HDAC
Histone deacetylases (HDACs), known for their ability to target histones as well as more than 50 non-histone proteins, are classified into three major classes according to their homology to yeast proteins, including class I (HDAC1, HDAC2, HDAC3 and HDAC8), class II (HDAC4, HDAC5, HDAC7 and HDAC9) and class IV (HDAC11). HDAC inhibitors, a large group of compounds that is able to induce the accumulation of acetylated histones as well as non-histone proteins, are divided into several structural classes including hydroxamates, cyclic peptides, aliphatic acids and benzamides. HDAC inhibitors have been investigated for their efficacy as anticancer agents in the treatment of a wild range of cancers.
Products for HDAC
- Cat.No. Product Name Information
-
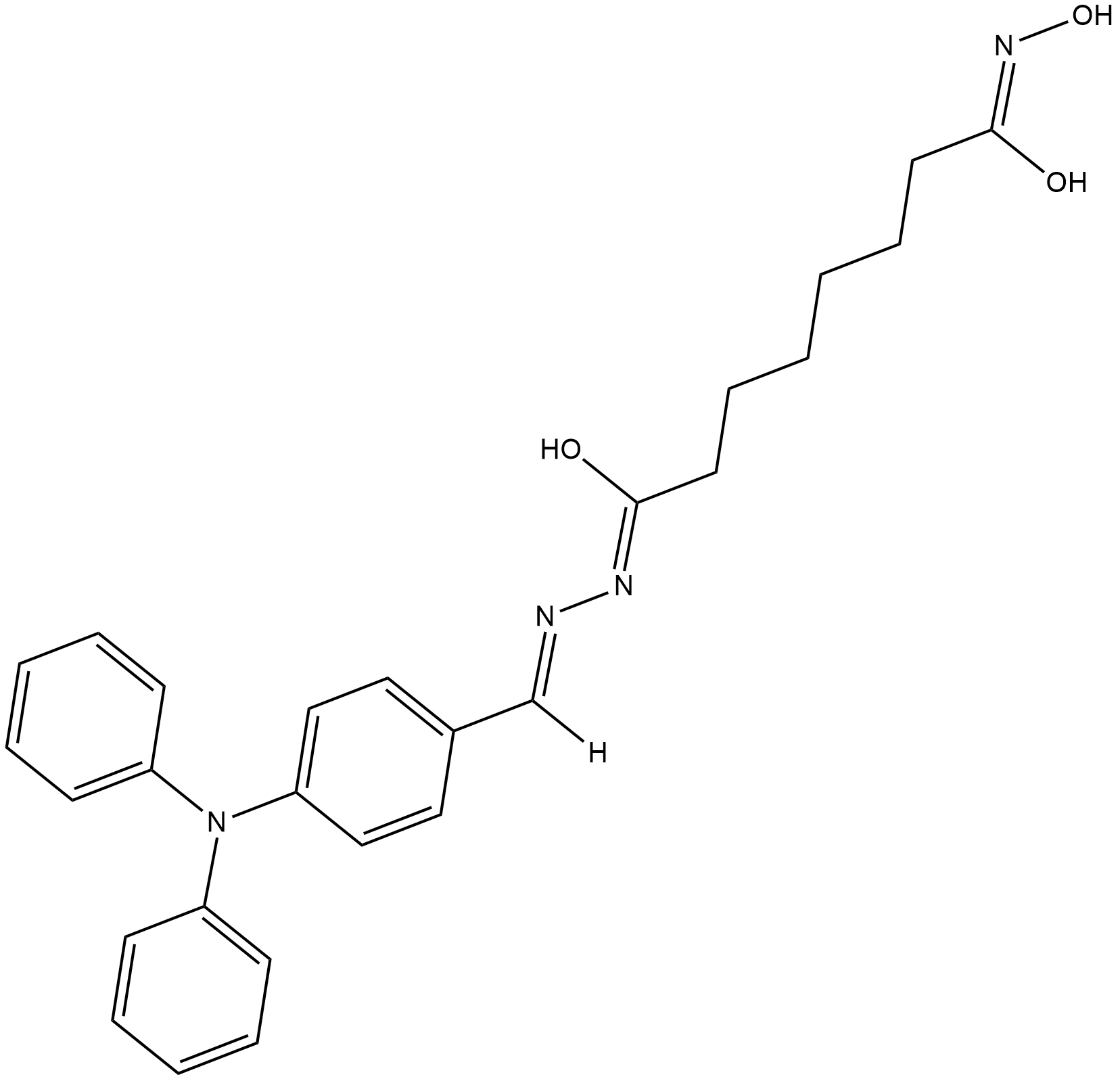
GC12667
4SC-202
4SC-202 (4SC-202) is a selective class I HDAC inhibitor with IC50 of 1.20 μM, 1.12 μM, and 0.57 μM for HDAC1, HDAC2, and HDAC3, respectively. It also displays inhibitory activity against Lysine specific demethylase 1 (LSD1).
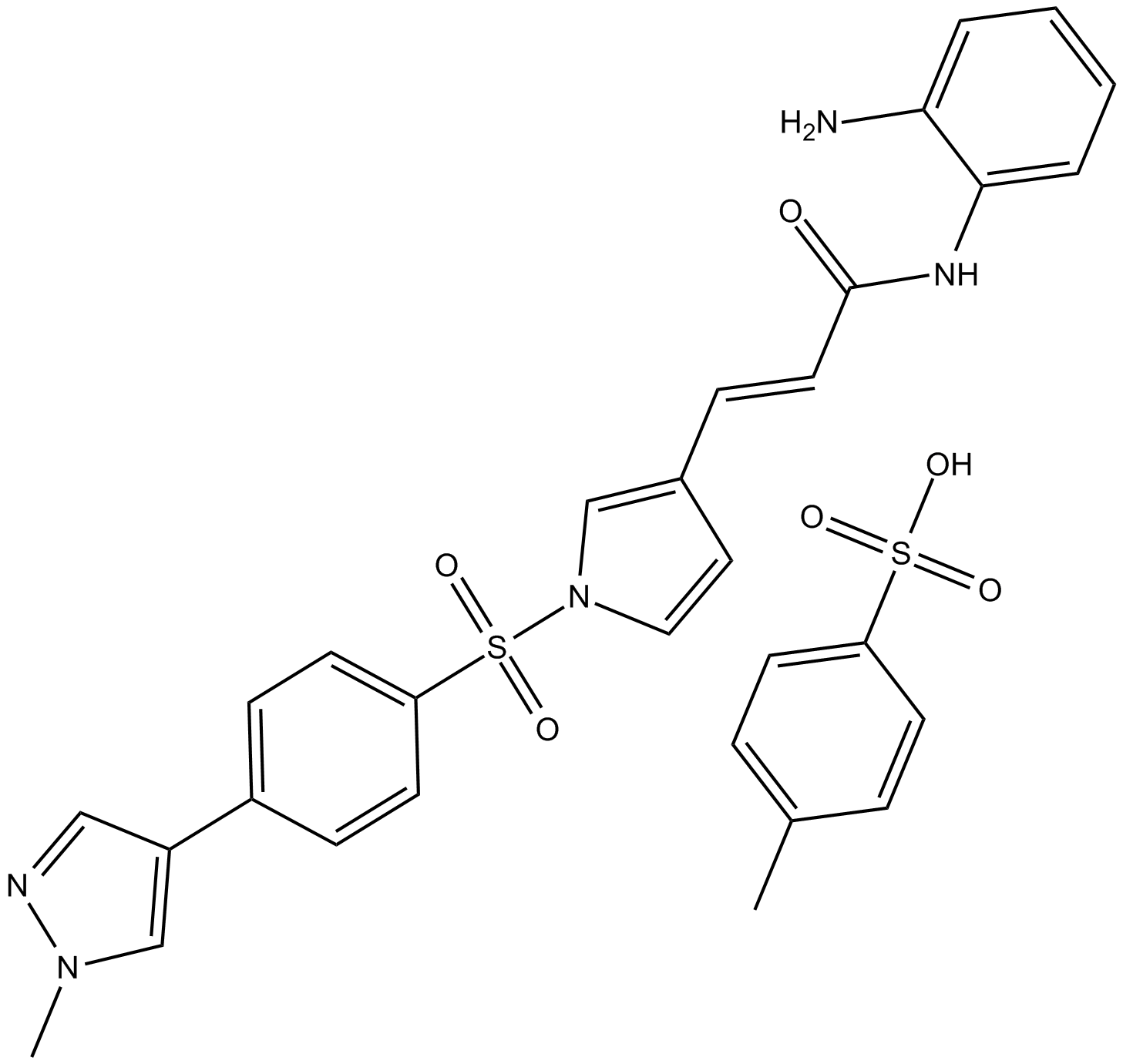
-
GC66055
5-Phenylpentan-2-one
5-Phenylpentan-2-one is a potent histone deacetylases (HDACs) inhibitor. 5-Phenylpentan-2-one can be used for urea cycle disorder research.
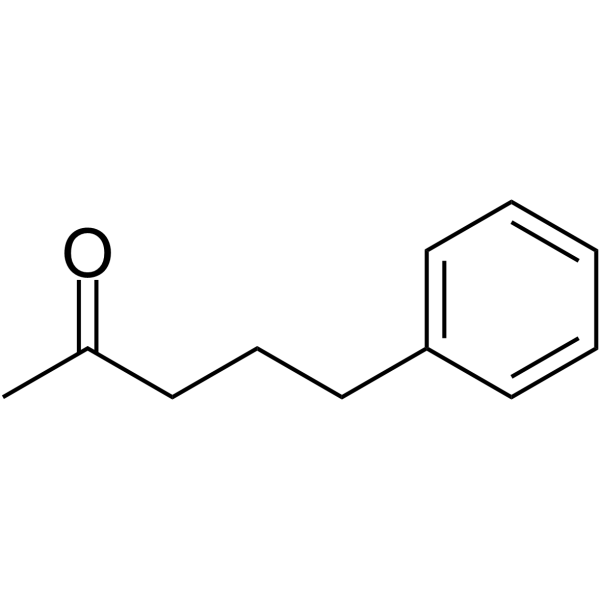
-
GA20605
Ac-Lys-AMC
Ac-Lys-AMC is cleaved by trypsin.

-
GC34458
ACY-1083
ACY-1083 is a selective and brain-penetrating HDAC6 inhibitor with an IC50 of 3 nM and is 260-fold more selective for HDAC6 than all other classes of HDAC isoforms. ACY-1083 effectively reverses chemotherapy-induced peripheral neuropathy.
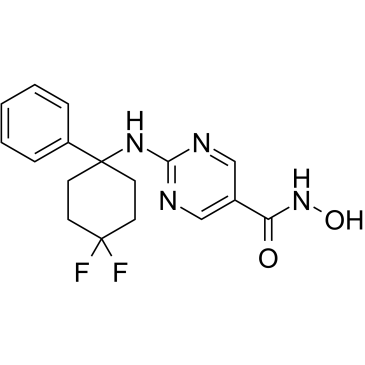
-
GC10417
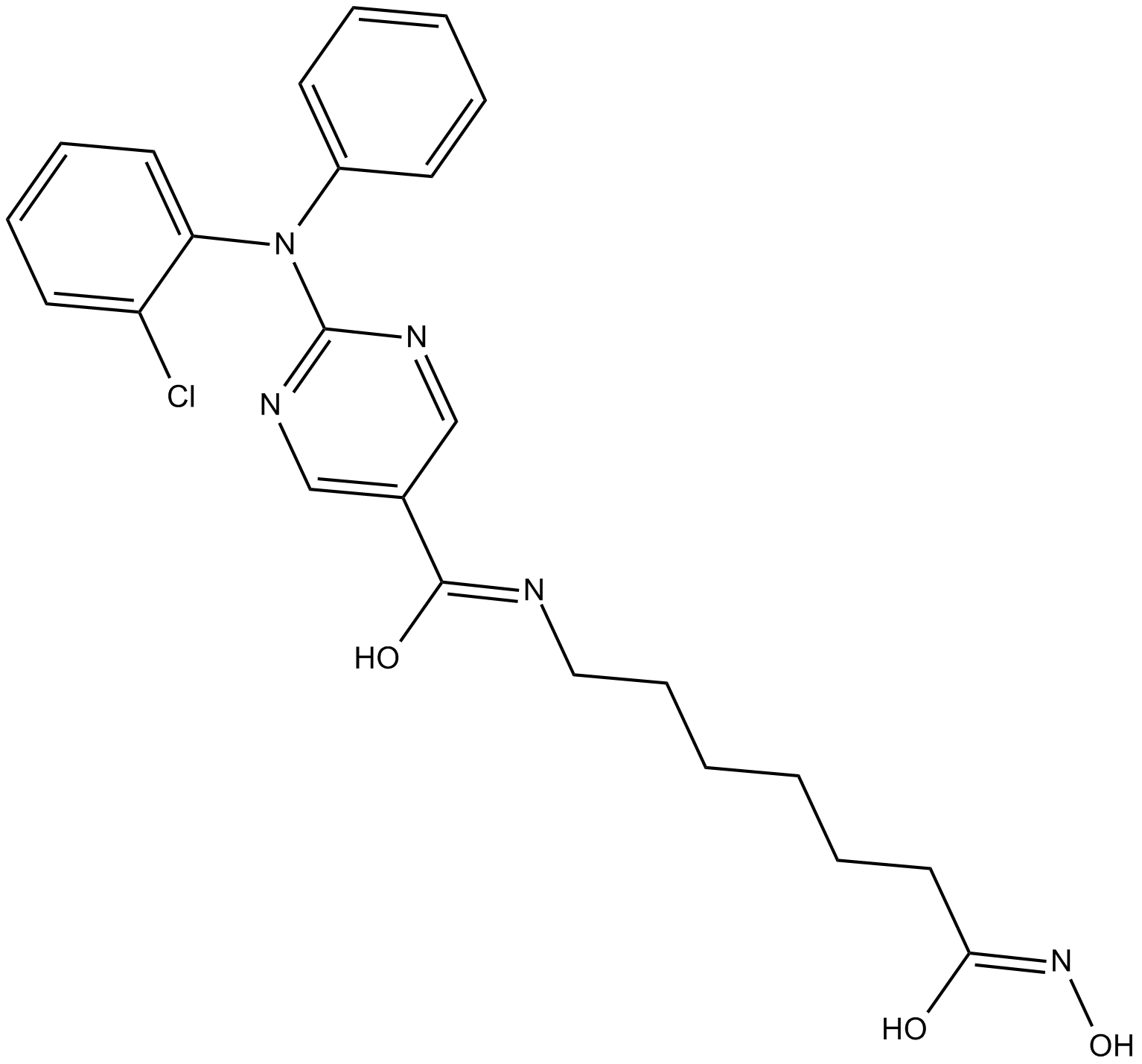
ACY-241
ACY-241 (ACY241) is a second generation potent, orally active and high-selective HDAC6 inhibitor with an IC50 of 2.6 nM (IC50s of 35 nM, 45 nM, 46 nM and 137 nM for HDAC1, HDAC2, HDAC3 and HDAC8, respectively). ACY-241 has anticancer effects.
-
GC19020
ACY-738
ACY-738 is a potent, selective and orally-bioavailable HDAC6 inhibitor, with an IC50s of 1.7 nM; ACY-738 also inhibits HDAC1, HDAC2, and HDAC3, with IC50s of 94, 128, and 218 nM.

-
GC30782
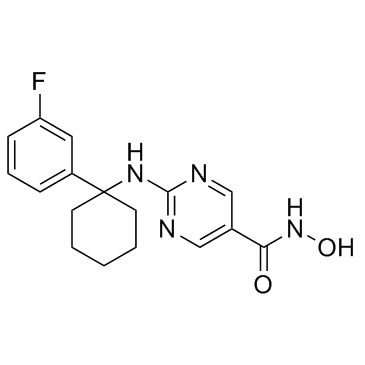
ACY-775
An HDAC6 inhibitor
-
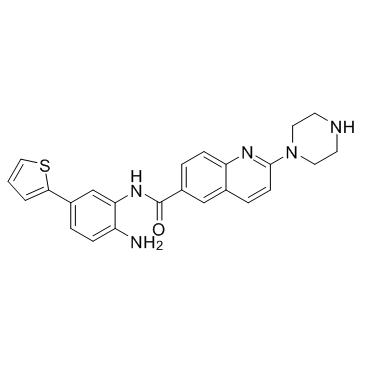
GC30526
ACY-957
ACY-957 is an orally active and selective inhibitor of HDAC1 and HDAC2, with IC50s of 7 nM, 18 nM, and 1300 nM against HDAC1/2/3, respectively, and shows no inhibition on HDAC4/5/6/7/8/9.
-
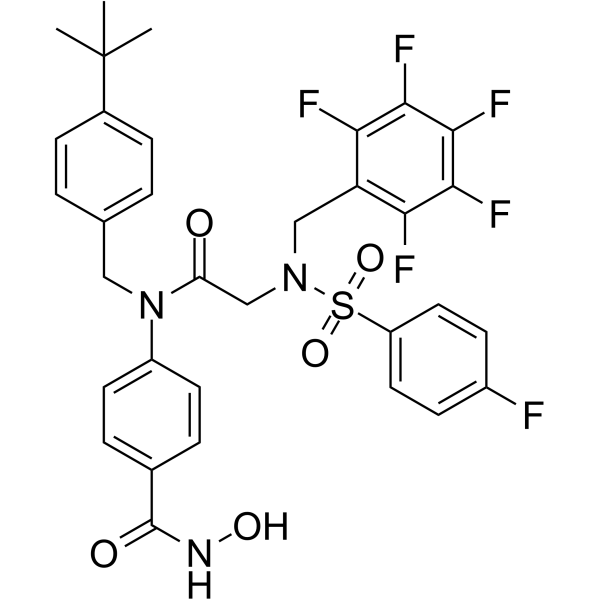
GC65330
AES-135
AES-135, a hydroxamic acid-based pan-HDAC inhibitor, prolongs survival in an orthotopic mouse model of pancreatic cancer. AES-135 inhibits HDAC3, HDAC6, HDAC8, and HDAC11 with IC50s ranging from 190-1100 nM.
