P2X purinergic receptor
P2X purinergic receptor is a famliy of ATP-gated cation channels. It is widely distributed in the body and paly an important role in regulation of synaptic transmission, vascular endothelium and nociception etc.
Products for P2X purinergic receptor
- Cat.No. Product Name Information
-
GC62560
(E/Z)-Sivopixant
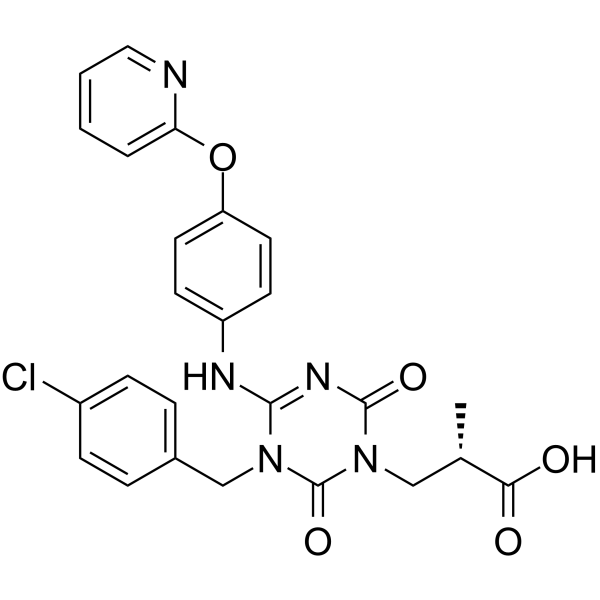
(E/Z)-Sivopixant ((E/Z)-S-600918) is a potent P2X3 receptor antagonist with an IC50 of 4 nM.
-
GC15416
2-Methylthioadenosine triphosphate tetrasodium salt
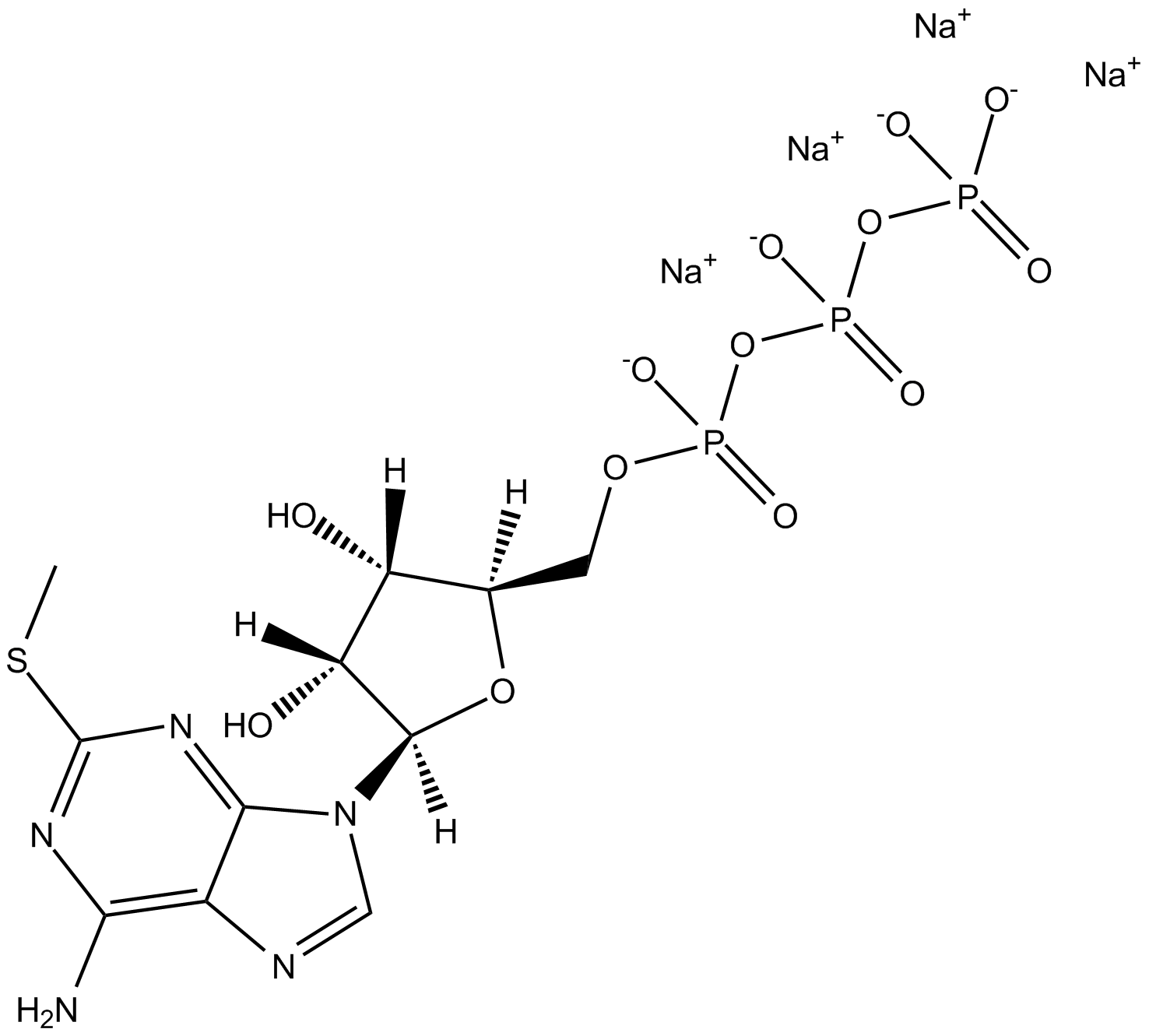
2-Methylthioadenosine triphosphate tetrasodium salt is a non-specific P2-receptor agonist.
-
GC13943
5-BDBD
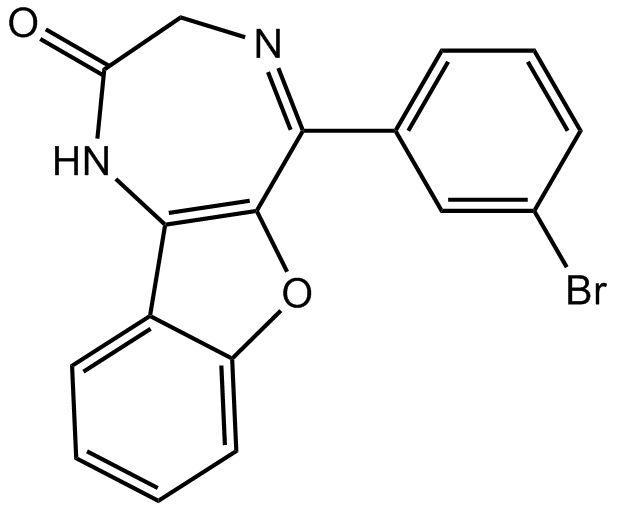
P2X4 receptor antagonist
-
GC50474
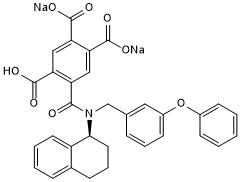
A 317491 sodium salt
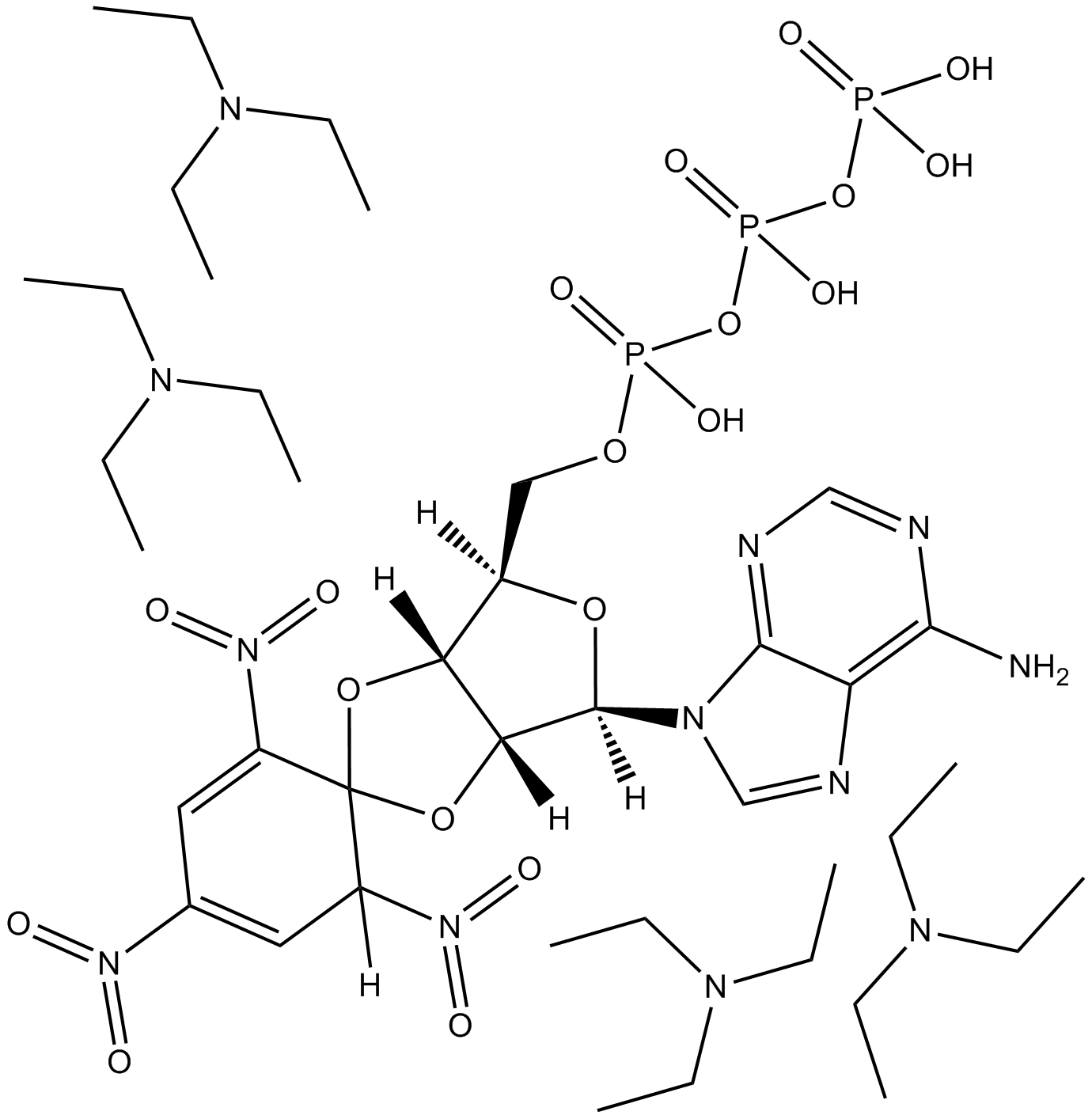
Selective, high affinity P2X3 and P2X2/3 receptor antagonist; antinociceptive
-
GC17212
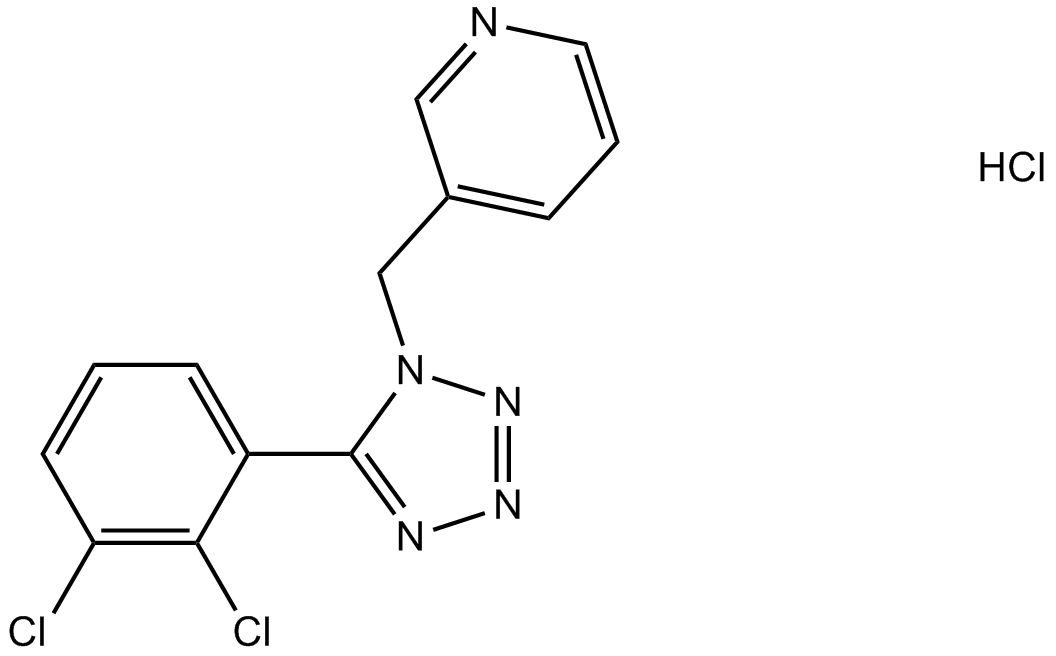
A 438079
An antagonist of the nucleotide receptor P2X7
-
GC13733
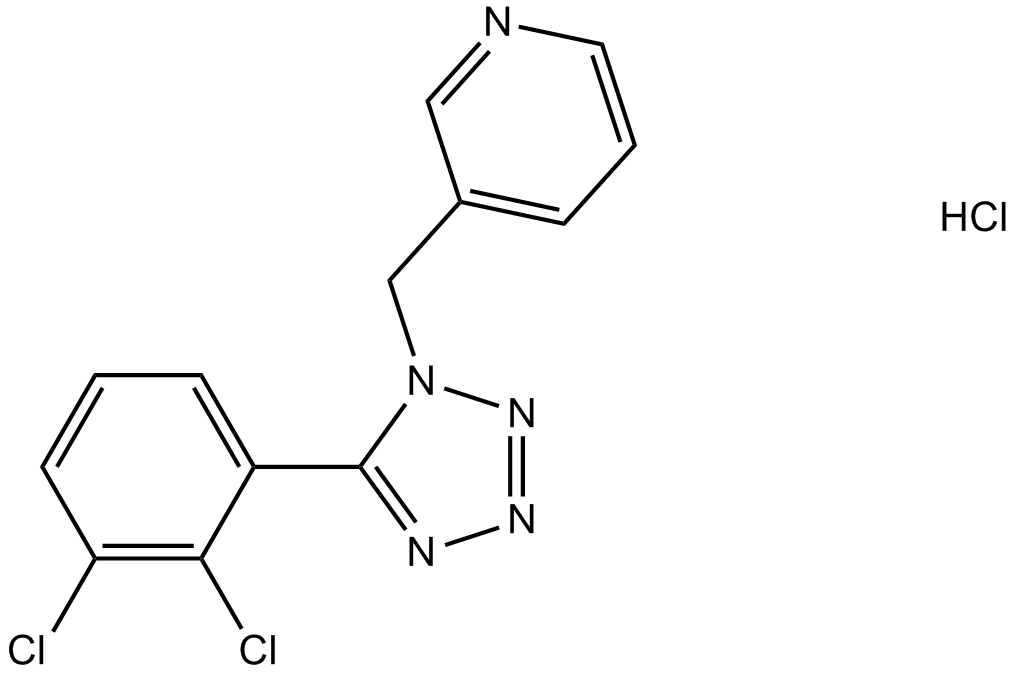
A 438079 hydrochloride
An antagonist of the nucleotide receptor P2X7
-
GC17870
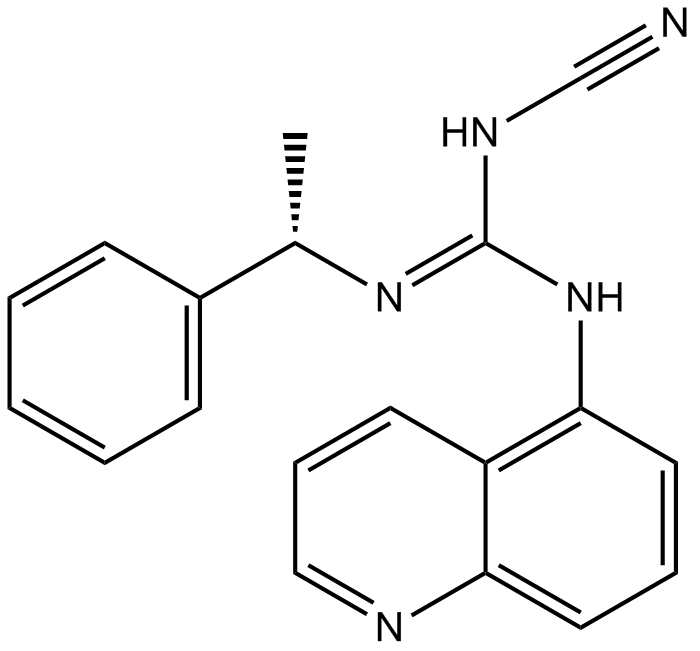
A 804598
P2X7 antagonist,potent and selective
-
GC15434
A 839977
P2X7 antagonist,potent and selective
-
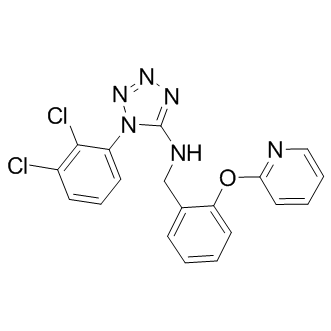
GC17710
A-317491
A P2X3 and P2X2/3 receptor antagonist
-
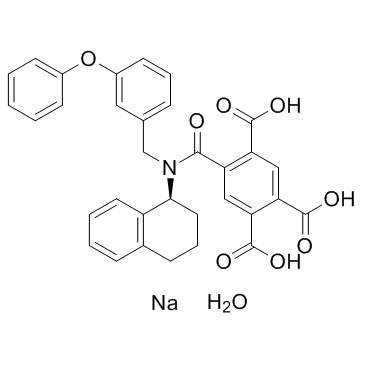
GC35211
A-317491 sodium salt hydrate
A-317491 sodium salt hydrate is a potent, selective and non-nucleotide antagonist of P2X3 and P2X2/3 receptors, with Kis of 22, 22, 9, and 92 nM for hP2X3, rP2X3, hP2X2/3, and rP2X2/3, respectively.
