Cancer Biology
- Cancer Biology Peptides(84)
- Ras-like GTPases(11)
- Leukemia(1)
- Cell Proliferation, Viability & Cytotoxicity(0)
- Others(7)
- Tumor Microenvironment(30)
- Cell Migration & Metastasis(19)
Products for Cancer Biology
- Cat.No. Product Name Information
-
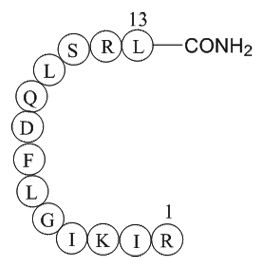
GC65288
Resibufagin
Resibufagin is a kind of bufadienolide isolated from the venom of Bufo bufo gargarizans, has anti-tumor activities.
-
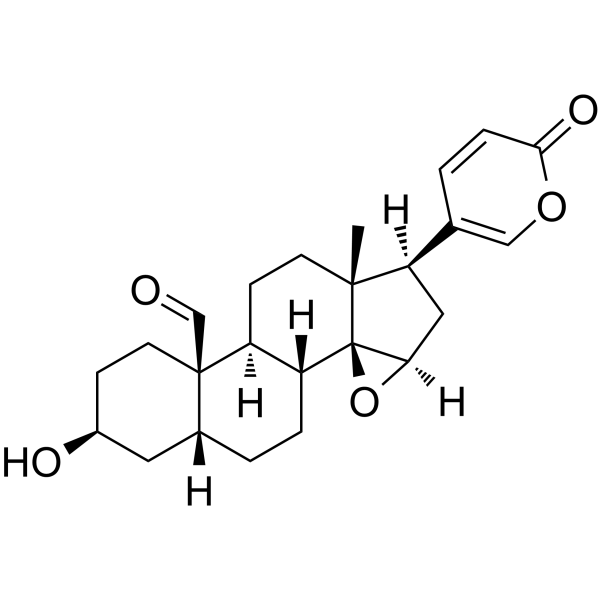
GC18955
Reveromycin C
Reveromycin C is a polyketide originally isolated from Streptomyces that has antifungal activity against C.
-
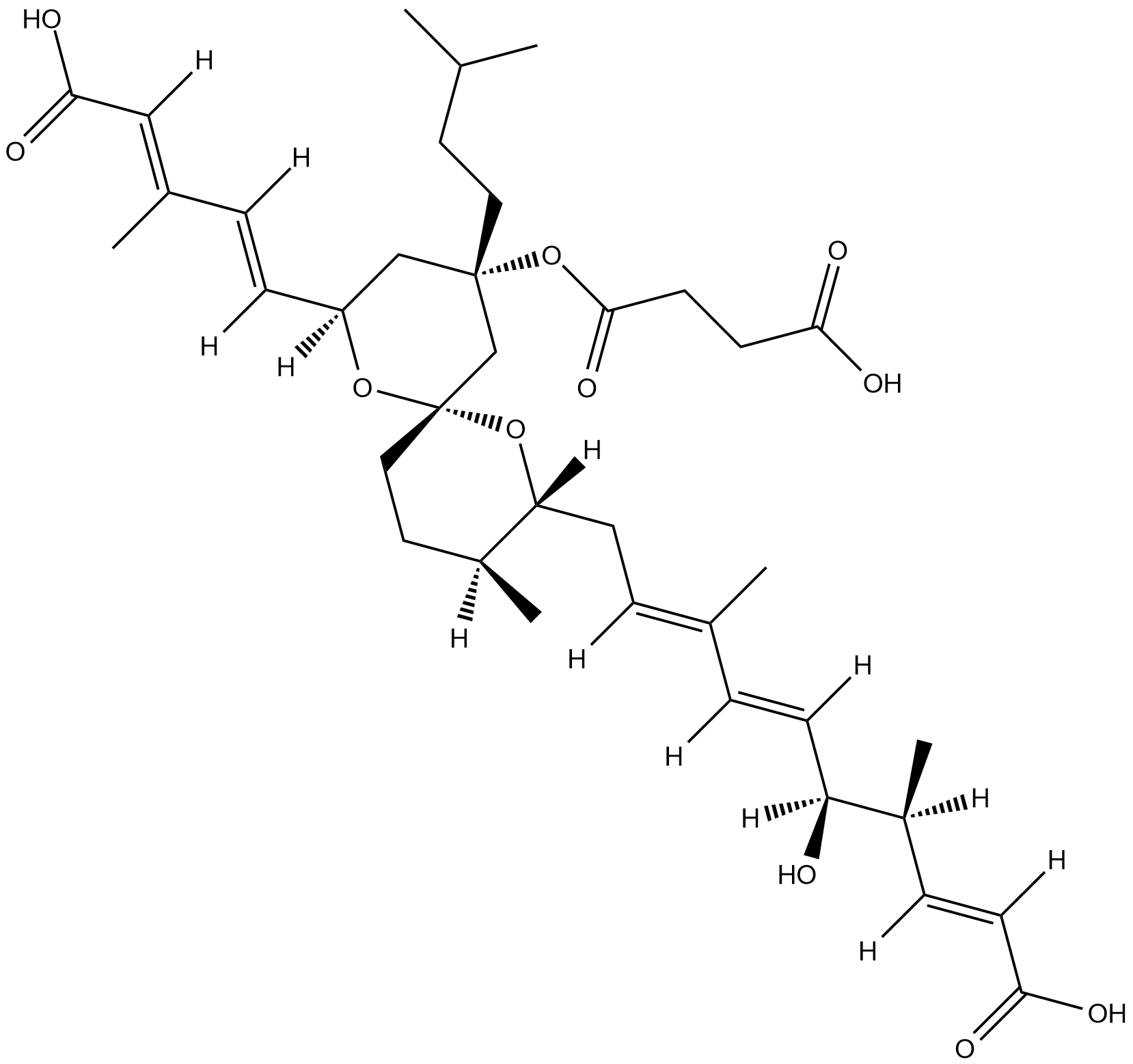
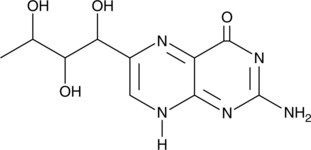
GC52067
Rhamnopterin