-
GC67984
Alteminostat
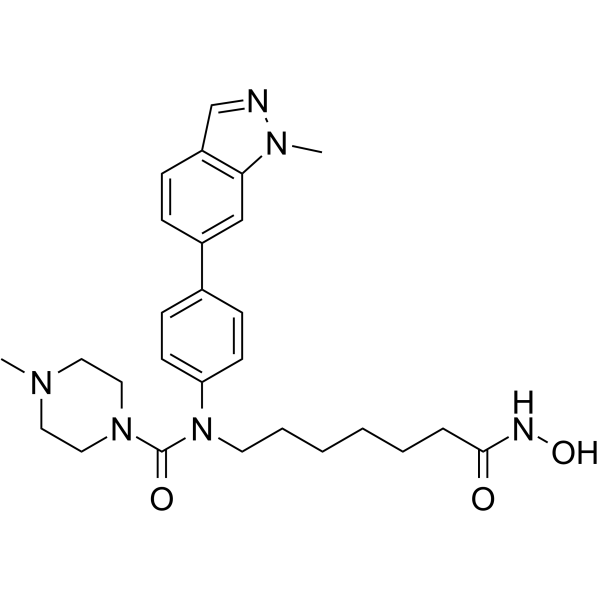
-
GC30763
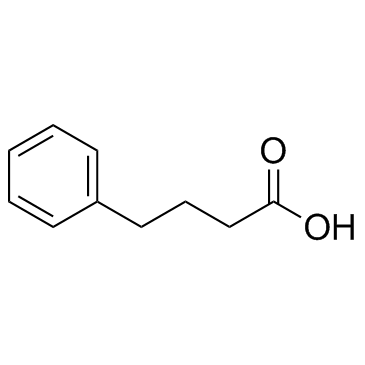
Benzenebutyric acid (4-Phenylbutyric acid)
Benzenebutyric acid (4-Phenylbutyric acid) (4-PBA) is an inhibitor of HDAC and endoplasmic reticulum (ER) stress, used in cancer and infection research.
-
GC12074
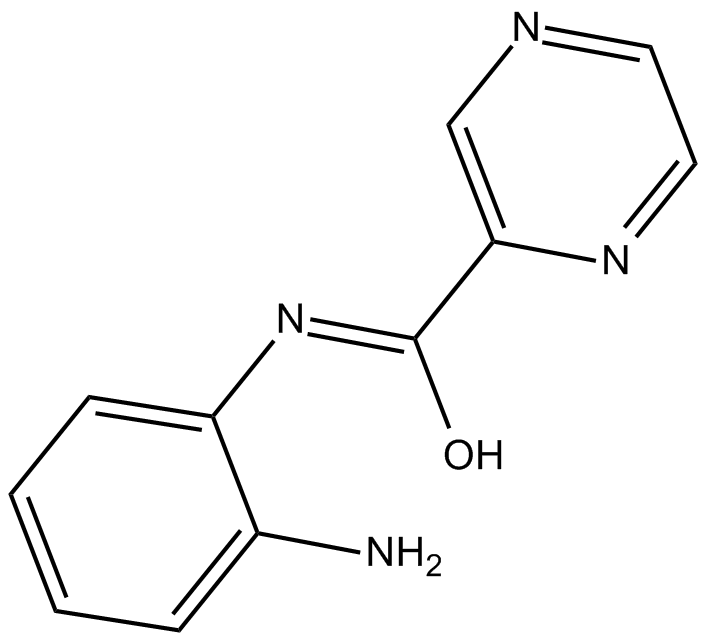
BG45
Novel HDAC3-selective inhibitor
-
GC25139
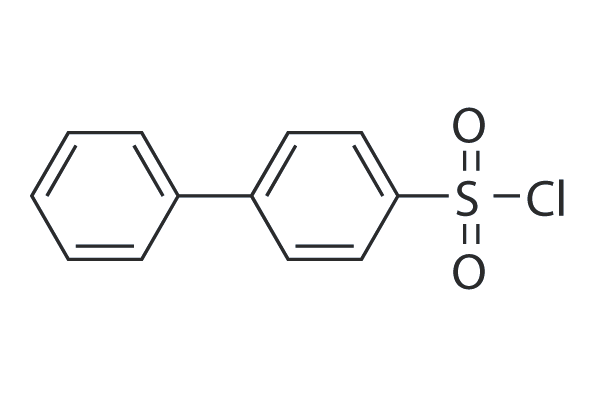
Biphenyl-4-sulfonyl chloride
Biphenyl-4-sulfonyl chloride (p-Phenylbenzenesulfonyl, 4-Phenylbenzenesulfonyl, p-Biphenylsulfonyl) is a HDAC inhibitor with synthetic applications in palladium-catalyzed desulfitative C-arylation.
-
GC12822
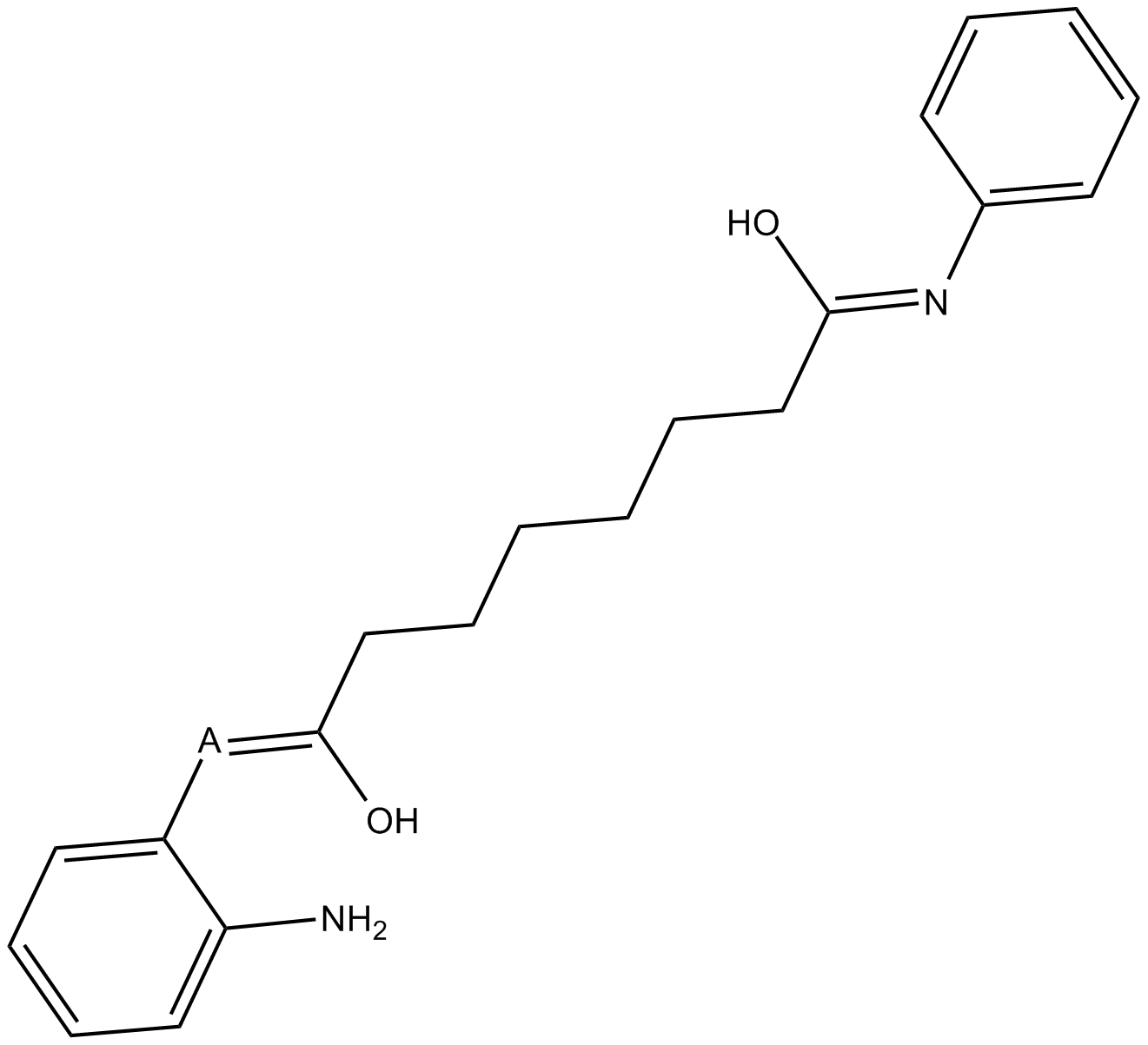
BML-210(CAY10433)
BML-210(CAY10433) is a novel HDAC inhibitor, and its mechanism of action has not been characterized.
-
GC33331
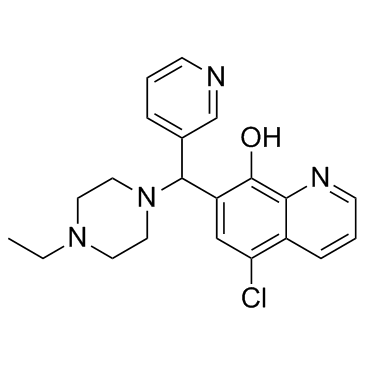
BRD 4354
BRD 4354 is a moderately potent inhibitor of HDAC5 and HDAC9, with IC50s of 0.85 and 1.88 μM, respectively.
-
GC35551
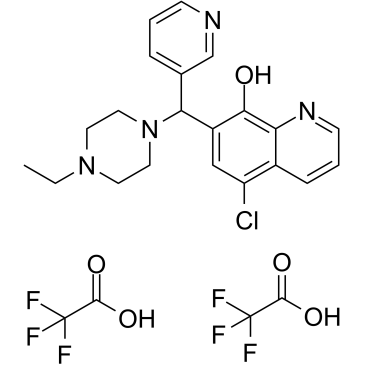
BRD 4354 ditrifluoroacetate
BRD 4354 (ditrifluoroacetate) is a moderately potent inhibitor of HDAC5 and HDAC9, with IC50s of 0.85 and 1.88 μM, respectively.
-
GC38742
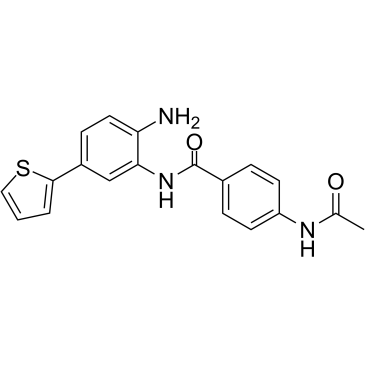
BRD-6929
An HDAC1 and HDAC2 inhibitor
-
GC12484
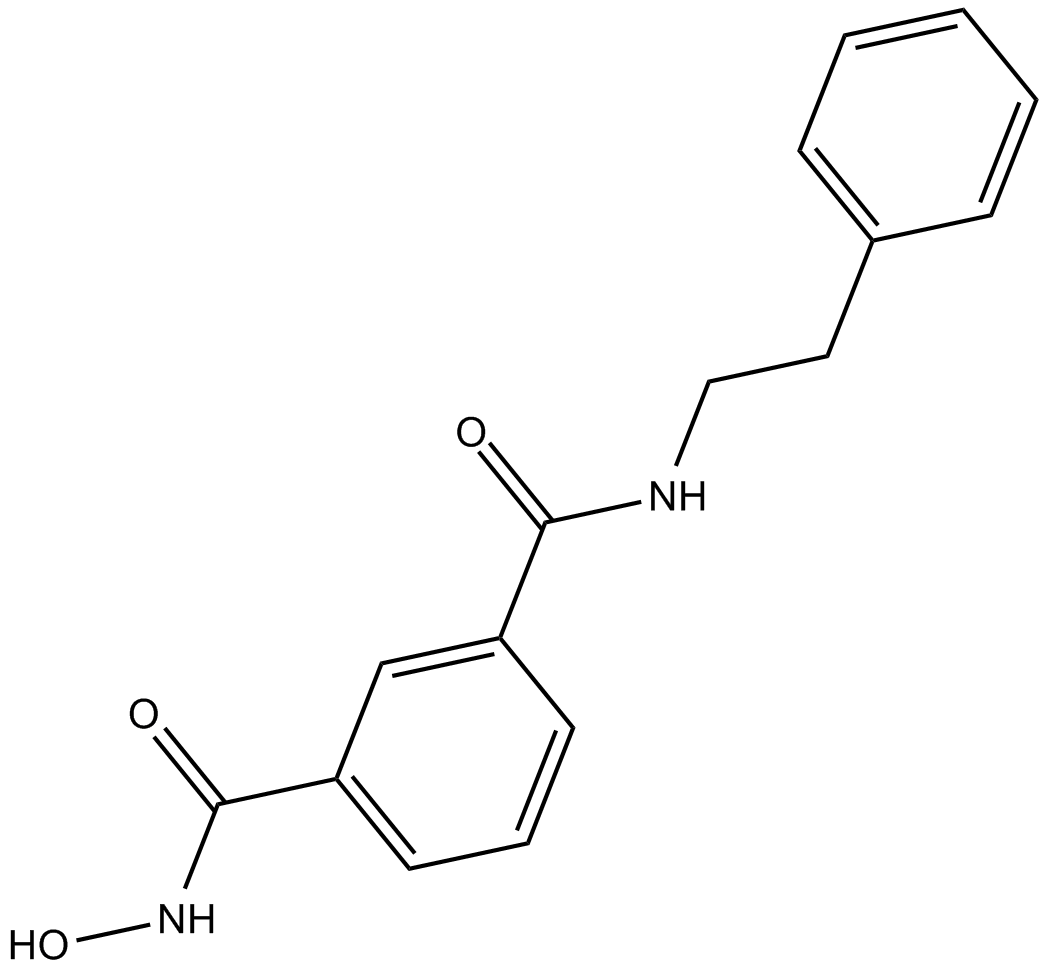
BRD73954
potent and selective HDAC inhibitor
-
GC15410
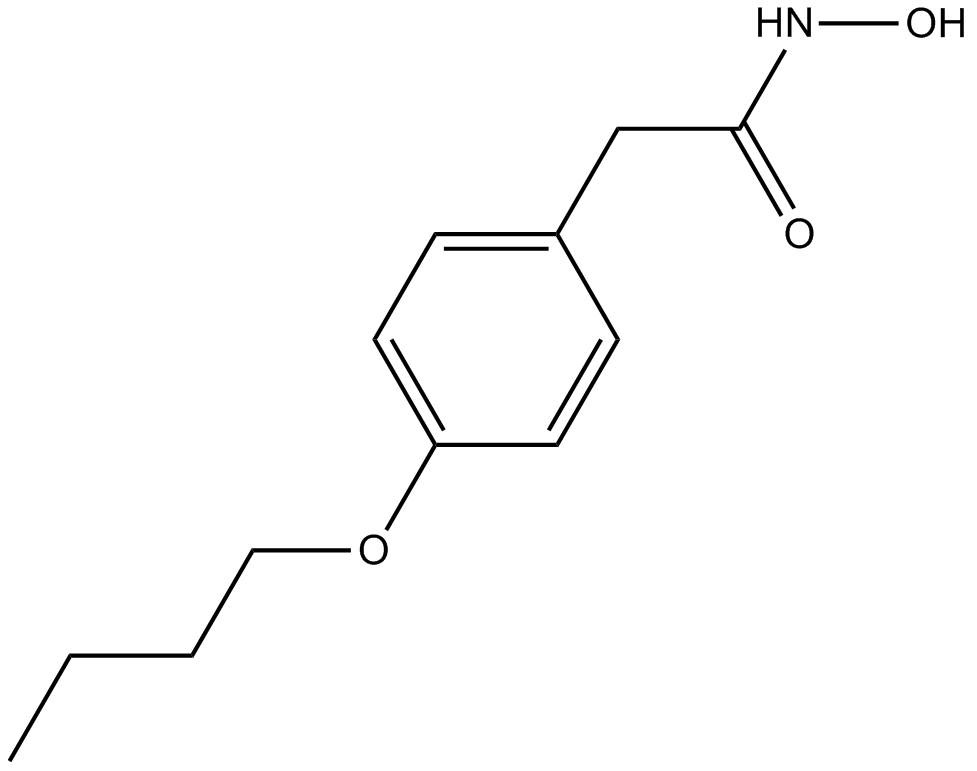
Bufexamac
COX inhibitor
-
GC64761
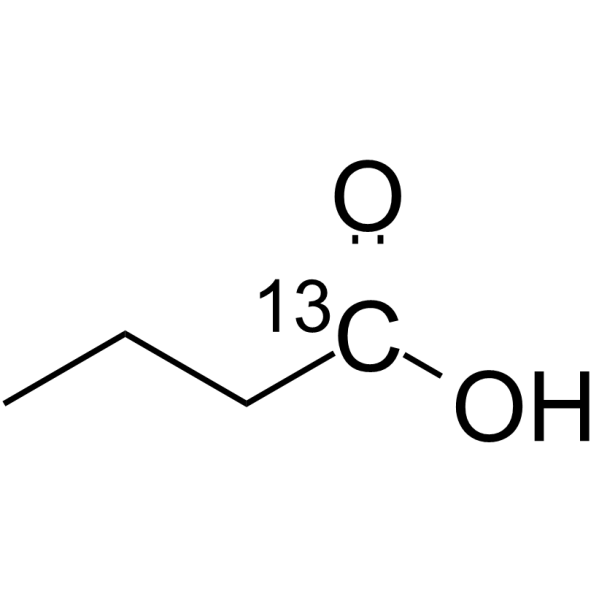
Butyric acid-13C1
-
GC12971
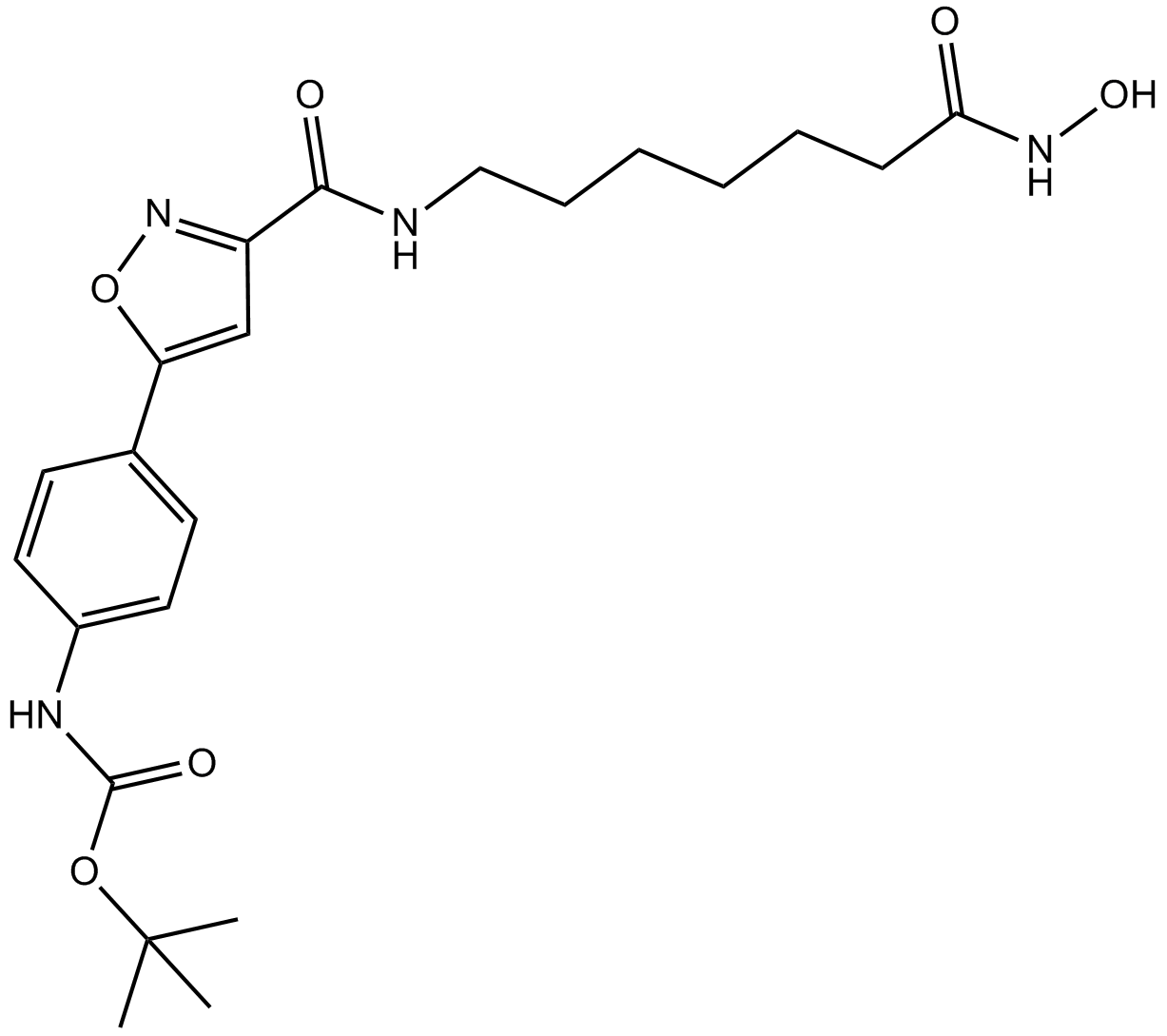
CAY10603
potent and selective inhibitor of HDAC6
-
GC14058
CBHA
CBHA is a potent HDAC inhibitor, exhibiting ID50 values of 10 and 70 nM in vitro for HDAC1 and HDAC3, respectively. CBHA also induces apoptosis and suppresses tumor growth.
