Signaling Pathways
The antibody-drug conjugate (ADC), a humanized or human monoclonal antibody conjugated with highly cytotoxic small molecules (payloads) through chemical linkers, is a novel therapeutic format and has great potential to make a paradigm shift in cancer chemotherapy. The three components of the ADC together give rise to a powerful oncolytic agent capable of delivering normally intolerable cytotoxins directly to cancer cells, which then internalize and release the cell-destroying drugs. At present, two ADCs, Adcetris and Kadcyla, have received regulatory approval with >40 others in clinical development.
ADCs are administered intravenously in order to prevent the mAb from being destroyed by gastric acids and proteolytic enzymes. The mAb component of the ADC enables it to circulate in the bloodstream until it finds and binds to tumor-specific cell surface antigens present on target cancer cells. Linker chemistry is an important determinant of the safety, specificity, potency and activity of ADCs. Linkers are designed to be stable in the blood stream (to conform to the increased circulation time of mAbs) and labile at the cancer site to allow rapid release of the cytotoxic drug. First generation ADCs made use of early cytotoxins such as the anthracycline, doxorubicin or the anti-metabolite/antifolate agent, methotrexate. Current cytotoxins have far greater potency and can be divided into three main groups: auristatins, maytansines and calicheamicins.
The development of site-specific conjugation methodologies for constructing homogeneous ADCs is an especially promising path to improving ADC design, which will open the way for novel cancer therapeutics.
References:
[1] Tsuchikama K, et al. Protein Cell. 2016 Oct 14. DOI:10.1007/s13238-016-0323-0.
[2] Peters C, et al. Biosci Rep. 2015 Jun 12;35(4). pii: e00225. doi: 10.1042/BSR20150089.
Targets for Signaling Pathways
- Proteases(29)
- Apoptosis(824)
- Chromatin/Epigenetics(15)
- Metabolism(336)
- MAPK Signaling(26)
- Tyrosine Kinase(73)
- DNA Damage/DNA Repair(49)
- PI3K/Akt/mTOR Signaling(38)
- Microbiology & Virology(42)
- Cell Cycle/Checkpoint(168)
- Ubiquitination/ Proteasome(24)
- JAK/STAT Signaling(10)
- TGF-β / Smad Signaling(21)
- Angiogenesis(59)
- GPCR/G protein(3)
- Stem Cell(19)
- Membrane Transporter/Ion Channel(161)
- Cancer Biology(366)
- Endocrinology and Hormones(152)
- Neuroscience(367)
- Obesity, Appetite Control & Diabetes(17)
- Peptide Inhibitors and Substrate(2)
- Other Signal Transduction(156)
- Immunology/Inflammation(934)
- Cardiovascular(98)
- Vitamin D Related(0)
- Antibody-drug Conjugate/ADC Related(0)
- PROTAC(212)
- Ox Stress Reagents(24)
- Others(4010)
- Antiparasitics(6)
- Toxins(62)
Products for Signaling Pathways
- Cat.No. Product Name Information
-
GC14729
(R)-Crizotinib
A c-MET and ALK receptor tyrosine kinase inhibitor
-
GC62738
(R)-eIF4A3-IN-2
(R)-eIF4A3-IN-2 is a less active enantiomer of eIF4A3-IN-2.
-
GC69812
(R)-Elexacaftor
(R)-Elexacaftor is the enantiomer of Elexacaftor (Compound 1) and comes from Compound 37 in patent WO2018107100A1. It is a corrector of cystic fibrosis transmembrane conductance regulator (CFTR), with an EC50 value of 0.29 uM for CFTR dF508.
-
GC67857
(R)-Elsubrutinib

-
GC64179
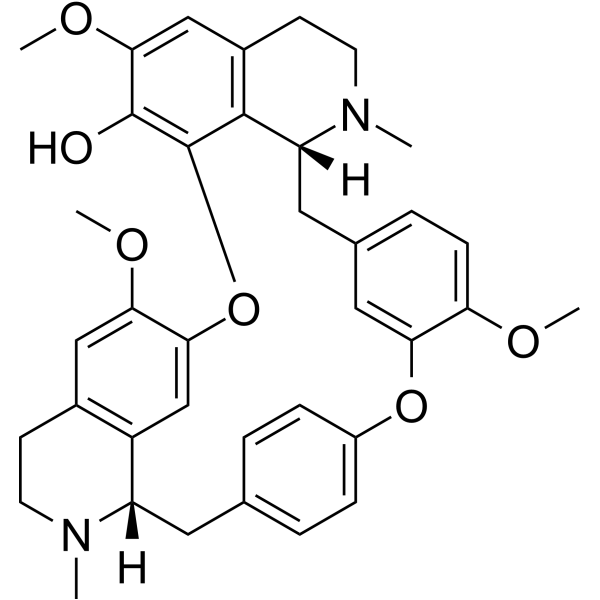
(R)-Fangchinoline
(R)-Fangchinoline (Thalrugosine), a alkaloids from genus Stephania,exhibits antimicrobial and hypotensive activity.
-
GC65384
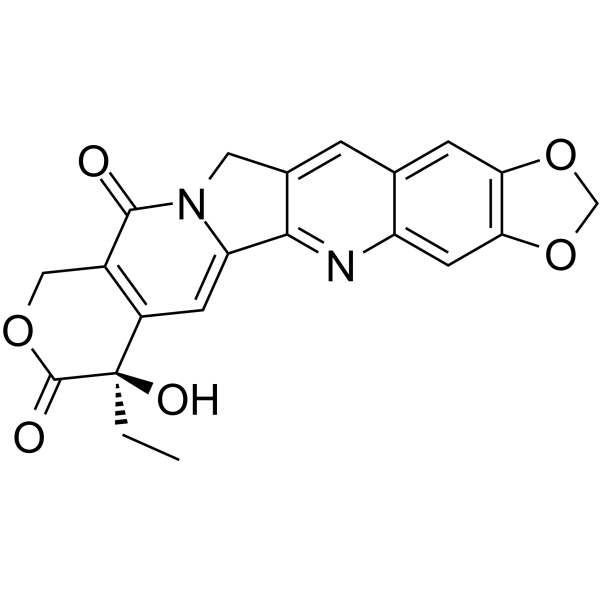
(R)-FL118
(R)-FL118 (10,11-(Methylenedioxy)-20(R)-camptothecin) is the R-enantiomer of FL118. (R)-FL118 shows anticancer activity.
-
GC62739
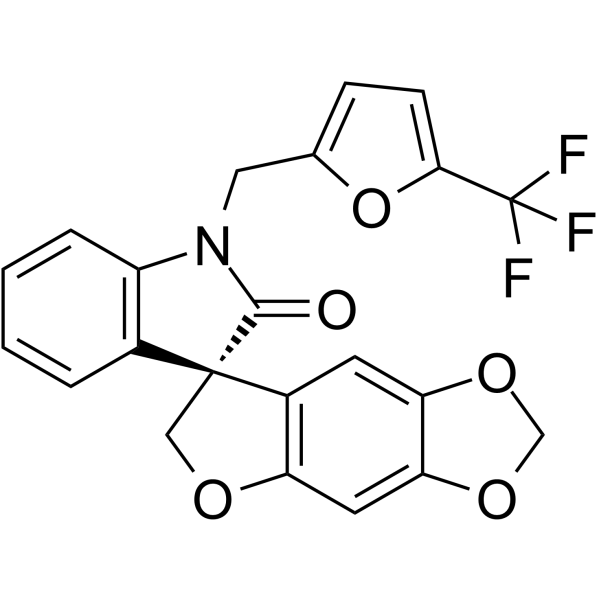
(R)-Funapide
(R)-Funapide ((R)-TV 45070) is the less active R-enantiomer of Funapide.
-
GC64210
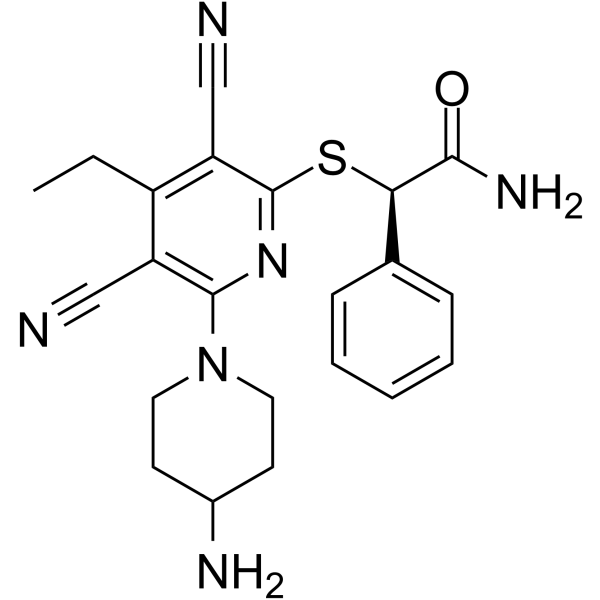
(R)-GSK-3685032
(R)-GSK-3685032 is the R-enantiomer of GSK-3685032. GSK-3685032 is a non-time-dependent, noncovalently, first-in-class reversible DNMT1-selective inhibitor, with an IC50 of 0.036 μM. GSK-3685032 induces robust loss of DNA methylation, transcriptional activation, and cancer cell growth inhibition.
-
GC52290
(R)-HTS-3
An inhibitor of LPCAT3
-
GC49066
(R)-Hydroxychloroquine (sulfate)
An isomer of hydroxychloroquine
-
GC65373
(R)-IL-17 modulator 4
(R)-IL-17 modulator 4 is the R-configure of IL-17 modulator 4.

-
GC67864
(R)-Irsenontrine

-
GC61847
(R)-Lanicemine
(R)-Lanicemine ((R)-AZD6765) is the less active R-enantiomer of Lanicemine.
-
GC66419
(R)-Lercanidipine-d3 hydrochloride
(R)-lercanidipine D3 (hydrochloride) is a deuterium labeled (R)-Lercanidipine hydrochloride. (R)-Lercanidipine D3 (hydrochloride), the R-enantiomer of Lercanidipine, is a calcium channel blocker.
-
GC62740
(R)-M8891
(R)-M8891 (compound R-9) is a less active isomer of M8891.
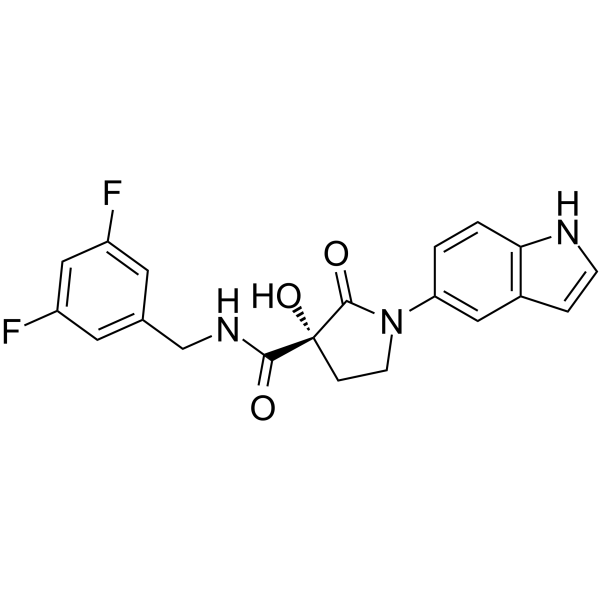
-
GC67937
(R)-Mirtazapine

-
GC61858
(R)-MLN-4760
(R)-MLN-4760, the R-enantiomer of MLN-4760, is an ACE2 inhibitor, with an IC50 of 8.4 μM.

