Cancer Biology
- Cancer Biology Peptides(84)
- Ras-like GTPases(11)
- Leukemia(1)
- Cell Proliferation, Viability & Cytotoxicity(0)
- Others(7)
- Tumor Microenvironment(30)
- Cell Migration & Metastasis(19)
Products for Cancer Biology
- Cat.No. Product Name Information
-
GP10141
Cytochrome P450 CYP1B1 (190-198) [Homo sapiens]
![Cytochrome P450 CYP1B1 (190-198) [Homo sapiens] Chemical Structure Cytochrome P450 CYP1B1 (190-198) [Homo sapiens] Chemical Structure](/media/struct/GP1/GP10141.png)
-
GC43361
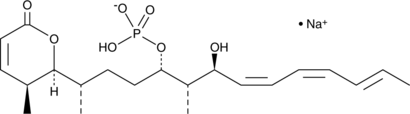
Cytostatin (sodium salt)
Cytostatin is a natural antitumor inhibitor of cell adhesion to extracellular matrix, blocking adhesion of B16 melanoma cells to laminin and collagen type IV in vitro (IC50s = 1.3 and 1.4 μg/ml, respectively) and B16 cells metastatic activity in mice.
-
GC43368
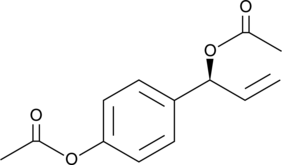
D,L-1′-Acetoxychavicol Acetate
D,L-1′-Acetoxychavicol acetate is a natural compound first isolated from the rhizomes of ginger-like plants.
-
GC43504
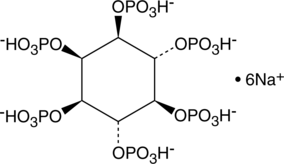
D-myo-Inositol-1,2,3,4,5,6-hexaphosphate (sodium salt)
Phytic acid (Inositol hexaphosphate) hexasodium is a phosphorus storage compound of seeds and cereal grains.
-
GC47168
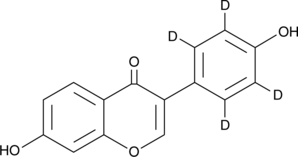
Daidzein-d4
An internal standard for the quantification of daidzein
-
GC60121
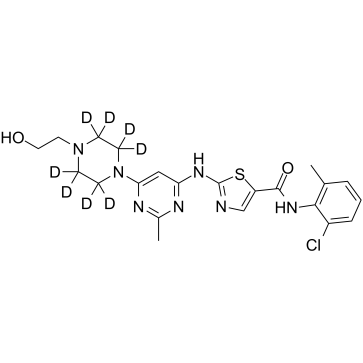
Dasatinib D8
An internal standard for the quantification of GC- or LC-MS
-
GC48581
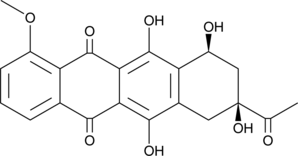
Daunorubicinone
An aglycone form of daunorubicin
-
GC40697
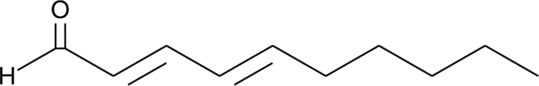
DDA
DDA is a lipid peroxidation product of linolieic acid.
-
GC43390
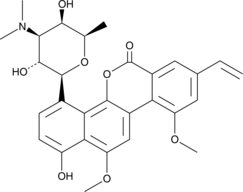
Deacetylravidomycin
Deacetylravidomycin is a microbial metabolite that has been found in Streptomyces and has light-dependent antibiotic and anticancer activities.
-
GC43391
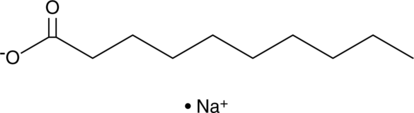
Decanoic Acid (sodium salt)
Decanoic acid is a saturated medium-chain fatty acid that contains 10 carbons.
-
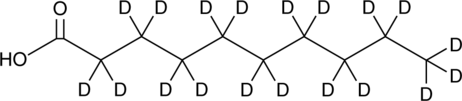
GC45815
Decanoic Acid-d19
An internal standard for the quantification of decanoic acid
-
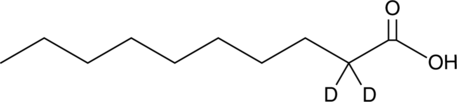
GC46127
Decanoic Acid-d2
An internal standard for the quantification of decanoic acid
-
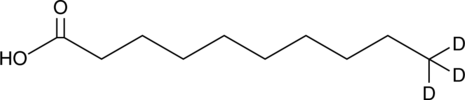
GC45722
Decanoic Acid-d3
An internal standard for the quantification of decanoic acid
-
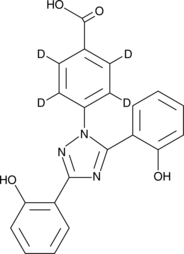
GC40230
Deferasirox-d4
Deferasirox-d4 is intended for use as an internal standard for the quantification of deferasirox by GC- or LC-MS.
-
GC52316
Deferitrin
An iron chelator and derivative of desferrithiocin

-
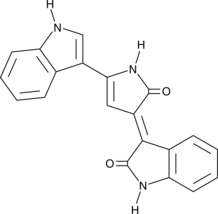
GC40654
Deoxyviolacein
Deoxyviolacein is a bacterial metabolite and byproduct in the biosynthesis of the bisindole alkaloid violacein that has anticancer, antibacterial, and antifungal properties.
