Cancer Biology
- Cancer Biology Peptides(84)
- Ras-like GTPases(11)
- Leukemia(1)
- Cell Proliferation, Viability & Cytotoxicity(0)
- Others(7)
- Tumor Microenvironment(30)
- Cell Migration & Metastasis(19)
Products for Cancer Biology
- Cat.No. Product Name Information
-
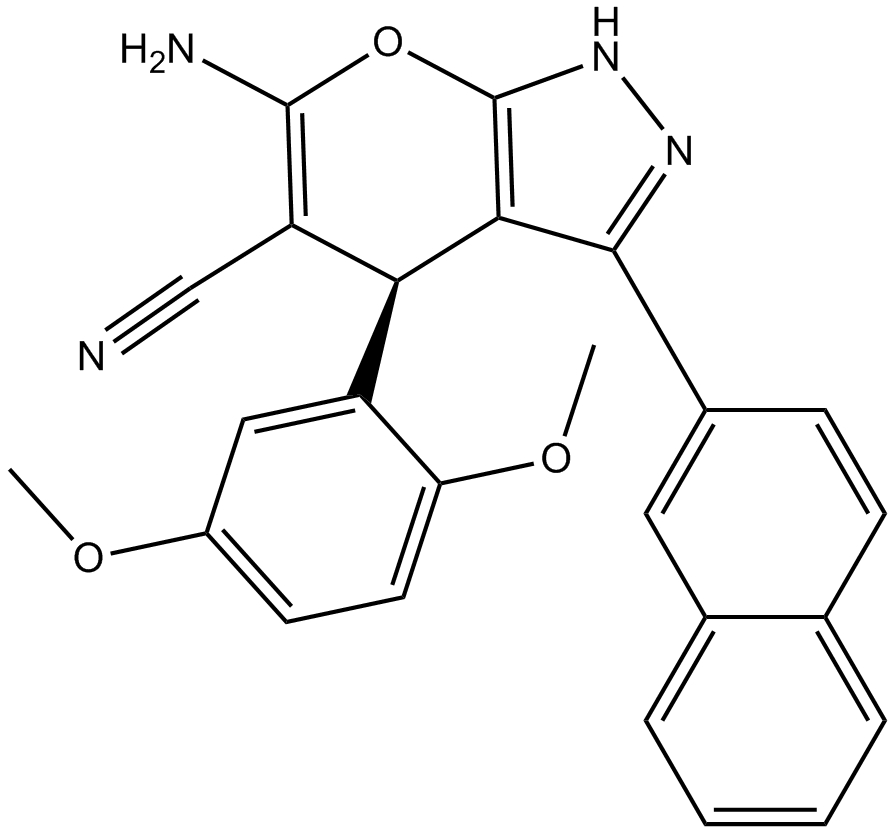
GC46163
IT-143B
A bacterial metabolite
-
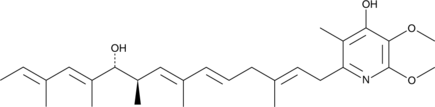
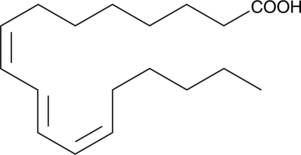
GC41645
Jacaric Acid
Jacaric acid is a conjugated polyunsaturated fatty acid first isolated from seeds of Jacaranda plants.
-
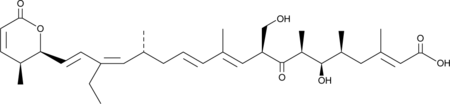
GC41236
Kazusamycin A
Kazusamycin A is an antibiotic from Streptomyces and a hydroxy analog of Leptomycin B that demonstrates cytotoxic activity against various human and mouse tumor lines.
-
GC44009
KM 233
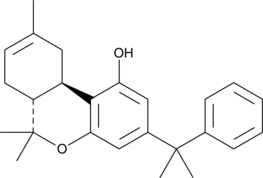
Because selective activation of peripheral cannabinoid (CB2) receptors in rat C-6 glioma cells has been shown to induce apoptosis through enhanced ceramide synthesis de novo, CB2 agonists present potential as anti glioma agents.
-
GC41627
Kumbicin C
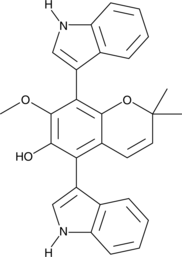
Kumbicin C is a bis-indolyl benzenoid fungal metabolite produced by A.
-
GC47533
L-778,123
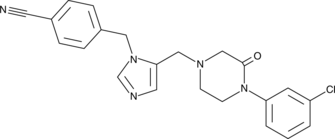
A dual inhibitor of FTase and GGTase I
-
GC47572
L-Lysine lactam (hydrochloride)
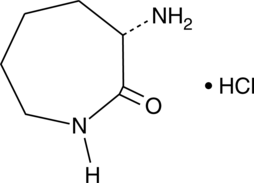
A building block
-
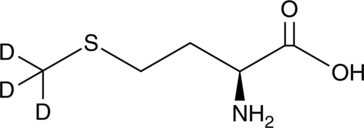
GC49368
L-Methionine-d3
An internal standard for the quantification of L-methionine
-
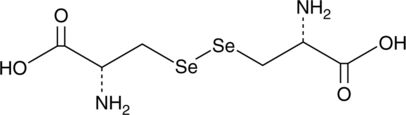
GC44084
L-Selenocystine
L-Selenocystine is a diselenide-bridged amino acid that may be confused with selenocysteine (Sec), which is a rare amino acid featuring a single selenium atom.
-
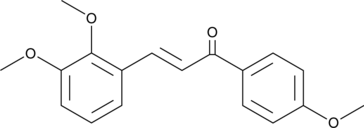
GC49560
L6H21
L6H21, a Chalcone derivative, is an orally active, potent and specific myeloid differentiation 2 (MD-2) inhibitor.
-
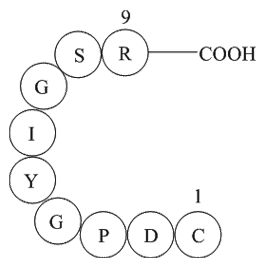
GP10103
Laminin (925-933)
Laminin (925-933)(CDPGYIGSR), is the sequence of Laminin on the B1 chain.
-
GC66014
Lepidiline A
Lepidiline A shows cytotoxic activity against HL-60 cells with an IC50 of 32.3 μM.

-
GC66012
Lepidiline C
Lepidiline C shows cytotoxic activity against HL-60 cells with an IC50 of 27.7 μM.
