PROTAC
PROTACs or Proteolysis Targeting Chimeric Molecules are heterobifunctional nanomolecules that theoretically target any protein for ubiquitination and degradation. In terms of the structure, PROTACs consist of one moiety which is recognized by the E3 ligase. This moiety is then chemically and covalently linked to a small molecule or a protein that recognizes the target protein. The trimeric complex formation leads to the transfer of ubiquitins to the target protein.
By removing target proteins directly rather than merely blocking them, PROTACs can provide multiple advantages over small molecule inhibitors, which can require high systemic exposure to achieve sufficient inhibition, often resulting in toxic side effects and eventual drug resistance. PROTAC molecules possess good tissue distribution and the ability to target intracellular proteins, thus can be directly applied to cells or injected into animals without the use of vectors.
Targeted protein degradation using the PROTAC technology is emerging as a novel therapeutic method to address diseases, such as cancer, driven by the aberrant expression of a disease-causing protein. In addition to the use of PROTACs for the treatment of human disease, these molecules provide a chemical genetic approach to “knock down” proteins to study their function. Currently, there are several small molecule inhibitors that have been found to show good biological activity by specifically targeting BET, estrogen receptor (ER), androgen receptor, etc.
References:
[1] Sakamoto KM. Pediatr Res. 2010 May;67(5):505-8.
[2] Neklesa TK, et al. Pharmacol Ther. 2017 Jun;174:138-144.
Targets for PROTAC
Products for PROTAC
- Cat.No. Product Name Information
-
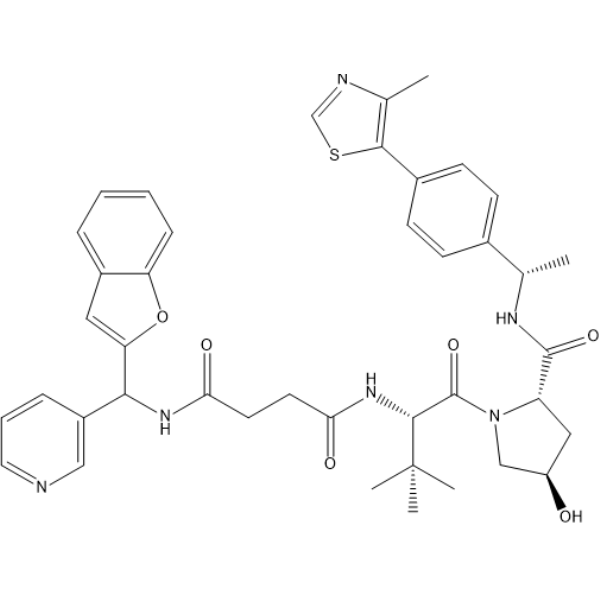
GC68352
(S,R,S)-AHPC-C2-amide-benzofuranylmethyl-pyridine
-
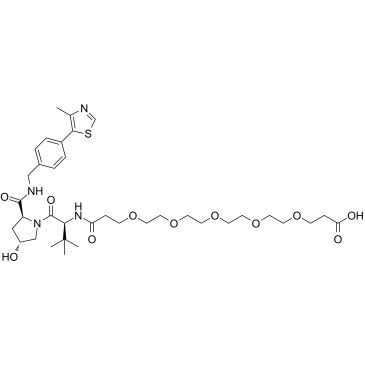
GC60009
(S,R,S)-AHPC-PEG5-COOH
(S,R,S)-AHPC-PEG5-COOH (VH032-PEG5-COOH) is a synthesized E3 ligase ligand-linker conjugate that incorporates the (S,R,S)-AHPC based VHL ligand and 5-unit PEG linker used in PROTAC technology.
-
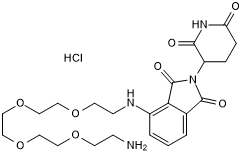
GC50469
A 410099.1 amide-alkylC4-amine
A 410099.1 amide-alkylC4-amine
-
GC50468
A 410099.1 amide-PEG2-amine
A 410099.1 amide-PEG2-amine
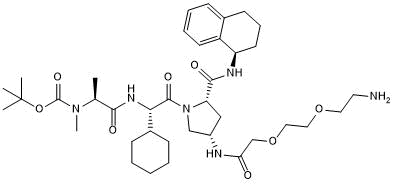
-
GC50467
A 410099.1 amide-PEG3-amine
A 410099.1 amide-PEG3-amine
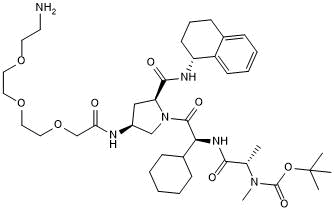
-
GC50737
A 410099.1 amide-PEG4-amine
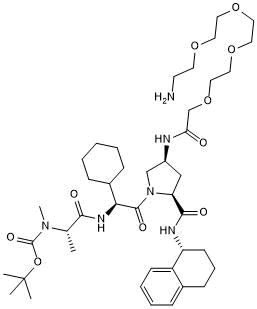
-
GC50739
A 410099.1 amide-PEG5-amine
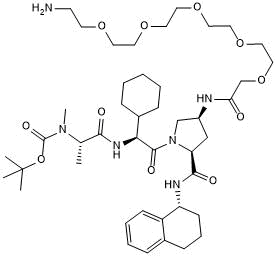
-
GC33280
A1874
A1874 is a nutlin-based (MDM2 ligand) and BRD4-degrading PROTAC with a DC50 of 32 nM (induce BRD4 degradation in cells). Effective in inhibiting many cancer cell lines proliferation.
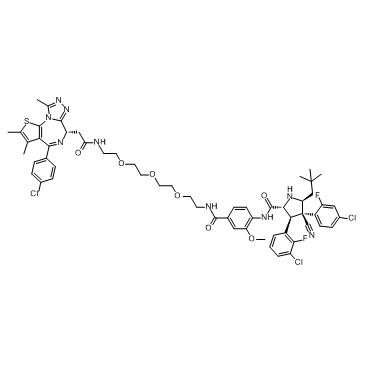
-
GC35227
ACBI1
ACBI1 is a potent and cooperative SMARCA2, SMARCA4 and PBRM1 degrader with DC50s of 6, 11 and 32 nM, respectively. ACBI1 is a PROTAC degrader. ACBI1 shows anti-proliferative activity. ACBI1 induces apoptosis.
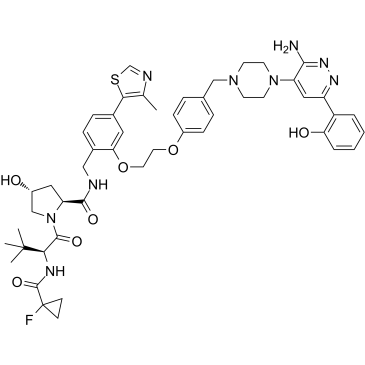
-
GC50744
ARCC 4 negative control
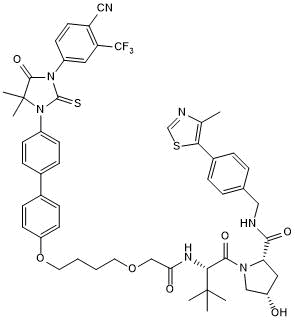
-
GC60594
ARCC-4
ARCC-4 is a low-nanomolar Androgen Receptor (AR) degrader based on PROTAC, with a DC50 of 5?nM. ARCC-4 is an enzalutamide-based von Hippel-Lindau (VHL)-recruiting AR PROTAC and outperforms enzalutamide. ARCC-4 effectively degrades clinically relevant AR mutants associated with antiandrogen therapy.
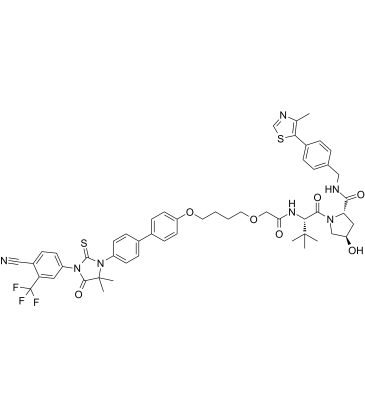
-
GC63492
ARD-2128
ARD-2128 is a highly potent, orally bioavailable PROTAC androgen receptor (AR) degrader. ARD-2128 effectively reduces AR protein, suppresses AR-regulated genes in tumor tissues, and inhibits growth of tumor without signs of toxicity. ARD-2128 has the potential for the research of the prostate cancer.
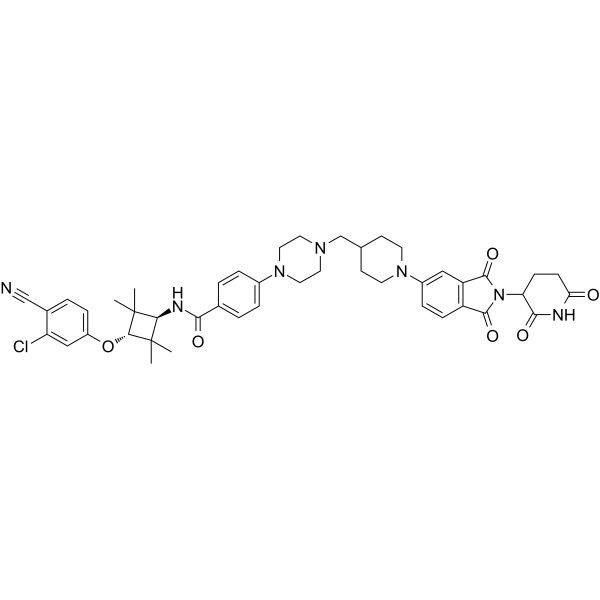
-
GC63704
ARD-2585
ARD-2585 is an exceptionally potent and orally active PROTAC degrader of androgen receptor.
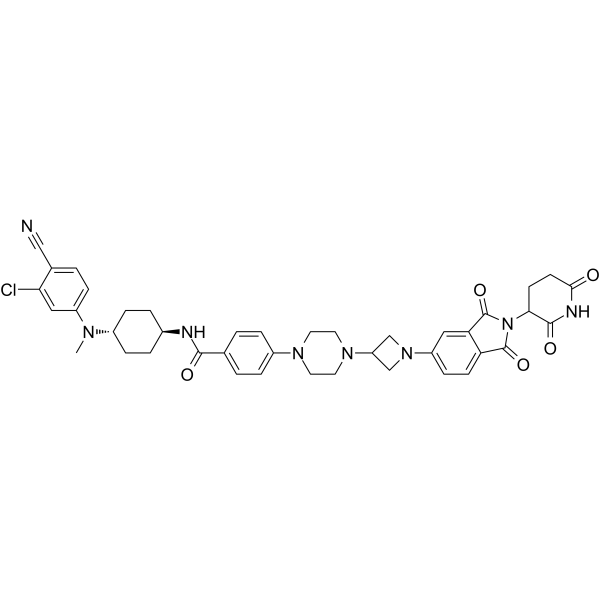
-
GC32685
ARV-771
ARV-771 is a potent BET PROTAC based on E3 ligase von Hippel-Lindau with Kds of 34 nM, 4.7 nM, 8.3 nM, 7.6 nM, 9.6 nM, and 7.6 nM for BRD2(1), BRD2(2), BRD3(1), BRD3(2), BRD4(1), and BRD4(2), respectively.
