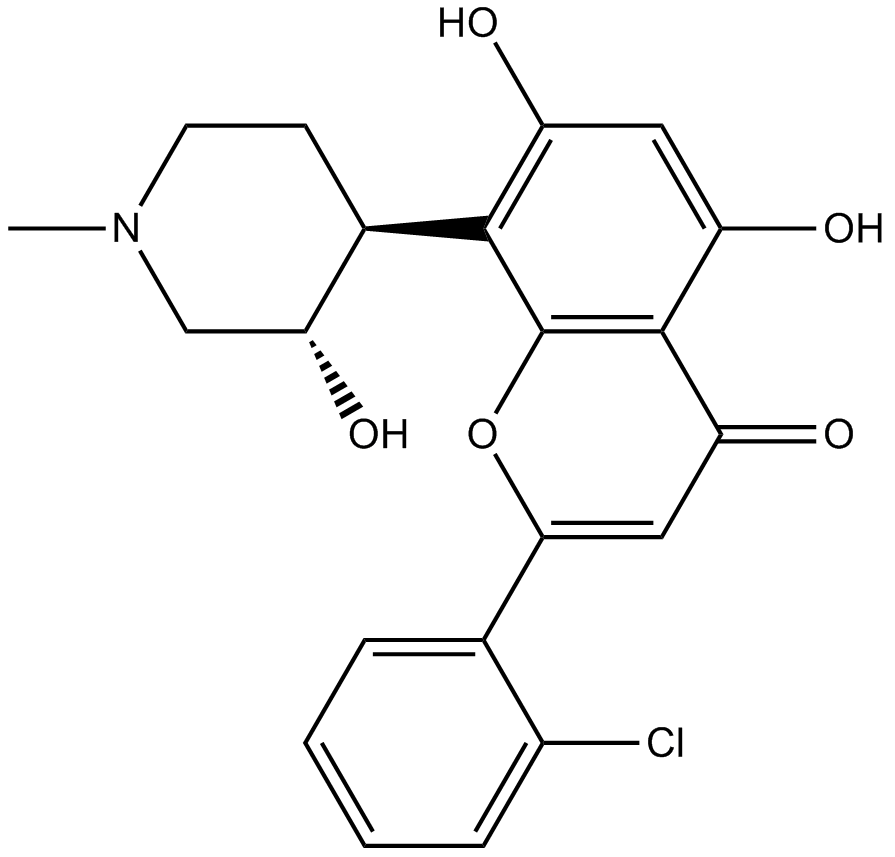
Flavopiridol
An inhibitor of cyclin-dependent kinases
-
GC16425
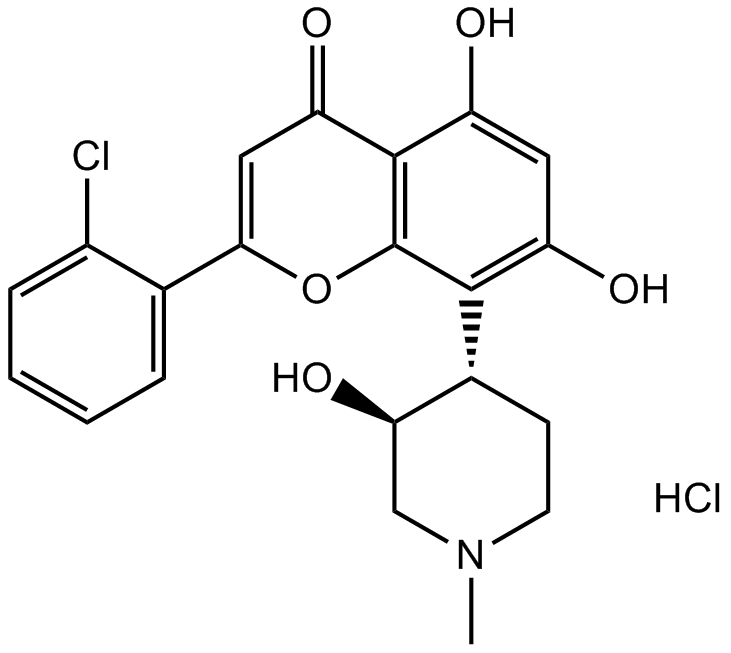
Flavopiridol hydrochloride
CDK inhibitor, potent and selective
-
GC14466
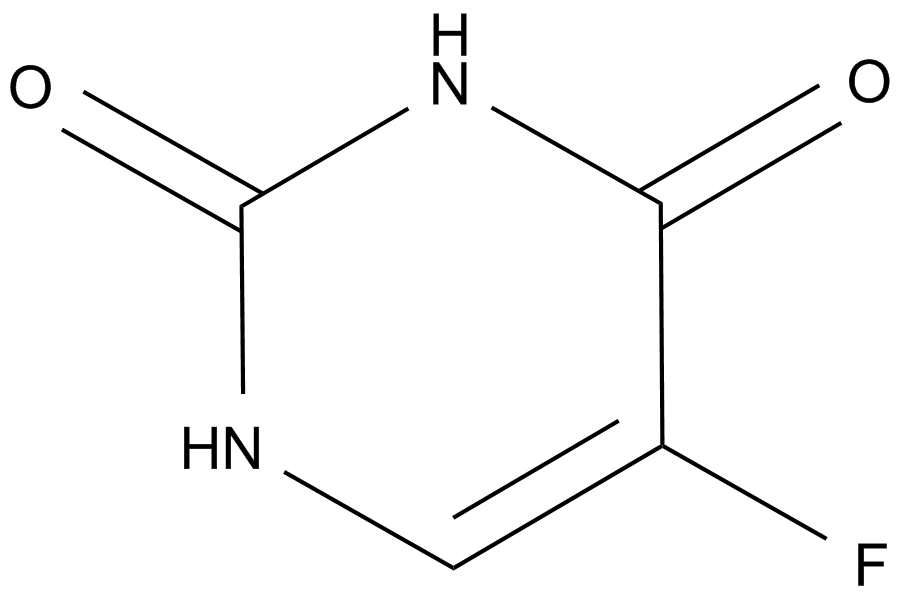
Fluorouracil (Adrucil)
A prodrug form of FdUMP
-
GC63646
Formycin A
Formycin A (NSC 102811), a purine nucleoside antibiotic, is a potent human immunodeficiency virus type 1 (HIV-1) inhibitor with an EC50 of 10 μM.
