Signaling Pathways
The antibody-drug conjugate (ADC), a humanized or human monoclonal antibody conjugated with highly cytotoxic small molecules (payloads) through chemical linkers, is a novel therapeutic format and has great potential to make a paradigm shift in cancer chemotherapy. The three components of the ADC together give rise to a powerful oncolytic agent capable of delivering normally intolerable cytotoxins directly to cancer cells, which then internalize and release the cell-destroying drugs. At present, two ADCs, Adcetris and Kadcyla, have received regulatory approval with >40 others in clinical development.
ADCs are administered intravenously in order to prevent the mAb from being destroyed by gastric acids and proteolytic enzymes. The mAb component of the ADC enables it to circulate in the bloodstream until it finds and binds to tumor-specific cell surface antigens present on target cancer cells. Linker chemistry is an important determinant of the safety, specificity, potency and activity of ADCs. Linkers are designed to be stable in the blood stream (to conform to the increased circulation time of mAbs) and labile at the cancer site to allow rapid release of the cytotoxic drug. First generation ADCs made use of early cytotoxins such as the anthracycline, doxorubicin or the anti-metabolite/antifolate agent, methotrexate. Current cytotoxins have far greater potency and can be divided into three main groups: auristatins, maytansines and calicheamicins.
The development of site-specific conjugation methodologies for constructing homogeneous ADCs is an especially promising path to improving ADC design, which will open the way for novel cancer therapeutics.
References:
[1] Tsuchikama K, et al. Protein Cell. 2016 Oct 14. DOI:10.1007/s13238-016-0323-0.
[2] Peters C, et al. Biosci Rep. 2015 Jun 12;35(4). pii: e00225. doi: 10.1042/BSR20150089.
Targets for Signaling Pathways
- Proteases(29)
- Apoptosis(824)
- Chromatin/Epigenetics(15)
- Metabolism(336)
- MAPK Signaling(26)
- Tyrosine Kinase(73)
- DNA Damage/DNA Repair(49)
- PI3K/Akt/mTOR Signaling(38)
- Microbiology & Virology(42)
- Cell Cycle/Checkpoint(168)
- Ubiquitination/ Proteasome(24)
- JAK/STAT Signaling(10)
- TGF-β / Smad Signaling(21)
- Angiogenesis(59)
- GPCR/G protein(3)
- Stem Cell(19)
- Membrane Transporter/Ion Channel(161)
- Cancer Biology(366)
- Endocrinology and Hormones(152)
- Neuroscience(367)
- Obesity, Appetite Control & Diabetes(17)
- Peptide Inhibitors and Substrate(2)
- Other Signal Transduction(156)
- Immunology/Inflammation(934)
- Cardiovascular(98)
- Vitamin D Related(0)
- Antibody-drug Conjugate/ADC Related(0)
- PROTAC(212)
- Ox Stress Reagents(24)
- Others(4010)
- Antiparasitics(6)
- Toxins(62)
Products for Signaling Pathways
- Cat.No. Product Name Information
-
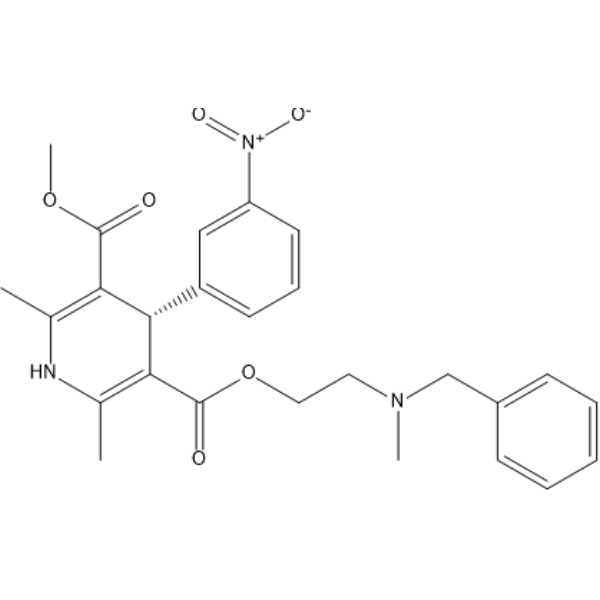
GC68476
(S)-Nicardipine
-
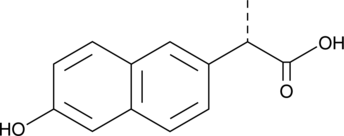
GC49179
(S)-O-Desmethyl Naproxen
A metabolite of (S)-naproxen
-
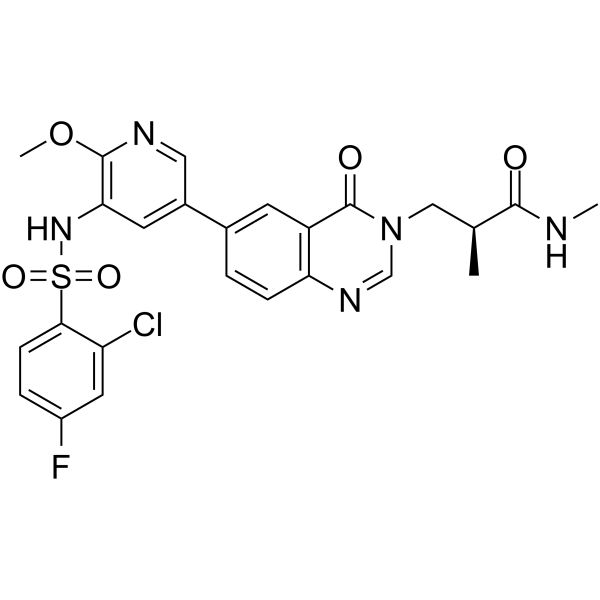
GC62450
(S)-PI3Kα-IN-4
(S)-PI3Kα-IN-4 is a potent inhibitor of PI3Kα, with an IC50 of 2.3 nM. (S)-PI3Kα-IN-4 shows 38.3-, 4.25-, and 4.93-fold selectivity for PI3Kα over PI3Kβ, PI3Kδ, and PI3Kγ, respectively. (S)-PI3Kα-IN-4 can be used for the research of cancer.
-
GC69941
(S)-Renzapride
(S)-Renzapride ((S)-BRL 24924) is an isomer of Renzapride. Renzapride is a 5-HT4 receptor agonist with a Ki value of 115 nM. It also acts as an antagonist for the 5HT2b and 5HT3 receptors. Renzapride can be used in research on constipation-predominant irritable bowel syndrome (C-IBS).
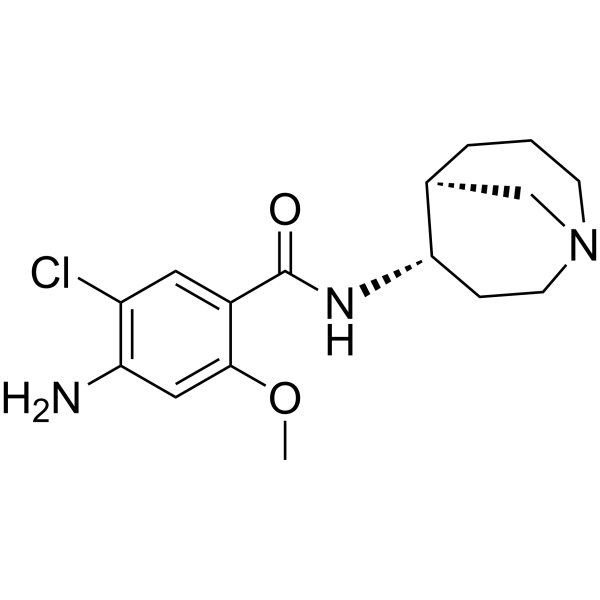
-
GC63797
(S)-Sunvozertinib
(S)-Sunvozertinib ((S)-DZD9008), the S-enantiomer of Sunvozertinib, shows inhibitory activity against EGFR exon 20 NPH and ASV insertions, EGFR L858R/T790M mutation and Her2 exon20 YVMA insertion (IC50=51.2 nM, 51.9 nM, 1 nM, and 21.2 nM, respectively). (S)-Sunvozertinib also inhibits BTK.
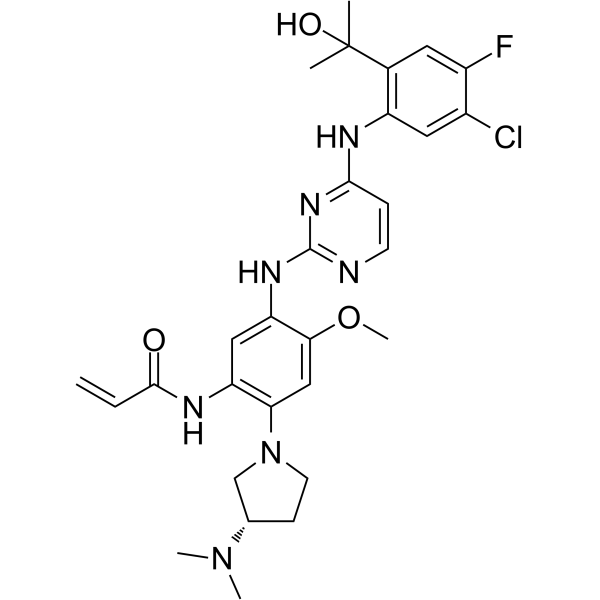
-
GC66064
(S)-Thalidomide
(S)-Thalidomide ((S)-(-)-Thalidomide) is the S-enantiomer of Thalidomide. (S)-Thalidomide has immunomodulatory, anti-inflammatory, antiangiogenic and pro-apoptotic effects. (S)-Thalidomide induces teratogenic effects by binding to cereblon (CRBN) .
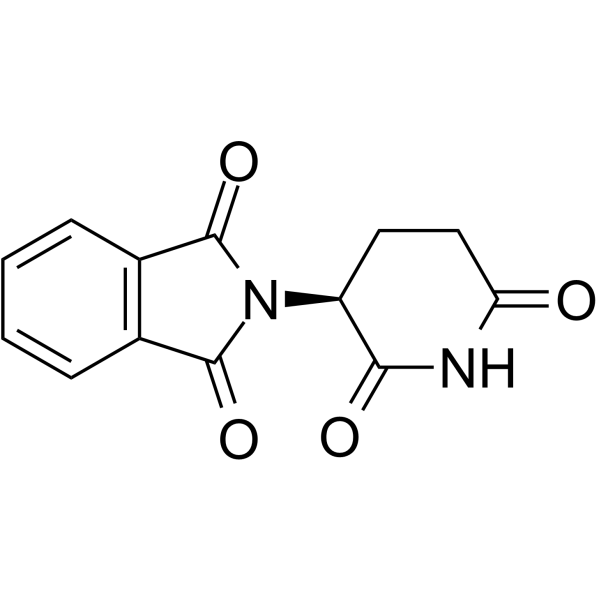
-
GC69977
(S)-Veliflapon
(S)-Veliflapon ((S)-BAY X 1005) is an orally active leukotriene biosynthesis and 5-lipoxygenase activating protein (FLAP) inhibitor. It inhibits the formation of leukotriene B4 (LTB4) in rat, mouse, and human white blood cells with IC50 values of 0.026 μM, 0.039 μM, and 0.22 μM respectively. (S)-Veliflapon exhibits enantioselectivity in human whole blood.
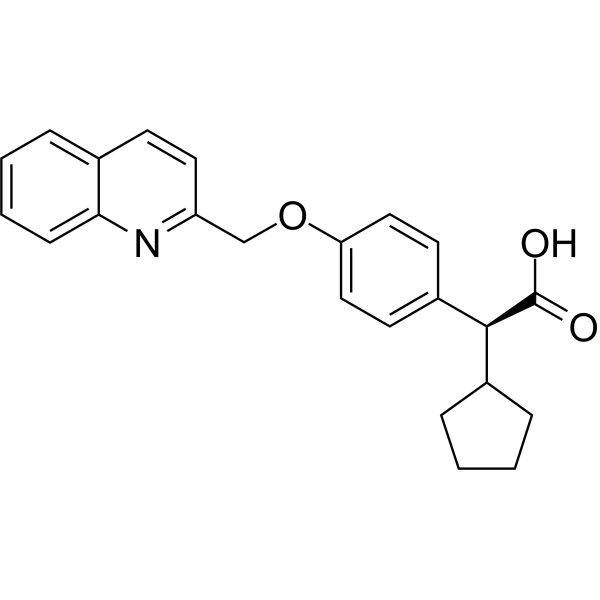
-
GC69979
(S)-VQW-765
(S)-VQW-765 ((S)-AQW-051) is a partial agonist of the α7 nicotinic acetylcholine receptor (nAChR) with oral activity, selectivity and efficacy. It has potential applications in cognitive impairments associated with neurological diseases such as Alzheimer's disease or schizophrenia.
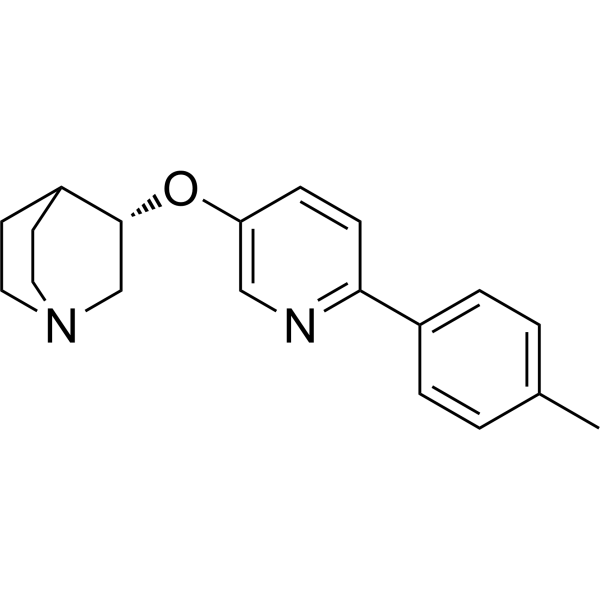
-
GC65883
(S,R)-WT IDH1 Inhibitor 2
(S,R)-WT IDH1 Inhibitor 2 (GSK321) is a potent, selective mutant IDH1 inhibitor with IC50 values of 2.9, 3.8, 4.6 and 46 nM for R132G, R132C, R132H and WT IDH1, respectively, and >100-fold selectivity over IDH2. (S,R)-WT IDH1 Inhibitor 2 induces decrease in intracellular 2-HG, abrogation of the myeloid differentiation block and induction of granulocytic differentiation at the level of leukemic blasts and more immature stem-like cells. (S,R)-WT IDH1 Inhibitor 2 can be used for research of acute myeloid leukemia (AML) and other cancers.