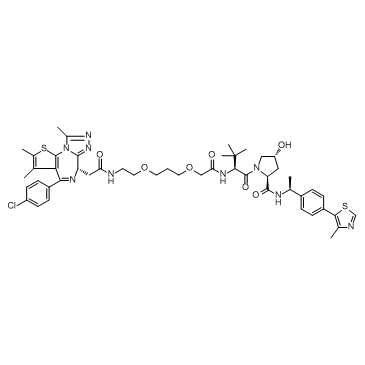
-
GC19038
ARV-825
ARV-825 is a BRD4 Inhibitor based on PROTAC technology.
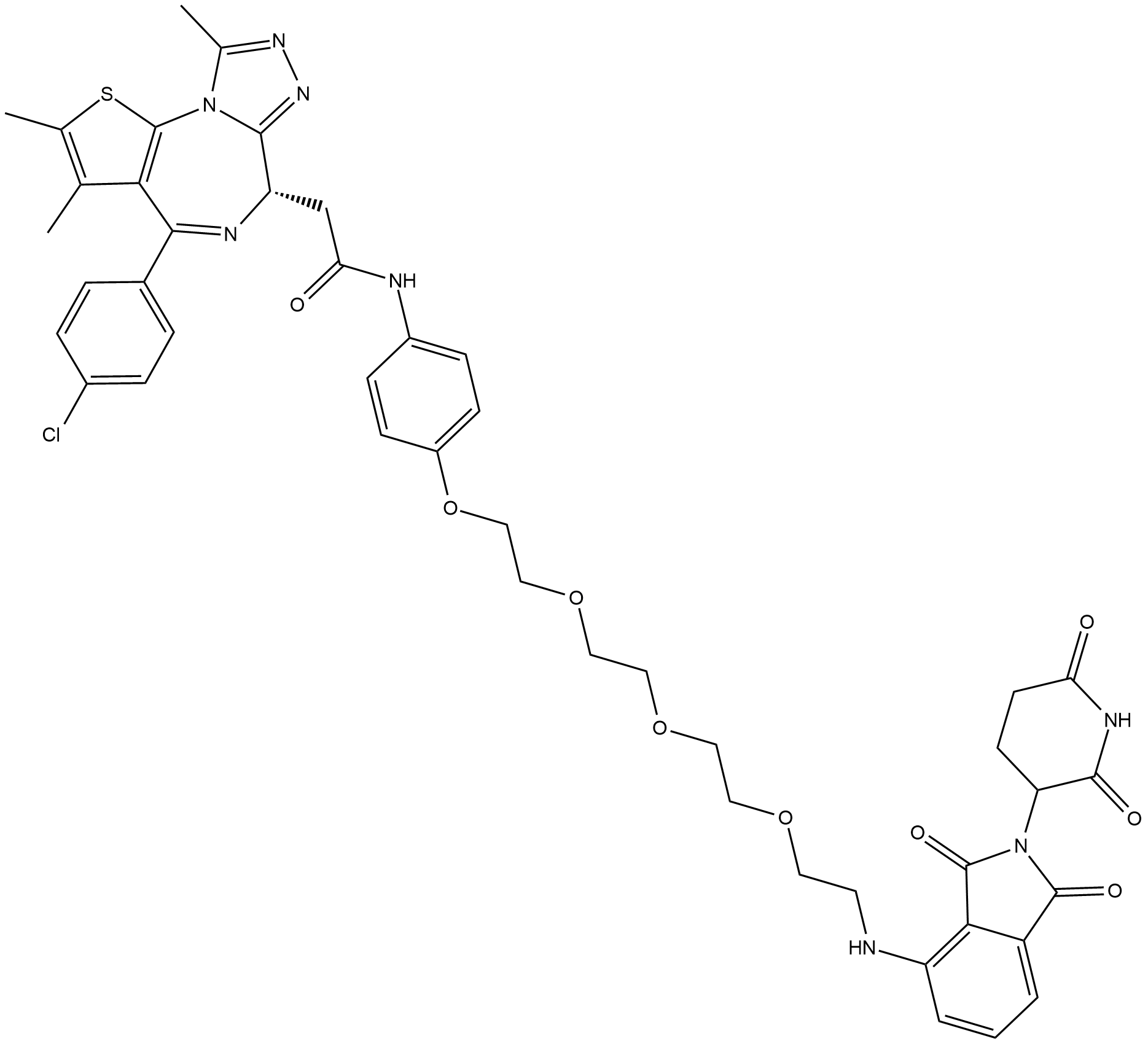
-
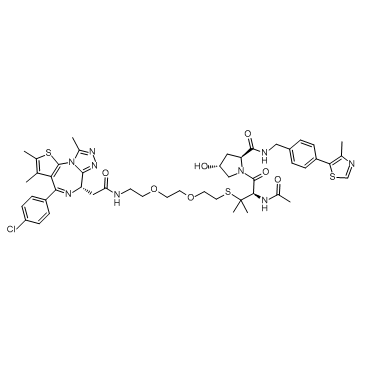
GC33354
AT6
AT6 is a PROTAC AT1 analogue, which is a PROTAC connected by ligands for von Hippel-Lindau and BRD4 with highly selectivity to bromodomain (Brd4).
-
GC50630
aTAG 2139
Degrader of MTH1 fusion proteins for use within the aTAG system

-
GC50631
aTAG 4531
Degrader of MTH1 fusion proteins for use within the aTAG system
-
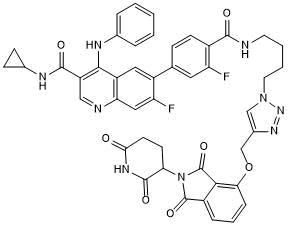
GC65506
BETd-246
BETd-246 is a second-generation and PROTAC-based BET bromodomain (BRD) inhibitor connected by ligands for Cereblon and BET, exhibiting superior selectivity, potency and antitumor activity.
-
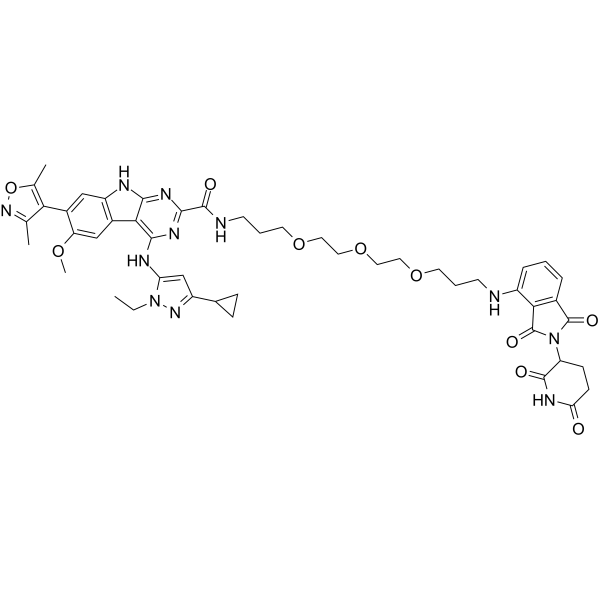
GC32791
BETd-260 (ZBC 260)
BETd-260 (ZBC 260) (ZBC 260) is a PROTAC connected by ligands for Cereblon and BET, with as low as 30 pM against BRD4 protein in RS4;11 leukemia cell line. BETd-260 (ZBC 260) potently suppresses cell viability and robustly induces apoptosis in hepatocellular carcinoma (HCC) cells.
-
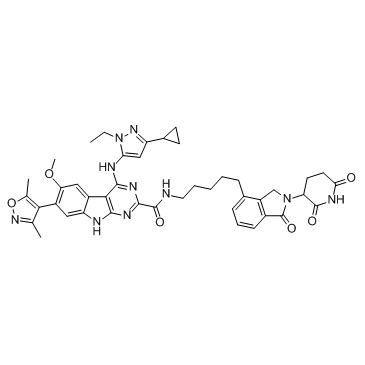
GC65457
BI-3663
BI-3663 is a highly selective PTK2/FAK PROTAC (DC50=30 nM), with Cereblon ligands to hijack E3 ligases for PTK2 degradation. BI-3663 inhibits PTK2 with an IC50 of 18 nM. BI-3663 is a PROTAC that composes of BI-4464 linked to Pomalidomide with a linker. Anti-cancer activity.
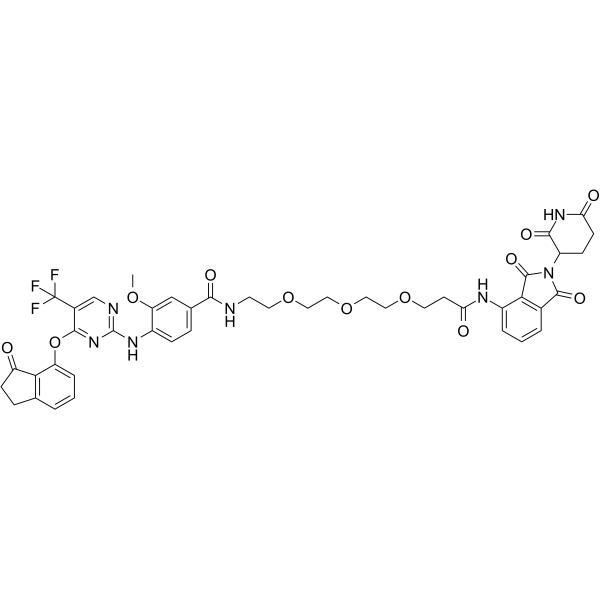
-
GC33017
BRD4 degrader AT1
BRD4 degrader AT1 is a PROTAC connected by ligands for von Hippel-Lindau and BRD4 as a highly selective Brd4 degrader, with a Kd of 44 nM for Brd4BD2 in cells.
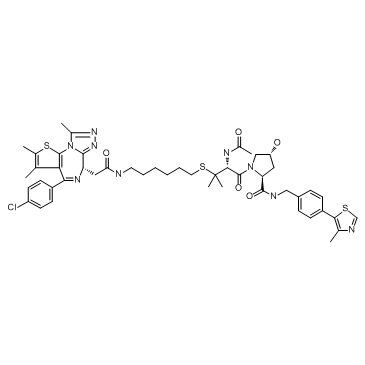
-
GC65128
BSJ-03-123
BSJ-03-123 is a PROTAC connected by ligands for Cereblon and CDK as a potent and novel CDK6-selective small-molecule degrader.