-
GC68475
(R)-Nicardipine

-
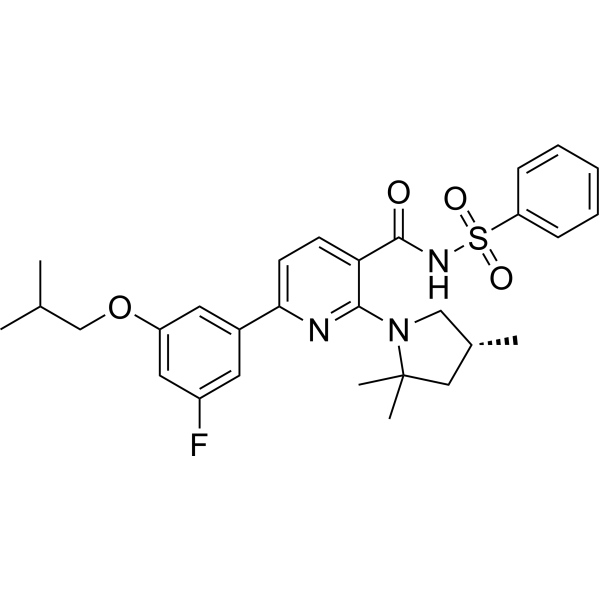
GC67767
(R)-Olacaftor
-
GC64366
(R)-ONO-2952
(R)-ONO-2952 is a R-enantiomer of ONO-2952.
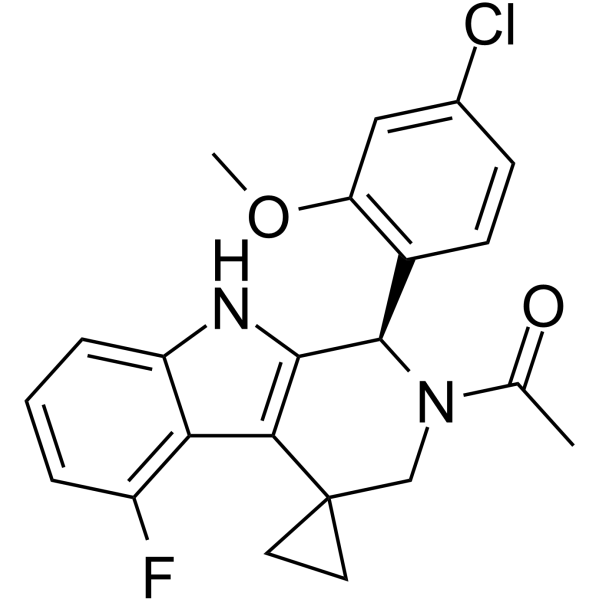
-
GC49253
(R)-P7C3-Ome
An analog of P7C3-A20
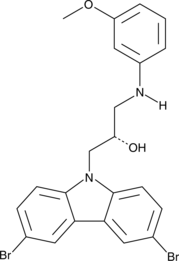
-
GC67717
(R)-PF-04991532
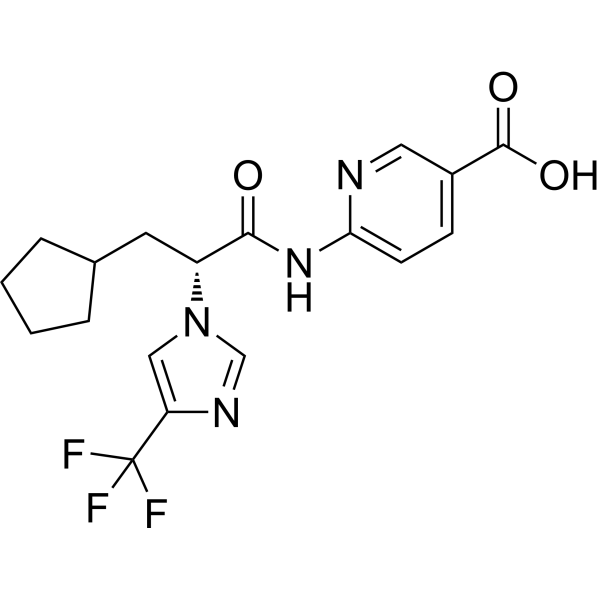
-
GC62741
(R)-PF-06256142
(R)-PF-06256142 is the R enantiomer of PF-06256142 with low active.
-
GC62742
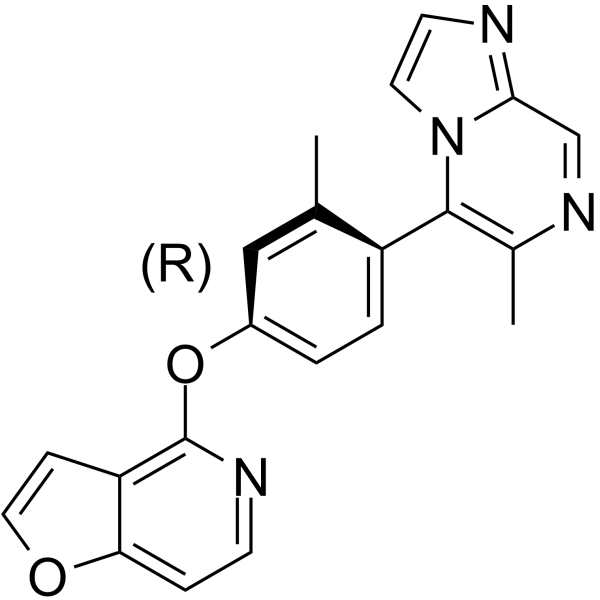
(R)-Pirtobrutinib
(R)-Pirtobrutinib ((R)-LOXO-305) is a less active enantiomer of Pirtobrutinib.
-
GC62513
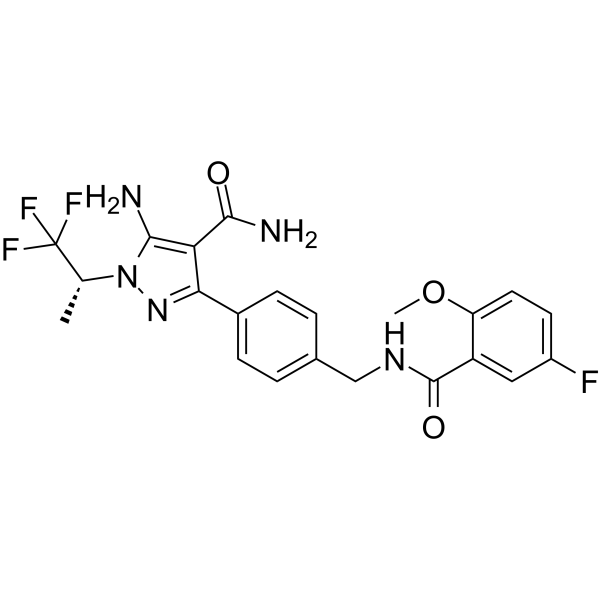
(R)-Posenacaftor sodium
(R)-Posenacaftor (R)-PTI-801) sodium is the R enantiomer of Posenacaftor.
-
GC68408
(R)-Praziquantel-d11
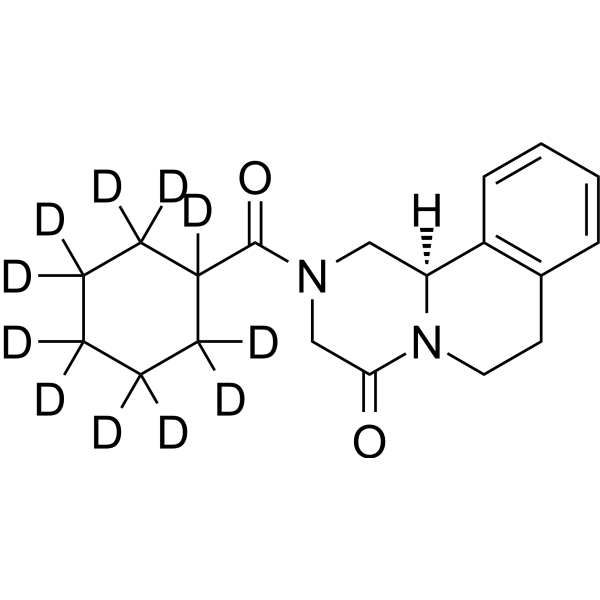
-
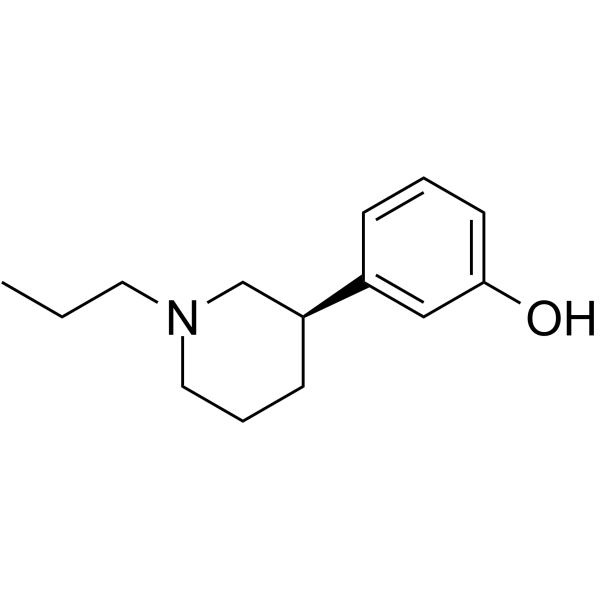
GC68383
(R)-Preclamol
-
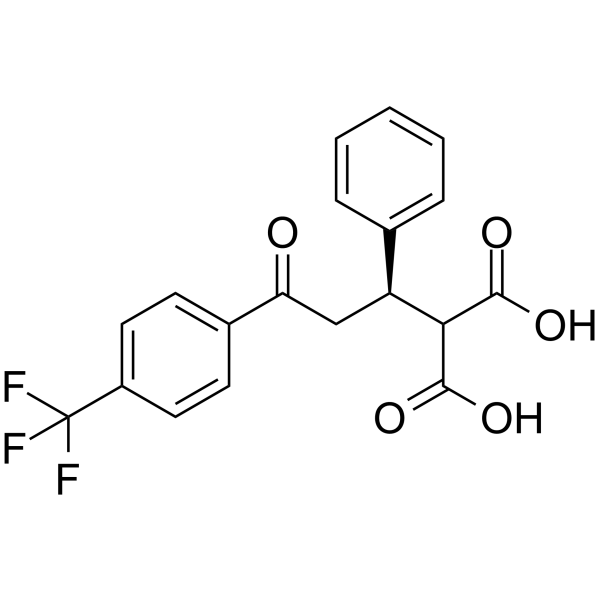
GC63802
(R)-PS210
(R)-PS210, the R enantiomer of PS210 (compound 4h-eutomer), is a substrate-selective allosteric activator of PDK1 with an AC50 value of 1.8 μM. (R)-PS210 targets to the PIF-binding pocket of?PDK1. PIF: The protein kinase C-related kinase 2 (PRK2)-interacting fragment.