-
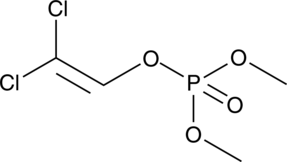
GC47211
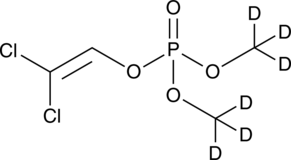
Dichlorvos
An organophosphate insecticide
-
GC49293
Dichlorvos-d6
An internal standard for the quantification of dichlorvos
-
GC47215
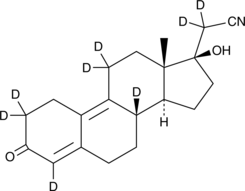
Dienogest-d8
An internal standard for the quantification of dienogest
-
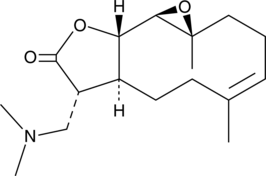
GC52194
Dimethylamino Parthenolide
-
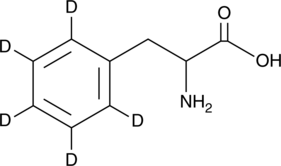
GC49383
DL-Phenyl-d5-alanine
An internal standard for the quantification of DL-phenylalanine
-
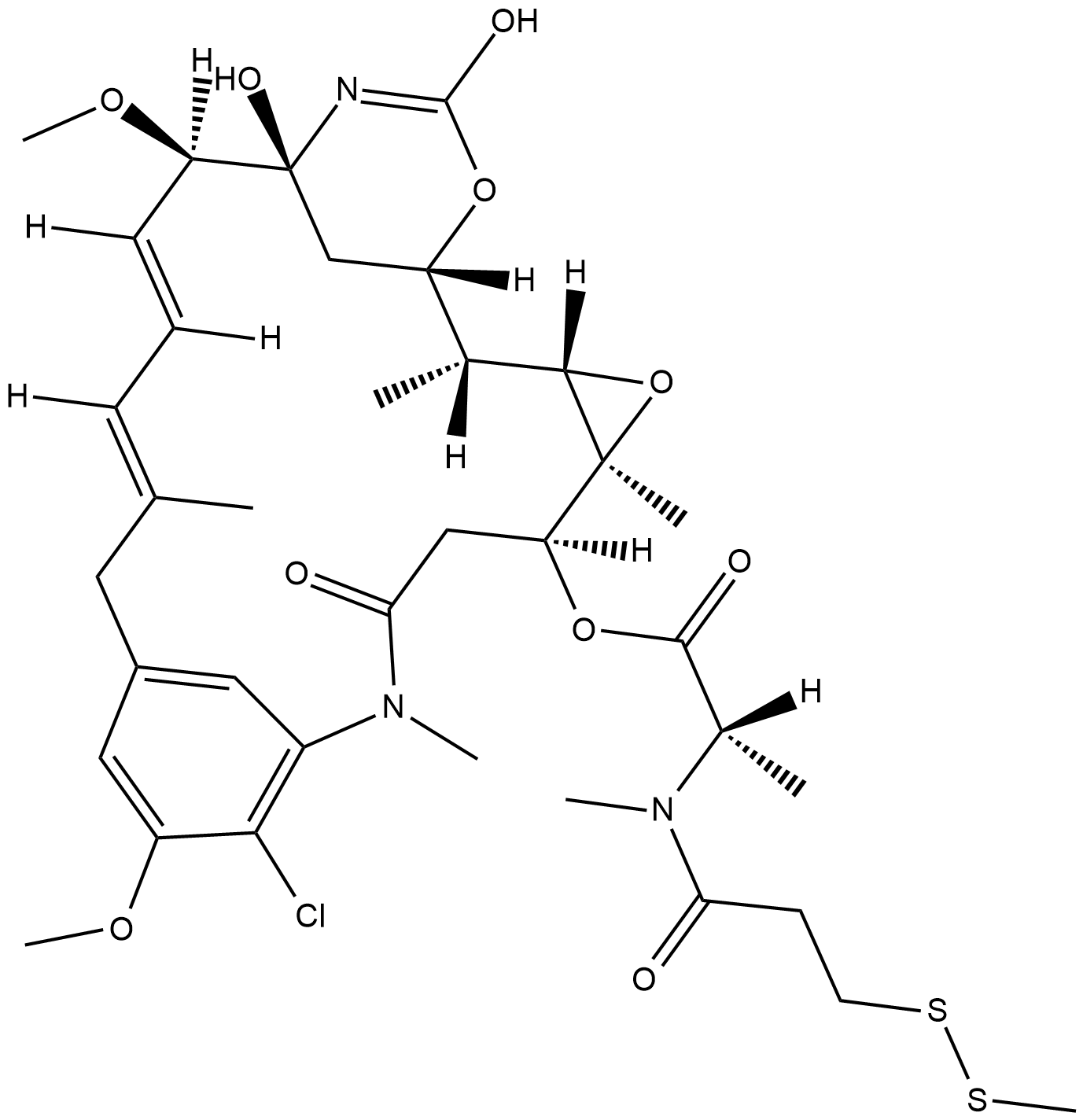
GC18254
DM1-SMe
DM1-SMe is an analog of mertansine with a thiomethane cap attached to the sulfhydryl group.
-
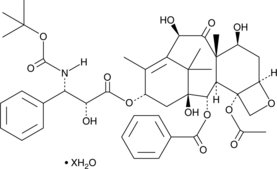
GC48968
Docetaxel (hydrate)
An analog of taxol with antitumor properties
-
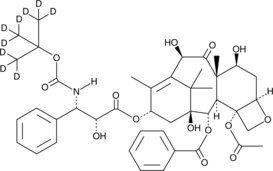
GC47250
Docetaxel-d9
An internal standard for the quantification of docetaxel
-
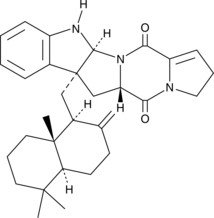
GC40082
Drimentine B
Drimentine B is a terpenylated diketopiperazine antibiotic originally isolated from Actinomycete bacteria.
-
GC43573
Drimentine C
Drimentine C is a terpenylated diketopiperazine antibiotic originally isolated from Actinomycete bacteria.
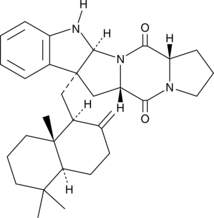
-
GC49001
DSPE-PEG(2000)-amine (sodium salt)
A PEGylated derivative of DSPE
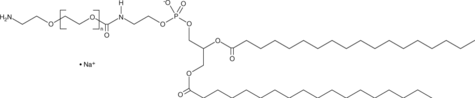
-
GC52333
DSPE-PEG(2000)-NHS
A PEGylated derivative of DSPE
-
GC49058
DTPA
DTPA is a compound used to construct magnetic adsorbent, which can simultaneously remove heavy metal and dye from complex wastewater.