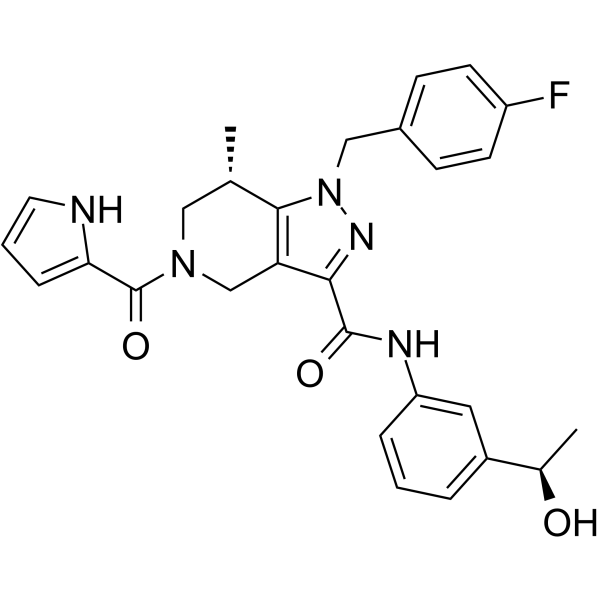
-
GC62283
(S,R,S)-AHPC
(S,R,S)-AHPC (VH032-NH2) is the VH032-based VHL ligand used in the recruitment of the von Hippel-Lindau (VHL) protein. (S,R,S)-AHPC can be connected to the ligand for protein (e.g., BCR-ABL1) by a linker to form PROTACs (e.g., GMB-475). GMB-475 induces the degradation of BCR-ABL1 with an IC50 of 1.11 μM in Ba/F3 cells.
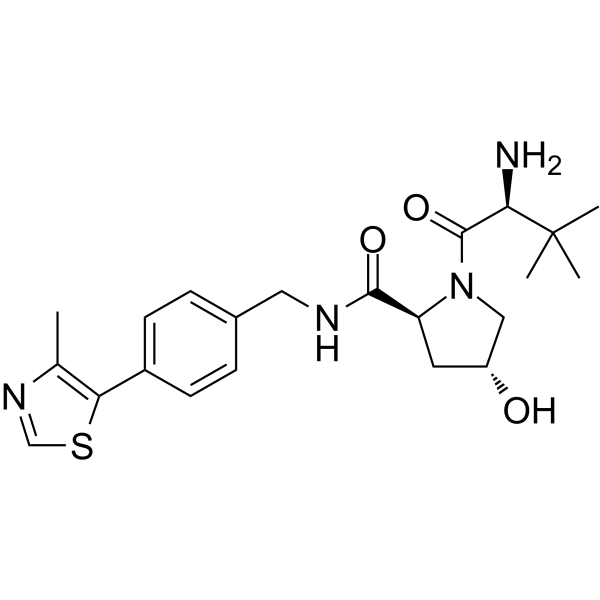
-
GC69944
(S,R,S)-AHPC-3-methylbutanyl acetate-methanesulfonothioate-Me-C10-NH2 TFA
(S,R,S)-AHPC-3-methylbutanyl acetate-methanesulfonothioate-Me-C10-NH2 TFA is a synthetic E3 ligase ligand-linker conjugate containing the (S,R,S)-AHPC ligand, which can be used in PROTAC research.
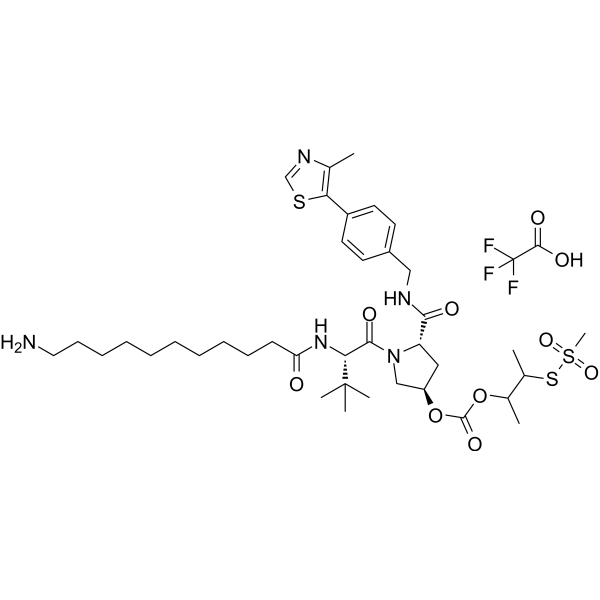
-
GC69945
(S,R,S)-AHPC-3-methylbutanyl acetate-methanesulfonothioate-PEG3-NH2 TFA
"(S,R,S)-AHPC-3-methylbutanyl acetate-methanesulfonothioate-PEG3-NH2 TFA" is a synthetic E3 ligase ligand-linker conjugate, which includes a ligand based on (S,R,S)-AHPC and a three-unit PEG linker."
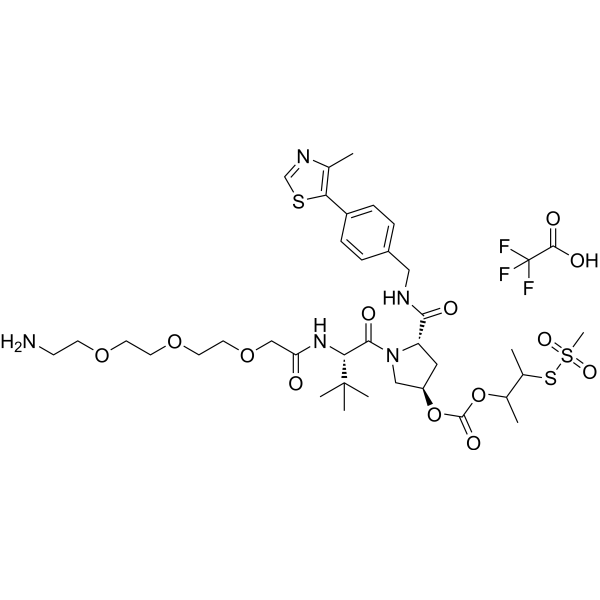
-
GC62148
(S,R,S)-AHPC-amido-C5-acid
(S,R,S)-AHPC-amido-C5-acid incorporates a VHL ligand for the E3 ubiquitin ligase, and a PROTAC linker. (S,R,S)-AHPC-amido-C5-acid can be used to design XY028-133.
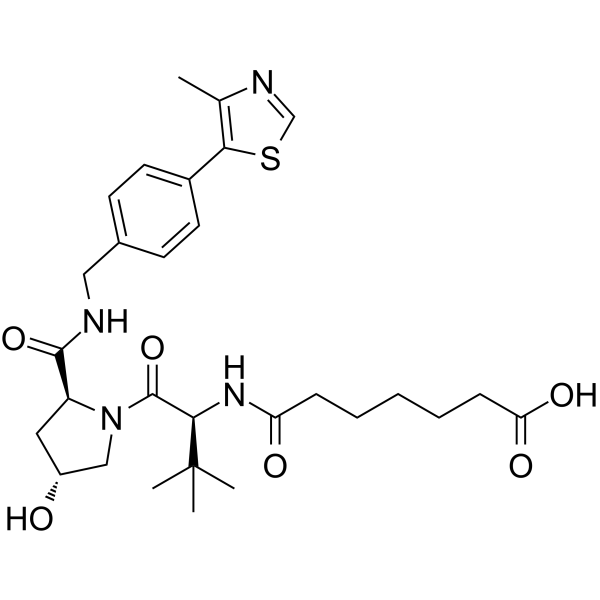
-
GC69946
(S,R,S)-AHPC-amido-C7-acid
(S,R,S)-AHPC-amido-C7-acid contains a VHL ligand for recruiting E3 ubiquitin ligases and a PROTAC linker. (S,R,S)-AHPC-amido-C5-acid can be used for designing PROTACs.
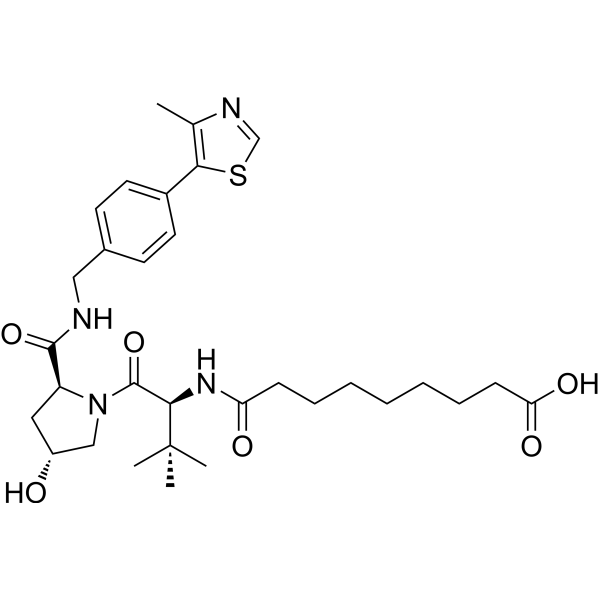
-
GC62684
(S,R,S)-AHPC-Boc-trans-3-aminocyclobutanol-Pip-CH2COOH
(S,R,S)-AHPC-Boc-trans-3-aminocyclobutanol-Pip-CH2COOH (VH032-Boc-trans-3-aminocyclobutanol-Pip-CH2COOH) is a E3 ligase ligand-linker conjugate that contains on one end a VHL ligand. (S,R,S)-AHPC-Boc-trans-3-aminocyclobutanol-Pip-CH2COOH is used in PROTAC technology.
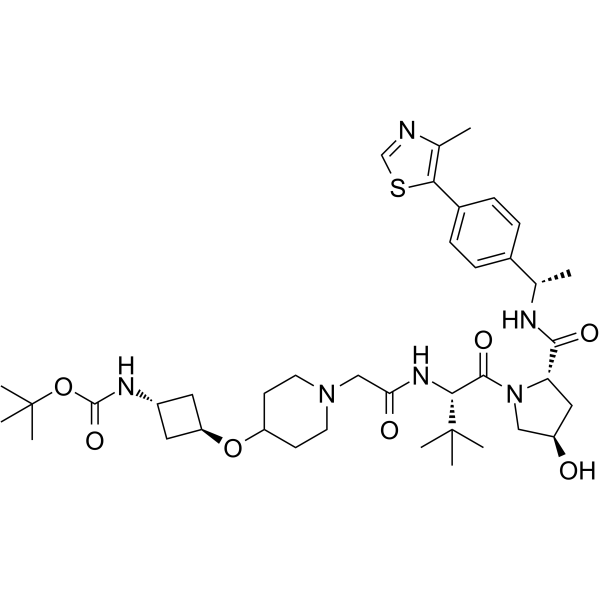
-
GC69947
(S,R,S)-AHPC-C10-NHBoc
(S,R,S)-AHPC-C10-NHBoc is a synthetic E3 ligase ligand-linker conjugate, containing a VHL ligand based on (S,R,S)-AHPC and one linker. It can be used to synthesize targeted PROTACs for BET.
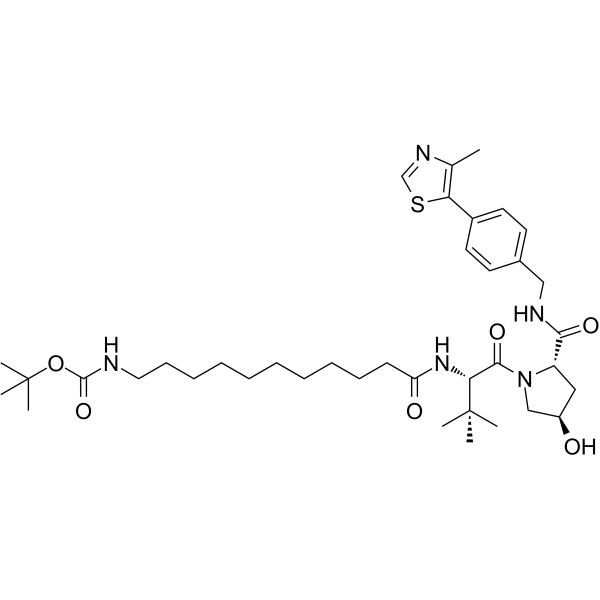
-
GC68352
(S,R,S)-AHPC-C2-amide-benzofuranylmethyl-pyridine
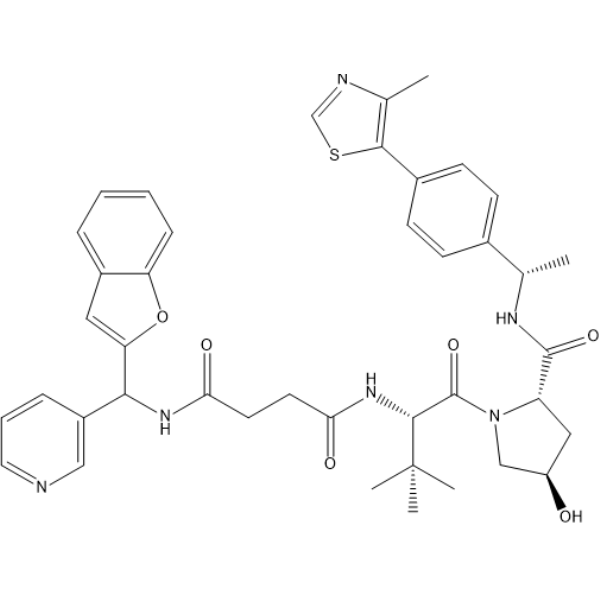
-
GC69743
(S,R,S)-AHPC-C4-NH2 hydrochloride
(S,R,S)-AHPC-C4-NH2 hydrochloride is a synthetic E3 ligase ligand-linker conjugate, containing a VHL ligand based on (S,R,S)-AHPC and one linker. It is used for the synthesis of targeted PROTACs for EED.
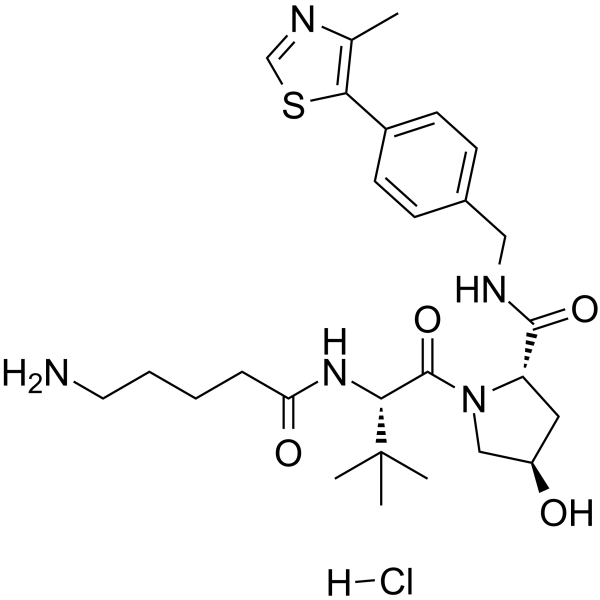
-
GC65133
(S,R,S)-AHPC-C5-COOH
(S,R,S)-AHPC-C5-COOH (VH032-C5-COOH) is a synthesized?E3 ligase ligand-linker conjugate, contains the VH032 VHL-based ligand and a linker to form PROTACs. VH-032 is a selective and potent inhibitor of VHL/HIF-1α interaction with a Kd of 185 nM, has the potential for the study of anemia and ischemic diseases.