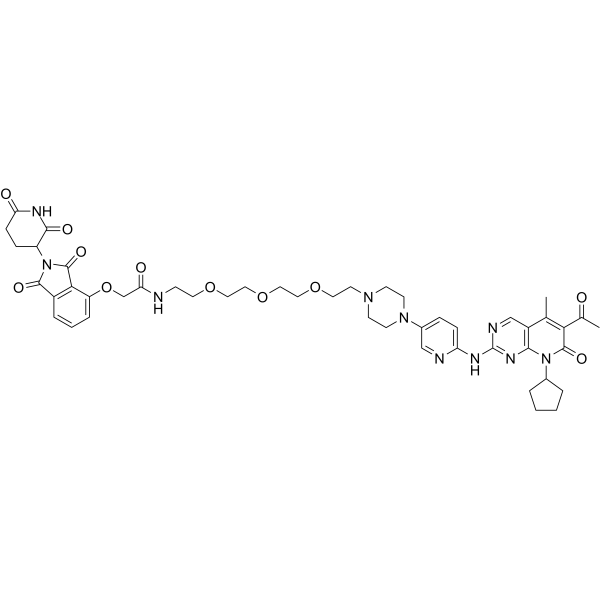
-
GC50615
BSJ-03-204
Selective Cdk4/6 degrader
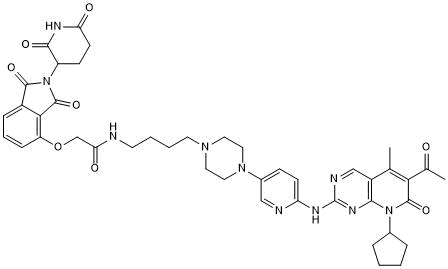
-
GC50614
BSJ-04-132
Selective Cdk4 degrader
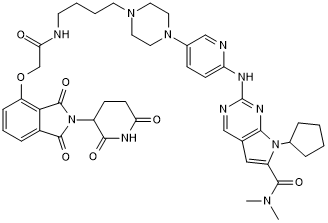
-
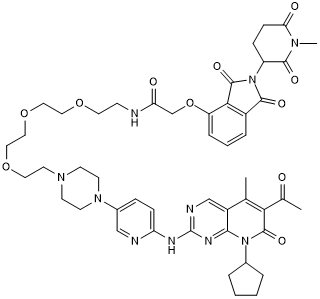
GC50610
BSJ-Bump
Negative control for BSJ-03-123
-
GC67861
CCT367766 formic
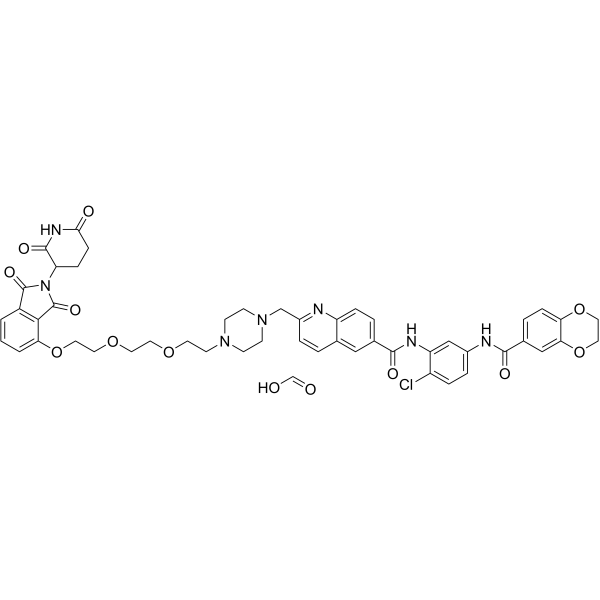
-
GC35635
CDK9 Antagonist-1
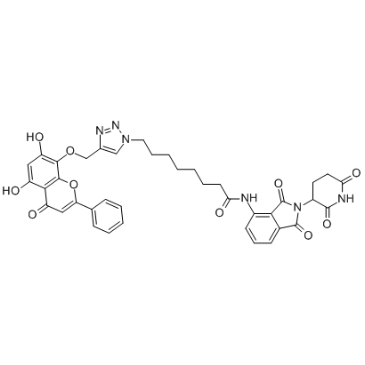
-
GC50363
cis MZ 1
Negative Control for MZ 1
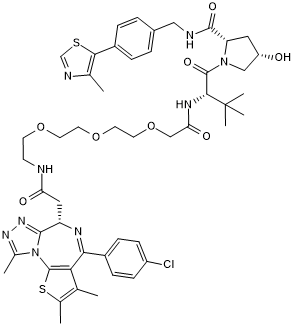
-
GC50629
cis VH 032, amine dihydrochloride
Negative control for VH 032, amine
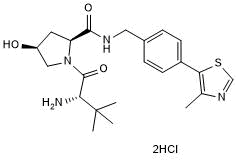
-
GC50616
cis-VZ 185
Negative control for VZ 185
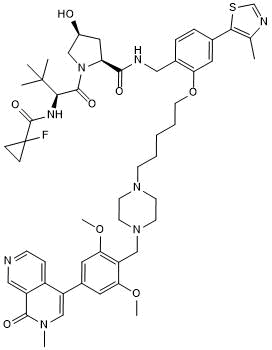
-
GC35739
CP-10
CP-10 is a PROTAC connected by ligands for Cereblon and CDK, with highly selective, specific, and remarkable CDK6 degradation (DC50=2.1 nM). It inhibits proliferation of several haematopoietic cancer cells with impressive potency including multiple myeloma, and can still degrades mutated and overexpressed CDK6.
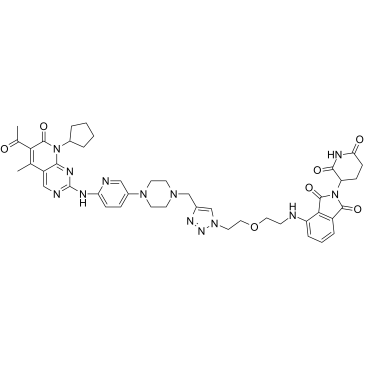
-
GC50619
CRBN-6-5-5-VHL
Potent and selective cereblon degrader; cell-permeable
-
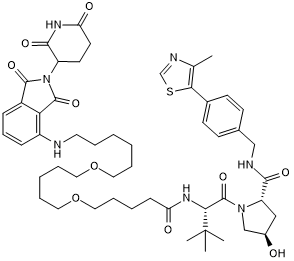
GC66361
DB-0646
DB-0646, a PROTAC, is a multi-kinase degrader.
-
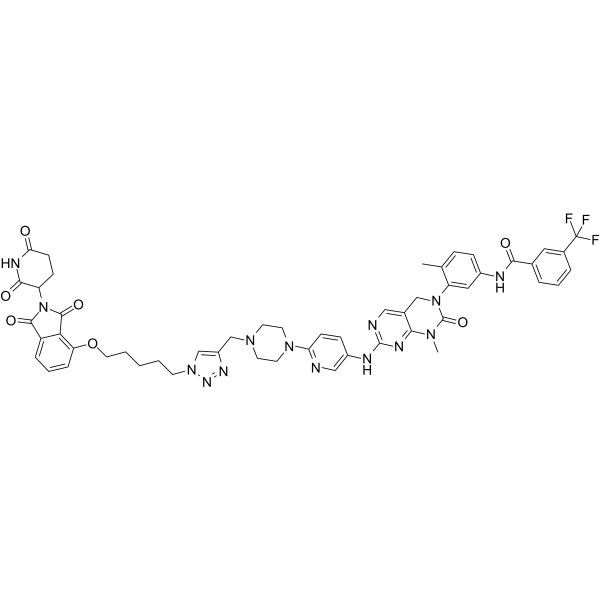
GC19119
dBET1
dBET1 is a potent BRD4 protein degrader based on PROTAC technology with an EC50 of 430 nM.
-
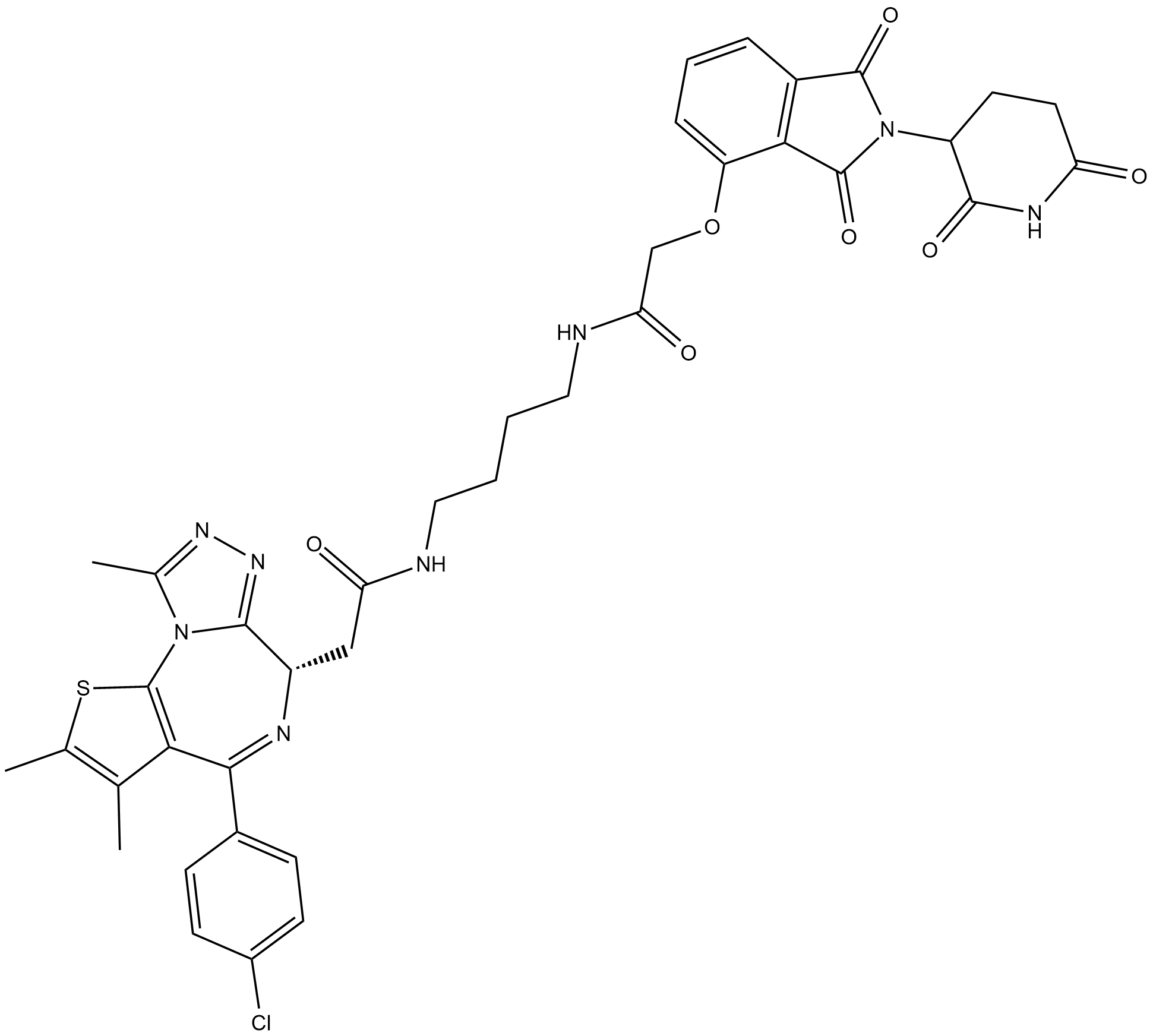
GC63445
dBET23
dBET23 is a highly effective and selective PROTAC BRD4 degrader with a DC50/5h of ~ 50 nM for BRD4BD1 protein.