-
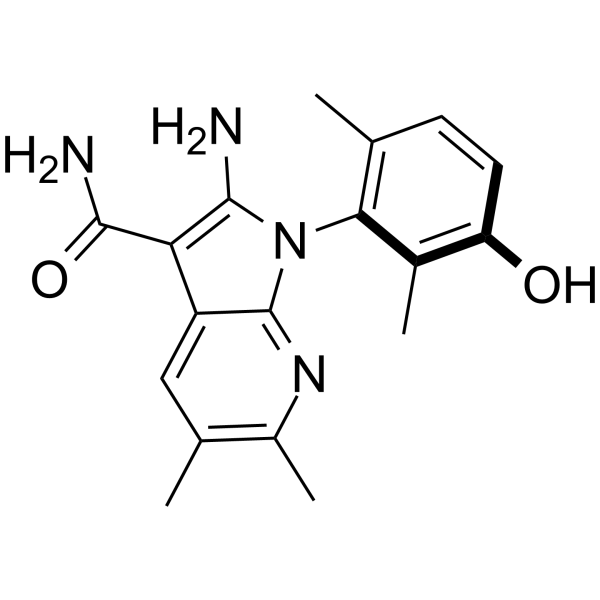
GC65591
(R)-RP-6306
(R)-RP-6306 (183) can be used for the research of Myt1 mediated diseases and kinds of cancer for slowing the progression of cancer.
-
GC69839
(R)-Tegoprazan
(R)-Tegoprazan ((R)-CJ-12420; example 3) is a benzimidazole derivative and an effective inhibitor of gastric H+/K+-ATPase. Its IC50 for canine kidney Na+/K+-ATPase is 98 nM. (R)-Tegoprazan has potential in the research of gastrointestinal diseases.
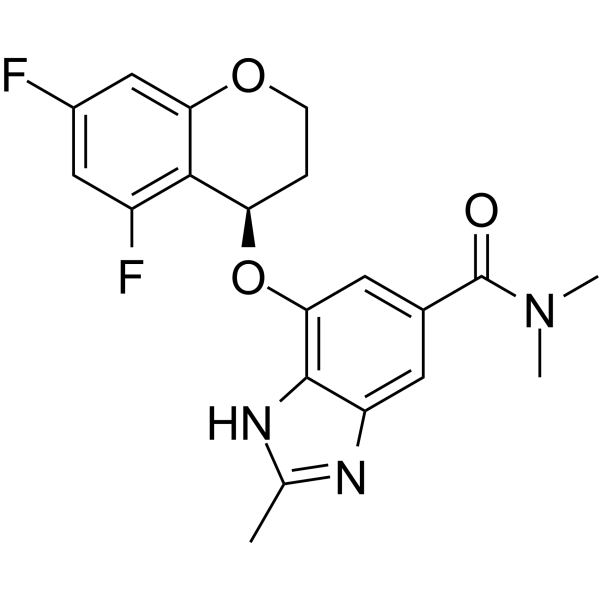
-
GC69847
(R)-VT104
(R)-VT104 is the R-enantiomer of VT104. The IC50 value of (R)-VT104 for firefly luciferase is 0.1-1 μμ. VT104 is an orally active pan-TEAD deacetylase inhibitor.
-
GC65884
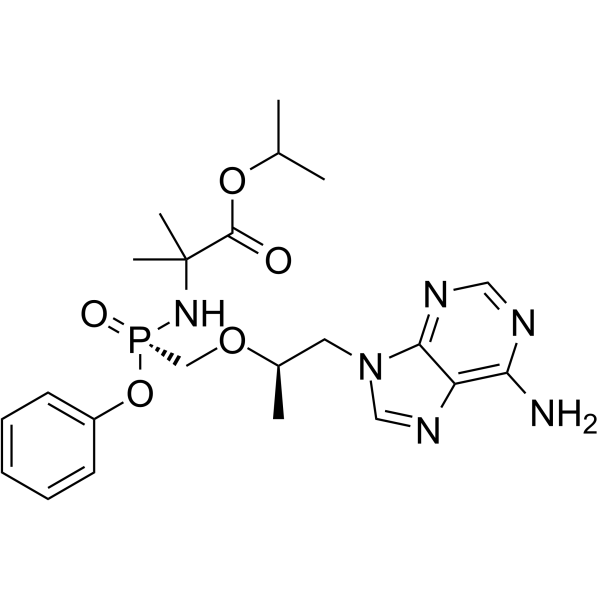
(R,1R)-Tenofovir amibufenamide
(R,1R)-Tenofovir amibufenamide ((R,1R)-HS-10234) can be used for the purifying a tenofovir prodrug. Tynofovir (tenofovir) is a nucleoside acids reverse transcriptase inhibitors.
-
GC63983
(R,R)-BAY-Y 3118
(R,R)-BAY-Y 3118 is the R-enantiomer of BAY-Y 3118.
-
GC62588
(R,R)-CXCR2-IN-2
(R,R)-CXCR2-IN-2, diastereoisomer of CXCR2-IN-2 (compound 68), is a brain penetrant CXCR2 antagonist with a pIC50 of 9 and 6.8 in the Tango assay and d in the HWB Gro-α induced CD11b expression assay, respectively.
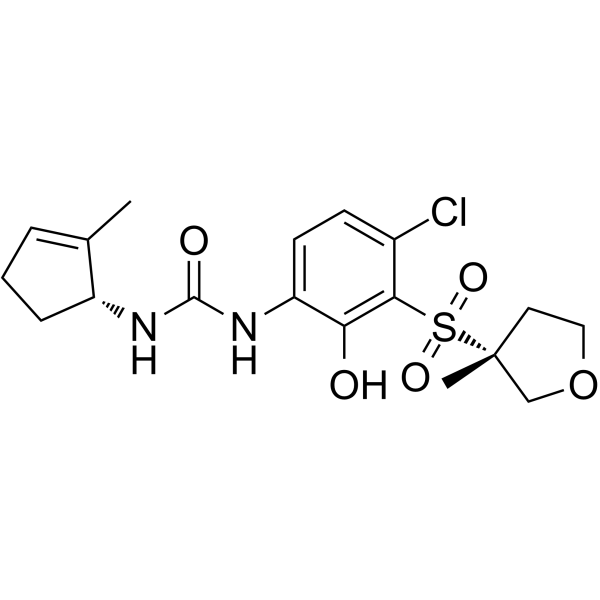
-
GC14145
(R,R)-Formoterol
β2-selective adrenergic agonist
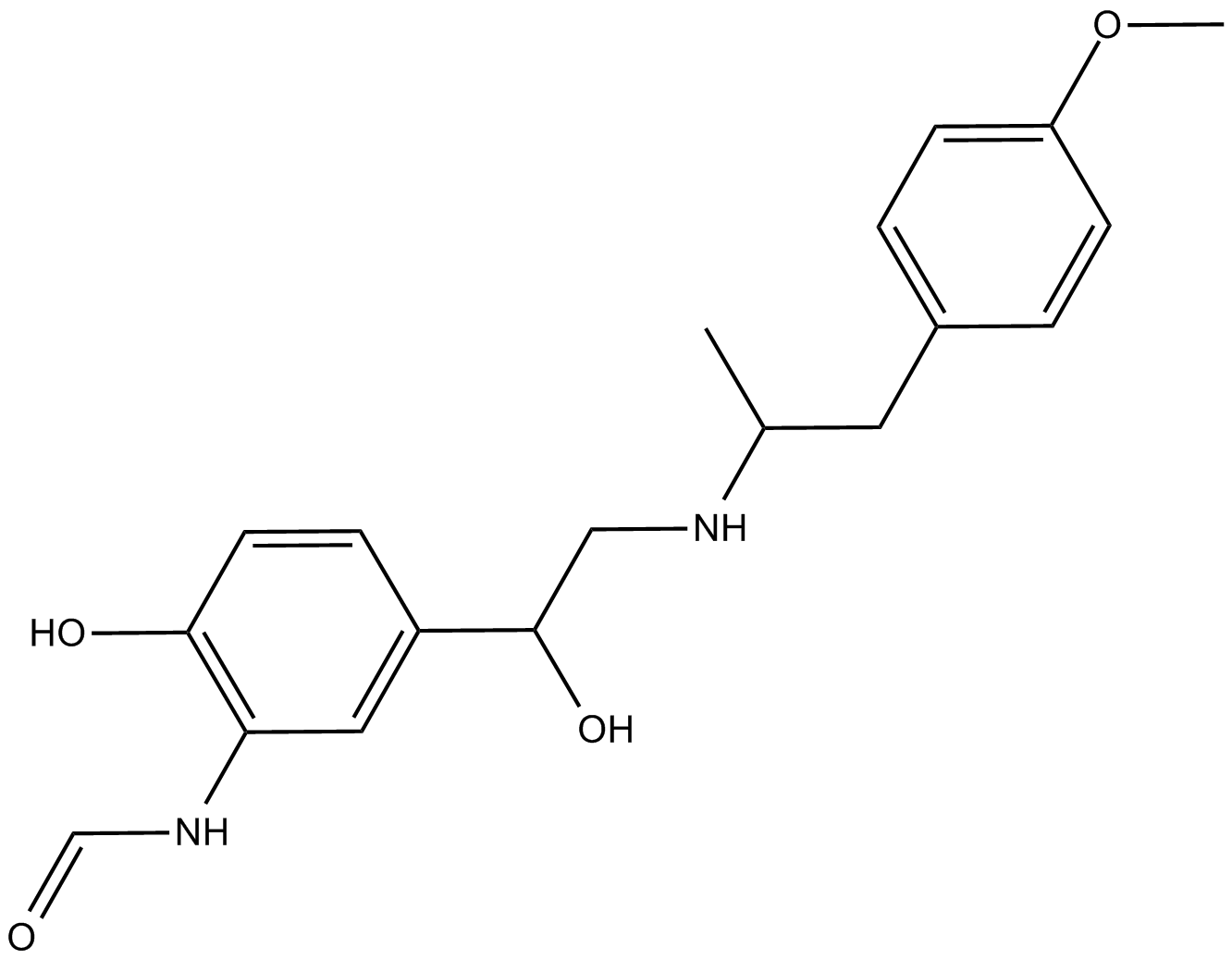
-
GC69836
(R,R)-VVD-118313
(R,R)-VVD-118313 is an isomer of VVD-118313. VVD-118313 is a selective JAK1 inhibitor that can block JAK1-dependent phosphorylation and cytokine signaling. VVD-118313 can be used for cancer research.

-
GC52185
(R,S)-Anatabine-d4

-
GC69837
(R/S)-Alicaforsen
(R/S)-Alicaforsen is the racemic form of Alicaforsen, which consists of both R and S configurations. Alicaforsen is a 20-base length antisense oligonucleotide that inhibits the production of ICAM-1, an important adhesion molecule involved in the migration and transport process of white blood cells to inflammatory sites.

-
GC63452
(Rac)-5-Hydroxymethyl Tolterodine
(Rac)-5-Hydroxymethyl Tolterodine ((Rac)-Desfesoterodine), an active metabolite of Tolterodine, is a mAChR antagonist (Ki values of 2.3 nM, 2 nM, 2.5 nM, 2.8 nM, and 2.9 nM for M1, M2, M3, M4, and M5 receptors, respectively).