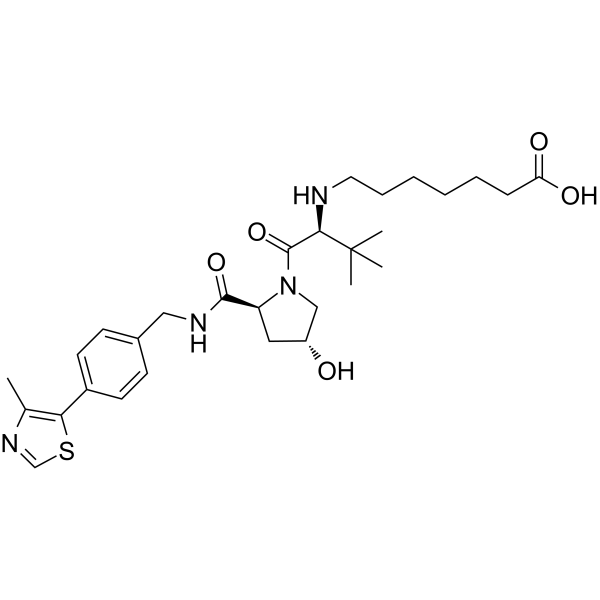
-
GC69948
(S,R,S)-AHPC-CO-C9-acid
"(S,R,S)-AHPC-CO-C9-acid" is a ligand-linker complex of E3 ligase, which can be attached to the ligand of a protein to form a PROTAC."
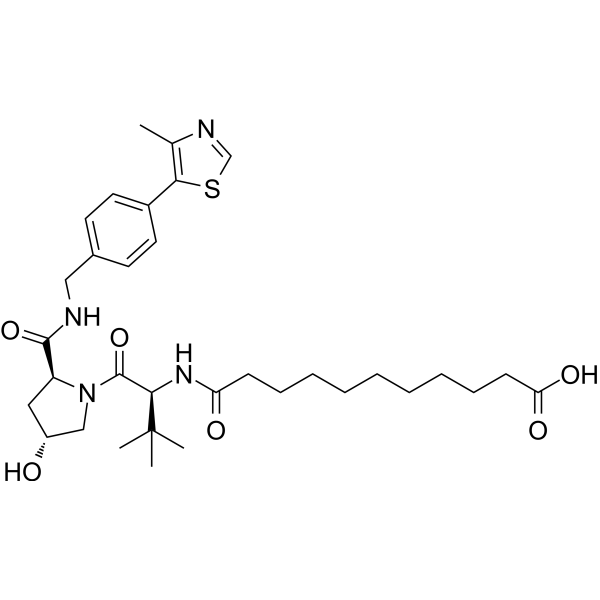
-
GC69949
(S,R,S)-AHPC-isobutyl acetate-methanesulfonothioate-Me-C10-NH2 TFA
(Please note that this is a chemical name and may not have a direct translation into English) (Please also note that the use of chemicals should only be done by trained professionals in appropriate settings with proper safety measures.) (Please consult with a professional chemist for accurate information regarding this compound.) The (S,R,S)-AHPC-isobutyl acetate-methanesulfonothioate-Me-C10-NH2 TFA is part of PROTAC BRD4 Degrader-12.
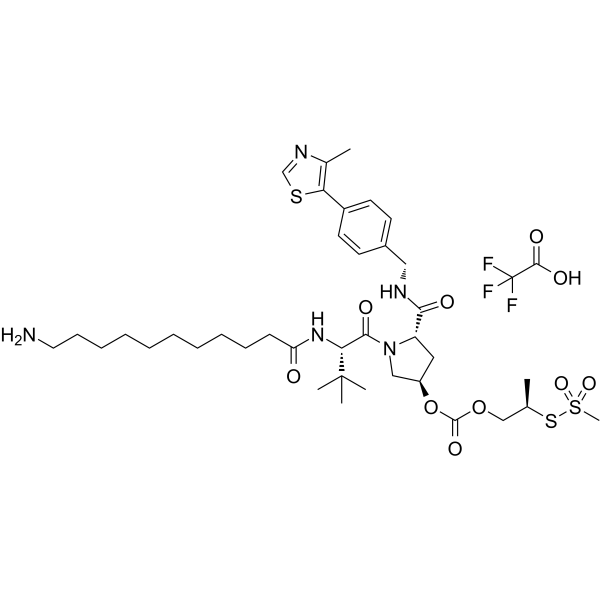
-
GC67831
(S,R,S)-AHPC-Me-C10-Br
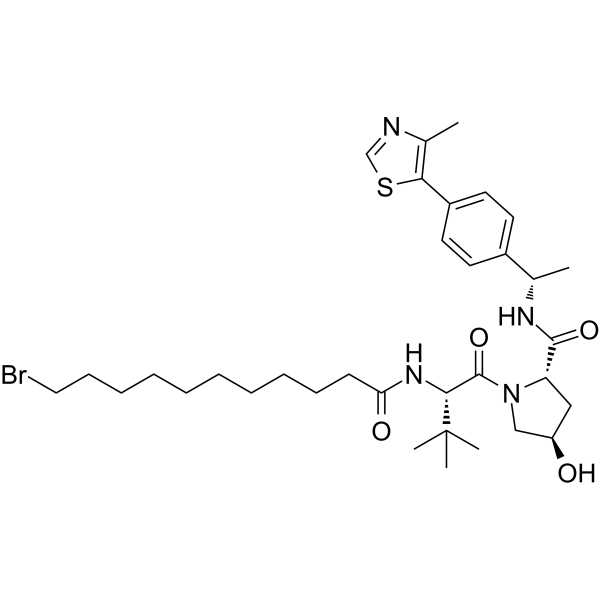
-
GC64371
(S,R,S)-AHPC-Me-C10-NH2
(S,R,S)-AHPC-Me-C10-NH2 is a synthesized E3 ligase ligand-linker conjugate that incorporates the a VHL ligand and a linker. (S,R,S)-AHPC-Me-C10-NH2 can be used in PROTAC MS432.
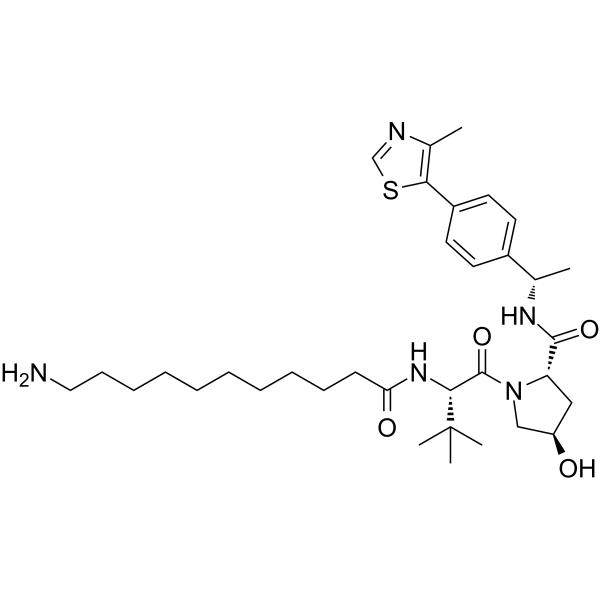
-
GC63494
(S,R,S)-AHPC-Me-C5-COOH
(S,R,S)-AHPC-Me-C5-COOH is a synthesized E3 ligase ligand-linker conjugate that incorporates the a VHL ligand and a linker. (S,R,S)-AHPC-Me-C5-COOH can be used in PROTAC DT2216.
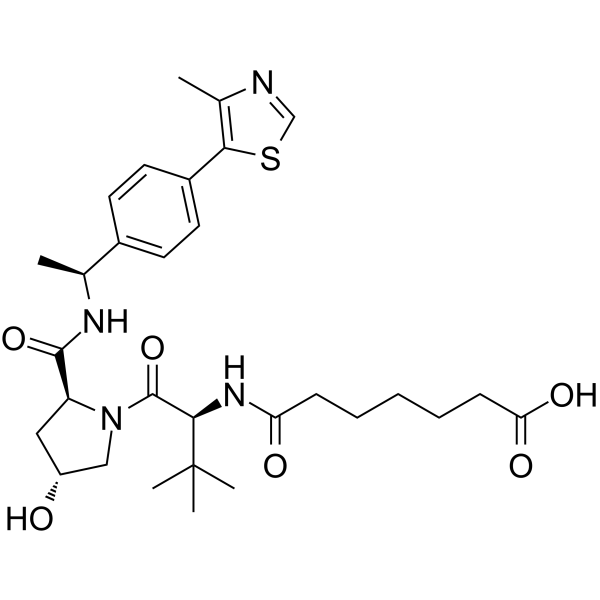
-
GC67729
(S,R,S)-AHPC-Me-C6-NH2
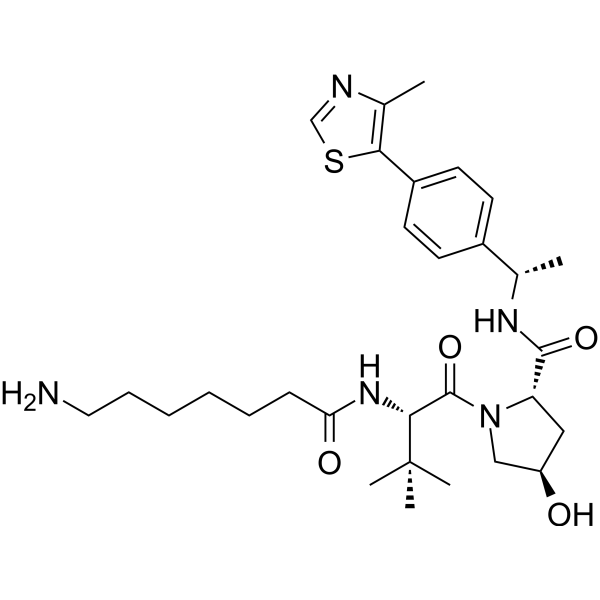
-
GC63561
(S,R,S)-AHPC-Me-C7 ester
(S,R,S)-AHPC-Me-C7 ester is a E3 ligase ligand-linker conjugate used to synthesise BCL-XL PROTAC degraders.
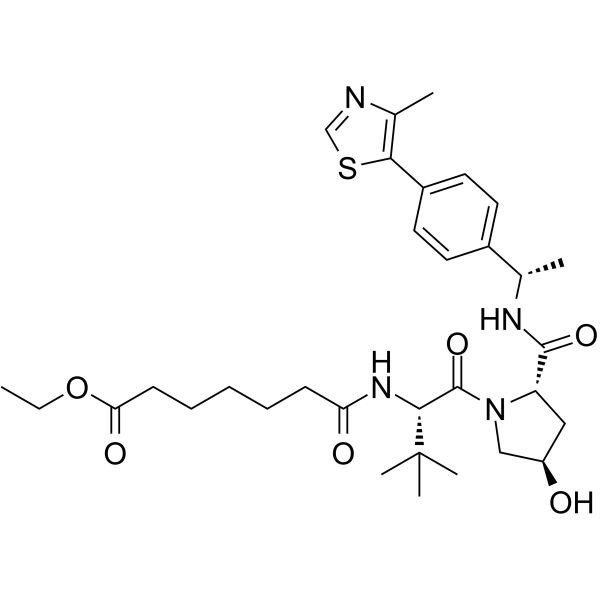
-
GC62585
(S,R,S)-AHPC-Me-CO-CH2-PEG3-NH2
(S,R,S)-AHPC-Me-CO-CH2-PEG3-NH2 is a synthesized E3 ligase ligand-linker conjugate that incorporates the a VHL ligand and a linker. (S,R,S)-AHPC-Me-CO-CH2-PEG3-NH2 can be used in PROTAC BRD4 Degrader-5 and PROTAC BRD4 Degrader-5-CO-PEG3-N3.
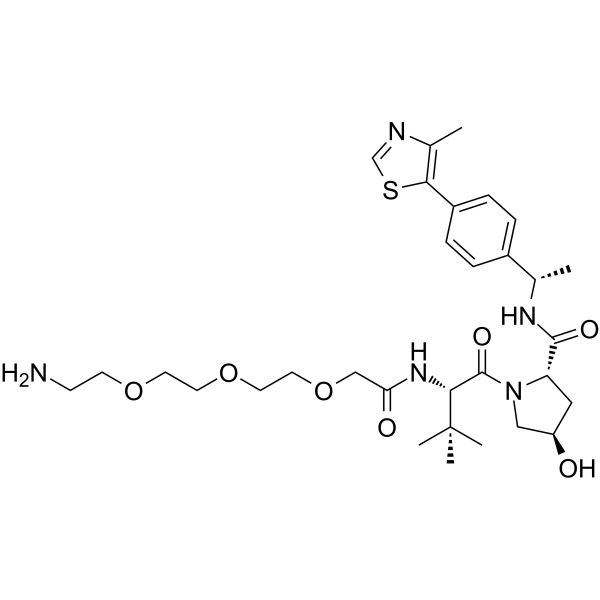
-
GC62183
(S,R,S)-AHPC-O-Ph-PEG1-NH2
(S,R,S)-AHPC-O-Ph-PEG1-NH2 (VH032-O-Ph-PEG1-NH2) is E3 ligase ligand-linker conjugate and incorporates a VHL ligand for the E3 ubiquitin ligase, and a PROTAC linker. (S,R,S)-AHPC-O-Ph-PEG1-NH2 is used in PROTAC EED degrader-1. PROTAC EED degrader-1 is a PROTAC targeting EED with a pKD of 9.02.
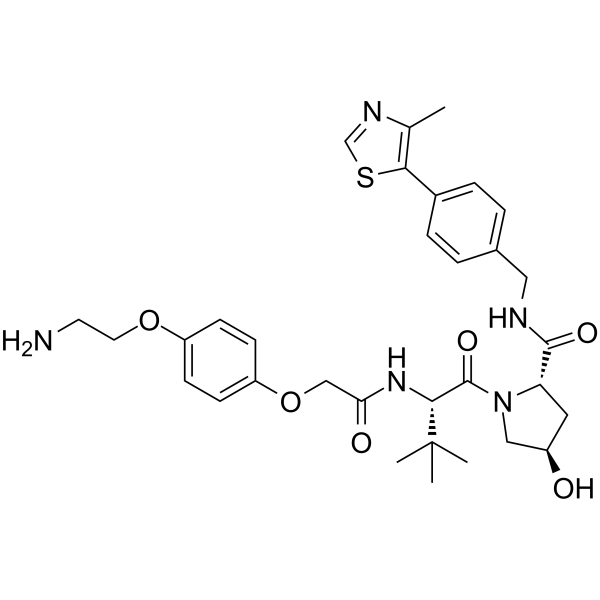
-
GC66630
(S,R,S)-AHPC-PEG2-acid
(S,R,S)-AHPC-PEG2-acid is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
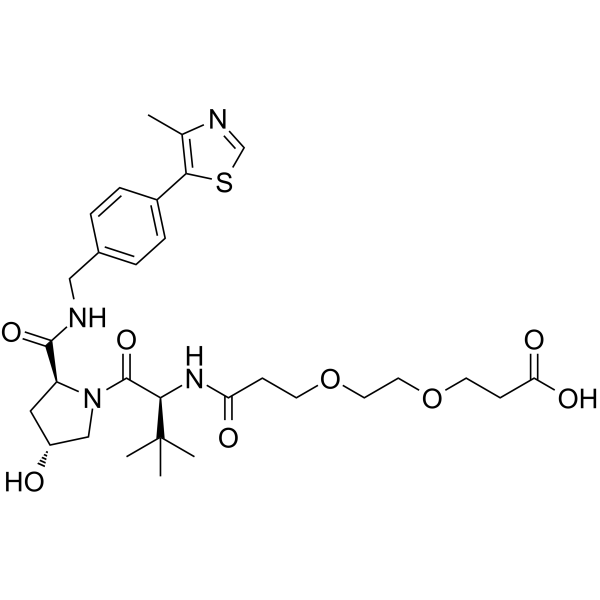
-
GC61774
(S,R,S)-AHPC-PEG5-Boc
(S,R,S)-AHPC-PEG5-Boc is a E3 ligase ligand-linker conjugate that incorporates the (S,R,S)-AHPC based VHL ligand and a linker used for Cdc20 degrader CP5V.