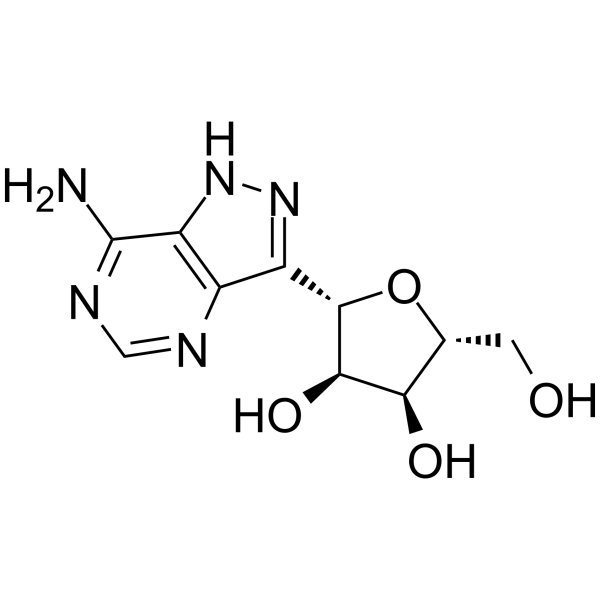
-
GC32362
Fosamprenavir (Amprenavir phosphate)
Fosamprenavir (Amprenavir phosphate) (Amprenavir phosphate;GW 433908) is a phosphate ester prodrug of the antiretroviral protease inhibitor Amprenavir, with improved solubility.
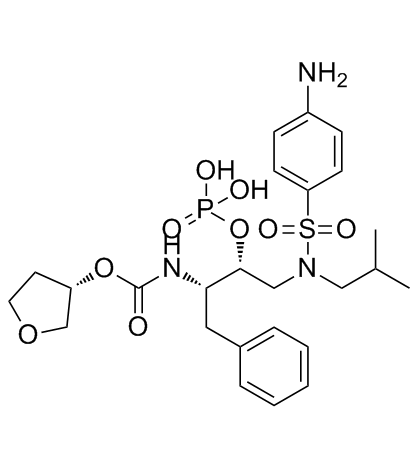
-
GC11300
Fosamprenavir Calcium Salt
A prodrug form of amprenavir
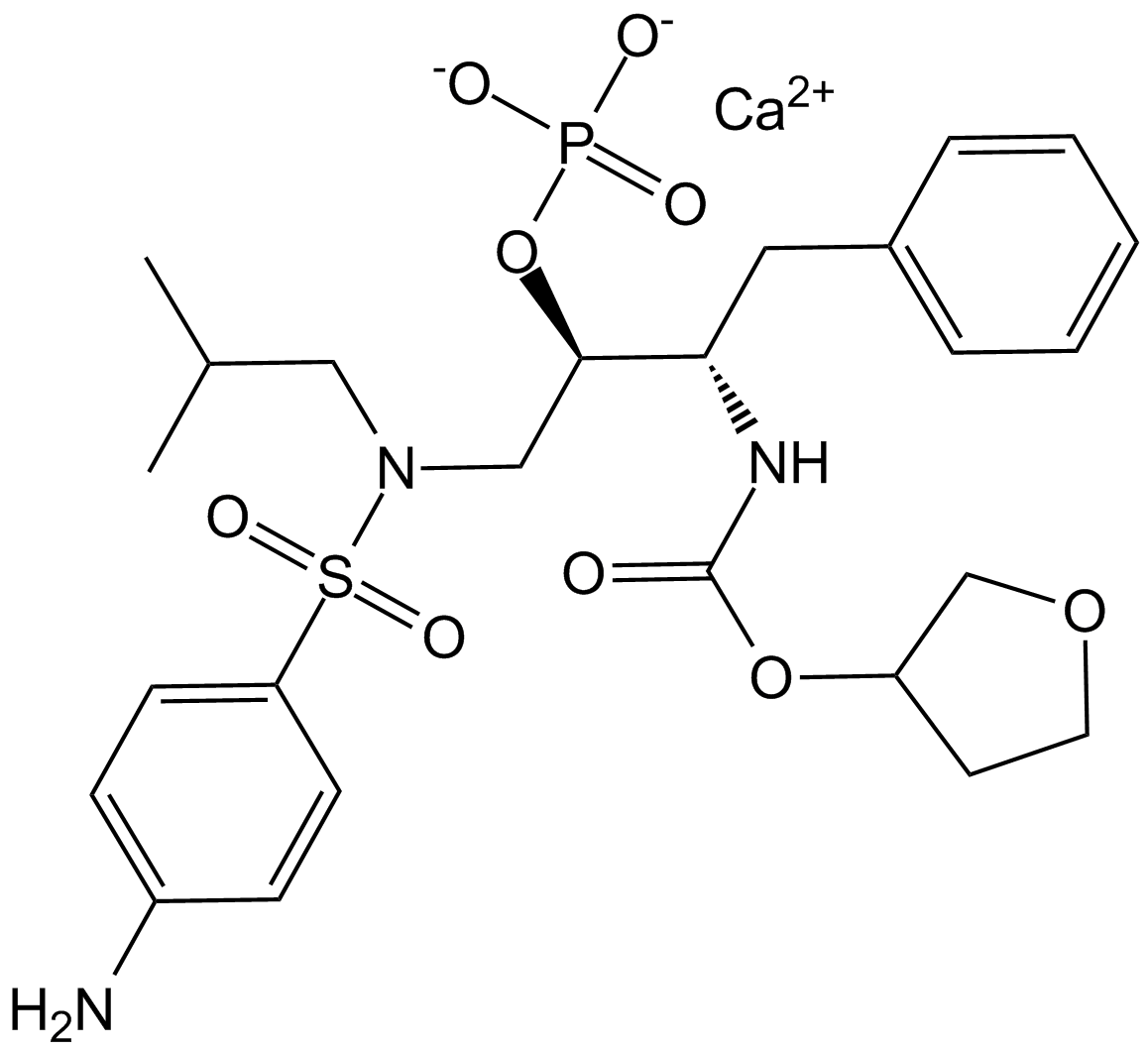
-
GC36072
Fostemsavir Tris
Fostemsavir Tris (BMS-663068 (Tris)) is the phosphonooxymethyl prodrug of BMS-626529.
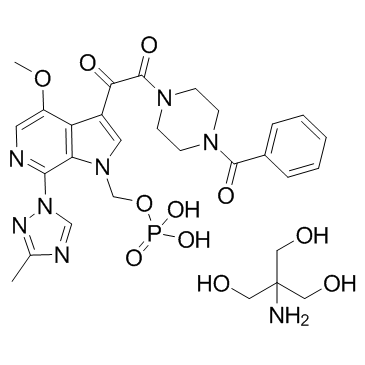
-
GC40849
Funalenone
Funalenone is a phenalenone originally isolated from A.
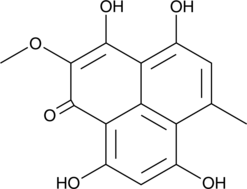
-
GC49229
Ganodermanontriol
Ganodermanontriol, a sterol isolated from Ganoderma lucidum, induces anti-inflammatory activity in tert-butyl hydroperoxide (t-BHP)-damaged hepatic cells through the expression of HO-1. Ganodermanontriol exhibits hepatoprotective activity.
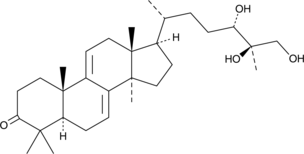
-
GC33910
Gardiquimod trifluoroacetate
Gardiquimod trifluoroacetate, an imidazoquinoline analog, is a TLR7/8 agonist.