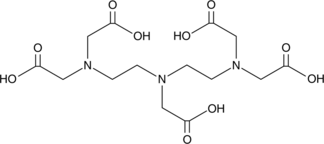
-
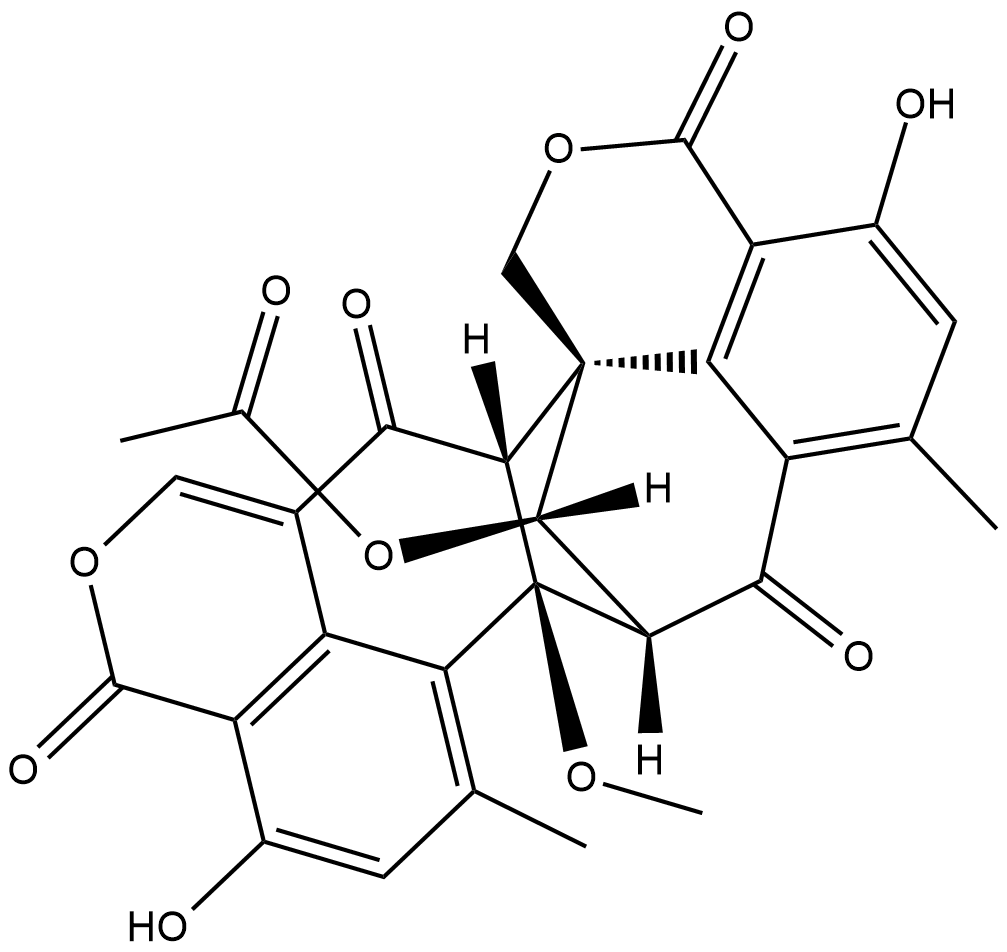
GC18992
Duclauxin
Duclauxin decreases proliferation of tumor cells in vitro and increases the lifespan of mice innoculated with Ehrlich ascitic tumor cells.
-
GP10022
Dynamin inhibitory peptide
-
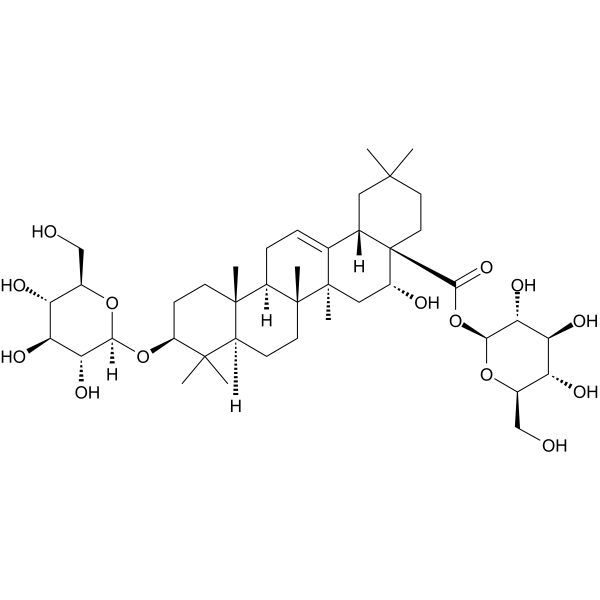
GC65123
Eclalbasaponin I
Eclalbasaponin I is isolated from Eclipta prostrata L with antitumor activity. Eclalbasaponin I inhibits the proliferation of hepatoma cell smmc-7721 with an IC50 value of 111.1703 μg/ml.
-
GP10030
EGF receptor substrate eps15 acetyl - [Mus musculus]/[Homo sapiens]
EGF receptor substrate eps15 (Eps15) has been identified as a 142-kDa substrate of the EGF receptor1.
![EGF receptor substrate eps15 acetyl - [Mus musculus]/[Homo sapiens] Chemical Structure EGF receptor substrate eps15 acetyl - [Mus musculus]/[Homo sapiens] Chemical Structure](/media/struct/GP1/GP10030.png)
-
GP10108
EGF-R (661-681) T669 Peptide
-
GC43591
Eicosapentaenoyl 1-propanol-2-amide
Monoacylglycerols (MAGs) of ω-3 polyunsaturated fatty acids have diverse physiological and health effects.
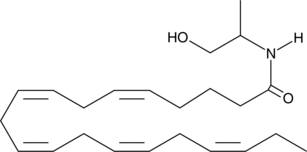
-
GC49522
EM-12
EM-12 (EM-12), a teratogenic Thalidomide analogue, is more active than Thalidomide and is much more stable for hydrolysis. EM-12 enhances 1,2-dimethylhydrazine-induction of rat colon adenocarcinomas.