-
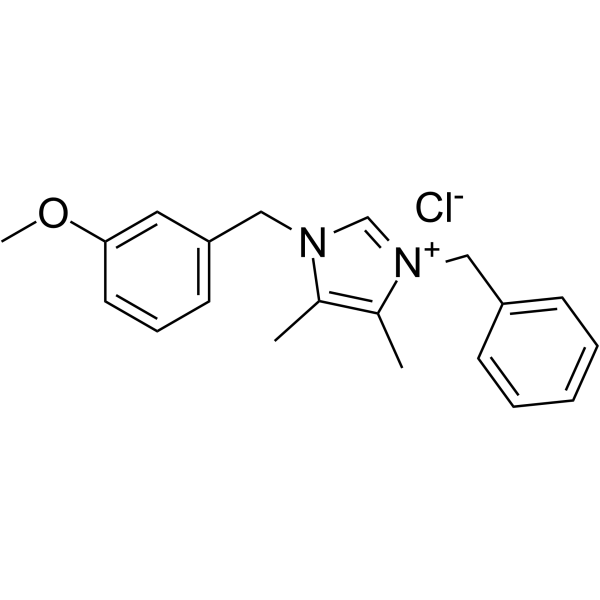
GC47554
Letrozole-d4
An internal standard for the quantification of letrozole
-
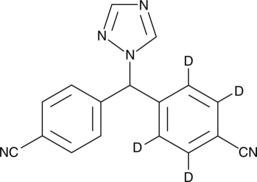
GC44050
Leucinostatin A
Leucinostatin A is a fungal metabolite originally isolated from P.
-
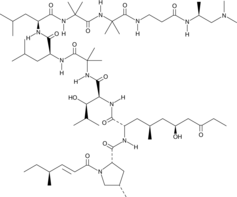
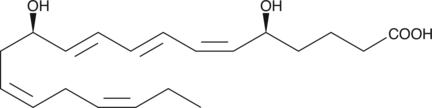
GC41107
Leukotriene B5
Leukotriene B5 (LTB5) is a leukotriene with diverse biological activities.
-
GC49020
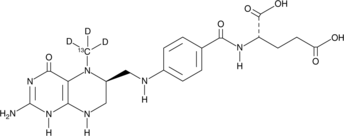
Levomefolate-13C-d3
An internal standard for the quantification of levomefolate
-
GC18789
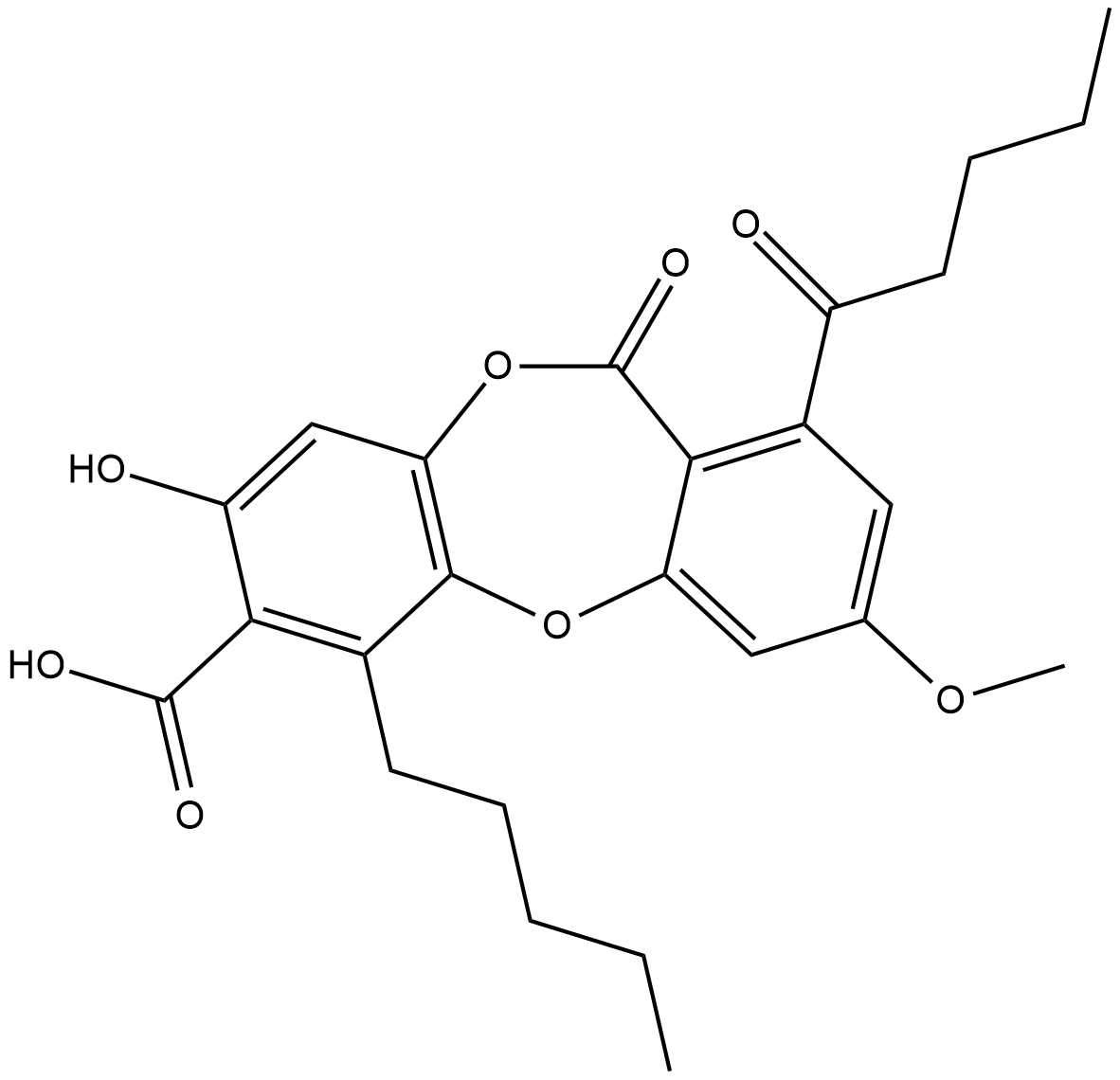
Lobaric Acid
Lobaric acid is a depsidone metabolite that has been isolated from Stereocaulon lichen species with antioxidant, antiproliferative, antiviral, and enzyme inhibitory activites.
-
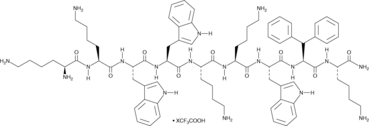
GC48989
LTX-315 (trifluoroacetate salt)
A synthetic cationic amphiphilic peptide
-
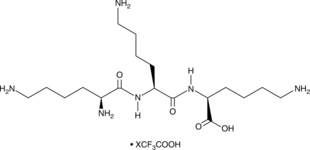
GC52314
Lysyllysyllysine (trifluoroacetate salt)
A tripeptide
-
GC41185
MAGL Inhibitor Compound 23
MAGL Inhibitor Compound 23 is a potent, selective, reversible and competitive inhibitor of MAGL, with an IC50 of 80 nM. MAGL Inhibitor Compound 23 exhibits anti-proliferative effects against human breast, colorectal, and ovarian cancer cells. MAGL Inhibitor Compound 23 blocks MAGL in cell-based as well as in vivo assays.