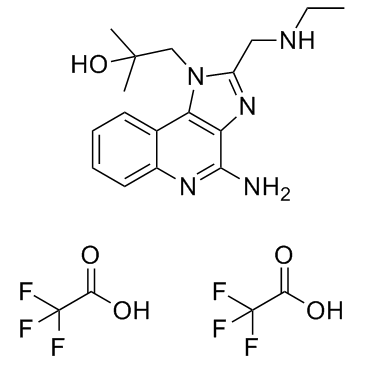
-
GC47398
Genistein-d4
An internal standard for the quantification of genistein
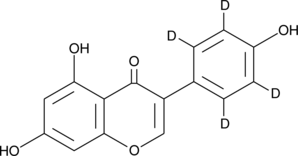
-
GC36143
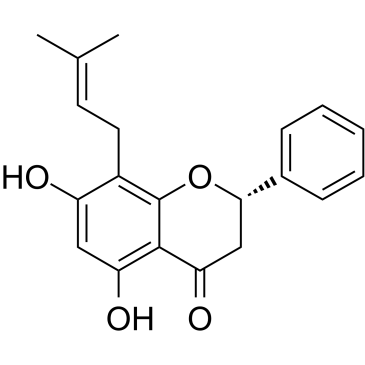
Glabranine
Glabranine, an flavonoid, is isolated from Tephrosia s.
-
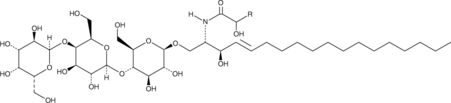
GC52492
Globotriaosylceramide (hydroxy) (porcine RBC)
A sphingolipid
-
GC52491
Globotriaosylceramide (non-hydroxy) (porcine RBC)
A sphingolipid

-
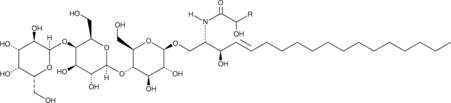
GC43758
Globotriaosylceramides (hydroxy) (porcine)
Globotriaosycleramides are glycosphingolipids found in mammalian cell membranes that are synthesized from lactosylceramides.
-
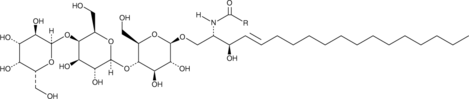
GC43760
Globotriaosylceramides (porcine)
Globotriaosycleramides are glycosphingolipids found in mammalian cell membranes that are synthesized from lactosylceramides.
-
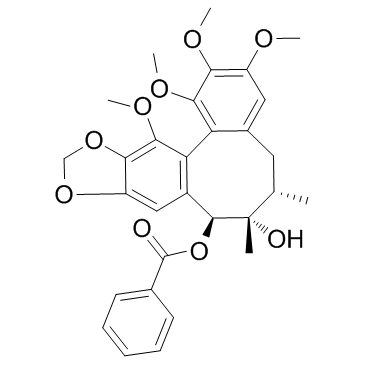
GC32210
Gomisin G
Gomisin G is an ethanolic extract of the stems of Kadsura interior; exhibits potent anti-HIV activity with EC50 and therapeutic index (TI) values of 0.006 microgram/mL and 300, respectively.
-
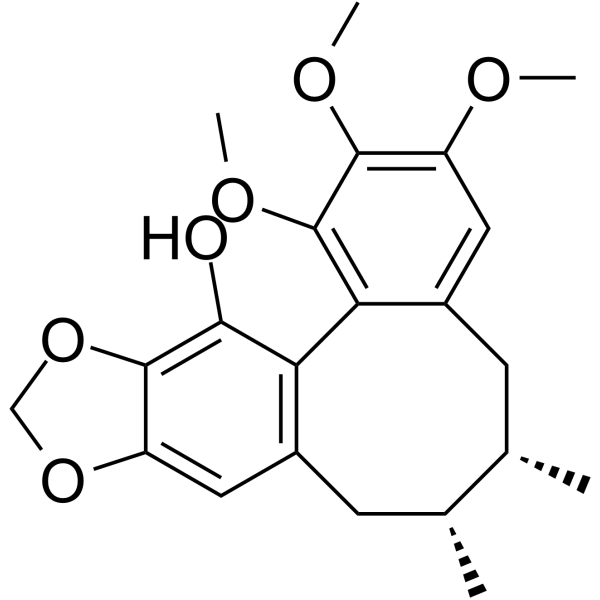
GC64233
Gomisin M2
Gomisin M2 ((+)-Gomisin M2) is a lignan isolated from the fruits of Schisandra rubriflora with anti-HIV activity (EC50 of 2.4 μM).
-
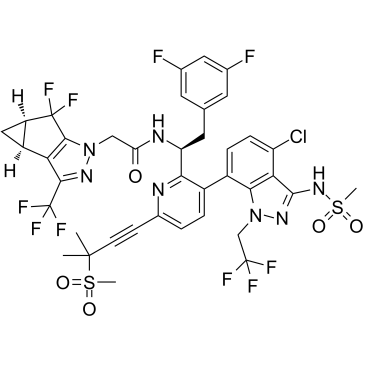
GC38790
GS-6207
GS-6207 (GS-6207) is a HIV-1 capsid inhibitor.