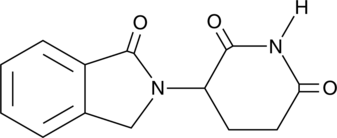
-
GC46137
Emestrin
A mycotoxin
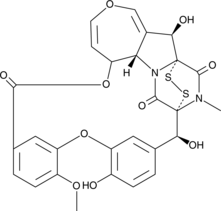
-
GC49124
EN219
EN219 is a moderately selective synthetic covalent ligand against an N-terminal cysteine (C8) of RNF114 with an IC50 of 470 nM. EN219 inhibits RNF114-mediated autoubiquitination and p21 ubiquitination.
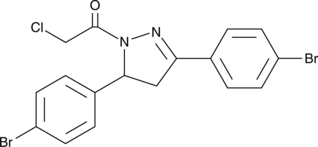
-
GP10117
Endostatin (84-114)-NH2 (JKC367)
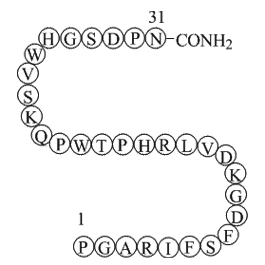
-
GC49648
Enobosarm-d4
An internal standard for the quantification of enobosarm
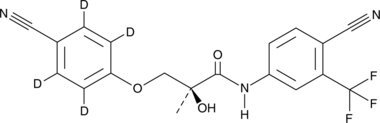
-
GC47297
EP4 Receptor Antagonist 1
An EP4 receptor antagonist
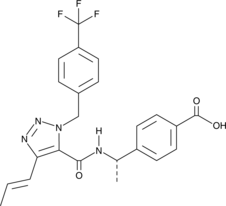
-
GP10005
Epidermal growth factor receptor (994-1002) acetyl/amide
EGF-family receptor
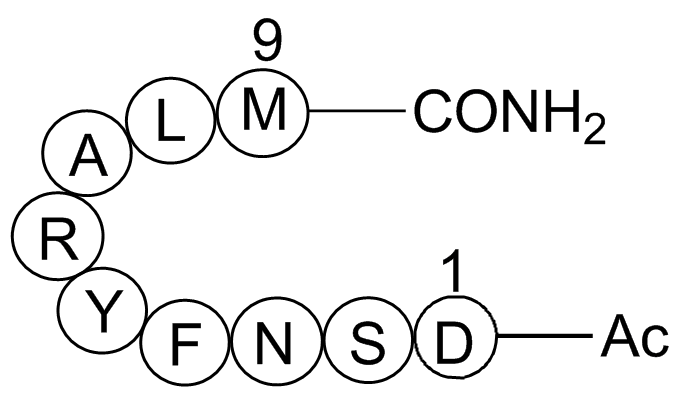
-
GP10095
Epidermal Growth Factor Receptor Peptide (985-996)
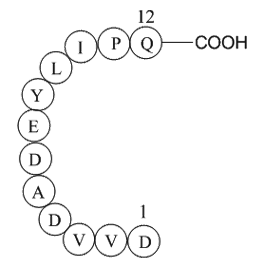
-
GP10132
erbB-2
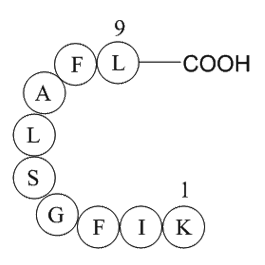
-
GC43631
Estrone 3-sulfate (sodium salt)
Estrone sulfate, a biologically inactive form of estrogen, is a major circulating plasma estrogen that is converted into the biologically active estrogen, estrone (E1) by steroid sulfatase (STS). strone sulfate can be used for the research of breast cancer.
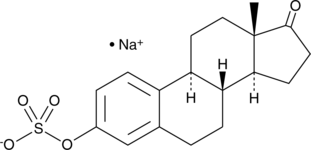
-
GC52303
Ethyl Mycophenolate
A potential impurity found in commercial preparations of mycophenolate mofetil
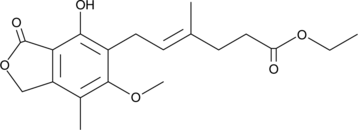
-
GC15555
Farnesyl Thiosalicylic Acid Amide
inhibits tumor growth
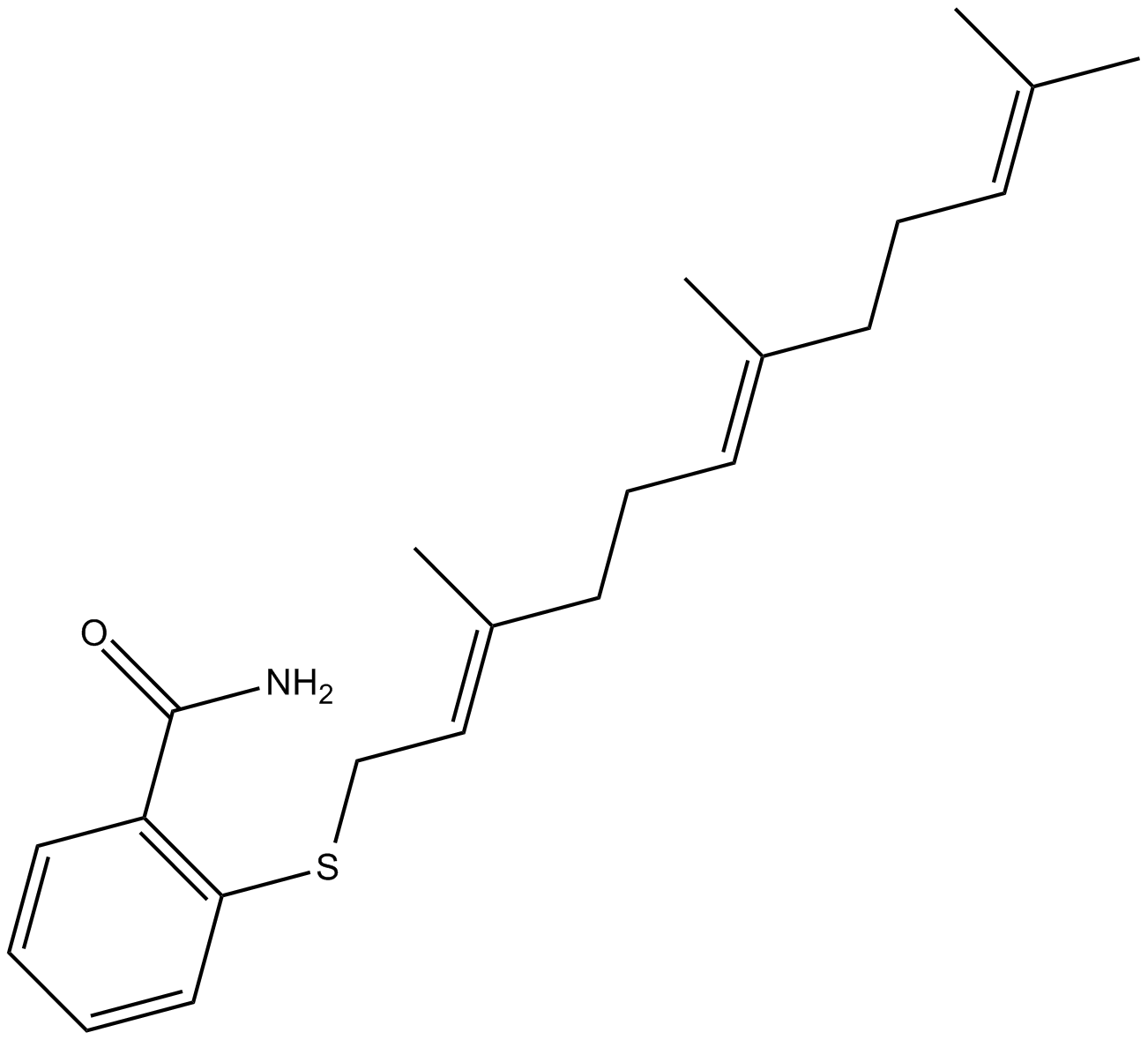
-
GP10123