-
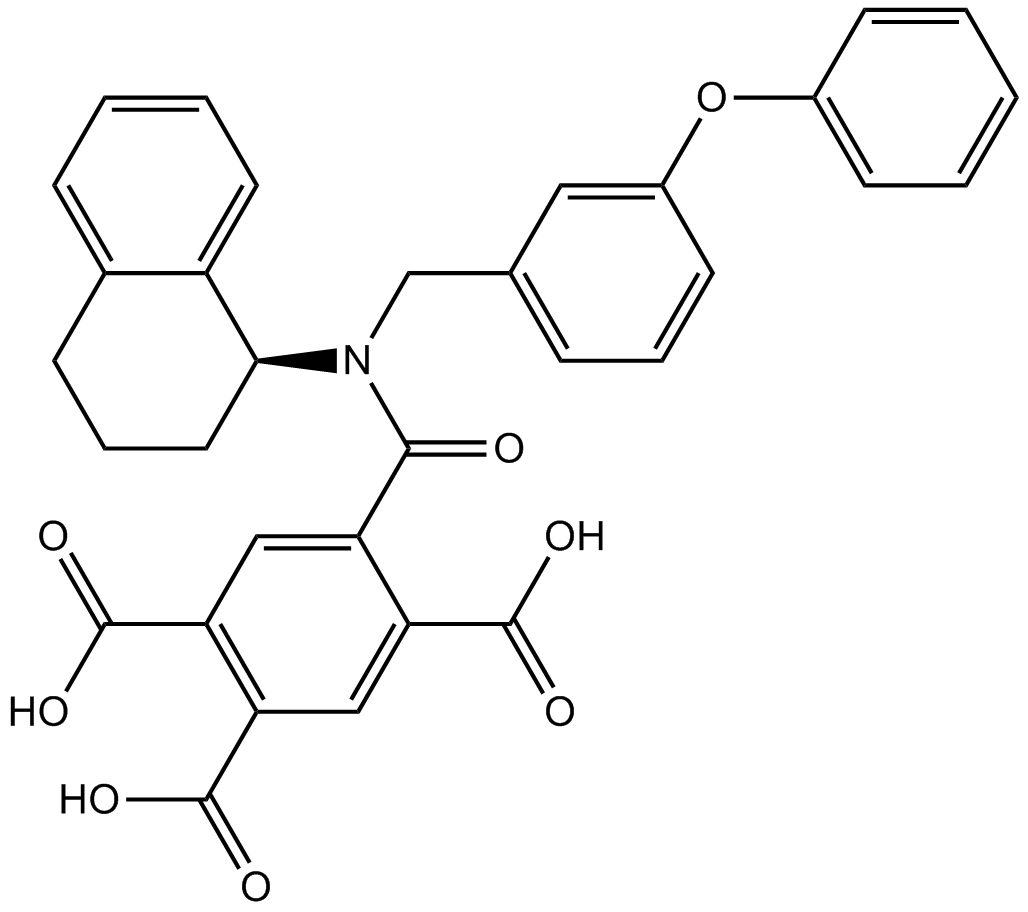
GC11842
A-740003
A selective P2X7 antagonist
-
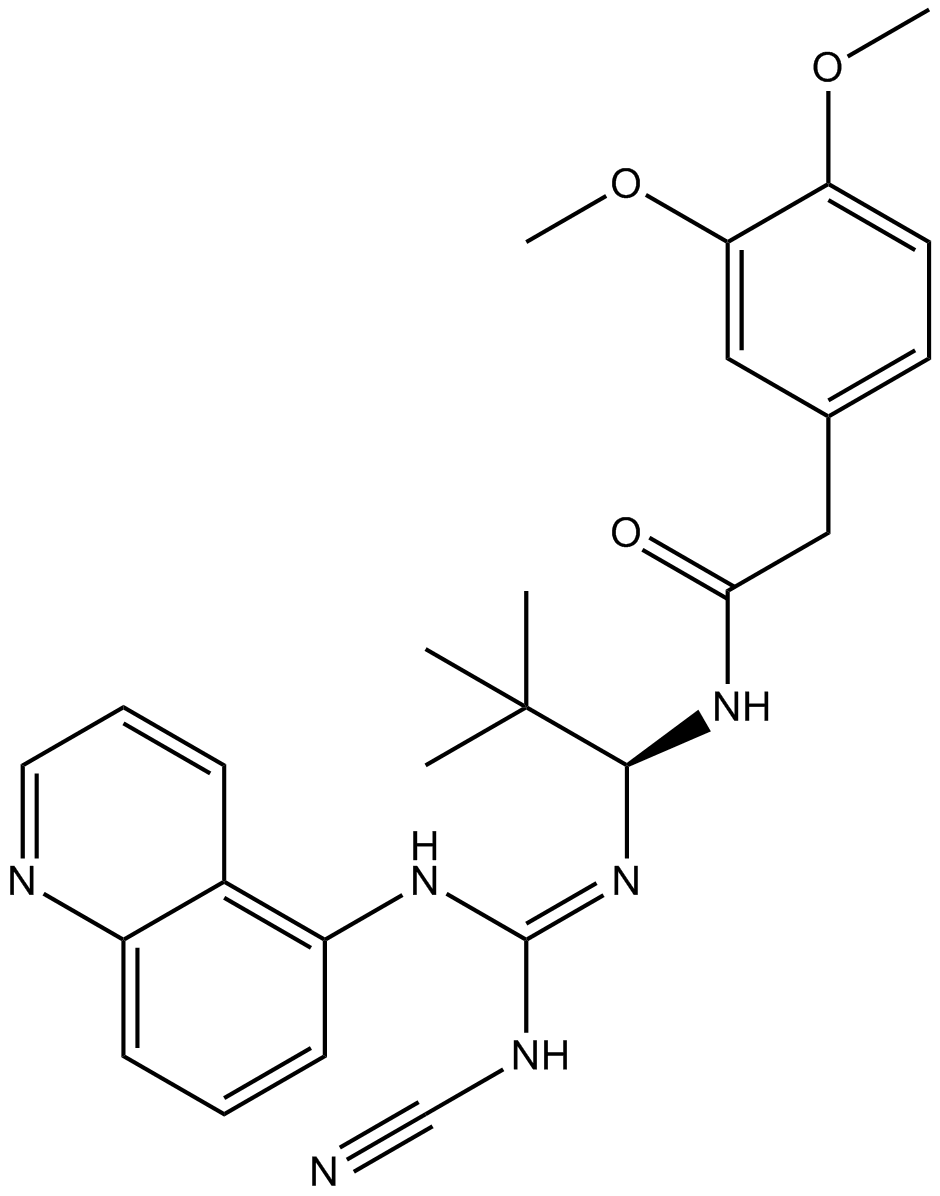
GC19022
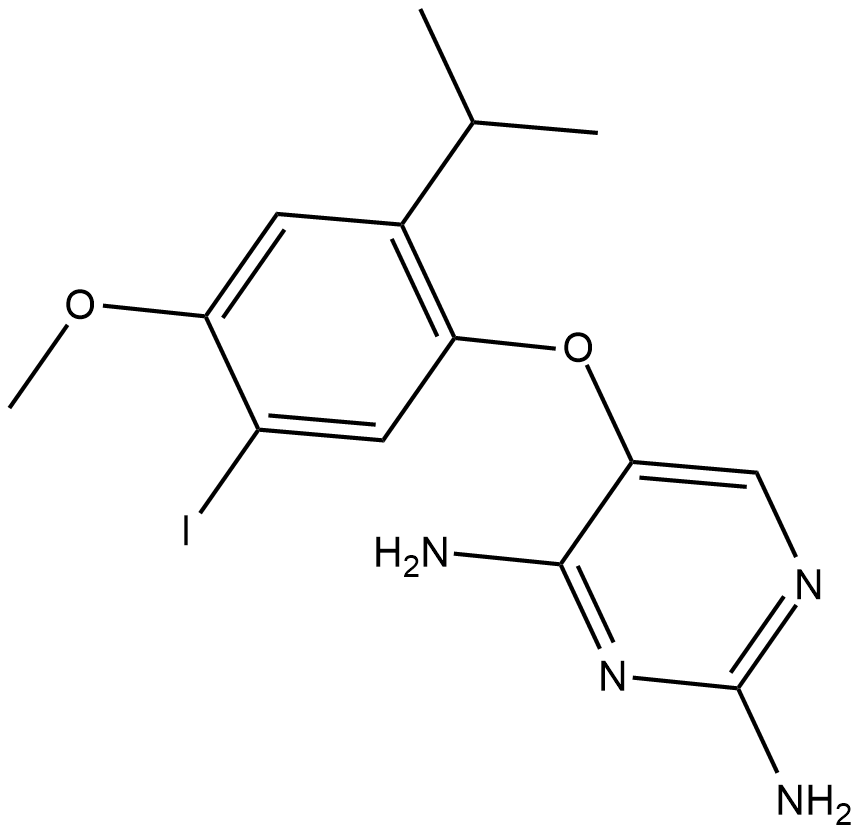
AF-353
AF-353 is a novel, potent and orally bioavailable P2X3/P2X2/3 receptor antagonist, inhibits human and rat P2X3 (pIC50= 8.0).
-
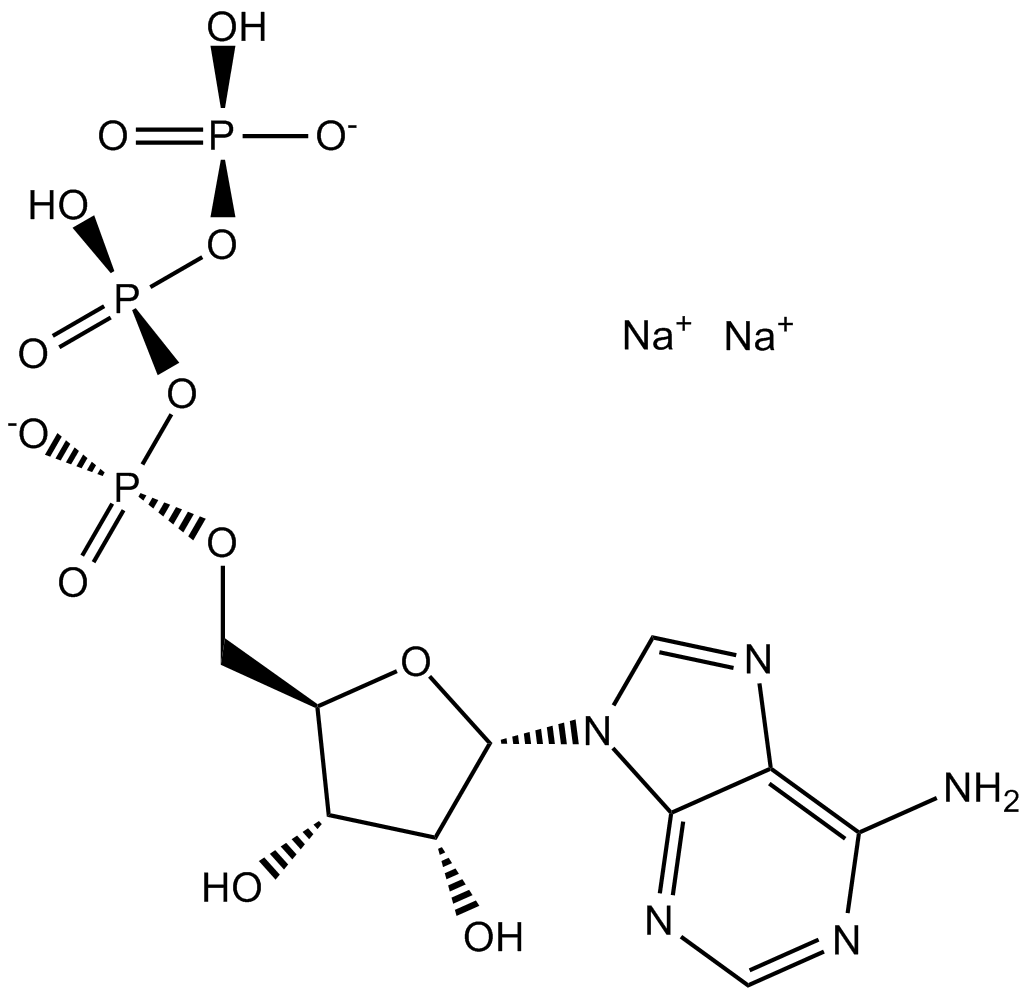
GC11109
ATP disodium salt
P2 purinoceptor agonist
-
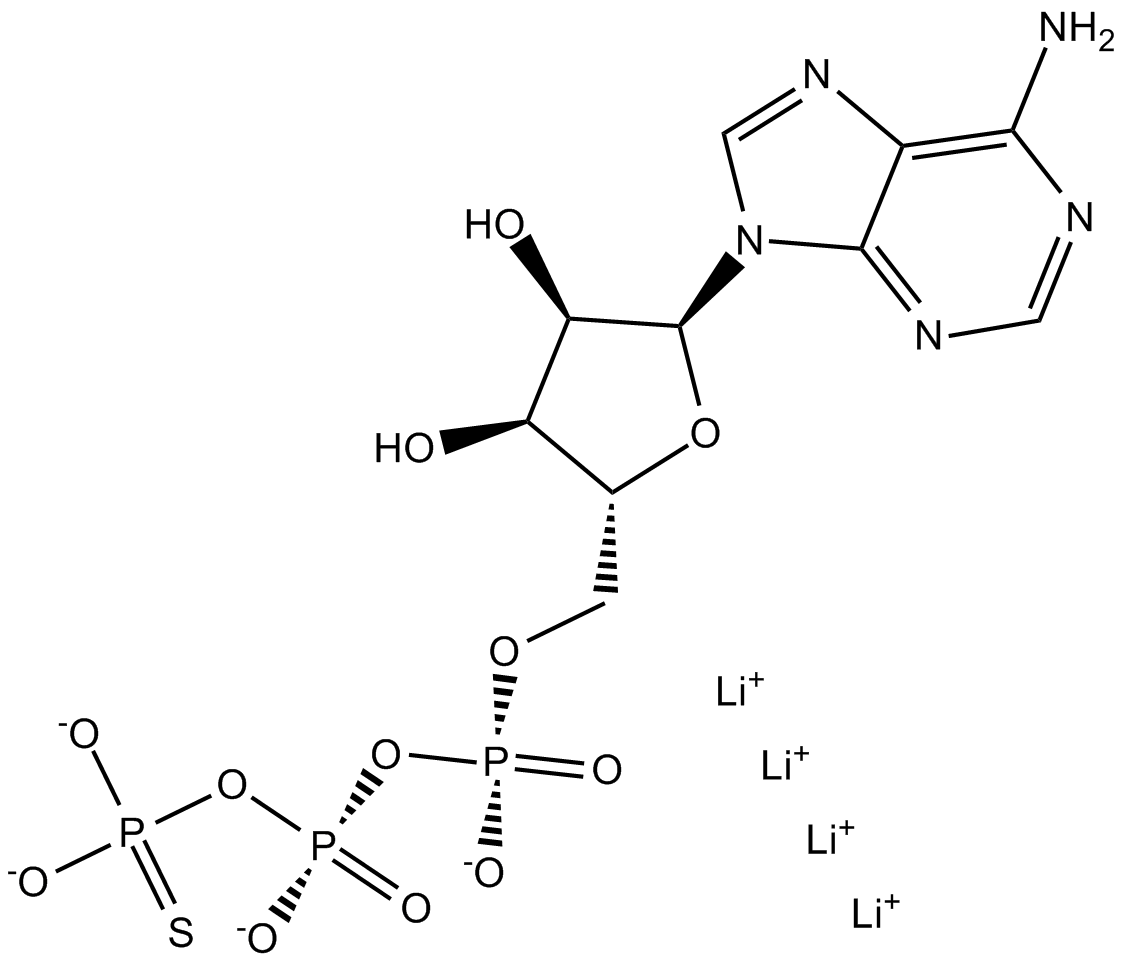
GC12574
ATPγS tetralithium salt
ATPγS (tetralithium salt) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A.