-
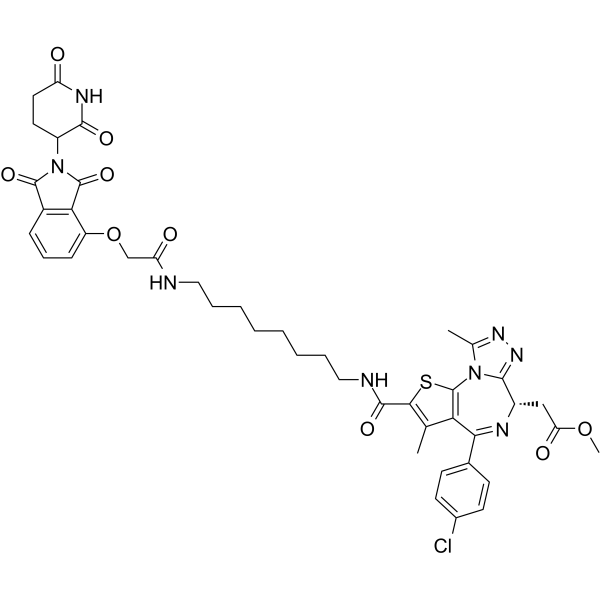
GC35815
dBET57
dBET57 is a potent and selective degrader of BRD4BD1 based on the PROTAC technology. dBET57 mediates recruitment to the CRL4Cereblon E3 ubiquitin ligase, with a DC50/5h of 500 nM for BRD4BD1, and is inactive on BRD4BD2.
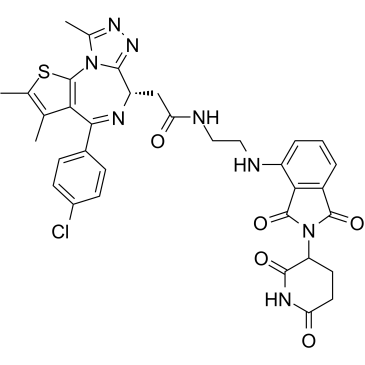
-
GC32719
dBET6
dBET6 is a highly potent, selective and cell-permeable PROTAC connected by ligands for Cereblon and BET, with an IC50 of 14 nM, and has antitumor activity.
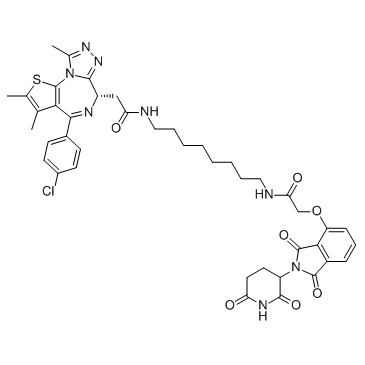
-
GC50518
dBRD9
Potent and selective BRD9 degrading PROTAC
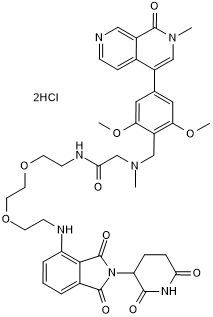
-
GC50736
dBRD9-A
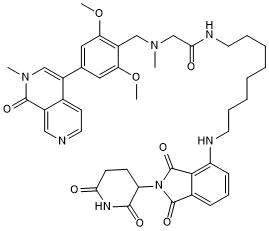
-
GC50749
DD 03-171
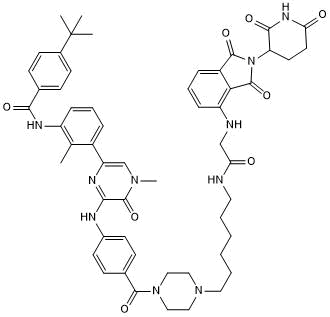
-
GC35882
dMCL1-2
dMCL1-2 is a potent and selective PROTAC of myeloid cell leukemia 1 (MCL1) (Bcl-2 family member) based on Cereblon, which binds to MCL1 with a KD of 30 nM. dMCL1-2 activats the cellular apoptosis machinery by degradation of MCL1.
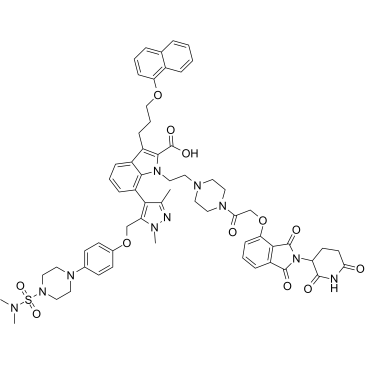
-
GC62942
DP-C-4
DP-C-4 is a Cereblon-based dual PROTAC for simultaneous degradation of EGFR and PARP.
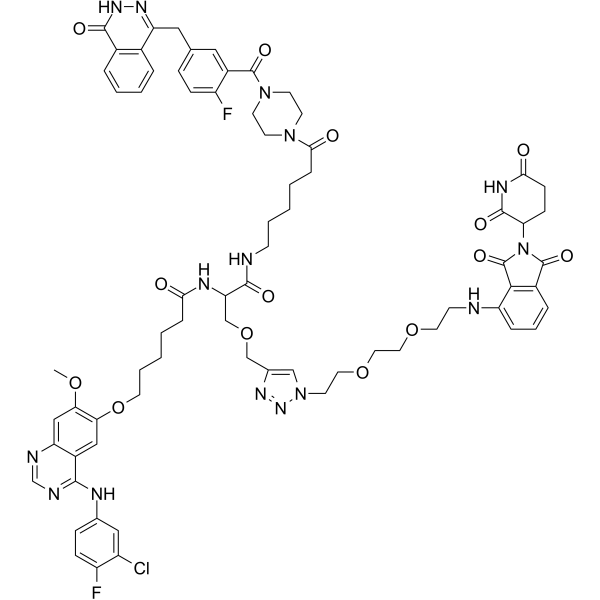
-
GC50754
dTAG-13-NEG
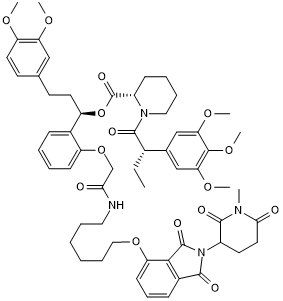
-
GC50756
dTAGV-1
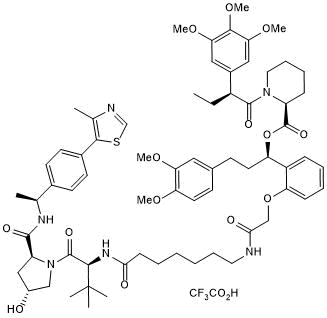
-
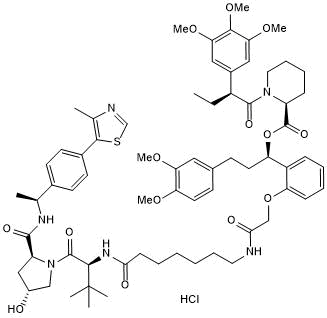
GC50757
dTAGV-1 hydrochloride
-
GC67914
dTAGV-1 TFA

-
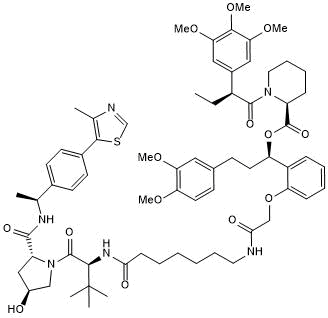
GC50755
dTAGV-1-NEG
dTAGV-1-NEG is a diastereomer and as a heterobifunctional negative control of dTAGV-1. dTAGV-1 is an FKBP12F36V-selective degrader.
-
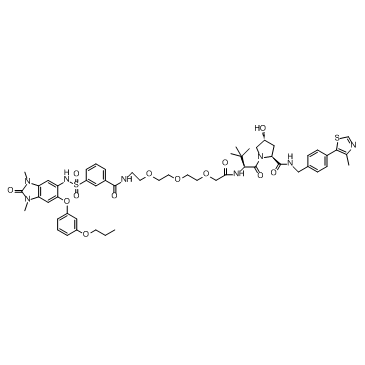
GC35904
dTRIM24
dTRIM24 is a selective bifunctional degrader of TRIM24 based on PROTAC, consists of ligands for von Hippel-Lindau and TRIM24.
-
GC32902
E3 ligase Ligand-Linker Conjugates 10
E3 ligase Ligand-Linker Conjugates 10 (VH032-PEG2-C4-Cl) is a conjugate of ligands for E3 and 13-atom-length linker. The connector of linker is Halogen group. E3 ligase Ligand-Linker Conjugates 10 incorporates the (S,R,S)-AHPC based VHL ligand and an alkyl/ether-based linker. E3 ligase Ligand-Linker Conjugates 10 is capable of inducing the degradation of GFP-HaloTag7 in cell-based assays.
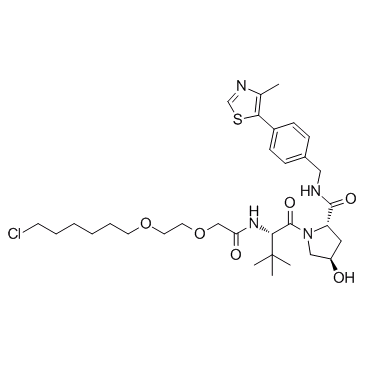
-
GC32987
E3 ligase Ligand-Linker Conjugates 8
E3 ligase Ligand-Linker Conjugates 8 (VH032-C6-PEG3-C4-Cl) is a conjugate of ligands for E3 and 20-atom-length linker. The connector of linker is Halogen group. E3 ligase Ligand-Linker Conjugates 8 incorporates the (S,R,S)-AHPC based VHL ligand and an alkyl/ether-based linker. E3 ligase Ligand-Linker Conjugates 8 is capable of inducing the degradation of GFP-HaloTag7 in cell-based assays.
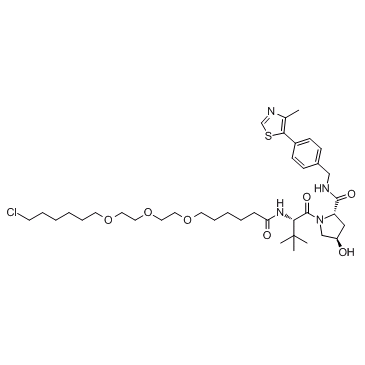
-
GC32976
E3 ligase Ligand-Linker Conjugates 9
E3 ligase Ligand-Linker Conjugates 9 is a conjugate of ligands for E3 and 25-atom-length linker. The connector of linker is Halogen group. E3 ligase Ligand-Linker Conjugates 9 incorporates the (S,R,S)-AHPC based VHL ligand and 6-unit PEG linker. E3 ligase Ligand-Linker Conjugates 9 is capable of inducing the degradation of GFP-HaloTag7 in cell-based assays.
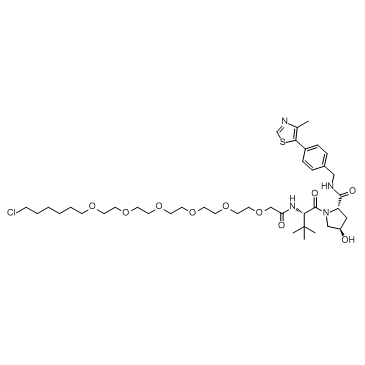
-
GC36001
ERD-308
ERD-308 is a highly potent von Hippel-Lindau-based PROTAC degrader of estrogen receptor (ER) for ER positive breast cancer treatment. ERD-308 induces >95% of ER degradation at concentrations as low as 5 nM in both cell lines (DC50 (concentration causing 50% of protein degradation) of 0.17 nM and 0.43 nM in MCF-7 and T47D ER+ cells, respectively).