-
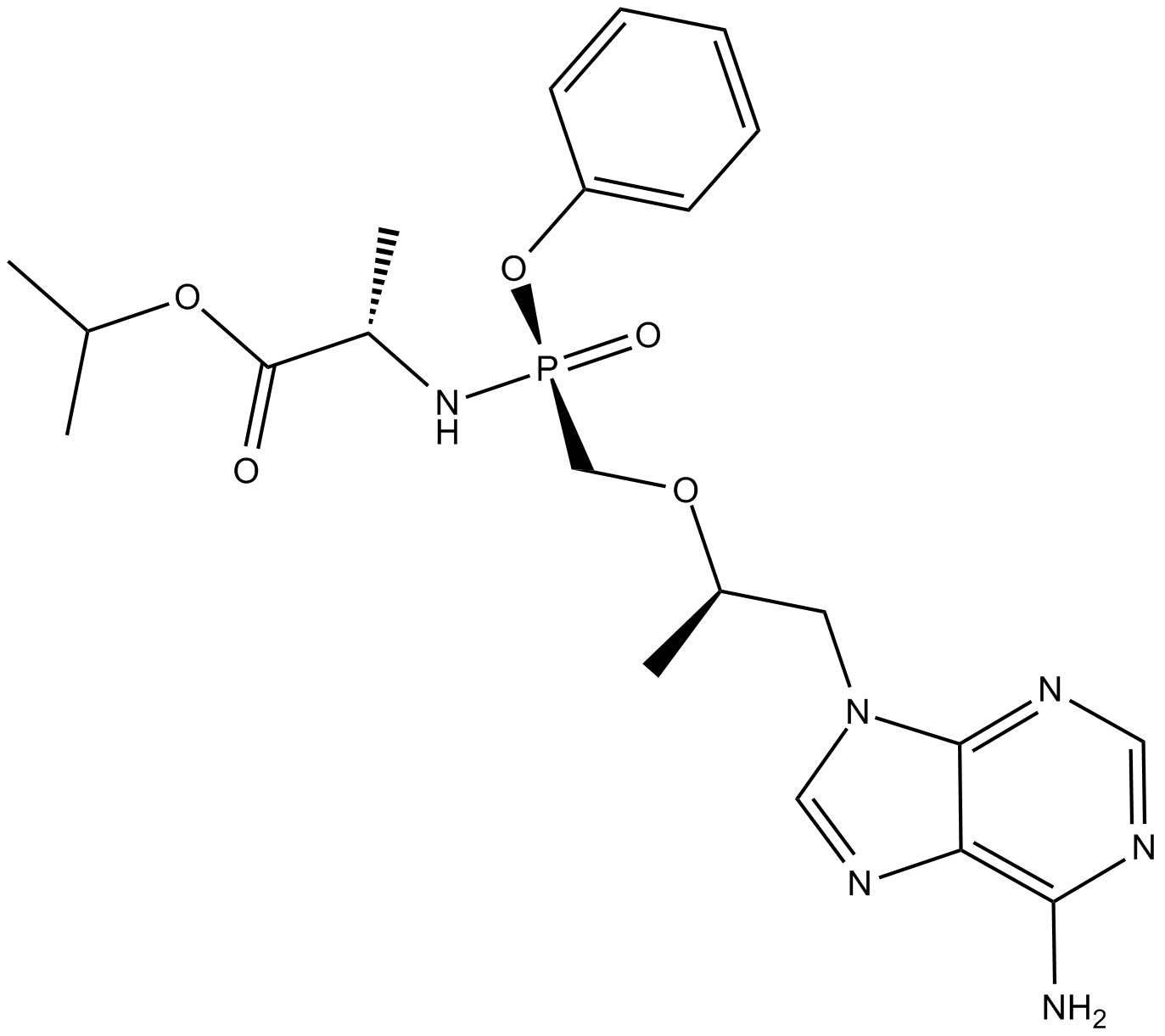
GC12679
GS-7340
GS-7340 (GS-7340) is an investigational oral prodrug of Tenofovir.
-
GC12555
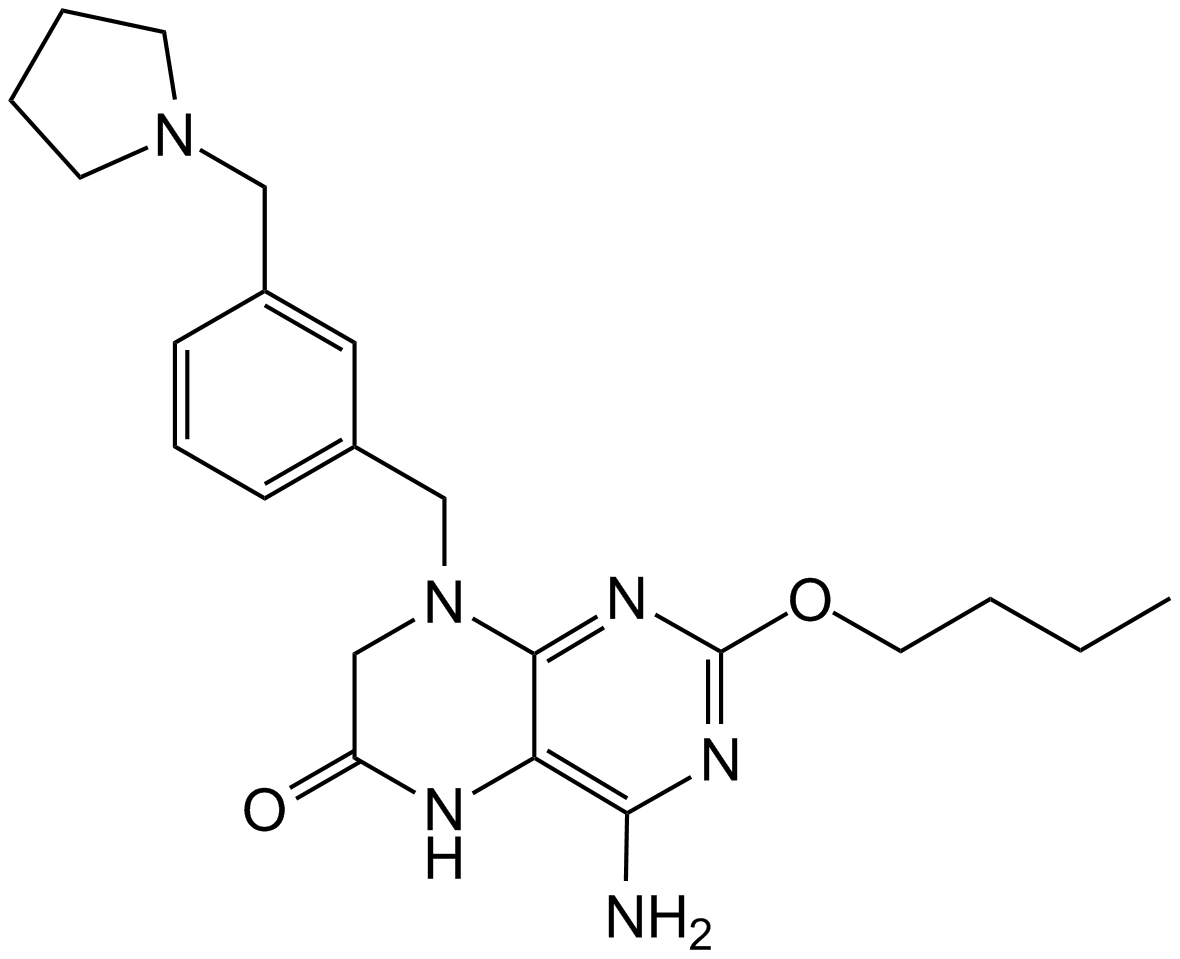
GS-9620
A TLR7 agonist
-
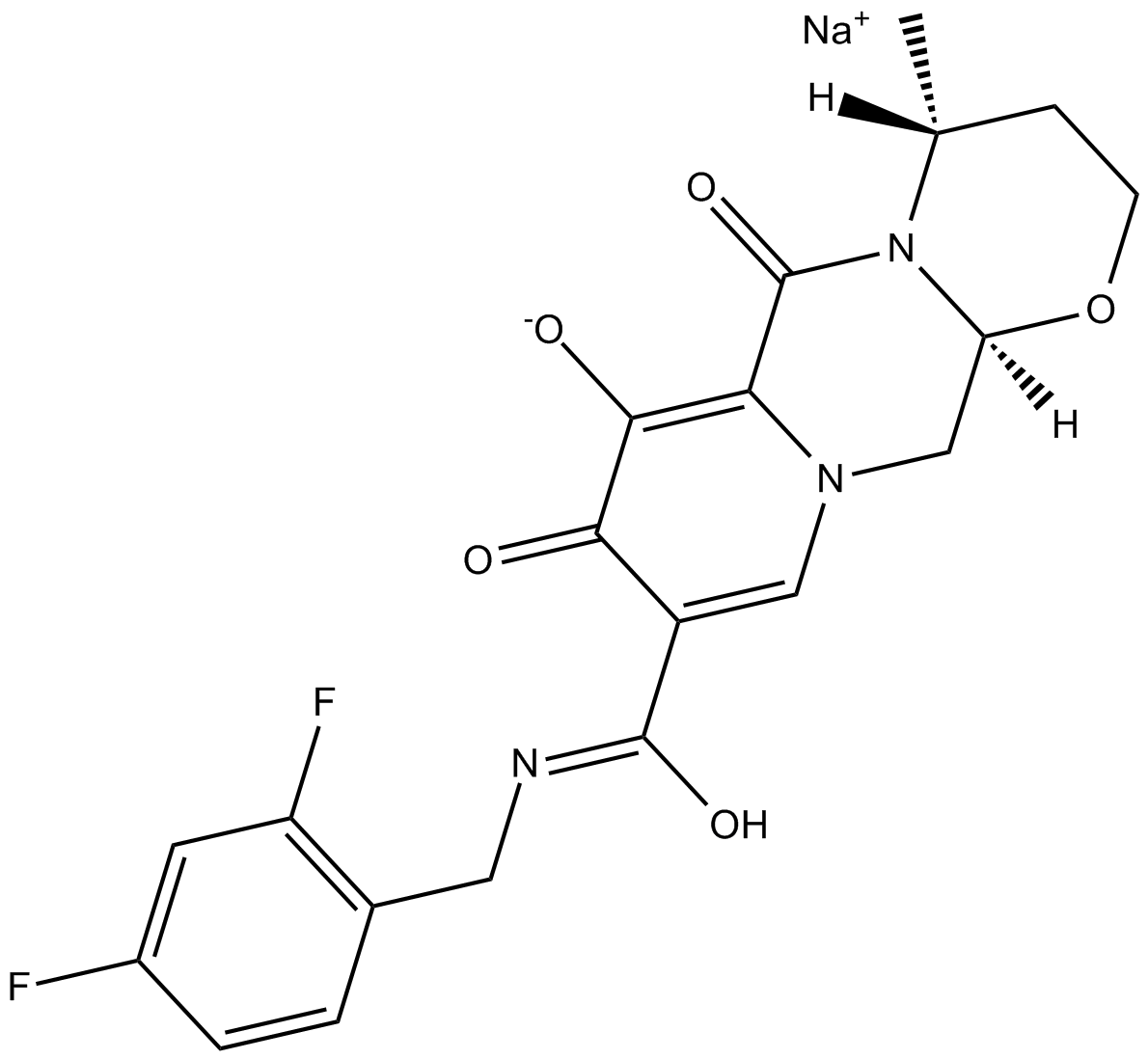
GC14057
GSK1349572 sodiuM salt