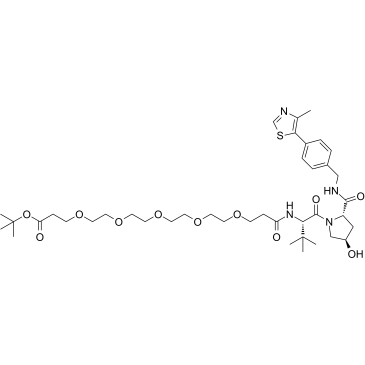
-
GC67672
(S,S)-GNE 5729
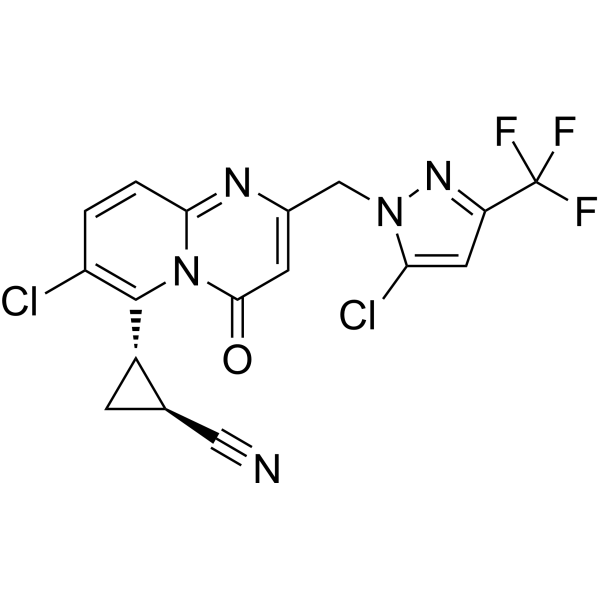
-
GC67682
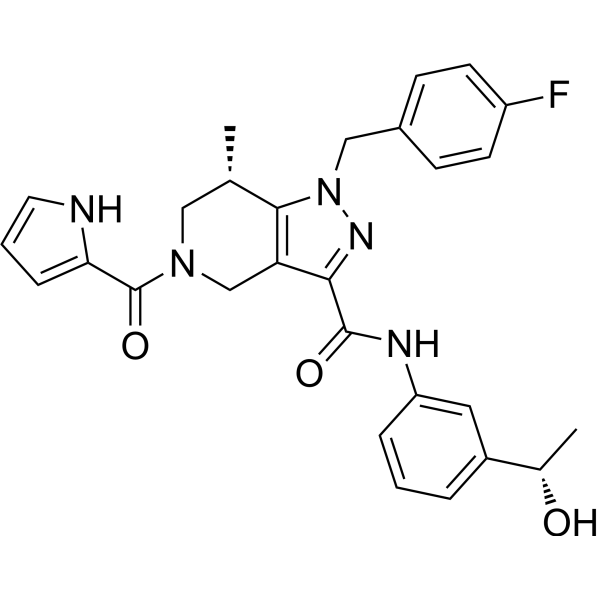
(S,S)-GSK321
-
GC64535
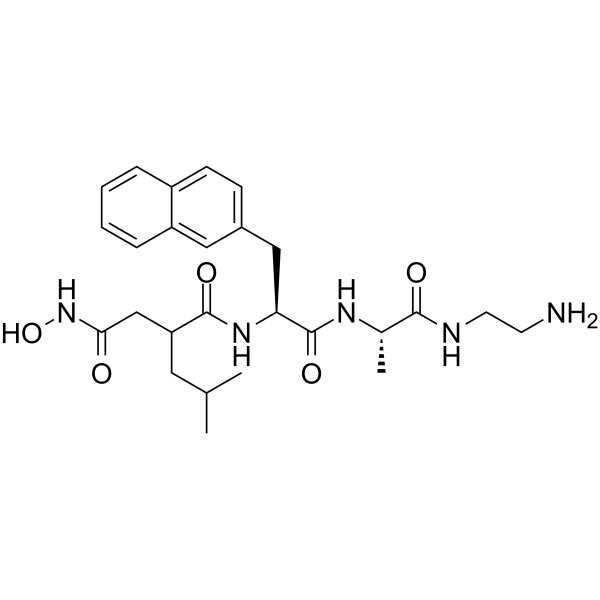
(S,S)-TAPI-1
(S,S)-TAPI-1 is an isomer of TAPI-1.
-
GC67761
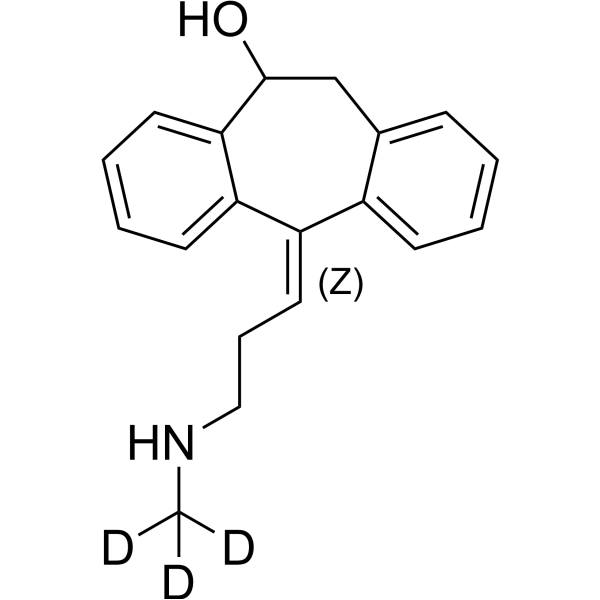
(Z)-10-Hydroxynortriptyline-d3
-
GC62291
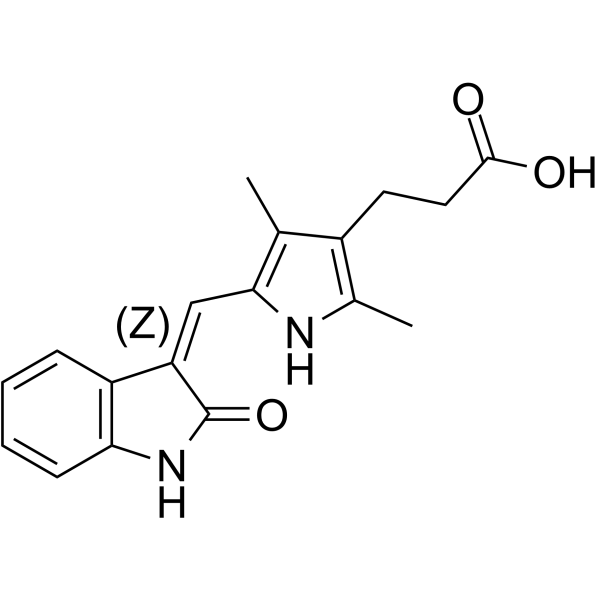
(Z)-Orantinib
(Z)-Orantinib ((Z)-SU6668) is a potent, selective, orally active and ATP competitive inhibitor of Flk‐1/KDR, PDGFRβ, and FGFR1, with IC50s of 2.1, 0.008, and 1.2 ?M, respectively. (Z)-Orantinib is a potent antiangiogenic and antitumor agent that induces regression of established tumors.
-
GC11965
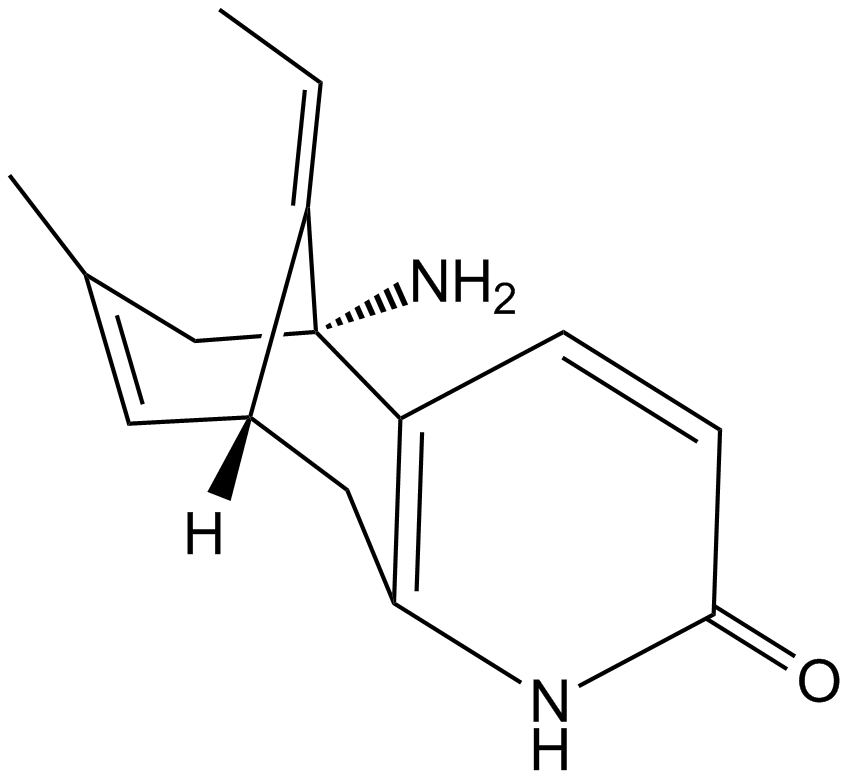
(±)-Huperzine A
A neuroprotective AChE inhibitor
-
GC14252
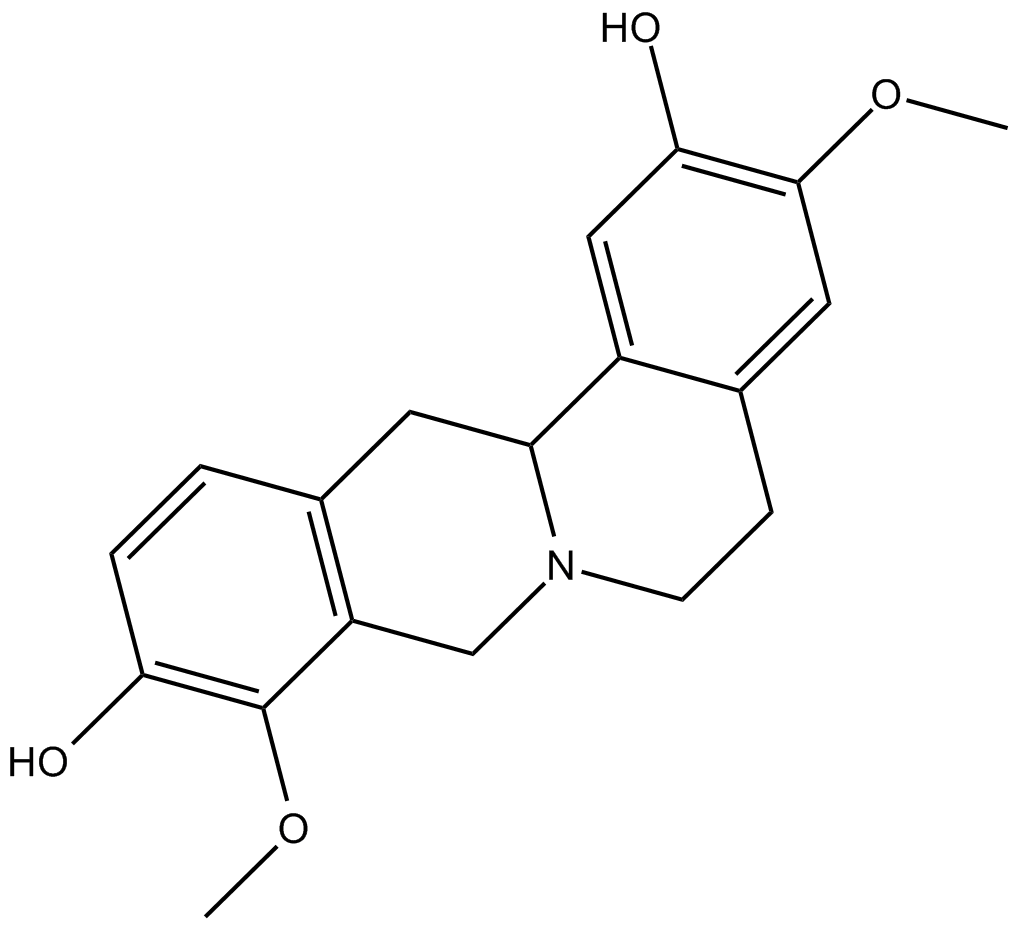
(–)-Stepholidine
A dopamine receptor antagonist and 5-HT1A partial agonist
-
GC63946
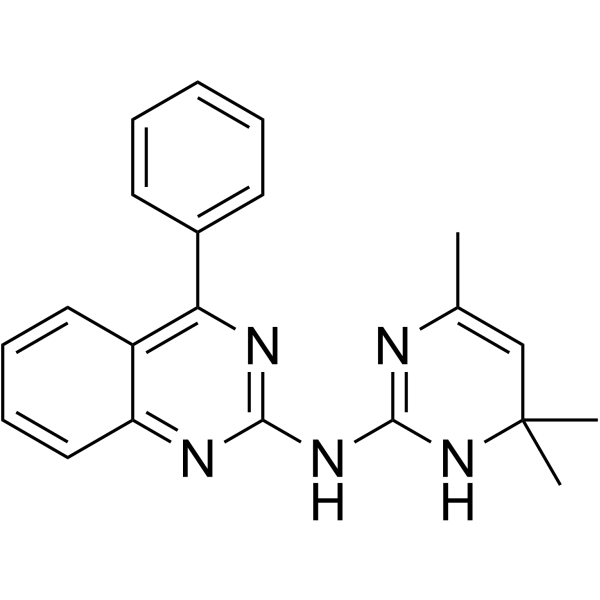
0990CL
0990CL is a?specific heterotrimeric Gαi subunit inhibitor by direct interaction with Gαi.
-
GC62761
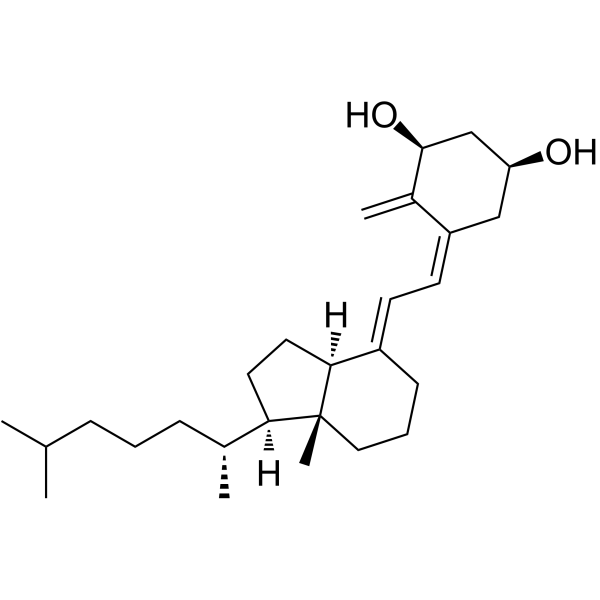
1α-Hydroxy-3-epi-vitamin D3
1α-Hydroxy-3-epi-vitamin D3, a natural metabolite of 1alpha,25-dihydroxyvitamin D3, is a potent suppressor of parathyroid hormone (PTH) secretion.