-
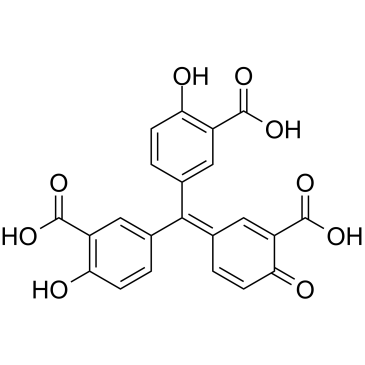
GC39699
Aurintricarboxylic acid
Aurintricarboxylic acid is a nanomolar-potency, allosteric antagonist with selectivity towards αβ-methylene-ATP-sensitive P2X1Rs and P2X3Rs, with IC50s of 8.6 nM and 72.9 nM for rP2X1R and rP2X3R, respectively.
-
GC17375
AZ 10606120 dihydrochloride
Potent P2X7 receptor antagonist
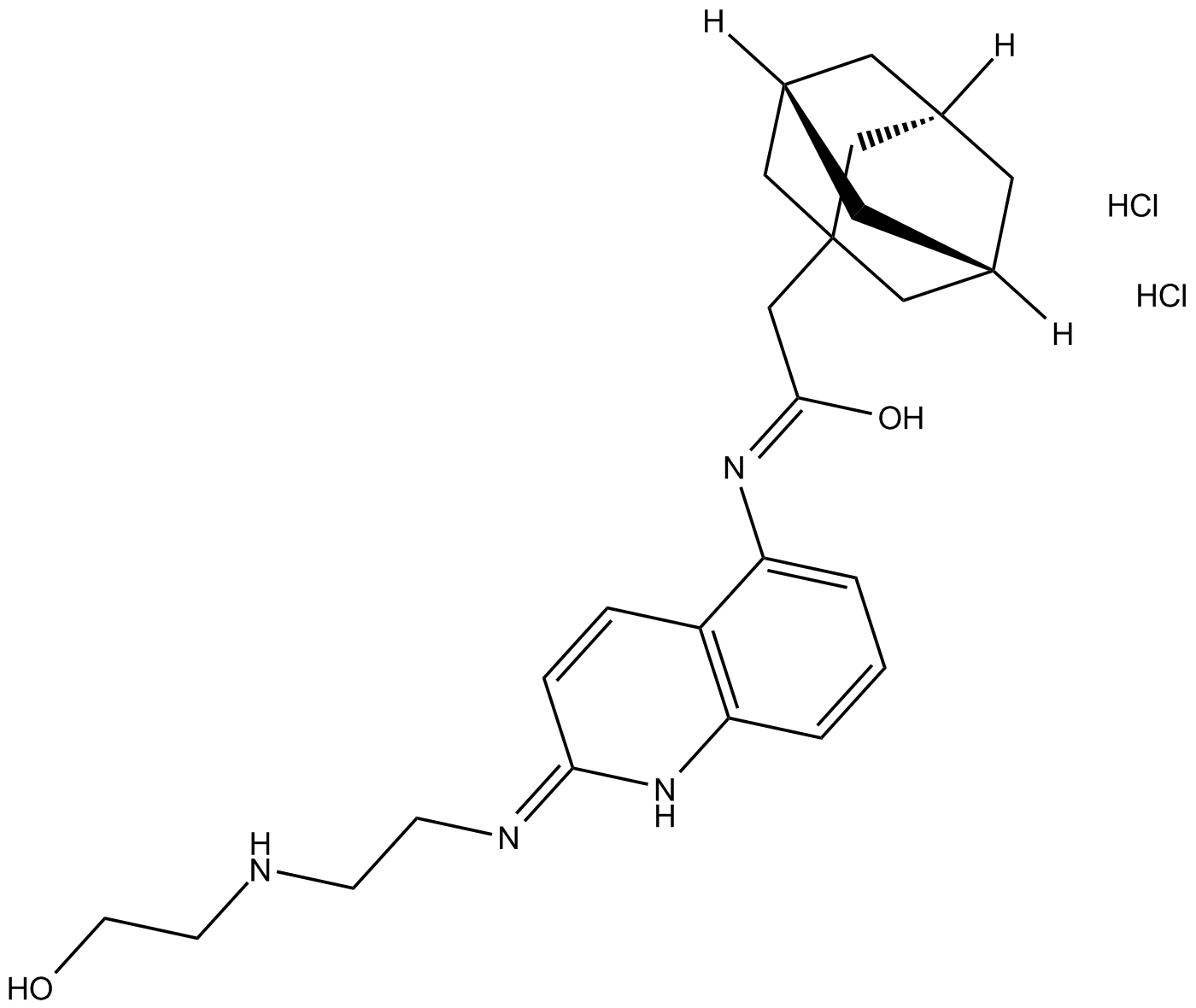
-
GC18052
AZ 11645373
Human P2X7 antagonist,potent and selective
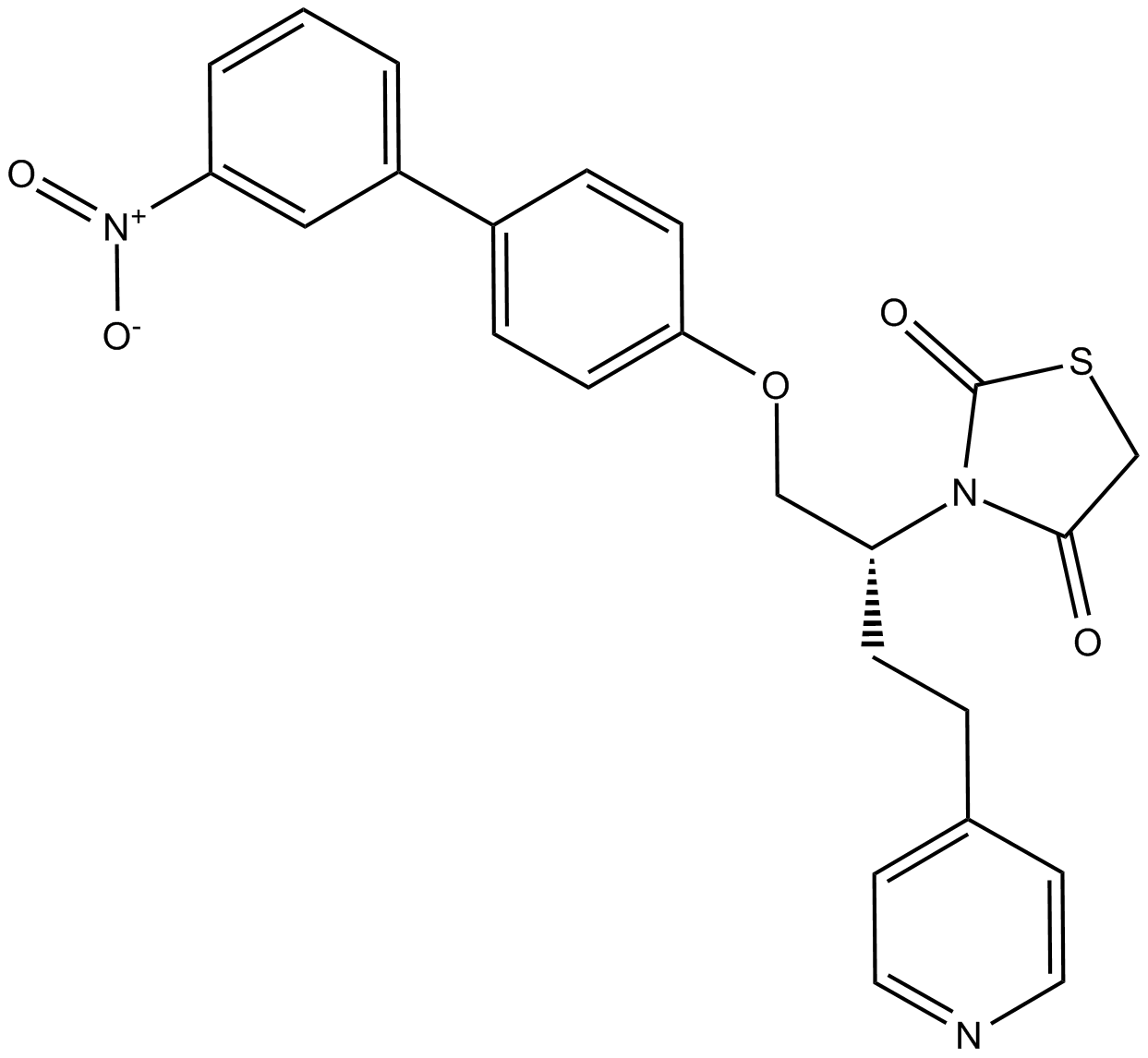
-
GC31674
AZD9056 hydrochloride
AZD9056 hydrochloride is a selective orally active inhibitor of P2X7 which plays a significant role in inflammation and pain-causing diseases.
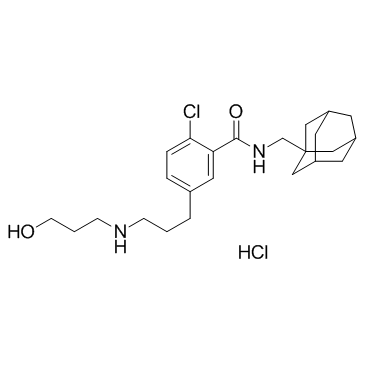
-
GC60090
BLU-5937
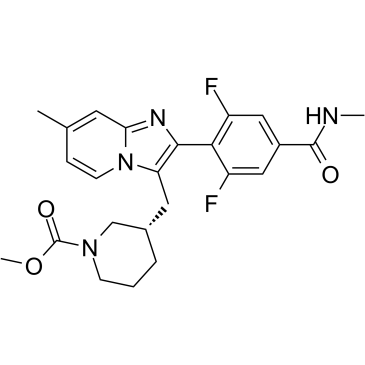
-
GC12421
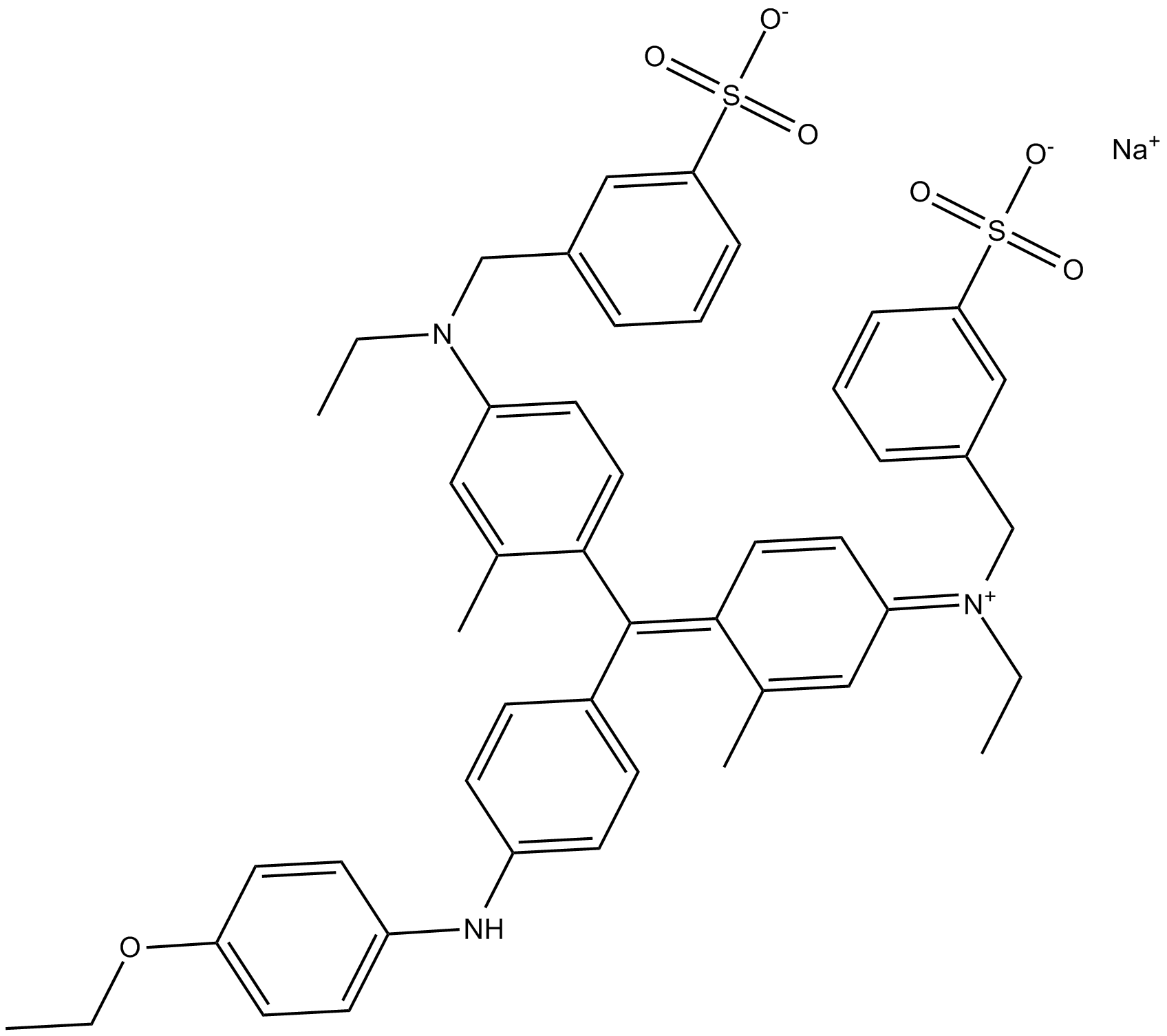
Brilliant blue G-250
Brilliant Blue G-250 is a dye commonly used for the visualization of proteins separated by SDS-PAGE, offering a simple staining procedure and high quantitation.
-
GC35567
Bullatine A
Bullatine A, a diterpenoid alkaloid of the genus Aconitum, possesses anti-rheumatic, anti-inflammatory and anti-nociceptive effects.