GSK1349572 sodiuM salt (S/GSK1349572 sodium) is a highly potent and orally bioavailable HIV integrase strand transfer inhibitor with an IC50 of 2.7 nM for HIV-1 integrase-catalyzed strand transfer.
-
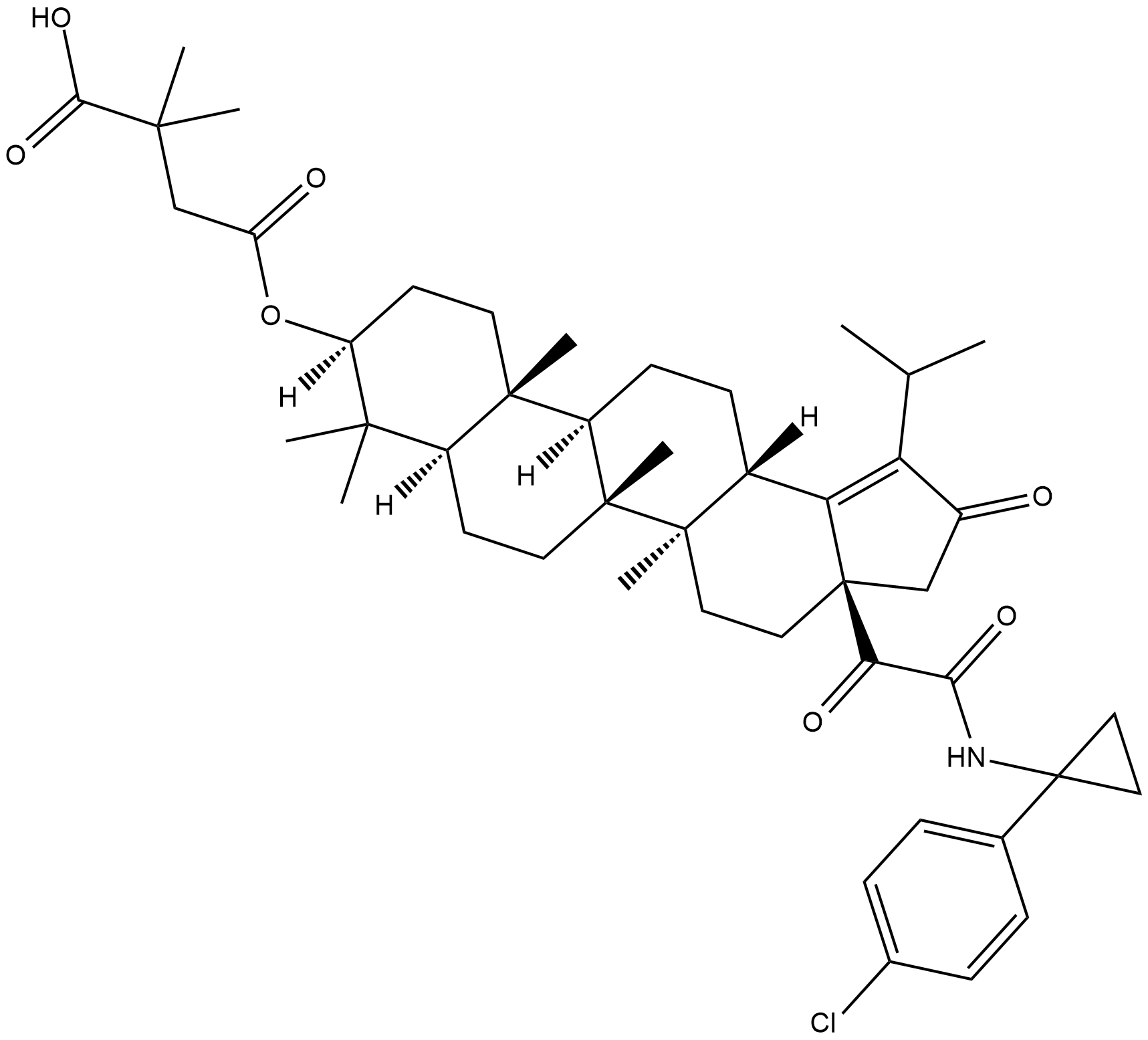
GC18484
GSK2578999A
-
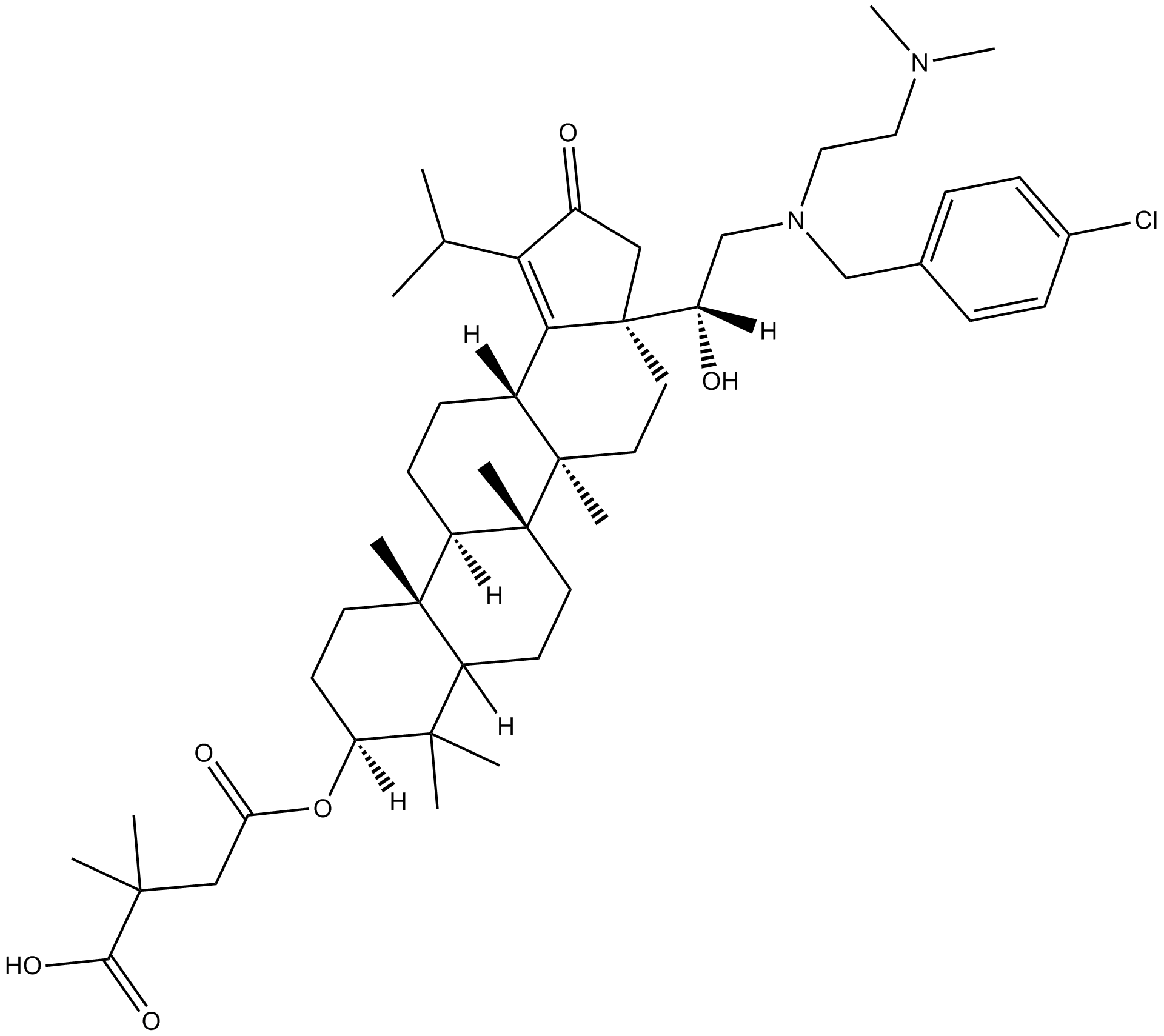
GC12329
GSK2838232
HIV-1 maturation inhibitor