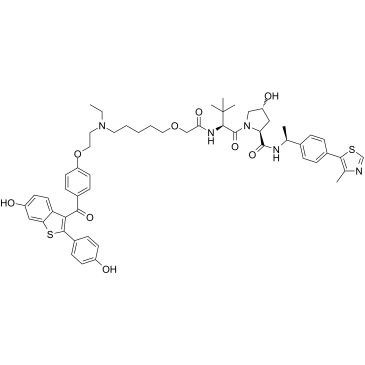
-
GC39264
FKBP12 PROTAC dTAG-13
FKBP12 PROTAC dTAG-13 (dTAG-13), a PROTAC-based heterobifunctional degrader, is a selective degrader of FKBP12F36V with expression of FKBP12F36V in-frame with a protein of interest. FKBP12 PROTAC dTAG-13 effectively engages FKBP12F36V and CRBN, thereby selectively degrading FKBP12F36V.
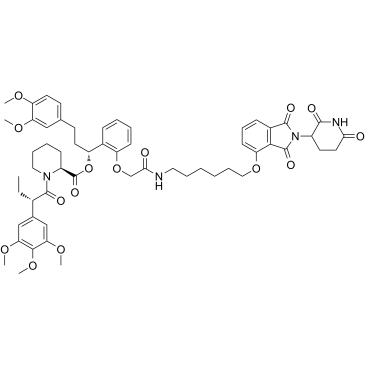
-
GC39289
FKBP12 PROTAC dTAG-7
FKBP12 PROTAC dTAG-7 (dTAG-7) is a heterobifunctional degrader. FKBP12 PROTAC dTAG-7 (dTAG-7) is a degrader of FKBP12F36V with expression of FKBP12F36V in-frame with a protein of interest. FKBP12 PROTAC dTAG-7 (dTAG-7) also is a selective degrader of BET bromodomain transcriptional co-activator BRD4 by bridging BET bromodomains to an E3 ubiquitin ligase CRBN.
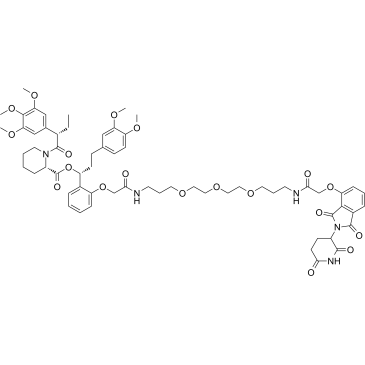
-
GC19509
Gefitinib-based PROTAC 3
A VHL-recruiting PROTAC
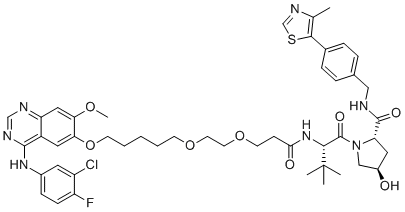
-
GC36167
GMB-475
GMB-475 is a degrader of BCR-ABL1 tyrosine kinase based on PROTAC, overcoming BCR-ABL1-dependent drug resistance. GMB-475 targets BCR-ABL1 protein and recruits the E3 ligase Von Hippel Lindau (VHL), resulting in ubiquitination and subsequent degradation of the oncogenic fusion protein.
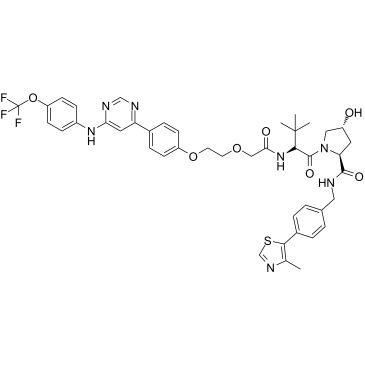
-
GC32831
HaloPROTAC 2
HaloPROTAC 2 (HaloPROTAC 2) is a conjugate of ligands for E3 and 21-atom-length linker. The connector of linker is Halogen group. HaloPROTAC 2 incorporates the VH032 based VHL ligand and 5-unit PEG linker. HaloPROTAC 2 is capable of inducing the degradation of GFP-HaloTag7 in cell-based assays.
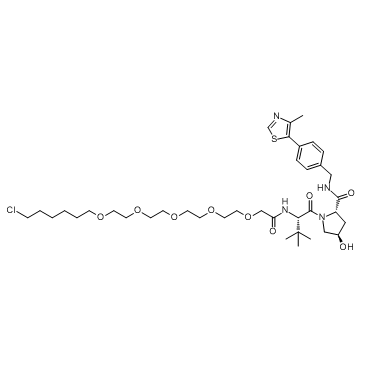
-
GC40877
Heclin
Heclin is an inhibitor of HECT E3 ubiquitin ligases (IC50s = 6.8, 6.3, and 6.9 μM for Smurf2, Nedd4, and WWP1 HECT ligase domains, respectively).
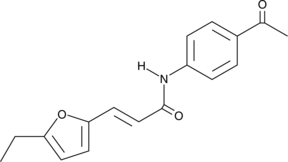
-
GC33391
Homo-PROTAC cereblon degrader 1
Homo-PROTAC cereblon degrader 1 (compound 15a) is a highly potent and efficient Cereblon (CRBN) degrader with only minimal effects on IKZF1 and IKZF3.
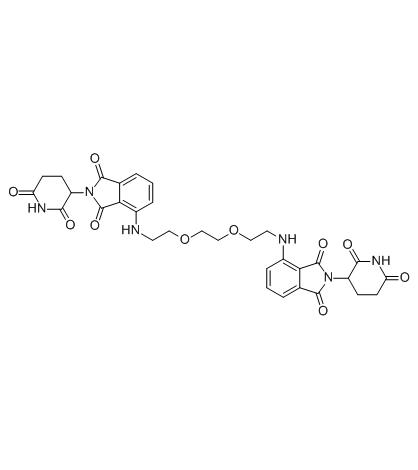
-
GC65285
Homo-PROTAC pVHL30 degrader 1
Homo-PROTAC pVHL30 degrader 1 is a potent pVHL30 degrader based on PROTAC, consists of two ligands of von Hippel-Lindau.
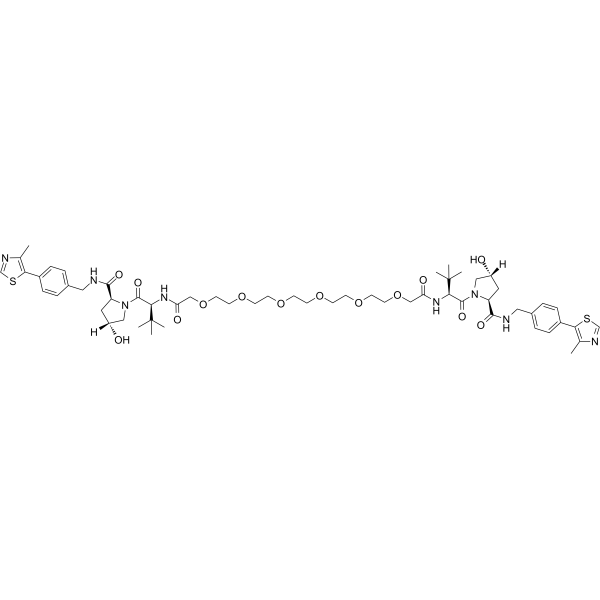
-
GC65559
INY-03-041
INY-03-041 is a potent, highly selective and PROTAC-based pan-AKT degrader consisting of the ATP-competitive AKT inhibitor GDC-0068 conjugated to Lenalidomide (Cereblon ligand). INY-03-041 inhibits AKT1, AKT2 and AKT3 with IC50s of 2.0 nM, 6.8 nM and 3.5 nM, respectively.
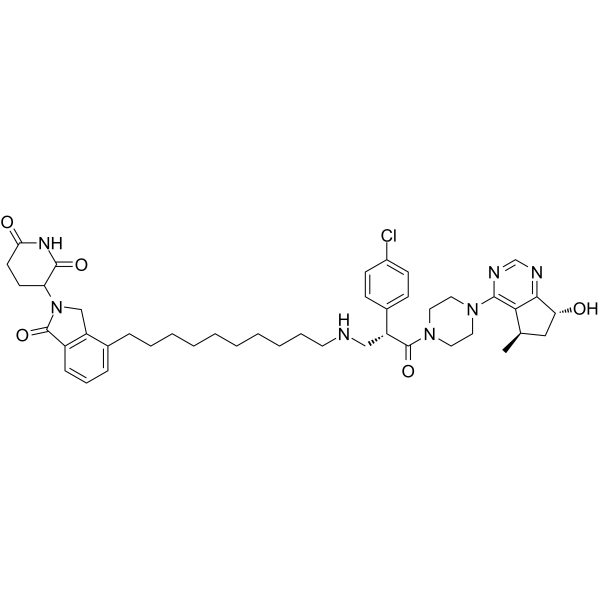
-
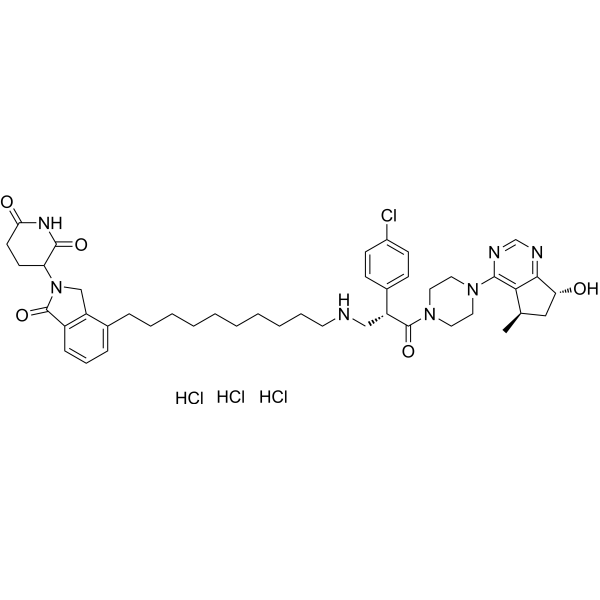
GC67757
INY-03-041 trihydrochloride
-
GC63707
JB170
JB170 is a potent and highly specific PROTAC-mediated AURORA-A (Aurora Kinase) degrader (DC50=28 nM) by linking Alisertib, to the Cereblon-binding molecule Thalidomide. JB170 preferentially binds AURORA-A (EC50=193 nM) over AURORA-B (EC50=1.4 ?M). JB170-mediated S-phase arrest is caused specifically by AURORA-A depletion. JB170 has excellent ability to inhibit non-catalytic function of AURORA-A kinase.