-
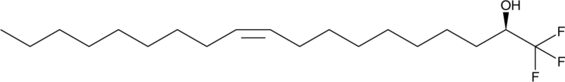
GC49034
1(R)-(Trifluoromethyl)oleyl alcohol
An oleic acid analog
-
GC65171
1,1′,1′′,1′′′-[1,4-Piperazinediylbis(2,1-ethanediylnitrilo)]tetrakis[2-dodecanol]
1,1′,1′′,1′′′-[1,4-Piperazinediylbis(2,1-ethanediylnitrilo)]tetrakis[2-dodecanol] is a lipid/lipidoid used in preparation of lipid-based or lipidoid nanoparticles.
![1,1′,1′′,1′′′-[1,4-Piperazinediylbis(2,1-ethanediylnitrilo)]tetrakis[2-dodecanol] Chemical Structure 1,1′,1′′,1′′′-[1,4-Piperazinediylbis(2,1-ethanediylnitrilo)]tetrakis[2-dodecanol] Chemical Structure](/media/struct/GC6/GC65171.png)
-
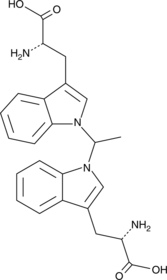
GC49768
1,1’-Ethylidene-bis-(L-tryptophan)
A potential impurity found in commercial preparations of L-tryptophan
-
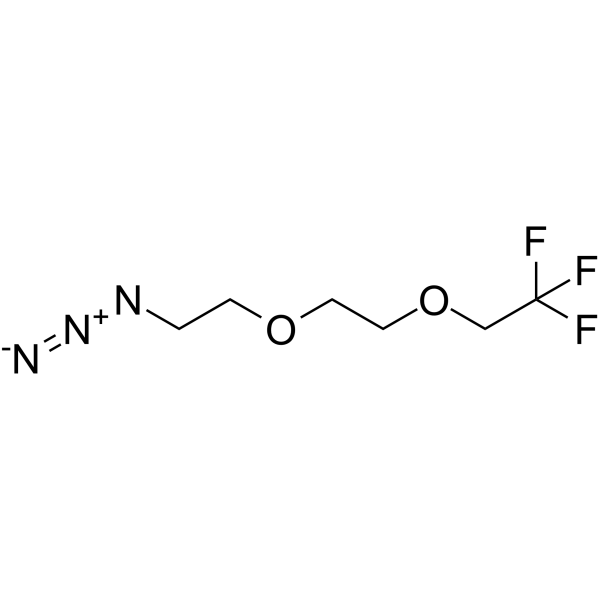
GC65729
1,1,1-Trifluoroethyl-PEG2-azide
1,1,1-Trifluoroethyl-PEG2-azide is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
-
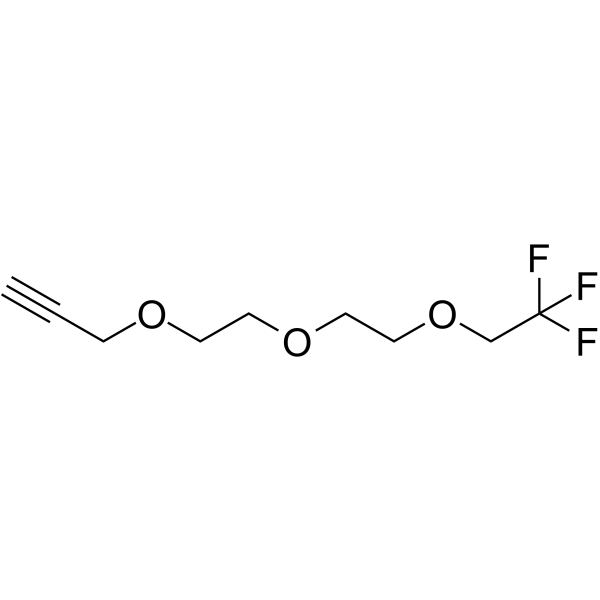
GC65715
1,1,1-Trifluoroethyl-PEG2-propargyl
1,1,1-Trifluoroethyl-PEG2-propargyl is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
-
GC66567
1,1,1-Trifluoroethyl-PEG4-alcohol
1,1,1-Trifluoroethyl-PEG4-alcohol is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
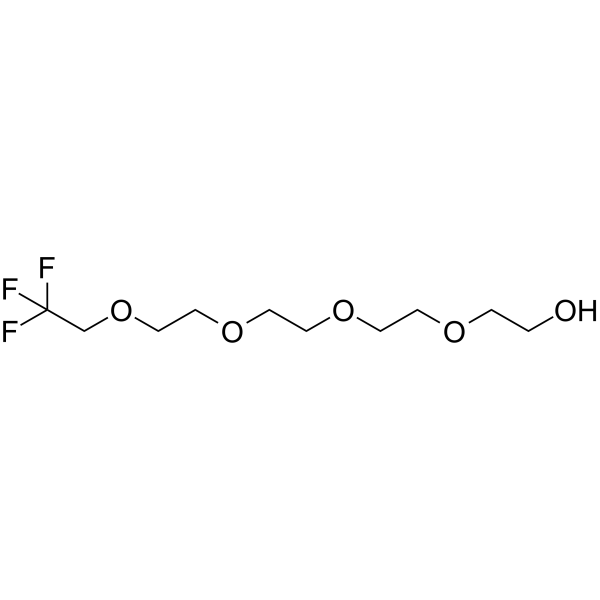
-
GC67011
1,1,1-Trifluoroethyl-PEG4-amine
111-Trifluoroethyl-PEG4-amine is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
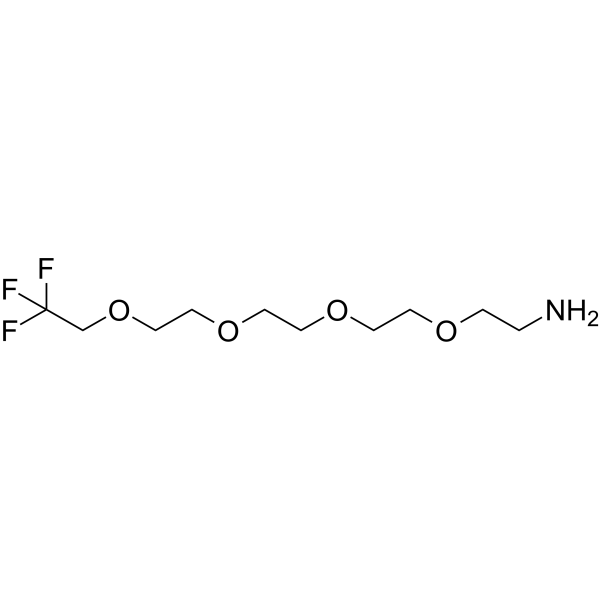
-
GC67295
1,1,1-Trifluoroethyl-PEG4-aminooxy
1,1,1-Trifluoroethyl-PEG4-aminooxy is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
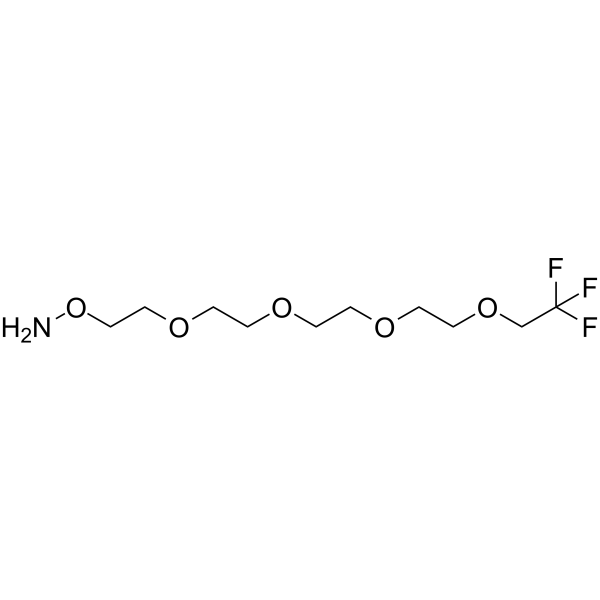
-
GC67595
1,1,1-Trifluoroethyl-PEG4-azide
111-Trifluoroethyl-PEG4-azide is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
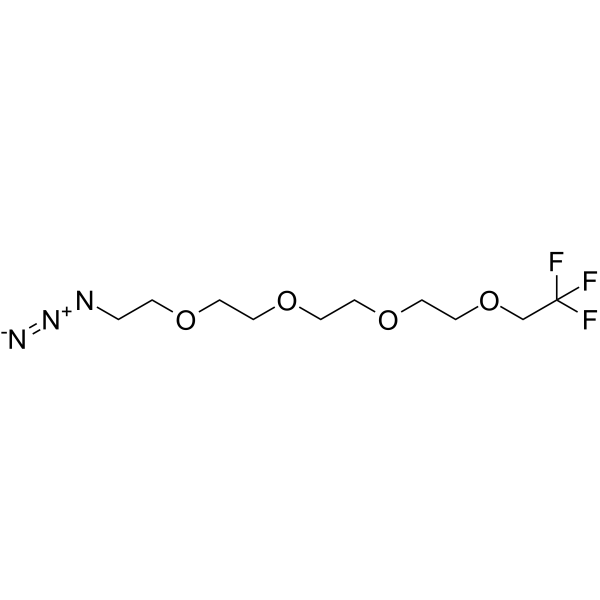
-
GC66971
1,1,1-Trifluoroethyl-PEG4-propargyl
1,1,1-Trifluoroethyl-PEG4-propargyl is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
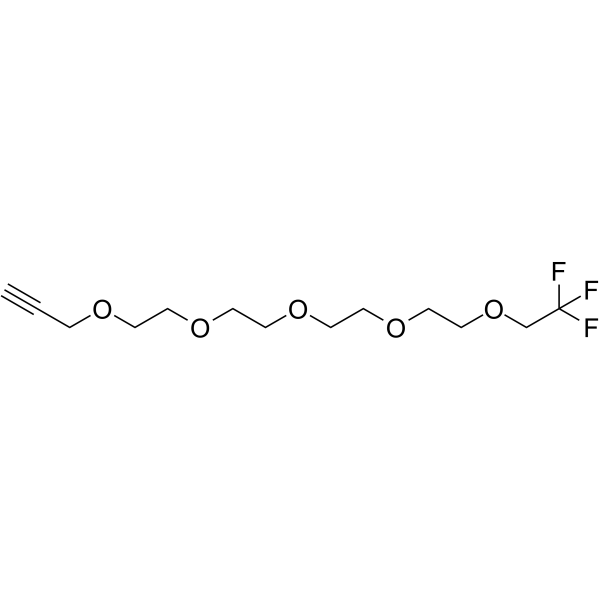
-
GC65718
1,1,1-Trifluoroethyl-PEG4-Tos
1,1,1-Trifluoroethyl-PEG4-Tos is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
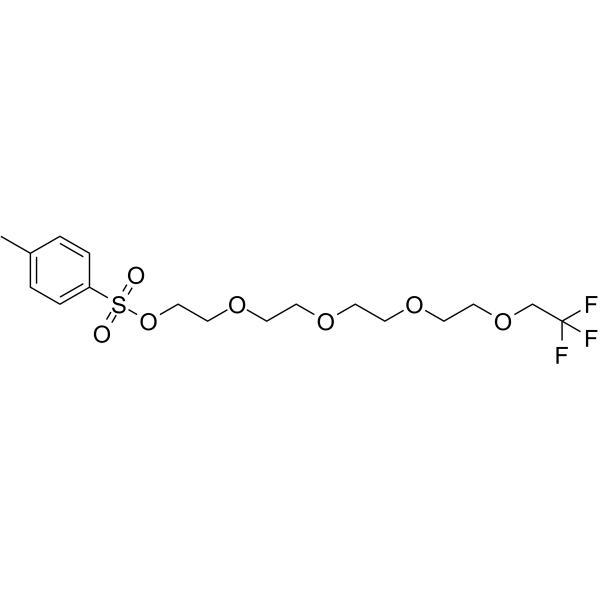
-
GC67833
1,1,3-Tribromoacetone

-
GC17055
1,2,3,4,5,6-Hexabromocyclohexane
1,2,3,4,5,6-六溴环己烷(1,2,3,4,5,6-Hexabromocyclohexane)直接抑制JAK2激酶结构域内的口袋,抑制自身磷酸化。
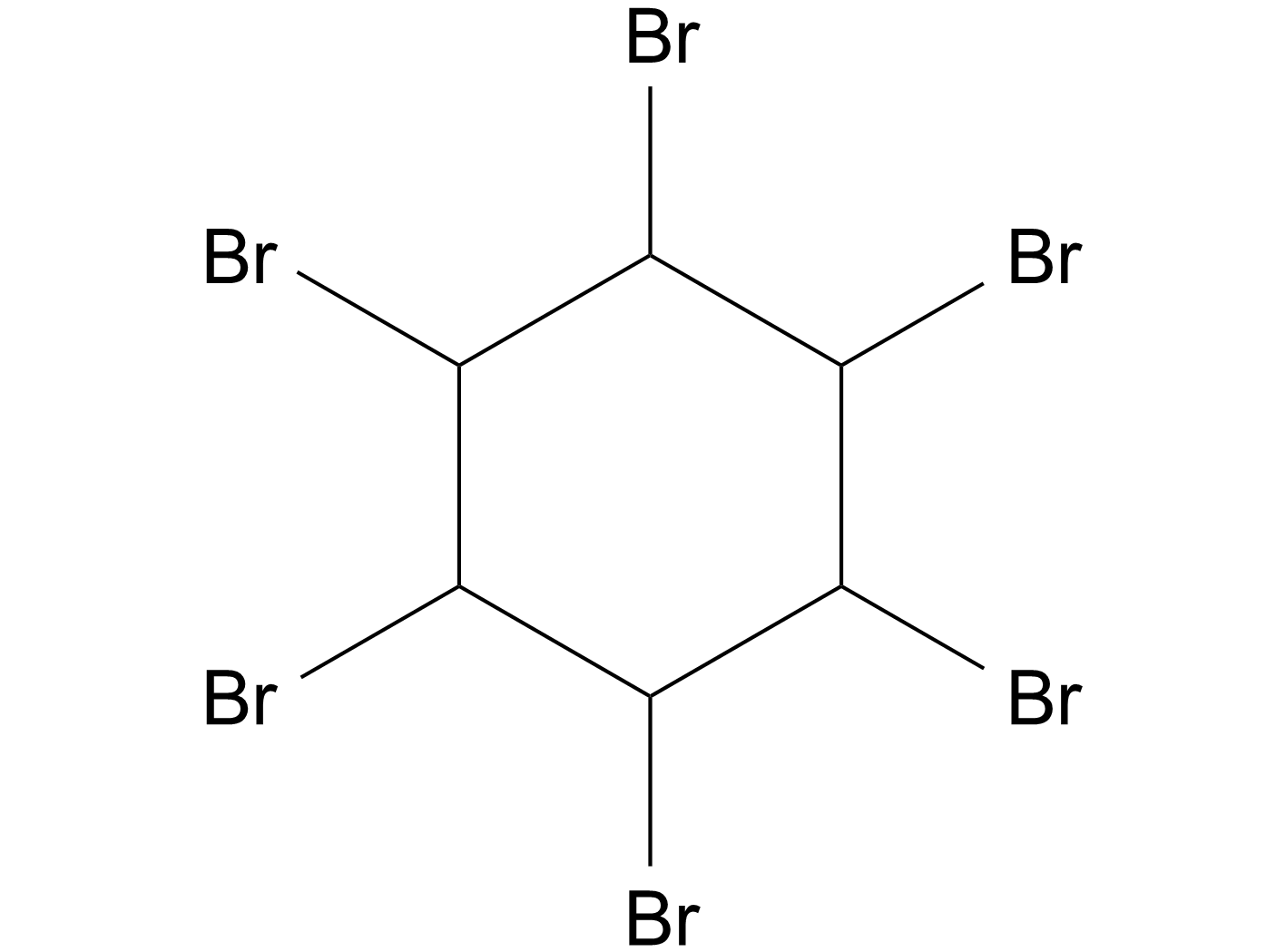
-
GC61719
1,2,3,4-Tetramethylbenzene
1,2,3,4-Tetramethylbenzene consists of a benzene ring with four methyl groups (-CH3) as a substituent.