Fas C- Terminal Tripeptide

-
GC46146
FD-211
A δ lactone with anticancer activity
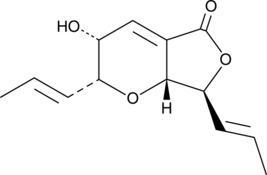
-
GC49563
Fenhexamid
Fenhexamid, a botryticide, is a sterol biosynthesis inhibitor.
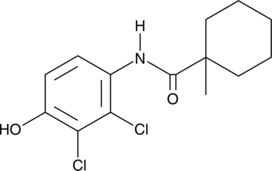
-
GP10146
ferritin heavy chain fragment [Multiple species]
![ferritin heavy chain fragment [Multiple species] Chemical Structure ferritin heavy chain fragment [Multiple species] Chemical Structure](/media/struct/GP1/GP10146.png)
-
GC47361
Fludioxonil
A fungicide
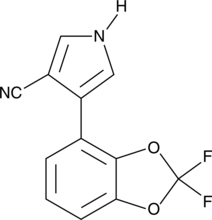
-
GC49074
Forphenicine
A bacterial metabolite and an inhibitor of alkaline phosphatase
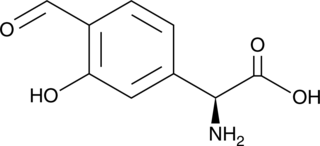
-
GC52014
Forphenicinol
An immunomodulator and a derivative of forphenicine
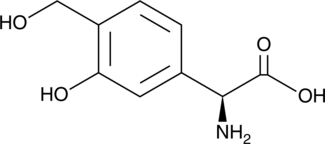
-
GC49089
FR900359
A cyclic depsipeptide and an inhibitor of Gαq, Gα11, and Gα14
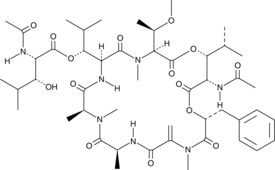
-
GC16236
Fucoidan
anticancer, antiviral, neuroprotective, immune-modulating
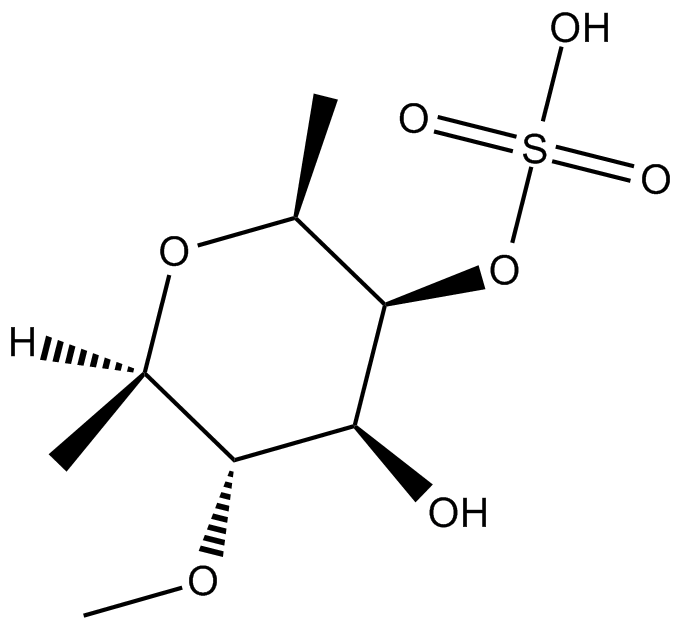
-
GC52483
Fucosylated Ganglioside GM1 (porcine) (ammonium salt)
A sphingolipid
-
GC47377
Fumitremorgin B
A mycotoxin
-
GC52288
Fumonisin B1-13C34
An internal standard for the quantification of fumonisin B1
-
GC47378
Fumonisin B2-13C34
An internal standard for the quantification of fumonisin B2
-
GC52047
Fustin
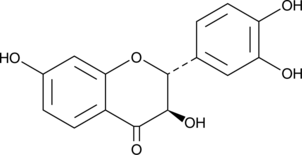
-
GP10092
G protein coupled receptor [Homo sapiens]
G protein coupled receptor [Homo sapiens], (C110H168N38O45), a peptide with the sequence H2N-QESHNSGNRSDGPGKNTTLHNEFDT-OH, MW= 2742.74.