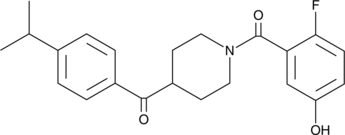
-
GC46031
Malabaricone B
A diarylnonanoid with diverse biological activities
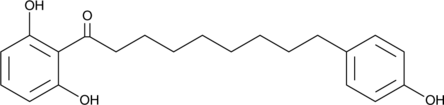
-
GC44118
Malvidin (chloride)
Malvidin is an O-methylated anthocyanidin responsible for the pigments in grapes and blueberries.
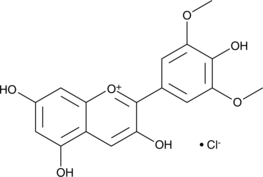
-
GC44126
Manzamine A
Manzamine A is a β-carboline alkaloid with diverse activities that has been found in marine sponges, including X.
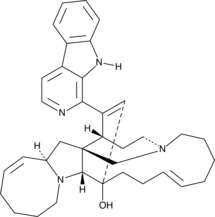
-
GP10140
MAP kinase fragment [Multiple species]
![MAP kinase fragment [Multiple species] Chemical Structure MAP kinase fragment [Multiple species] Chemical Structure](/media/struct/GP1/GP10140.png)
-
GC44168
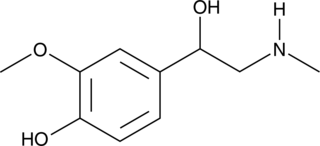
Metanephrine
Metanephrine is a metabolite of epinephrine produced by the action of catechol-O-methyl transferase on epinephrine.
-
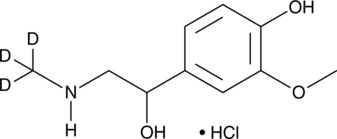
GC49423
Metanephrine-d3 (hydrochloride)
An internal standard for the quantification of metanephrine
-
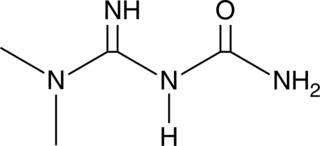
GC49879
Metformin hydroxy analog 2
An oxidation product of metformin
-
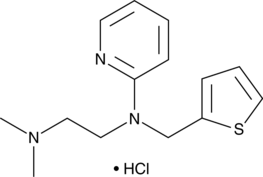
GC48805
Methapyrilene (hydrochloride)
An H1 receptor antagonist and non-genotoxic carcinogen
-
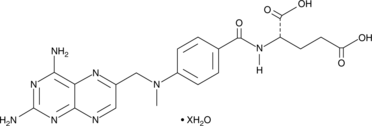
GC47649
Methotrexate (hydrate)
A folic acid and aminopterin derivative
-
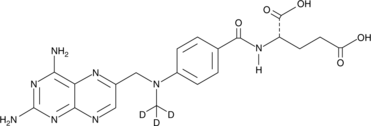
GC47650
Methotrexate-d3
An internal standard for the quantification of methotrexate
-
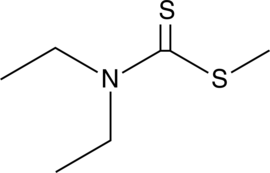
GC49241
Methyl Diethyldithiocarbamate
An active metabolite of disulfiram
-
GC50686
Miro1 Reducer
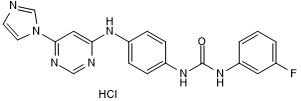
-
GC16331
Misonidazole
nitroimidazole with radiosensitizing and antineoplastic properties
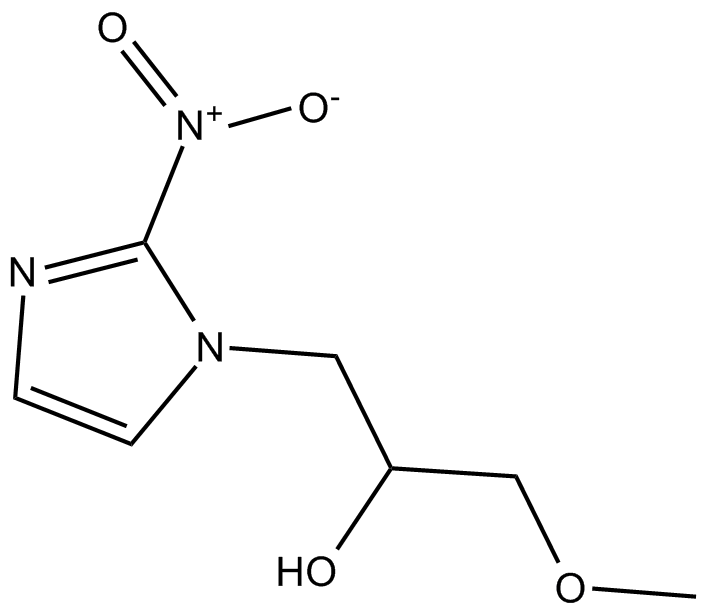
-
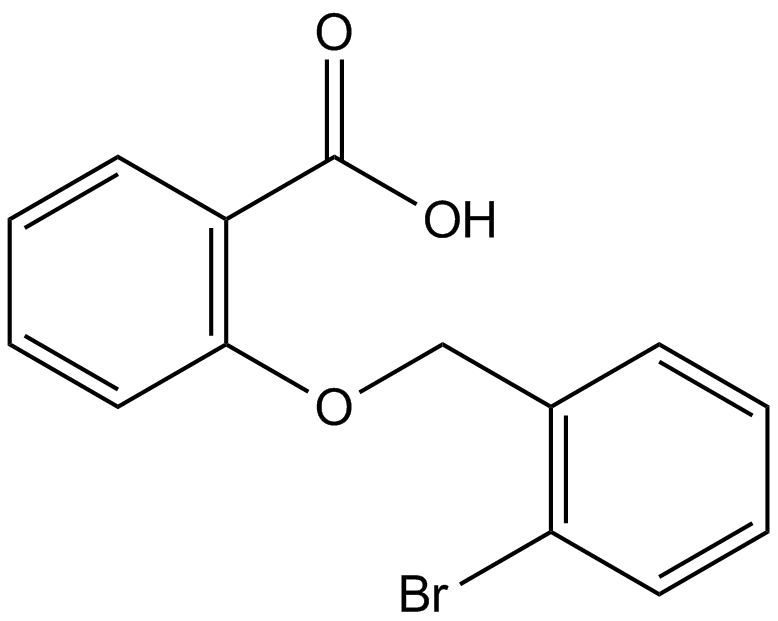
GC12876
ML-097
ML-097 (CID-2160985) is a pan Ras-related GTPases activator that can activate Rac1, cell division cycle 42, Ras, and Rab7.