A derivative of biopterin
-
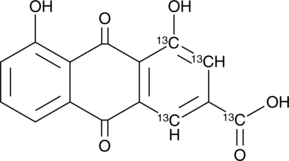
GC45798
Rhein-13C4
An internal standard for the quantification of rhein
-
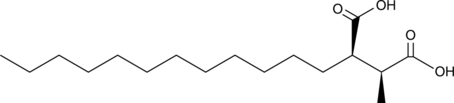
GC46214
Roccellic Acid
A lichen secondary metabolite
-
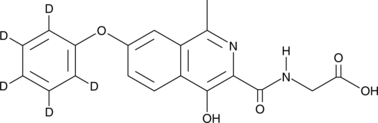
GC49190
Roxadustat-d5
An internal standard for the quantification of roxadustat
-
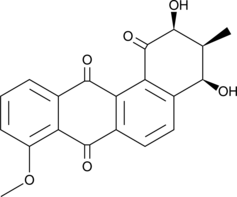
GC48693
Rubiginone D2
A polyketide with antibacterial and anticancer activities
-
GC46215
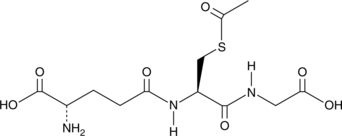
S-Acetyl-L-glutathione
A derivative of glutathione
-
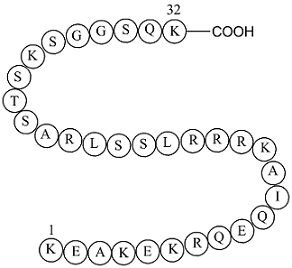
GP10104
S6 Kinase Substrate Peptide 32
Measures the activity of kinases that phosphorylate ribosomal protein S6.