![G protein coupled receptor [Homo sapiens] Chemical Structure G protein coupled receptor [Homo sapiens] Chemical Structure](/media/struct/GP1/GP10092.png)
-
GC49420
Gallic Acid-d2
An internal standard for the quantification of gallic acid
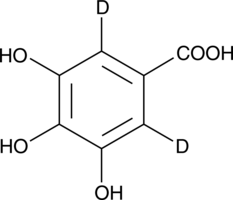
-
GC43727
Ganglioside GD1a mixture (sodium salt)
Ganglioside GD1a is a sialic acid-containing glycosphingolipid found in brain, erythrocytes, bone marrow, testis, spleen, and liver.
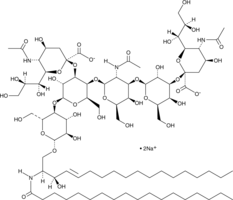
-
GC43728
Ganglioside GD1b Mixture (sodium salt)
Ganglioside GD1b is an acidic glycosphingolipid that contains two sialic acid residues linked to an inner galactose unit.
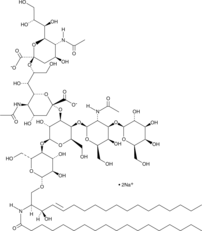
-
GC49490
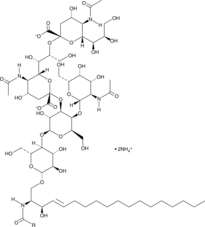
Ganglioside GD2 Mixture (bovine buttermilk) (ammonium salt)
A semi-synthetic mixture of ganglioside GD2
-
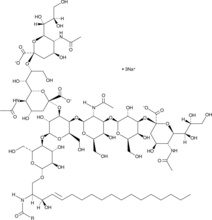
GC52505
Ganglioside GT1b (bovine) (sodium salt)
A sphingolipid
-
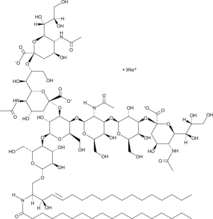
GC43735
Ganglioside GT1b Mixture (sodium salt)
Ganglioside GT1b is a trisialoganglioside that is characterized by having two sialic residues linked to the inner galactose unit.
-
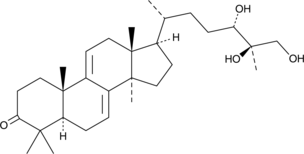
GC49229
Ganodermanontriol
Ganodermanontriol, a sterol isolated from Ganoderma lucidum, induces anti-inflammatory activity in tert-butyl hydroperoxide (t-BHP)-damaged hepatic cells through the expression of HO-1. Ganodermanontriol exhibits hepatoprotective activity.
-
GP10107
Gap 26
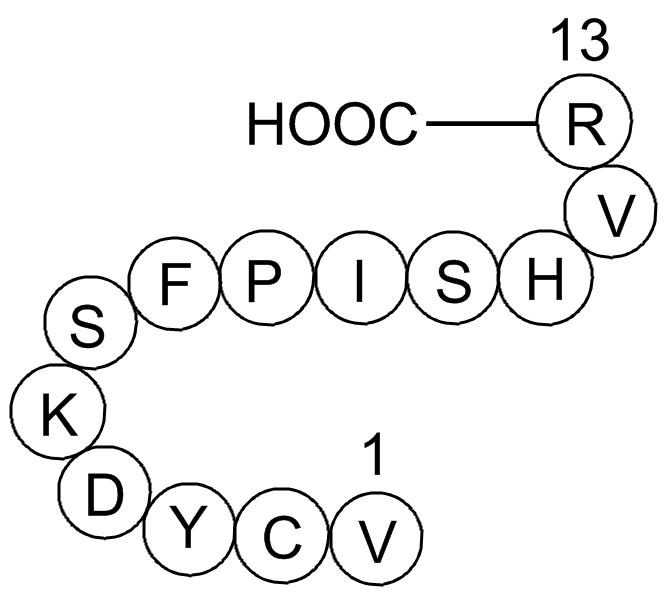
-
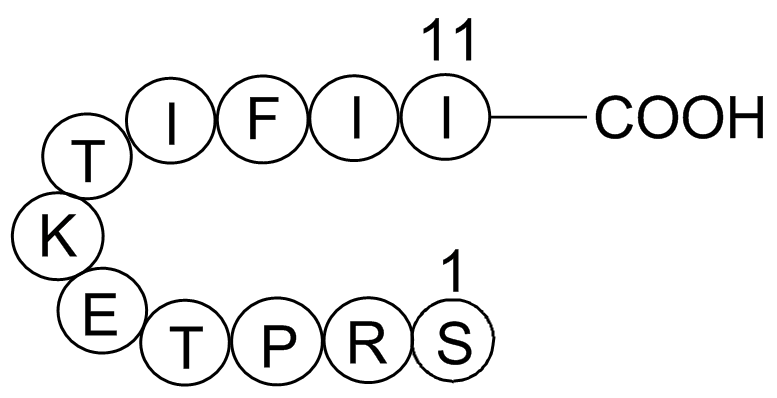
GP10119
Gap 27
A connexin-mimetic peptide
-
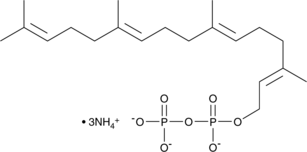
GC43747
Geranylgeranyl Pyrophosphate (ammonium salt)
Geranylgeranyl pyrophosphate is an intermediate in the HMG-CoA reductase pathway derived directly from farnesyl pyrophosphate and used in the biosynthesis of terpenes and terpenoids.
-
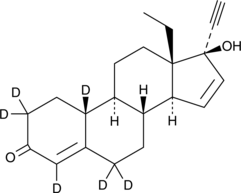
GC48763
Gestodene-d6
An internal standard for the quantification of gestodene
-
GC18840
Gilvocarcin V
Gilvocarcin V is an antitumor antibiotic with a coumarin-based aromatic structure that was orignally isolated from the culture broth of S.