-
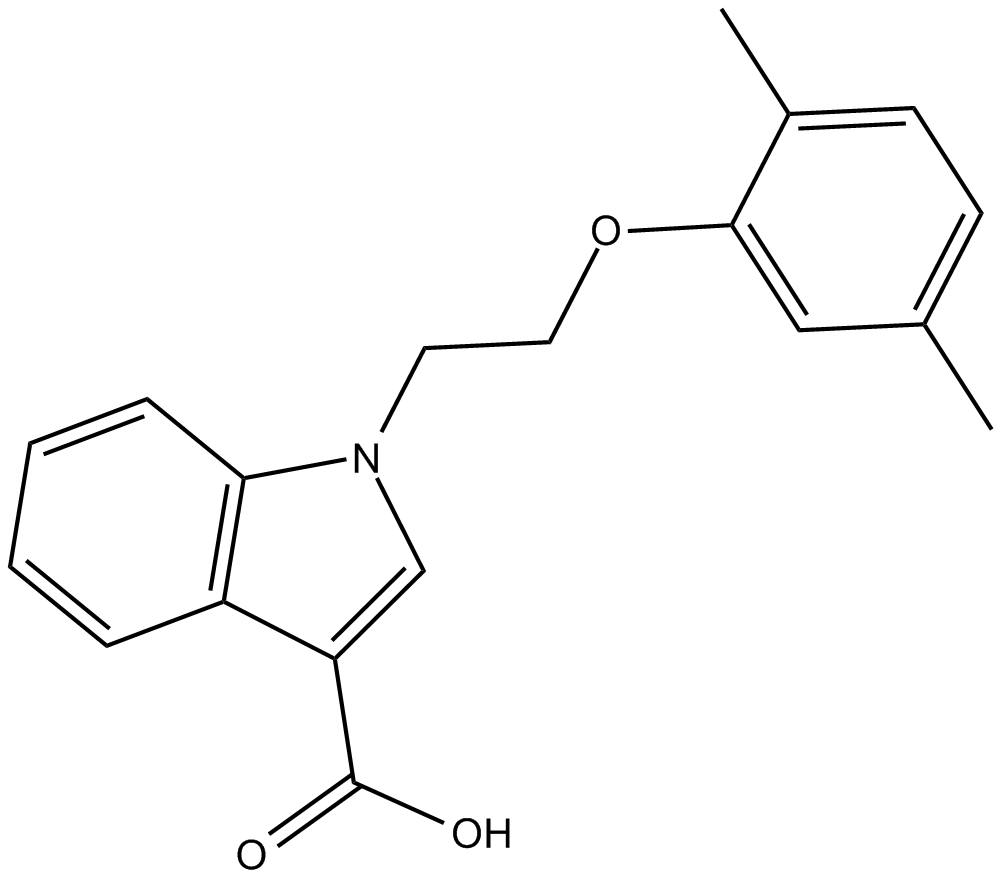
GC13315
ML-098
activator of the GTP-binding protein Rab7
-
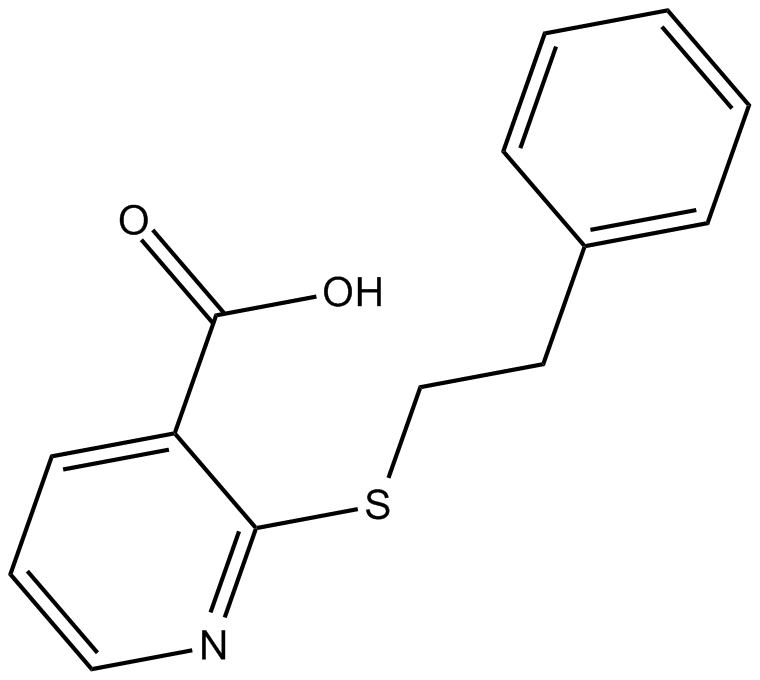
GC16765
ML-099
pan activator of Ras-related GTPases
-
GC18616
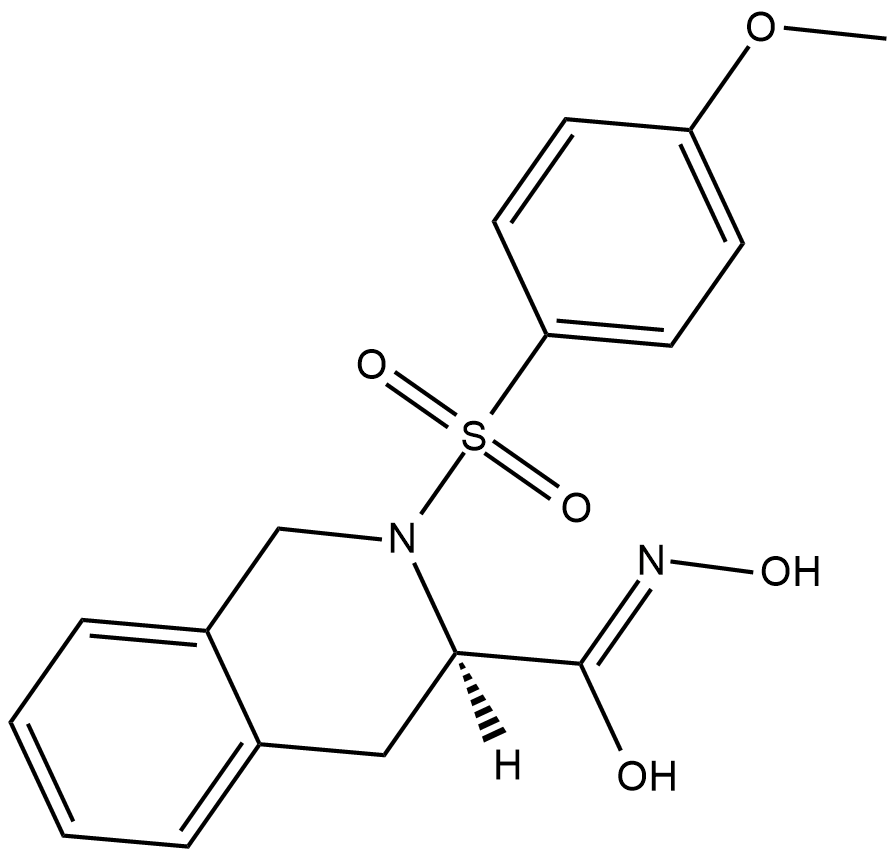
MMP-8 Inhibitor I
MMP-8 Inhibitor I is a selective inhibitor of the neutrophil collagenase matrix metalloproteinase-8 (MMP-8) with an IC50 value of 4 nM.