-
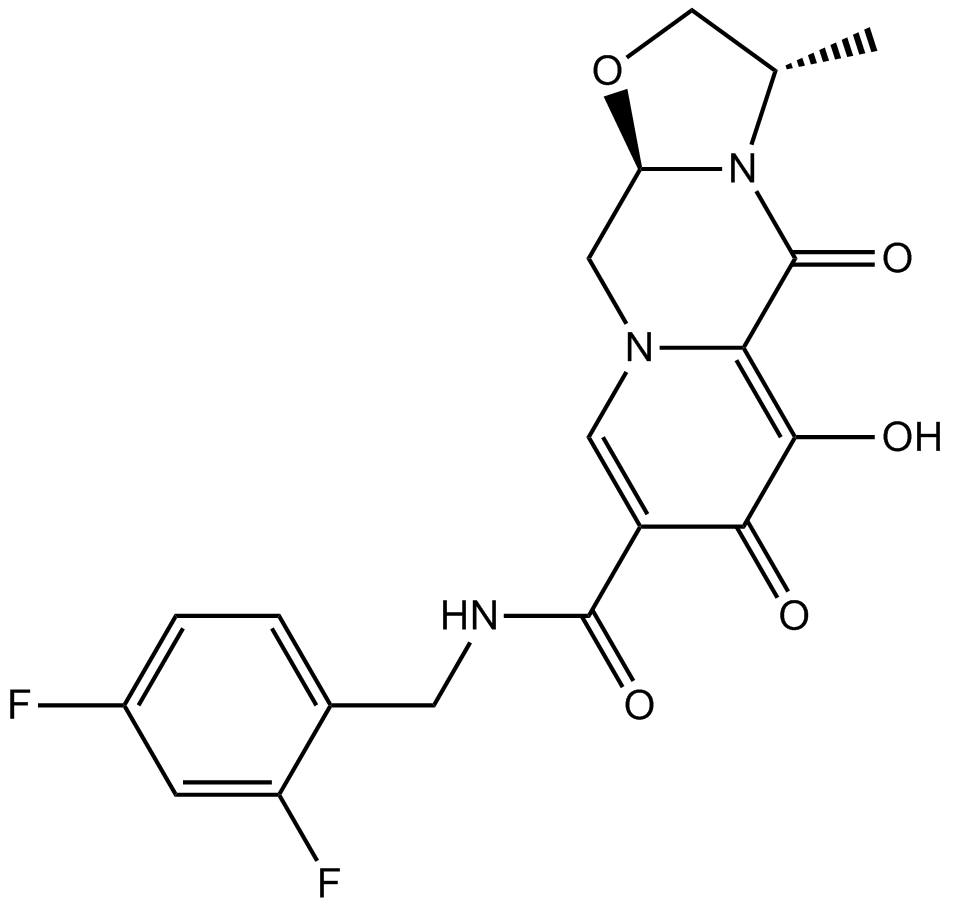
GC16215
GSK744 (S/GSK1265744)
GSK744 (S/GSK1265744) (GSK-1265744) is a orally active and long-acting HIV integrase inhibitor and organic anion transporter 1/3 (OAT1/OAT3) inhibitor with IC50 values of 2.5 nM, 0.41 μM and 0.81 μM for HIVADA, OAT3 and OAT1, respectively.