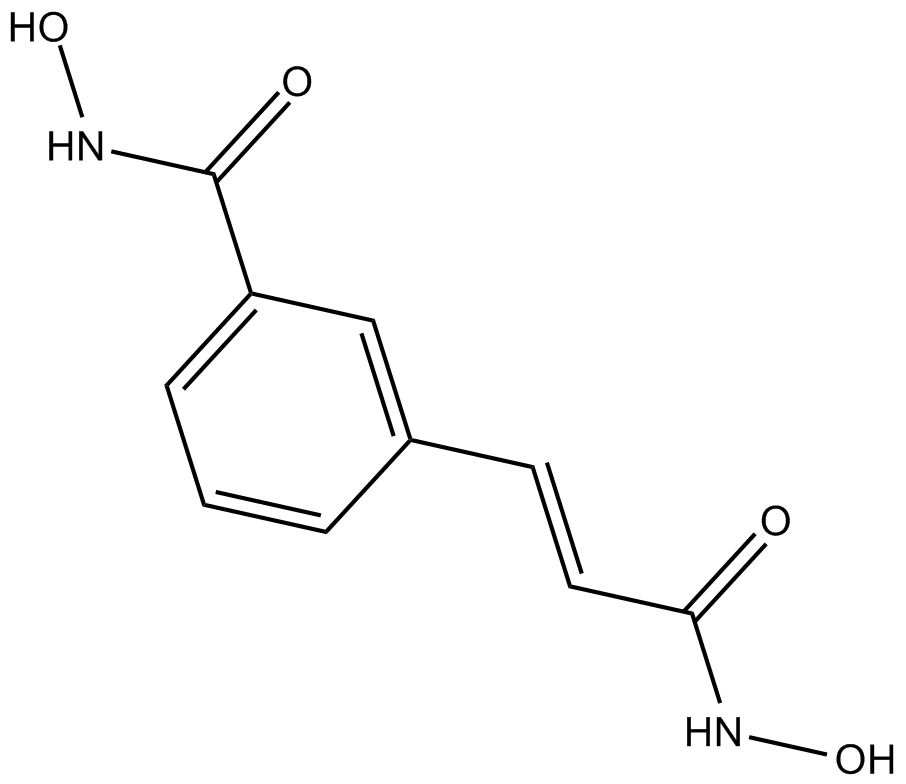
-
GC35668
CG-200745
CG-200745 (CG-200745) is an orally active, potent pan-HDAC inhibitor which has the hydroxamic acid moiety to bind zinc at the bottom of catalytic pocket. CG-200745 inhibits deacetylation of histone H3 and tubulin. CG-200745 induces the accumulation of p53, promotes p53-dependent transactivation, and enhances the expression of MDM2 and p21 (Waf1/Cip1) proteins. CG-200745 enhances the sensitivity of Gemcitabine-resistant cells to Gemcitabine and 5-Fluorouracil (5-FU; ). CG-200745 induces apoptosis and has anti-tumour effects.
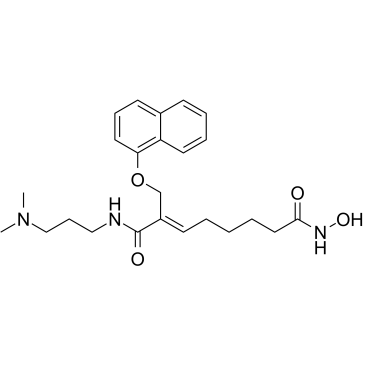
-
GC64942
CHDI-390576
CHDI-390576, a potent, cell permeable and CNS penetrant class IIa histone deacetylase (HDAC) inhibitor with IC50s of 54 nM, 60 nM, 31 nM, 50 nM for class IIa HDAC4, HDAC5, HDAC7, HDAC9, respectively, shows >500-fold selectivity over class I HDACs (1, 2, 3) and ~150-fold selectivity over HDAC8 and the class IIb HDAC6 isoform.
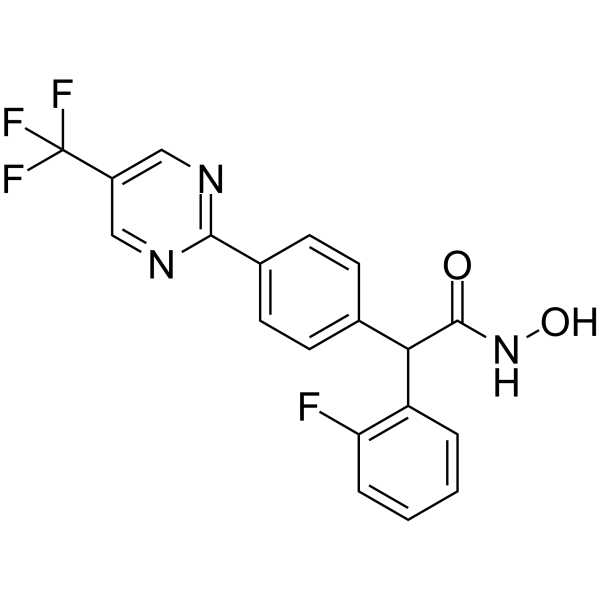
-
GC16042
Chidamide
Chidamide (Chidamide impurity) is an impurity of Chidamide. Chidamide is a potent and orally bioavailable HDAC enzymes class I (HDAC1/2/3) and class IIb (HDAC10) inhibitor.
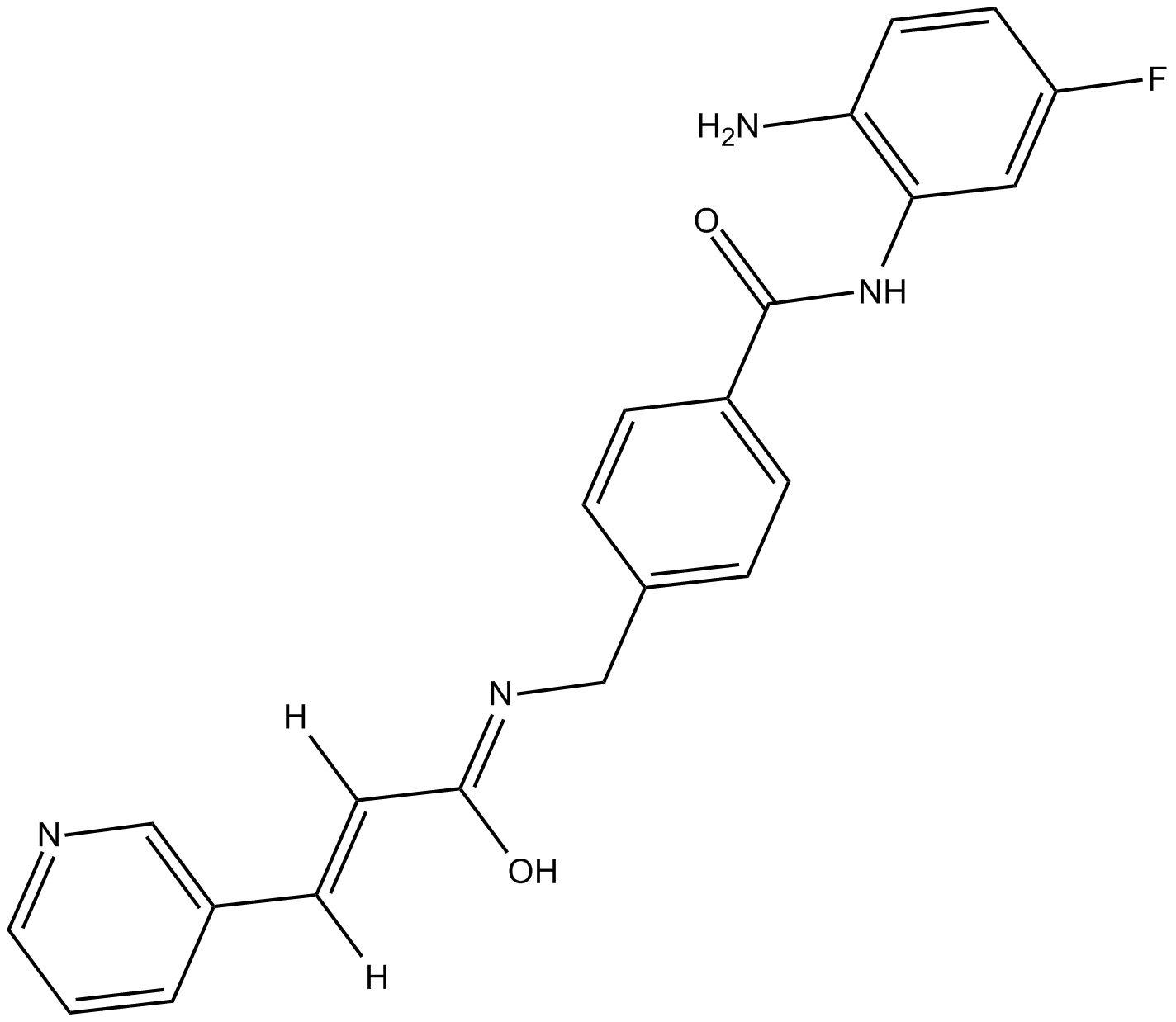
-
GC65426
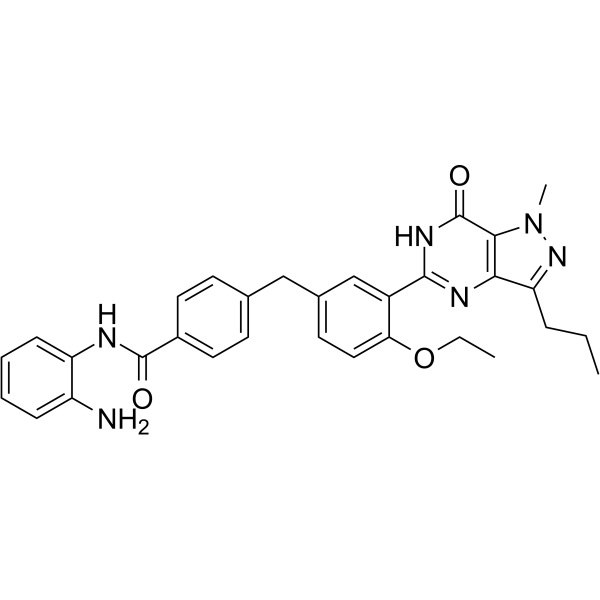
CM-675
CM-675 is a dual phosphodiesterase 5 (PDE5) and class I histone deacetylases-selective inhibitor, with IC50 values of 114 nM and 673 nM for PDE5 and HDAC1, respectively.
-
GC34165
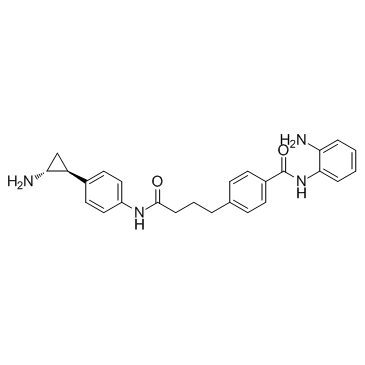
Corin
Corin is a dual inhibitor of histone lysine specific demethylase (LSD1) and histone deacetylase (HDAC), with a Ki(inact) of 110 nM for LSD1 and an IC50 of 147 nM for HDAC1.
-
GC32565
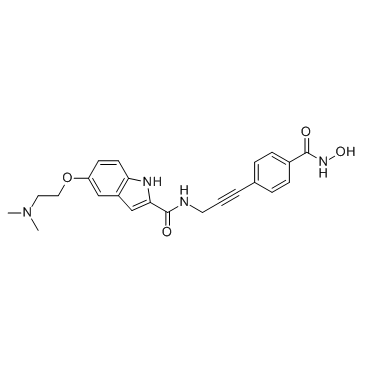
CRA-026440
CRA-026440 is a potent, broad-spectrum HDAC inhibitor.
-
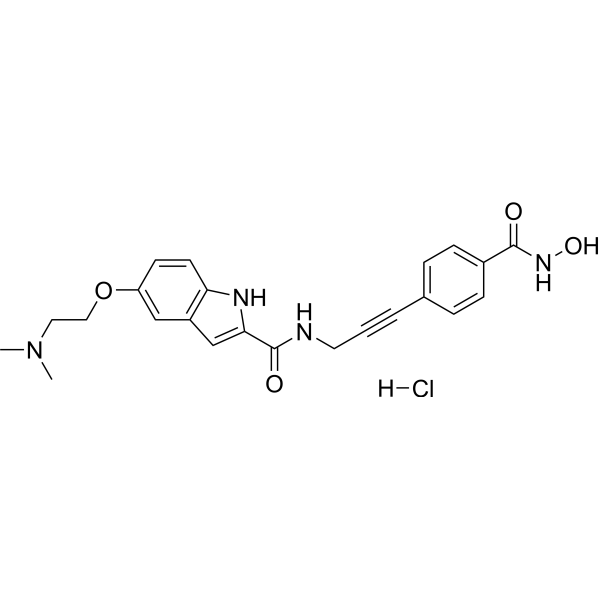
GC67674
CRA-026440 hydrochloride
-
GC38412
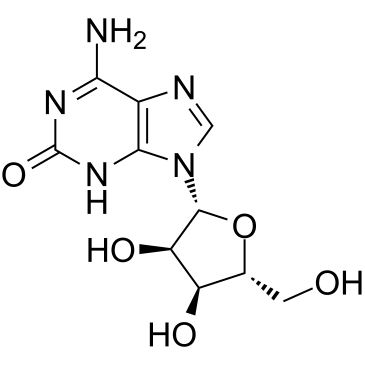
Crotonoside
A guanosine analog with diverse biological activities
-
GC35893
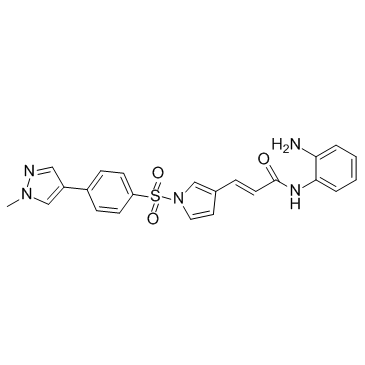
Domatinostat
Domatinostat (4SC-202 free base) is a selective class I HDAC inhibitor with IC50 of 1.20 μM, 1.12 μM, and 0.57 μM for HDAC1, HDAC2, and HDAC3, respectively. It also displays inhibitory activity against Lysine specific demethylase 1 (LSD1).