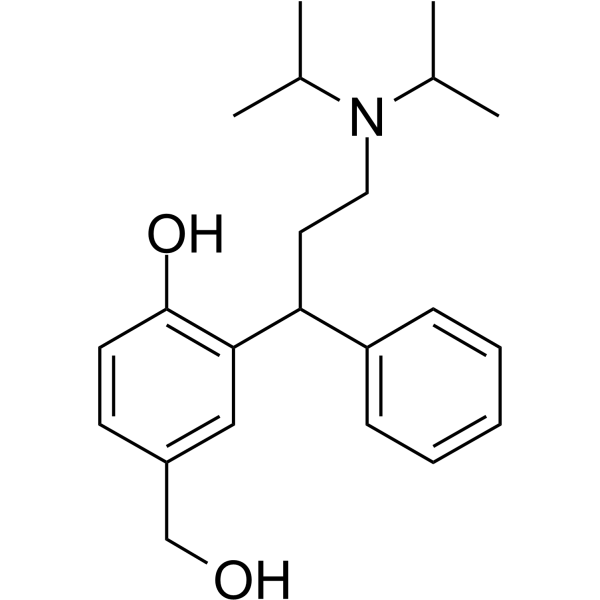
-
GC61548
(Rac)-5-Keto Fluvastatin
(Rac)-5-Keto Fluvastatin (3-Hydroxy-5-Keto Fluvastatin) is an impurity of Fluvastatin (XU 62320).
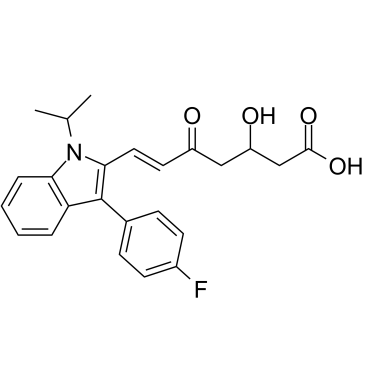
-
GC61488
(Rac)-Acolbifene
(Rac)-Acolbifene (EM-343; (Rac)-EM-652) is the racemic form of EM652 (estrogen receptor?antagonist), has anti-estrogenic and estrogenic activities. (Rac)-Acolbifene (EM-343; (Rac)-EM-652) contains a piperidine ring, shows good pharmacological profile,relative binding affinity (RBA)=380.
-
GC69794
(Rac)-Arnebin 1
(Rac)-Arnebin 1 is the racemic form of β,β-Dimethylacrylalkannin and/or β,β-Dimethylacrylshikonin. These compounds are naphthoquinones isolated from plants in the Lithospermum genus, with β,β-Dimethylacrylshikonin exhibiting anti-tumor activity.
-
GC68414
(Rac)-Atropine-d3
-
GC69795
(Rac)-BIO8898
(Rac)-BIO8898 is a CD40-CD154 co-stimulatory interaction inhibitor. (Rac)-BIO8898 inhibits the binding of CD154 to CD40-Ig, with an IC50 of 25 μM.
-
GC65477
(Rac)-CFT7455
(Rac)-CFT7455 is a zinc finger transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) degrader, acting via the ubiquitin proteasome pathway, with a GI50 of 0.05 nM for NCIH929.1 cells. (Rac)-CFT7455 is the racemic isomer of CFT7455 which is a IKZF1/IKZF3 degrader with anticancer activity.
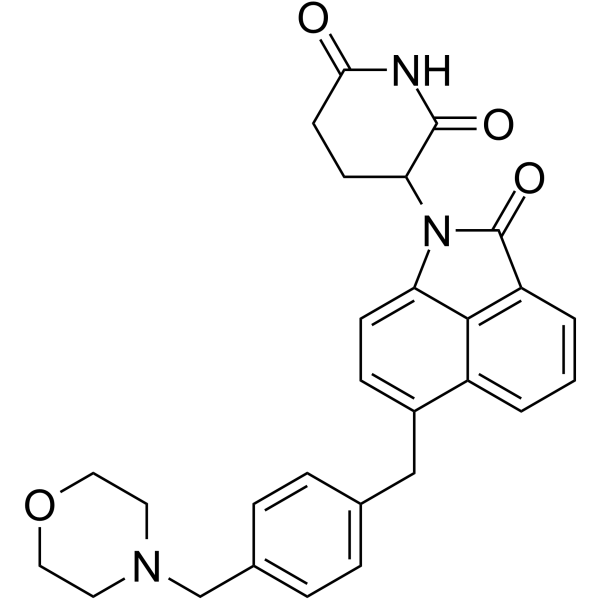
-
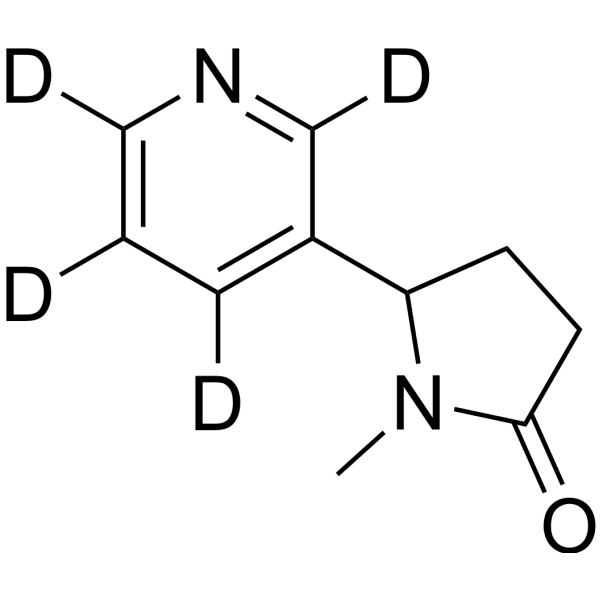
GC67993
(Rac)-Cotinine-d4
-
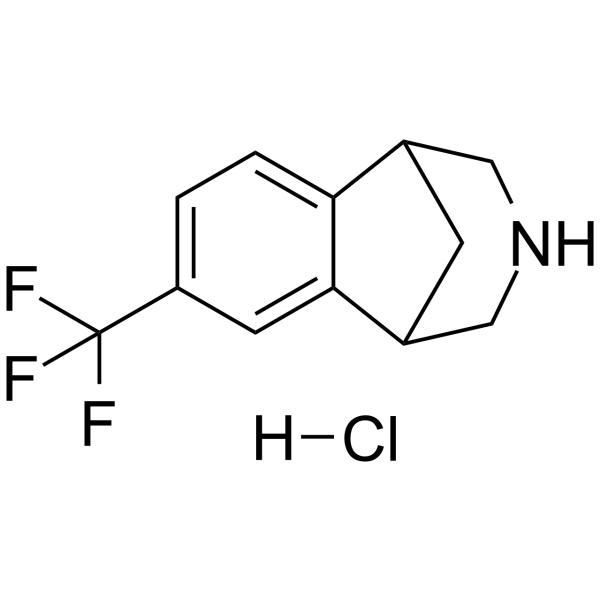
GC63511
(Rac)-CP-601927 hydrochloride
(Rac)-CP-601927 hydrochloride is the racemate of CP-601927.
-
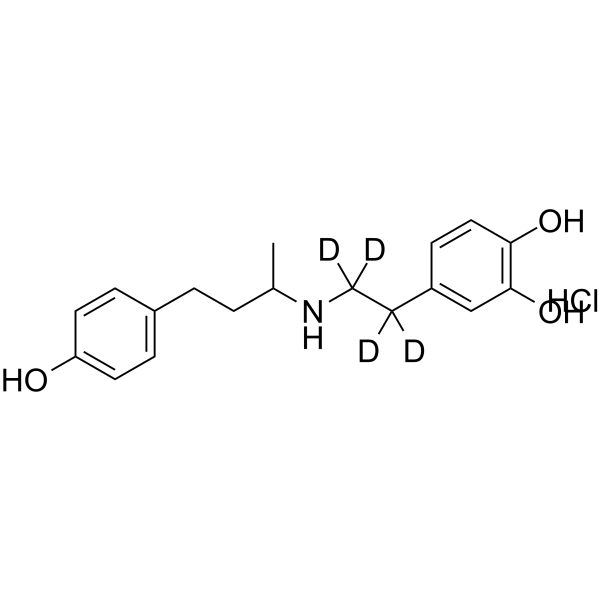
GC64481
(rac)-Dobutamine-d4 hydrochloride
-
GC62642
(rac)-Dobutamine-d6 hydrochloride
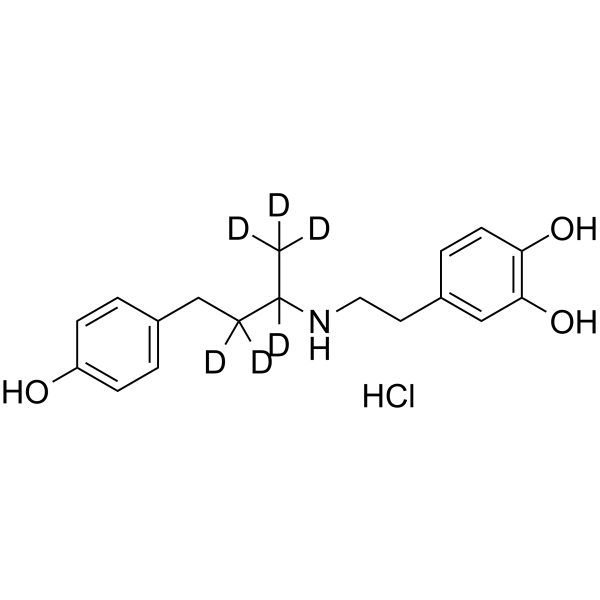
-
GC62743
(Rac)-EC5026
(Rac)-EC5026 ((Rac)-BPN-19186) is a potent piperidine inhibitor of soluble epoxide hydrolase (sEH) extracted from patent WO2019156991A1, page 39, has a Ki of 0.06 nM.
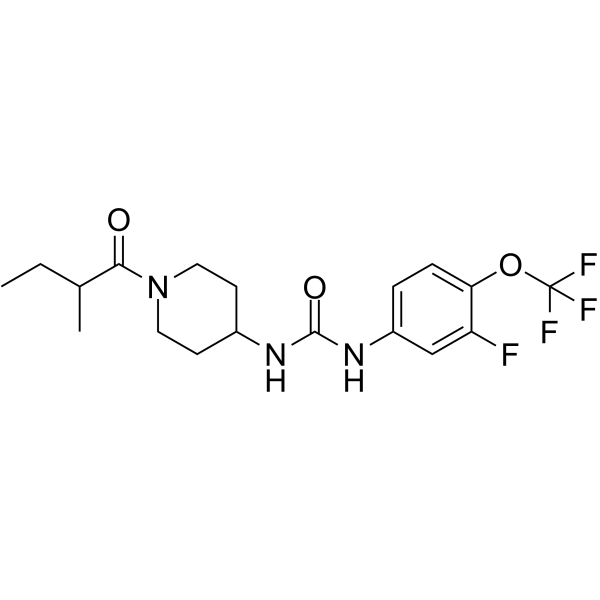
-
GC69796
(Rac)-Etavopivat
(Rac)-Etavopivat ((Rac)-FT-4202) is an isomer of Etavopivat. Etavopivat is an orally active activator of red blood cell pyruvate kinase-R (PKR), which can be used for research on sickle cell disease and other hemoglobin disorders.