-
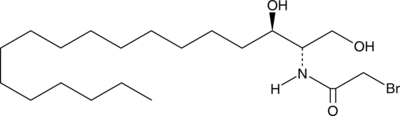
GC49668
SABRAC
An inhibitor of acid ceramidase
-
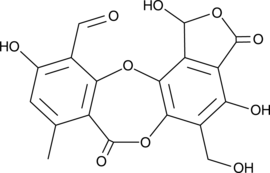
GC46216
Salazinic Acid
A depsidone lichen metabolite
-
GC40653
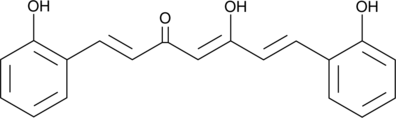
Salicylcurcumin
Curcuminoids are natural and synthetic analogs of curcumin, a natural polyphenol with modulating effects in inflammation, cancer, and immunity.
-
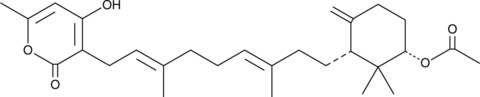
GC44873
Sartorypyrone A
Sartorypyrone A is a meroditerpene metabolite produced by Neosartorya fungal species.
-
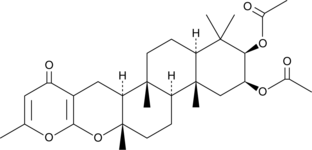
GC46217
Sartorypyrone B
A fungal metabolite
-
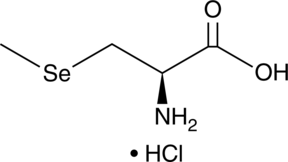
GC48914
Se-Methylselenocysteine (hydrochloride)
Se-methylselenocysteine (hydrochloride) (MSC), a derivative of selenocysteine methylation, is a natural monomethylated selenoamino acid.
-
GC49723
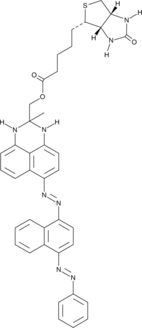
SenTraGor™ Cell Senescence Reagent
A cellular senescence detection reagent
-
GC49592
Sermorelin (acetate)
A growth hormone-releasing hormone analog

-
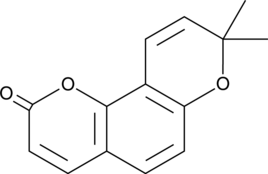
GC46221
Seselin
An angular pyranocoumarin with diverse biological activities
-
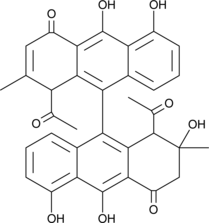
GC44884
Setomimycin
Setomimycin is a pre-anthraquinone originally isolated from S.