-
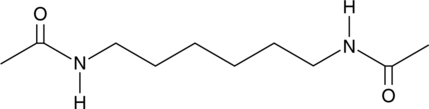
GC49027
Hexamethylene bisacetamide
Hexamethylene bisacetamide is a tumor cell-differentiating agent.
-
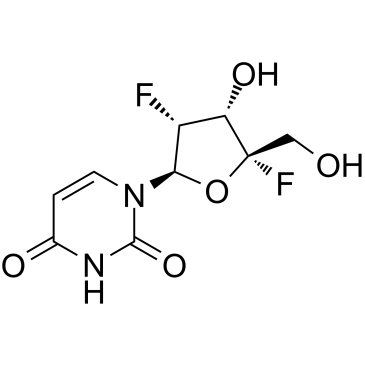
GC36232
HIV-1 inhibitor-3
HIV-1 inhibitor-3 is a HIV infection inhibitor extracted from patent US2018360927.
-
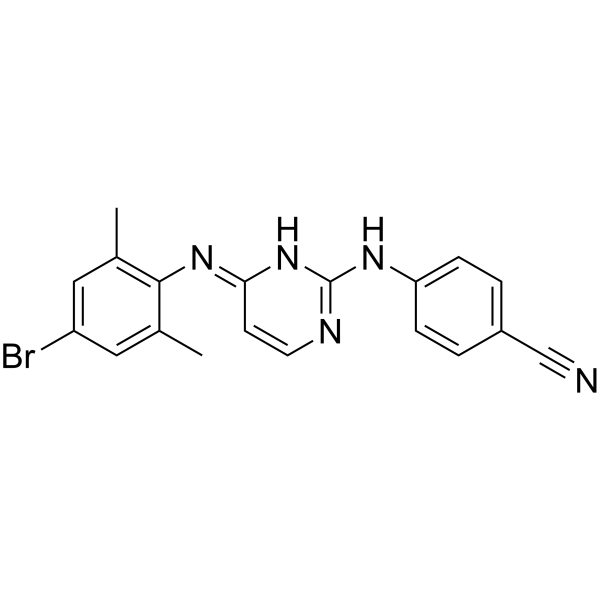
GC68023
HIV-1 inhibitor-48
-
GC65256
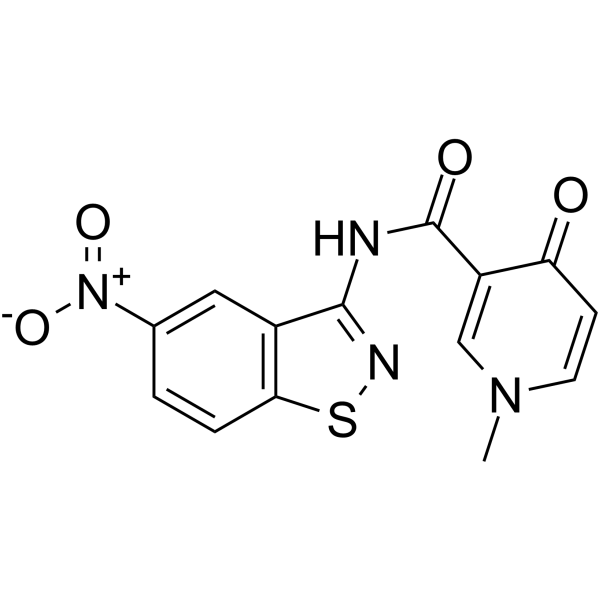
HIV-1 inhibitor-6
HIV-1 inhibitor-6 (compound 9), a diheteroarylamide-based compound, is a potent HIV-1 pre-mRNA alternative splicing inhibitor.
-
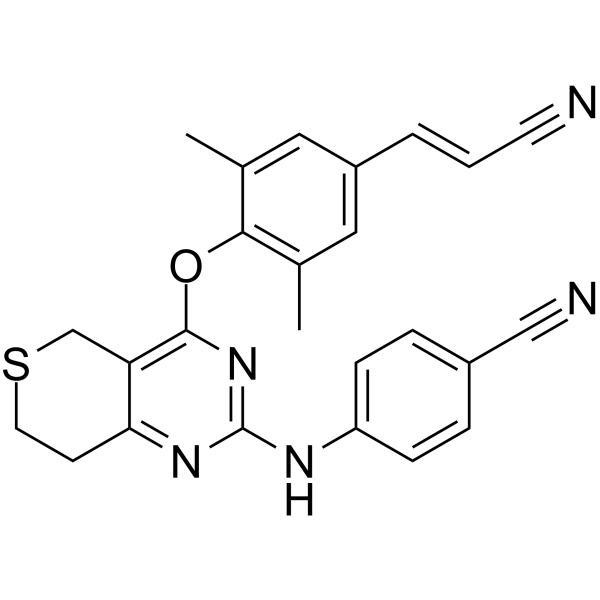
GC65158
HIV-1 inhibitor-8
HIV-1 inhibitor-8 is an orally active, low-toxicity and potent HIV?1 non-nucleoside reverse transcriptase inhibitor (NNRTI).
-
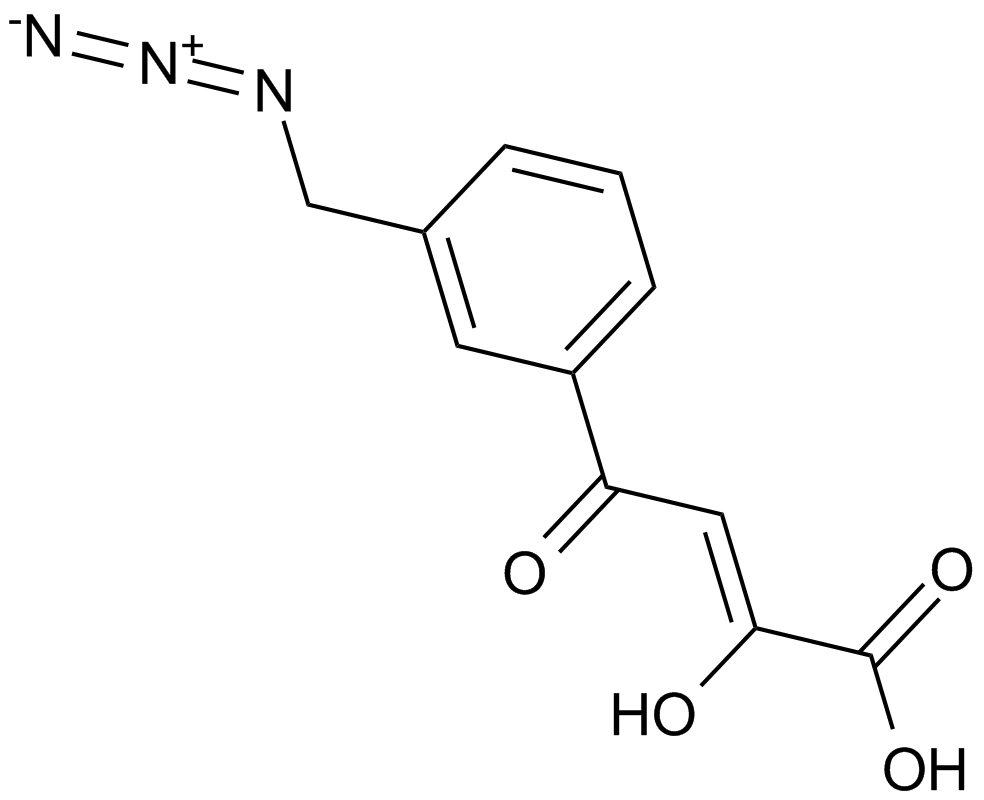
GC12998
HIV-1 integrase inhibitor
-
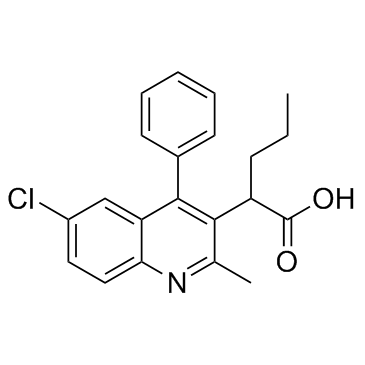
GC36233
HIV-1 integrase inhibitor 2
HIV-1 integrase inhibitor 2 (CX05168) is a quinoline-based protein-protein interaction inhibitor of LEDGF/p75 and HIV integrase.