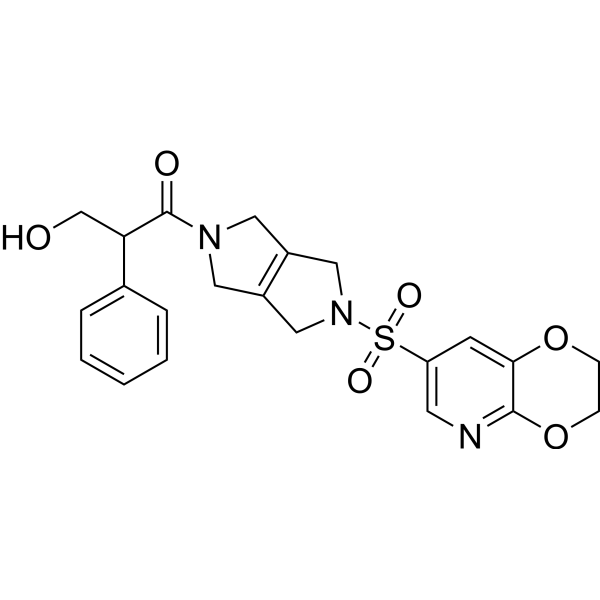
-
GC62332
(Rac)-Finerenone
(Rac)-Finerenone ((Rac)-BAY 94-8862) is the racemate of Finerenone.
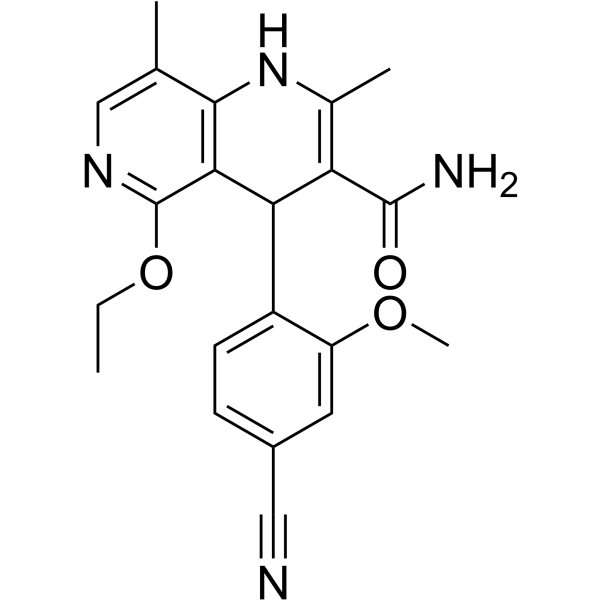
-
GC62528
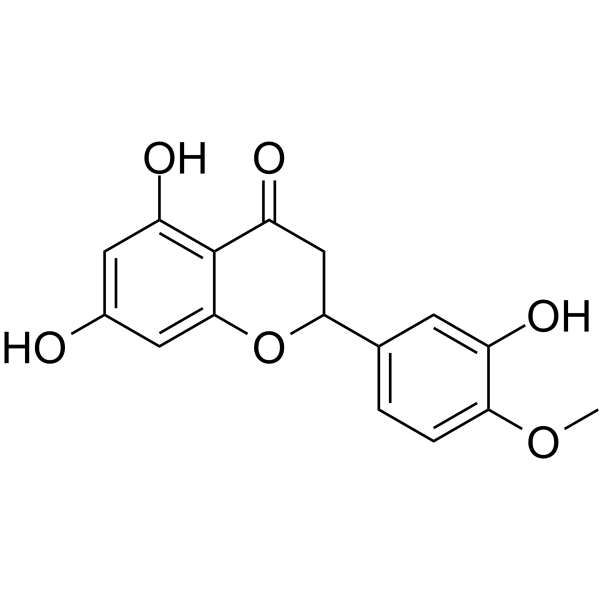
(Rac)-Hesperetin
(Rac)-Hesperetin is the racemate of Hesperetin.
-
GC61750
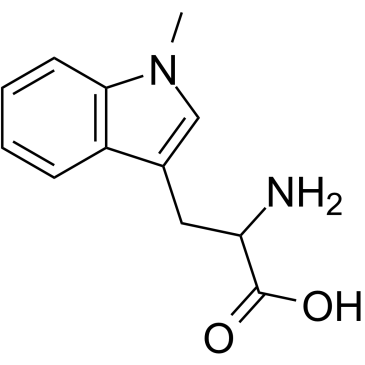
(Rac)-Indoximod
(Rac)-Indoximod (1-Methyl-DL-tryptophan) is an indoleamine 2,3-dioxygenase (IDO) inhibitor.
-
GC61446
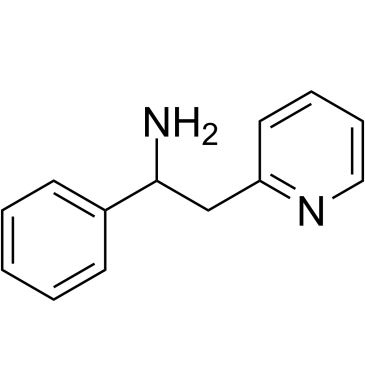
(Rac)-Lanicemine
(Rac)-Lanicemine ((Rac)-AZD6765) is the racemate of Lanicemine.
-
GC63632
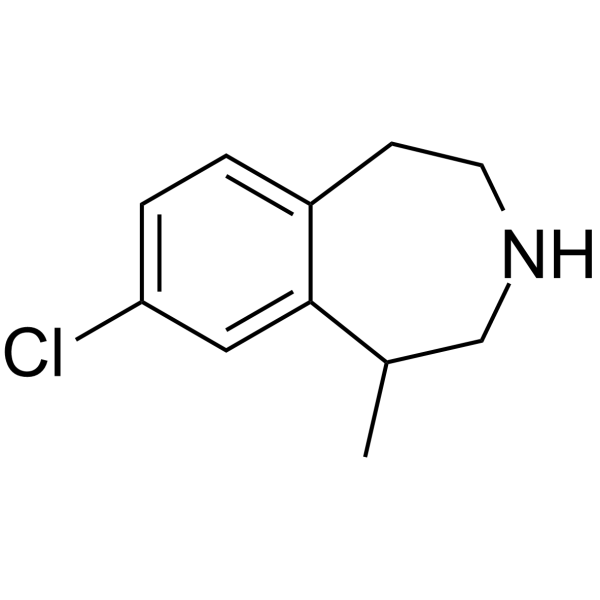
(Rac)-Lorcaserin
-
GC63382
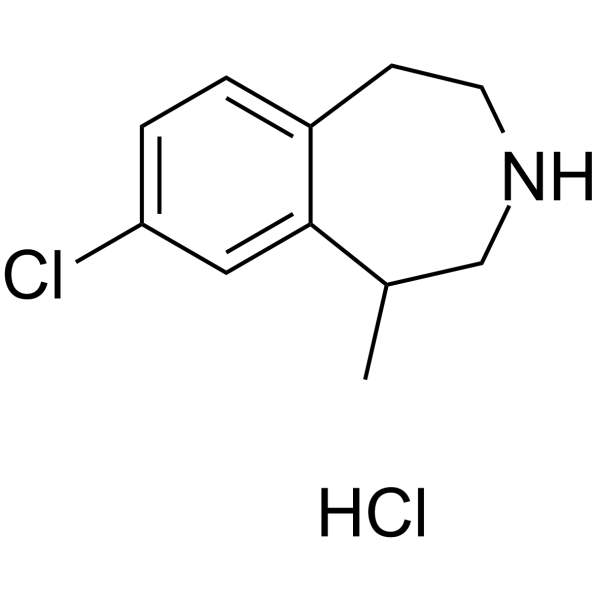
(Rac)-Lorcaserin hydrochloride
-
GC64069
(Rac)-Lys-SMCC-DM1
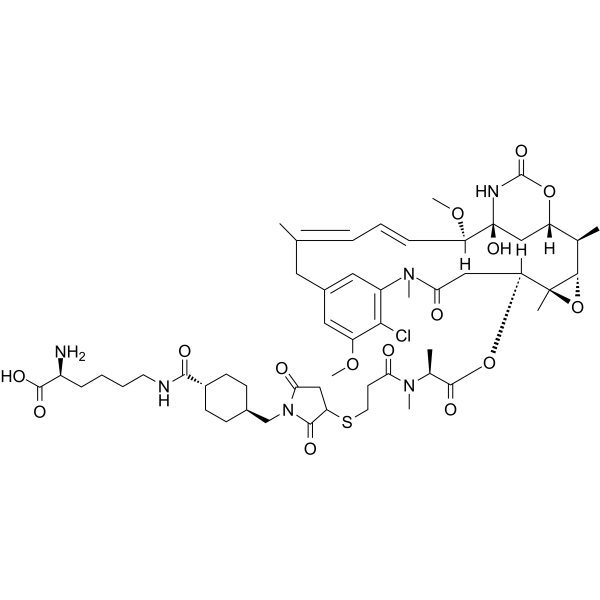
(Rac)-Lys-SMCC-DM1 ((Rac)-Lys-Nε-MCC-DM1) is the racemate of Lys-SMCC-DM1. Lys-SMCC-DM1 is a linker-payload component that has the potential to inhibit tubulin polymerization. Lys-SMCC-DM1 is the active metabolite of T-DM1.
-
GC61873
(Rac)-MEM 1003
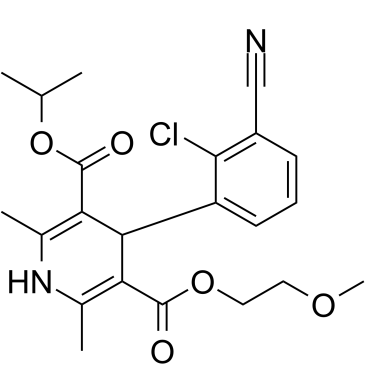
(Rac)-MEM 1003 is the racemate of MEM 1003.
-
GC64490
(Rac)-MGV354
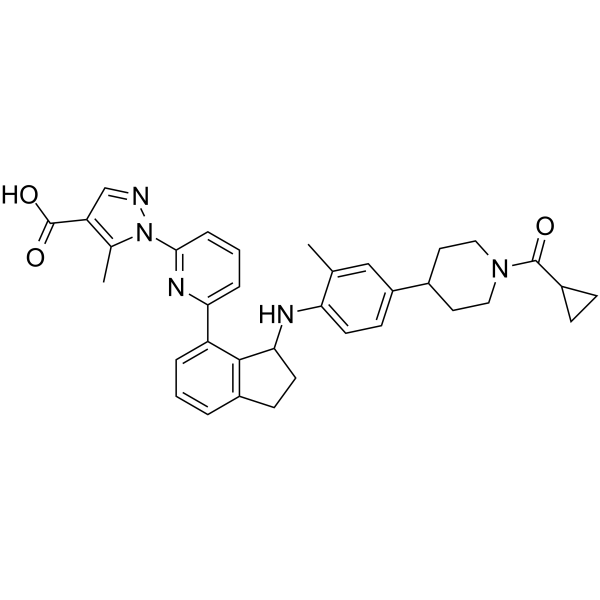
(Rac)-MGV354 is the racemate of MGV354.
-
GC63410
(Rac)-MRI-1867
(Rac)-MRI-1867 ((Rac)-MRI-1867, compound 6b) is a cannabinoid receptor type 1 (CB1R)/iNOS antagonist, with a Ki of 5.7 nM for CB1R.