-
GC50200
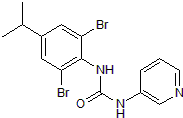
BX 430
Selective P2X4 allosteric antagonist
-
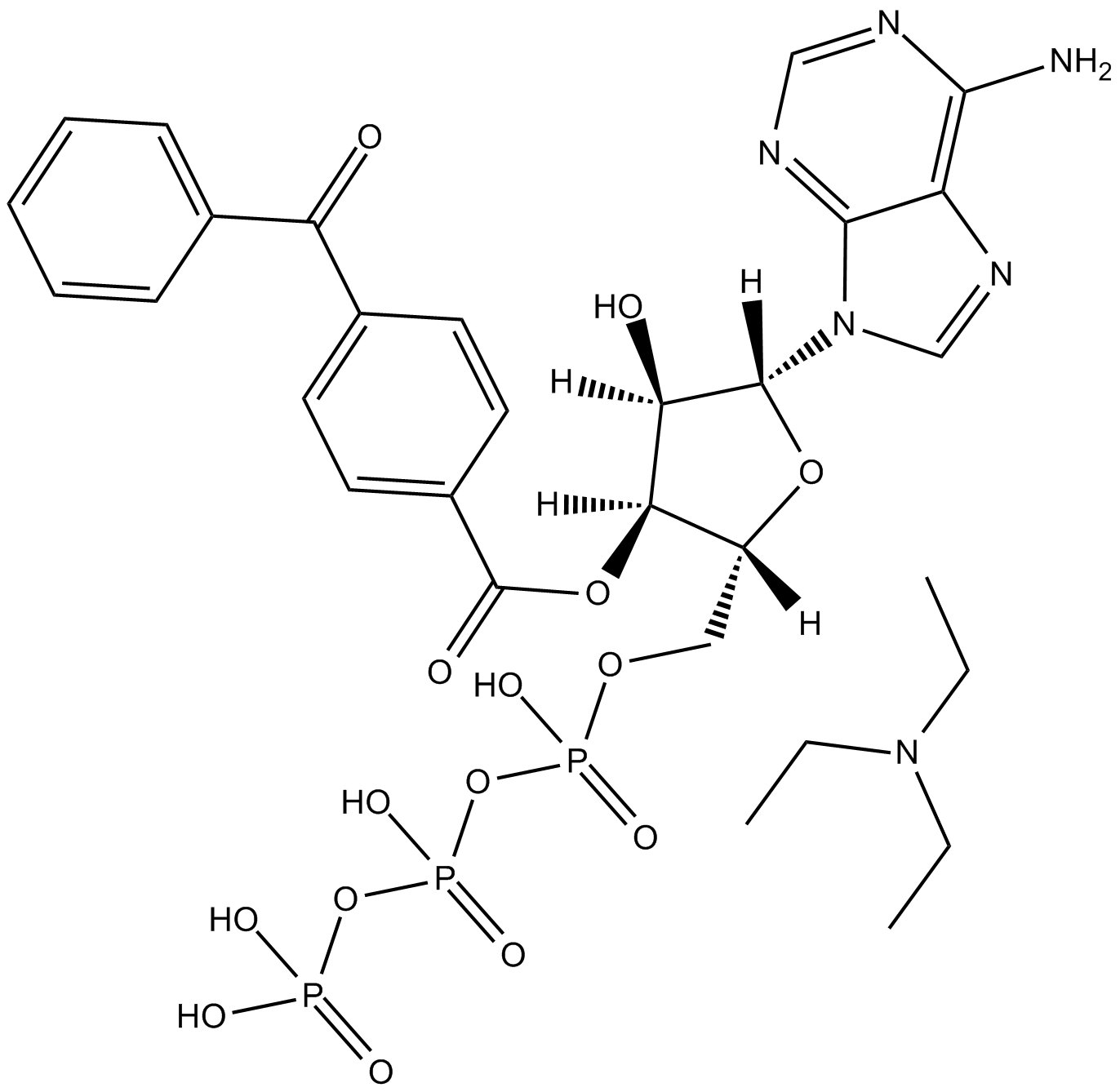
GC15898
BzATP triethylammonium salt
P2X7 receptor agonist
-
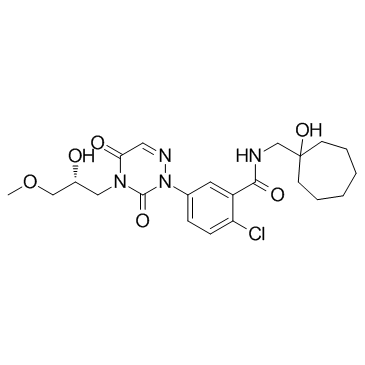
GC31157
CE-224535 (PF-04905428)
CE-224535 (PF-04905428) is a selective P2X7 receptor antagonist.
-
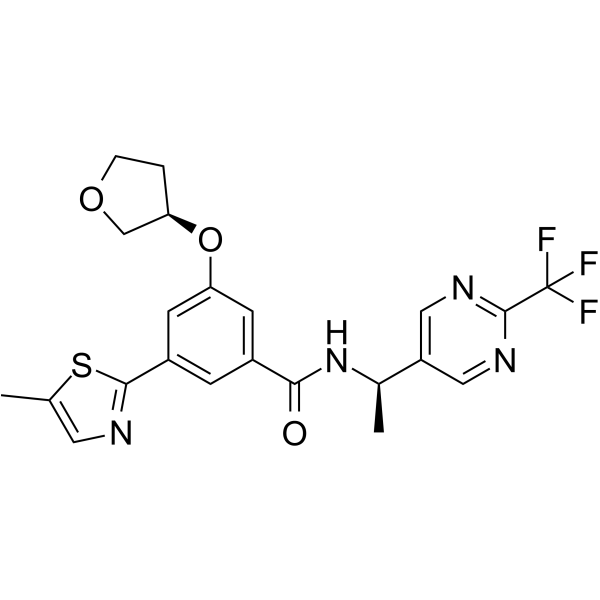
GC62950
Eliapixant
Eliapixant (BAY 1817080) is a potent and selective antagonist of P2X3 receptor, with an IC50 of 8 nM.
-
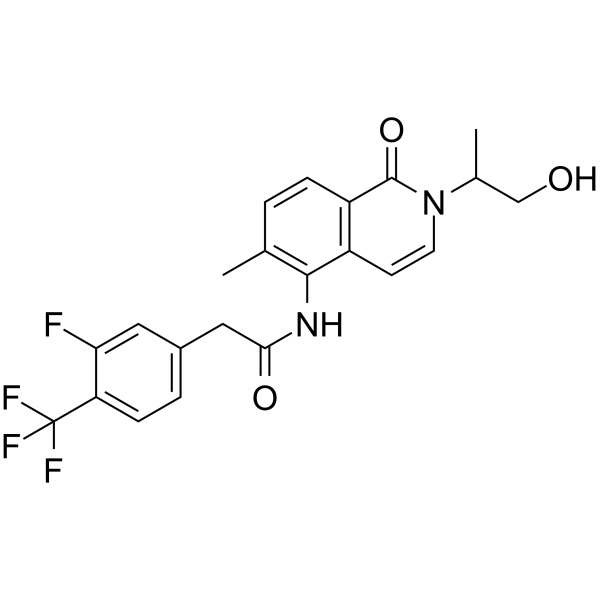
GC67895
EVT-401
-
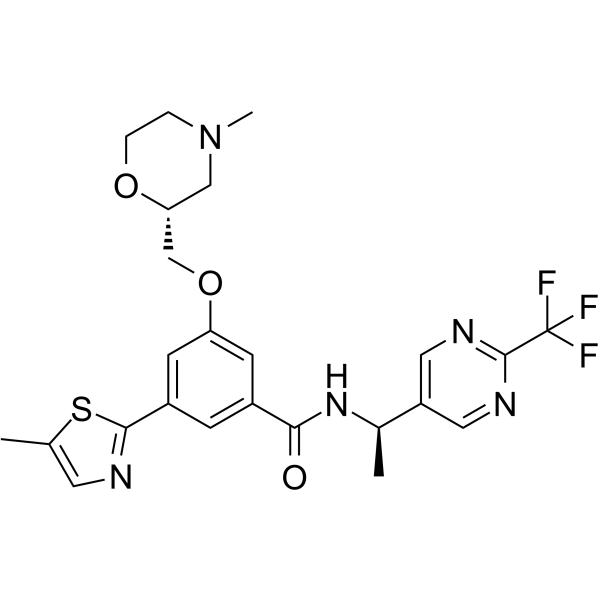
GC63444
Filapixant
Filapixant is a purinoreceptor antagonist extracted from patent WO2016091776A1, example 348.
-
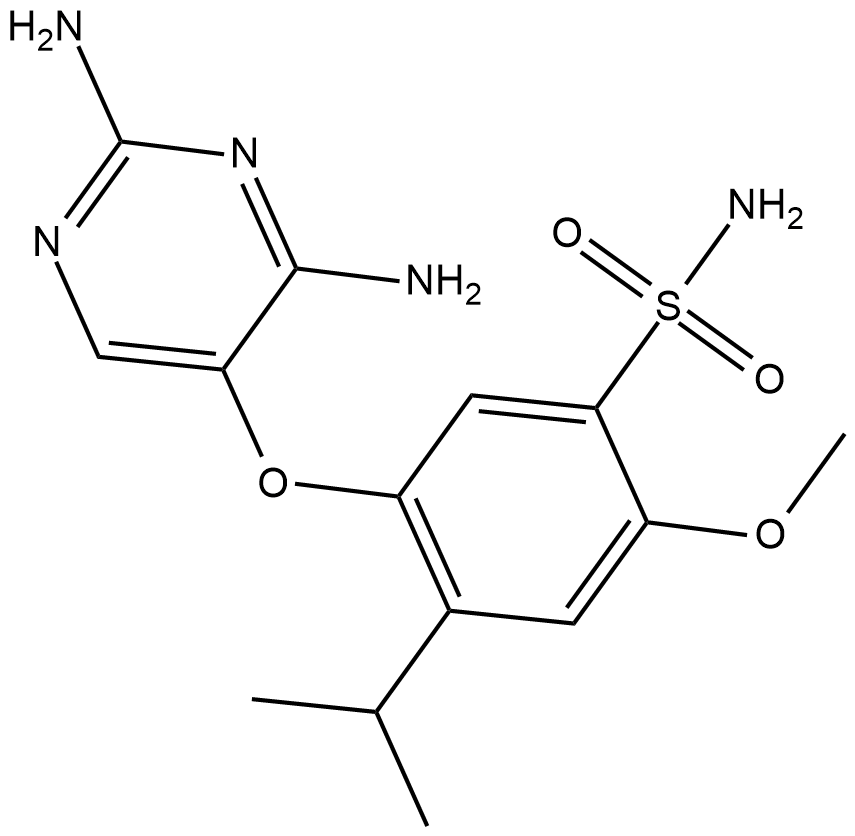
GC19422
Gefapixant
Gefapixant is a P2X3 receptor antagonist
-
GC63838
GSK-1482160
GSK-1482160 is an orally available negative allosteric modulator of the P2X7 receptor.
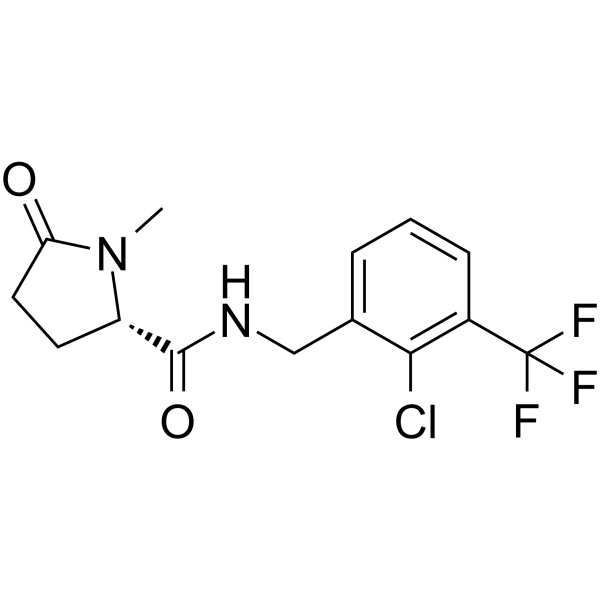
-
GC36202
GW791343 dihydrochloride
GW791343 dihydrochloride is a potent human P2X7 receptor negative allosteric modulator (exhibits species-specific activity), produces a non-competitive antagonist effect on human P2X7 receptor, with a pIC50 of 6.9-7.2.
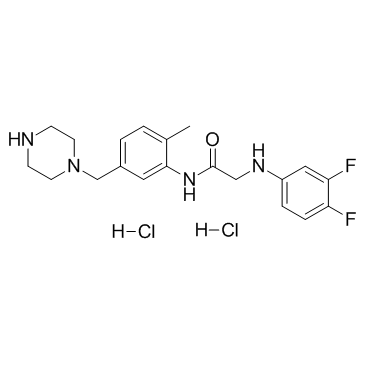
-
GC14207
GW791343 HCl
GW791343 HCl is a potent human P2X7 receptor negative allosteric modulator (exhibits species-specific activity), produces a non-competitive antagonist effect on human P2X7 receptor, with a pIC50 of 6.9-7.2.
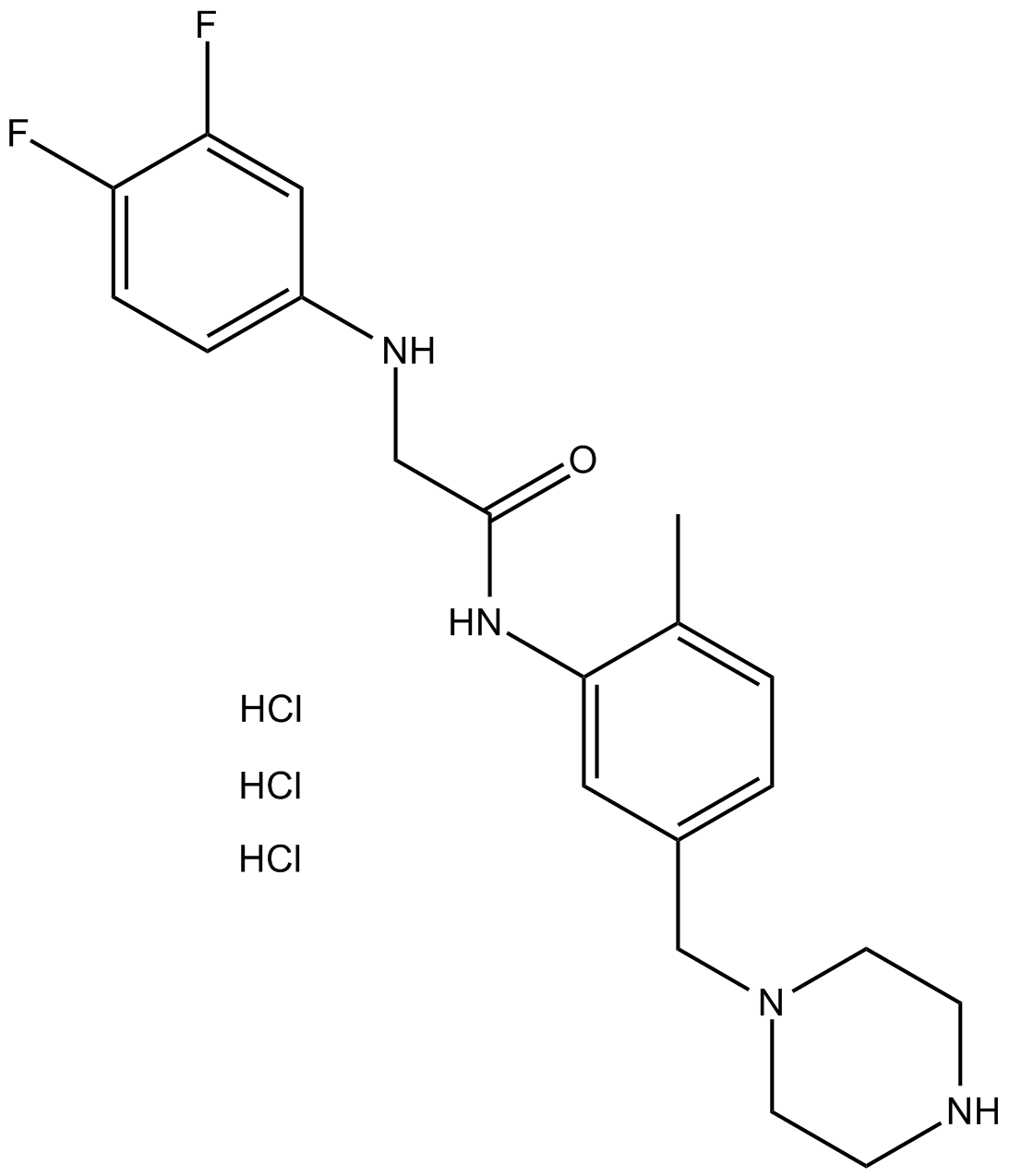
-
GC61565
Indophagolin
Indophagolin is a potent, indoline-containing autophagy inhibitor (IC50=140 nM). Indophagolin antagonizes the purinergic receptor P2X4 as well as P2X1 and P2X3 with IC50s of 2.71, 2.40 and 3.49 μM, respectively. Indophagolin also antagonizes the Gq-protein-coupled P2Y4, P2Y6, and P2Y11 receptors (IC50s =3.4~15.4 μM). Indophagolin has a strong antagonistic effect on serotonin receptor 5-HT6 (IC50=1.0 μM) and a moderate effect on receptors 5-HT1B, 5-HT2B, 5-HT4e, and 5-HT7.