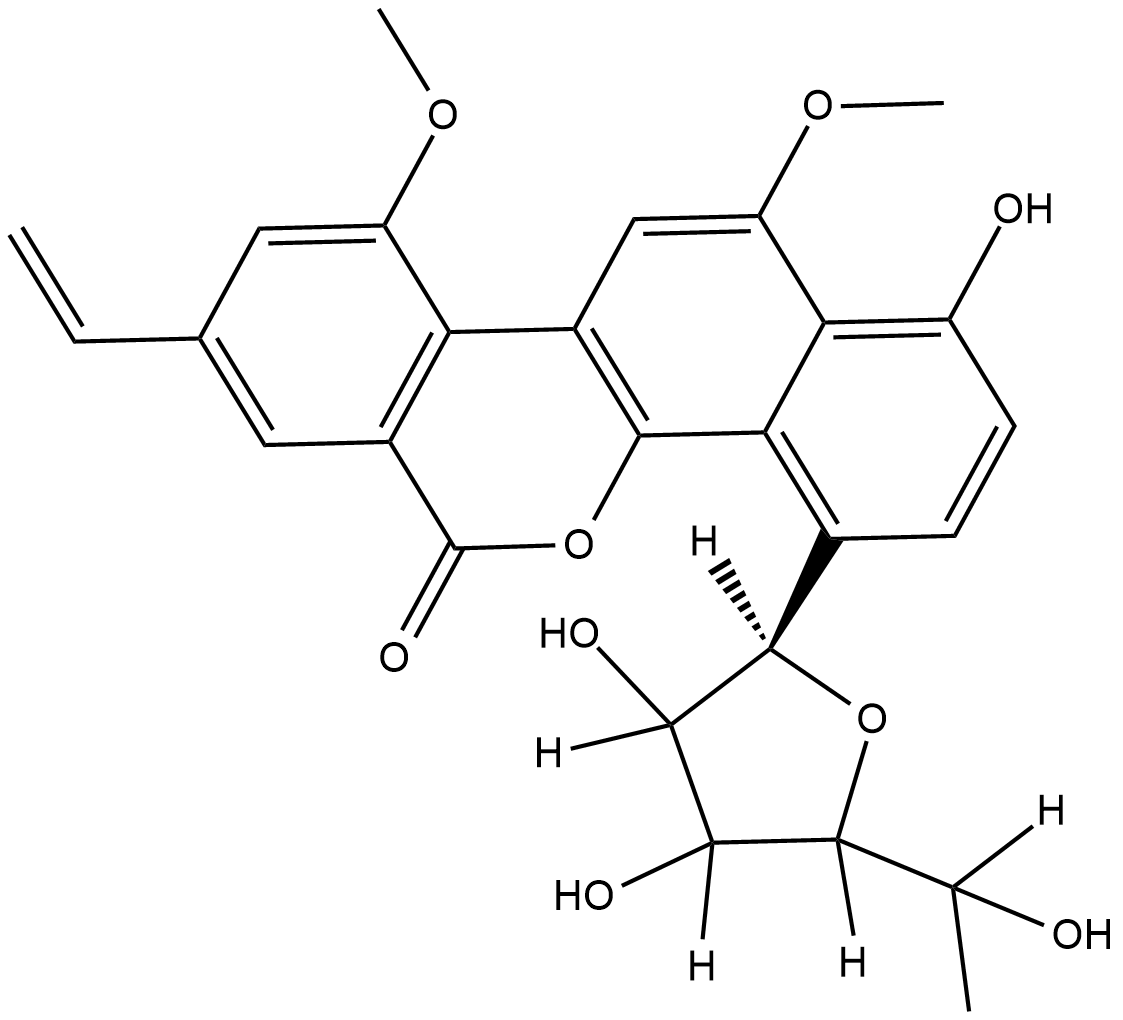
-
GC45811
GKK1032B
GKK1032B is an alkaloid compound that can be found in endophytic fungus Penicillium sp. GKK1032B can induce the apoptosis of human osteosarcoma MG63 cells through caspase pathway activation.
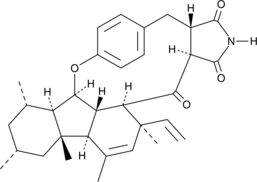
-
GC40049
Glucocorticoid Resistance Inhibitor
Glucocorticoid resistance inhibitor is an inhibitor of glucocorticoid resistance.
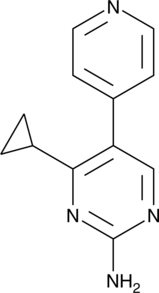
-
GC48720
Griseofulvic Acid
A metabolite of griseofulvin
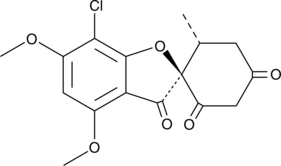
-
GC43784
GRP (human) (trifluoroacetate salt)
Gastrin-releasing peptide (GRP) is a neuropeptide that stimulates gastrin release.

-
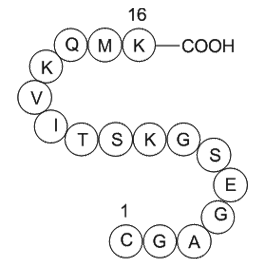
GP10115
GTP-Binding Protein Fragment, G alpha
-
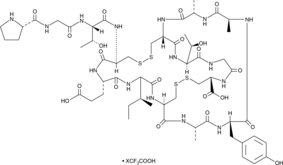
GC49048
Guanylin (human) (trifluoroacetate salt)
A peptide hormone activator of GC-C
-
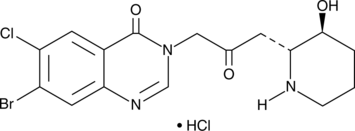
GC43804
Halofuginone (hydrochloride)
Halofuginone is a halogenated derivative of febrifugine, a natural quinazolinone-containing compound found in the Chinese herb D.