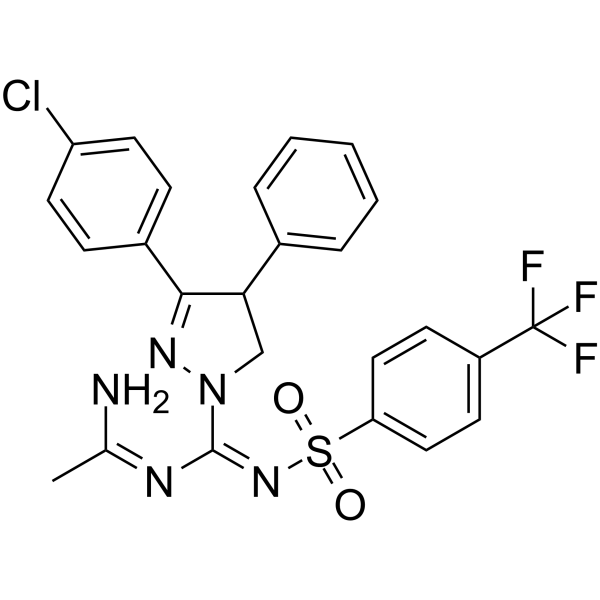
-
GC62228
(rac)-Nebivolol-d4
(Rac)-Nebivolol-d4 ((Rac)-R 065824-d4) is a labelled racemic Nebivolol.
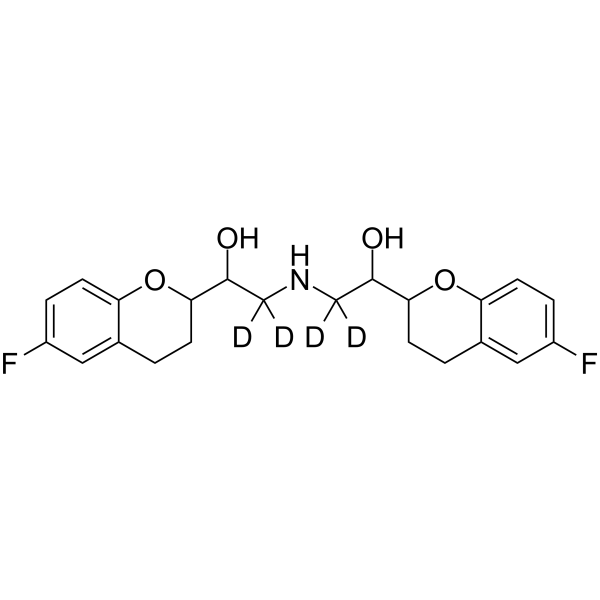
-
GC62744
(Rac)-OSMI-1
(Rac)-OSMI-1 is the racemate of OSMI-1.
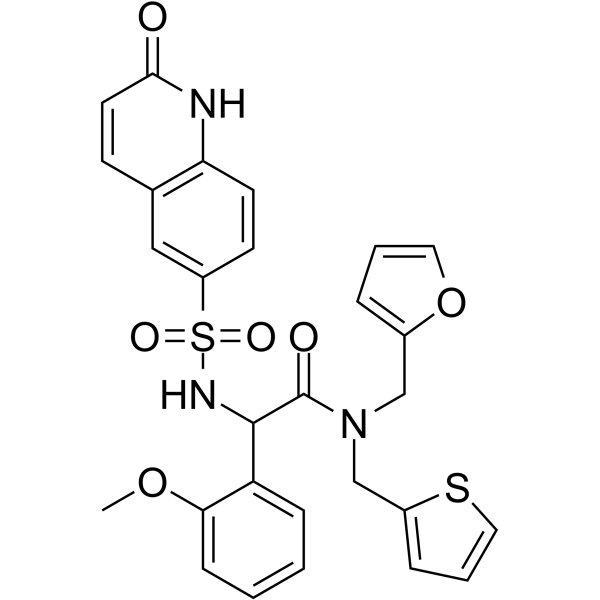
-
GC62745
(Rac)-PF-06256142
(Rac)-PF-06256142 is the less effective enantiomer of PF-06256142.
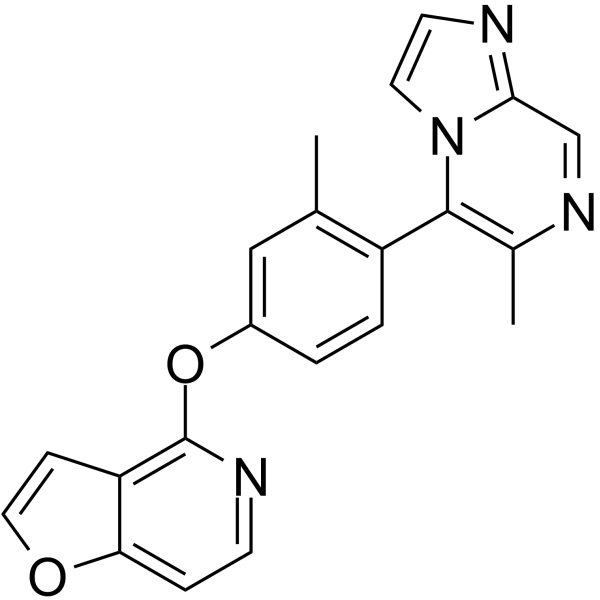
-
GC66334
(Rac)-PF-184 hydrate
(Rac)-PF-184 hydrate is a potent inhibitory factor-κB kinase 2 (IKK-2) inhibitor with an IC50 of 37 nM. (Rac)-PF-184 hydrate has anti-inflammatory effects.
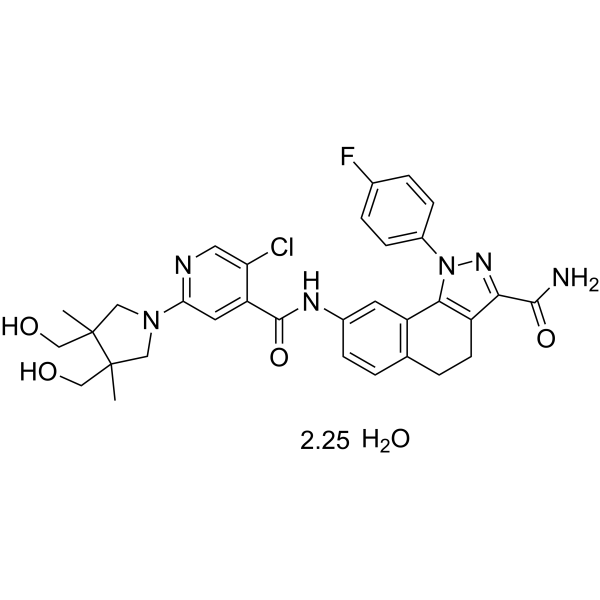
-
GC63292
(rac)-PF-998425
(Rac)-PF-998425 is a potent, selective, nonsteroidal androgen receptor (AR) antagonist.
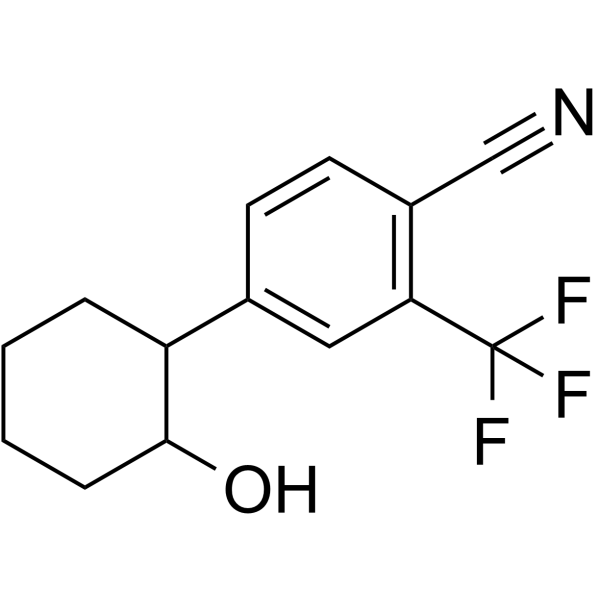
-
GC64123
(Rac)-RP-6306
(Rac)-RP-6306 (compound 181) is a potent Myt1 inhibitor with an IC50 of <10 nM. (Rac)-RP-6306 (compound 181) has anticancer effects.
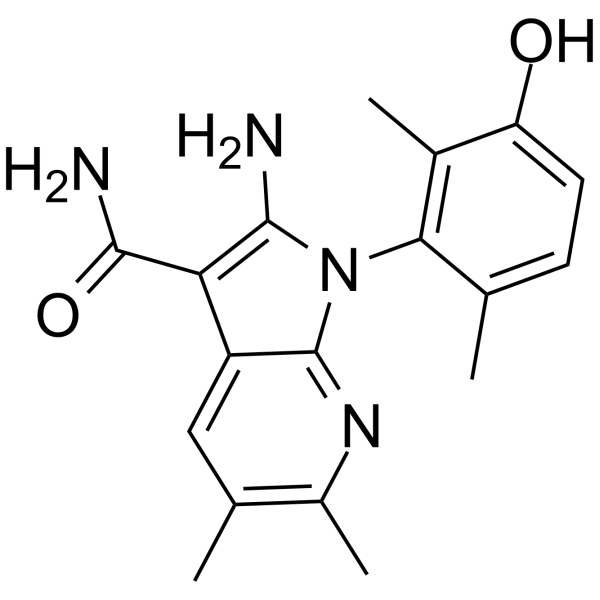
-
GC69797
(Rac)-SHIN2
(Rac)-SHIN2 is a serine hydroxymethyltransferase (SHMT) inhibitor with a 1,4-dihydropyran[2,3-c]pyrazole structure. It is involved in folate or one-carbon metabolism pathways and prevents viral infections. SHMT1 and SHMT2 are cytosolic and/or mitochondrial isoforms of serine hydroxymethyltransferase respectively.
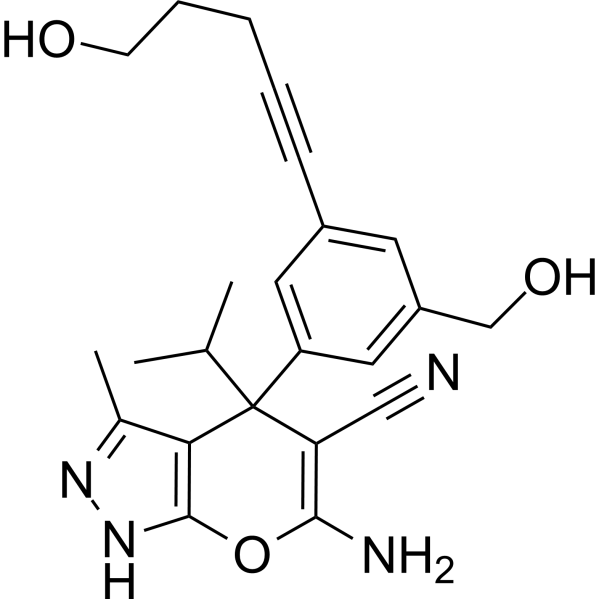
-
GC69798
(Rac)-SNC80
(Rac)-SNC80 is the racemic form of SNC80. SNC80 (NIH 10815) is an effective and highly selective non-peptide delta-opioid receptor agonist with a Ki of 1.78 nM and IC50 of 2.73 nM. SNC80 has anti-nociceptive, anti-hyperalgesic, and antidepressant-like effects, and can be used in research on various types of headaches.
-
GC64905
(Rac)-Upacicalcet
(Rac)-Upacicalcet is the racemate of Upacicalcet.
-
GC69799
(Rac)-ZLc-002
(Rac)-ZLc-002 is an inhibitor that interacts with the binding protein between nNOS and nitric oxide synthase 1 (NOS1AP). It inhibits inflammatory pain and chemotherapy-induced neuropathic pain, and synergistically reduces tumor cell viability with Paclitaxel.