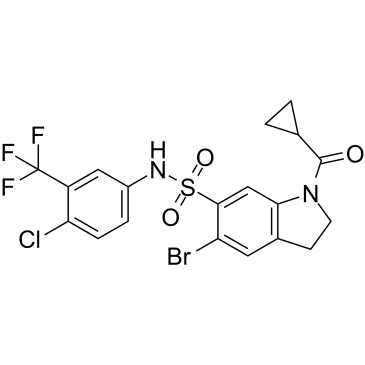
-
GC10990
iso-PPADS tetrasodium salt
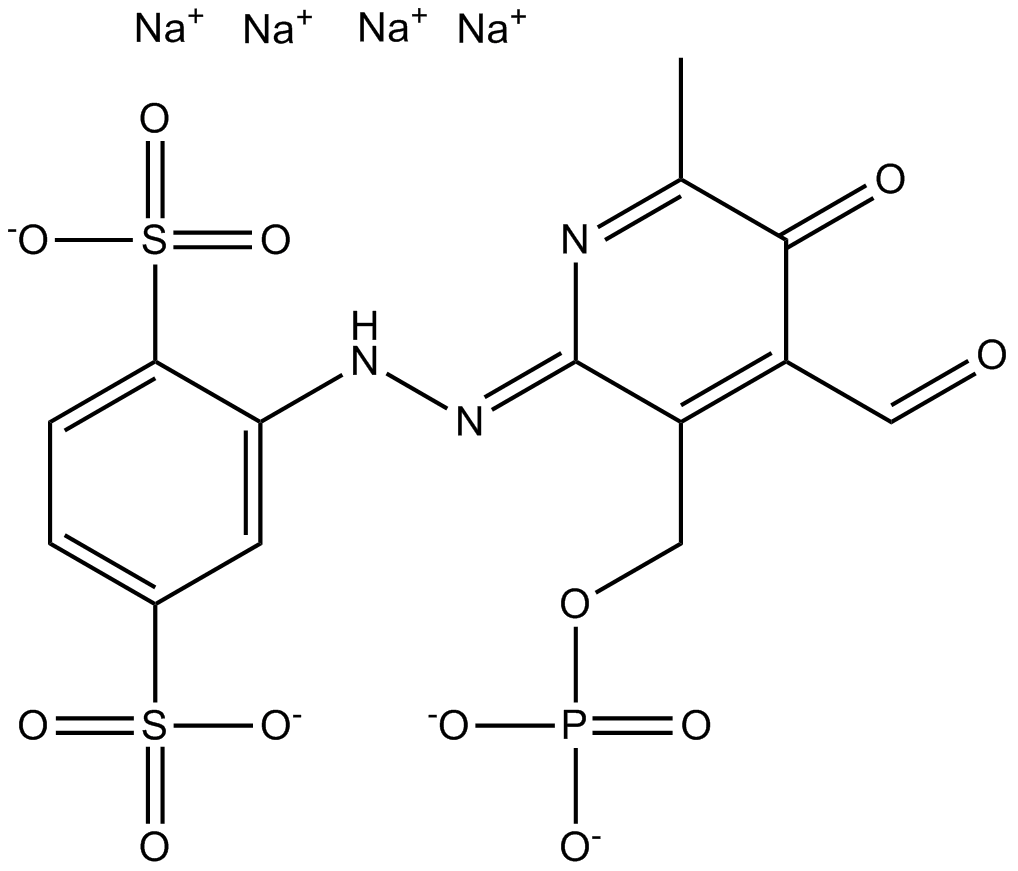
P2 purinoceptors antagonist, specific
-
GC12339
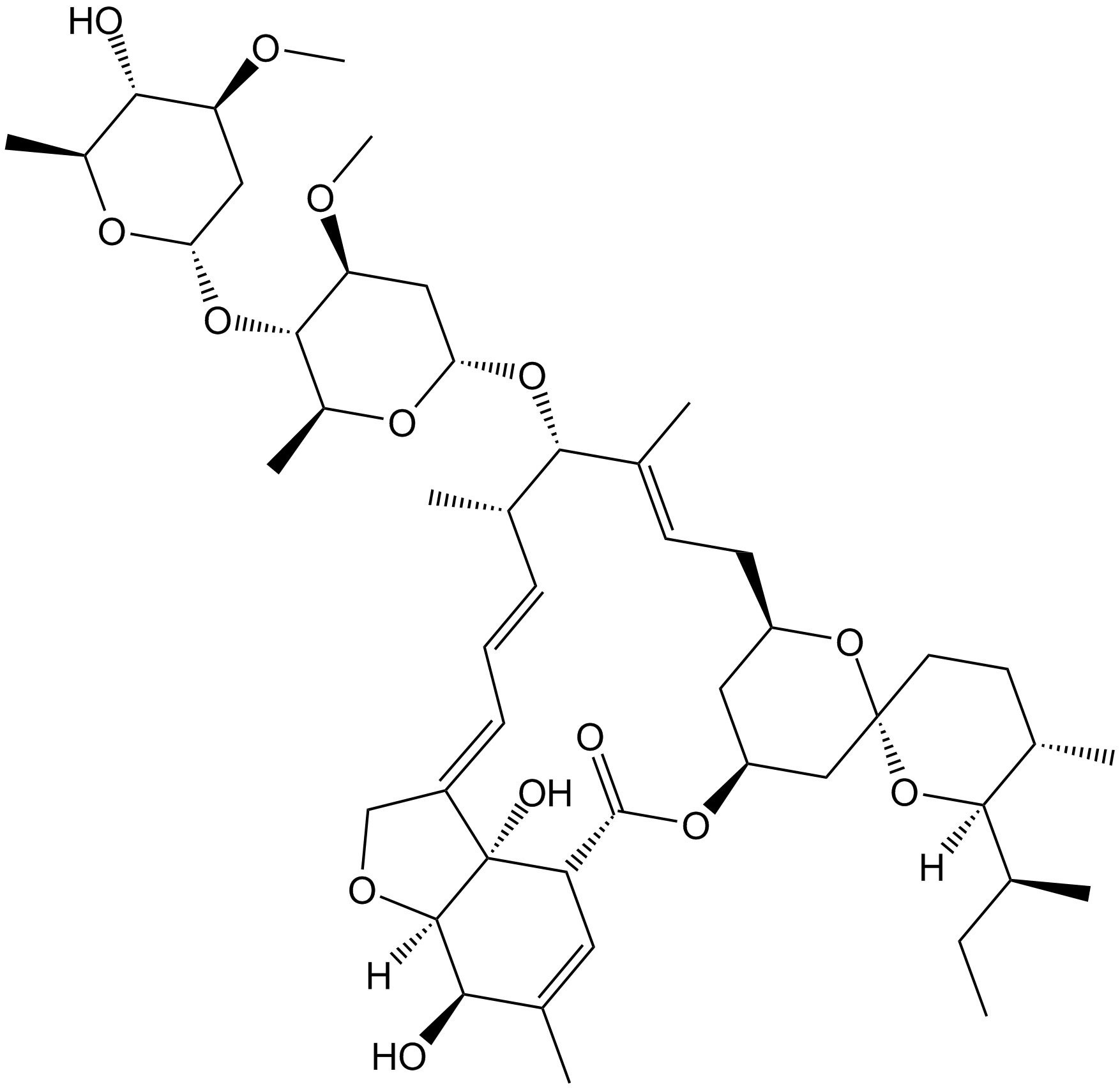
Ivermectin
NAChR/purinergic P2X4 receptor modulator
-
GC64822
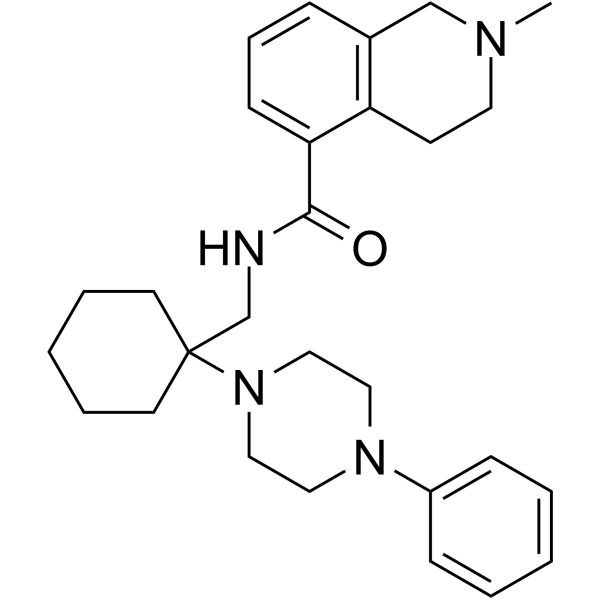
JNJ-42253432
JNJ-42253432 is a CNS-penetrant, high-affinity and orally active P2X7 antagonist, with pKi values of 9.1 and 7.9 for rat and human P2X7 channels, respectively.
-
GC43931
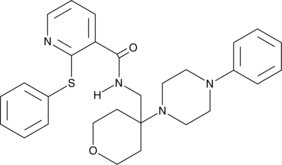
JNJ-47965567
JNJ-47965567 is a selective antagonist of the purinergic receptor P2X subtype 7 (P2X7), a ligand-gated ion channel.
-
GC62284
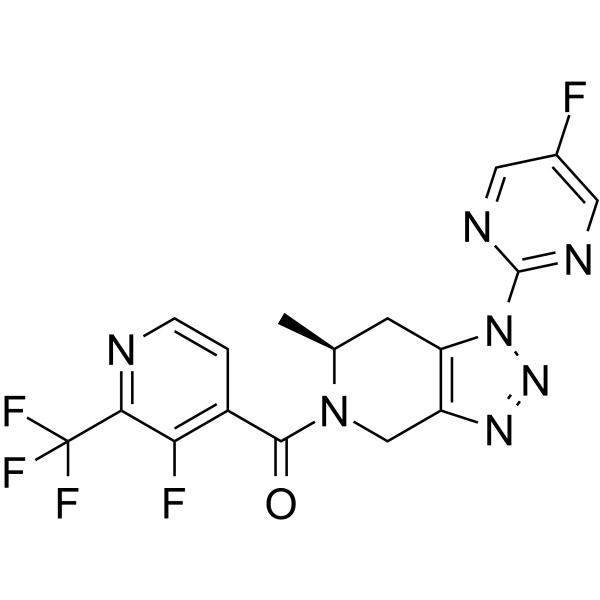
JNJ-55308942
JNJ-55308942 is a high-affinity, selective, brain-penetrant P2X7 functional antagonist (hP2X7: IC50=10 nM, Ki=7.1 nM; rP2X7: IC50=15 nM, Ki=2.9 nM).