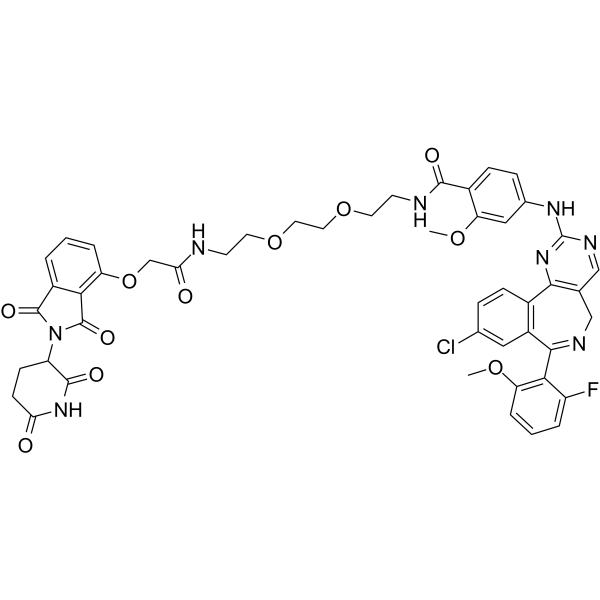
-
GC65471
JH-XI-10-02
JH-XI-10-02 is a PROTAC connected by ligands for Cereblon and CDK. JH-XI-10-02 is a highly potent and selective PROTAC CDK8 degrader, with an IC50 of 159 nM. JH-XI-10-02 causes proteasomal degradation, does not affect CDK8 mRNA levels. JH-XI-10-02 shows no effect on CDK19.
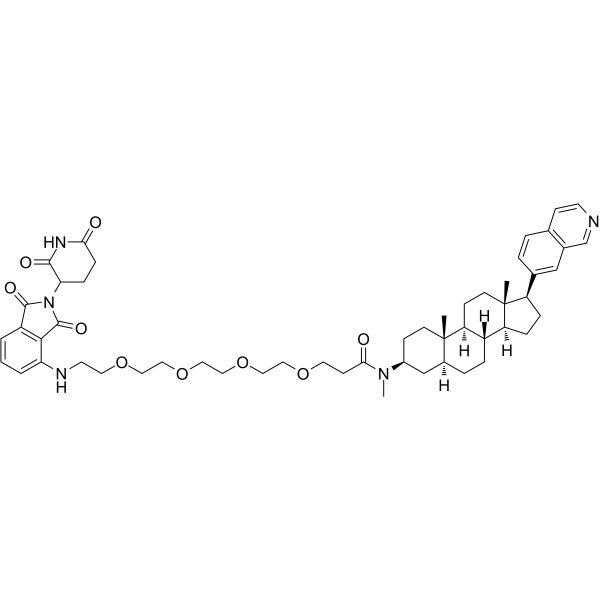
-
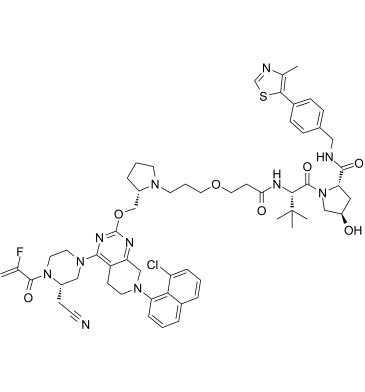
GC61766
LC-2
LC-2 is a potent and first-in-class von Hippel-Lindau-based PROTAC capable of degrading endogenous KRAS G12C, with DC50s between 0.25 and 0.76 μM. LC-2 covalently binds KRAS G12C with a MRTX849 warhead and recruits the E3 ligase VHL, inducing rapid and sustained KRAS G12C degradation leading to suppression of MAPK signaling in both homozygous and heterozygous KRAS G12C cell lines.
-
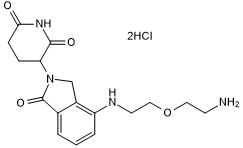
GC50669
Lenalidomide 4'-PEG1-amine
-
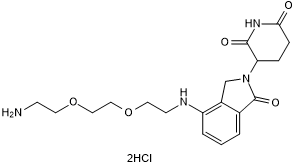
GC50684
Lenalidomide 4'-PEG2-amine
-
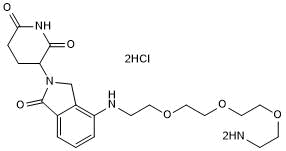
GC50694
Lenalidomide 4'-PEG3-amine
-
GC38812
MD-224
MD-224 is a first-in-class and highly potent small-molecule human murine double minute 2 (MDM2) degrader based on the proteolysistargeting chimera (PROTAC) concept. MD-224 consists of ligands for Cereblon and MDM2. MD-224 induces rapid degradation of MDM2 at concentrations <1 nM in human leukemia cells, and achieves an IC50 value of 1.5 nM in inhibition of growth of RS4;11 cells. MD-224 has the potential to be a new class of anticancer agent.
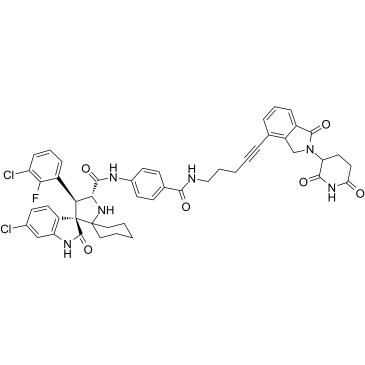
-
GC64651
MI-389
MI-389 is a PROTAC translation termination factor GSPT1 degrader. MI-389 disrupts a target that is a shared dependency in different AML and ALL cell lines, and that MI-389 action is dependent on the CRL4CRBN E3 ligase.
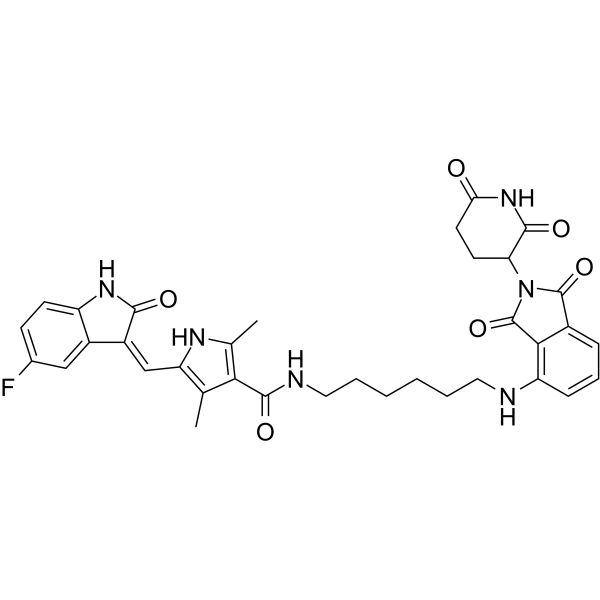
-
GC69501
MS159
MS159 is an effective nuclear receptor binding SET domain protein 2 (NSD2) PROTAC degrader. MS159 can inhibit the growth of tumor cells. MS159 is an effective chemical tool for exploring the role of NSD2 in health and disease.
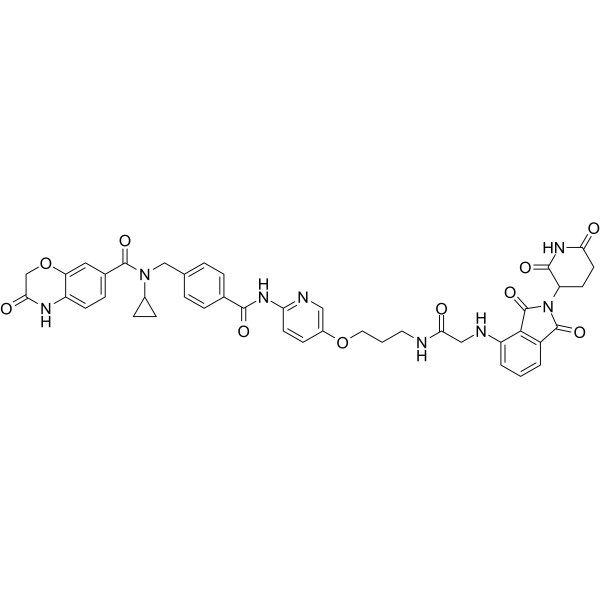
-
GC65466
MS170
MS170 is a potent and selective PROTAC AKT degrader. MS170 depletes cellular total AKT (T-AKT) with the DC50 value of 32 nM. MS170 binds to AKT1, AKT2, and AKT3 with Kds of 1.3 nM, 77 nM, and 6.5 nM, respectively.
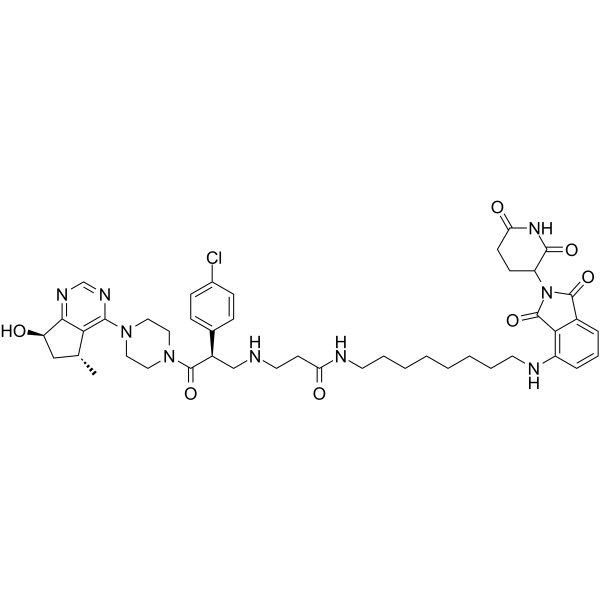
-
GC67710
MS177
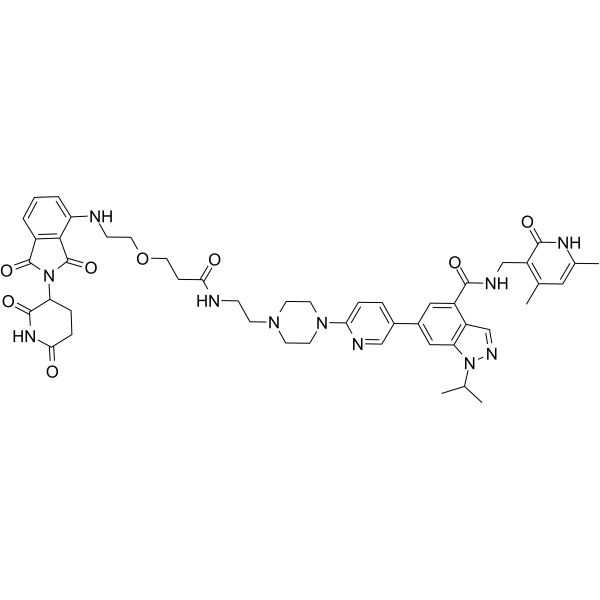
-
GC69502
MS21
MS21 is a novel AKT degrader that selectively inhibits the growth of mutant cancers through the PI3K/PTEN pathway.