-
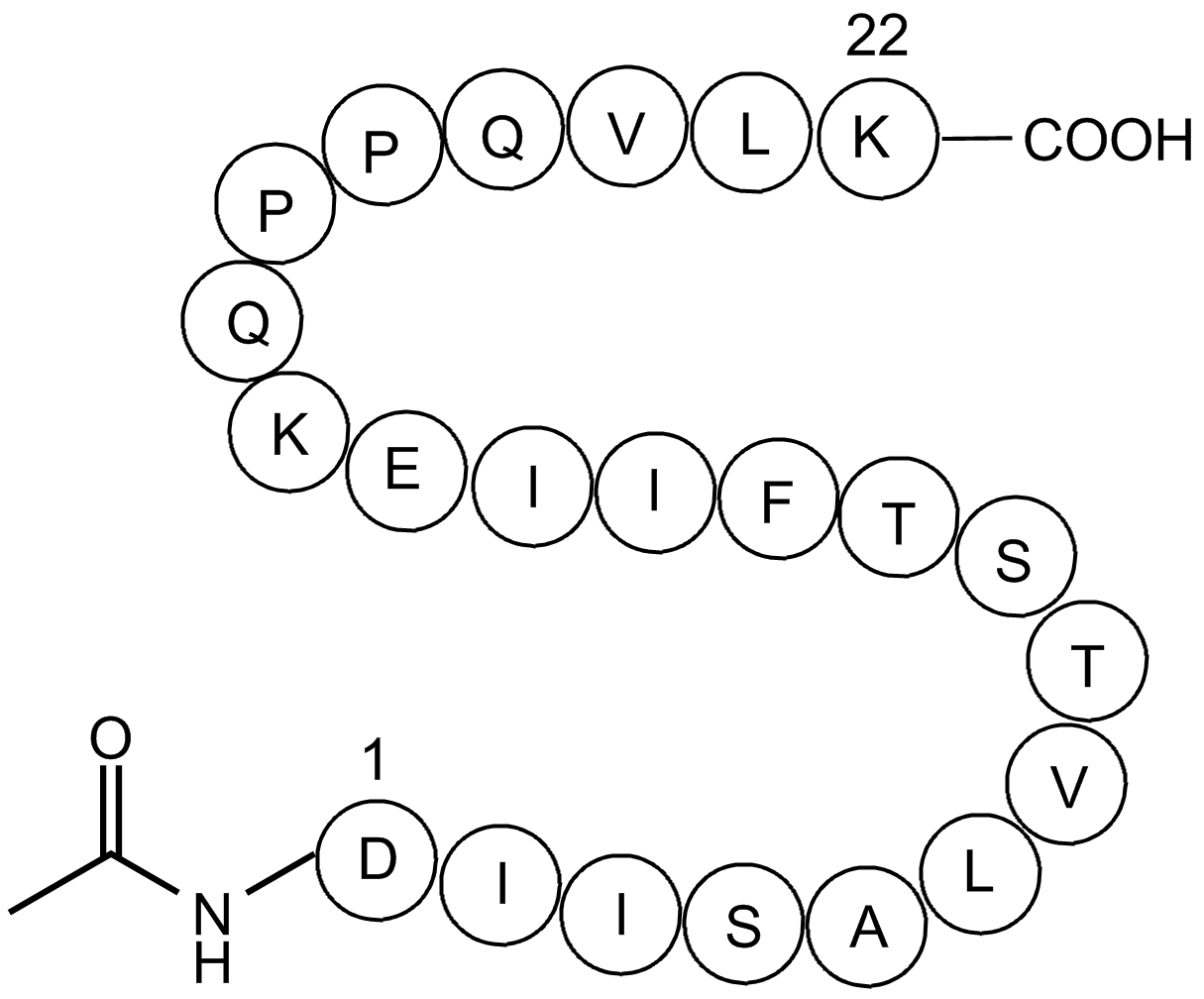
GP10093
signal transducer and activator of transcription 5 (322-343) acetyl/amide
signal transducer and activator of transcription 5 (322-343) acetylamide, (C115H193N27O33), a peptide with the sequence Ac-DIISALVTSTFIIEKQPPQVLK-amide, MW= 2481.92.
-
GP10145
signal transducer and activator of transcription 6 fragment
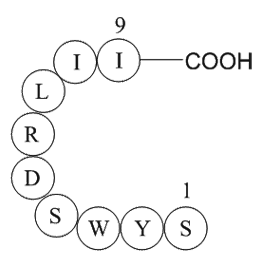
-
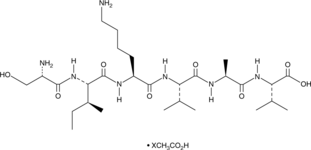
GC49713
SIKVAV (acetate)
A laminin α1-derived peptide
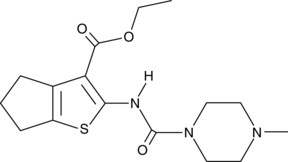
-
GC49049
SMU127
An agonist of TLR1/2
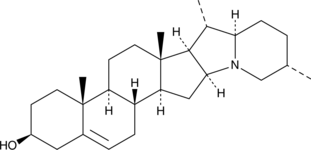
-
GC52075
Solanidine
Solanidine is a cholestane alkaloid isolated from several potato species including Solanum demissum, Solanum acaule, and Solanum tuberosum. Solanidine can inhibit proliferation and exhibit obvious antitumor effect.