-
GC14202
KN-62
Inhibitor of Ca2+/calmodulin-dependent kinase type II

-
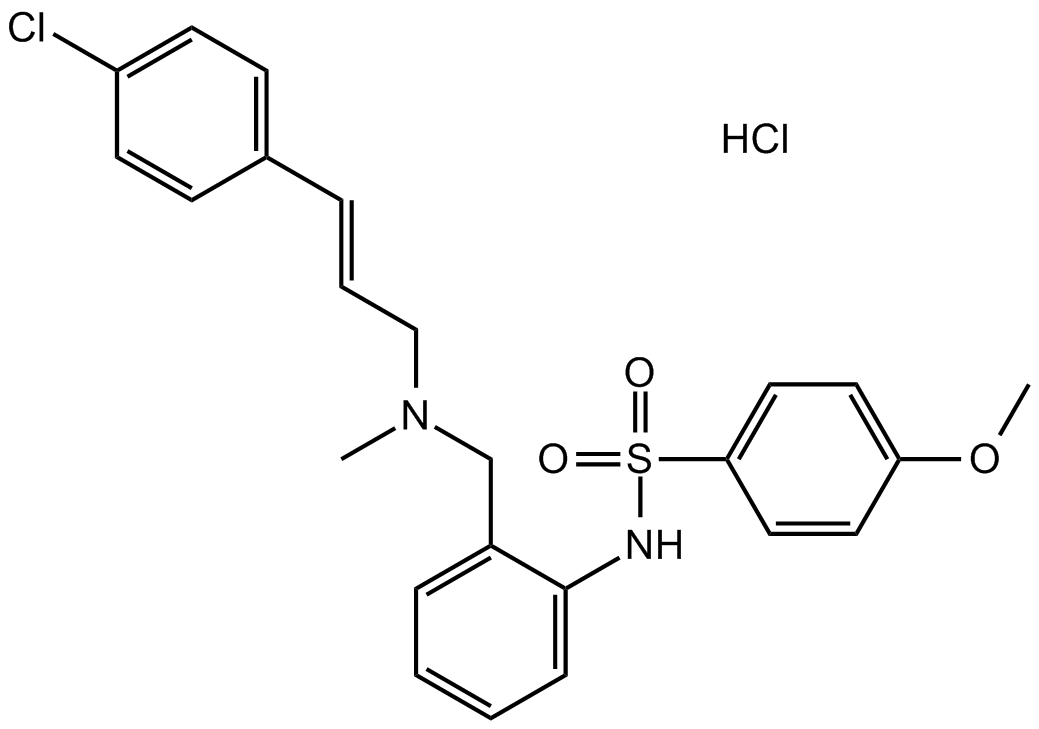
GC15988
KN-92 hydrochloride
An inactive control compound for a CaMKII inhibitor
-
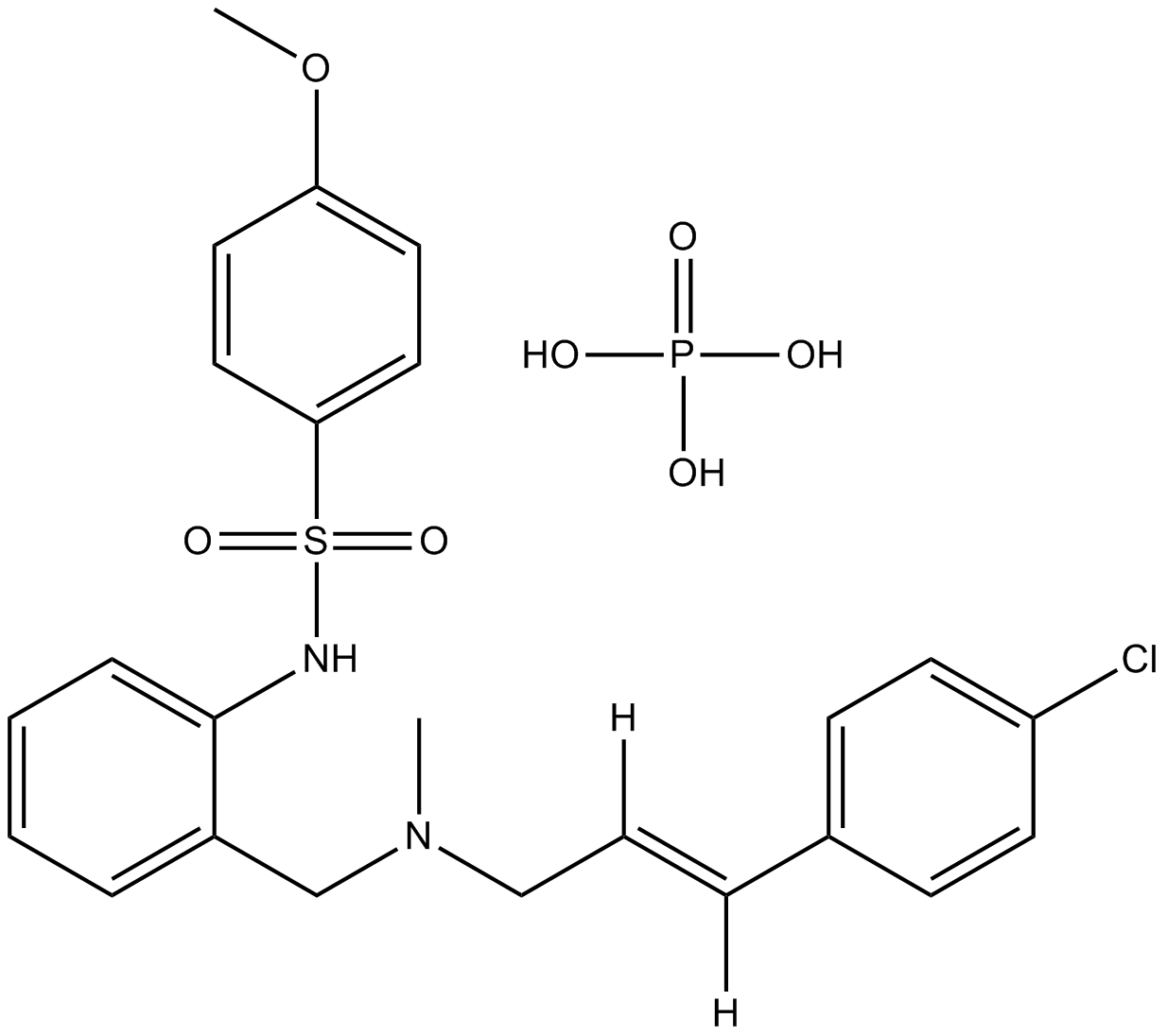
GC16981
KN-92 phosphate
-
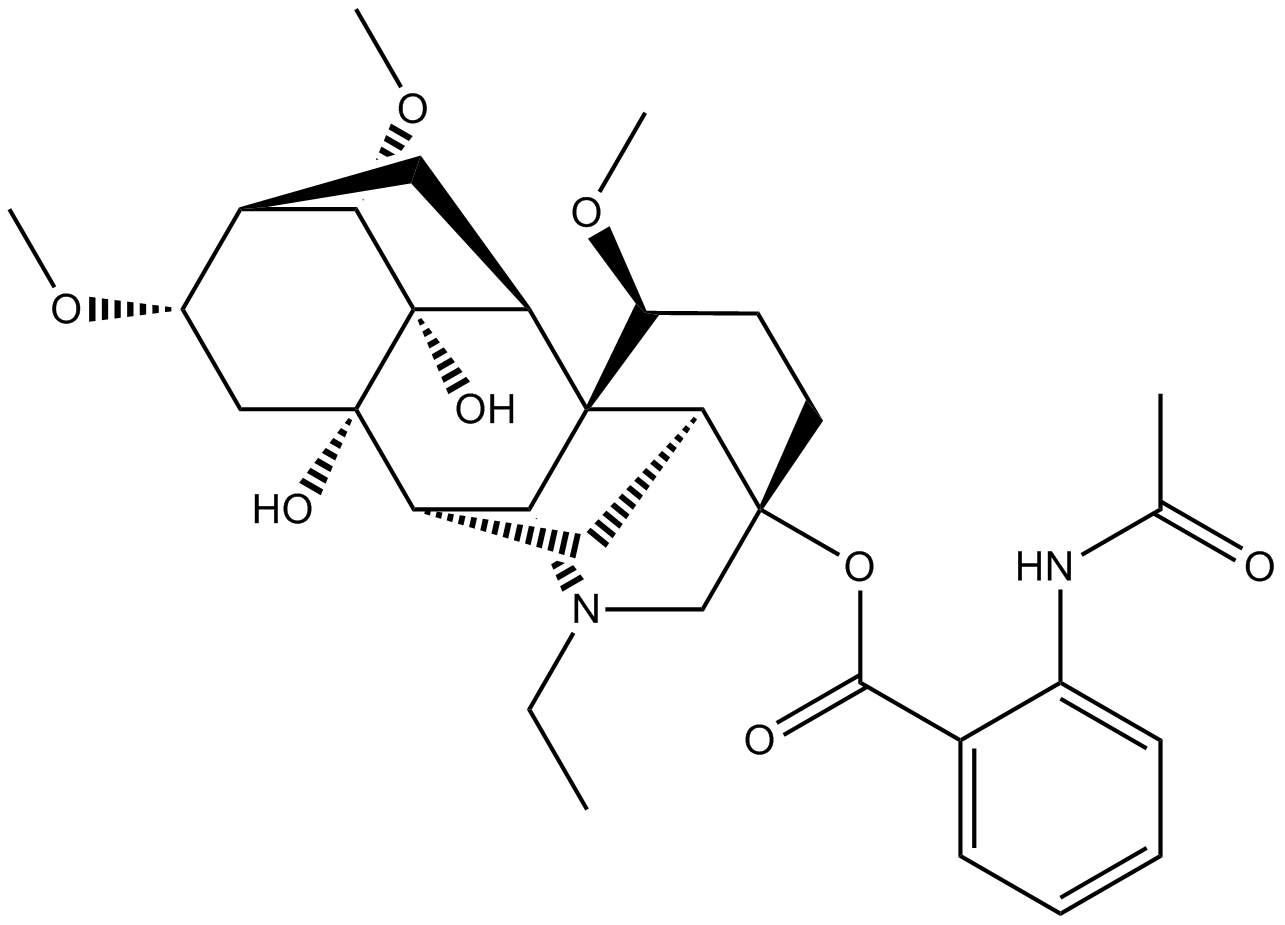
GN10747
Lannaconitine
-
GC63996
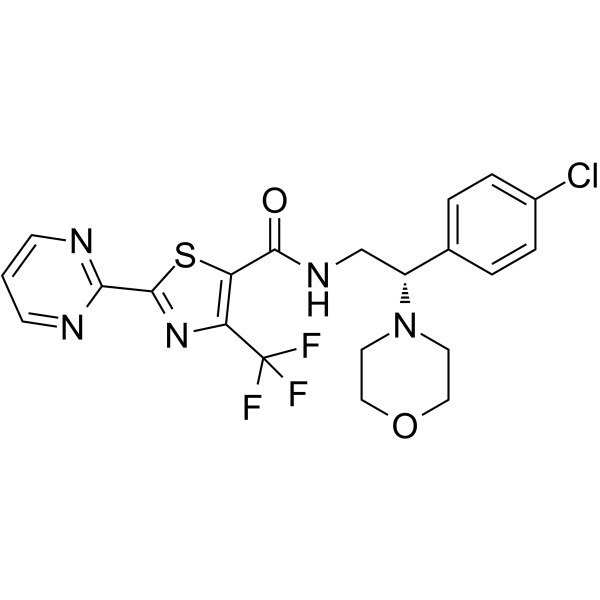
Lu AF27139
Lu AF27139 is a potent, selective, and orally active antagonist of P2X7 receptor (IC50s of 12 and 2.4 nM for human and rat, Kis of 22, 54, and 13 nM for mouse, human, and rat, respectively).
-
GC36613
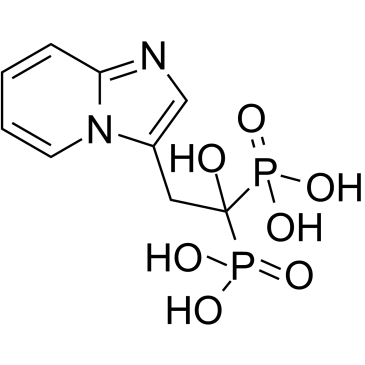
Minodronic acid
Minodronic acid (YM-529) is a third-generation bisphosphonate that directly and indirectly prevents proliferation, induces apoptosis, and inhibits metastasis of various types of cancer cells. Minodronic acid (YM-529) is an antagonist of purinergic P2X2/3 receptors involved in pain.
-
GC10411
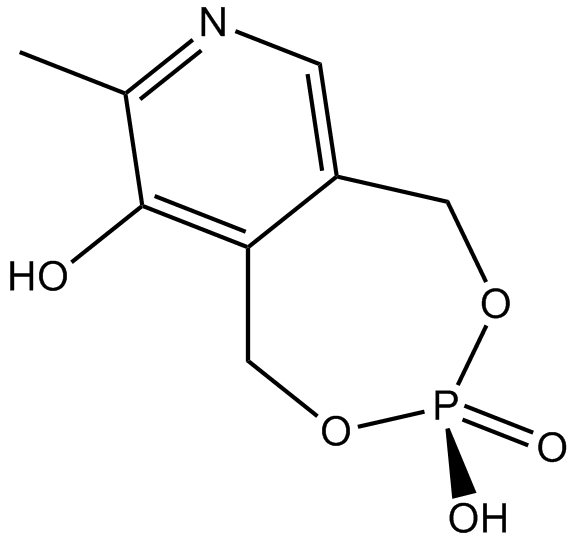
MRS 2219
P2X1 receptor potentiator
-
GC11289
NF 023
NF 023 is a selective and competitive P2X1 receptor antagonist, with IC50 values of 0.21 μM, 28.9 μM, > 50 μM and > 100 μM for human P2X1, P2X3, P2X2, and P2X4-mediated responses respectively.

-
GC13224
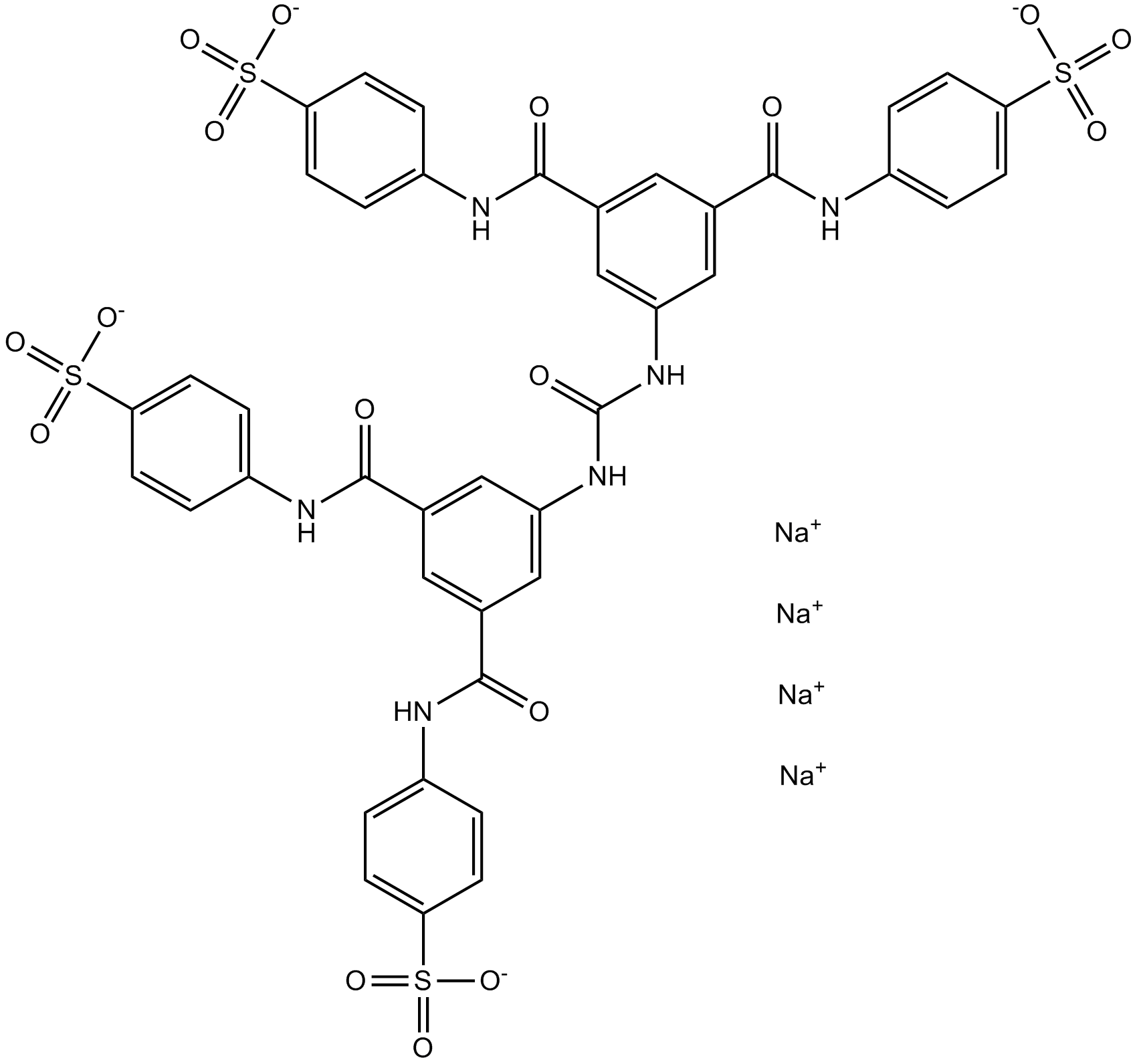
NF 110
P2X3 antagonist
-
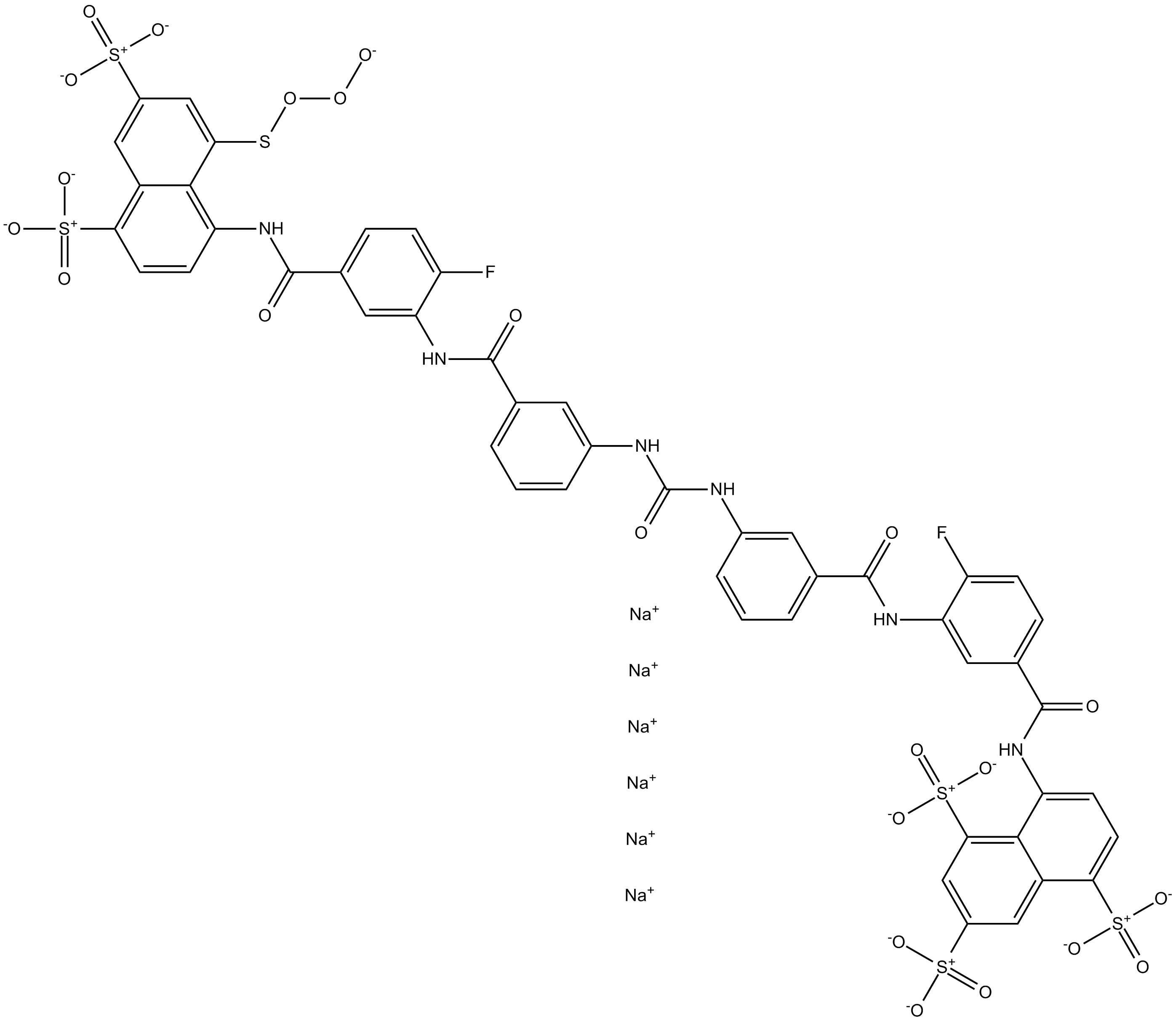
GC17320
NF 157
P2Y11/P2X1 antagonist
-
GC14992
NF 279
P2X1 antagonist
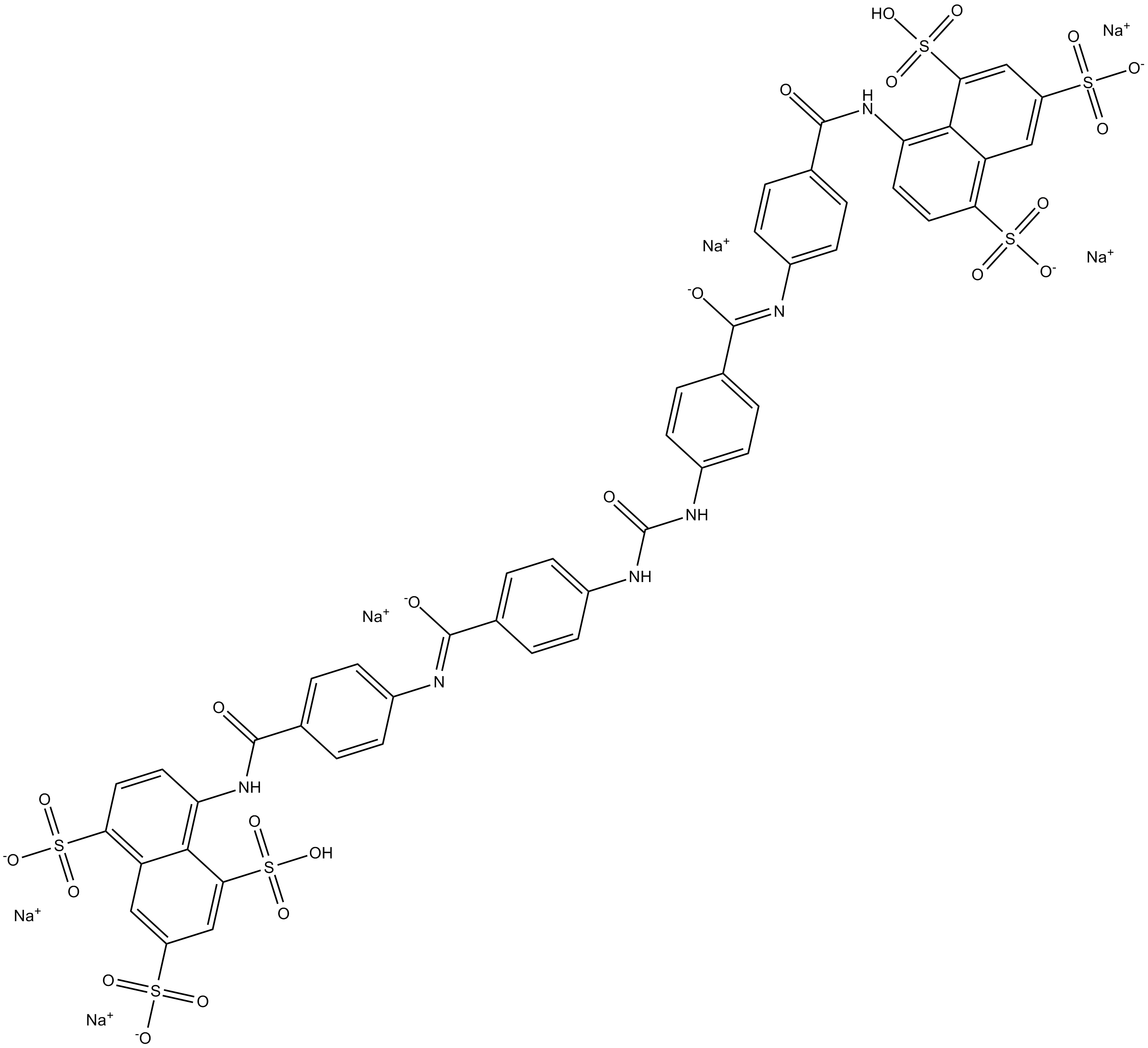