-
GC19129
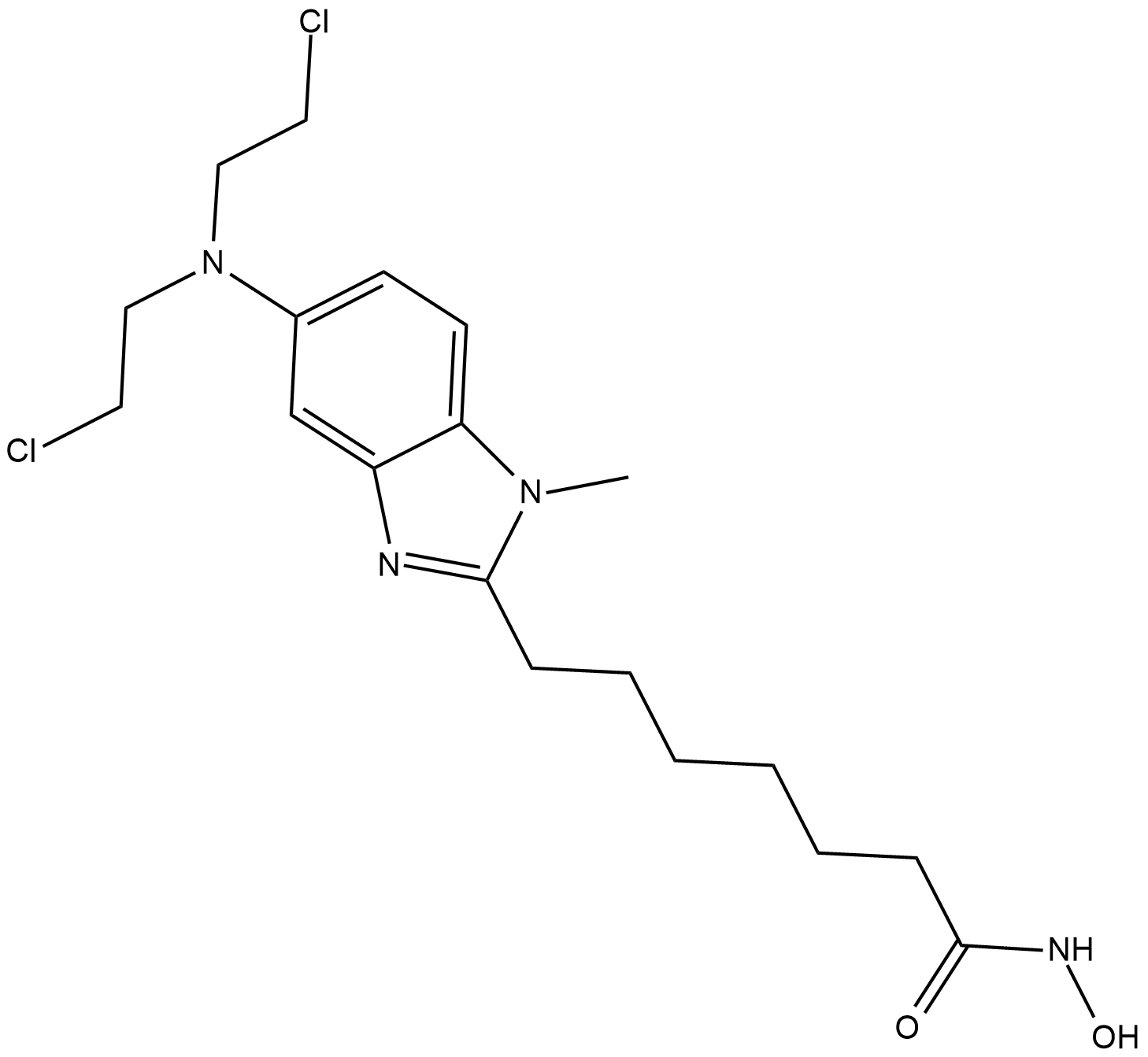
EDO-S101
EDO-S101 is a pan HDAC inhibitor; inhibits HDAC1, HDAC2 and HDAC3 with IC50 values of 9, 9 and 25 nM, respectively.
-
GC64191
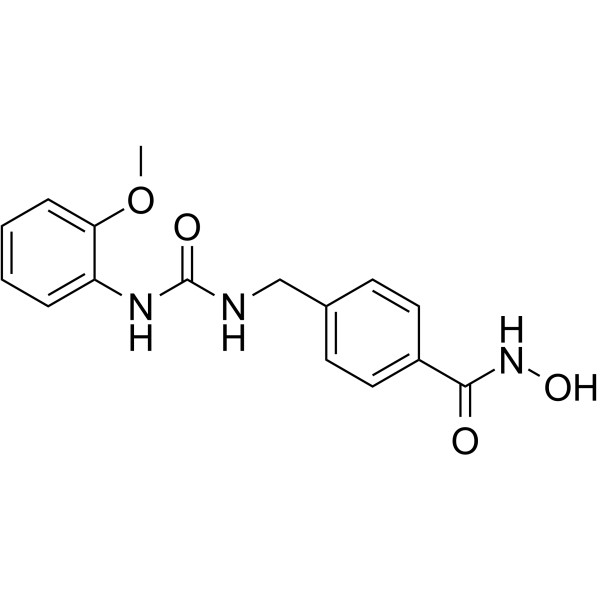
Elevenostat
Elevenostat (JB3-22) is a selective HDAC11 inhibitor (IC50=0.235??M). Anti-multiple myeloma (MM) activity.
-
GC65206
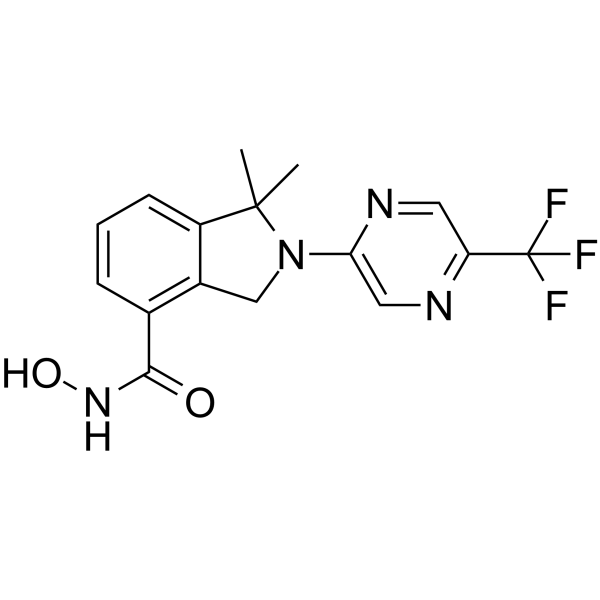
FT895
FT895 is a potent and selective HDAC11 inhibitor with an IC50 of 3 nM.
-
GC33050
Givinostat (ITF-2357)
Givinostat (ITF-2357) (ITF-2357) is a HDAC inhibitor with an IC50 of 198 and 157 nM for HDAC1 and HDAC3, respectively.

-
GC33159
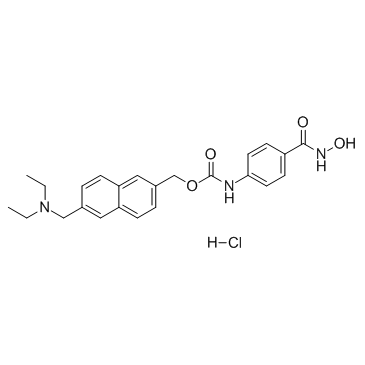
Givinostat hydrochloride (ITF-2357 hydrochloride)
Givinostat (ITF-2357) hydrochloride is a HDAC inhibitor with an IC50 of 198 and 157 nM for HDAC1 and HDAC3, respectively.
-
GC18402
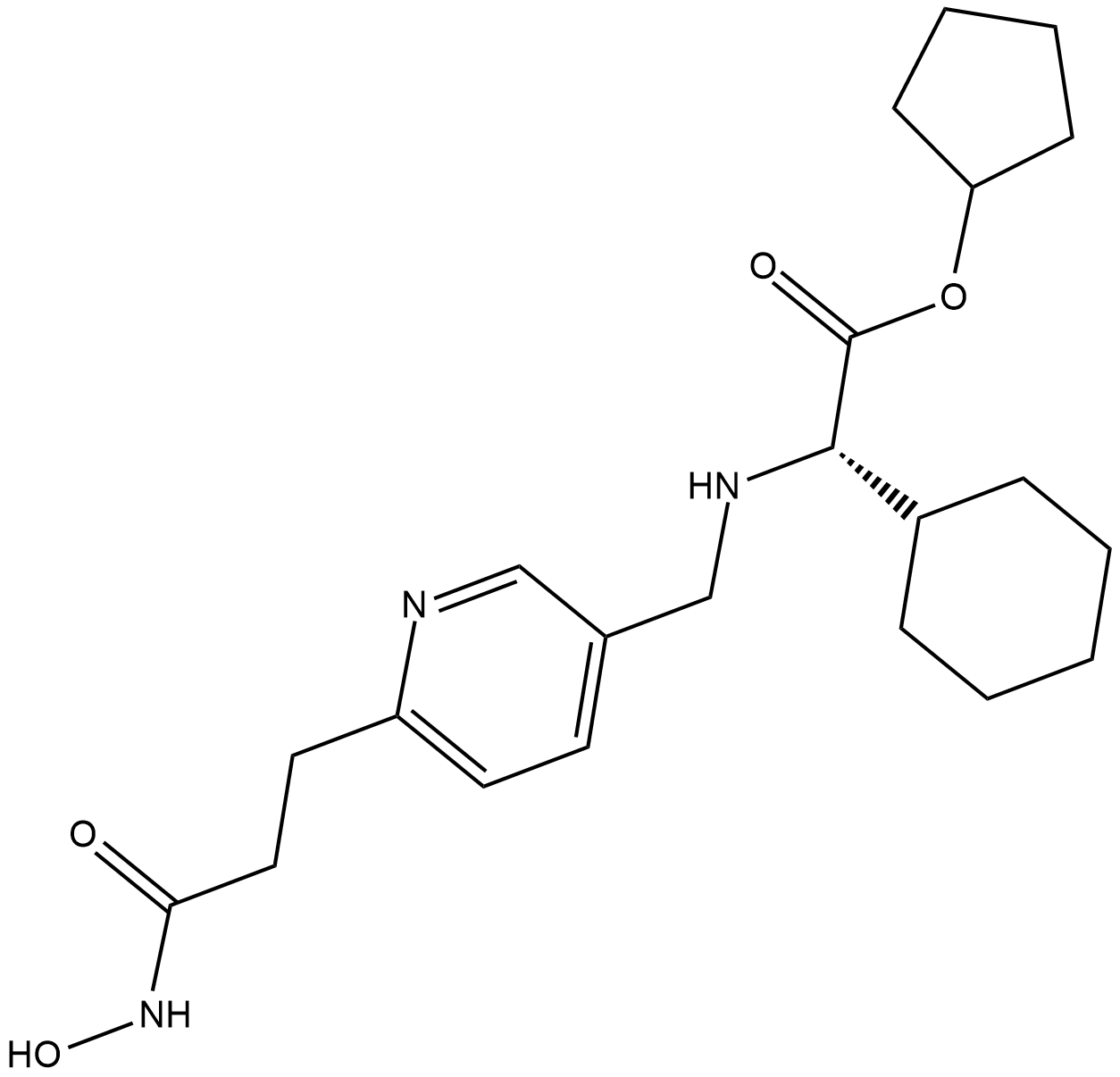
GSK3117391
GSK3117391 is a histone deacetylase (HDAC) inhibitor, extracted from patent WO/2008040934 A1.
-
GC19190
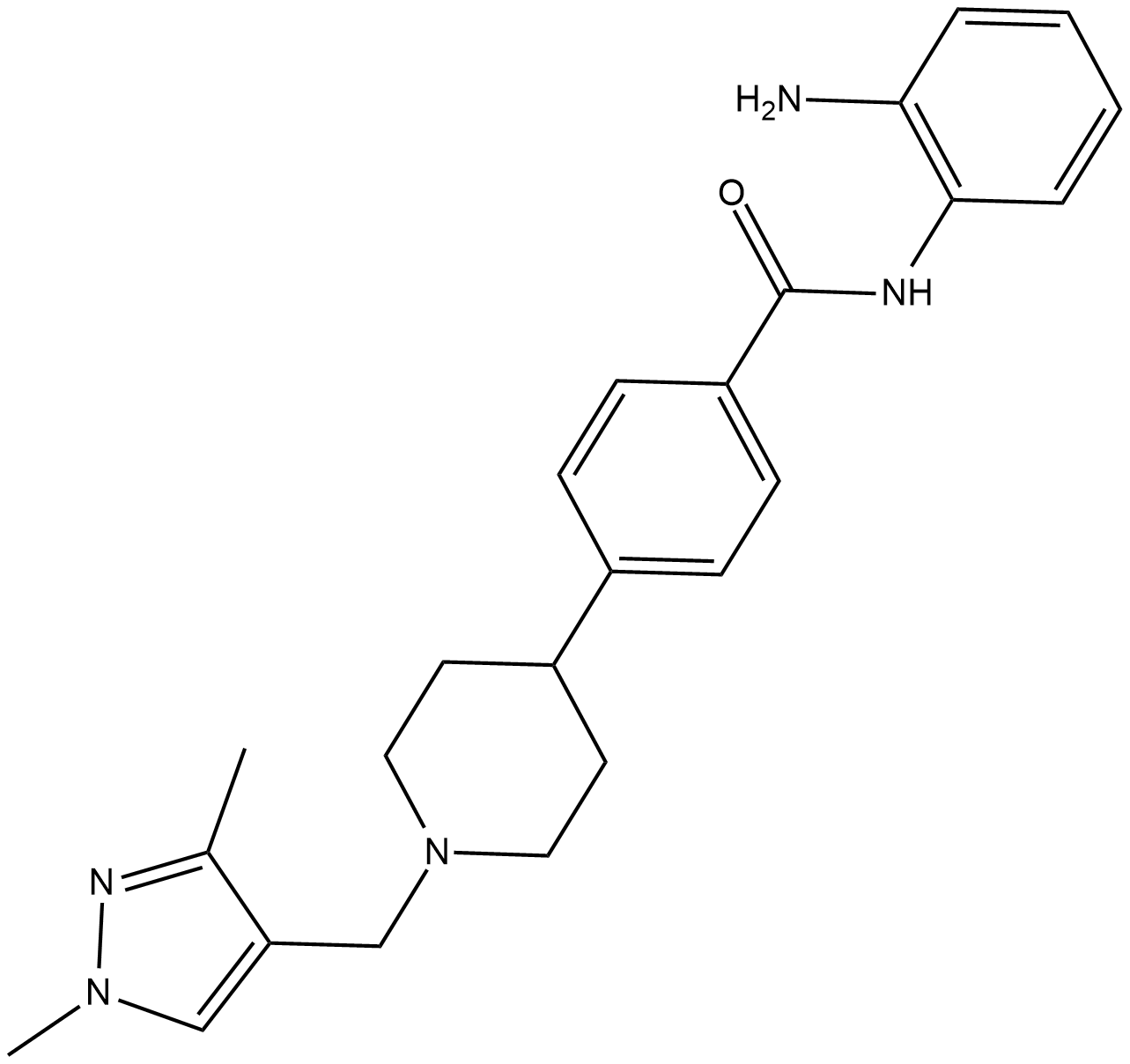
HDAC-IN-4
HDAC-IN-4 is a potent, selective and orally active class I HDAC inhibitor with IC50s of 63 nM, 570 nM and 550 nM for HDAC1, HDAC2 and HDAC3, respectively. HDAC-IN-4 has no activity against HDAC class II. HDAC-IN-4 has antitumor activity.
-
GC66052
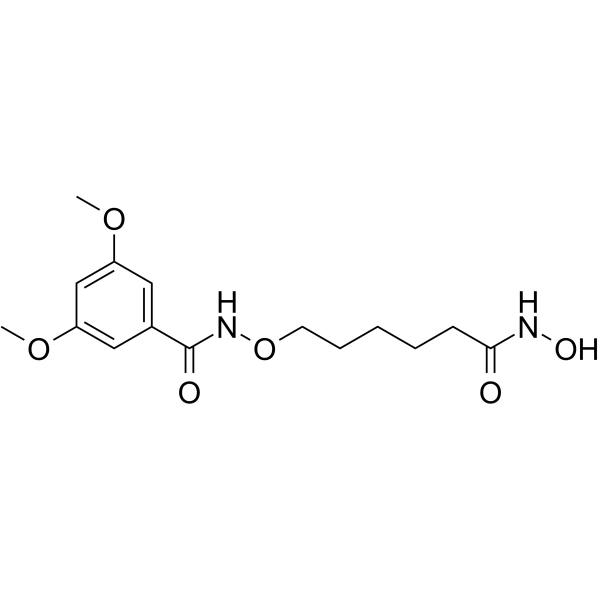
HDAC-IN-40
HDAC-IN-40 is a potent alkoxyamide-based HDAC inhibitor with Ki values of 60 nM and 30 nM for HDAC2 and HDAC6, respectively. HDAC-IN-40 had antitumor effects.
-
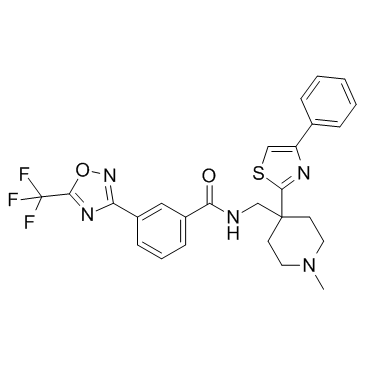
GC33395
HDAC-IN-5
HDAC-IN-5 is a histone deacetylase (HDAC) inhibitor.
-
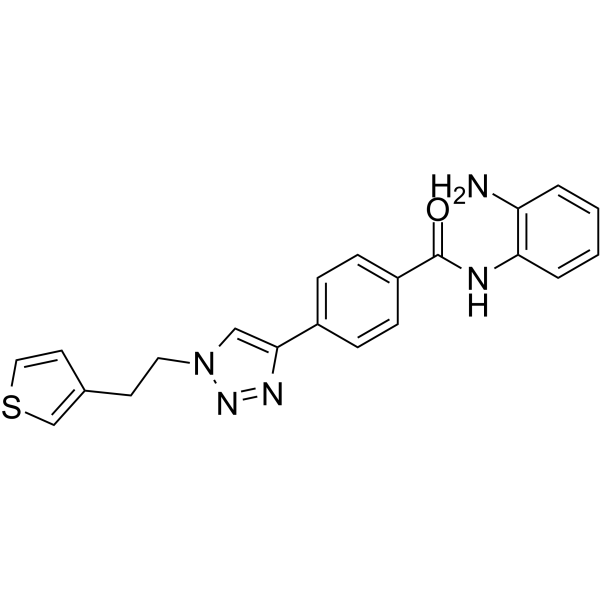
GC68010
HDAC3-IN-T247
-
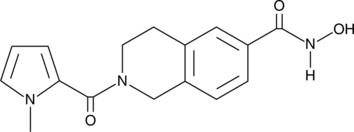
GC41085
HDAC6 Inhibitor
HDAC6 Inhibitor is a potent and selective HDAC6 inhibitor (IC50=36 nM). HDAC6 Inhibitor weakly inhibits other HDAC isoforms. HDAC6 Inhibitor inhibits acyl-tubulin accumulation in cells with an IC50 value of 210 nM.