-
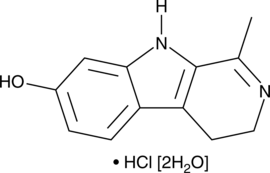
GC49855
Harmalol (hydrochloride hydrate)
A β-carboline alkaloid and an active metabolite of harmaline
-
GC40724
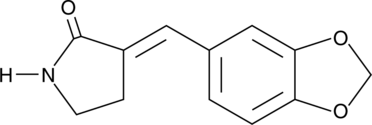
Heat Shock Protein Inhibitor II
Heat shock protein (Hsp) inhibitor II is the active form of Hsp inhibitor I and a benzylidene lactam compound that prevents the synthesis of inducible Hsps, such as Hsp105, Hsp72, and Hsp40.
-
GC48388
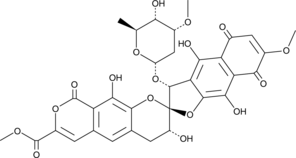
Heliquinomycin
A bacterial metabolite with diverse biological activities
-
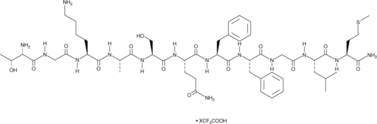
GC49882
Hemokinin 1 (human) (trifluoroacetate salt)
A peptide agonist of NK1 receptors
-
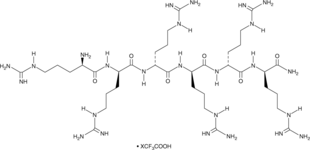
GC49915
Hexa-D-Arginine (trifluoroacetate salt)
A furin inhibitor
-
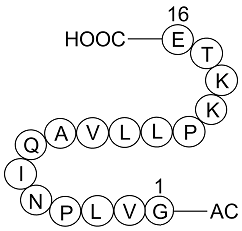
GP10020
Histone-H2A-(107-122)-Ac-OH
Histone-H2A peptide
-
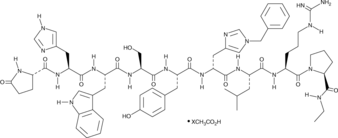
GC43866
Histrelin (acetate)
Histrelin is a synthetic gonadotropin-releasing hormone (GNRH) agonist that binds to the GNRH receptor (GNRHR; Ki = 0.2 nM in CHO cells expressing the human receptor).
-
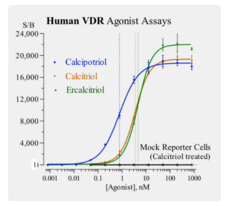
GC52504
Human Vitamin D Receptor Reporter Assay System
A nuclear receptor cell-based reporter assay
-
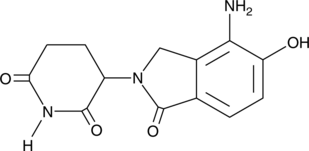
GC49282
Hydroxy Lenalidomide
A metabolite of lenalidomide
-
GC45959
IKD-8344
A macrocyclic dilactone with diverse biological activities
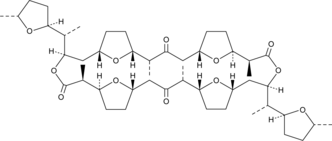
-
GC49359
IM-156 (acetate)
A mitochondrial complex I inhibitor and an AMPKα activator
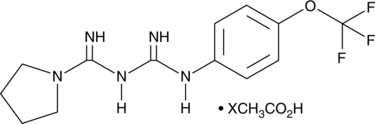
-
GC52190
Imidazole Ketone Erastin
Imidazole Ketone Erastin is a ferroptosis inducer.
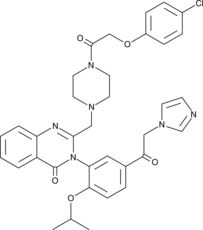
-
GC47454
IMS 2186
An anti-choroidal neovascularization agent
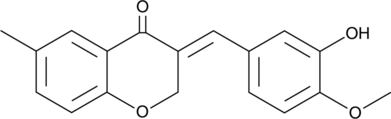
-
GC52472
Inostamycin A (sodium salt)
A bacterial metabolite with anticancer activity
-
GP10041
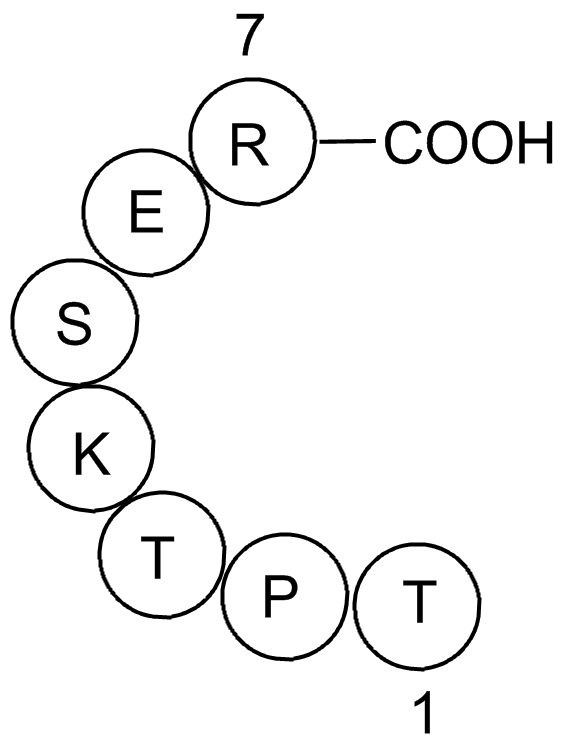
Insulin like growth factor II fragment variant
Insulin like growth factor II fragment variant has a sequence of Thr-Pro-Thr-Lys-Ser-Glu-Arg.
-
GC48396
IR 780
A fluorescent probe for in vivo tumor cell imaging
-
GC48378
IR 783
A fluorescent probe for in vivo tumor cell imaging
-
GC49634
Isoflupredone
Isoflupredone belongs to the class of corticosteroids and exerts its effect by binding to glucocorticoid and mineralocorticoid receptors of animals, such as horses.
-
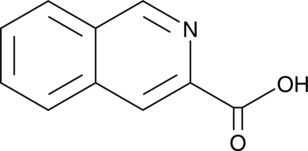
GC43920
Isoquinoline-3-carboxylic Acid
Isoquinoline-3-carboxylic acid is a synthetic intermediate that has been used to design novel antitumor compounds.