-
GC62746
(RS)-AMPA monohydrate
(RS)-AMPA ((±)-AMPA) monohydrate is a glutamate analogue and a potent and selective excitatory neurotransmitter L-glutamic acid agonist.
-
GC64804
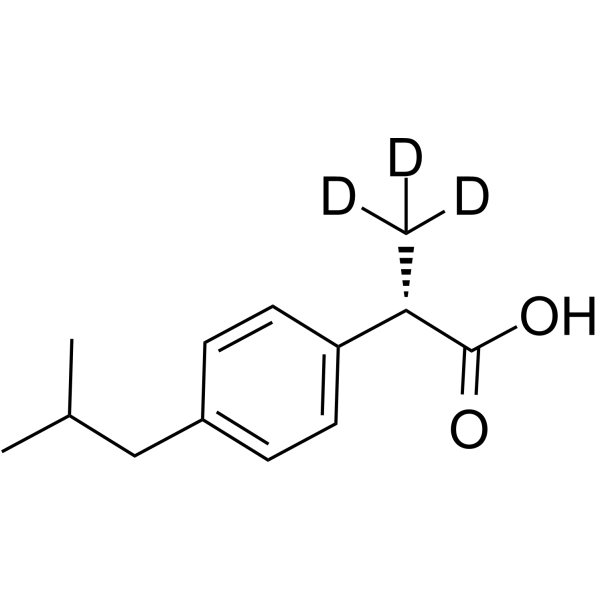
(S)-(+)-Ibuprofen D3
(S)-(+)-Ibuprofen D3 ((S)-Ibuprofen D3) is a deuterium labeled (S)-(+)-Ibuprofen.
-
GC25002
(S)-(-)-α-Methylbenzylamine
(S)-(-)-α-Methylbenzylamine is a chiral auxiliary in the enantiodivergent synthesis of simple isoquinoline alkaloids.

-
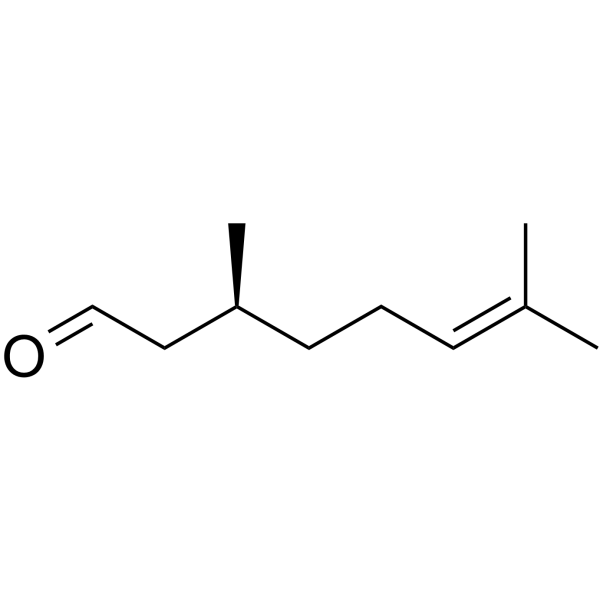
GC62747
(S)-(-)-Citronellal
-
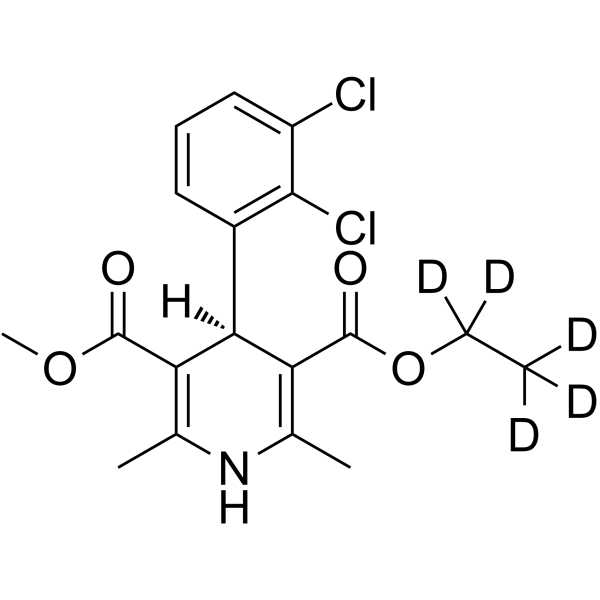
GC63321
(S)-(-)-Felodipine-d5
-
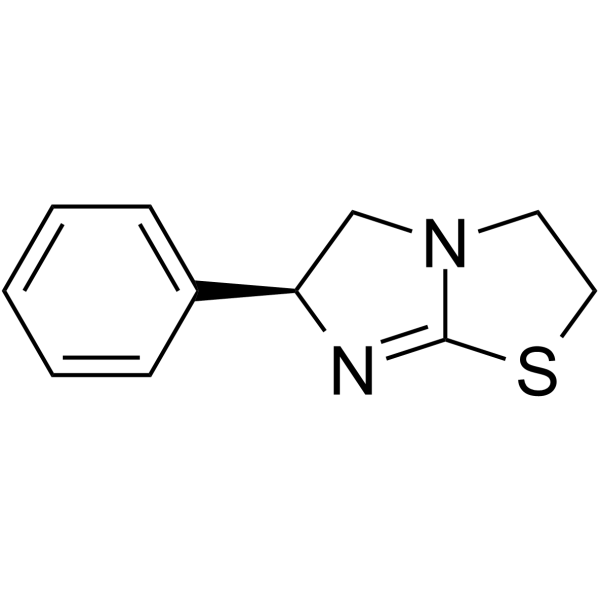
GC64735
(S)-(-)-Levamisole
(S)-(-)-Levamisole (Levamisole), an anthelmintic agent with immunomodulatory properties.
-
GC66076
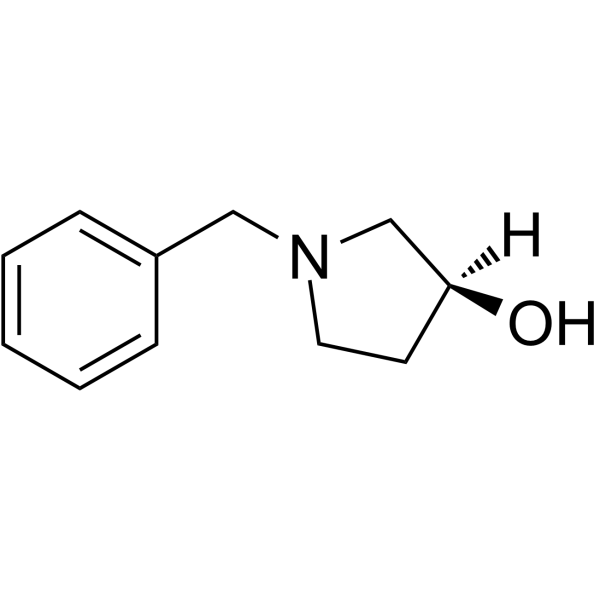
(S)-1-Benzylpyrrolidin-3-ol
(S)-1-Benzylpyrrolidin-3-ol (Compound 14) can be used to synthesis impurities of the anti-hypertensive drug Barnidipine hydrochloride (HY-107322).
-
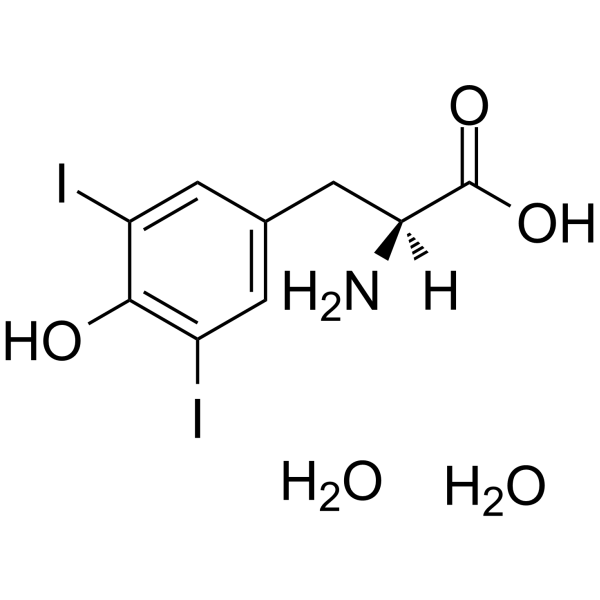
GC62748
(S)-2-Amino-3-(4-hydroxy-3,5-diiodophenyl)propanoic acid dihydrate
(S)-2-Amino-3-(4-hydroxy-3,5-diiodophenyl)propanoic acid dihydrate is an endogenous metabolite.
-
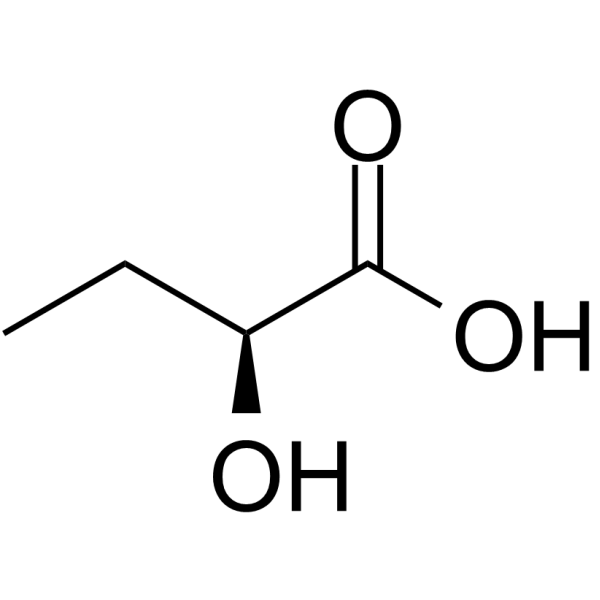
GC64746
(S)-2-Hydroxybutanoic acid
(S)-2-Hydroxybutanoic acid is the S-enantiomer of?2-Hydroxybutanoic acid.
-
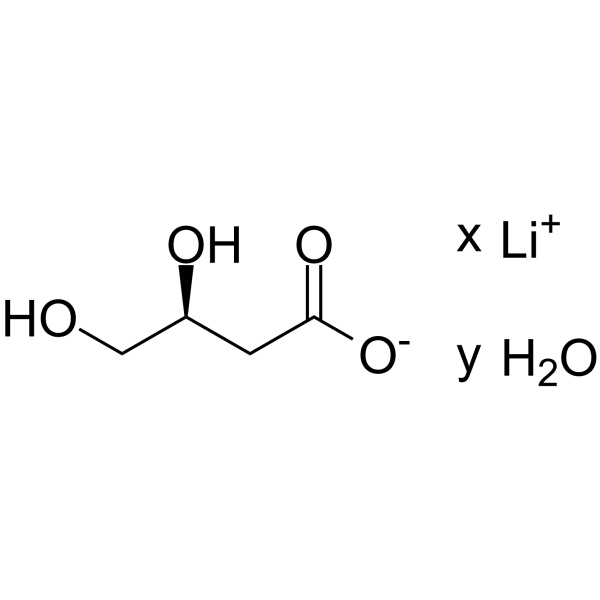
GC64473
(S)-3,4-Dihydroxybutyric acid lithium hydrate
(S)-3,4-Dihydroxybutyric acid (lithium hydrate) is a normal human urinary metabolite that is excreted in increased concentration in patients with succinic semialdehyde dehydrogenase (SSADH) deficiency.