-
GC33317
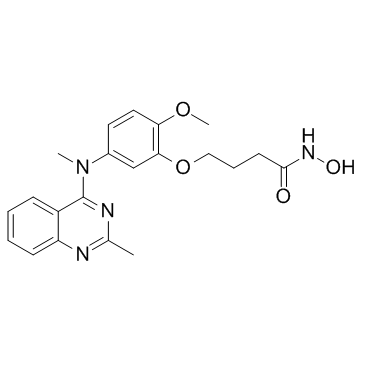
HDAC6-IN-1
HDAC6-IN-1 is a potent and selective inhibitor for HDAC6 with an IC50 of 17 nM and shows 25-fold and 200-fold selectivity relative to HDAC1 (IC50=422 nM) and HDAC8 (IC50=3398 nM), respectively.
-
GC19189
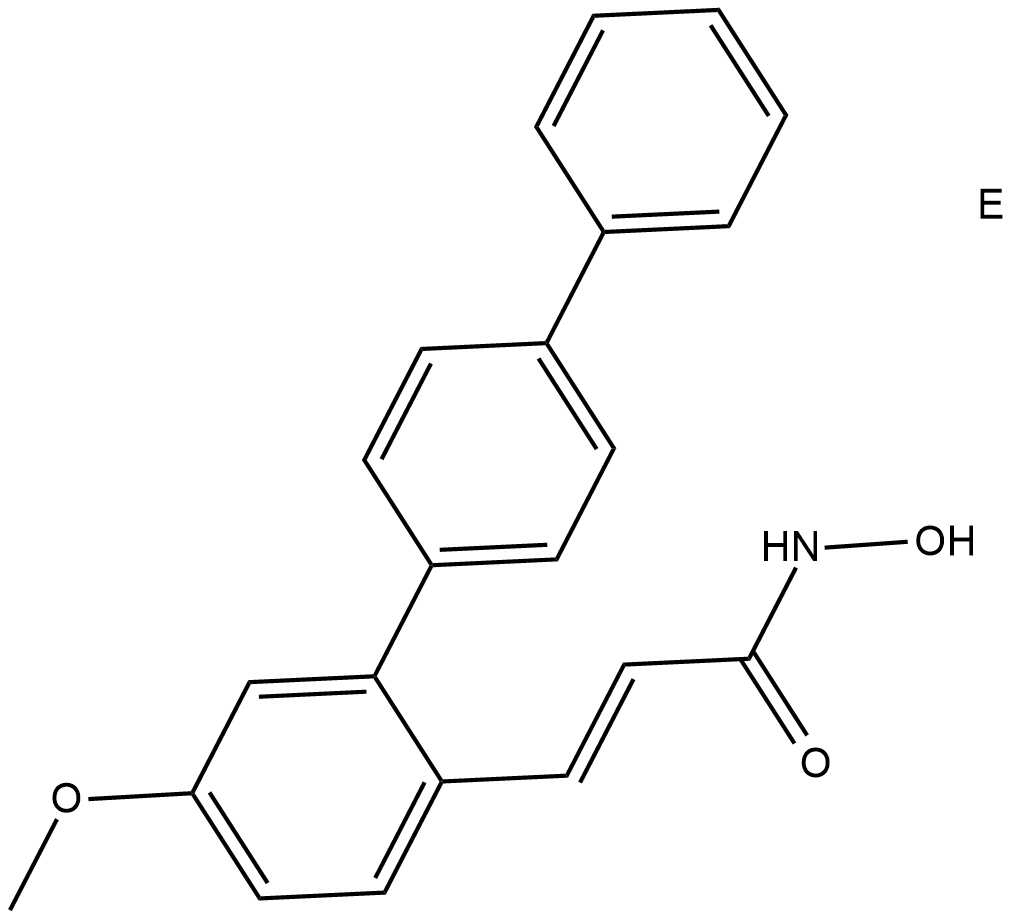
HDAC8-IN-1
HDAC8-IN-1 is a HDAC8 inhibitor with an IC50 of 27.2 nM.
-
GC65460
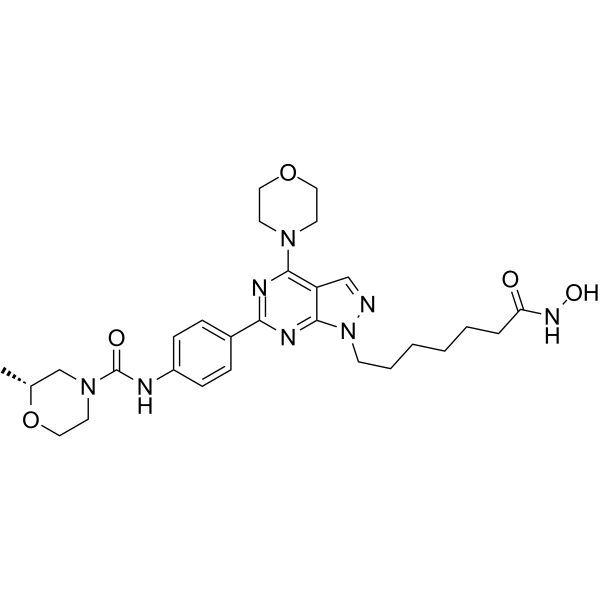
HDACs/mTOR Inhibitor 1
HDACs/mTOR Inhibitor 1 is a dual Histone Deacetylases (HDACs) and mammalian target of Rapamycin (mTOR) target inhibitor for treating hematologic malignancies, with IC50s of 0.19 nM, 1.8 nM, 1.2 nM and >500 nM for HDAC1, HDAC6, mTOR and PI3Kα, respectively. HDACs/mTOR Inhibitor 1 stimulates cell cycle arrest in G0/G1 phase and induce tumor cell apoptosis with low toxicity in vivo.
-
GC12334
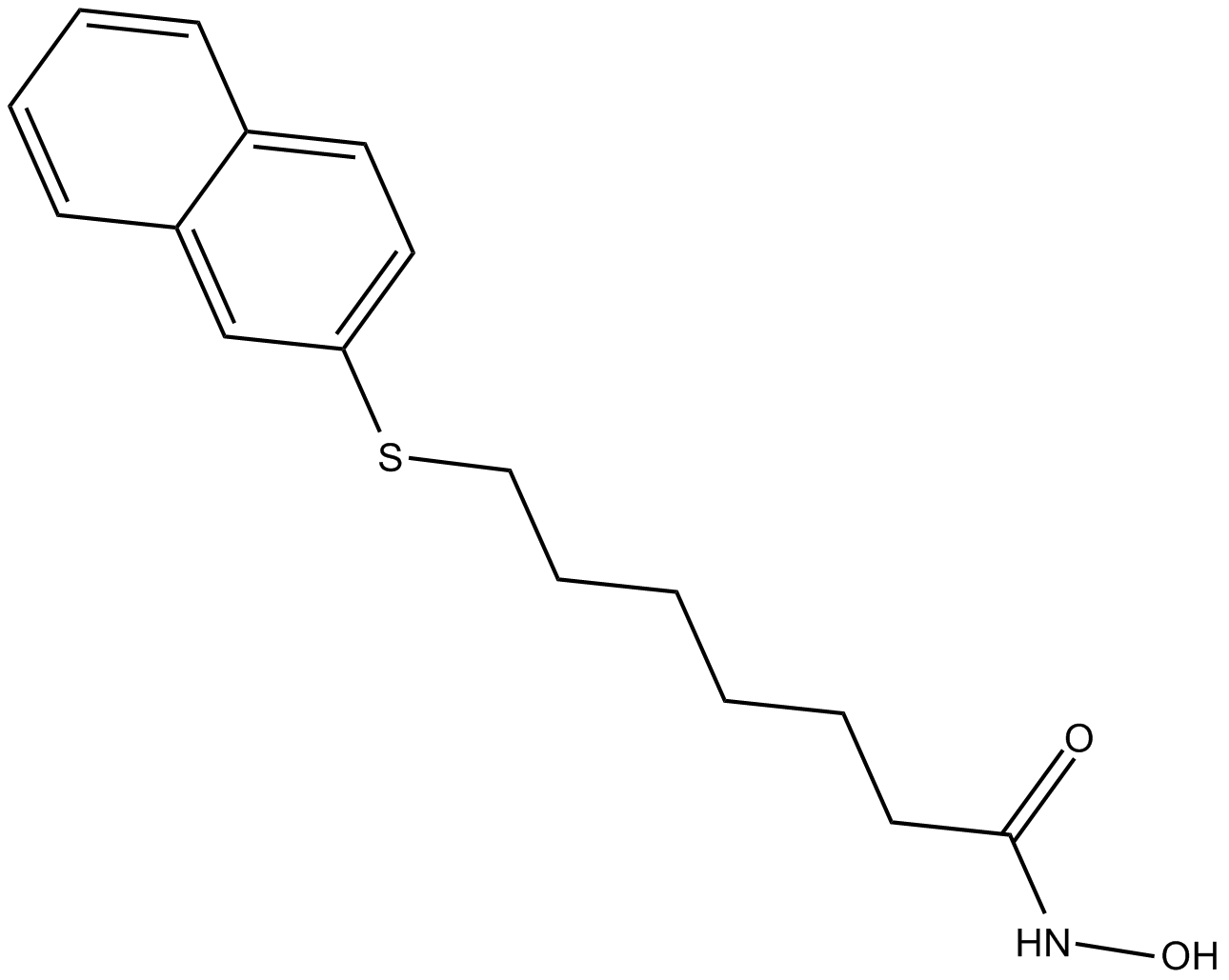
HNHA
HDAC inhibitor
-
GC11574
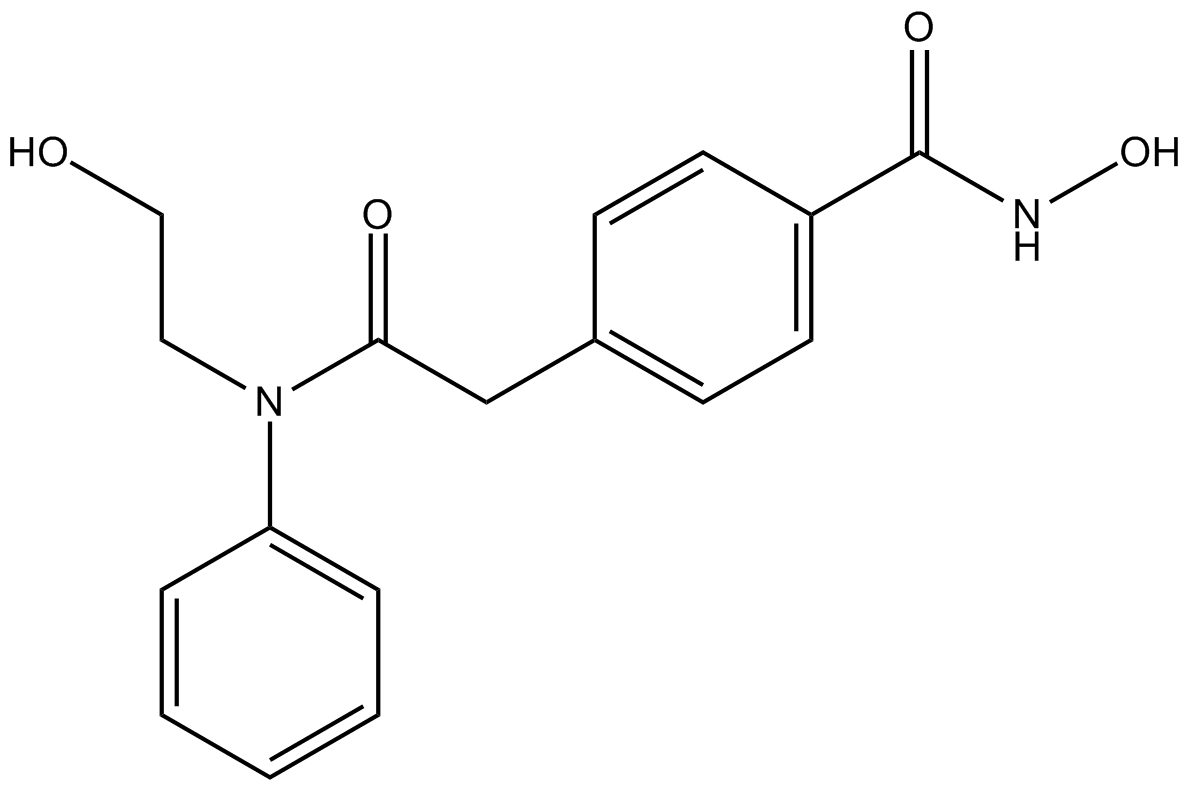
HPOB
HDAC6 inhibitor, potent and selective
-
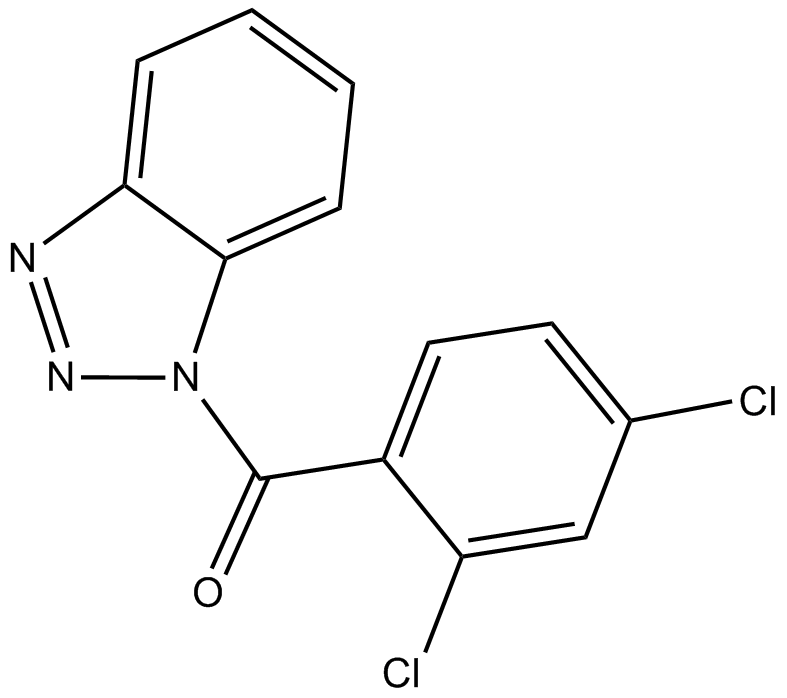
GC14597
ITSA-1 (ITSA1)
ITSA-1 (ITSA1) is an activator of histone deacetylase (HDAC), and counteract trichostatin A (TSA)-induced cell cycle arrest, histone acetylation, and transcriptional activation.
-
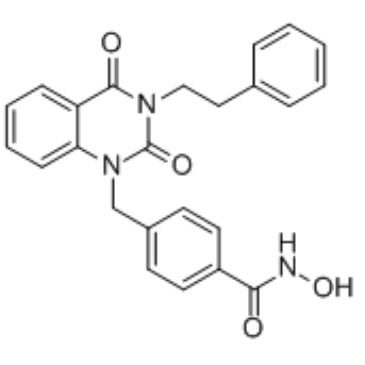
GC34629
J22352
J22352 is a PROTAC (proteolysis-targeting chimeras)-like and highly selective HDAC6 inhibitor with an IC50 value of 4.7 nM. J22352 promotes HDAC6 degradation and induces anticancer effects by inhibiting autophagy and eliciting the antitumor immune response in glioblastoma cancers, and leading to the restoration of host antitumor activity by reducing the immunosuppressive activity of PD-L1.
-
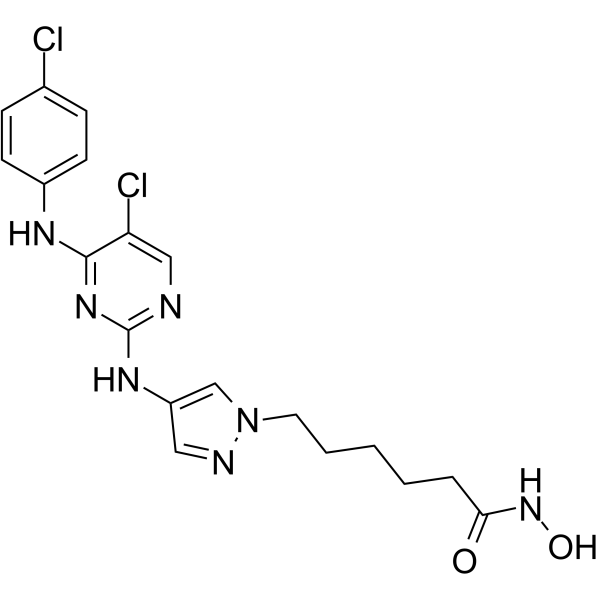
GC64959
JAK/HDAC-IN-1
JAK/HDAC-IN-1 is a potent JAK2/HDAC dual inhibitor, exhibits antiproliferative and proapoptotic activities in several hematological cell lines. JAK/HDAC-IN-1 shows IC50s of 4 and 2 nM for JAK2 and HDAC, respectively.
-
GC25552
KT-531
KT-531 (KT531) is a potent, selective HDAC6 inhibitor with IC50 of 8.5 nM, displays 39-fold selectivity over other HDAC isoforms.