-
GC46176
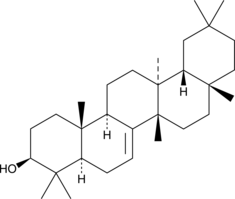
Multiflorenol
A triterpene
-
GC41325
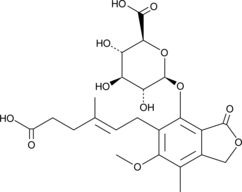
Mycophenolic Acid β-D-Glucuronide
Mycophenolic Acid β-D-Glucuronide is a metabolite of the immunosuppressant mycophenolic acid (MPA). Mycophenolic Acid β-D-Glucuronide shows anti-tumor activity and can be used in adenocarcinoma research.
-
GP10114
Myelopeptide-2 (MP-2)

-
GC49256
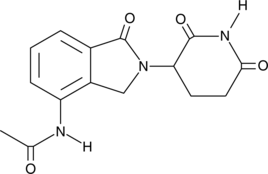
N-acetyl Lenalidomide
A metabolite of lenalidomide
-
GC18506
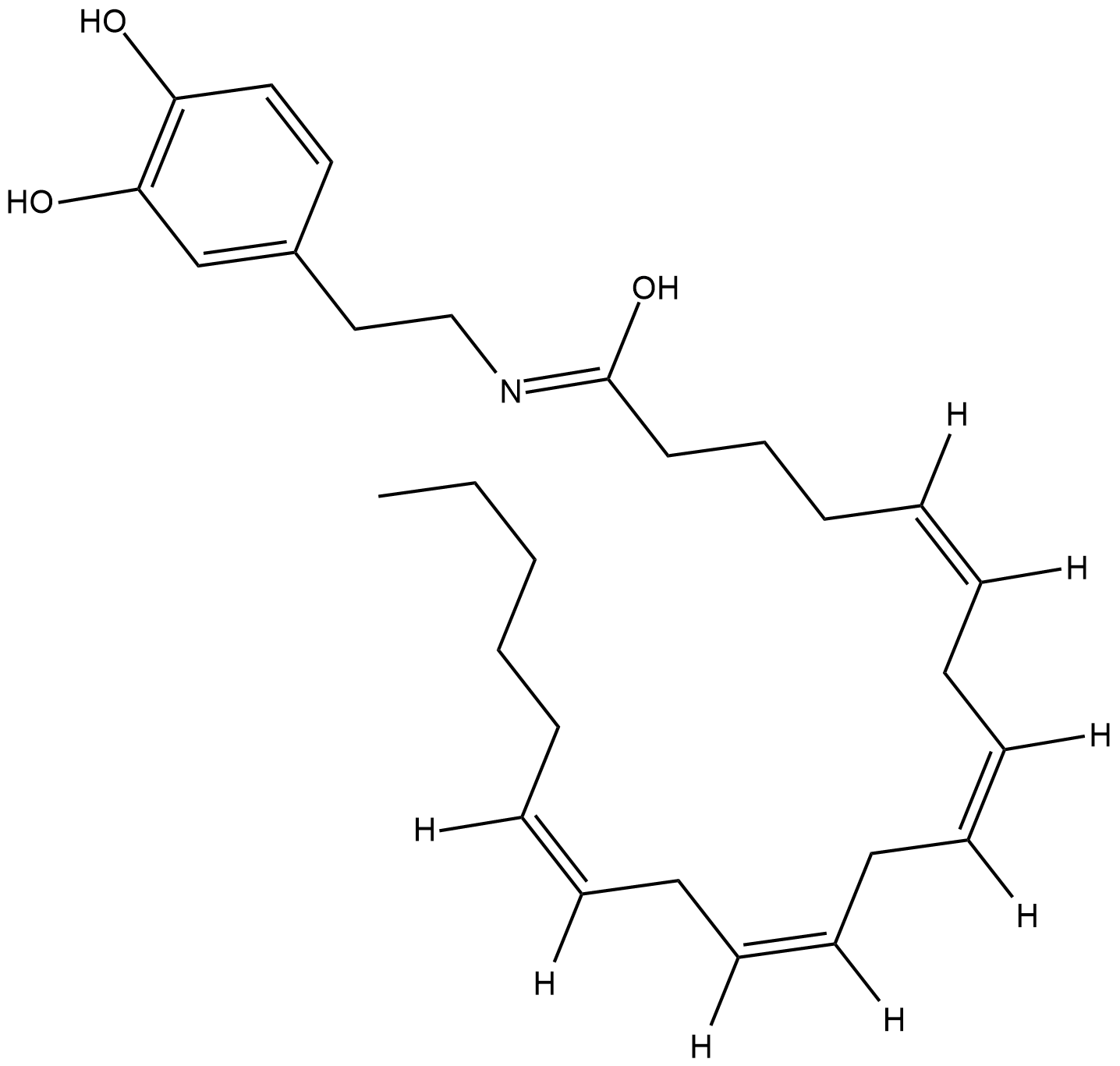
N-Arachidonoyl Dopamine
N-Arachidonoyl Dopamine is a potent and selective endogenous CB1 receptor agonist with a Ki of 250 nM.
-
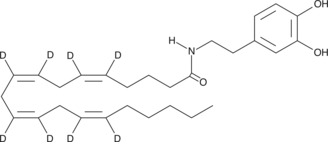
GC47748
N-Arachidonoyl Dopamine-d8
An internal standard for the quantification of Narachidonoyl dopamine
-
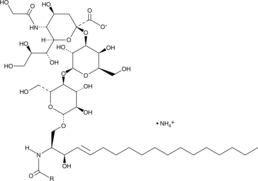
GC48335
N-glycolyl-Ganglioside GM3 Mixture (ammonium salt)
A mixture of N-glycolyl-ganglioside GM3
-
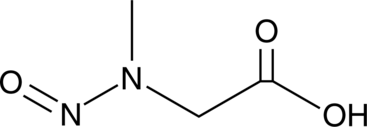
GC49350
N-Nitroso Sarcosine
A carcinogen
-
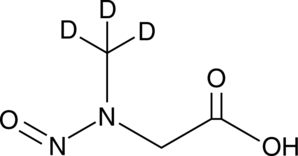
GC49354
N-Nitroso Sarcosine-d3
An internal standard for the quantification of nitroso sarcosine
-
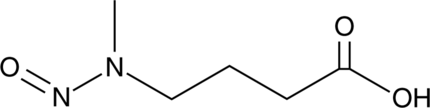
GC47786
N-Nitroso-N-methyl-4-Aminobutyric Acid
A tobacco-specific nitrosamine carcinogen
-
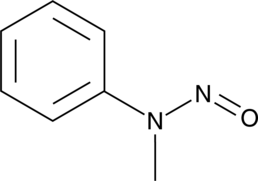
GC49859
N-Nitroso-N-methylaniline
An N-nitrosamine
-
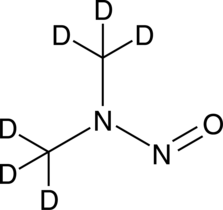
GC49731
N-Nitrosodimethylamine-d6
An internal standard for the quantification of N-nitrosodimethylamine
-
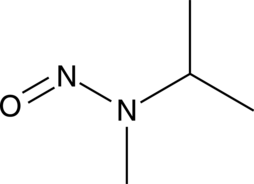
GC49814
N-Nitrosomethylisopropylamine
An N-nitrosamine
-
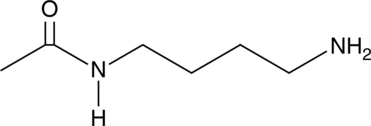
GC52162
N1-Acetylputrescine
-
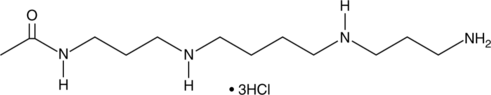
GC44285
N1-Acetylspermine (hydrochloride)
N1-Acetylspermine is a monoacetylated derivative of spermine, an endogenous polyamine synthesized from spermidine, that displays lower Km and higher Vmax values than spermine, making it a better substrate of polyamine oxidase than the non-acetylated polyamine.