-
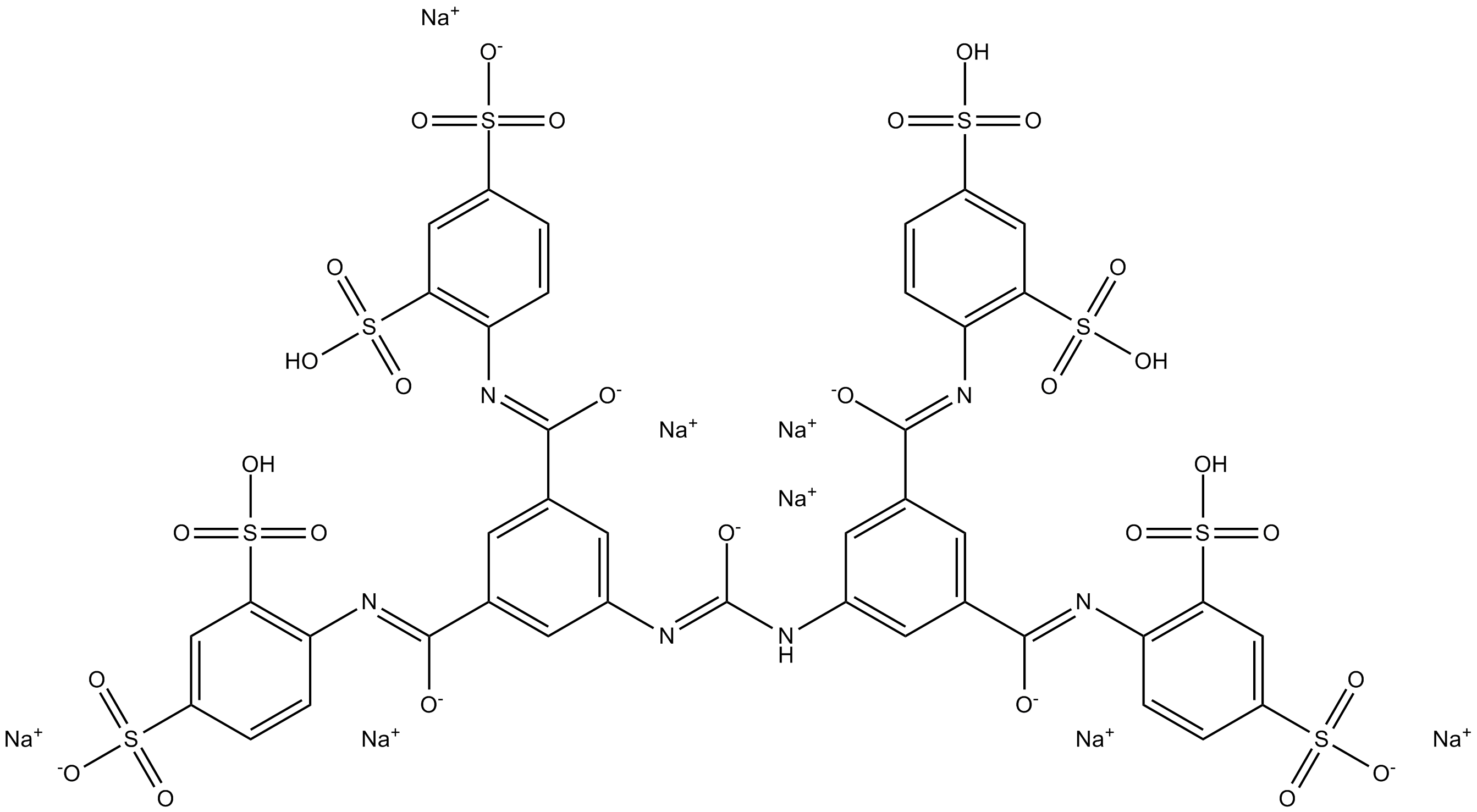
GC11326
NF 449
NF 449 is a highly potent P2X1 receptor antagonist, with IC50s of 0.28, 0.69, and 120 nM for rP2X1, rP2X1+5, P2X2+3, respectively.
-
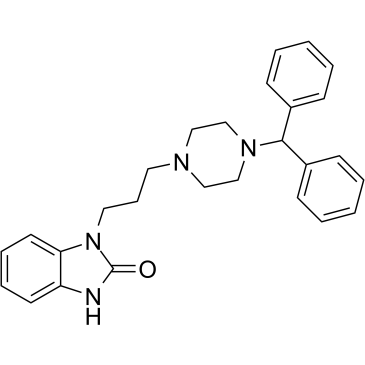
GC61401
Oxatomide
Oxatomide is a potent and orally active dual H1-histamine receptor and P2X7 receptor antagonist with antihistamine and anti-allergic activity.
-
GC60281
P2X3 antagonist 34
P2X3 antagonist 34 is a potent, selective and orally active P2X3 homotrimeric receptor antagonist with IC50s of 25?nM, 92?nM and 126?nM for human P2X3, rat P2X3 and guinea pig P2X3 receptors, respectively.
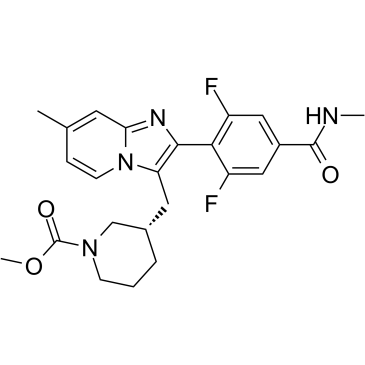
-
GC69647
P2X7-IN-2 TFA
P2X7-IN-2 TFA (compound 58) is an inhibitor of the P2X7 receptor. It inhibits the release of IL-Iβ with an IC50 value of 0.01 nM. P2X7-IN-2 TFA can be used for research on autoimmune diseases, inflammation, and cardiovascular diseases.

-
GC16659
PPADS tetrasodium salt
PPADS tetrasodium salta is a non-selective P2X receptor antagonist.
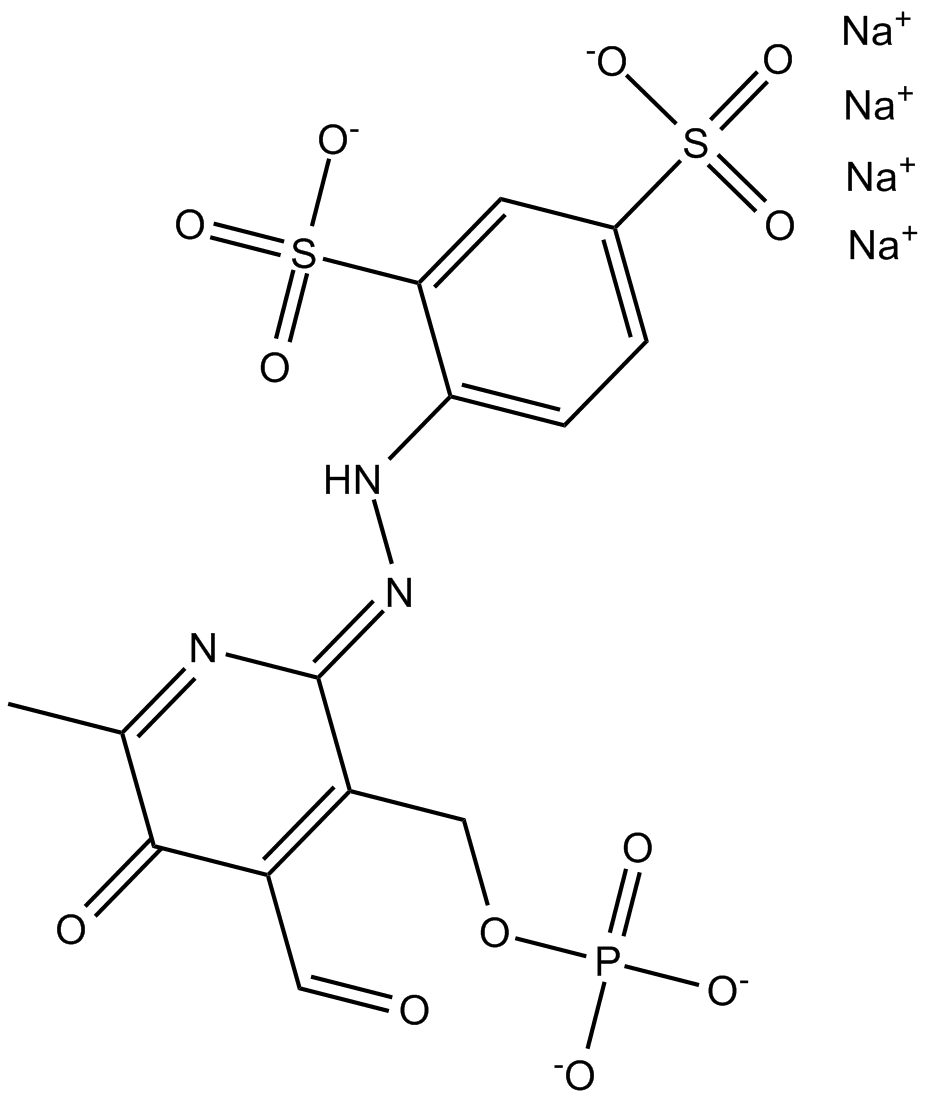
-
GC16342
PPNDS
PPNDS is a selective and competitive meprin β inhibitor (IC50: 80 nM, Ki: 8 nM), and also inhibits ADAM10 (IC50: 1.2 μM).
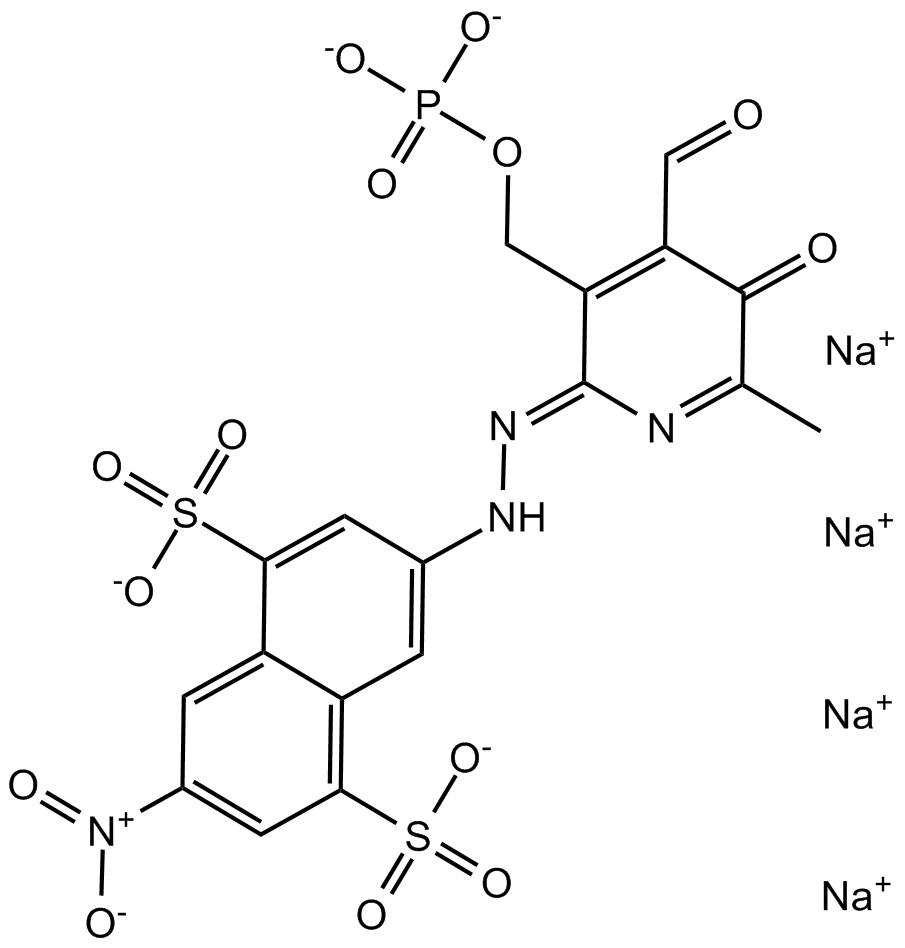
-
GC30805
PSB-12062 (N-(p-Methylphenylsulfonyl)phenoxazine)
PSB-12062 (N-(p-Methylphenylsulfonyl)phenoxazine) is a potent and selective P2X4 antagonist with an IC50 of 1.38 μM for human P2X4.