-
GC11949
MI-192
inhibitor of histone deacetylases (HDACs)
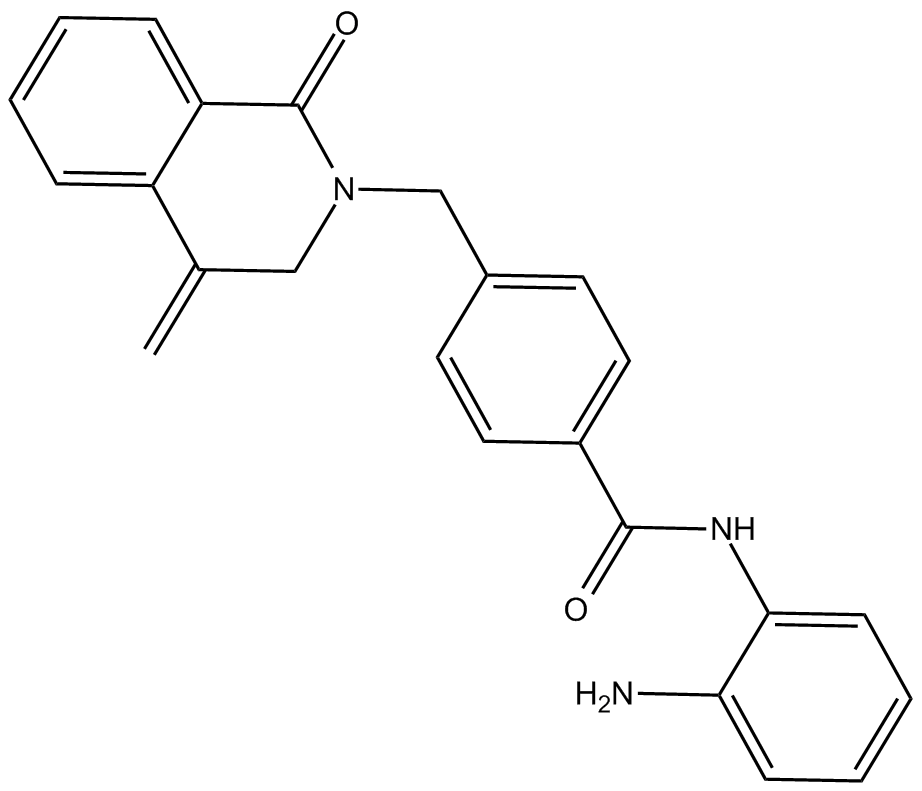
-
GC64729
MPT0B390
MPT0B390 is an arylsulfonamide-based derivative with potent HDAC inhibitory ability. MPT0B390, TIMP3 inducer, inhibits tumor growth, metastasis and angiogenesis. MPT0B390 shows antiproliferative activity against human colon cancer cell line HCT116 with the GI50 of 0.03 μM.
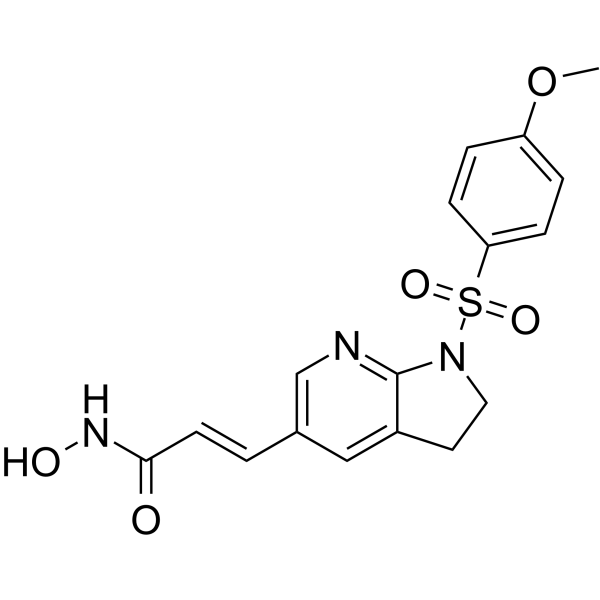
-
GC65965
MPT0E028
MPT0E028 is an orally active and selective HDAC inhibitor with IC50s of 53.0 nM, 106.2 nM, 29.5 nM for HDAC1, HDAC2 and HDAC6, respectively. MPT0E028 reduces the viability of B-cell lymphomas by inducing apoptosis and possesses potent direct Akt targeting ability and reduces Akt phosphorylation in B-cell lymphoma. MPT0E028 has good anticancer activity.
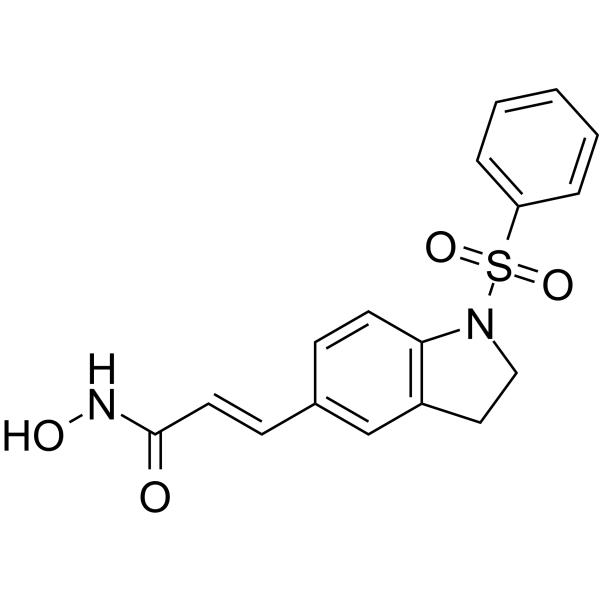
-
GC36690
Nampt-IN-3
Nampt-IN-3 (Compound 35) simultaneously inhibit nicotinamide phosphoribosyltransferase (NAMPT) and HDAC with IC50s of 31 nM and 55 nM, respectively. Nampt-IN-3 effectively induces cell apoptosis and autophagy and ultimately leads to cell death.
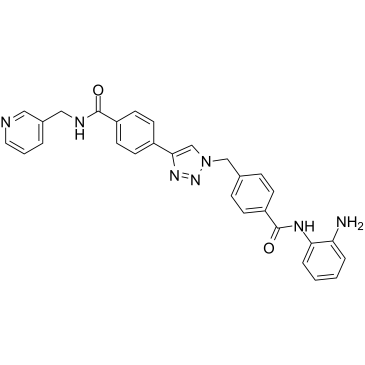
-
GC36691
Nanatinostat
Nanatinostat (CHR-3996) is a potent, class I selective and orally active histone deacetylase (HDAC) inhibitor with an IC50 of 8 nM.
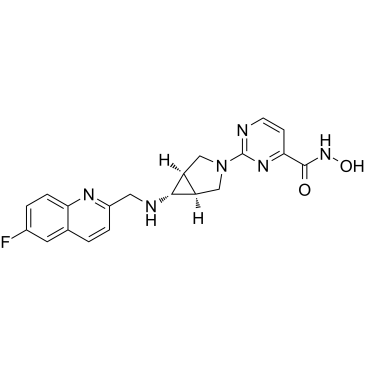
-
GC13764
Nexturastat A
HDAC6 inhibitor,highly potent and selective
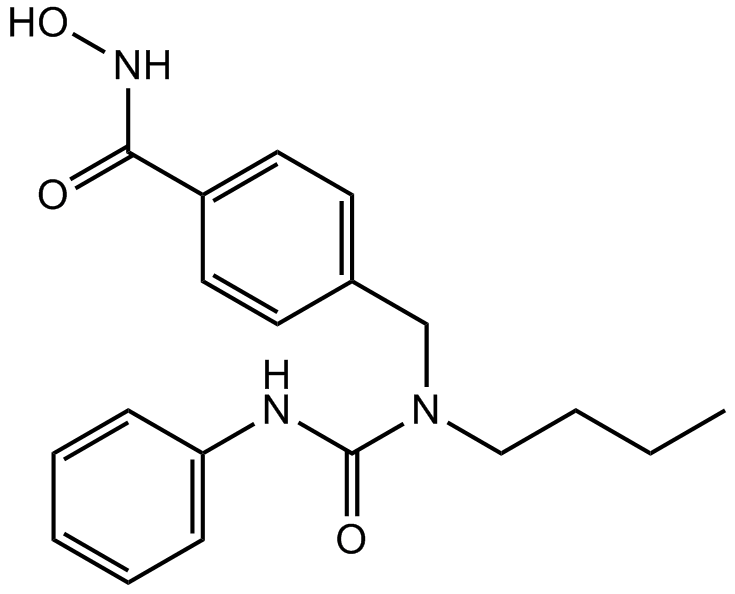
-
GC32964
NKL 22
A selective HDAC1/3 inhibitor
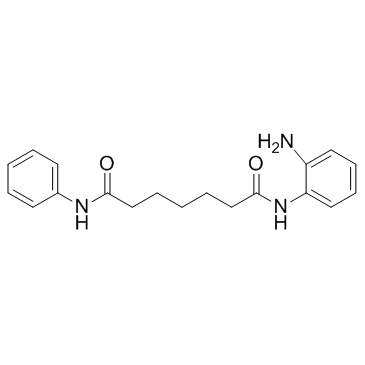
-
GC13925
Nullscript
negative control of scriptaid, HDAC inhibitor
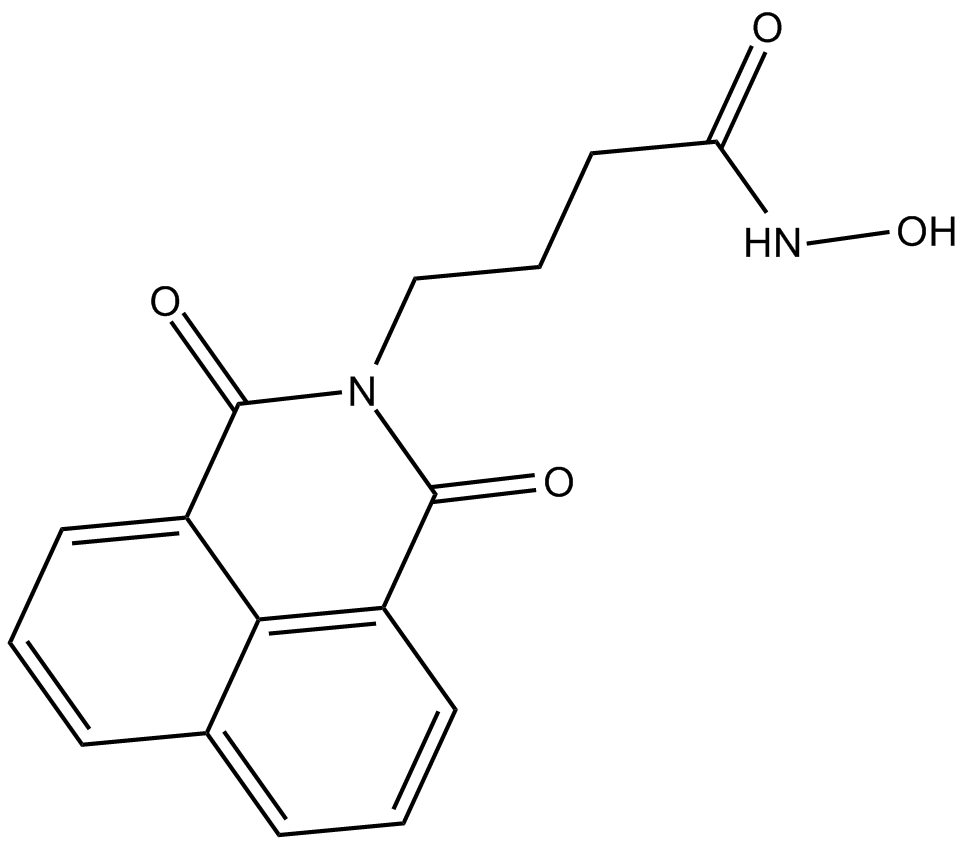
-
GC11360
ORY-1001
Selective inhibitor of KDM1A.
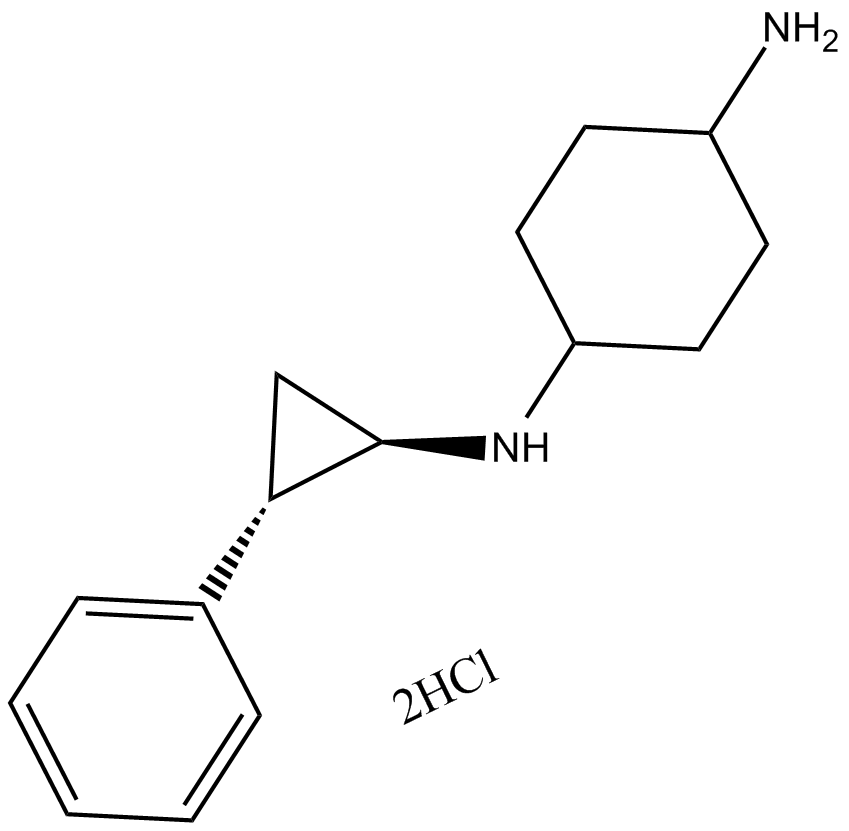
-
GC17472
Oxamflatin
HADC inhibitor
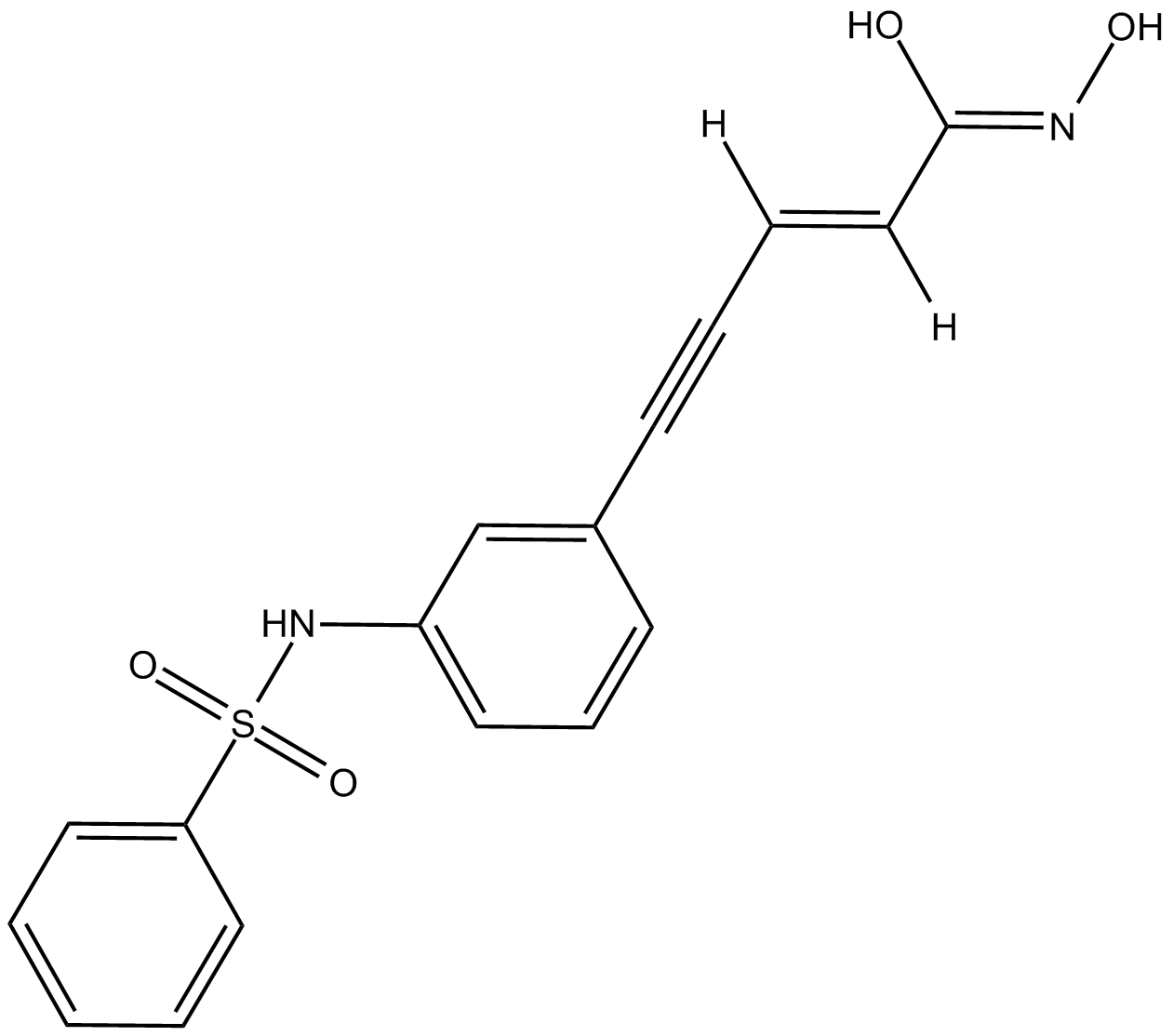
-
GC36905
PI3K/HDAC-IN-1
PI3K/HDAC-IN-1 is a potent dual inhibitor of PI3K/HDAC, potently inhibits PI3Kδ and HDAC1 with IC50s of 8.1 nM and 1.4 nM, respectively.