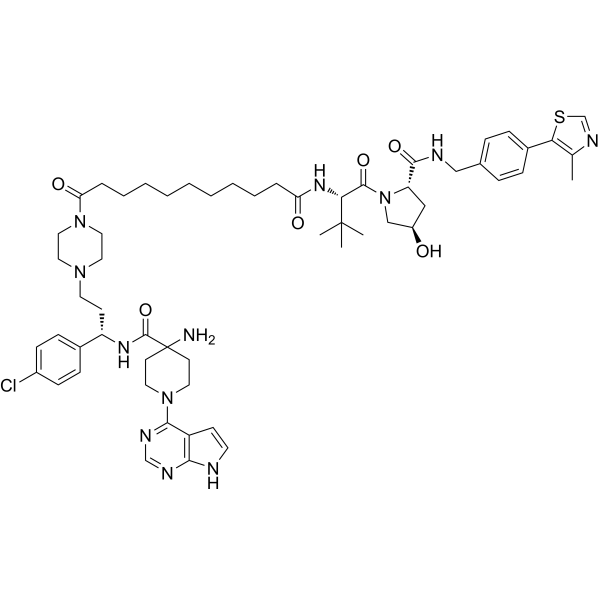
-
GC65243
MS4077
MS4077 is an anaplastic lymphoma kinase (ALK) PROTAC (degrader) based on Cereblon ligand, with a Kd of 37?nM for binding affinity to ALK.
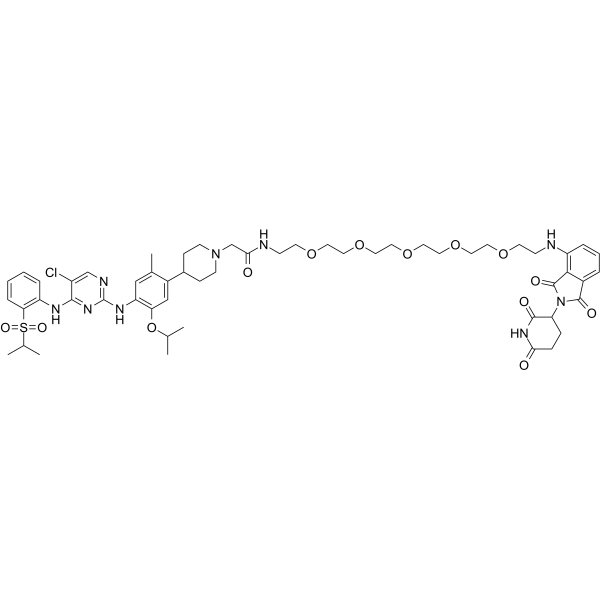
-
GC64966
MS4078
MS4078 is an anaplastic lymphoma kinase (ALK) PROTAC (degrader) based on Cereblon ligand, with a Kd of 19?nM for binding affinity to ALK.
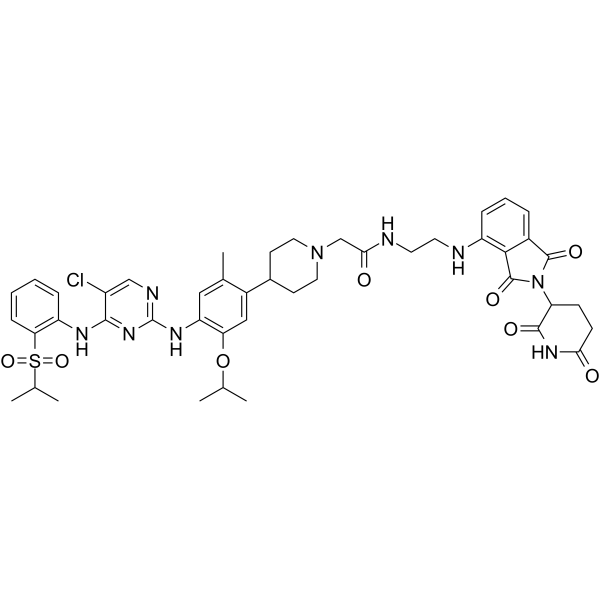
-
GC65052
MS4322
MS4322 (YS43-22) is a first-in-class PRMT5 degrader and a valuable chemical tool (PROTAC) for exploring the PRMT5 functions in health and disease.
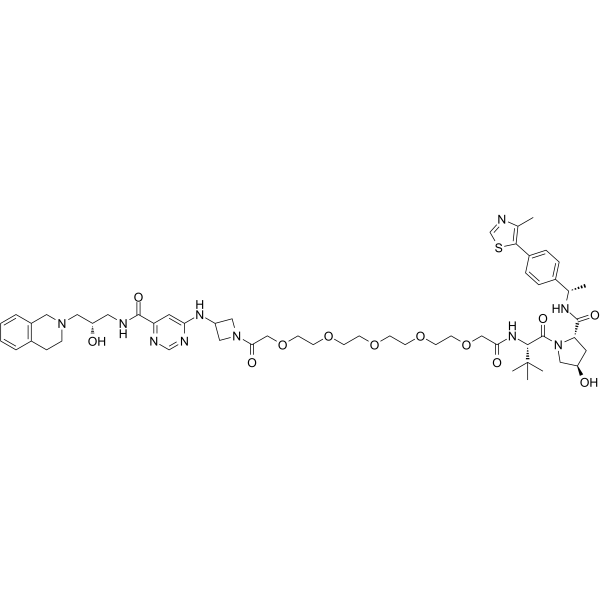
-
GC65876
MS4322 (isomer)
MS4322 (YS43-22) isomer is an isomer of MS4322. MS4322 is a potent and selective PRMT5 (protein arginine methyltransferase 5) degrader, and inhibits growth of multiple cancer cell lines.
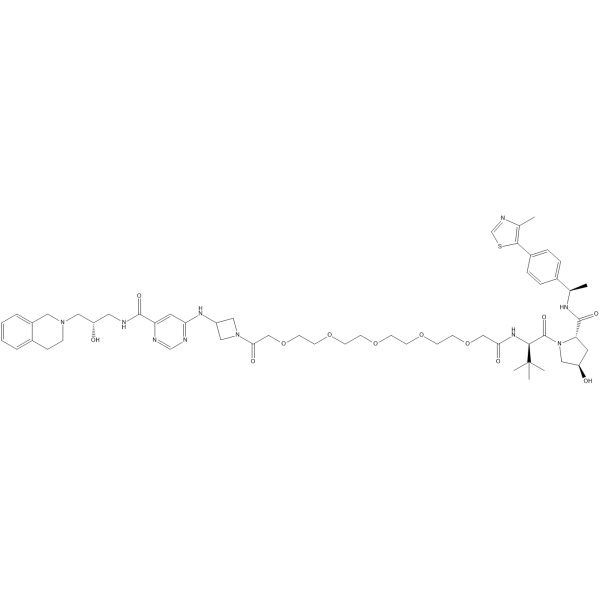
-
GC69505
MS9427
MS9427 is an effective PROTAC EGFR degrader, with Kd values of 7.1 nM and 4.3 nM for wild-type EGFR and mutant EGFR L858R, respectively. MS9427 selectively degrades mutant variants through the ubiquitin/proteasome system (UPS) and autophagy/lysosome pathway, but does not degrade WT EGFR. MS9427 has a strong inhibitory effect on NSCLC cell proliferation and can be used in cancer research.
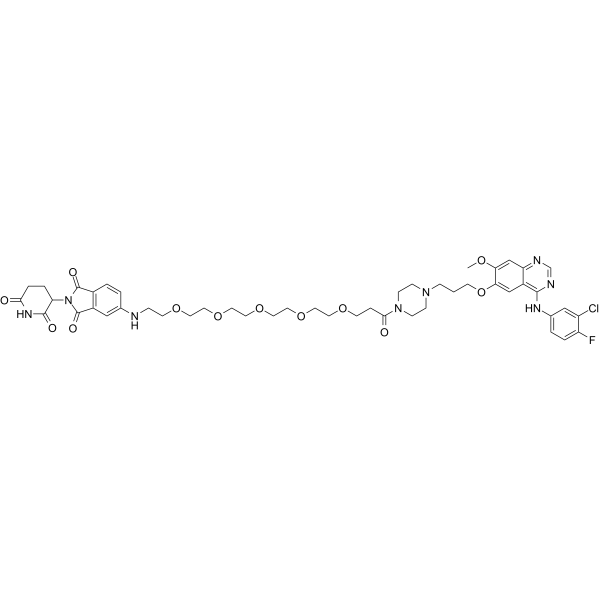
-
GC69506
MS9427 TFA
MS9427 TFA is an effective PROTAC EGFR degrader, with Kd values of 7.1 nM and 4.3 nM for wild-type EGFR and mutant EGFR L858R, respectively. MS9427 TFA selectively degrades mutant variants through the ubiquitin/proteasome system (UPS) and autophagy/lysosome pathway, but not WT EGFR. MS9427 TFA strongly inhibits NSCLC cell proliferation and can be used in cancer research.
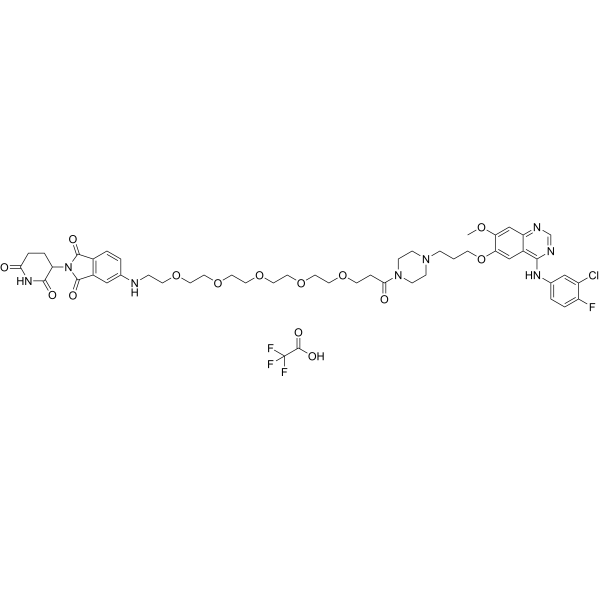
-
GC36661
MT-802
MT-802 is a potent BTK degrader based on Cereblon ligand, with a DC50 of 1 nM.
-
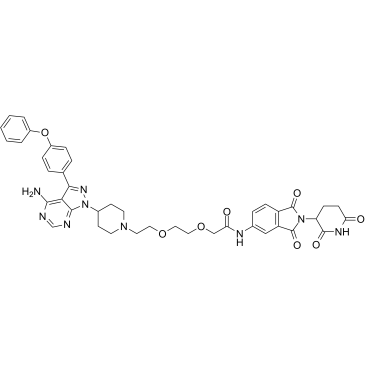
GC69744
MTX-23
MTX-23 is an AR-based PROTAC. MTX-23 inhibits CaP cell proliferation by degrading AR-V7 and AR-FL. MTX-23 induces apoptosis in cells.
-
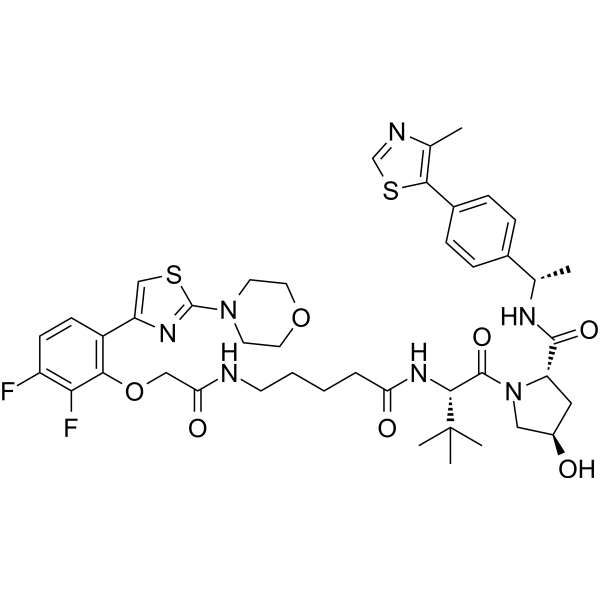
GC18729
MZ1
MZ1 is a hybrid compound that drives the selective proteasomal degradation of bromodomain-containing protein 4 (BRD4).