-
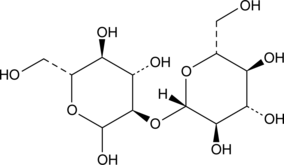
GC41090
Sophorose
Sophorose is a disaccharide component of the microbial glycolipids produced by yeast termed sophorolipids.
-
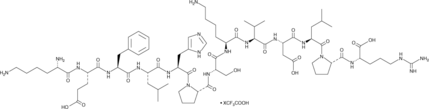
GC52397
SOR-C13 (trifluoroacetate salt)
A peptide TRPV6 antagonist
-
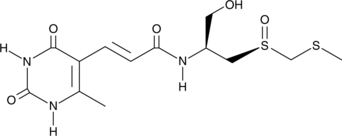
GC46223
Sparsomycin
A bacterial metabolite with diverse biological activities
-
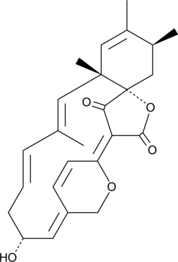
GC44941
Spirohexenolide A
Spirohexenolide A is a metabolite originally isolated from S.
-
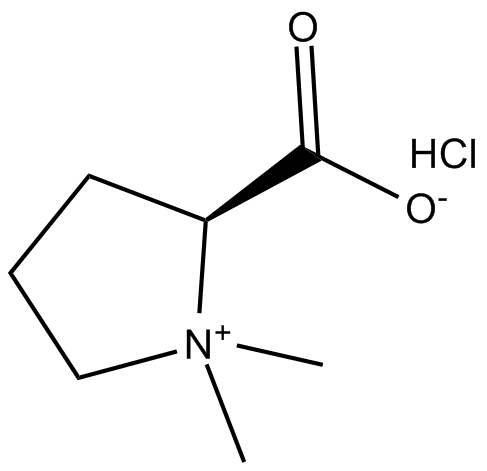
GC13840
Stachydrine (hydrochloride)
anti-metastatic agent
-
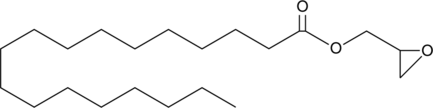
GC52011
Stearic Acid glycidyl ester
An esterified form of stearic acid
-
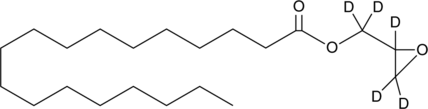
GC49770
Stearic Acid glycidyl ester-d5
An internal standard for the quantification of stearic acid glycidyl ester
-
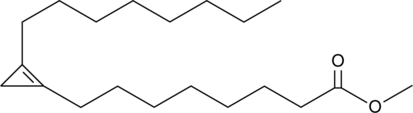
GC41518
Sterculic Acid methyl ester
Sterculic acid methyl ester is an ester form of sterculic acid, which is an inhibitor of δ9 desaturase.
-
GC44955
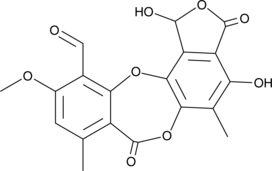
Stictic Acid
Stictic acid is a depsidone found in lichen.