-
GC62749
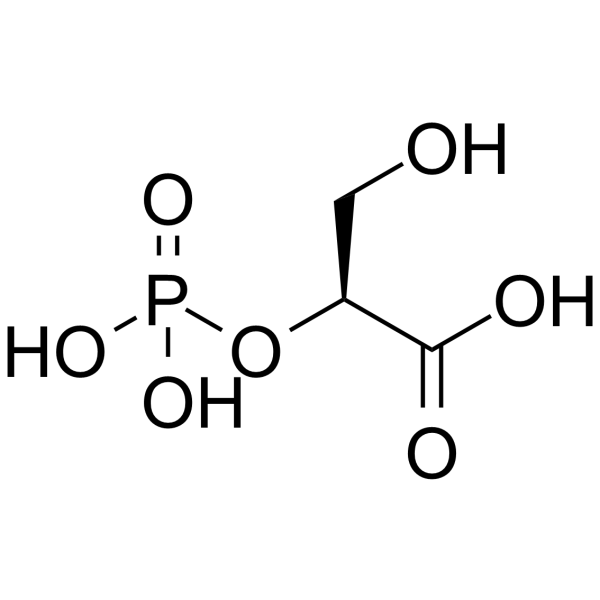
(S)-3-Hydroxy-2-(Phosphonooxy)Propanoic Acid
(S)-3-Hydroxy-2-(Phosphonooxy)Propanoic Acid is an endogenous metabolite.
-
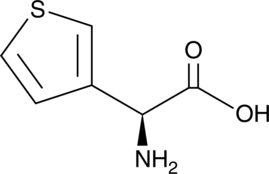
GC49028
(S)-3-Thienylglycine
A thienyl-containing amino acid
-
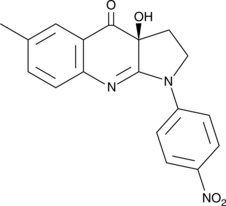
GC52192
(S)-4'-nitro-Blebbistatin
(S)-4'-nitro-Blebbistatin is a non-cytotoxic, photostable, fluorescent and specific Myosin II inhibitor, usd in the study of the specific role of myosin II in physiological, developmental, and cell biological studies.
-
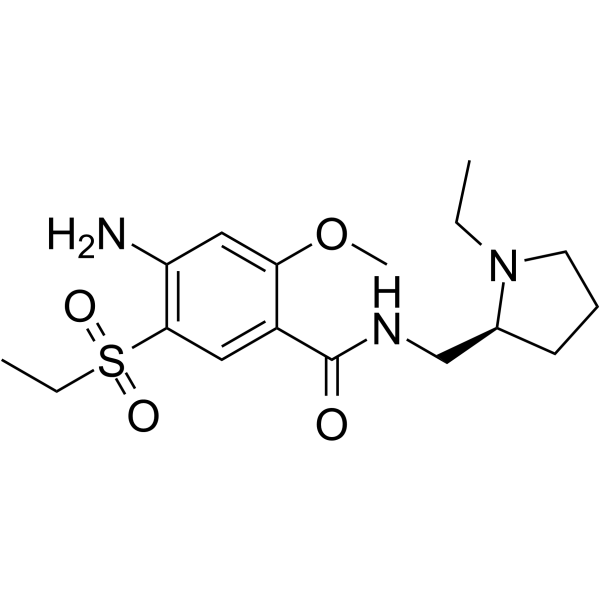
GC63655
(S)-Amisulpride
(S)-Amisulpride (Esamisulpride) is a potent dopamine D2/D3 receptor antagonist.
-
GC64594
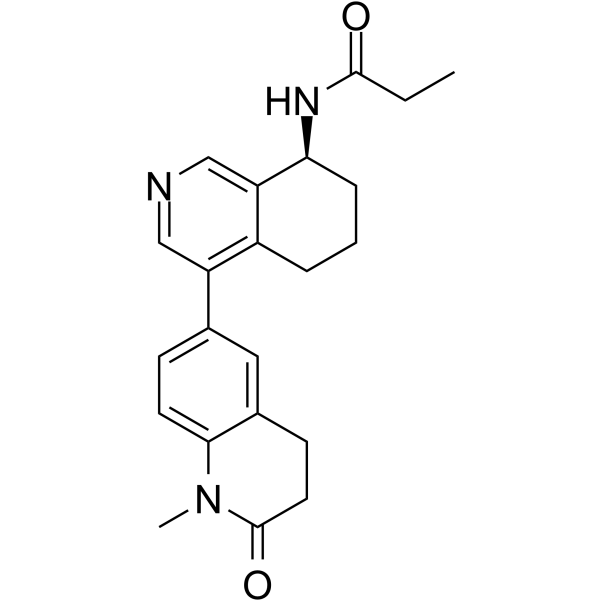
(S)-Baxdrostat
(S)-Baxdrostat is the S-enantiomer of Baxdrostat.
-
GC48719
(S)-Canadine
(S)-Canadine is an alkaloid and intermediate in the biosynthesis of berberine with insecticidal activity.

-
GC62750
(S)-Enzaplatovir
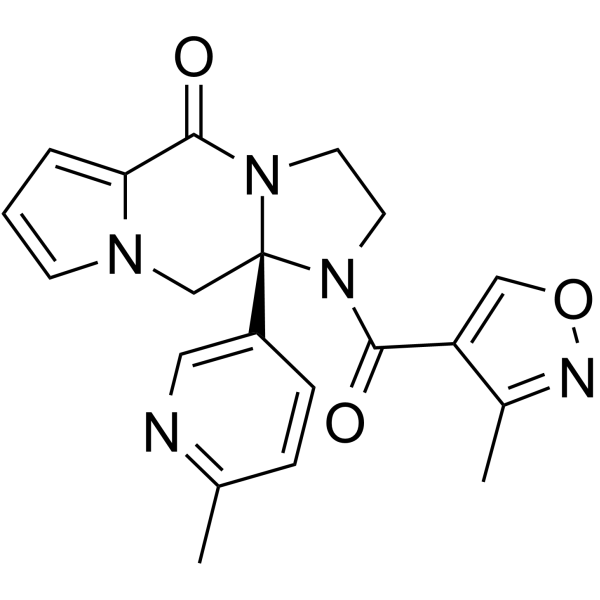
(S)-Enzaplatovir ((S)-BTA-C585) is the S-enantiomer of Enzaplatovir. (S)-Enzaplatovir shows antiviral activities with an EC50 of 56 nM for respiratory syncytial viral (RSV) (patent WO2011094823A1 compound 77).
-
GC49520
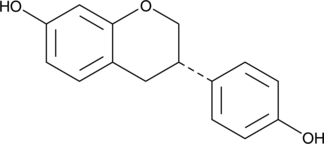
(S)-Equol
An estrogen receptor β agonist
-
GC63526
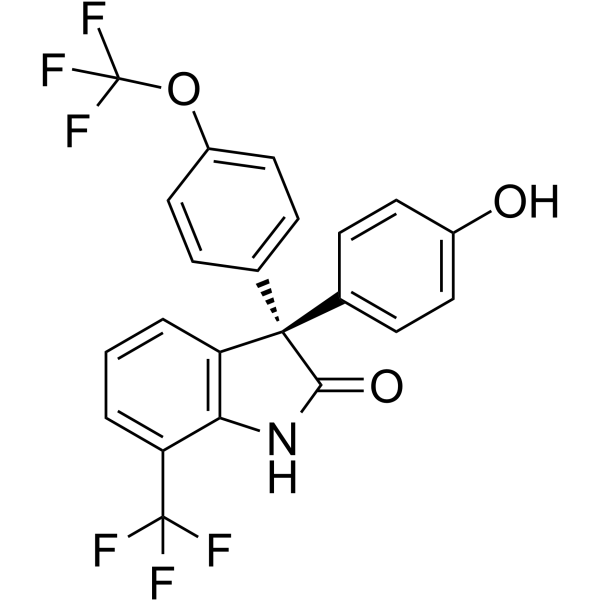
(S)-ErSO
(S)-ErSO is the dextrorotatory enantiomer of ErSO. (S)-ErSO is inactive in MCF-7 cells (from patent WO2020009958A1, compound (s)-105).
-
GC65877
(S)-GFB-12811
(S)-GFB-12811 (compound 596) is a potent and selective CDK5 inhibitor, with an IC50 value less than 10 nM. (S)-GFB-12811 can be used in the research of cell cycle progression, neuronal development, tumorigenesis.
-
GC62751
(S)-Higenamine hydrobromide
(S)-Higenamine ((S)-Norcoclaurine) hydrobromide, a S-enantiomer of Higenamine, is the entry compound in benzylisoquinoline alkaloid biosynthesis.
-
GC49067
(S)-Hydroxychloroquine (sulfate)
An isomer of hydroxychloroquine
-
GC69890
(S)-Imlunestrant tosylate
(S)-Imlunestrant tosylate ((S)-LY-3484356) is the (S)-enantiomer of Imlunestrant. It can be used for cancer research.
-
GC69891
(S)-Indoximod-d3
(S)-Indoximod-d3 is the deuterated form of (S)-Indoximod. (S)-Indoximod (1-Methyl-L-tryptophan) is an inhibitor of indoleamine 2,3-dioxygenase (IDO). (S)-Indoximod can be used for cancer research.
-
GC69904
(S)-JDQ-443
(S)-JDQ-443 is an isomer of JDQ-443. JDQ-443 is an orally effective and selective covalent KRAS G12C inhibitor. JDQ-443 has anti-tumor activity.
-
GC65997
(S)-LY3177833 hydrate
(S)-LY3177833 ((S)-Example 2) hydrate is an orally active CDC7 kinase inhibitor. (S)-LY3177833 hydrate shows broad in vitro anticancer activity.
-
GC69914
(S)-Malic acid-d3
(S)-Malic acid-d3 is the deuterated form of (S)-Malic acid. (S)-Malic acid ((S)-2-Hydroxysuccinic acid) is a naturally occurring dicarboxylic acid and a source of fruity sour taste, commonly used as a food additive.
-
GC67989
(S)-Mirtazapine
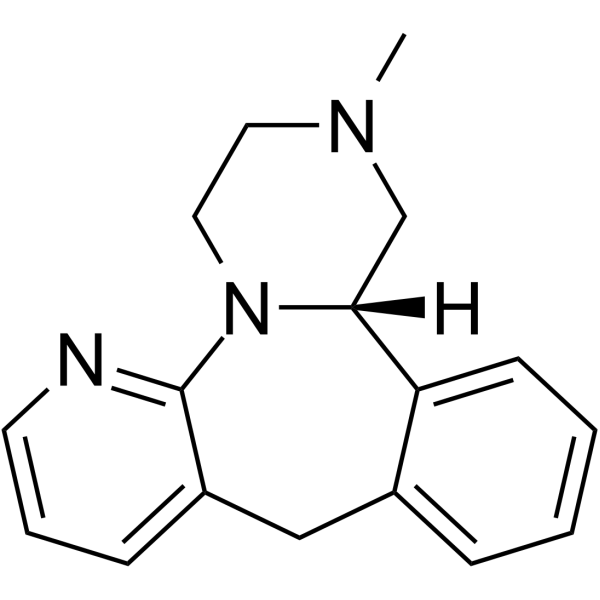
-
GC68410
(S)-Mirtazapine-d3