-
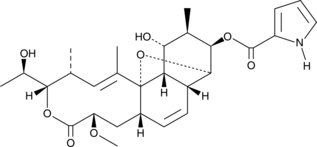
GC40081
Nargenicin
Nargenicin is a macrolide antibiotic that selectively inhibits the growth of S.
-
GC45926
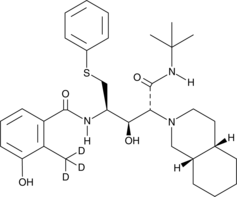
Nelfinavir-d3
An internal standard for the quantification of nelfinavir
-
GC47768
Neuromedin U-25 (human) (trifluoroacetate salt)
A neuropeptide with diverse roles

-
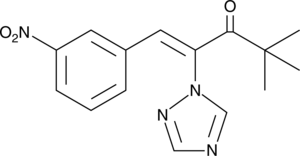
GC46184
Nexinhib20
An inhibitor of Rab27a-JFC1 protein-protein interactions
-
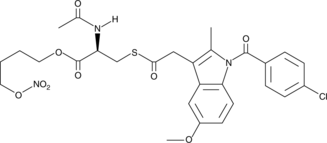
GC44439
NO-Indomethacin
NO-indomethacin is a hybrid molecule of indomethacin and a nitric oxide (NO) donor.
-
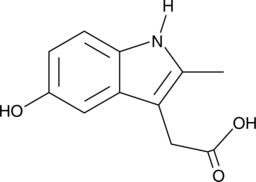
GC49316
O-Desmethyl-N-deschlorobenzoyl Indomethacin
A metabolite of indomethacin
-
GC47814
Ochratoxin A-13C20
An internal standard for the quantification of ochratoxin A
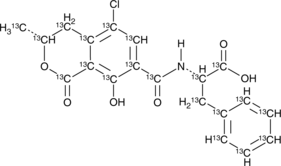
-
GC44507
Oligomycin E
Oligomycin E is a minor metabolite of the oligomycin complex produced by several species of Streptomyces.
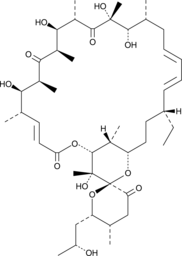
-
GC49236
Orlistat-d3
An internal standard for the quantification of orlistat
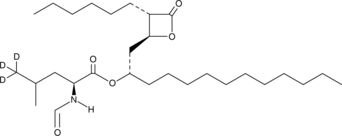
-
GC49073
Oxanosine
A guanosine analog with diverse biological activities
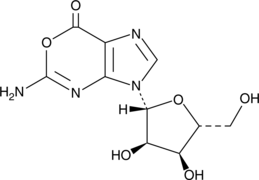
-
GP10036
p53 tumor suppressor fragment
Regulates cell cycle
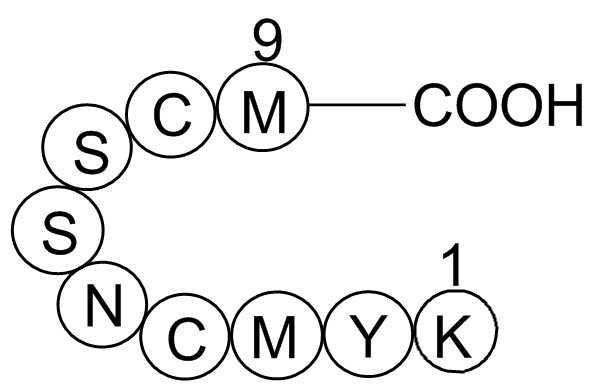
-
GC44531
PACAP (1-27) (human, mouse, ovine, porcine, rat) (trifluoroacetate salt)
Pituitary adenylate cyclase-activating peptide (PACAP) (1-27) is a PACAP receptor agonist with IC50 values of 3, 2, and 5 nM, respectively, for rat PAC1, rat VPAC1, and human VPAC2 recombinant receptors expressed in CHO cells.
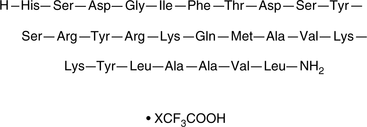
-
GC44533
PACAP (6-27) (human, chicken, mouse, ovine, porcine, rat) (trifluoroacetate salt)
Pituitary adenylate cyclase-activating peptide (PACAP) (6-27) is a PACAP receptor antagonist with IC50 values of 1,500, 600, and 300 nM, respectively, for rat PAC1, rat VPAC1, and human VPAC2 recombinant receptors expressed in CHO cells.

-
GC44535
PACAP-related Peptide (human) (trifluoroacetate salt)
PACAP-related peptide (PRP) is an endogenous 29-amino acid peptide that belongs to the secretin/glucagon superfamily of peptides, which includes secretin, glucagon, glucagon-like peptide-1, GLP-2, and pituitary adenylate cyclase-activating polypeptide.

-
GC49024
Palmitic Acid MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
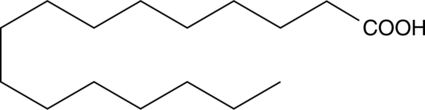
-
GC49023
Palmitic Acid-d9 MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
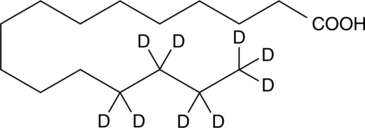
-
GC49422
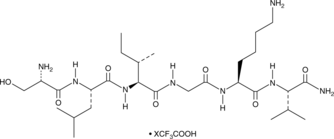
PAR2 (1-6) amide (human) (trifluoroacetate salt)
A peptide agonist of PAR2
-
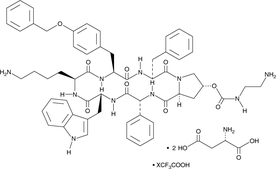
GC47925
Pasireotide (aspartate) (trifluoroacetate salt)
A somatostatin receptor agonist
-
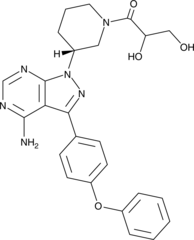
GC48493
PCI 45227
PCI 45227 (PCI-45227) is a dihydrodiolactive metabolite of Ibrutinib, has inhibitory activity towards BTK approximately 15 times lower than that of ibrutinib.