-
GC46228
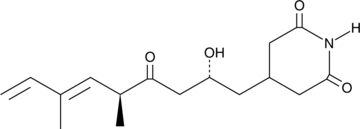
Streptimidone
A bacterial metabolite with diverse biological activities
-
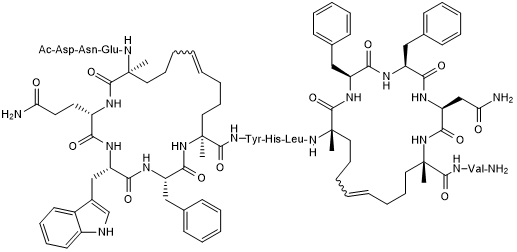
GC50507
StRIP16
Rab8a GTPase-binding stapled peptide
-
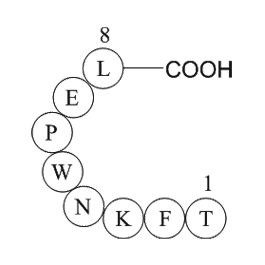
GP10034
survivin (baculoviral IAP repeat-containing protein 5) (21-28)
Cancer Biology Peptides
-
GC44975
SW203668 (trifluoroacetate salt)
SW203668 is an irreversible inhibitor of stearoyl-CoA desaturase (IC50 = 54 nM).

-
GC44988
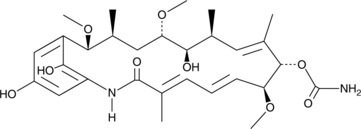
TAN 420C
TAN 420C is a hydroquinone ansamycin antibiotic originally isolated from Streptomyces.
-
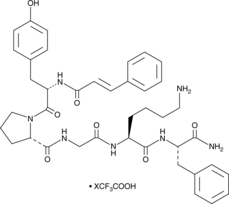
GC52460
tcY-NH2 (trifluoroacetate salt)
A peptide antagonist of PAR4
-
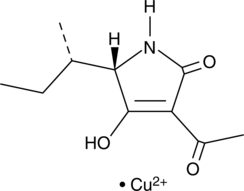
GC45013
Tenuazonic Acid (copper salt)
Tenuazonic acid is one of the major Alternaria mycotoxins commonly found as a natural contaminant in food (LD50 = 548 μg/egg in the chicken embryo toxicity assay).
-
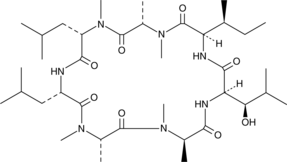
GC45016
Ternatin
Ternatin is a cyclic heptapeptide first isolated from the mushroom C.
-
GC41342
Terrecyclic Acid
Terrecyclic acid is a sesquiterpene originally isolated from A.
-
GC40341
Tetrahydro-11-deoxy Cortisol
Tetrahydro-11-deoxy cortisol (THS) is the primary urinary metabolite of 11-deoxycortisol.
-
GC14778
tetramethyl Nordihydroguaiaretic Acid
tetramethyl Nordihydroguaiaretic Acid is a synthetic derivative of Nordihydroguaiaretic acid and a non-selective lipoxygenase inhibitor.
-
GC45578
Thymohydroquinone
-
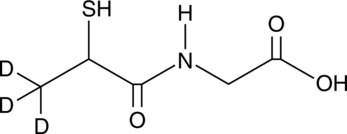
GC49062
Tiopronin-d3
An internal standard for the quantification of tiopronin
-
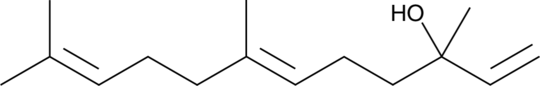
GC48193
trans-Nerolidol
A sesquiterpene with diverse biological activities
-
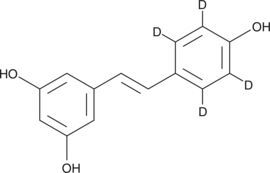
GC48194