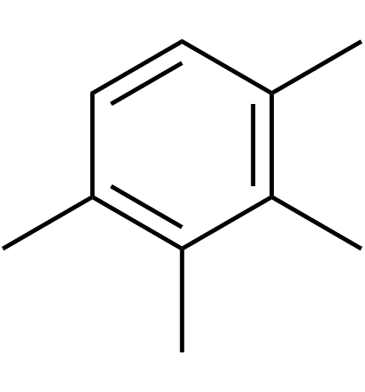
-
GC61729
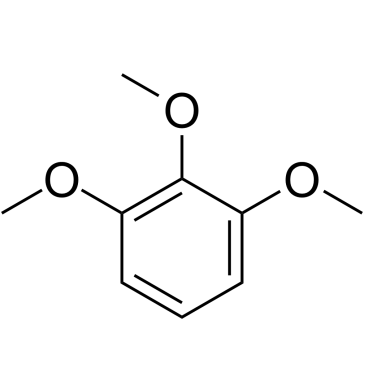
1,2,3-Trimethoxybenzene
1,2,3-Trimethoxybenzene is a member of the class of compounds known as anisoles.
-
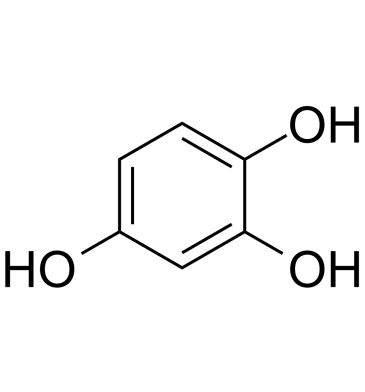
GC61898
1,2,4-Trihydroxybenzene
1,2,4-Trihydroxybenzene (Hydroxyhydroquinone), a by-product of coffee bean roasting, increases intracellular Ca2+ concentration in rat thymic lymphocytes.
-
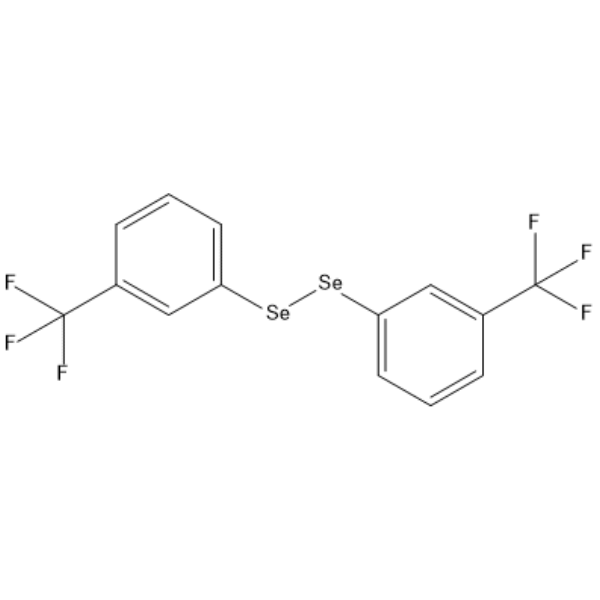
GC66452
1,2-Bis(3-(trifluoromethyl)phenyl)diselane
1,2-Bis(3-(trifluoromethyl)phenyl)diselane is an active compound and can be used for research.
-
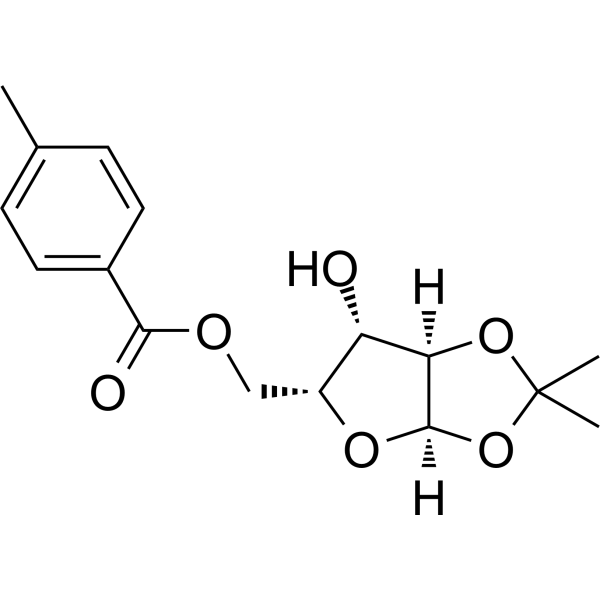
GC68484
1,2-O-Isopropylidene-5-O-p-toluoyl-a-D-xylofuranose
1,2-O-Isopropylidene-5-O-p-toluoyl-a-D-xylofuranose is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity and target malignant tumors in the inert lymphatic system. The anticancer mechanism in this process depends on inhibiting DNA synthesis, inducing apoptosis (cell death), etc.
-
GC61731
1,3,5-Triisopropylbenzene
1,3,5-Triisopropylbenzene acts as a fuel and fuel additive.
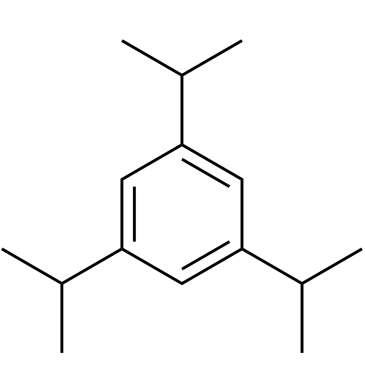
-
GC61399
1,3,6,8-Tetrahydroxynaphthalene
1,3,6,8-Tetrahydroxynaphthalene (T4HN) is an indispensable precursor to DHN (1,8-Dihydroxynaphthalene) melanin and is an unique symmetrical compound of polyketide origin.
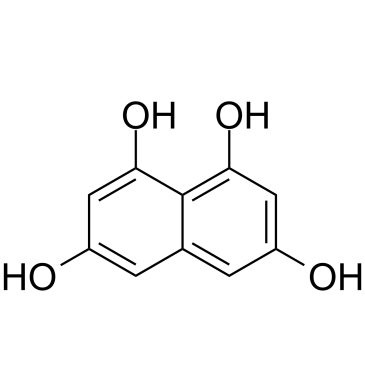
-
GC49220
1,3-Benzodioxole-5,6-diamine (hydrochloride)
A fluorescent derivatization reagent
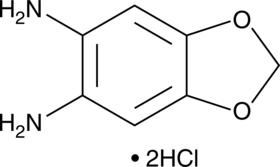
-
GC66949
1,3-bis(carboxyethoxy)-2,2-bis(carboxyethoxy)propane
1,3-bis(carboxyethoxy)-2,2-bis(carboxyethoxy)propane is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
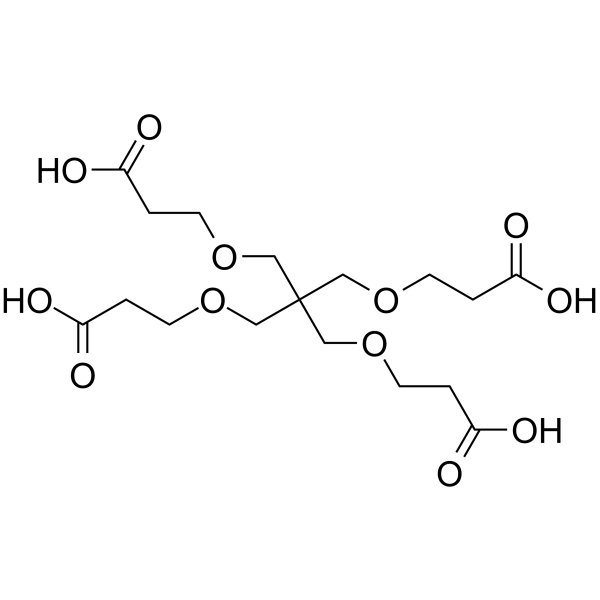
-
GC67054
1,3-Bis-aminooxy propane
1,3-Bis-aminooxy propane is an alkyl chain-based PROTAC linker that can be used in the synthesis of PROTACs.
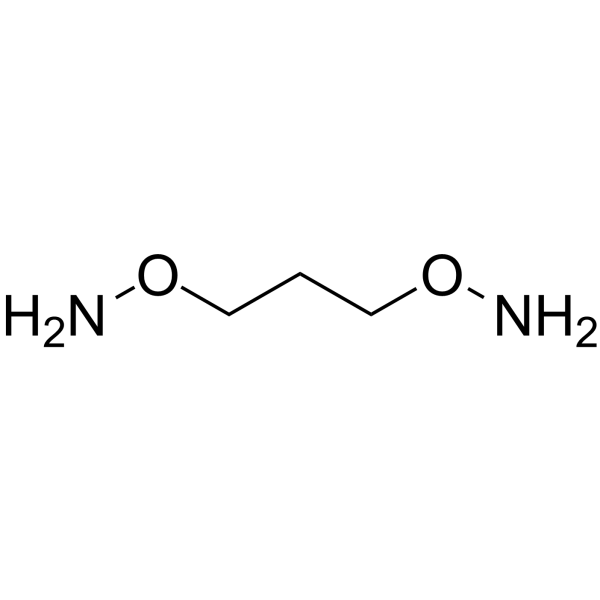
-
GC62352
1,3-Butanediol
1,3-Butanediol, an ethanol dimer providing a source of calories for human nutrition.
-
GC19661
1,3-Dihydroxyacetone
1,3-Dihydroxyacetone (DHA), the main active ingredient in sunless tanning skin-care preparations and an important precursor for the synthesis of various fine chemicals, is produced on an industrial scale by microbial fermentation of glycerol over Gluconobacter oxydans.

-
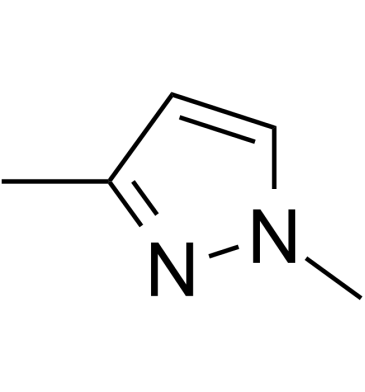
GC61827
1,3-Dimethylpyrazole
1,3-Dimethylpyrazole is a bioactive compound isolated from Moso Bamboo Root.
-
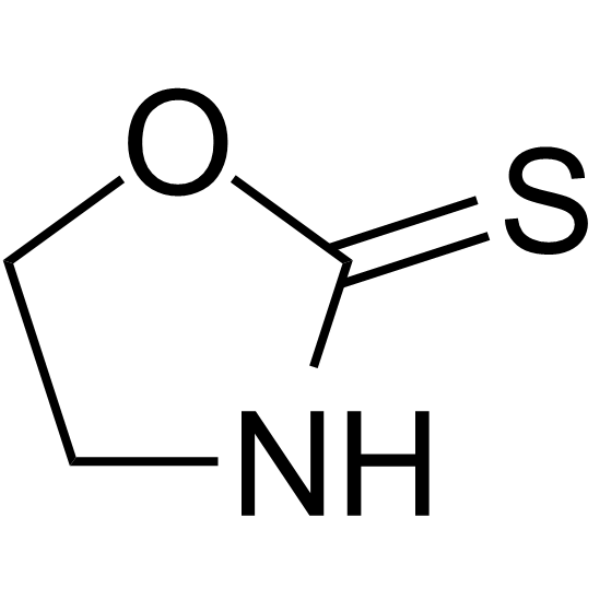
GC67985
1,3-Oxazolidine-2-thione
-
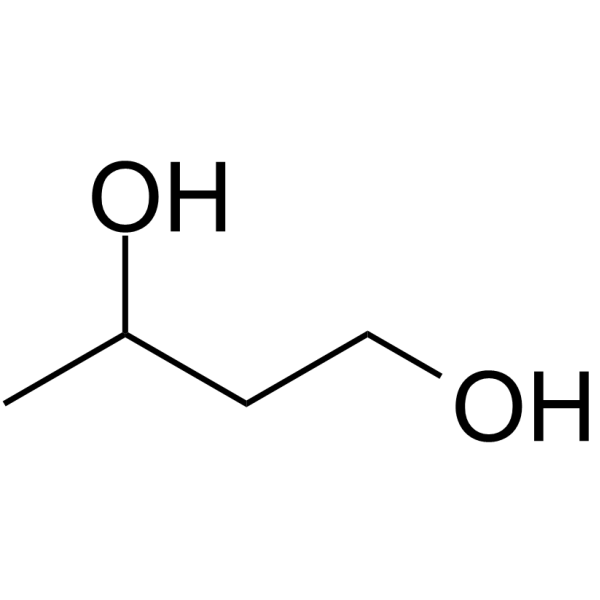
GC61709
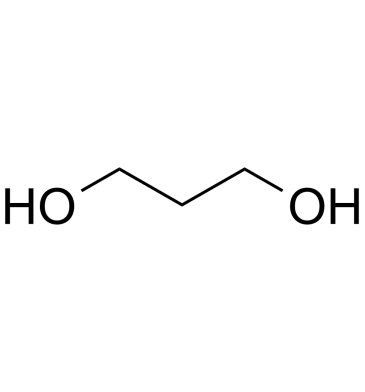
1,3-Propanediol
1,3-Propanediol is produced in nature by the fermentation of glycerol in microorganism.