-
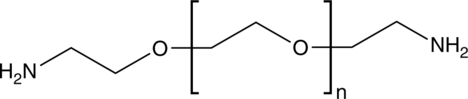
GC52107
PEG diamine
A PEGylated building block
-
GC44590
Pellitorine
An amide alkaloid with diverse biological activities
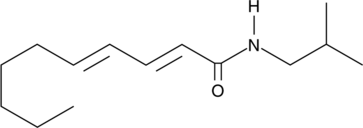
-
GC44591
Pemetrexed (sodium salt hydrate)
Pemetrexed (sodium salt hydrate) is a novel antifolate, the Ki values of the pentaglutamate of LY231514 are 1.3, 7.2, and 65 nM for inhibits thymidylate synthase (TS), dihydrofolate reductase (DHFR), and glycinamide ribonucleotide formyltransferase (GARFT), respectively.
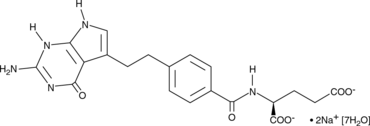
-
GC46198
Penicinoline
An alkaloid
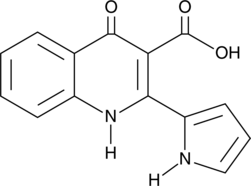
-
GC45814
Pentanoic Acid-d9
An internal standard for the quantification of pentanoic acid
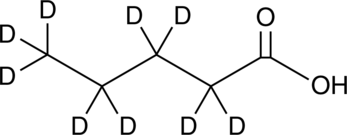
-
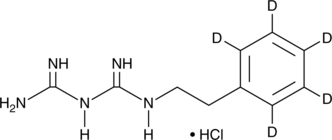
GC49530
Phenformin-d5 (hydrochloride)
An internal standard for the quantification of phenformin
-
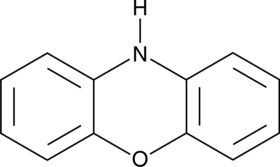
GC47948
Phenoxazine
A heterocyclic building block
-
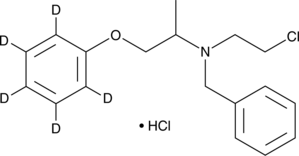
GC46202
Phenoxybenzamine-d5 (hydrochloride)
An internal standard for the quantification of phenoxybenzamine
-
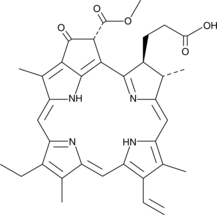
GC44629
Pheophorbide a
Pheophorbide a is a product of chlorophyll breakdown that has been used as a photosensitizer in photodynamic therapy for the treatment of cancer.
-
GC44637
PHPS1
PHPS1 is a cell-permeable, phosphotyrosine mimetic that inhibits the Src homology region 2 domain-containing phosphatase (SHP)-2 (IC50 = 2.1 μM; Ki = 0.73 μM).
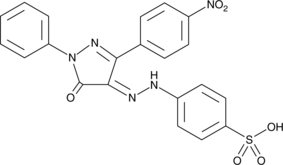
-
GC49441
Plerixafor (hydrochloride hydrate)
An antagonist of CXCR4
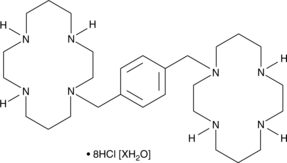
-
GC49148
Plitidepsin
A depsipeptide with diverse biological activities
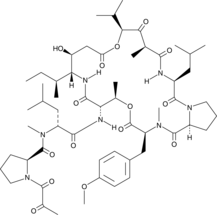
-
GC18271
Polmacoxib
Polmacoxib is an inhibitor of cyclooxygenase 2 (COX-2) and the carbonic anhydrase subtypes I (CAI) and CAII.
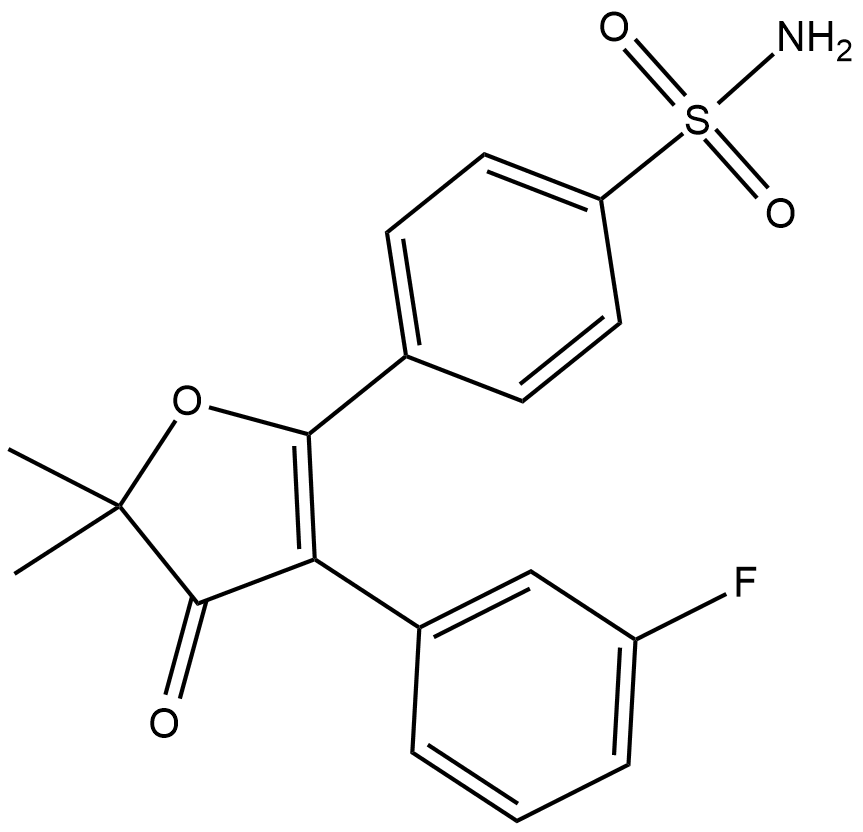
-
GC46207
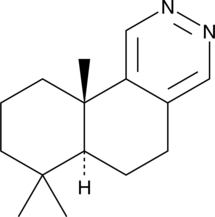
Polygodial pyridazine
A derivative of polygodial
-
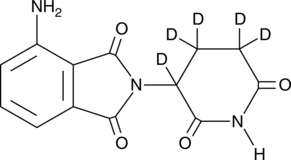
GC47965
Pomalidomide-d5
An internal standard for the quantification of pomalidomide
-
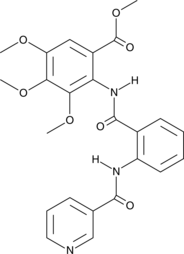
GC48501
Preterramide C
Preterramide C is a fungal metabolite.