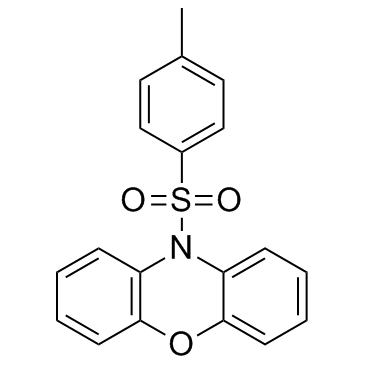
-
GC13584
Purotoxin 1
P2X3 receptor modulator
-
GC11216
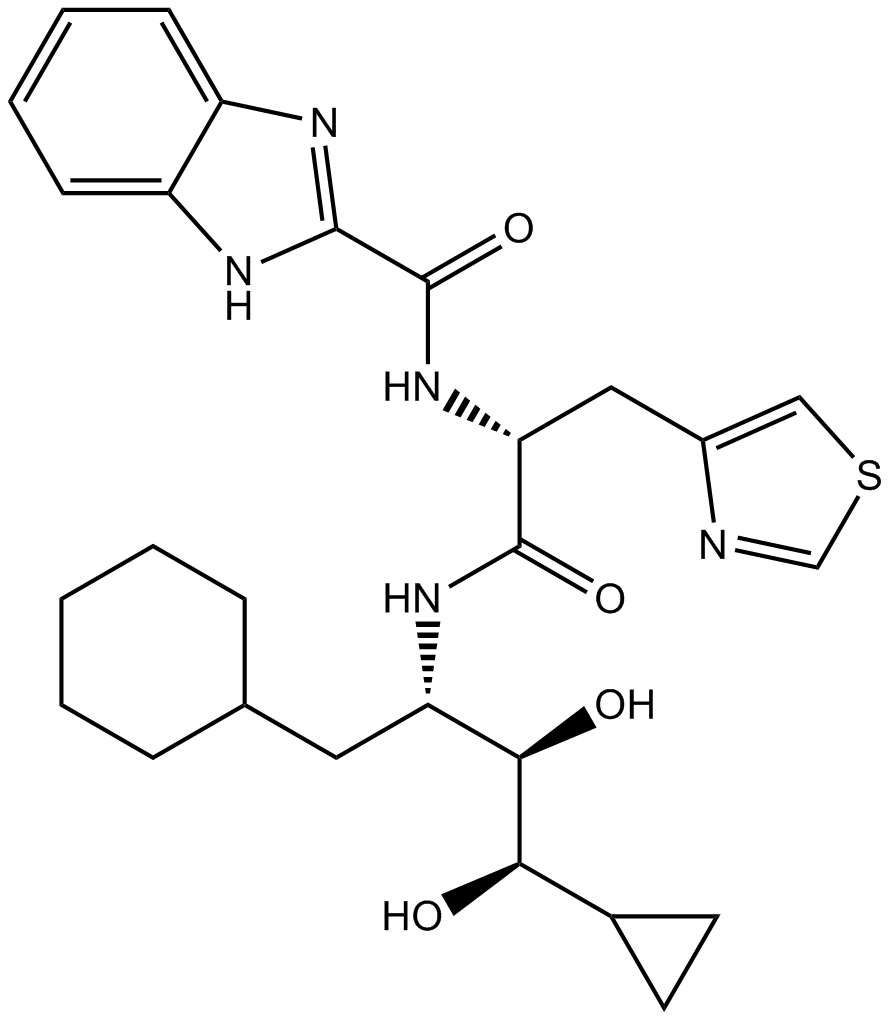
Ro 0437626
P2X1 purinergic receptor antagonist
-
GC16330
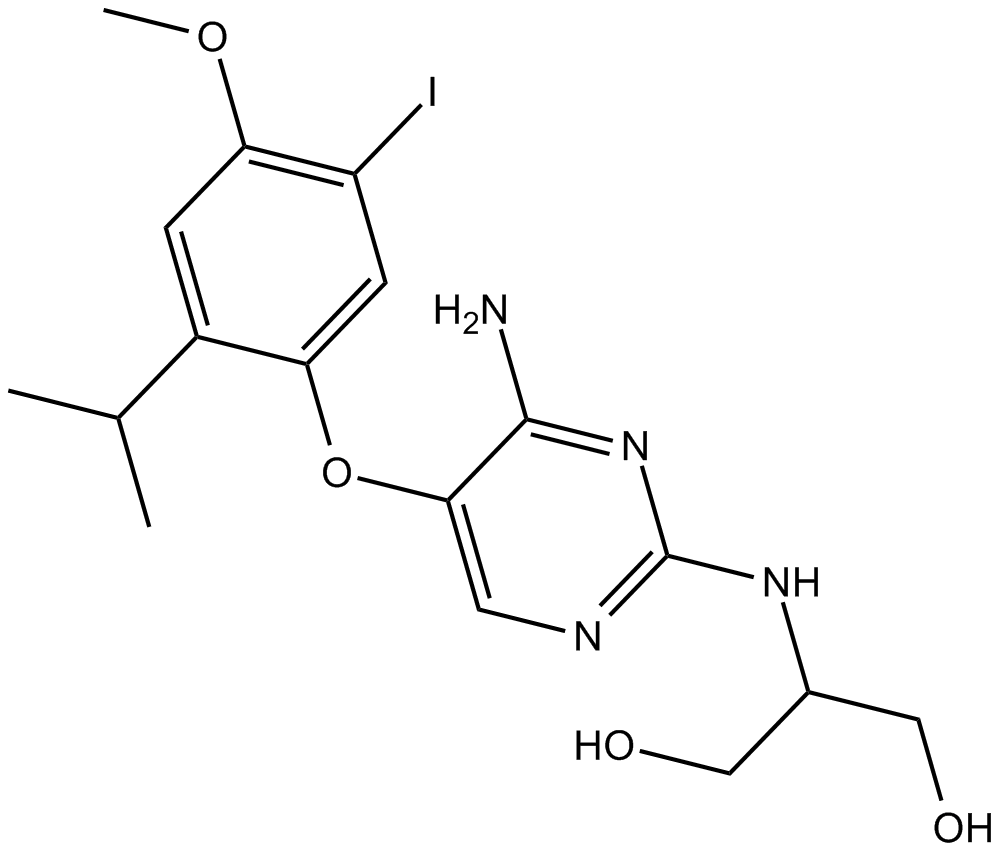
Ro 51
dual P2X3 and P2X2/3 antagonist
-
GC10152
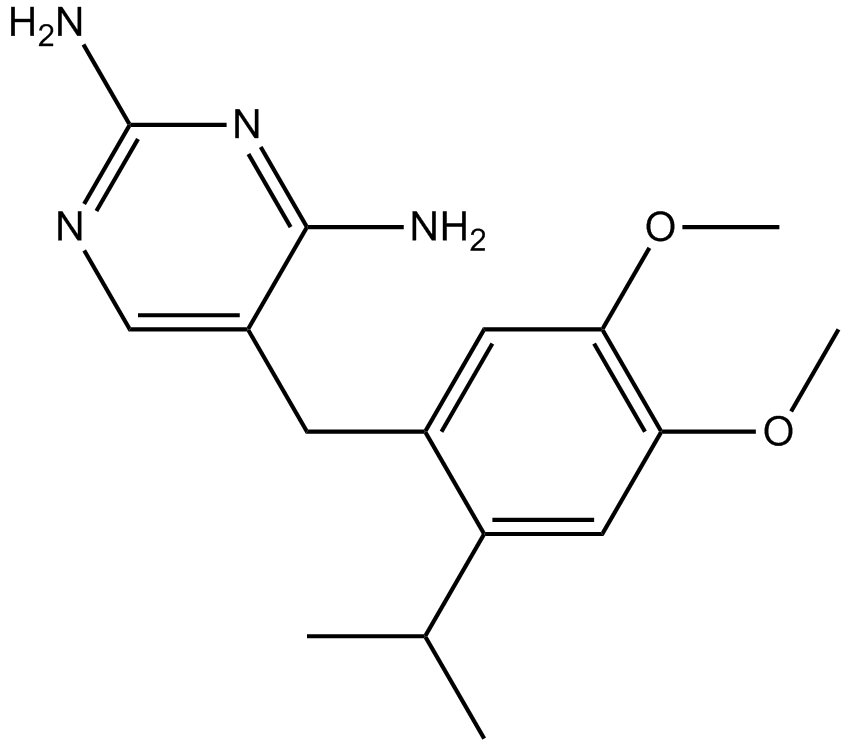
RO-3
homomeric P2X3 and heteromeric P2X2/3 receptor antagonist
-
GC16832
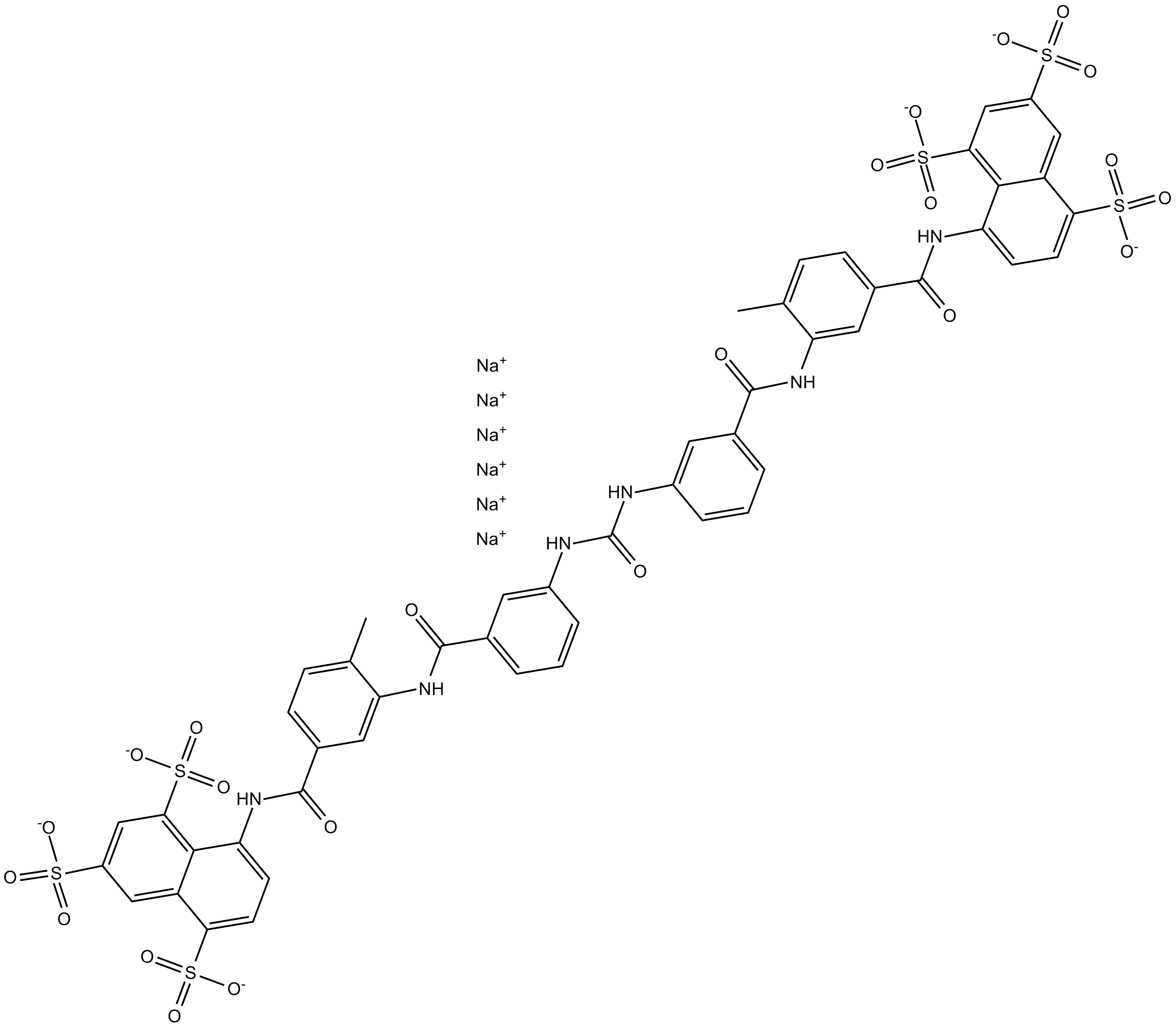
Suramin hexasodium salt
Suramin hexasodium salt(Suramin hexasodium salt) is a reversible and competitive protein-tyrosine phosphatases (PTPases) inhibitor.
-
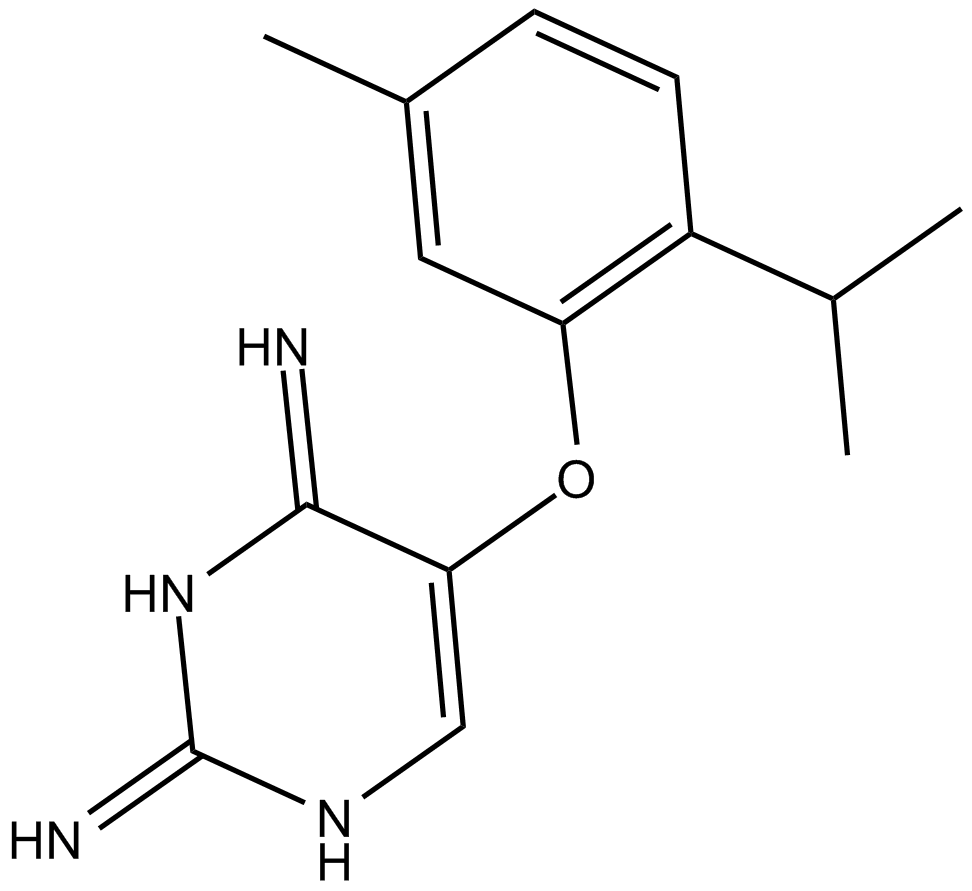
GC11682
TC-P 262
P2X3 and P2X2/3 receptor antagonist
-
GC17355
TNP-ATP triethylammonium salt
P2X receptor antagonist