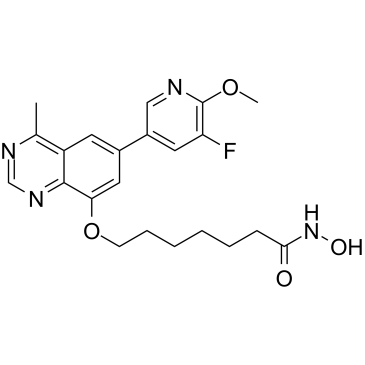
-
GC68474
Pivanex
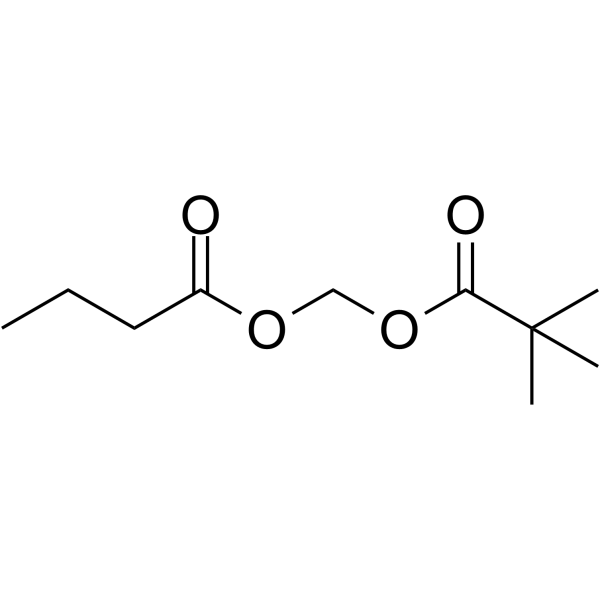
-
GC33115
Pomiferin (NSC 5113)
Pomiferin (NSC 5113) (NSC 5113) acts as an potential inhibitor of HDAC, with an IC50 of 1.05 μM, and also potently inhibits mTOR (IC50, 6.2 μM).
-
GC33290
Remetinostat (SHP-141)
Remetinostat (SHP-141) (SHP-141) is a hydroxamic acid-based inhibitor of histone deacetylase enzymes (HDAC) which is under development for the treatment of cutaneous T-cell lymphoma.
-
GC14285
RGFP966
RGFP966 is HDAC3 inhibitor with an IC50 of 0.08 μM. RGFP966 can suppress inflammatory responses.

-
GC14439
Santacruzamate A (CAY10683)
Santacruzamate A (CAY10683) (CAY-10683) is a potent and selective HDAC2 inhibitor with an IC50 of 119 pM.
-
GC64684
SB-429201
SB-429201 is a potent and selective HDAC1 (IC50~1.5 μM). SB-429201 displays at least a 20-fold preference for HDAC1 inhibition over HDAC3 and HDAC8.
-
GC31511
Sinapinic acid (Sinapic acid)
Sinapinic acid (Sinapic acid) (Sinapic acid) is a phenolic compound isolated from Hydnophytum formicarum Jack.
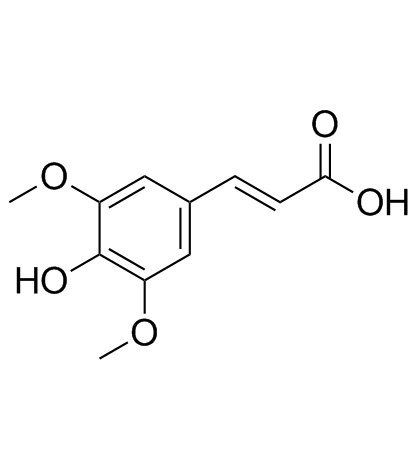
-
GC37643
SIS17
SIS17 is a mammalian histone deacetylase 11 (HDAC 11) inhibitor with an IC50 value of 0.83 μM, inhibits the demyristoylation HDAC11 substrate, serine hydroxymethyl transferase 2, without inhibiting other HDACs.
-
GC69917
Snail/HDAC-IN-1
Snail/HDAC-IN-1 is an effective dual-target inhibitor of Snail/HDAC. It has strong inhibitory activity against HDAC1 with an IC50 of 0.405 μM and a strong inhibitory effect on Snail with a Kd of 0.18 μM. Snail/HDAC-IN-1 increases histone H4 acetylation in HCT-116 cells and reduces the expression of Snail protein, thereby inducing apoptosis in cells.
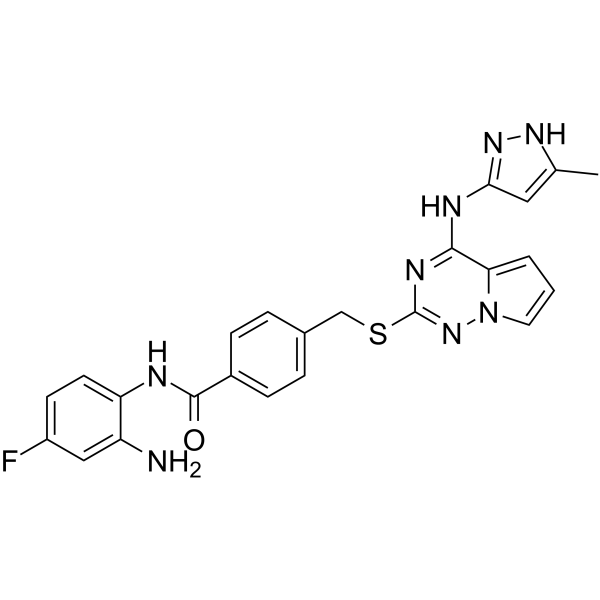
-
GC15857
Sodium butyrate
Histone deacetylase inhibitor
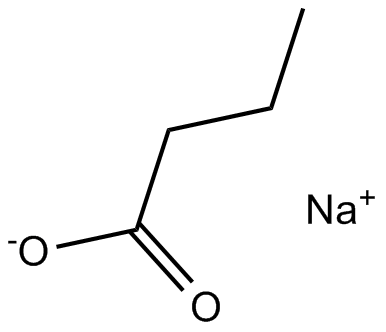
-
GC38137
Splitomicin
Inhibitor of yeast Sir2p
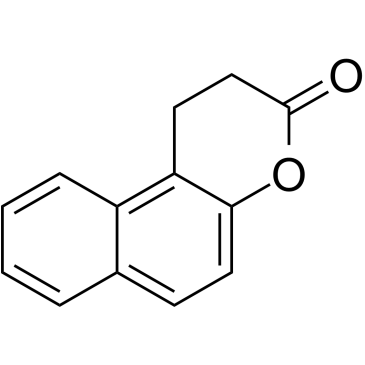
-
GC19337
SR-4370
SR-4370 is an inhibitor of HDAC, with IC50s of 0.13 uM, 0.58 uM, 0.006 uM, 2.3 uM, and 3.4 uM for HDAC1, HDAC2, HDAC3, HDAC8, and HDAC6, respectively.
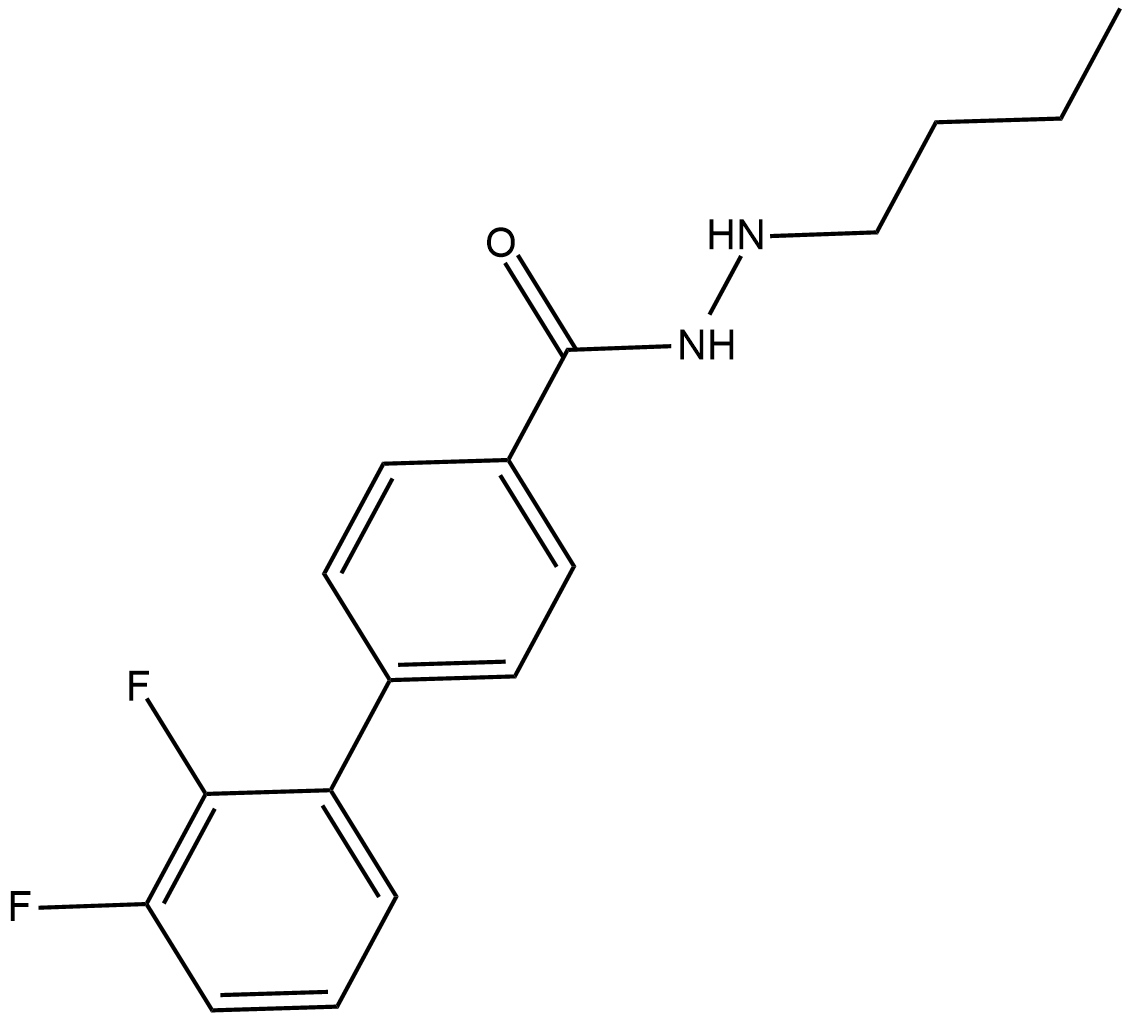
-
GC14016
Sulforaphane
Sulforaphane (SFN) known as [1-isothiocyanato-4-(methylsulfinyl)butane].
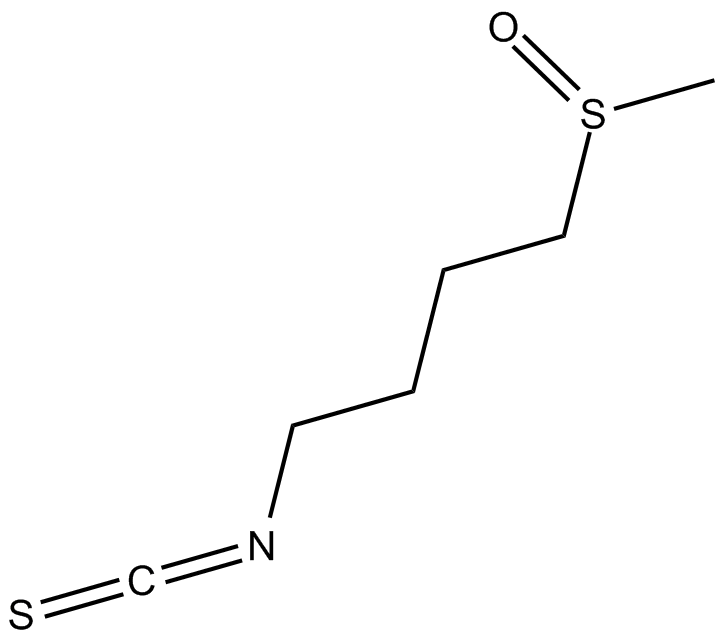
-
GC64968
SW-100
SW-100, a selective histone deacetylase 6 (HDAC6) inhibitor with an IC50 of 2.3 nM, shows at least 1000-fold selectivity for HDAC6 relative to all other HDAC isozymes.

-
GC37753
Tefinostat
Tefinostat (CHR-2845) is a monocyte/macrophage targeted histone deacetylase (HDAC) inhibitor. Tefinostat can be cleaved into active acid CHR-2847 by the intracellular esterase human carboxylesterase-1 (hCE-1). Tefinostat can be used for the research of leukaemias.
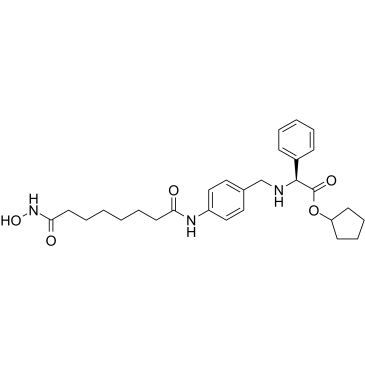
-
GC19406
TH34
TH34 is a potent HDAC6/8/10 inhibitor
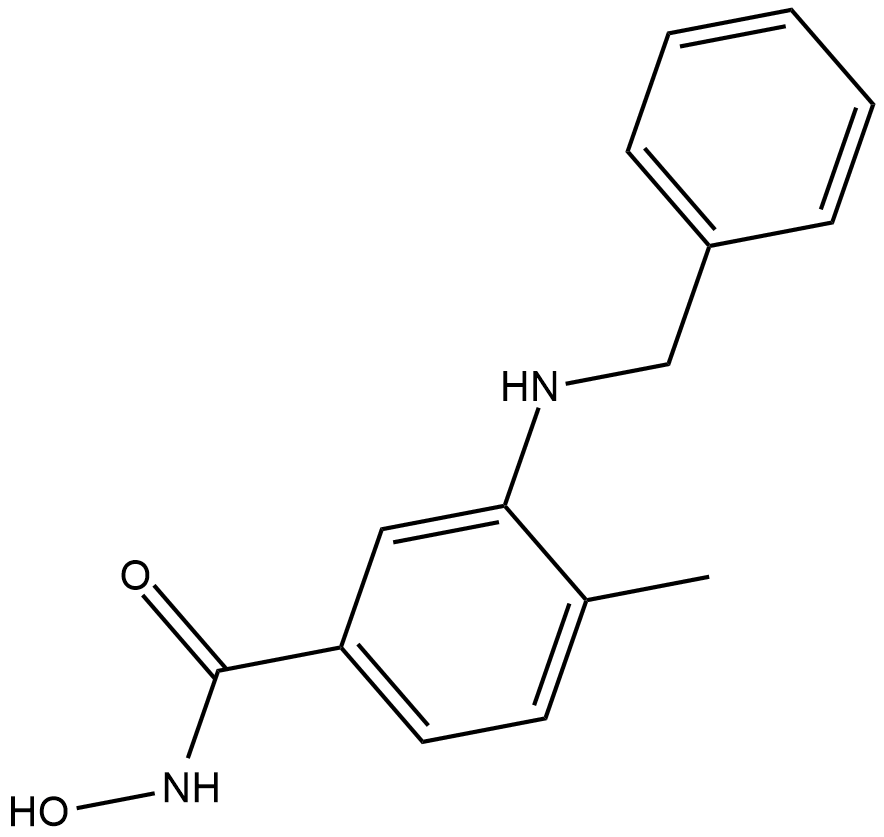
-
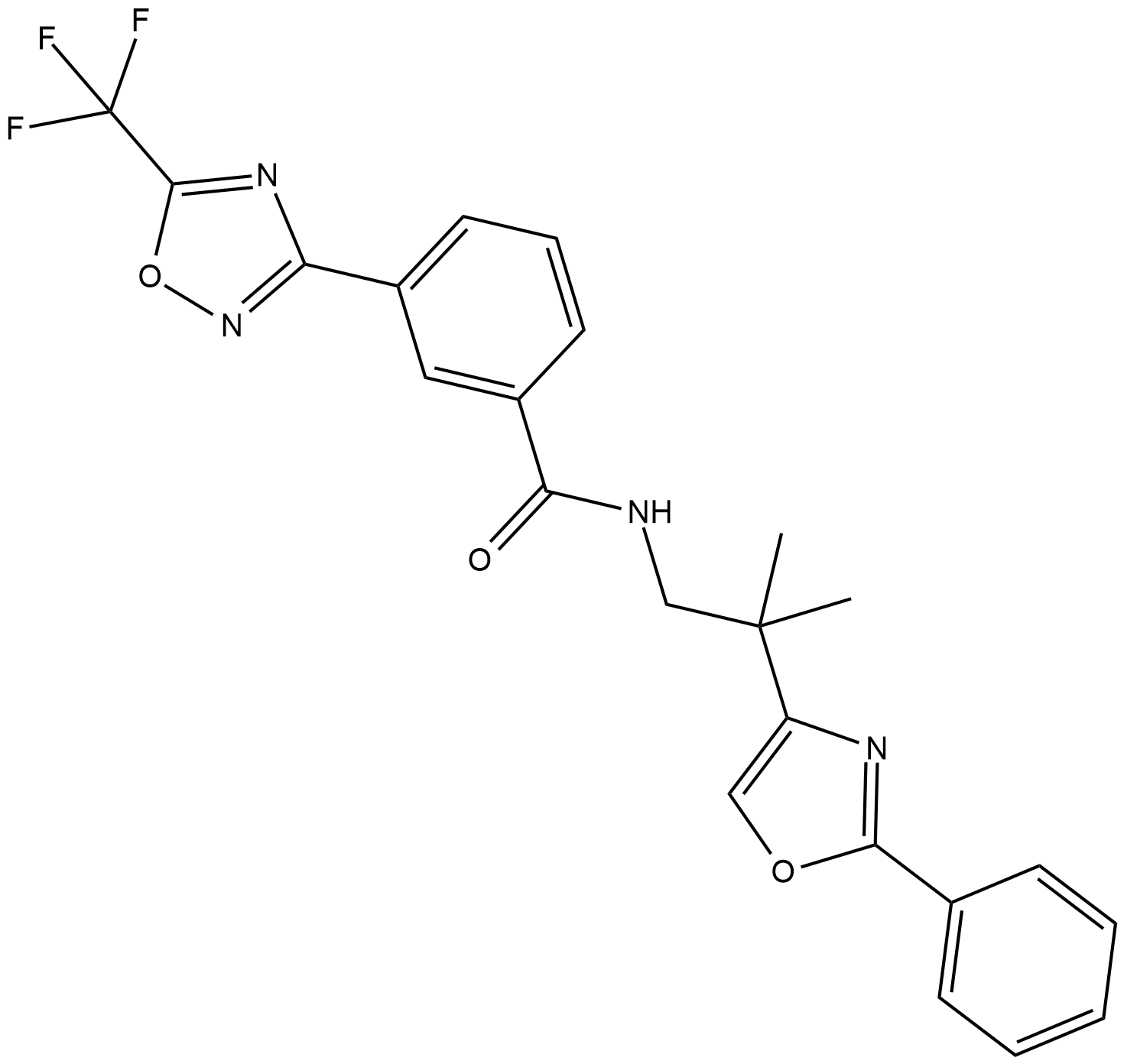
GC19360
TMP195
TMP195 is a potent and selective class IIa HDAC inhibitor with IC50s of 59 nM, 60 nM, 26 nM and 15 nM for HDAC4, HDAC5, HDAC7 and HDAC9, respectively.