-
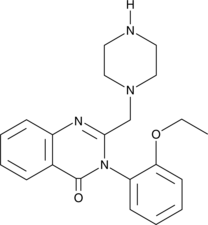
GC41572
PRLX-93936
PRLX-93936 is an analog of erastin that has antitumor activity.
-
GP10072
Pro-Adrenomedullin (153-185), human
Vasodilator
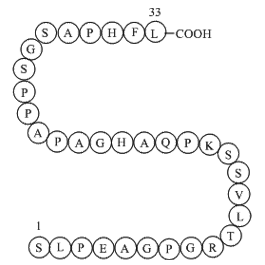
-
GC18310
Prostaglandin A1
Prostaglandin A1 (PGA1) was first isolated as a dehydration product of the PGE1 compounds found in human semen.
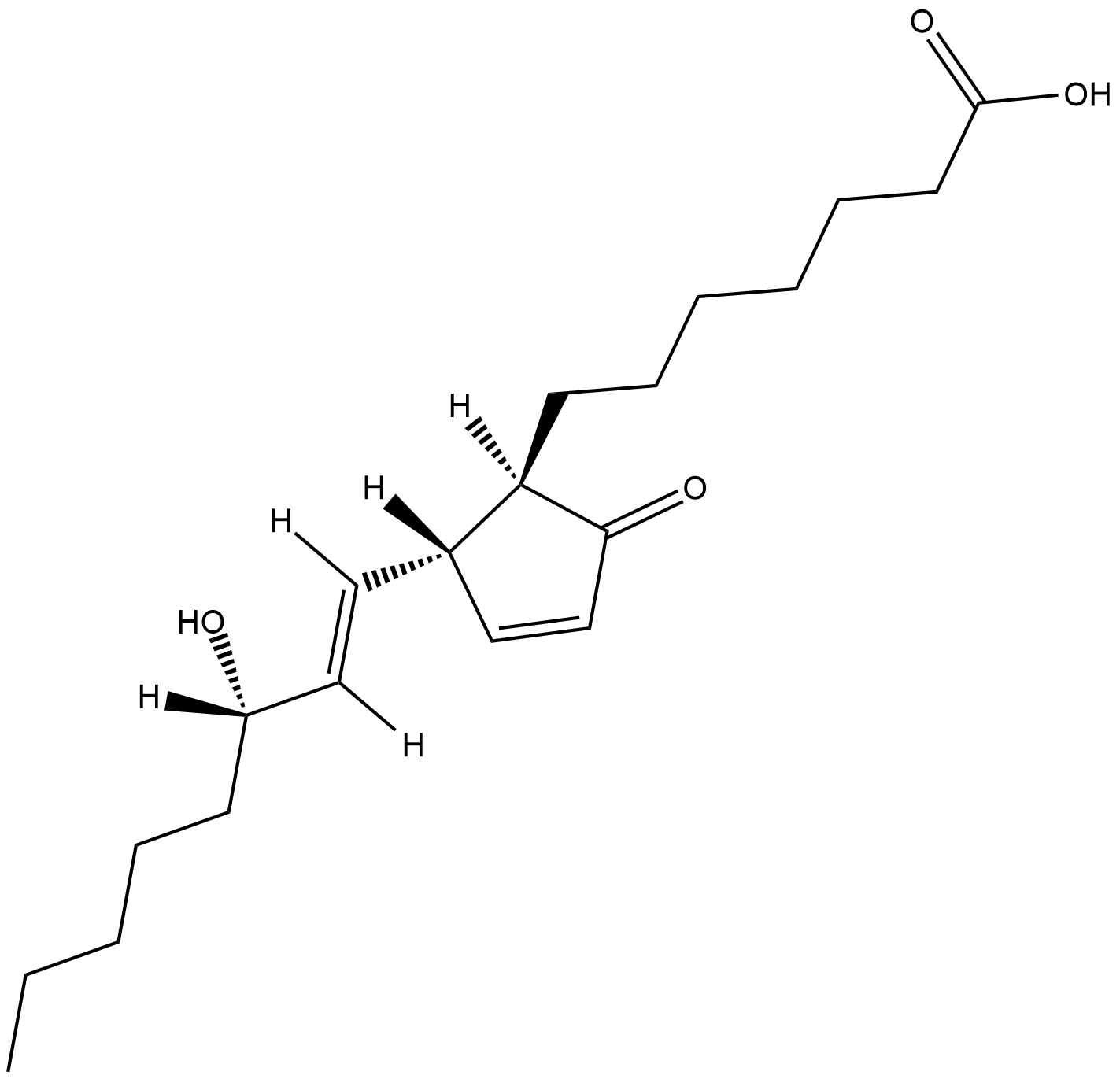
-
GP10002
prostate apoptosis response protein PAR-4 (2-7) [Homo sapiens]
Transcriptional repressor
![prostate apoptosis response protein PAR-4 (2-7) [Homo sapiens] Chemical Structure prostate apoptosis response protein PAR-4 (2-7) [Homo sapiens] Chemical Structure](/media/struct/GP1/GP10002.png)
-
GC48406
PSI (trifluoroacetate salt)
A proteasome inhibitor
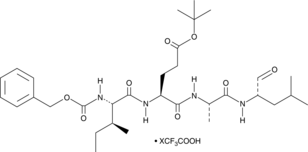
-
GC49914
Pt(II)-NHC Complex 2C
An inducer of immunogenic cancer cell death
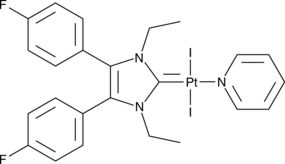
-
GC52484
Purified Ganglioside Mixture (bovine) (ammonium salt)
A mixture of purified bovine gangliosides

-
GC40568
Pyrocoll
Pyrocoll is a bacterial metabolite originally isolated from Streptomyces.
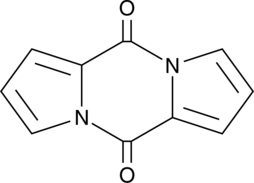
-
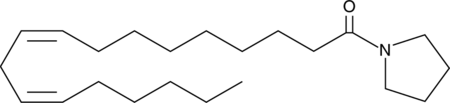
GC44791
Pyrrolidine Linoleamide
Linoleic acid is an essential fatty acid and one of the most abundant polyunsaturated fatty acids in the western diet.
-
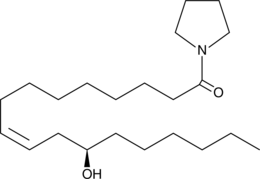
GC44792
Pyrrolidine Ricinoleamide
Ricinoleic acid is a naturally occurring 12-hydroxy fatty acid.
-
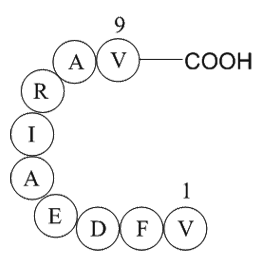
GP10032
Rac GTPase fragment
Fragment of small signaling G proteins
-
GC10906
RBC8
Ral GTPase inhibitor