-
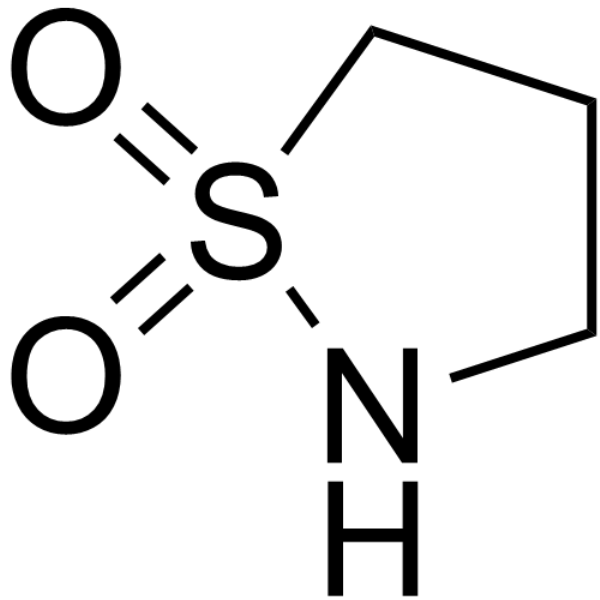
GC68134
1,3-Propanesultam
-
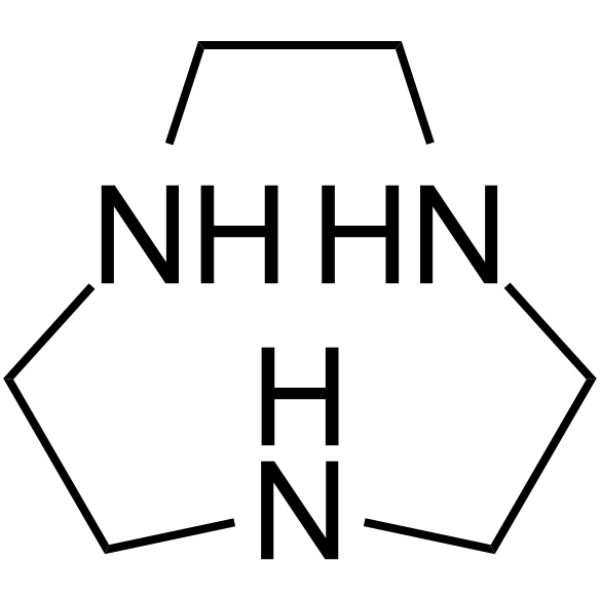
GC64247
1,4,7-Triazonane
1,4,7-Triazonane (1,4,7-Triazacyclononane), an intermediate in the synthesis of 1,4,7-trifunctionalized derivatives, is a possible reagent for compleximetric titrations with high cation-binding selectivity.
-
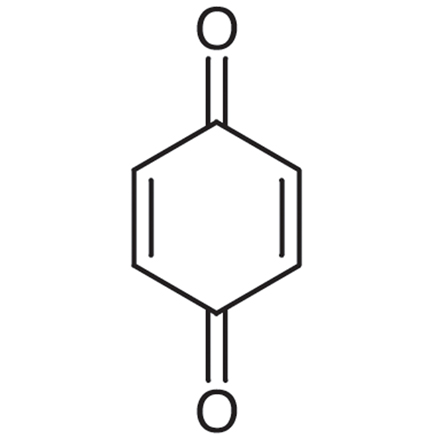
GC19528
1,4-Benzoquinone
A toxic metabolite of benzene
-
GC62755
1,4-D-Gulonolactone
1,4-D-Gulonolactone is an endogenous metabolite.
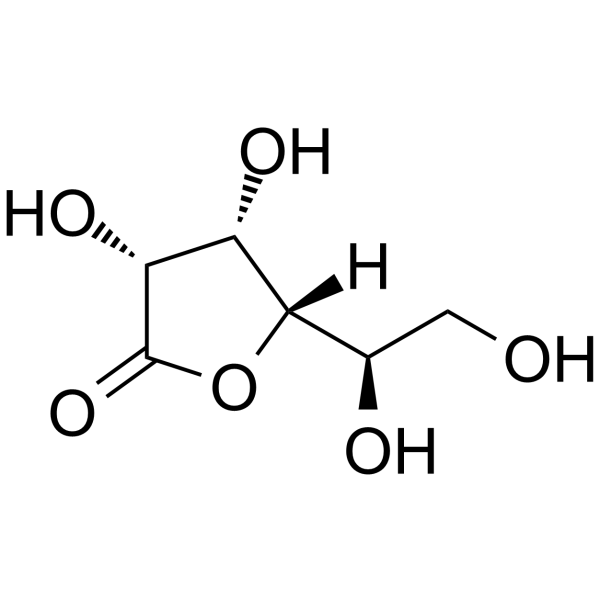
-
GC68486
1,4-Diaminobutane-d8 dihydrochloride
1,4-Diaminobutane-d8 (dihydrochloride) is the deuterated form of 1,4-Diaminobutane dihydrochloride. 1,4-Diaminobutane (Putrescine) dihydrochloride is an endogenous metabolite that can serve as an indicator of pollution caused by Cr(III) or Cr(VI) stress in higher plants such as barley and rapeseed.
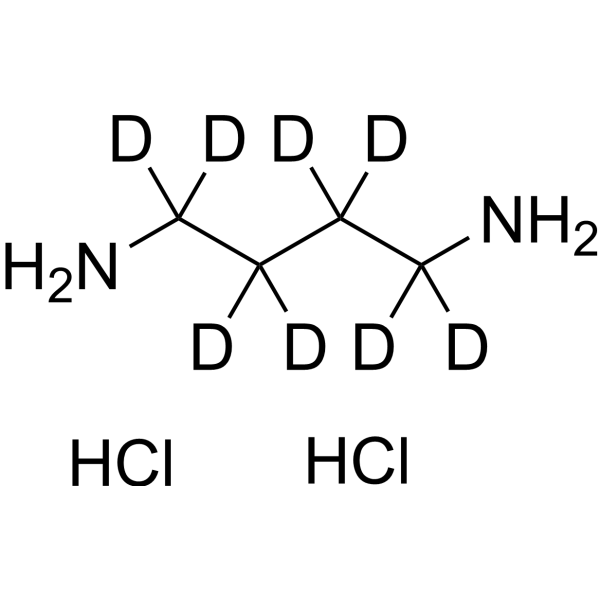
-
GC19683
1,4-Dichlorobenzene
1,4-Dichlorobenzene is used as an intermediate product in the manufacture of pigments, pesticides and disinfectants.

-
GC61949
1,4-DPCA ethyl ester
1,4-DPCA ethyl ester is the ethyl ester of 1,4-DPCA and can inhibit factor inhibiting HIF (FIH).
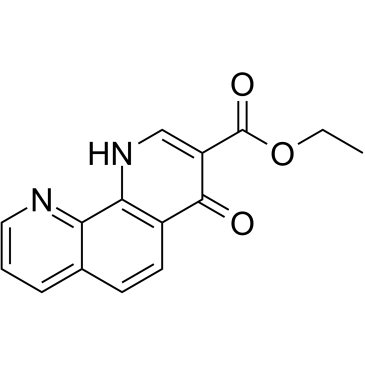
-
GC19717
1,6-anhydroglucose
1,6-anhydroglucose (1,6-Anhydro-β-D-glucopyranose) is an anhydrosugar produced through glucan pyrolysis and is widely found in nature.
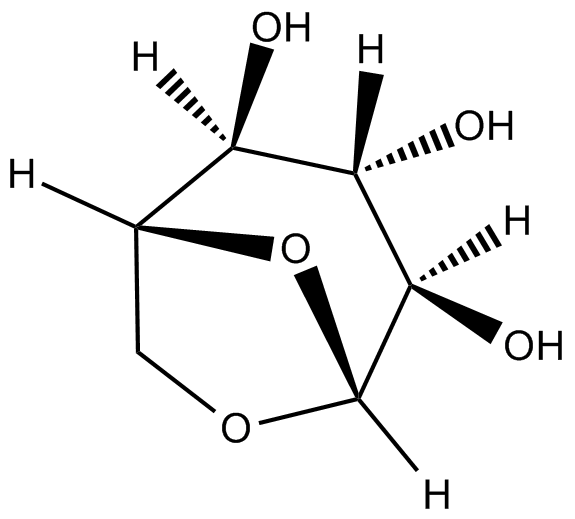
-
GC67200
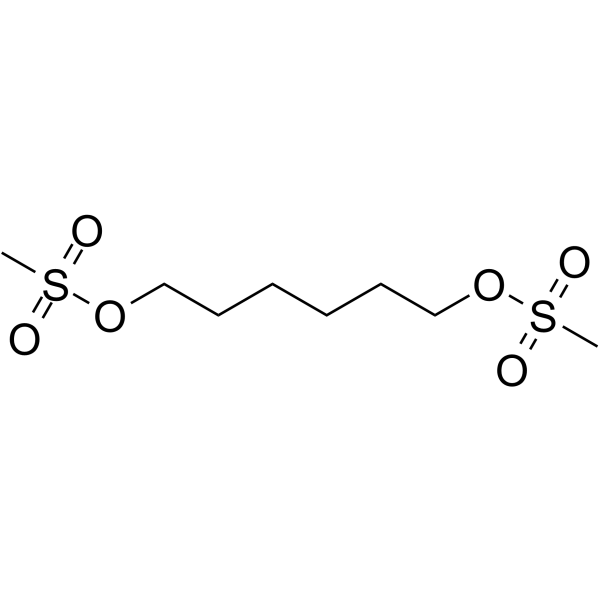
1,6-Bis(mesyloxy)hexane
16-Bismesyloxyhexane is a cleavable ADC linker used in the synthesis of antibody-drug conjugates (ADCs).
-
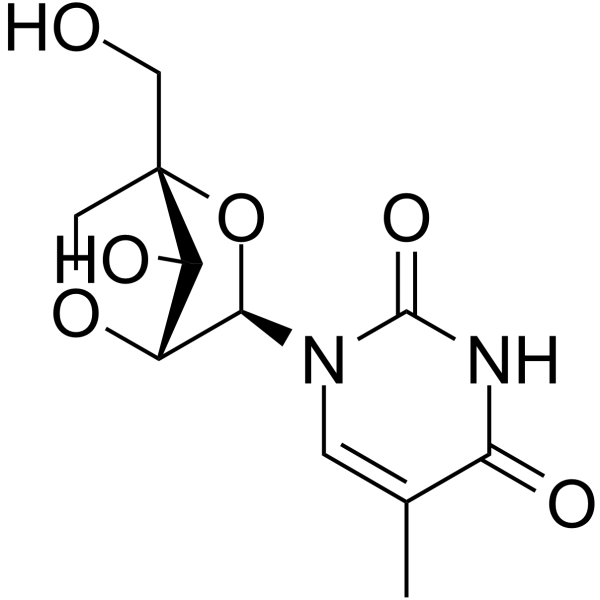
GC65551
1-(2'-O-4-C-Methylene-beta-D-ribofuranosyl)thymine
1-(2'-O-4-C-Methylene-beta-D-ribofuranosyl)thymine is a bicyclic nucleoside.
-
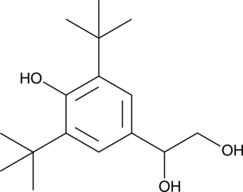
GC49537
1-(3,5-Di-tert-butyl-4-hydroxyphenyl)-1,2-ethanediol
A sterically hindered para-alkylphenol
-
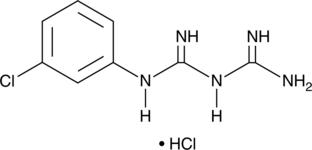
GC49346
1-(3-Chlorophenyl)biguanide (hydrochloride)
A 5-HT3 receptor agonist
-
GC49294
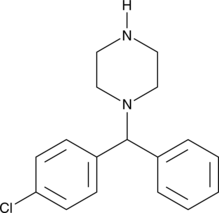
1-(4-Chlorobenzhydryl)piperazine
An inactive metabolite of meclizine and chlorcyclizine
-
GC52356
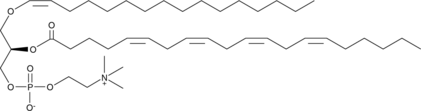
1-1(Z)-Hexadecenyl-2-Arachidonoyl-sn-glycero-3-PC
A plasmalogen
-
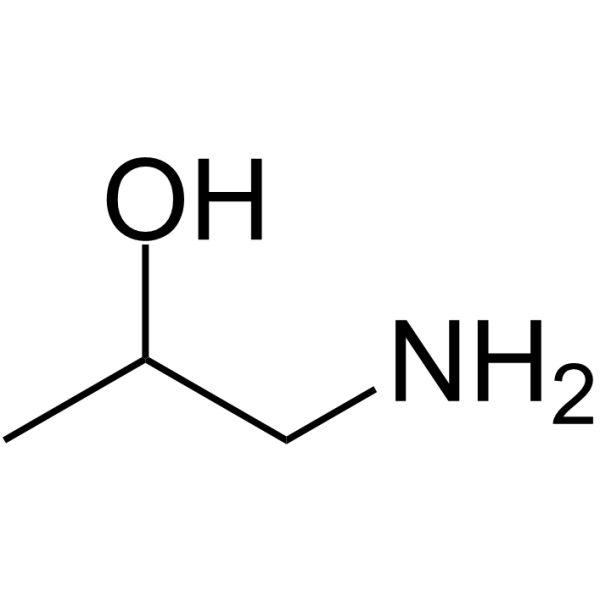
GC64404
1-Aminopropan-2-ol
1-Aminopropan-2-ol is a microbial metabolism of amino alcohol metabolism via propionaldehyde and acetaldehyde in a species of Pseudomonas.
-
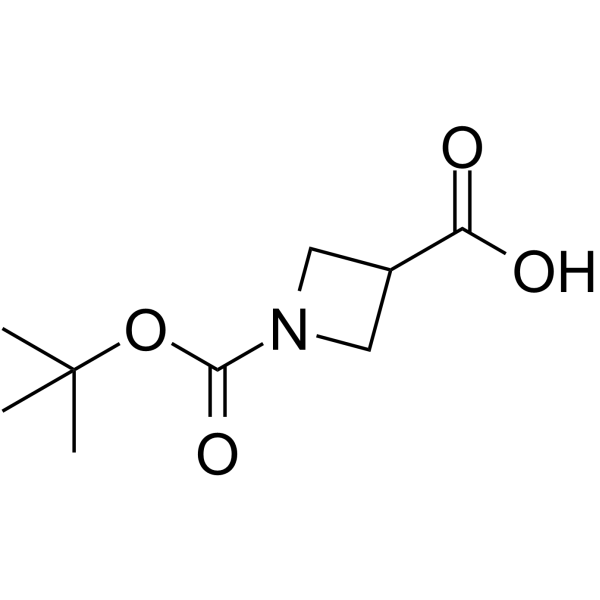
GC64176
1-Boc-azetidine-3-carboxylic acid
1-Boc-azetidine-3-carboxylic acid is a non-cleavable?ADC linker?used in the synthesis of antibody-drug conjugates (ADCs). 1-Boc-azetidine-3-carboxylic acid is also a alkyl chain-based PROTAC linker?that can be used in the synthesis of PROTACsup
-
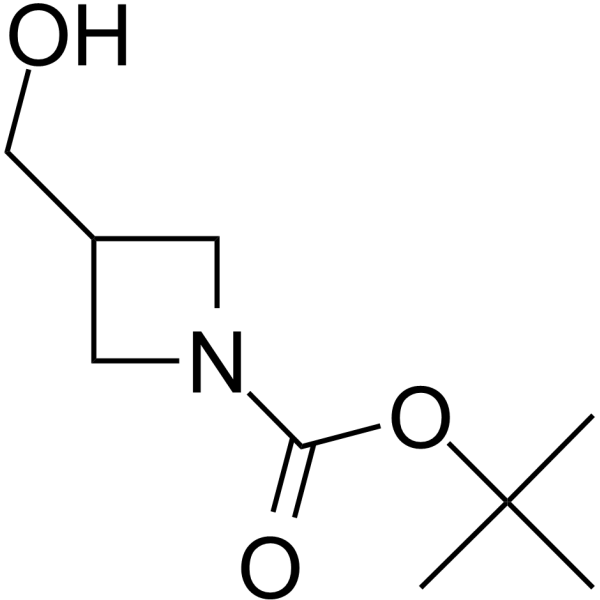
GC64159
1-Boc-azetidine-3-yl-methanol
1-Boc-azetidine-3-yl-methanol is a non-cleavable?ADC linker?used in the synthesis of antibody-drug conjugates (ADCs). 1-Boc-azetidine-3-yl-methanol is also a alkyl chain-based PROTAC linker?that can be used in the synthesis of PROTACs
-
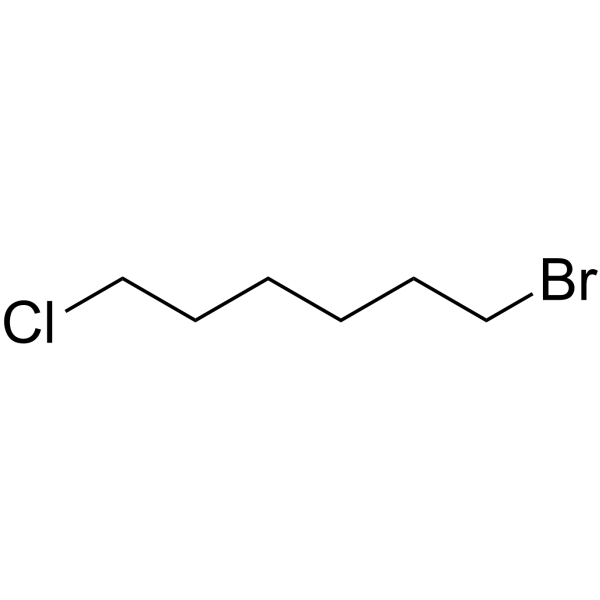
GC65223
1-Bromo-6-chlorohexane
1-Bromo-6-chlorohexane is a PROTAC linker can be used in the synthesis of PROTACs.