-
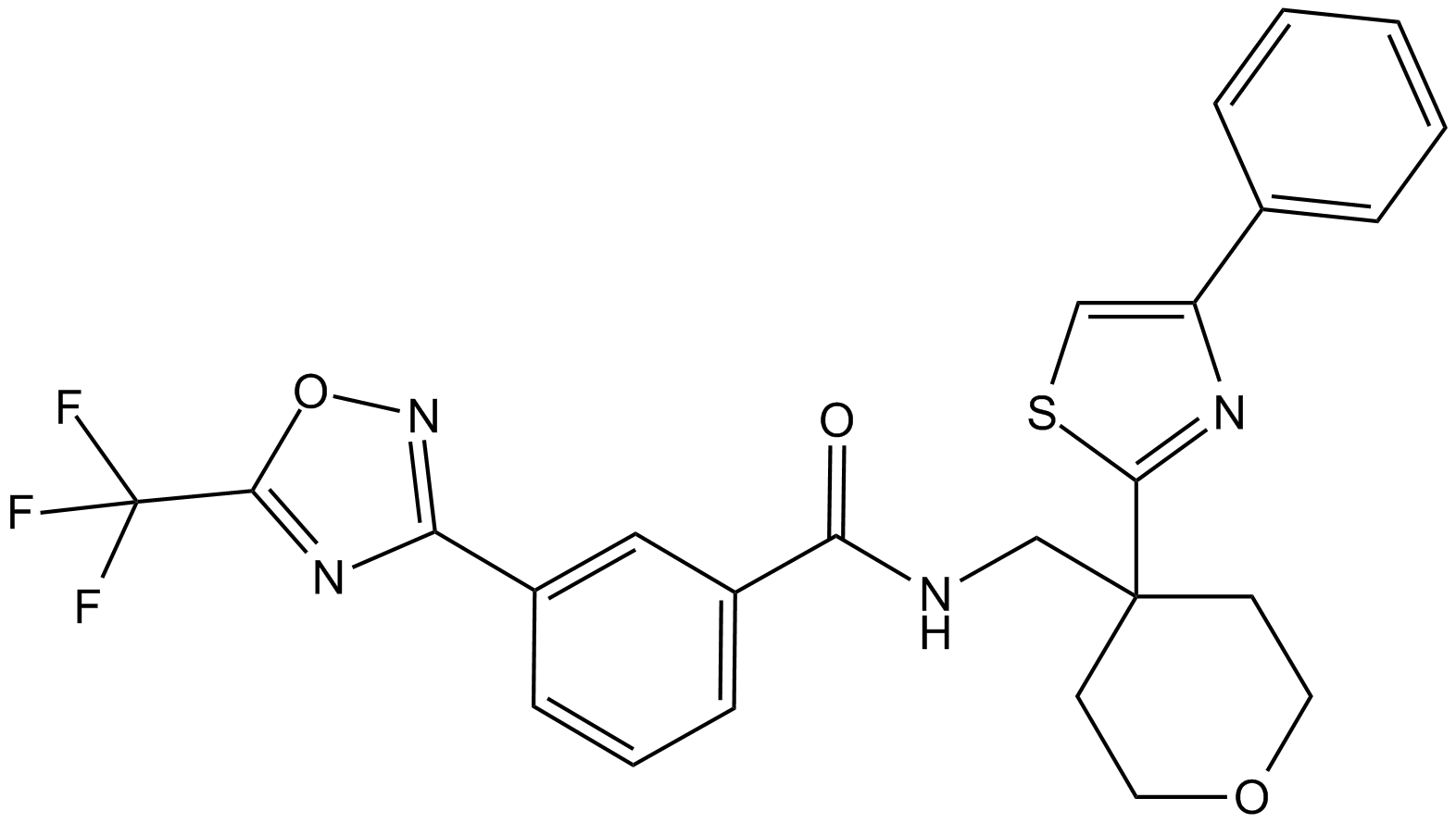
GC17703
TMP269
HDAC 4/5/7/9 inhibitor
-
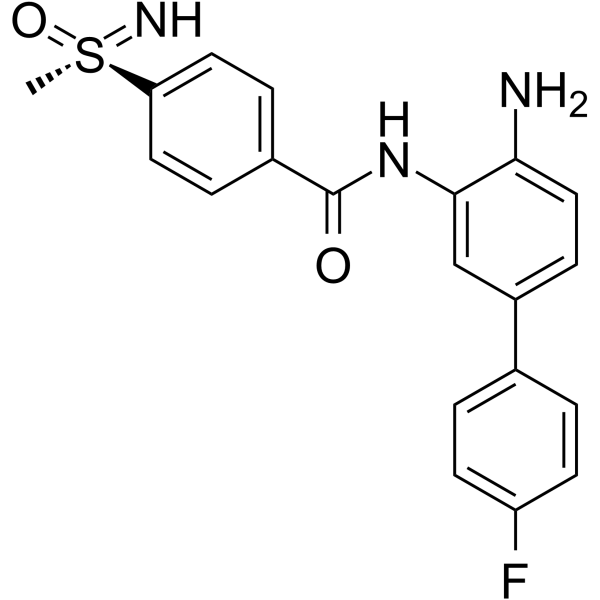
GC70040
TNG260
TNG260 is a selective CoREST deacetylase (CoreDAC) inhibitor. TNG260 selectively inhibits HDAC1 10 times more than HDAC3. TNG260 leads to the inhibition of HDAC1 and reverses anti-PD1 resistance driven by STK11 deficiency. TNG260 reduces tumor infiltration of neutrophils. TNG260 exhibits immune-mediated cell killing.
-
GC10322
Tubastatin A HCl
Tubastatin A HCl (Tubastatin A HCl) is a potent and selectiveHDAC6inhibitor withIC50of 15 nM in a cell-free assay, and is selective (1000-fold more) against all other isozymes except HDAC8 (57-fold more). Tubastatin A HCl also inhibits HDAC10 and metallo-β-lactamase domain-containing protein2 (MBLAC2).
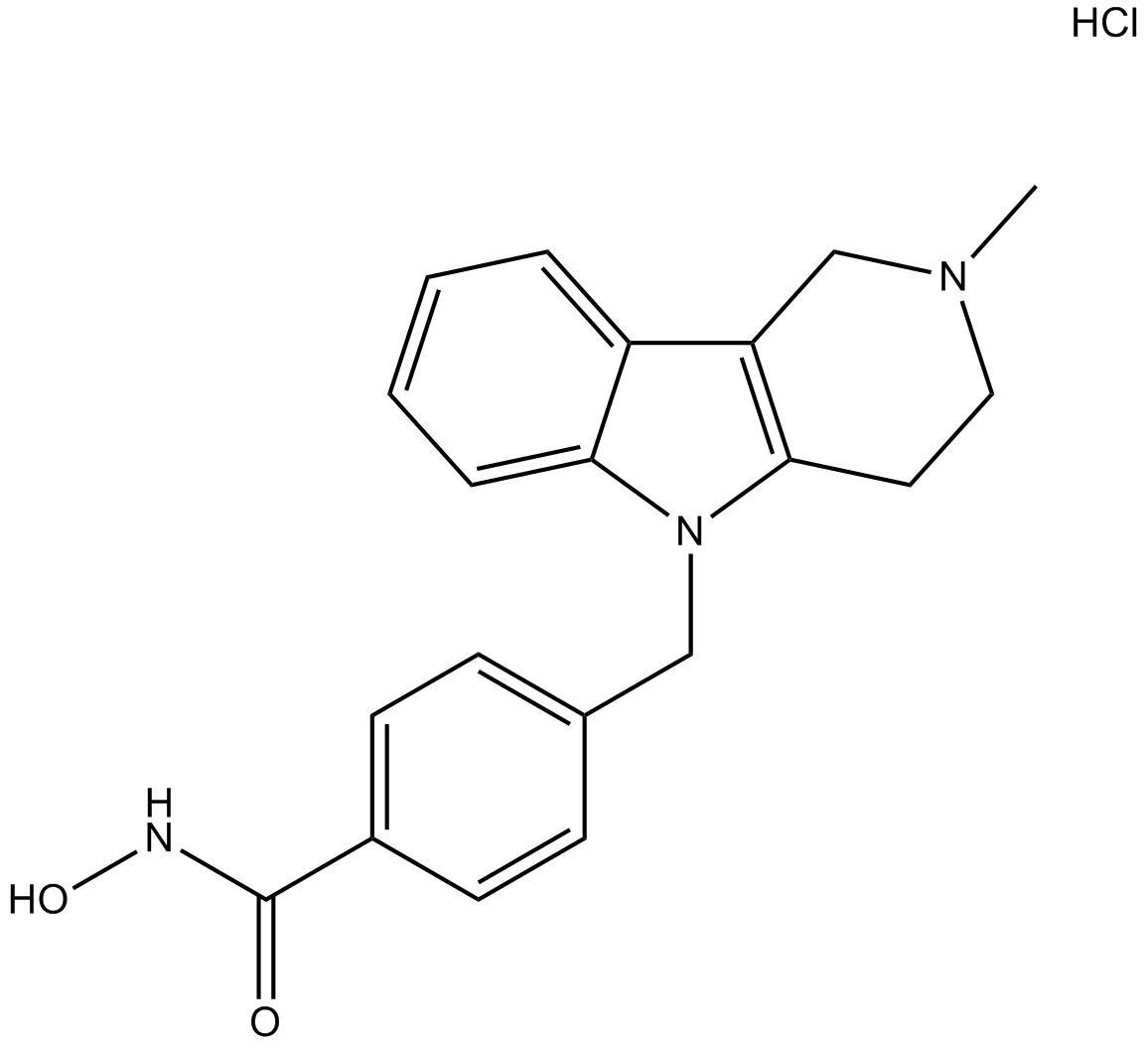
-
GC37845
Tucidinostat
An HDAC inhibitor
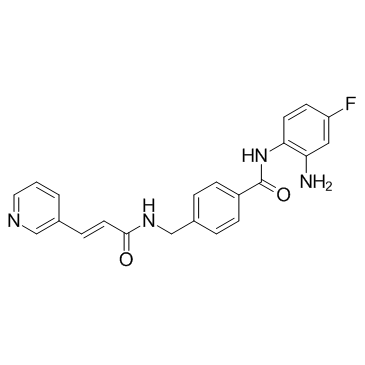
-
GC14367
UF 010
Novel and selective class I HDAC inhibitor
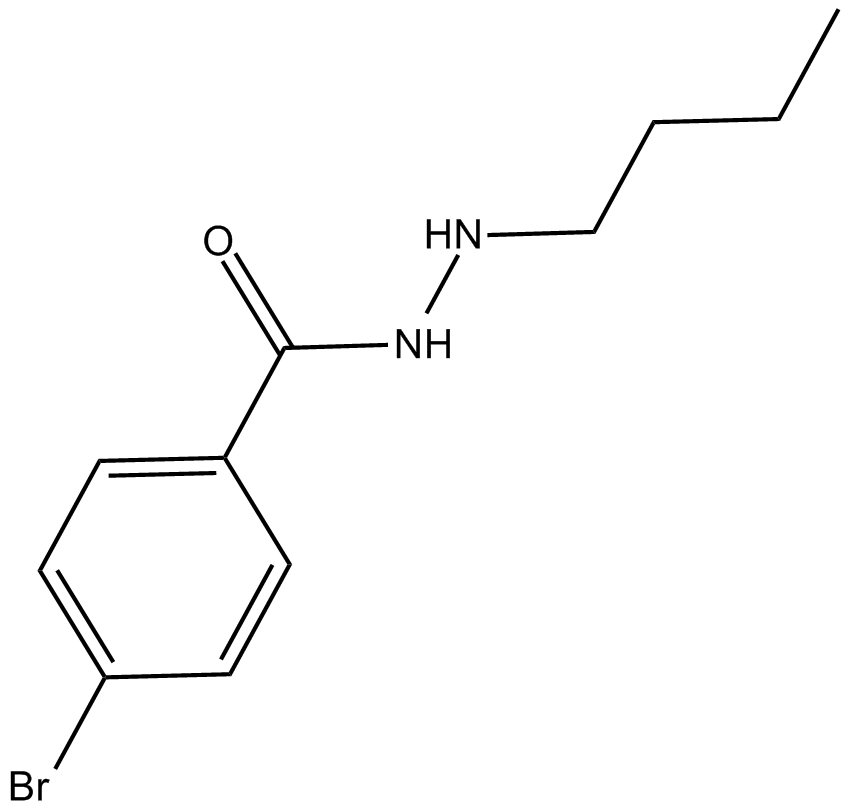
-
GC11424
Valproic acid
HDAC1 inhibitor
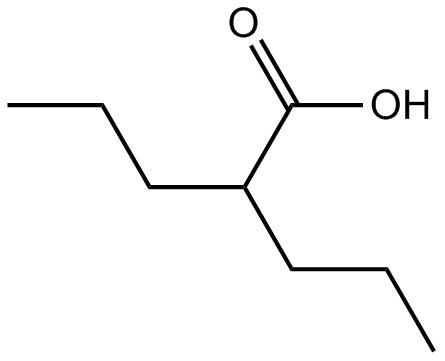
-
GC70109
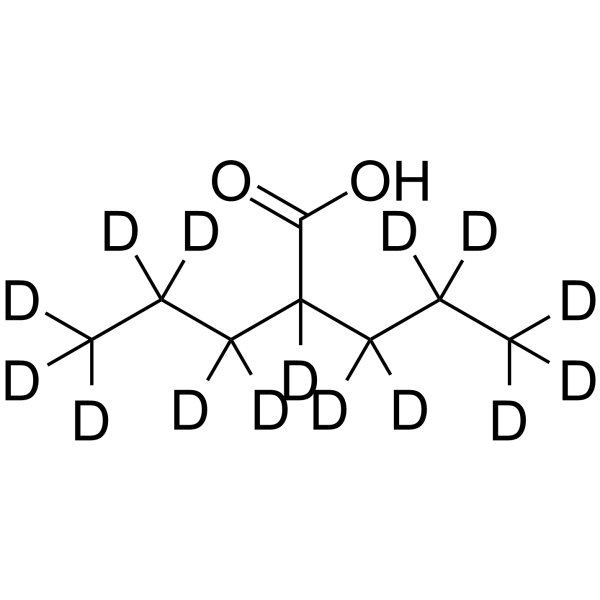
Valproic acid-d15
Valproic acid-d15 is the deuterated form of Valproic acid. Valproic acid (VPA; 2-Propylpentanoic Acid) is an HDAC inhibitor with an IC50 value of 0.5-2 mM, inhibiting the activity of HDAC1 (IC50, 400 μM), while inducing the degradation of HDAC2. Valproic acid activates Notch1 signaling and inhibits proliferation in small cell lung cancer (SCLC) cells. Valproic acid can be used for research on epilepsy, bipolar disorder, and migraines.
-
GC64378
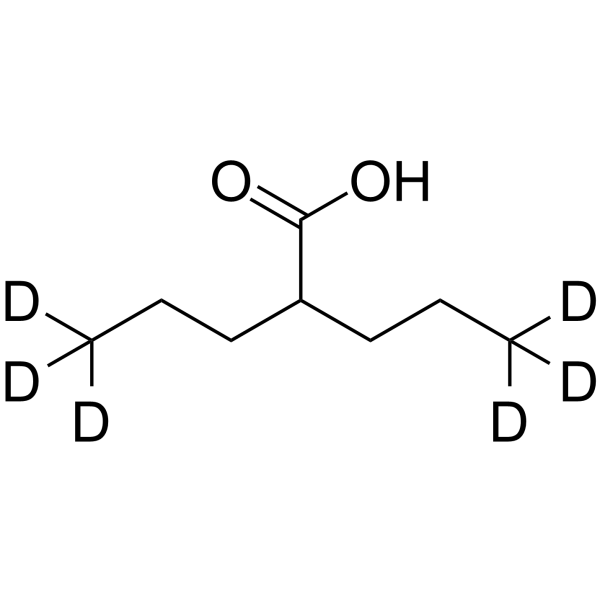
Valproic acid-d6
Valproic acid-d6 (VPA-d6) is the deuterium labeled Valproic acid. Valproic acid (VPA; 2-Propylpentanoic Acid) is an HDAC inhibitor, with IC50 in the range of 0.5 and 2 mM, also inhibits HDAC1 (IC50, 400 μM), and induces proteasomal degradation of HDAC2. Valproic acid activates Notch1 signaling and inhibits proliferation in small cell lung cancer (SCLC) cells. Valproic acid sodium salt is used in the treatment of epilepsy, bipolar disorder and prevention of migraine headaches.
-
GC16143
Varenicline Hydrochloride
α4β2 nicotinic receptor agonist

-
GC18206
WT161
WT161 is a potent inhibitor of HDAC6 with an IC50 value of 0.40 nM.