-
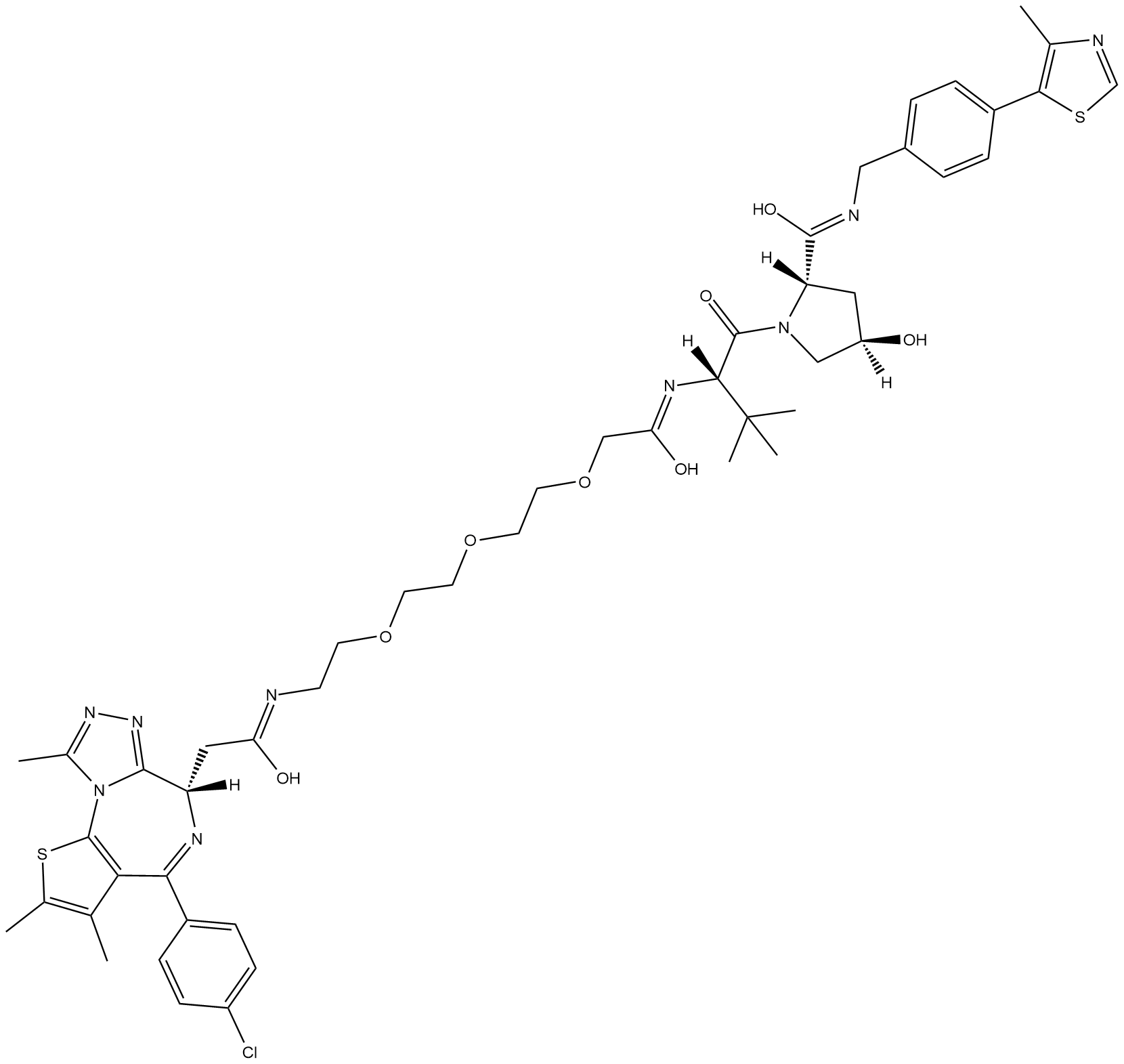
GC33102
MZP-54
MZP-54 is a PROTAC connected by ligands for von Hippel-Lindau and BRD3/4, with a Kd of 4 nM for Brd4BD2.
-
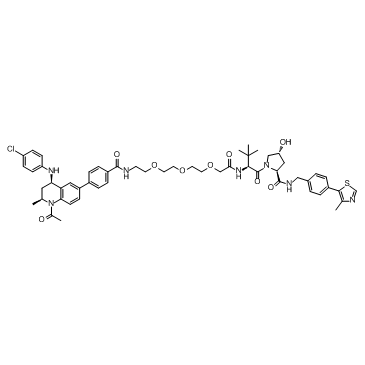
GC33363
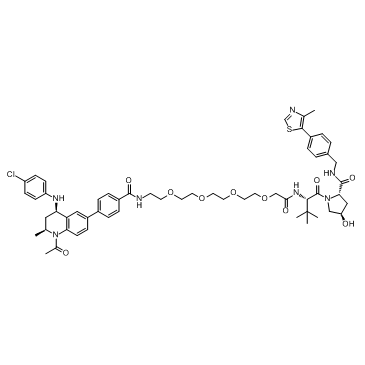
MZP-55
MZP-55 is a PROTAC connected by ligands for von Hippel-Lindau and BRD3/4, with a Kd of 8 nM for Brd4BD2.
-
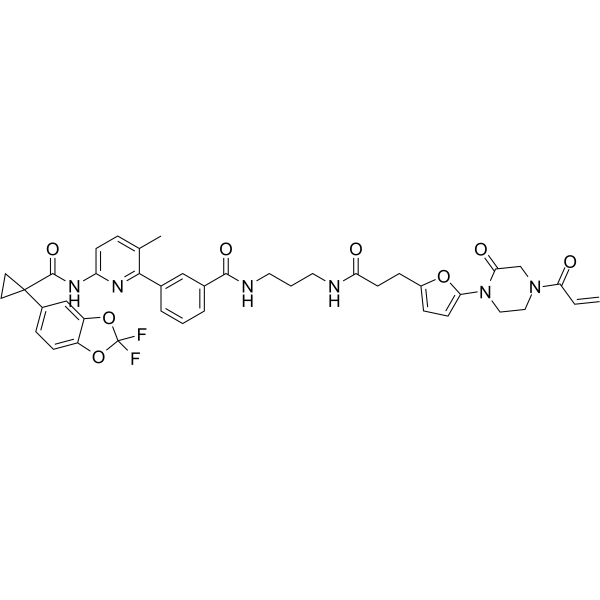
GC67889
NJH-2-056
-
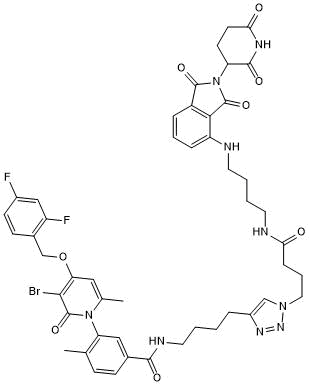
GC50740
NR 7h
-
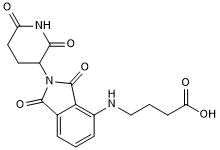
GC50665
Pomalidomide 4'-alkylC3-acid
-
GC50658
Pomalidomide 4'-alkylC3-amine
Pomalidomide 4'-alkylC3-amine is a synthesized E3 ligase ligand-linker conjugate that incorporates the Pomalidomide based cereblon ligand and a linker used in PROTAC technology.
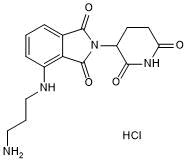
-
GC50674
Pomalidomide 4'-alkylC4-acid
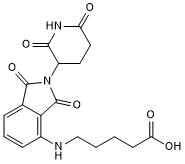
-
GC50546
Pomalidomide 4'-alkylC5-acid
Cereblon ligand with alkyl linker and terminal acid for onward chemistry
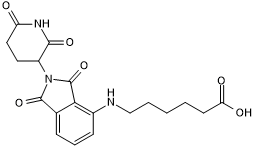
-
GC50554
Pomalidomide 4'-alkylC5-amine
Pomalidomide 4'-alkylC5-amine is a synthesized E3 ligase ligand-linker conjugate that incorporates the Thalidomide based cereblon ligand and a linker used in PROTAC technology.
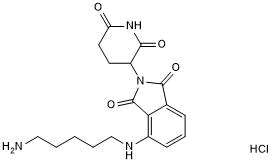
-
GC50689
Pomalidomide 4'-alkylC6-acid
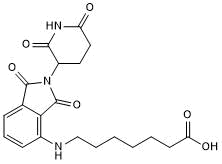
-
GC50696
Pomalidomide 4'-alkylC8-acid
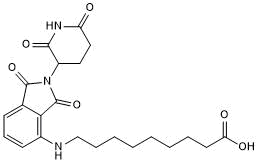
-
GC50675
Pomalidomide 4'-PEG1-acid
Pomalidomide 4'-PEG1-acid is a synthesized E3 ligase ligand-linker conjugate that incorporates the Pomalidomide based cereblon ligand and 1-unit PEG linker used in PROTAC technology.
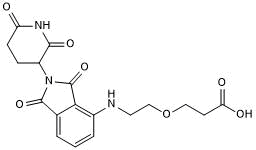
-
GC50690
Pomalidomide 4'-PEG2-acid
Pomalidomide 4'-PEG2-acid (Pomalidomide 4'-PEG2-acid) is a synthesized E3 ligase ligand-linker conjugate that incorporates the Pomalidomide based cereblon ligand and 2-unit PEG linker used in PROTAC technology.
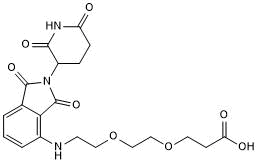
-
GC50530
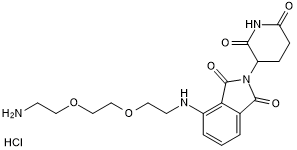
Pomalidomide 4'-PEG2-amine
Pomalidomide 4'-PEG2-amine is a synthesized E3 ligase ligand-linker conjugate that incorporates the Thalidomide based cereblon ligand and 2-unit PEG linker used in PROTAC technology.
-
GC50705
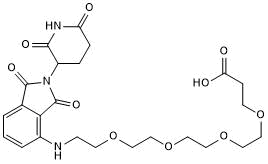
Pomalidomide 4'-PEG4-acid
Pomalidomide 4'-PEG4-acid is a E3 ligase ligand-linker conjugate.
-
GC50625
Pomalidomide 4'-PEG4-amine
Pomalidomide 4'-PEG4-amine is a synthesized E3 ligase ligand-linker conjugate that incorporates the Pomalidomide based cereblon ligand and 4-unit PEG linker used in the synthesis of PROTACs..