trans-Resveratrol-d4
An internal standard for the quantification of trans-resveratrol
-
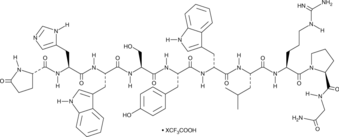
GC49475
Triptorelin (trifluoroacetate salt)
A synthetic GNRH peptide agonist
-
GC32772
Triptorelin ([DTrp6]-LH-RH)
A synthetic GNRH peptide agonist
![Triptorelin ([DTrp6]-LH-RH) Chemical Structure Triptorelin ([DTrp6]-LH-RH) Chemical Structure](/media/struct/GC3/GC32772.png)
-
GC52412
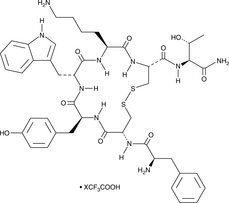
TT-232 (trifluoroacetate salt)
A synthetic peptide derivative of somatostatin
-
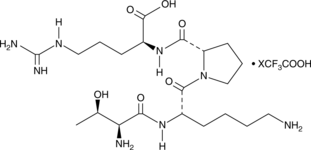
GC49099
Tuftsin (human) (trifluoroacetate salt)
A phagocytosis-stimulating peptide
-
GP10028
tumor protein p53 binding protein fragment [Homo sapiens]/[Mus musculus]
P53 binding protein fragment
![tumor protein p53 binding protein fragment [Homo sapiens]/[Mus musculus] Chemical Structure tumor protein p53 binding protein fragment [Homo sapiens]/[Mus musculus] Chemical Structure](/media/struct/GP1/GP10028.png)
-
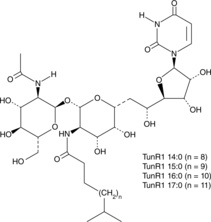
GC48213
TunR1
An antibiotic and derivative of tunicamycin
-
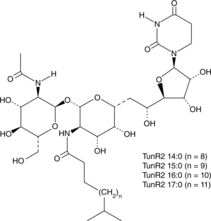
GC48214
TunR2
An antibiotic and derivative of tunicamycin
-
GP10142
ubiquitin specific protease 3 fragment
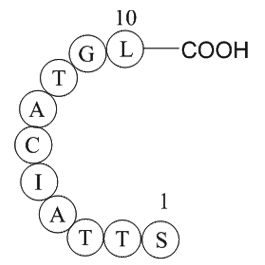
-
GC45110
UCM53
An inhibitor of bacterial cell division
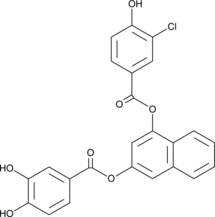
-
GC48226
Urdamycin A
A bacterial metabolite
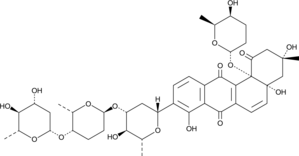
-
GC48227
Urdamycin B
A bacterial metabolite with antibacterial and anticancer activities
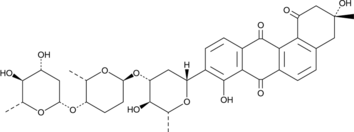
-
GC52242
V-125
An RXR agonist
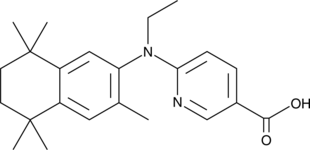
-
GC45139
Vapreotide (trifluoroacetate salt)
Vapreotide is a peptide neurokinin-1 receptor (NK1) antagonist and analog of somatostatin (IC50 = 330 nM in a radioligand binding assay).