-
GC36234
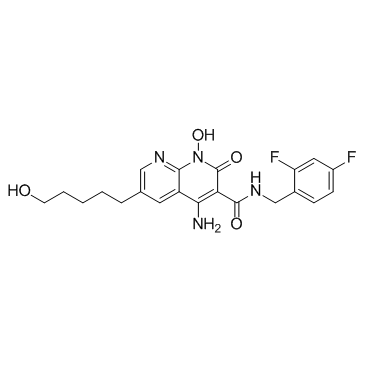
HIV-1 integrase inhibitor 3
HIV-1 integrase inhibitor 3 is a HIV-1 integrase strand transfer (INST) inhibitor with an IC50 of 2.7 nM.
-
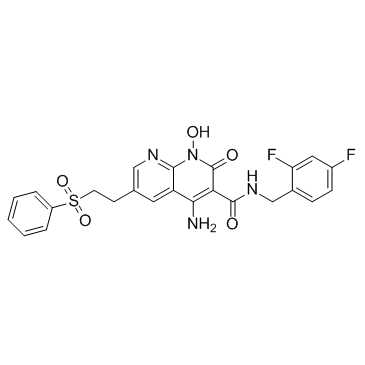
GC36235
HIV-1 integrase inhibitor 4
HIV-1 integrase inhibitor 4 is a HIV-1 integrase strand transfer (INST) inhibitor with an IC50 of 3.7 nM.
-
GC32342
HIV-1 Rev 34-50 (HIV-1 rev Protein (34-50))
HIV-1 Rev 34-50 (HIV-1 rev Protein (34-50)) is a 17-aa peptide derived from the Rev-responsive element (RRE)-binding domains of Rev in HIV-1, with anti-HIV-1 activity.

-
GC65134
HIV-IN-6
HIV-IN-6 is a HIV-Ⅰ viral replication inhibitor by targeting Src family kinases (SFK) that interact with Nef protein of the virus, such as Hck.

-
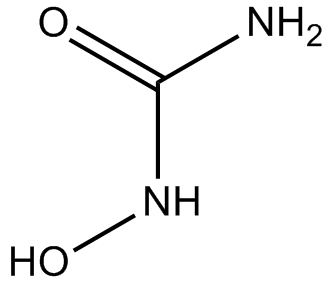
GC16843
Hydroxyurea
DNA synthesis inhibitor
-
GC65034
Ibalizumab
Ibalizumab (TMB-355) is a humanised IgG4 monoclonal antibody that prevents HIV cell entry by binding to CD4 receptor.

-
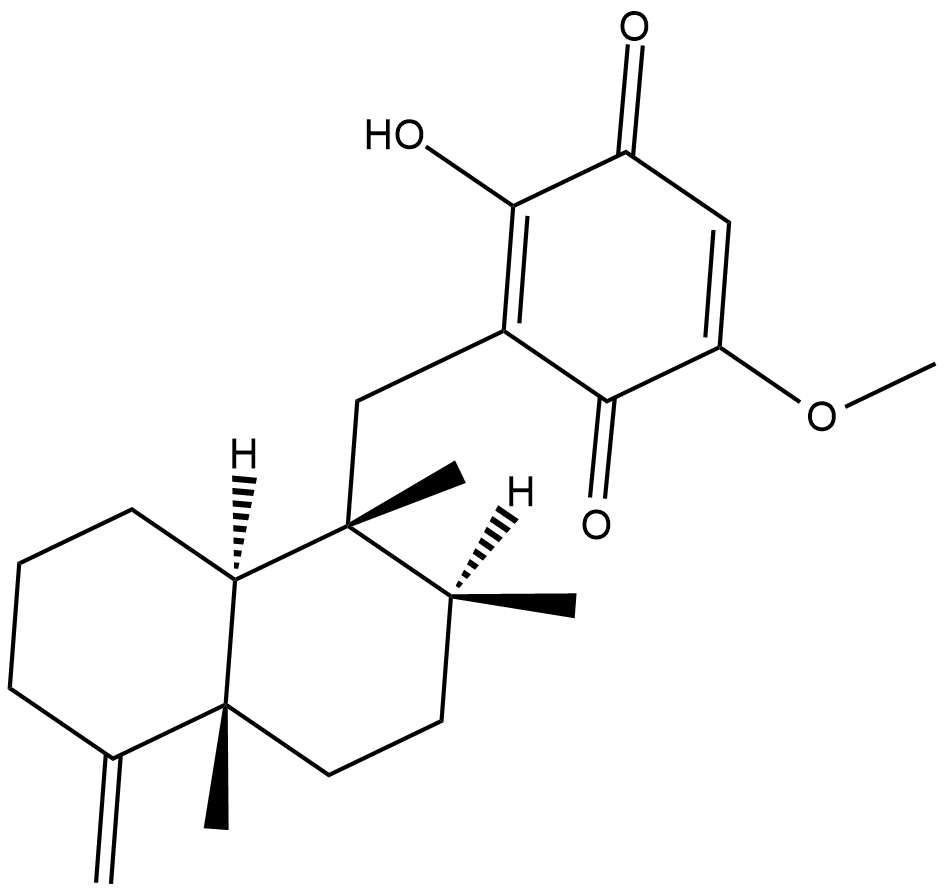
GC18545
Ilimaquinone
Ilimaquinone is a natural sesquiterpene quinone that has antimicrobial, anti-HIV, anti-mitotic, and anti-inflammatory properties.
-
GC61820
IMB-301
IMB-301 is a specific HIV-1 replication inhibitor that binds to hA3G (human APOBEC3G), interrupts the hA3G-Vif interaction and inhibits Vif-mediated degradation of hA3G.
-
GC36309
Indinavir
Indinavir (MK-639 free base) is an orally active and selective HIV-1 protease inhibitor with a Ki of 0.54 nM for PR.