-
GC49861
1-Carboxycyclohexaneacetic Acid
A potential impurity in commercial preparations of gabapentin

-
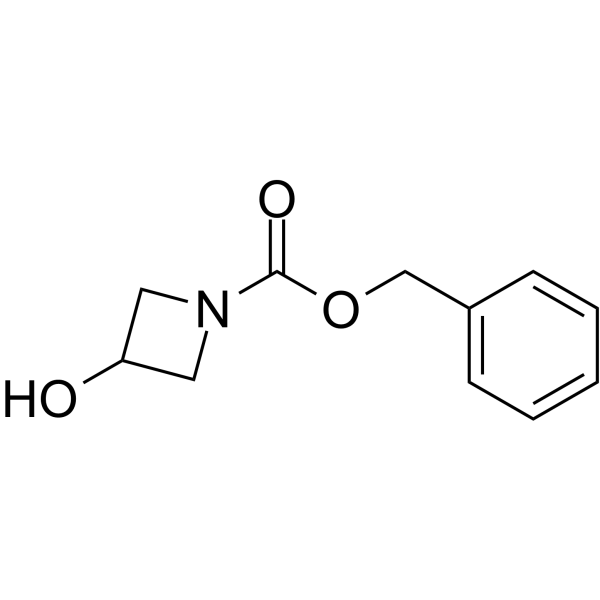
GC64145
1-Cbz-3-Hydroxyazetidine
1-Cbz-3-Hydroxyazetidine is a non-cleavable?ADC linker?used in the synthesis of antibody-drug conjugates (ADCs). 1-Cbz-3-Hydroxyazetidine is also a alkyl chain-based PROTAC linker?that can be used in the synthesis of PROTACs.
-
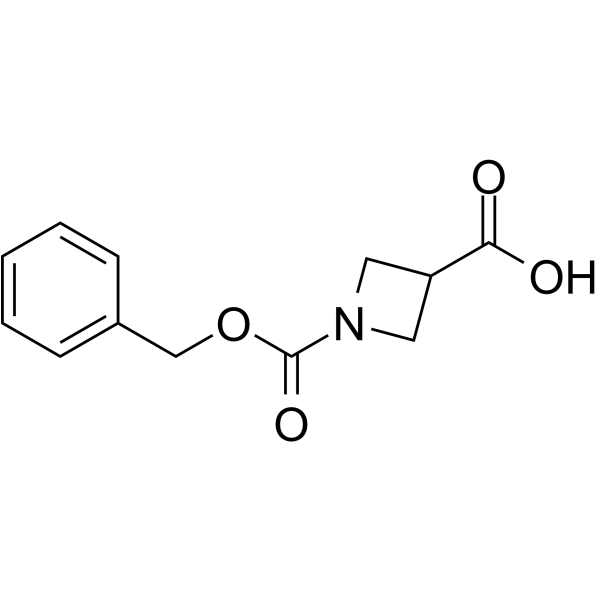
GC64047
1-Cbz-azetidine-3-carboxylic acid
1-Cbz-azetidine-3-carboxylic acid is a non-cleavable?ADC linker?used in the synthesis of antibody-drug conjugates (ADCs). 1-Cbz-azetidine-3-carboxylic acid is also a alkyl chain-based PROTAC linker?that can be used in the synthesis of PROTACs
-
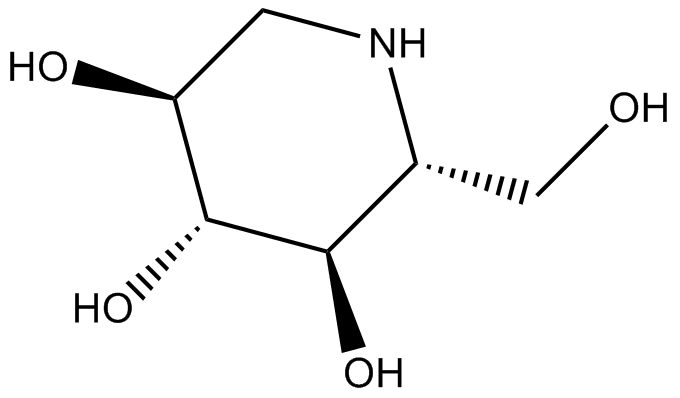
GN10317
1-Deoxynojirimycin
-
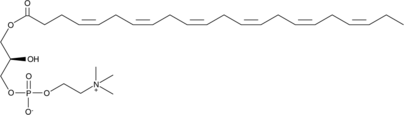
GC52186
1-Docosahexaenoyl-2-hydroxy-sn-glycero-3-PC
-
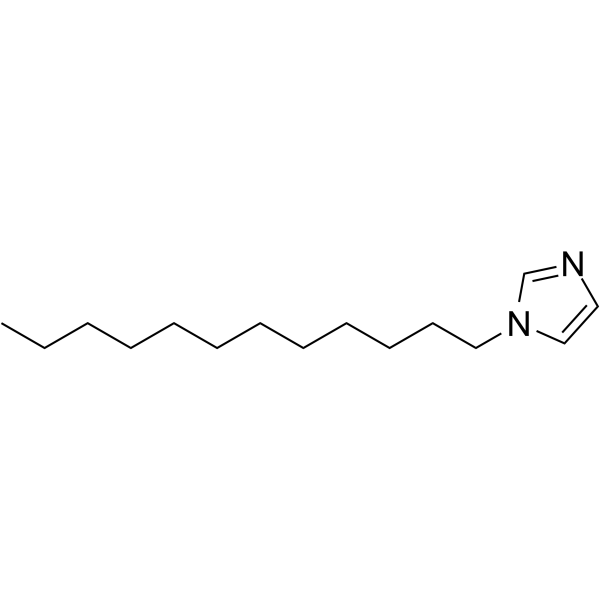
GC62760
1-Dodecylimidazole
1-Dodecylimidazole (N-Dodecylimidazole) is a lysosomotropic detergent and a cytotoxic agent. 1-Dodecylimidazole causes cell death by its acid-dependent accumulation in lysosomes, disruption of the lysosomal membrane, and releaseof cysteine proteases into the cytoplasm. 1-Dodecylimidazole has hypocholesterolaemic activity and broad-spectrum antifungal activity.
-
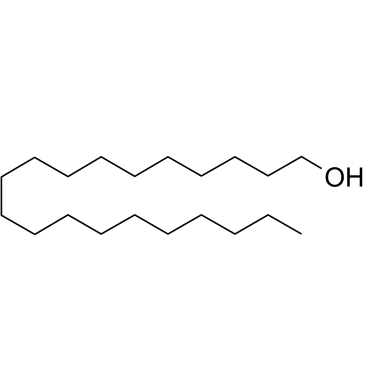
GC61690
1-Eicosanol
1-Eicosanol is a natural compound with antioxidant activity isolated from Hypericum carinatum.
-
GC49470
1-Ethyl-1-nitrosourea (hydrate)
A DNA alkylating agent
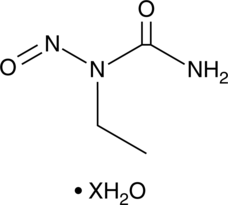
-
GC67844
1-Fluoronaphthalene

-
GC62754
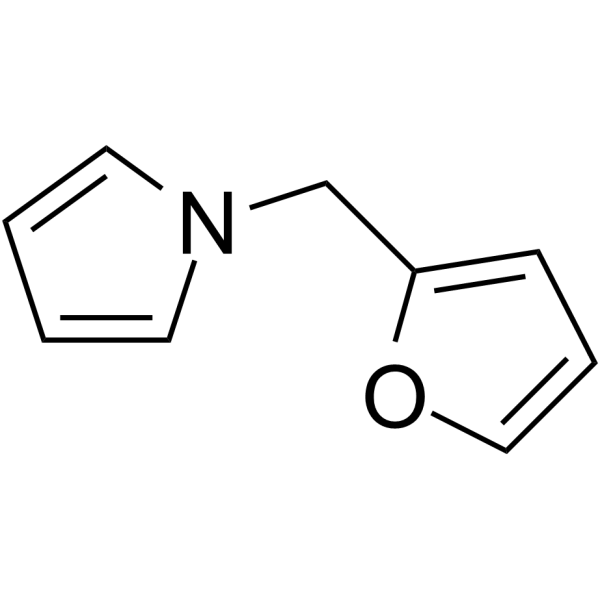
1-Furfurylpyrrole
1-Furfurylpyrrole has been identified as a potential contributor of flavor and aroma to popcorn.
-
GC61642
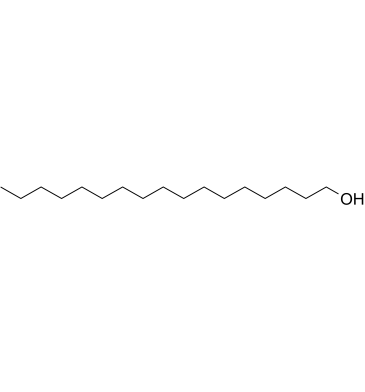
1-Heptadecanol
1-Heptadecanol is a long-chain primary alcohol with antibacterial activity from Solena amplexicaulis leaves.
-
GC68495
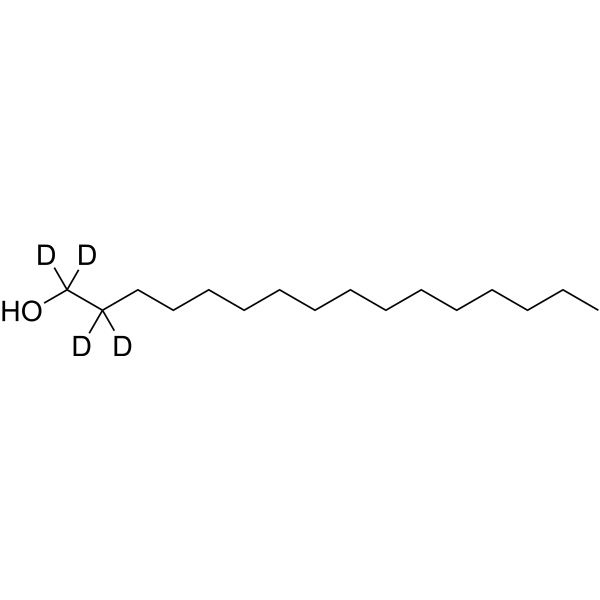
1-Hexadecanol-d4
1-Hexadecanol-d4 is the deuterated form of 1-hexadecanol. 1-Hexadecanol is a fatty alcohol, which is a hydrophobic substrate.
-
GC68496
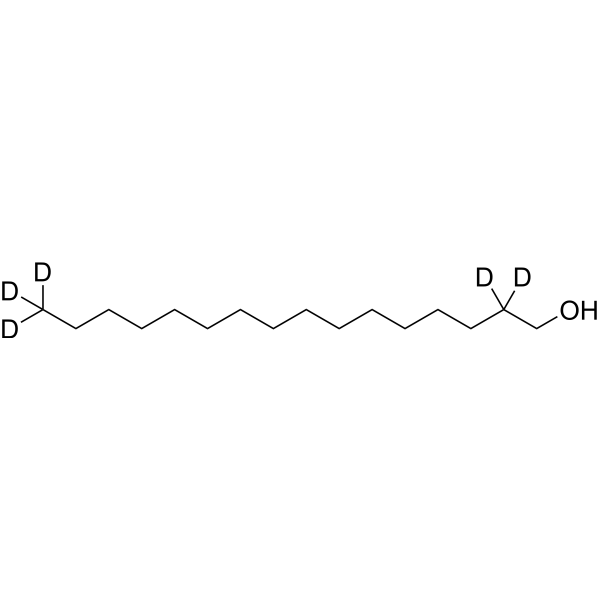
1-Hexadecanol-d5
1-Hexadecanol-d5 is the deuterated form of 1-hexadecanol. 1-Hexadecanol is a fatty alcohol, which is a hydrophobic substrate.
-
GC61723
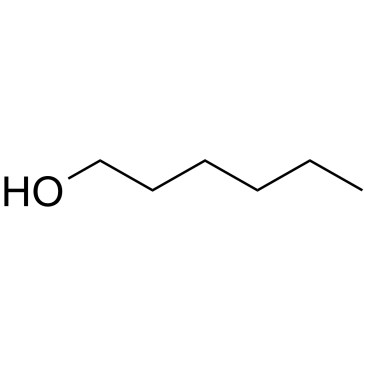
1-Hexanol
1-Hexanol, a primary alcohol, is a surfactant that can be employed in industrial processes to enhance interfacial properties.
-
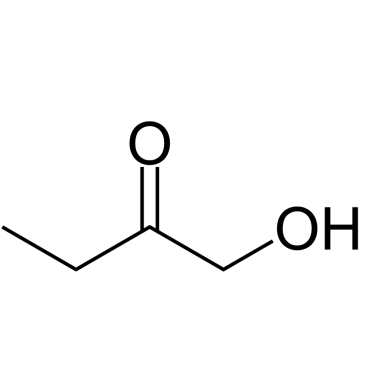
GC61643
1-Hydroxy-2-butanone
1-Hydroxy-2-butanone is a natural compound isolated from Bomboo Juice with antitubercular activity.