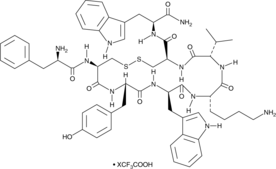
-
GC52364
Vimentin (139-159)-biotin Peptide
A biotinylated vimentin peptide

-
GC52371
Vimentin (G146R) (139-159)-biotin Peptide
A biotinylated mutant vimentin peptide

-
GC46235
Vitamin K1-d7
An internal standard for the quantification of vitamin K1
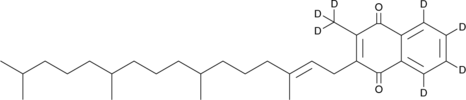
-
GC17093
VLX600
shows selective cytotoxicity against quiescent cancer cells
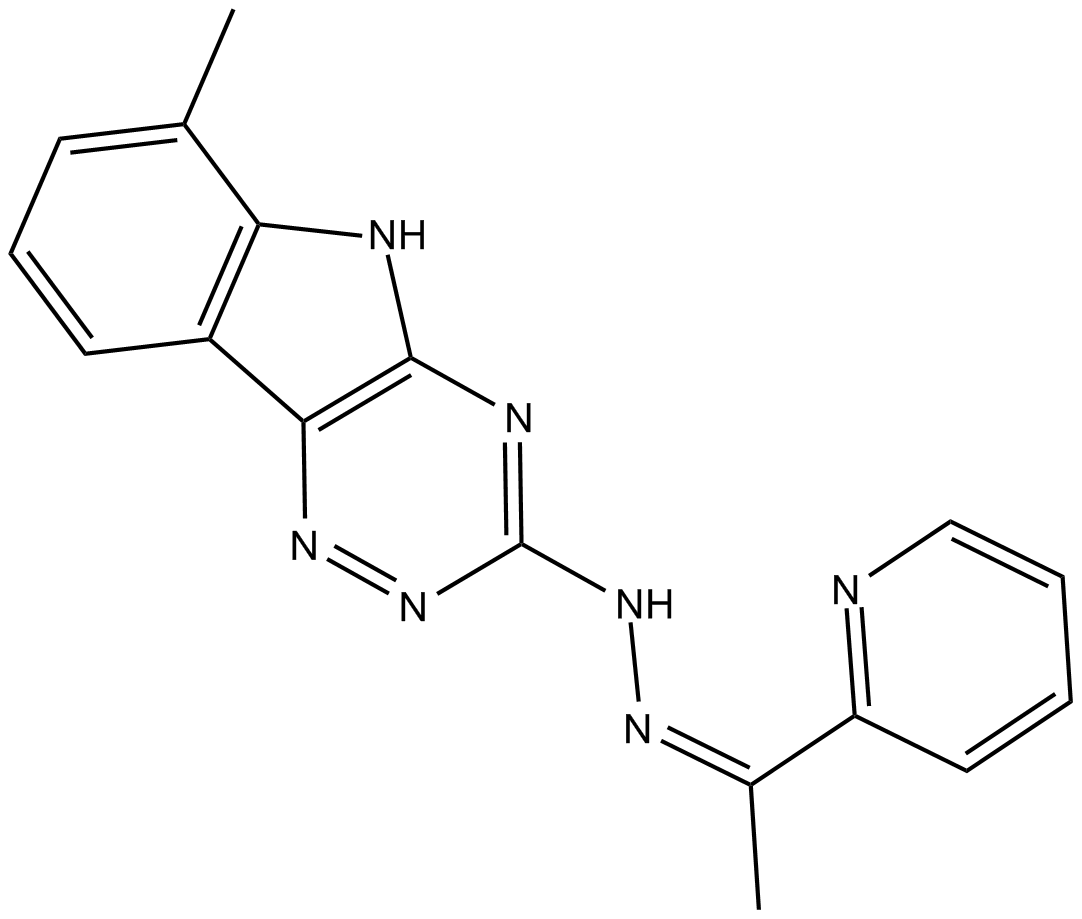
-
GC52084
YSK05
YSK05 is a pH-sensitive cationic lipid. YSK05 improves the intracellular trafficking of non-viral vectors. YSK05-MEND shows significantly good gene silencing activity and hemolytic activity. YSK05 overcomes the suppression of endosomal escape by PEGylation. YSK05 effectively enhances siRNA delivery both in vitro and in vivo.
-
GP10017
[Ser25] Protein Kinase C (19-31)
PKC substrate
![[Ser25] Protein Kinase C (19-31) Chemical Structure [Ser25] Protein Kinase C (19-31) Chemical Structure](/media/struct/GP1/GP10017.png)
-
GP10111
β-Pompilidotoxin